Open Access Article
Open Access ArticleDouble layer 3D codes: fluorescent supramolecular polymeric gels allowing direct recognition of the chloride anion using a smart phone†
Xiaofan
Ji
 a,
Wei
Chen
a,
Wei
Chen
 a,
Lingliang
Long
a,
Lingliang
Long
 *ad,
Feihe
Huang
*ad,
Feihe
Huang
 *c and
Jonathan L.
Sessler
*c and
Jonathan L.
Sessler
 *ab
*ab
aDepartment of Chemistry, The University of Texas at Austin, 105 East 24th Street, Stop A5300, Austin, Texas 78712, USA. E-mail: sessler@cm.utexas.edu
bInstitute for Supramolecular and Catalytic Chemistry, Shanghai University, Shanghai 200444, China
cState Key Laboratory of Chemical Engineering, Center for Chemistry of High-Performance & Novel Materials, Department of Chemistry, Zhejiang University, Hangzhou 310027, P. R. China. E-mail: fhuang@zju.edu.cn; Fax: +86-571-8795-3189; Tel: +86-571-8795-3189
dSchool of Chemistry and Chemical Engineering, Jiangsu University, Zhenjiang, Jiangsu 212013, P. R. China. E-mail: longlingliang@ujs.edu.cn
First published on 30th August 2018
Abstract
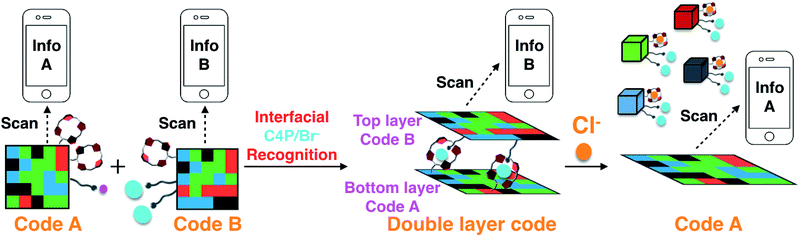
Reported here is a two-layer gel construct that permits the on-site detection of the chloride anion by means of a hand-held UV-vis lamp and a smart phone “app”. To prepare this system, eight fluorescent polymeric gels G1–G8 were synthesized. Gels G1–G4 containing C4P/imidazolium-F− anion recognition motifs were used to construct a fluorescent pattern (Code A) that may be read out by a smart phone as a 3D color code (Info A). Gels G5–G8, which are based on C4P/imidazolium-Br− anion interactions, were used to construct an array (Code B) that may be read as Info B. Code B was then adhered to the surface of Code A to produce an adhered double layer code system stabilized by interfacial C4P/imidazolium-Br− anion interactions. Readout of the top layer (Code B) could be achieved without interference from Code A. Treatment with a Cl− anion source was found to delaminate the top layer and leave intact the more robust bottom layer, allowing the bottom layer (Code A) to be read out. The disparate information content associated with Codes A and B thus permits the on-site recognition of Cl−. The present sensing strategy does not rely on elaborate instrumentation and could provide a new application for elaborated soft materials.
Introduction
Considerable progress has been made in the recognition of anionic analytes in recent years. Typically, detection is effected by means of spectroscopic means. For example, changes in NMR chemical shifts have been used to recognize inter alia chloride and fluoride ions.1–3 Separately, differences in UV/vis spectral intensity have been exploited to analyse a variety of anions.4–7 Several analyte-sensors have been reported that rely on changes in the fluorescence emission intensity.8–14 Anion complexation has also been monitored via mass spectrometry.15,16 However, most of these strategies rely on the availability of laboratory instrumentation to achieve good sensitivity. This can be problematic in terms of cost and accessibility. This provides an incentive to create anion detection platforms that might be amenable for use under field-type conditions. Here we report a new approach towards addressing this latter need namely a two layer polymeric gel system that acts as a two-stage bar code detection system for chloride anions.Barcodes are a widespread feature of modern life. One-dimensional (1D),17 two-dimensional (2D),18 and three-dimensional (3D) codes19,20 rely on lines and spaces, arrays and matrices, and colour, respectively, to provide encoded information. In spite of their ubiquity, the use of bar code approaches to sense targeted chemical analytes has not been widely explored and, to our knowledge, never been applied to the problem of anion sensing. In principle, however, it should be possible to produce a bar code system that would allow for the on-site detection of an anionic analyte using a barcode reader, such as incorporated in, e.g., a smart phone “app”.
Recently, we demonstrated that a “Texas-sized” tetraimidazolium receptor21,22 can be used to create soft materials that interact with anionic components built into the gels.20 When combined with fluorophore-containing domains, these hydrogels could be mechanically assembled into fluorescent 3D colour codes whose encoded information could be read out by a smart phone. However, these first generation materials did not allow for any function other than information coding and read-out. As a first test of whether fluorescent-based multidomain soft materials might have applications utility, we sought to prepare a more complex information encoding system that might function as an anion sensor. We thought this could be achieved by creating a stacked double layer of two different 3D codes, where the adhesion between the constituent layers could be modified by exposure to an analyte. Reading the top code layer, treating with an appropriate chemical stimulus (analyte), and then reading out the bottom code would allow for chemical sensing of the analyte in question (cf.Scheme 1). The targeted analyte would thus play the role of an information transducer, while the disparate information codes scanned in sequence using, e.g., a smart phone “app” (code reader) would allow direct on-site detection of the analyte. The net result would be a functional system that would not only respond to the specific environment, but provide sensing-type information as to what that might be. In the present proof-of-principle study this latter environmental input is a non-aqueous chloride anion source. The net result is a bar code-based chloride anion sensor.
To achieve the function set out in Scheme 1, we appreciated that we would not be able to use simply our previously reported code material.20 Rather the system would need to be fully redesigned. This is because the nature of the interaction between the two putative code layers is crucial to success: an overly strong or overly weak layer-to-layer adhesion could move the response dynamics outside the window where useful sensing is possible. With such a consideration in mind, a new set of gel-based 3D codes was prepared. These are based on calix[4]pyrroles (C4Ps) as the anion recognition element23,24 and halide anion salts of an imidazolium cation as the recognition partners (Scheme 2).25 Simple C4Ps are neutral systems and as a general rule considerably weaker anion receptors than the Texas-sized box used in our first generation system. In organic media, the anion affinities of C4Ps generally follow the trend F− > Cl− > Br− > I−.26 This ordering makes the chloride anion1,2,27,28 attractive for use as a test analyte since it has an intermediate affinity and might be able to promote the delamination of two layer gel system were it appropriately designed. The chloride anion is also an analyte of considerable interest in its own right. It is an “essential anion” that plays a key role in maintaining cellular osmotic pressure and pH-balance across a wide range of species, including humans. It is also highly corrosive. New methods for detecting and sensing Cl− are thus of interest.
The bromide and fluoride anions are arguably of lesser general interest; however, both represent counter anions that can be used in conjunction with imidazolium cations. The stronger affinity of F− for C4P leads us to propose that C4P/imidazolium-F− anion interactions can be used to prepare a robust bottom layer (Code A; Scheme 2a and b) that would be unresponsive when exposed to a chloride anion source.25 In contrast, C4P/imidazolium-Br− anion recognition could be used to stabilize a top layer (Code B; Scheme 2a and c) and promote inter-layer adhesion. In this case, exposure to a chloride anion source would be expected to compete with the C4P-bromide interactions leading to breakup of the layer and delamination of the two-layer system. Such a stimulus-induced loss of the top code layer would expose the lower code layer as shown in Scheme 1.
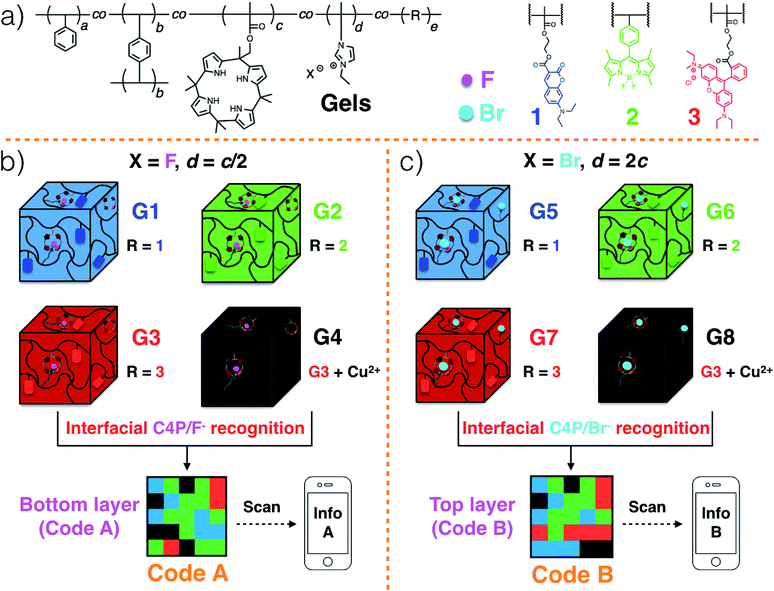
Polymeric gels29–33 consisting of a polystyrene (PS) network cross-linked by p-divinylbenzene,34 as well as tethered C4P anion receptors and pendant imidazolium-X− subunits, where X is either F or Br, were considered attractive for code construction.25 Various blue, green, and red fluorescent elements can be incorporated into the polymers. In accord with our previous work,20 this could be done by adding domains containing coumarin,35 BODIPY,36 and rhodamine B,37 dyes, respectively. To create a non-fluorescent (“black”) gel element, the red gel could be covered with excess Cu(CH3COO)2 powder.20 Individual gel elements could then be pushed together to create multi-colour fluorescent patterns, namely Code A and Code B in the case of the lower and upper layers, respectively (Scheme 2b and c).20 For the lower layer, the gel elements can be prepared so as to contain roughly 2× the number of C4P receptors as imidazolium-F− anion donor subunits. In contrast, a corresponding excess of imidazolium-Br− elements could be incorporated into the top layer. This stoichiometric mismatch should allow the two layers to adhere to one another through relatively weak C4P-bromide anion interactions. The resulting sandwich structure is expected to delaminate when exposed to a chloride anion source.
As true to date, read-out of the top layer (Code B) should be readily achievable using a commercially available smart phone “app”. To read Code A, excess competitive anion (Cl−) would be added to disrupt the adhesion between the two layers. This will leave intact the more robust bottom layer (Code A) stabilized by C4P/imidazolium-F− anion interactions as noted above (Scheme 1). Information encoded in this lower layer (Code A) will then be exposed in a way it can be read out. The chloride anion will thus play the role of an information transducer. As importantly, the system as a whole could act as a Cl− anion sensor. Evidence in support of these design expectations is provided below.
Results and discussion
The fluorescent polymeric gels of the present study were prepared via the free radical copolymerization of styrene, p-divinylbenzene (a covalent cross-linker), methacrylate-functionalized C4P anion monomer,38 imidazolium-anion monomer,39 and a fluorescent monomer containing either coumarin,35 BODIPY,36 or rhodamine B,37 in DMSO using azobisisobutyronitrile (AIBN) as the initiator (Scheme S1, ESI†). Imidazolium subunits bearing F− and Br− anions were used to prepare polymers G1–G4 and G5–G8, respectively. Once prepared, these gels were characterized by fluorescence measurements and rheological experiments (ESI, Fig. S1 and S2†). When illuminated by a handheld UV lamp, blue, green, red, and no fluorescence were seen for G1, G2, G3 and G4, respectively (Fig. 1a). When excited at 365 nm, emission maxima at 469 (coumarin)35 528 (BODIPY),36 and 609 (rhodamine B ester)37 nm were observed for G1, G2, and G3, respectively, whereas no obvious peak was seen for G4 (ESI, Fig. S1a–d†). Similar colour and fluorescent spectral profiles were seen for G5, G6, G7 and G8 (Fig. 1b, and ESI, Fig. S1e–h†). Rheological experiments were then used to investigate the viscoelasticity of gels G1–G8.32,33 The resulting data (ESI, Fig. S2†) proved consistent with dominant elastic behaviour, as would be expected for a gel state.Strain-dependent oscillatory rheological experiments (ESI, Fig. S3†), step-strain rheological measurements (ESI, Fig. S4†) and simple cut/heal tests (ESI, Fig. S5†) provided evidence that these gels would self-heal.32,33 For example, after cutting into two pieces, gels G1–G4 could be re-adhered to provide materials analogous to the original; presumably, this finding reflects the strength of the C4P/imidazolium-F− interactions within the materials (ESI Movie S1 and Fig. S5†). Based on the observed self-healing and the presence of similar C4P/imidazolium-F− recognition motifs in gels G1, G2, G3, and G4, we anticipated that it would be possible to prepare adhesive gels by contacting individual gels with one another.
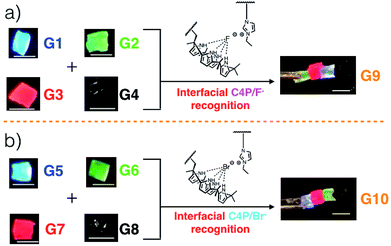
Gels G1, G2, G3, and G4 are each characterized by distinctive fluorescence emission features (ESI, Fig. S1†). Thus, in analogy to what proved true for the first generation gel system based on the “Texas-sized” tetraimidazolium box, it was expected that simple visual monitoring could be used to test whether the proposed hetero-healing would take place. Individual gel elements were thus contacted with one another by pushing together manually on a black nitrile substrate in the absence of any additional solvent. This contact led to formation of an integrated gel G9 within roughly one minute under ambient conditions. After setting aside for about 5 seconds, this newly formed patchwork gel could be lifted up off the nitrile substrate without disintegration (Fig. 1a and ESI Movie S1†).
When excited at 365 nm using a UV lamp, G9 displayed four distinct fluorescent emission features corresponding to the individual gel blocks from which it is derived. Such behaviour is taken as evidence that G9 is a macroscopic gel with blue, green, red, and non-fluorescent domains. Similar homo- and hetero-healing was seen in the case of gels G5–G8 with a specific hetero-healed gel G10 being produced through physical assembly of the individual gel elements. In this case, stabilization was ascribe to the presence of interfacial C4P/imidazolium-Br− anion interactions (Fig. 1b, and ESI, Movie S1†). As with G9, array G10 gives rise to individual and distinct fluorescent domains upon illumination with a hand held UV lamp. In both cases, stretching studies32,33 (ESI Fig. S6a–l†) provided support for the notion that the underlying C4P-imidazolium-X− (X = F or Br) interactions could be used to promote interfacial adhesion.
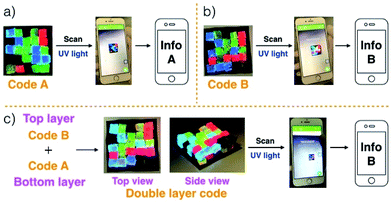
Gels G1–G4 and G5–G8 were then used to prepare two different 3D fluorescent code layers. First, blocks of G1–G4 were cut and arranged on a black nitrile substrate. Contact between the blocks produced a multi-block pattern corresponding to Code A. This pattern proved stable, a finding ascribed to the presence of stabilizing interfacial C4P/imidazolium-F− anion interactions. The colours of this pattern are dim under natural light; however, when placed under a UV lamp (λex = 365 nm), the blue, green, red, and non fluorescent features of the constituent G1, G2, G3, and G4 blocks were readily apparent. The pattern, Code A, was easily recognized by the freely available application software (“app”) COLORCODE™ downloaded onto a smart phone. The net result of this readout is the production of specific information (Info A, Fig. 2a; ESI Movie S3†). Using the same basic approach, G1–G4 blocks were used to form Code B stabilized through presumed interfacial C4P/imidazolium-Br− anion interactions. Again, the encoded information (Info B) could be read out under UV light (Fig. 2b; ESI Movie S4†).
Layering Code B (top layer) on to the surface of Code A (bottom layer) then produces a “sandwich structure” held together by interfacial interactions between the relative excess of C4P receptors in the bottom layer and the excess imidazolium-Br− anion units present in the top layer (Fig. 2c; ESI Movie S5†). A model stretching study (ESI, Fig. S6m†), involving a selected adhesive gel pair corresponding to the two layers (i.e., G1 and G5), served to confirm that the two layers would likely adhere to one another. Indeed, the assembled double layer system could be lifted up intact and proved capable of supporting its own weight soon after the adhesion process was complete (ESI Movie S5†). Most importantly, after production of the two-layer multi-block get array, only the top layer (Code B) could be readout using the code reading app (giving Info A) under conditions of UV illumination, while that of the bottom layer (Code A) was hidden.
Ensembles G9 and G10 were used, respectively, to test the anion-responsiveness of the C4P/imidazolium-F− and C4P/imidazolium-Br− anion interactions present in the construct produced by layering Code B over Code A (Fig. 3). In a first experiment, G9 was immersed in chloroform solutions containing TBAF, TBACl, TBABr, and TBAI, respectively. The same concentrations (20 mM) and volumes (20 mL) were used in all four cases. Initially, G9 proved stable in all four solutions. In the case of the TBAF shaking with immersion for 20 min lead to loss of integrity and dissociation into the four different gel blocks used to prepare G9. No disassembly was observed in the case of the other three solutions, as evidenced by the fact that G9 remained intact and could be lifted up from the corresponding solutions. These observations are in accord with the affinity trend for C4P noted above, which provides support for the conclusion that the Cl−, Br−, and I− anions would be too weak to interfere appreciably with the relatively strong C4P-to-F− interactions present in G9. The fluoride anion concentration in the TBAF solution used in these dissociation tests is high compared to the number of imidazolium-F− subunits present in the gels making up G9 (i.e., G1–G4). Since free F− anion can compete with the imidazolium-F− C4P interactions,40 a decrease in interfacial adhesion among gels G1–G4 was expected, as indeed was seen by experiment.
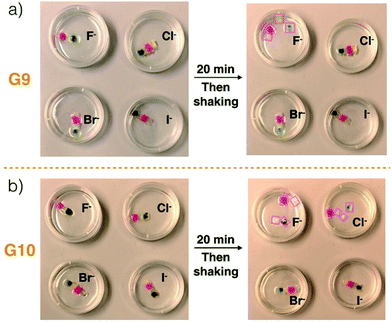 | ||
| Fig. 3 Photographs of (a) G9 and (b) G10 before and after immersion into chloroform solutions containing various tetrabutylammonium halide anion salts (viz. TBAF, TBACl, TBABr, and TBAI) for 20 min. The same concentrations (20 mM) and volumes (20 mL) were used in all cases. The size scale for constructs G9 and G10 is provided in Fig. 1. Disassembled gel blocks are shown using boxes. | ||
Construct G10, which relies on imidazolium-Br− C4P interactions, was subject to identical immersion-based competition experiments. In contrast to what was found for G9, disassembly was observed in the case of both TBAF and TBACl, while no disassembly occurred in otherwise identical tests involving the other two solutions (Fig. 3b and ESI Movie S7†). The disassembly is ascribed to the fact that both free F− and Cl− anions should be able to out-compete the imidazolium-Br− subunits for the C4P anion recognition sites present in the individual gel blocks G5–G8. It is important to note that these latter gels are not fully analogous to gels G1–G4 in that the number of imidazolium-Br− subunits exceeds the concentration of bromide anion provided by the TBABr solution used for the integrity studies. Therefore, in contrast to what was seen for TBAF and G9, treatment of G10 with a free Br− anion source does not lead to discernible levels of break up. Based on these results, we conclude that G9 will disassemble upon exposure to an F− anion source, while G10 will prove responsive to both the F− and Cl− anions. Because of their specific compositions, we expected that these predictions would hold true in the case of Code A and Code B, respectively.
To test the responsiveness of the double layer made up by adhering Code B on Code A, the two layer ensemble was immersed into chloroform solutions containing TBAF, TBACl, TBABr, and TBAI, respectively. The same concentrations (20 mM) and volumes (20 mL) were used in all four test studies. In accord with what was noted above, only the information corresponding to the top layer (Info B) could be read out by a smart phone. This proved true after initial exposure of Code B to all four solutions (ESI Movies S8–S11†). In the specific case of the TBAF solution. Shaking with immersion for 20 min led to totally disassembly of the construct and dispersion of the individual gels into the solvent medium (Fig. 4a, ESI Movie S8†). We conclude that under these conditions, neither the top layer nor the bottom layer remained intact.
An exactly analogous experiment was carried out using a solution of TBACl instead of TBAF. In this case, exposure led only to disassembly of the top layer, while the bottom layer remained intact (Fig. 4b, ESI Movie S9†). The bottom layer exposed in this way could be lifted out from the solution and its information (Info A) could be read out by means of a smart phone. No evidence of disassembly was seen in the case of the TBABr and TBAI solutions, and the original construct could be lifted out from the solution and read again as Info B (Fig. 4c and d, ESI Movies S10 and S11†).
As in the case of the test studies involving G9 and G10, the differential anion-responsiveness of the interfacial C4P/imidazolium-F− and C4P/imidazolium-Br− complexes (Fig. 3) is thought to account for these observations. Free F− anions can outcompete both the C4P/imidazolium-F− and C4P/imidazolium-Br− interactions under the conditions of the experiment (i.e., it can disassemble both Code A and Code B). In contrast, free Cl− anions can only breakup the C4P/imidazolium-Br− interactions that give rise to the integrity of the top layer but not outcompete the C4P/imidazolium-F− interactions present in the lower layer. Thus, only the top layer (Code B) is broken up upon exposure to the TBACl solutions. Neither the Br− and I− anions at the concentrations used can outcompete the imidazolium-Br− C4P interactions present in the gels. These weakly coordinating anions have no appreciable effect on the integrity of the two layers construct. In other words, only upon exposure to Cl− anions is the encoded information transformed (from Info B to Info A). The net result, we propose, is a new approach to the on-site recognition and sensing of the Cl− anion.27,28
It is important to appreciate that this function is made possible by the specific choice of anion receptor, C4P, whose inherent affinities impart the observed analyte specificity. A strongly binding system, such as the tetraimidazolium receptor used in our first generation system20 would not be expected to allow such an anion-based environmental response.
Because it relies on competitive interactions between a C4P host and a negatively charged guest (Cl−), the present system was expected to respond not just to the choice of anion (chloride vs. bromide or iodide), but also the specific Cl− concentration. Below a certain level the competition-induced loss in integrity would not suffice to effect delamination of the layers and little response would be seen; above what is expected to be a sharp cut-off concentration, good sensing would be expected. To test this proposition, the responsiveness of the double layer code was examined in chloroform solutions containing different concentrations of TBACl (ESI, Table S1†). These studies revealed that the information transformation phenomenon leading to differential code-based sensing was effective at Cl− anion concentrations ≥6 mM. This level of detection (LOD) corresponds to the range of chloride anion found within human red blood cells and is much lower than the >100 mM extracellular Cl− concentrations observed for healthy cells.41
Conclusions
In summary, a set of polymeric gels consisting of a polystyrene network cross-linked by p-divinylbenzene and a complex of tethered C4P anion receptors and pendant imidazolium-X− subunits (X = F, Cl, Br, and I) have been prepared. Fluorescent coumarin, BODIPY, and rhodamine subunits were incorporated, respectively, into the polymer backbones of the gels; this allowed production of individual gel blocks displaying blue, green, and red fluorescence upon UV irradiation. Covering a red gel block with excess Cu(CH3COO)2 powder produced a non-fluorescent (“black”) gel. The four fluorescent gels based on imidazolium-F− C4P interactions G1–G4 could be physically assembled on a black nitrile substrate to create a multi-color fluorescent gel array (Code A) that gave rise to a 3D information response (Info A) upon UV irradiation and reading with a smart phone. A complementary set of four fluorescent gels G5–G8 were produced by replacing the pendant imidazolium-F− subunits with imidazolium-Br− moieties. In this case, adhesion between individual gel blocks produced an array (Code B) that produced Info B upon UV illumination. A double layer construct, stabilized by means of interfacial C4P anion interactions, was then constructed by adhering Code B on to the surface of Code A. Only the encoded information of the top layer (Code B) could be read out using a smart phone, while that of the bottom layer (Code A) was hidden. Immersing the double layer code in a solution of TBACl served to remove the top layer and leave intact the more robust bottom layer. This was not true when the corresponding bromide or iodide anion salts were used. These anions bind weakly to the C4P receptor and are not able to compete effectively with the host–guest interactions that stabilize the present gel system.Since the bottom layer encodes for different information than the top layer (Info A vs. Info B), the system functions as a chloride anion sensor that operates by what we believe is a hitherto unexplored recognition and readout mechanism, namely selective layer-by-layer delamination. Furthermore, the top layer can work as a protective (or masking) layer for the bottom layer. This is because the “hidden information” stored in the bottom layer can be read out only if the right reading method (here chloride anion recognition) is applied. The approach reported here thus differs from more classic chloride detection strategies in that it is not predicated on the use of laboratory instrumentation. Rather, and in direct contrast to existing chemosensor-type approaches, it permits the detection of an anionic analyte using a barcode reader. This could prove advantageous in terms of cost, accessibility, and downstream information processing. The present system thus illustrates what might be a new applications area for soft materials.
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
Acknowledgment is made to the donors of The American Chemical Society Petroleum Research Fund for partial support of this research (grant no. 56925-ND7 to J. L. S.). Further support from the Robert A. Welch Foundation (F-0018 to J. L. S.) and Shanghai University is acknowledged. F. H. thanks the National Natural Science Foundation of China (21434005, 91527301) for financial support.Notes and references
- K. P. McDonald, B. Qiao, E. B. Twum, S. Lee, P. J. Gamache, C. H. Chen, Y. Yi and A. H. Flood, Chem. Commun., 2014, 50, 13285–13288 RSC.
- Y. Li and A. H. Flood, Angew. Chem., Int. Ed., 2008, 47, 2649–2652 CrossRef PubMed.
- H. Aboubakr, H. Brisset, O. Siri and J. M. Raimundo, Anal. Chem., 2013, 85, 9968–9974 CrossRef PubMed.
- R. Nishiyabu and P. Anzenbacher Jr, Org. Lett., 2006, 8, 359–362 CrossRef PubMed.
- R. Nishiyabu and P. Anzenbacher Jr, J. Am. Chem. Soc., 2005, 127, 8270–8271 CrossRef PubMed.
- H. Miyaji, W. Sato and J. L. Sessler, Angew. Chem., Int. Ed., 2000, 39, 1777–1780 CrossRef PubMed.
- P. A. Gale and C. Caltagirone, Chem. Soc. Rev., 2015, 44, 4212–4227 RSC.
- H. S. Jung, P. S. Kwon, J. W. Lee, J. I. Kim, C. S. Hong, J. W. Kim, S. Yan, J. Y. Lee, J. H. Lee, T. Joo and J. S. Kim, J. Am. Chem. Soc., 2009, 131, 2008–2012 CrossRef PubMed.
- M. H. Lee, J. H. Han, J. H. Lee, N. Park, R. Kumar, C. Kang and J. S. Kim, Angew. Chem., Int. Ed., 2013, 52, 6206–6209 CrossRef PubMed.
- L. J. Chen, Y. Y. Ren, N. W. Wu, B. Sun, J. Q. Ma, L. Zhang, H. Tan, M. Liu, X. Li and H. B. Yang, J. Am. Chem. Soc., 2015, 137, 11725–11735 CrossRef PubMed.
- R. M. Duke, E. B. Veale, F. M. Pfeffer, P. E. Kruger and T. Gunnlaugsson, Chem. Soc. Rev., 2010, 39, 3936–3953 RSC.
- Z. Guo, W. Zhu and H. Tian, Macromolecules, 2010, 43, 739–744 CrossRef.
- E. Mulugeta, Q. He, D. Sareen, S.-J. Hong, J. H. Oh, V. M. Lynch, J. L. Sessler, S. K. Kim and C.-H. Lee, Chem, 2017, 3, 1008–1020 Search PubMed.
- B. Sui, B. Kim, Y. Zhang, A. Frazer and K. D. Belfield, ACS Appl. Mater. Interfaces, 2013, 5, 2920–2923 CrossRef PubMed.
- S. Chakraborty, S. Saha, L. M. P. Lima, U. Warzok, S. Sarkar, B. Akhuli, M. Nandi, S. Bej, N. N. Adarsh, C. A. Schalley, R. Delgado and P. Ghosh, J. Org. Chem., 2017, 82, 10007–10014 CrossRef PubMed.
- O. Dumele, B. Schreib, U. Warzok, N. Trapp, C. A. Schalley and F. Diederich, Angew. Chem., Int. Ed., 2017, 56, 1152–1157 CrossRef PubMed.
- J. M. Meruga, C. Fountain, J. Kellar, G. Crawford, A. Baride, P. S. May, W. Cross and R. Hoover, Int. J. Comput. Appl., 2015, 37, 17–27 Search PubMed.
- S. Han, H. J. Bae, J. Kim, S. Shin, S. E. Choi, S. H. Lee, S. Kwon and W. Park, Adv. Mater., 2012, 24, 5924–5929 CrossRef PubMed.
- D. H. Park, C. J. Han, Y. G. Shul and J. H. Choy, Sci. Rep., 2014, 4, 4879 CrossRef PubMed.
- X. Ji, R. T. Wu, L. Long, X. S. Ke, C. Guo, Y. J. Ghang, V. M. Lynch, F. Huang and J. L. Sessler, Adv. Mater., 2018, 30, 1705480 CrossRef PubMed.
- H. Y. Gong, B. M. Rambo, E. Karnas, V. M. Lynch and J. L. Sessler, Nat. Chem., 2010, 2, 406–409 CrossRef PubMed.
- Y. D. Yang, J. L. Sessler and H. Y. Gong, Chem. Commun., 2017, 53, 9684–9696 RSC.
- D. S. Kim and J. L. Sessler, Chem. Soc. Rev., 2015, 44, 532–546 RSC.
- S. K. Kim and J. L. Sessler, Acc. Chem. Res., 2014, 47, 2525–2536 CrossRef PubMed.
- A. Aydogan and J. L. Sessler, Chem. Commun., 2014, 50, 13600–13603 RSC.
- P. A. Gale, J. L. Sessler, V. Kral and V. Lynch, J. Am. Chem. Soc., 1996, 118, 5140–5141 CrossRef.
- J. J. Gassensmith, S. Matthys, J. J. Lee, A. Wojcik, P. V. Kamat and B. D. Smith, Chem.–Eur. J., 2010, 16, 2916–2921 CrossRef PubMed.
- R. B. Elmes, P. Turner and K. A. Jolliffe, Org. Lett., 2013, 15, 5638–5641 CrossRef PubMed.
- E. A. Appel, F. Biedermann, D. Hoogland, J. Del Barrio, M. D. Driscoll, S. Hay, D. J. Wales and O. A. Scherman, J. Am. Chem. Soc., 2017, 139, 12985–12993 CrossRef PubMed.
- W. Zheng, G. Yang, N. Shao, L. J. Chen, B. Ou, S. T. Jiang, G. Chen and H. B. Yang, J. Am. Chem. Soc., 2017, 139, 13811–13820 CrossRef PubMed.
- H. Chen, X. Ma, S. Wu and H. Tian, Angew. Chem., Int. Ed., 2014, 53, 14149–14152 CrossRef PubMed.
- J. Liu, C. S. Tan, Z. Yu, Y. Lan, C. Abell and O. A. Scherman, Adv. Mater., 2017, 29, 1604951 CrossRef PubMed.
- J. Liu, C. S. Y. Tan, Z. Yu, N. Li, C. Abell and O. A. Scherman, Adv. Mater., 2017, 29, 1605325 CrossRef PubMed.
- T. Sekine, Y. Takashima, H. Yamaguchi and A. Harada, Chem. Lett., 2016, 45, 1250–1252 CrossRef.
- T. Werther, S. Wahlefeld, J. Salewski, U. Kuhlmann, I. Zebger, P. Hildebrandt and H. Dobbek, Nat. Commun., 2017, 8, 16084 CrossRef.
- X. Wang, D. S. Miller, E. Bukusoglu, J. J. de Pablo and N. L. Abbott, Nat. Mater., 2016, 15, 106–112 CrossRef PubMed.
- M. Beija, C. A. Afonso and J. M. Martinho, Chem. Soc. Rev., 2009, 38, 2410–2433 RSC.
- E. S. Silver, B. M. Rambo, C. W. Bielawski and J. L. Sessler, J. Am. Chem. Soc., 2014, 136, 2252–2255 CrossRef PubMed.
- L. Xia, Q. Cui, X. Suo, Y. Li, X. Cui, Q. Yang, J. Xu, Y. Yang and H. Xing, Adv. Funct. Mater., 2018, 28, 1704292 CrossRef.
- M. Nakahata, Y. Takashima, A. Hashidzume and A. Harada, Angew. Chem., Int. Ed., 2013, 52, 5731–5735 CrossRef PubMed.
- N. Busschaert, S. H. Park, K. H. Baek, Y. P. Choi, J. Park, E. N. W. Howe, J. R. Hiscock, L. E. Karagiannidis, I. Marques, V. Felix, W. Namkung, J. L. Sessler, P. A. Gale and I. Shin, Nat. Chem., 2017, 9, 667–675 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc03463j |
| This journal is © The Royal Society of Chemistry 2018 |