Understanding spatiotemporal mechanical behavior, viscoelasticity, and functions of stem cell-derived cardiomyocytes†
Lihua
Lou
 a,
Alberto
Sesena Rubfiaro
a,
Alberto
Sesena Rubfiaro
 b,
Jin
He
b,
Jin
He
 b and
Arvind
Agarwal
b and
Arvind
Agarwal
 *a
*a
aMechanical and Materials Engineering, School of Biomedical, Materials and Mechanical Engineering (SBMME), College of Engineering and Computing, Florida International University, 10555 West Flagler Street, Miami, FL 33174, USA. E-mail: agarwala@fiu.edu
bDepartment of Physics, Florida International University, Miami, FL 33174, USA
First published on 5th June 2023
Abstract
Understanding myocytes' spatiotemporal mechanical behavior and viscoelasticity is a long-standing challenge as it plays a critical role in regulating structural and functional homeostasis. To probe the time-dependent viscoelastic behaviors of cardiomyocytes with cross-linked polymer networks, we measure stem cell-derived cardiomyocyte's (hiPSC-CM) deformation, adhesion, and contractility using atomic force microscopy (AFM) nanoindentation, fluidic micropipette, and digital image correlation (DIC). Our results show a cytoplasm load of 7–14 nN, a de-adhesion force of 0.1–1 nN, and an adhesion force between two hiPSC-CMs of 50–100 nN with an interface energy of 0.45 pJ. Based on the load–displacement curve, we model its dynamic viscoelasticity and discover its intimate associations with physiological properties. Cell detaching and contractile modeling demonstrate cell–cell adhesion and beating related strains manifesting viscoelastic behavior, highlighting viscoelasticity plays the primary role in governing hiPSC-CM spatiotemporal mechanics and functions. Overall, this study provides valuable information about the mechanical properties, adhesion behaviors, and viscoelasticity of single hiPSC-CM, shedding light on mechanical–structure relationships and their dynamic responses to mechanical stimuli and spontaneous contraction.
Introduction
The capability to quantify a single cardiomyocyte's mechanical behavior1,2 is critical for elucidating cardiac biological and pharmaceutical questions.3,4 It promises to decipher myocyte mechanotransduction,5 physiological development,6 and pathological mechanism.3In vitro mechanical test platform allows measuring the influences of bio-environment factors, cardiovascular drugs,7 substrate topology/stiffness,8–11 aging, and pH on the viscoelasticity, adhesion,12 and beating mechanics13,14 of hiPSC-CM. Compared to 2D or 3D cardiac microbundles or patches,15–18 this platform is simple and has data precision due to controllable cell quality and technical maturity.Despite the significant advances in single-cell mechanics measuring tools, the studies are limited to its force and strain responses,1 deformation–disease relationship,7 substrate effects,8,9 differentiation-mechanics association,19 mutation-adhesion mechanism,20 and disease detection.21 Moreover, most of the studies focused on semi-quantitative or quantitative tools6,22,23 in detecting cell mechanical properties and intercellular adhesions or the functions of genes,24 proteins/receptors,25 and growth factor signaling26 from the biological and chemical perspectives. However, the studies of single cardiomyocytes' mechanics and viscoelasticity are limited. Viscoelasticity indicates instantaneous responsiveness with velocity dependency behavior when faced with external deformation.27,28 In theory, cardiomyocytes exhibit unique viscoelasticity and adaptability attributed to high contractility and arrangement of myofilaments and cytoskeleton.8 Therefore, fundamental research of mechanics and viscoelasticity of single cardiomyocyte with complex structure and functional behaviors can provide fundamental insights into (1) the structure–contractility–viscoelasticity relationship; (2) the underlying disease mechanism; (3) developing novel therapies; (4) fabricating fully functional cardiac patches.
Existing studies of single cardiomyocyte mechanics can be categorized into two branches: (1) contractile mechanics; (2) mechanics measurement and modeling. Iribe et al.29 studied the effects of external stretching load on a single isolated cardiomyocyte end-systolic force. The results demonstrated thin filaments' (in)activation exhibiting velocity- and load-dependent. Ribeiro et al.8 investigated the influences of substrate stiffness and physiological shape on single hiPSC-CM contractility, demonstrating associations between contractility and myofibril maturity metrics. However, the contractility–viscoelasticity relationship remains unclear. As for cardiomyocyte mechanics measurement and modeling, Lieber et al.30 reported the effects of aging on cardiac myocytes' stiffness and its relationship with left ventricular diastolic dysfunction. Deitch et al.31 explored substrate-induced nanomechanical and viscoelastic changes of 2D cultured neonatal rat cardiomyocytes. Lanzicher et al.32 explored the linkages between neonatal rat cardiomyocytes' membrane adhesion and cytoskeletal viscoelastic with mutated lamin A/C gene (LMNA). Zhang et al.33 characterized a single cardiomyocyte cell contractile behavior using a linear dynamic model to explore effects of subcellular structure on cytoskeleton mechanics. However, there is little knowledge regarding the uniqueness of cardiomyocyte viscoelasticity, mechanical, and adhesion roles and mechanisms in maintaining its morphogenesis and regulating functional behaviors. Moreover, the structural–mechanical–functional associations at a single-cell level and the potential application of cell mechanics and related technologies in biological and clinical studies remain unexplored.
This study aimed to explore force-controlled single hiPSC-CM mechanical performance, revealing its dynamic changes in architecture and compositions, including cell membrane, cytoskeleton, and cytoplasm physiological status. We seek to provide insights into hiPSC-CM viscoelasticity, adhesion, cell–cell/cell–substrate communication, spatiotemporal displacements/strains, and mechanotransduction at cell junctions using AFM nanoindentation, fluidic micropipette, and DIC technique. More importantly, we intended to discover the interrelationship among viscoelastic behaviors, surface mechanical properties, adhesion, and beating mechanics. Overall, we targeted to explore fundamental hiPSC-CM mechanical behaviors, structure–contractility–viscoelasticity relationships, and the consequences of these properties for physiological functions. This work has potential practical applications for cardiovascular drug screening and mechanics-disease mechanisms research. Beyond that, the unique structural, mechanical, and adhesion properties of hiPSC-CM are critical for practical applications for cardiovascular drug screening and mechanics-disease mechanisms research.
Results and discussion
Single hiPSC-CM mechanical properties
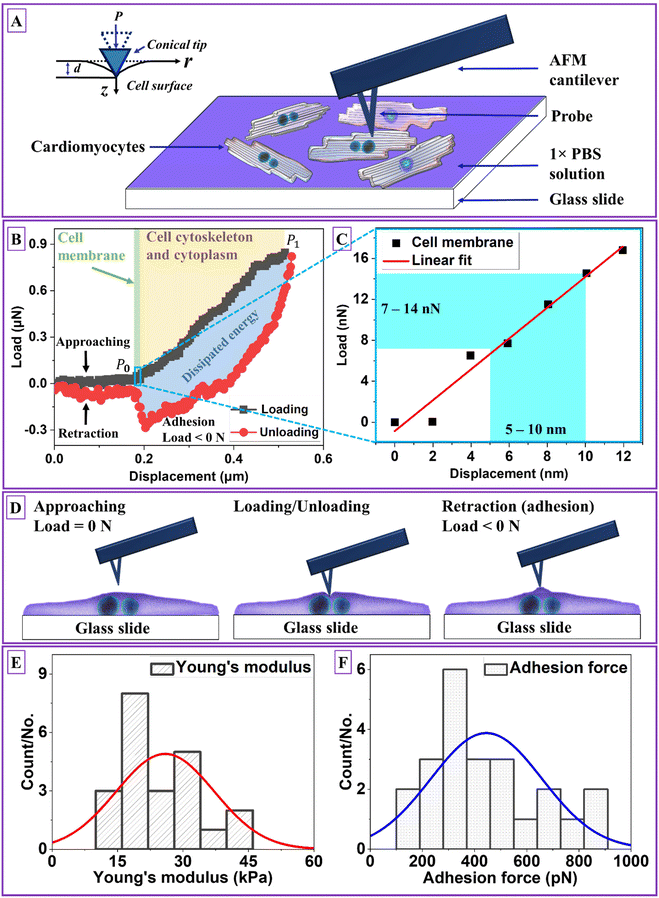
During AFM nanoindentation on living hiPSC-CMs seeded on a glass slide without fixation (Fig. 1A), the probe exerts a force on the cell membrane, cytoskeleton, and cytoplasm to reveal the mechanical response that is illustrated as the load–displacement curve in Fig. 1B. The load–displacement curve consisted of the approaching–loading–unloading–retraction process, where P0 and P1 are the loading curve's start and endpoints. The average indentation depth was ∼300 nm, corresponding to a deformation strain (ε) of 0.075–0.12. ε is calculated using eqn (1):| ε = d/thiPSC-CM | (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 34). According to Lulevich35et al.'s study, 0.2 < ε < 0.3 and 0.3 < ε < 0.8 suggested cell membrane remain impermeable and bursting with stress peaks, respectively. The stress peak(s) displayed on the loading curve, indicating rupture of membrane, a break of cytoskeleton crosslinkers, leakage of cytoplasm, and drop of cell pressure. In this study, there was no stress peak on the loading curve, suggesting hiPSC-CM remained undamaged during and after AFM nanoindentation testing. The loading section includes two regions: (1) an initial contact and bending of the cell membrane (green color) and (2) cytoskeleton and cytoplasm deformation (yellow color). The initial linear stage (Fig. 1C) with a displacement within 10 nm corresponds to the elastic lipid bilayer of 5–10 nm in thickness2 with a detected cell membrane load of 7–14 nN.
34). According to Lulevich35et al.'s study, 0.2 < ε < 0.3 and 0.3 < ε < 0.8 suggested cell membrane remain impermeable and bursting with stress peaks, respectively. The stress peak(s) displayed on the loading curve, indicating rupture of membrane, a break of cytoskeleton crosslinkers, leakage of cytoplasm, and drop of cell pressure. In this study, there was no stress peak on the loading curve, suggesting hiPSC-CM remained undamaged during and after AFM nanoindentation testing. The loading section includes two regions: (1) an initial contact and bending of the cell membrane (green color) and (2) cytoskeleton and cytoplasm deformation (yellow color). The initial linear stage (Fig. 1C) with a displacement within 10 nm corresponds to the elastic lipid bilayer of 5–10 nm in thickness2 with a detected cell membrane load of 7–14 nN.
The nonlinear region gradually manifested with displacement higher than 10 nm, attributing to a composite character of sarcomeric cytoskeleton and cytoplasm. Cytoskeletons of intertwined and interlinked actin, microtubules, and intermediate filaments demonstrate nonlinear stiffness and viscoelastic characteristics.3 Cytoplasm constitutes ∼80% water and is treated as a viscoplastic fluid.2 The combined elastic cell membrane, viscoelastic cytoskeleton, and viscoplastic cytoplasm characteristics were manifested with dominated viscoelasticity with a hysteresis phenomenon, showing as a blue interspace within loading and unloading curves. The dissipation loop, the interspace, shows a dissipated energy of 55.75 ± 7.98% due to heat loss trigged by internal composite macro-polymers friction. The dissipated energy was calculated using eqn (2):
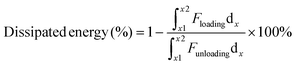 | (2) |
Fig. 1D illustrates three probe–sample interaction stages corresponding to the load–displacement curve: (1) approaching stage related to the process of that probe moving toward the sample surface (P0) with a load of 0 N, (2) the loading–unloading stage depicting the direct mechanical interaction of the probe–sample, and (3) the retraction stage tracking adhesion of the probe–sample by detaching the probe from the sample with a load < 0 N.
The reduced elastic modulus (E′) of hiPSC-CMs were calculated based on the loading curve using the Derjaguin–Muller–Toporov (DMT) model28,36 (eqn (3)). According to Garcia et al.,27,37 the force equation for a conical probe of half-angle θ of 10° is eqn (4):
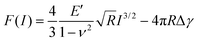 | (3) |
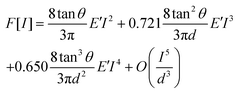 | (4) |
Garcia et al.28 differentiated viscoelastic descriptions into two steps: (1) mechanical-equivalent models or continuum mechanics theories, and (2) transform deflection into tip–sample distance relationships. Using the acquired elastic modulus and viscoelasticity of hiPSC-CMs, we modeled the von Mises stress distribution near the indentation region using the finite element method. The Mooney–Rivlin two-parameter model and two-generalized Maxwell branches were applied to simulate hiPSC hyperelasticity and viscoelasticity, respectively. The stress and strain were computed using a strain energy density function Ws (eqn (5)):
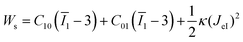 | (5) |
HiPSC-CM adhesion behaviors
The fluidic micropipette technique22 combines AFM with micro-channeled cantilevers with tunable nano- or micro-sized apertures (Fig. 2A), allowing single-cell isolation, injection or deposition materials, and adhesion characterization. The test was performed by approaching a targeted hiPSC-CM with a 1× PBS-filled cantilever (Fig. 2B). Sucking was applied under negative pressure of ≤1 bar upon aperture reaching and right above the cell surface, corresponding to a load of 0 N (Fig. 2C). After the hiPSC-CM membrane was tightly contacted within the aperture, a small load of 15–45 nN was observed. The targeted cell was detached by retracting the cantilever, correlating to the isolating process with a load < 0 N (red curves). The minimum force during the isolating process was pull-off or adhesion force. The load–displacement curve represented the intermediate force changing and deformation mechanism corresponding to the measurement process, as shown in Fig. 2D and E. Video V1† shows the hiPSC-CM pull-off process.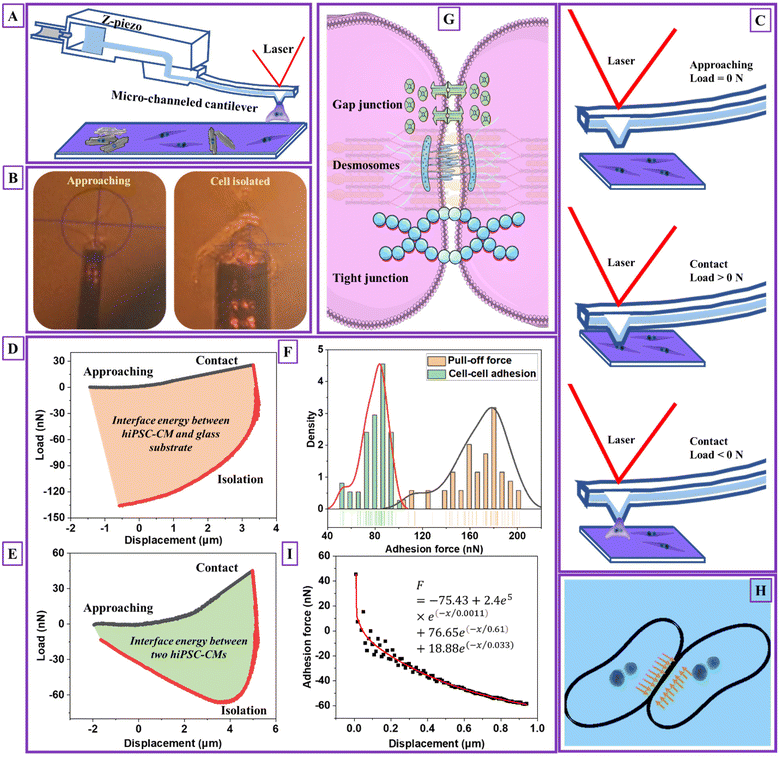 | ||
| Fig. 2 Cardiomyocyte–cardiomyocyte (D4) adhesion measurement. (A) Schematic images of detaching one single hiPSC-CM and measuring adhesion between two hiPSC-CMs' process, (B) the images of practical detaching one single hiPSC-CM and measuring adhesion between two hiPSC-CMs' process, (C) a schematic of hiPSC-CM adhesion detection process and the corresponding cell–micropipette interaction statuses, (D) load–displacement curve of detaching one single hiPSC-CM process (n = 50, 25 cells), (E) load–displacement curve of measuring adhesion between two hiPSC-CMs' process (n = 50, 25 cells), (F) the detected cardiomyocyte–cardiomyocyte adhesion force distribution and pull-off force distribution when detaching one single hiPSC-CM from the glass substrate, (G) a schematic of cardiomyocyte–cardiomyocyte adhesion mechanism, (H) a schematic of cardiomyocyte–cardiomyocyte adhesion and compression distribution at the contact region, and (I) the fitted cell–cell adhesion force during the initial detaching process using the Prony series model (the black scatters are extracted from the initial unloading curve from Fig. 2E with unchanged load and modified displacement values. The modification considers the start unloading point with a displacement of 0. The data from the initial 0–1 μm unloading section is used for data modeling. The raw and treated data is attached in an Excel sheet, see ESI†). Note: D4 means hiPSC-CMs were measured after they started beating for four days. | ||
The pull-off force between hiPSC-CM and glass substrate (Fig. 2F) was 100–210 nN with an average value of 167.58 ± 22.56 nN. The adhesion force between two hiPSC-CMs was 50–100 nN with an average value of 79.22 ± 10.80 nN, attributed to gap junctions, desmosomes, and tight junctions (Fig. 2G). The gap and tight junctions are strong mechanical connections linked to the cytoskeleton, regulating intercellular force and signal transmission and influencing cell regulatory and migratory behaviors.3 Additionally, cadherins,39 integrins, nectins, and junctional adhesive molecules are in charge of the physical cell–cell linkage. The functions of these molecules are regulating intercellular communication and signaling.39 It indicates that single-cell mechanics and cell–cell or cell–matrix adhesion might be directly correlated. Due to adhesive contact, there is pressure at the interface (Fig. 2H). The interface energy (γ) is the dissipation energy during the approaching–contact–detaching process, which is calculated by eqn (6):
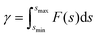 | (6) |
The cell–cell adhesion force during the detaching process was nonlinear, with a rapid decrease at the initial stage, as plotted in Fig. 2I. We hypothesized that cell–cell adhesion exhibits viscoelastic behavior due to structure connections with cell structural building blocks. To prove that we extracted the data for viscoelastic modeling using eqn (7):
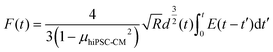 | (7) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) e(−x/0.61) + 18.88
e(−x/0.61) + 18.88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) e(−x/0.033). The corresponding viscosity coefficients η1, η2 and η3 are 2.4 × 105, 76.65, and 18.88 Pa s, respectively, determining the time-dependent energy dissipation rate. Specifically, η1 associated with the first Maxwell unit, characterizing the material's short-term (high frequency) viscosity. Corresponding, η2 and η3 associated with the second and third Maxwell units, representing intermediate-term (middle frequency) and long-term (low-frequency) materials viscosity. A lower value of η1 compared to η2 and η3 indicates less resistance to deformation due to elasticity of cell membrane during the initial indentation process. Moreover, the inequality η1 < η3 < η2 suggests the frequency-dependent viscoelastic behavior28 with a stronger viscous response at higher frequencies and a relatively weaker viscous response at intermediate frequencies. The viscoelastic behavior of cell–cell adhesion verifies our hypothesis that its intimate association with single-cell mechanics is due to linkages between cell–cell junctions and the cytoskeletons. The adhesion and viscoelasticity research highlighted the importance of fabricating biomaterials with adhesive molecules and proteins to support cardiomyocytes' electrical and chemical coupling. It is worth noting that the geometry of the micropipette is circular with a flat aperture of 4 μm in diameter. The limitation of this modeling is not considering the effect of micropipette geometry and contact area.28
e(−x/0.033). The corresponding viscosity coefficients η1, η2 and η3 are 2.4 × 105, 76.65, and 18.88 Pa s, respectively, determining the time-dependent energy dissipation rate. Specifically, η1 associated with the first Maxwell unit, characterizing the material's short-term (high frequency) viscosity. Corresponding, η2 and η3 associated with the second and third Maxwell units, representing intermediate-term (middle frequency) and long-term (low-frequency) materials viscosity. A lower value of η1 compared to η2 and η3 indicates less resistance to deformation due to elasticity of cell membrane during the initial indentation process. Moreover, the inequality η1 < η3 < η2 suggests the frequency-dependent viscoelastic behavior28 with a stronger viscous response at higher frequencies and a relatively weaker viscous response at intermediate frequencies. The viscoelastic behavior of cell–cell adhesion verifies our hypothesis that its intimate association with single-cell mechanics is due to linkages between cell–cell junctions and the cytoskeletons. The adhesion and viscoelasticity research highlighted the importance of fabricating biomaterials with adhesive molecules and proteins to support cardiomyocytes' electrical and chemical coupling. It is worth noting that the geometry of the micropipette is circular with a flat aperture of 4 μm in diameter. The limitation of this modeling is not considering the effect of micropipette geometry and contact area.28
Single hiPSC-CM contractility
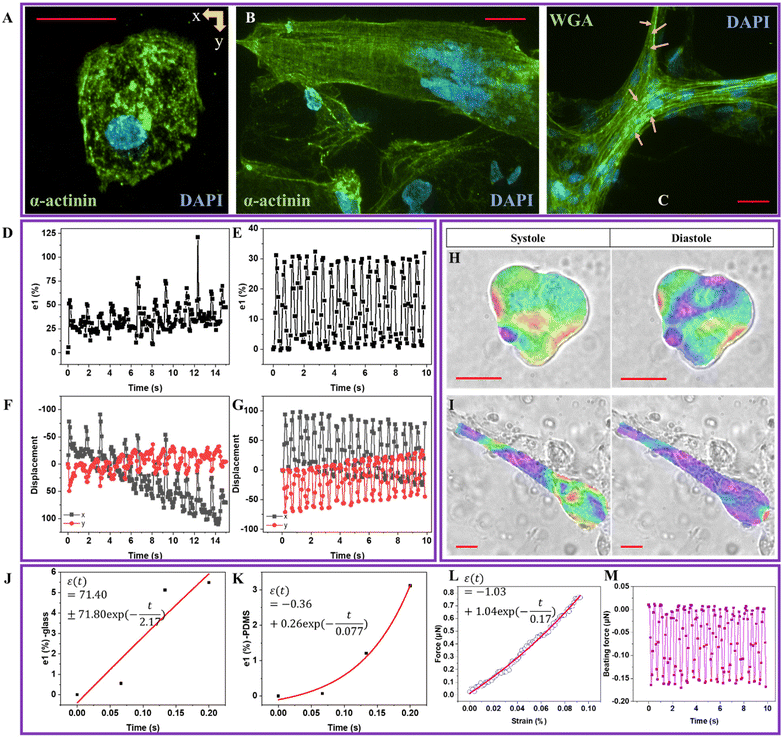
Spontaneous cardiomyocyte (CM) contractility41 is the chemical–electrical–mechanical composite process controlled through the gap junctions of intercalated discs via excitation–contraction coupling. Therefore, contractility behavior, including beating rate, rhythm, and full-field strain map, directly reflects CM functions and maturation, determined by physiological structure, mechanics, gene expression, and metabolism.13 A dimensionless factor defines cell shape sc (0 ≤ sc ≤ 1) using eqn (S3) (in the ESI†). sc closes to 0 and 1 representing circle and rectangular. As shown, hiPSC-CMs with rounded (Fig. 3A) and elongated rectangular shapes (Fig. 3B) were observed on glass and PDMS substrates, where the corresponding sc was 0.25 and 0.75, respectively. Compared to a randomly distributed structure, the elongated shape exhibited a highly aligned myofibrillar cytoskeleton and T-tubule (Fig. 3C), contributing to maturation, calcium metabolism, and contractile anisotropy.41 T-tubules and spontaneous rhythmic beating are indirect evidence of energy metabolism, electrophysiological maturation, and gene expression of Ca2+ channels relating to its physiological condition.The DIC technique characterized the full-field contractile-related deformation, revealing spatiotemporal beating frequency, rhythm, and magnitude. The time-dependent principal strains (e1) calculated from tracked displacements were shown in Fig. 3D and E, which play a critical role in showing CM homeostasis.13 HiPSC-CMs seeded on the glass substrate displayed an inhomogeneous beating rhythm. The time-dependent displacements (Fig. 3F and G) along the x- and y-direction were analyzed to compare the contractile anisotropy. Nearly 2–3 times lower in y-direction displacement magnitude than that of x-direction was observed with a consistent abnormal rhythm similar to strain. The time-dependent e1 color maps at the systole and diastole phases are plotted in Fig. 3H and I, where blue/purple and yellow/red are denoted as contraction and dilation, respectively. HiPSC-CMs seeded on the PDMS substrate demonstrated more uniformity in the diastole phase than in glass. Videos V2 and V3† show the bright-field beating and DIC analyzed the major principal strain (e1) videos of hiPSC-CM seeded on cover glass (250×), respectively. Videos V4 and V5† show hiPSC-CM seeded on PDMS substrate (250×). We observed substrate-stimulated hiPSC-CM shape differences, inhomogeneous/homogenous beating rhythm, and contractile anisotropy. Myocyte is the major contributor to heart contractility, where spontaneous beating is directly associated with heart abnormalities. For example, cardiac electrophysiologists monitor heart rhythm using a 12-lead electrocardiogram to check heart attack, failure, ischemia, myocardial infarction, and arrhythmia. It indicated that hiPSC-CM morphology–contractile-pathology is tightly interrelated.
We hypothesized that contractile-related e1 could be linked to the detected viscoelasticity measured by AFM We analyzed the left half cycle of e1, which fitted with the Kelvin–Voigt viscoelastic model42 using eqn (S4) (in the ESI†). Due to the spontaneous contractile being recorded and not detected by the AFM probe, there is no need to introduce tip geometry. The fitting curve for hiPSC-CM seeded on the glass substrate (Fig. 3J) exhibited an R2 of 60% and an equation of 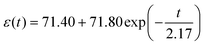 . The fitting results for hiPSC-CM seeded on the PDMS substrate (Fig. 3K) demonstrated an R2 of 97.40% with the equation of
. The fitting results for hiPSC-CM seeded on the PDMS substrate (Fig. 3K) demonstrated an R2 of 97.40% with the equation of 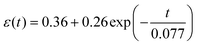 . The viscosity coefficients of hiPSC-CM seeded on glass and PDMS substrates are 71.80 and 0.26 Pa s, respectively. This suggests that hiPSC-CM seeded on the glass substrate is more resistant to spontaneous contractile deformation with prominent viscous behavior. The data proved our hypothesis and suggested that substrate stiffness8 influences hiPSC-CM viscoelasticity. Typically, hiPSC-CM maturation on soft substrates reduces viscosity, attributed to alterations in cytoskeletal organization, expressions of proteins involved in cell adhesion and mechanotransduction, and contractile–structure associations. Actin, myosin, troponin, tropomyosin, nebulin, and titin are major proteins involved in the contractile of cardiomyocytes.43 These proteins are the building blocks of thin and thick myofilaments, intertwining with cytoskeletal networks to support contractility.43 To calculate the strain-related beating force, we fitted the force–strain curve (Fig. 3L), exhibiting an R2 of 99.46% with the equation of
. The viscosity coefficients of hiPSC-CM seeded on glass and PDMS substrates are 71.80 and 0.26 Pa s, respectively. This suggests that hiPSC-CM seeded on the glass substrate is more resistant to spontaneous contractile deformation with prominent viscous behavior. The data proved our hypothesis and suggested that substrate stiffness8 influences hiPSC-CM viscoelasticity. Typically, hiPSC-CM maturation on soft substrates reduces viscosity, attributed to alterations in cytoskeletal organization, expressions of proteins involved in cell adhesion and mechanotransduction, and contractile–structure associations. Actin, myosin, troponin, tropomyosin, nebulin, and titin are major proteins involved in the contractile of cardiomyocytes.43 These proteins are the building blocks of thin and thick myofilaments, intertwining with cytoskeletal networks to support contractility.43 To calculate the strain-related beating force, we fitted the force–strain curve (Fig. 3L), exhibiting an R2 of 99.46% with the equation of 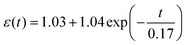 . The calculated time-dependent beating force for hiPSC-CM seeded on the PDMS substrate is plotted in Fig. 3M, revealing a maximum force at systole and diastole phases of ∼0.01 and ∼0.15 ± 0.02 μN. Therefore, we successfully connected hiPSC-CM anamorphism with contractile forces using non-invasive DIC and AFM nanoindentation techniques, which quantitatively characterize a biomechanical phenomenon using force rather than biological descriptive, modeling or “biased” invasive methods.1,2 The importance of hiPSC-CM contractility is the guidance of designing nN–μN force responsive biomaterials for cardiac bioengineering.
. The calculated time-dependent beating force for hiPSC-CM seeded on the PDMS substrate is plotted in Fig. 3M, revealing a maximum force at systole and diastole phases of ∼0.01 and ∼0.15 ± 0.02 μN. Therefore, we successfully connected hiPSC-CM anamorphism with contractile forces using non-invasive DIC and AFM nanoindentation techniques, which quantitatively characterize a biomechanical phenomenon using force rather than biological descriptive, modeling or “biased” invasive methods.1,2 The importance of hiPSC-CM contractility is the guidance of designing nN–μN force responsive biomaterials for cardiac bioengineering.
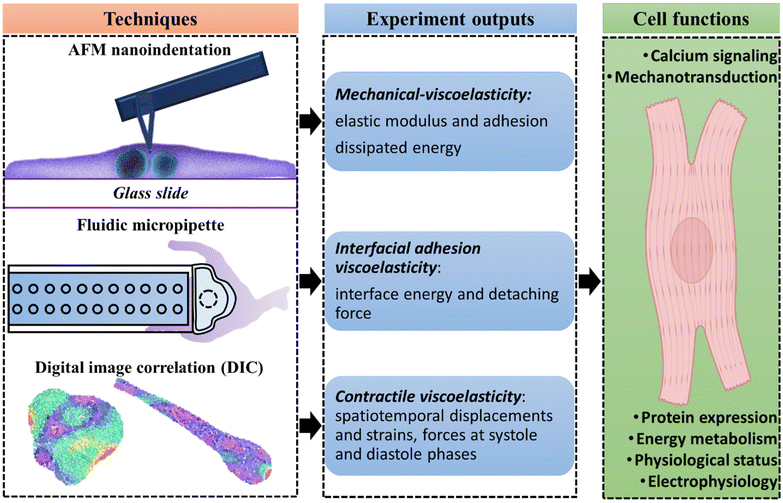
In summary, we endeavor to test the feasibility of constructing a hiPSC-CM mechanic's testbed to address numerous myocardial physiology and pathophysiology issues. Three technologies, including AFM nanoindentation, fluidic micropipette, and DIC technique, were employed to study single hiPSC-CM Young's modulus, model deformation mechanism, probe cell adhesion and interface energy, and measure beating mechanics. The results demonstrated that the cytoskeleton's viscoelasticity dominated single hiPSC-CM mechanics with a visible energy dissipation characteristic. The adhesion force between two hiPSC-CMs was primarily governed by gap and tight junctions that directly link with their cytoskeletons and single-cell mechanics, manifesting by its viscoelastic behavior that fits perfectly with the Prony series model. Distinct morphology and time-dependent beating mechanics were observed for hiPSC-CM seeded on glass and PDMS substrate. The time-dependent contractile strain for hiPSC-CM seeded on PDMS substrate also exhibited viscoelasticity, indicating substrate stiffness is essential in maintaining hiPSC-CM functions. Contractile associated with electrical and mechanical stimuli are determined by beating-related proteins' expression, maturation of ion channels, calcium signaling, and energy utilization.41 These factors are also critical for the functional characteristics of cardiomyocytes, including propagating electrical signaling and performing structural and mechanical integrity.44 Therefore, this study disclosed associations between cardiomyocyte contractile and functional properties. Most importantly, our study revealed robust structural–mechanical–viscoelasticity, demonstrating the potential practicality of our mechanical platform conception, as illustrated in Fig. 4.
Conclusions
The viscoelasticity of cardiomyocytes and the underlying structural–mechanical–functional mechanism lack systematic study, although it is critical for cardiac functions. This study investigated a single cardiomyocyte's spatiotemporal mechanical and viscoelastic behaviors using three cutting-edge technologies: AFM nanoindentation, fluidic micropipette, and DIC Our results showed a detected cytoplasm load and dissipated energy of 7–14 nN and 55.75 ± 7.98%, indicating hiPSC-CM's mechanics and viscoelasticity is intimately linked with cytoskeleton network, intracellular structures, and extracellular matrix. The detected adhesion force between two hiPSC-CMs was 50–100 nN with an interface energy of 0.58 ± 0.11 pJ, attributing to cell interfacial molecular connections, including cadherins and gap/tight junctions. Based on the acquired data, we modeled cell–cell adhesion and contractility, demonstrating the fundamental role of cardiomyocyte mechanics and time-dependent viscoelasticity in organizing and maintaining its electrical and mechanical integrations with the surrounding environment. Overall, this study highlights the viscoelasticity of cardiomyocytes and its essential inter-relationship with internal structure, contractile, and functions, contributing to designing biomaterials with mimicked force response mechanisms as that of cardiomyocytes.Materials and methods
HiPSC culture and cardiomyocyte differentiation
HiPSCs (human iPSCs from reprogrammed fibroblasts, GM23338) were purchased from Coriell Institute for Medical Research and grown on Matrigel-coated plates (B.D. Biosciences) in stem basal medium (mTeSR1, STEMCELL Technologies) for about 7 days to reach 80–90% of confluence. On day 0, hiPSCs were treated with CHIR99021 (12 μM, Tocris, 4423) diluted in RPMI/B27 – insulin for 24 hours, and the media changed every day until day 3. On day 3, hiPSCs were treated with IWP4 (5 μM, Tocris, 5214) mixed with RPMI/B27 – insulin, which was removed on day 4. The medium was changed every other day. From day 9, hiPSCs were maintained in RPMI/B27 containing insulin with regular medium change. Spontaneous contractions were observed between 10 to 12 days.Seeding hiPSC-CM
The cover glass was incubated with 10 μg ml−1 of human fibronectin (Corning, 356008) in sterile phosphate-buffer solution (1× PBS, Gibco) for 1 hour at 37 °C and rinsed with sterile 1× PBS. HiPSC-CMs were digested from tissue culture plates by adding TrypLE express enzyme (Gibco, 12605010) at 37 °C for 15 minutes. The detached cells were resuspended in RPMI/B27 containing insulin. HiPSC-CMs were replanted on cover glass-fibronectin coated at a density of 3 × 105 cells with RPMI/B27 containing insulin, fetal bovine serum (10%, Gibco, 10099141), pen strep (1%, Gibco, 15140148) and Y-27632 (5 μM, Tocris, 1254) for 48 hours. On day 3, hiPSC-CMs were maintained in RPMI/B27 containing insulin. Spontaneous contractions were observed between 7 to 9 days.Immunostaining and immunofluorescence imaging
HiPSC-CMs seeded on the cover glass were prepared for immunocytochemical analysis on day 4 after the spontaneous beating started. The cells were immersed in paraformaldehyde (4%) for 20 minutes, blocked, and permeabilized with Triton X-100 (0.2%) and bovine serum albumin (1%) in 1× PBS for 20 minutes at room temperature. Cells were stained overnight with the primary antibody, anti-sarcomeric α-actinin (Abcam, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 dilution). Samples were incubated with goat anti-mouse secondary antibody Alexa-488 for 40 minutes (Abcam, 1
300 dilution). Samples were incubated with goat anti-mouse secondary antibody Alexa-488 for 40 minutes (Abcam, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 dilution), followed by 10 minutes incubation with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, 30 nM). For T-tubule labeling, the samples were incubated with WGA-Alexa Fluor 488 (Life Technologies, W11261) for 10 min and rinsed with 1× PBS. Fluorescence images were acquired using the confocal microscope (Nikon ECLIPSE Ti2) with a 60× objective.
300 dilution), followed by 10 minutes incubation with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, 30 nM). For T-tubule labeling, the samples were incubated with WGA-Alexa Fluor 488 (Life Technologies, W11261) for 10 min and rinsed with 1× PBS. Fluorescence images were acquired using the confocal microscope (Nikon ECLIPSE Ti2) with a 60× objective.
Mechanics of hiPSC-CM measured by AFM
HiPSC-CM cell mechanics were measured using AFM (Nanosurf FlexAFM, Liestal, Switzerland). HiPSC-CMs were seeded on cover glasses with a diameter of 15 mm and incubated at 37 °C. Before the measurements, the cover glass was transferred to a thin flat microscope slide. Cantilever Tap150Al-G (NanoANDMore, Watsonville, CA) with length, width, resonant frequency, stiffness, half-cone angle, and tip radius of 125 μm, 25 μm, 150 kHz, 5 N m−1, 10° at the apex, and <10° was selected for single cell mechanic measurements. The spring constant was calibrated using the frequency sweep method using Nanosurf C3000 software before deflection and crosstalk calibrations. The nanoindentation test was performed at the Spectroscopy mode using static force, force–distance spectroscopy grid, and data point 1024. The load–displacement curve was analyzed by Nanosurf ANA software, where Derjaguin–Muller–Toporov (DMT) model was applied for Young's modulus calculation. The minimum backward force was the adhesion force between the probe and hiPSC-CM, verifying contact. Fifty tests on 25 cells were performed, where the data was denoted as mean ± standard deviation.Adhesion force measurement
The adhesion forces between hiPSC-CM and glass slide or between two hiPSC-CMs were measured using micropipette cantilevers (Cytosurge AG, Opfikon, Switzerland) with a circular and flat aperture of 4 μm in diameter and spring constant of 2 N m−1. The fluidic channel of the micropipette cantilever was filled with 1× PBS (5 μl) using a hand-held pipette (2–20 μl). It was mounted to the head of the AFM, whereas the Nanosurf AFM system was mounted on an inverted microscope (Carl Zeiss, Germany). After manual laser alignment, the micropipette cantilever's spring constant and deflection sensitivity were calibrated using the thermal tuning and Cytosurge software. Before the measurement, the hiPSC-CM seeded glass was transferred to a Petri dish filled with 1× PBS. The test was completed within 1.5 h to avoid cell death. A hiPSC-CM was selected, approached, and sucked under a pressure of 600–800 mbar for 10 s to ensure the sealing of the cell to the cantilever. The cantilever is retracted using a speed of 1 μm s−1 to isolate hiPSC-CM. The load–displacement curve was analyzed by Nanosurf ANA software, where the adhesion force and interface energy were analyzed. Fifty tests on 25 cells were performed, where the data was denoted as mean ± standard deviation.Bright-field imaging and DIC analysis
HiPSC-CMs seeded on cover glass or PDMS substrate were removed from the incubator and placed on the inverted microscope with 40× objective and a C.C.D. camera (FLIR, Grasshopper3 GS3-U3-15S5M) for bright-field beating video recording. The video with a frame rate of 15. The video was converted into sequential images for DIC analysis using VIC-2D software (Correlated Solutions, Irmo, SC). The initial image was selected as the reference, with all others as deformed images. The applied subset and step size were 51 and 8, where the corresponding displacements as of the reference image were analyzed for each frame. The time-dependent x- and y-directional displacement, principal strain, and strain maps were acquired for beating mechanics analysis.Author contributions
A. A. and L. L. designed the concept of the work. A. S. R. and J. H. performed hiPSC-CM differentiation, culture, staining, imaging, and corresponding protocol writing. L. L. performed AFM nanoindentation, micropipette, and DIC analysis. The manuscript was written by L. L. and A. A. All authors have approved the final version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the Engineering Research Centers Program of the National Science Foundation under N.S.F. Cooperative Agreement no. EEC-1647837. The authors acknowledge Mr B. Aguiar's support in supplementary Video V1† editing.References
- S. R. Heidemann and D. Wirtz, Trends Cell Biol., 2004, 14, 160–166 CrossRef CAS PubMed
.
- D. A. Fletcher and R. D. Mullins, Nature, 2010, 463, 485–492 CrossRef CAS PubMed
.
- M. A. Caporizzo and B. L. Prosser, Nat. Rev. Cardiol., 2022, 19, 364–378 CrossRef PubMed
.
- L. Lou, K. O. Lopez, P. Nautiyal and A. Agarwal, Adv. NanoBiomed Res., 2021, 1, 2100075 CrossRef CAS
.
- C. A. Blair and B. L. Pruitt, Adv. Healthcare Mater., 2020, 9, 1901656 CrossRef CAS PubMed
.
- M. Mathelié-Guinlet, F. Viela, J. Dehullu, S. Filimonava, J. M. Rauceo, P. N. Lipke and Y. F. Dufrêne, Commun. Biol., 2021, 4, 1–8 CrossRef PubMed
.
- L. Wang, W. Dou, M. Malhi, M. Zhu, H. Liu, J. Plakhotnik, Z. Xu, Q. Zhao, J. Chen and S. Chen, ACS Appl. Mater. Interfaces, 2018, 10, 21173–21183 CrossRef CAS PubMed
.
- A. J. Ribeiro, Y.-S. Ang, J.-D. Fu, R. N. Rivas, T. M. Mohamed, G. C. Higgs, D. Srivastava and B. L. Pruitt, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 12705–12710 CrossRef CAS PubMed
.
- K. Dasbiswas, S. Majkut, D. Discher and S. A. Safran, Nat. Commun., 2015, 6, 1–8 Search PubMed
.
- L. Lou, T. Paul, B. A. Aguiar, T. Dolmetsch, C. Zhang and A. Agarwal, ACS Appl. Mater. Interfaces, 2022, 14, 42876–42886 CrossRef CAS PubMed
.
- L. Lou, L. Paolino and A. Agarwal, ACS Appl. Mater. Interfaces, 2023, 24197–24208 CrossRef CAS PubMed
.
- A. Shinde, K. Illath, P. Gupta, P. Shinde, K.-T. Lim, M. Nagai and T. S. Santra, Cells, 2021, 10, 577 CrossRef CAS PubMed
.
- D. J. Shiwarski, J. W. Tashman, A. Tsamis, J. M. Bliley, M. A. Blundon, E. Aranda-Michel, Q. Jallerat, J. M. Szymanski, B. M. McCartney and A. W. Feinberg, Nat. Commun., 2020, 11, 1–15 CrossRef PubMed
.
- Q. Zheng, M. Peng, Z. Liu, S. Li, R. Han, H. Ouyang, Y. Fan, C. Pan, W. Hu and J. Zhai, Sci. Adv., 2021, 7, eabe7738 CrossRef CAS PubMed
.
- M. S. Ma, S. Sundaram, L. Lou, A. Agarwal, C. S. Chen and T. G. Bifano, Front. Bioeng. Biotechnol., 2023, 11, 703 Search PubMed
.
- A. Sesena-Rubfiaro, N. J. Prajapati, L. Paolino, L. Lou, D. Cotayo, P. Pandey, M. Shaver, J. D. Hutcheson, A. Agarwal and J. He, ACS Biomater. Sci. Eng., 2023, 1644–1655 CrossRef CAS PubMed
.
- L. Lou, A. S. Rubfiaro, J. He and A. Agarwal, Adv. Mater. Technol., 2021, 6, 2100669 CrossRef
.
- H. Li, S. Sundaram, R. Hu, L. Lou, F. Sanchez, W. McDonald, A. Agarwal, C. S. Chen and T. G. Bifano, IEEE Trans. Biomed. Eng., 2023, 1–12 Search PubMed
.
- K. H. Vining and D. J. Mooney, Nat. Rev. Mol. Cell Biol., 2017, 18, 728–742 CrossRef CAS PubMed
.
- T. Panciera, A. Citron, D. Di Biagio, G. Battilana, A. Gandin, S. Giulitti, M. Forcato, S. Bicciato, V. Panzetta and S. Fusco, Nat. Mater., 2020, 19, 797–806 CrossRef CAS PubMed
.
- T. Chowdhury, B. Cressiot, C. Parisi, G. Smolyakov, B. Thiébot, L. Trichet, F. M. Fernandes, J. Pelta and P. Manivet, ACS Sens., 2023, 406–426 CrossRef CAS PubMed
.
- O. Guillaume-Gentil, E. Potthoff, D. Ossola, C. M. Franz, T. Zambelli and J. A. Vorholt, Trends Biotechnol., 2014, 32, 381–388 CrossRef CAS PubMed
.
- L. Lou, A. S. Rubfiaro, J. He and A. Agarwal, Adv. Mater. Technol., 2021, 2100669 CrossRef
.
- T. Hattori, S. Shimizu, Y. Koyama, K. Yamada, R. Kuwahara, N. Kumamoto, S. Matsuzaki, A. Ito, T. Katayama and M. Tohyama, Mol. Psychiatry, 2010, 15, 798–809 CrossRef CAS PubMed
.
- E. Kardash, M. Reichman-Fried, J.-L. Maître, B. Boldajipour, E. Papusheva, E.-M. Messerschmidt, C.-P. Heisenberg and E. Raz, Nat. Cell Biol., 2010, 12, 47–53 CrossRef CAS PubMed
.
- E. Kim, A. Lisby, C. Ma, N. Lo, U. Ehmer, K. E. Hayer, E. E. Furth and P. Viatour, Nat. Commun., 2019, 10, 1–17 CrossRef CAS PubMed
.
- P. D. Garcia and R. Garcia, Nanoscale, 2018, 10, 19799–19809 RSC
.
- R. Garcia, Chem. Soc. Rev., 2020, 49, 5850–5884 RSC
.
- G. Iribe, T. Kaneko, Y. Yamaguchi and K. Naruse, Prog. Biophys. Mol. Biol., 2014, 115, 103–114 CrossRef PubMed
.
- S. C. Lieber, N. Aubry, J. Pain, G. Diaz, S.-J. Kim and S. F. Vatner, Am. J. Physiol.: Heart Circ. Physiol., 2004, 287, H645–H651 CrossRef CAS PubMed
.
- S. Deitch, B. Z. Gao and D. Dean, MCB Mol. Cell. Biomech., 2012, 9, 227 Search PubMed
.
- T. Lanzicher, V. Martinelli, C. S. Long, G. Del Favero, L. Puzzi, M. Borelli, L. Mestroni, M. R. Taylor and O. Sbaizero, Nucleus, 2015, 6, 394–407 CrossRef CAS PubMed
.
- C. Zhang, W. Wang, W. He, N. Xi, Y. Wang and L. Liu, Biophys. J., 2018, 114, 188–200 CrossRef CAS PubMed
.
- S. Kadota, L. Pabon, H. Reinecke and C. E. Murry, Stem Cell Rep., 2017, 8, 278–289 CrossRef CAS PubMed
.
- V. Lulevich, T. Zink, H.-Y. Chen, F.-T. Liu and G.-y. Liu, Langmuir, 2006, 22, 8151–8155 CrossRef CAS PubMed
.
- B. V. Derjaguin, V. M. Muller and Y. P. Toporov, J. Colloid Interface Sci., 1975, 53, 314–326 CrossRef CAS
.
- P. D. Garcia and R. Garcia, Biophys. J., 2018, 114, 2923–2932 CrossRef CAS PubMed
.
- A. Viljoen, M. Mathelié-Guinlet, A. Ray, N. Strohmeyer, Y. J. Oh, P. Hinterdorfer, D. J. Müller, D. Alsteens and Y. F. Dufrêne, Nat. Rev. Methods Primers, 2021, 1, 1–24 CrossRef
.
- B. M. Gumbiner, Nat. Rev. Mol. Cell Biol., 2005, 6, 622–634 CrossRef CAS PubMed
.
-
D. Gutierrez-Lemini, Engineering Viscoelasticity, Springer, Chambersburg, PA, 2014 Search PubMed
.
- K. Ronaldson-Bouchard, S. P. Ma, K. Yeager, T. Chen, L. Song, D. Sirabella, K. Morikawa, D. Teles, M. Yazawa and G. Vunjak-Novakovic, Nature, 2018, 556, 239–243 CrossRef CAS PubMed
.
- M. Backholm, W. S. Ryu and K. Dalnoki-Veress, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4528–4533 CrossRef CAS PubMed
.
- B. L. Lin, T. Song and S. Sadayappan, Compr. Physiol., 2011, 675–692 Search PubMed
.
- K. Ronaldson-Bouchard, K. Yeager, D. Teles, T. Chen, S. Ma, L. Song, K. Morikawa, H. M. Wobma, A. Vasciaveo and E. C. Ruiz, Nat. Protoc., 2019, 14, 2781–2817 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr01553j |
| This journal is © The Royal Society of Chemistry 2023 |