Hierarchical assembly and modeling of DNA nanotube networks using Y-shaped DNA origami seeds†
Yanqi
Jiang‡
 a,
Michael S.
Pacella‡
a,
Michael S.
Pacella‡
 a,
Sojeong
Lee
b,
Jasen
Zhang
a,
Sojeong
Lee
b,
Jasen
Zhang
 a,
Jonathan A.
Gunn
a,
Jonathan A.
Gunn
 a,
Paul
Vallejo
a,
Pragya
Singh
a,
Tiffany
Hou
a,
Evan
Liu
a,
Paul
Vallejo
a,
Pragya
Singh
a,
Tiffany
Hou
a,
Evan
Liu
 a and
Rebecca
Schulman
a and
Rebecca
Schulman
 *ac
*ac
aDepartment of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, Maryland 21218, USA. E-mail: rschulm3@jhu.edu
bDepartment of Biomedical Engineering, Johns Hopkins University, Baltimore, Maryland 21218, USA
cDepartment of Computer Science, Johns Hopkins University, Baltimore, Maryland 21218, USA
First published on 29th May 2024
Abstract
DNA nanotechnology offers many means to synthesize custom nanostructured materials from the ground up in a hierarchical fashion. While the assembly of DNA nanostructures from small (nanometer-scale) monomeric components has been studied extensively, how the hierarchical assembly of rigid or semi-flexible units produces multi-micron scale structures is less understood. Here we demonstrate a mechanism for assembling micron-scale semi-flexible DNA nanotubes into extended structures. These nanotubes assemble from nanometer-scale tile monomers into materials via heterogeneous nucleation from rigid, Y-shaped DNA origami seeds to form Y-seeded nanotube architectures. These structures then assemble into networks via nanotube end-to-end joining. We measure the kinetics of network growth and find that the assembly of networks can be approximated by a model of hierarchical assembly that assumes a single joining rate between DNA nanotube ends. Because the number of nucleation sites on Y-seeds and their spatial arrangement can be systematically varied by design, this hierarchical assembly process could be used to form a wide variety of networks and to understand the assembly mechanisms that lead to different types of material architectures at length scales of tens to hundreds of microns.
1 Introduction
A longstanding goal of nanotechnology is the development of methods for synthesizing custom matter from the ground up in a hierarchical fashion. Hierarchical assembly, in which nanoscale components are organized across size scales of microns and beyond, controls processes as diverse as inorganic crystal growth1 and the formation of complex biological superstructures such as the cytoskeleton.2 Cells exploit organization at the angstrom and nanometer scales to achieve specific chemical functions and then assemble and organize organelles,3 filaments,4 and other substructures to optimize transport, mechanics, and chemical synthesis at the 100 nm to micron scale. Architected materials ranging from insect shells, biominerals, and soft tissues likewise use hierarchical organization and dynamic behavior to optimize performance. Hierarchical self-assembly offers the ability not only to inexpensively assemble synthetic structures with a range of feature sizes but also to create materials that are capable of autonomous reconfiguration in response to stimuli through dynamic self-assembly.While there are clear advantages to using hierarchical self-assembly for creating structures and devices, it has been difficult to develop comprehensive rational design strategies. A central challenge is that the results of hierarchical assembly can be hard to predict: the products are orders of magnitude larger than their smallest feature sizes or molecular components. As a result, the kinetics of assembly is controlled by multiple reactions that occur over vastly different size and time scales. Models of self-assembly must therefore incorporate information about the different mechanisms and rates of these processes.
In this paper, we sought to develop a hierarchically assembled DNA architecture in which semi-flexible filaments – DNA nanotubes – are organized into specific extended network geometries, and to understand their assembly mechanism and kinetics using models. The highly programmable base pairing and well-defined secondary structures of DNA offer routes for constructing a wide range of DNA-based devices, circuits, and biomaterials, but the mechanisms of assembly are not yet well understood beyond the nanoscale.
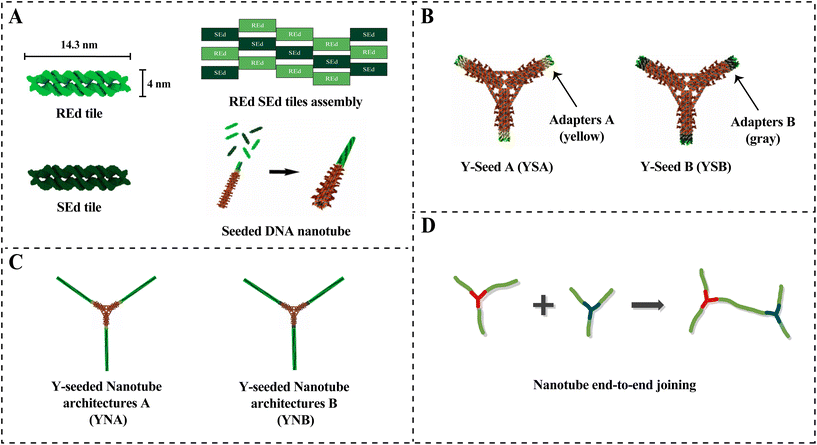
In our system, two types of DAE-E double crossover REd and SEd tiles with four sticky ends were used (Fig. 1A and ESI section 1.1†) to form lattices that cyclize into DNA nanotubes. Tile monomers nucleate from nanometer-scale scaffolded DNA origami5 seeds with a Y-shaped geometry,6 termed YSA and YSB (Fig. 1B). Each arm of a Y-seed contains a set of single-stranded DNA adapter strands at its ends, which nucleate the growth of DNA nanotubes from REd SEd tiles. However, the adapters of YSA and YSB differ in their sequences (ESI section 1.2†). The two types of adapters are specifically engineered so that the resulting structures – termed Y-seeded nanotube architectures types A and B (YNA and YNB) (Fig. 1C) – will have complementary, rather than identical, sticky ends (see Fig. S3† for detailed mechanistic basis). These complementary ends enable end-to-end joining between YNA and YNB, while YNA–YNA and YNB–YNB connections cannot form due to their identical sticky ends. Similar to the end-to-end joining of two seeded DNA nanotubes,7 these Y-seeded nanotube architectures then hierarchically assemble into micrometer-scale extended networks over a period of around 24 hours via an end-to-end joining process (Fig. 1D).
While predictive kinetic models exist for the assembly of DNA nanotubes from small DNA tiles,8,9 a more general and comprehensive model for the hierarchical assembly of DNA networks from larger and more complicated components such as Y-seeded nanotube architectures nucleated from Y-shaped DNA origami seeds remains elusive.7,10,11 Here we tracked and quantified the hierarchical assembly of such structures and developed a simple explanatory model that predicts their growth into large networks. The model is consistent with a single joining rate governing the hierarchical assembly across all scales and may provide a basis for systematically varying components to rationally design a variety of large-scale DNA materials.
2 Results and discussion
A and B Y-shaped DNA origami seeds can nucleate and grow Y-seeded nanotube architectures with high yield
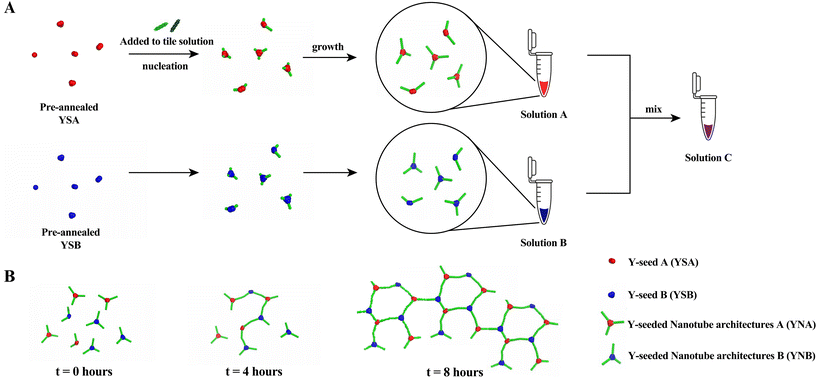
Jorgenson et al.6 demonstrated that nanotubes could grow efficiently from YSA with a yield of 45 ± 3%. To enable the formation of networks from Y-seeded nanotubes through end-to-end joining of nanotubes, we designed a new DNA origami, YSB, which has the same structure as YSA but different sequences in the adapters so that the nanotubes on each Y-arm of a YNA and a YNB can join at their ends to form large networks.To determine the yields of Y-seeded nanotubes that nucleated on YSA and the new YSB, we nucleated and grew Y-seeded nanotubes in separate tubes by adding pre-annealed Y-seeds (final concentration 6 pM) to 25 nM DNA tile solutions (Fig. 2A). These solutions were each incubated at 32 °C and 20 micrographs were taken at 0, 4, and 8 hours (Fig. 2B) to characterize the progress of network formation. We used an automated image processing algorithm (ESI section 3.1†) to count the number of Y-seeded nanotube architectures with 1, 2, and 3 arms. The yields of 3-armed nanotubes for both YSA and YSB (Fig. 3) are consistent with the yields observed in Jorgenson et al.6 for YSA. Fig. 3 suggests that approximately 46% of YNA and 54%of YNB possess three binding sites, while 42% of YNA and 36% of YNB have two, and 12% of YNA and 10% of YNB have only one. YNAs and YNBs with two or three binding sites facilitate network growth and expansion without impeding it, although their growth efficiency varies. For instance, a three-armed YNA attaching to a YNB within a large network leaves two arms free to connect to additional YNBs. In contrast, a two-armed YNA would have only one free arm. However, a one-armed YNA acts as a cap when joining a network, blocking further extensions from its attachment point.
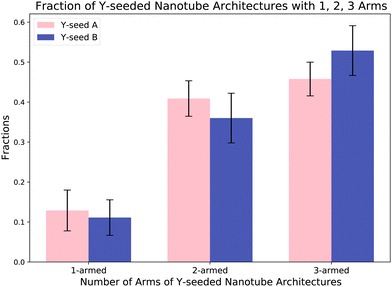 | ||
| Fig. 3 The fractions of Y-seeded nanotube architectures with 1, 2 or 3 arms observed in 20 fluorescence micrographs of assembled Y-seeded nanotube architectures after 24 hours of assembly. Each micrograph contained around 50 structures, so a total of about 1000 structures was observed. Error bars represent the standard deviation of the mean over the 20 images. The number of Y-seeds with no arms (only Y-seeds observed) was insignificant and not included in the analysis. The fractions of Y-seeded nanotube architectures observed with each number of arms are consistent with Jorgenson et al.6 | ||
Y-seeded nanotube architectures form networks by end-to-end joining
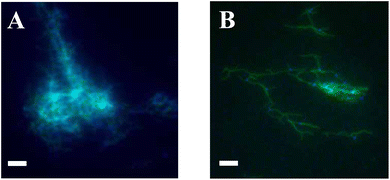
We next sought to test whether the two types of Y-seeded nanotube architectures would form networks via an end-to-end joining process after mixing. We mixed YNA with YNB as described in Fig. 2A to create solution C, which was aliquoted into multiple tubes for characterization. All tubes were then incubated at 32 °C. At 0, 4 and 8 hours after mixing, 6 μL from each aliquot was deposited onto a glass slide for imaging (Fig. 2B). To characterize the progress of the assembly reaction, we took micrographs of each slide at 15 random locations. As expected, at 0 hours the structures observed were still primarily individual Y-seeded nanotube architectures. 4 hours after mixing, we began to see small networks with at least one YNA joined with one YNB. 8 hours after mixing, we saw large networks, a few of which contained around 50 Y-seeds. Examples of these structures are shown in Fig. 4. The structures of the networks clearly indicated that end-to-end joining between individual Y-seeded nanotube architectures occurs and results in the formation of networks. The micrographs also indicated that networks can grow to a size incorporating around 50 Y-seeds in as few as 8 hours.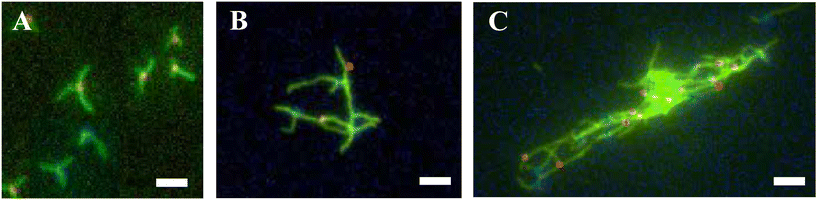 | ||
| Fig. 4 Networks observed after different durations of hierarchical self-assembly. Fluorescence micrographs were taken 0 h, 4 h and 8 h after solution C was mixed as described in Fig. 2. Here YSA was labeled with ATTO 647N (red), YSB was labeled with ATTO 488 (blue), and the nanotube tile monomers were labeled with Cy3 (green). (See Materials and methods, ESI section 1.1 and 1.3.†) (A) Right after mixing (0 h), mainly individual Y-seeded nanotube architectures are observed. (B) Four hours after mixing, small networks have begun to form. (C) Eight hours after mixing, large networks incorporating around 50 Y-seeds (either YSA or YSB) can be observed. Scale bars: 5 μm. | ||
Blob and edge detection enable identification and the counting of labeled Y-seeds in a network
To further investigate the kinetics of the network growth, we sought to measure the sizes of the networks produced during the hierarchical self-assembly process by measuring the total number of Y-seeds (YSA + YSB) incorporated in each network (see Materials and methods). Here we met two challenges. First, during the long time period required for the network growth, the ATTO 647N dye became less visible as the reaction proceeded, possibly due to the adsorption of the dye to the test tube walls. To address this issue, after using ATTO 647N to label YSA in preliminary experiments and confirming the successful synthesis of YSA and YNA, we switched to using ATTO 488 to label both YSA and YSB to measure the size of networks. The other challenge was that as the networks grew, individual seeds within structures became difficult to discern (Fig. 4B and C). In some networks, the density of ATTO 488 labeled Y-seeds was so high that it was impossible to properly distinguish individual Y-seeds (Fig. 5A). To address this problem, we altered the protocol so that only 25% of the Y-seeds (both YSA and YSB) were labeled. In the resulting images, individual seeds could be discerned even in larger networks (Fig. 5B).We next developed automated image processing techniques to rapidly measure the sizes of networks on slides (see Materials and methods, ESI section 3.2†). We first performed edge detection on the Cy3 channel to outline each nanotube network. We then used blob detection on the corresponding ATTO 488 channel image to identify the centers of each labeled Y-seed. Finally, we combined the results of edge detection, which outlined the areas within a micrograph corresponding to networks, and blob detection, which identified individual Y-seeds, by overlaying the network outlines with the Y-seed locations and counting the number of Y-seeds inside each network.
The distribution of network sizes formed at later stages is bimodal
To model the relationship between the true number of seeds in a network n and the number of labeled seeds k, we used the binomial probability distribution: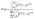 | (1) |
We used this equation to create a probability distribution over the true number of seeds for each network and used these distributions to estimate the number of seeds in networks at 3 time points during the reaction: 1 hour, 8 hours, and 23 hours. Example distributions are shown in Fig. 6 (gray, dotted). The distributions have two interesting features. First, large networks form after about 8 hours, as indicated by the peak that appears for a size of about 50 seeds after 8 hours (Fig. 6, middle row). Second, once large networks begin to form, they dominate the statistics at later time points (bottom row). The distributions at 23 hours are extremely bimodal; either a Y-seed is part of a relatively small network or it is part of one of several large networks that dominate the distributions.
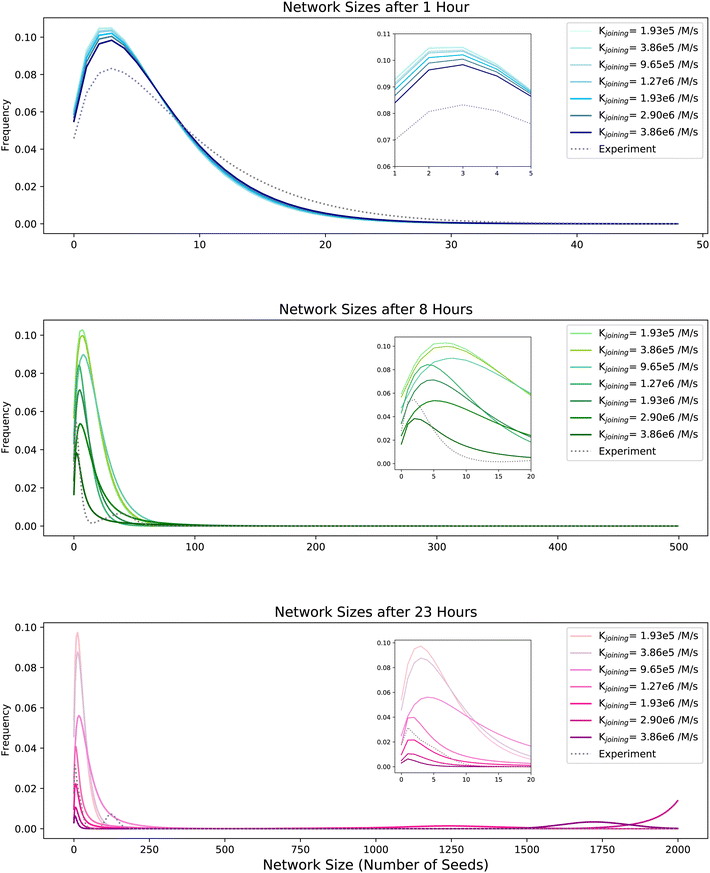 | ||
| Fig. 6 Network size distributions as measured from experiments and predicted by simulation. The plots show the frequencies of an individual nanotube being present in networks of given sizes, as measured from experiments (gray, dotted) and sampled from simulations (blue, green, and pink solid lines respectively) after different reaction times: 1 hour (top), 8 hours (middle) and 23 hours (bottom). The solid lines indicate different reaction rate constants for the joining reaction used in the simulations. For experimental measurements, 25 separate fluorescence microscopy images were analyzed for each time point. 25% of all seeds were fluorescently labeled to optimize the resolution of individual seeds, and the true number of seeds in each network was determined as described in the text. 1000 YSA and 1000 YSB were simulated here with 50 iterations. The number of arms of the YSA and YSB were set using the fractions of 1-armed, 2-armed, and 3-armed Y-seeds measured in Fig. 2. The insets in each panel provide zoomed-in views of the regions inside the gray-dotted rectangles. | ||
A simple model of hierarchical assembly is consistent with measured network sizes
To gain more insight into the mechanism of hierarchical assembly, we developed a computational model of network assembly that assumes that free nanotubes of complementary types can join at a reaction rate Kjoining. The model also incorporates the yields of Y-seeds with different numbers of arms, which will influence the efficiency of network formation. We used a value of Kjoining measured previously as the rate of joining between two types of single nanotubes,12 3.86 × 106 M−1 s−1 (referred to as “the measured value of Kjoining”). We used the Gillespie algorithm to sample joining reactions (ESI section 3.1†). We then compared the network sizes predicted using this model with the predictions of models in which the joining rate was 0.05-, 0.10-, 0.25-, 0.33-, 0.50-, or 0.75-fold the measured value of Kjoining of 3.86 × 106 M−1 s−1. Each of these predictions was then used to calculate the corresponding distribution of network sizes that might be inferred in a simulation in which only 25% of the seeds observed were labeled (Fig. 6). After 1 hour of growth, the simulation using the measured joining rate predicted that most species were already in networks of sizes 2 to 3 seeds. At 8 hours and 23 hours, there were more ‘peaks’ with higher seed counts, indicating that more Y-seeds were incorporated and larger networks formed over time. Overall, the comparison shows that 1.93 × 106 M−1 s−1 (0.5-fold the measured value of Kjoining), 2.90 × 106 M−1 s−1 (0.75-fold the measured value of Kjoining), and 3.86 × 106 M−1 s−1 (the measured value of Kjoining) best fit the network sizes estimated from experiment. The simulations qualitatively fit the observed results, suggesting that this simple model may have value for predicting the progress of the hierarchical assembly of DNA nanotube networks.Large networks grow rapidly by incorporating medium-sized networks and then begin to plateau in size as networks are depleted
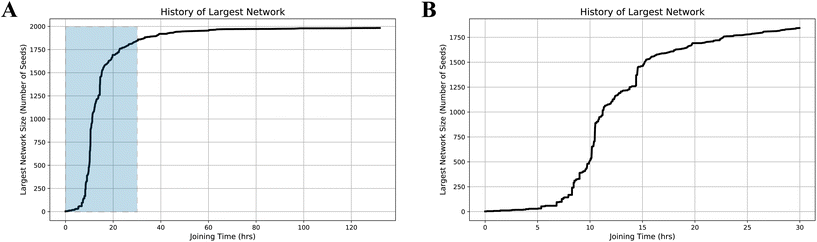
To further explore the mechanism by which large networks grow, we used simulations to follow example trajectories by which a large nanotube network would form. To do so, we first identified the largest network in the system at the end of a simulation trajectory. We then tracked the sizes of all networks at each time point in the simulation and recorded the largest network (Fig. 7A). The detailed development of the largest network during the first 30 hours, after most joining events have already taken place, is shown in Fig. 7B. Using recursion, the series of end-to-end joining events that led to the formation of the single largest network was then analyzed. The size of the largest network, as measured by the number of Y-seeds incorporated, was tracked until all 2000 Y-seeds were incorporated into one single network. As more and more joining events happen, the size of the precursors of this largest network first increases gradually, indicating that the growth occurs mainly via the attachment of individual Y-seeded nanotube architectures. At intermediate times, the network grows rapidly and large vertical jumps occur, indicating that the network grows by joining another large network containing many seeds, significantly increasing its size after a single event. Interestingly, at later stages, the rate of network size increase slows down again, indicating that the growth occurs via the addition of the remaining individual Y-seeded nanotube architectures and small networks to existing large-sized networks. This change likely occurs because most of the medium-sized networks have been depleted by this point, having been incorporated into larger networks. Since the larger networks have a lower diffusion rate than the small structures, the joining events at the later stages primarily depend on the incorporation of single nanotubes or small networks.3 Conclusions
In this study, we experimentally characterized the hierarchical assembly of DNA nanotube networks from Y-shaped DNA origami seeds by quantifying network sizes measured as the number of Y-seeds incorporated in a network at different time points during the assembly process. We then tested the ability of a simple size-independent model of nanotube joining to qualitatively reproduce our experimental kinetic data using different rate constants for the end-to-end joining reaction. Our simple but comprehensive model successfully reproduces the trends seen in the experimental results, specifically, the bimodal nature of the network size distribution during joining. Both the experimental and model distributions are primarily dominated by a single, large network that forms after roughly 8 hours and continues to increase in size until eventually plateauing as the number of components available for joining continues to decrease.The general model we developed for the hierarchical assembly of DNA nanostructures from larger components, like Y-shaped seeds instead of regular one-arm seeds,8 to form extensive DNA networks confirms the similarity between the mechanism of single nanotube end-to-end joining and that of the growth of DNA nanotubes into extended networks. The model connects (1) the assembly of individual DNA staple and scaffold strands to form nanometer-scale Y-seeds, (2) the nucleation and growth of DNA nanotubes from DAE-E tiles at the adapter sites of the Y-seeds (1–10 micrometer scale), and (3) the hierarchical assembly of Y-seeded nanotube architectures into extended networks via end-to-end joining (10–100 micrometer scale). It may enable the prediction of super-large networks that incorporate thousands of seeds and provide the flexibility of changing different parameters.
A better understanding of what controls the rate of hierarchical assembly will be critical for the design of novel hierarchical nanostructures. Factors that potentially affect the rates of hierarchical assembly include the diffusion rate of large networks, the differences in the diffusion rate of networks of different sizes, and how a YNA–YNB joining event between two different networks impacts the potential for another joining event to occur nearby. The rigidity of the Y network, the length of sticky ends of the nanotube tile monomers, the temperature, and the salt composition of the solution may also affect the joining rates. More accurate and detailed models taking into account more parameters could be developed through further study to help predict the growth of DNA nanotube-based structures at different scales.
4 Materials and methods
Design and self-assembly of Y-seeds A and B
DNA origami Y-seeds were formed based on the protocol outlined by Jorgenson.6 The sequences used in this study are listed in ESI section 1.† In this work, we directly adapted the Y-seeds from Jorgenson's work as YSA and labeled these structures with ATTO 647N dye. We revised the design of the YSA by replacing some strands (ESI section 1.2†) in the adapters to create YSB and labeled YSB with ATTO 488 dye. Then in long-time imaging, we switched to using ATTO 488 for labeling both Y-seeds. Adapters, seed dye strands, dye attachment strands, struts, and staple strands were synthesized by Integrated DNA Technologies, Inc. M13mp18 scaffold strand was purchased from Bayou Biolabs. Adapter strands were PAGE purified. All samples were prepared in TAE buffer (40 mM Tris-Acetate, 1 mM EDTA) to which 12.5 mM magnesium acetate was added. After mixing the scaffold strand, staples, struts, adapters, dyes, and dye attachment strands together with buffer, the mixture was heated in an Eppendorf Mastercycler to 65 °C for 15 minutes and then immediately lowered to 47 °C for 48 hours, after which the temperature was decreased by 1 °C per minute until the thermocycler reached room temperature (20 °C). YSA and YSB were annealed separately, after which the seeds were purified using centrifugal filtration (100 kDa) to remove excess staples and adapters not incorporated into seeds13 (ESI Section 2.1†).Self-assembly of DNA nanotubes on Y-seeds
DAE-E DNA tiles consisting of five DNA strands were used in this study to grow DNA nanotubes on Y-seeds. The DNA nanotube tiles and adapters were PAGE purified from IDT. The fluorescent labeling strands were HPLC purified. To grow nanotubes on Y-seeds, we first annealed the Y-seeds of two types as described above. Next, two mixtures each containing 50 nM DNA tiles, 4 nM adapter strands, and TAE-Mg2+ buffer were annealed from 90 to 45 °C at 1 °C per minute, held at 45 °C for 1 h, and then annealed from 45 to 32 °C at 0.1 °C per minute. Once the tile mixtures reached 45 °C, pre-annealed Y-seeds A and B were heated to 45 °C and then added to the tile mixtures at a final concentration of 6 pM each to create solution A and solution B. The two solutions were incubated at 32 °C for at least 15 h to allow nanotubes to nucleate and grow. The two solutions were then mixed at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, and the incubation was continued at 32 °C to form networks and to be characterized at different times (Fig. 1).
1 ratio, and the incubation was continued at 32 °C to form networks and to be characterized at different times (Fig. 1).
Fluorescence microscopy
Fluorescence microscopy images were taken at 1, 8, and 23 hours after the Y-seeded nanotube architectures YNA and YNB were mixed. At each time point, 6 μL of solution C was transferred to an 18 mm by 18 mm glass coverslip for fluorescence imaging. The samples were imaged on an inverted microscope (Olympus IX71) using a 60×/1.45 NA oil immersion objective and Olympus Cy3 and ATTO 488 filter cube sets. Images were captured on a cooled CCD camera (iXon3, Andor). At each time point, 15 images were captured at random locations to ensure that products were sampled without bias. All captured images were used in the analysis except those where background noise was too high to allow for the reliable measurement of product sizes.Nanotube image processing
Fluorescence micrographs of nanotubes were processed in an automated fashion using the scikit-image14 library available for Python. First, edge detection was performed using the Canny algorithm.15 Small gaps in the detected edges were closed by applying a dilation (to join adjacent edges) followed by an erosion (to restore a single pixel-width edge). Finally, artifacts were removed by applying a filter to remove detected objects below a threshold size. Because we expect nanotubes to be of uniform width, nanotube length (in pixels) was calculated by dividing the total number of pixels in a tube by the mean width of the tubes in a given image. The length in pixels was then multiplied by 0.17 μm per pixel to calculate the final length in microns. Blob detection was used to locate the positions of the ATTO 488 seeds in each image. The number of Y-seeds in each network was then determined by counting the number of ATTO 488 seeds contained within the Cy3 boundaries of each distinct network. A Python script to perform the described nanotube image processing is available by a shared link at the beginning of ESI section 3.†Author contributions
Y. Jiang, M. S. Pacella, and R. Schulman designed the experiments. Y. Jiang, M. S. Pacella, J. Zhang, J. Gunn, P. Vallejo, P. Singh, T. Hou and E. Liu conducted the experiments. Y. Jiang, M. S. Pacella, S. Lee, and R. Schulman ran the simulation and analyzed the data. All the authors discussed the results, and Y. Jiang, M. S. Pacella, S. Lee, and R. Schulman wrote the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was supported by DOE award DE-SC0010426 to R. S. We expressed our thanks to Ruby Liu for her contributions in conducting the experiments on building DNA nanotube networks and imaging analysis, both of which were crucial to the advancements of our study.References
- B. Cai, Z. Li and C. L. Chen, Acc. Chem. Res., 2021, 54, 81–91 CrossRef CAS PubMed.
- C. R. Safinya, U. Raviv, D. J. Needleman, A. Zidovska, M. C. Choi, M. A. Ojeda-Lopez, K. K. Ewert, Y. Li, H. P. Miller, J. Quispe, B. Carragher, C. S. Potter, M. W. Kim, S. C. Feinstein and L. Wilson, Adv. Mater., 2011, 23, 2260–2270 CrossRef CAS PubMed.
- E. Gomes and J. Shorter, J. Biol. Chem., 2019, 294, 7115–7127 CrossRef CAS PubMed.
- H. Herrmann, H. Bär, L. Kreplak, S. V. Strelkov and U. Aebi, Nat. Rev. Mol. Cell Biol., 2007, 8, 562–573 CrossRef CAS PubMed.
- P. W. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- T. D. Jorgenson, A. M. Mohammed, D. K. Agrawal and R. Schulman, ACS Nano, 2017, 11, 1927–1936 CrossRef CAS PubMed.
- A. M. Mohammed, P. Šulc, J. Zenk and R. Schulman, Nat. Nanotechnol., 2017, 12, 312–316 CrossRef CAS PubMed.
- A. M. Mohammed and R. Schulman, Nano Lett., 2013, 13, 4006–4013 CrossRef CAS PubMed.
- S. W. Schaffter, D. Scalise, T. M. Murphy, A. Patel and R. Schulman, Nat. Commun., 2020, 11, 1–25 CrossRef PubMed.
- G. Tikhomirov, P. Petersen and L. Qian, Nature, 2017, 552, 67–71 CrossRef CAS PubMed.
- P. Petersen, G. Tikhomirov and L. Qian, Nat. Commun., 2018, 9, 5362 CrossRef CAS PubMed.
- M. S. Pacella, V. Mardanlou, S. Agarwal, A. Patel, E. Jelezniakov, A. M. Mohammed, E. Franco and R. Schulman, Mol. Syst. Des. Eng., 2020, 5, 544–558 RSC.
- D. K. Agrawal, R. Jiang, S. Reinhart, A. M. Mohammed, T. D. Jorgenson and R. Schulman, ACS Nano, 2017, 11, 9770–9779 CrossRef CAS PubMed.
- S. Van Der Walt, J. L. Schönberger, J. Nunez-Iglesias, F. Boulogne, J. D. Warner, N. Yager, E. Gouillart and T. Yu, PeerJ, 2014, 2, e453 CrossRef PubMed.
- J. Canny, IEEE Trans. Pattern Anal. Mach. Intell., 1986, PAMI-8, 679–698 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: All designs of sequences, experiment protocols and data analysis methods. See DOI: https://doi.org/10.1039/d4nr01066c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |