Open Access Article
Open Access ArticleIntegrated therapy platform of exosomal system: hybrid inorganic/organic nanoparticles with exosomes for cancer treatment
Ming-Hsien
Chan†
 a,
Zhi-Xuan
Chang†
b,
Chi-Ying F.
Huang
a,
Zhi-Xuan
Chang†
b,
Chi-Ying F.
Huang
 bc,
L. James
Lee
bd,
Ru-Shi
Liu
bc,
L. James
Lee
bd,
Ru-Shi
Liu
 *ae and
Michael
Hsiao
*ae and
Michael
Hsiao
 *af
*af
aGenomics Research Center, Academia Sinica, Taipei, 115, Taiwan. E-mail: mhsiao@gate.sinica.edu.tw
bInstitute of Biopharmaceutical Sciences, National Yang Ming Chiao Tung University, Taipei, Taiwan
cInstitute of Clinical Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan
dDepartment of Chemical and Biomolecular Engineering, Ohio State University, Columbus, 43210, Ohio, USA
eDepartment of Chemistry, National Taiwan University, Taipei 106, Taiwan. E-mail: rsliu@ntu.edu.tw
fDepartment of Biochemistry, College of Medicine, Kaohsiung Medical University, Kaohsiung 807, Taiwan. E-mail: mhsiao@gate.sinica.edu.tw
First published on 18th January 2022
Abstract
Recent studies have found that exosomes or extracellular vehicles (EVs) are associated with cancer metastasis, disease progression, diagnosis, and treatment, leading to a rapidly emerging area of exocrine vesicle research. Relying on the superior targeting function and bio-compatibility of exosomes, researchers have been able to deliver drugs to cancer stem cells deep within tumors in mouse models. Despite significant efforts made in this relatively new field of exosome research, progress has been held back by challenges such as inefficient separation methods, difficulties in characterization/tracking, and a lack of specific biomarkers. Therefore, current researches are devoted to combining nanomaterials with exosomes to improve these shortcomings. Adding inorganic/organic nanoparticles such as artificial liposomes and iron oxide can bring more drug options and various fluorescent or magnetic diagnostic possibilities to the exosome system. Moreover, the applications of exosomes need to be further evaluated under actual physiological conditions. This review article highlights the potential of exosome-biomimetic nanoparticles for their use as drug carriers to improve the efficacy of anticancer therapy.
1. Introduction
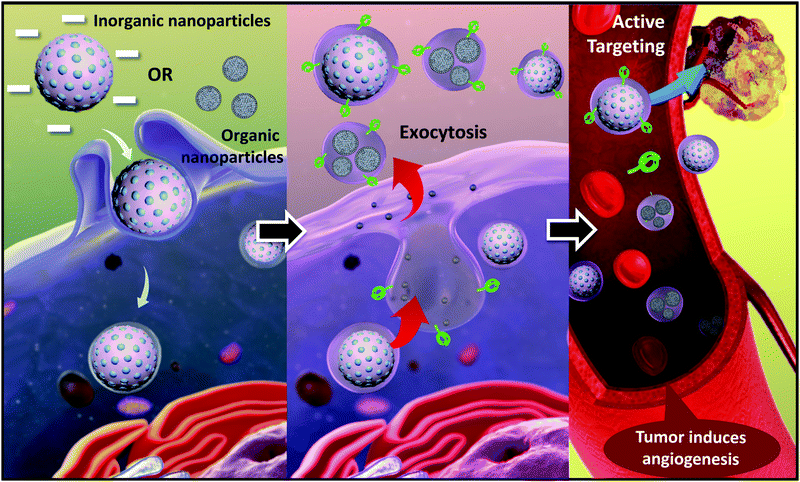
Cancer is characterized by the US National Cancer Institute-National Institutes of Health (NIH NCI) as a significant disease that requires several therapeutic strategies to reduce its current high mortality. Highly aggressive cancer types such as lung adenocarcinoma (LUAD), glioblastoma (GBM), and pancreatic ductal adenocarcinoma (PDAC) originate from organs that are difficult to diagnose and trace. One of the primary reasons these cancers have high mortality rates is the lack of treatment means and diagnosis methods. In an effort to prevent the life-threatening scenarios caused by these high-fatality cancers, exosomes are considered as a new generation of nano drugs with both therapeutic and diagnostic functions.1–3 Many studies have found that exosomes derived from tumor cells contain an abundance of specific microRNAs, and their biochemical properties render them stable and easy to store.4–6 When exosomes are designed or selectively encapsulated with unique biologically active molecules, they can be used to deliver tumor suppressor molecules or chemotherapy drugs.7 Moreover, exosomes are considered an excellent candidate for GBM treatment since they can be developed as a multifunctional drug system that can penetrate the blood-brain tumor barrier.8 Exosomes have many functions and can penetrate physiological barriers in the brain and pancreas, and their therapeutic efficacy has been successfully demonstrated in previous research works.9Currently, many researchers are developing new technologies that can produce therapeutic exosomes (tExos) containing multiple therapeutics and targeting ligands as a new drug platform to treat cancers.10 Although exosomes have many advantages, many challenges remain that need to be overcome for their use in disease treatment and diagnosis. These problems can be solved by combining exosomes with inorganic/organic nanomaterials to optimize the medical application value of exosomes. The following are several essential issues that such hybrid nanomaterials can address. (1) The total number of exosomes consumed in the treatment is too high. Based on current research, exosomes carrying genes or proteins require a dose of >1010 particles per mL to achieve a therapeutic effect. On the other hand, when combined with inorganic/organic nanomaterials, only 10 ng mL−1 of the drug is necessary to produce the same therapeutic effect.11 In detail, compared with other nanomaterials, the drug-carrying capacity of exosomes is challenging o control. The drugs needed to be sent into the cell first, then collected and purified the exosome with the drug in the cell supernatant.
However, inorganic/organic nanomaterials can preferentially carry and pack medicines with known concentrations in bench synthesis, protecting the drug molecules that won’t react with the unpredictable cellular environment, such as protein or nucleic acid substance and decreasing the drug dose. (2) Exosomes in circulation are quickly taken up by immune cells in the body and are difficult to detect.12 After modification with specific ligands, inorganic/organic nanomaterials can prevent the exosome uptake by immune cells. Moreover, these particles can be detected using luminescence, ultrasonic signaling, and magnetic properties for diagnosis. (3) The central core of the exosome is hydrophilic. As a result, exosomes cannot load hydrophobic drugs quickly. Since the center of some inorganic/organic nanomaterials has hydrophobic pores or spaces, they thereby allow the particles to carry hydrophobic drugs, which can then be enveloped with exosomes for delivery to cancer cells.13,14 Inorganic/organic nanoparticles hybridized with exosomes have unique properties, such as enhanced permeability and retention (EPR) effect, modifiability, intelligent responsiveness, and co-delivery of multiple drugs. These materials play an essential role in the precise and synergistic treatment of tumors by enhancing efficacy and reducing toxicity and side effects. Moreover, hybrid nanomaterials are often coupled with targeting molecules to achieve the targeted delivery of anti-tumor drugs.15 These biomimetic nanoplatforms combine the unique functions of natural biomaterials and the versatility of artificial nanomaterials, resulting in anti-tumor nanomedicines with five essential characteristics: long circulation time, tumor targeting, deep tumor penetration, high cellular internalization, and strong drug release (Fig. 1). In this review, we first introduce the properties of exosomes, followed by the description of the cancer applications of exosomes with a deep focus on how exosomes are combined with inorganic/organic nanomaterials to provide multiple and beneficial therapeutic and diagnostic results.
2. Exosomes
2.1. The background of exosomes
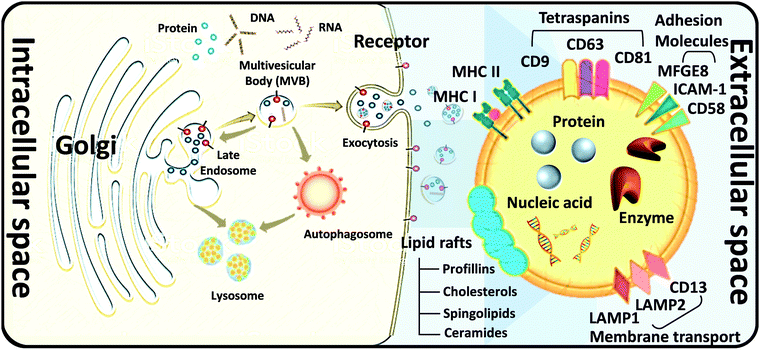
Cell-secreted extracellular vehicles (EVs), particularly exosomes with sizes of 30–150 nm in diameter, encapsulating various active biomolecules, including nucleic acids, lipids, and proteins, have emerged as promising therapeutic agents. Unpurified exosomes belong to a larger group of cell-derived heterogeneous vesicles defined as EVs produced and released by cells.EVs are distinct in size and biogenesis, and there are three main types of double-layer vesicles: exosomes, apoptotic bodies, and microvesicles. Apoptotic bodies are 0.5–5 μm in diameter; they are produced and released by dying cells that undergo apoptosis. Microvesicles are 50–1000 nm in diameter and are formed by “pinching off” from the plasma membrane. Exosomes are 30–200 nm in diameter. They are intraluminal vesicles (ILVs) in multivesicular bodies (MVBs), which are organelles involved in the intracellular traffic of biomolecules and released via inward budding at the plasma membrane (Fig. 2).16 Exosomes were first reported 30 years ago. In the 1980s, exosomes were discovered by researchers working with maturing reticulocytes.17 They observed that small vesicles created by inward budding in the intracellular endosome would form a multivesicular body (MVB). The MVB would then fuse with the cell plasma membrane (PM). The internal vesicles would be secreted from cells as exosomes.18,19 In the past, exosomes were considered to be the dustbins of cells, but due to recent discoveries of their desirable features for use in cell communication, they are expected to become a new type of biomarker for disease diagnosis and drug nanocarriers. In addition to their biocompatibility (due to their presence in many body fluids and tissues), exosomes have intrinsic transmembrane and membrane-anchored proteins that provide prolonged blood circulation and promote tissue-directed delivery and cellular uptake of their internal contents. Exosomes are usually formed in three steps: (1) endocytosis of extracellular material, (2) maturation, and (3) exocytosis.
2.2. Isolation, purification, and analysis of exosomes
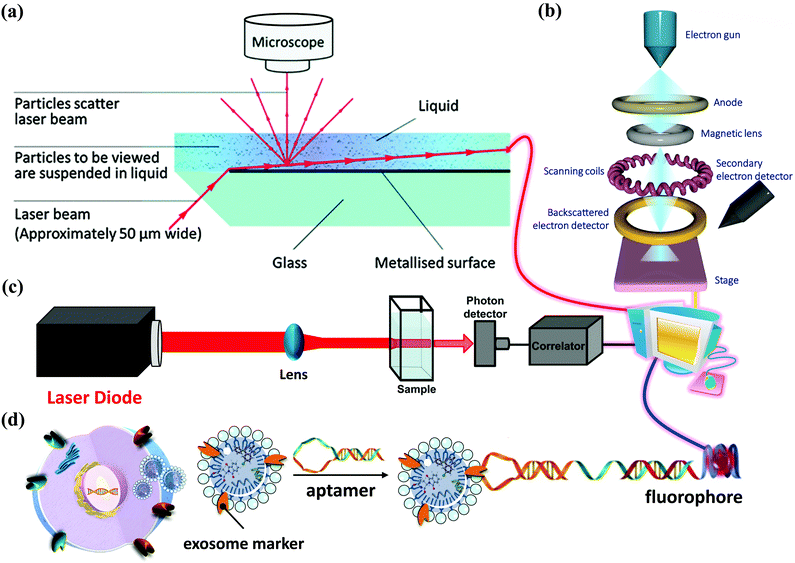
How to obtain and confirm high-purity exosomes is currently a hot research topic. Most of the current methods of combining nanoparticles with exosomes as a composite nano-platform are first to use cells as providers of exosomes, then add target nanomaterials to the cells for cell uptake, and finally release the package through exocytosis. After these procedures, a composite carrier covered with exosomes and target nanomaterials was born. Therefore, isolation, purification, and analysis of exosomes are also important topics. Here is a brief description of several popular ways to connect the hybrid inorganic/organic nanoparticles with exosomes. High-speed UC, which is regarded as the most common isolation method, mainly depends on vesicle size and density. Gradient-based UC can also be applied further to purify the exosomes to a narrower distributed population.20 Ultrafiltration membranes, including tangential flow filtration (TFF), can also be used to isolate exosomes based on vesicle sizes.21 Depending on the size of the membrane, this method allows for the separation of exosomes from proteins and other macromolecules. Immunoaffinity methods utilize interactions between antibodies and selective exosome surface proteins to isolate the exosomes or exosome subpopulations.21 Size exclusion chromatography (SEC) relies on resin-filled porous columns to sort EVs and exosomes into fractions based on their different retention times in the column.20 In the precipitation method, exosomes can be isolated from biological fluids by changing their solubility.22Other techniques apply polymers to precipitate exosomes based on their surface features and polymer molecular weights.23 After exosomes are sorted and collected from a sample, and they are characterized by size, protein expression, RNA content, and lipid composition. Several techniques can be applied, such as nanoparticle tracking analysis (NTA), dynamic light scattering (DLS) and nanoscale flow cytometry for exosome size and number quantification, LC-MS, western blot, and ELISA for protein profiling and characterization, Next Generation Sequencing (NGS), microarray, and qPCR for RNA profiling and characterization, and several microscopy techniques for exosome morphology characterization.24–26 In the following, we briefly describe several widely used methods for exosome analysis. Nanoparticle Tracking Analyzer (NTA) uses laser light to irradiate a nanoparticle suspension. The particle size, scattered light intensity, quantity, concentration, and dynamic images of moving particles under Brownian motion can be obtained at the same time (Fig. 3a).27,28 The working principle of the Transmission/Scanning Electron Microscope (TEM/SEM) is the use of an electron beam with an energy of 1–30 kV to irradiate the surface of the analyzed sample in the raster scan mode.29 Secondary electrons and backscattered electrons are imaged to obtain information on the microstructure and morphology of the sample surface and have high resolution (Fig. 3b). Dynamic light scattering (DLS) is a technique that collects fluctuations in the intensity of the scattered light of particles undergoing Brownian motion in the solution.30 These light intensity fluctuations are converted into a correlation curve through an appropriate instrument, thereby obtaining their speed (Fig. 3c).
In addition, in 1989, Sidney Altman and Thomas Cech discovered that small RNA or DNA has functions such as ligand binding, activation, and gene regulation. They can be screened for secondary structure RNA or single-stranded DNA that specifically binds to the target protein. These specialized small molecule nucleic acids are called aptamers. Aptamers can also be used for exosome detection, such as the fluorescence method for exosome competitive detection of CD63-specific aptamers (Fig. 3d). In the presence of exosomes, CD63 on the exosomes binds to the aptamer, resulting in the shedding of short sequences into the supernatant. The number of exosomes can be estimated by detecting the fluorescence intensity in the supernatant. Compared with conventional methods, the detection cost and difficulty are significantly reduced, while the sensitivity is guaranteed, which provides a basis for subsequent detection of specific cancer cell exosomes.
These standard analysis methods have their advantages and drawbacks. Therefore, evaluating exosomes from multiple perspectives is necessary, especially for hybrid exosomes where inorganic/organic nanoparticles may lead to significant exosome size and morphology changes.
3. The cancer applications of exosomes and hybrid with inorganic/organic nanoparticles
At first, exosomes were considered excess membrane proteins released by the regulating membrane function during cell maturation. They were characterized as organelles that remove cell debris and eliminate cell surface molecules. Later, an increasing number of studies found that exosomes play essential roles in mediating inflammation and cell proliferation, regulating the extracellular microenvironment, and inducing the body's immune response.32 At present, exosomes are considered biomarkers and prognostic factors of diseases, and they have the potential to serve as carriers for gene and drug delivery, which have important clinical significance.33–353.1. Diagnostic and prognostic markers of cancer
Because exosomes can be detected in body fluids (blood, urine, saliva, and cerebrospinal fluid), they are considered non-invasive or minimally invasive biomarkers for disease diagnosis. They can detect many pathological conditions, including cancer potential (Fig. 4a).36–39 The current focus of genomic medicine is to develop tests based on personalized and precision medicine. Therefore, a liquid biopsy that uses body fluids (mainly blood and urine) instead of tissue samples obtained through surgery for diagnosis is rapidly attracting attention. This method is convenient for patients with minimal harm. The liquid biopsy covers three significant types, including circulating tumor cells (CTCs), free DNA (cfDNA), and exosomes (or EVs). Exosomes are the most attractive type in liquid biopsy due to their high stability and ease of collection from body fluids such as urine and blood. miRNAs are considered potential diagnostic biomarkers because they play a regulatory role in the body. Changes in exosomal miRNA enrichment can reflect physiological and pathological changes, indicating their possible use as diagnostic biomarkers. According to reports, miR-23b-3p, miR-10b-5p, and miR-21-5p are promising prognostic biomarkers for non-small cell lung cancer (NSCLC) patients.40–42 In addition to miRNA, exosomes also contain proteins from host cells, so they have potential functions as biomarkers. Analysis of the entire proteome of exosomes by mass spectrometry has highlighted various biomarkers in prostate and bladder cancer.43 A study involving 276 NSCLC patients identified NY-ESO-1, EGFR, PLAP, EpCAM, and Alix in exosomes as protein biomarkers for overall survival.44 Plasma-derived exosomes have been successfully used to evaluate multi-label models using the EV Array system, consisting of 37 antibodies against lung cancer-related proteins.45 This method uses only 10 μL of plasma samples for analysis. Exosomal lncRNA is also defined as a new tumor biomarker.46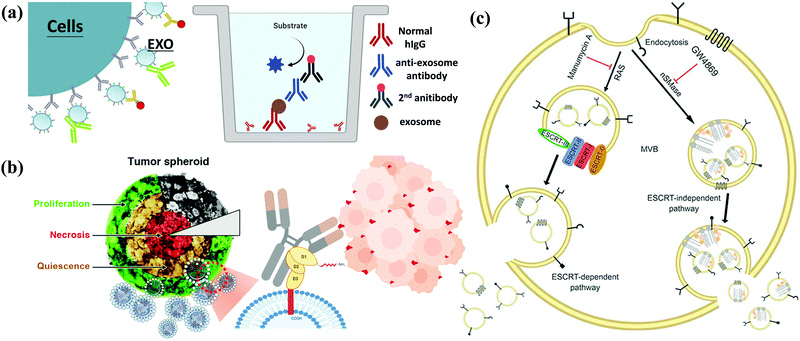 | ||
| Fig. 4 Applications of exosomes. (a) Diagnosis: exosomes from patients can be used for disease diagnosis. (b) Drug delivery: therapeutic drugs such as chemicals, peptides, and siRNA can be delivered to patients. (c) Therapy: exosomes from MSCs can be used to treat various diseases. (c) Reproduced with permission.31 Copyright 2020, Wiley Online Library. | ||
3.2. Drug delivery vehicles
One of the most promising drug delivery systems is liposomes. Liposomes are artificial vesicles with a phospholipid membrane self-assembled into different sizes and forms in an aqueous milieu. Liposomes allow for long circulation and a wide range of techniques for high concentration drug loading, and they can be prepared easily. However, the immunocompatibility of liposomes is debatable, and their targeting capability in vivo is often insufficient.47 Compared to liposomes, exosomes are more stable in circulation because of their endogenous nature and unique surface content.48 Due to their small size and bubble shape, exosomes can move from one cell to another, release their contents through the cell membrane. Due to their protein content and genetic material, biological and small drugs can be incorporated into exosomes. An example of such a system is doxorubicin-loaded exosomes.49 Once exosomes have been internalized into recipient cells, and their cargo can exert its effects. In a 2007 study, the authors prove that exosome mRNA can be translated into recipient cells.50 They first collected exosomes from mice and then incubated them with human cells. After incubation, three different mouse proteins were found in human recipient cells, indicating that exosome cargos can be transferred into cells.51 Exosome-based therapy has several significant advantages: (1) low immunogenicity; (2) no vascular occlusion; and (3) systemic injection of exosomes can cross the blood–brain barrier and enter the brain parenchyma.52Recently, post-insertion of siRNA into exosomes by conventional bulk electroporation has been developed and demonstrated success; the performance was superior to that of synthetic nanocarriers in several mouse models for central neuron and cancer treatment.53–55 Many clinical trials on exosome therapy have also been launched worldwide.56 Despite their many advantages, the application of exosomes in gene delivery for clinical use has been slow because producing sufficient quantities for large-scale in vivo use is technically challenging. Even though the exosome dose (∼50 μg kg−1 or ∼1 μg exosomal RNA from ∼109 exosomes in one injection per mouse, similar to an adeno-associated virus dose) is lower than using synthetic RNA and nanocarriers (∼1 mg kg−1), large numbers of cell cultures are required. It takes days of incubation, followed by purification and nuclei acid loading, to generate gene-containing exosomes in sufficient quantities for repeated use in human therapy because 150–200 times more exosomes are needed for a patient than for a mouse. Only a few cell types, such as mesenchymal stem cells (MSCs) and dendritic cells (DCs), are known to secrete relatively high numbers of exosomes, e.g., >5 × 104 per cell in 1–2 days, but the strong electric field required to insert RNAs would damage many exosomes, leading to a low yield. Post-insertion of large biomolecules into nano-sized exosomes is also inefficient. New technologies to generate many exosomes with a high loading of RNAs and targeting peptides in exosomes are essential for nucleic acid-based exosome therapy.
3.3. Anticancer vaccine for cancer immunotherapy
Recently, the success of immune checkpoint therapy in several cancer types has increased people's interest in the further exploration of immune disorders in tumors, including exosomal tumor immunotherapy, which is expected to produce anticancer vaccines (Fig. 4b).57 Tumor-derived exosomes carry antigens and have been used as a source of specific stimulation of the immune response against tumors. One study found that B cells transformed with Epstein–Barr virus released exosomes containing major histocompatibility complex (MHC)-II, thereby activating CD4+ T cells and the exosomes secreted by dendritic revealed to express MHC-I, which activates CD8+ T cells.58 Subsequent studies found that those exosomes can induce anti-tumor immunity in experimental animal models and human clinical trials, including colorectal cancer, metastatic skin cancer, and non-small cell lung cancer.59–61 In mice, tumor-derived exosomes can serve as an antigen delivery system and prevent the development of autologous tumors in a CD4+ and CD8+ T cell-dependent manner.62 In phase I clinical trial, patients with advanced non-small-cell lung cancer or metastatic melanoma were vaccinated with dendritic cell-derived exosomes and showed an anti-tumor immune response and tumor regression. Recent evidence showed that dendritic cells loaded with tumor exosomes effectively produce an anti-tumor response against autologous tumor cells in patients with malignant glioma.63Researchers believe ascites-derived exosomes are as effective as dendritic cells or tumor-derived exosomes in sensitizing dendritic cells and the primary cytotoxic T lymphocytes, thereby killing autologous tumor cells in vitro.64 Moreover, exosomes derived from the malignant effusion of ovarian cancer patients have been prepared and will undergo clinical trials.65 It was found that the combination of ascites-derived exosomes from colorectal cancer patients and granulocyte-macrophage colony-stimulating factors can effectively induce effective carcinoembryonic antigen-specific anti-tumor immunity in patients with advanced colorectal cancer.66
3.4. Inhibition of exosomes
Elevated levels of exosomes are usually associated with the progression of different types of cancer. Some researchers hope to prevent poor patient prognosis by reducing tumor exosomes to normal levels (Fig. 4c).31,67 From this perspective, many ongoing studies aim to regulate the release of exosomes by modulating the process of exosome secretion or by explicitly targeting their components to inhibit their interaction with target cells. Currently, our understanding of exosomal mechanisms and functions under different physiological and pathological conditions is increasing. Although its biological functions have not yet been fully resolved, researchers have conducted in-depth explorations in many fields. Exosomes are not waste particles but a key medium for cell-to-cell communication. As the satellites of host cells, exosomes contain a large amount of biological information, and their functions exceed initial expectations. Host cells control the contents of exosomes, thus changing their fate or that of other cells. Secondly, exosomes have a strong influence on tumor progression and metastasis, and they can be used to predict the location of metastasis and to establish a pre-metastasis niche. Additionally, the exosomes secreted by tumor cells can either block tumor growth by promoting the anti-tumor immune response or induce tumor growth by weakening anti-tumor immunity or promoting angiogenesis.68In the late stage, cancer cell-derived exosomes produce much stronger immunosuppressive effects than immune activation, and vaccines based on antigen-carrying exosomes are also in clinical trials.69
Although the use of exosomes for diagnosis and treatment is still in its infancy, their use as a carrier combined with gene therapy has excellent clinical application potential with the deeper understanding and systematic evaluation of exosomes.
3.5. Advantages of exosomes-inorganic/organic nanoparticles hybrid system
Organic/inorganic nanocomposites are usually organic polymer composites with inorganic nanoscale structural units. They combine the advantages of inorganic materials (such as rigidity, thermal stability) and organic polymers (such as flexibility, dielectric properties, elasticity, and processability). Exosomes themselves is multifunctional organic carrier, and their lipid bilayer structure is like liposomes. It has excellent compatibility in a biological environment. Combining inorganic materials can significantly improve the biocompatibility of inorganic materials and reduce the release of metal ions generated when inorganic materials are degraded into the biological environment to cause physical toxicity. In addition, the inherent characteristics of inorganic materials, such as fluorescent development, magnetic tracking, etc., also provide exosomes with tracking and monitoring effects. In addition to combining with inorganic nanomaterials, Exosomes can also be easily integrated with organic nanoparticles such as liposomes for membrane fusion. Liposomes are preparations formed by encapsulating drug molecules in vesicles formed by phospholipid bilayer membranes. Since the basic structure of the biological plasma membrane is also a phospholipid bilayer membrane, liposomes have a design similar to exosomes. They both have good biocompatibility that can be easy for the bioapplications. By incorporating targeted substances or chemotherapeutic drugs into the liposome membrane, this artificial organic nanoparticle can deliver the cargos in the cavity to the exosome.87 The modified exosome can also carry medicines and post-modified targeted substances.Mixed functional materials composed of inorganic/organic ingredients are considered a potential platform for expanding the diversified fields of medicine. Combining exosome and organic/organic nanoparticles can produce innovative materials by modifying the composition on the molecular scale. And balance the advantages between the two to adjust the size, design, function, and form. The following chapters are the critical part of this manuscript. We will describe and discuss the contributions of various exosome hybrid organic–inorganic composite materials in the past five years.
4. Hybrid inorganic/organic nanoparticles with exosomes
Due to their unique properties, hybrid nanomaterials have recently been studied as promising platforms for imaging and therapeutic applications (Table 1). Since the hybridization of inorganic/organic nanoparticles with exosomes is still a relatively novel technology, many novel types of research have been developed in 2021. This table has sorted out the relevant literature published in the past five years and selected several representative works for in-depth review. Nanotechnology offers great potential for future biomedical applications.88 Such nanoparticles can retain the valuable features of their inorganic and organic parts and systematically modify the properties of hybrid exosomes through the combination of functional elements. Furthermore, the conjugation of targeting moieties on the surface of these nanomaterials or exosomes provides a nanoplatform with specifically targeted imaging and therapeutic properties. This section summarizes the latest studies on synthesizing inorganic/organic nanoparticles with exosomes and their applications in the biomedical field. This paragraph integrates some relevant references that point out those specific nanoparticles can be used to optimize the functions of exosomes and improve their shortcomings to obtain a more complete clinically applicable nanotherapeutic theranostic platform.| Combination nanoparticles | Biological target (source of exosomes) | Theranostic methods | Cancer type | Ref. | |
|---|---|---|---|---|---|
| Liposome | Exosome (HEK293T) | RT-PCR | Mesenchymal stem cell | 2018, Lin et al.70 | |
| Runx2 expression (cell) | |||||
| Exosome (Fibroblasts) | NBD (Liposome) | Metastatic peritoneal cancer | 2020, Lv et al.71 | ||
| CD47-SIRPa | CD9 antibody (Exosome) | ||||
| DIR (gETL) | |||||
| Exosome (HT29) | DOX | Colorectal cancer | 2021, Kwon et al.72 | ||
| Exosome (CT26) | CD9 | Colorectal cancer | 2021, L. Cheng et al.73 | ||
| CD47-SIRPa | Photothermal therapy | ||||
| ICG/R837 | |||||
| Exosome (Fibroblast) | CD9, Calcein | Pulmonary fibrosis | 2021, L. Sun et al.74 | ||
| Clodronate | |||||
| CXCR4, miRNA, Antagomir-188 | CXCR4, CD9 | Osteolytic cancer | 2021, Y. Hu et al.75 | ||
| Exosome (NIH-3T3) | |||||
| Metal–organic framework (MOF) | Zn | Exosome (MDA-MB-231) | FluoBSA | Breast cancer | 2018, Cheng et al.76 |
| pH change (cell) | |||||
| Fe | Exosome (HeLa) | Calcein | Cervical cancer | 2017, Illes et al.77 | |
| Folate | |||||
| Inorganic nanoparticles | Porous silicon | Exosome (H22 or Bel7402) | DOX | Liver cancer | 2019, Yong et al.78 |
| Lung cancer | |||||
| CD54 | Breast cancer | ||||
| Melanoma | |||||
| Quantum dots (black phosphorus) | Exosome (LLC cells or Hepa1-6) tumor antigen | DIR | Lung cancer | 2020, Liu et al.79 | |
| Liver cancer | |||||
| MnCO | Exosome (4T1) | CD63 antibody | Breast cancer | 2021, Zhu et al.80 | |
| CD9 antibody | |||||
| Fe3O4 | Exosome (Rat) | RhB | Myocardial infarction | 2020, Liu et al.81 | |
| MLC and CD63 antibody | PKH26 | ||||
| Urinary exosome | UV-Vis | Prostate cancer | 2021, Pan et al.82 | ||
| DOX | |||||
| Gold (Au) | Exosome (HEK293T) | Dil | Brain cancer | 2019, Khongkow et al.83 | |
| RVG Lamp2b | |||||
| Exosome (MSC) | CD63 antibody | Mesenchymal stem cell | 2019, Sancho-Albero et al.84 | ||
| Exosome (B16-F10) | PKH67 | Melanoma | 2019, Sancho-Albero et al.85 | ||
| Temperature (cell) | |||||
| Exosome (FBS) | pH change (cell) | Colon cancer | 2021, Lee et al.86 | ||
| Breast cancer | |||||
| Cervical cancer | |||||
4.1. Exosome–liposome hybrid nanoparticles
Liposomes are formed by encapsulating drug molecules in vesicles formed by a phospholipid bilayer membrane.89 Since the basic structure of the biological plasma membrane is also a phospholipid bilayer membrane, and liposomes have a structure similar to biological cells, so they have good biocompatibility (Fig. 5a).90 After entering the human body, liposomes act as an “invader” to activate the immune mechanism. They are swallowed by the reticuloendothelial system, thereby causing targeted enrichment in tissues such as the liver, spleen, lung, and bone marrow. This phenomenon is known as the passive targeting of liposomes. Certain targeting substances can be incorporated into the liposome membrane; in this way, liposomes can be targeted to a specific site through biological or physical factors. The adding ligands provide the active targeting of liposomes, and it has emerged as an essential mechanism in drug delivery.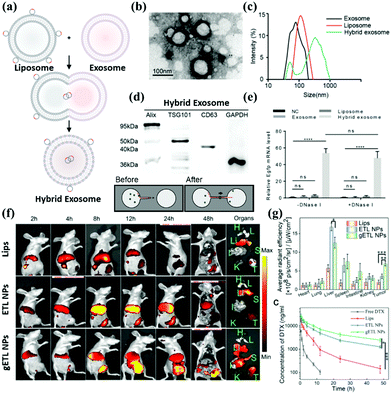 | ||
| Fig. 5 Assessment of hybrid exosomes encapsulating large plasmids. (a) Illustration of the procedure to produce hybrid exosomes. The isolated exosomes were incubated with liposomes for 12 h at 37 °C to induce fusion. (b) The combination of exosome and liposome under SEM. (c) DLS measures exosome and liposome and hybrid exosome, respectively. (d) Hybrid Exosome and Exosome have the same surface integral protein. (e) The cargo in the Liposome can be fused with the Exosome so that the Hybrid Exosome contains the target drug or gene. (f) In addition to carrying cargo, the surface modification can also deliver the target molecule or antibody into the hybrid exosome through lipid fusion. (g) Target modification enhances metabolism and enhances therapeutic efficacy. Reproduced with permission.70,71 Copyright 2018 and 2019, Wiley Online Library. | ||
Techniques include heat-sensitive liposomes, magnetically guided liposomes, and antibody-guided liposomes (Fig. 5b and c).91–93 However, at present, several limitations remain. First, its preparation technology introduces specific difficulties to industrial production; additionally, the encapsulation rate is low for some water-insoluble drugs, which easily leak from liposomes. Poor stability is also a problem in the commercialization of liposomes. The current freeze-drying method may be an effective way to extend the storage period of liposomes. Identification by the immune system is also a challenge encountered by liposomes. The exosome itself can evade immune identification, combined with the active targeting of liposomes to create a new drug hybrid carrier system. Exosomes are a promising drug delivery vehicle. However, it is challenging to encapsulate large nucleic acids in exosomes due to their small sizes. The current reports on exosomes as drug delivery vehicles are mainly related to small nucleic acids such as miRNA, siRNA, or low-molecular-weight drugs, which are much smaller than large plasmids, such as those expressing Cas9, with a minimum size of 5–6 kb. Therefore, there is an urgent need to find a suitable method to encapsulate the CRISPR/Cas9 system into exosomes. Lin et al. successfully encapsulated large nucleic acids, including CRISPR/Cas9 expression vectors, into hybrid exosomes produced by incubating primitive exosomes with liposomes (Fig. 5d and e).70 The loaded hybrid exosomes can be endocytosed by mesenchymal stem cells (MSCs) and release cargo.
The hybrid exosome delivery CRISPR/Cas9 system has been verified to be capable of expression in MSCs and regulate the expression of target genes. These target genes provide a new strategy for delivering the CRISPR-Cas9 system to MSCs for gene manipulation. Further research has shown that liposomes can offer additional advantages to exosomes. To improve the therapeutic effect of exosome–liposome hybrid nanoparticles, hyperthermic intraperitoneal chemotherapy (HIPEC) was recently developed. Lv et al. synthesized a genetically engineered exosome–thermosensitive liposome hybrid (gETL). They loaded it with docetaxel (DTX) to analyze its characteristics in cell line-derived (CDX) and patient-derived tumor xenografts (PDXs) for MPC treatment.87 They found that when gETL was loaded with DTX, the results revealed significantly inhibited tumor progression. The anti-tumor effect of HIPEC was enhanced when combined with the treatment (Fig. 5f and g).
In terms of the mechanism, treatment with therapeutic gETL could induce macrophage polarization, enhance macrophage-mediated tumor cell phagocytosis, and induce apoptosis. This study provides a new direction for treating metastatic peritoneal cancer through the combination of drug-loaded gETL and HIPEC.
4.2. Exosome-coated metal–organic framework nanoparticles
Metal–organic framework (MOF) materials are an essential classification of new metal–organic materials (MOMs). MOF is one of the most popular research fields in new inorganic–organic materials science because it combines two chemical disciplines, inorganic chemistry and organic chemistry, which are often separated.MOF consists of metal atoms or clusters of atoms coordinated by organic ligands to form a one-, two-, or three-dimensional structure.94 MOF can be used in gas adsorption, gas storage, gas separation, catalysts, and other fields. MOF nanoparticles can form a porous three-dimensional framework. Due to this construction principle, the composition and structure of these materials are highly adjustable. Additionally, the inner and outer surfaces can be independently functionalized. With such characteristics, MOF nanoparticles (NPs) can be designed to meet the specific requirements of the desired application. In the field of drug delivery, a so-called “composite material” with exosomes has been synthesized (Fig. 6a).95
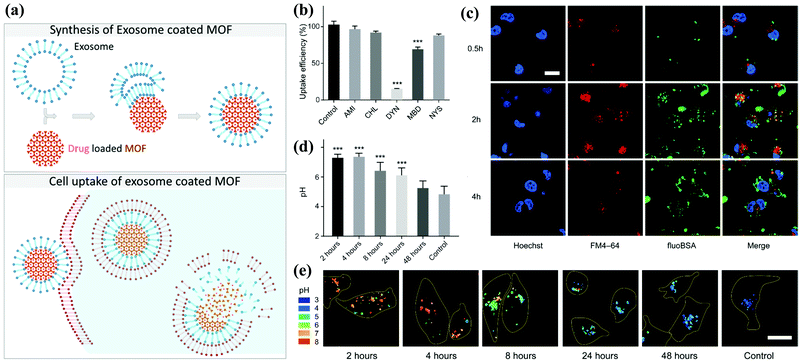 | ||
| Fig. 6 Schematic illustration of the synthesis of the exosome-coated MOF. (a) The subsequent cell uptake and proposed release mechanism of the cargo. (b) Analysis of the degree of phagocytosis and (c) the accumulation effect under confocal images. (d and e) The decomposition state of exosome-coated MOF under different pH environments. Reproduced with permission.76,77 Copyright 2017 and 2018, American Chemical Society. | ||
Compared with the artificial lipid layer as a closed system, exosomes provide significant advantages: for example, they are non-immunogenic and used by cells for communication purposes. Combined with the high porosity provided by MOF, the composite material can achieve a high drug loading. They are also designed to be biodegradable because they are coated with exosomes. Illes et al. were the first to report that MOF nanoparticles encapsulated by exosomes are an efficient drug delivery system with an “onboard trigger” function (Fig. 6b–e). This approach synergistically combines the characteristics of MOF NPs and exosomes to produce a promising system that promotes easy and efficient loading and sealing. Moreover, it has high treatment efficiency, and medicine does not leak prematurely.77 Subsequent articles related to exosome-coated MOF have been published in recent years.96–99
A biomimetic nanosystem was designed to assemble cargo proteins in the pH-responsive metal–organic framework (MOF) nanoparticles and use the extracellular vesicle membrane (EVM) to camouflage them.100–102 The protein is immobilized in the MOF (ZIF-8) matrix through the self-assembly of the metal node (Zn2+) and the blocking of the organic ligand. MOF has a large internal surface area and non-covalent affinity, significantly improving the protein loading efficiency and loading capacity. After the exosome-coated MOF is internalized by the recipient cell, the pH responsiveness of the metal–ligand bond causes the release of the protein in acidic endosomes and lysosomes. Additionally, the exosome may contain the integrin-related transmembrane protein CD47, which mediates a “do not eat me” signal to avoid phagocytosis, allowing the composite material to undergo specific endocytosis.103
4.3. Exosome-coated inorganic nanoparticles
The development of inorganic nanotechnology has diversified research in chemistry, physics, materials science, engineering, biology, and medicine. Taking biomedicine research as an example, nanomaterials exhibit unique physical and chemical properties at the nanoscale, such as magnetic, optical, thermal, and biomolecule-like properties, which are widely used in biomedical imaging research and development to assist nanomedicine.104 Nanomaterials have also been used for diagnosis, treatment, and prevention purposes. In recent years, combining two (or more) functional nanocomposites (bi-functional or multifunctional nanocomposites) has advanced the development of nanomedicine platforms, such as those used to study the growth and aging of the human body, and biochemical manipulation of mechanism and scale has become the driving force for accelerating multimodal imaging.105 Next, we enumerate the combinations of common inorganic nanomaterials and exosomes and their related applications in the biomedical field. We briefly describe their applications' current status and prospects in biomedical imaging.At present, many published reports describe using mesoporous silicon materials or modified mesoporous silicon materials as carriers, along with their applications in drug delivery systems. Yong et al. proposed using porous silicon nanoparticles (PSiNPs) as drug carriers to deliver into tumor cells and generate PSiNPs (E-PSiNPs) through exosomes as a drug carrier for targeted cancer chemotherapy. When tumor cells are incubated with doxorubicin-loaded PSiNPs (DOX@PSiNPs), they will pass by the exocytotic pathway to generate exosome-encapsulated DOX-loaded PSiNPs (DOX@E-PSiNPs). DOX@E-PSiNPs also possess the characteristics of exosomal targeting, drug loading, and biological diagnostics, which leads to applying biomimetic nanoparticles as drug carriers to effectively deliver anticancer drugs (Fig. 7a). The biodistribution evaluation also demonstrates E-PSiNPs can be used as a targeting flatform for drug accumulation (Fig. 7b).108,109 In addition to their drug delivery functions, mesoporous materials can be modified through functional groups on their surfaces to provide them with different properties, which can be applied to various types of biomedicine and biotechnology. For example, cell type identification, disease diagnosis, intracellular visualization, and drug and gene delivery have been researched, and results confirm that mesoporous materials can enter cells through endocytosis (Fig. 7c). The authors also revealed that E-PSiNPs could send the whole nanosystem to enter the core of cells (Fig. 7d). These grafted molecules can use various environmental factors, such as light-cutting effects, redox reactions, the presence of enzymes, or different pH values, to produce different chemical reactions, and then control the opening and closing of holes to achieve the effect of controlled drug release (Fig. 7e and f).
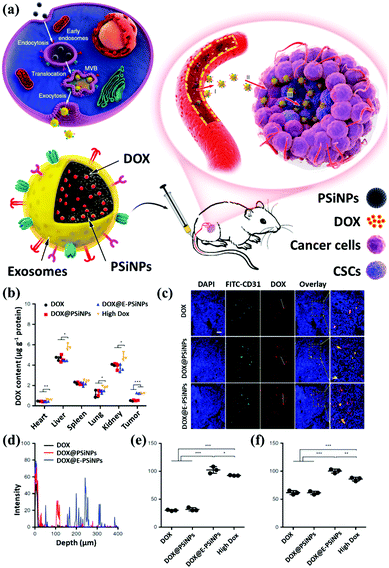 | ||
| Fig. 7 Schematic illustration of E-PSiNPs as drug carriers for targeted cancer chemotherapy. (a) Schematic illustration of the preparation of DOX@E-PSiNPs. DOX@PSiNPs are endocytosed into cancer cells after incubation and localized in multivesicular bodies (MVBs) and autophagosomes. After MVBs or amphisomes fuse with the cell membrane, DOX@E-PSiNPs are exocytosed into extracellular space. (b) Schematics showing how DOX@E-PSiNPs efficiently target tumor cells after intravenous injection into tumor-bearing mice. (c) The confocal images and (d) signal quantification of DOX@E-PSiNPs accumulation. The DOX accumulation under (e) neutral and (f) low pH values. Reproduced with permission.78 Copyright 2019, Springer Nature Limited. | ||
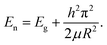 | (1-1) |
As illustrated in the above formula, the energy gap of nanomaterials is inversely proportional to their size. In modern biological analysis, various kinds of organic dyes are used. However, as technology advances, greater flexibility in these dyes is being sought.
To this end, quantum dots have quickly filled this role, as they are superior to traditional organic dyes on several counts, with brightness being among the most immediately obvious (owing to the high extinction coefficient combined with a comparable quantum yield to fluorescent dyes) as well as their stability (leading to much less photobleaching). It has been estimated that quantum dots are 20 times brighter and 100 times more stable than traditional fluorescent reporters. For single-particle tracking, the irregular blinking of quantum dots is a minor drawback. However, several groups have developed quantum dots that are essentially nonblinking. Their utility in single-molecule tracking experiments has been demonstrated.114,115 However, the toxicity and stability of quantum dots are still shortcomings that need to be overcome in current research. Liu et al. proposed a method of coating exosomes on the periphery of black phosphorus quantum dots (BPQDs). The author first took out the mouse blood to purify the exosome, and then added BPQDs to the exosome (Fig. 8a). Quantum dots with exosomes on the surface have better dispersion than simple quantum dots (Fig. 8b and f). Next, the thickness and element distribution of the material was analyzed through an atomic force microscope (Fig. 8c–e), confirming that the size of the material is close to the same, reflecting the potential strength of exosome to optimize nanomaterials. Then analyze the material size (Fig. 8g) and the total BPQDs carried by the exosome (Fig. 8h). Finally, to confirm that the outer layer is an exosome membrane, a western blot was used to analyze the markers of the composite material (Fig. 8i).
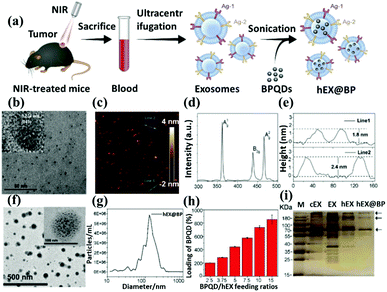 | ||
| Fig. 8 Schematic representation of black phosphorus quantum dots through exosome coating. (a) Graphic illustration describes the structure of exosome-encapsulated black phosphorus quantum dots (hEX@BP) and their roles in photothermal immunotherapy. Ag: antigen; NIR: near-infrared BPQDs: black phosphorus quantum dots. Transmission electron microscopy images of (b) BPQDs and (f) hEX@BP. The atomic force microscope indicated the (c) linear depth, (d) Raman shift, and (e) height of BPs. The (g) size distribution, (h) BPQDs loading ratio, and (i) western blot of hEX@BP. Reproduced with permission.79 Copyright 2020, Royal Society of Chemistry. | ||
In addition, the electron–ion pairs in these elements can correspond to the absorption of photons in the visible electromagnetic spectrum, resulting in colored compounds. Due to these behaviors, transition metals exhibit rich and fascinating chemical properties. In 2021, Pan et al. designed a nanocarrier based on the exosome-embedded and dox-loaded phosphomolybdic acid/Fe3O4 nanoparticles (Exo-PMA/Fe-HSA@DOX, Fig. 9a).80 They use the urinary exosomal membrane as a nano-platform for chemical/chemokinetic therapy and targeted homogenous prostate cancer diagnosis. The nanocarriers of urinary exosomes do not contain intracellular substances such as histone 3 and COX IV (Fig. 9b). Successfully achieved excellent synergistic low-dose chemical/chemical kinetic performance in vivo. In addition, the increased intracellular PMA/Fe-HSA@DOX nanoparticles (NPs) greatly enhance the bypass of the EGFR/AKT/NF-kB/IkB signaling pathway. The in vivo mice model is used to verify its therapeutic effect, which shows that the targeted homologous cancer therapy based on the urine exosomes of cancer patients embodies a new type of targeted anticancer program in an efficient and straightforward method (Fig. 9c). In the same year, Zhu et al. developed exosomal nanovesicles (MMV) loaded with manganese carbonyl (MnCO) to overcome this tumor treatment problem (Fig. 9d).82 MMV has a uniform size and good dispersion (Fig. 9e and f). After administration, MMV has shown admirable performance in actively targeting tumors, mitochondrial damage, and radiosensitization therapy and can promote 90% tumor growth inhibition under very low dose (only 2Gy) radiotherapy (Fig. 9g). Overall, this study emphasizes a novel and practical way to improve the efficacy of tumor radiotherapy.
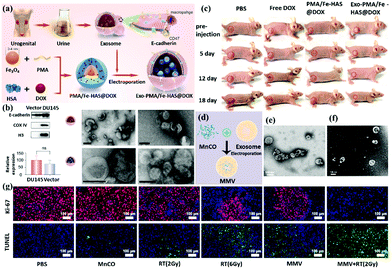 | ||
| Fig. 9 Schematic representation of transition elements nanoparticles (Fe3O4 and MnCO) functionalization through exosome coating. (a) Preparation method and synthesis schematic diagram of Exo-PMA/Fe3O4. (b) The performance of the vector in exosomes and the changes in pH environment regulation morphology of Exo-PMA/Fe3O4. (c) Analysis of mouse treatment data. (d) Schematic diagram of MnCO@Exosome (MMV) preparation. (e) TEM and (f) SEM topography analysis. (g) Tissue staining analysis and use radiotherapy together with MMV for treatment. Reproduced with permission.80,82 Copyright 2021, Elsevier Ltd. All. | ||
This unique physical phenomenon occurs only for metallic elements with an abundance of free conductive electrons that are easily affected by irradiating light, for example, Au, Ag, and Cu. The SPR effect for metallic colloid nanoparticles displays a unique physical impact and corresponds to the color of the solution. For example, 13 nm gold nanoparticles exhibit a red color in solution because their LSPR peak is 520 nm. The wavelength of SPR in the previously mentioned study can be derived from eqn (1-2), as shown below.118 The SPR frequency ωsp of metal nanoparticles is related to their geometry factor κ and the dielectric constant εm of the surrounding medium. Wavelength and frequency are reciprocally related.
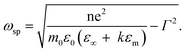 | (1-2) |
From this equation and the literature, the most convenient factor for changing the peak site of SPR is the geometry factor.119 For gold nanoparticles of various sizes, the SPR peak slightly shifts from about 520 to 540 nm as the particle size increases from 5 to 100 nm.
Therefore, the geometric structures of gold nanorods, gold nanoshells, and gold nanocages are commonly used to determine the difference in SPR peaks. Based on the SPR effect of the gold nanomaterial, Sancho-Albero et al. proposed a technique of exosome embedding gold nanoparticles. In Fig. 10a, the exosome is first released from the cells. The medium is extracted to purify the exosome, which is mixed with gold nanoparticles and fabricated to produce a gold nanoparticle surface-functionalized exosome. Moreover, the authors tested different cell lines and confirmed that exosomes could lead gold nanoparticles to target and accumulate in the cancer cells after pumping the exosome/AuNPs (Fig. 10b). To quantify the accumulation effect of exosome/AuNPs, they chose different cell lines to show that exosomes can actively carry AuNPs into the cancer cells (Fig. 10c). The gold nanoparticles are sent into the cells through transwell co-culture at different time points for Confocal (Fig. 10d) and Time course (Fig. 10e) analysis.85
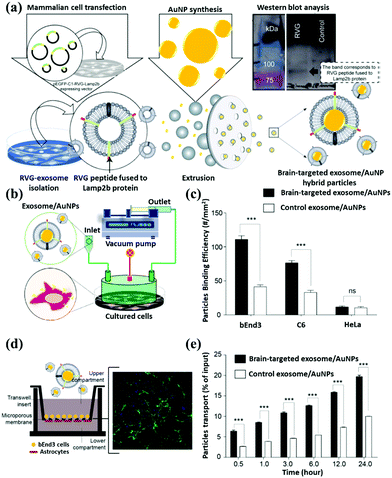 | ||
| Fig. 10 Schematic representation of gold nanoparticle surface functionalization through exosome coating. (a) The brain-targeting property of AuNPs after fabrication with neuron-targeted exosomes. Signals of fluorescently labeled nanoparticles in the basal chamber were measured in 0.5 mL aliquots at different time points. (b) A microinjection device was used to deliver exosome/AuNPs systematically. (c) Quantification of exosome/AuNPs and determining the degree of phagocytosis of the material by various cell lines. Confocal microscope (d) image analysis of cell uptake and (e) quantifying the endocytosis effect of cells on exosome/AuNPs under transwell incubation. Reproduced with permission.85 Copyright 2018, Royal Society of Chemistry. | ||
5. Conclusions and future perspectives
Recent studies have found that exosomes or EVs are associated with cancer metastasis, disease progression, diagnosis, and treatment, leading to a rapidly emerging cell research area. Cells secrete exosomes via exocytosis. This has also been observed for other rigid nanoparticles, such as silica and carbon. Relying on the superior targeting function and biocompatibility of cancer exosomes, researchers have been able to deliver drugs to cancer stem cells deep within tumors in mouse models. Despite significant efforts made in this relatively new field of exosome research, progress has been held back by challenges such as inefficient separation methods, difficulties in characterization, and a lack of specific biomarkers.Moreover, the applications of exosomes need to be further evaluated under actual physiological conditions. Inorganic/organic nanoparticles, on the other hand, produce powerful in vivo near-infrared fluorescence, thus generating remarkable tumor imaging tracking ability in subcutaneous, orthotopic, and metastatic tumors. This review describes an approach to cancer therapy that uses exosome-biomimetic nanoparticles. Hybrid inorganic/organic nanoparticles with exosomes can be applied as drug carriers to deliver anticancer drugs to the centers of cancer cells efficiently. Biocompatible hybrid inorganic/organic nanoparticles with exosomes have been synthesized for targeted cancer chemotherapy. Following intravenous injection, nanocomposites actively accumulate in the tumor site. They are subject to cross-reactive cellular uptake by cancer cells, resulting in augmented in vivo drug delivery enrichment in tumor cells. At present, diversified hybrid inorganic/organic nanoparticles with exosomes for both therapy and diagnosis are still in urgent need of development.
This combination can achieve theranostics on the same platform and provide advantages such as targeting, improved dispersion, and avoiding immune system clearance. However, as in most clinical applications, the biological application of nanomaterials still needs more research to optimize and improve their performance in vivo. The complexity of hybrid inorganic/organic nanoparticles with exosomes will require more attention to their safety and biocompatibility, even though they may provide excellent therapeutic and diagnostic functions. In addition, its biodegradability also needs to be evaluated to ensure that the inorganic/organic materials will not accumulate in the body. Therefore, it is believed that hybrid inorganic/organic nanoparticles with exosomes will become a hot topic for future research after being baptized by scientists' brainstorming.
Author contributions
Conceptualization, M. H., R. S. L., L. J. L., and C. Y. H.; writing – original draft preparation, M.-H. C. and Z.-X. C.; writing – review and editing, M. H. C. and Z. X. C.; supervision, R. S. L. and M. H. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by Genomics Research Center, Academia Sinica, Taiwan to M. H. and by Ministry of Science and Technology (MOST 109-2113-M-002-020-MY3) to R. S. L. In addition, Ming-Hsien Chan is supported by the Academia Sinica Outstanding Postdoctoral Fellowship.References
- A. V. Vlassov, S. Magdaleno, R. Setterquist and R. Conrad, Biochim. Biophys. Acta, Gen. Subj., 2012, 1820, 940–948 CrossRef CAS PubMed.
- S. A. A. Kooijmans, P. Vader, S. M. van Dommelen, W. W. van Solinge and R. M. Schiffelers, Int. J. Nanomed., 2012, 7, 1525–1541 CAS.
- D. Ha, N. N. Yang and V. Nadithe, Acta Pharm. Sin. B, 2016, 6, 287–296 CrossRef PubMed.
- H. Valadi, K. Ekstrom, A. Bossios, M. Sjostrand, J. J. Lee and J. O. Lotvall, Nat. Cell Biol., 2007, 9, 654–672 CrossRef CAS PubMed.
- H. Q. Xin, Y. Li and M. Chopp, Front. Cell. Neurosci., 2014, 8, 377 Search PubMed.
- P. A. Steck, M. A. Pershouse, S. A. Jasser, W. K. A. Yung, H. Lin, A. H. Ligon, L. A. Langford, M. L. Baumgard, T. Hattier, T. Davis, C. Frye, R. Hu, B. Swedlund, D. H. F. Teng and S. V. Tavtigian, Nat. Genet., 1997, 15, 356–362 CrossRef CAS PubMed.
- D. Szkutnik-Fiedler, Pharmaceutics, 2020, 12, 1180 CrossRef CAS PubMed.
- C. C. Chen, L. N. Liu, F. X. Ma, C. W. Wong, X. N. E. Guo, J. V. Chacko, H. P. Farhoodi, S. X. Zhang, J. Zimak, A. Segaliny, M. Riazifar, V. Pham, M. A. Digman, E. J. Pone and W. A. Zhao, Cell. Mol. Bioeng., 2016, 9, 509–529 CrossRef CAS PubMed.
- S. Kamerkar, V. S. LeBleu, H. Sugimoto, S. J. Yang, C. F. Ruivo, S. A. Melo, J. J. Lee and R. Kalluri, Nature, 2017, 546, 498–499 CrossRef CAS PubMed.
- S. G. Antimisiaris, S. Mourtas and A. Marazioti, Pharmaceutics, 2018, 10, 218 CrossRef CAS PubMed.
- L. Song, P. P. Li, W. Yang, X. H. Lin, H. Liang, X. F. Chen, G. Liu, J. Li and H. H. Yang, Adv. Funct. Mater., 2018, 28, 1707496 CrossRef.
- J. Y. Ha, S. Y. Choi, J. H. Lee, S. H. Hong and H. J. Lee, Front Mol. Biosci., 2020, 7, 596366 CrossRef CAS PubMed.
- R. C. Lai, R. W. Y. Yeo, K. H. Tan and S. K. Lim, Biotechnol. Adv., 2013, 31, 543–551 CrossRef CAS PubMed.
- C. Schindler, A. Collinson, C. Matthews, A. Pointon, L. Jenkinson, R. R. Minter, T. J. Vaughan and N. J. Tigue, PLoS One, 2019, 14, e0214545 CrossRef CAS PubMed.
- M. P. Stewart, A. Sharei, X. Y. Ding, G. Sahay, R. Langer and K. F. Jensen, Nature, 2016, 538, 183–192 CrossRef CAS PubMed.
- A. Calistri, P. Sette, C. Salata, E. Cancellotti, C. Forghieri, A. Cornin, H. Gottlinger, G. Campadelli-Fiume, G. Palu and C. Parolin, J. Virol., 2007, 81, 11468–11478 CrossRef CAS PubMed.
- R. M. Johnstone, M. Adam, J. R. Hammond, L. Orr and C. Turbide, J. Biol. Chem., 1987, 262, 9412–9420 CrossRef CAS.
- J. Kowal, M. Tkach and C. Thery, Curr. Opin. Cell Biol., 2014, 29, 116–125 CrossRef CAS PubMed.
- N. P. Hessvik and A. Llorente, Cell. Mol. Life Sci., 2018, 75, 193–208 CrossRef CAS PubMed.
- Y. Q. Koh, F. B. Almughlliq, K. Vaswani, H. N. Peiris and M. D. Mitchell, Front. Biosci.-Landmark., 2018, 23, 865–874 CrossRef CAS PubMed.
- F. Yang, X. Z. Liao, Y. Tian and G. Y. Li, Biotechnol. J., 2017, 12, 1600699 CrossRef PubMed.
- J. Lee, H. Kim, Y. Heo, Y. K. Yoo, S. I. Han, C. Kim, D. Hur, H. Kim, J. Y. Kang and J. H. Lee, Analyst, 2020, 145, 157–164 RSC.
- S. S. Huang, L. Wang, T. F. Bruce and R. K. Marcus, Biotechnol. Prog., 2020, 36, e2998 CrossRef CAS PubMed.
- P. Saravanan, J. Gu, X. Wang, P. Nguyen, G. Yoshimatsu, M. Lawrence and B. Naziruddin, Am. J. Transplant., 2017, 17, 723 Search PubMed.
- Y. A. Bai, Y. X. Lu, K. Wang, Z. L. Cheng, Y. L. Qu, S. H. Qiu, L. Zhou, Z. H. Wu, H. Y. Liu, J. L. Zhao and H. L. Mao, Nano-Micro Lett., 2019, 11, 59 CrossRef CAS PubMed.
- Y. Z. Zhu, S. Lee, X. R. Zheng, G. J. Zheng, H. R. Tseng and S. Hou, Mod. Pathol., 2017, 30, 538a Search PubMed.
- C. Y. Soo, Y. Q. Song, Y. Zheng, E. C. Campbell, A. C. Riches, F. Gunn-Moore and S. J. Powis, Immunology, 2012, 136, 192–197 CrossRef CAS PubMed.
- C. Gardiner, R. Dragovic, D. Tannetta, P. Harrison, C. Redman and I. Sargent, Int. J. Lab. Hematol., 2012, 34, 87 Search PubMed.
- E. van der Pol, F. A. W. Coumans, A. E. Grootemaat, C. Gardiner, I. L. Sargent, P. Harrison, A. Sturk, T. G. van Leeuwen and R. Nieuwland, J. Thromb. Haemostasis, 2014, 12, 1182–1192 CrossRef CAS PubMed.
- K. Kluszczynska, L. Czernek, W. Cypryk, L. Peczek and M. Duchler, Curr. Pharm. Des., 2019, 25, 4464–4485 CrossRef CAS PubMed.
- M. Catalano and L. O'Driscoll, J. Extracell. Vesicles, 2020, 9, 1703244 CrossRef CAS PubMed.
- Y. Zheng, A. Hasan, M. M. Nejadi Babadaei, E. Behzadi, M. Nouri, M. Sharifi and M. Falahati, Biomed. Pharmacother., 2020, 129, 110442 CrossRef CAS PubMed.
- X. Y. Zhao, D. L. Wu, X. D. Ma, J. L. Wang, W. J. Hou and W. Zhang, Biomed. Pharmacother., 2020, 128, 110237 CrossRef CAS PubMed.
- Z. Wan, Y. Dong, M. Y. Wei, X. T. Gao, G. D. Yang, J. Zhang and L. Liu, Adv. Biosyst., 2020, 4, e2000061 CrossRef PubMed.
- J. E. Pullan, M. I. Confeld, J. K. Osborn, J. Kim, K. Sarkar and S. Mallik, Mol. Pharmaceutics, 2019, 16, 1789–1798 CrossRef CAS PubMed.
- G. Morales, D. Cardenas, C. Arroyo, L. Ayala, R. Seqqat and M. Torres, Trop. Med. Int. Health, 2017, 22, 129–130 Search PubMed.
- Y. H. Chang, K. C. Wu, H. J. Harn, S. Z. Lin and D. C. Ding, Cell Transplant., 2018, 27, 349–363 Search PubMed.
- R. Pluta and M. Ułamek-Kozioł, Adv. Exp. Med. Biol., 2019, 1118, 71–82 CrossRef CAS PubMed.
- L. Q. Jiang, H. J. Dong, H. Cao, X. F. Ji, S. Y. Luan and L. Liu, Med. Sci. Monit., 2019, 25, 3329–3335 CrossRef CAS PubMed.
- X. B. Qin, S. R. Yu, L. L. Zhou, M. Q. Shi, Y. Hu, X. Y. Xu, B. Shen, S. W. Liu, D. L. Yan and J. F. Feng, Int. J. Nanomed., 2017, 12, 3721–3733 CrossRef CAS PubMed.
- D. L. Yuwen, B. B. Sheng, J. Liu, W. Wenyu and Y. Q. Shu, Eur. Rev. Med. Pharmaco., 2017, 21, 2650–2658 Search PubMed.
- W. J. Huang, Y. Y. Yan, Y. Liu, M. T. Lin, J. X. Ma, W. Zhang, J. W. Dai, J. J. Li, Q. R. Guo, H. B. Chen, B. Makabel, H. Liu, C. Y. Su, H. Bi and J. Y. Zhang, Signal Transduction Targeted Ther., 2020, 5, 39 CrossRef CAS PubMed.
- Y. T. Wang, T. J. Shi, S. Srivastava, J. Kagan, T. Liu and K. D. Rodland, Cancers, 2020, 12, 2335 CrossRef CAS PubMed.
- N. Wang, X. G. Song, L. S. Liu, L. M. Niu, X. W. Wang, X. R. Song and L. Xie, Cancer Sci., 2018, 109, 1701–1709 CrossRef CAS PubMed.
- D. J. Clark, W. E. Fondrie, A. Yang and L. Mao, J. Proteomics, 2016, 133, 161–169 CrossRef CAS PubMed.
- K. O'Brien, K. Breyne, S. Ughetto, L. C. Laurent and X. O. Breakefield, Nat. Rev. Mol. Cell Biol., 2020, 21, 585–606 CrossRef PubMed.
- J. Guan, Q. Shen, Z. Zhang, Z. X. Jiang, Y. Yang, M. Q. Lou, J. Qian, W. Y. Lu and C. Y. Zhan, Nat. Commun., 2018, 9, 2982 CrossRef PubMed.
- T. Yamashita, Y. Takahashi, M. Nishikawa and Y. Takakura, Eur. J. Pharm. Biopharm., 2016, 98, 1–8 CrossRef CAS PubMed.
- A. Thakur, R. K. Sidu, H. Zou, M. K. Alam, M. S. Yang and Y. J. Lee, Int. J. Nanomed., 2020, 15, 8331–8343 CrossRef PubMed.
- X. M. Guo, J. Ma, J. Sun and G. X. Gao, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 151–156 CrossRef CAS PubMed.
- D. Gallego-Perez, D. Pal, S. Ghatak, V. Malkoc, N. Higuita-Castro, S. Gnyawali, L. Q. Chang, W. C. Liao, J. F. Shi, M. Sinha, K. Singh, E. Steen, A. Sunyecz, R. Stewart, J. Moore, T. Ziebro, R. G. Northcutt, M. Homsy, P. Bertani, W. Lu, S. Roy, S. Khanna, C. Rink, V. B. Sundaresan, J. J. Otero, L. J. Lee and C. K. Sen, Nat. Nanotechnol., 2017, 12, 974–979 CrossRef CAS PubMed.
- L. A. Ambattu, S. Ramesan, C. Dekiwadia, E. Hanssen, H. Y. Li and L. S. Y. Yeo, Commun. Biol., 2020, 3, 553 CrossRef CAS PubMed.
- S. W. Guo, N. Perets, O. Betzer, S. Ben-Shaul, A. Sheinin, I. Michaelevski, R. Popovtzer, D. Offen and S. Levenberg, ACS Nano, 2019, 13, 10015–10028 CrossRef CAS PubMed.
- M. Kim, G. Kim, D. W. Hwang and M. Lee, J. Biomed. Nanotechnol., 2019, 15, 2401–2412 CrossRef CAS PubMed.
- H. Y. Tao, H. L. Xu, L. Zuo, C. Li, G. Qiao, M. Y. Guo, L. H. Zheng, M. Leitgeb and X. K. Lin, Int. J. Biol. Macromol., 2020, 161, 470–480 CrossRef CAS PubMed.
- Y. H. Cao, E. B. Ma, S. Cestellos-Blanco, B. Zhang, R. Y. Qiu, Y. D. Su, J. A. Doudna and P. D. Yang, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 7899–7904 CrossRef CAS PubMed.
- Z. J. Xu, S. S. Zeng, Z. C. Gong and Y. L. Yan, Mol. Cancer, 2020, 19, 160 CrossRef CAS PubMed.
- A. Gallo, S. Vella, M. Miele, F. Timoneri, M. Di Bella, S. Bosi, M. Sciveres and P. G. Conaldi, Cancer Lett., 2017, 388, 334–343 CrossRef CAS PubMed.
- M. Iero, R. Valenti, V. Huber, P. Filipazzi, G. Parmiani, S. Fais and L. Rivoltini, Cell Death Differ., 2008, 15, 80–88 CrossRef CAS PubMed.
- H. Y. Xu, N. Li, N. Yao, X. F. Xu, H. X. Wang, X. Y. Liu and Y. Zhang, Oncol. Rep., 2019, 42, 866–879 CAS.
- L. H. Lv, Y. L. Wan, Y. Lin, W. Zhang, M. Yang, G. L. Li, H. M. Lin, C. Z. Shang, Y. J. Chen and J. Min, J. Biol. Chem., 2012, 287, 15874–15885 CrossRef CAS PubMed.
- H. Samaha, A. Pignata, K. Fousek, J. Ren, F. W. Lam, F. Stossi, J. Dubrulle, V. S. Salsman, S. Krishnan, S. H. Hong, M. L. Baker, A. Shree, A. Z. Gad, T. Shum, D. Fukumura, T. T. Byrd, M. Mukherjee, S. P. Marrelli, J. S. Orange, S. K. Joseph, P. H. Sorensen, M. D. Taylor, M. Hegde, M. Mamonkin, R. K. Jain, S. El-Naggar and N. Ahmed, Nature, 2018, 561, 331–332 CrossRef CAS PubMed.
- X. J. Gao, N. Ran, X. Dong, B. F. Zuo, R. Yang, Q. B. Zhou, H. M. Moulton, Y. Q. Seow and H. F. Yin, Sci. Transl. Med., 2018, 10, eaat0195 CrossRef PubMed.
- B. H. Liang, X. M. Hu, Y. H. Ding and M. J. Liu, J. Cell. Physiol., 2021, 236, 4138–4151 CrossRef CAS PubMed.
- M. Naseri, M. Bozorgmehr, M. Zoller, E. R. Pirmardan and Z. Madjd, OncoImmunology, 2020, 9, 1779991 CrossRef PubMed.
- N. Abu, N. Othman, N. S. Ab Razak, S. N. Nasir and R. Jamal, Gut, 2019, 68, A14–A15 Search PubMed.
- V. S. LeBleu and R. Kalluri, Immunity, 2019, 50, 547–549 CrossRef CAS PubMed.
- X. Y. Qiu, J. Liu, C. X. Zheng, Y. T. Su, L. L. Bao, B. Zhu, S. Y. Liu, L. L. Wang, X. Wang, Y. R. Wang, W. M. Zhao, J. Zhou, Z. H. Deng, S. Y. Liu and Y. Jin, Cell Proliferation, 2020, 53, e12830 CrossRef CAS PubMed.
- S. Dusoswa, S. Horrevorts, S. Schetters, T. Wurdinger, Y. Van Kooyk and J. Garcia-Vallejo, Neuro-Oncology, 2018, 20, 89 CrossRef.
- Y. Lin, J. H. Wu, W. H. Gu, Y. L. Huang, Z. C. Tong, L. J. Huang and J. L. Tan, Adv. Sci., 2018, 5, 1700611 CrossRef PubMed.
- Q. J. Lv, L. L. Cheng, Y. Lu, X. G. Zhang, Y. Z. Wang, J. F. Deng, J. B. Zhou, B. Liu and J. Liu, Adv. Sci., 2020, 7, 2000515 CrossRef CAS PubMed.
- S.-H. Kwon, H. Al Faruque, H. Kee, E. Kim and S. Park, Colloids Surf., B, 2021, 205, 111915 CrossRef CAS PubMed.
- L. Cheng, X. Zhang, J. Tang, Q. Lv and J. Liu, Biomaterials, 2021, 275, 120964 CrossRef CAS PubMed.
- L. Sun, M. Fan, D. Huang, B. Li, R. Xu, F. Gao and Y. Chen, Biomaterials, 2021, 271, 120761 CrossRef CAS PubMed.
- Y. Hu, X. Li, Q. Zhang, Z. Gu, Y. Luo, J. Guo, X. Wang, Y. Jing, X. Chen and J. Su, Bioact. Mater., 2021, 6, 2905–2913 CrossRef CAS PubMed.
- G. Cheng, W. Q. Li, L. Ha, X. H. Han, S. J. Hao, Y. Wan, Z. G. Wang, F. P. Dong, X. Zou, Y. W. Mao and S. Y. Zheng, J. Am. Chem. Soc., 2018, 140, 7282–7291 CrossRef CAS PubMed.
- B. Illes, P. Hirschle, S. Baenert, V. Cauda, S. Wuttke and H. Engelke, Chem. Mater., 2017, 29, 8042–8046 CrossRef CAS.
- T. Y. Yong, X. Q. Zhang, N. N. Bie, H. B. Zhang, X. T. Zhang, F. Y. Li, A. Hakeem, J. Hu, L. Gan, H. A. Santos and X. L. Yang, Nat. Commun., 2019, 10, 3838 CrossRef PubMed.
- Q. Liu, T. J. Fan, Y. Y. Zheng, S. L. Yang, Z. Q. Yu, Y. H. Duo, Y. H. Zhang, D. Adah, L. L. Shi, Z. Sun, D. Wang, J. L. Xie, H. Wu, Z. Z. Wu, C. C. Ge, L. J. Qiao, C. Y. Wei, L. D. Huang, Q. T. Yan, Q. H. Yang, S. Y. Bao, L. P. Liu and H. Zhang, Nanoscale, 2020, 12, 19939–19952 RSC.
- D. Zhu, Z. Liu, Y. Li, Q. Huang, L. Xia and K. Li, Biomaterials, 2021, 274, 120894 CrossRef CAS PubMed.
- S. Y. Liu, X. Chen, L. L. Bao, T. Liu, P. Y. Yuan, X. S. Yang, X. Y. Qiu, J. J. Gooding, Y. K. Bai, J. J. Xiao, F. X. Pu and Y. Jin, Nat. Biomed. Eng., 2020, 4, 1063–1064 CrossRef CAS PubMed.
- S. Pan, Y. Zhang, M. Huang, Z. Deng, A. Zhang, L. Pei, L. Wang, W. Zhao, L. Ma, Q. Zhang and D. Cui, Biomaterials, 2021, 275, 120946 CrossRef CAS PubMed.
- M. Khongkow, T. Yata, S. Boonrungsiman, U. R. Ruktanonchai, D. Graham and K. Namdeel, Sci. Rep., 2019, 9, 8278 CrossRef PubMed.
- M. Sancho-Albero, N. Navascues, G. Mendoza, V. Sebastian, M. Arruebo, P. Martin-Duque and J. Santamaria, J. Nanobiotechnol., 2019, 17, 16 CrossRef PubMed.
- M. Sancho-Albero, M. D. Encabo-Berzosa, M. Beltran-Visiedo, L. Fernandez-Messina, V. Sebastian, F. Sanchez-Madrid, M. Arruebo, J. Santamaria and P. Martin-Duque, Nanoscale, 2019, 11, 18825–18836 RSC.
- E. S. Lee, B. S. Cha, S. Kim and K. S. Park, Int. J. Mol. Sci., 2021, 22, 4433 CrossRef CAS PubMed.
- G. H. Nam, Y. Choi, G. B. Kim, S. Kim, S. A. Kim and I. S. Kim, Adv. Mater., 2020, 32, e2002440 CrossRef PubMed.
- M. S. Yoon, Y. J. Lee, H. J. Shin, C. W. Park, S. B. Han, J. K. Jung, J. S. Kim and D. H. Shin, Pharmaceutics, 2020, 12, 1156 CrossRef CAS PubMed.
- L. Sercombe, T. Veerati, F. Moheimani, S. Y. Wu, A. K. Sood and S. Hua, Front. Pharmacol., 2015, 6, 286 Search PubMed.
- K. B. Knudsen, H. Northeved, P. E. K. Kumar, A. Permin, T. Gjetting, T. L. Andresen, S. Larsen, K. M. Wegener, J. Lykkesfeldt, K. Jantzen, S. Loft, P. Moller and M. Roursgaard, Nanomed. Nanotechnol., 2015, 11, 467–477 CrossRef CAS PubMed.
- H. Yang, Q. V. Le, G. Shim, Y. K. Oh and Y. K. Shin, Acta Pharm. Sin. B, 2020, 10, 2212–2226 CrossRef CAS PubMed.
- C. G. Yan, Y. K. Wu, J. Feng, W. F. Chen, X. Liu, P. Hao, R. M. Yang, J. Zhang, B. Q. Lin, Y. K. Xu and R. Y. Liu, Int. J. Nanomed., 2013, 8, 245–255 Search PubMed.
- D. S. Dimitrov, Y. Feng and P. Prabakaran, J. Comput. Theor. Nanosci., 2008, 5, 751–759 CrossRef CAS.
- L. Cheng, Q. S. Zhang, M. Xu, Q. C. Zhai and C. L. Zhang, J. Colloid Interface Sci., 2021, 583, 299–309 CrossRef CAS PubMed.
- W. Q. Wang, L. Wang, S. Liu and Z. G. Xie, Bioconjugate Chem., 2017, 28, 2784–2793 CrossRef CAS PubMed.
- Y. Cao, Y. Wang, X. M. Yu, X. H. Jiang, G. Li and J. Zhao, Biosens. Bioelectron., 2020, 166, 112452 CrossRef CAS PubMed.
- Y. T. Fu and Q. Ma, Nanoscale, 2020, 12, 13879–13898 RSC.
- Y. Wang, Curr. Gene Ther., 2020, 20, 356–372 CrossRef CAS PubMed.
- E. J. Bunggulawa, W. Wang, T. Y. Yin, N. Wang, C. Durkan, Y. Z. Wang and G. X. Wang, J. Nanobiotechnol., 2018, 16, 81 CrossRef CAS PubMed.
- E. van der Pol, A. N. Boing, P. Harrison, A. Sturk and R. Nieuwland, Pharmacol. Rev., 2012, 64, 676–705 CrossRef CAS PubMed.
- S. El Andaloussi, I. Maeger, X. O. Breakefield and M. J. A. Wood, Nat. Rev. Drug Discovery, 2013, 12, 348–358 CrossRef PubMed.
- M. Yang, X. Y. Liu, Q. Q. Luo, L. L. Xu and F. X. Chen, J. Nanobiotechnol., 2020, 18, 100 CrossRef CAS PubMed.
- E. Koh, E. J. Lee, G. H. Nam, Y. Hong, E. Cho, Y. Yang and I. S. Kim, Biomaterials, 2017, 121, 121–129 CrossRef CAS PubMed.
- E. Giacomini, S. Makieva, V. Murdica, R. Vago and P. Vigano, Curr. Opin. Obstet. Gyn., 2020, 32, 179–184 CrossRef PubMed.
- K. O. Jung, Y. H. Kim, S. J. Chung, C. H. Lee, S. Rhee, G. Pratx, J. K. Chung and H. Youn, Int. J. Mol. Sci., 2020, 21, 7850 CrossRef CAS PubMed.
- H. M. Saleh, F. A. El-Saied, T. A. Salaheldin and A. A. Hezo, Constr. Build. Mater., 2019, 218, 556–567 CrossRef CAS.
- P. Singh, S. Srivastava and S. K. Singh, ACS Biomater. Sci. Eng., 2019, 5, 4882–4898 CrossRef CAS PubMed.
- A. Tamayo, M. A. Mazo, R. Ruiz-Caro, A. Martin-Illana, L. M. Bedoya, M. D. Veiga-Ochoa and J. Rubio, Chem. Eng. J., 2015, 280, 165–174 CrossRef CAS.
- K. K. Qian and R. H. Bogner, J. Pharm. Sci., 2012, 101, 444–463 CrossRef CAS PubMed.
- L. Walekar, T. Dutta, P. Kumar, Y. S. Ok, S. Pawar, A. Deep and K. H. Kim, TrAC, Trends Anal. Chem., 2017, 97, 458–467 CrossRef CAS.
- P. Reineck, A. Francis, A. Orth, D. W. M. Lau, R. D. V. Nixon-Luke, I. Das Rastogi, W. A. W. Razali, N. M. Cordina, L. M. Parker, V. K. A. Sreenivasan, L. J. Brown and B. C. Gibson, Adv. Opt. Mater., 2016, 4, 1549–1557 CrossRef CAS.
- M. L. Lu, C. W. Lai, H. J. Pan, C. T. Chen, P. T. Chou and Y. F. Chen, Nano Lett., 2013, 13, 1920–1927 CrossRef CAS PubMed.
- M. Lannoo, C. Delerue, G. Allan and Y. M. Niquet, Philos. Trans. R. Soc., A, 2003, 361, 259–272 CrossRef CAS PubMed.
- F. Y. Song, P. S. Tang, H. Durst, D. T. Cramb and W. C. W. Chan, Angew. Chem., Int. Ed., 2012, 51, 8773–8777 CrossRef CAS PubMed.
- X. S. Tang, J. Yang, S. Q. Li, Z. Z. Liu, Z. P. Hu, J. Y. Hao, J. Du, Y. X. Leng, H. Y. Qin, X. Lin, Y. Lin, Y. X. Tian, M. Zhou and Q. H. Xiong, Adv. Sci., 2019, 6, 1900412 CrossRef CAS PubMed.
- G. Palui, S. Ray and A. Banerjee, J. Mater. Chem., 2009, 19, 3457–3468 RSC.
- R. E. Messersmith, G. J. Nusz and S. M. Reed, J. Phys. Chem. C, 2013, 117, 26725–26733 CrossRef CAS PubMed.
- W. B. Ewe, H. S. Chu and E. P. Li, Opt. Express, 2007, 15, 18200–18208 CrossRef PubMed.
- R. Rodriguez-Oliveros and J. A. Sanchez-Gil, Opt. Express, 2011, 19, 12208–12219 CrossRef CAS PubMed.
Footnote |
| † Ming-Hsien Chan and Zhi-Xuan Chang contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |