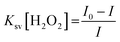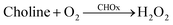DOI:
10.1039/D0RA03055D
(Paper)
RSC Adv., 2020,
10, 24190-24202
CdTe QD-based inhibition and reactivation assay of acetylcholinesterase for the detection of organophosphorus pesticides†
Received
5th April 2020
, Accepted 8th June 2020
First published on 25th June 2020
Abstract
An enzyme immobilized glutathione (GSH)-capped CdTe quantum dot (QD)-based fluorescence assay has been developed for monitoring organophosphate pesticides. In principle, GSH-capped CdTe QDs exhibit higher sensitivity towards H2O2 produced from the active enzymatic reaction of acetylcholinesterase (AChE) and choline oxidase (CHOx), which results in the fluorescence (FL) “turn-off” of the GSH-capped CdTe QDs. A “turn-on” FL of the CdTe QDs at 520 nm was recovered in the presence of organophosphate (OP). The FL changes of the GSH-capped CdTe QD/AChE/CHOx biosensor reasonably correspond to the amount of OP pesticides. The detection limit of the CdTe/AChE/CHOx biosensor towards paraoxon, dichlorvos, malathion and triazophos was 1.62 × 10−15 M, 75.3 × 10−15 M, 0.23 × 10−9 M and 10.6 × 10−12 M, respectively. The GSH-capped CdTe QDs/AChE/CHOx biosensor was applied as a FL nanoprobe for assaying the enzymatic activity of AChE. The inhibited AChE was reactivated up to 94% using pyridine oximate (2-PyOx−), and functionalized pyridinium oximates (4-C12PyOx− and 4-C18PyOx−) of varying chain lengths. It was found that the reactivation potency of the tested oximes varied with the chain length of the oximes. This biosensing system offers the promising benefit for the determination of the OP pesticides in food, water and environmental samples.
Introduction
In the current scenario, the determination of acetylcholinesterase (AChE) activity is of paramount importance due to pharmacological and toxicological concerns.1 AChE is the major enzyme that hydrolyzes acetylcholine, a key neurotransmitter for synaptic transmission, into acetic acid and choline. Mild inhibition has been shown to have therapeutic relevance in Alzheimer's disease (AD), myasthenia gravis, and glaucoma among others.2,3 In contrast, the strong inhibition of AChE can lead to cholinergic poisoning.4 To combat this, AChE reactivators have been developed to remove the offending AChE inhibitors, restoring acetylcholine levels to normal.5,6 It is really challenging to design a nanoprobe for monitoring AChE activity in the presence of reversible (carbamate, acridine) and irreversible (organophosphorus) inhibitors. The probe reported thus far can determine the inhibition7 of AChE, and is limited to monitoring percentage reactivation of organophosphorus-inhibited enzyme.
With high sensitivity and simplification, fluorescence-based sensors have been widely applied as one of the most commonly used sensing candidates for environmental monitoring,8 food safety9 and quality control.10 Quite recently, many optical11–13 and electrochemical14–16 methods/biosensors have been applied to determine pesticide residues in food samples. Various kinds of materials have been widely employed for the fabrication of a fluorescence (FL) sensing platform, including fluorescent dyes,17 semiconductor nanomaterials,18 metal nanomaterials,19,20 carbon quantum dots (CQD),21 and rare earth materials.22 It is also very critical to design a proper recognition unit that can be combined with the FL probe for responding to the fluorescent “turn-off”, “turn-on”, or “ratiometric” signal. Carbon quantum dots have been extensively investigated for probing AChE and the detection of organophosphorus (OP) pesticides.23–25 We have developed a FRET-based CQD–AuNP system for the detection of pesticides, along with monitoring the inhibition and reactivation of AChE.26
Biosensors based on CdTe QDs have been developed due to their unique electronic and optical properties, such as their broad absorption spectra, narrow and symmetric emission bands, less environmental sensitivity, and high quantum yield.27–32 These advantages of the CdTe QDs in the narrow emission band have enabled the sensitive detection of trace elements without spectral interferences. Over the past decades, quantum dots (QDs) have been widely studied and applied for the detection of trace amounts of hazardous substances, including pesticides and heavy metals. The Willner group33 proposed that controlling the photophysical properties of QDs by H2O2 might provide a versatile method to develop CdSe/ZnS QDs-based sensors. Sahoo et al.34 reported the detection of pesticides at the ppm level in water through an optical and electrochemical approach using zinc oxide quantum dots as the nanosensor. An in situ growth of CdTe QDs was shown to be a novel route to the label-free and visual detection of butyrylcholinesterase (BChE), an important enzyme in human bodies.35 The concentration of thiocholine produced by the enzymolysis of butyrylcholine (BTCh) was found to be proportional to the FL intensity of QD. A fluorescence turn-on microfluidic sensor composed of a QD-AChE aerogel has been demonstrated for the detection of the OP pesticides.36 The FL intensity of the QD-AChE aerogel is quenched by thiocholine (generated from the AChE-catalyzed hydrolysis of ATCh), which is recovered (turn-on) by the addition of OPs. A fluorescent nanocomposite was fabricated using a simple supramolecular method to coat mercaptopropionic acid-capped CdTe QDs with surfactant–cyclodextrin shells. The strong fluorescence of the nanocomposite is effectively quenched by the addition of paraoxon due to the host–guest complexation between the β-CD cavity and paraoxon degradation product. The potential of the CdTe/CTAB/β-CD composite sensor was also tested with blood samples of paraoxon-poisoned rats.37 Moreover, a TGA-capped CdTe QDs paper-based device was developed for a simple multi-step bi-enzyme (AChE and choline oxidase) assay for OP and carbamate insecticides detection. The assay is based on the changing of the fluorescence intensity of TGA-capped CdTe QDs due to the reduction of H2O2 produced by the bi-enzyme reaction in the presence of insecticides.38 Likewise, the GSH-capped Mn-doped ZnS quantum dots were applied as a phosphorescent probe for the bi-enzymatic determination of organophosphorus pesticides (paraoxon) with an ∼0.1 pM limit of detection.39 For most of these methods, the inhibitory protocols have been applied in which AChE is deactivated and cannot be further used. This cholinesterase reactivator is used as an antidote for poisoning due to organophosphate pesticides/nerve agents or other chemicals that have anticholinesterase activity.40 It is also of value in the control of an overdosage by anticholinesterase agents used in the treatment of myasthenia gravis.41 Therefore, the highly sensitive nanoprobe is essential for monitoring the inhibition and reactivation of AChE.
In this work, a simple, rapid, and sensitive method for the optical detection of organophosphorus pesticides (paraoxon, dichlorvos, malathion and triazophos), which is based on the inhibition mechanism of AChE (Scheme 1) has been developed. This detection method of OPs is mainly composed of one step, which involves blending the sample with AChE and the solution containing ACh, CHOx and QDs (Scheme 1), without the complex process of an assembly for the biosensor. The principle of the method is the inhibition efficiency of the pesticide to AChE activity could be evaluated by measuring the fluorescence changes of the CdTe QDs. Systematic studies on the different inhibition effects of the oxo form and thio form of OPs have been performed. The matrix effect often occurs in the analysis of real samples. Therefore, the interference of metal ions and selected organic compounds has been investigated. The results exhibited a good linear relationship in the range of 1.6 × 10−15 M to 3.02 × 10−12 M (paraoxon), 75.3 × 10−15 M to 2.26 × 10−9 M (dichlorvos), 0.23 × 10−9 M to 3.54 × 10−9 M (malathion) and 10.6 × 10−12 M to 1.59 × 10−9 M (triazophos), respectively.
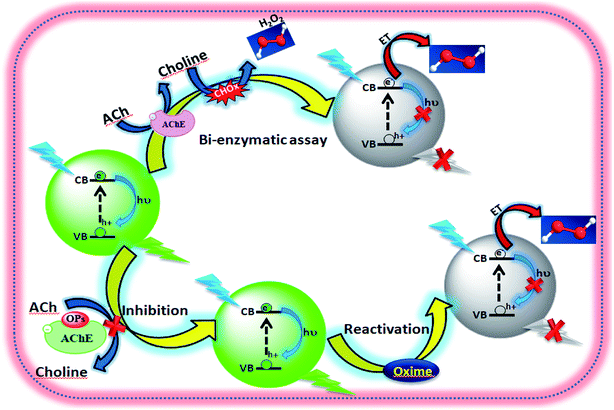 |
| | Scheme 1 Schematic presentation of fluorescence probing inhibition and reactivation of AChE. | |
The biosensor showed a low detection limit of 1.6 × 10−15 M, 75.3 × 10−15 M, 0.23 × 10−9 M and 10.6 × 10−12 M for paraoxon, dichlorvos, malathion and triazophos, respectively. Furthermore, a possible mechanism for the fluorescence quenching of CdTe QDs in the presence of H2O2 has been proposed. At the initial stage, the detachment of thiol (–SH) from the surface of the QDs caused quenching of the FL intensity. When the QDs were seriously quenched, the quenching of the QDs was due to the combination of the detachment of a thiol at the ion from the surface of the QDs and the oxidation of tellurium (Te). The results also reveal that the proposed biosensor can be applied in real samples.
Similarly, for the recovery of the inhibited enzyme activity, oxime reactivators (4-C18PyOx−, 4-C12PyOx− and 2-PyOx−) are used as atropine. The recovery percentages, reactivation rate constant and kinetic parameter (namely kr, KD, kr2) of different oximes have been reported.
Experimental
Synthesis of GSH-capped CdTe QDs
Briefly, a NaHTe solution was prepared by dissolving Te powder (1 mM) and NaBH4 (4 mM) in 5 mL of nitrogen bubbled Millipore water. The solution was ultrasonicated for 1 h to produce a pink colored NaHTe solution. In a three-necked flask with an attached condenser, Cd2+ precursor solutions were prepared by mixing a solution of Cd(CH3COO)2 (0.2280 g, 0.0127 M) in the presence of GSH (0.2768 g, 0.03 M) as a stabilizing agent, and the solutions were then adjusted to pH 11.0 using 1 M NaOH. The resulting solution was despoiled by N2 for 30 min. Under vigorous stirring, the oxygen-free NaHTe solution, as prepared above, was injected. In our experiments, the typical molar ratio of Cd2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaHTe
NaHTe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GSH was 1
GSH was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2
0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.4. The resulting colloidal CdTe was refluxed under nitrogen flow at 100 °C for different times to obtain the GSH-capped CdTe QDs of different sizes. The aliquots of the reaction solution were pipetted out at different intervals of time (10 min interval) for the optical measurements.
2.4. The resulting colloidal CdTe was refluxed under nitrogen flow at 100 °C for different times to obtain the GSH-capped CdTe QDs of different sizes. The aliquots of the reaction solution were pipetted out at different intervals of time (10 min interval) for the optical measurements.
FL quantum yield of GSH-capped CdTe QDs
The fluorescence quantum yields (QYs) of the samples were determined relative to fluorescein (in 0.1 M NaOH) as a standard using the gradient method.29 Eqn (1) was followed for the calculation of QY:| |
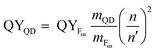 | (1) |
where QYFsn = 0.95 is the QY of fluorescein dissolved in 0.1 M NaOH, mFsn and mQD are the slopes of curves acquired by plotting the integrated fluorescence intensity versus the optical density of solutions of different concentrations of the fluorescein and QDs, respectively. n = 1.32 and n′ = 1.35 are the refractive indices of the solvents in which the QDs and the Fsn dye were dissolved, respectively.
Time-resolved measurements
For the time-resolved fluorescence measurements, the samples were excited at 380 nm using an Edinburg FLS-980 time-resolved fluorescence spectrometer with a 450 W Xe arc continuous lamp as a light source and cooled PMT tube as the detector. A Xe pulse lamp with 60 W power and 1 μs pulse width was used for the lifetime measurements.
Fluorescence assay for the inhibition of AChE
The GSH-capped CdTe QDs were reasonably utilized to quantify the organophosphate pesticide (OP) by following the fluorescence response of CdTe QDs upon the reaction with H2O2 generated from acetylcholinesterase (AChE) and choline oxidase (CHOx). Every portion of the proposed biosensors was prepared in the same stock solution. Under experiments, 10 mL of AChE (10 mU mL−1) and 10 mL of CHOx (1.25 U mL−1) in tris buffer were first added to 3 mL of CdTe QDs solution, and then the solution was added to 25 mL of tris buffer solution (50 mM, pH 8). The solution was stirred for 5 min prior to incubation at 5 °C for 24 h. Stocks of the CdTe quantum dot-immobilized enzymes (CdTe QDs/AChE/CHOx) were kept at 5 °C prior to use.
Inhibition of AChE and detection of organophosphate
The solution of 0.7 mL of Milli-Q water and 0.2 mL of different concentrations of organophosphate pesticide was added to the biosensor solution, and incubated for 15 min in a water bath at 37 °C. The mixture solution was added to 0.2 mL acetylcholine (10 mg mL−1), and incubated by stirring for 30 min. The PL signals were monitored by fluorescence spectroscopy, and the measurements were repeated 3 times. The percentage of the enzyme inhibition (I%) was plotted against the concentration of the organophosphate pesticide to obtain the calibration curve. The percentage of the enzyme inhibition (I%) was calculated using eqn (2), where F0 and FI are the fluorescence quenching without and with organophosphate pesticide after incubation for 30 min.| |
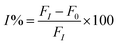 | (2) |
Reactivation of inhibited AChE
The degree of reactivated AChE was measured as an increase of the FL intensity at 520 nm. Stock solutions (10 mM) of the reactivators, viz., quaternized oxime (4-C18PyOx−, 4-C12PyOx−), and syn-pyridine aldoxime (2-PyOx−) were prepared. The stock solution of 4-C12PyOx− was diluted to 10–1000 μM, and was subsequently reacted with the inhibited AChE, following the addition of ATChI and the CdTe QDs solution, and incubated for 5–10 min. The FL spectra of the solution was recorded at 520 nm. The inhibition reactivation efficiency (R%) was taken as the output parameter and calculated using eqn (3), where FI is from the activity of the controlled intact enzyme (relative to the activity of the inhibited enzyme), and Fr is from the activity of the reactivated enzyme.| |
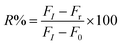 | (3) |
During the experiment process, the control batch was set up in which the activity of the enzyme was kept constant (completely inhibited). The values of the reactivation study of the inhibited enzyme were evaluated by calculating the ratio of the recovered enzyme activity and control activity. The optimal parameters, viz., kobs, KD, kr, were calculated by a non-linear fit (eqn (4) derived from the following Scheme 2). Here, kobs is the apparent reactivation rate constant of the oxime, KD is the dissociation constant of the inhibited enzyme–oxime complex [EPOX], and kr is the maximal reactivation rate constant. A calculation of the constant values was performed by a non-linear regression analysis using the curve fitting program Prism™ Ver. 6.1 (Graph Pad Software, San Diego, USA).
| |
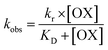 | (4) |
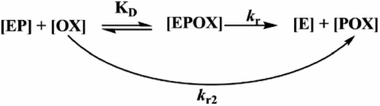 |
| | Scheme 2 Schematic representation of the reactivation of the OP-inhibited AChE by oxime. | |
Organophosphate detection in real samples
The organophosphate detection in real samples was carried out as follows: (1) apple, tomato juice, field water, and sample; all were kept at room temperature for a day. The spiked organophosphate pesticide with different concentrations were injected in the CdTe QDs/enzyme biosensor, and then the mixture was incubated at 37 °C for 15 min; (2) the CdTe QDs/enzyme biosensor was added by acetylcholine, and then incubated at 37 °C for 30 min; (3) the FL spectrum was recorded and the FL intensity was plotted as a calibration curve. The organophosphate pesticide in the real sample water was determined from this calibration curve.
Results and discussion
Synthesis and characterization of probe
The CdTe quantum dots were synthesized using Cd(CH3COO)2 and NaHTe as the Cd and Te sources, respectively, and glutathione as a capping agent. Aliquots withdrawn at the indicated times showed absorbance and fluorescence spectra, which exhibited the characteristic CdTe QDs absorption and fluorescence peaks at 360 nm and 520 nm, respectively (Fig. S1a†), with a high quantum yield (41%). The absorption spectra of CdTe QDs with different sizes (Fig. 1a) were used to calculate the optical bandgap energy (Table S1†). In addition, fractions exposed to UV light (365 nm) showed highly green fluorescence with an absolute intensity that was proportional to the incubation time, shifting from green to yellow, and then to red after 1 h (Fig. 1b and S1b†). Absorption and emission peaks were very narrow (with a full bandwidth at half maximum of 50 nm), supporting the possibility of using these QDs as cellular probes because of their tunable fluorescence (Fig. 1b). Fig. S1b† displayed the normalized FL intensities of the CdTe QDs with different excitation wavelengths from 310 nm to 410 nm. Similarly, we have also studied fluorescence decay parameters (Table S2†), and lifetime measurements of the CdTe QDs solution with a refluxing time interval (different sized QD in Fig. 1c). It was observed that the average FL lifetime increased with increasing size of CdTe QDs. Fig. 1g shows the cyclic voltammogram of the GSH-capped CdTe quantum dots with increasing size. This data gives knowledge about the formation of different sized CdTe QDs. Size is an important factor that affects the potential biological applications of QDs. CdTe QDs ranged from 3–6 nm in size with green and red QDs (Fig S1c†). As expected, yellow QDs exhibited intermediate sizes. A homogeneous, narrow size distribution was observed for all QDs with a distribution range of 2–3 nm, which favors their use in fluorescence applications.
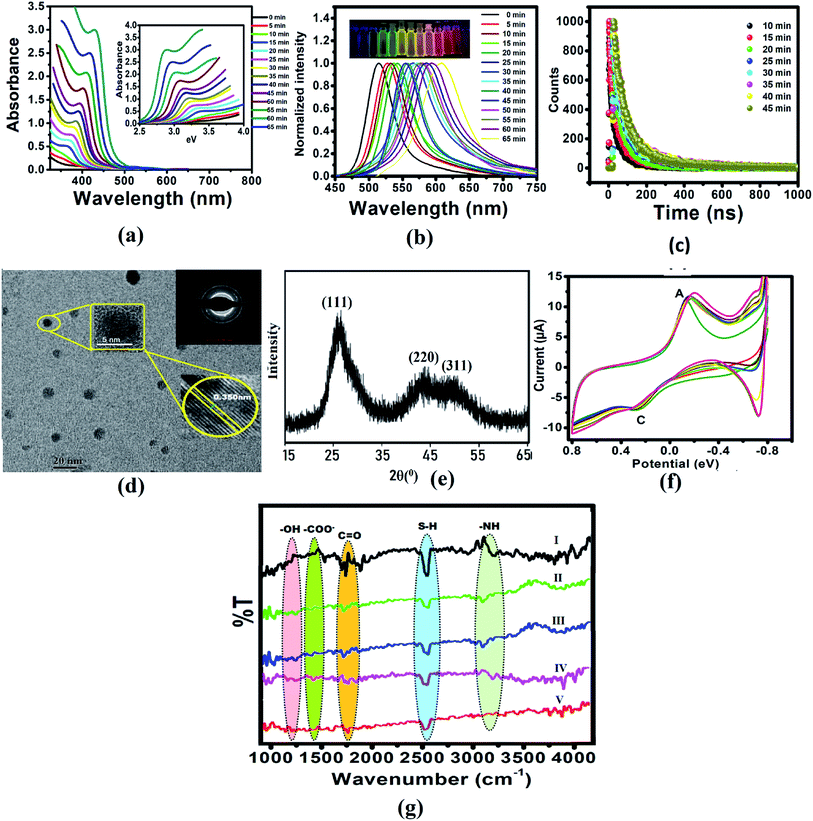 |
| | Fig. 1 (a) Absorption spectra of CdTe QDs with different sizes and refluxing time. The inset shows the relationship between the absorbance and energy gap. (b) Emission spectra of CdTe QDs with different sizes, inset: photographic image observed by the naked eyes under 365 nm UV lamp illumination. (c) Lifetime of CdTe QDs solution with refluxing time interval (different sized QD). (d) TEM image, (e) XRD pattern, (f) FTIR spectrum of CdTe QDs, and (g) cyclic voltammograms of CdTe QDs at different time intervals. | |
Additionally, the QDs nanometric size was confirmed by TEM analysis, and most QDs were average sized (3–5 nm). High-resolution transmission electron microscopy (HRTEM) clearly illustrated the crystallinity and high quality of the CdTe nanocrystals (Fig. 1d). The X-ray diffraction (XRD) pattern (Fig. 1e) of the vacuum-dried CdTe powder showed a peak at 26° (111) and a broad band at ca. 45° due to the overlap of the (220) and (311) diffractions, confirming that the CdTe QDs have a zinc blended crystal structure.
To confirm the presence of GSH in QDs, FTIR studies were conducted for GSH, CdTe-GSH QDs, and CdTe-GSH in buffer, CdTe-GSH/AChE in buffer and CdTe-GSH/AChE/ChOx (Fig. 1f(I–V)). The IR bands around 1713–1602 cm−1 (symmetric ν-COO−), 1397 cm−1 (asymmetric ν-COO−), 1713 cm−1 (antisymmetric ν-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 1280 cm−1 (–OH) indicate the presence of a –COOH group, while the band at 1075 cm−1 can be assigned to a stretching ν-C–N. Characteristic ν-N–H stretching modes observed at 3346 cm−1 and 3250 cm−1 provide evidence of a –NH group. The characteristic –SH stretching mode is clearly seen at 2526 cm−1. As expected, the IR bands of the main functional groups, –COOH, NH–, and –SH are detected in the neat GSH. In the CdTe-GSH QDs, the disappearance of the S–H group vibration at 2526 cm−1 (ν-S–H) is likely the result of a covalent bond formation between the thiol and the Cd atom at the CdTe QD surface, suggesting the oxidation of cysteine residues. These IR results are identical to those described for other CdTe-GSH QDs. The thiol capping of the CdTe QD is further validated by cyclic voltammetry (CV). Fig. 1g shows a cyclic voltammogram of the GSH-capped CdTe quantum dots with increasing size. The cathodic peak (C2) (PGSSG ∼ −0.72 V) was obtained due to the oxidation of the thiol to GSSG (2GSH → GSSG + 2e− + 2H+), which is shifted towards a lower negative potential with increasing quantum dot size. A size dependence CV behavior has also been observed for the thioglycolic acid (TGA) capped-CdTe QDs (Fig. S2†).
O) and 1280 cm−1 (–OH) indicate the presence of a –COOH group, while the band at 1075 cm−1 can be assigned to a stretching ν-C–N. Characteristic ν-N–H stretching modes observed at 3346 cm−1 and 3250 cm−1 provide evidence of a –NH group. The characteristic –SH stretching mode is clearly seen at 2526 cm−1. As expected, the IR bands of the main functional groups, –COOH, NH–, and –SH are detected in the neat GSH. In the CdTe-GSH QDs, the disappearance of the S–H group vibration at 2526 cm−1 (ν-S–H) is likely the result of a covalent bond formation between the thiol and the Cd atom at the CdTe QD surface, suggesting the oxidation of cysteine residues. These IR results are identical to those described for other CdTe-GSH QDs. The thiol capping of the CdTe QD is further validated by cyclic voltammetry (CV). Fig. 1g shows a cyclic voltammogram of the GSH-capped CdTe quantum dots with increasing size. The cathodic peak (C2) (PGSSG ∼ −0.72 V) was obtained due to the oxidation of the thiol to GSSG (2GSH → GSSG + 2e− + 2H+), which is shifted towards a lower negative potential with increasing quantum dot size. A size dependence CV behavior has also been observed for the thioglycolic acid (TGA) capped-CdTe QDs (Fig. S2†).
Principle of catalytic reaction of H2O2
To study the reaction time of H2O2 with CdTe QDs, the fluorescence spectra of the CdTe QDs were estimated within the presence of H2O2 at various time intervals (Fig. 2a). The emission spectra at 520 nm showed a large drop off in the array of 0–35 min. At times greater than 15 min, the emission spectra of CdTe QDs showed a very slight decrease (Fig. 2b). For the time effectiveness for the sensing ability, 30 min was chosen as the ideal synergist time of H2O2. The catalytic activity of H2O2 within CdTe QDs was explored by varying the amount of H2O2. Upon the addition of H2O2, the intensity of FL spectra was decreased as a function of the concentration of H2O2 (Fig. 2c). The great linearity in the H2O2 concentration array of 1–125 mM with R2 = 0.9989 is shown in Fig. 2b. Further, the Stern–Volmer eqn (5), was used to represent the affinity between CdTe QDs and the H2O2 quencher, where I0 and I are the FL-intensity of CdTe QDs in the presence and absence of H2O2 (ranging from 0 to 50 mM), and Ksv is the Stern–Volmer constant. The Stern–Volmer plots (Fig. 2d) demonstrated that the relative FL intensity of the CdTe QDs depends on the amount of H2O2, and the Stern–Volmer constant Ksv is 3.8 mM.| |
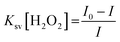 | (5) |
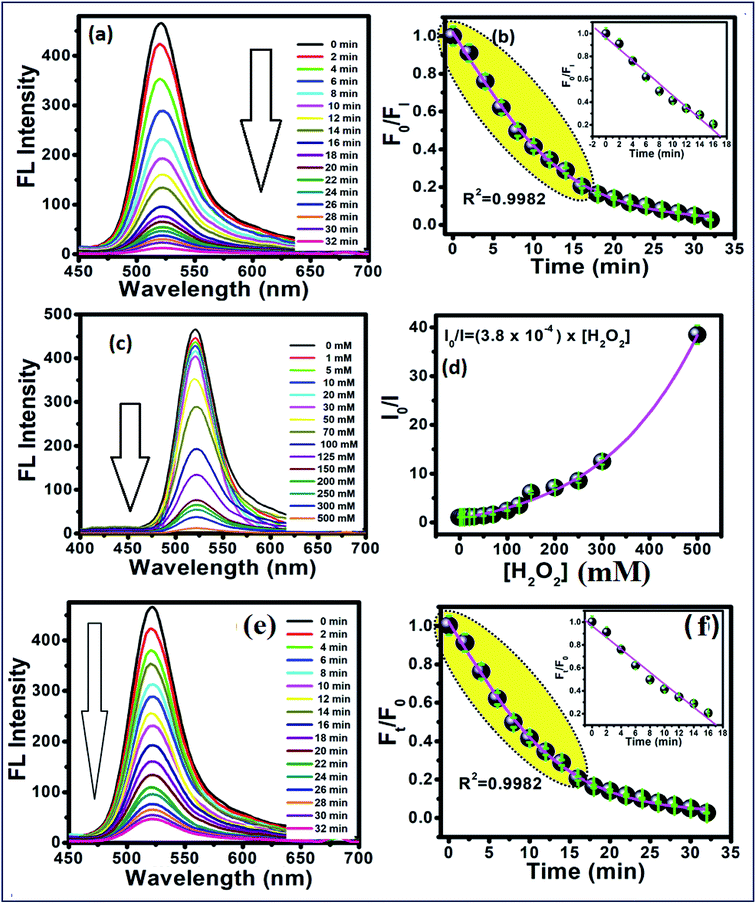 |
| | Fig. 2 (a) Time-dependent fluorescence changes of 5 mM of the CdTe QDs after interacting with 0.1 M H2O2, and (b) calibration curve between time and F0/FI. (c) Concentration-dependent fluorescence changes at 30 min after interacted with different concentration of H2O2. (d) Plots between the concentrations of H2O2 and I0/I. (e) Time-dependent fluorescence changes of CdTe QDs/AChE/CHOx after interaction with acetylcholine (ACh) in tris buffer pH 8, and (f) shows the relationship between Ft/F0 and time. The inset shows the linear calibration curve. | |
As a notable perspective, the CdTe QDs displayed the peroxidase-like catalytic action. The catalytic activity was proposed to happen via the electron transfer process.42 Thus, it was suggested that the small size of the CdTe QDs could facilitate this without much of a stretch and legitimately move the bottomless electron on the surface of CdTe QDs to the LUMO of H2O2, as follows in eqn (6)–(10), bringing about the PL extinguishing of the CdTe-GSH QDs:
| | |
O2 + 2GSH + 4OH− → 2O2− + 2GSSG + 2H2O
| (6) |
| | |
O2− + H2O2 → 4OH* + O2
| (7) |
| | |
O2− + CdTe → CdTe (eisc−)
| (8) |
| | |
OH* + CdTe → CdTe (hisc+)
| (9) |
| | |
CdTe (eisc−) + CdTe (hisc+) → (CdTe QDs)* → hν
| (10) |
Responses to acetylcholine in enzyme immobilized nanoprobe
The immobilized enzyme on the nanomaterials is significant because of the fact that they take advantage of promoting the stability and sensitivity of the enzyme.43 In this work, it is normal that the CdTe-GSH QDs with the edging of the carboxyl, amine and hydroxyl groups could effortlessly interact with acetylcholinesterase (AChE) and choline oxidase (CHOx) through hydrogen bonding interactions. The presence of AChE and CHOx in the framework demonstrated the noteworthy decrease of the fluorescence quenching of CdTe upon increment of time (Fig. 2e). This awesome sight is analogous to the fluorescence quenching of the directly included H2O2. Then again, no such fluorescent changes have been noticed for the other system without both enzymes of AChE and CHOx fused on CdTe QDs. It was observed that the fluorescent quenching of the CdTe QDs occurred as a result of H2O2 created from the enzymatic response, as shown in eqn (11) and (12). Moreover, the H+ particle from the dissociation of acetate and an electron from CdTe QDs could catalyze the turnover of H2O2 to water and oxygen, as appears in eqn (9) and (10), bringing about the perception of little bubbles around the reaction vessel.| |
 | (11) |
| |
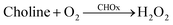 | (12) |
As shown in Fig. 2e, the time-dependent enzymatic reaction was explored in tris buffer at pH 8 on the CdTe QDs/AChE/CHOx biosensor in the presence of acetylcholine. Decreasing the emission spectra at 520 nm was allied with the amount of H2O2 generated by an enzymatic response as a function of the reaction time. To confirm the best possible amount of enzymes, the different concentrations of AChE and CHOx in the CdTe QDs/AChE/CHOx biosensor under time-span was observed in a tris-buffer solution pH 8.0. The fluorescence spectra at 520 nm was essentially decreased in the presence of AChE (10 mU mL−1) and CHOx (1.25 mU mL−1) under the response time of 32 min. On the ground of the material cost utilized, the optimized condition for employing the amount of enzyme in this investigation was chosen as 10 mU mL−1 (AChE) and 1.25 mU mL−1 (CHOx). The incubation time was examined from 0 to 32 min after the addition of acetylcholine. The normalized FL intensity at 520 nm was completely decreased upon the incubation time of 32 min. However, the incubation time of 32 min has not been effective for the valid time detection test. The incubation time at 15 min of the CdTe within the presence of 10 mU mL−1 (AChE) and 1.25 mU mL−1 (CHOx) showed over half (50%) quenching of the emission intensity. After that, incubation times that were greater than 15 min showed a small quenching of the emission intensity. Consequently, the incubation time of the CdTe QDs/AChE/CHOx biosensor with acetylcholine at 15 min (Fig. 2f) was chosen for all experiments. The pH impact on this biosensor plays a significant role for this analysis. When the pH is increased, the FL intensity of the CdTe QDs/AChE/CHOx biosensor (Fig. 2) demonstrated increased affinity up to pH 7–8. At pH > 9, the maximum emission intensity was slightly decreased. Moreover, the FL manner within the presence of ACh with different pH values was inspected in the range of pH 5–9. As in Fig. 3, an enormous decrease of the emission intensity at 520 nm was observed between pH 7–9. As practical consistency with over a comparative fluorescent quenching of CdTe QDs/AChE/CHOx biosensor at pH 7–9, all the experiments for the detection of organophosphates were studied at pH 8.
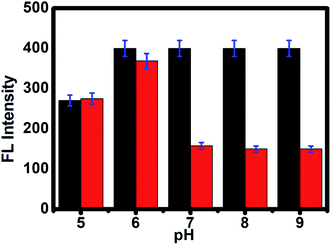 |
| | Fig. 3 The PL intensity of CdTe QDs/AChE/CHOx before (black bar) and after incubation in ACh for 30 min (red bar) at various pH values from 5 to 9. | |
CdTeQDs/AChE/CHOx nanoprobe used for the detection of OPs
The inhibition of AChE by an organophosphorus ester takes place via a chemical reaction in which the serine hydroxyl moiety in the enzyme active site is phosphorylated in a manner analogous to the acetylation of AChE. In contrast to the acetylated enzyme, which rapidly breaks down to give acetic acid and the regenerated enzyme, the phosphorylated enzyme is highly stable. In some cases, depending on the groups attached to the phosphorus atom (R and R′), it is irreversibly inhibited. The serine hydroxyl group, blocked by a phosphoryl moiety, is no longer able to participate in the hydrolysis of ACh. The FL intensity of this CdTe QDs/AChE/CHOx nanoprobe toward organophosphate is shown in Fig. 4a. The FL intensity of the CdTe QDs was gradually increased upon the increment of the paraoxon concentration. This showed that the organophosphate pesticides (OPs) actually inhibited the activity of the enzyme acetylcholinesterase (AChE), and prompted the decrease of H2O2, bringing about the turn-on FL intensity of CdTe QDs within the presence of OPs. Collectively, these outcomes exhibited that the CdTe QDs/AChE/CHOx biosensor can indirectly recognize the amount of OPs through the inhibition of the AChE enzymatic reaction, which induces a fluorescence recovery. Four selected organophosphates (OPs), including paraoxon, dichlorvos, malathion and triazophos at concentrations of 10 ng L−1 to 500 ng L−1, were also analyzed with respect to an inhibition efficiency of the AChE enzymatic response, as demonstrated in Fig. 4b. At 500 ng L−1, the concentration of the OPs tended to be the % inhibition at 90 ± 2% (paraoxon), 74 ± 2%, (dichlorvos), 58 ± 3% (malathion) and 50 ± 1% (triazophos). However, the % inhibition potency in both concentrations of OPs demonstrated the comparable tendency change for all OPs. Taking such previous work in the inhibition capacity of OPs toward AChE, a likely effect of the electrophilicity at a phosphorus molecule has been dependably considered because the inhibition capacity of the oxo-structures is more efficient than the thio-structure.19,26 Another factor is the steric effect of the OPs to the active site of AChE, which contains a profound and tapered crevasse.44,45 In such matters, the inhibition capacity of paraoxon and dichlorvos is more capable than that of malathion and triazophos. Concerning the various inhibition efficiencies of OPs, it possibly contributed to the corresponding absorption capacity of OPs on the surface of the nanoprobe. As per the substance structure of the OPs, dichlorvos pesticide has a more hydrophobic structure than paraoxon. This was potentially excused because the hydrophobic dichlorvos did not favor going through the hydrophilic regions of the CdTeQDs/AChE/CHOx biosensor, bringing about a lower inhibition capacity.44 For example, the inhibition of paraoxon and dichlorvos has been mostly studied in detecting applications. The % inhibition efficiency (I%) of the enzyme was plotted as a function of the concentration of OPs, as appeared in Fig. 4b. The appropriately linear calibration curve was plotted between the logarithm concentration of the OPs and the FL intensity ratio (F0/FI), R2 = 0.99. Fig. 4c shows that the maximum % inhibition efficiency (I%) of paraoxon, dichlorvos, malathion and triazophos is 90 ± 2%, 74 ± 2%, 58 ± 3% and 50 ± 1%, respectively.
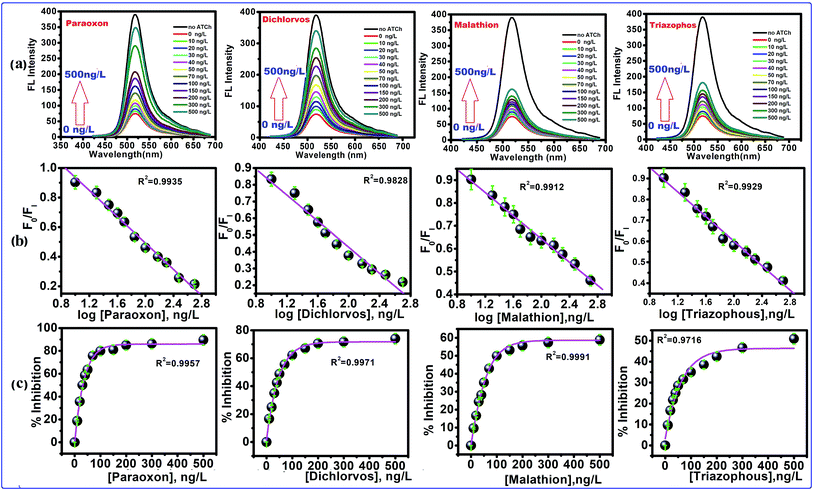 |
| | Fig. 4 The FL quenching of CdTe QDs excited at 380 nm with different concentrations of pesticides. (a) FL spectra of CdTe QDs with increasing concentration of pesticides, (b) the calibration curve of FI/FI0 and concentration of pesticides (FI = FL of intensity of CdTe QDs with inhibited AChE, and FI0 is the FL intensity of CdTe QDs with active AChE), the calibration curve FI/FI0 and logarithm concentration of pesticides, (c) concentration dependent % inhibition of AChE. | |
Often, the detection limit of a pesticide by a sensing framework is tolerable at 10% inhibition, depending on the AChE activity.46–49 Subsequently, the recognition concentration of dichlorvos by the CdTe QDs/AChE/CHOx biosensor was 75.3 × 10−15 M. In correlation, this worth is lower than a potentiometric detection,50 the enzymatic immobilization on a working electrode for an amperometric detection,51 the fluorescence quenching sensor for the Tb3+ complex52 and an optical mode.53 On account of the linear range of 75.3 × 10−15 M to 2.26 × 10−9 M (10–500 ng L−1) (Fig. 4b) with the eq. of I% = 74 ± 2% log[dichlorvos], R2 = 0.9828 exhibited the LOD of 1.6 × 10−15 M. This value is lower than the spectrophotometric cutinized assay54 and conductometric detection.55 The naked eye intensity upon the various concentrations of paraoxon with ACh under a UV light has been shown in Fig. S1d.† Under the UV light, the sensing platform observed the strong green brightness, and a nonbright solution was observed after introducing ACh without OPs. For the remaining solution in the center containers of Fig. S1d,† the tendency of the green brightness of this sensing platform has been raised, leading to an increment of the OPs concentration. These results suggested that the CdTe QDs/AChE/CHOx biosensor was capable of detecting OPs by quantitative visual interpretation.
Determination of organophosphate in real samples
To investigate the sensibleness of the system, the CdTe QDs/AChE/CHOx sensing system was applied to the detection of pesticides in field water, tomato juice and apple juice samples. The calibration curve of OPs was achieved from the standard addition experiment. Table 1 demonstrates the correlation of the concentration of organophosphate that was determined by the CdTeQDs/AChE/CHOx biosensor. The satisfactory accuracy in the amount of OPs at 0.5 to 1.5 (ng mL−1) spiked in the solution system was obtained with the % RSD of 1.23–7.19% and recovery of 82–110.2% for the CdTeQDs/AChE/CHOx biosensor. The developed CdTeQDs/AChE/CHOx biosensor can be used with acceptable accuracy and precision in the quantitative analysis of OPs in various real samples.
Table 1 Detection of organophosphorus pesticides in spiked real samples
| Real samples |
Spiked (ng mL−1) |
Paraoxon |
Dichlorvos |
Malathion |
Triazophos |
| Found (ng mL−1) |
Recovery (%) |
RSD (%) (n = 6) |
Found (ng mL−1) |
Recovery (%) |
RSD (%) (n = 6) |
Found (ng mL−1) |
Recovery (%) |
RSD (%) (n = 6) |
Found (ng mL−1) |
Recovery (%) |
RSD (%) (n = 6) |
| Field water |
0.5 |
0.54 |
110.2 |
2.73 |
0.49 |
97.3 |
3.24 |
0.54 |
104.1 |
3.33 |
0.56 |
110 |
2.72 |
| 1.0 |
0.98 |
97.6 |
6.16 |
1.10 |
101.3 |
1.23 |
0.56 |
93.3 |
1.63 |
0.95 |
95.3 |
1.64 |
| 1.5 |
1.47 |
99.2 |
4.12 |
1.41 |
98.91 |
2.27 |
0.96 |
102.2 |
2.56 |
1.49 |
98.5 |
3.74 |
| Tomato juice |
0.5 |
0.52 |
106.6 |
4.22 |
0.40 |
82.0 |
2.23 |
0.43 |
93.6 |
3.10 |
0.43 |
90.2 |
3.33 |
| 1.0 |
0.97 |
95.4 |
3.91 |
0.97 |
97.0 |
1.76 |
0.98 |
97.6 |
1.55 |
1.42 |
86.6 |
1.90 |
| 1.5 |
1.53 |
103.1 |
3.86 |
1.52 |
102.3 |
4.21 |
1.46 |
96.3 |
2.30 |
1.53 |
105.1 |
4.12 |
| Apple juice |
0.5 |
0.56 |
105.6 |
7.19 |
0.44 |
93.3 |
3.57 |
0.49 |
99.1 |
3.44 |
0.35 |
84.2 |
3.72 |
| 1.0 |
0.94 |
95.4 |
1.35 |
0.95 |
91.6 |
1.81 |
1.11 |
101.6 |
1.43 |
0.99 |
99.1 |
1.53 |
| 1.5 |
1.59 |
107.1 |
4.13 |
1.42 |
98.01 |
3.31 |
1.46 |
92.2 |
2.56 |
1.39 |
91.5 |
4.71 |
Reactivation of inhibited AChE by oximes
After the addition of oximates, such as 4-C18PyOx−, 4-C12PyOx− and 2-PyOx−, to the inhibited AChE for 60–120 min, the observed FL intensity regained almost all of its capability as shown in Fig. 5a, S3(a) and S4(a),† which indicated that the inhibited AChE is reactivated by 4-C18PyOx−, 4-C12PyOx− and 2-PyOx−. The reactivities of AChE by oximes with organophosphate compounds are fairly closely related to the dissociation constants (pKa) of the oximes, although other factors do have some influence, particularly with oximes. When the abilities of these compounds to reactivate the inhibited AChE were compared, marked differences were found. 4-C18PyOx− and 4-C12PyOx− are much more effective in reactivating AChE inhibited with OPs than 2-PyOx− of comparable reactivity with organophosphate compounds.
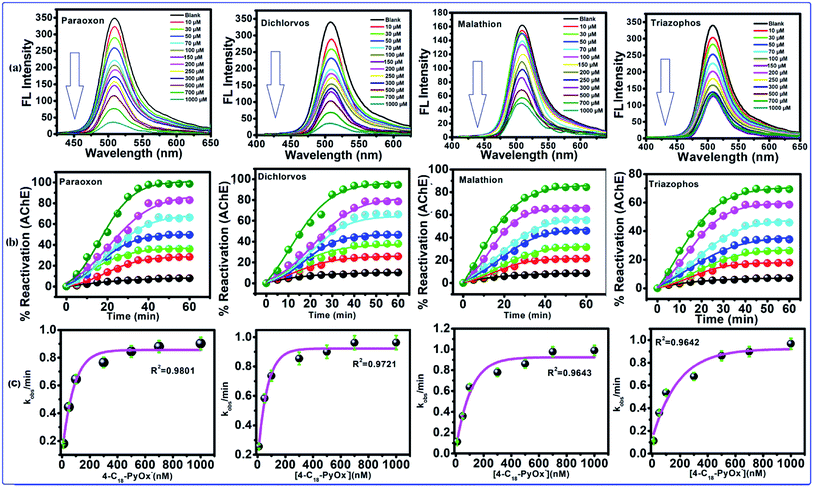 |
| | Fig. 5 Reactivation of inhibited AChE through the OPs using 4-C18-PyOx−. (a) The fluorescence spectra of CdTeQD, in the presence of 4-C18-PyOx−, inhibited AChE by paraoxon, dichlorvos, malathion, triazophos. (b) Plot between the % reactivation of AChE and time/min, and the (c) plot between the kobs/min and concentration of oxime enabled the calculation of KD and kr. | |
The 4-C18PyOx− showed 94.37, 88.12, 84.18 and 73.62% maximum reactivation of inhibited AChE by paraoxon, dichlorvos, malathion and triazophos, respectively (Fig. 5b). It is evident from the plots that maximum reactivation was obtained with respect to the time interval. The maximum concentration of 4-C18PyOx−, i.e., 1000 μM L−1 was obtained at which the intensity was quenched at 520 nm (Fig. 5b). The % reactivation was also calculated and plotted with increasing time interval. The kobs/min was plotted with the concentration of 4-C18PyOx−, and enabled the calculation of KD and kr (Fig. 5c). Likewise, Fig. S3† showed the relationship between the % reactivation and time. At 1000 μM L−1 concentration, 4-C12PyOx− showed 90.21, 83.42, 77.25 and 63.79% maximum reactivation against paraoxon, dichlorvos, malathion and triazophos, respectively (Fig. S3†). On the other hand, at 1000 μM L−1 of 2-PyOx−, 87.92, 72.53, 64.20 and 55.98% maximum reactivation was obtained against paraoxon, dichlorvos, malathion and triazophos, respectively. As a general trend, 2-PyOx− showed the largest reactivation against paraoxon (87%) and lowest against triazophos (64%) (Fig. S4†).
The phosphorylation of the enzyme depends on the alkyl groups associated with the phosphorus atom.56 The reactivation parameters supported that the 4-C18PyOx− and 4-C18PyOx− showed larger reactivation potency than the 2-PyOx− oxime (Fig. 6) because the strength of the complex was increased by the electrostatic attraction between the cationic site in the reactivator and the anionic site in the inhibited enzyme. Furthermore, the reactivation rate constant and kinetic parameters (namely, kr, KD, kr2) of different oximes on OPs that inhibited AChE are summarized in Table 2. The minimum value of the dissociation constant KD was obtained for 4-C18PyOx− for all pesticides, showing that the 4-C18PyOx− is the best reactivator among the tested oximes. Also, for the quaternized oxime, the values of the first order rate constant kr and the second order rate constant kr2 are the maximum values, which provide further evidence that 4-C18PyOx− is the best reactivator for the OPs-inhibited enzyme. Similarly, we have also studied the fluorescence decay parameter, as shown in Table S3 and Fig. S3.† These results give information about the average lifetime of the nanoprobe in the presence and absence of the pesticides and oximes. Sample A has an average lifetime of the nanoprobe that is very low, as compared to the presence of OPs and oximes, i.e., 5.65 ns, 76.05 ns and 37.35 ns, respectively (Fig. S5 and Table S3†). An electronegative hydroxyl group of a serine at the active site of AChE reacted with the relatively electropositive phosphorous atom by the loss of one of the side groups on the phosphorous atom was designated the leaving group.57,58
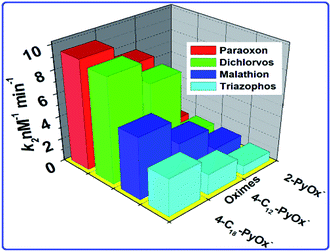 |
| | Fig. 6 The relationship between the oximes and the rate constant k2. | |
Table 2 Reactivation rate constants for the oxime-induced reactivation of Ops-inhibited AChE
| Reactivator |
Paraoxon |
Dichlorvos |
Malathion |
Triazophos |
| KD (μM) |
kr (min−1) |
kr2 (mM−1 min−1) |
KD (μM) |
kr (min−1) |
kr2 (mM−1 min−1) |
KD (μM) |
kr (min−1) |
kr2 (mM−1 min−1) |
KD (μM) |
kr (min−1) |
kr2 (mM−1 min−1) |
| 4-C18PyOx− |
9.71 ± 0.05 |
0.09 ± 0.01 |
6.10 ± 0.02 |
10.30 ± 0.05 |
0.10 ± 0.24 |
9.80 ± 0.15 |
27.60 ± 0.03 |
0.11 ± 0.02 |
3.18 ± 0.07 |
21.68 ± 0.21 |
0.96 ± 0.10 |
5.39 ± 0.05 |
| 4-C12PyOx− |
14.50 ± 0.09 |
0.11 ± 0.09 |
3.73 ± 0.14 |
18.70 ± 0.02 |
0.13 ± 0.04 |
7.21 ± 0.18 |
54.48 ± 0.11 |
0.05 ± 0.04 |
1.81 ± 0.14 |
49.59 ± 0.12 |
0.11 ± 0.05 |
3.24 ± 0.06 |
| 2-PyOx− |
77.60 ± 0.04 |
0.04 ± 0.03 |
2.05 ± 0.16 |
91.5 ± 0.07 |
0.07 ± 0.03 |
1.09 ± 0.07 |
116.3 ± 0.08 |
0.08 ± 0.01 |
0.81 ± 0.02 |
98.37 ± 0.07 |
0.10 ± 0.26 |
1.12 ± 0.20 |
Impact of interfering ions
Because of the high wealth of metal ions in soil and water, the interference of metal ions for the OPs detection by this framework was explored. The selectivity of this strategy was analyzed within the presence of metal ions. Fig. 7a shows that the results showed that most metal ions exhibited no interference to the OPs detection. Therefore, in the presence of metal ions, no changes occurred in the FL of the CdTeQDs/AChE/ChOx system. Moreover, the possible interference of different substances, such as sugars (glucose, mannose and maltose), amino acids (Phe, Glu, Asp, Trp, Leu, His, Gln, Arg, Met and Cys), vitamins (biotin, folic acid, niacin, thiamine and pyridoxine), were explored in the detection system. As displayed in Fig. 7b, the interference of the above-mentioned substances was negligible. The results proved that a sensitive and selective enzymatic assay method has been developed based on the CdTeQDs/AChE/ChOx system.
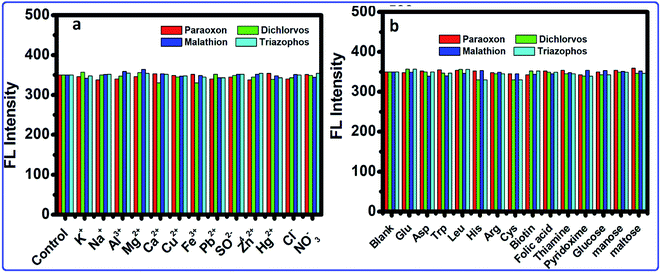 |
| | Fig. 7 The selectivity and interference of the CdTeQDs/AChE/ChOx detection system. (a) FL intensity of the system in the presence of 10 nM mL−1 of metal ions. (b) FL intensity of the system in the presence of different substances at the 10 nM mL−1 level. | |
Conclusion
In summing up, the coincident enzyme-based sensors of CdTe quantum dots (CdTeQDs/AChE/CHOx) for the detection of organophosphate pesticides (OPs) have been successfully developed. The developed CdTe QDs peroxidase-catalytic assay is more sensitive when compared with the traditional enzyme activity-based methods because of the highly effective catalytic activity of the QDs. More importantly, the developed CdTe QDs enzyme assay is convenient to implement and requires only a simple, inexpensive detection apparatus. Thus, it is practical for rapid field detection. With further development, the assay can be configured into an array system for the rapid, low-cost, and large-scale field screening of OPs neurotoxins. Taking an advantage, the CdTeQDs/AChE/CHOx biosensor would be easily oxidized by H2O2 at the pH value of 8, inducing the fluorescence quenching. Upon the addition of OPs, it will react with acetylcholinesterase (AChE) to inhibit the generation of H2O2 in the system, resulting in a strong fluorescence response. Instead, the reactivation of AChE gives a turn-off response of the probe. The versatile biosensor thus developed can be applied for the detection of H2O2, acetylcholine, choline oxidase activity, AChE activity and OP pesticides. This biosensor highlighted the sensitive and rapid detection of paraoxon with a detection limit of 1.6 × 10−15 M that was lower than the earlier reported method. Moreover, this methodology offered the advantage for determining the OPs in real samples.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
Financial assistance from SERB-EEQ/2017 (DST), New Delhi (2018–2020) grant (No. EEQ/2017/000331) University Grants Commission (U.G.C.), New Delhi, Research award (2016–18) grant (No. 30-1/2015) (SA-II), DST-FIST and UGC-SAP (SR/FST/CSI-259/2014(c) and F-540/7/DRS-II/2016 (SAP-I)) is gratefully acknowledged. We are thankful to the National Chemical Laboratory, Pune for TEM analysis. The help provided by Prof. M. K. Deb School of Studies in Chemistry, Pt. Ravishankar Shukla University is gratefully acknowledged. We are also grateful to Dr Bhabani Shankar Panigrahi, IGCAR, Chennai for TCSPC analysis.
References
- T. Zorbaz, P. Mišetić, N. Probst, S. Žunec, A. Zandona, G. Mendaš, V. Micek, N. M. Hrvat, M. Katalinić, A. Braïki, L. Jean, P.-Y. Renard, V. G. Marković and Z. Kovarik, ACS Chem. Neurosci., 2020, 11, 1072–1084 CrossRef CAS PubMed.
- V. K. Meena, S. Chaturvedi, R. K. Sharma, A. K. Mishra and P. P. Hazari, Mol. Pharm., 2019, 16, 2296–2308 CrossRef CAS PubMed.
- S. V. R. Jonnalagadda, A. J. Gerace, K. Thai, J. Johnson, K. Tsimenidis, J. M. Jakubowski, C. Shen, K. J. Henderson, P. Tamamis and M. Gkikas, J. Phys. Chem. B, 2020, 124, 487–503 CrossRef CAS PubMed.
- V. S. Lin, R. F. Volk, A. J. DeLeon, L. N. Anderson, S. O. Purvine, A. K. Shukla, H. C. Bernstein, J. N. Smith and A. T. Wright, Chem. Res. Toxicol., 2020, 33, 414–425 Search PubMed.
- S. Li, X. Liu, Q. Liu and Z. Chen, Anal. Chem., 2020, 92, 3361–3365 CrossRef CAS PubMed.
- F.-L. Hsu, S. Y. Bae, J. McGuire, D. R. Anderson, S. M. Bester, J. J. Height, S. D. Pegan and A. J. Walz, ACS Med. Chem. Lett., 2019, 10, 761–766 CrossRef CAS PubMed.
- J. Ma, T. Si, C. Yan, Y. Li, Q. Li, X. Lu and Y. Guo, ACS Sens., 2020, 5, 83–92 CrossRef CAS PubMed.
- C. Dincer, R. Bruch, E. Costa-Rama, M. T. Fernández-Abedul, A. Merkoçi, A. Manz, G. A. Urban and F. Güder, Adv. Mater., 2019, 31, 1806739 CrossRef PubMed.
- X. Shi, W. Wei, Z. Fu, W. Gao, C. Zhang, Q. Zhao, F. Deng and X. Lu, Talanta, 2019, 194, 809–821 CrossRef CAS PubMed.
- K. D. Mahajan, G. Ruan, G. Vieira, T. Porter, J. J. Chalmers, R. Sooryakumar and J. O. Winter, J. Mater. Chem. B, 2020, 8, 3534–3541 RSC.
- S. Chu, W. Huang, F. Shen, T. Li, S. Li, W. Xu, C. Lv, Q. Luo and J. Liu, Nanoscale, 2020, 12, 5829–5833 RSC.
- Y. Wu, J. Wu, L. Jiao, W. Xu, H. Wang, X. Wei, W. Gu, G. Ren, N. Zhang, Q. Zhang, L. Huang, L. Gu and C. Zhu, Anal. Chem., 2020, 92, 3373–3379 CrossRef CAS PubMed.
- R. Singh, N. Kumar, R. Mehta, H. Kumar and V. P. Singh, Trends Environ. Anal. Chem., 2020 DOI:10.1016/j.teac.2020.e.00086.
- Z. Liu, X. Xia, G. Zhou, L. Ge and F. Li, Analyst, 2020, 145, 2339–2344 RSC.
- J. Zhang, B. Wang, Y. Li, W. Shu, H. Hu and L. Yang, RSC Adv., 2019, 9, 25248–25256 RSC.
- S. Yang, J. Liu, H. Zheng, J. Zhong and J. Zhou, Nanoscale, 2020, 12, 3701–3714 RSC.
- S. Wu, Y. Li, T. Deng, X. Wang, S. Hu, G. Peng, X. Huang, Y. Ling and F. Liu, Org. Biomol. Chem., 2020, 18, 2468–2474 RSC.
- M. K. Bera and S. Mohapatra, Colloid. Surface. Physicochem. Eng. Aspect., 2020, 596, 124710 CrossRef CAS.
- M. L. Satnami, J. Korram, R. Nagwanshi, S. K. Vaishnav, I. Karbhal, H. K. Dewangan and K. K. Ghosh, Sens. Actuators, B, 2018, 267, 155–164 CrossRef CAS.
- I. S. Che Sulaiman, B. W. Chieng, M. J. Osman, K. K. Ong, J. I. A. Rashid, W. M. Z. Wan Yunus, S. A. M. Noor, N. A. M. Kasim, N. A. Halim and A. Mohamad, Microchim. Acta, 2020, 187, 131 CrossRef CAS PubMed.
- H. Li, D. Su, H. Gao, X. Yan, D. Kong, R. Jin, X. Liu, C. Wang and G. Lu, Anal. Chem., 2020, 92(4), 3198–3205 CrossRef CAS PubMed.
- J. Perisa, Z. Antic, C.-G. Ma, J. Papan, D. Jovanovic and M. D. Dramicanin, J. Mater. Sci. Technol., 2020, 38, 197–204 CrossRef.
- P. Wang, H. Li, M. M. Hassan, Z. Guo, Z.-Z. Zhang and Q. Chen, J. Agric. Food Chem., 2019, 67(14), 4071–4079 CrossRef CAS PubMed.
- H. Li, X. Yan, G. Lu and X. Su, Sens. Actuators, B, 2018, 260, 563–570 CrossRef CAS.
- M. Wang, M. Li, J. Lu, B. Fan, Y. He, Y. Huang and F. Wang, RSC Adv., 2018, 8, 11551–11556 RSC.
- J. Korram, L. Dewangan, R. Nagwanshi, I. Karbhal, K. K. Ghosh and M. L. Satnami, New J. Chem., 2019, 43, 6874–6882 RSC.
- S. K. Vaishanav, J. Korram, R. Nagwanshi, K. K. Ghosh and M. L. Satnami, Sens. Actuators, B, 2017, 245, 196–206 CrossRef CAS.
- N. Huang, Y. Qin, M. Li, T. Chen, M. Lu and J. Zhao, Analyst, 2019, 144, 3436–3441 RSC.
- T. Han and G. Wang, J. Mater. Chem. B, 2019, 7, 2613–2618 RSC.
- Y. Wang, R. Hu, G. Lin, I. Roy and K. T. Yong, ACS Appl. Mater. Interfaces, 2013, 5, 2786–2799 CrossRef CAS PubMed.
- C. Mongin, S. Garakyaraghi, N. Razgoniaeva, M. Zamkov and F. N. Castellano, Science, 2016, 351, 369–372 CrossRef CAS PubMed.
- J. J. Lia and J. J. Zhu, Analyst, 2013, 138, 2506–2515 RSC.
- R. Gill, L. Bahshi, R. Freeman and I. Willner, Angew. Chem., Int. Ed., 2008, 47, 1676–1679 CrossRef CAS PubMed.
- D. Sahoo, A. Mandal, T. Mitra, K. Chakraborty, M. Bardhan and A. K. Dasgupta, J. Agric. Food Chem., 2018, 66, 414–423 CrossRef CAS PubMed.
- W. Denga, P. Chen, P. Hua, Z. Hea, M. Zhang, X. Yuan and K. Huang, Sens. Actuators, B, 2019, 292, 180–186 CrossRef.
- T. Hu, J. Xu, Y. Ye, Y. Han, X. Li, Z. Wang, D. Sun, Y. Zhou and Z. Ni, Biosens. Bioelectron., 2019, 136, 112–117 CrossRef CAS PubMed.
- R. R. Kashapov, A. M. Bekmukhametova, K. A. Petrov, I. R. Nizammev, M. K. Kadirov and L. Y. Zakharova, Sens. Actuators, B, 2018, 273, 592–599 CrossRef CAS.
- A. Apilux, W. Siangproh, N. Insin, O. Chailapakul and V. Prachayasittikul, Anal. Methods, 2017, 9, 519–527 RSC.
- R. Ban, J.-J. Zhu and J. Zhang, Microchim. Acta, 2014, 181, 1591–1599 CrossRef CAS.
- G. Mercey, T. Verdelet, J. Renou, M. Kliachyna, R. Baati, F. Nachon, L. Jean and P.-Y. Renard, Acc. Chem. Res., 2012, 45(5), 756–766 CrossRef CAS PubMed.
- C. Galdeano, N. Coquelle, M. Cieslikiewicz-Bouet, M. Bartolini, B. Pérez, M. Vi. Clos, I. Silman, L. Jean, J.-P. Colletier, P.-Y. Renard and D. Muñoz-Torrero, Molecules, 2018, 23(1–19), 634 CrossRef PubMed.
- Y. Dong, J. Shao, C. Chen, H. Li, R. Wang, Y. Chi, X. Lin and G. Chen, Carbon, 2012, 50, 4738–4743 CrossRef CAS.
- Z. B. Qu, X. Zhou, L. Gu, R. Lan, D. Sun, D. Yu and G. Shi, Chem. Commun., 2013, 49, 9830–9832 RSC.
- Y. He, J. Sun, D. Feng, H. Chen, F. Gao and L. Wang, Biosens. Bioelectron., 2015, 74, 418–422 CrossRef CAS PubMed.
- S. Umrao, M. H. Jang, J. H. Oh, G. Kim, S. Sahoo, Y. H. Cho and A. Srivastva, Carbon, 2015, 81, 514–524 CrossRef CAS.
- T. D. Lazarevic-Pasti, A. M. Bondzic, I. A. Pasti and V. M. Vasic, Pestic. Biochem. Physiol., 2012, 104, 236–242 CrossRef CAS.
- S. Jin, Z. Xu, J. Chen, X. Liang, Y. Wu and X. Qian, Anal. Chim. Acta, 2004, 523, 117–123 CrossRef CAS.
- R. Pimsen, A. Khumsri, S. Wacharasindhu, G. Tumcharern and M. Sukwattanasinitt, Biosens. Bioelectron., 2014, 62, 8–12 CrossRef CAS PubMed.
- Z. Zheng, Y. Zhou, X. Li, S. Liu and Z. Tang, Biosens. Bioelectron., 2011, 26, 3081–3085 CrossRef CAS PubMed.
- A. T. Lawal and S. B. Adeloju, Biosens. Bioelectron., 2009, 25, 406–410 CrossRef CAS PubMed.
- H. A. Azab, A. S. Orabi and A. M. Abbas, J. Lumin., 2015, 167, 360–370 CrossRef CAS.
- F. C. Wong, M. L. Ahmad, Y. Heng and L. B. Peng, Talanta, 2006, 69, 888–893 CrossRef CAS PubMed.
- I. Walz and W. Schwack, Eur. Food Res. Technol., 2007, 226, 1135–1143 CrossRef.
- S. V. Dzyadevych, A. P. Soldatkin, V. N. Arkhypova, A. V. Elskaya, J. M. Chovelon, C. A. Georgiou, C. Martelet and N. Jaffrezic-Renault, Sens. Actuators, B, 2005, 105, 81–87 CrossRef CAS.
- I. H. Um, K. Akhtar, Y. H. Shin and J. Y. Han, J. Org. Chem., 2007, 72, 3823–3829 CrossRef CAS PubMed.
- N. Singh, Y. Karpichev, B. Gupta, M. L. Satnami, J. Marek, K. Kuca and K. K. Ghosh, J. Phys. Chem. B, 2013, 117, 3806–3817 CrossRef CAS PubMed.
- M. L. Satnami, S. Dhritlahre, R. Nagwanshi, I. Karbhal, K. K. Ghosh and F. Nome, J. Phys. Chem. B, 2010, 114, 16759–16765 CrossRef CAS PubMed.
- L. Wong, Z. Radic, R. J. M. Bruggemann, N. Hosea, H. A. Berman and P. Taylor, Biochemistry, 2000, 39, 5750–5757 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Data relating to the optical properties, fluorescence lifetimes, and reactivation of AChE are available. See DOI: 10.1039/d0ra03055d |
|
| This journal is © The Royal Society of Chemistry 2020 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article a and
Manmohan L. Satnami
a and
Manmohan L. Satnami *a
*a
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaHTe
NaHTe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GSH was 1
GSH was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2
0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.4. The resulting colloidal CdTe was refluxed under nitrogen flow at 100 °C for different times to obtain the GSH-capped CdTe QDs of different sizes. The aliquots of the reaction solution were pipetted out at different intervals of time (10 min interval) for the optical measurements.
2.4. The resulting colloidal CdTe was refluxed under nitrogen flow at 100 °C for different times to obtain the GSH-capped CdTe QDs of different sizes. The aliquots of the reaction solution were pipetted out at different intervals of time (10 min interval) for the optical measurements.
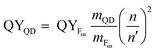
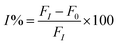
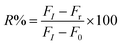
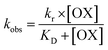
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 1280 cm−1 (–OH) indicate the presence of a –COOH group, while the band at 1075 cm−1 can be assigned to a stretching ν-C–N. Characteristic ν-N–H stretching modes observed at 3346 cm−1 and 3250 cm−1 provide evidence of a –NH group. The characteristic –SH stretching mode is clearly seen at 2526 cm−1. As expected, the IR bands of the main functional groups, –COOH, NH–, and –SH are detected in the neat GSH. In the CdTe-GSH QDs, the disappearance of the S–H group vibration at 2526 cm−1 (ν-S–H) is likely the result of a covalent bond formation between the thiol and the Cd atom at the CdTe QD surface, suggesting the oxidation of cysteine residues. These IR results are identical to those described for other CdTe-GSH QDs. The thiol capping of the CdTe QD is further validated by cyclic voltammetry (CV). Fig. 1g shows a cyclic voltammogram of the GSH-capped CdTe quantum dots with increasing size. The cathodic peak (C2) (PGSSG ∼ −0.72 V) was obtained due to the oxidation of the thiol to GSSG (2GSH → GSSG + 2e− + 2H+), which is shifted towards a lower negative potential with increasing quantum dot size. A size dependence CV behavior has also been observed for the thioglycolic acid (TGA) capped-CdTe QDs (Fig. S2†).
O) and 1280 cm−1 (–OH) indicate the presence of a –COOH group, while the band at 1075 cm−1 can be assigned to a stretching ν-C–N. Characteristic ν-N–H stretching modes observed at 3346 cm−1 and 3250 cm−1 provide evidence of a –NH group. The characteristic –SH stretching mode is clearly seen at 2526 cm−1. As expected, the IR bands of the main functional groups, –COOH, NH–, and –SH are detected in the neat GSH. In the CdTe-GSH QDs, the disappearance of the S–H group vibration at 2526 cm−1 (ν-S–H) is likely the result of a covalent bond formation between the thiol and the Cd atom at the CdTe QD surface, suggesting the oxidation of cysteine residues. These IR results are identical to those described for other CdTe-GSH QDs. The thiol capping of the CdTe QD is further validated by cyclic voltammetry (CV). Fig. 1g shows a cyclic voltammogram of the GSH-capped CdTe quantum dots with increasing size. The cathodic peak (C2) (PGSSG ∼ −0.72 V) was obtained due to the oxidation of the thiol to GSSG (2GSH → GSSG + 2e− + 2H+), which is shifted towards a lower negative potential with increasing quantum dot size. A size dependence CV behavior has also been observed for the thioglycolic acid (TGA) capped-CdTe QDs (Fig. S2†).