Open Access Article
Open Access ArticleDual-signal lateral flow assay using vancomycin-modified nanotags for rapid and sensitive detection of Staphylococcus aureus†
Shu Wang‡
ab,
Wanzhu Shen‡cd,
Shuai Zhengcd,
Zhigang Lia,
Chongwen Wang *cd,
Long Zhang*ab and
Yong Liu*ab
*cd,
Long Zhang*ab and
Yong Liu*ab
aHefei Institute of Physical Science, Chinese Academy of Sciences, Hefei 230036, PR China. E-mail: liuyong@aiofm.ac.cn; zhanglong@aiofm.ac.cn; wangchongwen1987@126.com
bUniversity of Science and Technology of China, Hefei 230036, PR China
cAnhui Agricultural University, Hefei 230036, PR China
dBeijing Institute of Radiation Medicine, Beijing 100850, PR China
First published on 9th April 2021
Abstract
This paper reports a colorimetric-fluorescent dual-signal lateral flow assay (LFA) based on vancomycin (Van)-modified SiO2–Au-QD tags for sensitive and quantitative detection of Staphylococcus aureus (S. aureus). The combination of high-performance Van-tags and detection antibodies integrated into the LFA system produced assays with high sensitivity and specificity. The visualization limit of the colorimetric signal and the detection limit of the fluorescence signal of the proposed method for S. aureus can reach 104 and 100 cells mL−1, respectively.
Staphylococcus aureus (S. aureus) is one of the major iatrogenic and foodborne bacteria that cause a wide range of infectious diseases, such as skin infection, sepsis, pneumonia, and osteomyelitis.1,2 Accurate detection of S. aureus in food samples and clinical specimens is crucial to guarantee food safety and guide effective treatment. Current diagnostic technologies, mainly conventional blood cultures or plate cultures, polymerase chain reaction (PCR), mass spectrometry, and DNA sequencing-based methods, require long testing time (several hours to several days), complicated equipment, and tedious procedures, thus limiting their applications to point-of-care testing (POCT).3–5
Lateral Flow Assay (LFA) has become the most mature and widely used POCT method in recent years because of its rapidity, portability, low cost, and simple operation.6–9 However, utilizing LFA for sensitive detection of S. aureus in complex samples is still a challenge. Limited research on LFA-based methods for detection of S. aureus has been reported, and the difficulty lies in three aspects. First, traditional LFA strip is based on colloidal gold (Au NPs) as reporters to provide colorimetric signal, which result in limited sensitivity.10 Second, complex matrix interferences in real samples adversely affect the stability and practical application of LFA system for bacteria detection.11 Third, a pair of antibodies to S. aureus with high affinity and no cross-reactivity is needed for LFA detection; such antibodies are difficult to screen and expensive with poor reproducibility.12,13 To address these problems, scholars should focus on developing and integrating novel nanotags with high signal intensity and good stability and new biorecognition molecules integrated into LFA system for S. aureus detection.
Vancomycin (Van) is a broad-spectrum glycopeptide antibiotic that can specifically bind to D-Ala–D-Ala moieties on the cell wall of most Gram-positive bacteria via five-point hydrogen bonds.14,15 Due to its high stability, low cost, and easy preparation, vancomycin has attracted wide attention to construct biosensors for detection of Gram-positive bacteria.16–18 Our previous works demonstrated that Van-modified magnetic nanoparticles (e.g., Fe3O4@Au, and Fe3O4@Ag) could rapidly recognize and capture S. aureus in complex samples.19,20
In this work, a sensitive colorimetric-fluorescent dual-signal LFA strip for S. aureus detection was proposed on the basis of rapid and strong binding capability between vancomycin molecule-modified nanotags and S. aureus. A novel Van-modified dual-signal tag (SiO2–Au-QD–Van) was fabricated via polyethyleneimine (PEI)-mediated assembly strategy.21–23 The tag consisted of a monodisperse SiO2 core (∼200 nm) as supporter, a layer of Au NPs (3 nm) to provide colorimetric signal, a layer of carboxylated CdSe/ZnS QDs to provide strong and stable fluorescence signal, and surface-modified vancomycin to effectively bind to S. aureus (Scheme 1a). The high-performance SiO2–Au-QD–Van tag was introduced into a simple LFA strip to rapidly capture S. aureus and provide two signal modes. A detection antibody for S. aureus was coated onto the test line of the strip to immobilize the formed Van-tags-S. aureus complexes. The colorimetric intensity and fluorescence intensity of test line supported the rapid detection and high-sensitivity quantitative analysis of S. aureus, respectively. As far as we know, this work is the first to construct LFA strip by using the combination of vancomycin and specific antibody for bacteria detection.
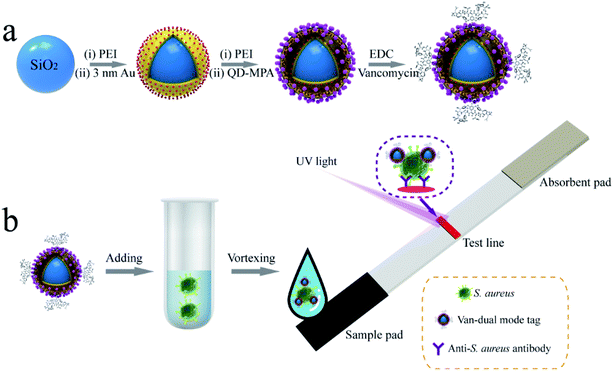 | ||
| Scheme 1 Schematic of (a) synthesis of vancomycin-modified dual-signal tag and (b) mechanisms for rapid detection of S. aureus based on dual-signal tag-based LFA strip. | ||
Scheme 1b shows a simple LFA strip designed for rapid detection of S. aureus. The strip consists of a sample pad for Van tags/bacterial sample loading, an absorbent pad to provide capillary force, and a test line to coat S. aureus detection antibody. In test samples containing target S. aureus, the added SiO2–Au-QD–Van tags bind tightly to S. aureus due to the high affinity of vancomycin toward the target bacteria. The formed SiO2–Au-QD–Van–S. aureus complexes migrate along the nitrocellulose (NC) membrane under the capillary forces of absorbent pad and are caught by the test line that coated the S. aureus antibody. Considering the high price of vancomycin antibody, we do not set a control zone on the LFA strip. Thus, only one test line shows colorimetric/fluorescence signal in the presence of S. aureus. The visible/fluorescence intensity of the LFA strip is derived from the amount of SiO2–Au-QD–Van–S. aureus complexes immobilized on the test line. The amount is proportional to the bacterial concentrations in the samples. Finally, quantitative analysis is performed in fluorescence mode because the fluorescence intensity of the strip can be easily read.
High-performance SiO2–Au-QD–Van tags are the key to building dual-signal LFA strip. As illustrated in Scheme 1a, the dual-signal core of SiO2–Au-QD–Van tag was fabricated through our previously reported LBL assembly method followed by modification of vancomycin on its surface. Firstly, monodispersed SiO2 (∼200 nm) spheres were reacted with aqueous PEI solution under intense sonication to form PEI-coated SiO2 NPs (SiO2@PEI).24 Secondly, the SiO2–Au NPs were prepared by adsorbing numerous small Au NPs (∼3 nm) onto the SiO2@PEI surface through electrostatic interaction between positively charged PEI layer and negatively charged Au NPs. Thirdly, the prepared SiO2–Au NPs were vigorously interacted with PEI solution again to coat the second PEI layer, thereby forming SiO2–Au–PEI nanostructure. Finally, SiO2–Au-QD NPs were obtained by electrostatic adsorption of dense CdSe/ZnS–MPA QDs onto the SiO2–Au–PEI surface. ESI S1† shows the detailed fabrication process of dual-signal SiO2–Au-QD NPs.
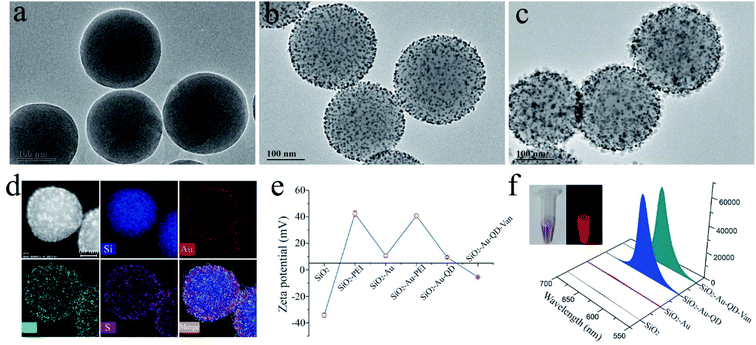
Transmission Electron Microscopy (TEM) was employed to verify the morphology of the dual-signal tags. Fig. 1a, b, and c display the typical TEM images of 200 nm SiO2 core, SiO2–Au, and SiO2–Au-QD NPs, respectively. All the prepared NPs show good dispersivity and homogeneous nanostructure. After the successive coating of 3 nm Au NPs layer and QDs layer, the diameter of SiO2–Au-QD NPs increased to approximately 240 nm. The EDS elemental mapping results further demonstrated the dense and uniform distribution of Au NPs (red signal) on the SiO2 surface (blue signal) and CdSe/ZnS QDs (azure and purple signal) on the SiO2–Au surface (Fig. 1d). The abundant CdSe/ZnS-MPA QDs on the outermost layer of the dual-signal tags not only generated high luminescence for quantitative analysis but also provided numerous carboxyl groups for subsequent modification of vancomycin. The zeta potential of SiO2, SiO2@PEI, SiO2–Au, SiO2–Au–PEI, and SiO2–Au-QD NPs were measured to be −34.4, +42.3, +10.6, +40.7, and +9.4 mV, respectively (Fig. 1e). The zeta potential of the products increased significantly when the PEI layer was formed and decreased after the adsorption of negatively charged Au NPs and QDs. This finding indicated that the formation of dual-signal tags was driven by electrostatic interaction.
Vancomycin molecules were modified onto the surface of SiO2–Au-QD NPs according to our previously reported approach.25 Scheme 1a shows the binding of the surface carboxyl groups of SiO2–Au-QD NPs to vancomycin molecules through carbodiimide activation to form SiO2–Au-QD–Van NPs. Fig. 1e and S1a† show that the zeta potential of SiO2–Au-QD decreased to −5.5 mV after vancomycin modification. This finding demonstrated the successful preparation of SiO2–Au-QD–Van tags. In addition, Fig. 1f (inset) presents the photographs of SiO2–Au-QD–Van tags under visible and UV light, indicating the good colorimetric signal and luminescence of the dual-signal tags. The figure also shows the fluorescence spectra of SiO2, SiO2–Au, SiO2–Au-QD, and SiO2–Au-QD–Van tags. SiO2–Au-QD–Van NPs as well as SiO2–Au-QD NPs exhibited strong fluorescence signal at 625 nm by excitation with 360 nm UV light, indicating that fluorescence quenching did not occur after vancomycin modification. The SiO2–Au-QD–Van tags showed excellent optical/chemical stability in the sample solution within a wide pH range of 4–13 due to the high stability of CdSe/ZnS core–shell QDs and SiO2 core used (Fig. S1b†). These results suggested the potential of SiO2–Au-QD–Van tags for bacteria detection in complex samples.
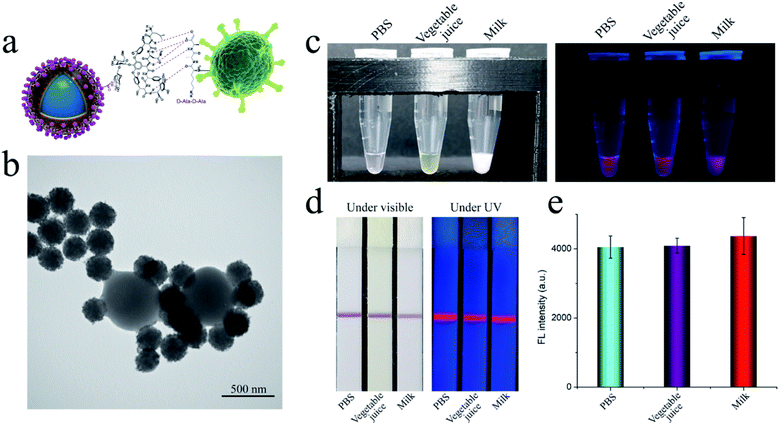
Vancomycin binds to the cell wall of S. aureus though hydrogen bonding in the terminal peptide unit (D-Ala–D-Ala) (Fig. 2a). The TEM study confirmed the effective binding of the fabricated SiO2–Au-QD–Van tags to S. aureus in aqueous solution (Fig. 2b). The S. aureus used in this work was verified by PCR (Fig. S2†), indicating the amplification of the specific gene of S. aureus.26 To further study the bind ability of SiO2–Au-QD–Van tags in complex samples, we spiked 50 μL of high-salt solution (0.1 M PBS), vegetable juice, and milk samples with S. aureus (106 cells mL−1) and mixed them with Van nanotags (2 μL). As shown in Fig. 2c, the Van-tags were dispersed well in these complex samples with stable fluorescence intensity. The three samples containing Van tags were mixed with 25 μL of PBST buffer (1% Tween) and then tested by S. aureus antibody-modified LFA strip. Fig. 2d reveals the photographs and fluorescence images of dual-signal LFA strips with the test line for S. aureus detection. All the test lines showed a distinct purple color band and bright fluorescence band at 106 cells mL−1 of S. aureus under visible light and UV light, respectively. The detailed fluorescence signal value of the test lines was measured by a commercial fluorescent reader (Fig. 2e). The fluorescence intensity only slightly differed among the three tested strips, indicating that the SiO2–Au-QD–Van based LFA strip can work well in real samples.
After the feasibility of the proposed method was confirmed, some important experimental conditions were optimized to ensure the high performance of the LFA system. The NC membrane type of the LFA strip was first investigated, considering the big size of S. aureus (∼600 nm) and SiO2–Au-QD–Van tags (∼250 nm). The suitable NC membrane allowed the good transport of S. aureus and nanotags, and generated the best immune binding efficiency on the test line.27 As revealed in Fig. S3a,† we achieved the highest signal-to-noise ratio (SNR) for S. aureus detection using CN95 membrane with 15 μm pore size. Our previous works systematically investigated the running buffer for big nanotags.28–30 We made slight adjustments and found that the PBS solution (10 mM, pH 7.4)-based running buffer containing 1% Tween 20 and 1% BSA can reduce the nonspecific adsorption of SiO2–Au-QD–Van tags on the NC membrane and generate the highest fluorescence intensity on the test line (Fig. S3b†). In addition, 0.8 mg mL−1 of the detection antibody coated on the test line is sufficient for S. aureus detection (Fig. S3c†).
In this study, SiO2–Au-QD–Van tags were directly added into 50 μL of the test sample to bind to S. aureus. The mixture was then mixed with 25 μL of running buffer for LFA detection. As mentioned in ESI S1.3,† the concentration of SiO2–Au-QD–Van solution was fixed at 2 mg mL−1. Notably, the concentration of SiO2–Au-QD–Van tags used for LFA detection seriously influence the final fluorescence signal at the test line on the strip. As shown in Fig. S4,† excessive amounts of SiO2–Au-QD–Van tags (4 μL) used reduce the detection sensitivity and increase the fluorescence background of LFA strip. This phenomenon can be attributed to the dense SiO2–Au-QD–Van tags attached on the bacterial surface will affect the formation of nanotags S. aureus-antibody sandwich complexes on the test line. Thus, the optimized dosage of SiO2–Au-QD–Van tags was chosen 3 μL (2 mg mL−1) for LFA-based S. aureus detection.
The incubation time of SiO2–Au-QD–Van tags (3 μL) with S. aureus was next optimized to achieve the highest sensitivity. The TEM images in Fig. S5a† reveal that the number of SiO2–Au-QD–Van tags bound to S. aureus increased with increasing incubation time (2–8 min). This finding suggested the high affinity of Van-tags toward S. aureus. However, we found that excessive Van-tags conjugated outside the bacterial surface had adverse effect on the immunoreaction between the bacteria and test line (Fig. S5b†). Incubation for 2 min is suitable for Van-tags-based LFA detection. The proposed LFA strip only needs 12 min for one testing with the addition of 10 min for chromatography.
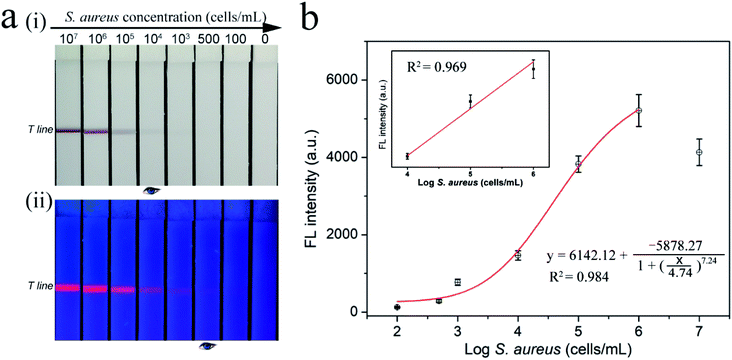
After the optimization of the experimental details, the sensitivity of the SiO2–Au-QD–Van tags-based LFA strip was verified. We tested a series of samples containing different concentrations (107 to 0 cell mL−1) of S. aureus. Fig. 3a shows the photographs (i) and fluorescence images (ii) of the test strips for S. aureus detection. The visible purple color band and red fluorescence band of the test line were observed at high bacteria concentrations, and their signal intensity decreased gradually with decreasing concentrations of S. aureus. As shown in Fig. 3a(i), the visible test line of the tested strips for S. aureus was observed at 104 cells mL−1 by the naked eye. Under UV light, the fluorescence signal of the SiO2–Au-QD–Van tags can significantly improve the detection sensitivity of LFA strip and can be applied to quantitative analysis. Based on the fluorescence images in Fig. 3a(ii), the fluorescence signal on the test line was visible in as low as 500 cells mL−1 of S. aureus. Fig. 3b shows the corresponding calibration curve of the recorded fluorescence intensity. The calibration curve is fitted by a four-parameter logistic equation as shown in inset of Fig. 3b, where y is the fluorescence intensity at test line, and x is the logarithmic concentration of S. aureus. We estimated the limit of detection (LOD) by the fluorescence signal for S. aureus to be 100 cells mL−1 at an SNR of 3 by using the IUPAC protocol.31,32 Thus, the fluorescence detection limit of our proposed dual-signal strip is 100 times more sensitive than that acquired from the colorimetric signal. Moreover, the detection performance (sensitivity and dynamic range) of SiO2–Au-QD–Van tags based-LFA in PBS buffer and vegetable juice were equal to that of in milk samples (Fig. S6†). These results indicated the good stability of the proposed method in different samples. We also employed traditional plate counting to verify the accuracy of our method. As shown in Fig. S7,† the results of plate counting method are consistent with the those of Van-tags based LFA.
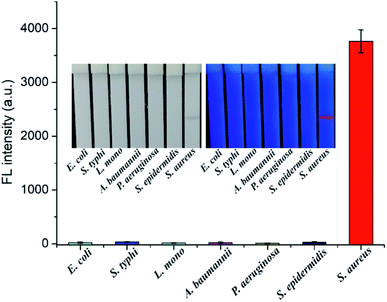
We determined the reproducibility of our method by testing four batches of dual-signal LFA strips with 106 and 104 cells mL−1 S. aureus. As shown in Fig. S8,† all the LFA strips exhibited homogeneous and stable visible/fluorescence signal on the test lines, with relative standard deviation (RSD) less than 6.8%. This finding demonstrated the good reproducibility of SiO2–Au-QD–Van-based LFA. We further assessed the specificity of the proposed method by testing various common pathogenic bacteria including Escherichia coli (E. coli), Salmonella typhimurium (S. typhi), L. monocytogenes (L. mono), Acinetobacter baumannii (A. baumannii), Pseudomonas aeruginosa (P. aeruginosa), and Staphylococcus epidermidis (S. epidermidis) as interferents. All the bacterial samples were prepared at a concentration of 105 cells mL−1 and detected by SiO2–Au-QD–Van-based LFA. As shown in Fig. 4, the positive group (S. aureus) exhibited distinct visible/fluorescence band on the tested strip, whereas no test line was observed for the six other bacteria. These results indicated the good specificity of our method. Notably, several previous studies have shown that the vancomycin-modified magnetic beads can effectively and electively capture multidrug-resistant S. aureus including methicillin-resistant S. aureus (MRSA) and vancomycin-resistant bacteria.15,33,34 Theoretically speaking, the SiO2–Au-QD–Van tags have the ability to bind multidrug-resistant S. aureus, including MRSA and vancomycin-resistant S. aureus. Hence, the combination of Van-nanotags and detection antibody-modified LFA strip allows the accurate detection of S. aureus between other common bacteria. In addition, the SiO2–Au-QD–Van-based LFA strips exhibited no obvious fluorescence signal attenuation of the test zone after storage for 30 days (Fig. S9†), which confirms the good stability of our proposed method.
The excellent detection performance (e.g., sensitivity, stability, specificity) of the proposed method for S. aureus could be attributed to the high-powered SiO2–Au-QD–Van used in this study. The proposed SiO2–Au-QD–Van tags combined the advantages of the stable SiO2† core, a layer of Au NPs, and a layer of carboxylated QDs. The tags cannot only have monodispersity and excellent stability in complex solution but also provides strong colorimetric and luminescence signal for rapid and sensitive determination of S. aureus. Moreover, Van-modified tags with low price but high affinity and S. aureus antibody with specificity were conjunct to improve the application of our method in POCT. The successfully developed rapid and sensitive LFA strip can ensure the detection assay to be completed within 12 min. The proposed SiO2–Au-QD–Van tag-based method exhibits better sensitivity and more functions than other LFA-based bacteria detection approaches (Table 1). The colorimetric mode of our method can be used for rapid identification of S. aureus-contaminated food or S. aureus infection samples, whereas the fluorescent mode is suitable for sensitive and quantitative determination of bacterial concentration in these samples.
| Detection method | Bacteria | LOD (cells mL−1) | Assay time (min) | Reference |
|---|---|---|---|---|
| Colorimetric LFA | S. aureus | 1 × 106 | 15 | Wiriyachaiporn, 2013 (ref. 35) |
| Colorimetric LFA | E. coli O157 | 4.5 × 103 | 20 | Zhu, 2018 (ref. 36) |
| Fluorescent LFA | E. coli O157 | 3 × 103 | 120 | Li, 2019 (ref. 37) |
| Magnetic-QD LFA | S. typhi | 3.75 × 103 | 35 | Hu, 2019 (ref. 38) |
| Magnetic-QD LFA | E. coli O157 | 2.39 × 102 | 74 | Huang, 2019 (ref. 39) |
| Fluorescent LFA | S. aureus | 1.4 × 103 | 20 | Shi, 2019 (ref. 40) |
| Fluorescent LFA | S. typhi | 5 × 102 | 15 | Zhang, 2020 (ref. 41) |
| Fe3O4@CuS LFA | E. coli O157 | 102 | 15 | Zhang, 2020 (ref. 42) |
| Nanozyme LFA | E. coli O157 | 102 | 25 | Wang, 2020 (ref. 43) |
| Colorimetric LFA | S. aureus | 1 × 103 | >20 | Zhao, 2021 (ref. 44) |
| Dual-signal LFA | S. aureus | 100 by fluorescence signal | 12 | This work |
| 104 by colorimetric signal |
In summary, we propose a simple and sensitive LFA method for S. aureus detection by using vancomycin-modified SiO2–Au-QD NPs as dual-signal tags. We fabricated novel dual-signal tags by PEI-mediated electrostatic adsorption of a layer of 3 nm Au NPs and a layer of QDs on the 200 nm SiO2 core. The tags possess excellent stability and high colorimetric/fluorescence signal. Vancomycin molecules were conjugated onto the surface of SiO2–Au-QD NPs to rapidly bind to target S. aureus with high affinity. By introducing Van-tags into LFA strip, we were able to rapidly and sensitively detect S. aureus in complex samples within 12 min via observation of colorimetric or fluorescence signal. In fluorescence quantitative mode, the LOD of SiO2–Au-QD–Van-based LFA for S. aureus is as low as 100 cells/mL. Considering the other advantages including high stability, specificity, low cost, and easy to operate, we believe that the proposed method will become a promising tool for S. aureus detection in the POCT field.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant no. 81830101), and the Natural Science Foundation of Anhui Province (grant no. 1908085QB85).References
- S. Khoshnood, M. Heidary, A. Asadi, S. Soleimani, M. Motahar, M. Savari, M. Saki and M. Abdi, Biomed. Pharmacother., 2019, 109, 1809–1818 CrossRef CAS PubMed.
- M. Shahdordizadeh, S. M. Taghdisi, N. Ansari, F. Alebooye Langroodi, K. Abnous and M. Ramezani, Sens. Actuators, B, 2017, 241, 619–635 CrossRef CAS.
- C. Wang, M. M. Meloni, X. Wu, M. Zhuo, T. He, J. Wang, C. Wang and P. Dong, AIP Adv., 2019, 9, 010701 CrossRef.
- J. Chen, S. M. Andler, J. M. Goddard, S. R. Nugen and V. M. Rotello, Chem. Soc. Rev., 2017, 46, 1272–1283 RSC.
- J. Luo, M. Jiang, J. Xiong, J. Li, X. Zhang, H. Wei and J. Yu, Anal. Chim. Acta, 2018, 1044, 147–153 CrossRef CAS PubMed.
- C. Parolo, A. Sena-Torralba, J. F. Bergua, E. Calucho, C. Fuentes-Chust, L. Hu, L. Rivas, R. Álvarez-Diduk, E. P. Nguyen, S. Cinti, D. Quesada-González and A. Merkoçi, Nat. Protoc., 2020, 15, 3788–3816 CrossRef.
- W. Shen, C. Wang, X. Yang, C. Wang, Z. Zhou, X. Liu, R. Xiao, B. Gu and S. Wang, J. Mater. Chem. C, 2020, 8, 12854 RSC.
- C. Wang, C. Wang, X. Wang, K. Wang, Y. Zhu, Z. Rong, W. Wang, R. Xiao and S. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 19495–19505 CrossRef CAS PubMed.
- L. Huang, S. Tian, W. Zhao, K. Liu, X. Ma and J. Guo, Analyst, 2020, 145, 2828–2840 RSC.
- X. Liu, X. Yang, K. Li, H. Liu, R. Xiao, W. Wang, C. Wang and S. Wang, Sens. Actuators, B, 2020, 320, 128350 CrossRef CAS.
- L. Guo, Y. Shao, H. Duan, W. Ma, Y. Leng, X. Huang and Y. Xiong, Anal. Chem., 2019, 91, 4727–4734 CrossRef CAS PubMed.
- D. Cheng, M. Yu, F. Fu, W. Han, G. Li, J. Xie, Y. Song, M. T. Swihart and E. Song, Anal. Chem., 2016, 88, 820–825 CrossRef CAS PubMed.
- J. Wang, X. Wu, C. Wang, N. Shao, P. Dong, R. Xiao and S. Wang, ACS Appl. Mater. Interfaces, 2015, 7, 20919–20929 CrossRef.
- S. Yang, H. Ouyang, X. Su, H. Gao, W. Kong, M. Wang, Q. Shu and Z. Fu, Biosens. Bioelectron., 2016, 78, 174–180 CrossRef CAS PubMed.
- T. Y. Liu, K. T. Tsai, H. H. Wang, Y. Chen, Y. H. Chen, Y. C. Chao, H. H. Chang, C. H. Lin, J. K. Wang and Y. L. Wang, Nat. Commun., 2011, 2, 538 CrossRef.
- Y. Pang, N. Wan, L. Shi, C. Wang, Z. Sun, R. Xiao and S. Wang, Anal. Chim. Acta, 2019, 1077, 288–296 CrossRef CAS PubMed.
- X. Meng, F. Li, F. Li, Y. Xiong and H. Xu, Sens. Actuators, B, 2017, 247, 546–555 CrossRef CAS.
- M. Yu, H. Wang, F. Fu, L. Li, J. Li, G. Li, Y. Song, M. T. Swihart and E. Song, Anal. Chem., 2017, 89, 4085–4090 CrossRef CAS PubMed.
- C. Zhang, C. Wang, R. Xiao, L. Tang, J. Huang, D. Wu, S. Liu, Y. Wang, D. Zhang, S. Wang and X. Chen, J. Mater. Chem. B, 2018, 6, 3751–3761 RSC.
- C. Wang, B. Gu, Q. Liu, Y. Pang, R. Xiao and S. Wang, Int. J. Nanomed., 2018, 13, 1159–1178 CrossRef CAS PubMed.
- C. Wang, X. Yang, B. Gu, H. Liu, Z. Zhou, L. Shi, X. Cheng and S. Wang, Anal. Chem., 2020, 92, 15542–15549 CrossRef CAS PubMed.
- C. Wang, W. Shen, Z. Rong, X. Liu, B. Gu, R. Xiao and S. Wang, Nanoscale, 2020, 12, 795–807 RSC.
- C. Wang, M. Li, Q. Li, K. Zhang, C. Wang, R. Xiao and S. Wang, RSC Adv., 2017, 7, 13138–13148 RSC.
- K. Wang, Y. Wang, C. Wang, X. Jia, J. Li, R. Xiao and S. Wang, RSC Adv., 2018, 8, 30825–30831 RSC.
- C. Wang, K. Zhang, Z. Zhou, Q. Li, L. Shao, R. Z. Hao, R. Xiao and S. Wang, Int. J. Nanomed., 2017, 12, 3077–3094 CrossRef CAS PubMed.
- J. Štěpán, R. Pantůček, V. Růžičková, S. Rosypal, V. Hájek and J. Doškař, Mol. Cell. Probes, 2001, 15, 249–257 CrossRef PubMed.
- R. Xiao, L. Lu, Z. Rong, C. Wang, Y. Peng, F. Wang, J. Wang, M. Sun, J. Dong, D. Wang, L. Wang, N. Sun and S. Wang, Biosens. Bioelectron., 2020, 168, 112524 CrossRef CAS PubMed.
- C. Wang, R. Xiao, S. Wang, X. Yang, Z. Bai, X. Li, Z. Rong, B. Shen and S. Wang, Biosens. Bioelectron., 2019, 146, 111754 CrossRef CAS PubMed.
- H. Liu, E. Dai, R. Xiao, Z. Zhou, M. Zhang, Z. Bai, Y. Shao, K. Qi, J. Tu, C. Wang and S. Wang, Sens. Actuators, B, 2021, 329, 129196 CrossRef CAS PubMed.
- X. Yang, X. Liu, B. Gu, H. Liu, R. Xiao, C. Wang and S. Wang, Mikrochim. Acta, 2020, 187, 570 CrossRef CAS PubMed.
- X. Wang, N. Choi, Z. Cheng, J. Ko, L. Chen and J. Choo, Anal. Chem., 2017, 89, 1163–1169 CrossRef CAS PubMed.
- X. Fu, J. Wen, J. Li, H. Lin, Y. Liu, X. Zhuang, C. Tian and L. Chen, Nanoscale, 2019, 11, 15530–15536 RSC.
- A. J. Kell, G. Stewart, S. Ryan, R. Peytavi, M. Boissinot, A. Huletsky, M. G. Bergeron and B. Simard, ACS Nano, 2008, 2, 1777–1788 CrossRef CAS PubMed.
- H. Gu, P. L. Ho, K. W. Tsang, L. Wang and B. Xu, J. Am. Chem. Soc., 2003, 125, 15702–15703 CrossRef CAS PubMed.
- S. Wiriyachaiporn, P. H. Howarth, K. D. Bruce and L. A. Dailey, Diagn. Microbiol. Infect. Dis., 2013, 75, 28–36 CrossRef CAS PubMed.
- C. Zhu, G. Zhao and W. Dou, Anal. Chim. Acta, 2018, 1038, 97–104 CrossRef CAS PubMed.
- Q. Li, Y. Yang, F. Hu, Y. Cai, X. Liu and X. He, Anal. Biochem., 2019, 564–565, 32–39 CrossRef CAS PubMed.
- J. Hu, Y. Z. Jiang, M. Tang, L. L. Wu, H. Y. Xie, Z. L. Zhang and D. W. Pang, Anal. Chem., 2019, 91, 1178–1184 CrossRef CAS PubMed.
- Z. Huang, J. Peng, J. Han, G. Zhang, Y. Huang, M. Duan, D. Liu, Y. Xiong, S. Xia and W. Lai, Food Chem., 2019, 276, 333–341 CrossRef CAS PubMed.
- Y. Shi, H. Yang, Y. He and Z. Fu, Talanta, 2019, 205, 120130 CrossRef CAS PubMed.
- B. Zhang, X. Yang, X. Liu, J. Li, C. Wang and S. Wang, RSC Adv., 2020, 10, 2483–2489 RSC.
- M. Zhang, T. Bu, Y. Tian, X. Sun, Q. Wang, Y. Liu, F. Bai, S. Zhao and L. Wang, Food Chem., 2020, 332, 127398 CrossRef CAS PubMed.
- Z. Wang, X. Yao, Y. Zhang, R. Wang, Y. Ji, J. Sun, D. Zhang and J. Wang, Food Chem., 2020, 329, 127224 CrossRef CAS PubMed.
- M. Zhao, X. Yao, S. Liu, H. Zhang, L. Wang, X. Yin, L. Su, B. Xu, J. Wang, Q. Lan and D. Zhang, Food Chem., 2021, 339, 127955 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01085a |
| ‡ The authors S. Wang and W. Shen contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |