Recent progress in nanomaterials for bacteria-related tumor therapy
Fuping
Zhang
a,
Shuyu
Wang
*b,
Shuo
Yang
c,
Feihe
Ma
 *ad and
Hui
Gao
*a
*ad and
Hui
Gao
*a
aSchool of Materials Science and Engineering, Tiangong University, Tianjin 300387, China. E-mail: feihema@tiangong.edu.cn; huigao@tiangong.edu.cn
bSchool of Environmental Science and Engineering, Tiangong University, Tianjin 300387, China. E-mail: wangshuyu@tiangong.edu.cn
cDepartment of Medical Statistics, School of Public Health, Sun Yat-sen University, Guangzhou 510080, China
dKey Laboratory of Functional Polymer Materials of Ministry of Education, Institute of Polymer Chemistry and College of Chemistry, Nankai University, Tianjin, 300071, P.R. China
First published on 24th February 2024
Abstract
Many studies suggest that tumor microbiome closely relates to the oncogenesis and anti-tumor responses in multiple cancer types (e.g., colorectal cancer (CRC), breast cancer, lung cancer and pancreatic cancer), thereby raising an emerging research area of bacteria-related tumor therapy. Nanomaterials have long been used for both cancer and bacterial infection treatment, holding great potential for bacteria-related tumor therapy. In this review, we summarized recent progress in nanomaterials for bacteria-related tumor therapy. We focus on the types and mechanisms of pathogenic bacteria in the development and promotion of cancers and emphasize how nanomaterials work. We also briefly discuss the design principles and challenges of nanomaterials for bacteria-related tumor therapy. We hope this review can provide some insights into this emerging and rapidly growing research area.
1. Introduction
Increasing evidence suggests that there are various bacteria existing in tumor tissues, and the intratumoral microbiome plays important roles in tumor genesis, progression, metastasis, immune response and chemosensitivity.1–3 For example, intratumoral bacteria can directly cause DNA damage, leading to increased gene mutations, causing cancer and promoting the growth of cancer cells.4,5 The intratumoral bacteria can also convert chemotherapy drugs into inactive forms6 or upregulate tumor cell autophagy,7 causing chemotherapy resistance. Moreover, intratumoral bacteria can affect the tumor immune microenvironment, inhibiting anti-tumor immune responses and making the tumors more aggressive.8To date, the use of antibiotics remains the most effective and widely used strategy against bacterial infections, which has shown benefits for bacteria-related tumor therapy.9 However, systemic or oral administration of antibiotics lacks anti-bacterial specificity, causing undesired dysbiosis of gut microbiota.10,11 Moreover, the poor accumulation of free antibiotics in the infected tumor tissues requires a large dose and repeated administration of antibiotics to efficiently kill bacteria, which can enhance gut dysbiosis and develop drug resistance.12 The rapid developments in nanotechnology and materials science provide advanced strategies against both bacterial and cancer.13,14 In comparison with small molecules, nanomaterials have a higher surface-to-volume ratio, providing a good tailorable surface for functionality modifications.15 Nanomaterials themselves can directly exhibit antibacterial or antitumor activities, such as silver nanoparticles,16 carbon-based nanomaterials,17 nanozymes18 and polycationic nanomaterials.19 Meanwhile, nanomaterials including mesoporous silica nanomaterials,20 liposomes21 and polymeric nanoparticles22 can also be used as drug carriers to effectively deliver antibacterial and anti-tumor drugs to infection or tumor sites, improving the therapeutic efficacy and reducing side effects of the free drugs.23,24 Therefore, nanomaterials certainly hold great potential for bacteria-related tumor therapy.
In this review, the recent progress in nanomaterials for bacteria-related tumor therapy is summarized. The main types and mechanisms of pathogenic bacteria in the development and promotion of cancers are first introduced. Then, we summarize the current nanomaterials for the treatment of bacteria-related tumors including CRC, breast cancer, lung cancer and pancreatic cancer, and discuss the mechanisms. Finally, we briefly discussed the challenges in the area of development of nanomaterials for bacteria-related tumor therapy. We hope this review will provide some insights into this emerging and rapidly growing research area.
2. Pathogenic bacteria in tumors
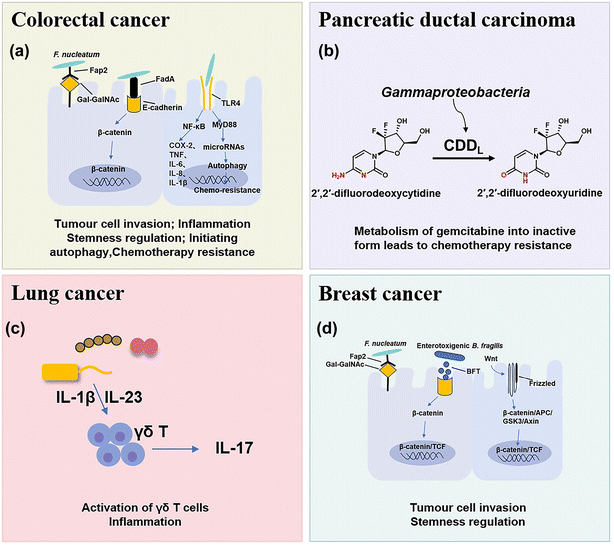
The microbial composition in tumors shows cases distinct patterns (Fig. 1). For example, rectal tumors predominantly harbor Firmicutes and Bacteroidetes.25 Pancreatic cancer exhibits a dominance of Proteus akin to the normal duodenal microbiome, possibly indicating bacterial migration from the duodenum through the pancreatic duct.3,26 Proteobacteria and Firmicutes are present across all cancer types, while their ratio (P/F) varies among tumors, potentially holding significance in understanding individual cancer microbiomes.27 The non-gastrointestinal tumors such as breast, lung, and ovarian cancers display the presence of actinomycetes such as corynebacteriaceae and micrococcaceae.1 These microbial distinctions in different cancers provide insights into potential relationships between microbial presence and tumor development. Here, we briefly discuss the main types of pathogenic bacteria and their mechanisms in the oncogenesis of cancers including CRC, pancreatic cancer, lung cancer and breast cancer.2.1 CRC
CRC is one of the most common causes of cancer death in the world, and chemotherapy and immunotherapy are most used clinically for the treatment of CRC.28 The intestine contains approximately 3 × 1013 bacterial cells that are symbiotic with the host, which play important roles in the occurrence and treatment of CRC and have attracted increasing interest in recent years.29In mouse models of CRC, F. nucleatum can drive a pro-inflammatory tumor microenvironment and suppress anti-cancer immune responses. F. nucleatum can activate the NF-κB pathway, and upregulate pro-inflammatory factors (COX-2, TNF, IL-6, IL-8, and IL-1β),35 thus promoting chronic inflammation and infiltrating intra-tumoural myeloid cells to enhance tumorigenesis.36 In addition, some studies have preliminarily found that F. nucleatum may activate TLR4 innate immune signaling, subsequently downregulation certain microRNAs to initiate the autophagy pathway, and reduce the sensitive response of F. nucleatum-treated CRC cells to 5-FU and oxaliplatin chemotherapy through autophagy.7,37 Moreover, it was found that short-chain fatty acids (SCFAs), a derivative metabolite of F. nucleatum, can further promote its role in carcinogenesis by changing the function of the immune system.38 SCFAs can cause numerous changes in different types of immune cells such as Treg cells, innate lymphoid cells type 3 (ILC3s), neutrophils and dendritic cells, and usually affect the immune system in a receptor-mediated way.39
2.2 Pancreatic cancer
Pancreatic ductal carcinoma (PDAC) is a highly malignant tumor with a hidden location, low early diagnosis rate, poor efficacy of various treatments, and extremely poor prognosis.42 The survival rate within five years after diagnosis is less than 10%.43 Many studies have found that the intratumoral microbial composition of long-term survival (LTS) patients and short-term survival (STS) patients with PDAC is varied, and regulation of the tumor microbiome can affect tumor growth and immune infiltration.442.3 Lung cancer
Lung cancer is the most common primary malignant tumor and the 1st cause of cancer deaths worldwide.46 Because of the presence of mucosa, lung tissue attracts a large number of microorganisms to colonize, such as Staphylococcus aureus, Streptococcus pneumoniae, and Pseudomonas aeruginosa.47 Due to the limitation of technology, few viable microbial cells can be isolated from healthy lungs, so the intervention mechanism of lung microflora is rarely studied.Tsay et al.48 demonstrated that Streptococcus and Veillonella enriched in the lower respiratory tract of lung cancer patients affect the proliferation, survival and tissue invasion of lung cancer cells by up-regulating RK/MAPK and PI3K/AKT pathways. Jin et al.49 used the KP mouse model of lung adenocarcinoma (LUAD) driven by Kras activation point mutation and p53 deletion, which clearly proved that the increase of lung bacteria and ecological imbalance would accelerate LUAD by producing γδT cells and IL-17. The evidence is mounting that bacterial microbe-associated molecular patterns (MAMPs) and lipoteichoic acid (LTAs) will lead to the activation of the transcription factors, such as NF-κB and AP-1, thus inducing inflammation and promoting lung cancer.49
2.4 Breast cancer
Breast cancer is the most common type of cancer and the leading cause of cancer death in women.50 Fu et al.51 reported that the tumors of spontaneous mouse mammary tumor (BT) model mice contained a large number of intracellular bacteria, similar to human breast cancer. Under the physiological conditions, these intracellular bacteria can move with cancer cells in the circulation and play a key role in tumor metastasis and colonization.513. Nanomaterials for bacteria-related tumor therapy
As discussed above, tumor-associated bacteria can metabolize drugs into inactive forms, induce drug resistance, and regulate the response of the immune system, thus having a significant impact on the therapeutic effect.54 With the continuous development and perfection of nano-materials, nano-drug delivery systems including liposomes, polymeric micelles, and inorganic nanoparticles provide new possibilities for both bacterial infection and cancer therapies by improving the pharmacokinetic behavior of drugs, increasing the stability of drugs, and realizing targeted drug delivery and controlled release.55 Liposomes are spherical nanoparticles composed of phospholipid bilayer membranes, and their size is usually between 50 and 500 nanometers. Liposome nanoparticles can encapsulate hydrophilic and hydrophobic drugs and release drugs in tumor cells, thus achieving targeted drug delivery and controlled release.56 A polymeric micelle is a kind of nanoparticle formed by polymer self-assembly, which can encapsulate and release chemotherapy drugs and has good biocompatibility.57 Mesoporous silica nanoparticles are considered one of the most potential nano-carriers of antibacterial drugs because of their porosity, large specific surface area and easy modification. Gold nanoparticles have also been studied for cancer treatment because of their excellent antibacterial mechanism.58 Nanozymes with oxidase activity can selectively catalyze the tumor microenvironment and generate a large amount of ROS, so it is widely used in the treatment of tumors.593.1 Chemotherapy
Chemotherapy is one of the most commonly used and effective means to treat cancer. The basic principle is that chemical drugs enter the tumors to inhibit and kill cancer cells, thus achieving the purpose of treating cancer.55 Previous studies have shown that tumor-related bacteria can lead to chemotherapy resistance by activating autophagy or inducing drug inactivation.54 Therefore, the use of nano-drugs to regulate bacteria closely related to cancer progression and chemotherapy resistance can be used as an important method to improve the efficacy of chemotherapy.Conventional liposome generally lacks self-adaptability which is important for the tumor accumulation of nanomedicines. Shi et al.60 synthesized a new type of pH-responsive liposome 2-(4-((1,5-bis(octadecyloxy)-1,5-dioxopentan-2-yl) carbamoyl) pyridin-1-ium-1-yl) acetate, abbreviated DCPA in 2021, which can complex with water molecules through hydrogen bonds (DCPA-H2O). Due to its high HOMO binding energy, it can be rapidly protonated in acidic environments. Therefore, DCPA liposomes circulating in the blood can quickly target the surface of negatively charged bacteria or tumor cells and release the loaded drugs. The strong self-targeting properties make DCPA-H2O liposomes a very promising nanocarrier for the treatment of bacteria-related tumors. Recently, they used DCPA liposomes to co-deliver the antibiotic ciprofloxacin and the chemotherapy drug gemcitabine to bacteria-related tumors (Fig. 2a and b).61In vivo and in vitro results show that GC-DCPA-H2O can accumulate in tumor tissue very fast and eliminate tumor bacteria with high efficiency, thereby preventing the inactivation of gemcitabine and significantly inhibiting tumor growth.
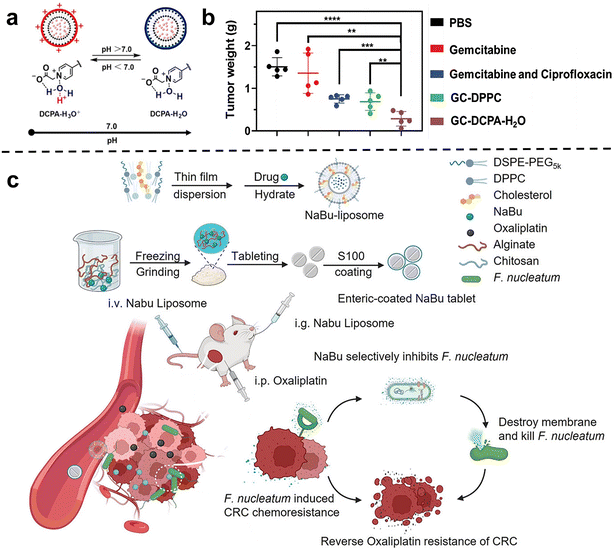 | ||
| Fig. 2 (a) DCPA-H2O has the function of pH response and is negatively charged in a normal physiological environment, and will be protonated rapidly in a tumor or infected site (pH < 6.8). Reproduced with permission from ref. 60. Copyright 2021, Wiley. (b) Tumor weight at sacrifice. Treatment either consisted of tail-vein injections of PBS, free gemcitabine or gemcitabine combined with ciprofloxacin in solution, GC-DPPC or self-targeting GC-DCPA-H2O liposomes. Reproduced with permission from ref. 61. Copyright 2023, Wiley. (c) Schematic illustration of the NaBu-liposome to regulate F. nucleatum-induced oxaliplatin chemoresistance in colorectal cancer. Reproduced with permission from ref. 62. Copyright 2023, Elsevierc. | ||
Chen et al.62 tried to use sodium butyrate (NaBu), the main product of intestinal microbial fermentation, to treat bacterially infected colorectal cancer (Fig. 2c). However, since sodium butyrate is a small hydrophilic molecule, it is easily excreted in the body. In order to improve bioavailability, sodium butyrate was embedded in liposomes and injected intravenously into subcutaneous colorectal tumors, orthotopic colorectal tumors, and spontaneously formed colorectal tumors in F. nucleatum-infected mice. They showed that the liposome-embedded sodium butyrate can effectively inhibit the growth of F. nucleatum in colorectal tumors and reduce F. nucleatum-induced chemotherapy resistance, achieving effective tumor chemotherapy, and prolonging the overall survival of mice.
Zhang et al. 63 isolated a phage strain from human saliva that can specifically lyse F. nucleatum. This phage was used to design a bio-non-biological hybrid nanomaterial, that is, the phage was modified in dextran nanoparticles (DNPs) encapsulating the anti-colorectal cancer drug irinotecan (IRT)(Fig. 3a). The results showed that in mice with orthotopic colorectal tumors or spontaneously formed colorectal tumors, oral or intravenous administration could inhibit the proliferation of F. nucleatum and significantly improve the efficiency of CRC chemotherapy treatment. Chen et al.64 designed nitroreductase-mediated supramolecular self-assembly. After enzyme catalysis, the precursor undergoes a self-elimination reaction to form a gel, which self-assembles into supramolecular nanofibers. The supramolecular nanofibers then target and capture F. nucleatum to inhibit intratumoral bacteria and enhance subsequent chemotherapy.
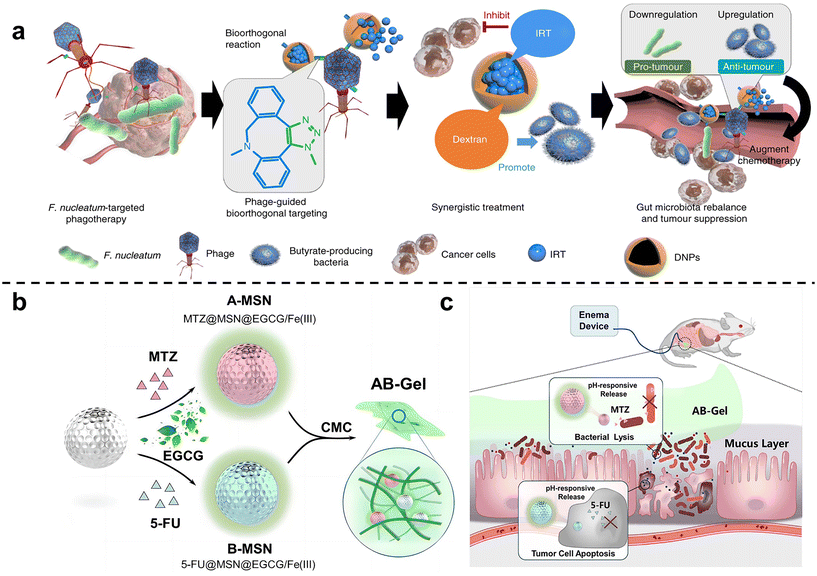 | ||
| Fig. 3 (a) Illustration of the phage-guided biotic-abiotic hybrid nanosystem and its mechanism. Reproduced with permission from ref. 63. Copyright 2019, Springer Nature. (b) EGCG/Fe(III) network encapsulated MSN loaded with MTZ and 5-FU, mixing into CMC, constructed as the AB-Gel. (c) AB-Gel was directly perfused into the colon to the tumor site, which not only realized the rapid and accurate release of drugs but also reduced the side effects, thus increasing the chemotherapy effect. Reproduced with permission from ref. 66. Copyright 2021, Elsevier. | ||
Chen et al.65 used dendritic mesoporous silicas (DMSNs) to load silver nanoparticles (Ag NPs), and combined with chemotherapy to treat F. nucleatum-infected colorectal cancer. DMSN was firstly modified with Ag+ through sulfhydryl bonds, and then Ag NPs were grown in situ in the mesopores of SH-DMSNs through the sodium borohydride rapid reduction method. Finally, the chemotherapy drug epirubicin (EPI) was loaded to form antibacterial and antitumor Ag@DMSNs-EPI NPs (ADEN for short). The designed antibacterial-antitumor nanomedicines can effectively treat colorectal cancer by clearing F. nucleatum within tumors and reshaping the tumor microenvironment. Chen et al.66 used mesoporous silica nanoparticles to prepare a drug delivery gel (Fig. 3b and c). The antibiotic metronidazole and the chemotherapy drug 5-fluorouracil (5-FU) were loaded in MSN and then blended with carboxymethylcellulose (CMC) to obtain the anti-CRC gel AB-Gel. In vitro experiments showed that antibiotic-loaded A-MSN has strong antibacterial activity, and 5-FU-loaded B-MSN has strong anti-cancer ability. In vivo studies further confirmed that combined microbiota modulation can significantly improve the efficacy of colorectal cancer treatment.
In order to inhibit chemotherapy resistance caused by commensal bacteria in lung tumors, Han et al.67 constructed an inhalable gallium-based metal nanoparticle coated with capsular polysaccharidecp (CP), defined as GaTa-cpNPs (Fig. 4a). Tannin (Ta) is a natural polyphenol with many phenolic hydroxyl groups. GaTa NPs were successfully synthesized by self-assembly through a coordination reaction between tannin and gallium metal ions. The Streptococcus pneumoniae capsular polysaccharide coating can enable GaTa-cp NPs to effectively adhere to lung tissue and masquerade as normal tissue components to reduce immune clearance in the body. In addition, they also included the chemotherapy drug epoposide to verify the drug delivery and tumor treatment capabilities of GaTa-cp NPs. In the acidic environment of the tumor, Ga3+ and the drug Eto are released. In multiple Escherichia coli Nissle 1917 (EcN)-infected mouse lung cancer models, local lung microbiota can be effectively depleted to inhibit bacterial-induced chemotherapy resistance, thereby enhancing lung cancer treatment. Recently, Qiu et al.68 used micelles to jointly deliver antibiotics and anticancer drugs, called colistin cross-linked gemcitabine micelles (CCGM) for EcN-infected breast cancer therapy (Fig. 4b). Once CCGM is delivered into the tumor microenvironment, it triggers a glutathione response, and the self-immolating linker is disconnected, resulting in the release of colistin and gemcitabine, thereby inhibiting the growth of intratumoral bacteria and eliminating bacterial-induced tumor resistance to enhance tumor chemotherapy.
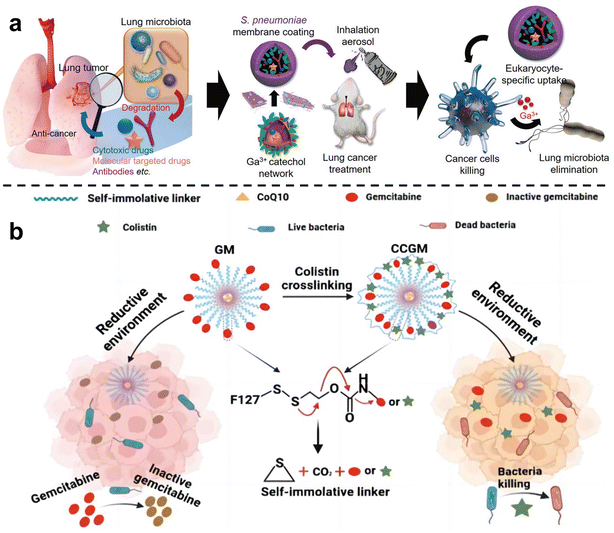 | ||
| Fig. 4 (a) Scheme for the synthesis of the inhalable capsular polysaccharide (CP)-camouflaged gallium-based metal–organic network (MON). It can effectively eliminate various microorganisms in the lung tumor model, thus reducing the drug resistance induced by bacteria. Reproduced with permission from ref. 67. Copyright 2023, Wiley. (b) Schematic illustration of CCGM eliminating bacterial-induced cancer drug resistance. Reproduced with permission from ref. 68. Copyright 2022, American Chemical Society. | ||
Recently, our group has developed a series of polymeric nanosystems to overcome F. nucleatum caused issues including chemotherapy resistance and immunosuppression in CRC therapy. Based on our decadal works on supramolecular assembly, we rationally designed a size-switchable supramolecular nanomedicine (PG-Pt-LA/CB[7]) to effectively reverse F. nucleatum-induced chemotherapy resistance of CRC to oxaliplatin (Fig. 5a–f).69 This was the first report on using a natural fatty acid lauric acid (LA) as an antibacterial agent to selectively kill F. nucleatum, which overcomes the drawbacks of broad-spectrum antibiotics. In detail, LA and platinum(IV) oxaliplatin prodrug (OxPt-COOH) were coupled with hyperbranched poly(glycidol) (PG) and self-assembled to form PG-Pt-LA nanoparticles, and then cucurbit[7]uril (CB[7]) was added to induce supramolecular assembly. The PG-Pt-LA/CB[7] (∼210 nm) is stable in blood with a long half-life, while disassembles in response to the CRC microenvironment and turns into small PG-Pt-LA NPs (∼10 nm) to penetrate the tumor deeply. In vivo results confirmed that LA has a significant effect in eliminating F. nucleatum present in solid CRC tumors and can reduce chemotherapy resistance, thereby increasing the therapeutic effect of chemotherapy drugs on tumors.
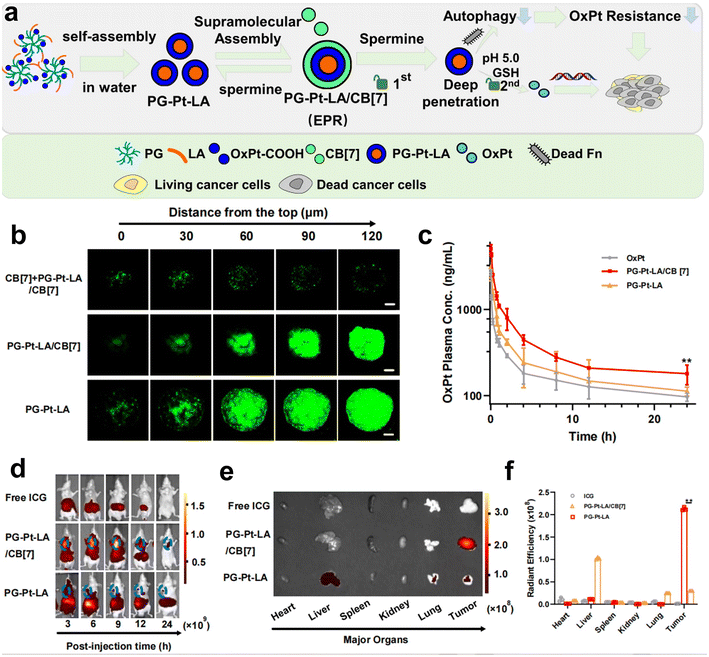 | ||
| Fig. 5 (a) The assembly diagram of supramolecular nanomedicine. (b) CLSM observation on the HT29 3D multicellular spheroids subjected to prior incubation of CB[7] with the aim of depletion of spermine, followed by different treatments. (c) Pharmacokinetics of OxPt, PG-Pt-LA/CB[7] and PG-Pt-LA upon intravenous dosage. (d) Change of biodistributions of free ICG, ICG-labeled PG-Pt-LA/CB[7] and ICG-labeled PG-Pt-LA with time. (e) Fluorescence imaging and (f) quantitative analysis of the dissected organs and tumors after different treatments. Reproduced with permission from ref. 69. Copyright 2022, Elsevier. | ||
Intracellular F. nucleatum was hard to eliminate using free antibiotics due to its inability to traverse cell membranes. In a recent study, Li et al.70 fabricated an acidity-responsive nanomaterial that can deliver LA and OxPt-COOH into tumor cells to kill the intratumoral bacteria and treat cancer (Fig. 6a–d). OLP and PP can self-assemble in water to form stable nanoassembly, in which the PBA can specifically bind to tumor cells and facilitate receptor-mediated endocytosis. After cellular uptake, the OLP/PP nanoassembly can disassemble in response to the tumor's acidic microenvironment (pH 5.0) to trigger the release of drugs. The results showed that the constructed OLP/PP nanoassembly can effectively eliminate both extracellular and intracellular F. nucleatum and inhibit tumor growth.
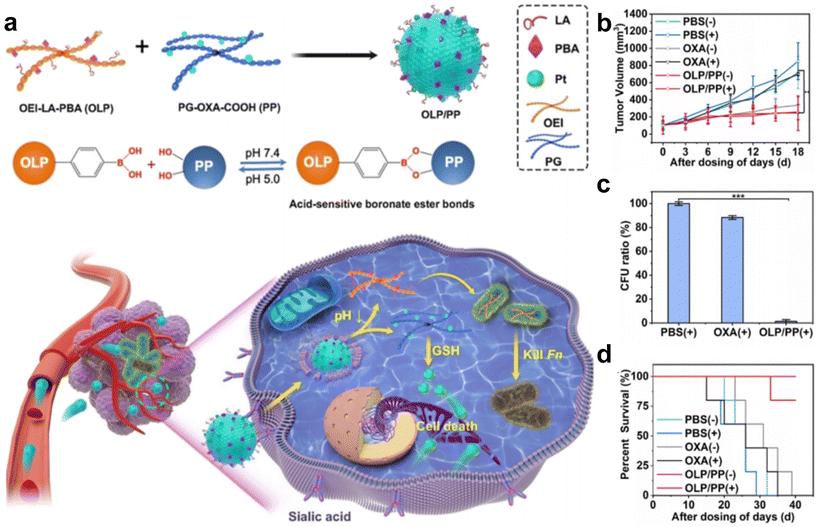 | ||
| Fig. 6 (a) Schematic illustration of OLP/PP for the treatment of CRC tumors colonized with F. nucleatum. (b) Tumor volume, (c) quantification of F. nucleatum in tumors and (d) survival fractions of the F. nucleatum-infected HT29 tumor-bearing mice under diverse treatments. Reproduced with permission from ref. 70. Copyright 2023, American Chemical Society. | ||
3.2 Immunotherapy
Cancer immunotherapy aims to activate the immune system of patients and kill cancer cells by their own immune function.55 This new treatment method is expected to achieve the goal of long-term survival with a tumor or a complete cure of tumor and is becoming one of the important development directions in the field of cancer treatment. Recent studies have shown that the enrichment of F. nucleatum in colorectal cancer can drive the formation of tumor microenvironment in immunosuppression.71 As such, eliminating tumor-associated bacteria may be an effective strategy to improve the effect of tumor immunotherapy.Chen et al.72 designed a liposome-based nanomedicine that mimics F. nucleatum to reverse immunotherapy resistance for the infected CRC treatment (Fig. 7a and b). The antibiotic colistin is loaded into liposomes, and then fused with the cytoplasmic membrane FM of F. nucleatum to obtain colistin-loaded FM fusion liposomes (colistin-LipoFM). Since the surface of the nuclear-cytoplasmic membrane (FM) contains Fap-2, it can specifically target Gal-GalNAc that was overexpressed on the surface of colorectal tumor cells, allowing colistin to effectively accumulate at the site of the tumor infection without affecting intestinal microorganisms and selectively kill tumor-colonizing F. nucleatum. Recently, Huang et al.73 also designed an antibiotic silver-tinidazole complex encapsulated liposomes (LipoAgTNZ) (Fig. 7c and d). This LipoAgTNZ can accurately target anaerobic bacteria in infected tumors, release silver particles and tinidazole in the acidic tumor microenvironment, and kill F. nucleatum in the tumor. Interestingly, they found that eliminating bacteria in colorectal cancer tumors can expose microbial epitopes, thus activating immune responses to enhance tumor treatment efficacy.
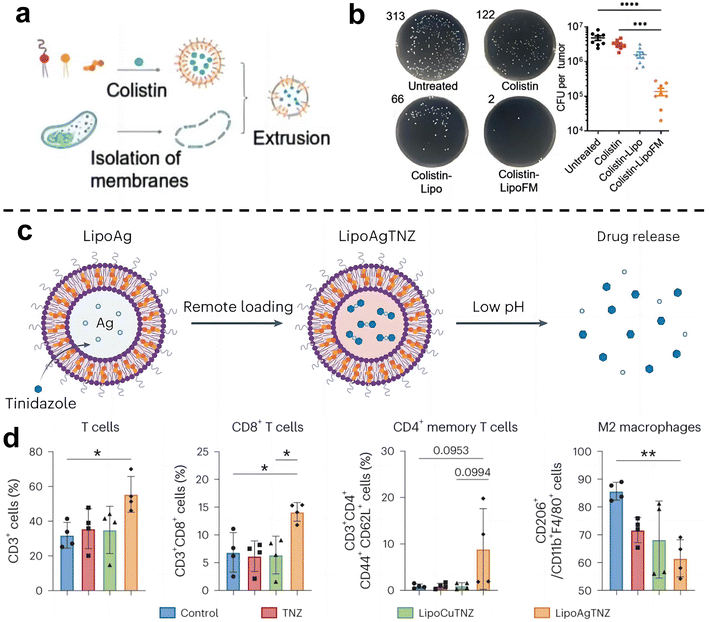 | ||
| Fig. 7 (a) Synthetic legend of Colistin-LipoFM. (b) Colony plate images (left) and the colony-forming units (CFUs) (right) showing the F. nucleatum abundance in homogenized tumors of mice treated with colistin-LipoFM, colistin-Lipo and free colistin. Reproduced with permission from ref. 72. Copyright 2023, Wiley. (c) Illustration of remote loading by a silver nitrate gradient and drug release in response to low pH. (d) Analyses of the immune cell population in the tumor microenvironment after treatment. Reproduced with permission from ref. 73. Copyright 2023, Springer Nature. | ||
Zhang et al.74 used hyaluronic acid (HA)-modified photoluminescent organosilica nanodots (OSiNDs) to deliver gemcitabine (Gem) and the antibiotic ciprofloxacin (Cip) into bacterially infected mouse colon tumors (Fig. 8a). After treatment with Gem/Cip@SiPNG@HA, the killed bacteria promoted the maturation of antigen-presenting dendritic cells (DCs), while Gem reduced the levels of MDSCs, thereby triggering T cells enhanced cancer immunotherapy. Song et al.75 designed a bacteria-targeting nanomaterial to target bacteria living in tumor sites. Bacterial lipophosphocholic acid antibodies extracted from bacterial walls were modified on mesoporous silica nanoparticles, which could precisely target LTA on the bacterial surface within tumors and deliver anti-tumor drugs. Ma et al.76 also designed an inhalable, dual-loaded mesoporous silica nanoparticle (MSN@DOX-AMP) to treat lung cancer (Fig. 8b). Efficient loading of anti-tumor antibiotic doxorubicin (DOX) and antimicrobial peptide HHC36 using physical adsorption and thiol–ene click chemistry. In mice with lung cancer with Staphylococcus aureus as a commensal bacterium, it was demonstrated that MSN@DOX-AMP could strongly destroy the tumor/bacteria symbiosis system of lung cancer, achieving better therapeutic effects that compared with treating tumors or bacteria alone.
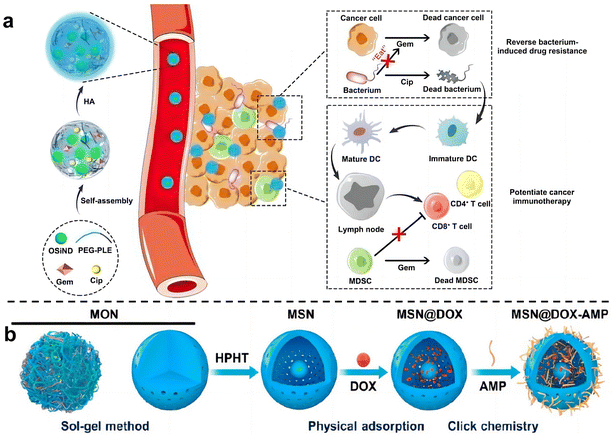 | ||
| Fig. 8 (a) Schematic diagram illustrates the preparation and mechanism of Gem/Cip@SiPNG, which can reverse bacterial-induced chemotherapy resistance and potentiate cancer immunotherapy. Reproduced with permission from ref. 74. Copyright 2021, Wiley. (b) Preparation of the dual-drug-delivery mesoporous silica nanoparticles (MSNs). Reproduced with permission from ref. 76. Copyright 2023, American Chemical Society. | ||
3.3 Synergetic therapy
After carefully designed nano-system with two or more treatments can be skillfully combined to act on tumors together, thus producing remarkable synergistic effects. This combination therapy strategy can not only significantly improve the therapeutic effect, make the tumor inhibition rate reach more than 90% within 20 days, but also greatly reduce the toxic and side effects so that patients can bear fewer side effects during the treatment.77Qu et al.78 developed a gold nanoparticle-based antibacterial nanoplatform by using bovine serum albumin (BSA) as a carrier to improve biocompatibility and stability (Au@BSA-CuPpIX) (Fig. 9a). Gold nanoparticles (Au NPs) are captured through biomimetic mineralization and then modified with an antibacterial metalloporphyrin (CuPpIX) sonosensitizer to synthesize nanoparticles with ultra-small diameters. Reactive oxygen species (ROS) are generated under the action of ultrasound, and in vivo experiments showed that Au@BSA-CuPpIX can effectively kill bacteria in tumors and improve the therapeutic effect of colorectal cancer. In addition, the wrapped gold nanoparticles reduce the phototoxicity of metalloporphyrins and avoid severe inflammation and metal damage.
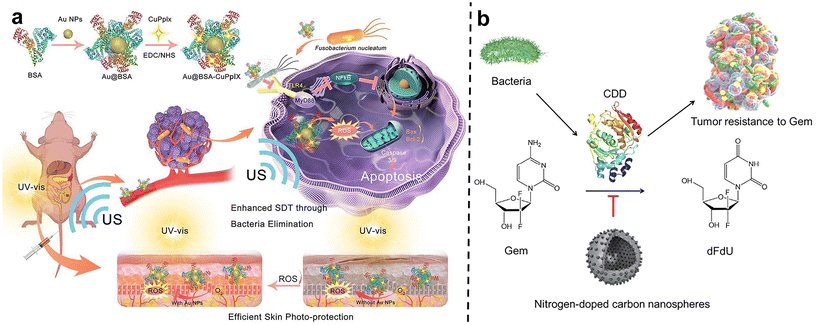 | ||
| Fig. 9 (a) The scheme describes the synthesis and mechanism of Au@BSA-CuPpIX. Au@BSA-CuPpIX, which can effectively eliminate F. nucleatum to reduce the level of apoptosis inhibitor protein in cancer cells, and enhance the tumor treatment effect of SDT. Au NPs can reduce the phototoxicity of CuPpIX in the skin, thus reducing skin damage. Reproduced with permission from ref. 78. Copyright 2023, American Chemical Society. (b) Schematic of N-CS-induced inhibitory effects against CDD expressed by bacteria to metabolize Gem into dFdU. Reproduced with permission from ref. 80. Copyright 2022, Elsevier. | ||
Wang et al.79 pioneered the construction of a highly biomimetic artificial protein-supported copper single-atom nanozyme (BSA-Cu SAN). Due to the multivalent nature of copper, BSA-Cu SAN can produce catalytic H2O2 to generate ROS, consume glutathione, kill intratumoral F. nucleatum, and effectively restore the autophagy level of tumor cells elevated by F. nucleatum. At the same time, bovine serum albumin is loaded on the nanozyme, which gives the nanozyme high hydrophilicity and biocompatibility, solving the problem of high metal ion toxicity, and achieving satisfactory tumor treatment effects. Xi et al.80 used N-doped carbon spheres (N-CSs) as a colorectal cancer treatment system and found that they have the dual functions of nanozymes and nanoinhibitors, capable of reversing bacterial-induced tumor drug resistance and improving immunity in pathogen-symbiotic tumors (Fig. 9b). First, N-CSs, as a peroxidase-like nanozyme, can catalyze H2O2 in the tumor microenvironment to produce highly cytotoxic hydroxyl radicals (˙OH) to kill cancer cells. Secondly, N-CSs, as nanoinhibitors, inhibited bacteria at tumor sites, successfully reversing bacterial-induced gemcitabine resistance and restoring tumor susceptibility to gemcitabine. It provides an effective way to overcome tumor resistance induced by intratumoral bacteria and to synergize chemotherapy with catalytic therapy.
Wang et al.81 proposed and synthesized a dual-cascade response nanoparticle (sNP@G/IR) with a shell-core structure that can eliminate intra-tumor bacteria and achieve effective drug delivery for EcN-infected pancreatic cancer therapy (Fig. 10a and b). Firstly, an amphiphilic GSH-responsive polymer (SGP) with good antibacterial activity and biocompatibility was synthesized, which contained the chemotherapy drug Gem and the photothermal agent IR1048 and was self-assembled to form NP@G/IR. It was further coated with hyaluronic acid (HA) with a tumor-targeting effect to obtain dual cascade reaction nanoparticles (sNP@G/IR). This inhibits the growth of bacterially colonized tumors by eliminating tumor-resident intracellular bacteria and killing cancer cells with precise delivery. Liu et al.82 synthesized an amphiphilic polymer by coupling hydrophilic polyethylene glycol (PEG) with the poorly water-soluble photosensitizer pyropheophoride-a (Ppa) and the antimicrobial peptide ubiquicidin (UBI) (Fig. 10c), and then self-assembled into polymer micelles (UPPM) in the aqueous solution. UBI peptides endow UPPM with excellent bacterial targeting capabilities. Under light, UPPM can kill bacteria by inducing bacterial membrane damage, thereby effectively inhibiting Gem metabolism. In subcutaneous PDAC animal models injected with bacteria, UPPM can eradicate intratumoral bacteria via photodynamic therapy without affecting the intestinal microbiota.
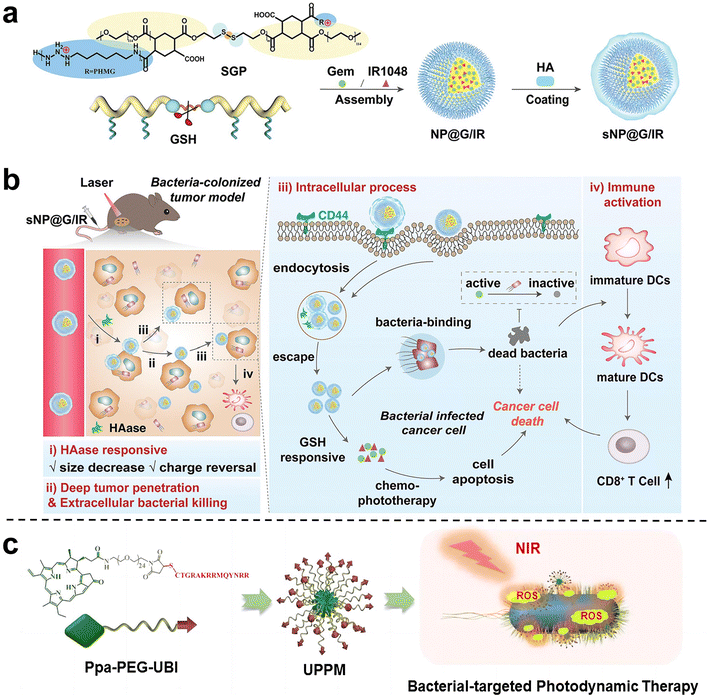 | ||
| Fig. 10 (a) Preparation process of sNP@G/IR. (b) Schematic diagram illustrating that sNP@G/IR can kill bacteria and activate immunity. After the HAase response, the particle size of sNP@G/IR decreases and the charge is reversed, which can achieve deep penetration of the tumor and bacterial killing. After rapid endocytosis, NP@G/IR is beneficial for the clearance of intracellular bacteria and immune activation. Reproduced with permission from ref. 81. Copyright 2023, Wiley. (c) Schematic illustration of the preparation process of UPPM micelles. Reproduced with permission from ref. 82. Copyright 2023, American Chemical Society. | ||
Immunotherapy is promising for the treatment of CRC, however, the therapeutic efficiency is also affected by F. nucleatum. Recently, Xin et al.83 developed an in situ activated anti-tumor and antibacterial nanoplatform (Cu2O/BNN6@MSN-Dex) that allows combined photothermal and gas therapy to enhance F. nucleatum-infected colorectal cancer immunotherapy (Fig. 11a–f). Boronized mesoporous silica nanoparticles MSN-PBA were first synthesized, and then cuprous oxide (Cu2O) and N,N′-di-sec-butyl-N,N′-dinitroso-1,4-phenylene diamine (BNN6) were encapsulated in MSN-PBA. Finally, polyhydroxydextran (Dex) was coated through dynamic cross-linking boronic acid bonds to obtain Cu2O/BNN6@MSN-Dex. Cu2O/BNN6@MSN-Dex can accumulate into tumor tissues and target tumor cells through the PBA ligand. After penetrating into the tumor, the loaded Cu2O will be sulfated by endogenous H2S overexpressed in tumors, resulting in copper sulfide with significant photoacoustic (PA) and photothermal (PT) properties. Under 808 nm laser irradiation, non-invasive PTT was performed with in situ local hyperthermia and induced BNN6 to produce NO. Dextran makes Cu2O/BNN6@MSN-Dex have good biocompatibility, responding in the tumor microenvironment, and is decomposed by glucanase, which is beneficial to the release of NO. In vivo and in vitro experiments demonstrated that combined photothermal and NO gas treatment can specifically eliminate F. nucleatum within tumors and significantly inhibit tumor growth, providing an effective strategy to enhance CRC treatment.
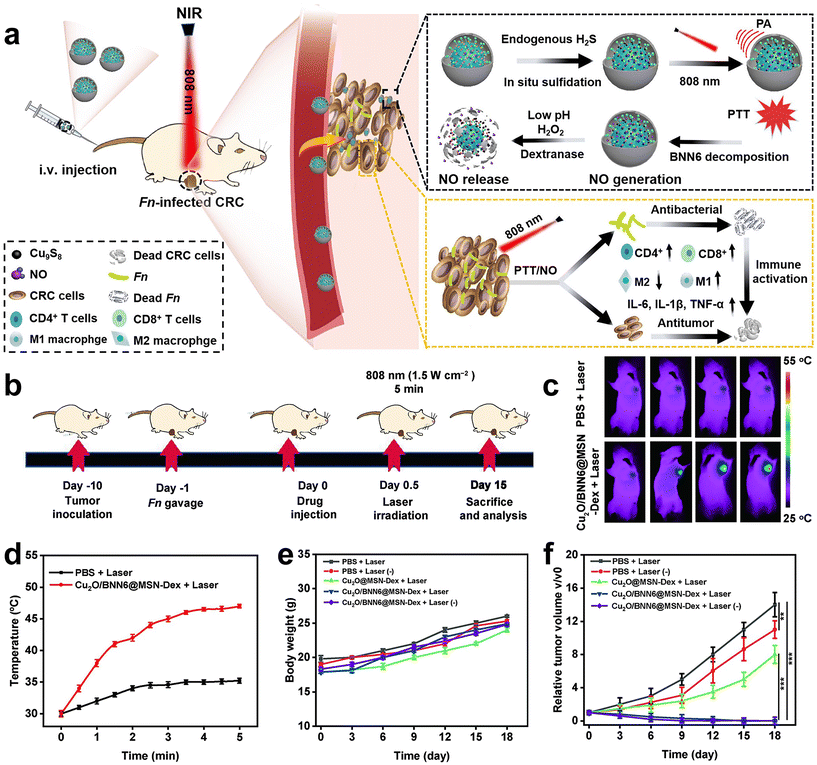 | ||
| Fig. 11 (a) Schematic diagram of the photothermal and gas combined therapy with Cu2O/BNN6@MSN-Dex. (b) Scheme showing the experimental schedule. (c) In vivo fluorescence imaging and (d) temperature versus time curves of F. nucleatum-infected HT29 tumor mice treated with PBS or Cu2O/BNN6@MSN-Dex under 808 nm laser irradiation. (e) Body weight and (f) tumor growth curves of F. nucleatum-infected mice bearing HT29 tumors after different treatments. Reproduced with permission from ref. 83. Copyright 2023, Elsevier. | ||
4. Conclusions and perspective
In this review, we summarized the key research progress in the field of nanomaterials for bacteria-related tumor therapy. Current studies mostly focus on targeting the tumor-commensal bacteria model and using nanomaterials to eradicate cancer-associated microorganisms to enhance tumor treatment effects. It is obvious that this strategy has shown prosperous significance. One key challenge for the construction of such nanomaterials is how to effectively integrate antibacterial and antitumor functions into a single system. More importantly, it is optimum for antibacterial agents to selectively exert antibacterial effects on intratumoral bacteria to prevent microbiome imbalance caused by unselective sterilization.Currently, there is relatively little research on the mechanism of the progression of bacteria colonization in tumor tissue. This should be a long process with many pathological changes. However, current animal models are too artificial. For example, when building a bacterial related tumor model, researchers usually use intratumoral injection or intravenous injection of a single bacterial species. Although this method is simple and feasible, it cannot truly simulate the diversity of the intratumoral microbiota and the complex bacteria-tumor interaction. Therefore, more attention needs to be paid to building a more authentic bacterial related tumor model in order to obtain more accurate and in-depth research results. Moreover, the effectiveness of such a therapeutic strategy should be further evaluated using the above models. Overall, this is an emerging research area which is worthy of attention. We believe more innovative methods and strategies will emerge in the near future, which will be helpful in improving tumor treatment effects and improving patients’ quality of life.
Author contributions
Fuping Zhang: writing–original draft, figures designing and investigation. Shuo Yang: investigation. Shuyu Wang, Feihe Ma and Hui Gao: writing–reviewing and editing, supervision, and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by The National Natural Science Foundation of China (U20A20260, 22075209, 52103180), a distinguished professor of Tianjin, the Training Project of Innovation Team of Colleges and Universities in Tianjin (TD13-5020), the Tianjin Enterprise Key Laboratory for Application Research of Hyaluronic Acid (KTRDHA-Z201901), and the Key Program of Tianjin Municipal Natural Science Foundation (22JCZDJC00570).References
- D. Nejman, I. Livyatan, G. Fuks, N. Gavert, Y. Zwang, L. T. Geller, A. Rotter-Maskowitz, R. Weiser, G. Mallel, E. Gigi, A. Meltser, G. M. Douglas, I. Kamer, V. Gopalakrishnan, T. Dadosh, S. Levin-Zaidman, S. Avnet, T. Atlan, Z. A. Cooper, R. Arora, A. P. Cogdill, Md A. W. Khan, G. Ologun, Y. Bussi, A. Weinberger, M. Lotan-Pompan, O. Golani, G. Perry, M. Rokah, K. Bahar-Shany, E. A. Rozeman, C. U. Blank, A. Ronai, R. Shaoul, A. Amit, T. Dorfman, R. Kremer, Z. R. Cohen, S. Harnof, T. Siegal, E. Yehuda-Shnaidman, E. N. Gal-Yam, H. Shapira, N. Baldini, M. G. I. Langille, A. Ben-Nun, B. Kaufman, A. Nissan, T. Golan, M. Dadiani, K. Levanon, J. Bar, S. Yust-Katz, I. Barshack, D. S. Peeper, D. J. Raz, E. Segal, J. A. Wargo, J. Sandbank, N. Shental and R. Straussman, Science, 2020, 368, 973–980 CrossRef CAS PubMed
.
- G. El Tekle and W. S. Garrett, Nat. Rev. Cancer, 2023, 23, 600–618 CrossRef CAS PubMed
.
- R. J. Knippel, J. L. Drewes and C. L. Sears, Cancer Discovery, 2021, 11, 2378–2395 CrossRef CAS PubMed
.
- P. J. Dziubańska-Kusibab, H. Berger, F. Battistini, B. A. M. Bouwman, A. Iftekhar, R. Katainen, T. Cajuso, N. Crosetto, M. Orozco, L. A. Aaltonen and T. F. Meyer, Nat. Med., 2020, 26, 1063–1069 CrossRef PubMed
.
- C. Pleguezuelos-Manzano, J. Puschhof, R. Bonnet, P. Quirke, M. Meyerson, E. Cuppen, R. van Boxtel and H. Clevers, Nature, 2020, 580, 269–273 CrossRef CAS PubMed
.
- L. T. Geller, M. Barzily-Rokni, T. Danino, O. H. Jonas, N. Shental, D. Nejman, N. Gavert, Y. Zwang and Z. A. Cooper, Science, 2017, 357, 1156–1160 CrossRef CAS PubMed
.
- T. Yu, F. Guo, Y. Yu, T. Sun, D. Ma, J. Han, Y. Qian, I. Kryczek, D. Sun, N. Nagarsheth, Y. Chen, H. Chen, J. Hong, W. Zou and J.-Y. Fang, Cell, 2017, 170, 548–563 CrossRef CAS PubMed
.
- C. Gur, Y. Ibrahim, B. Isaacson, R. Yamin, J. Abed, M. Gamliel, J. Enk, Y. Bar-On, N. Stanietsky-Kaynan, S. Coppenhagen-Glazer, N. Shussman, G. Almogy, A. Cuapio, E. Hofer, D. Mevorach, A. Tabib, R. Ortenberg, G. Markel, K. Miklić, S. Jonjic, C. A. Brennan, W. S. Garrett, G. Bachrach and O. Mandelboim, Immunity, 2015, 42, 344–355 CrossRef CAS PubMed
.
- F. Baquero and B. R. Levin, Nat. Rev. Microbiol., 2020, 19, 123–132 CrossRef PubMed
.
- F. Peyrusson, H. Varet, T. K. Nguyen, R. Legendre, O. Sismeiro, J.-Y. Coppée, C. Wolz, T. Tenson and F. Van Bambeke, Nat. Commun., 2020, 11, 2200 CrossRef CAS PubMed
.
- L. Maier, C. V. Goemans, J. Wirbel, M. Kuhn, C. Eberl, M. Pruteanu, P. Müller, S. Garcia-Santamarina, E. Cacace, B. Zhang, C. Gekeler, T. Banerjee, E. E. Anderson, A. Milanese, U. Löber, S. K. Forslund, K. R. Patil, M. Zimmermann, B. Stecher, G. Zeller, P. Bork and A. Typas, Nature, 2021, 599, 120–124 CrossRef CAS PubMed
.
- J. G. Liu, O. Gefen, I. Ronin, M. Bar-Meir and N. Balaban, Science, 2020, 367, 200–204 CrossRef CAS PubMed
.
- Y. Liu, L. Shi, L. Su, H. C. van der Mei, P. C. Jutte, Y. Ren and H. J. Busscher, Chem. Soc. Rev., 2019, 48, 428–446 RSC
.
- X. Kong, Y. Qi, X. Wang, R. Jiang, J. Wang, Y. Fang, J. Gao and K. C. Hwang, Prog. Mater. Sci., 2023, 134, 101070 CrossRef CAS
.
- A. Gupta, S. Mumtaz, C.-H. Li, I. Hussain and V. M. Rotello, Chem. Soc. Rev., 2019, 48, 415–427 RSC
.
- N. Dasgupta and C. Ramalingam, Environ. Chem. Lett., 2016, 14, 477–485 CrossRef CAS
.
- H. Sun, N. Gao, K. Dong, J. Ren and X. Qu, ACS Nano, 2014, 8, 6202–6210 CrossRef CAS PubMed
.
- M. Liang and X. Yan, Acc. Chem. Res., 2019, 52, 2190–2200 CrossRef CAS PubMed
.
- Y. Zhu, C. Xu, N. Zhang, X. Ding, B. Yu and F. J. Xu, Adv. Funct. Mater., 2018, 28, 1706709 CrossRef
.
- B. Li, Y. Liao, X. Su, S. Chen, X. Wang, B. Shen, H. Song and P. Yue, J. Nanobiotechnol., 2023, 21, 325 CrossRef PubMed
.
- S. Dong, Y. Bi, X. Sun, Y. Zhao, R. Sun, F. Hao, Y. Sun, Y. Wang, X. Li, W. Deng, X. Liu, J. Ha, L. Teng, P. Gong, J. Xie, B. Y. S. Kim, Z. Yang, W. Jiang and L. Teng, Small, 2022, 18, 2107690 CrossRef CAS PubMed
.
- M. Li, Y. Wang, L. Zhang, Q. Liu, F. Jiang, W. Hou, Y. Wang, H. Fang and Y. Zhang, ACS Nano, 2023, 17, 16703–16714 CrossRef CAS PubMed
.
- P. P. Kalelkar, M. Riddick and A. J. García, Nat. Rev. Mater., 2021, 7, 39–54 CrossRef PubMed
.
- M. Bar-Zeev, Y. D. Livney and Y. G. Assaraf, Drug Resistance Updates, 2017, 31, 15–30 CrossRef PubMed
.
- C. C. Wong and J. Yu, Nat. Rev. Clin. Oncol., 2023, 20, 429–452 CrossRef PubMed
.
- V. Chandra and F. McAllister, Gut, 2021, 70, 1419–1425 CrossRef PubMed
.
- J. An, H. Kwon and Y. J. Kim, J. Clin. Med., 2023, 12, 2216 CrossRef CAS PubMed
.
- R. L. Siegel, N. S. Wagle, A. Cercek, R. A. Smith and A. Jemal, Ca-Cancer J. Clin., 2023, 73, 233–254 CrossRef PubMed
.
- M. S. R. Rajoka, H. M. Mehwish, Y. Xiong, X. Song, N. Hussain, Q. Zhu and Z. He, Trends Food Sci. Technol., 2021, 107, 240–251 CrossRef
.
- N. Wang and J.-Y. Fang, Trends Microbiol., 2023, 31, 159–172 CrossRef CAS PubMed
.
- J. Abed, J. E. M. Emgård, G. Zamir, M. Faroja, G. Almogy, A. Grenov, A. Sol, R. Naor, E. Pikarsky, K. A. Atlan, A. Mellul, S. Chaushu, A. L. Manson, A. M. Earl, N. Ou, C. A. Brennan, W. S. Garrett and G. Bachrach, Cell Host Microbe, 2016, 20, 215–225 CrossRef CAS PubMed
.
- M. Xu, M. Yamada, M. Li, H. Liu, S. G. Chen and Y. W. Han, J. Biol. Chem., 2007, 282, 25000–25009 CrossRef CAS PubMed
.
- M. R. Rubinstein, X. Wang, W. Liu, Y. Hao, G. Cai and Y. W. Han, Cell Host Microbe, 2013, 14, 195–206 CrossRef CAS PubMed
.
- Q. Meng, Q. Gao, S. Mehrazarin, K. Tangwanichgapong, Y. Wang, Y. Huang, Y. Pan, S. Robinson, Z. Liu, A. Zangiabadi, R. Lux, P. N. Papapanou, X. E. Guo, H. Wang, L. E. Berchowitz and Y. W. Han, EMBO Rep., 2021, 22, 52891 CrossRef PubMed
.
- A. Janney, F. Powrie and E. H. Mann, Nature, 2020, 585, 509–517 CrossRef CAS PubMed
.
- A. D. Kostic, E. Chun, L. Robertson, J. N. Glickman, C. A. Gallini, M. Michaud, T. E. Clancy, D. C. Chung, P. Lochhead, G. L. Hold, E. M. El-Omar, D. Brenner, C. S. Fuchs, M. Meyerson and W. S. Garrett, Cell Host Microbe, 2013, 14, 207–215 CrossRef CAS PubMed
.
- S. Zhang, Y. Yang, W. Weng, B. Guo, G. Cai, Y. Ma and S. Cai, J. Exp. Clin. Cancer Res., 2019, 38, 14 CrossRef PubMed
.
- M. Nomura, R. Nagatomo, K. Inoue, K. Doi, J. Shimizu, K. Baba, T. Saito, S. Matsumoto and M. Muto, Ann. Oncol., 2019, 30, 509 CrossRef
.
- G. Li, J. Lin, C. Zhang, H. Gao, H. Lu, X. Gao, R. Zhu, Z. Li, M. Li and Z. Liu, Gut Microbes, 2021, 13, 1968257 CrossRef PubMed
.
- W. T. Cheng, H. K. Kantilal and F. Davamani, Malays. J. Med. Sci., 2020, 27, 9–21 CrossRef PubMed
.
- C. M. Dejea, P. Fathi, J. M. Craig, A. Boleij, R. Taddese, A. L. Geis, X. Wu, C. E. D. Shields, E. M. Hechenbleikner, D. L. Huso, R. A. Anders, F. M. Giardiello, E. C. Wick, H. Wang, S. Wu, D. M. Pardoll, F. Housseau and C. L. Sears, Science, 2018, 359, 592–597 CrossRef CAS PubMed
.
- Y. Wang, G. Yang, L. You, J. Yang, M. Feng, J. Qiu, F. Zhao, Y. Liu, Z. Cao, L. Zheng, T. Zhang and Y. Zhao, Mol. Cancer, 2019, 18, 173 CrossRef PubMed
.
- D. Iain, Nat. Rev. Gastroenterol. Hepatol., 2018, 15, 328 Search PubMed
.
- E. Riquelme, Y. Zhang, L. Zhang, M. Montiel, M. Zoltan, W. Dong, P. Quesada, I. Sahin, V. Chandra, A. S. Lucas, P. Scheet, H. Xu, S. M. Hanash, L. Feng, J. K. Burks, K. Do, C. B. Peterson, D. Nejman and F. McAllister, Cell, 2019, 178, 795–806 CrossRef CAS PubMed
.
- S. Pushalkar, M. Hundeyin, D. Daley, C. P. Zambirinis, E. Kurz, A. Mishra, N. Mohan, B. Aykut, M. Usyk, L. E. Torres, G. Werba, K. Zhang, Y. Guo, Q. Li, N. Akkad, S. Lall, B. Wadowski, J. Gutierrez, M. Janal, A. Saxena, X. Li, D. Cohen, R. B. Sartor, D. Saxena and G. Miller, Cancer Discovery, 2018, 8, 403–416 CrossRef CAS PubMed
.
- A. Leiter, R. R. Veluswamy and J. P. Wisnivesky, Nat. Rev. Clin. Oncol., 2023, 20, 624–639 CrossRef PubMed
.
- W. Barcik, R. C. T. Boutin, M. Sokolowska and B. B. Finlay, Immunity, 2020, 52, 241–255 CrossRef CAS PubMed
.
- J. J. Tsay, B. G. Wu, I. Sulaiman, K. Gershner, R. Schluger, Y. Li, T. Yie, P. Meyn, E. Olsen, L. Perez, B. Franca, J. Carpenito, T. Iizumi, M. El-Ashmawy, M. Badri, J. T. Morton, N. Shen, L. He, G. Michaud, S. Rafeq, J. L. Bessich, R. L. Smith, H. Sauthoff, K. Felner, R. Pillai, A. Zavitsanou, S. B. Koralov, V. Mezzano, C. A. Loomis, A. L. Moreira, W. Moore, A. Tsirigos, A. Heguy, W. N. Rom, D. H. Sterman, H. I. Pass, J. C. Clemente, H. Li, R. Bonneau, K. Wong, T. Papagiannakopoulos and L. N. Segal, Cancer Discovery, 2021, 11, 293–307 CrossRef CAS PubMed
.
- C. Jin, G. K. Lagoudas, C. Zhao, S. Bullman, A. Bhutkar, B. Hu, S. Ameh, D. Sandel, X. S. Liang, S. Mazzilli, M. T. Whary, M. Meyerson, R. Germain, P. C. Blainey, J. G. Fox and T. Jacks, Cell, 2019, 176, 998–1013 CrossRef CAS PubMed
.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed
.
- A. Fu, B. Yao, T. Dong, Y. Chen, J. Yao, Y. Liu, H. Li, H. Bai, X. Liu, Y. Zhang, C. Wang, Y. Guo, N. Li and S. Cai, Cell, 2022, 185, 1356–1372 CrossRef CAS PubMed
.
- L. Parhi, T. Alon-Maimon, A. Sol, D. Nejman, A. Shhadeh, T. Fainsod-Levi, O. Yajuk, B. Isaacson, J. Abed, N. Maalouf, A. Nissan, J. Sandbank, E. Yehuda-Shnaidman, F. Ponath, J. Vogel, O. Mandelboim, Z. Granot, R. Straussman and G. Bachrach, Nat. Commun., 2020, 11, 3259 CrossRef CAS PubMed
.
- S. Parida, S. Wu, S. Siddharth, G. Wang, N. Muniraj, A. Nagalingam, C. Hum, P. Mistriotis, H. Hao, C. C. Talbot, K. Konstantopoulos, K. L. Gabrielson, C. L. Sears and D. Sharma, Cancer Discovery, 2021, 11, 1138–1157 CrossRef CAS PubMed
.
- S. Bullman, C. S. Pedamallu, E. Sicinska, T. E. Clancy, X. Zhang, D. Cai, D. Neuberg, K. Huang, F. Guevara, T. Nelson, O. Chipashvili, T. Hagan, M. Walker, A. Ramachandran, B. Diosdado, G. Serna, N. Mulet, S. Landolfi, S. R. Y. Cajal, R. Fasani, A. J. Aguirre, K. Ng, E. Élez, S. Ogino, J. Tabernero, C. S. Fuchs, W. C. Hahn, P. Nuciforo and M. Meyerson, Science, 2017, 358, 1443–1448 CrossRef CAS PubMed
.
- W. Mu, Q. Chu, Y. Liu and N. Zhang, Nano-Micro Lett., 2022, 12, 142 CrossRef PubMed
.
- K. Forier, K. Raemdonck, S. C. D. Smedt, J. Demeester, T. Coenye and K. Braeckmans, J. Controlled Release, 2014, 190, 607–623 CrossRef CAS PubMed
.
- X. Sun, G. Wang, H. Zhang, S. Hu, X. Liu, J. Tang and Y. Shen, ACS Nano, 2018, 12, 6179–6192 CrossRef CAS PubMed
.
- K. Zheng, M. I. Setyawati, D. T. Leong and J. Xie, ACS Nano, 2017, 11, 6904–6910 CrossRef CAS PubMed
.
- L. Zhang, H. Wang and X. Qu, Adv. Mater., 2023, 2211147 Search PubMed
.
- D. Wang, G. Yang, H. C. van der Mei, Y. Ren, H. J. Busscher and L. Shi, Angew. Chem., Int. Ed., 2021, 60, 17714–17719 CrossRef CAS PubMed
.
- D. Wang, Y. Cao, G. Yang, S. Zhang, H. C. van der Mei, Y. Ren, T. G. van Kooten, D. A. de Groot, J. J. de Haan, L. Shi and H. J. Busscher, Adv. Funct. Mater., 2023, 33, 2215153 CrossRef CAS
.
- L. Chen, R. Zhao, Z. Kang, Z. Cao, N. Liu, J. Shen, C. Wang, F. Pan, X. Zhou, Z. Liu, Y. Yang and Q. Chen, J. Controlled Release, 2023, 363, 43–56 CrossRef CAS PubMed
.
- D. Zheng, X. Dong, P. Pan, K. Chen, J. Fan, S. Cheng and X. Zhang, Nat. Biomed. Eng., 2019, 3, 717–728 CrossRef CAS PubMed
.
- J. Chen, P. Zhang, Y. Zhao, J. Zhao, X. Wu, R. Zhang, R. Cha, Q. Yao and Y. Gao, Sci. Adv., 2022, 8, 2789 CrossRef PubMed
.
- Q. Chen, Y. Zhu, K. Wang, X. Wang, Y. Zhang, M. Zhou, J. Du, X. Qu, Z. Shi, Y. Zhang, Y. Chen and H. Qin, Nano Today, 2023, 52, 101994 CrossRef CAS
.
- Z. Chen, J. Li, P. Pan, P. Bao, X. Zeng and X. Zhang, Nano Today, 2021, 41, 101329 CrossRef CAS
.
- Z. Han, Q. Chen, D. Zheng, K. Chen, Q. Huang, Z. Zhuang and X. Zhang, Adv. Mater., 2023, 35, 2302551 CrossRef CAS PubMed
.
- Q. Qiu, D. Lu, G. Liu, X. Yang, J. Li, H. Ren, J. Liu, B. Sun and Y. Zhang, Bioconjugate Chem., 2022, 33, 1944–1952 CrossRef CAS PubMed
.
- X. Yan, F. Ma, Q. Chen, X. Gou, X. Li, L. Zhang and H. Gao, Chem. Eng. J., 2022, 450, 137605 CrossRef CAS
.
- X. Li, Y. Ma, Y. Xin, F. Ma and H. Gao, ACS Appl. Mater. Interfaces, 2023, 15, 14164–14172 CAS
.
- X. Zheng, G. Zhu, D. Pan, H. Li, K. Xiao, Q. Gong, Z. Gu, K. Luo and W. Li, Adv. Mater., 2024, 36, 2308977 CrossRef CAS PubMed
.
- L. Chen, R. Zhao, J. Shen, N. Liu, Z. Zheng, Y. Miao, J. Zhu, L. Zhang, Y. Wang, H. Fang, J. Zhou, M. Li, Y. Yang, Z. Liu and Q. Chen, Adv. Mater., 2023, 35, 2306281 CrossRef CAS PubMed
.
- M. Wang, B. Rousseau, K. Qiu, G. Huang, Y. Zhang, H. Su, C. Le Bihan-Benjamin, I. Khati, O. Artz, M. B. Foote, Y. Cheng, K. Lee, M. Z. Miao, Y. Sun, P. Bousquet, M. Hilmi, E. Dumas, A. Hamy, F. Reyal, L. Lin, P. M. Armistead, W. Song, A. Vargason, J. C. Arthur, Y. Liu, J. Guo, X. Zhou, J. Nguyen, Y. He, J. P.-Y. Ting, A. C. Anselmo and L. Huang, Nat. Biotechnol., 2023 DOI:10.1038/s41587-023-01957-8
.
- X. Zhang, X. Chen, Y. Guo, G. Gao, D. Wang, Y. Wu, J. Liu, G. Liang, Y. Zhao and F. Wu, Angew. Chem., Int. Ed., 2021, 60, 14013–14021 CrossRef CAS PubMed
.
- W. Song, D. Zheng, S. Zeng, X. Zeng and X. Zhang, ACS Nano, 2022, 16, 17402–17413 CrossRef CAS PubMed
.
- Z. Ma, H. Wang, Z. Shi, F. Yan, Q. Li, J. Chen, Z. Cui, Y. Zhang, X. Jin, Y. Jia and L. Wang, ACS Nano, 2023, 17, 5740–5756 CrossRef CAS PubMed
.
- W. Fan, B. Yung, P. Huang and X. Chen, Chem. Rev., 2017, 117, 13566–13638 CrossRef CAS PubMed
.
- X. Qu, F. Yin, M. Pei, Q. Chen, Y. Zhang, S. Lu, X. Zhang, Z. Liu, X. Li, H. Chen, Y. Zhang and H. Qin, ACS Nano, 2023, 17, 11466–11480 CrossRef CAS PubMed
.
- X. Wang, Q. Chen, Y. Zhu, K. Wang, Y. Chang, X. Wu, W. Bao, T. Cao, H. Chen, Y. Zhang and H. Qin, Signal Transduction Targeted Ther., 2023, 8, 277 CrossRef CAS PubMed
.
- J. Xi, Y. Wang, X. Gao, Y. Huang, J. Chen, Y. Chen, L. Fan and L. Gao, Nano Today, 2022, 43, 101395 CrossRef CAS
.
- X. Kang, F. Bu, W. Feng, F. Liu, X. Yang, H. Li, Y. Yu, G. Li, H. Xiao and X. Wang, Adv. Mater., 2022, 34, 2206765 CrossRef CAS PubMed
.
- R. Liu, H. Yang, S. Qu, P. Yang, X. Zhi, Y. Xu, Z. Dai and L. Qian, Aggregate, 2023, 5, e423 CrossRef
.
- Y. Xin, Y. Yu, M. Su, X. Li, M. Elsabahy and H. Gao, J. Controlled Release, 2023, 359, 69–84 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2024 |