Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Programmable short peptides for modulating stem cell fate in tissue engineering and regenerative medicine
Rohan
Vishwanath
a,
Abhijit
Biswas
b,
Unnati
Modi
b,
Sharad
Gupta
 b,
Dhiraj
Bhatia
b,
Dhiraj
Bhatia
 *b and
Raghu
Solanki
*b and
Raghu
Solanki
 *b
*b
aSchool of Life Science, Central University of Gujarat, Gandhinagar-382030, India
bDepartment of Biological Sciences and Engineering, Indian Institute of Technology Gandhinagar, Palaj, Gujarat 382355, India. E-mail: raghu.solanki@iitgn.ac.in; dhiraj.bhatia@iitgn.ac.in
First published on 14th January 2025
Abstract
Recent advancements in tissue engineering and regenerative medicine have introduced promising strategies to address tissue and organ deficiencies. This review highlights the critical role of short peptides, particularly their ability to self-assemble into matrices that mimic the extracellular matrix (ECM). These low molecular weight peptides exhibit target-specific activities, modulate gene expression, and influence cell differentiation pathways. They are stable, programmable, non-cytotoxic, biocompatible, biodegradable, capable of crossing the cell membrane and easy to synthesize. This review underscores the importance of peptide structure and concentration in directing stem cell differentiation and explores their diverse biomedical applications. Peptides such as Aβ1–40, Aβ1–42, RADA16, A13 and KEDW are discussed for their roles in modulating stem cell differentiation into neuronal, glial, myocardial, osteogenic, hepatocyte and pancreatic lineages. Furthermore, this review delves into the underlying signaling mechanisms, the chemistry and design of short peptides and their potential for engineering biocompatible materials that mimic stem cell microenvironments. Short peptide-based biomaterials and scaffolds represent a promising avenue in stem cell therapy, tissue engineering, and regenerative medicine.
1. Introduction
Stem cell research investigates the fundamental properties of stem cells and their potential applications in regenerative medicine.1,2 Current treatment strategies focus on stem cell-based therapies, aiming to replace damaged or diseased tissues with healthy cells derived from stem cells.3 In the 1960s, Friedenstein pioneered stem cell research by isolating and characterizing bone marrow-derived cells from guinea pigs, demonstrating their ability to differentiate into various cell types.4,5 For more than six decades, transplantation of hematopoietic stem cells (HSCs) has remained the primary curative therapy for a wide range of genetic and hematological disorders.6 Approximately half a century ago, Till and McCulloch identified a distinct progenitor cell type within the bone marrow capable of clonal expansion and differentiation into all hematopoietic cell lineages, marking a pivotal advancement in HSC research.7 The discovery of mouse embryonic stem cells (mESCs) revolutionized developmental biology by establishing mice as invaluable models for studying mammalian stem cell biology.8,9 Despite their affordability and genetic manipulability, the utility of mESCs in regenerative medicine remains limited due to ethical and technical challenges. In 2006, Yamanaka and Takahashi demonstrated that multipotent adult stem cells could be reprogrammed into induced pluripotent stem cells (iPSCs).10 Over time, these breakthroughs have paved the way for developing biocompatible cells for therapeutic applications.Stem cells play a pivotal role in biomedicine by advancing regenerative therapies and improving our understanding of complex human biological processes.11 They serve as a robust and cost-effective tool for investing disease mechanisms and discovering new treatments for diseases such as sickle cell anemia, liver disease and COVID-19 infection.12 However, stem-cell based approaches face limitations such as immune rejection, tumorigenicity, and ethical concerns associated with embryonic stem cells.13
Short peptides have emerged as promising tools in stem cell research, offering precise control over stem cell behaviour through modulation of signaling pathways, promoting cell adhesion, cell growth, and enabling targeted delivery of therapeutic agents.14 Their versatility and ease of synthesis make peptides valuable in developing safer and more effective stem cell-based therapies for various diseases and injuries.15 Short peptides are fundamental biomolecules that constitute proteins and possess unique advantages; rendering them critical in stem cell-based regenerative biomedicine.16 They can traverse cell membranes and be easily modified to perform specific biological functions. Short peptides enhance the adhesion, growth, and proliferation of stem cells and utilize various mechanisms to penetrate cell membranes and activate signaling pathways that regulate gene expression and differentiation.17
The concentration and structure of short peptides influence the direction of stem cell differentiation. The highly cationic nature of short polypeptides enables them to penetrate cell membranes and facilitate the delivery of other molecules, such as biological cargos.18 In addition to enhancing the production of neural differentiation markers, the short peptides including KE (Lys-Glu), AED (Ala-Glu-Asp), KED (Lys-Glu-Asp), and AEDG (Ala-Glu-Asp-Gly) peptides can stimulate neural differentiation in stem cells.19 The KED peptide has the ability to promote neuronal development, which may contribute to its neuroprotective effects.20 Research indicates that the KED peptide regulates the expression of genes related to cell aging and death (p16, p21), neural development (NES, GAP43), and Alzheimer's disease pathogenesis (SUMO, APOE, and IGF1).21
Self-assembling peptides (SAPs) are favoured for their nanoscale porosity and fibrous topographies, which resemble the natural ECM. In particular, RADA16 has been shown to promote proliferation and differentiation across various cell types, including stem cells, progenitors, and established cell lines from humans, mice, chickens and other animals.14 The self-assembling property of these peptides and their adaptable molecular design offer advantages in managing scaffold degradation, enhancing cell adhesion, and controlled delivery of growth factors within encapsulated cells. These characteristics make SAPs versatile for improving scaffold–cell interactions in tissue regenerative applications such as cartilage repair.22
In this review article, we provide a comprehensive overview of the role of short peptides in stem cell research. We discuss the applications of various short peptides in modulating stem cell fate, with a focus on recent evidence supporting their roles in neuronal differentiation, epithelial differentiation, myocardial differentiation, osteogenic differentiation, pancreatic differentiation, and hepatocyte differentiation. By delving into these diverse applications, we highlight the significance of short peptides in influencing stem cell differentiation pathways and their potential for advancing the field of regenerative medicine. This review aims to offer insights into the chemistry, design, and functional applications of short peptides in creating biocompatible microenvironments for effective stem cell therapy and tissue engineering.
2. Short peptides: characteristics and advantages as a modulator for stem cell fate
Peptides are short chains of amino acids, consisting of two to fifty residues, linked by covalent bonds formed through condensation reactions.23 These peptides are fundamental biomolecules that play critical roles in various biological processes and are integral components in fields such as regenerative medicine and biotechnology.24 One of the many advantages of short peptides is their ease of synthesis, development, and analysis.25 They offer a range of possibilities for incorporating functionality, and exhibit excellent biocompatibility and biodegradability.26,27 Short peptides have demonstrated neuroprotective and anti-carcinogenic properties in various experimental models, and they are considered safe for extended administration in animals.28Short peptides can be categorized based on their biological functions (Fig. 1). Antimicrobial peptides (AMPs) help to fight against microbial infections by disrupting microbial membranes.29–32 Bioactive peptides exhibit therapeutic properties including anti-cancer, immunomodulatory, anti-hyperuricemia, anti-inflammatory and other human health promoting properties.33–36 Molecular self-assembly of peptides refers to the process by which individual peptide molecules spontaneously organize into ordered structures through non-covalent weak interactions.26,37 Hydrophobic and hydrophilic amino acids are present in the structure of small peptides, which have ability to self-organize into 3D nanofiber structures such as a stable hydrogel. Self-assembling peptides interact with stem cells to improve their activity or survival, resulting in intended outcomes. They help alleviate symptoms of arthritis, heart dysfunction, and neurodegenerative diseases (like Parkinson's and Alzheimer's).38 They help to enhance angiogenesis in the case of vascular disorders. Neuroplasticity processes can be promoted by neural differentiation of stem cells. Protein and peptide self-assembly play a role in both biological function and malfunction. Collagen fibril networks serve as a biochemical framework, controlling the mechanical characteristics and shape of biological tissue in a variety of ways.39 The pathological hallmarks of catastrophic degenerative diseases have long been thought to be protein misfolding and the production of amyloid fibrils.40
There are several factors that can affect peptide self-assembly such as pH and temperature.41 Changes in pH result in H-bond and salt bridges, which disrupt the structure of peptides. Protonation and deprotonation can occur due to changes in pH in the amino acid side chains. pH-responsive amino acids and chemical linkers can help to modify various nanostructures by peptides that can undergo self-assembly.42 These systems are therefore highly biocompatible and easily adjustable, making them excellent options for drug release as well as fibrous materials that can imitate the natural ECM.43 Peptides exist as a monomer initially, but upon heating, they can convert into various forms such as nanofibers, micelles and other peptide structures including non-specific aggregate-like networks depending on the buffer's pH and calcium content.44 The decrease in temperature promotes the formation of hydrogen bonds through constant increasing ionization, leading to the formation of a nanowire structure and facilitating self-assembly.45
Experiments on synthetic peptides and complex peptide bioregulators have revealed their specific effects and proposed major molecular mechanisms underlying their biological activity.46 Research has shown that short peptides influence gene expression, cell division, apoptosis, differentiation, and organ function.47,48 Short peptides have been shown to be crucial for restoring age-related genetically conditioned changes, modulating transcription and facilitating biological information transfer.49
Reported studies suggest that peptides also play crucial roles in stem cell research, particularly in reprogramming and differentiation processes50,51 (discussed in Section 4 in-depth). They can act as signaling molecules to induce specific differentiation pathways, regulating stem cells towards certain cell types such as neurons, cardiomyocytes, or other specific cells.52 Peptides also enhance the efficiency of reprogramming techniques by influencing gene expression patterns and epigenetic modifications, thereby promoting the development of functional and specialized cell populations in regenerative medicine and disease modeling.50,53 These peptides are integral components of proteins, offering significant advantages for stem cell-based regenerative therapy.54 They have very low molecular weight and target specific activity and they are stable, versatile, cost-effective, non-cytotoxic, biodegradable, biocompatible, capable of crossing cell membranes and easy to modify for specific applications.55
3. Stem cell niche and the role of short peptides in modulating signaling pathways
Stem cells reside in a unique tissue specific microenvironment known as a niche; a phenomenon originally proposed by Schofield in 1978 based on studies of hematopoietic stem cells produced from bone marrow.56 Schofield's hypothesis emphasized the environment created by the cells nearby, which allows stem cells to continue self-renewal and maintain their individuality. The stem cell niche encompasses a dynamic ensemble of physicochemical and biological stimuli that influence their differentiation, fate and self-renewal potential.57 Key events related to stem cell survival, proliferation, self-renewal and differentiation are highly regulated in these niches. The stem cell niche comprises three main components: cell–cell interactions, cell–ECM interactions and cell-soluble factor interactions.58 These components provide critical decision-making information to the cell. For instance, the ECM supplies topographical, mechanical and biochemical signals while cell–cell communication can be either heterologous (i.e. signals from the brain) or homologous as seen with daughter cells. In the clinical context, alterations in the stem cell niche can lead to changes in stem cell activity, contributing to the pathophysiology of various disorders including cancer,59 gliomas,60 Crohn's disease61 and leukemia.62Short peptides can alter stem cell differentiation by interacting with their microenvironment or stem cell niche. To successfully use short peptides in stem cell therapy, understanding the complex interaction of essential constituents such as transcription factors, epigenetic regulation, microRNAs, and signaling pathways that facilitate self-renewal, differentiation, and pluripotency is important. These interactions involve various signals like growth factors, cell adhesion molecules and mechanical signals; all of which can influence stem cell development. Short peptides can deliver these signals that promote desired cellular differentiation by activating the signaling mechanism that controls the expression of differentiation genetic information.63 During development, progenitors respond to specific morphogen regimens that activate transcriptional networks to define cell fates. Morphogens and signaling pathways such as TGF-β superfamily members, ERKs and canonical WNTs, which are crucial for lineage specification and differentiation are also necessary for cultured ESCs to maintain or self-renew. Recent research explores replicating these signaling pathways in vitro to determine whether ESCs should self-renew or differentiate. For example, the W9 peptide stimulated the differentiation to osteoblasts in MC3T3-E1 cells and MSCs.64 W9 peptide promoted the mineralization of stem cells derived from human adipose tissues more effectively than BMP2 or saline treatments in a dose dependent manner. W9 peptide induced higher expression of osteogenesis-related genes COL1A1, RUNX2 and TGFR2 while decreasing BMPR1B and BMPR2 expression in adipose stem cells. Study indicates that W9-induced osteogenic induction was caused by the stimulation of the TGF and PI3K/Akt signaling pathway in human adipose-derived stem cells.64 Evidence also suggests that short peptides trigger cell differentiation by modulating the Wnt/β-catenin,65 TGF-β,66,67 Notch,68 BMP,69 MAPK70 and PI3K-Akt71 signaling pathways.
3.1 Wnt/β-catenin signalling pathway
Wnt peptides are secreted morphogens crucial for embryonic development, stem cell proliferation, self-renewal of cells, tissue regeneration and adult tissue remodelling. The aberrant Wnt signalling pathway contributes to solid tumors and haematological malignancies.72 Therefore, activation of the Wnt/β-catenin protein signalling pathway is of significant use in stem cell therapy and regenerative medicine.72 The Wnt signaling pathway is categorized into canonical and non-canonical pathways that are β catenin dependent and β catenin independent, respectively.73 The canonical pathway predominantly regulates cell migration, differentiation and proliferation. Wnt family ligands transmit signals via Frizzled (FZD) receptors and LRP5/6 co-receptors to activate the Wnt/β-catenin protein signaling cascade (Fig. 2). | ||
| Fig. 2 Peptides with therapeutic effects targeting the Wnt signaling pathway (figure reprinted from ref. 74 with permission, Copyright© 2023). | ||
There are 19 Wnt genes, separated in 20 conserved Wnt subfamilies. Canonical signaling is typically activated by WNT1, WNT2, WNT3, WNT8 and WNT19 ligands while WNT4, WNT5a, WNT7 and WNT11 typically activate non-canonical pathways.74 The transmembrane protein Klotho is crucial in the cellular process. Chen et al. synthesized Klotho-derived peptide 6 (KP6; QPVVTLYHWDLPQRLQDAYGGWANRALADH) based on the human Klotho KL1 domain sequence.75 KP6, a 30 amino acid peptide, contains a Wnt-binding motif within the KL1 domain (Gln 186 to His215) and mimics Klotho's function. KP6 binds to Wnt, inhibiting its interaction with LRP6 and thereby blocking the Wnt/β-catenin signaling pathway. KP6 inhibits the expression of its downstream target genes in tubular epithelial cells and glomerular podocytes, and it also reduces β-catenin activation in vivo. Additionally, KP6 promotes survival and recovery from kidney damage in diabetic mice.75
Low-molecular-mass peptide 2 (LMP2),76 atrial natriuretic peptide (ANP),77 amyloid β peptide 42 (Aβ42),78 P182–195 (sequence: MEENAYQVFLTSDI)79 and C-peptide80 are other examples of peptides that can modulate the Wnt/β-catenin signaling pathway. Dietrich L. et al. synthesized a cell penetrating peptide (NLS-StAx-h), stapled peptide inhibitor of oncogenic Wnt signaling that effectively disrupts β-catenin-transcription factor interactions.81 In summary, Wnt proteins are pivotal in development and regeneration, with aberrant signaling linked to diseases such as cancers. The Wnt/β-catenin pathway's modulation is crucial in stem cell therapy. Peptides like KP6, ANP, C peptide and NLS-StAx-h can modulate this pathway, offering therapeutic avenues for medical interventions.
3.2 TGF-β signalling pathway
The transforming growth factor-β (TGF-β) is a multifunctional superfamily of cytokines that can both promote and inhibit cancer progression.82 It regulates cell division, invasion, migration, apoptosis and proliferation through suppressor of mother against decapentaplegic (Smad) and non-Smad pathways. TGF-β, Activin, Inhibins, Nodal, bone morphogenetic proteins (BMPs), anti-Mullerian hormone (AMH), and growth and differentiation factors (GDFs) are all members of the TGF-β superfamily, which is highly conserved across species.83 These cytokines are essential for diverse biological processes such as growth inhibition, cell migration, ECM remodelling and immune regulation. Dysregulated TGF-β signaling contributes to diseases like cancer, fibrosis and inflammation where it promotes aberrant cell behaviours.83 Peptide-based drug development efforts are increasingly targeting the TGF-β pathway to mitigate its pathological effects.TGF-β receptors are categorized into three types: type I (e.g. TGFRI) also known as activin-like kinases, ALKs, type II (e.g. TGFRII) and type III (e.g. TGFRIIII). Receptor types I and II possess an intracellular domain with serine/threonine kinase activity. TGF-β signals primarily through the type I receptor, specifically ALK5, and the type II receptor, such as TβRII, both necessary for signalling pathway activation. Ligand binding to the membrane receptor and receptor phosphorylation triggers the SMAD signaling pathway.84 TGF-β family ligands activate these receptors, leading to the phosphorylation of Smad proteins in the cell cytoplasm which then translocate to inside the nucleus.85 In the nucleus, Smad proteins act as transcriptional co-factors, activating target genes and determining cell fate in response to external stimuli. Smad proteins are categorized into three classes based on their structure and function; receptor-regulated “R-Smad” such as Smad1, Smad2, Smad3, Smad5 and Smad8, “Co-Smad” like Smad4 and inhibitory “I-Smad” including Smad6 and Smad7.86 The TGF-β superfamily comprises two main branches; one includes Activin, Nodal, myostatin, Leffty and TGF-β, while the other consists of BMPs (Fig. 3).
 | ||
| Fig. 3 The TGF-β/SMAD protein signaling pathway involves ligands like TGF-β, myostatin, and GDF11 binding to cell membrane receptors to phosphorylate downstream SMAD2/3 (R-SMADs), while BMPs like BMP2, BMP4, and BMP7 phosphorylate SMAD1/5/8 (R-SMADs). Figure reprinted from ref. 85 with permission, Copyright© 2020. | ||
According to Jienny Lee et al., soy peptides activate the mTOR signalling pathway, leading to the synthesis of TGF-β1 and cytokines like VEGF and IL-6 in serum-free conditions.87 In another study, P. Yu et al. synthesized the RADA-16 peptide using solid-phase peptide synthesis, generating a well-defined hydrogel via supramolecular peptide self-assembly.88 TGF-β1 (10 ng mL−1) injected in the RADA-16 hydrogel then known as TGF-β1/RADA-16 hydrogel. This designed hydrogel promoted chondrogenic development and sGAG formation in BMSCs while decreasing the production of pro-inflammatory factors. Additionally, the hydrogel significantly activated the Smad and ERK/MAPK pathways in BMSCs. M. Pitou et al. synthesized peptides 90-YYVGRKPK-97 (peptide 8) and 91-YVGRKP-96 (peptide 6) which favor the ERK1/2 and Smad2 pathways, leading to articular extracellular matrix formation at concentrations of 789.06 pg ml−1 and 562.5 pg ml−1 respectively in hDPSCs (human dental pulp stem cells).89 TGF-β1 also favors the Smad1/5/8 pathway, resulting in the expression of metalloproteinases ADAMTS-5 and MMP13, leading to a hypertrophic chondrocyte phenotype. Additionally, TGF-β1 promotes the Smad1/5/8 pathway, which in turn triggers the expression of metalloproteinases ADAMTS-5 and MMP13, which in turn produces a hypertrophic chondrocyte phenotype. Taken together, these two short peptides- especially peptide 8 could be used in conjunction with a scaffold to induce in vivo chondrogenesis in injured articular cartilage, offering an alternative for treating osteoarthritis.89 Efforts in peptide-based drug development are increasingly targeting the TGF-β pathway, crucial in diverse biological processes and implicated in regenerative medicine, tissue engineering, cancer, fibrosis, and inflammation.
3.3 Notch signalling pathways
Thomas Hunt Morgan first identified Notch mutations in Drosophila over 100 years ago.90 Notch signaling is a highly conserved pathway that coordinates neighbouring cells during development and homeostasis.91 In the past few years, Notch signaling has been majorly recognized as a major pathway coordinating developmental processes in most organs and tissues of metazoans. Notch signaling is crucial during both embryonic development of the CNS and postnatally, as it contributes to the maintenance of the stem cell pool and regulates proliferation, differentiation, migration, and apoptosis.92The Notch pathway consists of five ligands such as Delta-like 1, Delta-like 3, Delta-like 4, Jagged 1, and Jagged 2 and four receptors like Notch1, Notch2, Notch3, and Notch4.93 The connection between a ligand and a receptor initiates cleavage of the Notch intracellular domain (NICD) by gamma-secretase, followed by Notch1 intracellular domain (NICD) binding to the transcriptional factor CSL expressed by the RBPJ gene to control downstream Notch-dependent genes such as HEY, HES and SNAIL (Fig. 4).92 Notch receptors are synthesized as 300–350 kDa precursors and processed into two non-covalently linked subunits. The resulting mature heterodimeric structure of the protein is displayed on the outer cell surface. The cell extracellular domain with 29–36 N-terminal EGF-like repeats that are available for ligand interaction. The activation of Notch receptors is further regulated by O-fucose, fringe and Rumi glycosyltransferases in response to certain ligands.93
 | ||
| Fig. 4 Schematic representation of the Notch signaling pathway (figure reprinted from ref. 93 with permission, Copyright© 2019 Elsevier B.V.). | ||
Molecular studies performed by S. R. Jung et al. demonstrated that silk peptides inhibit the expression of adipocyte-specific genes [peroxisome proliferator-activated receptor γ (PPARγ)] and its targets [aP2, Cd36 and CCAAT enhancer binding protein α (C/EBPα)]. Silk peptides reduce adipose synthesis by suppressing the Notch signalling pathway and silencing Notch target genes (Hes-1 and Hey-1). Y. Itokazu et al. investigated the impact of various bioactive amyloid β-peptides (Aβs) on neural stem cell (NSC) proliferation. Aβ1–42 was found to activate the NICD, leading to increased expression of Musashi-1 and Pax6, a paired box protein. Aβ1–42 stimulates NSC proliferation by influencing the expression of glycoproteins involved in Notch signaling. Another Jagged1-mimicking peptide, J1, composed of 17 amino acids (CDDYYYGFGCNKFCRPR), was synthesized by G. Ferrari-Toninelli et al. The results demonstrated that the J1 peptide induced Notch activation, significantly increasing NICD and Hes1 levels, and resulting in a dose-dependent increase in SH-SY5Y cell proliferation.94 This evidence suggests that short peptides have a pleiotropic effect on stem cell fate, including cell growth arrest, induction of differentiation, and reduction of cell motility, which may offer a therapeutic advantage for brain tumor treatment.
3.4 Bone morphogenetic protein (BMP) signalling pathway
Bone morphogenetic proteins (BMPs) were discovered to promote ectopic bone growth in the mid-1960s.95 Much research has demonstrated that BMPs may induce MSCs to differentiate into bone, supporting their significance in bone and cartilage production. BMP signals play crucial roles during embryogenesis, from early pattern related events to tissue specification, organ development, and germ cell differentiation. BMPs are members of the TGF-β protein superfamily.96When homo- or heterodimeric BMP ligands attach to type I receptors and form a heteromeric complex with type II receptors, the BMP signalling pathway can initiate. Type II receptors phosphorylate type I receptors, which then activate R-Smads (Smads 1, 5, and 8 for BMP ligands) via serine-threonine phosphorylation.97 Activated R-Smads multimerize and accumulate in the nucleus to function as transcription factors. BMP receptors transmit signals not only through Smad phosphorylation but also via p38 activation.98 Signaling can also occur through repressor-Smads and Smads specialized for TGF-β signals, such as Nodal.99 BMP signals are modulated by factors such as receptor oligomerization, degradation, dephosphorylation, endocytosis, interaction with other pathways, and pseudo-receptor expression. Downstream Smad proteins are further regulated by interactions with transcriptional modifiers, nuclear import/export, and dephosphorylation.97
Tong et al. designed a BMP9 peptide mimic, P3, constructed from the type I receptor binding region, with millimolar binding affinities for the receptors activin protein receptor-like kinase 1 such as ALK1, ALK2 and ALK3.100 They discovered that this BMP9-derived peptide can selectively increase ALK1 mediated BMP9 signaling in pulmonary artery endothelial cells (hPAECs) while modulating BMP9 and BMP4 signaling in a cell type specific manner.100 In another study, M. Yang et al. examined an osteogenic peptide (GETNPADSKPGSIR, P-GM-2) derived from Gadus morhua peptides.101 The cell-based studies showed that P-GM-2 enhanced osteoblast proliferation, differentiation and mineralization by increasing osteogenic markers. P-GM-2 phosphorylates GSK-3β, stabilizing β-catenin protein and activating the Wnt/β-catenin protein pathway. It also activates the BMP/Smad and MAPK pathways. Validation with specific inhibitors confirmed P-GM-2's activation of BMP as well as Wnt/β-catenin pathways, highlighting its potential as an antiosteoporosis agent.101 Recent studies on BMP9 and osteogenic peptides highlight their therapeutic potential in enhancing bone formation and treating osteoporosis.102 Additionally, BMP-based therapeutic treatments are being developed for cardiovascular and kidney diseases.96
3.5 Other signaling pathways
In addition to the discussed signalling pathways such as Notch, Wnt/β-catenin, TGF-β, and BMPs, other pathways such as MAPK,103 PI3K,104 Akt,105 and mTOR106 are well-documented for their roles in peptide-based therapeutic applications including tissue defects, wounds, cancer, cardiovascular disorders, and neurodegenerative diseases. Collectively, the evidence highlights the potential of short peptides in regenerative medicine and therapeutic applications through various signaling pathway cross-talks.1074. Biomedical applications of short peptides in stem cell differentiation
Peptide form and concentration determine the path of cell differentiation.20 In neural tissue, epidermis and mesenchyme, pluripotent cells undergo differentiation in response to peptides such as AEDG and AEDP.19 For example, AEDP (cortagen) at a dose of 10 ng ml−1 induced 80% of the cells in culture to mesenchyme and epidermis via differentiation. Doses of 2, 20, and 100 ng ml−1 resulted in a 30–48% differentiation rate, whereas doses of 50 and 200 ng ml−1 had no effect on pluripotent cell differentiation.108 Peptides exert their influence by connecting with histone proteins, thereby modulating gene accessibility for transcription. They can also directly bind with DNA, regulating gene expression either by activation or inhibition. These mechanisms enable peptides to activate signaling pathways that regulate gene expression during the process of differentiation. Peptides such as AEDL and KEDW induce differentiation in pancreatic and lung cells, while KE, AED, and KED activate neural differentiation. Immune cells differentiate through KE, KED, or EDA peptides. Table 1 summarizes a list of short peptides and their mechanisms for stem cell differentiation.| Sr. no. | Peptide | Sequence | Differentiation | In vitro/in vivo model | Brief findings | Ref. |
|---|---|---|---|---|---|---|
| 1 | RAD-RGI | Ac-(RADA)4-G2-RGIDKRHWNSQ-NH2 | Neuronal | Rat Schwann cells and neurites | Peptides provide 3D environment and enhanced RSC adhesion, myelination, and neurotrophic production in vitro and in vivo | 109 |
| RAD-IKV | Ac-(RADA)4-G2-IKVAV-NH2 | |||||
| 2 | RADA 16-I | RADARADARADARADA | Cardiac | Human iPSCs | In the absence of RAD16-I, ascorbic acid enhances hiPSC cardiac differentiation by upregulating the expression of specific cardiac genes and protein | 110 |
| 3 | RGD+ | GRGDS | Cardiomyocyte | Myocardial cells from Wistar rat hPSC cell line | RGD peptides increased cardiomyocyte cell viability and differentiation of scaffolds | 111 |
| 4 | IVFK, | Ac-Ile-Val-Phe-Lys-NH2 | Osteogenic | hMSC cell lines, HUVEC cells | Peptides promote proliferation and osteogenic differentiation of hMSCs, and promote angiogenesis of HUVECs | 112 |
| IVZK | Ac-Ile-Val-Cha-Lys-NH2 | |||||
| 5 | BFP-1 | GQGFSYPYKAVFSTQ | Osteogenic | MBSCs or D1 cells | Compared to BMP-7, BFP-1 exhibits greater osteogenic differentiation activity | 113 |
| 6 | A13 peptide | RQVFQVAYIIIKA | Hepatocyte | Primary rat hepatocytes, male Sprague Dawley rats | Primary hepatocytes cultured on the A13 peptide continued to express hepatic differentiation markers | 114 |
| 7 | KEDW | Lys-Glu-Asp-Trp-NH2 | Pancreatic | Endocrine pancreatic cell line | Upregulated the expression of PDX1, NGN3, PAX6, FOXA2, NKX2-2, NKX6.1, and PAX4 genes, while downregulating the expression of MNX1 and HOXA3 | 115 |
| 8 | Noggin | MQQAALLLGLSLADA | Endogenous progenitors | Mouse spinal cord injury (SCI) model | Combination of noggin and PDGF enhanced overall myelination of regenerated axons and, significantly improved functional recovery | 116 |
| 9 | DEGA | Asp-Gly-Glu-Ala | Osteogenic | hMSCs | DGEA peptide facilitates adhesion, spreading, and early commitment to osteogenic differentiation, while EEE peptide enhances mineralization capabilities | 117 |
| EEE | Glu-Glu-Glu | |||||
| 10 | ADEL | Ala-Asp-Glu-Leu | Bronchial epithelium | Bronchoepithelial human embryonic cell culture | ADEL protects the bronchial epithelium against pulmonary diseases | 118 |
| 11 | OGP or | ALKRQGRTLYGFGG | Osteogenic | Female hMSCs | The incorporation of either OGP or BMP-2 significantly enhanced the osteogenic differentiation of hMSCs | 119 |
| BMP-2 | KIPKASSVPTELSAISTLYL |
The detailed mechanisms of these peptides modulating stem cell fate to specific differentiation are discussed below:
4.1 Ectodermal differentiation
The ectoderm is one of the three primary germ layers formed during embryogenesis, with the other two being the mesoderm and endoderm. Each germ layer gives rise to specific tissues and organs in the developing embryo. Focusing on the ectoderm, it can differentiate into two main subdivisions: the surface ectoderm and neuroectoderm. The surface ectoderm further differentiates into the epidermis and exocrine glands. The epidermis forms the outermost layer of the skin and gives rise to the lining of the mouth, nostril, and anus. The exocrine gland, derived from the surface ectoderm, includes the mammary glands, sweat glands, and salivary glands. The neuro-ectoderm, on the other hand, differentiates into components of the central and peripheral nervous system.120 We have highlighted the role of peptides in neuronal and epithelial differentiation by summarizing recent advances.Maria B. Fonseca et al. examined the role of various Aβ fragments in NSC proliferation and differentiation, also investigating if autophagy is involved in Aβ-induced alterations of neural fate.124 The findings show that both Aβ1–40 and Aβ1–42 significantly induce neuronal and glial-specific protein markers; however, Aβ1–40 preferentially enhances the neurogenesis of NSCs while Aβ1–42 appears to favour gliogenesis. These results support various important roles for different Aβ peptides in NSC fate decisions and emphasize the significance of autophagy in controlling this activity. S. Yang et al. designed a self-assembling peptide (SAP) hydrogel nanofiber for peripheral nerve regeneration.109 The hydrogel was based on the self-assembling backbone Ac-(RADA)4-NH2 (RAD), dual-functionalized with a BDNF-mimetic peptide epitope RGIDKRHWNSQ (RGI) and a laminin-derived motif IKVAV (IKV) (Fig. 5A).
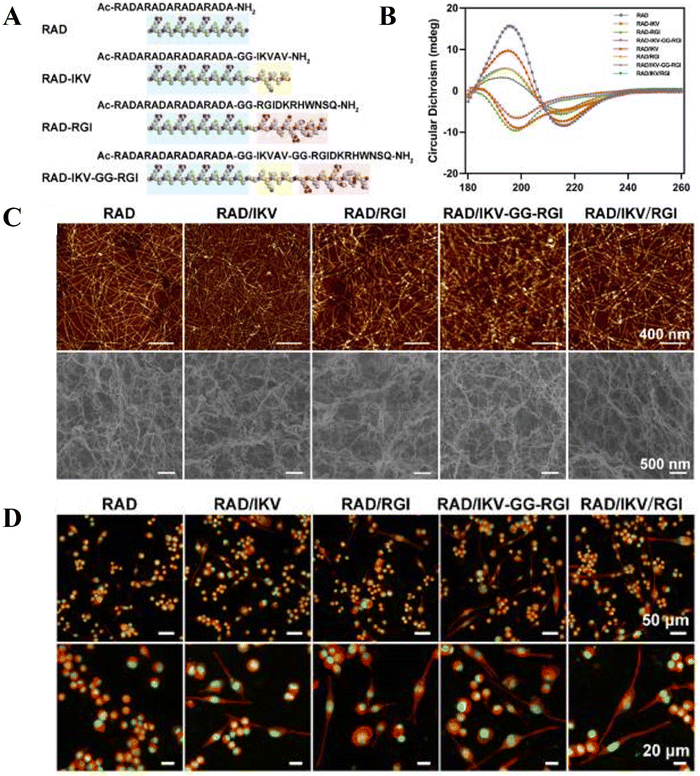 | ||
| Fig. 5 Characterization of SAP and hydrogels with rat Schwann cell (RSC) adhesion and spreading behavior on the surface of self-assembling peptides (SAP) and hydrogels: the sequences of the functionalized SAP (A). CD spectra of SAP solutions (B), atomic force microscopy photographs of peptide solutions and scanning electron microscopy morphologies of hydrogels (C), representative fluorescence microscopy images of rat Schwann cells on various peptide scaffolds (D). Rhodamine–phalloidin was used to stain β-actin (red) in the cells, while SYTOX Green was used to stain the nuclei (green) (figures reprinted from ref. 109 with permission, Copyright© 2020). | ||
The formerly mentioned hydrogel provided a three-dimensional (3D) microenvironment for rat Schwann cells (RSCs) and neurites. The researchers found that the dual-functionalized SAP hydrogels effectively filled a 10 mm gap representing a sciatic nerve lesion in rats in vivo and enhanced RSC adhesion, myelination, and neurotrophin production in vitro (Fig. 5D). The outcomes demonstrated how IKVAV and RGI work synergistically to promote axonal regeneration and functional recovery following peripheral nerve damage. In another study, short peptide PepB (AAAAEK) was designed to deliver retinoic acid (RA) to human neural stem cells (hNSCs) and enhance neuronal development.125 Treatment with 0.1 μM RA or 0.1 μM RA-PepB3 resulted in a 2-2.5-fold increase in β-III tubulin and a 2.5-fold increase in MAP2 expression compared to the control, with no significant difference between the RA and RA-PepB3 treatments. Overall, this evidence suggests that bioactive scaffolds and peptides can enhance cell viability, proliferation, and neuronal differentiation, aiming to create functional tissue grafts for in vivo applications, highlighting the potential role of peptides in the treatment of neurodegenerative diseases.
Due to dysfunction in epithelial barriers, the chance of developing allergic diseases and other disorders including oral cancer, celiac disease, type 2 inflammatory diseases, asthma, allergic rhinitis, chronic rhinosinusitis and eosinophilic esophagitis increases.129 S. Raghupathy et al. explored the potential of the short amino acid containing peptide L-R5 such as partial inhibitor of protein kinase C zeta to improve epithelial paracellular uptake in a reversible as well as non-toxic manner.130 The L-R5 structure is a pentapeptide containing a cell membrane-penetrating group at its N-terminus and the sequence myristoyl-ARRWR. When administered apically in vitro, L-R5 improved epithelial uptake ability within minutes enhancing apical-to-basolateral (AB) transport of 4 kDa dextran (use as model of peptide and protein) and naloxone (BSC class III drug) in nasal human primary epithelial cells. Several investigations have shown that the tetrapeptide Ala-Asp-Glu-Leu (ADEL) is effective in models of acute bacterial lung inflammation, fibrosis, and toxic lung injury. V. K. Khavinson et al. reported that tetrapeptide (ADEL) modulates Ki67, Mcl-1, p53, CD79, and NOS-3 protein levels in human bronchial epithelial cell cultures at various passages.118 The findings demonstrated that the ADEL peptide may provide a defense mechanism to the bronchial epithelium from pulmonary disease. Additionally, ADEL may protect bronchial tissue against aging. Furthermore, enhancing intestinal repair mechanisms using regulatory peptides or other modulatory factors may provide future options for treating disorders characterized by epithelial surface damage.131
4.2 Mesodermal differentiation
Human pluripotent stem cells exhibit characteristics similar to the epiblast, which forms the three germ layers. Using human induced iPSCs, mesoderm and endoderm were produced with over 75% purity. The single-cell dynamics of iPSCs derived mesoderm as well as endoderm cells were studied using time-lapse imaging for visualization.132 Both mesoderm and endoderm cells exhibited random migration with short-term directional persistence, and no significant differences were observed in their migration patterns. A. Rodaway and R. Patient discovered the existence of mesendoderm in C. elegans to Xenopus, capable of giving rise to both endoderm and mesoderm.133 In early zebrafish development, the marginal zone surrounding the vegetal edge contains both endodermal and mesodermal precursor cells. This marginal zone co-expresses brachyury and Gata5, which are distinct markers for mesoderm and endoderm progenitors, respectively. Mesodermal cells are the most prevalent in the human body, contributing to a wide range of cell types including the musculoskeletal system such as bone, cartilage, and muscle, cardiovascular systems like the heart, blood, and blood vessels, and connective tissues found throughout the body.134Self-assembling peptides have shown considerable promise for tissue engineering, particularly in healing wounded myocardium. Numerous protein and cellular treatments have been successfully tested in preclinical animal models.139 A novel SAP was designed by modifying a peptide such as RADA16 with a cell-adhesive motif as well as a BMP-2 binding motif, as synthesized by H. D. Guo et al.69 The effects of this functionalized SAP on c-kit+ MSC adherence, survival and differentiation were further investigated. Heart function, neovascularization, and myocardial regeneration were assessed after transplanting SAP containing BMP-2 and c-kit+ MSCs in rat MI models. The cells adhered and dispersed readily on the well-organized nanofibrous scaffolds that the SAP spontaneously created. The SAP protected the cells from hypoxia and serum deprivation. Following SAP breakdown, BMP-2 was released continuously, inducing c-kit+ MSCs to differentiate into cardiomyocytes. These findings imply that functionalized SAP effectively increases stem cell engraftment, survival, and differentiation. The local continuous release of BMP-2 with SAP is a promising method for enhancing engrafted stem cell differentiation and healing of infarcted myocardium.
In another study, Y. Zhang et al. demonstrated the potential of ECM-derived collagen I hydrogel loaded with molecules such as histone deacetylase 7 (HDAC7)-derived-phosphorylated 7-amino-acid peptide (7Ap) as a novel candidate for MI treatment.140 The 7Ap-collagen increased neo-micro vessel formation, improved stem cell antigen-1 positive (Sca-1+) stem cell recruitment and differentiation, reduced cellular apoptosis, and promoted cardiomyocyte cycle progression. Additionally, 7Ap-collagen reduced LV wall fibrosis, decreased infarct wall thinning, and significantly enhanced cardiac function at 2 weeks after-MI.
O. Schussler et al. proposed a novel approach to develop a highly effective collagen-cell scaffold using arginine–glycine–aspartic acid–serine (RGD+) adhesion peptides cross-linked to a therapeutically approved collagen matrix.111 The primary advantages were improved cell contractility, cardiomyocyte viability, and differentiation. V. A. Puig-Sanvicens et al. studied the cardiac differentiation properties of hiPSCs when cultivated with and without ascorbic acid (AA) and embedded in peptides like RAD16-I.110 In adherent cultures and in the absence of RAD16-I, AA promotes hiPSC cardiac cell differentiation by upregulating the expression of specific cardiac genes and proteins, increasing the number of contracting clusters during differentiation. These findings indicate the potential application of SAP and peptide-based scaffolds as a medium for enhancing cellular transplantation in the heart or designing a true myocardial equivalent.
G. Gulseren et al. reported increased bone regeneration efficiency using alkaline phosphatase (ALP) mimetic peptide nanofibers that express both activity such as catalytic and matrix-regulatory capabilities of alkaline phosphatase, which is an enzyme that regulates phosphate homeostasis and the synthesis of calcifiable bone matrix.146 Histidine-presenting peptide nanostructures were designed to act as phosphatases, catalyzing phosphate hydrolysis while also serving as a scaffold for the formation of bone-like nodules. These mesenchymal and osteoblast-like cell lines were stimulated to undergo osteogenesis by these alkaline phosphatase-like peptide nanofibers.
Zhou et al. developed peptides derived from the bioactive domains of BMP-2 (residues 73–92) that were covalently conjugated to the surface of mesoporous silica nanoparticles (MSNs) using an aminosilane linker.147 These peptides were loaded with dexamethasone to promote osteogenic differentiation of rBM-MSCs, mitigating the high-dose negative effects of BMP-2. After 48 hours of incubation, MSNs-pep showed significantly higher cell viability than MSNs at doses of 50 and 100 μg mL−1 (P < 0.01), with a cell viability of approximately 93.7% at 100 μg mL−1. The results demonstrated that compared to ungrafted MSNs, MSN-pep exhibited improved cellular uptake, biocompatibility, and dispersibility. After three weeks, ectopic bone growth was observed in 4-week-old male SD rats implanted with this construct. In another study, S. Alshehri et al. designed SAP-scaffolds to promote the proliferation and osteogenic differentiation of hMSCs.112 These rationally designed peptides are synthetic and amphiphilic, containing self-assembling tetrapeptides composed of four non-toxic amino acids. They synthesized two self-assembling tetrapeptides, aromatic (IVFK) and nonaromatic (IVZK), which formed hydrogels upon PBS addition (Fig. 6A). SEM micrographs confirmed porous fiber networks from entangled peptide nanofibers, with nanometer-scale pores smaller than cell nuclei, potentially limiting passive cell mobility. Hydrogel stiffness increased with peptide concentration. Osteogenic differentiation of bone-marrow derived mesenchymal stem cells (BM-MSCs) in these hydrogels was observed through bright-field microscopic images after 7 and 14 days, showing cell shape changes and visible mineralization after 14 days (Fig. 6B). Cells exhibited a branching “osteocyte-like” structure, confirmed by confocal fluorescence microscopic images of the F-actin cytoskeleton, indicating successful differentiation. Alizarin red staining validated the mineralization process during differentiation. These tetrapeptide hydrogels promote both osteogenic differentiation and angiogenesis, making them suitable scaffolds for bone tissue engineering.
 | ||
| Fig. 6 Self-assembling peptide (IVFK and IVZK) based hydrogels (A), self-assembled peptide in hydrogel in 1× PBS (a). The morphology of IVFK (b) and IVZK (c) nanofibrous hydrogels using SEM. Porosity of the peptide hydrogels was determined using SEM images (d). Mechanical stiffness values (e); osteogenic differentiation of MSCs (B), phase contrast images of MSCs cultured in different hydrogels in osteogenic media after 7 and 14 days (a), Alizarin red-S staining (b) and In vitro morphology of hMSCs after 3 weeks of culturing. Figures are reprinted from ref. 112 with permission, Copyright© 2021. | ||
H. Hosseinkhani et al. designed a three-dimensional network of nanofibers generated by the self-assembly activity of peptide-amphiphile (PA) molecules.148 When rat MSCs were seeded into PA nanofibers with or without RGD, the PA nanofibers containing RGD showed a higher number of adhered cells. In measuring MSC osteogenic differentiation, the PA nanofibers with RGD exhibited the highest alkaline phosphatase (ALP) activity and osteocalcin content compared to those having absence of RGD. Both values were remarkably higher than those observed in static tissue culture plates in two-dimensional culture. The study concluded that PA nanofibers served as a cell scaffold, influencing MSC adhesion, proliferation, and osteogenic differentiation. Peptide-based fibrillar hydrogels have been shown to resemble the structure of the extracellular matrix, providing a niche for cells to perform their physiological functions.149
A study done by L. A. Castillo Diaz et al. demonstrated that an ionic-complementary peptide hydrogel, FEFEFKFK (comprising phenylalanine, glutamic acid, and lysine), can host hMSCs in 3D and stimulate osteogenic development.150 Over 12 days of growth, hMSCs remained viable and proliferated in the hydrogel. With osteogenic stimulation, they differentiated into osteoblasts. These findings indicate that a biodegradable octapeptide hydrogel can host and stimulate stem cell differentiation, with potential applications in regenerating hard tissues like alveolar bone.
Apart from these discussed self-assembled peptides, BMP-2 derived KIPKASSVPTELSAISTLYL151 and γEPRRγEVCγEL,152 peptide amphiphile (PA) with RGDS/DGEA,153 RGD,154 VN and BFP-1 peptide,155 hydroxyapatite (HA)-binding peptides,156 bone marrow homing peptide 1 (BMHP1),157 RGDS containing biomimetic peptide amphiphile (PA)158 and tenascin-C mimetic peptide159 have also been reported as modulators of osteogenic differentiation. Short peptides may trigger osteogenic cell development by activating pathways such as p38/MAPK and PI3K-Akt. These pathways can be activated either by peptides binding to signalling cascade components or by regulating the gene expression code for the p38/MAPK as well as PI3K-Akt proteins.
4.3 Endodermal differentiation
During gastrulation, the endoderm germ layer emerges from the epiblast. After that, endoderm cells differentiate to form the embryonic gut's epithelial lining, which organogenesis develops into the complete gastrointestinal tract, the glandular structures of the respiratory system, lungs, and other organs such as the pancreas and liver.Yamato Kikkawa et al. screened hepatocyte attachment peptides using 25 physiologically active peptides from laminin α1 and assessed the preservation of hepatic function with primary rat hepatocytes.114 Peptide A13 contains amino acid sequence RQVFQVAYIIIKA, which includes mice laminin α1 chain residues 121 to 133, demonstrated the highest activity. Additionally, hepatic differentiation markers such as tyrosine aminotransferase, tryptophan-2,3-dioxygenase, and cytochrome P450D remained present in primary hepatocytes treated with A13 peptides. C. E. Semino et al. designed a self-assembling peptide scaffold in a putative adult rat hepatocyte progenitor cell line-Lig-8 to enhance tissue-like function.163 Differentiated progeny cells exhibit hepatocyte maturation markers such as binucleation, up-regulation of albumin, and expression of cytochrome P450s CYP1A1, CYP1A2, and CYP2E1. They also express the transcription factor C/EBPα. Other peptides, such as myristoylated HBV preS1-peptides,164 RAD16-I combined with RGD (integrin-binding sequence) and YIG (laminin receptor binding sequence),165 corn peptides and the synthetic pentapeptide (QLLPF),166 were designed and their effects on hepatocytes were examined. Based on these studies, short peptides show promise in liver disease therapy as modulators of hepatocyte differentiation and function.
Glucagon-like peptide 1 (GLP-1) activates pancreatic β-cells to produce glucose-dependent insulin, suppresses glucagon secretion, delays stomach emptying, reduces food intake, and promotes glucose clearance, aiding in type 2 diabetes management.172 GLP-1 agonists enhance β-cell proliferation and differentiation. Recent research suggests that they stimulate pancreatic stem cells to develop into β-cells. The gut's endocrine cells release the glucagon-like peptides GLP-1 and GLP-2 in response to food consumption. These peptides control energy absorption and storage as well as cell survival and proliferation.173 GLP-1 increases islet mass by promoting pancreatic β-cell proliferation, neogenesis, and the differentiation of exocrine cells into β-cells. GLP-2 enhances cell proliferation in the gastrointestinal mucosa, aiding in mucosal growth and reducing intestinal damage. Both peptides inhibit apoptosis, preserving β-cell mass and gut epithelium.
Apart from GLPs, peptide KEDW (Lys-Glu-Asp-Trp-NH2) has been shown to lower blood glucose levels in both in vitro and in vivo models. V. Kh. Khavinson et al. discovered that the KEDW peptide stimulates the expression of genes crucial for the functional activity of endocrine pancreas cells, likely by binding to DNA and activating gene expression.115 Due to its effects on the differentiation and function of pancreatic cells, the peptide could be used to correct pancreatic cell function, aiding in developing better treatments for diabetes and other pancreatic diseases. Pancreatic cells show decreased expression of differentiation markers as they age. The tetrapeptide pancragen induces differentiation factors in acinar (Pdx1, Ptfl a) and islet of Langerhans (Pdx1, Pax6, Pax4, Foxa2, NKx2.2) in both “young” and “aged” cultures.174 Additionally, two synthetic peptides, Spadin and Mini-Spadin, derived from the sortilin propeptide (PE), replicate the actions of PE, enhance insulin secretion in response to glucose and protect β cells from cytokine-induced apoptosis.175 This evidence suggests that researchers are increasingly focused on developing short peptides that specifically target pancreatic β cells to enhance their survival and regeneration for diabetes management.
5. Challenges and future directions
Despite their potential, short peptides in stem cell research face several limitations. One major challenge is their susceptibility to enzymatic degradation, leading to a loss of bioavailability and effectiveness.176 This degradation is primarily due to the presence of numerous proteases that break down peptides or peptide-conjugated drugs. Additionally, short peptides can sometimes change conformation and bind to undesirable sites during stem cell growth, causing unwanted side effects. The large size of many peptides can also hinder their uptake by stem cells, complicating the differentiation process. Maintaining cell viability and metabolic activity is another significant hurdle, as short peptide conjugation can be rejected by the host immune system, leading to cell death.177 To address these issues, peptides are often combined with polymers or scaffolds to prevent early degradation.178 Improving peptide delivery systems, creating more stable and bioavailable peptide models, and understanding the exact mechanisms underlying peptide-mediated signaling pathways in stem cells are crucial areas for future research. Advances in peptide engineering, such as the development of peptide mimetics and peptide conjugates, may help overcome these challenges and enhance the therapeutic potential of short peptides in stem cell research.6. Conclusion
Short peptides have demonstrated significant potential in stem cell research and regenerative medicine. They can effectively modulate stem cell activity, enhance cell viability, and direct differentiation. In cardiovascular diseases, SAP and peptide-based scaffolds have shown promise in improving heart function and cardiac regeneration. Similarly, tetrapeptide hydrogels have been successful in promoting osteogenic differentiation and angiogenesis for bone tissue engineering. In liver disease therapy, peptides such as A13 and myristoylated HBV preS1-peptides have shown potential in enhancing hepatocyte function and differentiation. Overall, short peptides are promising modulators of cellular behaviour, offering new therapeutic avenues for treating a variety of diseases and injuries. Our review provides a comprehensive introduction to stem cell fate and differentiation induced through short peptides, emphasizing the pivotal role of short peptides. Hence, we present extensive discussion about how short peptides can modulate critical signalling pathways, such as Notch, Wnt-β-catenin, TGF-β and BMP to facilitate stem cell differentiation. Additionally, we explore their potential applications in neuronal, epithelial, cardiac, hepatocyte, pancreatic and osteogenic differentiation, offering significant insights into advancements in stem cell therapy and tissue engineering.Author contributions
Rohan Vishwanath: literature review and writing – original draft preparation, Raghu Solanki: conceptualization, literature review and writing – original draft preparation, writing – review and editing, Abhijit Biswas: writing – review and editing, Unnati Modi: writing – review and editing, Sharad Gupta: writing – review and editing Dhiraj Bhatia: conceptualization, writing – review and editing, and supervision. All authors confirmed the final version of the manuscript.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
The authors would like to thank IIT, Gandhinagar for providing the necessary facilities. R. S. acknowledges the Science and Engineering Research Board (SERB), Government of India, for financial support through the National Post-Doctoral Fellowship (NPDF). DB thanks SERB-DST, GUJCOST, GSBTM, STARS-MoES for research grants.References
- J. Li, Z. Wu, L. Zhao, Y. Liu, Y. Su, X. Gong, F. Liu and L. Zhang, Stem Cell Res. Ther., 2023, 14, 381 Search PubMed.
- Y. Jin, S. Li, Q. Yu, T. Chen and D. Liu, MedComm, 2023, 4, e291 Search PubMed.
- H. Han, B.-T. Chen, Y. Liu, Y. Wang, L. Xing, H. Wang, T.-J. Zhou and H.-L. Jiang, J. Controlled Release, 2024, 365, 981–1003 CrossRef PubMed.
- A. J. Friedenstein, R. K. Chailakhyan, N. V. Latsinik, A. F. Panasyuk and I. V. Keiliss-Borok, Transplantation, 1974, 17, 331–340 CrossRef PubMed.
- J. T. Triffitt, Biomater. Transl., 2021, 2, 287 Search PubMed.
- M. Chivu-Economescu and M. Rubach, Curr. Stem Cell Res. Ther., 2017, 12, 124–133 CrossRef PubMed.
- A. J. Becker, E. A. McCulloch and J. E. Till.
- C. Eguizabal, B. Aran, S. M. Chuva de Sousa Lopes, M. Geens, B. Heindryckx, S. Panula, M. Popovic, R. Vassena and A. Veiga, Hum. Reprod. Open, 2019, 2019, hoy024 Search PubMed.
- P. Lotfinegad, A. Movassaghpour, J. Majidi and B. Baradaran, Adv. Pharm. Bull., 2014, 4, 5 Search PubMed.
- K. Takahashi and S. Yamanaka, Cell, 2006, 126, 663–676 CrossRef PubMed.
- S. Bharadwaj, N. Kirtipal, M. Bharti and R. C. Sobti, Biomedical Research, Medicine, and Disease, CRC Press, 2023, pp. 503–520 Search PubMed.
- A. Mahmoudi, P. Meidany, W. Almahmeed, T. Jamialahmadi and A. Sahebkar, Curr. Stem Cell Rep., 2024, 1–23 Search PubMed.
- P. Yadav, R. Vats, A. Bano and R. Bhardwaj, Stem Cells, Elsevier, 2024, pp. 355–362 Search PubMed.
- A. A. Dayem, S. Bin Lee, K. M. Lim, A. Kim, H. J. Shin, B. Vellingiri, Y. B. Kim and S.-G. Cho, Biomed. Pharmacother., 2023, 160, 114376 CrossRef PubMed.
- X. Zhang, T. Lei and H. Du, Stem Cell Res. Ther., 2021, 12, 1–10 CrossRef PubMed.
- L. P. Datta, S. Manchineella and T. Govindaraju, Biomaterials, 2020, 230, 119633 CrossRef PubMed.
- T.-C. Sung, T. Wang, Q. Liu, Q.-D. Ling, S. K. Subbiah, R. R. Renuka, S.-T. Hsu, A. Umezawa and A. Higuchi, J. Mater. Chem. B, 2023, 11, 1389–1415 RSC.
- M. C. Morris, S. Deshayes, F. Heitz and G. Divita, Biol. Cell., 2008, 100, 201–217 CrossRef CAS.
- S. Caputi, O. Trubiani, B. Sinjari, S. Trofimova, F. Diomede, N. Linkova, A. Diatlova and V. Khavinson, Int. J. Immunopathol. Pharmacol., 2019, 33, 2058738419828613 CrossRef CAS PubMed.
- V. Khavinson, N. Linkova, A. Diatlova and S. Trofimova, Stem Cell Rev. Rep., 2020, 16, 118–125 CrossRef PubMed.
- V. K. Khavinson, N. S. Lin’kova and R. S. Umnov, Bull. Exp. Biol. Med., 2021, 171, 190–193 CrossRef.
- J. Kisiday, M. Jin, B. Kurz, H. Hung, C. Semino, S. Zhang and A. J. Grodzinsky, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9996–10001 CrossRef PubMed.
- S. Yang, M. Wang, T. Wang, M. Sun, H. Huang, X. Shi, S. Duan, Y. Wu, J. Zhu and F. Liu, Mater. Today Bio, 2023, 100644 CrossRef.
- A. Hernandez, J. D. Hartgerink and S. Young, Front. Bioeng. Biotechnol., 2023, 11, 1139782 CrossRef PubMed.
- K. Sharma, K. K. Sharma, A. Sharma and R. Jain, Drug Discovery Today, 2023, 28, 103464 CrossRef PubMed.
- S.-Y. Qin, J.-Q. Feng, Y.-J. Cheng, W.-L. Liu, A.-Q. Zhang, L. Wang, H. Wang and X.-Z. Zhang, Coord. Chem. Rev., 2024, 502, 215600 CrossRef.
- R. Das, B. Gayakvad, S. D. Shinde, J. Rani, A. Jain and B. Sahu, ACS Appl. Bio Mater., 2020, 3, 5474–5499 Search PubMed.
- E. V. Prazdnova, M. S. Mazanko, V. A. Chistyakov, A. A. Bogdanova, A. G. Refeld, E. Y. Kharchenko and M. L. Chikindas, Probiotics Antimicrob. Proteins, 2022, 1–16 Search PubMed.
- H. Gong, X. Hu, L. Zhang, K. Fa, M. Liao, H. Liu, G. Fragneto, M. Campana and J. R. Lu, J. Colloid Interface Sci., 2023, 637, 182–192 CrossRef.
- N. Yadav and V. S. Chauhan, Adv. Colloid Interface Sci., 2024, 103282 CrossRef PubMed.
- Y. Wang, Y. Zhang, R. Su, Y. Wang and W. Qi, J. Mater. Chem. B, 2024, 12, 5061–5075 RSC.
- R. Solanki, N. Makwana, R. Kumar, M. Joshi, A. Patel, D. Bhatia and D. K. Sahoo, RSC Adv., 2024, 14, 33568–33586 RSC.
- K. Wang and Y. Wang, Food Funct., 2024, 15, 5714–5736 RSC.
- T. Zhao, G. Su, L. Zhang, J. Chen, Y. Zhang, W. Liu, M. Zhao, J. Zhang and Q. Huang, Food Front., 2024, 5, 1145–1165 CrossRef CAS.
- T. P. C. Ezeorba, A. L. Ezugwu, I. F. Chukwuma, E. G. Anaduaka and C. C. Udenigwe, Food Chem., 2024, 435, 137632 CrossRef PubMed.
- Y. Atma, B. S. Murray, A. Sadeghpour and F. M. Goycoolea, Food Funct., 2024, 15, 3959–3979 RSC.
- S. Yadav, A. K. Sharma and P. Kumar, Front. Bioeng. Biotechnol., 2020, 8, 127 CrossRef.
- S. Toksoz, H. Acar and M. O. Guler, Soft Matter, 2010, 6, 5839–5849 RSC.
- Y. Loo, M. Goktas, A. B. Tekinay, M. O. Guler, C. A. E. Hauser and A. Mitraki, Adv. Healthcare Mater., 2015, 4, 2557–2586 CrossRef CAS.
- S. Bashir, A. Aiman, M. Shahid, A. A. Chaudhary, N. Sami, S. F. Basir, I. Hassan and A. Islam, Ageing Res. Rev., 2024, 102276 CrossRef.
- Y. Zhou, Q. Li, Y. Wu, X. Li, Y. Zhou, Z. Wang, H. Liang, F. Ding, S. Hong and N. F. Steinmetz, ACS Nano, 2023, 17, 8004–8025 CrossRef CAS PubMed.
- X. Yang, L. Ma, K. Lu and D. Zhao, Protein J., 2024, 1–13 Search PubMed.
- P. Shi, X. He, H. Cong, B. Yu and Y. Shen, ACS Mater. Lett., 2024, 6, 1649–1677 CrossRef CAS.
- T. Li, X.-M. Lu, M.-R. Zhang, K. Hu and Z. Li, Bioact. Mater., 2022, 11, 268–282 CAS.
- W. Zhou, Y. Li and D. Zhu, Chem. – Asian J., 2007, 2, 222–229 CrossRef CAS PubMed.
- V. K. Khavinson, I. G. Popovich, N. S. Linkova, E. S. Mironova and A. R. Ilina, Molecules, 2021, 26, 7053 CrossRef CAS PubMed.
- M. Petkovic, M. V. Mouritzen, B. Mojsoska and H. Jenssen, Biomolecules, 2021, 11, 952 CrossRef PubMed.
- R. Lv, Y. Dong, Z. Bao, S. Zhang, S. Lin and N. Sun, Trends Food Sci. Technol., 2022, 122, 171–186 CrossRef.
- A. Ilina, V. Khavinson, N. Linkova and M. Petukhov, Int. J. Mol. Sci., 2022, 23, 4259 CrossRef PubMed.
- H. Wang, Y. Yang, J. Liu and L. Qian, Nat. Rev. Mol. Cell Biol., 2021, 22, 410–424 CrossRef PubMed.
- N. L. Francis, N. K. Bennett, A. Halikere, Z. P. Pang and P. V. Moghe, ACS Biomater. Sci. Eng., 2016, 2, 1030–1038 CrossRef.
- F. Yu, Y. Chen, M. Zhou, L. Liu, B. Liu, J. Liu, T. Pan, Y. Luo, X. Zhang and H. Ou, Cell Death Discovery, 2024, 10, 51 CrossRef PubMed.
- S. Nemec and K. A. Kilian, Nat. Rev. Mater., 2021, 6, 69–83 CrossRef.
- M. Liu, K. Pan, Z. Guo and Z. Li, Regenerative Medicine and Brain Repair, Springer, 2024, pp. 199–217 Search PubMed.
- K. Klimek and G. Ginalska, Polymers, 2020, 12, 844 CrossRef PubMed.
- R. Schofield, Blood Cells, 1978, 4, 7–25 Search PubMed.
- I. A. Khlusov and M. Y. Khlusova, Bioceram. Biocompos., 2019, 291–321 Search PubMed.
- M. Willadsen, M. Chaise, I. Yarovoy, A. Q. Zhang and N. Parashurama, Bioeng. Transl. Med., 2018, 3, 232–255 CrossRef.
- F. Ju, M. M. Atyah, N. Horstmann, S. Gul, R. Vago, C. J. Bruns, Y. Zhao, Q.-Z. Dong and N. Ren, Stem Cell Res. Ther., 2022, 13, 233 CrossRef PubMed.
- A. A. Brockman, B. C. Mobley and R. A. Ihrie, J. Histochem. Cytochem., 2021, 69, 819–834 CrossRef PubMed.
- L. F. Buttó, A. Pelletier, S. K. More, N. Zhao, A. Osme, C. L. Hager, M. A. Ghannoum, R.-P. Sekaly, F. Cominelli and M. Dave, Stem Cell Rep., 2020, 15, 389–407 CrossRef.
- T. Marchand and S. Pinho, Front. Immunol., 2021, 12, 775128 CrossRef.
- A. Ellert-Miklaszewska, K. Poleszak and B. Kaminska, Future Med. Chem., 2017, 9, 199–221 CrossRef PubMed.
- Y. Otsuki, M. Ii, K. Moriwaki, M. Okada, K. Ueda and M. Asahi, Biochem. Biophys. Res. Commun., 2018, 495, 904–910 CrossRef PubMed.
- S. M. Sundaram, L. Varier, K. Z. Fathima, A. Dharmarajan and S. Warrier, Life Sci., 2023, 316, 121384 CrossRef PubMed.
- M. S. Rahman, N. Akhtar, H. M. Jamil, R. S. Banik and S. M. Asaduzzaman, Bone Res., 2015, 3, 1–20 Search PubMed.
- H. Zhang, Z. Liao, W. Wang, Y. Liu, H. Zhu, H. Liang, B. Zhang and X. Chen, Oncogene, 2023, 42, 113–123 CrossRef.
- S. Zhai, J. Lin, Y. Ji, R. Zhang, Z. Zhang, Y. Cao, Y. Liu, X. Tang, J. Liu and P. Liu, Cell Discovery, 2023, 9, 95 CrossRef PubMed.
- H. Guo, J. Wu, H. Wang and Y. Tan, Stem Cell Rev. Rep., 2024, 1–15 Search PubMed.
- Y. Li, T. Jin, N. Liu, J. Wang, Z. Qin, S. Yin, Y. Zhang, Z. Fu, Y. Wu and Y. Wang, J. Neuroinflammation, 2023, 20, 53 CrossRef PubMed.
- C. Di and W. Jia, Crit. Rev. Food Sci. Nutr., 2024, 64, 9210–9227 Search PubMed.
- C. Bonnet, A. Brahmbhatt, S. X. Deng and J. J. Zheng, RSC Chem. Biol., 2021, 2, 1144–1157 RSC.
- M. Holzem, M. Boutros and T. W. Holstein, Nat. Rev. Genet., 2024, 1–13 Search PubMed.
- N. Zhang, H. Shen, B. Chen, H. Hu, C. Liu, Y. Chen and W. Cong, Front. Med., 2023, 10, 1164656 CrossRef.
- X. Chen, H. Tan, J. Xu, Y. Tian, Q. Yuan, Y. Zuo, Q. Chen, X. Hong, H. Fu and F. F. Hou, Kidney Int., 2022, 102, 506–520 Search PubMed.
- X.-Y. Chen, S.-F. Wan, N.-N. Yao, Z.-J. Lin, Y.-G. Mao, X.-H. Yu and Y.-Z. Wang, Mil. Med. Res., 2021, 8, 1–16 Search PubMed.
- A. Colini Baldeschi, E. Pittaluga, F. Andreola, S. Rossi, M. Cozzolino, G. Nicotera, G. Sferrazza, P. Pierimarchi and A. Serafino, Front. Aging Neurosci., 2018, 10, 20 Search PubMed.
- B. Yang, S. Li, Z. Chen, F. Feng, L. He, B. Liu, T. He, X. Wang, R. Chen and Z. Chen, FASEB J., 2020, 34, 3583–3593 CrossRef.
- D. B. Bernkopf, M. Brückner, M. V. Hadjihannas and J. Behrens, Nat. Commun., 2019, 10, 4251 CrossRef PubMed.
- S. A. Khaliq, M.-O. Baek, H.-J. Cho, S. J. Chon and M.-S. Yoon, Front. Cell Dev. Biol., 2020, 8, 609551 CrossRef PubMed.
- L. Dietrich, B. Rathmer, K. Ewan, T. Bange, S. Heinrichs, T. C. Dale, D. Schade and T. N. Grossmann, Cell Chem. Biol., 2017, 24, 958–968 CrossRef PubMed.
- S. Ali, M. U. Rehman, A. M. Yatoo, A. Arafah, A. Khan, S. Rashid, S. Majid, A. Ali and M. N. Ali, Eur. J. Pharmacol., 2023, 947, 175678 CrossRef.
- R. J. Akhurst and A. Hata, Nat. Rev. Drug Discovery, 2012, 11, 790–811 CrossRef PubMed.
- U. Blank and S. Karlsson, Blood, J. Am. Soc. Hematol., 2015, 125, 3542–3550 Search PubMed.
- S.-N. Li and J.-F. Wu, Stem Cell Res. Ther., 2020, 11, 41 CrossRef PubMed.
- K.-S. Park, Int. J. Stem Cells, 2011, 4, 18 CrossRef.
- J. Lee, K.-B. Roh, S.-C. Kim, J. Lee and D. Park, J. Nutr. Biochem., 2012, 23, 1341–1351 CrossRef.
- P. Yu, L. Duan, Z. Yan, J. Li and D.-Z. Cai, Curr. Stem Cell Res. Ther., 2024, 19, 257–266 CrossRef.
- M. Pitou, E. Papachristou, D. Bratsios, G.-M. Kefala, A. S. Tsagkarakou, D. D. Leonidas, A. Aggeli, G. E. Papadopoulos, R. M. Papi and T. Choli-Papadopoulou, Biomedicines, 2023, 11, 3182 CrossRef PubMed.
- T. H. Morgan, Am. Nat., 1917, 51, 513–544 CrossRef.
- S. J. Bray, Nat. Rev. Mol. Cell Biol., 2006, 7, 678–689 CrossRef CAS.
- N. V. Katolikova, A. A. Khudiakov, D. D. Shafranskaya, A. D. Prjibelski, A. E. Masharskiy, M. S. Mor, A. S. Golovkin, A. K. Zaytseva, I. E. Neganova and E. V. Efimova, Int. J. Mol. Sci., 2023, 24, 1429 CrossRef CAS PubMed.
- A. Malashicheva, A. Kostina, A. Kostareva, O. Irtyuga, M. Gordeev and V. Uspensky, Biochim. Biophys. Acta, Mol. Basis Dis., 2020, 1866, 165631 CrossRef CAS.
- G. Ferrari-Toninelli, S. A. Bonini, D. Uberti, L. Buizza, P. Bettinsoli, P. L. Poliani, F. Facchetti and M. Memo, Neuro-Oncology, 2010, 12, 1231–1243 CrossRef CAS PubMed.
- M. R. Urist, Science, 1965, 150, 893–899 CrossRef CAS.
- R. N. Wang, J. Green, Z. Wang, Y. Deng, M. Qiao, M. Peabody, Q. Zhang, J. Ye, Z. Yan and S. Denduluri, Genes Dis., 2014, 1, 87–105 CrossRef PubMed.
- T. U. Wagner, FEBS J., 2007, 274, 2968–2976 CrossRef CAS.
- U. Nöth, R. Tuli, R. Seghatoleslami, M. Howard, A. Shah, D. J. Hall, N. J. Hickok and R. S. Tuan, Exp. Cell Res., 2003, 291, 201–211 CrossRef.
- X. Guo and X.-F. Wang, Cell Res., 2009, 19, 71–88 CrossRef CAS.
- Z. Tong, J. Guo, R. C. Glen, N. W. Morrell and W. Li, Sci. Rep., 2019, 9, 13446 CrossRef PubMed.
- M. Yang, Z. Gao, S. Cheng, Z. Wang, H. Ei-Seedi and M. Du, J. Agric. Food Chem., 2024, 72, 9691–9702 CrossRef CAS PubMed.
- Q. Liu, X. Peng, X. Liu, X. Mou, Y. Guo, L. Yang, Y. Chen, Y. Zhou, Z. Shi and Z. Yang, Composites, Part B, 2023, 262, 110805 CrossRef CAS.
- F. Xu, F. Yang, Y. Qiu, C. Wang, Q. Zou, L. Wang, X. Li, M. Jin, K. Liu and S. Zhang, Fish Shellfish Immunol., 2024, 145, 109351 CrossRef CAS.
- Q. Hu, C. Chen, Z. Lin, L. Zhang, S. Guan, X. Zhuang, G. Dong and J. Shen, Biol. Pharm. Bull., 2023, 46, 382–393 CrossRef CAS.
- K. Zhang, S. Li, J. Hou, Y. Hong, X. Chen, C. Zhou, H. Wu, G. Zheng, C. Zeng and H. Wu, Cell. Mol. Neurobiol., 2023, 43, 2989–3003 CrossRef CAS PubMed.
- J. Liu, L. Zhu, Y. Bao, Z. Du, L. Shi, X. Hong, Z. Zou and G. Peng, J. Controlled Release, 2024, 365, 729–743 CrossRef CAS PubMed.
- E. Y. Plotnikov, D. N. Silachev, V. A. Popkov, L. D. Zorova, I. B. Pevzner, S. D. Zorov, S. S. Jankauskas, V. A. Babenko, G. T. Sukhikh and D. B. Zorov, Heart, Lung Circ., 2017, 26, 648–659 CrossRef.
- N. S. Lin’Kova, A. V. Trofimov and A. V. Dudkov, Bull. Exp. Biol. Med., 2011, 151, 530–531 CrossRef.
- S. Yang, C. Wang, J. Zhu, C. Lu, H. Li, F. Chen, J. Lu, Z. Zhang, X. Yan and H. Zhao, Theranostics, 2020, 10, 8227 CrossRef PubMed.
- V. A. C. Puig-Sanvicens, C. E. Semino and N. I. Zur Nieden, Differentiation, 2015, 90, 101–110 CrossRef.
- O. Schussler, C. Coirault, M. Louis-Tisserand, W. Al-Chare, P. Oliviero, C. Menard, R. J. Michelot, P. Bochet, D. R. Salomon and J. C. Chachques, Nat. Rev. Cardiol., 2009, 6, 240–249 CrossRef.
- S. Alshehri, H. H. Susapto and C. A. E. Hauser, Biomacromolecules, 2021, 22, 2094–2106 CrossRef PubMed.
- H. K. Kim, J. H. Kim, D. S. Park, K. S. Park, S. S. Kang, J. S. Lee, M. H. Jeong and T. R. Yoon, Biomaterials, 2012, 33, 7057–7063 CrossRef.
- Y. Kikkawa, N. Takahashi, Y. Matsuda, T. Miwa, T. Akizuki, A. Kataoka and M. Nomizu, Biomaterials, 2009, 30, 6888–6895 CrossRef PubMed.
- V. K. Khavinson, S. M. Tendler, N. A. Kasyanenko, S. I. Tarnovskaya, N. S. Linkova, V. V. Ashapkin, P. P. Yakutseni and B. F. Vanyushin, Am. J. Biomed. Sci., 2015, 7, 156–169 Search PubMed.
- D. R. Smith, D. J. Margul, C. M. Dumont, M. A. Carlson, M. K. Munsell, M. Johnson, B. J. Cummings, A. J. Anderson and L. D. Shea, Biotechnol. Bioeng., 2019, 116, 155–167 Search PubMed.
- H. Ceylan, S. Kocabey, H. Unal Gulsuner, O. S. Balcik, M. O. Guler and A. B. Tekinay, Biomacromolecules, 2014, 15, 2407–2418 CrossRef PubMed.
- V. K. Khavinson, S. M. Tendler, B. F. Vanyushin, N. A. Kasyanenko, I. M. Kvetnoy, N. S. Linkova, V. V. Ashapkin, V. O. Polyakova, V. S. Basharina and A. Bernadotte, Lung, 2014, 192, 781–791 Search PubMed.
- S. Li, Y. Xu, J. Yu and M. L. Becker, Biomaterials, 2017, 141, 176–187 Search PubMed.
- S. Jadalannagari and O. S. Aljitawi, Tissue Eng., Part B, 2015, 21, 314–322 CrossRef PubMed.
- T. Yasuhara, S. Kawauchi, K. Kin, J. Morimoto, M. Kameda, T. Sasaki, B. Bonsack, C. Kingsbury, N. Tajiri and C. V. Borlongan, CNS Neurosci. Ther., 2020, 26, 595–602 CrossRef PubMed.
- J. A. Zimmermann and D. V. Schaffer, Brain Res. Bull., 2019, 150, 50–60 CrossRef PubMed.
- R. Patel, M. Santhosh, J. K. Dash, R. Karpoormath, A. Jha, J. Kwak, M. Patel and J. H. Kim, Polym. Adv. Technol., 2019, 30, 4–12 CrossRef CAS.
- M. B. Fonseca, S. Solá, J. M. Xavier, P. A. Dionísio and C. M. P. Rodrigues, Mol. Neurobiol., 2013, 48, 829–840 CrossRef CAS PubMed.
- W. Ma, G. Jin, P. M. Gehret, N. C. Chada and W. H. Suh, Biomolecules, 2018, 8, 48 CrossRef.
- H. Zhao, K. B. Adler, C. Bai, F. Tang and X. Wang, J. Proteome Res., 2006, 5, 743–755 CrossRef CAS PubMed.
- R. P. Schleimer and S. Berdnikovs, J. Allergy Clin. Immunol., 2017, 139, 1752–1761 CrossRef CAS PubMed.
- S. Fleming, J. Pathol., 1991, 164, 95–100 CrossRef CAS PubMed.
- Y. Bai, J. Boath, G. R. White, U. G. I. U. Kariyawasam, C. S. Farah and C. Darido, Cancers, 2021, 13, 5123 CrossRef CAS PubMed.
- S. Ragupathy, J. Brunner and G. Borchard, Eur. J. Pharm. Sci., 2021, 160, 105747 CrossRef CAS PubMed.
- A. U. Dignass, Inflammatory Bowel Dis., 2001, 7, 68–77 CrossRef CAS PubMed.
- K. Maruyama, S. Miyazaki, R. Kobayashi, H. Hikita, T. Tsubone and K. Ohnuma, In Vitro Cell. Dev. Biol.: Anim., 2024, 1–9 Search PubMed.
- A. Rodaway and R. Patient, Cell, 2001, 105, 169–172 CrossRef CAS PubMed.
- E. Ferretti and A.-K. Hadjantonakis, Curr. Opin. Cell Biol., 2019, 61, 110–116 CrossRef CAS.
- Y. Kalou, A. M. Al-Khani and K. H. Haider, Heart, Lung Circ., 2023, 32, 870–880 CrossRef PubMed.
- N. H. Goradel, F. G. Hour, B. Negahdari, Z. V. Malekshahi, M. Hashemzehi, A. Masoudifar and H. Mirzaei, J. Cell. Biochem., 2018, 119, 95–104 CrossRef CAS PubMed.
- C. W. Tsao, A. W. Aday, Z. I. Almarzooq, C. A. M. Anderson, P. Arora, C. L. Avery, C. M. Baker-Smith, A. Z. Beaton, A. K. Boehme and A. E. Buxton, Circulation, 2023, 147, e93–e621 CrossRef.
- D. M. Nelson, Z. Ma, K. L. Fujimoto, R. Hashizume and W. R. Wagner, Acta Biomater., 2011, 7, 1–15 CrossRef CAS PubMed.
- K. M. French, I. Somasuntharam and M. E. Davis, Adv. Drug Delivery Rev., 2016, 96, 40–53 CrossRef PubMed.
- Y. Zhang, D. Zhu, Y. Wei, Y. Wu, W. Cui, L. Liuqin, G. Fan, Q. Yang, Z. Wang and Z. Xu, Acta Biomater., 2019, 86, 223–234 CrossRef.
- H. Gao, J. Huang, Q. Wei and C. He, Bioengineering, 2023, 10, 201 CrossRef PubMed.
- N. Varo, Advances in Laboratory Medicine/Avances en Medicina de Laboratorio, 2024, vol. 5, pp. 1–3 Search PubMed.
- G. B. M. Maciel, R. M. Maciel and C. C. Danesi, Mol. Biol. Rep., 2023, 50, 2857–2863 CrossRef PubMed.
- M. T. Valenti, L. Dalle Carbonare and M. Mottes, Int. J. Mol. Sci., 2016, 18, 41 CrossRef PubMed.
- Y. Bai, X. Li, K. Wu, B. C. Heng, X. Zhang and X. Deng, Med. Rev., 2024 DOI:10.1515/mr-2024-0023.
- G. Gulseren, I. C. Yasa, O. Ustahuseyin, E. D. Tekin, A. B. Tekinay and M. O. Guler, Biomacromolecules, 2015, 16, 2198–2208 CrossRef.
- X. Zhou, W. Feng, K. Qiu, L. Chen, W. Wang, W. Nie, X. Mo and C. He, ACS Appl. Mater. Interfaces, 2015, 7, 15777–15789 CrossRef.
- H. Hosseinkhani, M. Hosseinkhani, F. Tian, H. Kobayashi and Y. Tabata, Biomaterials, 2006, 27, 4079–4086 CrossRef.
- J. E. Gough, A. Saiani and A. F. Miller, Bioinspired, Biomimetic Nanobiomater., 2012, 1, 4–12 CrossRef.
- L. A. Castillo Diaz, M. Elsawy, A. Saiani, J. E. Gough and A. F. Miller, J. Tissue Eng., 2016, 7, 2041731416649789 CrossRef PubMed.
- V. Lukasova, M. Buzgo, V. Sovkova, J. Dankova, M. Rampichova and E. Amler, Cell Proliferation, 2017, 50, e12357 CrossRef PubMed.
- J. S. Lee, J. S. Lee and W. L. Murphy, Acta Biomater., 2010, 6, 21–28 CrossRef PubMed.
- J. M. Anderson, J. B. Vines, J. L. Patterson, H. Chen, A. Javed and H.-W. Jun, Acta Biomater., 2011, 7, 675–682 CrossRef CAS PubMed.
- S. X. Hsiong, T. Boontheekul, N. Huebsch and D. J. Mooney, Tissue Eng., Part A, 2009, 15, 263–272 CrossRef CAS PubMed.
- M. Wang, Y. Deng, P. Zhou, Z. Luo, Q. Li, B. Xie, X. Zhang, T. Chen, D. Pei and Z. Tang, ACS Appl. Mater. Interfaces, 2015, 7, 4560–4572 CrossRef CAS PubMed.
- A. Polini, J. Wang, H. Bai, Y. Zhu, A. P. Tomsia and C. Mao, Biomater. Sci., 2014, 2, 1779–1786 RSC.
- F.-Y. Cao, W.-N. Yin, J.-X. Fan, R.-X. Zhuo and X.-Z. Zhang, Biomater. Sci., 2015, 3, 345–351 RSC.
- J. M. Anderson, M. Kushwaha, A. Tambralli, S. L. Bellis, R. P. Camata and H.-W. Jun, Biomacromolecules, 2009, 10, 2935–2944 CrossRef CAS.
- M. Sever, B. Mammadov, M. O. Guler and A. B. Tekinay, Biomacromolecules, 2014, 15, 4480–4487 CrossRef CAS PubMed.
- M. Balakrishnan and J. Rehm, Hepatology, 2024, 79, 451–459 CrossRef PubMed.
- X. Lu, H. Guo, X. Wei, D. Lu, W. Shu, Y. Song, N. Qiu and X. Xu, Int. J. Nanomed., 2023, 2873–2890 CrossRef CAS.
- Y. Tanaka, E. Furuhata, S. Maeda, M. Kishima, H. Suzuki and T. Suzuki, Sci. Data, 2023, 10, 93 CrossRef CAS.
- C. E. Semino, J. R. Merok, G. G. Crane, G. Panagiotakos and S. Zhang, Differentiation, 2003, 71, 262–270 CrossRef CAS.
- A. Meier, S. Mehrle, T. S. Weiss, W. Mier and S. Urban, Hepatology, 2013, 58, 31–42 CrossRef CAS.
- J. Wu, N. Marí-Buyé, T. F. Muiños, S. Borrós, P. Favia and C. E. Semino, J. Nanobiotechnol., 2010, 8, 1–15 CrossRef.
- Z. Ma, T. Hou, W. Shi, W. Liu and H. He, Int. J. Mol. Sci., 2015, 16, 22062–22080 CrossRef CAS.
- K. Piper, S. Brickwood, L. W. Turnpenny, I. T. Cameron, S. G. Ball, D. I. Wilson and N. A. Hanley, J. Endocrinol., 2004, 181, 11–24 Search PubMed.
- H. Edlund, Diabetologia, 2001, 44, 1071–1079 CrossRef PubMed.
- L. Sui, R. L. Leibel and D. Egli, Curr. Protoc. Hum. Genet., 2018, 99, e68 CrossRef PubMed.
- J. K. Mfopou, B. Chen, L. Sui, K. Sermon and L. Bouwens, Diabetes, 2010, 59, 2094–2101 CrossRef PubMed.
- B. Soria, Differentiation, 2001, 68, 205–219 CrossRef.
- J. F. List and J. F. Habener, Am. J. Physiol.: Endocrinol. Metab., 2004, 286, E875–E881 CrossRef.
- D. J. Drucker, Mol. Endocrinol., 2003, 17, 161–171 CrossRef.
- V. K. Khavinson, A. O. Durnova, V. O. Polyakova, G. H. Tolibova, N. S. Linkova, I. M. Kvetnoy, T. V. Kvetnaia and S. I. Tarnovskaya, Bull. Exp. Biol. Med., 2013, 154, 501–504 CrossRef PubMed.
- G. Daziano, N. Blondeau, S. Béraud-Dufour, A. Abderrahmani, C. Rovère, C. Heurteaux, J. Mazella, P. Lebrun and T. Coppola, Pharmacol. Res., 2021, 167, 105539 CrossRef.
- N. Yadav, M. K. Chauhan and V. S. Chauhan, Biomater. Sci., 2020, 8, 84–100 RSC.
- K. E. Eckhart, S. J. Schmidt, F. A. Starvaggi, M. E. Wolf, W. M. Vickery and S. A. Sydlik, Regener. Eng. Transl. Med., 2021, 7, 460–484 CrossRef.
- J. Bojarska, Int. J. Nutr. Sci., 2021, 6, 1046 Search PubMed.
| This journal is © The Royal Society of Chemistry 2025 |