Gold nanoclusters Au25AcCys18 normalize intracellular ROS without increasing cytoplasmic alarmin acHMGB1 abundance in human microglia and neurons†
Issan
Zhang
 a,
Dusica
Maysinger
a,
Dusica
Maysinger
 *a,
Maja
Beus
*a,
Maja
Beus
 a,
Antonija
Mravak
a,
Antonija
Mravak
 b,
Ziqi
Yu
cd,
Martina
Perić Bakulić
b,
Ziqi
Yu
cd,
Martina
Perić Bakulić
 ef,
Patrick A.
Dion
d,
Guy A.
Rouleau
d,
Vlasta
Bonačić-Koutecký
ef,
Patrick A.
Dion
d,
Guy A.
Rouleau
d,
Vlasta
Bonačić-Koutecký
 fg,
Rodolphe
Antoine
fg,
Rodolphe
Antoine
 h and
Željka
Sanader Maršić
h and
Željka
Sanader Maršić
 *bf
*bf
aDepartment of Pharmacology and Therapeutics, McGill University, 3655 Promenade Sir-William-Osler, H3G 1Y6 Montreal, Canada. E-mail: dusica.maysinger@mcgill.ca
bFaculty of Science, University of Split, Ruđera Boškovića 33, 21000 Split, Croatia. E-mail: zsm@pmfst.hr
cDivision of Experimental Medicine, Faculty of Medicine, McGill University, Montreal, QC, Canada
dMontreal Neurological Institute, Department of Neurology and Neurosurgery, McGill University, Montreal, QC, Canada
eFaculty of Chemistry and Technology, University of Split, Ruđera Boškovića 35, 21000 Split, Croatia
fCenter of Excellence for Science and Technology, Integration of Mediterranean Region (STIM), Faculty of Science, University of Split, Ruđera Boškovića 33, 21000 Split, Croatia
gChemistry Department, Humboldt University of Berlin, Brook-Taylor-Strasse 2, 12489 Berlin, Germany
hInstitut Lumière Matière, CNRS UMR 5306, Université Claude Bernard Lyon 1, Univ. Lyon, 69622 Villeurbanne Cedex, France
First published on 28th November 2024
Abstract
This study focuses on the modulatory effects of gold nanoclusters with 25 gold atoms and 18 acetyl cysteines (Au25AcCys18) in human microglia, human iPSC-derived neurons and SH-SY5Y differentiated human neuronal cells. The combination of chemical, biological, and computational methods shows the well-retained viability of these human cells treated with Au25AcCys18, interactions between Au25AcCys18 and transcription factor TFEB (computational approach), interactions between TFEB and HMGB1 (proximity ligation assay and molecular modeling using AlphaFold), modulation of the abundance and location of acHMGB1 by Au25AcCys18 (immunocytochemistry), and the reduction of ROS in cells treated with Au25AcCys18 (CellROX live imaging). These novel findings in human neural cells, particularly neurons, encourage further studies in experimental animal models of neurological disorders and/or human organoids to exploit the unique structural and photophysical properties of gold nanoclusters and to better understand their ability to modulate molecular mechanisms in human cells.
Introduction
The meteoric rise of nanomaterials for therapeutic purposes in the last few decades raised important questions at the cellular and subcellular levels. Gold nanoclusters (AuNCs) can be atomically precise, giving them molecule-like identities with highly predictable physicochemical properties.1,2 Biological applications of AuNCs (e.g. photodynamic therapy and bioimaging) benefit from their size and ligand tunability.2–4 Bare metal clusters are unstable in solution and need protection. The combination of metal nanoclusters with peptides or proteins brings together the unique optical, electronic, and catalytic properties of metal nanoclusters with the biological functions of biomolecules. Thiol-containing ligands such as glutathione,5 mercaptoundecanoic acid,6 and captopril7 are used for specific functionalities and enhance the solubility of AuNCs in solvents. These ligands allow for precise control over the size, shape, and surface chemistry of AuNCs, facilitating their use in catalysis,8 sensing,9 and drug delivery.10 Cysteine ligands notably stabilize AuNCs and facilitate interactions with cells. N-Acetyl-cysteine is water-soluble, endogenous and non-toxic at low concentrations. Au25 nanoclusters have been thoroughly characterized using high-resolution mass spectrometry. Their size strikes a balance, being small enough for bioactivity and avoiding the disruption of cellular proteins, yet large enough to avoid toxicity due to their excessive surface area, as seen in smaller nanoclusters.11,12 Au25AcCys18 (Au25: 25 gold atoms; AcCys: N-acetyl-cysteine) has a long lifetime and optical properties attractive for photosensitization.3,13–15 Its early discovery, easy preparation, high stability, and excitation in the ultraviolet-visible and near infrared regions (tunable photoluminescence and large Stokes shift) make it advantageous for biomedical applications.16In the dynamic cellular environment, rapidly shifting oxidative states under physiological and pathological conditions can significantly affect Au25AcCys18 interactions. The brain maintains tight control of oxidative processes due to the sensitivity of neural cells and high energy demands.17 Dysregulation and/or excessive stress associated with hypoxia or inflammation are hallmarks of many neurological disorders18,19 and would affect the impact of Au25AcCys18 in cells. We have previously shown that human astrocytes respond to AuNCs in a size- and ligand-dependent manner at the level of organelles.4 The present work focuses on microglia and neurons, which are particularly sensitive to reactive oxygen species (ROS) and redox imbalance. Activated microglia release ROS and soluble factors such as high mobility group box 1 (HMGB1), a multifaceted protein with emerging binding partners and functions under physiological and pathological conditions.20,21
We hypothesize that Au25AcCys18 can modulate cellular ROS under hypoxic and normoxic conditions. Au25AcCys18 could also modulate the localization of redox-responsive proteins such as HMGB1 and TFEB, as well as their protein–protein interactions, resulting in changes in lysosomal cathepsin B activity. The results show reduced ROS abundance and maintenance of low nuclear acetylated HMGB1 (acHMGB1) abundance both in human microglia and human iPSC-derived neurons. Collectively, the data suggest context-dependent biological effects of Au25AcCys18 as related to its nano-bio interactome.
Experimental
Gold nanocluster synthesis
Au25AcCys18 and AcCys18 were provided by Dr Rodolphe Antoine from the Institut Lumière Matière, University of Lyon, France. The synthesis and characterization were described by Fakhouri et al.3 A summary of their structures is provided in Fig. 1.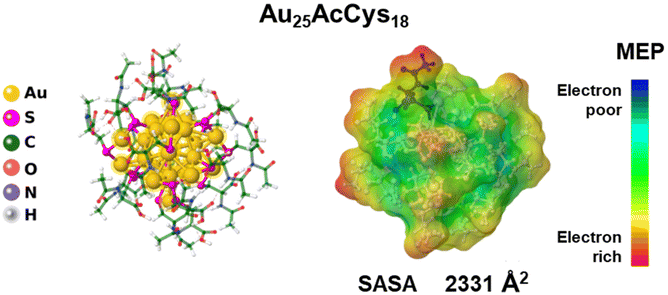 | ||
| Fig. 1 Structure of the gold nanocluster Au25AcCys18 and atomic representation with a colour scheme (left). The icosahedral gold core and S–Au–S–Au–S staple motifs are highlighted, whereas acetyl cysteine (AcCys) ligands are presented as sticks.3 Au = gold, S = sulfur, C = carbon, O = oxygen, N = nitrogen, and H = hydrogen. (Right) The solvent-accessible surface area (SASA) colored by molecular electrostatic potentials (MEPs) indicates the interacting surface of the nanocluster with the potential for electrostatic interactions. | ||
In silico modeling and analyses
The protein structures for HMGB1 (pdb: 2YRQ) and TFEB (pdb: AF_AFA0A024RCY3F1) were taken from the Protein Data Bank.22 Interactions between TFEB and Au25AcCys18 or HMGB1 were predicted using AlphaFold.23,24 The calculation of solvent accessible surface area (SASA) and molecular electrostatic potential (MEP) was performed in Jmol,25 whereas the analysis of TFEB–{Cys9} contact interactions was carried out in Chimerax.26The density functional theory-optimized structure of Au25AcCys18 was taken from our previous study.3 To calculate the interaction energy, we performed single point calculations using the Perdew–Burke–Ernzerhof functional27,28 within Gaussian software. The split valence polarization (SVP) atomic orbital (AO) basis set29 was used for gold and sulphur, and the 3-21G AO basis set30,31 was used for all other atoms. For gold atoms, a relativistic effective core potential (19-e− RECP) was employed.32
Solvent-Accessible Surface Area (SASA) represents the portion of a molecule that is available for contact with solvent (as opposed to the part that is buried inside). It is a key factor in understanding molecular interactions and behavior in biological environments. SASA is calculated by “rolling” a spherical probe (representing a solvent molecule) over the surface of the molecule we are investigating. The radius of this probe is usually set to the radius of a water molecule (∼1.4 Å). The resulting surface area represents the part of the molecule that could interact with solvent molecules.
Cell line culture
The human microglia (HMC3) and human neuroblastoma (SH-SY5Y) cell lines were originally obtained from the American Type Culture Collection. HMC3 cells were kept in Dulbecco's Modified Eagle's Medium (DMEM, Thermo Fisher Scientific) with 5% (v/v) fetal bovine serum (FBS, Wisent) and 1% (v/v) penicillin–streptomycin (Thermo Fisher Scientific). SH-SY5Y cells were maintained in F12/DMEM (Thermo Fisher Scientific) supplemented with 10% FBS and 1% penicillin–streptomycin. The cells were kept at 37 °C with 5% CO2 and 95% relative humidity.iPSC culture
Human iPSCs were generated from the lymphocytes of consenting individuals. The iPSC line SBP012 was reprogrammed as described previously.33 iPSCs were cultured on a Matrigel (Corning) coated plate with mTeSR1 medium (STEMCELL Technologies) in a 100% humidity incubator at 37 °C and 5% CO2.Neuronal differentiation
Neural progenitor cells (NPCs) were induced using a STEMdiff™ SMADi Neural Induction kit (STEMCELL Technologies) according to the manufacturer's embryoid body (EB) protocol. On day 0, a single-cell suspension of iPSCs was generated using Gentle Cell Dissociation reagent and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well were seeded in an ultra-low attachment 96-well plate (Corning) to form EBs. The cells were cultured in STEMdiff™ Neural Induction Medium with SMADi and 10 μM Y-27632. A partial medium change was performed daily from Day 1 to Day 4 using STEMdiff™ Neural Induction Medium with SMADi. On Day 5, EBs were collected using a 1 mL wide-bore serological pipette and a 40 μm strainer. EBs were then replated in a poly-L-ornithine hydrobromide (PLO) and laminin-coated 6-well plate (Corning). A full medium change was performed daily from day 6 to day 11. When the neural induction efficiency exceeds 75%, neural rosettes were manually selected on day 12 using the STEMdiff™ Neural Rosette Selection Reagent and replated onto a PLO/laminin-coated 6-well plate. Daily full medium changes were performed until the NPCs were ready for the first passage when cultures reached approximately 80–90% confluency. NPCs were maintained in STEMdiff™ Neural Progenitor Medium until ready for differentiation after two passages using Accutase (STEMCELL Technologies). For the final differentiation into forebrain neurons, NPCs were digested with Accutase and seeded onto PLO/laminin-coated glass coverslips (Mandel Scientific) with a STEMdiff™ Forebrain Neuron Differentiation Kit. Daily full medium changes were performed for 6 days before switching to the STEMdiff™ Forebrain Neuron Maturation Kit. Maturation media were changed twice a week until the neurons were ready for use.
000 cells per well were seeded in an ultra-low attachment 96-well plate (Corning) to form EBs. The cells were cultured in STEMdiff™ Neural Induction Medium with SMADi and 10 μM Y-27632. A partial medium change was performed daily from Day 1 to Day 4 using STEMdiff™ Neural Induction Medium with SMADi. On Day 5, EBs were collected using a 1 mL wide-bore serological pipette and a 40 μm strainer. EBs were then replated in a poly-L-ornithine hydrobromide (PLO) and laminin-coated 6-well plate (Corning). A full medium change was performed daily from day 6 to day 11. When the neural induction efficiency exceeds 75%, neural rosettes were manually selected on day 12 using the STEMdiff™ Neural Rosette Selection Reagent and replated onto a PLO/laminin-coated 6-well plate. Daily full medium changes were performed until the NPCs were ready for the first passage when cultures reached approximately 80–90% confluency. NPCs were maintained in STEMdiff™ Neural Progenitor Medium until ready for differentiation after two passages using Accutase (STEMCELL Technologies). For the final differentiation into forebrain neurons, NPCs were digested with Accutase and seeded onto PLO/laminin-coated glass coverslips (Mandel Scientific) with a STEMdiff™ Forebrain Neuron Differentiation Kit. Daily full medium changes were performed for 6 days before switching to the STEMdiff™ Forebrain Neuron Maturation Kit. Maturation media were changed twice a week until the neurons were ready for use.
Cell viability
Cells were seeded at 7000 cells (24 h treatment) or 5000 cells (72 h treatment) per well in 96-well plates (Sarstedt) and cultured overnight before treatment. For cell counting, the cells were incubated after treatment for 15 min with Hoechst 33342 (Millipore-Sigma, 10 μM) and imaged using a fluorescence microscope (Leica DMI4000B). Cell numbers were counted using the Cell Counter function in ImageJ (version 1.53t). For the MTT assay, the culture media were removed and the cells were incubated with 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT, 0.5 mg mL−1, Millipore-Sigma) dissolved in culture medium, for 30 min at 37 °C. The MTT media were removed and the cells were lysed in dimethyl sulfoxide (100 μL per well). The released formazan was measured colorimetrically at 595 nm using a microplate reader (Spark, Tecan).Oxidative stress
Cells were seeded onto glass coverslips (12 mm diameter, Assistent) at 7000 cells per coverslip and cultured for 24 h. Hypoxic conditions (4 h) were achieved using a BioSpherix XvivoSystem model X3 Hypoxia Hood with a glovebox. Nitrogen gas was used to flush oxygen, and the conditions were set to 37 °C, 5% CO2, and 1.0% O2.At the end of the treatment, the cells were incubated with CellROX Deep Red (5 μM, Thermo Fisher Scientific) and Hoechst 33342 (10 μM) for 30 min at 37 °C, in phenol- and serum-free DMEM (Thermo Fisher Scientific). The cells were washed in phenol- and serum-free DMEM before imaging with a fluorescence microscope. Fluorescence was analyzed in ImageJ.
Immunocytochemistry in cell lines
Cells were seeded onto glass coverslips at 7000 cells per coverslip and incubated for 24 h before treatment. The cells were washed twice with phosphate-buffered saline (PBS) and treated under serum-deprived conditions. Following treatment, the cells were fixed (4% (w/v) with paraformaldehyde, BDH, 10 min) and permeabilized (0.1% Triton X-100 (v/v), Millipore-Sigma, 10 min). The samples were incubated with primary antibodies (rabbit anti-acetylated HMGB1, 1/1000, MyBioSource, MBS9404216) overnight in 10% (v/v) goat serum (Thermo Fisher Scientific). The samples were washed thrice for 5 min with PBS and then incubated with secondary antibodies (goat anti-rabbit Alexa Fluor 647, 1/500, Thermo Fisher Scientific) for 1 h at room temperature, along with Hoechst 33342 (10 μM). The cells were washed thrice for 5 min with PBS and then mounted onto microscope slides using Aqua-Poly/Mount (Polysciences). Samples were imaged using a fluorescence microscope and fluorescence was analyzed in ImageJ.Immunocytochemistry for iPSCs
iPSC-differentiated neurons were washed twice with PBS and fixed for 1 h with 5% (w/v) sucrose and 3.7% (w/v) formaldehyde in PBS at room temperature. Following fixation, neurons were permeabilized using the same buffer with 0.2% (v/v) Triton X-100 (2 min). The neurons were rinsed twice in PBS and incubated for 10 min in 50 mM NH4CI in PBS for 10 min, washed twice with PBS before blocking with 10% (v/v) donkey serum (DS) in PBS for 20 min, and immunolabelled with primary antibodies diluted in 5% DS and 0.05% Triton X-100 in PBS overnight at 4 °C. The neurons were washed thrice with PBS for 5 min at room temperature and then incubated with secondary antibodies conjugated to Alexa 647 or Alexa 488 diluted in 5% DS and 0.05% Triton X-100 in PBS for 1 h. Neurons were washed thrice in PBS for 5 min. Hoechst 33342 (10 μM) was used to label nuclei for 10 min. Then the neurons were washed thrice with PBS and once in double distilled water, and were mounted with ProLong™ Diamond Antifade Mountant (Thermo Fisher Scientific). Images were acquired with a 63X lens on an SP8 confocal microscope (Leica Microsystems) and analyzed with ImageJ.Proximity ligation assay
Cells were seeded onto glass coverslips for immunocytochemistry experiments. Following treatment, the cells were fixed (4% paraformaldehyde, 10 min) and permeabilized (0.1% Triton X-100, 10 min). The samples were blocked in Duolink blocking solution and incubated with primary antibodies overnight (rabbit anti-TFEB, 1/500, Sigma-Aldrich, SAB4503154; mouse anti-HMGB1, 1/500, Abcam, ab190377). The proximity ligation assay was then performed following the manufacturer's instructions for Duolink (Millipore-Sigma). In brief, the samples were washed twice in buffer A (5 minand then incubated with Plus and Minus probes for 1 h at 37 °C. The cells were washed twice and then incubated with ligase (30 min, 37 °C). The cells were washed again twice and then incubated with polymerase for signal amplification (100 min, 37 °C). For actin labelling, the cells were incubated with Alexa Fluor 488 Phalloidin (1/400, 20 min, Thermo Fisher Scientific). The samples were mounted onto glass slides with Duolink in situ mounting medium with DAPI and imaged using a fluorescence microscope.Statistics
Statistical significance was calculated using one-way ANOVA followed by Dunnett's test. When the homogeneity of variance or sample size is not met, Welch's ANOVA followed by the Games-Howell test was performed. Experiments were performed independently at least three times. p-Values smaller than 0.05 were considered statistically significant.Results
Structure and properties of Au25AcCys18
Au25AcCys18 exhibits unique structural properties that influence its behavior and functionality. The core consists of gold atoms arranged in an icosahedral Au13 kernel with a central Au atom, protected by six Au2(SR)3 staple motifs.34,35 The ligands can significantly influence the AuNC properties. Both ligands and the nanocluster size play crucial roles in determining their activity and biodistribution in vivo. The choice of acetyl-cysteine (AcCys) as a protecting ligand for AuNCs was primarily for its biocompatibility and endogenous antioxidant properties.36 AcCys at concentrations used in this study does not interfere with cellular functions, and its flexibility is influenced by H-bond networks. AcCys provides greater flexibility for AuNCs compared to larger ligands like glutathione,3 owing to fewer H-bonds and allowing for structural flexibility and performance. The overall size of the nanocluster is approximately 2 nm, with a solvent-accessible surface area of 2331 Å2, making it small enough for cellular uptake and interaction with biomolecules (Fig. 1).Biological evaluation in human neural cells: viability and modulation of ROS
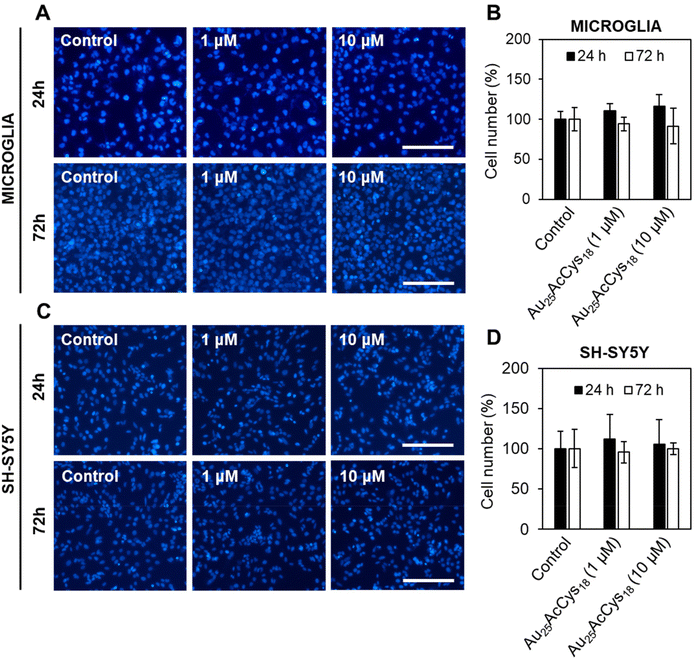
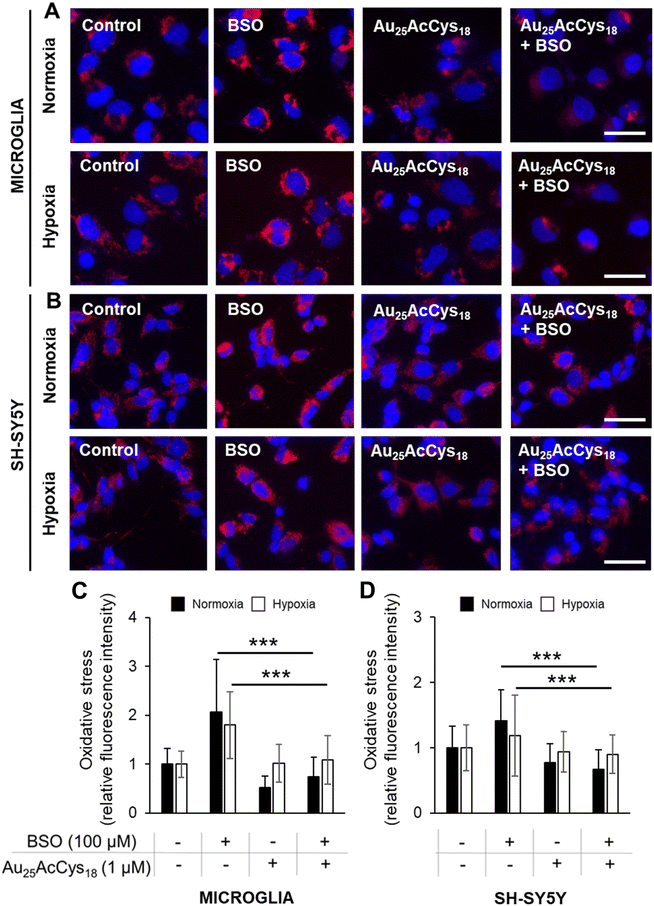
We first assessed the viability of human microglia and neurons in response to Au25AcCys18. These neural cell types play critical roles in the central nervous system under physiological and pathological conditions. Microglia are sensitive to foreign materials as they constantly survey the brain. Their response to nanostructures and other compounds sets the stage for therapeutic applications, as they directly and indirectly influence other neural cells. Neurons do not have the same ability as microglia to proliferate, and their loss is critical to brain health. We examined the potential toxicity of Au25AcCys18 by establishing time- and concentration-dependent effects on cell viability (cell count) after 24 h and 72 h (Fig. 2 and Fig. S1†). The results showed that Au25AcCys18 was well tolerated at the concentration used throughout this manuscript (1 μM) in both cell types.Cellular redox is a fast-changing state that participates in cellular signalling. Altered redox as a result of changing energy expenditure or production influences the functionality of redox-sensitive proteins and pathways. A cellular response to Au25AcCys18 under conditions of oxidative stress was simulated chemically using the GSH inhibitor buthionine sulfoximine (BSO), as well as physically using a hypoxic chamber (Fig. 3). Depletion of antioxidant defences with BSO causes an increase in ROS, as detected with the fluorescent probe CellROX. Neuronal cells were more sensitive to hypoxia with GSH depletion, and Au25AcCys18 brought down ROS to near-control levels. This suggests a protective effect of Au25AcCys18 against excessive cellular ROS.
Modulation of HMGB1 localization
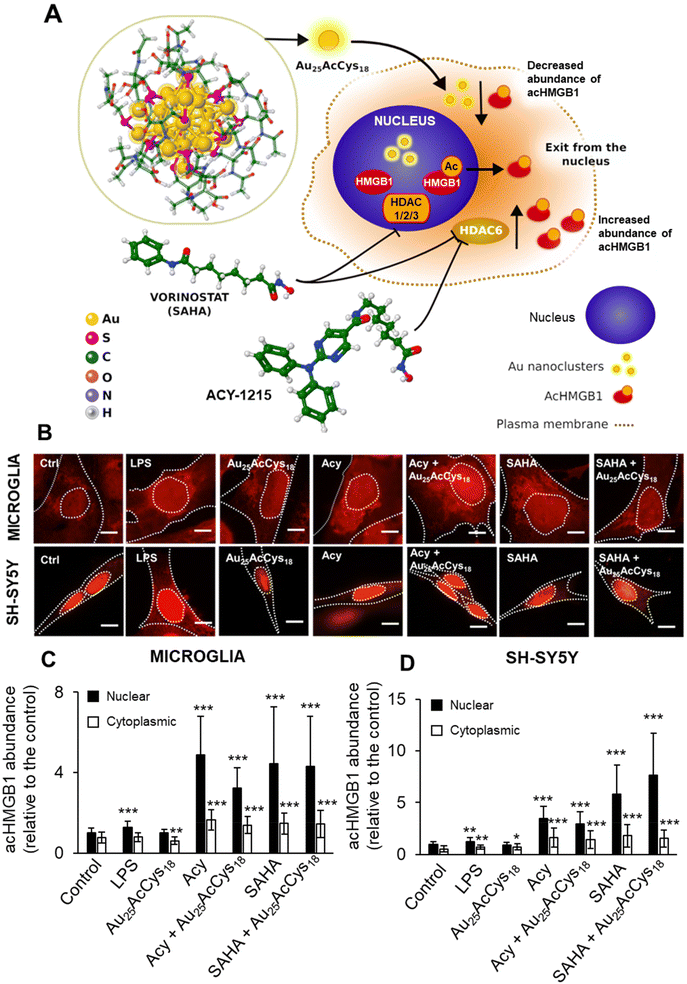
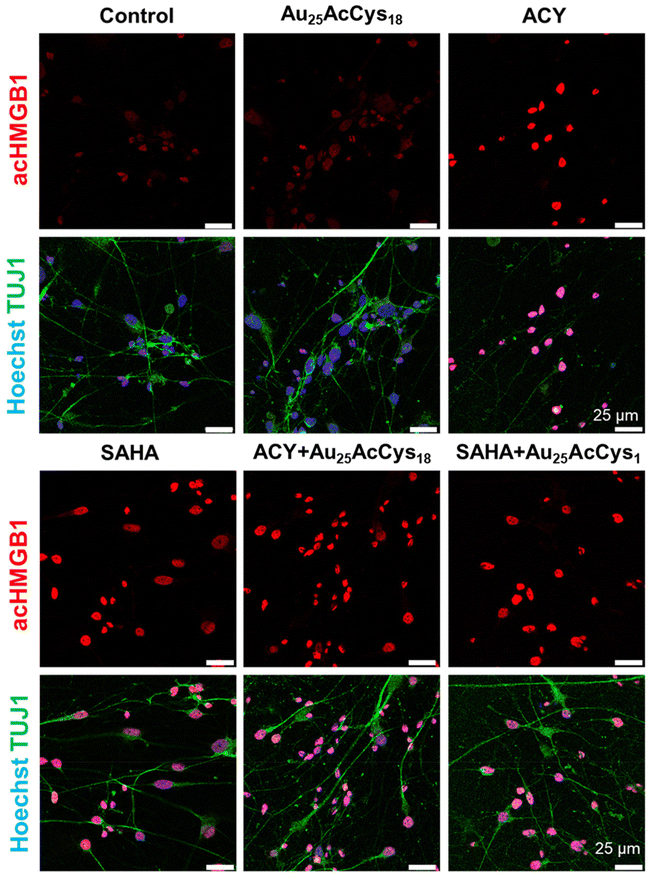
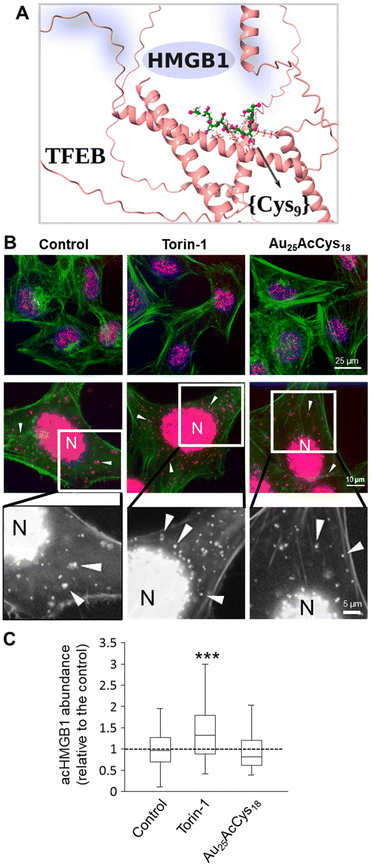
Microglial and neuronal cells are affected by pro-inflammagens such as lipopolysaccharides (LPSs), which activate a cascade of intracellular pathways culminating in the release of soluble factors such as HMGB1. HMGB1 is mostly concentrated in the nucleus, where it serves as a DNA-associated protein facilitating transcription. Inflammation and ROS induce post-translational modifications (e.g. acetylation) that relocate HMGB1 to the cytosol and extracellular space as a signalling protein, thus affecting neuron–neuron and neuron–glia interactions. Pharmacological manipulation using histone deacetylase inhibitors (ACY-1215, SAHA) increased the cellular level of acHMGB1 (Fig. 4 and 5). The concept is illustrated in Fig. 4a. We show the morphology of human microglia and neuron-like cells (differentiated SH-SY5Y), and the abundance of acHMGB1 under hypoxic and normoxic conditions (Fig. 4b and d). Au25AcCys18 itself maintains cytoplasmic acHMGB1 to a low, barely detectable level both in differentiated SH-SY5Y (Fig. 4d) and human neurons, comparable to untreated controls (Fig. 5). Au25AcCys18 does not significantly change the subcellular distribution of acHMGB1, as the effect of histone deacetylase inhibitors is very strong. Considering that HMGB1 translocates from the nucleus to the cytosol under cellular stress, we next investigated if cytosolic HMGB1 will associate with another stress-responsive transcription factor: TFEB.Interaction with transcription factors TFEB and HMGB1
HMGB1 is a co-factor of numerous transcription factors. Its association with lysosomal compartments for cellular release suggests a potential interaction with the key lysosomal regulator TFEB.37 Little is known about TFEB and HMGB1 interactions. We aimed to investigate the sites and characteristics of their interactions and compare them with the putative interactions of TFEB and Au25AcCys18. We employed AlphaFold (v.2.0), which uses an end-to-end deep neural network to predict protein structures from sequences, including protein complexes.38 It has achieved unprecedented accuracy, significantly outperforming other methods in the latest Critical Assessment of Protein Structure Prediction (CASP14) round.23Fig. 6a presents the interaction sites for HMGB1 and Au25AcCys18 at the surface of TFEB, whereas Fig. S2† shows details that include TFEB amino acids interacting with HMGB1 versus {Cys9}. Prediction of the TFEB and HMGB1 complex was done using their primary sequences, and AlphaFold provided their three-dimensional complex model, together with the predicted local distance difference test (pLDDT). The pLDDT is a per-residue confidence measure for structural predictions, scaled from 0 to 100. Higher scores indicate higher confidence and better prediction accuracy. A pLDDT above 90 signifies high accuracy for both backbone and side chains, whereas a score above 70 indicates accurate backbone prediction with some side chain errors. The pLDDT score can vary across a protein, indicating regions of differing prediction reliability and identifying which parts of the structure are likely accurate. Comparison of the pLDDT plots in Fig. S2a† shows that interacting regions of TFEB and HMGB1 are dark and light blue (pLDDT > 90 and 90 > pLLDT > 70, respectively), indicating high accuracy for the backbone, and slightly less for side chains. On the other hand, the interaction of TFEB and Au25AcCys18 was modelled using a sequence of 9 Cys residues due to the need for a flexible model of TFEB, and because only surface ligands of Au25NCs can interact with TFEB. AlphaFold effectively models the flexibility of TFEB and the {Cys9} ligand, which is crucial for their strong interaction. The model shows the flexibility of TFEB, consistent with its varying NMR structures and shapes {Cys9} for optimal interactions. We also tested a {Cys18} model which forms a hairpin and interacts with TFEB similarly to {Cys9}. Other docking tools might be less reliable as they use a fixed TFEB structure (which is very flexible in solution), leading to results that heavily depend on the initial TFEB configuration and are unsuitable for large molecules like Au25AcCys18. On the other hand, the accuracy of AlphaFold2 can be improved by including the training data and code.39 Fig. S2b† shows the 3D structure of the TFEB and {Cys9} complex. Comparing the positions of HMGB1 and {Cys9} with TFEB (Fig. 6a), and the interacting amino acids (Fig. S2c and d†) it is apparent that they occupy different parts of TFEB, with different amino acids involved.The biological association between HMGB1 and TFEB was detected in neural cells using a proximity ligation assay (Fig. 6). Most interactions occur in the nucleus, whereas an increased association in the cytosol is associated with autophagy, as shown in cells treated with the mTOR inhibitor Torin-1 (Fig. 6b and c). Au25AcCys18 maintained a level of HMGB1–TFEB association comparable to that of the control condition. An overview of interaction sites between HMGB1 and TFEB is given (Fig. S2a and c†). Additionally, the {Cys9} model for Au25AcCys18 and TFEB interaction sites is shown (Fig. S2b and d†).
Discussion
This study addresses the impact and mechanism of atomically defined AuNCs in human neural cells. The rationale for such investigations is a paucity of data for Au25AcCys18 in brain cells and a need for a better understanding of intracellular events in human microglia and neurons. So far, Au25AcCys18 has not been tested in human neurons derived from iPSCs. This is a close model of human neurons useful for drug and nanostructure screening.40,41It is well established that many stressors, including cerebral hypoxia, lead to hyperactivation of microglia, astrocytes, and deleterious effects in neurons.42–46 Considering that AuNCs are promising nanotools for imaging and eventual therapeutic intervention, it is a prerequisite to improve our understanding of their deleterious and beneficial effects. Our results show that Au25AcCys18 exerts distinct modulatory effects in human microglia, neuron-like cells, and neurons. A decrease in the abundance of ROS under hypoxic and normoxic conditions, maintenance of HMGB1–TFEB association at control levels and low toxicity in human neural cells are novel findings reported herein. Further studies at the level of protein–protein interactions are warranted, particularly in disease models.
Neurological disorders have consistently challenged therapeutic modalities for several reasons; brain accessibility is limited by the size and properties of compounds, and the delicate balance between neural cells does not accommodate drastic manipulations without significant side effects and sequelae. Yet, the demand for better diagnostic solutions and brain drugs continues to grow globally. Nanotechnological approaches to the brain offer several advantages. AuNCs in this study are atomically defined and can be as small as a few nanometers, with tunable surface properties. These features can facilitate blood–brain barrier entry and discourage unwanted immune reactivity. Microglia are often the first responders in the brain; by consuming and interacting with AuNCs, they can directly or indirectly influence other neural cells.3,4,47,48 AuNCs seem to have a range of effects relevant to neurological disorders, particularly when functionalized with AcCys, which is key to antioxidant defenses. By altering ROS levels, Au25AcCys18 modulates hypoxia-driven pathways, which would otherwise exacerbate microenvironmental damage in brain cancer and stroke.49–51 Microglial hyperactivation is also a source of excessive ROS, and when unopposed due to ineffective endogenous antioxidant molecules or enzymatic function, it results in impaired cellular functions. Our studies show remarkable effects of Au25AcCys18 in normalizing ROS concentrations under normoxic and hypoxic conditions.
We investigated if the AuNCs could have subtle effects on protein–protein interactions, particularly with redox-responsive transcription factors TFEB and HMGB1. The AuNC interactome is only beginning to be studied. Interaction partners can be partially surmised based on subcellular localization. We investigated lysosomal proteins because HMGB1 is trafficked through lysosomal compartments upon its acetylation and extranuclear release.52,53 Its alarmin function is enhanced by pro-inflammagens, making it a pharmacological target.20,54 Although Au25AcCys18 does not directly affect HMGB1 acetylation (e.g. by modulating acetylases or deacetylases), it attenuates the effect of deacetylase inhibitors in neuron-like cells. This might indicate that Au25AcCys18 exerts mainly cytosolic effects, as pan- (vorinostat, SAHA) and cytosolic (ACY-1215) histone deacetylase inhibitors had similar results. In addition, ROS can increase extranuclear HMGB1 and TFEB activation through the lysosomal sensor TRPML1.55 Thus, AuNC-mediated normalization of ROS could also normalize HMGB1 localization and benefit inflammatory resolution.
TFEB participates in the regulation of lysosomal function and biogenesis as well as cellular adaptation under stress.56 Effects of Au25AcCys18 on lysosomes were investigated through TFEB activation, cathepsin activity, and TFEB interactions. TFEB–HMGB1 interactions detected by the proximity ligation assay indicate that most interactions are nuclear, as both proteins are transcription regulators abundant in the nucleus.37,57,58 Cytosolic interactions increased upon mTOR inhibition with Torin-1, indicating that TFEB and HMGB1 interaction could participate in autophagy.59 TFEB and HMGB1 coincide in their interaction with autophagic flux regulator Beclin-1, thus contributing to the same protein complexes and pathways.60,61 TFEB acetylation at several lysine residues is also a determinant of its dimerization and transcriptional activity, sometimes with opposing effects.37,62 TFEB and HMGB1 can undergo several types of post-translational modifications (PTMs), including phosphorylation and oxidation.37,52,62,63 Such modifications under oxidative stress could alter their interaction affinity for other redox-responsive proteins, and variable adaptability could make neurons particularly vulnerable. Under oxidative stress, TFEB can be oxidized and prone to dimerization. AcHMGB1 has a limited ability to interact with AuNCs,4 suggesting that PTMs contribute to altered protein–protein interactions. It remains to be examined how phosphorylated HMGB1 and TFEB differ in their interaction, and how other PTMs influence their interaction in models of neurological disorders.
The models of TFEB–HMGB1 and TFEB–Au25AcCys18 interactions were studied using AlphaFold as implemented in Chimerax. Thus, the flexibility of all interacting partners was considered. The results clearly show that critical sites for the TFEB–HMGB1 interaction are TFEB residues Y75 to Q86 and S209 to K219, whereas a distinct set of TFEB residues (K237 to K274) is involved in interactions with the {Cys9} model of Au25AcCys18. Interactions involve both charged and hydrophobic amino acids, which form ion–ion and hydrophobic connections between TFEB and HMGB1, or the {Cys9} model. Advantages of AlphaFold2 predictions include the possibility to accurately model multiunit protein–protein complexes from their primary sequences without the need for resolved crystal structures, whereas the pLDDT metric offers a quantitative assessment of the models. As only the protein parts of the Protein Data Bank structures were used as the AlphaFold2 training set, this method cannot model small molecules, DNA or post-translational modifications, and it struggles to predict the impact of point mutations.
In summary, results from our studies suggest that Au25AcCys18 (i) is only toxic at high concentrations in human neurons and glia; (ii) reduces oxidative stress under hypoxic and normoxic conditions and (iii) maintains HMGB1–TFEB association in human neural cells at the control level. These novel findings provide a first step in future connectome investigations in human neural cells exposed to or treated with AuNCs. They also point towards studies on how AuNCs could affect other transcription factor complexes under physiological and pathological conditions in cell models, particularly human organoids.
Conclusions
The motivation for this study stems from seminal discoveries highlighting the unique properties of AuNCs using several complementary experimental techniques and computational methods.3,64,65 Au25AcCys18 is particularly interesting because it does not seem harmful to human cells within the concentration range and models proposed. They were tested for bioimaging,15,66–68 and our recent studies explain the uniqueness of Au25 with 16 ligands, supporting the notion that this type of nanocluster is a “captain of the big ship”.69 We provide new insight into some biological effects in neural cells and show that Au25AcCys18 could interact with the transcription factor TFEB (computational model), modulate HMGB1 localization without markedly increasing its abundance, and normalize excessive levels of ROS under hypoxic and normoxic conditions. Considering that so far Au25AcCys18 has not been tested in human neurons derived from iPSCs, we aim to stimulate further preclinical investigations on AuNCs in human brain models (e.g. organoids). As our studies are limited to HMGB1–TFEB association in the presence of Au25AcCys18, this is a first step in future connectome investigations in human neural cells exposed to or treated with AuNCs. Further investigations on the likely modulatory effects of different protein–protein associations are warranted.Author contributions
DM, IZ, and ZSM conceptualized the studies and wrote the manuscript drafts and revisions. IZ and MB performed all experiments in cells, and ZQY performed experiments in iPSC-derived neurons. PD and GR supervised experiments in iPSC-derived neurons. ZSM, AM, and MPB carried out modeling, analyzed and prepared the results in publishable format. VBK and AR participated in the initial experimental plan and provided feedback on the text.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
DM acknowledges funding from FRQ-S (#294233) and NSERC (RGPIN 2020-07011). ZSM and VBK were supported by the project STIM-REI (contract number: KK.01.1.1.01.0003), funded by the European Union through the European Regional Development Fund—the Operational Programme Competitiveness and Cohesion 2014-2020 (KK.01.1.1.01). GR is funded by the ALS-Canada-Brain Canada Arthur J. Hudson Translation Team Grant. VBK, ZSM, MPB and AM acknowledge funding from “Confined molecular systems: from a new generation of materials to the stars” (COSY) supported by COST (European Cooperation in Science and Technology).References
- V. Bonačić-Koutecký and R. Antoine, Nanoscale, 2019, 11, 12436–12448 RSC.
- S. M. van de Looij, E. R. Hebels, M. Viola, M. Hembury, S. Oliveira and T. Vermonden, Bioconjugate Chem., 2022, 33, 4–23 CrossRef CAS PubMed.
- H. Fakhouri, M. P. Bakulić, I. Zhang, H. Yuan, D. Bain, F. Rondepierre, P.-F. Brevet, Ž. S. Maršić, R. Antoine, V. Bonačić-Koutecký and D. Maysinger, Commun. Chem., 2023, 6, 1–10 CrossRef PubMed.
- D. Maysinger, Ž. Sanader Maršić, E. R. Gran, A. Shobo, J.-R. Macairan, I. Zhang, M. Perić Bakulić, R. Antoine, G. Multhaup and V. Bonačić-Kouteckỳ, ACS Chem. Neurosci., 2022, 13, 464–476 CrossRef CAS PubMed.
- Y. Negishi, K. Nobusada and T. Tsukuda, J. Am. Chem. Soc., 2005, 127, 5261–5270 CrossRef CAS PubMed.
- L. Zhu, Y. Zeng, M. Teubner, B. Grimm-Lebsanft, A. R. Ziefuß, C. Rehbock, M. A. Rübhausen, S. Barcikowski, W. J. Parak and I. Chakraborty, ACS Appl. Nano Mater., 2021, 4, 3197–3203 CrossRef CAS.
- S. R. Bhattacharya, K. Bhattacharya, V. J. Xavier, A. Ziarati, D. Picard and T. Bürgi, ACS Appl. Mater. Interfaces, 2022, 14, 29521–29536 CrossRef CAS PubMed.
- S. Tian, Y. Cao, T. Chen, S. Zang and J. Xie, Chem. Commun., 2020, 56, 1163–1174 RSC.
- G. F. Combes, A.-M. Vučković, M. Perić Bakulić, R. Antoine, V. Bonačić-Koutecky and K. Trajković, Cancers, 2021, 13, 4206 CrossRef CAS PubMed.
- H. Huang, R. Liu, J. Yang, J. Dai, S. Fan, J. Pi, Y. Wei and X. Guo, Pharmaceutics, 2023, 15, 1868 CrossRef CAS PubMed.
- J. Yang, F. Yang, C. Zhang, X. He and R. Jin, ACS Mater. Lett., 2022, 4, 1279–1296 CrossRef CAS.
- J. Peng and X. Liang, Medicine, 2019, 98, e15311 CrossRef CAS PubMed.
- A.-M. Hada, A.-M. Craciun, M. Focsan, A. Vulpoi, E.-L. Borcan and S. Astilean, Microchim. Acta, 2022, 189, 337 CrossRef CAS PubMed.
- M. Hembury, N. Beztsinna, H. Asadi, J. B. van den Dikkenberg, J. D. Meeldijk, W. E. Hennink and T. Vermonden, Biomacromolecules, 2018, 19, 2841–2848 CrossRef CAS PubMed.
- C. Zhang, X. Gao, W. Chen, M. He, Y. Yu, G. Gao and T. Sun, iScience, 2022, 25, 105022 CrossRef CAS PubMed.
- X. Yuan, Y. Yu, Q. Yao, Q. Zhang and J. Xie, J. Phys. Chem. Lett., 2012, 3, 2310–2314 CrossRef CAS PubMed.
- L.-P. Bernier, E. M. York and B. A. MacVicar, Trends Neurosci., 2020, 43, 854–869 CrossRef CAS PubMed.
- T. F. Outeiro, P. S. Brocardo and D. P. Gelain, J. Neurochem., 2024, 168, 1423–1425 CrossRef CAS PubMed.
- C. Pallarés-Moratalla and G. Bergers, Front. Immunol., 2024, 15, 1305087 CrossRef PubMed.
- U. Andersson, H. Yang and H. Harris, Expert Opin. Ther. Targets, 2018, 22, 263–277 CrossRef CAS PubMed.
- U. Andersson and H. Yang, J. Intensive Care Med. DOI:10.1016/j.jointm.2022.02.001.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov and M. Steinegger, Nat. Methods, 2022, 19, 679–682 CrossRef CAS PubMed.
- Jmol: an open-source Java viewer for chemical structures in 3D. https://www.jmol.org/.
- E. C. Meng, T. D. Goddard, E. F. Pettersen, G. S. Couch, Z. J. Pearson, J. H. Morris and T. E. Ferrin, Protein Sci., 2023, 32, e4792 CrossRef CAS PubMed.
- J. P. Perdew, J. A. Chevary, S. H. Vosko, K. A. Jackson, M. R. Pederson, D. J. Singh and C. Fiolhais, Phys. Rev. B: Condens. Matter Mater. Phys., 1992, 46, 6671–6687 CrossRef CAS PubMed.
- J. P. Perdew, M. Ernzerhof and K. Burke, J. Chem. Phys., 1996, 105, 9982–9985 CrossRef CAS.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC.
- J. S. Binkley, J. A. Pople and W. J. Hehre, J. Am. Chem. Soc., 1980, 102, 939–947 CrossRef CAS.
- K. D. Dobbs and W. J. Hehre, J. Comput. Chem., 1987, 8, 880–893 CrossRef CAS.
- M. Dolg, H. Stoll and H. Preuss, J. Chem. Phys., 1989, 90, 1730–1734 CrossRef CAS.
- S. Stern, R. Santos, M. C. Marchetto, A. P. D. Mendes, G. A. Rouleau, S. Biesmans, Q.-W. Wang, J. Yao, P. Charnay, A. G. Bang, M. Alda and F. H. Gage, Mol. Psychiatry, 2018, 23, 1453–1465 CrossRef CAS PubMed.
- M. W. Heaven, A. Dass, P. S. White, K. M. Holt and R. W. Murray, J. Am. Chem. Soc., 2008, 130, 3754–3755 CrossRef CAS PubMed.
- M. Zhu, C. M. Aikens, F. J. Hollander, G. C. Schatz and R. Jin, J. Am. Chem. Soc., 2008, 130, 5883–5885 CrossRef CAS PubMed.
- B. Pedre, U. Barayeu, D. Ezeriņa and T. P. Dick, Pharmacol. Ther., 2021, 228, 107916 CrossRef CAS PubMed.
- H. Chen, S. Gong, H. Zhang, Y. Chen, Y. Liu, J. Hao, H. Liu and X. Li, Cell Death Discovery, 2024, 10, 84 CrossRef CAS PubMed.
- R. Yin, B. Y. Feng, A. Varshney and B. G. Pierce, Protein Sci., 2022, 31, e4379 CrossRef CAS PubMed.
- G. Ahdritz, N. Bouatta, C. Floristean, S. Kadyan, Q. Xia, W. Gerecke, T. J. O'Donnell, D. Berenberg, I. Fisk, N. Zanichelli, B. Zhang, A. Nowaczynski, B. Wang, M. M. Stepniewska-Dziubinska, S. Zhang, A. Ojewole, M. E. Guney, S. Biderman, A. M. Watkins, S. Ra, P. R. Lorenzo, L. Nivon, B. Weitzner, Y.-E. A. Ban, S. Chen, M. Zhang, C. Li, S. L. Song, Y. He, P. K. Sorger, E. Mostaque, Z. Zhang, R. Bonneau and M. AlQuraishi, Nat. Methods, 2024, 21, 1514–1524 CrossRef CAS PubMed.
- R. Dolmetsch and D. H. Geschwind, Cell, 2011, 145, 831–834 CrossRef CAS PubMed.
- K. Takahashi, K. Tanabe, M. Ohnuki, M. Narita, T. Ichisaka, K. Tomoda and S. Yamanaka, Cell, 2007, 131, 861–872 CrossRef CAS PubMed.
- P.-B. Bielawski, I. Zhang, C. Correa-Paz, F. Campos, M. Migliavacca, E. Polo, P. Del Pino, B. Pelaz, D. Vivien and D. Maysinger, ACS Pharmacol. Transl. Sci., 2024, 7, 680–692 CrossRef CAS PubMed.
- S. Y. Lee and W.-S. Chung, Curr. Opin. Neurobiol., 2024, 84, 102840 CrossRef CAS PubMed.
- S. A. Liddelow, K. A. Guttenplan, L. E. Clarke, F. C. Bennett, C. J. Bohlen, L. Schirmer, M. L. Bennett, A. E. Münch, W.-S. Chung, T. C. Peterson, D. K. Wilton, A. Frouin, B. A. Napier, N. Panicker, M. Kumar, M. S. Buckwalter, D. H. Rowitch, V. L. Dawson, T. M. Dawson, B. Stevens and B. A. Barres, Nature, 2017, 541, 481–487 CrossRef CAS PubMed.
- W. Lu and J. Wen, Mol. Neurobiol., 2024, 61, 6161–6174 CrossRef CAS PubMed.
- G. Nzou, R. T. Wicks, N. R. VanOstrand, G. A. Mekky, S. A. Seale, A. El-Taibany, E. E. Wicks, C. M. Nechtman, E. J. Marrotte, V. S. Makani, S. V. Murphy, M. C. Seeds, J. D. Jackson and A. J. Atala, Sci. Rep., 2020, 10, 9766 CrossRef CAS PubMed.
- C. C. Escoubas and A. V. Molofsky, Trends Immunol., 2024, 45, 358–370 CrossRef CAS PubMed.
- S. Zhao, A. D. Umpierre and L.-J. Wu, Trends Neurosci., 2024, 47, 181–194 CrossRef CAS PubMed.
- N. Colwell, M. Larion, A. J. Giles, A. N. Seldomridge, S. Sizdahkhani, M. R. Gilbert and D. M. Park, Neuro-Oncology, 2017, 19, 887–896 CrossRef CAS PubMed.
- N. Lénárt, C. Cserép, E. Császár, B. Pósfai and Á. Dénes, Glia, 2024, 72, 833–856 CrossRef PubMed.
- A. M. Planas, Glia, 2024, 72, 1016–1053 CrossRef CAS PubMed.
- U. Andersson, D. J. Antoine and K. J. Tracey, J. Intern. Med., 2014, 276, 420–424 CrossRef CAS PubMed.
- R. Chen, R. Kang and D. Tang, Exp. Mol. Med., 2022, 54, 91–102 CrossRef CAS PubMed.
- M. E. Street, Horm. Res. Paediatr., 2020, 93, 73–75 CrossRef CAS PubMed.
- X. Zhang, X. Cheng, L. Yu, J. Yang, R. Calvo, S. Patnaik, X. Hu, Q. Gao, M. Yang, M. Lawas, M. Delling, J. Marugan, M. Ferrer and H. Xu, Nat. Commun., 2016, 7, 12109 CrossRef CAS PubMed.
- N. Raben and R. Puertollano, Annu. Rev. Cell Dev. Biol., 2016, 32, 255–278 CrossRef CAS PubMed.
- Y. Feng, Y. Chen, X. Wu, J. Chen, Q. Zhou, B. Liu, L. Zhang and C. Yi, Autophagy, 2024, 20, 4–14 CrossRef CAS PubMed.
- M. T. Lotze and K. J. Tracey, Nat. Rev. Immunol., 2005, 5, 331–342 CrossRef CAS PubMed.
- D. Tang, R. Kang, K. M. Livesey, C.-W. Cheh, A. Farkas, P. Loughran, G. Hoppe, M. E. Bianchi, K. J. Tracey, H. J. Zeh and M. T. Lotze, J. Cell Biol., 2010, 190, 881–892 CrossRef CAS PubMed.
- X. Ma, H. Liu, J. T. Murphy, S. R. Foyil, R. J. Godar, H. Abuirqeba, C. J. Weinheimer, P. M. Barger and A. Diwan, Mol. Cell. Biol., 2015, 35, 956–976 CrossRef PubMed.
- M. Palmieri, S. Impey, H. Kang, A. di Ronza, C. Pelz, M. Sardiello and A. Ballabio, Hum. Mol. Genet., 2011, 20, 3852–3866 CrossRef CAS PubMed.
- J. A. Martina, D. Guerrero-Gómez, E. Gómez-Orte, J. Antonio Bárcena, J. Cabello, A. Miranda-Vizuete and R. Puertollano, EMBO J., 2021, 40, e105793 CrossRef CAS PubMed.
- M. Takla, S. Keshri and D. C. Rubinsztein, EMBO Rep., 2023, 24, e57574 CrossRef CAS PubMed.
- M. Perić, Ž. S. Maršić, I. Russier-Antoine, H. Fakhouri, F. Bertorelle, P.-F. Brevet, X. le Guével, R. Antoine and V. Bonačić-Koutecký, Phys. Chem. Chem. Phys., 2019, 21, 23916–23921 RSC.
- H. Qian, M. Zhu, Z. Wu and R. Jin, Acc. Chem. Res., 2012, 45, 1470–1479 CrossRef CAS PubMed.
- J. Ma, M. Yang, B. Zhang and M. Niu, Nanoscale, 2024, 16, 7287–7306 RSC.
- K. Tan, H. Ma, X. Mu, Z. Wang, Q. Wang, H. Wang and X.-D. Zhang, Anal. Bioanal. Chem., 2024, 416, 5871–5891 CrossRef CAS PubMed.
- H. Yang, Y. Wu, H. Ruan, F. Guo, Y. Liang, G. Qin, X. Liu, Z. Zhang, J. Yuan and X. Fang, Anal. Chem., 2022, 94, 3056–3064 CrossRef CAS PubMed.
- X. Kang, H. Chong and M. Zhu, Nanoscale, 2018, 10, 10758–10834 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr03512g |
| This journal is © The Royal Society of Chemistry 2025 |