Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
ProFA – valorization of macroalgae biomass as a source of proteins and formic acid†
Stefanie Wesinger a,
Keerthana Erattemparambil
a,
Keerthana Erattemparambil b,
Andreas Liese
b,
Andreas Liese b,
Maximilian J. Poller
b,
Maximilian J. Poller a,
Ana Malvis Romerob and
Jakob Albert
a,
Ana Malvis Romerob and
Jakob Albert *a
*a
aInstitute of Technical and Macromolecular Chemistry, University of Hamburg, Bundesstrasse 45, 20146 Hamburg, Germany. E-mail: jakob.albert@uni-hamburg.de
bInstitute of Technical Biocatalysis, Hamburg University of Technology, Denickestr. 15, 21073 Hamburg, Germany
First published on 26th May 2025
Abstract
Macroalgae are promising feedstocks for the sustainable production of bioplastic films from proteins or platform chemicals such as formic acid (FA) as they are valuable sources of proteins and carbohydrates. This study explores the possibility of using macroalgae as biomass for biogenic FA production using the OxFA process in combination with the extraction of proteins from macroalgae with a focus on resource utilisation and innovation. Herein, the extraction of proteins and the utilisation of the other components of macroalgae to produce FA are linked for the first time. The aim was to find out which macroalgae is best suited to produce FA and protein-rich solids. For this purpose, three different algae were tested: the brown alga Fucus vesiculosus, the green alga Ulva fenestrata and the red alga Porphyra dioica. In addition, the most suitable catalyst for this study was selected from the two polyoxometalates H5PV2Mo10O40 (HPA-2) and H8PV5Mo7O40 (HPA-5) known for their suitability in the OxFA process. After Porphyra dioica proved to be a promising substrate, the parameters such as temperature (80–120 °C), reaction time (18–30 h) and catalyst/substrate ratio (0.05 to 0.5) were evaluated for their statistical influence using a Box–Behnken design of experiments. The resulting model was then used to optimise the protein content and FA yield. The optimal conditions were determined to be 80 °C, 30 hours and a catalyst-to-substrate ratio of 0.5 resulting in a protein yield of 59.5% and a formic acid yield of 16.4%. For protein extraction from the solid residues, three different methods such as alkaline hydrolysis, ultrasound-assisted extraction (UAE) and ionic liquid extraction (ILE) were investigated. All extraction methods resulted in a protein recovery of more than 40% dw, with UAE yielding the highest protein recovery of 87.2% dw (at 100% sonication amplitude) showing 30% higher protein recovery than alkaline hydrolysis and 40% higher protein recovery than ILE. It turned out that the OxFA process followed by protein extraction using UAE gave a high protein recovery and a promising yield of formic acid.
Sustainability spotlightTo realize the goal of a carbon-neutral society, the closing of carbon and nitrogen cycles in platform chemicals would be extremely important. Macroalgae are promising feedstocks for the sustainable production of bioplastic films from proteins or platform chemicals such as formic acid as they are valuable sources of proteins and carbohydrates. This study explores the possibility of using macroalgae as biomass for biogenic formic acid production with the OxFA process in combination with the extraction of proteins from macroalgae with a focus on resource utilisation and innovation. |
Introduction
Biomass is an important feedstock to target the problem of shifting the chemical industry to more sustainable processes. Algal biomass is an abundant renewable source of carbon (C) and nitrogen (N) combined within one species. Specifically, macroalgae represent a vast and underexplored resource for protein extraction, with benefits such as rapid growth rates and minimal environmental footprint. Recently, macroalgae proteins have gained significant scientific interest for application in non-food industries such as cosmetic and pharmaceutical sectors.1Macroalgae are, on the one hand, important sources of proteins, lipids, vitamins, and other biologically active compounds like phycobilin and carotenoids.2 On the other hand, using the carbohydrates from macroalgal biomass led to products like biogas,3 bioethanol or biobutanol.4 In addition, the proteins, polysaccharides and lipids found in macroalgae can be utilised for the production of biopolymers.5 The utilisation of macroalgae not only meets the growing demand for alternative protein sources, but also does so with minimal impact on the environment associated with the cultivation of macroalgae.1,6,7 Therefore, protein extraction from macroalgae is an important process for sustainable resource utilisation and innovative industrial application of products. Macroalgae are divided into three different types: red, brown and green macroalgae. One example for a red alga is the species Porphyra dioica, which is known for its high protein content (25–30% dw). These species are found especially in warm waters like in Portugal, New Zealand, South Africa, etc. and in freshwater and saltwater.8–10 One example for a brown alga is the species Fucus vesiculosus also known as Bladderwrack mainly present in cold-temperature water regions. These algae have a protein content of around 6% dw to 10% dw.11,12 For a green alga Ulva fenestrata is an example which can be found in many parts of the world in polluted and pure water as well as open or protected areas. This species is considered an important protein source with a protein content of 15% dw to 20% dw and a wide range of polysaccharides like cellulose, starch, glucan and ulvan.13–15
Various protein extraction techniques such as liquid extraction and ultrasonic extraction as well as novel methods such as ionic liquid extraction and two-phase separation have been investigated for macroalgae proteins.1 In general, a combination of extraction techniques together with a pre-treatment increases the protein yield. The fundamental principle of extraction techniques is the disruption of the cell wall. Alkaline hydrolysis is a conventional extraction technique for algal protein extraction.1 The application of acid before alkali extraction enhances the liberation of polysaccharides and proteins within the cell wall matrix and can increase the solubilization of proteins.16 Ultrasound-assisted extraction (UAE) is a technology that utilizes sound waves at frequencies higher than 20 kHz to destroy the cell wall matrices of the macroalgae.17 During the treatment, smaller cavitation bubbles within the liquid surrounding the macroalgae cell collapse which results in higher shear stress and eventually break down of the cell wall.18 UAE results in lower solvent consumption and is in general a simple, efficient and inexpensive extraction method. Nevertheless, using ultrasound could lead to excessive heat generation which might interfere with the protein structure.19 A more novel protein extraction approach is Ionic Liquid Extraction (ILE). Ionic liquids are known to be efficient extraction media for complex biomolecules especially lignin or hemicellulose in lignocellulosic biomass.20 These ILs interact with the macroalgae cell walls while disrupting intermolecular H-bonds in the cellulose present in the cell wall causing their dissolution. This results in partial or complete damage of the cell wall.1 Utilizing ILs synthesized from natural sources can increase the biocompatible nature and sustainability by using ILs with less toxic environmental impacts.21,22
Different extraction approaches can be combined to improve the extraction of valuable components and the remaining fractions can be used as raw materials to produce other compounds, thus minimising waste streams. This concept increases the value of macroalgae as a bioresource and promotes sustainable development. Therefore, the selection of the most effective technology for the separation of proteins from macroalgae is crucial.1,19
If proteins are extracted from the macroalgae, carbohydrates are usually waste products. In order to increase the total added value of macroalgae, these carbohydrates can be converted into platform chemicals. One possible value product is formic acid (FA). FA is widely used in various industries such as the chemical, agricultural, leather and rubber industries.23 The latter is also discussed as a hydrogen carrier and as a liquid syngas equivalent.24,25 Biogenic formic acid can be produced from biomass especially carbohydrates under mild conditions (<100 °C) using the OxFA process.26 Herein, biomass is selectively oxidised using a polyoxometalate (POM) catalyst to produce FA and carbon dioxide. Macroalgae, which are rich in polysaccharides, can also be used to produce biogenic FA using the OxFA process and therefore represent a sustainable alternative to fossil fuels.27–29
This study focuses on combining the OxFA process with protein extraction methods for the complete valorization of macroalgae. In the OxFA process, carbohydrates are oxidized to FA, leaving a solid residue ideally consisting of mainly non-water-soluble proteins in unreacted macroalgae remains. We investigated various methods to extract these proteins therefore increasing the utilization of the algae. Three different macroalgae species, namely Porphyra dioica, Ulva fenestrata and Fucus vesiculosus were tested for their use in this biorefinery concept. By applying a Box–Behnken design of experiments (DoE), the statistical influences on the OxFA process with the most promising macroalgae species are analyzed. Following the optimization, the solid residue obtained is utilized for protein extraction. This study also explores various protein extraction methods, emphasizing both traditional and novel techniques such as solid–liquid extraction, ultrasound-assisted extraction (UAE) and ionic-liquid based extraction (ILE). Additionally, it investigates the factors influencing protein recovery for each extraction method. Understanding these aspects is crucial for optimizing protein extraction protocols with maximized protein recovery. A simplified process scheme is shown in Fig. 1.
 | ||
| Fig. 1 Schematic flow-chart of the ProFA process. The unit operations are marked in blue and the reactants and products are colored greenish. | ||
Experimental
Substrates and catalysts
The macroalgae used for the study, the red alga Porphyra dioica, the green alga Ulva fenestrata and the brown alga Fucus vesiculosus, were cultivated by the company ALGAplus in the coastal lagoon of Ria de Aveiro (40°38′ N, 8°44′ W) based on an open Integrated Multi-Tropic Aquaculture system. After harvesting, the macroalgae biomass was washed with seawater, centrifuged to remove the excess water, and dried in an air tunnel at low temperature by ALGAplus. To make sure that different particle sizes have no influence on the outcome of the reaction, the samples were milled to 250 μm using a Retsch ZM 200 (Retsch, Germany) at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm in Hamburg.
000 rpm in Hamburg.
The used catalysts HPA-2 (H5PV2Mo10O40) and HPA-5 (H8PV5Mo7O40) were synthesized according to a procedure described by Albert28 and Raabe et al.30 Further information about the synthesis and characterization of the catalysts is given in the ESI (Table S1 and Fig. S1 to S4).†
Catalytic oxidation experiments
All catalytic oxidation experiments were performed in a 600 mL Hastelloy C276 autoclave (Parr Instruments, USA) equipped with a gas entrainment stirrer (Cemp International, Germany). The reactor was filled with 10 g of algae and 200 g of water. The amount of catalysts varied between 0.5 g and 5 g according to the desired catalyst/substrate ratio of 0.05 and 0.5 gcatalyst gsubstrate−1. The reactor was closed and purged three times with molecular oxygen (30 bar). To set up the reaction conditions the reactor was pre-pressurized with ca. 20 bar of oxygen, heated up to the desired reaction temperature (80 °C or 120 °C) and a stirrer speed of 300 rpm was set. The screening experiments for the different algal substrates were conducted non-isothermally and isochore, and therefore the reaction temperature is the starting temperature of the experiment. The DoE experiments are carried out in an identical, but isothermal, set-up. At the reaction temperature, the desired pressure of 30 bar oxygen was adjusted and the stirrer speed was increased to 1000 rpm. After the reaction time (between 18 h and 30), the stirrer speed was set to 300 rpm and the reactor was allowed to cool down to room temperature. A gas sample was taken from the cooled down gas phase and analyzed via gas chromatography (GC). The reactor was depressurized. The solid residue was filtered and washed with deionized water and dried overnight at 40 °C. The dry solid residue was weight and analyzed via organic elemental (CHNS) analysis for protein content. The liquid phase was analyzed via High Performance Liquid Chromatography (HPLC). The benchmark experiments were conducted as described with a temperature of 90 °C, a reaction time of 24 h and an amount of catalyst of 1 mmol (1.67 g (HPA-5) and 1.88 g (HPA-2)).Protein extraction methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) volume ratio of 1
volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, containing 0.2 g/100 mL of N-acetyl-L-cysteine (NAC). NAC is a reducing agent and as reported by Harnedy et al.31 its addition can increase the yield of alkaline soluble protein during extraction. Alkaline extraction was performed in a thermoshaker (Grant Instruments, United Kingdom) at 750 rpm and 20 °C for 4 h. The extract was separated by centrifugation at 10
15, containing 0.2 g/100 mL of N-acetyl-L-cysteine (NAC). NAC is a reducing agent and as reported by Harnedy et al.31 its addition can increase the yield of alkaline soluble protein during extraction. Alkaline extraction was performed in a thermoshaker (Grant Instruments, United Kingdom) at 750 rpm and 20 °C for 4 h. The extract was separated by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 minutes and was analyzed for protein content. The extraction was performed in duplicate (n = 2).
000 rpm for 20 minutes and was analyzed for protein content. The extraction was performed in duplicate (n = 2).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15. An HD 2700 ultrasound machine (Bandelin, Germany) with an MS 73 probe was used. The samples were kept in an ice bath throughout the experiment to prevent heating up. An extraction time of 10 min and frequency of 20 kHz was used, and the amplitude was regulated between 20–100%. The highest frequency (100%) is a power of 70 W. The resulting solution was centrifuged at 5000 rpm for 15 min at 4 °C. The extract was separated and analyzed for protein content.
15. An HD 2700 ultrasound machine (Bandelin, Germany) with an MS 73 probe was used. The samples were kept in an ice bath throughout the experiment to prevent heating up. An extraction time of 10 min and frequency of 20 kHz was used, and the amplitude was regulated between 20–100%. The highest frequency (100%) is a power of 70 W. The resulting solution was centrifuged at 5000 rpm for 15 min at 4 °C. The extract was separated and analyzed for protein content.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15). The solution was mixed using a Vortex mini (Janke and Kunkel, Germany) and extracted for 10 min at 25 °C in a thermoshaker (Grant Instruments, United Kingdom) at 1000 rpm. The mixture was then centrifuged at 3000 rpm for 10 minutes at 4 °C. The extract was separated and analyzed for protein content.
15). The solution was mixed using a Vortex mini (Janke and Kunkel, Germany) and extracted for 10 min at 25 °C in a thermoshaker (Grant Instruments, United Kingdom) at 1000 rpm. The mixture was then centrifuged at 3000 rpm for 10 minutes at 4 °C. The extract was separated and analyzed for protein content.Analytic methods
For the pre-characterization of the algal biomass, the protein, amino acid, carbohydrate, lipid, moisture and ash contents and the elemental composition were analyzed. For product analysis GC was used for the gas phase and HPLC for the liquid phase. The solid residue was analyzed for proteins and elemental composition.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 45 s. Around 5 μL of the samples were run through an SDS gel (SERVAGelTM TG PRiMETM 12% gel – 12 sample wells gel) alongside a standard marker ranging from 11 to 180 kDa. After completion of electrophoresis, the gel was removed from the gel tray and stained with a protein stain (SERVA Quick Coomassie Stain) for about 24 h. The gel was washed with water to remove excess stain for another 24 h and was used to obtain and compare the results.
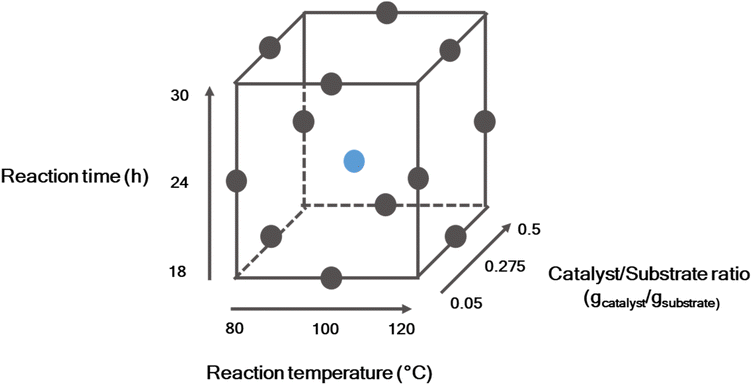
000 rpm for 45 s. Around 5 μL of the samples were run through an SDS gel (SERVAGelTM TG PRiMETM 12% gel – 12 sample wells gel) alongside a standard marker ranging from 11 to 180 kDa. After completion of electrophoresis, the gel was removed from the gel tray and stained with a protein stain (SERVA Quick Coomassie Stain) for about 24 h. The gel was washed with water to remove excess stain for another 24 h and was used to obtain and compare the results.The parameter room in which the statistical influences are determined is shown in Fig. 2. The dots represent the screened experiments.
Results and discussion
Macroalgae characterization
In order to assess the potential of different types of macroalgae for valorization i.e. production of FA and extraction of proteins, they were first characterized in detail. The protein, lipid, carbohydrate, and moisture contents as well as the ash content were analyzed. These values can be seen in the diagram in Fig. 3. | ||
| Fig. 3 Characterization of different macroalgae – Fucus vesiculosus (brown alga), Ulva fenestrata (green alga), and Porphyra dioica (red alga). | ||
The ash content is very high in all samples. For the algae Porphyra dioica and Fucus vesiculosus it is 43.4% (±1.0%) and 43.1% (±1%) respectively. Only the tested green alga Ulva fenestrata has an ash content of 27.3% (±0.2%) below 30%. The lowest percentage in the composition of the macroalgae is the lipid content. For all substrates tested, this is less than 10%, for the red alga Porphyra dioica it is even less than 1%. The moisture content before drying of the algae is only above 10% (11.2%) (±0.2%) for the green alga Ulva fenestrata and otherwise below 10%. The most interesting values for this study are the protein content and the carbohydrate content, as both have an influence on the later value products of the study. With a protein content of just under 30% (29.8%), the red alga Porphyra dioica has the highest tested value in the study. The other algae have a protein content of 16.0% for the green alga Ulva fenestrata and 6.8% for the brown alga Fucus vesiculosus. The higher the carbohydrate content, the higher the expected yield of the target product FA can be. Therefore, the macroalgae Ulva fenestrata and Fucus vesiculosus are good candidates for the production of FA with around 45% of carbohydrates (46.9% and 45.3%). All these characterization data lay well in line with other studies of these three macroalgae.6,37–47 The characterization of Porphyra dioica in this study shows a 5% higher protein content and a relatively low carbohydrate content compared to that in other studies by Pimental et al.6 and Teles et al.38 It is known that the growth phase at harvest and the growth conditions have a significant influence on the composition of the macroalgae. The location and climatic conditions at the cultivation area also have an impact on the composition of the biomass.10 Therefore, the deviations of the values from the cited studies are not surprising. Some reports for Porphyra dioica show even higher protein contents.10 On the one hand, with all the data of the three macroalgae, the reported protein content of Porphyra dioica in this study has the potential to result in high amounts of protein extracted from these macroalgae increasing the potential usage for a protein source. On the other hand, FA as the second target product is derived through the carbohydrates existing in the macroalgae samples. Therefore, Fucus vesiculosus and Ulva fenestrata with high carbohydrate content are the most promising substrates for the production of FA.
To complete the characterization of the three different macroalgae species, the amino acid profile and the CHNS-elemental composition results are shown in Fig. S2–S4 and Table S1.† The amino acid profile helps to understand the distribution of various amino acids present in the biomass, which is important for potential further use as a substrate for biopolymer film development. There are two groups of amino acids present in the macroalgae samples – hydrophobic and hydrophilic amino acids. The hydrophilic amino acids restrict protein-based biopolymer film applications in packaging of wet materials. Hence, it is beneficial to have a higher distribution of hydrophobic amino acids in the extracted proteins. Hydrophobic amino acids are Val (valine), Met (methionine), Ile (isoleucine), Leu (leucine), Phe (Phenylalanine) Gly (glycine), and Ala (alanine).48 The amino acid profile of the used algae and a short description can be found in Fig. S6–S8.† By using the OxFA process as a pre-treatment method, the amino acid distribution can change significantly. The next step will therefore focus on the catalytic oxidation of the three macroalgae substrates introduced.
Catalytic oxidation of macroalgae using the OxFA process
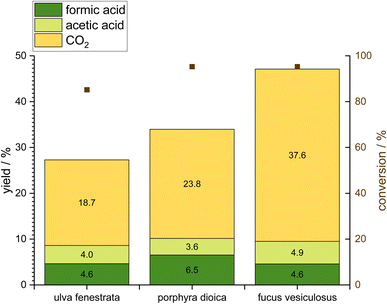
The catalytic conversion of the three macroalgae is aimed at pretreating these algae for protein extraction and obtaining FA as a valuable product. Therefore, all macroalgae were oxidized under similar reaction conditions at a temperature of 90 °C, a pressure of 30 bar oxygen, a stirrer speed of 1000 rpm, and a reaction time of 24 h using 10 g macroalgae together with a catalyst amount of 1 mmol. The used catalyst is the well-established HPA-5 which has been proven to be an efficient catalyst for biomass oxidation especially the OxFA process.26 Fig. 4 shows the OxFA-reaction produced FA (dark green) as well as the by-products CO2 (yellow) and acetic acid (light green). Overall, a high conversion was observed for all macroalgae with 95.3% for Fucus vesiculosus and Porphyra dioica and 85.2% for Ulva fenestrata. However, FA-yields of 4.6% for Ulva fenestrata and Fucus vesiculosus as well as 6.5% for Porphyra dioica are comparatively low compared to that using sugars or lignocellulosic biomass as substrates for the OxFA process. The FA-yield depends on the substrate and the number of oxygen functionalities in the carbon framework.27 The most favorable substrate in the study of Albert et al.27 is pomace. Remarkably, the two macroalgae with the highest carbohydrate content Fucus vesiculosus and Ulva fenestrata (Fig. 3) result in the lowest FA-yield. For the brown alga Fucus vesiculosus the oxidation reaction results in a high overoxidation to CO2 of 37.6%. With FA and acetic acid as valuable products, this leads to a value to waste ratio of only 0.25. With a value to waste ratio below 1, the waste product CO2 is more likely to be produced than valuable products. For the other two screened algae, the value to waste ratio lies under 1 as well, with a value of 0.42 for Porphyra dioica and 0.46 for Ulva fenestrata. In terms of the value to waste ratio, Ulva fenestrata is the best starting material, but regarding FA-yield Porphyra dioica leads to the highest yield amongst the screened macroalgae.As the idea of this study is to generate two valuable products, namely FA and proteins, we decided to maximize the amount of solid residue, i.e. lower the conversion to preserve the protein content. The analysis of proteins in the reaction broth is difficult as the homogeneous POM catalyst interferes with the usual analytical methods. Esser et al.49 showed the possibility of membrane separation through a nanofiltration membrane for the POM catalyst. However, this works because the target product FA or other small compounds can diffuse through the membrane and the catalyst is retained. Since proteins are much larger molecules, a new process has to be established for the separation of the proteins or amino acids from the catalyst, which warrants a study of its own. Therefore, this study focuses on the proteins left in the solid residue after the reaction as these proteins might be more hydrophobic and therefore a good feedstock for the production of biopolymer films. Since the initial experiment showed that the conversion with the five-times vanadium substituted Keggin-type POM-catalyst HPA-5 is too high for an investigation of the solid residue, we chose a less active catalyst for the next experiments. The next catalyst tested was the two-times vanadium substituted Keggin-type POM HPA-2. Raabe et al.50 and Esser et al.51 showed in different studies the variable redox-activity of different vanadium-substituted POMs. Due to the different V-contents (5 vanadium atoms in HPA-5 and 2 vanadium atoms in HPA-2), the redox-activity could be tuned to lower the redox-potential with HPA-2 (Fig. S9 & S10†). As shown by Krueger et al.,52 the requirement of having V atoms adjacent to each other leads to an activity difference as well. The trends of the three macroalgae are similar, as basically the same chemistry is applied, just with a lower redox potential. The conversion of the three tested macroalgae could be reduced to 93.3% for Fucus vesiculosus, 87.3% for Porphyra dioica and 70% for Ulva fenestrata (Fig. 5). The highest reduction of conversion with nearly 15% could be achieved with Ulva fenestrata. A significant reduction of 8% was observed for Porphyra dioica; however with Fucus vesiculosus a reduction of just 2% could be achieved. Interestingly, the HPA-2-catalyst suppresses the overoxidation to CO2 and reduces the yield of CO2 to about 7% or 8% for Ulva fenestrata, and Fucus vesiculosus, with just 2% yield of CO2 from Porphyra dioica. Another noteworthy point is the FA-yield, which increases in the case of Porphyra dioica up to 8.6% (before 6.5%) and Fucus vesiculosus up to 5.4% (before 4.6%) and decreases in the case of Ulva fenestrate to 3.9% (before 4.6%).
To obtain a complete picture of the process, the solid residue after the reaction was characterized for protein content by HPLC and for the amino acid profile. Exemplarily, the amino acid profiles for the red alga Porphyra dioica and its solid residue after the OxFA process are shown in Fig. 6 and all other comparison diagrams are shown in Fig. S11 and S12.† The comparison between the solid residue and the neat Porphyra dioica shows that in general the relative content of most amino acids is increasing. Specifically, the hydrophobic amino acids (Val, Met, Ile, Leu, Phe, Gly and Ala) are more abundant, which is desirable for the production of biopolymer films. The highest increase among these was noticed for leucine (13%) and valine (16%). The other hydrophobic amino acids increase with 8% Phe, 4% Gly, 7% Ala, and 9% Ile, respectively. The only hydrophobic amino acid which decreases is Met from 1.7% to 0.9%. A similar trend can be seen in the other macroalgae samples. For Ulva fenestrata (Fig. S11†) the highest increase of hydrophobic amino acids was found for Ala (17%) and for Leu (12%). For the amino acid methionine, the same decreasing trend is found.
For Fucus vesiculosus, valine (14%) and leucine (19%) have the highest increase in hydrophobic amino acids. Remarkably, in the solid residue of the Fucus vesiculosus sample methionine is increasing compared to that in the raw macroalgae sample from 1.6% to 2.1%. The protein content between the macroalgae samples and the solid residue samples is depicted in Fig. 7. The elemental composition of the solid residues is shown in Table S3 in the ESI.† Clearly, the amount of protein increases after the pretreatment with the OxFA process in the solid residue. The highest total amount of protein content is with the red macroalga Porphyra dioica and also its solid residue. For the alga Fucus vesiculosus an increase of the protein content by about 11% from 6% initially to 17% was achieved after the pretreatment.
Therefore, the initial proof of concept was successful: the OxFA process can serve as a useful pretreatment for protein extraction from macroalgae. It increases the protein content in the sample by converting the carbohydrates to a second valuable product FA. Nevertheless, to valorize the proteins in the solid residue an extraction method has to be applied on the solid residue. The standard method for protein extraction from biogenic solids is alkaline hydrolysis, which was also applied to this solid residue. Table 1 shows the obtained protein quantity for all three macro algae solid residues after alkali extraction, which was measured via SDS-PAGE gel electrophoresis. The same trend that has already been shown for neat algae and solid residues can also be observed here, namely that Porphyra dioica provides the highest protein yield and is therefore the best source of protein.
| Alkaline extract | Protein quantity (g L−1) |
|---|---|
| Porphyra dioica solid residue | 18.3 |
| Ulva fenestrata solid residue | 10.0 |
| Fucus vesiculosus solid residue | 3.88 |
These results provide a promising proof-of-concept for the combination of the OxFA process and subsequent protein extraction from algae. Nevertheless, a FA-yield of 8.6% and a protein quantity of 18.3 g L−1 require further improvement for an economic process. The same applies for the high yield of CO2 waste, which limits the valorization of the carbon content. For this reason, the next section identifies the statistical influence of various parameters on the OxFA process for valorization of macroalgae via a Box–Behnken design of experiments and provides a further optimization of the protein recovery and FA-yield.
Optimizing protein recovery and FA-yield via Box–Behnken DoE
For the DoE, a new batch of Porphyra dioica from the same supplier and region was used. To account for the significant differences in the composition, the new batch has also been fully characterized using the above-described methods, as shown in Fig. S13–S15 and Table S3.† The second batch used has a high carbohydrate content of 48% dw which is on the upper end of the reported literature values between 46.4% dw by Noda et al.53 and 45.40% dw reported by Smith et al.54 The other valuable product in this study is proteins. Here, the second batch of Porphyra dioica has a 3% higher protein content compared to the first batch (Fig. S13†). The value of 33% dw of proteins is well in line with the literature value with a reported range of 25–47% dw.1,55,56 As already discussed, the variations in the biomass composition can be attributed to various factors like environmental conditions such as light, temperature, nutritional availability, and the harvesting seasons as well as the harvesting method. Another influential factor is the life cycle stage in which the macroalgae are harvested.10,52,53 Hence, understanding the detailed composition of the biomass is essential for exploring its potential for utilizing the macroalgae in this process.The sensitivity analysis was performed using the statistical method of Box–Behnken DoE with the help of the software Design Expert.36 The influence of three parameters was investigated on three levels. Furthermore, two-dimensional interactions between these factors were determined.57 The Box–Behnken design was also used for response surface methodology for optimization, because the parameters are estimated using quadratic models, a sequential design is built and the detection of the lack of fit is possible for the model.33,57 For this purpose the temperature was varied between 80 °C and 120 °C where 80 °C is the lowest temperature reported in the literature where a catalytic reaction is activated.58 The expectation for the lowest temperature is a high solid residue and therefore high protein content. The highest temperature of 120 °C is chosen because of the expected high FA-yield and a temperature above 120 °C would lead to the denaturation of proteins. Time as a parameter was ranged between 18 h and 30 h because it is known that for complex water-insoluble biomass like beech wood an initial time is needed to get significant high yields of FA.58 The screening time of 24 h was therefore set to the middle point and it was varied from around 6 h to 18 as shown in Fig. 2.
The response surface obtained is displayed in Fig. 9. The model indicates that protein recovery is only dependent on temperature which is clearly seen in Fig. 8. Decreasing the temperature increases the protein recovery especially when the temperature decreases below 80 °C. This is mainly due to the low conversion of the macroalgae as well as less denaturation of the proteins due to lower temperature. However, the OxFA process requires a minimum temperature of 80 °C for an effective activity of the POM catalyst. Even if a large amount of solid residue and therefore a large amount of protein in the solid residue is favorable, the OxFA process helps as a pre-treatment method for the proteins. This fact will be addressed later in detail with the screening of different further protein extraction methods.
 | ||
| Fig. 9 The 3D response surface for protein recovery as result of the Box–Behnken DoE. The red dots depict the run points above the surface. | ||
The same Box–Behnken design was also used to analyze significant parameters for FA production. The suggested model here is a quadratic one with a sequential p-value of 0.0026. The statistical evaluation ANOVA is shown in Table S6.† The model is significant with a p-value < 0.0001. The lack of fit with a p-value of 0.0096 is significant, which means that the model does not fit well enough to the data. To get a non-significant lack of fit a p-value higher than 0.05 is required. To verify this, two verification runs were performed. Fig. S17† displays the parameter room for the verification points as green dots. The experiments are run with the following parameters: for the first verification run a reaction time of 30 h, a reaction temperature of 80 °C and a catalyst/substrate ratio of 0.5 were chosen. For the second verification run a temperature of 120 °C, a reaction time of 30 h and a catalyst/substrate ratio of 0.5 were picked. The other reaction parameters were kept constant. The points were set to the edges of the parameter space. Fig. S18† shows the model predicted values and the actual experimental values. These experiments are well in line and the two verification points (marked with black circles) fit very well to the model and the model is validated. Therefore, the model was used for further interpretation and optimization.
The model found the following parameters to be significant: the temperature A, the catalyst/substrate ratio (C) and the quadratic interactions of these two A2 and C2 as well as the quadratic interactions between the two parameters AC. This is visualized in the Pareto chart (Fig. 10) where every parameter which is below the red line (significance limit) is deemed statistically significant.
The response surface for the FA-yield is depicted in Fig. 11. From the model, the temperature and the catalyst/substrate ratio are found to be the most influential factors. The best FA-yield was obtained for a catalyst/substrate ratio of 0.5. This can be seen by the red colored region in the FA-yield (FA) – temperature-catalyst ratio plot in Fig. 11 on the right side. The model indicates that a temperature value between 90 °C and 110 °C produces the highest FA-yield. Interestingly, time is found to be an insignificant factor. Reichert et al.58 reported that in the OxFA process, the formation of formic acid is much higher within the first 14 h of the reaction. After that the carbon dioxide formation reduces the formic acid selectivity.59 This effect is probably the increasing amount of formic acid coupled with the decreasing pH that reduces the catalyst activity and thereby the FA production.60,61 To obtain a maximum conversion a reaction time of more than 18 h was initially chosen. Therefore, the influence of time is found to be an insignificant factor. The diagram on the left side of Fig. 11 illustrates this very well.
 | ||
| Fig. 11 The 3D response surfaces for FA-yield as result of the Box–Behnken DoE. The red dots depict the run points above the surface and the pink dots depict the run points below the surface. | ||
The analysis of the influence parameters on the FA-yield shows a big contrast to the best parameters for protein recovery. As the latter favors a low temperature of 80 °C, high temperatures are more favorable for a high FA-yield. On the other hand, the second significant parameter for FA production, the catalyst/substrate ratio has no significant influence on protein recovery. Nevertheless, optimizing this process is a challenge due to the contrary influences of the temperature. It requires a compromise between FA-yield and protein recovery. The Design Expert software36 can provide optimized conditions finding a compromise between FA-yield and protein recovery, these are shown in Fig. 12. The red dots show the suggested reaction conditions which are the following: 80 °C, 30 h, 0.5 gcatalyst g−1substrate with the fixed reaction parameters of 1000 rpm stirrer speed, 30 bar oxygen pressure, 10 g of Porphyra dioica, and 200 g of water as solvent. The blue dots visualize the actual values and these are also shown in Table 2.
 | ||
| Fig. 12 The optimized conditions resulting from the Box–Behnken DoE maximizing FA-yield as well as protein recovery. | ||
| Predicted | Actual | |
|---|---|---|
| a Reaction conditions: 30 bar oxygen pressure, 1000 rpm stirrer speed, 30 h reaction time, 80 °C, 200 g of water with 10 g suspended macroalgae and a catalyst/substrate ratio of 0.5, catalyst: HPA-2. | ||
| FA-yield (%) | 19.13 | 16.41 |
| Protein recovery (% dw) | 50.40 | 59.55 |
The experiment resulted in 59.55% protein recovery and 16.41% FA-yield which is a 7.8% higher FA-yield as that after the screening of different macroalgae. A comparison of the optimized FA-yield with Porphyra dioica as the substrate is shown in Fig. 13. In general, algal biomass results in FA-yields less than 30%.27 The FA-yield depends on the number of oxygen functionalities in the substrate which is higher for lignocellulosic biomass like pomace.23 Lignocellulosic biomass consists mainly of cellulose, hemicellulose, and lignin which also affect the FA-yield. Algae reportedly have negligible amounts of lignin and low carbohydrate (48%) compared to these biomasses (over 60%). The data shown in Fig. 13 indicate that only algae Chlorella sp. results in a higher FA-yield of 21.6% compared to the Porphyra dioica in this study which resulted in a yield of 16.41%. Based on the biomass characterization, Porphyra dioica has a higher protein content of 33% dw compared to Chlorella sp. with a protein content of 19.3%dw.62 Subsequently, the overall carbohydrate content of Chlorella sp. might be higher than that of Porphyra dioica (49.9% dw). Since the aim of the study was to obtain a maximum FA-yield along with maximum recovery of proteins, Porphyra dioica with its high protein content is a better substrate for the aim of this study. Furthermore, since proteins can also be extracted from the remaining solid residue the usage of macroalgae as biomass is advantageous even with low FA-yields.
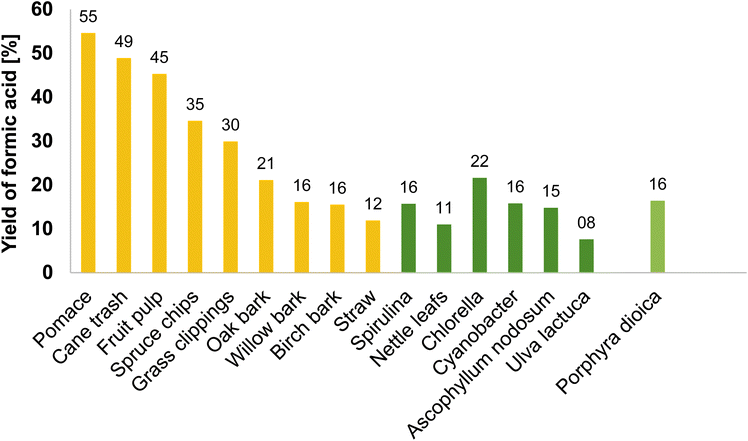 | ||
| Fig. 13 Graph for comparing the FA-yield for different biomass substrates for the OxFA process (yellow and dark green): reaction conditions: 3.3 g substrate, 1.9 g (11 mmol) additive para-toluene sulfonic acid and 1.74 g (0.9 mmol) HPA-2 catalyst dissolved in 100.0 mL H2O, 30 bar O2, 90 °C, 24 h, 1000 rpm;27 light green: optimized reaction conditions for protein and FA production from Porphyra dioica: 80 °C, 30 h, 0.5 gcatalyst gsubstrate−1, 30 bar oxygen pressure, 1000 rpm stirrer speed, 10 g of Porphyra dioica as the substrate, 200 mL of water as the solvent. | ||
Protein extraction from the solid residue of the OxFA process
The solid residue after the reaction is most likely unreacted algae and potentially lipids, and not utilized carbohydrates and therefore further extraction is necessary. To investigate the best extraction approach for the solid residue three extraction methods were tested: alkaline hydrolysis as a state-of-the-art method,55 and ultrasound-assisted extraction and ionic liquid extraction as novel approaches. Since the solid residue was already exposed to acidic conditions during the OxFA process, the choice of the tested extraction method excludes an acid extraction. It is important to keep in mind that every extraction method is the second part of a two-step extraction of proteins from a macroalga with the first step of an acid treatment with the valuable by-product FA.For the first approach, the alkaline hydrolysis, the solid samples were extracted using NaOH based on the study by Harnedy et al.31 which reported that alkaline solutions can help in the extraction of non-water-soluble proteins of macroalgae. According to Kadam et al.,63 the percentage of the extracted protein could be enhanced from 16.90% to 56.35% using 0.4 M NaOH instead of 0.4 M HCl as the extraction medium. According to a study by Harnedy et al.,31 the concentration of NaOH is a critical parameter in alkali-based extraction. Therefore, the concentration of NaOH in this study varied between 0.08 M and 5 M NaOH. The yield of alkali-soluble nitrogen can be increased by adding a reducing agent like NAC (N-acetyl-L-cysteine). Hence, 2 g per L NAC was used for the extraction as well. The extracts were analyzed for protein content and the protein recovery was calculated (equation 10) with respect to the protein content in the solid residue. To confirm the presence of proteins in the extract an initial SDS PAGE gel electrophoresis was conducted (Fig. S19†). A qualitative assessment of the protein profile shows the presence of protein with the range of 11 to 180 kDa with a regular intensity throughout the range. The intensity of the bands increases in the low molecular weight regions, and this indicates large amounts of smaller-sized proteins possibly in the form of peptides. The profile indicates higher amounts of protein content for 0.14 M NaOH concentration visible as a darker band. In contrast to the study of Harnedy et al.31 where the protein recovery remained constant when the NaOH concentration was increased from 0.12 M to 0.14 M, this study observed an increasing trend in the protein recovery. This trend was also confirmed in our experiments (Fig. 13). The protein recovery is increasing with an increase in the NaOH concentration up to 1 M NaOH (57.8%) with a slight stagnation between 0.4 M and 0.8 M NaOH (47%). The maximum value of 57.8% protein recovery is similar to the value of 59.75% for Ascophyllum nodosum obtained by Kadam et al.63 A further increase in the NaOH concentration to 5 M decreases the protein recovery to 35.8%. Lu et al.64 reported a similar trend for protein recovery with NaOH concentration, wherein the protein recovery initially increases with increasing concentration, reaching a peak, and subsequently decreases. The reason for the increasing effect is that high alkali concentrations can promote a breakdown of hydrogen bonds and dissociation of hydrogen from proteins, enhancing protein solubility. The decreasing effect after a concentration of 1 M NaOH could be explained by the fact that strong alkali solutions and therefore elevated pH-values can disrupt the protein structure and cause denaturation leading to a loss of solubility of proteins.64
The second approach tested was the Ultrasound-Assisted Extraction (UAE). This UAE was applied to the solid residue in an alkali medium of 0.4 M NaOH. Hildebrand et al.65 reported this method to be an efficient approach for protein extraction if it is combined with an alkali medium. A study by Garcia-Vaquero et al.66 analyzed the influential parameters for UAE and reported that the sonication amplitude impacts the amount of protein extracted into the medium. Therefore, this study analyses the influence of this parameter as well which is shown in Fig. 14. In line with the study by Garcia-Vaquero et al.66 the sonication amplitude in our study was varied between 20% and 100% with a fixed extraction time of 10 minutes in an alkali medium with a concentration of 0.4 M NaOH. The time was fixed to 10 min as a study from Hildebrand et al.65 showed that a longer duration of ultrasonic treatment has a negative effect on the protein recovery (73.6% after 18 h compared to 76.6% after 10 min). The extracted samples were analyzed via SDS-PAGE gel electrophoresis to confirm the presence of proteins, and these results are shown in Fig. S20.† The SDS-PAGE results show increasing protein content with an increase in the sonication amplitude. Like the alkali extraction approach the bands in the SDS-PAGE gel electrophoresis are spread throughout a region between 11 to 180 kDa. The regions with lower molecular weights show a stronger intensity and therefore indicate a high number of smaller-sized proteins. Fig. 14 shows the comparison between the sonication amplitudes of 20%, 50% and 100%, respectively. The sonication amplitude of 100% clearly shows the best protein recovery of 87.2%. Ultrasound treatment induces cavitation within the liquid medium which leads to cell wall damage within the structure and an increased surface area enhancing the protein recovery. Hence, alkali extraction could be boosted with ultrasound. For comparison, non-ultrasound assisted alkali extraction with 0.4 M NaOH leads to a protein recovery of 47.8% and ultrasound assisted alkali extraction has a protein recovery of 68.2% with the lowest tested sonication amplitude of 20%. The maximum protein recovery in this study (87.2%) was even higher than previous reported values by Hildebrand et al. (76.6%)62 and Kadam et al. (57.2%)63 with ultrasonic assisted extraction in an alkali medium. This can be attributed to the OxFA process, which works as an acidic pre-treatment of the macroalgae sample, resulting in an overall higher value for the protein extraction in this study.
The third extraction approach is ionic liquid extraction. The IL used in this study was choline chloride (ChCl), a green, cheap and environmentally friendly solvent.21 To test the influence of the concentration of ChCl in the extraction media, IL/water concentrations of 10–50 wt% of IL are tested. Higher concentrations of IL pose challenges in handling and processability as well as cause interference in analytical measurements.21 The nitrogen atoms in the ChCl solution affected the protein content estimation by the nitrogen content method, which was applied in the other tested extraction methods, and therefore a Bradford assay was used to quantify the proteins extracted by the IL. It is reported in a study by Suarez Garcia et al. that the Bradford assay method for quantification of the proteins is not impacted by ChCl.21 The resulting protein recovery with this approach is shown in Fig. 15. A clear trend is visible throughout an increase in the ChCl content. The higher the IL content in the extraction media, the higher the protein recovery. This could achieve an increase in the recovery from 5.0% at 10 wt% of ChCl to 47.6% at 50 wt% of ChCl, which is a 9.5-fold increase. Similar trends were observed by Suarez Garcia et al.21 and Martins et al.67 ChCl acts as a solubilizing agent helping the protein release from the macroalgae matrix and stabilizes the protein during extraction preventing denaturation. According to studies by Martins et al.,67 the extraction of protein from macroalgae is also due to the salting-in effect caused by the addition of ChCl, especially in the case of proteins such as phycobiliproteins whose solubility increases with even a slight addition of CCl. However, the protein recovery obtained in this study was higher than the values reported in these previous studies. This is likely due to the OxFA process causing a pretreatment effect as mentioned above.
After investigating three different extraction techniques for the solid residue of the red macroalga Porphyra dioica after the OxFA process, Fig. 16 shows the comparison of these extraction techniques. It can be clearly seen that the best protein recovery approach with the highest value for protein recovery of 87.2% is UAE. Since the proteins are extracted into an alkali medium further purification is needed to utilize these proteins for applications like biofilm production. Ultrafiltration might be the best possible purification technique as other techniques like chromatography would need further treatment of the extraction broth (Fig. 17).
The presented results clearly show that the combination of the OxFA process and UAE is a promising approach for the valorisation of macroalgae, specifically Porphyra dioica, producing proteins and FA as valuable products. While the OxFA process is obviously an efficient pre-treatment method for protein extraction, some protein is lost during this treatment.
Conclusions
This study investigated the combined use of the OxFA process and different protein extraction methods for the production of proteins and formic acid from macroalgae (ProFA process). Three different subtypes of algae, specifically a red alga (Porphyra dioica), a brown alga (Fucus vesiculosus), and a green alga (Ulva fenestrata) have been tested. Out of these screened algae the red alga Porphyra dioica provided the best results. By tuning the redox-activity of the OxFA catalyst, the amount of protein-rich solid residue could be increased to 13.7% of the total algae with a protein content of 36.4% dw of protein, while achieving a FA yield of 8.6%.After the identification of the most promising substrate, the OxFA process (in combination with an alkali extraction method) was further enhanced to optimize protein recovery next to FA yield. For this purpose, a Box–Behnken-DoE was applied and identified the temperature to be a significant parameter for protein recovery, while both the temperature and catalyst/substrate ratio had a significant influence on the FA yield. Using the optimized reaction parameters (80 °C, 30 h, 0.5 gcatalyst gsubstrate−1) Porphyra dioica as the substrate, and 200 mL of water as the solvent a yield of 16.4% with a protein recovery of 59.6% was achieved.
Following optimization of the OxFA process, the next step was the identification of the best protein extraction technique. Since the OxFA process already serves as acidic pre-treatment, three non-acidic methods were examined: alkali extraction (AE), ultrasound-assisted alkali-based extraction (UAE), and a method using choline chloride as ionic liquid (ILE). UAE was found to be the most effective protein extraction technique with a protein recovery of 87.2%. Therefore, the combination of the OxFA process with UAE was found to be the most efficient way to obtain protein and formic acid from macroalgae. Analysis of the extracted proteins showed that their composition is suitable for applications such as the production of biopolymer films.
In summary, this process provides a possibility for the sustainable and holistic utilization of algal biomass producing valuable proteins and formic acid.
Data availability
Various additional results and analytical data of catalysts and reaction solutions are included in the ESI.† Further raw measurement data and calculation files are available at Zenodo at https://doi.org/10.5281/zenodo.15605969.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This project was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), SFB 1615-503850735. The authors thank the Central Elemental Analysis Service of the Department of Chemistry at the University of Hamburg as well as the division for central element analytics of the Hamburg University of Technology conducting ICP as well as CHNS measurements.References
- M. Gordalina, H. M. Pinheiro, M. Mateus, M. M. R. da Fonseca and M. T. Cesário, Macroalgae as Protein Sources—A Review on Protein Bioactivity, Extraction, Purification and Characterization, Appl. Sci., 2021, 11, 7969 CrossRef CAS.
- G. Jiao, G. Yu, J. Zhang and H. S. Ewart, Chemical structures and bioactivities of sulfated polysaccharides from marine algae, Mar. Drugs, 2011, 9, 196–223 CrossRef CAS PubMed.
- J. C. Costa, P. R. Gonçalves, A. Nobre and M. M. Alves, Biomethanation potential of macroalgae Ulva spp. and Gracilaria spp. and in co-digestion with waste activated sludge, Bioresour. Technol., 2012, 114, 320–326 CrossRef CAS PubMed.
- K. A. Jung, S.-R. Lim, Y. Kim and J. M. Park, Potentials of macroalgae as feedstocks for biorefinery, Bioresour. Technol., 2013, 135, 182–190 CrossRef CAS PubMed.
- A. Aswathi Mohan, A. Robert Antony, K. Greeshma, J.-H. Yun, R. Ramanan and H.-S. Kim, Algal biopolymers as sustainable resources for a net-zero carbon bioeconomy, Bioresour. Technol., 2022, 344, 126397 CrossRef CAS PubMed.
- F. B. Pimentel, M. Cermeño, T. Kleekayai, P. A. Harnedy-Rothwell, E. Fernandes, R. C. Alves, M. B. P. P. Oliveira and R. J. FitzGerald, Enzymatic Modification of Porphyra dioica-Derived Proteins to Improve their Antioxidant Potential, Molecules, 2020, 25(12), 2838 CrossRef CAS PubMed.
- A. Naseri, G. S. Marinho, S. L. Holdt, J. M. Bartela and C. Jacobsen, Enzyme-assisted extraction and characterization of protein from red seaweed Palmaria palmata, Algal Res., 2020, 47, 101849 CrossRef.
- M. J. Holmes and J. Brodie, Morphology, seasonal phenology and observations on some aspects of the life history in culture of Porphyra dioica (Bangiales, Rhodophyta) from Devon, UK, Phycologia, 2004, 43, 176–188 CrossRef.
- P. Rawiwan, Y. Peng, I. G. P. B. Paramayuda and S. Y. Quek, Red seaweed: A promising alternative protein source for global food sustainability, Trends Food Sci. Technol., 2022, 123, 37–56 CrossRef CAS.
- F. B. Pimentel, M. Cermeño, T. Kleekayai, P. A. Harnedy, R. J. FitzGerald, R. C. Alves and M. B. P. P. Oliveira, Effect of in vitro simulated gastrointestinal digestion on the antioxidant activity of the red seaweed Porphyra dioica, Food Res. Int., 2020, 136, 109309 CrossRef CAS PubMed.
- L. Buedenbender, F. A. Astone and D. Tasdemir, Bioactive Molecular Networking for Mapping the Antimicrobial Constituents of the Baltic Brown Alga Fucus vesiculosus, Mar. Drugs, 2020, 18(6), 311 CrossRef CAS PubMed.
- A. R. Ribeiro, T. Madeira, G. Botelho, D. Martins, R. M. Ferreira, A. M. S. Silva, S. M. Cardoso and R. Costa, Brown Algae Fucus vesiculosus in Pasta: Effects on Textural Quality, Cooking Properties, and Sensorial Traits, Foods, 2022, 11(11), 1561 CrossRef CAS PubMed.
- J. Olsson, G. B. Toth, A. Oerbekke, S. Cvijetinovic, N. Wahlström, H. Harrysson, S. Steinhagen, A. Kinnby, J. White, U. Edlund, I. Undeland, H. Pavia and E. Albers, Cultivation conditions affect the monosaccharide composition in Ulva fenestrata, J. Appl. Phycol., 2020, 32, 3255–3263 CrossRef CAS.
- S. I. Kozhenkova, E. N. Chernova and V. M. Shulkin, Microelement composition of the green alga Ulva fenestrata from Peter the Great Bay, Sea of Japan, Russ. J. Mar. Biol., 2006, 32, 289–296 CrossRef CAS.
- L. Juul, S. Steinhagen, A. Bruhn, S. K. Jensen, I. Undeland and T. K. Dalsgaard, Combining pressing and alkaline extraction to increase protein yield from Ulva fenestrata biomass, Food Bioprod. Process., 2022, 134, 80–85 CrossRef CAS.
- M. Cermeño, T. Kleekayai, M. Amigo-Benavent, P. Harnedy-Rothwell and R. J. FitzGerald, Current knowledge on the extraction, purification, identification, and validation of bioactive peptides from seaweed, Electrophoresis, 2020, 41, 1694–1717 CrossRef PubMed.
- R. Mittal, H. A. Tavanandi, V. A. Mantri and K. S. M. S. Raghavarao, Ultrasound assisted methods for enhanced extraction of phycobiliproteins from marine macro-algae, Gelidium pusillum (Rhodophyta), Ultrason. Sonochem., 2017, 38, 92–103 CrossRef CAS PubMed.
- J. R. McMillan, I. A. Watson, M. Ali and W. Jaafar, Evaluation and comparison of algal cell disruption methods: Microwave, waterbath, blender, ultrasonic and laser treatment, Appl. Energy, 2013, 103, 128–134 CrossRef.
- J. Echave, M. Fraga-Corral, P. Garcia-Perez, J. Popović-Djordjević, E. H Avdović, M. Radulović, J. Xiao, M. A Prieto and J. Simal-Gandara, Seaweed Protein Hydrolysates and Bioactive Peptides: Extraction, Purification, and Applications, Mar. Drugs, 2021, 19(9), 500 CrossRef CAS PubMed.
- Z. Lei, C. Dai, J. Hallett and M. Shiflett, Introduction: Ionic Liquids for Diverse Applications, Chem. Rev., 2024, 124, 7533–7535 CrossRef CAS PubMed.
- E. Suarez Garcia, C. F. Miranda, M. T. Cesario, R. H. Wijffels, C. van den Berg and M. H. M. Eppink, Ionic Liquid-Assisted Selective Extraction and Partitioning of Biomolecules from Macroalgae, ACS Sustain. Chem. Eng., 2023, 11, 1752–1762 CrossRef CAS PubMed.
- M. M. Pereira, S. N. Pedro, M. V. Quental, A. Mohamadou, J. A. P. Coutinho and M. G. Freire, Integrated Approach to Extract and Purify Proteins from Honey by Ionic Liquid-Based Three-Phase Partitioning, ACS Sustain. Chem. Eng., 2022, 10, 9275–9281 CrossRef CAS PubMed.
- P. Preuster and J. Albert, Biogenic formic acid as a green hydrogen carrier, Energy Technol., 2018, 6(3), 501–509 CrossRef CAS.
- F. Kroll, M. Schörner, M. Schmidt, F. T. U. Kohler, J. Albert and P. Schühle, Hydrogen production from wet biomass via a formic acid route under mild conditions, Int. J. Hydrogen Energy, 2024, 62, 959–968 CrossRef CAS.
- J. Albert, A. Jess, C. Kern, F. Pöhlmann, K. Glowienka and P. Wasserscheid, Formic Acid-Based Fischer-Tropsch Synthesis for Green Fuel Production from Wet Waste Biomass and Renewable Excess Energy, ACS Sustainable Chem. Eng., 2016, 4(9), 5078–5086 CrossRef CAS.
- J. Albert, R. Wölfel, A. Bösmann and P. Wasserscheid, Selective oxidation of complex, water-insoluble biomass to formic acid using additives as reaction accelerators, Energy Environ. Sci., 2012, 5(7), 7956–7962 RSC.
- J. Albert and P. Wasserscheid, Expanding the scope of biogenic substrates for the selective production of formic acid from water-insoluble and wet waste biomass, Green Chem., 2015, 17, 5164–5171 RSC.
- J. Albert, D. Lüders, A. Bösmann, D. M. Guldi and P. Wasserscheid, Spectroscopic and electrochemical characterization of heteropoly acids for their optimized application in selective biomass oxidation to formic acid, Green Chem., 2014, 16, 226–237 RSC.
- J.-C. Raabe, M. J. Poller, D. Voß and J. Albert, H8 PV5 Mo7 O40 - A Unique Polyoxometalate for Acid and RedOx Catalysis: Synthesis, Characterization, and Modern Applications in Green Chemical Processes, ChemSusChem, 2023, 16, e202300072 CrossRef CAS PubMed.
- J.-C. Raabe, J. Aceituno Cruz, J. Albert and M. J. Poller, Comparative Spectroscopic and Electrochemical Study of V(V)-Substituted Keggin-Type Phosphomolybdates and -Tungstates, Inorganics, 2023, 11, 138 CrossRef CAS.
- P. A. Harnedy and R. J. FitzGerald, Extraction of protein from the macroalga Palmaria palmata, LWT–Food Sci. Technol., 2013, 51, 375–382 CrossRef CAS.
- J. Kjeldahl, Neue Methode zur Bestimmung des Stickstoffs in organischen Körpern, Z. für Anal. Chem., 1883, 22, 366–382 CrossRef.
- A. Lamp, M. Kaltschmitt and O. Lüdtke, Improved HPLC-method for estimation and correction of amino acid losses during hydrolysis of unknown samples, Anal. Biochem., 2018, 543, 140–145 CrossRef CAS PubMed.
- A. Sluiter, B. Hames, R. Ruiz, C. Scarlata, J. Sluiter, D. Templeton and D. Crocker, Determination of Structural Carbohydrates and Lignin in Biomass, 2012 Search PubMed.
- M. M. Bradford, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 1976, 248–254 CrossRef CAS PubMed.
- Design-Expert® Software, Stat Ease Inc., Minneapolis, MN, USA Search PubMed.
- L. Juul, M. Danielsen, C. Nebel, S. Steinhagen, A. Bruhn, S. K. Jensen, I. Undeland and T. K. Dalsgaard, Ulva fenestrata protein – Comparison of three extraction methods with respect to protein yield and protein quality, Algal Res., 2021, 60, 102496 CrossRef.
- M. Teles, P. Adão, C. Afonso, R. Bernardino, M. Guedes, R. Baptista and S. Bernardino, Development and Characterization of Films for Food Application Incorporating Porphyran Extracted from Porphyra dioica, Coatings, 2022, 12, 148 CrossRef CAS.
- S. Steinhagen, K. Larsson, J. Olsson, E. Albers, I. Undeland, H. Pavia and G. B. Toth, Closed life-cycle aquaculture of sea lettuce (Ulva fenestrata): performance and biochemical profile differ in early developmental stages, Front. Mar. Sci., 2022, 9 DOI:10.3389/fmars.2022.942679.
- S. Steinhagen, S. Enge, K. Larsson, J. Olsson, G. M. Nylund, E. Albers, H. Pavia, I. Undeland and G. B. Toth, Sustainable Large-Scale Aquaculture of the Northern Hemisphere Sea Lettuce, Ulva fenestrata, in an Off-Shore Seafarm, J. Mar. Sci. Eng., 2021, 9, 615 CrossRef.
- A. B. Ross, J. M. Jones, M. L. Kubacki and T. Bridgeman, Classification of macroalgae as fuel and its thermochemical behaviour, Bioresour. Technol., 2008, 99, 6494–6504 CrossRef CAS PubMed.
- I. Peinado, J. Girón, G. Koutsidis and J. M. Ames, Chemical composition, antioxidant activity and sensory evaluation of five different species of brown edible seaweeds, Food Res. Int., 2014, 66, 36–44 CrossRef CAS.
- L. Paiva, E. Lima, A. I. Neto and J. Baptista, Seasonal Variability of the Biochemical Composition and Antioxidant Properties of Fucus spiralis at Two Azorean Islands, Mar. Drugs, 2018, 16(8), 248 CrossRef PubMed.
- A. Malvis Romero, F. Brozio, S. Kammler, C. Burkhardt, L. Baruth, M. Kaltschmitt, G. Antranikian and A. Liese, Enzyme-Assisted Extraction of Alginate from Beach Wrack Fucus vesiculosus, Chem. Ing. Tech., 2023, 95, 549–556 CrossRef CAS.
- F. Herbreteau, L. J. M. Coiffard, A. Derrien and Y. de Roeck-Holtzhauer, The Fatty Acid Composition of Five Species of Macroalgae, Bot. Mar., 1997, 40, 25–28 CAS.
- J. Fleurence, M. Morançais and J. Dumay, in Proteins in Food Processing, Elsevier, 2018, pp. 245–262 Search PubMed.
- A. R. Circuncisão, M. D. Catarino, S. M. Cardoso and A. M. S. Silva, Minerals from Macroalgae Origin: Health Benefits and Risks for Consumers, Mar. Drugs, 2018, 16(11), 400 CrossRef PubMed.
- M. R. Barnes and I. C. Gray, Bioinformatics for Geneticists, John Wiley, Chichester, 2005 Search PubMed.
- T. Esser, M. Huber, D. Voß and J. Albert, Development of an efficient downstream process for product separation and catalyst recycling of a homogeneous polyoxometalate catalyst by means of nanofiltration membranes and design of experiments, Chem. Eng. Res. Des., 2022, 185, 37–50 CrossRef CAS.
- J.-C. Raabe, J. Albert and M. J. Poller, Spectroscopic, Crystallographic, and Electrochemical Study of Different Manganese(II)-Substituted Keggin-Type Phosphomolybdates, Chem.–Eur. J., 2022, 28, e202201084 CrossRef CAS PubMed.
- T. Esser, A. Wassenberg, J.-C. Raabe, D. Voß and J. Albert, Catalytic Valorization of Humins by Selective Oxidation Using Transition-Metal-Substituted Keggin-Type Polyoxometalate Catalysts, ACS Sustain. Chem. Eng., 2024, 12, 543–560 CrossRef CAS.
- J.-D. H. Krueger, M. J. Poller, C. Lyall, J. Lowe, U. Hintermair and J. Albert, In-Situ Investigations of Polyoxometalate-Catalysed Biomass Oxidation to Formic Acid by Using Multinuclear High Resolution Flow NMR Spectroscopy, ChemCatChem, 2024, 16(16) DOI:10.1002/cctc.202400402.
- H. Noda, Health benefits and nutritional properties of nori, J. Appl. Phycol., 1993, 5, 255–258 CrossRef.
- J. L. Smith, G. Summers and R. Wong, Nutrient and heavy metal content of edible seaweeds in New Zealand, N. Z. J. Crop Hortic. Sci., 2010, 38, 19–28 CrossRef CAS.
- R. O’ Brien, M. Hayes, G. Sheldrake, B. Tiwari and P. Walsh, Macroalgal Proteins: A Review, Foods, 2022, 11(4), 571 CrossRef PubMed.
- M. Øverland, L. T. Mydland and A. Skrede, Marine macroalgae as sources of protein and bioactive compounds in feed for monogastric animals, J. Sci. Food Agric., 2019, 99, 13–24 CrossRef PubMed.
- K. Siebertz, Statistische Versuchsplanung, Springer Berlin Heidelberg, Berlin, Heidelberg, 2017 Search PubMed.
- J. Reichert and J. Albert, Detailed Kinetic Investigations on the Selective Oxidation of Biomass to Formic Acid (OxFA Process) Using Model Substrates and Real Biomass, ACS Sustain. Chem. Eng., 2017, 5, 7383–7392 CrossRef CAS.
- A. Bukowski, K. Schnepf, S. Wesinger, A. Brandt-Talbot and J. Albert, Sensitivity Analysis and Parameter Optimization for the Fractionative Catalytic Conversion of Lignocellulosic Biomass in the Polyoxometalate–Ionosolv Concept, ACS Sustain. Chem. Eng., 2022, 10, 8474–8483 CrossRef CAS.
- J. Reichert, B. Brunner, A. Jess, P. Wasserscheid and J. Albert, Biomass oxidation to formic acid in aqueous media using polyoxometalate catalysts – boosting FA selectivity by in-situ extraction, Energy Environ. Sci., 2015, 8, 2985–2990 RSC.
- I. V. Kozhevnikov, Catalysis by Heteropoly Acids and Multicomponent Polyoxometalates in Liquid-Phase Reactions, Chem. Rev., 1998, 98, 171–198 CrossRef CAS PubMed.
- G. F. Ferreira, L. F. Ríos Pinto, P. O. Carvalho, M. B. Coelho, M. N. Eberlin, R. Maciel Filho and L. V. Fregolente, Biomass and lipid characterization of microalgae genera Botryococcus, Chlorella, and Desmodesmus aiming high-value fatty acid production, Biomass Convers. Biorefin., 2021, 11, 1675–1689 CrossRef CAS.
- S. U. Kadam, C. Álvarez, B. K. Tiwari and C. P. O'Donnell, Extraction and characterization of protein from Irish brown seaweed Ascophyllum nodosum, Food Res. Int., 2017, 99, 1021–1027 CrossRef CAS PubMed.
- K. Lu, X. Zhao, S.-H. Ho, R. Ma, Y. Xie and J. Chen, Biorefining and the Functional Properties of Proteins from Lipid and Pigment Extract Residue of Chlorella pyrenoidosa, Mar. Drugs, 2019, 17(8), 454 CrossRef CAS PubMed.
- G. Hildebrand, M. M. Poojary, C. O'Donnell, M. N. Lund, M. Garcia-Vaquero and B. K. Tiwari, Ultrasound-assisted processing of Chlorella vulgaris for enhanced protein extraction, J. Appl. Phycol., 2020, 32, 1709–1718 CrossRef CAS.
- M. Garcia-Vaquero, V. Ummat, B. Tiwari and G. Rajauria, Exploring Ultrasound, Microwave and Ultrasound-Microwave Assisted Extraction Technologies to Increase the Extraction of Bioactive Compounds and Antioxidants from Brown Macroalgae, Mar. Drugs, 2020, 18(3), 172 CrossRef CAS PubMed.
- M. Martins, F. A. Vieira, I. Correia, R. A. S. Ferreira, H. Abreu, J. A. P. Coutinho and S. P. M. Ventura, Recovery of phycobiliproteins from the red macroalga Gracilaria sp. using ionic liquid aqueous solutions, Green Chem., 2016, 18, 4287–4296 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Various additional results and analytical data of catalysts and reaction solutions. See DOI: https://doi.org/10.1039/d5su00012b |
| This journal is © The Royal Society of Chemistry 2025 |