Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bioinspired design of DNA in aqueous ionic liquid media for sustainable packaging of horseradish peroxidase under biotic stress†
Diksha Dhiman‡
a,
Aaftaab Sethi‡ b,
Rakesh Sinhaa,
Sagar Biswasa,
Gregory Franklin
b,
Rakesh Sinhaa,
Sagar Biswasa,
Gregory Franklin a and
Dibyendu Mondal
a and
Dibyendu Mondal *ac
*ac
aInstitute of Plant Genetics (IPG), Polish Academy of Sciences, Strzeszyńska 34, 60-479 Poznań, Poland. E-mail: m.dibyendu@jainuniversity.ac.in; dmon@igr.poznan.pl
bLaboratory of Biomolecular Interactions and Transport, Department of Gene Expression, Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University, Uniwersytetu Poznanskiego 6, Poznan 61-614, Poland
cCentre for Nano and Material Sciences, Jain (Deemed-to-be University), Jain Global Campus, Kanakapura, Bangalore, Karnataka 562112, India
First published on 23rd December 2024
Abstract
We show that a combination of DNA and ionic liquid significantly increases the stability and activity of HRP and achieves a 4.8-fold higher peroxidase activity than PBS buffer. Also, HRP retains 84% of its activity in IL+DNA compared to 24% in PBS against trypsin digestion. Molecular modeling and spectroscopic studies reveal a protective microenvironment.
In biological systems, macromolecular crowding is a natural phenomenon in which proteins exist in a densely packed environment within cells.1,2 To simulate these crowded conditions in vitro, researchers often use polyethylene glycol, dextran, Ficoll, bovine serum albumin and hemoglobin as crowding agents.3–5 DNA, a natural biopolymer, also replicates the crowded intracellular environment and provides a stabilizing effect that allows enzymes to adopt their folded conformations. Recently, we have shown that B-DNA can act as a crowding agent, increasing both the activity and stability of cytochrome c.6 Previous studies have highlighted the significant effects of macromolecular crowding on various biological functions, including protein folding,7,8 thermal stability,9 and enzyme kinetics.4 The effects of crowding on enzyme activity are diverse. While some enzymes may exhibit increased activity due to conformational changes, the efficiency of other enzymes may be reduced due to diffusion limitations.5,10 Furthermore, crowding can induce liquid–liquid phase separation in proteins, suggesting that soft interactions play a crucial, yet unexplored role in enzymatic behavior under crowded conditions.5 Interestingly, ionic liquids (ILs) offer the possibility of enhancing protein interactions through softer mechanisms.11–13 ILs are characterized by their high thermal stability, tunable viscosity and ability to solubilize various biomacromolecules.14–16 These solvents have been extensively used to purify, stabilize and activate proteins by modifying their microenvironment through ionic interactions, hydrogen bonding and hydrophobic interactions.15,16 Our recent work has shown that protein-compatible ILs can effectively modulate the activity and stability of enzymes during their immobilization with nanomaterials.13
Despite the promising results of using ILs and DNA, no studies have yet explored the combined effects of DNA as a crowding agent alongside ILs on enzyme activity and stability. The use of DNA in IL media offers additional advantages as DNA is a renewable resource and well-designed ILs can be environmentally friendly. This combination is particularly advantageous for applications in green chemistry and sustainable protein packaging. In this study, we aim to increase the activity and stability of horseradish peroxidase (HRP) by using a biological fluid-inspired solvent system consisting of B-DNA and cholinium phosphonoacetate ([Ch]2[PAA]) IL (Fig. S1, ESI†). HRP is a widely used enzyme in biocatalysis and diagnostics due to its ability to catalyze oxidative reactions.17 Although HRP has been successfully used in various processes, it is sensitive to abiotic and biotic stress. Various methods, including covalent immobilization on functionalized graphene oxide and bacterial cellulose, have been tried to improve the stability and performance of HRP,18,19 but the full potential of HRP remains largely untapped. This study demonstrates the potential of using DNA in IL media for improved packaging of HRP with increased peroxidase activity and stability. The effects of trypsin protease as a biotic stressor on HRP activity were also investigated. Various spectroscopic techniques, including UV-visible spectroscopy and circular dichroism (CD), were used to assess the structural stability of the enzyme. Molecular docking studies were also performed to explore the interactions between HRP, DNA and IL. Overall, this research suggests that DNA–IL media can serve as a sustainable, biologically inspired, molecularly crowded system for packaging proteins under biotic stress and overcome traditional challenges in biocatalysis.
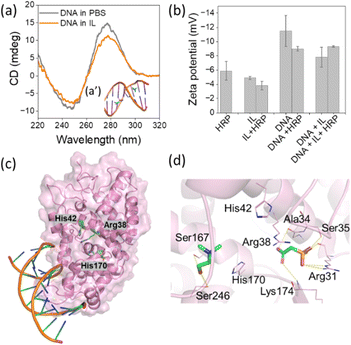
To design bioinspired media, firstly DNA interaction with the IL was evaluated. Fig. 1a presents the CD spectrum of DNA in the presence of phosphate buffer (pH 7.4) and 15 wt% [Ch]2[PAA]. B-DNA in PBS exhibits a prominent positive band at 277 nm due to π–π base stacking and a less intense negative band is observed at 246 nm corresponds to the helicity.14 A marginal shift of the negative band in the presence of IL indicates an interaction of the DNA with IL counterions. Docking of IL counterions to DNA was performed to assess their interactions. The anion of IL showed binding to the major groove of DNA (−5.844 kcal mol−1) and thus exhibited a stronger affinity than the cation (−5.159 kcal mol−1), which was localised in the minor groove (Fig. 1a′ inset). Zeta (ζ)-potential studies were also performed to evaluate the propensity to form HRP–DNA, HRP–IL and HRP–IL + DNA complexes. As shown in Fig. 1b, the ζ-potential of HRP in phosphate buffer (pH = 7.4) was −5.9 mV, while the ζ-potential of HRP decreased slightly in the presence of IL. However, due to the very high ζ-potential of DNA in water, HRP–DNA exhibited a relatively high ζ-potential, indicating greater dispersion stability of HRP in the presence of DNA.20,21 Similarly, HRP exhibited a high ζ-potential in the presence of IL + DNA, emphasising the increasing colloidal stability of HRP in the presence of a bioinspired designer solvent system. In addition, the interaction of HRP with DNA, IL and IL + DNA was predicted with HDock.22 More than half of the top ten predicted poses showed DNA binding in a similar region (Fig. S2, ESI†), predominantly near the heme-binding site of HRP but, crucially, without blocking it (Fig. 1c). The DNA bases showed several polar contacts with HRP residues such as Arg178, Gly191 and Arg206. The localization of DNA at this site suggests that it can stabilise HRP in its active state by reducing conformational flexibility and improving its stability. This is consistent with previous studies showing that DNA increases protein stability.6 Next, IL docking was extended to HRP, with the anion again showing a better binding value (−5.328 kcal mol−1) than the cation (−3.504 kcal mol−1). Both ions were found to bind in close proximity to the heme-binding site (Fig. 1d). Importantly, neither the anion nor the cation blocked access to the active site, so that the catalytic pocket remained accessible. The anion exhibited an extensive interaction network consisting of multiple hydrogen bonds with Arg31, Ala34, Ser35 and Lys174. The cation bound to a diagonally opposite site and formed hydrogen bonds with Ser167 and Ser246 (Fig. 1d). Finally, docking simulations were performed for the IL and HRP–DNA complexes to ascertain whether the IL preferentially binds to the protein, the DNA or the interface between them. The results showed that the IL was localized in the same region as observed in the HRP-only docking experiments (Fig. 1d), namely near the heme-binding site. This indicates that the IL has a strong preference for binding near the heme-binding site of HRP. The complementary binding of DNA and IL to key regions of HRP suggests that together they may enhance HRP activity. Accordingly, we investigated the peroxidase activity of HRP in the presence of different concentrations of IL, DNA and DNA + IL. A detailed protocol for the activity assays can be found in the ESI.†
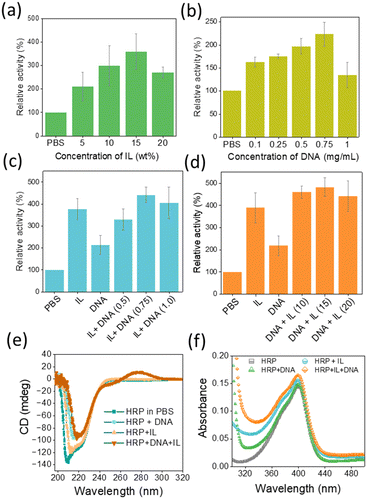
The peroxidase activity of HRP was investigated in the presence of H2O2 using 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS).6 Fig. 2a–d show the peroxidase activity of HRP in the presence of IL, DNA and DNA + IL systems. The corresponding UV-Vis absorbance (at 420 nm) vs. time plots are provided in Fig. S3 (ESI†). The activity of HRP in PBS was considered to be 100%. To optimize the concentration of IL, the activity of HRP was investigated in different concentrations of [Ch]2[PAA] IL from 0 to 20 wt% (Fig. 2a). The relative activity of HRP increased with increasing IL concentration, with the maximum relative activity recorded at 15 wt% IL (3.58-fold). A further increase in IL concentration led to a decrease in peroxidase activity. Similarly, the DNA concentration was varied from 0 to 0.75 mg mL−1 and it was found that the optimum concentration was 0.75 mg mL−1 DNA, resulting in a 2.22-fold increase in the relative activity of HP compared to PBS (Fig. 2b). To understand the effect of DNA in the presence of IL, the DNA concentration was varied, keeping the IL concentration at 15 wt%. As can be seen in Fig. 2c, HRP activity was maximal (∼4.4-fold) in the presence of IL (15 wt%) + DNA (0.75 mg mL−1), i.e. stronger than in a system with only DNA and only IL. Similarly, peroxidase activity was analysed by fixing the DNA concentration at 0.75 mg mL−1 and varying the IL concentration (Fig. 2d). When the relative HRP activity was evaluated in the presence of DNA + IL formulations (Fig. 2d), a 4.8-fold increase in activity was observed at the optimal concentrations of IL (15%) and DNA (0.75 mg mL−1). When evaluating the HRP activity from the two series of experiments in Fig. 2c and d, it becomes clear that the incubation of HRP with DNA followed by IL is more favourable than IL followed by DNA. Overall, the trend of activity follows the order DNA + IL > IL > DNA > PBS. Hasan et al.23 investigated the stability and activity of HRP in the presence of molecular crowders, dextran 70 and PEG-4000. Their results show that PEG-4000 increases the stability of HRP but at the same time impairs its enzymatic activity. Through our work, in which we developed a formulation containing both molecular crowders and IL, the synergy between the structural stability and activity of HRP was achieved. The results obtained in the present study thus confirm the fact that the activity and stability of enzymes can be remarkably tuned by a carefully designed solvent system.
To further investigate the influence of the bioinspired solvents, the structural stability of HRP was examined in the presence of optimized IL, DNA and IL + DNA concentrations (Fig. 2e and f). The characteristic absorption peak of HRP at 403 nm24 in the UV region was preserved in the presence of all four solvent systems, indicating the retention of its native structure. In addition, the CD spectra showed the preservation of the secondary structure of the enzyme, as evidenced by the retention of the peaks at 222 nm and 208 nm, indicating a stable α-helix content.25 The positive CD band around 280 nm in Fig. 2e for the HRP + DNA and HRP + IL + DNA system arises due to π–π base stacking induced by the DNA in the aqueous medium.
The binding patterns in IL docking suggest that the interactions (Fig. 1d), particularly that of the anion, may play a crucial role in stabilizing the more active conformation of the enzyme. By establishing a strong network of hydrogen bonds near the heme-binding site, the IL may help to maintain the structural integrity of the enzyme and prevent unwanted conformational changes that could otherwise reduce catalytic efficiency. This improved stabilisation likely increases the enzyme's ability to bind and process substrates more effectively, which may lead to increased catalytic activity. The complementary binding of DNA and IL to key regions of HRP suggests that together they increase the stability of HRP in its more active conformation (Fig. 1c and d). Moreover, the ability of IL and DNA to form multiple hydrogen bonds with residues directly involved in maintaining the architecture of the active site and side residues, respectively, suggests a mechanism by which IL + DNA can preserve the enzyme in its catalytically active form against various stress conditions. Accordingly, the effects of biotic stress such as trypsin digestion on the activity and stability of HRP in the presence of different solvent systems were investigated by peroxidase activity assays and SDS-PAGE analysis. Detailed protocols for the activity of HRP in the presence of trypsin and SDS-PAGE can be found in the ESI.†
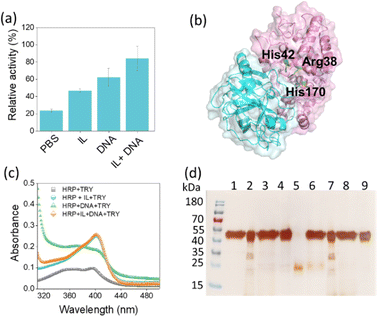
Trypsin (TRY) hydrolyses the peptide bond on basic amino acids and thereby denatures the target protein.26 To investigate the resistance of the developed solvent to proteolytic digestion of HRP, the system consisting of HRP in buffer, HRP in IL, HRP in DNA and HRP in IL + DNA was incubated with 6 μM trypsin at 37 °C for 24 h. After interaction, only 24% of the activity in PBS was retained compared to the initial activity of HRP in PBS buffer (pH = 7.4) (Fig. 3a), while HRP in the presence of IL and DNA maintained a decent performance with 46% and 62% of activity, respectively. Notably, HRP exhibited the highest relative activity in the presence of the IL + DNA solvent system, retaining approximately 84% of its initial activity against TRY digestion (Fig. 3a). In addition, a molecular docking study with HDock was performed to predict how TRY binds to HRP, potentially providing clues to the likely reason for the reduced protease digestion of HRP by TRY. Here, the AlphaFold predictions could not be used to validate the HRP/TRY complex as the ipTM score was 0.38, which is also below the threshold for reliable predictions (Fig. S4, ESI†). Therefore, the model with the highest score in HDock was used for further analysis. The results show that both DNA and TRY bind to a similar region on HRP (Fig. 1c and 3b), especially near the catalytic site. This overlap suggests that DNA may hinder the access of TRY to HRP, ultimately leading to slower degradation of HRP by TRY. The results obtained with HDock was also validated from docking results performed using GRAMM (Fig. S5, ESI†).27 Although there was some variability in the top 5 poses generated by GRAMM and HDock, with two poses in the top 5 for each of them binding to the region displayed (Fig. S5, ESI†). It is important to note that such variability in binding regions is anticipated. Nonetheless, these results support the experimental findings.
In addition, the effect of TRY on HRP structure was analysed by incubating HRP in the presence of buffer, IL, DNA and IL + DNA for 24 h using the UV-Vis spectra of HRP in the Soret region (Fig. 3c). As can be seen in Fig. 3c, in the presence of DNA and buffer, there is a slow blue shift in the UV spectrum of HRP due to changes in the protein structure around the heme, especially alterations in critical amino acids such as histidine.28 Moreover, there is a marked drop in the absorbance of HRP at 403 nm, indicating greater degradation of HRP by TRY in PBS. These structural stability results are consistent with the HRP activity result (Fig. 3a), which shows that HRP has the lowest activity after trypsin digestion in the presence of PBS buffer than in DNA solution. However, the 403 nm peak of HRP was maintained in the presence of IL and IL + DNA. SDS-PAGE analysis of HRP before and after digestion with trypsin was also performed (Fig. 3d). Most of the native HRP in the presence of trypsin was degraded in the presence of PBS buffer. However, degradation of the enzyme was slower when digested in the presence of DNA and IL. The SDS-PAGE band for HRP after digestion with TRY in the presence of IL + DNA was almost intact, indicating the combined effect of DNA and IL to protect HRP from biotic stress such as protease. Furthermore, we examined TRY activity in these solvent systems using N-α-benzoyl-DL-arginine 4-nitroanilide as the substrate. The results showed that TRY activity in the presence of DNA was higher than in the control (Tris–HCl pH 7.8), whereas in the presence of IL was significantly lower than the control (Fig. S6, ESI†). However, CD studies revealed that the overall secondary structure of TRY remains stable in the presence of both IL and DNA solutions (Fig. S7, ESI†). Overall, the combination of DNA and IL creates a unique microenvironment that stabilizes HRP and improves its structural integrity and activity. This molecularly crowded system not only prevents TRY access to HRP, but also promotes a protective barrier that effectively reduces the rate of enzymatic degradation.
In summary, DNA and IL can effectively increase the stability and activity of HRP, resulting in a 4.8-fold higher peroxidase activity than the control. The interaction of IL and DNA with HRP creates a protective environment that preserves the structural integrity of HRP and increases its catalytic activity. According to docking experiments, both DNA and IL preferentially bind near the heme-binding site without blocking the active site, indicating a synergistic effect that keeps HRP in its highly active conformation. Experimental results show that HRP exhibits remarkable resistance to proteolytic degradation and retains significantly higher activity when exposed to the DNA + IL system compared to conventional buffers. This innovative, sustainable formulation addresses biotic stress challenges and demonstrates potential for developing effective biocatalytic systems in crowded environments.
This work was supported by the National Science Centre (NCN), SONATA project no. Reg. UMO-2021/43/D/ST4/00699. D. M. and G. F. acknowledge the NANOPLANT project, which received funding from the European Union's Horizon 2020 research and innovation program under grant agreement no. 856961.
Data availability
The study data are included in the article and ESI;† raw data are available upon request.Conflicts of interest
There are no conflicts to declare.References
- F.-X. Theillet, et al., Chem. Rev., 2014, 114(13), 6661–6714 CrossRef.
- A. R. Subramanya and C. R. Boyd-Shiwarski, Annu. Rev. Physiol., 2024, 86, 429–452 CrossRef PubMed.
- C. Alfano, et al., Chem. Rev., 2024, 124(6), 3186–3219 CrossRef PubMed.
- R. Chapanian, et al., Nat. Commun., 2014, 5, 4683 CrossRef PubMed.
- G. Rivas and P. A. Minton, Annu. Rev. Biochem., 2022, 91, 321–351 CrossRef PubMed.
- S. M. Shet, et al., Int. J. Biol. Macromol., 2022, 215, 184–191 CrossRef PubMed.
- D. Gomez, et al., J. Phys. Chem. Lett., 2019, 10(24), 7650–7656 CrossRef PubMed.
- J. Peters, et al., Chem. Rev., 2023, 123(23), 13441–13488 CrossRef CAS PubMed.
- L. Stagg, et al., Proc. Natl. Acad. Sci. U. S. A., 2007, 104(48), 18976–18981 CrossRef CAS PubMed.
- A. Pastore and A. P. Temussi, Trends Biochem. Sci., 2022, 47(12), 1048–1058 CrossRef CAS.
- K. Bhakuni, et al., Chem. Commun., 2019, 55(40), 5747–5750 RSC.
- P. Bharadwaj, et al., Green Chem., 2023, 25(17), 6666–6676 RSC.
- S. K. Thayallath, et al., Chem. Commun., 2023, 59(39), 5894–5897 RSC.
- C. Mukesh, et al., Chem. Commun., 2013, 49(61), 6849–6851 RSC.
- S. P. M. Ventura, et al., Chem. Rev., 2017, 117(10), 6984–7052 CrossRef PubMed.
- P. Bharmoria, et al., Chem. Rev., 2024, 124(6), 3037–3084 CrossRef PubMed.
- G. R. Lopes, et al., RSC Adv., 2014, 4, 37244–37265 RSC.
- M. Besharati, et al., Int. J. Biol. Macromol., 2018, 106, 1314–1322 CrossRef.
- B. Yu, et al., Process Biochem., 2019, 79, 40–48 CrossRef.
- T. A. Shmool, et al., JACS Au, 2022, 2, 2068–2080 CrossRef PubMed.
- D. Dhiman, et al., Mol. Pharmaceutics, 2023, 20, 3150–3159 CrossRef PubMed.
- Y. Yan, et al., Nat. Protoc., 2020, 15(5), 1829–1852 CrossRef.
- S. Hasan and A. Naeem, J. Mol. Recognit., 2021, 34, e2902, DOI:10.1002/jmr.2902.
- D. Morales-Urrea, et al., Sci. Rep., 2023, 13, 13363 CrossRef CAS PubMed.
- N. J. Greenfield, Nat. Protoc., 2006, 1, 2876–2890 CrossRef CAS PubMed.
- M. Krieger, et al., J. Mol. Biol., 1974, 83, 209–230 CrossRef CAS PubMed.
- A. Singh, et al., Methods Mol. Biol., 2024, 2714, 101–112 CrossRef CAS PubMed.
- T. Li, et al., BMC Struct. Biol., 2011, 11, 13 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details and characterization. See DOI: https://doi.org/10.1039/d4cc05803h |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2025 |