Open Access Article
Open Access ArticlePyridine appended 2-hydrazinylthiazole derivatives: design, synthesis, in vitro and in silico antimycobacterial studies†
Ramkishore Matsaa,
Parameshwar Makambe,
Guneswar Sethic,
Ahammed Ameen Thottasseri a,
Aswani Raj Kizhakkandiyila,
Krishna Ramadasc,
Vignesh Mariappand,
Agieshkumar Balakrishna Pillaid and
Tharanikkarasu Kannan
a,
Aswani Raj Kizhakkandiyila,
Krishna Ramadasc,
Vignesh Mariappand,
Agieshkumar Balakrishna Pillaid and
Tharanikkarasu Kannan *a
*a
aDepartment of Chemistry, Pondicherry University, Kalapet, Puducherry 605 014, India. E-mail: tharani.che@pondiuni.edu.in
bDr Param Laboratories, Plot No. 478, BN. Reddy Nagar, Cherlapally, Hyderabad, Telangana 500 051, India
cCentre for Bioinformatics, Pondicherry University, Puducherry 605 014, India
dCentral Inter-Disciplinary Research Facility (CIDRF), Sri Balaji Vidyapeeth (Deemed to be University), Puducherry 607 402, India
eDivision of Research and Innovation, Department of Chemistry, Uttaranchal University, Arcadia Grant, P.O. Chandanwari, Premnagar, Dehradun, Uttarakhand 248007, India
First published on 22nd June 2022
Abstract
An array of pyridine appended 2-hydrazinylthiazole derivatives has been synthesized to discover novel chemotherapeutic agents for Mycobacterium tuberculosis (Mtb). The drug-likeness of pyridine appended 2-hydrazinylthiazole derivatives was validated using the Lipinski and Veber rules. The designed thiazole molecules have been synthesized through Hantzsch thiazole methodologies. The in vitro antimycobacterial studies have been conducted using Luciferase reporter phage (LRP) assay. Out of thirty pyridine appended 2-hydrazinylthiazole derivatives, the compounds 2b, 3b, 5b, and 8b have exhibited good antimycobacterial activity against Mtb, an H37Rv strain with the minimum inhibitory concentration in the range of 6.40–7.14 μM. In addition, in vitro cytotoxicity of active molecules has been observed against Human Embryonic Kidney Cell lines (HEK293t) using MTT assay. The compounds 3b and 8b are nontoxic and their cell viability is 87% and 96.71% respectively. The in silico analyses of the pyridine appended 2-hydrazinylthiazole derivatives have been studied to find the mode of binding of the active compounds with KasA protein of Mtb. The active compounds showed a strong binding score (−5.27 to −6.23 kcal mol−1).
1. Introduction
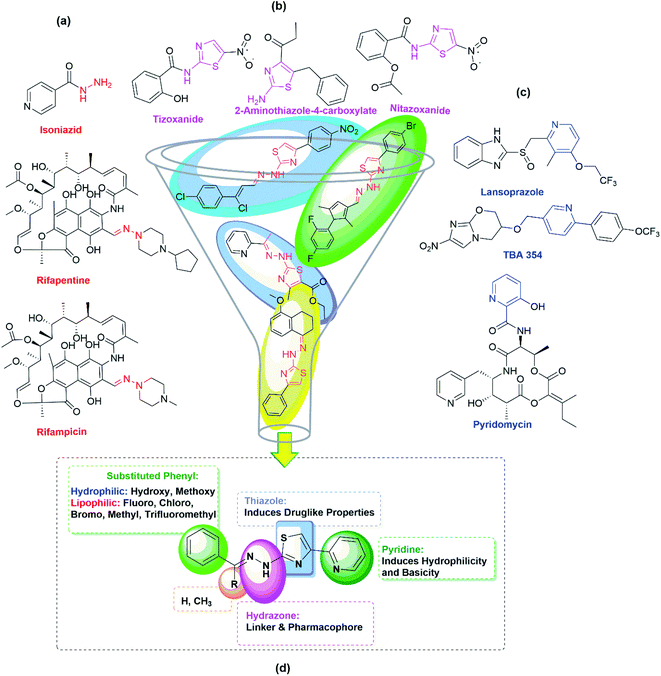
Tuberculosis (TB) is an infectious airborne disease caused by Mycobacterium tuberculosis (Mtb). TB remained the topmost infectious disease before the coronavirus (COVID-19) pandemic. According to the Global tuberculosis report 2021, there were 9.9 million infections and 1.3 million deaths. The COVID-19 pandemic has halted years of global progress in reducing TB deaths.1 The majority of existing TB drugs were discovered in the late 1950s, and due to the extensive usage of these drugs for the past seventy years, these drugs are less effective in controlling the disease as Mycobacterium has developed resistance to them. Bedaquiline and delamanid are used in conjunction with other TB medicines for curing MDR-TB.2–4 Pretomanid has most recently entered into the market for treating XDR-TB, and it is used in combination with bedaquiline, and linezolid.5 Developing new and effective TB drugs for the treatment of XDR-TB and MDR-TB with shorter and simpler regimens with negligible side effects is the need of the hour.A broad range of established and new scaffolds were tested for their antitubercular activity over the last decade to identify novel anti-TB drugs. Thiazole derivatives are promising compounds to act as antitubercular agents as they are target-specific. Besides, thiazole compounds are known to act as the anticancer,6 antitumor,7,8 antimalarial,9 antimicrobial,10 anti-inflammatory,11 and anti-hypolipidemic12 agents. Moreover, the thiazole moiety is structurally analogous to thiolactomycin, an antibiotic that exists naturally but with synthetic challenges.13 During the biosynthesis of the Mtb cell wall, thiolactomycin inhibits β-ketoacyl-ACP synthase (KasA), leading to the death of Mtb.14 Nitazoxanide (NTZ), a thiazole ring containing oral FDA-approved drug to treat protozoal infections, significantly inhibits intracellular Mtb development.15,16 Similarly, tizoxanide, a metabolite of NTZ has also been reported to inhibit non-replicative and replicative Mtb strains.17 Interestingly, derivatives of 2-aminothiazole-4-carboxylate are known to be potent inhibitors of Mtb's KasA protein.18 Due to the wide range of biological activity of 2-amino thiazole derivatives, in our research lab, we synthesized novel and effective 2-amino thiazole derivatives with antitubercular activity.19,20 We found 2-(2-hydrazinyl)thiazole derivatives showed good antitubercular activity when the 2-pyridyl group was introduced at imine carbon of the thiazole ring.21 To fine-tune and check the effect of substituents on the antitubercular activity of the derivatives of the pyridine group introduced at the 4th place of the thiazole ring, different functional groups on imine carbon have been incorporated and evaluated in the present investigation for their anti-TB activities. Interestingly, all the pyridine appended 2-hydrazinylthiazole derivatives discussed in the present report show better antitubercular activity than the previously reported derivatives of 2-hydrazinylthiazoles. The detailed results are presented in this paper. The thiazole derivatives discussed in the present investigation have been synthesized easily through classical Hantzsch thiazole methodologies. The advantage of this method is that there is much scope for generating a class of novel thiazole derivatives.22
2. Results and discussion
2.1. Designing of pyridine appended 2-hydrazinylthiazole through molecular hybridization approach
In the first step of the present investigation, pyridine appended 2-hydrazinylthiazole derivatives have been designed, and molecular hybridization23,24 approach was used for this purpose. As given in Fig. 1, the thiazole scaffold is linked to pyridine on one side and the hydrazine group on the other side for better pharmacological properties during the inhibition of TB.Pyridine is one of the essential heterocyclic scaffolds found in natural substances, such as alkaloids (trigonelline), vitamins (vitamin B3 and B6), coenzymes (nicotinamide adenine dinucleotide), etc. Because of its versatile properties, such as water solubility, good chemical stability, and hydrogen bond-forming capability, pyridine moiety plays a vital role in medicinal chemistry. Pyridine derivatives show excellent antitubercular activity. For example, lansoprazole, a pyridine moiety-containing drug that inhibits gastric acid secretion by binding to the proton-pump receptor, has an intracellular effect against Mtb. Lansoprazole kills Mtb by attacking the cytochrome bc1-complex, according to ex vivo pharmacokinetic studies.25 Pyridomycin is another pyridine-containing and natural antibiotic drug that demonstrates intense activity against various Mycobacteria, including Mtb and M. smegmatis.26 TBA-354 is another pyridine-containing drug that shows promising antitubercular activity with replicative and static action against Mtb.27 The antitubercular activity of pyridine-containing drugs and pyridine-containing derivatives synthesized in our lab previously21 prompted us to include a 2-pyridyl scaffold at the 4th place of the thiazole ring in the present investigation. Fig. 1(c) gives the structure of some of the pyridine-containing antitubercular drugs.
The hydrazone (R1R2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NNH2) is another essential and promising scaffold in medicinal chemistry. The lone pair electrons of amine nitrogen are conjugated with the imine group present in hydrazone compounds. The nitrogen atom of the hydrazone is nucleophilic, while the carbon is both electrophilic and nucleophilic.28 These functional features make hydrazone a versatile chemical entity with antimalarial, antiviral, anti-HIV, anti-schistosomiasis, antimicrobial, anthelmintic, anticancer, antiplatelet, antidiabetic, antidepressant, anticonvulsant, anti-inflammatory, analgesic, and antioxidant properties.29,30 Moreover, the most promising antitubercular drugs such as rifampicin, isoniazid, and rifapentine contain hydrazone scaffold (see Fig. 1(a)) in their structures. With unique hydrogen bonding donor and receiving regions, hydrazones have gained much attention as potent anti-TB molecules.31 The combination of 2-aminothiazole and hydrazone scaffolds in the derivatives of 2-(2-hydrazinyl)thiazole exhibited good inhibitory potentials against the strain of Mtb, H37Rv.19,32–34 The encouraging results of the in vitro studies and the higher possibility of synthesizing different types of substitutions motivated us to synthesize derivatives of pyridine appended 2-hydrazinylthiazole in finding out anti-TB agents with improved inhibitory potentials.
NNH2) is another essential and promising scaffold in medicinal chemistry. The lone pair electrons of amine nitrogen are conjugated with the imine group present in hydrazone compounds. The nitrogen atom of the hydrazone is nucleophilic, while the carbon is both electrophilic and nucleophilic.28 These functional features make hydrazone a versatile chemical entity with antimalarial, antiviral, anti-HIV, anti-schistosomiasis, antimicrobial, anthelmintic, anticancer, antiplatelet, antidiabetic, antidepressant, anticonvulsant, anti-inflammatory, analgesic, and antioxidant properties.29,30 Moreover, the most promising antitubercular drugs such as rifampicin, isoniazid, and rifapentine contain hydrazone scaffold (see Fig. 1(a)) in their structures. With unique hydrogen bonding donor and receiving regions, hydrazones have gained much attention as potent anti-TB molecules.31 The combination of 2-aminothiazole and hydrazone scaffolds in the derivatives of 2-(2-hydrazinyl)thiazole exhibited good inhibitory potentials against the strain of Mtb, H37Rv.19,32–34 The encouraging results of the in vitro studies and the higher possibility of synthesizing different types of substitutions motivated us to synthesize derivatives of pyridine appended 2-hydrazinylthiazole in finding out anti-TB agents with improved inhibitory potentials.
In the second step of this research, Lipinski and Veber rules were used to evaluate the drug-like molecule (DLM) nature of pyridine appended 2-hydrazinylthiazoles.35,36 The DLM nature can be found out using molecular weight, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value, number of hydrogen bond acceptors, and donors mentioned in the Lipinski rule. Besides, total polar surface area (TPSA) and the number of rotatable bonds (RBs) mentioned in the Veber rule can also be used to understand the DLM nature of the pyridine appended 2-hydrazinylthiazoles. Molinspiration server has been used to deduce the physicochemical properties of designed pyridine appended 2-hydrazinylthiazoles, and the deduced data are tabulated in Table 1. The results indicate that the molecular weights are in the range of 232.31–370.43, which is less than Lipinski's recommended value of 500. The log
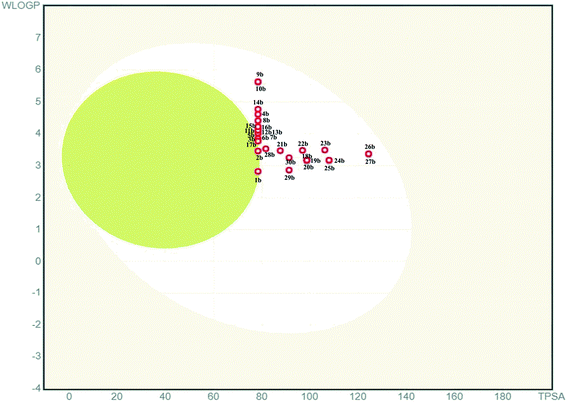
P value, number of hydrogen bond acceptors, and donors mentioned in the Lipinski rule. Besides, total polar surface area (TPSA) and the number of rotatable bonds (RBs) mentioned in the Veber rule can also be used to understand the DLM nature of the pyridine appended 2-hydrazinylthiazoles. Molinspiration server has been used to deduce the physicochemical properties of designed pyridine appended 2-hydrazinylthiazoles, and the deduced data are tabulated in Table 1. The results indicate that the molecular weights are in the range of 232.31–370.43, which is less than Lipinski's recommended value of 500. The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values are between 1.64 and 4.10, and these values are less than 5 which is suggested by Lipinski rule. Likewise, the hydrogen bond acceptors of all compounds are between 4 to 7, and hydrogen bond donors are less than two that are far below the Lipinski rule's recommended values. The total number of RBs of all the compounds is in the range of 4–7, and the TPSA of the compounds is in the range of 50.17–96.00 Å2, which is less than the recommended values. Therefore, all these compounds have not violated the Lipinski and Veber rules and have DLM characteristics. The boiled egg diagram of the pyridine appended 2-hydrazinylthiazoles has been predicted through the Swiss ADME web-based tool,37 and the graph is shown in Fig. 2. A compound should have high gastrointestinal absorption to act as an orally active drug (under white region). Most of the pyridine appended 2-hydrazinylthiazoles exhibited high gastrointestinal absorption except compounds 1b and 2b, and these compounds have exhibited blood–brain barrier permeation (under yellow region).
P values are between 1.64 and 4.10, and these values are less than 5 which is suggested by Lipinski rule. Likewise, the hydrogen bond acceptors of all compounds are between 4 to 7, and hydrogen bond donors are less than two that are far below the Lipinski rule's recommended values. The total number of RBs of all the compounds is in the range of 4–7, and the TPSA of the compounds is in the range of 50.17–96.00 Å2, which is less than the recommended values. Therefore, all these compounds have not violated the Lipinski and Veber rules and have DLM characteristics. The boiled egg diagram of the pyridine appended 2-hydrazinylthiazoles has been predicted through the Swiss ADME web-based tool,37 and the graph is shown in Fig. 2. A compound should have high gastrointestinal absorption to act as an orally active drug (under white region). Most of the pyridine appended 2-hydrazinylthiazoles exhibited high gastrointestinal absorption except compounds 1b and 2b, and these compounds have exhibited blood–brain barrier permeation (under yellow region).
| Structure and code | Lipinski rule | Veber rule | No. of violations | In vitro | In silico | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MW | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
HAs | HDs | RBs | TPSA | MIC against Mtb, H37Rv (μM) | Cytotoxicity% against HEK 293t at 6.5 μM | Glide score (kcal mol−1) | No. of H-bond (interacting residue with distance) | ||
| a Calculated from molinspiration online server (https://www.molinspiration.com/cgi-bin/properties).b NA = not analysed. | |||||||||||
 |
232.31 | 2.04 | 4 | 1 | 3 | 50.17 | 0 | 430.46 | NA | −5.804 | 1 (Thr 315, 2.80 Å) |
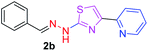 |
280.36 | 2.81 | 4 | 1 | 4 | 50.17 | 0 | 7.13 | 38.23 | −6.233 | 1 (Thr 315, 2.94 Å) |
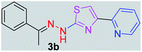 |
294.38 | 3.26 | 4 | 1 | 4 | 50.17 | 0 | 6.79 | 87.47 | −5.277 | 1 (Val 278, 3.19 Å) |
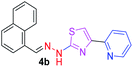 |
330.42 | 3.97 | 4 | 1 | 4 | 50.17 | 0 | 75.66 | NA | −5.015 | 1 (Thr 315, 2.81 Å) |
 |
298.35 | 2.93 | 4 | 1 | 4 | 50.17 | 0 | 6.70 | 41.58 | −6.095 | 2 (Thr 315, 3.29 Å and 2.93 Å) |
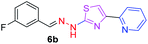 |
298.35 | 2.95 | 4 | 1 | 4 | 50.17 | 0 | 83.79 | NA | −6.165 | 1 (Thr 315, 2.93 Å) |
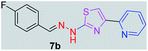 |
298.35 | 2.98 | 4 | 1 | 4 | 50.17 | 0 | 83.79 | NA | −6.186 | 1 (Thr 315, 2.93 Å) |
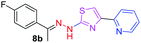 |
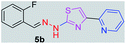
312.37 | 3.42 | 4 | 1 | 4 | 50.17 | 0 | 6.40 | 92.17 | −5.482 | 1 (Thr 315, 2.90 Å) |
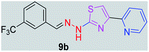 |
348.35 | 3.68 | 4 | 1 | 5 | 50.17 | 0 | 71.77 | NA | −5.016 | 2 (Thr 315, 3.25 Å, 2.86 Å) |
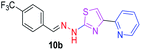 |
348.35 | 3.71 | 4 | 1 | 5 | 50.17 | 0 | 71.77 | NA | −5.039 | 2 (Thr 315, 3.26 Å, 2.86 Å) |
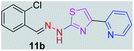 |
314.80 | 3.44 | 4 | 1 | 4 | 50.17 | 0 | 79.42 | NA | −5.932 | 2 (Thr 315, 3.30 Å, 2.81 Å) |
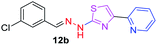 |
314.80 | 3.47 | 4 | 1 | 4 | 50.17 | 0 | 317.66 | NA | −6.111 | 2 (Thr 315, 3.34 Å, 2.95 Å) |
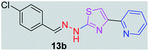 |
314.80 | 3.49 | 4 | 1 | 4 | 50.17 | 0 | 79.42 | NA | −5.708 | 2 (Thr 315, 3.28 Å, 2.86 Å) |
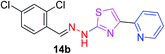 |
349.25 | 4.10 | 4 | 1 | 4 | 50.17 | 0 | 286.33 | NA | −5.932 | 4 (Thr 315, 3.22 Å, 2.87 Å) (Arg 214, 3.15 Å, 3.25 Å) |
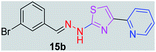 |
359.25 | 3.60 | 4 | 1 | 4 | 50.17 | 0 | 278.36 | NA | −5.621 | 1 (Thr 315, 2.80 Å) |
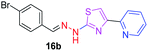 |
359.25 | 3.62 | 4 | 1 | 4 | 50.17 | 0 | 278.36 | NA | −6.008 | 2 (Thr 315, 3.22 Å, 2.92 Å) |
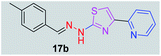 |
294.38 | 3.26 | 4 | 1 | 4 | 50.17 | 0 | 339.70 | NA | −5.035 | 1 (Thr 315, 2.88 Å) |
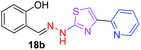 |
296.36 | 2.75 | 5 | 2 | 4 | 70.40 | 0 | 337.43 | NA | −5.707 | 4 (Thr 315, 3.23 Å, 2.86 Å) (Arg 214, 3.25 Å, 3.19 Å) |
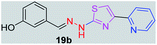 |
296.36 | 2.31 | 5 | 2 | 4 | 70.40 | 0 | 337.43 | NA | −5.056 | 2 (Thr 315, 3.21 Å, 2.93 Å) |
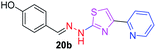 |
296.36 | 2.33 | 5 | 2 | 4 | 70.40 | 0 | 84.36 | NA | −4.685 | 1 (Thr 315, 3.00 Å) |
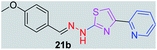 |
310.38 | 2.87 | 5 | 1 | 5 | 59.41 | 0 | 80.55 | NA | −4.949 | 1 (Thr 315, 2.78 Å) |
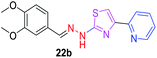 |
340.41 | 2.46 | 6 | 1 | 6 | 68.64 | 0 | 293.76 | NA | −5.019 | 2 (Thr 315, 3.30 Å, 2.84 Å) |
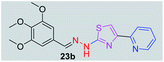 |
370.43 | 2.44 | 7 | 1 | 7 | 77.88 | 0 | 269.96 | NA | −4.937 | 2 (Thr 315, 3.22 Å, 2.86 Å) |
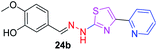 |
326.38 | 2.15 | 6 | 2 | 5 | 79.64 | 0 | 306.39 | NA | −5.001 | 2 (Thr 315, 3.21 Å, 2.92 Å) |
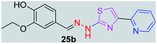 |
340.41 | 2.53 | 6 | 2 | 6 | 79.64 | 0 | 73.44 | NA | −4.985 | 1 (Thr 315, 2.95 Å) |
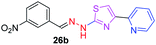 |
325.35 | 2.75 | 7 | 1 | 5 | 96.00 | 0 | 307.36 | NA | −4.517 | 1 (Met 213, 3.07 Å) |
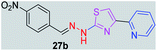 |
325.35 | 2.77 | 7 | 1 | 5 | 96.00 | 0 | >307.36 | NA | −4.87 | — |
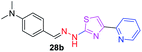 |
323.43 | 2.91 | 5 | 1 | 5 | 53.41 | 0 | 309.19 | NA | −4.609 | 2 (Thr 315, 3.22 Å, 2.91 Å) |
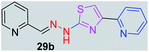 |
281.34 | 1.64 | 5 | 1 | 4 | 63.07 | 0 | 355.44 | NA | −6.013 | 2 (Thr 315, 3.29 Å, 2.89 Å) |
 |
295.37 | 2.02 | 5 | 1 | 4 | 63.07 | 0 | 338.56 | NA | −5.640 | 1 (Thr 315, 2.80 Å) |
| Rifampicin | — | — | — | — | — | — | — | 2.4 | NA | NA | NA |
| Isoniazid | — | — | — | — | — | — | — | NA | NA | −5.830 | 1 (Thr 315, 2.89 Å) |
2.2. Synthesis of pyridine appended 2-hydrazinylthiazoles
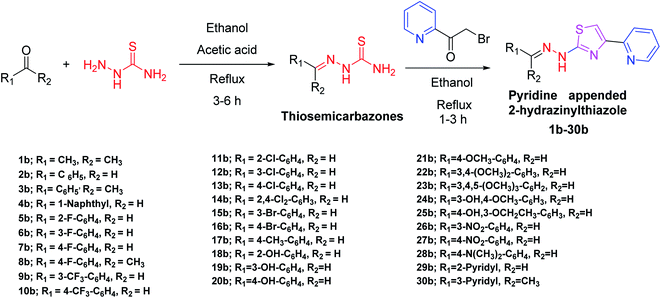
In the third step of this research, as given in Scheme 1, all the designed pyridine appended 2-hydrazinylthiazoles with DLM nature have been synthesized. To synthesize pyridine appended 2-hydrazinylthiazole derivatives, first, thiosemicarbazones were synthesized from thiosemicarbazide and the respective carbonyl compounds using the literature method.19,38The structure of thiosemicarbazones has been confirmed using different spectroscopic techniques. The characteristic imine proton present in thiosemicarbazones appears as a single peak between 8.5 ppm and 9.0 ppm in the 1H nuclear magnetic resonance (NMR) spectra, and methyl protons at the imine group resonate between 2.28 ppm to 2.37 ppm. The proton present in –NH resonates between 10.05 ppm and 12.47 ppm, whereas protons present in aromatic groups resonate between 7.00 ppm and 8.4 ppm. In 13C NMR spectra, the characteristic imine carbon resonates around 150 ppm, and in the infrared spectra, the stretching vibrations of the characteristic imine group appear in between 1542 and 1579 cm−1. The spectral data of thiosemicarbazones presented here are similar to the already published data.38 In the next step, as shown in Scheme 1, novel pyridine appended 2-hydrazinylthiazole derivatives have been synthesized from 2-(bromoacetyl)pyridine and corresponding thiosemicarbazones using the classical Hanzch-thiazole synthesis method. All the structures of the novel pyridine appended 2-hydrazinylthiazole derivatives have been confirmed using spectroscopic techniques. The characteristic thiazole proton resonates between 7.5–8.5 ppm, and the methyl protons attached to the imine group resonates around 2–2.5 ppm in 1H NMR spectra. The –NH protons of thiosemicarbazones resonate at 9.0–9.5 ppm, whereas the same protons resonate downfield at 10.5–12.5 ppm in the derivatives of pyridine appended 2-hydrazinylthiazoles, as a result of aromaticity present in the thiazole ring. The spectral data and spectra of all the novel compounds are given in ESI.† Structures of all the synthesized derivatives of pyridine appended 2-hydrazinylthiazole are given in Table 1.
2.3. Single crystal X-ray studies
In the next step, the single crystals of some of the pyridine appended 2-hydrazinylthiazole derivatives have been grown to understand the crystal packing and spatial orientation. As a representative structure, the crystal packing of a single crystal of 29b is given in Fig. 3, and the crystal parameters of 29b are tabulated in Table S1 (see ESI†). The compound 29b has intermolecular hydrogen bonding with another three molecules of the same compound. The nitrogen atom in a 2-pyridyl ring connected to the imine group forms H-bonding with the N–H group present in another molecule with a distance of 2.197 Å. The N–H group present in the same molecule forms H-bonding with imine substituted 2-pyridyl nitrogen with a distance of 2.063 Å. Likewise, the 2-pyridyl nitrogen, attached to the 4th position of the thiazole ring, interacts with another molecule's N–H group through a hydrogen bonding with a distance of 2.063 Å.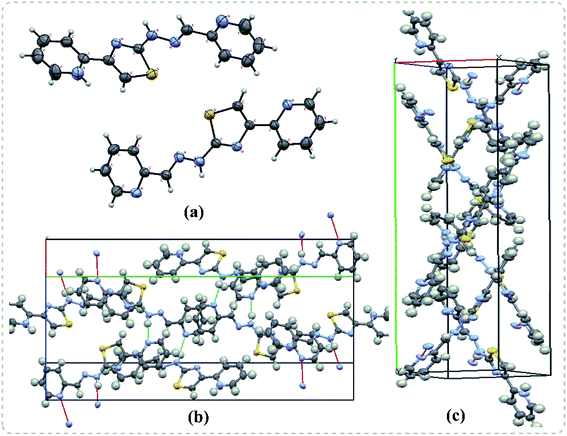 | ||
| Fig. 3 Single crystal X-ray diffraction results of 29b. (a) The ORTEP diagram (b) H-bonding interactions (c) double helical shape packing diagram in the crystal lattice. | ||
2.4. In vitro antitubercular screening of pyridine appended 2-hydrazinylthiazole derivatives
After successful synthesis and characterization, the antitubercular activity of pyridine appended 2-hydrazinylthiazole derivatives have been evaluated against Mtb, H37Rv strain using Luciferase reporter phage (LRP) assay. Most of the pyridine appended 2-hydrazinylthiazole derivatives have shown good antitubercular activity than the reported 2-hydrazinyl thiazole derivatives.19,39–41 The substitution effect on pyridine appended 2-hydrazinylthiazole derivatives on Mtb, H37Rv strain at the imine carbon was evaluated, and results are given in Table 1. The detailed in vitro antitubercular activity of pyridine appended 2-hydrazinylthiazole derivatives against Mtb, H37Rv are given in Table S2 (see ESI†). The pyridine appended 2-hydrazinylthiazole derivatives are categorized into three groups based on imine carbon substitution. The pyridine appended 2-hydrazinylthiazole derivatives substituted by aliphatic, phenyl, and pyridyl groups are given in one category. The halo and other substituted pyridine appended 2-hydrazinylthiazole derivatives are the other two categories. In the first category, the antitubercular activity of pyridine appended 2-hydrazinylthiazoles substituted with aliphatic, phenyl, and pyridyl groups show the MIC values in the range of 6–431 μM. Among these, phenyl substituted derivatives have shown excellent antitubercular activities. The compound 3b, phenyl and methyl substitution on the imine carbon of pyridine appended 2-hydrazinylthiazole has shown antitubercular activity with MIC value 6.79 μM. When the methyl group is substituted by the hydrogen in compound 3b, compound 2b has shown antitubercular activity with MIC value 7.13 μM. In the case of compound 4b where the phenyl group is substituted by a naphthyl group in compound 2b, the antitubercular activity is decreased with the MIC value 75.66 μM. In the case of compounds 30b and 29b where pyridyl substitution on the imine carbon of pyridine appended 2-hydrazinylthiazole derivatives have shown insufficient activity with the MIC values 338.56 μM and 355.44 μM, respectively. When two methyl groups are introduced at imine carbon of pyridine appended 2-hydrazinylthiazole, the resulting compound 1b has shown insufficient antitubercular activity with MIC value 430.46 μM.In the second category, halogen groups have been introduced on the phenyl ring of the pyridine appended 2-hydrazinylthiazole derivatives. All these compounds showed antitubercular activity with MIC values in the range of 6–318 μM. When the fluorine atom is substituted on the different positions at the phenyl ring of compound 2b, the antitubercular activity show MIC values in the range of 6–84 μM. In the case of compound 5b where fluorine substitution on the 2nd position of the phenyl group present in pyridine appended 2-hydrazinylthiazole, it shows excellent antitubercular activity with the MIC value 6.7 μM. When the fluorine was introduced on third and fourth positions of the phenyl group present in pyridine appended 2-hydrazinylthiazole, the resulting compounds 6b and 7b have shown similar antitubercular activity with MIC value 83.79 μM. When the 4-fluorophenyl and a methyl group are substituted at the imine carbon of pyridine appended 2-hydrazinylthiazole, the resulting compound 8b has shown excellent antitubercular activity with MIC value 6.4 μM. When chlorine atom is introduced on the different positions of phenyl ring of pyridine appended 2-hydrazinylthiazole, the compounds have shown moderate antitubercular activity. In compounds 11b and 13b, the chlorine is substituted at second and fourth positions on the phenyl ring of pyridine appended 2-hydrazinylthiazole, and these compounds have shown similar antitubercular activity with MIC value 79.42 μM. When the chlorine atom is substituted at second and fourth positions on the phenyl ring of pyridine appended 2-hydrazinylthiazole, the resulting compound 14b has shown insufficient antitubercular activity with MIC value 286.33 μM. In the case of compound 12b where chlorine is substituted at the third position on phenyl ring of pyridine appended 2-hydrazinylthiazole, it has shown insufficient antitubercular activity with MIC value 317.66 μM. In the case of bromine substitution at third and fourth positions on phenyl ring of pyridine appended 2-hydrazinylthiazoles, the resulting compounds 15b and 16b have shown similar antitubercular activity with MIC value of 278.36 μM. When the trifluoromethyl group is introduced at third and fourth positions of phenyl ring of pyridine appended 2-hydrazinylthiazole, the resulting compounds 9b and 10b have shown similar antitubercular activity with MIC value 71.77 μM.
In the third category, pyridine appended 2-hydrazinylthiazole derivatives substituted with methyl, methoxy, hydroxy, nitro, and diamine groups on the phenyl ring are considered. The antitubercular activity of these compounds shows the MIC values in the range of 73–340 μM. When a methyl group is substituted at the fourth position on the phenyl ring of pyridine appended 2-hydrazinylthiazole, the resulting compound 17b has shown insufficient antitubercular activity with MIC value 339.7 μM. When a hydroxy group is substituted at different positions on the phenyl ring of pyridine appended 2-hydrazinylthiazole derivatives, the antitubercular activity was in the range of 84 to 338 μM. In the case of compound 20b where the 4-hydroxy substituted phenyl ring of pyridine appended 2-hydrazinylthiazole, moderate antitubercular activity was observed with MIC value 84.36 μM. In the case of 18b and 19b where –OH group is attached at second and third positions on phenyl ring of pyridine appended 2-hydrazinylthiazole, both the molecules have shown similar antitubercular activity with MIC value 337.43 μM. In the case of compound 21b where the 4-methoxy group is substituted on the phenyl ring of pyridine appended 2-hydrazinylthiazole, it has shown moderate antitubercular activity with MIC value 80.55 μM. Further increasing the number of methoxy groups on phenyl ring of pyridine appended 2-hydrazinylthiazole, the resulting compounds 22b and 23b have shown insufficient antitubercular activity with the MIC values 293.76 μM and 269.96 μM, respectively. In the case of compound 24b where the –OH group at 3rd position, and methoxy group at the 4th position on the phenyl ring of pyridine appended 2-hydrazinylthiazole insufficient antitubercular activity was observed with MIC value 306.39 μM. When the hydroxy group is substituted at fourth and ethoxy group at third positions on phenyl ring of pyridine appended 2-hydrazinylthiazole, the resulting compound 25b showed moderate antitubercular activity with MIC value of 73.44 μM. In nitro substitution at third and fourth positions on the phenyl ring, the resulting compounds 26b and 27b have shown insufficient antitubercular activity with MIC value 307.36 μM. When the 4-dimethylamine is substituted on the phenyl ring, the resulting compound 28b has shown poor antitubercular activity with MIC value 309.19 μM. Overall, the compounds 2b, 3b, 5b, and 8b have shown excellent antitubercular activity with the MIC values range of 6–8 μM. These values are near to the standard rifampicin MIC value, i.e., 2.4 μM. The comparison of MIC values of different pyridine appended 2-hydrazinylthiazole derivatives is shown in Fig. 4.
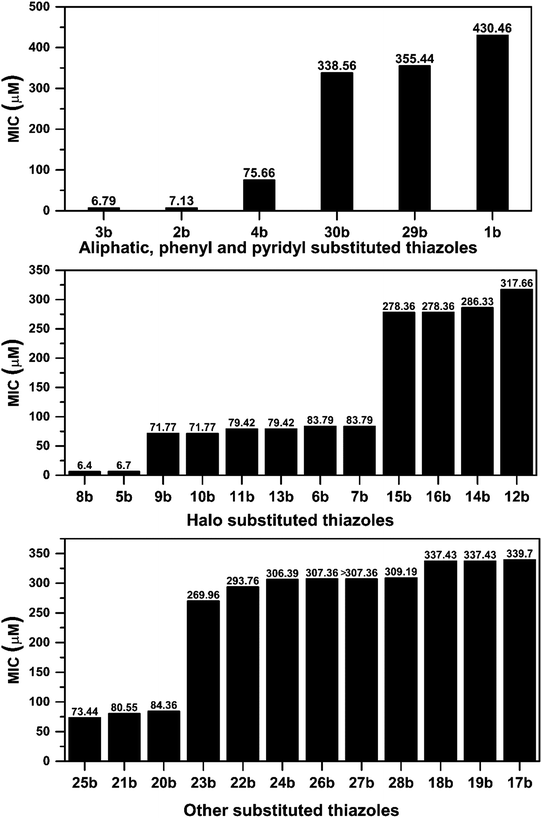 | ||
| Fig. 4 Correlation between antitubercular activity and substitution on the imine group of pyridine appended 2-hydrazinylthiazoles. | ||
Similarly, the correlation between log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values and their antitubercular activity of pyridine appended 2-hydrazinylthiazole derivatives is shown in Fig. 5. The log
P values and their antitubercular activity of pyridine appended 2-hydrazinylthiazole derivatives is shown in Fig. 5. The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value is gradually increased by introducing lipophilic groups at imine carbon of pyridine appended 2-hydrazinylthiazole. In the first category, the log
P value is gradually increased by introducing lipophilic groups at imine carbon of pyridine appended 2-hydrazinylthiazole. In the first category, the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of pyridine appended 2-hydrazinylthiazoles are in the range of 1.64 to 3.97, and their MIC values are in the range of 6–431 μM. In the case of pyridyl substituted pyridine appended 2-hydrazinylthiazole derivatives, as in the case of compounds 29b and 30b, least log
P values of pyridine appended 2-hydrazinylthiazoles are in the range of 1.64 to 3.97, and their MIC values are in the range of 6–431 μM. In the case of pyridyl substituted pyridine appended 2-hydrazinylthiazole derivatives, as in the case of compounds 29b and 30b, least log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of 1.64 and 2.02, respectively was observed, and antitubercular activity of these compounds was also poor. When the log
P values of 1.64 and 2.02, respectively was observed, and antitubercular activity of these compounds was also poor. When the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value was increased to 2.04, the antitubercular activity decreased for the compound 30b with the MIC value 430.46 μM. Further, an increase in log
P value was increased to 2.04, the antitubercular activity decreased for the compound 30b with the MIC value 430.46 μM. Further, an increase in log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value to 3.26 leads to the increased antitubercular activity of the pyridine appended 2-hydrazinylthiazole derivatives. The compounds 2b and 3b with the log
P value to 3.26 leads to the increased antitubercular activity of the pyridine appended 2-hydrazinylthiazole derivatives. The compounds 2b and 3b with the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values 2.81 and 3.26 show good antitubercular activity with MIC values 7.13 μM and 6.79 μM, respectively. When the log
P values 2.81 and 3.26 show good antitubercular activity with MIC values 7.13 μM and 6.79 μM, respectively. When the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value is increased to 3.97, as in compound 4b, it has shown moderate antitubercular activity with a MIC value of 75.66 μM.
P value is increased to 3.97, as in compound 4b, it has shown moderate antitubercular activity with a MIC value of 75.66 μM.
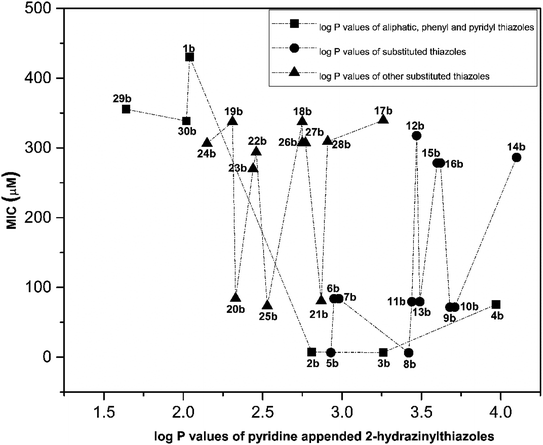 | ||
Fig. 5 Correlation between log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of pyridine appended 2-hydrazinylthiazoles and their antitubercular activity. P values of pyridine appended 2-hydrazinylthiazoles and their antitubercular activity. | ||
In the second category, the halogen-substituted pyridine appended 2-hydrazinylthiazole derivatives have shown the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value in the range of 2.93–4.1. Compound 5b has the log
P value in the range of 2.93–4.1. Compound 5b has the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of 2.93 and showed excellent antitubercular activity with a MIC value of 6.7 μM. When the log
P value of 2.93 and showed excellent antitubercular activity with a MIC value of 6.7 μM. When the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value is increased to 2.98, the compounds 6b and 7b have shown moderate antitubercular activity with MIC value 83.79 μM. Interestingly, a further increase in log
P value is increased to 2.98, the compounds 6b and 7b have shown moderate antitubercular activity with MIC value 83.79 μM. Interestingly, a further increase in log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P to 3.42, compound 8b has shown excellent antitubercular activity with MIC value 6.4 μM. When the log
P to 3.42, compound 8b has shown excellent antitubercular activity with MIC value 6.4 μM. When the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value is increased to 3.47, the antitubercular activity is decreased for 11b and 12b with MIC values 79.42 μM and 317.66 μM, respectively. In the case of compound 13b, which has the log
P value is increased to 3.47, the antitubercular activity is decreased for 11b and 12b with MIC values 79.42 μM and 317.66 μM, respectively. In the case of compound 13b, which has the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of 3.49, it showed moderate antitubercular activity with a MIC value of 79.42 μM. When the log
P value of 3.49, it showed moderate antitubercular activity with a MIC value of 79.42 μM. When the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value is increased to 3.62, the compounds 15b and 16b have shown similar and inferior antitubercular activity with MIC value 278.36 μM. The compounds 9b and 10b have the log
P value is increased to 3.62, the compounds 15b and 16b have shown similar and inferior antitubercular activity with MIC value 278.36 μM. The compounds 9b and 10b have the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values 3.62 and 3.68 and showed similar and moderate antitubercular activity with MIC value 71.77 μM. Further increase in the log
P values 3.62 and 3.68 and showed similar and moderate antitubercular activity with MIC value 71.77 μM. Further increase in the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value to 4.1, the compound 14b has shown poor antitubercular activity with MIC value 286.33 μM. In the third category, the pyridine appended 2-hydrazinylthiazole derivatives have log
P value to 4.1, the compound 14b has shown poor antitubercular activity with MIC value 286.33 μM. In the third category, the pyridine appended 2-hydrazinylthiazole derivatives have log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values in the range of 2.15–3.26 and showed moderate to poor antitubercular activity. The compounds 24b and 19b have log
P values in the range of 2.15–3.26 and showed moderate to poor antitubercular activity. The compounds 24b and 19b have log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values 2.15 and 2.31 and showed poor antitubercular activity with MIC values 306.39 μM and 337.43 μM, respectively.
P values 2.15 and 2.31 and showed poor antitubercular activity with MIC values 306.39 μM and 337.43 μM, respectively.
Further increasing the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value to 2.33, the compound 20b has shown antitubercular activity with MIC value 84.36 μM. When the log
P value to 2.33, the compound 20b has shown antitubercular activity with MIC value 84.36 μM. When the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value increased to 2.46, the compounds 23b and 22b have shown moderate antitubercular activity with MIC values 269.96 μM and 293.76 μM, respectively. Compounds 25b and 18b with log
P value increased to 2.46, the compounds 23b and 22b have shown moderate antitubercular activity with MIC values 269.96 μM and 293.76 μM, respectively. Compounds 25b and 18b with log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of 2.53 and 2.75 respectively showed antitubercular activity 73.44 μM and 337.43 μM, respectively. When the log
P values of 2.53 and 2.75 respectively showed antitubercular activity 73.44 μM and 337.43 μM, respectively. When the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value is increased to 2.77, the compounds 26b and 27b have shown similar antitubercular activity with MIC value 307.36 μM. Further increasing the log
P value is increased to 2.77, the compounds 26b and 27b have shown similar antitubercular activity with MIC value 307.36 μM. Further increasing the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value to 2.87, the compound 21b has shown moderate antitubercular activity with a MIC value of 80.55 μM. When the log
P value to 2.87, the compound 21b has shown moderate antitubercular activity with a MIC value of 80.55 μM. When the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value increased further to 3.26, the antitubercular activity decreased for the compounds 28b and 17b with MIC values 309.19 μM and 339.7 μM, respectively. Overall, most of the active pyridine appended 2-hydrazinylthiazole derivatives have shown good antitubercular activity when the log
P value increased further to 3.26, the antitubercular activity decreased for the compounds 28b and 17b with MIC values 309.19 μM and 339.7 μM, respectively. Overall, most of the active pyridine appended 2-hydrazinylthiazole derivatives have shown good antitubercular activity when the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value in the range of 2.81–3.42.
P value in the range of 2.81–3.42.
2.5. In vitro cytotoxicity studies of pyridine appended 2-hydrazinylthiazole derivatives
After in vitro antitubercular evaluation, the active pyridine appended 2-hydrazinylthiazoles have been tested against Human Embryonic Kidney Cell lines (HEK 293t) to understand the cytotoxicity of the target thiazole derivatives. The active compounds 2b, 3b, 5b, and 8b were treated with HEK 293t cells at 6.5 μM concentrations, and MTT assay was used to analyse the cell growth. The cell viability of the compounds is in the range of 38–97%, and the results are presented in Fig. 6. Pyridine appended 2-hydrazinylthiazoles have shown more than 50% cell viability considered as nontoxic. The compounds 3b and 8b have shown cell viability at 87.52% and 96.71%, and hence, these compounds are termed to be nontoxic.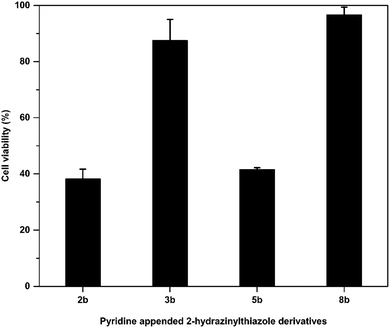 | ||
| Fig. 6 Cytotoxicity of pyridine appended 2-hydrazinylthiazole derivatives against human embryonic kidney cell lines. | ||
2.6. Molecular docking studies
Docking studies for pyridine appended 2-hydrazinylthiazole derivatives against Mtb, KasA protein was carried out in order to determine the mode of action because the 2-aminothiazole pharmacopore is structurally similar to the thiolactomycin (TLM) which is naturally occurring antibiotic.13,19 TLM inhibits β-ketoacyl-ACP synthase (KasA) in mtFabH fatty acid synthesis, which in turn inhibits cell wall biosynthesis, resulting in Mtb death.14 In the mycolic acid biosynthesis,42 the long-chain fatty acid, KasA protein, plays an important role, thus, KasA protein is selected as a target for docking studies in the present investigation. All the active antitubercular agents have shown good interaction with KasA protein by forming a strong hydrogen bonding with the pyridine appended 2-hydrazinylthiazole molecules. The binding score of the complex was found to be in the range of −4.517 kcal mol−1 to −6.233 kcal mol−1. Some molecules showed a better binding score than standard drug isoniazid, i.e., −5.830 kcal mol−1. The in silico results and interaction diagram with Mtb, KasA protein are given in Table 1 and Fig. 7, respectively. The hydrophobic interacting residues of the KasA protein of Mtb are shown in Table S2 (see ESI†). The active pyridine appended 2-hydrazinylthiazoles have strong interaction with the KasA protein of Mtb. The pyridine nitrogen in compound 2b has hydrogen bonding with Thr 135 residue of KasA protein with a distance of 2.94 Å. This compound has several hydrophobic interactions with Arg 214, Met 213, Ala 215, His 311, Gly 318, Phe 402, Thr 313, Pro 280, Val 278, Phe 404, Ile 317, Ala 279 of KasA protein. In the case of compound 3b, the hydrazone N–H has hydrogen bonding with KasA protein through Val 278 residue with a distance of 3.19 Å and has several hydrophobic interactions such as Met 213, Arg 214, Ala 215, Ile 317, Ala 279, Pro 280, Thr 313, Thr 315, Gly 318, Asp 319, Glu 322, Phe 402, Gly 403, Phe 404 of KasA protein. The halo substituted active compounds have interaction (s) with Thr 315 residue of KasA protein. Compound 5b has two hydrogen-bonding interactions with Thr 315. The nitrogen atom in the thiazole ring has one hydrogen bonding with Thr 315 with a distance of 3.29 Å. The nitrogen atom in the pyridine ring has another hydrogen bonding with Thr 315 with a distance of 2.93 Å. Compounds 6b, 7b, and 8b have hydrogen bonds with KasA protein through Thr 135 residue with a distance of 2.93 Å, 2.93 Å, and 2.90 Å, respectively.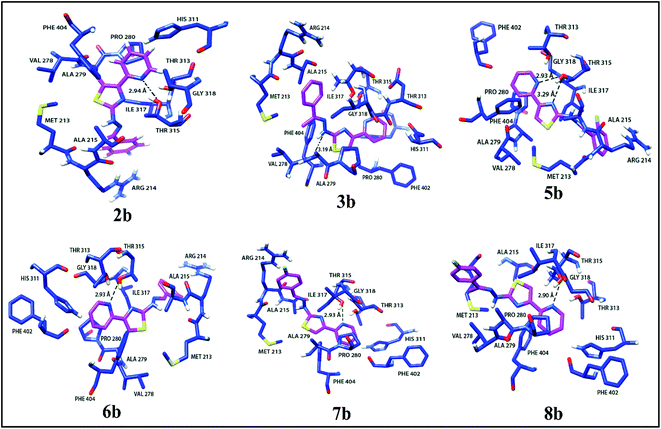 | ||
| Fig. 7 Interaction diagrams of active pyridine appended 2-hydrazinylthiazole derivatives with KasA protein of Mtb. | ||
3. Conclusion
The derivatives of pyridine appended 2-hydrazinylthiazole have been designed based on Lipinski and Veber rules. The designed thiazole derivatives were synthesized from 2-(bromoacetyl)pyridine and a variety of thiosemicarbazones. The resulting compounds have been evaluated against the H37Rv strain of Mtb to understand their antitubercular activity. Pyridine appended 2-hydrazinylthiazole derivatives with phenyl (2b and 3b), phenyl substitution with 2- and 4-fluoro (5b and 8b) groups at imine carbon have shown excellent inhibitory potential with MIC values of 6–8 μM. Human embryonic kidney cell lines were used to determine the in vitro cytotoxicity properties of active pyridine appended 2-hydrazinylthiazoles at 6.5 μM. The active compounds 3b and 8b show cell viability of more than 87.52% and 96.71%, respectively, and are nontoxic. The docking studies of pyridine appended 2-hydrazinylthiazoles have been done against Mtb, KasA protein, and the molecules showed a binding score in the range of −5.27 to −6.23 kcal mol−1.4. Experimental section
4.1. Materials and methods
Bruker Avance-II NMR spectrometer (400 MHz for 1H nucleus and 101 MHz for 13C nucleus) was used to get NMR spectra. Thermo Nicolet 6700 Fourier transform infrared (FT-IR) spectrometer was used to get FT-IR spectra. Thermo Scientific High-Resolution Magnetic Sector MS DFS mass spectrometer was used to record mass spectra (MS). Rigaku-Oxford Xcalibur Eos single-crystal X-ray diffractometer with Mo-Kα radiation (λ = 0.71073 Å) was used to obtain single crystal X-ray data. For empirical absorption correction, the implemented SCALE3 ABSPACK scaling algorithm was used. The XL in the Olex 2-1.2 package was used for the refinement and SHELXS-97 was used for the structural solution.4.2. Synthesis of thiosemicarbazone derivatives
Carbonyl compounds (1.1 mmol) and appropriate thiosemicarbazide (1 mmol) were dissolved in ethanol. To this solution, acetic acid (glacial) (a catalytic amount) was added. The resulting solution was refluxed for 3–6 hours and then cooled to room temperature. The resulting precipitate was filtered to obtain the corresponding thiosemicarbazone.4.3. General procedure for the synthesis of pyridine appended 2-hydrazinylthiazole derivatives
2-(Bromoacetyl)pyridine (1.1 mmol) and appropriate thiosemicarbazone (1.0 mmol) were dissolved in ethyl alcohol. The resulting solution was refluxed for 30 min–6 h and after cooling to room temperature, the reaction mixture was neutralized with an aqueous NaHCO3 solution. Finally, the solid obtained was filtered and recrystallized from ethyl alcohol to get the final products.4.4. In vitro antimycobacterial assay
The antimycobacterial activity of pyridine appended 2-hydrazinylthiazole derivatives was evaluated using Luciferase Reporter Phage (LRP) assay method with modification.43 The brief procedure as follows. About 400 μL of sterile Middlebrook 7H9 broth containing pyridine appended 2-hydrazinylthiazole derivatives at concentrations of 100 μg mL−1 were aliquoted into test cryovials (T) and 400 μL of Middlebrook 7H9 broth containing rifampicin (2 μg mL−1) was transferred to drug control cryovial (D). Next, the control cryovial (C) was aliquoted with 400 μL of sterile below-mentioned 7H9 broth (Himedia). Mtb H37Rv cell suspension (100 μL) (McFarland no. 2) were used to inoculate all cryovials and incubated at 37 °C for 72 h. Then, 40 μL of 0.1 M CaCl2 and 50 μL of mycobacteriophage (phAE202) were added into all the cryovials (cell-phage mixture) and incubated for 4 h at 37 °C. Then, 100 μL of the cell-phage mixture was transferred to a luminometer cuvette and added with 100 μL of D-luciferin substrate (Cayman chemicals, USA). The relative light unit (RLU) was measured at 10 s integration time in a luminometer (Lumat 9508, Berthold, Germany). The compounds considered as an antitubercular agent if test compounds showed a 50% reduction in RLU when compared to the control RLU. The % of RLU reduction was calculated by using the below-mentioned formula.The active compounds were further tested for their antitubercular activities at 25 μg mL−1 and 2 μg mL−1 concentrations by LRP assay, as mentioned above.
4.5. In vitro cytotoxicity assay
4.6. Molecular docking
To perform the molecular docking study, the protein was retrieved from the RCSB protein data bank (https://www.rcsb.org/pdb/home/home.do). Before the docking study, the target protein was prepared using the protein preparation wizard, in which the protein was pre-processed, optimized, and minimized. Similarly, to prepare the ligands for docking in maestro format, Schrodinger v11.5 (LigPrep module) was used. A grid box was generated around the protein in which the ligands bind to the protein's active site. The prepared protein and ligands were docked using Schrodinger v11.5. The glide score was calculated within the Schrodinger v11.5 software. The active site prediction was made using the COACH server,44 which can be accessed freely. The COACH server is generally used to predict protein–ligand binding sites. The LIGPLOT (version 4.5.3) program was used to generate schematic representations of protein-ligand complexes. UCSF Chimera (version 1.5.3) has been used for 3-D visualization and structural analysis of protein-ligand complex.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to Mr Anbarasu Sivaraj, Sathyabama Institute of Science and Technology, Chennai, India for the generation of the antitubercular data against Mtb, H37Rv strain. University Research Fellowship (URF) to Mr Ramkishore Matsa by Pondicherry University is gratefully acknowledged. Funding by University Grants Commission (UGC), New Delhi (No. F.540/6/DSA-1/2016/(SAP-1) dated 31-10-2018) to Department of Chemistry, Pondicherry University is gratefully acknowledged. We acknowledge Pondicherry University's Central Instrumentation Facility (CIF) for their support in characterizing pyridine appended 2-hydrazinylthiazole derivatives.References
- World Health Organization, Global Tuberculosis Report 2021, 2021 Search PubMed
.
- C. R. Horsburgh, C. E. Barry and C. Lange, N. Engl. J. Med., 2015, 373, 2149–2160 CrossRef CAS PubMed
.
- L. G. Dover and G. D. Coxon, J. Med. Chem., 2011, 54, 6157–6165 CrossRef CAS PubMed
.
- B. Liu, F. Li, T. Zhou, X.-Q. Tang and G.-W. Hu, J. Heterocycl. Chem., 2018, 55, 1863–1873 CrossRef CAS
.
- S. J. Keam, Drugs, 2019, 79, 1797–1803 CrossRef PubMed
.
- E. L. Luzina and A. V Popov, Eur. J. Med. Chem., 2009, 44, 4944–4953 CrossRef CAS PubMed
.
- S. M. El-Messery, G. S. Hassan, F. A. M. Al-Omary and H. I. El-Subbagh, Eur. J. Med. Chem., 2012, 54, 615–625 CrossRef CAS PubMed
.
- J. Cai, M. Sun, X. Wu, J. Chen, P. Wang, X. Zong and M. Ji, Eur. J. Med. Chem., 2013, 63, 702–712 CrossRef CAS PubMed
.
- P. Makam, P. K. Thakur and T. Kannan, Eur. J. Pharm. Sci., 2014, 52, 138–145 CrossRef CAS PubMed
.
- R. P. Karuvalam, K. R. Haridas, S. K. Nayak, T. N. Guru Row, P. Rajeesh, R. Rishikesan and N. S. Kumari, Eur. J. Med. Chem., 2012, 49, 172–182 CrossRef CAS PubMed
.
- M. H. M. Helal, M. A. Salem, M. S. A. El-Gaby and M. Aljahdali, Eur. J. Med. Chem., 2013, 65, 517–526 CrossRef CAS PubMed
.
- S. N. Mokale, P. T. Sanap and D. B. Shinde, Eur. J. Med. Chem., 2010, 45, 3096–3100 CrossRef CAS PubMed
.
- G. Pappenberger, T. Schulz-Gasch, E. Kusznir, F. Müller and M. Hennig, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2007, 63, 1208–1216 CrossRef CAS PubMed
.
- S. R. Luckner, C. A. Machutta, P. J. Tonge and C. Kisker, Structure, 2009, 17, 1004–1013 CrossRef CAS PubMed
.
- S. Ranjbar, V. Haridas, A. Nambu, L. D. Jasenosky, S. Sadhukhan, T. S. Ebert, V. Hornung, G. H. Cassell, J. V Falvo and A. E. Goldfeld, iScience, 2019, 22, 299–313 CrossRef CAS PubMed
.
- L. M. Fox and L. D. Saravolatz, Clin. Infect. Dis., 2005, 40, 1173–1180 CrossRef CAS PubMed
.
- E. P. Harausz, K. A. Chervenak, C. E. Good, M. R. Jacobs, R. S. Wallis, M. Sanchez-Felix and W. H. Boom, Tuberculosis, 2016, 98, 92–96 CrossRef CAS PubMed
.
- Q. Al-Balas, N. G. Anthony, B. Al-Jaidi, A. Alnimr, G. Abbott, A. K. Brown, R. C. Taylor, G. S. Besra, T. D. McHugh, S. H. Gillespie, B. F. Johnston, S. P. Mackay and G. D. Coxon, PLoS One, 2009, 4, e5617 CrossRef PubMed
.
- P. Makam, R. Kankanala, A. Prakash and T. Kannan, Eur. J. Med. Chem., 2013, 69, 564–576 CrossRef CAS PubMed
.
- P. Makam and T. Kannan, Eur. J. Med. Chem., 2014, 87, 643–656 CrossRef CAS PubMed
.
- P. Makam, R. Kankanala, A. Prakash and T. Kannan, Eur. J. Med. Chem., 2013, 69, 564–576 CrossRef CAS PubMed
.
- M. A. Metwally, E. Abdel-Latif, F. A. Amer and G. Kaupp, J. Sulfur Chem., 2004, 25, 63–85 CrossRef CAS
.
- C. Viegas-Junior, A. Danuello, V. D. S. Bolzani, E. J. Barreiro and C. A. M. Fraga, Curr. Med. Chem., 2007, 14, 1829–1852 CrossRef CAS PubMed
.
- C. A. M. Fraga, Expert Opin. Drug Discovery, 2009, 4, 605–609 CrossRef CAS PubMed
.
- J. Rybniker, A. Vocat, C. Sala, P. Busso, F. Pojer, A. Benjak and S. T. Cole, Nat. Commun., 2015, 6, 7659 CrossRef PubMed
.
- R. C. Hartkoorn, C. Sala, J. Neres, F. Pojer, S. Magnet, R. Mukherjee, S. Uplekar, S. Boy-Röttger, K.-H. Altmann and S. T. Cole, EMBO Mol. Med., 2012, 4, 1032–1042 CrossRef CAS PubMed
.
- S. Ntshangase, A. Shobo, H. G. Kruger, A. Asperger, D. Niemeyer, P. I. Arvidsson, T. Govender and S. Baijnath, Xenobiotica, 2018, 48, 938–944 CrossRef CAS PubMed
.
- E. J. Corey and D. Enders, Tetrahedron Lett., 1976, 17, 11–14 CrossRef
.
- S. Rollas and G. S. Küçükgüzel, Molecules, 2007, 12 Search PubMed
.
- Ł. Popiołek, Med. Chem. Res., 2017, 26, 287–301 CrossRef PubMed
.
- B. Mathew, J. Suresh, M. J. Ahsan, G. E. Mathew, D. Usman, P. N. S. Subramanyan, K. F. Safna and S. Maddela, Infect. Disord.: Drug Targets, 2015, 15, 76–88 CAS
.
- P. Makam, P. K. Thakur and T. Kannan, Eur. J. Pharm. Sci., 2014, 52, 138–145 CrossRef CAS PubMed
.
- A. Arshad, H. Osman, M. C. Bagley, C. K. Lam, S. Mohamad and A. S. M. Zahariluddin, Eur. J. Med. Chem., 2011, 46, 3788–3794 CrossRef CAS PubMed
.
- G. Turan-Zitouni, Z. A. Kaplancıklı and A. Özdemir, Eur. J. Med. Chem., 2010, 45, 2085–2088 CrossRef CAS PubMed
.
- C. A. Lipinski, Drug Discovery Today: Technol., 2004, 1, 337–341 CrossRef CAS PubMed
.
- D. F. Veber, S. R. Johnson, H. Y. Cheng, B. R. Smith, K. W. Ward and K. D. Kopple, J. Med. Chem., 2002, 45, 2615–2623 CrossRef CAS PubMed
.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef PubMed
.
- R. Matsa, P. Makam, M. Kaushik, S. L. Hoti and T. Kannan, Eur. J. Pharm. Sci., 2019, 104986 CrossRef CAS PubMed
.
- K. K. Roy, S. Singh, S. K. Sharma, R. Srivastava, V. Chaturvedi and A. K. Saxena, Bioorg. Med. Chem. Lett., 2011, 21, 5589–5593 CrossRef CAS PubMed
.
- P. V Sowmya, B. Poojary, V. Kumar, U. Vishwanatha and P. Shetty, Arch. Pharmacal Res., 2017 DOI:10.1007/s12272-017-0967-1
.
- M. Hublikar, V. Kadu, J. K. Dublad, D. Raut, S. Shirame, P. Makam and R. Bhosale, Arch. Pharm., 2020, 353, 2000003 CrossRef PubMed
.
- R. Veyron-Churlet, S. Bigot, O. Guerrini, S. Verdoux, W. Malaga, M. Daffé and D. Zerbib, J. Mol. Biol., 2005, 353, 847–858 CrossRef CAS PubMed
.
- A. Sivaraj, V. Kumar, R. Sunder, K. Parthasarathy and G. Kasivelu, J. Cluster Sci., 2020,(31), 287–291 CrossRef CAS
.
- J. Yang, A. Roy and Y. Zhang, Bioinformatics, 2013, 29, 2588–2595 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02163c |
| This journal is © The Royal Society of Chemistry 2022 |