Open Access Article
Open Access ArticleAutonomous optimisation of a nanoparticle catalysed reduction reaction in continuous flow†
Brendan L.
Hall
 ,
Connor J.
Taylor
,
Connor J.
Taylor
 ,
Ricardo
Labes
,
Ricardo
Labes
 ,
Alexander F.
Massey
,
Alexander F.
Massey
 ,
Robert
Menzel
,
Robert
Menzel
 ,
Richard A.
Bourne
,
Richard A.
Bourne
 and
Thomas W.
Chamberlain
and
Thomas W.
Chamberlain
 *
*
Institute for Process Research and Development, School of Chemistry, University of Leeds, Leeds, LS2 9JT, UK. E-mail: T.W.Chamberlain@Leeds.ac.uk; Tel: +44 (0)113 343 6468
First published on 13th April 2021
Abstract
An automated continuous flow reactor system equipped with inline analysis, was developed and applied in the self-optimisation of a nanoparticle catalysed reaction. The system was used to optimise the experimental conditions of a gold nanoparticle catalysed 4-nitrophenol reduction reaction, towards maximum conversion in under 2.5 hours. The data obtained from this optimisation was then used to generate a kinetic model, allowing us to predict the outcome of the reaction under different conditions. By combining continuous flow nanoparticle synthesis with this approach, the development timeline for these emerging catalysts could be significantly accelerated.
Due to their high energy surfaces and extremely high surface atom to volume ratios,1 nanoparticles have been demonstrated as excellent catalysts in a wide variety of chemical transformations, such as: photocatalytic water splitting,2 hydrogenations,3 oxidations4 and cross-coupling reactions.5 Over the past two decades there has been an increasing focus on further optimising the performance of catalytic nanoparticles.6 These reports often investigate the effects of nanoparticle properties such as size, shape or composition on their performance as catalysts.7–9
Nanoparticle catalysed reaction optimisation can be a challenging task. Unlike homogeneous organic or metal-complex based catalysts, nanoparticle catalysts typically present as a non-discrete distribution of structures. These structures can be almost infinitely modified, providing additional variables to explore. The outcome of these reactions depends on both the intrinsic properties of the nanoparticles and the experimental conditions under which the reactions are performed.10 Therefore, it is important to compare each discrete catalyst system under a range of different reaction conditions. The difficulty of implementing such an approach arises from the sheer number of experiments required to thoroughly explore the reaction conditions in this way.
Recently, chemists have adopted a range of enabling technologies to assist with repetitive tasks such as reaction analysis and screening. This has led to the development of systems that utilise algorithms for the automated optimisation of reactions, particularly in the area of continuous flow research.11 Automated continuous flow systems have been developed for the self-optimisation of a wide variety of reactions12,13 and have led to accelerated chemical process development timelines.14
One of the first reported uses of a self-optimising continuous flow system was for the synthesis of cadmium–selenium quantum dot nanoparticles. Where a quantum dot synthesis method was optimised to produce nanoparticles with a maximum fluorescence intensity for a selected emission wavelength.15 Continuous flow reactors more generally, have become popular platforms for the synthesis of materials and nanoparticles in particular owing to their superior heat and mass transfer properties.16–19 Examples include the synthesis of metal,20 bimetallic,21 quantum dot22 and metal oxide23 nanoparticles.
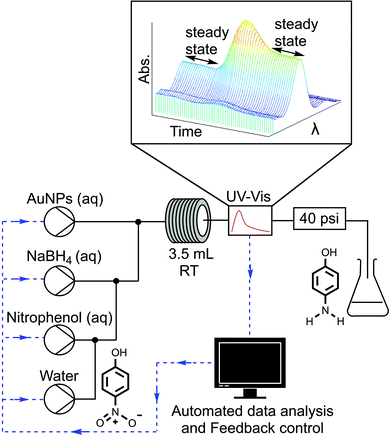
In this communication we report what is, to the best of our knowledge, the first example of a self-optimising continuous flow reactor for the optimisation of a nanoparticle catalysed reaction. The general workflow of this approach begins as a window of operating conditions is defined for the optimisation. Automated HPLC pumps then pump nanoparticles and reagents into a tubular reactor. The reactor outlet stream is analysed with an inline analytical technique, in this case UV-vis. The resulting analysis data is then interpreted to determine, for example, conversion or selectivity and the results are fed to an optimisation algorithm that determines the next array of experimental conditions to be set by the system. This automated process is repeated iteratively until an optimum solution is obtained.
To demonstrate our system, the conditions of a gold nanoparticle (AuNP) catalysed 4-nitrophenol (4-NP) reduction reaction were optimised for maximum conversion. Reduction reactions are an important class of reactions with special relevance in synthetic drug development.24 4-NP reductions are commonly used as a benchmark reaction for comparing the catalytic performance of different nanoparticle catalysts.25 The AuNPs in this study were synthesised using a Turkevich protocol26 and confirmed by TEM to have an average diameter of 15.7 ± 5.5 nm (ESI,† Section 2.4).
The closed loop optimisation system is shown in Fig. 1. Automated pumps were used to flow aqueous solutions of the AuNPs, reducing agent NaBH4 and 4-NP into a tubular flow reactor (PTFE, 0.8 mm ID, 3.5 mL volume). Altering the pump flow rates allowed different concentrations of reagents as well as different residence times to be explored. The active surface area of the nanoparticles present in the reaction was correlated to the concentration of Au present using a geometric approximation (ESI,† Section 2.2), based on the shape and average nanoparticle size determined by TEM analysis. The AuNPs were characterised with XPS, revealing the surface AuNP atoms to be present in the Au(0) oxidation state, both before and after the reaction (ESI,† Section 2.5). Further TEM characterization was performed to confirm there was no change in nanoparticle size after the nanoparticles were used to catalyse the reaction (ESI,† Section 2.4). A water dilution pump was also included to increase the range of conditions that could be attained by the system, as not all combinations of factors could be achieved without a diluent i.e. particular concentrations and residence times. The global optimisation algorithm Stable Noisy Optimisation by Branch and Fit (SNOBFIT)27 was used to optimise the reaction, allowing us to demonstrate the platforms ability to explore a wide range of reaction variables within the design space shown in Table 1.
| AuNP SA (m2 L−1) | NaBH4 conc. (mM) | Res. time (min) | |
|---|---|---|---|
| Upper bound | 0.16 | 2.5 | 3 |
| Lower bound | 0.04 | 0.5 | 1 |
The NaBH4 solution was kept in an ice bath and adjusted to pH 10 with a solution of sodium hydroxide (0.1 M), to prevent hydrolysis when stored in the reactor reservoir. The initial concentration of 4-NP (0.06 mM) was kept the same for each experiment to simplify analysis. The outlet of the reactor was monitored with an inline UV-vis absorption spectrometer, this allowed real time monitoring of 4-NP concentration and verification of steady state conditions.
Reaction conversion was determined by integrating the UV-vis absorption profile of 4-NP between 350–450 nm. As the AuNP and 4-NP absorption bands overlap within this range, the measured UV-vis spectra were deconvoluted by subtracting an AuNP reference band that was scaled to match the AuNP peak intensity in the measured spectrum between 500–560 nm (ESI,† Section 5).
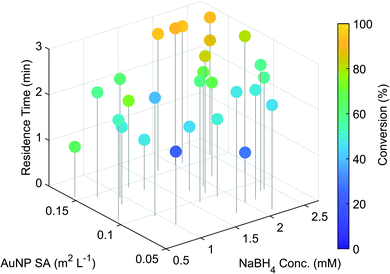
The optimisation was performed in under 2.5 hours and the outcome of the individual experiments are displayed in Fig. 2. A trend towards the highest conversion (95%) was found at longer residence times, with higher AuNP surface areas and NaBH4 concentrations. These results fit well with the findings of previously reported studies,28,29 demonstrating clearly the systems capability for nanoparticle catalysed reaction optimisation.
The global optimisation algorithm used in this study explored a wide range of conditions within the experimental design space. Therefore, it was possible to use this data set to identify kinetic parameters and obtain a high degree of understanding of the reaction system. Previous studies have shown how kinetic models can be generated using structured data from similar automated flow systems,30,31 but kinetic models built from data obtained during a self-optimisation experiment have not been reported prior to this work. Ordinary differential equation (ODE) solvers were used to predict reaction progression under different conditions using a commonly accepted kinetic model for bi-molecular surface catalysed reactions, the Langmuir Hinshelwood (LH) model.32 This model is shown in (eqn (1)), where S is nanoparticle surface area per unit volume, k is the overall rate constant for the reaction, CNit/CBH4 represent the concentrations and KNit/KBH4 are the absorption coefficients of 4-NP and sodium borohydride respectively (ESI,† Section 6).
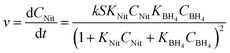 | (1) |
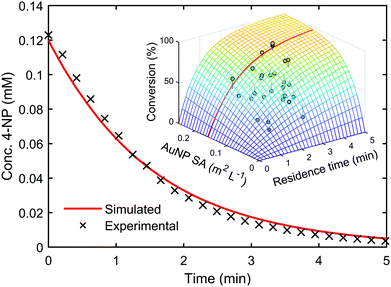
Kinetic models such as this can be used to predict the outcome of reactions under different conditions, even when alternative reactor configurations are used.33 A genetic algorithm34 was used to determine the kinetic parameters within the LH model by maximising the convergence of the predicted reaction outcomes to experimental data. Upon identifying this kinetic information, it was possible to use the model to predict reaction conversion under different experimental conditions. The model was used to predict the reaction kinetics of a AuNP catalysed 4-NP reduction reaction performed in a batch reactor with a two-fold increase in 4-NP concentration, compared to the flow optimisation study. The resulting extrapolated reaction kinetics showed good agreement with the model prediction, with a residual error of only ±1.85%, see Fig. 3 (ESI,† Section 6.1).
In summary, a continuous flow reactor platform has been developed for the efficient optimisation of nanoparticle catalysed reactions. Automated reactor controls and a self-optimising feedback loop were integrated into an automated flow reactor system. The effect of changing residence times, sodium borohydride concentrations and AuNP surface area were explored using the global optimisation algorithm SNOBFIT. The system was able to identify the optimum reaction conditions for maximum 4-NP conversion within a set operating window. Furthermore, the data gained during this optimisation process was used to obtain a kinetic model by employing ODE solvers and a genetic algorithm to fit kinetic parameters to the experimental data, thereby generating a response surface for the explored design space. The generated model was then validated by accurately predicting the outcome of a nanoparticle catalysed reduction reaction performed in batch. The system presented herein allows efficient determination of the optimal conditions for nanoparticle catalysed reactions. Integrating this system into automated flow reactors for nanoparticle synthesis, could also enable rapid determination of the conditions required for producing optimal catalytic nanoparticles as well as the best conditions for their respective catalysed reaction.
This work was supported by Zabeeda P. Aslam at the Leeds Electron Microscopy and Spectroscopy Centre (LEMAS). RAB was supported by the Royal Academy of Engineering under the Research Chairs and Senior Research Fellowships scheme (RCSRF1920\9\38). We also acknowledge support from the Henry Royce Institute (EPSRC grants: EP/P022464/1, EP/R00661X/1), which funded the VXSF Facilities within the Bragg Centre for Materials Research at Leeds, the EPSRC via the Complex Particulates and Processes CDT program (BH, CP3-CDT), the Cognitive Chemical Manufacturing project (TWC, RAB, RL, EP/R032807/1) and the University of Leeds.
Conflicts of interest
There are no conflicts to declare.Notes and references
- Z.-Y. Zhou, N. Tian, J.-T. Li, I. Broadwell and S.-G. Sun, Chem. Soc. Rev., 2011, 40, 4167–4185 RSC.
- Q. Wang and K. Domen, Chem. Rev., 2019, 120, 919–985 CrossRef PubMed.
- T. Mitsudome and K. Kaneda, Green Chem., 2013, 15, 2636–2654 RSC.
- T. Mallat and A. Baiker, Annu. Rev. Chem. Biomol. Eng., 2012, 3, 11–28 CrossRef CAS PubMed.
- A. Fihri, M. Bouhrara, B. Nekoueishahraki, J. M. Basset and V. Polshettiwar, Chem. Soc. Rev., 2011, 40, 5181–5203 RSC.
- D. Astruc, Chem. Rev., 2020, 120, 461–463 CrossRef CAS PubMed.
- K.-J. Wu and L. Torrente-Murciano, React. Chem. Eng., 2018, 3, 267–276 RSC.
- A. Monga and B. Pal, New J. Chem., 2015, 39, 304–313 RSC.
- B. Roldan Cuenya, Thin Solid Films, 2010, 518, 3127–3150 CrossRef.
- N. Narayan, A. Meiyazhagan and R. Vajtai, Materials, 2019, 12, 1–12 CrossRef PubMed.
- A. D. Clayton, J. A. Manson, C. J. Taylor, T. W. Chamberlain, B. A. Taylor, G. Clemens and R. A. Bourne, React. Chem. Eng., 2019, 4, 1545–1554 RSC.
- B. J. Reizman, Y.-M. Wang, S. L. Buchwald and K. F. Jensen, React. Chem. Eng., 2016, 1, 658–666 RSC.
- L. M. Baumgartner, C. W. Coley, B. J. Reizman, K. W. Gao Ab and K. F. Jensen, React. Chem. Eng., 2018, 3, 301–311 RSC.
- N. Holmes, G. R. Akien, A. J. Blacker, R. L. Woodward, R. E. Meadows and R. A. Bourne, React. Chem. Eng., 2016, 1, 366–371 RSC.
- S. Krishnadasan, R. J. C. Brown, J. deMello and A. J. deMello, Lab Chip, 2007, 7, 1377–1612 RSC.
- O. Długosz and M. Banach, React. Chem. Eng., 2020, 5, 1619–1641 RSC.
- E. J. Roberts, L. R. Karadaghi, L. Wang, N. Malmstadt and R. L. Brutchey, ACS Appl. Mater. Interfaces, 2019, 11, 27479–27502 CrossRef CAS PubMed.
- A. Sivo, R. d. S. Galaverna, G. R. Gomes, J. C. Pastre and G. Vilé, React. Chem. Eng., 2021 10.1039/D0RE00411A.
- V. Sebastian, S. Khan and A. Kulkarni, J. Flow Chem., 2017, 7, 96–105 CrossRef CAS.
- K.-J. Wu, G. M. De Varine Bohan and L. Torrente-Murciano, React. Chem. Eng., 2017, 2, 116–128 RSC.
- H. Liu, J. Huang, D. Sun, O.-W. Tareque, J. Li and Q. Li, J. Nanopart. Res., 2014, 16, 1–9 Search PubMed.
- R. W. Epps, M. S. Bowen, A. A. Volk, K. Abdel-Latif, S. Han, K. G. Reyes, A. Amassian and M. Abolhasani, Adv. Mater., 2020, 32, 2001626 CrossRef CAS PubMed.
- E. Lester, P. Blood, J. Denyer, D. Giddings, B. Azzopardi and M. Poliakoff, J. Supercrit. Fluids, 2006, 37, 209–214 CrossRef CAS.
- J. S. Carey, D. Laffan, C. Thomson and M. T. Williams, Org. Biomol. Chem., 2006, 4, 2337–2347 RSC.
- T. Aditya, A. Pal and T. Pal, Chem. Commun., 2015, 51, 9410–9431 RSC.
- N. G. Bastus, J. Comenge and V. V. Puntes, Langmuir, 2011, 27, 11098–11105 CrossRef CAS PubMed.
- W. Huyer and A. Neumaier, ACM Trans. Math. Softw., 2008, 35, 20575–20587 CrossRef.
- S. Wunder, F. Polzer, Y. Lu, Y. Mei and M. Ballauff, J. Phys. Chem. C, 2010, 114, 8814–8820 CrossRef CAS.
- P. Hervés, M. Pérez-Lorenzo, L. M. Liz-Marzán, J. Dzubiella, Y. Lu and M. Ballauff, Chem. Soc. Rev., 2012, 41, 5577–5587 RSC.
- C. A. Hone, A. Boyd, A. O'Kearney-McMullan, R. A. Bourne and F. L. Muller, React. Chem. Eng., 2019, 4, 1565–1570 RSC.
- C. J. Taylor, M. Booth, J. A. Manson, M. J. W. Willis, G. Clemens, B. A. Taylor, T. W. Chamberlain and R. A. Bourne, Chem. Eng. J., 2020, 127017 Search PubMed.
- W. H. Kang and H. Chuan Weinberg, Chem. Rev., 1995, 95, 667–676 CrossRef.
- N. Zaborenko, M. W. Bedore, T. F. Jamison and K. F. Jensen, Org. Process Res. Dev., 2011, 15, 131–139 CrossRef CAS.
- D. E. Goldberg, Genetic Algorithms in Search, Optimization and Machine Learning, Addison-Wesley Longman Publishing Co., Inc., USA, 1st edn, 1989 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methodology, automation code, XPS and TEM characterisation. See DOI: 10.1039/d1cc00859e |
| This journal is © The Royal Society of Chemistry 2021 |