Open Access Article
Open Access Article3(2H)-pyridazinone derivatives: a new scaffold for novel plant activators†
Qinjie Shia,
Yanxia Shib,
Kang Changa,
Jianqin Chena,
Zhenjiang Zhaoc,
Weiping Zhua,
Yufang Xu *a,
BaoJu Li*b and
Xuhong Qian*a
*a,
BaoJu Li*b and
Xuhong Qian*a
aState Key Laboratory of Bioreactor Engineering, Shanghai Key Laboratory of Chemical Biology, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China. E-mail: yfxu@ecust.edu.cn
bInstitute of Vegetables and Flowers, Chinese Academy of Agricultural Science, Beijing 100081, China. E-mail: libj@mail.caas.net.cn
cState Key Laboratory of Bioreactor Engineering, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China
First published on 6th November 2019
Abstract
Due to the emergence of drug resistance, pesticide residue and environmental contamination, it is important to develop novel eco-friendly strategies to protect plants. Among them, plant activators have been gaining more and more attention. Herein, based on SHAFTS method, a new scaffold for novel plant activators was predicted and the discovery and structure–activity relationships of a series of 3(2H)-pyridazinone derivatives as novel plant activators were elucidated in detail. The vast majority of compounds exhibited excellent broad-spectrum induced resistance activity against tested diseases in vivo but no direct antimicrobial activity in vitro. Among them, compound 32 showed excellent efficacy against four pathogens and great potential as new plant activators in crop protection.
Threatened by a large quantity of pathogens, such as fungi, bacteria, and viruses, the production of agricultural crops is still facing a big challenge.1,2 Although the use of many traditional pesticides could directly control the diseases and improve the productivity, it simultaneously created serious problems, including pesticide residue, environmental contamination, and drug resistance.3
In order to deal with these issues, plant activator, a novel kind of agrochemical, has been gaining more and more attention.4 It can protect crops from diseases by inducing plant immune responses without direct toxicity to pathogens and shows high efficacy, low toxicity, and no drug resistance.5
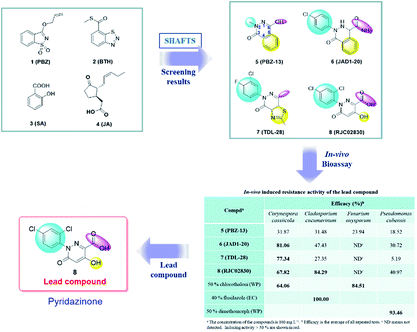
Several plant activators, such as PBZ,6 BTH,7,8 TDL,9 and ISOTIANIL10 that molecularly mimic salicylic acid (SA), could induce the plant's systemic acquired resistance4 and have been commercially applied in crop protection.11–16 Owing to the great advantages of plant activators, it is necessary to develop more efficient and eco-friendly plant activators to protect plants. In our previous work, a novel approach called SHApe-FeaTure Similarity (SHAFTS),17,18 which is a hybrid 3D similarity calculation algorithm considering both the molecular shape and pharmacophore features, was used for virtual screening to find novel scaffolds of plant activators.19,20
Pyridazinone scaffolds are a series of potent bioactive molecules applied in medicine and agriculture. Among them, 3(2H)-pyridazinone derivatives are extensively developed in crop protection, such as insecticide/acaricide,21 herbicide,22 plant growth regulator,23 and fungicides,24 while there is no application as plant activators.
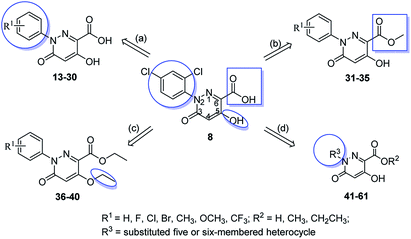
In this communication, based on the structures of PBZ, BTH and plant hormones SA and jasmonic acid (JA), several 3(2H)-pyridazinone compounds with high similarity scores were discovered as promising plant activators by SHAFTS method against the MayBridge database. In order to verify the lead compound, we bought them from the commercial library and tested the in vivo induced resistance activity against four pathogens (Fig. 1, Table S1†). Results displayed that compound 8 had induced resistance against two pathogens. Therefore, compound 8 was chosen as the lead compound for the derivation of the new plant activator (Fig. 1). By comparing the activities of the four compounds, the preliminary structure–activity relationship was obtained as follows. The large steric hindrance in the 2-position and the electron-withdrawing property in the 6-position of compound 5 may be beneficial to the bioactivity (5 vs. 6). The aromatic ring in the position [4,5-d] maybe optional and the electron-donating group in the 6-position of compound 7 may reduce the bioactivity (6 vs. 7); hydrogen bond donor –OH and acceptor –COOH in the 5 and 6 positions of compound 8 may be necessary for broad-spectrum antimicrobial activity (7 vs. 8). Therefore, the optimization of the lead compound was carried out through four aspects. (a) Introducing the substituents to the benzene ring; (b) changing the acid to ester; (c) modifying the hydroxyl group; (d) replacement of benzene ring by a five- or six-membered heterocyclic ring (Fig. 2). Hence, a series of 5-hydroxyl-2-aryl-3-oxo-2,3-dihydropyridazine-6-carboxylic acid and its derivatives with broad-spectrum induced antimicrobial activity were designed and synthesized, and their structure–activity relationship was studied.
Initially, we introduced different substituents into ortho-, meta-, and para-positions of the phenyl ring to investigate the electronic and steric effect on the activity. Compounds 13–30 were synthesized, and their in vivo induced resistance activities are shown in Tables 1 and 2. According to the results, most of the compounds showed a strong induced resistance activity against two or more test pathogens. Comparing the compounds 13–18 (Table 1) and 19–24 (Table 2), when a mono-substituent was introduced to the para-position of the benzene ring, all compounds exhibited improved activities compared to the none substituted compound 13, which simultaneously showed resistance against two or three pathogens (compounds 14–18). The general trend in induced activity was as follows: 4-CF3 ≥ 4-F ≥ 4-Cl ≥ 4-OCF3 ≥ 4-OCH3.
| Compound | R1 | Efficacy (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MM | CC | RS | BC | FO | PI | PT | PL | ||
| a ND-not detected; inducing activity > 50% are shown in bold. | |||||||||
| 13 | H | 84.44 | 82.62 | −2.86 | −8.40 | 5.94 | 33.17 | ND | 26.95 |
| 14 | 4-F | 27.78 | 75.41 | 50.05 | −1.41 | −7.5 | 7.09 | ND | 52.77 |
| 15 | 4-Cl | 58.33 | 50.82 | −4.04 | −9.06 | 11.31 | 32.46 | ND | 69.58 |
| 16 | 4-CF3 | 17.5 | 82.3 | 6.01 | 11.78 | −2.13 | 54.62 | ND | 64.98 |
| 17 | 4-OCH3 | 58.61 | 44.26 | 46.35 | 24.19 | 2.58 | 2.08 | ND | 51.89 |
| 18 | 4-OCF3 | ND | ND | 59.05 | ND | 76.92 | 16.67 | 66.15 | ND |
| 25 | 3,5-Di-Cl | 91.67 | 72.31 | −2.86 | −10.4 | −1.46 | 23.53 | ND | 60.38 |
| 26 | 2,4-Di-Cl | 39.17 | 81.31 | 12.51 | −0.91 | 11.31 | 26.38 | ND | 48.7 |
| 27 | 3,4-Di-Cl | 43.33 | 65.66 | 3.74 | −15.1 | 1.9 | 19.95 | ND | 38.44 |
| 28 | 3,4-Di-F | ND | ND | 53.12 | ND | 82.05 | −9.72 | 100 | ND |
| 29 | 4-F-3-CF3 | ND | ND | 53.12 | ND | 10.26 | 2.78 | 100 | ND |
| 30 | 3,5-Di-OCH3 | ND | ND | 59.05 | ND | 92.31 | −5.56 | 44.62 | ND |
| BTH | 88.06 | −8.2 | 16.06 | 46.8 | 19.37 | 64.98 | ND | 75.59 | |
| 50% kresoximmethyl (WG) | 98.43 | ||||||||
| 75% chlorothalonil (WP) | 90.16 | ||||||||
| 5% validamycin A (WP) | 83.38 | ||||||||
| 50% procymidone (WP) | 41.87 | ||||||||
| 70% mildothane (WP) | 75.14 | ||||||||
| 50% dimethomorph (WP) | 96.78 | ||||||||
| 20% bismerthlazol (WP) | 68.55 | 67.8 | |||||||
| Compound | R1 | Efficacy (%) | |||
|---|---|---|---|---|---|
| RS | FO | PI | PT | ||
| a ND-not detected; inducing activity > 50% are shown in bold. | |||||
| 19 | 2-F | 40.36 | 71.79 | 13.89 | 4.62 |
| 20 | 2-CF3 | 54.9 | 23.08 | 25 | 44.62 |
| 21 | 2-OCH3 | 32.34 | 38.46 | 22.22 | ND |
| 22 | 3-F | 30.17 | 35.9 | 6.94 | 4.62 |
| 23 | 3-CF3 | 61.42 | −23.08 | 11.11 | 38.46 |
| 24 | 3-OCH3 | 73 | 58.97 | 0 | 78.46 |
| 5% validamycin A (WP) | 83.38 | ||||
| 70% mildothane (WP) | 75.14 | ||||
| 50% dimethomorph (WP) | 96.78 | ||||
| 20% bismerthlazol (WP) | 68.55 | ||||
The results indicated that the activity would be better with the introduction of a fluorine-containing group in the para-position of the benzene ring due to the electron absorption effect. However, when these groups were introduced into the ortho- or meta-position (compounds 19–24), the activity almost disappeared except for compound 24. We believe that there are two reasons for this. One is that the dihedral angle between the benzene ring and the pyridazinone ring became larger when the groups were introduced from the para-position to the ortho-position, leading to a reduction in the activity. The other is that the larger steric hindrance at the para-position was better than the meta-position for the activity. On the other hand, when two-substituents were introduced to the benzene ring, the induced activities of compounds against test pathogens were in the following order: 3,4-di-F ≥ 3,5-di-Cl ≥ 4-F-3-CF3 ≥ 3,5-di-OCH3 » 2,4-di-Cl, 3,4-di-Cl. It implied that the fluorine-containing group introduced to the 4-position or the 3,5-di-substituent of the benzene ring was beneficial to broad-spectrum disease resistance. To sum up, the stronger the electron absorption effect of the phenyl group, the higher the activity.
Next, we explored the importance of the withdrawing effect of the carboxylic group through esterification and compounds 31–35 were derived. The results showed that these compounds had an improved broad-spectrum activity (Table 3). Surprisingly, compound 32 showed excellent activity against four pathogens. Hence, the withdrawing effect of the carboxylic group is not necessary in the existence of the hydrogen bond acceptor for broad-spectrum disease resistance.
| Compound | R1 | Efficacy (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MM | CC | RS | BC | FO | PI | PT | PL | ||
| a ND-not detected; inducing activity > 50% are shown in bold. | |||||||||
| 31 | 4-F | 71.53 | 62.55 | 32.02 | 20.2 | −2.8 | 9.23 | ND | 50.83 |
| 32 | 4-OCH3 | 55.83 | 67.9 | 64.53 | 29.18 | −5.49 | 22.1 | ND | 54.36 |
| 33 | 2-OCH3 | ND | ND | 68.55 | ND | 87.18 | 5.56 | 63.08 | ND |
| 34 | 3-F | ND | ND | 24.04 | ND | 17.95 | 29.86 | −66.2 | ND |
| 35 | 3,4-Di-Cl | 10 | 74.43 | 39.7 | −11.9 | −0.11 | 14.95 | ND | 71.61 |
| BTH | 88.06 | −8.2 | 16.06 | 46.8 | 19.37 | 64.98 | ND | 75.59 | |
| 50% kresoximmethyl (WG) | 98.43 | ||||||||
| 75% chlorothalonil (WP) | 90.16 | ||||||||
| 5% validamycin A (WP) | 83.38 | ||||||||
| 50% procymidone (WP) | 41.87 | ||||||||
| 70% mildothane (WP) | 75.14 | ||||||||
| 50% dimethomorph (WP) | 96.78 | ||||||||
| 20% bismerthlazol (WP) | 68.55 | 67.8 | |||||||
Then, we investigated the necessity of the hydrogen bond donor –OH. The results are shown in Table S3;† when the hydroxyl group was replaced by ether, the activity was lost. It implied that the hydrogen bond donor hydroxyl group is very important for the induced activity.
Based on the above research and the structures of the commercial plant activators, a strategy of bioisosteric replacement of benzene ring with a heterocyclic ring was carried out for the discussion of the effect of aromaticity and polarity of 2-substituent of the pyridazinone ring on activity. The results are shown in Table S4;† when the benzene ring was replaced by a five- or six-membered heterocyclic ring with a nitrogen atom, the induced resistance activity was weakened. When the benzene ring was replaced by a pyridin-4-yl group, it was helpful to maintain the activity by introducing one electron-withdrawing group to the pyridine ring than an electron-donating group, and the higher the polarity of the substituents, the better the activity (compounds 41–45). Whereas, the ester compounds exhibited a mild decrease in the activity than their acid products (compounds 47–51). A similar conclusion could be obtained when introducing other five-membered heterocycles with a nitrogen atom into the pyridazione ring (compounds 53–61).
For plant activators, one of the remarkable characteristics is that they are effective in vivo but not in vitro. Therefore, some representative compounds with good in vivo activity were chosen to test the direct antimicrobial activity. As we can see (Table 4), all compounds had no direct antimicrobial activity in vitro compared to the positive controls. The results demonstrated their potency to be used as novel plant activators. Therefore, 3(2H)-pyridazinone derivatives have great potential as new plant activators in crop protection.
| Compound | Efficacy (%) | |||
|---|---|---|---|---|
| MM | CC | RS | PI | |
| 14 | 0.00 | 9.80 | 4.75 | 5.96 |
| 15 | 15.12 | 13.14 | 0.59 | 1.99 |
| 16 | 4.26 | 11.58 | 4.15 | 7.62 |
| 17 | 5.62 | 10.24 | 0.89 | 6.29 |
| 25 | 10.47 | 9.35 | 0.89 | 3.64 |
| 31 | 1.36 | 18.26 | 0.00 | 2.98 |
| 32 | 9.69 | 11.58 | 8.90 | 0.33 |
| 35 | 4.56 | 20.04 | −1.48 | 7.28 |
| CK | 10 | 12 | 19 | 11 |
| 50% kresoximmethyl (WG) | 71 | |||
| 75% chlorothalonil (WP) | 21 | |||
| 5% validamycin A (WP) | 50 | |||
| 50% dimethomorph (WP) | 80 | |||
In summary, based on the SHAFTS method, a new scaffold for novel plant activators was predicted and a series of 3(2H)-pyridazinone derivatives as novel plant activators were designed and synthesized. Their induced resistance activities were evaluated through in vivo and in vitro assays using a broad spectrum of pathogens, including fungi, bacteria, and oomycete. The results showed that most 2-phenyl or heterocyclic rings with electron-withdrawing group substituted 3(2H)-pyridazinone derivatives are potent to be plant activators, which exhibited high efficacy in vivo inducing activity but no direct antibiotic activity in vitro. The hydrogen bond donor –OH in the 3-position and hydrogen bond acceptor groups in the 6-position of 3(2H)-pyridazinone may be vital for broad-spectrum disease resistance. Among them, compound 32 showed good broad-spectrum efficacy against four pathogens. Altogether, 3(2H)-pyridazinone derivatives have great potential as new plant activators in crop protection.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Research and Development Program of China (2018YFD0200100), and the Fundamental Research Funds for the Central Universities.Notes and references
- M. Narusaka, T. Minami, C. Iwabuchi, T. Hamasaki, S. Takasaki, K. Kawamura and Y. Narusaka, PLoS One, 2015, 10, e0115864 CrossRef.
- K. Jiang, T. Kurimoto, E. K. Seo, S. Miyazaki, M. Nakajima, H. Nakamura and T. Asami, J. Agric. Food Chem., 2015, 63, 7124–7133 CrossRef CAS.
- A. L. S. Rangel de Souza, S. A. De Souza, M. V. V. De Oliveira, T. M. Ferraz, F. A. M. M. A. Figueiredo, N. D. Da Silva, P. L. Rangel, C. R. S. Panisset, F. L. Olivares, E. Campostrini and G. A. De Souza Filho, Plant Soil, 2015, 399, 257–270 CrossRef.
- Y. Bektas and T. Eulgem, Front. Plant Sci., 2014, 5, 804 Search PubMed.
- H. J. Lee, Y. J. Park, P. J. Seo, J. H. Kim, H. J. Sim, S. G. Kim and C. M. Park, Plant Cell, 2015, 27, 3425–3438 CrossRef CAS PubMed.
- M. Uchiyama, H. Abe, R. Sato, M. Shimura and T. Watanabe, Agric. Biol. Chem., 1973, 37, 737–745 CrossRef CAS.
- L. Friedrich, K. Lawton, W. Ruess, P. Masner, N. Specker, M. G. Rella, B. Meier, S. Dincher, T. Staub, S. Uknes, J.-P. Métraux, H. Kessmann and J. Ryals, Plant J., 1996, 10, 61–70 CrossRef CAS.
- W. Kunz, R. Schurter and T. Maetzke, Pestic. Sci., 1997, 50, 275–282 CrossRef CAS.
- K. Tsubata, K. Kuroda, Y. Yamamoto and N. Yasokawa, J. Pestic. Sci., 2006, 31, 161–162 CrossRef CAS.
- M. Ogava, A. Kadowaki, T. Yamada, and O. Kadooka, R&D Report, 2011, 1, 1–16.
- Y. Xu, Z. Zhao, X. Qian, Z. Qian, W. Tian and J. Zhong, J. Agric. Food Chem., 2006, 54, 8793–8798 CrossRef CAS.
- Q. Du, W. Zhu, Z. Zhao, X. Qian and Y. Xu, J. Agric. Food Chem., 2012, 60, 346–353 CrossRef CAS.
- M. Yasuda, H. Nakashita, S. Hasegawa, M. Nishioka, Y. Arai, M. Uramoto, I. Yamaguchi and S. Yoshida, Biosci., Biotechnol., Biochem., 2003, 67, 322–328 CrossRef CAS.
- M. Nishioka, H. Nakashita, H. Suzuki, S. Akiyama, S. Yoshida and I. Yamaguchi, J. Pestic. Sci., 2003, 28, 416–421 CrossRef CAS.
- P. Stanetty, M. Turner and M. D. Mihovilovic, Journal, 2005, 10, 367–375 CAS.
- Z. Fan, Z. Shi, H. Zhang, X. Liu, L. Bao, L. Ma, X. Zuo, Q. Zheng and N. Mi, J. Agric. Food Chem., 2009, 57, 4279–4286 CrossRef CAS.
- X. Liu, H. Jiang and H. Li, J. Chem. Inf. Model., 2011, 51, 2372–2385 CrossRef CAS.
- W. Lu, X. Liu, X. Cao, M. Xue, K. Liu, Z. Zhao, X. Shen, H. Jiang, Y. Xu, J. Huang and H. Li, J. Med. Chem., 2011, 54, 3564–3574 CrossRef CAS.
- K. Chang, Y. Shi, J. Chen, Z. He, Z. Xu, Z. Zhao, W. Zhu, H. Li, Y. Xu, B. Li and X. Qian, MedChemComm, 2016, 7, 1849–1857 RSC.
- K. Chang, J. Q. Chen, Y. X. Shi, M. J. Sun, P. F. Li, Z. J. Zhao, W. P. Zhu, H. L. Li, Y. F. Xu, B. J. Li and X. H. Qian, Chin. Chem. Lett., 2017, 28, 919–926 CrossRef CAS.
- K. Hirata, Y. Kawamura, M. Kudo and H. Igarashi, J. Pestic. Sci., 1995, 20, 213–221 CrossRef.
- B. Riggle and B. Dunbar, J. Agric. Food Chem., 1990, 38, 1922–1925 CrossRef CAS.
- D. H. Schoene and O. L. Hoffmann, Science, 1949, 109, 588–590 CrossRef CAS.
- S. Tsuboi, S. Sasaki and Y. Hattori, US Pat., 4731385, 1988.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06892a |
| This journal is © The Royal Society of Chemistry 2019 |