Metal-phenolic self-assembly shielded probiotics in hydrogel reinforced wound healing with antibiotic treatment†
Chen
Zhou‡
ab,
Yaping
Zou‡
ab,
Ruiling
Xu
ab,
Xiaowen
Han
ab,
Zhen
Xiang
ab,
Hao
Guo
ab,
Xing
Li
ab,
Jie
Liang
abc,
Xingdong
Zhang
ab,
Yujiang
Fan
 *ab and
Yong
Sun
*ab and
Yong
Sun
 *ab
*ab
aNational Engineering Research Center for Biomaterials, Sichuan University, 29# Wangjiang Road, Chengdu, China. E-mail: fan_yujiang@scu.edu.cn; sunyong8702@scu.edu.cn
bCollege of Biomedical Engineering, Sichuan University, 29# Wangjiang Road, Chengdu, China
cSichuan Testing Center for Biomaterials and Medical Devices, Sichuan University, 29# Wangjiang Road, Chengdu, China
First published on 12th May 2023
Abstract
Living probiotics secrete bioactive substances to accelerate wound healing, but the clinical application of antibiotics inhibits the survival of probiotics. Inspired by the chelation of tannic acid and ferric ions, we developed a metal-phenolic self-assembly shielded probiotic (Lactobacillus reuteri, L. reuteri@FeTA) to prevent interference from antibiotics. Here, a superimposing layer was formed on the surface of L. reuteri to adsorb and inactivate antibiotics. These shielded probiotics were loaded into an injectable hydrogel (Gel/L@FeTA) formed by carboxylated chitosan and oxidized hyaluronan. The Gel/L@FeTA aided the survival of probiotics and supported the continuous secretion of lactic acid to perform biological functions in an environment containing gentamicin. Furthermore, the Gel/L@FeTA hydrogels presented a better performance than the Gel/L in inflammatory regulation, angiogenesis, and tissue regeneration both in vitro and in vivo in the presence of antibiotics. Hence, a new method for designing probiotic-based biomaterials for clinical wound management is provided.
New conceptsCompared with other advanced materials, living probiotics could continue to secrete bioactive substances until the wound heals, but their survival is crucial for their efficacy. Thus, the application of antibiotics during the treatment should be carefully considered. Here, we developed a novel probiotic hydrogel system with double-layer protection through a metal-polyphenol complex covering the surface of the probiotics layer-by-layer and a hydrogel network formed by a Schiff base reaction. The hydrogel network provided a space for the normal physiological activities of probiotics and achieved superior performance including water absorption, injectability, and antibacterial properties. Moreover, probiotics survived even in an environment containing antibiotics (gentamicin, penicillin, and cephalosporin) because of the adsorption and isolation behavior of the supramolecular structure, which substantially improves the efficacy of probiotic-based wound treatment. In a full-thickness round skin wound model, using antibiotics in vivo, the self-assembly shielded probiotic hydrogel played a role in accelerating wound healing and facilitated a series of beneficial processes such as inflammatory regulation, angiogenesis, and tissue regeneration. Thus, this study provides a simple strategy for enhancing the therapeutic effect of probiotic-based biomaterials in tissue repair. |
Introduction
As a member of the normal human microbiota, probiotics show various biological activities and functions, which play an important role in maintaining the homeostasis of the internal environment and fighting against diseases.1–3 Using probiotics in clinical settings to improve the curative effect is an attractive and innovative strategy, which has attracted increased attention in recent years.4–6 In previous studies, probiotics have been reported to prevent pathogenic infection, regulate inflammation, and promote tissue regeneration and remodeling by continuously releasing bioactive substances such as organic acids, bacteriocins, and enzymes, which have a positive effect on wound healing.7–9 In contrast with other advanced materials, including bioactive ceramics, natural extracts, metal nanoparticles, and exosomes, the continuous release of bioactive substances by probiotics is not affected by the consumption of raw materials during the therapy. The multi-function advantages of probiotics demonstrate their potential value in wound treatment.10Normally, using antibiotics as adjuvant therapy in serious wounds is necessary.11–13 However, because of the nonspecific killing action of antibiotics, their use leads to a drastic depletion of probiotics and a major setback for the curative effect. Hence, improving the survival ability of probiotics in trauma sites is crucial for achieving a curative effect.14 According to previous studies,15,16 the physical encapsulation of probiotics in a biocompatible hydrogel network could provide a space for their normal physiological activities while avoiding attack by the immune system. In addition, the inherent absorbability, high porosity, and stable physical structure of hydrogels provide a favorable environment for wound healing.17–19 Nevertheless, due to the permeability of the barrier, antibiotics as part of the treatment, still have an inevitable impact on the biological activity of probiotics.
Layer-by-layer self-assembly technology is safe and easy and provides effective protective packaging for the molecular surface. This technology is widely used in food engineering, environmental science, and medicine.20 Recent studies21–23 have shown that the self-assembled nano-package achieves the adsorption, blocking, and inactivation of foreign material through intermolecular interactions, thus attaining optimal protection. Moreover, the composition of these nanocarriers is usually based on rich plant extracts with high biocompatibility and biosafety. As a common food additive and functional substance applied in biomedical engineering, tannic acid (TA) forms a supramolecular layer-by-layer structure by chelation with ferric ions (Fe3+). This structure could be adsorbed on the surface of bacteria and could achieve the isolation of antibiotics through molecular interactions and precise adsorption.24,25 This strategy has also been reported for the surface modification of probiotics by oral delivery to avoid the influence of antibiotics and has produced surprising results.26 Therefore, this strategy has the potential to enhance the therapeutic effect of probiotic-based biomaterials and improve the multiple functional effects of probiotics.
As one of the most common probiotics, Lactobacillus reuteri (L. reuteri) shows biosafety, which allows its wide application in food engineering and medicine. L. reuteri produces bioactive substances (e.g., antibacterial agents) to inhibit pathogens and boost immune regulation, as well as to promote wound repair.27–29 In this study, based on the chelation of tannic acid and ferric ions, a nano-protective layer was assembled layer-by-layer on the surface of L. reuteri (L. reuteri@FeTA) and then wrapped into an injectable hydrogel formed by carboxylated chitosan (CCS) and oxidized hyaluronic acid (OHA) based on a Schiff base reaction (Gel/L@FeTA). The properties of these hydrogels were characterized in vitro in the presence of antibiotics. Furthermore, a full-thickness skin injury model of BALB/c mice treated with/without antibiotics was established, and immunohistochemistry analysis and western blots were performed to evaluate the healing properties of the hydrogels in vivo. We found that Gel/L@FeTA could effectively shield the antibiotic to maintain the viability of L. reuteri, which in turn promoted a series of processes, such as immune regulation and angiogenesis, to accelerate wound healing. These results indicate that Gel/L@FeTA may be a potential candidate for clinical wound treatment.
Results and discussion
Synthesis and structural characterization of L. reuteri@FeTA and the hydrogels
The scheme of the synthesis of L. reuteri@FeTA and the hydrogels is shown in Fig. 1A and Fig. S1 (ESI†). L. reuteri was evenly dispersed in a TA solution and formed a dense packaging in a layer-by-layer self-assembly by complexation with FeCl3. Due to the adsorption of the polyphenols and simultaneous cross-linking, this supramolecular structure could stably depose on the surface of bacteria.20 The scanning electron microscope (SEM) image revealed a uniform nano-packaging on the surface of L. reuteri@FeTA compared to the L. reuteri without any modification (Fig. 1B). In addition, measurement of the diameter of L. reuteri and L. reuteri@FeTA revealed that the thickness of L. reuteri@FeTA increased by about 15 nm compared to the unmodified probiotic, which was similar to previous reports26 (Fig. 1C). These results confirm that the Fe3+-TA chelate successfully enclosed the surface of L. reuteri.The hydrogels were synthesized according to our previous study.30 As a hydrogel precursor, OHA polymer was synthesized to obtain aldehydes by the oxidation of NaIO4, and the hydrogel was prepared by a Schiff base reaction between the amino group from CCS and the aldehyde group from OHA at 37 °C. The 1H NMR spectra of OHA revealed that new peaks appeared at 4.90, 5.00, and 5.10 ppm, which corresponded to the hemiacetalic proton between the aldehydes and the neighboring hydroxyl groups. The proton signal at 9.50 ppm belonged to the proton in the aldehyde31 (Fig. S2A, ESI†). The ATR-FTIR spectrum also exhibited a new absorption peak at 1730 cm−1, which corresponded to the stretching vibration of –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and the typical peaks of the Schiff base at 1645 cm−1 confirmed that the hydrogel was successfully synthesized32 (Fig. S2B, ESI†).
O, and the typical peaks of the Schiff base at 1645 cm−1 confirmed that the hydrogel was successfully synthesized32 (Fig. S2B, ESI†).
To obtain the hydrogels with probiotics, 106 CFU mL−1 of L. reuteri and L. reuteri@FeTA were mixed in the OHA solution before gelation. SEM analysis of the microstructure of each sample revealed that the hydrogels had an evenly porous structure and the scattered bacteria exhibited a representative rod-like shape in the hydrogel network (Fig. 1D). This suggests that the probiotic was uniformly loaded within the hydrogel, and both the morphology of the hydrogel and probiotic was not affected. Furthermore, the transmission electron microscope (TEM) images also exhibited that the Fe3+-TA chelate, as a distinct extra shell,33 showed stable adhesion to the surface of L. reuteri after loading in the network of the hydrogel, and the elemental maps of Fe in the Gel/L@FeTA proved the existence of a ferric ion complex from L. reuteri@FeTA (Fig. 1E). In addition, the water absorption capacity, degradation behavior, and mechanical properties of the hydrogels were also investigated because they were related to the wound-dressing requirements. The water absorption of the hydrogels was not affected after loading with probiotics. The hydrogels also exhibited a stable degradation behavior, without notable differences under physiological conditions (Fig. S3, ESI†). On the other hand, the modulus and compression behavior of all hydrogels showed a regular and unified change under the same conditions (Fig. S4, ESI†). Thus, encapsulation of probiotics had no substantial effect on the performance of the hydrogel, which is consistent with the results of the previous study.27,34
The antibiotic shielding behavior of the hydrogels
Living bacterial probiotics can secrete large amounts of metabolites and organic acids, which have been used for inflammation regulation and improvement of the wound microenvironment.15 Therefore, avoiding the interference of antibiotics and maintaining the activity of probiotics is essential during the treatment. To observe the activity of L. reuteri under the antibiotic conditions before loading into Gel, the bacterial growth curve and spread plate method were employed in this study. As shown in Fig. S5 and S6 (ESI†), the normal growth of bacteria was exhibited both in L. reuteri and L. reuteri@FeTA without antibiotics. After treatment with antibiotics, L. reuteri@FeTA still maintains stable bacterial vitality, while the L. reuteri was significantly inhibited. These results indicated that the Fe3+-TA chelation could not only isolate and adsorb the antibiotics but also did not affect the normal proliferation of bacteria. Similar results were also reflected in L. reuteri@FeTA after loading into Gel. Under conditions without antibiotics (Fig. 1F1), L. reuteri exhibited normal proliferative activity both in Gel/L and Gel/L@FeTA (p > 0.05). However, because of antibiotic interference (Fig. 1F2–4), the proliferation behavior of L. reuteri in Gel/L was significantly decreased. By contrast, the L. reuteri in Gel/L@FeTA could proliferate continuously even in the presence of common antibiotics (gentamicin, penicillin, and cephalosporin) (Fig. S7, ESI†). This suggests that the Fe3+-TA chelate through layers of packaging provides a tight coating—like a suit of armor—to reduce the harmful effects on probiotics, without affecting their vitality and proliferation.35In addition, the survivability of L. reuteri in hydrogels was evaluated using LIVE/DEAD BacLight Bacterial Viability kits with gentamicin as the representative antibiotic. In the CLSM images (Fig. 1G and Fig. S8, ESI†), more green signals (representing living bacteria) were observed in both Gel-L and Gel-L@FeTA under normal physiological conditions without the antibiotic, while red signals (representing dead bacteria) were less abundant. When the hydrogels were applied to the physiological environment containing gentamicin, red (death) signals spread all over the Gel-L and few bacteria survived. By contrast, numerous green signals (representing survival) could still be observed in Gel-L@FeTA. Similar phenomena were also observed in the bacterial growth plate (Fig. 1H and Fig. S9, ESI†).
As an effective secretion from living probiotics, suitable lactic acid has a positive effect to regulate physiological activity through immunoregulation, antibacterial, and other properties.36,37 As it had not affected the normal proliferation of bacteria, the proliferative clones from L. reuteri@FeTA could stably secrete lactic acid even under the condition of gentamicin (Fig. S10, ESI†). Therefore, the cumulative release of lactic acid in the hydrogels under different conditions was also analyzed. As shown in Fig. 1I, lactic acid was secreted both in Gel-L and Gel-L@FeTA because the bacterial activity was undisturbed and the probiotics survived in the hydrogels under normal physiological conditions. Under the influence of gentamicin, less lactic acid was secreted in Gel-L. Nevertheless, Gel-L@FeTA could still release lactic acid continuously for 48 h. Although hydrogels provided a suitable living space for probiotics, the vitality and function of bacteria were severely damaged once the antibiotic was immersed in the hydrogel's network. These results suggest that the superimposed layer not only played a role in reducing the mortality of the probiotics by adsorbing and isolating the antibiotic, but also did not affect the proliferation of probiotics, and thus supported the sustained secretion of bioactive substances like lactic acid by the probiotics.21,26
In vitro antibacterial activity, inflammatory regulation capacity and biocompatibility of the hydrogels
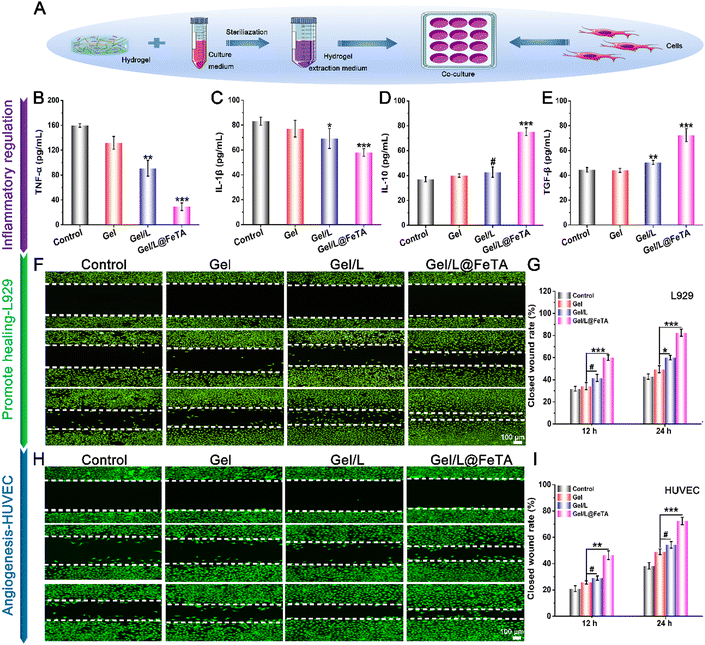
High antibacterial performance is imperative in the design of wound dressings.38,39 To eliminate the disturbance from the probiotic itself, a sterilized hydrogel extract was used to evaluate the antibacterial capacity. The images of the spread plates of two common pathogens – Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus) revealed that fewer bacteria appeared in the hydrogel groups compared with the control, especially since no obvious bacterial clones were found in the Gel/L and Gel/L@FeTA groups (Fig. S11, ESI†). As a hydrogel component, CCS shows excellent antibacterial capacity by continuously releasing cations to destroy bacterial cell walls and also could provide a suitable space for the survival of probiotics by adjusting the concentration.40 On the other hand, L. reuteri also could release antimicrobial agents such as organic acids and bacteriocins.15 Therefore, the gel synthesized with CCS as a precursor not only exhibited high antibacterial behavior but also improved the sterilization effect by synergism with probiotics.Furthermore, a co-culture system of a hydrogel extract and cells was employed to evaluate the effect of the hydrogel on cell behavior including inflammatory regulation, cell proliferation, spreading, and migration (Fig. 2A). The levels of inflammatory cytokines were investigated using an ELISA kit after 24 h of treatment (Fig. 2B–E). As representative pro-inflammatory factors, TNF-α and IL-1β showed lower concentrations in Gel/L@FeTA than in the gel and control group (p < 0.001). By contrast, the hydrogel extract of Gel/L@FeTA promoted the expression of IL-10 and TGF-β (which play a substantial role in the anti-inflammatory process) compared with the gel and the control group (p < 0.001). Nonetheless, despite the regulatory trend of Gel/L being similar to that of Gel/L@FeTA, the effect was not substantial. This may be due to the influence of antibodies in the cell culture medium, which has a negative effect on the activity of the probiotic. The secretory level of organic substances by living probiotics has a considerable influence on the expression of inflammatory factors.
In addition, a highly biocompatible dressing material is desirable.41,42 Compared with L. reuteri, the L. reuteri@FeTA presented better cell proliferation capacity because of the shielding effect of Fe-TA to reduce the interference of antibiotics (Fig. S12, ESI†). Thus, the biocompatibility of the hydrogel loaded with probiotic should be further tested with different types of cells. In this study, L929 cells and HUVECs were used to test the biocompatibility through the live/dead cell, cytoskeleton/nuclear, and CCK8 assays. For L929, as shown in Fig. S13 (ESI†), numerous fluorescent green signals (representing living cells) were observed in each group, while the expression of red fluorescence (representing dead cells) could hardly be observed. The morphology of the cytoskeleton and nucleus in the hydrogel groups presented normal structural characteristics like those of the control group. The CCK8 assay further evaluated cell proliferation in the hydrogel extract. No significant difference was observed in the cell vitality of each hydrogel group compared to the control at 1 day. Nonetheless, compared with other groups, the cell vitality in Gel/L@FeTA had significantly increased at 3 days and 5 days (p < 0.001). By comparison, the proliferation rate of cells in Gel/L was not faster than in Gel/L@FeTA, even though Gel/L still showed a significant potential for promoting cell proliferation (p < 0.001). The same results were also reflected in HUVECs (Fig. S14, ESI†), in which the cells were related to angiogenesis. And the proliferation, migration, and arrangement of HUVECs had a positive effect on vascularization during the process of wound healing.43 Overall, all of these hydrogels exhibited high biocompatibility, and Gel/L@FeTA showed the optimal performance to promote cell proliferation.
Furthermore, the cell scratch model was employed in this study to evaluate the intuitive cell migration effect of hydrogels in vitro. For L929 cells (Fig. 2F and G), the closed wound rate of Gel/L@FeTA exceeded 50% at 12 h. A large number of living cells densely spread in the scratch was observed at 24 h and the highest wound-healing rate (82.6 ± 3.26%) was achieved (p < 0.001). Regarding the performance of Gel/L, no significant differences in wound closure were observed compared with the gel group at 12 h (p > 0.05). Similar results were also shown in HUVECs (Fig. 2H and I), where the closed wound rate of Gel/L@FeTA reached 46.3 ± 3.29% in 12 h and 72.3 ± 2.87% in 24 h, respectively, which had a significant promotion effect compared to Gel/L (p < 0.01 for 12 h, and p < 0.01 for 24 h). However, the cell migration effect had no clear difference between the Gel/L and Gel both in two time points (p > 0.05). Hence, Gel/L@FeTA exhibited a higher capacity to promote cell migration compared with other samples.
These results indicate that the hydrogel loaded with probiotics showed high biocompatibility to promote cell proliferation and migration, which are related to wound tissue repair and healing. In particular, the Gel/L@FeTA loaded with probiotics exhibited optimal performance in vitro, which may be attributed to the effective barriers that protected L. reuteri@FeTA to avoid the lethality of antibiotics in the culture medium. Previous studies34 have shown that the appropriate probiotic loading in a hydrogel had a positive effect on the cells, but it should also be noted that the excessive addition of probiotics can cause negative effects on the cells and affect their normal biological behavior because of the excessive secretion of active substances, bacterial auto-immunogenicity, and large-scale bacterial proliferation. Therefore, a suitable concentration of probiotics and an effective protection mechanism resulted in the high antibacterial effects, inflammatory regulation, and biocompatibility of Gel/L@FeTA.
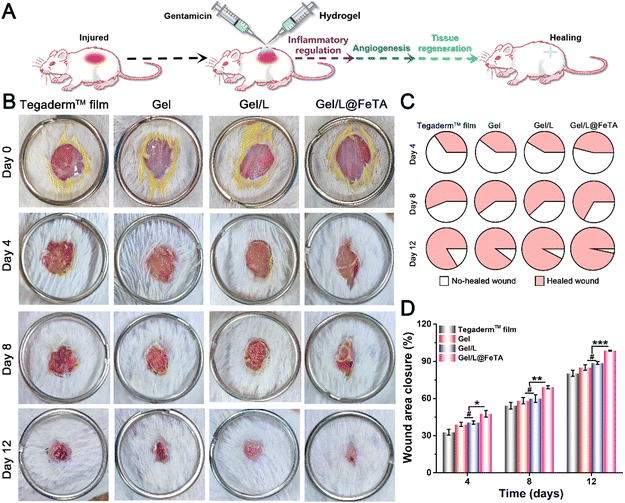
In vivo wound healing ability of the hydrogels
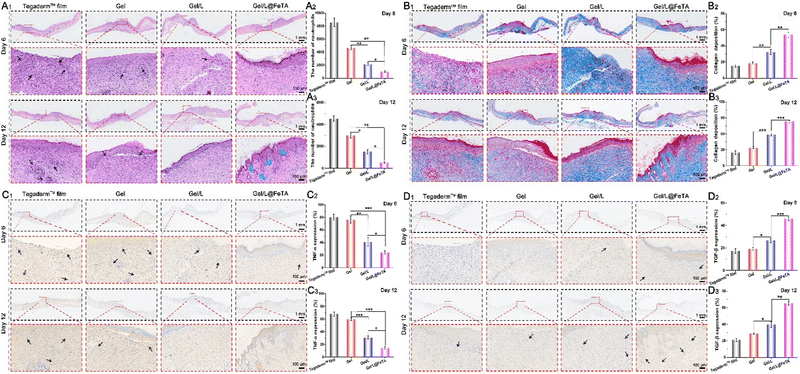
As a common medicine applied in wound management, antibiotics are widely used to prevent infection and purulent discharge in some injuries, such as extensive burns, diabetic ulcers, and postoperative wounds.44 However, treatment with antibiotics also leads to side effects, especially the loss of the normal microbiota in the human body. At present, the therapeutic efficacy of using exogenous probiotics in therapy should be considered with caution. Therefore, a full-thickness round skin wound model was employed to evaluate the healing-promoting properties of hydrogels during antibiotic therapy in vivo (Fig. 3A). The images of the wound site at a fixed time (Fig. 3B) revealed that the Gel/L@FeTA group exhibited the fastest healing speed and the highest healing quality compared with other samples. Gel/L@FeTA also performed well to promote wound repair (Fig. 3C and D), and the closure rate reached 98% at 12 days, which was significantly higher than other groups (p < 0.001). Although the Gel/L was also loaded with probiotics under the same conditions as Gel/L@FeTA, the appearance and closure rate of the wound site did not show advantages compared with the gel during the treatment (p > 0.05). It is worth noting that the weak healing effects of L. reuteri@FeTA without Gel might be due to immune system clearance,15 despite it being better than that of L. reuteri treatment (Fig. S15 and S16, ESI†). Therefore, using probiotics alone for wound treatment might not achieve a satisfactory therapeutic effect.45The same result was also reflected in the appearance and analysis of the tissue in HE staining. In addition, more hair follicles and thicker neo-granulation tissue were observed in the Gel/L@FeTA group both at 6 days and 12 days (Fig. 4A and Fig. S17, and Table S1 and S3, ESI†). By comparison, fewer neutrophils were observed in the Gel/L@FeTA groups, and their number gradually decreased during the healing process. Nonetheless, the expression of collagen deposition of Gel/L@FeTA in Masson's trichrome staining (Fig. 4B) was significantly higher than that of the Gel/L both at 6 days (p < 0.01) and 12 days (p < 0.001). In addition, Gel/L@FeTA also played a role in immunomodulation to significantly inhibit the expression of TNF-α and increase the concentration of TGF-β compared to Gel/L (Fig. 4C and D). These good regulatory functions of Gel/L@FeTA were beneficial to promote wound healing. Although Gel/L showed significant facilitation in tissue regeneration, collagen deposition and immunomodulation compared with Gel, the efficacy is still obviously interfered with by antibiotics, opposite to Gel/L@FeTA. In addition, the concentrations of CD31 and VEGF (two proteins related to angiogenesis),46,47 in the wound site were evaluated by western blot assays. As shown in Fig. 5A–C, the wound tissue treated with Gel/L@FeTA exhibited a higher expression of CD31 and VEGF both at 6 days and 12 days, in sharp contrast to the others. On the other hand, whether the hydrogel was loaded with pure probiotics or not, the expression of CD31 and VEGF exhibited no clear differences in the wound site, in terms of the intervention with antibiotics (p > 0.05).
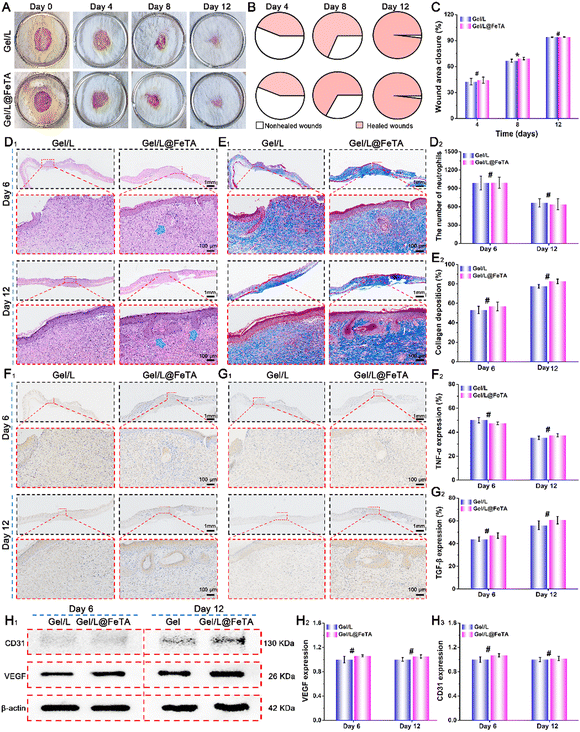
To further investigate the differences in the properties of Gel/L and Gel/L@FeTA in vivo, the above measures were repeated once more without the application of antibiotics. As shown in Fig. 6A–C, near-uniform healing states were observed in these two groups, and the closure rate reached a high level at 12 days for both (93.8% for Gel/L and 94.1% for Gel/L@FeTA, p > 0.05). Moreover, no obvious differences were observed in the histology and histochemistry of these two groups (Fig. 6D–I and Fig. S18, and Table S2, S4, ESI†), including the size of the wound tissue, collagen deposition, and expression of biotic factors (TNF-α, TGF-β, CD31, and VEGF). These results suggest that Gel/L and Gel/L@FeTA had the same functional effect and quality to accelerate wound healing during the treatment period, without the use of antibiotics.
The in vivo results confirmed that the hydrogel loaded with L. reuteri improved the quality of wound healing through collagen deposition, immune regulation, and vascular regeneration. The antibiotics commonly used in clinical treatment often affect the activity of probiotics. Thus, the biological properties of probiotics in the hydrogel are greatly reduced, and their efficacy also becomes unsatisfactory. For instance, the performance of Gel/L was lost once antibiotics were injected. By comparison, because of the protective enclosure, the bioactivity of L. reuteri remained stable in Gel/L@FeTA and thus played a role to accelerate wound healing and facilitate a series of beneficial processes, even in the presence of antibiotics. These results are consistent with the characterization in vitro.
Conclusions
In conclusion, Gel/L@FeTA hydrogels were successfully designed and prepared in this study. Through layer-by-layer self-assembly, L. reuteri@FeTA was complexed into the precursor of OHA and CCS by gelation with a Schiff base. These biocompatible hydrogels exhibited high water absorption, degradation, and mechanical performance, and improved the survival of L. reuteri even in the presence of antibiotics. Thus, the hydrogel loaded with encapsulated probiotics boosted immune regulation, angiogenesis, and other beneficial processes for wound healing in vitro. The in vivo experiment confirmed that L@FeTA hydrogels effectively promoted collagen deposition around the wound, regulated inflammatory factors, and promoted the expression of angiogenesis-related proteins to accelerate wound healing in comparison with Gel/L hydrogels during treatment with gentamicin. Therefore, this work provides a simple strategy for enhancing the therapeutic effect of probiotic-based biomaterials in tissue repair.Materials and methods
The detailed materials and methods are presented in the supporting information.Author contributions
All authors contributed to this work. Chen Zhou: conceptualization, investigation, writing – original draft, formal analysis. Yaping Zou: investigation, writing – review & editing. Ruiling Xu: software, investigation. Xiaowen Han: software, investigation. Zhen Xiang: investigation. Hao Guo: investigation. Xing Li: investigation. Jie Liang: validation, resources. Xingdong Zhang: funding acquisition, project administration, writing – review & editing. Yujiang Fan: funding acquisition, project administration, writing – review & editing. Yong Sun: funding acquisition, project administration, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the National Key R&D Project of China (No. 2022YFC2401800) and the National Natural Science Foundation of China (No. 32071352 and 32271419).Notes and references
- C. Bárcena, R. Valdés-Mas, P. Mayoral, C. Garabaya, S. Durand, F. Rodriguez, M. T. Fernandez-Garcia, N. Salazar, A. M. Nogacka, N. Garatachea, N. Bossut, F. Aprahamian, A. Lucia, G. Kroemer, J. M. P. Freije, P. M. Quiros and C. López-Otín, Nat. Med., 2019, 25, 1234–1242 CrossRef PubMed.
- P. Piewngam, Y. Zheng, T. H. Nguyen, S. W. Dickey, H. S. Joo, A. E. Villaruz, K. A. Glose, E. L. Fisher, R. L. Hunt, B. Li, J. Chiou, S. Pharkjaksu, S. Khongthong, G. Y. C. Cheung, P. Kiratisin and M. Otto, Nature, 2018, 562, 532–537 CrossRef CAS PubMed.
- N. Kamada, G. Y. Chen, N. Inohara and G. Nunez, Nat. Immunol., 2013, 14, 685–690 CrossRef CAS PubMed.
- B. Álvarez and L. Á. Fernández, Microb. Biotechnol., 2017, 10, 1057–1061 CrossRef PubMed.
- W. Hou, J. Li, Z. Cao, S. Lin, C. Pan, Y. Pang and J. Liu, Small, 2021, 17, e2101810 CrossRef PubMed.
- S. Messaoudi, M. Manai, G. Kergourlay, H. Prevost, N. Connil, J. M. Chobert and X. Dousset, Food Microbiol., 2013, 36, 296–304 CrossRef CAS PubMed.
- Y. Lu, H. Li, J. Wang, M. Yao, Y. Peng, T. Liu, Z. Li, G. Luo and J. Deng, Adv. Funct. Mater., 2021, 31, 2105749 CrossRef CAS.
- J. Lukic, V. Chen, I. Strahinic, J. Begovic, H. Lev-Tov, S. C. Davis, M. Tomic-Canic and I. Pastar, Wound Repair Regen., 2017, 25, 912–922 CrossRef PubMed.
- W. Mohammedsaeed, S. Cruickshank, A. J. McBain and C. A. O'Neill, Sci. Rep., 2015, 5, 16147 CrossRef CAS PubMed.
- R. I. Jari Litany and P. K. Praseetha, J. Controlled Release, 2022, 349, 443–459 CrossRef CAS PubMed.
- M. Muhrbeck, A. Wladis, M. Lampi, P. Andersson and J. P. E. Junker, Injury, 2022, 53, 381–392 CrossRef PubMed.
- M. A. S. Buijs, J. van den Kieboom, J. Sliepen, K. L. H. Wever, J. M. van Breugel, F. Hietbrink, F. F. A. IJpma and G. A. M. Govaert, Injury, 2022, 53, 3930–3937 CrossRef CAS PubMed.
- M. Mobayen, M. Hedayati Ch, M. J. Ghazanfari, M. Sadeghi, S. S. Mirmasoudi, A. Feizkhah, M. Mobayen, P. Bagheri Toolaroud and S. Karkhah, Burns, 2022, 48, 730–731 CrossRef PubMed.
- S. Aghamohammad and M. Rohani, Microbiol. Res., 2023, 267, 127275 CrossRef PubMed.
- Z. Ming, L. Han, M. Bao, H. Zhu, S. Qiang, S. Xue and W. Liu, Adv. Sci., 2021, 8, e2102545 CrossRef PubMed.
- R. Wang, K. Guo, W. zhang, Y. He, K. Yang, Q. Chen, L. Yang, Z. Di, J. Qiu, P. Lei, Y. Gu, Z. Luo, X. Xu, Z. Xu, X. Feng, S. Li, Z. Yu and H. Xu, Adv. Funct. Mater., 2022, 32, 2113034 CrossRef CAS.
- S. Han, Q. Wu, J. Zhu, J. Zhang, A. Chen, S. Su, J. Liu, J. Huang, X. Yang and L. Guan, Mater. Horiz., 2023, 10, 1012–1019 RSC.
- L. Wu, Y. Chen, G. Zeng, N. Mao, N. Li, L. Li, X. Xu and L. Yan, Chem. Eng. J., 2023, 457, 141244 CrossRef CAS.
- T. Xie, J. Ding, X. Han, H. Jia, Y. Yang, S. Liang, W. Wang, W. Liu and W. Wang, Mater. Horiz., 2020, 7, 605–614 RSC.
- H. Ejima, J. J. Richardson and F. Caruso, Nano Today, 2017, 12, 136–148 CrossRef CAS.
- G. Fan, P. Wasuwanich, M. R. Rodriguez-Otero and A. L. Furst, J. Am. Chem. Soc., 2022, 144, 2438–2443 CrossRef CAS PubMed.
- Z. Zhao, D. C. Pan, Q. M. Qi, J. Kim, N. Kapate, T. Sun, C. W. T. Shields, L. L. Wang, D. Wu, C. J. Kwon, W. He, J. Guo and S. Mitragotri, Adv. Mater., 2020, 32, e2003492 CrossRef PubMed.
- J. Ma, Z. Zhou, K. Li, K. Li, L. Liu, W. Zhang, J. Xu, X. Tu, L. Du and H. Zhang, Food Chem., 2021, 354, 129510 CrossRef CAS PubMed.
- J. Guo, Y. Ping, H. Ejima, K. Alt, M. Meissner, J. J. Richardson, Y. Yan, K. Peter, D. von Elverfeldt, C. E. Hagemeyer and F. Caruso, Angew. Chem., Int. Ed., 2014, 53, 5546–5551 CrossRef CAS PubMed.
- H. Ejima, J. J. Richardson, K. Liang, J. P. Best, M. P. van Koeverden, G. K. Such, J. Cui and F. Caruso, Science, 2013, 341, 154–157 CrossRef CAS PubMed.
- J. Pan, G. Gong, Q. Wang, J. Shang, Y. He, C. Catania, D. Birnbaum, Y. Li, Z. Jia, Y. Zhang, N. S. Joshi and J. Guo, Nat. Commun., 2022, 13, 2117 CrossRef CAS PubMed.
- Z. Ming, L. Han, M. Bao, H. Zhu, S. Qiang, S. Xue and W. Liu, Adv. Sci., 2021, 8, 2102545 CrossRef CAS PubMed.
- R. Francavilla, L. Polimeno, A. Demichina, G. Maurogiovanni, B. Principi, G. Scaccianoce, E. Ierardi, F. Russo, G. Riezzo, A. Di Leo, L. Cavallo, A. Francavilla and J. Versalovic, J. Clin. Gastroenterol., 2014, 48, 407–413 CrossRef PubMed.
- F. Meng, F. Zhang, Q. Chen, M. Yang, Y. Yang, X. Li, W. Gu and J. Yu, Biomed. Pharmacother., 2022, 147, 112521 CrossRef CAS PubMed.
- C. Zhou, R. Xu, X. Han, L. Tong, L. Xiong, J. Liang, Y. Sun, X. Zhang and Y. Fan, Composites, Part B, 2023, 250, 110451 CrossRef CAS.
- X. Yi, Z. Xu, Q. Liu, H. Zhou, L. Yuan, D. Li, L. Zhao, C. Mu and L. Ge, Biomater. Adv., 2022, 137, 212804 CrossRef CAS PubMed.
- J. Qu, X. Zhao, Y. Liang, Y. Xu, P. X. Ma and B. Guo, Chem. Eng. J., 2019, 362, 548–560 CrossRef CAS.
- X. Yang, J. Yang, Z. Ye, G. Zhang, W. Nie, H. Cheng, M. Peng, K. Zhang, J. Liu, Z. Zhang and J. Shi, ACS Nano, 2022, 16, 4041–4058 CrossRef CAS PubMed.
- L. Mei, D. Zhang, H. Shao, Y. Hao, T. Zhang, W. Zheng, Y. Ji, P. Ling, Y. Lu and Q. Zhou, ACS Appl. Mater. Interfaces, 2022, 14, 20538–20550 CrossRef CAS PubMed.
- P. Ding, Z. Wang, Z. Wu, M. Hu, W. Zhu, N. Sun and R. Pei, ACS Appl. Mater. Interfaces, 2021, 13, 3694–3700 CrossRef CAS PubMed.
- F. M. Davis and K. Gallagher, Cell Host Microbe, 2018, 23, 432–434 CrossRef CAS PubMed.
- W. Liu, Y. Wang, L. H. M. Bozi, P. Fischer, M. P. Jedrychowski, H. Xiao, T. Wu, N. Darabedian, X. He, E. L. Mills, N. Burger, S. Shin, A. Reddy, H. G. Sprenger, N. Tran, S. Winther, S. M. Hinshaw, J. Shen, H. S. Seo, K. Song, A. Z. Xu, L. Sebastian, J. Zhao, S. Dhe-Paganon, J. Che, S. P. Gygi, H. Arthanari and E. T. Chouchani, Nature, 2023, 616, 790–797 CrossRef CAS PubMed.
- C. Wang, X. Jiang, H. J. Kim, S. Zhang, X. Zhou, Y. Chen, H. Ling, Y. Xue, Z. Chen, M. Qu, L. Ren, J. Zhu, A. Libanori, Y. Zhu, H. Kang, S. Ahadian, M. R. Dokmeci, P. Servati, X. He, Z. Gu, W. Sun and A. Khademhosseini, Biomaterials, 2022, 285, 121479 CrossRef CAS PubMed.
- C. Zhou, Z. Yang, X. Xun, L. Ma, Z. Chen, X. Hu, X. Wu, Y. Wan and H. Ao, Bioact Mater, 2022, 13, 212–222 CrossRef CAS PubMed.
- L. Yang, Z. Han, C. Chen, Z. Li, S. Yu, Y. Qu and R. Zeng, Mater. Sci. Eng., C, 2020, 117, 111265 CrossRef CAS PubMed.
- J. Wang, H. Cheng, W. Chen, P. Han, X. Yao, B. Tang, W. Duan, P. Li, X. Wei, P. K. Chu and X. Zhang, Chem. Eng. J., 2023, 452, 139474 CrossRef CAS.
- G. Theocharidis, S. Rahmani, S. Lee, Z. Li, A. Lobao, K. Kounas, X. L. Katopodi, P. Wang, S. Moon, I. S. Vlachos, M. Niewczas, D. Mooney and A. Veves, Biomaterials, 2022, 288, 121692 CrossRef CAS PubMed.
- C. Wu, L. Long, Y. Zhang, Y. Xu, Y. Lu, Z. Yang, Y. Guo, J. Zhang, X. Hu and Y. Wang, J. Controlled Release, 2022, 344, 249–260 CrossRef CAS PubMed.
- G. H. R. Vale de Macedo, V. L. Chagas, M. H. Cruz dos Santos, G. D. Costa dos Santos, J. M. N. Bazán, A. Zagmignan, E. M. Carvalho, R. D. C. Mendonça de Miranda, C. S. Teixeira and L. C. Nascimento da Silva, Biochem. Eng. J., 2022, 187, 108664 CrossRef CAS.
- C. Togo, A. P. Zidorio, V. Gonçalves, P. Botelho, K. de Carvalho and E. Dutra, Nutrients, 2021, 14, 111 CrossRef PubMed.
- J. He, Y. Liang, M. Shi and B. Guo, Chem. Eng. J., 2020, 385, 123464 CrossRef CAS.
- Z. Yang, H. Chen, P. Yang, X. Shen, Y. Hu, Y. Cheng, H. Yao and Z. Zhang, Biomaterials, 2022, 282, 121401 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3mh00033h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |