Open Access Article
Open Access ArticleUnravelling complex mechanisms in materials processes with cryogenic electron microscopy
Minyoung
Lee
ab,
Yonggoon
Jeon
 abc,
Sungin
Kim
abd,
Ihnkyung
Jung
ab,
Sungsu
Kang
abe,
Seol-Ha
Jeong
*a and
Jungwon
Park
abc,
Sungin
Kim
abd,
Ihnkyung
Jung
ab,
Sungsu
Kang
abe,
Seol-Ha
Jeong
*a and
Jungwon
Park
 *abfg
*abfg
aDepartment of Chemical and Biological Engineering, and Institute of Chemical Processes, Seoul National University, Seoul 08826, Republic of Korea. E-mail: jungwonpark@snu.ac.kr; jsh528@snu.ac.kr
bCenter for Nanoparticle Research, Institute for Basic Science (IBS), Seoul, 08826, Republic of Korea
cDepartment of Physics and Chemistry, Korea Military Academy (KMA), Seoul 01805, Republic of Korea
dDepartment of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA
eDepartment of Chemistry, University of Chicago, Chicago, IL 60637, USA
fInstitute of Engineering Research, Seoul National University, Seoul, 08826, Republic of Korea
gAdvanced Institute of Convergence Technology, Seoul National University, Suwon, 16229, Republic of Korea
First published on 17th December 2024
Abstract
Investigating nanoscale structural variations, including heterogeneities, defects, and interfacial characteristics, is crucial for gaining insight into material properties and functionalities. Cryogenic electron microscopy (cryo-EM) is developing as a powerful tool in materials science particularly for non-invasively understanding nanoscale structures of materials. These advancements bring us closer to the ultimate goal of correlating nanoscale structures to bulk functional outcomes. However, while understanding mechanisms from structural information requires analysis that closely mimics operation conditions, current challenges in cryo-EM imaging and sample preparation hinder the extraction of detailed mechanistic insights. In this Perspective, we discuss the innovative strategies and the potential for using cryo-EM for revealing mechanisms in materials science, with examples from high-resolution imaging, correlative elemental analysis, and three-dimensional and time-resolved analysis. Furthermore, we propose improvements in cryo-sample preparation, optimized instrumentation setup for imaging, and data interpretation techniques to enable the wider use of cryo-EM and achieve deeper context into materials to bridge structural observations with mechanistic understanding.
Introduction
Materials at the nanoscale often exhibit unique properties driven by surface1–5 or quantum effects6–8 that influence the macroscopic properties of chemistry-based processes, such as those in energy storage systems,9,10 catalysis,11–15 biochemistry,16 self-assembly17 or crystallization.18–22 In solid materials, heterogeneities in morphology, presence of defects,10,23 and interfacial characteristics24–26 contribute to bulk properties in complex ways. Examples include nanoscale crystallinity affecting diffusive properties27,28 or molecular structural variations giving rise to macroscale morphologies.20,29,30 Gaining insight into how these unique nanoscale structures affect macroscopic characteristics allows for designing materials with tailored functionalities, enhancing materials performance across various fields.Investigation of such nanoscale structural variations requires characterization techniques capable of sub-nanometer resolution. Among such tools is cryogenic electron microscopy (cryo-EM), recognized by the Nobel Prize in Chemistry for its application in determining structures of proteins and biological organelles in their vitrified native states.31 The technique involves rapidly freezing samples near liquid nitrogen temperature, which fixes the samples of interest within a thin layer of vitreous ice. This allows for the 3D structure analysis of proteins based on numerous solvated particle projections acquired by transmission electron microscopy (TEM) maintained at cryogenic temperatures. Cryo-EM have been widely utilized for elucidating mechanisms of biomolecular interactions through single particle analysis with 2D projections,31 and even with in situ visualization of cellular processes enabled by cryo-electron tomography.20 Despite the sensitivity of biomaterials to environmental conditions and the electron beam in electron microscopy, cryo-EM had enabled their imaging at unprecedented resolution. With the extensive use of cryo-EM for structural biology, the automation of sample preparation,32–34 advancements in instrumentation,35,36 and algorithms for image processing37–40 have led to improvements in image quality, and as a result, deepened the level of structural investigation. These methods are currently well-established into a pipeline for efficient data acquisition.
In materials science, imaging vitrified samples using cryo-EM had been initially utilized for the observation of the structures of solvated soft matter such as block copolymers,41–43 surfactant micelles,44 and single-walled carbon nanotubes.45 These previous observations of assembled structures using cryo-EM span scales of hundreds of nanometers down to sub-nanometer scales,46 allowing spatial resolutions sufficient for revealing individual assembly components at the nanoscale.46 While cryo-EM initially focused on structures of soft matter, cryo-EM later expanded to observing a wider range of materials including inorganic materials and composites,20,46,47 naturally expanding into the mechanistic studies of these materials in context of their operation in applications. Imaging using the TEM or scanning TEM (STEM) mode inherently provides structural information, and while structures of materials are closely tied to their functionalities and mechanisms in processes, bridging the gap between structural information and mechanisms of action is a prevalent goal. Therefore, there has been a growing need for obtaining more context beyond nanoscale structural information, such as higher resolutions down to atomic scale or multi-dimensional information such as correlative spectroscopic analysis or time-resolved information. The process of interpreting data is also crucial for deriving mechanistic understanding from structural data.
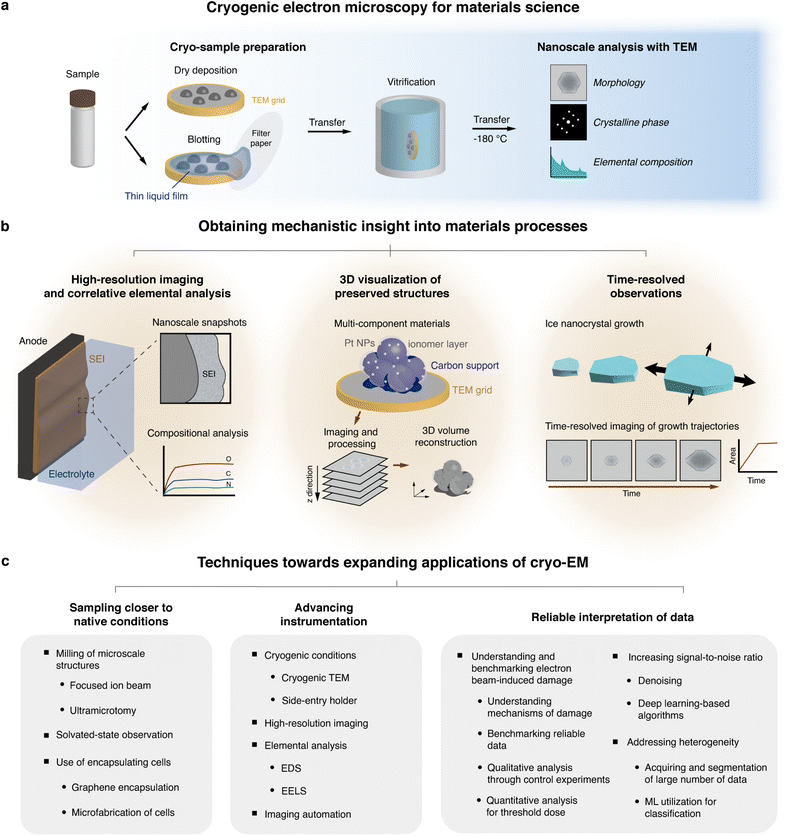
High-resolution analysis of inorganic materials has been performed with dry-state, conventional aberration-corrected TEM or STEM, which can be combined with energy-dispersive X-ray spectroscopy (EDS) and electron energy loss spectroscopy (EELS) to simultaneously retrieve elemental and chemical bonding information. Hence, various mechanisms in materials processes have been unveiled by investigating structures of functional materials at various length scales.9,48,49 However, a large portion of materials processes operate in interface with solutions,50,51 which may be altered by the ultrahigh vacuum conditions in electron microscopes when investigated using EM.52 In materials that exhibit weaker chemical bonds, the effects from the electron beam can deteriorate sample integrity.53,54 The key difference between cryo-EM and conventional EM is that cryo-EM techniques involve freezing samples and preserving their native states,18 while minimizing electron beam-induced reactions.26,47,55 This enables more accurate data acquisition regarding material structures in their operational conditions. Samples for cryo-EM analysis are normally deposited or transferred to a grid, which are then cooled by plunging into a cryogen at liquid nitrogen temperature and transferred to a side-entry cryo-transfer holder (Fig. 1a).26,56
As such, cryo-EM had broadened our understanding of material structures in near-native conditions. Currently, the field is moving towards gaining fundamental understanding of nanoscale structures and linking these insights to material mechanisms that are important in real-world applications47 (Fig. 1b). However, this task is complicated by the extensive sample preparation requirements for cryo-EM, such as thinning, which may inadvertently alter the native state of the samples. Additionally, deeper understanding on the interaction between the electron beam and low-temperature, solution phase samples, the accurate interpretation of the data,47 as well as a current lack of widespread instrumental capabilities that enable high-resolution imaging or elemental analysis are key limitations that must be improved57 (Fig. 1c).
In this Perspective, we highlight the advantages of cryo-EM for its potential in elucidating operational mechanisms of specific material systems at the nanoscale by utilizing high-resolution imaging, correlative elemental analysis, three-dimensional and time-resolved analysis to further bridge the gap between structural observations and mechanistic understanding. We additionally discuss innovations needed in cryo-sample preparation, data interpretation, and instrumentation. These enhancements aim to allow for standardized use of cryo-EM in materials science, improving the applicability of cryo-EM for unveiling unknown mechanisms in materials science.
Obtaining mechanistic insight into materials using cryo-EM imaging methods
Utilizing cryo-EM has enabled imaging intricate nanoscale structures of a variety of materials by mitigating two issues associated with TEM imaging – (1) reducing structural changes due to the imaging electron beam and (2) maintaining the sample in solution state under the high vacuum of the TEM. The first challenge is associated with the inevitable knock-on, radiolysis or thermal damage from the electron beam.58 Through multiple studies on different materials, especially organic materials59,60 or Li dendrites,26 it was found that cooling the specimen near liquid nitrogen temperature reduces structural damage during imaging. The second challenge of conventional TEM is from the vacuum environment, which limits imaging to dry samples. The advantages of cryo-EM, which involves obtaining “snapshots” of solution-based processes in frozen-state, allows for us to obtain structural insights with the use of higher electron doses for high-resolution imaging, as well as longer electron exposure times for elemental mapping of beam-sensitive or metastable regions of the sample.61,62 Cryo-EM has been applied to observing the sub-nanometer-scale structures of various beam-sensitive materials, including metal organic frameworks (MOFs),54,63,64 zeolites,65 or perovskites,66–68 which have exhibited significantly less structural change arising from electron beam-induced damage compared to using conventional TEM. These results provide opportunities for the structures of fragile materials to be characterized with TEM.Beyond the structural information provided by projection images, there has been a growing need for obtaining more comprehensive data to derive meaningful mechanistic insights. Higher resolutions down to atomic scale, multi-dimensional information such as correlative spectroscopic analysis or time-resolved information offer promising methods for obtaining more context into materials. However, these methods are also currently associated with challenges in sample preparation, data interpretation, and instrumentation. In this section, we discuss the recent advancements and the opportunities for overcoming these challenges, with the overarching goal of enhancing mechanistic understanding in materials sciences through cryo-EM.
Cryogenic high-resolution TEM (HRTEM) and correlative elemental analysis
A crucial aspect to consider in analyzing materials processes is understanding not only the structure, but also the atomic components and their spatial arrangements in a material. In this aspect, HRTEM enables the visualization of materials at atomic scales, including phases or defects that contribute to the bulk properties of a material. The incorporation of cryogenic temperatures to these high-resolution techniques is critical because the method utilizes high electron doses that damage many fragile samples. The damage mitigation effects of cryo-EM also bring opportunities for the use of energy dispersive X-ray spectroscopy (EDS) and electron energy loss spectroscopy (EELS) for elemental analysis. EDS is used to obtain information on the elemental composition of the samples, while a region of the sample can be selected to produce a map of elements present. However, EDS normally require relatively higher electron doses and repeated exposures to collect signal from sample, and these limitations could benefit from the use of cryo-EM. For systems that exhibit inelastic scattering of electrons at low energies, EELS elemental analysis can be performed for materials that exhibit energy loss at low energy loss regions, and can be a viable option that is more dose-efficient.69 Additionally, EELS vibrational spectroscopy is sensitive to the chemical states or the phases for certain materials,70 enabling their observation correlatively with the structure images obtained with TEM. We specifically highlight the opportunities for mechanistic understanding with cryo-EM by using cases from two fields – battery chemistry and quantum materials, as examples for high-resolution observations and correlative spectroscopic analysis.Analysis for battery mechanisms
A prominent field that utilizes high resolution and elemental analysis with cryo-EM is for elucidating operation mechanisms in batteries. Batteries, which work through transport of ions and electrons in an assembly of the anode, cathode, electrolyte, and separator, contain highly-reactive components that give rise to unique interfaces in the regions of contact between those compartments.71,72 Particularly, Li plating at the anode is affected by the presence of the solid electrolyte interphase (SEI), a nanoscale passivation layer formed from the reaction between surface Li and electrolytes. The SEI governs the transport of Li from the electrolyte to the Li metal surface,73–75 and also affects the resultant structures of Li dendrites, which are main causes of battery failure and safety concerns.72 Despite this significance, observation of SEI has been limited using conventional TEM because Li metals and the SEI are severely fragile toward the electron beam. The issue becomes worse as higher electron dose rates are used for high magnification imaging. In pioneering work by Li et. al., the incorporation of cryogenic temperatures has enabled Li dendrites to be observed with high-resolution TEM, revealing the lattice structure of Li and the presence of defects in areas with kinks (Fig. 2a).26 In addition, as the Li dendrites can be deposited directly from a coin cell,78,79 the structures of the SEI that has arisen from the operating conditions could be obtained. SEI interfaces, which are generally amorphous with nanoscale inorganic domains such as Li2O or Li carbonates, can be characterized with imaging (Fig. 2b) and EELS (Fig. 2c).78,80,81 With these methods, insights into the structures and compositions of the SEI could be obtained (Fig. 2d), which also influence the shapes of Li dendrites.73,76,82 SEI properties have been found to change with different types of electrolytes used (namely ether-based81,83 and carbonate-based81,84 electrolytes), at the presence of electrolyte additives,76,85 or different current densities.73,86 Specifically, the distribution of organic and inorganic domains, thickness, and layered structures of the SEI can affect its mechanical properties and the diffusion dynamics of Li within the SEI, influencing the Li plating process during deposition. This insight is relevant to enhancing the performance of Li metal batteries by improving function through SEI engineering.87–89 In one example, Gao et. al. demonstrates such SEI engineering that resulted in a high-performance Li metal battery under low temperature at high rate-charging conditions (Fig. 2e).77 Cryo-EM images of the SEI reveal a multilayer SEI that effectively seals the Li surface at low temperatures, improving its performance (Fig. 2f).77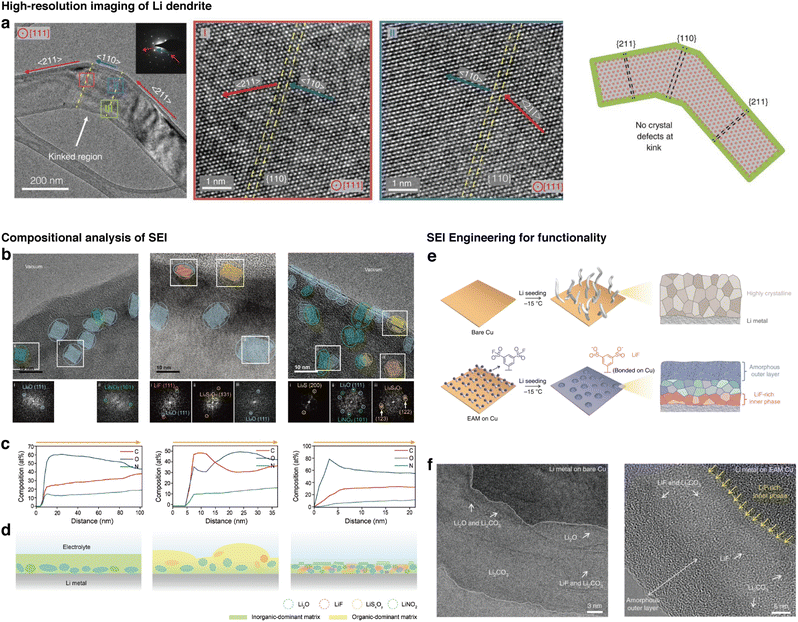 | ||
| Fig. 2 High-resolution cryo-EM and correlative elemental analysis of battery materials for understanding operation mechanisms. (a) High-resolution imaging of a Li dendrite at the region of a kink, revealing local crystallographic properties of the Li dendrite (reproduced with permission from ref. 26, copyright 2017 AAAS). (b) Images of the SEI revealing areas with local crystallinity, in which the crystallographic phases are determined using fast Fourier transform patterns. (c) Composition profile of C, O, N along the depth of the SEI. (d) Predicted morphologies and composition of the SEI based on cryo-EM analysis (reproduced from ref. 76, copyright 2024 ACS Publications). (e) Scheme for an example of SEI engineering for improving battery performance. (f) Characterization of the SEI for revealing the role of SEI on performance (reproduced from ref. 77, copyright 2020 Nature Publishing Group). | ||
As such, cryo-EM provides opportunities for evaluation of SEI structures in various cycling conditions. Beyond Li deposition in battery chemistry, cryo-EM may also be used to study calendar aging,90 a significant failure mode where an active material left in contact with the electrolyte degrades over time. Another less-understood structure to visualize with the cryo-EM is the cathode electrolyte interface, which has been shown to contribute to stable cycling in a Li-sulfurized polyacrylonitrile cathode.91
Elucidating atomic origins of properties in quantum materials
High-resolution observations are crucial for understanding the atomic-scale mechanisms behind the unique properties of quantum materials. These properties, which include superconductivity, topological states or colossal magnetoresistivity,92 are primarily governed by the electronic, magnetic, or lattice order within the materials.93 At the nanoscale, many of these states coexist, yet the complex interplay between them remains elusive. Observing these materials at cryogenic temperatures is crucial as it preserves the phases and properties that only manifest at low temperatures, and suppresses thermal vibrations of atoms. Liquid N2 and liquid He holders, developed decades ago, enable these low temperature observations within electron microscopes.92 In one of the earlier works, tetragonal and orthorhombic phases of a La2−xBaxCuO4 cuprate were investigated at temperatures as low as 20 K with a liquid He holder, using electron diffraction and dark-field imaging to reveal the formation of twin boundaries at low temperatures.94The demand for higher resolutions in cryo-EM for quantum materials is driven by the need to observe fine structural features down to the picometer-scale displacements of individual atoms that give rise to the unique properties.95 Recent advances in aberration-corrected cryo-STEM have enabled significant improvements in spatial and spectral resolution. A major challenge behind high resolution imaging is specimen drift caused by the cooling holder or the sample stage, which must be addressed as it leads to a loss in resolution. One approach demonstrated by Savitzky et. al. proposed imaging with fast frame rates and employing a post-imaging drift correction algorithm, which performs cross-correlation analysis of low signal images for accurate frame alignment.96 The picometer-scale resolution achieved with this method has particularly beneficial for observing one class of quantum material phases, known as charge-ordered phases, in which electrons and the atomic lattice form periodic patterns that break the translational symmetries of the crystal.93 These phenomena are closely linked to the material's properties that may influence superconductivity or other electronic behaviors such as colossal magnetoresistance and metal–insulator transitions.
The states of a transition metal dichalcogenide 1T′-TaTe2 (Fig. 3a–c) was imaged with high-resolution cryo-STEM to reveal the presence of Ta trimer states at 93 K, which are not present at room temperature (Fig. 3d and e). Fourier transforms were used to compare images obtained in the two temperature regimes and reveal the presence of a modulated structure in the lattice at low temperatures (Fig. 3c and d).97 In a different example, periodic lattice displacements of a charge-ordered manganite, Nd1/2Sr1/2Mn2O3 was mapped to reveal the existence of an intermediate state of electrons that contain both site and bond order and breaks inversion symmetry. The images of atoms in their intra-unit-cell arrangements enable the understanding of charge ordering through displacement pattern analysis with group theory.98
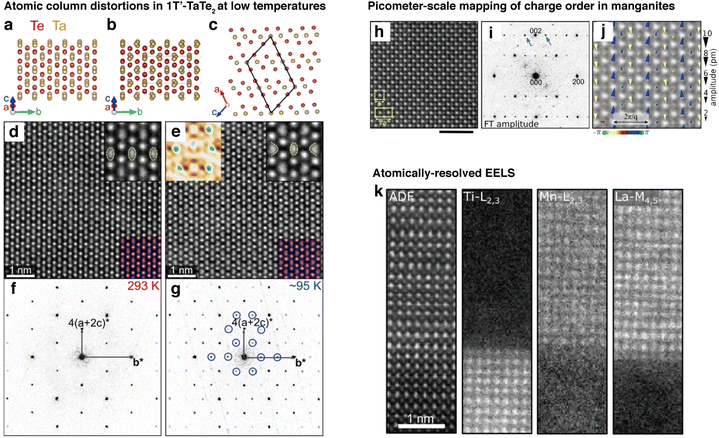 | ||
| Fig. 3 High-resolution, low temperature imaging of atomic columns of quantum materials. (a–c) Models of the structure of 1T′-TaTe2 at (a) room temperature and (b) low temperatures. (c) Lattice model indicating the monoclinic unit cell. (d and e) STEM image at (d) 293 K and (e) 95 K. The insets at the top right are magnified views of images, and the insets at the bottom right are multislice simulations of HAADF-STEM images. The inset at the top left of (e) is the intensity difference map. (f and g) Fourier transforms of the STEM images at (f) 293 K and (g) 95 K. Blue circles represent superlattice peaks. (h–j) Picometer-scale mapping of charge order of Nd1/2Sr1/2Mn2O3 with (h) HAADF-STEM image at 95 K, (i) corresponding fast Fourier transform image, and (j) map of the periodic lattice displacements. (k) Atomically-resolved EELS of La0.8Sr0.2MnO3/SrTiO3 (reproduced with permission from ref. 93, copyright 2021 ACS Publications). | ||
While structures at very high resolutions were observed for different temperatures of quantum materials so far, the quest for leveraging cryo-EM in elucidating mechanisms for a variety of quantum material systems necessitates increasing the versatility of the imaging technique. For instance, finer control of temperature of the sample would be essential for investigating the dynamics of electronic phases at multiple different temperatures.47,93,99 Moreover, correlative spectroscopic analysis such as EELS at atomic scales could provide additional insight into electronic structures beyond the images from STEM (Fig. 3k).93,100 However, a major challenge is the additional drift101 that would be introduced with these two techniques due to the heating process of side-entry holders for variable temperature experiments, and the longer frame dwell times required for EELS.102 While direct electron detectors have mitigated some of these effects with faster readout times, further advancements in drift correction will be crucial for high-resolution imaging of a wide variety of quantum materials.
The capabilities offered by cryogenic conditions in high-resolution imaging and correlative spectroscopic analysis are pivotal not only for battery chemistry and quantum materials, as discussed, but also for a broader range of systems in materials science. High-resolution cryo-EM is beneficial for imaging crystalline materials, as it reveals interfaces, defects, or structural modulations at atomic levels that dictate material properties. Correlative spectroscopic analysis further enhances this by localizing elements that are present for multicomponent systems, revealing how different components interact and influence material behavior during operation.
Visualizing structural heterogeneities in three dimensions
Heterogeneity inherent in nanoscale structures of materials plays crucial roles in determining the mechanical103 and mass transport104 properties. Local enhancements or depletions of specific properties combine to influence the overall behaviors and mechanisms. Thus, probing the 3D structures of various material components is essential for understanding bulk chemistries. For example, in the context of designing catalysts, key questions include how nanoparticles are dispersed on the substrate105 and whether they are embedded within the pores,106 because the performance of the catalyst rely on the spatial distribution of catalytically active nanoparticles within the support. However, retrieving information along the depth direction parallel to the electron beam path is challenging with two-dimensional projections of the TEM. To accurately understand these 3D properties, additional cryo-EM sample preparation techniques and selecting suitable imaging modalities for visualizing materials in three dimensions are important.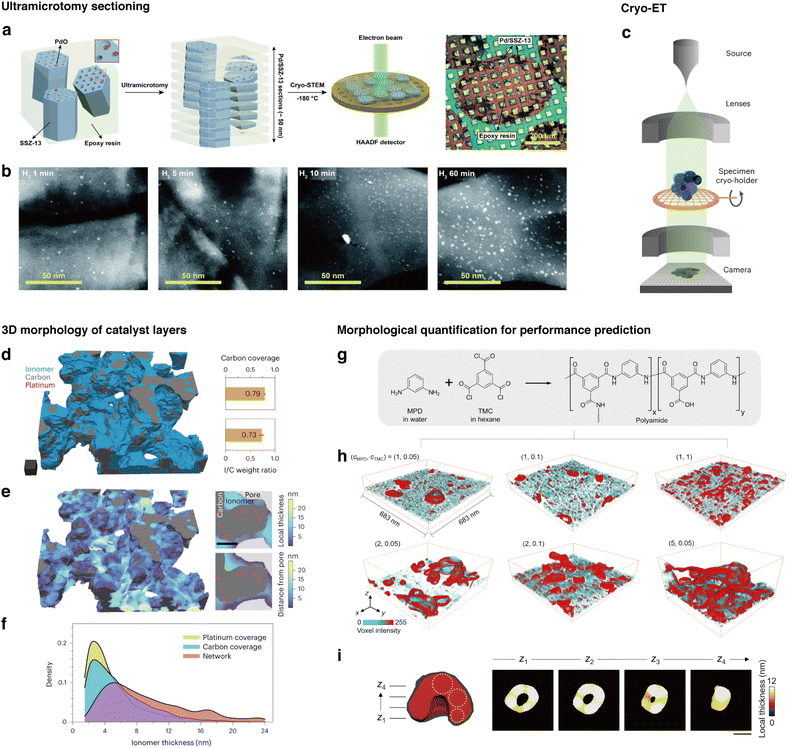 | ||
| Fig. 4 Techniques for associating 3D morphology with functional outcomes for preserved structures enabled by cryo-EM. (a) Schematic of the ultramicrotomy sectioning process of SSZ-13 to visualize PdO clusters embedded within the pores of the support. (b) Cryo-STEM images of sections according to different H2 treatment times (reproduced with permission from ref. 65, copyright 2021 Royal Society of Chemistry). (c) Schematic of a cryo-ET experiment. (d) 3D map of a catalyst obtained using cryo-ET, and analysis of 3D distribution of ionomers enabled by cryo-ET. (e) Map of local thickness of ionomers on the catalyst. (f) Distribution of the ionomer thickness plotted for different sampling and calculation methods. (Reproduced with permission from ref. 107, copyright 2023 Nature Publishing Group). (g) Schematic for production of the polyamide membranes. (h) 3D maps of polyamide membranes with different monomer concentrations obtained with cryo-ET. (i) Schematic for quantification of 3D morphologies of crumples (reproduced with permission from ref. 108, copyright 2022 AAAS). | ||
3D volumes obtained from TEM could also be used to predict performance outcomes based on shape factors. In the work by An et. al., polyamide membranes used for commercial molecular separation were imaged with cryo-ET to observe membrane crumpling morphologies with varying monomer concentrations (Fig. 4g and h).108 Furthermore, membrane microenvironments were quantitatively measured with nanoscale morphological parameters to predict methanol permeability within the membranes (Fig. 4i).108 In subsequent work by Kalutantirige et. al., it was discovered that regions where two or more crumples meet exhibited thicker membrane walls, suggesting that crumples form through a coalescence mechanism where crumple walls connect and merge.113 These observations were correlated with coarse-grained molecular dynamics (MD) simulations to verify the proposed membrane formation mechanisms. Furthermore, by using graph theory as a basis for quantifying interconnected crumples, the study proposes that structural rigidity trends monotonically with the mechanical robustness of the membranes, providing insights into the nano-structural architecture that influences performance at the bulk level.113
Determining the 3D morphology of materials provides another dimension for quantitative analysis for obtaining insights into structures that affect macroscopic properties. Techniques that acquire information in the depth direction can be adapted for imaging the interior of microscale particles, particularly those with complex internal architectures such as pores, or those composed of multiple components. Furthermore, 3D imaging can enhance the analysis of amorphous or anisotropically shaped materials by revealing the three-dimensional shapes that affect their bulk properties. By conducting correlative analysis of material performance and strategically integrating quantification parameters for analyzing nanoscale morphologies, the gap between structural data and mechanistic insights can be bridged.
Time-resolved observations of materials processes
Along with the nanoscale high spatial resolution cryo-EM offers, obtaining data along the temporal dimension is also crucial for developing a mechanistic understanding of how material structures form and evolve over time. Tracking changes over time enables understanding of nucleation mechanisms, important structural intermediates, and how final products are ultimately shaped. Time-resolved studies also provide insight into the kinetics and stability of phases at the nanometer scale, achieving deeper understanding of the dynamic processes that ultimately govern the behavior and functionality of complex materials.The intermediate stages of a self-assembly process occurring in solution can be probed using cryo-EM by producing aliquots of samples at different stages of assembly. For producing cryogenic samples, a few microliters of solution are extracted from each aliquot and deposited onto a TEM grid. The grid is then blotted with filter paper to produce a thin layer of liquid, and plunge-frozen in liquid ethane to produce a layer of frozen solution containing the structures to be observed. Sequentially imaging different time frames of the assembly process reveals the nanoscale structures in various intermediate stages. Owing to the versatility of the technique, formation mechanisms of a variety of materials systems have been revealed to obtain insights into synthetic mechanisms. In one example, the oriented attachment mechanism during self-assembly was revealed by cryo-EM imaging of cyclosporin A, which shows the intermediate structures of small crystals undergoing attachment to form a large mesocrystal (Fig. 5a–e).115 In another example by Tsarfati et. al., perylene diimide crystallization was observed, where densified spheres formed initially as shown in Fig. 5f and g. These spheres undergo an intermediate phase exhibiting faceted intermediates in Fig. 5h and i, and evolve into crystalline fibrillar structures in Fig. 5j and k. The scheme of the crystallization process in Fig. 5l suggests a densification process followed by crystallization that occurs within the dense phases, ultimately forming ordered fibrillar structures.117 Other examples that use time-resolved cryo-EM analysis include early-stage biomimetic mineralization,118–121 formation of MOFs,122 hierarchical structural evolution,123–125 oriented attachment processes,126 chiral self-assembly,127 and amorphous-to-ordered-phase transitions18 by enabling time-resolved imaging for processes at time scales of minutes, hours, or even days.
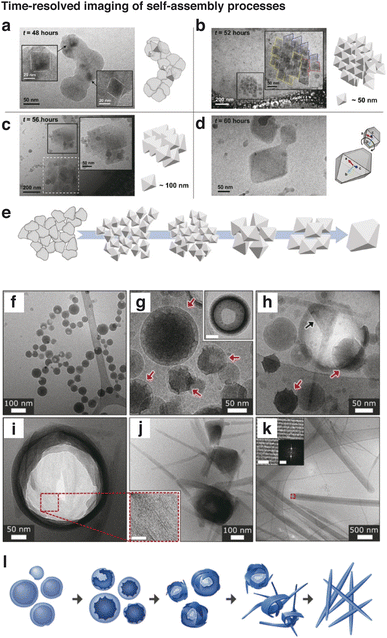 | ||
| Fig. 5 Time-resolved imaging of self-assembly processes. (a–d) Cryo-EM images of structures observed during self-assembly of cyclosporin A nanoparticles after (a) 48 h, (b) 52 h, (c) 56 h, and (d) 60 h after initiation (reproduced with permission from ref. 115, copyright 2022 ACS Publications). (e) Schematic of the proposed crystallization pathway. (f–k) Crystallization of perylene diimides (f) observed immediately, (g) observed after 30–50 min, (h) exhibiting faceted intermediate showing crystallinity, (i) with ruptured aggregate, (j) showing growth of fibrous crystals, and (k) fully developed. (l) Schematic of the proposed crystallization pathway (reproduced with permission from ref. 117, copyright 2018 ACS Publications). | ||
In many self-assembly systems, however, cryo-EM observation of these intermediate structures faces challenges associated with the coexistence of multiple mechanisms at the nanoscale, obscuring the dominant mechanistic picture.128 A wide range of heterogeneity of morphologies formed at the nanoscale complicates the interpretation of the assembly process. In addition, blotting of samples for thinning the solution can lead to damage of structures of embedded materials,129 especially for larger, microscale assemblies. These limitations hinder the reliable observation of macroscale structures. To overcome these challenges, correlation of structures obtained from imaging with microscale analysis, such as with diffraction or scattering techniques are essential.130 Particularly for larger structures, it is important to perform additional imaging analysis with larger-scale techniques, such as scanning electron microscopy (SEM)131 or optical microscopy132 to ensure that samples are minimally affected by the sample preparation process. The blotting process of samples is difficult to avoid during conventional cryo-EM sampling procedures, but developments in sampling procedures that do not require blotting could improve the structural stability of these assemblies.
Cryo-EM is therefore a valuable tool for bridging this resolution gap by allowing direct imaging at the nanoscale. Currently, ice nanocrystal growth has been visualized at cryogenic conditions by deposition of residual water vapor onto a graphene grid maintained at liquid nitrogen temperature, which induces a vapor-to-solid transition for observation. This approach has enabled molecular-scale visualization of ice growth processes on 2D materials substrate in situ (Fig. 6a and b).135 In another study, amorphous ice was prepared by vitrifying a thin liquid film in liquid ethane, and ice crystallization was induced by heating amorphous ice to 143 K. This resulted in an amorphous-to-crystalline transition in a thin amorphous ice film (Fig. 6c). Snapshots of different regions of the ice film at different time periods were obtained, as shown in Fig. 6d, in order to mitigate electron beam damage. This study also tracks the growth of individual ice nanocrystals and correlates the results with MD simulations, revealing the growth anisotropy of heterocrystalline ice (Fig. 6e and f).136 Specialized in situ cryo-EM techniques, such as the incorporation of laser-induced heating have also been used to observe various forms of H2O, such as detecting the properties of deeply supercooled water before crystallization.137,138
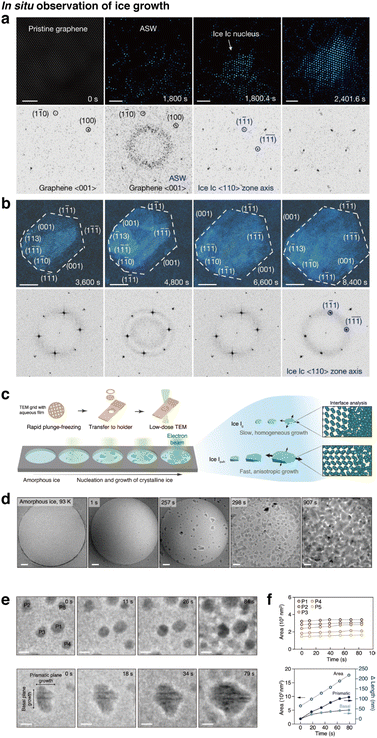 | ||
| Fig. 6 Direct observation of in situ ice growth via individual particle tracking. (a and b) High-resolution TEM images and the corresponding FFTs for (a) nucleation of cubic ice from residual water vapor within the TEM column on a graphene surface and (b) a cubic ice nanocrystal with defined facets. (Reproduced with permission from ref. 135, copyright 2023 Nature Publishing Group). (c) Schematic for the observation of ice crystallization in amorphous ice films. (d) Snapshots of ice crystallization of a free-standing amorphous ice film at different annealing time periods at different locations. (e) Individual particle tracking of growth over time and (f) plots of particle growth over time (reproduced with permission from ref. 136, copyright 2023 Nature Publishing Group). | ||
Nano-to-molecular-scale observation of ice within the TEM is important in applications for ice growth optimization in various low-temperature systems such as cryopreservation materials. Whereas the energetics and the formation mechanisms of cubic and hexagonal ice remain poorly understood,133,134,139,140 cryo-EM is emerging as a valuable tool for being able to track the structures and growth of ice crystals. This provides crucial insights into ice physics, where nanoscale ice crystals shape the formation of larger, microscale ice structures. Furthermore, studying the interaction between ice crystals and nanoscale materials using cryo-EM, particularly within biological macromolecules or cell membranes, is highly valuable. The formation of crystalline ice within cells that have been treated with different antifreezing agents have been achieved using cryo-EM, offering significant insights into antifreezing processes.141–143
Time-resolved analysis enables the direct monitoring of the structural changes over time to reveal process mechanisms or kinetics. Observing aliquots of samples undergoing operation can be widely used because of its applicability to many different materials systems. However, these observations do not capture the evolution of individual structural units over time. Additionally, the heterogeneity within a sample, such as particles being at different stages of evolution within the same aliquot, complicates data interpretation. Tracking of in situ processes of individual units has been demonstrated, for instance, with the crystallization of ice, where the process temperature corresponds to temperatures compatible with cryo-EM. However, many materials processes do not undergo dynamics at these very low temperatures. One proposed technique for observing transient dynamics within the TEM is by melting the ice to allow configurations of molecules to change before the sample is revitrified again.144 A technique for melting ice in the TEM using a laser beam has been demonstrated for viruses to observe their microsecond-scale structural dynamics.145 Another method could involve freezing and thawing samples by increasing temperature, while maintaining the liquid and the sample encapsulated to prevent drying.146 The development of new in situ techniques and directly tracking individual structural units over time will be beneficial for achieving mechanistic studies.
Technical developments for expanding applications of cryo-EM
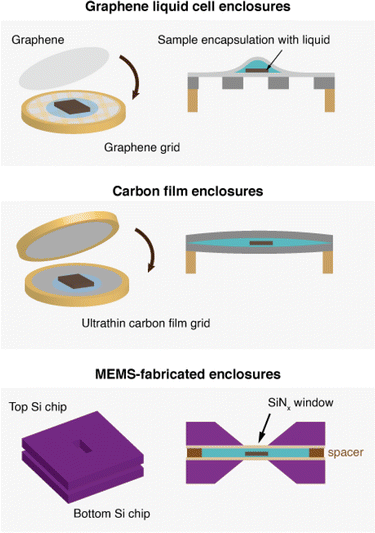
Interpreting the effect of nanoscale structures on macroscopic properties have long been of interest for many materials science disciplines, which provide fundamental insight into how materials should be designed for tailored functionalities.30 The implementation of cryo-EM has brought promising developments for the observation of materials with reduced structural changes that may have occurred from drying or electron beam effects. On another note, as demonstrated by An et. al., nanoscale structural morphologies, even for highly amorphous shapes, can be quantified for correlation with their expected performances.108 Cryo-EM has vast opportunities in materials science for obtaining images of nanoscale structures for this purpose, especially in their metastable states. However, cryo-EM use in materials science poses challenges due to the ultrathin sampling requirements, the small visualization area of the TEM, the static nature of cryogenic imaging, and beam-induced effects. Fortunately, advancements in instrumentation and pipelines for image processing for interpretation has been significantly developed for purposes in reconstructing 3D volumes of proteins for structural biology.31 Such techniques can be modified and implemented to adapt to the benchmarking of cryo-EM analysis for materials science as well. However, there are other developments that are required specifically for materials science that we will also discuss in this section. These developments will allow the acquisition of image data under conditions that closely mirror the native environment, determining structure–function relationships for materials. Specifically, carefully designed model systems during sample preparation, as well as consideration in data interpretation and acquisition are necessary to realize data collection representative of realistic conditions using cryo-EM.Sample preparation
Optimizing instrumentation for materials science
For cryo-EM in structural biology, a revolutionary advancement that enabled the reconstruction of sub-nanometer-resolution 3D structures of proteins is the implementation of instruments dedicated to imaging biological materials. Examples include the direct electron detector,163 the use of 300 kV acceleration voltage,164 and the implementation of energy filters165 and phase plates166 to increase SNR and contrast for proteins embedded in vitreous ice. Additionally, developments in algorithms for dose-weighting167 for extracting reliable data and pipelines for high-throughput data acquisition have made cryo-EM a standardized and widely-used technique in structural biology. The instrumentation available for imaging biological systems may be utilized for imaging materials with cryo-EM. Incorporating cold field emission guns for high brightness, and direct electron detectors for increased SNR is beneficial for imaging beam-sensitive materials, retrieving maximum information from small electron doses. However, many cryo-TEM setups for structural biology are not optimized to meet the specific requirements of materials scientists.57 One example is the need for spherical aberration (Cs) correction48 which is crucial for high-resolution imaging for nanoscale materials—a feature typically not necessary for biological applications and thus often lacking in many cryo-TEM setups. Spherical aberration arises from inherent limitations in electron lenses, resulting in a significant reduction in point resolution. In images captured with an uncorrected microscope, spherical aberration causes a reversal in contrast beyond the resolution limit,168 making accurate interpretation of the images challenging. For the resolution range used for images in high-resolution EM, Cs correction drastically improves the achievable resolution of the images.169 Unfortunately, incorporating Cs correction into an existing uncorrected cryo-EM setup is challenging as it requires substantial modifications to the electron column as well as specific operation protocols. To expand the use of cryo-EM in materials sciences, it would be highly beneficial to develop more cryo-EM setups specifically tailored for materials research. These setups could include conventional Cs-corrected microscopes with compatible cryo-transfer holders, or dedicated cryo-EMs incorporated with Cs correction. Additionally, as discussed with examples from mechanistic studies in battery chemistry and quantum materials, electron diffraction, STEM imaging and correlative elemental analysis techniques such as EDS or EELS are necessary in materials science but are not commonly used in structural biology. STEM, EDS, and EELS require installation of dedicated detectors. In addition to adding these detectors, techniques such as EELS can benefit from incorporating a monochromator to produce highly coherent electron beams to achieve higher spectral resolution.170 Depending on the spectral resolution needed for imaging fine electronic structures with EELS, a monochromator for the electron beam may be useful.TEM observation at cryogenic temperatures can be performed in two ways: by utilizing a dedicated cryogenic TEM equipped with a cryo-stage and an autoloader, or by using a cryogenic holder on a conventional TEM. A dedicated cryo-TEM is less versatile in terms of instrumentation but provides higher imaging stability, reducing drift or vibration from the external environment. For materials science purposes, using a cryogenic TEM holder may be a compelling option in many cases because it is versatile, economical, and more suitable for samples that undergo changes in ambient conditions. Additionally, commercial cryogenic holders have dual-axis tilt, which is crucial for aligning the desired crystallographic orientations in HRTEM. However, cryogenic holders suffer from more drift during imaging compared to a dedicated cryogenic stage, causing blurred images due to the boiling of the liquid nitrogen or vibrations from the external environment.93,167 In addition, contaminants may be deposited during sample transfer within the cryogenic stations,171 and automated imaging is more difficult to perform with the side-entry holder setup.
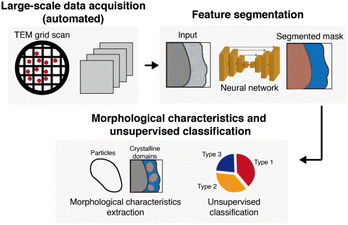
Software and hardware development for automated imaging for materials science may also be beneficial, especially for large-scale imaging to address heterogeneity.172 This process involves acquiring images at low magnifications with minimal dose, and then designating locations to automatically image at high resolution, so the electron dose that the sample is exposed to is reduced. Currently, software is optimized to locate holes of holey support grids for data acquisition. Optimizing software to locate areas for imaging in materials systems using feature or contrast analysis could be implemented to achieve automated imaging for nanoscale materials.
In terms of sample preparation, setups that provide air-tight environments are beneficial for imaging air-sensitive materials such as Li. Sampling procedures within an Ar environment in a glove box and methods to transport samples from one area to the next have been developed to minimize the exposure of sensitive samples to air. These are crucial developments to maintain the integrity of the samples being imaged under cryo-EM.163
Interpretation of data for obtaining mechanistic insights
A crucial aspect for utilizing cryo-EM for making conclusions in terms of mechanisms is to provide a reliable interpretation of data, mainly images, obtained from samples of interest.47 A few necessary considerations before drawing conclusions from TEM data include understanding electron beam-induced effects, increasing signal-to-noise ratio (SNR), and implementing techniques to address data heterogeneity.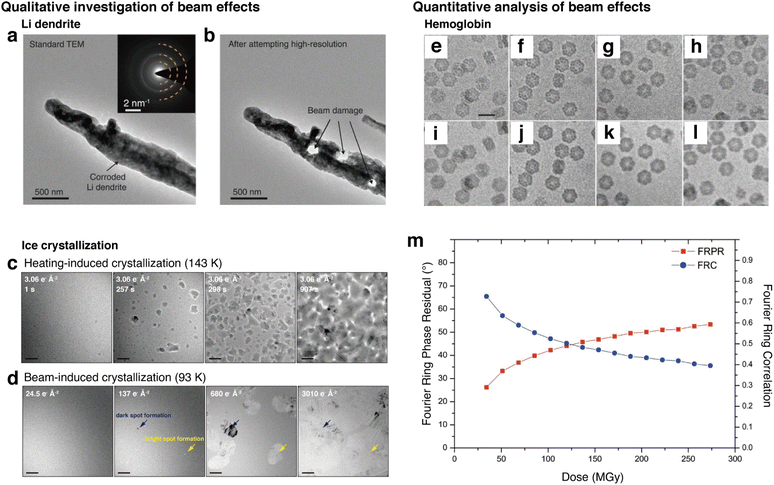 | ||
| Fig. 8 Investigation and benchmarking of electron-beam effects on different samples. (a) Li dendrite image before high-resolution imaging attempt using conventional, room temperature TEM. (b) Li dendrite after beam damage caused by high electron doses after attempting high-resolution (reproduced with permission from ref. 26, copyright 2017 AAAS). (c) Example of heating-induced crystallization of ice, which is the intended process to be observed. (d) Example of beam-induced crystallization of ice from continuous irradiation, which produces artifacts that are different from the crystallization process observed by heating (reproduced with permission from ref. 136, copyright 2023 Nature Publishing Group). (e–l) Aligned and summed images of the protein hemoglobin embedded in vitreous ice, in which (a and e) are low-flux, (b and f) are medium-flux, (c and g) are high-flux, and (d and h) are high-flux short-exposure. (a–d) are 50 e Å−2 and (e–h) are 250 e Å−2. (m) Plots of FRPR and FRC according to different dose rates calculated in MGy, providing information on the changes in low-resolution information according to accumulated dose. The threshold value of 45 deg for FRPR and 0.5 for FRC can be used to benchmark imaging dose rates (reproduced with permission from ref. 178, copyright 2011 IUCr Journals). | ||
One way to benchmark electron beam effects quantitatively is to utilize the intensity of crystalline peaks in the Fast Fourier Transform (FFT) of the images.179 By obtaining sequential images of a material while undergoing electron beam-induced changes, one can gain insight into the effects that the beam poses on the crystallinity of the sample. Additionally, parameters such as the Fourier ring correlation (FRC), which indicates the highest reliable resolution of an image,178 can be quantified as a function of cumulative dose. In an experiment for imaging the protein hemoglobin embedded in vitreous ice, the effects of cumulative dose and dose rates on the protein images have been shown (Fig. 8e–l). This data was quantified using the Fourier Ring Phase Residual (FRPR) and the FRC of images with different cumulative doses (Fig. 8m).180 These parameters can be used to set a beam dose threshold with respect to the preservation of low-resolution structures. A value of 45° for the FRPR, and 0.5 for the FRC was used to benchmark the threshold of dose and to determine which images obtained are reliable for use. These dose-based image quantification methods have been utilized in structural biology to fully utilize information from images, giving more weight for reliability to the images that have had less exposure to the beam.181
Because of the heterogeneous properties of nanoscale materials, imaging materials within their operational environment is particularly beneficial. 3D imaging with strategic sample preparation procedures, as demonstrated by An et. al. and Kalutantirige et. al., have revealed how interactions with surrounding environments affect the structures and mechanisms within materials.108,113 Tracking individual particles, as performed in ice crystallization observations,135,136 is also beneficial for interpretation of data, as it directly visualizes structural effects within observed processes.
Conclusion and future outlook
In this Perspective, we have outlined the uses of cryo-EM for materials science, particularly in its role in revealing nanoscale mechanisms of complicated processes. In addition to the two-dimensional imaging of projections using the cryo-TEM and STEM modes, high-resolution imaging, correlative elemental analysis, three-dimensional imaging and time-resolved analysis have contributed vastly to providing more context behind mechanistic understanding of processes. Moreover, cryo-EM enables the observation of materials processes under conditions that reflect the native states, and with its use in conjunction with other correlative analytical techniques, it can reveal relationships between structural observations and mechanistic understanding. Currently, however, cryo-EM in use for elucidating mechanisms still require optimization in benchmarking such as quantification of beam-damage, or new approaches for producing thin samples. These challenges require additional improvements in cryo-sample preparation, instrumentation, and data interpretation, as we have outlined in this Perspective. These enhancements will enable the novel studies of materials processes, while standardizing the use of cryo-EM as a tool for general use in materials science.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
M. L., Y. J., S. K., I. J., S. K., S.-H. J. and J. P. conceived and collaboratively drafted the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean government (RS-2024-00408823, RS-2023-00283902, and RS-2024-00421181). The authors acknowledge financial support from the Samsung Science and Technology Foundation (SSTF-BA-2302-06). The authors also acknowledge the financial support from the Institute for Basic Science (IBS-R006-D1).Notes and references
- M. A. Pfeifer, G. J. Williams, I. A. Vartanyants, R. Harder and I. K. Robinson, Nature, 2006, 442, 63–66 CrossRef CAS
.
- M. Azubel, J. Koivisto, S. Malola, D. Bushnell, G. L. Hura, A. L. Koh, H. Tsunoyama, T. Tsukuda, M. Pettersson, H. Häkkinen and R. D. Kornberg, Science, 2014, 345, 909–912 CrossRef PubMed
.
- B. H. Kim, J. Heo, S. Kim, C. F. Reboul, H. Chun, D. Kang, H. Bae, H. Hyun, J. Lim, H. Lee, B. Han, T. Hyeon, A. P. Alivisatos, P. Ercius, H. Elmlund and J. Park, Science, 2020, 368, 60–67 CrossRef
.
- S. Jeon, T. Heo, S.-Y. Hwang, J. Ciston, K. C. Bustillo, B. W. Reed, J. Ham, S. Kang, S. Kim, J. Lim, K. Lim, J. S. Kim, M.-H. Kang, R. S. Bloom, S. Hong, K. Kim, A. Zettl, W. Y. Kim, P. Ercius, J. Park and W. C. Lee, Science, 2021, 371, 498–503 CrossRef PubMed
.
- H. Wietfeldt, R. Meana-Pañeda, C. Machello, C. F. Reboul, C. T. S. Van, S. Kim, J. Heo, B. H. Kim, S. Kang, P. Ercius, J. Park and H. Elmlund, Commun. Chem., 2024, 7, 4 CrossRef
.
- M. A. Boles, D. Ling, T. Hyeon and D. V. Talapin, Nat. Mater., 2016, 15, 141–153 CrossRef PubMed
.
- A. H. Ip, S. M. Thon, S. Hoogland, O. Voznyy, D. Zhitomirsky, R. Debnath, L. Levina, L. R. Rollny, G. H. Carey, A. Fischer, K. W. Kemp, I. J. Kramer, Z. Ning, A. J. Labelle, K. W. Chou, A. Amassian and E. H. Sargent, Nat. Nanotechnol., 2012, 7, 577–582 CrossRef PubMed
.
- N. Kholmicheva, P. Moroz, H. Eckard, G. Jensen and M. Zamkov, ACS Energy Lett., 2017, 2, 154–160 CrossRef
.
- H. Park, H. Park, K. Song, S. H. Song, S. Kang, K. H. Ko, D. Eum, Y. Jeon, J. Kim, W. M. Seong, H. Kim, J. Park and K. Kang, Nat. Chem., 2022, 14, 614–622 CrossRef PubMed
.
- D. Eum, S. O. Park, H. Y. Jang, Y. Jeon, J. H. Song, S. Han, K. Kim and K. Kang, Nat. Mater., 2024, 23, 1093–1099 CrossRef PubMed
.
- M. Tang, H. Yan, X. Zhang, Z. Zheng and S. Chen, Adv. Mater., 2023, 2306387 CrossRef PubMed
.
- R. Sun, Z. Xia, X. Xu, R. Deng, S. Wang and G. Sun, Nano Energy, 2020, 75, 104919 CrossRef
.
- F. C. Cetinbas, R. K. Ahluwalia, N. N. Kariuki and D. J. Myers, J. Electrochem. Soc., 2018, 165, F1051–F1058 CrossRef CAS
.
- S. Kim, J. Kwag, C. Machello, S. Kang, J. Heo, C. F. Reboul, D. Kang, S. Kang, S. Shim, S. J. Park, B. H. Kim, T. Hyeon, P. Ercius, H. Elmlund and J. Park, Nano Lett., 2021, 21, 1175–1183 CrossRef CAS PubMed
.
- J. Kim, A. Park, J. Kim, S. J. Kwak, J. Y. Lee, D. Lee, S. Kim, B. K. Choi, S. Kim, J. Kwag, Y. Kim, S. Jeon, W. C. Lee, T. Hyeon, C. H. Lee, W. B. Lee and J. Park, Adv. Mater., 2022, 34, 2206066 CrossRef CAS
.
- M. H. Kang, J. Park, S. Kang, S. Jeon, M. Lee, J. Y. Shim, J. Lee, T. J. Jeon, M. K. Ahn, S. M. Lee, O. Kwon, B. H. Kim, J. R. Meyerson, M. J. Lee, K. Il Lim, S. H. Roh, W. C. Lee and J. Park, Adv. Mater., 2021, 33, 2102991 CrossRef CAS
.
- M. C. Weidman, D. M. Smilgies and W. A. Tisdale, Nat. Mater., 2016, 15, 775–781 CrossRef CAS
.
- L. Houben, H. Weissman, S. G. Wolf and B. Rybtchinski, Nature, 2020, 579, 540–543 CrossRef CAS PubMed
.
- A. McPherson, Methods Mol. Biol., 2017, 1607, 17–50 CrossRef CAS PubMed
.
- J. S. Du, Y. Bae and J. J. De Yoreo, Nat. Rev. Mater., 2024, 9, 229–248 CrossRef CAS
.
- E. B. Moore and V. Molinero, Phys. Chem. Chem. Phys., 2011, 13, 20008–20016 RSC
.
- R. S. Smith, C. Huang, E. K. L. Wong and B. D. Kay, Surf. Sci. Lett., 1996, 367, L13–L18 CrossRef
.
- H. Baek, S. Kang, J. Heo, S. Choi, R. Kim, K. Kim, N. Ahn, Y. G. Yoon, T. Lee, J. B. Chang, K. S. Lee, Y. G. Park and J. Park, Nat. Commun., 2024, 15, 1671 CrossRef
.
- G. Zhu, M. L. Sushko, J. S. Loring, B. A. Legg, M. Song, J. A. Soltis, X. Huang, K. M. Rosso and J. J. De Yoreo, Nature, 2021, 590, 416–422 CrossRef PubMed
.
- D. Jacobsson, F. Panciera, J. Tersoff, M. C. Reuter, S. Lehmann, S. Hofmann, K. A. Dick and F. M. Ross, Nature, 2016, 531, 317–322 CrossRef PubMed
.
- Y. Li, Y. Li, A. Pei, K. Yan, Y. Sun, C.-L. Wu, L.-M. Joubert, R. Chin, A. L. Koh, Y. Yu, J. Perrino, B. Butz, S. Chu and Y. Cui, Science, 2017, 358, 506–510 CrossRef PubMed
.
- E. Trevisanello, R. Ruess, G. Conforto, F. H. Richter and J. Janek, Adv. Energy Mater., 2021, 11, 2003400 CrossRef
.
- C. Kirsch, C. Dreßler and D. Sebastiani, J. Phys. Chem. C, 2022, 126, 12136–12149 CrossRef
.
- X. Wang, J. Li and Q. Chen, Matter, 2023, 6, 2488–2612 CrossRef
.
- D. Li, Q. Chen, J. Chun, K. Fichthorn, J. De Yoreo and H. Zheng, Chem. Rev., 2023, 123, 3127–3159 CrossRef PubMed
.
- Y. Cheng, Cell, 2015, 161, 450–457 CrossRef PubMed
.
- M. C. Darrow, J. P. Moore, R. J. Walker, K. Doering and R. S. King, Microsc. Microanal., 2019, 25, 994–995 CrossRef
.
- G. Weissenberger, R. J. M. Henderikx and P. J. Peters, Nat. Methods, 2021, 18, 463–471 CrossRef
.
- V. P. Dandey, W. C. Budell, H. Wei, D. Bobe, K. Maruthi, M. Kopylov, E. T. Eng, P. A. Kahn, J. E. Hinshaw, N. Kundu, C. M. Nimigean, C. Fan, N. Sukomon, S. A. Darst, R. M. Saecker, J. Chen, B. Malone, C. S. Potter and B. Carragher, Nat. Methods, 2020, 17, 897–900 CrossRef
.
- S. Wu, J. P. Armache and Y. Cheng, Microscopy, 2016, 65, 35–41 CrossRef
.
- X. Li, P. Mooney, S. Zheng, C. R. Booth, M. B. Braunfeld, S. Gubbens, D. A. Agard and Y. Cheng, Nat. Methods, 2013, 10, 584–590 CrossRef
.
- A. Punjani, J. L. Rubinstein, D. J. Fleet and M. A. Brubaker, Nat. Methods, 2017, 14, 290–296 CrossRef PubMed
.
- E. D. Zhong, T. Bepler, B. Berger and J. H. Davis, Nat. Methods, 2021, 18, 176–185 CrossRef PubMed
.
- J. L. Vilas, J. M. Carazo and C. O. S. Sorzano, Chem. Rev., 2022, 122, 13915–13951 CrossRef PubMed
.
- S. Q. Zheng, E. Palovcak, J. P. Armache, K. A. Verba, Y. Cheng and D. A. Agard, Nat. Methods, 2017, 14, 331–332 CrossRef PubMed
.
- A. L. Parry, P. H. H. Bomans, S. J. Holder, N. A. J. M. Sommerdijk and S. C. G. Biagini, Angew. Chem., Int. Ed., 2008, 47, 8859–8862 CrossRef PubMed
.
- K. Mortensenf and Y. Talmon, Macromolecules, 1995, 28, 8829–8834 CrossRef
.
- Y. He, Z. Li, P. Simone and T. P. Lodge, J. Am. Chem. Soc., 2006, 128, 2745–2750 CrossRef PubMed
.
- R. Zana and Y. Talmon, Nature, 1993, 362, 228–230 CrossRef
.
- V. A. Davis, A. N. G. Parra-Vasquez, M. J. Green, P. K. Rai, N. Behabtu, V. Prieto, R. D. Booker, J. Schmidt, E. Kesselman, W. Zhou, H. Fan, W. W. Adams, R. H. Hauge, J. E. Fischer, Y. Cohen, Y. Talmon, R. E. Smalley and M. Pasquali, Nat. Nanotechnol., 2009, 4, 830–834 CrossRef PubMed
.
- X. C. Ren, X. Q. Zhang, R. Xu, J. Q. Huang and Q. Zhang, Adv. Mater., 2020, 32, 1908293 CrossRef
.
- Y. Li, W. Huang, Y. Li, W. Chiu and Y. Cui, ACS Nano, 2020, 14, 9263–9276 CrossRef
.
- S. Kang, J. Cha, Y. S. Jo, Y. J. Lee, H. Sohn, Y. Kim, C. K. Song, Y. Kim, D. H. Lim, J. Park and C. W. Yoon, Adv. Mater., 2023, 35, 2203364 CrossRef
.
- Y. Kim, S. Kang, D. Kang, K. R. Lee, C. K. Song, J. Sung, J. S. Kim, H. Lee, J. Park and J. Yi, Angew. Chem., Int. Ed., 2021, 60, 25411–25418 CrossRef PubMed
.
- M. V. Kovalenko, L. Manna, A. Cabot, Z. Hens, D. V. Talapin, C. R. Kagan, V. I. Klimov, A. L. Rogach, P. Reiss, D. J. Milliron, P. Guyot-Sionnnest, G. Konstantatos, W. J. Parak, T. Hyeon, B. A. Korgel, C. B. Murray and W. Heiss, ACS Nano, 2015, 9, 1012–1057 CrossRef PubMed
.
- J. Kim, S. Kang, F. Cheng, Y. Wang, X. Ye and J. Park, MRS Bull., 2024, 49, 365–376 CrossRef
.
- H. Wu, H. Friedrich, J. P. Patterson, N. A. J. M. Sommerdijk and N. de Jonge, Adv. Mater., 2020, 32, 2001582 CrossRef PubMed
.
- R. F. Egerton, P. Li and M. Malac, Micron, 2004, 35, 399–409 CrossRef PubMed
.
- L. Liu, D. Zhang, Y. Zhu and Y. Han, Commun. Chem., 2020, 3, 99 CrossRef CAS
.
- Y. Li, W. Huang, Y. Li, A. Pei, D. T. Boyle and Y. Cui, Joule, 2018, 2, 2167–2177 CrossRef
.
- X. Wang, Y. Li and Y. S. Meng, Joule, 2018, 2, 2225–2234 CrossRef
.
- R. E. A. Williams, D. W. Mccomb and S. Subramaniam, MRS Bull., 2019, 44, 929–934 CrossRef
.
- R. F. Egerton, Micron, 2019, 119, 72–87 CrossRef PubMed
.
- G. T. Oostergetel, F. J. Esselink and G. Hadziioannou, Langmuir, 1995, 11, 3721–3724 CrossRef
.
- M. J. M. Wirix, P. H. H. Bomans, H. Friedrich, N. A. J. M. Sommerdijk and G. De With, Nano Lett., 2014, 14, 2033–2038 CrossRef PubMed
.
- K. Steinberg, X. Yuan, C. K. Klein, N. Lazouski, M. Mecklenburg, K. Manthiram and Y. Li, Nat. Energy, 2023, 8, 138–148 CrossRef
.
- E. Zhang, M. Mecklenburg, X. Yuan, C. Wang, B. Liu and Y. Li, iScience, 2022, 22, 105689 CrossRef
.
- F. Banihashemi, G. Bu, A. Thaker, D. Williams, J. Y. S. Lin and B. L. Nannenga, Ultramicroscopy, 2020, 216, 113048 CrossRef PubMed
.
- Y. Li, K. Wang, W. Zhou, Y. Li, R. Vila, W. Huang, H. Wang, G. Chen, G. H. Wu, Y. Tsao, H. Wang, R. Sinclair, W. Chiu and Y. Cui, Matter, 2019, 1, 428–438 CrossRef PubMed
.
- Y. Kim, J. Sung, S. Kang, J. Lee, M. H. Kang, S. Hwang, H. Park, J. Kim, Y. Kim, E. Lee, G. S. Park, D. H. Kim and J. Park, J. Mater. Chem. A, 2021, 9, 19796–19806 RSC
.
- E. J. Vandenbussche, C. P. Clark, R. J. Holmes and D. J. Flannigan, ACS Omega, 2020, 5, 31867–31871 Search PubMed
.
- L. Bao, P. Gao, T. Song, F. Xu, Z. Li and G. Xu, Crystals, 2023, 13, 973 CrossRef CAS
.
- B. Yuan and Y. Yu, Chem, 2022, 8, 327–339 CAS
.
- N. Dumaresq, N. Brodusch, S. Bessette and R. Gauvin, Ultramicroscopy, 2024, 262, 113977 CrossRef CAS
.
- H. Tan, J. Verbeeck, A. Abakumov and G. Van Tendeloo, Ultramicroscopy, 2012, 116, 24–33 CrossRef CAS
.
- Z. Luo, X. Qiu, C. Liu, S. Li, C. Wang, G. Zou, H. Hou and X. Ji, Nano Energy, 2021, 79, 105507 CrossRef
.
- X. B. Cheng, R. Zhang, C. Z. Zhao and Q. Zhang, Chem. Rev., 2017, 117, 10403–10473 CrossRef PubMed
.
- X. Yuan, B. Liu, M. Mecklenburg and Y. Li, Nature, 2023, 620, 86–91 CrossRef PubMed
.
- W. Liu, P. Liu and D. Mitlin, Adv. Energy Mater., 2020, 10, 2002297 CrossRef
.
- E. Peled and S. Menkin, J. Electrochem. Soc., 2017, 164, A1703–A1719 CrossRef
.
- H. Park, Y. Jeon, M. Park, I. Jung, J. Shin, Y. Kim, W. K. Kim, K. H. Ryu, W. B. Lee and J. Park, ACS Nano, 2024, 18, 12885–12896 CrossRef PubMed
.
- Y. Gao, T. Rojas, K. Wang, S. Liu, D. Wang, T. Chen, H. Wang, A. T. Ngo and D. Wang, Nat. Energy, 2020, 5, 534–542 CrossRef
.
- M. J. Zachman, Z. Tu, S. Choudhury, L. A. Archer and L. F. Kourkoutis, Nature, 2018, 560, 345–349 CrossRef PubMed
.
- N. S. Dutta, P. J. Weddle, O. Hathaway, M. Al-Jassim and K. Jungjohann, ACS Energy Lett., 2024, 9, 2464–2471 CrossRef PubMed
.
- J. Quinn, B. Wu, Y. Xu, M. H. Engelhard, J. Xiao and C. Wang, ACS Nano, 2022, 16, 21063–21070 CrossRef PubMed
.
- H. Park, Y. Jeon, W. J. Chung, Y. Bae, J. Kim, H. Baek and J. Park, ACS Energy Lett., 2023, 8, 715–721 CrossRef
.
- C. Fang, J. Li, M. Zhang, Y. Zhang, F. Yang, J. Z. Lee, M. H. Lee, J. Alvarado, M. A. Schroeder, Y. Yang, B. Lu, N. Williams, M. Ceja, L. Yang, M. Cai, J. Gu, K. Xu, X. Wang and Y. S. Meng, Nature, 2019, 572, 511–515 CrossRef PubMed
.
- B. Han, X. Li, Q. Wang, Y. Zou, G. Xu, Y. Cheng, Z. Zhang, Y. Zhao, Y. Deng, J. Li and M. Gu, Adv. Mater., 2021, 34, 2108252 CrossRef
.
- X. Wang, M. Zhang, J. Alvarado, S. Wang, M. Sina, B. Lu, J. Bouwer, W. Xu, J. Xiao, J. G. Zhang, J. Liu and Y. S. Meng, Nano Lett., 2017, 17, 7606–7612 CrossRef CAS
.
- Y. Liu, D. Lin, Y. Li, G. Chen, A. Pei, O. Nix, Y. Li and Y. Cui, Nat. Commun., 2018, 9, 3656 CrossRef PubMed
.
- K. Dong, Y. Xu, J. Tan, M. Osenberg, F. Sun, Z. Kochovski, D. T. Pham, S. Mei, A. Hilger, E. Ryan, Y. Lu, J. Banhart and I. Manke, ACS Energy Lett., 2021, 6, 1719–1728 CrossRef CAS
.
- B. Zhang, H. Shi, Z. Ju, K. Huang, C. Lian, Y. Wang, O. Sheng, J. Zheng, J. Nai, T. Liu, Y. Jin, Y. Liu, C. J. Zhang and X. Tao, J. Mater. Chem. A, 2020, 8, 26045–26054 RSC
.
- Y. Xu, H. Jia, P. Gao, D. E. Galvez-Aranda, S. P. Beltran, X. Cao, P. M. L. Le, J. Liu, M. H. Engelhard, S. Li, G. Ren, J. M. Seminario, P. B. Balbuena, J. G. Zhang, W. Xu and C. Wang, Nat. Energy, 2023, 8, 1345–1354 CrossRef CAS
.
- Y. Yang, Y. Yin, D. M. Davies, M. Zhang, M. Mayer, Y. Zhang, E. S. Sablina, S. Wang, J. Z. Lee, O. Borodin, C. S. Rustomji and Y. S. Meng, Energy Environ. Sci., 2020, 13, 2209–2219 RSC
.
- D. T. Boyle, W. Huang, H. Wang, Y. Li, H. Chen, Z. Yu, W. Zhang, Z. Bao and Y. Cui, Nat. Energy, 2021, 6, 487–494 CrossRef CAS
.
- X. Xing, Y. Li, X. Wang, V. Petrova, H. Liu and P. Liu, Energy Storage Mater., 2019, 21, 474–480 CrossRef
.
- Y. Zhu, Acc. Chem. Res., 2021, 54, 3518–3528 CrossRef CAS PubMed
.
- E. Bianco and L. F. Kourkoutis, Acc. Chem. Res., 2021, 54, 3277–3287 CrossRef CAS PubMed
.
- Y. Zhu, A. R. Moodenbaugh, Z. X. Cai, J. Tafto, M. Suenaga and D. O. Welch, Phys. Rev. Lett., 1994, 73, 3026 CrossRef CAS PubMed
.
- P. Moradifar, Y. Liu, J. Shi, M. L. Siukola Thurston, H. Utzat, T. B. van Driel, A. M. Lindenberg and J. A. Dionne, Chem. Rev., 2023, 123, 12757–12794 CrossRef CAS
.
- B. H. Savitzky, I. El Baggari, C. B. Clement, E. Waite, B. H. Goodge, D. J. Baek, J. P. Sheckelton, C. Pasco, H. Nair, N. J. Schreiber, J. Hoffman, A. S. Admasu, J. Kim, S. W. Cheong, A. Bhattacharya, D. G. Schlom, T. M. McQueen, R. Hovden and L. F. Kourkoutis, Ultramicroscopy, 2018, 191, 56–65 CrossRef CAS PubMed
.
- I. El Baggari, N. Sivadas, G. M. Stiehl, J. Waelder, D. C. Ralph, C. J. Fennie and L. F. Kourkoutis, Phys. Rev. Lett., 2020, 125, 165302 CrossRef CAS PubMed
.
- I. El Baggari, D. J. Baek, M. J. Zachman, D. Lu, Y. Hikita, H. Y. Hwang, E. A. Nowadnick and L. F. Kourkoutis, Nat. Commun., 2021, 12, 3747 CrossRef CAS PubMed
.
- B. H. Goodge, B. H. Goodge, E. Bianco, N. Schnitzer, H. W. Zandbergen, H. W. Zandbergen, L. F. Kourkoutis and L. F. Kourkoutis, Microsc. Microanal., 2020, 26, 439–446 CrossRef CAS PubMed
.
- B. H. Goodge, D. J. Baek and L. F. Kourkoutis, arXiv, 2020, preprint arxiv: 2007.09747, DOI:10.48550/arXiv.2007.09747.
- X. He, R. Kostin, E. Knight, M. G. Han, J. Mun, I. Bozovic, C. Jing and Y. Zhu, Ultramicroscopy, 2024, 267, 114037 CrossRef CAS
.
- B. Plotkin-Swing, A. Mittelberger, B. Haas, J. C. Idrobo, B. Graner, N. Dellby, M. T. Hotz, C. E. Meyer, S. C. Quillin, O. L. Krivanek and T. C. Lovejoy, Microsc. Microanal., 2023, 29, 1698–1699 CrossRef
.
- A. J. Silvaroli, T. R. Heyl, M. Chyasnavichyus, J. M. Beebe, D. Ahn, S. Mangold, Q. Chen, M. Wang and K. R. Shull, Macromolecules, 2023, 56, 4075–4086 CrossRef
.
- C. A. Ron, K. Vanness, A. Rasmuson and W. P. Johnson, Environ. Sci.:Nano, 2019, 6, 1921–1931 RSC
.
- J. Lee, S. Kang, E. Lee, M. Kang, J. Sung, T. J. Kim, P. Christopher, J. Park and D. H. Kim, J. Mater. Chem. A, 2022, 10, 7029–7035 RSC
.
- C. Galeano, J. C. Meier, V. Peinecke, H. Bongard, I. Katsounaros, A. A. Topalov, A. Lu, K. J. J. Mayrhofer and F. Schüth, J. Am. Chem. Soc., 2012, 134, 20457–20465 CrossRef
.
- R. Girod, T. Lazaridis, H. A. Gasteiger and V. Tileli, Nat. Catal., 2023, 6, 383–391 CrossRef
.
- H. An, J. W. Smith, B. Ji, S. Cotty, S. Zhou, L. Yao, F. C. Kalutantirige, W. Chen, Z. Ou, X. Su, J. Feng and Q. Chen, Sci. Adv., 2022, 8, eabk1888 CrossRef PubMed
.
- Y. Li, Z. Wu, C. Wang, X. Yu, W. Gao, B. Wang, C. Wu, Y. Yao, J. Yang and Z. Zou, Adv. Funct. Mater., 2024, 34, 2310428 CrossRef
.
- S. Ott, A. Orfanidi, H. Schmies, B. Anke, H. N. Nong, J. Hübner, U. Gernert, M. Gliech, M. Lerch and P. Strasser, Nat. Mater., 2020, 19, 77–85 CrossRef PubMed
.
- M. Lopez-Haro, L. Guétaz, T. Printemps, A. Morin, S. Escribano, P. H. Jouneau, P. Bayle-Guillemaud, F. Chandezon and G. Gebel, Nat. Commun., 2014, 5, 5229 CrossRef PubMed
.
- K. Yu, J. L. Hart, J. Xie, M. L. Taheri and P. Ferreira, Nano Energy, 2023, 111, 108393 CrossRef
.
- F. C. Kalutantirige, J. He, L. Yao, S. Cotty, S. Zhou, J. W. Smith, E. Tajkhorshid, C. M. Schroeder, J. S. Moore, H. An, X. Su, Y. Li and Q. Chen, Nat. Commun., 2024, 15, 2852 CrossRef CAS PubMed
.
- J. P. Patterson, Y. Xu, M. A. Moradi, N. A. J. M. Sommerdijk and H. Friedrich, Acc. Chem. Res., 2017, 50, 1495–1501 CrossRef CAS
.
- Z. Chen, K. Higashi, K. Ueda and K. Moribe, Nano Lett., 2022, 22, 6841–6846 CrossRef CAS PubMed
.
- J. J. De Yoreo, P. U. P. A. Gilbert, N. A. J. M. Sommerdijk, R. L. Penn, S. Whitelam, D. Joester, H. Zhang, J. D. Rimer, A. Navrotsky, J. F. Banfield, A. F. Wallace, F. M. Michel, F. C. Meldrum, H. Cölfen and P. M. Dove, Science, 2015, 349, aaa6760 CrossRef PubMed
.
- Y. Tsarfati, S. Rosenne, H. Weissman, L. J. W. Shimon, D. Gur, B. A. Palmer and B. Rybtchinski, ACS Cent. Sci., 2018, 4, 1031–1036 CrossRef CAS
.
- A. Dey, P. H. H. Bomans, F. A. Müller, J. Will, P. M. Frederik, G. De With and N. A. J. M. Sommerdijk, Nat. Mater., 2010, 9, 1010–1014 CrossRef
.
- B. P. Pichon, P. H. H. Bomans, P. M. Frederik and N. A. J. M. Sommerdijk, J. Am. Chem. Soc., 2008, 130, 4034–4040 CrossRef PubMed
.
- E. M. Pouget, P. H. H. Bomans, J. A. C. M. Goos, P. M. Frederik, G. De With and N. A. J. M. Sommerdijk, Science, 2009, 323, 1455–1458 CrossRef PubMed
.
- J. J. De Yoreo and N. A. J. M. Sommerdijk, Nat. Rev. Mater., 2016, 1, 16035 CrossRef
.
- A. F. Ogata, A. M. Rakowski, B. P. Carpenter, D. A. Fishman, J. G. Merham, P. J. Hurst and J. P. Patterson, J. Am. Chem. Soc., 2020, 142, 1433–1442 CrossRef PubMed
.
- L. He, H. K. Hsu, L. Li, L. T. Lin, T. H. Tu, T. G. Ong, G. G. Liou and Y. T. Chan, Chem, 2022, 8, 494–507 Search PubMed
.
- F. Wang, O. Gnewou, A. Solemanifar, V. P. Conticello and E. H. Egelman, Chem. Rev., 2022, 122, 14055–14065 CrossRef
.
- L. Kang, A. Chao, M. Zhang, T. Yu, J. Wang, Q. Wang, H. Yu, N. Jiang and D. Zhang, J. Am. Chem. Soc., 2021, 143, 5890–5902 CrossRef
.
- A. E. S. Van Driessche, N. Van Gerven, R. R. M. Joosten, W. L. Ling, M. Bacia, N. Sommerdijk and M. Sleutel, Nat. Commun., 2021, 12, 3902 CrossRef PubMed
.
- L. Ziserman, H. Y. Lee, S. R. Raghavan, A. Mor and D. Danino, J. Am. Chem. Soc., 2011, 133, 2511–2517 CrossRef PubMed
.
- C. Lei, Y. H. Wang, P. X. Zhuang, Y. T. Li, Q. Q. Wan, Y. X. Ma, F. R. Tay and L. N. Niu, J. Dent. Res., 2022, 101, 505–514 CrossRef
.
- M. Armstrong, B.-G. Han, S. Gomez, J. Turner, D. A. Fletcher and R. M. Glaeser, Biophys. J., 2020, 118, 708–719 CrossRef PubMed
.
- S. Y. Schmid, K. Lachowski, H. T. Chiang, L. Pozzo, J. De Yoreo and S. Zhang, Angew. Chem., Int. Ed., 2023, 62, e202309725 CrossRef PubMed
.
- J. P. Patterson, M. P. Robin, C. Chassenieux, O. Colombani and R. K. O'Reilly, Chem. Soc. Rev., 2014, 43, 2412–2425 RSC
.
- E. Sutter, P. Sutter, A. V. Tkachenko, R. Krahne, J. De Graaf, M. Arciniegas and L. Manna, Nat. Commun., 2016, 7, 11213 CrossRef
.
- G. P. Johari, Philos. Mag. B, 1998, 78, 375–383 CrossRef
.
- L. Lupi, A. Hudait, B. Peters, M. Grünwald, R. Gotchy Mullen, A. H. Nguyen and V. Molinero, Nature, 2017, 551, 218–222 CrossRef PubMed
.
- X. Huang, L. Wang, K. Liu, L. Liao, H. Sun, J. Wang, X. Tian, Z. Xu, W. Wang, L. Liu, Y. Jiang, J. Chen, E. Wang and X. Bai, Nature, 2023, 617, 86–91 CrossRef PubMed
.
- M. Lee, S. Y. Lee, M. H. Kang, T. K. Won, S. Kang, J. Kim, J. Park and D. J. Ahn, Nat. Commun., 2024, 15, 908 CrossRef
.
- C. R. Krüger, N. J. Mowry, G. Bongiovanni, M. Drabbels and U. J. Lorenz, Nat. Commun., 2023, 14, 2812 CrossRef
.
- C. R. Krüger, N. J. Mowry, M. Drabbels and U. J. Lorenz, J. Phys. Chem. Lett., 2024, 15, 4244–4248 CrossRef PubMed
.
- T. L. Malkin, B. J. Murray, C. G. Salzmann, V. Molinero, S. J. Pickering and T. F. Whale, Phys. Chem. Chem. Phys., 2015, 17, 60–76 RSC
.
- W. F. Kuhs, C. Sippel, A. Falenty and T. C. Hansen, Proc. Natl. Acad. Sci. U.S.A., 2012, 109, 21259–21264 CrossRef
.
- J. Lee, S. Y. Lee, D. K. Lim, D. J. Ahn and S. Lee, J. Am. Chem. Soc., 2019, 141, 18682–18693 CrossRef PubMed
.
- C. Lee, Y. Lee, W. H. Jung, T.-Y. Kim, T. Kim, D.-N. Kim and D. J. Ahn, Sci. Adv., 2022, 8, 185 Search PubMed
.
- S. Y. Lee, M. Kim, T. K. Won, S. H. Back, Y. Hong, B. Kim and D. J. Ahn, Nat. Commun., 2022, 13, 6532 CrossRef PubMed
.
- U. J. Lorenz, Curr. Opin. Struct. Biol., 2024, 87, 102840 CrossRef PubMed
.
- O. F. Harder, S. V. Barrass, M. Drabbels and U. J. Lorenz, Nat. Commun., 2023, 14, 5649 CrossRef PubMed
.
- L. Rutten, M. de Beer, R. Roverts, E. M. Sánchez and N. Sommerdijk, Microsc. Microanal., 2023, 29, 1935–1936 CrossRef
.
- A. P. Conlan, E. Tillotson, A. Rakowski, D. Cooper and S. J. Haigh, J. Microsc., 2020, 279, 168–176 CrossRef PubMed
.
-
V. Lam and E. Villa, in Methods in Molecular Biology, Humana Press Inc., USA, 2021, vol. 2215, pp. 49–82 Search PubMed
.
- X. Zhang, Z. Guo, X. Li, Q. Liu, H. Hu, F. Li, Q. Huang, L. Zhang, Y. Tang and J. Huang, Energy Environ. Sci., 2024, 17, 1436–1447 RSC
.
- D. Chmielewski, M. F. Schmid, G. Simmons, J. Jin and W. Chiu, Nat. Microbiol., 2022, 7, 1270–1279 CrossRef PubMed
.
- R. A. Woldeyes, M. Nishiga, A. S. Vander Roest, L. Engel, P. Giri, G. C. Montenegro, A. C. Wu, A. R. Dunn, J. A. Spudich, D. Bernstein, M. F. Schmid, J. C. Wu and W. Chiu, bioRxiv, 2023 DOI:10.1101/2023.10.26.564098
.
- A. Matatyaho Ya'Akobi and Y. Talmon, Acc. Chem. Res., 2021, 54, 2100–2109 CrossRef PubMed
.
- H. Koh, E. Detsi and E. A. Stach, Microsc. Microanal., 2023, 29, 1350–1356 CrossRef
.
- Z. Zhang, Y. Li, R. Xu, W. Zhou, Y. Li, S. T. Oyakhire, Y. Wu, J. Xu, H. Wang, Z. Yu, D. T. Boyle, W. Huang, Y. Ye, H. Chen, J. Wan, Z. Bao, W. Chiu and Y. Cui, Science, 2022, 375, 66–70 CrossRef
.
- R. B. G. Ravelli, F. J. T. Nijpels, R. J. M. Henderikx, G. Weissenberger, S. Thewessem, A. Gijsbers, B. Beulen, C. López-Iglesias and P. J. Peters, Nat. Commun., 2020, 11, 2563 CrossRef PubMed
.
- M. R. Vos, P. H. H. Bomans, P. M. Frederik and N. A. J. M. Sommerdijk, Ultramicroscopy, 2008, 108, 1478–1483 CrossRef
.
- J. Dubochet, J. Lepault, R. Freeman, J. A. Berriman and J. -C. Homo, J. Microsc., 1982, 128, 219–237 CrossRef
.
- P. Zhang, H. Du, S. Cui, P. Zhou and Y. Xu, Responsive Mater., 2023, 1, e20230025 CrossRef
.
- J. Xu, X. Gao, L. Zheng, X. Jia, K. Xu, Y. Ma, X. Wei, N. Liu, H. Peng and H. W. Wang, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2309384121 CrossRef PubMed
.
- L. Kong, J. Liu, M. Zhang, Z. Lu, H. Xue, A. Ren, J. Liu, J. Li, W. L. Ling and G. Ren, Nat. Commun., 2023, 14, 5641 CrossRef CAS PubMed
.
- Y. Xie, J. Wang, B. H. Savitzky, Z. Chen, Y. Wang, S. Betzler, K. Bustillo, K. Persson, Y. Cui, L.-W. Wang, C. Ophus, P. Ercius and H. Zheng, Sci. Adv., 2023, 9, eadc9721 CrossRef PubMed
.
- Y. Son, B. H. Kim, B. K. Choi, Z. Luo, J. Kim, G. H. Kim, S. J. Park, T. Hyeon, S. Mehraeen and J. Park, ACS Appl. Mater. Interfaces, 2022, 14, 22810–22817 CrossRef CAS PubMed
.
-
G. McMullan, A. R. Faruqi and R. Henderson, in Methods Enzymol., Academic Press Inc., USA, 2016, vol. 579, pp. 1–17 Search PubMed
.
- K. Neselu, B. Wang, W. J. Rice, C. S. Potter, B. Carragher and E. Y. D. Chua, J. Struct. Biol. X, 2023, 7, 100085 CAS
.
- T. Nakane, A. Kotecha, A. Sente, G. McMullan, S. Masiulis, P. M. G. E. Brown, I. T. Grigoras, L. Malinauskaite, T. Malinauskas, J. Miehling, T. Uchański, L. Yu, D. Karia, E. V. Pechnikova, E. de Jong, J. Keizer, M. Bischoff, J. McCormack, P. Tiemeijer, S. W. Hardwick, D. Y. Chirgadze, G. Murshudov, A. R. Aricescu and S. H. W. Scheres, Nature, 2020, 587, 152–156 CrossRef CAS PubMed
.
- R. Danev and W. Baumeister, Curr. Opin. Struct. Biol., 2017, 46, 87–94 CrossRef CAS
.
- Y. Kayama, R. N. Burton-Smith, C. Song, N. Terahara, T. Kato and K. Murata, Sci. Rep., 2021, 11, 8395 CrossRef CAS PubMed
.
- C. Hetherington, Mater. Today, 2004, 7, 50–55 CrossRef
.
- N. Tanaka, Sci. Technol. Adv. Mater., 2008, 9, 014111 CrossRef PubMed
.
- N. D. Browning, I. Arslan, R. Erni, J. C. Idrobo, A. Ziegler, J. Bradley, Z. Dai, E. A. Stach and A. Bleloch, J. Phys.: Conf. Ser., 2006, 26, 59 CrossRef
.
- K. Sader, R. Matadeen, P. Castro Hartmann, T. Halsan and C. Schlichten, Acta Crystallogr., Sect. D:Struct. Biol., 2020, 76, 313–325 CrossRef CAS
.
- K. S. Kapoor, S. Kong, H. Sugimoto, W. Guo, V. Boominathan, Y. L. Chen, S. L. Biswal, T. Terlier, K. M. McAndrews and R. Kalluri, ACS Nano, 2024, 18, 11717–11731 CrossRef CAS
.
- D. Cheng, B. Lu, G. Raghavendran, M. Zhang and Y. S. Meng, Matter, 2022, 5, 26–42 CrossRef CAS
.
- H. Ma, D. Kim, S. I. Park, B. K. Choi, G. Park, H. Baek, H. Lee, H. Kim, J. S. Yu, W. C. Lee, J. Park and J. Yang, Adv. Sci., 2023, 10, 2205690 CrossRef CAS
.
- M. U. Rothmann, W. Li, Y. Zhu, A. Liu, Z. Ku, U. Bach, J. Etheridge and Y. B. Cheng, Adv. Mater., 2018, 30, 1800629 CrossRef
.
- O. Dyck, S. Kim, S. V. Kalinin and S. Jesse, Appl. Phys. Lett., 2017, 111, 113104 CrossRef
.
- H. Malekpour, P. Ramnani, S. Srinivasan, G. Balasubramanian, D. L. Nika, A. Mulchandani, R. K. Lake and A. A. Balandin, Nanoscale, 2016, 8, 14608–14616 RSC
.
- W. Zhao, X. Huang, J. Yang, G. Qiu, L. Qu, S. Zhao, Z. Luo, X. Wang, Y. Jiu, H. Mao, X. Ding, J. Tan, Y. Hu, L. Pan, L. Chen and H. Li, bioRxiv, 2022 DOI:10.1101/2022.12.01.518675
.
-
M. S'Ari, J. Cattle, N. Hondow, H. Blade, S. Cosgrove, R. M. Brydson and A. P. Brown, in Journal of Physics: Conference Series, Institute of Physics Publishing, UK, 2015, vol. 644, p. 012038 Search PubMed
.
- M. Karuppasamy, F. Karimi Nejadasl, M. Vulovic, A. J. Koster and R. B. G. Ravelli, J. Synchrotron Radiat., 2011, 18, 398–412 CrossRef PubMed
.
- J. Cheng and X. Zhang, Biophys. Rep., 2021, 7, 152–158 Search PubMed
.
- J. Lv, H. Zhang, D. Zhang, L. Liu and Y. Han, Acc. Mater. Res., 2022, 3, 552–564 CrossRef
.
- A. S. Frangakis, J. Struct. Biol., 2021, 213, 107804 CrossRef
.
-
B. Salmon and A. Krull, in Lecture Notes in Computer Science, Springer, Cham, Switzerland, 2023, p. 13804 Search PubMed
.
-
A. Krull, T.-O. Buchholz and F. Jug, in 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Institute of Electrical and Electronics Engineers, USA, 2019, pp. 2124–2132 Search PubMed
.
- T. Bepler, K. Kelley, A. J. Noble and B. Berger, Nat. Commun., 2020, 11, 5208 CrossRef PubMed
.
- A. Cheng and Y. Yu, Curr. Opin. Struct. Biol., 2024, 86, 102795 CrossRef PubMed
.
- A. Shah, J. A. Schiller, I. Ramos, J. Serrano, D. K. Adams, S. Tawfick and E. Ertekin, Mater. Today Commun., 2023, 35, 106127 CrossRef
.
- L. Yao, H. An, S. Zhou, A. Kim, E. Luijten and Q. Chen, Nanoscale, 2022, 14, 16479–16489 RSC
.
| This journal is © The Royal Society of Chemistry 2025 |