Point-of-care diagnostics for SARS-CoV-2 wastewater-based epidemiology: a big leap toward miniaturization
Ahmed
Donia
 a,
Nonsikelelo Precios
Mthethwa-Hlongwa
a,
Isaac Dennis
Amoah
ab,
Sheena
Kumari
a and
Faizal
Bux
a,
Nonsikelelo Precios
Mthethwa-Hlongwa
a,
Isaac Dennis
Amoah
ab,
Sheena
Kumari
a and
Faizal
Bux
 *a
*a
aInstitute for Water and Wastewater Technology, Durban University of Technology, Durban, South Africa. E-mail: faizalb@dut.ac.za
bDepartment of Environmental Science, University of Arizona, Tucson, AZ, USA
First published on 15th July 2024
Abstract
The integration of point-of-care diagnostics in SARS-CoV-2 wastewater-based epidemiology signifies a substantial leap forward in disease surveillance and monitoring. Various innovative methods have been explored for the detection of SARS-CoV-2 in wastewater samples, each with its unique advantages and applications. Loop-mediated isothermal amplification (LAMP) has emerged as a prominent technique due to its sensitivity, rapidity, and simplicity. It amplifies pathogen genetic material without the need for a thermal cycler, making it suitable for point-of-care detection. Pairing microfluidic technology with LAMP enables swift and automated analysis directly on a chip. Additionally, paper-based devices offered a cost-effective and straightforward approach for LAMP-based detection, particularly beneficial in resource-limited settings. Combining LAMP with CRISPR/Cas technology enhances specificity and sensitivity, crucial for variant-specific detection like VarLOCK (variant-specific SHERLOCK). Aptamer-based electrochemical chips offer high specificity and stability, making them suitable for wastewater analysis. By integrating aptamer technology with filtration and purification systems, detecting SARS-CoV-2 on-site in wastewater becomes feasible, offering a practical solution for monitoring viral transmission. These methods showcase the diverse approaches in SARS-CoV-2 detection through wastewater-based epidemiology, promising effective disease surveillance. In this review, we will summarize these recent advancements in point-of-care diagnostics for SARS-CoV-2 wastewater-based epidemiology and explore their potential applications beyond SARS-CoV-2.
Water impactIntegrating point-of-care diagnostics into SARS-CoV-2 wastewater surveillance is a novel method to enhance disease monitoring with rapid on-site detection, vital for early outbreak identification and timely public health responses. This innovation not only broadens its applications beyond COVID-19 surveillance but also offers potential for detecting other emerging and re-emerging pathogens in wastewater, bolstering public health efforts. |
I. Introduction
A. Overview of wastewater-based epidemiology
Wastewater-based epidemiology (WBE) analyzes pollutants and biomarkers in untreated wastewater to gain insights into community behavior, substance use, and disease prevalence, offering real-time understanding of health conditions and environmental exposure. Initially introduced by Christian Daughton, WBE assesses drug residues in wastewater as indicators of population-level drug consumption.1 Amidst the COVID-19 pandemic, WBE identifies SARS-CoV-2 RNA in wastewater, aiding global pandemic surveillance and reinforcing adherence to World Health Organization (WHO) guidelines for wastewater management.2 Wastewater monitoring of SARS-CoV-2 demonstrates superior cost-effectiveness compared to clinical testing, providing comprehensive population-level surveillance at a fraction of the cost. Wright and colleagues conducted a comparative cost analysis and found that wastewater-based epidemiology was more cost-effective, with data obtained at only 1.7% of the total cost of clinical testing ($6042 for wastewater-based epidemiology versus $338![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for clinical testing).3
000 for clinical testing).3
B. Importance of point-of-care diagnostics in wastewater surveillance
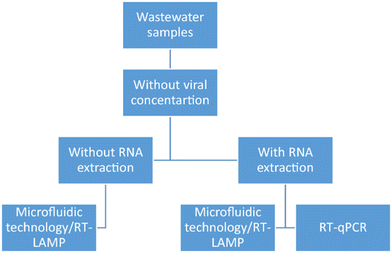
In areas with limited resources, conventional laboratory methods such as PCR and ELISA are often inaccessible or costly. Point-of-care (POC) devices offer a solution, providing on-site detection that replaces expensive methods with accessible, rapid, and cost-effective alternatives.4Microfluidic devices, especially, excel at detecting contaminants in water and food. They are valued for their affordability, simplicity, minimal sample requirements, speed, and on-site usability.5,6 Different approaches have been explored, including PCR, loop-mediated isothermal amplification (LAMP), mass spectrometry, and electrochemistry. A microfluidic quantitative PCR (MFQPCR) system has been used to detect human viral and enteric bacterial pathogens in wastewater treatment plants, reaching concentrations as low as 2 copies per μl of cDNA/DNA7 and 100 cells per liter,8 respectively. For on-chip nucleic acid testing, chips are typically divided into three sections: sampling, sensing, and signaling. Essential components of microfluidic devices include micropumps, microvalves, micromixers, droplet generators, separators, traps, cell cultures, and gradient generators.9 Despite these advancements, challenges such as bubble generation and reagent evaporation in microfluidic PCR need to be addressed through improved modeling of microfluidic platforms for controlled fluid transfer.10
Electrochemistry emerges as a promising point-of-care method for various targets in wastewater, offering rapid, selective, and reproducible responses.11 This integrates sensor electrodes from electrochemical biosensors with sustainable polymers and microfluidics for affordable sensing options. These devices, employing cost-effective polymers like plastics and thin-layer metal electrodes, inherently reduce production costs, which are further diminished through miniaturization. Biofuel cells, notably powered by enzymes or microbes mediating glucose, oxygen, and other digestate conversions, have shown promise. Manufacturing plastic-based sensing platforms, utilizing printed circuit board technology, offers a feasible route. Leveraging this established industrial process, single board costs ranging from USD 0.20–0.52 per cm2, depending on layer complexity, contrast significantly with USD 10–20 per cm2 for glass-based chips. Mass production can substantially lower these costs, promising the creation of genuinely cost-effective devices.12
The optical analytical technique encompasses methodologies like ultra-violet visible, fluorescence, chemiluminescence, or Raman spectroscopy, employed to monitor signal alterations. It includes a recognition component that engages with the target analyte and a transducer element that signals the interaction.13 Immunoprobes, typically colloidal gold, carbon, or colloidal selenium nanoparticles, are pivotal in colorimetric assays for detecting target analytes.14,15 They interact specifically with targets, inducing observable color changes, enabling qualitative or quantitative analysis. Advancements in nanotechnology have led to the widespread adoption of luminescent nanoparticles in point-of-care testing due to their optical stability and suitability for immunoassays, including quantum dots,16 dye-doped nanoparticles,17 and up-converting nanoparticles.18
C. Rationale for using wastewater surveillance for SARS-CoV-2 monitoring
SARS-CoV-2 may integrate into wastewater treatment plants due to its potential to adhere to fecal particles and subsequently infiltrate these facilities.19 Multiple studies from various countries have confirmed the presence of SARS-CoV-2 in sewage samples, with viral RNA levels showing an upward trajectory over time.20–23 A notable finding is the prolonged shedding of SARS-CoV-2 RNA via the digestive system compared to the respiratory tract, lasting up to 27.9 days from the onset of symptoms.23 The presence of SARS-CoV-2 viral load in wastewater reflects both asymptomatic and symptomatic shedding, providing a more accurate representation of community infection levels.24 Current surveillance primarily relies on diagnostic testing, which is limited in its ability to provide timely warnings for surges and is susceptible to various biases. In contrast, wastewater surveillance has emerged as a reliable, less biased, and predictive tool for monitoring SARS-CoV-2 infection rates in near-real-time.25–27 Wastewater surveillance has demonstrated its effectiveness by identifying lower-than-expected viral circulation in central Florida during the 2022/23 winter. Moreover, its capability to detect subtle changes in infection rates 30–46 days before clinical tests, even at low baseline levels of circulation, enables communities to confidently resume normal activities during low-risk periods while promptly implementing mitigation measures when the risk escalates.28Wastewater surveillance has been used for monitoring viruses other than SARS-CoV-2 such as poliovirus, norovirus, hepatitis A virus, influenza, respiratory syncytial virus (RSV) and enteroviruses. In efforts toward poliomyelitis eradication, Greece undertook enhanced laboratory surveillance of enteroviruses between 2008 and 2014. The Hellenic Polioviruses/Enteroviruses Reference Laboratory rigorously monitored high-risk populations, immigrants from poliovirus-endemic regions, and environmental samples. Out of 722 stool and 179 sewage water samples examined, no wild-type polioviruses were detected; however, two vaccine-derived strains were identified. Enteroviruses were found in 25.3% of stool and 25.1% of sewage samples, predominantly species A, B, or C. Genetic analysis revealed close relationships between sewage and stool isolates, underscoring the importance of monitoring asymptomatic carriers to preserve Greece's polio-free status amid ongoing population movements.29
Environmental surveillance in London, UK, was conducted to test sewage samples for investigation of the molecular properties of type 2 poliovirus isolates found in sewage to detect virus transmission in the community.30 The findings revealed 118 genetically linked poliovirus isolates related to the serotype 2 Sabin vaccine strain in 21 of 52 sequential sewage samples collected in London between February 8 and July 4, 2022. Expansion of surveillance sites helped localize transmission to several boroughs in north and east London. All isolates lacked two key attenuating mutations, were recombinants with a species C enterovirus, and a subset (20 of 118) met the criterion for a vaccine-derived poliovirus, displaying six to ten nucleotide changes in the gene encoding the VP1 capsid protein. Environmental surveillance facilitated early detection of poliovirus importation and circulation in London, enabling a rapid public health response, including enhanced surveillance and an inactivated polio vaccine campaign among children aged 1–9 years. Whole-genome sequencing established linkage of isolates and confirmed transmission of a unique recombinant poliovirus lineage detected in Israel and the USA.30 The detection of poliovirus in London's wastewater has sparked considerable public interest and media coverage. It underscores the ongoing risk of resurgence globally until the virus is eradicated. The simultaneous identification of genetically linked poliovirus strains in New York, USA, and Israel suggests the potential spread of the virus through international travel, reaching geographically distant regions.31
In response to a foodborne hepatitis A outbreak in Italy, urban sewages at wastewater treatment plants were monitored to investigate the spread of hepatitis A virus. From July 2012 to September 2013, 38 out of 157 sewage samples (24.2%) tested positive for Hepatitis A virus, including the IA variant linked to the outbreak. Notably, while clinical surveillance identified variants not found in wastewater, the majority of sequences belonged to genotype IB, indicating low/intermediate endemicity.32
A study demonstrated the efficacy of wastewater-based surveillance as a valuable tool for monitoring viral circulation and serving as an early warning system, particularly for respiratory viruses like SARS-CoV-2, influenza, and RSV.33 Over a 15 month period from September 2021 to November 2022, weekly sampling campaigns were conducted at two wastewater treatment plants in Barcelona, Spain, serving the entire population. The samples underwent concentration using the aluminum hydroxide adsorption-precipitation method and subsequent analysis through RNA extraction and RT-qPCR. In all cases, SARS-CoV-2 was detected, whereas the positivity rates for influenza virus and RSV were notably lower (10.65% for influenza A (IAV), 0.82% for influenza B (IBV), 37.70% for RSV-A, and 34.43% for RSV-B). SARS-CoV-2 gene copy concentrations were consistently higher, typically ranging approximately 1 to 2 logarithmic units above those of the other respiratory viruses. Distinct spikes in influenza A virus H3:N2 during February and March 2022, as well as RSV occurrences in the winter of 2021, were noted, aligning with the chronological pattern of infections documented in the clinical database of the Catalan Government. Overall, the findings underscore the utility of wastewater surveillance in providing novel insights into the abundance of respiratory viruses in the Barcelona area, demonstrating a favorable correlation with clinical data.33
The importance of wastewater surveillance is extended to include pharmaceuticals/drugs.34 Water pollution due to pharmaceuticals is an escalating concern, necessitating water quality monitoring for public health protection. Particularly, antidepressants, benzodiazepines, antiepileptics, and antipsychotics pose risks to aquatic life. A study developed a comprehensive method for detecting 105 pharmaceutical residues in small water samples, applied to screen samples from four Wastewater Treatment Plants in northern Italy. Samples were filtered, extracted, and analysed by Ultra-High-Performance Liquid Chromatography-Quadrupole Time-of-Flight High-Resolution Mass Spectrometry (UHPLC-QTOF-HRMS). Adequate sensitivity was achieved, detecting 23 of 105 targeted drugs in all samples. Retrospective analysis identified carbamazepine metabolites, including carbamazepine-10,11-epoxide, posing neurotoxic risks.35
II. Point-of-care diagnostics: an overview
Point-of-care testing, which refers to diagnostic testing conducted directly at or near the patient's location, offers the advantage of providing rapid results to healthcare providers without the delay associated with transporting samples to a central laboratory.36 The inception of point-of-care testing dates to 1962 with the development of a quick method to measure blood glucose levels.37 However, it wasn't until the early 1990s that point-of-care testing gained significant momentum with the introduction of portable devices capable of measuring electrolytes in emergency departments.38Biosensors, acting as analytical tools, detect specific targets by converting biomolecular recognition events into measurable physicochemical signals. Developing simple, rapid, affordable yet highly sensitive biosensors for POC testing remains challenging. Comprising biological recognition and transducer components, biosensors interact with targets through various molecular interactions like antibody/antigen or enzyme/substrate binding, amplifying them into detectable signals. Transducers—electrochemical, optical, electronic, and mass-sensitive devices—convert these interactions into measurable outputs proportional to the target concentration.39 Microfluidic systems, akin to miniaturized laboratories, streamline analytical processes and provide portability for POC testing. Crucial to developing miniaturized biosensors for POC testing are sensor specificity, driven by target recognition, and sensitivity, influenced by transducer and amplifier performance. These aspects enable the detection of diverse targets with high accuracy and efficiency.39–41
Electrochemical devices can also be miniaturized. Miniaturized electrochemical sensors are employed for detecting trace amounts of targets such as small organic molecules, metal ions, and biomolecules. This is achieved by measuring changes in electrochemical signals (e.g., current, voltage, potential, or impedance) arising from the oxidation/reduction of chemical/biological molecules using electrodes and electrochemical units. Typically, electrodes are modified to enhance sensor selectivity primarily through the attachment of specific recognition elements like aptamers, antibodies, and receptors. Miniaturized electrochemical sensors sensor systems offer several advantages including simple instrumentation, user-friendliness, high sensitivity and selectivity, minimal sample pretreatment, short analysis time, portability, and cost-effectiveness.42
Miniaturization technologies have been also applied for optical biosensor design.43 Certain optical detection techniques, which are not readily adaptable to conventional sensing systems, such as fluorescence lifetime imaging, time-lapse microscopy, and multicolor analysis, have been effectively integrated as measurement methods in microfluidic sensors.44,45 Additionally, surface plasmon resonance, mass spectrometry, and surface acoustic wave are among the other methods successfully employed in the design of microfluidic sensors.43,46,47
III. Recent advancements in point-of-care diagnostics for SARS-CoV-2 in wastewater
The target genes for SARS-CoV-2 are based on the conserved and specific open reading frame 1ab (ORF1ab), spike (S), RNA-dependent RNA polymerase (RdRp), envelope (E), and nucleocapsid (N) genes48 (Fig. 1). The nucleic acid-based detection method, notably reverse transcription-PCR (RT-PCR), offers high sensitivity and specificity, making it the gold standard for surveillance of SARS-CoV-2.49 However, RT-PCR's reliance on expensive equipment, specialized reagents, and trained personnel in biosafety-compliant laboratories hinders its suitability for timely detection and surveillance. Hence, there's an urgent need for rapid and accessible SARS-CoV-2 detection and surveillance methods, leading to the emergence of real-time nucleic acid detection, particularly in the form of point-of-care testing. The point-of-care testing, performed at the sampling site using portable instruments, offers a valuable complement to RT-PCR in standard laboratories.50 It provides quick results, enabling faster clinical decision-making and epidemic screening.51–54 Although real-time detection presents advantages such as accuracy, rapidity, portability, simplicity, and cost-effectiveness, it also introduces new challenges for clinical laboratories.55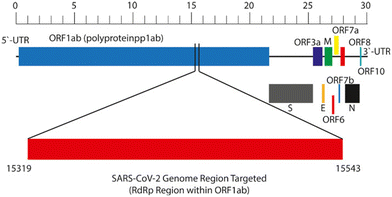 | ||
| Fig. 1 Genomic location of the target region within the SARS-CoV-2 genome.56 | ||
This leap toward miniaturization not only facilitates rapid results but also introduces the prospect of compact, portable diagnostic tools that can be easily deployed to wastewater treatment plants or community collection points. The miniaturization of point-of-care diagnostics for SARS-CoV-2 in wastewater-based epidemiology offers numerous advantages beyond speed and portability. By reducing the size of diagnostic devices, researchers can enhance cost-effectiveness, making these technologies more accessible for widespread use. This leap toward miniaturization also aligns with the growing trend of decentralized healthcare solutions, empowering communities to take a more active role in monitoring and managing public health.57 Overall, the applying of point-of-care diagnostics in wastewater-based epidemiology marks a transformative step forward, not only in the battle against the current pandemic but also in the development of agile, community-centric approaches to infectious disease control.58
Table 1 summarized the current studies about point-of-care diagnostics for SARS-CoV-2 wastewater-based surveillance.
| Point-of-care diagnostics principle | Target gene/sequence/protein | Ref. |
|---|---|---|
a VarLOCK assays (variant-specific SHERLOCK): SHERLOCK: (![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) pecific pecific ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) igh-sensitivity igh-sensitivity ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) nzymatic nzymatic ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) eporter unLOCKing). eporter unLOCKing).
|
||
| LAMP/microfluidic technology | N/ORF1a | 59 |
| LAMP | N | 60 |
| LAMP | N and E | 61 |
| LAMP | N and E | 62 |
| LAMP | RdRp | 56 |
| LAMP/electrochemical sensor | N and ORF1ab | 63 |
| Isothermal amplification/microfluidic technology/colorimetric sensor | N gene, E gene and Orf1a | 64 |
| Field-effect transistor | N and ORF1ab | 65 |
| Paper-based device (CRISPR/Cas12a and LAMP) | N, E, and S | 66 |
| VarLOCK (variant-specific SHERLOCK)a | S | 67 |
| Aptamer-based electrochemical chip | Spike protein | 68 |
A. LAMP
LAMP stands out as a contemporary isothermal nucleic acid amplification technique, gaining traction in point-of-care detection, especially during COVID-19 pandemic.59,60 It has several notable advantages, including sensitivity, rapidness, the absence of a need for a thermal cycler, and tolerance against sample inhibitors. Consequently, it emerges as a promising alternative to quantitative polymerase chain reaction (qPCR). It can amplify pathogen genetic material in under an hour, requiring a specific set of four to six primers to ensure precision and specificity.69 To confirm the success of LAMP, various approaches can be employed, such as monitoring changes in fluorescence using intercalating dyes, employing DNA probes combined with gold nanoparticles, observing alterations in turbidity due to the formation of magnesium pyrophosphate precipitates, utilizing pH indicators, or conducting gel electrophoresis followed by UV detection.59 We presented a broad insight into published point-of-care diagnostics used for SARS-CoV-2 wastewater-based epidemiology and the possibility to be used against other environmental pathogens.Amoah and colleagues explored two optimized RT-LAMP protocols targeting N gene utilizing color change and fluorescence detection.60 These protocols demonstrated a limit of detection of 10 copies/25 μl reaction with positive amplification within 35 minutes. Fig. 2 showed that the limit of detection assessed using the fluorescence method is 10 copies/25 μl reaction. Over a 4 week monitoring period of wastewater from four treatment plants, the colorimetric protocol identified a prevalence of 12.5%, increasing to 44% when the RNA template concentration was raised fivefold. The limit of RT-LAMP detection was high so this was one of the major limitations compared to droplet digit PCR and qPCR. It means when there is low concentration of virus in the samples, the LAMP was not effective as opposed to droplet digit PCR. The prevalence of the fluorescent RT-LAMP was 31% and 47% when utilizing initial templates with concentrations of 92.7 (±28.2) ng μl−1 and 480 (±134.5) ng μl−1 of the extracted RNA, respectively. They recommended its utilization as a rapid molecular analysis tool to for COVID-19 detection within the catchment areas of wastewater treatment plants. This adaptable approach could be instrumental in future outbreaks, offering a swifter, more cost-effective, and less complex means of monitoring infections through wastewater-based epidemiology.60
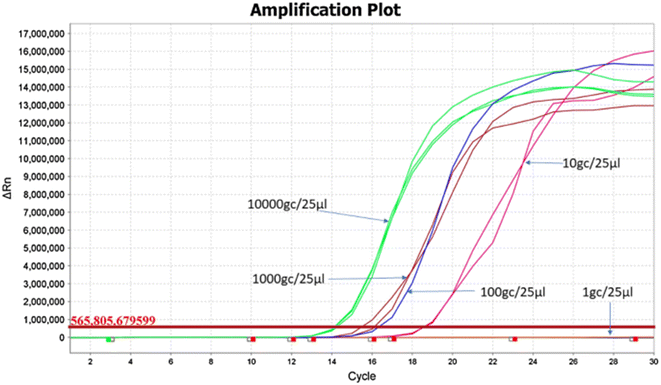 | ||
| Fig. 2 Amplification plot for limit of detection assessment using the fluorescence method.60 | ||
A recent study explored the use of tampons as passive swabs for sample collection and RT-LAMP to targeting the N and E genes in wastewater.61 The results of this workflow were available within three hours of sample collection. While this assay is approximately 20 times less analytically sensitive than RT-droplet digital PCR, during a building-level wastewater surveillance campaign conducted alongside independent weekly clinical testing of all students, the method demonstrated a three-day positive predictive value (PPV) of 75% (excluding convalescent cases) and a same-day negative predictive value (NPV) of 80% for incident COVID-19 cases (Fig. 3). These predictive values are comparable to those reported by wastewater monitoring using RT-qPCR. Despite lower analytical sensitivity, these findings suggest that the tampon swab and RT-LAMP workflow provides a cost-effective and rapid approach that could be effectively employed for scalable building-level wastewater surveillance for COVID-19, even in low-resource settings.61
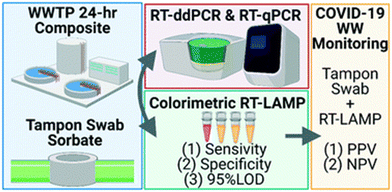 | ||
| Fig. 3 Schematic representation of building-level wastewater surveillance using tampon swabs and RT-LAMP for rapid SARS-CoV-2 RNA detection.61 | ||
A recent study compared the efficacy of RT-LAMP with RT-qPCR assays targeting the N and E genes of the SARS-CoV-2 genome in detecting RNA in untreated wastewater samples (Fig. 4).62 The RT-qPCR assays demonstrated consistent amplification down to 2 × 102 GC per reaction, with greater sensitivity at 2 × 101 GC per reaction for the US CDC N1 and N2 assays. In contrast, RT-LAMP exhibited lower sensitivity, detecting SARS-CoV-2 only at or above 2 × 103 GC per reaction. While RT-qPCR identified SARS-CoV-2 RNA in 29 out of 30 wastewater samples, RT-LAMP identified 27 positive samples, showing strong concordance with the US CDC N1 and E_Sarbeco RT-qPCR assays but lower agreement with the US CDC N2 assay. Despite lower sensitivity, RT-LAMP may offer advantages for wastewater surveillance, such as rapid results and simplicity. Logistic regression analysis indicated that RT-LAMP results correlated with RNA quantified by the US CDC N1 and E_Sarbeco assays, supporting the use of RT-LAMP as a specific and efficient method for screening wastewater samples for SARS-CoV-2, particularly in resource-limited settings.62
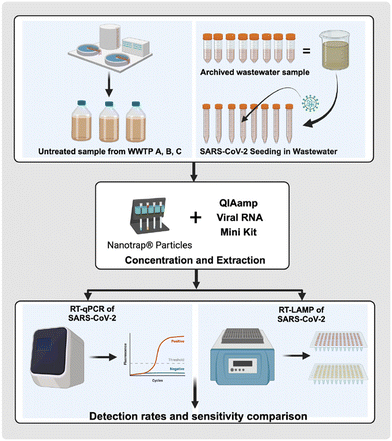 | ||
| Fig. 4 Graphical abstract of comparison of RT-LAMP and RT-qPCR assays for detecting SARS-CoV-2 in wastewater.62 | ||
A study introduced a colorimetric RT-LAMP assay for the visual detection of SARS-CoV-2 in clinical and sewage samples.56 The assay, based on the RNA dependent RNA polymerase (RdRp) gene, demonstrated a high success rate in detecting virus RNA in clinical samples and exhibited specificity in a short timeframe of 20 minutes, and its sensitivity was further enhanced by adding bovine serum albumin (BSA) (Fig. 5). This optimized assay was successfully applied to detect SARS-CoV-2 in sewage waters from virus hotspots in Lahore, Pakistan. To evaluate the specificity of the LAMP assay for SARS-CoV-2, they aligned each of the eight regions targeted by the primers with the reference genome sequences of six related coronaviruses. They conducted an in-silico analysis to assess the specificity of primers. The analysis revealed that most of the primer sequences exhibited high specificity for SARS-CoV-2, displaying no significant similarity to any of the six related coronaviruses included in the study. Although the F1c and LB regions displayed some similarity to the Tor2 and HUK1 viruses, these similarities were determined to be insufficient to cause nonspecific amplifications. Therefore, owing to the inherent specificity of the LAMP assay—utilizing six primers targeting eight distinct regions in the target—and the meticulous design of the primers, they demonstrated that LAMP assay, used in this study, has the potential to differentiate SARS-CoV-2 from various related coronaviruses. Additionally, it can detect most common SARS-CoV-2 variants with equal efficiency.56
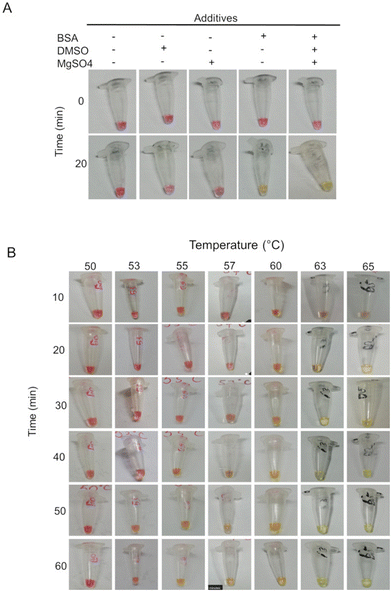 | ||
| Fig. 5 Optimization of the LAMP assay:56 The LAMP assay utilized a 225 nt synthetic RdRp gene fragment. (A) Results of the RdRp gene-based LAMP assay are shown with and without amplification-enhancing additives (BSA, MgSO4, and/or DMSO). (B) Images display the RdRp gene-based LAMP assay results across different temperatures and time points. Pink indicates negative results, while yellow indicates positive results. | ||
 | ||
| Fig. 6 Graphical abstract of integration of RT-LAMP and microfluidic technology for detection of SARS-CoV-2 in wastewater.59 | ||
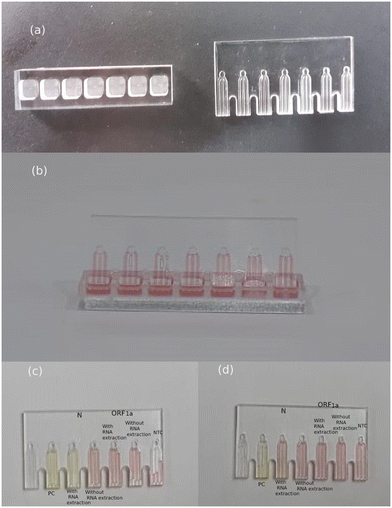 | ||
| Fig. 7 Integration of microfluidic devices with RT-LAMP for the detection of SARS-CoV-2 in wastewater.59 (a) Microfluidic device and wells. (b) Successful loading of RT-LAMP mixtures into the microfluidic device. (c) Sample ID 1 displayed a color change to yellow in the microchannels of the N gene after RNA extraction. (d) Sample ID 3 did not exhibit a color change, remaining red in the microchannels of the N gene after RNA extraction. NTC: negative control. PC: positive control. | ||
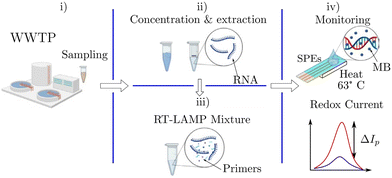 | ||
| Fig. 8 Workflow of an RT-LAMP-based electrochemical sensor for wastewater samples:63 i) sampling from the wastewater treatment plant. ii) Nucleic acid extraction and concentration. iii) RT-LAMP mixtures for genetic amplification. iv) Electrochemical monitoring of RT-LAMP products through redox current. | ||
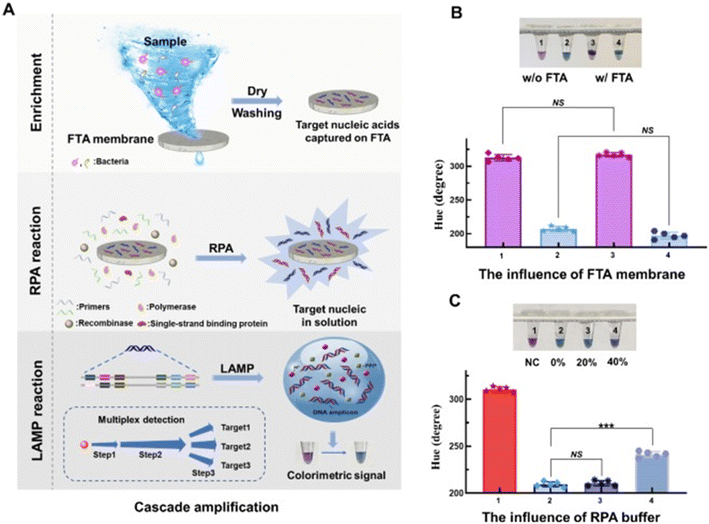 | ||
| Fig. 9 Design and optimization of a Flinders Technology Associates (FTA)-based multiplexed colorimetric assay for sensitive pathogen detection in wastewater.64 (A) Design of the FTA-based multiplexed colorimetric assay for detecting various pathogens. (B) Impact of the FTA membrane on the SEC-LAMP assay. Tubes 1–4: negative control without the FTA membrane, positive control (500 CFU per reaction, Salmonella) without the FTA membrane, negative control with the FTA membrane, and positive control (500 CFU per reaction, Salmonella) with the FTA membrane, respectively. (C) Influence of RPA reaction solution volume on the SEC-LAMP assay. Tubes 1–4: negative control, positive control (500 CFU per reaction, Salmonella), positive control with 20% (v/v) RPA reaction solution, and positive control with 40% (v/v) RPA reaction solution, respectively. | ||
B. Field-effect transistor
Field-effect transistor (FET) biosensors present a promising avenue due to their low-cost fabrication, miniaturization, reliability, sensitivity, and label-free detection capabilities.72 Extended-gate FET (EGFET) devices, in particular, offer a straightforward and cost-effective option for biosensing.73 Despite technological progress, EGFET sensors face limitations in portable applications due to challenges in electrode integration and the need for labeled receptors for specificity.74 To address these challenges, Alvarez-Serna and colleagues used an EGFET device for nucleic acid detection without electrode modification.65 This device detected two segments of the SARS-CoV-2 genome (N and ORF1ab genes) through quantifying pH changes resulting from the isothermal amplification process, leveraging the selectivity of EGFET-based sensors to measure pH (Fig. 10). This sensor measured real wastewater samples for detection of SARS-CoV-2, with a limit of detection of 0.31 × 10−3 ng μL−1 for end-point measurement. Furthermore, the sensor can provide real-time-like results, with a robust response within just 15 minutes, even at concentrations as low as 0.37 ng μL−1.65 Firstly, they established the sensor's capability to detect pH fluctuations within the range where both negative and positive samples fall. Then, using the pH value of the negative template control (NTC) sample as a reference, they observed pH shifts in positive samples due to isothermal amplification. Sensor characterization was conducted using SARS-CoV-2 positive samples across seven concentrations via serial dilution. The calibration curve depicted a linear relationship between drain-to-source current (IDS) and sample concentration. There is a straightforward relationship among the current IDS and changes in pH; which in turn, are promoted by the RT-LAMP reaction.65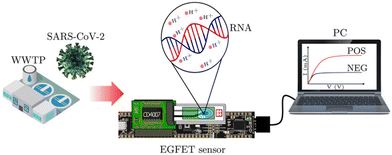 | ||
| Fig. 10 Portable sensor monitors RT-LAMP for SARS-CoV-2 in wastewater.65 Wastewater samples are collected from wastewater treatment plants. Next, the genetic material is extracted and concentrated. Then, the RNA is combined with RT-LAMP primers, and a 50 μL drop is applied onto the electrodes of the EGFET. The RT-LAMP reaction takes place at the electrode surface, with temperature control at 63 °C for approximately 30 minutes. Finally, the RT-LAMP products are monitored by observing changes in drain-to-source current (IDS), which occur due to pH alterations as the reaction progresses. The pH value of the negative template control sample serves as the baseline value. Consequently, in the positive samples, one can observe pH changes attributed to the isothermal amplification process. | ||
C. Paper-based device
There has been growing interest in enhancing the specificity of analytical methods using CRISPR/Cas-based gene editing techniques. CRISPR/Cas technology offers high-fidelity recognition down to single-base resolution and efficient signal transduction through trans-cleavage, making it a promising detection method.75,76 However, despite its advantages, the sensitivity of CRISPR/Cas alone may not be sufficient.77 To address this, some studies have explored combining CRISPR/Cas with isothermal amplification methods.75,78 This combination utilizes the specificity of CRISPR/Cas to mitigate false positives in isothermal amplification while leveraging the high sensitivity of isothermal amplification to enhance the limit of detection (LOD) of CRISPR/Cas. One such combination is Cas12a-RT-LAMP, which has been employed for SARS-CoV-2 detection.79,80 For instance, Broughton and colleagues developed a lateral flow assay integrating Cas12a and RT-LAMP, achieving detection as low as 10 copies per μL in clinical samples.79 Other approaches include visual detection methods utilizing the fluorescence characteristics of FAM, where Wang et al.81 and Pang et al.78 integrated Cas12a trans-cleavage reaction and isothermal amplification into a single tube by creating two temperature regions in one device. Despite these advancements, the combination of CRISPR/Cas and isothermal amplification methods still requires further exploration and optimization.Cao and colleagues introduced a new paper-based device that combines RT-LAMP with CRISPR/Cas12a for analyzing wastewater (Fig. 11).66 Wastewater samples undergo filtration and lysis on a microporous membrane, after which they are transferred to a paper-based device for RNA purification and distribution. The device utilizes high-fidelity RT-LAMP to detect specific gene fragments (N, E, and S) of SARS-CoV-2. Results are observable by the naked eye following excitation by 480 nm light or lateral flow dipsticks. When SARS-CoV-2 is present, RT-LAMP produces amplicons that activate Cas12a-gRNA, resulting in non-specific cleavage of single-stranded DNA probes. Fluorescence is observed through the separation of FAM-BHQ1 in fluorescence-based methods, while the lateral flow method shows a distinct test-stained band due to the separation of FAM-biotin. In the absence of SARS-CoV-2, the probe remains intact, resulting in no fluorescence. The device's design and components are illustrated in Fig. 11. Testing involved using SARS-CoV-2 spiked wastewater to determine limits of detection (LODs) and quantitative capabilities, followed by blind testing to confirm reliability. This paper-based device allowed for the simultaneous detection of the N, E, and S genes, having limit of detection 25, 310, and 10 copies per mL, respectively. Furthermore, the device enabled semiquantitative analysis within the range of 0 to 310 copies per mL, owing to the varying LODs of the three genes. Blind experiments confirmed the device's suitability for wastewater analysis, showing 97.7% sensitivity and 82% accuracy in semiquantitative assessment.66
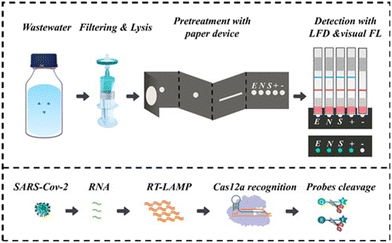 | ||
| Fig. 11 Paper device for detection of SARS-CoV-2 in wastewater using CRISPR/Cas12a and RT-LAMP.66 Wastewater samples are filtered and lysed on a microporous membrane, then transferred to a paper device for RNA purification and distribution. High-fidelity RT-LAMP detects SARS-CoV-2, which is visible by the naked eye following excitation by 480 nm light or through the use of lateral flow dipsticks. In the presence of SARS-CoV-2, RT-LAMP generates amplicons activating Cas12a-gRNA, leading to unspecific ssDNA probe cleavage. Fluorescence arises from FAM-BHQ1 separation in fluorescence method, while lateral flow method exhibits strong test-stained band due to FAM-biotin separation. Without SARS-CoV-2, probe remains intact, yielding no fluorescence. | ||
The device underwent a six-month application period from June to November 2021 for analyzing wastewater, aimed at further validating its performance. The Erqiao wastewater treatment plant in Huaguoyuan, among the largest communities in Asia, and the Jinyang wastewater treatment plant, situated in a new urban area in Guiyang, were selected as the designated locations for this evaluation. Following the analysis, no viruses were detected in the sewage over the course of six months, indicating that the communities were experiencing a low-risk status, consistent with the observations of the Centers for Disease Control and Prevention (CDC). Consequently, the innovative device exhibits promising potential for sewage detection.66
D. VarLOCK (variant-specific SHERLOCK)
CRISPR-based diagnostic tests offer a new platform for detecting pathogens. Techniques like SHERLOCK (![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) pecific
pecific ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) igh-sensitivity
igh-sensitivity ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) nzymatic
nzymatic ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) eporter unLOCKing) uses a two-step process involving target amplification and CRISPR-based nucleic acid detection,75,77 successfully applied to detect SARS-CoV-2 in clinical samples.79 However, these methods are more complex than point-of-care tests due to RNA extraction and multiple liquid-handling steps, increasing the risk of sample cross-contamination. DETECTR, a recently emerging molecular diagnostic method, utilizes CRISPR-Cas12a along with isothermal nucleic acid amplification.79
eporter unLOCKing) uses a two-step process involving target amplification and CRISPR-based nucleic acid detection,75,77 successfully applied to detect SARS-CoV-2 in clinical samples.79 However, these methods are more complex than point-of-care tests due to RNA extraction and multiple liquid-handling steps, increasing the risk of sample cross-contamination. DETECTR, a recently emerging molecular diagnostic method, utilizes CRISPR-Cas12a along with isothermal nucleic acid amplification.79
In 2017, Jennifer Doudna's team unveiled the CRISPR-Cas diagnostic tool named DNA endonuclease-targeted CRISPR trans reporter (DETECTR).75 This technique leverages the collateral activity of the Cas12a protein, which is activated upon recognition of target RNA. Specifically, the LbCas12a variant of Cas12a demonstrates nonspecific collateral activity, breaking down neighboring DNA molecules after identifying target RNA. When utilized alongside single-stranded DNA reporters (probes) within a biological sample, Cas12a's detection of pathogenic nucleic acids triggers collateral activity, resulting in the destruction of DNA probes. These probes, resembling conventional TaqMan probes, emit a fluorescent signal upon degradation, which can be detected using a fluorimeter. To further enhance DETECTR's capabilities, an isothermal preamplification step (RPA) has been incorporated, improving analytical sensitivity, and obviating the necessity for costly equipment. SHERLOCK operates on similar principles to DETECTR but relies on the activity of the Cas13 nuclease from Leptotrichia wadei (Fig. 12). Unlike Cas12a, Cas13 exclusively identifies and cleaves RNA rather than DNA.82
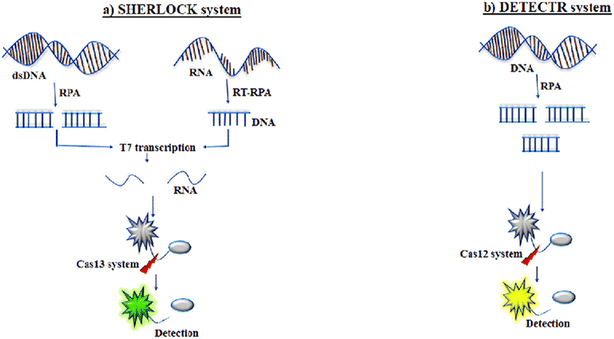 | ||
| Fig. 12 Mechanism of SHERLOCK and DETECTR system. (a) SHERLOCK system. (b) DETECTR system.83 (a) Recombinase polymerase amplification (RPA) or reverse transcription (RT)-RPA amplifies targeted double-stranded DNA (dsDNA) or RNA, respectively. This is followed by coupling RPA with T7 transcription to convert targeted RNA, allowing detection by the Cas13 system. The amplification steps, along with the reporter probe, facilitate specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) to identify the targeted sequence. (B) DNA is amplified with RPA in the DNA endonuclease-targeted CRISPR trans reporter (DETECTR). The Cas12 system associates with the single-stranded DNA (ssDNA) of interest, initiating the DNase activity of the Cas12 system. This amplification step, in conjunction with the reporter probe, enables DETECTR to detect the targeted sequence. | ||
Joung et al.84 introduced STOP (SHERLOCK Testing in One Pot), a simple method for detecting SARS-CoV-2 with sensitivity comparable to RT-qPCR assays. STOP combines simplified RNA extraction, isothermal amplification, and CRISPR-based detection from nasopharyngeal or anterior nasal swab samples. Operating at a single temperature, STOP provides results in under an hour with minimal equipment.
Nan et al.67 expanded on the CRISPR-Cas assay SHERLOCK to develop VarLOCK (variant-specific SHERLOCK), an innovative molecular diagnostic approach suitable for known variants of concern (VOCs) and newly detected variants (Fig. 13). VarLOCK swiftly and sensitively detects SARS-CoV-2 VOCs, applicable at point of care or in testing facilities of any size. It also estimates VOC proportions in pooled wastewater samples, complementing sequencing efforts for rapid identification of infected individuals and breaking infection chains. They optimized VarLOCK assays for various spike gene mutations of SARS-CoV-2 and validated them with samples from the Cardiff University Testing Service. VarLOCK's utility extends to national wastewater surveillance of SARS-CoV-2 variants, demonstrating adaptability to new and emerging VOCs.
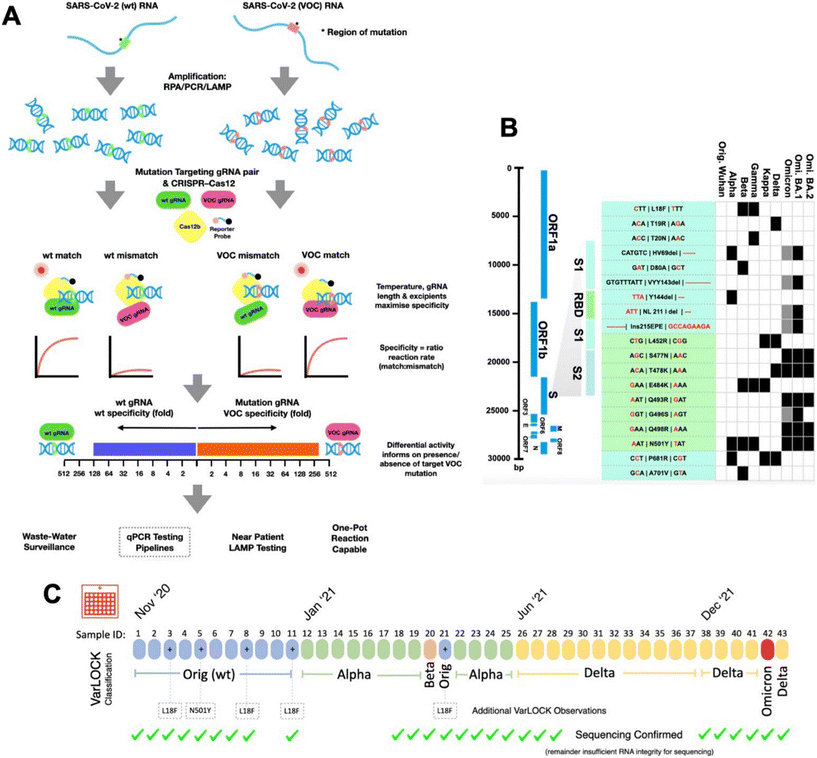 | ||
| Fig. 13 SARS-CoV-2 variant of concern identification using the VarLOCK assay.67 (A) The VarLOCK assay is illustrated as follows: SARS-CoV-2 RNA regions, whether containing the wildtype (green) or mutant (pink) sequences, are amplified using RPA, LAMP, or PCR. The resulting amplified DNA fragments are then subjected to the SHERLOCK assay with a pair of gRNAs specific to either the wildtype sequence (wt gRNA, green) or the mutant sequence (VOC gRNA, pink). The goal is to establish conditions where only a perfect match between the gRNA and the amplified DNA will activate the Cas12b protein's collateral nuclease activity, leading to the cleavage of a quenched fluorescent reporter. This cleavage results in an increase in fluorescence, allowing the nuclease activity to be monitored. The ratiometric response of an unknown SARS-CoV-2 variant sample indicates the presence or absence of the targeted mutation, thus enabling the identification of the VOC. (B) The schematic on the left shows the genome of SARS-CoV-2, with an expanded view of the spike protein encoding region, highlighting the locations of the RBD, S1, and S2. Nineteen mutation sites were selected for the VarLOCK assay, for which gRNA pairs were designed and optimized. The table displays the nucleotide sequences encoding the relevant amino acids, with nucleotide substitutions/deletions marked in red. The association of these mutations with various variants of concern is also indicated, creating a barcode-like identification matrix. (C) This approach, when applied to saliva samples, successfully identifies the original Wuhan, Alpha, Beta, Delta, and Omicron variants, with results confirmed by sequencing. | ||
They validated and implemented VarLOCK using patient samples from the Cardiff University COVID19 Testing Service. These assays can also be employed in national wastewater surveillance, facilitating identification and mapping of variant prevalence across different times and locations. Coupled with LAMP amplification, VarLOCK assays offer potential for widespread use in near-patient variant-specific diagnostic testing. They showcased VarLOCK's rapid adaptability to new variants, exemplified by the development of an Omicron-specific assay within two weeks of its classification by the WHO. This advancement could significantly enhance public health initiatives by enabling swifter identification of Omicron infections and providing a scalable approach for future variants of concern.67
E. Aptamer-based electrochemical chip
Aptamers, highly specific nucleic acid-based recognition elements, are selected in labs to target molecules, including viral proteins.85 They possess exceptional specificity, stability, and versatility for surface attachment and signal transmission, making them ideal for biosensors. During the COVID-19 pandemic, aptamers have been engineered to target various parts of the SARS-CoV-2 virus, such as the spike protein, nucleocapsid protein, and the whole virus, with very low dissociation constants. A universal aptamer has been developed capable of detecting not only the original SARS-CoV-2 strain but also several variants, including alpha, beta, gamma, epsilon, kappa, delta, and omicron. A dimeric aptamer, DSA1N5, was created by combining two previously selected aptamers, demonstrating a detection sensitivity of 100 copies per mL of SARS-CoV-2. Despite evaluating these aptamers in saliva and nasopharyngeal fluid, their potential for detecting SARS-CoV-2 in wastewater remains unexplored. While there have been advancements in biosensing technologies for clinical samples, their adaptation to wastewater analysis is challenging due to low virus levels and matrix interference.86–88Facing this challenge, Sen and colleagues aimed to assess the suitability of aptamer technology for wastewater analysis and develop a prototype for aptamer-based wastewater monitoring. They began by examining the stability and specificity of their universal aptamer in wastewater using a dot blot method. Then, they integrated an electrochemical aptamer test with a water filtration, purification, and enrichment system to determine if this setup could achieve the required limit of detection and specificity for wastewater analysis. Finally, they used the integrated system to analyze actual wastewater samples and compared its performance with that of RT-qPCR.68
They combined an aptamer-based electrochemical chip with a filtration, purification, and extraction system to create an on-site solution for wastewater analysis (Fig. 14). Using a dimeric aptamer effective against wild-type and variants like alpha, delta, and omicron, they confirmed its stability in wastewater (diluted to 50%) with minimal impact on binding affinity. The sensing chip demonstrates a detection limit of 1000 copies per L (1 copy per mL), enhanced by the filtration, purification, and extraction system. This integrated setup can detect trace virus levels (<10 copies per mL), moderate contamination (10–1000 copies per mL), or high levels (>1000 copies per mL) in native wastewater, offering a practical in-field wastewater analysis solution.68
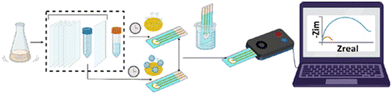 | ||
| Fig. 14 Graphical abstract of aptamer-based electrochemical chip with a filtration, purification, and extraction system to create an on-site solution for wastewater analysis.68 Samples undergo filtration, purification, and extraction to concentrate nucleic acids, including SARS-CoV-2 RNA. Dimeric aptamers on an electrochemical chip bind specifically to SARS-CoV-2 RNA or proteins, enhancing binding strength. Binding causes a structural change, altering electrochemical properties detected as redox current. Signal amplification by filtration, purification, and extraction precedes quantification, categorizing viral load levels. | ||
Microfluidics provides precise control over experimental environments using laminar flow, fluid mixing, and broader fluid behavior concepts. Microfluidic technology, enabling rapid on-site analysis with minimal reagent usage, reduces time, sample loss, contamination risks, and costs, while enhancing automation and reliability.92 However, microfluidic devices, which predominantly made from polydimethylsiloxane (PDMS), face challenges despite their advantages. PDMS processing is time-consuming, prone to swelling in certain solvents, difficult to seal, and exhibits selective molecule extraction. Finding affordable, solvent-compatible materials with versatile processing methods remains a significant obstacle for scaling up production for both research and commercial use. Alternative materials like thermoplastics and processing techniques such as hot embossing and 3D printing require further exploration to meet application requirements. Moreover, microfluidic devices often rely on external equipment, hindering accessibility and scalability. Integrating smaller, cost-effective components into machines could alleviate this issue. Streamlining sample preparation procedures and developing programmable microfluidic chips would further enhance accessibility and versatility, overcoming current limitations in reliability and compatibility between component chips.93 Enhancing microfluidic accessibility hinges on developing cost-effective materials compatible with various solvents, seamlessly transitioning from research to commercialization. Additionally, devising fluid movement techniques devoid of external pumps and microfluidic chips performing specific sample preparations is imperative.94
CRISPR-Cas offers significant benefits for point-of-care testing in resource-limited environments. It provides real-time patient information, aiding in better treatment decisions and improving outcomes. CRISPR-based tests are affordable, sensitive, user-friendly, and independent of complex equipment, making them ideal for on-site point-of-care testing. With short detection cycles, low cost, high sensitivity, and versatile readout methods, CRISPR-Cas systems reduce dependency on PCR-based instruments, promising effective and accessible diagnostics.95 Its distinctive ability to precisely target and cleave DNA sequences makes CRISPR-Cas technology a ground-breaking tool for diagnostics, genomic and molecular editing. Despite the notable progress, most CRISPR-Cas sensors primarily detect nucleic acids, limiting their broader applicability. Challenges include oversensitivity, off-target recognition, and false positives due to the system's tolerance for mismatched nucleotides, hindering widespread adoption.96 Despite hurdles, CRISPR technology promises to revolutionize sensing and enhance POCT globally.97
Aptamers, folded into 3D nanostructures via specific base pairing, offer increased sensitivity and selectivity. Their feasibility for chemical synthesis allows sequence-specific modifications to enhance binding affinity. Aptamers' self-amplification enhances biosensor sensitivity, while their binding-induced structure-switching improves selectivity. Adaptability to evolving biosensor requirements, such as real-time and in-field detection, is highlighted. Their modular nature facilitates multifunctional biosensor development, promising advancements beyond mere binding strength enhancement.98 However, commercialization of aptamers and aptameric biosensors faces several challenges. Aptamer generation via SELEX technologies is time-consuming and labor-intensive, with a low success rate.98 PCR during SELEX technology presents challenges due to the complexity of the reaction mixture and the generation of biased and artifactual products. Factors such as unequal primer hybridization, varying polymerase efficiency, and intertemplate hybridization contribute to this bias.99 Addressing these challenges is crucial for the advancement of biosensors utilizing aptamers.
Point-of-care diagnostics represent a promising avenue for rapid and on-site analysis, particularly in the context of wastewater management. However, several limitations persist in their application to wastewater analysis, necessitating improvements to enhance their efficacy and reliability. One significant limitation lies in the complexity and heterogeneity of wastewater samples.100 Unlike clinical samples, which are relatively standardized, wastewater can contain a myriad of substances, including organic matter, chemicals, and microbial contaminants.
Wastewater samples may contain compounds that cross-react with assay reagents or interfere with detection mechanisms, leading to false-positive or false-negative outcomes.101 The presence of interfering substances can compromise the accuracy of results obtained from point-of-care diagnostics systems. Moreover, the current lack of standardization and validation protocols specific to point-of-care diagnostics in wastewater analysis hinders their widespread adoption. Establishing standardized procedures for calibrating, validating, and quality-assuring point-of-care diagnostics platforms is imperative for ensuring the accuracy and comparability of results across different settings.102
In summary, while point-of-care diagnostics holds immense potential for wastewater analysis, overcoming current limitations requires concerted efforts towards technological innovation, methodological refinement, and standardization initiatives. By addressing these challenges, point-of-care diagnostics can become indispensable tools for monitoring and managing wastewater resources effectively.
Conclusion
The integration of point-of-care diagnostics into SARS-CoV-2 wastewater-based epidemiology marks a notable advancement in COVID-19 surveillance. While RT-PCR has demonstrated high sensitivity, its complexity and reliance on specialized equipment pose challenges. Point-of-care testing methods, such as LAMP, microfluidic technology, field-effect transistors, biosensors, and CRISPR/Cas, offer rapid and accessible alternatives crucial for timely detection and surveillance efforts. Combining LAMP with microfluidic technology and paper-based devices further enhances their utility for on-site analysis. Furthermore, the development of aptamer-based electrochemical chips provides solutions with high specificity and stability, facilitating practical monitoring of viral transmission in wastewater. Despite current challenges associated with sample complexity and standardization, ongoing research aims to refine these point-of-care diagnostics to improve their effectiveness and reliability. In summary, point-of-care diagnostics offer a revolutionary approach to wastewater-based epidemiology, providing promising prospects for efficient disease surveillance and management beyond the scope of SARS-CoV-2.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This research received no external funding.References
- C. Daughton, The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends, Sci. Total Environ., 2020, 726, 138149 CrossRef CAS.
- M. Mousazadeh, R. Ashoori, B. Paital, I. Kabdaşlı, Z. Frontistis and M. Hashemi, et al., Wastewater based epidemiology perspective as a faster protocol for detecting coronavirus RNA in human populations: a review with specific reference to SARS-CoV-2 virus, Pathogens, 2021, 10(8), 1008 CrossRef CAS.
- J. Wright, E. M. Driver, D. A. Bowes, B. Johnston and R. U. Halden, Comparison of high-frequency in-pipe SARS-CoV-2 wastewater-based surveillance to concurrent COVID-19 random clinical testing on a public US university campus, Sci. Total Environ., 2022, 820, 152877 CrossRef CAS.
- S. Kumar, M. Nehra, J. Mehta, N. Dilbaghi, G. Marrazza and A. J. S. Kaushik, Point-of-care strategies for detection of waterborne pathogens, Sensors, 2019, 19(20), 4476 CrossRef CAS PubMed.
- L. S. A. Busa, S. Mohammadi, M. Maeki, A. Ishida, H. Tani and M. J. M. Tokeshi, Advances in microfluidic paper-based analytical devices for food and water analysis, Micromachines, 2016, 7(5), 86 CrossRef PubMed.
- S.-U. Hassan, A. Tariq, Z. Noreen, A. Donia, S. Z. Zaidi and H. Bokhari, et al., Capillary-driven flow microfluidics combined with smartphone detection: an emerging tool for point-of-care diagnostics, Diagnostics, 2020, 10(8), 509 CrossRef CAS.
- S. Ishii, G. Kitamura, T. Segawa, A. Kobayashi, T. Miura and D. Sano, et al., Microfluidic quantitative PCR for simultaneous quantification of multiple viruses in environmental water samples, Appl. Environ. Microbiol., 2014, 80(24), 7505–7511 CrossRef PubMed.
- S. Ishii, T. Segawa and S. Okabe, Simultaneous quantification of multiple food-and waterborne pathogens by use of microfluidic quantitative PCR, Appl. Environ. Microbiol., 2013, 79(9), 2891–2898 CrossRef CAS PubMed.
- Z. Mumtaz, Z. Rashid, A. Ali, A. Arif, F. Ameen and M. S. AlTami, et al., Prospects of microfluidic technology in nucleic acid detection approaches, Biosensors, 2023, 13(6), 584 CrossRef CAS PubMed.
- S. H. Lee, J. Song, B. Cho, S. Hong, O. Hoxha and T. Kang, et al., Bubble-free rapid microfluidic PCR, 2019, vol. 126, pp. 725–733.
- A. D. da Silva, W. J. Paschoalino, R. C. Neto and L. T. Kubota, Electrochemical point-of-care devices for monitoring waterborne pathogens: Protozoa, bacteria, and viruses–An overview, Case Stud. Chem. Environ. Eng., 2022, 5, 100182 CrossRef CAS.
- J. Rainbow, E. Sedlackova, S. Jiang, G. Maxted, D. Moschou and L. Richtera, et al., Integrated electrochemical biosensors for detection of waterborne pathogens in low-resource settings, Biosensors, 2020, 10(4), 36 CrossRef CAS PubMed.
- N. Kalyani, S. Goel and S. Jaiswal, On-site sensing of pesticides using point-of-care biosensors: a review, Environ. Chem. Lett., 2021, 19(1), 345–354 CrossRef CAS.
- P. D. Howes, R. Chandrawati and M. M. Stevens, Colloidal nanoparticles as advanced biological sensors, Science, 2014, 346(6205), 1247390 CrossRef PubMed.
- J. Yang, K. Wang, H. Xu, W. Yan, Q. Jin and D. Cui, Detection platforms for point-of-care testing based on colorimetric, luminescent and magnetic assays: A review, Talanta, 2019, 202, 96–110 CrossRef CAS PubMed.
- C. Wang, F. Hou and Y. Ma, Simultaneous quantitative detection of multiple tumor markers with a rapid and sensitive multicolor quantum dots based immunochromatographic test strip, Biosens. Bioelectron., 2015, 68, 156–162 CrossRef CAS.
- M.-L. Järvenpää, K. Kuningas, I. Niemi, P. Hedberg, N. Ristiniemi and K. Pettersson, et al., Rapid and sensitive cardiac troponin I immunoassay based on fluorescent europium (III)-chelate-dyed nanoparticles, Clin. Chim. Acta, 2012, 414, 70–75 CrossRef.
- C. Liu, W. Ma, Z. Gao, J. Huang, Y. Hou and C. Xu, et al., Upconversion luminescence nanoparticles-based lateral flow immunochromatographic assay for cephalexin detection, J. Mater. Chem. C, 2014, 2(45), 9637–9642 RSC.
- L. Heller, C. R. Mota and D. B. Greco, COVID-19 faecal-oral transmission: Are we asking the right questions?, Sci. Total Environ., 2020, 729, 138919 CrossRef CAS PubMed.
- W. Ahmed, N. Angel, J. Edson, K. Bibby, A. Bivins and J. W. O'Brien, et al., First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community, Sci. Total Environ., 2020, 728, 138764 CrossRef CAS PubMed.
- S. J. N. Mallapaty, How sewage could reveal true scale of coronavirus outbreak, Nature, 2020, 580(7802), 176–177 CrossRef CAS PubMed.
- G. Medema, L. Heijnen, G. Elsinga, R. Italiaander and A. Brouwer, Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands, Environ. Sci. Technol. Lett., 2020, 7(7), 511–516 CrossRef CAS PubMed.
- Y. Wu, C. Guo, L. Tang, Z. Hong, J. Zhou and X. Dong, et al., Prolonged presence of SARS-CoV-2 viral RNA in faecal samples, The Lancet Gastroenterology & Hepatology, 2020, 5(5), 434–435 Search PubMed.
- J. R. Thompson, Y. V. Nancharaiah, X. Gu, W. L. Lee, V. B. Rajal and M. B. Haines, et al., Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management, Water Res., 2020, 184, 116181 CrossRef CAS PubMed.
- A. Donia, S.-U. Hassan, X. Zhang, L. Al-Madboly and H. Bokhari, COVID-19 crisis creates opportunity towards global monitoring & surveillance, Pathogens, 2021, 10(3), 256 CrossRef CAS PubMed.
- N. W. West, J. Hartrick, M. Alamin, A. A. Vasquez, A. Bahmani and C. L. Turner, et al., Passive swab versus grab sampling for detection of SARS-CoV-2 markers in wastewater, Sci. Total Environ., 2023, 889, 164180 CrossRef CAS.
- N. W. West, A. A. Vasquez, A. Bahmani, M. F. Khan, J. Hartrick and C. L. Turner, et al., Sensitive detection of SARS-CoV-2 molecular markers in urban community sewersheds using automated viral RNA purification and digital droplet PCR, Sci. Total Environ., 2022, 847, 157547 CrossRef CAS PubMed.
- B. S. Jimenez, T. Sterling, A. Brown, B. Modica, K. Gibson and H. Collins, et al., Wastewater surveillance in the COVID-19 post-emergency pandemic period: A promising approach to monitor and predict SARS-CoV-2 surges and evolution, Heliyon, 2023, 9(11), e22356 CrossRef PubMed.
- V. Pogka, S. Labropoulou, M. Emmanouil, A. Voulgari-Kokota, A. Vernardaki and T. Georgakopoulou, et al., Laboratory surveillance of polio and other enteroviruses in high-risk populations and environmental samples, Appl. Environ. Microbiol., 2017, 83(5), e02872-16 CrossRef PubMed.
- D. Klapsa, T. Wilton, A. Zealand, E. Bujaki, E. Saxentoff and C. Troman, et al., Sustained detection of type 2 poliovirus in London sewage between February and July, 2022, by enhanced environmental surveillance, Lancet, 2022, 400(10362), 1531–1538 CrossRef CAS PubMed.
- M. Hill and A. J. Pollard, Detection of poliovirus in London highlights the value of sewage surveillance, Lancet, 2022, 400(10362), 1491–1492 CrossRef PubMed.
- G. La Rosa, S. Della Libera, M. Iaconelli, A. R. Ciccaglione, R. Bruni and S. Taffon, et al., Surveillance of hepatitis A virus in urban sewages and comparison with cases notified in the course of an outbreak, Italy 2013, BMC Infect. Dis., 2014, 14, 1–11 CrossRef PubMed.
- D. Toribio-Avedillo, C. Gómez-Gómez, L. Sala-Comorera, L. Rodríguez-Rubio, A. Carcereny and D. García-Pedemonte, et al., Monitoring influenza and respiratory syncytial virus in wastewater. Beyond COVID-19, Sci. Total Environ., 2023, 892, 164495 CrossRef CAS.
- M. Papageorgiou, I. Zioris, T. Danis, D. Bikiaris and D. Lambropoulou, Comprehensive investigation of a wide range of pharmaceuticals and personal care products in urban and hospital wastewaters in Greece, Sci. Total Environ., 2019, 694, 133565 CrossRef CAS PubMed.
- M. Massano, A. Salomone, E. Gerace, E. Alladio, M. Vincenti and M. Minella, Wastewater surveillance of 105 pharmaceutical drugs and metabolites by means of ultra-high-performance liquid-chromatography-tandem high resolution mass spectrometry, J. Chromatogr. A, 2023, 1693, 463896 CrossRef CAS.
- K. Gj, Principles and practice of point-of-care testing, Lippincott Williams & Wilkins, Philadelphia, PA, USA, 2002 Search PubMed.
- L. C. Clark and C. Lyons, Electrode systems for continuous monitoring in cardiovascular surgery, Ann. N. Y. Acad. Sci., 1962, 102(1), 29–45 CrossRef CAS PubMed.
- K. A. Erickson and P. Wilding, Evaluation of a novel point-of-care system, the i-STAT portable clinical analyzer, Clin. Chem., 1993, 39(2), 283–287 CrossRef CAS.
- D. Liu, J. Wang, L. Wu, Y. Huang, Y. Zhang and M. Zhu, et al., Trends in miniaturized biosensors for point-of-care testing, TrAC, Trends Anal. Chem., 2020, 122, 115701 CrossRef CAS.
- Z. Zhu, Z. Guan, D. Liu, S. Jia, J. Li and Z. Lei, et al., Translating molecular recognition into a pressure signal to enable rapid, sensitive, and portable biomedical analysis, Angew. Chem., 2015, 127(36), 10594–10599 CrossRef.
- D. Liu, X. Li, J. Zhou, S. Liu, T. Tian and Y. Song, et al., A fully integrated distance readout ELISA-Chip for point-of-care testing with sample-in-answer-out capability, Biosens. Bioelectron., 2017, 96, 332–338 CrossRef CAS.
- W. Zhang, R. Wang, F. Luo, P. Wang and Z. Lin, Miniaturized electrochemical sensors and their point-of-care applications, Chin. Chem. Lett., 2020, 31(3), 589–600 CrossRef CAS.
- B. Derkus, Applying the miniaturization technologies for biosensor design, Biosens. Bioelectron., 2016, 79, 901–913 CrossRef CAS.
- R. K. Benninger, O. Hofmann, B. Önfelt, I. Munro, C. Dunsby and D. M. Davis, et al., Fluorescence-Lifetime Imaging of DNA–Dye Interactions within Continuous-Flow Microfluidic Systems, Angew. Chem., Int. Ed., 2007, 46(13), 2228–2231 CrossRef CAS.
- A. S. Hansen, N. Hao and E. K. O'shea, High-throughput microfluidics to control and measure signaling dynamics in single yeast cells, Nat. Protoc., 2015, 10(8), 1181–1197 CrossRef CAS PubMed.
- E. Galopin, M. Beaugeois, B. Pinchemel, J.-C. Camart, M. Bouazaoui and V. Thomy, SPR biosensing coupled to a digital microfluidic microstreaming system, Biosens. Bioelectron., 2007, 23(5), 746–750 CrossRef CAS PubMed.
- A. Pohl and I. M. Weiss, Real-time monitoring of calcium carbonate and cationic peptide deposition on carboxylate-SAM using a microfluidic SAW biosensor, Beilstein J. Nanotechnol., 2014, 5(1), 1823–1835 CrossRef PubMed.
- Y. Zhou, F. Pei, M. Ji, L. Wang, H. Zhao and H. Li, et al., Sensitivity evaluation of 2019 novel coronavirus (SARS-CoV-2) RT-PCR detection kits and strategy to reduce false negative, PLoS One, 2020, 15(11), e0241469 CrossRef CAS PubMed.
- W. Ahmed, S. L. Simpson, P. M. Bertsch, K. Bibby, A. Bivins and L. L. Blackall, et al., Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance, 2022, vol. 805, p. 149877.
- S. Petralia and S. Conoci, PCR technologies for point of care testing: progress and perspectives, ACS Sens., 2017, 2(7), 876–891 CrossRef CAS PubMed.
- F. S. Iliescu, A. M. Ionescu, L. Gogianu, M. Simion, V. Dediu and M. C. Chifiriuc, et al., Point-of-care testing—the key in the battle against SARS-CoV-2 pandemic, Micromachines, 2021, 12(12), 1464 CrossRef PubMed.
- S. Chakraborty, Democratizing Nucleic Acid based Molecular Diagnostic tests for Infectious Diseases at Resource-Limited Settings–from Point of Care to Extreme Point of Care, Sens. Diagn., 2024, 3(4), 536–561 RSC.
- Z. Zhang, P. Ma, R. Ahmed, J. Wang, D. Akin and F. Soto, et al., Advanced point-of-care testing technologies for human acute respiratory virus detection, Adv. Mater., 2022, 34(1), 2103646 CrossRef CAS PubMed.
- S. Jain, M. Nehra, R. Kumar, N. Dilbaghi, T. Hu and S. Kumar, et al., Internet of medical things (IoMT)-integrated biosensors for point-of-care testing of infectious diseases, Biosens. Bioelectron., 2021, 179, 113074 CrossRef CAS PubMed.
- Q. Ye, D. Lu, T. Zhang, J. Mao and S. Shang, Recent advances and clinical application in point-of-care testing of SARS-CoV-2, J. Med. Virol., 2022, 94(5), 1866–1875 CrossRef CAS PubMed.
- M. F. U. Haque, S. S. Bukhari, R. Ejaz, F. U. Zaman, K. R. Sreejith and N. Rashid, et al., A novel RdRp-based colorimetric RT-LAMP assay for rapid and sensitive detection of SARS-CoV-2 in clinical and sewage samples from Pakistan, Virus Res., 2021, 302, 198484 CrossRef CAS PubMed.
- S. K. Vashist, J. H. Luong, S. K. Vashist and J. H. Luong, An overview of point-of-care technologies enabling next-generation healthcare monitoring and management, Springer, 2019 Search PubMed.
- T. Mackuľak, M. Gal, V. Špalková, M. Feher, K. Briestenska and M. Mikušová, et al., Wastewater-based epidemiology as an early warning system for the spreading of SARS-CoV-2 and its mutations in the population, Int. J. Environ. Res. Public Health, 2021, 18(11), 5629 CrossRef PubMed.
- A. Donia, M. Furqan Shahid, S.-U. Hassan, R. Shahid, A. Ahmad and A. Javed, et al., Integration of RT-LAMP and Microfluidic Technology for Detection of SARS-CoV-2 in Wastewater as an Advanced Point-of-care Platform, Food Environ. Virol., 2022, 14(4), 364–373 CrossRef CAS PubMed.
- I. D. Amoah, N. P. Mthethwa, L. Pillay, N. Deepnarain, K. Pillay and O. O. Awolusi, et al., RT-LAMP: a cheaper, simpler and faster alternative for the detection of SARS-CoV-2 in wastewater, Food Environ. Virol., 2021, 13, 447–456 CrossRef CAS PubMed.
- A. Bivins, M. Lott, M. Shaffer, Z. Wu, D. North and E. K. Lipp, et al., Building-level wastewater surveillance using tampon swabs and RT-LAMP for rapid SARS-CoV-2 RNA detection, Environ. Sci.: Water Res. Technol., 2022, 8(1), 173–183 RSC.
- J. Akter, W. J. Smith, M. Gebrewold, I. Kim, S. L. Simpson and A. Bivins, et al., Evaluation of colorimetric RT-LAMP for screening of SARS-CoV-2 in untreated wastewater, Sci. Total Environ., 2023, 167964 Search PubMed.
- R. G. Ramírez-Chavarría, E. Castillo-Villanueva, B. E. Alvarez-Serna, J. Carrillo-Reyes, R. M. Ramírez-Zamora and G. Buitrón, et al., Loop-mediated isothermal amplification-based electrochemical sensor for detecting SARS-CoV-2 in wastewater samples, J. Environ. Chem. Eng., 2022, 10(3), 107488 CrossRef PubMed.
- K. Yin, X. Ding, Z. Xu, Z. Li, X. Wang and H. Zhao, et al., Multiplexed colorimetric detection of SARS-CoV-2 and other pathogens in wastewater on a 3D printed integrated microfluidic chip, Sens. Actuators, B, 2021, 344, 130242 CrossRef CAS PubMed.
- B. E. Alvarez-Serna, R. G. Ramírez-Chavarría, E. Castillo-Villanueva, J. Carrillo-Reyes, R. M. Ramírez-Zamora and G. Buitrón, et al., Label-free and portable field-effect sensor for monitoring rt-lamp products to detect SARS-CoV-2 in wastewater, Talanta, 2023, 253, 124060 CrossRef CAS.
- H. Cao, K. Mao, F. Ran, P. Xu, Y. Zhao and X. Zhang, et al., Paper device combining CRISPR/Cas12a and reverse-transcription loop-mediated isothermal amplification for SARS-CoV-2 detection in wastewater, Environ. Sci. Technol., 2022, 56(18), 13245–13253 CrossRef CAS PubMed.
- X. Nan, P. Hardinge, S. Hoehn, S. N. Dighe, J. Ukeri and D. F. Pease, et al., VarLOCK: sequencing-independent, rapid detection of SARS-CoV-2 variants of concern for point-of-care testing, qPCR pipelines and national wastewater surveillance, Sci. Rep., 2023, 13(1), 20832 CrossRef CAS PubMed.
- P. Sen, Z. Zhang, P. Li, B. R. Adhikari, T. Guo and J. Gu, et al., Integrating Water Purification with Electrochemical Aptamer Sensing for Detecting SARS-CoV-2 in Wastewater, ACS Sens., 2023, 8(4), 1558–1567 CrossRef CAS PubMed.
- Y. H. Baek, J. Um, K. J. C. Antigua, J.-H. Park, Y. Kim and S. Oh, et al., Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2, Emerging Microbes Infect., 2020, 9(1), 998–1007 CrossRef CAS PubMed.
- L. Mautner, C.-K. Baillie, H. M. Herold, W. Volkwein, P. Guertler and U. Eberle, et al., Rapid point-of-care detection of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification (RT-LAMP), Virol. J., 2020, 17, 1–14 CrossRef PubMed.
- S. Wei, E. Kohl, A. Djandji, S. Morgan, S. Whittier and M. Mansukhani, et al., Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction, Sci. Rep., 2021, 11, 2402 CrossRef CAS PubMed.
- D. Sadighbayan, M. Hasanzadeh and E. Ghafar-Zadeh, Biosensing based on field-effect transistors (FET): Recent progress and challenges, TrAC, Trends Anal. Chem., 2020, 133, 116067 CrossRef CAS PubMed.
- S. A. Pullano, C. D. Critello, I. Mahbub, N. T. Tasneem, S. Shamsir and S. K. Islam, et al., EGFET-based sensors for bioanalytical applications: A review, Sensors, 2018, 18(11), 4042 CrossRef PubMed.
- P. Fathi-Hafshejani, N. Azam, L. Wang, M. A. Kuroda, M. C. Hamilton and S. Hasim, et al., Two-dimensional-material-based field-effect transistor biosensor for detecting COVID-19 virus (SARS-CoV-2), ACS Nano, 2021, 15(7), 11461–11469 CrossRef CAS PubMed.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian and J. M. Palefsky, et al., CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity, Science, 2018, 360(6387), 436–469 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6, Science, 2018, 360(6387), 439–444 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy and J. Joung, et al., Nucleic acid detection with CRISPR-Cas13a/C2c2, Science, 2017, 356(6336), 438–442 CrossRef CAS PubMed.
- B. Pang, J. Xu, Y. Liu, H. Peng, W. Feng and Y. Cao, et al., Isothermal amplification and ambient visualization in a single tube for the detection of SARS-CoV-2 using loop-mediated amplification and CRISPR technology, Anal. Chem., 2020, 92(24), 16204–16212 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita and J. Singh, et al., CRISPR–Cas12-based detection of SARS-CoV-2, Nat. Biotechnol., 2020, 38(7), 870–874 CrossRef CAS PubMed.
- Y. Zhao, X. Fang, H. Yu, Y. Fu and Y. Zhao, Universal Exponential Amplification Confers Multilocus Detection of Mutation-Prone Virus, Anal. Chem., 2022, 94(2), 927–933 CrossRef CAS PubMed.
- R. Wang, C. Qian, Y. Pang, M. Li, Y. Yang and H. Ma, et al., opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection, Biosens. Bioelectron., 2021, 172, 112766 CrossRef CAS PubMed.
- M. I. Mustafa and A. M. Makhawi, SHERLOCK and DETECTR: CRISPR-Cas systems as potential rapid diagnostic tools for emerging infectious diseases, J. Clin. Microbiol., 2021, 59(3), e00745-20 CrossRef PubMed.
- V. E. Hillary, S. Ignacimuthu and S. A. Ceasar, Potential of CRISPR/Cas system in the diagnosis of COVID-19 infection, Expert Rev. Mol. Diagn., 2021, 21(11), 1179–1189 CrossRef CAS PubMed.
- J. Joung, A. Ladha, M. Saito, N.-G. Kim, A. E. Woolley and M. Segel, et al., Detection of SARS-CoV-2 with SHERLOCK one-pot testing, N. Engl. J. Med., 2020, 383(15), 1492–1494 CrossRef CAS PubMed.
- B. Santosh and P. K. Yadava, Nucleic acid aptamers: research tools in disease diagnostics and therapeutics, BioMed Res. Int., 2014, 2014, 540451 Search PubMed.
- Z. Zhang, R. Pandey, J. Li, J. Gu, D. White and H. D. Stacey, et al., High-Affinity Dimeric Aptamers Enable the Rapid Electrochemical Detection of Wild-Type and B.1.1.7 SARS-CoV-2 in Unprocessed Saliva, Angew. Chem., Int. Ed., 2021, 60(45), 24266–24274 CrossRef CAS PubMed.
- J. Li, Z. Zhang, J. Gu, H. D. Stacey, J. C. Ang and A. Capretta, et al., Diverse high-affinity DNA aptamers for wild-type and B.1.1.7 SARS-CoV-2 spike proteins from a pre-structured DNA library, Nucleic Acids Res., 2021, 49(13), 7267–7279 CrossRef CAS PubMed.
- Z. Zhang, J. Li, J. Gu, R. Amini, H. D. Stacey and J. C. Ang, et al., A Universal DNA Aptamer that Recognizes Spike Proteins of Diverse SARS-CoV-2 Variants of Concern, Chemistry, 2022, 28(15), e202200524 CrossRef PubMed.
- T. J. Moehling, G. Choi, L. C. Dugan, M. Salit and R. J. Meagher, LAMP diagnostics at the point-of-care: emerging trends and perspectives for the developer community, Expert Rev. Mol. Diagn., 2021, 21(1), 43–61 CrossRef CAS PubMed.
- Y. Yu, J. X. Zhou, B. Li, M. Ji, Y. Wang and E. Carnaby, et al., A quantitative RT-qLAMP for the detection of SARS-CoV-2 and human gene in clinical application, Microb. Biotechnol., 2022, 15(10), 2619–2630 CrossRef CAS PubMed.
- B. A. Lakshmi and S. Kim, Recent trends in the utilization of LAMP for the diagnosis of viruses, bacteria, and allergens in food, Recent developments in applied microbiology and biochemistry, Elsevier, 2021, pp. 291–297 Search PubMed.
- J. Saez, R. Catalan-Carrio, R. M. Owens, L. Basabe-Desmonts and F. Benito-Lopez, Microfluidics and materials for smart water monitoring: A review, Anal. Chim. Acta, 2021, 1186, 338392 CrossRef CAS PubMed.
- D. R. Reyes, H. van Heeren, S. Guha, L. Herbertson, A. P. Tzannis and J. Ducrée, et al., Accelerating innovation and commercialization through standardization of microfluidic-based medical devices, Lab Chip, 2021, 21(1), 9–21 RSC.
- S. Battat, D. A. Weitz and G. M. Whitesides, An outlook on microfluidics: the promise and the challenge, Lab Chip, 2022, 22(3), 530–536 RSC.
- T. Huang, R. Zhang and J. Li, CRISPR-Cas-based techniques for pathogen detection: Retrospect, recent advances, and future perspectives, J. Adv. Res., 2023, 50, 69–82 CrossRef CAS PubMed.
- X.-H. Zhang, L. Y. Tee, X.-G. Wang, Q.-S. Huang and S.-H. Yang, Off-target effects in CRISPR/Cas9-mediated genome engineering, Mol. Ther.–Nucleic Acids, 2015, 4, e264 CrossRef CAS PubMed.
- A. Kumaran, N. Jude Serpes, T. Gupta, A. James, A. Sharma and D. Kumar, et al., Advancements in CRISPR-based biosensing for next-gen point of care diagnostic application, Biosensors, 2023, 13(2), 202 CrossRef CAS PubMed.
- H. Yoo, H. Jo and S. S. Oh, Detection and beyond: Challenges and advances in aptamer-based biosensors, Mater. Adv., 2020, 1(8), 2663–2687 RSC.
- G. T. Rozenblum, V. G. Lopez, A. D. Vitullo and M. Radrizzani, Aptamers: current challenges and future prospects, Expert Opin. Drug Discovery, 2016, 11(2), 127–135 CrossRef CAS PubMed.
- K. Obaideen, N. Shehata, E. T. Sayed, M. A. Abdelkareem, M. S. Mahmoud and A. Olabi, The role of wastewater treatment in achieving sustainable development goals (SDGs) and sustainability guideline, Energy Nexus, 2022, 7, 100112 CrossRef.
- K. A. Vu, T. A. Nguyen and T. P. Nguyen, Advances in Pretreatment Methods for Free Nucleic Acid Removal in Wastewater Samples: Enhancing Accuracy in Pathogenic Detection and Future Directions, Appl. Microbiol., 2023, 4(1), 1–15 CrossRef.
- I. Michael-Kordatou, P. Karaolia and D. Fatta-Kassinos, Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification, J. Environ. Chem. Eng., 2020, 8(5), 104306 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |





