Open Access Article
Open Access ArticleCatch-enrich-release approach for amine-containing natural products†
Michelle Jessy
Müller
 ,
Andrea
Dorst
,
Constanze
Paulus
,
Imran
Khan
and
Simon
Sieber
,
Andrea
Dorst
,
Constanze
Paulus
,
Imran
Khan
and
Simon
Sieber
 *
*
Department of Chemistry, University of Zurich, 8057 Zurich, Switzerland. E-mail: simon.sieber@uzh.ch
First published on 17th October 2022
Abstract
Amine-containing natural products are an important class of therapeutic compounds. Herein, we report a chemoselective approach to catch and enrich amine-containing natural products, and release them as underivatized compounds. The strategy exploits the selectivity of the enzyme legumain for the specific release of amine-containing natural products.
For centuries, natural products (NPs) have been exploited for their therapeutic properties and have had a crucial impact on human health.1,2 Their success is related to their structural diversity and the large chemical space that they occupy compared to synthetic compounds.3–6 The discovery of novel bioactive NPs is and will continue to be essential for the treatment of current diseases7–9
A phenotypic screening followed by a bioactivity-guided fractionation represents the classical approach to identifying active NPs. The process includes (1) the selection of an organism, (2) the extraction of the compounds from the biomass, (3) the separation of the active compound by the selection of active fraction(s), and (4) the structure elucidation of the isolated compound.10 During the process, several challenges might be encountered, such as the accessibility of the biomass, the isolation/exclusion of known NPs, the low production of the active compound, and the cost of the isolation and structure elucidation procedure.3,11,12
Novel strategies have been developed recently to facilitate the identification of known NPs (dereplication). One consists of genome mining (e.g. antiSMASH) to predict the presence of NPs in a crude extract.13–16 Other approaches are based on metabolomics and use tandem mass spectrometry analysis or high-resolution mass spectrometry (HRMS) to identify NPs using platforms such as the Global Natural Products Social Molecular Networking (GNPS).17–20 Those powerful approaches have the advantage of building upon growing databases while requiring only small amounts of analytical samples.
Another emerging technology consists of chemoselective approaches for the enrichment of NPs possessing a specific functional group.21–25 This concept is based on a selective catching via a derivatisation agent25,26 attached to a solid support and the release of the (un)derivatised NPs triggered by an external stimulus.27 Currently, several methods have been developed for the catch and release of NPs possessing amine,28–32 hydroxy,33–36 carbonyl,28,29,37–40 carboxylic acid,28,29,34 thiol,28,29,41 and alkyne42,43 functional groups. Only a few strategies afforded an underivatized NP as the released compound by using siloxyl-functionalized resin33–36 or by utilizing a pyridyl disulfide activation group.41
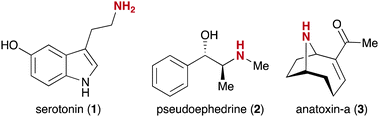
Herein, we are reporting a chemoselective catch-enrich-release approach to obtain underivatized amine-containing NPs. Our methodology exploits the advantage of streptavidin-coated magnetic beads and the high specificity of the enzyme legumain.44 The amine functional group is highly abundant in natural products45 including many bioactive molecules.46 The importance of this functional group is highlighted by the discovery of the neurotransmitter serotonin (1, Fig. 1),47 the stimulant pseudoephedrine (2),48 and the neurotoxin anatoxin-a (3).49
Unlike other chemoselective approaches for amine-containing compounds, we envisioned that the chemoselectivity of our enrichment strategy would arise from the releasing step instead of focusing on the initial catch.28,29,31,40 To obtain an underivatized compound, we sought to use an enzymatic cleavage as the last step of our protocol. The asparaginyl endopeptidase legumain was selected since it is commercially available, has high tolerance at amino acid P1′50 and has a selective recognition for the residues at P1–P4.51
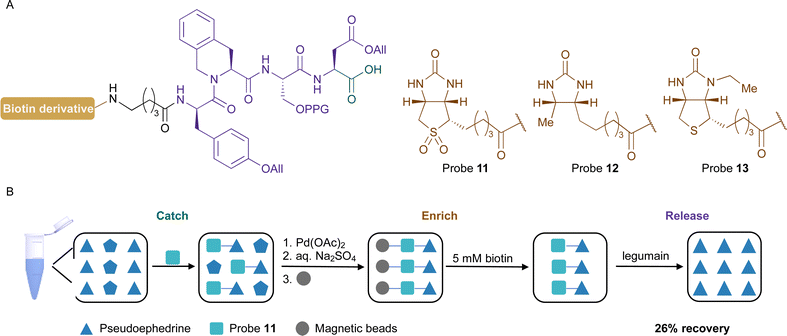
The design of the desired probe includes a biotin anchor for the enrichment process, an amino acid sequence for the selective enzymatic release, and a carboxylic acid group for the catch of the amine-containing compounds (Fig. 2A). The first approach was based on (D)-biotin and used L-Ala-L-Ala-L-Ala-L-Asn recognition sequence for legumain. The probe 4 was synthesised by solid-phase peptide synthesis (SPPS) and its reactivity towards amine-containing compounds was investigated. The carboxylic acid was activated using N-hydroxysuccinimide and, unfortunately, the product was not stable and several by-products were observed. We hypothesised that the amide of Asn side chain might react with the activated electrophile. Similar results were observed in a previous study.52
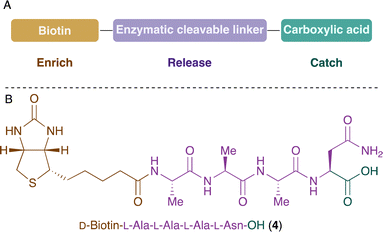 | ||
| Fig. 2 (A) Design of the probe for the catch, enrich and release of amine-containing NPs. (B) Structure of the probe 4 with asparagine in position P1. | ||
To circumvent the undesired reactivity of the Asn side chain, we turned our attention to an approach using Asp instead. The design of the second approach was inspired by a recent study from Drag and co-workers, where the legumain recognition sequence was optimized for Asp.53 The new probe 5 consisted of biotin followed by the linker 6-aminocaproic acid (6-Ahx) and the amino sequence D-Tyr-L-Tic-L-Ser-L-Asp(OAll). The probe 5 was synthesized by SPPS and was converted to the switchable fluorescent probe 6 using HATU and 7-amino-4-methylcoumarin (AMC, 7) followed by removing the allyl group using the Tsuji–Trost condition (Fig. 3A).54 The efficacy of legumain-mediated cleavage was investigated by treatment of probe 6 with 15 ng mL−1 of the enzyme legumain. We were pleased to observe a release of 34% of the fluorophore after 130 minutes (Fig. 3B and Fig. S1, ESI†). Furthermore, the efficacy of legumain was also evaluated with the commercially available substrate Z-L-Ala-L-Ala-L-Asn-AMC, which yielded a recovery 84% (Fig. S1, ESI†). The amino acid at position P1 is hypothesized to account for the difference in yield between both compounds.
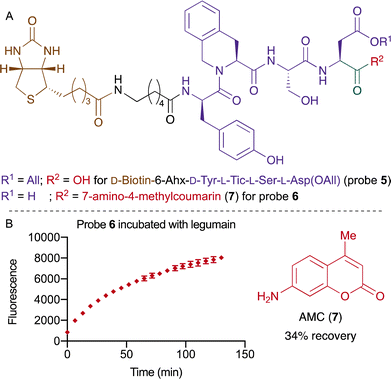 | ||
| Fig. 3 (A) Structure of probe 5 and fluorescent probe 6. (B) Kinetic release of AMC (7) while incubating probe 6 with legumain at a concentration of 15 ng mL−1. | ||
With this encouraging result, the enrichment strategy was tested by incubating probe 6 with streptavidin-coated magnetic beads followed by enzymatic cleavage. The release of AMC (7) was not observed, and we concluded that the proximity of the streptavidin hindered legumain from accessing the recognition sequence. Therefore, it was necessary to release the probe from the solid support before performing the enzymatic cleavage.
It was crucial to establish a protocol using mild conditions for the release of the probe from the solid support to preserve the structural integrity of natural products during the procedure. Therefore, a recent method from Bearden and co-workers was adapted with the use of a mixture of phenol and chloroform for the release of biotinylated compounds from streptavidin-coated beads (Fig. S2, ESI†).55 The conditions were applied to our system and an efficient release of probe 6 was observed. However, the release of AMC (7) was not observed after attempting enzymatic cleavage. We hypothesized that the remaining traces of phenol might alter the structure of legumain,55 therefore, we stopped investigating this approach.
To encourage the competitive elution of biotinylated compounds from the streptavidin beads, probe analogues containing biotin derivatives, which have a weaker affinity towards streptavidin as compared with biotin, can be applied. Using this approach, the compounds can be eluted with a biotin solution. Biotin sulfone (8), desthiobiotin (9), and N′3-ethyl biotin (10) were selected as biotin derivatives and the probes bearing these anchors were synthesized by SPPS (Fig. 4A and Fig. S3A, ESI†). Both derivatives 8 and 9 were synthesized following the reported procedures26g,56 while the tyrosine and the serine were protected to avoid side reactions. The attachment and the release from the solid support were performed with the three probes, and their concentrations after their release from the beads with a 5 mM biotin solution were analysed by UHPLC-MS. The biotin sulfone probe 11 showed the most promising result with a 27% recovery after a 10-minute incubation. The calculated recovery yield for the desthiobiotin analogues 12 and the N′3-ethyl biotin probe 13 were 12% and 8%, respectively(Fig. 4 and Fig. S3B, ESI†).
Then, we investigated the deprotection of the propargyl and the two allyl groups in the recognition sequence of legumain. It was crucial to perform this reaction before the release of the probe from the solid support to render the recognition sequence accessible to legumain. Preliminary results indicated that the deprotection was more efficient when performed prior to the attachment of the probe to the beads. The optimal reaction conditions include the catalyst Pd(OAc)2, the water-soluble ligand trisodium 3,3′,3′′-phosphanetriyltris(benzene-1-sulfonate (TPPTS), morpholine as a scavenger, and the addition of MgCl2 to prevent the unprotected peptide from reacting with the catalyst.57 The reaction was monitored by UHPLC-MS and full conversion was observed after 2 hours. It was necessary to include a washing step with a saturated aqueous Na2SO4 solution to remove excess of MgCl2.
With the optimized conditions in hand, the procedure (Fig. 4B and Fig. S4, ESI†) with probe 11 was applied to the catch, enrichment, and release of the natural product pseudoephedrine (2). For the catching step, compound 2 was coupled to probe 11 with HATU and the protection groups were removed. The enrichment was performed by incubating the modified probe with magnetic beads to remove unreacted reagents and side products. The release from the solid support was then achieved with a 5 mM biotin solution. The natural product 2 was recovered in a 26% yield after treatment with legumain. We hypothesised that this recovery is mainly due to the efficacy of the enzymatic cleavage due, most likely, to having aspartic acid at position P1. To further demonstrate the chemoselectivity of our novel protocol, probe 11 was examined in a controlled complex matrix containing the following compounds at an equimolar concentration: serotonin (1), pseudoephedrine (2), 1-deoxynojirimycin, isopropanol, prop-2-en-1-ol, 4-methylbenzenethiol, histamine, acarbose, and vancomycin. We were glad to obtain 1 and 2 and 1-deoxynojirimycin as the underivatized natural products with a recovery yield of 9%, 3%, and <1%, respectively. Furthermore, the masses corresponding to the probe reacted with histamine, and acarbose were also observed after the release from the solid support.
In addition, we tested our protocol with a sample collected from cyanobacteria mats detected in Neuchâtel, Switzerland. The biomass was extracted and submitted to the catch-enrich-release strategy using probe 11. The extract and released product were analysed by UHPLC-HRMS/HRMS to identify compounds present in both samples. A comparison with a cyanobacteria metabolites database (CyanMetDB)58 led to the identification of two potent neurotoxins: anatoxin-a (3) and dihydroanatoxin-a (Table S1 and Fig. S5, ESI†). The structure of both compounds was confirmed by HRMS/HRMS analysis and comparison with analytical standard (Fig. S6 and S7, ESI†).59
In conclusion, we developed the first catch-enrich-release approach that results underivatized amine-containing compounds as products. The robustness of the strategy was demonstrated with the natural products serotonin (1), pseudoephedrine (2), 1-deoxynojirimycin, anatoxin-a (3), and dihydroanatoxin-a. Furthermore, the optimized probe 11 is quickly accessible via SPPS and can be readily used to improve the discovery and identification of amine-containing natural products.
The authors thank professors Karl Gademann, Nina Hartrampf and Jason Holland, and Dr Ektoras Yiannakas for excellent advice and discussions. Jiajun Ren and Erik Jung are thanked for their critical advice on the manuscript. The authors thank Professor Pilar Junier and Sami Zhioua for providing the biomass sample. The authors are grateful for the financial support of the Swiss National Science Foundation (CRSK-2_190556), the Dr. Helmut Legerlotz Stiftung, and the University of Zurich. Timea Szabo is thanked for the technical assistance.
Conflicts of interest
There are no conflicts to declare.Notes and references
- J. K. Borchardt, Drug News Perspect., 2002, 15, 187 CrossRef PubMed.
- A. G. Atanasov, et al. , Biotechnol. Adv., 2015, 33, 1582–1614 CrossRef CAS PubMed.
- A. G. Atanasov, et al. , Nat. Rev. Drug Discovery, 2021, 20, 200–216 CrossRef CAS PubMed.
- H. Lachance, S. Wetzel, K. Kumar and H. Waldmann, J. Med. Chem., 2012, 55, 5989–6001 CrossRef CAS PubMed.
- S. Stone, D. J. Newman, S. L. Colletti and D. S. Tan, Nat. Prod. Rep., 2021, 39, 20–32 RSC.
- M. Feher and J. M. Schmidt, J. Chem. Inf. Comput. Sci., 2003, 43, 218–227 CrossRef CAS PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2016, 79, 629–661 CrossRef CAS PubMed.
- M. I. Hutchings, A. W. Truman and B. Wilkinson, Curr. Opin. Microbiol., 2019, 51, 72–80 CrossRef CAS PubMed.
- F. Bucar, A. Wube and M. Schmid, Nat. Prod. Rep., 2013, 30, 525–545 RSC.
- S. Bernardini, A. Tiezzi, V. L. Masci and E. Ovidi, Nat. Prod. Res., 2017, 32, 1926–1950 CrossRef PubMed.
- J. Clardy and C. Walsh, Nature, 2004, 432, 829–837 CrossRef CAS PubMed.
- M. H. Medema, et al. , Nucleic Acids Res., 2011, 39, W339–46 CrossRef CAS PubMed.
- K. Blin, M. H. Medema, D. Kazempour, M. A. Fischbach, R. Breitling, E. Takano and T. Weber, Nucleic Acids Res., 2013, 41, W204–12 CrossRef PubMed.
- T. Weber, et al. , Nucleic Acids Res., 2015, 43, W237–43 CrossRef CAS PubMed.
- K. Blin, et al. , Nucleic Acids Res., 2017, 45, W36–41 CrossRef CAS PubMed.
- M. Wang, et al. , Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- H. Mohimani, et al. , Nat. Chem. Biol., 2017, 13, 30–37 CrossRef CAS PubMed.
- A. Gurevich, A. Mikheenko, A. Shlemov, A. Korobeynikov, H. Mohimani and P. A. Pevzner, Nat. Microbiol., 2018, 3, 319–327 CrossRef CAS PubMed.
- Example: S. Sieber, S. M. Grendelmeier, L. A. Harris, D. A. Mitchell and K. Gademann, J. Nat. Prod., 2020, 83, 438–446 CrossRef CAS PubMed.
- T. Böttcher, M. Pitscheider and S. A. Sieber, Angew. Chem., Int. Ed., 2010, 49, 2680–2698 CrossRef PubMed.
- E. J. Tollefson and E. E. Carlson, in Mass Spectrometry-Based Chemical Proteomics, ed. W. A. Tao and Y. Zhang, John Wiley & Sons Inc., United States of America, 2019, ch. 7, pp. 187–206 Search PubMed.
- J. Chen, Y. Tian, Y. Zhang and F. Xu, J. Anal. Test., 2020, 4, 175–182 CrossRef.
- H. Li, T. Li, X. Shi and G. Xu, J. Chromatogr. A, 2021, 1636, 461785 CrossRef CAS PubMed.
- C. C. Hughes, Nat. Prod. Rep., 2021, 38, 1684–1705 RSC.
- Recent examples of derivatisation agent: (a) D. Reimer and C. C. Hughes, J. Nat. Prod., 2017, 80, 126–133 CrossRef CAS PubMed; (b) G. Castro-Falcón, D. Hahn, D. Reimer and C. C. Hughes, ACS Chem. Biol., 2016, 11, 2328–2336 CrossRef PubMed; (c) G. Castro-Falcón, N. Millán-Aguiñaga, C. Roullier, P. R. Jensen and C. C. Hughes, ACS Chem. Biol., 2018, 13, 3097–3106 CrossRef PubMed; (d) L. A. Harris, P. M. B. Saint-Vincent, X. Guo, G. A. Hudson, A. J. DiCaprio, L. Zhu and D. A. Mitchell, ACS Chem. Biol., 2020, 15, 3167–3175 CrossRef CAS PubMed; (e) C. L. Cox, J. I. Tietz, K. Sokolowski, J. O. Melby, J. R. Doroghazi and D. A. Mitchell, ACS Chem. Biol., 2014, 9, 2014–2022 CrossRef CAS PubMed; (f) T. Maxson, J. I. Tietz, G. A. Hudson, X. R. Guo, H.-C. Tai and D. A. Mitchell, J. Am. Chem. Soc., 2016, 138, 15157–15166 CrossRef CAS PubMed; (g) R. J. B. Schäfer, M. R. Monaco, M. Li, A. Tirla, P. Rivera-Fuentes and H. Wennemers, J. Am. Chem. Soc., 2019, 141, 18644–18648 CrossRef PubMed.
- G. Leriche, L. Chisholm and A. Wagner, Bioorg. Med. Chem., 2012, 20, 571–582 CrossRef CAS PubMed.
- E. E. Carlson and B. F. Cravatt, Nat. Methods, 2007, 4, 429–435 CrossRef CAS PubMed.
- E. E. Carlson and B. F. Cravatt, J. Am. Chem. Soc., 2007, 129, 15780–15782 CrossRef CAS PubMed.
- H. Li, Q. Qin, L. Qiao, X. Shi and G. Xu, Chem. Commun., 2015, 51, 11321–11324 RSC.
- N. Garg, L. P. Conway, C. Ballet, M. S. P. Correia, F. K. S. Olsson, M. Vujasinovic, J.-M. Löhr and D. Globisch, Angew. Chem., Int. Ed., 2018, 57, 13805–13809 CrossRef CAS PubMed.
- W. Lin, Z. Yang, A. Kaur, A. Block, M. Vujasinovic, J.-M. Löhr and D. Globisch, RSC Chem. Biol., 2021, 2, 1479–1483 RSC.
- A. Y. Odendaal, D. J. Trader and E. E. Carlson, Chem. Sci., 2011, 2, 760–764 RSC.
- D. J. Trader and E. E. Carlson, Org. Lett., 2011, 13, 5652–5655 CrossRef CAS PubMed.
- D. J. Trader and E. E. Carlson, J. Org. Chem., 2013, 78, 7349–7355 CrossRef CAS PubMed.
- D. J. Trader and E. E. Carlson, Tetrahedron, 2014, 70, 4191–4196 CrossRef CAS.
- W. Yuan, J. L. Edwards and S. Li, Chem. Commun., 2013, 49, 11080–11082 RSC.
- L. P. Conway, N. Garg, W. Lin, M. Vujasinovic, J.-M. Löhr and D. Globisch, Chem. Commun., 2019, 155, 9080–9083 RSC.
- W. Lin, L. P. Conway, A. Block, G. Sommi, M. Vujasinovic, J.-M. Löhr and D. Globisch, Analyst, 2020, 145, 3822–3831 RSC.
- W. Lin, L. P. Conway, M. Vujasinovic, J. Matthias Löhr and D. Globisch, Angew. Chem., Int. Ed., 2021, 60, 23232–23240 CrossRef CAS PubMed.
- S. L. Capehart and E. E. Carlson, Chem. Commun., 2016, 52, 13229–13232 RSC.
- R. Orth and S. A. Sieber, J. Org. Chem., 2009, 74, 8476–8479 CrossRef CAS PubMed.
- H. Jeon, C. Lim, J. M. Lee and S. Kim, Chem. Sci., 2015, 6, 2806–2811 RSC.
- J.-M. Chen, P. M. Dando, N. D. Rawlings, M. A. Brown, N. E. Young, R. A. Stevens, E. Hewitt, C. Watts and A. J. Barrett, J. Biol. Chem., 1997, 272, 8090–8098 CrossRef CAS PubMed.
- P. Ertl and T. Schuhmann, J. Nat. Prod., 2019, 82, 1258–1263 CrossRef CAS PubMed.
- P. Ertl, E. Altmann and J. M. McKenna, J. Med. Chem., 2020, 63, 8408–8418 CrossRef CAS PubMed.
- M. Vialli and V. Erspamer, Boll. d. Soc. Med-chir. Pavia, 1937, 357–363 Search PubMed.
- A. Ladenburg and C. Oelschlägel, Ber. Dtsch. Chem. Ges., 1889, 22, 1823–1827 CrossRef.
- J. P. Devlin, O. E. Edwards, P. R. Gorham, N. R. Hunter, R. K. Pike and B. Stavric, Can. J. Chem., 1977, 55, 1367–1371 CrossRef CAS.
- G. Schwarz, J. Brandenburg, M. Reich, T. Burster, C. Driessen and H. Kalbacher, Biol. Chem., 2002, 383, 1813–1816 CAS.
- M. A. Mathieu, M. Bogyo, C. R. Caffrey, Y. Choe, J. Lee, H. Chapman, M. Sajid, C. S. Craik and J. H. McKerrow, Mol. Biochem. Parasitol., 2002, 121, 99–105 CrossRef CAS.
- S. Mojsov, A. R. Mitchell and R. B. Merrifield, J. Org. Chem., 1980, 45, 555–560 CrossRef CAS.
- M. Poreba, R. Solberg, W. Rut, N. N. Lunde, P. Kasperkiewicz, S. J. Snipas, M. Mihelic, D. Turk, B. Turk, G. S. Salvesen and M. Drag, Cell Chem. Biol., 2016, 23, 1023–1035 CrossRef CAS PubMed.
- H. Kunz and H. Waldmann, Angew. Chem., Int. Ed. Engl., 1984, 23, 71–72 CrossRef.
- S. Bearden, F. Wang and A. R. Hall, Anal. Chem., 2019, 91, 7996–8001 CrossRef CAS PubMed.
- L.-Q. Ying and B. P. Branchaud, Chem. Commun., 2011, 47, 8593–8595 RSC.
- N. Kamo, G. Hayashi and A. Okamoto, Chem. Commun., 2018, 54, 4337–4340 RSC.
- M. R. Jones, et al. , Water Res., 2021, 196, 117017 CrossRef CAS PubMed.
- K. J. James, J. Crowley, B. Hamilton, M. Lehane, O. Skulberg and A. Furey, Rapid Commun. Mass Spectrom., 2005, 19, 1167–1175 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Containing the synthetic and analytical procedures as well as the spectra of the synthetic compounds. See DOI: https://doi.org/10.1039/d2cc04905h |
| This journal is © The Royal Society of Chemistry 2022 |