Open Access Article
Open Access ArticleFabrication of photoluminescent nanoparticles from carbazole-derived chalcones: a study of optical properties, cell biomarking, and metabolism†
Susana Lucía
Estrada-Flores
a,
Cesar
Garcias-Morales
 *a,
Catalina M.
Perez-Berumen
a,
Arxel
de León-Santillán
b,
Mario
Rodríguez
c,
Juan Pablo
García-Merinos
*a,
Catalina M.
Perez-Berumen
a,
Arxel
de León-Santillán
b,
Mario
Rodríguez
c,
Juan Pablo
García-Merinos
 d,
Jesús A.
Claudio-Rizo
d,
Jesús A.
Claudio-Rizo
 a and
Eder Iván
Martínez-Mora
a
a and
Eder Iván
Martínez-Mora
a
aFacultad de Ciencias Químicas, Universidad Autónoma de Coahuila, Ing. J. Cárdenas Valdez S/N, República, 25280 Saltillo, Coahuila, Mexico. E-mail: cgarcias@uadec.edu.mx
bConacyt-Centro de Investigación en Química Aplicada, Blvd. Enrique Reyna H. 140, 25294 Saltillo, Coah, Mexico
cResearch Group of Optical Properties of Materials (GPOM), Centro de Investigaciones en Óptica, A.P. 1-948, 37000 León, Guanajuato, Mexico
dInstituto de Investigaciones Químico Biológicas, Universidad Michoacana de San Nicolás de Hidalgo, Ed.B-1, C.U., Morelia, Michoacán 58030, Mexico
First published on 22nd June 2023
Abstract
In this work, the synthesis by Claisen–Schmidt condensation of two new chalcones: (2E)-3-[4-(9H-carbazol-9-yl)phenyl]-1-(2,4-difluorophenyl)prop-2-en-1-one (M1) and (2E)-3-[4-(9H-carbazol-9-yl)phenyl]-1-(3,4-difluorophenyl)prop-2-en-1-one (M2), with a donor–acceptor structure for photonic applications is reported. The characterization of the optoelectronic properties is evaluated, the two chalcones have photoluminescence in both solution and in the solid state, however, the intensity of the photoluminescence is higher in the solid state (I/I0 = 2). The electronic distribution of M1 and M2 was studied by the solvatocromismo and acidochromism experiment, where it was determined that the absorption and emission of the molecule change according to the polarity of the solvent. The D–A structure was corroborated by the data obtained by DFT analysis using the B3LYP functional and the 6-31G(d) base. Silica precursor-coated nanoparticles were fabricated by the microemulsion method, obtaining spherical biocompatible nanoparticles with a size distribution of 42 nm and ΦF of 8% and 18% for M1 and M2 respectively. Cytotoxicity tests were carried out on different cell lines, healthy cells, colon cancer cells, and breast cancer cells, finding that the NPs coated with silica were not cytotoxic, but when irradiated at 366 nm for 1 h, the population of diseased cells is reduced almost entirely.
Introduction
Due to the optical properties of organic fluorophores, the scientific community has chosen to design and synthesize new molecules based on this type of compound to evaluate their luminescent properties for application in optoelectronics and biomedicine areas.1–6Chalcones are interesting molecules owing to their non-linear optical properties resulting from their design with electron-donating and electro-attracting groups, obtaining a Donor–π–Acceptor system (D–π–A),7–11 this leads to structures with the push–pull effect that promotes internal charge transfer (ICT) complexes with interesting photophysical properties, which allow their application as biomarkers, organic field-effect transistors (OFETs), organic light emitting diodes (OLEDs) and organic photovoltaics (OPVs).12–16
Molecules with the D–A electronic structure are used as chemosensors has been reported, it is due to optical properties such as colour change or fluorescence since this electronic structure can present absorption bands caused by internal charge transfers ICT, which modify the optical properties of molecules through the electron density modification and these properties can be evaluated by computational calculations using DFT.17–19
In the design of D–A molecules, carbazole is highlighted as a donor fragment, which has been reported as part of the molecular structure of fluorophores with dual state emission (DSE),20–22 as well as systems with aggregation induced emission enhancement (AIEE)23–25 with high fluorescence quantum yields in the solid state (ΦF = 14–59%) and in the liquid state (ΦF = 73–100%).26 It is possible to take advantage of the optical properties of carbazole by including it in the chalcone structure, resulting in band gap values of ∼3.25 eV and an increasing in the ICT.27,28 The emission of the molecule in solution will depend on the solvent in which it is dissolved due to the change in electron density; where the fluorescence quantum yield can decrease with the increasing of the solvent polarity from 53 to 8%.29–31
The synthesis of molecules derived from carbazole under D–D–A structure has been reported, these compounds have emission in the infrared region in the solid state since carbazole derivatives are good fluorophores they can be used in the development of potential molecules for cell imaging and photodynamic therapy (PDT).32,33 Carbazole derivatives in which an H atom has been replaced by iodine have also been reported, where it improved the molar absorption coefficient and red-shifted displacement of absorption and emission; additionally, iodine improves the formation of reactive species (ROS) that lead to cell apoptosis, making them good candidates for photodynamic therapy.34,35 Recently, nanoparticles of compounds derived from carbazole have been manufactured, which have allowed for improving selectivity and biocompatibility with the biological system.36,37
The use of organic fluorophores for photodynamic therapy involves the evaluation of the optical and toxicological properties. For this reason, manufacturing methods of luminescent organic nanoparticles coated with a biocompatible material have been reported using techniques such as microemulsion and reprecipitation, where the optical properties have been preserved, and biocompatibility has improved. The use of silica allows preserving the optoelectronic properties such as the maximum wavelength of absorption (λa) and emission (λe), as well as the fluorescence intensity increase with the formation of aggregates. Molecules coated with silica nanoparticles can also be internalized at the cellular level (fixation in the cell membrane or nucleus) adapting new ways of cell staining and control of cell metabolism.38–42
Nanoparticles sizes used in this application have average around 25 to 82 nm that in dark cytotoxicity assays (MTT assay, olive moment, neutral comet assay, lactate dehydrogenase assay) prove to be biocompatible by showing cell viabilities after 48 hours of incubation of 80% in cells of the immune system, cells of the HeLa type, A375, HGF, HepG2, and fibroblasts.43–49
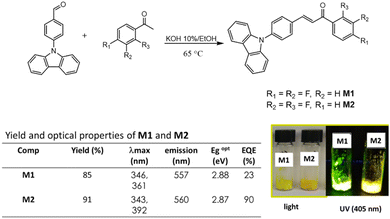
This work reports the synthesis of the new chalcones (2E)-3-[4-(9H-carbazol-9-yl)phenyl]-1-(2,4-difluorophenyl)prop-2-en-1-one (M1) and (2E)-3-[4-(9H-carbazol-9-yl)phenyl]-1-(3,4-difluorophenyl)prop-2-en-1-one (M2) derived from carbazole under a D–A architecture and the fabrication of nanoparticles using the microemulsion method; the aggregation induced emission enhancement, absorption, emission and fluorescence quantum yield of the material is evaluated to determine its possible application as cell biomarkers in photodynamic therapy (PDT).
Experimental
Materials and methods
The reagents and solvents: N-(4-formylphenyl)carbazole, 2,4-difluoroacetophenone, 3,4-difluoroacetophenone, Aerosol-OT, dioxane, ethyl ether, aluminium trichloride, triethoxyvinylsilane (VTES), aminopropyltrietosisilane (APTES), were obtained from Sigma-Aldrich and used without purification. Reagent grade solvents; toluene, hexane, chloroform, chlorobenzene, ethyl acetate, and hydrochloric acid were obtained from Jalmek Cientifica. Tetrahydrofuran and sodium hydroxide were obtained from CTR, Mexico. Triethylamine, acetonitrile, acetone, methanol, methylene chloride, and propanol were obtained from Fisher Scientific reagent grade. 3-(4,5-dimethylthiazolyl)-2,5-diphenyltetrazolium bromide (MTT), Alsever's solution, rhodamine-b and live/dead cell proliferation kit was purchased from ThermoFisher Scientific.The Edinburg FS5 Fluorometer with an integrating sphere was used to determine the fluorescence quantum yields. The SYNERGY H1 microplate reader was used to obtain the emission spectra, and the absorption spectra were obtained using a JENWAY 7315 Spectrophotometer. The morphological study of the coated nanoparticles was performed by Transmission Electron Microscopy (TEM) with the FEI Company equipment (Titan 300 KV). The particle size distribution was determined in the Nanotrac Wave III Q equipment from the Microtrac® commercial house.
Synthesis of chalcones
The chalcones were synthesized by the Claisen–Schmidt condensation reaction (Fig. 1). The structure characterization was carried out by FT-IR where a band at 1663 cm−1 is observed in M1 and 1666 cm−1 for M2 for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. By NMR, two signals as double were observed at 7.45 and 7.71 ppm with a coupling constant of 3JH,H = 15.4 Hz, which correspond to the hydrogens in the α,β-system of M1 while for M2 these hydrogens were observed as double at 7.53 and 7.92 ppm, with a 3JH,H = 15.4 Hz, the complete assignment of the protons and carbons can be shown in the ESI.†
O. By NMR, two signals as double were observed at 7.45 and 7.71 ppm with a coupling constant of 3JH,H = 15.4 Hz, which correspond to the hydrogens in the α,β-system of M1 while for M2 these hydrogens were observed as double at 7.53 and 7.92 ppm, with a 3JH,H = 15.4 Hz, the complete assignment of the protons and carbons can be shown in the ESI.†
In the absorption spectrum in the solid state, two bands were observed for M1, at 346 nm and 361 nm, the first one is attributed to nN → π* transition, and the second band is attributed to the internal charge transfer (ICT) complex due to the D–A structure. For M2, two bands were also observed, at 343 nm attributed to the nN → π* transition and 392 nm attributed to the ICT. Finally, the quantum yield (ΦF) was determined, M2, reached a ΦF of 90%, and for M1 the ΦF is 23%.
Results and discussion
Photophysical properties
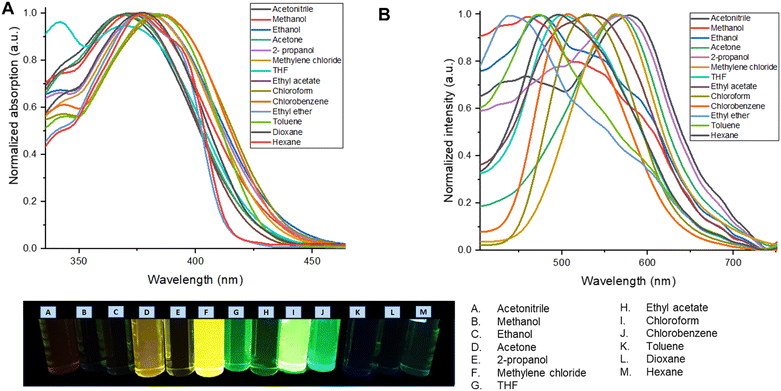
To study the photophysical properties of M1 and M2, the solvatochromism analysis was carried out, which consisted in obtaining the emission and absorption spectra in solvents with different polarities. The study of solvatochromism was mainly focused on the ICT band. The interaction of the solvent with the molecule and its effect on photoluminescence was studied in terms of the Lippert–Mataga equation (eqn (1)).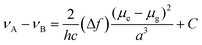 | (1) |
In this νA and νB are the wavenumber in cm−1 of the maximum absorption and emission, h is Planck's constant, c is the speed of light, Δf is the polarity of the solvent obtained from eqn (2); a corresponds to the Onsager radius of each molecule calculated by DFT, being 16.25 Å for the M1 and 16.27 Å for M2; and finally, Δμ is the dipole moment existing between the ground state and the excited state.
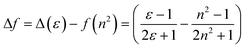 | (2) |
Eqn (2) shows the polarity of a solvent, where Δ(ε) is the difference of the dielectric constant in each solvent and n is the refractive index of each solvent.
Table 1 shows the photophysical properties of M1, when the molecule is in aprotic polar solvents, for example in acetonitrile the emission band shifts towards the red region (573 nm), and in apolar solvents as hexane, the emission shifts towards UV-vis (419 nm) with a difference of 154 nm. However, quenching of fluorescence in acetonitrile, acetone as well as in nonpolar solvents (ethyl ether, toluene, and hexane) was observed, meanwhile, in methylene chloride and THF the fluorescence intensity of M1 is higher (Fig. 2). THF is considered the best solvent to carry out the manufacture of nanoparticles by the reprecipitation and microemulsion method, as well as the AIEE due to the good photophysical properties that M1 presents when it is dissolved in this.
| Solvent | ε | n | Δƒ | λ a (nm) | νa (cm−1) | λ e (nm) | νe (cm−1) | Δν (cm−1) |
|---|---|---|---|---|---|---|---|---|
| a ε is the dielectric constant, n is the refractive index of each one of the solvents, λa is the maximum absorption, νa is the maximum absorption in cm−1, λe is the maximum emission, νe is the maximum emission in cm−1, Δν is the difference between maximum emission and absorption. | ||||||||
| Acetonitrile | 37 | 1.346 | 0.305 | 368 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 173 173 |
573 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 452 452 |
9721 |
| Methanol | 33 | 1.3288 | 0.308 | 378 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 455 455 |
526 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 011 011 |
7443 |
| Ethanol | 24 | 1.3611 | 0.289 | 368 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 173 173 |
530 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 867 867 |
8305 |
| Acetone | 21 | 1.3588 | 0.285 | 368 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 173 173 |
568 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 605 605 |
9568 |
| 2-Propanol | 18 | 1.3756 | 0.277 | 376 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 595 595 |
530 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 867 867 |
7727 |
| Methylene chloride | 9.1 | 1.424 | 0.223 | 376 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 595 595 |
559 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 889 889 |
8706 |
| THF | 7.5 | 1.405 | 0.21 | 368 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 173 173 |
506 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 762 762 |
7411 |
| Ethyl acetate | 6 | 1.3723 | 0.201 | 370 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027 027 |
537 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 621 621 |
8405 |
| Chloroform | 4.8 | 1.4459 | 0.148 | 382 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 178 178 |
529 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 903 903 |
7274 |
| Chlorobenzene | 4.7 | 1.5248 | 0.12 | 382 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 178 178 |
502 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 920 920 |
6257 |
| Ethyl ether | 4.3 | 1.3526 | 0.165 | 374 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 737 737 |
504 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 999 999 |
6738 |
| Toluene | 2.4 | 1.4961 | 0.013 | 382 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 178 178 |
476 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 361 361 |
8816 |
| 1,4-Dioxane | 2.25 | 1.4224 | 0.021 | 372 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 881 881 |
481 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 211 211 |
9670 |
| Hexane | 2 | 1.38 | 0.0012 | 374 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 737 737 |
419 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 993 993 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 744 744 |
 | ||
| Fig. 2 (A) Normalized UV-vis absorption spectrum of the M1 solvatochromism study. (B) Normalized emission spectrum of the M1 in the solvatochromism study. | ||
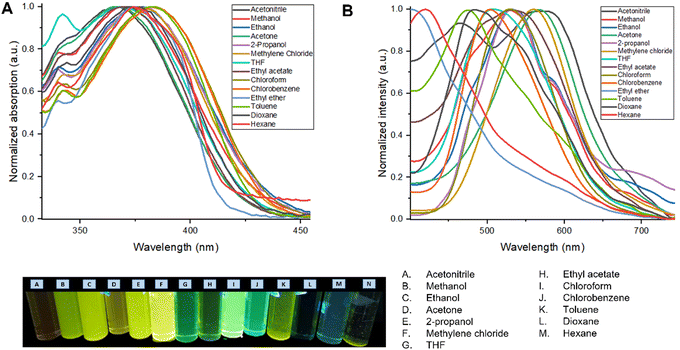
Table 2 shows the photophysical properties of M2. It is observed that in aprotic polar solvents the emission band shifts towards the red region (574 nm, in acetonitrile), and in less polar solvents as toluene the emission shifts towards UV-vis (443 nm) with a difference of 131 nm. In solvents with higher polarity such as acetonitrile, methanol, ethanol, and isopropanol, quenching of fluorescence was observed. This effect is also observed in nonpolar solvents such as toluene, dioxane, and hexane, while the fluorescence intensity is higher when M2 is dissolved in methylene chloride and THF (Fig. 3).
| Solvent | ε | n | Δƒ | λ a (nm) | ν a (cm−1) | λ e (nm) | ν e (cm−1) | Δν (cm−1) |
|---|---|---|---|---|---|---|---|---|
| a ε is the dielectric constant, n is the refractive index of each one of the solvents, λa is the maximum absorption, νa is the maximum absorption in cm−1, λe is the maximum emission, νe is the maximum emission in cm−1, Δν is the difference between maximum emission and absorption. | ||||||||
| Acetonitrile | 37 | 1.346 | 0.305 | 368 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 173 173 |
574 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 421 421 |
9752 |
| Methanol | 33 | 1.3288 | 0.308 | 374 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 737 737 |
461 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 691 691 |
5046 |
| Ethanol | 24 | 1.3611 | 0.289 | 382 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 178 178 |
476 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 008 008 |
5169 |
| Acetone | 21 | 1.3588 | 0.285 | 370 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027 027 |
568 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 605 605 |
9421 |
| 2-propanol | 18 | 1.3756 | 0.277 | 378 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 455 455 |
568 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 605 605 |
8849 |
| Methylene chloride | 9.1 | 1.424 | 0.223 | 378 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 455 455 |
561 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128 128 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 326 326 |
| THF | 7.5 | 1.405 | 0.21 | 342 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 239 239 |
498 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080 080 |
9159 |
| Ethyl acetate | 6 | 1.3723 | 0.201 | 370 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027 027 |
557 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 621 621 |
8405 |
| Chloroform | 4.8 | 1.4459 | 0.148 | 384 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 041 041 |
530 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 867 867 |
7173 |
| Chlorobenzene | 4.7 | 1.5248 | 0.12 | 384 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 041 041 |
529 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 903 903 |
7138 |
| Ethyl ether | 4.3 | 1.3526 | 0.165 | 374 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 737 737 |
506 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 762 762 |
6975 |
| Toluene | 2.4 | 1.4961 | 0.013 | 384 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 041 041 |
443 | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 573 573 |
3468 |
| 1,4-dioxane | 2.25 | 1.4224 | 0.021 | 382 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 178 178 |
476 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 008 008 |
5169 |
| Hexane | 2 | 1.38 | 0.0012 | 376 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 595 595 |
498 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080 080 |
6515 |
 | ||
| Fig. 3 (A) Normalized UV-vis absorption spectrum of the M2 solvatochromism study. (B) Normalized emission spectrum of the M2 in the solvatochromism study. | ||
According to the value of Δν presented in Tables 1 and 2 it is possible to identify three different regions for M1 and M2 depending on the dipole moment that they have: one for aprotic polar solvents like acetonitrile, acetone, methylene chloride, THF, and ethyl acetate; the protic polar for methanol, ethanol, 2-propanol, and finally, the non-polar one for chloroform, chlorobenzene, ethyl ether, toluene, dioxane, and hexane.
The bathochromic shift, is accompanied by an increase in the dipole moment, and for this reason there is a shift towards the infrared region. The last occurs in polar solvents because the excited states are more polar than the ground states. An increase in the polarity of the solvent causes a dipole–dipole interaction to decrease the energy of the excited state more than that of the ground state, causing radiation of lower energy to be absorbed.
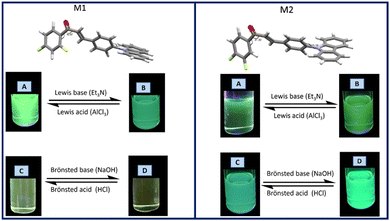
On the other hand, since acidity could significantly influence the photophysical properties of a fluorophore in solution, the effect of acidochromism on the solution absorption spectra of M1 and M2 were studied using a Lewis acid and base (AlCl3 and Et3N), as well as a Brønsted acid and base (HCl y NaOH) Fig. 4.
In the M1 titration using AlCl3 (Fig. S1, ESI†) an isosbestic point is observed at 316 nm indicating an equilibrium between two species, whereas the acid concentration increases, the π → π* transition band is more intense, while the intensity of the band attributed to ICT decreases. On the other hand, when M1 was titrated using a Lewis base (Fig. S1, ESI†), a band intensity decreases both in the band attributed to the π → π* transition, as well as in the band attributed to the ICT of the molecule, with a decrease in fluorescence intensity. The titration of M1 with a Brønsted acid (HCl) does not lead to any significant change in the absorption bands, while when using NaOH the band corresponding to ICT tends to disappear.
In the titration of M2 with AlCl3 a decrease in the intensity of the bands for the electronic transitions π → π* and ICT was observed (Fig. S2, ESI†). The absorption intensity decreases as the concentration of the acid increases, causing quenching of the fluorescence. In the titration spectrum using triethyl amine (Et3N), a decrease in intensity of the ICT band is observed at 366 nm as the Et3N concentration increases, and at 290 nm an isosbestic point is observed, indicating an equilibrium between two species. When M2 is titrated with HCl, it is observed that there is a decrease in the intensity of the ICT band with increasing acid concentration, as well as a 6 nm hypsochromic shift of the π → π* transition band. The titration of M2 with NaOH (Fig. S2, ESI†) shows an isosbestic point at 290 nm as the NaOH concentration increases, with the disappearance of the ICT band.
Computational modelling
A computational analysis of the excited and basal state of the M1 and M2 molecules was carried out to understand their photophysical properties; the calculations were carried out using density functional theory (DFT) in the Gaussian 09 platform with the B3LYP functional50,51 and the base 6-31G(d).52–54 This theory level was chosen because they have been used to study the linear and non-linear optical properties of chalcones, where the experimental data and those obtained by theoretical calculations are agree.55–57 First, the geometry of the molecule was optimized and then the energies of the ground and excited states (S0 → S1) were obtained. Table 3 shows the HOMO and LUMO values for M1 and M2, comparing the energy of the optical band gap (Egop) obtained from the absorption spectrum of M1 and M2; 3.35 and 3.38 eV respectively, with the calculated band gap 3.23 eV for M1 and 3.27 eV for M2, we conclude that they agree. These values give us the zone of extraction of the molecules corresponding to the ultraviolet region.In Fig. 5, the electronic distribution of M1 and M2 is shown both in the ground state S0 and in the excited state S1. It is observed that in the ground state, in the HOMO orbital, the electron density is found mainly on the carbazole, which is rich in electrons and therefore in a donor fraction (D), while, in the excited state S1 in the orbital LUMO, the electron density mainly in the α,β-unsaturated ketone which functions as acceptor (A), this corroborates that the electronic distribution of M1 and M2 is donor–acceptor and that it favors an ICT.
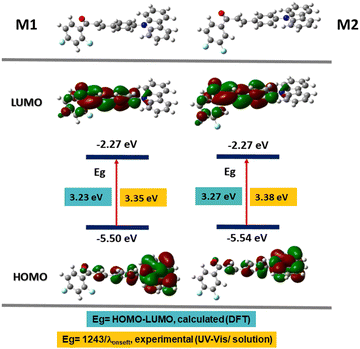 | ||
| Fig. 5 Energy values of the HOMO and LUMO orbitals calculated by DFT and electron density distribution. | ||
Aggregation induced emission enhancement
To evaluate the AIEE phenomenon in the photoluminescent properties of M1 and M2, fluorescence spectra were obtained in a THF/water mixture at different ratios v/v%. Fig. 6 shows the photoluminescence obtained for M1 in different THF/Water ratios in a range from 100/0 to 10/90. In the THF fraction was from 90 to 40%, there is a quenching of the fluorescence, on the other hand when the water fraction was from 70 to 90% the formation of aggregates is observed, and the fluorescence of the molecule becomes visible, registering an intensity I/I0 = 2.07 in the ratio 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80.
80.
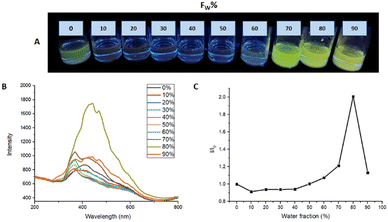 | ||
| Fig. 6 (A) AIEE M1 samples under 366 nm to different THF/water ratios. (B) AIEE test emission spectrum. (C) plot of water fraction vs. I/I0. | ||
Fig. 7 shows the emission spectra of M2 in different THF/water ratios. The analysis revealed that the THF/water ratio where M2 presented the highest photoluminescence intensity is 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70, reaching a ratio I/I0 = 1.526. The increment in photoluminescence is due to the formation of microaggregates characteristic of AIEE behavior. When the molecule is dissolved it can easily rotate around a bond and when it is excited the absorbed energy is dissipated by this rotation used as a non-radiative relaxation channel. On the other hand, when water is added to the solution, microaggregates are formed where the lattice energy and packing restrict the rotation of the molecule, and the relaxation channels are not efficient, so the energy absorbed by the molecule is emitted in the form of light.
70, reaching a ratio I/I0 = 1.526. The increment in photoluminescence is due to the formation of microaggregates characteristic of AIEE behavior. When the molecule is dissolved it can easily rotate around a bond and when it is excited the absorbed energy is dissipated by this rotation used as a non-radiative relaxation channel. On the other hand, when water is added to the solution, microaggregates are formed where the lattice energy and packing restrict the rotation of the molecule, and the relaxation channels are not efficient, so the energy absorbed by the molecule is emitted in the form of light.
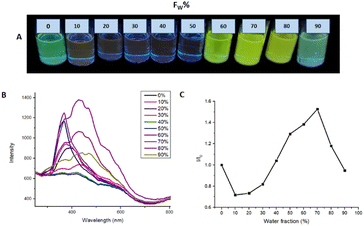 | ||
| Fig. 7 (A) AIEE M2 samples under 366 nm to different THF/water ratios. (B) AIEE test emission spectrum. (C) plot of water fraction vs. I/I0. | ||
Nanoparticles fabrication
To increase the biocompatibility of M1 and M2, the materials were coated with silica forming nanoparticles (NPs) by the microemulsion method. For which the methodology described in the ESI† was followed, as emulsifying agents Aerosol-OT and Triton X100 were used in different concentrations (0.11, 0.22, and 0.44 g).The optical properties of the NPs were evaluated by UV-vis and fluorescence spectroscopy. Fig. 8 shows the absorption and emission spectra of the nanoparticle emulsions obtained from M1.
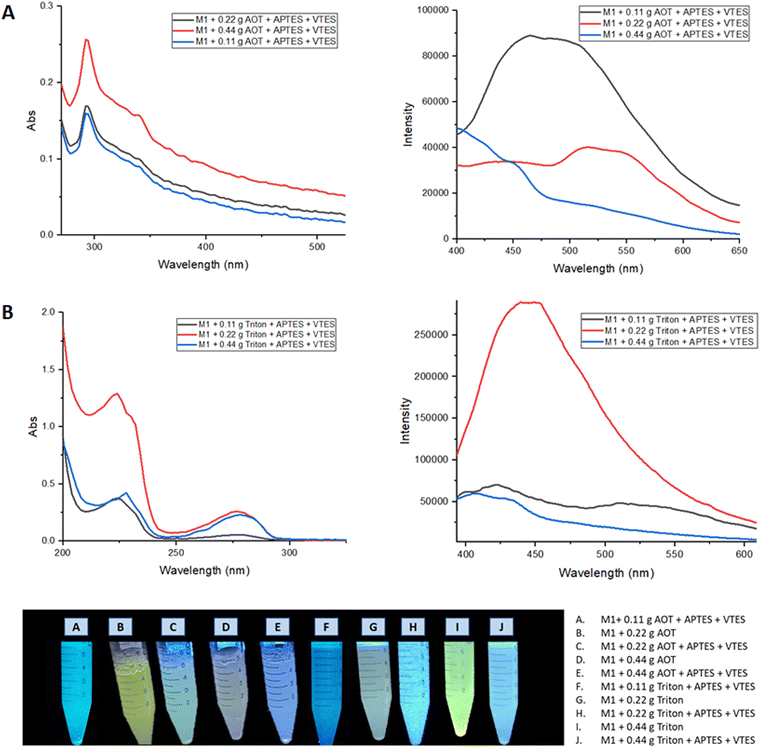 | ||
| Fig. 8 (A) Absorption and emission spectra of nanoemulsion of M1 NPs with Aerosol-OT. (B) Absorption and emission spectra of nanoemulsion of M1 NPs with Triton X-100. | ||
When nanoparticles are coated using APTES and VTES, the fluorescence intensity is higher when 0.22 g of Aerosol-OT is used as well as Triton. The highest quantum yield obtained for these NPs was when Triton was used as a surfactant, registering ΦNPs = 8.3% compared to that obtained in solid with Φsol = 23% a considerable decrease is observed, showing that it would not be a good staining agent.
Fig. 9 shows the absorption and emission spectra of the nanoemulsions of M2, the best optical properties are obtained when 0.22 g of surfactant was used; where the nanoparticles manufactured in Aerosol-OT present greater intensity in the fluorescence spectrum, observing a band at 520 nm. The highest quantum yield obtained for these nanoparticles was when Aerosol-OT was used as a surfactant agent, registering ΦNPs = 18.5% compared to that obtained in solid of Φsol = 90%, a considerable decrease is observed. When compared to the fluorescence quantum yields of the most common porphyrin-based photosensitizers,58–60 which are between 10 and 20%, it can be concluded that the M2 nanoparticles with 0.22 g of Aerosol-OT comply with the photophysical properties for cell markers. During the NPs fabrication process, the polycondensation and hydrolysis reactions are responsible for the fact that the chalcone molecules are covered in the generated polysiloxane network, and these organic molecules are physically embedded since they do not have free –OH or –NH2 groups that can participate in the growth process of siloxane chains. This ensures that the chemical structure of the chalcone is free and bioactive once the physical interactions with the polysiloxane (silica) matrix are broken, benefiting from its properties both as biomarkers and dynamic phototherapy.
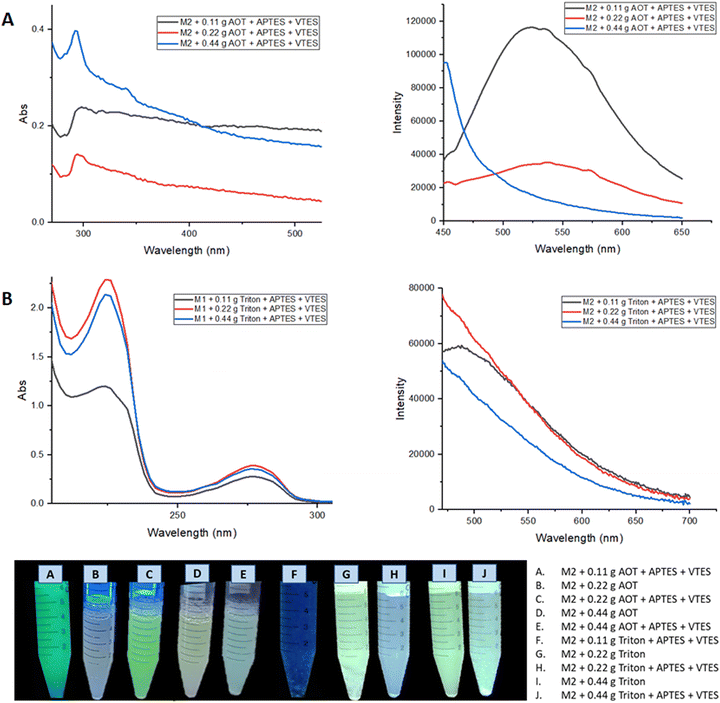 | ||
| Fig. 9 (A) Absorption and emission spectra of nanoemulsion of M2 NPs with Aerosol-OT. (B) Absorption and emission spectra of nanoemulsion of M2 NPs with Triton X-100. | ||
NPs morphology and size distribution
The morphology of the NPs fabricated by the microemulsion method of M1 and M2 + Aerosol OT at a concentration of 0.22 g were studied using transmission electron microscopy (TEM) using equipment FEI Company equipment (Titan 300 KV); the suspensions were placed on a Lacey Carbon Film 300 Mesh Copper grid, finally, they were left to dry for 2 hours.Fig. 10 shows the micrographs obtained where it can be seen that spherical NPs are obtained and that are not agglomerated. The microemulsion method allows obtained spherical nanoparticles where M1 and M2 are into the nucleus and coated with silica, which makes the NP highly dispersible in water and biocompatible with the human body, without the loss of optical properties of the chalcones under study.
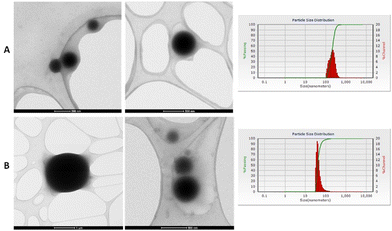 | ||
| Fig. 10 (A) NPs micrographs of M1 with 0.22 g of Aerosol-OT. (B) NP's micrographs of M2 with 0.22 g of Aerosol-OT. | ||
Dynamic light scattering (DLS) measurements of the NPs were performed to determine the particle size distribution. Measurements were made at room temperature with a laser incidence angle of 90°. Table 4 shows the average size obtained for NPs of chalcones, which were prepared using different concentrations of surfactant. When 0.11 g of surfactant was used, the NPs size obtained for M1 are 102–132 nm, for M2, particles that were out of the nanometric scale were obtained, so the amount of surfactant used is not enough. This is attributed to the fact that the surfactant agent is not enough to maintain the microemulsion and the NP's agglomerate, increasing the size which is 132 nm for the NPs of M1 and 588 nm for the NPs of M2. On the other hand, when 0.44 g of surfactant was used, an average size between 60 and 90 nm was obtained, however, a large amount of surfactant caused the quenching of the chalcone fluorescence. The nanoparticles with the best optical properties (M1 + 0.22 g Triton and M2 + 0.22 g Aerosol-OT) presented mean diameters of 5.86 and 42.8 nm, respectively.
| Sample | Size of M1 (nm) | Size of M2 (nm) |
|---|---|---|
| 0.11 g Triton + APTES + VTES | 132.5 | 588 |
| 0.11 g Aerosol-OT + APTES + VTES | 102.2 | 409 |
| 0.22 g Triton + APTES + VTES | 5.86 | 6.96 |
| 0.22 g Aerosol-OT + APTES + VTES | 403 | 42.8 |
| 0.44 g Triton + APTES + VTES | 93.7 | 1.34 |
| 0.44 g Aerosol-OT + APTES + VTES | 60.5 | 87.8 |
The NPs show a variable average diameter, this size is suitable enough to undergo cellular internalization, and they can be transported at the cell membrane level as well as at the nucleus level. The size of the nanoparticle determines cytotoxic effects in different cell models, nanoparticles smaller than 10 nm can cause a variation in cell metabolism and/or cell death due to rapid internalization and subsequent rapid degradation, affecting fundamental biological processes. For biomarker purposes, it is necessary that the particle size can interact with the membrane proteins to be able to detect them after the biochemical assembly that occurs; while for dynamic phototherapy purposes, it is also, desired that the NPs be fixed in the membranes so that the emitted light quickly reaches the cell nucleus and stops the growth of mutated cells such as cancer cells.
Cytotoxicity assays of NPs
Hemocompatibility was determined through the hemolysis test of the NPs of M1 (0.22 g Triton + APTES + VTES) and M2 (0.22 g Aerosol-OT + APTES + VTES). An analysis of variance (ANVA) was carried out and through a comparison of means test by Tukey, it was determined that all the treatments are statistically different from each other (Table 5). According to the ASTM F 756-08 standard, if a material has a hemolytic capacity greater than 5%, it is considered a hemolytic or non-hemocompatible material, causing lysis of red blood cells and therefore cytotoxic.61 Analyzing the results obtained, we can say that the only non-hemolytic NPs is the M2 (0.22 g AOT + APTES + VTES). This is associated with the larger size of the NPs, which prevents rapid internalization and does not promote severe degradation effects that tend to lyse the erythrocyte membrane. The effect of the destruction of the erythrocyte membrane is easily observed by decreasing the size of the NPs, which is associated with rapid internalization and destructive association with membrane proteins, producing cell lysis and death.| Sample | %Hemolysis | |
|---|---|---|
| a Capital letters different from each other indicate a significant difference (with a significance level of p = 0.05). | ||
| M1 + 0.22 g Triton | 26.33 ± 1.6 | A |
| M1 + 0.22 g Triton + APTES + VTES | 20.68 ± 0.8 | B |
| M2 + 0.22 g Aerosol-OT | 7.69 ± 1.3 | C |
| M2 + 0.22 g Aerosol-OT + APTES+ VTES | 1.87 ± 1.4 | D |
To evaluate the effect of the NPs of M1 and M2 on the metabolism of monocytes, fibroblasts, colon cancer cells, and breast cancer cells, an MTT assay was performed. Fig. 11 shows the results obtained from the metabolic activity of monocytes, fibroblasts, colon cancer cells, and mama cancer cells in the NPs presence of the M1+ 0.22 g Triton, M1 + 0.22 g Triton + APTES + VTES, M2 + 0.22 g Aerosol-OT, M2 + 0.22 g Aerosol-OT + APTES + VTES; the samples were evaluated at 24 and 72 hours of incubation. In the samples incubated for 24 hours with monocytes, metabolic activity percentages of 18.05%, 36.8%, 70.2%, and 57.6% were obtained, respectively, indicating that the cells presented a decrease in their metabolic activity. While at 72 hours the metabolic activity was 84.6%, 180.8%, 120.5%, and 272.8%; This could be due that after the first 24 hours, the monocytes underwent a process of adaptation to the NPs, since cell viability decreased in all cases; however, after 72 hours, viability increased in all cases even more than the control sample, indicating that the composition of the nanoparticles with chalcones increases the metabolism of monocytes at 72 h, representing a biocompatible character with these cells of the immune system.
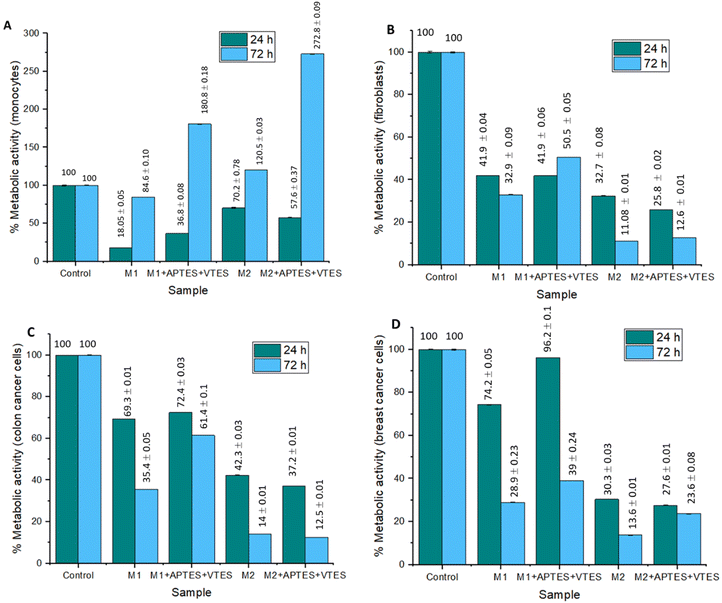 | ||
| Fig. 11 Graphs of the effect of M1 and M2 NPs on metabolic activity after 24 and 72 hours of incubation in (A) monocytes, (B) fibroblasts, (C) colon cancer cells, and (D) Breast cancer cells. | ||
The metabolic activity of the fibroblasts is less than 49.1% in all cases, so it can be determined that the NPs of M1 and M2 are cytotoxic. In this case, with this type of cells that build the extracellular matrix, the nanoparticle size and composition decrease the activity of their mitochondrial dehydrogenases, decreasing the reduction of MTT to insoluble formazan; this could be associated with the fact that M1 and M2 NPs associate with membrane proteins, preventing the diffusion of gases and nutrients, effects that decrease the metabolic activity of these cells. In colon and breast cancer cells, the NPs of M2 turned out to be cytotoxic, having metabolic activities lower than 37.2%, while the NPs of M1 presented percentages of up to 96.2%, demonstrating no cytotoxicity to this type of cells. These results demonstrate that the size of the NPs is important in the metabolic activity of cancer cells, indicating that the larger the nanoparticles, the metabolic activity decreases in cancer cells. This is associated with the effects of interaction with membrane proteins during the internalization process, limiting the respiratory metabolism of cells due to the effects of diffusion of gases, nutrients, metabolites, or waste products. In the case of M1 nanoparticles with a smaller molecular size, there is no loss of diffusion of biochemical components, so the metabolic activity of the cells is not altered.
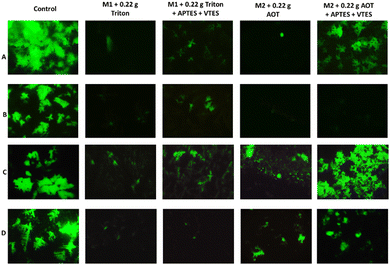
A cell proliferation assay was performed to determine whether colon and breast cancer cells died when are in contact with the NPs of M1 and M2. For this, cell staining of living cells with the live/dead fluorescent agent was performed. Fig. 12 A and C show the live colon cancer cell and breast cancer cell populations stained with calcein in the presence of the suspensions M1+ 0.22 g Triton (1), M1 + 0.22 g Triton + APTES + VTES (2), M2 + 0.22 g Aerosol-OT (3) and M2 + 0.22 g Aerosol-OT + APTES + VTES (4). In both cases (A and C) trials 1, 2, and 3 show fewer cell populations than the control since the nanomaterials are shown to be cytotoxic, while the NPs of trial 4 maintain cell populations like the control. For both M1 and M2, it can be observed that there is less cell death when the nanoparticles are coated with silica. These results are by the cellular metabolism assay; since if there is a decrease in metabolism, cell proliferation must also decrease. In this test, highly fluorescent calcein is produced by the action of esterases of cells with active metabolism (alive), and the higher the content of cell populations, the greater the proliferation and production of calcein. The short-range interactions that chalcones produce with the polysiloxane matrix (silica) are associated with a decrease in the inhibitory biological response to proliferation.
Fig. 12B and D show the colon cancer and breast cancer cell populations in contact with the NPs based on: M1+ 0.22 g Triton, M1 + 0.22 g Triton + APTES + VTES, M2 + 0.22 g Aerosol-OT and M2 + 0.22 g Aerosol-OT + APTES + VTES irradiated at 366 nm for 1 hour. The irradiation of the control sample (B and D) does not show cell death, on the other hand when colon cancer cells were irradiated for one hour at 366 nm in the presence of NPs of both M1 + 0.22 g Triton + APTES + VTES and M2 + 0.22 g Aerosol-OT + APTES + VTES, cell death was observed, obtaining better results of inhibition of cancer cell proliferation when using M2-based NPs, which demonstrates that for anticancer phototherapy strategies with colon cells, cell death is linked by interaction phenomena with membrane proteins during internalization, preventing fundamental biological functions such as respiration, preventing their proliferation. When breast cancer cells were irradiated for one hour at 366 nm, live cell populations decreased considerably compared to control, with better results being obtained when NPs based on M1 + 0.22 g Triton + APTES + VTES were used. In this case, the inhibition of cell proliferation occurs with smaller nanoparticles, which is associated with degradation products that are generated at the cytosol level and that can reach the cell nucleus to inactivate the cell proliferation mechanisms; Interestingly, the results of this work demonstrate two interaction effects of the nanoparticle structure associated with chalcones for the inhibition of cell proliferation, representing potential biomedical application in anticancer dynamic phototherapy. The chalcone structure determines the size of the silica nanoparticle and its biological response to anticancer phototherapy strategies. In the case of chalcone M1 NPs of smaller molecular size are obtained, probably due to the repulsive effect of the two nearby fluorines, proving to decrease the growth and proliferation of breast cancer cells. In the case of chalcone M2, it can be seen that the chemical environment of the aromatic rings favors greater growth of the polysiloxane (silica) matrix, generating a larger particle size and evidencing an inhibition effect of proliferation by effects at the membrane protein level with colon cancer cells.
It can be concluded that there is greater cell death when the cells come into contact with the material and it is additionally irradiated, which is associated with the fact that chalcones emit UV light that reaches the cell nucleus and slows down the growth of cancer cells, which is fundamental in anticancer photodynamic therapies. By modifying the chemical structure of the chalcone, higher energy can be generated, which has a greater impact on the inhibition of cancer cell proliferation.
Cellular staining tests using M1 and M2 NPs
M1 + 0.22 g Triton + APTES + VTES and M2 + 0.22 g Aerosol-OT + APTES + VTES nanoparticles were internalized into colon cancer cells to test whether they can function as a staining agent for colon contrast images. To identify the part of the cell where the nanoparticles were, rhodamine was used as a cell staining agent, this stains the cell membrane red due to the affinity of the cell wall for the fluorophore.62The membranes of the cells stained by rhodamine in red can be identified both in the images obtained from M1 and M2 (Fig. 13), the internal part of the cell is stained in yellow color, indicating that the nanoparticles that they emit yellow fluorescence when excited at 360 nm and may be the cause of cellular staining inside the cell. These results demonstrate that NPs stained with rhodamine can be used as biomarkers of cancer cells at the cellular and nuclear membrane level, since when interacting with membrane proteins, physicochemical associations are carried out between NPs that contain chalcones and membrane proteins, producing a variation in the native photoluminescence of the chalcone, and allowing the identification of the nanoparticle and its cellular fate. Cell biomarking effect is attributed to the fact that the size of the fluorescent NPs is adequate for internalization to the nucleus. In this sense, this system under study represents a potential biomarking agent for the identification of cancer cells.
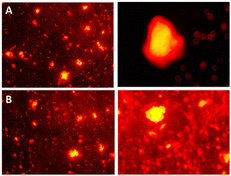 | ||
| Fig. 13 Cell staining of colon cancer cells viewed under 360 nm using: (A) M1 + 0.22 g Triton + APTES + VTES. (B) M2 + 0.22 g Aerosol-OT + APTES + VTES. | ||
Conclusions
In conclusion, the synthesis of two new chalcones M1 and M2 with D–A electronic structure was carried out, these molecules are fluorescent both in solid state and in solution. Through AIEE experiments using THF:H2O solvent mixture, it was observed that by increasing the volume of water the chalcones presented higher fluorescence intensity for M1 and M2. With this information it was possible to manufacture biocompatible NPs of M1 and M2 using the microemulsion method under a structure M+ surfactant + APTES + VTES, obtaining spherical nanoparticles with a size of 5.86 nm for M1 and 42.8 nm for M2. The quantum yields of NPs for M1 are ΦNPs = 8.3 and 18.5% for M2.The molecule with the best optical properties is M2, using 0.22 g of Aerosol-OT for the fabrication of coated nanoparticles, with a quantum yield ΦNPs = 18.5%; these NPs have better inhibition on the metabolic activity of cancer cells, preventing their proliferation, and these effects are increased when irradiated by UV light of 366 nm, besides they do not present hemolytic activity and has percentages of metabolic activity above 70% for healthy cells; they are highly capable of staining colon cancer cells in the presence of rhodamine-b, generating stained nanoparticles that can be internalized within colon cancer cells that can be identified acting at the cell membrane and nucleus level. With this in mind, these innovative NPs with chalcones could be successfully employed as cancer cell biomarkers and in dynamic anticancer phototherapies.
Author contributions
Estrada-Flores, S. L., Garcias-Morales, C., and Martínez-Mora, E. I., contributed to the design, synthesis, purification, and optical characterization of the chalcones, as well as to nanoparticle fabrication. García Merinos, J. P., contributed to the structural characterization by NMR and FT-IR. Pérez Berumen, C. M., De León-Santillán, A. and Rodríguez, M., contributed to the characterization of morphology and nanoparticles size, as well as the photophysical properties. Estrada-Flores, S. L. and Claudio-Rizo, J. A., carried out the cytotoxicity and cell staining studies. All authors participated in the discussion and writing of the article.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to the Consejo Nacional de Ciencia y Tecnología (CONACyT) for the financial support that has been provided with Scholarship number 787927 for the master's degree training of Q. Susana Lucía Estrada Flores in the master's program in Chemical Science and Technology at the Universidad Autónoma de Coahuila. The authors thank also CONACyT for supporting the FORDECYT-PRONACES/6660/2020 project.References
- Y. Cheng, Y. Luo, R. Peng, Q. Cao, Q. Wu and Q. Cui, Colloids Surf., A, 2021, 613, 126096 CrossRef CAS.
- M. Korzec, S. Kotowicz, R. Gawecki, K. Malarz, A. Mrozek-Wilczkiewicz, M. Siwy, E. Schab-Balcerzak, J. Grzelak and S. Maćkowski, Dyes Pigm., 2021, 193, 109508 CrossRef CAS.
- J. Maillard, K. Klehs, C. Rumble, E. Vauthey, M. Heilemann and A. Fürstenberg, Chem. Sci., 2021, 12, 1352–1362 RSC.
- F. Shahrokhi and Y. Zhao, Org. Lett., 2019, 21, 9306–9310 CrossRef CAS PubMed.
- Q. Shen, S. Wang, N.-D. Yang, C. Zhang, Q. Wu and C. Yu, J. Lumin., 2020, 225, 117338 CrossRef CAS.
- C. Teng, S. Zhang, Y. Tian, Q. Cheng, H. Dang, D. Yin and L. Yan, Nanomedicine, 2022, 44, 102574 CrossRef CAS PubMed.
- M. N. El-Nahass, T. A. Fayed, S. A. Elazim, M. M. El-Gamil, D. F. Draz and F. Hassan, J. Mol. Struct., 2021, 1240, 130581 CrossRef CAS.
- N. Luo, J. Liu, S. Wang and C. Wang, Org. Biomol. Chem., 2020, 18, 9210–9215 RSC.
- P. S. Patil, N. B. Gummagol, A. Ekbote, Q. A. Wong, C. K. Quah, M. Shkir, S. R. Maidur and S. V. Rao, J. Mol. Struct., 2020, 1219, 128523 CrossRef CAS.
- M. Shkir, A. Irfan, S. AlFaify, P. Shankaragouda Patil and A. G. Al-Sehemi, Optik, 2019, 199, 163354 CrossRef CAS.
- Q. A. Wong, C. K. Quah, X. A. Wong, S. R. Maidur, H. C. Kwong, Y.-F. Win, P. S. Patil and N. B. Gummagol, J. Mol. Struct., 2022, 1267, 133584 CrossRef CAS.
- T. Andrade-Filho, T. Silva, E. Belo, A. Raiol, R. V. S. de Oliveira, P. S. B. Marinho, H. R. Bitencourt, A. M. R. Marinho, A. R. da Cunha and R. Gester, J. Mol. Struct., 2021, 1246, 131182 CrossRef CAS.
- A. H. Anizaim, M. F. Zaini, I. A. Razak and S. Arshad, J. Solid State Chem., 2021, 304, 122551 CrossRef CAS.
- D. Haleshappa, R. Bairy, A. Jayarama, C. K. Quah, H. C. Kwong and P. S. Patil, J. Mol. Struct., 2021, 1232, 130053 CrossRef.
- S. Omar, M. Shkir, M. Ajmal Khan, Z. Ahmad and S. AlFaify, Optik, 2020, 204, 164172 CrossRef CAS.
- Z. Yang, Y. Guo, S.-L. Ai, S.-X. Wang, J.-Z. Zhang, Y.-X. Zhang, Q.-C. Zou and H.-X. Wang, Mater. Chem. Front., 2019, 3, 571–578 RSC.
- M. Khalid, I. Shafiq, Umm-e-Hani, K. Mahmood, R. Hussain, M. F. ur Rehman, M. A. Assiri, M. Imran and M. S. Akram, Sci. Rep., 2023, 13, 1395 CrossRef CAS PubMed.
- T. Saleem, S. Khan, M. Yaqub, M. Khalid, M. Islam, M. Yousaf ur Rehman, M. Rashid, I. Shafiq, A. A. C. Braga, A. Syed, A. H. Bahkali, J. F. Trant and Z. Shafiq, New J. Chem., 2022, 46, 18233–18243 RSC.
- M. Khalid, M. U. Khan, I. Shafiq, R. Hussain, K. Mahmood, A. Hussain, R. Jawaria, A. Hussain, M. Imran, M. A. Assiri, A. Ali, M. F. ur Rehman, K. Sun and Y. Li, Arabian J. Chem., 2021, 14, 103295 CrossRef CAS.
- N. Altinolcek, A. Battal, C. N. Vardalli, M. Tavasli, H. A. Yu, W. J. Peveler and P. J. Skabara, J. Mol. Struct., 2021, 1239, 130494 CrossRef CAS.
- Z. Heydari and P. Rashidi-Ranjbar, J. Photochem. Photobiol., A, 2019, 377, 8–13 CrossRef CAS.
- P. S. Singh, P. M. Badani and R. M. Kamble, J. Photochem. Photobiol., A, 2021, 419, 113457 CrossRef CAS.
- M. Pervez, A. K. Pearce, J. T. Husband, L. Male, M. Torrent-Sucarrat and R. K. O’Reilly, Chem. – Eur. J., 2022, 28, e202201877 CrossRef CAS PubMed.
- Y.-W. Tu, C.-C. Wang, A. S. Godana and C.-Y. Yu, Eur. Polym. J., 2019, 112, 283–290 CrossRef CAS.
- L. Yan, Y. Zhang, B. Xu and W. Tian, Nanoscale, 2016, 8, 2471–2487 RSC.
- J. L. Belmonte-Vázquez, E. A. Hernández-Morales, F. Hernández, M. C. García-González, L. D. Miranda, R. Crespo-Otero and B. Rodríguez-Molina, Eur. J. Org. Chem., 2022, e202200372 Search PubMed.
- M. Kazmi, I. Khan, A. Khan, S. A. Halim, A. Saeed, S. Mehsud, A. Al-Harrasi and A. Ibrar, Bioorg. Med. Chem., 2019, 27, 115123 CrossRef CAS PubMed.
- L.-A. Lozano-Hernández, J.-L. Maldonado, C. Garcias-Morales, A. Espinosa Roa, O. Barbosa-García, M. Rodríguez and E. Pérez-Gutiérrez, Molecules, 2018, 23(2), 280 CrossRef PubMed.
- H. Chen, S. Fang, L. Wang, X. Liu, J. Yan, N. Zhang and K. Zheng, J. Mol. Liq., 2021, 321, 114913 CrossRef CAS.
- D. B. Christopher Leslee and S. Karuppannan, J. Photochem. Photobiol., A, 2022, 429, 113937 CrossRef CAS.
- M. F. Zaini, I. A. Razak, W. M. Khairul and S. Arshad, Acta Crystallogr., Sect. E: Crystallogr. Commun., 2020, 76, 387–391 CrossRef CAS PubMed.
- Z. Liu, Q. Wang, W. Qiu, Y. Lyu, Z. Zhu, X. Zhao and W. Zhu, Chem. Sci., 2022, 13, 3599–3608 RSC.
- Z. Chen, H. Qin, Y. Yin, D. Deng, S.-Y. Qin, N. Li, K. Wang and Y. Sun, Chem. – Eur. J., 2023, 29, e202203797 CAS.
- W. Du, X. Lu, T. Yuan, Z. Sun, X. Li, S. Li, Q. Zhang, X. Tian, D. Li and Y. Tian, Analyst, 2022, 147, 66–71 RSC.
- T. C. Pham, D. J. Lee, D. H. Kim, J. Yoon, T. D. Lam, H. M. Kim and S. Lee, Chem. Commun., 2023, 59, 4503–4506 RSC.
- T. Mallick, A. Karmakar, J. Bag, S. Sahu, M. Mishra and N. A. Begum, Dyes Pigm., 2020, 173, 107994 CrossRef CAS.
- H. Shen, Y. Li, X. Kang, J. Wu, R. Chen, X. Wei, X. Zhang, J. Qi and Q. Liu, ACS Appl. Nano Mater., 2023, 6, 6056–6065 CrossRef CAS.
- L. Aparicio-Ixta, G. Ramos-Ortiz, J. L. Pichardo-Molina, J. L. Maldonado, M. Rodríguez, V. M. Tellez-Lopez, D. Martinez-Fong, M. G. Zolotukhin, S. Fomine, M. A. Meneses-Nava and O. Barbosa-García, Nanoscale, 2012, 4, 7751–7759 RSC.
- B. Chen, G. Feng, B. He, C. Goh, S. Xu, G. Ramos-Ortiz, L. Aparicio-Ixta, J. Zhou, L. Ng, Z. Zhao, B. Liu and B. Z. Tang, Small, 2016, 12, 782–792 CrossRef CAS PubMed.
- X. Lou, Z. Zhao and B. Z. Tang, Small, 2016, 12, 6429 CrossRef CAS.
- Z. Zhao, B. Chen, J. Geng, Z. Chang, L. Aparicio-Ixta, H. Nie, C. C. Goh, L. G. Ng, A. Qin, G. Ramos-Ortiz, B. Liu and B. Z. Tang, Part. Part. Syst. Charact., 2014, 31, 481–491 CrossRef CAS.
- Z. Zhao, B. Chen, J. Geng, Z. Chang, L. Aparicio-Ixta, H. Nie, C. C. Goh, L. G. Ng, A. Qin, G. Ramos-Ortiz, B. Liu and B. Z. Tang, Part. Part. Syst. Charact., 2014, 31, 409 CrossRef.
- Z. Huang, D. Li, F. Guo, T. Xian, H.-S. Hu, J. Xu, Y.-F. Luo, Z.-Z. Chen, B.-C. Wang and Y.-M. Zhang, Colloids Surf., B, 2022, 220, 112956 CrossRef CAS.
- T. Jin, D. Cheng, G. Jiang, W. Xing, P. Liu, B. Wang, W. Zhu, H. Sun, Z. Sun, Y. Xu and X. Qian, Bioact. Mater., 2022, 14, 42–51 CrossRef CAS PubMed.
- S. K. Rout, V. Priya, A. Setia, A. K. Mehata, S. Mohan, M. Albratty, A. Najmi, A. M. Meraya, H. A. Makeen, M. M. Tambuwala and M. S. Muthu, Biomed. Pharmacother., 2022, 153, 113451 CrossRef CAS PubMed.
- N. Singh, J. Kim, J. Kim, K. Lee, Z. Zunbul, I. Lee, E. Kim, S.-G. Chi and J. S. Kim, Bioact. Mater., 2023, 21, 358–380 CrossRef PubMed.
- W. Sun, X. Wang, Z. Cheng, X. Wang, N. Fan, P. Dong, M. Qiong Tong, Y. Liu and W. Sun, Biomed. Pharmacother., 2023, 158, 114071 CrossRef CAS PubMed.
- S. Wang, X. Liu, M. Yang, L. Ouyang, J. Ding, S. Wang and W. Zhou, Asian J. Pharm. Sci., 2022, 17, 557–570 CrossRef PubMed.
- Q. Zhang, X. Wang, G. Kuang and Y. Zhao, Bioact. Mater., 2023, 24, 185–196 CrossRef CAS PubMed.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS PubMed.
- W. J. Hehre, R. Ditchfield and J. A. Pople, J. Chem. Phys., 1972, 56, 2257–2261 CrossRef CAS.
- P. C. Hariharan and J. A. Pople, Theor. Chim. Acta, 1973, 28, 213–222 CrossRef CAS.
- M. M. Francl, W. J. Pietro, W. J. Hehre, J. S. Binkley, M. S. Gordon, D. J. DeFrees and J. A. Pople, J. Chem. Phys., 1982, 77, 3654–3665 CrossRef CAS.
- C. Garcias-Morales, J. Espinosa Maldonado, A. Ariza Castolo, C. M. Perez Berumen, M. A. Lobato, M. A. Rodriguez and A. Espinoza Roa, J. Mol. Struct., 2021, 1228, 129444 CrossRef CAS.
- B. Kosar and C. Albayrak, Spectrochim. Acta, Part A, 2011, 78, 160–167 CrossRef PubMed.
- E. Mathew, V. V. Salian, I. Hubert Joe and B. Narayana, Opt. Laser Technol., 2019, 120, 105697 CrossRef CAS.
- D. A. Caminos and E. N. Durantini, J. Porphyrins phthalocyanines, 2005, 09, 334–342 CrossRef CAS.
- D. D. Ferreyra, M. B. Spesia, M. E. Milanesio and E. N. Durantini, J. Photochem. Photobiol., A, 2014, 282, 16–24 CrossRef CAS.
- S. J. Mora, M. E. Milanesio and E. N. Durantini, J. Photochem. Photobiol., A, 2013, 270, 75–84 CrossRef CAS.
- S. Henkelman, G. Rakhorst, J. Blanton and W. van Oeveren, Mater. Sci. Eng.: C, 2009, 29, 1650–1654 CrossRef CAS.
- A. T. Jacobs, D. Martinez Castaneda-Cruz, M. M. Rose and L. Connelly, Biochem. Pharmacol., 2022, 204, 115209 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma00108c |
| This journal is © The Royal Society of Chemistry 2023 |