Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Preparation of bovine serum albumin nanospheres via desolvation: a study of synthesis, characterization, and aging†
Blake A.
Bartlett
 ,
John
Klier
* and
Sepideh
Razavi
,
John
Klier
* and
Sepideh
Razavi
 *
*
School of Sustainable Chemical, Biological and Materials Engineering, University of Oklahoma, Norman, OK 73019, USA. E-mail: srazavi@ou.edu
First published on 10th January 2025
Abstract
Serum albumin has myriad uses in biotechnology, but its value as a nanocarrier or nanoplatform for therapeutics is becoming increasingly important, notably with albumin-bound chemotherapeutics. Another emerging field is the fabrication of biopolymeric nanoparticles using albumin as a building block to achieve highly-tunable nonimmunogenic capsules or scaffolds that may be cheaply and reliably produced. The aim of this study was to characterize and optimize the desolvation process used for fabrication of albumin nanoparticles under ambient conditions, studying both glutaraldehyde (GT) and glucose (GLU) as crosslinking agents and the effect of various synthesis conditions including pH, electrolyte concentration, and rate of desolvation on particle size and stability. Particle size, polydispersity index, and zeta potential were investigated, morphology was examined using scanning electron microscopy (SEM), and long-term stability and degradation modes were studied using dynamic light scattering (DLS) and transmission electron microscopy (TEM). It was determined that the optimized synthesis procedure for synthesis of Bovine Serum Albumin (BSA) nanoparticles at the investigated scale under ambient conditions was addition of ethanol at a rate of 0.625 mL min−1via infusion against the vial wall and a pH of 9 with the addition of no other electrolytes. Optimized BSA nanoparticles were synthesized at a size of 86 ± 3.7 nm (σ = 1.85) using glutaraldehyde as a crosslinker and a size of 92 ± 1.9 nm (σ = 0.95) using glucose as a crosslinker with polydispersity indices of 0.08 and 0.05, respectively. Nanoparticles synthesized via the optimized procedure, using both crosslinkers, were found to maintain colloidal stability significantly longer than cases previously reported in the literature, with insignificant changes in hydrodynamic size many months after synthesis.
1 Introduction
Advances in immunotherapy research have shown antibody-based therapies to be potent in the treatment of cancer.1 However, existing small-molecule therapeutics often face significant challenges in engineering and suffer from poor biological half-lives, which necessitates larger doses or extended treatment times that strain what limited production infrastructure exists. These shortcomings have emphasized the importance of nanoparticle-based delivery systems, which aim to improve drug distribution in the body. Both for existing therapies and for those that continue to be developed, there is a need for stable, biocompatible nanocarriers that are easy to produce. Biopolymeric nanoparticles produced with native or homologous proteins can be nonimmunogenic, nontoxic, biodegradable, biocompatible, and may have native biological functions that complement the nanocarrier system if the protein building block is chosen carefully, making these types of particles an attractive option. Serum albumin is abundant in nearly all biological fluids, most notably blood, and as an endogenous protein has all of these desired traits.2Albumin is essential for the maintenance of oncotic blood pressure and pH in the body, so it is slowly metabolized by the body and enjoys a sizable half-life of around 19 days.2,3 Albumin has no enzymatic function, but acts as a transport molecule for lipids, fatty acids, and hormones, making it important in the circulation of hydrophobic small-molecule drugs like ibuprofen, naproxen, warfarin, and many more.4–6 The critical role of albumin in small-molecule transport has a strong positive correlation with the growth and survival of eukaryotic cell cultures,7 and albumin-bound therapeutics have exploited this role to great effect. The most famous example of the benefits of albumin as a nanocarrier is Abraxane®, an albumin-bound formulation of paclitaxel used for treatment of several types of cancer.2,8 In fact, for chemotherapeutic drugs that are intended to enter the circulatory system, albumin is particularly well-suited for protein-drug conjugates due to its ability to rapidly diffuse across leaky blood vessels in tumors7 and tendency to concentrate in some cancerous tissues due to a combination of binding to upregulated proteins2,8 and upregulation of amino acid metabolism within cancer cells.9,10 While albumin does not cross the blood–brain barrier in healthy individuals, upregulation of Gp60 (60 kDa glycoprotein, also known as albondin) in many brain cancers (such as gliomas) enables transport into tumor tissue via transcytosis.11 Further, from a practical economic perspective, both bovine serum albumin (BSA) and human serum albumin (HSA) are commercially available in large, pure quantities for relatively small costs. These two proteins have 75.8% amino acid identity and 88% amino acid similarity, with residues important for tertiary structure highly conserved.4 As a result, the proteins may be used interchangeably in laboratory experiments. HSA is generally preferred in actual nanomedicine formulations, but there is some precedent for the proteins still being interchangeable in these situations.3
Synthesis of albumin nanospheres as nanovectors is well documented in the literature,11 with synthesis primarily being through one of two methods: high-pressure homogenization (HPH) and desolvation, although other methods also exist.2,12 Particles formed by high-pressure homogenization are nanoemulsions (also called miniemulsions) formed by strong shear forces temporarily altering the tertiary structure of the protein,13 and the method is popular for the industrial preparation of protein or protein-conjugate drugs. These emulsions rapidly solubilize upon injection or other mixture with protein solutions, so the nanoparticle character of the formulation is really only true when the drug is first formulated. In fact, Abraxane®, which is formulated using high-pressure homogenization, is typically supplied to clinics as a lyophilized powder that is reconstituted when needed, with the resulting dispersion only stable for around 24 hours.14 These extreme conditions can also result in a permanent partial denaturation of the protein that potentially decreases biological activity.15 There are some examples of albumin-based HPH particles used for drug loading,16 as well as some research demonstrating crosslinking of particles following HPH to improve stability,17 but research being done in these areas account for a small subset of overall HPH investigations relative to the “traditional” method of relying on steric entanglements to preserve nanoemulsion structure. The desolvation method involves mixing an aqueous protein solution with a desolvating agent, a water-miscible solvent (typically an alcohol, acetone, or similar organic solvent) in which the protein is insoluble. This results in nucleation and growth of small protein aggregates. Done properly, this process results in a highly uniform distribution of spherical particles whose size and polydispersity may be tuned via a handful of variables including temperature, protein concentration, desolvating agent identity, desolvating agent addition rate (and mode), stir rate, and stir time before crosslinker addition.11,12 The desolvation method is used in this study, with major motivations being investigation of nanoparticle long-term stability and size uniformity using that method of particle formation, as well as minimization of size using ambient conditions. This study will also investigate the use of both glutaraldehyde and glucose as crosslinkers (further discussed below). The goal of this work is to investigate the synthesis procedure using nontoxic alternative chemistries competitive with glutaraldehyde in terms of size, stability, reproducibility, and morphology.
The addition of desolvating agent causes coacervation of solid protein particles from an aqueous solution via replacement of adsorbed water molecules, which denatures the protein in the process.18 However, this denaturation is reversible, and it is often this phenomenon that is exploited to fractionate albumin (sometimes called “Fraction V”) from blood samples at industrial scale.6 The ideal procedure yields small, monodisperse colloidal particles, and the conditions where this occurs are similar to those used to synthesize albumin hydrogels (differing primarily in concentrations of solutes and desolvating agent),19 so resultant particles exhibit certain gel-like properties. To achieve long-term stability, addition of a crosslinker is necessary, as the basic desolvated gel-like particles are soft and tend to agglomerate when they collide in solution, eventually resulting in flocculation and sedimentation. In previous studies, a wide variety of crosslinker molecules have been studied, most commonly glutaraldehyde,18,20–23 but also glucose,24 sorbitol, ascorbic acid, citric acid, and tannic acid,20 as well as 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and similar carbodiimides.25 While the exact chemical mechanism of crosslinking varies (and in some cases is not fully understood26–28), the end result is generally a linkage of two basic amino acids (with the protein N-terminus also available if accessible). Aldehyde groups, like those in glutaraldehyde or in aldoses like D-glucose, rapidly form imines with accessible lysine residues, particularly deprotonated lysines. In the case of glutaraldehyde, the final crosslink is simply a diimine between two lysine residues (see Fig. 1A). In crosslinking with glucose, the final crosslink may include many advanced glycation end products (AGEs), or nonenzymatic glycosylation products, but the most common by an order of magnitude is glucosepane, a lysine-arginine linkage.27 This linkage begins with an imine forming between a lysine and the terminal aldehyde of D-glucose, forming a Schiff base. This intermediate rearranges to an enaminol and then an Amadori product. At the time of writing, it is not conclusively known how the reaction proceeds from this point to the ring closure and reaction with arginine to form glucosepane, but it likely involves a carbonyl shift down the length of the carbohydrate to eliminate the C-4 hydroxyl group and a series of nucleophilic attacks by an arginine resulting in the formation of the bicylic structure and elimination of three water molecules (see Fig. 1B).27 In both cases, the reaction proceeds spontaneously at ambient temperature, albeit slowly enough that an overnight equilibration step is necessary.
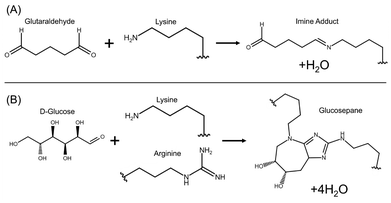 | ||
| Fig. 1 Overview of chemical reactions for the two crosslinkers studied. (A) Glutaraldehyde crosslinking involves formation of an imine from an aldehyde and a primary amine. The crosslink is formed once both terminal aldehydes have reacted, forming a diimine. For a detailed mechanism of this reaction and overview of rare side reactions, see Farris et al. 2010.29 (B) Glucose crosslinking begins with a lysine residue reacting with the the terminal aldehyde of D-glucose forming an imine, but the following linkage formed with an arginine residue is more complicated and not fully understood. However, the glucosepane product is by far the most plentiful glycation adduct. For more information on glycation reactions and the mechanism of this reaction, see Biemel et al. 2002.27 | ||
There is little published information regarding how long desolvated and crosslinked particles remain stable, which is necessary to determine any sort of shelf stability. Specific applications, such as nanoencapsulation and artificial antigen-presenting cells, also necessitate a high degree of particle surface stability to maintain efficacy.30–33 Jun et al. 201123 report 100 nm BSA nanoparticles crosslinked with GT aggregating into a solid cake within three months of synthesis. In their study, particles were synthesized via desolvation using acetone followed by GT crosslinking, washing, and redispersion in absolute ethanol using a probe sonicator. Amighi et al. 202020 report significant increases to both average size and polydispersity (recorded using DLS) of BSA nanoparticles crosslinked with GT and GLU within 28 days of synthesis. For example, average size of GT-crosslinked particles is reported to change from 121 ± 2.6 nm at the time of synthesis to 148 ± 3.1 nm at 28 days after synthesis, over a 20% increase. No numerical data for any time thereafter is provided, only stating that samples “were found stable at pH 9 for 3 months”. In their study, particles were synthesized via desolvation using ethanol followed by GT crosslinking, washing, and redispersion in ultrapure water using a probe sonicator. Thalhammer-Thurner & Debbage 202334 report a “minimal tendency to aggregate” after three years, but support this claim with only a single DLS measurement with high polydispersity. At the time of writing, we are unaware of any other publication that quantitatively studies long-term stability of albumin nanoparticles. These previous studies imply a relatively short period of stability, and quantitative data beyond 28 days after synthesis is extremely limited. As the applications of albumin-based nanoparticles moves beyond controlled-release drug formulations, there is a critical need to improve and characterize the long-term stability of these particles that could serve as a platform for cell-targeting therapies.
The usefulness of albumin as a nanocarrier is well established in the literature,2,3,7,8 as is its versatility as a building block for the synthesis of biopolymeric nanoparticles.35,36 However, there has been ongoing discussion regarding the optimization of a nanoparticle synthesis procedure, including what it means to be “optimized”. Generally, smaller particles are more desirable because they have more potential pathways for interaction with cells and a lower chance of being filtered from the bloodstream by the kidneys or by phagocytes.21 Much of the precedent in synthesis of albumin nanoparticles has had the drug loading application in mind (taking advantage of those gel-like properties previously mentioned),2,3,10,21,22,35 but innovations in immunotherapy demonstrate the need for scaffolds to help these new therapeutics circulate.37–39 Depending on the desired application, there may be a lower bound for particle size, as particularly small nanoparticles may not be capable of sufficient drug loading for controlled release applications. Likewise, antibody-based immunotherapy applications often benefit from multifunctional antigen binding that necessitates room for multiple ligands to be bound. Regardless, minimizing size is often the primary goal of the optimization process, but increasing scrutiny has been given to simplifying the synthesis procedure and eliminating toxic reagents. This is the primary motivation for studying glucose as an alternative to glutaraldehyde. While there has been some success using glucose as a crosslinker, reported particles were significantly larger and disintegrated far faster than otherwise identical particles that used glutaraldehyde as a crosslinker.20
Size, yield, and polydispersity of nanoparticles are highly influenced by numerous operating conditions, with the “ideal” conditions often within a narrow range. Because so many variables are at play, what is “optimum” under one set of conditions is often different than those reported elsewhere, and some trends can even reverse when different variables are controlled. Langer et al. 200321 and Tarhini et al. 201840 both reported that protein concentration has little effect on size, whereas Amighi et al. 202020 and Kaur & Singh 201541 reported the opposite. Amighi et al. 202020 reported that the relationship between temperature and particle size depends on the desolvating agent, with higher temperatures being associated with smaller particles when acetone is used, but with larger particles when ethanol is used. von Storp et al. 201218 reported that particle diameter generally increases with desolvating agent concentration, but the trend is inverted if methanol is used. Despite the constantly shifting landscape of “optimized” procedures, some parameters have been well-described. For example, Weber et al. 200042 reported that nanoparticle size is associated with parameters such as initial pH and amount of desolvation agent, and these findings have been largely included in following reported procedures. However, many of these reported procedures have optimized for only one or two parameters, and often involve harsh conditions and toxic chemistries. Tanjung et al. 202412 provides an overview of 23 experimental studies of albumin nanoparticle synthesis using desolvation and categorizes them based on the studied variables. Of the 23 articles, only 5 studied the effect of pH, only 3 studied the effect of alternative crosslinkers, none studied the effect of desolvating agent addition rate, and none studied long-term stability. Our own literature review has identified only a handful of other articles in these areas, indicating a significant gap in our understanding of particle synthesis. We have studied the effects of various important parameters including initial pH, desolvating agent addition rate, and crosslinker type on particle size with an emphasis on greener nontoxic chemistries and conditions, namely ambient temperatures and glucose as an alternative crosslinker. Additionally, since it is desired that the size and surface properties of nanoparticles, especially those used as nanovectors, be tunable, syntheses were performed in a variety of electrolyte concentrations to determine any permanent effects on particle size, surface attributes, and stability. These added electrolytes are in addition to the ions added by pH equilibration. Further, samples were also stored in (that is to say, redispersed in during the washing process) a variety of electrolyte concentrations to determine whether biosimilar electrolytic strength is sufficient to induce aggregation, with the goal of verifying whether BSA nanoparticles are colloidally stable and suitable for in vivo use.
2 Materials and methods
2.1 Materials
Bovine serum albumin (Fraction V, purity ≥98%) was obtained from Fisher. A 50 wt% aqueous glutaraldehyde solution was obtained from TCI America and diluted to an 8 wt% solution in deionized water for use as a crosslinker. α-D-Glucose (anhydrous, purity ≥96%) was obtained from Sigma-Aldrich and dissolved in deionized water to make a 15 wt% solution for use as a crosslinker. Potassium nitrate (purity ≥99%) was obtained from Sigma-Aldrich, potassium hydroxide was obtained from Macron (purity ≥85%), phosphate-buffered saline (PBS) was obtained from Thermo Scientific (10× concentration, pH 7.4), and absolute ethanol was obtained from Fisher. Uranyl acetate (purity ≥98%) was obtained from Sigma-Aldrich. All reagents were of analytical grade and were used as received. Ultrapure deionized water was dispensed using a Milli-Q® benchtop water purification system (resistivity of 18.2 MΩ cm). SEM pin stub specimen mounts were obtained from Ted Pella. Carbon-coated copper TEM grids were obtained from Sigma-Aldrich.2.2 Synthesis procedure
The procedure for fabrication of nanoparticles was adapted from the work of Amighi et al. 202020 and Jun et al. 201123 with modifications resulting from the optimization studies described in section 3. First, 40 mg of BSA was dissolved in 2 mL of ultrapure water (or electrolyte solutions ranging from 1 mM KNO3 to 1× PBS, see section 3.4) to make a 20 mg mL−1 or 2 wt% protein solution. Solution was stirred until all BSA was fully dissolved, then equilibrated to pH of 9 using 1 M KOH (pH values ranging from 6.5–10.5 were studied, with further discussion in section 3.1). While stirring at a rate of 600 rpm, a New Era Pump Systems NE-1000 syringe pump was used to infuse 8 mL of absolute ethanol at a rate of 0.625 mL min−1 (varied between 0.25 and 1.0 mL min−1 to determine optimum rate, discussed further in section 3.2). Ethanol was infused against the glass, as opposed to dropwise addition. Care was taken to ensure that the syringe tubing was never submerged in the bulk solution. In cases of dropwise addition and submerged tubing, the solution would have localized high concentrations of ethanol resulting in protein aggregation and sedimentation, high polydispersity, and lower particle yield. The solution became slightly turbid shortly before ethanol addition was complete, visually appearing as the clear solution becoming white and cloudy (see ESI Video S1†). Immediately after ethanol addition, 120 μL of a 8 wt% glutaraldehyde solution or 15 wt% glucose solution were added. The vial was then covered to prevent evaporation and the dispersion was allowed to stir for at least 12 hours, preferably overnight. All syntheses were performed at ambient temperature ≈25 °C.Samples were washed to remove unreacted components and ethanol via centrifugation (Thermo Scientific Fresco 21 microcentrifuge) at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 50 minutes at a temperature of 10 °C, a common practice for high spin speeds and protein-based samples, which tend to be heat-sensitive. Supernatant was disposed and replaced with an equivalent volume of 1 mM KNO3 or ultrapure water (or electrolyte solutions up to 1 M KNO3, see section 3.7). Pelletized particles were redispersed using a VWR Analog Vortex Mixer. Centrifugation, washing, and redispersal steps were repeated two additional times for a total of three (3) cycles.
000g for 50 minutes at a temperature of 10 °C, a common practice for high spin speeds and protein-based samples, which tend to be heat-sensitive. Supernatant was disposed and replaced with an equivalent volume of 1 mM KNO3 or ultrapure water (or electrolyte solutions up to 1 M KNO3, see section 3.7). Pelletized particles were redispersed using a VWR Analog Vortex Mixer. Centrifugation, washing, and redispersal steps were repeated two additional times for a total of three (3) cycles.
2.3 Storage
Following synthesis, washing, and redispersal steps, nanoparticle dispersions were transferred by pipette to a clean vial. Dispersions were labeled and stored in a refrigerator at 5 °C. Aliquots removed for DLS measurements were retained for future measurements and returned to storage, but aliquots removed for zeta potential measurements were discarded.2.4 Characterization
![[X with combining overline]](https://www.rsc.org/images/entities/i_char_0058_0305.gif) , where PDI = σ2/
, where PDI = σ2/![[X with combining overline]](https://www.rsc.org/images/entities/i_char_0058_0305.gif) 2. The value describes the distribution of sizes within the sample, with smaller values indicating more homogenous (or monodisperse) populations. Values smaller than 0.05 indicate a highly monodisperse sample, whereas values greater than 0.7 indicate a broad size distribution not suitable for DLS analysis.43 DLS measurements of average size were rounded to the nearest whole number, with error bars rounded to the nearest tenth of a nanometer. All measurements were performed in quintuplicate, and error bars in figures and tables represent two standard deviations (2σ) unless stated otherwise. For each reported variation in synthesis procedure, experiments were independently repeated n times, with the specific sample sizes reported in relevant figures below.
2. The value describes the distribution of sizes within the sample, with smaller values indicating more homogenous (or monodisperse) populations. Values smaller than 0.05 indicate a highly monodisperse sample, whereas values greater than 0.7 indicate a broad size distribution not suitable for DLS analysis.43 DLS measurements of average size were rounded to the nearest whole number, with error bars rounded to the nearest tenth of a nanometer. All measurements were performed in quintuplicate, and error bars in figures and tables represent two standard deviations (2σ) unless stated otherwise. For each reported variation in synthesis procedure, experiments were independently repeated n times, with the specific sample sizes reported in relevant figures below.
Statistical analysis of DLS measurements between variations in synthesis procedure was performed to verify statistical significance between data sets. Independent two-sample Student's t-tests were performed on aggregate data from any two selected sets, and the corresponding p-value determined. The p-value is the probability that the two data sets are not significantly different, so a p-value p less than 0.05 indicates that the observed difference is unlikely to have occurred by random chance, suggesting with over 95% confidence that the difference between the two data sets is statistically significant.44 Where relevant, p-values between data sets are labeled in figures, with sets where p > 0.05 labeled “n.s.” meaning “not significant”.
Zeta potential values were calculated according to the methods described in Leunissen 2007,45 which we summarize below. Effective ionic concentration (ceff) was determined using the conductance values measured and reported by the zeta potential instrument:
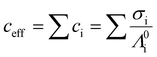 | (1) |
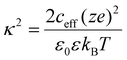 | (2) |
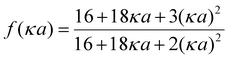 | (3) |
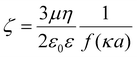 | (4) |
BSA nanoparticle samples were also imaged using a JEOL 2000-FX Scanning Transmission Electron Miscroscope (STEM) under a 200 kV accelerating voltage. Samples were prepared on a glow discharged copper grid with either a carbon film or holey carbon film. Grids were immersed in a droplet of sample before wicking away excess fluid and similarly washing in deionized water. The grid was negative stained by immersing in a droplet of 2 wt% uranyl acetate solution before wicking and washing as before. Grids were allowed to air dry prior to imaging.
3 Results & discussion
3.1 Effect of pH during synthesis
Serum albumin, like many proteins, undergoes certain conformational changes in response to the pH of its environment. There are five different pH-dependent conformation states described in the literature, with the pH values at which these transitions occur also relatively well-known.49 The “normal” conformer is the state at physiological pH, often described as “heart-shaped”. The transition between the “normal” and “fast” conformations (occurring when pH is lowered below 4.3), or the N–F transition, is particularly well-described due to the role of the F conformer in certain tissue transport mechanisms50 and amyloid formation in certain diseases.15,51 However, it has been demonstrated that the ideal pH for the desolvation of albumin nanoparticles is roughly pH = 9,20,21,23 with this pH yielding the smallest particles. Albumin nanospheres were synthesized across a range of initial pH from 6.5–10.5 and the resulting particle sizes were found to be minimized around pH = 9 in agreement with previous literature (see Table 1).| pH | Average particle size (nm) | PDI |
|---|---|---|
| 6.7 | 170 ± 8.4 | 0.15 |
| 7.2 | 163 ± 8.2 | 0.14 |
| 9.1 | 94 ± 1.1 | 0.04 |
| 10.5 | 549 ± 37 | 0.23 |
The transition to the “basic” conformer, or the N–B transition, occurs around pH = 8, and the transition to the “aged” conformer, or the B–A transition, occurs around pH = 10, so the ideal conformation for this method of nanoparticle synthesis seems to be the B conformer, irrespective of crosslinker. At the time of writing, very little information is known conclusively about the B conformer,50 although it is believed to mainly involve changes in domains I and II of albumin tertiary structure,52 a slight decrease in helical content,53 and may depend on resonance structures of histidine residues in domain I.54 Without more structural information about the B-form, any attempts to explain the physical role of pH in the optimization of the synthesis procedure are largely speculative. It is clear that extremely alkaline or acidic conditions will disrupt the secondary structure of albumin, resulting in denaturation and precipitation. Further, the isoelectric point of BSA is roughly 4.7, so most mild acidic conditions result in reduced surface charge, and the decrease in electrostatic repulsion results in larger particles.21 In the remaining range of pH where particle synthesis is reasonably reproducible, around 7–10, BSA has a sizable net negative charge, with electrostatic repulsion undoubtedly playing a significant role in decreasing particle size. However, we observe a local minimum in synthesized particle size with pH, so net charge (which continues to decrease as pH increases) is not the only factor. Some have hypothesized that the B-form is capable of forming intermolecular β-sheets20 that allow for more efficient packing during self-assembly, but often this seems to be based on the misconception that native albumin includes β-strands. Many early predictions of albumin structure based on indirect methods mistakenly identified the extended-chain and turn components of albumin as β-strands,50,55 although it is now known that the only organized secondary structure of albumin is in α-helices and turns.4,56 While it is possible that the N–B transition involves changes in secondary structure, we are not aware of any experiment that has demonstrated this behavior, and the continued confusion in the literature makes it necessary to emphasize that no β-strand structures exist in the N-form. Further research is necessary in this area to properly describe the N–B transition and its role in the synthesis of albumin nanospheres. Regardless, our findings concur with previous publications that the ideal initial pH for minimization of nanoparticle size (and maximization of reproducibility) is pH = 9, which will be used for the remainder of this study.
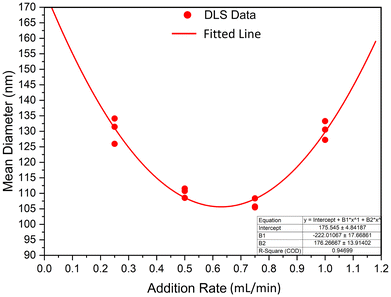
3.2 Effect of ethanol addition rate
The rate of desolvating agent addition was found to impact particle size and polydispersity, and the mode of addition was also found to impact these variables. It was found that dropwise ethanol addition resulted in a larger size distribution compared to infusion against the glass walls, including the formation of sediment in solution. Infusion allowed for a more homogeneous change in ethanol concentration with time (i.e., a well-mixed system), where dropwise addition likely suffered from transient areas of high concentration. The impact of addition mode on the results of synthesis may be visually observed at its most dramatic in Fig. 2, where dropwise ethanol addition resulted in a significant amount of aggregation, a significant increase in average size, and a significant increase in polydispersity index. Sample A, synthesized using dropwise addition, had an average size of 549 ± 37 nm (PDI = 0.23), whereas sample B, synthesized using infusion against the glass, had an average size of 109 ± 1.3 nm (PDI = 0.08). With respect to minimization of size and polydispersity as well as maximizing reproducibility of the synthesis procedure, infusion of ethanol against the glass was the superior desolvation method.Particle formation is governed by desolvation, namely solvent replacement in protein structure resulting in denaturation. When this phenomenon is carefully controlled, nanoparticles form. Rapid addition of the desolvating agent results in gelation,19 and extremely rapid addition may result in protein simply “crashing” from solution as an amorphous aggregate. Using a 2 wt% protein solution with an initial pH of 9 (further discussed in section 3.1) and GT as crosslinker, the rate of ethanol addition was varied between 0.25 and 1 mL min−1 and its effect on particle size studied. The findings are summarized in Fig. 3. It was found that both slow and rapid ethanol addition was associated with larger particle sizes, with a local minimum in between. We believe that this trend arises from two competing phenomena, namely rate of protein desolvation and agglomeration. Before crosslinker addition, particles formed from the desolvation of the protein are soft and gel-like, with a tendency to agglomerate upon collision. At particularly low ethanol addition rates, nucleating particles have more time to collide before the crosslinking reaction solidifies their structure and prevents further agglomeration. In these conditions, particle growth occurs through coalescence in addition to monomer migration (where BSA is the monomer), resulting in a larger average size and polydispersity. At particularly high addition rates, the time frame for nucleation is short, resulting in fewer nuclei and thus larger particle size as desolvated BSA is distributed between fewer growing seeds. The ideal system maximizes nucleation sites while minimizing coalescence (as well as other growth mechanisms like Ostwald ripening). For the investigated scale, where 8 mL of absolute ethanol is infused to 2 mL of aqueous albumin solution for a total volume of 10 mL, minimum size and size distribution was achieved at an ethanol addition rate of ≈0.625 mL min−1, with crosslinker added immediately afterwards. Unless stated otherwise, all other syntheses were performed using this optimal ethanol addition rate.
3.3 Effect of crosslinker
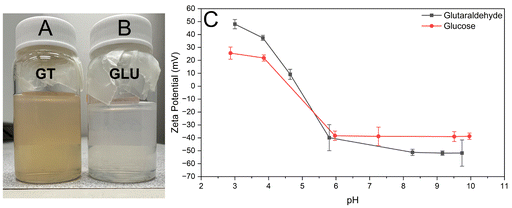
The specific crosslinker used during synthesis, either GT or GLU, was found to only have immediate effects on the size and optical properties of resultant particles. Differences in long-term stability are discussed further in section 3.7. Nanoparticles crosslinked with GLU were found to have a larger average size than particles crosslinked with GT in otherwise identical conditions. However, this difference in the optimized procedure is generally less than 30 nm, and many samples are within 10 nm. This may be attributed to the slower kinetics of GLU crosslinking forming fewer total crosslinks26 and larger physical size of the GLU crosslink making it easier for the particle to swell.27 The other main difference between crosslinkers comes from optical properties. GT crosslinking is associated with a color change, with the dispersion appearing a reddish-yellow color (see ESI Video S2†), a phenomenon reported in previous literature arising from the crosslink molecule itself and possibly from certain aldol condensation oligomer products.57,58 GLU crosslinking, on the other hand, exhibits no clear color change, with the dispersion appearing a cloudy white (see Fig. 4). While the glucose crosslink molecule is more poorly understood, previous literature indicates that advanced glycation end products (AGEs) primarily absorb light in near-UV and blue wavelengths, although the blue absorbance peak is much lower in magnitude than that of UV.59–61 At the concentrations studied in this work, glucose-crosslinked samples appear colorless or white in bulk media. Zeta potential of the synthesized particles using GT and GLU as crosslinkers are provided in Fig. 4C as a function of pH. GT-crosslinked particles exhibit a larger magnitude of zeta potential at all pH values studied.3.4 Effect of electrolyte addition during synthesis
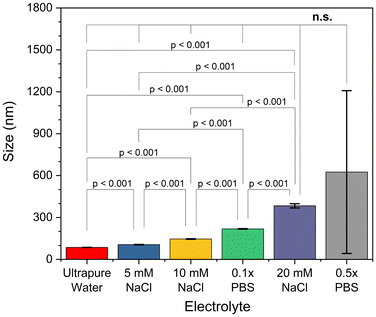
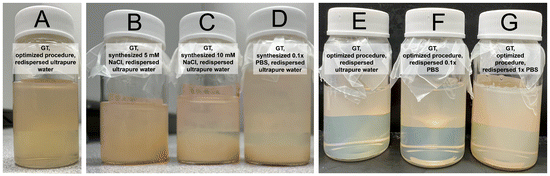
Nanoparticles were synthesized using the optimized procedure (ethanol infused against glass at 0.625 mL min−1, pH of 9) and GT as crosslinker, varying only the concentration of electrolytes in the reaction medium or in the redispersal medium. The effect of increasing electrolyte concentration could be visually observed during synthesis. It was found that, as electrolyte concentration in the reaction medium increased, both size and polydispersity of the resulting particles increased (see Fig. 5). Syntheses in media without additional electrolytes were found to corroborate the predictions made by the trendline of ethanol addition rate (compare Fig. 3 to the first column of Fig. 5). Weak electrolyte solutions yielded larger particles in a slightly more turbid suspension compared to syntheses in ultrapure water, whereas strong electrolyte solutions (exceeding around 10 mM) produced inherently unstable and murky particle dispersions, with some aggregates settling out immediately (see Fig. 5 and 8). In these cases, the aggregates were recovered and included in later DLS measurements to the best of our ability. Beyond 20 mM electrolyte concentration, the synthesis would effectively fail, with protein crashing out of solution and forming a thick aggregate on the glass during the reaction step. Recovery of these aggregates was not feasible, so the DLS data for these samples likely underestimates average particle size. This trend is in agreement with observations previously made in the literature.21 In the optimized procedure in pH-equilibrated ultrapure water, turbidity slowly increased as ethanol was infused, eventually yielding a translucent dispersion. In syntheses of increasing electrolyte concentration (up to 0.5× PBS), the transition between clear solution and turbid dispersion became both faster and more dramatic. In syntheses using strong electrolyte concentrations (20 mM NaCl or higher), the solution transitioned from clear to nearly opaque in a span of seconds, indicative of weak colloidal stability of the protein, which resulted in rapid formation of larger, more polydisperse, and less stable particles. The size was indeed tunable, but control was very sensitive, and came at the cost of larger size distributions.In terms of zeta potential, the presence of electrolytes during the synthesis procedure had no permanent effect on protein structure or charge, only serving to mask charges during the synthesis procedure. As the electrolytes were removed during the washing procedure, shielding of the charges by the ionic cloud was consequently removed. As a result, the particle zeta potential values exhibited similar values across the pH spectrum studied, and were independent of the initial electrolytic strength (see Fig. 6). Thus, we can think of reaction electrolytic strength as a tool to tune the final size of synthesized nanoparticles without altering zeta potential, although this comes at the cost of increased polydispersity and is limited in scope (as size begins to diverge at electrolytic strengths beyond about 20 mM of a monovalent salt, κ−1 ≈ 2.1 nm).
3.5 Effect of pH following synthesis
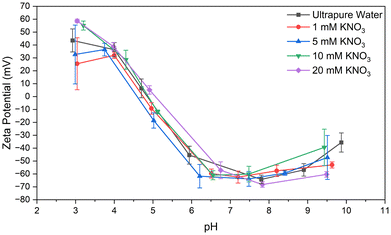
Analysis of zeta potential of synthesized particles across a range of pH from 3–10 showed that the synthesis and crosslinking of particles does not significantly shift the isoelectric point (see Fig. 6). Further, at pH values exceeding 6, the zeta potential roughly plateaus at a value greater than 30 mV in magnitude, indicating a sizable range of pH where particles would remain colloidally stable (neglecting the effect of other chemical or environmental factors).62 Notably, at physiological pH (around 7.4), zeta potential measurements indicate high stability, which along with the mildness of these conditions contributes to the observed slow aging of particles synthesized by the optimized procedure. We suspect that, since the particles are synthesized and crosslinked under alkaline conditions, where the B-form is most stable, but stored under neutral conditions where the N-form is most stable, that the tendency of the protein molecules to revert to the N-form may introduce an entropic strain within the nanoparticles. Further research into the morphological change associated with the N–B transition is necessary for this to be anything more than hypothetical. This strain is likely small, as the B-form does not appear to undergo a dramatic morphological transformation like that seen in the F-form,50 and may increase the porosity of the particles or their ability to bind certain ligands. Such strain may also increase the kinetics of crosslink breakage or other mechanisms of particle degradation.63 However, storage in alkalaine conditions pH > 8 would not be advantageous regardless of the most favorable morphological state of BSA. Prolonged exposure to alkaline conditions could result in alkaline hydrolysis of structurally-important disulfide bonds and peptide bonds of the protein backbone within BSA molecules, degrading the nanoparticle structure.64,65 This effect was visually observed as an increase in turbidity of dispersion aliquots that were held at pH ≥ 9 for periods exceeding around an hour. Further, this would introduce the need to equilibrate pH before a stored dispersion could be utilized in cellular applications.3.6 Morphology analysis
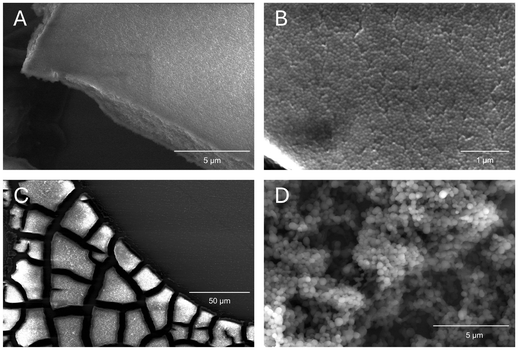
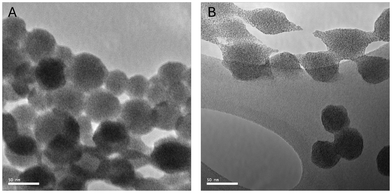
BSA nanoparticles were analyzed to study their morphology using SEM and TEM. It was desired for nanoparticles to exist as single spheres in dispersion, only forming multisphere complexes in the event of aggregation due to aging or drying (due to Marangoni flows). It was also desired to view nanoparticles in as close to their native state as possible, so no sputter coating was performed for SEM samples. These naked nanoparticles are relatively poor conductors, so samples, particularly of smaller nanoparticles, suffered from charging artifacts and progressive radiation damage. It was found that particles crosslinked with glutaraldehyde were more resistant to electron microscopy operating conditions, which follows from glutaraldehyde commonly being used as a fixative for biological samples. Samples for SEM were prepared by allowing a droplet of aqueous dispersion to air-dry, so samples visually displayed the coffee ring effect under low magnification even when diluted. Magnifying the “coffee ring” region revealed a layer of aggregated spherical nanoparticles (see Fig. 7). Parts B and D of Fig. 7 show the nanostructure of these aggregates, representing an optimized and unoptimized synthesis, respectively. It can be seen that generally particles are spherical, but the optimized procedure yields highly spherical and monodisperse nanoparticles, whereas unoptimized synthesis procedures yield particles of a wide size range. In short, SEM sample preparation rendered samples prone to damage over time, but imaging yielded useful information related to physical size and monodispersity, as well as some surface characteristics. The individual particles measured by SEM are within a few nanometers of the average size reported by DLS. The two results are in agreement as DLS measures the particles’ size in their hydrated state. TEM imaging enabled us to capture higher-resolution images of particles and provided a longer imaging window; however, uranyl acetate staining is known to cause contraction of biological materials, so TEM micrographs were unreliable for determining average size of particles.66,67 However, TEM imaging also confirmed the spherical morphology and monodisperse size distribution of nanoparticle dispersions.3.7 Long-term stability analysis
Colloidal stability was periodically assessed by tracking the hydrodynamic size and comparing it to the initial size at the time of synthesis. Further, particle morphology was investigated over time to help elucidate failure modes of the albumin nanospheres as they aged. It was found that, regardless of crosslinker, particles synthesized using the optimized procedure and redispersed in ultrapure water or a weak electrolyte solution (such as 1 mM KNO3) remained colloidally stable for over one year to date, exhibiting no visible flocculation (see Fig. 8 and Tables 3, 4). Notably, we see a larger increase in average size in GLU-crosslinked particles than GT-crosslinked particles over the same time spans. We speculate that this is due to three factors: slower kinetics of the GLU crosslinking reaction (therefore fewer crosslinks formed) and larger physical size of the crosslink (therefore more subject to swelling), and lower colloidal stability rendering GLU-crosslinked particles more susceptible to aggregation (see Fig. 4C). The sedimentation behavior observed over time for samples synthesized in concentrated electrolyte solution was positively correlated to the electrolyte concentration used during the synthesis but not strongly trending with redispersal medium for the time period and concentrations studied (see Fig. 8).From zeta potential measurements we determine that particles synthesized in a range of electrolytic strengths should maintain colloidal stability (see Fig. 6); however, we document sedimentation of particle dispersions synthesized in electrolyte solutions ≥10 mM of a monovalent salt. This indicates gravitational instability of larger particles, where increased size and mass decreases the gravitational length (the ratio between the thermal energy and buoyant force acting on a particle), resulting in sedimentation.68 We see this phenomenon trend with particle size when comparing Fig. 5 and 8, as the sediment layer of sample C is taller than the sediment layer of sample D.
As illustrated in Fig. 8, the long-term stability of BSA nanosphere samples was strongly influenced by the reaction conditions, as average size trends positively with reaction electrolytic strength and larger particles are inherently less gravitationally stable. Zeta potential measurements indicate that these particles are still colloidally stable, but the density mismatch with solution causes them to sediment out over time. However, it was found that flocs formed in these samples, synthesized in media with increased electrolytic strengths, were often of a loose physical association that could be reversed with vortexting, dilution, sonication, or even simple stirring. For example, a sample synthesized in 10 mM NaCl (labeled as sample D in Fig. 8) had aggregates that were visible with the naked eye after aging for roughly 8 months, but after vortexing the bulk sample and diluting an aliquot for DLS analysis, average size was reported as 148 ± 2.8 nm with a PDI of 0.06. When comparing to the size reported by DLS when the particles were first synthesized, which was 146 ± 3.8 nm with a PDI of 0.06, we may conclude that flocculation of these BSA nanoparticles is largely reversible if the particles within the floc are not significantly degraded. This does not apply generally, however, as samples synthesized in strong electrolytic strengths (i.e. ≥20 mM of a monovalent salt) had large aggregates that persisted even when sonicated.
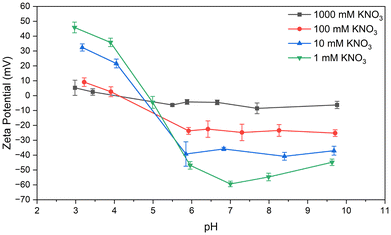
Particles were also synthesized in ultrapure water equilibrated to a pH of 9 per the optimized procedure before washing and redispersal in a variety of electrolyte solutions. BSA nanoparticles dispersed in electrolytic strengths up to 1× PBS behaved, visually appeared, and aged nearly indistinguishably from samples redispersed in ultrapure water or 1 mM KNO3. As expected, zeta potential measurements indicate decreased colloidal stability as electrolytic strength increases (see Fig. 9), with electrolytic strength of 100 mM KNO3 resulting in “incipient” stability (−15 ≤ ζ ≤ −30 mV).62,69 100 mM KNO3 is of comparable electrolytic strength to 1× PBS, which is designed to emulate physiological conditions. The two solutions have Debye lengths (κ−1) of ≈0.96 and ≈0.70 nm, respectively,70 whereas in 1 M KNO3 it shrinks to ≈0.3 nm. Storage in electrolytic solutions more concentrated than around 1× PBS will likely result in agglomeration and precipitation, but dispersions in solutions of 1× PBS or weaker have remained stable one year after initial synthesis (see Table 2). In summary, when BSA nanoparticles were synthesized via the optimized procedure, redispersal media with an electrolytic strength comparable to physiological conditions or weaker had a minor effect for the time period studied (see Table 2).
| Sample | Age (days) | Size (nm) | PDI |
|---|---|---|---|
| 0.1× PBS | 0 | 87 ± 2.8 | 0.10 |
| 92 | 90 ± 1.4 | 0.07 | |
| 346 | 91 ± 4.0 | 0.10 | |
| 0.5× PBS | 0 | 97 ± 0.7 | 0.11 |
| 91 | 96 ± 1.6 | 0.11 | |
| 345 | 102 ± 7.2 | 0.14 | |
| 1.0× PBS | 0 | 101 ± 1.5 | 0.06 |
| 88 | 102 ± 1.7 | 0.06 | |
| 341 | 107 ± 9.2 | 0.09 |
In stable dispersions produced via the optimized procedure, no visible flocs have yet formed at the time scale of around a year. Even with glucose-crosslinked particles, which we would generally assume to be less stable due to the slower kinetics of the crosslinking reaction, larger physical size of the crosslink bridge, and less favorable equilibrium, it takes many months before we see a statistically significant change in average particle size (see Tables 3 and 4), with polydispersity remaining small as well. With initial average particle size typically just below 100 nm, size changes of this magnitude still represent a quite small change in diameter. Regardless, we find that GLU-crosslinked particles exhibit a larger increase in size compared to GT-crosslinked particles. It is notable that these changes in dispersion character are significantly smaller than those reported by Amighi et al. 202020 and Jun et al. 2011,23 which do not report even “optimized” dispersions remaining stable beyond three months after synthesis. Both of the aforementioned studies utilized a probe sonicator for particle redispersion, whereas this study utilized a vortexer. We speculate that the shear forces particles were exposed to during probe sonication may have destabilized them, where the gentler treatment of a vortexer preserved their structural integrity. The differences in reported stability make it clear that optimization of washing steps is also important. Regardless, further information about particle conformation at these points in the aging time frame where degradation becomes relevant is necessary, which is where electron microscopy comes in.
| Glutaraldehyde crosslinker | |||
|---|---|---|---|
| Sample | Age (days) | Size (nm) | PDI |
| 1 | 0 | 108 ± 1.3 | 0.10 |
| 108 | 108 ± 2.7 | 0.08 | |
| 282 | 109 ± 1.3 | 0.08 | |
| 2 | 0 | 96 ± 3.1 | 0.08 |
| 74 | 90 ± 2.7 | 0.09 | |
| 253 | 93 ± 4.2 | 0.07 | |
| 3 | 0 | 95 ± 6.2 | 0.05 |
| 53 | 95 ± 1.7 | 0.05 | |
| 306 | 99 ± 6.0 | 0.07 | |
| 4 | 0 | 112 ± 1.6 | 0.08 |
| 43 | 109 ± 2.7 | 0.04 | |
| 296 | 112 ± 8.3 | 0.08 | |
| 5 | 0 | 86 ± 1.6 | 0.10 |
| 47 | 88 ± 2.3 | 0.10 | |
| 121 | 88 ± 6.0 | 0.08 | |
| 6 | 0 | 86 ± 0.6 | 0.11 |
| 7 | 87 ± 2.3 | 0.11 | |
| 103 | 90 ± 6.0 | 0.10 | |
| Glucose crosslinker | |||
|---|---|---|---|
| Sample | Age (days) | Size (nm) | PDI |
| 1 | 0 | 110 ± 0.6 | 0.03 |
| 73 | 118 ± 2.2 | 0.06 | |
| 246 | 129 ± 3.8 | 0.07 | |
| 2 | 0 | 94 ± 1.1 | 0.04 |
| 59 | 99 ± 1.5 | 0.05 | |
| 312 | 108 ± 5.7 | 0.06 | |
| 3 | 0 | 100 ± 4.4 | 0.04 |
| 62 | 101 ± 3.9 | 0.04 | |
| 311 | 108 ± 7.9 | 0.04 | |
| 4 | 0 | 92 ± 1.9 | 0.05 |
| 30 | 98 ± 2.2 | 0.07 | |
| 271 | 98 ± 8.2 | 0.04 | |
| 5 | 0 | 96 ± 2.3 | 0.08 |
| 175 | 104 ± 3.5 | 0.04 | |
| 249 | 109 ± 5.5 | 0.08 | |
| 6 | 0 | 108 ± 3.9 | 0.05 |
| 127 | 116 ± 2.9 | 0.06 | |
| 201 | 121 ± 8.2 | 0.06 | |
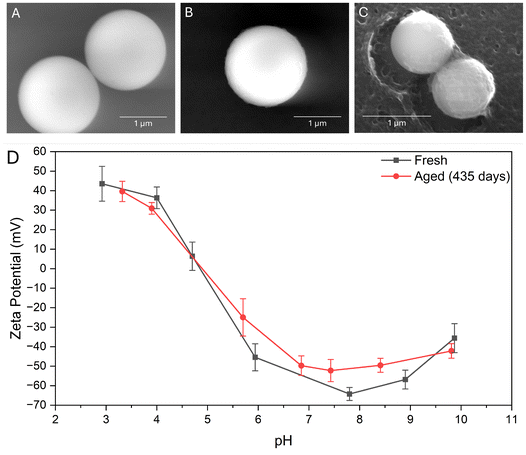
When imaging using SEM, it was found that particles were damaged by the electron beam. It was infeasible to capture high-resolution surface characteristics of small particles before they were significantly damaged, and sputter coating would conceal any of these surface details. However, it was found that larger spheres produced via an intentionally unoptimized procedure could be imaged without issue, so these images were used for Fig. 10. By imaging the same sample of particles at several different points in time, we observed “fraying” around the edges as they aged (see Fig. 10B), adding a surface roughness not observed in fresh particles regardless of crosslinker. While this does not compromise the particle's stability (see Fig. 10D) or bulk characteristics at this stage, it indicates that the gradual failure mechanism that the particles undergo primarily occurs from the outside in. Bonds break over time and the “solid” character of fresh nanoparticles is progressively diminished. We may observe this in highly aged samples, such as in Fig. 10C, where some particles have degraded to the point of losing surface material to the bulk media. These results indicate that drug formulations involving immobilization of surface ligands would have a shorter efficacy lifetime relative to simpler drug-loading applications. However, zeta potential measurements indicate that this slow degradation process does not significantly reduce colloidal stability for the time period studied, with even highly aged samples exhibiting zeta potentials ≥30 mV in magnitude at physiological pH. Studies by Michnik et al. 200571 and Madeira et al. 202172 indicate that albumin protein tends to aggregate and eventually thermally denature when stored in aqueous conditions for long periods of time. Neither of these phenomena result in changes in the chemical composition of the protein, so considering the zeta potential data we believe that the observed degradation occurs via breaking of crosslink bonds.
We gain further insight into this process in TEM when samples are prepared on holey carbon grids. Irradiation with the electron beam causes beam-induced motion of the holes of the holey carbon grid, pulling nanoparticles that aggregated near the edge apart. When this happens, we see that some material pulls away from the particle surface, as though the particles were sticky (see Fig. 11). This process occurred over the course of imaging, and may represent an accelerated version of the degradation imaged in SEM. As electrons bombard the sample, bonds are broken and the sample decays from the outside in, exacerbating the “fraying” degradation seen in particles of similar age in SEM. In dispersion, the nanoparticles likely fail via an analogous long-term mechanism (see Fig. 10). The outermost layers of crosslinks or intramolecular bonds progressively break, but the particle remains nearly constant in size due to the physical interactions between protein monomers (van der Waals forces, steric hindrance, etc.), resulting in a sort of soft, gel-like layer similar to a corona. Over time, failure occurs via sloughing of this soft material into solution as well as collisions between “sticky” or “frayed” particles resulting in aggregation, whereas fresh particles are more likely to elastically collide or form small aggregates that can be broken apart with gentle shear forces. The polydispersity index also trends very weakly with age, but has been observed to decrease slightly with many samples. We suspect that these less predictable changes in PDI could be partially due to the Vroman effect, with free albumin that has resolubilized before adsorbing to other particles as a protein corona.11
4 Conclusion
Bovine serum albumin nanospheres were produced via desolvation procedure optimized to minimize size and maximize stability at room temperature. In agreement with previous studies, this work finds desolvation to be an effective and consistent method of nanoparticle synthesis. It was found that both pH and desolvating agent addition rate have optimum values, following approximately parabolic trends with local minima in particle size for the ranges studied. To our knowledge, this work is the first to optimize the parameter of desolvating agent addition. Optimized values for these two parameters were pH = 9 and ethanol addition rate of 0.625 mL min−1, with the ethanol added via infusion against the wall of the synthesis vial. The effect of two crosslinkers, glutaraldehyde and glucose, were studied for their effect on particle size and stability. The optimized synthesis procedure yielded average particle sizes of 86 ± 3.8 nm using glutaraldehyde as a crosslinker and a size of 92 ± 1.8 using glucose as a crosslinker with polydispersity indices of 0.08 and 0.05, respectively. This indicates a high degree of monodispersity for both crosslinkers, exceeding that reported in previous literature. At physiological pH (7.0–7.4) and dispersal medium of 1 mM KNO3, these optimized nanoparticle samples were found to have zeta potentials of −32 ± 2.3 mV for glutaraldehyde-crosslinked particles and −39 ± 7.1 mV for glucose-crosslinked particles, indicating high colloidal stability.62,69 Particles maintained long-term colloidal stability in physiological electrolytic conditions, and the small Debye length in these conditions (κ−1 ≤ 1 nm) is capable of accommodating small-molecule ligands or antibodies.73 The optimization steps taken by this study yielded glucose-crosslinked BSA nanoparticles with a stability exceeding that reported in previous literature. This improvement in stability is especially relevant in cell targeting and artificial antigen presentation applications.30,74Nanoparticle morphology was confirmed using electron microscopy imaging, providing insight into how BSA nanoparticles age. DLS and zeta potential analyses were performed to study the long-term stability of particle suspensions. It was found that the optimized procedure yields small, highly monodisperse particles that “fray” around the edges as they age, progressively losing their originally smooth surface character. For both studied crosslinkers, particles were found to exhibit superior stability compared to what is reported in the literature.20,23 Using the optimized procedure, particles of similar size and zeta potential were achieved when glutaraldehyde was substituted with glucose. These findings will be beneficial in applications where BSA nanoparticles will be used in biological systems and glutaraldehyde should be avoided.
Several areas for future research are also identified by this work. The usefulness of protein-based particles as nanocarriers for controlled release of drugs has been studied in many publications, but fewer have investigated protein-drug conjugates (Abraxane® and similar therapies being exceptions), inviting further research into conjugation of therapeutics for both controlled-release and cell-targeting applications. Additional research is necessary to understand the role of the B-form of BSA in the optimized desolvation procedure, as well as the chemical mechanism behind GLU crosslinking (specifically the formation of glucosepane). Further inquiry in these areas would make for a more holistic understanding of the nanoparticle synthesis process, which would aid in both fine-tuning and scaling of that process. The desolvation procedure used in this study requires less specialized equipment and less hazardous conditions compared to HPH, but the appeal of continuous production invites further research guided by the findings of this work.
Author contributions
Blake Bartlett was responsible for data collection, formal analysis, investigation, methodology, validation, and writing. Sepideh Razavi and John Klier were responsible for administration, resources, supervision, and editing.Data availability
The data supporting the findings of this study are available within the article, and supplementary video is provided in the supplementary materials. Raw data is available upon request. S1 timelapse of desolvation under optimized conditions and addition of glutaraldehyde crosslinker, S2 timelapse of the glutaraldehyde crosslinking reaction.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the National Science Foundation Graduate Research Fellowship under Grant No. 10557390. S. R. acknowledges support by the National Science Foundation through CAREER grant (CBET-2144020). Financial support provided by the University of Oklahoma Libraries ‘Open Access Fund’ is acknowledged.References
- L. A. Emens, P. A. Ascierto, P. K. Darcy, S. Demaria, A. M. M. Eggermont, W. L. Redmonds, B. Seliger and F. M. Marincola, Cancer Immunotherapy: Opportunities and Challenges in the Rapidly Evolving Clinical Landscape, Eur. J. Cancer, 2017, 81, 116–129 CrossRef CAS PubMed.
- A. Spada, J. Emami, J. A. Tuszynski and A. Lavasanifar, The Uniqueness of Albumin as a Carrier in Nanodrug Delivery, Mol. Pharmaceutics, 2021, 18, 1862–1894 CrossRef CAS PubMed.
- F. An and X. Zhang, Strategies for Preparing Albumin-Based Nanoparticles for Multifunctional Bioimaging and Drug Delivery, Theranostics, 2017, 7, 3667–3689 CrossRef CAS PubMed.
- A. Bujacz, Structures of Bovine, Equine, and Leporine Serum Albumin, Acta Crystallogr., 2012, 68, 1278–1289 CrossRef CAS PubMed.
- A. Bujacz, K. Zielinski and B. Sekula, Structural Studies of Bovine, Equine, and Leporine Serum Albumin Complexes with Naproxen, Proteins, 2014, 82, 2199–2208 CrossRef CAS PubMed.
- R. Raoufinia, A. Mota, N. Keyhanvar, F. Safari, S. Shamekhi and J. Abdolalizadeh, Overview of Albumin and Its Purification Methods, Adv. Pharm. Bull., 2016, 6, 495–507 CrossRef CAS PubMed.
- V. Mishra and R. J. Heath, Structural and Biochemical Features of Human Serum Albumin Essential for Eukaryotic Cell Culture, Int. J. Mol. Sci., 2021, 22, 8411 CrossRef CAS PubMed.
- E. Miele, G. P. Spinelli, E. Miele, F. Tomao and S. Tomao, Albumin-Bound Formulation of Paclitaxel (Abraxane® ABI-007) in the Treatment of Breast Cancer, Int. J. Nanomed., 2009, 4, 99–105 CAS.
- Z. Wei, X. Liu, C. Cheng, W. Yu and P. Yi, Metabolism of Amino Acids in Cancer, Front. Cell Dev. Biol., 2021, 8, 603837 CrossRef PubMed.
- E. N. Hoogenboezem and C. L. Duvall, Harnessing Albumin as a Carrier for Cancer Therapies, Adv. Drug Delivery Rev., 2018, 130, 73–89 CrossRef CAS PubMed.
- A. Parodi, J. Miao, S. M. Soond, M. Rudzińska and A. A. Zamyatnin Jr., Albumin Nanovectors in Cancer Therapy and Imaging, Biomolecules, 2019, 9, 218 CrossRef PubMed.
- Y. P. Tanjung, M. K. Dewi, V. A. Gatera, M. I. Barliana, I. M. Joni and A. Y. Chaerunisaa, Factors Affecting the Synthesis of Bovine Serum Albumin Nanoparticles Using the Desolvation Method, Nanotechnol., Sci. Appl., 2024, 17, 21–40 CrossRef CAS PubMed.
- J. Floury, A. Desrumaux and J. Legrand, Effect of Ultra-high-pressure Homogenization on Structure and on Rheological Properties of Soy Protein-stabilized Emulsions, J. Food Sci., 2002, 67, 9 CrossRef.
- Food and D. Administration, ABRAXANE (paclitaxel) Label, 2020, https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/021660s047lbl.pdf Search PubMed.
- Y. Jia, C. Cocker and J. Sampath, Insights into Protein Unfolding under pH, Temperature, and Shear using Molecular Dynamics Simulations, arXiv Preprint, 2024.
- A. D. Putri, P. Chen, Y. Su, J. Lin, J. Liou and C. Hsieh, Optimization and Development of Selective Histone Deacetylase Inhibitor (MPT0B291)-Loaded Albumin Nanoparticles for Anticancer Therapy, Pharmaceutics, 2021, 13, 1728 CrossRef CAS PubMed.
- B. R. Riegger, R. Kowalski, L. Hilfert, G. E. M. Tovar and M. Bach, Chitosan Nanoparticles via High-Pressure Homogenization-Assisted Miniemulsion Crosslinking for Mixed-Matrix Membrane Adsorbers, Carbohydr. Polym., 2018, 201, 172–181 CrossRef CAS PubMed.
- B. von Storp, A. Boeker, M. Ploeger and K. Langer, Albumin Nanoparticles with Predictable Size by Desolvation Procedure, J Microencapsulation, 2012, 29, 138–146 CrossRef CAS PubMed.
- S. H. Arabi, D. Haselberger and D. Hinderberger, The Effect of Ethanol on Gelation, Nanoscopic, and Macroscopic Properties of Serum Albumin Hydrogels, Molecules, 2020, 25, 1927 CrossRef CAS PubMed.
- F. Amighi, Z. Emam-Djomeh and M. Labbafi-Mazraeh-Shahi, Effect of Different Cross-Linking Agents on the Preparation of Bovine Serum Albumin Nanoparticles, J. Iran. Chem. Soc., 2020, 17, 1223–1235 CrossRef CAS.
- K. Langer, S. Balthasar, V. Vogel, N. Dinauer, H. von Briesen and D. Schubert, Optimization of the Preparation Process for Human Serum Albumin (HSA) Nanoparticles, Int. J. Pharm., 2003, 257, 169–180 CrossRef CAS PubMed.
- O. P. Rubino, R. Kowalsky and J. Swarbrick, Albumin Microspheres as a Drug Delivery System: Relation Among Turbidity Ratio, Degree of Cross-Linking, and Drug Release, Pharm. Res., 1993, 10, 1059–1065 CrossRef CAS PubMed.
- J. Y. Jun, H. H. Nguyen, S. Paik, H. Chun, B. Kang and S. Ko, Preparation of Size-Controlled Bovine Serum Albumin (BSA) Nanoparticles by a Modified Desolvation Method, Food Chem., 2011, 127, 1892–1898 CrossRef CAS.
- H. Niknejad and R. Mahmoudzadeh, Comparison of Different Crosslinking Methods for Preparation of Docetaxel-loaded Albumin Nanoparticles, Iran. J. Pharm. Res., 2015, 14, 385–394 CAS.
- S. Ghosh, P. Das and B. Nayak, High Potency EDC-crosslinked Bovine Serum Albumin Nanoencapsulation of Berberine Enhances in vitro Anticancer Efficacy Against Glioblastoma by Inducing ROS Mediated Cell Apoptosis, New J. Chem., 2022, 46, 23254–23267 RSC.
- J. Anguizola, R. Matsuda, O. S. Barnaby, K. S. Joseph, C. Wa, E. DeBolt, M. Koke and D. S. Hage, Review: Glycation of Human Serum Albumin, Clin. Chim. Acta, 2013, 64–76 CrossRef CAS PubMed.
- K. M. Biemel, D. A. Friedl and M. O. Lederer, Identification and Quantification of Major Maillard Cross-Links in Human Serum Albumin and Lens Protein, J. Biol. Chem., 2002, 277, 24907–24915 CrossRef CAS PubMed.
- A. Twarda-Clapa, A. Olczak, A. M. Białkowska and M. Koziołkiewicz, Advanced Glycation End-Products (AGEs): Formation, Chemistry, Classification, Receptors, and Diseases Related to AGEs, Cells, 2022, 11, 1312 CrossRef CAS PubMed.
- S. Farris, J. Song and Q. Huang, Alternative Reaction Mechanism for the Cross-Linking of Gelatin with Glutaraldehyde, J. Agric. Food Chem., 2010, 58, 998–1003 CrossRef CAS PubMed.
- F. Hou, Z. Guo, M. T. Ho, Y. Hui and C. Zhao, Particle-Based Artificial Antigen-Presenting Cell Systems for T Cell Activation in Adoptive T Cell Therapy, ACS Nano, 2024, 18, 8571–8599 CrossRef CAS.
- K. Alhallak, J. Sun, K. Wasden, N. Guenthner, J. O'Neal, B. Muz, J. King, D. Kohnen, R. Vij, S. Achilefu, J. F. DiPersio and A. K. Azab, Nanoparticle T-cell Engagers as a Modular Platform for Cancer Immunotherapy, Leukemia, 2021, 35, 2346–2357 CrossRef CAS PubMed.
- H. Yuan, W. Jiang, C. A. von Roemeling, Y. Qie, X. Liu, Y. Chen, Y. Wang, R. E. Wharen, K. Yun, G. Bu, K. L. Knutson and B. Y. S. Kim, Multivalent Bi-Specific Nanobioconjugate Engager for Targeted Cancer Immunotherapy, Nat. Nanotechnol., 2017, 12, 763–769 CrossRef CAS PubMed.
- D. Schmid, C. G. Park, C. A. Hartl, N. Subedi, A. N. Cartwright, R. B. Puerto, Y. Zheng, J. Maiarana, G. J. Freeman, K. W. Wucherpfennig, D. J. Irvine and M. S. Goldberg, T Cell-Targeting Nanoparticles Focus Delivery of Immunotherapy to Improve Antitumor Immunity, Nat. Commun., 2017, 8, 1747 CrossRef.
- G. C. Thalhammer-Thurner and P. Debbage, Albumin-based Nanoparticles: Small, Uniform, and Reproducible, Nanoscale Adv., 2023, 5, 503–512 RSC.
- Y. Jiang and M. Stenzel, Drug Delivery Vehicles Based on Albumin-Polymer Conjugates, Macromol. Biosci., 2016, 16, 791–802 CrossRef CAS PubMed.
- M. L. Verma, B. S. Dhanya, Sukriti, V. Rani, M. Thakur, J. Jeslin and R. Kushwaha, Carbohydrate and Protein Based Biopolymeric Nanoparticles: Current Status and Biotechnological Applications, Int. J. Biol. Macromol., 2020, 154, 390–412 CrossRef CAS PubMed.
- A. Brouillard, N. Deshpande and A. A. Kulkarni, Engineered Multifunctional Nano- and Biological Materials for Cancer Immunotherapy, Adv. Healthcare Mater., 2021, 10, 2001680 CrossRef CAS PubMed.
- M. S. Kang, T. W. S. Kong, J. Y. X. Khoo and T. Loh, Recent Developments in Chemical Conjugation Strategies Targeting Native Amino Acids in Proteins and their Applications in Antibody-Drug Conjugates, Chem. Sci., 2021, 12, 13613 RSC.
- N. Dinauer, S. Balthasar, C. Weber, J. Kreuter, K. Langer and H. von Briesen, Selective Targeting of Antibody-Conjugated Nanoparticles to Leukemic Cells and Primary T-Lymphocytes, Biomaterials, 2005, 26, 5898–5906 CrossRef CAS PubMed.
- M. Tarhini, I. Benlyamani, S. Hamdani, G. Agusti, H. Fessi, H. Greige-Gerges, A. Bentaher and A. Elaissari, Protein-Based Nanoparticle Preparation via Nanoprecipitation Method, Materials, 2018, 11, 394 CrossRef PubMed.
- H. Kaur and A. Singh, Design, Development and Characterization of Serratiopeptidase Loaded Albumin Nanoparticles, J. Appl. Pharm. Sci., 2015, 5, 103–109 CrossRef.
- C. Weber, J. Kreuter and K. Langer, Desolvation Process and Surface Characteristics of HSA-nanoparticles, Int. J. Pharm., 2000, 196, 197–200 CrossRef CAS PubMed.
- M. Danaei, M. Deghankhold, S. Ataei, F. H. Davarani, R. Javanmard, A. Dokhani, S. Khorasani and M. R. Mozafari, Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems, Pharmaceutics, 2018, 10, 57 CrossRef PubMed.
- T. K. Kim, T Test as a Parametric Statistic, Korean J. Anesthesiol., 2015, 68, 540–546 CrossRef PubMed.
- M. E. Leunissen, Ph.D. thesis, Utrecht University, 2007.
- E. L. Correia, S. Thakur, A. Ervin, E. Shields and S. Razavi, Adsorption of Surfactant Molecules onto the Surface of Colloidal Particles: Case of Like-Charged Species, Colloids Surf., A, 2023, 676, 132142 CrossRef CAS.
- J. W. Swan and E. M. Furst, A Simpler Expression for Henry's Function Describing the Electrophoretic Mobility of Spherical Colloids, J. Colloid Interface Sci., 2012, 388, 92–94 CrossRef CAS PubMed.
- D. Velegol, Colloidal Systems, CreateSpace Independent Publishing Platform, Scotts Valley, California, 1993 Search PubMed.
- P. J. Sadler and A. Tucker, pH-Induced Structural Transitions of Bovine Serum Albumin, Eur. J. Biochem., 1993, 212, 811–817 CrossRef CAS PubMed.
- D. C. Carter and J. X. Ho, Structure of Serum Albumin, Adv. Protein. Chem., 1994, 45, 153–203 CrossRef CAS PubMed.
- K. Baler, O. A. Martin, M. A. Carignano, G. A. Ameer, J. A. Vila and I. Szleifer, Electrostatic Unfolding and Interactions of Albumin Driven by pH Changes: A Molecular Dynamics Study, J. Phys. Chem. B, 2014, 118, 921–930 CrossRef CAS PubMed.
- M. Dockal, D. C. Carter and F. Rüker, Conformational Transitions of the Three Recombinant Domains of Human Serum Albumin Depending on pH, J. Biol. Chem., 2000, 275, 3042–3050 CrossRef CAS PubMed.
- S. Era, K. B. Itoh, M. Sogami, K. Kuwata, T. Iwama, H. Yamada and H. Watari, Structural Transition of Bovine Plasma Albumin in the Alkaline Region–the N-B Transition, Int. J. Pept. Protein Res., 1990, 35, 1–11 CrossRef CAS PubMed.
- O. J. M. Bos, J. F. A. Labro, M. J. E. Fischer, J. Wilting and L. H. M. Janssen, The Molecular Mechanism of the Neutral-to-Base Transition of Human Serum Albumin, J. Biol. Chem., 1989, 264, 953–959 CrossRef CAS PubMed.
- S. Lin, M. Li and Y. Wei, Ethanol or/and Captopril-induced Precipitation and Secondary Conformational Changes of Human Serum Albumin, Spectrochim. Acta, Part A, 2004, 60, 3107–3111 CrossRef PubMed.
- S. Sugio, A. Kashima, S. Mochizuki, M. Noda and K. Kobayashi, Crystal Structure of Human Serum Albumin at 2.5 Å Resolution, Protein Eng., 1999, 12, 439–446 CrossRef CAS PubMed.
- D. Hopwood, C. R. Allen and M. McCabe, The Reactions Between Glutaraldehyde and Various Proteins. An Investigation of their Kinetics, Histochem. J., 1970, 2, 137–150 CrossRef CAS PubMed.
- X. Ma, X. Sun, D. Hargrove, J. Chen, D. Song, Q. Dong, X. Lu, T. Fan, Y. Fu and Y. Lei, A Biocompatible and Biodegradable Protein Hydrogel with Green and Red Autofluorescence: Preparation, Characterization, and In Vivo Biodegradation Tracking and Modeling, Sci. Rep., 2016, 6, 19370 CrossRef CAS PubMed.
- K. Yanagisawa, Z. Makita, K. Shiroshita, T. Ueda, T. Fusegawa, S. Kuwajima, M. Takeuchi and T. Koike, Specific Fluorescence Assay for Advanced Glycation End Products in Blood and Urine of Diabetic Patients, Metabolism, 1998, 47, 1348–1353 CrossRef CAS PubMed.
- M. Larsson, R. Favilla and T. Strömberg, Assessment of Advanced Glycated End Product Accumulation in Skin using Auto Fluorescence Multispectral Imaging, Comput. Biol. Med., 2017, 85, 106–111 CrossRef CAS PubMed.
- A. Perrone, A. Giovino, J. Benny and F. Martinelli, Advanced Glycation End Products (AGEs): Biochemistry, Signaling, Analytical Methods, and Epigenetic Effects, Oxid. Med. Cell. Longevity, 2020, 2020, 3818196 Search PubMed.
- A. Kumar and C. K. Dixit, in Methods for Characterization of Nanoparticles, Elsevier Ltd, 2017, ch. 3, pp. 43–58 Search PubMed.
- B. Ahmad, M. Z. Kamal and R. H. Khan, Alkali-Induced Conformational Transition in Different Domains of Bovine Serum Albumin, Protein Pept. Lett., 2004, 11, 307–315 CrossRef CAS PubMed.
- T. M. Florence, Degradation of Protein Disulphide Bonds in Dilute Alkali, Biochem. J., 1980, 189, 507–520 CrossRef CAS PubMed.
- M. Kluz, H. Nieznańska, R. Dec, I. DziÈ©cielewski, B. Niżyński, G. Ścibisz, W. Puławski, G. Staszczak, E. Klein, J. Smalc-Koziorowska and W. Dzwolak, Revisiting the Conformational State of Albumin Conjugated to Gold Nanoclusters: A Self-Assembly Pathway to Giant Superstructures Unraveleed, PLoS One, 2019, 14, e0218975 CrossRef CAS PubMed.
- Y. Zhang, T. Huang, D. M. Jorgens, A. Nickerson, L. Jin and J. Pelz, et al., Quantitating Morphological Changes in Biological Samples During Scanning Electron Microscopy Sample Preparation with Correlative Super-Resolution Microscopy, PLoS One, 2017, 12, e0176839 CrossRef PubMed.
- P. Serwer, Flattening and Shrinkage of Bacteriophage T7 After Preparation for Electron Microscopy by Negative Staining, J. Ultrastruct. Res., 1977, 58, 235–243 CrossRef CAS PubMed.
- T. Eckert, M. Schmidt and D. de las Heras, Gravity-induced Phase Phenomena in Plate-Rod Colloidal Mixtures, Commun. Phys., 2021, 4, 202 CrossRef CAS.
- T. M. Riddick, Control of Colloid Stability through Zeta Potential, Livingston Publishing Company, Wynnewood, Pennsylvania, 1968 Search PubMed.
- C. Chu, I. Sarangadharan, A. Regmi, Y. Chen, C. Hsu, W. Chang, G. Lee, J. Chyi, C. Chen, S. Shiesh, G. Lee and Y. Wang, Beyond the Debye Length in High Ionic Strength Solution: Direct Protein Detection with Field-Effect Transistors (FETs) in Human Serum, Sci. Rep., 2017, 7, 5256 CrossRef PubMed.
- A. Michnik, K. Michalik and Z. Drzazga, Stability of Bovine Serum Albumin at Different pH, J. Therm. Anal. Calorim., 2005, 80, 399–406 CrossRef CAS.
- P. P. Madeira, I. L. D. Rocha, M. E. Rosa, M. G. Freira and J. A. P. Coutinho, On the Aggregation of Bovine Serum Albumin, J. Mol. Liq., 2021, 349, 118183 CrossRef.
- V. Kesler, B. Murmann and H. T. Soh, Going Beyond the Debye Length: Overcoming Charge Screening Limitations in Next-Generation Bioelectronic Sensors, ACS Nano, 2020, 14, 16194–16201 CrossRef CAS PubMed.
- J. V. Kim, J. Latouche, I. Rivière and M. Sadelain, The ABCs of Artificial Antigen Presentation, Nat. Biotechnol., 2004, 22, 403–410 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr04682j |
| This journal is © The Royal Society of Chemistry 2025 |