Development of an engineered thermostable amine dehydrogenase for the synthesis of structurally diverse chiral amines†
Lei
Liu
a,
Dong-Hao
Wang
a,
Fei-Fei
Chen
 a,
Zhi-Jun
Zhang
a,
Qi
Chen
a,
Jian-He
Xu
a,
Zhi-Long
Wang
a,
Zhi-Jun
Zhang
a,
Qi
Chen
a,
Jian-He
Xu
a,
Zhi-Long
Wang
 b and
Gao-Wei
Zheng
b and
Gao-Wei
Zheng
 *a
*a
aState Key Laboratory of Bioreactor Engineering, Shanghai Collaborative Innovation Center for Biomanufacturing, East China University of Science and Technology, 130 Meilong Road, Shanghai 200237, P. R. China. E-mail: gaoweizheng@ecust.edu.cn; Fax: (+86) 21 6425 0840
bState Key Laboratory of Microbial Metabolism, School of Pharmacy, Shanghai Jiao Tong University, Shanghai 200240, P. R. China
First published on 12th February 2020
Abstract
Amine dehydrogenases (AmDHs) are emerging as a class of attractive biocatalysts for synthesizing chiral amines via asymmetric reductive amination of ketones with inexpensive ammonia as an amino donor. However, the AmDHs developed to date exhibit limited substrate scope. Here, using directed evolution, we engineered a GkAmDH based on a thermostable phenylalanine dehydrogenase from Geobacillus kaustophilus. The newly developed AmDH is able to catalyze reductive amination of a diverse set of ketones and functionalized hydroxy ketones with ammonia or primary amines with up to >99% conversion, thus accessing structurally diverse chiral primary and secondary amines and chiral vicinal amino alcohols, with excellent enantioselectivity (up to >99% ee) and releasing water as the sole by-product.
Chiral amines are an important class of intermediates for the synthesis of numerous pharmaceutical drugs, bioactive compounds, and agrochemicals.1 They are also extensively used as resolving agents in kinetic resolution reactions.2 Therefore, the synthesis of such molecules continues to be of great interest for chemists. Traditional synthesis methods, however, rely heavily on precious transition metal catalysts and other reagents well known to be environmentally harmful, and have the additional downside of often failing to generate optically pure reaction products.
Advances in protein engineering have enabled the emergence of biocatalysis as a powerful alternative method for the synthesis of chiral compounds in the chemical and pharmaceutical industries.3–8 To date, numerous biocatalytic processes have been developed for the synthesis of chiral amines; for example, the (dynamic) kinetic resolution of racemic amines catalyzed by amine oxidases9–12 and lipases,13,14 asymmetric reduction of imines catalyzed by (artificial) imine reductases,15–20 (reductive) amination of ketones catalyzed by transaminases,21–24 reductive aminases20,25,26 and amine dehydrogenases,27–33 direct C–H amination of alkane by cytochrome P450 monooxygenases,34,35 and a series of elegant multi-enzymatic cascade reactions.36–41
In particular, a recently developed class of engineered enzymes, amine dehydrogenases (AmDHs), are now appreciated as attractive biocatalysts for the synthesis of chiral primary amines using simple and green approaches; they can use inexpensive free ammonia as an amino donor for the reductive amination of ketones. Moreover, only water is generated as a by-product in this process. Three AmDHs were initially created by Bommarius and co-workers through directed evolution of naturally occurring leucine dehydrogenases and phenylalanine dehydrogenases.32,42 Using the same approach, Li and co-workers subsequently developed an AmDH based on Rhodococcus phenylalanine dehydrogenase, which displayed detectable activity towards the bulky 4-phenyl-2-butanone, a challenging substrate for these enzymes.31 More recently, Schell and co-workers also developed a thermostable CtAmDH by directed evolution of a Caldalkalibacillus thermarum phenylalanine dehydrogenase, and demonstrated an efficient biphasic system for the amination of phenoxy-2-propanone.30 In addition, Vergne-Vaxelaire and Grogan identified a family of native AmDHs for reduction amination of ketones.27
Notably, by coupling the engineered AmDHs with alcohol dehydrogenases (ADHs), both the Turner group and our group constructed elegant dual-enzyme ‘hydrogen-borrowing’ cascade systems for the production of chiral amines from available racemic alcohols in a green manner.41,43 Mutti and coworkers performed a detailed investigation of AmDH-catalyzed reductive amination that considered substrate acceptance, optimal reaction conditions, and stereoselectivity.44 Very recently, we further engineered AmDHs through mutation of two key residues surrounding the substrate-binding pocket which affect the binding of bulky aliphatic ketones.29 The resultant AmDH mutants displayed broad substrate scopes, and are capable of accepting previously inaccessible bulky substrate ketones like 2-heptanone and 2-octanone.
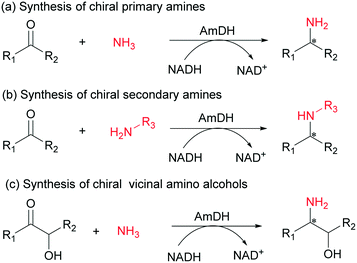
Despite recent progress with AmDHs, the enzymes were mainly used for the synthesis of chiral primary amines via reductive amination of ketones with ammonia. Until recently, Mutti and co-workers reported the synthesis of chiral secondary amines from ketones and achiral organic amino donors using AmDHs.28,45 However, few examples showed that AmDHs could have been utilized for asymmetric reductive amination of functionalized aromatic ketones (e.g., aromatic hydroxy ketones).46 Herein, we performed directed evolution of a thermostable phenylalanine dehydrogenase from Geobacillus kaustophilus to develop an AmDH that possesses a broad substrate scope. The resulting GkAmDH can catalyze the synthesis of structurally diverse chiral primary and secondary amines from ketones with ammonia or organic amines and chiral vicinal amino alcohols through amination of hydroxy ketones with ammonia (Scheme 1).
Initially, four phenylalanine dehydrogenases, which were identified from different microorganisms, were used as starting templates for the development of AmDHs by introducing the corresponding double mutations of K78S/N276L to alter substrate specificity.27–33 Among the engineered AmDHs, GkAmDH (based on Geobacillus kaustophilus phenylalanine dehydrogenase) displayed the highest amination activity (6.0 U mg−1 protein) towards the model substrate 4-fluorophenylacetone (1) (Table S2†). The optimal pH and temperature of GkAmDH were 9.0 and 55 °C, respectively (Fig. S1†). Analysis of kinetic parameters showed that GkAmDH gave a KM of 7.8 mM and kcat of 6.0 s−1 towards ketone 1, values comparable to those reported for BbAmDH and CtAmDH.30,32 Its melting temperature (Tm) reached 71.5 °C (Table S3†), suggesting that it is a thermostable biocatalyst for reductive amination. Moreover, we observed that its amination activity increased linearly with increases in the NH3/NH4+ concentration ranging from 0 to 5.0 M, and reached 18.0 U mg−1 towards substrate 1 at 5.0 M (Fig. S2†).
With this promising enzyme in hand, we subsequently explored its amination activity towards a series of ketones using ammonia as an amino donor. As shown in Fig. 1, GkAmDH exhibited amination activities towards all tested substrates 1–12, including aromatic ketones with a range of substitutions on their aromatic rings and aliphatic ketones with different carbon chain lengths, as well as the challenging 4-phenyl-2-butanone (8). GkAmDH exhibited higher activities towards 1–4, which bear electron-withdrawing substituents on their benzene rings, than towards 6 and 7 with electron-donating substituents, clearly suggesting that electron-withdrawing substituents may activate the carbonyl carbon. Moreover, GkAmDH displayed similar activities towards 1–3, suggesting that the substituent's position on the benzene ring had no apparent effect on the enzyme's activity.
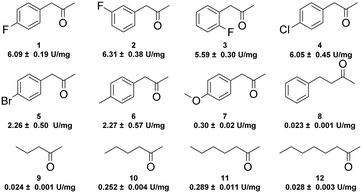 | ||
| Fig. 1 Specific activities (U mg−1 protein) of GkAmDH towards various ketones using ammonia as an amino donor. | ||
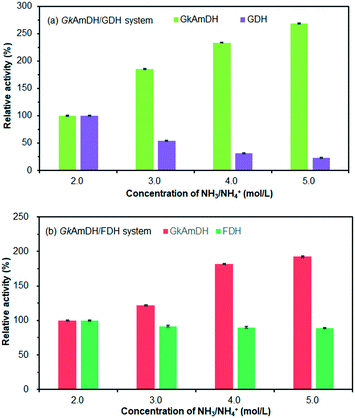
We next attempted the reductive amination of ketones 1–12 in 5 M NH3·H2O/NH4Cl buffer (pH 9.0) using GkAmDH combined with glucose dehydrogenase (GDH) used for the regeneration of NADH. However, we found that high concentrations of NH4+ caused poor GDH activity (Fig. 2a). We therefore examined another system based on formate dehydrogenase (FDH) with NH3·H2O/NH4COOH buffer (Fig. 2b). Consequently, use of a high concentration of NH3·H2O/NH4COOH buffer had no apparent deleterious effects on FDH activity. We therefore used an optimized FDH-based regeneration system (5 M, pH 9.0) in subsequent reductive amination reactions.
Using the optimized reaction system, analytical scale reductive amination reactions of various ketones 1–12 with ammonia were carried out using the GkAmDH/FDH coupling system in NH3·H2O/NH4COOH buffer (5 M, pH 9.0): GkAmDH exhibited great reactivity towards ketones 1–6 (Table 1), findings consistent with our results for specific activity (Fig. 1). 50 mM substrates 1–6 were converted with up to 99% conversions with low enzyme loading. For the challenging substrates 7–11, good-to-excellent conversions were also achieved using 10 g L−1GkAmDH. Only for substrate 12, GkAmDH exhibited poor conversion (<5%), suggesting that excessively long alkyl chains can hinder substrate access to the GkAmDH active site. Furthermore, GkAmDH displayed excellent stereoselectivity for all the tested substrates, and yielded (R)-configuration chiral primary amines with up to 99% enantiomeric excess (ee). These results highlight the strong potential of GkAmDH as a biocatalyst for the synthesis of chiral primary amines.
| Substrate | Substrate concn. (mM) | GkAmDH (mg mL−1) | Conv. (%) | ee (%) |
|---|---|---|---|---|
| a Reaction conditions: 0.5 mL reaction mixture containing NH3·H2O/NH4COOH (5 M NH4+, pH 9.0), 10–50 mM substrate, 1 mM NAD+, 10 mg mL−1 FDH (lyophilized cell-free extract) and GkAmDH (lyophilized cell-free extract) shaken at 1000 rpm at 40 °C for 24 h. Conversions and enantiomeric excess were determined via GC and chiral GC, respectively. | ||||
| 1 | 50 | 1 | 99 | >99 |
| 2 | 50 | 1 | 98 | >99 |
| 3 | 50 | 1 | 98 | >99 |
| 4 | 50 | 1 | 91 | >99 |
| 5 | 50 | 3 | 98 | >99 |
| 6 | 50 | 3 | 99 | >99 |
| 7 | 50 | 10 | 99 | >99 |
| 8 | 50 | 10 | 85 | 98 |
| 9 | 10 | 10 | 77 | >99 |
| 10 | 10 | 10 | 99 | >99 |
| 11 | 10 | 10 | 98 | >99 |
| 12 | 10 | 10 | <5 | >99 |
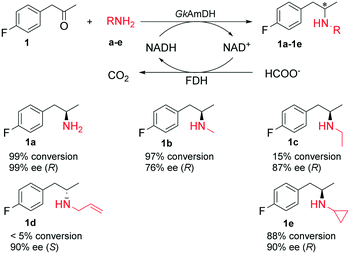
Recently, a reductive aminase AspRedAm and several imine reductases have been described for the synthesis of chiral secondary amines via reductive amination of ketones with primary amines.19,26,47,48 To explore the possibility of using AmDHs for the synthesis of chiral secondary amines, reductive amination of the model substrate 1 with four primary amines b–e was attempted with GkAmDH. Consequently, GkAmDH can reduce the imines formed with primary amines b and e with 1 at 97% conversion and 88% conversion, respectively. In contrast, GkAmDH only provided 15% and <5% conversions in the reduction of 1 with ethylamine c and allylamine d, highlighting the substantial impact of the amino donors on GkAmDH's activity (Scheme 2). The corresponding chiral secondary amines were produced with good stereoselectivity (78–90% ee). Interestingly, a reversed stereoselectivity was observed for 1d, perhaps owing to the rotation of the carbon–carbon bond connecting the phenyl and carbon of the carbonyl group. Recently, Mutti and co-workers reported AmDH-catalyzed coupling amination of ketones with primary amines in which up to 43% of the substrate was converted into the (R)-selective secondary amines with up to 72% ee.45 Taken together, these results demonstrate that AmDHs possess the unique ability to reduce the imines formed from both inorganic ammonia and organic amines with ketones for the preparation of optically pure primary and secondary amines.
Encouragingly, we observed that GkAmDH showed amination activity towards aromatic hydroxy ketones 13 and 14 (3.50 U mg−1 and 2.06 U mg−1, respectively). Docking analysis showed that the hydroxy ketone 14 is accommodated into the binding pocket of GkAmDH in a reasonable conformation in which the terminal hydroxyl group points towards the binding residues S78 and L276, while the carbonyl group points to the two catalytic residues K90 and D125, thereby generating satisfactory amination activity (Fig. 3).
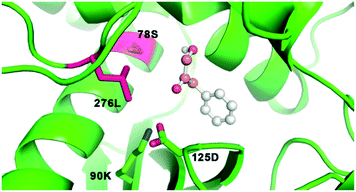 | ||
| Fig. 3 The docking model of substrate 14 into the binding pocket of GkAmDH. S78 and L276 are two key residues for altering substrate specificity and K90 and D125 are the catalytic residues. | ||
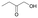
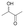
We also detected the activity of GkAmDH for aliphatic hydroxy ketones 15 and 16 (0.39 U mg−1 and 0.04 U mg−1, respectively) (Table 2). The results indicate that AmDHs are able to catalyze the reduction of imines formed from hydroxy ketones with inexpensive ammonia to access vicinal amino alcohols. These amino alcohols are extensively exploited as privileged scaffolds in pharmaceutically active molecules and natural products,49 such as norephedrine and norpseudoephedrine, which are used as amphetamine pharmaceuticals in most countries.50 In addition, they also serve as important chiral auxiliaries or ligands in asymmetric synthesis reactions.51,52
| Substrate | Specific activity of GkAmDHK78S/N276L | The best AmDH | Specific activity (U mg−1) |
|---|---|---|---|
| a Activity was measured in NH3·H2O/NH4COOH buffer (2 M, pH 9.0) containing 0.2 mM NADH and 10 mM substrate (except 1 mM for substrate 14) at 40 °C. | |||
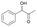 13
13
|
3.50 ± 0.01 | AmDHK78S/N276C | 9.35 ± 0.08 |
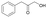 14
14
|
2.06 ± 0.08 | AmDHK78S/N276T | 4.59 ± 0.37 |
 15
15
|
0.39 ± 0.01 | AmDHK78T/N276L | 0.43 ± 0.01 |
 16
16
|
0.04 ± 0.01 | AmDHK78T/N276C | 0.07 ± 0.01 |
Seeking to develop AmDHs with higher activity towards hydroxy ketones, we undertook double site combinational mutagenesis at two key residues S78 and L276 affecting substrate specificity using GkAmDH as the template. After screening separately using hydroxy ketones 13–16, the best mutants displayed further increased activity towards the corresponding substrate (Table 2). To investigate whether chiral vicinal amino alcohols could be achieved using the newly developed AmDHs, we next performed the reductive amination of 14 on a preparative scale as a representative example. Using the best AmDHK78S/N276T, 35 mM 14 was smoothly converted to (S)-2-amino-3-phenylpropanol ((S)-14a) in 41% isolated yield and >99% ee. This result showcases the potential of the newly developed AmDH enzymes for the preparation of chiral vicinal amino alcohols, especially for aromatic chiral vicinal amino alcohols.
Finally, ketone 7 was aminated with ammonia on a 100 mL scale (Scheme 3). The preparative reductive amination of 7 (200 mM, 5% (v/v) DMSO) was performed in NH3·H2O/NH4COOH buffer (5 M, pH 9.0) and the pH was maintained at 9.0 by titrating 5 M NH3·H2O during the reaction course. Consequently, 99% of the substrate was converted into the corresponding chiral primary amine product (R)-4-methoxyamphetamine ((R)-7a) within 6 h, corresponding to a calculated space–time yield of 130.9 g L−1 d−1. After normal workup, the product was prepared with 43% isolated yield and >99% ee.
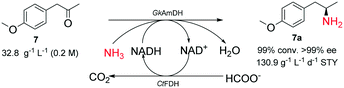 | ||
| Scheme 3 Synthesis of 4-methoxyamphetamine (7a) catalyzed by the GkAmDH/FDH system on a 100 mL scale. | ||
We also performed the reductive coupling of ketone 1 (10 mM, 1% (v/v) DMSO) with methylamine b on a 50 mL scale. The 98% conversion was reached in 12 h, and the corresponding chiral secondary amine product 1b was obtained with 45% yield and 70% ee.
In summary, the present study developed a thermostable GkAmDH by engineering a phenylalanine dehydrogenase from Geobacillus kaustophilus. The newly developed GkAmDH not only catalyzes the reductive amination of ketones with inexpensive ammonia and primary amines to chiral primary and secondary amines, but also can mediate the bioamination of ketones 13–16 with ammonia to access chiral vicinal amino alcohols. Using GkAmDH, a wide range of chiral primary and secondary amines and chiral vicinal amino alcohol 14a were synthesized with up to >99% conversion and up to >99% ee, generating water as the sole by-product. These results demonstrate extensive applications of AmDHs for the synthesis of structurally diverse chiral amines using a simple and green approach.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 21878085, 21536004 and 21871085), the Fundamental Research Funds for the Central Universities (22221818014), and the State Key Laboratory of Microbial Metabolism (MMLKF18-09).Notes and references
- M. E. Welsch, S. A. Snyder and B. R. Stockwell, Curr. Opin. Chem. Biol., 2010, 14, 347–361 CrossRef CAS PubMed.
- T. C. Nugent, Chiral amine synthesis: methods, developments and applications, John Wiley & Sons, Weinheim, Germany, 2010 Search PubMed.
- O. Kuchner and F. H. Arnold, Trends Biotechnol., 1997, 15, 523–530 CrossRef CAS PubMed.
- U. T. Bornscheuer, G. W. Huisman, R. J. Kazlauskas, S. Lutz, J. C. Moore and K. Robins, Nature, 2012, 485, 185–194 CrossRef CAS PubMed.
- J. M. Woodley, Curr. Opin. Chem. Biol., 2013, 17, 310–316 CrossRef CAS PubMed.
- Z. Chen and A. P. Zeng, Curr. Opin. Biotechnol., 2016, 42, 198–205 CrossRef CAS PubMed.
- G. W. Zheng and J. H. Xu, Curr. Opin. Biotechnol., 2011, 22, 784–792 CrossRef CAS PubMed.
- G. Grogan, Curr. Opin. Chem. Biol., 2018, 43, 15–22 CrossRef CAS PubMed.
- D. Ghislieri, A. P. Green, M. Pontini, S. C. Willies, I. Rowles, A. Frank, G. Grogan and N. J. Turner, J. Am. Chem. Soc., 2013, 135, 10863–10869 CrossRef CAS PubMed.
- T. Li, J. Liang, A. Ambrogelly, T. Brennan, G. Gloor, G. Huisman, J. Lalonde, A. Lekhal, B. Mijts, S. Muley, L. Newman, M. Tobin, G. Wong, A. Zaks and X. Y. Zhang, J. Am. Chem. Soc., 2012, 134, 6467–6472 CrossRef CAS PubMed.
- G. Y. Li, P. Y. Yao, R. Gong, J. L. Li, P. Liu, R. Lonsdale, Q. Q. Wu, J. P. Lin, D. M. Zhu and M. T. Reetz, Chem. Sci., 2017, 8, 4666 RSC.
- S. Nakano, K. Yasukawa, T. Tokiwa, T. Ishikawa, E. Ishitsubo, N. Matsuo, S. Ito, H. Tokiwa and Y. Asano, J. Phys. Chem. B, 2016, 120, 10736–10743 CrossRef CAS PubMed.
- L. Munoz, A. M. Rodriguez, G. Rosell, M. P. Bosch and A. Guerrero, Org. Biomol. Chem., 2011, 9, 8171–8177 RSC.
- M. T. Reetz and K. Schimossek, Chimia, 1996, 50, 668–669 CAS.
- J. Mangas-Sanchez, S. P. France, S. L. Montgomery, G. A. Aleku, H. Man, M. Sharma, J. I. Ramsden, G. Grogan and N. J. Turner, Curr. Opin. Chem. Biol., 2017, 37, 19–25 CrossRef CAS PubMed.
- H. Li, P. Tian, J. H. Xu and G. W. Zheng, Org. Lett., 2017, 19, 3151–3154 CrossRef CAS PubMed.
- H. Li, G. X. Zhang, L. M. Li, Y. S. Ou, M. Y. Wang, C. X. Li, G. W. Zheng and J. H. Xu, ChemCatChem, 2016, 8, 724–727 CrossRef CAS.
- H. Li, Z. J. Luan, G. W. Zheng and J. H. Xu, Adv. Synth. Catal., 2015, 357, 1692–1696 CrossRef CAS.
- P. Matzel, M. Gand and M. Höhne, Green Chem., 2017, 19, 385–389 RSC.
- S. C. Cosgrove, A. Brzezniak, S. P. France, J. I. Ramsden, J. Mangas-Sanchez, S. L. Montgomery, R. S. Heath and N. J. Turner, Methods Enzymol., 2018, 608, 131–149 Search PubMed.
- A. Gomm and E. O'Reilly, Curr. Opin. Chem. Biol., 2018, 43, 106–112 CrossRef CAS PubMed.
- I. Slabu, J. L. Galman, R. C. Lloyd and N. J. Turner, ACS Catal., 2017, 7, 8263–8284 CrossRef CAS.
- C. E. Paul, M. Rodriguez-Mata, E. Busto, I. Lavandera, V. Gotor-Fernandez, V. Gotor, S. Garcia-Cerrada, J. Mendiola, O. de Frutos and I. Collado, Org. Process Res. Dev., 2014, 18, 788–792 CrossRef CAS.
- J. Rudat, B. R. Brucher and C. Syldatk, AMB Express, 2012, 2, 11 CrossRef PubMed.
- G. A. Aleku, J. Mangas-Sanchez, J. Citoler, S. P. France, S. L. Montgomery, R. S. Heath, M. P. Thompson and N. J. Turner, ChemCatChem, 2018, 10, 515–519 CrossRef CAS.
- G. A. Aleku, S. P. France, H. Man, J. Mangas-Sanchez, S. L. Montgomery, M. Sharma, F. Leipold, S. Hussain, G. Grogan and N. J. Turner, Nat. Chem., 2017, 9, 961–969 CrossRef CAS PubMed.
- O. Mayol, K. Bastard, L. Beloti, A. Frese, J. P. Turkenburg, J. L. Petit, A. Mariage, A. Debard, V. Pellouin, A. Perret, V. de Berardinis, A. Zaparucha, G. Grogan and C. Vergne-Vaxelaire, Nat. Catal., 2019, 2, 324–333 CrossRef CAS.
- V. Tseliou, T. Knaus, M. F. Masman, M. L. Corrado and F. G. Mutti, Nat. Commun., 2019, 10, 3717 CrossRef PubMed.
- F. F. Chen, G. W. Zheng, L. Liu, H. Li, Q. Chen, F. L. Li, C. X. Li and J. H. Xu, ACS Catal., 2018, 8, 2622–2628 CrossRef CAS.
- A. Pushpanath, E. Siirola, A. Bornadel, D. Woodlock and U. E. Schell, ACS Catal., 2017, 7, 3204–3209 CrossRef CAS.
- L. J. Ye, H. H. Toh, Y. Yang, J. P. Adams, R. Snajdrova and Z. Li, ACS Catal., 2015, 5, 1119–1122 CrossRef CAS.
- M. J. Abrahamson, J. W. Wong and A. S. Bommarius, Adv. Synth. Catal., 2013, 355, 1780–1786 CrossRef CAS.
- M. J. Abrahamson, E. Vazquez-Figueroa, N. B. Woodall, J. C. Moore and A. S. Bommarius, Angew. Chem., Int. Ed., 2012, 51, 3969–3972 CrossRef CAS PubMed.
- C. K. Prier, R. J. K. Zhang, A. R. Buller, S. Brinkmann-Chen and F. H. Arnold, Nat. Chem., 2017, 9, 629–634 CrossRef CAS PubMed.
- P. Dydio, H. M. Key, H. Hayashi, D. S. Clark and J. F. Hartwig, J. Am. Chem. Soc., 2017, 139, 1750–1753 CrossRef CAS PubMed.
- J. H. Schrittwieser, S. Velikogne, M. Hall and W. Kroutil, Chem. Rev., 2017, 118, 270–348 CrossRef PubMed.
- S. P. France, S. Hussain, A. M. Hill, L. J. Hepworth, R. M. Howard, K. R. Mulholland, S. L. Flitsch and N. J. Turner, ACS Catal., 2016, 6, 3753–3759 CrossRef CAS.
- H. D. Peng, E. M. Wei, J. L. Wang, Y. N. Zhang, L. Cheng, H. M. Ma, Z. X. Deng and X. D. Qu, ACS Chem. Biol., 2016, 11, 3278–3283 CrossRef CAS PubMed.
- F. Parmeggiani, S. L. Lovelock, N. J. Weise, S. T. Ahmed and N. J. Turner, Angew. Chem., Int. Ed., 2015, 54, 4608–4611 CrossRef CAS PubMed.
- S. L. Montgomery, J. Mangas-Sanchez, M. P. Thompson, G. A. Aleku, B. Dominguez and N. J. Turner, Angew. Chem., Int. Ed., 2017, 56, 10491–10494 CrossRef CAS PubMed.
- F. F. Chen, Y. Y. Liu, G. W. Zheng and J. H. Xu, ChemCatChem, 2015, 7, 3838–3841 CrossRef CAS.
- B. R. Bommarius, M. Schurmann and A. S. Bommarius, Chem. Commun., 2014, 50, 14953–14955 RSC.
- F. G. Mutti, T. Knaus, N. S. Scrutton, M. Breuer and N. J. Turner, Science, 2015, 349, 1525–1529 CrossRef CAS PubMed.
- T. Knaus, W. Bohmer and F. G. Mutti, Green Chem., 2017, 19, 453–463 RSC.
- V. Tseliou, M. F. Masman, W. Bohmer, T. Knaus and F. G. Mutti, ChemBioChem, 2019, 20, 800–812 CrossRef CAS PubMed.
- F. F. Chen, S. C. Cosgrove, W. R. Birmingham, J. Mangas-Sanchez, J. Citoler, M. P. Thompson, G.-W. Zheng, J.-H. Xu and N. J. Turner, ACS Catal., 2019, 9, 11813–11818 CrossRef CAS.
- P. N. Scheller, M. Lenz, S. C. Hammer, B. Hauer and B. M. Nestl, ChemCatChem, 2015, 7, 3239–3242 CrossRef CAS.
- T. Huber, L. Schneider, A. Präg, S. Gerhardt, O. Einsle and M. Müller, ChemCatChem, 2014, 6, 2248–2252 CrossRef CAS.
- S. C. Bergmeier, Tetrahedron, 2000, 56, 2561–2576 CrossRef CAS.
- M. Yakoot, J. Pharmacol. Pharmacother., 2012, 3, 4–6 CrossRef CAS PubMed.
- D. J. Ager, I. Prakash and D. R. Schaad, Chem. Rev., 1996, 96, 835–875 CrossRef CAS PubMed.
- Q. Y. Tan, X. Q. Wang, Y. Xiong, Z. M. Zhao, L. Li, P. Tang and M. Zhang, Angew. Chem., Int. Ed., 2017, 56, 4829–4833 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cy00071j |
| This journal is © The Royal Society of Chemistry 2020 |