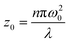Mitochondria-localized dinuclear iridium(III) complexes for two-photon photodynamic therapy†
Zanru
Tan
a,
Jiang
Feng
b,
Jiangping
Liu
*c,
Taihong
Liu
 *b,
Huihui
Wu
*d and
Hui
Chao
*b,
Huihui
Wu
*d and
Hui
Chao
 *a
*a
aMOE Key Laboratory of Bioinorganic and Synthetic Chemistry, Guangdong Basic Research Center of Excellence for Functional Molecular Engineering, School of Chemistry, Sun Yat-sen University, Guangzhou, 510275, P. R. China. E-mail: ceschh@mail.sysu.edu.cn
bKey Laboratory of Applied Surface and Colloid Chemistry of Ministry of Education, School of Chemistry and Chemical Engineering, Shaanxi Normal University, Xi'an, Shaanxi 710119, P. R. China. E-mail: liuth121@snnu.edu.cn
cInstitute of Flexible Electronics (IFE, Future Technologies), Xiamen University, Xiamen, 361102, P. R. China. E-mail: liujping@xmu.edu.cn
dDepartment of Dermatology, The East Division of the First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, 510700, P. R. China. E-mail: cometvivi@126.com
First published on 21st January 2025
Abstract
Photodynamic therapy (PDT), as a non-invasive cancer treatment, offers significant advantages including high temporal–spatial selectivity, minimal surgical intervention, and low toxicity, thereby garnering considerable research interest from across the world. In this study, we have developed a series of dinuclear cyclometalated Ir(III) complexes as potential two-photon photodynamic anticancer agents. These Ir(III) complexes demonstrate significant two-photon absorption (2PA) cross-sections (σ2 = 66–166 GM) and specifically target mitochondria. Amongst them, N-Ir4 manifests an IC50 value of 2.0 μM and a phototoxicity index (PI) of 24. Under two-photon excitation, N-Ir4 efficiently generates reactive oxygen species (ROS), leading to mitochondrial damage and cell death. Our study reveals drastically enhanced optical properties forged by forming a dinuclear complex bridged by two conjugated rigid planar moieties and sheds light on a potential paradigm to boost 2PA cross-sections.
Introduction
Cancer has evolved as a major threat to human health and has escalated to a leading cause of mortality worldwide. In 2020, there were 19.3 million newly diagnosed cases and 10 million cancer-related deaths reported. It is predicted that by 2040 the incidence of cancer will rise to 29 million.1,2 Unfortunately, traditional cancer treatment modalities, including chemotherapy, radiotherapy, and surgical resection, continue to exhibit numerous limitations.PDT has emerged as a promising alternative in recent decades.3–9 In a PDT regimen, a photosensitizer (PS) absorbs light energy and transits to the excited state by which this energy (type II PDT) or the excited electron (type I PDT) is further shifted to the neighboring oxygen molecules, generating reactive oxygen species (ROS). These ROS oxidize pivotal macro-biomolecules, such as lipids, proteins, and DNA, triggering deeper biological responses and ultimately inducing cell death.10–12 Nevertheless, current commercially available PSs, such as porphyrin and chlorin derivatives, suffer from various limitations, including tedious preparation, poor photostability, and inadequate selectivity.13–15 Therefore, a novel class of PSs with bold structural novelty is highly demanded to push the envelope of this field.
Many Ir(III) complexes have been considered candidate drugs for anticancer therapy due to their favorable photophysical attributes and unique physiological properties, including strong photostability, ease of synthesis, and modifications.16–18 Besides, phosphorescent cyclometalated Ir(III) complexes often specifically accumulate in various cellular compartments such as mitochondria, endoplasmic reticulum, lysosomes, Golgi apparatus, and nuclei, providing a convenient way to induce cytotoxicity through the generation of ROS.19,20 Whilst mononuclear Ir(III) complexes have been extensively used as PSs for anticancer therapy, their dinuclear counterparts remain largely underexplored. A recent study suggested that a dinuclear cyclometalated Ir(III) complex in which the monomeric units were simply bridged by a flexible alkyl chain outperformed its mononuclear form in both photoluminescence quantum yield and singlet oxygen quantum yield.21 Indeed, homonuclear bimetallic complexes are often paired with interesting properties that are beyond the reach of their monomers.22–25 These advances prompt us to scrutinize the optical properties of a novel series of dinuclear Ir(III) complexes with rigid bridging ligands in which the optimized planar configuration may stand a chance to largely reshape its 2PA transition probabilities. The 2PA process involves the simultaneous absorption of two photons. Compared to single-photon absorption, 2PA offers an elegant way to excite Ir(III) complexes in the near-infrared regime and allows deeper tissue penetration for PDT applications.26–28 Indeed, barely any small molecular PSs could possess viable 2PA cross-sections to deliver efficient 2PA PDT. For example, the classical PS, Photofrin, only exhibits a σ2 value of 10 GM, and Visudyne exhibits a σ2 value of 50 GM.29,30 Hence, there remains an urgent need to validate the structural design know-how of endowing PSs with significantly higher σ2 values.
Herein, a series of novel binuclear Ir(III) complexes were synthesized (shown in Scheme 1). In contrast to the routine multinuclear framework, the Ir(III) monomers were interfaced from the flank in an asymmetric manner via two large conjugate planes. Their photophysical properties and photocytotoxicity were investigated. Interestingly, benefiting from the potentially extended planar configuration, the dinuclear complexes feature significant enhancement in 2PA absorption compared to the reported mononuclear complexes.31
 | ||
| Scheme 1 Chemical structure and the putative mechanism of action of the Ir(III) complexes in 2PA PDT. | ||
Results and discussion
Synthesis and characterization
The synthetic route to Ir(III) complexes is illustrated in Fig. S1.† The ligand was synthesized using glacial acetic acid as a solvent at 120 °C. The subsequent Ir(III) complexes were prepared according to ref. 31 and purification was done through column chromatography in dichloromethane/methanol. Characterization of the final products was performed using mass spectrometry and nuclear magnetic resonance techniques.The UV-vis absorption spectrum of Ir1–4 (10 μM) in dichloromethane is shown in Fig. 1(b). The intense absorption bands at below 300 nm can be assigned to the absorption of intra-ligand charge transfer (ILCT) and the moderate shoulder bands between 370 and 420 nm can be attributed to the absorption of metal–ligand charge transfer (MLCT). The photophysical properties of the Ir1–4 complex are summarized in Table S1.† Upon excitation at 405 nm, the complex exhibits a maximum emission peak at approximately 570 nm. The luminescence quantum yield is observed to be in the range of 0.01–0.02, and the lifetimes of these complexes span the range of 200–203 ns. The photophysical properties of the four Ir(III) complexes are remarkably similar, indicating the substitution of a single atom does not significantly affect their photophysical characteristics. The UV-vis absorption spectrum of Ir1–4 (10 μM) in dichloromethane is shown in Fig. 1(b). The intense absorption bands at below 300 nm can be assigned to the absorption of intra-ligand charge transfer (ILCT) and the moderate shoulder bands between 370 and 420 nm can be attributed to the absorption of metal–ligand charge transfer (MLCT). The photophysical properties of the Ir1–4 complex are summarized in Table S1.† Upon excitation at 405 nm, the complex exhibits a maximum emission peak at approximately 570 nm. The luminescence quantum yield is observed to be in the range of 0.01–0.02, and the lifetimes of these complexes span the range of 200–203 ns. The photophysical properties of the four Ir(III) complexes are remarkably similar, indicating that the substitution of a single atom does not significantly affect their photophysical characteristics.
The complexes exhibit high solubility in organic solvents, such as dichloromethane, but demonstrate poor solubility in aqueous solutions. To enhance the solubility of these complexes in aqueous environments, nanoparticles were synthesized through co-assembly with the commercial amphiphilic polymer DSPE-PEG2000. The resulting encapsulated complexes were named N-Ir1–4. Characterization of the N-Ir4 nanoparticles using transmission electron microscopy (TEM) revealed a spherical morphology with an average diameter of approximately 170 nm (Fig. 1(a)). The average particle sizes of N-Ir1–4 were measured using dynamic light scattering (DLS) are 203, 198, 175, and 176 nm, respectively (Fig. 1(a), and Fig. S2†). The particle size of N-Ir1–4 did not change significantly over 7 days (Fig. S3†), demonstrating good stability in phosphate-buffered saline (PBS) for subsequent PDT research. The stability of N-Ir1–4 at physiological pH was also demonstrated using zeta potential (Fig. S4†) and UV–Vis spectra (Fig. S5†) measurements.
2PA cross-sections
The 2PA cross-sections of Ir1–4 were determined using the open-aperture z-scan technique. The results shown in Fig. S6 and Table S2† indicate that the highest σ2 values of Ir2–4 at 720 nm and were determined to be 166, 66, and 107 GM, respectively (1 GM = 1 × 10−50 cm4 s per photon per molecule). While the maximum σ2 value for Ir1 was found to be 135 GM at 730 nm. In our group's prior research, we investigated mononuclear Ir(III) complexes with analogous structures, which exhibited the highest σ2 values of 85 GM at 750 nm.32 Besides, these σ2 values are higher than those of many other mononuclear Ir(III) complexes.31,32ROS generation yield
Reactive oxygen species (ROSs) exhibit significant reactivity and selectivity, playing crucial roles in cellular processes such as growth and proliferation, development and differentiation, aging, and apoptosis, as well as in numerous physiological and pathological mechanisms.33 Electron spin resonance (ESR) is a robust technique employed to detect, identify, and quantify reactive oxygen species (ROS) in various systems. Spin-trapping techniques using ESR allow for the measurement of free radical species generated in different chemical systems.In this study, 2,2,6,6-tetramethylpiperidine (TEMP) was employed as a singlet oxygen trapping agent, and the singlet oxygen generated by the complexes was determined in solution. The data revealed a characteristic triplet peak (Fig. S7†), indicative of the production of 1O2. These results imply that the complexes operate via type II PDT mechanisms. Although electron spin resonance (ESR) is a robust technique for ROS detection, it is crucial to account for its limitations and potential sources of error, particularly regarding the stability of radical traps and the impact of environmental factors.
To further quantify the yield of singlet oxygen, an indirect measurement was performed using 9,10-anthracenediyl-bis(methylene) dimalonic acid (ABDA) as a 1O2 scavenger and Ru(bpy)32+ (ΦΔ = 0.81) as a standard to assess the complexes’ ability to generate ROSs under light.34 The oxidation of ABDA by photogenerated 1O2 leads to a decrease of optical density at 378 nm in a stoichiometric manner and follows a zero-order kinetics process. ΦΔ (1O2) values were determined by comparison of the slope of the OD378nm attenuation curve with a standard compound, as is shown in Fig. S8 and Table S3.† The ΦΔ values of Ir1 (0.31), Ir3 (0.32), and Ir4 (0.31) are comparable to that of the Ir(III) complex [Ir(ppy)2(bpy)]+ (0.35).35 However, the quantum yield of Ir2 is slightly lower, which may be attributed to the influence of substituent groups.
Lipophilicity and cellular uptake
The lipophilicity of a compound is intricately linked to its physicochemical and physiological properties, particularly in terms of cellular uptake and intracellular distribution. The lipid–water distribution coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w) for the n-octanol/water system complexes were evaluated, and the findings are illustrated in Fig. S9.† The lipid–water distribution coefficients for the complexes are 1.11, 1.01, 0.81, and 1.69, respectively, suggesting that these complexes exhibit the ability to traverse the cell membrane. Due to the limited solubility of the complex in aqueous solution, we employed the commercial amphiphilic polymer DSPE-PEG2000 to encapsulate the complex, forming nanoparticles designated as N-Ir1–4. As illustrated in Fig. S10,† the intracellular uptake of both unencapsulated and encapsulated Ir(III) complexes was quantified using ICP-MS. The data indicate that encapsulation with DSPE-PEG2000 significantly enhances the cellular uptake of the Ir(III) complexes, with the maximum accumulation observed at 8 hours.
Po/w) for the n-octanol/water system complexes were evaluated, and the findings are illustrated in Fig. S9.† The lipid–water distribution coefficients for the complexes are 1.11, 1.01, 0.81, and 1.69, respectively, suggesting that these complexes exhibit the ability to traverse the cell membrane. Due to the limited solubility of the complex in aqueous solution, we employed the commercial amphiphilic polymer DSPE-PEG2000 to encapsulate the complex, forming nanoparticles designated as N-Ir1–4. As illustrated in Fig. S10,† the intracellular uptake of both unencapsulated and encapsulated Ir(III) complexes was quantified using ICP-MS. The data indicate that encapsulation with DSPE-PEG2000 significantly enhances the cellular uptake of the Ir(III) complexes, with the maximum accumulation observed at 8 hours.
Intracellular distribution
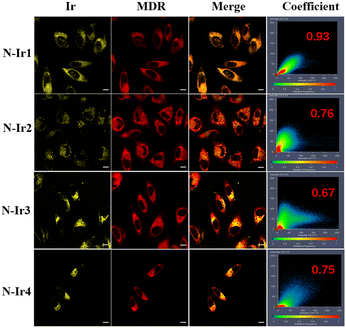
Colocalization experiments utilizing commercially available dyes were conducted to examine the intracellular distribution of complexes. Specifically, the colocalization of mitochondrial channel luminescent patterns was investigated using MitoTracker Deep Red FM and Ir(III) complexes. High colocalization coefficients were observed, indicating effective mitochondria-targeted accumulation.36–39 The colocalization coefficients of the complex and MitoTracker Deep Red FM are presented in Fig. 2. The results demonstrated that the complexes specifically targeted mitochondria. The mitochondrion, a double-membraned organelle, plays a crucial role in apoptosis, calcium regulation, and ROS management. It is an important anti-tumor mechanism to destroy the redox balance of mitochondria by producing reactive oxygen species in situ under illumination, thus affecting the function and morphology of cells.40Cytotoxicity assay
The evaluation of the (photo-)toxicities of Ir(III) complexes in A375 cancer cells and HLF cells using the MTT assay provides substantial insights into verifying the PDT effect. The IC50 values are summarized in Table 1. N-Ir4 exhibited an IC50 value of 2 μM towards A375 cells, with a PI of 24, which is comparable to most of the reported cyclometalated Ir(III) PSs.| Cell line | Ir1 | Ir2 | Ir3 | Ir4 | Cisplatin | |
|---|---|---|---|---|---|---|
| a Irradiated with an LED area light source (405 nm, 12 J cm−2). b PI = IC50 (dark)/IC50 (light). | ||||||
| A375 | Dark | 11.0 ± 1.2 | 47 ± 6.3 | 28.1 ± 3.6 | 48.0 ± 6.3 | >100 |
| Lighta | 1.1 ± 0.4 | 6.9 ± 1.6 | 4.7 ± 1.1 | 2 ± 0.4 | >100 | |
| PIb | 10.0 | 6.8 | 6.0 | 24 | ||
| HLF | Dark | >100 | >100 | >100 | >100 | 69.0 ± 10.2 |
| Lighta | 12.4 ± 2.5 | 11.0 ± 3.1 | 18.2 ± 3.6 | 15.1 ± 3.7 | 57.2 ± 8.4 | |
| PIb | >8.1 | >9.1 | >5.5 | >6.62 | 1.1 | |
ROS generation under two-photon excitation
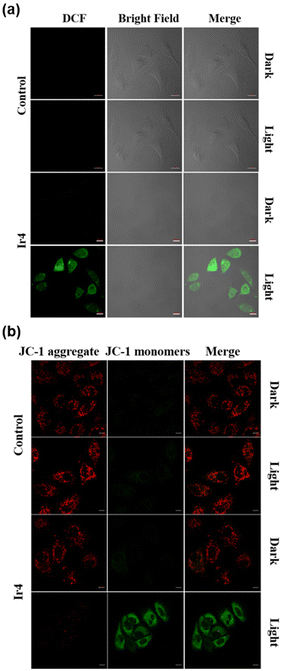
The utilization of DCFH-DA as a fluorescent probe for the detection of reactive oxygen species (ROS) within cells is a well-established technique. DCFH-DA is a non-fluorescent compound capable of readily traversing the cell membrane to enter live cells. Once inside, cellular esterases catalyze the hydrolysis of DCFH-DA, converting it into DCFH. Upon interaction with ROS, DCFH is oxidized to form the highly fluorescent compound 2,7-dichlorofluorescein (DCF). Consequently, the fluorescence intensity of intracellular DCF serves as an indirect measure of intracellular ROS levels.41 Confocal imaging in Fig. 3(a) showed negligible green fluorescence in the control group and the sample group in the dark, while the sample group showed obvious green fluorescence after two-photon laser irradiation (720 nm, 15 s). According to these results, N-Ir4 can generate significant ROS when exposed to two-photon irradiation.Mitochondrial membrane potential (MMP) assessment
Given that mitochondria have been identified as the primary sites for the internalization of Ir(III) complexes, it is reasonable to anticipate mitochondrial dysfunction in cells post-PDT. MMP analysis serves as an early indicator of mitochondrial activity. To assess the extent of mitochondrial damage, JC-1, a widely utilized commercial dye, is particularly well-suited for the analysis of the MMP. In mitochondria exhibiting high MMPs, JC-1 forms aggregates that emit red fluorescence. Conversely, in dysfunctional or damaged mitochondria characterized by low MMP, JC-1 remains in its monomeric form, emitting green fluorescence. Therefore, mitochondrial damage can be detected through this colorimetric shift from red to green. As shown in Fig. 3(b), following two-photon laser irradiation at 720 nm, the cells treated with N-Ir4 exhibit green fluorescence of monomer JC-1, indicating mitochondrial dysfunction. Conversely, the control group did not demonstrate any changes in the MMP despite undergoing two-photon laser irradiation.Live/dead dual staining
In the investigation of 2PA PDT, the calcein AM (live cell)/EthD-1 (dead cell) double staining technique is frequently employed to assess cell viability. This method enables the visual differentiation of live and dead cells through fluorescence labeling, thereby facilitating the evaluation of PDT's impact on cell survival. As illustrated in Fig. 4, cells were incubated with 2 μM N-Ir4. Prior to two-photon laser irradiation at 720 nm, both the experimental and control groups exhibited green fluorescence indicative of live cells. However, following two-photon laser irradiation at 720 nm, the control group continued to display green fluorescence. Following two-photon laser irradiation at 720 nm, the control group exhibited persistent green fluorescence with only negligible red fluorescence. In contrast, the cells treated with N-Ir4 demonstrated substantial red fluorescence and a markedly reduced green fluorescence. These observations indicate that the N-Ir4 complex induces cellular damage under two-photon irradiation.Two-photon laser penetrating ability and ROS generation in 3D multicellular tumor spheroids (MCTSs)
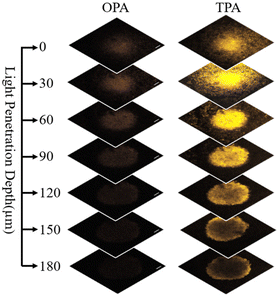
Three-dimensional (3D) cell spheres more accurately simulate tumor behavior compared to traditional two-dimensional (2D) cell cultures. Due to their structural similarity to actual tumors, 3D cell spheres encompass cells located on the spherical surface as well as cells embedded deep within the sphere. These spheres contain both proliferating and non-proliferating cells, with the central region often characterized by an anoxic environment. This structural configuration renders 3D cell spheres an optimal model for the investigation of tumor biology, drug screening, and drug delivery. Within the 3D cell sphere, intercellular interactions are more intimate, thereby emulating the intricate relationships observed among cells in vivo. This characteristic facilitates a more precise assessment of the comprehensive effects of pharmacological agents on cellular communities, rather than on individual cells.Using confocal laser scanning microscopy (CLSM) along the Z-axis, complexes can be visualized intuitively in sections at varying depths within the spheroids. As illustrated in Fig. 5, two-photon irradiation at 720 nm achieves a penetration depth of 180 μm, whereas one-photon irradiation at 405 nm is restricted to a depth of 60 μm. 2PA PDT thus demonstrates significant advantages in achieving deeper tissue penetration, which is crucial for the treatment of conditions such as deep-seated tumors.
The production of ROSs by N-Ir4 within 3D MCTSs was similar to the detection of ROS production in a monolayer cell model using DCFH-DA (Fig. 6). The cells were incubated with N-Ir4 (2 μM) for 8 h, followed by an additional 1 h of incubation with 1 μM DCFH-DA. Under dark conditions, the cells treated with N-Ir4 did not exhibit green fluorescence. However, upon two-photon irradiation, a green fluorescence signal was observed on the surface of the cell spheroid. This phenomenon is attributed to the requirement of oxygen for the production of 1O2 resulting in the absence of green fluorescence in the anoxic core of the spheroid.
Conclusions
In this study, we synthesized and fully characterized four novel binuclear Ir(III) complexes. We evaluated their UV-vis absorption spectra and fluorescence spectra and assessed the PDT efficacy in the A375 cell line. Based on the cellular uptake and MTT profile, complex N-Ir4 was chosen for further investigation. This complex specifically targets mitochondria and exhibits an IC50 value of 2.0 μM and a PI value of 24. In monolayer cells, it produces ROS in a 2PA manner, leading to MMP loss and cell death. Similar 2PA PDT therapeutic outcomes were observed in MCTSs without any appreciable compromise caused by the penetration depth of 180 nm. This study highlights a potential strategy to strengthen 2PA cross-sections via a simple multinuclear reconstruction process and may serve as a paradigm for the fast-developing field of PDT agents.Experimental section
General information
Unless otherwise indicated, all chemicals were purchased from commercial sources and were of analytical reagent grade. The materials 1,10-phenanthroline-5,6-dione, 4-tert-butyl benzenamine, 4-nitrobenzaldehyde, benzaldehyde, 4-fluoro-benzaldehyde, 4-methylbenzaldehyde, 4-cyanobenzaldehyde, and IrCl3·3H2O were purchased from Energy Chemical company. Palladium 10% on carbon, phosphate buffered saline (PBS), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were obtained from Sigma Aldrich Trading Co., Ltd (USA). Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) with 1% penicillin/streptomycin and 10% fetal bovine serum (FBS) at 37 °C under 5% CO2 and 95% air, among which DMEM, FBS, and penicillin/streptomycin were purchased from Gibco (USA). MitoTracker Deep Red and 2′,7′-dichlorofluorescein diacetate (DCFH-DA) were purchased from Thermo Fisher (USA). Calcein AM/PI cell viability/cytotoxicity assay kits were purchased from Beyotime. All buffer components were of biological grade, obtained from commercial suppliers, and used as received.1H spectra were recorded using a Bruker AVANCE III 400 MHz NMR spectrometer with DMSO-d6 as a deuterated solvent. Tetramethylsilane (TMS; δ = 0 ppm) was used as an internal standard. MALDI-TOF MS analysis was performed using a Bruker Autoflex mass spectrometer. The ultraviolet and visible (UV-Vis) absorption spectra were recorded using a PerkinElmer Lambda 950 spectrometer. Luminescence emission spectra were recorded using a Hitachi F-4500 luminescence spectrometer. The EPR measurements were determined using a Bruker Model EMXPlus-10/12 spectrometer at 298 K. Inductively coupled plasma mass spectrometry (ICP-MS) experiments were conducted using an iCAP RQ (ThermoFisher) spectrometer. Confocal fluorescence imaging was performed using an LSM-880 laser scanning confocal microscope (Carl Zeiss Co., Ltd, Germany) with an oil-immersion objective lens. Time-resolved emission measurements were performed using an SR-500I-D1 (femtosecond fluorescence spectrum measurement system, Coherent Inc.). 2PA cross-section measurements were performed using a mode-locked Yb:KGW laser (Light Conversion, Pharos) with a pulse width of 200 fs and a repetition rate of 10 kHz. Cell viability was measured using a SpectraMax CMax Plus (Molecular Devices) absorbance microplate reader.
Synthesis of ligand tbpip-NO2
The ligand tbpip-NO2 was synthesized following the synthetic method as described in our previous work: 1,10-phenanthroline-5,6-dione (420 mg, 0.2 mM), 4-tert-butylaniline (298 mg, 0.2 mmol), 4-nitrobenzaldehyde (302 mg, 0.2 mmol), and ammonium acetate (1.078 g, 1.4 mmol) were dissolved in 30 mL of acetic acid in a round bottom flask, purged with Ar, and refluxed at 120 °C for 12 h. The crude product was purified by silica gel column chromatography (dichloromethane/acetonitrile, v/v = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and the solvent was removed by rotary evaporation to obtain 857 mg of a pale yellow solid. Yield: 860 mg, 91%. ES-MS (CH3OH): m/z = 474.44 [M + H+]+.
1), and the solvent was removed by rotary evaporation to obtain 857 mg of a pale yellow solid. Yield: 860 mg, 91%. ES-MS (CH3OH): m/z = 474.44 [M + H+]+.
Synthesis of ligand tbpip-NH2
The ligand tbpip-NO2 (857 mg, 0.18 mmol) and Pd/C (212 mg, 0.2 mmol) were dissolved in 40 mL of ethanol and subjected to reflux at 65 °C for 1 h. Subsequently, a diluted solution of H2O2 in ethanol was added dropwise, and the reaction was allowed to proceed for an additional 12 h. The mixture was then subjected to filtration while hot, and the solvent was removed via rotary evaporation. The resulting product was recrystallized from ethanol. Yield: 651 mg, 82%. ES-MS (CH3OH): m/z = 444.42 [M + H+]+.Synthesis of Ir-tbpip-NH2
A mixture comprising the ligand tbpip-NH2 (620 mg, 0.14 mmol) and [Ir(ppy)2Cl]2 (749 mg, 0.07 mmol) in a solvent system of dichloromethane and methanol (5 mL each) was subjected to reflux under an argon atmosphere at 85 °C for a duration of 6 h. Upon completion of the reaction, the solvent was removed by evaporation to yield a solid residue. This residue was subsequently purified via column chromatography on alumina using a dichloromethane/methanol eluent to obtain a pure yellow product. Yield: 1003 mg, 73%. ES-MS (CH3OH): m/z = 944.58 [M − Cl−]+.Synthesis of the complex Ir1
A mixture comprising the complex Ir-tbpip-NH2 (240 mg, 0.0245 mmol) and the ligand 1,10-phenanthroline-5,6-dione (53 mg, 0.025 mmol), benzaldehyde (27 mg, 0.025 mmol), and ammonium acetate (100 mg, 0.13 mmol) was dissolved in 10 mL of acetic acid in a round bottom flask, purged with Ar, and refluxed at 135 °C for 12 h. The crude product was purified by silica gel column chromatography (dichloromethane/methanol, v/v = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and the solvent was removed by rotary evaporation to obtain 194 mg of crude product. Then the crude product (194 mg, 0.015 mmol) and [Ir(ppy)2Cl]2 (86 mg, 0.008 mmol) in a solvent system of dichloromethane and methanol (5 mL each) were subjected to reflux under an argon atmosphere at 85 °C for a duration of 6 h. Upon completion of the reaction, the solvent was removed by evaporation to yield a solid residue. This residue was subsequently purified via column chromatography on alumina using a dichloromethane/methanol eluent to obtain a pure yellow product, which recrystallized with toluene and dichloromethane. Yield: 125 mg, 46%. 1H NMR (400 MHz, d6-DMSO) δ 9.38–9.29 (m, 2H), 8.33–8.23 (m, 5H), 8.22–8.10 (m, 4H), 8.00–7.86 (m, 10H), 7.86–7.69 (m, 8H), 7.65–7.46 (m, 9H), 7.42 (dd, J = 8.2, 6.6 Hz, 2H), 7.27 (d, J = 8.6 Hz, 2H), 7.11–6.93 (m, 11H), 6.34–6.25 (m, 4H), 1.42–1.28 (m, 9H). ES-MS (CH3OH): m/z = 862.31 [M − 2Cl−]2+.
1), and the solvent was removed by rotary evaporation to obtain 194 mg of crude product. Then the crude product (194 mg, 0.015 mmol) and [Ir(ppy)2Cl]2 (86 mg, 0.008 mmol) in a solvent system of dichloromethane and methanol (5 mL each) were subjected to reflux under an argon atmosphere at 85 °C for a duration of 6 h. Upon completion of the reaction, the solvent was removed by evaporation to yield a solid residue. This residue was subsequently purified via column chromatography on alumina using a dichloromethane/methanol eluent to obtain a pure yellow product, which recrystallized with toluene and dichloromethane. Yield: 125 mg, 46%. 1H NMR (400 MHz, d6-DMSO) δ 9.38–9.29 (m, 2H), 8.33–8.23 (m, 5H), 8.22–8.10 (m, 4H), 8.00–7.86 (m, 10H), 7.86–7.69 (m, 8H), 7.65–7.46 (m, 9H), 7.42 (dd, J = 8.2, 6.6 Hz, 2H), 7.27 (d, J = 8.6 Hz, 2H), 7.11–6.93 (m, 11H), 6.34–6.25 (m, 4H), 1.42–1.28 (m, 9H). ES-MS (CH3OH): m/z = 862.31 [M − 2Cl−]2+.
Synthesis of the complex Ir2
The complex Ir2 was synthesized using a method analogous to that employed for the synthesis of complex Ir1, with the substitution of 4-methyl benzaldehyde in place of benzaldehyde. Yield: 162 mg, 60%. 1H NMR (400 MHz, DMSO-d6) δ 9.34 (dd, J = 13.3, 8.3, 1.5 Hz, 2H), 8.33–8.24 (m, 6H), 8.20–8.09 (m, 4H), 8.00–7.87 (m, 10H), 7.85–7.68 (m, 8H), 7.64 (dd, J = 8.6, 1.4 Hz, 1H), 7.54–7.41 (m, 7H), 7.23 (d, J = 8.1 Hz, 2H), 7.11–6.93 (m,12H),6.34–6.25 (m, 4H), 2.36 (s, 3H), 1.28 (d, J = 2.0 Hz, 9H). ES-MS (CH3OH): m/z = 869.98 [M − 2Cl− + H+]2+.Synthesis of the complex Ir3
The complex Ir3 was synthesized using a method analogous to that employed for the synthesis of complex Ir1, with the substitution of 4-cyanobenzaldehyde in place of benzaldehyde. Yield: 117 mg, 43%. 1H NMR (400 MHz, DMSO-d6) δ 9.35 (t, J = 8.3, 1.5 Hz,2H), 8.33–8.25 (m, 6H), 8.22–8.12 (m, 4H), 8.00–7.85 (m, 16H), 7.81–7.70 (m, 6H), 7.67–7.62 (m, 1H), 7.52 (m, 5H), 7.11–6.94 (m, 12H), 6.34–6.26 (m, 4H), 1.31–1.19 (m, 9H). m/z = 875.21 [M − 2Cl− + H+]2+.Synthesis of the complex Ir4
The complex Ir4 was synthesized using a method analogous to that employed for the synthesis of complex Ir1, with the substitution of 4-fluoro-benzaldehyde in place of benzaldehyde. Yield: 136 mg, 50%. 1H NMR (400 MHz, DMSO-d6) δ 9.34 (dd, J = 12.2, 8.3, 1.5 Hz, 2H), 8.32–8.24 (m, 6H), 8.21–8.11 (m, 4H), 8.00–7.88 (m, 10H), 7.86–7.79 (m, 4H), 7.74–7.68 (m, 3H), 7.65–7.58 (m, 3H), 7.57–7.47 (m, 5H), 7.31–7.22 (m, 3H), 7.12–6.94 (m, 12H), 6.34–6.26 (m, 4H), 1.28 (d, J = 1.6 Hz, 9H). m/z = 871.35 [M − 2Cl−]2+.log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) P measurement
P measurement
The lipid–water distribution coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PO/W) is frequently employed to assess the hydrophilicity or lipophilicity of compounds, as determined by the standard “shake-flask” method. The n-octanol/water partition coefficient (log
PO/W) is frequently employed to assess the hydrophilicity or lipophilicity of compounds, as determined by the standard “shake-flask” method. The n-octanol/water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) was calculated using eqn (1).
P) was calculated using eqn (1).log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = lg(C[solute]octanol/C[solute]water) P = lg(C[solute]octanol/C[solute]water) | (1) |
The concentrations of the solute in n-octanol and water, [solute]octanol and [solute]water, were measured through UV absorption. Equal volumes of n-octanol and water were combined in a centrifuge tube, and the mixture was agitated at 37 °C in a shaker incubator overnight. Subsequently, the mixture was allowed to stir for 24 hours before being left to stand for phase separation. The upper phase consisted of an n-octanol solution saturated with water, while the lower phase comprised an aqueous solution saturated with n-octanol. These two solutions were meticulously separated, and the complex (5 μM) was subsequently dissolved in the n-octanol solution saturated with water. An equivalent volume of the saturated n-octanol aqueous solution was then introduced, and the resultant mixture was agitated overnight, followed by continuous stirring for 24 hours. Subsequently, the aqueous and organic phases were separated, and the two phases were assayed at a wavelength of 290 nm with a UV/Vis spectrophotometer, namely [solute]Octanol and [solute]Water. The experimental group was parallel three times.
2PA cross-section measurements
| q0(0) = βI0Leff |
| Leff = (1 − e−α0L)/α0 |
The NLO and δ2PA coefficient uncertainties are estimated to be commonly ∼15% arising predominantly from (a) estimation errors of the beam waist (at the focus) and thereby leading to the peak intensity errors (which is a crucial parameter), (b) the input laser power fluctuation, (c) fitting errors, etc. We have attempted to compare the NLO coefficients obtained for the present set of molecules with literature reports. Pure solvent samples under the same experimental conditions show no significant results.
Singlet oxygen detection
The singlet oxygen yield of the complex was determined by an indirect method with the use of UV-Vis spectroscopy. ABDA was used as a 1O2 detection probe, while [Ru(bpy)3]2+ was used as the standard. The complex whose absorbance at 405 nm was adjusted to the same value of 0.25, and then mixed ABDA (100 μM) in methanol at quartz cuvettes. The above solution was irradiated with 405 nm light (10.9 mW cm−2) every 10 s, and the absorbance at 378 nm was measured. The ΦΔ value was determined using the following equations:| ΦsΔ = ΦrΔ × (ss × Fr)/(sr × Fs) |
| F = (1 − OD405s)/(1 − OD405r) |
Cell culture
A375 cell lines were procured from the American Type Culture Collection (ATCC). The cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 100 units per mL penicillin, and 50 μg mL−1 streptomycin. All cell cultures were maintained under conditions of 37 °C, 5% CO2, and 95% humidity. The cells were subcultured through successive trypsinization and re-seeding.Monolayer cell cytotoxicity tests
Cytotoxicity was assessed utilizing the MTT assay. A375 cell lines were seeded in 96-well plates at a density of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and incubated for 24 hours in DMEM supplemented with 10% FBS. Ir(III) complexes at various concentrations were introduced to the plates containing adherent cell lines and subsequently incubated for an additional 8 hours, after which the medium was replaced with fresh medium. Following the necessary incubation period, cells in the light treatment group were exposed to irradiation using a lamp with a 405 nm bandpass for 600 seconds (20 mW cm−2), while the dark treatment group was incubated for an additional 48 hours without irradiation. Subsequently, the MTT assay was performed. Subsequently, the MTT assay was conducted to assess cell viability. The culture medium was aspirated and replaced with 150 μL per well of DMSO. After shaking for 2 min, the OD595nm values were measured with a SpectraMax CMax Plus, Molecular Devices. The experimental group is parallel three times.
000 cells per well and incubated for 24 hours in DMEM supplemented with 10% FBS. Ir(III) complexes at various concentrations were introduced to the plates containing adherent cell lines and subsequently incubated for an additional 8 hours, after which the medium was replaced with fresh medium. Following the necessary incubation period, cells in the light treatment group were exposed to irradiation using a lamp with a 405 nm bandpass for 600 seconds (20 mW cm−2), while the dark treatment group was incubated for an additional 48 hours without irradiation. Subsequently, the MTT assay was performed. Subsequently, the MTT assay was conducted to assess cell viability. The culture medium was aspirated and replaced with 150 μL per well of DMSO. After shaking for 2 min, the OD595nm values were measured with a SpectraMax CMax Plus, Molecular Devices. The experimental group is parallel three times.
Cellular localization
An overnight culture of A375 cells at a concentration of 1 × 105cells per mL was established in Corning confocal dishes at 37 °C. Following an 8-hour treatment with the complexes at a concentration of 2.0 μM, the cells were rinsed with PBS and subsequently stained with the commercially available MitoTracker Deep Red at a concentration of 50 nM for 30 minutes. The cells were then washed with PBS to remove any excess dye. Confocal fluorescence microscopy was performed using a Zeiss LSM 880 laser microscopy system to investigate colocalization.Cell uptake
A375 cancer cells were seeded into a 6-well plate for 24 h. The medium was removed and replaced with Ir1–4 and N-Ir1–4 in plain DMEM. After different times (2–10 h) of incubation, the cells were scraped out, collected in PBS (3 mL), and counted. The cells were pelletized, and HNO3 (65%, 1 mL) was added to these cells, and the mixture was incubated at room temperature for 24 h to digest entirely. The solution was then diluted to a final volume of 10 mL with Milli-Q water, and the cellular concentration of iridium was measured using ICP-MS.ROS level detection
The DCFH-DA assay was employed to measure intracellular ROS production by the complexes under two-photon excitation. A375 cells were seeded in a confocal laser dish at a density of 1 × 105 cells per mL and subsequently incubated with the complex (2.0 μM) for 8 h. Following this, the culture medium was replaced, and the cells were incubated with the fluorescent probe DCFH-DA (1 μM) for 20 minutes. The experimental group was subjected to two-photon irradiation (6 J, 300 s), while the dark group was without treatment. Fluorescent images were gathered using an excitation wavelength of 488 nm, and the emission was recorded between 510 and 540 nm. The production of ROSs was assessed by using the same protocol in 3D multicellular spheroids.Mitochondrial membrane potential detection
The A375 cells were seeded in a confocal laser dish at a density of 1 × 105 cells per mL and subsequently incubated with the complex (2.0 μM) for 8 hours. Following incubation, the culture medium was replaced, and the cells were treated with JC-1 according to the manufacturer's instructions. Imaging of the samples was performed using a confocal laser scanning microscope. Fluorescence images of the JC-1 aggregates were acquired with an excitation wavelength of 543 nm, and emissions were recorded between 580 and 610 nm. For the JC-1 monomer, the images were captured using an excitation wavelength of 488 nm, with emissions recorded between 510 and 540 nm. The experimental groups subjected to light exposure underwent two-photon irradiation at 720 nm with an energy dose of 6 J, followed by a 20-minute incubation period. Subsequently, these samples were imaged using a confocal microscope, ensuring consistent imaging conditions across all samples. Control groups were maintained under identical conditions to the experimental groups, with the exception that they were not incubated with Ir(III) complexes.Live/dead viability assay
A375 cells were seeded in 96-well plates at a density of 1 × 104 cells per well and incubated with complexes at a concentration of 2.0 μM for 8 hours. Subsequently, the culture medium was replaced, and calcein AM along with ethidium homodimer-1 (EthD-1) was added according to the manufacturer's instructions. Following additional incubation, the cells were imaged using a confocal microscope. For the dark groups, fluorescence images of calcein AM were acquired using an excitation wavelength of 488 nm, with emission recorded between 510 and 540 nm. For EthD-1, excitation was performed at a wavelength of 530 nm, and emission was recorded between 640 and 670 nm. In light groups, the cells were exposed to two-photon irradiation (720 nm, 6 J) and photographed under the same conditions. Controls were managed under the same conditions as the experimental group except for the incubation of Ir(III) complexes.Z-Axis scanning of 3D MCSs
The culture of 3D multicellular spheroids was prepared according to a previously reported method. A suspension of 1.5% agarose in fresh PBS and A375 cells was added to a 96-well microassay culture plate until the total volume was 200 μL per well. A375 3D MCSs with an average diameter of 800 μm were cultured in a 37 °C incubator for 3 days. At the same time, the culture medium was replaced every two days. Then the A375 3D MCSs were incubated with Ir(III) complexes (2 μM) for 8 h. Then the cells were imaged using a confocal microscope. For one-photon imaging, the excitation wavelength was 405 nm, while it was 720 nm for two-photon imaging.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22120102002, 92353301, and 22477105), the Science and Technology Innovation Program of the Hunan Province of China (2021RC5028), the Natural Science Foundation of Xiamen, China (3502Z202471010), the Chancellor's Fund for Xiamen University (20720240078), and the AMS Newton International Fellowship (NIFR7/1063). Scheme 1 was drawn using Fig. 2.References
- P. Agostinis, K. Berg, K. A. Cengel, T. H. Foster, A. W. Girotti, S. O. Gollnick, S. M. Hahn, M. R. Hamblin, A. Juzeniene, D. Kessel, M. Korbelik, J. Moan, P. Mroz, D. Nowis, J. Piette, B. C. Wilson and J. Golab, Ca-Cancer J. Clin., 2011, 61, 250–281 CrossRef PubMed.
- K. Yeoh and A. Gray, Clin. Oncol., 2022, 34(9), e377–e382 CrossRef CAS PubMed.
- Y. A. Hagyousif, B. M. Sharaf, R. A. Zenati, W. El-Huneidi, Y. Bustanji, E. Abu-Gharbieh, M.AY. Alqudah, A. D. Giddey, A. Y. Abuhelwa, K. H. Alzoubi, N. C. Soares and M. H. Semreen, Int. J. Mol. Sci., 2023, 13, 1604 CrossRef PubMed.
- P. Agostinis, K. Berg, K. A. Cengel, T. H. Foster, A. W. Girotti, S. O. Gollnick, S. M. Hahn, M. R. Hamblin, A. Juzeniene, D. Kessel, M. Korbelik, J. Moan, P. Mroz, D. Nowis, J. Piette, B. C. Wilson and J. Golab, Ca-Cancer J. Clin., 2011, 61, 250–281 CrossRef PubMed.
- X. Zhao, J. Liu, J. Fan, H. Chao and X. Peng, Chem. Soc. Rev., 2021, 50, 4185–4219 RSC.
- Z. Xie, T. Fan, J. An, W. Choi, Y. Duo, Y. Ge, B. Zhang, G. Nie, N. Xie, T. Zheng, Y. Chen, H. Zhang and J. S. Kim, Chem. Soc. Rev., 2020, 49, 8065 RSC.
- D. Dolmans, D. Fukumura and R. K. Jain, Nat. Rev. Cancer, 2003, 3, 380–387 CrossRef CAS PubMed.
- X. Wang, J. Peng, C. Meng and F. Feng, Chem. Sci., 2024, 15, 12234–12257 RSC.
- R. L. Siegel, K. D. Miller, N. S. Wagle and A. Jemal, CA Cancer J. Clin., 2023, 73, 17–48 CrossRef PubMed.
- J. P. Celli, B. Q. Spring, I. Rizvi, C. L. Evans, K. S. Samkoe, S. Verma, B. W. Pogue and T. Hasan, Chem. Rev., 2010, 110, 2795–2838 CrossRef CAS PubMed.
- T. C. Pham, V. N. Nguyen, Y. Choi, S. Lee and J. Yoon, Chem. Rev., 2021, 121, 13454–13619 CrossRef CAS PubMed.
- W. Fan, P. Huang and X. Chen, Chem. Soc. Rev., 2016, 45, 6488–6519 RSC.
- M. Ethirajan, Y. Chen, P. Joshi and R. K. Pandey, Chem. Soc. Rev., 2011, 40, 340–362 RSC.
- N. Patel, P. Pera, P. Joshi, M. Dukh, W. A. Tabaczynski, K. E. Siters, M. Kryman, R. R. Cheruku, F. Durrani, J. R. Missert, R. Watson, T. Y. Ohulchanskyy, E. C. Tracy, H. Baumann and R. K. Pandey, J. Med. Chem., 2016, 59, 9774–9787 CrossRef CAS PubMed.
- M. R. Hamblin, Photochem. Photobiol., 2020, 96, 506–516 CrossRef CAS PubMed.
- D. Tang, M. Cui, B. Wang, C. Xu, Z. Cao, J. Guo, H. Xiao and K. Shang, Adv. Mater., 2024, 36, 2406815 CrossRef CAS PubMed.
- J. S. Nam, M. Kang, J. Kang, S. Park, S. J. Lee, H. Kim, J. Seo, O. Kwon, M. H. Lim, H. Rhee and T. Kwon, J. Am. Chem. Soc., 2016, 138, 10968–10977 CrossRef CAS PubMed.
- W. Li, T. Li, Y. Pan, S. Li, G. Xu, Z. Zhang, H. Liang and F. Yang, J. Med. Chem., 2024, 67, 3843–3859 CrossRef CAS PubMed.
- W. Lin, Y. Liu, J. Wang, Z. Zhao, K. Lu, H. Meng, R. Luoliu, X. He, J. Shen, Z. Mao and W. Xia, Angew. Chem., Int. Ed., 2023, 62, e202310158 CrossRef CAS PubMed.
- L. Wang, R. Guan, L. Xie, X. Liao, K. Xiong, T. W. Rees, Y. Chen, L. Ji and H. Chao, Angew. Chem., Int. Ed., 2021, 60, 4657–4665 CrossRef CAS PubMed.
- N. Neelambaran, S. Shamjith, V. P. Murali, K. K. Maiti and J. Joseph, ACS Appl. Bio. Mater., 2023, 6(12), 5776–5788 CrossRef CAS PubMed.
- G. Xu, Q. Liang, L. Gao, S. Xu, W. Luo, Q. Wu, J. Li, Z. Zhang, H. Liang and F. Yang, J. Med. Chem., 2024, 67, 19573–19585 CrossRef CAS PubMed.
- W. Li, T. Li, Y. Pan, S. Li, G. Xu, Z. Zhang, H. Liang and F. Yang, J. Med. Chem., 2024, 67, 3843–3859 CrossRef CAS PubMed.
- M. Huang, J. Cui, Q. Wu, S. Liu, D. Zhu, G. Li, M. R. Bryce, D. Wang and B. Tang, Inorg. Chem., 2024, 63, 24030–24040 CrossRef CAS PubMed.
- L. Wei, R. Kushwaha, T. Sadhukhan, H. Wu, A. Dao, Z. Zhang, H. Zhu, Q. Gong, J. Ru, C. Liang, P. Zhang, S. Banerjee and H. Huang, J. Med. Chem., 2024, 67, 11125–11137 CrossRef CAS PubMed.
- F. Helmchen and W. Denk, Nat. Methods, 2005, 2, 932–940 CrossRef CAS PubMed.
- A. Soleimany, D. K. Aghmiouni, M. Amirikhah, M. A. Shokrgozar, S. Khoee and B. Sarmento, Adv. Funct. Mater., 2024, 7, 2408594 CrossRef.
- Y. Chen, R. Guan, C. Zhang, J. Huang, L. Ji and H. Chao, Coord. Chem. Rev., 2016, 310, 16–40 CrossRef CAS.
- S. Golovynskyi, I. Golovynska, L. I. Stepanova, O. I. Datsenko, L. Liu, J. Qu and T. Y. Ohulchanskyy, J. Biophotonics, 2018, 11, 201800141 CrossRef PubMed.
- J. Tian, B. Huang, M. H. Nawaz and W. Zhang, Coord. Chem. Rev., 2020, 420, 213410 CrossRef CAS.
- C. Jin, J. Liu, Y. Chen, L. Zeng, R. Guan, C. Ouyang, L. Ji and H. Chao, Chem. – Eur. J., 2015, 21, 12000–12010 CrossRef CAS PubMed.
- K. Qiu, H. Huang, B. Liu, Y. Liu, Z. Huang, Y. Chen, L. Ji and H. Chao, ACS Appl. Mater. Interfaces, 2016, 8, 12702–12710 CrossRef CAS PubMed.
- W. Dröge, Physiol. Rev., 2002, 82, 47–95 CrossRef PubMed.
- M. M. Kim, R. Penjweini and T. C. Zhu, J. Biomed. Opt., 2017, 22, 028002 CrossRef PubMed.
- Z. Tan, M. Lin, J. Liu, H. Wu and H. Chao, Dalton Trans., 2024, 53, 12917–12926 RSC.
- Y. Chen, T. W. Rees, L. Ji and H. Chao, Curr. Opin. Chem. Biol., 2018, 43, 51–57 CrossRef CAS PubMed.
- C. Jin, F. Liang, J. Wang, L. Wang, J. Liu, X. Liao, T. W. Rees, B. Yuan, H. Wang, Y. Shen, Z. Pei, L. Ji and H. Chao, Angew. Chem., Int. Ed., 2020, 59, 15987–15991 CrossRef CAS PubMed.
- H. Yuan, Z. Han, Y. Chen, F. Qi, H. Fang, Z. Guo, S. Zhang and W. He, Angew. Chem., Int. Ed., 2021, 60, 8174–8181 CrossRef CAS PubMed.
- H. Huang, S. Banerjee, K. Qiu, P. Zhang, O. Blacque, T. Malcomson, M. J. Paterson, G. J. Clarkson, M. Staniforth, V. G. Stavros, G. Gasser, H. Chao and P. J. Sadler, Nat. Chem., 2019, 11, 1041–1048 CrossRef CAS PubMed.
- R. Guan, L. Xie, T. W. Rees, L. Ji and H. Chao, J. Inorg. Biochem., 2019, 204, 110985 CrossRef PubMed.
- D. Yu, Y. Zha, Z. Zhong, Y. Ruan, Z. Li, L. Sun and S. Hou, Sens. Actuators, B, 2021, 339, 129878 CrossRef CAS.
- M. Sheik-Bahae, A. A. Said, T. H. Wei, D. J. Hagan and E. W. V. Stryland, IEEE J. Quantum Electron., 1990, 26, 760–769 CrossRef CAS.
- M. Nikolay, D. Mikhail and R. Aleksander, Opt. Express, 2008, 16, 4029–4047 CrossRef PubMed.
- W. Feng, K. Liu, J. Y. Zang, G. Wang, R. Miao, L. Ding, T. Liu, J. Kong and Y. Fang, ACS Appl. Mater. Interfaces, 2021, 13, 28985–28995 CrossRef CAS PubMed.
- N. Zhang, L. Liu, H. X. Chang, K. Liu, T. Liu, L. Ding and Y. Fang, J. Phys. Chem. Lett., 2023, 14, 7283–7289 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dt03426k |
| This journal is © The Royal Society of Chemistry 2025 |