Aptamer recognition-trigged label-free homogeneous electrochemical strategy for an ultrasensitive cancer-derived exosome assay†
Xuehan
Yin
,
Ting
Hou
 ,
Bingzheng
Huang
,
Limin
Yang
* and
Feng
Li
,
Bingzheng
Huang
,
Limin
Yang
* and
Feng
Li
 *
*
College of Chemistry and Pharmaceutical Sciences, Qingdao Agricultural University, Qingdao, 266109, People's Republic of China. E-mail: lifeng@qau.edu.cn
First published on 21st October 2019
Abstract
We developed an aptamer recognition-trigged label-free homogeneous electrochemical biosensing method for highly selective and ultrasensitive detection of cancer-derived exosomes.
Exosomes, a kind of small extracellular vesicle (30–150 nm), are generated by a multitude of cell types and excreted into saliva, serum, urine, and plasma.1,2 Exosomes can act as a multi-component delivery vehicle for carrying a variety of cargoes, such as specific proteins, DNA, and RNA (mRNAs and microRNAs), thus serving as a means of mediating cell–cell communication.3,4 In recent years, with the growing interest in exploring their functions in physiology and pathology, cancer-derived exosomes have been found to play a vital role in cancer occurrence and progression, and can regulate the tumor microenvironment and promote tumor migration and invasion.5,6 In view of these features, cancer-specific exosomes are ideal candidates to serve as a new and promising cancer biomarker.7 Therefore, developing facile, highly sensitive and cost-effective strategies for cancer-derived exosome detection is highly desired in noninvasive early cancer diagnosis.
Although many qualitative studies have been reported, the detection of cancer-specific exosomes remains a great challenge. Traditional characterization techniques, including transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), atomic force microscopy and dynamic light scattering have been used for direct analysis of exosomes,8–10 but these methods fail to detect cancerous exosomes with specificity. Other methods, such as western blotting, flow cytometry or enzyme-linked immunosorbent methods, need expensive equipment or unstable antibodies.11–13 To overcome the drawbacks of these approaches, great efforts have been devoted to developing new methods for exosome detection, such as surface plasmon resonance, colorimetric methods, fluorescence spectroscopy, and surface-enhanced Raman scattering.14–18 These methods have made impressive progress, but several major limitations in terms of sophisticated technical skills and low sensitivity have impeded their widespread applications. Thus, there is a growing and urgent demand for constructing simple, ultrasensitive and easier-to-use strategies to detect cancer-derived exosomes rapidly and specifically.
Compared with antibodies, aptamers possess unique advantages such as chemical stability, easy production, ready modification and low cost. Therefore, aptamer-based strategies have been widely used for quantitative determination of a wide range of analytes.19,20 In particular, electrochemical aptamer-based exosome assays have attracted significant attention in cancer-derived exosome analysis owing to the intrinsic advantages of low cost, simple instrumentation, excellent portability, and easy-operation.21–23 Generally, aptamer probes are usually directly assembled on the electrode surface in conventional electrochemical methods,24 which inevitably hinders the effective recognition between the exosome and aptamer because of the spatial hindrance effect. To solve this problem, Tan's group proposed a DNA nanotetrahedron-based electrochemical method for enhancing the exosome capturing efficiency,25 which greatly improved the detection sensitivity of exosomes. However, the recognition and binding efficiency of the aptamer toward exosomes on the solution-electrode interface is also relatively low compared with that in homogeneous solution, which unavoidably limits the further improvement of the detection sensitivity. More importantly, in all these reported electrode-based electrochemical approaches for exosome assays, the modification of probes on the electrode surface is sophisticated and time-consuming. Inspiringly, an immobilization-free electrochemical protocol opens a promising way for quantitatively analyzing different types of targets, which not only diminishes the experimental protocols and reduces the test cost but also eliminates local steric hindrance.26–30 In homogeneous electrochemical sensing strategies, all the target recognition and signal-amplification procedures occur in homogeneous solution, which are very beneficial for increasing the detection sensitivity. In particular, the label-free homogeneous electrochemical methods circumvented expensive signal probe labeling as well.
Herein, we reported a label-free homogeneous electrochemical sensing platform for an ultrasensitive cancer-derived exosome assay based on Exonuclease III (Exo III)-assisted recycling amplification. The nucleic acids used in this detection system typically contained an aptamer probe P1, a trigger DNA probe P2, and a hairpin DNA probe HP (Scheme 1). The aptamer probe was used to target CD63 protein, which was enriched in most cancer-derived exosome surfaces. The partially complementary P1–P2 duplex DNA probe was synthesized with a blunt 3′-terminus that can resist the Exo III digestion. The designed HP probe contained a loop region I, a stem region II that included rich –GC– sequences and a DNA fragment III that was complementary to the part of the P2 probe. Moreover, the HP probe could form a stable stem-loop structure including a 3′-protruding DNA fragment. The anthracycline doxorubicin (DOX) was selected as an excellent electrochemical redox-active indicator, which showed a stronger binding affinity for double-stranded –GC– sequences than –AT– sequences. In the absence of cancer-derived exosomes, both P1–P2 and HP probes maintained their conformations. When DOX was added into the detection system, most of the DOX would be intercalated into P1–P2 and HP, whose diffusivity was greatly decreased. Furthermore, due to the electrostatic repulsion of P1–P2 and HP probes toward the indium tin oxide (ITO) electrode, the intercalated DOX molecules could not easily reach the electrode surface. Thus, a significant decrease of the electrochemical signal was obtained. In contrast, in the presence of cancer-derived exosomes, the aptamer recognized and bonded exosomes with high affinity, leading to the release of P2. When the released P2 probe hybridized with the HP probe and formed a P2-HP duplex, the Exo III cleavage process was triggered following with the release of intact P2. Then, the released P2 hybridized with another HP probe to trigger the next digestion procedure. In this way, a single exosome can initiate the continual digestion of HP probes, which ultimately lead to less HP probes remaining in the solution. Under these conditions, a great many DOX molecules would be free in solution, which showed strong diffusivity toward the ITO electrode, leading to a great current signal. Therefore, this label-free homogeneous electrochemical platform with the outstanding virtues of simplicity, versatility and excellent sensitivity, can be readily applied in cancer-derived exosome detection.
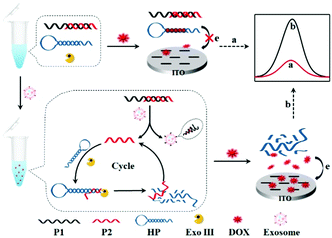 | ||
| Scheme 1 Principle of the aptamer recognition-trigged label-free homogeneous electrochemical strategy for ultrasensitive detection of cancer-derived exosomes. | ||
The electrochemical behaviors of DOX were first investigated using ITO electrodes as the working electrode. Owing to the unique merits of easy preparation and stable structure of hairpin DNA structures, HP1 with rich –GC– sequences and HP2 with rich –AT– sequences were designed respectively. There was a highly differential pulse voltammogram (DPV) signal for free DOX in solution (Fig. S1, curve a, ESI†). However, significantly reduced DPV signals were observed when DOX molecules were added into the HP1 and HP2 solution (Fig. S1, curve b and c, ESI†), respectively. Moreover, it is worth noting that the signal of DOX in the presence of HP1 was lower than that in the presence of HP2. This definitely indicated that DOX showed a higher affinity with –GC– compared to –AT– sequences, resulting in more DOX molecules within HP1, thus inducing a more obviously reduced electrochemical signal. Next, we compared the electrochemical behaviors of DOX with that of methylene blue (MB), a common electrochemical indicator used in electrochemical biosensors.31 More interestingly, we found that the DPV signals of the DOX were much higher than that of the MB under the same concentrations (Fig. S2, ESI†). The well-defined DPV signals of DOX were also detected in different kinds of buffer solutions (Fig. S3, ESI†). In addition, DOX displayed stable signals at different temperatures (Fig. S4, ESI†) and had a high current signal at pH 7.4 (Fig. S5, ESI†). Therefore, all these results demonstrated that DOX was very suitable as a highly effective electrochemical signal reporter for electrochemical assays.
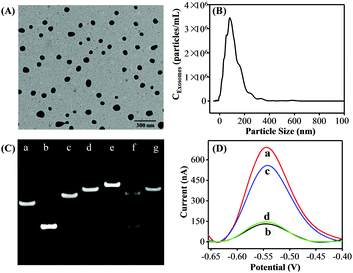
Exosomes were prepared from the cell culture supernatant of human breast cancer cell line MCF-7 cells, which were cultured in exosome-free fetal bovine serum (FBS) and purified by a commercial exosome purification kit. TEM analysis was first employed to investigate the morphology and size of the purified exosomes. The MCF-7 cell-derived exosomes showed spherical shape with a diameter from 50 to 120 nm (Fig. 1A). Then, the concentration and size distribution of the obtained exosomes were characterized by the NTA technique (Fig. 1B) and the results were well consistent with the TEM results and previous reports.21
To confirm the feasibility of the proposed strategy, we performed native polyacrylamide gel electrophoresis (PAGE) experiments and the DPV responses. As shown in Fig. 1C, compared with P1 (lane a) and P2 (lane b) only, the P1 + P2 (lane c) migrated slower, indicating the successful formation of P1–P2 duplex DNA probes. A slower moving electrophoresis band (line e) than that of HP (line d) was observed when adding P2 into the HP solution, demonstrating the hybridization product between HP and P2. Whereas, the duplex P2–HP complex treated by Exo III showed two bands in line f. In addition, the band of the HP + Exo III system (lane g) was similar to that of HP only (lane d). According to the assay principle, after P2 hybridized with the HP, they would be completely digested by Exo III, generating a new ssDNA and releasing the intact P2. Therefore, the results were well consistent with the as-proposed strategy. Inspired by the above results, the DPV responses of the proposed strategy under different conditions were performed to further examine the feasibility of this assay. As a proof of concept, the MCF-7 cell-derived exosomes were selected as the target. As shown in Fig. 1D, a great DPV signal was detected in the solution of DOX. However, a dramatic decrease in the electrochemical signal was observed after both HP and P1–P2 probes were introduced into the DOX solution. In the presence of the MCF-7 cell-derived exosomes, the DPV signal was significantly increased. Also, a negligible peak current change was observed when aptamer P1 was displaced with a random sequence P3. Therefore, all the above results strongly demonstrated the aptamer recognition-induced DNA release and the Exo III-assisted signal amplification for exosome quantitation was feasible.
Appropriate concentrations of HP and Exo III are critical to the detection of the exosome. In the case of fixing 0.02 μM P1–P2, the HP concentration was optimized by measuring the concentration ratio between P1–P2 and HP. The optimal amounts of DOX were 0.08, 0.1, and 0.12 μM when the concentration ratios of P1–P2 and HP were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, 1
0.5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 1
1, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (Fig. S6A–C, ESI†), respectively. Fig. S6D (ESI†) showed that when the concentration ratio between P1–P2 and HP was 1
2 (Fig. S6A–C, ESI†), respectively. Fig. S6D (ESI†) showed that when the concentration ratio between P1–P2 and HP was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, the DPV peak current change Δip showed a similar value but higher than that of 1
2, the DPV peak current change Δip showed a similar value but higher than that of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
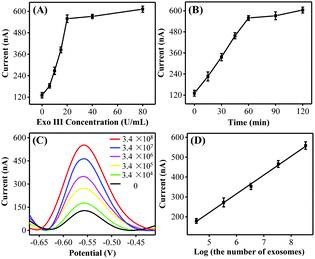
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5. Taking into consideration the DNA consumption, 0.02 μM was selected as the optimal HP amount for the subsequent exosome detection. In addition, the peak currents gradually increased and reached a plateau at 20 U mL−1 and 60 min (Fig. 2B), respectively. Thus, the optimal Exo III amount and the reaction time were selected as 20 U mL−1 and 60 min, respectively.
0.5. Taking into consideration the DNA consumption, 0.02 μM was selected as the optimal HP amount for the subsequent exosome detection. In addition, the peak currents gradually increased and reached a plateau at 20 U mL−1 and 60 min (Fig. 2B), respectively. Thus, the optimal Exo III amount and the reaction time were selected as 20 U mL−1 and 60 min, respectively.
To further confirm whether the as-proposed label-free homogeneous electrochemical platform could be used for the detection of exosomes, the DPV responses were tested with a series of concentrations of MCF-7 cell-derived exosomes. As shown in Fig. 2C, the DPV signals increased successively with the exosome concentration. The DPV peak current displayed a good linear relationship with the logarithm of the number of target exosomes in the range from 3.4 × 104 to 3.4 × 108 particles per mL (Fig. 2D). The limit of detection was estimated to be 1.2 × 104 particles per mL by using the 3σ/slope method (σ is the standard deviation of the blank samples), which was superior to those of the most currently available methods for exosome detection (Table S2, ESI†). Such high sensitivity may be attributed to the improved recognition efficiency of the aptamer toward exosomes and the high Exo III-assisted amplification efficiency in the homogeneous solution. We next investigated the repeatability and the reproducibility of the proposed biosensing strategy. The relative standard deviation (RSD) values were 4.1% and 3.7% respectively, confirming that this approach displayed satisfactory repeatability and reproducibility for exosome assay.
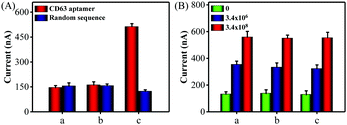
Next, a series of contrast experiments were performed to explore the specificity of the biosensing platform. It has been reported that the CD63 expression of cancer-derived exosomes was higher than that of normal cell-derived exosomes. A significant current change was observed when MCF-7 cell-derived exosomes were added into the detection system (Fig. S7, ESI†). However, the signal change was negligible when human hepatocyte cell line HL-7702 cell-derived exosomes existed. Meanwhile, almost no signal changes were detected when the CD63 aptamer was substituted by a random sequence P3. Consequently, all the above results confirmed that our method was capable of discriminating cancer cell-derived exosomes from the normal cell-derived exosomes with excellent selectivity.
Encouraged by satisfactory sensitivity and selectivity, the detection platform was expected to detect tumor exosomes in complex biological samples. To confirm this, we first assessed the performance of the method for direct detection of exosomes in the culture medium. When CD 63 aptamer was used in the detection system, a significant electrochemical signal was observed in the presence of MCF-7 cells culture medium, while the cell culture supernatant filtrate with a 10 kDa filter showed negligible signal changes (Fig. 3A). At the same time, almost no current signal changes were detected in all of the groups using a random sequence. Also, Fig. S8 (ESI†) showed that the proposed method could detect HL-7702 cell-derived exosomes in the culture medium. Next, we spiked MCF-7 cell-derived exosomes with a known concentration in diluted exosome-free FBS to simulate the clinical system. As shown in Fig. 3B, the current signals obtained from the buffer solution and FBS (5% and 20%) exhibited no obvious difference. Therefore, all of these data clearly demonstrate that the proposed method holds great promise in biological and clinical samples.
In conclusion, we have established a simple and label-free homogeneous electrochemical platform for ultrasensitive detection of cancer-derived exosomes. By virtue of DOX as an excellent electrochemical signal reporter and Exo III-assisted signal amplification, the detection limit for MCF-7 cell-derived exosomes was down to 1.2 × 104 particles per mL, which was much lower than those of most previous reports. Moreover, the proposed strategy demonstrated excellent selectivity to distinguish the cancer cell-derived exosomes from normal cell-derived exosomes and has been effectively applied to detect the target exosomes spiked in biological samples. This sensing platform effectively circumvented the expensive signal molecular labeling and complicated probe immobilization processes, thus showing the outstanding features of simplicity, rapidness, cost-efficiency, and easy manipulation. More importantly, this “signal-on” strategy can quantify a wide range of analytes by choosing the corresponding aptamers, thus holding much promise in biochemical research and disease diagnosis.
This work was financially supported by the National Natural Science Foundation of China (No. 21804077, 21675095 and 21775082), the Special Foundation for Distinguished Taishan Scholars of Shandong Province (ts201511052), and the Research Foundation for Distinguished Scholars of Qingdao Agricultural University (665-1119006).
Conflicts of interest
There are no conflicts to declare.Notes and references
- S. Wan, L. Zhang, S. Wang, Y. Liu, C. Wu, C. Cui, H. Sun, M. Shi, Y. Jiang, L. Li, L. Qiu and W. Tan, J. Am. Chem. Soc., 2017, 139, 5289–5292 CrossRef CAS PubMed
.
- W. Wang, J. Luo and S. Wang, Adv. Healthcare Mater., 2018, 7, 1800484 CrossRef PubMed
.
- M. Tkach and C. Théry, Cell, 2016, 164, 1226–1232 CrossRef CAS PubMed
.
- L. Balaj, R. Lessard, L. Dai, Y. J. Cho, S. L. Pomeroy, X. O. Breakefield and J. Skog, Nat. Commun., 2011, 2, 180 CrossRef PubMed
.
- I. Lazar, E. Clement, S. Dauvillier, D. Milhas, M. Ducoux-Petit, S. LeGonidec, C. Moro, V. Soldan, S. Dalle, S. Balor, M. Golzio, O. Burlet-Schiltz, P. Valet, C. Muller and L. Nieto, Cancer Res., 2016, 76, 4051–4057 CrossRef CAS PubMed
.
- V. Luga, L. Zhang, A. M. Viloria-Petit, A. A. Ogunjimi, M. R. Inanlou, E. Chiu, M. Buchanan, A. N. Hosein, M. Basik and J. L. Wrana, Cell, 2012, 151, 1542–1556 CrossRef CAS PubMed
.
- M. R. Speicher and K. Pantel, Nat. Biotechnol., 2014, 32, 441–443 CrossRef CAS PubMed
.
- E. Van der Pol, F. A. W. Coumans, A. E. Grootemaat, C. Gardiner, I. L. Sargent, P. Harrison, A. Sturk, G. van Leeuwen and R. Nieuwl, J. Thromb. Haemostasis, 2014, 12, 1182–1192 CrossRef CAS PubMed
.
- R. A. Dragovic, C. Gardiner, A. S. Brooks, D. S. Tannetta, D. J. P. Ferguson, P. Hole, B. Carr, C. W. G. Redman, A. L. Harris, P. J. Dobson, P. Harrison and I. L. Sargent, Nanomedicine, 2011, 7, 780–788 CrossRef CAS PubMed
.
- S. D. Ibsen, J. Wright, J. M. Lewis, S. Kim, S. Y. Ko, J. Ong, S. Manouchehri, A. Vyas, J. Akers, C. C. Chen, B. S. Carter, S. C. Esener and M. J. Heller, ACS Nano, 2017, 11, 6641–6651 CrossRef CAS PubMed
.
- H. Zhou, P. S. Yuen, T. Pisitkun, P. A. Gonzales, H. Yasuda, J. W. Dear, P. Gross, M. A. Knepper and R. A. Star, Kidney Int., 2006, 69, 1471–1476 CrossRef CAS PubMed
.
- A. Clayton, J. Court, H. Navabi, M. Adams, M. D. Mason, J. A. Hobot, G. R. Newman and B. Jasani, J. Immunol. Methods, 2001, 247, 163–174 CrossRef CAS PubMed
.
- C. Liu, X. Xu, B. Li, B. Situ, W. Pan, Y. Hu, T. An, S. Yao and L. Zheng, Nano Lett., 2018, 18, 4226–4232 CrossRef CAS PubMed
.
- D. L. M. Rupert, C. Lasser, M. Eldh, S. Block, V. P. Zhdanov, J. O. Lotvall, M. Bally and F. Höök, Anal. Chem., 2014, 86, 5929–5936 CrossRef CAS PubMed
.
- F. He, J. Wang, B.-C. Yin and B.-C. Ye, Anal. Chem., 2018, 90, 8072–8079 CrossRef CAS PubMed
.
- X. Yu, L. He, M. Pentok, H. Yang, Y. Yang, Z. Li, N. He, Y. Deng, L. Song, T. Liu, X. Chen and H. Luo, Nanoscale, 2019, 11, 15589–15595 RSC
.
- Y. M. Wang, J. W. Liu, G. B. Adkins, W. Shen, M. P. Trinh, L. Y. Duan, J. H. Jiang and W. Zhong, Anal. Chem., 2017, 89, 12327–12333 CrossRef CAS PubMed
.
- Z. Wang, S. Zong, Y. Wang, N. Li, L. Li, J. Lu, Z. Wang, B. Chen and Y. Cui, Nanoscale, 2018, 10, 9053–9062 RSC
.
- B. Liu, B. Zhang, G. Chen and D. Tang, Chem. Commun., 2014, 50, 1900–1902 RSC
.
- Y. Nie, X. Yuan, P. Zhang, Y. Q. Chai and R. Yuan, Anal. Chem., 2019, 91, 3452–3458 CrossRef CAS PubMed
.
- R. Huang, L. He, Y. Xia, H. Xu, C. Liu, H. Xie, S. Wang, L. Peng, Y. Liu, N. He and Z. Li, Small, 2019, 15, 1900735 CrossRef PubMed
.
- H. Zhang, Z. Wang, Q. Zhang, F. Wang and Y. Liu, Biosens. Bioelectron., 2019, 124, 184–190 CrossRef PubMed
.
- H. Dong, H. Chen, J. Jiang, H. Zhang, C. Cai and Q. Shen, Anal. Chem., 2018, 90, 4507–4513 CrossRef CAS PubMed
.
- Q. Zhou, A. Rahimian, K. Son, D. S. Shin, T. Patel and A. Revzin, Methods, 2016, 97, 88–93 CrossRef CAS PubMed
.
- S. Wang, L. Zhang, S. Wan, S. Cansiz, C. Cui, Y. Liu, R. Cai, C. Hong, I. T. Teng, M. Shi, Y. Wu, Y. Dong and W. Tan, ACS Nano, 2017, 11, 3943–3949 CrossRef CAS PubMed
.
- F. Xuan, T. W. Fan and I. M. Hsing, ACS Nano, 2015, 9, 5027–5033 CrossRef CAS PubMed
.
- F. Xuan, X. Luo and I. M. Hsing, Anal. Chem., 2013, 85, 4586–4593 CrossRef CAS PubMed
.
- P. Gai, C. Gu, H. Li, X. Sun and F. Li, Anal. Chem., 2017, 89, 12293–12298 CrossRef CAS PubMed
.
- F. Liu, L. Yang, X. Yin, X. Liu, L. Ge and F. Li, Biosens. Bioelectron., 2019, 141, 111446 CrossRef CAS PubMed
.
- J. Zhuang, D. Tang, W. Lai, G. Chen and H. Yang, Anal. Chem., 2014, 86, 8400–8407 CrossRef CAS PubMed
.
- X. Lin, Y. Ni and S. Kokot, Anal. Chim. Acta, 2015, 867, 29–37 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and supplementary figures. See DOI: 10.1039/c9cc07253e |
| This journal is © The Royal Society of Chemistry 2019 |