Development of a two-dimensional peptide functionalized-reduced graphene oxide biomaterial for wound care applications†
Shubhi
Joshi
 ab,
Panchali
Barman
c,
Mayank
Maan
ab,
Panchali
Barman
c,
Mayank
Maan
 a,
Hemant
Goyal
a,
Sheetal
Sharma
a,
Rajesh
Kumar
a,
Hemant
Goyal
a,
Sheetal
Sharma
a,
Rajesh
Kumar
 d,
Gaurav
Verma
ef and
Avneet
Saini
d,
Gaurav
Verma
ef and
Avneet
Saini
 *a
*a
aDepartment of Biophysics, Panjab University, Chandigarh, U.T. 160014, India. E-mail: avneet@pu.ac.in; Tel: +91 9876442266
bEnergy Research Centre, Panjab University, Chandigarh, U.T. 160014, India
cInstitute of Forensic Science & Criminology, Panjab University, Chandigarh, U.T. 160014, India
dDepartment of Physics, Panjab University, Chandigarh, U.T. 160014, India
eDr Shanti Swarup Bhatnagar University Institute of Chemical Engineering & Technology, Panjab University, Chandigarh, U.T. 160014, India
fCentre for Nanoscience & Nanotechnology (U.I.E.A.S.T), Panjab University, Chandigarh, U.T. 160014, India
First published on 11th October 2024
Abstract
Increased incidences of antibiotic resistance have necessitated the development of novel wound disinfection strategies with minimal risk of resistance development. This study aimed at developing a biocompatible wound dressing biomaterial with the potential to treat acute and chronic wounds infected with multidrug-resistant Pseudomonas aeruginosa. A multifunctional antibacterial nanoconjugate was synthesized by covalently coupling a synthetically designed peptide (DP1, i.e., RFGRFLRKILRFLKK) with reduced graphene oxide (rGO). The conjugate displayed antibacterial and antibiofilm activities against multidrug-resistant Pseudomonas aeruginosa. In vitro studies demonstrated 94% hemocompatibility of the nanoconjugate even at concentrations as high as 512 μg mL−1. Cytotoxicity studies on 3T3-L1 cells showed 95% cell viability, signifying biocompatibility. Owing to these properties, the biomedical applicability of the nanoconjugate was assessed as an antibacterial wound dressing agent. rGO-DP1-loaded wound dressing exhibited enhanced reduction in bacterial bioburden (6 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 CFU) with potential for wound re-epithelization (77.3%) compared to the uncoated bandage. Moreover, an improvement in the material properties of the bandage was observed in terms of enhanced tensile strength and decreased elongation at break (%). Collectively, these findings suggest that rGO-DP1 is an effective biomaterial that, when loaded on wound dressings, has the potential to be used as a facile, sustainable and progressive agent for bacterial wound disinfection as well as healing.
10 CFU) with potential for wound re-epithelization (77.3%) compared to the uncoated bandage. Moreover, an improvement in the material properties of the bandage was observed in terms of enhanced tensile strength and decreased elongation at break (%). Collectively, these findings suggest that rGO-DP1 is an effective biomaterial that, when loaded on wound dressings, has the potential to be used as a facile, sustainable and progressive agent for bacterial wound disinfection as well as healing.
1. Introduction
Skin may appear to be nothing more than a protective coating around the bones and tissues, but it is actually an exceptionally complicated and sophisticated structure that has the ability to heal itself upon damage.1 It serves as the first line of defence against any potentially harmful external influences as well as pathogenic invasion. Factors such as surgery, injury, burn, radiation, infection and underlying diseases often result in the formation of chronic or acute wounds, thereby disturbing the histological structure and function of the skin.2,3 Wounds have a tendency to heal within a predictable period depending on the severity of the lesion. Risk of microbial infection is considered as the most severe complication for any type of wound.4 Wounds infected by pathogenic organisms are difficult to treat owing to formation of a biofilm, which results in delayed healing, causing the risk of soft tissue damage.2,5 Moreover, if the infected wound is left untreated, it results in serious complications, often resulting in morbidity and mortality. Pathogenic bacteria, such as Staphylococcus aureus, Enterococcus faecalis, Escherichia coli and Pseudomonas aeruginosa (P. aeruginosa), tend to colonize chronic lesions.6–8 Among all the pathogens associated with wound-related infections and delayed healing, P. aeruginosa is particularly considered an opportunistic pathogen because of its ability to express virulence factors and surface proteins that influence the rate of wound healing.9P. aeruginosa causes infection in burn wounds, skin and soft tissue, leg ulcers, surgical wounds and diabetic foot ulcers.10,11 Despite scientific advances, disinfection and treatment of P. aeruginosa-infected wounds continue to be a great challenge due to a variety of complications appearing during the complex wound-healing process. Wound management approaches, such as antibiotics, acetic acid and negative-pressure wound therapy, are used to treat wounds infected with P. aeruginosa.6,7,12,13 Amongst the different therapeutic approaches, antibiotics are considered the first-line therapy to control the spread of infection at the wound site. However, because of its ability to form protective bacterial communities in the form of biofilms, P. aeruginosa exhibits resistance towards conventional antibiotics, thereby aggravating the severity of the infection. Also, the sustained emergence of antibiotic resistance (IMCJ2.S1, JNQH-PA57, UY1PSABAL, UY1PSABAL2, PA154197, etc.) complicates and impedes the treatment of pathogenic bacterial infections.14–16 Amidst the COVID-19 pandemic, antibiotics were utilized as a therapeutic measure for addressing secondary bacterial infections. However, the excessive and improper utilization of antibiotics contributed to the emergence and dissemination of antimicrobial resistance.Therefore, development of novel antimicrobial agents with low susceptibility to developing antimicrobial resistance, along with the ability to efficiently reduce P. aeruginosa infection in wounds, is an absolute necessity. Lately, nano-sized biomaterials such as silver, zinc oxide, curcumin, ferric chloride and gentamicin-based complexes are being evaluated for their effectiveness towards controlling P. aeruginosa infection in wounds.17,18 Amongst the different nano-sized antimicrobial agents, cationic antimicrobial peptides (AMPs) are also considered an effective alternative to conventional antibiotics owing to their broad-spectrum antibacterial activity, distinctive mechanism of action and low propensity to develop antimicrobial resistance.19–24 DP1 is an amphipathic synthetic antimicrobial peptide (sAMP) that displays helical characteristics as well as stability in membrane-mimicking environments. DP1 possesses a net positive charge of +7 and a negative GRAVY (grand average of hydropathy) score, indicating its hydrophilic nature.25 It displays broad-spectrum antimicrobial activity against Gram-negative, Gram-positive and multidrug-resistant bacterial strains. High cytocompatibility and hemocompatibility make it a prospective therapeutic agent.26,27 However, the biomedical applicability of the novel peptide is limited due to its short half-life and proteolytic degradation.28 It has been reported that the conjugation of peptides with nanoparticles helps overcome these intrinsic limitations, thereby enhancing their applicability.29–31 Despite promising research on peptides and their nanoconjugates with silver, gold, zinc and quantum dots, only a few studies reporting their use as anti-pseudomonal wound dressing either as hydrogels or as electrospun nanofibers have been reported.32–34
To exploit the antimicrobial property of these peptides for wound healing, a two-dimensional (2D) material to enhance its bioavailability was required and reduced graphene oxide (rGO), in particular, provided large surface area and abundant oxygen-containing moieties that enable the ease of peptide binding, enhanced stability, hemocompatibility and improved permeability of bacterial membranes.35,36 This study reports the synthesis of a peptide-functionalized rGO nanoconjugate using a covalent conjugation approach, where an in-house designed peptide (DP1) RFGRFLRKILRFLKK is conjugated with rGO. In vitro biocompatibility of the nanoconjugate was assessed by determining its hemocompatibility towards erythrocytes, cell viability towards 3T3-L1 cells and anti-pseudomonal potential. To control and manage wounds infected with P. aeruginosa, the rGO-DP1 nanoconjugate was used to develop an anti-pseudomonal wound dressing. The murine wound excision model was used to study the effect of the designed dressing on the antibacterial efficacy, and wound healing potential was assessed over eight days. Generally, in vitro models that do not mimic real-time wound biofilms and complexities of clinical wounds are used to assess the antibiofilm activity of antibacterial materials.37,38 Also, evaluation of the antibiofilm activity is frequently done with antibacterial solutions that tend to display dissimilar dispersal and permeation in comparison to wound dressings.39 This is the first time that the efficacy of a novel wound dressing embedded with a peptide-functionalized reduced graphene oxide nanoformulation is being reported.
2. Experimental section: materials
All reagents used during the course of this study were of analytical grade. Graphite nanopowder (98% purity), L-ascorbic acid (99.7% purity), phosphate-buffered saline (PBS) tablets and sodium hydroxide pellets were procured from Sisco Research Laboratories Pvt. Ltd (SRL, India). Design peptide DP1 (RFGRFLRKILRFLKK) was bought from GL Biochem (Shanghai) Ltd (China). 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride, cetrimide agar and Luria–Bertani broth were purchased from HiMedia Laboratories Pvt. Ltd (India). N-Hydroxysuccinimide was obtained from Avra Synthesis Pvt. Ltd (India). Ethanol was bought from Changshu Yangyuan Chemical Co., Ltd (China). Triton X-100 was procured from Sigma-Aldrich (USA).2.1. Synthesis and validation of a two-dimensional peptide functionalized-reduced graphene oxide-based wound dressing
This work was chiefly divided into two parts. The first part involved the synthesis, characterization and in vitro evaluation of a sustainable peptide-functionalized nanoconjugate (section 2.2–2.9). In the second part, the biomedical applicability of the synthesized nanoconjugate was determined by developing an antibacterial wound dressing wherein the efficacy of the wound dressing was evaluated in a P. aeruginosa infected murine wound excision model (section 2.10–2.19).2.2. Synthesis of rGO-DP1 nanoconjugate
Sustainable reduced graphene oxide (rGO) was synthesized by the application of L-ascorbic acid using a green synthesis approach.40 rGO nanopowder was synthesized by adding 1 g graphite flakes to 50 mL concentrated H2SO4 with constant stirring in an ice-water bath. Further, 3 g KMnO4 was gradually added to the graphite–H2SO4 mixture. Since the reaction was exothermic in nature, the temperature was maintained below 10 °C. The resultant suspension was constantly stirred for 25 minutes at 35 °C, followed by 15 minutes of sonication in an ultrasonic bath sonicator. After sonication, the reaction was stopped by the addition of deionised (DI) water. The pH of the suspension was adjusted to ∼6 by 1 M sodium hydroxide (NaOH), followed by sonication for 1 hour. L-Ascorbic acid (10 g) was dissolved in 100 mL DI water and the resultant solution was slowly added to the obtained suspension by maintaining the temperature at 35 °C. Reduction was performed by heating the solution at 95 °C for 1 hour in a hot water bath. Black precipitates were obtained after the completion of the reduction reaction. These precipitates were allowed to settle overnight, followed by a series of washings alternately using 1 M HCl and DI water along with filtrations until a supernatant with neutral pH was obtained. The pellet was freeze-dried to obtain a fine rGO powder. Further, peptide-functionalized nanoconjugate was synthesized by conjugating the peptide (DP1) with rGO by applying the covalent conjugation approach.41 During this protocol, 5 mg of finely powdered rGO was dissolved in 10 mL DI water. The rGO solution was sonicated at 20 kHz for 30 minutes. To prevent heat-induced damage, sonication was performed by placing the sonication vessel in an ice water bath (0 °C). The obtained solution was then centrifuged at 630g for 30 minutes to obtain a supernatant containing a clear solution with uniform-sized particles. In the next step, cross-linkers, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS), were successively added into the graphitic solution and sonicated for 1 hour to generate semi-stable NHS ester linkages with the graphitic derivatives. Then, the pH of the mixture was adjusted to 8 by using 1 mM sodium hydroxide (NaOH). The peptide (dissolved in PBS, 0.145 mM, 0.1 mL) was added to the obtained mixture, followed by incubation at 4 °C in the dark for 12 hours to enable cross-linking of the NHS esters with peptides. Unbound peptides were removed from the final solution via centrifugation at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 750g for 30 minutes. The obtained sediment was then lyophilized and stored for future use. The number of peptide molecules bound with graphitic nanomaterials was calculated using eqn (1).
750g for 30 minutes. The obtained sediment was then lyophilized and stored for future use. The number of peptide molecules bound with graphitic nanomaterials was calculated using eqn (1).| Npep = 0.65(Rconjugate3 − RrGO3)/Rpep3 | (1) |
2.3. Characterization
The physico-chemical properties of the nanoconjugates were evaluated by the application of different analytical techniques. The chemical composition of the synthesized specimen was characterized by conducting Fourier-transform infrared (FT-IR) spectroscopy using a PerkinElmer-400 FT-IR spectrophotometer. The identification and determination of elemental composition were conducted by an FESEM (HITACHI SU8010) equipped energy dispersive X-ray spectrophotometer (EDS). Morphology, crystallographic structure and particle size of the synthesized samples were evaluated using JEOL JEM-2100 Plus high-resolution transmission electron microscopy (HR-TEM). Zeta potential was determined using Malvern Nano-S90 Zetasizer. Wetting properties were evaluated by conducting water contact angle measurements using a KRÜSS-DSA25E drop shape analyser.2.4. In vitro hemocompatibility assessment
The hemolytic assay of rGO and rGO-DP1 was carried out according to a previously reported protocol.42 Hemocompatibility of the specimen was assessed by evaluating hemolytic toxicity towards murine erythrocytes. Fresh blood from mice was obtained in heparin-treated tubes. Further, erythrocytes were separated from blood by mixing the collected blood with PBS and centrifuged for 10 minutes at 3000 rpm at 5 °C. The collected erythrocytes were washed with PBS 4 times. Triton X-100 mixed with RBCs was used as a positive control, while PBS mixed with RBCs was used as a negative control. The synthesized formulations were added to the RBC suspension at different concentrations (2–512 μg mL−1) and incubated for 3 hours at 37 °C with gentle shaking. Following incubation, the samples were centrifuged at 10,016g for 3 minutes and the supernatants were carefully collected for measurements. The degree of hemolysis induced by the samples was determined by measuring the absorption of the supernatants at 540 nm using a UV spectrophotometer.33,40 The percent hemolysis was calculated using eqn (2).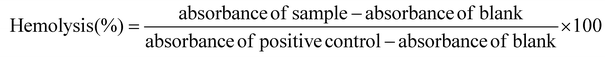 | (2) |
2.5. Cell viability assay
The effect of the synthesized graphitic material rGO and its DP1-functionalized nanoconjugate on the 3T3-L1 cells was evaluated by conducting the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. The cell lines were obtained from the National Centre for Cell Science, Pune, India. Cells were cultured and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) bovine serum, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin in a 5% carbon dioxide (CO2) incubator with 98% humidity at 37 °C. Before experimentation, the cells were harvested by trypsinization and seeded in a 96-well plate. Further, upon reaching 75–80% confluency, the cells were treated with the specimen at a concentration of 5–500 μg mL−1 for 24 hours at 37 °C in 5% CO2. After 24 hours, the cells were treated with MTT (5 mg ml−1) for a period of 4 hours. Then, cells were lysed with molecular-grade dimethyl sulfoxide (DMSO). Absorbance was taken at 570 nm and compared with the wells containing untreated cells. The percentage of cell viability was calculated using eqn (3).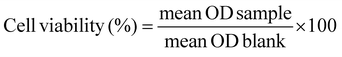 | (3) |
2.6. Determination of antibacterial and antibiofilm activity
The minimum inhibitory concentration (MIC) of rGO and rGO-DP1 was determined by the broth micro-dilution method using a previously described protocol.43 The bacterial cultures were inoculated in Luria–Bertani (LB) broth. The inoculated broth was incubated overnight at 37 °C. The following day, fresh LB broth was inoculated using the overnight culture and incubated at 37 °C to obtain cells in the mid-exponential growth phase. The respective bacterial cultures were diluted with sterile broth to obtain 105 CFU per mL. Using a multichannel pipette, sterile broth was dispensed into the wells of a microtiter plate. Stock solution of the test samples was prepared (1 mg mL−1). The specimens were further dispensed into the wells and serially diluted (two-folds) till the desired concentration (2–512 μg mL−1) was achieved. The standardized bacterial cultures were then added to each well and the plates were incubated at 37 °C for 24 hours in a static incubator. Wells with only sterile broth and broth-containing bacterial culture (without agent) served as negative and positive controls, respectively. Post incubation, absorbance values were recorded at 600 nm. To avoid any contamination, all the experimental work was performed in a biosafety level 2 (BSL-2) laboratory.2.7. Quantitative biofilm inhibition assay
Biofilm inhibition assay was performed in the presence of rGO, DP1 and rGO-DP1 at sub-MICs. 1 ml of the LB medium supplemented with sub-MICs of the specimen were inoculated with a diluted culture of P. aeruginosa (1 × 106 CFU per ml). The plate was incubated at 37 °C under static conditions. Sterile LB was used as a negative control, while PAO1 culture without any drug treatment served as a positive control. The media containing planktonic cells was discarded; wells were washed thrice with PBS (pH 7.2) and the adherent biofilms were stained with 0.1% crystal violet (CV) dye. After staining, the dye was removed and wells were washed thrice with PBS (pH 7.2) to remove excess CV. The plate was air-dried at 37 °C, followed by adding 33% glacial acetic acid to each well to elute the bound dye. Absorbance values were measured using an ELISA plate reader (Bio-Rad, Hercules, CA) at 595 nm. All sets were processed similarly for up to 7 days.442.8. Biofilm eradication assay
The overnight culture of PAO1 was diluted using sterile LB to attain 1 × 106 CFU per ml, and 200 ml of the adjusted culture was dispensed into microcentrifuge tubes to allow biofilm formation. After every 24 hours, the spent culture medium was discarded and fresh LB was replenished in the wells. Cultures were allowed to grow at 37 °C under static conditions for 1 day (young biofilm), 4 days (peak biofilm) and 7 days (mature biofilm). On completion of the designated time, preformed biofilms were challenged with sub-MICs of rGO, DP1 and rGO-DP1 for 24 hours at 37 °C. Tubes lacking the specimen served as a positive control, while sterile LB was used as a negative control. The contents were aspirated to remove any planktonic cells from the treated and untreated wells, followed by washing thrice with PBS. After washing, adherent biofilms were stained with 0.1% CV, followed by washing with PBS. The bound CV was then eluted from adherent biofilms using 33% acetic acid, and the absorbance values were recorded at 595 nm.45,462.9. Microscopic examination of P. aeruginosa biofilms
FESEM was employed to visually examine the biofilm formation in PAO1 following drug treatment. PAO1 biofilms were allowed to form over sterile coverslips placed in 5 ml of the LB medium supplemented with sub-MICs of rGO, DP1 and rGO-DP1 in a six-well culture dish at 37 °C for 4 days. The PAO1 culture without any drug specimen served as a positive control. After every 24 hours, the culture medium was discarded and fresh LB containing the specimen was added to the wells. Post incubation, the spent media was aspirated and coverslips were washed with PBS (pH 7.2) to remove any unbound cells. Peak-day biofilms were fixed with 2% glutaraldehyde for 2 hours at 37 °C, followed by three rounds of washing with PBS. Biofilms were then sequentially dehydrated using different concentrations of EtOH, including 30, 50, 70, 90 and 100%. Coverslips were air-dried, gold-coated and viewed under the field emission scanning electron microscope (FESEM; SU8010, Hitachi, Tokyo, Japan) to analyse PAO1 biofilms.2.10. Preparation of the peptide-functionalized rGO-based wound dressing
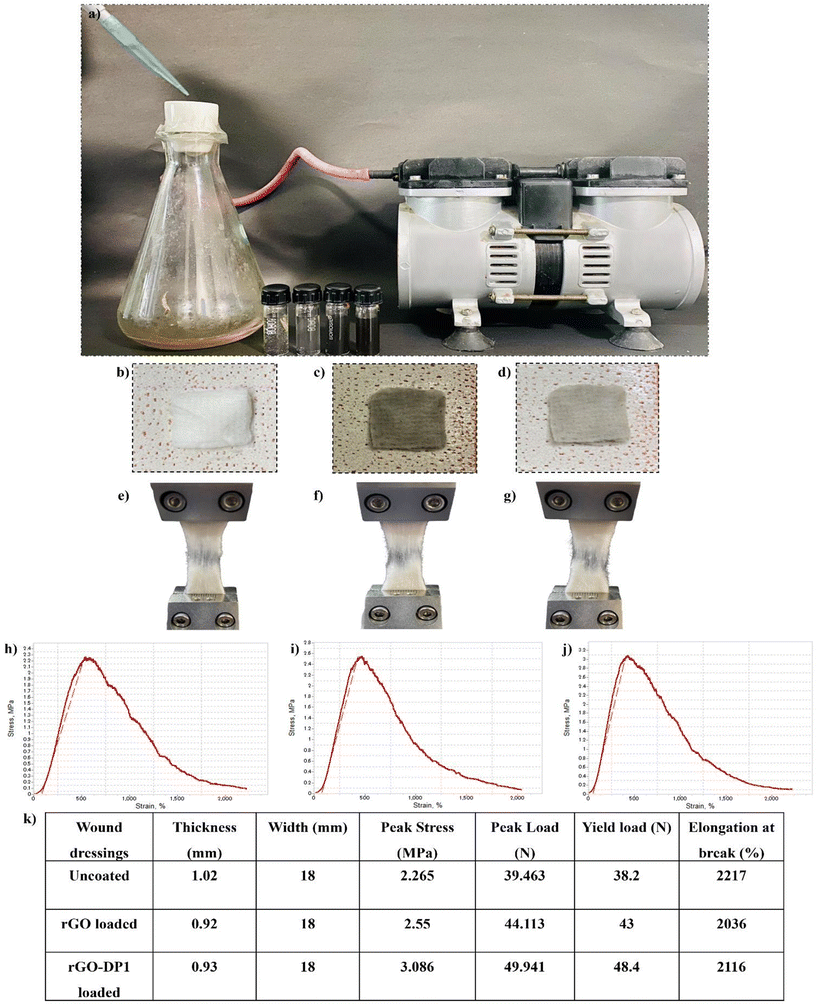
The peptide-functionalized rGO-based wound dressing was synthesized by passing 0.5 ml of the rGO-DP1 solution (1 mg mL−1) through a sterile cotton fabric (1 cm2). Fig. 1 displays a schematic illustration describing the green synthesis of the peptide-functionalized nanoconjugate-based wound dressing. The cotton substrate was placed on a porous ceramic crucible attached to a negative-pressure suction filter. The rGO-DP1 solution was slowly added to the cotton fabric until the complete solution passed through it. This filtration procedure was repeated until the solution was completely absorbed by the cotton fabric. Further, the rGO-DP1-loaded fabric was mounted on an elastic adhesive wound dressing.47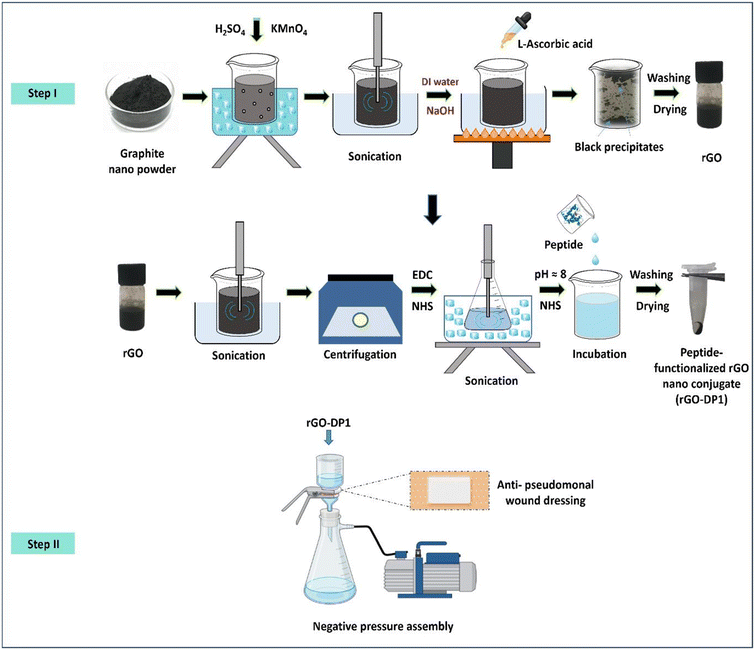 | ||
| Fig. 1 Schematic describing the two-step process for developing the rGO-DP1-loaded anti-pseudomonal wound dressing. | ||
2.11. Growth curve experiment
To conduct the growth curve experiment, an isolated colony of P. aeruginosa (ATCC 15692) was inoculated into 15 ml nutrient broth and incubated overnight. The following day, the optical density (OD) of this culture was measured using a UV-vis spectrophotometer. OD of the inoculum was adjusted by following the dilution formula (eqn (4).| (OD1)V1 = (OD2)V2 | (4) |
Where, OD1: initial OD of the primary culture after overnight incubation, V1: volume of the primary culture to be added to the secondary culture, OD2: desired OD of the inoculum (as a standard, this value was adjusted to 0.05), V2: volume of the secondary culture.
The calculated volume V1 was inoculated from the primary culture into the secondary culture, confirming the OD to be 0.05 at 600 nm. The OD of the secondary culture was checked at every 60 minute interval and recorded, followed by plotting a standard growth curve of the organism. Simultaneously, the culture was plated on cetrimide agar plates after every reading in order to calculate the bacterial load (IBSC-PUNANO-071-2020).
2.12. In vivo wound excision model animals
All animal experiments performed in the present study were compiled with the guidelines of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) and were approved by the Institutional Animal Ethical Committee (IAEC) of Panjab University, Chandigarh, India (PU/45/99/CPCSEA/IEAC/2021/467). To determine the in vivo antibacterial activity of the rGO-DP1-loaded wound dressing against P. aeruginosa, a wound infection model was established on female Balb/C mice at the age of 6–8 weeks, weighing between 20 to 30 g. Animals were procured from the Central Animal House facility of Panjab University, Chandigarh, India. They were housed in polypropylene cages bedded with rice husk at the animal house facility at the Department of Biophysics, Panjab University, Chandigarh, India. The animals were provided ad libitum access to clean drinking water and a standard animal pellet diet throughout the course of this experiment. Prior to the commencement of the experiment, animals were kept for acclimatization to experimental conditions for a period of 7 days.2.13 Experimental design
Animals were randomly divided into six experimental groups and each group contained 6 female BALB/c mice. The mice were anesthetized by ketamine (90 mg kg−1 of body weight) and xylazine (10 mg kg−1 of body weight) intraperitoneally as an analgesic before wound generation. The dorsal fur of the mice was removed with clippers and depilatory cream 24 hours prior to inducing the wound. A full-thickness excisional wound was made on the dorsal surface of each mouse by using a 6 mm biopsy punch. Further, the infection was established by administering P. aeruginosa topically at the site of the wound. Group I (G1) animals served as a control, which were given only the wound. Animals in group II (G2) were wounded with a wound punch and then the wound site was infected with P. aeruginosa. Group III (G3) animals were wounded, infected with bacteria P. aeruginosa, and the wound was treated by the application of rGO-loaded wound dressing. Animals belonging to experimental group IV (G4) were wounded, infected and treated with DP1-loaded wound dressing. Group V (G5) mice were also wounded and infected with P. aeruginosa and treated by covering the wound with the rGO-DP1-loaded wound dressing. Animals of experimental group VI (G6) were wounded, infected and treated by covering the wound with antibiotic (azithromycin) loaded wound dressing. During the course of the study, animals were given standard laboratory feed and normal drinking water. After application of the wound dressing, treatment was given for a period of 7 days, following which the mice were sacrificed on the 8th day of the treatment.472.14. Wound infection
Five minutes after the wound was induced, the wounds were inoculated with 20 μl of P. aeruginosa (104 CFU per mL) using a micropipette. Mice were individually housed after the establishment of wounds and infection. They received buprenorphine (0.03 mg kg−1 of body weight) subcutaneously as an analgesic post-wound generation.482.15. Assessment of body weight
The body weights of the mice in all the groups were recorded throughout the study every day until sacrificing them. The daily changes in percent body weights were calculated and plotted.2.16. Evaluation of wound contraction rate
The rate of wound healing was determined by taking photographs of the wounds at days 0 and 8 to follow the visual wound healing through assessment of the diameter of the wound.49,50 The percentage of wound closure was evaluated using eqn (5). | (5) |
2.17. Qualitative evaluation of wound bioburden
To determine the amount of bacteria (P. aeruginosa) in the tissues, wounds from mice were collected and homogenized in sterile saline. Serially diluted aliquots of the homogenate were cultured on cetrimide agar and incubated for 18 hours at 37 °C. After incubation, the colonies were counted and CFU per g tissue was subsequently calculated.262.18. Histopathological assay
To evaluate granulation tissue deposition during the wound healing process, tissue samples were fixed in 10% formalin and embedded in paraffin wax. According to standard protocol, the vertical sections were stained with hematoxylin–eosin stains. The stained sections were viewed under a Leica DFC295 light microscope.512.19. Statistical analysis
Data have been reported as mean ± standard deviation (SD). To check the normal distribution of the data, Shapiro–Wilk test and Q–Q plot were performed. Skewness and kurtosis values were calculated. One-way ANOVA and two-way ANOVA were employed for the analysis of the data using (GraphPad Prism 6.01) software.3. Results and discussion
Development of the peptide-functionalized rGO-loaded wound dressing comprised two steps (Fig. 1). In the first step, the peptide-functionalized nanoconjugate, i.e., rGO-DP1, was synthesized by applying a “green-chemistry approach” wherein rGO was prepared by using L-ascorbic acid as a reducing agent.35 The peptide DP1 was conjugated with rGO using covalent interactions by the application of EDC and NHS as cross-linkers. In the second step, the antibacterial wound dressing was synthesized by loading rGO-DP1 on a sterile wound dressing material using a negative pressure vacuum pump assembly.3.1. Characterization of the rGO-DP1 nanoconjugate
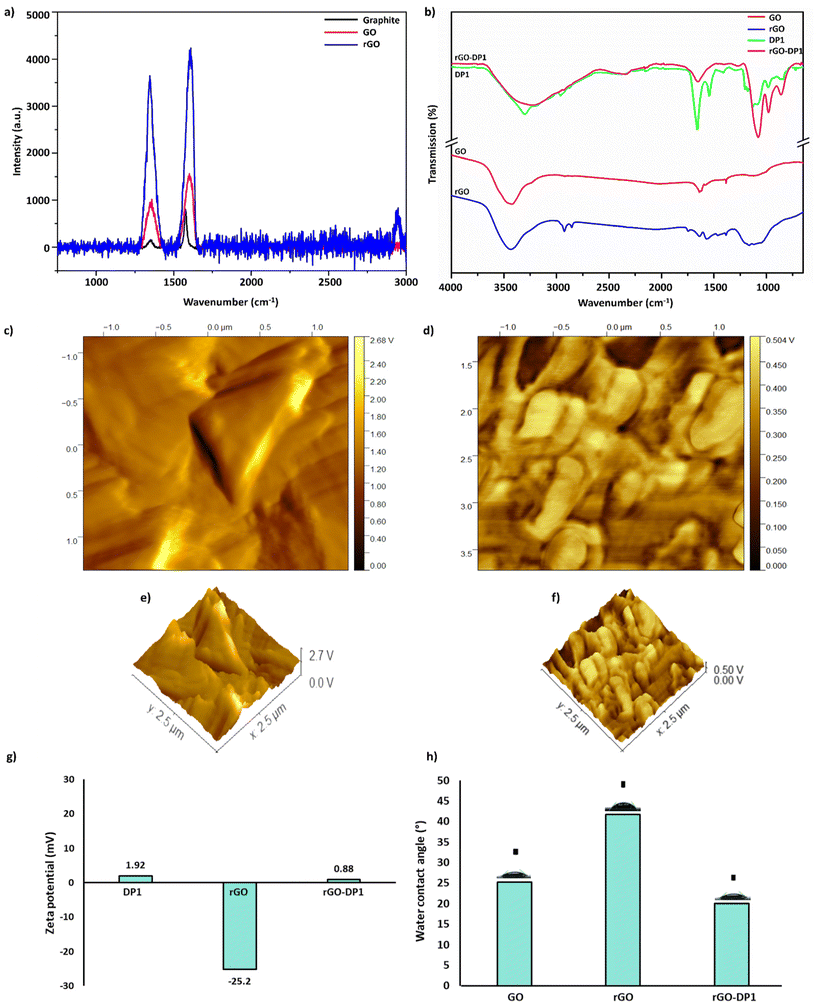
Raman spectroscopy was conducted to evaluate the physico-chemical properties of the graphitic materials and to confirm the reduction of GO to form rGO (Fig. 2a). It was observed that graphite displayed a D-band at 178 cm−1 and a G-band at 804 cm−1, respectively. The G-band at 2439 cm−1 and D-band at 2984 cm−1 were observed in the Raman spectra of GO. In comparison to GO, rGO spectra displayed a shift and intensification of the D band (7655 cm−1) and G-band at (8182 cm−1). In the rGO spectra, sharpness of the D peak was also observed, which is attributed to the removal of amorphous components during the reduction reaction. In addition to this, a less intense peak characteristic of the defect-activated band was observed at 2900 cm−1. The results suggested a successful reduction of GO to form rGO.1 Further, synthesis of the rGO-DP1 nanoconjugate was affirmed by conducting spectroscopy and microscopy-based analyses. Using eqn (1), it was estimated that approximately 37 peptide molecules were bound to the rGO sheet that was ∼50 nm in size. Comparative Fourier transform infrared (FT-IR) spectra of GO, rGO, DP1 and rGO-DP1 have been displayed in Fig. 2b. The FT-IR spectra of rGO-DP1 showed a broad peak between 3500–3100 cm−1, characteristic of N–H stretching and corresponding to secondary amide groups. It also represents the presence of intramolecular O–H stretching vibrations characteristic of graphitic materials. The presence of these peaks is an indicator of active binding between rGO and DP1.35,45 Energy-dispersive X-ray spectroscopy (EDS) and elemental mapping of rGO-DP1 revealed the presence of carbon (57.36%), oxygen (19.46%), chlorine (2.27%), sodium (5.88%) and nitrogen (15.02%) (ESI Fig. 1(a and b)).† Conjugation of DP1 with rGO is responsible for increasing the oxygen content, thereby decreasing the C/O ratio. Furthermore, the increased oxygen content of the conjugate can also be attributed to the amino acid side chains of the peptide.52 These results corroborated the FT-IR data where the presence of N–H, O–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and N–O spectral bands indicated the successful synthesis of peptide-functionalized rGO nanoconjugate.
C and N–O spectral bands indicated the successful synthesis of peptide-functionalized rGO nanoconjugate.
Atomic force microscopy (AFM) and high-resolution transmission electron microscopy (HR-TEM) provided a visual representation of rGO and the synthesized conjugate. AFM micrographs of rGO displayed the occurrence of flake-like morphology with the presence of sharp edges (Fig. 2c and e). Upon conjugation with the peptide, a change in morphology and height was observed (Fig. 2d and f). Peptide binding often results in the formation of clusters on the rGO surface. HR-TEM images of rGO-DP1 revealed a uniform deposition of peptide throughout the surface of the rGO sheet (Fig. S2a–c†). Conjugation of DP1 was affirmed by increased thickness and the presence of a honeycomb pattern on the rGO sheet. As shown in Fig. 2(g), the zeta potential of DP1, rGO and rGO-DP1 was observed to be −25.2 mV, +1.92 mV and +0.88 mV, respectively.
DP1 adorned a net positive charge of +1.92 mV and rGO possessed a negative charge of −25.2 mV. However, upon covalent interaction, the conjugate exhibited a positive charge of 0.88 mV. The change in charge upon conjugation indicated interfacial adhesion between the reduced graphitic derivative and DP1. A similar trend has been reported previously for rGO that depicted a net negative charge of −59.7 mV, but upon keratin conjugation, a positive relationship between the decrease in negative values occurred.53 The change in charge is attributed to the conjugation reaction that exposes the primary amines present on the peptide side chains, which subsequently results in the occurrence of variable degrees of charge depending on the length of the coupled peptide.54
Wettability is an important parameter that needs to be deliberated during the synthesis of biomaterials with antimicrobial properties. The wettability of a wound dressing material, as measured by the water contact angle, is a critical factor in the material's ability to absorb biomaterials, enable wound healing and facilitate the removal of the dressing from the wound site.55–57 GO and rGO displayed a water contact angle of 25° and 44.2°, respectively. However, upon coupling with DP1, the synthesized conjugate displayed a decrease in water contact angle to 20° (Fig. 2g). The smaller water contact angle values indicate increased hydrophilicity of the formulated conjugate. The increase in the conjugate's hydrophilic characteristic is attributed to the adsorption of the peptide on rGO.58 DP1 has a hydrophobicity value of 0.351, and the sum of hydropathy values obtained by conducting the grand average of hydropathicity index (GRAVY) affirms the hydrophilic characteristics of DP1.25 In addition to this, FT-IR analysis of the conjugate also revealed the presence of OH moieties upon conjugation, and these interactions may also be considered responsible for the decrease in water contact angle values.59
3.2. Antibacterial and antibiofilm properties
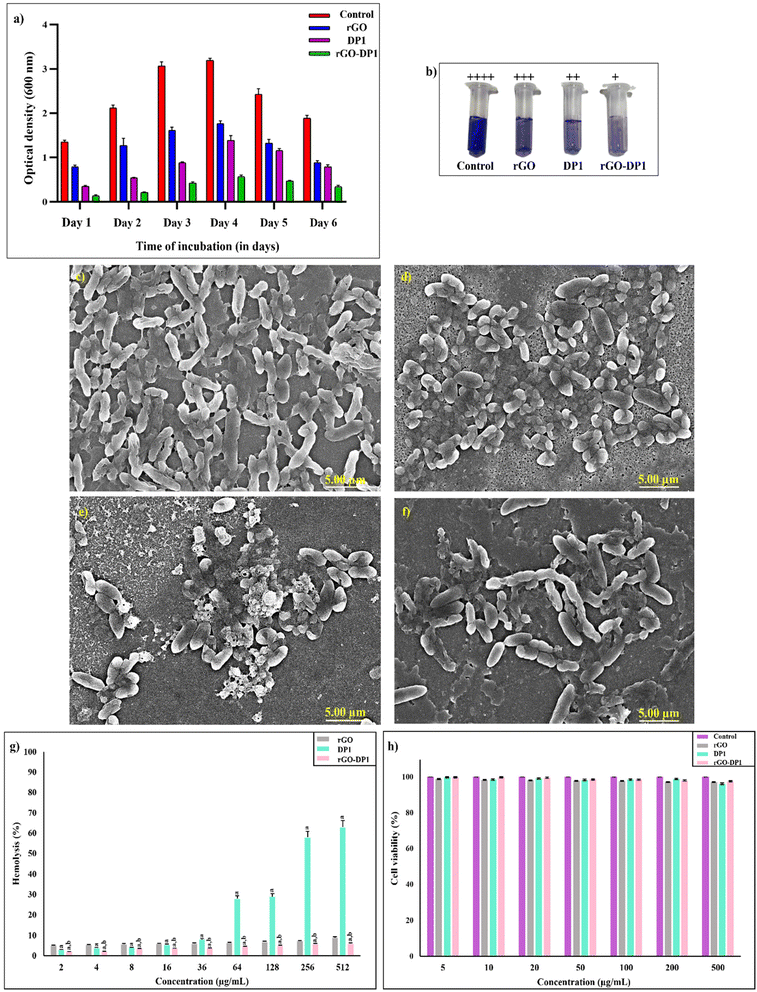
The antibacterial efficiency of the nanoconjugate rGO-DP1 was assessed against multidrug-resistant Pseudomonas aeruginosa (PAO1), a nosocomial bacterium infamous for causing infections in surgical wounds. MIC of rGO-DP1 against P. aeruginosa was observed to be 128 μg mL−1. The antibiofilm activity of DP1 functionalized rGO conjugate was evaluated by conducting a quantitative biofilm inhibition assay supplemented by FESEM-based microscopic analysis. Fig. 3(a) displays the time-dependent biofilm eradication after incubating the P. aeruginosa biofilm with DP1, rGO and rGO-DP1. Changes in the optical density were measured over a duration of 6 days to evaluate the effect of the incubated agents on the biofilm. The formation of a P. aeruginosa biofilm is majorly classified into 5 stages, where 2 hours after culture inoculation, P. aeruginosa enters stage I of the biofilm formation. During this stage, the bacterial cells initiate attachment towards the abiotic substrate. After 8 hours of incubation, the biofilm formation enters stage II, during which the attachment of cells becomes irreversible. During stage III, i.e., after 14 hours, microcolony formation takes place. During the initial 4 days, the biofilm remains in stage IV, where maturation of biofilm takes place and a three-dimensional community of bacterial cells is formed. After 5 days of incubation, the biofilm enters stage V and the bacterial cells are released from the mature biofilm with the aim to colonize surrounding surfaces.57 Therefore, by observing the optical density on different days, it was inferred that rGO-DP1 displayed a five-fold reduction in the P. aeruginosa biofilm. Biofilm inhibition was further assessed by conducting the FESEM analysis. As displayed in Fig. 3(c), untreated P. aeruginosa biofilms displayed a densely packed network of bacterial cells rooted in extracellular polymeric substances. rGO-treated P. aeruginosa induced alteration in the morphology of the biofilm by inhibiting the formation of extracellular polymeric substances.The micrograph of the rGO-DP1-treated P. aeruginosa displayed inflated bacterial cells with an apparent alteration of the cell wall along with negligible production of extracellular polymeric substances. There are numerous factors responsible for the antibiofilm activity exhibited by the rGO-DP1 nanoconjugate. Firstly, the role of the peptide DP1 is considered of utmost importance as amongst both the conjugated materials and it exhibits broad-spectrum antibacterial as well as antibiofilm activity. Upon conjugation with the large surface area of rGO, the number of peptides increased, which enhanced the activity of the formulated conjugate. Considering the significance of the graphitic material, it was observed that the morphology of rGO also plays an important factor in the inhibition of the P. aeruginosa biofilm. It has been reported previously that the sharp edges of rGO sheets pierce into the cell wall, inhibit cell growth and subsequently affect the rate of biofilm formation by exerting oxidative stress.35 Upon the application of rGO-DP1 as an antimicrobial and antibiofilm agent, combined defensive mechanisms involving antimicrobial activity imparted by coupled cationic DP1 along with the blade-like structure of rGO exhibited a lethal depolarization effect on the cell membrane, thereby rupturing the P. aeruginosa biofilm structure. These results established rGO-DP1 nanoconjugate as an effective antimicrobial agent with biofilm inhibitory properties as well.
3.3. In vitro biocompatibility assessment
Applicability of the nanoconjugate as an antibacterial agent necessitates comprehensive evaluation of its effect on erythrocytes and healthy cells. Hemocompatibility of rGO, DP1 and rGO-DP1 was evaluated by determining the hemolytic toxicity upon incubation with erythrocytes at different concentrations (2 to 512 μg mL−1) (Fig. 3g). It was observed that rGO displayed 90% hemocompatibility at all tested concentrations. DP1 displayed dose-dependent hemolytic toxicity,24–26 while the DP1-functionalized rGO conjugate displayed >90% hemocompatibility at all tested concentrations. It was interesting to note that rGO-DP1 showed 94% hemocompatibility even at the highest tested concentration of 512 μg mL−1. Overall, the synthesized nanoconjugate showed higher compatibility in comparison to DP1 and rGO. Further, the cytocompatibility of rGO and its peptide-functionalized conjugate was evaluated by cell viability studies of 3T3-L1 cells using a dye-based MTT assay (Fig. 3h). It turns out that no significant decrease in cell viability was observed. More than 95% of cells were alive upon incubation with rGO, DP1 and rGO-DP1, thereby indicating good cytocompatibility. These results prove the viability of using rGO-DP1 as a non-toxic material for wound dressing applications.3.4. Synthesis and mechanical properties of rGO-DP1-based antibacterial wound dressing
The antibacterial wound dressing was synthesized by loading rGO-DP1 on a 1 cm2 sterile wound dressing material (Fig. 4a–d). Fig. 4(a) displays the assembly used for the synthesis of wound dressing and the synthesized wound dressings (b–d). The effect of specimen loading on the mechanical properties was observed by conducting tensile strength studies (Fig. 4e–k). In comparison to the uncoated wound dressing, the highest peak load was observed in the rGO-DP1-loaded wound dressing. Peak load is referred to as the maximum stress a material can endure before being completely deformed. The significant increase in tensile strength is accredited to the homogenous dispersion of rGO into the porous wound dressing structure. In addition to this, rGO is a two-dimensional material with a large surface-to-volume ratio and high adsorption capacity. This property enables them to form strong interfacial interactions with polymer matrices, which results in the enhancement of mechanical properties. Owing to these properties, graphitic materials are considered as nanofillers that have the potential to improve the mechanical properties of polymeric materials even at low loading capacities.60 In addition to the current approach, rGO-DP1 can also be incorporated into the wound dressings using methods such as spray coating, dip coating, layer-by-layer assembly and electrospinning. Each technique has its own advantages and limitations. A common issue with these methods is the potential for excessive deposition of the nano-formulation. However, applying a negative pressure suction filter addresses this problem by removing unabsorbed formulations, thereby resulting in the formation of a uniformly distributed wound dressing.3.5. In vitro and in vivo antibacterial activity of the wound dressing
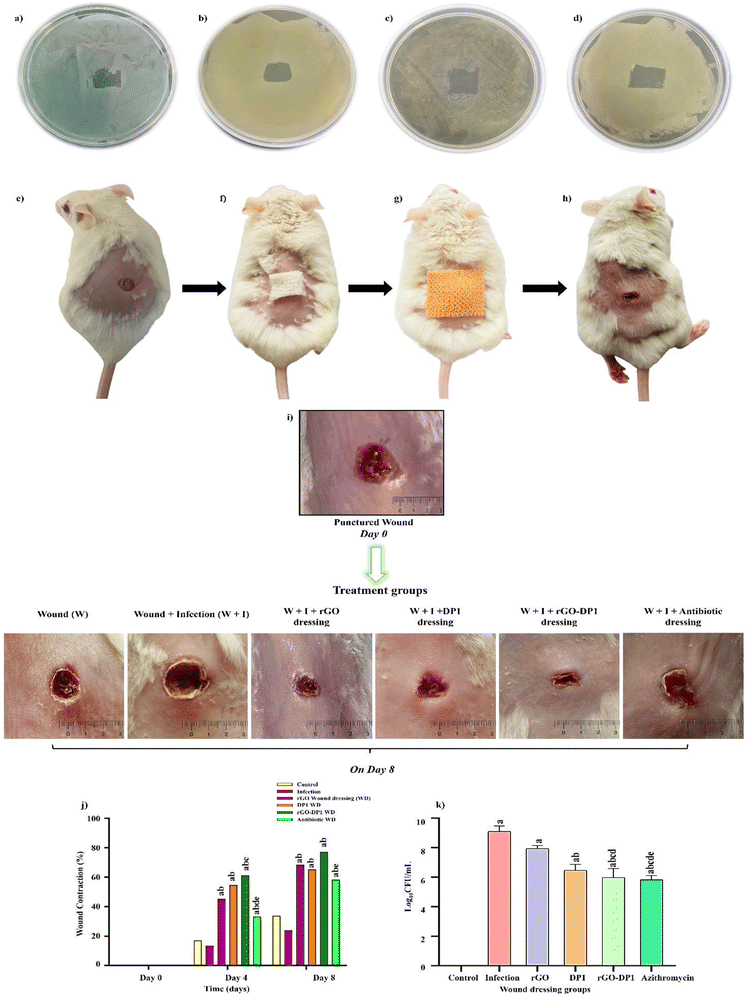
The antibacterial efficacy of the dressing against P. aeruginosa was assessed by conducting the zone of inhibition assay. A standard antibiotic, i.e., azithromycin, was also loaded on the sterile dressing and evaluated for its anti-pseudomonal efficacy. Bacterial inhibition was evaluated by determining the square zone formed by the wound dressings. As seen in Fig. 5(a–d) rGO, DP1, rGO-DP1 and azithromycin-loaded wound dressings displayed 6.2 mm, 8 mm, 8.4 mm and 9.5 mm inhibition area, respectively. Further, the wound dressings were evaluated for antibacterial efficacy in the bacteria-infected murine excision model. P. aeruginosa present in the exponential log phase was used to initiate bacterial infection in the murine model. Fig. 5(e–h) displays different stages of the in vivo model wherein after acclimatization (72 hours) of mice, the first step involved the depilation of the back hair and waiting for 24 hours. Next, anesthesia was given, and a wound of approximately 6 mm, resembling clinically relevant acute wounds, was made using a biopsy punch. Further, P. aeruginosa was inoculated on the wound site to induce infection and after 30 minutes, the wound dressing was applied. Throughout the course of the treatment, all mice were observed for any changes in behavior and weight. On the 8th day, after sacrificing, the tissues were collected from the wounds, homogenized and plated for evaluation of bacterial bioburden at the site of the wound. The effect of different wound dressings on the mice in the excision wound model was evaluated by determining the decrease in the wound area compared to day 0 on which the wound was perforated and infected and on the day of sacrifice, i.e., 8th day, to determine the wound closure percentage (Fig. 5i). It was observed that the rGO-DP1 dressing significantly reduced the wound size owing to the fast rate of re-epithelization in comparison to the P. aeruginosa-infected experimental group with rGO, DP1 or even the antibiotic-loaded wound dressing material. The lowest rate of wound healing was observed in the control group with no treatment or infection, followed by the P. aeruginosa infected group, antibiotic, peptide and rGO-DP1-loaded dressing group. This, in turn, led us to a very interesting observation regarding the wound-healing property of the peptide, which needs to be explored further. The wound contraction rate was determined by measuring the area of the wound on day 0 and day 8, respectively (Fig. 5j). The size of the healed area was compared to the size of the initial wound to assess the wound closure rate in all the experimental groups. The lowest wound contraction was observed in the infection group (24%), followed by the antibiotic group (58.3%) on the final day before surgery. In the rGO-DP1 and antibiotic dressing groups, wound contraction was observed to be 77.3% and 58.3%, respectively. DP1 and rGO-loaded wound dressings displayed promising wound healing characteristics by displaying 64% and 69% wound closure, respectively. The highest wound contraction was observed in the rGO-DP1 wound dressing with 78% closure. The healing power of DP1 has also been demonstrated in S. aureus-infected murine wound studies.26 Graphitic derivatives such as GO and rGO have also been shown to enhance tissue regeneration.61The wound scab was carefully plucked and examined for bacterial bioburden, with the intention to analyse the effect of an 8-day wound dressing course on P. aeruginosa (Fig. 5k). The control group did not show the presence of P. aeruginosa. However, in the infection group, highest presence of pathogen load was detected. Upon application of rGO and DP1-loaded wound dressing, bacterial load of 8 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 CFU and 7 log
10 CFU and 7 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 CFU was observed, respectively. The group treated with rGO-DP1-loaded wound dressing exhibited 6 log
10 CFU was observed, respectively. The group treated with rGO-DP1-loaded wound dressing exhibited 6 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 CFU bacterial bioburden. This reduction in bacterial load was comparable to that obtained with antibiotic-containing wound dressing. Thus, it may be inferred that the rGO-DP1-loaded wound dressing has the ability to combat drug-resistant bacterial infections, inhibit biofilm formation and accelerate wound healing.
10 CFU bacterial bioburden. This reduction in bacterial load was comparable to that obtained with antibiotic-containing wound dressing. Thus, it may be inferred that the rGO-DP1-loaded wound dressing has the ability to combat drug-resistant bacterial infections, inhibit biofilm formation and accelerate wound healing.
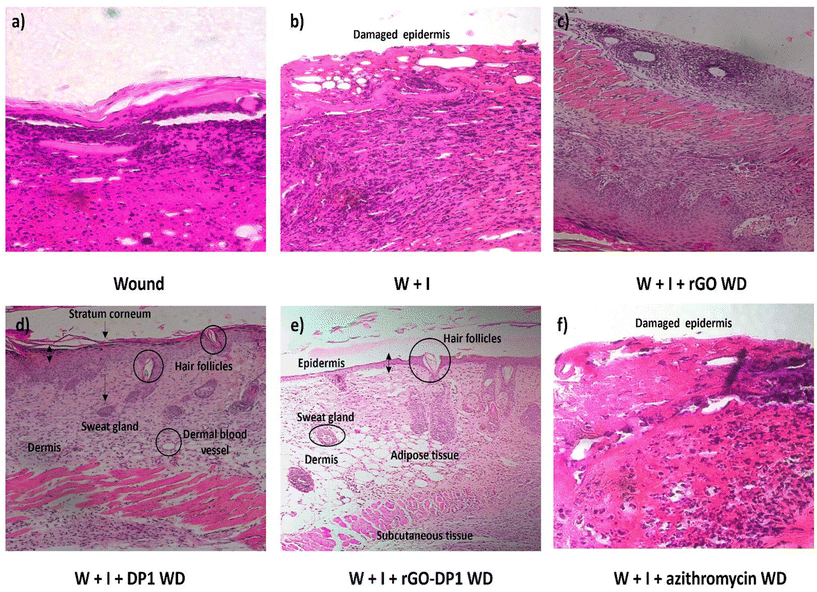
3.6. Histological analysis
Anti-pseudomonal activity and wound healing progression were further assessed using histopathological analysis. Upon completion of the 8th day, the wound tissues were carefully extracted and preserved in 10% formalin solution before conducting the hematoxylin and eosin (H&E) staining (Fig. 6). The control group exhibited normal skin histoarchitecture without any discernible abnormalities. In comparison to the control group, the P. aeruginosa-infected group displayed a damaged ulcerative surface. Since the infection was at an early stage, the occurrence of monocytes was also evident. They are usually present during the preliminary phase of wound infection.62 Further, the application of the rGO-loaded wound dressing resulted in the development of the stratum granulosum and stratum basale layers, which are part of the epidermis layer of the skin. Overall, epidermal remodelling was present in the rGO dressing group. These findings were consistent with the wound contraction rate pictographs, wherein the graphitic derivative displayed modest skin healing (Fig. 6c). The peptide and rGO-DP1-loaded wound dressing resulted in the regeneration of the epidermal layer representing wound remodelling (Fig. 6d and e). In comparison to the infected group, the regeneration of stratum corneum, stratum granulosum and stratum basale was observed in the peptide wound dressing group. In the basal layer, keratinocytes that are primarily responsible for the establishment of an epidermal water barrier by synthesizing and secreting phospholipids were also observed.The histology of skin treated with rGO-DP1-loaded wound dressing displayed healed skin structures with typical epithelization, reestablishment of adnexa and fibrosis within the epidermis layer. The presence of hair follicles is considered as a characteristic trait of wound repair and skin regeneration. Amongst rGO, DP1 and rGO-DP1-treated groups, the conjugate treated group showed faster re-epithelization of the skin tissue. However, the azithromycin-treated group did not show improvement in the wound histoarchitecture, even though it decreased the bacterial bioburden (Fig. 6f). Reduction in the bacterial bioburden in the rGO-DP1-loaded wound dressing may be attributed to a number of factors. Firstly, the antibacterial activity of DP1, even in its conjugate form, is attributed as a significant factor. In addition, the presence of rGO's nano-knife morphology along with oxygen-containing functionalities, contributes to the disruption of bacterial cell walls. Charge and hydrophilicity of the loaded nanoconjugate also plays a crucial role in reducing P. aeruginosa infestation. The hydrophilicity of the conjugate provides hydrophilic groups on the dressing material surface, which in turn enables enhanced interaction with the wound, thereby providing enhanced cellular compatibility. Upon comparing the bacterial bioburden data with the healing capacity, it may be stated that although the antibiotic was effective in reducing the infection caused by P. aeruginosa, it had no effect on the wound contraction rate.
4. Conclusion
For the very first time, a synthetically designed peptide-functionalized rGO nanoconjugate has been developed as an effective biomaterial for wound care. The antibacterial dressing was developed using a two-step process involving the synthesis of a rGO-DP1 nanoconjugate, followed by its incorporation into the wound dressing material. Comprehensive physico-chemical analyses, including Raman spectroscopy, Fourier-transform infrared spectroscopy and atomic force microscopy, confirmed the reduction of graphene oxide to reduced graphene oxide and subsequent covalent binding of the peptide to rGO. The synthesized conjugate displayed antibacterial properties and was effective against multidrug-resistant Pseudomonas aeruginosa as well. It also showed hemocompatibility, cell viability and antibiofilm potency, collectively highlighting its potential as a safe and effective biomaterial for wound disinfection and management. The rGO-DP1 was uniformly loaded onto the sterile wound dressing, resulting in improved mechanical strength and increased hydrophilicity. In vivo studies demonstrated a significant reduction in bacterial bioburden and enhanced wound closure compared to standard treatments, highlighting the wound healing potential of the wound dressing. Overall, the peptide functionalized-rGO biomaterial presents a promising strategy for developing sustainable wound dressings with the potential to open new avenues for the treatment of acute and chronic wounds infested with bacterial infections.Author contributions
Shubhi Joshi: writing – review & editing, writing – original draft, visualization, software, methodology, investigation, formal analysis, conceptualization. Panchali Barman: software, formal analysis, investigation. Mayank Maan: software, formal analysis, investigation. Hemant Goyal: methodology, investigation. Sheetal Sharma: methodology, validation. Rajesh Kumar: software, investigation, Gaurav Verma: software, investigation Avneet Saini: writing – review & editing, validation, supervision, resources, project administration, methodology, conceptualization.Data availability
Data will be made available on request.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
SJ and AS are thankful to the Department of Science and Technology (DST)-Women Scientist Scheme-B (WOS-B) (DST/WOS-B/HN-6/2021) for providing financial support to carry out this research work.References
- S. X. Chen, L. J. Zhang and R. L. Gallo, J. Invest. Dermatol., 2019, 139, 1002–1009 CrossRef CAS.
- X. Ding, Q. Tang, Z. Xu, Y. Xu, H. Zhang, D. Zheng, S. Wang, Q. Tan, J. Maitz, P. K. Maitz, S. Yin, Y. Wang and J. Chen, Burns Trauma, 2022, 10, tkac014 CrossRef PubMed.
- A. Rezaei and H. Ehtesabi, Mater. Today Chem., 2022, 24, 100910 CrossRef CAS.
- S. Homaeigohar and A. R. Boccaccini, Acta Biomater., 2020, 107, 25–49 CrossRef CAS.
- K. Razdan, J. Garcia-Lara, V. R. Sinha and K. K. Singh, Drug Discovery Today, 2022, 27, 2137–2150 CrossRef CAS PubMed.
- R. Jamaledin, C. K. Y. Yiu, E. N. Zare, L. N. Niu, R. Vecchione, G. Chen, Z. Gu, F. R. Tay and P. Makvandi, Adv. Mater., 2020, 32, 1–29 CrossRef.
- M. Bassetti, M. Peghin, N. Castaldo and D. R. Giacobbe, Expert Opin. Drug Saf., 2019, 18, 635–650 CrossRef.
- B. Short, A. Bakri, A. Baz, C. Williams, J. Brown and G. Ramage, Curr. Clin. Microbiol. Rep., 2023, 10, 9–16 CrossRef.
- C. Liao, X. Huang, Q. Wang, D. Yao and W. Lu, Front. Cell. Infect. Microbiol., 2022, 12, 926758 CrossRef CAS PubMed.
- K. H. Turner, J. Everett, U. Trivedi, K. P. Rumbaugh and M. Whiteley, PLoS Genet., 2014, 10(7), 1004518 CrossRef.
- S. Phan, C. H. Feng, R. Huang, Z. X. Lee, Y. Moua, O. J. Phung and J. R. Lenhard, Microorganisms, 2023, 11(5), 1210 CrossRef PubMed.
- V. L. Madhusudhan, Int. Wound J., 2016, 13, 1129–1136 CrossRef CAS PubMed.
- G. Wang, Z. Li, T. Li, S. Wang, L. Zheng, L. Zhang and P. Tang, BioMed Res. Int., 2018, 8(1), 41 Search PubMed.
- H. Cao, T. Xia, Y. Li, Z. Xu, S. Bougouffa, Y. K. Lo, V. B. Bajic, H. Luo, P. C. Y. Woo and A. Yan, Antimicrob. Agents Chemother., 2019, 63(4), 10–1128 CrossRef.
- E. L. Madaha, C. Mienie, H. K. Gonsu, R. N. Bughe, M. C. Fonkoua, W. F. Mbacham, K. A. Alayande, C. C. Bezuidenhout and C. N. Ateba, PLoS One, 2020, 15, 1–16 CrossRef PubMed.
- M. Hao, W. Ma, X. Dong, X. Li, F. Cheng and Y. Wang, BMC Microbiol., 2021, 21, 1–16 CrossRef PubMed.
- M. Khatami, R. S. Varma, N. Zafarnia, H. Yaghoobi, M. Sarani and V. G. Kumar, Sustainable Chem. Pharm., 2018, 10, 9–15 CrossRef.
- S. Alven, X. Nqoro and B. A. Aderibigbe, Polymers, 2020, 12, 1–25 CrossRef CAS; J. Xuan, W. Feng, J. Wang, R. Wang, B. Zhang, L. Bo, Z.-S. Chen, H. Yang and L. Sun, Drug Resistance Updates, 2023, 68, 100954 CrossRef PubMed.
- H. Shao, J. Zhou, X. Lin, Y. Zhou, Y. Xue, W. Hong, X. Lin, X. Jia and Y. Fan, Biomaterials, 2023, 300, 122183 CrossRef CAS.
- Z. Chen, C. Zhou, Y. Xu, K. Wen, J. Song, S. Bai, C. Wu, W. Huang, Q. Cai, K. Zhou, H. Wang, Y. Wang, X. Feng and Y. Bai, Biomaterials, 2021, 275, 120858 CrossRef CAS.
- J. Shi, C. Chen, D. Wang, Z. Wang and Y. Liu, Commun. Biol., 2022, 5, 1–14 CrossRef.
- J. Talapko, T. Meštrović, M. Juzbašić, M. Tomas, S. Erić, L. H. Aleksijević, S. Bekić, D. Schwarz, S. Matić, M. Neuberg and I. Škrlec, Antibiotics, 2022, 11(10), 1417 CrossRef CAS PubMed.
- S. Sharma, P. Barman, S. Joshi, S. Preet and A. Saini, Colloids Surf., B, 2022, 211, 112303 CrossRef CAS.
- P. Sharma, S. Sharma, S. Joshi, P. Barman and A. Bhatt, Sci. Rep., 2022, 12(1), 1–14 CrossRef PubMed.
- P. Barman, C. Sharma, S. Joshi, S. Sharma, M. Maan, P. Rishi and N. Singla, Probiotics Antimicrob. Proteins, 2023, 1–14 Search PubMed.
- M. Maan, H. Goyal, S. Joshi, P. Barman and S. Sharma, Life Sci., 2024, 340, 122458 CrossRef CAS.
- Z. Lai, X. Yuan, H. Chen, Y. Zhu, N. Dong and A. Shan, Biotechnol. Adv., 2022, 59, 107962 CrossRef CAS.
- V. Del Genio, R. Bellavita, A. Falanga, K. Hervé-Aubert, I. Chourpa and S. Galdiero, Pharmaceutics, 2022, 14, 1–21 Search PubMed.
- F. Xu, Y. Yuan, Y. Wang and Q. Yin, Biomed. Pharmacother., 2023, 158, 114117 CrossRef CAS PubMed.
- A. R. P. Silva, M. S. Guimaraes, J. Rabelo, L. H. Belen, C. J. Perecin, J. G. Farias, J. H. P. M. Santos and C. O. Rangel-Yagui, J. Mater. Chem. B, 2022, 10(19), 3587–3600 RSC.
- S. Atefyekta, E. Blomstrand, A. K. Rajasekharan, S. Svensson, M. Trobos, J. Hong, T. J. Webster, P. Thomsen and M. Andersson, ACS Biomater. Sci. Eng., 2021, 7, 1693–1702 CrossRef CAS PubMed.
- R. Weishaupt, J. N. Zünd, L. Heuberger, F. Zuber, G. Faccio, F. Robotti, A. Ferrari, G. Fortunato, Q. Ren, K. Maniura-Weber and A. G. Guex, Adv. Healthcare Mater., 2020, 9, 1–13 Search PubMed.
- S. Joshi, R. Siddiqui, P. Sharma, R. Kumar, G. Verma and A. Saini, Sci. Rep., 2020, 10, 1–11 CrossRef PubMed.
- W. J. Jeong, J. Bu, L. J. Kubiatowicz, S. S. Chen, Y. S. Kim and S. Hong, Nano Converg., 2018, 5, 1–18 CrossRef PubMed.
- I. Guzmán-Soto, C. McTiernan, M. Gonzalez-Gomez, A. Ross, K. Gupta, E. J. Suuronen, T. F. Mah, M. Griffith and E. I. Alarcon, iScience, 2021, 24(5), 102443 CrossRef PubMed.
- G. Brackman and T. Coenye, in Advances in Microbiology, Infectious Diseases and Public Health. Advances in Experimental Medicine and Biology, 2015, pp. 15–32 Search PubMed.
- E. K. Stuermer, I. Plattfaut, M. Dietrich, F. Brill, A. Kampe, V. Wiencke, A. Ulatowski, M. Geffken, J. D. Rembe, E. A. Naumova, S. E. Debus and R. Smeets, Front. Microbiol., 2021, 12, 1–10 Search PubMed.
- S. Abdolhosseinzadeh, H. Asgharzadeh and H. S. Kim, Sci. Rep., 2015, 5, 1–7 Search PubMed.
- H. Wang, Q. Zhang, X. Chu, T. Chen, J. Ge and R. Yu, Angew. Chem., Int. Ed., 2011, 50, 7065–7069 CrossRef CAS PubMed.
- K. H. Liao, Y. S. Lin, C. W. MacOsko and C. L. Haynes, ACS Appl. Mater. Interfaces, 2011, 3, 2607–2615 CrossRef CAS PubMed.
- I. Pal, D. Bhattacharyya, R. K. Kar, D. Zarena, A. Bhunia and H. S. Atreya, Sci. Rep., 2019, 9, 1–11 CrossRef CAS.
- S. Rajamani, R. Sandy, K. Kota, L. Lundh, G. Gomba, K. Recabo, A. Duplantier and R. G. Panchal, J. Microbiol. Methods, 2019, 159, 179–185 CrossRef CAS PubMed.
- S. Joshi, J. Chadha, K. Harjai, G. Verma and A. Saini, FlatChem, 2024, 44, 100626 CrossRef CAS.
- X. Chen, T. R. Thomsen, H. Winkler and Y. Xu, BMC Microbiol., 2020, 20, 1–11 CrossRef.
- H. Sun, N. Gao, K. Dong, J. Ren and X. Qu, ACS Nano, 2014, 8, 6202–6210 CrossRef CAS.
- M. Cao, W. Zhao, L. Wang, R. Li, H. Gong, Y. Zhang, H. Xu and J. R. Lu, ACS Appl. Mater. Interfaces, 2018, 10, 24937–24946 CrossRef CAS.
- X. Kang, P. Guan, C. Xiao, C. Liu, Y. Guan, Y. Lin, Y. Tian, K. Ren, Y. Huang, R. Fu, C. Ning, L. Fan, G. Tan and L. Zhou, Adv. Healthcare Mater., 2023, 12, 1–16 Search PubMed.
- C. F. Moreira, P. Cassini-Vieira, M. C. C. Canesso, M. Felipetto, H. Ranfley, M. M. Teixeira, J. R. Nicoli, F. S. Martins and L. S. Barcelos, Probiotics Antimicrob. Proteins, 2021, 13, 709–719 CrossRef CAS.
- M. Das, S. Mondal, R. Ghosh, S. Darbar, L. Roy, A. Kumar, D. Debasish and A. K. Mallick, J. Biomed. Mater. Res. A., 2024, 1–18 CAS.
- S. V. Otari, M. Kumar, M. Z. Anwar, N. D. Thorat, S. K. S. Patel, D. Lee, J. H. Lee, J. K. Lee, Y. C. Kang and L. Zhang, Sci. Rep., 2017, 7, 1–14 CrossRef CAS.
- E. Jimenez-Cervantes Amieva, R. Fuentes-Ramirez, A. L. Martinez-Hernandez, B. Millan-Chiu, L. M. Lopez-Marin, V. M. Castaño and C. Velasco-Santos, J. Alloys Compd., 2015, 643, S137–S143 CrossRef CAS.
- M. Q. Khan, D. Kharaghani, Sanaullah, A. Shahzad, Y. Saito, T. Yamamoto, H. Ogasawara and I. S. Kim, Polym. Test., 2019, 74, 39–44 CrossRef CAS.
- T. T. Li, Y. Zhong, H. K. Peng, H. T. Ren, H. Chen, J. H. Lin and C. W. Lou, J. Mater. Sci., 2021, 56, 4407–4419 CrossRef CAS.
- A. Awadhiya, S. Tyeb, K. Rathore and V. Verma, Eng. Life Sci., 2017, 17, 204–214 CrossRef CAS PubMed.
- Z. Li, Y. Wang, A. Kozbial, G. Shenoy, F. Zhou, R. McGinley, P. Ireland, B. Morganstein, A. Kunkel, S. P. Surwade, L. Li and H. Liu, Nat. Mater., 2013, 12, 925–931 CrossRef CAS PubMed.
- Y. Wang, W. C. Lee, K. K. Manga, P. K. Ang, J. Lu, Y. P. Liu, C. T. Lim and K. P. Loh, Adv. Mater., 2012, 24, 4285–4290 CrossRef CAS PubMed.
- T. Rasamiravaka, Q. Labtani, P. Duez and M. El Jaziri, BioMed Res. Int., 2015, 759348 Search PubMed.
- S. Joshi, D. Sharma, A. Saini and G. Verma, Prog. Org. Coat., 2024, 195, 108636 CrossRef CAS.
- J. Du and H. M. Cheng, Macromol. Chem. Phys., 2012, 213, 1060–1077 CrossRef CAS.
- K. E. Eckhart, S. J. Schmidt, F. A. Starvaggi, M. E. Wolf, W. M. Vickery and S. A. Sydlik, Regener. Eng. Transl. Med., 2021, 7, 460–484 CrossRef CAS.
- P. Rishi, T. Vashist, A. Sharma, A. Kaur, A. Kaur, N. Kaur, I. P. Kaur and R. Tewari, Pathog. Dis., 2018, 76, 1–9 Search PubMed.
- M. Zhang and C. Zhang, Regener. Biomater., 2023, 10 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr02233e |
| This journal is © The Royal Society of Chemistry 2024 |