 Open Access Article
Open Access ArticleMorphogenesis of confined biofilms: how mechanical interactions determine cellular patterning and global geometry
Kee-Myoung
Nam
 a and
Jing
Yan
a and
Jing
Yan
 *ab
*ab
aDepartment of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06511, USA
bQuantitative Biology Institute, Yale University, New Haven, CT 06511, USA. E-mail: jing.yan@yale.edu
First published on 27th January 2025
Abstract
Biofilms are surface-attached bacterial communities encased within extracellular matrices that play significant roles in health and society and serve as prototypical examples of proliferating active nematics. Recent advances in fluorescence microscopy have facilitated an unprecedented view of biofilm development at the single-cell level, thus providing the opportunity to develop a mechanistic understanding of how biofilm development is influenced by mechanical interactions with the environment. Here, we review recent studies that examined the morphogenesis of Vibrio cholerae biofilms under confinement at both single-cell and continuum levels. We describe how biofilms under different confinement modes—embedded and interstitial—can acquire various global geometries and forms of cell orientational ordering different from those in unconfined biofilms, and we demonstrate how these properties arise from the mechanical interplay between the biofilm and its confining medium. We also discuss how this interplay is fundamentally governed by the extracellular matrix, which facilitates the transmission of mechanical stress from the medium into the biofilm via adhesion and friction, and serves as the key feature that distinguishes biofilms from classical bacterial colonies. These studies lay the groundwork for many potential future directions, all of which will contribute to the establishment of a new “developmental biology” of biofilms.
1. Introduction
Biofilms are surface-attached communities of bacteria, encased within extracellular matrices (ECMs) of biopolymers (Fig. 1A and B). Biofilms play many significant roles in health, nature, and industry,1 ranging from gut microbiome homeostasis2 to biogeochemical cycling3 to water and soil bioremediation.4 Pathogenic biofilms constitute a major threat to public health, as they exhibit heightened tolerance to antibiotics and the host immune response, in part due to the physicochemical properties of the ECM.5–7 From a physical perspective, biofilms are also notable examples of active collectives with non-conserved particle number: the cells within the biofilm metabolize nutrients in the environment to drive growth and proliferation, from which the resulting collective motion can enable the biofilm to do mechanical work on its surroundings.8–10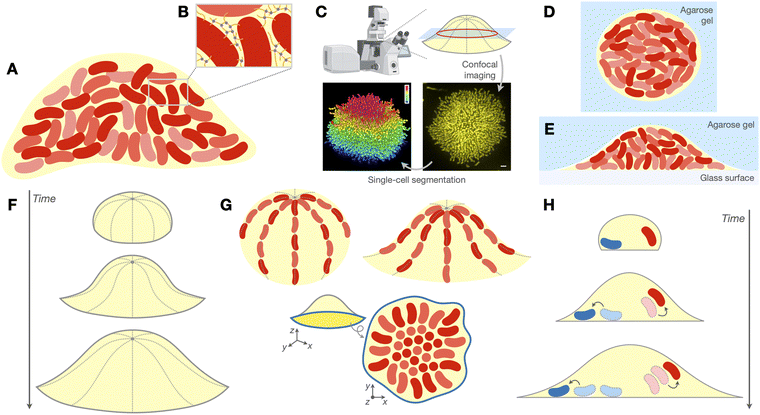 | ||
| Fig. 1 Biofilms and their morphogenesis under soft confinement. (A) Biofilms are surface-attached communities of bacteria (red) embedded in an extracellular matrix (ECM, yellow) of biopolymers. (B) The ECM consists of various polymeric macromolecules, such as polysaccharides, proteins, and/or extracellular nucleic acids, that form a mechanically resilient polymer network through crosslinking and other interactions. (C) Top: Our research group and others have established a live imaging platform, based on spinning disk confocal microscopy, that can image growing biofilms containing tens of thousands of cells at single-cell resolution. Bottom: Example cross-sectional view of the basal layer of a V. cholerae biofilm expressing mKO after 18 h of growth obtained from this imaging platform (right). Upon processing these images with an adaptive single-cell segmentation algorithm, the position and orientation of every cell within the biofilm can be reconstructed (left). The cells in the segmentation are coloured by their z-positions. Scale bar, 3 μm. Graphic at top left was created using BioRender; images in bottom row were adapted with permission from ref. 11. (D) Schematic of embedded biofilms, which are entirely surrounded by an agarose hydrogel (blue). (E) Schematic of interstitial biofilms, which grow within the interstitial space between a glass surface (gray) and an overlaid agarose hydrogel (blue). (F)–(H) We consider three features of biofilm organization in this perspective: the evolution of the biofilm's global geometry over time (F; see Section 3), the local arrangement or ordering of cells within the biofilm (G; see Section 4), and the spatiotemporal trajectories taken by the cells throughout biofilm development (H; see Section 5). In G, we show three examples of local cellular patterning that we cover: the bipolar alignment of cells along the biofilm–gel interface in embedded biofilms (top left; see Section 4.2), a similar form of longitudinal alignment of cells along the biofilm–gel interface in interstitial biofilms (top right; see Section 4.3), and the “aster patterning” of cells within the basal layer (blue outline) of cells in unconfined and interstitial biofilms (bottom; see Section 4.1). | ||
When the constituent cells are rod-shaped (as in Escherichia coli, Vibrio cholerae, Pseudomonas aeruginosa and many other species), the biofilm can also be described as an active nematic, in which the alignment of neighbouring cells can dynamically shape the biofilm's higher-order architecture.8,9,12,13 The most prototypical examples of active nematics are composed of self-propelled, or motile, particles that engage in collective motion, not unlike the motion of macroscopic active collectives like airborne flocks of starlings.14 Examples include communities of swimming bacteria, epithelial layers, mixtures of microtubules and motor proteins, and synthetic self-propelled colloids. Such active nematics exhibit an extraordinary diversity of collective behaviours, many of which are manifestations of active turbulence,12,13,15 and further intriguing modes of collective motion are continually being discovered.16–18 Biofilms, however, are fundamentally different in that the constituent cells are not motile in most cases, but rather are physically pushed by the growth and division of neighbouring cells. As such, biofilms fall within the bracket of “proliferating active matter”,10 whose collective properties are not as well-characterized. Therefore, we envision that biofilms serve as a paradigmatic model system for revealing these properties.
Given the broad importance of biofilms to health and society and their significance as model active nematics, it is natural to ask: How are the cells within a biofilm organized, and how does this organization evolve over time? How does the ECM influence this organization? And how does this organization influence the behaviours of cells within the biofilm? These types of questions have long been at the forefront of eukaryotic developmental biology, and technical advances in light-sheet microscopy enabled the single-cell mapping of early zebrafish embryos as early as 2008;19 subsequent advances facilitated the single-cell mapping of fruit-fly and mouse embryos,20,21 as well as other embryos and complex tissues (for a recent history, see ref. 22). However, the same questions have been more challenging to address in biofilms. The small size of bacterial cells within a densely packed biofilm makes them more difficult to resolve than their larger eukaryotic counterparts; this difficulty is compounded by their lack of membrane-bound nuclei, which serve as useful landmarks for tracking eukaryotic cells. Fluorescence microscopy is also fundamentally constrained by the fluorescent proteins used to visualize the cells, which undergo photobleaching and induce phototoxicity under prolonged excitation. As such, for a long time, high-resolution biofilm imaging was mostly limited to live imaging of immature biofilms23 or to imaging of processed samples using scanning electron microscopy,24 neither of which were sufficient to characterize the full biofilm developmental program at the single-cell level.
Several groups, including us, have pioneered live imaging of bacterial biofilms at single-cell resolution, starting from the mid-2010's.11,25–28 In our laboratory, we have established an imaging platform that can image growing biofilms containing on the order of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells and resolve them with single-cell resolution (Fig. 1C). This platform, which bears similarities to earlier platforms that were designed to resolve individual cells in fixed biofilms,25,26 takes advantage of several technical advances (see Section 2) to enable the tracking and analysis of the full spatiotemporal organization of growing biofilms at single-cell resolution.11 More recently, several groups have employed light-sheet microscopy to further reduce phototoxicity and photobleaching and push the temporal resolution to minutes, enabling the tracking of cell positional fates.29,30
000 cells and resolve them with single-cell resolution (Fig. 1C). This platform, which bears similarities to earlier platforms that were designed to resolve individual cells in fixed biofilms,25,26 takes advantage of several technical advances (see Section 2) to enable the tracking and analysis of the full spatiotemporal organization of growing biofilms at single-cell resolution.11 More recently, several groups have employed light-sheet microscopy to further reduce phototoxicity and photobleaching and push the temporal resolution to minutes, enabling the tracking of cell positional fates.29,30
Most in vitro studies of biofilms—including the study that introduced our imaging platform11—have focused on unconfined biofilms, which grow freely on solid substrates without mechanical obstruction. But there are also notable examples of biofilms in nature that grow under mechanical confinement. For instance, pathogenic bacteria such as V. cholerae and P. aeruginosa often form biofilms within mucosal layers that line a wide range of host tissues.31–33 Other pathogens, such as uropathogenic E. coli, can invade host cells and form intracellular biofilm-like aggregates, or “pods,” that can evade the host immune response.6,34–36 Intracellular bacteria also feature in a wide range of tumours,37,38 and have been implicated in the initiation, promotion, and/or progression of tumorigenesis39 and cancer metastasis.40 Beyond the human body, a remarkable panoply of bacterial species form biofilms in a wide range of natural habitats, many of which lie within confined niches such as the soil and the Earth's subsurface.3,41 Indeed, the significance of such confined environments as bacterial habitats is further underscored by recent studies of bacterial motility42,43 and colony morphogenesis44,45 in in vitro analogues of these niches. The mechanical stresses that these environments exert upon biofilms growing within them can influence their internal organization and growth dynamics; in turn, growing biofilms can respond to these stresses by re-shaping their environments. As such, confined biofilms are expected to exhibit fundamentally different physical features from their unconfined counterparts.11,26,27
In this perspective, we describe a series of studies46–48 from our group that utilized our imaging platform to elucidate the effects of soft confinement (i.e., confinement by a soft, deformable material, such as a polymeric hydrogel) on biofilm development at single-cell resolution, using the rod-shaped bacterium V. cholerae as a model organism. Though V. cholerae cells are known to assume a characteristic helical shape owing to the periplasmic protein CrvA,49 due to their short length (1.5–3 μm), they can be treated as straight rods to a first-order approximation. These studies focused on two types of confined biofilms: biofilms embedded within an agarose hydrogel with no surface present (Fig. 1D), and biofilms grown within the interstitial space between a glass surface and an overlaid agarose gel (Fig. 1E). The overarching themes that emerge from these studies are twofold: (1) the geometry and internal organization of confined biofilms are controlled by the mechanical interplay between the biofilm and its confining medium; and (2) this interplay critically depends on the ECM, which facilitates the transmission of mechanical stress from the medium into the biofilm. We shall see that this interplay controls both the cellular patterning—in the form of orientational ordering and the emergence of topological defects—and the global geometry of the confined biofilm, echoing related observations that have been made in other multicellular collectives,50,51 such as eukaryotic tissues.52–54
The structure of this paper is as follows. We first provide a brief introduction to biofilms and the imaging platform used in the above studies to visualize live biofilms at single-cell resolution (Section 2; Fig. 1A–C). We then discuss the consequences of soft confinement, as elucidated in the above studies, on three fundamental aspects of the organization of a developing biofilm: the biofilm's global geometry (Section 3; Fig. 1F), the local arrangement and ordering exhibited by the biofilm's constituent cells (Section 4; Fig. 1G), and the spatiotemporal trajectories that cells take within the biofilm (Section 5; Fig. 1H). In each of these three sections, we first consider the unconfined case as a point of departure, then consider how each aspect of biofilm organization changes with the introduction of soft confinement (Fig. 1D and E). Finally, we summarize the key themes that emerge from these findings and the many future directions that remain to be explored in the realm of biofilm development.
2. A brief introduction to biofilms
Free-swimming, or “planktonic,” bacteria form biofilms by first attaching to a hospitable surface, transitioning to a sessile state, and proliferating while producing the ECM that binds the cells to each other and to the surface (Fig. 1A). This phenotypic transition is realized by intracellular signalling pathways that engender large-scale changes in gene expression, thus suppressing behaviours related to the planktonic state, such as flagellar assembly, while promoting behaviours related to the biofilm state, such as ECM production.28,55–59 Subsequently, nutrient depletion or other environmental perturbations may then cause the cells within the biofilm to revert to the planktonic phenotype, degrade the ECM, and swim away to colonize other niches, in a process known as dispersal.60,61As we alluded to previously, the key feature that distinguishes biofilms from classical bacterial colonies is the ECM, which consists of a complex assortment of polymeric macromolecules that often includes polysaccharides, proteins, and/or extracellular nucleic acids62–64 (Fig. 1B). The key function of the ECM is to adhere the cells within the biofilm to each other and to the surface; it is through this function that the ECM shields biofilm-dwelling cells from various external perturbations, including hydrodynamic shear stress, chemical insults, the host immune response, and grazing by eukaryotic predators.5,7,64 The ECMs of different bacterial species vary enormously in molecular composition, as do their mechanisms of production and degradation.62–64 Importantly, these molecular components do not exist as a simple mixture, but rather form a mechanically resilient polymer network through crosslinking and other attractive interactions, thus giving rise to the ECM's unique material properties62,64–67 (Fig. 1B). For instance, the major components of the V. cholerae biofilm ECM are Vibrio polysaccharide (VPS), which is a polymer composed of unique repeating tetrasaccharide units,68,69 and three adhesion proteins: RbmA, which facilitates cell–cell adhesion;70,71 and Bap1 and RbmC, which facilitate cell–surface adhesion and, together with VPS, form envelope structures that group cells into small clusters72–74 (Fig. 2A; for a recent review, see ref. 58). These three adhesins all rely on VPS for proper function,71–74 which means that eliminating VPS has the effect of simultaneously abrogating cell–cell and cell–surface adhesion.
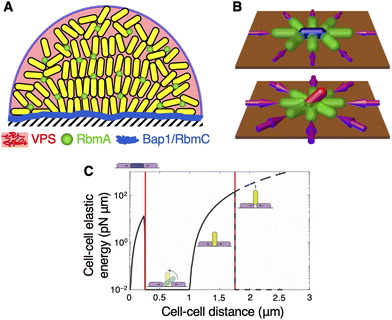 | ||
| Fig. 2 Geometry and cellular organization in unconfined biofilms. (A) Schematic of unconfined V. cholerae biofilm. The cells are surrounded by Vibrio polysaccharide (VPS, pink), which constitutes most of the ECM; cell–cell adhesion is facilitated by RbmA (green); and cell–surface adhesion is facilitated by Bap1 and RbmC (blue). Bap1 and RbmC also form envelopes that surround small clusters of cells. (B) Schematic of cell verticalization. A horizontal cell (blue) is compressed due to the growth of neighbouring cells (green), which gives rise to a mechanical instability that causes the cell to rotate out of the plane (red). Adapted with permission from ref. 75. (C) Cell–cell elastic energy due to growth-induced compression of the central cell (yellow) by neighbouring cells (purple), as a function of cell–cell distance. The energy landscape exhibits four phases: the system (1) accumulates energy while maintaining a horizontal configuration, (2) releases the energy by verticalizing the central cell, (3) accumulates energy from further compression, and (4) again releases the energy by ejecting the central cell from the surface (“pinch-off”). Adapted with permission from ref. 47. | ||
Our aforementioned protocol for imaging live biofilms at single-cell resolution11,28,46–48 uses a customized spinning disk confocal microscope to visualize biofilms in which the cells constitutively express a fluorescent protein (Fig. 1C). The various components of this platform were configured to strike a balance between maximizing optical resolution and minimizing spherical aberrations in 3D imaging; a time-adaptive z-range that increases over time was also incorporated to mitigate photobleaching and phototoxicity due to wasteful illumination.11 The resulting z-slices are then processed with a custom 3D adaptive local thresholding algorithm for single-cell segmentation, through which we can reconstruct the position and orientation of each cell within the biofilm (Fig. 1C). It is precisely through this imaging platform—and others like it26,27,29—that our research group and others have been able to work towards building a rigorous understanding of how cellular organization emerges within a developing biofilm, the details of which we spell out in the rest of this perspective.
2.1. A note on terminology
In the studies that we focus on in this perspective,46–48 the “reference” V. cholerae strain that was used as the genetic background for all mutants was the rugose strain with an rbmA deletion. This strain harbours a point mutation that causes robust biofilm formation, but its biofilms lack cell–cell adhesion and therefore are less densely packed when grown without confinement. We call this strain the WT* strain.To avoid having to continually refer to gene names, we call the mutant strain in which the cell–surface adhesin genes, bap1 and rbmC, have also been deleted (ΔrbmAΔbap1ΔrbmC) the non-surface-adherent strain. We will also consider colonies formed by a mutant strain that lacks vpsL, which is a key biogenesis gene for VPS. This strain is incapable of both cell–surface and cell–gel adhesion and does not produce VPS, and therefore we refer to this strain as the non-biofilm-forming strain. This strain essentially behaves like other non-biofilm-forming bacteria such as lab-domesticated E. coli, and therefore allows for direct comparison between our work and the vast literature on the biophysics of classical bacterial colonies.76–84 We will mostly focus on these three strains throughout the rest of this paper, save for some occasional exceptions that we will point out as we encounter them.
3. How soft confinement shapes biofilm geometry
3.1. Unconfined biofilms
We now turn to how soft confinement affects biofilm geometry. As a point of departure, we first consider the geometry of unconfined biofilms. Using the aforementioned imaging platform, Yan et al.11 found that unconfined biofilms formed by the well-characterised rugose V. cholerae strain85 grow from single founder cells to planar branched structures to dense hemispherical domes (Fig. 1C), in agreement with similar observations made with fixed V. cholerae biofilms.26 This bears some similarities to classical bacterial colonies, which also undergo a layering transition to form a “wedding-cake-like” architecture,76–78 but tend to form more conical structures within the same timeframe.77 The full biofilm growth program was later imaged and modelled using an anisotropic cell–cell interaction potential by the Drescher lab, who also demonstrated how biofilm architecture can be shaped by fluid flow.27,86Yan et al. observed that, within the core of these growing biofilms, the initially horizontal cells first verticalize and, with each subsequent cell division, project their offspring into the third dimension. Surrounding this verticalized core is a subpopulation of horizontal cells that align radially outward; we shall revisit this radial alignment in Section 4.1. First, how does this verticalization occur? To resolve this question, Beroz et al.75 implemented an agent-based model (ABM) to identify the mechanism underlying cell verticalization in silico. In this ABM, cells were modelled as spherocylinders that grow in length, divide at a critical length, and physically push against each other. The model also incorporated the effects of the ECM, which was assumed to form a soft, elastic shell around each cell, cause the cells to adhere to the surface, and incur a drag against each cell's motion due to bulk viscosity and surface friction. This ABM revealed that verticalization is primarily a consequence of a mechanical instability at the single-cell level that arises when a surface-adhered cell is subject to sufficient in-plane compression from the growth of neighbouring cells (Fig. 2B). This explains why the innermost cells within an early-stage, planar biofilm, which experience sustained compression from the cells on the periphery, verticalize first and seed the dome shape that emerges as the biofilm grows. A subsequent analysis of the same model by Nijjer et al.47 found that this verticalization instability is followed by a pinch-off instability, in which further accumulation of elastic potential energy causes the cell to be ejected from the surface (Fig. 2C). Crucially, the energy barrier to the pinch-off instability, which determines the duration for which the cell remains stably verticalized, was found to scale with the strength of cell–surface adhesion. In other words, strong cell–surface adhesion—which requires the ECM—gives rise to a stably verticalized core, as observed in the dome-shaped biofilm in Fig. 1C (see also Fig. 5A).
To investigate the role of the ECM on biofilm development, Yan et al. imaged biofilms formed by mutant strains in which the genes responsible for facilitating cell–cell adhesion, cell–surface adhesion, or both were deleted. Removing cell–cell adhesion resulted in a much less densely packed biofilm, with a commensurately larger volume; removing cell–surface adhesion resulted in a free-floating, disordered biofilm that is detached from the surface. As might be expected from these two phenotypes, removing both cell–cell and cell–surface adhesion resulted in a highly diffuse, disordered “web” of cells that are detached from the surface.
3.2. A spherical-to-ellipsoidal bifurcation in embedded biofilms
Zhang et al.46 examined V. cholerae biofilms embedded in agarose gels (Fig. 1D), which undergo a morphogenetic process that, unlike surface-attached biofilms, is more akin to the expansion of a viscoelastic inclusion within an elastic medium. The gel stiffness, Ga, was systematically varied over three orders of magnitude, between ∼300 Pa to ∼70 kPa, by tuning the agarose concentration (0.3% to 2%), while the biofilm stiffness, Gb, was varied between ∼100 Pa and ∼2 kPa by deleting various combinations of ECM components.66 This dual approach allowed for a detailed analysis of how the stiffness of the gel and biofilm together dictate the geometry and internal organization of a growing embedded biofilm over time.Using this approach, we observed an intriguing growth profile for biofilms under stiff confinement (Fig. 3A). Namely, the biofilm initially grows into a prolate ellipsoidal geometry, due to the division of intrinsically anisotropic cells along their long axes; then transitions into a nearly spherical geometry; then finally transitions into an oblate ellipsoidal geometry (Fig. 3B). By systematically varying the gel and biofilm stiffness, we found that this growth profile is controlled by the stiffness contrast, Gb/Ga (Fig. 3C): when the biofilm is stiffer than the gel (Gb/Ga > 1), the biofilm retains its spherical geometry as it matures, but when the gel is stiffer than the biofilm (Gb/Ga < 1), the biofilm exhibits the two-stage growth profile described above, resulting in an oblate geometry.
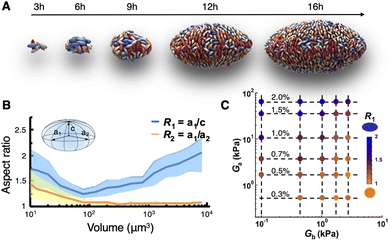 | ||
| Fig. 3 Geometry of embedded biofilms. (A) Growth profile of a WT* biofilm embedded in a 2% agarose gel over 16 hours. Each image is shown to a different scale for clarity. (B) Evolution of the two aspect ratios of WT* biofilms embedded within a 2% gel, as indicated in the inset schematic, as a function of biofilm volume. The two curves show that the biofilm quickly approaches a nearly spherical geometry, then assumes an increasingly oblate ellipsoidal geometry. (C) Phase diagram of embedded WT* biofilm shape, quantified here as the aspect ratio of the longest and shortest axes, as a function of the gel stiffness, Ga, and biofilm stiffness, Gb. All panels were adapted with permission from ref. 46. | ||
We attributed this unusual two-stage growth profile to the mechanical confinement exerted by the gel, which imposes an external constraint upon the biofilm; the biofilm–gel system seeks to minimize the total mechanical energy by choosing its preferred shape at each point along the developmental trajectory. This intuition was formalized with a continuum model, which described the biofilm as a solid inclusion that grows within a void in an elastic medium.87 According to this model, the growing biofilm first deforms the gel elastically, so that the stress-free shape of the void is preserved (Stage I). Then, after the biofilm reaches a certain critical volume, further growth deforms the gel beyond its elastic limit, damaging the gel and altering the void's stress-free shape (Stage II). Analysis results from this model recapitulated the key dichotomy observed in the phase diagram in Fig. 3C: a biofilm that is stiffer than the gel will tend towards a spherical geometry, whereas a biofilm that is softer than the gel will assume an oblate geometry.
3.3. A dome-to-lens bifurcation in interstitial biofilms
In a parallel line of work,48 Nijjer et al. examined the same questions in interstitial V. cholerae biofilms sandwiched between a gel and a glass surface (Fig. 1E). The presence of the glass surface introduces cell–surface friction as well as gel–surface adhesion; in order for the biofilm to grow, it needs to work against both effects. When confined with a soft gel, e.g., a gel with concentration c = 0.2% (corresponding to a gel stiffness of Ga ≈ 150 Pa), interstitial WT* biofilms were observed to assume a hemispherical dome shape, much like unconfined biofilms. However, as c was increased from 0.2% to 2.5% (Ga ≈ 150 kPa), the biofilms were observed to undergo a geometric bifurcation, from a dome shape for c < 1.5% to a flatter lens shape for c > 1.5%. These geometries were distinguished in terms of the biofilm's contact angle, ψ, with the surface at the gel–surface interface, which was as large as ∼120° in dome-shaped biofilms and as small as ∼20° in lens-shaped biofilms (Fig. 4A); and the biofilm's height and radius, which both scaled as n1/3 in dome-shaped biofilms (where n is the number of cells), but rather scaled as n1/5 and n2/5, respectively, in lens-shaped biofilms. These latter power law exponents are reminiscent of those seen in hydraulic fracture.88,89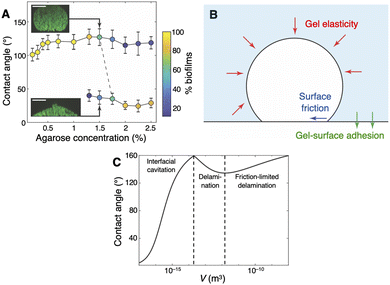 | ||
| Fig. 4 Geometry of interstitial biofilms. (A) Contact angle made by interstitial WT* biofilms at the glass surface, as a function of gel concentration. The insets show two example biofilms, one dome-shaped and one lens-shaped, under 1.5% gel confinement. Scale bars, 10 μm. (B) Schematic of energetic model of interstitial biofilm growth, showing the competing contributions of gel elasticity, biofilm–surface friction, and gel–surface adhesion. (C) An example solution of the energetic model, showing the three stages of interstitial biofilm growth and the corresponding contact angle evolution: the biofilm first grows exclusively into the gel (Stage I), then delaminates the gel from the surface (Stage II), until the delamination becomes limited by surface friction (Stage III). All panels were adapted with permission from ref. 48. | ||
Hypothesizing that cell–surface adhesion can modulate this geometric bifurcation, we found that non-surface-adherent biofilms undergo the bifurcation at a lower gel stiffness; subsequently, imaging biofilms with a titratable level of cell–surface adhesion revealed that the critical gel stiffness increases with the level of cell–surface adhesion. This inspired a mathematical model of interstitial biofilm growth that integrated the contributions of gel stiffness, gel–surface adhesion, and cell–surface friction (Fig. 4B). In this model, the biofilm was described as a volumetrically expanding ideal liquid between a semi-infinite elastic medium and a rigid surface that are bonded at their interface, and the system's total potential energy was defined as the sum of two parts: (1) the gel–surface adhesion energy, which increases as the biofilm delaminates the gel from the surface; and (2) the elastic energy stored in the gel, which increases as the biofilm expands and deforms the gel. An additional dissipative term was introduced to account for the biofilm–surface friction that arises upon gel–surface delamination and biofilm expansion along the surface. The competition between these three terms determines the energetically least expensive growth pathway of the biofilm, which in turn dictates its shape evolution.
We found that this model predicts a three-stage growth profile, with a corresponding non-monotonic evolution in the contact angle, ψ (Fig. 4C). In Stage I, the biofilm exclusively grows into—and deforms—the gel, resulting in increasing ψ; this continues until the deformation reaches the point where gel–surface delamination becomes energetically favorable, which causes the biofilm to expand along the surface and results in decreasing ψ (Stage II); then, finally, the delamination becomes limited by biofilm–surface friction and ψ increases again (Stage III). According to this model, the biofilm–surface friction coefficient, η, controls the durations of the latter two stages: if η is small, then delamination continues unhindered by friction and ψ asymptotically decays to a minimum value90 (i.e., the system remains in Stage II indefinitely); but if η is large, then delamination quickly enters the friction-limited regime and ψ continues to increase (i.e., Stage II quickly gives way to Stage III). This model complements the two-stage growth profile of embedded biofilms (Fig. 3A and B), by showing how gel confinement and the presence of a rigid surface can jointly shape a growing interstitial biofilm.
4. How cell–surface adhesion and soft confinement determine cellular patterning
4.1. Aster patterning in the basal layer of unconfined and interstitial biofilms
Classical colonies of rod-shaped bacteria—which do not produce ECM—are globally disordered when growing radially outward: they exhibit patches, or “microdomains,” of nematically aligned cells, whose local order is destroyed within relatively short length-scales by topological defects.80,81,83,91 So it was especially striking that, in submerged V. cholerae biofilms grown on a rigid substrate, the basal layer of cells was found to exhibit a remarkable degree of cellular ordering, comprising a core of verticalized cells and a periphery of radially aligned, horizontal cells (Fig. 5A).11,26 Later, Nijjer et al.47 observed the same “aster” pattern within the basal layer of interstitial WT* biofilms, with even stronger radial alignment in the periphery (Fig. 5B).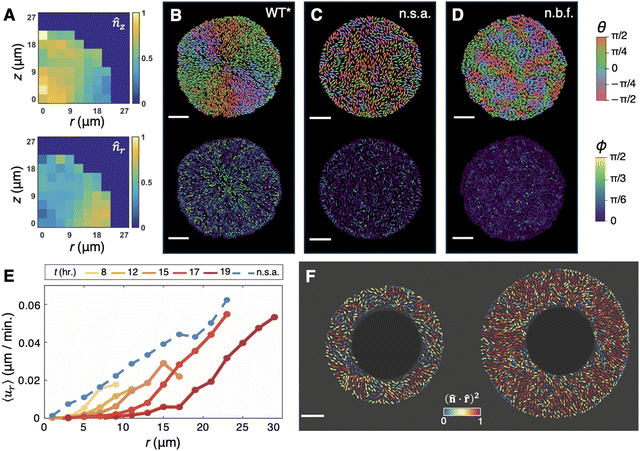 | ||
Fig. 5 Aster patterning in the basal layer of unconfined and interstitial biofilms. (A) Histograms of the vertical component, ![[n with combining circumflex]](https://www.rsc.org/images/entities/i_char_006e_0302.gif) z, and radial component, z, and radial component, ![[n with combining circumflex]](https://www.rsc.org/images/entities/i_char_006e_0302.gif) r, of each cell within a collection of unconfined rugose biofilms. Adapted with permission from ref. 11. (B)–(D) Basal layers of an interstitial WT* biofilm (B), an interstitial non-surface-adherent (n.s.a.) biofilm (C), and an interstitial colony formed by the non-biofilm-forming (n.b.f.) strain (D), with each cell coloured according to its in-plane orientation (θ) and its angular deviation from the glass surface (ϕ). Scale bars, 10 μm. (E) The average radial velocity, ur, as a function of radial position, r, within interstitial WT* biofilms over time (solid lines), as well as interstitial non-surface-adherent biofilms (blue dashed line). (F) A non-surface-adherent biofilm that was irradiated with a 405 nm laser within a circle of radius 15 μm at the center, 0 hours (left) and 4 hours (right) after irradiation. Each cell is coloured according to its degree of radial alignment. Panels B–F were adapted with permission from ref. 47. r, of each cell within a collection of unconfined rugose biofilms. Adapted with permission from ref. 11. (B)–(D) Basal layers of an interstitial WT* biofilm (B), an interstitial non-surface-adherent (n.s.a.) biofilm (C), and an interstitial colony formed by the non-biofilm-forming (n.b.f.) strain (D), with each cell coloured according to its in-plane orientation (θ) and its angular deviation from the glass surface (ϕ). Scale bars, 10 μm. (E) The average radial velocity, ur, as a function of radial position, r, within interstitial WT* biofilms over time (solid lines), as well as interstitial non-surface-adherent biofilms (blue dashed line). (F) A non-surface-adherent biofilm that was irradiated with a 405 nm laser within a circle of radius 15 μm at the center, 0 hours (left) and 4 hours (right) after irradiation. Each cell is coloured according to its degree of radial alignment. Panels B–F were adapted with permission from ref. 47. | ||
Given that the key discrepancy between colonies and biofilms is the ECM, Yan et al.11 hypothesized that the ECM is primarily responsible for this aster patterning. Indeed, Yan et al. found that, without confinement, Δbap1ΔrbmC biofilms exhibit a completely disordered cellular organization, with negligible correlations between neighbouring cell orientations. In other words, the long-range organization within wild-type biofilms is not a consequence of inherent nematic interactions between individual cells, but rather depends critically on cell–surface interactions and on the growth history. However, the free-floating, spherical geometry of these mutant biofilms, and especially their lack of a “basal layer,” made mechanistic study difficult. Hence, Nijjer et al. exploited the similar geometries of the WT* and nonadherent mutants when grown as interstitial biofilms, and found that, indeed, interstitial colonies formed by either the non-surface-adherent biofilm mutant or the non-biofilm-forming strain contain basal layers of mostly horizontal cells with no radial alignment (Fig. 5C and D). Moreover, we found that, in interstitial biofilms with a titratable level of cell–surface adhesion, the aster patterning grew stronger as the level of cell–surface adhesion was increased, illustrating a direct, quantitative link between cell–surface adhesion and aster patterning. ABMs recapitulated this effect in silico: with or without confinement, surface-adherent biofilms formed the aster pattern but nonadherent biofilms did not (see also ref. 75). Altogether, these observations attested to the fundamental role of the ECM—in particular, the role of VPS, Bap1, and RbmC, which together confer cell–surface adhesion—in establishing both components of this pattern.
To investigate how the two components of the aster pattern arise from cell–surface adhesion, we turned to mathematical modelling. As described in Section 3.1, we and others75–77 established that the innermost cells in the basal layer verticalize due to mechanical instabilities that occur with the accumulation of growth-induced pressure. In particular, we found that the stability of the verticalized configuration prior to pinch-off (Fig. 2C) scales with the cell–surface adhesion energy. This explains why cell–surface adhesion is necessary for the formation of a stably verticalized core in the basal layer: without it, the cells are pinched off too readily, resulting in a basal layer of mostly horizontal cells.
Now, to explain the radial alignment along the periphery, we turned to continuum modelling.8,12,84 Biofilms are communities of non-motile but growing cells; this growth, which is an energy-intensive process, generates a flow field that dictates the motion of each cell throughout biofilm growth. In other words, biofilms are an example of “proliferating active matter”.10 But once the verticalized core is established, the verticalized cells elongate and project their offspring into the third dimension, which translates into a zero-velocity core in the corresponding in-plane flow field. Indeed, by imaging biofilms in which each cell expresses a bright punctum that can be tracked with higher temporal resolution29 (see Section 5), we were able to observe such a zero-velocity core in WT* biofilms but not in non-surface-adherent biofilms (Fig. 5E).
Inspired by work by Basaran et al.84 on the inward growth of bacterial colonies, we formalized these notions in a continuum model, in which the basal layer is approximated as an axisymmetric population of cells whose growth gives rise to a radial flow field, u(r, t), where r denotes radial distance from the biofilm centre; and an “orientation field,” Θ(r, t), which measures the angular deviation of a cell's director with respect to the radial direction. According to this model, Θ(r, t) is determined by u(r, t) through a partial differential equation,
| ∂tΘ + ur∂rΘ = −f(r, t)sin(2Θ), | (1) |
As a direct test of this model, we showed that simply irradiating the cells in the centre of a non-surface-adherent biofilm—thus killing them and introducing a growth void in the otherwise disordered biofilm—was sufficient to induce radial alignment in the basal layer (Fig. 5F). But, in allowing for an arbitrary flow field, eqn (1) paved the way towards more novel predictions of cellular ordering induced by differential growth. For instance, if a biofilm is seeded such that the cells in the core grow faster within the basal layer than those in the periphery, the model predicts a strictly negative cell alignment torque, f < 0, which would result in circumferential alignment within the periphery. This prediction was indeed experimentally confirmed.
In all, these models suggest a two-step mechanism for aster patterning due to cell–surface adhesion: (1) accumulation of growth-induced pressure within the biofilm core causes verticalization of the innermost cells within the basal layer, which is stabilized by cell–surface adhesion; and (2) the verticalized cells impose a zero-velocity core on the growth-induced flow field within the basal layer, which drives radial alignment in the periphery. The radial alignment then directs further growth-induced compression into the core, thus increasing the radius of the verticalized core—r0(t) in the above model—over time. The ensuing “reorientation cascade” results in the robust propagation of the aster pattern throughout biofilm development. This is a striking illustration of how the ECM can guide biofilm organization, and how proliferating active matter can differ from their nongrowing counterparts.
4.2. Bipolar alignment in embedded biofilms
In the absence of a rigid substrate, Zhang et al. also found that embedded biofilms under stiff confinement—which grow to exhibit an oblate ellipsoidal geometry, as described in Section 3.2 (Fig. 3)—exhibit bipolar alignment of cells along the biofilm–gel interface (Fig. 6A), in a manner reminiscent of the molecular ordering found on the surface of nematic liquid crystal droplets.92 In particular, the cells along the biofilm–gel interface were found to align along lines of constant longitude (meridians) between the two poles of the short principal axis of the biofilm. The two poles therefore constitute +1 topological defects, or “boojums,” from which the cells emanate radially. Increasing the gel stiffness served to increase this interfacial ordering.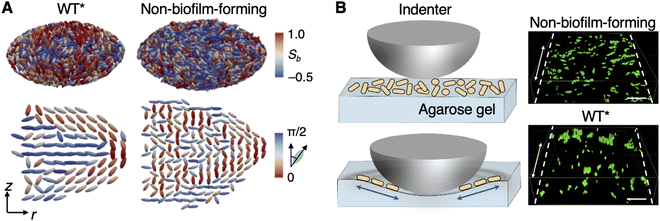 | ||
| Fig. 6 Bipolar alignment in embedded biofilms. (A) Architectures of a WT* biofilm and a colony formed by the non-biofilm-forming strain, each embedded within a 2% gel. The top row shows a bipolar order parameter, Sb, that was measured for each cell; the bottom row shows the azimuthally averaged cell orientations and their angular deviations from the z-axis. (B) Left: Schematic of the gel indentation experiment. The double-headed arrows indicate the direction of gel stretching. Right: Images of non-biofilm-forming and WT* cells on a deformed gel surface that was stretched along the dashed line. Scale bar, 10 μm. All panels were adapted with permission from ref. 46. | ||
What are the origins of this bipolar alignment? Unlike the rigid, nondeformable glass substrate on which interstitial biofilms are grown, the gel surrounding an embedded biofilm undergoes considerable deformation as the biofilm grows; this, in turn, causes the biofilm–gel interface to dilate significantly. Therefore, we hypothesized that the bipolar alignment arises from the mechanical interplay between the biofilm and the gel. As cells proliferate and push against the gel, the gel experiences tensile stresses that align with the long axes of the oblate biofilm. In turn, these stresses are transmitted back to the cells, which adhere to the gel through the ECM, causing the cells to align with the axis of maximum tensile stress. Crucially, we observed that embedded colonies formed by the non-biofilm-forming strain did not exhibit bipolar alignment (Fig. 6A), highlighting the key role of the ECM in establishing cellular ordering within biofilms. These observations were further supported with an in vitro indentation experiment, in which a spherical indenter was used to deform a gel sample in which cells have been dispersed in random orientations (Fig. 6B). Upon indentation, non-biofilm-forming cells maintained random orientations, whereas biofilm-forming cells readily aligned with the direction of the tensile stress in the gel. These observations collectively demonstrate that mechanical stress transmission from the gel to the biofilm via cell–gel adhesion is both necessary and sufficient for bipolar alignment.
4.3. Bulk orientational ordering in interstitial biofilms
Coming back to interstitial biofilms, we also found that the biofilm shape, which is jointly determined by cell–surface friction, gel elasticity, and gel–surface adhesion (Section 3.3), has a profound effect on the 3D organization of cells within the biofilm interior. In particular, Nijjer et al. observed that, as the gel concentration, c, was increased from 0.2% to 1.5%, the dome-shaped biofilm exhibited increasing local nematic order; at c = 1% and c = 1.5%, we also observed the emergence of two boojums, situated at the origin and the apex of the biofilm (Fig. 7A), not unlike those observed in biofilms embedded within stiff gels (Fig. 6A). The dome-to-lens bifurcation near c = 1.5% was accompanied by a topological transition in the cellular orientation field: while the two boojums remained and were smoothly connected by cells in the biofilm core, an additional −1/2 disclination ring was observed in the biofilm interior (Fig. 7B).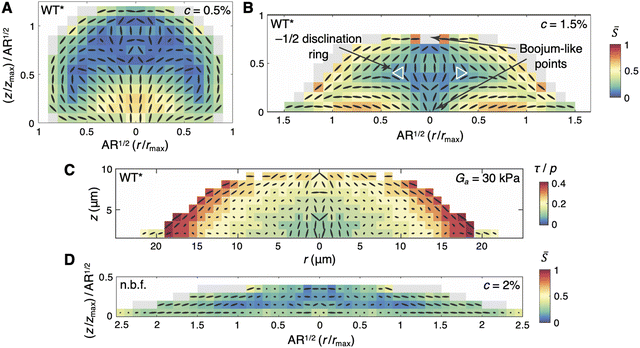 | ||
Fig. 7 Bulk orientational ordering in interstitial biofilms. (A) and (B) Azimuthally averaged cell orientations (ovals) for interstitial WT* biofilms grown under 0.5% (A) and 1.5% (B) gel confinement. Each box is coloured according to the average scalar nematic order parameter, ![[S with combining macron]](https://www.rsc.org/images/entities/i_char_0053_0304.gif) , over cells within the box (see ref. 48 for details). The dimensions of each biofilm were rescaled by their maximum values and further rescaled by the average aspect ratio (AR) over all replicates. Both biofilms exhibit two boojums, one at the apex and another at the origin; the lens-shaped biofilm in panel B additionally exhibits a −1/2 disclination ring within the biofilm interior. (C) Azimuthally averaged first principal stress directions (ovals) for a simulated interstitial WT* biofilm under a gel of stiffness 30 kPa, which corresponds to a 1.5% gel (Fig. 3C). Each box is coloured according to the average stress anisotropy, τ/p, over cells within the box (see ref. 48 for details). (D) Azimuthally averaged cell orientations (ovals) for interstitial colonies formed by the non-biofilm-forming mutant, grown under 2% gel confinement. As in panels A and B, each box is coloured according to the average scalar nematic order parameter, , over cells within the box (see ref. 48 for details). The dimensions of each biofilm were rescaled by their maximum values and further rescaled by the average aspect ratio (AR) over all replicates. Both biofilms exhibit two boojums, one at the apex and another at the origin; the lens-shaped biofilm in panel B additionally exhibits a −1/2 disclination ring within the biofilm interior. (C) Azimuthally averaged first principal stress directions (ovals) for a simulated interstitial WT* biofilm under a gel of stiffness 30 kPa, which corresponds to a 1.5% gel (Fig. 3C). Each box is coloured according to the average stress anisotropy, τ/p, over cells within the box (see ref. 48 for details). (D) Azimuthally averaged cell orientations (ovals) for interstitial colonies formed by the non-biofilm-forming mutant, grown under 2% gel confinement. As in panels A and B, each box is coloured according to the average scalar nematic order parameter, ![[S with combining macron]](https://www.rsc.org/images/entities/i_char_0053_0304.gif) , over cells within the box (see ref. 48 for details), and the biofilm dimensions were rescaled as described above. All panels were adapted with permission from ref. 48. , over cells within the box (see ref. 48 for details), and the biofilm dimensions were rescaled as described above. All panels were adapted with permission from ref. 48. | ||
To identify the mechanisms underlying this topological transition, we turned to ABMs, which allowed for the quantification of the mechanical stress experienced by each cell.48,93 This analysis revealed that the orientational ordering in interstitial biofilms is driven by stress anisotropy (Fig. 7C), due to shear stress that arises from growth-induced expansion of the biofilm–gel interface, and results in the alignment of cells parallel to the interface. This is in contrast with cells on the rigid substrate, which do not seek to align with the minimal compressive stress, but rather align radially due to growth-induced flow (as described in Section 4.1; Fig. 5B).
The emergence of the −1/2 disclination ring in lens-shaped biofilms can be explained by considering the contact angle, ψ, at the triple-contact point between the biofilm, gel, and surface. In dome-shaped biofilms, a large value for ψ allows for a smooth transition of cell orientations between the biofilm–gel and biofilm–surface interfaces. Therefore, no topological defects beyond the two boojums are necessary. However, in lens-shaped biofilms, smoothly transitioning between the biofilm–gel and biofilm–surface interfaces requires a splay conformation at the triple-contact point. Therefore, assuming axisymmetry, as we traverse the boundary of an azimuthal cross-section of the biofilm clockwise, the two boojums and two splay points together incur an angular change of −2π, which necessitates a defect of charge −1 in the biofilm interior. This constraint is satisfied by the −1/2 disclination ring. Indeed, in non-surface-adherent biofilms, the verticalized core and radial alignment in the basal layer are eliminated, resulting in the disappearance of the boojum at the origin and the −1/2 disclination ring. In interstitial colonies formed by the non-biofilm-forming strain, cells assume random orientations much like in classical bacterial colonies (Fig. 7D).
5. How soft confinement modulates cell trajectories
5.1. “Fountain-like” trajectories in unconfined biofilms
The experiments we have discussed thus far have mostly relied on imaging with a somewhat low acquisition rate (e.g., one acquisition every 30 minutes), which strikes an appropriate balance between the need to minimize phototoxicity and the need for sufficiently high temporal resolution to observe how biofilm organization evolves over time. However, this temporal resolution is not sufficient for tracking the motion of individual cells within the biofilm, which can reach velocities of tens of microns per hour for cells at the periphery; nor is it suitable for tracking cell division events within the biofilm, which is necessary if one is interested in assembling a lineage map for the biofilm.To resolve the incipient issue of phototoxicity while imaging V. cholerae biofilms at high temporal resolution (while maintaining high spatial resolution), the Bassler lab29 implemented an imaging setup based on dual-view light-sheet microscopy (DiSPIM), which accommodates high spatiotemporal resolution while incurring much less photobleaching than with spinning disk confocal microscopy,22,94 albeit at the expense of limited optical sectioning. To overcome this limitation—which is especially relevant in V. cholerae biofilms, due to their thickness and density—Qin et al. engineered a V. cholerae strain that constitutively expresses a fluorescently labelled viral protein, μNS, which self-assembles into a bright polar punctum in the cytoplasm.29,95 The punctum localizes to one of the two poles of the cell and, upon division, is inherited by the daughter cell corresponding to that pole. In this way, Qin et al. were able to track the trajectories of these puncta with high temporal resolution.
Using this approach, Qin et al. found that cell trajectories within unconfined wild-type V. cholerae biofilms exhibit two distinct stages. (Here, “wild-type” refers to a clinical V. cholerae isolate that retains the full complement of ECM components and their underlying regulatory networks.96) In the initial phase (<5 hours), during which cells within the biofilm core had not yet verticalized, the puncta followed in-plane trajectories that resembled Brownian motion, frequently and randomly changing their directions of motion. In particular, the mean squared displacement (MSD) of the puncta was found to roughly scale as τ1.2, where τ is the time between consecutive puncta position measurements. Subsequently, the puncta began to follow more directed trajectories that extended both along the surface and throughout the bulk of the biofilm, with the MSD scaling as τ1.8 by 7 hours. It was in this phase that Qin et al. observed a collective “fountain-like” flow comprising three classes of cell trajectories (Fig. 8A): (1) cells that remained trapped on the surface and remained close to the biofilm centre, due to cell–surface friction; (2) cells that extended radially outward from the biofilm centre; and (3) cells that extended radially outward, then arced downward towards the surface. The subset of the last group of cells that reached the surface again were able to “overtake” the cells in the first group.
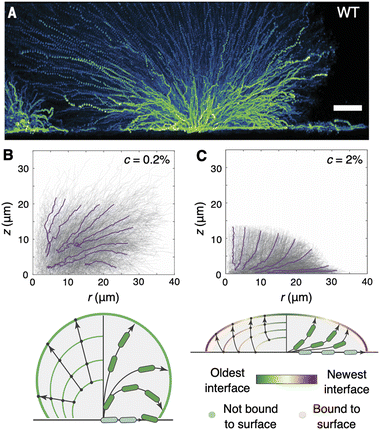 | ||
| Fig. 8 Cell trajectories in growing biofilms. (A) Side view projections of puncta trajectories within a growing wild-type V. cholerae biofilm. Colours indicate the fluorescence intensity of the puncta. Scale bar, 10 μm. Adapted from ref. 29 with permission from the American Association for the Advancement of Science, copyright 2020. (B) and (C) Side view projections of puncta trajectories within growing WT* biofilms under 0.2% (B) and 2% (C) gel confinement, and accompanying schematics of the fountain-like (B) and lotus-like (C) flows that arise under the two regimes. Purple lines denote averaged trajectories that reach the biofilm–gel interface. Panels B–C were adapted with permission from ref. 48. | ||
5.2. A cell trajectory bifurcation in interstitial biofilms
How does this picture change with confinement? Using the same puncta-tracking strategy, we found that cells under soft confinement depart the surface and arc downward, in the same “fountain-like” flow pattern seen in unconfined biofilms (Fig. 8B).48 In contrast, cells under stiff confinement were seen to arc upward, away from the surface, meaning that cells that lay within the basal layer at any given time had remained there for all preceding time (Fig. 8C)—no overtaking is happening on the basal layer.This bifurcation in the trajectory curvature was attributed to the differing progressions of the biofilm–gel interface, within which cells adhere to the gel through the ECM. Indeed, by visualizing the progression of the biofilm–gel interface with tracer particles in the gel (similar to traction force microscopy97), we found that, under soft confinement, the biofilm–gel interface expands in surface area purely by gel deformation, due to the low elastic modulus of the gel and the relative unfavorability of gel–surface delamination, consistent with our continuum theory (Section 3.3); therefore, little “new” biofilm–gel interface is created (Fig. 8B). In contrast, under stiff confinement, the system continually creates new biofilm–gel interface through gel–surface delamination, and “old” portions of the biofilm–gel interface are continually displaced upward from the surface (Fig. 8C). Cells are anchored at the upward-moving biofilm–gel interface through the ECM, and therefore cannot bend downward as in unconfined biofilms, or in softly confined biofilms for which the gel does not provide sufficiently strong anchoring. Consequently, the cell trajectories arc upward and create a “lotus-like” pattern instead of the fountain-like pattern. Indeed, when non-biofilm-forming cells were subjected to stiff confinement, the global lens-shaped geometry was maintained but the upward cell trajectories were not, providing further evidence that the evolution of the biofilm–gel interface, coupled with cell–gel adhesion, determines the positional fate of each cell within the biofilm.
6. Conclusions
In this perspective, we have outlined a series of recent studies, mostly from our group, that detail the spatiotemporal organization of biofilms under soft confinement. Each aspect of this organization—the global geometry of the biofilm, the orientational ordering of the constituent cells, and the trajectories that cells follow throughout biofilm growth—is fundamentally controlled by the mechanical interplay between the biofilm and its confining medium. We have seen how this interplay differentially manifests in embedded and interstitial biofilms, arising from the differing energetic costs of the several ways that the biofilm interacts with and transforms its environment: it deforms the gel (both embedded and interstitial), expands along the surface (interstitial only), and delaminates the gel from the surface (interstitial only). A vital ingredient for the realization of this interplay is the ECM, which is not only what limits biofilm expansion along the surface through cell–surface friction, but also what facilitates stress transmission from the gel into the biofilm through cell–gel adhesion. It is through these effects that the remarkable diversity of anisotropic geometries, cellular patterns, and topological defects we have described above come into being.These studies were made feasible by a customized confocal microscopy and analysis platform that was designed to overcome the challenges of resolving single bacterial cells within dense biofilms with high spatiotemporal resolution. But an equally significant aspect of the above studies is the usage of computational approaches, namely ABMs and continuum models, to pinpoint plausible physical mechanisms for the experimental observations and—perhaps more importantly—generate novel, quantitative predictions that can be tested with subsequent experiments. ABMs and continuum models have both been frequently wielded as tools in the search for mechanistic explanations for interesting phenomena in microbial communities27,75–78,80–84,86,91,98 (for a recent review, see ref. 99), and these studies demonstrate their continued importance to the field.
It is instructive to compare these confined biofilms to eukaryotic tissues, whose developmental trajectories are well-known to be influenced by mechanical interactions with their environments. Indeed, studies of villification in the chick gut52 and follicle patterning in the avian skin53 have revealed striking examples of intricate cellular patterns that arise from compressive stresses from neighbouring tissues (in the case of villi) or reciprocal interactions between the cells and the surrounding ECM (in the case of follicles). However, another hallmark of eukaryotic development is the specification of distinct cell fates, through modulation of molecular signalling, in response to these mechanical interactions.100,101 Such cell-fate specification is not only essential for establishing the diversity of cell-types that make up a multicellular organism, but also can drive higher-order morphogenesis through collective cell migration102 and sorting.103 Such effects likely do not exist in the confined biofilms examined in the above studies, as these biofilms were all formed from the rugose V. cholerae strain (or mutants thereof), which is “locked” in a biofilm-forming phenotype.85 However, phenotypic heterogeneity is widely appreciated to be common among microbial communities,104,105 and a growing body of evidence suggests that such phenotypic heterogeneity among biofilm-dwelling cells may further shape the organization of a developing biofilm.106 Much further work remains to be done to elucidate these effects.
The studies we have described here lay the groundwork for many future directions, including:
• Lineage tracking, which may be considered the “holy grail” of single-cell biofilm imaging. Lineage tracking poses an especially thorny technical challenge in 3D biofilms, as it will require imaging biofilms with sufficiently high spatiotemporal resolution to unambiguously resolve every division event and track the subsequent motion of the daughter cells.28
• Imaging of biofilms on surfaces or in environments that better mimic host tissues and mucosa,32 or natural habitats such as soil.42–45
• Gene expression profiling in live biofilms. A spatial transcriptomics protocol, based on a variant of single-molecule fluorescence in situ hybridization (FISH), was recently successfully repurposed for bacteria,107,108 revealing an unprecedented, spatially resolved view of gene expression in fixed biofilms. But our ability to simultaneously image the expression of multiple genes in live biofilms remains severely limited.
• Relating cellular and ECM organization within biofilms. As we have repeatedly seen throughout this perspective, the ECM plays a crucial role in dictating cellular organization within biofilms, but how the ECM—which is dynamically produced and secreted by the cells—is distributed throughout the biofilm as the cells proliferate remains technically difficult to elucidate. For example, recent work from the Drescher lab demonstrated that, by mapping cellular displacement upon shear, in situ microrheology can be performed on biofilms,109 whereby heterogeneous mechanical properties within the biofilm could be identified and correlated with the heterogeneous spatial distribution of the various ECM components.
• Expanding the single-cell approach to multispecies biofilms. Though we have made great strides towards understanding how single-species biofilms develop, biofilms in the wild are most often thought to be polymicrobial.110,111 The biofilms that constitute dental plaque, for example, contain hundreds of species with beautiful spatial organizations revealed by combinatorial labelling and spectral imaging FISH in fixed samples.112 A major challenge is therefore to apply the live single-cell imaging approach reviewed here to multispecies biofilms and reveal the contributions of interspecies interactions to the developmental process.
Fulfilling these future directions will require overcoming numerous technical challenges: photobleaching and phototoxicity remain ever-present issues in fluorescence microscopy; current live imaging platforms only allow for the simultaneous imaging of a few channels; and, despite recent advances, accurate cell segmentation of 3D microbial communities remains difficult. We hope that the steady introduction of novel tools and techniques, such as optical protocols for reducing photobleaching and phototoxicity,113 more photostable fluorescent proteins,114 multispectral imaging,115 and deep-learning-based methods for cell segmentation116,117 will incrementally pave the way towards achieving these goals in the long term.
Author contributions
K.-M. N. and J. Y. both wrote the paper.Data availability
This article does not contain original data.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to Jung-Shen B. Tai for helpful discussions. J. Y. acknowledges support from the National Institute of General Medical Sciences of the National Institutes of Health under Award Number DP2GM146253 and the Alfred P. Sloan Foundation (FG-2023-20857).Notes and references
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Nat. Rev. Microbiol., 2004, 2, 95–108 CrossRef CAS PubMed.
- J.-P. Motta, J. L. Wallace, A. G. Buret, C. Deraison and N. Vergnolle, Nat. Rev. Gastroenterol. Hepatol., 2021, 18, 314–334 CrossRef PubMed.
- H.-C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Nat. Rev. Microbiol., 2016, 14, 563–575 CrossRef CAS PubMed.
- S. Mishra, Y. Huang, J. Li, X. Wu, Z. Zhou, Q. Lei, P. Bhatt and S. Chen, Chemosphere, 2022, 294, 133609 CrossRef CAS PubMed.
- J. Yan and B. L. Bassler, Cell Host Microbe, 2019, 26, 15–21 CrossRef CAS PubMed.
- L. K. Vestby, T. Grønseth, R. Simm and L. L. Nesse, Antibiotics, 2020, 9, 59 CrossRef CAS PubMed.
- O. Ciofu, C. Moser, P. Ø. Jensen and N. Høiby, Nat. Rev. Microbiol., 2022, 20, 621–635 CrossRef CAS PubMed.
- M. C. Marchetti, J. F. Joanny, S. Ramaswamy, T. B. Liverpool, J. Prost, M. Rao and R. A. Simha, Rev. Mod. Phys., 2013, 85, 1143 CrossRef CAS.
- D. Needleman and Z. Dogic, Nat. Rev. Mater., 2017, 2, 17048 CrossRef CAS.
- O. Hallatschek, S. S. Datta, K. Drescher, J. Dunkel, J. Elgeti, B. Waclaw and N. S. Wingreen, Nat. Rev. Phys., 2023, 5, 407–419 CrossRef PubMed.
- J. Yan, A. G. Sharo, H. A. Stone, N. S. Wingreen and B. L. Bassler, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E5337–E5343 CAS.
- A. Doostmohammadi, J. Ignés-Mullol, J. M. Yeomans and F. Sagués, Nat. Commun., 2018, 9, 3246 CrossRef PubMed.
- M. Bär, R. Großmann, S. Heidenreich and F. Peruani, Annu. Rev. Condens. Matter Phys., 2020, 11, 441–466 CrossRef.
- M. Ballerini, N. Cabibbo, R. Candelier, A. Cavagna, E. Cisbani, I. Giardina, V. Lecomte, A. Orlandi, G. Parisi, A. Procaccini, M. Viale and V. Zdravkovic, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 1232–1237 CrossRef CAS PubMed.
- T. Sanchez, D. T. N. Chen, S. J. DeCamp, M. Heymann and Z. Dogic, Nature, 2012, 491, 431–434 CrossRef CAS PubMed.
- G. Duclos, C. Erlenkämper, J.-F. Joanny and P. Silberzan, Nat. Phys., 2017, 13, 58–62 Search PubMed.
- J. Zhang, R. Alert, J. Yan, N. S. Wingreen and S. Granick, Nat. Phys., 2021, 17, 961–967 Search PubMed.
- S. Das, M. Ciarchi, Z. Zhou, J. Yan, J. Zhang and R. Alert, Phys. Rev. X, 2024, 14, 031008 Search PubMed.
- P. J. Keller, A. D. Schmidt, J. Wittbrodt and E. H. K. Stelzer, Science, 2008, 322, 1065–1069 CrossRef CAS PubMed.
- L. A. Royer, W. C. Lemon, R. K. Chhetri, Y. Wan, M. Coleman, E. W. Myers and P. J. Keller, Nat. Biotechnol., 2016, 34, 1267–1278 CrossRef CAS PubMed.
- K. McDole, L. Guignard, F. Amat, A. Berger, G. Malandain, L. A. Royer, S. C. Turaga, K. Branson and P. J. Keller, Cell, 2018, 175, 859–876.E33 CrossRef CAS PubMed.
- Y. Wan, K. McDole and P. J. Keller, Annu. Rev. Cell Dev. Biol., 2019, 35, 655–681 CrossRef CAS PubMed.
- K. Zhao, B. S. Tseng, B. Beckerman, F. Jin, M. L. Gibiansky, J. J. Harrison, E. Luijten, M. R. Parsek and G. C. L. Wong, Nature, 2013, 497, 388–391 CrossRef CAS PubMed.
- D. O. Serra, A. M. Richter, G. Klauck, F. Mika and R. Hengge, mBio, 2013, 4, e00103-13 CrossRef PubMed.
- E. J. Stewart, A. E. Satorius, J. G. Younger and M. J. Solomon, Langmuir, 2013, 29, 7017–7024 CrossRef CAS PubMed.
- K. Drescher, J. Dunkel, C. D. Nadell, S. van Teeffelen, I. Grnja, N. S. Wingreen, H. A. Stone and B. L. Bassler, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E2066–E2072 CrossRef CAS PubMed.
- R. Hartmann, P. K. Singh, P. Pearce, R. Mok, B. Song, F. Díaz-Pascual, J. Dunkel and K. Drescher, Nat. Phys., 2019, 15, 251–256 Search PubMed.
- G. C. L. Wong, J. D. Antani, P. P. Lele, J. Chen, B. Nan, M. J. Kühn, A. Persat, J.-L. Bru, N. M. Høyland-Kroghsbo, A. Siryaporn, J. C. Conrad, F. Carrara, Y. Yawata, R. Stocker, Y. V. Brun, G. B. Whitfield, C. K. Lee, J. de Anda, W. C. Schmidt, R. Golestanian, G. A. O’Toole, K. A. Floyd, F. H. Yildiz, S. Yang, F. Jin, M. Toyofuku, L. Eberl, N. Nomura, L. A. Zacharoff, M. Y. El-Naggar, S. E. Yalcin, N. S. Malvankar, M. D. Rojas-Andrade, A. I. Hochbaum, J. Yan, H. A. Stone, N. S. Wingreen, B. L. Bassler, Y. Wu, H. Xu, K. Drescher and J. Dunkel, Phys. Biol., 2021, 18, 051501 Search PubMed.
- B. Qin, C. Fei, A. A. Bridges, A. A. Mashruwala, H. A. Stone, N. S. Wingreen and B. L. Bassler, Science, 2020, 369, 71–77 CrossRef CAS PubMed.
- J. Zhang, M. Zhang, Y. Wang, E. Donarski and A. Gahlmann, J. Phys. Chem. B, 2021, 125, 12187–12196 CrossRef CAS PubMed.
- T. Bjarnsholt, P. Ø. Jensen, M. J. Fiandaca, J. Pedersen, C. R. Hansen, C. B. Andersen, T. Pressler, M. Givskov and N. Høiby, Pediatr. Pulmonol., 2009, 44, 547–558 CrossRef PubMed.
- T. Bjarnsholt, M. Alhede, M. Alhede, S. R. Eickhardt-Sørensen, C. Moser, M. Kühl, P. Ø. Jensen and N. Høiby, Trends Microbiol., 2013, 21, 466–474 CrossRef CAS PubMed.
- A. Cont, T. Rossy, Z. Al-Mayyah and A. Persat, eLife, 2020, 9, e56533 CrossRef CAS PubMed.
- G. G. Anderson, J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser and S. J. Hultgren, Science, 2003, 301, 105–107 CrossRef CAS PubMed.
- M. A. Croxen and B. B. Finlay, Nat. Rev. Microbiol., 2010, 8, 26–38 CrossRef CAS PubMed.
- A. L. Flores-Mireles, J. N. Walker, M. Caparon and S. J. Hultgren, Nat. Rev. Microbiol., 2015, 13, 269–284 CrossRef CAS PubMed.
- D. Nejman, I. Livyatan, G. Fuks, N. Gavert, Y. Zwang, L. T. Geller, A. Rotter-Maskowitz, R. Weiser, G. Mallel, E. Gigi, A. Meltser, G. M. Douglas, I. Kamer, V. Gopalakrishnan, T. Dadosh, S. Levin-Zaidman, S. Avnet, T. Atlan, Z. A. Cooper, R. Arora, A. P. Cogdill, Md. A. W. Khan, G. Ologun, Y. Bussi, A. Weinberger, M. Lotan-Pompan, O. Golani, G. Perry, M. Rokah, K. Bahar-Shany, E. A. Rozeman, C. U. Blank, A. Ronai, R. Shaoul, A. Amit, T. Dorfman, R. Kremer, Z. R. Cohen, S. Harnof, T. Siegal, E. Yehuda-Shnaidman, E. N. Gal-Yam, H. Shapira, N. Baldini, M. G. I. Langille, A. Ben-Nun, B. Kaufman, A. Nissan, T. Golan, M. Dadiani, K. Levanon, J. Bar, S. Yust-Katz, I. Barshack, D. S. Peeper, D. J. Raz, E. Segal, J. A. Wargo, J. Sandbank, N. Shental and R. Straussman, Science, 2020, 368, 973–980 CrossRef CAS PubMed.
- J. L. Galeano Niño, H. Wu, K. D. LaCourse, A. G. Kempchinsky, A. Baryiames, B. Barber, N. Futran, J. Houlton, C. Sather, E. Sicinska, A. Taylor, S. S. Minot, C. D. Johnston and S. Bullman, Nature, 2022, 611, 810–817 CrossRef PubMed.
- G. El Tekle and W. S. Garrett, Nat. Rev. Cancer, 2023, 23, 600–618 CrossRef CAS PubMed.
- A. Fu, B. Yao, T. Dong, Y. Chen, J. Yao, Y. Liu, H. Li, H. Bai, X. Liu, Y. Zhang, C. Wang, Y. Guo, N. Li and S. Cai, Cell, 2022, 185, 1356–1372 CrossRef CAS PubMed.
- H.-C. Flemming and S. Wuertz, Nat. Rev. Microbiol., 2019, 17, 247–260 CrossRef CAS PubMed.
- T. Bhattacharjee and S. S. Datta, Nat. Commun., 2019, 10, 2075 CrossRef PubMed.
- T. Bhattacharjee and S. S. Datta, Soft Matter, 2019, 15, 9920 RSC.
- A. Martínez-Calvo, T. Bhattacharjee, R. K. Bay, H. N. Luu, A. M. Hancock, N. S. Wingreen and S. S. Datta, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2208019119 CrossRef PubMed.
- M. Sreepadmanabh, M. Ganesh, P. Sanjenbam, C. Kurzthaler, D. Agashe and T. Bhattacharjee, Nat. Commun., 2024, 15, 9561 CrossRef CAS PubMed.
- Q. Zhang, J. Li, J. Nijjer, H. Lu, M. Kothari, R. Alert, T. Cohen and J. Yan, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2107107118 CrossRef CAS PubMed.
- J. Nijjer, C. Li, Q. Zhang, H. Lu, S. Zhang and J. Yan, Nat. Commun., 2021, 12, 6632 CrossRef CAS PubMed.
- J. Nijjer, C. Li, M. Kothari, T. Henzel, Q. Zhang, J.-S. B. Tai, S. Zhou, T. Cohen, S. Zhang and J. Yan, Nat. Phys., 2023, 19, 1936–1944 Search PubMed.
- T. M. Bartlett, B. P. Bratton, A. Duvshani, A. Miguel, Y. Sheng, N. R. Martin, J. P. Nguyen, A. Persat, S. M. Desmarais, M. S. VanNieuwenhze, K. C. Huang, J. Zhu, J. W. Shaevitz and Z. Gitai, Cell, 2017, 168, 172–185.E15 CrossRef CAS PubMed.
- S. R. K. Vedula, M. C. Leong, T. L. Lai, P. Hersen, A. J. Kabla, C. T. Lim and B. Ladoux, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 12974–12979 CrossRef CAS PubMed.
- R. Ienaga, K. Beppu and Y. T. Maeda, Soft Matter, 2023, 19, 5016 RSC.
- A. E. Shyer, T. Tallinen, N. L. Nerurkar, Z. Wei, E. S. Gil, D. L. Kaplan, C. J. Tabin and L. Mahadevan, Science, 2013, 342, 212–218 CrossRef CAS PubMed.
- K. H. Palmquist, S. F. Tiemann, F. L. Ezzeddine, S. Yang, C. R. Pfeifer, A. Erzberger, A. R. Rodrigues and A. E. Shyer, Cell, 2022, 185, 1960–1973.e11 CrossRef CAS PubMed.
- C. R. Pfeifer, A. E. Shyer and A. R. Rodrigues, Curr. Opin. Cell Biol., 2024, 86, 102305 CrossRef CAS PubMed.
- J. K. Teschler, D. Zamorano-Sánchez, A. S. Utada, C. J. A. Warner, G. C. L. Wong, R. G. Linington and F. H. Yildiz, Nat. Rev. Microbiol., 2015, 13, 255–268 CrossRef CAS PubMed.
- U. Jenal, A. Reinders and C. Lori, Nat. Rev. Microbiol., 2017, 15, 271–284 CrossRef CAS PubMed.
- J. G. Conner, D. Zamorano-Sánchez, J. H. Park, H. Sondermann and F. H. Yildiz, Curr. Opin. Microbiol., 2017, 36, 20–29 CrossRef CAS PubMed.
- J. K. Teschler, C. D. Nadell, K. Drescher and F. H. Yildiz, Annu. Rev. Microbiol., 2022, 76, 503–532 CrossRef CAS PubMed.
- A. A. Bridges, J. A. Prentice, N. S. Wingreen and B. L. Bassler, Annu. Rev. Microbiol., 2022, 76, 235–257 CrossRef CAS PubMed.
- K. P. Rumbaugh and K. Sauer, Nat. Rev. Microbiol., 2020, 18, 571–586 CrossRef CAS PubMed.
- K. Sauer, P. Stoodley, D. M. Goeres, L. Hall-Stoodley, M. Burmølle, P. S. Stewart and T. Bjarnsholt, Nat. Rev. Microbiol., 2022, 20, 608–620 CrossRef CAS PubMed.
- M. F. Moradali and B. H. A. Rehm, Nat. Rev. Microbiol., 2020, 18, 195–210 CrossRef CAS PubMed.
- L. Karygianni, Z. Ren, H. Koo and T. Thurnheer, Trends Microbiol., 2020, 28, 668–681 CrossRef CAS PubMed.
- H.-C. Flemming, E. D. van Hullebusch, T. R. Neu, P. H. Nielsen, T. Seviour, P. Stoodley, J. Wingender and S. Wuertz, Nat. Rev. Microbiol., 2023, 21, 70–86 CrossRef CAS PubMed.
- J. Yan, C. D. Nadell, H. A. Stone, N. S. Wingreen and B. L. Bassler, Nat. Commun., 2017, 8, 327 CrossRef PubMed.
- J. Yan, A. Moreau, S. Khodaparast, A. Perazzo, J. Feng, C. Fei, S. Mao, S. Mukherjee, A. Košmrlj, N. S. Wingreen, B. L. Bassler and H. A. Stone, Adv. Mater., 2018, 30, e1804153 CrossRef PubMed.
- Q. Zhang, D. Nguyen, J.-S. B. Tai, X. Xu, J. Nijjer, X. Huang, Y. Li and J. Yan, Adv. Funct. Mater., 2022, 32, 2110699 CrossRef CAS.
- F. H. Yildiz and G. K. Schoolnik, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 4028–4033 CrossRef CAS PubMed.
- F. Yildiz, J. Fong, I. Sadovskaya, T. Grard and E. Vinogradov, PLoS One, 2014, 9, e86751 CrossRef PubMed.
- J. C. N. Fong, K. Karplus, G. K. Schoolnik and F. H. Yildiz, J. Bacteriol., 2006, 188, 1049–1059 CrossRef CAS PubMed.
- J. C. N. Fong, A. Rogers, A. K. Michael, N. C. Parsley, W.-C. Cornell, Y.-C. Lin, P. K. Singh, R. Hartmann, K. Drescher, E. Vinogradov, L. E. P. Dietrich, C. L. Partch and F. H. Yildiz, eLife, 2017, 6, e26163 CrossRef PubMed.
- C. Absalon, K. Van Dellen and P. I. Watnick, PLoS Pathog., 2011, 7, e1002210 CrossRef CAS PubMed.
- V. Berk, J. C. N. Fong, G. T. Dempsey, O. N. Develioglu, X. Zhuang, J. Liphardt, F. H. Yildiz and S. Chu, Science, 2012, 337, 236–239 CrossRef CAS PubMed.
- X. Huang, T. Nero, R. Weerasekera, K. H. Matej, A. Hinbest, Z. Jiang, R. F. Lee, L. Wu, C. Chak, J. Nijjer, I. Gibaldi, H. Yang, N. Gamble, W.-L. Ng, S. A. Malaker, K. Sumigray, R. Olson and J. Yan, Nat. Commun., 2023, 14, 2104 CrossRef CAS PubMed.
- F. Beroz, J. Yan, Y. Meir, B. Sabass, H. A. Stone, B. L. Bassler and N. S. Wingreen, Nat. Phys., 2018, 14, 954–960 Search PubMed.
- M. A. A. Grant, B. Wacław, R. J. Allen and P. Cicuta, J. R. Soc., Interface, 2014, 11, 20140400 CrossRef PubMed.
- M. R. Warren, H. Sun, Y. Yan, J. Cremer, B. Li and T. Hwa, eLife, 2019, 8, e41093 CrossRef PubMed.
- Z. You, D. J. G. Pearce, A. Sengupta and L. Giomi, Phys. Rev. Lett., 2019, 123, 178001 CrossRef CAS PubMed.
- F. D. C. Farrell, O. Hallatschek, D. Marenduzzo and B. Waclaw, Phys. Rev. Lett., 2013, 111, 168101 CrossRef CAS PubMed.
- A. Doostmohammadi, S. P. Thampi and J. M. Yeomans, Phys. Rev. Lett., 2016, 117, 048102 CrossRef PubMed.
- Z. You, D. J. G. Pearce, A. Sengupta and L. Giomi, Phys. Rev. X, 2018, 8, 031065 CAS.
- Z. You, D. J. G. Pearce and L. Giomi, Sci. Adv., 2021, 7, eabc8685 CrossRef CAS PubMed.
- D. Dell’Arciprete, M. L. Blow, A. T. Brown, F. D. C. Farrell, J. S. Lintuvuori, A. F. McVey, D. Marenduzzo and W. C. K. Poon, Nat. Commun., 2018, 9, 4190 CrossRef PubMed.
- M. Basaran, Y. I. Yaman, T. C. Yüce, R. Vetter and A. Kocabas, eLife, 2022, 11, e72187 CrossRef CAS PubMed.
- S. Beyhan and F. H. Yildiz, Mol. Microbiol., 2007, 63, 995–1007 CrossRef CAS PubMed.
- P. Pearce, B. Song, D. J. Skinner, R. Mok, R. Hartmann, P. K. Singh, H. Jeckel, J. S. Oishi, K. Drescher and J. Dunkel, Phys. Rev. Lett., 2019, 123, 258101 CrossRef CAS PubMed.
- J. Li, M. Kothari, S. Chockalingam, T. Henzel, Q. Zhang, X. Li, J. Yan and T. Cohen, J. Mech. Phys. Solids, 2022, 159, 104709 CrossRef.
- C.-Y. Lai, Z. Zheng, E. Dressaire and H. A. Stone, Philos. Trans. R. Soc., A, 2016, 374, 20150425 CrossRef PubMed.
- N. J. O’Keeffe, H. E. Huppert and P. F. Linden, J. Fluid Mech., 2018, 844, 435–458 CrossRef.
- T. Henzel, J. Nijjer, S. Chockalingam, H. Wahdat, A. J. Crosby, J. Yan and T. Cohen, PNAS Nexus, 2022, 1, 1–8 CrossRef PubMed.
- D. Volfson, S. Cookson, J. Hasty and L. S. Tsimring, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15346–15351 CrossRef PubMed.
- D. Andrienko, J. Mol. Liq., 2018, 267, 520–541 CrossRef CAS.
- C. Li, J. Nijjer, L. Feng, Q. Zhang, J. Yan and S. Zhang, Soft Matter, 2024, 20, 3401 RSC.
- Y. Wu, P. Wawrzusin, J. Senseney, R. S. Fischer, R. Christensen, A. Santella, A. G. York, P. W. Winter, C. M. Waterman, Z. Bao, D. A. Colón-Ramos, M. McAuliffe and H. Shroff, Nat. Biotechnol., 2013, 31, 1032–1038 CrossRef CAS PubMed.
- B. R. Parry, I. V. Surovtsev, M. T. Cabeen, C. S. O’Hern, E. R. Dufresne and C. Jacobs-Wagner, Cell, 2014, 156, 183–194 CrossRef CAS PubMed.
- K. H. Thelin and R. K. Taylor, Infect. Immun., 1996, 64, 2853–2856 CrossRef CAS PubMed.
- R. W. Style, R. Boltyanskiy, G. K. German, C. Hyland, C. W. MacMinn, A. F. Mertz, L. A. Wilen, Y. Xu and E. R. Dufresne, Soft Matter, 2014, 10, 4047–4055 RSC.
- B. J. R. Cockx, T. Foster, R. J. Clegg, K. Alden, S. Arya, D. J. Stekel, B. F. Smets and J.-U. Kreft, PLoS Comput. Biol., 2024, 20, e1011303 CrossRef CAS PubMed.
- K. Nagarajan, C. Ni and T. Lu, ACS Synth. Biol., 2022, 11, 3564–3574 CrossRef CAS PubMed.
- Y. Maroudas-Sacks and K. Keren, Annu. Rev. Cell Dev. Biol., 2021, 37, 469–493 CrossRef CAS PubMed.
- H. De Belly, E. K. Paluch and K. J. Chalut, Nat. Rev. Mol. Cell Biol., 2022, 23, 465–480 CrossRef CAS PubMed.
- E. H. Barriga, K. Franze, G. Charras and R. Mayor, Nature, 2018, 554, 523–527 CrossRef CAS PubMed.
- J.-L. Maître, H. Berthoumieux, S. F. G. Krens, G. Salbreux, F. Jülicher, E. Paluch and C.-P. Heisenberg, Science, 2012, 338, 253–256 CrossRef PubMed.
- J.-W. Veening, W. K. Smits and O. P. Kuipers, Annu. Rev. Microbiol., 2008, 62, 193–210 CrossRef CAS PubMed.
- J. Jo, A. Price-Whelan and L. E. P. Dietrich, Nat. Rev. Microbiol., 2022, 20, 593–607 CrossRef CAS PubMed.
- G. R. Squyres and D. K. Newman, Curr. Opin. Microbiol., 2024, 82, 102537 CrossRef CAS PubMed.
- D. Dar, N. Dar, L. Cai and D. K. Newman, Science, 2021, 373, eabi4882 CrossRef CAS PubMed.
- G. E. Johnson, C. Fei, N. S. Wingreen and B. L. Bassler, bioRxiv, 2024, preprint, bioRxiv:2024.07.17.603784 DOI:10.1101/2024.07.17.603784.
- T. Ohmura, D. J. Skinner, K. Neuhaus, G. P. T. Choi, J. Dunkel and K. Drescher, Adv. Mater., 2024, 36, 2314059 CrossRef CAS PubMed.
- R. Wolcott, J. W. Costerton, D. Raoult and S. J. Cutler, Clin. Microbiol. Infect., 2013, 19, 107–112 CrossRef CAS PubMed.
- W. H. Bowen, R. A. Burne, H. Wu and H. Koo, Trends Microbiol., 2018, 26, 229–242 CrossRef CAS PubMed.
- J. L. Mark Welch, B. J. Rossetti, C. W. Rieken, F. E. Dewhirst and G. G. Borisy, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E791–E800 CrossRef CAS PubMed.
- L. Ludvikova, E. Simon, M. Deygas, T. Panier, M.-A. Plamont, J. Ollion, A. Tebo, M. Piel, L. Jullien, L. Robert, T. Le Saux and A. Espagne, Nat. Biotechnol., 2024, 42, 872–876 CrossRef CAS PubMed.
- M. Hirano, R. Ando, S. Shimozono, M. Sugiyama, N. Takeda, H. Kurokawa, R. Deguchi, K. Endo, K. Haga, R. Takai-Todaka, S. Inaura, Y. Matsumura, H. Hama, Y. Okada, T. Fujiwara, T. Morimoto, K. Katayama and A. Miyawaki, Nat. Biotechnol., 2022, 40, 1132–1142 CrossRef CAS PubMed.
- A. Kumar, K. E. McNally, Y. Zhang, A. Haslett-Saunders, X. Wang, J. Guillem-Marti, D. Lee, B. Huang, R. R. Kay, D. Baker, E. Derivery and J. D. Manton, bioRxiv, 2024, preprint, bioRxiv:2024.06.12.597784 DOI:10.1101/2024.06.12.597784.
- H. Jeckel and K. Drescher, FEMS Microbiol. Rev., 2021, 45, fuaa062 CrossRef CAS PubMed.
- E. Jelli, T. Ohmura, N. Netter, M. Abt, E. Jiménez-Siebert, K. Neuhaus, D. K. H. Rode, C. D. Nadell and K. Drescher, Mol. Microbiol., 2023, 119, 659–676 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |


