Exploring the entropy-driven amplification reaction and trans-cleavage activity of CRISPR-Cas12a for the development of an electrochemiluminescence biosensor for the detection of the SARS-CoV-2 RdRp gene in real samples and environmental surveillance†
Kai
Zhang
 *a,
Zhenqiang
Fan
*a,
Zhenqiang
Fan
 a,
Yuedi
Ding
a,
Sha
Zhu
b,
Minhao
Xie
a,
Yuedi
Ding
a,
Sha
Zhu
b,
Minhao
Xie
 a and
Nan
Hao
*c
a and
Nan
Hao
*c
aNHC Key Laboratory of Nuclear Medicine, Jiangsu Key Laboratory of Molecular Nuclear Medicine, Jiangsu Institute of Nuclear Medicine, Wuxi, Jiangsu 214063, China. E-mail: zhangkai@jsinm.org
bDepartment of Oncology, The Affiliated Wuxi No.2 People's Hospital of Nanjing Medical University, Wuxi, 214000, PR China
cSchool of Chemistry and Chemical Engineering, Jiangsu University, Zhenjiang, Jiangsu 212013, PR China. E-mail: hn@ujs.edu.cn
First published on 25th November 2021
Abstract
In this paper, we present the first idea of using a DNA triple helix structure to inhibit CRISPR-Cas12a activity and apply it to the design of an electrochemiluminescent biosensor for the detection of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA-dependent RNA polymerase (RdRp) gene in real samples and environmental surveillance. We employed a segment from the RdRp gene of SARS-CoV-2 by an entropy-driven reaction, which was paired with double-stranded DNA that can activate CRISPR-Cas12a activity by Hoogsteen pairing to form triple-stranded DNA, thereby inhibiting the binding interaction of the double-stranded DNA with CRISPR-Cas12a, which in turn inhibits the trans cleavage activity of CRISPR-Cas12a. The inhibited CRISPR-Cas12a is unable to cut the nucleic acid modified on the electrode surface, resulting in the inability of the ferrocene (Fc) modified on the other end of the nucleic acid to move away from the electrode surface, and thus failing to cause electrochemiluminescence changes in GOAu–Ru modified on the electrode surface. The extent of the electrogenic chemiluminescence change can reflect the concentration of the gene to be tested. Using this system, we achieved the detection of the SARS-CoV-2 RdRp gene with a detection limit of 32.80 aM.
Environmental significanceWe have proposed the first idea of using a DNA triple helix structure to inhibit CRISPR-Cas12a activity and applied it to design an ECL biosensor for detecting the SARS-CoV-2 RdRp gene in real samples and environmental surveillance. We employed an entropy-driven reaction to amplify a segment of nucleic acid that can activate the activity of CRISPR-Cas12a by Hoogsteen pairing to form triple-stranded DNA, thereby inhibiting the trans cleavage activity of CRISPR-Cas12a. This method may provide a potential platform for the diagnosis and treatment of coronavirus disease 2019 (COVID-19). It plays a key role in the environmental monitoring of COVID-19. |
1. Introduction
DNA machines are a class of molecular machines built from DNA. The programmable, predictive, logical, and self-assembling properties of DNA, due to its adherence to the Watson–Crick base pairing principle, allow this class of machines to be rationally designed.1,2 Various DNA machines have been constructed and synthesized in highly programmable ways, such as DNA tweezers3,4 DNA motors,5–7 DNA walking machines,8–11 and DNA robots.12 Among these designs, DNA walking machines can precisely control programmed oligonucleotides at the micron or nanometer scale, exhibiting the most prolific and abundant designs with broad application potential in areas such as biosensors,13,14 biocomputing,15 and drug delivery.16,17 After being driven by toehold strand replacement reactions, enzymes, and environmental stimuli, DNA walkers can move along designed trajectories, which include one-dimensional (1D) planar trajectories,18 two-dimensional (2D) DNA origami,19–21 or three-dimensional stereoscopic tracks.22 In particular, among them, 3D DNA walking machines possess a more powerful signal amplification capability. This is attributed to the fact that all the reaction components are concentrated in micron- or nanoscale 3D orbitals, resulting in a higher local effective concentration and stable signal output. Therefore, the application of 3D-DNA walking machines has attracted a lot of attention. For example, in biosensing, Li and his colleagues designed an enzyme-driven DNA walking sensor with 3D orbitals that enables isothermal homogeneous signal amplification for the detection of specific nucleic acids.21 Fan et al. also programmed a random DNA walker driven by nucleic acid exonuclease III that generates cascaded signal amplification for ultrasensitive bioanalysis.17 Although these methods show ultra-high sensitivity, they are all enzyme-dependent methods that have some inherent enzymatic drawbacks: susceptibility to complex experimental conditions (e.g., buffer and temperature), high detection costs, and unsuitability for long-term storage. Therefore, the development of an enzyme-free amplification strategy has aroused great interest among researchers.In contrast to signal amplification systems that require enzymes, the entropy-driven reaction is a catalytic signal amplification strategy that does not rely on enzymes or precise complicated temperature cycling.23–25 Strand replacement reactions mediated by a toehold are driven by entropy-increasing facilitation, applying a series of single nucleic acid chains that play multiple roles in the catalytic circuit.26 Due to its unique and extraordinary driving force, the total number of base pairs in the catalytic circuit remains unchanged during signal amplification. As a result, this strategy has less circuit leakage and a low background signal, providing a more reliable method for accurate detection. Therefore, entropy-driven reactions may be an important tool for enzymatic-free ultrasensitive target assays.
CRISPR-Cas is a bacterial defense system that cleaves and eliminates the threat of viruses and foreign plasmids. Translated from this system,27,28 CRISPR-Cas technology has become a powerful gene editing tool that is widely used in gene function research and gene modification therapy.29–32 In addition, scientists have found that some class II Cas proteins bind and cleave target sequences under the guidance of sgRNA, while activating the auxiliary cleavage activity of Cas proteins, i.e., the ability to rapidly and efficiently degrade random single-stranded DNA or RNA sequences. The above features make the CRISPR-Cas system suitable for both molecular recognition (accurate identification of nucleic acid sequences) and signal output amplification (catalytic activity of auxiliary cleavage enzymes), which has great potential in molecular diagnostics.27,33 Therefore, CRISPR detection technology is called the “next-generation molecular detection technology”. Among them, the CRISPR-Cas12a system has the activity of targeting DNA targets and non-specific cleavage of ssDNA, and CRISPR-Cas12a-based bioanalytical methods have been widely used in genetic testing and protein analysis.29–31,34,35 However, since the activator of CRISPR-Cas12a is either double-stranded DNA or single-stranded DNA generated in the sensing system, and the activated CRISPR-Cas12a can indiscriminately cleave single-stranded DNA, traditional biosensor designs often have the disadvantage that the activated CRISPR-Cas12a cleaves the original single-stranded nucleic acid in the sensing system. To address this, we propose a pioneering strategy to inhibit the activation of DNA (single- or double-stranded DNA) by CRISPR-Cas12a intermediates and thus inhibit CRISPR-Cas12a activity, and then design and construct a biosensing system for the detection of the target.
In the double helix structure of DNA, two hydrogen bonds can be formed between bases A and T, and three hydrogen bonds can be formed between G and C. This results in the formation of a complementary double helix between two polydeoxyribonucleotides, which is mainly characterized by hydrogen bonds, base stacking forces, and electrostatic attraction between the double-stranded DNA molecules. At the same time, since the paired bases attached to the glycosyl groups of the two reverse-parallel main chains are not directly opposite each other, an asymmetric double helix structure is formed, which results in the formation of large and small grooves with varying gaps along the helix between the main chains. Felsenfeld et al. first discovered and proved the structural configuration of triple-helical DNA (THD) in 1957, and defined the basic structure of triple-stranded DNA: triple-stranded DNA is based on Watson–Crick base pairing, and the third stranded base can form a Hoogsteen pairing with the purines in the Watson–Crick base pairing.36 The large grooves of DNA are twisted together in parallel or anti-parallel to form a triple helix structure. The basic building block of triple-helical DNA is the double-base system, and triple-helical DNA is similar to it in that it is a three-base system. There are two main types of triple-stranded DNA: purine type, purine–purine–pyrimidine (Pu–Pu–Py) type, and 2-pyrimidine type, pyrimidine–purine–pyrimidine (Py–Pu–Py) type.37 The purine type is a type of triple helix in which the third strand of purine DNA is inserted into a large groove in double-helical DNA to counter parallel the direction of the purine strand in Watson–Crick double-stranded DNA and specifically binds to the purine DNA strand.38 Similar to the pyrimidine type, a third pyrimidine DNA strand is inserted into a large groove in double-helical DNA to counter parallel the orientation of the purine strand in Watson–Crick double-stranded DNA and specifically binds to the purine DNA strand, a type of triple helix structure. Due to the specificity of the triple DNA structure, the biological significance of these systems and their potential clinical applications as well as the construction of biosensing systems have also attracted widespread interest.39–41
Electrochemiluminescence (ECL) is an analytical technique that is increasingly being used in bioassay and sensing technologies.23,25,42–47 ECL is a type of chemiluminescence caused by the migration of unstable excited states back to the ground state when certain substances with electrochemiluminescence activity are at a certain potential and interact with redox substances in solution. It has high sensitivity, good reproducibility, continuous detection, fast detection, and simple device characteristics towards the commonly used fluorescence methods and the most studied ECL active substances including trichloropyridine ruthenium (Ru(bpy)32+), luminol, and so on.48 The relatively small number of ECL active substances has led to limitations in the application of ECL technology. The discovery of electro-luminescent functional composites has effectively solved this problem. Electro-luminescent functional composites are composites that can emit light under the effect of electrical excitation.23,25,29,42,45,49 ECLs generally belong to activated doped luminescent materials, i.e., luminescent materials with selective doping of activators in the matrix, which have the advantages of low cost, non-toxicity, chemical inertness, and easy modification, and have great potential for biosensing and bioimaging applications. Graphene oxide (GO), obtained by chemical modification of graphene, is a graphene derivative that contains a broad specific surface area as well as oxygen functional groups such as hydroxyl, carboxyl and epoxy groups.50 It has significant applications in energy storage, electronic devices, and sensors.51–53
In this work, we propose for the first time the idea of using a DNA triple helix structure to inhibit the activity of CRISPR-Cas12a for the design of an ECL biosensor. Since CRISPR-Cas12a activity requires double-stranded DNA (dsDNA) for activation, we designed Hoogsteen-paired triple-stranded DNA based on Hoogsteen pairing to inhibit the binding of double-stranded DNA (dsDNA) to CRISPR-Cas12a, thus achieving inhibition of CRISPR-Cas12a trans cleavage activity, in combination with an entropy-driven isothermal amplification strategy without the use of an isothermal amplification enzyme for detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) related RNA-dependent RNA polymerase (RdRp) gene (SARS-CoV-2 RdRp gene) sequences in real samples and environmental surveillance. This method has high specificity as it can clearly distinguish a base change. We anticipate that this method may provide a potential platform for the diagnosis and treatment of SARS-CoV-2.
2. Experimental section
2.1 Materials and reagents
The oligonucleotides purified by high-performance liquid chromatography (HPLC) used in our detection strategy were purchased from Genscript Biotech (Nanjing, China), and their sequences are listed in Table S1.† Tris(4,4′-dicarboxylic acid-2,2′-bipyridyl)ruthenium(II) dichloride (Ru(dcbpy)32+) and polyethyleneimine (PEI) modified graphene oxide nanofilms (GO-PEI) were obtained from XFNANO (Nanjing, China). In addition, other common analytical chemicals and reagents used in this study were purchased from Aladdin Reagents (Shanghai, China), and ultrapure water was processed through a Milli-Q purification system, unless otherwise noted.2.2 Synthesis of GOAu–Ru
The synthesis of GOAu–Ru was conducted according to the work of Ye et al.54 and our previous work.23 First, Ru(dbpy)32+ aqueous solution (5 mL, 2 mg mL−1) was prepared in advance. Afterward, 5 mL of EDC (2 mg mL−1) and NHS (1 mg mL−1) were added to the Ru(dbpy)32+ aqueous solution with continuous stirring for 2 h to activate the –COOH of Ru(dbpy)32+. 10 mL of GO-PEI solution (1 mg mL−1) was then added to the above activation solution under magnetic stirring for 2 h. Next, 10 mL of 13 nm-sized Au nanoparticles were added to the prepared GOAu–Ru hybrid solution and stirred continuously for 2 h to form an electrostatic interaction based on Au–N bonds, thus immobilizing Au on the GO surface. These samples were then centrifuged and washed several times to remove excess unreacted reagents. Finally, the GOAu–Ru was diluted in 4 mL of purified water.2.3 Fabrication of the ECL biosensor
A suspension of 8 μL of GOAu–Ru was added dropwise to a GCE that had been treated to a smooth interface and dried under nitrogen to prepare GOAu–Ru/GCE. DNA7 at a concentration of 1 μM was added to PBS buffer (pH 7.4) containing 5.0 mM MgCl2 and 0.1 mM TCEP and incubated at 25 °C for 1.5 h to break the disulfide bonds. The GOAu–Ru/GCE electrode was immersed in DNA7 PBS buffer overnight to allow DNA7 to modify the surface of the gold electrode through the Au–S bonds. The hybrid DNA/GOAu–Ru/GCE was immersed in 60 μL of 100 μM 6-mercaptohexanol (MCH) PBS solution (0.1 M, pH 7.4) for passivating the unoccupied sites outside of the modified GCE-specific hybrid DNA probe to introduce DNA7 perpendicular to the GCE surface. Besides, after each modification, the GCE needs to be rinsed with PBS solution (20 mM) to remove substances that are not explicitly adsorbed. Thus, the construction of an ECL biosensor (MCH/DNA7/GOAu–Ru/GCE) was achieved.2.4 SARS-CoV-2 RdRp gene assay
100 μL of DNA1, DNA2 and DNA3 (200 nM each) were annealed at 95 °C for 5 min and allowed to cool slowly to room temperature for approximately 4 h to form a DNA1/DNA2/DNA3 complex. Meanwhile, 200 nM DNA5 and DNA6 were annealed to form a DNA5/DNA6 duplex in the same process. For the target detection, the DNA1/DNA2/DNA3 complex, DNA4 and DNA5/DNA6 duplex (200 nM each) in a tube were incubated with different concentrations of the target (SARs-CoV-2 RdRp gene) at 37 °C in Tris-HCl buffer (10 mM Tris, 12.5 mM MgCl2, pH 8.0) for 2 h to form a DNA2/DNA5/DNA6 complex. Next, 50 μL of the above DNA2/DNA5/DNA6 complex solution (final concentration was 50 nM) was added into the cleavage buffer containing 50 nM CRISPR-Cas12a/gRNA, with a final volume of 200 μL. The DNA7/GOAu–Ru/GCE based biosensor was immersed in the above solution for 1 h before the ECL assay.2.5 Gel electrophoresis
Non-denaturing PAGE was selected for PAGE gels (20%) to characterize the entropy-driven reaction process and the formation of DNA2/DNA5/DNA6 complexes. Tris-boric acid-EDTA (TBE) was used as the electrophoresis buffer (89 mM Tris, 89 mM boric acid and 2 mM EDTA, pH 8.0–8.5) for electrophoresis experiments of nucleic acids. Pre-prepared DNA samples (20 μL) were mixed with 1× loading buffer containing 0.25% bromophenol blue and 60% glycerol, loaded into the wells of the gel, and run at 115 V for 2.5 h, respectively. At the end of the run, the gels were stained with ethidium bromide (EB) for 20 min under shaking and photographed with a gel imaging system (ChemiDoc MP, Bio-Rad).2.6 Instruments and measurements
Transmission electron microscopy (TEM) measurements were conducted on a 1400 PLUS (Nippon Electronics Co.). X-ray photoelectron spectroscopy (XPS) and Fourier transform infrared spectroscopy (FT-IR) in the spectral range of 4000–500 cm−1 were carried out on a Thermo Fisher K-Alpha and Thermo Fisher Nicolet iS5, respectively. UV-vis absorption spectroscopy was performed using a Spectra Max M5e. The ECL experiments were conducted with an ECL-6B instrument built by the State Key Laboratory of Analytical Chemistry for Life Sciences, Nanjing University. The voltage sweep range was 0–1.3 V and the sweep rate was moderate (0.1 V s−1). A three-electrode system was used, which consisted of a 3 mm diameter glassy carbon electrode (GCE), an Ag/AgCl reference electrode and a platinum pair electrode. The electrolyte used for ECL quantification was phosphate-buffered saline (PBS) (0.1 M, pH 7.4). The energy-dispersive X-ray spectroscopy (EDS) patterns and elemental mapping images were collected using an Oxford X-max50 microscope.3. Results and discussion
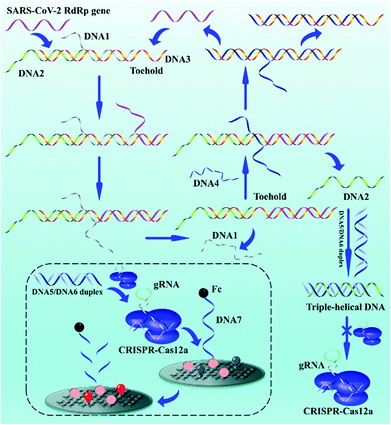
3.1 The working principle
The biosensor based on entropy-driven amplification and CRISPR-Cas shear for the detection of the SARS-CoV-2 RdRp gene consists of four parts (Scheme 1): (1) a trigger system containing an entropy-driven reaction system. (2) A CRISPR-Cas12a activity inhibition system. The entropy-driven reaction produces DNA2 that binds to the activator of CRISPR-Cas12a (DNA5/DNA6 duplex) to form triple helix DNA, inhibiting the binding of the DNA5/DNA6 duplex to CRISPR-Cas12a and thus the trans-activity of CRISPR-Cas12a. (3) A CRISPR-Cas12a shear system, including the CRISPR-Cas12a/gRNA complex and DNA5/DNA6 duplex. (4) A sensing system, based on the ferrocene (Fc)-quenched GOAu–Ru electrochemiluminescent biosensor.In the presence of the SARS-CoV-2 RdRp gene sequence in the detection system, the SARS-CoV-2 RdRp gene can form the target DNA/DNA2/DNA3 complex by hybridizing with a toehold in the DNA1/DNA2/DNA3 complex and further binding to the DNA1/DNA2/DNA3 complex, which in turn replaces DNA1. DNA4 in the reaction system further binds to a toehold in the DNA3 intermediate, displacing DNA2 and the SARS-CoV-2 RdRp gene and hybridizing with DNA3 to form the DNA3/DNA4 duplex. DNA2 becomes free. The free DNA2 further forms triple-stranded DNA with the DNA5/DNA6 duplex, resulting in the inability of the DNA5/DNA6 duplex to bind to the CRISPR-Cas12a/gRNA complex, further inhibiting the shearing activity of CRISPR-Cas12a. The key parts of the system design are the formation of the triple-helix complex and the inhibition of Crisper-Cas12a activity. We selected sequences that could form a triple-helix complex structure (part of DNA2: 5′-TTCCTTTCTCCTTCTTT-3′; part of DNA5: 5′-TTTTCTTCCTCTTTCCTT-3′, and part of DNA6: 5′-AAGGAAAGAGGAAGAAA-3′) based on previous reports.55 The DNA5/DNA6 duplex contained a PAM sequence of 5′-TTTN-3′ (N is any DNA oligonucleotide, C in this work) within the complementary DNA region. The formation of the triple-helix complex not only inhibits the binding of DNA5/DNA6 to gRNA, but also inhibits the PAM region. Therefore, the ferrocene (Fc), modified at the 3′ end of DNA7, quenches the ECL signal of GOAu–Ru on the electrode surface.56 If the SARS-CoV-2 RdRp gene is not present in the system, the entropy-driven reaction is not activated, and therefore the shearing activity of CRISPR-Cas12a is not inhibited. This leads to CRISPR-Cas12a cutting the DNA7 modified on the electrode surface, resulting in the Fc moving away from the electrode surface and thus the enhanced ECL signal of GOAu–Ru. The modification characterization data for the GCE are in the ESI† (Fig. S1).
Therefore, we successfully developed a system to convert the concentration of nucleic acid to be measured, through entropy-driven reactions and the trans-activity of CRISPR-Cas12a, into ECL signal changes at the electrode surface. The presence of the SARS-CoV-2 RdRp gene in the system can then be detected by collecting the ECL signal changes.
3.2 Characterization of GOAu–Ru composites
The TEM image shows Au nanoparticles (AuNPs) dispersed in the GO–Ru hybrid (Fig. 1A) is about 13 nm. In addition, O, N, Au, Ru, and C were uniformly distributed in the GOAu–Ru composites by EDS elemental analysis (Fig. 1B). In addition, the EDS pattern, likewise, shows the presence of the O, N, Au, Ru, and C elements (Fig. 1C). In addition, the percentages of the different elements in GOAu–Ru are shown in Table S2.† Both the EDS mapping and EDS spectral characterization verified the formation of GOAu–Ru.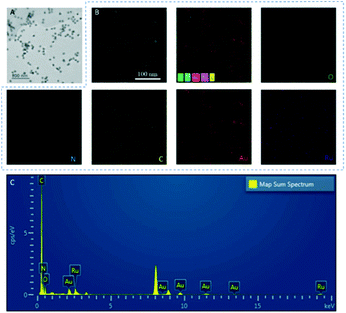 | ||
| Fig. 1 (A) TEM characterization of GOAu–Ru. (B) Electron images of GOAu–Ru, EDS overlays, and elemental mapping of elements in GOAu–Ru. (C) The EDS spectrum of GOAu–Ru nanocomposites. | ||
3.3 The ECL mechanism
Next, to clarify the ECL mechanism of the emitter composite GOAu–Ru, the CV and ECL signal responses under different detection conditions are presented in Fig. 3C and D.23 The CV curve of GOAu–Ru exhibits an inadequate signal response at 1.05 V, indicating that GOAu–Ru loses electrons to produce [GOAu–Ru]3+. In general, the ECL intensity of trichloropyridine ruthenium and its derivatives increases dramatically with the participation of co-reactants.57 The common co-reactants mainly include tri-n-propylamine (TPrA), triethylamine (TEA), diethylamine (DEA), etc., in which Ru can obtain a stable and strong ECL signal with their participation.58,59 For the detection solution with the co-reactant TEA, the electro-oxidation of TEA showed a clear oxidation peak at 0.87 V. A weak ECL signal was observed when GOAu–Ru/GCE was detected under the conditions without TEA. This is due to the fact that the TEA in the solution acts as a co-reactant in the emission process, and thus the ECL intensity is much higher than that of the assay solution without a co-reactant. The whole evidence suggests that GOAu–Ru as an ECL emitter and TEA as a co-reactant improve the ECL efficiency. Based on the above experimental phenomena, the anodic mechanism of GOAu–Ru and the co-reactant TEA is explained as follows. First, TEA and [GOAu–Ru]2+ are electrochemically oxidized to TEA+˙ and [GOAu–Ru]3+ (eqn (1)–(3)). Then, TEA+˙ loses its hydrogen cation and is converted to the strongly reducing TEA˙, which further reacts with [GOAu–Ru]3+ to form the excited state [GOAu–Ru]2+* (eqn (4)). Finally, the non-stationary excited state energy dissipates, producing optical light back to the ground state (eqn (5)).| TEA-e− → TEA+˙ | (1) |
| TEA+˙ → TEA˙ + H+ | (2) |
| [GOAu–Ru(II)]-e− → [GOAu–Ru(III)] | (3) |
| [GOAu–Ru(III)] + TEA˙ → [GOAu–Ru(II)*] + product | (4) |
| [GOAu–Ru(II)*] → [GOAu–Ru(II)] + hv | (5) |
3.4 Feasibility of the experimental conditions
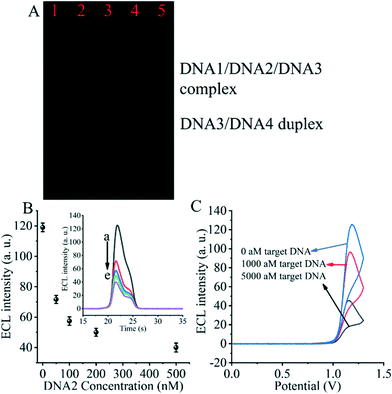
The entropy-driven reaction was first verified using electrophoresis experiments (Fig. 2A). The hybridization of DNA1, DNA2 and DNA3 (lane 1) resulted in a single bright band, indicating that the hybridization reaction was fully achieved and the DNA1/DNA2/DNA3 complex was formed. Afterwards, due to the incubation of the hybridized DNA/DNA2/DNA3 complex with the SARS-CoV-2 RdRp gene (lane 2), a decrease in the brightness of the DNA/DNA2/DNA3 complex was observed. The DNA3/DNA4 duplex control appears in the third lane. The fourth lane shows a plot of the product of the DNA1/DNA2/DNA3 complex after incubation with the SARS-CoV-2 RdRp gene and DNA4. We can clearly see that after the entropy-driven reaction, a bright DNA3/DNA4 duplex is produced and there is a faint short band at the front, which is probably the disengaged DNA1 and DNA2. Meanwhile, when the DNA1/DNA2/DNA3 complex was not incubated with the SARS-CoV-2 RdRp gene and DNA4 together, but only after DNA4 incubation, no obvious DNA3/DNA4 duplex was produced (lane 5). Therefore, we can judge that after triggering by the SARS-CoV-2 RdRp gene and with the help of DNA4, the entropy-driven reaction proceeded smoothly.In addition, the most critical issue is the formation of the DNA2/DNA5/DNA6 complex, which needs to be verified with electrophoretic experiments. We can find by the electrophoresis figure (Fig. S2†) that DNA2 and the DNA5/DNA6 duplex (ratio 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) form a triple-helix structure after co-incubation, and bright DNA2/DNA5/DNA6 complex bands can be found by electrophoresis, and the DNA5/DNA6 duplex (lane 2) was also present. In addition, DNA2 and the DNA5/DNA6 duplex (1
1) form a triple-helix structure after co-incubation, and bright DNA2/DNA5/DNA6 complex bands can be found by electrophoresis, and the DNA5/DNA6 duplex (lane 2) was also present. In addition, DNA2 and the DNA5/DNA6 duplex (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) can form the DNA2/DNA5/DNA6 complex after co-incubation, and bright bands are obtained (lane 3). Therefore, the results of this experiment demonstrated the formation of the DNA2/DNA5/DNA6 triple-helix complex.
1 ratio) can form the DNA2/DNA5/DNA6 complex after co-incubation, and bright bands are obtained (lane 3). Therefore, the results of this experiment demonstrated the formation of the DNA2/DNA5/DNA6 triple-helix complex.
For the critical element of the triple-helix-based system we designed to inhibit CRISPR-Cas12a activity for detecting the RdRp gene of SARS-CoV-2, the core issue is the reaction of DNA2 with the DNA5/DNA6 duplex to generate the DNA2/DNA5/DNA6 complex. Therefore, we next validated the effect of DNA2 on the ECL signal (Fig. 2B). When the DNA5/DNA6 duplex was pre-reacted with the CRISPR-Cas12a/gRNA complex and further processed the biosensor, we obtained the highest ECL signal value (Fig. 2B insert, curve a), indicating that the activity of the CRISPR-Cas12a/gRNA complex was activated by the DNA5/DNA6 duplex. Meanwhile, when the DNA5/DNA6 duplex was pre-reacted with DNA2 and further treated with the DNA5/DNA6 duplex and CRISPR-Cas12a/gRNA complex, no significant changes in the ECL signal values were found (Fig. 2B, curve e). Moreover, the ECL signal values decreased as the concentration of DNA2 increased (Fig. 2B and curves a to e in the inset). This indicates that the shearing activity of the CRISPR-Cas12a/gRNA complex was inhibited. Since the only difference between these two sets of experiments was the presence or absence of DNA2, we can conclude that DNA2 inhibited the activity of the CRISPR-Cas12a/gRNA complex.
Then, we investigated the changes of the ECL signal in the biosensor under different conditions. If there was no SARS-CoV-2 RdRp gene triggering the entropy-driven response and directly treated with CRISPR-Cas12a/gRNA, an extreme ECL signal value could be clearly observed (Fig. 2C). In contrast, the system caused more ECL signal decrease if the RdRp gene of SARS-CoV-2 was present in the design and triggered the entropy-driven response by incubation with 100 aM and 5000 aM SARS-CoV-2 RdRp gene. This is because the entropy-driven reaction triggered by the RdRp gene of SARS-CoV-2 leads to the release of DNA2 and binds to the activatable DNA5/DNA6 duplex, which in turn inhibits the activity of CRISPR-Cas12a, ultimately leading to the inability of DNA7 to be cleaved, and thus the change in the ECL signal is not significant. The above data suggest that the entropy-driven reaction triggers the formation of the triple-helix DNA, which plays a crucial role in the change of the signal. Therefore, we can announce that an ECL biosensor based on CRISPR-Cas12a-triggered triple-helix formation for SARS-CoV-2 RdRp gene analysis has been developed successfully.
3.5 Optimization of the experimental conditions
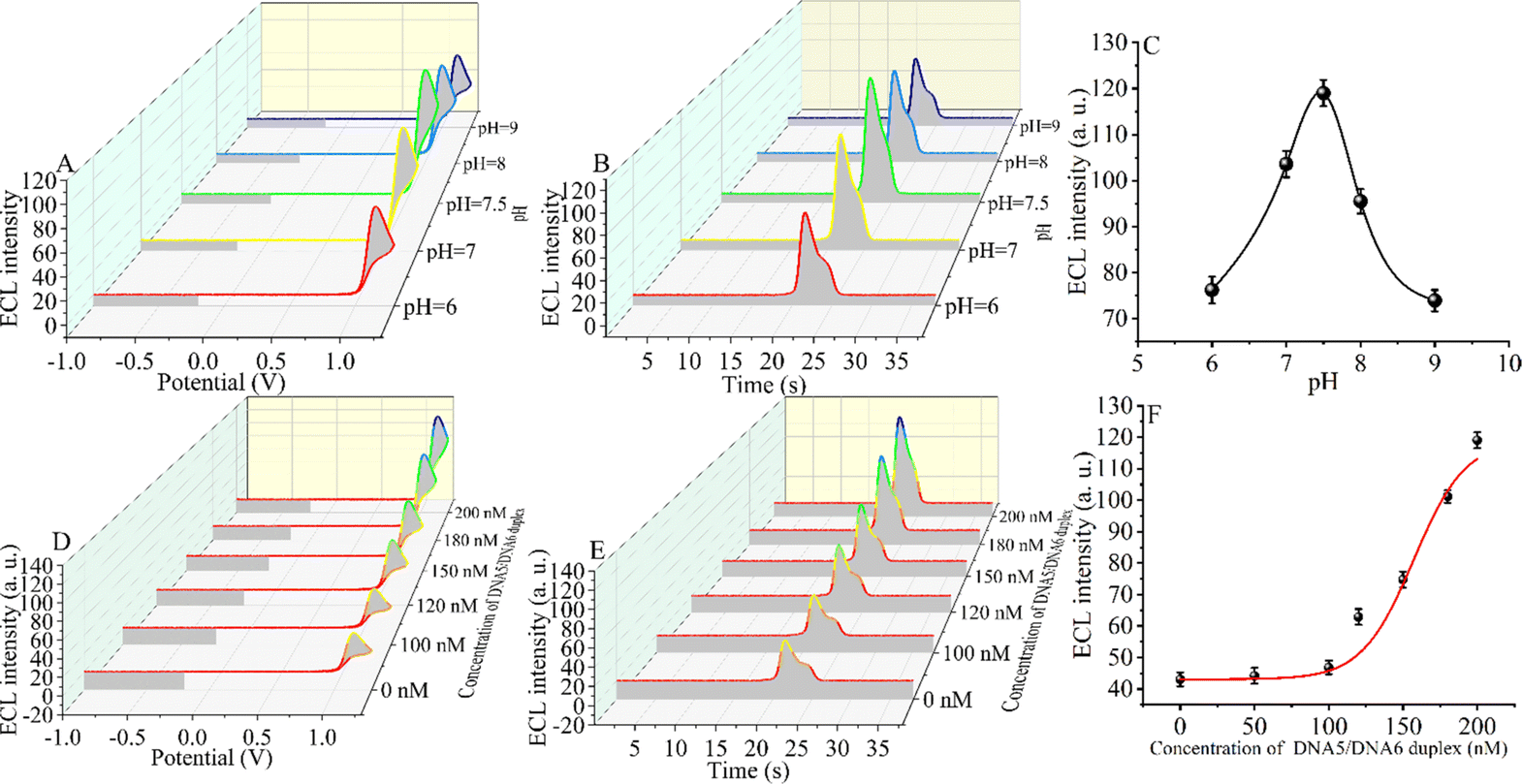
An appropriate pH environment is critical for the sensitivity of the constructed CRISPR-Cas12a-based biosensor. The ECL signal intensity was tested under weakly acidic, neutral, and weakly basic conditions. When the ECL intensity was in the range of pH 6.0 to pH 9.0 (Fig. 3A–C), the signal first increased and then decreased slightly, reaching the maximum ECL intensity at pH = 7.5 (the target DNA concentration is 5 fM). The reason for the maximum signal at pH = 7.5 may be that CRISPR-Cas12a has the highest shear activity at this pH.60,61 The optimal environment for the formation of the triple-helix complex is alkaline, and the melting temperature (Tm) of the triple-helix complex at pH = 7.5 is only 56 °C.55 However, considering that the optimal pH for enzymatic activity of CRISPR-Cas12a is 7.5 and that the reaction temperature (Tr) of our measurements is 37 °C, at which the triple-helix is still stable (Tm > Tr), we chose pH 7.5 as the optimal reaction condition.Next, we optimized the concentration of the DNA5/DNA6 duplex, and we selected different concentrations of the DNA5/DNA6 duplex to be added to the reaction system after the entropy-driven reaction (Fig. 3D–F). Other conditions, including the nucleic acid concentration and reaction time, were kept constant. The results revealed that the ECL signal decreased with the change of the DNA5/DNA6 concentration. The ECL signal reached a minimum when the concentration of the DNA5/DNA6 duplex coincided with that of DNA2. The reason for this phenomenon may be that in the reaction system we designed, below 100 nM DNA5/DNA6 duplex, DNA2 binds completely to the DNA5/DNA6 duplex and in turn completely inhibits the shearing activity of CRISPR-Cas12a. When the concentration of the DNA5/DNA6 duplex is further increased and exceeds that of DNA2, the higher concentration of the DNA5/DNA6 duplex also activates the shearing activity of CRISPR-Cas12a and therefore leads to an increase in the ECL signal. Consequently, we chose 100 nM for the next step of the SARS-CoV-2 RdRp gene detection experiment because, at this concentration, the DNA5/DNA6 duplex can be entirely bound to DNA2 completely.
3.6 Assay performance and accurate evaluation of the ECL biosensor
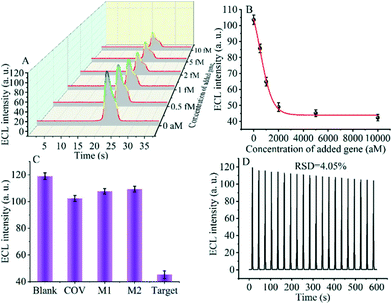
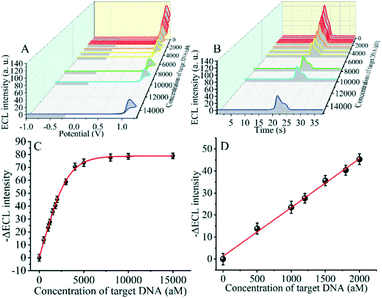
The detection performance of the biosensor used to detect different concentrations of the SARS-CoV-2 RdRp gene was investigated, as shown in Fig. 4A and B. The concentration of the target gene was analyzed by calculating the degree of change of the ECL signal. As shown in Fig. 4B, the ΔECL intensity increased rapidly with increasing concentration of the SARS-CoV-2 RdRp gene, ranging from 0 aM to 1 fM. In addition, the relationship between the ΔECL intensity and the target gene was also investigated (Fig. 4C). The linear equation is: Y = 1.289 + 0.022X (R2 = 0.9952), where Y represents the difference between the ECL values of the sample and the blank sample at a specific concentration and X represents the concentration of the target gene (Fig. 4D). Using this biosensor, we obtained a limit of detection (LOD) of 32.80 aM using the 3σ/slope method. The σ is the standard deviation when measuring 0 aM SARS-CoV-2 RdRp gene, and the slope is obtained from the linear equation. The excellent detection sensitivity is attributed to the amplification effect of the entropy-driven reaction and the efficient activation of double-stranded nucleic acids for CRISPR-Cas12a shearing activity. Thus, our design provides an efficient and ultra-sensitive detection solution for analyzing COVID-19 nucleic acid markers. The LOD was comparable to the work shown in Table S3† in the past few years.3.7 Applicability of the biosensor in real samples
To further investigate whether the proposed SARS-CoV-2 RdRp gene detection strategy can be applied to real samples and meet clinical needs, the biosensor was applied to the detection of the SARS-CoV-2 RdRp gene in tenfold diluted serum. Firstly, different concentrations of the SARS-CoV-2 RdRp gene were added to the tenfold diluted serum, and then the prepared samples were used instead of standard samples for the detection of the SARS-CoV-2 RdRp gene by using the CRISPR-Cas12a-based GOAu–Ru biosensor. The assay results showed that the intensity of ECL decreased gradually with the increase of SARS-CoV-2 RdRp gene addition (Fig. 5A), and the change of ECL reached a more stable value at the added amount of 5000 aM (Fig. 5B). This result fits well with the detection curve of the SARS-CoV-2 RdRp gene in buffer solution. The results demonstrate that our designed biosensing system can be applied for detecting the SARS-CoV-2 RdRp gene in real samples. Therefore, our proposed detection strategy for this analytical platform has the potential to be widely used for clinical applications in detecting COVID-19 related nucleic acid markers in complex biological samples.3.8 Selectivity and reproducibility of the CRISPR-Cas12a-based GOAu–Ru biosensor
Specificity and stability are essential metrics for evaluating the CRISPR-Cas12a-based GOAu–Ru biosensor. Therefore, next, we tested various targets to evaluate the specificity and stability of our constructed biosensing system. We selected a blank solution (without target DNA), the SARS-COV RdRp gene (COV, 5 fM), mismatched DNA1 (M1, 5 fM), and mismatched DNA2 (M2, 5 fM) for the control group. As shown in Fig. 5C, there was no significant difference in the electrochemiluminescence signal after using the blank solution, SARS-COV RdRp gene, mismatched DNA1, and mismatched DNA2 acting on the system. In contrast, the electrogenerated chemiluminescence signal changed more after the SARS-CoV-2 RdRp gene (5 fM) was applied to the CRISPR-Cas12a-based GOAu–Ru biosensor. The results indicate that our constructed CRISPR-Cas12a-based GOAu–Ru has good selectivity for the RdRp gene of SARS-COV-2.Stability is also an important performance indicator for the success of CRISPR-Cas12a-based GOAu–Ru biosensor construction. We measured the stability of the electrochemiluminescence signal and the results are shown in Fig. 5D. It can be seen from the figure that the electrochemiluminescence signal of the biosensor has a good stability (RSD = 4.05%) despite performing for three minutes and twenty scans. Thus, this indicates that our constructed CRISPR-based GOAu–Ru biosensor is expected to be used for the detection of the SARS-COV-2 RdRp gene.
3.9 Applicability of the biosensor in environmental surveillance
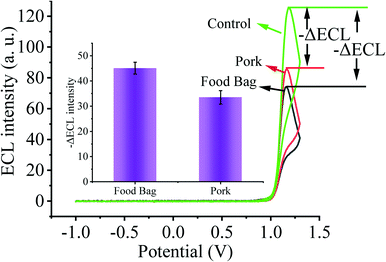
As the primary route of infection, healthy individuals can be directly infected by SARS-CoV-2 through respiratory droplets from inhalation and contact with mucous membranes, aerosols, and food surfaces. Moreover, contaminated environmental matrices also play a key role in the transmission of SARS-CoV-2. Next, in order to validate the prospective application of our designed biosensing system in environmental detection, we selected several environmental matrices as the study objects to verify the capability of our constructed biosensor for SARS-CoV-2 environmental detection (Fig. 6). We sprayed 2 fM SARS-CoV-2 nucleic acid buffer on the surface of food bags as well as pork and stored them at 4 °C for one week, and then collected SARS-CoV-2 nucleic acid on the surface of the bags and pork, and used the substrate without SARS-CoV-2 nucleic acid spraying as the control. By testing with our constructed sensing system, we clearly observed that the solution collected on the surface of the substrate sprayed with SARS-CoV-2 nucleic acid caused a higher ECL signal change. Thus, our sensor has potential applications in monitoring the spread of SARS-CoV-2 nucleic acids in the environment.4. Conclusions
In summary, we have proposed the first idea of using a DNA triple helix structure to inhibit CRISPR-Cas12a activity and applied it to design an electrochemiluminescent biosensor for detecting the SARS-CoV-2 RdRp gene. We employed an entropy-driven reaction to amplify a segment of nucleic acid that pairs with double-stranded DNA that can activate the activity of CRISPR-Cas12a by Hoogsteen pairing to form triple-stranded DNA, thereby inhibiting the binding of double-stranded DNA to CRISPR-Cas12a and thus the trans cleavage activity of CRISPR-Cas12a. Adopting this new concept, we built a novel biosensor to detect the SARS-CoV-2 RdRp gene by combining GOAu–Ru as the ECL luminous source. The inability of the inhibited activity of CRISPR-Cas12a to cleave the nucleic acid modified on the electrode surface results in the inability of the Fc modified at the other end of the nucleic acid to move away from the electrode surface, thus failing to induce electrochemiluminescence changes in the GOAu–Ru modified on the electrode surface. In contrast, when the activity of CRISPR-Cas12a cannot be inhibited, the electrochemiluminescence of GOAu–Ru changes. Using this system, we detected the SARS-CoV-2 RdRp gene with a detection limit of 32.80 aM. Moreover, the detection system has good stability and reproducibility, and we expect that this method may provide a potential platform for the diagnosis and treatment of coronavirus disease 2019 (COVID-19). In particular, it plays a key role in the environmental monitoring of COVID-19.Author contributions
Kai Zhang: conceptualization, writing – review and editing. Zhenqiang Fan: data curation, characterization, software. Yuedi Ding: data curation, characterization, software. Sha Zhu: scheme revision, discussion. Minhao Xie: reviewing, discussion Nan Hao: reviewing, conceptualization.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21705061), the Jiangsu Provincial Key Medical Discipline (Laboratory) (ZDXKA2016017), and the Innovation Capacity Development Plan of Jiangsu Province (BM2018023).Notes and references
- Y. Wang, L. Xia, C. Wei, H. Wang, H. Wang, R. Yuan and S. Wei, Ultrasensitive photoelectrochemical microRNA biosensor based on doxorubicin sensitized graphitic carbon nitride assisted by a target-activated enzyme-free DNA walker, Chem. Commun., 2019, 55, 13082–13084 RSC.
- N. Zhang, X.-M. Shi, H.-Q. Guo, X.-Z. Zhao, W.-W. Zhao, J.-J. Xu and H.-Y. Chen, Gold Nanoparticle Couples with Entropy-Driven Toehold-Mediated DNA Strand Displacement Reaction on Magnetic Beads: Toward Ultrasensitive Energy-Transfer-Based Photoelectrochemical Detection of miRNA-141 in Real Blood Sample, Anal. Chem., 2018, 90, 11892–11898 CrossRef CAS.
- X.-M. Li, W. Li, A.-Q. Ge and H.-Y. Chen, Logic-Based Dual-Functional DNA Tweezers with Protein and Small Molecule as Mechanical Activators, J. Phys. Chem. C, 2010, 114, 21948–21952 CrossRef CAS.
- M. H. Liu, J. L. Fu, C. Hejesen, Y. H. Yang, N. W. Woodbury, K. Gothelf, Y. Liu and H. Yan, A DNA tweezer-actuated enzyme nanoreactor, Nat. Commun., 2013, 4, 5 Search PubMed.
- Y. Yu, W. S. Zhang, Y. Guo, H. Peng, M. Zhu, D. Miao and G. Su, Engineering of exosome-triggered enzyme-powered DNA motors for highly sensitive fluorescence detection of tumor-derived exosomes, Biosens. Bioelectron., 2020, 167, 112482 CrossRef CAS PubMed.
- O. J. Wilkinson, C. Carrasco, C. Aicart-Ramos, F. Moreno-Herrero and M. S. Dillingham, Bulk and single-molecule analysis of a bacterial DNA2-like helicase nuclease reveals a single-stranded DNA looping motor, Nucleic Acids Res., 2020, 48, 7991–8005 CrossRef CAS PubMed.
- L. Li, Y. Zhang, Z. Yan, M. Chen, L. Zhang, P. Zhao and J. Yu, Ultrasensitive Photoelectrochemical Detection of MicroRNA on Paper by Combining a Cascade Nanozyme-Engineered Biocatalytic Precipitation Reaction and Target-Triggerable DNA Motor, ACS Sens., 2020, 5, 1482–1490 CrossRef CAS.
- X. Zhou, W. Zhang, Z. Wang, J. Han, G. Xie and S. Chen, Ultrasensitive aptasensing of insulin based on hollow porous C3N4/S2O82−/AuPtAg ECL ternary system and DNA walker amplification, Biosens. Bioelectron., 2020, 148, 111795 CrossRef CAS.
- K. Zhang, K. Wang, Y. Huang, X. Zhu, M. Xie and J. Wang, Sensitive detection of cytokine in complex biological samples by using MB track mediated DNA walker and nicking enzyme assisted signal amplification method combined biosensor, Talanta, 2018, 189, 122–128 CrossRef PubMed.
- Y. Zhang, L. Wang, F. Luo, B. Qiu, L. Guo, Z. Weng, Z. Lin and G. Chen, An electrochemiluminescence biosensor for Kras mutations based on locked nucleic acid functionalized DNA walkers and hyperbranched rolling circle amplification, Chem. Commun., 2017, 53, 2910–2913 RSC.
- K. Zhang, W. Huang, H. Li, M. Xie and J. Wang, Ultrasensitive detection of hERG potassium channel in single-cell with photocleavable and entropy-driven reactions by using an electrochemical biosensor, Biosens. Bioelectron., 2019, 132, 310–318 CrossRef PubMed.
- Y. Chang, Z. Wu, Q. Sun, Y. Zhuo, Y. Chai and R. Yuan, Simply Constructed and Highly Efficient Classified Cargo-Discharge DNA Robot: A DNA Walking Nanomachine Platform for Ultrasensitive Multiplexed Sensing, Anal. Chem., 2019, 91, 8123–8128 CrossRef PubMed.
- Y. Yin, G. Chen, L. Gong, K. Ge, W. Pan, N. Li, J. O. a. Machuki, Y. Yu, D. Geng, H. Dong and F. Gao, DNAzyme-Powered Three-Dimensional DNA Walker Nanoprobe for Detection Amyloid β-Peptide Oligomer in Living Cells and in Vivo, Anal. Chem., 2020, 92, 9247–9256 CrossRef PubMed.
- N. Wu, K. Wang, Y.-T. Wang, M.-L. Chen, X.-W. Chen, T. Yang and J.-H. Wang, Three-Dimensional DNA Nanomachine Biosensor by Integrating DNA Walker and Rolling Machine Cascade Amplification for Ultrasensitive Detection of Cancer-Related Gene, Anal. Chem., 2020, 92, 11111–11118 CrossRef PubMed.
- D. Gilbert, M. Heiner and C. Rohr, Petri-net-based 2D design of DNA walker circuits, Nat. Comput., 2018, 17, 161–182 CrossRef PubMed.
- K. Yehl, A. Mugler, S. Vivek, Y. Liu, Y. Zhang, M. Fan, E. R. Weeks and K. Salaita, High-speed DNA-based rolling motors powered by RNase H, Nat. Nanotechnol., 2016, 11, 184–190 CrossRef CAS PubMed.
- X. Qu, D. Zhu, G. Yao, S. Su, J. Chao, H. Liu, X. Zuo, L. Wang, J. Shi, L. Wang, W. Huang, H. Pei and C. Fan, An Exonuclease III-Powered, On-Particle Stochastic DNA Walker, Angew. Chem., Int. Ed., 2017, 56, 1855–1858 CrossRef CAS PubMed.
- J. Bath, S. J. Green and A. J. Turberfield, A Free-Running DNA Motor Powered by a Nicking Enzyme, Angew. Chem., Int. Ed., 2005, 44, 4358–4361 CrossRef CAS PubMed.
- K. Wang, M. Feng, M.-Q. He, F.-H. Zhai, Y. Dai, R.-H. He and Y.-L. Yu, DNA-fueled target recycling-induced two-leg DNA walker for amplified electrochemical detection of nucleic acid, Talanta, 2018, 188, 685–690 CrossRef PubMed.
- K. Wang, M.-Q. He, F.-H. Zhai, J. Wang, R.-H. He and Y.-L. Yu, Autonomous DNA nanomachine based on cascade amplification of strand displacement and DNA walker for detection of multiple DNAs, Biosens. Bioelectron., 2018, 105, 159–165 CrossRef PubMed.
- X. Yang, Y. Tang, S. D. Mason, J. Chen and F. Li, Enzyme-Powered Three-Dimensional DNA Nanomachine for DNA Walking, Payload Release, and Biosensing, ACS Nano, 2016, 10, 2324–2330 CrossRef PubMed.
- W. Li, L. Wang and W. Jiang, A catalytic assembled enzyme-free three-dimensional DNA walker and its sensing application, Chem. Commun., 2017, 53, 5527–5530 RSC.
- K. Zhang, Z. Fan, B. Yao, T. Zhang, Y. Ding, S. Zhu and M. Xie, Entropy-driven electrochemiluminescence ultra-sensitive detection strategy of NF-κB p50 as the regulator of cytokine storm, Biosens. Bioelectron., 2021, 176, 112942 CrossRef PubMed.
- D. Y. Zhang, A. J. Turberfield, B. Yurke and E. Winfree, Engineering Entropy-Driven Reactions and Networks Catalyzed by DNA, Science, 2007, 318, 1121–1125 CrossRef.
- Z. Fan, B. Yao, Y. Ding, J. Zhao, M. Xie and K. Zhang, Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds, Biosens. Bioelectron., 2021, 178, 113015 CrossRef PubMed.
- C.-P. Liang, P.-Q. Ma, H. Liu, X. Guo, B.-C. Yin and B.-C. Ye, Rational Engineering of a Dynamic, Entropy-Driven DNA Nanomachine for Intracellular MicroRNA Imaging, Angew. Chem., Int. Ed., 2017, 56, 9077–9081 CrossRef CAS PubMed.
- R. Zeng, W. Wang, M. Chen, Q. Wan, C. Wang, D. Knopp and D. Tang, CRISPR-Cas12a-driven MXene-PEDOT:PSS piezoresistive wireless biosensor, Nano Energy, 2021, 82, 105711 CrossRef CAS.
- K. Hsieh, G. Zhao and T.-H. Wang, Applying biosensor development concepts to improve preamplification-free CRISPR/Cas12a-Dx, Analyst, 2020, 145, 4880–4888 RSC.
- K. Zhang, Z. Fan, B. Yao, Y. Ding, J. Zhao, M. Xie and J. Pan, Exploring the trans-cleavage activity of CRISPR-Cas12a for the development of a Mxene based electrochemiluminescence biosensor for the detection of Siglec-5, Biosens. Bioelectron., 2021, 178, 113019 CrossRef CAS.
- Z. Fan, Y. Ding, B. Yao, J. Wang and K. Zhang, Electrochemiluminescence platform for transcription factor diagnosis by using CRISPR–Cas12a trans-cleavage activity, Chem. Commun., 2021, 57, 8015–8018 RSC.
- K. Zhang, Z. Fan, Y. Ding and M. Xie, A pH-engineering regenerative DNA tetrahedron ECL biosensor for the assay of SARS-CoV-2 RdRp gene based on CRISPR/Cas12a trans-activity, Chem. Eng. J., 2022, 429, 132472 CrossRef CAS.
- K. Zhang, Z. Fan, Y. Huang, Y. Ding and M. Xie, A strategy combining 3D-DNA Walker and CRISPR-Cas12a trans-cleavage activity applied to MXene based electrochemiluminescent sensor for SARS-CoV-2 RdRp gene detection, Talanta, 2022, 236, 122868 CrossRef CAS PubMed.
- R. Nouri, Z. Tang, M. Dong, T. Liu, A. Kshirsagar and W. Guan, CRISPR-based detection of SARS-CoV-2: A review from sample to result, Biosens. Bioelectron., 2021, 178, 113012 CrossRef CAS.
- Y. Jiang, M. Hu, A.-A. Liu, Y. Lin, L. Liu, B. Yu, X. Zhou and D.-W. Pang, Detection of SARS-CoV-2 by CRISPR/Cas12a-Enhanced Colorimetry, ACS Sens., 2021, 6, 1086–1093 CrossRef CAS PubMed.
- J. Li, S. Yang, C. Zuo, L. Dai, Y. Guo and G. Xie, Applying CRISPR-Cas12a as a Signal Amplifier to Construct Biosensors for Non-DNA Targets in Ultralow Concentrations, Acs Sensors, 2020, 5, 970–977 CrossRef CAS.
- G. Felsenfeld, D. R. Davies and A. Rich, FORMATION OF A THREE-STRANDED POLYNUCLEOTIDE MOLECULE, J. Am. Chem. Soc., 1957, 79, 2023–2024 CrossRef CAS.
- M. R. Kiran and M. Bansal, Structural Polymorphism in d(T)12.d(A)12*d(T)12 Triple Helices, J. Biomol. Struct. Dyn., 1995, 13, 493–505 CrossRef CAS.
- Y. Hu, A. Cecconello, A. Idili, F. Ricci and I. Willner, Triplex DNA Nanostructures: From Basic Properties to Applications, Angew. Chem., Int. Ed., 2017, 56, 15210–15233 CrossRef CAS.
- D. Zhu, J. Zhu, Y. Zhu, L. Wang and W. Jiang, Sensitive detection of transcription factors using an Ag+−stabilized self-assembly triplex DNA molecular switch, Chem. Commun., 2014, 50, 14987–14990 RSC.
- J. Zheng, J. Li, Y. Jiang, J. Jin, K. Wang, R. Yang and W. Tan, Design of Aptamer-Based Sensing Platform Using Triple-Helix Molecular Switch, Anal. Chem., 2011, 83, 6586–6592 CrossRef CAS.
- F. Iacovelli, A. Idili, A. Benincasa, D. Mariottini, A. Ottaviani, M. Falconi, F. Ricci and A. Desideri, Simulative and Experimental Characterization of a pH-Dependent Clamp-like DNA Triple-Helix Nanoswitch, J. Am. Chem. Soc., 2017, 139, 5321–5329 CrossRef CAS.
- Z. Fan, B. Yao, Y. Ding, D. Xu, J. Zhao and K. Zhang, Rational engineering the DNA tetrahedrons of dual wavelength ratiometric electrochemiluminescence biosensor for high efficient detection of SARS-CoV-2 RdRp gene by using entropy-driven and bipedal DNA walker amplification strategy, Chem. Eng. J., 2022, 427, 131686 CrossRef CAS PubMed.
- L. Zhu, J. Ye, M. Yan, L. Yu, Y. Peng, J. Huang and X. Yang, Sensitive and Programmable “Signal-Off” Electrochemiluminescence Sensing Platform Based on Cascade Amplification and Multiple Quenching Mechanisms, Anal. Chem., 2021, 93, 2644–2651 CrossRef CAS PubMed.
- T. Zhou, R. Huang, M. Huang, J. Shen, Y. Shan and D. Xing, CRISPR/Cas13a Powered Portable Electrochemiluminescence Chip for Ultrasensitive and Specific MiRNA Detection, Adv. Sci., 2020, 7, 1903661 CrossRef.
- Z. Fan, Z. Lin, Z. Wang, J. Wang, M. Xie, J. Zhao, K. Zhang and W. Huang, Dual-Wavelength Electrochemiluminescence Ratiometric Biosensor for NF-κB p50 Detection with Dimethylthiodiaminoterephthalate Fluorophore and Self-Assembled DNA Tetrahedron Nanostructures Probe, ACS Appl. Mater. Interfaces, 2020, 12, 11409–11418 CrossRef.
- Y. Liu, J. Lei, Y. Huang and H. Ju, Off-On Electrochemiluminescence System for Sensitive Detection of ATP via Target-Induced Structure Switching, Anal. Chem., 2014, 86, 8735–8741 CrossRef PubMed.
- Y. He, Y. Chai, R. Yuan, H. Wang, L. Bai and N. Liao, A supersandwich electrochemiluminescence immunosensor based on mimic-intramolecular interaction for sensitive detection of proteins, Analyst, 2014, 139, 5209–5214 RSC.
- J. Ge, Y. Zhao, C. Li and G. Jie, Versatile Electrochemiluminescence and Electrochemical “On–Off” Assays of Methyltransferases and Aflatoxin B1 Based on a Novel Multifunctional DNA Nanotube, Anal. Chem., 2019, 91, 3546–3554 CrossRef PubMed.
- Z. Fan, B. Yao, Y. Ding, M. Xie, J. Zhao, K. Zhang and W. Huang, Electrochemiluminescence aptasensor for Siglec-5 detection based on MoS2@Au nanocomposites emitter and exonuclease III-powered DNA walker, Sens. Actuators, B, 2021, 334, 129592 CrossRef PubMed.
- D. D. Kulkarni, I. Choi, S. S. Singamaneni and V. V. Tsukruk, Graphene Oxide−Polyelectrolyte Nanomembranes, ACS Nano, 2010, 4, 4667–4676 CrossRef PubMed.
- B. Genorio, K. L. Harrison, J. G. Connell, G. Dražić, K. R. Zavadil, N. M. Markovic and D. Strmcnik, Tuning the Selectivity and Activity of Electrochemical Interfaces with Defective Graphene Oxide and Reduced Graphene Oxide, ACS Appl. Mater. Interfaces, 2019, 11, 34517–34525 CrossRef CAS PubMed.
- Z. Xu, Y. Bando, L. Liu, W. Wang, X. Bai and D. Golberg, Electrical Conductivity, Chemistry, and Bonding Alternations under Graphene Oxide to Graphene Transition As Revealed by In Situ TEM, ACS Nano, 2011, 5, 4401–4406 CrossRef CAS PubMed.
- Y. V. Stebunov, O. A. Aftenieva, A. V. Arsenin and V. S. Volkov, Highly Sensitive and Selective Sensor Chips with Graphene-Oxide Linking Layer, ACS Appl. Mater. Interfaces, 2015, 7, 21727–21734 CrossRef CAS.
- J. Ye, L. Zhu, M. Yan, Q. Zhu, Q. Lu, J. Huang, H. Cui and X. Yang, Dual-Wavelength Ratiometric Electrochemiluminescence Immunosensor for Cardiac Troponin I Detection, Anal. Chem., 2019, 91, 1524–1531 CrossRef CAS.
- A. Amodio, B. Zhao, A. Porchetta, A. Idili, M. Castronovo, C. Fan and F. Ricci, Rational Design of pH-Controlled DNA Strand Displacement, J. Am. Chem. Soc., 2014, 136, 16469–16472 CrossRef CAS PubMed.
- H. Lei, C. Niu, T. Li, Y. Wan, W.-B. Liang, R. Yuan and P. Liao, A Novel Electrochemiluminescent Immunoassay based on Target Transformation Assisted with Catalyzed Hairpin Assembly Amplification for the Ultrasensitive Bioassay, ACS Appl. Mater. Interfaces, 2019, 11, 31427–31433 CrossRef CAS.
- P. J. Taverna, H. Mayfield and A. R. J. Andrews, Determination of cadmium ions in water by a novel electrochemiluminescence method, Anal. Chim. Acta, 1998, 373, 111–117 CrossRef CAS.
- Y. Yuan, S. Han, L. Hu, S. Parveen and G. Xu, Coreactants of tris(2,2′-bipyridyl)ruthenium(II) Electrogenerated Chemiluminescence, Electrochim. Acta, 2012, 82, 484–492 CrossRef CAS.
- H. Zheng and Y. Zu, Emission of Tris(2,2′-bipyridine)ruthenium(II) by Coreactant Electrogenerated Chemiluminescence: From O2-Insensitive to Highly O2-Sensitive, J. Phys. Chem. B, 2005, 109, 12049–12053 CrossRef CAS.
- R. Zhou, Y. Li, T. Dong, Y. Tang and F. Li, A sequence-specific plasmonic loop-mediated isothermal amplification assay with orthogonal color readouts enabled by CRISPR Cas12a, Chem. Commun., 2020, 56, 3536–3538 RSC.
- T. Yuan, O. Mukama, Z. Li, W. Chen, Y. Zhang, J. de Dieu Habimana, Y. Zhang, R. Zeng, C. Nie, Z. He and L. Zeng, A rapid and sensitive CRISPR/Cas12a based lateral flow biosensor for the detection of Epstein-Barr virus, Analyst, 2020, 145, 6388–6394 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1en00645b |
| This journal is © The Royal Society of Chemistry 2022 |