 Open Access Article
Open Access ArticleTargeted DNP for biomolecular solid-state NMR
Diego
Gauto†
 a,
Ons
Dakhlaoui
a,
Ons
Dakhlaoui
 ab,
Ildefonso
Marin-Montesinos
ab,
Ildefonso
Marin-Montesinos
 ac,
Sabine
Hediger
ac,
Sabine
Hediger
 *a and
Gaël
De Paëpe
*a and
Gaël
De Paëpe
 *a
*a
aUniv. Grenoble Alpes, CEA, CNRS, IRIG-MEM, Grenoble, France. E-mail: sabine.hediger@cea.fr; gael.depaepe@cea.fr
bUniv. Grenoble Alpes, CNRS, CERMAV, Grenoble, France
cUniversity of Aveiro, CICECO Chem. Dept., Aveiro, Portugal
First published on 23rd March 2021
Abstract
High-field dynamic nuclear polarization is revolutionizing the scope of solid-state NMR with new applications in surface chemistry, materials science and structural biology. In this perspective article, we focus on a specific DNP approach, called targeted DNP, in which the paramagnets introduced to polarize are not uniformly distributed in the sample but site-specifically located on the biomolecular system. After reviewing the various targeting strategies reported to date, including a bio-orthogonal chemistry-based approach, we discuss the potential of targeted DNP to improve the overall NMR sensitivity while avoiding the use of glass-forming DNP matrix. This is especially relevant to the study of diluted biomolecular systems such as, for instance, membrane proteins within their lipidic environment. We also discuss routes towards extracting structural information from paramagnetic relaxation enhancement (PRE) induced by targeted DNP at cryogenic temperature, and the possibility to recover site-specific information in the vicinity of the paramagnetic moieties using high-resolution selective DNP spectra. Finally, we review the potential of targeted DNP for in-cell NMR studies and how it can be used to extract a given protein NMR signal from a complex cellular background.
Introduction
The function of biological systems, e.g. proteins, nucleic acids, lipid membranes and carbohydrates is rooted in the details of their dynamical structures. Thus, structural insights in terms of stability, conformational dynamics and intra-/inter-molecular interactions can not only provide answers regarding why and how challenging biological systems perform their function, but also reveal the reasons for a loss of functionality (inhibitor binding, aggregation, etc.).For this reason, different methods were developed to elucidate the structure of biomolecules, including X-ray crystallography, cryo-electron microscopy (Cryo-EM), neutron diffraction, small-angle X-ray scattering (SAXS), and nuclear magnetic resonance (NMR). While all these methods complement each other, the specific strength of NMR is its capability to provide site-specific structural details, to reveal molecular interactions, and also to give insights into conformational dynamics, particularly relevant for the study of low-populated excited states.1–7
Despite its uniqueness, specificity and versatility, the Achilles' heel of NMR lies in its inherent low sensitivity that comes from the low interaction energies involved and the resulting small nuclear spin population differences, which are directly proportional to the NMR signal. In addition, most of the NMR active isotopes, except for 1H and 31P, are only present at very low natural abundance (e.g. 1.108% for 13C, 0.365% for 15N). As a result, advances in molecular biology to produce isotopically labelled systems have led to a major leap in the capability of NMR to provide structural insights on biomolecules. This triggered the development of innovative multidimensional NMR methods making use of the multiple spin interactions present in isotopically labeled systems. Over the years, improving sensitivity and resolution has been a constant motivation for biomolecular NMR, necessary for the structural study of ever larger and more complex systems.8
In this battle for more sensitivity, we have witnessed, for instance, the successful development of intense magnetic fields, cryogenic solution-state probes and ultra-fast magic angle sample-spinning (MAS) solid-state probes, as well as advanced pulse sequence engineering and optimized sample preparation. Finally, the successful development of a hyperpolarization approach called high field Dynamic Nuclear Polarization (DNP), pioneered by the Griffin group, is currently revolutionizing MAS solid-state NMR (ssNMR) enabling several orders of magnitude of sensitivity gains.9–11 Such unprecedented improvement in sensitivity enabled data to be recorded using experiments that were previously deemed infeasible. In addition, MAS-DNP can be applied to a broad range of systems with emerging new applications in surface chemistry, materials science and structural biology.11–21
In this perspective article, we will discuss a specific DNP approach, called targeted DNP (tDNP), in the context of biomolecular applications. We will first explain why it differs from conventional DNP experiments. We will then review various strategies to implement tDNP experiments and discuss foreseen benefits in terms of sensitivity and selectivity improvement. Next, we will report on the feasibility of obtaining structural information through paramagnetic relaxation enhancement (PRE) induced by tDNP at cryogenic temperature, and the possibility to recover high-resolution information using the recently introduced selective DNP (SelDNP) methodology. Finally, the development of tDNP in the context of in-cell DNP-enhanced NMR will be discussed.
Targeted DNP versus conventional DNP
In MAS-DNP experiments, the NMR sensitivity enhancement is achieved by transferring the polarization from unpaired electrons, contained in dopant molecules called Polarizing Agents (PAs), to the surrounding nuclear spins of the system of interest. PAs are typically composed of stable organic mono- or bi-radicals, although DNP with metals has also been demonstrated.22–37 The transfer of polarization is driven by high-power high-frequency microwave (μw) irradiation while spinning the sample at the magic angle at cryogenic temperature, usually around 100 K using cold nitrogen gas38 or at lower temperature using closed-loop cryogenic helium spinning systems.39–41In conventional biomolecular MAS-DNP experiments, PAs are typically dissolved in a glass-forming matrix (usually a mixture of glycerol or DMSO, D2O and H2O) and randomly distributed around the sample of interest. The polarization is transferred from the electron spin system to surrounding nuclei (typically 1H, 13C, 15N in the case of biomolecules), then to more distant nuclei, either directly through-space using hyperfine couplings to the electrons, and/or relying on spin-diffusion mechanism among nuclear spins.
In tDNP approaches, on the contrary, the PAs are not randomly distributed, but located or anchored at specific molecular sites. It is important to note that the use of localized paramagnetic species is not new to the field of biomolecular magnetic resonance. A large part of proteins naturally bears paramagnetic centers in the form of metal ion binding sites or naturally occurring radical moieties. These endogenous paramagnetic centers can directly be observed by electron paramagnetic resonance (EPR),42–44 whereas NMR can be used to indirectly detect the influence of the paramagnets through resonance broadening and shifting.45–47 Extension of this methodology to diamagnetic systems has led to the development of various spin-labeling strategies,48–50 enabling the use of EPR51,52 and paramagnetic NMR53–56 to study structure and dynamics of biomolecules.
Making use of spin labels in the context of MAS-DNP was thus a natural next step in the development of the technique, and several groups in the last years have investigated the benefits of tagging their biomolecular system with the PA required for hyperpolarization.57,58 The motivation and objectives of the different approaches were diverse, going from optimization of the sample preparation for best sensitivity59,60 and biocompatibility,61–63 investigation of the bleaching/quenching effect of the paramagnetic tag64–66 under DNP conditions for its potential use as a structural probe,67–69 selection of a particular protein in a complex environment,62,63,70–75 to the detection of protein–protein interactions,76 and the development of high-resolution localized techniques through selective DNP.77
Targeting strategies
Covalent binding and site-directed spin labeling of PAs
Considering that the replacement of monomeric TEMPO by the bis-nitroxide biradical TOTAPOL has been one of the major milestones in the development of high-field MAS-DNP,23 it is not surprising that the first attempt towards tDNP in 2011 has been conducted using a TOTAPOL moiety covalently linked to the system of interest, a decapeptide in this case (see Fig. 1a).59 More recently, in the context of antimicrobial peptides, Sani et al. introduced by solid-phase synthesis one or two spin label amino acid TOAC (2,2,6,6-tetramethyl-N-oxyl-4-amino-4-carboxylic acid, see Fig. 1b) at the N-terminus of a maculatin mutant in order to investigate with MAS-DNP the interaction of this antimicrobial peptide with the bacterial cell membrane of E. coli.74 Although neither the monoradical TOAC nor the biradical TOAC–TOAC are optimized PAs for MAS-DNP, DNP enhancement factors of 10 and 13, respectively, were obtained on cell samples.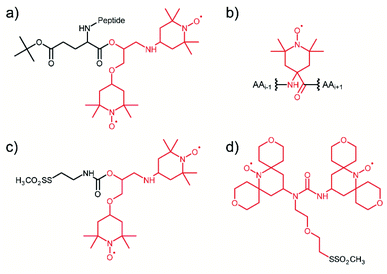 | ||
| Fig. 1 Illustration of the several tDNP spin labeling approaches based on the covalent binding of the PA onto the targeted system: (a) TOTAPOL is covalently attached to the terminal CO of a decapeptide.59 (b) Spin-label amino acid TOAC compatible with solid-phase peptide synthesis.74 (c) and (d) MTS derivatives of TOTAPOL and AMUPol, respectively, which reacts with accessible cysteine thiol groups of proteins.64,67 The original radical or biradical moiety is highlighted in red throughout the figure. | ||
Targeted DNP was also implemented on proteins using the site-directed spin-labeling (SDSL) strategy, combining site-directed mutagenesis and sulfhydryl chemistry. For this purpose, Voinov et al.64 and van der Cruijsen et al.67 synthesized methanethiosulfonate (MTS) derivatives of the DNP biradicals TOTAPOL23 and AMUPol31 (Fig. 1c and d), respectively, which can react selectively with thiol groups of surface exposed cysteines through the formation of a covalent disulfide bond. This approach is rather robust, with efficient coupling efficiencies, and also versatile, since the position of the tagged PA can be chosen using cysteine mutagenesis. In addition to MTS, protein thiol groups are known to react specifically as well with the maleimide and iodoacetamide functional groups, which can broaden the chemical possibilities for targeting PAs in the future. When used in the context of large protein complexes and membrane proteins in their lipidic environment, this strategy definitively leads to a non-uniform dispersion of PA, ensuring proximity between the diluted biomolecule of interest and the hyperpolarizing agent.
Endogenous metal DNP and site-directed spin labeling of metal PAs
Paramagnetic metal ions are ubiquitous in biology and can thus be leveraged for tDNP experiments. In particular, Mn2+ and Gd3+ form high-spin complexes (S = 5/2 for Mn2+ and S = 7/2 for Gd3+) whose magnetic properties are compatible with DNP at high magnetic fields.29 While Mn2+ has biological roles enabled by its redox reactivity, Gd3+ is interesting due to its high redox stability which makes it a favorable candidate for in-cell DNP. The first demonstration of their use as PA has been published in 2011.28 In contrast to bis-nitroxide radicals, MAS-DNP with paramagnetic metal ions rely on the solid-effect (SE) mechanism. Irradiation on the narrow central EPR transition of the half-integer electronic spin of the metal ion leads to polarization transfer to the hyperfine coupled nuclear spins.29The use of paramagnetic metal ions for tDNP has been developed so far by the Corzilius group. A first study investigated the use of endogenous Mn2+, whose presence is essential in hammerhead ribozymes for their self-cleavage function (see Fig. 2a).65,69 Extension of this approach for diamagnetic biomolecules was obtained via chelator ligands compatible with site-directed spin-labeling of cysteines, like DOTA-M or 4MMDPA (see Fig. 2b and c).68
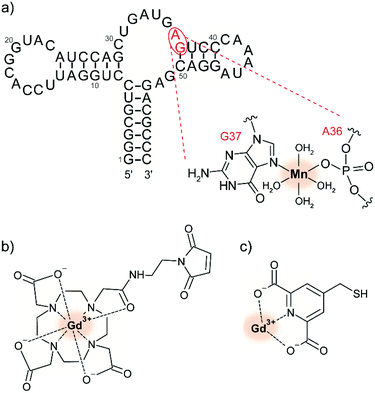 | ||
| Fig. 2 (a) Predicted secondary structure of the full-length hammerhead ribozyme with the proposed selective binding site for divalent ions. Adapted by permission from Springer Nature Customer Service Centre GmbH: Springer Nature, Journal of Biomolecular NMR, dynamic nuclear polarization of nucleic acid with endogenously bound manganese, P. Wenk et al., Copyright Springer Science + Business Media Dordrecht (2015).65 Chemical structure of (b) maleimide derivatized Gd-DOTA and (c) Gd-4MMDPA spin labels, which can react with the thiol group of exposed cysteines, forming either a carbon–sulfur or a disulfide bond, respectively.68 The metal ion used as PA is highlighted with a red shadowed circle. | ||
Ligand-binding PAs
Most of the tDNP spin-labeling approaches presented so far are based on a covalent bond attaching the PA to the protein of interest, with the exact position of the spin label being chemically engineered. Alternatives have been proposed where the location of the spin label is determined through protein–ligand interactions, with the PA being covalently attached to the ligand. In that case, the position of the PA is determined by the ligand-binding site. In addition, some selectivity is introduced through the specificity of the interaction, which might allow the targeting of a specific protein in the presence of other biomolecules.A first example in that direction was proposed by the Etzkorn group in 2016. With the aim to selectively enhance the 20 kDa Bcl-xL protein in cell lysates, they covalently attached a TOTAPOL moiety to the Bak peptide, the binding partner of Bcl-xL (see Fig. 3a).71 A similar approach was used by the group of McDermott who tagged a TOTAPOL moiety (or a derivative) to trimethoprim (TMP), which has a high affinity to E. coli dihydrofolate reductase (DHFR) (Fig. 3b), in order to investigate the signal attenuation induced by the presence of the paramagnetic spin-label66 and the possibility to maintain effective DNP efficiencies in the case of dilute samples.60 Finally, our group also reported the use of a galactose ligand tethered to a TOTAPOL moiety with the goal to selectively probe the galactose binding site of the lectin LecA with MAS-DNP (Fig. 3c).77
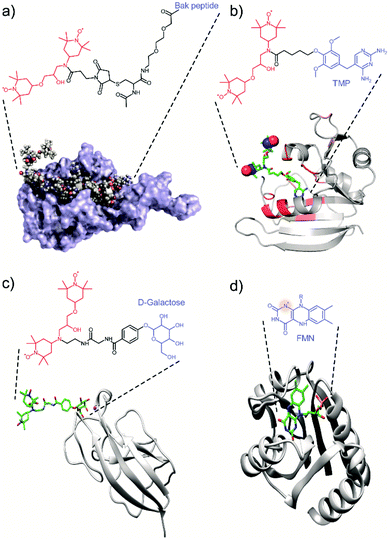 | ||
| Fig. 3 Examples of tDNP approaches that uses (a–c) the specific affinity of a ligand-derivatized PA with the protein of interest, and (d) an endogenous paramagnetic cofactor. (a) Protein Bcl-xL interacting with the Bak peptide tethered to TOTAPOL. Adapted with permission from ref. 71. Copyright 2016 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (b) DHFR protein interacting with TMP tethered to TOTAPOL. Adapted with permission from ref. 66. Copyright (2017) American Chemical Society. (c) LecA interacting with galactose tethered to TOTAPOL.77 (d) Protein flavodoxin with its endogenous stable semiquinone radical (flavin mononucleotide).61 The biradical and the ligand moiety are highlighted respectively in red and blue throughout the figure. The nitrogen radical in (d) is highlighted with a red shadowed circle. | ||
Whereas all these examples made use of exogenous PA tethered to protein ligands, some biomolecular systems naturally possess endogenous transient or stable organic radical like flavins or quinones, which can be used for tDNP. Such an example has been shown by Maly et al.61 who used the semiquinone (SQ) form of the flavin cofactor in the protein flavodoxin to obtain DNP sensitivity enhancement of proton spins by MAS-DNP (Fig. 3d). The appeal of this approach is that it allows the study of an intact biomolecular system. However, it is also limited since monomeric flavodoxins are far from being competitive in terms of DNP efficiency compared to optimized PAs, especially at high magnetic fields and fast sample spinning.
PAs with unspecific affinity: the case of lipid-anchored PAs
Unspecific affinity of the PAs with the sample of interest have been reported in several studies.78–82 In the context of matrix-free (MF) sample preparation, which circumvents the use of a glass-forming matrix, we showed that unspecific affinity of the PA for the system of interest can be used to preserve a uniform distribution of PA molecules.78,83In the context of tDNP, this MF approach is particularly interesting in the case of heterogeneous biomolecular samples with domains of different polarities like liposomes or cells. For instance, the use of DMSO or glycerol in higher concentrations, usually required for DNP samples, is known to perturb the lipid bilayer structure. In addition, it is not straightforward to predict the location of water-soluble PAs such as TOTAPOL,23 AMUPol31 or AsymPolPOK25 in such multi-component biomolecular systems. This was recently illustrated in the work by the Hong group that uses paramagnetic relaxation enhancement of lipid signals to investigate the partitioning of AMUPol and TOTAPOL in the lipid bilayer.84
To avoid perturbation of such complex and fragile biomolecular systems, and to control the location of the PA, it was relevant to investigate alternative protocols and to design new specific PAs. Thus, a lipid-anchored biradical, N-propyl-PALMIPOL, was developed in our group (see Fig. 4).62 It consists of a TOTAPOL moiety functionalized with a palmitate chain, which is able to localize to the lipidic membrane, keeping the biradical function at the surface of the liposomes (see Fig. 4). This leads to appreciable DNP enhancement despite the absence of excess cryoprotectant (DMSO/glycerol) as usually introduced in DNP samples. A similar idea was also proposed by the Long group who compared different commercial spin-labeled phospholipids, with the TEMPO moiety located either on the phosphate head group (see Fig. 4), in the middle or at the end of the lipid choline chain.63,70 Subsequently, other groups have further explored this direction and reported improved matrix-free DNP efficiency using other lipid-anchored biradicals.72,73
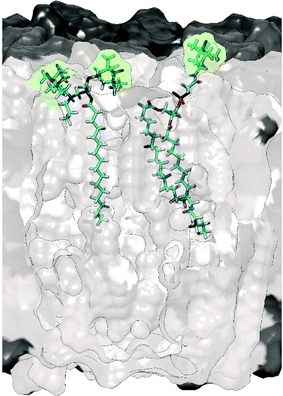 | ||
| Fig. 4 Molecular model illustrating the embedding of the biradical N-propyl-PALMIPOL62 (on the left) and the TEMPO-functionalized phosphatidylcholine TEMPO-PC70 (on the right) in the phospholipid bilayer. The green shadow highlights the location of the radical moieties. | ||
Bio-orthogonal chemistry for in-cell tDNP applications
The extension of the tDNP approach to in-cell NMR applications is highly desirable since the main limitation of such studies lies in the difficulty of detecting an “endogenous” amount of protein within a large cellular background. tDNP could certainly be useful to preferentially enhance the protein of interest over the cellular background. However, it is not straightforward to specifically target a given protein inside the cellular environment. Approaches based on cysteine chemistry are limited because of their lack of selectivity, and methods relying on protein–ligand interactions are only possible for systems with known and very specific ligands.Alternative strategies require the incorporation of an unnatural amino acid (UAA)85 and the development of bio-orthogonal chemistry.86 This path has proven very successful for the efficient and specific incorporation of optical probes into proteins of the cellular milieu.87,88 In the context of DNP experiments, Debelouchina and coworkers have reported the first successful example of targetable tetrazine-based PAs for biological systems.75 In their work, the unnatural amino acid norbornene-lysine is introduced in the protein sequence through amber suppression.85 DNP spin labeling is then obtained through biorthogonal conjugation of a TOTAPOL-tetrazine radical (TTz) as illustrated in Fig. 5. The approach has been demonstrated on several proteins and in cell lysates.75
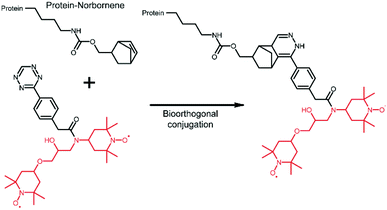 | ||
| Fig. 5 Tetrazine-derivatized TOTAPOL designed to perform a biorthogonal conjugation with the unnatural amino acid norbornene-lysine genetically introduced in the mutant protein.75 The TOTAPOL moiety is highlighted in red. | ||
Targeted DNP for improved sensitivity
As stated before, a conventional MAS-DNP experiment requires the PA to be distributed as homogeneously as possible inside the sample. This has been conventionally obtained by co-dissolving the PA and the system of interest in a glass-forming solvent mixture commonly dubbed DNP juice (for biomolecules, often a glycerol– or DMSO–water mixture).18,89 The analyte is then studied under DNP conditions in the form of a frozen solution. The major drawback of this glassy matrix approach lies in the fact that the solvent occupies a non-negligible part of the fixed and limited volume of the ssNMR sample rotor. This impacts significantly the amount of analyte inside the rotor, and consequently the sensitivity of its signal. In most cases, simply reducing the amount of solvent lead to PA aggregation and its phase separation from the system of interest with detrimental effects on the DNP efficiency. For organic and biomolecules, this phase separation can be avoided when some affinity of the PA for the analyte is present, allowing thus the preparation of matrix-free samples.78 This kind of non-specific affinity with commonly-used PAs has been described for several biomaterials and biomolecules like cellulose,78 proteins,79,80 fibrils,81 and bacterial cell wall polymers.82 In the absence of such an affinity, Takahashi et al. has shown that an intermediate molecule like trehalose can be used to bridge the affinity between the PA and the investigated system (see Fig. 6).83 | ||
| Fig. 6 Schematic illustration of the matrix-free approach developed by Takahashi et al.,83 using either direct (a) or indirect (b) affinity of the PA with the system of interest. Reproduced from ref. 83 with permission from The Royal Society of Chemistry, copyright 2013. | ||
As the PA is always in close proximity to the system of interest in MF samples and in tDNP applications, its concentration should be reoptimized, rather than relying on the same concentration as commonly used for frozen solution preparations. For instance, the supernatant solution of only 1 mM in TOTAPOL was found to be optimum for the study of extracted bacterial cell wall polymers due to the affinity of the PA for the peptidoglycan,82 whereas standard concentrations of about 10 mM are usually required in samples prepared with the conventional frozen solution method.
Similar observations were reported in the work from Voinov et al.,64 where the biradical TOTAPOL is directly attached to a solvent exposed cysteine of the heptahelical transmembrane protein, Anabaena sensory rhodopsin (ASR) embedded in a multilamellar liposome mixture (DMPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMPA 9
DMPA 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). For a given mass of protein sample, tDNP with a matrix-free deuterated buffer provided similar DNP sensitivity as the conventional frozen solution DNP buffer with TOTAPOL dissolved into a glycerol-d8/D2O/H2O matrix. Interestingly, removing the glassy matrix via the tDNP approach enabled more sample to be packed inside the rotor, which translated into a 4-fold sensitivity increase.
1). For a given mass of protein sample, tDNP with a matrix-free deuterated buffer provided similar DNP sensitivity as the conventional frozen solution DNP buffer with TOTAPOL dissolved into a glycerol-d8/D2O/H2O matrix. Interestingly, removing the glassy matrix via the tDNP approach enabled more sample to be packed inside the rotor, which translated into a 4-fold sensitivity increase.
Consistent observations were recently made in the work from Good et al. aiming at optimizing matrix-free sample preparation for membrane proteins with a biradical-tagged phospholipid as PA.73 The ability to maximize the NMR sensitivity of membrane proteins reconstituted in lipids by tDNP is an important direction since the very small amount of protein present in lipid sample (to mimic relevant physiological condition) severely limits sensitivity for conventional NMR approaches. Nevertheless, the loss of NMR resolution induced by sample freezing typically requires multiple isotopic labeling schemes to be used for obtaining intermolecular information.
Another advantage of the tDNP approach, recently highlighted by Rogawski et al., lies in the fact that sub-millimolar concentration of bound PA can be used. For instance, the authors use a biradical-derivatized trimethoprim ligand (TMP-T) of dihydrofolate reductase (DHFR) to report 2D correlation experiments of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DHFR
1 DHFR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TMP-T complexes at 0.5 mM, allowing characterization of DHFR with only ∼15 nmol of proteins.60
TMP-T complexes at 0.5 mM, allowing characterization of DHFR with only ∼15 nmol of proteins.60
Targeted DNP and paramagnetic enhanced relaxation
Introduction of paramagnetic tags to the protein of interest, covalently bonded or not, induces paramagnetic effects that can translate into contact shifts, pseudo-contact shifts and paramagnetic relaxation enhancement (PRE) effects.47,53 These effects are well-known in solution-state NMR46,90,91 and have recently been extended to biomolecular solid samples at room temperature.92,93 In the case of nitroxide radicals, the main contribution is PRE. It translates into a variation of the longitudinal and transverse coherence nuclear decay times that can be modelled to extract distance information. Even though the usual solution state PRE theory has not yet been adapted and validated for MAS NMR at cryogenic temperature, it is directly linked to the bleaching phenomenon observed in DNP, where signal from spins close to the PA are strongly attenuated and/or broadened beyond detection.78,94,95Recently the Baldus and McDermott groups reported evidence of site-specific signal bleaching for residues close to the tagged PA (Fig. 7).66,67 In the latter, the PRE induced signal attenuation of a PA-functionalized ligand was investigated with solution-state NMR and compared to 100 K DNP-enhanced data.66 Using specifically-labeled proteins, the authors measured the bleaching of the NMR signal intensity for numerous sites in the protein and showed a correlation with the distance to the tagged PA. They discuss three cases: (i) residues at 10 Å or closer to the unpaired electrons are fully bleached; (ii) residues which are 10 to 25 Å in distance are strongly affected by the presence of the paramagnets; (iii) above 30 Å, much less bleaching effect is observed.66 A more precise quantification of the distance-dependency of the bleaching effect is not straightforward, mostly because of the heterogeneous broadening at 100 K, which limits site-specific information, and the conformational disorder of the paramagnetic tag.
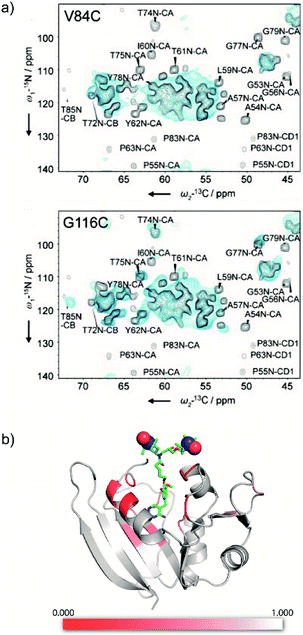 | ||
| Fig. 7 (a) 800 MHz low-temperature DNP 2D NCA of U-[13C, 15N] spectra (blue) of KcsA mutants V84C (top) and G116C (bottom), spin labeled with AMUPol-MTSSL (see Fig. 1d), compared to room temperature 700 MHz solid-state NMR spectra (black) of wild-type KcsA (black). The presence of the spin label in the mutants leads to paramagnetic bleaching at cryogenic temperature. Reproduced with permission from ref. 67. Copyright 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (b) Model of the protein DHFR complexed with the high affinity ligand TMP derivatized with TOTAPOL (see as well Fig. 3b), with heat map showing the bleaching information obtained from low temperature DNP spectra. The strongest reduction in signal intensity (in red) is observed for residues proximal to the TMP-radical binding pocket. Reproduced with permission from ref. 66. Copyright (2017) American Chemical Society. | ||
It is also worth pointing out that Corzilius and co-workers have also investigated similar questions in the context of paramagnetic metal centers. They notably reported signal attenuation of the full-length hammerhead ribozyme (HHRz) complexed with the paramagnetic Mn2+ divalent ion, when compared to the diamagnetic reference with Mg2+.65 Nevertheless, access to site-specific information was limited due to significant spectral overlap of RNA resonances. An approach to recover resolution in the proximity of the paramagnetic tag while indirectly detecting the PRE effect is described in the following section.
Targeted DNP with SelDNP: improving sensitivity and resolution
In 2019, our group reported a novel approach that combines the use of localized PRE and difference spectroscopy in order to recover high-resolution DNP spectra from uniformly labeled biomolecules by means of spectroscopic selection.77 This approach, called SelDNP, requires the introduction of a Paramagnetic Tag (PT) on the protein of interest and was first demonstrated to reveal the carbohydrate-binding site of the protein LecA (see Fig. 3c). It enabled recovery of site-specific information from the interaction site thanks to high-quality multidimensional data. It nicely complements other NMR methodologies that can be used to locate binding sites and solve their structures.96–98 Very importantly, SelDNP does not require the use of advanced non-uniform isotopic labeling strategies to produce resolved data from residues located within a 30 Å sphere from the PT. In addition, the method has no inherent limitations with respect to the size of the biomolecular system.High-resolution DNP-enhanced SelDNP spectra of protein binding sites
SelDNP requires the acquisition of the same NMR experiment on two samples. The first sample is used to produce the reference dataset, for which it is assumed that all resonances are uniformly polarized by DNP. The sample can be prepared using standard DNP sample preparation protocols where the PA is randomly distributed around the protein molecules. The second sample is prepared following a tDNP approach. For instance, Marin-Montesinos et al. have used a functionalized ligand tethered to a PA moiety as PT in order to selectively introduce PRE effects around the binding site and to polarize the rest of the protein. SelDNP spectra are obtained by subtracting the two datasets. Resonances from residues which are close to the PT are more bleached in the tDNP spectrum while they are of similar intensity to the rest of the residues in the reference spectrum; those resonances are therefore revealed by taking the difference of the two spectra. In contrast, resonances from residues that are far enough to be unaffected by the presence of the PT will appear similarly in both datasets and will thus be minimized or even suppressed by the difference spectroscopy. SelDNP results in high-resolution spectra with a reduced number of peaks, all originating from sites close to the PT, and thus to the binding site. Examples of SelDNP spectra can be found in Fig. 8. For each sample, a 13C–13C double-quantum (DQ) dipolar correlation spectrum was acquired with a mixing time set to produce one-bond correlations. Note that only positive intensities are retained in the 2D SelDNP spectra, and those positive peaks originate from the protein with its native ligand.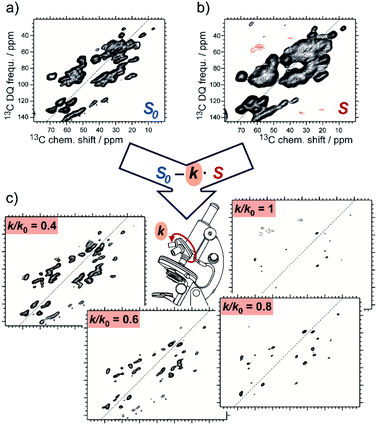 | ||
| Fig. 8 Schematic principle of the SelDNP approach. (a) Reference dataset S0 recorded on the sample prepared with the protein and its native ligand together with a non-specific PA. (b) tDNP dataset S recorded on the sample prepared with the protein and the ligand functionalized with the PA (acting as PT). The difference of the two datasets using a variable relative scaling factor k (selectivity factor) produces a set of SelDNP spectra given in (c) for k = 1, 0.8, 0.6, and 0.4, with increasing number of peaks for lower values of k. These peaks correspond to residues at increasing distances from the PT position. Spectra (a) and (b) are 2D 13C–13C double-quantum single-quantum correlation spectra. They were acquired on the protein LecA as described in ref. 77. | ||
Tuning the spectral selectivity and its relationship to the distance to the paramagnetic tag
By varying the scaling or weight factor k between the two spectra, one can tune the number of positive cross-peaks displayed in the SelDNP spectra. This corresponds to adjusting the focus around the PT by selecting resonances as a function of the PRE magnitude, which is related to the distance between the corresponding nuclei and the electron spins of the PT. As discussed in the work from Rogawski et al.,66 the direct analysis of this relationship requires the use of selectively labeled proteins. In the work from Marin-Montesinos et al.,77 a solution to this problem is proposed. Not only can one recover spectral resolution by selecting residues close to the PT, but it is also possible to relate the selectivity factor k to an approximate distance cut-off from the PT (Fig. 9d), and thus to introduce long-range constraints with respect to the PT.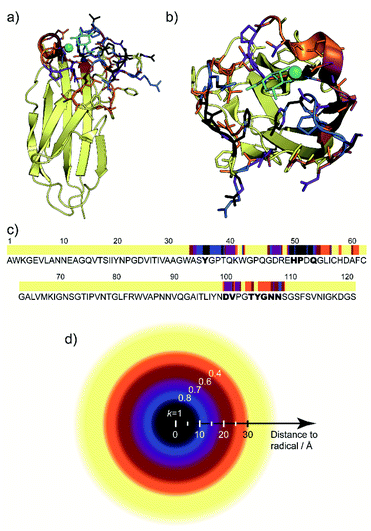 | ||
| Fig. 9 (a) and (b) Lateral and top views of the protein LecA with residues present in the SelDNP spectra of Fig. 8c) highlighted in colour, depending in which spectrum they first appear: black for k = 1, blue for k = 0.8, purple for k = 0.7, burgundy for k = 0.6, orange for k = 0.4. The galactose ligand is in cyan and the calcium ion in green. (c) LecA protein sequence with the residues assigned from SelDNP spectra highlighted using the same colour code as in (a). Residues identified in the crystal structure to be in interaction with the ligand are identified in bold. (d) Illustration of the cut off distance to PT of residues appearing in the SelDNP spectrum for a given selectivity factor k. Adapted from ref. 77 with permission from the Royal Society of Chemistry, copyright 2019. | ||
Resonance assignment of SelDNP spectra: locating the binding site
The one-bond 2D 13C–13C SelDNP correlation spectra reported in Fig. 8c are 13C NMR fingerprints of the residues around the binding site. A residue-type assignment can be obtained by comparing the resonances to tabulated values for the different amino acids in proteins. Using minimal prior information known about the protein, i.e., that the detected residues should be grouped in only a few stretches of the protein sequence and that 51Pro and 50His are involved in the galactose binding of LecA, Marin-Montesinos et al. were able to propose a sequence assignment of the residues contained in the set of SelDNP spectra from LecA, in agreement with location of the binding site as determined in the crystal structure of the protein (Fig. 9).77SelDNP can, in principle, also be applied to 15N–13C correlation experiments in order to extract sequential information. Nevertheless, the poor resolution in the 15N dimension might make it challenging. Therefore, it is a good strategy to favor multidimensional experiments that provide side-chain 13C correlations, even for sequential assignment, due to the wider inherent spectral distribution of such correlations.99 We are currently developing computational algorithms for guided resonance assignment to extend SelDNP to more complex biomolecular assemblies.Targeted DNP for in-cell NMR: improving selectivity in complex environments
NMR applications related to structural biology have mostly been performed so far under in vitro conditions. Although there are growing evidences that atomic-scale studies should also be performed under conditions closer to the cellular environment,100–102 it has proven very challenging. Examples of NMR studies on proteins embedded in their natural membrane or surrounded by their natural cellular environment have clearly highlighted many challenges ahead, from the production of isotopically labeled samples to the limited sensitivity resulting from the diluted conditions. As a consequence, background signal rapidly becomes a major obstacle to extract site-specific information from the biomolecular system of interest.71The field of in-cell NMR, pioneered in the liquid-state by Selenko and coworkers,100 is now expanding to solid-state NMR103,104 with the main argument being that large biomolecular assemblies become impractical for solution-state NMR due to resonance broadening. In addition, studying proteins at their physiological concentration strongly limits the amount of active material. For instance, in a recent work, Narasimhan and Scherpe et al. estimated that only 10–30 μg of labeled ubiquitin is contained in a 3.2 mm rotor filled with cells, when the protein is present at physiological concentrations.105 The resonances of such diluted amounts of protein, even when labeled, face significant overlapping and crowding from signals contributed by the cellular background, which constitutes a tremendous limitation. Therefore, a hyperpolarization approach such as DNP is necessary and potentially more suitable for these complex biological environments, especially if combined with innovative on-going methodological developments.101,102,105–107
Introducing selectivity using tDNP for in-cell NMR studies
The first attempt towards in-cell DNP uses methodology that was developed in standard in-cell solution and solid-state NMR. The 13C, 15N isotopically labeled protein is introduced into unlabeled or even isotopically depleted cells (i.e.2H, 12C labeled) using for example electroporation or pore-forming peptides. The DNP PA is then added to the cells in order to record DNP-enhanced NMR data. More details can be found in the recent review from the Frederick group.108 DNP experiments need to be performed relatively rapidly after addition of the PA in order to limit radical reduction.109 In this approach, the challenge is to detect the labeled protein at μM concentration, which can be overcome using multiple-quantum filtered pulse sequences that only select NMR-active nuclei that are close in space. This works well for the study of soluble proteins (e.g. ubiquitin, alpha-synuclein) inside cells, to track e.g. structural changes or complex formation.A second approach requires producing the protein of interest directly inside the cells (E. coli, insect cells, and more recently mammalian cells) and is thus more suited for the study of non-soluble proteins such as membrane proteins, or proteins that require post translational modification in order to be functional (e.g. glycosylation or phosphorylation).110 Even though the labeled cell medium is usually introduced just before triggering the protein overexpression, partial isotopic enrichment of the rest of the cell cannot be avoided. In this second approach, selecting the protein signal out of the partly isotopically labeled background can be problematic even when employing multiple-quantum filters.
For this reason, DNP methods that can selectively hyperpolarize low amounts of a target protein in a large cellular background were recently introduced. The first demonstration of such a background suppression approach was presented by Viennet et al.71 It relies on a tDNP approach combined with deuteration of the DNP matrix. Specifically, the authors demonstrated first that 50 μM of a TOTAPOL-modified binding peptide was enough to selectively hyperpolarize about 50 μM of a 20 kDa protein (Bcl-xL, see Fig. 3a) dissolved into a deuterated buffer, that was essential to prevent 1H–1H spin diffusion whereby hyperpolarization would be lost to the solvent. In a second step, they validated their approach on non-purified cell lysates, which were previously suspended in D2O buffer and then concentrated. The end pellet was estimated to contain a total protein concentration of ∼35 mg ml−1, including about 10% of Bcl-xL. tDNP experiments performed with the TOTAPOL-modified binding peptide revealed a non-uniform protein-specific DNP enhancement compared to conventional DNP experiment with a non-specific PA. Assuming that the binding peptide only interacts with Bcl-xL, the cellular background contribution could be removed by subtracting a scaled uniformly polarized spectrum (see Fig. 10a).71
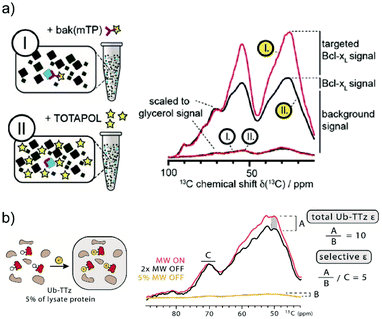 | ||
| Fig. 10 Examples of tDNP applied to selectively enhance proteins at close to endogenous concentration in isotopically labeled bacterial lysates. (a) In the work from Viennet et al., two samples are prepared (left panel) using two PAs: either Bak-mTP, a ligand functionalized with TOTAPOL (sample I), or “free” TOTAPOL (sample II). The corresponding 13C CPMAS spectra (red and black lines, respectively) are shown on the right-hand side. Spectra with white and yellow labels correspond to data obtained without and with microwave irradiation, respectively. Normalizing the background signal from the two on-spectra using the glycerol resonance reveals a selective enhancement of the targeted protein when using the functionalized PA. The different signal contributions are indicated next to the scaled spectra. Adapted with permission from ref. 71. Copyright 2016 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (b) In the work from Lim et al., the tetrazine-functionalized PA reacts directly in the isotopically labeled bacterial lysate with the overexpressed ubiquitin mutant containing the UAA norbornene-lysine (left panel). Normalizing the spectra obtained with and without microwave irradiation according to the glycerol resonance (right panel) allows the estimation of left-over enhancement denoted as A. Considering that ubiquitin accounts for 5% of the protein signal (estimated using biochemical assays), the total DNP enhancement for ubiquitin is obtained by scaling A with 5% of the microwave off signal intensity (denoted B). The resulting DNP enhancement for the targeted ubiquitin is about 10, much larger than the background signal enhancement of ∼2 (denoted C). Adapted with permission from ref. 75. Copyright 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
More recently, the Debelouchina group demonstrated the first use of bioorthogonal chemistry in the context of tDNP experiments as described above.75 Their approach was first tested on three purified mutant proteins (ubiquitin, heterochromatin protein HP1α, and the structural maintenance of chromosomes protein SMC) at concentrations of only tens of μM. As a proof of concept, tDNP using the tetrazine-based PA was then applied to isotopically labeled bacterial lysates containing overexpressed ubiquitin mutants without purification. Similar to the work from Etzkorn and co-workers, the data show a clear selectivity towards the targeted protein (see Fig. 10b).75 Both approaches show clear promises for future in-cell tDNP applications, if sufficient spectral resolution can be obtained.
Perspectives and conclusions
tDNP is a rapidly developing approach which show great promises for the study of biomolecules by DNP-enhanced NMR. As detailed in this review, reasons to further develop the tDNP methodology are multiple, both for in vitro and in-cell structural studies. The combination of hyperpolarization and PRE measurements enables residue-specific structural characterization of challenging biomolecular complexes. This is clearly an appealing approach to combat the limited sensitivity of NMR while accessing long range distance constraints (10 to 30 Å or more). Unfortunately, the low temperature required for DNP often induces significant spectral resolution degradation which is usually overcome with samples comprising of multiple isotopic labeling schemes. Nevertheless, as shown in this review, this resolution limitation can be mitigated by the SelDNP approach. This promising method can deliver resolved site-specific information for resonance assignment and also can provide distance information (including PRE-type constraints). Finally, we envisage that the tDNP approach will be useful for the emerging field of in-cell solid-state NMR by selectively improving the sensitivity of the targeted protein over the large cellular background.Of course, many new developments are still necessary to move from the proof-of-concept stage to a robust and widely applicable solution. This includes developing DNP at higher magnetic fields, combining DNP-NMR data with computational approaches,111 but also accessing sustainable helium spinning at ultra-low temperature (<100 K), and developing chemically persistent radicals for in-cell tDNP applications.
Author contributions
Diego Gauto contributed to the outline and wrote the first draft of the manuscript. Ons Dakhlaoui and Ildefonso Marin-Montesinos participated in the writing of the following versions of the document. Diego Gauto and Ons Dakhlaoui designed and produced the figures. Sabine Hediger and Gaël De Paëpe designed the outline of the review and were involved in all stages of the writing process. All authors contributed to the editing of the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Wing Ying Chow for stimulating discussions and proof reading the manuscript. This work was supported by the French National Research Agency (CBH-EUR-GS and ARCANE ANR-17-EURE-0003, Glyco@Alps ANR-15-IDEX-02, and ANR-16-CE11-0030-03) and the European Research Council Grant ERC-CoG-2015 No. 682895 (to G. D. P.).References
- I. Bertini, K. S. McGreevy and G. Parigi, NMR of Biomolecules, NMR of Biomolecules: Towards Mechanistic Systems Biology, Wiley-VCH Verlag GmbH & Co. KGaA, 2012, DOI:10.1002/9783527644506.
- R. Tycko, Solid-State NMR Studies of Amyloid Fibril Structure, Annu. Rev. Phys. Chem., 2011, 62, 279–299 CrossRef CAS PubMed.
- W. T. Franks, A. H. Linden, B. Kunert, B.-J. van Rossum and H. Oschkinat, Solid-state magic-angle spinning NMR of membrane proteins and protein–ligand interactions, Eur. J. Cell Biol., 2012, 91, 340–348 CrossRef CAS.
- L. E. Kay, New Views of Functionally Dynamic Proteins by Solution NMR Spectroscopy, J. Mol. Biol., 2016, 428, 323–331 CrossRef CAS PubMed.
- P. Schanda and M. Ernst, Studying dynamics by magic-angle spinning solid-state NMR spectroscopy: principles and applications to biomolecules, Prog. Nucl. Magn. Reson. Spectrosc., 2016, 96, 1–46 CrossRef CAS PubMed.
- V. S. Mandala, J. K. Williams and M. Hong, Structure and Dynamics of Membrane Proteins from Solid-State NMR, Annu. Rev. Biophys., 2018, 47, 201–222 CrossRef CAS PubMed.
- A. Sekhar and L. E. Kay, An NMR View of Protein Dynamics in Health and Disease, Annu. Rev. Biophys., 2019, 48, 297–319 CrossRef CAS PubMed.
- J.-H. H. Ardenkjaer-Larsen, G. S. Boebinger, A. Comment, S. Duckett, A. S. Edison, F. Engelke, C. Griesinger, R. G. Griffin, C. Hilty, H. Maeda, G. Parigi, T. Prisner, E. Ravera, J. van Bentum, S. Vega, A. Webb, C. Luchinat, H. Schwalbe and L. Frydman, Facing and Overcoming Sensitivity Challenges in Biomolecular NMR Spectroscopy, Angew. Chem., Int. Ed., 2015, 54, 9162–9185 CrossRef CAS.
- Q. Z. Ni, E. Daviso, T. V. Can, E. Markhasin, S. K. Jawla, T. M. Swager, R. J. Temkin, J. Herzfeld and R. G. Griffin, High Frequency Dynamic Nuclear Polarization, Acc. Chem. Res., 2013, 46, 1933–1941 CrossRef CAS PubMed.
- C. P. Slichter, The discovery and renaissance of dynamic nuclear polarization, Rep. Prog. Phys., 2014, 77, 072501 CrossRef PubMed.
- A. B. Barnes, G. De Paëpe, P. C. A. van der Wel, K.-N. Hu, C.-G. Joo, V. S. Bajaj, M. L. Mak-Jurkauskas, J. R. Sirigiri, J. Herzfeld, R. J. Temkin and R. G. Griffin, High-Field Dynamic Nuclear Polarization for Solid and Solution Biological NMR, Appl. Magn. Reson., 2008, 34, 237–263 CrossRef CAS PubMed.
- A. J. Rossini, A. Zagdoun, M. Lelli, A. Lesage, C. Cop, L. Emsley, C. Copéret and L. Emsley, Dynamic Nuclear Polarization Surface Enhanced NMR Spectroscopy, Acc. Chem. Res., 2013, 46, 1942–1951 CrossRef CAS PubMed.
- A. G. M. Rankin, J. Trébosc, F. Pourpoint, J.-P. Amoureux and O. Lafon, Recent developments in MAS DNP-NMR of materials, Solid State Nucl. Magn. Reson., 2019, 101, 116–143 CrossRef CAS PubMed.
- Y. Su, L. Andreas and R. G. Griffin, Magic Angle Spinning NMR of Proteins: High-Frequency Dynamic Nuclear Polarization and 1 H Detection, Annu. Rev. Biochem., 2015, 84, 465–497 CrossRef CAS PubMed.
- D. Lee, S. Hediger and G. De Paëpe, Is solid-state NMR enhanced by dynamic nuclear polarization?, Solid State Nucl. Magn. Reson., 2015, 66–67, 6–20 CrossRef CAS PubMed.
- A. N. Smith and J. R. Long, Dynamic Nuclear Polarization as an Enabling Technology for Solid State Nuclear Magnetic Resonance Spectroscopy, Anal. Chem., 2016, 88, 122–132 CrossRef CAS PubMed.
- Ü. Akbey and H. Oschkinat, Structural biology applications of solid state MAS DNP NMR, J. Magn. Reson., 2016, 269, 213–224 CrossRef PubMed.
- A. S. Lilly Thankamony, J. J. Wittmann, M. Kaushik and B. Corzilius, Dynamic nuclear polarization for sensitivity enhancement in modern solid-state NMR, Prog. Nucl. Magn. Reson. Spectrosc., 2017, 102–103, 120–195 CrossRef CAS PubMed.
- D. Lee, S. Hediger and G. De Paëpe, High-Field Solid-State NMR with Dynamic Nuclear Polarization, in Modern Magnetic Resonance, ed. G. A. Webb, Springer International Publishing, 2018, pp. 861–877, at http://link.springer.com/10.1007/978-3-319-28275-6_73-1 Search PubMed.
- L. Zhao, A. C. Pinon, L. Emsley and A. J. Rossini, DNP-enhanced solid-state NMR spectroscopy of active pharmaceutical ingredients, Magn. Reson. Chem., 2018, 56, 583–609 CrossRef CAS.
- K. Jaudzems, T. Polenova, G. Pintacuda, H. Oschkinat and A. Lesage, DNP NMR of biomolecular assemblies, J. Struct. Biol., 2019, 206, 90–98 CrossRef CAS PubMed.
- T. V. Can, M. A. Caporini, F. Mentink-Vigier, B. Corzilius, J. J. Walish, M. Rosay, W. E. Maas, M. Baldus, S. Vega, T. M. Swager and R. G. Griffin, Overhauser effects in insulating solids, J. Chem. Phys., 2014, 141, 064202 CrossRef CAS PubMed.
- C. Song, K. N. Hu, C. G. Joo, T. M. Swager and R. G. Griffin, TOTAPOL: a biradical polarizing agent for dynamic nuclear polarization experiments in aqueous media, J. Am. Chem. Soc., 2006, 128, 11385–11390 CrossRef CAS PubMed.
- F. Mentink-Vigier, G. Mathies, Y. Liu, A.-L. Barra, M. A. Caporini, D. Lee, S. Hediger, R. G. Griffin and G. De Paëpe, Efficient cross-effect dynamic nuclear polarization without depolarization in high-resolution MAS NMR, Chem. Sci., 2017, 8, 8150–8163 RSC.
- F. Mentink-Vigier, I. Marin-Montesinos, A. P. Jagtap, T. Halbritter, J. van Tol, S. Hediger, D. Lee, S. T. Sigurdsson and G. De Paëpe, Computationally Assisted Design of Polarizing Agents for Dynamic Nuclear Polarization Enhanced NMR: The AsymPol Family, J. Am. Chem. Soc., 2018, 140, 11013–11019 CrossRef CAS PubMed.
- D. Wisser, G. Karthikeyan, A. Lund, G. Casano, H. Karoui, M. Yulikov, G. Menzildjian, A. C. Pinon, A. Purea, F. Engelke, S. R. Chaudhari, D. Kubicki, A. J. Rossini, I. B. Moroz, D. Gajan, C. Copéret, G. Jeschke, M. Lelli and L. Emsley, et al., BDPA-Nitroxide Biradicals Tailored for Efficient Dynamic Nuclear Polarization Enhanced Solid-State NMR at Magnetic Fields up to 21.1 T, J. Am. Chem. Soc., 2018, 140, 13340–13349 CrossRef CAS PubMed.
- A. Lund, G. Casano, G. Menzildjian, M. Kaushik, G. Stevanato, M. Yulikov, R. Jabbour, D. Wisser, M. Renom-Carrasco, C. Thieuleux, F. Bernada, H. Karoui, D. Siri, M. Rosay, I. V. Sergeyev, D. Gajan, M. Lelli, L. Emsley and O. Ouari, et al., TinyPols: a family of water-soluble binitroxides tailored for dynamic nuclear polarization enhanced NMR spectroscopy at 18.8 and 21.1 T, Chem. Sci., 2020, 11, 2810–2818 RSC.
- B. Corzilius, A. A. Smith, A. B. Barnes, C. Luchinat, I. Bertini and R. G. Griffin, High-Field Dynamic Nuclear Polarization with High-Spin Transition Metal Ions, J. Am. Chem. Soc., 2011, 133, 5648–5651 CrossRef CAS PubMed.
- B. Corzilius, Paramagnetic metal ions for dynamic nuclear polarization, eMagRes, 2018, 7, 179–194 CAS.
- Y. Matsuki, T. Maly, O. Ouari, H. Karoui, F. Le Moigne, E. Rizzato, S. Lyubenova, J. Herzfeld, T. Prisner, P. Tordo and R. G. Griffin, Dynamic Nuclear Polarization with a Rigid Biradical, Angew. Chem., Int. Ed., 2009, 48, 4996–5000 CrossRef CAS PubMed.
- C. Sauvée, M. Rosay, G. Casano, F. Aussenac, R. T. Weber, O. Ouari and P. Tordo, Highly efficient, water-soluble polarizing agents for dynamic nuclear polarization at high frequency, Angew. Chem., Int. Ed., 2013, 52, 10858–10861 CrossRef PubMed.
- A. Zagdoun, G. Casano, O. Ouari, M. Schwarzwälder, A. J. Rossini, F. Aussenac, M. Yulikov, G. Jeschke, C. Copéret, A. Lesage, P. Tordo and L. Emsley, Large Molecular Weight Nitroxide Biradicals Providing Efficient Dynamic Nuclear Polarization at Temperatures up to 200 K, J. Am. Chem. Soc., 2013, 135, 12790–12797 CrossRef CAS PubMed.
- W.-M. Yau, K. R. Thurber and R. Tycko, Synthesis and evaluation of nitroxide-based oligoradicals for low-temperature dynamic nuclear polarization in solid state NMR, J. Magn. Reson., 2014, 244, 98–106 CrossRef CAS PubMed.
- G. Mathies, M. A. Caporini, V. K. Michaelis, Y. Liu, K.-N. Hu, D. Mance, J. L. Zweier, M. Rosay, M. Baldus and R. G. Griffin, Efficient Dynamic Nuclear Polarization at 800 MHz/527 GHz with Trityl-Nitroxide Biradicals, Angew. Chem., 2015, 127, 11936–11940 CrossRef.
- D. J. Kubicki, G. Casano, M. Schwarzwälder, S. Abel, C. Sauvée, K. Ganesan, M. Yulikov, A. J. Rossini, G. Jeschke, C. Copéret, A. Lesage, P. Tordo, O. Ouari and L. Emsley, Rational design of dinitroxide biradicals for efficient cross-effect dynamic nuclear polarization, Chem. Sci., 2016, 7, 550–558 RSC.
- C. Sauvée, G. Casano, S. Abel, A. Rockenbauer, D. Akhmetzyanov, H. Karoui, D. Siri, F. Aussenac, W. Maas, R. T. Weber, T. Prisner, M. Rosay, P. Tordo and O. Ouari, Tailoring of Polarizing Agents in the bTurea Series for Cross-Effect Dynamic Nuclear Polarization in Aqueous Media, Chem.–Eur. J., 2016, 22, 5598–5606 CrossRef PubMed.
- A. P. Jagtap, M. A. Geiger, D. Stöppler, M. Orwick-Rydmark, H. Oschkinat and S. T. Sigurdsson, BcTol: a highly water-soluble biradical for efficient dynamic nuclear polarization of biomolecules, Chem. Commun., 2016, 52, 7020–7023 RSC.
- M. Rosay, L. Tometich, S. Pawsey, R. Bader, R. Schauwecker, M. Blank, P. M. Borchard, S. R. Cauffman, K. L. Felch, R. T. Weber, R. J. Temkin, R. G. Griffin and W. E. Maas, Solid-state dynamic nuclear polarization at 263 GHz: spectrometer design and experimental results, Phys. Chem. Chem. Phys., 2010, 12, 5850–5860 RSC.
- Y. Matsuki, S. Nakamura, S. Fukui, H. Suematsu and T. Fujiwara, Closed-cycle cold helium magic-angle spinning for sensitivity-enhanced multi-dimensional solid-state NMR, J. Magn. Reson., 2015, 259, 76–81 CrossRef CAS PubMed.
- E. Bouleau, P. Saint-Bonnet, F. Mentink-Vigier, H. Takahashi, J.-F. Jacquot, M. Bardet, F. Aussenac, A. Purea, F. Engelke, S. Hediger, D. Lee and G. De Paëpe, Pushing NMR sensitivity limits using dynamic nuclear polarization with closed-loop cryogenic helium sample spinning, Chem. Sci., 2015, 6, 6806–6812 RSC.
- D. Lee, E. Bouleau, P. Saint-Bonnet, S. Hediger and G. De Paëpe, Ultra-low temperature MAS-DNP, J. Magn. Reson., 2016, 264, 116–124 CrossRef CAS.
- T. Prisner, M. Rohrer and F. MacMillan, Pulsed EPR Spectroscopy: Biological Applications, Annu. Rev. Phys. Chem., 2001, 52, 279–313 CrossRef CAS PubMed.
- M. Bennati and T. F. Prisner, New developments in high field electron paramagnetic resonance with applications in structural biology, Rep. Prog. Phys., 2005, 68, 411–448 CrossRef CAS.
- M. J. Davies and C. L. Hawkins, EPR spin trapping of protein radicals, Free Radicals Biol. Med., 2004, 36, 1072–1086 CrossRef CAS PubMed.
- F. Arnesano, L. Banci and M. Piccioli, NMR structures of paramagnetic metalloproteins, Q. Rev. Biophys., 2005, 38, 167–219 CrossRef CAS.
- G. Otting, Protein NMR Using Paramagnetic Ions, Annu. Rev. Biophys., 2010, 39, 387–405 CrossRef CAS PubMed.
- A. J. Pell, G. Pintacuda and C. P. Grey, Paramagnetic NMR in solution and the solid state, Prog. Nucl. Magn. Reson. Spectrosc., 2019, 111, 1–271 CrossRef CAS.
- X. C. Su and J. L. Chen, Site-Specific Tagging of Proteins with Paramagnetic Ions for Determination of Protein Structures in Solution and in Cells, Acc. Chem. Res., 2019, 52, 1675–1686 CrossRef CAS.
- S. A. Shelke and S. T. Sigurdsson, Site-directed spin labelling of nucleic acids, Eur. J. Org. Chem., 2012, 2291–2301, DOI:10.1002/ejoc.201101434.
- M. Brodrecht, K. Herr, S. Bothe, M. de Oliveira, T. Gutmann and G. Buntkowsky, Efficient Building Blocks for Solid-Phase Peptide Synthesis of Spin Labeled Peptides for Electron Paramagnetic Resonance and Dynamic Nuclear Polarization Applications, ChemPhysChem, 2019, 20, 1475–1487 CAS.
- G. Jeschke and Y. Polyhach, Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance, Phys. Chem. Chem. Phys., 2007, 9, 1895 RSC.
- J. P. Klare and H. J. Steinhoff, Spin labeling EPR, Photosynth. Res., 2009, 102, 377–390 CrossRef CAS PubMed.
- G. M. Clore, Practical Aspects of Paramagnetic Relaxation Enhancement in Biological Macromolecules, in Methods in Enzymology, ed. P. Z. Qin and K. Warncke, Academic Press Inc., 2015, vol. 564, pp. 485–497 Search PubMed.
- C. Nitsche and G. Otting, Pseudocontact shifts in biomolecular NMR using paramagnetic metal tags, Prog. Nucl. Magn. Reson. Spectrosc., 2017, 98–99, 20–49 CrossRef CAS PubMed.
- C. P. Jaroniec, Structural studies of proteins by paramagnetic solid-state NMR spectroscopy, J. Magn. Reson., 2015, 253, 50–59 CrossRef CAS PubMed.
- M. Tang and D. Lam, Paramagnetic solid-state NMR of proteins, Solid State Nucl. Magn. Reson., 2019, 103, 9–16 CrossRef CAS PubMed.
- R. Rogawski and A. E. McDermott, New NMR tools for protein structure and function: spin tags for dynamic nuclear polarization solid state NMR, Arch. Biochem. Biophys., 2017, 628, 102–113 CrossRef CAS PubMed.
- A. Chakraborty, F. Deligey, J. Quach, F. Mentink-Vigier, P. Wang and T. Wang, Biomolecular complex viewed by dynamic nuclear polarization solid-state NMR spectroscopy, Biochem. Soc. Trans., 2020, 48, 1089–1099 CrossRef CAS.
- V. Vitzthum, F. Borcard, S. Jannin, M. Morin, P. Miéville, M. A. Caporini, A. Sienkiewicz, S. Gerber-Lemaire and G. Bodenhausen, Fractional spin-labeling of polymers for enhancing NMR sensitivity by solvent-free dynamic nuclear polarization, ChemPhysChem, 2011, 12, 2929–2932 CrossRef CAS.
- R. Rogawski, I. V. Sergeyev, Y. Li, M. F. Ottaviani, V. Cornish and A. E. McDermott, Dynamic Nuclear Polarization Signal Enhancement with High-Affinity Biradical Tags, J. Phys. Chem. B, 2017, 121, 1169–1175 CrossRef CAS.
- T. Maly, D. Cui, R. G. Griffin and A. F. Miller, 1H dynamic nuclear polarization based on an endogenous radical, J. Phys. Chem. B, 2012, 116, 7055–7065 CrossRef CAS PubMed.
- C. Fernández-de-Alba, H. Takahashi, A. Richard, Y. Chenavier, L. Dubois, V. Maurel, D. Lee, S. Hediger, G. De Paëpe and G. De Paëpe, Matrix-Free DNP-Enhanced NMR Spectroscopy of Liposomes Using a Lipid-Anchored Biradical, Chem.–Eur. J., 2015, 21, 4512–4517 CrossRef PubMed.
- A. N. Smith, M. A. Caporini, G. E. Fanucci and J. R. Long, A Method for Dynamic Nuclear Polarization Enhancement of Membrane Proteins, Angew. Chem., Int. Ed., 2015, 54, 1542–1546 CrossRef CAS PubMed.
- M. A. Voinov, D. B. Good, M. E. Ward, S. Milikisiyants, A. Marek, M. A. Caporini, M. Rosay, R. A. Munro, M. Ljumovic, L. S. Brown, V. Ladizhansky and A. I. Smirnov, Cysteine-Specific Labeling of Proteins with a Nitroxide Biradical for Dynamic Nuclear Polarization NMR, J. Phys. Chem. B, 2015, 119, 10180–10190 CrossRef CAS PubMed.
- P. Wenk, M. Kaushik, D. Richter, M. Vogel, B. Suess and B. Corzilius, Dynamic nuclear polarization of nucleic acid with endogenously bound manganese, J. Biomol. NMR, 2015, 63, 97–109 CrossRef CAS PubMed.
- R. Rogawski, I. V. Sergeyev, Y. Zhang, T. H. Tran, Y. Li, L. Tong and A. E. McDermott, NMR Signal Quenching from Bound Biradical Affinity Reagents in DNP Samples, J. Phys. Chem. B, 2017, 121, 10770–10781 CrossRef CAS PubMed.
- E. A. W. van der Cruijsen, E. J. Koers, C. Sauvée, R. E. Hulse, M. Weingarth, O. Ouari, E. Perozo, P. Tordo and M. Baldus, Biomolecular DNP-Supported NMR Spectroscopy using Site-Directed Spin Labeling, Chem.–Eur. J., 2015, 21, 12971–12977 CrossRef CAS PubMed.
- M. Kaushik, T. Bahrenberg, T. V. Can, M. A. Caporini, R. Silvers, J. Heiliger, A. A. Smith, H. Schwalbe, R. G. Griffin and B. Corzilius, Gd(III) and Mn(II) complexes for dynamic nuclear polarization: small molecular chelate polarizing agents and applications with site-directed spin labeling of proteins, Phys. Chem. Chem. Phys., 2016, 18, 27205–27218 RSC.
- D. Daube, M. Vogel, B. Suess and B. Corzilius, Dynamic nuclear polarization on a hybridized hammerhead ribozyme: an explorative study of RNA folding and direct DNP with a paramagnetic metal ion cofactor, Solid State Nucl. Magn. Reson., 2019, 101, 21–30 CrossRef CAS.
- A. N. Smith, U. T. Twahir, T. Dubroca, G. E. Fanucci and J. R. Long, Molecular Rationale for Improved Dynamic Nuclear Polarization of Biomembranes, J. Phys. Chem. B, 2016, 120, 7880–7888 CrossRef CAS PubMed.
- T. Viennet, A. Viegas, A. Kuepper, S. Arens, V. Gelev, O. Petrov, T. N. Grossmann, H. Heise and M. Etzkorn, Selective Protein Hyperpolarization in Cell Lysates Using Targeted Dynamic Nuclear Polarization, Angew. Chem., Int. Ed., 2016, 55, 10746–10750 CrossRef CAS.
- E. S. Salnikov, S. Abel, G. Karthikeyan, H. Karoui, F. Aussenac, P. Tordo, B. Bechinger and O. Ouari, Dynamic Nuclear Polarization/Solid-State NMR Spectroscopy of Membrane Polypeptides: Free-Radical Optimization for Matrix-Free Lipid Bilayer Samples, ChemPhysChem, 2017, 18, 2103–2113 CrossRef CAS.
- D. B. Good, M. A. Voinov, D. Bolton, M. E. Ward, I. V. Sergeyev, M. Caporini, P. Scheffer, A. Lo, M. Rosay, A. Marek, L. S. Brown, A. I. Smirnov and V. Ladizhansky, A biradical-tagged phospholipid as a polarizing agent for solid-state MAS dynamic nuclear polarization NMR of membrane proteins, Solid State Nucl. Magn. Reson., 2019, 100, 92–101 CrossRef CAS PubMed.
- M.-A. Sani, S. Zhu, V. Hofferek and F. Separovic, Nitroxide spin-labeled peptides for DNP-NMR in-cell studies, FASEB J., 2019, 33, 11021–11027 CrossRef CAS PubMed.
- B. J. Lim, B. E. Ackermann and G. T. Debelouchina, Targetable Tetrazine-Based Dynamic Nuclear Polarization Agents for Biological Systems, ChemBioChem, 2020, 21, 1315–1319 CrossRef CAS PubMed.
- B. J. Wylie, B. G. Dzikovski, S. Pawsey, M. Caporini, M. Rosay, J. H. Freed and A. E. McDermott, Dynamic nuclear polarization of membrane proteins: covalently bound spin-labels at protein-protein interfaces, J. Biomol. NMR, 2015, 61, 361–367 CrossRef CAS PubMed.
- I. Marin-Montesinos, D. Goyard, E. Gillon, O. Renaudet, A. Imberty, S. Hediger and G. De Paëpe, Selective high-resolution DNP-enhanced NMR of biomolecular binding sites, Chem. Sci., 2019, 10, 3366–3374 RSC.
- H. Takahashi, D. Lee, L. Dubois, M. Bardet, S. Hediger and G. De Paëpe, Rapid Natural-Abundance 2D 13C-13C Correlation Spectroscopy Using Dynamic Nuclear Polarization Enhanced Solid-State NMR and Matrix-Free Sample Preparation, Angew. Chem., Int. Ed., 2012, 51, 11766–11769 CrossRef CAS PubMed.
- E. Ravera, B. Corzilius, V. K. Michaelis, C. Rosa, R. G. Griffin, C. Luchinat and I. Bertini, Dynamic nuclear polarization of sedimented solutes, J. Am. Chem. Soc., 2013, 135, 1641–1644 CrossRef CAS PubMed.
- E. Ravera, B. Corzilius, V. K. Michaelis, C. Luchinat, R. G. Griffin and I. Bertini, DNP-Enhanced MAS NMR of Bovine Serum Albumin Sediments and Solutions, J. Phys. Chem. B, 2014, 118, 2957–2965 CrossRef CAS PubMed.
- M. Nagaraj, T. W. Franks, S. Saeidpour, T. Schubeis, H. Oschkinat, C. Ritter and B. J. van Rossum, Surface Binding of TOTAPOL Assists Structural Investigations of Amyloid Fibrils by Dynamic Nuclear Polarization NMR Spectroscopy, ChemBioChem, 2016, 1308–1311, DOI:10.1002/cbic.201600185.
- H. Takahashi, I. Ayala, M. Bardet, G. De Paëpe, J.-P. Simorre and S. Hediger, Solid-state NMR on bacterial cells: selective cell wall signal enhancement and resolution improvement using dynamic nuclear polarization, J. Am. Chem. Soc., 2013, 135, 5105–5110 CrossRef CAS.
- H. Takahashi, S. Hediger and G. De Paëpe, Matrix-free dynamic nuclear polarization enables solid-state NMR 13C–13C correlation spectroscopy of proteins at natural isotopic abundance, Chem. Commun., 2013, 49, 9479–9481 RSC.
- S. Y. Liao, M. Lee, T. Wang, I. V. Sergeyev and M. Hong, Efficient DNP NMR of membrane proteins: sample preparation protocols, sensitivity, and radical location, J. Biomol. NMR, 2016, 64, 223–237 CrossRef CAS PubMed.
- L. Wang, A. Brock, B. Herberich and P. G. Schultz, Expanding the Genetic Code of Escherichia coli, Science, 2001, 292, 498–500 CrossRef CAS PubMed.
- J. A. Prescher and C. R. Bertozzi, Chemistry in Living Systems, Nature Chemical Biology, Nature Publishing Group, 2005, vol. 1, pp. 13–21 Search PubMed.
- K. Lang, L. Davis, J. Torres-Kolbus, C. Chou, A. Deiters and J. W. Chin, Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction, Nat. Chem., 2012, 4, 298–304 CrossRef CAS PubMed.
- T. Plass, S. Milles, C. Koehler, J. Szymański, R. Mueller, M. Wießler, C. Schultz and E. A. Lemke, Amino acids for diels-alder reactions in living cells, Angew. Chem., Int. Ed., 2012, 51, 4166–4170 CrossRef CAS PubMed.
- D. A. Hall, D. C. Maus, G. J. Gerfen, S. J. Inati, L. R. Becerra, F. W. Dahlquist and R. G. Griffin, Polarization-Enhanced NMR Spectroscopy of Biomolecules in Frozen Solution, Science, 1997, 276, 930–932 CrossRef CAS PubMed.
- I. Bertini, C. Luchinat and G. Parigi, Solution NMR of Paramagnetic Molecules: Applications to Metallobiomolecules and Models, 2001, at https://www.elsevier.com/books/solution-nmr-of-paramagnetic-molecules/bertini/978-0-444-20529-2 Search PubMed.
- L. Banci, I. Bertini, C. Luchinat and M. Mori, NMR in structural proteomics and beyond, Prog. Nucl. Magn. Reson. Spectrosc., 2010, 56, 247–266 CrossRef CAS.
- I. Sengupta, P. S. Nadaud and C. P. Jaroniec, Protein Structure Determination with Paramagnetic Solid-State NMR Spectroscopy, Acc. Chem. Res., 2013, 46, 2117–2126 CrossRef CAS.
- M. J. Knight, I. C. Felli, R. Pierattelli, L. Emsley and G. Pintacuda, Magic Angle Spinning NMR of Paramagnetic Proteins, Acc. Chem. Res., 2013, 46, 2108–2116 CrossRef CAS PubMed.
- A. J. Rossini, A. Zagdoun, M. Lelli, D. Gajan, F. Rascón, M. Rosay, W. E. Maas, C. Copéret, A. Lesage and L. Emsley, One hundred fold overall sensitivity enhancements for silicon-29 NMR spectroscopy of surfaces by dynamic nuclear polarization with CPMG acquisition, Chem. Sci., 2012, 3, 108–115 RSC.
- B. Corzilius, L. B. Andreas, A. A. Smith, Q. Z. Ni and R. G. Griffin, Paramagnet induced signal quenching in MAS–DNP experiments in frozen homogeneous solutions, J. Magn. Reson., 2014, 240, 113–123 CrossRef CAS PubMed.
- J. Orts, M. A. Wälti, M. Marsh, L. Vera, A. D. Gossert, P. Güntert and R. Riek, NMR-Based Determination of the 3D Structure of the Ligand–Protein Interaction Site without Protein Resonance Assignment, J. Am. Chem. Soc., 2016, 138, 4393–4400 CrossRef CAS.
- F. Torres, D. Ghosh, D. Strotz, C. N. Chi, B. Davis and J. Orts, Protein–fragment complex structures derived by NMR molecular replacement, RSC Med. Chem., 2020, 11, 591–596 RSC.
- A.-H. Emwas, K. Szczepski, B. G. Poulson, K. Chandra, R. T. McKay, M. Dhahri, F. Alahmari, L. Jaremko, J. I. Lachowicz and M. Jaremko, NMR as a “Gold Standard” Method in Drug Design and Discovery, Molecules, 2020, 25, 4597 CrossRef CAS PubMed.
- I. V. Sergeyev, B. Itin, R. Rogawski, L. A. Day and A. E. McDermott, Efficient assignment and NMR analysis of an intact virus using sequential side-chain correlations and DNP sensitization, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5171–5176 CrossRef CAS PubMed.
- F. X. Theillet, A. Binolfi, B. Bekei, A. Martorana, H. M. Rose, M. Stuiver, S. Verzini, D. Lorenz, M. Van Rossum, D. Goldfarb and P. Selenko, Structural disorder of monomeric α-synuclein persists in mammalian cells, Nature, 2016, 530, 45–50 CrossRef CAS PubMed.
- M. Kaplan, A. Cukkemane, G. C. P. Van Zundert, S. Narasimhan, M. Daniëls, D. Mance, G. Waksman, A. M. J. J. Bonvin, R. Fronzes, G. E. Folkers and M. Baldus, Probing a cell-embedded megadalton protein complex by DNP-supported solid-state NMR, Nat. Methods, 2015, 12, 649–652 CrossRef CAS PubMed.
- K. K. Frederick, V. K. Michaelis, B. Corzilius, T. C. Ong, A. C. Jacavone, R. G. Griffin and S. Lindquist, Sensitivity-Enhanced NMR Reveals Alterations in Protein Structure by Cellular Milieus, Cell, 2015, 163, 620–628 CrossRef CAS PubMed.
- M. Renault, R. Tommassen-van Boxtel, M. P. Bos, J. A. Post, J. Tommassen and M. Baldus, Cellular solid-state nuclear magnetic resonance spectroscopy, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 4863–4868 CrossRef CAS PubMed.
- S. Reckel, J. J. Lopez, F. Löhr, C. Glaubitz and V. Dötsch, In-Cell Solid-State NMR as a Tool to Study Proteins in Large Complexes, ChemBioChem, 2012, 13, 534–537 CrossRef CAS PubMed.
- S. Narasimhan, S. Scherpe, A. Lucini Paioni, J. van der Zwan, G. E. Folkers, H. Ovaa and M. Baldus, DNP-Supported Solid-State NMR Spectroscopy of Proteins Inside Mammalian Cells, Angew. Chem., Int. Ed., 2019, 58, 12969–12973 CrossRef CAS.
- K. Yamamoto, M. A. Caporini, S.-C. Im, L. Waskell and A. Ramamoorthy, Cellular solid-state NMR investigation of a membrane protein using dynamic nuclear polarization, Biochim. Biophys. Acta, Biomembr., 2015, 1848, 342–349 CrossRef CAS PubMed.
- B. J. Albert, C. Gao, E. L. Sesti, E. P. Saliba, N. Alaniva, F. J. Scott, S. T. Sigurdsson and A. B. Barnes, Dynamic Nuclear Polarization Nuclear Magnetic Resonance in Human Cells Using Fluorescent Polarizing Agents, Biochemistry, 2018, 57, 4741–4746 CrossRef CAS PubMed.
- W. N. Costello, Y. Xiao and K. K. Frederick, DNP-Assisted NMR Investigation of Proteins at Endogenous Levels in Cellular Milieu, in Methods in Enzymology, Academic Press Inc., 2019, vol. 615, pp. 373–406 Search PubMed.
- K. M. McCoy, R. Rogawski, O. Stovicek and A. E. McDermott, Stability of nitroxide biradical TOTAPOL in biological samples, J. Magn. Reson., 2019, 303, 115–120 CrossRef CAS PubMed.
- E. Luchinat and L. Banci, In-Cell NMR in Human Cells: Direct Protein Expression Allows Structural Studies of Protein Folding and Maturation, Acc. Chem. Res., 2018, 51, 1550–1557 CrossRef CAS.
- J. Heiliger, T. Matzel, E. C. Çetiner, H. Schwalbe, G. Kuenze and B. Corzilius, Site-specific dynamic nuclear polarization in a Gd(III)-labeled protein, Phys. Chem. Chem. Phys., 2020, 22, 25455–25466 RSC.
Footnote |
| † Present address: ICSN, CNRS UPR 2301, Univ. Paris-Saclay, Gif-sur-Yvette, France. |
| This journal is © The Royal Society of Chemistry 2021 |





