DNA functionalized programmable hybrid biomaterials for targeted multiplexed applications
Nihal
Singh
 ,
Ankur
Singh
,
Ankur
Singh
 ,
Mukesh
Dhanka
and
Dhiraj
Bhatia
,
Mukesh
Dhanka
and
Dhiraj
Bhatia
 *
*
Discipline of Bioengineering, Indian Institute of Technology Gandhinagar, Gujarat, India 382355. E-mail: dhiraj.bhatia@iitgn.ac.in
First published on 14th June 2024
Abstract
With the advent of DNA nanotechnology, DNA-based biomaterials have emerged as a unique class of materials at the center of various biological advances. Owing to DNA's high modification capacity via programmable Watson–Crick base-pairing, DNA structures of desired design with increased complexity have been developed. However, the limited scalability, along with poor mechanical properties, high synthesis costs, and poor stability, reduced the adaptability of DNA-based materials to complex biological applications. DNA-based hybrid biomaterials were designed to overcome these limitations by conjugating DNA with functional materials. Today, DNA-based hybrid materials have attracted significant attention in biological engineering with broad application prospects in biomedicine, clinical diagnosis, and nanodevices. Here, we summarize the recent advances in DNA-based hybrid materials with an in-depth understanding of general molecular design principles, functionalities, and applications. Finally, the challenges and prospects associated with DNA-based hybrid materials are discussed at the end of this review.
1. Introduction
DNA, a material chosen by nature as a carrier of genetic information, is now considered an important structural material owing to its high modification capacity both at the genomic and at the structural level. Seeman's proposal of using DNA as a building block for generation of self-assembled nanostructures utilizing Watson–Crick base-pairing has seen significant advances over the past 40 years (Fig. 1).1,2 Today, with the advent of DNA nanotechnology with nanomolecular structural modifications using DNA origami, DNA is at the center of emerging research not only in fundamental biology but also in pharmaceutical and industrial fields.3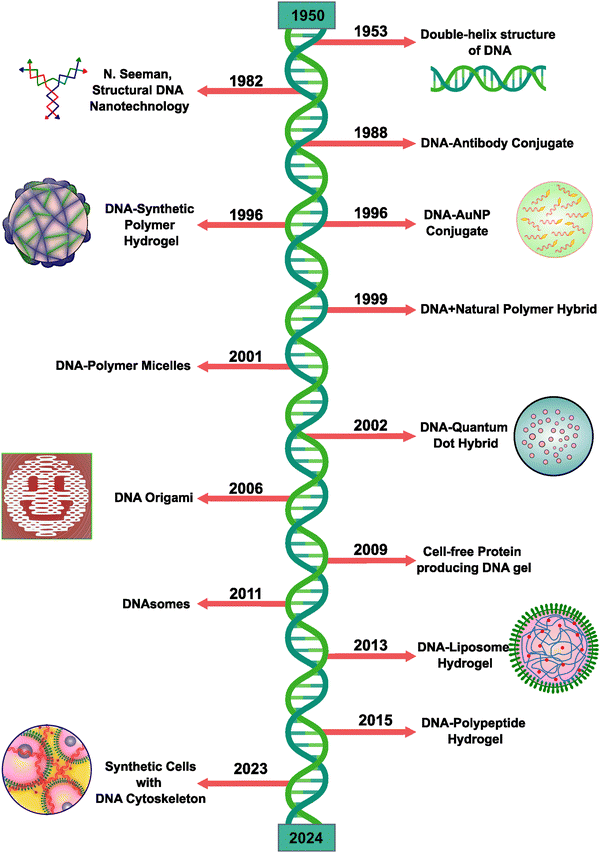 | ||
| Fig. 1 Development roadmap of the major milestones in DNA-based hybrid materials over the past several decades: N. Seeman, structural DNA nanotechnology;4 DNA–antibody conjugate;5 DNA–synthetic polymer hydrogel;6 DNA–AuNP conjugate;7 DNA–natural polymer hybrid;8 DNA–polymer micelles;9 DNA–quantum dot hybrid;10 DNA origami;11 cell-free protein-producing DNA gel;12 DNAsomes;13 DNA–liposome hydrogel;14 DNA–polypeptide hydrogel;15 and synthetic cells with the DNA cytoskeleton.16 | ||
The ease of synthesis of a desired DNA sequence, formation of superstructures such as i-motifs, triple helices, and quadruplexes along with various 2D and 3D structures of any desired shape utilizing simple DNA hybridization, and site-specific modification of the DNA strand by various enzymes have made DNA an attractive material for various biological applications.17,18 However, although DNA-based materials are highly designable, they are prone to hydrolysis and enzymatic degradation.19 Apart from their limited stability in living systems, their limited scalability, immunogenicity, and high synthesis cost make DNA-based materials unattractive candidates for clinical therapeutics.20,21 Today, extensive research is being done towards the development of biomaterials that can not only act as scaffolds but also possess multiple functionalities such as superior physical properties, stimuli-responsiveness, higher adhesion strength, self-proliferative and differentiation potential, etc. Hence, to overcome the limitations associated with DNA and to introduce new functionalities not obtained by DNA alone, the researchers conjugated DNA with functional materials with the aim of developing hybrids, thus bringing the synergistic or contrasting functional properties of different components into one system.
DNA-based hybrid materials constructed by combining DNA with other materials, such as polymers, metallic nanoparticles, peptides, and inorganic compounds, have attracted significant attention in the field of biological engineering.22–24 Combining DNA with other materials confers additional functionalities to the already large repertoire of capabilities that DNA demonstrates.25–27 The advancement of DNA nanotechnology and the limitless possibility to combine DNA with a wide array of functional materials have opened a broad and innovative area of research toward developing novel multi-functional DNA-based hybrid materials. Although the development of functional DNA-based hybrid materials is in a dynamic state, there is an unmet need for a comprehensive and timely review of functional DNA-based hybrid materials right now.
We provide here a comprehensive introduction to the current status of the functional DNA-based hybrid materials with a specific focus on: first, the present preparation strategies of the DNA-based hybrid materials; second, the principle and design criterion for the introduction of different functionalities in pure DNA-based materials; third, the current application of functional DNA-based hybrid materials; and lastly, the challenges and prospects of the functional DNA-based hybrid materials. We hope this work will provide a comprehensive idea of the current state of functional DNA-based hybrid materials in research and contribute to further stimulating innovation and development in this area.
2. Preparation of DNA-based hybrid materials
Considering the variability introduced by the type of DNA used (ssDNA, dsDNA, linear, and cyclic) as well as by the chemical nature of hybrid materials (polymers, lipids, inorganic compounds, metals, and peptides), it is difficult to standardize any particular method for preparation of DNA-based hybrid materials. Subsequently, it is also challenging to classify DNA-based hybrid materials based on their mode of synthesis. Additionally, due to the conjugation component of synthesis, some materials are developed by combining two or more methods. Therefore, in this review, we have broadly classified the synthesis of DNA-based hybrid materials based on the crosslinking method used to conjugate DNA and the hybrid materials. Two crosslinking methods for preparing DNA-based hybrid materials presented in this work are (i) physical crosslinking and (ii) chemical crosslinking.2.1 Physical crosslinking
In the physical crosslinking method, DNA and the hybrid materials are combined to form functional biohybrids through non-covalent physical interactions such as hydrophobic interactions, ionic interactions, electrostatic interactions, hydrogen bonding, and physical entanglement.20,22,28 The commonly used methods for forming DNA biohybrids include self-assembly, electrostatic interactions, and pH-dependent complexation.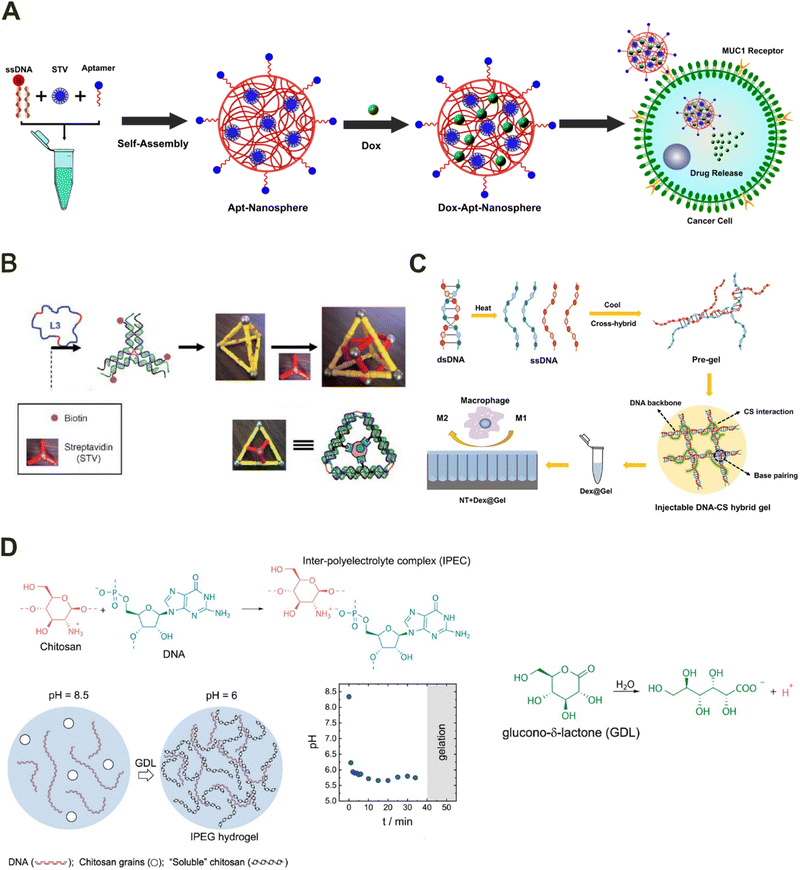 | ||
| Fig. 2 Different routes for DNA-based hybrid material formation by physical cross-linking. (A) Self-assembly of aptamer-conjugated hybrid nanospheres via step-growth polymerization and the biotin–streptavidin conjugate system. Drafted and adapted from ref. 32. Copyright 2022, American Chemical Society. (B) Self-assembly of DNA polyhedra to organize proteins in three-dimensional space. Reproduced with permission from ref. 33. Copyright 2012, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (C) DNA–chitosan injectable hydrogel synthesized via electrostatic interactions for delivery of dexamethasone (Dex) to induce macrophages M2 polarization. Reproduced with permission from ref.37. Copyright 2021, Elsevier. (D) DNA–chitosan hydrogel synthesized utilizing pH-dependent modulation. Reproduced with permission from ref.38. Copyright 2021 American Chemical Society. | ||
Recently utilizing a similar principle, Lee and colleagues developed a polymer/DNA complex for gene delivery.45 The developed polymer/DNA complex was made by complexing DNA with polyethylene glycol (PEG) grafted poly(L-lysine) (PLL). The developed complex demonstrated high transfection efficiency with lower cytotoxicity than other polymer/DNA complexes. Apart from utilizing electrostatically conjugated DNA hybrid materials for gene delivery, they have also been used to form matrix networks such as hydrogels. Chen and colleagues developed a DNA–chitosan injectable hydrogel for dexamethasone (Dex) delivery to induce macrophage M2 polarization.37 The hydrogel was synthesized by simply mixing chitosan with the DNA pre-gel formed through heating and cooling treatments (Fig. 2(C)). In another study, Singh and colleagues investigated the effect of DNA macrostructure on collagen fibrillogenesis and the effect of different mass fractions of DNA and collagen on DNA/collagen film formation.46 The films formed had a dense fibrous network with variable size fibrils formed due to the electrostatic interaction between DNA macrostructure and collagen. The films demonstrated aligned cellular growth along larger fibrils and promoted cellular uptake and neuronal differentiation. The study demonstrates that not only the type of DNA (linear, cyclic, ssDNA, and dsDNA) but also the shape, size, and 3D architecture of DNA-based materials can lead to variable interactions with other materials and can result in the development of new functionalities. Hence, understanding the effect of DNA on cationic materials can be used for the development of different types of hybrid materials. Overall, the formation of various biomaterials via electrostatic interaction with DNA has garnered significant attention in recent years primarily due to the ease of synthesis and robust mechanical properties of the developed network materials.
2.2 Chemical cross-linking
As a ubiquitous and robust molecule, DNA can form a wide variety of bonds/chemical interactions with either self- or non-self-molecules. The smallest functional unit of a DNA strand is a single nucleotide, which in turn is formed from three components: a nitrogen-containing cyclic base, a deoxyribose (pentose) sugar, and a phosphate group. The phosphate groups form a phosphor-diester linkage between the 3′ C-atom of the preceding sugar group and the 5′ C-atom of the former sugar group, forming a sugar-phosphate backbone and providing directionality to the DNA strands. The nitrogenous bases can be categorized into bicyclic purines (adenine and guanine) and homocyclic pyrimidines (cytosine and thymine). These nitrogenous bases extend outwards from the backbone and establish H-bonding with the intra- and inter-strand nitrogenous bases in an anti-parallel orientation, thereby stabilizing the duplex strands.1 This nitrogenous base-pairing is possible due to the unique molecular blueprint of ribonucleotides. The nucleic acids, because of their unique structural components, can form hydrogen bonds, covalent bonds (phosphodiester and thiol bonds), polar–non-polar interactions, metal–ligand complexes, and host–guest complexes with other polymers including peptides, amphiphiles, metal ions, lipids, and others.54,55 These bonds allow the DNA molecules to easily form physical (reversible) bonds and/or chemical (usually nonreversible) bonds with other biomolecules, opening new avenues for development of DNA-based hybrid biomaterials. The physical methods of assembly of DNA hybrid materials are easy to perform, require less time for synthesis, and often lead to poor mechano-physical properties.56,57 Most of the physically crosslinked DNA materials show a lack of functional diversity in terms of the availability of non-DNA functional moieties and are usually weak in terms of mechanical stiffness. To strengthen these major caveats of DNA-based biomaterials, a vast number of research groups have focused on chemically modifying the DNA molecule.The smallest customizable element of a covalently joined DNA hybrid composite material is called a block copolymer and is made up of one or more homopolymer subunits (blocks). In the case of hybrid DNA molecules, any polymer could be covalently coupled with the DNA backbone, thereby forming a DNA block copolymer. Utilizing this scheme various biomaterials can be chemically cross-linked with the DNA to form the intended DNA-block copolymer chains. These hybrid DNA block copolymers can be further used to form higher molecular complexes via non-covalent interactions. The DNA block copolymer can be synthesized by two schemes: the first scheme of modifying ssDNA units is by chemically conjugating organic compounds containing amino, thiol, carboxylic, and other groups in an aqueous phase.28 The amine group can be made to react with organic polymers having carboxylic groups (–COOH) such as PEG and pNIPAM (Fig. 3(A)).58–61 Additionally, the thiol group modified nucleotides can react with materials having either a 2-pyridyl disulfide group (–SH), a maleimide, or an acrylic acid group (undergoing Michael addition reaction).62,63 Rationally, the generation of water-soluble DNA block copolymers is actively promoted by the aqueous method of chemical modifications, but the limitation to this scheme lies in incorporating non-polar hydrophobic compounds into the DNA backbone.
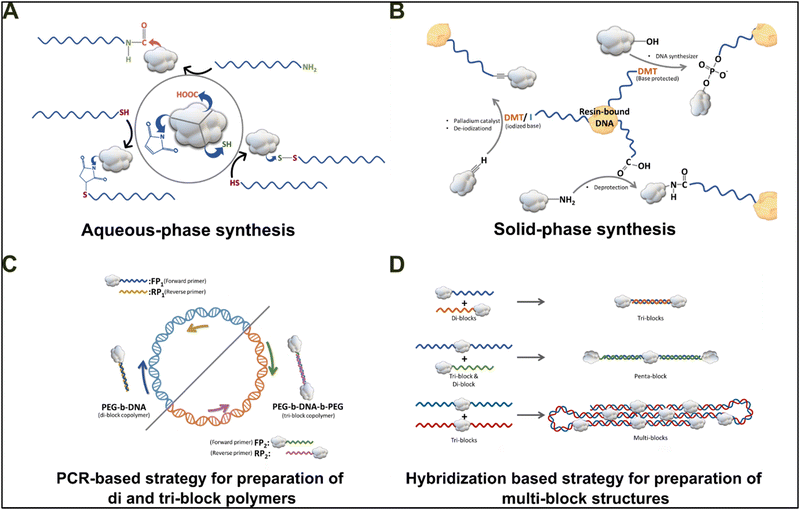 | ||
| Fig. 3 Different routes for DNA-based hybrid material formation via chemically crosslinking organic molecules with blocks of DNA. (A) Conjugation of amphiphilic or hydrophilic moieties to DNA blocks by aqueous phase synthesis methods. (B) Solid-state-based coupling schemes incorporating hydrophobic, organic, and amphiphilic molecules into DNA blocks. The lower panel shows efficient strategies for the addition/extension of block copolymer chains by (C) PCR and (D) HCR to form higher-order structures using DNA-hybridization strategies. Using these strategies, we can form DNA–hybrid biomaterials of any other non-DNA polymers, metals, micelles, lipids, amphiphiles, and others. Drafted and adapted from ref. 28. | ||
The second scheme, i.e., solid phase synthesis, offers an opportunity to incorporate an amphiphilic or a hydrophobic polymer into the DNA backbone by blocking the DNA strand on a solid substrate. In this scheme, the 3′-OH group of the ssDNA strand is anchored to an activated resin, while the 5′-phosphorylated end is accessible for undergoing detritylation by treatment with dimethoxytrityl (DMT). Then, the organic or amphiphilic moiety is modified to yield a phosphoramide terminal that can form a covalent linkage with the 5′-detritylated end of the resin-bound DNA strand. Next, the DNA–polymer blockchain is freed from the resin by treating the sample with concentrated ammonia (Fig. 3(B)).64,65 The DNA block copolymers synthesized by the solid phase synthesis method include DNA-b-polystyrene (DNA/PS), DNA-b-poly(propylene oxide) (DNA/PPO), and several other di-block polymer chains. To form a triblock assembly, the organic component can be modified to represent two phosphoramide linkage ends, thereby forming DNA–polymer–DNA blockchains.66 A carboxylated DNA strand is needed to form a block copolymer with amine-containing polybutadiene (PB) (Fig. 3(B)).67 Also, organic compounds with a terminal ethynyl group (such as poly(p-phenyleneethynylene), PPE) can be coupled with DNA by iodizing the 5′-detritylated nucleobase followed by the Sonogashira–Hagihara coupling reaction catalyzed by palladium (Fig. 3(B)).68
However, chemical methods of synthesis of block copolymer chains are limited by the degree of polymerization (∼50 nucleotides). Exceeding this limit, the specificity of the addition of residues deteriorates rapidly. To circumvent the above limitation, methods involving the use of the chemically synthesized DNA–block polymer chains to generate multiblock architectures using either the PCR (Fig. 3(C)) or hybridization chain reaction (Fig. 3(D)) have been developed.66,69–71 These methods incorporating the PCR and HCR provide an added level of synthesis for forming higher-order complexes using DNA-based hybridization.
3. Functional DNA-based hybrid materials
DNA-based hybrid materials represent an important class of materials with applications in various fields of science and technology. The core aim of developing a hybrid is to combine and bring different functional properties of two or more components into one system. The type of functionality introduced mainly depends on the end-stage application; for example, materials with adhesive functionality are preferred for wound healing, materials developed for minimally invasive surgery are usually injectable, and stimuli responsiveness is a critical functionality for drug delivery and sensor-based applications. Considering the flexibility of DNA at the structural and functional levels, DNA-based hybrid materials with different functionalities have been developed. In this section, we briefly describe several important functionalities that have been introduced in DNA-based materials for specific applications.3.1 Enhanced mechanical stiffness
The characterization of the stiffness of the DNA as a molecule is quite tough using conventional approaches. Nonetheless, researchers showed tremendous interest in estimating the elastic modulus of the DNA by stretching the molecules before they reach their breaking point. To estimate the mechanical properties of DNA, Nguyen and colleagues demonstrated a straightforward end-to-end pulling experiment using AFM and a 30-nucleotide-long DNA strand sandwiched between a gold surface and a streptavidin-coated AFM cantilever tip. The authors discovered that the mechanical stiffness of ssDNA strands (G′ = 18.0 ± 0.5 MPa) was much lower in comparison to that of dsDNA strands (G′ = 55.2 ± 3.2 MPa).72 The results were obvious as the ssDNA strand lacks stability provided by sequence-based hybridization, as compared to dsDNA. Building up on this logic, the DNA strands utilized in DNA origami structures may vary in their mechanical stiffness ranges, depending upon sequence content (GC%), the number of base pairs in the strands, stacking of the DNA strands (parallel or antiparallel), twist during the stacking of the strands, etc. In another study to determine the mechanical properties of DNA, Rief and colleagues calculated the unzipping force needed to separate DNA duplexes. The authors identified that the force needed to unzip a poly(dG–dC) duplex was around 20 pN, whereas the force required to unzip the poly(dA–dT) strands was half of the former.73 Another research group shed light on the mechanical stiffness of the DNA origami structures in 2016. They tethered various origami structures between a gold substrate and a cantilever tip in either horizontal or longitudinal orientations and reported the dissociation forces needed to disassemble the various DNA origami structures (DNA nanotiles (28 pN), nanopyramids (35 pN), nanotubes (8 columns −49.4 pN and 6 columns −48.5 pN).74,75 In summary, we can infer that the GC% content, length of the DNA strands, alignment of these strands in 3D, the number of Holliday junctions, etc. enhance the stiffness of DNA-based structures. However, utilization of DNA-based materials for applications such as tissue engineering would require higher mechanical strength, which can be achieved by conjugating the DNA-based materials with polymers having high mechanical strength.Pure DNA-based materials offer programmability, reproducibility, and biological inertness; however, the inherent tensile strength of DNA-based materials pales in comparison with other synthetic/natural polymers.57 Linear, unbranched strands of DNA in the hydrogels make them mechanically weak as compared to the similar-sized branched polymeric hydrogels. Also, pure-DNA hydrogels exhibit tensile strength with the magnitude of 103 Pa, which is considerably lower than those of other polymeric hydrogels.56
Therefore, extensive research is being done towards the development of materials that exhibit high mechanical strength and have all the functionalities offered by DNA. The doping of other non-DNA polymers within DNA-based materials enables the formation of additional chemical and physical bonds, which help increase the mechanical stiffness of the DNA hybrid systems. These mechanically reinforced hybrid DNA materials can be aptly implemented in the fabrication of scaffolds for tissue engineering and 3D-tissue model systems.76 The added non-DNA polymers can either form interpenetrating networks within DNA scaffolds or form double network structures.
In principle, any polymer that may interact with DNA strands via polar interactions, van der Waals forces, ionic, H-bonding, or host–guest interactions may contribute towards an increment in the mechanical stiffness of the hybrid materials. The choice of polymers could be based on the putative application, stability, robustness, cytocompatibility, etc.
A classical approach utilized to modulate the mechanical strength of a polymeric hydrogel is the introduction of a second monomer that can polymerize within the existing polymer network to form a hybrid hydrogel with interpenetrating networks. Utilizing a similar principle, Cao and colleagues demonstrated the formation of a 3D interpenetrating DNA/AAm–BIS hydrogel system for cell culture and imaging (Fig. 4(A)).77 The authors first developed a DNA hydrogel system using Y-shaped DNA arms and crosslinking them utilizing a linker DNA. Next, a secondary network was developed by polymerization of homogenously diffused molecules of monomeric acrylamide (AAm) and bisacrylamide (BIS). The polymerization of AAm/BIS forms interpenetrating networks of a highly branched acrylamide gel within a preexisting DNA hydrogel. The subsequent addition of AAm/BIS polymers within DNA hydrogels increased the polymeric concentrations per unit area, thereby increasing the polymeric network density of the hybrid hydrogel and resulting in enhanced mechanical properties compared to the DNA hydrogel alone. The enhancement of the mechanical properties could also be attributed to an increase in non-covalent interactions between these two distinct polymers, forming a physically entangled, interpenetrating hybrid hydrogel system. Also, the hybrid interpenetrating network system was effective in trapping cells in their 3D orientations, which is necessary for studying cellular behavior in a 3D scaffold system.
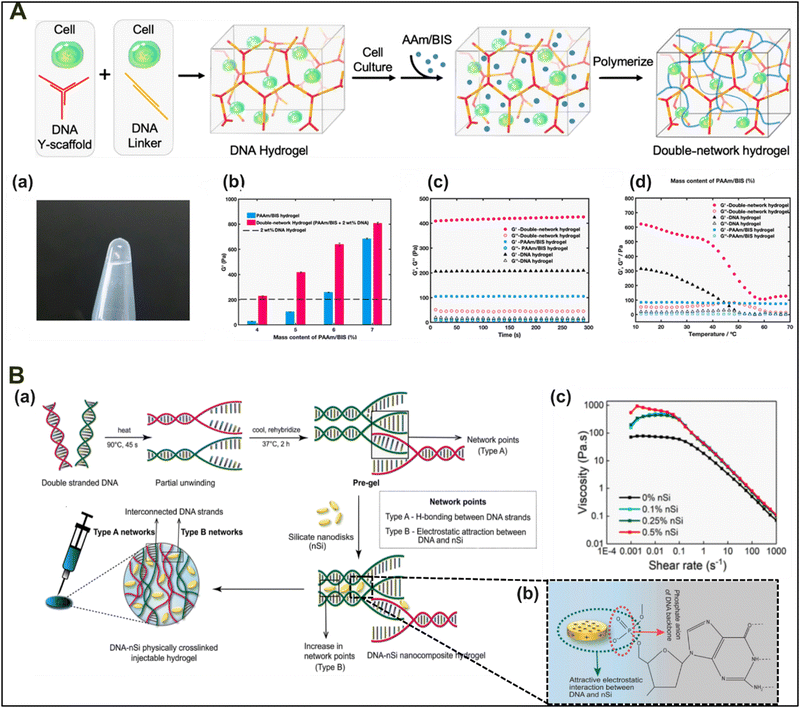 | ||
| Fig. 4 Schematic of an (A) interpenetrating DNA/PAAm network for 3D cell culture and imaging. (a) Gross image of a solidified interpenetrating DNA/PAAm–BIS hydrogel. (b) Modulus values of the gel at different percentages reflect mechanical strength enhancement by increasing polymeric concentrations. Rheology of the hydrogels was studied using (c) the time sweep test [at 1 Hz and 1% strain, at 25 °C] and (d) the temperature-ramp test. Reproduced with permission from ref. 77 Copyright 2020, American Chemical Society. (B) Schematic representation of (a) a double-network hybrid hydrogel formed by DNA–nanosilicate disks, (b) with the underlying mechanism of interaction and (c) demonstrating that nSi concentration is directly related to the hydrogel mechanical stability under introduced deforming stress. Reproduced with permission from ref. 78 Copyright 2018, American Chemical Society. | ||
In another study, Basu and colleagues demonstrated the fabrication of an injectable double network hydrogel system with a DNA network forming electrostatic interactions with 2D silicate nanodisks (nSi) (Fig. 4(B)).78 The authors demonstrated that the developed hybrid material enhanced mechanical resilience compared to the DNA hydrogel alone. The added mechanical resilience comes from the reversible electrostatic interactions of nSi disks with the negatively charged phosphate backbone of the DNA strands, which provides additional support to the network structure. The doping levels of nSi were in direct correlation with an increment in elasticity and stress endurance of this double-network hydrogel system. Increasing the concentrations of nSi within DNA hydrogels resulted in a concentration-dependent increase in the yield stress. The increment in the mechanical stiffness was attributed to the reinforcement of the hybrid hydrogel system by dispersed nSi disks. Similar studies were carried out, where gelatin hydrogels were doped with synthetic silicate disks, thereby improving the thermal and physiological stability of the hybrid gels by enhancing their mechanical stiffness.79
3.2 Promoting cellular adhesion
DNA-based materials can be used to enhance cellular adhesion by integrating copolymers with which cells adhere, namely fibronectin, hyaluronic acid, collagen, and others. Oftentimes, in order to make a bio-inspired hybrid material pro-adhesive, the goal is to select materials that are chemically inert with minimum cytotoxicity. Utilizing appropriate polymers with their unique properties brings synergistic effects. For example, conjugating hyaluronic acid, a major component of the bone tissue, covalently to the DNA strands results in the formation of a fibrous material with adhesive properties and mineralization similar to the bone tissue.80 Today, the aim of biomaterial science engineering is to fabricate bio-inspired structures with 3D architecture and functionality similar to native tissue, which can serve as templates for appropriate cellular growth. Towards this aim, an earlier research work performed by Faisal and colleagues reported a structurally tunable peptide-conjugated DNA-based matrix (ECMDP) for cellular adhesion, viability, and growth (Fig. 5(A)).81 Taking inspiration from collagen structure, they formed a DNA ribbon utilizing five strands with one exposed overhang for conjugating a human fibronectin-derived RGD peptide using biotin–streptavidin binding. The ECMDP showed adhesion levels compared to commercially available cell adherence substrates, including poly-L-lysine and fibronectin. Their findings indicate that pure DNA or protein-based materials are not sufficient enough to efficiently aid in cellular adhesion, whereas the ECMDP is able to. The authors identified that the cellular adhesion was dependent on the persistence length of the DNA, with decrease in the persistence length by increasing the length of DNA strands resulting in lower expression of focal adhesion kinase (FAK-a signal transduction protein involved in cellular development, migration, and tumor suppression) and localization of FOXO1a (transcription factor) levels within the nucleus and cytoplasm. The subsequent covalent binding of a ligand (RGD peptide) onto a pentameric repeating unit of DNA enables the structure to promote active binding with the cells cultured over it. The author identified that the spatial localization and firm anchorage of the RGD peptide provided by the ribbon backbone of the DNA/peptide hybrid material resulted in enhanced cellular adhesion. Overall, the controlled and precise fibrillar organization provided by the DNA, along with the incorporation of cellular adhesion functionality by the RGD peptide, resulted in the development of materials with increased cellular adhesiveness and bioactivity.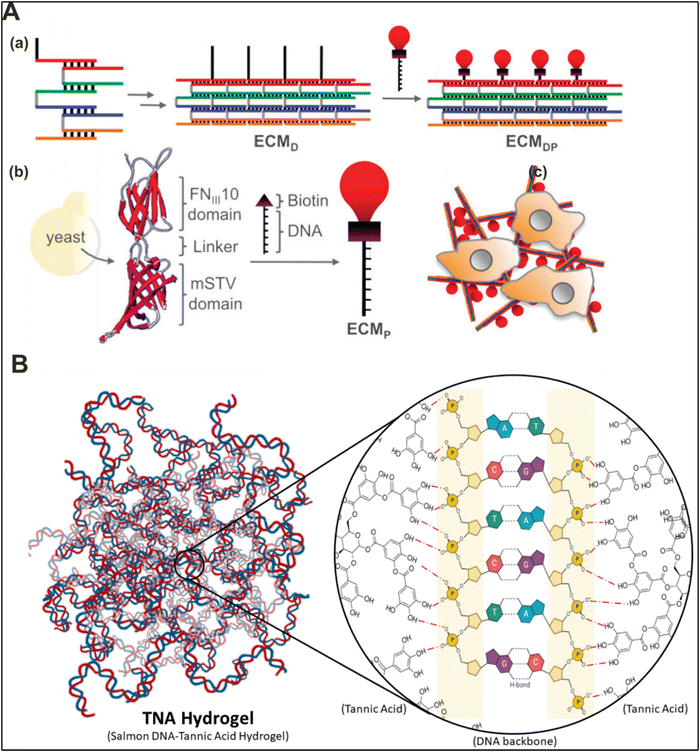 | ||
| Fig. 5 Adhesive functional DNA-based hybrid biomaterials. (A) Schematic representation of the formation of a collagen-inspired DNA ribbon scaffold (ECMDP) with a conjugated functionalized peptide for efficient cellular growth and adhesion. Reproduced with permission from ref. 81 Copyright 2010, American Chemical Society. (B) Schematic depicting the chemical interaction between tannic acid and DNA forming a TNA hydrogel. The ratio of DNA to TA is critical for inducing spontaneous gelation and improving mechanical stiffness. Drafted and adapted from ref. 82. | ||
DNA-hybrid materials have also been explored to develop strong bio-inspired adhesives for clinical applications. In 2015, Shin and colleagues demonstrated the formation of a hybrid hydrogel by mixing tannic acid (TA), a plant-derived polyphenolic compound, and DNA to form TNA hydrogels (TA + DNA) (Fig. 5(B)).82 TA is known to bind with macromolecules via non-covalent interactions, namely thrombin, elastin, and gelatin. The availability of multivalent hydroxyl groups (catechols and gallols) in TA facilitates efficient H-bond paired complexation with nucleic acids, thereby acting as a molecular glue for both the DNA and the proteins of interest.83–85 The addition of TA to DNA resulted in spontaneous gelation of the hydrogel at the stoichiometric ratio of 1.3 [DNA/TA (D/T)], whereas no gelation was observed at the D/T ratio of 2.6, and gels with higher TA constituents showed an increased stiffness. The TNA hydrogel (D/T = 1.3) had lower mechanical stiffness and, therefore, showed more extensibility (∼30 mm; 10.5 ± 0.2 kPa), as compared to the hydrogel with D/T = 0.9 (extension ∼15 mm; 33.9 ± 2.1 kPa). The work demonstrated that the TNA hydrogel can lift the subcutaneous tissue with a weight 1.6 times higher than the TNA hydrogel against gravity, demonstrating its strong adhesive properties. The extensibility might be caused by several factors, including the ultra-long strands of the DNA template (∼20 kbps) and the molecular-like interaction between TA and the DNA strands. Furthermore, the high programmability of DNA is a boon for exploring further growth in the development of TNA hydrogels.
3.3 Injectability
Self-assembled DNA strand materials consist mainly of non-covalent interactions between their neighboring strands. These weak yet abundant forces make the DNA hydrogels suitable for development of therapeutically relevant injectable hydrogel systems by virtue of their low viscosity and high water content with resistance against shearing stress. The clinical importance of injectable hybrid DNA hydrogels lies in minimally invasive procedures for target delivery, sustained release of the therapeutic compounds, improved bioavailability of the drugs, and safe and efficacious personalized therapy for patients. As a result, the realm of smart biomaterials for therapeutic and clinical applications often demands materials to be injectable (thixotropic). The presence of rigid chemical bonds, such as covalent bonds, makes the interconnections between the polymers stiff and thus possesses reduced elasticity. In contrast, the presence of physical bonds makes the polymers slide over each other upon application of a shearing force, without breakage of polymeric strands. For a hybrid biomaterial, the ratio of physical to chemical bonds should be optimized to make them not only injectable but also resilient to mechanical stress with high plasticity. Based on this rationale, various researchers have embarked on the journey to develop novel injectable platforms for therapeutic applications.In 2017, Gacacin and colleagues reported the preparation of a multifunctional protein–DNA hybrid hydrogel to slow down the process of osteoporosis by actively inhibiting the resorption by osteoclasts (Fig. 6(A)).86 The developed hydrogel included the clever design of incorporating a triblock template of DNA–PEG–denatured HSA (human serum albumin), where the DNA strands offer network formation and denatured HSA provides the site for branching using PEG as a linker between the two. The HSA protein is modified in a series of steps to yield a helical denatured PEGylated–polypeptide backbone, with 18 PEG molecules conjugated over the HSA surface. The PEG group on the HSA couples with the alkyne-modified ssDNA strands by following a Huisgen cycloaddition reaction. Furthermore, the hybrid hydrogel was also loaded with a fusion protein that can selectively inhibit Rho-signaling processes in osteoclasts, a repertoire of bone cells responsible for causing osteoporosis by resorbing bone minerals. The hydrogel formed using a triblock architecture demonstrated stress-induced deformation (>100%) as well as self-healing properties under reduced strain (0.1%). The thixotropic behavior of the developed injectable protein–DNA hydrogel makes it extremely desirable for clinical evaluation of osteoporosis.
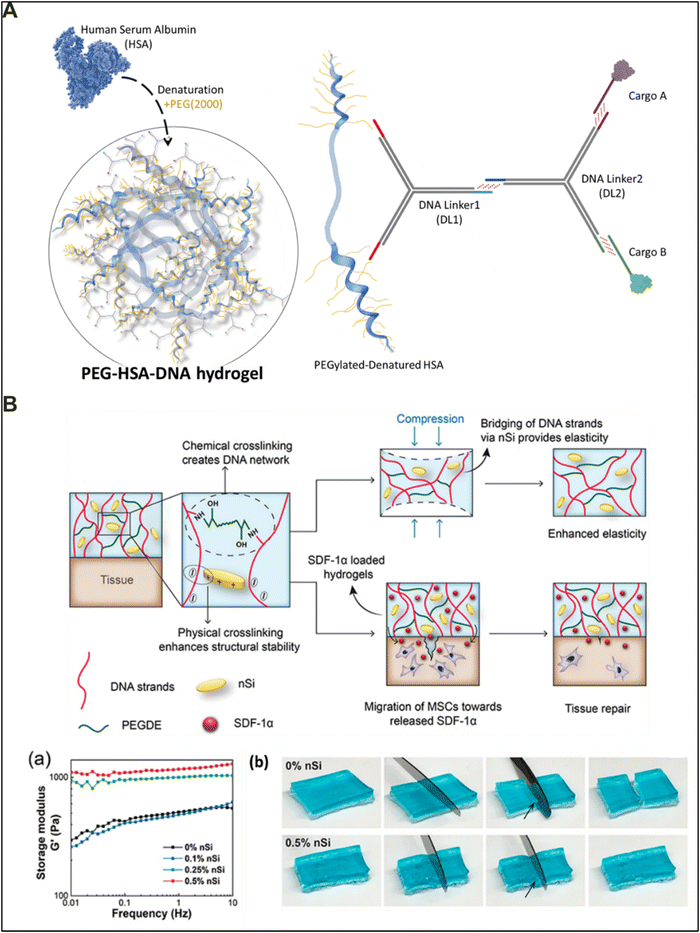 | ||
| Fig. 6 Injectable DNA-based hybrid materials. (A) Schematic representation of the PEG–HAS–DNA hydrogel synthesized by chemically conjugating denatured HAS with PEG, resulting in high branching nodes and target sites for cargo A and B associated DNA crosslinkers for drug delivery applications. Drafted and adapted from ref. 86. (B) Schematic representation of the DNA/nSi nanocomposite double interpenetrating network systems based on electrostatic interactions. The SDF-1α loaded nanocomposite hydrogel system mediates cellular migration and promotes the tissue repair response. (a) Rheological characterization showing the amplitude-sweep test performed at 0.01 Hz to 10 Hz. (b) Images showing self-healing properties of the nanocomposite material at varying nSi levels. Reproduced with permission from ref. 87. Copyright 2019, American Chemical Society. | ||
Another well-known example of an injectable hybrid DNA material includes the double cross-linked DNA/nSi network structures (Fig. 6(B)).87 Basu and colleagues formed a physically crosslinked DNA hydrogel in which, after adding cationic silicate nanodisks (nSi), the emergent electrostatic interaction between these components gave rise to an interpenetrating DNA/nSi hybrid nanocomposite hydrogel. Because of the non-covalent nature of the second interpenetrating network, it shows low viscosity and higher elasticity. The nanocomposite gel system can undergo shear-thinning without undergoing deformity, and the viscosity increases with an increase in nSi concentrations. The storage modulus values also increase with an increase in the nSi concentrations (Fig. 6(B)-a). Overall, DNA-based materials can be the principal materials for the development of injectable systems because of their low viscosity and stiffness. However, in recent years the introduction of a second polymer providing increased plasticity and a high fracture point has advanced the functionality of the DNA-based material to be utilized for various tissue engineering applications. Additionally, developing DNA-based hybrid materials with natural polymers like collagen, fibronectin, silk, and chitosan can create biomaterials that not only have high plasticity and injectability but also offer increased bioactivity, stability, and non-immunogenicity.
3.4 Stimuli responsiveness
The foundation of DNA nanotechnology lies upon various non-covalent and covalent interactions with the DNA strands, enabling them to form novel conserved domains with functionality. These include i-motifs, G-quadruplexes, cytosine-rich regions with affinity for silver ions, Holliday junctions, tetraplexes, and others. These 3D functional motifs impart selective responsiveness against pH, temperature, metal ions, templated DNA, and molecules of interest.88–93 However, apart from DNA's inherent ability to respond to various stimuli, the introduction of other functional agents has increased the functionality of an already large repertoire of DNA-based sensors. Today, a range of multi-stimuli-responsive DNA-based hybrid biomaterials that can be used for different biological applications have been developed.94–97 For example, the addition of an azobenzene moiety can render the DNA hydrogels photoresponsive, and simultaneously, the incorporation of aptamers can make the hybrid system responsive to multiple targeted stimuli through aptamer recognition.98–101 These modifications add to the ability of DNA-based materials to respond to various stimuli, which has opened doors for the design and fabrication of smart stimuli-responsive materials that can offer excellent clinical and therapeutic properties.Concerning another application of DNA-based hybrid materials, Huang and colleagues demonstrated the clever utilization of an aptamer-functionalized DNAzyme-based hydrogel system for detecting heavy metal ions quantitatively along with visual output.102 The developed material system demonstrated high sensitivity with the ability to correctly quantify Pb2+ ion concentration up to 10 nM. The strategy included the hybridization of two strands of DNA-coupled acrylamide chains using a lead-ion-specific aptamer as a cross-linker. The designed model entrapped gold–platinum core–shell nanoparticles (Au@PtNPs) within the DNA–acrylamide hydrogel system, using a lead-sensitive aptamer as a crosslinker. They placed the developed hydrogel system in a microfluidic volumetric graduated bar chart chip (V-chip) (Fig. 7(A)-a), with a colorful indicator dye and hydrogen peroxide in different wells of the V-chip. The hydrogel undergoes gel-to-sol conversion in the presence of Pb2+ ions and releases encapsulated Au@PtNPs in the vicinity, which catalyzes the conversion of H2O2 into O2 gas and water. The built-up O2 gas is diverted to pass through the dye well, thereby raising the level of the indicator dye in proportion to the O2 produced, which in turn depends on Pb2+ ion levels in the sample (Fig. 7(A)-a–c). The versatility of the developed system lies in its ability to program the biosensor to detect any molecule by simply swapping the target-specific DNAzyme from the system.
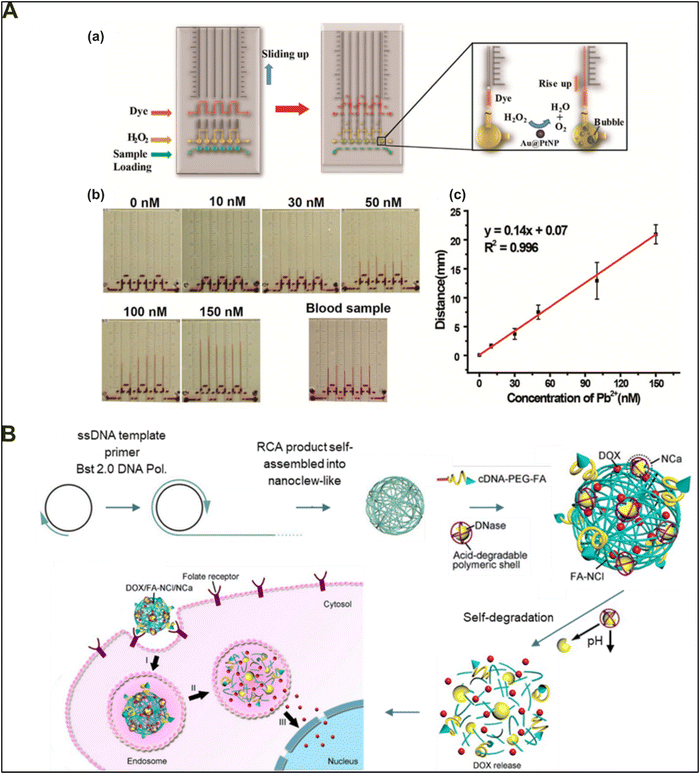 | ||
| Fig. 7 Stimuli-responsive DNA-based hybrid materials. (A) Illustration of a biosensor device based on stimuli-triggered release of Au@PtNPs within a DNAzyme cross-linked hydrogel for quantitative detection of lead ions on a V-chip device. (b) Working of the device in response to various levels of Pb2+ ions and (c) the linear relationship between the distance travelled by the ink dye and the Pb2+ concentration levels. Reproduced with permission from ref. 102. Copyright 2014, American Chemical Society. (B) Workflow of a pH-sensitive DNA nanoclew hydrogel system for tumor-targeted drug delivery applications. The released doxorubicin drug targets the nucleus and shows its intended therapeutic anti-cancer efficacy at a whopping IC50 value of 0.9 μM drug concentration. Reproduced with permission from ref. 103. Copyright 2014, American Chemical Society. | ||
In another work, Sun and colleagues demonstrated the advantages of a DNA-based hybrid material by engineering an intracellular pH-responsive self-degradable DNA hydrogel for anticancer drug delivery applications.103 The developed hydrogel constitutes three major components: a cocoon-like DNA hydrogel core, also known as a DNA nanoclew (NCl); a pH-responsive DNase I nanocapsule (NCa); and finally, a tumor-cell targeting folic acid (FA) ligand conjugated over a PEGylated complementary DNA (PEG-cDNA) to the DNA nanoclew (Fig. 7(B)). The NCl core contains GC-rich DNA strands for enhanced loading of doxorubicin and is synthesized by the RCA mechanism. The DNase I enzyme was encapsulated in an acid-responsive protein-based nanocapsule forming NCa. When the complex is administered to the tumor microenvironment, the folic acid ligand, a well-known biomarker of cancer cells, facilitates cellular entry through endosomal pathways. In the late endosomal cycle, the formation of an acidic endosome triggers the degradation of the pH-sensitive polymeric shell of NCa, thereby releasing the DNase I enzyme. The released DNase I enzyme chops off the NCl to release the intercalated dox for enhanced antitumor efficacy. The drug-delivery system was pre-programmed to deliver the anti-tumor cargo by sensing the intracellular environmental cue, which highlights the practical application of DNA hybrid materials (DNA/protein in this instance). The utilized strategy offers insights into the formulation and fabrication of derivatized programmable drug-delivery systems with the capabilities of sensing their macro-/micro-environments.
DNA-based hybrid materials have also been utilized for the development of stimuli-responsive robotic systems. In a recent work performed by Fern and colleagues, a DNA strand displacement-mediated soft robotic system was developed with the ability to swell upon applying controlled input.104 The authors developed a hydrogel system made up of polyacrylamide chains crosslinked using a DNA linker. Modulation of the length of DNA crosslinks using strand displacement was utilized to control the shape of the hydrogel. Also, the developed system utilized the catalytic amplification of input DNA strands to enable stimuli responsiveness at very low concentrations of signal. Overall, the authors demonstrated the potential application of DNA-based hybrid materials for the development of soft robotic systems with biomolecular circuits.
Another study performed by Hu and colleagues showed an amalgamation of various stimuli-responsive hybrid DNA materials that underwent stimuli-triggered reversible shape transformations (Fig. 8).105 In their study, they reported a bilayer sandwich design of DNA–acrylamide hybrid hydrogel systems with differentially-stimuli-responsive hydrogel layers within the sandwich. System I entails a pH-responsive TA T cross-linked hybrid DNA/acrylamide hydrogel system (hydrogel I). System II is slightly modified to incorporate N-isopropyl acrylamide (for thermal responsiveness) in place of acrylamide (hydrogel II) (Fig. 8(a)). A sandwich of hydrogel-I/hydrogel-II shows excellent acidic and thermo-responsive behavior in a reversible manner (Fig. 8(b)).
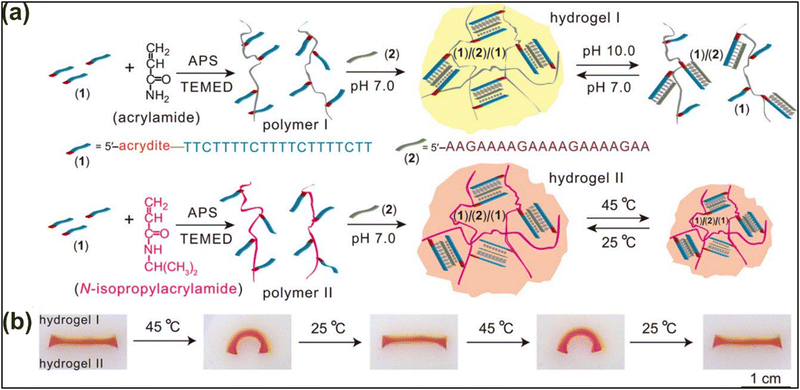 | ||
| Fig. 8 Stimuli-/thermo-responsive DNA hybrid hydrogels showing reversible and stimuli-triggered shape-switchable smart materials. (a) The preparation of hydrogel I and hydrogel II systems. Hydrogel I is formed by TA T-based crosslinking between acrylamide, strand 1 and strand 2; whereas hydrogel II shows TA T stabilized cross-linking between strand 1 polymerized with N-isopropylacrylamide and strand 2. (b) A bilayer composite of hydrogel I/II shows thermo-responsive and reversible shape-modulating properties. Reproduced with permission from ref. 105 Copyright 2016, American Chemical Society. | ||
4. Applications of DNA-based hybrid materials
As discussed above, DNA-based hybrid materials with different functionalities, including high mechanical strength, adhesion, injectability, stimuli responsiveness, etc., have been made in recent years. Most of the functional materials were designed for a specific application, with their functionality dictating their biological applications. The following section briefly describes the most recent applications of DNA-based hybrid materials with specific examples.4.1 Gene therapy
Gene therapy is emerging as a prime candidate for treating various diseases, including cytological, immunological, and genetic illnesses.106,107 The basic principle of gene therapy is to deliver the exogenous gene segments to the target cells. However, naked nucleic acid is readily degraded in the extracellular environment and cannot cross the biological cell membrane because of its negative charge. To overcome the barrier, various delivery agents were developed over the years.108,109 As one of the initial aims of developing DNA-based materials was to utilize the DNA employed as both the building block and functional component for gene therapy, DNA-based materials are considered the ideal carriers for the delivery of nucleic acid.110,111 However, to impart additional properties, including increased stability, gene regulation, stimuli responsiveness, enhanced targeting, and biosafety, DNA-based hybrid materials were developed.112One common approach towards gene therapy is the delivery of antisense oligonucleotides to suppress the corresponding protein expression by targeting respective heterogenous nuclear RNA (hnRNA) or mRNA. Towards this approach, Zhang and colleagues developed a novel siRNA delivery system via nucleic acid hybridization.113 DNA-g-PCL brushes were developed to serve as the framework of the nanogel with siRNA crosslinking the DNA-g-PCL brushes together (Fig. 9(A)). The developed nanogel effectively delivers siRNA to different cells both in vitro and in vivo without the need for transfection agents. Apart from suppressing gene expression via antisense oligonucleotide delivery, gene editing is another promising approach for gene therapy.114 Moreover, with the advent of CRISPR/Cas9 technology, limitations in regard to inefficient gene transfer and off-target expression have been severely reduced.115 For efficient gene therapy, the Cas9 protein and guide RNA (gRNA), two main components of the CRISPR/Cas system with gRNA recognizing the targeted DNA region of interest and directing Cas9 nuclease for editing, need to be delivered inside the cell. However, targeted delivery of Cas9 protein and guide RNA (gRNA) without degradation poses a significant challenge towards therapeutic development. DNA-based hybrid materials have been shown to be resistant to enzymatic degradation and allow targeted cargo delivery, making them appropriate materials for CRISPR/Cas9-based gene editing. Towards this aim, Ding and colleagues developed a non-cationic nanogel, wherein Cas9/sgRNA-loaded DNA-g-PCL brushes were crosslinked via linker DNA to form a nanogel via nucleic acid hybridization.116 The developed nanogel demonstrated excellent physiological stability with increased tolerance towards nuclease digestion. The nanogel was readily taken up by HeLA cells and actively suppressed the expression of EGFP, making it a promising material for targeted gene editing (Fig. 9(B)). In a similar work, Sun and colleagues delivered CRISPR/Cas9 using a yarn-like DNA nanoclew.117 The DNA nanoclew was formed via the self-assembly of palindromic sequences synthesized using rolling circle amplification (Fig. 9(C)). To enable endosomal escape, the DNA nanoclew was hybridized with the cationic polymer polyethyleneimine (PEI). The developed DNA nanoclew demonstrated significant disruption of EGFP expression both in vitro and in vivo. Overall, DNA-based hybrid materials have been extensively applied for gene therapy because of the substantial advantage offered via the use of highly programmable sequences, which can be utilized for targeting as well as for therapeutics while being the building block of the materials.
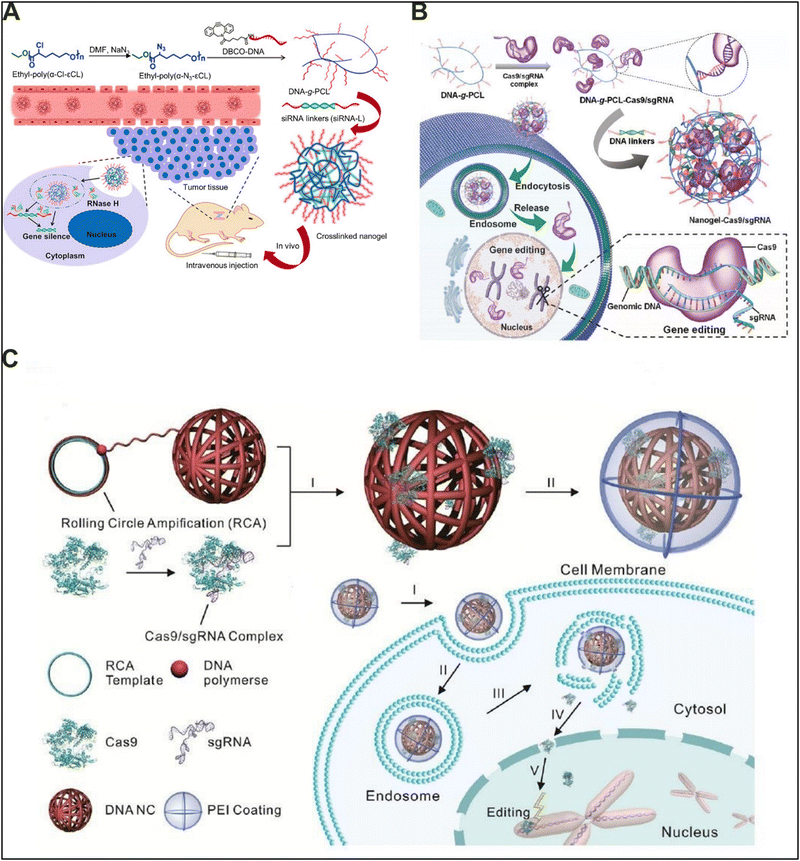 | ||
| Fig. 9 Gene therapy applications of DNA-based hybrid materials. (A) siRNA delivery via a crosslinked nanogel formed through nucleic acid hybridization between RNA and DNA. Reproduced with permission from ref. 113. Copyright 2018, John Wiley and Sons. (B) Non-cationic nanogel of DNA-g-PCL brushes for CRISPR/Cas9 delivery synthesized via nucleic acid hybridization between the linker and PCL grafted DNA. Reproduced with permission from ref. 116. Copyright 2019, Royal Society of Chemistry. (C) Yarn-like DNA nanoclew for delivery of CRISPR/Cas9, formed via self-assembly of palindromic sequences, synthesized using rolling circle amplification. Reproduced with permission from ref. 117. Copyright 2015, Wiley Journal. | ||
4.2 Drug delivery
Compared to conventional materials, DNA-based hybrid materials, with precise control over the structure using highly programmable DNA sequences with the flexibility to incorporate even the most inert chemical substance using appropriate hybrid materials, have surfaced as the ideal carriers for the delivery of both biological and chemical substances in recent years.118,119 Utilizing the highly programmable sequence, various DNA-based hybrid materials with high binding affinity and specificity towards a target molecule have been designed. In addition, considering their significant advantages over other drug carriers, extensive research has been done towards developing DNA-based hybrid materials with additional functionalities, including injectability, in situ gelation, and self-healing.78,120,121A simple hybrid design that has been extensively studied for the delivery of hydrophobic drugs involves hybridization of DNA with a hydrophobic polymer. The DNA acts as a shell and enables enhanced cellular uptake with active targeting, while the polymer acts as a hydrophobic core to carry hydrophobic drugs. Towards this design, one of the first works was demonstrated by Hermann and coworkers.122 They developed a DNA–polymer micelle utilizing the polypropylene oxide (PPO) polymer as the hydrophobic core to carry the anticancer drug doxorubicin (DOX). At the same time, DNA was used to form the shell with a sequence complementary to the folate receptor targeting unit (Fig. 10(A)). The hybrid material demonstrated the efficacy of the synergistic effect of active targeting with the chemotherapeutic drug in killing cancer cells. Since then, considerable research has utilized the above system for targeted drug delivery.
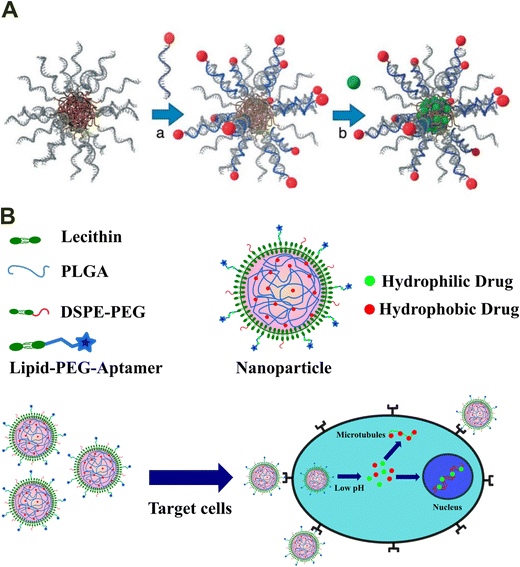 | ||
| Fig. 10 Drug delivery applications of DNA-based hybrid materials. (A) Doxorubicin (green dots) loaded DNA–polymer micelle synthesized using ssDNA (green) complementary to the folate receptor ligand (red dot) and the polypropylene oxide (PPO) (brown) polymer. Reproduced with permission from ref. 122. Copyright 2008, Wiley online library. (B) DNA–lipid–poly(lactide-co-glycolic acid) based hybrid nanoparticles for targeted co-delivery of two chemotherapeutic drugs, PTX and DOX, into cancer cells. Drafted and adapted from ref. 123. | ||
In recent years, significant research involving biomaterials utilizing aptamers as functional units for targeted drug delivery has been done. Aptamers are short ssDNAs capable of recognizing specific cellular receptors and thus offer a significant advantage for drug delivery into targeted cells. Tan and colleagues utilized sgc8 aptamers for the targeted delivery of lipid–poly(lactide-co-glycolic acid) hybrid nanoparticles into cancer cells.123 The dsDNA shell of the nanoparticle formed through the base pairing of the sgc8 aptamer with ssDNA attached with the PLGA core was used for DOX loading through intercalation. In contrast, the PLGA core encapsulated paclitaxel (PTX) via hydrophobic interactions (Fig. 10(B)). The developed system successfully co-delivered two anti-tumor drugs inside cancer cells.
Apart from delivering chemical compounds, DNA-based hybrid materials have also been utilized to deliver biological compounds, including nucleic acids, enzymes, and proteins, under controlled physiological conditions without alteration in their native properties. Apart from immobilizing or encapsulating proteins in the materials, DNA-based materials have also been utilized for cell-free protein synthesis, circumventing the need to immobilize and encapsulate proteins.124 Overall, DNA-based hybrid materials are promising drug delivery systems with critical characteristics, including high biocompatibility and loading efficiency, triggered stimuli responsiveness, controllable retention kinetics, and effective active targeting ability.
4.3 Sensing
Through precise Watson–Crick base pairing, DNA-based materials with specific size, shape, and conformation have been designed. In particular, due to their excellent sensitivity and specificity, DNA structure-based biosensing systems have attracted considerable attention with applications ranging from medical diagnosis to the quantification of heavy metal contamination in water.125–130 The development of DNA-based sensors, especially for detecting cationic pollutants, proteins, small molecules, and nucleic acid, has received considerable support from both industry and clinical research. However, the application of pure DNA sensor systems is heavily limited because of their instability and rapid enzymatic degradation in vivo.129,131 Moreover, pure DNA sensors lack intrinsic optical, electrical, and mechanical properties, significantly limiting their biological applications. To overcome these limitations, DNA hybridized with other materials or technologies was soon realized, resulting in the development of novel hybrid nanostructures and devices.One of the early and highly utilized design principles on which many DNA-based biosensors were based was nanobeacons.132–136 Nanobeacon technology is based on the conformational shift that DNA undergoes during appropriate base pairing. Briefly, a system can be designed with a fluorophore and quencher-associated DNA, which, upon hybridization, will bring the fluorophore and the quencher close to each other and thus will result in no fluorescence. However, dehybridization will result in a signal readout. Mirkin and colleagues utilized this principle to design a novel oligonucleotide-modified gold nanoparticle to detect specific mRNA in cells.137 They utilized the excellent quenching properties of gold nanoparticles and higher cellular uptake kinetics of the DNA oligonucleotide to develop a sensor with high signal output, low background fluorescence, and excellent sensitivity towards mRNA (Fig. 11(A)). Over the years, due to the simplicity and sensitivity of nanobeacon technology, DNA-based sensors have been heavily utilized for the detection of nucleic acids inside the cells. However, the extraordinary recognition ability of DNA-based sensors is not limited to nucleic acids only, but they have also been utilized for detection of various macromolecules, small molecules, and cationic pollutants.138–140
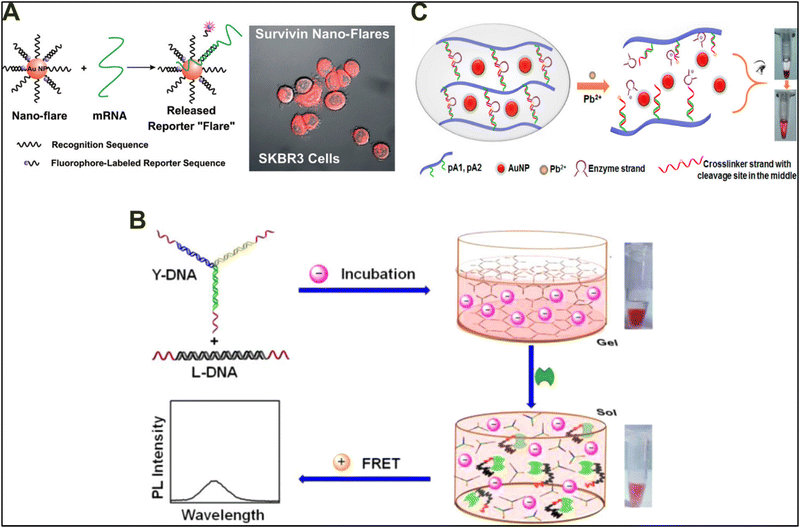 | ||
| Fig. 11 Biosensing applications of DNA-based hybrid materials. (A) Oligonucleotide-modified gold nanoparticles functionalized with an mRNA recognition sequence, capable of recognizing and reporting the presence of specific mRNA within cells. Reproduced with permission from ref. 137. Copyright 2007, American Chemical Society. (B) Self-assembled aptamer-functionalized DNA hydrogel for colorimetric and fluorescence detection of thrombin protein. Reproduced with permission from ref. 141. Copyright 2013, American Chemical Society. (C) DNA–AuNPs hybrid hydrogel for visual detection of lead ions via DNAzyme-mediated cleavage of the crosslinking substrate and subsequent release of encapsulated AuNPs. Reproduced with permission from ref. 139 Copyright 2014, American Chemical Society. | ||
Some of the important biomarkers that are used for the diagnosis of diseases are proteins. Hence, sensitive detection of proteins is of critical importance in clinical diagnosis. DNA-based sensors utilizing aptamers for the detection of proteins have been realized. Lei and colleagues designed an aptamer-functionalized DNA hydrogel for the detection of thrombin.141 The DNA hydrogel was developed using a Y-shaped DNA with the thrombin aptamer as a linker (Fig. 11(B)). In the presence of thrombin, the aptamer undergoes strand displacement and results in the dissociation of the hydrogel, releasing the AuNPs stored within the hydrogel. Furthermore, polyethyleneimine (PEI) wrapped positively charged quantum dots (PEI-QDs) were synthesized to act as the fluorophore. The released negatively charged AuNPs interact with PEI-QDs, and the fluorescence resonance energy transfer between PEI-QDs and AuNPs quenched the fluorescence signal, indicating the presence of thrombin.
Apart from detecting biomolecules, DNA-based hybrid materials have also been applied to detect inorganic substances such as metal ions. Estimation of the amount of heavy metals is needed both in environmental pollution studies and to monitor diseases associated with metal poisoning in humans. The ability of DNA to interact with metal ions comes by virtue of DNA being a polyanionic polymer with multiple functional groups. Huang and colleagues developed a DNA–AuNP hybrid hydrogel that can detect lead ions with concentrations as low as 10 nM.139 The hydrogel was synthesized by functionalizing the polyacrylamide backbone with ssDNA (pA1,pA2) (Fig. 11(C)). A single-stranded DNA with a sequence complementary to pA1 and pA2 was used as a crosslinker. The presence of lead ions activates the DNAzyme and induces the cleavage of the crosslinking strand, resulting in hydrogel degradation. Hydrogel degradation results in the release of AuNPs, which are then used for colorimetric detection of the amount of lead ions in the sample. Apart from lead, DNA-based hybrid materials have also been designed to detect different metal ions.142,143 In addition to the sensors introduced above, a variety of DNA-based hybrid materials as sensors have been developed and applied in biomedicine, clinical diagnosis, and environmental safety. With the advancement in DNA nanotechnology and an increased understanding of materials and chemical science, more complex and sensitive sensors will continue to develop in the future.
4.4 Cell culture and tissue engineering
Cell culture is a crucial cellular and molecular biology technique allowing researchers to study the physiology, biochemistry, and metabolism of wild-type and pathogenic cells. Research involving the development of a biomaterial matrix that can support, promote, and induce cellular growth is of critical importance, especially for cells with limited proliferative and differentiation potential (hepatocytes and neurites). Therefore, mimicking the native extracellular matrix's mechanical and functional components is necessary for a bioactive scaffold. DNA, a negative polyelectrolyte, mimics the negative charge density of most extracellular matrices. Therefore, developing DNA-based biohybrids with DNA mimicking the ECM and the second component acting as a ligand for induction of proliferation and differentiation may circumvent the limitation associated with cell culture and can result in novel biohybrids with more effective and broader applications in tissue engineering. Hivare and colleagues designed a peptide-functionalized DNA hydrogel to enhance neuroblastoma cell growth and differentiation.144 The developed DNA hydrogel was functionalized with the IKVAV peptide, which is known to be responsible for enhanced attachment and differentiation of neural cells (Fig. 12(A)). The group observed that the developed hybrid hydrogel enhanced neural differentiation, prolonged neurite length, and altered endocytic mechanisms in neuroblastoma stem cells. The developed hydrogel demonstrated the advantages of DNA-based hybrid materials for cell culture applications and paved the way for the development of materials for different cells utilizing a similar design principle.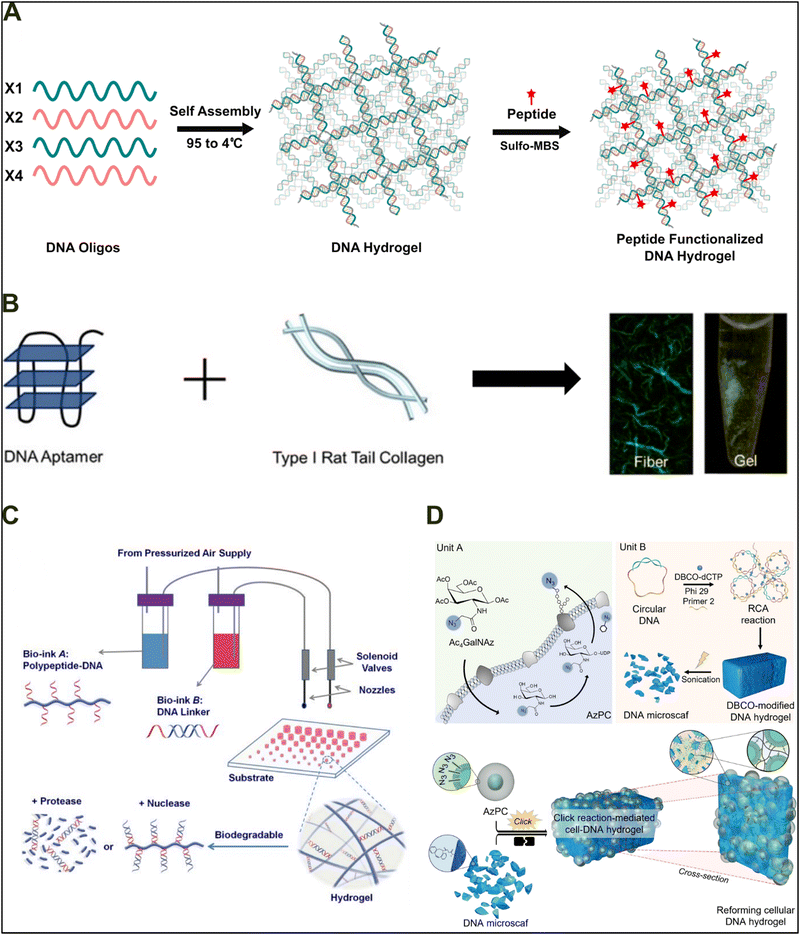 | ||
| Fig. 12 Cell culture and tissue engineering applications of DNA-based hybrid materials. (A) Self-assembled 4-way junction DNA hydrogel functionalized with the IKVAV peptide through sulfo-MBS chemistry for attachment and differentiation of neural cells. Drafted and adapted from ref. 144. Copyright 2022, Royal Society of Chemistry. (B) Nucleic acid collagen complex (NACC) fibers and gel for cell culture application formed by conjugating an aptamer and type I rat tail collagen at different mass percent ratios. Reproduced with permission from ref. 145. Copyright 2020, Elsevier. (C) A bio-printable DNA-based hybrid 3D hydrogel synthesized using two different bio inks (bio-ink A and bio-ink b) with excellent biocompatibility and controllable degradation kinetics via nucleases and proteases. Reproduced with permission from ref. 146. Copyright 2015, Wiley. (D) A DNA hybrid hydrogel synthesized via covalent assembly of modified mammalian cells (Unit A) and DNA microscafs (Unit B) through click chemistry. Reproduced with permission from ref. 147. Copyright 2023, Elsevier. | ||
A field that is closely associated with cell culture and derived from its application is regenerative medicine/tissue engineering. The core principle of regenerative medicine is to either ‘regenerate’ or ‘replace’ the damaged tissue/organ to achieve its normal function. Although significant research towards improving body's own regenerative potential is being done, a greater focus is directed towards replacing the damaged tissues/organs with engineered functional tissues directly. The first generation of engineered biomaterials mainly focused on mimicking the mechanical properties of the native tissues (hips, inter-vertebral disc, teeth and heart valve); however, they were limited in their ability to allow repair and regeneration while mimicking the native architecture of the host tissue. The neo-generation biomaterials are developed with functionalities such as cell specific interaction, growth factor delivery, stimuli responsiveness etc., allowing precise control over tissue repair. Towards this aim, DNA-based hybrid materials have emerged as a novel class of biomaterials integrating unique functionality offered by conjugation of different components/materials as well as precise structural regulation via programmable DNA sequences.
In a recent work performed by James and colleagues, a ssDNA and collagen-based hybrid biomaterial formed via electrostatic interaction was utilized for growing primary human osteocytes for bone tissue engineering.145 Complex fibers were synthesized using the specific ratio of ssDNA to collagen, which were then combined with hydroxyapatite to form mineralized microfibers (Fig. 12(B)). At the nanoscale, the mineralized microfibers resemble the native bone extracellular matrix, which was remodeled by cultured cells into densified, tissue-like structures within 3 days. The developed fibers show the promise to be utilized as coatings and scaffolds for tissues within the human body.
With the recent advancements in 3D bioprinting and its substantial implementation in designing and developing tissue-like structures, Lie and colleagues explored the possibility of developing a bio-printable DNA-based hybrid scaffold material.146,148,149 To develop a multi-layer 3D hydrogel, they utilized two different bio inks, ‘bio-ink A’ containing a DNA–polypeptide conjugate synthesized via click chemistry and ‘bio-ink B’ containing a dsDNA based crosslinking agent (Fig. 12(C)). By programming alternate deposition of bio-ink A and bio-ink B at a defined molar ratio, rapid (within seconds) in situ hydrogel formation occurred under physiological conditions. The printed hydrogel demonstrated excellent merging and healing properties while maintaining 3D structure at the millimeter level. The hydrogel demonstrated excellent biocompatibility and allowed long term growth of AtT-20 cells. The developed hydrogel established that DNA-based hybrid materials can be utilized for development of bioprintable materials allowing the development of complex 3D tissue constructs for tissue engineering. Apart from utilizing hybrid tissue constructs made up of non-biological components, cells can themselves be modified and used as conjugate materials for biological hybrid materials. Biomaterials with cells covalently embedded to the artificial matrix circumvent the major limitation of cellular washing from the implanted site associated with autologous transplantation.150,151 Nam and colleagues addressed this problem by developing a hydrogel utilizing the covalent assembly between DNA and modified therapeutic cells.147 The mammalian cells were modified by introduction of an azide group on the cell surface via metabolic engineering (Unit A), allowing cells to act as building units themselves (Fig. 12(D)). The modified cells were covalently attached to a DNA micro-scaffold (Unit B - DNA microscafs) synthesized via rolling circle amplification through a clickable moiety. DNA microscafs provided a porous matrix for nutrient flow and ultrasoft mechanical properties to the final construct. The developed construct was injectable, biocompatible, and demonstrated therapeutic effects in vivo. Overall, the work demonstrated the flexibility of DNA-based materials to be conjugated with living biological components and paved a way for utilization of DNA-based hybrids for cell replacement therapy.
5. Conclusion and perspectives
The past three decades have witnessed the growth and development of the domain of DNA nanotechnology. The main focus revolved around the hidden potential of DNA as a polymeric customizable “LEGO” block for creating predictable shapes and structures of interest. However, the DNA strand has its own limitations owing to its weak mechanical strength, limited stability, and poor scalability, which other polymers do not have. To resolve these caveats, the integration of DNA with other biologically relevant biopolymers is a must, resulting in the generation of hybrid DNA biomaterials.The introduction of hybrid materials is essential to broaden the scope of DNA-based materials in clinical, biomedical, research, and therapeutic areas. Conjugation of pure DNA materials with different materials imparts necessary functionalities such as improved mechanical strength, enhanced adhesion, injectability, self-healing, conductivity, and responsiveness to environmental stimuli. The scope of this review is to lay down key discoveries and directions in which hybrid-DNA biomaterials have shown growth and improvements in the context of their biological applications, including but not limited to cell culture, biosensing, clinical assays, stimuli-responsive smart device fabrication, etc. These compiled research ideas can be taken as a road map to study the girth and depth of this field. Even though the hybrid DNA smart materials show enhanced and obvious advantages over classical pure DNA structures, these hybrid materials nonetheless show some unavoidable pitfalls: (1) the greatest concern is the cytotoxic or immunosafety standards of the putative hybrid polymeric materials being utilized for biological applications. (2) The synthesis and standardization of two or more component systems have more often proven to be more expensive than the system involving only one variable. (3) Biocompatibility of the hybrid system must be checked before randomly mixing one or more polymeric compounds, which may prove to be unrewarding. (4) Simple chemical reactions with limited steps are a must to address the problem of scalability. (5) Biodegradation and pharmaco-kinetics of hybrid DNA materials within the body have rarely been reported.
The above-stated limitations could be solved mostly, if not all by rationally selecting synergistic candidate materials that can be conjugated to DNA. Selection of appropriate materials can therefore overcome the limitation of cytotoxicity or immunogenicity. The cost of synthesis and scaling-up issues can also be resolved by selecting materials that are present in abundance in nature and simultaneously designing more efficient methods and techniques to gain a higher yield of the biomaterials needed for the fabrication of hybrid systems. Integration and exploration of basic organic/inorganic chemistry could be a key to devising and implementing novel yet simple chemical conjugation steps, for example, conjugating diblock components using a single-step click chemistry. These steps could cut down the cost and increase the speed of the synthesis process while also minimizing exposure to harsh and rigorous chemical treatments, which could solve the cytotoxicity and biocompatibility issues. More in-depth understanding and exploration will be needed to thoroughly improve our understanding of the pharmacokinetics and pharmacodynamics of the developed hybrid DNA systems for therapeutic drug-related biological applications. The demand for standard protocols or methods is highly appreciated as the kinetic profile of different materials will vary depending on the type of material conjugated with DNA. Overall, we hope this review provides a deeper and comprehensive understanding of the current state of DNA-based hybrid materials and their significance in tissue engineering, drug delivery, gene therapy, sensing, etc.
Data availability
This is a review article. So no new data were collected for writing this manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors also thank the researchers working on different aspects of DNA-based hybrid materials. N. S. thanks IITGN-MoE, GoI for a PhD fellowship. A. S. thanks the GoI for the Prime Minister's Research Fellowship. D. B. and M. D. acknowledge the Indian Institute of Technology, Gandhinagar for the infrastructure and financial support.References
- N. C. Seeman, Nucleic acid junctions and lattices, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed.
- F. Zhang, J. Nangreave, Y. Liu and H. Yan, Structural DNA Nanotechnology: State of the Art and Future Perspective, J. Am. Chem. Soc., 2014, 136, 11198–11211 CrossRef CAS PubMed.
- P. Chidchob and H. F. Sleiman, Recent advances in DNA nanotechnology, Curr. Opin. Chem. Biol., 2018, 46, 63–70 CrossRef CAS PubMed.
- N. C. Seeman, Nucleic acid junctions and lattices, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed.
- N. Dattagupta, W. J. Knowles, V. T. Marchesi and D. M. Crothers, Nucleic acid-protein conjugate used in immunoassay, 1988 Search PubMed.
- S. Nagahara and T. Matsuda, Hydrogel formation via hybridization of oligonucleotides derivatized in water-soluble vinyl polymers, Polym. Gels Networks, 1996, 4, 111–127 CrossRef CAS.
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, A DNA-based method for rationally assembling nanoparticles into macroscopic materials, Nature, 1996, 382, 607–609 CrossRef CAS PubMed.
- T. Ochiya, et al., New delivery system for plasmid DNA in vivo using atelocollagen as a carrier material: the Minipellet, Nat. Med., 1999, 5, 707–710 CrossRef CAS PubMed.
- J. H. Jeong and T. G. Park, Novel Polymer−DNA Hybrid Polymeric Micelles Composed of Hydrophobic Poly(D,L-lactic-co-glycolic Acid) and Hydrophilic Oligonucleotides, Bioconjugate Chem., 2001, 12, 917–923 CrossRef CAS PubMed.
- W. J. Parak, et al., Conjugation of DNA to Silanized Colloidal Semiconductor Nanocrystalline Quantum Dots, Chem. Mater., 2002, 14, 2113–2119 CrossRef CAS.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- N. Park, S. H. Um, H. Funabashi, J. Xu and D. Luo, A cell-free protein-producing gel, Nat. Mater., 2009, 8, 432–437 CrossRef CAS.
- Y. H. Roh, et al., DNAsomes: Multifunctional DNA-based nanocarriers, Small Weinh. Bergstr. Ger., 2011, 7, 74–78 CrossRef CAS PubMed.
- Y. Dayani and N. Malmstadt, Liposomes with Double-Stranded DNA Anchoring the Bilayer to a Hydrogel Core, Biomacromolecules, 2013, 14, 3380–3385 CrossRef CAS.
- C. Li, et al., Rapid Formation of a Supramolecular Polypeptide–DNA Hydrogel for In Situ Three-Dimensional Multilayer Bioprinting, Angew. Chem., Int. Ed., 2015, 54, 3957–3961 CrossRef CAS PubMed.
- P. Zhan, K. Jahnke, N. Liu and K. Göpfrich, Functional DNA-based cytoskeletons for synthetic cells, Nat. Chem., 2022, 14, 958–963 CrossRef CAS PubMed.
- V. Morya, S. Walia, B. B. Mandal, C. Ghoroi and D. Bhatia, Functional DNA Based Hydrogels: Development, Properties and Biological Applications, ACS Biomater. Sci. Eng., 2020, 6, 6021–6035 CrossRef CAS PubMed.
- Y. Zhang, et al., Dynamic DNA Structures, Small Weinh. Bergstr. Ger., 2019, 15, e1900228 CrossRef PubMed.
- S. I. S. Hendrikse, S. L. Gras and A. V. Ellis, Opportunities and Challenges in DNA-Hybrid Nanomaterials, ACS Nano, 2019, 13, 8512–8516 CrossRef CAS PubMed.
- C. J. Whitfield, et al., Functional DNA–Polymer Conjugates, Chem. Rev., 2021, 121, 11030–11084 CrossRef CAS PubMed.
- A. Keller and V. Linko, Challenges and Perspectives of DNA Nanostructures in Biomedicine, Angew. Chem., Int. Ed., 2020, 59, 15818–15833 CrossRef CAS PubMed.
- D. Menon, R. Singh, K. B. Joshi, S. Gupta and D. Bhatia, Designer, Programmable DNA-peptide hybrid materials with emergent properties to probe and modulate biological systems, ChemBioChem, 2023, 24, e202200580 CrossRef CAS.
- S. W. Shin, J. S. Yuk, S. H. Chun, Y. T. Lim and S. H. Um, Hybrid material of structural DNA with inorganic compound: synthesis, applications, and perspective, Nano Convergence, 2020, 7, 2 CrossRef CAS PubMed.
- J. Zimmermann, M. Kwak, A. J. Musser and A. Herrmann, Amphiphilic DNA block copolymers: nucleic acid-polymer hybrid materials for diagnostics and biomedicine, Methods Mol. Biol., 2011, 751, 239–266 CrossRef CAS.
- D. Wang, S. Li, Z. Zhao, X. Zhang and W. Tan, Engineering a Second-Order DNA Logic-Gated Nanorobot to Sense and Release on Live Cell Membranes for Multiplexed Diagnosis and Synergistic Therapy, Angew. Chem., Int. Ed., 2021, 60, 15816–15820 CrossRef CAS.
- H. Li, et al., A DNA Molecular Robot that Autonomously Walks on the Cell Membrane to Drive Cell Motility, Angew. Chem., Int. Ed., 2021, 60, 26087–26095 CrossRef CAS.
- A. E. Marras, L. Zhou, H.-J. Su and C. E. Castro, Programmable motion of DNA origami mechanisms, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 713–718 CrossRef CAS PubMed.
- M. Kwak and A. Herrmann, Nucleic Acid/Organic Polymer Hybrid Materials: Synthesis, Superstructures, and Applications, Angew. Chem., Int. Ed., 2010, 49, 8574–8587 CrossRef CAS PubMed.
- N. Stephanopoulos, J. H. Ortony and S. I. Stupp, Self-Assembly for the Synthesis of Functional Biomaterials, Acta Mater., 2013, 61, 912–930 CrossRef CAS PubMed.
- N. Stephanopoulos, Hybrid Nanostructures from the Self-Assembly of Proteins and DNA, Chem, 2020, 6, 364–405 CAS.
- X. Yan, S. Huang, Y. Wang, Y. Tang and Y. Tian, Bottom-Up Self-Assembly Based on DNA Nanotechnology, Nanomaterials, 2020, 10, 2047 CrossRef CAS PubMed.
- D. Lee, et al., Self-Assembled DNA–Protein Hybrid Nanospheres: Biocompatible Nano-Drug-Carriers for Targeted Cancer Therapy, ACS Appl. Mater. Interfaces, 2022, 14, 37493–37503 CrossRef CAS.
- C. Zhang, et al., DNA-Directed Three-Dimensional Protein Organization, Angew. Chem., Int. Ed., 2012, 51, 3382–3385 CrossRef CAS PubMed.
- Y. Tian, et al., Ordered three-dimensional nanomaterials using DNA-prescribed and valence-controlled material voxels, Nat. Mater., 2020, 19, 789–796 CrossRef CAS PubMed.
- A. Hernandez-Garcia, Strategies to Build Hybrid Protein–DNA Nanostructures, Nanomaterials, 2021, 11, 1332 CrossRef CAS PubMed.
- H. Li, J. D. Carter and T. H. LaBean, Nanofabrication by DNA self-assembly, Mater. Today, 2009, 12, 24–32 CrossRef CAS.
- F. Chen, et al., A novel tunable, highly biocompatible and injectable DNA-chitosan hybrid hydrogel fabricated by electrostatic interaction between chitosan and DNA backbone, Int. J. Pharm., 2021, 606, 120938 CrossRef CAS PubMed.
- K. Morikawa, Y. Masubuchi, Y. Shchipunov and A. Zinchenko, DNA-Chitosan Hydrogels: Formation, Properties, and Functionalization with Catalytic Nanoparticles, ACS Appl. Bio Mater., 2021, 4, 1823–1832 CrossRef CAS.
- A. G. Cherstvy, Electrostatic interactions in biological DNA-related systems, Phys. Chem. Chem. Phys., 2011, 13, 9942–9968 RSC.
- U. Rungsardthong, et al., Effect of Polymer Ionization on the Interaction with DNA in Nonviral Gene Delivery Systems, Biomacromolecules, 2003, 4, 683–690 CrossRef CAS PubMed.
- S. Izui, P. H. Lambert and P. A. Miescher, In vitro demonstration of a particular affinity of glomerular basement membrane and collagen for DNA. A possible basis for a local formation of DNA-anti-DNA complexes in systemic lupus erythematosus, J. Exp. Med., 1976, 144, 428–443 CrossRef CAS.
- T. J. Thomas, H.-A. Tajmir-Riahi and C. K. S. Pillai, Biodegradable Polymers for Gene Delivery, Molecules, 2019, 24, 3744 CrossRef CAS PubMed.
- S. Barua, et al., Discovery of Cationic Polymers for Non-viral Gene Delivery using Combinatorial Approaches, Comb. Chem. High Throughput Screening, 2011, 14, 908–924 CrossRef CAS PubMed.
- X. Gao and L. Huang, Potentiation of cationic liposome-mediated gene delivery by polycations, Biochemistry, 1996, 35, 1027–1036 CrossRef CAS PubMed.
- H. Lee, J. H. Jeong and T. G. Park, PEG grafted polylysine with fusogenic peptide for gene delivery: high transfection efficiency with low cytotoxicity, J. Controlled Release, 2002, 79, 283–291 CrossRef CAS PubMed.
- N. Singh, A. Singh and D. Bhatia, Self-assembled DNA-collagen bioactive scaffolds promote cellular uptake and neuronal differentiation, bioRxiv, 2024, preprint, DOI:10.1101/2024.05.24.595848.
- V. A. Bloomfield, DNA condensation, Curr. Opin. Struct. Biol., 1996, 6, 334–341 CrossRef CAS.
- A. Mann, R. Richa and M. Ganguli, DNA condensation by poly-L-lysine at the single molecule level: role of DNA concentration and polymer length, J. Controlled Release, 2008, 125, 252–262 CrossRef CAS PubMed.
- D. A. Kondinskaia and A. A. Gurtovenko, Supramolecular complexes of DNA with cationic polymers: The effect of polymer concentration, Polymer, 2018, 142, 277–284 CrossRef CAS.
- C. M. Jewell and D. M. Lynn, Surface-Mediated Delivery of DNA: Cationic Polymers Take Charge, Curr. Opin. Colloid Interface Sci., 2008, 13, 395–402 CrossRef CAS PubMed.
- S. Barua, et al., Discovery of Cationic Polymers for Non-viral Gene Delivery using Combinatorial Approaches, Comb. Chem. High Throughput Screening, 2011, 14, 908–924 CrossRef CAS PubMed.
- S. P. Strand, S. Danielsen, B. E. Christensen and K. M. Vårum, Influence of chitosan structure on the formation and stability of DNA-chitosan polyelectrolyte complexes, Biomacromolecules, 2005, 6, 3357–3366 CrossRef CAS PubMed.
- P. Pakornpadungsit, T. Prasopdee, N. M. Swainson, A. Chworos and W. Smitthipong, DNA:chitosan complex, known as a drug delivery system, can create a porous scaffold, Polym. Test., 2020, 83, 106333 CrossRef CAS.
- Z. Shen, et al., Polymer–Nucleic Acid Interactions, Top. Curr. Chem., 2017, 375, 44 CrossRef PubMed.
- F. Cozzolino, I. Iacobucci, V. Monaco and M. Monti, Protein–DNA/RNA Interactions: An Overview of Investigation Methods in the -Omics Era, J. Proteome Res., 2021, 20, 3018–3030 CrossRef CAS PubMed.
- C. Lachance-Brais, et al., Small Molecule-Templated DNA Hydrogel with Record Stiffness Integrates and Releases DNA Nanostructures and Gene Silencing Nucleic Acids, Adv. Sci., 2023, 2205713, DOI:10.1002/advs.202205713.
- D. Cao, Y. Xie and J. Song, DNA Hydrogels in the Perspective of Mechanical Properties, Macromol. Rapid Commun., 2022, 43, 2200281 CrossRef CAS.
- J. H. Jeong, S. H. Kim, S. W. Kim and T. G. Park, Polyelectrolyte Complex Micelles Composed of c-raf Antisense Oligodeoxynucleotide−Poly(ethylene glycol) Conjugate and Poly(ethylenimine): Effect of Systemic Administration on Tumor Growth, Bioconjugate Chem., 2005, 16, 1034–1037 CrossRef CAS PubMed.
- M. Oishi, Y. Nagasaki, K. Itaka, N. Nishiyama and K. Kataoka, Lactosylated Poly(ethylene glycol)-siRNA Conjugate through Acid-Labile β-Thiopropionate Linkage to Construct pH-Sensitive Polyion Complex Micelles Achieving Enhanced Gene Silencing in Hepatoma Cells, J. Am. Chem. Soc., 2005, 127, 1624–1625 CrossRef CAS PubMed.
- Y. G. Takei, et al., Temperature-responsive bioconjugates. 1. Synthesis of temperature-responsive oligomers with reactive end groups and their coupling to biomolecules, Bioconjugate Chem., 1993, 4, 42–46 CrossRef CAS PubMed.
- R. B. Fong, Z. Ding, C. J. Long, A. S. Hoffman and P. S. Stayton, Thermoprecipitation of Streptavidin via Oligonucleotide-Mediated Self-Assembly with Poly(N-isopropylacrylamide), Bioconjugate Chem., 1999, 10, 720–725 CrossRef CAS PubMed.
- M. Oishi, et al., Supramolecular Assemblies for the Cytoplasmic Delivery of Antisense Oligodeoxynucleotide: Polyion Complex (PIC) Micelles Based on Poly(ethylene glycol)-SS-Oligodeoxynucleotide Conjugate, Biomacromolecules, 2005, 6, 2449–2454 CrossRef CAS PubMed.
- M. Oishi, F. Nagatsugi, S. Sasaki, Y. Nagasaki and K. Kataoka, Smart Polyion Complex Micelles for Targeted Intracellular Delivery of PEGylated Antisense Oligonucleotides Containing Acid-Labile Linkages, ChemBioChem, 2005, 6, 718–725 CrossRef CAS PubMed.
- F. E. Alemdaroglu, K. Ding, R. Berger and A. Herrmann, DNA-Templated Synthesis in Three Dimensions: Introducing a Micellar Scaffold for Organic Reactions, Angew. Chem., Int. Ed., 2006, 45, 4206–4210 CrossRef CAS PubMed.
- Z. Li, Y. Zhang, P. Fullhart and C. A. Mirkin, Reversible and Chemically Programmable Micelle Assembly with DNA Block-Copolymer Amphiphiles, Nano Lett., 2004, 4, 1055–1058 CrossRef CAS.
- F. E. Alemdaroglu, M. Safak, J. Wang, R. Berger and A. Herrmann, DNA multiblock copolymers, Chem. Commun., 2007,(13), 1358–1359 RSC.
- F. Teixeira Jr., P. Rigler and C. Vebert-Nardin, Nucleo-copolymers: oligonucleotide-based amphiphilic diblock copolymers, Chem. Commun., 2007,(11), 1130–1132 RSC.
- C. J. Yang, M. Pinto, K. Schanze and W. Tan, Direct Synthesis of an Oligonucleotide–Poly(phenylene ethynylene) Conjugate with a Precise One-to-One Molecular Ratio, Angew. Chem., Int. Ed., 2005, 117, 2628–2632 CrossRef.
- T. F. A. de Greef and E. W. Meijer, Supramolecular polymers, Nature, 2008, 453, 171–173 CrossRef CAS PubMed.
- E. A. Fogleman, W. C. Yount, J. Xu and S. L. Craig, Modular, Well-Behaved Reversible Polymers from DNA-Based Monomers, Angew. Chem., Int. Ed., 2002, 41, 4026–4028 CrossRef CAS PubMed.
- M. Safak, F. E. Alemdaroglu, Y. Li, E. Ergen and A. Herrmann, Polymerase Chain Reaction as an Efficient Tool for the Preparation of Block Copolymers, Adv. Mater., 2007, 19, 1499–1505 CrossRef CAS.
- T.-H. Nguyen, et al., An improved measurement of dsDNA elasticity using AFM, Nanotechnology, 2010, 21, 075101 CrossRef PubMed.
- M. Rief, H. Clausen-Schaumann and H. E. Gaub, Sequence-dependent mechanics of single DNA molecules, Nat. Struct. Biol., 1999, 6, 346–349 CrossRef CAS PubMed.
- P. Shrestha, et al., Mechanical properties of DNA origami nanoassemblies are determined by Holliday junction mechanophores, Nucleic Acids Res., 2016, 44, 6574–6582 CrossRef PubMed.
- J. Ji, D. Karna and H. Mao, DNA origami nano-mechanics, Chem. Soc. Rev., 2021, 50, 11966–11978 RSC.
- F. X. Jiang, B. Yurke, B. L. Firestein and N. A. Langrana, Neurite outgrowth on a DNA crosslinked hydrogel with tunable stiffnesses, Ann. Biomed. Eng., 2008, 36, 1565–1579 CrossRef PubMed.
- T. Cao, H. Jia, Y. Dong, S. Gui and D. Liu, In Situ Formation of Covalent Second Network in a DNA Supramolecular Hydrogel and Its Application for 3D Cell Imaging, ACS Appl. Mater. Interfaces, 2020, 12, 4185–4192 CrossRef CAS PubMed.
- S. Basu, et al., Harnessing the Noncovalent Interactions of DNA Backbone with 2D Silicate Nanodisks To Fabricate Injectable Therapeutic Hydrogels, ACS Nano, 2018, 12, 9866–9880 CrossRef CAS PubMed.
- A. K. Gaharwar, et al., Shear-thinning nanocomposite hydrogels for the treatment of hemorrhage, ACS Nano, 2014, 8, 9833–9842 CrossRef CAS PubMed.
- C. Qi, et al., Tetrahedral framework nucleic acids/hyaluronic acid-methacrylic anhydride hybrid hydrogel with antimicrobial and anti-inflammatory properties for infected wound healing, Int. J. Oral Sci., 2024, 16, 30 CrossRef CAS PubMed.
- F. A. Aldaye, W. T. Senapedis, P. A. Silver and J. C. Way, A Structurally Tunable DNA-Based Extracellular Matrix, J. Am. Chem. Soc., 2010, 132, 14727–14729 CrossRef CAS PubMed.
- M. Shin, et al., DNA/Tannic Acid Hybrid Gel Exhibiting Biodegradability, Extensibility, Tissue Adhesiveness, and Hemostatic Ability, Adv. Funct. Mater., 2015, 25, 1270–1278 CrossRef CAS.
- A. Shukla, J. C. Fang, S. Puranam, F. R. Jensen and P. T. Hammond, Hemostatic Multilayer Coatings, Adv. Mater., 2012, 24, 492–496 CrossRef CAS PubMed.
- J. C. Isenburg, D. T. Simionescu and N. R. Vyavahare, Elastin stabilization in cardiovascular implants: improved resistance to enzymatic degradation by treatment with tannic acid, Biomaterials, 2004, 25, 3293–3302 CrossRef CAS PubMed.
- J. P. Van Buren and W. B. Robinson, Formation of complexes between protein and tannic acid, J. Agric. Food Chem., 1969, 17, 772–777 CrossRef CAS.
- J. Gačanin, et al., Spatiotemporally Controlled Release of Rho-Inhibiting C3 Toxin from a Protein–DNA Hybrid Hydrogel for Targeted Inhibition of Osteoclast Formation and Activity, Adv. Healthcare Mater., 2017, 6, 1700392 CrossRef PubMed.
- S. Basu, A.-R. Alkiswani, S. Pacelli and A. Paul, Nucleic Acid-Based Dual Cross-Linked Hydrogels for in Situ Tissue Repair via Directional Stem Cell Migration, ACS Appl. Mater. Interfaces, 2019, 11, 34621–34633 CrossRef CAS PubMed.
- C.-H. Lu, et al., Switchable Catalytic Acrylamide Hydrogels Cross-Linked by Hemin/G-Quadruplexes, Nano Lett., 2013, 13, 1298–1302 CrossRef CAS PubMed.
- C. Wang, M. Fadeev, M. Vázquez-González and I. Willner, Stimuli-Responsive Donor–Acceptor and DNA-Crosslinked Hydrogels: Application as Shape-Memory and Self-Healing Materials, Adv. Funct. Mater., 2018, 28, 1803111 CrossRef.
- Y. Wu, D. Wang, I. Willner, Y. Tian and L. Jiang, Smart DNA Hydrogel Integrated Nanochannels with High Ion Flux and Adjustable Selective Ionic Transport, Angew. Chem., Int. Ed., 2018, 57, 7790–7794 CrossRef CAS PubMed.
- W. Guo, et al., Switchable Bifunctional Stimuli-Triggered Poly-N-Isopropylacrylamide/DNA Hydrogels, Angew. Chem., Int. Ed., 2014, 53, 10134–10138 CrossRef CAS PubMed.
- W. Guo, et al., Reversible Ag+-crosslinked DNA hydrogels, Chem. Commun., 2014, 50, 4065–4068 RSC.
- X. Yu, Y. Hu, J. S. Kahn, A. Cecconello and I. Willner, Orthogonal Dual-Triggered Shape-Memory DNA-Based Hydrogels, Chem. – Eur. J., 2016, 22, 14504–14507 CrossRef CAS PubMed.
- V. Kumar and P. Guleria, Application of DNA–Nanosensor for Environmental Monitoring: Recent Advances and Perspectives, Curr. Pollut. Rep., 2020 DOI:10.1007/s40726-020-00165-1.
- M. Z. Quazi, J. H. Choi, M. Kim and N. Park, DNA and Nanomaterials: A Functional Combination for DNA Sensing, ACS Appl. Bio Mater., 2024, 7, 778–786 CrossRef CAS PubMed.
- Y. Hua, J. Ma, D. Li and R. Wang, DNA-Based Biosensors for the Biochemical Analysis: A Review, Biosensors, 2022, 12, 183 CrossRef CAS PubMed.
- H. C. Budnikov, G. A. Evtugyn and A. V. Porfireva, Electrochemical DNA sensors based on electropolymerized materials, Talanta, 2012, 102, 137–155 CrossRef CAS PubMed.
- L. Peng, et al., Macroscopic Volume Change of Dynamic Hydrogels Induced by Reversible DNA Hybridization, J. Am. Chem. Soc., 2012, 134, 12302–12307 CrossRef CAS PubMed.
- M. Oishi and K. Nakatani, Dynamically Programmed Switchable DNA Hydrogels Based on a DNA Circuit Mechanism, Small, 2019, 15, 1900490 CrossRef PubMed.
- A. J. Simon, L. T. Walls-Smith and K. W. Plaxco, Exploiting the conformational-selection mechanism to control the response kinetics of a “smart” DNA hydrogel, Analyst, 2018, 143, 2531–2538 RSC.
- L. Yan, et al., Target-Responsive “Sweet” Hydrogel with Glucometer Readout for Portable and Quantitative Detection of Non-Glucose Targets, J. Am. Chem. Soc., 2013, 135, 3748–3751 CrossRef CAS PubMed.
- Y. Huang, et al., Target-Responsive DNAzyme Cross-Linked Hydrogel for Visual Quantitative Detection of Lead, Anal. Chem., 2014, 86, 11434–11439 CrossRef CAS PubMed.
- W. Sun, et al., Cocoon-Like Self-Degradable DNA Nanoclew for Anticancer Drug Delivery, J. Am. Chem. Soc., 2014, 136, 14722–14725 CrossRef CAS PubMed.
- J. Fern and R. Schulman, Modular DNA strand-displacement controllers for directing material expansion, Nat. Commun., 2018, 9, 3766 CrossRef PubMed.
- Y. Hu, et al., Reversible Modulation of DNA-Based Hydrogel Shapes by Internal Stress Interactions, J. Am. Chem. Soc., 2016, 138, 16112–16119 CrossRef CAS.
- G. A. R. Gonçalves and R. M. A. Paiva, Gene therapy: advances, challenges and perspectives, Einstein, 2017, 15, 369–375 CrossRef PubMed.
- E. Papanikolaou and A. Bosio, The Promise and the Hope of Gene Therapy, Front. Genome Ed., 2021, 3, 618346 CrossRef PubMed.
- Y. Sung and S. Kim, Recent advances in the development of gene delivery systems, Biomater. Res., 2019, 23, 8 CrossRef CAS PubMed.
- H. Xu, Z. Li and J. Si, Nanocarriers in gene therapy: a review, J. Biomed. Nanotechnol., 2014, 10, 3483–3507 CrossRef CAS PubMed.
- H. Lee, et al., Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery, Nat. Nanotechnol., 2012, 7, 389–393 CrossRef CAS PubMed.
- W. Tang, et al., An Aptamer-Modified DNA Tetrahedron-Based Nanogel for Combined Chemo/Gene Therapy of Multidrug-Resistant Tumors, ACS Appl. Bio Mater., 2021, 4, 7701–7707 CrossRef CAS PubMed.
- J. Liu, et al., A DNA-Based Nanocarrier for Efficient Gene Delivery and Combined Cancer Therapy, Nano Lett., 2018, 18, 3328–3334 CrossRef CAS.
- F. Ding, et al., A Crosslinked Nucleic Acid Nanogel for Effective siRNA Delivery and Antitumor Therapy, Angew. Chem., Int. Ed., 2018, 57, 3064–3068 CrossRef CAS PubMed.
- H. Li, et al., Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects, Signal Transduction Targeted Ther., 2020, 5, 1–23 CrossRef.
- A. V. Anzalone, L. W. Koblan and D. R. Liu, Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors, Nat. Biotechnol., 2020, 38, 824–844 CrossRef CAS PubMed.
- F. Ding, et al., A non-cationic nucleic acid nanogel for the delivery of the CRISPR/Cas9 gene editing tool, Nanoscale, 2019, 11, 17211–17215 RSC.
- W. Sun, et al., Self-Assembled DNA Nanoclews for the Efficient Delivery of CRISPR–Cas9 for Genome Editing, Angew. Chem., Int. Ed., 2015, 54, 12029–12033 CrossRef CAS.
- Y. Wu, et al., Programmable protein–DNA hybrid hydrogels for the immobilization and release of functional proteins, Chem. Commun., 2014, 50, 14620–14622 RSC.
- C. Wang, et al., Rational Design of DNA Framework-Based Hybrid Nanomaterials for Anticancer Drug Delivery, Small Weinh. Bergstr. Ger., 2020, 16, e2002578 CrossRef.
- M. Shin, et al., DNA/Tannic Acid Hybrid Gel Exhibiting Biodegradability, Extensibility, Tissue Adhesiveness, and Hemostatic Ability, Adv. Funct. Mater., 2015, 25, 1270–1278 CrossRef CAS.
- C. Li, et al., A Writable Polypeptide–DNA Hydrogel with Rationally Designed Multi-modification Sites, Small, 2015, 11, 1138–1143 CrossRef CAS.
- F. E. Alemdaroglu, N. C. Alemdaroglu, P. Langguth and A. Herrmann, DNA Block Copolymer Micelles – A Combinatorial Tool for Cancer Nanotechnology, Adv. Mater., 2008, 20, 899–902 CrossRef CAS.
- F. Huang, et al., Self-assembled hybrid nanoparticles for targeted co-delivery of two drugs into cancer cells, Chem. Commun., 2014, 50, 3103–3105 RSC.
- N. Park, S. H. Um, H. Funabashi, J. Xu and D. Luo, A cell-free protein-producing gel, Nat. Mater., 2009, 8, 432–437 CrossRef CAS PubMed.
- A. Azadbakht, M. Roushani, A. R. Abbasi and Z. Derikvand, A novel impedimetric aptasensor, based on functionalized carbon nanotubes and prussian blue as labels, Anal. Biochem., 2016, 512, 58–69 CrossRef CAS.
- S. Saha, V. Prakash, S. Halder, K. Chakraborty and Y. Krishnan, A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells, Nat. Nanotechnol., 2015, 10, 645–651 CrossRef CAS PubMed.
- M. J. Campolongo, et al., Adaptive DNA-based materials for switching, sensing, and logic devices, J. Mater. Chem., 2011, 21, 6113–6121 RSC.
- A. Cecconello, C.-H. Lu, J. Elbaz and I. Willner, Au nanoparticle/DNA rotaxane hybrid nanostructures exhibiting switchable fluorescence properties, Nano Lett., 2013, 13, 6275–6280 CrossRef CAS PubMed.
- K. M. Abu-Salah, et al., DNA-Based Nanobiosensors as an Emerging Platform for Detection of Disease, Sensors, 2015, 15, 14539–14568 CrossRef CAS PubMed.
- J. Cao, C. Feng, Y. Liu, S. Wang and F. Liu, Highly sensitive and rapid bacteria detection using molecular beacon-Au nanoparticles hybrid nanoprobes, Biosens. Bioelectron., 2014, 57, 133–138 CrossRef CAS PubMed.
- Y. Hua, J. Ma, D. Li and R. Wang, DNA-Based Biosensors for the Biochemical Analysis: A Review, Biosensors, 2022, 12, 183 CrossRef CAS PubMed.
- R. A. Reynolds, C. A. Mirkin and R. L. Letsinger, Homogeneous, Nanoparticle-Based Quantitative Colorimetric Detection of Oligonucleotides, J. Am. Chem. Soc., 2000, 122, 3795–3796 CrossRef CAS.
- C. Bao, et al., Bioresponsive antisense DNA gold nanobeacons as a hybrid in vivo theranostics platform for the inhibition of cancer cells and metastasis, Sci. Rep., 2015, 5, 12297 CrossRef CAS PubMed.
- Z. Chen, et al., Dendrimer-Functionalized Superparamagnetic Nanobeacons for Real-Time Detection and Depletion of HSP90α mRNA and MR Imaging, Theranostics, 2019, 9, 5784–5796 CrossRef CAS PubMed.
- J. Conde, J. Rosa, J. M. de la Fuente and P. V. Baptista, Gold-nanobeacons for simultaneous gene specific silencing and intracellular tracking of the silencing events, Biomaterials, 2013, 34, 2516–2523 CrossRef CAS PubMed.
- S. Song, et al., Gold-nanoparticle-based multicolor nanobeacons for sequence-specific DNA analysis, Angew. Chem., Int. Ed., 2009, 48, 8670–8674 CrossRef CAS PubMed.
- D. S. Seferos, D. A. Giljohann, H. D. Hill, A. E. Prigodich and C. A. Mirkin, Nano-Flares:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Probes for Transfection and mRNA Detection in Living Cells, J. Am. Chem. Soc., 2007, 129, 15477–15479 CrossRef CAS.
Probes for Transfection and mRNA Detection in Living Cells, J. Am. Chem. Soc., 2007, 129, 15477–15479 CrossRef CAS. - D. Yu, Z. Zha, S. Tang, Y. Qiu and D. Liu, Modification-Free Fluorescent Biosensor for CEA Based on Polydopamine-Coated Upconversion Nanoparticles, J. Fluoresc., 2022, 32, 1289–1297 CrossRef CAS.
- Y. Huang, et al., Target-Responsive DNAzyme Cross-Linked Hydrogel for Visual Quantitative Detection of Lead, Anal. Chem., 2014, 86, 11434–11439 CrossRef CAS PubMed.
- S. Kim and H. J. Lee, Gold Nanostar Enhanced Surface Plasmon Resonance Detection of an Antibiotic at Attomolar Concentrations via an Aptamer-Antibody Sandwich Assay, Anal. Chem., 2017, 89, 6624–6630 CrossRef CAS PubMed.
- L. Zhang, J. Lei, L. Liu, C. Li and H. Ju, Self-Assembled DNA Hydrogel as Switchable Material for Aptamer-Based Fluorescent Detection of Protein, Anal. Chem., 2013, 85, 11077–11082 CrossRef CAS.
- M. Yamada and H. Aono, DNA–inorganic hybrid material as selective absorbent for harmful compounds, Polymer, 2008, 49, 4658–4665 CrossRef CAS.
- K. M. Abu-Salah, A. A. Ansari and S. A. Alrokayan, DNA-Based Applications in Nanobiotechnology, J. Biomed. Biotechnol., 2010, 2010, 715295 Search PubMed.
- P. Hivare, et al., Peptide functionalized DNA hydrogel enhances neuroblastoma cell growth and differentiation, Nanoscale, 2022, 14, 8611–8620 RSC.
- B. D. James, P. Guerin, Z. Iverson and J. B. Allen, Mineralized DNA-collagen complex-based biomaterials for bone tissue engineering, Int. J. Biol. Macromol., 2020, 161, 1127–1139 CrossRef CAS PubMed.
- C. Li, et al., Rapid formation of a supramolecular polypeptide-DNA hydrogel for in situ three-dimensional multilayer bioprinting, Angew. Chem., Int. Ed., 2015, 54, 3957–3961 CrossRef CAS PubMed.
- H. Nam, et al., Module-assembly of injectable cellular DNA hydrogel via clickable cells and DNA scaffolds, Chem. Eng. J., 2023, 452, 139492 CAS.
- S. V. Murphy and A. Atala, 3D bioprinting of tissues and organs, Nat. Biotechnol., 2014, 32, 773–785 CrossRef CAS PubMed.
- Y. Luo, X. Lin and P. Huang, 3D Bioprinting of Artificial Tissues: Construction of Biomimetic Microstructures, Macromol. Biosci., 2018, 18, 1800034 CrossRef.
- R. Al Hamed, A. H. Bazarbachi, F. Malard, J.-L. Harousseau and M. Mohty, Current status of autologous stem cell transplantation for multiple myeloma, Blood Cancer J., 2019, 9, 44 CrossRef PubMed.
- J. V. Terrovitis, R. R. Smith and E. Marbán, Assessment and Optimization of Cell Engraftment after Transplantation into the Heart, Circ. Res., 2010, 106, 479–494 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |
