Open Access Article
Open Access ArticleWatasemycin biosynthesis in Streptomyces venezuelae: thiazoline C-methylation by a type B radical-SAM methylase homologue†
Yuki
Inahashi‡
ab,
Shanshan
Zhou‡
a,
Maureen J.
Bibb
c,
Lijiang
Song
a,
Mahmoud M.
Al-Bassam§
c,
Mervyn J.
Bibb
*c and
Gregory L.
Challis
 *a
*a
aDepartment of Chemistry, University of Warwick, Coventry, CV4 7AL, UK. E-mail: g.l.challis@warwick.ac.uk
bKitasato Institute for Life Sciences, Kitasato University, 5-9-1, Shirokane, Minato-ku, Tokyo, Japan
cDepartment of Molecular Microbiology, John Innes Centre, Norwich, NR4 7UH, UK. E-mail: mervyn.bibb@jic.ac.uk
First published on 19th January 2017
Abstract
2-Hydroxyphenylthiazolines are a family of iron-chelating nonribosomal peptide natural products that function as virulence-conferring siderophores in various Gram-negative bacteria. They have also been reported as metabolites of Gram-positive Streptomyces species. Transcriptional analyses of Streptomyces venezuelae ATCC 10712 revealed that its genome contains a putative 2-hydroxyphenylthiazoline biosynthetic gene cluster. Heterologous expression of the gene cluster in Streptomyces coelicolor M1152 showed that the mono- and dimethylated derivatives, thiazostatin and watasemycin, respectively, of the 2-hydroxyphenylthiazoline enantiopyochelin are two of its metabolic products. In addition, isopyochelin, a novel isomer of pyochelin containing a C-methylated thiazolidine, was identified as a third metabolic product of the cluster. Metabolites with molecular formulae corresponding to aerugine and pulicatins A/B were also detected. The structure and stereochemistry of isopyochelin were confirmed by comparison with synthetic standards. The role of two genes in the cluster encoding homologues of PchK, which is proposed to catalyse thiazoline reduction in the biosynthesis of enantiopyochelin in Pseudomonas protegens, was investigated. One was required for the production of all the metabolic products of the cluster, whereas the other appears not to be involved in the biosynthesis of any of them. Deletion of a gene in the cluster encoding a type B radical-SAM methylase homologue abolished the production of watasemycin, but not thiazostatin or isopyochelin. Feeding of thiazostatin to the mutant lacking the functional PchK homologue resulted in complete conversion to watasemycin, demonstrating that thiazoline C-methylation by the type B radical-SAM methylase homologue is the final step in watasemycin biosynthesis.
Introduction
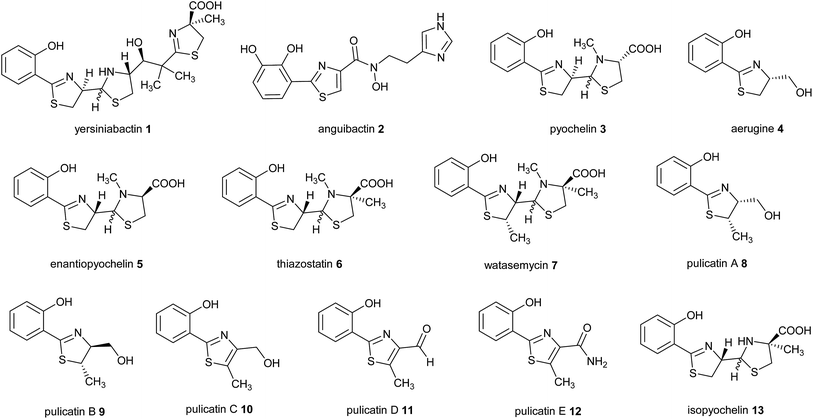
2-Hydroxyphenylthiazolines are a family of bacterial natural products with a variety of biological activities. Some members of this family function as siderophores, iron chelators produced for iron uptake that are virulence factors in numerous pathogens.1–3 Examples include yersiniabactin 1 produced by enteropathogenic Yersinia species,4 anguibactin 2 from Vibrio anguillarum 775, which causes hemorrhagic septicemia in fish,5,6 and pyochelin 3 and aerugine 4 produced by Pseudomonas species including Pseudomonas aeruginosa, an opportunistic human pathogen (Fig. 1).7,8Intriguingly, the non-pathogenic pseudomonad Pseudomonas protegens produces enantiopyochelin 5, which has also been shown to function as a siderophore (Fig. 1).9 Other members of the 2-hydroxyphenylthiazoline family are produced by non-pathogenic Actinobacteria. These include thiazostatin 6, watasemycin 7 and the pulicatins 8–12 (Fig. 1), produced by Streptomyces species, which have been reported to have antioxidant, antibacterial and neuroactive properties, respectively.10–12 However, it is not currently known whether these compounds are also able to function as siderophores.
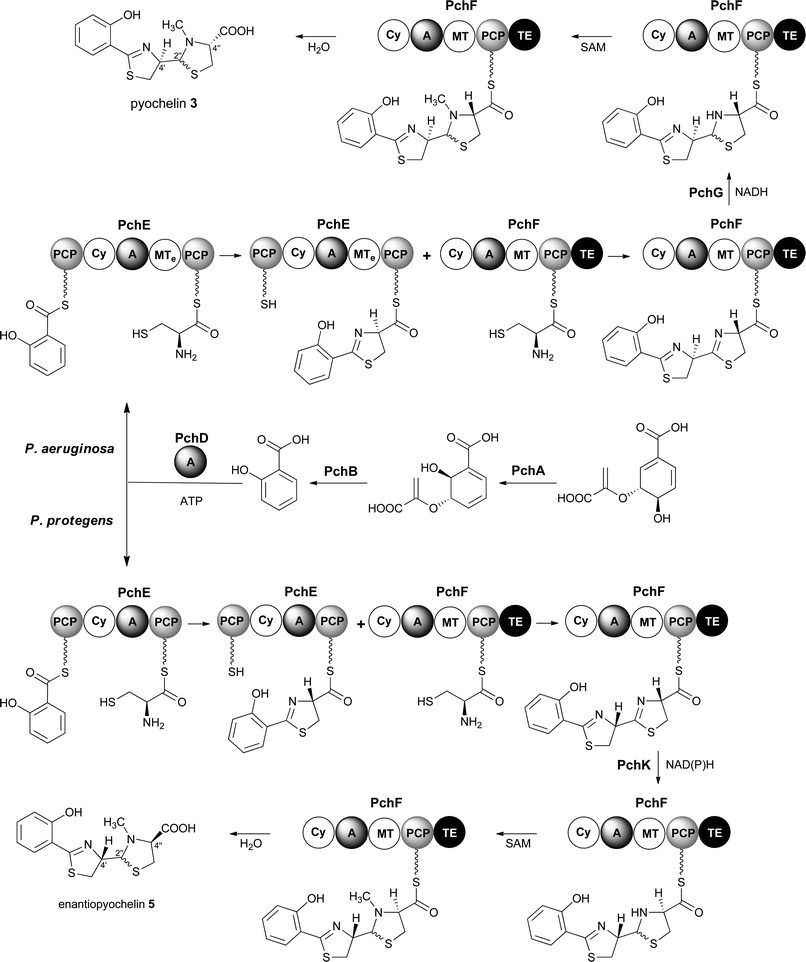
Pyochelin 3 biosynthesis in P. aeruginosa is well characterized.13–18 Seven proteins (PchABCDEFG) assemble pyochelin 3 from salicylate and two molecules of L-cysteine (Fig. 2). Salicylate is generated from chorismate by PchA and PchB. It is then activated by reaction with ATP, catalysed by the standalone adenylation (A) domain PchD. The resulting salicyl adenylate reacts with the phosphopantetheine thiol of the N-terminal peptidyl carrier protein (PCP) domain of the PchE nonribosomal peptide synthetase (NRPS) to form the corresponding salicyl thioester. The A domain of PchE similarly loads L-cysteine onto the C-terminal PCP domain of PchE and the heterocyclisation (Cy) domain catalyses the condensation of the salicyl thioester with the cysteinyl thioester to form a 2-hydroxyphenylthiazolinyl thioester. Epimerisation of the cysteine-derived stereocentre in this intermediate is catalysed by the methyl transferase-like (MTe) domain of PchE. The A domain of PchF then loads a second molecule of L-cysteine onto its PCP domain and the Cy domain catalyses condensation of the resulting cysteinyl thioester with the PchE-bound 2-hydroxyphenylthiazolinyl thioester to generate a second thiazoline, which is reduced to the corresponding thiazolidine by PchG and N-methylated by the MT domain of PchF. Hydrolytic release of pyochelin 3 from PchF is catalysed by the C-terminal thioesterase (TE) domain. PchC is a type II TE that removes incorrectly loaded molecules from the PCP domain of PchE and PchF.18
Enantiopyochelin 5 biosynthesis in P. protegens involves homologues of PchABCDEF (Fig. 2).9 The PchE homologue lacks the MTe domain responsible for epimerisation of the stereocentre in the first thiazoline ring and a homologue of pchG, which encodes the thiazoline reductase utilized by P. aeruginosa, is not present in the enantiopyochelin biosynthetic gene cluster. Instead, the product of pchK is proposed to catalyse reduction of the second thiazoline after the configuration of the C-4′′ stereocentre has been inverted by a hitherto unidentified enzyme. The absence of the MTe domain in the PchE homologue and the substitution of PchG with PchK are together able to account for the production of enantiopyochelin 5 by P. protegens.
The plant pathogen Streptomyces scabies 87.22 has also been shown to produce pyochelin 3 in an iron-deficient medium.19 The gene cluster responsible for its biosynthesis encodes homologues of PchCDEFG (a single enzyme encoded by the scab1381 gene is proposed to be responsible for salicylate biosynthesis in S. scabies) (Fig. 2).19 Transcription of the gene cluster is repressed by a TetR-family protein encoded by scab1401 and activated by an AfsR-family protein encoded by scab1371.19
Here we report the identification of a putative 2-hydroxyphenylthiazoline biosynthetic gene cluster in the genome of Streptomyces venezuelae ATCC 10712 by comparative transcriptional analyses of the wild type strain and a bldM mutant. The main metabolic products of this cluster were identified as the known 2-hydroxyphenyl-thiazolines thiazostatin 6 and watasemycin 7, and a novel metabolite isopyochelin 13via a heterologous expression approach. Gene deletion experiments defined an essential role for a PchK homologue in the biosynthesis of all metabolic products of the cluster, and showed that a type B radical-SAM methylase homologue is responsible for methylation of the thiazoline ring in thiazostatin 6 to form watasemycin 7.
Results and discussion
Microarray analysis of S. venezuelae bldM mutant
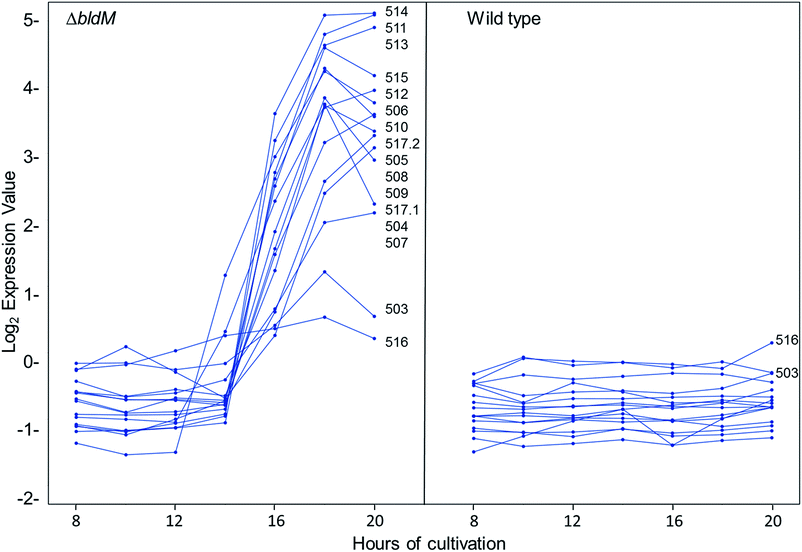
We recently reported the results of DNA microarray analyses of S. venezuelae, which showed that the transcription of the chloramphenicol biosynthetic gene cluster is markedly increased in a bldM mutant relative to the wild type strain.20 The bldM gene encodes an atypical orphan response regulator required for morphological differentiation.21 Analysis of the same microarray data set revealed another cluster of co-ordinately regulated genes, sven0503–sven0517, that is up-regulated in the bldM mutant (Fig. 3). The transcriptional profiles of the flanking genes sven0498–sven0502 and sven0519–sven0527 (sven0518 was not represented on the microarray), which showed basal levels of expression throughout growth in both the wild type strain and the bldM mutant (see ESI†), indicated that they are unlikely to be functionally related to sven0503–sven0517 (note also that a leucyl-tRNA gene, currently lacking a sven designation, lies between sven0517 and sven0518, further suggesting that sven0517 lies at the right hand end of the gene cluster).Sequence analysis of the sven0503–sven0517 gene cluster
Most of the sven0503–sven0517 genes encode proteins with >50% similarity to those encoded by genes within the S. scabies pyochelin biosynthetic gene cluster (Fig. 4 and ESI†). Sven0510, Sven0512 and Sven0517 are homologues of Scab1411 (PchD), Scab1481 (PchE) and Scab1471 (PchF), respectively. Sven0511 is a homologue of the proofreading type II TE Scab1421 (PchC) and Sven0506 is homologous to the putative salicylate synthase encoded by scab1381. No homologues of PchG can be found in the S. venezuelae genome. However, Sven0516 is 47% similar to PchK from P. protegens CHA0, which has been proposed to function as a thiazoline reductase in enantiopyochelin biosynthesis.9 Moreover, Sven0508 is 48% similar to Sven0516, suggesting it may also be able to function as a thiazoline reductase.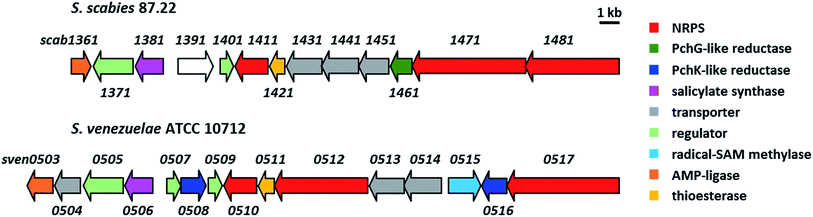 | ||
| Fig. 4 Comparison of the organisation of the watasemycin and pyochelin biosynthetic gene clusters in S. venezuelae ATCC 10712 and S. scabies 87.22, respectively. | ||
Interestingly, a homologue of sven0515, which encodes a putative type B radical-SAM methylase, cannot be found in any 2-hydroxyphenylthiazoline biosynthetic gene cluster reported to date. Such enzymes are known to catalyse the methylation of unactivated carbon centres in the biosynthesis of a variety of specialised metabolites.22 Taken together, these analyses suggested that the sven0503–sven0517 gene cluster may direct the biosynthesis of a C-methylated pyochelin derivative.
Sven0507/Sven0509 and Sven0505 are highly similar to the TetR-like Scab1401 repressor and the AfsR family Scab1371 activator, respectively, of pyochelin biosynthesis in S. scabies. Similarly, Sven0513 and Sven0514 are homologues of the putative ABC transporter ATPase/permease fusions encoded by scab1431 and scab1441, respectively, which are likely responsible for pyochelin export. No obvious role can be postulated for the Scab1361 homologue Sven0503, a putative AMP ligase, or Sven0504, a putative Na+/H+ antiporter that is not homologous to any of the proteins encoded by the S. scabies pyochelin biosynthetic gene cluster.
Expression of the gene cluster in S. coelicolor M1152
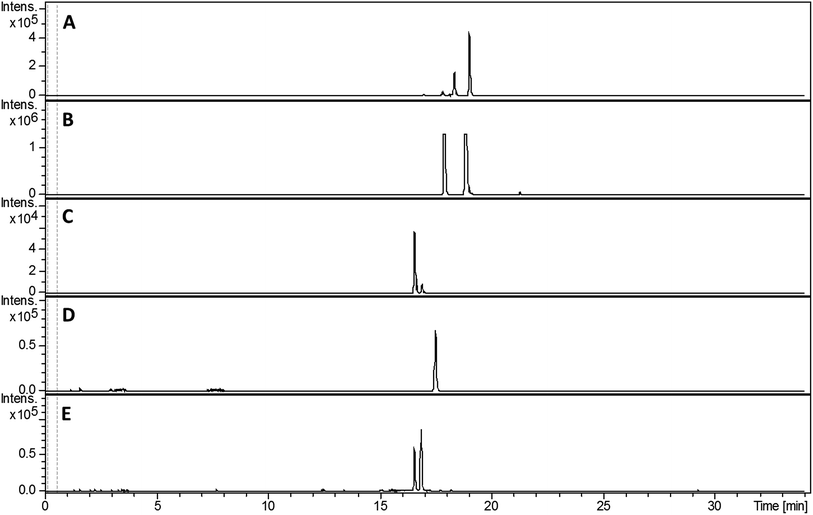
Despite screening various growth media, we failed to detect the production of any 2-hydroxyphenylthiazoline-containing metabolites by S. venezuelae. This is potentially explained by the low levels of sven0516 expression in the bldM mutant (Fig. 3), which is surprising given that sven0517 is likely to be in the same operon, and possibly reflects differential mRNA stability for the two genes. We therefore elected to express the sven0503–sven0517 gene cluster in the engineered host S. coelicolor M1152.23 A clone (SV-2_E03) from an ordered genomic cosmid library of the S. venezuelae chromosome containing a segment extending from sven0496 to sven0518 was PCR-targeted in Escherichia coli with a 5.2 kb SspI fragment from pIJ10702 that contains oriT, and the øC31 integrase gene and phage attachment site (attP). The resulting cosmid, SV-2_E03::SspI, was introduced into S. coelicolor M1152 by conjugation, whereupon it integrated into the chromosomal øC31 attB site. Wild type S. coelicolor M1152 and the SV-2_E03::SspI derivative were incubated in YD medium for five days and the culture broths were extracted with ethanol. UHPLC-ESI-Q-TOF-MS analyses identified metabolites giving rise to ions with m/z = 339.0840, 353.0983, 325.0675, 210.0588 and 224.0736, corresponding to [M + H]+ for the known 2-hydrophenylthiazolines thiazostatin 6 (m/z calculated for C15H19N2O3S2+: 339.0832), watasemycin 7 (m/z calculated for C16H21N2O3S2+: 353.0988), pyochelin 3 (m/z calculated for C14H17N2O3S2+: 325.0675) aerugine 4 (m/z calculated for C10H12NO2S+: 210.0583) and pulicatins A/B 8/9 (m/z calculated for C11H14NO2S+: 224.0740), respectively, in the culture extract of S. coelicolor M1152/SV-2_E03::SspI (Fig. 5). These metabolites were not present in the culture extracts of the unmodified host. Several other liquid growth media, including TSB, ISP2 and GSP, were also found to support production of these compounds.Structure elucidation of metabolic products
Metabolites were isolated from the culture broths using Diaion HP-20 resin, eluting with methanol. After ethyl acetate extraction and HPLC separation, the fractions containing the compounds with molecular formulae corresponding to thiazostatin and watasemycin were collected and analysed by 1H NMR spectroscopy (see ESI†). The spectroscopic data were identical to those previously reported for thiazostatins A and B 6, and watasemycins A and B 7, isolated from Streptomyces tolurosus and Streptomyces sp. TP-A0597, respectively.A mixture of pyochelin 3 and neopyochelin (the C-4′ epimer of pyochelin) was synthesised according to a literature procedure.24 Surprisingly, none of these four diastereomeric compounds had the same retention time as the metabolites with molecular formulae corresponding to pyochelin 3 (see ESI†). We thus hypothesised that these metabolites are isomers of pyochelin 3 in which C-4′′ is methylated instead of the nitrogen atom of the thiazolidine. To test this hypothesis, an authentic standard of the C-4′′ methylated isomer of pyochelin (hereafter referred to as isopyochelin) was synthesised as a mixture of four diastereomers via condensation of the known 2-hydroxyphenylthiazoline aldehyde 14 with L-2-methylcysteine (Scheme 1).
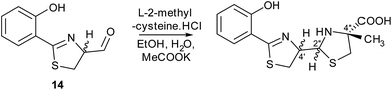 | ||
| Scheme 1 Synthesis of an authentic standard of (4′′R)-isopyochelin as a mixture of stereoisomers at C-4′ and C-2′′. | ||
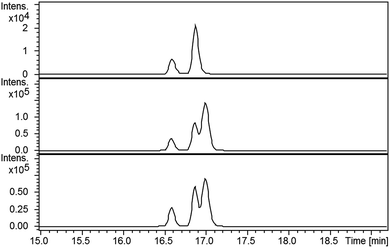
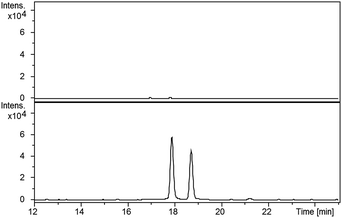
LC-MS comparisons showed that two of the synthetic isopyochelin diastereomers had the same retention time as the metabolites in the culture extract (Fig. 6). The absolute configuration of C-4′′ in the natural products was established as S by condensing 2-hydroxyphenylthiazoline aldehyde 14 with D-2-methylcysteine and comparing the retention times of the metabolites in the extract with synthetic (4′′R) and (4′′S)-isopyochelins derived from L- and D-2-methylcysteine, respectively (see ESI†). Based on the assumption that natural isopyochelin and thiazostatin 6 have the same relative stereochemistry (see below), the structure of the former was assigned as 13 (Fig. 1).
Biosynthetic role of sven0508 and sven0516
The observation that the sven0503–sven0517 gene cluster contains two genes encoding PchK-like putative thiazoline reductases intrigued us, because only a single thiazoline reduction appears to be involved in the biosynthesis of thiazostatin 6, watasemycin 7 and isopyochelin 13. To investigate the role played by sven0508 and sven0516 in the biosynthesis of 6, 7 and 13, individual in-frame deletions of these genes were created in SV-2_E03 by PCR targeting. The mutagenized cosmids were subsequently targeted with the 5.2 kb SspI fragment from pIJ10702 and then introduced into S. coelicolor M1152 from E. coli ET12567/pUZ8002 by conjugation.Ethanol extracts of culture broths from the sven0508 and sven0516 mutants were analysed by LC-MS. Aerugine 4, thiazostatin 6, watasemycin 7, pulicatins A/B 8/9 and isopyochelin 13 were all still observed in the extract from the sven0508 mutant (see ESI†). On the other hand, none of these metabolites could be detected in the extract from the sven0516 mutant (see ESI†). Thus we conclude that Sven0516 plays an essential role in the biosynthesis of all the metabolic products of the sven0503–sven0517 gene cluster.
Revised stereochemistry of watasemycin and thiazostatin
The finding that a PchK homologue plays an essential role in assembling the metabolic products of the sven0503–sven0517 gene cluster is in accord with our assignment of the C-4′′ configuration of isopyochelin 13 as S. Assuming isopyochelin 13 and watasemycin 7 have the same C-4′′ absolute configuration, the relative stereochemistry previously assigned to watasemycin on the basis of NOE NMR studies suggests its C-5′ and C-4′ stereocentres should both be R-configured.However, the 5′R stereochemical assignment conflicts with the absolute stereochemistry proposed, on the basis of NOESY, Mosher's ester and CD studies, for the corresponding stereogenic centre in pulicatins A and B 8 and 9.12 The C4′ hydrogen substituent and the C5′ methyl group in watasemycin 7 are proposed to be anti to each other on the basis of NOE studies.11 We thus suspect that the relative configuration of watasemycin 7 has been misassigned and hypothesize that the correct stereochemical assignment is 4′S, 5′S, 4′′S (as shown in Fig. 1). By analogy, we propose that the stereochemistry of thiazostatin 6 is 4′S, 4′′S. Although further experiments will be required to confirm these stereochemical reassignments, it is noteworthy that the MTe domain of the PchE homologue Sven0512 appears, on the basis of conserved domain searches,25 to be non-functional. This is consistent with the 4′S configuration, resulting from net incorporation of L-cysteine into the thiazoline ring of thiazostatin 6, watasemycin 7 and isopyochelin 13, as in enantiopyochelin 5 biosynthesis (Fig. 2).9
Sven0515 C-methylates the thiazoline of thiazostatin
Compared with pyochelin 3, watasemycin 7 contains two additional methyl groups at C-5′ and C-4′′ (Fig. 1). Based on its similarity to type B radical-SAM methylases,21 Sven0515 could be responsible for introducing one or both of these. We therefore deleted sven0515 from SV-2_E03, introduced the 5.2 kb SspI fragment from pIJ10702 and transferred the resulting construct into S. coelicolor M1152, as described above. LC-MS analysis of ethanol extracts from the culture broth of this strain showed that it is unable to produce watasemycin 7 or pulicatins A/B 8/9 (Fig. 7). Thus, Sven0515 appears to be involved in the methylation of C-5′, but not C-4′′. To investigate the timing of C-5′ methylation, we fed the metabolites in the extract of the sven0515 mutant to the sven0516 mutant. This resulted in complete conversion of the thiazostatin 6 in the extract to watasemycin 7 (Fig. 8), indicating that thiazostatin is the substrate of Sven0515.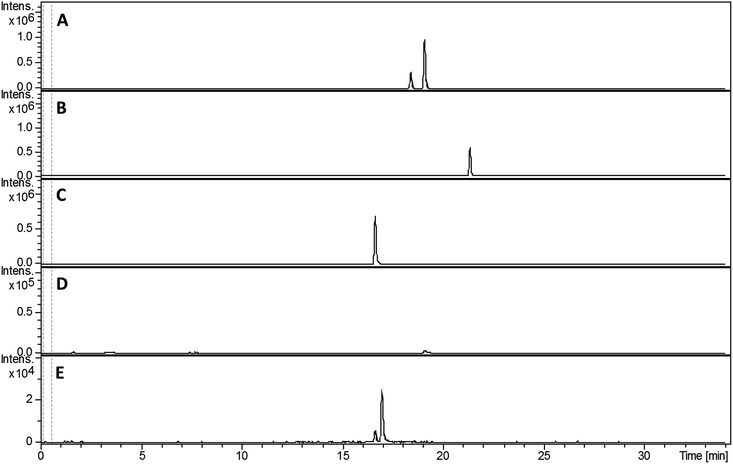 | ||
| Fig. 7 Extracted ion chromatograms at m/z = 339.0832 (A), 353.0988 (B), 210.0583 (C), 224.0740 (D) and 325.0675 (E), corresponding to [M + H]+ for thiazostatin, watasemycin, aerugine, pulicatin and isopyochelin, respectively, from LC-MS analyses of the ethanol extract of S. coelicolor M1152/SV-2_E03::SspI/Δsven0515 culture broth. Two peaks are observed for thiazostatin and isopyochelin because they exist as a mixture of diastereomers resulting from epimerisation at C-2′′. The peak with a retention time of approximately 21.5 minutes in the m/z = 353.0988 chromatogram (B) appears, on the basis of LC-MS/MS analyses, to be due to the methyl ester of thiazostatin (see ESI†). | ||
Proposed pathway for watasemycin biosynthesis
Taken together, the above data lead us to propose a pathway for the biosynthesis of watasemycin 7 in S. venezuelae (Fig. 9). The early stages of this pathway, up to the formation of the 2-hydroxyphenyl-bis-thiazolinyl thioester intermediate attached to the PCP domain of Sven0517, mirror the biosynthesis of enantiopyochelin 5 (Fig. 2). At this point the MT domain of Sven0517 appears to catalyse methylation of C-4′′ (Fig. 9). An analogous transformation has been shown to occur in the biosynthesis of yersiniabactin.26 Sven0516 catalyses thiazoline reduction and the MT domain of Sven0517 then methylates the nitrogen atom of the resulting thiazolidine, as in pyochelin biosynthesis.16 Hydrolysis of the thioester by the TE domain affords thiazostatin 6, which is methylated at C-5′ by Sven0515 to give watasemycin 7. Isopyochelin 13 presumably results from TE-catalysed thioester hydrolysis prior to the N-methylation reaction, suggesting that this may be a slow step in thiazostatin 6 biosynthesis. The biosynthetic origins of aerugine 4 and pulicatins A/B 8/9 are unclear, but it seems likely that they arise from hydrolytic cleavage and subsequent reduction of thiazostatin 6/isopyochelin 13 and watasemycin 7, respectively.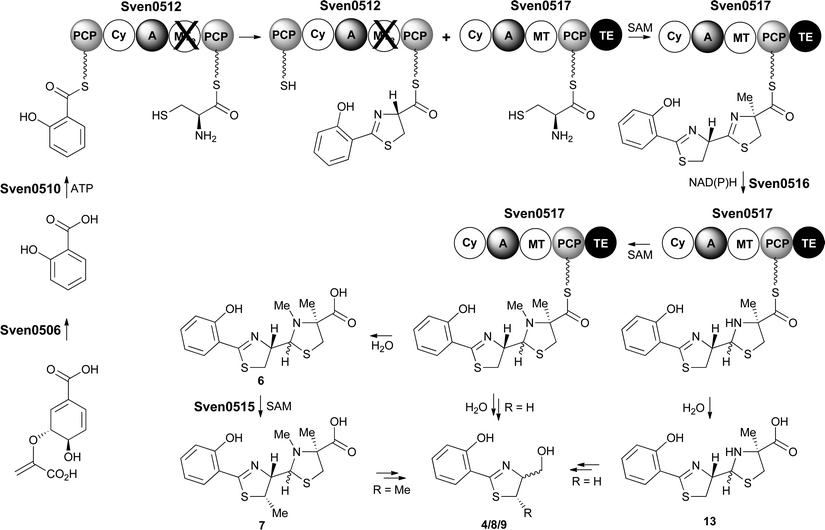 | ||
| Fig. 9 Proposed pathway for the biosynthesis of thiazostatin 6, watasemycin 7, isopyochelin 13, aerugine 4 and pulicatins A/B 8/9 in S. venezuelae. | ||
Conclusions
We have identified the known Streptomyces metabolites aerugine 4, thiazostatin 6, watasemycin 7 and pulicatins A/B 8/9, and the novel natural product isopyochelin 13 as the metabolic products of a cryptic 2-hydroxyphenylthiazoline biosynthetic gene cluster in S. venezuelae ATCC10712 using a heterologous expression approach. The absolute stereochemistry of isopyochelin 13 was assigned as 4′′S by comparison with synthetic standards. In combination with the absolute and relative stereochemistry reported for pulicatins A and B 8 and 9,12 previous investigations of the relative stereochemistry of watasemycin 7,11 and biosynthetic considerations, this prompted us to assign the stereochemistry of watasemycin 7 and thiazostatin 6 as 4′S, 5′S, 4′′S and 4′S, 4′′S, respectively. Gene deletion experiments established an essential role for the PchK homologue Sven0516 in the biosynthesis of all the metabolic products of the gene cluster and showed that the type B radical-SAM methylase homologue encoded by sven0515 is responsible for the conversion of thiazostatin 6 to watasemycin 7. Type B radical-SAM methylases are known to catalyse methylation of unactivated carbon centres in the biosynthesis of several different classes of natural products, including aminoglycosides, β-lactams, phosphonates, and ribosomally biosynthesised and posttranslationally-modified peptides.22 However, to the best of our knowledge the Sven0515-catalysed methylation of thiazostatin 6 is the first experimentally-validated example of such a reaction in the biosynthesis of a nonribosomal peptide.Acknowledgements
This research was supported by a Chancellor's International Scholarship from the University of Warwick (to S. Z.), and the BBSRC through the MIBTP doctoral training partnership (BB/J014532/1) and an Institute Strategic Programme Grant “Understanding and Exploiting Plant and Microbial Secondary Metabolism” (BB/J004561/1). The Dionex 3000RS/Bruker MaXis Impact instrument used in this work was purchased with a grant from the BBSRC (BB/K002341/1).Notes and references
- M. L. Guerinot, Annu. Rev. Microbiol., 1994, 48, 743–772 CrossRef CAS PubMed.
- J. H. Crosa and C. T. Walsh, Microbiol. Mol. Biol. Rev., 2002, 66, 223–249 CrossRef CAS PubMed.
- M. Miethke and M. A. Marahiel, Microbiol. Mol. Biol. Rev., 2007, 71, 413–451 CrossRef CAS PubMed.
- H. Drechsel, H. Stephan, R. Lotz, H. Haag, H. Zähner, K. Hantke and G. Jung, Liebigs Ann., 1995, 10, 1727–1733 CrossRef.
- L. a. Actis, W. Fish, J. H. Crosa, K. Kellerman, S. R. Ellenberger, F. M. Hauser and J. Sanders-Loehr, J. Bacteriol., 1986, 167, 57–65 CrossRef CAS PubMed.
- M. A. F. Jalal, M. B. Hossain, D. van der Helm, J. Sanders-Loehr, L. A. Actis and J. H. Crosa, J. Am. Chem. Soc., 1989, 111, 292–296 CrossRef CAS.
- C. D. Cox, K. L. Rinehart, M. L. Moore and J. C. Cook, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 4256–4260 CrossRef CAS.
- A. Zunnundzhanov, I. A. Bessonova, N. D. Abdullaev and D. K. Ogai, Chem. Nat. Compd., 1987, 23, 461–465 CrossRef.
- Z. A. Youard, G. L. A. Mislin, P. A. Majcherczyk, I. J. Schalk and C. Reimmann, J. Biol. Chem., 2007, 282, 35546–35553 CrossRef CAS PubMed.
- K. Shindo, A. Takenaka, T. Noguchi, Y. Hayakawa and H. Seto, J. Antibiot., 1989, 42, 1526–1529 CrossRef CAS PubMed.
- T. Sasaki, Y. Igarashi and T. Furumai, J. Antibiot., 2002, 55, 249–255 CrossRef CAS PubMed.
- Z. Lin, R. R. Antemano, R. W. Hughen, M. D. B. Tianero, O. Peraud, M. G. Haygood, G. P. Concepcion, B. M. Olivera, A. Light and E. W. Schmidt, J. Nat. Prod., 2010, 73, 1922–1926 CrossRef CAS PubMed.
- L. Serino, C. Reimmann, H. Baur, M. Beyeler, P. Visca and D. Haas, Mol. Gen. Genet., 1995, 249, 217–228 CrossRef CAS PubMed.
- L. Serino, C. Reimmann, P. Visca, M. Beyeler, V. D. Chiesa and D. Haas, J. Bacteriol., 1997, 179, 248–257 CrossRef CAS PubMed.
- L. E. N. Quadri, T. A. Keating, H. M. Patel and C. T. Walsh, Biochemistry, 1999, 38, 14941–14954 CrossRef CAS PubMed.
- H. M. Patel and C. T. Walsh, Biochemistry, 2001, 40, 9023–9031 CrossRef CAS PubMed.
- C. Reimmann, H. M. Patel, L. Serino, M. Barone, C. T. Walsh and D. Haas, J. Bacteriol., 2001, 183, 813–820 CrossRef CAS PubMed.
- C. Reimmann, H. M. Patel, C. T. Walsh and D. Haas, J. Bacteriol., 2004, 186, 6367–6373 CrossRef CAS PubMed.
- R. F. Seipke, L. Song, J. Bicz, P. Laskaris, A. M. Yaxley, G. L. Challis and R. Loria, Microbiology, 2011, 157, 2681–2693 CrossRef CAS PubMed.
- L. T. Fernández-Martínez, C. Borsetto, J. P. Gomez-Escribano, M. J. Bibb, M. M. Al-Bassam, G. Chandra and M. J. Bibb, Antimicrob. Agents Chemother., 2014, 58, 7441–7450 CrossRef PubMed.
- M. M. Al-Bassam, M. J. Bibb, M. J. Bush, G. Chandra and M. J. Buttner, PLoS Genet., 2014, 10, e1004554 Search PubMed.
- S. Zhou, L. M. Alkhalaf and G. L. Challis, Curr. Opin. Chem. Biol., 2016, 35, 73–79 CrossRef CAS PubMed.
- J. P. Gomez-Escribano and M. J. Bibb, Microb. Biotechnol., 2011, 4, 207–215 CrossRef CAS PubMed.
- A. Zamri and M. A. Abdallah, Tetrahedron, 2000, 56, 249–256 CrossRef CAS.
- A. Marchler-Bauer, M. K. Derbyshire, N. R. Gonzales, S. Lu, F. Chitsaz, L. Y. Geer, R. C. Geer, J. He, M. Gwadz, D. I. Hurwitz, C. J. Lanczycki, F. Lu, G. H. Marchler, J. S. Song, N. Thanki, Z. Wang, R. A. Yamashita, D. Zhang, C. Zheng and S. H. Bryant, Nucleic Acids Res., 2015, 43, D222–D226 CrossRef PubMed.
- D. A. Miller, C. T. Walsh and L. Luo, J. Am. Chem. Soc., 2001, 123, 8434–8435 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures and additional tables, figures, chromatograms and results from biological assays. See DOI: 10.1039/c6sc03533g |
| ‡ These authors contributed equally. |
| § Current address: Department of Bioengineering, University of California, San Diego, USA. |
| This journal is © The Royal Society of Chemistry 2017 |