NIR-activated quercetin-based nanogels embedded with CuS nanoclusters for the treatment of drug-resistant biofilms and accelerated chronic wound healing†
Amit
Nain
a,
Yu-Ting
Tseng
b,
Akash
Gupta
c,
Yu-Feng
Lin
b,
Sangili
Arumugam
 d,
Yu-Fen
Huang
d,
Yu-Fen
Huang
 e,
Chih-Ching
Huang
e,
Chih-Ching
Huang
 *fg and
Huan-Tsung
Chang
*fg and
Huan-Tsung
Chang
 *dhij
*dhij
aDepartment of Material Engineering, Indian Institute of Science, Bangalore, 560012, Karnataka, India
bDepartment of Chemistry, National Taiwan University, Taipei, 10617, Taiwan
cDavid H Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
dDepartment of Biomedical Sciences, Chang Gung University, Taoyuan, 33302, Taiwan. E-mail: changht@mail.cgu.edu.tw
eInstitute of Analytical and Environmental Sciences, National Tsing Hua University, Hsinchu, 30013, Taiwan
fDepartment of Bioscience and Biotechnology and Center of Excellence for the Oceans, National Taiwan Ocean University, Keelung, 202301, Taiwan. E-mail: huanging@ntou.edu.tw
gSchool of Pharmacy, College of Pharmacy, Kaohsiung Medical University, Kaohsiung, 80708, Taiwan
hGraduate Institute of Biomedical Sciences, Chang Gung University, Taoyuan, 33302, Taiwan
iCenter for Advanced Biomaterials and Technology Innovation, Chang Gung University, Taoyuan, 33302, Taiwan
jDivision of Breast Surgery, Department of General Surgery, Chang Gung Memorial Hospital, Linkou, Taoyuan 33305, Taiwan
First published on 19th September 2023
Abstract
We have developed multifunctional nanogels with antimicrobial, antioxidant, and anti-inflammatory properties, facilitating rapid wound healing. To prepare the multifunctional nanogels, we utilized quercetin (Qu) and a mild carbonization process to form carbonized nanogels (CNGs). These CNGs possess excellent antioxidative and bacterial targeting properties. Subsequently, we utilized the Qu–CNGs as templates to prepare nanogels incorporating copper sulfide (CuS) nanoclusters, further enhancing their functionality. Notably, the CuS/Qu–CNGs nanocomposites demonstrated an exceptional minimum inhibitory concentration against tested bacteria, approximately 125-fold lower than monomeric Qu or Qu–CNGs. This enhanced antimicrobial effect was achieved by leveraging near-infrared II (NIR-II) light irradiation. Additionally, the CuS/Qu–CNGs exhibited efficient penetration into the extracellular biofilm matrix, eradicating methicillin-resistant Staphylococcus aureus-associated biofilms in diabetic mice wounds. Furthermore, the nanocomposites were found to suppress proinflammatory cytokines, such as IL-1β, at the wound sites while regulating the expression of anti-inflammatory factors, including IL-10 and TGF-β1, throughout the recovery process. The presence of CuS/Qu–CNGs promoted angiogenesis, epithelialization, and collagen synthesis, thereby accelerating wound healing. Our developed CuS/Qu–CNGs nanocomposites have great potential in addressing the challenges associated with delayed wound healing caused by microbial pathogenesis.
New conceptsThis study presents the development of multifunctional quercetin carbonized nanogels embedded with copper sulfide nanoclusters (CuS/Qu–CNGs) with remarkable antimicrobial, antioxidative, and anti-inflammatory properties. A unique feature of this research is near-infrared II (NIR-II) responsiveness, which amplifies the antimicrobial efficacy of the nanogels. The nanocomposites adeptly permeate the extracellular biofilm matrix, enabling successful eradication of formidable biofilms of methicillin-resistant Staphylococcus aureus (MRSA) in wounds of diabetic mice. Moreover, the presence of CuS/Qu–CNGs mitigates the production of pro-inflammatory cytokines and modulates the expression of anti-inflammatory factors. This immunomodulation promotes key steps in a wound healing process such as angiogenesis, epithelialization, and collagen synthesis. Consequently, the CuS/Qu–CNGs showcase vast potential as a skin disinfectant for treating bacterial infections and hastening the healing process of chronic wounds. |
Introduction
Wound healing requires the tissue to undergo successive hemostasis, inflammation, proliferation, and maturation phases.1 The process is more complex in chronic wounds, owing to long-term infection and/or suppressed immune response, resulting in slower wound healing.2 In particular, exudative fluid and necrotic tissues in the superficial wounds provide a favorable environment for bacteria to form biofilms, causing chronic infections with an increased mortality risk.3 Moreover, multidrug-resistant (MDR) bacteria develop efflux pumps, produce hydrolytic enzymes, modify the target, and block binding sites and entry ports to withstand antibiotics.4 Current approaches to combat microbial pathogenesis include antibiotics, skin disinfectants, and hydrogels. However, their clinical indications in wound healing are not fully understood.5 Conventional broad-spectrum antibiotics are very effective but have no role in wound healing.6 In addition, continuous and rapidly growing antimicrobial resistance has further reduced the efficacy of conventional antibiotics.7 Clinically used skin disinfectants such as triclosan, triclocarban, and benzalkonium chloride often lead to contact dermatitis, mucous membrane irritation, and in some cases, interfere with thyroid metabolism or act as endocrine disruptors, mainly due to lower immunocompatibility and severe toxicity at a higher dosage.8 Although hydrogels are the next-best alternative in wound healing, faster dehydration rate in exposed wounds, poor mechanical stability in the swollen state, and ability to cause skin maceration have restricted their clinical use.9 Nevertheless, altered levels of protein glycosylation in the wound site impair the ability of macrophages and neutrophils to phagocytose bacteria, further prolonging the inflammatory phase.10 Non-regulated expression of cytokines in the inflammatory phase can cause the production of cytotoxic enzymes and elevated oxidative stress leading to severe tissue damage.11 Thus, antimicrobial activity combined with optimized antioxidant and/or anti-inflammatory response is critical for developing therapeutics with high wound healing potency.11Nanomaterial-based antimicrobials have been developed to disintegrate bacteria, primarily by releasing metal ions and elevated oxidative stress.12–14 However, their stability in biofluids and non-selective toxicity is still a major pursuit.15 Several polymers and biocompatible organic molecules also exhibit broad-spectrum antibacterial activity but possess intrinsic toxicity and are insoluble in aqueous.16 Alternatively, catalytic and near-infrared (NIR) irradiation-based treatments feature stimuli-triggered controlled elevation of oxidative stress (photodynamic and catalytic therapy) and hyperthermia (photothermal therapy;) to eradicate bacteria.17 For example, Xiao et al. demonstrated ultrafast and broad-spectrum antimicrobial response through a combined effect from photo- and chemo-dynamic therapy.18 Recently, Li et al. reported NIR-II (1060 nm) activated hydrogel for photothermal/thermodynamic synergistic antimicrobial therapy.19 In another study, Tian et al. investigated perylene and tetracyanoquinodimethane-based organic nanoparticles with intermolecular charge-transfer complexes having programmable optical absorption (1000–1350 nm) for photothermal (1064 nm) antibacterial therapy.20 Although the authors achieved deeper tissue penetration for localized photothermia against planktonic cells, antimicrobial activity against biofilms was not investigated. Furthermore, additional in vivo studies are necessary to fully elucidate the wound-healing capabilities of these newly developed biomaterials.
Our previous work showed that bovine serum albumin (BSA) stabilized catalytic and photoresponsive copper sulfide (CuS) nanoassemblies can effectively eliminate bacteria from the wound site under NIR-II irradiation.21 These BSA-templated CuS nanoassemblies, featuring variable oxidation states of copper, offer excellent water dispersibility, efficient light-harvesting capabilities, and robust reactive oxygen species (ROS) generation. Additionally, their low cytotoxicity positions them as a promising antimicrobial treatment option.21 However, the nanoassemblies did not exhibit antioxidant properties, and their efficacy in wound healing was not investigated. Toward this end, we employed quercetin (Qu), a polyphenol that belongs to a family of flavonoid compounds with strong antioxidative properties, as a template for metal nanoclusters.22 Through a process of moderate carbonization and polymerization, quercetin (Qu) was transformed into carbonized nanogels (Qu–CNGs). These nanogels subsequently acted as a scaffold for the nucleation and growth of minuscule copper sulfide (CuS) nanoclusters, culminating in the formation of CuS/Qu–CNGs nanocomposites. The Qu–CNGs possess intrinsic antioxidant and catalytic (oxidase- and peroxidase-like) activities and photoresponsive properties. The as-prepared CuS/Qu–CNGs with enhanced physical interactions with bacterial membrane amplify the synergistic bactericidal response from localized catalytic and photoresponsive therapies. Relative to Qu–CNGs, CuS/Qu–CNGs showed negligible cytotoxicity towards human skin cells (HaCaT cells) and red blood cells (RBCs). Furthermore, surgical wounds on diabetic mice infected with methicillin-resistant Staphylococcus aureus (MRSA) were efficiently treated with CuS/Qu–CNGs through destroying mature biofilm under NIR-II (1064 nm) irradiation via enhanced contact with bacterium membrane. The CuS/Qu–CNGs stimulated angiogenesis, epithelialization, and collagen deposition by modulating pro- and/or anti-inflammatory cytokines during the inflammatory phase, which accelerated wound healing (Fig. 1). Biocompatible CuS/Qu–CNGs with antimicrobial/oxidative/inflammatory activities hold great potential as an effective therapeutic for the treatment of bacterial infections and rapid healing of chronic wounds.
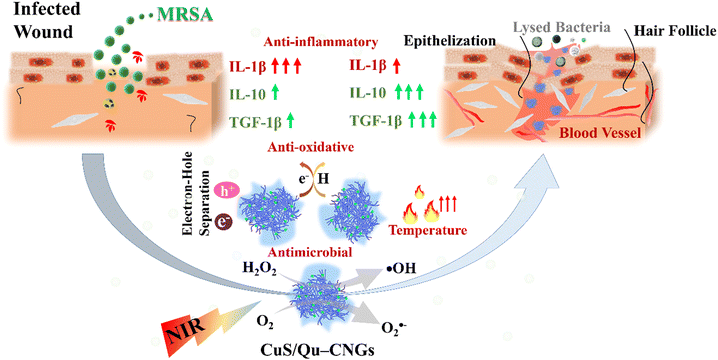 | ||
| Fig. 1 Schematic representation of the properties of CuS/Qu–CNGs and their role in the wound healing process. | ||
Results and discussion
Intrinsic and photoresponsive characteristics of CuS/Qu–CNGs
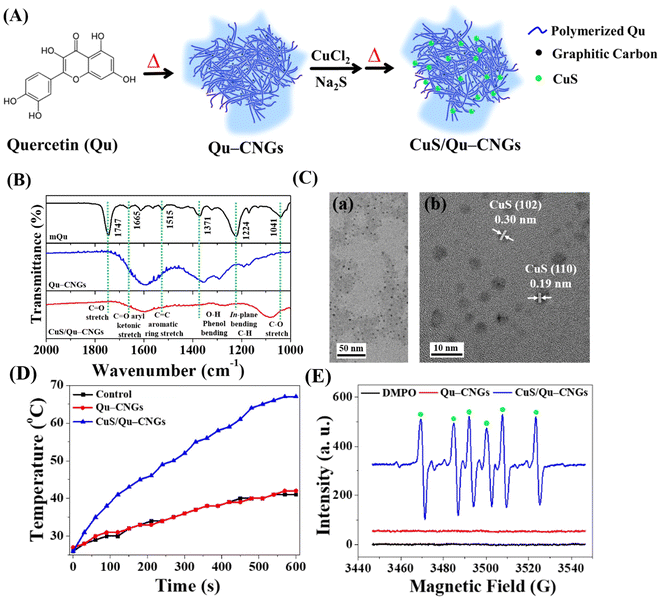
Upon pyrolyzing Qu at a temperature of 270 °C for a duration of 2 h, a brown residue is obtained, as depicted in Fig. S1A (ESI†). A subsequent alkaline treatment further refines this residue, resulting in the generation of highly dispersible Qu–CNGs. Briefly, CNGs were synthesized through dehydration, polymerization, crosslinking, and partial carbonization.23 During heating, polymers were formed through the dehydration-mediated condensation of Qu, and their further crosslinking generated micro-sized supramolecular structures.24 The gel-like structures undergo fragmentation upon prolonged heating, resulting in the assembly of irregular nanostructures (i.e., Qu–CNGs). Meanwhile, partially carbonized graphene dots are formed in situ within close-knit polymeric nanonetworks during heating, as shown in Fig. S1B (ESI†). Insufficient carbonization endows CNGs with the functional groups from precursor molecules. To clarify, Qu–CNGs, even at concentrations as high as 10 mg mL−1, do not form a macroscopic hydrogel. Rather, they exist as a colloidal solution with a viscosity of approximately 20 cP. We refer to these as “nanogels” due to their polymeric nanonetwork structure comprised of carbonized quercetin, observable at the nanoscale (Fig. S1B, ESI†). The Fourier transform infrared (FT-IR) spectra of monomeric Qu (mQu) revealed the characteristic bands at 1665, 1515–1614, and 1360 cm−1 corresponding to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O aryl ketonic stretch (weak; submerged), C
O aryl ketonic stretch (weak; submerged), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic ring stretch, and –O–H phenol bending, respectively, whereas the peaks were broadened after the formation of Qu–CNGs (Fig. 2B). Further, the as-formed Qu–CNGs were incubated with CuCl2 and Na2S at 90 °C for 15 min to prepare CuS nanoclusters in the GNCs (CuS/Qu–CNGs) (Fig. 2A). Transmission electron microscope (TEM) images show tiny CuS nanoclusters (∼2 nm) are uniformly embedded on Qu–CNGs templates (Fig. 2C(a)). The CuS/Qu–CNGs are highly crystalline, with d-spacing of 0.30 and 0.19 nm assigned to the (102) and (110) crystal planes of covellite-phase hexagonal CuS (Fig. 2C-b), respectively.21
C aromatic ring stretch, and –O–H phenol bending, respectively, whereas the peaks were broadened after the formation of Qu–CNGs (Fig. 2B). Further, the as-formed Qu–CNGs were incubated with CuCl2 and Na2S at 90 °C for 15 min to prepare CuS nanoclusters in the GNCs (CuS/Qu–CNGs) (Fig. 2A). Transmission electron microscope (TEM) images show tiny CuS nanoclusters (∼2 nm) are uniformly embedded on Qu–CNGs templates (Fig. 2C(a)). The CuS/Qu–CNGs are highly crystalline, with d-spacing of 0.30 and 0.19 nm assigned to the (102) and (110) crystal planes of covellite-phase hexagonal CuS (Fig. 2C-b), respectively.21
X-ray diffraction (XRD) pattern of CuS/Qu–CNGs exhibits (101), (102), (103), (105), (106), and (110) planes, which are also attributed to pure covellite-phase hexagonal CuS nanocrystals (Fig. S2, ESI†).25 Scanning transmission electron microscope coupled energy-dispersive X-ray spectroscopy (STEM/EDS) elemental mapping of CuS/Qu–CNGs indicates the coexistence of Cu and S within networks of Qu–CNGs (Fig. S3, ESI†). After the CuS nanoclusters deposited on Qu–CNGs, the zeta potential (ζ) of Qu–CNGs changes from −11.6 ± 2.9 mV (n = 5) to −40.2 ± 0.8 mV (n = 5). The negative charge is mainly attributed to the phenolic and ketonic groups in Qu. The UV-Vis absorption spectrum of mQu show characteristic bands centered at 256 and 370 nm, attributed to A–C benzoyl and B-ring cinnamoyl systems, respectively (Fig. S4, ESI†). In contrast to mQu, Qu–CNGs exhibit a distinct peak at 320 nm, probably due to the formation of the phenolic oxide o-quinone structure.26 Gradual blue shift and a weakened shoulder (∼370 nm) after carbonization can be ascribed to the π → π* and n → π* transitions of conjugated C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C
O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bonds, respectively, supporting the formation of graphitic multilayer carbon clusters within CNGs.27 Moreover, additional characteristic NIR broadband (620–1060 nm) was observed in CuS/Qu–CNGs, attributed to the formation of tiny-sized CuS nanoclusters within the Qu–CNGs matrix.28 X-ray photoelectron spectroscopy (XPS) analysis of C 1s reveals ∼79% of C
N bonds, respectively, supporting the formation of graphitic multilayer carbon clusters within CNGs.27 Moreover, additional characteristic NIR broadband (620–1060 nm) was observed in CuS/Qu–CNGs, attributed to the formation of tiny-sized CuS nanoclusters within the Qu–CNGs matrix.28 X-ray photoelectron spectroscopy (XPS) analysis of C 1s reveals ∼79% of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C groups in CNGs, relative to mQu (50.9%) (Fig. S5, ESI†). In contrast to mQu (C–O 42%), the abundance of the C–O bond was significantly reduced to 6.23% after pyrolysis to form Qu–CNGs. Deconvoluted O 1s XPS spectra exhibit a much higher O
C groups in CNGs, relative to mQu (50.9%) (Fig. S5, ESI†). In contrast to mQu (C–O 42%), the abundance of the C–O bond was significantly reduced to 6.23% after pyrolysis to form Qu–CNGs. Deconvoluted O 1s XPS spectra exhibit a much higher O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C in Qu–CNGs (∼33%), relative to mQu (∼7%). Moreover, characteristic peaks of 2p orbitals of Cu (Cu 2p1/2 and 2p3/2) and S (chemisorbed sulfur) in CuS/Qu–CNGs confirm the formation of CuS nanoclusters.
C in Qu–CNGs (∼33%), relative to mQu (∼7%). Moreover, characteristic peaks of 2p orbitals of Cu (Cu 2p1/2 and 2p3/2) and S (chemisorbed sulfur) in CuS/Qu–CNGs confirm the formation of CuS nanoclusters.
Our results suggest that Qu–CNGs are formed by partial carbonization and subsequent polymerization. Hence, several essential functional groups from the precursor with reducing abilities are preserved (Fig. S6, ESI†). To substantiate the mild carbonization, we employed the Folin-Ciocâlteu reagent to estimate the total phenolic content in the monomeric and pyrolytic Qu products (Fig. S6B, ESI†).29 In contrast to mQu, the phenolic content (reducing capability) present in Qu–CNGs was slightly lower, which supports mild carbonization of Qu at 270 °C. However, CuS/Qu–CNGs (Abs765 ∼ 0.77) exhibited comparable antioxidant properties to that of ascorbic acid (Abs765 ∼ 0.94) or mQu (Abs76 ∼ 0.73), which is higher than those produced by Qu–CNGs (Abs765 ∼ 0.60). Due to CuS, Qu–CNG's structure becomes looser, which results in higher scavenging activity, as more phenols are exposed and available to undergo electron transfer reaction. Although polyphenols are directly correlated with antioxidant properties, we evaluated the radical scavenging activity of CNGs through a well-known 2,2-diphenyl-1-picryl-hydrazyl-hydrate (DPPH) assay.30 The DPPH scavenging assay further reveals the CuS/Qu–CNGs (5.0 μg mL−1, in terms of Qu–CNGs) have more excellent radical scavenging capability mainly through electron and proton transfer processes.31 Contrastingly, ascorbic acid (positive control) at the same concentration, i.e., 5.0 μg mL−1, could not facilitate the electron transfer. The superior antioxidant property of CuS/Qu–CNGs is mainly attributed to the self-preserved phenolic groups within CNGs and the variable oxidation states of Cu.
Intrinsic catalytic activities of CNGs dispersed in sodium phosphate buffer (10 mM, pH 7.4) were investigated using amplex red (AR).32,33 The CuS/Qu–CNGs catalyzed the efficient O2-mediated oxidation of AR to produce highly fluorescent resorufin (7-hydroxy-3H-phenoxazin-3-one; λex/λem: 540/585 nm) (Fig. S7, ESI†). The oxidase-like activity of CuS/Qu–CNGs is 5-fold higher than that of the mQu and Qu–CNGs. Tiny CuS nanoclusters embedded in Qu–CNGs offer high specific surface area, which endows abundant catalytically active reaction centers to boost the intrinsic oxidase-like activity of CuS/Qu–CNGs. Further, CuS/Qu–CNGs can effectively catalyze AR to yield resorufin in the presence of H2O2 (1.0 mM) (Fig. S8, ESI†). The CuS/Qu–CNGs exhibit comparable fluorescence to that provided by HRP (0.1 unit mL−1) and ∼600-fold higher than the Qu–CNGs. The stronger peroxidase-like activity is attributed to the ability of Cu ions dissipated from the CuS surface to participate in Fenton-like reactions (H2O2 + Cu2+ → ˙OH + OH + Cu+).34
We then investigated the photoresponsive properties of CuS/Qu–CNGs. Fig. 2D shows a quick rise in the temperature profile of CuS/Qu–CNGs (∼68 °C) within 10 min of NIR-II irradiation (1064 nm, 0.94 W cm−2), which is drastically higher (< 40 °C) than that of the control (DI water) or Qu–CNGs, attributed to their characteristic and broad NIR photoabsorption (Fig. S4, ESI†). In addition, CuS/Qu–CNGs manifest an apparent concentration- and power density-dependent elevation in temperature (Fig. S9, ESI†). Excellent photothermal performance of CuS/Qu–CNGs is ascribed to intrinsic overlapping plasmon frequency (1064 nm), which engendered a near-field enhancement effect.35 The increased temperature is associated with the kinetic energy of the electron after collective and coherent oscillation and collisions in space during vibrational transitions.36 Furthermore, the photodynamic activity of CuS/Qu–CNGs was evaluated using AR as a substrate under NIR-II laser (1064 nm, 0.94 W cm−2) irradiation (Fig. S10, ESI†). The CuS/Qu–CNGs over Qu–CNGs displayed significantly higher activity (>tenfold) towards oxygen (O2)-mediated oxidation of AR after 10 min of NIR-II exposure. The amount of ROS may be even higher; the presence of the scavenger, i.e., Qu–CNGs in the same system, probably lowered the actual intensity. In control experiments, only AR or Cu2+ ions (data not shown) were incapable of AR oxidation. In addition, electron spin resonance (ESR) measurement with DMPO revealed a distinctive sextet due to hydroperoxyl radicals (˙OOH) generated by CuS/Qu–CNGs (Fig. 2E). Under NIR-II irradiation, CuS/Qu–CNGs facilitate the generation of superoxide (O2−˙) induced ˙OOH formation (O2 + e− → O2−; O2−˙ + H2O → ˙OOH + OH−) via type I photo-oxidative reaction.37 A negligible signal was obtained from CNGs without NIR-II laser (Fig. S11, ESI†). The amplified catalytic response mainly originated from NIR-II irradiation-assisted faster electron transfer Cu2+/Cu+/Cu0 and O2/H2O on the surface of CuS nanoclusters on Qu–CNGs. Moreover, the indirect formation of ˙OOH through the Fenton-like reaction of Cu2+/Cu+ cannot be excluded.38
Superior antimicrobial activity of CuS/Qu–CNGs coupled with NIR-II irradiation and H2O2
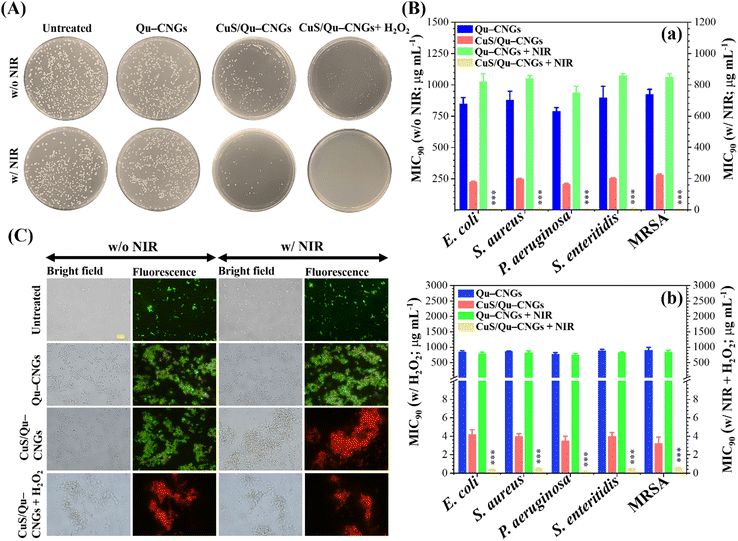
By taking advantage of antioxidant, enzyme-like, and photoresponsive characteristics, in vitro antibacterial performance of CuS/Qu–CNGs was evaluated against S. aureus, S. enteritidis, E. coli, and P. aeruginosa, and methicillin-resistant S. aureus (MRSA) bacterial strains without and with NIR-II laser irradiation (1064 nm, 0.94 W cm−2) (Fig. 3). Colony formation assays of E. coli treated with CuS/Qu–CNGs (209 ± 12/45 ± 3 CFU, n = 4) revealed a significant reduction in bacterial colonies in the absence/presence of NIR-II irradiation, as compared to those of untreated (475 ± 27/403 ± 16 CFU, n = 4) and Qu–CNGs (413 ± 31/390 ± 15 CFU, n = 4) treated groups (Fig. 3A). Bactericidal effect of CuS/Qu–CNGs (71 ± 11 CFU, n = 4) was amplified in the presence of H2O2 (10 μM), due to their intrinsic peroxidase-like activity to transform H2O2 into severely cytotoxic ˙OH. Moreover, negligible colonies of E. coli were counted when co-treated with CuS/Qu–CNGs and H2O2 under NIR-II irradiation. Contrastingly, in control experiments, only H2O2 (up to 500 μM) slightly inhibits bacteria (Fig. S12, ESI†). The MIC90 (minimal inhibitory concentration required to inhibit >90% of the isolates) values were determined by a standard serial dilution method. As shown in Fig. 3B, the MIC90 values of Qu–CNGs against tested bacterial strains were estimated to be >750 μg mL−1 in the absence and/or presence of NIR-II irradiation and/or H2O2. As expected, NIR-II irradiation reduced the MIC90 values of CuS/Qu–CNGs to 6–9 μg mL−1 (in terms of Qu–CNGs), which is ∼35-fold lower than those treated in the absence of photoirradiation (Fig. 3B(a)). H2O2 coupled treatment further decreased the MIC90 values of CuS/Qu–CNGs to ca. 4 and 0.2 μg mL−1 (in terms of Qu–CNGs) in the absence and presence of NIR-II irradiation, respectively. Moreover, the bacteriostatic activity of CuS/Qu–CNGs against E. coli was determined through a well-known live/dead SYTO 9 (green (G) fluorescence)/propidium iodide (PI; red (R)) staining assay (Fig. 3C).39 The results indicate that CuS/Qu–CNGs cause the most effective disintegration (relative red-to-green fluorescence (R/G) ∼ 4.1) of bacterial cell membrane upon co-treatment with photoirradiation and H2O2, relative to untreated (R/G ∼ 0.1) or Qu–CNGs (R/G ∼ 0.3) treated groups (Fig. S13, ESI†). The R/G vale of CuS/Qu–CNGs coupled with photoirradiation and H2O2 alone is 3.5 and 3.2, respectively.The extent of bacteria aggregation demonstrates the strong physical interaction between bacterial membrane and Qu–CNGs, ascribed to polyphenolic and other self-preserved pyrolytic products of Qu (Fig. 3C).23 Literature suggested that phenolic compounds exhibit a higher tendency to bind at the cytoplasmic membrane of bacteria.40 The broad-spectrum antimicrobial activity of CuS/Qu–CNGs is at least 3500-fold higher than Qu–CNGs. The synergistic response from photodynamic (˙OOH) and photothermal (>45 °C) effects, along with the strong intrinsic peroxidase-like ability to decompose H2O2 into hydroxyl radical (˙OH) in situ to induce oxidative stress, are probably the main contributors for membrane disruption. ROS generating capacity of the CuS/Qu–CNGs was estimated through 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) acetylation and further oxidation to produce a fluorescent DCF, and the fluorescence intensity accounts for the intracellular ROS levels (Fig. S14A, ESI†).41E. coli co-treated with CuS/Qu–CNGs and H2O2 under NIR-II irradiation (1064 nm, 0.94 W cm−2, 10 min) exhibit comparable fluorescent intensity to that of H2O2 (positive control, 1.0 mM) and 20-fold higher than that of Qu–CNGs treated bacteria (Fig. S14B, ESI†). A similar trend of bacteria aggregation confirms that CuS/Qu–CNGs facilitate their strong physical interactions with the cell membrane. These results are consistent with the ESR measurements, which also indicated the formation of ROS upon NIR-II irradiation (Fig. 2E).
Subsequently, the bacterium membrane potential was assessed using o-nitrophenyl-β-D-galactopyranoside (ONPG) as a substrate.42 The CuS/Qu–CNGs over Qu–CNGs induced more release of cytoplasmic β-galactosidase from E. coli to form o-nitrophenol (ONP; yellow color) (Fig. S15, ESI†). In addition, TEM images of E. coli treated with both CuS/Qu–CNGs and H2O2 under NIR-II irradiation show disrupted bacterial membranes; however, untreated bacteria exhibit intact morphology with well-defined cell membranes (Fig. S16, ESI†). Polyphenolic Qu promotes cellular attachment, whereas, in the presence of H2O2 and NIR treatment, it causes irreversible bacterial damage by the continuous and localized heat and bombardment of ROS. Moreover, the biocompatibility of CNGs was evaluated through in vitro cytotoxicity (Fig. S17, ESI†) and hemolysis (Fig. S18, ESI†) assay against HaCaT skin cell lines and erythrocytes, respectively. In contrast to mQu (∼15%), CuS/Qu–CNGs or Qu–CNGs exhibit ∼100% cell viability up to 200 μg mL−1, which is ∼2500-fold higher than respective MIC90 values. The higher toxicity of mQu is ascribed to its poor aqueous solubility.43 It is reported that mQu often exhibits severe cytotoxic effects, including proinflammatory properties, mitochondrial toxicity, potential mutagenicity, and inhibition of essential enzymes involved in hormonal metabolism, and also associated with food safety concerns.44,45
Wound healing application of CuS/Qu–CNGs
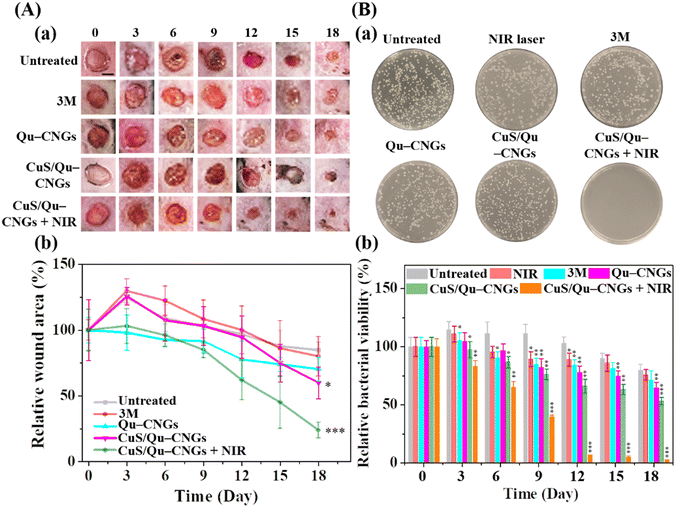
The host's immune system generally destroys pathogens on the exposed wound at the early stage of the wound-healing process.46 However, in a chronic wound, microbes attach to the wound surface and proliferate to develop a biofilm which becomes mature over time due to immunodeficiency of the diabetic disease.47 To address this issue, Qu–CNGs and CuS/Qu–CNGs were employed both to inhibit and eradicate MRSA biofilms. The effectiveness of these treatments was assessed through crystal violet staining assays, both with and without NIR-II irradiation (Fig. S19, ESI†).48 Compared to CNGs (<5%), CuS/Qu–CNGs (1.0 μg mL−1, in terms of Qu) coupled with NIR-II laser irradiation (1064 nm, 1.20 W cm−2, 10 min) suppressed ∼95% of biofilm formation (Fig. S19A, ESI†) and eradicated 82% of full-grown biofilm (Fig. S19B, ESI†), while negligible antibiofilm response was observed without NIR-II irradiation. Interestingly, vancomycin (100 μg mL−1) completely inhibited the biofilm formation of bacterial suspensions of MRSA, whereas it eradicated only ca. 50% of mature biofilm, even at 20-fold higher dosage than that of CuS/Qu–CNGs. Drug-resistant bacteria tend to hinder or neutralize traditional antibiotics by activating intrinsic self-defense barriers, which engenders acquired mutation.49 The superior antimicrobial response can be ascribed to unique polymeric structures of CNGs that facilitated substantial contact-mediated biofilm disintegration, followed by localized catalytic and NIR-assisted phototherapeutic action.50Owing to strong antibiofilm activity, we next investigated the in vivo efficacy of CuS/Qu–CNGs using a mouse model of Type 2 diabetes mellitus (BKS.Cg-Dock7m+/+Leprdb/J) infected with MRSA. Contrary to in vitro study (externally supplied H2O2), the H2O2 burst generated around wounded tissue was employed as a substrate for peroxidase-like activity. Fig. 4A shows the photographs (Fig. 4A(a)) and relative wound closure area (Fig. 4A(b)) of postoperative wounds (∼10 mm) untreated and treated separately with 3 M bandage, Qu–CNGs or CuS/Qu–CNGs (w/o and w/NIR-II; 1064 nm, 1.32 W cm−2). The inability of the control group to inhibit biofilm formation can be attributed to the compromised immune response commonly seen in diabetic mice. As a result, wound sizes in the control group were observed to be slightly larger on the third day compared to their initial sizes on day 0.51 Contrary to CuS/Qu–CNGs, the untreated, 3 M bandage and Qu–CNGs treated groups developed ulcers and edema on wound sites after 6 days. On the 15th day, the relative wound area of CuS/Qu–CNGs treated groups without and with NIR-II irradiation were 80% and 52%, respectively, while control groups did not exhibit any signs of recovery (Fig. 4A). Biofluids collected from CuS/Qu–CNGs (w/NIR-II) treated wounds on day 12 show no conspicuous bacterial colonies, relative to those of untreated (850 ± 36 CFU) and 3 M bandage (721 ± 51 CFU), Qu–CNGs (695 ± 62 CFU), and CuS/Qu–CNGs (w/o NIR-II; 515 ± 45 CFU) treated groups (Fig. 4B(a)). A further antimicrobial assessment revealed that CuS/Qu–CNGs (w/NIR-II) significantly inhibited MRSA from the 12th day onwards, while >90% and >50% bacteria viabilities were recorded for untreated and other treated groups, respectively, till the 18th day (Fig. 4B(b)). The CuS/Qu–CNGs treated group displayed higher epithelialization (∼55% wound closure) after day 15 and healed completely on day 18 (Fig. 4A). Uncured MRSA infection in the control groups caused severe inflammation and tissue damage and resulted in delayed healing.52
The wound healing process is comprised of hemostasis (blood clotting), inflammatory, proliferative (granulation), and maturation (remodeling) phases.53 Fig. S20 (ESI†) shows the histological evaluation of tissue specimens collected on postoperative day 15 using H&E and Masson's trichrome staining. Antimicrobial CuS/Qu–CNGs mitigated the MRSA infection-induced inflammation, which was quite prominent in the untreated group, as indicated by the immune cells (yellow arrow). In addition to the efficient bactericidal effect, the CuS/Qu–CNGs with NIR-II irradiation also accelerated wound healing by promoting angiogenesis, epithelialization, and collagen deposition. Erythrocytes are less pliable to deliver oxygen to the chronic wound for tissue metabolism and collagen synthesis. The blue color represents collagen fibers in Masson's trichrome staining, and its intensity corresponds to the relative collagen content in the tissue sections.52 Our results indicate that more collagen is formed in the recovered dermal layers after treatment with the CuS/Qu–CNGs under NIR-II irradiation, and hence a more intense blue color compared to that in the untreated or without NIR-II laser-treated groups (Fig. S20, ESI†). We also noted CuS/Qu–CNGs assisted in the development of blood vessels (cyan arrow) and hair follicles (black arrow) when coupled with NIR-II laser treatment (Fig. S20, ESI†). The formation of a new network of blood vessels allows an adequate supply of oxygen and nutrients for the stimulated growth of granulation tissue.53 Moreover, we also observed higher epithelialization (green arrow) in the CuS/Qu–CNGs treated group under NIR-II irradiation relative to other groups, which implies that the wound is completely recovered (Fig. S20, ESI†).54
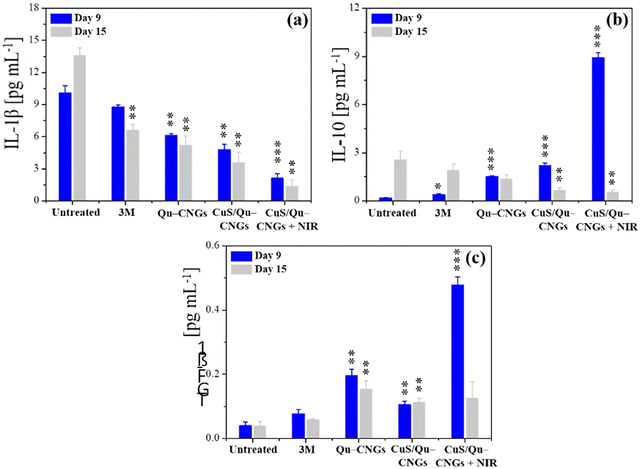
Prolonging the inflammatory phase due to microbial pathogenesis and suppressed immune response of diabetic mice significantly contribute to delayed healing.55 The inflammation phase begins immediately after injury and induces localized swelling to prevent bleeding and infection. During this phase, compromised cells, invaded pathogens etc., are removed from the wound site.56 Inflammation is a natural part of the wound healing process, however, impaired neutrophil and macrophage phagocytosis of bacteria in diabetic wounds, due to abnormal protein glycosylation may protract the inflammatory phase.57 Moreover, higher consumption of complement proteins in this phase due to bacterial infection may lead to restrained chemotaxis.58 Thus, controlled regulation and modulation of pro- and/or anti-inflammatory cytokine levels is vital.59 Tissue specimens from the infectious wound sites were extracted on days 9 and 15 to quantify the levels of Interleukin 1 Beta (IL-1β), IL-10, and transforming growth factor-beta 1 (TGF-β1) using a standard enzyme-linked immunosorbent assay (ELISA) (Fig. 5).59 The CuS/Qu–CNGs under NIR-II irradiation show reduced levels of a pro-inflammatory cytokine, i.e., IL-1β on day 9 (2.12 pg mL−1), which were highly expressed in the untreated group (control; 10.1 pg mL−1) due to bacterial pathogenesis (Fig. 5a and 3B(b)). We observed that CuS/Qu–CNGs not only suppressed proinflammatory IL-1β but also up- and down-regulated anti-inflammatory IL-10 on day 9 and 15, respectively, which probably contributed to faster healing (Fig. 5b). TGF-β1, a multifunctional housekeeping gene, belongs to a class of anti-inflammatory cytokines.60Fig. 5c indicates CuS/Qu–CNGs under NIR-II irradiation spiked the expression of TGF-β1 while recovering from infection (day 9), and the same levels were maintained as those in control on day 15 (after recovery). The anti-inflammatory response of CuS/Qu–CNGs mainly originates from the intrinsic radical scavenging capacity of self-preserved polyphenolic groups within Qu–CNGs.61 Chronic wounds are mainly stalled due to the extended stay of proinflammatory macrophages (M1) in the inflammatory phase, restricting subsequent proliferation and remodeling processes vital for systematic tissue regeneration and wound closure.62 M1 macrophages promote the secretion of proinflammatory cytokines, including IL-6, 8, etc., and tumor necrosis factor α (TNF-α), in response to which M2-like anti-inflammatory phenotype is stimulated.63 Our results indicated high-level expression of IL-10 during infection in CuS/Qu–CNGs treated groups (w/NIR-II), typically associated with accumulation of M2 macrophages (Fig. 5b).64 Further, M2 macrophages induce the production of fibroblast and vascular endothelial growth factors to enhance angiogenesis and tissue regeneration.65 Therefore, therapeutic methods that facilitate the transition from M1 to M2 macrophages hold great potential to stimulate the healing process in chronic wounds.66 We further employed DCFH-DA staining assays to determine the lipopolysaccharide (LPS) induced oxidative stress in RAW264.7 macrophages using two different sets of experiments.67 Firstly, RAW264.7 macrophages co-incubated with LPS for 18 h in the absence and presence of Qu–CNGs or CuS/Qu–CNGs revealed anti-inflammatory action of as-prepared CNGs (Fig. S21A, ESI†). In another study, LPS pre-incubated with Qu–CNGs or CuS/Qu–CNGs for 6 h exhibit similar fluorescence to those of untreated macrophage, suggesting that the unique assembly of CNGs facilitates the LPS-binding to inhibit oxidative stress-induced inflammation (Fig. S21B, ESI†). Superior in vivo antimicrobial/oxidative/inflammatory properties of CuS/Qu–CNGs effectively suppressed inflammation and promoted wound healing in diabetic mice.
Furthermore, we have evaluated the potential in vivo toxicity of CuS/Qu–CNGs by monitoring mice body weights, biochemical levels in the bloodstream, and histological evaluation of vital organs using H&E staining (Fig. S22, ESI†). The body weights of mice before and after administration of CuS/Qu–CNGs (∼25 mg kg−1) displayed no significant difference compared to control groups (untreated ones) up to 18 days (Fig. S22A, ESI†). In addition, the collected blood samples from untreated and CuS/Qu–CNGs treated mice were analyzed for several vital biochemicals responsible for inflammation in the liver [aspartate transaminase (AST), alanine transaminase (ALT), gamma-glutamyl transferase (GGT), and total bilirubin (T-BIL)] and kidney (blood urea nitrogen (BUN). As shown in Fig. S22B (ESI†), the levels of those indicators of treated groups exhibit no significant change relative to the untreated ones. Moreover, we also analyzed H&E stained tissue sections of various important organs, including the heart, spleen, lung, kidney, and liver (Fig. S22C, ESI†). The stained tissue slices from CuS/Qu–CNGs treated mice at day 18 indicated no distinguishable tissue damage. Collectively, our results suggest that CuS/Qu–CNGs exhibit substantial in vivo biocompatibility and thus hold great potential as a safe antimicrobial agent.
In this study, we successfully used Qu–CNGs as a template to incorporate CuS nanoclusters, introducing additional photoresponsive properties to the nanogels. The resultant CuS/Qu–CNGs nanocomposites have significantly amplified their antimicrobial effects when exposed to NIR-II light irradiation. Besides superior antimicrobial response, the nanocomposites demonstrated an exceptional ability to penetrate extracellular biofilm matrices, leading to the eradication of formidable MRSA biofilms in wounds inflicted on diabetic mice. Notably, the nanocomposites suppressed proinflammatory cytokines such as IL-1β while simultaneously boosting the expression of anti-inflammatory factors, including IL-10 and TGF-β1. This duality aided the wound recovery process. Furthermore, CuS/Qu–CNGs encouraged crucial wound healing processes, including angiogenesis, epithelialization, and collagen synthesis, thus accelerating overall wound healing. The CuS/Qu–CNGs nanocomposites show immense potential in treating bacterial infections and enhancing the healing process of chronic wounds, especially those complicated by microbial pathogenesis. However, we acknowledge several areas that future work can address to refine the clarity and applicability of CuS/Qu–CNGs nanocomposites.
Our current findings suggest that Qu–CNGs enhance physical interactions with bacterial membranes, inflicting irreversible damage due to the synergistic catalytic and photoresponsive properties of CuS. Our future work will focus on gaining deeper insight into the precise mechanisms of these interactions and resultant damage. We plan to further examine the potential induction of oxidative stress to directly disrupt the cell membrane. A comprehensive understanding of this mechanism will enable better design and the use of similar materials. While our study indicates that CuS/Qu–CNGs have higher biocompatibility with HaCaT skin cells and erythrocytes compared to mQu, we will further investigate potential cytotoxic effects on various cell types and possible systemic toxicity following application. Additionally, we believe there is room to optimize the preparation process of CuS/Qu–CNGs for improved efficacy or different applications. Determining particle concentration, size, and surface properties could yield substantial improvements. Simultaneously, efforts to scale the production process for CuS/Qu–CNGs could enhance the research's applicability to real-world scenarios.
Conclusions
We prepared Qu–CNGs through polymerization and subsequent mild carbonization. Further, CuS nanoclusters were grown in situ to prepare CuS/Qu–CNGs. The Qu–CNGs inherit antioxidant and anti-inflammatory activities, and CuS possess peroxidase-like and photoresponsive properties. MIC90 values of CuS/Qu–CNGs (6–9 μg mL−1) is ∼125-fold lower than mQu or Qu–CNGs under NIR-II irradiation and were further improved by ∼30-fold (ca. 0.2 μg mL−1) in the presence of H2O2. Qu–CNGs enhanced physical interaction with bacteria membrane, causing irreversible damage to the bacteria through synergistic catalytic and photoresponsive properties of CuS. Over mQu, CuS/Qu–CNGs showed higher biocompatibility towards HaCaT skin cells and erythrocytes. Furthermore, administration of CuS/Qu–CNGs under NIR-II irradiation eradicated MRSA biofilms from diabetic mice wounds due to their unique polymeric structure, facilitating direct contact with bacterium membrane. The CuS/Qu–CNGs not only suppress proinflammatory cytokines (IL-1β) but also modulate the expression of anti-inflammatory IL-10 and TGF-β1 during different stages of recovery from infection. Remarkable in vivo antimicrobial/oxidative/inflammatory properties of CuS/Qu–CNGs accelerate chronic wound healing. Our results revealed that CuS/Qu–CNGs hold great potential as a skin disinfectant for treating bacterial infections and rapid healing of chronic wounds.Experimental section
Synthesis of quercetin carbonized nanogels (Qu–CNGs)
A glass vial containing 50 mg of quercetin dehydrate (Qu) was heated at 270 °C for 2 h. The solid dark brown residue after the thermal treatment was cooled to room temperature (25 °C) and dispersed in 5.0 mL of NaOH (0.1 M) through sonication (1 h). The solution was centrifuged at a relative centrifugal force (RCF) of 5000g for 30 min to remove larger aggregates. The supernatant containing Qu–CNGs was stored at −20 °C for further experiments.Synthesis of CuS/Qu–CNGs
Qu–CNGs (100 μL, 10 mg mL−1) dispersed in 10 mM sodium phosphate buffer (808 μL, pH 7.4) were incubated with CuCl2 (32 μL, 100 mM) for 10 min, to which Na2S (32 μL, 0.5 M) was spiked, resulting in an immediate color change from brown to black. The resulting solution was then heated at 90 °C for 15 min and cooled to room temperature. The as-formed CuS/Qu–CNGs were purified through dialysis (molecular weight cut off of membrane; 1.0 kD) against sodium phosphate buffer (10 mM, pH 7.4) for 3 h, with the buffer being replaced every 30 min and stored at 4 °C when not in use.Antioxidant assays
Radical scavenging abilities of Qu–CNGs and CuS/Qu–CNGs were evaluated using a facile colorimetric-based 2,2-diphenyl-1-picryl-hydrazyl-hydrate (DPPH) assay. 5.0 μg mL−1 (in terms of Qu) of mQu, Qu–CNGs, CuS/Qu–CNGs, or ascorbic acid (positive control) dispersed in 10 mM sodium phosphate buffer was directly reacted with DPPH (200 μM) at ambient temperature under orbital shaking (150 rpm) for 1 h. Then, 200 μL from each mixture was transferred to a 96-well flat bottom plate to record the UV-Vis absorption spectra in the range of 300–900 nm.Photoresponsive properties of CuS/Qu–CNGs
A continuous near-infrared laser (NIR-II, 1064 nm, 0.94 W cm−2) with DPSSL driver II (Tangyu Precision Machinery Industry Co., Ltd, Taipei, Taiwan) coupled with a light guide was used to investigate the photothermal and photodynamic activities of CNGs. 50 μg mL−1 (in terms of Qu) of Qu–CNGs or CuS/Qu–CNGs dispersed in 10 mM sodium phosphate buffer (0.5 mL, pH 7.4) was irradiated with the NIR-II laser, and the temperature change was recorded over time using a TFC-305A Type K single-input thermocouple thermometer (Yi Chun Electrics Co., Ltd, Taipei, Taiwan).Typically, the photodynamic performances of Qu–CNGs and CuS/Qu–CNGs were evaluated through the reaction of AR (10 μM) with 50 μg mL−1 (in terms of Qu) of Qu–CNGs or CuS/Qu–CNGs dispersed in 10 mM sodium phosphate buffer (0.5 mL, pH 7.4) in the dark and under photoirradiation at ambient temperature for 10 min. Then, 200 μL from each sample was poured into 96-well microtiter plates to record the fluorescence intensity at 590 nm upon excitation at 540 nm. 5,5-Dimethyl-1-pyrroline-N-oxide (DMPO) was employed as a spin trap to perform electron spin resonance (ESR) spectroscopy analysis to detect the produced free radicals. The ESR parameters are as follows: modulation frequency, 100 kHz; g-factor, 2.00627; modulation amplitude, 0.1 mT; microwave power, 15 mW; microwave frequency, 9.8 gHz; scan range, 100 G; time constant, 0.328 s; sampling time, 20 ms; receiver gain, 30.
Bacterial cultures and antibacterial assays
Colonies of P. aeruginosa (ATCC 27853), S. aureus (BCRC10781), E. coli (BRBC 12438), S. enteritidis (BRBC 10744), and MRSA (ATCC 43300) were plucked and grown in Luria Bertani (LB) growth media. Each bacterial suspension was incubated at 37 °C until the absorbance at 600 nm reached 1.0 (optical path length = 1 cm). The bacterial suspension (1.0 mL) was purified by centrifugation (5000g, 27 °C, 10 min) and washed thrice with phosphate-buffered saline (PBS, pH 7.4, containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2.0 mM KH2PO4; 1 mL). Then, 200 μL of the bacterial suspension [1.0 × 104 colony-forming unit (CFU) mL−1] was spread over solidified LB agar plates and allowed to grow overnight at 37 °C for the colony formation assay.The MIC90 (minimal inhibitory concentration required to kill >90% of the bacteria population) values were obtained using the broth microdilution method. The bacterial suspensions [1.0 × 103 CFU mL−1] were first incubated with Qu–CNGs and CuS/Qu–CNGs in the presence and absence of H2O2 (10 μM) in PBS solution at room temperature for one hour. Then, each of the mixtures was further incubated with or without NIR-II laser (1064 nm, 0.94 W cm−2) irradiation at room temperature for 10 min. Finally, 200 μL from each bacterial solution was spread onto the solidified LB agar plates at 37 °C, and CFUs were counted manually after incubating overnight.
Antibiofilm activities of CuS/Qu–CNGs
For biofilm inhibition assay, MRSA suspensions (1 × 106 CFU mL−1) dispersed in Tryptic Soy Broth (TSB) were individually incubated with vancomycin (100 μg mL−1; positive control), Qu–CNGs, and CuS/Qu–CNGs (1.0 μg mL−1, in terms of Qu) without and with NIR-II laser irradiation (1064 nm, 0.94 W Cm−2; slightly higher power density was used due to apparent potency of biofilms) for 10 min in a sterile 96-well flat bottom plate at 37 °C for 24 h under mild shaking (100 rpm). After washing three times with 5.0 mM sodium phosphate buffer (0.2 mL, pH 7.4), as-formed biofilms were fixed using paraformaldehyde (∼4%) for 20 min before staining with crystal violet (0.1%, 10 min). Lastly, biofilms were then solubilized in acetic acid (0.2 mL, 30%) to measure absorbance values at 595 nm.On the other hand, biofilms of MRSA bacterial cultures (1 × 106 CFU mL−1) dispersed in TSB were formed first in a sterile 96-well flat bottom plate (37 °C, 48 h). Mature biofilms were washed thrice with 5.0 mM sodium phosphate buffer (0.2 mL, pH 7.4) and then treated with vancomycin (100 μg mL−1; positive control) and 5.0 μg mL−1 (in terms of Qu) of Qu–CNGs and CuS/Qu–CNGs without and with NIR-II laser irradiation (1064 nm, 0.94 W cm−2). Post-treatment biofilms were further incubated for 24 h at 37 °C. After washing three times with the same buffer, the biofilms were fixed by paraformaldehyde (∼4%) for 20 min, followed by staining with crystal violet (0.1%, 10 min). Finally, the biofilms were dissolved in acetic acid (0.2 mL, 30%) and subjected to absorbance measurement at 595 nm.
Wound healing and histology
The wound healing capacity of the CNGs was investigated using an MRSA-infected diabetic mouse model. All the animal-related experimental procedures were in agreement and approved by the Institutional Animal Care and Use Committee of the National Laboratory Animal Center (NLAC; Permit No. IACUC2012-037). A gas mixture of Zoletil (5 mg kg−1) and Xylazine (117 mg kg−1) was employed to anesthetize the BKS.Cg-7m+/+Leprdb/J (db/db) male mice weighing (ca. ∼35 g). The skin around the dorsal area was trimmed and disinfected using 70% ethanol before aseptic surgery. Circular-shaped wounds of diameter ∼10 mm were created through incision of the dorsal skin with sterile surgical scissors. Then, 50 μL of MRSA suspension [1 × 108 CFU mL−1] dispersed in sterile PBS solution was transferred onto the wound site to induce the infection, followed by dressing in a sterilized cotton gauze moistened in PBS to avoid cross-contamination. After 24 h, the wound site was smeared using a sterile swab and cultured on LB agar plates for 24 h at 37 °C to confirm the MRSA infection. To evaluate the in vivo antimicrobial action of as-prepared CNGs, biofluid from infected wound site untreated and treated with 25 μg mL−1 (in terms of Qu) of Qu–CNGs or CuS/Qu–CNGs in the absence and presence of NIR-II laser (1064 nm, 1.32 W Cm−2) was swabbed on day 0, 3, 6, 9, 12, 15, and 18 and cultured on LB agar plates to monitor the bacteria growth through colony formation assay. In addition, the infection site covered with a conventional antimicrobial bandage, i.e., 3 M, was used as a control group to determine the relative efficacy of CNGs. Altogether, wound healing was also monitored by capturing the digital photographs of real-time wound size on days 0, 3, 6, 9, 12, 15, and 18 post-surgeries. The relative wound size was calculated using the following equation: Relative wound area (%) = An/A0 × 100, where A0 and An are the wound area on day 0 and on the nth day, respectively.Skin histology and collagen formation in the infectious wounded tissue were examined by Hematoxylin and eosin (H&E) and Masson's trichrome staining, respectively. The tissue specimen containing the entire wound, including surrounding healthy skin (dermis and subcutaneous tissue), was surgically removed and fixed with 10% formaldehyde solution.
Determination of cytokine levels using ELISA
To quantify interleukins (ILs) on the infectious wound site to monitor inflammatory response, the overdose of anesthesia euthanized animals and the tissue specimens containing wounded and surrounding skin area were collected and frozen in liquid nitrogen. Upon thawing, the tissue samples (100 mg mL−1) were homogenized in a solution containing NaCl (400 mM), Tween 20 (0.05%), BSA (0.5%), phenylmethylsulfonyl fluoride (PMSF; 0.1 mM), benzethonium chloride (0.1 mM), EDTA (10 mM), and aprotinin (20 KI; ∼33 μg mL−1) through bath sonication for 30 min. Each of the suspensions was then centrifuged (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 10 min, 4 °C), and supernatants were collected to estimate the concentrations of IL-1β, IL-10, and TGF-β1 using ELISA kits as per the manufacturer's standard protocols. All samples were assayed in triplicates.
000g, 10 min, 4 °C), and supernatants were collected to estimate the concentrations of IL-1β, IL-10, and TGF-β1 using ELISA kits as per the manufacturer's standard protocols. All samples were assayed in triplicates.
In vitro anti-inflammatory response
Lipopolysaccharides (LPS) induced inflammation in RAW264.7 was employed to investigate the anti-inflammatory response of as-prepared CNGs through two different sets of experiments. Briefly, RAW264.7 macrophages (1 × 105 cells per well) in DMEM (5% FBS) were seeded in a 96-well plate in 5% CO2 at 37 °C for 24 h. Then, LPS (1.0 μg mL−1) was incubated with the macrophages for 18 h at 37 °C in the absence and presence of 25 μg mL−1 (in terms of Qu) of Qu–CNGs and CuS/Qu–CNGs. Subsequently, growth media was replaced with DCFH-DA (100 μM) dye solution to capture bright field and fluorescence images of cells.In another study, 25 μg mL−1 (in terms of Qu) of Qu–CNGs or CuS/Qu–CNGs pre-incubated with LPS (1.0 μg mL−1) at room temperature for 6 h were purified using Amicon Ultra Centrifugal Filters (UFC800308, Merck KGaA, Darmstadt, Germany) with a molecular weight cutoff (MWCO) of 10 K. Solutions at the top were collected after centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 10 min, 4 °C) includes LPS, LPS/Qu–CNGs and LPS/CuS/Qu–CNGs, which were incubated with the RAW264.7 cells for 18 h 5% CO2 at 37 °C to induce inflammation, then replaced with the fresh DMEM (10% FBS) to further grow for 24 h. Eventually, the growth media were removed, and DCFH-DA (100 μM) solution was added into each well and further incubated at 37 °C for 30 min before fluorescence observation. Fluorescence intensities at 530 nm were recorded with a fixed excitation at 490 nm using a monochromatic microplate spectrophotometer.
000g, 10 min, 4 °C) includes LPS, LPS/Qu–CNGs and LPS/CuS/Qu–CNGs, which were incubated with the RAW264.7 cells for 18 h 5% CO2 at 37 °C to induce inflammation, then replaced with the fresh DMEM (10% FBS) to further grow for 24 h. Eventually, the growth media were removed, and DCFH-DA (100 μM) solution was added into each well and further incubated at 37 °C for 30 min before fluorescence observation. Fluorescence intensities at 530 nm were recorded with a fixed excitation at 490 nm using a monochromatic microplate spectrophotometer.
Please see the ESI† for the details on the following contents: materials, characterization of CNGs, determination of total phenolic content, enzyme-like activities of CuS/Qu–CNGs, bacterial viability assays, ROS assays, bacterium membrane potential assays, TEM images of bacteria, in vitro cytotoxicity assays, hemolysis assays, and in vivo biocompatibility.
Author contributions
Amit Nain: carried out the experiments and wrote the original draft. Yu-Ting Tseng: assisted with resources, software, and validation. Akash Gupta: revised the manuscript. Yu-Feng Lin: interpreted the results and revised the original manuscript. Arumugam Sangili: provided feedback and interpreted the results and the mechanism. Yu-Fen Huang: provided feedback and interpreted the results and the mechanism. Ching Huang: conceived the conceptualization supervised the project from the beginning to the end, and prepared the manuscript. Huan-Tsung Chang: Funding acquisition and revised the manuscript.Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
This study was supported by the Ministry of Science and Technology, Taiwan under the contracts 110-2113-M-002-005-MY3, 110-2622-M-002-001, and 108-2638-M-002-001-MY2 and by the Chang Gung University under the contracts UMRPD1N0051 and UMRPD1N0141. Thanks to Ms Chia-Ying Chien of the Ministry of Science and Technology (National Taiwan University) for the assistance in TEM and EDS experiments. The authors gratefully acknowledge Mr Jui-Chin Lee for using electron spectroscopy for chemical analysis (PHI-5000) provided by the Instrument Center of National Cheng Kung University. The authors thank the Instrumentation Center at National Tsing Hua University (NTHU) for providing the EPR analysis.References
- P. Martin and R. Nunan, Br. J. Dermatol., 2015, 173, 370–378 CrossRef CAS PubMed.
- A. Figueiredo, E. C. Leal and E. Carvalho, Pharmacol. Res., 2020, 159, 104977 CrossRef CAS PubMed.
- E. M. Tottoli, R. Dorati, I. Genta, E. Chiesa, S. Pisani and B. Conti, Pharmaceutics, 2020, 12, 735 CrossRef CAS PubMed.
- M. H. Kollef, M. Bassetti, B. Francois, J. Burnham, G. Dimopoulos, J. G. Montero, J. Lipman, C. E. Luyt, D. P. Nicolau and M. J. Postma, Intensive Care Med., 2017, 43, 1187–1197 CrossRef PubMed.
- M. Zhao, J. Wu, H. Wu, A. H. Sawalha and Q. Lu, Clin. Rev. Allergy Immunol., 2021, 62, 1–19 Search PubMed.
- S. L. Percival, C. Emanuel, K. F. Cutting and D. W. Williams, Int. Wound J., 2012, 9, 14–32 CrossRef PubMed.
- P. Durão, R. Balbontín and I. Gordo, Trends Microbiol., 2018, 26, 677–691 CrossRef PubMed.
- D. W. Lachenmeier, Side Eff. Drugs Annu., 2019, 41, 251–256 Search PubMed.
- L. S. G. Aparicio, J. B. Sarabia, T. A. C. Villegas, P. H. L. Fabres, N. E. D. Martínez, E. P. Camberos, A. L. Navarro and A. B. C. Ceseña, Biomater. Sci., 2021, 9, 726–744 RSC.
- L. P. Erwig and N. A. R. Gow, Nat. Rev. Microbiol., 2016, 14, 163–176 CrossRef CAS PubMed.
- S. Zhang, J. Hou, Q. Yuan, P. Xin, H. Cheng, Z. Gu and J. Wu, Chem. Eng. J., 2020, 392, 123775 CrossRef CAS.
- A. Nain, Y. T. Tseng, S. C. Wei, A. P. Periasamy, C. C. Huang, F. G. Tseng and H. T. Chang, J. Hazard. Mater., 2020, 389, 121821 CrossRef CAS PubMed.
- K. Zheng and J. Xie, ACS Nano, 2020, 14, 11533–11541 CrossRef CAS PubMed.
- S. C. Wei, A. Nain, Y. F. Lin, R. S. Wu, P. Srivastava, L. Chang, Y. F. Huang, H. T. Chang, K. T. Chuang and C. C. Huang, Carbon, 2023, 201, 952–961 CrossRef CAS.
- M. Lakemeyer, W. Zhao, F. A. Mandl, P. Hammann and S. A. Sieber, Angew. Chem., Int. Ed., 2018, 57, 14440–14475 CrossRef CAS PubMed.
- C. Yang, X. Ding, R. J. Ono, H. Lee, L. Y. Hsu, Y. W. Tong, J. Hedrick and Y. Y. Yang, Adv. Mater., 2014, 26, 7346–7351 CrossRef CAS PubMed.
- G. Qing, X. Zhao, N. Gong, J. Chen, X. Li, Y. Gan, Y. Wang, Z. Zhang, Y. Zhang, W. Guo, Y. Luo and X. Liang, Nat. Commun., 2019, 10, 4336 CrossRef PubMed.
- Q. Xiao, B. Mai, Y. Nie, C. Yuan, M. Xiang, Z. Shi, J. Wu, W. Leung, C. Xu, S. Q. Yao, P. Wang and L. Gao, ACS Appl. Mater. Interface, 2021, 13, 11588–11596 CrossRef CAS PubMed.
- Q. Li, Y. Zhang, X. Huang, D. Yang, L. Weng, C. Ou, X. Song and X. Dong, Chem. Eng. J., 2021, 407, 127200 CrossRef CAS.
- S. Tian, H. Bai, S. Li, Y. Xiao, X. Cui, X. Li, J. Tan, Z. Huang, D. Shen and W. Liu, Angew. Chem., Int. Ed., 2021, 60, 11758–11762 CrossRef CAS PubMed.
- A. Nain, S. C. Wei, Y. F. Lin, Y. T. Tseng, R. P. Mandal, Y. F. Huang, C. C. Huang, F. G. Tseng and H. T. Chang, ACS Appl. Mater. Interfaces, 2021, 13, 7865–7878 CrossRef CAS PubMed.
- M. A. Rahim, S. L. Kristufek, S. Pan, J. J. Richardson and F. Caruso, Angew. Chem., Int. Ed., 2019, 58, 1904–1927 CrossRef CAS PubMed.
- Y. J. Li, S. C. Wei, H. W. Chu, H. J. Jian, A. Anand, A. Nain, Y. F. Huang, H. T. Chang, C. C. Huang and J. Y. Lai, Chem. Eng. J., 2022, 437, 135315 CrossRef CAS.
- Y. R. Chiou, C. J. Lin, S. G. Harroun, Y. R. Chen, L. Chang, A. T. Wu, F. C. Chang, Y. W. Lin, H. J. Lin, A. Anand, B. Unnikrishnan, A. Nain and C. C. Huang, Nanoscale, 2022, 14, 11719–11730 RSC.
- Y. Liu, M. Liu and M. T. Swihart, Chem. Mater., 2017, 29, 4783–4791 CrossRef CAS.
- M. Salomäki, L. Marttila, H. Kivelä, T. Ouvinen and J. Lukkari, J. Phys. Chem. B, 2018, 122, 6314–6327 CrossRef PubMed.
- C. Dhanush and M. G. Sethuraman, Spectrochim. Acta, Part A, 2021, 260, 119920 CrossRef CAS PubMed.
- H. Y. Wang, X. W. Hua, F. G. Wu, B. Li, P. Liu, N. Gu, Z. Wang and Z. Chen, ACS Appl. Mater. Interfaces, 2015, 7, 7082–7092 CrossRef CAS PubMed.
- T. Margraf, A. R. Karnopp, N. D. Rosso and D. Granato, J. Food Sci., 2015, 80, C2397–C2403 CrossRef CAS PubMed.
- B. Payet, A. S. C. Sing and J. Smadja, J. Agric. Food Chem., 2005, 53, 10074–10079 CrossRef CAS PubMed.
- A. Barzegar, PLoS One, 2012, 7, e39660 CrossRef PubMed.
- M. Raineri, E. L. Winkler, T. E. Torres, M. V. Mansilla, M. S. Nadal, R. D. Zysler and E. Lima, Nanoscale, 2019, 11, 18393–18406 RSC.
- D. Debski, R. Smulik, J. Zielonka, B. Michałowski, M. Jakubowska, K. Debowska, J. Adamus, A. Marcinek, B. Kalyanaraman and A. Sikora, Free Radical Biol. Med., 2016, 95, 323–332 CrossRef CAS PubMed.
- A. N. Pham, G. Xing, C. J. Miller and T. D. Waite, J. Catal., 2013, 301(116), 54–64 CrossRef CAS.
- L. Mascaretti, A. Dutta, Š. Kment, V. M. Shalaev, A. Boltasseva, R. Zbořil and A. Naldoni, Adv. Mater., 2019, 31, 1805513 CrossRef PubMed.
- J. Huang, J. Zhou, J. Zhuang, H. Gao, D. Huang, L. Wang, W. Wu, Q. Li, D. P. Yang and M. Y. Han, ACS Appl. Mater. Interfaces, 2017, 9, 36606–36614 CrossRef CAS PubMed.
- X. Zhang, Y. Guo, J. Tian, B. Sun, Z. Liang, X. Xu and H. Cui, Appl. Catal., B, 2018, 232, 355–364 CrossRef CAS.
- Y. Wang, Z. Li, Y. Hu, J. Liu, M. Guo, H. Wei, S. Zheng, T. Jiang, X. Sun, Z. Ma, Y. Sun, F. Besenbacher, C. Chen and M. Yu, Biomaterials, 2020, 255, 120167 CrossRef CAS PubMed.
- P. Stiefel, S. S. Emrich, K. M. Weber and Q. Ren, BMC Microbiol., 2015, 15, 36 CrossRef PubMed.
- A. H. Beckett, S. J. Patki and A. E. Robinson, Nature, 1958, 181, 712 CrossRef CAS PubMed.
- S. Nakamura, M. Shibuya, H. Nakashima, R. Hisamura, N. Masuda, T. Imagawa, M. Uehara and K. Tsubota, Invest. Ophthalmol. Visual Sci., 2007, 48, 1552–1558 CrossRef PubMed.
- F. F. Han, Y. H. Gao, C. Luan, Y. G. Xie, Y. F. Liu and Y. Z. Wang, J. Dairy Sci., 2013, 96, 3471–3487 CrossRef CAS PubMed.
- M. H. Abraham and W. E. Acree, J. Mol. Liq., 2014, 197, 157–159 CrossRef CAS.
- T. Okamoto, Int. J. Mol. Med., 2005, 16, 275–278 CAS.
- Q. Zhang, X.-H. Zhao and Z.-J. Wang, Toxicol. In Vitro, 2009, 23, 797–807 CrossRef CAS PubMed.
- N. Strbo and N. Yin, Adv. Wound Care, 2014, 3, 492–501 CrossRef PubMed.
- S. L. Percival, S. M. McCarty and B. Lipsky, Adv. Wound Care, 2015, 4, 373–381 CrossRef PubMed.
- A. Nain, H. H. Huang, D. M. Chevrier, Y. T. Tseng, A. Sangili, Y. F. Lin, Y. F. Huang, L. Chang, F. C. Chang, C. C. Huang, F. G. Tseng and H. T. Chang, Nanoscale, 2021, 13, 18632–18646 RSC.
- J. M. Munita and C. A. Arias, Microbiol. Spectrum, 2016, 4, 2–4 Search PubMed.
- G. Y. Huang, W. J. Chang, T. W. Lu, I. L. Tsai, S. J. Wu, M. H. Ho and F. L. Mi, Chem. Eng., 2022, 431, 134059 CrossRef CAS.
- Y. Lu, H. Li, J. Wang, M. Yao, Y. Peng, T. Liu, Z. Li, G. Luo and J. Deng, Adv. Funct. Mater., 2021, 31, 2105749 CrossRef CAS.
- A. S. Eming, T. Krieg and J. M. Davidson, J. Dermatol., 2007, 127, 514–525 Search PubMed.
- R. S. Kirsner and W. H. Eaglstein, Dermatol. Clin., 1993, 11, 629–640 CrossRef CAS.
- Y. Fu, J. Zhang, Y. Wang, J. Li, J. Bao, X. Xu, C. Zhang, Y. Li, H. Wu and Z. Gu, Carbohydr. Polym., 2021, 257, 117598 CrossRef CAS PubMed.
- N. X. Landén, D. Li and M. Ståhle, Cell. Mol. Life Sci., 2016, 73, 3861–3885 CrossRef PubMed.
- J. B. Acosta, D. G. D. Barco, D. C. Vera, W. Savigne, P. L. Saura, G. G. Nieto and G. S. Schultz, Int. Wound J., 2008, 5, 530–539 CrossRef PubMed.
- N. Bryan, H. Ahswin, N. Smart, Y. Bayon, S. Wohlert and J. A. Hunt, Eur. Cell Mater., 2012, 24, e65 CrossRef PubMed.
- A. Kitani, I. Fuss, K. Nakamura, F. Kumaki, T. Usui and W. Strober, J. Exp. Med., 2003, 198, 1179–1188 CrossRef CAS PubMed.
- M. L. Usui, J. N. Mansbridge, W. G. Carter, M. Fujita and J. E. Olerud, J. Histochem. Cytochem., 2008, 56, 687–696 CrossRef CAS PubMed.
- S. Sanjabi, L. A. Zenewicz, M. Kamanaka and R. A. Flavell, Curr. Opin. Pharmacol., 2009, 9, 447–453 CrossRef CAS PubMed.
- S. Kumar and A. K. Pandey, Sci. World J., 2013, 162750 Search PubMed.
- A. Sindrilaru, T. Peters, S. Wieschalka, C. Baican, A. Baican, H. Peter, A. Hainzl, S. Schatz, Y. Qi and A. Schlecht, J. Clin. Invest., 2011, 121, 985–997 CrossRef CAS PubMed.
- F. O. Martinez and S. Gordon, F1000Prime Rep., 2014, 6, 13 Search PubMed.
- L. Qi, H. Yu, Y. Zhang, D. Zhao, P. Lv, Y. Zhong and Y. Xu, Oncotarget, 2016, 7, 71673–71685 CrossRef PubMed.
- K. E. Johnson and T. A. Wilgus, Adv. Wound care, 2014, 3, 647–661 CrossRef PubMed.
- H. Kim, S. Y. Wang, G. Kwak, Y. Yang, I. C. Kwon and S. H. Kim, Adv. Sci., 2019, 6, 1900513 CrossRef CAS PubMed.
- H. Wang, H. Wang, J. Guan, W. Guan and Z. Liu, Sci. Rep., 2023, 13, 9670 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of the materials, experimental methods, and Fig. S1–S22. See DOI: https://doi.org/10.1039/d3nh00275f |
| This journal is © The Royal Society of Chemistry 2023 |