Tapping into the natural aromatic potential of microbial lignin valorization towards aromatic fine chemicals†
Xiao-Lei
Zhang
ab,
Zhi-Hua
Liu
*ab,
Bing-Zhi
Li
 *ab and
Ying-Jin
Yuan
*ab and
Ying-Jin
Yuan
 ab
ab
aFrontiers Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education), School of Chemical Engineering and Technology, Tianjin University, Tianjin 300072, China. E-mail: zhliu@tju.edu.cn
bFrontiers Research Institute for Synthetic Biology, Tianjin University, China. E-mail: bzli@tju.edu.cn
First published on 17th October 2024
Abstract
Lignin valorization presents significant opportunities for advancing a sustainable bioeconomy and carbon neutrality goals. In this case, biological conversion of lignin offers promising pathways for the production of aromatic fine chemicals, effectively addressing the challenges associated with the heterogeneous and macromolecular structure of lignin. However, the pathways for converting lignin derivatives into aromatic chemicals remain underdeveloped, and their conversion efficiency remains unsatisfactory. This work aims to prospect the bioconversion pathways of lignin toward valuable aromatics, which aligns with the principles of the atom economy concept. A comprehensive overview of critical pathways for converting lignin into aromatic chemicals is presented together with a thorough discussion on advanced technologies essential for enhancing lignin bioconversion and aromatic synthesis. Furthermore, existing challenges and emerging strategies are explored. The findings of this study are expected to offer valuable insights into recent advancements and future directions in the biological conversion of lignin. These insights can facilitate lignin valorization and promote the development of a lignin-based bioeconomy.
1 Introduction
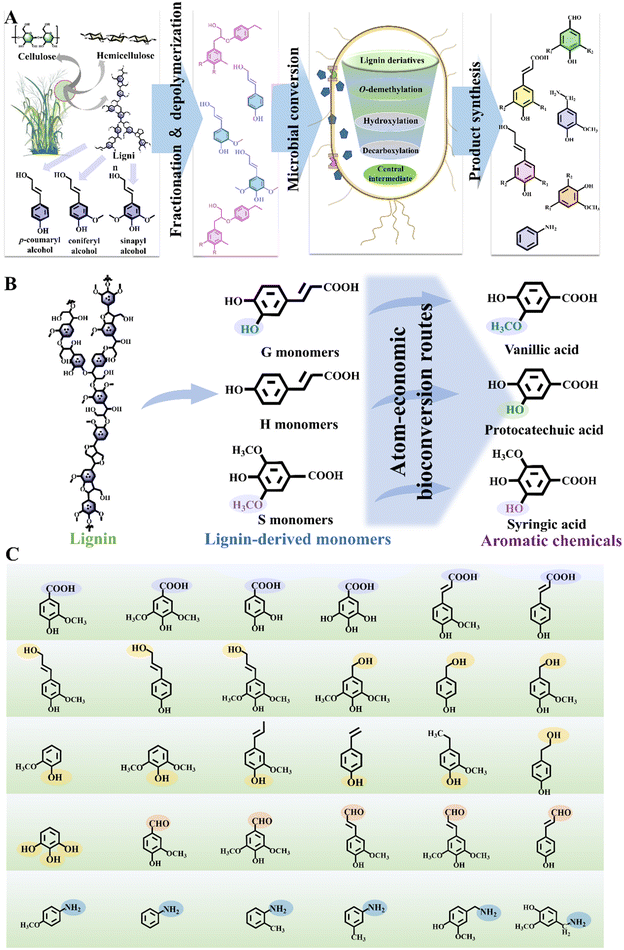
Lignocellulosic biomass refineries produce a range of biofuels, chemicals and materials, reducing regional fossil energy consumption and greenhouse gas emissions while contributing to carbon neutrality.1–3 Lignin, one of the three major components of lignocellulosic biomass, is globally produced in vast quantities, exceeding 300 million tons annually.1,2,4 Thus, it can serve as an exceptional raw material for producing high-value products, where lignin valorization plays a crucial role in promoting feasible biorefineries and fostering the growth of a sustainable biomass-based economy.5Lignin has a complex and irregular structure, featuring an amorphous and disordered arrangement of its constituent units.6 It is formed through the bonding of three methoxylated phenylpropane units, namely, p-coumaryl alcohol (H), coniferyl alcohol (G) and sinapyl alcohol (S). Approximately two-thirds of the chemical bonds connecting these basic structural units are C–O bonds, while the remaining one-third consists of C–C bonds.7 These linkages include β-O-4, α-O-4, 4-O-5, β–β, and β-1 connections. Furthermore, lignin is abundant in functional groups, including benzene rings, methoxy groups, phenolic hydroxyl groups, carboxyl groups, and carbonyl groups.8 The diverse array of aromatic units, functional groups, and linkages in lignin contributes to its structural complexity and recalcitrance, presenting both challenges and opportunities for lignin valorization and highlighting the need to explore effective utilization strategies.9
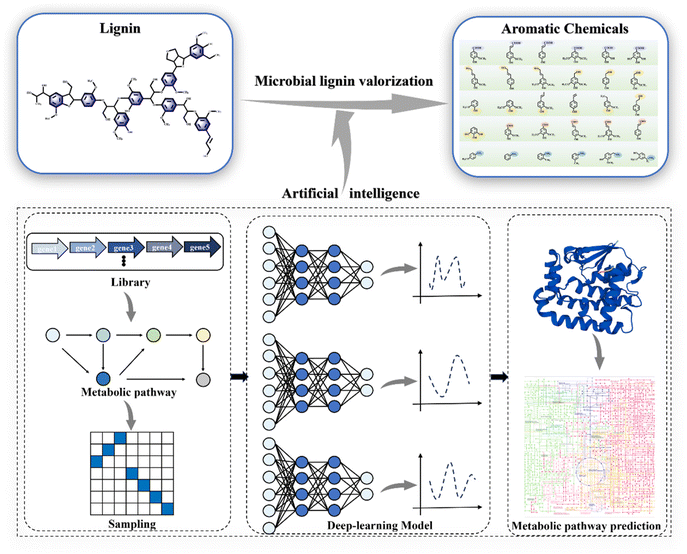
Developing innovative strategies for lignin conversion is crucial to address the challenges posed by its heterogeneity.10 Microbial conversion holds the potential to transform the promising renewable lignin source into bioproducts, serving as an inevitable requirement to advance the industrialization of lignin valorization and biorefineries.11,12 Microbial lignin conversion aligns with natural development principles and supports environmental sustainability. Systems biology tools have helped identify many ligninolytic genes, enzymes, and metabolic pathways, leading to the engineering of efficient bacterial strains for lignin bioconversion. Technological advancements in artificial intelligence and directed evolution have also facilitated the design of key ligninolytic enzymes and the successful synthesis of diverse lignin-related products.13 Consequently, these innovative approaches have enabled the assimilation of lignin and the synthesis of valuable products (Fig. 1A–C).14
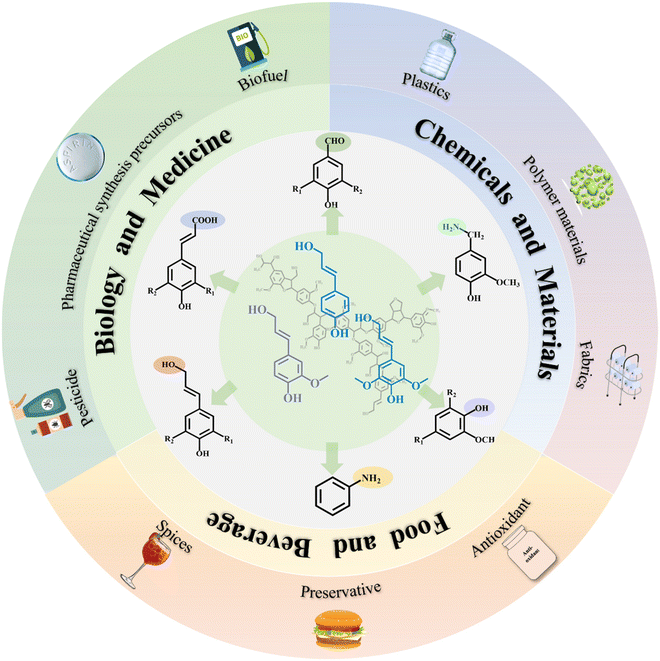
Aromatic chemicals are predominantly sourced from botanical extracts or synthetic processes.13 Generally, these compounds exhibit a variety of bioactivities, such as antibacterial, anti-inflammatory, analgesic, antioxidant, and antitumor effects. They also find extensive applications across various industries, including medicine, food, materials, and energy (Fig. 2). Common industrial methods such as pressing and solvent leaching are employed for extracting aromatic natural products from plants. However, the limited availability and low stability of aromatics in plants and challenges related to large-scale production can be restrictive. Moreover, the chemical synthesis of aromatic chemicals often requires organic solvents and harsh conditions, which can negatively impact the environment and human health.15
Microbial lignin valorization has opened new avenues for the sustainable and efficient production of aromatic fine chemicals. Inexpensive lignin presents a promising aromatic carbon source for the production of aromatic fine chemicals. The generation of benzene rings in lignin is a complex and energy-intensive aspect throughout its life cycle. The substantial aromaticity of lignin has spurred the exploration of efficient conversion pathways that yield valuable natural aromatic products. Microbial synthesis can efficiently leverage the aromatic structure of lignin for producing these chemicals. Especially, various ligninolytic microorganisms have developed a ‘biological funnel’ pathway for converting heterogeneous lignin into valuable aromatic platform chemicals, which aligns with the atom economy concept.16 The microbial conversion strategy offers a practical approach for transforming complex lignin resources into valuable bioproducts, providing a potential solution for lignin valorization.17,18
However, although microbial conversion presents significant advantages, challenges and issues persist in fully elucidating the bioconversion of lignin into fine aromatic chemicals.19 The structural complexity of lignin contributes to its recalcitrance, creating challenges in its microbial valorization into valuable aromatic fine chemicals. Firstly, the inherent heterogeneity and macromolecular nature of lignin contribute to its poor bioaccessibility, making its direct conversion challenging.20 Secondly, lignin bioconversion involves the utilization of living cells, constituting a more intricate system that requires deeper comprehension and enhancement.21 Thirdly, the current bioconversion efficiency is unsatisfactory for the microbial conversion of lignin into aromatic derivatives. Thus, it is necessary to further exploit systematic technical solutions to enhance the synthesis efficiency of aromatic products from lignin bioconversion.
This work provides a comprehensive review of the microbial conversion of lignin into aromatic fine chemicals, in line with atom-economic and sustainable green concepts. It emphasizes the strategic use of the inherent aromatic features of lignin to maximize the production of valuable aromatic chemicals. Initially, this review presents a summary of the critical biological pathways for converting lignin to aromatic chemicals, followed by an exploration of microbial conversion processes, including ligninolytic and non-ligninolytic strain chassis, key enzymes, and conversion strategies. Subsequently, cutting-edge technologies are identified for microbial lignin valorization. Finally, the challenges and prospects for both lignin valorization and aromatic biosynthesis are discussed. This comprehensive work aims to pave the way for sustainable lignin valorization and the economically viable synthesis of aromatic fine chemicals.
2 Critical pathways for the microbial conversion of lignin derivatives into aromatic fine chemicals
Natural bioconversion pathways for lignin involve several key steps, including macromolecular lignin depolymerization, transmembrane transport of aromatic molecules, biological funnel pathways, and subsequent synthesis of target products (Fig. 1A).22 The depolymerization of lignin aims to break down the intricate structure of lignin, releasing lignin-derived aromatic compounds for subsequent utilization. The efficient transportation of these compounds is required for the intracellular utilization of lignin derivatives. Within the cells, lignin assimilation and product biosynthesis pathways play a crucial role in channeling lignin-derived compounds towards the desired aromatic chemicals.2.1 Lignin depolymerization for obtaining lignin derivatives
One of the primary obstacles in lignin bioconversion is acquiring lignin derivatives suitable for microbial assimilation. The structural complexity of lignin surpasses that of cellulose and hemicellulose, leading to fundamental differences in the structure and properties of the products derived from depolymerization.23 Generally, lignin depolymerization involves thermal, chemical and biological methods.24 The biological depolymerization of lignin relies on key ligninolytic enzymes, including laccase, manganese peroxidase (MnP), lignin peroxidase (LiP), versatile peroxidase (VP), and dye-decolorizing peroxidase (DyP).25Numerous microorganisms have been found to possess efficient ligninolytic capabilities.26Bacillus subtilis S11Y showed remarkable lignin-depolymerizing capabilities.27 The draft genome sequences of B. subtilis S11Y revealed the absence of conventional dye-decolorizing peroxidase or catalase-peroxidase genes commonly associated with bacterial lignin depolymerization. In B. subtilis S11Y, potential genes encoded ligninolytic enzymes were identified as oxidative stress-related enzymes, which included Cu/Zn type-superoxide dismutase and a heme-containing monofunctional catalase.27 Five native ligninolytic bacterial strains were isolated from agricultural soil via the serial dilution method.28 A mixed bacterial culture was constructed to promote the biodepolymerization of kraft lignin. By the sixth day of incubation, it achieved peak levels of kraft lignin degradation and vanillin production, reaching a titer of 72.5 mg L−1.28 It was further confirmed that B. subtilis played a crucial role in the oxidation, depolymerization and repolymerization of soluble aromatic substrates. This ability was facilitated by the highly active enzyme bacterial laccase CotABsu, particularly due to its twelve active site residues.29
It was found that the synergistic effects of laccases and TiO2-assisted photocatalytic treatment significantly enhanced the depolymerization of lignin.30In vitro studies confirmed that Schizophyllum commune, a white-rot fungus known for its laccase production, effectively degraded agricultural residual lignin and provided a safe, affordable and environmentally compatible depolymerization route.30 The ligninolytic ability of Mycobacterium smegmatis was found to be better than that of other strains.31 Lignin was preferentially degraded over cellulose with the degradation rate of 50% on the eighth day. The maximum activities of laccase, lignin peroxidase and manganese peroxidase were 0.18, 0.12, and 0.52 U mL−1, respectively.31 Among the bacteria isolated from Peruvian rainforest soils, B. pumilus and B. atrophaeus exhibited laccase activities of about 0.14 U mL−1 and 0.055 U mL−1, respectively. Unlike fungi and most other ligninolytic bacteria, these strains exhibited both intracellular and extracellular laccase activities, specifically targeting the β-O-4 bond of lignin.32 Two bacterial consortia, sourced from proximate sediments of the East and South China Seas and possessing alkali lignin degradation capabilities, demonstrated maximum lignin degradation rates of 57% and 18%, respectively. Both consortia also actively produced ligninolytic enzymes laccase, manganese peroxidase, and lignin peroxidase, facilitating the degradation of lignin into smaller constituents.33
The N246A variant of the dye decolorizing peroxidase of type B (DypB) from Rhodococcus jostii was overexpressed in Escherichia coli to enhance the catalysis performance of DypB.34 The enzyme exhibited broad dye-decolorization activity, showing increased effectiveness especially in the presence of Mn2+. It also exhibited the ability to oxidize a wide variety of aromatic monomers derived from lignin. Notably, it can degrade the guaiacylglycerol-β-guaiacyl ether, a dimer model compound of lignin, producing guaiacol at a rate of 12.5 μmol min−1 mg enzyme within 10 min.34 The ligninolytic bacteria P. putida KT2440 was confirmed to be capable of producing a ligninolytic secretome containing active oxidase and peroxidase enzymes, which exhibited lignin degradation capabilities.35 This secretome facilitated a series of extracellular oxidative cleavage reactions of lignin, leading to the degradation of approximately 8.6% of alkali lignin. Moreover, the lignin degradation rate increased to 14.5% with the addition of H2O2 and Mn2+.35 These studies suggested that employing a variety of ligninolytic enzymes effectively enhanced the lignin depolymerization. Additionally, the synergistic effects of diverse ligninolytic microbial strains and enzymes play a vital role in the depolymerization process. The above-mentioned findings indicated that the successful assessment of ligninolytic enzymes holds promise for their application in lignin depolymerization for generating valuable lignin derivatives.
Overall, biological depolymerization is an optimal choice for obtaining lignin derivatives suitable for downstream upgrading. However, it is still necessary to screen and identify the ligninolytic hosts and enzymes. The extracellular lignin depolymerization mechanism is also required for understanding lignin structural alterations and degradation pathways. For example, considering the unpredictable reactions of lignin catalyzed by laccases, it is still necessary to uncover the molecular mechanisms and optimal conditions for the depolymerization and polymerization of lignin for laccase. To date, the depolymerization efficiency of lignin is still unsatisfactory and the uncontrollable side reactions still significantly affect the purity of lignin derivatives. The depolymerization process will yield a complex mixture of aromatic chemicals, posing challenges in converting them into a singular product. Therefore, the integration of computer-aided design, co-expression of proteins, multi-omics analysis, and screening is essential for developing highly active ligninolytic enzymes under industrial conditions. Also, the further development of coupled chemical and biological approaches is needed to promote the depolymerization efficiency. Combining these strategies can greatly enhance the depolymerization performance of lignin, resulting in lignin derivatives suitable for microbial conversion.
2.2 The uptake of lignin derivatives through microbial cell membranes
Following lignin depolymerization, lignin derivatives will be transported across microbial cell membranes for intracellular metabolism.36 Two transportation modes have been identified for lignin derivatives, including active membrane transporters and transporter-assisted membrane crossing. It has been confirmed that active transport serves as the primary uptake mode for lignin derivatives.36 In comparison, passive diffusion act as the transport mechanism for uncharged monomers and dimers in a computational model of a bacterial membrane.37 Predictions suggest that both lignin biosynthesis and catabolism are probably facilitated by passive transport processes for uncharged aromatic chemicals in bacteria and plants. This involves gradients in membrane concentration, compound delivery and utilization rates, which regulate the membrane translocation rates.38 The inner membrane transporters in bacteria responsible for the production of lignin derivatives can be classified into three categories including ATP-binding cassette transporters, major facilitator superfamily transporters, and tripartite ATP-independent periplasmic transporters.36The efficient uptake of lignin derivatives relies on specific aromatic transporters for cellular entry. Three aromatic-associated transporters had been identified, which are responsible for transporting phenol, vanillate, benzoate and guaiacol.39 It was also observed that the upregulation of ATP-binding cassette transporters in Burkholderia sp. ISTR5 and P. putida KT2440 enhanced the efficient transport of lignin derivatives.40 In P. putida KT2440, the transporters for lignin-related monomers and their substrate selectivity were investigated, emphasizing five genes that encode aromatic acid/H+ symporter family transporters. The findings indicated that at physiological pH levels, PcaK and HcnK are primarily responsible for the uptake of protocatechuic acid/4-hydroxybenzoic acid and ferulic acid/4-coumaric acid, respectively, whereas VanK functions as a vanillic acid/protocatechuic acid transporter.40,41 Furthermore, it was confirmed that T. versicolor and G. subvermispora takes up 4-hydroxybenzoic acid and vanillic acid and employ intracellular catabolic pathways to channel these lignin derivatives into central carbon metabolism.42
The transporter gene responsible for the uptake of 5,5′-dehydrodivanillate was identified in Sphingobium sp. SYK-6.43 This gene encoded a putative major facilitator superfamily transporter (SLG_07710) and MarR-type transcriptional regulator (SLG_07780). Besides characterizing its functionality, the transporter gene was utilized for transporting the aromatic substrate and biosynthesizing a value-added metabolite. Furthermore, SLG_07710 overexpression significantly facilitated the production of 2-pyrone-4,6-dicarboxylate, a crucial metabolite from 5,5′-dehydrodivanillate for functional polymers.43 A mutant strain of Sphingobium sp. SYK-6 lacking the 2-pyrone-4,6-dicarboxylate hydrolase gene ligI was cultivated in 5,5′-dehydrodivanillate, leading to the accumulation of 2-pyrone-4,6-dicarboxylate. Upon the introduction of a SLG_07710-expression plasmid into the ligI mutant, the conversion of 5,5′-dehydrodivanillate and subsequent production of 2-pyrone-4,6-dicarboxylate after 48 h increased by 1.35 and 1.34 times, respectively.43 The results highlighted the critical importance of a comprehensive understanding of the transport and catabolic pathways for efficient lignin utilization. In this case, enhancing the expression of transporter genes has the potential to boost the biosynthesis of lignin-derived metabolites.
R. opacus underwent adaptive evolution with phenol as its sole carbon source, resulting in a remarkable 373% growth improvement, with the phenol consumption rate reaching approximately 20 mg L−1 h−1 and nearly doubling lipid production from phenol.44 This enhancement correlated with the upregulation of relevant genes involved in lignin uptake and metabolism.44R. palustris, equipped with the TarPQM system, significantly enhanced the uptake and conversion of aromatics produced from lignin, such as coumarate, ferulic acid, and caffeate.45 It was confirmed that R. palustris possessed two primary and secondary transporters with distinct energetic profiles, both utilizing high-affinity periplasmic binding proteins.45
Overall, it has been demonstrated that inner membrane transporters are essential. These transporter proteins should exhibit high affinity and specificity towards aromatics to facilitate the uptake of these derivatives. Overexpressing these transporter proteins in bacteria has been shown to improve the uptake of lignin-derived aromatics, thereby aiding in the production of value-added products. Despite this, the structure and function of these transporter protein membranes remain unclear. Identifying and characterizing specific transporters is still crucial for the efficient and flexible transportation of diverse aromatics. A comprehensive understanding of the structural and functional aspects of transporters is also vital for elucidating the underlying mechanisms for the uptake of aromatic chemicals. Furthermore, to enhance the uptake and conversion of lignin derivatives, it is necessary to engineer the transporters within the aromatic uptake systems. This knowledge can offer valuable insights for engineering and designing transporters to improve bacterial uptake efficiency of lignin derivatives.
2.3 Intracellular catabolism pathways of lignin derivatives
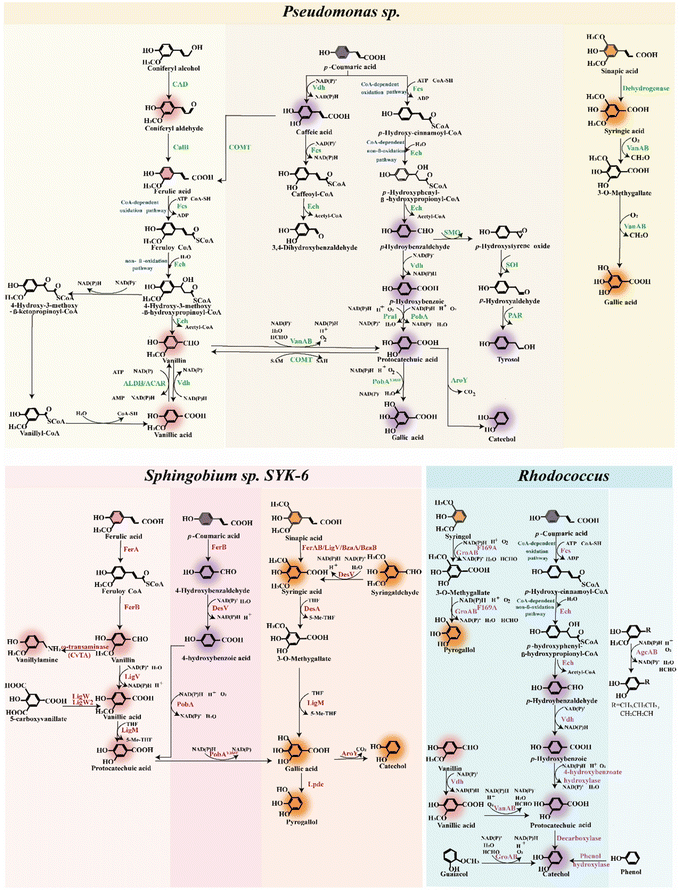
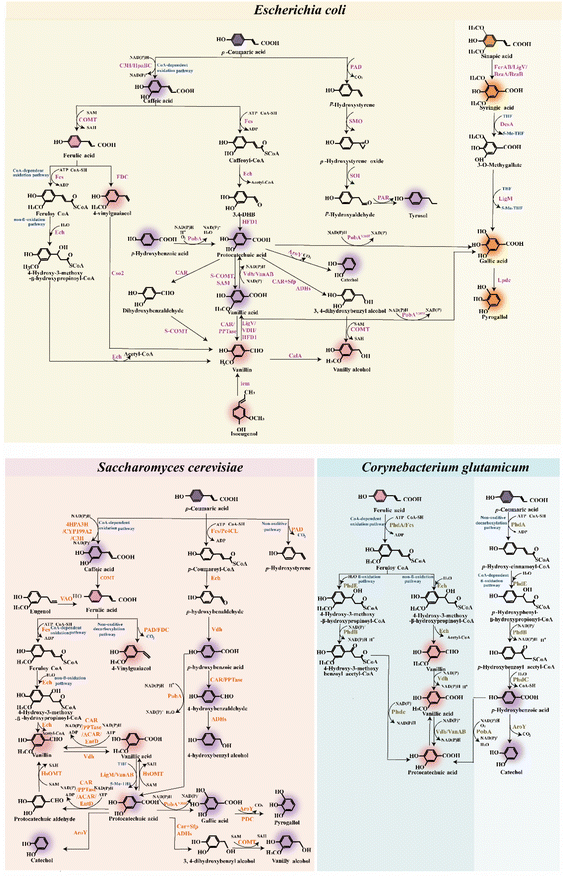
Ligninolytic strains have evolved the intracellular catabolism pathways to convert lignin derivatives into valuable products with high atom efficiency (Fig. 1B and C).20 The conversion of diverse lignin feedstocks typically involves multiple engineered microorganisms, each specialized for different substrate compositions and product outputs. An ideal microbial host should possess the ability to simultaneously assimilate diverse lignin derivatives, demonstrate resilience to high levels of toxic compounds, and efficiently produce value-added products.46 Achieving these traits in non-model microbes requires modifications and transformations of varying complexity. Optimizing non-model strains necessitates advancements in genetic tool development, robustness engineering, and overall pathway integration. Microbial strain design efforts must align with feedstock engineering, innovative depolymerization strategies, process engineering, and potential downstream applications.Certain ligninolytic microbes possess natural catabolism pathways for heterogeneous lignin derivatives, potentially addressing the challenges associated with the heterogeneity of lignin (Fig. 3).47 Through engineering strategies, non-ligninolytic strains also exhibit excellent lignin degradation characteristics (Fig. 4). Producing desirable aromatic chemicals from various underexploited lignin derivatives requires high specificity in side-chain catabolism.48 A critical bottleneck and rate-limiting step in the biological conversion of lignin involves key nodes within the biological funneling pathway. For example, starting with three fundamental lignin building blocks, varying in aromatic ring methylation, several enzymatic reactions occur in the upper pathways of aromatic catabolism. These reactions include aromatic O-demethylation, hydroxylation and decarboxylation.18 In the upper pathways, the O-demethylation of methoxy-containing aromatics is essential for their conversion into important central intermediates, such as catechol or protocatechuic acid.49
Lignin hydrolysate obtained after depolymerization contains various lignin-derived aromatic monomers, which serve as precursors for aromatic fine chemicals through biological funneling pathways. Bacteria employ three pathways for the degradation of an H-type representative aromatic p-coumaric acid including CoA-dependent oxidation, CoA-independent pathways and decarboxylation pathway.48 Ferulic acid, a representative G-type aromatic derivative, undergoes metabolism through four pathways, including CoA-dependent oxidation pathway, non-oxidative decarboxylation pathway and side-chain reduction pathway. Syringic acid is commonly regarded as a representative of S-type aromatics, which bears a higher number of methoxy groups on its aromatic ring. However, the microbial metabolism of S-type aromatics has not yet reached optimal performance levels.36
Recently, novel assimilation pathways for lignin derivatives have been identified in Sphingobium sp. SYK-6. Using multiplexed fitness profiling, a randomly barcoded transposon insertion library was developed, and the catabolism pathways for lignin derivatives were determined.50 This result not only confirmed several known aromatic catabolic pathways, but also established new catabolism pathways. The illumination of novel aromatic catabolism features in this model bacterium presents targets for future metabolic engineering.50 Additionally, R. opacus has been adaptively developed to assimilate various aromatic chemicals. The evolved mutants exhibited growth improvements of up to 1900% over the wild-type strain.39 Whole-genome sequencing revealed redox-related gene mutations shared by several adapted mutants. Comparative transcriptomics identified five aromatic funneling pathways and by-product detoxification pathways, which were upregulated in response to specific aromatic chemicals. Gene knockout experiments verified two β-ketoadipate pathways for the degradation of five aromatics.39 Numerous modified GcoA variants (a cytochrome P450 protein from the family CYP255A) were evaluated for their ability to catalyze aromatic O-demethylation.51 GcoA requires two strategically planned single amino acid substitutions, F169S and T296S, to effectively catalyze the o- and p-isomers of vanillin, respectively. In vivo demethylation was demonstrated using the T296S variation, employing P. putida KT2440 strains previously optimized for synthesizing p-vanillin from ferulate. This expands the demethylate capabilities of cytochrome P450 enzymes.51 These findings highlight the key reactions in aromatic utilization, enhancing the understanding of aromatic bioconversion.
Furthermore, 15 bacterial species showing lignin bioconversion ability were identified, while three promising bacterial strains, including P. putida KT2440, Rhodococcus jostii RHA1, Amycolatopsis sp. ATCC 39116, were intensively investigated for their lignin decomposition properties.52P. putida KT2440 exhibited accelerated growth in lignin-rich medium and released outer membrane vesicles of two sizes. The extracellular proteome of the bacterial cultures was found to be significantly enriched with enzymes involved in the metabolic pathways of lignin monomers, specifically p-coumaric acid and ferulic acid catabolism. It was demonstrated that various enzymes within the secreted outer membrane vesicles of P. putida are essential for lignin biodepolymerization and bioconversion.52
Overall, various ligninolytic strains possess intracellular aromatic metabolism pathways to assimilate lignin derivatives toward targeted products. However, the heterogeneity of lignin necessitates versatile enzymes with specific substrate selectivity for complex metabolic pathways. The availability and activity of key enzymes in intracellular metabolism are crucial for pathway efficiency and product yield. Certain enzymes have specific requirements for reaction conditions such as temperature and pH, which complicates pathway regulation and optimization. There may be limitations in the supply of key enzymes, necessitating strategies such as genetic and protein engineering to elevate enzyme expression and catalytic efficiency. Increasing the expression of crucial enzymes and supplying necessary aromatic precursors will enhance the biosynthesis of aromatic fine chemicals. Furthermore, prospecting novel lignin catabolism pathways and screening effective ligninolytic enzymes are still required to funnel lignin derivatives into targeted aromatic products.
Additionally, toxicity and inhibition by aromatic intermediates can impair microbial growth and metabolism, necessitating adaptive evolution strategies. Moreover, redox balance and energy input are critical in lignin bioconversion, with metabolic engineering, cofactor manipulation, and process optimization as potential solutions. Therefore, a comprehensive understanding of lignin metabolism within cellular systems is essential to unlock the full potential of lignin as a valuable resource.
3 Funneling lignin derivatives into aromatic acids via biological funnel pathway
Ligninolytic microorganisms exhibit a unique ability to enzymatically convert a wide range of heterogeneous lignin derivatives into various aromatic acids through a range of enzyme-mediated reactions, including oxidation, carboxylation, dehydrogenation via biological funnel pathways.18 Aromatic acids, characterized by a carboxylic acid group linked to an aromatic ring, include compounds such as protocatechuic acid, gallic acid, and vanillin acid. These chemicals are widely utilized across industries, such as pharmaceuticals, food additives, fragrances, and polymer synthesis, due to their biological activities, flavoring attributes, and role as foundational precursors for generating valuable chemicals (Table 1).53| Aromatic chemicals | Host strains | Substrates | Gene origin | Engineering strategies | Performance (titer) | Performance (yield) | Ref. |
|---|---|---|---|---|---|---|---|
| Protocatechuic acid | P. putida KT2440 | Lignin-derived monomers in hydrolysates | pcaHG; vanAB | Knocking out protocatechuate 3,4-dioxygenase and overexpressing vanillate-O-demethylase | 253.88 mg L−1 | 70.85 % | 54 |
| Protocatechuic acid | C. glutamicum | Ferulic acid | CeVanAB | Heterologous expressing vanAB | 1.064 g L−1 | 62.8% | 55 |
| Protocatechuic acid | P. putida KT2440 | Ethanol depolymerized lignin derivatives | — | Deleting pcaGH | 6.73 mg L−1 | 17.5% | 56 |
| Protocatechuic acid | P. putida KT2440 | Syringic acid | vanAB | Overexpressing vanAB | — | — | 132 |
| Protocatechuic acid | P. putida KT2440 | p-Coumaric acid and ferulic acid | pobA; vanAB; HcnK | Co-expressing pobA, vanAB, and HcnK | 12.7 g L−1 | — | 98 |
| Protocatechuic acid | P. putida KT2440 | Lignin-derived p-coumaric acid and vanillic acid | pcaHG; vanAB | Deleting pcaHG and overexpressing vanAB | — | >90% (p-coumaric acid), >50% (vanillic acid) | 58 |
| Protocatechuic acid | S. cerevisiae | Alkaline pretreated lignin liquor | RdMetf1; Pc4CL; PpECH; PpVDH; PdVanA; PdVanB | Deleting ADH6, ADH7, BDH2, FDC1, MHT1 and SAM4; overexpressing MET6, metf1 and PodA; heterologous expressing 4CL, ech, vdh, vanA, vanB | 810 mg L−1 | — | 59 |
| Protocatechuic acid | Aspergillus niger | Multiple benzoates and cinnamic acids | phyA; prcA | Double deleting phyA and prcA | — | ≥80% | 60 |
| Gallic acid | E. coli | Lignin-derived syringate | desA; ligM; lpdc | Co-expressing desA, ligM and lpdc | 59.6 mg L−1 | — | 63 |
| Gallic acid | E. coli | Ferulic acid | PpFCS; PpECH; ScHFD11; PpVanAB; PpPobA | Overexpressing fcs, ech, hdf1, vanAB and PobAY385F | 3.3 g L−1 | 97.9% | 65 |
| Gallic acid | E. coli | p-Coumaric acid | PpFCS; PpECH; ScHFD1; PpPobA | Overexpressing fcs, ech, hdf1, vanAB and PobAY385F | 3.4 g L−1 | 99.8% | 65 |
| Gallic acid | R. opacus PD630 | Alkaline-pretreated lignin stream | pobA*; pobA**; pobA | Heterologous expressing pobA*, pobA**, and wild pobA; integrating hydroxylation, O-demethylation, and aryl side-chain oxidation systems | 0.407 g g−1 lignin | — | 64 |
| p-Hydroxybenzoic acid | Burkholderia glumae BGR1 | p-Coumaric acid | phb3h; bcl; phcs II | Deleting phb3h and bcl; overexpressing phcs II | 9.27 mg L−1 h−1 | 99.0% | 106 |
| 4-Hydroxy-phenylacetic acid | E. coli | p-Coumaric acid | pad; styAB; styC; feaB | Introducing pad, styAB, styC, and feaB genes | 2.08 g L−1 | 91.3% | 67 |
| 3,4-Dihydroxy-phenylacetic acid | E. coli | p-Coumaric acid | pad; styAB; styC; feaB | Expressing pad, styAB, styC, and feaB genes | 2.27 g L−1 | 90% | 67 |
| Homovanillic acid | E. coli | Ferulic acid | pad; styAB; styC; feaB | Co-expressing pad, styAB, and styC with feaB genes | 692 mg L−1 | 76.2% | 67 |
| Vanillic acid | P. putida KT2440 | Maize bran hydrolysate derived ferulic acid | CatA/A2; pcaHG | Knocking out catA/A2 and pcaHG | 0.507 g g−1 | 90% | 77 |
| Caffeic acid | E. coli | p-Coumaric acid | hpaB; hpaC | Substrate delay; expressing hpaB and hpaC | 3.46 g L−1 | 86.5% | 133 |
| Caffeic acid | E. coli | p-Coumaric acid | hpaBC | Expressing 4-hydroxyphenylacetate 3-hydroxylase (4HP3H) | 3.82 g L−1 | 53.0% | 134 |
Protocatechuic acid, recognized for its antihyperglycemic and neuroprotective effects, demonstrates notable antioxidant and anti-inflammatory properties. With its active acid and hydroxyl group, it serves as a valuable precursor for the synthesis of numerous valuable compounds, including adipic acid, gallic acid and pyrogallol.54p-Coumaric acid undergoes conversion to caffeic acid via the CoA-dependent oxidation pathway, followed by subsequent reactions, leading to the formation of protocatechuic acid. Protocatechuic acid can also be produced through the CoA-independent pathway and non-oxidative decarboxylation pathway.36 For example, p-hydroxybenzaldehyde served as a precursor of protocatechuic acid within a biological funneling pathway. Some ligninolytic strains also have the ability to metabolize G-type aromatic monomers, such as ferulic acid and vanillic acid, into protocatechuic acid.55
In the case of ligninolytic strains, an engineered P. putida KT2440 strain achieved promising biological co-upgrading of ethanol-assisted depolymerized lignin with a final titer of 6.73 mg L−1 protocatechuic acid and a 17.5% (w/w) yield of total lignin monomers.56 The engineered strain also effectively accumulated 253.88 mg L−1 protocatechuic acid by knocking out protocatechuate 3,4-dioxygenase and overexpressing vanillate-O-demethylase, with a remarkable conversion yield of 70.85%.54 The engineered strain also effectively accumulated protocatechuic acid by inhibiting the degradation pathway and regulating the catabolite repression control protein. The chromosomally integrated strain accumulated 22.68 mM protocatechuic acid from a mixture of 20 mM p-coumaric acid and 4 mM ferulic acid, with a remarkable molar conversion yield of 94.5%.54 Furthermore, in Sphingomonas sp., a mutant ligAB gene encoding protocatechuate 4,5-dioxygenase enabled growth in a substrate mixture of 5 mM butyric acid and 5 mM vanillic acid, leading to the accumulation of approximately 4 mM protocatechuic acid.57 Genetically, P. putida KT2440, with a deletion in the protocatechuate 3,4-dioxygenase and overexpression of the vanillate-O-demethylase gene, converted p-coumaric acid and vanillic acid into protocatechuic acid with yields exceeding 90% and 50%, respectively.58
In the case of non-ligninolytic strains, S. cerevisiae was engineered with an artificial biological funnel pathway, efficiently converting p-coumaric acid and ferulic acid into protocatechuic acid.16 The deletion and overexpression of specific genes, combined with the integration of the multi-copy heterologous pobA gene and optimized fermentation strategies, resulted in the highest yield of 810 mg L−1 protocatechuic acid.59 The identified protocatechuate hydroxylase (decarboxylating) (PhyA) enabled the development of a fungal cell factory capable of accumulating protocatechuic acid from various lignin derivatives. This engineered pathway effectively yielded 10.32 mM protocatechuic acid, with a conversion rate of over 99%, demonstrating a promising and sustainable approach for lignin valorization.60
Gallic acid, which is well-known for its potent antioxidant activity, is generally extracted from various fruits and plants.61,62 Alternatively, the biological conversion of lignin derivatives provides a promising route for the production of gallic acid. For example, the S-type lignin monomer syringic acid can be converted into gallic acid by the enzymes of O-demethylase (DesA) and vanillate/3-O-methygallate O-demethylase (LigM). DesA and LigM were confirmed to catalyze syringate to gallate with a titer of 59.6 mg L−1.63 In the case of ligninolytic strains, by deleting the degradation pathways for protocatechuic acid and gallic acid and introducing the genes responsible for the main reactions, Rhodococcus erythropolis achieved the one-pot conversion of H-, G- and S-type lignin and yielded a gallic acid titer of 0.42 g L−1 from an alkaline-pretreated lignin stream.64 This engineered biocatalyst offered a sustainable and facile strategy for the production of gallic acid from lignin with a yield of 91.59% and 0.630 g g−1 lignin.64
In the case of non-ligninolytic strains, a whole-cell biocatalytic system in E. coli was designed, incorporating enzymes such as acyl-CoA synthetase, enoyl-CoA hydratase, hexadecenal dehydrogenase, vanillic acid O-demethylase and p-hydroxybenzoate hydroxylase, or 4-hydroxyphenylacetate 3-monooxygenase oxygenase. Engineered recombinant E. coli expressed feruloyl-CoA synthetase, enoyl-CoA hydratase/aldolase, vanillate O-demethylase oxygenase, and a mutant form of PobAY385F from P. putida. Employing a fed-batch approach, the strain yielded 19.57 mM gallic acid from 20 mM ferulic acid, achieving a conversion efficiency of 97.9%.65 These findings highlighted the significant advancements in efficiently converting lignin into gallic acid, facilitated by the synergistic integration of multiple technologies.
Vanillic acid serves as a precursor of vanillin, which is extensively utilized in diets, cosmetics and pharmaceuticals, showcasing multifunctional effects including antimutagenic, antiangiogenetic and anti-sickling.66 Ferulic acid can be converted into vanillic acid via the CoA-dependent oxidation pathway. Regarding non-ligninolytic strains, Corynebacterium glutamicum employed p-hydroxycinnamoyl-CoA synthetase (PhdA) to catalyze ferulic acid, yielding feruloyl-CoA. This intermediate was further processed by enoyl-CoA hydrase/aldolase (PhdE), leading to the formation of an intermediate compound, 4-hydroxy-3-methoxybenzoyl acetyl-CoA. Subsequent oxidation by 3-hydroxyacyl-CoA dehydrogenase (PhdB) transformed the hydroxy group into a keto group, resulting in the production of vanillic acid through the catalysis of 3-oxoacyl-CoA ketohydrolase (PhdC) and vanillin dehydrogenase (Vdh). For biocatalytic conversion using p-coumaric acid, three different expression plasmids were introduced into E. coli, enabling the efficient conversion of syringaldehyde and syringic acid via Burkholderia sp. ISTR5.40 Moreover, an engineered E. coli BL21 (DE3) strain, specialized in reducing aromatic aldehydes, was developed.67 This strain facilitated the production of aromatic aldehyde precursors, leading to the increased synthesis of hydroxyphenylacetic acids through a one-pot bioconversion process. This process involved two distinct pathways, yielding 4-hydroxyphenylacetic acid, homovanillic acid, and 3,4-dihydroxyphenylacetic acid. Additionally, a modeling strategy was utilized to assess the production of hydroxyphenylacetic acids from lignocellulosic biomass hydrolysate, resulting in the addition of 5.2 mM 4-hydroxyphenylacetic acid.67 Furthermore, the psychrotolerant strain Paraburkholderia aromaticivorans AR20-38 showed efficient catalysis of ferulic acid into vanillic acid, with the optimized culture conditions leading to molar vanillic acid yields exceeding 90%.68 The findings highlighted the potential of lignin as a renewable resource for valuable aromatic acids sustainably.
Overall, the engineered strains exhibited the successful synthesis of diverse aromatic acids, showcasing promising potential for converting lignin into valuable aromatic fine chemicals. However, to fully harness this potential for microbial production, challenges such as substrate diversity, intricate structures, pathway inefficiencies, enzyme limitations, and the accumulation of metabolic byproducts must be addressed. Firstly, the metabolic pathways of representative aromatic acids remain lengthy, complex and energy intensive, partly due to the limited heterologous expression and constrained carbon metabolism. The development and optimization of effective metabolic pathways are still required to funnel lignin derivatives into aromatic acids. Secondly, a significant knowledge gap exists regarding the enzymatic catalysis mechanisms and pathway refinement. Thus, it is crucial to deeply analyze the molecular mechanism of the catalytic role of the related enzymes and continuously refine the known metabolic pathways. Additionally, the synthesis efficiency of aromatic acids remains inadequate due to the limited steps in the metabolic pathways, requiring protein engineering and screening of ligninolytic strains and key enzymes to enhance catalytic functions.
4 Converting lignin derivatives into aromatic alcohols
The hydroxyl-rich benzene ring in lignin monomers enables their bioconversion into various aromatic alcohols or phenols. Aromatic alcohols and phenols, characterized by a hydroxyl group attached to an aromatic ring structure, include benzyl alcohol, eugenol and phenol. Due to their antimicrobial properties, antioxidant activity and aromaticity, they are valuable components in pharmaceuticals, cosmetics, flavors, and polymers. The lignin-derived G-type monomer ferulic acid can be converted into vanillin, and then turned into vanillyl alcohol. Alternatively, the lignin-derived H-type monomer p-coumaric acid undergoes non-oxidative decarboxylation to yield tyrosol (Table 2).| Aromatic chemicals | Host strains | Substrates | Gene origin | Engineering strategies | Performance (titer) | Performance (yield) | Ref. |
|---|---|---|---|---|---|---|---|
| Pyrogallol | E. coli | Syringic acid | desA; ligM; lpdc | Co-expressing desA, ligM, and lpdc genes | 7.3 mg L−1 | — | 63 |
| Vanillyl alcohol | Cystobasidium laryngis FMYD002 | Vanillin | — | Isolating and identifying a yeast from decaying wood | — | — | 71 |
| 4-Hydroxybenzyl alcohol, vanillyl alcohol, and syringyl alcohol | L. starkeyi | 4-Hydroxybenzaldehyde, vanillin, and syringaldehyde | — | Converting three representatives of lignin derivative aldehydes into their alcohol forms | 122.12, 152.15, and 182.17 g mol−1, respectively | 90.1%, 97.6%, 97.5% | 72 |
| Coniferyl alcohol | Amycolatopsis sp. HR167 | Eugenol | vaoA | Heterologous expressing vaoA | 4.7 g L−1 | — | 73 |
| Tyrosol | E. coli | p-Coumaric acid | fdc1; smo; soi; par | Co-expressing fdc1, smo, soi, and par genes | 2.07 g L−1 | over 90% | 74 |
| Catechol | P. putida KT2440 | Maize bran hydrolysate derived p-coumaric acid | catA/A2; pcaHG | Knocking out catA/A2 and pcaHG | — | 58% | 77 |
| Catechol | E. coli | Vanillin | — | Co-expressing a catechol biosynthesis pathway with an active aromatic transporter CouP under induction by a vanillin self-inducible promoter, ADH7 | — | Improved about 30% and 40% under promoter pTrc and ADH7. | 102 |
| 4-Hydrostyrene | C. glutamicum | Lignin-extracted p-coumaric acid | BaPAD | Deleting phdA; heterologous expressing pad | 1.7 g L−1 | 73% | 135 |
| 4-Hydrostyrene | E. coli | p-Coumaric acid | BaPAD | Heterologous expressing pad | 31.9 g L−1 | 88.7% | 136 |
| 4-Hydrostyrene | E. coli | Alkaline hydrolyzate of corn cob | LpPDC | Heterologous expressing pdc | 1.0035 g L−1 | 137 | |
| Hydroxytyrosol | E. coli | p-Coumaric acid | pad; styAB; styC; adh; par1; yqhD; hpaBC | Introducing pad, styAB, styC, adh, par1, yqhD, and hpaBC genes | 1.83 g L−1 | 97.5% | 138 |
| Tyrosol | E. coli | p-Coumaric acid | pad; styAB; styC; adh; par1; yqhD | Co-expressing pad, styAB, styC, adh, par1 with yqhD genes | 2.046 g L−1 | 97.4% | 138 |
| Homovanillyl alcohol | E. coli | Ferulic acid | pad; styAB; styC; adh; par1; yqhD | Co-expressing pad, styAB, styC, adh, par1, and yqhD genes | 1.189 g L−1 | 91.9% | 138 |
Vanillyl alcohol, a phenolic compound, features a hydroxyl group attached to an aromatic ring. It finds notable applications in fragrances, flavoring, pharmaceuticals and polymer development.69 In the case of ligninolytic strains, such as R. opacus PD630, the metabolic distribution of vanillin was elucidated by integrating intermediate identification, putative gene prediction, and target gene verification. The results indicated that a substantial amount of vanillin consumed was converted into vanillyl alcohol in R. opacus PD630, with a yield of 1.69%.70 Additionally, Cystobasidium laryngis, isolated from decaying wood, was demonstrated to catalyze the reduction of vanillin to alcohol. This conversion significantly decreased the toxicity of lignin, effectively safeguarding the microbes from harm.71
A novel approach was devised for synthesizing aromatic alcohols and lipids using L. starkeyi from lignin derivatives.72 This process effectively converted three representative lignin-derived aldehydes into their corresponding aromatic alcohols, including 4-hydroxybenzyl alcohol, vanillyl alcohol, and syringyl alcohol, with a remarkable yield of 90.1%, 97.6%, and 97.5%, respectively. Remarkably, doubling the concentration of syringaldehyde in individual lignin fermentation enhanced cell growth to 17.01 g L−1 and lipid accumulation to 30.72%. This study introduced an innovative approach for producing aromatic alcohols from lignin derivatives.72 The vanillyl alcohol oxidase gene vaoA from Penicillium simplicissimum CBS 170.90 was expressed in the vanillin-tolerant Gram-positive strain Amycolatopsis sp. HR167. Utilizing eugenol as a cost-effective substrate, the biotechnological production of coniferyl alcohol achieved a peak concentration of 4.7 g L−1 after a 16 h bioconversion process.73E. coli harbored ferulic acid decarboxylase from S. cerevisiae, styrene monooxygenase, styrene oxide isomerase from P. putida, and phenylacetaldehyde reductase from Solanum lycopersicum, which converted over 90% p-coumaric acid to tyrosol, reaching a titer of 545.51 mg L−1.74
The bioconversion of lignin derivatives into valuable phenols also shows the potential to promote lignin valorization. Catechol is a phenolic compound and exhibits antioxidant properties and metal-chelating abilities, which are valuable in pharmaceuticals, cosmetics, and materials science. Catechol is also a precursor for vanillin or eugenol synthesis, making it useful in the food, perfume, and personal care product industries.75 Numerous lignin-derived aromatic precursors, such as phenol, guaiacol, benzoic acid, and protocatechuic acid, have been identified for catechol biosynthesis.16 In the case of ligninolytic strains, it is possible that when protocatechuic acid accumulates, R. jostii can further decarboxylate it into catechol, thereby preventing higher concentrations of protocatechuic acid from building up.76P. putida KT2440 was engineered through knocking out the genes catA/A2 encoding catechol 1,2-dioxygenase and pcaHG encoding protocatechuate 3,4-dioxygenase, inhibiting unwanted metabolic reactions.77 To overcome metabolic obstacles such as vanillic acid accumulation, the codon-optimized vanAB gene that encodes vanillate-O-demethylase was overexpressed toward catechol biosynthesis. The engineered strain could survive on the medium of lignin-derived model compounds and biomass hydrolysate composed of different lignin monomers to produce catechol, with a yield higher than 60%.77
Pyrogallol, characterized by a benzene ring substituted with three hydroxyl groups, is a polyphenol of significant value in pharmaceuticals, photography, and antioxidants. The lignin-derived S-type aromatic syringic acid can be converted into gallic acid, which was subsequently converted into pyrogallol. Alkali lignin underwent a depolymerization process to produce vanillin and syringate as the main aromatic monomers.63 A new synthetic pathway consisted of vanillin degradation (ligV and ligM), a protocatechuate decarboxylase (aroY) and a catechol dioxygenase (CatA) in Sphingomonas SYK-6. Subsequently, engineered E. coli can convert syringate into pyrogallol by co-expressing desA, ligM, and Lpdc, with the yield of 7.3 mg L−1.63 A coenzyme-free biocatalyst converted lignin-derived aromatics into a range of valuable polymers and beneficial building blocks.78 By combining newly discovered phenolic acid decarboxylase with aromatic dioxygenase, the conversion of primary lignin monomers into 4-vinylphenol was accomplished without requiring coenzymes. The yield of 4-vinylphenol achieved 20.5 g L−1.78 Additionally, isoeugenol, a propenyl-substituted guaiacol, is also utilized in flavor formulations, essential oils, and vanillin synthesis. An engineered bacterial eugenol oxidase was optimized through computational prediction-guided mutations to selectively catalyze the dehydrogenation of 4-npropylguaiacol to isoeugenol.79
Overall, engineered strains demonstrate potential to valorize lignin derivatives into aromatic alcohols and phenols. However, the yields of high-value aromatic alcohols and phenols from lignin derivatives remain relatively limited, posing challenges for large-scale industrial production. Achieving precise control and high selectivity in synthesizing specific products is often difficult due to the limited selectivity of bioconversion processes, which involve multiple enzyme-catalyzed reactions. For example, to enhance catechol production, it is necessary to effectively convert protocatechuic acid, while hindering its degradation in the TCA cycle. Therefore, efforts focusing on enzyme mining and protein engineering strategies are crucial to enhance the enzymatic cascade efficiency. Additionally, the bioconversion can often generate impurities, and cross-reactions may occur in the various steps. Thus, it is necessary to identify and block the branching pathways of lignin derivatives and products. Moreover, designing artificial pathways is limited by a lack of comprehensive genetic information about certain aromatic alcohols. In this case, to expand the range of aromatic alcohols, innovative reaction pathways and engineered gene expression in heterologous synthesis pathways must be developed through microbial lignin conversion. For example, the synthesis pathway of aromatic alcohols from S-type monomer remains unclear. Prospecting novel conversion pathway and key enzymes can facilitate the conversion of S-type monomers into aromatic alcohols.
5 Transforming lignin derivatives into aromatic aldehydes
Aromatic aldehydes are characterized by an aldehyde group linked to an aromatic ring structure. They are widely utilized in the fragrance, flavor, pharmaceutical, and fine chemical industries due to their distinctive aromas and flavor enhancement properties. The notable compounds in this class include benzaldehyde, vanillin and cinnamaldehyde. Particularly, vanillin is a commercially available product widely used across various industries, such as food, cosmetic and pharmaceuticals.24 The microbial production of vanillin from lignin has emerged as a sustainable alternative, achievable through lignin depolymerization or the bioconversion of aromatic substrates. The bioconversion of aromatic substrates is the primary pathway attracting significant attention, particularly in converting the lignin-derived H- and G-type monomers into vanillin. This conversion can occur via the CoA-dependent β-oxidation pathway, CoA-dependent non-β-oxidation pathway and non-oxidative decarboxylation pathway (Table 3).16| Aromatic chemicals | Host strains | Substrates | Gene origin | Engineering strategies | Performance (titer) | Performance (yield) | Ref. |
|---|---|---|---|---|---|---|---|
| Vanillin | R. jostii RHA1 | Minimal medium containing 2.5% wheat straw lignocellulose and 0.05% glucose | vdh | Deleting vdh | 96 mg L−1 | — | 80 |
| Vanillin | P. putida KT2440 | Ferulic acid | fcs; ech; vdh | Overexpressing fcs and ech; deleting vdh | — | 86% | 81 |
| Vanillin | Bacillus ligniniphilus L1 | Wheat straw | vdh | Deleting vdh | 352 mg L−1 | — | 82 |
| Vanillin | Bacillus pumilus ZB1 | Isoeugenol, eugenol, vanillyl alcohol | — | Incubating B. pumilus ZB1 in the basal medium for 24 h to achieve the bioconversion of guaiacyl lignin monomers to vanillin. | 71.99 mg L−1 | 61.1% | 83 |
| Vanillin | Bacillus subtilis | Ferulic acid | — | Packing a stirring bioreactor with a carbon fiber textiles (FT) biofilm | — | 57.42% | 84 |
| Vanillin | Streptomyces sannanensis MTCC 6637 | De-starched wheat bran | — | Using Streptomyces sannanensis MTCC 6637 for biotransformation of ferulic acid esters | 0.708 g L−1 | — | 85 |
| Vanillin | E. coli | Ferulic acid | PfECH; PfFCS | Heterologous expressing fcs and ech | 4.28 g L−1 | — | 86 |
| Vanillin | E. coli | Ferulic acid | FCS; ECH | Heterologous expressing fcs and ech, overexpressing gltA and deleting icdA | 5.14 g L−1 | 86.6% | 87 |
| Vanillin | Pseudomonas fluorescens BF13 | Ferulic acid | vdh | Inactivating vdh | 1.28 g L−1 | — | 139 |
| Vanillin | Lactobacillus plantarum CECT 748(T) | Ferulic acid | — | Screening a panel of LAB isolates for releasing phenolics from agrowaste materials like rice bran and their biotransformation to industrially important compounds | — | 5.01% | 140 |
| Vanillin | Amycolatopsis sp. strain ATCC 39116 | Ferulic acid | vdh | Deleting vdh | 1.03 g L−1 | — | 141 |
| Vanillin | Bacillus pumilus S-1 | Isoeugenol | — | Identifying a Bacillus pumilus strain based on biochemical tests, cellular fatty acid composition, RiboPrint pattern and 16S rRNA gene sequence analyses | 3.75 g L−1 | 40.5% | 142 |
| Vanillin | E. coli | Vanillic acid | — | Identifying a novel type III fungal CAR and expressing CAR using the pETDuet-1 plasmid system in combination with an autoinduction protocol | 1.4 g L−1, 0.029 g L−1 h−1 | — | 143 |
| Vanillin | Mycobacterium abscessus | Vanillic acid | Mycobacterium abscessus B1MLD7 | Testing fourteen CARs heterologously expressed in E coli | 2.86 g L−1 | — | 144 |
| Vanillin | Streptomyces sp. ssr-198 | Wheat bran ferulic acid | — | Using FAE of E. lactis SR1 and Streptomyces sp. ssr-198 | 685 mg L−1 | — | 145 |
| Vanillin | Bacillus safensis SMS1003 | Eugenol | — | Identifying bacterial strain SMS1003 using biochemical tests and molecular phylogenic analysis | 0.12 g L−1 | 26% | 146 |
| Vanillin | Trichosporon asahii | Isoeugenol | — | Isolating 40 isoeugenol-tolerant yeasts and collecting them from aromatic plants | 2.4 g L−1 | 52.5% | 147 |
| Vanillin | Pycnoporus cinnabarinus | Ferulic acid | — | High-density incubating Pycnoporus cinnabarinus in glucose phospholipid medium | 1.575 g L−1 | — | 148 |
| Vanillin | Lactic acid bacteria | Ferulic acid | — | Using Oenococcus oeni or Lactobacillus sp. | — | 0–1% | 149 |
| Vanillin | Streptomyces sp. strain V-1 | Ferulic acid | — | Using several macroporous adsorbent resins to adsorb vanillin in situ during the bioconversion | 19.2 g L−1 | — | 150 |
| Vanillin | E. coli JM109 | Ferulic acid | fcs; ech | Co-expressing fcs and ech genes from Pseudomonas; using response surface methodology | 2.52 g L−1 | 70.6% | 151 |
| Vanillin | Aspergillus niger CGMCC0774 and Pycnoporus cinnabarinus CGMCC1115 | Waste residue of rice bran oil derived ferulic acid | — | Developing a new technology of transforming ferulic acid into vanillin by combining fungal strains Aspergillus niger CGMCC0774 and Pycnoporus cinnabarinus CGMCC1115 | 2.2 g L−1 | — | 152 |
| Vanillin | Aspergillus niger, Pycnoporus cinnabarinus | Ferulic acid | — | Biotransforming ferulic acid by Aspergillus niger, Pycnoporus cinnabarinus and resin | 2.8 g L−1 | — | 152 |
| Vanillin | Enterobacter hormaechei | Pomegranate peel-derived ferulic acid | — | Optimizing different process parameters and examining their effect on biovanillin production using central composite design of response surface methodology | 4.2 g L−1 | — | 153 |
| Vanillin | Amycolatopsis sp. ATCC 39116 | Ferulic acid | — | Using a solid–liquid two-phase fed-batch partitioning bioreactor (TPPB) system to remove in situ product, enhancing transformation productivity by this strain. | 9.18 g L−1 | — | 154 |
| Vanillin | E. coli | Isoeugenol | Fdc; Cso2 | Using immobilized enzymes Fdc and Cso2 | 1.2 g L−1 | — | 155 |
| Vanillin | Pseudomonas nitroreducens Jin1 | Isoeugenol | — | Constructing recombinant E. coli over-expressing isoeugenol monooxygenase; optimizing the culture conditions for enzyme production and reaction process for vanillin | 252 mM, 115 g L−1 d−1 | 82.3% | 156 |
| Vanillin | Bacillus fusiformis CGMCC1347 | Isoeugenol | — | Isolating a novel strain Bacillus fusiformis CGMCC1347 from soil; avoiding the product inhibition by adding resin HD-8 | 8.10 g L−1 | 78.4% | 157 |
| Vanillin | Pseudomonas resinovorans SPR1 | Eugenol | — | Isolating a novel strain with the capability to grow on eugenol | 240 mg L−1 | 10% | 158 |
| Vanillin | Candida galli strain PGO6 | Eugenol | — | Isolating a novel strain with the capability to grow on eugenol | 1.12 g L−1 | 44% | 158 |
| Vanillin | Psychrobacter sp. strain CSW | Isoeugenol | — | Screening a moderately halotolerant Gram-negative coccobacilli to convert isoeugenol to vanillin | 1.28 g L−1 | 10.2% | 159 |
| Vanillin | P. putida | Isoeugenol | — | Incubating Pseudomonas putida (HUT 8100) in usage a. | 11.95 g L−1 | 6.2% | 160 |
| Syringaldehyde | R. jostii | Organosolv lignin | vdh | Deleting vdh | 0.243 g L−1 | — | 161 |
| Syringaldehyde | E. coli | Syringic acid | — | Screening fourteen microbial carboxylic acid reductases (CARs). | 1.63 g L−1 | 90% | 88 |
In the case of ligninolytic strains, for example, R. jostii RHA1 demonstrated the efficient conversion of lignin into vanillin. Genome sequencing facilitated metabolic pathway engineering for this conversion. Deleting vanillin dehydrogenase gene in the R. jostii RHA1 strain resulted in the accumulation of vanillin with a titer of 96 mg L−1 on a minimal medium containing 2.5% wheat straw lignocellulose and 0.05% glucose.80P. putida successfully produced vanillin from ferulic acid by inactivating the vdh gene, molybdate transporter, and overexpression of the acyl-CoA synthetase and enoyl-CoA hydratase genes.81 Moreover, knocking down the vanilloid oxidase in P. putida demonstrated efficient conversion of ferulic acid to vanillin, achieving a molar yield of up to 86%.81 In the case of non-ligninolytic strains, the system biology-guided design of extremophilic B. ligniniphilus L1 enabled the biosynthesis of vanillin after lignin depolymerization. Deleting the vanillin dehydrogenase gene in B. ligniniphilus L1 resulted in a substantial increase in the vanillin titer, reaching 352 mg L−1, which represented a 28-fold enhancement. This research established a robust biosynthetic framework for vanillin synthesis from lignin, making a significant contribution to a more environmentally friendly approach.82B. pumilus ZB1 exhibited excellent capacity for converting various substrates such as guaiacyl, isoeugenol, eugenol, and vanillyl alcohol into vanillin. Moreover, it showed the capability to convert 61.1% of the isoeugenol and eugenol found in pyrolyzed bio-oil obtained from Masson's pine into vanillin precursors.83 A stirred bioreactor was used to produce vanillin from ferulic acid by employing a B. subtilis biofilm grown on carbon fiber textiles. The molar yield of vanillin from ferulic acid reached 57.4%, with a conversion efficiency of 93.5%.84Streptomyces sannanensis MTCC 6637 utilized its esterase to catalyze ferulic acid esters into ferulic acid, which was subsequently transformed into vanillin via feruloyl-CoA synthetase and enoyl-CoA hydratase/aldolase, achieving a titer of 708 mg L−1.85
Vanillin can also be synthesized using engineered E. coli, which possesses the Pseudomonas genes encoding feruloyl-CoA synthetase and feruloyl-CoA hydratase/aldolase. Furthermore, fermentation optimization significantly increased the titer of vanillin to 28 mM.86E. coli JM109/pBB1 used the hydrolysates of corn cob containing 1.17 g L−1 ferulic acid and 2.16 g L−1p-coumaric acid to produce vanillin. E. coli was engineered by heterologously expressing acyl-CoA synthetase, enoyl-CoA hydratase, citrate synthase and deleting isocitrate dehydrogenase, yielding vanillin with a titer of 5.14 g L−1 from ferulic acid.87S. cerevisiae utilized a combination of engineering strategies involving enzyme fusion and metabolism regulation in the heterologous rate-limiting enzyme. This approach successfully led to the accumulation of 1.97 mmol L−1 vanillin from ferulic acid and p-coumaric acid substrates.17 These results demonstrated that the synthesis of vanillin from lignin derivatives offers promising possibilities to promote both lignin valorization and vanillin production.
Syringaldehyde is highly valuable as a versatile building block in the synthesis of various high-value products, including pharmaceuticals, flavors, fragrances, and antioxidants. The biocatalytic reduction of syringic acid to syringaldehyde, an energetically disfavored reaction, was evaluated using fourteen microbial carboxylic acid reductases.88 Among them, nine exhibited positive syringic acid reduction, while carboxylic acid reductase from Mycobacterium abscessus demonstrated the highest analytical yield. A whole-cell biocatalyst consisting of recombinant E. coli was designed by expressing the carboxylic acid reductase gene, which successfully achieved 90% conversion of syringic acid into syringaldehyde.88 The results indicated that effective engineering approaches facilitated the microbial synthesis of aromatic aldehydes from lignin derivatives.
Overall, these findings underscore the significant progress in elucidating the conversion pathways, enhancing the conversion efficiency, and increasing the product yield of aromatic aldehydes. Nevertheless, there remains ample room for improvement in producing high-value aromatic aldehydes such as vanillin. Firstly, the yield of syringaldehyde, for instance, is low in lignin bioconversion, possibly due to inefficient enzymes, competing reactions, and side reactions. In addition, the downstream reaction of vanillin should be inhibited, which accelerates its degradation. The identification and heterologous expression of key enzymes are essential for increased aromatic aldehyde production. Secondly, precision control is necessary to knock out genes associated with the downstream aromatic aldehyde degradation pathway. Besides, aromatic aldehydes can have cytotoxic effects on the host cells. Metabolic engineering techniques, such as rational design and cofactor engineering, should be utilized to systematically modify the host cells, thereby improving the robustness of the host bacteria. Furthermore, effective fermentation strategies have the potential to enhance the biosynthesis performance of aromatic aldehydes. Considering all these factors, the bioconversion of lignin derivatives to aromatic aldehydes offers a sustainable and viable alternative for lignin biomanufacturing.
6 Functionalizing lignin derivatives into aromatic amines
Aromatic amines are vital organic compounds distinguished by an amino group attached to an aromatic ring structure. They are widely used as antioxidants, in photography, and as pharmaceutical intermediates. Also, their global consumption surpasses 4 million tons annually, valuing this market at around $10 billion.89 Notably, lignin has demonstrated significant potential for effective bioconversion, enabling the synthesis of diverse aromatic amines (Table 4).90| Aromatic chemicals | Host strains | Substrates | Gene origin | Engineering strategies | Performance (titer) | Performance (yield) | Ref. |
|---|---|---|---|---|---|---|---|
| Vanillylamine | E. coli CV | Lignin-derived vanillin | — | Establishing a two-phase reaction medium composed of organic solvent–water; applying Escherichia coli CV as ω-transaminase biocatalyst to the biosynthesis of vanillylamine from lignin-derived vanillin. | — | 100% | 91 |
| Vanillylamine | P. putida KT2440 | Vanillic acid | cvTA; aladh; catA; vdh; ald; bdh | Overexpressing cvTA and aladh genes; delete catA, vdh, ald, and bdh genes; L-alanine and/or NH4Cl as the amine donor | 174 mg L−1 | — | 93 |
| Vanillylamine | E. coli | Ferulic acid | PpFCS; PpECH; CvTA; BsAlaDH | Heterologous expressing fcs, ech, cvTA and aladh | — | 71.5% | 94 |
| Vanillylamine | P. putida KT2440 | Ferulic acid | CvATA; BsAlaDH; | Expressing ω-transaminase and L-alanine dehydrogenase. Overexpression of fcs, ech, ATA and aladh; deleting pcaGH, area, vdh, ald and bdh | — | 90.0% | 95 |
| Vanillylamine | E. coli BL21 (DE3) | Vanillin | CV2025 w-TAm | Heterologous expressing CV2025 w-TAm; (S)-α-methylbenzylamine as the amine donor | — | 100% | 96 |
| Vanillylamine | E. coli | Vanillic acid | CAR/PPTase; cvTA; aladh | Introducing CAR/PPTase, CvTA and AlaDH; NH4Cl as the amine donor | 3.58 g L−1 | 94.5% | 162 |
| Vanillylamine | E. coli | Ferulic acid | fcs; ech; cvTA; aladh | Introducing fcs, ech, cvTA, aladh genes; NH4Cl as the amine donor | 3.64 g L−1 | 96.1% | 162 |
| Cinnamylamine | E. coli | Cinnamic acid | CAR/PPTase; CvTA | Co-expressing CAR/PPTase with CvTA | 523.15 mg L−1 | — | 97 |
Vanillylamine, as an important drug precursor and fine chemical intermediate, possesses significant economic value.91 The synthesis of vanillylamine has garnered significant attention, particularly due to its crucial role as a precursor for capsaicin, a physiologically active alkaloid.92 It was reported that the biosynthesis of vanillylamine from the G-type monomer, ferulic acid, can be achieved by amine transaminases from Chromobacterium violaceum.16 In the case of ligninolytic strains, for example, a novel screening method based on the cell growth of P. putida KT2440 was developed to assess the in vivo activity of recombinant aromatic amine transferases towards vanillylamine.93 This approach led to the identification of the highly active native enzyme Pp-SpuC-II and aromatic amine transferases from C. violaceum in vivo. This whole-cell bioconversion of vanillin yielded about 0.70 mM and 0.92 mM vanillylamine, respectively. The concept of double enzyme co-expression was introduced using a relatively low dosage of amine donors, including vanillin, L-alanine and isopropylamine.93 Regarding non-ligninolytic strains, a recombinant E. coli HNIQLE-AlaDH, expressing co-transaminase from Aspergillus terreus and alanine dehydrogenase from B. subtilis, achieved a 71.5% yield of vanillylamine from vanillin.94 An effective whole-cell-catalyzed bioconversion enabled the conversion of vanillin into vanillylamine.95 A recently recombinant E. coli 30CA cell, expressing L-alanine dehydrogenase and ω-transaminase, successfully converted 50 mM and 60 mM vanillin into vanillylamine, achieving a maximum yield of 90.0% from 60 mM vanillin. This highlighted its eco-friendly characteristics and potential for lignin valorization into valuable chemicals.95
Vanillin was identified as a suitable substrate for vanillylamine biosynthesis. The ω-transaminase from C. violaceum was employed as an omega-transaminase biocatalyst to biosynthesize vanillylamine from lignin-derived vanillin in a two-phase reaction medium, consisting of an organic solvent and water.91 In a dibutyl phthalate-water medium, a C. violaceum whole-cell biocatalyst efficiently converted 50 mM vanillin into vanillylamine with 100% yield. The complete conversion of benzaldehyde, anisaldehyde and veratraldehyde into their respective amines was demonstrated.91E. coli harbored CV2025 ω-transaminase from C. violaceum DSM30191 and showed the selective amination of lignin-derived vanillin into vanillylamine, combining with an excess of the amine donor (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 mol mol−1).96 The ω-aminotransferase Cv-ωTA from C. violaceum, the carboxylic acid reductase from Neurospora crassa and phosphopantetheinyl transferase from E. coli were co-expressed in E. coli. This engineered strain promoted the conversion of cinnamic acid to cinnamylamine, with a titer of 523.2 mg L−1.97 These findings underscore the need for developing enzymes with a broader substrate range and engineering strategies to functionalize lignin derivatives into aromatic amines.
4 mol mol−1).96 The ω-aminotransferase Cv-ωTA from C. violaceum, the carboxylic acid reductase from Neurospora crassa and phosphopantetheinyl transferase from E. coli were co-expressed in E. coli. This engineered strain promoted the conversion of cinnamic acid to cinnamylamine, with a titer of 523.2 mg L−1.97 These findings underscore the need for developing enzymes with a broader substrate range and engineering strategies to functionalize lignin derivatives into aromatic amines.
Overall, these findings significantly advance the understanding and utilization of lignin-derived aromatic amine resources. Microbial conversion has enabled the functionalization of lignin derivatives for the biosynthesis of aromatic amines. However, the range of aromatic amines synthesized through lignin bioconversion is currently limited, and the specific pathways for their synthesis remain unclear. Specific aminopeptidases need precise targeting and modification for lignin conversion toward aromatic amines. Additionally, the bioconversion of lignin to aromatic amines results in the generation of diverse products, with low selectivity towards the desired target amines. Therefore, screening and modification of specific ammonia-related enzymes are required to promote the efficient synthesis of target aromatic amines. Besides, the high reactivity and volatility of aromatic amines predispose them to oxidation and decomposition, requiring the precise control of biological pathways. Furthermore, the potential toxicity of amine products to enzymes necessitates improved enzyme stability, possibly through immobilization for activity under extreme conditions. Additionally, challenges in the storage and utilization of aromatic amines pose hurdles in large-scale industrial production. Addressing these issues requires further in-depth research and technological improvements to enhance the efficiency and selectivity of bioconversion.
7 Emerging technologies boost microbial lignin valorization toward aromatic fine chemicals
7.1 Synthetic biology facilitates the construction of aromatic synthesis pathway
Synthetic biology, a rising interdisciplinary field, holds significant potential for reprogramming and controlling biological functions.26 When integrated with metabolic engineering, it can significantly boost lignin valorization by exploring microbial metabolism and designing novel enzyme systems. Synthetic biology allows the controlled synthesis of targeted aromatic fine chemicals by modulating the expression of crucial enzymes and regulatory genes (Fig. 5A–C).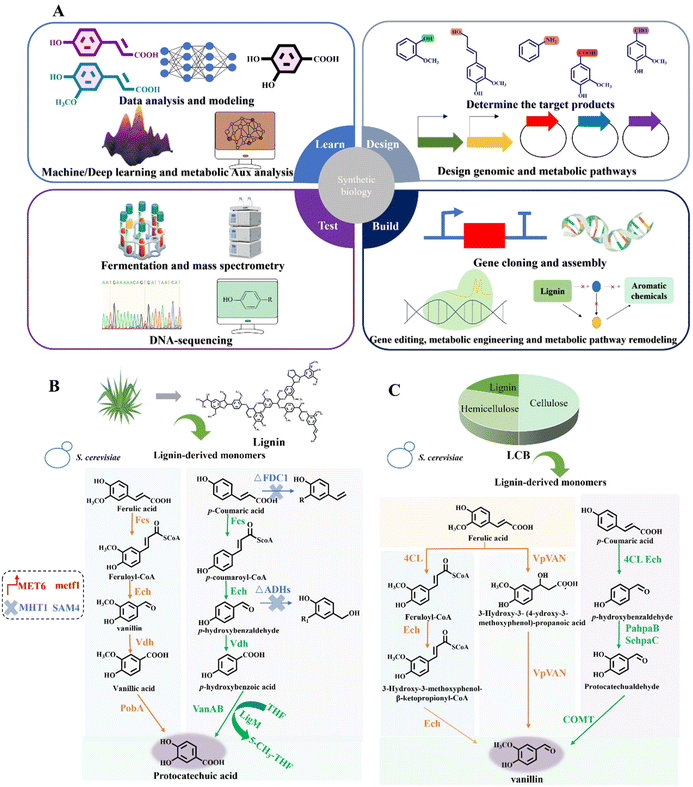 | ||
| Fig. 5 Synthetic biology facilitates the construction of aromatic synthesis pathway. (A) Depolymerized lignin monomers can be valorized into a variety of target aromatic chemicals through the “Design–Build–Test–Learn” cycle of synthetic biology. Specifically, “Design” means determining the target aromatic products and specific metabolic pathways for lignin bioconversion. “Build” means constructing a new system capable of bio-converting lignin into high-value aromatic chemicals via genetic engineering. “Test” means detection technologies such as mass spectrometry and DNA-sequencing, which play an important role in ensuring product alignment with system design specifications and identifying defects for further improvement after fermentation. “Learn” signifies the application of machine learning and deep learning in data analysis, modeling and metabolic aux analysis, which provides feedback and improvements for the design and construction of desired lignin bioconversion process. (B) Typical application of synthetic biology in the construction of lignin aromatic biosynthesis pathway.59 Lignin-derived monomers ferulic acid and p-coumaric acid can be valorized into protocatechuic acid through biological funnel pathway in S. cerevisiae, with a molar conversion yield of 94.5%. The synthetic biology methods include hydroxylase expression enhancement, demethylase enhancement and tetrahydrofolate regulation.59 (C) Application of synthetic biology for lignin bioconversion into value-added chemicals.17 Lignin derivatives can be transformed into vanillin through biological funneling in S. cerevisiae, with the yield of 10.5 mg per g carbon source. Engineering strategies in the bioconversion process encompass hydroxylase fusion and S-adenosylmethionine synthesis.17 | ||
Three of the most important reactions, namely O-demethylation, hydroxylation and decarboxylation, bridge the bioconversion from lignin to aromatic chemicals. In the case of O-demethylation, S. cerevisiae was engineered for stable vanillin production from ferulic acid and p-coumaric acid.17 Hydroxylase expression enhancement, demethylase fusion and S-adenosylmethionine synthesis were utilized for pathway construction and metabolism regulation. The final yield reached 10.5 mg vanillin per g carbon source (Fig. 5C).17 Moreover, by co-expressing the key rate-limiting enzymes of 4-hydroxybenzoate hydroxylase, vanillate-O-demethylase and transporter HcnK in P. putida KT2440, it was possible to convert p-coumaric acid and ferulic acid into protocatechuic acid with a titer of 12.7 g L−1.98 Besides, the catabolite repression control protein serves as a pivotal global regulator that modulates carbon catabolite repression in microbial systems. 4-Hydroxybenzoate hydroxylase PobA and vanillate demethylase VanAB were confirmed to be the targets of the catabolite repression control protein. Deleting the gene encoding the catabolite repression control protein significantly decreased 4-hydroxybenzoate and vanillate accumulation, boosting muconate production by about 70%.99 A full-length Class I electron transfer system from Rhodopseudomonas palustris HaA2 was expressed and characterized. The key enzyme in this system, CYP199A4, selectively demethylated veratric acid at the para position, yielding 1.2 g L−1 of vanillic acid.100 The tetrahydrofolate (THF)-dependent O-demethylase LigM from Sphingobium sp. SYK-6 was investigated, which converted vanillic acid into protocatechuic acid. The substrate selectivity was elucidated rationally and ligM was successfully overexpressed in E. coli, achieving up to 80 mg L−1 in the fermentation broth. The plant methionine synthase MetE was co-expressed in E. coli and combined with LigM to mitigate cofactor consumption.101
Regarding hydroxylation, ferulic acid and p-coumaric acid could be converted through the biological funnel pathway in Saccharomyces cerevisiae. Synthetic biology strategies promoted the synthesis of protocatechuic acid, including hydroxylase expression enhancement, demethylase enhancement and tetrahydrofolate regulation. The molar conversion yield of protocatechuic acid reached 94.5% (Fig. 5B).59 Synthetic biology also helped to construct numerous bacterial biosensors capable of detecting aromatic chemicals and assessing lignin conversion. For example, genes of CouP, LigV, LigM and Aro were overexpressed under a vanillin self-inducible promoter, ADH7, using E. coli, facilitating the transport of vanillin across the cell membrane and its conversion into catechol without additional inducers.102 The strain with couP integrated into the catechol operon showed a 40% increase in catechol and a 10% increase in vanillic acid production compared to the strain without the transporter.102
In the case of decarboxylation, introducing a microbial phenolic acid decarboxylase with Gibson assembly successfully achieved the bioconversion of p-coumaric acid into 4-vinylphenol in C. glutamicum, E. coli, and B. subtilis. Among them, C. glutamicum showed the best performance, with a titer of 187 g L−1 in a biphasic system.103 The heterologous expression of the vanillin synthase gene from Vanilla planifolia in S. cerevisiae facilitated the catalysis of ferulic acid into vanillin. Vanillin was further aminated to vanillylamine with the conversion rate of 100% through the heterologous expression of CV2025 w-transaminase in E. coli.96 A cascade of coenzyme-independent ferulic acid decarboxylase and Caulobacter segnis dioxygenase was utilized to enhance an enzymatic vanillin synthesis system.104 The highest yield of 35.0% was obtained with immobilized enzymes using ferulic acid. The site-directed mutagenesis further improved Caulobacter segnis dioxygenase mutants to achieve a yield of 58.4%.104Paraburkholderia aromaticivorans AR20-38 effectively converted vanillic acid from ferulic acid at low and moderate temperatures. Transcriptome analysis revealed the characterization of transcriptional factors, cellular transporters and differentially expressed genes, suggesting a plausible membrane vesicle mechanism.105
A synthetic biology strategy was also conducted by deleting two genes associated with p-hydroxybenzoic acid degradation in Burkholderia glumae strain BGR, which efficiently produced p-hydroxybenzoic acid from p-coumaric acid. The overexpression of phcs II, which encodes p-hydroxycinnamoyl-CoA synthetase II, yielded an impressive 99.0% of p-hydroxybenzoic acid from 20 mM p-coumaric acid, marking the remarkable titer of p-hydroxybenzoic acid produced biologically.106 These studies highlighted the potential of two-component systems as a valuable tool in the critical phase of microbial lignin conversion.
Furthermore, the representative ligninolytic strain, R. opacus PD630, with its inherent “biological funneling” pathways, holds significant potential as a promising platform for lignin valorization. Employing green fluorescent protein-based sensors and deletion analysis, the transcription factors controlling aromatic degradation were identified, highlighting the application potential of synthetic biology.107 This study revealed the regulatory role of transcriptional activators in phenol, guaiacol, and other aromatic metabolism, advancing R. opacus as a lignin valorization platform.107
In summary, advancements in synthetic biology have provided a promising framework for efficiently converting lignin into valuable aromatic chemicals, paving the way for engineering microbial cell factories. However, the significant challenges in lignin bioconversion call for more research on synthetic biology. Advancing lignin valorization requires developing new strains through the Design–Build–Test–Learn cycle of biofoundries (Fig. 5A). Integrating advanced DNA sequencing, genome editing, and transposon libraries will deepen our understanding of the lignin bioconversion genetic mechanisms, boosting the design of genetic circuits for the effective utilization of lignin-derived monomers.
Secondly, screening microorganisms with high bioconversion potential should be a priority, enabling them to digest and convert lignin into specific building block chemicals. Ideal host strains should be engineered through synthetic biology to integrate depolymerization and conversion pathways, addressing the heterogeneity of lignin and enhancing its bioconversion efficiency. Multi-level omics technology can assist in accumulating biological information, screening potential ligninolytic host strains, and deducing related pathways.
Thirdly, advancing genetic tools and manipulation systems for ligninolytic microorganisms is necessary, leveraging insights from systems biology regarding essential pathways and enzymes in lignin degradation. Innovative systems that synergistically combine ligninolytic microorganisms and enzymes should also be developed to expedite lignin processing and improve its catalytic properties. Additionally, synthetic biology has played a crucial role in enhancing the solvent tolerance of host bacteria, countering product and substrate toxicity. These approaches can lead to the development of better industrial strains, ultimately contributing to optimizing lignin valorization processes.
7.2 Artificial intelligence guides the design of lignin valorization process
Artificial intelligence (AI) encompasses the ability of a system to adapt flexibly to achieve goals, which includes interpreting data, learning from it, and applying insights effectively.108,109 Data-driven models leverage accumulated data to contribute to the improvement of biorefineries.110 Pertinent data for machine learning applications may encompass biomass images, lignin quality data, bioconversion parameters, desired product yield, and other relevant characteristics.110 The classification of extensive data sets from lignocellulosic biomass processes will aid in devising cost-effective bioconversion for generating high-value products.111,112AI also facilitates the screening and engineering of microbial strains to achieve efficient lignin depolymerization and bioconversion. Machine learning algorithms analyze vast experimental data and structural information to forecast the types and yields of lignin depolymerized products.113 AI aids the analysis of genomics and metabolomics data, the metabolic pathways of potential lignin bioconversion strains, and the predicted product yields. Furthermore, AI can be utilized to devise and refine genetic engineering strategies aimed at improving the microbial degradation and conversion efficiency of lignin. AI plays a pivotal role in uncovering new biocatalysts and targets for metabolic engineering, thereby fostering research and practical applications in lignin bioconversion. Moreover, AI also contributes to the enhancement of lignin bioconversion enzymes by optimizing their activity, stability, and specificity.114 Through structure prediction and simulation, strategies for enzyme mutation can be designed and fine-tuned, together with forecasting the effects of mutations on the performance and catalytic efficacy of enzymes. This facilitates the design of more effective ligninolytic enzymes, driving lignin bioconversion forward.
AI has significantly advanced lignin depolymerization. The integration of synthetic biology tools with deep learning methodologies shows promising potential for boosting the product yields.115 Utilizing artificial neural networks and the Taguchi method as dynamic statistical techniques, the alkali sodium hydroxide pretreatment of hybrid Napier and Deenanath grasses was optimized, with the experimental results closely matching the model predictions.116
AI plays a crucial role in the selection and modification of ligninolytic enzymes. Previous studies have dedicated extensive efforts to leveraging microbes as biocatalysts for producing a diverse array of aromatics and derivatives. Drawing on data from existing literature, a novel predictive model was developed to estimate the bio-oil yield, char yield, and reaction time. This model included factors such as the surface properties of the catalyst and the weight-averaged molecular weight of the lignin used in the reaction.117 AI-based methods facilitate the efficient and precise prediction of enzyme properties, encompassing substrate specificity, catalytic activity, and stability. These methodologies aid in identifying promising ligninolytic enzymes and informing the rational design of enzyme variants with enhanced efficiency and specificity. This accelerates the progress in developing biocatalysts for lignin valorization.
Comamonas testosteroni FJ17 exhibited the enzymatic activity of laccase and the delignification of rice straw. Molecular docking and AI modeling revealed the interaction features between laccase and lignin, achieving a peak activity of 2016.7 U L−1.118 White-rot fungi can release versatile peroxidases to effectively degrade lignin. The model structures obtained through deep-learning-based ab initio structure prediction methods were validated as reliable foundations for one-shot Protein Repair One Stop Shop (PROSS) stability-design computations.119 Engineered versatile peroxidases with up to 43 mutations were successfully expressed in S. cerevisiae, a feat not achieved by its wildtype counterparts. This computational optimization enhanced the functional diversity of natural enzymes, directly leveraging genomic databases.119 A novel unsupervised learning approach was devised for categorizing laccases independent of homology. This study emphasized that physicochemical properties showed superior performance compared to other descriptors, offering a valuable method for categorizing laccases and gaining deeper insights into their physicochemical characteristics.120 These findings contribute to a better understanding and utilization of laccases and versatile peroxidases for lignin valorization.
AI, particularly machine learning, is instrumental in devising efficient synthetic strategies for value-added products, enhancing the predictability of metabolic engineering. This capability empowers researchers to improve their ability to forecast, optimize, and manipulate metabolic pathways in designing value-added products.112 For instance, the synthesis of isoeugenol, a valuable fragrance compound, was achieved by genetically modifying a bacterial eugenol oxidase to convert 4-n-propylguaiacol. Initially, guided by computational predictions, five mutations were introduced to boost the thermal stability of the enzyme. Subsequent iterations involved incorporating and assessing additional mutations to improve chemoselectivity and catalytic activity. This study showcases the potential to reengineer a native enzyme into a tailored biocatalyst for valorizing lignin-derived monophenols.79
In summary, AI enables the analysis of complex biological data to discover new enzymes and metabolic pathways in microbial lignin valorization. This not only aids in predicting the physicochemical and chemical properties of lignin but also facilitates the identification of new lignin-derived compounds. Furthermore, AI algorithms can assist in the rational design of ligninolytic enzymes with improved catalytic properties. Moreover, AI-based models can optimize the process conditions, forecast product yields, and guide the selection of optimal microbial hosts for lignin bioconversion. AI is anticipated to further enhance the exploration and modification of carrier proteins and intracellular proteins involved in transmembrane transport. This cutting-edge technology also holds potential for accelerating the development of sustainable and efficient strategies for producing valuable aromatic chemicals (Fig. 6).
7.3 Directed evolution for the engineering of ligninolytic enzymes
Directed evolution is deeply rooted in both adaptive evolution and classical strain engineering, emerging as one of the most effective and popular methods for generating new or improved functionality in proteins, metabolic processes, and even entire genomes (Fig. 7A).121 The ligninolytic enzymatic consortium, primarily composed of nonspecific oxidoreductases such as laccases, peroxidases, and H2O2-producing oxidases, stands as a versatile tool in biotechnology.122 Directed evolution strategies have been employed to adapt these enzymes to the harsh industrial conditions, such as high temperatures, organic solvents, extreme pH, and inhibitors. This approach has been effective in enhancing the enzyme activity.122 A protein engineering toolkit was developed for ferulic acid decarboxylase from S. cerevisiae, utilizing directed evolution and/or structure-guided methodologies.123 The uniqueness of this tool stems from its capability to screen ferulic acid decarboxylase activity within a mutant library, employing a newly developed fluorescent plate-assay in a high-throughput manner. Evaluating the variant library with diverse substitutions unveiled several variants with enhanced activity. Computational analysis confirmed the assay results, supporting the proposed molecular model. This study offers vital insights into improving the substrate-specific ferulic acid decarboxylase activity, paving the way for customizing its substrates.123 | ||
| Fig. 7 Directed evolution empowers the engineering of ligninolytic enzymes. (A) Key steps of funneling lignin derivatives into aromatic chemicals with directed evolution. Firstly, DNA variants library can be generated through random mutation, followed by gene evolution via screening and selection. This results in evolved enzymes capable of catalyzing the bioconversion of lignin into valuable aromatic chemicals, including O-demethylation, hydroxylation and decarboxylation. (B) Typical research on directed evolution for lignin bioconversion utilizes flow cytometry to select mutants of lignin peroxidase with enhanced oxidative stability.124 To enhance the oxidative stability of lignin peroxidase, a random mutagenesis library consisting of 106 lignin peroxidase genes was established, and then flow cytometry was employed to identify mutants outperforming the wild-type. After two rounds of sorting, the proportion of variants exhibiting enhanced stability increased from 1.4% to 52.3%.124 (C) Another significant application of directed evolution in lignin bioconversion, which enhanced the oxidizing efficiency of dye-decolorizing peroxidases under alkaline pH conditions.129 Firstly, random mutation was introduced via error-prone PCR to create a DNA variant library, followed by high-throughput screening to identify and select the desired enzyme, 6E10 variant, which exhibited a 100-fold increase in catalytic efficiency for 2,6-dimethoxyphenol. This variant also demonstrated improved enzyme yield, stability, ABTS activity, redox potential, and catalytic efficiencies for phenolic and aromatic amines, with the optimal pH at 8.5.129 | ||
To enhance the oxidative stability of lignin peroxidase, a random mutagenesis library consisting of 106 mutated lignin peroxidase genes was constructed.124 Flow cytometry was employed to screen the expressed enzymes with increased activity. Two rounds of sorting significantly increased the population of stable variants from 1.4% to 52.3%. The most stable variants displayed two to four mutations and retained up to 80% of initial activity (Fig. 7B).124 A colorimetric screening method enhanced the functionality of lignin peroxidases via directed evolution, resulting in the identification of three different types of lignin peroxidase mutants. The Kcat/Km values for 2,4-DCP and H2O2 substrates in the mutant enzymes increased approximately 4-fold and 89-fold, respectively.125 Directed evolution, combined with a fast colorimetric screening strategy, identified mutant genes with enhanced polychlorinated phenol degradability and H2O2 stability. Subsequently, these genes were successfully expressed as secretive lignin peroxidases in recombinant S. cerevisiae, demonstrating about 1.6-fold increased stability against H2O2 and enhanced degradation activity towards 2,4-dichlorophenol compared to the parent strain.126
Combining directed evolution with rational design enhanced the catalytic activity of laccase towards lignin phenolics. Although natural laccases have evolved to oxidize lignin under acidic conditions and mild temperatures, the modified enzyme, capable of oxidizing lignin at pH 10, demonstrated optimal activity at 70 °C and exhibited a significant improvement in thermal resistance. This advancement was realized through the introduction of eight mutations in the protein sequence, resulting in increased heat resistance of the enzyme.127 Additionally, directed evolution was employed to modify a thermostable versatile peroxidase with high-redox potential but limited activity at basic pH.128 Combining directed evolution with hybrid techniques, a basic pH-active versatile peroxidase was generated. The evolutionary pathway was precisely managed using minimal mutational loads of 1–3 nucleotide changes per evolutionary round, complemented by in vivo recombination. After three rounds of directed evolution with error-prone PCR, a variant with the beneficial E140G, P182S and Q229P mutations was isolated, doubling the activity of the enzyme at pH 8.0 compared to the previous evolution stage.128
Directed evolution also enhanced the oxidizing efficiency of dye-decolorizing peroxidases from P. putida MET94 at alkaline pH (Fig. 7C).129 Three rounds of random mutagenesis with high-throughput screening led to the identification of the desired variant. The laccase variant showed enhanced alkali tolerance and heat resistance for lignin degradation. Epistasis between A142V and E188K mutations was proven to be the crucial site determining the substrate specificity, particular for 2,6-dimethoxyphenol with a 100-fold catalytic efficiency increase. The variant also showed improved yield, stability, ABTS activity, and redox potential, with an optimal pH of 8.5.129 A laboratory evolution approach enhanced the selectivity of the metallo-oxidase McoA from Aquifex aeolicus towards aromatic chemicals.130 Four rounds of random mutagenesis of the mcoA gene and subsequent high-throughput screening obtained the variant. This variant demonstrated a 100-fold increase in catalytic efficiency compared to the wild-type enzyme for the common laccase substrate ABTS. It also enhanced activity towards phenolics and synthetic aromatic dyes.130 Under the optimal enzymatic conditions, the mutation resulted in a 10% enhancement in the oxidative degradation of the lignin model compound compared to the wild-type enzyme. Additionally, the variant exhibited a 5–30% increase in the production of high-value aldehydes.131 These findings demonstrate the effectiveness of directed evolution in enhancing enzyme activity and target product yields.
Therefore, directed evolution serves as a powerful engineering tool for the development and modification of effective ligninolytic enzymes. However, several limitations must be addressed to further enhance their activity. Firstly, natural ligninolytic enzymes or catalysts are sensitive to harsh conditions, frequently leading to their inactivation. Thus, the main challenge involves identifying enzymes that can withstand extreme environmental conditions, a task often achieved through directed evolution. Secondly, the structural diversity and heterogeneity of lignin make it challenging to identify enzymes or catalysts with broad substrate specificity. Therefore, an effective directed evolution strategy is necessary. Thirdly, ligninolytic enzymes often result in a wide range of products, making it challenging to achieve high selectivity for the target aromatic chemicals. An effective directed evolution strategy is still required to enhance the performance of enzyme cascades. Moreover, although directed evolution can improve the performance of enzymes, it is crucial to carefully evaluate the scalability and economic feasibility of the resulting biocatalysts. Additionally, the integration of directed evolution, computational biology and other methodologies can further enhance the performance of enzymes, paving the way for sustainable lignin valorization.
8 Conclusions and outlooks
The findings show that the microbial conversion of lignin holds promising potential to enhance both lignin valorization and aromatic fine chemical synthesis, aligning with atom-economy and sustainable principles. Deeper insights into bioconversion pathways and various types of aromatic products have been gained by leveraging microbial lignin valorization to unlock its inherent aromatic potential. This progress is anticipated to stimulate market expansion and render the lignin valorization process economically viable and sustainable.However, various challenges and obstacles in lignin valorization still exist. The heterogeneous structure of lignin presents a notable challenge to conversion efficiency. To generate lignin derivatives suitable for bioconversion, it is crucial to gain a more comprehensive understanding of the lignin depolymerization mechanism and the processability of lignin derivatives. Although the significant cleavage of inter-unit linkages and molecular weight reduction have been achieved, the production yields of bioavailable lignin derivatives and subsequent bioconversion have not reached ideal levels. Additionally, the complete conversion of all heterogeneous components of lignin into targeted aromatics remains elusive. Although naturally occurring ligninolytic microorganisms exist, they often lack the desired properties for industrial applications, such as high yields and efficient production capacities. Therefore, the industrial application of wild ligninolytic microorganisms remains challenging.
Accordingly, to address these challenges, the implementation of advanced technologies is necessary. Firstly, the development of innovative and environmentally friendly lignin depolymerization technologies should be prioritized. Additionally, it is crucial to focus on technologies for mixed depolymerization products to enable easier subsequent upgrading and bioconversion. Furthermore, it is necessary to further optimize the lignin conversion pathways by redesigning the natural lignin conversion pathways and establishing new artificial routes in model microorganisms. In this process, the integration of advanced technologies can facilitate the modification of key enzymes and improve their expression efficiency. Moreover, screening dominant ligninolytic microorganisms is vital to expand bioconversion resources. Exploring novel catabolic pathways and genes in the metabolism of lignin derivatives is also necessary. These efforts will collectively contribute to establishing a comprehensive lignin metabolism network. Overcoming the bottleneck in lignin bioconversion holds great promise for promoting the bioeconomy, achieving carbon neutrality, and making industrially feasible lignin bioconversion a reality.
Data availability
No primary research results, software or code has been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2023YFC3403500) and the Key Research and Development Program of Ningxia Hui Autonomous Region (2024BEE02005).References
- Z. Luo, C. Liu, A. Radu, D. F. De Waard, Y. Wang, J. T. Behaghel De Bueren, P. D. Kouris, M. D. Boot, J. Xiao, H. Zhang, R. Xiao, J. S. Luterbacher and E. J. M. Hensen, Nat. Chem. Eng., 2024, 1, 61–72 CrossRef.
- Q. Liu, T. Kawai, Y. Inukai, D. Aoki, Z. Feng, Y. Xiao, K. Fukushima, X. Lin, W. Shi, W. Busch, Y. Matsushita and B. Li, Nat. Commun., 2023, 14, 4866 CrossRef CAS PubMed.
- D. M. Alonso, S. H. Hakim, S. Zhou, W. Won, O. Hosseinaei, J. Tao, V. Garcia-Negron, A. H. Motagamwala, M. A. Mellmer, K. Huang, C. J. Houtman, N. Labbé, D. P. Harper, C. T. Maravelias, T. Runge and J. A. Dumesic, Sci. Adv., 2017, 3, e1603301 CrossRef.
- A. L. Yaguchi, S. J. Lee and M. A. Blenner, Trends Biotechnol., 2021, 39, 1037–1064 CrossRef CAS PubMed.
- Z.-H. Liu, N. Hao, Y.-Y. Wang, C. Dou, F. Lin, R. Shen, R. Bura, D. B. Hodge, B. E. Dale, A. J. Ragauskas, B. Yang and J. S. Yuan, Nat. Commun., 2021, 12, 3912 CrossRef CAS PubMed.
- J. Guo, D. Liu and Y. Xu, Sustainable Energy Fuels, 2024, 8, 1153–1184 RSC.
- L. Lin, Y. Cheng, Y. Pu, S. Sun, X. Li, M. Jin, E. A. Pierson, D. C. Gross, B. E. Dale, S. Y. Dai, A. J. Ragauskas and J. S. Yuan, Green Chem., 2016, 18, 5536–5547 RSC.
- P. Li, J. Ren, Z. Jiang, L. Huang, C. Wu and W. Wu, RSC Adv., 2022, 12, 10289–10305 RSC.
- S.-Y. Zhu, S.-C. Liu, C.-X. Zhang, X. Xin, Z.-H. Liu, L.-J. Zhang, B.-Z. Li and Y.-J. Yuan, Green Chem., 2024, 26, 5260–5272 RSC.
- R.-Y. Liu, Z.-H. Liu, B.-Z. Li and Y.-J. Yuan, Green Chem., 2024, 26, 1770–1789 RSC.
- H. Liu, X. Tao, S. Ntakirutimana, Z.-H. Liu, B.-Z. Li and Y.-J. Yuan, Chem. Eng. J., 2024, 495, 153375 CrossRef CAS.
- S. J. B. Mallinson, M. M. Machovina, R. L. Silveira, M. Garcia-Borràs, N. Gallup, C. W. Johnson, M. D. Allen, M. S. Skaf, M. F. Crowley, E. L. Neidle, K. N. Houk, G. T. Beckham, J. L. DuBois and J. E. McGeehan, Nat. Commun., 2018, 9, 2487 CrossRef.
- F. Wu, P. Cao, G. Song, W. Chen and Q. Wang, J. Chem. Technol. Biotechnol., 2018, 93, 2804–2816 CrossRef CAS.
- J. M. Perez, C. Sener, S. Misra, G. E. Umana, J. Coplien, D. Haak, Y. Li, C. T. Maravelias, S. D. Karlen, J. Ralph, T. J. Donohue and D. R. Noguera, Green Chem., 2022, 24, 2795–2811 RSC.
- S. Wang and G. Song, Nat. Sustainability, 2023, 6, 1295–1296 CrossRef.
- Z.-H. Liu, B.-Z. Li, J. S. Yuan and Y.-J. Yuan, Trends Biotechnol., 2022, 40, 1550–1566 CrossRef CAS PubMed.
- X. Xin, R.-K. Zhang, S.-C. Liu, Z.-J. He, R.-Y. Liu, H.-N. Lan, Z.-H. Liu, B.-Z. Li and Y.-J. Yuan, Chem. Eng. J., 2024, 149815 CrossRef CAS.
- E. Erickson, A. Bleem, E. Kuatsjah, A. Z. Werner, J. L. DuBois, J. E. McGeehan, L. D. Eltis and G. T. Beckham, Nat. Catal., 2022, 5, 86–98 CrossRef CAS.
- H.-N. Lan, R.-Y. Liu, Z.-H. Liu, X. Li, B.-Z. Li and Y.-J. Yuan, Biotechnol. Adv., 2023, 64, 108107 CrossRef CAS PubMed.
- J. Becker and C. Wittmann, Biotechnol. Adv., 2019, 37, 107360 CrossRef CAS PubMed.
- Z.-M. Zhao, Z.-H. Liu, T. Zhang, R. Meng, Z. Gong, Y. Li, J. Hu, A. J. Ragauskas, B.-Z. Li and Y.-J. Yuan, Biotechnol. Adv., 2024, 70, 108274 CrossRef CAS PubMed.
- L. Zhao, J. Zhang, D. Zhao, L. Jia, B. Qin, X. Cao, L. Zang, F. Lu and F. Liu, Ind. Crops Prod., 2022, 188, 115715 CrossRef CAS.
- T. D. H. Bugg, Chem. Commun., 2024, 60, 804–814 RSC.
- N. Zhou, W. P. D. W. Thilakarathna, Q. S. He and H. P. V. Rupasinghe, Front. Energy Res., 2022, 9, 758744 CrossRef.
- Z. Chen and C. Wan, Renewable Sustainable Energy Rev., 2017, 73, 610–621 CrossRef CAS.
- Z.-H. Liu, R. K. Le, M. Kosa, B. Yang, J. Yuan and A. J. Ragauskas, Renewable Sustainable Energy Rev., 2019, 105, 349–362 CrossRef CAS.
- F. A. Riyadi, N. F. Azman, F. Nadia Md Akhir, N. Othman and H. Hara, J. Gen. Appl. Microbiol., 2023, 69, 278–286 CrossRef CAS PubMed.
- S. Baghel and J. Anandkumar, Bioresour. Technol. Rep., 2019, 8, 100335 CrossRef.
- Z. Choolaei, R. Flick, A. N. Khusnutdinova, E. A. Edwards and A. F. Yakunin, J. Biotechnol., 2021, 325, 128–137 CrossRef CAS PubMed.
- V. P. Kumar, M. Sridhar and R. G. Rao, Sci. Rep., 2022, 12, 11170 CrossRef CAS PubMed.
- K. Zhang, R. Xu, A. E.-F. Abomohra, S. Xie, Z. Yu, Q. Guo, P. Liu, L. Peng and X. Li, Energy Convers. Manage., 2019, 199, 111928 CrossRef CAS.
- X. Huang, N. Santhanam, D. V. Badri, W. J. Hunter, D. K. Manter, S. R. Decker, J. M. Vivanco and K. F. Reardon, Biotechnol. Bioeng., 2013, 110, 1616–1626 CrossRef CAS PubMed.
- Y. Ley, X.-Y. Cheng, Z.-Y. Ying, N.-Y. Zhou and Y. Xu, Microbiol. Spectr., 2023, 11, e04424–e04422 Search PubMed.
- E. Vignali, F. Tonin, L. Pollegioni and E. Rosini, Appl. Microbiol. Biotechnol., 2018, 102, 10579–10588 CrossRef CAS PubMed.
- Z. Xu, B. Peng, R. B. Kitata, C. D. Nicora, K. K. Weitz, Y. Pu, T. Shi, J. R. Cort, A. J. Ragauskas and B. Yang, Biotechnol. Biofuels, 2022, 15, 117 CrossRef CAS PubMed.
- H. Liu, Z.-H. Liu, R.-K. Zhang, J. S. Yuan, B.-Z. Li and Y.-J. Yuan, Biotechnol. Adv., 2022, 60, 108000 CrossRef CAS PubMed.
- B. E. Blass, in Basic Principles of Drug Discovery and Development, Elsevier, 2015, pp. 245–306 Search PubMed.
- J. V. Vermaas, R. A. Dixon, F. Chen, S. D. Mansfield, W. Boerjan, J. Ralph, M. F. Crowley and G. T. Beckham, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 23117–23123 CrossRef CAS PubMed.
- W. R. Henson, T. Campbell, D. M. DeLorenzo, Y. Gao, B. Berla, S. J. Kim, M. Foston, T. S. Moon and G. Dantas, Metab. Eng., 2018, 49, 69–83 CrossRef CAS PubMed.
- R. Morya, M. Kumar and I. Shekhar Thakur, Bioresour. Technol., 2021, 330, 124981 CrossRef CAS PubMed.
- A. Wada, É. T. Prates, R. Hirano, A. Z. Werner, N. Kamimura, D. A. Jacobson, G. T. Beckham and E. Masai, Metab. Eng., 2021, 64, 167–179 CrossRef CAS PubMed.
- C. Del Cerro, E. Erickson, T. Dong, A. R. Wong, E. K. Eder, S. O. Purvine, H. D. Mitchell, K. K. Weitz, L. M. Markillie, M. C. Burnet, D. W. Hoyt, R. K. Chu, J.-F. Cheng, K. J. Ramirez, R. Katahira, W. Xiong, M. E. Himmel, V. Subramanian, J. G. Linger and D. Salvachúa, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2017381118 CrossRef PubMed.
- K. Mori, K. Niinuma, M. Fujita, N. Kamimura and E. Masai, Appl. Environ. Microbiol., 2018, 84, e01314–e01318 CrossRef CAS PubMed.
- A. Yoneda, W. R. Henson, N. K. Goldner, K. J. Park, K. J. Forsberg, S. J. Kim, M. W. Pesesky, M. Foston, G. Dantas and T. S. Moon, Nucleic Acids Res., 2016, 44, 2240–2254 CrossRef CAS PubMed.
- R. C. Salmon, M. J. Cliff, J. B. Rafferty and D. J. Kelly, PLoS One, 2013, 8, e59844 CrossRef CAS PubMed.
- C. C. Azubuike, M. N. Allemann and J. K. Michener, Curr. Opin. Microbiol., 2022, 65, 64–72 CrossRef CAS PubMed.
- R. Weiss, G. M. Guebitz, A. Pellis and G. S. Nyanhongo, Trends Biotechnol., 2020, 38, 1215–1231 CrossRef CAS PubMed.
- Z. Zhao, Z. Liu, Y. Pu, X. Meng, J. Xu, J. S. Yuan and A. J. Ragauskas, ChemSusChem, 2020, 13, 5423–5432 CrossRef CAS PubMed.
- F. Li, Y. Zhao, L. Xue, F. Ma, S. Y. Dai and S. Xie, Trends Biotechnol., 2022, 40, 1469–1487 CrossRef CAS PubMed.
- A. Bleem, R. Kato, Z. A. Kellermyer, R. Katahira, M. Miyamoto, K. Niinuma, N. Kamimura, E. Masai and G. T. Beckham, Cell Rep., 2023, 42, 112847 CrossRef CAS PubMed.
- E. S. Ellis, D. J. Hinchen, A. Bleem, L. Bu, S. J. B. Mallinson, M. D. Allen, B. R. Streit, M. M. Machovina, Q. V. Doolin, W. E. Michener, C. W. Johnson, B. C. Knott, G. T. Beckham, J. E. McGeehan and J. L. DuBois, JACS Au, 2021, 1, 252–261 CrossRef CAS PubMed.
- D. Salvachúa, A. Z. Werner, I. Pardo, M. Michalska, B. A. Black, B. S. Donohoe, S. J. Haugen, R. Katahira, S. Notonier, K. J. Ramirez, A. Amore, S. O. Purvine, E. M. Zink, P. E. Abraham, R. J. Giannone, S. Poudel, P. D. Laible, R. L. Hettich and G. T. Beckham, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 9302–9310 CrossRef PubMed.
- J. Wang, X. Shen, J. Rey, Q. Yuan and Y. Yan, Appl. Microbiol. Biotechnol., 2018, 102, 47–61 CrossRef CAS PubMed.
- X. Jin, X. Li, L. Zou, Z. Zheng and J. Ouyang, Molecules, 2024, 29, 1555 CrossRef CAS PubMed.
- N. Okai, T. Masuda, Y. Takeshima, K. Tanaka, K. Yoshida, M. Miyamoto, C. Ogino and A. Kondo, AMB Express, 2017, 7, 130 CrossRef PubMed.
- L. T. Nguyen, M. H. Tran and E. Y. Lee, Bioresour. Technol., 2021, 338, 125563 CrossRef CAS PubMed.
- T. Sonoki, K. Takahashi, H. Sugita, M. Hatamura, Y. Azuma, T. Sato, S. Suzuki, N. Kamimura and E. Masai, ACS Sustainable Chem. Eng., 2018, 6, 1256–1264 CrossRef CAS.
- P. Upadhyay and A. Lali, Prep. Biochem. Biotechnol., 2021, 51, 979–984 CrossRef CAS PubMed.
- R.-K. Zhang, Y.-S. Tan, Y.-Z. Cui, X. Xin, Z.-H. Liu, B.-Z. Li and Y.-J. Yuan, Green Chem., 2021, 23, 6515–6526 RSC.
- R. J. M. Lubbers and R. P. De Vries, mBio, 2021, 12, e00391–e00321 CrossRef CAS PubMed.
- J. Bai, Y. Zhang, C. Tang, Y. Hou, X. Ai, X. Chen, Y. Zhang, X. Wang and X. Meng, Biomed. Pharmacother., 2021, 133, 110985 CrossRef CAS PubMed.
- N. A. Al Zahrani, R. M. El-Shishtawy and A. M. Asiri, Eur. J. Med. Chem., 2020, 204, 112609 CrossRef CAS PubMed.
- W. Wu, T. Dutta, A. M. Varman, A. Eudes, B. Manalansan, D. Loqué and S. Singh, Sci. Rep., 2017, 7, 8420 CrossRef PubMed.
- C. Cai, Z. Xu, H. Zhou, S. Chen and M. Jin, Sci. Adv., 2021, 7, eabg4585 CrossRef CAS PubMed.
- B. Fu, G. Xiao, Y. Zhang and J. Yuan, J. Agric. Food Chem., 2021, 69, 11336–11341 CrossRef CAS PubMed.
- L. Xu, F. Liaqat, J. Sun, M. I. Khazi, R. Xie and D. Zhu, Renewable Sustainable Energy Rev., 2024, 189, 113905 CrossRef CAS.
- M. Zhao, Y. Tao, X. Wu and Y. Xiao, ACS Sustainable Chem. Eng., 2021, 9, 6400–6409 CrossRef CAS.
- T. M. Ludwikowski, A. O. Wagner and R. Margesin, Ann. Microbiol., 2023, 73, 11 CrossRef CAS.
- Z. Chen, X. Shen, J. Wang, J. Wang, R. Zhang, J. F. Rey, Q. Yuan and Y. Yan, ACS Synth. Biol., 2017, 6, 1784–1792 CrossRef CAS PubMed.
- H. Zhou, Z. Xu, C. Cai, J. Li and M. Jin, Bioresour. Technol., 2022, 347, 126348 CrossRef CAS PubMed.
- J. Rönnander, J. Ljunggren, E. Hedenström and S. A. I. Wright, AMB Express, 2018, 8, 137 CrossRef PubMed.
- F. J. N. Putra, P. Kahar, A. Kondo and C. Ogino, Biochem. Eng. J., 2023, 200, 109065 CrossRef CAS.
- J. Overhage, A. Steinbüchel and H. Priefert, J. Biotechnol., 2006, 125, 369–376 CrossRef CAS PubMed.
- Y. Lai, H. Chen, L. Liu, B. Fu, P. Wu, W. Li, J. Hu and J. Yuan, ACS Synth. Biol., 2022, 11, 441–447 CrossRef CAS PubMed.
- S. Jeenpadiphat, I. Mongkolpichayarak and D. N. Tungasmita, J. Anal. Appl. Pyrolysis, 2016, 121, 318–328 CrossRef CAS.
- H. Hara, L. D. Eltis, J. E. Davies and W. W. Mohn, J. Bacteriol., 2007, 189, 1641–1647 CrossRef CAS PubMed.
- P. Upadhyay and A. Lali, Prep. Biochem. Biotechnol., 2022, 52, 80–88 CrossRef CAS PubMed.
- J. Ni, Y.-T. Wu, F. Tao, Y. Peng and P. Xu, J. Am. Chem. Soc., 2018, 140, 16001–16005 CrossRef CAS PubMed.
- Y. Guo, L. Alvigini, M. Trajkovic, L. Alonso-Cotchico, E. Monza, S. Savino, I. Marić, A. Mattevi and M. W. Fraaije, Nat. Commun., 2022, 13, 7195 CrossRef CAS PubMed.
- P. D. Sainsbury, E. M. Hardiman, M. Ahmad, H. Otani, N. Seghezzi, L. D. Eltis and T. D. H. Bugg, ACS Chem. Biol., 2013, 8, 2151–2156 CrossRef CAS PubMed.
- N. Graf and J. Altenbuchner, Appl. Microbiol. Biotechnol., 2014, 98, 137–149 CrossRef CAS PubMed.
- D. Zhu, L. Xu, S. Sethupathy, H. Si, F. Ahmad, R. Zhang, W. Zhang, B. Yang and J. Sun, Green Chem., 2021, 23, 9554–9570 RSC.
- K. Zuo, H. Li, J. Chen, Q. Ran, M. Huang, X. Cui, L. He, J. Liu and Z. Jiang, Front. Microbiol., 2022, 13, 901690 CrossRef PubMed.
- P. Chen, L. Yan, S. Zhang, Z. Wu, S. Li, X. Yan, N. Wang, N. Liang and H. Li, Food Sci. Biotechnol., 2017, 26, 143–152 CrossRef CAS PubMed.
- P. Chattopadhyay, G. Banerjee and S. K. Sen, J. Cleaner Prod., 2018, 182, 272–279 CrossRef CAS.
- F. Luziatelli, L. Brunetti, A. G. Ficca and M. Ruzzi, Front. Bioeng. Biotechnol., 2019, 7, 279 CrossRef PubMed.
- E. Lee, S. Yoon, A. Das, S. Lee, C. Li, J. Kim, M. Choi, D. Oh and S. Kim, Biotechnol. Bioeng., 2009, 102, 200–208 CrossRef CAS PubMed.
- H.-S. Lee, J. Park and Y. J. Yeon, Enzyme Microb. Technol., 2022, 161, 110099 CrossRef CAS PubMed.
- A. M. Tafesh and J. Weiguny, Chem. Rev., 1996, 96, 2035–2052 CrossRef CAS PubMed.
- L. Xu, Z. He, H. Zhang, S. Wu, C. Dong and Z. Fang, Bioresour. Technol., 2021, 320, 124252 CrossRef CAS PubMed.
- Y. Li, B. Fan, L. Yang, C. Ma and Y.-C. He, Ind. Crops Prod., 2023, 205, 117550 CrossRef CAS.
- J. Zhao, Y. Li, L. Yi, G. Zhang and B. Ye, ACS Sustainable Chem. Eng., 2023, 11, 7683–7691 CrossRef CAS.
- J. H. C. Manfrão-Netto, F. Lund, N. Muratovska, E. M. Larsson, N. S. Parachin and M. Carlquist, Microb. Biotechnol., 2021, 14, 2448–2462 CrossRef PubMed.
- L. Li, C. Ma, H. Chai and Y.-C. He, Bioresour. Technol., 2023, 385, 129453 CrossRef CAS PubMed.
- Q. Li, R. Gao, Y. Li, B. Fan, C. Ma and Y.-C. He, Bioresour. Technol., 2023, 384, 129292 CrossRef CAS PubMed.
- C. J. Du, L. Rios-Solis, J. M. Ward, P. A. Dalby and G. J. Lye, Biocatal. Biotransform., 2014, 32, 302–313 CrossRef CAS.
- Q. Wang, L. Ma, Z. Wang, Q. Chen, Q. Wang and Q. Qi, Biotechnol. Biofuels, 2022, 15, 100 CrossRef CAS PubMed.
- J. Li, C. Yue, W. Wei, Y. Shang, P. Zhang and B.-C. Ye, Bioresour. Technol., 2022, 344, 126221 CrossRef CAS PubMed.
- C. W. Johnson, P. E. Abraham, J. G. Linger, P. Khanna, R. L. Hettich and G. T. Beckham, Metab. Eng. Commun., 2017, 5, 19–25 CrossRef PubMed.
- S. G. Bell, A. B. H. Tan, E. O. D. Johnson and L.-L. Wong, Mol. BioSyst., 2009, 6, 206–214 RSC.
- E. Rosini, P. D'Arrigo and L. Pollegioni, Catal. Sci. Technol., 2016, 6, 7729–7737 RSC.
- W. Wu, F. Liu and S. Singh, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 2970–2975 CrossRef CAS PubMed.
- R.-Y. Liu, C. Wang, B.-Z. Li, Z.-H. Liu and Y.-J. Yuan, Chem. Eng. J., 2024, 499, 156286 CrossRef CAS.
- X. Yao, Y. Lv, H. Yu, H. Cao, L. Wang, B. Wen, T. Gu, F. Wang, L. Sun and F. Xin, Appl. Microbiol. Biotechnol., 2020, 104, 3897–3907 CrossRef CAS PubMed.
- C. Poyntner, T. M. Ludwikowski, A. O. Wagner and R. Margesin, AMB Express, 2022, 12, 148 CrossRef CAS PubMed.
- D.-H. Jung, E.-J. Kim, E. Jung, R. J. Kazlauskas, K.-Y. Choi and B.-G. Kim, Biotechnol. Bioeng., 2016, 113, 1493–1503 CrossRef CAS PubMed.
- J. Diao, R. Carr and T. S. Moon, Commun. Biol., 2022, 5, 1109 CrossRef CAS PubMed.
- A. Kaplan and M. Haenlein, Bus. Horiz., 2019, 62, 15–25 CrossRef.
- J. S. Cho, G. B. Kim, H. Eun, C. W. Moon and S. Y. Lee, JACS Au, 2022, 2, 1781–1799 CrossRef CAS PubMed.
- A. B. Culaba, A. P. Mayol, J. L. G. San Juan, C. L. Vinoya, R. S. Concepcion, A. A. Bandala, R. R. P. Vicerra, A. T. Ubando, W.-H. Chen and J.-S. Chang, Bioresour. Technol., 2022, 344, 126215 CrossRef CAS PubMed.
- L. König-Mattern, E. I. Sanchez Medina, A. O. Komarova, S. Linke, L. Rihko-Struckmann, J. S. Luterbacher and K. Sundmacher, Chem. Eng. J., 2024, 495, 153524 CrossRef.
- C. E. Lawson, J. M. Martí, T. Radivojevic, S. V. R. Jonnalagadda, R. Gentz, N. J. Hillson, S. Peisert, J. Kim, B. A. Simmons, C. J. Petzold, S. W. Singer, A. Mukhopadhyay, D. Tanjore, J. G. Dunn and H. Garcia Martin, Metab. Eng., 2021, 63, 34–60 CrossRef CAS PubMed.
- A. Castro Garcia, S. Cheng, S. E. McGlynn and J. S. Cross, ACS Omega, 2023, 8, 32078–32089 CrossRef CAS PubMed.
- E. Orsi, L. Schada Von Borzyskowski, S. Noack, P. I. Nikel and S. N. Lindner, Nat. Commun., 2024, 15, 3447 CrossRef CAS PubMed.
- S. Sethupathy, G. Murillo Morales, L. Gao, H. Wang, B. Yang, J. Jiang, J. Sun and D. Zhu, Bioresour. Technol., 2022, 347, 126696 CrossRef CAS PubMed.
- S. Mohaptra, P. K. Dash, S. S. Behera and H. Thatoi, Environ. Technol., 2016, 37, 940–951 CrossRef CAS PubMed.
- A. Castro Garcia, C. Shuo and J. S. Cross, Bioresour. Technol., 2022, 345, 126503 CrossRef CAS PubMed.
- L. Wang, C. Xue, G. Owens and Z. Chen, Bioresour. Technol., 2022, 345, 126565 CrossRef CAS PubMed.
- S. Barber-Zucker, V. Mindel, E. Garcia-Ruiz, J. J. Weinstein, M. Alcalde and S. J. Fleishman, J. Am. Chem. Soc., 2022, 144, 3564–3571 CrossRef CAS PubMed.
- T. Weirick, S. S. Sahu, R. Mahalingam and R. Kaundal, BMC Bioinf., 2014, 15, S15 CrossRef PubMed.
- Y. Wang, P. Xue, M. Cao, T. Yu, S. T. Lane and H. Zhao, Chem. Rev., 2021, 121, 12384–12444 CrossRef CAS PubMed.
- E. Garcia-Ruiz, D. M. Mate, D. Gonzalez-Perez, P. Molina-Espeja, S. Camarero, A. T. Martínez, A. O. Ballesteros and M. Alcalde, in Cascade Biocatalysis, ed. S. Riva and W. Fessner, Wiley, 1st edn, 2014, pp. 1–22 Search PubMed.
- H. Duţă, A. Filip, L. C. Nagy, E. Z. A. Nagy, R. Tőtős and L. C. Bencze, Sci. Rep., 2022, 12, 3347 CrossRef PubMed.
- K. Ilić Đurđić, S. Ece, R. Ostafe, S. Vogel, A. M. Balaž, S. Schillberg, R. Fischer and R. Prodanović, J. Biosci. Bioeng., 2020, 129, 664–671 CrossRef PubMed.
- K. Ryu, S. Y. Hwang, K. H. Kim, J. H. Kang and E. K. Lee, J. Biotechnol., 2008, 133, 110–115 CrossRef CAS PubMed.
- K. Ryu, J. H. Kang, L. Wang and E. K. Lee, J. Biotechnol., 2008, 135, 241–246 CrossRef CAS PubMed.
- D. Rodríguez-Escribano, R. Pliego-Magán, F. De Salas, P. Aza, P. Gentili, P. Ihalainen, T. Levée, V. Meyer, M. Petit-Conil, S. Tapin-Lingua, M. Lecourt and S. Camarero, Biotechnol. Biofuels, 2022, 15, 149 CrossRef PubMed.
- D. Gonzalez-Perez, I. Mateljak, E. Garcia-Ruiz, F. J. Ruiz-Dueñas, A. T. Martinez and M. Alcalde, Catal. Sci. Technol., 2016, 6, 6625–6636 RSC.
- V. Brissos, D. Tavares, A. C. Sousa, M. P. Robalo and L. O. Martins, ACS Catal., 2017, 7, 3454–3465 CrossRef CAS.
- V. Brissos, M. Ferreira, G. Grass and L. O. Martins, ACS Catal., 2015, 5, 4932–4941 CrossRef CAS.
- Y. Yang, S. Ghatge and H.-G. Hur, Appl. Microbiol. Biotechnol., 2023, 107, 273–286 CrossRef CAS PubMed.
- J. Mueller, H. Willett, A. M. Feist and W. Niu, Biotechnol. Bioeng., 2022, 119, 2541–2550 CrossRef CAS PubMed.
- J. A. Jones, S. M. Collins, V. R. Vernacchio, D. M. Lachance and M. A. G. Koffas, Biotechnol. Prog., 2016, 32, 21–25 CrossRef CAS PubMed.
- Q. Huang, Y. Lin and Y. Yan, Biotechnol. Bioeng., 2013, 110, 3188–3196 CrossRef CAS PubMed.
- A. Rodriguez, J. A. Meadows, N. Sun, B. A. Simmons and J. M. Gladden, Microb. Cell Fact., 2021, 20, 181 CrossRef CAS PubMed.
- D.-H. Jung, W. Choi, K.-Y. Choi, E. Jung, H. Yun, R. J. Kazlauskas and B.-G. Kim, Appl. Microbiol. Biotechnol., 2013, 97, 1501–1511 CrossRef CAS PubMed.
- J. M. Salgado, R. Rodríguez-Solana, J. A. Curiel, B. De Las Rivas, R. Muñoz and J. M. Domínguez, Enzyme Microb. Technol., 2014, 58–59, 22–28 CrossRef CAS PubMed.
- M. Zhao, X. Hong, Abdullah, R. Yao and Y. Xiao, Green Chem., 2021, 23, 838–847 RSC.
- D. Di Gioia, F. Luziatelli, A. Negroni, A. G. Ficca, F. Fava and M. Ruzzi, J. Biotechnol., 2011, 156, 309–316 CrossRef CAS PubMed.
- B. Kaur, D. Chakraborty and B. Kumar, BioMed Res. Int., 2013, 2013, 1–6 Search PubMed.
- C. Fleige, G. Hansen, J. Kroll and A. Steinbüchel, Appl. Environ. Microbiol., 2013, 79, 81–90 CrossRef CAS PubMed.
- D. Hua, C. Ma, S. Lin, L. Song, Z. Deng, Z. Maomy, Z. Zhang, B. Yu and P. Xu, J. Biotechnol., 2007, 130, 463–470 CrossRef CAS PubMed.
- M. Horvat, G. Fiume, S. Fritsche and M. Winkler, J. Biotechnol., 2019, 304, 44–51 CrossRef CAS PubMed.
- J. Park, H.-S. Lee, J. Oh, J. C. Joo and Y. J. Yeon, Biochem. Eng. J., 2020, 161, 107683 CrossRef CAS.
- A. Sharma, J. Singh, P. Sharma, G. S. Tomar, S. Singh, M. Grover and L. Nain, 3 Biotech, 2021, 11, 462 CrossRef PubMed.
- A. Singh, K. Mukhopadhyay and S. Ghosh Sachan, Biocatal. Biotransform., 2019, 37, 291–303 CrossRef CAS.
- M. Ashengroph and J. Amini, 3 Biotech, 2017, 7, 358 CrossRef PubMed.
- C. Stentelaire, L. Lesage-Meessen, J. Oddou, O. Bernard, G. Bastin, B. C. Ceccaldi and M. Asther, J. Biosci. Bioeng., 2000, 89, 223–230 CrossRef CAS PubMed.
- A. Bloem, A. Bertrand, A. Lonvaud-Funel and G. De Revel, Lett. Appl. Microbiol., 2006, 44, 62–67 CrossRef PubMed.
- D. Hua, C. Ma, L. Song, S. Lin, Z. Zhang, Z. Deng and P. Xu, Appl. Microbiol. Biotechnol., 2007, 74, 783–790 CrossRef CAS PubMed.
- P. Barghini, D. Di Gioia, F. Fava and M. Ruzzi, Microb. Cell Fact., 2007, 6, 13 CrossRef PubMed.
- L. Zheng, Pu. Zheng, Z. Sun, Y. Bai, J. Wang and X. Guo, Bioresour. Technol., 2007, 98, 1115–1119 CrossRef CAS PubMed.
- S. Saeed, S. Q. Raza, S. S. Zafar, H. Mujahid, M. Irfan and T. Mehmood, Biomass Convers. Biorefin., 2024, 14, 679–688 CrossRef.
- X. Ma and A. J. Daugulis, Biotechnol. Prog., 2014, 30, 207–214 CrossRef CAS PubMed.
- T. Furuya, M. Kuroiwa and K. Kino, J. Biotechnol., 2017, 243, 25–28 CrossRef CAS PubMed.
- Q. Wang, X. Wu, X. Lu, Y. He, B. Ma and Y. Xu, Appl. Biochem. Biotechnol., 2021, 193, 1116–1128 CrossRef CAS PubMed.
- L.-Q. Zhao, Z.-H. Sun, P. Zheng and J.-Y. He, Process Biochem., 2006, 41, 1673–1676 CrossRef CAS.
- L.-Q. Zhao, Z.-H. Sun, P. Zheng and L.-L. Zhu, Biotechnol. Lett., 2005, 27, 1505–1509 CrossRef CAS PubMed.
- M. Ashengroph, I. Nahvi, H. Zarkesh-Esfahani and F. Momenbeik, Appl. Biochem. Biotechnol., 2012, 166, 1–12 CrossRef CAS PubMed.
- H. Karakaya and M. Yilmaztekin, J. Agric. Sci., 2022, 28, 423–429 Search PubMed.
- C. S. Lancefield, G. M. M. Rashid, F. Bouxin, A. Wasak, W.-C. Tu, J. Hallett, S. Zein, J. Rodríguez, S. D. Jackson, N. J. Westwood and T. D. H. Bugg, ACS Sustainable Chem. Eng., 2016, 4, 6921–6930 CrossRef CAS.
- B. Fu, G. Xiao, Y. Zhu, Y. Chen, Y. Lai and J. Yuan, ACS Agric. Sci. Technol., 2021, 1, 566–571 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4gc03567d |
| This journal is © The Royal Society of Chemistry 2024 |