Amplification-free detection of Mycobacterium tuberculosis using CRISPR-Cas12a and graphene field-effect transistors†
Weiqi
Wang‡
ab,
Huanyu
Du‡
ab,
Changhao
Dai
ab,
Hongwenjie
Ma
ab,
Shi
Luo
ab,
Xuejun
Wang
 ab,
Mingquan
Guo
*c,
Derong
Kong
*ab and
Dacheng
Wei
ab,
Mingquan
Guo
*c,
Derong
Kong
*ab and
Dacheng
Wei
 *ab
*ab
aState Key Laboratory of Molecular Engineering of Polymers, Department of Macromolecular Science, Fudan University, Shanghai 200433, China. E-mail: weidc@fudan.edu.cn; drkong@fudan.edu.cn
bInstitute of Molecular Materials and Devices, Fudan University, Shanghai 200433, China
cDepartment of Laboratory Medicine, Shanghai Public Health Clinical Center, Fudan University, Shanghai 201508, China. E-mail: gmqjiandan@163.com
First published on 11th January 2025
Abstract
Current molecular tests for tuberculosis (TB), such as whole genome sequencing and Xpert Mycobacterium tuberculosis/rifampicin resistance assay, exhibit limited sensitivity and necessitate the pre-amplification step of target DNA. This limitation greatly increases detection time and poses an increased risk of infection. Here, we present a graphene field-effect transistor (GFET) based on the CRISPR/Cas system for detecting Mycobacterium tuberculosis. The CRISPR/Cas12a system has the ability to specifically recognize and cleave target DNA. By integrating the system onto the FET platform and utilizing its electrical amplification capability, we achieve rapid and sensitive detection without requiring sample pre-amplification, with a limit of detection (LoD) as low as 2.42 × 10−18 M. Cas12a-GFET devices can differentiate 30 positive cases from 56 serum samples within 5 minutes. These findings highlight its immense potential in future biological analysis and clinical diagnosis.
1. Introduction
Tuberculosis (TB), caused by the bacterium Mycobacterium tuberculosis (MTB), is a chronic contagious disease, affecting mainly the human respiratory system.1 In 2023, approximately 7.5 million patients were newly diagnosed worldwide, and it is the highest figure since monitoring began.2 The challenge of curbing the high mortality rate of tuberculosis requires early, rapid and accurate detection. Currently, positive culture of MTB is the gold standard for diagnosis,3 but it requires biosafety laboratory infrastructure and well-trained personnel, making it time-consuming and costly. Other commonly used diagnostic methods for MTB include acid-fast bacilli (AFB) smear microscopy,4 molecular biology techniques (e.g., Xpert MTB/RIF),5 and immunologic methods (e.g., interferon gamma (IFN-γ) release assay (IGRAs)).6,7 Although sputum AFB smear is widely used for TB diagnosis, it has low sensitivity and specificity. Xpert MTB/RIF, a polymerase chain reaction (PCR)-based assay, simultaneously detects MTB and rifampicin resistance, but it is complicated to perform and has high equipment costs. IGRAs can detect IFN-γ release in response to MTB-specific antigens, but it is time-consuming (12–18 h).8 Therefore, there is an urgent need to develop a rapid diagnosis method for MTB that is sensitive, specific and easy to perform, so that it can be effectively deployed in areas with limited medical resources.Field effect transistors (FETs) can be used as a promising detection platform due to their advantages such as no labeling and fast response.9–11 They detect trace biological analytes by monitoring changes in the conductivity of semiconductor channels, which originate from chemical or physical perturbations of specific interactions between the recognition element and the analyte.12,13 At present, FETs are widely used in the detection of RNA,14 DNA,15 lactose,16 hydrogen peroxide,17 and acetylcholine.18 However, the application of FET biosensors in nucleic acid detection of MTB has not been reported to date. Due to the stable double-stranded DNA structure of the MTB nucleic acid, conventional probes are unable to directly detect DNA targets without undergoing a necessary pre-treatment process.19 To solve these problems, Kiana Aran's team introduced clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) 9 into a GFET for the first time. Clinical DNA samples from patients with Duchenne muscular dystrophy can be detected within 15 minutes, with a limit of detection (LoD) of 1.7 × 10−15 M.20 Most clinical applications, however, require the detection of nucleic acid concentrations down to the attomolar level, and the limited sensitivity does not meet the high standard of nucleic acid samples for infectious disease diagnostics.21 Combining nucleic acid amplification techniques (such as RT-qPCR, RT-RPA, and HCR) with CRISPR-Cas technology facilitates the highly specific detection of extremely low concentrations of target nucleic acids.22–25 However, the amplification process can extend the detection time. A sensitive detection platform for unamplified samples can reduce the detection time and minimize errors introduced by the amplification process.26,27
In this study, we combine the amplification-free CRISPR/Cas12a system with a GFET. The unamplified nucleic acid detection method is a promising nucleic acid detection technology, which simplifies the detection process, shortens the detection time, and may reduce the detection cost.28–30 Cas12a exhibits a smaller size compared to Cas9 and demonstrates a higher modification density on the graphene surface, thereby offering promising prospects for reducing detection limits and shortening detection time in biological assays.31 When the target DNA is added, CRISPR RNA (crRNA) that hybridizes to a complementary sequence of a target DNA or RNA guides the Cas12a protein to recognize nucleic acids with high specificity, thus promoting the application of CRISPR/Cas systems in molecular diagnosis.32 The GFET can enhance compromised signals of the CRISPR/Cas detection system and relieve it from dependence on expensive fluorescence-based instruments.33,34 Meanwhile the CRISPR/Cas system is capable of supplementing the limited specificity of GFET sensors.35 This platform is able to effectively distinguish the target DNA sequence from human DNA, the Escherichia coli gene and the Streptococcus agalactiae sip gene. With a limit of detection of 2.42 × 10−18 M, our device can accurately differentiate DNA extracted from serum samples of TB patients and healthy individuals within 5 minutes with 98.6% accuracy. This strategy enables rapid molecular assays to be a widely applicable tool for fast, cheap, and precise detection of low-concentration presumptive MTB samples, which holds great promise in high-quality screening of tuberculosis worldwide.
2. Methods
2.1. Fabrication of the GFET biosensors
The preparation of the device generally has the following steps: electrode mask lithography and gold plating, graphene transfer and patterning (Fig. S1†). The source electrode and drain electrode of the device are first lithographically patterned on a silicon wafer with 300 nm thermal silicon oxide (SiO2/Si), and then Cr/Au is adhered to the wafer by means of metal evaporation (Fig. S2†). As the adhesive layer, the thickness of chromium is 5 nm, and 38 nm gold is deposited on the surface of chromium metal. Graphene is synthesized on copper foil by vapor deposition and is directly removed from the copper foil by the electrochemical bubbling method and transferred to the connected circuit between the source and drain. Then, we pattern the graphene by standard photolithography and plasma etching (ZEPTO RIE) to define the sensor area (W × L: 30 × 100 μm2). An Ag/AgCl reference electrode is used to apply gate pressure to the solution in the PDMS well. Finally, the device is connected to the PCB through wire bonding, and the external semiconductor tester is connected to test and analyze the target.2.2. Cas12a modification
After the transfer and patterning of graphene, the device is immersed in a 5 mM PASE solution overnight. It is then washed with ethanol and water before placing the PDMS well above the sensor channel. The crRNA sequence is purchased from BiOligo Biotechnology and sequenced (Table S1†). Cas12a (purchased from the company) is stored at −20 °C. The Cas12a protein is diluted to a concentration of 5 nM using 1× NE buffer, while crRNA is diluted to a concentration of 2 nM using TE buffer (BiOligo Biotechnology). Cas12a (5 nM, 35 μL) and crRNA (2 nM, 50 μL) are mixed and incubated at 37 °C for 30 minutes in a polymerase chain reaction (PCR) thermal cycler. After complete binding between Cas12a and crRNA, the mixture is added to the PDMS well and kept at room temperature for at least 4 hours to allow fixation of the Cas12a-crRNA RNP complex on the graphene surface.2.3. Characterization
Raman spectrometers (LabRam HR Evolution, Horiba Jobin Yvon, 532 nm Ar ion laser) are used for Raman spectroscopy. X-ray photoelectron spectroscopy (XPS) analysis is carried out on a Thermo Scientific K-Alpha+ instrument, and the incident beam is produced by Al-X-rays with an energy of 1486.6 eV. The air morphology of GFET devices is analyzed before and after Cas12a modification using an atomic force microscope (AFM) in tapping mode with a tip radius of 20–25 nm.2.4. Device measurement
Electrical measurements are made using a B1500A semiconductor analyzer (Santa Clara, California B1500A semiconductor analyzer, USA). The test solution is stored in the PDMS well and the Ag/AgCl electrode is inserted into the solution to apply a gate pressure. The drain–source voltage (Vds) is maintained at 50 mV, and a gate voltage from −400 mV to 400 mV is applied, resulting in the Ids–Vgs (drain–source current versus gate voltage) curve. Before the test, 1× NE buffer (100 μL) is added into the PDMS well and then stored for 5 minutes. After the measured stable transfer curve is obtained (the Dirac point is basically unchanged), 10 μL solution is extracted and an equal amount of solution to be measured is added. The Ids–Vgs curve is obtained after incubation for 15 min. For the measurement of real-time Ids, a voltage of 50 mV is applied between the source and the drain, and Ids is read in real time. After obtaining a stable current, the sample is added to read the channel current change.2.5. Preparation of test samples
Synthetic DNA samples, including the specific sequences Is1081, 16S rDNA, and sipDNA, are obtained from BiOligo Biotechnology. Prior to detection, the samples are centrifuged at 4 °C (4000 rpm, 5 min) and dissolved in 1× NE buffer (BiOligo), followed by gradient dilution to the target concentration. Nanodrop (NanoDrop Lite Plus, Thermo Fisher) is used to test the concentration of the gradient dilution sample. The test starts when the theoretical calibration value matches the test value.2.6. Clinical samples
Serum samples are collected from 30 patients with TB or latent TB and 26 healthy people from the Shanghai Public Health Clinical Center (Table S3†). The MagMAX™ DNA Multi-Copy Ultra 2.0 kit (Thermo Fisher) is used to achieve high throughput separation of DNA from serum. Protease K is added to the serum sample and heated at 65 °C for 20 minutes to destroy the bacterial structure and release DNA. The DNA binding bead mixture consists of a binding solution and DNA binding beads, which are introduced into treated serum samples for the purpose of adsorbing the DNA released through cleavage. The mixture is shaken at a speed of 800 revolutions per minute (rpm) for 5 minutes and allowed to settle, and magnets are employed to facilitate the precipitation of magnetic beads, followed by decanting the supernatant. The magnetic beads are subsequently subjected to multiple rinses with a cleaning solution, followed by the elution of DNA from the beads and its subsequent storage in the DNA eluent. The study has received ethical approval by the Ethics Committee of Shanghai Public Health Clinical Center (approval ID: #2021-S057-03).3. Results and discussion
3.1. Cas12a-GFET biosensor
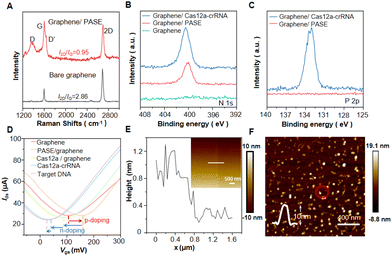
The configuration of the Cas12a-GFET for the detection of MTB is shown in Fig. 1A. The sensor is a liquid-gated FET, and graphene prepared by chemical vapor deposition is attached to the Au electrode to connect the source and drain. A polydimethylsiloxane (PDMS) chamber is placed on the sensing channel and an Ag/AgCl reference electrode is inserted into the solution in the chamber to provide the gate voltage. The sensing area is designed to be 100 × 30 μm2 (W × L). Finally, the device is connected to the printed circuit board (PCB) by wire-bonding (Fig. S3†), and an external semiconductor analyzer is connected to test the target.The Cas12a protein is immobilized onto the GFET through molecule linkers: 1-pyrenebutanoic acid succinimidyl ester (PASE). The aromatic pyrene group at one end of PASE is π–π stacked with the graphene surface, and the other end is N-hydroxysuccinimide (NHS), which can bind to amine groups present in biomolecules (Fig. S4†).36,37 Cas12a anchored on the graphene recognizes the hairpin structure of crRNA to form a functional ribonucleoprotein (RNP).38 Cas12a subsequently adopts a conformation that is initiated for recognition of target DNA, and the seed sequence of the bound crRNA is pre-sequenced to mediate base pairing with the target DNA strand. When MTB DNA is added, Cas12a-crRNA RNP specifically identifies the protospacer-adjacent motif (PAM) sequences, repositioning the target DNA sequence in alignment with the crRNA and facilitating localized strand separation of the DNA in close proximity to the PAM. This allows the target DNA strand to be captured by the pre-ordered seed sequence of the crRNA, resulting in crRNA-target DNA hybridization.39,40 In contrast, the non-target DNA remains unrecognized (Fig. 1B). The transfer curve of a GFET device is obtained by sweeping the gate–source voltage (Vgs) at a constant drain voltage, and the Dirac point (VDirac) can be read in the transfer curve.41VDirac is the grid voltage corresponding to the minimum current where the electron transport is almost equal to the hole transport, at which point the graphene is charge neutral. After the GFET identifies the negatively charged analyte, the charge is transferred to the graphene and n-doping occurs. Consequently, the Fermi level of graphene changes, and VDirac shifts towards a negative potential (Fig. S5†). Upon introduction of MTB DNA into the sensing system, the Cas12a protein specifically recognizes and captures the DNA, causing the shift of VDirac (ΔVDirac). Therefore, the presence of target DNA can be detected in external semiconductor devices.
3.2. Modification and characterization of Cas12a-GFET
To verify the successful modification of PASE, the Raman spectra of graphene are tested before and after PASE modification (Fig. 2A). Raman spectroscopy of pristine graphene exhibits two distinct peaks, namely the 2D peak at ∼2683 cm−1 and the G peak at ∼1598 cm−1. The 2D peak represents the vibration mode of the two photonic lattices, while the G peak corresponds to the vibration mode of the sp2 hybrid carbon atoms in graphene.42,43 The intensity ratio of the 2D peak to the G peak (I2D/IG) is ∼2.86, indicating the high quality of pristine graphene. After PASE modification, the I2D/IG decreases to 0.95, indicating the presence of some disordered states on graphene.44,45 D peaks (∼1367 cm−1) and D′ peaks (∼1632 cm−1) emerge due to sp3 bonding, orbital hybridization, and relative resonance.Compared with the pristine graphene, X-ray photoelectron spectroscopy (XPS) of modified graphene shows a more pronounced N 1s peak, further verifying the modification of PASE (Fig. 2B). Due to the presence of phosphate groups within the RNA skeleton, graphene functionalized with Cas-crRNA RNP demonstrates an additional P 2p peak at a binding energy of ∼133 eV (Fig. 2C). In addition, the functionalization of graphene is monitored through transfer curves after each modification step (Fig. 2D). The positive shift of VDirac is caused by p-doping after the modification of PASE. When the Cas12a protein is anchored on the graphene, VDirac shifts from 202 to 162 mV, indicating that the strong negative charge of the protein causes an n-doping effect on graphene.46 The change of VDirac following the binding of Cas12a-crRNA and target DNA can also be attributed to this reason. The atomic force microscopy (AFM) characterization shows the pristine graphene with an average height of 0.6 nm (Fig. 2E). The Cas12a protein with a height of about 10 nm is uniformly distributed on the graphene surface (Fig. 2F).
3.3. Detection performance of MTB
As a specific DNA sequence of Mycobacterium tuberculosis, the presence of MTB can be accurately determined by detecting Is1081.47 When the Is1081 concentration in the NE buffer increases from 6 × 10−20 M to 6 × 10−15 M, the Dirac point shows a continuous negative shift (Fig. 3A), which is attributed to non-electrostatic stacking and doping effects between graphene and target DNA.48 Cas12a-GFET produces a |ΔVDirac| response of up to ∼30 mV upon addition of 6 × 10−15 M Is1081, while the response of a bare GFET is negligible (|ΔVDirac| = 3.13 mV). The |ΔVDirac| response value of Cas12a-GFET is linearly correlated with the concentration of Is1081 (Fig. 3B). The regression equation is determined to be |ΔVDirac| = 2.445![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ctarget + 52.46 with a correlation coefficient value (R2) of 0.972, where Ctarget refers to the Is1081 concentration. The limit of detection, defined as the concentration at which |ΔVDirac| reaches 3 times the control result, is calculated to be ∼2.42 × 10−18 M (Fig. S6 and S7†). Upon addition of Is1081 at a concentration of 6 × 10−16 M, |ΔVDirac| gradually increases over a period of 30 minutes as the reaction progresses (Fig. 3C and S8†).
Ctarget + 52.46 with a correlation coefficient value (R2) of 0.972, where Ctarget refers to the Is1081 concentration. The limit of detection, defined as the concentration at which |ΔVDirac| reaches 3 times the control result, is calculated to be ∼2.42 × 10−18 M (Fig. S6 and S7†). Upon addition of Is1081 at a concentration of 6 × 10−16 M, |ΔVDirac| gradually increases over a period of 30 minutes as the reaction progresses (Fig. 3C and S8†).
We also evaluate the dynamic response of the Cas12a-GFET biosensor to Is1081. The real-time current response of the Cas12a-GFET biosensor is measured after successive additions of Is1081 at concentrations ranging from 6 × 10−20 M to 6 × 10−15 M (Fig. 3D). The current response is normalized to ΔI/I0 = (I − I0)/I0, where I is the drain–source current and I0 is the initial value of I. |ΔI/I0| of Cas12a-GFET shows a linear correlation with the Is1081 concentration (Fig. 3E). The |ΔI/I0| response to 6 × 10−15 M Is1081 is approximately 17.3%. Meanwhile, upon the addition of 1× NE buffer to the device modified with Cas12a-crRNA RPN, there is no significant current change (Fig. 3C and S9†).
The cellular structure and dsDNA genetic material of Escherichia coli and Streptococcus agalactiae are similar to those of MTB.49,50 Therefore, the Escherichia coli specific sequence sipDNA and Streptococcus agalactiae specific sequence 16S rDNA are selected as control samples. An appreciable |ΔVDirac| response of up to ∼14.0 mV is observed for Is1081 at a concentration of 6 × 10−16 M, and a negligible |ΔVDirac| response smaller than 4.0 mV for 16S rDNA and sipDNA at a concentration of 6 × 10−15 M (Fig. 3F). This indicates that Cas12a-GFET is able to distinguish MTB nucleic acids from those of other bacteria and has high specificity in MTB nucleic acid detection.
We evaluate the performance of the Cas12a-GFETs at various temperatures by adding the target sample Is1081 at a concentration of 6 × 10−16 M (Fig. S10†). The recovery rate is 80% (Table S2†). As a semi-quantitative detection method, Cas12a-GFET exhibits relatively good reliability and accuracy. The devices demonstrate a robust response to the target DNA at 24 °C and 44 °C, but they fail to detect the target at 4 °C, attributed to protein inactivation at low temperature. In reproducibility tests, different devices show high consistency in |ΔVDirac| response after the addition of Is1081 (Fig. S11†). Moreover, Cas12a-GFETs maintain good detection capabilities after being stored at room temperature for seven days, showcasing their remarkable stability (Fig. S12†).
3.4. Tests of clinical samples
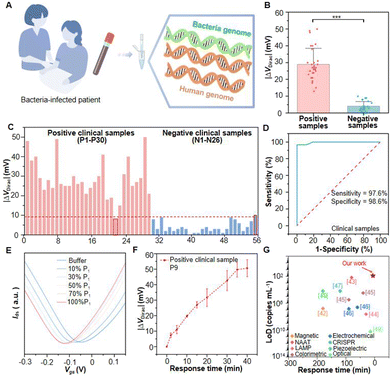
To further validate the practical application of the device in TB detection, DNA is extracted from the serum by the DNA magnetic bead method and added to the device to read the Dirac point offset (Fig. 4A and S13†). We test 30 serum samples (P1–P30) collected from TB patients and 26 samples (N1–N26) from healthy people (Fig. 4B). Cas12a GFETs correctly distinguish patient samples from healthy samples. The |ΔVDirac| signal of positive samples reaches more than 30 mV while a negligible |ΔVDirac| signal of about 10 mV is detected upon the addition of negative clinical DNA samples to the Cas12a-FET (Fig. 4C). Among them, 54 samples show correct results, demonstrating high accuracy. However, the Cas12a-GFETs produce incorrect results when testing samples P22 and N26, potentially attributable to differences between devices or discrepancies in DNA concentrations resulting from sample pretreatment.As can be seen from the receiver operating characteristic (ROC) curve analysis performed on the clinical test results, the AUC (area under the curve) in the curve is 0.995, with a sensitivity of 97.6% and a specificity of 98.6% (Fig. 4D). As an ultrasensitive detection method, the Cas12a-GFET is capable of discriminating the diluted TB serum sample as low as 10% of the pristine sample (Fig. 4E) and reaches a saturated |ΔVDirac| signal at a dilution level of 50%. With increasing diluted concentrations of P1 dilution, VDirac exhibited a significant negative shift, indicating n-type doping effects on graphene. After the introduction of clinical sample P9, Dirac points are measured multiple times within 40 minutes. |ΔVDirac| increases continuously within 40 minutes, which is linearly correlated with time (Fig. 4F). The calculated reaction time is 3.6 minutes (Fig. S14†). In clinical trials, the proposed Cas12a-GFET demonstrates good accuracy and rapid detection capability.
4. Conclusion
In summary, we develop a GFET sensor modified with a CRISPR-Cas12a system that achieves unamplified detection of serum samples from TB patients. Cas12a-crRNA RNP specifically recognizes the PAM sequence and initiates the complementary base pairing of crRNA and target DNA, thereby conferring excellent specificity to Cas12a-GFETs. Utilizing the inherent signal sensitization of graphene transistors, our sensors achieve an ultra-low LoD of ∼2.42 × 10−18 M. Compared with other detection methods, Cas12a-GFET exhibits the shortest detection time and demonstrates good performance in terms of the LoD (Fig. 4G and Table S4†). Notably, the proposed sensors can provide a universal platform for nucleic acid detection, requiring only the design of crRNA corresponding to the target DNA sequence. Cas12a-GFET is a candidate sensor for the rapid and non-invasive detection of various biomarkers by liquid biopsy. Currently, there remain instances of misdiagnosis when using GFETs for the detection of MTB. Subsequent work may integrate multi-channel nucleic acid detection technology into the platform, enabling the simultaneous identification of multiple biomarkers.51 Future research will increasingly concentrate on improving the adaptability of GFETs in complex environments and investigating their potential for reusability to lower operational costs. Furthermore, the sensors can be integrated with microelectronic systems for home testing, alleviating the burden on public healthcare services and mitigating the risk of viral transmission.Data availability
The datasets supporting this article have been uploaded as part of the ESI. Data will be made available on request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the National Natural Science Foundation of China (22304031 and 61890940), National Science Fund for Distinguished Young Scholars (T2425006), Shanghai Natural Science Foundation (24CL2901100), the National Key R&D Program of China (2021YFC2301100), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB30000000), the Program of Shanghai Academic Research Leaders (23XD1420200), the Chongqing Bayu Scholar Program (DP2020036), the China Postdoctoral Science Foundation (2023M730635) and Fudan University.References
- K. C. Rahlwes, B. R. S. Dias, P. C. Campos, S. Alvarez-Arguedas and M. U. Shiloh, Virulence, 2023, 14, 2150449 CrossRef PubMed.
- World Health Organization, Global tuberculosis report, Geneva 2023, Global report, 2023 Search PubMed.
- N. Alonso, N. Griffa, R. D. Moyano, M. L. Mon, M. A. C. Olivieri, S. Barandiaran, M. M. Vivot, G. Fiorini, A. M. Canal, M. P. Santangelo, M. Singh and M. I. Romano, Immunol. Methods, 2021, 491, 112941 CrossRef CAS PubMed.
- J. B. Ma, G. Y. Jiang, Q. Q. Ma, M. M. Du, H. Wang, J. G. Wu, C. Wang, X. W. Xie, T. Li, S. X. Chen, L. X. Zhang and M. Wu, Anal. Methods, 2022, 14, 438–448 RSC.
- H. Zhang, X. Dai, P. Hu, L. Tian, C. Li, B. Ding and X. He, Microbiol. Spectrum, 2024, 12, e04098–e04023 Search PubMed.
- S. Chandrashekara, R. Panchagnula and Y. Chennupati, Rheumatology, 2023, 62, 3952–3956 CrossRef CAS PubMed.
- Y. Yang, H. J. Wang, W. L. Hu, G. N. Bai and C. Z. Hua, Diagnostics, 2022, 12, 453 CrossRef CAS PubMed.
- P. Ogongo, A. Tran, F. Marzan, D. Gingrich, M. Krone, F. Aweeka and J. D. Ernst, Front. Immunol., 2024, 15, 1378040 CrossRef CAS PubMed.
- Y. T. Yang, D. R. Kong, Y. G. Wu, Y. H. Chen, C. Dai, C. Chen, J. H. Zhao, S. Luo, W. T. Liu, Y. Q. Liu and D. C. Wei, Anal. Chem., 2023, 95, 13281–13288 CrossRef CAS PubMed.
- A. Hassibi, A. Manickam, R. Singh, S. Bolouki, R. Sinha, K. B. Jirage, M. W. McDermott, B. Hassibi, H. Vikalo, G. Mazarei, L. Pei, L. Bousse, M. Miller, M. Heshami, M. P. Savage, M. T. Taylor, N. Gamini, N. Wood, P. Mantina, P. Grogan, P. Kuimelis, P. Savalia, S. Conradson, Y. Li, R. B. Meyer, E. Ku, J. Ebert, B. A. Pinsky, G. Dolganov, T. Van, K. A. Johnson, P. Naraghi-Arani, R. G. Kuimelis and G. Schoolnik, Nat. Biotechnol., 2018, 36, 738–745 CrossRef CAS PubMed.
- L. Q. Wang, X. J. Wang, Y. G. Wu, M. Q. Guo, C. J. Gu, C. J. Dai, D. R. Kong, Y. Wang, C. Zhang, D. Qu, C. H. Fan, Y. H. Xie, Z. Q. Zhu, Y. Q. Liu and D. C. Wei, Nat. Biomed. Eng., 2022, 6, 276–285 CrossRef CAS PubMed.
- J. C. Shu, W. Q. Cao and M. S. Cao, Adv. Funct. Mater., 2021, 31, 2100470 CrossRef CAS.
- L. Laxmi, B. Mahapatra, R. V. Krishna and P. K. Patel, AIP Conf. Proc., 2021, 2327, 020011 CrossRef.
- M. Tian, M. Qiao, C. Shen, F. Meng, L. A. Frank, V. V. Krasitskaya, T. Wang, X. Zhang, R. Song, Y. Li, J. Liu, S. Xu and J. Wang, Appl. Surf. Sci., 2020, 527, 1468390 Search PubMed.
- S. Chen, Y. Sun, Y. Xia, K. Lv, B. Man and C. Yang, Biosens. Bioelectron., 2020, 156, 112128 CrossRef CAS PubMed.
- E. Danielson, M. Dindo, A. J. Porkovich, P. Kumar, Z. Wang, P. Jain, T. Mete, Z. Ziadi, R. Kikkeri, P. Laurino and M. Sowwan, Biosens. Bioelectron., 2020, 165, 112419 CrossRef CAS PubMed.
- S. H. Lee, K. H. Kim, S. E. Seo, M. Kim, J. Park and O. S. Kwon, J. Ind. Eng. Chem., 2020, 83, 29–34 CrossRef CAS.
- G. E. Fenoy, W. A. Marmisollé, O. Azzaroni and W. Knoll, Biosens. Bioelectron., 2019, 148, 111796 CrossRef PubMed.
- C. R. Sun, C. H. Wang, F. Xiao, N. Jia, X. L. Huang, J. Fu, Y. Zhang, J. Zhou, G. R. Wang and Y. Wang, Heliyon, 2024, 10, 31901 Search PubMed.
- R. Hajian, S. Balderston, T. Tran, T. DeBoer, J. Etienne, M. Sandhu, N. Wauford, J. Chung, J. Nokes, M. Athaiya, J. Paredes, R. Peytavi, B. Goldsmith, N. Murthy, I. M. Conboy and K. Aran, Nat. Biomed. Eng., 2019, 3, 427–437 CrossRef CAS PubMed.
- R. Arnaout, R. A. Lee, G. R. Lee, C. Callahan, A. Cheng, C. F. Yen, K. P. Smith, R. Arora and J. E. Kirby, Clin. Infect. Dis., 2021, 73, 3042–3046 CrossRef PubMed.
- J. Yang, Z. Xu, L. Yu, B. Wang, R. Hu, J. Tang, J. Lv, H. Xiao, X. Tan, G. Wang, J. X. Li, Y. Liu, P. L. Shao and B. Zhang, Angew. Chem., 2024, 136, 202318800 Search PubMed.
- Y. H. Yang, J. C. Liu and X. H. Zhou, Biosens. Bioelectron., 2021, 190, 113418 CrossRef CAS PubMed.
- Y. H. Yang, F. Wang, B. Y. Xue and X. H. Zhou, J. Hazard. Mater., 2023, 459, 132077 CrossRef CAS PubMed.
- Y. H. Yang, L. L. Sun, J. H. Zhao, Y. Jian, T. L. Han and X. H. Zhou, Biosens. Bioelectron., 2024, 255, 116239 CrossRef CAS PubMed.
- Y. H. Yang, J. S. Tan, F. Wang, W. M. Sun, H. C. Shi, Z. Cheng, Y. C. Xie and X. H. Zhou, Biosens. Bioelectron., 2024, 263, 116617 CrossRef CAS PubMed.
- J. B. Ma, M. M. Du, C. Wang, X. W. Xie, H. Wang, T. Li, S. X. Chen, L. X. Zhang, S. Mao, X. H. Zhou and M. Wu, ACS Sens., 2021, 6, 3367–3376 CrossRef CAS PubMed.
- R. Hu, Y. Liu, G. Wang, J. Lv, J. Yang, Y. Liu and B. Zhang, Anal. Chim. Acta, 2023, 1280, 341870 CrossRef CAS PubMed.
- Y. Liu, Y. Yang, G. Wang, D. Wang, P. L. Shao, J. Tang and B. Zhang, Nat. Biomed. Eng., 2023, 7, 1636–1648 CrossRef CAS PubMed.
- J. Yang, Z. Xu, L. Yu, B. Wang, R. Hu, J. Tang, J. Lv, H. Xiao, X. Tan, G. Wang, J.-X. Li, Y. Liu, P.-L. Shao and B. Zhang, Angew. Chem., Int. Ed., 2024, 63, e202318800 CrossRef CAS PubMed.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Science, 2018, 360, 439–444 CrossRef CAS PubMed.
- H. M. Li, Y. Xie, F. M. Chen, H. W. Bai, L. S. Xiu, X. N. Zhou, X. K. Guo, Q. Q. Hu and K. Yin, Chem. Soc. Rev., 2023, 52, 361–382 RSC.
- F. Teng, L. Guo, T. Cui, X. G. Wang, K. Xu, Q. Q. Gao, Q. Zhou and W. Li, Genome Biol., 2019, 20, 1–7 CrossRef CAS PubMed.
- H. J. Li, J. Yang, G. F. Wu, Z. Y. Weng, Y. Song, Y. X. Zhang, J. A. Vanegas, L. Avery, Z. Gao, H. Sun, Y. P. Chen, K. D. Dieckhaus, X. Gao and Y. Zhang, Angew. Chem., Int. Ed., 2022, 61, e202203826 CrossRef CAS PubMed.
- X. Zhang, Q. Jing, S. Ao, G. F. Schneider, D. Kireev, Z. Zhang and W. Fu, Small, 2020, 16, 1902820 CrossRef CAS PubMed.
- S. Xu, J. Zhan, B. Man, S. Jiang, W. Yue, S. Gao, C. Guo, H. Liu, Z. Li, J. Wang and Y. Zhou, Nat. Commun., 2017, 8, 14902 CrossRef CAS PubMed.
- J. Xu, Z. J. Liu, Z. Zhang and T. B. Wu, Anal. Chem., 2023, 95, 10664–10669 CrossRef CAS PubMed.
- Z. Wang, K. Y. Yi, Q. Y. Lin, L. Yang, X. S. Chen, H. Chen, Y. Liu and D. C. Wei, Nat. Commun., 2019, 10, 1544 CrossRef PubMed.
- D. C. Swarts, Biochem. Soc. Trans., 2019, 47, 1499–1510 CrossRef CAS PubMed.
- W. Fu, L. Feng, G. Panaitov, D. Kireev, D. Mayer, A. Offenhausser and H. J. Krause, Sci. Adv., 2017, 3, e1701247 CrossRef PubMed.
- A. C. Ferrari, J. C. Meyer, V. Scardaci, C. Casiraghi, M. Lazzeri, F. Mauri, S. Piscanec, D. Jiang, K. S. Novoselov, S. Roth and A. K. Geim, Rev. Lett., 2006, 97, 187401 CrossRef CAS PubMed.
- S. Niyogi, E. Bekyarova, M. E. Itkis, H. Zhang, K. Shepperd, J. Hicks, M. Sprinkle, C. Berger, C. N. Lau, W. A. deHeer, E. H. Conrad and R. C. Haddon, Nano Lett., 2010, 10, 4061 CrossRef CAS PubMed.
- G. Wu, X. Tang, M. Meyyappan and K. W. C. Lai, Appl. Surf. Sci., 2017, 425, 713–721 CrossRef CAS.
- D. R. Kong, S. Zhang, M. Q. Guo, S. Li, Q. Wang, J. Gou, Y. Wu, Y. Chen, Y. Yang, C. Dai, Z. Tian, A. T. S. Wee, Y. Liu and D. Wei, Adv. Mater., 2023, 2307366 Search PubMed.
- C. Dai, M. Guo, Y. Wu, B. P. Cao, X. Wang, Y. Wu, H. Kang, D. Kong, Z. Zhu, T. Ying, Y. Liu and D. Wei, J. Am. Chem. Soc., 2021, 143, 19794–19801 CrossRef CAS.
- D. S. Boyle, R. McNerney, H. Teng Low, B. T. Leader and A. C. Pérez-Osorio, PLoS One, 2014, 9, e103091 CrossRef PubMed.
- H. Kang, X. Wang, M. Guo, C. Dai, R. Chen, L. Yang, Y. Wu, T. Ying, Z. Zhu, D. Wei, Y. Liu and D. Wei, Nano Lett., 2021, 21, 7897–7904 CrossRef CAS PubMed.
- W. Jiang, D. Bikard and D. Cox, Nat. Biotechnol., 2013, 31, 233–239 CrossRef CAS PubMed.
- S. J. Wang, Y. Q. Liu, R. F. Liu, L. Xie, H. M. Yang, S. G. Ge and J. H. Yu, Anal. Chim. Acta, 2024, 1291, 342213 CrossRef CAS PubMed.
- A. Ahmad, M. Imran and H. Ahsan, Pharmaceutics, 2023, 15, 1630 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr03852e |
| ‡ The authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2025 |