Optical nano-biosensing interface via nucleic acid amplification strategy: construction and application
Hong
Zhou†
a,
Jing
Liu†
a,
Jing-Juan
Xu
 *b,
Shu-Sheng
Zhang
*b,
Shu-Sheng
Zhang
 *a and
Hong-Yuan
Chen
b
*a and
Hong-Yuan
Chen
b
aShandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi 276005, China. E-mail: shushzhang@126.com
bKey Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, China. E-mail: xujj@nju.edu.cn
First published on 15th February 2018
Abstract
Modern optical detection technology plays a critical role in current clinical detection due to its high sensitivity and accuracy. However, higher requirements such as extremely high detection sensitivity have been put forward due to the clinical needs for the early finding and diagnosing of malignant tumors which are significant for tumor therapy. The technology of isothermal amplification with nucleic acids opens up avenues for meeting this requirement. Recent reports have shown that a nucleic acid amplification-assisted modern optical sensing interface has achieved satisfactory sensitivity and accuracy, high speed and specificity. Compared with isothermal amplification technology designed to work completely in a solution system, solid biosensing interfaces demonstrated better performances in stability and sensitivity due to their ease of separation from the reaction mixture and the better signal transduction on these optical nano-biosensing interfaces. Also the flexibility and designability during the construction of these nano-biosensing interfaces provided a promising research topic for the ultrasensitive detection of cancer diseases. In this review, we describe the construction of the burgeoning number of optical nano-biosensing interfaces assisted by a nucleic acid amplification strategy, and provide insightful views on: (1) approaches to the smart fabrication of an optical nano-biosensing interface, (2) biosensing mechanisms via the nucleic acid amplification method, (3) the newest strategies and future perspectives.
1. Introduction
The ultrasensitive detection of biomolecules (e.g. DNA or proteins) or cells has become increasingly important in the fields of food detection, clinical diagnostics, environmental monitoring, and anti-bioterrorism.1 In recent years, with the development of interface modification technology, nanotechnology, molecule assembly techniques, and molecular recognition technology, a variety of detection technologies have been developed and used in the detection of biological molecules. Modern optical-detection techniques with their inherent advantages of simplicity, convenience and sensitivity have been widely applied to detect biomolecules. A variety of optical-detection methods, including fluorescence,2 surface-enhanced Raman scattering,3 chemiluminescence,4 colorimetric assays,5 surface plasmon resonance (SPR),6 and electrochemiluminescence (ECL),7 have been designed for biosensing with a wide dynamic range and high sensitivity. In particular, enormous attention has been paid to the development of signal amplification strategies using optical signal probes in bioassays in order to realize excellent performance for the detection of low-abundance analytes. A series of amplification strategies or principles have been developed, such as bio-functional nanomaterials8 or the isothermal amplification of nucleic acids.9 Here, an amplification strategy assisted by isothermal amplification of oligonucleotides has already become a powerful tool for efficient and smart signal amplified detection under gentle and constant temperature, such as the hybridization chain reaction (HCR), rolling-circle amplification (RCA), the strand displacement reaction (SDR), loop-mediated amplification (LAMP), and target-recycling amplification with endonuclease, exonuclease and polymerase. The combination of nucleic acid isothermal amplification and optical-detection technology has also attracted intense attention. For example, nucleic acid isothermal amplification-assisted optical interfaces have been widely employed in recent biosensing due to their excellent stability, sensitivity, and designability, all of which demonstrate good prospects for clinical applications.Nucleic acid amplification methods commonly occur in a homogeneous system. However, in the face of complex detection samples, including human serum, saliva and urine, it is easy to be influenced by cross-contamination during the detection. Thus the accuracy of these methods becomes a real problem through enhanced sensitivity via signal amplification. Moreover, in situ detection of intracellular target analytes is also a big challenge because it is hard for nucleic acids to enter into cells without transfection reagents or nanomaterials. To obtain highly selective and sensitive tests for biomolecules in complex biological microenvironments, various reaction interfaces based on nucleic acid amplification reactions, including macro-reaction and micro-reaction interfaces, were combined with an optical system for fabricating a series of novel detection methods. Macro-reaction interfaces, like electrodes, glass slides, microwell plates, all kinds of chips, etc., as general substrates for all kinds of amplification reactions, have been applied in optics-based nucleic acid isothermal amplification reactions. These macro-reaction interfaces, demonstrating many unique advantages, such as visual inspection, ease of operation, simple separation, as well as sensitivity and specificity, have been widely developed for in vitro detection. Their only deficiency is their larger size that limits their application in intracellular or in vivo analysis and detection. Compared with macro-reaction interfaces, micro-reaction interfaces like noble metal nanoparticles, inorganic nanomaterials, magnetic nanoparticles, graphene and novel two-dimensional (2D) layered nanomaterials analogous to graphene, etc., with large surface areas, good biocompatibility, large loading capacity and high stability, have also been widely applied in optics-based nucleic acid isothermal amplification reactions. Benefiting from these advantages, in situ sensing detection was successfully realized, which greatly extended its clinical application. Up to now, numerous studies related to nucleic acid assisted isothermal amplification have been published. However, the recent smart construction, application and development of nucleic acid amplification technology on modern optical nano-biosensing interfaces have not been comprehensively reviewed.
In this review, as described in Fig. 1, we first discuss recent representative work on the construction and application of macro-reaction interfaces for optical biosensing methods, including fluorescence, surface-enhanced Raman scattering, chemiluminescence, colorimetric assays, electrochemiluminescence (ECL) and surface plasmon resonance (SPR) assisted by diverse isothermal amplification strategies. Then we summarize advances in nanomaterial based micro-reaction interfaces via a nucleic acid amplification strategy for the biosensing of various clinical analytes. These are classified into six sections: five different types of nanomaterial supported micro-reaction interfaces and special situation-cell surface interfaces. Finally, the challenges and outlook of this field for clinical detection will be discussed.
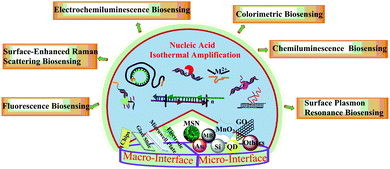 | ||
| Fig. 1 Schematic illustration of various nucleic acid isothermal amplification assisted optical biosensing methods on macro-reaction interfaces and micro-reaction interfaces. | ||
2. Nucleic acid amplification assisted optical macro-reaction interfaces for biosensing
Recently, optical detection methods have been integrated with various isothermal amplification reactions to construct a series of analysis interfaces and to fabricate novel analysis strategies, which have achieved remarkably sensitive and selective detection of tumor markers.10–13 Some optical analysis systems, such as surface-enhanced Raman scattering (SERS), electrochemiluminescence (ECL), colorimetric assays, fluorescence and surface plasmon resonance (SPR), have usually used the macro-reaction interfaces as carriers for biosensing assisted by all kinds of isothermal amplification reactions. These macro-reaction interfaces include electrodes, glass slides, microwell plates, and all kinds of chips. Here we review different types of nucleic acid amplification strategies for optical biosensing based on macro-reaction interfaces.2.1 Rolling-circle amplification (RCA) assisted optical macro-reaction interfaces for biosensing
Rolling circle amplification (RCA) is a simple and efficient in vitro DNA amplification technology, imitating internal rolling-circle replication and generating a large number of repetitive sequences of target sequences.14 In the presence of DNA polymerase, the primer would extend and generate a long ssDNA product with thousands of repeated complementary sequences on the circular template. Linear RCA could generally achieve approximately 103-fold amplification in a short period, and the number could be further increased with the addition of a co-reagent DNA or protein. Therefore, it has been widely utilized in the diagnostic field for the detection of various DNA, RNA, or proteins combined with electrochemical or optical testing techniques.9Fig. 2A illuminates the principle of the classical RCA reaction. Both ends of a single-stranded probe DNA (green) are potentially complementary to a target DNA sequence (orange). With the target DNA and a DNA ligase, the two ends are connected to form circular DNA, which then serves as the template for an RCA reaction to produce a long ssDNA. This long RCA product as an amplification transducer can amplify an output signal over thousands of times and has become an ideal analytical method for the ultrasensitive and selective optical detection of various targets.
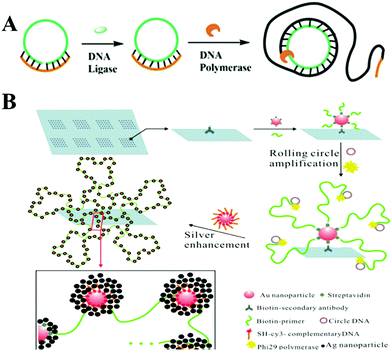 | ||
| Fig. 2 Schematic illustration of RCA strategy and RCA-based sensitive SERS assays. (A) Schematic illustration of RCA strategy. (B) SERS assays based on nano rolling-circle amplification (left) and nano hyperbranched rolling circle amplification (right) for protein microarrays. Reprinted with permission from ref. 20. Copyright (2012) American Chemical Society. | ||
In SERS assays, gold–silver staining methods were usually used for Raman scattering enhancement, attributed to the electromagnetic effect (“hot spots”) arising from the Ag nanoparticles grown around the fluorophore-labeled Au NPs.19 In order to increase these “hot spot” groups, Yan and co-workers reported a SERS sensing method employing an aldehyde-activated chip for protein microarrays with the help of nano RCA and nano hyperbranched rolling circle amplification (HRCA). As depicted in Fig. 2B, streptavidin-combined Au NPs (SA–Au NPs) were previously bound to the protein, then biotin modified primers bonded to SA–Au NPs and simultaneously initiated the RCA reaction. Thus, the intensity of the Raman signal was detected for target protein analysis, with a detection limit lower than 10 zeptomolar.20 Considering that HRCA could create longer ssDNA and thus be more efficient than RCA, they further designed a nanoHRCA strategy to achieve a much stronger SERS signal for protein microarrays than nanoRCA. Besides nucleic acids and proteins, some pathogenic microorganisms could also be sensitively detected using this RCA-assisted SERS assay method. For example, Chen's group immobilized antibodies on a 96-well microplate to capture target V. parahaemolyticus. Then an aptamer ssDNA also acting as the primer for the RCA process was recognized via the binding effect. A single-strand DNA probe labeled with silver coated gold nanoparticles (Au@Ag) hybridizes with the RCA products and forms a long Au@Ag/RCA product structure for an extraordinarily enhanced Raman response for V. parahaemolyticus in food samples.21 There are also some problems with the SERS method, including difficulty in quantification and the low reproducibility of SERS substrates. There is a long way to go to develop highly reproducible SERS platforms for commercially available analysis.
In addition to the above-mentioned microplate slides used in colorimetric biosensing platforms, portable microchips were further developed to satisfy the requirement for rapid and effective point-of-care (POC) diagnostics. RCA based colorimetric assays could also be realized for the highly sensitive detection of proteins or bacterial pathogens on portable microchips, as reported by Lin's group25 and Lee's group.26 Recently, Li's group designed a portable, low-cost, low-volume, disposable, but more robust and sensitive “all-in-one” paper-based POC diagnostic system for the detection of DNA and microRNA combined with isothermal RCA techniques.27 As Fig. 3A describes, a preformed 5′-biotinylated DNA–streptavidin conjugate was printed onto a nitrocellulose membrane surface which successfully performed the RCA reaction on a patterned paper device. Distinctive features of this work were the printing of DNA primers efficiently onto a macro-paper surface and the production of a 96-microzone paper device, providing high potential for high-throughput assays. This work greatly exploited application fields of POC testing in resource-limited settings, which was particularly suitable for early clinical diagnosis with simplified steps, and no additional reagent handling. Colorimetric assays, with their advantages of simplicity, speediness and perceptual intuition, showed excellent prospects for rapid and effective clinical point-of-care (POC) diagnostics. Two problems with this method are listed below. One is how to further improve detecting sensitivity so it is high enough for the naked eye to distinguish targets of low concentrations. The second problem is how to retain the catalytic activity of enzymes used in colorimetric assays under the unstable situation of transportation and complex real samples. Further studies of novel artificial nanoenzymes and signal amplification strategies may promote its development in POC diagnostics.28
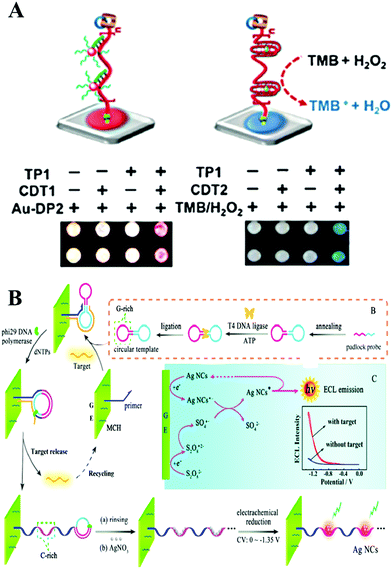 | ||
| Fig. 3 (A) Performing RCA with paper-bound primer colorimetric assay using Au NP–DNA conjugates (left), and colorimetric assay mediated by a peroxidase-mimicking DNAzyme (right). Reprinted with permission from ref. 27. Copyright (2016) Wiley-VCH. (B) Principle of target-cycling synchronized RCA and in situ electrochemical generation of Ag NCs, preparation of the circular template, and ECL mechanism of Ag NCs/S2O82−-based ECL system. Reprinted with permission from ref. 39. Copyright (2016) American Chemical Society. | ||
Further combined with a nucleic acid amplification strategy, ECL has attracted intense attention and led to simpler and more sensitive biomolecule analysis. And different sensor interfaces were built to detect different molecules, based on the combination of ECL and nucleic acid isothermal amplification. DNA was popular in constructing the biosensing interface due to its easy modification and electrical conductivity. The ECL signal was induced by the hybridization between DNA modified on ECL materials and DNA modified on the electrode. And the signal could be further amplified by nucleic acid isothermal amplification. Taking RCA amplification as an example, Su and co-workers reported a highly sensitive RCA-ECL biosensor for point mutation.36 In this work, an allele-discriminating padlock probe was designed for targeting the sequence in the p53 oncogene locus. Streptavidin-coated magnetic beads were employed as an immobilization matrix as well as a separation tool at the surface of a platinum electrode. Ru(bpy)32+ (TBR)-tagged probes were hybridized with hundreds of copies of the circular DNA template sequence, generating an enhanced ECL signal. A low to 2 amol detection limit of mutated strands was obtained by this assay. Due to antibody or aptamer technology, this powerful amplification strategy was applied in the detection of 0.15 fM H-IgG37 and 16 Ramos cells,38 respectively, with excellent selectivity, and also showed good performance in complex samples and whole blood samples. In a further combination with in situ electrochemical generation of silver nanoclusters as a novel ECL probe, Yuan's group subtly designed a circular template containing a guanine-rich (G-rich) region and a binding region for realizing target miR-21-cycling synchronized RCA (Fig. 3B). The DNA product of the RCA, which possessed cytosine-rich (C-rich) sequences, further formed a stable cytosine–Ag+–cytosine structure after incubation with Ag NO3 solution. Finally, in situ electrochemical generation of silver nanoclusters was realized in ECL signal probes for the rapid and ultrasensitive detection of miR-21 with a detection limit down to 22 aM.39 ECL is a burgeoning tool for biosensing. However, up to now, most nanoemitters still show a lower ECL efficiency compared with Ru(bpy)32+. Therefore, further studies of highly efficient, stable, and low-cost ECL luminophores are essential to the development of the ECL technique.35
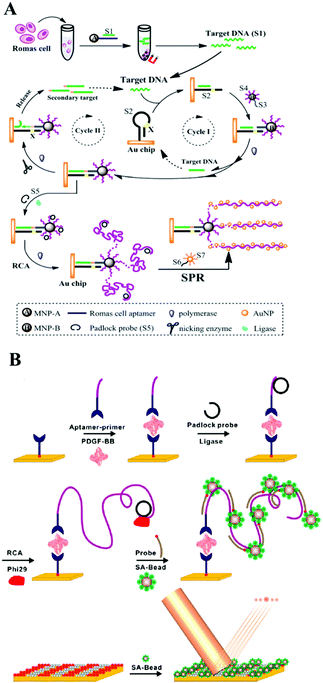 | ||
| Fig. 4 (A) Multiple signal amplification SPR assay for DNA and cancer cells. Reprinted with permission from ref. 52. Copyright (2014) Royal Society of Chemistry. (B) Schematic of RCA-based microbead diffractometric detection assay of PDGF-BB. Reprinted with permission from ref. 56. Copyright (2010) American Chemical Society. | ||
Generally, we find that optical approaches showed excellent performance as powerful tools for remarkably sensitive and selective detection of various samples, including DNA or RNA, proteins, and cancer cells with the assistance of nucleic acid isothermal amplification strategies. Further simple but powerful optical biosensors which meet the demands for clinical cost-effectiveness, and on-site and timely diagnosis will be established, combined with the already developed optical methods and novel nucleic acid amplification strategies.
A comparison of different RCA based optical sensing assays is listed in Table 1. Although all the strategies are based on RCA, they demonstrate different detection limits. Generally, the sensitivity of an optical sensing assay is related not only to optical methods but also to the protocols that integrate with other isothermal amplification techniques. Moreover, the sensing interfaces and modification conditions also greatly influence the detection limit.
| Target | Method | Sensing interface | Detection limit | Ref. |
|---|---|---|---|---|
| DNA | SERS | Probe-modified gold electrode | 10.0 pM | 17 |
| DNA | SERS | Gold slide | 6.3 fM | 18 |
| Protein | SERS | Aldehyde-activated chip | 10 zM | 20 |
| Vp | SERS | 96-well microplate | 1 cfu mL−1 | 21 |
| DNA methylation | Colorimetric | Microplate slide | 93 fM | 22 |
| Thrombin | Colorimetric | Microtiter plate | 1 fM | 23 |
| α-Fetoprotein | Colorimetric | 96-well microplate | 33.45 pg mL−1 | 24 |
| Thrombin | Colorimetric | Microchip | 0.083 pg mL−1 | 25 |
| Pathogen | Colorimetric | Microchip | 6 × 107 copies mL−1 | 26 |
| DNA | Colorimetric | 96-microzone paper device | 10 pM | 27 |
| DNA | ECL | Platinum electrode | 2 amol | 36 |
| Human IgG | ECL | Macroporous Au-paper electrode | 25 pM | 37 |
| Cell | ECL | 96 microtiter plates | 16 cells | 38 |
| MicroRNA | ECL | Gold electrode | 22 aM | 39 |
| PDGF | Fluorescence | Glass slides | 0.3 nM | 44 |
| ATP | Fluorescence | Glass slide | 1 μM | 45 |
| DNA, cells | SPR | Gold chip | 10 aM | 52 |
| DNA | SPR | Gold electrode | 44 aM | 53 |
| Thrombin | SPR | Gold chip | 0.78 aM | 54 |
| Cell | Chemiluminescence imaging | 96-well microtiter plat | 163 cells | 55 |
2.2 Nucleic acid-cleaving enzyme assisted optical macro-reaction interfaces for biosensing
Nucleic acid-cleaving enzymes, including endonuclease and exonuclease, could cleave the phosphodiester bonds of nucleic acids. In this part we will summarize nucleic acid-cleaving enzyme assisted optical biosensing methods on macro-reaction interfaces classified by different cleavage mechanisms. Traditional nucleic acid-cleaving enzymes could be divided into two types on their cleaving mechanism. One is exonucleases (Exos) and the other is endonucleases.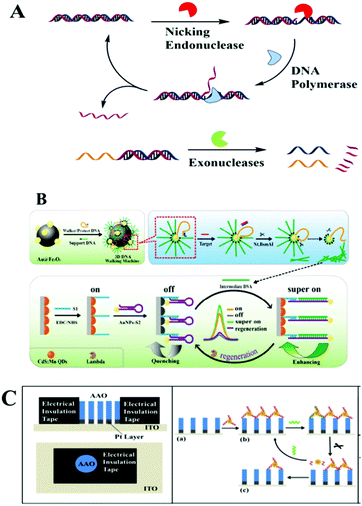 | ||
| Fig. 5 (A) Schematic illustration of the nucleic acid-cleaving enzymes strategy. (B) “On–off–super on” ECL biosensor. Reprinted with permission from ref. 61. Copyright (2017) American Chemical Society. (C) Nanopore-based ECL platform on nanopore-based electrode. Reprinted with permission from ref. 85. Copyright (2017) American Chemical Society. | ||
Benefiting from the unique character of NEase, which only catalyzes the hydrolysis of one strand of a duplex at specific recognition sites, many works further integrated it with strand displacement amplification (SDA) to further amplify the detection signals. The SDA isothermal amplification process needs a strand-displacing DNA polymerase and a nicking endonuclease. It starts with a polymerase extension reaction and creates a new complementary strand with a special nicking site, which would be cleaved by a NEase. With the help of DNA polymerase, the 3′-end of the nick was further extended, thus displacing the downstream strand and regenerating the nicking site. After the unidirectional amplification process of SDA, thousands of target copies were accumulated. For example, after a circular strand displacement polymerization reaction, a detection limit of 6.4 × 10−15 M for ssDNA targets could be achieved within 1000 s by designing target-specific hairpin fluorescent probes.63 SDA as an isothermal amplification approach is widely used for modern optical biosensing. For example, Xu's group employed this strategy in combination with energy transfer from CdS NCs to Au NPs and CdTe NPs, or Au NP enhanced ECL. Highly selective detection of DNA was presented with a low detection limit at aM level.64,65 Besides this ECL method, other methods including SERS, fluorescence detection, or chemiluminescence imaging arrays were also enhanced by an SDA cascade isothermal amplification strategy for the detection of DNA, RNA or protein.66–74
2.3 DNAzyme assisted optical macro-reaction interfaces for biosensing
DNAzymes are DNA molecules generally containing a substrate strand and an enzyme strand. The substrate strand contains a single RNA linkage (rA) serving as a cleavage site, while the enzyme strand has distinct primary and secondary structures. In the presence of metal ions or amino acids acting as catalytic cofactors, the enzyme strand cleaved the substrate strand into two parts.87–89 Due to metal ions being required to activate DNAzymes, DNAzymes are very suitable for metal sensing using various types of DNAzymes.90 As the study progressed, the high recognition function and catalytic activity of DNAzyme were verified, and DNAzymes have been successfully developed in the construction of functional molecular probes for signal amplification. Compared to aptamers, DNAzymes often show higher selection efficiency. Therefore, more and more DNAzyme assisted molecular probes have been used in various optical methods, with significantly improved selectivity and simplicity. Generally, the reports of DNAzyme assisted optical biosensors on macro-reaction interfaces could be divided into two types. One is the construction of DNAzymes directly on the macro-reaction interface, including electrodes and chips. The other is the construction of target-induced DNAzymes in solution, when the product of the DNAzyme reaction was introduced onto the surface of the biosensing interfaces, generating different signal changes. Ju's group employed DNAzyme strand functionalized QDs acting as a capture probe and a reductive-oxidative ECL signal probe, luminol-reduced gold nanoparticles (Au@luminol) as an oxidative-reductive ECL signal probe, and a Cy5 modified Mg2+ substrate strand as the quencher, and they constructed an Mg2+-dependent DNAzyme-triggered ratiometric ECL biosensor for Mg2+ detection in HeLa cell extract.91 Gao and co-workers further introduced this DNAzyme assisted DNA recycling strategy into SERS cycling detection of thrombin combined with RCA.92 Although it is easy to operate, there are many suppression factors during this direct DNAzyme modification and reaction on biosensing interfaces, such as limits from the DNA sequence design. Two examples from Chai's group showed another DNAzyme based approach for troponin I and microRNA, which realized a target-induced cycling DNAzyme reaction in the solution, and then introduced the product of the DNAzyme reaction onto the electrode with enhanced ECL readout.93,94 Just recently work by Jie and co-workers presented another ECL biosensor with a multiple signal amplification strategy and in situ silver nanocluster synthesis as a versatile probe for ECL and traditional electrochemical methods.95 DNAzymes, as an enzyme-free technique with specificity, could also be applied for the construction of other optical biosensing platforms, including fluorescence or SRP. Li and co-workers constructed an SPR biosensor for microRNA based on multicomponent DNAzyme mediated catalysis.96 A multicomponent DNAzyme catalyzed hairpin is assembled on the target, and a large number of streptavidin are generated, immobilized on the gold chip, obtaining a high SPR signal. Among a great many studies on DNAzymes, Tan's group have shown us a cycling Zn2+–DNAzyme-based ELISA (DLISA) strategy for multiple protein analytes.972.4 Toehold-mediated DNA strand displacement assisted optical macro-reaction interfaces for biosensing
During toehold-mediated DNA strand displacement, the prehybridized strand is displaced from a DNA duplex containing a “toehold” overhang. The toehold refers to a single-stranded domain with 5–8 bases where strand displacement could be initiated through branched chain migration. The kinetics of the strand displacement process is determined and regulated by the situation of the bases in the toehold region, including the number and species of the bases.98 Theoretically, the rate of strand displacement can be accelerated 106-fold using a toehold-mediated DNA strand displacement strategy.99 Two types of classical strand displacement reactions are the hybridization chain reaction (HCR) and catalyzed hairpin assembly (CHA), which have attracted much attention for improving the performance of enzyme-free optical biosensors for the amplified detection of various targets.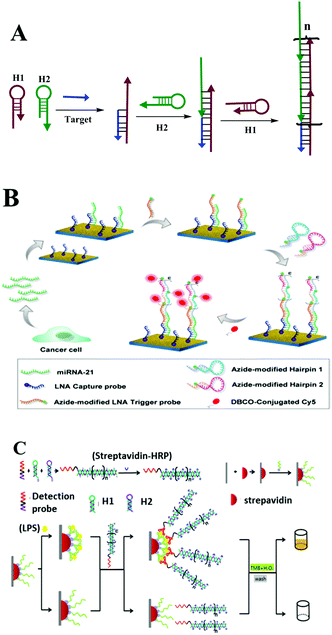 | ||
| Fig. 6 HCR based optical biosensing. (A) Schematic illustration of HCR strategy. (B) MiRNA determination by means of a PE-HCR assay on a plasmonic chip spotted with LNA capture probes. Reprinted with permission from ref. 101. Copyright (2016) American Chemical Society. (C) HCR-based aptasensor for the sensitive detection of LPS. Reprinted with permission from ref. 102. Copyright (2016) Nature group. | ||
This method introduces triggered DNA self-assembly into nanostructures and exploits DNA as an amplifying transducer for biosensing and bioimaging. They introduced a novel strategy in which DNA can accomplish the roles of recognition and signal amplification simultaneously by binding to a substrate. Since this discovery of HCR, many efforts have been made to apply it as a signal amplification strategy for optical biosensing assays. Here, we take macro-reaction interfaces as an example. Yin and co-workers recently reported a plasmon-enhanced HCR strategy for low-abundance microRNA quantification. As described in Fig. 6B, they initially prepared a roughened plasmonic substrate by seeding gold nanoclusters on a glass surface as the fluorescence enhancer. The captured target miRNAs were then recognized by an azide moiety-tagged LNA trigger probe, which acts as the initiator for the following HCR reaction. After a series of reactions, the final resulting nicked double helix was conjugated with fluorescent dye Cy5 via copper-free click chemistry. Cy5 fluorophores showed an overlapping of the fluorescence emission wavelength with the plasmonic resonance peak of the gold substrate.101 Thus, significantly enhanced fluorescence detection at the femtomolar level (1.0 fM–1.0 pM) was obtained on this plasmonic chip, which is 2 orders of magnitude lower than that of RTPCR (0.1 pM–1.0 nM). Besides cancer biomarkers, the HCR strategy has also performed well in food safety analysis. For example, there is a just published report on lipopolysaccharide detection (Fig. 6C). Lipopolysaccharides (LPS), integral components of the outer membrane of all Gram-negative bacteria, are closely associated with foodborne diseases. Xie and co-workers developed a novel HCR-based signal amplification assay for LPS from E. coli O111:B4 with low detection limits of 1.73 ng mL−1.102 By changing the modification of the interface or signal probe molecules, this simple enzyme-free HCR technique has been widely used for the detection of DNA, proteins, bacteria and cancer cells using various optical methods, including traditional fluorescence, ECL, ELISA, SERS or SPR.103–113
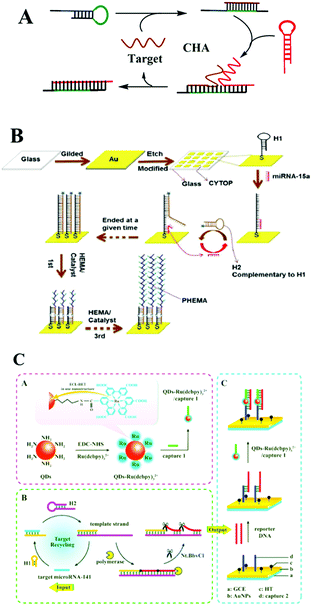 | ||
| Fig. 7 CHA based optical biosensing. (A) Schematic illustration of CHA strategy. (B) CHA-based orthogonal signal amplification for SPRi detection of miRNA-15a. (C) ECL-RET based dual amplification assay including target recycling and double-output conversion strategies. Reprinted with permission from ref. 114 and 115. Copyright (2017) American Chemical Society. | ||
3. Nucleic acid amplification assisted optical micro-reaction interfaces for biosensing
Compared with macro-reaction interfaces, micro-reaction interfaces like noble metal nanoparticles, inorganic nanomaterials, magnetic nanoparticles, graphene and novel two-dimensional (2D) layered nanomaterials analogous to graphene, etc., with large surface areas, good biocompatibility, large loading capacity and high stability, have been widely applied in optics-based nucleic acid isothermal amplification reactions. And thanks to these advantages, the micro-interface as a good cellular transporter has also been widely used for the biosensing analysis of targets inside cells.3.1 Noble metal nanoparticle interfaces for optical biosensing based on nucleic acid amplification
Recently, remarkable advances in the detection of tumor markers have been achieved by constructing highly sensitive isothermal amplification assays based on nanomaterial reaction interfaces. In this regard, noble metal nanoparticles have attracted substantial attention because of their distinct physical and optical properties, including localized surface plasmon resonance (LSPR), fluorescence resonance energy transfer (FRET), and SERS. In addition, noble metal nanoparticles have several advantages, such as easy synthesis and diverse functionalization options for surface modification, which make them particularly suitable for optical biosensing. The modification of nucleic acid on an Au NP surface is usually based on the formation of Au–S bonds which provide stronger and more stable surface binding compared to most physisorption processes.119 Moreover, the size, density, aggregation state, surface charge and other surface properties of an Au NP–DNA composite could be finely tuned to provide diverse biological functions which are very important during the fabrication of chemical and biological sensors.120 In further combination with a nucleic acid isothermal amplification reaction, noble metal nanoparticles showed important effects during the detection of various biomolecules.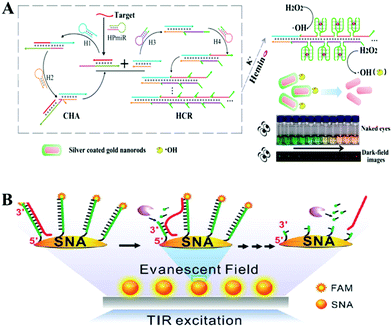 | ||
| Fig. 8 Noble metal nanoparticle-based in vitro assays assisted by nucleic acid amplification. (A) CHA and HCR assisted amplification detection of miR-141 based on silver coated gold nanorods. Reprinted with permission from ref. 121. Copyright (2017) American Chemical Society. (B) Single-particle fluorescence analysis for the operation of an Exo III-powered stochastic DNA walker moving on the SNA surface using TIRF microscopy. Reprinted with permission from ref. 122. Copyright (2017) Wiley-VCH. | ||
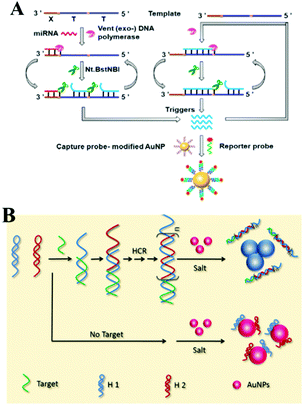
For the aspect of fluorescence biosensing, Qu and co-workers used 15 nm sized Au NPs functionalized with thiol tagged oligonucleotides and developed a 3D track (Fig. 8B). A stochastic DNA walker was presented which could autonomously move on this 3D track. The driving force was derived from Exo III assisted unidirectional digestion of DNA tracks.122 Zhang and co-workers also integrated DNA modified Au NPs with an enzyme-assisted isothermal amplification reaction to construct a novel and smart binding-induced DNA nanomachine. Based on the specific reaction trigger-target binding and unique three-dimensional DNA tracked Au NP probes, the nanomachines have achieved sensitive and selective detection of many kinds of biomolecules: for example proteins and nucleic acid.123 By incorporating the duplex-specific nuclease (DSN) enzyme and DNA-functionalized Au NP probes, Degliangeli and co-workers developed a sensitive and direct assay for the quantification of cancer-related miRNAs.124 The target-miRNA hybridized with the unique fluorescence quenched DNA–Au NP probes, forming DNA–RNA heteroduplexes which can be hydrolyzed by DSN enzyme. Then the target-miRNA, which remained intact during the hydrolyzation process, induced target-recycling amplification and significant signal enhancement. Low to 104 copies per ng RNA can be detected by this method. Besides Au NPs, other noble metal nanoparticles have also been utilized in combination with an enzyme-assisted nucleic acid isothermal amplification reaction for sensitive fluorescence biosensing.125,126 For example, Cui and co-workers utilized C-rich hairpin DNA probes for silver nanocluster Ag NCs nucleation and primers with a G-rich sequence to induce fluorescence enhancement. This fluorescence enhancement was realized by converting the excitation/emission pair of Ag NCs/hairpin DNAs, then obtaining fluorescent enhancement at a longer wavelength. Further benefiting from a target miRNAs cycling SDA reaction, sensitive detection of miR-16-3p and miR-19b-5p was realized in an easy and rapid way.125 Also using Ag NCs as a fluorescent reporter, Zhang's group further integrated an Hg2+-meditated conformational molecular beacon and endonuclease-assisted target recycling amplification to develop a label-free strategy for ultrasensitive miRNA detection.126 In that work, in the presence of a target, the hybridization product could be recognized by the endonuclease Nt.BbvCI, resulting in the cleavage of the probe DNA and release of Hg2+, which could efficiently quench the fluorescence of Ag NCs.
SERS, a modern and useful optical technique, has recently been employed for biomolecular analysis through isothermal amplification strategies reacting on noble metal nanoparticle constructed micro-reaction interfaces.127,128 For example, Li and co-workers have just built a SERS biosensing protocol for the selective and sensitive assay of miRNA with the help of target-triggered rolling circle polymerization (RCP) and rolling circle transcription (RCT).127 Also using the SERS technique, Ye and co-author designed a circular exponential amplification reaction to develop a direct sensing system for the sensitive detection of multiple microRNAs in lung cancer cells with a limit of 0.5 fM, which could discriminate target miRNA from among a variety of RNA mixtures with similar sequences (Fig. 9A). That means great potential for further clinical application in early diagnosis.128
 | ||
| Fig. 9 (A) Multiple signal amplification SPR assay for DNA and cancer cells. Reprinted with permission from ref. 128. Copyright (2014) Royal Society of Chemistry. (B) Schematic of highly sensitive colorimetric detection of DNA by the use of gold nanoparticles and HCA amplification. Reprinted with permission from ref. 140. Copyright (2013) American Chemical Society. | ||
The Au NP based nucleic acid amplification technique has also been integrated in a colorimetric assay to develop amplified colorimetric assays for the ultrasensitive detection of significant biomolecules.129,130 Tan and co-workers took advantage of the isothermal amplification method, SDA, to design a simple, rapid colorimetric method for detecting short DNA sequences. The isothermal amplification produces oligonucleotide X as a trigger, which was converted to a reporter oligonucleotide Y, which influenced the colorimetric readout by mediating DNA nanosphere aggregation. This approach increased the sensitivity to nucleic acids to 100 fM in 10 min or less.131 Similarly, Zhang et al. also developed an ultrasensitive colorimetric detection system for transcription factor NF-κB p50 by employing the SDA amplification reaction. The incorporation of SDA into a colorimetric assay successfully achieved target analysis in direct visualization with the naked eye.132 Moreover, with the development of NEase,133–135 exonuclease III aided amplification, loop-mediated isothermal amplification (LAMP), and RCA have been inserted into colorimetric assays for the sensitive analysis of proteins and nucleic acids.136–139 In addition to the above-mentioned enzyme assisted amplification strategies, enzyme-free nucleic acid isothermal amplification methods were also constructed on Au NP-based micro-reaction interfaces to obtain amplified colorimetric strategies for significant biomolecules assays.140–142 For example, Liu and co-workers demonstrated a highly specific and sensitive colorimetric DNA assay with the HCR technique. As illustrated in Fig. 9B, target DNA hybridization with hairpin auxiliary probes increased the HCR amplification reaction, resulting in a decrease in the stabilizer ssDNA and the aggregation of Au NPs. Taking advantage of HCR and Au NPs, the detection of the target can be seen with the naked eye in this method.140 Additionally, the high selectivity of this protocol, which can identify target sequences from that with a single base-pair mismatch, and relatively quick detection time (60 min) without expensive equipment make it a potential approach for applications in the fields of environmental or clinical monitoring.
 | ||
| Fig. 10 (A) HCR based ultrasensitive mRNA imaging in living cells. Reprinted with permission from ref. 143. Copyright (2013) American Chemical Society. (B) Walking motor on the Au NP surface for real-time RNA imaging inside the cells. Reprinted with permission from ref. 149. Copyright (2017) Nature group. | ||
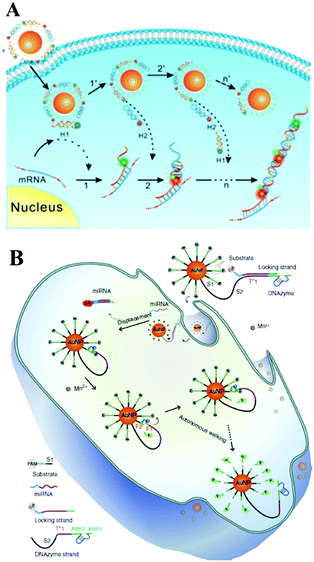
With the rapid development of ion-dependent DNAzyme, many researchers have focused on studies of a DNAzyme biosensing system on the surface of Au NPs. For example, Lu's group first demonstrated a DNAzyme–Au NP probe which could serve as an intracellular metal ion sensor inside cells. Such a method can be generally applied to the detection of other metal ions or DNA by changing DNAzymes.147 Subsequent studies from Tang's group showed a DNAzyme-based two-color fluorescence nanoprobe for the simultaneous imaging of intracellular Zn2+ and Cu2+.148 Very recently, Le's group presented a satisfying DNAzyme motor sensing strategy on Au NPs. In Fig. 10B, we can see DNAzyme molecules silenced by a locking strand were both modified on an Au NP surface. With the target miRNA inside the cells, the hybridization reaction with target miRNA unlocked the foregoing locking strand from the DNAzyme through a strand displacement reaction. The unlocked DNAzyme strand could thus subsequently hybridize with its surrounding substrate. With the assistance of cofactor Mn2+, the activated DNAzyme would cleave the substrate and generate two DNA segments (F1 and F2). Therefore, FAM modified on the F1 segment recovered its fluorescence due to being away from the Au NP surface. Meanwhile, the DNAzyme separated from F2 could subsequently react circularly with substrate strands with an enhanced fluorescence signal. This process is just like the walking motor from one substrate strand to the next around the Au NP surface and meantime it realizes real-time RNA imaging inside the cells.149 Similarly, a recent report from Wang's group designed a DNAzyme biosensing system also on the surface of Au NPs. Here, the DNAzyme substrate sequence and target-binding sequence are linked together as an intact hairpin structure.150 Significant signal enhancement was also realized during this target-recycling strategy for an amplified sensing strategy in living cells.151 Furthermore, Zhu and co-workers designed near-infrared (NIR) photo-thermally activated DNAzymes conjugated to gold nanoshells and constructed a sensing method for the detection of intracellular metal ions. The presented novel NIR activated DNAzyme-based sensing system provided a new efficient tool for target cellular imaging.152 The same group also demonstrated a dual-functional probe composed of Au NPs, DNAzyme and ZnO quantum dots (QDs) for intracellular gene regulation and drug delivery in a controlled manner.153
3.2 Two-dimensional (2D) layered nanomaterial interfaces for optical biosensing based on nucleic acid amplification
Thanks to its unique low-dimensional physical properties and high quenching efficiency, graphene is a promising two-dimensional (2D) layered nanomaterial which could be integrated with isothermal amplification techniques for the amplified detection of biomolecules.154–156 DNA, with a regular sequence of deoxyribose sugars and phosphate groups as its backbone was modified on a graphene surface mainly due to π–π stacking of the DNA bases. Different nucleobases have differential interaction strength with graphene, and guanine (G) is regarded as having the highest binding energy.157 As for graphene oxide (GO), the situation is more complicated. GO contains many functional groups, such as carboxyls, hydroxyls, esters and epoxides, so hydrogen bonding and electrostatic repulsion with the oxygen-rich domains show important attraction in addition to DNA base stacking. Generally, due to the double helical structure of dsDNA which could shield the π–π interactions from the bases, GO showed more highly efficient adsorption of single-stranded DNA in comparison with double-stranded DNA.158,159 Liu and co-workers designed a graphene oxide based Exo III-mediated recycling cleavage reaction to analyze multi-biomolecules. As illustrated in Fig. 11A, due to non-covalent π–π stacking interactions between DNA and graphene, the nucleic acid fluorescent probes 1 and 2 were adsorbed on the quencher graphene oxide. Then, analytes 3 and 4 hybridized with the corresponding probes 1 and 2, forming non-GO adsorbed duplex regions and triggered Exo III-assisted signal amplification for the fluorescence analysis of multi-targets. The multiplexed analysis of biomolecules and the operation of logic gates were also evaluated by this method, indicating that this method offered great potential applications in biological analysis.160 Chen and co-workers also used Exo III and GO-assisted signal amplification during the construction of a CL biosensor for ultrasensitive DNA detection.161 Besides Exo III, T7 exonuclease, DNA polymerase and nicking endonuclease assisted nucleic acid isothermal amplification reactions with nanomaterials–graphene oxide have also been developed for the fast and accurate detection of biomarker-miRNAs.162–164 GO could also be integrated with HCR165–168 and RCA169–173 for the optical biosensing of various biomarkers. For example in fluorescence analysis, Tang's group employed GO as a nanocarrier for the assay of two types of intracellular microRNA167 and intracellular telomerase activity.168 Zhang's group utilized self-assembled nucleic acid molecules on graphene oxide nanoplates and an RCA strategy to realize target miRNA imaging in single cells.171 Besides this, many recent studies have proved that an RCA assisted GO substrate provided excellent performance in the field of fluorescence biosensing of miRNA.172,173 With the help of metal ion-dependent DNAzymes, various metal ions, including Cu2+, Pb2+ or Mg2+, in environmental and biological samples could be simultaneously detected based on a DNAzyme branched junction structure and GO.174 Target-triggered HCR or CHA have also become useful tools for the construction of enzyme-free nucleic acid isothermal amplification on GO micro-reaction interfaces in biomolecule assays. For example, Zhen and co-workers recently employed target-triggered CHA on GO and presented the amplified fluorescence anisotropy detection of microRNA-21.175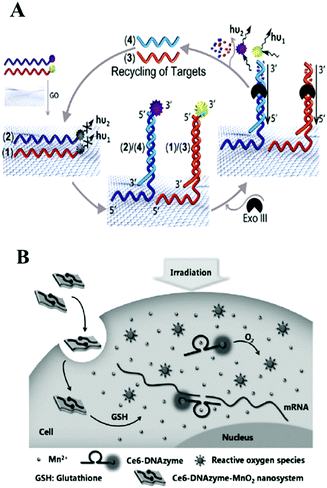 | ||
| Fig. 11 (A) Amplified multiplexed analysis of DNA using graphene oxide as a support and the Exo III-triggered recycling of the analytes. Reprinted with permission from ref. 160. Copyright (2012) American Chemical Society. (B) DNAzyme–MnO2 nanosystem was reported for cellular imaging and gene-silencing therapy. Reprinted with permission from ref. 181. Copyright (2015) Wiley-VCH. | ||
Additionally, WS2 and MnO2 nanosheets, as novel 2D layered nanomaterials analogous to graphene, have become very promising nanoplatforms in bioanalytical applications because of their excellent light absorption capacity and fast electron-transfer rate. Single-strand DNA labeled with a fluorophore can be adsorbed on the surface of WS2 or MnO2 nanosheets through a van der Waals interaction between nucleobases and the basal plane of the nanosheets, resulting in fluorescence quenching.176 For example, inspired by the unique properties of transition metal dichalcogenides as nanoquenchers, Xi and co-workers first combined super quencher-WS2 nanosheets with duplex-specific nuclease signal amplification for the simple and sensitive detection of miRNAs, achieving the ultrasensitive detection of miR-21 as low as 300 fM.177 Biodegradable MnO2 nanosheets also exhibited excellent application in the detection of intracellular targets in recent reports.178 Combined with DNAzyme, MnO2 nanosheet/DNAzyme could also act as a catalytic DNA circuit generator for intracellular signal amplification, monitoring DNA base-excision repairs in living cells. In this work, MnO2 nanosheets served as DNA nanocarriers as well as cofactor suppliers of DNAzyme. In the cellular environment, MnO2 nanosheets were reduced to Mn2+ acting as DNAzyme cofactor. After many cycling reactions, this method offered at least 40-fold amplified signals during the monitoring of apurinic/apyrimidinic endonuclease-initiated and DNA glycosylase-initiated BER pathways.179 Similarly, employing MnO2 nanosheets as DNA carriers and DNAzyme cofactor sources, the above group also assembled enzyme-initiated DNAzyme nanodevices which provided new ideas for monitoring enzyme catalysis in situ.180 A DNAzyme–MnO2 nanosystem is also an efficient tool not only for cellular imaging but also for gene-silencing therapy. For example, in Tan's work, shown in Fig. 11B, a smart DNAzyme–MnO2 nanosystem was reported for cellular imaging and gene-silencing therapy. Here MnO2 nanosheets could protect DNAzyme from endogenous nuclease digestion during the endocytosis process. Inside the cells, self-generated Mn2+ ions could activate DNAzyme for RNA cleavage.181
3.3 Magnetic nanoparticle interfaces for optical biosensing based on nucleic acid amplification
Magnetic nanoparticles with excellent separation ability have also been widely used in constructing micro-reaction interfaces for amplified DNA, RNA and protein detection.182–186 Magnetic nanoparticles could be functionalized with hydroxyl, amine, carboxyl, aldehyde, and epoxy groups, to bind target DNA through stable covalent attachment.187 Also, molecular recognition pairs are another popular strategy to attach DNA to surfaces which could be employed by affinity coupling, for example, a streptavidin–biotin pair.188 Up to now, a large number of magnetic nanoparticle interface based SERS sensing approaches have been reported. For example, researchers have combined magnetic beads or an Fe3O4@Ag NPs composite with nuclease, such as DSN, nicking endonuclease or polymerases, for signal amplified detection of various targets containing ATP, lysozyme, proteins, DNA methyltransferase, DNA and RNA with a low detection limit.189–196 Without using nuclease, Ye et al. proposed a SERS signal amplification strategy based on the combination of a magnetic bead based bifunctional probe, biobarcode probes and HCR, achieving the simultaneous detection of multiple cancer biomarkers.197 Additionally, some new types of magnetic materials have also been developed in order to meet the current requirements for detection. For example, Chai's group used a magnetic SERS substrate (Co@C/PEI/Ag) and exponential RCA strategy to design an “OFF–ON” surface enhancement SERS sensing system for miRNA detection.198 Thiol-hairpin DNA (H1) modified with Cy5 as a Raman label was immobilized on the surface of Co@C/PEI/Ag with a strong Raman signal. Another DNA P2 as placeholder DNA could weaken the Raman signal via a hybridization event, while the target miRNA 155 could recover the biosensor's “ON” status after a series of hybridization and exponential amplification reactions.Besides the SERS sensing approach, there are many new ideas being brought forward using magnetic nanoparticle interfaces for colorimetric biosensing, ECL or CL analysis, and fluorescence detection. For example, using a DNA enzyme-catalyzed colorimetric reaction, homogeneous PDGF-BB detection of low to 2 pg mL−1 in 2 h was obtained in an aptamer-functionalized magnetic microbead-based colorimetric assay which is more competitive than traditional assays such as ELISA.199 Apart from nucleic acids,200–202 and proteins assays,203 further employing selective binding of the aptamer to the receptors, low to 10 HL-60 cells could be successfully distinguished by the naked eye via a color change in colorimetric biosensing on messenger DNA (mDNA)–aptamer modified magnetic nanoparticle interfaces.204
As to magnetic nanoparticle based ECL analysis, more sensitive detection results were obtained generally compared with the colorimetric method, mainly due to the uniquely low background and high sensitivity of ECL. For example, assisted by magnesphere paramagnetic particles, HRCA and NEase-assisted target recycling amplification, Lin and co-workers designed an ultrasensitive ECL biosensing approach for p53 gene (0.02 fM) detection.205 Similarly, in recent work, Zhang and co-author also employed HRCA based dual signal amplification techniques on magnesphere paramagnetic particles and developed an ultrasensitive ECL biosensor for Kras mutant genes with an essential application in diagnostics and individualized patients’ therapeutic strategies for colorectal cancer.206 Furthermore, Jiang and co-workers found that a novel magnetic nanoparticle CoFe2O4 could enhance the ECL intensity of N-(aminobutyl)-N-(ethylisoluminol) (ABEI)–H2O2 due to its peroxidase-like activity. They then designed a convenient ECL biosensor for MUC1 detection with good efficiency and sensitivity with a detection limit at fM level.207 In addition, CL incorporating all kinds of amplification strategies based on magnetic nanoparticle micro-reaction interfaces have been proposed by different groups and have achieved the ultrasensitive detection of various analytes, including nucleic acids, proteins and small molecules.208–210
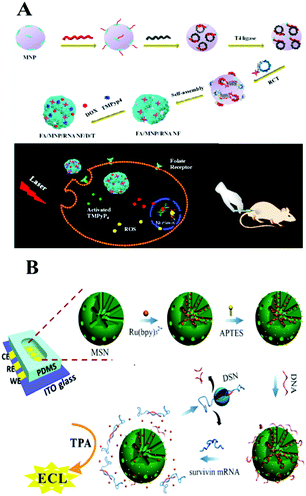
Considering the requirement for integration of clinical diagnosis and treatment, Guo and co-workers recently developed a magnetic nanoparticle based delivery system.211 As shown in Fig. 12A, magnetic nanoparticles were modified with RNA nanoflowers by biotin–avidin conjugation. The anticancer drug doxorubicin (DOX) as well as the photosensitizer TMPyP4 which could generate reactive oxygen species with 650 nm light were both introduced onto this nanocarrier. It showed better treatment efficacy than a single-drug delivery platform. Additionally, the prepared magnetic composite could also be used as a probe for detection of low to 50 HeLa cells.212 Thus, we found that the nucleic acid amplification technique assisted by magnetic nanoparticles holds great promise for potential applications in cancer risk assessment and molecular diagnostics and therapeutics.
 | ||
| Fig. 12 (A) Magnetic nanoparticle based co-delivery system. Reprinted with permission from ref. 211. Copyright (2017) American Chemical Society. (B) ECL microreactor for detection of survivin mRNA in cells based on DNA bio-gates of MSNs and the DSN amplification mechanism. Reprinted with permission from ref. 222. Copyright (2015) Royal Society of Chemistry. | ||
3.4 Silica nanoparticle interfaces for optical biosensing based on nucleic acid amplification
Silica nanoparticles (SNPs), possessing the good properties of a hydrophilic surface, high stability, ease of modification and surface functionalization, have become an important nanomaterial for biosensing. There are many types of interaction between SNPs and DNA: for example, the electrostatic interaction between negatively charged ssDNA and positively charged functionalized SNPs (such as NH2-SNPs), or the covalent attachment between NH2-DNA and a carboxylated or aminopropyl modified SNP surface. Also, the click reaction of copper-catalyzed azidealkyne cycloaddition was employed to link DNA on N3-SNPs. Generally, SNPs used in biosensors can be divided into two categories: nonporous (solid) NPs and mesoporous NPs, both of which bear an amorphous silica structure. Mesoporous silica NPs possess 2–50 nm meso-pores and thus have been widely used for the delivery of small molecules or drugs through physical or chemical adsorption. While nonporous SNPs acted as nanocarriers through encapsulation or the conjugation effect. For example, on the surface of silicon microbeads, Yang's group developed a target miRNA-induced HCR amplification reaction to produce long self-assembled DNA polymers and to further fabricate an Ag NP-conjugation on this long DNA polymer via an Ag–S bond. Then the further dissolved Ag NP loaded long DNA polymer generated a large number of silver ions which could cause the aggregation of previously prepared Au NPs@4-ABT (4-aminobenzenethiol) solution via forming N–Ag–N coordination compounds. Thus, a “hot-spot” was subsequently formed and showed an obviously enhanced SERS signal.213 Very recently, Wang's group fabricated a DNA walker on substrate-modified silica microspheres for l exonuclease (Exo) III-assisted target recycling amplification (ERA) detection of carcinoembryonic antigen (CEA).214 With smart modification of nucleic acids, nucleic acid functionalized silica-nanoflower biopolymers could be prepared as cellular transporters for cancer cell recognition and target drug delivery.215Recently, mesoporous silica which not only shows the character of SNPs, but also shows the advantages of a mesoporous material has been used in the field of controlled drug/DNA modification and delivery. Many recent studies focus on mesoporous silica nanoparticles (MSN) and develop a series of sensing approaches using MSN as a nano-carrier/amplifier for bioanalysis. Willner's group did a lot of MSN related research, some of which employed ion-dependent DNAzyme or exonuclease for signal amplification. The reported DNAzyme-capped mesoporous SiO2 nanoparticles provide stimuli-responsive containers for the biocatalytic release of an anti-cancer drug or the assay of various targets, such as ion ATP, or nucleotides.216–219 Besides fluorescent methods, Ju's group combined target-induced proximity hybridization with an MSN probe and the cleavage effect of endonuclease Nt.BbvCl, and developed an electrochemical DNA biogate for highly sensitive electrochemical immunoassay.220 Zhang's group employed the SERS technique and a nicking endonuclease based amplification reaction on MSN to build a nanocarrier for the sensitive detection of DNA methyltransferases.221 ECL, as a modern electrochemical approach, also demonstrated good performance with the assistance of an amplification reaction on MSN. For example, as shown in Fig. 12B, Chen's group employed a DNA bio-gate and DSN assisted target recycling and developed a facile ECL assay for the sensitive detection of survivin mRNA.222 They first encapsulated a mass of Ru(bpy)32+ into the MSNs via physical absorption, and then functionalized the external surface with 3-aminopropyltriethoxysilane (APTES) to form the NH2-MSNs. Then the negatively charged ssDNA could be electrostatically adsorbed onto the positively charged NH2-MSNs as a DNA bio-gate by sealing of the mesopores. Target survivin mRNA would trigger the release of the entrapped Ru(bpy)32+ by hybridization with ssDNA, thus generating an ECL signal with coreactant TPA in the solution. This facile method enables the accurate quantification of survivin mRNA detection in cancer cells with a low detection limit of 0.1 fM.
Facing the requirements of in situ detection of intracellular analytes, including mRNA or other micromolecules, MSN-based in situ amplification attracted researchers' attention due to the unique advantages of MSN in a target-stimulation induced release process. For example, Zhu's group assembled multicomponent nucleic acid enzymes on the surface of silica-coated gold nanorods for in situ multiplexed logic imaging of miR-21 and miR-145 and controlled drug release. This work provided a general strategy for disease diagnosis, prognosis, and combination treatment with chemotherapy and gene therapy.223 Another example of in situ detection using MSN-based nucleic acid amplification is from Zhang's group. They proposed a core–shell DNA self-assembly and target-initiated HCR for reduced glutathione detection in single living cells which has potential for pathological variations or biological processes in live cells.224
3.5 Other nanoparticle interfaces for optical biosensing based on nucleic acid amplification
In addition to the above-mentioned common nanoparticles, some other kinds of nanoparticles, including nanomaterial composites, polystyrene nanoparticles, sepharose beads, metal–organic frameworks, and semiconductor quantum dots have also been successfully employed for the construction of sensitive bioanalytical methods for biomarkers. For example, very recently, Li's group reported a functional nanosystem (ZnO@PDA-hpDNAs) containing hairpin DNA strands and polydopamine-coated ZnO nanoparticles. As described in Fig. 13A, the system consists of four functional hairpin DNA probes (H0, H1, H2 and H3) on ZnO@polydopamine (PDA) nanoparticles, named a ZnO@PDA-hpDNAs nanosystem. Among these, H1 and H2 possess two separate single-strand tails with inactive Mg2+-dependent DNAzyme sequences at their 5′ and 3′ ends, respectively. During the HCR reaction inside living cells, this nanosystem could form target-triggered, wire-shaped DNAzyme active DNAzyme nanostructures and showed the sensitive fluorescence detection of survivin mRNA as a tumor-related biomarker.225 Another good example using a fluorescence approach is a polystyrene nanoparticle and SDA amplification enhanced fluorescence aptasensor for the detection of attomolar proteins in biological samples, as reported by Huang and co-workers.226 These various nanoparticle-based interfaces provided an excellent sensing platform for some label-free, enzyme-dependent or enzyme-free detections of cancer biomarkers inside or outside cells via chemiluminescence or other optical methods.227–231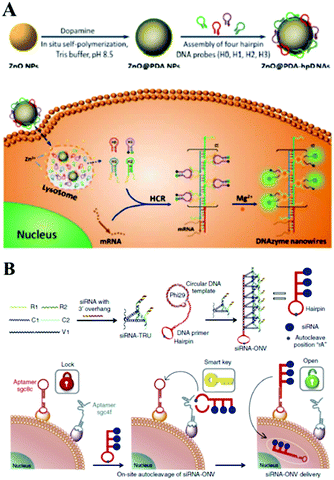 | ||
| Fig. 13 (A) ZnO@PDA-hpDNAs nanosystem based optical biosensing for live cell mRNA imaging via the target-triggered HCR mediated intracellular self-assembly of wire-shaped active DNAzyme nanostructures. Reprinted with permission from ref. 225. Copyright (2017) Royal Society of Chemistry. (B) Schematic illustration of ‘dual lock-and-key’-controlled cell-subtype-specific siRNA delivery. Reprinted with permission from ref. 239. Copyright (2017) Nature group. | ||
3.6 Cell surface-interfaces for optical biosensing based on nucleic acid amplification
In addition, a cell-assisted reaction interface has also been utilized to develop amplified bioanalytical methods for various biomolecules. For example, previously mentioned nucleic acid amplification reactions, including HCR, RCA, or NEase, could occur on the cell surface to sensitively monitor the amount of target cells, to deliver RNA or for molecular imaging. In the past few years, NEase or DNAzyme based nucleic acid amplification reactions on the cell surface were reported by Tan's or Kong's group for cell membrane-anchored biosensing.232,233 Recently enzyme-free methods, such as HCR, RCA have attracted researchers' attention. For example, Wang and co-worker first proposed aptamer structure switching triggered cell surface HCR isothermal amplification strategies for fluorescence detection and targeting therapy.234 Also using an aptamer structure, Tan's group established a DNA nanodevice tethered by fluorescent aptamer (aptNDs) on the surface of a target living cell by anchoring and an in situ self-assembly approach which showed significant potential for the construction of nanodevices applied to bioimaging on cell surfaces via specific interactions.235 Further combined with inorganic materials such as nanocrystal QDs or Cu NPs, Ma's group and Qu's group respectively proposed a DNA-programmed QD polymerization strategy assisted by a hybridization chain reaction (HCR) for molecular imaging or fluorescence of cancer cells.236,237 Benefiting from HCR amplification and the formation of linear QD–aptamer polymers (QAPs), the photoluminescence signals were significantly improved compared with QD–aptamer monomer (QAM)-treated target cancer cells. Also in a very recent study depicted in Fig. 14, Song and co-workers designed a DNA hydrogel with an aptamer-toehold for the recognition, cloaking and decloaking of circulating tumor cells. Initially, MCF-7 cells are dispersed in solution, and then the aptamer–initiator bi-blocks specifically bind to the EpCAM on the cell surface which could then trigger the HCR reaction to form a DNA hydrogel. Confocal images of aptamer–initiator bi-blocks (red) colocalized with DiO stained lipid on the cell membrane (green) could be obviously observed. Additionally, due to the designed ATP responsive region in the DNA hydrogel, ATP can be used to destroy the DNA hydrogel on the cell surface. This liquid biopsy tool is expected to be an effective route to gathering faithful information about primary tumors from captured CTCs during clinical cancer diagnostics and therapeutics.238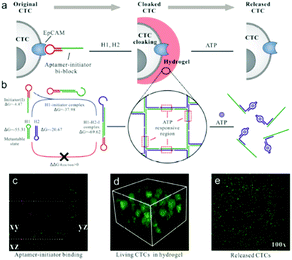 | ||
| Fig. 14 An aptamer-trigger clamped hybridization chain reaction (atcHCR) method for in situ identification and subsequent cloaking/decloaking of CTCs by porous DNA hydrogels. Reprinted with permission from ref. 238. Copyright (2017) American Chemical Society. | ||
Apart from the above-mentioned HCR based amplification reaction on the cell surface, RCA is another powerful tool on the cell surface-interfaces for optical biosensing. For example, in Fig. 13B, Ju's group successfully constructed an RCA based DNA dual lock-and-key strategy for cell-subtype-specific siRNA delivery, which provided a new method for the application of RNAi in accurate diagnosis and intervention.239 Here, an RCA assisted oligonucleotide nano-vehicle was designed to load siRNA with a higher payload capacity and enhanced stability against nuclease degradation for controllable siRNA delivery. Zhang and co-workers recently reported another metabolic method for cell surface enhanced glycan imaging via an RCA reaction. This work could further be used for monitoring the glycosylation status on a cell's surface and extend the applications of RCA in metabolic systems.240 Recently Wang's group integrated the RCA technique into a split aptamer design, which produced a long ssDNA as an RCA product with repeated Split-b supplying polyvalent binding sites and Poly T regions for assembling signal output, and they constructed a novel strategy of polyvalent aptameric DNA nanoensembles for SMMC-7721 cancer cell detection and manipulation.241
4. Conclusions and perspectives
In this review, we have summarized remarkable advances in the development of novel ultrasensitive modern optical nano-biosensing interfaces based on isothermal amplification of nucleic acid. Macro-reaction interfaces and nanoparticle-based micro-reaction interfaces each offer different features and benefits in biosensing. Further construction of isothermal amplification reactions on smartly designed modern optical nano-biosensing interfaces has been widely applied in the recent biosensing field due to their excellent designability and stability, and demonstrates great promise for clinical application. First, burgeoning modern optical biosensing technology and newly-designed interface assembly approaches provided excellent methodologies and stable photoelectric features. Moreover, further ingenious combination with isothermal amplification strategies greatly enhanced the biosensing sensitivity and also improved its performance in the clinical detection of cancer disease-related nucleic acid, proteins and cells. Besides this, further attempt ay intracellular isothermal amplification and portable modern optical devices have achieved preliminary success which were a big step towards in situ amplified tests of cancer cells.Although nucleic acid isothermal amplification-based methods have exhibited good performances in ultrasensitive optical biosensing for extensive analytes, further development and routine applications still face many challenges. The first challenge is the limited types of analytes. Nucleic acid-mediated application reactions, including nucleic acid-cleaving enzymes or polymerase assisted RCA, SDA, etc. or enzyme-free HCR, CHA were all well designed and optimized for nucleic acid analyses. Only a few proteins could be adapted to be successfully detected using these cycling signal amplification strategies with the benefit of well-understood aptamer structures. These proteins are just a minority in clinical cancer biomarkers, including thrombin, human IgE, PDGF-BB. The development of broadly suitable molecular machine or translators which could be universal for general clinical cancer relevant proteins biomarkers is urgently needed. Recently developed modified SELEX techniques, which have the potential to generate aptamers for nearly any protein target, present good opportunities for further research and development on nucleic acid assisted isothermal amplification assays.
Second, the requirement for well-controlled buffer systems in nucleic acid isothermal amplification is another challenge for clinical applications. Although nucleic acid isothermal amplification strategies which break the rules of programed heating have pushed us a big step closer to addressing a central problem in precise and effective clinical cancer diagnosis, the limiting factors from buffers still exist. Different polymerase or nucleic acid-cleaving enzymes have their own optimal buffer systems. And when the sensing system was used in biological fluids, for example samples from serum or plasma, urine, or saliva, most of these sensors may perform unsatisfactorily with poor sensitivity. Although the utilization of optical sensing interfaces could greatly reduce the influence from complex sample solutions, via a simple separation process, interfering biomolecules or proteins in biological samples may be more or less attached to the surface of the interface non-specifically, affecting the subsequent reactions. In addition, in situ sensitive detection of intracellular target analytes is also a difficult problem due to the complex environment inside the cell which could greatly influence the effect of nuclease. In this aspect, DNA-NP nanostructures show great promise to greatly enhance the stability and sensitivity of optical biosensors, as well as providing excellent opportunities for in vivo imaging, diagnosis or therapy of tumor cells. For example, some noble NPs, such as gold or silver NPs, have been found to possess excellent biocompatibility and stability, as well as being able to provide various enhanced specific SERS or LSPR signals for amplification strategies for detecting multiple clinic targets.120 Moreover, these kinds of NPs could also protect nucleic acids from enzymatic degradation in biofluids which is very essential for the detection of real clinical samples or in vivo cell assay.242,243
Third, in order to satisfy the demand for personalized clinical diagnostics, rapid, low-cost and multiplex detection is an opportunity as well as a challenge for optical biosensors. By good design of multichannel or multicultural sensing platforms on optical interfaces, such as chips, or microwell plates, it could realize the simultaneous detection of multiple analytes. However, current multiplex detections are mainly obtained on macro-sensing interfaces, and only a few studies of multiplex assays on nanoparticle-based micro-sensing interfaces have been reported, though they generally showed more flexibility and sensitivity in biosensing compared with micro-sensing interfaces. It will be of great significance to widely construct multiplex detection on powerful micro-sensing interfaces which hold many advantages in real-time monitoring within a very short time. In addition, the rapid development of optical instruments and new optical techniques, such as near infrared (NIR), dark-field imaging techniques and two-photon techniques provide excellent and promising approaches for higher-performance imaging with reduced phototoxicity and higher temporal and spatial resolution.244–246
These current and oncoming challenges provide new opportunities for further research and development of improved analytical methods for all aspects of detection abilities, including accuracy, selectivity, efficiency and sensitivity. Moreover, further efficient multimode clinical detection and in vivo 3D imaging become a promising trend. If we can overcome the stability problem of probes in enzyme resistance inside cells, an isothermal amplification strategy assisted modern optical method could be a strong tool in understanding life processes, and then help us to diagnose and treat cancers at a higher level. With the rapid development of nanomaterials and analytical methodology, it is expected that more excellent and simple-to-handle gentle amplification strategies could emerge brilliantly to build smart optical biosensing platforms for the sensitive assay of many malignant diseases with low cost, less time, ease of operation and high sensitivity. Therefore, real clinical applications for accurate and minimally invasive diagnosis even with portable POC devices, precise and efficient drug delivery and disease therapeutics could be realized.
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
Financial support was provided by the National Natural Science Foundation of China (Grant No. 21327902, 21505065, 21535002, 21535003, 21675074, 21775063), the “Innovation Team Development Plan” of the Ministry of Education Rolling Support (IRT_15R31).Notes and references
- L. Wu and X. G. Qu, Chem. Soc. Rev., 2015, 44, 2963 RSC.
- L. M. Lu, X. B. Zhang, R. M. Kong, B. Yang and W. H. Tan, J. Am. Chem. Soc., 2011, 133, 11686 CrossRef CAS PubMed.
- J. Zheng, A. L. Jiao, R. H. Yang, H. M. Li, J. S. Li, M. L. Shi, C. Ma, Y. Jiang, L. Deng and W. H. Tan, J. Am. Chem. Soc., 2012, 134, 19957 CrossRef CAS PubMed.
- R. Freeman, X. Q. Liu and I. Willner, J. Am. Chem. Soc., 2011, 133, 11597 CrossRef CAS PubMed.
- L. H. Guo, Y. Xu, A. R. Ferhan, G. N. Chen and D. H. Kim, J. Am. Chem. Soc., 2013, 135, 12338 CrossRef CAS PubMed.
- T. H. Seefeld, A. R. Halpern and R. M. Corn, J. Am. Chem. Soc., 2012, 134, 12358 CrossRef CAS PubMed.
- Z. Y. Liu, W. J. Qi and G. B. Xu, Chem. Soc. Rev., 2015, 44, 3117 RSC.
- J. P. Lei and H. X. Ju, Chem. Soc. Rev., 2012, 41, 2122 RSC.
- Y. X. Zhao, F. Chen, Q. Li, L. H. Wang and C. H. Fan, Chem. Rev., 2015, 115, 12491 CrossRef CAS PubMed.
- R. Deng, L. Tang, Q. Tian, Y. Wang, L. Lin and L. Li, Angew. Chem., Int. Ed., 2014, 53, 2389 CrossRef CAS PubMed.
- R. X. Duan, X. D. Lou and F. Xia, Chem. Soc. Rev., 2016, 45, 1738 RSC.
- Z. Cheglakov, T. M. Cronin, C. He and Y. Weizmann, J. Am. Chem. Soc., 2015, 137, 6116 CrossRef CAS PubMed.
- R. Duan, X. Zuo, S. Wang, X. Quan, D. Chen, Z. Chen, L. Jiang, C. Fan and F. Xia, J. Am. Chem. Soc., 2013, 135, 4604 CrossRef CAS PubMed.
- M. M. Ali, F. Li, Z. Q. Zhang, K. X. Zhang, D.-K. Kang, J. A. Ankrum, X. C. Le and W. A. Zhao, Chem. Soc. Rev., 2014, 43, 3324 RSC.
- Z. Y. Wang, S. F. Zong, L. Wu, D. Zhu and Y. P. Cui, Chem. Rev., 2017, 117, 7910 CrossRef PubMed.
- J.-F. Li, Y.-J. Zhang, S.-Y. Ding, R. Panneerselvam and Z.-Q. Tian, Chem. Rev., 2017, 117, 5002 CrossRef CAS PubMed.
- J. Hu and C. Y. Zhang, Anal. Chem., 2010, 82, 8991 CrossRef CAS PubMed.
- B. Guven, I. H. Boyaci, U. Tamer, E. Acar Soykut and U. Dogan, Talanta, 2015, 136, 68 CrossRef CAS PubMed.
- Y. C. Cao, R. Jin and C. A. Mirkin, Science, 2002, 297, 1536 CrossRef CAS PubMed.
- J. Yan, S. Su, S. He, Y. He, B. Zhao, D. Wang, H. Zhang, Q. Huang, S. P. Song and C. H. Fan, Anal. Chem., 2012, 84, 9139 CrossRef CAS PubMed.
- L. Yao, Y. W. Ye, J. Teng, F. Xue, D. D. Pan, B. G. Li and W. Chen, Anal. Chem., 2017, 89, 9775 CrossRef CAS PubMed.
- Y. Geng, J. Wu, L. Shao, F. Yan and H. Ju, Biosens. Bioelectron., 2014, 61, 593 CrossRef CAS PubMed.
- S. Wang, S. Bi, Z. H. Wang, J. F. Xia, F. F. Zhang, M. Yang, R. J. Gui, Y. H. Li and Y. Z. Xia, Chem. Commun., 2015, 51, 7927 RSC.
- C. Chen, M. Luo, T. Ye, N. Li, X. Ji and Z. He, Analyst, 2015, 140, 4515 RSC.
- X. Lin, Q. Chen, W. Liu, H. Li and J. M. Lin, Biosens. Bioelectron., 2014, 56, 71 CrossRef CAS PubMed.
- H. Y. Lee, H. Jeong, Y. Jung, B. Jang, Y. Chang Seo, H. Lee and H. Lee, Adv. Mater., 2015, 17, 3513 CrossRef PubMed.
- M. Liu, C. Y. Hui, Q. Zhang, J. Gu, B. Kannan, S. Jahanshahi-Anbuhi, C. M. Filipe, J. D. Brennan and Y. Li, Angew. Chem., Int. Ed., 2016, 55, 2709 CrossRef PubMed.
- L. H. Tang and J. H. Li, ACS Sens., 2017, 2, 857 CrossRef CAS PubMed.
- W. j. Miao, Chem. Rev., 2008, 108, 2506 CrossRef CAS PubMed.
- M. M. Richter, Chem. Rev., 2004, 104, 3003 CrossRef CAS PubMed.
- P. Wu, X. D. Hou, J. J. Xu and H. Y. Chen, Chem. Rev., 2014, 114, 11027 CrossRef CAS PubMed.
- L. Z. Hu and G. B. Xu, Chem. Soc. Rev., 2010, 39, 3275 RSC.
- Z. F. Ding, B. M. Quinn, S. K. Haram, L. E. Pell, B. A. Korgel and A. J. Bard, Science, 2002, 296, 1293 CrossRef CAS PubMed.
- Y. Chen, S. W. Zhou, L. L. Li and J. J. Zhu, Nano Today, 2017, 12, 98 CrossRef CAS.
- L. L. Li, Y. Chen and J. J. Zhu, Anal. Chem., 2017, 89, 358 CrossRef CAS PubMed.
- Q. Su, D. Xing and X. M. Zhou, Biosens. Bioelectron., 2010, 25, 1615 CrossRef CAS PubMed.
- L. D. Wu, C. Ma, X. X. Zheng, H. Y. Liu and J. H. Yu, Biosens. Bioelectron., 2015, 68, 413 CrossRef CAS PubMed.
- M. Chen, S. Bi, X. Q. Jia and P. He, Anal. Chim. Acta, 2014, 837, 44 CrossRef CAS PubMed.
- A. Y. Chen, S. Y. Ma, Y. Zhuo, Y. Q. Chai and R. Yuan, Anal. Chem., 2016, 88, 3203 CrossRef CAS PubMed.
- R. Deng, L. Tang, Q. Tian, Y. Wang, L. Lin and L. Li, Angew. Chem., Int. Ed., 2014, 53, 2389 CrossRef CAS PubMed.
- L. He, D. Q. Lu, H. Liang, S. T. Xie, X. B. Zhang, Q. L. Liu, Q. Yuan and W. H. Tan, J. Am. Chem. Soc., 2018, 140, 258 CrossRef CAS PubMed.
- Z. Cheglakov, T. M. Cronin, C. He and Y. Weizmann, J. Am. Chem. Soc., 2015, 137, 6116 CrossRef CAS PubMed.
- R. Duan, X. Zuo, S. Wang, X. Quan, D. Chen, Z. Chen, L. Jiang, C. Fan and F. Xia, J. Am. Chem. Soc., 2013, 135, 4604 CrossRef CAS PubMed.
- L. T. Yang, C. W. Fung, E. J. Cho and A. D. Ellington, Anal. Chem., 2007, 79, 3320 CrossRef CAS PubMed.
- E. J. Cho, L. T. Yang, M. Levy and A. D. Ellington, J. Am. Chem. Soc., 2005, 127, 2022 CrossRef CAS PubMed.
- W. A. Zhao, C. H. Cui, S. Bose, D. G. Guo, C. Shen, W. P. Wong, K. Halvorsen, O. C. Farokhzad, G. S. Teo, J. A. Phillips, D. M. Dorfman, R. Karnik and J. M. Karp, PNAS, 2012, 109, 19626 CrossRef CAS PubMed.
- S. Krishnan, V. Mani, D. Wasalathanthri, C. V. Kumar and J. F. Rusling, Angew. Chem., Int. Ed., 2011, 50, 1175 CrossRef CAS PubMed.
- E. J. Kim, B. H. Chung and H. J. Lee, Anal. Chem., 2012, 84, 10091 CrossRef CAS PubMed.
- J. Wang and H. S. Zhou, Anal. Chem., 2008, 80, 7174 CrossRef CAS PubMed.
- L. He, M. D. Musick, S. R. Nicewarner, F. G. Salinas, S. J. Benkovic, M. J. Natan and C. D. Keating, J. Am. Chem. Soc., 2000, 122, 9071 CrossRef CAS.
- Y. Uludag and I. E. Tothill, Anal. Chem., 2012, 84, 5898 CrossRef CAS PubMed.
- P. He, W. P. Qiao, L. J. Liu and S. S. Zhang, Chem. Commun., 2014, 50, 10718 RSC.
- W. B. Sun, W. L. Song, X. Y. Guo and Z. H. Wang, Anal. Chim. Acta, 2017, 978, 42 CrossRef CAS PubMed.
- P. He, L. J. Liu, W. P. Qiao and S. S. Zhang, Chem. Commun., 2014, 50, 1481 RSC.
- S. Bi, B. Ji, Z. Zhang and S. S. Zhang, Chem. Commun., 2013, 49, 3452 RSC.
- J. Lee, K. Icoz, A. Roberts, A. D. Ellington and C. A. Savran, Anal. Chem., 2010, 82, 197 CrossRef CAS PubMed.
- Y. L. Chen, L. Ding, T. T. Liu and H. X. Ju, Anal. Chem., 2013, 85, 11153 CrossRef CAS PubMed.
- H. Zhou, Y. Y. Zhang, J. Liu, J. J. Xu and H. Y. Chen, Chem. Commun., 2013, 49, 2246 RSC.
- G. F. Jie and J. X. Yuan, Anal. Chem., 2012, 84, 2811 CrossRef CAS PubMed.
- G. F. Jie, L. Wang, J. X. Yuan and S. S. Zhang, Anal. Chem., 2011, 83, 3873 CrossRef CAS PubMed.
- Z. Q. Xu, L. L. Liao, Y. Q. Chai, H. J. Wang and R. Yuan, Anal. Chem., 2017, 89, 8282 CrossRef CAS PubMed.
- C. Zong, J. Wu, M. M. Liu, F. Yan and H. X. Ju, Chem. Sci., 2015, 6, 2602 RSC.
- Q. Guo, X. Yang, K. Wang, W. Tan, W. Li, H. Tang and H. Li, Nucleic Acids Res., 2009, 37, 20 CrossRef PubMed.
- H. Zhou, J. Liu, J. J. Xu and H. Y. Chen, Anal. Chem., 2011, 83, 8320 CrossRef CAS PubMed.
- H. Zhou, J. Liu, J. J. Xu and H. Y. Chen, Chem. Commun., 2011, 47, 8358 RSC.
- J. Liu, X. Y. Xin, H. Zhou and S. S. Zhang, Chem. Commun., 2015, 51, 10843 RSC.
- F. L. Gao, J. P. Lei and H. X. Ju, RSC Adv., 2013, 3, 13163 RSC.
- S. J. Ye, X. M. Zhai, Y. Y. Wu and S. P. Kuang, Biosens. Bioelectron., 2016, 79, 130 CrossRef CAS PubMed.
- S. Z. Yue, T. T. Zhao, S. Bia and Z. P. Zhang, Biosens. Bioelectron., 2017, 98, 234 CrossRef CAS PubMed.
- C. X. Li, J. Lin, Y. S. Guo and S. S. Zhang, Chem. Commun., 2011, 47, 4442 RSC.
- A. Y. Chen, G. F. Gui, Y. Zhuo, Y. Q. Chai, Y. Xiang and R. Yuan, Anal. Chem., 2015, 87, 6328 CrossRef CAS PubMed.
- M. J. Niu, Y. J. Wang, S. Li and Y. S. Guo, Sens. Actuators, B, 2016, 223, 359 CrossRef CAS.
- M. Jiao, G. F. Jie, L. Tan and S. Y. Niu, Anal. Chim. Acta, 2017, 983, 166 CrossRef CAS PubMed.
- P. He, Y. Zhang, L. Liu, W. Qiao and S. S. Zhang, Chem. – Eur. J., 2013, 19, 7452 CrossRef CAS PubMed.
- D. Wu, X. Xin, X. H. Pang, M. Pietraszkiewicz, R. Hozyst, X. G. Sun and Q. Wei, ACS Appl. Mater. Interfaces, 2015, 7, 12663 CAS.
- P. Zhao, L. F. Zhou, Z. Nie, X. H. Xu, W. Li, Y. Huang, K. Y. He and S. Z. Yao, Anal. Chem., 2013, 85, 6279 CrossRef CAS PubMed.
- Z. Wu, Z. Zhen, J. H. Jiang, G. L. Shen and R. Q. Yu, J. Am. Chem. Soc., 2009, 131, 12325 CrossRef CAS PubMed.
- X. Y. Jiang, H. J. Wang, R. Yuan and Y. Q. Chai, Anal. Chem., 2016, 88, 9243 CrossRef CAS PubMed.
- X. Li, A. Y. Chen, Y. Q. Chai and R. Yuan, Analyst, 2017, 142, 2185 RSC.
- Y. Gao and X. B. Li, Anal. Chem., 2013, 85, 11494 CrossRef CAS PubMed.
- G. F. Jie, K. Chen, X. C. Wang and Z. K. Lu, RSC Adv., 2016, 6, 2065 RSC.
- N. Hao, P. P. Dai, T. Yu, J. J. Xu and H. Y. Chen, Chem. Commun., 2015, 51, 13504 RSC.
- N. Hao, X. L. Li, H. R. Zhang, J. J. Xu and H. Y. Chen, Chem. Commun., 2014, 50, 14828 RSC.
- Q.-M. Feng, Y.-Z. Shen, M.-X. Li, Z.-L. Zhang, W. Zhao, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2016, 88, 937 CrossRef CAS PubMed.
- X.-L. Huo, H. Yang, W. Zhao, J.-J. Xu and H.-Y. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 33360 CAS.
- Y. He, X. Yang, R. Yuan and Y. Q. Chai, Anal. Chem., 2017, 89, 8538 CrossRef CAS PubMed.
- J. W. Liu, Z. H. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948 CrossRef CAS PubMed.
- I. Willner, B. Shlyahovsky, M. Zayats and B. Willner, Chem. Soc. Rev., 2008, 37, 1153 RSC.
- L. Gong, Z. L. Zhao, Y.-F. Lv, S.-Y. Huan, T. Fu, X.-B. Zhang, G.-L. Shen and R.-Q. Yu, Chem. Commun., 2015, 51, 979 RSC.
- W. H. Zhou, R. Saran and J. W. Liu, Chem. Rev., 2017, 117, 8272 CrossRef CAS PubMed.
- Y. Cheng, Y. Huang, J. P. Lei, L. Zhang and H. X. Ju, Anal. Chem., 2014, 86, 5158 CrossRef CAS PubMed.
- F. L. Gao, L. L. Du, D. Q. Tang, Y. Lu, Y. Z. Zhang and L. X. Zhang, Biosens. Bioelectron., 2015, 66, 423 CrossRef CAS PubMed.
- Z. Q. Xu, Y. W. Dong, J. Y. Li and R. Yuan, Chem. Commun., 2015, 51, 14369 RSC.
- P. Zhang, X. Y. Wu, R. Yuan and Y. Q. Chai, Anal. Chem., 2015, 87, 3202 CrossRef CAS PubMed.
- G. F. Jie, L. Tan, Y. Zhao and X. C. Wang, Biosens. Bioelectron., 2017, 94, 243 CrossRef CAS PubMed.
- X. Li, W. Cheng, D. Li, J. Wu, X. Ding, Q. Cheng and S. Ding, Biosens. Bioelectron., 2016, 80, 98 CrossRef CAS PubMed.
- R. Hu, T. Liu, X.-B. Zhang, Y. H. Yang, T. Chen, C. C. Wu, Y. Liu, G. Z. Zhu, S. Y. Huan, T. Fu and W. H. Tan, Anal. Chem., 2015, 87, 7746 CrossRef CAS PubMed.
- D. Y. Zhang and G. Seelig, Nat. Chem., 2011, 3, 103 CrossRef CAS PubMed.
- A. J. Genot, D. Y. Zhang, J. Bath and A. J. Turberfield, J. Am. Chem. Soc., 2011, 133, 2177 CrossRef CAS PubMed.
- R. M. Dirks and N. A. Pierce, PNAS, 2004, 43, 15275 CrossRef PubMed.
- F. F. Yin, H. Q. Liu, Q. Li, X. Gao, Y. M. Yin and D. B. Liu, Anal. Chem., 2016, 88, 4600 CrossRef CAS PubMed.
- P. Y. Xie, L. J. Zhu, X. L. Shao, K. L. Huang, J. J. Tian and W. T. Xu, Sci. Rep., 2016, 6, 29524 CrossRef CAS PubMed.
- Q. Guo, J.-J. Han, S. Shan, D.-F. Liu, S.-S. Wu, Y.-H. Xiong and W.-H. Lai, Biosens. Bioelectron., 2016, 86, 990 CrossRef CAS PubMed.
- W. Q. Song, K. L. Zhu, Z. J. Cao, C. Lau and J. Z. Lu, Analyst, 2012, 137, 1396 RSC.
- J. Chao, Z. H. Li, J. Li, H. Z. Peng, S. Su, Q. Li, C. F. Zhu, X. L. Zuo, S. P. Song, L. H. Wang and L. H. Wang, Biosens. Bioelectron., 2016, 81, 92 CrossRef CAS PubMed.
- G. T. Jie and G. F. Jie, RSC Adv., 2016, 6, 24780 RSC.
- Y. Chen, J. Xu, J. Su, Y. Xiang, R. Yuan and Y. Q. Chai, Anal. Chem., 2012, 84, 7750 CrossRef CAS PubMed.
- L. J. Xiao, Y. Q. Chai, R. Yuan, Y. L. Cao, H. J. Wang and L. J. Bai, Talanta, 2013, 115, 577 CrossRef CAS PubMed.
- J. Choi, K. R. Love, Y. Gong, T. M. Gierahn and J. C. Love, Anal. Chem., 2011, 83, 6890 CrossRef CAS PubMed.
- F. L. Gao, J. P. Lei and H. X. Ju, Anal. Chem., 2013, 85, 11788 CrossRef CAS PubMed.
- S. Dai, Q. W. Xue, J. Zhu, Y. S. Ding, W. Jiang and L. Wang, Biosens. Bioelectron., 2014, 51, 421 CrossRef CAS PubMed.
- X. M. Li, Y. Wang, L. L. Wang and Q. L. Wei, Chem. Commun., 2014, 50, 5049 RSC.
- L. L. Liang, F. F. Lan, L. Li, S. G. Ge, J. H. Yu, N. Ren, H. Y. Liu and M. Yan, Biosens. Bioelectron., 2016, 86, 756 CrossRef CAS PubMed.
- F. C. Hu, J. Y. Xu and Y. Chen, Anal. Chem., 2017, 89, 10071 CrossRef CAS PubMed.
- Z. Y. Li, Z. F. Lin, X. Y. Wu, H. T. Chen, Y. Q. Chai and R. Yuan, Anal. Chem., 2017, 89, 6029 CrossRef CAS PubMed.
- Y.-Q. Yu, J.-P. Wang, M. Zhao, L.-R. Hong, Y.-Q. Chai, R. Yuan and Y. Zhuo, Biosens. Bioelectron., 2016, 77, 442 CrossRef CAS PubMed.
- Q.-M. Feng, Y.-H. Guo, J.-J. Xu and H.-Y. Chen, Biosens. Bioelectron., 2018, 100, 571 CrossRef CAS PubMed.
- Q.-M. Feng, Y.-H. Guo, J.-J. Xu and H.-Y. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 17637 CAS.
- D. Liu, W. Chen, K. Sun, K. Deng, W. Zhang, Z. Wang and X. Jiang, Angew. Chem., Int. Ed., 2011, 50, 4103 CrossRef CAS PubMed.
- W. Zhou, X. Gao, D. B. Liu and X. Y. Chen, Chem. Rev., 2015, 115, 10575 CrossRef CAS PubMed.
- Y. Gu, J. Song, M.-X. Li, T.-T. Zhang, W. Zhao, J.-J. Xu, M. L. Liu and H.-Y. Chen, Anal. Chem., 2017, 89, 10585 CrossRef CAS PubMed.
- X. M. Qu, D. Zhu, G. B. Yao, S. Su, J. Chao, H. J. Liu, X. L. Zuo, L. H. Wang, J. Y. Shi, L. H. Wang, W. Huang, H. Pei and C. H. Fan, Angew. Chem., Int. Ed., 2017, 56, 1855 CrossRef CAS PubMed.
- H. Q. Zhang, M. D. Lai, A. Zuehlke, H. Y. Peng, X. F. Li and X. C. Le, Angew. Chem., Int. Ed., 2015, 54, 14326 CrossRef CAS PubMed.
- F. Degliangeli, P. Kshirsagar, V. Brunetti, P. P. Pompa and R. Fiammengo, J. Am. Chem. Soc., 2014, 136, 2264 CrossRef CAS PubMed.
- J. Zhang, C. Li, X. Zhi, G. A. Ramon, Y. Liu, C. Zhang, F. Pan and D. Cui, Anal. Chem., 2016, 88, 1294 CrossRef CAS PubMed.
- H. Dong, K. Hao, Y. Tian, S. Jin, H. Lu, S.-F. Zhou and X. Zhang, Biosens. Bioelectron., 2014, 53, 377 CrossRef CAS PubMed.
- X. M. Li, F. W. Zheng and R. Ren, Chem. Commun., 2015, 51, 11976 RSC.
- L. P. Ye, J. Hu, L. Liang and C. Y. Zhang, Chem. Commun., 2014, 50, 11883 RSC.
- J. Van Ness, L. K. Van Ness and D. J. Galas, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4504 CrossRef CAS PubMed.
- M. D. Stone, M. Mihalusova, C. M. O'Connor, R. Prathapam, K. Collins and X. Zhuang, Nature, 2007, 446, 458 CrossRef CAS PubMed.
- E. Tan, J. Wong, D. Nguyen, Y. Zhang, B. Erwin, L. K. Van Ness, S. M. Baker, D. J. Galas and A. Niemz, Anal. Chem., 2005, 77, 7984 CrossRef CAS PubMed.
- Y. Zhang, J. Hu and C. Y. Zhang, Anal. Chem., 2012, 84, 9544 CrossRef CAS PubMed.
- J. Li, H. E. Fu, L. J. Wu, A. X. Zheng, G. N. Chen and H. H. Yang, Anal. Chem., 2012, 84, 5309 CrossRef CAS PubMed.
- T. Yu, Z.-F. Huang, W. Zhao, J.-J. Xu and H.-Y. Chen, Talanta, 2018, 178, 594 CrossRef CAS PubMed.
- W. Xu, X. J. Xue, T. H. Li, H. Q. Zeng and X. G. Liu, Angew. Chem., Int. Ed., 2009, 48, 6849 CrossRef CAS PubMed.
- Z. Zhou, W. Wei, Y. Zhang and S. Liu, J. Mater. Chem. B, 2013, 1, 2851 RSC.
- X. Yang and Z. Gao, Nanoscale, 2014, 6, 3055 RSC.
- J. K. Wong, S. P. Yip and T. M. Lee, Small, 2014, 10, 1495 CrossRef CAS PubMed.
- J. Li, T. Deng, X. Chu, R. Yang, J. Jiang, G. Shen and R. Yu, Rolling, Anal. Chem., 2010, 82, 28116 Search PubMed.
- P. Liu, X. Yang, S. Sun, Q. Wang, K. Wang, J. Huang, J. Liu and L. He, Anal. Chem., 2013, 85, 7689 CrossRef CAS PubMed.
- C. Ma, W. Wang, A. Mulchandani and C. Shi, Anal. Biochem., 2014, 457, 19 CrossRef CAS PubMed.
- C. Ma, W. Wang, Z. Li, L. Cao and Q. Wang, Anal. Biochem., 2012, 429, 99 CrossRef CAS PubMed.
- Z. Wu, G.-Q. Liu, X.-L. Yang and J.-H. Jiang, J. Am. Chem. Soc., 2015, 137, 6829 CrossRef CAS PubMed.
- D. X. Li, W. J. Zhou, R. Yuan and Y. Xiang, Anal. Chem., 2017, 89, 9934 CrossRef CAS PubMed.
- C.-P. Liang, P.-Q. Ma, H. Liu, X. G. Guo, B.-C. Yin and B.-C. Ye, Angew. Chem., Int. Ed., 2017, 56, 9077 CrossRef CAS PubMed.
- X. W. He, T. Zeng, Z. Li, G. L. Wang and N. Ma, Angew. Chem., Int. Ed., 2016, 55, 3073–3076 CrossRef CAS PubMed.
- P. W. Wu, K. Hwang, T. Lan and Y. Lu, J. Am. Chem. Soc., 2013, 135, 5254 CrossRef CAS PubMed.
- L. Li, J. Feng, Y. Y. Fan and B. Tang, Anal. Chem., 2015, 87, 4829 CrossRef CAS PubMed.
- H. Y. Peng, X.-F. Li, H. Q. Zhang and X. C. Le, Nat. Commun., 2017, 8, 14378 CrossRef PubMed.
- Y. J. Yang, J. Huang, X. H. Yang, X. X. He, K. Quan, N. L. Xie, M. Ou and K. M. Wang, Anal. Chem., 2017, 89, 5850 CrossRef CAS PubMed.
- Y. J. Yang, J. Huang, X. H. Yang, K. Quan, H. Wang, L. Ying, N. L. Xie, M. Ou and K. M. Wang, Anal. Chem., 2016, 88, 5981 CrossRef CAS PubMed.
- W. J. Wang, N. S. Reddy Satyavolu, Z. K. Wu, J.-R. Zhang, J.-J. Zhu and Y. Lu, Angew. Chem., Int. Ed., 2017, 6, 6798 CrossRef PubMed.
- Z.-M. He, P.-H. Zhang, X. Li, J.-R. Zhang and J.-J. Zhu, Sci. Rep., 2016, 6, 22737 CrossRef CAS PubMed.
- X. Li, X. Wang, L. Zhang, S. Lee and H. Dai, Science, 2008, 319, 1229 CrossRef CAS PubMed.
- Y. Wang, Z. Li, J. Wang, J. Li and Y. Lin, Trends Biotechnol., 2011, 29, 205 CrossRef CAS PubMed.
- Y. Liu, X. Dong and P. Chen, Chem. Soc. Rev., 2012, 41, 2283 RSC.
- H. Vovusha and B. Sanyal, RSC Adv., 2015, 5, 67427 RSC.
- B. W. Liu, S. Salgado, V. Maheshwari and J. W. Liu, Curr. Opin. Colloid Interface Sci., 2016, 26, 41 CrossRef CAS.
- S. J. Sowerby, C. A. Cohn, W. M. Heckl and N. G. Holm, PNAS, 2001, 98, 820 CrossRef CAS PubMed.
- X. Liu, R. Aizen, R. Freeman, O. Yehezkeli and I. Willner, ACS Nano, 2012, 6, 3553 CrossRef CAS PubMed.
- C. Chen and B. Li, J. Mater. Chem. B, 2013, 1, 6671 Search PubMed.
- Y. Ma, L. Chen, L. Zhang, S. Liao and J. Zhao, Analyst, 2015, 140, 4076 RSC.
- H. Liu, L. Li, Q. Wang, L. Duan and B. Tang, Anal. Chem., 2014, 86, 5487 CrossRef CAS PubMed.
- W. Li, T. Hou, M. Wu and F. Li, Talanta, 2016, 148, 116 CrossRef CAS PubMed.
- J. Ge, Z.-M. Huang, Q. Xi, R.-Q. Yu, J.-H. Jiang and X. Chu, Chem. Commun., 2014, 50, 11879 RSC.
- X. Wang, A. Jiang, T. Hou, H. Li and F. Li, Biosens. Bioelectron., 2015, 70, 324 CrossRef CAS PubMed.
- L. Li, J. Feng, H. Y. Liu, Q. L. Li, L. L. Tong and B. Tang, Chem. Sci., 2016, 7, 1940 RSC.
- M. Hong, L. D. Xu, Q. W. Xue, L. Li and B. Tang, Anal. Chem., 2016, 88, 12177 CrossRef CAS PubMed.
- M. Liu, J. Song, S. Shuang, C. Dong, J. D. Brennan and Y. Li, ACS Nano, 2014, 8, 5564 CrossRef CAS PubMed.
- X. Zhu, Y. Shen, J. Cao, L. Yin, F. Ban, Y. Shu and G. Li, Chem. Commun., 2015, 51, 10002 RSC.
- Z. Zhang, Y. Y. Wang, N. B. Zhang and S. S. Zhang, Chem. Sci., 2016, 7, 4184 RSC.
- C. Hong, A. Baek, S. S. Hah, W. Jung and D.-E. Kim, Anal. Chem., 2016, 88, 2999 CrossRef CAS PubMed.
- H. Yan, Y. C. Xu, Y. Lu and W. L. Xing, Anal. Chem., 2017, 89, 10137 CrossRef CAS PubMed.
- W. Yun, H. Wu, X. Y. Liu, M. Fu, J. L. Jiang, Y. F. Du, L. Z. Yang and Y. Huang, Anal. Chim. Acta, 2017, 986, 115 CrossRef CAS PubMed.
- S. J. Zhen, X. Xiao, C. H. Li and C. Z. Huang, Anal. Chem., 2017, 89, 8766 CrossRef CAS PubMed.
- X. L. Huang, Y. J. Liu, B. Yung, Y. H. Xiong and X. Y. Chen, ACS Nano, 2017, 11, 5238 CrossRef CAS PubMed.
- Q. Xi, D. M. Zhou, Y. Y. Kan, J. Ge, Z. K. Wu, R. Q. Yu and J. H. Jiang, Anal. Chem., 2014, 86, 1361 CrossRef CAS PubMed.
- J. Li, D. X. Li, R. Yuan and Y. Xiang, ACS Appl. Mater. Interfaces, 2017, 9, 5717 CAS.
- F. Chen, M. Bai, K. Cao, Y. Zhao, J. Wei and Y. X. Zhao, Adv. Funct. Mater., 2017, 27, 1702748 CrossRef.
- F. Chen, M. Bai, K. Cao, Y. Zhao, X. W. Cao, J. Wei, N. Wu, J. Li, L. H. Wang, C. H. Fan and Y. X. Zhao, ACS Nano, 2017, 11, 11908 CrossRef CAS PubMed.
- H. H. Fan, Z. L. Zhao, G. B. Yan, X. B. Zhang, C. Yang, H. M. Meng, Z. Chen, H. Liu and W. H. Tan, Angew. Chem., Int. Ed., 2015, 54, 4801 CrossRef CAS PubMed.
- G. Papadakis, A. Tsortos and E. Gizeli, Nano Lett., 2010, 10, 5093 CrossRef CAS PubMed.
- H. Brutzer, F. W. Schwarz and R. Seidel, Nano Lett., 2012, 12, 473 CrossRef CAS PubMed.
- Z. Zhang, Y. Wang, F. Zheng, R. Ren and S. S. Zhang, Chem. Commun., 2015, 51, 907 RSC.
- Y. Li, C. Lei, Y. Zeng, X. Ji and S. S. Zhang, Chem. Commun., 2012, 48, 10892 RSC.
- H. Zhang, Y. Liu, J. Gao and J. Zhen, Chem. Commun., 2015, 51, 16836 RSC.
- C. Heise and F. Bier, Immobilization of DNA on Micro-arrays: Immobilisation of DNA on Chips II, in Topics in Current Chemistry, ed. C. Wittmann, Springer Berlin/Heidelberg, 2005, vol. 261, pp. 1–25 Search PubMed.
- V. Tjong, L. Tang, S. Zauscher and A. Chilkoti, Chem. Soc. Rev., 2014, 43, 1612 RSC.
- Y. Pang, C. Wang, J. Wang, Z. Sun, R. Xiao and S. Wang, Biosens. Bioelectron., 2016, 79, 574 CrossRef CAS PubMed.
- S. Ye, Y. Yang, J. Xiao and S. S. Zhang, Chem. Commun., 2012, 48, 8535 RSC.
- Y. Li, C. Yu, H. Han, C. Zhao and X. Zhang, Biosens. Bioelectron., 2016, 81, 111 CrossRef CAS PubMed.
- X. M. Li, L. Wang and C. X. Li, Chem. – Eur. J., 2015, 21, 6817 CrossRef CAS PubMed.
- S. Ye, J. Xiao, Y. Guo and S. Zhang, Chem. – Eur. J., 2013, 19, 8111 CrossRef CAS PubMed.
- S. J. Ye, Y. Y. Wu, W. Zhang, N. Li and B. Tang, Chem. Commun., 2014, 50, 9409 RSC.
- P. P. Hu, H. Liu, S. J. Zhen, C. M. Li and C. Z. Huang, Biosens. Bioelectron., 2015, 73, 228 CrossRef CAS PubMed.
- F. L. Gao, Z. Zhu, J. P. Lei and H. X. Ju, Chem. Commun., 2012, 48, 10603 RSC.
- S. Ye, Y. Wu, X. Zhai and B. Tang, Anal. Chem., 2015, 87, 8242 CrossRef CAS PubMed.
- Y. He, X. Yang, R. Yuan and Y. Q. Chai, Anal. Chem., 2017, 89, 2866 CrossRef CAS PubMed.
- L. Tang, Y. Liu, M. M. Ali, D. K. Kang, W. Zhao and J. Li, Anal. Chem., 2012, 84, 4711 CrossRef CAS PubMed.
- C. Lin, Y. Zhang, X. Zhou, B. Yao and Q. Fang, Biosens. Bioelectron., 2013, 47, 515 CrossRef CAS PubMed.
- S. Bi, Y. Cui, Y. Dong and N. Zhang, Biosens. Bioelectron., 2014, 53, 207 CrossRef CAS PubMed.
- X. Yu, Z. L. Zhang and S. Y. Zheng, Biosens. Bioelectron., 2015, 66, 520 CrossRef CAS PubMed.
- Y. Zhang, W. Ren, H. Q. Luo and N. B. Li, Biosens. Bioelectron., 2016, 80, 463 CrossRef CAS PubMed.
- T. Yu, P. P. Dai, J. J. Xu and H. Y. Chen, ACS Appl. Mater. Interfaces, 2016, 8, 4434 CAS.
- L. L. Yang, Y. Z. Tao, G. Y. Yue, R. B. Li, B. Qiu, L. H. Guo, Z. Y. Lin and H. H. Yang, Anal. Chem., 2016, 88, 5097 CrossRef CAS PubMed.
- Y. Zhang, L. X. Wang, F. Luo, B. Qiu, L. H. Guo, Z. Q. Weng, Z. Y. Lin and G. N. Chen, Chem. Commun., 2017, 53, 2910 RSC.
- X. Y. Jiang, H. J. Wang, H. J. Wang, Y. Zhuo, R. Yuan and Y. Q. Chai, Anal. Chem., 2017, 89, 4280 CrossRef CAS PubMed.
- S. Bi, T. T. Zhao, B. Y. Luo and J.-J. Zhu, Chem. Commun., 2013, 49, 6906 RSC.
- C. Shi, Y. J. Ge, H. X. Gu and C. P. Ma, Biosens. Bioelectron., 2011, 26, 4697 CrossRef CAS PubMed.
- S. Bi, J. L. Zhang and S. S. Zhang, Chem. Commun., 2010, 46, 5509 RSC.
- Y. S. Guo, S. Li, Y. J. Wang and S. S. Zhang, Anal. Chem., 2017, 89, 2267 CrossRef CAS PubMed.
- Y. S. Guo, Y. J. Wang, S. Li, L. Niu, D. Wei and S. S. Zhang, Chem. Commun., 2017, 53, 4826 RSC.
- J. Zheng, Y. P. Hu, J. H. Bai, C. Ma, J. S. Li, Y. H. Li, M. L. Shi, W. H. Tan and R. H. Yang, Anal. Chem., 2014, 86, 2205 CrossRef CAS PubMed.
- M. Q. He, K. Wang, W.-J. Wang, Y.-L. Yu and J.-H. Wang, Anal. Chem., 2017, 89, 9292 CrossRef CAS PubMed.
- Z. Zhang, Y. T. Jiao, M. T. Zhu and S. S. Zhang, Anal. Chem., 2017, 89, 4320 CrossRef CAS PubMed.
- Z. X. Zhang, D. Balogh, F. A. Wang, S. Y. Sung, R. Nechushtai and I. Willner, ACS Nano, 2013, 7, 8455 CrossRef CAS PubMed.
- C.-H. Lu and I. Willner, Angew. Chem., Int. Ed., 2015, 54, 12212 CrossRef CAS PubMed.
- D. Balogh, M. A. Aleman Garcia, H. B. Albada and I. Willner, Angew. Chem., Int. Ed., 2015, 54, 11652 CrossRef CAS PubMed.
- Z. X. Zhang, D. Balogh, F. A. Wang and I. Willner, J. Am. Chem. Soc., 2013, 135, 1934 CrossRef CAS PubMed.
- K. W. Ren, J. Wu, Y. Zhang, F. Yan and H. X. Ju, Anal. Chem., 2014, 86, 7494 CrossRef CAS PubMed.
- X. M. Wang, M. R. Cui, H. Zhou and S. S. Zhang, Chem. Commun., 2015, 51, 13983 RSC.
- J. Wang, X.-L. Li, J.-D. Zhang, N. Hao, J.-J. Xu and H.-Y. Chen, Chem. Commun., 2015, 51, 11673 RSC.
- P. H. Zhang, Z. M. He, C. Wang, J. N. Chen, J. J. Zhao, X. N. Zhu, C.-Z. Li, Qi. H. Min and J.-J. Zhu, ACS Nano, 2015, 9, 789 CrossRef CAS PubMed.
- Z. Zhang, Y. T. Jiao, Y. Y. Wang and S. S. Zhang, Sci. Rep., 2016, 6, 29872 CrossRef CAS PubMed.
- D. G. He, X. He, X. Yang and H.-W. Li, Chem. Sci., 2017, 8, 2832 RSC.
- Y. Huang, X. Q. Liu, H. K. Huang, J. Qin, L. L. Zhang, S. L. Zhao, Z.-F. Chen and H. Liang, Anal. Chem., 2015, 87, 8107 CrossRef CAS PubMed.
- A. F. Jou, C.-H. Lu, Y.-C. Ou, S.-S. Wang, S.-L. Hsu, I. Willner and J. A. Ho, Chem. Sci., 2015, 6, 659 RSC.
- C. X. Song, X. H. Yang, K. M. Wang, Q. Wang, J. Huang, J. B. Liu, W. Liu and P. Liu, Anal. Chim. Acta, 2014, 827, 74 CrossRef CAS PubMed.
- T.-G. Cha, J. Pan, H. R. Chen, J. Salgado, X. Li, C. D. Mao and J. H. Choi, Nat. Nanotechnol., 2014, 9, 39 CrossRef CAS PubMed.
- Q. Wang, B.-C. Yin and B.-C. Ye, Biosens. Bioelectron., 2016, 80, 366 CrossRef CAS PubMed.
- W.-H. Chen, X. Yu, A. Cecconello, Y. S. Sohn, R. Nechushtai and I. Willner, Chem. Sci., 2017, 8, 5769 RSC.
- L. P. Qiu, T. Zhang, J. H. Jiang, C. C. Wu, G. Z. Zhu, M. X. You, X. G. Chen, L. Q. Zhang, C. Cui, R. Q. Yu and W. H. Tan, J. Am. Chem. Soc., 2014, 136, 13090 CrossRef CAS PubMed.
- X. X. Zhang, K. Y. Xiao, L. W. Cheng, H. Chen, B. H. Liu, S. Zhang and J. L. Kong, Anal. Chem., 2014, 86, 5567 CrossRef CAS PubMed.
- Y. M. Wang, Z. Wu, S. J. Liu and X. Chu, Anal. Chem., 2015, 87, 6470 CrossRef CAS PubMed.
- G. Z. Zhu, S. F. Zhang, E. Q. Song, J. Zheng, R. Hu, X. H. Fang and W. H. Tan, Angew. Chem., Int. Ed., 2013, 52, 5490 CrossRef CAS PubMed.
- Z. Li, X. W. He, X. C. Luo, L. Wang and N. Ma, Anal. Chem., 2016, 88, 9355 CrossRef CAS PubMed.
- Y. Zhang, Z. W. Chen, Y. Tao, Z. Z. Wang, J. S. Ren and X. G. Qu, Chem. Commun., 2015, 51, 11496 RSC.
- P. Song, D. K. Ye, X. L. Zuo, J. Li, J. B. Wang, H. J. Liu, M. T. Hwang, J. Chao, S. Su, L. H. Wang, J. Y. Shi, L. H. Wang, W. Huang, R. Lal and C. H. Fan, Nano Lett., 2017, 17, 5193 CrossRef CAS PubMed.
- K. W. Ren, Y. Liu, J. Wu, Y. Zhang, J. Zhu, M. Yang and H. X. Ju, Nat. Commun., 2016, 7, 13580 CrossRef CAS PubMed.
- X. R. Zhang, R. J. Li, Y. Y. Chen, S. S. Zhang, W. S. Wang and F. C. Li, Chem. Sci., 2016, 7, 6182 RSC.
- J. L. Tang, Y. R. Yu, H. Shi, X. X. He, Y. L. Lei, J. F. Shangguan, X. Yang, Z. Z. Qiao and K. M. Wang, Anal. Chem., 2017, 89, 6637 CrossRef CAS PubMed.
- D. S. Seferos, A. E. Prigodich, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Nano Lett., 2009, 9, 308 CrossRef CAS PubMed.
- H. Q. Zhang, F. Li, B. Dever, X.-F. Li and X. Chris Le, Chem. Rev., 2013, 113, 2812 CrossRef CAS PubMed.
- H. M. Meng, H. Liu, H. L. Kuai, R. Z. Peng, L. T. Mo and X.-B. Zhang, Chem. Soc. Rev., 2016, 45, 2583 RSC.
- Y. H. Peng, B. Xiong, L. Peng, H. Li, Y. He and E. S. Yeung, Anal. Chem., 2015, 87, 200 CrossRef CAS PubMed.
- B. Xiong, R. Zhou, J. R. Hao, Y. H. Jia, Y. He and E. S. Yeung, Nat. Commun., 2013, 4, 1708 CrossRef PubMed.
Footnote |
| † Hong Zhou and Jing Liu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |






![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 with H-index 84 (up to January, 2018), and several chapters and books. He has won lots of Advanced Achievement Awards in Science and Technology, and delivered numerous plenary and invited lectures at various conferences, universities and elsewhere. His research interests include electrochemical and photoelectrochemical sensing, bioelectrochemistry, nano and supermolecular electrochemistry, ultramicroelectrodes and biomolecular-electronic devices and Micro-Total Analysis System.
300 with H-index 84 (up to January, 2018), and several chapters and books. He has won lots of Advanced Achievement Awards in Science and Technology, and delivered numerous plenary and invited lectures at various conferences, universities and elsewhere. His research interests include electrochemical and photoelectrochemical sensing, bioelectrochemistry, nano and supermolecular electrochemistry, ultramicroelectrodes and biomolecular-electronic devices and Micro-Total Analysis System.