Multi-omics revealed the mechanisms of AgNP-priming enhanced rice salinity tolerance†
Si
Chen‡
a,
Zhengyan
Pan‡
b,
Jose R.
Peralta-Videa
 c and
Lijuan
Zhao
c and
Lijuan
Zhao
 *a
*a
aState Key Laboratory of Pollution Control and Resource Reuse, School of Environment, Nanjing University, Nanjing 210023, China. E-mail: jzhao@nju.edu.cn
bInstitute of Plant Protection, Liaoning Academy of Agricultural Sciences, Shenyang 110101, China
cDepartment of Chemistry and Biochemistry, The University of Texas at El Paso, 500 West University Avenue, El Paso, TX 79968, USA
First published on 25th October 2024
Abstract
Rice is highly susceptible to salt stress. Increasing the salt tolerance of rice is critical to reduce yield loss. Herein, we investigated the possibility of using an AgNP-based priming method (seed soaking (SP) and leaf spraying (LP)) to enhance rice salt tolerance. Under saline conditions, both SP (40 mg L−1) and LP (∼0.15 mg per plant) significantly increased the biomass (10.4–13.4%) and height (6.6–6.9%) of 6-week-old rice seedlings. In addition, SP significantly increased chlorophyll a (7.3%) and carotenoid (7.9%) content as well as total antioxidant capacity (10.5%), whereas it decreased malondialdehyde (MDA) content (16.9%) in rice leaves. These findings indicate that AgNP priming, especially SP, improved the salt tolerance of rice seedlings. A life cycle field study conducted in a real saline land revealed that SP significantly increased the rice grain yield by 25.8% compared to hydropriming. Multi-omics analyses demonstrated that AgNP priming induced metabolic and transcriptional reprogramming in both seeds and leaves. Notably, both SP and LP upregulated osmoprotectants in seeds and leaves. Furthermore, several transcriptional factors (TFs), such as WRKY and NAC, and salt-tolerance related genes, including the high-affinity K+ channel gene (OsHKT2;4, OsHAK5), the Ca2+/proton exchanger (CAX4), and the cation/Ca2+ exchanger (CCX4), were upregulated in leaves. Omics data provide a deep insight into the molecular mechanisms for enhanced salinity tolerance. Together, the results of this study suggest that seed priming with AgNPs can enhance the salt tolerance of rice and increase rice yield in saline soil, which provides an efficient and simple way to engineering salt-tolerant rice.
Environmental significanceGlobal food security is being threatened by soil salinization. Accelerating the adaptation of crops to salt stress is critical to reduce food insecurity. Herein, we propose an efficient and simple nano-enabled seed priming approach to enhance the salt tolerance of rice grown in a saline land. Using ROS-generating AgNPs as seed priming agents, rice seedlings' salt tolerance and grain yield were significantly increased. The results of this study provide a promising nano-enabled solution to utilize saline lands and increase crop production. |
Introduction
Salinity stress poses a significant threat to agricultural production as it impedes plant growth and reduces crop yield.1 With the ongoing climate change, the issue of global soil salinization (primarily caused by NaCl) is becoming increasingly severe.2 According to the Food and Agricultural Organization (FAO), approximately 7% of the world's land is affected by salinization.3 It is estimated that 46 million hectares of arable land worldwide are currently affected by salinization, and the area is increasing every year.4 Rice, an essential staple crop worldwide, is vulnerable to salt stress, especially at the seedling stage.5 Salt stress adversely affects rice by delaying seed germination, decreasing germination rate, inhibiting root growth, withering leaves, shortening plant height, and limiting tillering, consequently reducing the grain number per spike and seed setting rate.4 To mitigate yield loss and enhance food security, it is imperative to seek strategies to improve rice salt tolerance.Improving rice salt tolerance using traditional breeding methods is challenging, given that salt tolerance in rice is controlled by multiple genes and involves various physiological, biochemical, and molecular processes.6 Genetic engineering offers promising prospects; however, it still faces issues such as scarcity of key salt-tolerant genes, difficulty in cloning, and high cost.7 Thus, alternative approaches to enhance rice salt tolerance are urgently needed. Rice has evolved sophisticated signaling pathways to cope with salt stress.8 Reactive oxygen species (ROS), as key signaling molecules, have been found to play important roles in plants' response to stresses.9 Given this, we proposed a strategy of using ROS-generating nanoparticles (NPs) to activate defense responses and enhance the abiotic stress tolerance and immunity of plants.10 We hypothesized that NPs-generated ROS could act as a signaling molecule to trigger “stress memory”, alter molecular imprint, and enable the plants to be prepared for future stress. Our preliminary experiments demonstrated that rice (Oryza sativa L. subsp. Japonica Kato, cultivar Liaoxing 1) seeds pre-primed with ROS-generating silver nanoparticles (AgNPs) (dosing at 40 mg L−1, soaking for 24 h) exerted accelerated germination speed, improved germination rate and increased seedling vigor, indicating the enhanced salt tolerance of seeds and young seedlings (hydroponics).11 Omics data show that AgNPs seed priming induced the up-regulation of signaling metabolites (salicylic acid and nicotinamide) and signal transduction related pathways such as phytohormone signaling and mitogen-activated protein kinase (MAPK) pathways. Although these findings verified our hypothesis to some extent, some questions remain to be answered: (1) whether AgNPs seed priming can improve the salt tolerance of rice seedlings grown in real soil; (2) whether foliar priming can increase the salt tolerance of rice; (3) whether AgNPs seed priming could increase the yield of rice in a real saline field.
In this study, both a greenhouse pot trial and field trial were performed to address the above questions. Metabolomics and transcriptomics approaches were employed to elucidate the molecular changes induced by AgNPs priming. Greenhouse pot trial experiments revealed that both seed and leaf priming enhanced the salt tolerance of rice seedlings. A life cycle field trial indicated that AgNPs seed priming significantly increased the rice grain yield by 25.8%. Omics data showed that AgNPs priming induced considerable metabolic and transcriptomic reprogramming in rice seeds and leaves. Notably, the levels of osmoprotectant-related metabolites, such as sugars, and organic acids, were significantly upregulated in leaves upon priming. In addition, AgNPs priming triggered the upregulation of several transcription factors (TFs) and salt tolerance genes, such as WRKY, NAC, OsHKT2;4, OsHAK5, CAX4, and CCX4. These results provide a deep insight into the molecular mechanism for the enhanced salt tolerance.
Materials and methods
Characterization of the AgNPs
The AgNPs used in this study were purchased from Pantian Nano Material, Co., Ltd. in Shanghai, China. The AgNPs, characterized using transmission electron microscopy (TEM) (FEI, Tecnai F20, America), were spherical with an average size of about 20–37 nm. The mean hydrodynamic diameter (160 ± 8.7 nm) and zeta potential (−20.2 ± 0.4 mV) of the AgNPs (40 mg L−1) were determined by dynamic light scattering (Zetasizer Nano ZS, Malvern). The AgNPs had peroxidase-like catalytic activity and induced ·OH production in the presence of H2O2.11Application method of the AgNPs and salt stress
Rice seeds (Oryza sativa L. subsp. Japonica Kato, cultivar Liaoxing 1) were collected from Liaoning Rice Research Institute, China. The salt stress study had two phases: (1) seedling stage and (2) field stage (life cycle). The seedling stage study had two modes: (a) seed priming (SP) and (b) leaf priming (LP). The seed priming method was described in a previous publication.11 Briefly, sterilized rice seeds were soaked in an AgNP suspension (40 mg L−1, dosing according to the results of preliminary experiments) for 24 h, drained and rinsed with ultrapure water. The seeds were soaked in ultrapure water (hydropriming) as a control group. The AgNPs-primed and hydroprimed rice seeds were then germinated and planted in soil (the physiochemical characteristics of soils are listed in ESI†), and each pot (diameter: 8.5 cm, height: 11 cm, 200 g soil) contained five rice seedlings. For the leaf priming (leaf spraying) treatment, the hydroprimed seeds (24 h) were cultured in a greenhouse for 30 days. Then, 40 mg L−1 AgNPs (about 3–4 mL per plant) was sprayed on the leaf surface of 30-day-old rice seedlings. The control and SP groups were sprayed with the same volume of ultrapure water at the same time. After 7 days, salt stress was applied to 37-d-old rice seedlings. To avoid salinity shock, the NaCl solution (150 mM)12 was added into the pot three times, each time 50 mL, and irrigated once every 2 days, and the samples were collected one week after treatment. For the field test, AgNPs seed primed and leaf primed seedlings (37 d-old) were planted under natural field conditions at the research farm of Liaoning Rice Research Institute, Shenyang, China (salt-affected, salt concentration of 0.25%). Rice grains were collected from mature plants to determine yield, nutrients and mineral elements.Measurement of physiological responses
The leaf chlorophyll content was assessed according to Lichtenthaler.13 Briefly, fresh leaves (0.04 g) were extracted in 1.5 mL extraction solution (80% acetone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethanol = 1
ethanol = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) for 12 h. The chlorophyll content was estimated by measuring the absorbance of the supernatant at 470, 663 and 645 nm using a microplate reader (Varioskan LUX, Thermo Fisher Scientific, Vantaa, Finland).
1, v/v) for 12 h. The chlorophyll content was estimated by measuring the absorbance of the supernatant at 470, 663 and 645 nm using a microplate reader (Varioskan LUX, Thermo Fisher Scientific, Vantaa, Finland).
Lipid peroxidation was evaluated according to the method of thiobarbituric acid reactive substances (TBARS),14 as described in our previous publication.11 Briefly, after the samples (0.08 g) reacted with trichloroacetic acid (TCA) and 2-thiobarbituric acid (TBA) (0.5% in 20% TCA), the absorbance of the supernatant was measured at 450 nm, 532 nm and 600 nm using a microplate reader; the malondialdehyde (MDA) content was calculated as MDA (μmol L−1) = 6.45 × (A532–A600) − 0.56 × A450.
To determine the total antioxidant capacity (TAC) of leaves, iron-reducing antioxidant power (FRAP) was measured according to Benzie.15 Briefly, 1.5 mL of FRAP reagent was added to 50 μL extraction solution (obtained from the above chlorophyll extract). The reaction system was then incubated at 37 °C in the dark for approximately 1 h, and the supernatant was taken to measure the absorbance at 593 nm using a microplate reader.
The total phenol content was measured by mixing the 100 μL extraction solution with 400 μL water, 250 μL folin phenol reagent (2 M) and 1.25 mL Na2CO3 (20 g L−1). The mixture was placed in darkness at 37 °C for 4–5 h, and the absorbance of the supernatant was monitored at 735 nm using a microplate reader.16
Measurement of the total sugar content
The total sugar in rice grains was first extracted according to Verma's method.17 0.02 g of dried rice grains was taken in a 15 mL centrifuge tube, to which 2 mL of methanol (80%) was added, and the reaction system was heated in a water bath at 80 °C for 30 min, followed by centrifugation at 4500g for 20 min, and the supernatant was retained. The above steps were repeated three times, and the supernatant of the three times was mixed for the determination of total sugars. The total sugar content was determined according to the method of DuBois et al.18 1 mL of the supernatant was taken and mixed with 0.5 mL of phenol (5%) and 2.5 mL of H2SO4. After standing and cooling, the absorbance of the mixture was measured at 485 nm with a microplate reader.Measurement of mineral elements
Dry samples (0.1 g) of rice leaves or grains (homogenized and powdered) were digested with a mixture of HNO3 and H2O2 (v/v = 1/4) at 160 °C in a microwave digestion system (Milestone Ethos Up, Italy). The digested liquid was transferred to a 50 mL centrifuge tube and diluted to a constant volume with ultrapure water. The quantification of macro elements (Na, K, Ca, and Fe) and micro elements (Ag, Mn, Zn, and Mg) was performed using inductively coupled plasma–mass spectrometry (ICP-MS) (NexION-300, PerkinElmer, USA).Metabolome analysis
Metabolites in the samples were analyzed using gas chromatography-mass spectrometry (GC-MS). The samples comprised hydroprimed or AgNPs-primed rice seeds (24 h), and leaves from both control and treatment groups (SP and LP). Prior to analysis, the samples were thoroughly washed and quickly frozen in liquid nitrogen, and then ground to a fine powder using a pestle and mortar. A 60 mg sample was weighed into a 1.5 mL centrifuge tube, followed by the sequential addition of 360 μL methanol and 40 μL 2-chloro-L-phenylalanine (0.06 mg mL−1). The mixture was placed in a refrigerator at −20 °C for 5 min, and then homogenized with a 60 Hz bath-sonication for 30 min. Subsequently, 200 μL of chloroform and 400 μL of water were added and swirled for 2 min, followed by a bath-sonication at room temperature for 30 min. The mixture was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min (4 °C), and 150 μL of the supernatant was transferred to a glass derivatization vial, to which 80 μL of methoxylamine hydrochloride in pyridine (15 mg mL−1) was added. After vortexing for 2 min, the oximization reaction was conducted at 37 °C in a shaking incubator for 90 min. After removal from the incubator, 80 μL of BSTFA derivatization reagent and 20 μL of hexane were added, along with 10 μL of a mixed internal standard solution (C8/C9/C10/C12/C14/C16/C18/C20/C22/C24, all prepared with chloroform), and the reaction was carried out at 70 °C for 60 min after vortexing for 2 min. After derivatization, the samples were analyzed by GC-MS (7890B-5977A, Agilent, USA) for metabolomic analysis. Chromatographic conditions: the column was a DB-5MS capillary column (30 m × 0.25 mm × 0.25 μm, Agilent, USA). The carrier gas was high-purity helium (purity ≥ 99.999%) at a flow rate of 1.0 mL min−1. The temperature of the injection port was 260 °C, and the injection volume was 1 μL without shunt, with a solvent delay of 5 min. The column oven temperature was initially set at 60 °C and held for 0.5 minutes, followed by a ramp-up to 125 °C at 8 °C min−1, further increased to 210 °C at 8 °C min−1, 270 °C at 15 °C min−1, and ultimately to 305 °C at 20 °C min−1, and held for 5 min. The mass spectrometry (MS) conditions were as follows: the temperature of the electron bombardment ionization (EI) source was set at 230 °C, the temperature of the quadrupole was set at 150 °C, and the electron energy was set at 70 eV.
000 rpm for 10 min (4 °C), and 150 μL of the supernatant was transferred to a glass derivatization vial, to which 80 μL of methoxylamine hydrochloride in pyridine (15 mg mL−1) was added. After vortexing for 2 min, the oximization reaction was conducted at 37 °C in a shaking incubator for 90 min. After removal from the incubator, 80 μL of BSTFA derivatization reagent and 20 μL of hexane were added, along with 10 μL of a mixed internal standard solution (C8/C9/C10/C12/C14/C16/C18/C20/C22/C24, all prepared with chloroform), and the reaction was carried out at 70 °C for 60 min after vortexing for 2 min. After derivatization, the samples were analyzed by GC-MS (7890B-5977A, Agilent, USA) for metabolomic analysis. Chromatographic conditions: the column was a DB-5MS capillary column (30 m × 0.25 mm × 0.25 μm, Agilent, USA). The carrier gas was high-purity helium (purity ≥ 99.999%) at a flow rate of 1.0 mL min−1. The temperature of the injection port was 260 °C, and the injection volume was 1 μL without shunt, with a solvent delay of 5 min. The column oven temperature was initially set at 60 °C and held for 0.5 minutes, followed by a ramp-up to 125 °C at 8 °C min−1, further increased to 210 °C at 8 °C min−1, 270 °C at 15 °C min−1, and ultimately to 305 °C at 20 °C min−1, and held for 5 min. The mass spectrometry (MS) conditions were as follows: the temperature of the electron bombardment ionization (EI) source was set at 230 °C, the temperature of the quadrupole was set at 150 °C, and the electron energy was set at 70 eV.
Transcriptome analysis
For the RNA-seq analysis of seeds and leaves, total RNA was extracted using the RNAprep Pure Plant Kit (Tiangen, Beijing, China) according to the manufacturer's protocol. The purity and amount of RNA were analyzed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). The integrity of the RNA was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Following the manufacturer's instructions, the libraries were prepared using the VAHTS Universal V6 RNA-seq Library Prep Kit. The libraries were sequenced on an Illumina Novaseq 6000 platform and 150 bp paired-end reads. DESeq software was used to conduct differential expression analysis in each group.19 The threshold for significantly differentially expressed genes (DEGs) was set at FDR < 0.05 and |log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (foldchange)| ≥ 1. The transcriptome sequencing and analysis were conducted by OE Biotech, Inc., Shanghai, China.
2 (foldchange)| ≥ 1. The transcriptome sequencing and analysis were conducted by OE Biotech, Inc., Shanghai, China.
Statistical analysis
For metabolome analysis, four biological replicates were employed for each treatment. The online software OECloud tools (https://cloud.oebiotech.com) was used for the metabolites of KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis. For RNA-Seq analysis, four biological replicates were utilized for seed samples, and three replicates were used for leaf samples. The KEGG pathway enrichment analysis of genes was performed using KOBAS (http://kobas.cbi.pku.edu.cn/home.do).20 Additionally, five biological replicates were included in the greenhouse pot experiment, and three biological replicates were included in the field experiment. All data (except transcriptome data) were statistically analyzed using the Student's t-test, and p ≤ 0.05 indicates significant differences between treatment averages. The data are presented as mean ± SD in this study.Results and discussion
AgNPs priming enhanced the salt tolerance of rice seedlings
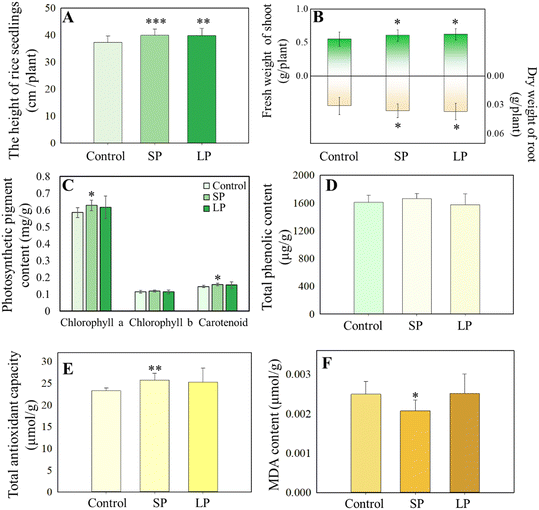
To test whether seed priming (SP) or leaf priming (LP) could enhance rice salt tolerance, 37-day old rice seedlings pre-treated by seed soaking (40 mg L−1) or foliar spraying (∼0.15 mg AgNPs per plant) were subjected to salt stress (irrigation of NaCl solution) for one week and growth performance was evaluated. Compared to the hydro-priming group (control), both SP and LP treatments significantly increased shoot height (p < 0.05) by 6.9% and 6.6%, respectively (Fig. 1A). Similarly, fresh shoot biomass was significantly increased (p < 0.05) by 10.0% and 13.1% under the SP and LP treatments, respectively (Fig. 1B). In addition, the root dry biomass was significantly increased (p < 0.05) by 16.7% and 18.4%, respectively, upon SP and LP treatments (Fig. 1B). These results indicate that AgNPs priming promoted both above-ground and below-ground tissues' growth under salt conditions. Khan et al.21 reported similar benefits of AgNPs-based seed priming (2.16 mg L−1) on pearl millet (Pennisetum glaucum L.) seedlings, including increased plant height (22%), fresh weight (58%), and dry weight (34%) under salt stress (150 mM). Furthermore, seed priming significantly increased the content of chlorophyll a (7.3%) and carotenoids (7.9%), compared to the control group (p ≤ 0.05), while chlorophyll b and total chlorophyll contents were unchanged (Fig. 1B). These results are consistent with previous studies in which AgNPs seed priming (0.2 mg L−1) was found to increase the chlorophyll content of wheat (Triticum aestivum L.) leaves under salt stress.22 TAC is an indicator of a plant's ability to mitigate oxidative damage.23 We found that SP significantly increased leaf TAC content by 10.5% (p ≤ 0.05) compared with the control group (Fig. 1E), while leaf TAC content remained unchanged upon LP treatment. Similarly, Khan et al.21 showed that seed priming with AgNPs alleviated the oxidative damage in pearl millet leaves under salt stress by enhancing the activity of antioxidant enzymes (catalase, superoxide dismutase, peroxidase, glutathione reductase, and glutathione peroxidase). In addition, SP significantly decreased leaf MDA content by 16.9% (p < 0.05), a marker of cell membrane lipid peroxidation, compared to the control (Fig. 1F). Again, LP did not change leaf MDA content. The increase of TAC content and decrease of MDA level indicate that seed priming resulted in a higher antioxidant capacity in rice leaves. These results also indicate that seed priming has a better performance in increasing rice salt tolerance as compared to leaf priming.A life cycle field study under a natural saline land
Under stress conditions, plants tend to allocate resources towards activating the defense system to endure challenges, even if it means compromising their normal growth and development.24 Since the seed priming and leaf priming data showed that AgNPs impart tolerance to salt stress to rice seedlings, we conducted a field trial in a natural saline land (0.25% NaCl) to investigate whether the AgNPs priming would impact the rice yield while improving the salt tolerance. Interestingly, the results showed that in the saline land, seed priming with AgNPs significantly increased grain yield by 25.8% (p < 0.05), compared to the control, whereas the leaf spray with AgNPs showed no significant change in yield (Fig. 2A). Similarly, it has been reported that ZnO NPs (25 mg L−1) seed priming significantly enhanced rice yield under drought stress.25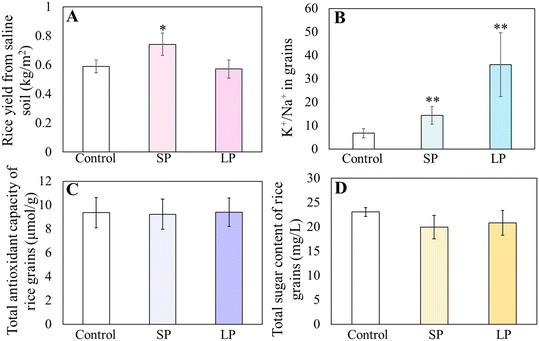 | ||
| Fig. 2 Effect of AgNPs priming on rice yield in saline soil (A). The K+/Na+ ratio (B), total antioxidant capacity (C) and total sugar content (D) of grains under salt stress after AgNP priming. | ||
Soil salinization will affect the nutritional quality of plant fruits,26 so we were interested to know whether AgNPs priming would affect the nutritional quality of the rice grains, particularly the mineral content. The nutritional quality of mature rice grains was determined through the content of macro-elements (K, Ca, Na, and Mg) and micro-elements (Ag, Fe, Mn, and Zn). In saline soil conditions, Na content in rice grains significantly decreased (p < 0.05) by 56% and 82% in SP and LP groups, respectively, while K was significantly reduced by 10% in the LP group (p < 0.05) and showed no change in the SP group (Table 1). However, both priming manners resulted in a significantly higher K+/Na+ ratio compared to the control (Fig. 2B). This interesting finding is significant for food safety, as diets high in Na can contribute to chronic diseases such as hypertension, cardiovascular diseases, and obesity.27 In addition, a higher K+/Na+ ratio also indicates improved salt stress tolerance in plants.28 Thus, AgNPs contribute to improve the nutritional quality of rice. The Ag content in grains was determined to evaluate the environmental impact of AgNPs. The results showed no significant difference in the Ag content of the treatment and the control groups (Table 1), indicating that the applied concentration of AgNPs (40 mg L−1) did not lead to Ag accumulation in the grain, thus posing no food safety concerns. Finally, the TAC and total sugar content in grains did not change in the AgNPs-primed plants (Fig. 2C and D), which is consistent with the report that AgNPs seed priming improves watermelon yield while maintaining fruit quality.29 Moreover, the increased yield observed with AgNPs seed priming holds promise for rice cultivation in saline soils.
| Ag | K | Ca | Na | |
|---|---|---|---|---|
| Asterisks indicate significant differences compared with a control (two-tailed Student's t-test, *P < 0.05, **P < 0.01). | ||||
| Control | 0.8 ± 0.71 | 2802 ± 149 | 129 ± 13 | 438 ± 171 |
| SP | 0.2 ± 0.03 | 2639 ± 184 | 134 ± 20 | 194 ± 53* |
| LP | 0.3 ± 0.08 | 2523 ± 98** | 122 ± 29 | 78 ± 25** |
| Mg | Fe | Mn | Zn | |
|---|---|---|---|---|
| Control | 1280 ± 44 | 13.9 ± 6.3 | 12.3 ± 1.4 | 39 ± 17 |
| SP | 1312 ± 81 | 12.7 ± 6.5 | 13.5 ± 2.3 | 36 ± 18 |
| LP | 1294 ± 40 | 9.0 ± 2.4 | 10.9 ± 1.2 | 35 ± 22 |
Seed metabolic reprogramming induced by AgNPs-seed priming
The primary hypothesis of this study was that seed priming or seedling leaf spraying with AgNPs confers tolerance to salt stress. This would be achieved through metabolism adjustment and transcriptional changes triggered by NPs. So, metabolomics and transcriptomics analyses were performed in treated and untreated seeds and leaves to characterize metabolite profiles and transcriptomes, aiming to elucidate the molecular mechanisms underlying enhanced salt tolerance.Metabolites are the products of metabolism that drive essential cellular functions, such as energy production and signal transduction.30 Therefore, we initially profiled the metabolites in rice seeds. By using GC-MS based non-target metabolomics, a total of 311 metabolites were identified and semi-quantified in rice seeds primed with AgNPs or water for 24 hours. A supervised multivariate analysis (orthogonal partial least-squares-discriminant analysis, OPLS-DA) model based on metabolomics datasets showed a clear separation between the two groups.11 This indicates that AgNPs seed priming induced metabolic reprogramming in rice seeds.
Differential metabolite screening using univariate t-test analysis revealed a significant increase (p < 0.05) in the content of glutamine and tryptophan (amino acids) in AgNPs-primed seeds (Fig. 3). The accumulation of these amino acids in plants plays a crucial role in maintaining nitrogen balance under salt stress conditions. Glutamine has been reported to improve the salt tolerance of salt-tolerant varieties,31 while tryptophan has shown similar beneficial effects in alleviating salt stress in various plant species, including potato (Solanum tuberosum L)32 and onion (Allium cepa).33 Interestingly, several antioxidant metabolites, including alpha-tocopherol, ascorbic acid, catechol, 1,2,4-benzenetriol, L-phenylalanine, isochlorogenic acid, 4-hydroxycinnamic acid, and trans-ferulic acid significantly decreased in AgNPs-primed seeds (p < 0.05), compared to hydroprimed seeds (Fig. 3). This metabolic imprint might be related to AgNPs-generated ROS during seed priming. Our previous study has shown that AgNPs catalyze the generation of ROS in rice seeds.11 The systematic reduction of these ROS-scavenging related metabolites may indicate that AgNPs triggered a disturbance of the reduction–oxidation (redox) homeostasis.
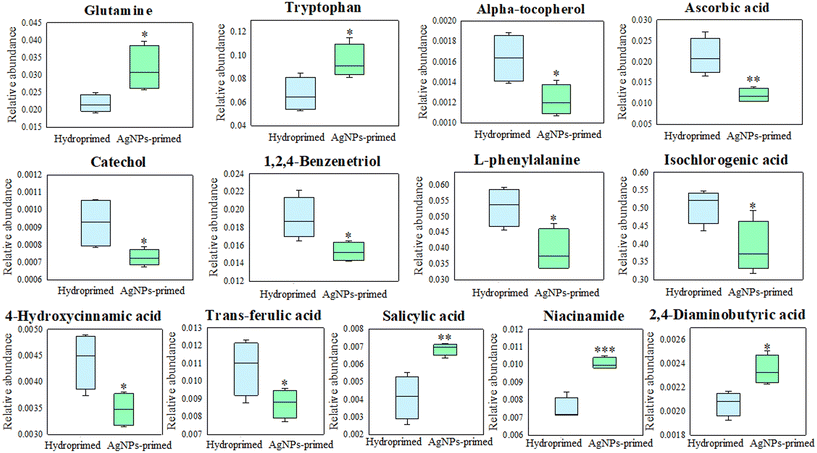 | ||
| Fig. 3 Significantly changed amino acids, antioxidants, and signaling transduction related metabolites in rice seeds. The rice seeds were soaked in the AgNP (40 mg L−1) solution or water for 24 h. | ||
ROS serve as pivotal regulators that mediate signaling pathways involved in plant responses to environmental fluctuations.34 Another noteworthy metabolic alteration is that AgNPs seed priming significantly increased the relative level of several signaling transduction-related metabolites in rice seeds (p < 0.05), including salicylic acid (SA, 67%), niacinamide (34%), and 2,4-diaminobutyric acid (DAB) (14%) (Fig. 3). Among them, SA is an important signaling molecule in enhancing plant resistance to abiotic and biotic stresses.35 Similarly, niacinamide, the amide derivative of nicotinic acid, is a critical signaling molecule associated with improving plant tolerance to diverse stresses.36 The upregulation of these signaling transduction-related metabolites suggests that the AgNPs trigger a defense response in the seeds, preparing plants to better withstand future salt stress challenges.
Conversely, the AgNPs-seed priming did not change the level of osmoprotectant-related metabolites that directly enhance rice tolerance to salt stress. For instance, the relative concentrations of soluble sugars (glucose, fructose, sucrose, and raffinose) and proline, crucial for salt tolerance, were comparable between AgNPs-primed and hydroprimed seeds. The decrease in antioxidant metabolites, increase in signaling metabolites, and no change in osmoprotectants suggest that ROS generated by AgNPs were recognized by rice seeds as a dangerous event and defense signaling pathways were activated for the subsequent salt stress.
Leaf metabolic reprogramming induced by AgNPs priming
To further investigate the specific metabolic changes related to salt tolerance in leaves induced by AgNPs priming, the metabolite data were analyzed by univariate t-test. The results showed that the AgNPs priming induced sugar accumulation in the leaves. Specifically, various saccharides, such as sucrose, altrose, D-fructose-6-phosphate, D-ribulose 5-phosphate, glucose-1-phosphate, maltotriose, and mannose 6-phosphate were significantly increased in the SP group (p < 0.05), as compared to the control group (Fig. 4 and S1†). The accumulation of sugar can be used as an osmotic substance to maintain cell homeostasis and as an energy source for plant growth and development.37 When compared to SP treatment, leaf spraying with AgNPs induced even greater accumulation of saccharides in the leaves, with the significant increase (p < 0.05) of the content of nine sugars (glucose, sucrose, 1-kestose, altrose, erythrotetrofuranose, glucose-6-phosphate, maltotriose, mannose 6-phosphate, and melezitose) (Fig. 4 and S1†). In addition, two sugar alcohols 1,5-anhydroglucitol and arabitol were significantly up-regulated in the SP group (p < 0.05) (Fig. S1†). These increases in sugars and sugar alcohols likely contributed to enhanced salt tolerance in rice.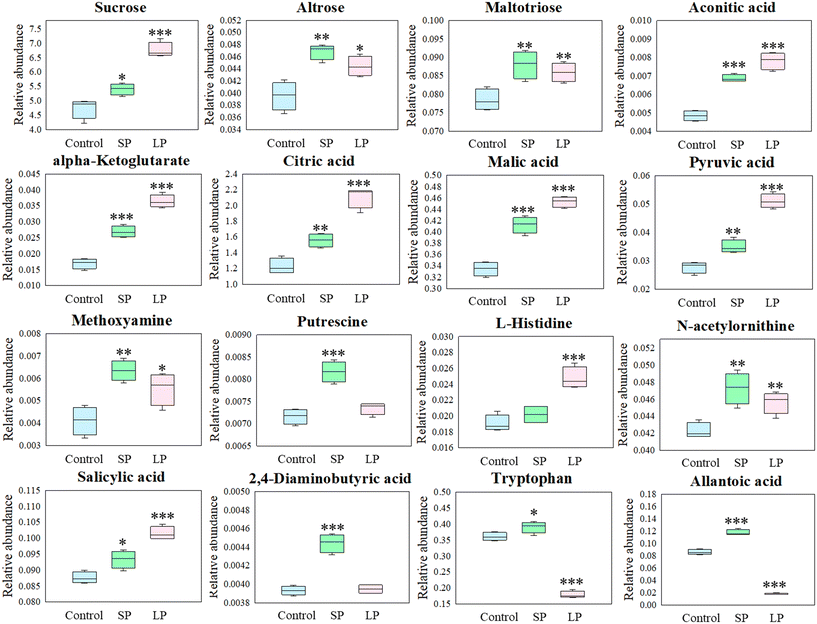 | ||
| Fig. 4 Significantly changed amino acids, antioxidants, and signaling transduction related metabolites in rice leaves. SP: seed priming and LP: leaf priming (leaf spraying). | ||
It has been shown that osmotic stress is accompanied by the accumulation of organic acids,38 as plants facing salt stress absorb excessive Na+, necessitating increased levels of organic acids to maintain intracellular pH stability and ion homeostasis.39 Several organic acids in TCA cycle intermediates increased significantly (p < 0.05) in the SP group, including aconitic acid, alpha-ketoglutarate, citric acid, malic acid and pyruvic acid (Fig. 4 and S2†). Tahjib-Ul-Arif et al.40 showed that exogenous application of citric acid promotes an increase in chlorophyll content by activating the antioxidant defense system and affecting secondary metabolism to improve crop growth and yield under various abiotic stress conditions. The TCA cycle also serves as a crucial source of metabolic energy for plants under salt stress. In addition, organic acids in Calvin cycle intermediates such as 3-phosphoglycerate, 3-phosphoglyceric acid, and glycerol 3-phosphate are also up-regulated in the SP group (Fig. S2†). Additional organic acid metabolic changes induced by AgNPs-seed priming include the up-regulation of glyceric acid, hydrocinnamic acid, oxalic acid, allantoic acid, isoferulic acid, quinic acid, shikimic acid, and 2,4-diaminobutyric acid in leaves (Fig. 4 and S2†). Similarly, leaf spraying AgNPs also caused a significant increase (p < 0.05) in the content of numerous organic acids in leaves, such as 3-phosphoglycerate, glyceric acid, alpha-aminoadipic acid, benzoic acid, cis-caffeic acid, hydrocinnamic acid, isoferulic acid, quinic acid, shikimic acid, and five TCA cycle intermediates (citric acid, aconitic acid, alpha-ketoglutarate, malic acid and pyruvic acid). Therefore, the increase in these organic acids likely contributed to enhanced salt tolerance in rice (Fig. 4 and S2†).
In addition, significant upregulation of certain amines was observed in the SP group (p < 0.05). Accumulation of amines within cells is known to enhance plant tolerance to salt stress.41 For example, putrescine, a new plant endogenous hormone type, is widely available in plants.37 Putrescine can act as an osmoprotectant to alleviate the damage of salt stress by reducing the accumulation of Na+ and Cl− in cells and the efflux of K+.42 Putrescine also functions as a signaling molecule to regulate stress responses in plants.43 Compared to the control group, the level of putrescine in the SP group was significantly increased by 14% (Fig. 4). Ghalati et al.44 showed that the exogenous foliar application of putrescine enhanced salt tolerance in guava (Psidium guajava L.) seedlings. Thus, the upregulation of putrescine induced by AgNPs seed priming likely contributes to enhanced salt tolerance in rice. In addition, the levels of ethanolamine, phosphoethanolamine, methoxyamine, N-acetylputrescine, and 2-monoolein were also significantly upregulated in the SP group (p < 0.05) (Fig. 4 and S3†). Whereas AgNPs leaf spraying resulted in few amines accumulating in leaves, only N-acetyl-D-hexosamine and the methoxyamine level were significantly up-regulated (p < 0.05) (Fig. 4 and S3†). Taken together, the up-regulated levels of these osmoprotectants likely contribute to the enhanced salt tolerance in rice stimulated by AgNPs.
The stimulation of AgNPs also induced the upregulation of several amino acid content in the leaves. For instance, AgNPs leaf spraying significantly increased the content of L-histidine in leaves by 30% (p < 0.05) compared to the control (Fig. 4). Research of histidine interactions with membranes and macromolecules has demonstrated its unique role in enhancing salt tolerance. Ji et al.45 showed that histidine enhances salt tolerance in maize (Zea mays) roots by modulating genes involved in phytohormone signaling, glycolysis and nitrogen metabolism pathways. Additionally, AgNPs leaf spraying induced metabolic changes include the upregulation of glutamyl-valine and N-acetylornithine, while AgNPs seed priming induced metabolic changes include the upregulation of N-acetylornithine, tryptophan and homoserine in leaves (Fig. 4 and S3†). These metabolites also appear to play an essential role in enhancing salt tolerance in rice.
Overlapping differentially accumulated metabolites (DAMs) in seeds and leaves induced by AgNPs priming
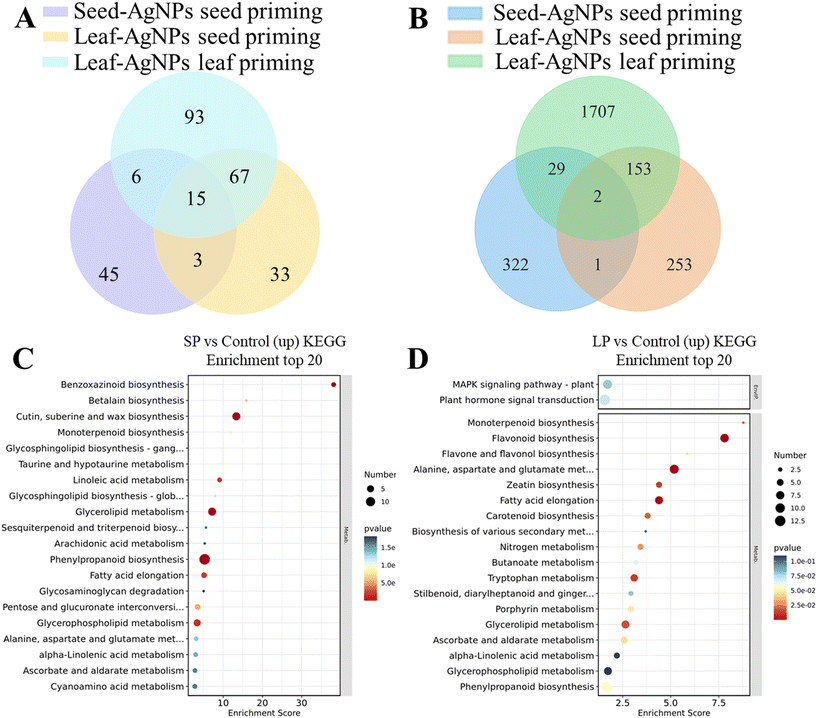
The Venn diagram (univariate t-test analysis) (Fig. 5A) showed that the number of DAMs in AgNPs-primed seeds, SP and LP groups was 70, 119 and 182, respectively. Notably, 18 DAMs were common between the AgNPs-primed seeds and the SP group, while 82 DAMs were common between the SP and LP groups. Among them, SA, DAB, allantoic acid and L-tryptophan metabolites were significantly up-regulated in both seeds and leaves induced by AgNPs seed priming (p < 0.05) (Fig. 3 and 4). As mentioned above, SA and DAB are crucial metabolites in plant stress response, potentially enhancing rice's ability to adjust and adapt to future stressful environments more efficiently. Tryptophan is known for its involvement in osmotic regulation, stomatal regulation and scavenging reactive oxygen species during stress responses.46 Allantoic acid plays a vital role in plant nitrogen metabolism, serving as a form of nitrogen storage and transport.47 In summary, these metabolites consistently showed up-regulation from the seed to the seedling stage following AgNPs treatment, suggesting their importance in enhancing rice's salt tolerance.Seed transcriptional reprogramming induced by AgNPs priming
Transcriptome sequencing of AgNPs primed seeds and leaves was carried out using RNA-Seq to dig out genes related to the enhancement of salt tolerance in rice. First, a total of 354 DEGs were detected in AgNPs-primed seeds, among which 318 were up-regulated and 36 down-regulated. Phytohormones are closely related to cell signaling pathways and are involved in plant response to salt stress by mediating complex signaling cascades.48 We found that AgNPs-primed seeds exhibited up-regulated expression of several genes related to hormone signal transduction, including ethylene-responsive transcription factor, auxin-responsive SAUR gene, and gibberellin receptor (Fig. 6A). Ethylene, auxin, gibberellin (GA), and cytokinin are key regulatory factors responding to salt stress.49 For instance, Iqbal et al.50 showed that the accumulation of auxin in wheat seeds enhances salt tolerance, thereby boosting wheat yield under saline conditions. GA also mitigates salt stress effects and enhances plant salt tolerance.51 Wang et al.52 showed that the accumulation of endogenous GA improved the salt tolerance of cucumber. In addition, the expression of cytokinin-o-glucosyltransferase, associated with cytokinin, was also up-regulated in the seeds (Fig. 6A). O-Glucosylation of cytokinin is considered to be a stable storage form of cytokinin.53 Overall, the activation of hormone signaling pathways and up-regulation of phytohormone-related genes in seeds by AgNPs priming likely contribute to enhancing salt tolerance in rice.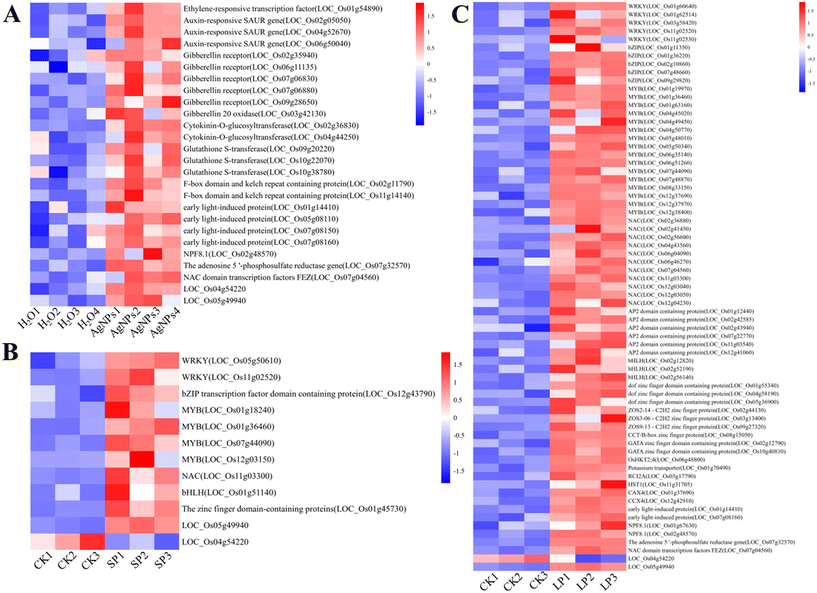 | ||
| Fig. 6 Heatmap of differentially expressed genes related to salt tolerance in rice seeds (A) and leaves (B and C) primed with AgNPs. | ||
Salt stress triggers the production of abundant harmful ROS. AgNPs priming induced significant up-regulation of several antioxidant-related genes in rice seeds. Notably, glutathione S-transferases (GSTs) are one of the main detoxification enzymes of plants, facilitating the conjugation of reduced glutathione with various electrophiles to form complexes that detoxify harmful substances.54 GSTs protect plants from oxidative stress mainly through detoxification.55 Compared with the control, the GST genes were notably up-regulated 2.1- to 2.3-fold in AgNPs-primed seeds (Fig. 6A). Qi et al.56 showed that Arabidopsis plants overexpressing GST possessed higher salt tolerance. The metabolic pathway of phenylpropane is the primary pathway for synthesizing secondary metabolites such as phenols, flavonoids and lignin, which constitute an important non-enzymatic antioxidant defense system in plants.57 The F-box domain and kelch repeat containing protein genes (OsFBK4 and OsFBK25) involved in the phenylpropanoid biosynthesis pathway were up-regulated 2.1- and 2.7- fold in the AgNPs-primed seeds (Fig. 6A). Zegeye et al.58 demonstrated that the OsFBK4 positively regulates rice plant height by promoting internode cell size and is involved in GA signaling and biosynthetic pathways. Increasing evidence underscores the role of F-box proteins in plants' response to salt stress. Zhao et al.59 found that wheat F-box gene TaFBA1 could improve the salt tolerance of transgenic tobacco by increasing the activity of antioxidant enzymes and decreasing MDA content and ROS accumulation. Therefore, AgNPs seed priming likely enhances rice's salt tolerance by activating its antioxidant defense system. In addition, matching the transcriptome and metabolome data with the KEGG database revealed that there were two common enrichment pathways: fatty acid biosynthesis and amino sugar and nucleotide sugar metabolism. This finding suggests that energy metabolism plays an important role in enhancing the salt tolerance of rice.
Leaf transcriptional reprogramming induced by AgNPs priming
First, a total of 409 DEGs were detected in rice leaves with AgNPs seed priming (SP), among which 319 were up-regulated and 90 were down-regulated. As for the rice leaves sprayed by AgNPs (LP), a total of 1891 DEGs were detected, of which 1023 were up-regulated and 868 were down-regulated. Transcription factors (TFs) constitute a diverse class of functional proteins ubiquitous in eukaryotes. They recognize and bind the cis-acting elements of target gene promoters, thereby activating or repressing gene expression, and are pivotal in plant responses to salt stress.60 Our results demonstrate that AgNPs priming significantly upregulated the expression of numerous TFs in the leaves compared with the control. Specifically, LP upregulated the TFs WRKY (2.1- to 20-fold), the basic leucine zipper (bZIP) (2.1- to 4-fold), the myeloblastosis (MYB) (2- to 5.6-fold), the NAC (2.1- to 26.9-fold), the APETALA2 (AP2) domain-containing proteins (1.9- to 2.7-fold), the basic helix–loop–helix (bHLH) (2.1- to 5.7-fold), and the zinc finger domain-containing proteins (2- to 49.6-fold) (Fig. 6C). WRKY TFs are involved in regulating abscisic acid (ABA), SA, and jasmonic acid (JA) signaling pathways crucial for plant stress responses. The OsWRKY53 has been reported as a key regulator of the salt tolerance of rice.8 Similarly, bZIP and NAC TFs participate in ABA signaling pathways essential for abiotic stress responses.61 Research by Liu et al.62 demonstrated the role of OsbZIP71 in enhancing rice salt tolerance. Hong et al.63 found that ONAC022 decreased the accumulation of Na+ in roots and shoots, increased protective substances such as proline and soluble sugar, and improved the salt tolerance of rice by regulating the ABA-mediated pathway. MYB proteins, prominent in plant TF families, enhance salt tolerance by mediating hormone synthesis and signal transduction.64 In our study, MYB80 in the SP group showed an 84.8-fold upregulation compared to the control (Fig. 6B). Tang et al.65 found that OsMYB6 transgenic rice plants had higher salt tolerance compared with wild-type plants. AP2 TFs also play an important role in improving plant salt tolerance. For example, the silencing of two AP2/ERF (ethylene response factor) transcription factors GhERF4L and GhERF54L in cotton can reduce the plant's salt tolerance.66 bHLH TFs act as positive regulators of salt stress. For example, in tobacco (Nicotiana tabacum) the TF NtbHLH123 enhances salt tolerance by activating the expression of NADPH oxidase NtRbohE.67 In our study, we found that the bHLH TF (LOC_Os08g42470) was upregulated by 56.6-fold in the SP group, compared to the control (Fig. 6B). Zinc finger proteins are involved in various RNA metabolism and play an important role in regulating gene expression and salt tolerance in plants.68 Among them, ZOS3-06-C2H2 zinc finger protein was up-regulated 49.6-fold in the LP group, compared to the control (Fig. 6C). Therefore, the upregulation of these TFs likely plays a beneficial role in the resistance of rice to salt stress.Maintaining ionic homeostasis and a balanced Na+/K+ ratio under salt stress is the key to enhancing plant salt tolerance. It has been reported that the main genes regulating Na+ in plants are vacuolar Na+/H+ antiporter (NHX) gene, plasma membrane Na+/H+ antiporter (SOS1) gene, and high-affinity K+ channel transporter (HKT) gene.2 In this study, we observed a 2.4-fold upregulation of OsHKT2;4 expression and a 3.6-fold upregulation of the potassium transporter gene OsHAK5 in the LP group (Fig. 6C). Members of the HKT family, such as OsHKT2;1, OsHKT2;3/OsHKT3, and OsHAK5, play critical roles in maintaining intracellular Na+/K+ homeostasis by translocating Na+ and K+, thereby regulating rice's response to salt stress.4 In addition, we found that the expression of two other Na+-related genes was up-regulated in the LP group: the salt-responsive protein genes RCI2A (increased by 2.8-fold) and HST1 (increased by 5.3-fold) (Fig. 6C). Overexpression of the RCI2A has been shown to mitigate salt stress-induced damage by reducing Na+ uptake in Arabidopsis thaliana.69 Aycan et al.70 demonstrated that the overexpression of the salt tolerance gene hst1 (hitomebore salt tolerant 1) reduces the accumulation of Na+, decreases lipid peroxidation and H2O2 content under salt stress, and increases the activity of proline and antioxidant enzymes, thereby improving salt tolerance in rice.
Calcium ion (Ca2+), as a second messenger, plays a regulatory role in cell signal transduction during plant growth and stress response. Under salt stress, plants first regulate Ca2+ concentrations through Ca2+ receptors, subsequently activating various protein kinases to mitigate salt-induced damage.71 Within the Ca2+/cation antiporter (CaCA) superfamily, Ca2+/proton exchangers (CAX) and cation/Ca2+ exchangers (CCX) mediate Ca2+ flux in plants.72 A family of capacity CAX transporters mediate part of high-capacity cation/H+ antiport across the tonoplast, and play an essential role in regulating the balance of Ca2+ and other cations in the cell.73 CAX also plays an important functional role in root growth under abiotic stress conditions.74 The product of the CCX4 gene acts as a cation/calcium exchanger and can help maintain cellular ion homeostasis during salt stress.75 In this study, we found that the expression of CAX4 gene and CCX4 gene was upregulated 2.2- and 6.0-fold in the LT group (Fig. 6C), respectively. Similarly, Bu et al.76 demonstrated that overexpression of CAX4 genes can significantly improve Arabidopsis tolerance to salt and ion stress. Cheng et al.77 found that CCX4 was up-regulated in the roots of cv. ‘Jinba’ under salt stress. The up-regulation of these genes may explain the enhanced salt tolerance observed in rice after AgNPs priming. In addition, by comparing the enriched KEGG pathways in metabolomics and transcriptomics, it was found that the pathways shared in the SP group were tyrosine metabolism, pentose and glucuronate interconversions, glycerolipid metabolism, and glycolysis/gluconeogenesis, while the pathways shared in the LP group were alanine, aspartate and glutamate metabolism (Fig. 5 and S4†). This finding suggests that AgNPs may enhance the salt tolerance of rice by promoting metabolic pathways such as energy metabolism, substance conversion and biosynthesis.
Overlapping DEGs in seeds and leaves induced by AgNPs priming
Venn analysis of the DEGs between AgNPs primed seeds vs. hydroprimed seeds, SP group vs. control, and LP group vs. control revealed commonalities and distinct patterns (Fig. 5B). Specifically, three DEGs (LOC_Os04g16722, LOC_Os04g54220 and LOC_Os05g49940) were identified in both AgNPs-primed seeds vs. hydroprimed seeds and SP group vs. control. Additionally, there were 31 common DEGs induced by SP and LP, of which 16 were up-regulated and only one was down-regulated in both groups. Among these up-regulated genes, several were newly discovered and may contribute to enhancing salt tolerance in rice. For instance, early light-induced protein (ELIP) accumulates in Tortula ruralis (Syntrichia ruralis)78 and halotolerant green alga Dunaliella79 under salt stress, indicating that ELIP is important in salt stress tolerance. We found that the expression of ELIP2 was up-regulated 2.3- to 2.7-fold in the AgNPs-primed seeds and 4.1- to 6.1-fold in the LP group (Fig. 6). Furthermore, the peptide transporter NPF8.1 of rice nitrate transporter 1/peptide transporter family (NPF) exhibited 2-fold and 3.7-fold up-regulation in the AgNPs-primed seeds and LP group, respectively (Fig. 6). Diyang et al.80 showed that OsNPF8.1 is a high-affinity rice polypeptide transporter, and the peptide transport mediated by OsNPF8.1 may contribute to the improvement of salt and drought tolerance of rice. Additionally, adenosine 5′-phosphosulfate reductase (APR) gene expression was up-regulated by 2.2- and 2.7-fold in the AgNPs-primed seeds and LP group, respectively (Fig. 6). APR mediated the production of cysteine in the sulfate assimilation pathway and is a key rate-limiting enzyme in the sulfur assimilation pathway. Sulfur compounds play an important role in plant stress defense.81 Koprivova et al.82 found that APR activity and mRNA levels of three APR isoforms were increased 3-fold in Arabidopsis (Arabidopsis thaliana) roots under salt stress, suggesting that APR accumulation plays a role in plant adaptation to salt stress. Moreover, the expression of NAC domain transcription factors FEZ was up-regulated 2.6- and 9.4-fold in the AgNPs-primed seeds and LP group, respectively (Fig. 6). KEGG enrichment analysis showed that the common enrichment pathway in both AgNPs-primed seeds and LP group were plant hormone signal transduction, flavone and flavonol biosynthesis, MAPK signaling pathway and zeatin biosynthesis (Fig. 5D). Notably, the SP and LP groups exhibited the highest number of common DEGs, totaling 155 genes (Fig. 5B), with 109 upregulated and 42 downregulated genes. KEGG enrichment analysis (Fig. 5C and D) showed eight common enrichment pathways in both SP and LP groups (monoterpenoid biosynthesis, glycerolipid metabolism, phenylpropanoid biosynthesis, fatty acid elongation, glycerophospholipid metabolism, alanine, aspartate and glutamate metabolism, alpha-linolenic acid metabolism and ascorbate and aldarate metabolism). In summary, AgNPs priming in different tissues (seed and leaf) and through different methods (seed priming and leaf priming) induces overlapping transcriptional responses, underscoring commonalities in rice plant stress adaptation. Interestingly, the consistent expression patterns of LOC_Os04g54220 and LOC_Os05g49940 across treatments suggest their potential significance in AgNPs-mediated responses.Conclusion
Overall, this study demonstrates that AgNPs priming alleviated the adverse effects of salt stress on rice growth. Seed priming with AgNPs increased rice plants' chlorophyll a, carotenoid content and total antioxidant capacity. Concurrently, it decreased the MDA content in salt-stressed rice seedlings and boosted the salt tolerance throughout the entire life cycle, resulting in increased yield. Therefore, AgNPs seed priming proved more effective in enhancing rice's salt tolerance compared to leaf priming. Omics studies provided insights into the molecular mechanisms underlying this enhancement. Metabolomics analysis revealed that AgNPs exposure either on seeds or seedling leaves promoted the accumulation of osmoprotectants, such as sugars, organic acids, amino acids, and amines. Transcriptome analysis demonstrated that AgNPs regulated the expression of salt stress response genes, limited the absorption of Na+, maintained ion homeostasis, and alleviated salt stress. In summary, AgNPs, whether used as a seed priming agent or sprayed on seedlings, effectively improved rice seedling growth under saline conditions, whereas AgNPs seed priming, instead of leaf priming, significantly increased the grain yield in a saline land. Lastly, given the high cost and uncertain environmental risk of AgNPs, other inexpensive and environmentally friendly nanomaterials are planned to be explored in future studies.Data availability
The data supporting the findings of this study are available within the paper. Should any additional raw data files be needed, they are available from the corresponding author upon reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by Liaoning Provincial Science and Technology Project of China under grant 2022JH2/101300161. Any opinions, finding, and conclusions or recommendations expressed in this material are those of authors and do not necessarily reflect the views of National Science Foundation of China.References
- Z. Maryum, T. Luqman, S. Nadeem, S. Khan, B. Wang, A. Ditta and M. K. R. Khan, An overview of salinity stress, mechanism of salinity tolerance and strategies for its management in cotton, Front. Plant Sci., 2022, 13, 907937 CrossRef.
- I. N. B. L. Reddy, B.-K. Kim, I.-S. Yoon, K.-H. Kim and T.-R. Kwon, Salt Tolerance in Rice: Focus on Mechanisms and Approaches, Rice Sci., 2017, 24(3), 123–144 CrossRef.
- T. Balasubramaniam, G. Shen, N. Esmaeili and H. Zhang, Plants' Response Mechanisms to Salinity Stress, Plants, 2023, 12(12), 2253 CrossRef CAS PubMed.
- C. Liu, B. Mao, D. Yuan, C. Chu and M. Duan, Salt tolerance in rice: Physiological responses and molecular mechanisms, Crop J., 2022, 10(1), 13–25 CrossRef.
- M. Hussain, S. Ahmad, S. Hussain, R. Lal, S. Ul-Allah and A. Nawaz, Chapter Six - Rice in Saline Soils: Physiology, Biochemistry, Genetics, and Management, in Advances in Agronomy, ed. D. L. Sparks, Academic Press, 2018, vol. 148, pp. 231–287 Search PubMed.
- R. Quan, J. Wang, J. Hui, H. Bai, X. Lyu, Y. Zhu, H. Zhang, Z. Zhang, S. Li and R. Huang, Improvement of Salt Tolerance Using Wild Rice Genes, Front. Plant Sci., 2017, 8, 2269 CrossRef.
- X. Sheng, Z. Ai, Y. Tan, Y. Hu, X. Guo, X. Liu, Z. Sun, D. Yu, J. Chen, N. Tang, M. Duan and D. Yuan, Novel Salinity-Tolerant Third-Generation Hybrid Rice Developed via CRISPR/Cas9-Mediated Gene Editing, Int. J. Mol. Sci., 2023, 24(9), 8025 CrossRef CAS.
- J. Yu, C. Zhu, W. Xuan, H. An, Y. Tian, B. Wang, W. Chi, G. Chen, Y. Ge, J. Li, Z. Dai, Y. Liu, Z. Sun, D. Xu, C. Wang and J. Wan, Genome-wide association studies identify OsWRKY53 as a key regulator of salt tolerance in rice, Nat. Commun., 2023, 14(1), 3550 CrossRef CAS PubMed.
- R. Mittler, S. I. Zandalinas, Y. Fichman and F. Van Breusegem, Reactive oxygen species signalling in plant stress responses, Nat. Rev. Mol. Cell Biol., 2022, 23(10), 663–679 CrossRef CAS PubMed.
- L. Zhao, T. Bai, H. Wei, J. L. Gardea-Torresdey, A. Keller and J. C. White, Nanobiotechnology-based strategies for enhanced crop stress resilience, Nat. Food, 2022, 3(10), 829–836 CrossRef.
- X. Yan, S. Chen, Z. Pan, W. Zhao, Y. Rui and L. Zhao, AgNPs-Triggered Seed Metabolic and Transcriptional Reprogramming Enhanced Rice Salt Tolerance and Blast Resistance, ACS Nano, 2023, 17(1), 492–504 CrossRef CAS.
- Y. C. Hariadi, A. Y. Nurhayati, S. Soeparjono and I. Arif, Screening Six Varieties of Rice (Oryzasativa) for Salinity Tolerance, Procedia Environ. Sci., 2015, 28, 78–87 CrossRef.
- H. K. Lichtenthaler, [34] Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes, in Methods in Enzymology, Academic Press, 1987, vol. 148, pp. 350–382 Search PubMed.
- N. Jambunathan, Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants, Methods Mol. Biol., 2010, 639, 292–298 CrossRef.
- I. F. Benzie and J. J. Strain, The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: the FRAP assay, Anal. Biochem., 1996, 239(1), 70–76 CrossRef CAS.
- V. L. Singleton and J. A. Rossi, Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents, Am. J. Enol. Vitic., 1965, 16(3), 144–158 CrossRef CAS.
- S. Verma and R. S. Dubey, Effect of cadmium on soluble sugars and enzymes of their metabolism in rice, Biol. Plant., 2001, 44(1), 117–123 CrossRef CAS.
- M. DuBois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Colorimetric Method for Determination of Sugars and Related Substances, Anal. Chem., 1956, 28(3), 350–356 CrossRef CAS.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15(12), 550 CrossRef PubMed.
- C. Xie, X. Mao, J. Huang, Y. Ding, J. Wu, S. Dong, L. Kong, G. Gao, C. Y. Li and L. Wei, KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases, Nucleic Acids Res., 2011, 39(Web Server issue), W316–W322 CrossRef CAS PubMed.
- I. Khan, M. A. Raza, S. A. Awan, G. A. Shah, M. Rizwan, B. Ali, R. Tariq, M. J. Hassan, M. N. Alyemeni, M. Brestic, X. Zhang, S. Ali and L. Huang, Amelioration of salt induced toxicity in pearl millet by seed priming with silver nanoparticles (AgNPs): The oxidative damage, antioxidant enzymes and ions uptake are major determinants of salt tolerant capacity, Plant Physiol. Biochem., 2020, 156, 221–232 CrossRef CAS PubMed.
- A. K. S. H. Mohamed, M. F. Qayyum, A. M. Abdel-Hadi, R. A. Rehman, S. Ali and M. Rizwan, Interactive effect of salinity and silver nanoparticles on photosynthetic and biochemical parameters of wheat, Arch. Agron. Soil Sci., 2017, 63(12), 1736–1747 CrossRef CAS.
- O. S. Singh, N. Pant, L. Laishram, M. M. Tewari, R. Dhoundiyal, K. Joshi and C. S. Pandey, Effect of CuO nanoparticles on polyphenols content and antioxidant activity in Ashwagandha (Withania somnifera L. Dunal), J. Pharmacogn. Phytochem., 2018, 7, 3433–3439 CAS.
- Y. Li, Y. Yang, Y. Hu, H. Liu, M. He, Z. Yang, F. Kong, X. Liu and X. Hou, DELLA and EDS1 Form a Feedback Regulatory Module to Fine-Tune Plant Growth–Defense Tradeoff in Arabidopsis, Mol. Plant, 2019, 12(11), 1485–1498 CrossRef CAS PubMed.
- M. Waqas Mazhar, M. Ishtiaq, I. Hussain, A. Parveen, K. Hayat Bhatti, M. Azeem, S. Thind, M. Ajaib, M. Maqbool, T. Sardar, K. Muzammil and N. Nasir, Seed nano-priming with Zinc Oxide nanoparticles in rice mitigates drought and enhances agronomic profile, PLoS One, 2022, 17(3), e0264967 CrossRef CAS.
- Q. Lu, G. Ge, D. Sa, Z. Wang, M. Hou and Y. S. Jia, Effects of salt stress levels on nutritional quality and microorganisms of alfalfa-influenced soil, PeerJ, 2021, 9, e11729 CrossRef PubMed.
- D. Mozaffarian, S. Fahimi and G. M. Singh, Global Sodium Consumption and Death From Cardiovascular Causes, J. Vasc. Surg., 2015, 61(2), 567 CrossRef.
- D. M. Almeida, M. M. Oliveira and N. J. M. Saibo, Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants, Genet. Mol. Biol., 2017, 40(1 suppl 1), 326–345 CrossRef CAS.
- P. Acharya, G. K. Jayaprakasha, K. M. Crosby, J. L. Jifon and B. S. Patil, Nanoparticle-Mediated Seed Priming Improves Germination, Growth, Yield, and Quality of Watermelons (Citrullus lanatus) at multi-locations in Texas, Sci. Rep., 2020, 10(1), 5037 CrossRef CAS PubMed.
- C. H. Johnson, J. Ivanisevic and G. Siuzdak, Metabolomics: beyond biomarkers and towards mechanisms, Nat. Rev. Mol. Cell Biol., 2016, 17(7), 451–459 CrossRef CAS PubMed.
- P. Kumar, M. Choudhary, T. Halder, N. R. Prakash, V. Singh, V. T. V., S. Sheoran, R. K. T., N. Longmei, S. Rakshit and K. H. M. Siddique, Salinity stress tolerance and omics approaches: revisiting the progress and achievements in major cereal crops, Heredity, 2022, 128(6), 497–518 CrossRef CAS PubMed.
- M. Gull, Z. A. Sajid and F. Aftab, Alleviation of Salt Stress in Solanum tuberosum L. by Exogenous Application of Indoleacetic acid and l-Tryptophan, J. Plant Growth Regul., 2023, 42(5), 3257–3273 CrossRef CAS.
- M. Hussein, S. Y. Faham and A. Kumar, Role of Foliar Application of Nicotinic Acid and Tryptophan on Onion Plants Response to Salinity Stress, J. Agric. Sci., 2014, 6, 41 Search PubMed.
- G. Decros, P. Baldet, B. Beauvoit, R. Stevens, A. Flandin, S. Colombié, Y. Gibon and P. Pétriacq, Get the Balance Right: ROS Homeostasis and Redox Signalling in Fruit, Front. Plant Sci., 2019, 10, 1091 CrossRef.
- M. I. R. Khan, P. Poor and T. Janda, Salicylic Acid: A Versatile Signaling Molecule in Plants, J. Plant Growth Regul., 2022, 41(5), 1887–1890 CrossRef CAS.
- T. Berglund, A. Wallström, T.-V. Nguyen, C. Laurell and A. B. Ohlsson, Nicotinamide; antioxidative and DNA hypomethylation effects in plant cells, Plant Physiol. Biochem., 2017, 118, 551–560 CrossRef CAS.
- N. Rajkumari, S. Chowrasia, J. Nishad, S. A. Ganie and T. K. Mondal, Metabolomics-mediated elucidation of rice responses to salt stress, Planta, 2023, 258(6), 111 CrossRef CAS PubMed.
- F. Kaplan, J. Kopka, D. W. Haskell, W. Zhao, K. C. Schiller, N. Gatzke, D. Y. Sung and C. L. Guy, Exploring the temperature-stress metabolome of Arabidopsis, Plant Physiol., 2004, 136(4), 4159–4168 CrossRef CAS.
- J. Chang, B. E. Cheong, S. Natera and U. Roessner, Morphological and metabolic responses to salt stress of rice (Oryza sativa L.) cultivars which differ in salinity tolerance, Plant Physiol. Biochem., 2019, 144, 427–435 CrossRef CAS.
- M. Tahjib-Ul-Arif, M. I. Zahan, M. M. Karim, S. Imran, C. T. Hunter, M. S. Islam, M. A. Mia, M. A. Hannan, M. S. Rhaman, M. A. Hossain, M. Brestic, M. Skalicky and Y. Murata, Citric Acid-Mediated Abiotic Stress Tolerance in Plants, Int. J. Mol. Sci., 2021, 22(13), 7235 CrossRef CAS PubMed.
- M. Sequera-Mutiozabal, C. Antoniou, A. F. Tiburcio, R. Alcázar and V. Fotopoulos, Polyamines: Emerging Hubs Promoting Drought and Salt Stress Tolerance in Plants, Curr. Mol. Biol. Rep., 2017, 3(1), 28–36 CrossRef.
- A. Ndayiragije and S. Lutts, Exogenous Putrescine Reduces Sodium and Chloride Accumulation in NaCl-Treated Calli of the Salt-Sensitive Rice Cultivar I Kong Pao, Plant Growth Regul., 2006, 48(1), 51–63 CrossRef CAS.
- A. Sharma and N. Garg, 7 - Polyamines: A promising strategy for imparting salinity stress tolerance in legumes, in Abiotic Stress and Legumes, ed. V. P. Singh, S. Singh, D. K. Tripathi, S. M. Prasad, R. Bhardwaj and D. K. Chauhan, Academic Press, 2021, pp. 137–174 Search PubMed.
- R. Esfandiari Ghalati, M. Shamili and A. Homaei, Effect of putrescine on biochemical and physiological characteristics of guava (Psidium guajava L.) seedlings under salt stress, Sci. Hortic., 2020, 261, 108961 CrossRef CAS.
- H. Ji, G. Yang, X. Zhang, Q. Zhong, Y. Qi, K. Wu and T. Shen, Regulation of salt tolerance in the roots of Zea mays by L-histidine through transcriptome analysis, Front. Plant Sci., 2022, 13, 1049954 CrossRef PubMed.
- F. J. Corpas, D. K. Gupta and J. M. Palma, Tryptophan: A Precursor of Signaling Molecules in Higher Plants, in Hormones and Plant Response, ed. D. K. Gupta and F. J. Corpas, Springer International Publishing, Cham, 2021, pp. 273–289 Search PubMed.
- A. M. Carter and M. Tegeder, Increasing Nitrogen Fixation and Seed Development in Soybean Requires Complex Adjustments of Nodule Nitrogen Metabolism and Partitioning Processes, Curr. Biol., 2016, 26(15), 2044–2051 CrossRef CAS PubMed.
- V. Verma, P. Ravindran and P. P. Kumar, Plant hormone-mediated regulation of stress responses, BMC Plant Biol., 2016, 16, 86 CrossRef.
- H. Ryu and Y.-G. Cho, Plant hormones in salt stress tolerance, J. Plant Biol., 2015, 58(3), 147–155 CrossRef CAS.
- M. Iqbal and S. Basra, Salt tolerance and regulation of gas exchange and hormonal homeostasis by auxin-priming in wheat, Pesqui. Agropecu. Bras., 2013, 48, 1210–1219 CrossRef.
- S. Hussain, M. B. Hafeez, R. Azam, K. Mehmood, M. Aziz, S. Ercisli, T. Javed, A. Raza, N. Zahra, S. Hussain and X. Ren, Deciphering the Role of Phytohormones and Osmolytes in Plant Tolerance Against Salt Stress: Implications, Possible Cross-Talk, and Prospects, J. Plant Growth Regul., 2023, 38–59 CAS.
- Y. Wang, X. Gong, W. Liu, L. Kong, X. Si, S. Guo and J. Sun, Gibberellin mediates spermidine-induced salt tolerance and the expression of GT-3b in cucumber, Plant Physiol. Biochem., 2020, 152, 147–156 CrossRef CAS PubMed.
- L. Chen, J. Zhao, J. Song and P. E. Jameson, Cytokinin glucosyl transferases, key regulators of cytokinin homeostasis, have potential value for wheat improvement, Plant Biotechnol. J., 2021, 19(5), 878–896 CrossRef CAS PubMed.
- I. Nianiou-Obeidat, P. Madesis, C. Kissoudis, G. Voulgari, E. Chronopoulou, A. Tsaftaris and N. E. Labrou, Plant glutathione transferase-mediated stress tolerance: functions and biotechnological applications, Plant Cell Rep., 2017, 36(6), 791–805 CrossRef CAS.
- B. Jia, M. Sun, X. Sun, R. Li, Z. Wang, J. Wu, Z. Wei, H. DuanMu, J. Xiao and Y. Zhu, Overexpression of GsGSTU13 and SCMRP in Medicago sativa confers increased salt–alkaline tolerance and methionine content, Physiol. Plant., 2016, 156(2), 176–189 CrossRef CAS PubMed.
- Y. C. Qi, W. Q. Liu, L. Y. Qiu, S. M. Zhang, L. Ma and H. Zhang, Overexpression of glutathione S-transferase gene increases salt tolerance of arabidopsis, Russ. J. Plant Physiol., 2010, 57(2), 233–240 CrossRef CAS.
- N. Q. Dong and H. X. Lin, Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions, J. Integr. Plant Biol., 2021, 63(1), 180–209 CrossRef CAS.
- W. A. Zegeye, D. Chen, M. Islam, H. Wang, A. Riaz, M. H. Rani, K. Hussain, Q. Liu, X. Zhan, S. Cheng, L. Cao and Y. Zhang, OsFBK4, a novel GA insensitive gene positively regulates plant height in rice (Oryza Sativa L.), Ecol. Genet. Genomics, 2022, 23, 100115 CAS.
- Z. Zhao, G. Zhang, S. Zhou, Y. Ren and W. Wang, The improvement of salt tolerance in transgenic tobacco by overexpression of wheat F-box gene TaFBA1, Plant Sci., 2017, 259, 71–85 CrossRef CAS PubMed.
- V. C. D. Fernando, Major transcription factor families involved in salinity stress tolerance in plants, in Transcription Factors for Abiotic Stress Tolerance in Plants, ed. S. H. Wani, Academic Press, ch. 7, 2020, pp. 99–109 Search PubMed.
- M. C. Baloglu, V. Eldem, M. Hajyzadeh and T. Unver, Genome-wide analysis of the bZIP transcription factors in cucumber, PLoS One, 2014, 9(4), e96014 CrossRef PubMed.
- C. Liu, B. Mao, S. Ou, W. Wang, L. Liu, Y. Wu, C. Chu and X. Wang, OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice, Plant Mol. Biol., 2014, 84(1), 19–36 CrossRef CAS PubMed.
- Y. Hong, H. Zhang, L. Huang, D. Li and F. Song, Overexpression of a Stress-Responsive NAC Transcription Factor Gene ONAC022 Improves Drought and Salt Tolerance in Rice, Front. Plant Sci., 2016, 7, 4 Search PubMed.
- P. S. Shukla, P. Agarwal, K. Gupta and P. K. Agarwal, Molecular characterization of an MYB transcription factor from a succulent halophyte involved in stress tolerance, AoB Plants, 2015, 7, plv054 CrossRef PubMed.
- Y. Tang, X. Bao, Y. Zhi, Q. Wu, Y. Guo, X. Yin, L. Zeng, J. Li, J. Zhang, W. He, W. Liu, Q. Wang, C. Jia, Z. Li and K. Liu, Overexpression of a MYB Family Gene, OsMYB6, Increases Drought and Salinity Stress Tolerance in Transgenic Rice, Front. Plant Sci., 2019, 10, 168 CrossRef PubMed.
- L. Long, W. W. Yang, P. Liao, Y. W. Guo, A. Kumar and W. Gao, Transcriptome analysis reveals differentially expressed ERF transcription factors associated with salt response in cotton, Plant Sci., 2019, 281, 72–81 CrossRef CAS.
- D. Liu, Y.-Y. Li, Z.-C. Zhou, X. Xiang, X. Liu, J. Wang, Z.-R. Hu, S.-P. Xiang, W. Li, Q.-Z. Xiao, Y. Wang, R.-S. Hu and Q. Zhao, Tobacco transcription factor bHLH123 improves salt tolerance by activating NADPH oxidase NtRbohE expression, Plant Physiol., 2021, 186(3), 1706–1720 CrossRef CAS PubMed.
- Z. Zhang, H. Liu, C. Sun, Q. Ma, H. Bu, K. Chong and Y. Xu, A C2H2 zinc-finger protein OsZFP213 interacts with OsMAPK3 to enhance salt tolerance in rice, J. Plant Physiol., 2018, 229, 100–110 CrossRef CAS PubMed.
- S. Mitsuya, M. Taniguchi, H. Miyake and T. Takabe, Overexpression of RCI2A decreases Na+ uptake and mitigates salinity-induced damages in Arabidopsis thaliana plants, Physiol. Plant., 2006, 128(1), 95–102 CrossRef CAS.
- M. Aycan, L. Nahar, M. Baslam and T. Mitsui, B-type response regulator hst1 controls salinity tolerance in rice by regulating transcription factors and antioxidant mechanisms, Plant Physiol. Biochem., 2023, 196, 542–555 CrossRef CAS PubMed.
- N. Tuteja and S. Mahajan, Calcium signaling network in plants: an overview, Plant Signaling Behav., 2007, 2(2), 79–85 CrossRef.
- M. Corso, F. G. Doccula, J. R. F. de Melo, A. Costa and N. Verbruggen, Endoplasmic reticulum-localized CCX2 is required for osmotolerance by regulating ER and cytosolic Ca2+ dynamics in Arabidopsis, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(15), 3966–3971 CrossRef CAS PubMed.
- N. Yamada, C. Theerawitaya, S. Cha-um, C. Kirdmanee and T. Takabe, Expression and functional analysis of putative vacuolar Ca2+−transporters (CAXs and ACAs) in roots of salt tolerant and sensitive rice cultivars, Protoplasma, 2014, 251(5), 1067–1075 CrossRef CAS.
- H. Mei, N. H. Cheng, J. Zhao, S. Park, R. A. Escareno, J. K. Pittman and K. D. Hirschi, Root development under metal stress in Arabidopsis thaliana requires the H+/cation antiporter CAX4, New Phytol., 2009, 183(1), 95–105 CrossRef CAS.
- A. Singh, P. Kanwar, A. K. Yadav, M. Mishra, S. K. Jha, V. Baranwal, A. Pandey, S. Kapoor, A. K. Tyagi and G. K. Pandey, Genome-wide expressional and functional analysis of calcium transport elements during abiotic stress and development in rice, FEBS J., 2014, 281(3), 894–915 CrossRef CAS.
- Y. Bu, W. Fu, J. Chen, T. Takano and S. Liu, Description of AtCAX4 in Response to Abiotic Stress in Arabidopsis, Int. J. Mol. Sci., 2021, 22(2), 856 CrossRef CAS PubMed.
- P. Cheng, J. Gao, Y. Feng, Z. Zhang, Y. Liu, W. Fang, S. Chen, F. Chen and J. Jiang, The chrysanthemum leaf and root transcript profiling in response to salinity stress, Gene, 2018, 674, 161–169 CrossRef CAS PubMed.
- Q. Zeng, X. Chen and A. J. Wood, Two early light-inducible protein (ELIP) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light1, J. Exp. Bot., 2002, 53(371), 1197–1205 CrossRef CAS.
- C. Chen, L. H. Bai, D. R. Qiao, H. Xu, G. L. Dong, K. Ruan, F. Huang and Y. Cao, Cloning and expression study of a putative carotene biosynthesis related (cbr) gene from the halotolerant green alga Dunaliella salina, Mol. Biol. Rep., 2008, 35(3), 321–327 CrossRef CAS.
- Q. Diyang, H. Rui, L. Ji, L. Ying, D. Jierong, X. Kuaifei, Z. Xuhua, F. Zhongming and Z. Mingyong, Peptide Transporter OsNPF8.1 Contributes to Sustainable Growth under Salt and Drought Stresses, and Grain Yield under Nitrogen Deficiency in Rice, Rice Sci., 2023, 30(2), 113–126 CrossRef.
- B.-R. Lee, A. Koprivova and S. Kopriva, The key enzyme of sulfate assimilation, adenosine 5′-phosphosulfate reductase, is regulated by HY5 in Arabidopsis, Plant J., 2011, 67(6), 1042–1054 CrossRef CAS PubMed.
- A. Koprivova, K. A. North and S. Kopriva, Complex Signaling Network in Regulation of Adenosine 5′-Phosphosulfate Reductase by Salt Stress in Arabidopsis Roots, Plant Physiol., 2008, 146(3), 1408–1420 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4en00685b |
| ‡ Si Chen and Zhengyan Pan contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |