Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multi-layered ZIF-coated cells for the release of bioactive molecules in hostile environments†
Lei
Gan
a,
Miriam de J.
Velásquez-Hernández
 a,
Anita
Emmerstorfer-Augustin
b,
Peter
Wied
a,
Anita
Emmerstorfer-Augustin
b,
Peter
Wied
 a,
Heimo
Wolinski
c,
Simone Dal
Zilio
d,
Marcello
Solomon
a,
Heimo
Wolinski
c,
Simone Dal
Zilio
d,
Marcello
Solomon
 a,
Weibin
Liang
e,
Christian
Doonan
a,
Weibin
Liang
e,
Christian
Doonan
 *e and
Paolo
Falcaro
*e and
Paolo
Falcaro
 *a
*a
aInstitute of Physical and Theoretical Chemistry, Graz University of Technology, Stremayrgasse 9, Graz, 8010, Austria. E-mail: paolo.falcaro@tugraz.at
bInstitute of Molecular Biotechnology, Graz University of Technology, NAWI Graz, BioTechMed-Graz,, Petergasse 14, Graz, 8010, Austria
cInstitute of Molecular Biosciences, BioTechMed-Graz, University of Graz, Graz, Austria
dIstituto Officina dei Materiali CNR, Basovizza, Edificio MM-SS, Trieste, Italy
eSchool of Physical Sciences, Faculty of Sciences, University of Adelaide, South Australia 5005, Australia. E-mail: christian.doonan@adelaide.edu.au
First published on 1st August 2022
Abstract
Metal–organic framework (MOF) coatings on cells enhance viability in cytotoxic environments. Here, we show how protective multi-layered MOF bio-composite shells on a model cell system (yeast) enhance the proliferation of living cells exposed to hostile protease-rich environments via the dissolution of the shells and release of a protease inhibitor (antitrypsin).
Molecular biology has demonstrated the importance of research on living cells and microorganisms for the advancement of biotechnological and biomedical applications.1–3 For example, Moorella thermoacetica bacterium was used for the reduction of CO2 to acetic acid,4 while stem cells have been applied to regenerative therapies, tissue engineering, and diagnosis.5 For these applications, cells are removed from their native environments and exposed to stressors (e.g., high temperature, enzymatic degradation).6 The relocation of cells in non-native environments often leads to loss of cell viability and limits the bioactivity of the system.7–10 To enhance the resistance of cells exposed to hostile conditions, methods for encapsulating cells within abiotic exoskeletons are being studied.7,9,11 To this end, metal–organic frameworks (MOFs), a class of materials synthesized from metal ions interconnected by multidentate organic linkers,12 have been used to prepare cytoprotective coatings on living cells.13 Zeolitic imidazolate framework-8 (ZIF-8)14—a MOF composed of tetrahedral Zn2+ ions linked by 2-methylimidazolate (mIM)—was successfully used for the fabrication of abiotic shells.15 ZIF-8, in its porous form, has a crystalline lattice with sodalite (sod) topology.14 However, by changing Zn2+
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HmIM, different crystalline phases (e.g., amorphous, diamondoid) with distinct mass transfer properties can be synthesized.16–18 ZIF-8 can be synthesized in water or buffer solutions13 and is known to self-assemble on naturally occurring bioentites, from proteins to bacteria.19,20 An additional feature of this chemistry is that the protective ZIF-8 coating can be simply removed from the cell by decreasing the pH,21 adding chelating agents (e.g., ethylenediaminetetraacetic acid, EDTA),22 or exposing ZIF-8 to some buffer solutions (e.g., phosphate-buffered saline, PBS).23–25 We have previously demonstrated the successful encapsulation of Saccharomyces cerevisiae (yeast cells) within sod ZIF-8 using a one-pot approach.26 In a few minutes, a ca. 100 nm thick ZIF-8 shell assembled on the yeast cell surface. This porous coating formed a cytoprotective barrier with perm-selective properties: i.e. it was permeable to nutrients (glucose) but not to larger cytotoxic molecules (e.g. lyticase).26 Thus, the yeast cell metabolic activity was maintained within the rigid abiotic shell even in the presence of cytotoxic molecules. Subsequent to the removal of the exoskeleton, the cells retained their biological functionality including reproduction.26 This ZIF-8-based coating strategy has been extended to viruses,27 bacteria26 and mammalian cells.28 Further research showed that yeast cells could be coated with a film of enzymes (β-galactosidase, β-gal) prior to the assembly of a protective ZIF-8 coating.29 The immobilized β-gal conferred biocatalytic properties to the exoskeleton by processing lactose into glucose (a cell nutrient). This study revealed that engineering abiotic shells with co-immobilized enzymes can enhance the viability of the encaged cells. To date, all the research on protective ZIF coatings has been focused on the shielding properties of ZIF-based exoskeletons as by using this cell@ZIF approach in an environment with overexpressed protease (e.g. inflammatory condition30 and some tumor environment31), cell death can be prevented. However, if this environment is the final destination of the cell, when the MOF shell is dissolved, exposure of the cell membrane to protease will cause cell death.32 To avoid degradation of the cell membrane subsequent to the removal of the ZIF-8 shell, one strategy could be to co-encapsulate a protease inhibitor that would be released during the MOF shell dissolution process. This would inactivate the cytotoxic enzyme in the surrounding environment and enable rapid cell proliferation.
HmIM, different crystalline phases (e.g., amorphous, diamondoid) with distinct mass transfer properties can be synthesized.16–18 ZIF-8 can be synthesized in water or buffer solutions13 and is known to self-assemble on naturally occurring bioentites, from proteins to bacteria.19,20 An additional feature of this chemistry is that the protective ZIF-8 coating can be simply removed from the cell by decreasing the pH,21 adding chelating agents (e.g., ethylenediaminetetraacetic acid, EDTA),22 or exposing ZIF-8 to some buffer solutions (e.g., phosphate-buffered saline, PBS).23–25 We have previously demonstrated the successful encapsulation of Saccharomyces cerevisiae (yeast cells) within sod ZIF-8 using a one-pot approach.26 In a few minutes, a ca. 100 nm thick ZIF-8 shell assembled on the yeast cell surface. This porous coating formed a cytoprotective barrier with perm-selective properties: i.e. it was permeable to nutrients (glucose) but not to larger cytotoxic molecules (e.g. lyticase).26 Thus, the yeast cell metabolic activity was maintained within the rigid abiotic shell even in the presence of cytotoxic molecules. Subsequent to the removal of the exoskeleton, the cells retained their biological functionality including reproduction.26 This ZIF-8-based coating strategy has been extended to viruses,27 bacteria26 and mammalian cells.28 Further research showed that yeast cells could be coated with a film of enzymes (β-galactosidase, β-gal) prior to the assembly of a protective ZIF-8 coating.29 The immobilized β-gal conferred biocatalytic properties to the exoskeleton by processing lactose into glucose (a cell nutrient). This study revealed that engineering abiotic shells with co-immobilized enzymes can enhance the viability of the encaged cells. To date, all the research on protective ZIF coatings has been focused on the shielding properties of ZIF-based exoskeletons as by using this cell@ZIF approach in an environment with overexpressed protease (e.g. inflammatory condition30 and some tumor environment31), cell death can be prevented. However, if this environment is the final destination of the cell, when the MOF shell is dissolved, exposure of the cell membrane to protease will cause cell death.32 To avoid degradation of the cell membrane subsequent to the removal of the ZIF-8 shell, one strategy could be to co-encapsulate a protease inhibitor that would be released during the MOF shell dissolution process. This would inactivate the cytotoxic enzyme in the surrounding environment and enable rapid cell proliferation.
In general, the potential of ZIF coatings to protect cells from environmental stressors and enhance cell thriving is still an undeveloped area of research. By showing that a biocomposite MOF shell could transform an environment from cytotoxic to biocompatible would further progress MOF materials for biotechnology and biomedicine. Here we demonstrate this proof-of-concept by using yeast as a model cell system, trypsin as a model protease enzyme, and Alpha-1-antitrypsin (AAT), a protein therapeutic, as a protease inhibitor.
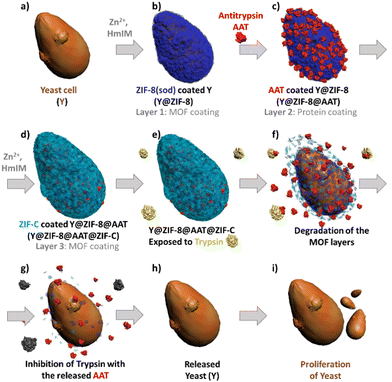
Inspired by the affinity of proteins for ZIFs33 and the ability of protein-functionalized surfaces to trigger the ZIF formation29 we developed a multilayer approach to control the AAT encapsulation within two ZIF layers. Firstly, a sod ZIF-8 layer is grown on yeast cells (Fig. 1a and b), followed by adsorption of AAT on the yeast@ZIF-8 biocomposite (Fig. 1c). Then a second ZIF shell is grown to cover the immobilized AAT (Fig. 1d). We note that, depending on synthesis conditions, either a phase pure ZIF-8 or ZIF-C (i.e. Zn2(mIM)2(CO3))34 shell can be deposited. ZIF-C was unexpected as it has not been reported to form a cell coating. This non-porous framework, which includes CO32− from atmospheric CO2, was observed as a product of biomimetic mineralization from ZIF-8 precursors and proteins.17 Compared to ZIF-8, ZIF-C shows different release kinetics of encapsulated molecules.17 Thus, this multilayered approach enables: (1) control over the spatial distribution of the AAT in the MOF exoskeleton, (2) the release profile of AAT to be modified via the selection of the ZIF phase for the outer layer (ZIF-8 – slow release, or ZIF-C – fast release) and, (3) artificial adaptability to protease-rich environments (Fig. 1e–i).
Saccharomyces cerevisiae (yeast cells; Y) was selected as a model organism as it is robust, non-pathogenic, easy to cultivate and divides similarly to human cells.35 The first protective ZIF-8 shell was synthesized by adding Y to an aqueous solution with the MOF precursors (Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HmIM = 1
HmIM = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
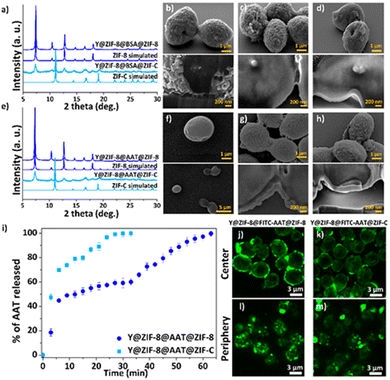
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 16, ESI†). When compared to previously reported one-pot protocols,26 the current higher ligand to metal ratio affords the rapid crystallization of Zn(mIM)2 into pure sod ZIF-8 as confirmed by the powder X-ray diffraction (PXRD) pattern (Fig. S1, ESI†) of the washed samples. Scanning electron microscopy (SEM) analyses reveal the formation of a homogenous ZIF-8 coating with an average shell thickness of ca. 60 ± 20 nm (Fig. 2 and Fig. S2 and S3, ESI†). The analysis of the dried Y@ZIF-8 with Fourier-transform infrared (FTIR) spectroscopy shows characteristic modes of the Zn–N bond of ZIF-8 (e.g. 421 cm−1, Fig. S4, ESI†). Next, Bovine Serum Albumin (BSA) was selected as an inexpensive model protein for the optimization of a protein layer on the ZIF-8 exoskeleton. By soaking 9 mg of Y@ZIF-8 in an aqueous solution of BSA, 0.05 mg of the protein were adsorbed on the Y@ZIF-8 surface (ESI†). Finally, Y@ZIF-8@BSA was exposed to the ZIF-8 precursors. Using a Zn2+
16, ESI†). When compared to previously reported one-pot protocols,26 the current higher ligand to metal ratio affords the rapid crystallization of Zn(mIM)2 into pure sod ZIF-8 as confirmed by the powder X-ray diffraction (PXRD) pattern (Fig. S1, ESI†) of the washed samples. Scanning electron microscopy (SEM) analyses reveal the formation of a homogenous ZIF-8 coating with an average shell thickness of ca. 60 ± 20 nm (Fig. 2 and Fig. S2 and S3, ESI†). The analysis of the dried Y@ZIF-8 with Fourier-transform infrared (FTIR) spectroscopy shows characteristic modes of the Zn–N bond of ZIF-8 (e.g. 421 cm−1, Fig. S4, ESI†). Next, Bovine Serum Albumin (BSA) was selected as an inexpensive model protein for the optimization of a protein layer on the ZIF-8 exoskeleton. By soaking 9 mg of Y@ZIF-8 in an aqueous solution of BSA, 0.05 mg of the protein were adsorbed on the Y@ZIF-8 surface (ESI†). Finally, Y@ZIF-8@BSA was exposed to the ZIF-8 precursors. Using a Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HmIM ratio of 1
HmIM ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 32, self-assembled shells of pure sod ZIF-8 (Fig. 2a and Table S1, ESI†) were achieved whereas a Zn2+
32, self-assembled shells of pure sod ZIF-8 (Fig. 2a and Table S1, ESI†) were achieved whereas a Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HmIM = 1
HmIM = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratio yielded a crystalline shell of pure ZIF-C (Fig. 2a and Table S1, see ESI† for full experimental procedure). A 100% adsorption efficiency (AE) of BSA was determined for both bio-composites (Fig. S5–S7, Table S2 and S3, ESI†). The morphology of Y@ZIF-8@BSA@ZIF-8 and Y@ZIF-8@BSA@ZIF-C were analyzed by SEM. Close inspection of the images clearly shows the formation of continuous ZIF coatings in both samples (SEM, Fig. 2c and d). An average shell thickness, determined from the cross-section (focused ion beam, FIB, ESI†), of the ZIF shells was ca. 102 nm and ca. 95 nm, for Y@ZIF-8@BSA@ZIF-8 and Y@ZIF-8@BSA@ZIF-C, respectively (Fig. 2c, 2d and Fig. S8, S9, ESI†). To ascertain the effect of the different crystalline phases on the outer ZIF-shells on the protein secretion, we measured the release profile of BSA upon the digestion of ZIF in an aqueous solution of EDTA (fast trigger release) and phosphate buffer (PB, 20 mM, pH 6.5, simulating acidic conditions associated with cancerous cells36 and inflammation37). Release tests triggered by EDTA show a 100% BSA release in ca. 15 min for Y@ZIF-8@BSA@ZIF-C, and ca. 30 min for Y@ZIF-8@BSA@ZIF-8 (Fig. S10, ESI†). The release profiles were then determined in PB (Fig. S11, ESI†). The profile measured from Y@ZIF-8@BSA@ZIF-C shows an 80% release of BSA in ca. 2 h, while for Y@ZIF-8@BSA@ZIF-8, 80% release was observed in ca. 15 h. In both media the release from ZIF-C was always faster than that from ZIF-8. These results indicate that, for the same digestion environment, the release kinetics of the immobilized protein can be tuned by engineering the crystalline phase of the outer ZIF shell.
4 ratio yielded a crystalline shell of pure ZIF-C (Fig. 2a and Table S1, see ESI† for full experimental procedure). A 100% adsorption efficiency (AE) of BSA was determined for both bio-composites (Fig. S5–S7, Table S2 and S3, ESI†). The morphology of Y@ZIF-8@BSA@ZIF-8 and Y@ZIF-8@BSA@ZIF-C were analyzed by SEM. Close inspection of the images clearly shows the formation of continuous ZIF coatings in both samples (SEM, Fig. 2c and d). An average shell thickness, determined from the cross-section (focused ion beam, FIB, ESI†), of the ZIF shells was ca. 102 nm and ca. 95 nm, for Y@ZIF-8@BSA@ZIF-8 and Y@ZIF-8@BSA@ZIF-C, respectively (Fig. 2c, 2d and Fig. S8, S9, ESI†). To ascertain the effect of the different crystalline phases on the outer ZIF-shells on the protein secretion, we measured the release profile of BSA upon the digestion of ZIF in an aqueous solution of EDTA (fast trigger release) and phosphate buffer (PB, 20 mM, pH 6.5, simulating acidic conditions associated with cancerous cells36 and inflammation37). Release tests triggered by EDTA show a 100% BSA release in ca. 15 min for Y@ZIF-8@BSA@ZIF-C, and ca. 30 min for Y@ZIF-8@BSA@ZIF-8 (Fig. S10, ESI†). The release profiles were then determined in PB (Fig. S11, ESI†). The profile measured from Y@ZIF-8@BSA@ZIF-C shows an 80% release of BSA in ca. 2 h, while for Y@ZIF-8@BSA@ZIF-8, 80% release was observed in ca. 15 h. In both media the release from ZIF-C was always faster than that from ZIF-8. These results indicate that, for the same digestion environment, the release kinetics of the immobilized protein can be tuned by engineering the crystalline phase of the outer ZIF shell.
Next, following the same protocols established with BSA, the fabrication of the multilayered system was tested with the clinical biotherapeutic AAT, and Y@ZIF-8@AAT@ZIF-8 and Y@ZIF-8@AAT@ZIF-C were prepared. The PXRD of Y@ZIF-8@AAT@ZIF-8 was analogous to pure ZIF-8 sod while for Y@ZIF-8@AAT@ZIF-C the pattern showed peaks attributed to both sod and ZIF-C (Fig. 2e). FTIR spectra of the biocomposites show modes attributed to the Zn–N bonds of the ZIF networks and the amide bonds from the proteins, 421 cm−1 and 1635 cm−1, respectively. Additionally, for the ZIF-C biocomposite, the 700–850 and 1300–1400 cm−1 bands were assigned to weak bending and asymmetric stretching of CO32− modes (Fig. S12, ESI†). The AAT adsorption efficiencies for Y@ZIF-8@AAT@ZIF-8 and Y@ZIF-8@AAT@ZIF-C were 100% (Fig. S7, Fig. S13–16, Table S4 and S5, ESI†). The morphology of the bio-composites was examined by SEM. Close inspection of the images revealed ellipsoidal monomodal particle distributions, suggesting that Y@ZIF-8@AAT particles act as seeds for the formation of the second ZIF-8 and ZIF-C layers. Furthermore, both ZIF-8 and ZIF-C coatings are continuous and possess a similar morphology to the BSA counterparts (rounded particle-like for ZIF-8 and plate-like for ZIF-C) (Fig. 2f–h). The average shell thicknesses obtained from Y@ZIF-8@AAT@ZIF-8 and Y@ZIF-8@AAT@ZIF-C are ca. 270 nm and ca. 125 nm, respectively (Fig. S17 and S18, ESI†). It is worth noting that the multilayer strategy inhibits the crystallization of independent AAT@ZIF-8 particles, which is not the case when yeast and AAT are simultaneously present during the ZIF formation (Fig. S19, ESI†).
The release profiles obtained from Y@ZIF-8@AAT@ZIF-8 and Y@ZIF-8@AAT@ZIF-C in EDTA and PB reveal trends similar to their BSA analogues (Fig. 2i and Fig. S20, ESI†). For example, in the digestion by EDTA, a 100% release of AAT is measured in 30 min for Y@ZIF-8@AAT@ZIF-C, while the full release takes 1 h for Y@ZIF-8@AAT@ZIF-8. For PB solution, a 50% release of AAT is measured in ca. 2 h for ZIF-C and again longer for ZIF-8 (i.e. ca. 18 h, ESI†). Confocal laser scanning microscopy (CLSM) was employed to assess the homogeneity and localization of the protein layer immobilized between the two ZIF layers. For this analysis, we used BSA and AAT tagged with fluorescein isothiocyanate (FITC-BSA, FITC-AAT) to synthesize the biocomposites. Fig. 2j and k show the center of Y@ZIF-8@FITC-AAT@ZIF-8 and Y@ZIF-8@FITC-AAT@ZIF-C, respectively. Fig. 2l and m show the periphery of Y@ZIF-8@FITC-AAT@ZIF-8 and Y@ZIF-8@FITC-AAT@ZIF-C composites (Videos S1, S2 and Fig. S21, ESI† for FITC-BSA samples, Fig. S22, ESI† for controls). In summary, the CLSM images confirm the successful immobilization of protein between the two ZIF layers on the yeast cells.
To evaluate whether AAT retains its protease inhibitor function after release from the ZIF shells, Y@ZIF-8@AAT@ZIF-8 and Y@ZIF-8@AAT@ZIF-C were digested in EDTA and PB, then the supernatant from the composite solutions were exposed to a trypsin solution. After 30 min storage at RT, the protease activity of trypsin was analyzed using a Trypsin Activity Assay (Fig. S23 and S24, ESI†). In PB, where the release is slower (Fig. S23 and S24, ESI†), we monitored the effect of AAT release from Y@ZIF-8@AAT@ZIF-8 on trypsin. Over the screened period, trypsin became increasingly inactivated by the release of AAT. For a 100% AAT release, trypsin become completely inhibited.
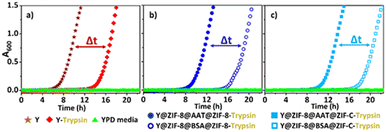
Finally, we ascertained the bio-protection functionality of AAT released from the ZIF coating on cells in a protease-rich environment. Y@ZIF-8@AAT@ZIF-8 and Y@ZIF-8@AAT@ZIF-C were directly exposed for 4 h to a trypsin solution (0.25 mg mL−1) with EDTA. The released yeast cells were washed with water, diluted, and resuspended in yeast growth medium (Yeast-extract-Peptone-Dextrose; YPD, ESI†).38 Cell proliferation was monitored by optical density measurements at 600 nm (OD600, ESI†).38 As control experiments, uncoated cells and the composites obtained with non-active protein (BSA) were exposed to trypsin under the same conditions used for AAT analogs. The OD600 measurements reveal that uncoated cells exposed to trypsin exhibit a longer lag phase (ca. 13 h) than the non-exposed cells (ca. 6 h, Fig. 3). The time difference (Δt = 7 h) to initiate the exponential growth of cells demonstrates the detrimental effect of trypsin on uncoated cells. When comparing the Y@ZIF-8@BSA@ZIF-C and Y@ZIF-8@AAT@ZIF-C systems exposed to trypsin, a Δt = 9 h was measured. Similarly, a Δt = 8 h was measured when evaluating the time difference in the lag phases of Y@ZIF-8@BSA@ZIF-8 and Y@ZIF-8@AAT@ZIF-8. Collectively, these data demonstrate that, in a trypsin-rich environment, yeast cells reproduce faster when AAT is released by ZIF coatings.
Overall, we demonstrate that a multilayer approach can be used to coat living cells and immobilize bioactive proteins (antitrypsin, AAT). While affording a homogeneous coating, the multistep process prevents the undesired formation of distinct AAT@ZIF-8 and Y@ZIF-8 particles. By tuning the crystalline phase of the outermost shell (i.e. ZIF-8 and ZIF-C), we could select two different release profiles for AAT from Y@ZIF-8@AAT@ZIF-8, Y@ZIF-8@AAT@ZIF-C. Finally, we demonstrated that the fast dissolution of a bio-composite ZIF shell triggers the release of cells while mitigating hostile extracellular conditions.
The authors acknowledge support from the Australian Research Council DP200102411, BioTechMed-Graz Young Researcher Group Program, TU Graz for the Lead Project (LP-03), the European Research Council under the European Union's Horizon 2020 Program (FP/2014–2020)/ERC Grant Agreement (771834—POPCRYSTAL).
Conflicts of interest
There are no conflicts to declare.Notes and references
- M. S. Vieira, A. K. Santos, R. Vasconcellos, V. A. M. Goulart, R. C. Parreira, A. H. Kihara, H. Ulrich and R. R. Resende, Biotechnol. Adv., 2018, 36, 1946–1970 CrossRef CAS PubMed.
- D. Hong, M. Park, S. H. Yang, J. Lee, Y.-G. Kim and I. S. Choi, Trends Biotechnol., 2013, 31, 442–447 CrossRef CAS PubMed.
- D. Carmona-Gutierrez, K. Kainz, A. Zimmermann, S. J. Hofer, M. A. Bauer, C. Ruckenstuhl, G. Kroemer and F. Madeo, Microb. Cell, 2022, 9, 72 CrossRef PubMed.
- K. K. Sakimoto, A. B. Wong and P. Yang, Science, 2016, 351, 74–77 CrossRef CAS PubMed.
- M. Cananzi, A. Atala and P. De Coppi, Reprod. Biomed. Online, 2009, 18, 17–27 CrossRef PubMed.
- J. Liu, J. Wen, Z. Zhang, H. Liu and Y. Sun, Microsyst. Nanoeng., 2015, 1, 1–15 Search PubMed.
- W. Youn, J. Y. Kim, J. Park, N. Kim, H. Choi, H. Cho and I. S. Choi, Adv. Mater., 2020, 32, 1907001 CrossRef CAS PubMed.
- W. Li, W. Bing, S. Huang, J. Ren and X. Qu, Adv. Funct. Mater., 2015, 25, 3775–3784 CrossRef CAS.
- W. Zhu, J. Guo, S. Amini, Y. Ju, J. O. Agola, A. Zimpel, J. Shang, A. Noureddine, F. Caruso, S. Wuttke, J. G. Croissant and C. J. Brinker, Adv. Mater., 2019, 31, 1900545 CrossRef PubMed.
- S. R. McCuskey, J. Chatsirisupachai, E. Zeglio, O. Parlak, P. Panoy, A. Herland, G. C. Bazan and T. Q. Nguyen, Chem. Rev., 2022, 122, 4791–4825 CrossRef CAS PubMed.
- M. de J. Velásquez-Hernández, M. Linares-Moreau, E. Astria, F. Carraro, M. Z. Alyami, N. M. Khashab, C. J. Sumby, C. J. Doonan and P. Falcaro, Coord. Chem. Rev., 2021, 429, 213651 CrossRef.
- H. Furukawa, K. E. Cordova, M. O’Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef PubMed.
- M. de J. Velásquez-Hernández, M. Linares-Moreau, E. Astria, F. Carraro, M. Z. Alyami, N. M. Khashab, C. J. Sumby, C. J. Doonan and P. Falcaro, Coord. Chem. Rev., 2020, 213651 Search PubMed.
- R. Banerjee, A. Phan, B. Wang, C. Knobler, H. Furukawa, M. O'Keeffe and O. M. Yaghi, Science, 2008, 319, 939–943 CrossRef CAS PubMed.
- R. Riccò, W. Liang, S. Li, J. J. Gassensmith, F. Caruso, C. Doonan and P. Falcaro, ACS Nano, 2018, 12, 13–23 CrossRef PubMed.
- Y. Chu, J. Hou, C. Boyer, J. J. Richardson, K. Liang and J. Xu, Appl. Mater. Today, 2018, 10, 93–105 CrossRef.
- F. Carraro, M. de J. Velásquez-Hernández, E. Astria, W. Liang, L. Twight, C. Parise, M. Ge, Z. Huang, R. Ricco and X. Zou, Chem. Sci., 2020, 11, 3397–3404 RSC.
- X. Wu, H. Yue, Y. Zhang, X. Gao, X. Li, L. Wang, Y. Cao, M. Hou, H. An and L. Zhang, Nat. Commun., 2019, 10, 1–8 Search PubMed.
- A. F. Ogata, A. M. Rakowski, B. P. Carpenter, D. A. Fishman, J. G. Merham, P. J. Hurst and J. P. Patterson, J. Am. Chem. Soc., 2020, 142, 1433–1442 CrossRef CAS PubMed.
- M. A. Luzuriaga, F. C. Herbert, O. R. Brohlin, J. Gadhvi, T. Howlett, A. Shahrivarkevishahi, Y. H. Wijesundara, S. Venkitapathi, K. Veera and R. Ehrman, ACS Nano, 2021, 15, 17426–17438 CrossRef CAS PubMed.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Nat. Commun., 2015, 6, 7240 CrossRef CAS PubMed.
- K. Liang, C. Carbonell, M. J. Styles, R. Ricco, J. Cui, J. J. Richardson, D. Maspoch, F. Caruso and P. Falcaro, Adv. Mater., 2015, 27, 7293–7298 CrossRef CAS PubMed.
- M. de J. Velásquez-Hernández, R. Ricco, F. Carraro, F. T. Limpoco, M. Linares-Moreau, E. Leitner, H. Wiltsche, J. Rattenberger, H. Schröttner, P. Frühwirt, E. M. Stadler, G. Gescheidt, H. Amenitsch, C. J. Doonan and P. Falcaro, CrystEngComm, 2019, 21, 4538–4544 RSC.
- M. A. Luzuriaga, C. E. Benjamin, M. W. Gaertner, H. Lee, F. C. Herbert, S. Mallick and J. J. Gassensmith, Supramol. Chem., 2019, 31, 485–490 CrossRef CAS PubMed.
- J. A. Allegretto, J. Dostalek, M. Rafti, B. Menges, O. Azzaroni and W. Knoll, J. Phys. Chem. A, 2018, 123, 1100–1109 CrossRef PubMed.
- K. Liang, J. J. Richardson, J. Cui, F. Caruso, C. J. Doonan and P. Falcaro, Adv. Mater., 2016, 28, 7910–7914 CrossRef CAS PubMed.
- S. Li, M. Dharmarwardana, R. P. Welch, C. E. Benjamin, A. M. Shamir, S. O. Nielsen and J. J. Gassensmith, ACS Appl. Mater. Interfaces, 2018, 10, 18161–18169 CrossRef CAS PubMed.
- L. Ha, U. Ryu, D.-C. Kang, J.-K. Kim, D. Sun, Y.-E. Kwon, K. M. Choi and D.-P. Kim, ACS Biomater. Sci. Eng., 2021, 7, 3075–3081 CrossRef CAS PubMed.
- K. Liang, J. J. Richardson, C. J. Doonan, X. Mulet, Y. Ju, J. Cui, F. Caruso and P. Falcaro, Angew. Chem., Int. Ed., 2017, 56, 8510–8515 CrossRef CAS PubMed.
- F. Schmidlin and N. W. Bunnett, Curr. Opin. Pharmacol., 2001, 1, 575–582 CrossRef CAS PubMed.
- S. Rakash, F. Rana, S. Rafiq, A. Masood and S. Amin, Biotechnol. Mol. Biol. Rev., 2012, 7, 90–101 CrossRef.
- L. Egger, J. Schneider, C. Rheme, M. Tapernoux, J. Häcki and C. Borner, Cell Death Differ., 2003, 10, 1188–1203 CrossRef CAS PubMed.
- J. G. Turner and C. J. Murphy, Langmuir, 2021, 37, 9910–9919 CrossRef CAS PubMed.
- S. A. Basnayake, J. Su, X. Zou and K. J. Balkus Jr., Inorg. Chem., 2015, 54, 1816–1821 CrossRef CAS PubMed.
- S. Mohammadi, B. Saberidokht, S. Subramaniam and A. Grama, BMC Syst. Biol., 2015, 9, 1–22 CrossRef PubMed.
- K. A. White, B. K. Grillo-Hill and D. L. Barber, J. Cell Sci., 2017, 130, 663–669 CrossRef CAS PubMed.
- A. Riemann, A. Ihling, J. Thomas, B. Schneider, O. Thews and M. Gekle, Biochim. Biophys. Acta, Mol. Cell Res., 2015, 1853, 299–307 CrossRef CAS PubMed.
- C.-W. Hung, J. Y. Martínez-Márquez, F. T. Javed and M. C. Duncan, Sci. Rep., 2018, 8, 1–16 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and additional plots. See DOI: https://doi.org/10.1039/d2cc03072a |
| This journal is © The Royal Society of Chemistry 2022 |