 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Magnetite-binding proteins from the magnetotactic bacterium Desulfamplus magnetovallimortis BW-1†
Anna
Pohl
 ab,
Sarah A. E.
Young
ab,
Sarah A. E.
Young
 ab,
Tara C.
Schmitz
ab,
Tara C.
Schmitz
 a,
Daniel
Farhadi
a,
Raz
Zarivach
a,
Daniel
Farhadi
a,
Raz
Zarivach
 c,
Damien
Faivre
c,
Damien
Faivre
 *bd and
Kerstin G.
Blank
*bd and
Kerstin G.
Blank
 *a
*a
aMax Planck Institute of Colloids and Interfaces, Mechano(bio)chemistry, Am Mühlenberg 1, 14476 Potsdam, Germany. E-mail: Kerstin.Blank@mpikg.mpg.de
bMax Planck Institute of Colloids and Interfaces, Department of Biomaterials, Am Mühlenberg 1, 14476 Potsdam, Germany. E-mail: Damien.Faivre@cea.fr
cDepartment of Life Sciences, The National Institute for Biotechnology in the Negev and Ilse Katz Institute for Nanoscale Science and Technology, Ben-Gurion University of the Negev, Beer Sheva 8410501, Israel
dAix-Marseille Université, CEA, CNRS, BIAM, 13108 Saint Paul lez Durance, France
First published on 25th November 2021
Abstract
Magnetite-binding proteins are in high demand for the functionalization of magnetic nanoparticles. Binding analysis of six previously uncharacterized proteins from the magnetotactic Deltaproteobacterium Desulfamplus magnetovallimortis BW-1 identified two new magnetite-binding proteins (Mad10, Mad11). These proteins can be utilized as affinity tags for the immobilization of recombinant fusion proteins to magnetite.
Magnetite (Fe3O4) nanoparticles with defined size, shape, crystallinity and magnetic properties are of great interest for a wide range of biomedical and biotechnological applications.1 Biofunctionalization of such nanoparticles with specific proteins alters particle surface properties, enhances biocompatibility and enables targeting to specific body parts of interest. In recent years, green synthetic routes towards biofunctionalized magnetite nanoparticles (MNPs) have moved into focus, inspired by the natural process of magnetite biomineralization in magnetotactic bacteria (MTB).1d Within specialized organelles, so-called magnetosomes, MTB mineralize monodisperse MNPs in the size range of a single magnetic domain.1d,2 The biomineralization process is genetically controlled and several proteins have been identified that strongly interact with the magnetite surface.2,3 Among others, these include Mms6,4 Mms7,4a Mms13 (MamC),4a,5 and MmsF.6 The identification and characterization of these proteins have provided important insights into the biomineralization process in MTB, including MNP size and shape control.5b,6b,7 At the same time, these proteins have started to serve as versatile domains for controlling in vitro MNP synthesis5b,7a,e,8 and/or for obtaining biofunctionalized MNPs.9
The search for magnetite-binding proteins was so far mainly restricted to the class of Alphaproteobacteria. It has recently been shown that MTB affiliated to the class of Deltaproteobacteria and the phylum of Nitrospirae possess additional genes essential for biomineralization.10 These magnetosome-associated deep-branching (mad) genes11 were first identified in Desulfamplus magnetovallimortis BW-1, which was found in a brackish water spring in Death Valley National Park, California, USA.12 Among the 30 encoded Mad proteins, Mad1–Mad11 are predicted to be involved in magnetite biomineralization.10a It has already been demonstrated that Mad1, Mad2 and Mad6 are necessary for the development of a magnetic phenotype in Desulfovibrio magneticus RS-1;13 however, no function could yet be assigned to most of these proteins except for Mad1, Mad6 and Mad9, which likely possess redox activity (Fig. S1–S11†).10a,13,14
In this work, we screened the proteins Mad1–Mad11 for magnetite binding, using a two-step approach. In the first step, we identified proteins possessing charged sequence motifs. Especially glutamic and aspartic acid display high affinity to magnetite, as evidenced when comparing the magnetite interaction of individual amino acids16 and when performing site-directed mutagenesis of magnetite-binding proteins.7b,17 While proteins with charged motifs were of high interest, we excluded proteins with predicted redox activity as well as proteins with a high fraction of transmembrane regions (Fig. S1–S11†). Considering these criteria, the proteins Mad3, Mad4, Mad5, Mad8, Mad10 and Mad11 were chosen for the second step. In this step, the selected proteins were recombinantly produced as fusion proteins. The DNA sequence of the respective Mad protein was genetically fused to the DNA sequence of superfolder green fluorescent protein (sfGFP18) to yield one polypeptide chain, consisting of both functional units. The resulting sfGFP-Mad-His fusion proteins (Fig. 1) allowed for visualizing the presence of these proteins during expression and purification. While Mad5, Mad10 and Mad11 are predicted to be soluble proteins, Mad3, Mad4 and Mad8 contain at least one transmembrane helix. Here, only the charged regions were used for fusion protein design (see ESI†). All proteins were expressed in the cytoplasm of E. coli in soluble form. Except for Mad11, all proteins could further be purified via immobilized metal ion affinity chromatography under native conditions (pH 8.0; see ESI†) and possessed the correct molecular weight and sufficient purity for further analysis (Fig. S12, S14–S20; Table S1†). The protein sfGFP-Mad11-His could only be purified when the pH was raised to 8.8; however, the amount and purity remained low (Fig. S12†). Despite the low purity of sfGFP-Mad11-His, we included this fusion protein in our subsequent screening for magnetite-binding proteins.
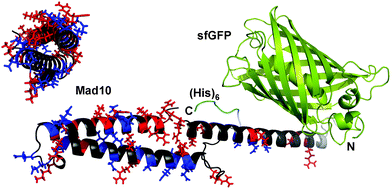 | ||
| Fig. 1 Fusion protein design. Each Mad protein was fused to the C-terminus of superfolder green fluorescent protein (sfGFP) via a short linker. For purification, a (His)6 tag was added at the C-terminus. The model shown is based on a 3D structure prediction of Mad10, performed with RaptorX.15 For Mad10, positively (His, Lys, Arg) and negatively (Glu, Asp) charged amino acids are highlighted in red and blue, respectively. | ||
All produced sfGFP-Mad-His fusion proteins were screened for their ability to interact with magnetite, using a quartz-crystal microbalance with dissipation (QCM-D). The fusion proteins, as well as a sfGFP-His control protein (see ESI†), were diluted in phosphate buffered saline pH 7.4 to match the pH value estimated for magnetosomes of Magnetospirillum magneticum AMB-1 (7.0–7.4).19 The diluted proteins (1 μM) were then exposed to commercially available magnetite-coated sensor chips. Changes in frequency and dissipation, caused by the protein–magnetite interaction on the sensor surface, were recorded in real-time. A decrease in the frequency signal originates from the adsorption of biomolecules onto the sensor surface. This decrease in the frequency signal is usually accompanied by an increase in the dissipation signal and correlates with the viscosity of the adsorbed biomolecule film. The fusion proteins sfGFP-Mad10-His and sfGFP-Mad11-His show fast binding to the magnetite sensor surface (Fig. 2; Fig. S21–S27†).
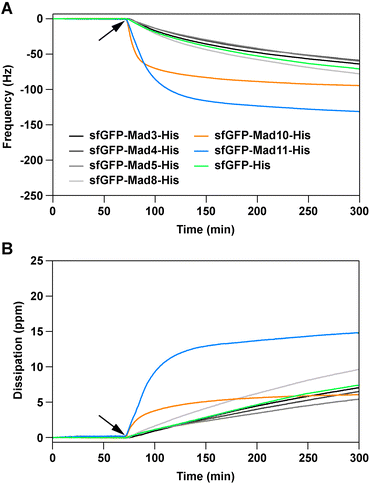 | ||
| Fig. 2 Magnetite-binding analysis, using a quartz-crystal microbalance with dissipation. Shown are the frequency (A) and dissipation (B) signals of the association phase. The protein solutions (1 μM in phosphate buffered saline) were flown over the magnetite sensor chip, pre-equilibrated with buffer. The time point where the proteins were added is indicated with an arrow. For both the frequency and dissipation signals, the 7th harmonic is displayed. For each protein, the data shows one example out of three independent repeats (see ESI† for all repeats, all harmonics and the dissociation phase). | ||
Importantly, the association is significantly faster than for the sfGFP-His control protein and a quasi-equilibrium state is reached within the timeframe of the experiment. For the other fusion proteins, the signal change is comparable to the control protein sfGFP-His. It has been reported that both GFP and the (His)6 tag weakly interact with magnetite.16,20 As the other Mad fusion proteins cause a similar response as sfGFP-His, we conclude that these proteins do not specifically bind to magnetite.
Strikingly, little dissociation from the sensor surface was observed for sfGFP-Mad10-His when flushing the sensor surface with a protein-free solution for >4 hours (Fig. S25†). This irreversible interaction could be caused by protein denaturation upon strong, non-specific adsorption to the magnetite surface. To investigate this possibility and to obtain additional proof for a specific protein–magnetite interaction, two sensor chips were exposed to sfGFP-His and sfGFP-Mad10-His sequentially (Fig. S28†). On the first sensor, sfGFP-His was bound first and then sfGFP-Mad10-His was added. For the second sensor, the order was reversed. Once sfGFP-Mad10-His was exposed to the sfGFP-His-coated sensor, the frequency signal decreased and reached a comparable minimum as measured when sfGFP-Mad10-His interacted with the bare magnetite sensor. In contrast, no change in signal was observed when sfGFP-His was added to a sensor previously exposed to sfGFP-Mad10-His. This is a strong indication that sfGFP-Mad10-His effectively competes with sfGFP-His and displaces sfGFP-His from the sensor surface in a specific manner. In addition, an experiment was performed with mixtures of fusion proteins (omitting the low purity sfGFP-Mad11-His sample). Only when sfGFP-Mad10-His was included in the mixture, a frequency change comparable to pure sfGFP-Mad10-His was observed (Fig. S29†). Overall, these experiments lead us to conclude that Mad10 specifically binds to magnetite in a fast and nearly irreversible manner. This confirms our earlier single-molecule force spectroscopy experiments with a Mad10-derived peptide that showed high-affinity binding to magnetite thin films.21
Mad10 is a unique protein as it also possesses a CXXC motif with putative redox or ion-binding function.22 To investigate the possible role of this motif on the protein–magnetite interaction and on MNP formation, we cloned an additional fusion protein where the CXXC motif was deleted (sfGFP-Mad10trunc-His; see ESI†). Furthermore, Mad10trunc was also cloned without the sfGFP fusion partner, placing the (His)6 tag either at the N-terminus (His-Mad10trunc) or the C-terminus (Mad10trunc-His). Only the fusion protein sfGFP-Mad10trunc-His could be purified in sufficient yield under native conditions (Fig. S12†). The sfGFP-free proteins largely remained in the flow-through of the Ni2+-NTA column (Fig. S13†), suggesting that the (His)6 tag is inaccessible and that these proteins oligomerize or aggregate.
To probe the influence of Mad10 on magnetite formation, sfGFP-Mad10-His and sfGFP-Mad10trunc-His were used in an in vitro MNP co-precipitation experiment, using sfGFP-His as a control. Desulfamplus magnetovallimortis BW-1 produces bullet-shaped MNPs. It has been hypothesized that magnetite-binding proteins are involved in MNP shape control, blocking the growth of certain crystal faces. Each synthesis was performed according to an established protocol23 for a total duration of 6 h, taking samples every hour. Transmission electron microscopy shows that all MNPs synthesized in the presence or absence of a Mad10 fusion protein exhibit high polydispersity. When compared to control particles, no morphological changes were observed (Fig. S30†). The crystal structure, the mean crystallite size and the growth kinetics were investigated using synchrotron-based powder X-ray diffraction (XRD). All samples show the characteristic peaks of the diffraction pattern of magnetite (Fig. 3A).24
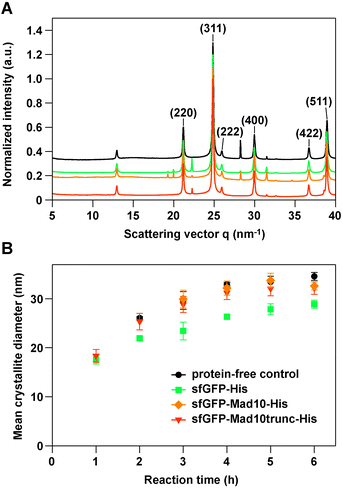 | ||
| Fig. 3 Synchrotron X-ray diffraction of MNPs synthesized with the co-precipitation method. (A) Representative 1D scattering profiles obtained after 6 h in the absence (protein-free control) or presence of 0.5 μM protein. The data was normalized to the (311) peak of magnetite. The different profiles are set off by 0.1 a.u. for better visualization. (B) Growth kinetics of MNPs in the different co-precipitation reactions. The data are the mean of 3–4 independent experiments and the error bars represent the standard error of the mean (see Table S2† for details). For the 1 h and 2 h time points, no magnetite was detected in some samples for the protein-free control reaction and the reaction with sfGFP-Mad10-His. | ||
The main peak of magnetite (311) was used to analyze the mean diameter of the crystallites. The mean diameter steadily increased and approached a plateau after approximately 4–5 h (Fig. 3A; Table S2†). The MNPs synthesized in the presence of the Mad10 fusion proteins follow the growth kinetics of the control particles and have approximately the same diameter after 6 h. MNPs synthesized in the presence of the sfGFP-His control show similar growth kinetics; however, the overall mean diameter is slightly smaller at nearly any time point. The final mean diameter of particles synthesized in the presence of sfGFP-His is only 29 ± 2 nm, while the particles synthesized in the presence of Mad10 fusion protein have a mean diameter of 32 ± 1 nm (sfGFP-Mad10-His) and 32 ± 3 nm (sfGFP-Mad10trunc-His). Mad10 thus has no measurable influence on the growth of MNPs synthesized in vitro. Also, under the conditions used, the CXXC motif does not seem to affect particle size and growth kinetics. To further investigate a possible redox function of this motif, pure Fe2+ or Fe3+ solutions can be used in future co-precipitation experiments instead of the current 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio.
2 ratio.
Other known magnetite-binding proteins, such as Mms6, MamC and MmsF, are bound to the magnetosome membrane4a,5b,6,17b and frequently form micelle-like assemblies when expressed recombinantly.6b,7e,17a It has further been shown that magnetite binding requires a helical structure.7b,d Mad10 is predicted to be helical21 (Fig. 1 and Fig. S10†) and appears to have a strong tendency to oligomerize or aggregate, as it was only possible to solubilize and purify His-Mad10trunc and Mad10trunc-His with high yield under denaturing conditions (8 M urea; Fig. S13†). Tertiary structure prediction suggests that two amphiphilic helices interact with each other in a coiled coil-like fashion (Fig. S10†). In the native environment, the hydrophobic faces of these helices may also interact with other proteins or the magnetosome membrane. We speculate that our inability to purify Mad11 may also be caused by oligomerization or aggregation of the sfGFP-Mad11-His fusion protein.
It has been proposed that a self-assembled array of acidic residues facilitates both magnetite binding and local supersaturation of iron ions, which is required for MNP nucleation.2,6b,7a,f The predicted tertiary structure of Mad10 does indeed show clustering of acidic residues on the protein surface (Fig. S32†). These are most likely responsible for magnetite binding in our experiment. The fact that we observe magnetite binding, but no effect on MNP morphology, may however indicate that the main natural role of Mad10 is iron binding and MNP nucleation. It has further been suggested that the presence and, possibly, the interaction of several different proteins may be required to control MNP size and shape.4b,6a,25 Mad10 alone may thus not be sufficient for exerting control over crystal size and/or morphology. Last but not least, steric hindrance imposed by the sfGFP fusion partner or the pH required for MNP co-precipitation (pH 9.0) may affect native Mad10 function or the assembly state of this highly charged protein. Future experiments, aimed at investigating the function of Mad10 (and Mad11), clearly need to focus on investigating the influence of the assembly state on biomineralization. Genetic modifications of MTB strains producing bullet-shaped MNPs will further elucidate the influence of these two proteins on magnetite crystal formation in vivo.13
From an application point of view, Mad10 or the previously characterized Mad10-derived peptide21 may serve as an affinity tag for the functionalization of MNPs, e.g. with fluorescent proteins. Labeling MNPs with fluorophores is reported in the literature but comprises a sequence of synthetic steps.26 Aiming at a bioinspired synthetic route towards fluorescently labelled MNPs, we thus tested whether our co-precipitation method can be used for the one-pot functionalization of MNPs with sfGFP-Mad10trunc-His. Particles synthesized in the presence of the fusion protein were imaged directly after synthesis (Fig. S31†). The overlay of bright-field and fluorescence images reveals the co-localization of sfGFP with the MNPs. Making use of the high-affinity Mad10-magnetite interaction, no additional steps were required. Mad10-derived binding domains can readily be fused to any other protein of interest, possibly providing a simple method for the green synthesis and functionalization of MNPs.
In summary, we identified two new magnetite-binding proteins (Mad10 and Mad11) from magnetotactic bacteria that produce bullet-shaped magnetite nanoparticles in vivo. These proteins are found in Deltaproteobacteria and Nitrospirae, suggesting a conserved role in magnetosome formation.10b,11,21 This is a first key step towards investigating currently unknown biomineralization processes leading to bullet-shaped particles. Moreover, these proteins can directly be fused to any protein of interest, opening up a wide range of applications in biomedicine and biotechnology, ranging from protein purification and biosensors to drug delivery and multimodal imaging.
Data availability
The raw data of this manuscript will be published on Zenodo and a doi has been reserved: doi: 10.5281/zenodo.5735991.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors thank Margit Rößner and Anna-Maria Tsirigoni for help with protein expression and characterization. They further thank the HZB (BESSY II, Helmholtz–Zentrum für Materialien und Energie, Berlin) for the allocation of X-ray synchrotron beam-time and Stefan Siegel and Chenghao Li for technical support. Funding was provided by the International Max Planck Research School on Multiscale Bio-Systems (AP), the Alexander von Humboldt Foundation (RZ) and the Max Planck Society. Open Access funding provided by the Max Planck Society.Notes and references
- (a) S. Laurent, D. Forge, M. Port, A. Roch, C. Robic, L. Van der Elst and R. N. Muller, Chem. Rev., 2008, 108, 2064 CrossRef CAS PubMed; (b) J. E. Rosen, L. Chan, D. B. Shieh and F. X. Gu, Nanomedicine, 2012, 8, 275 CrossRef CAS PubMed; (c) R. A. Revia and M. Zhang, Mater. Today, 2016, 19, 157 CrossRef CAS PubMed; (d) G. Mirabello, J. J. Lenders and N. A. Sommerdijk, Chem. Soc. Rev., 2016, 45, 5085 RSC.
- D. Faivre and D. Schüler, Chem. Rev., 2008, 108, 4875 CrossRef CAS PubMed.
- (a) C. Jogler and D. Schüler, Annu. Rev. Microbiol., 2009, 63, 501 CrossRef CAS PubMed; (b) R. Uebe and D. Schüler, Nat. Rev. Microbiol., 2016, 14, 621 CrossRef CAS PubMed.
- (a) A. Arakaki, J. Webb and T. Matsunaga, J. Biol. Chem., 2003, 278, 8745 CrossRef CAS PubMed; (b) M. Tanaka, E. Mazuyama, A. Arakaki and T. Matsunaga, J. Biol. Chem., 2011, 286, 6386 CrossRef CAS PubMed.
- (a) K. Grünberg, C. Wawer, B. M. Tebo and D. Schüler, Appl. Environ. Microbiol., 2001, 67, 4573 CrossRef PubMed; (b) C. Valverde-Tercedor, M. Montalbán-López, T. Perez-Gonzalez, M. S. Sanchez-Quesada, T. Prozorov, E. Pineda-Molina, M. A. Fernandez-Vivas, A. B. Rodriguez-Navarro, D. Trubitsyn, D. A. Bazylinski and C. Jimenez-Lopez, Appl. Microbiol. Biotechnol., 2015, 99, 5109 CrossRef CAS PubMed.
- (a) D. Murat, V. Falahati, L. Bertinetti, R. Csencsits, A. Körnig, K. Downing, D. Faivre and A. Komeili, Mol. Microbiol., 2012, 85, 684 CrossRef CAS PubMed; (b) A. E. Rawlings, J. P. Bramble, R. Walker, J. Bain, J. M. Galloway and S. S. Staniland, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 16094 CrossRef CAS PubMed.
- (a) S. M. Bird, A. E. Rawlings, J. M. Galloway and S. S. Staniland, RSC Adv., 2016, 6, 7356 RSC; (b) H. Nudelman, C. Valverde-Tercedor, S. Kolusheva, T. Perez Gonzalez, M. Widdrat, N. Grimberg, H. Levi, O. Nelkenbaum, G. Davidov, D. Faivre, C. Jimenez-Lopez and R. Zarivach, J. Struct. Biol., 2016, 194, 244 CrossRef CAS PubMed; (c) H. Nudelman, Y. Z. Lee, Y. L. Hung, S. Kolusheva, A. Upcher, Y. C. Chen, J. Y. Chen, S. C. Sue and R. Zarivach, Front. Microbiol., 2018, 9, 2480 CrossRef PubMed; (d) H. Nudelman, T. Perez Gonzalez, S. Kolushiva, M. Widdrat, V. Reichel, A. Peigneux, G. Davidov, R. Bitton, D. Faivre, C. Jimenez-Lopez and R. Zarivach, Acta Crystallogr., Sect. D: Struct. Biol., 2018, 74, 10 CrossRef CAS PubMed; (e) A. E. Rawlings, L. A. Somner, M. Fitzpatrick-Milton, T. P. Roebuck, C. Gwyn, P. Liravi, V. Seville, T. J. Neal, O. O. Mykhaylyk, S. A. Baldwin and S. S. Staniland, Nat. Commun., 2019, 10, 2873 CrossRef PubMed; (f) G. Davidov, G. Abelya, R. Zalk, B. Izbicki, S. Shaibi, L. Spektor, D. Shagidov, E. G. Meyron-Holtz, R. Zarivach and G. A. Frank, J. Am. Chem. Soc., 2020, 142, 19551 CrossRef CAS PubMed.
- T. Prozorov, S. K. Mallapragada, B. Narasimhan, L. Wang, P. Palo, M. Nilsen-Hamilton, T. J. Williams, D. A. Bazylinski, R. Prozorov and P. C. Canfield, Adv. Funct. Mater., 2007, 17, 951 CrossRef CAS.
- V. O. Shipunova, P. A. Kotelnikova, U. F. Aghayeva, O. A. Stremovskiy, I. A. Novikov, A. A. Schulga, M. P. Nikitin and S. M. Deyev, J. Magn. Magn. Mater., 2019, 469, 450 CrossRef CAS.
- (a) C. T. Lefèvre, D. Trubitsyn, F. Abreu, S. Kolinko, C. Jogler, L. G. P. de Almeida, A. T. R. de Vasconcelos, M. Kube, R. Reinhardt, U. Lins, D. Pignol, D. Schüler, D. A. Bazylinski and N. Ginet, Environ. Microbiol., 2013, 15, 2712 CrossRef PubMed; (b) W. Lin, A. Deng, Z. Wang, Y. Li, T. Wen, L.-F. Wu, M. Wu and Y. Pan, ISME J., 2014, 8, 2463 CrossRef CAS PubMed.
- S. Kolinko, M. Richter, F.-O. Glöckner, A. Brachmann and D. Schüler, Environ. Microbiol., 2016, 18, 21 CrossRef CAS PubMed.
- (a) C. T. Lefèvre, N. Menguy, F. Abreu, U. Lins, M. Pósfai, T. Prozorov, D. Pignol, R. B. Frankel and D. A. Bazylinski, Science, 2011, 334, 1720 CrossRef PubMed; (b) E. C. T. Descamps, C. L. Monteil, N. Menguy, N. Ginet, D. Pignol, D. A. Bazylinski and C. T. Lefèvre, Syst. Appl. Microbiol., 2017, 40, 280 CrossRef CAS PubMed.
- L. Rahn-Lee, M. E. Byrne, M. Zhang, D. Le Sage, D. R. Glenn, T. Milbourne, R. L. Walsworth, H. Vali and A. Komeili, PLoS Genet., 2015, 11, e1004811 CrossRef PubMed.
- M. I. Siponen, G. Adryanczyk, N. Ginet, P. Arnoux and D. Pignol, Biochem. Soc. Trans., 2012, 40, 1319 CrossRef CAS PubMed.
- M. Källberg, H. Wang, S. Wang, J. Peng, Z. Wang, H. Lu and J. Xu, Nat. Protoc., 2012, 7, 1511 CrossRef PubMed.
- S. P. Schwaminger, P. F. García, G. K. Merck, F. A. Bodensteiner, S. Heissler, S. Günther and S. Berensmeier, J. Phys. Chem. C, 2015, 119, 23032 CrossRef CAS.
- (a) L. Wang, T. Prozorov, P. E. Palo, X. Liu, D. Vaknin, R. Prozorov, S. Mallapragada and M. Nilsen-Hamilton, Biomacromolecules, 2012, 13, 98 CrossRef PubMed; (b) A. Yamagishi, K. Narumiya, M. Tanaka, T. Matsunaga and A. Arakaki, Sci. Rep., 2016, 6, 35670 CrossRef CAS PubMed.
- (a) X. Li, G. Zhang, N. Ngo, X. Zhao, S. R. Kain and C. C. Huang, J. Biol. Chem., 1997, 272, 28545 CrossRef CAS PubMed; (b) H. K. Kim and B. K. Kaang, Brain Res. Bull., 1998, 47, 35 CrossRef CAS PubMed; (c) J. D. Pédelacq, S. Cabantous, T. Tran, T. C. Terwilliger and G. S. Waldo, Nat. Biotechnol., 2006, 24, 79 CrossRef PubMed.
- Y. Eguchi, Y. Fukumori and A. Taoka, Biosci., Biotechnol., Biochem., 2018, 82, 1243 CrossRef CAS PubMed.
- S. P. Schwaminger, S. A. Blank-Shim, I. Scheifele, V. Pipich, P. Fraga-García and S. Berensmeier, Biotechnol. J., 2019, 14, e1800055 CrossRef PubMed.
- A. Pohl, F. Berger, R. M. A. Sullan, C. Valverde-Tercedor, K. Freindl, N. Spiridis, C. T. Lefèvre, N. Menguy, S. Klumpp, K. G. Blank and D. Faivre, Nano Lett., 2019, 19, 8207 CrossRef CAS PubMed.
- D. E. Fomenko, S. M. Marino and V. N. Gladyshev, Mol. Cells, 2008, 26, 228 CAS.
- J. Baumgartner, M. Antonietta Carillo, K. M. Eckes, P. Werner and D. Faivre, Langmuir, 2014, 30, 2129 CrossRef CAS PubMed.
- M. E. Fleet, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1984, 40, 1491 CrossRef.
- A. Arakaki, A. Yamagishi, A. Fukuyo, M. Tanaka and T. Matsunaga, Mol. Microbiol., 2014, 93, 554 CrossRef CAS PubMed.
- (a) Y. Sahoo, A. Goodarzi, M. T. Swihart, T. Y. Ohulchanskyy, N. Kaur, E. P. Furlani and P. N. Prasad, J. Phys. Chem. B, 2005, 109, 3879 CrossRef CAS PubMed; (b) Y. Zhang, N. Kohler and M. Zhang, Biomaterials, 2002, 23, 1553 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Bioinformatic analysis of Mad proteins; cloning, protein expression, purification and characterization; additional data of the QCM-D experiments and magnetic nanoparticle characterization. See DOI: 10.1039/d1nr04870h |
| This journal is © The Royal Society of Chemistry 2021 |
