Antibacterial supramolecular polymers constructed via self-sorting: promoting antibacterial performance and controllable degradation†
Yuchong
Yang
a,
Hao
Hu
a,
Linghui
Chen
a,
Haotian
Bai
b,
Shu
Wang
b,
Jiang-Fei
Xu
 *a and
Xi
Zhang
*a and
Xi
Zhang
 a
a
aKey Lab of Organic Optoelectronics and Molecular Engineering, Department of Chemistry, Tsinghua University, Beijing 100084, China. E-mail: xujf@mail.tsinghua.edu.cn
bKey Laboratory of Organic Solids, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China
First published on 19th March 2019
Abstract
An antibacterial supramolecular polymer is developed that exhibits enhanced antibacterial efficacy and controlled degradability. To this end, a water-soluble bifunctional monomer bearing two naphthalene moieties and a phenylbenzene moiety, as well as a small molecule with antibacterial activity were synthesized. The side-chain antibacterial supramolecular polymer was fabricated through self-sorting of the bifunctional monomer, the small-molecule antibacterial agent, and cucurbit[8]uril. It displayed enhanced antibacterial efficacy against ampicillin-resistant E. coli owing to the local enrichment effect. The controlled degradation of the antibacterial supramolecular polymer could well regulate its antibacterial performance from high to low activity. It is anticipated that this line of research may lead to novel antibacterial supramolecular materials for building degradable antibiotics with enhanced antibacterial efficacy to fight against drug-resistant bacteria.
Introduction
Infections caused by drug-resistant bacteria are a severe threat to human health, because of the long-term abuse of antibiotics in medical treatments, the agricultural field etc.1–7 Prolonged antibiotic treatment can lead to an increase of bacterial drug resistance.8 To tackle this severe threat, supramolecular antibiotics have been employed to combat antibiotic resistance, benefiting from the dynamicity, degradability and reversibility of supramolecular interactions.9–27 To avoid long-term exposure to the active antibiotics, the antibacterial performance can be switched “on” and “off” by a supramolecular strategy, and the emergence of drug-resistant bacteria could be slowed down. However, to further inhibit the emergence of drug-resistant bacteria, there is a need not only to construct degradable antibiotics, but also to promote the antibacterial activity of such antibacterial materials.We wondered if supramolecular polymers with controllable antibacterial efficacy could be fabricated. Supramolecular polymers refer to polymers in which monomers are connected with each other via non-covalent interactions.28–56 By construction of supramolecular polymers, functional monomers can be linked together through reversible non-covalent interactions. Therefore, we assume that if small-molecule antibiotics could be linked together to form supramolecular polymers, the antibacterial efficacy could be significantly enhanced owing to the local enrichment effect. Besides, benefiting from the dynamicity and reversibility of supramolecular interactions, such antibacterial supramolecular polymers can be controllably degraded. Thus, enhanced antibacterial efficacy and degradability of antibacterial materials could be realized by fabricating antibacterial supramolecular polymers.
Herein, we attempted to construct a side-chain antibacterial supramolecular polymer via a self-sorting strategy. Self-sorting is a self-assembly process in which molecules are endowed with the ability to selectively and specifically form complexes with their own recognition units within a complex mixture.57–64 It has been reported that supramolecular polymers with controlled structure and molecular weight can be constructed through the self-sorting strategy.65,66 By employing this strategy, we designed a side-chain antibacterial supramolecular polymer, by which antibacterial agents could be grafted to the supramolecular polymer backbone as the side chains. Then antibacterial agents could be locally enriched for promoting antibacterial performance. Moreover, the backbone of the antibacterial supramolecular polymer could be controllably degraded, thus reducing the antibacterial activity in case of need. The architecture of the antibacterial supramolecular polymer consisted of two parts, the supramolecular polymer backbone and the side-chain antibacterial agents. As shown in Scheme 1, a bifunctional monomer (Np–PhDi–Np), containing two naphthalene (Np) moieties and a phenylbenzene (PhDi) moiety, was designed and synthesized to construct the supramolecular polymer backbone through the host–guest interaction of Np–PhDi–Np and cucurbit[8]uril (CB[8]).
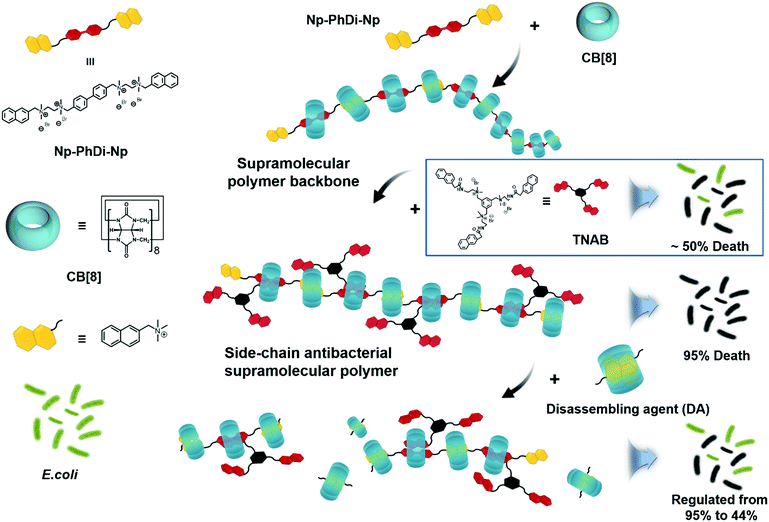 | ||
| Scheme 1 Chemical structures of building blocks and schematic illustration of the fabrication of side-chain antibacterial supramolecular polymer. | ||
A small molecule with antibacterial activity (TNAB) containing naphthalene acetamide (NpAM) moieties could be grafted to the supramolecular polymer backbone as side chains via self-sorting. The hydrophobic NpAM moieties of TNAB could insert into the membranes of bacteria for bacterial membranolysis. By locally enriching the TNAB on the supramolecular polymer backbone, the capacity for bacterial membranolysis may be remarkably increased. The degradation of the antibacterial supramolecular polymer could be realized by addition of a disassembling agent. Therefore, we may fabricate the antibacterial supramolecular polymer with tailor-made antibacterial efficacy and degradability.
Results and discussion
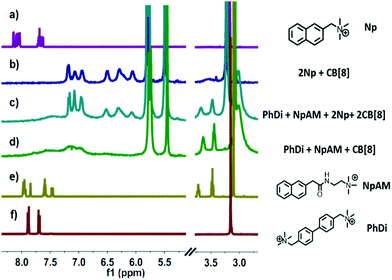
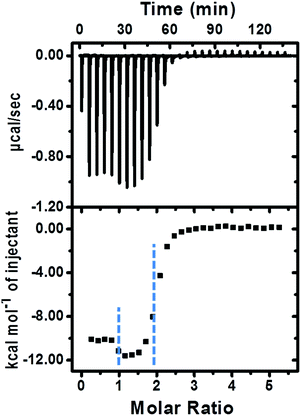
The fabrication of the supramolecular polymer backbone through the host–guest interaction of Np–PhDi–Np with CB[8] was demonstrated by isothermal titration calorimetry (ITC), 1H NMR and diffusion-ordered NMR spectroscopy (DOSY). As shown in the 1H NMR analysis in Fig. S1 (ESI†), when the molar ratio of CB[8] to Np–PhDi–Np increased from 0 to 1.0, the peaks of Np moiety shifted up-field, while the peaks of PhDi moiety shifted down-field. These shifting peaks indicated that the CB[8] combined with Np moieties of Np–PhDi–Np primarily, acting as the driving force for supramolecular polymerization. When the molar ratio of CB[8] to Np–PhDi–Np increased from 1.0 to 2.0, the peaks of PhDi moiety shifted up-field, which indicated that the CB[8] combined with the PhDi moiety of Np–PhDi–Np subsequently. The two-step host–guest complexations were also indicated by ITC data, as shown in Fig. 1. The first abrupt change at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 could be ascribed to the complexation between CB[8] and Np moieties, and the second abrupt change at a molar ratio of 2
1 could be ascribed to the complexation between CB[8] and Np moieties, and the second abrupt change at a molar ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 could be ascribed to the complexation between CB[8] and PhDi moiety. The two-step complexation may lead to the formation of the supramolecular polymer backbone.
1 could be ascribed to the complexation between CB[8] and PhDi moiety. The two-step complexation may lead to the formation of the supramolecular polymer backbone.
 | ||
| Fig. 1 ITC data (1× PBS buffer, pH = 7.0, 25.0 °C) of CB[8] (0.25 mM) titrated into Np–PhDi–Np (0.010 mM). | ||
Direct evidence for the formation of the supramolecular polymer backbone was demonstrated by DOSY. The diffusion coefficient of Np–PhDi–Np (0.25 mM) in water was 2.80 × 10−10 m2 s−1. After addition of 2 equivalents of CB[8], the diffusion coefficient was decreased to 1.10 × 10−10 m2 s−1, which indicated the formation of polymeric species. Taking the 1H NMR data, ITC data and DOSY measurements into account, the supramolecular polymer backbone is formed on the basis of the host–guest complexations between Np–PhDi–Np and CB[8].
To fabricate a side-chain antibacterial supramolecular polymer, TNAB containing three NpAM moieties was designed as the side chains to graft to the supramolecular polymer backbone through self-sorting. The self-sorting among Np, NpAM, PhDi model molecules and CB[8] was studied by 1H NMR, as demonstrated in Fig. 2. When Np, NpAM, PhDi model molecules and CB[8] were mixed at a ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, only the complexation between Np and CB[8] and the complexation of NpAM, PhDi and CB[8] were observed. The ITC data further demonstrated that Np could bind with CB[8] at a ratio of 2
2, only the complexation between Np and CB[8] and the complexation of NpAM, PhDi and CB[8] were observed. The ITC data further demonstrated that Np could bind with CB[8] at a ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with a binding constant of 2.25 × 1012 M−2. In addition, the NpAM moiety could insert into the supramolecular polymer backbone with a binding constant of 2.01 × 105 M−1, as shown in Fig. S4 (ESI†). Based on the self-sorting of the model molecules, TNAB, containing three NpAM moieties, could insert into the supramolecular polymer as side chains for constructing the antibacterial supramolecular polymer.
1 with a binding constant of 2.25 × 1012 M−2. In addition, the NpAM moiety could insert into the supramolecular polymer backbone with a binding constant of 2.01 × 105 M−1, as shown in Fig. S4 (ESI†). Based on the self-sorting of the model molecules, TNAB, containing three NpAM moieties, could insert into the supramolecular polymer as side chains for constructing the antibacterial supramolecular polymer.
The formation of the side-chain antibacterial supramolecular polymer was confirmed by analytical ultracentrifugation. As shown in Fig. S5a and b (ESI†), at a low concentration of 50 μM, the highest molecular weight of supramolecular polymer backbone was measured as 1.7 × 104 Da, corresponding to a degree of polymerization (DP) of 5. After addition of TNAB, the molecular weight of the antibacterial supramolecular polymer was increased up to 2.1 × 104 Da. The increased molecular weight indicated that 5 TNAB molecules were grafted to the supramolecular polymer backbone. Although there were three NpAM moieties on TNAB, the same DP of supramolecular backbone and antibacterial supramolecular polymer indicated that the crosslinking among the supramolecular polymer chains was negligible because of the low concentration. In addition, the DP of the antibacterial supramolecular polymer could be increased with increasing concentration of the building blocks. For example, the DP was measured to be 12 at a concentration of 0.50 mM, as shown in Fig. S5c (ESI†). Therefore, the side-chain antibacterial supramolecular polymer is successfully fabricated through self-sorting.
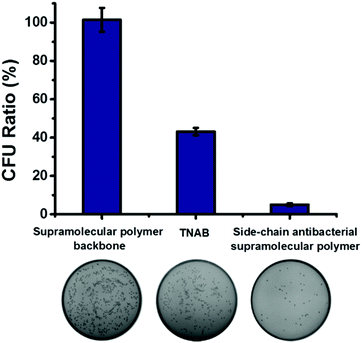
We have demonstrated successfully that antibacterial activity can be enhanced owing to the local enrichment of the antibacterial small molecules through the construction of the side-chain antibacterial supramolecular polymer. For comparison, antibacterial activity of the antibacterial supramolecular polymer, supramolecular polymer backbone and TNAB were investigated. As shown in Fig. 3, the supramolecular polymer backbone displayed almost no antibacterial activity, and the bacterial inhibition ratio of TNAB against ampicillin-resistant E. coli was calculated as 57%. When TNAB was grafted to the supramolecular backbone for forming the antibacterial supramolecular polymer, the bacterial inhibition ratio was remarkably increased up to 95%. Therefore, the antibacterial activity can be promoted by fabrication of the antibacterial supramolecular polymer.
The dynamic nature of supramolecular interactions endowed the supramolecular polymer with degradability, which could be used to turn off the antibacterial activity of the antibacterial supramolecular polymer for avoiding long-term drug exposure. The antibacterial supramolecular polymer could be degraded by addition of a disassembling agent (DA), naphthalene derivatives, through competing with the host–guest interaction between Np and CB[8], as indicated by DOSY shown in Table S1 (ESI†). Then the antibacterial efficacy could be regulated by the controllable degradation of the antibacterial supramolecular polymer. As shown in Fig. 4, by gradual addition of the DA to the antibacterial supramolecular polymer, the bacterial inhibition ratio was gradually decreased from 95% to 44%. The controllable degradation of the antibacterial supramolecular polymer could diminish the enrichment of TNAB, leading to the decrease of antibacterial activity. Therefore, the antibacterial efficacy of the antibacterial supramolecular polymer can be well regulated from high to low activity.
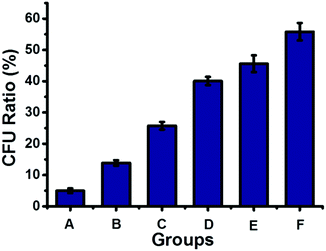 | ||
| Fig. 4 The antibacterial activity of the antibacterial supramolecular polymer with different amounts of a disassembling agent, naphthalene derivatives (DA shown in Scheme 1). (A) 50 μM antibacterial supramolecular polymer; (B) A + 10 μM DA; (C) A + 20 μM DA; (D) A + 30 μM DA; (E) A + 40 μM DA; (F) A + 50 μM DA. | ||
ζ-Potential measurements and scanning electron microscopy (SEM) were employed to understand the interactions between the antibacterial supramolecular polymer and bacteria. Upon addition of the antibacterial supramolecular polymer, the surface charge of E. coli was altered from −48.0 mV to 19.9 mV (Table S2, ESI†), indicating that the positively charged antibacterial supramolecular polymer was adsorbed onto the surface of E. coli. It was demonstrated by SEM that the antibacterial mechanism was membranolysis of the bacteria (Fig. S6, ESI†). As reported, the hydrophobic groups of antibacterial agents can insert into the membranes of bacteria, resulting in membranolysis.13,16,17 By locally enriching the hydrophobic groups of TNAB on the antibacterial supramolecular polymer, the membranolysis of bacteria can be enhanced, thus promoting the antibacterial efficacy.
The cytotoxicity of TNAB, supramolecular polymer backbone and antibacterial supramolecular polymer on mammalian cells was also investigated by MTT assay. A nonmalignant epithelial cell line, HaCaT, was chosen as representative cells. As shown in Fig. 5, no cytotoxicity was observed for the supramolecular antibacterial polymer, TNAB and supramolecular polymer backbone when their concentrations were lower than 0.125 mM. The antibacterial supramolecular polymer and TNAB exhibited cytotoxicity when their concentrations were higher than 0.125 mM. However, the supramolecular polymer backbone displayed no clear cytotoxicity even if the concentration was increased up to 0.50 mM. These results suggest that the fabrication of the antibacterial supramolecular polymer is a potential biocompatible strategy for further medical treatment.
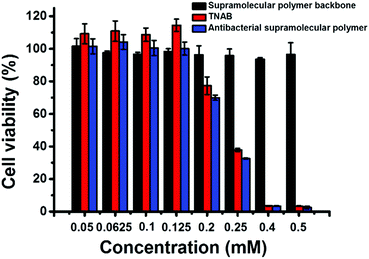 | ||
| Fig. 5 The cytotoxicity of supramolecular polymer backbone, TNAB and antibacterial supramolecular polymer on mammalian cells (HaCaT). | ||
Conclusions
In conclusion, we have successfully fabricated a side-chain antibacterial supramolecular polymer via the self-sorting strategy. The side-chain antibacterial supramolecular polymer exhibits enhanced antibacterial efficacy and controllable degradation. Besides E. coli, this kind of supramolecular polymeric antibacterial material could be used for killing other kinds of microbes. In addition, it is anticipated that other antibiotic reagents may also be supramolecularly polymerized through this method, thus providing a general strategy for building degradable polymer antibiotics with enhanced antibacterial efficacy to fight against drug-resistant bacteria.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (21434004, 21821001, 91527000) and the Tsinghua University Initiative Research Project (20181080084).Notes and references
- Y. Qian, D. Zhang, Y. Wu, Q. Chen and R. Liu, Acta Polym. Sin., 2016, 1300–1311 CAS.
- C. Zhu, L. Liu, Q. Yang, F. Lv and S. Wang, Chem. Rev., 2012, 112, 4687–4735 CrossRef CAS PubMed.
- Y. Yang, P. He, Y. Wang, H. Bai, S. Wang, J.-F. Xu and X. Zhang, Angew. Chem., Int. Ed., 2017, 56, 16239–16242 CrossRef CAS PubMed.
- S. E. Rossiter, M. H. Fletcher and W. M. Wuest, Chem. Rev., 2017, 117, 12415–12474 CrossRef CAS PubMed.
- G. D. Wright, ACS Infect. Dis., 2015, 1, 80–84 CrossRef CAS PubMed.
- I. B. Seiple, Z. Zhang, P. Jakubec, A. Langlois-Mercier, P. M. Wright, D. T. Hog, K. Yabu, S. R. Allu, T. Fukuzaki, P. N. Carlsen, Y. Kitamura, X. Zhou, M. L. Condakes, F. T. Szczypiński, W. D. Green and A. G. Myers, Nature, 2016, 533, 338–345 CrossRef CAS PubMed.
- D. J. Payne, M. N. Gwynn, D. J. Holmes and D. L. Pompliano, Nat. Rev. Drug Discovery, 2007, 6, 29–40 CrossRef CAS PubMed.
- H. Bai, X. Fu, Z. Huang, F. Lv, L. Liu, X. Zhang and S. Wang, ChemistrySelect, 2017, 2, 7940–7945 CrossRef CAS.
- W. A. Velema, J. P. van der Berg, M. J. Hansen, W. Szymanski, A. J. M. Driessen and B. L. Feringa, Nat. Chem., 2013, 5, 924–928 CrossRef CAS PubMed.
- W. Szymanski, J. M. Beierle, H. A. V. Kistemaker, W. A. Velema and B. L. Feringa, Chem. Rev., 2013, 113, 6114–6178 CrossRef CAS PubMed.
- H. Bai, F. Lv, L. Liu and S. Wang, Chem. – Eur. J., 2016, 22, 11114–11121 CrossRef CAS PubMed.
- Z. Huang, H. Zhang, H. Bai, Y. Bai, S. Wang and X. Zhang, ACS Macro Lett., 2016, 5, 1109–1113 CrossRef CAS.
- H. Bai, H. Yuan, C. Nie, B. Wang, F. Lv, L. Liu and S. Wang, Angew. Chem., Int. Ed., 2015, 54, 13208–13213 CrossRef CAS PubMed.
- X. Chen, F. Wang, J. Y. Hyun, T. Wei, J. Qiang, X. Ren, I. Shin and J. Yoon, Chem. Soc. Rev., 2016, 45, 2976–3016 RSC.
- X. Li, S. Lee and J. Yoon, Chem. Soc. Rev., 2018, 47, 1174–1188 RSC.
- L. Chen, H. Bai, J.-F. Xu, S. Wang and X. Zhang, ACS Appl. Mater. Interfaces, 2017, 9, 13950–13957 CrossRef CAS PubMed.
- Y. Yang, Z. Cai, Z. Huang, X. Tang and X. Zhang, Polym. J., 2018, 50, 33–44 CrossRef CAS.
- Y. Liu and J. A. Imlay, Science, 2013, 339, 1210–1213 CrossRef CAS PubMed.
- M. Garland, S. Loscher and M. Bogyo, Chem. Rev., 2017, 117, 4422–4461 CrossRef CAS PubMed.
- W. Kim, W. Zhu, G. L. Hendricks, D. Van Tyne, A. D. Steele, C. E. Keohane, N. Fricke, A. L. Conery, S. Shen, W. Pan, K. Lee, R. Rajamuthiah, B. B. Fuchs, P. M. Vlahovska, W. M. Wuest, M. S. Gilmore, H. Gao, F. M. Ausubel and E. Mylonakis, Nature, 2018, 556, 103–107 CrossRef CAS PubMed.
- B. S. Li, R. Wen, S. Xue, L. Shi, Z. Tang, Z. Wang and B. Z. Tang, Mater. Chem. Front., 2017, 1, 646–653 RSC.
- G. Huang, R. Wen, Z. Wang, B. S. Li and B. Z. Tang, Mater. Chem. Front., 2018, 2, 1884–1892 RSC.
- P. I. O’Daniel, Z. Peng, H. Pi, S. A. Testero, D. Ding, E. Spink, E. Leemans, M. A. Boudreau, T. Yamaguchi, V. A. Schroeder, W. R. Wolter, L. I. Llarrull, W. Song, E. Lastochkin, M. Kumarasiri, N. T. Antunes, M. Espahbodi, K. Lichtenwalter, M. A. Suckow, S. Vakulenko, S. Mobashery and M. Chang, J. Am. Chem. Soc., 2014, 136, 3664–3672 CrossRef PubMed.
- M. Zhang, P.-P. Zhu, P. Xin, W. Si, Z.-T. Li and J.-L. Hou, Angew. Chem., Int. Ed., 2017, 56, 2999–3003 CrossRef CAS PubMed.
- X. Li, D. Lee, J.-D. Huang and J. Yoon, Angew. Chem., Int. Ed., 2018, 57, 9885–9890 CrossRef CAS PubMed.
- X. Li, H. Bai, Y. Yang, J. Yoon, S. Wang and X. Zhang, Adv. Mater., 2019, 31, 1805092 Search PubMed.
- P.-Z. Chen, L.-Y. Niu, H. Zhang, Y.-Z. Chen and Q.-Z. Yang, Mater. Chem. Front., 2018, 2, 1323–1327 RSC.
- L. Yang, X. Tan, Z. Wang and X. Zhang, Chem. Rev., 2015, 115, 7196–7239 CrossRef CAS PubMed.
- J.-F. Xu, Y.-Z. Chen, D. Wu, L.-Z. Wu, C.-H. Tung and Q.-Z. Yang, Angew. Chem., Int. Ed., 2013, 52, 9738–9742 CrossRef CAS PubMed.
- X. Ma and H. Tian, Acc. Chem. Res., 2014, 47, 1971–1981 CrossRef CAS PubMed.
- R. Sun, C. Xue, X. Ma, M. Gao, H. Tian and Q. Li, J. Am. Chem. Soc., 2013, 135, 5990–5993 CrossRef CAS PubMed.
- H.-Q. Peng, X. Zheng, T. Han, R. T. K. Kwok, J. W. Y. Lam, X. Huang and B. Z. Tang, J. Am. Chem. Soc., 2017, 139, 10150–10156 CrossRef CAS PubMed.
- D.-S. Guo and Y. Liu, Chem. Soc. Rev., 2012, 41, 5907–5921 RSC.
- G. Yu, X. Zhao, J. Zhou, Z. Mao, X. Huang, Z. Wang, B. Hu, Y. Liu, F. Zhang, Z. He, O. Jacobson, C. Gao, W. Wang, C. Yu, X. Zhu, F. Huang and X. Chen, J. Am. Chem. Soc., 2018, 140, 8005–8019 CrossRef CAS PubMed.
- J. D. Barrio, P. N. Horton, D. Lairez, G. O. Lloyd, C. Toprakcioglu and O. A. Scherman, J. Am. Chem. Soc., 2013, 135, 11760–11763 CrossRef PubMed.
- C. Fouquey, J.-M. Lehn and A.-M. Levelut, Adv. Mater., 1990, 2, 254–257 CrossRef CAS.
- U. S. Schubert, O. Hien and C. Eschbaumer, Macromol. Rapid Commun., 2000, 21, 1156–1161 CrossRef CAS.
- M. Burnworth, L. Tang, J. R. Kumpfer, A. J. Duncan, F. L. Beyer, G. L. Fiore, S. J. Rowan and C. Weder, Nature, 2011, 472, 334–337 CrossRef CAS PubMed.
- J. Sautaux, L. M. Espinosa, S. Balog and C. Weder, Macromolecules, 2018, 51, 5867–5874 CrossRef CAS.
- T. Aida, E. W. Meijer and S. I. Stupp, Science, 2012, 335, 813–817 CrossRef CAS PubMed.
- J. Murray, K. Kim, T. Ogoshi, W. Yao and B. C. Gibb, Chem. Soc. Rev., 2017, 46, 2479–2496 RSC.
- A. Harada, Y. Takashima and H. Yamaguchi, Chem. Soc. Rev., 2009, 38, 875–882 RSC.
- S. Yang, A. V. Ambade and M. Weck, Chem. Soc. Rev., 2011, 40, 129–137 RSC.
- T. F. A. de Greef and E. W. Meijer, Nature, 2008, 453, 171–173 CrossRef CAS PubMed.
- X. Hou, C. Ke, Y. Zhou, Z. Xie, A. Alngadh, D. T. Keane, M. S. Nassar, Y. Y. Botros, C. A. Mirkin and J. F. Stoddart, Chem. – Eur. J., 2016, 22, 12301–12306 CrossRef CAS PubMed.
- J.-F. Xu and X. Zhang, Acta Polym. Sin., 2017, 37–49 Search PubMed.
- K.-D. Zhang, J. T. Hanifi, Y. Zhang, A. C.-H. Sue, T.-Y. Zhou, L. Zhang, X. Zhao, Y. Liu and Z.-T. Li, J. Am. Chem. Soc., 2013, 135, 17913–17918 CrossRef CAS PubMed.
- Z. Zhang, Y. Luo, J. Chen, S. Dong, Y. Yu, Z. Ma and F. Huang, Angew. Chem., Int. Ed., 2011, 50, 1397–1401 CrossRef CAS PubMed.
- H. Wang, X. Ji, Z. Li, C. N. Zhu, X. Yang, T. Li, Z. L. Wu and F. Huang, Mater. Chem. Front., 2017, 1, 167–171 RSC.
- Y. Liu, Y. Zhang, H. Zhu, H. Wang, W. Tian and B. Shi, Mater. Chem. Front., 2018, 2, 1568–1573 RSC.
- X. Yan, D. Xu, X. Chi, J. Chen, S. Dong, X. Ding, Y. Yu and F. Huang, Adv. Mater., 2012, 24, 362–369 CrossRef CAS PubMed.
- S. Dong, J. Leng, Y. Feng, M. Liu, C. J. Stackhouse, A. Schçnhals, L. Chiappisi, L. Gao, W. Chen, J. Shang, L. Jin, Z. Qi and C. A. Schalley, Sci. Adv., 2017, 3, eaao0900 CrossRef PubMed.
- X. Yan, Z. Liu, Q. Zhang, J. Lopez, H. Wang, H.-C. Wu, S. Niu, H. Yan, S. Wang, T. Lei, J. Li, D. Qi, P. Huang, J. Huang, Y. Zhang, Y. Wang, G. Li, J. B.-H. Tok, X. Chen and Z. Bao, J. Am. Chem. Soc., 2018, 140, 5280–5289 CrossRef CAS PubMed.
- Z. Huang, B. Qin, L. Chen, J.-F. Xu, C. F. J. Faul and X. Zhang, Macromol. Rapid Commun., 2017, 38, 1700312 CrossRef PubMed.
- Y.-Y. Huang, Y. Tian, X.-Q. Liu, Z. Niu, Q.-Z. Yang, V. Ramamurthy, C.-H. Tung, Y.-Z. Chen and L.-Z. Wu, Mater. Chem. Front., 2018, 2, 1893–1899 RSC.
- S. Ogi, V. Stepanenko, K. Sugiyasu, M. Takeuchi and F. Würthner, J. Am. Chem. Soc., 2015, 137, 3300–3307 CrossRef CAS PubMed.
- M. M. Safont-Sempere, G. Fernández and F. Würthner, Chem. Rev., 2011, 111, 5784–5814 CrossRef CAS PubMed.
- J.-M. Lehn, Science, 2002, 295, 2400–2403 CrossRef CAS PubMed.
- S. Liu, C. Ruspic, P. Mukhopadhyay, S. Chakrabarti, P. Y. Zavalij and L. Isaacs, J. Am. Chem. Soc., 2005, 127, 15959–15967 CrossRef CAS PubMed.
- M. Hutin, C. J. Cramer, L. Gagliardi, A. R. M. Shahi, G. Bernardinelli, R. Cerny and J. R. Nitschke, J. Am. Chem. Soc., 2007, 129, 8774–8780 CrossRef CAS PubMed.
- T. F. A. de Greef, G. Ercolani, G. B. W. L. Ligthart, E. W. Meijer and R. P. Sijbesma, J. Am. Chem. Soc., 2008, 130, 13755–13764 CrossRef CAS PubMed.
- B. H. Northrop, Y. Zheng, K. Chi and P. J. Stang, Acc. Chem. Res., 2009, 42, 1554–1563 CrossRef CAS PubMed.
- W. Jiang and C. A. Schalley, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10425–10429 CrossRef CAS PubMed.
- L. Cera and C. A. Schalley, Chem. Sci., 2014, 5, 2560–2567 RSC.
- Z. Huang, L. Yang, Y. Liu, Z. Wang, O. A. Scherman and X. Zhang, Angew. Chem., Int. Ed., 2014, 53, 5351–5355 CrossRef CAS PubMed.
- L. Chen, Z. Huang, J.-F. Xu, Z. Wang and X. Zhang, Polym. Chem., 2016, 7, 1397–1404 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9qm00028c |
| This journal is © the Partner Organisations 2019 |