Recent advances in nanotechnology-based COVID-19 vaccines and therapeutic antibodies
Lanying
Du
 *a,
Yang
Yang
b,
Xiujuan
Zhang
c and
Fang
Li
*de
*a,
Yang
Yang
b,
Xiujuan
Zhang
c and
Fang
Li
*de
aInstitute for Biomedical Sciences, Georgia State University, Atlanta, GA, USA. E-mail: ldu3@gsu.edu
bRoy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, IA, USA
cLindsley F. Kimball Research Institute, New York Blood Center, New York, New York, USA
dDepartment of Veterinary and Biomedical Sciences, University of Minnesota, Saint Paul, Minnesota, USA
eCenter for Coronavirus Research, University of Minnesota, Saint Paul, Minnesota, USA. E-mail: lifang@umn.edu
First published on 1st January 2022
Abstract
COVID-19 has caused a global pandemic and millions of deaths. It is imperative to develop effective countermeasures against the causative viral agent, SARS-CoV-2 and its many variants. Vaccines and therapeutic antibodies are the most effective approaches for preventing and treating COVID-19, respectively. SARS-CoV-2 enters host cells through the activities of the virus-surface spike (S) protein. Accordingly, the S protein is a prime target for vaccines and therapeutic antibodies. Dealing with particles with dimensions on the scale of nanometers, nanotechnology has emerged as a critical tool for rapidly designing and developing safe, effective, and urgently needed vaccines and therapeutics to control the COVID-19 pandemic. For example, nanotechnology was key to the fast-track approval of two mRNA vaccines for their wide use in human populations. In this review article, we first explore the roles of nanotechnology in battling COVID-19, including protein nanoparticles (for presentation of protein vaccines), lipid nanoparticles (for formulation with mRNAs), and nanobodies (as unique therapeutic antibodies). We then summarize the currently available COVID-19 vaccines and therapeutics based on nanotechnology.
Introduction
Nanotechnology is defined as the design, characterization, development, and employment of particles with dimensions on the scale of nanometers (1–100 nm).1,2 This technology has yielded nano-structures with unique properties, such as improved bioavailability, targeted drug delivery, and enhanced target-binding affinity.3 The development of nanotechnology has provided a cutting-edge platform for treating and preventing infectious diseases, including COVID-19 (caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)), Zika, dengue, influenza, Ebola, AIDS (caused by HIV-1), cancer caused by human papillomavirus, diseases associated with respiratory syncytial virus, and herpes simplex viral infections.3–8 One of the most notable and important applications of nanotechnology is to the design and development of effective vaccines and therapeutics to control viral diseases.COVID-19, a recently emerged respiratory infectious disease, was first reported in 2019 and has caused devastating damages to global health and economy.9 The causative agent SARS-CoV-2 is a novel beta-coronavirus.9 Two other beta-coronaviruses, SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV), caused outbreaks in 2002–2003 and have been infecting humans since 2012, respectively.10–18 SARS-CoV-2 spreads in human populations much more widely than both SARS-CoV-1 and MERS-CoV. Currently, the numbers of COVID-19 cases and fatalities continues to grow. Rapid development of a variety of effective and safe vaccines and therapeutic agents is a key strategy for stopping this global pandemic.
SARS-CoV-2 is a single-stranded and positive-sense RNA virus. The viral genome encodes four major structural proteins, including spike (S), envelope (E), nucleocapsid (N), and membrane (M), in addition to sixteen non-structural proteins (nsp1–16) and several accessory proteins (3a, 6, 7a, 7b, 8, and 10).19,20 The S protein plays an essential role in viral entry and is a critical determinant of viral infection and pathogenesis. Accordingly, it is an important target for vaccines and therapeutics.20–25 The S protein contains a receptor-binding S1 subunit and a membrane-fusion S2 subunit. The S1 subunit contains a N-terminal domain (NTD) and a C-terminal domain (CTD), also known as the receptor-binding domain (RBD) (Fig. 1a and b).21,26,27 The RBDs of both SARS-CoV-2 and SARS-CoV-1 recognize angiotensin-converting enzyme 2 (ACE2) as their cellular receptor for viral attachment to host cells.26,28–30 SARS-CoV-2 RBD binds to human ACE2 more tightly than SARS-CoV-1 RBD does (Fig. 1c).26,27 ACE2 binding by the RBD may trigger a conformational change in the S protein (i.e., transition of the S protein from its pre-fusion conformation to post-fusion conformation). The conformational change of the S protein leads to fusion of viral and cell membranes and SARS-CoV-2 entry into host cells.23,31–33 The RBD in the trimeric S protein of SARS-CoV-2 can exist in two states, “up” and “down”, but the binding of ACE2 to the RBD only occurs when the RBD in the “up” position (Fig. 1b and c).32,34 The strong ACE2 binding and potential hidden state of the RBD present challenges to the development of potent vaccines and therapeutics targeting SARS-CoV-2.
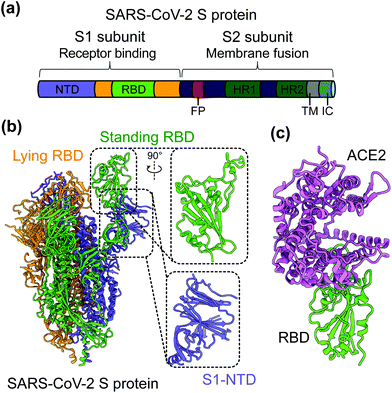 | ||
| Fig. 1 Structure of SARS-CoV-2 S protein. (a) Schematics of SARS-CoV-2 spike (S) protein domain organization. NTD, N-terminal domain; RBD, receptor-binding domain; FP, fusion peptide; HR1 and HR2, heptad region 1 and 2; TM: transmembrane domain; IC: intracellular tail. (b) Cryo-EM structure of SARS-CoV-2 S protein trimer (PDB 6VYB) with closed up views of the S1-NTD and RBD. Three copies of the S protein in the trimer are colored in green, orange and purple, respectively. (c) Crystal structure of SARS-CoV-2 RBD in complex with the host cell receptor angiotensin-converting enzyme 2 (ACE2) (PDB 6 M0J). SARS-CoV-2 RBD is colored in green and ACE2 is in orchid. | ||
In response to the rapid human-to-human spread of SARS-CoV-2 and the devastating pandemic, several nanotechnology-based tools, including protein nanoparticles (PNPs), lipid nanoparticles (LNPs), and nanobodies (Nbs), have been used to combat COVID-19 at a speed never before seen for other infectious diseases. In the remainder of this review, we will describe the roles of these nanotechnology-based tools in combating COVID-19, and will also summarize the currently available PNPs, NLPs, and Nbs targeting COVID-19.
Role of nanotechnology in the development of COVID-19 vaccines and therapeutic antibodies
Protein nanoparticles as a tool for delivery of SARS-CoV-2 vaccines
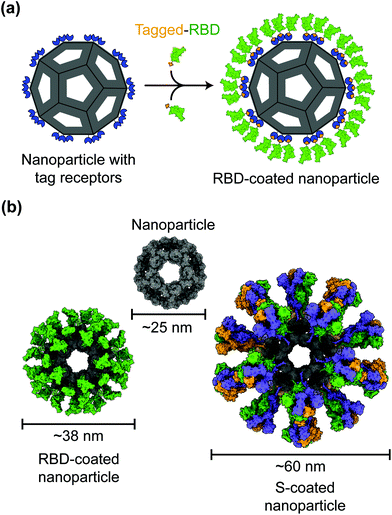
Protein nanoparticles are effective and safe delivery vehicles for protein-based vaccines. Nanoparticle-forming proteins are unusual in that they can spontaneously assemble into large multimers at nanometer sizes (such as 24-meric ferritin and 60-meric lumazine synthase). These virus particle-like multimers can then display many copies of target antigens (i.e., SARS-CoV-2 RBD or S protein) on their surface (Fig. 2).35–37 These viral antigens may be attached to nanoparticles via covalent or noncovalent interactions. Alternatively, nanoparticle proteins and viral antigens can be encoded in a single peptide chain that self assembles into antigen-presenting nanoparticles.38 In general, nanoparticles mimic the structures of virus particles and present antigens with high densities and in repetitive patterns; these features are what the mammalian immune systems have evolved to recognize.2 Hence, nanoparticle-based vaccines enhance antigen uptake by antigen-presenting cells (APCs) and stimulate the immune system to produce strong humoral (antibody) and cellular immune responses specific to the target antigens, conferring protection against SARS-CoV-2 infection.35,37,39Despite having the size and structure similar to virus particles, nanoparticles lack viral genetic material that could drive infection or replication. Hence in contrast to live attenuated virus vaccines, which may recover virulence, nanoparticle-based vaccines are relatively safe.40 Also, in contrast to inactivated virus vaccines whose antigens may have been destructed due to the inactivation procedures, nanoparticle-based vaccines present antigens in their original forms.41 Moreover, unlike recombinant protein subunit vaccines, which generally have low immunogenicity, nanoparticles have immunostimulatory functions that induce antigen-specific immune responses even in the absence of an adjuvant.42–44 The above being said, the inclusion of adjuvants in the vaccine components can further improve the immunogenicity and efficacy of nanoparticle-based vaccines. Indeed, antibodies induced by nanoparticle-based COVID-19 vaccines demonstrate significantly higher neutralization titers than those induced by recombinant proteins.37 Accordingly, nanoparticle-based COVID-19 vaccines are able to potently neutralize SARS-CoV-2 infection and protect immunized animals against SARS-CoV-2 challenge.37,45
Lipid nanoparticles as a tool for delivery of SARS-CoV-2 mRNA vaccines
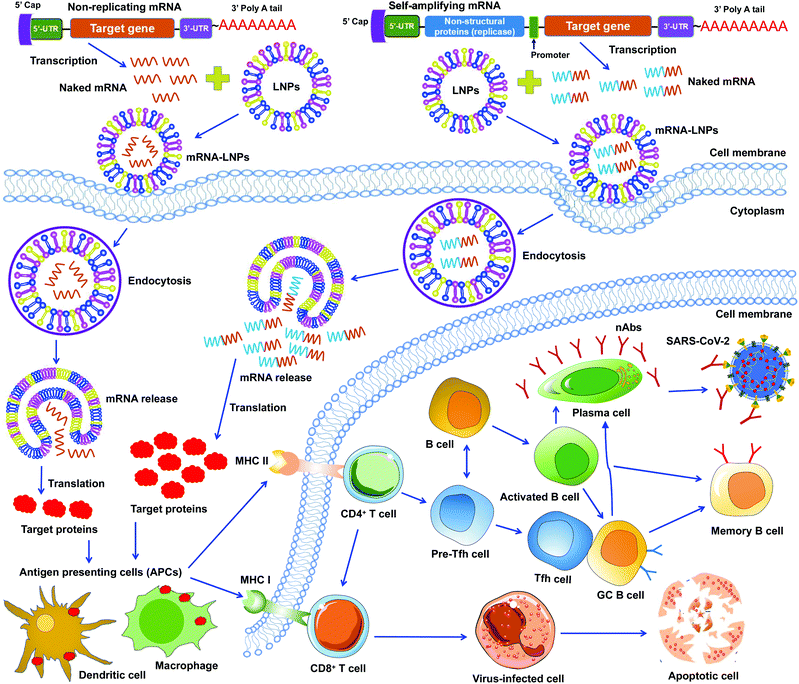
mRNA vaccines represent a new approach to protecting humans against viral infections. mRNAs can be rapidly synthesized at large scales. However, naked mRNAs are unstable and can be easily degraded under various physical, chemical, and enzymatic conditions. To improve mRNA vaccines’ stability, several strategies have been developed, including chemical modification of mRNAs, conjugation of mRNAs to other molecules, and encapsulation of mRNAs in lipid nanoparticles (LNPs).46–48 Among these approaches, LNPs demonstrate strong potentials in stabilizing mRNA vaccines and protecting them from degradation, ensuring effective and targeted delivery of mRNA vaccines.49To date, two types of mRNA vaccines have been reported against infectious diseases: non-replicating mRNAs and self-amplifying mRNAs. Non-replicating mRNAs contain the coding antigen sequence, 5′- and 3′-untranslated regions (UTRs), 5′-cap structure, and 3′-poly(A) tail for amplification.46,50 Self-amplifying mRNAs, also called replicons, are generated by replacing the structural proteins of an RNA virus (often an alphavirus or a flavivirus) with viral RNA-dependent RNA polymerases and antigen-encoding mRNAs.51,52 Both types of mRNAs can be delivered through LNPs. Like protein-based vaccines, non-replicating mRNA vaccines are safer because they do not contain any infectious viral components. If self-amplifying mRNAs are used, it is necessary to detect immune responses specific to both target proteins and RNA polymerases since the latter may induce an independent immune response. Both types of mRNA vaccines have been developed against SARS-CoV-2 infection (Fig. 3).47,53–55
Unlike DNA vaccines, mRNA vaccines do not need to cross nuclear membranes or integrate into cellular genomes, making them easier to be delivered.56 After LNP-mediated mRNAs are delivered into cells, they are internalized via endocytosis, released into the cytoplasm, and then translated to target proteins. Different from non-replicating mRNAs, the self-amplifying mRNAs can produce multiple copies of subgenomic mRNAs that express a higher level of target proteins.52,57 In either case, the target proteins are subsequently recognized by APCs (e.g., dendritic cells and macrophages), which subsequently activate T cells and B cells. Activated T cells will generate CD8+ and CD4+ T-cell responses to kill virus-infected cells and also help trigger B-cell responses. Some activated B cells differentiate into plasma cells to produce virus-neutralizing antibodies, whereas other activated B cells differentiate into germinal center B (GC B) cells, which may interact with T follicular helper cells (Tfh) cells or become memory B cells (Fig. 3).
A potential side effect of mRNA vaccines is that mRNAs can be recognized by pattern-recognition receptors (e.g., toll-like receptors: TLR3, TLR7, or TLR8) in endosomes. These receptors activate innate immune receptors, such as retinoic acid-inducible gene I (RIG-1), protein kinase RNA-activated (PKR), or melanoma differentiation-associated antigen 5 (MDA5), which in turn upregulate the secretion of proinflammatory cytokines and chemokines.56,58 To overcome this potential side effect, naturally-occurring modified nucleotides (e.g., pseudouridine-5′-triphosphate) can be introduced during mRNA synthesis to inhibit activation of these innate immune receptors.46,47,58
Nanobodies as unique SARS-CoV-2 therapeutics
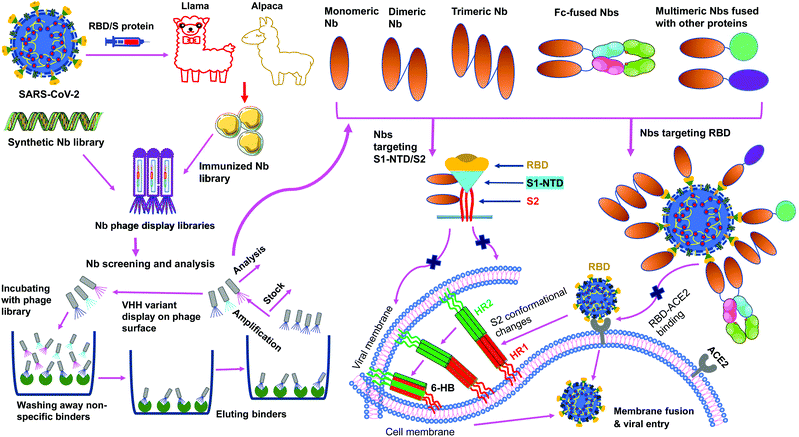
Nanobodies are single-domain antibodies derived from camelid heavy-chain-only antibodies.59,60 They are connected to nanotechnology due to their nano-scale size (about 4 nm by 2.5 nm). With a molecular weight of ∼12–14 kDa, nanobodies are among the smallest naturally occurring antibodies. Because of their small size, they can bind to hidden areas of antigens that are normally unreachable to conventional antibodies.59,60 Because of their elongated CDR3 loops, they are capable of binding to antigens with high affinities. Nanobodies can be expressed at high yield in a variety of expression systems (e.g., bacteria, yeasts and mammalian cells), allowing cheap and large-scale productions of nanobodies.59–63 They are stable under extreme conditions (e.g., extreme temperatures and extreme pH), making them easy to be stored and transported.62,63 Together, these features make nanobodies ideal prophylactic and therapeutic agents. They would be particularly suited to meet the urgent and large demands for battling the COVID-19 global pandemic.Monomeric nanobodies do not contain the Fc portion; consequently, they are unable to enter the Fc receptor-dependent pathway and generally lack cytotoxic activity arising from activation of the complement system and immune cells.64,65 Monomeric nanobodies generally have a short half-life because their size is below the renal filtration capacity, but this can be overcome by constructing Fc-tagged dimeric nanobodies or linking nanobodies to albumin to increase their size (Fig. 3).60,61,66 It is worth noting that the molecular weights of Fc-tagged dimeric nanobodies and albumin-linked nanobodies (both ∼75–80 kDa) are still much smaller than those of conventional antibodies (∼160 kDa), maintaining their small size advantage over conventional antibodies.61,66 Moreover, when Fc is fused with nanobodies, the amino acids of the Fc portion can be mutated to alleviate potential Fc-associated side effects.
SARS-CoV-2 nanobodies can be developed by immunizing camelids (llama or alpaca) with target antigens.67,68 Subsequently, immunized nanobody libraries can be established and screened. Alternatively, naïve nanobody libraries may be directly screened, skipping the immunization step.63,69,70 However, for nanobodies screened from naïve nanobody libraries, in vitro affinity maturation is usually needed to improve the nanobody's affinity for target antigens. Theoretically, neutralizing nanobodies function by binding to their target antigens (such as the RBD, S1, or S2 of SARS-CoV-2 S protein). Binding of nanobodies to the RBD may prevent the RBD from binding its receptor ACE2, thus blocking viral attachment and viral entry. Binding of nanobodies to the S1 or S2 may prevent the S protein from transitioning to its post-fusion conformation, thereby blocking membrane fusion and viral entry. The generation and potential mechanisms of SARS-CoV-2-targeting nanobodies are described in Fig. 4.
Currently developed COVID-19 vaccines based on protein nanoparticles
| Vaccine name | Target | Animals/humans | Ab responses and neutralizing Abs | Cellular and other responses | Protection and/or potential side effects | Ref. |
|---|---|---|---|---|---|---|
| Abs, antibodies; ACE2, angiotensin-converting enzyme 2; BAL, bronchoalveolar-lavage; COVID-19, Coronavirus disease 2019; GC B, germinal center B; hACE2-Tg, human ACE2-transgenic; HR2, heptad repeat region 2; LLPCs, long-lived plasma cells; LuS, lumazine synthase; MBCs, memory B cells; NHPs, non-human primates; RBD, receptor-binding domain; S, spike; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; S2P, S protein with two proline mutations in the S2 subunit; Tfh, T follicular helper cells; WT, wild-type. | ||||||
| Nanoparticle-based SARS-CoV-2 vaccines | ||||||
| RBD-NP | RBD | Mice | Induced SARS-CoV-2 S-specific Abs, with neutralizing Ab titers being 10-fold higher than a stabilized S protein against SARS-CoV-2 WT infection; neutralized SARS-CoV-2 B.1.1.7 variant | Elicited SARS-CoV-2 RBD-specific CD4+ T cell responses | Protected NHPs against SARS-CoV-2 infection | 72 and 73 |
| NHPs | ||||||
| VLP-RBD | RBD | Mice | Induced long-term RBD-specific Abs with neutralizing activity against live SARS-CoV-2 and pseudotyped SARS-CoV-2 WT and B.1.1.7, B.1.351, and P.1 variants, SARS-CoV-1, and SARS-related bat CoVs | N/A | Protected BALB/c mice against mouse-adapted SARS-CoV-2 challenge, with reduced clinical signs and pathological changes | 37 |
| RBD-SpyVLP | RBD | Mice | Induced SARS-CoV-2 RBD-specific Abs and neutralizing Abs against pseudotyped and/or live SARS-CoV-2 infection | Elicited SARS-CoV-2 RBD-specific B-cell responses and weak T-cell responses | N/A | 74 and 75 |
| RBD-mi3 | Pigs | |||||
| RBD-ferritin | RBD | Mice | Induced SARS-CoV-2 S/RBD-specific Abs and more potent neutralizing Abs than RBD alone or S2P protein against pseudotyped SARS-CoV-2 infection; elicited neutralizing Abs against pseudotyped SARS-CoV-2 B.1.1.7, B.1.351, and B.1.617 variants | Elicited SARS-CoV-2 S-specific T-cell immunity (S2GΔHR2), and high GC B cell responses | Protected NHPs from SARS-CoV-2 infection, with reduced virus replication and pathological damage | 35, 39 and 71 |
| S2GΔHR2 | S | NHPs | ||||
| S1-VLP | S1 | Mice | Induced SARS-CoV-2 S/RBD-specific Abs and neutralizing Abs against SARS-CoV-2 WT and B.1.1.7 variant | N/A | N/A | 76 |
| SpFN | S | Hamsters | Induced SARS-CoV-2 S/RBD-specific Abs and neutralizing Abs against pseudotyped SARS-CoV-2 WA1, B.1.1.7, and B.1.351 variants | N/A | Protected hamsters from SARS-CoV-2 (B.1.1.7 or B.1.351 variant) infection, with reduced virus replication and decreased weight loss, virus titers, and lung pathology | 45 |
| S-LuS | S | Mice | Induced higher SARS-CoV-2 S-specific Abs and pseudovirus neutralizing Abs than those by S2P protein | N/A | N/A | 77 |
| S-Fer | S | Mice | Induced higher SARS-CoV-2 S/RBD-specific Abs and pseudovirus neutralizing Abs than RBD monomer or S trimer protein | N/A | N/A | 36 |
| SΔC-Fer | S | |||||
| NVX-CoV2373 | S | Humans | Induced SARS-CoV-2 S-specific IgG Abs and neutralizing Abs against infection of live SARS-CoV-2 | Elicited SARS-CoV-2 S-specific CD4+ T-cell responses | Protected vaccinated people against SARS-CoV-2 infection, showing efficacy against B.1.1.7 and B.1.351 variants; no serious adverse effects were identified, except for mild fever | 38, 78–80 |
| Lipid nanoparticle-based SARS-CoV-2 mRNA vaccines | ||||||
| RBD-hFc | RBD | Mice | Induced SARS-CoV-2 S-specific IgG Abs and neutralizing Abs against pseudotyped SARS-CoV-2 infection | Elicited SARS-CoV-2 S-specific Th1 cell responses | Protected 70% of vaccinated hACE2-Tg mice from SARS-CoV-2 infection | 85 and 86 |
| mRNA-RBD | RBD | Mice | Induced SARS-CoV-2 RBD-specific IgG Abs and neutralizing Abs against pseudotyped and live SARS-CoV-2 infection | Elicited SARS-CoV-2 RBD-specific T-cell responses | Protected hACE2-Tg mice from SARS-CoV-2 infection | 83 |
| RBD-LNP | RBD | Mice | Induced SARS-CoV-2 and SARS-CoV-1 RBD-specific IgG Abs and neutralizing Abs against pseudotyped and live SARS-CoV-2 infection | Elicited SARS-CoV-2 RBD-specific T-cell, Tfh, GC B, and plasma cell responses | N/A | 47 |
| S1-LNP | S1 | |||||
| RBD | RBD | Mice | Induced SARS-CoV-2 S/RBD-specific IgG Abs and neutralizing Abs against pseudotyped and live SARS-CoV-2 infection | Elicited SARS-CoV-2 RBD-specific Tfh, GC B, T-cell, B-cell, LLPC and MBC responses | N/A | 81 and 82 |
| Full-length | S | |||||
| Δfurin | ||||||
| saRNA | S | Mice | Induced SARS-CoV-2 S-specific IgG Abs and neutralizing Abs against pseudotyped and live SARS-CoV-2 infection | Elicited SARS-CoV-2 S-specific T-cell responses | N/A | 89 |
| LION/repRNA-CoV2S | S | Mice | Induced SARS-CoV-2 S-specific IgG Abs and neutralizing Abs against pseudotyped and/or live SARS-CoV-2 infection | Elicited SARS-CoV-2 S1/RBD-specific T-cell responses (in spleen, lung (for mice) and PBMCs (for NHPs) | N/A | 53 |
| NHPs | ||||||
| CV07050101 | S | NHPs | Induced SARS-CoV-2 S-specific IgG Abs without neutralizing activity | Elicited SARS-CoV-2 S-specific T-cell responses | N/A | 87 |
| LUNAR-COV19 | S | Mice | Induced SARS-CoV-2 S-specific IgG and IgM Abs, S1/RBD-specific IgG Abs, and neutralizing Abs against live SARS-CoV-2 infection | Elicited SARS-CoV-2 S-specific CD8+ T and CD4+ T/Th1-dominant T-cell responses | Protected hACE2-Tg mice from SARS-CoV-2 infection | 90 |
| mRNA-LNP | S | NHPs | Induced SARS-CoV-2 S1/S2/RBD-specific Abs and neutralizing Abs against pseudotyped and live SARS-CoV-2 infection | Elicited SARS-CoV-2 S-specific memory B-cell, GC B cell, Tfh, and T-cell responses | Protected infant NHPs from SARS-CoV-2 infection | 94 |
| MRT5500 | S | Mice | Induced SARS-CoV-2 S-specific IgG Abs and neutralizing Abs against pseudotyped or live SARS-CoV-2 infection | Elicited SARS-CoV-2 S-specific T-cell responses in mice and NHPs | Protected hamsters from SARS-CoV-2 infection without weight loss and lung pathology | 92 |
| Hamsters | ||||||
| NHPs | ||||||
| mRNA-1273 | S | Mice | Induced SARS-CoV-2 S-specific IgG Abs and neutralizing Abs against pseudotyped (wild-type and D614 mutant) and live SARS-CoV-2 infection | Elicited SARS-CoV-2 S-specific CD4+ or CD8+ T-cell responses | Protected immunized mice and NHPs from SARS-CoV-2 infection without causing disease enhancement effect; had protective efficacy in preventing COVID-19 disease in human; elicited local and systemic adverse effects (e.g., fatigue, chills, headache, myalgia, pain), allergic reactions, etc. | 54, 88, 91, 97–100 and 138 |
| NHPs Humans | ||||||
| BNT162b1 | RBD | Mice | Induced dose-dependent, and SARS-CoV-2 RBD/S-specific Abs and neutralizing Abs against pseudotyped and live SARS-CoV-2 infection | Elicited SARS-CoV-2 RBD-specific CD4+ and CD8+ T-cell responses | Protected immunized mice and NHPs from SARS-CoV-2 infection; had protective efficacy in preventing COVID-19 disease in humans, with local and systemic reactions (fatigue, headache, myalgia, local pain, etc.), allergic reactions, and rare severe adverse events (for BNT162b2) | 55, 84, 93, 101–103, 131 and 140 |
| BNT162b2 | S | NHPs | ||||
| Humans | ||||||
| Lipid nanoparticles displaying SARS-CoV-2 therapeutic agents | ||||||
| hsACE2 | hACE2 | Mice | Secreted hACE2 in blood circulation and expressed hACE2 in bronchoalveolar lavage fluid; hACE2 bound SARS-CoV-2 RBD and neutralized pseudotyped SARS-CoV-2 infection | N/A | N/A | 96 |
Protein nanoparticle vaccines displaying SARS-CoV-2 RBD. Compared to recombinant SARS-CoV-2 RBD by itself or the full-length S protein, protein nanoparticle vaccines displaying SARS-CoV-2 RBD demonstrate good immunogenicity and/or protective effects in various animal models. First, 24-meric ferritin nanoparticles displaying 24 copies of SARS-CoV-2 RBD induced antibodies that potently neutralized pseudoviruses; they also protected non-human primates (NHPs) from SARS-CoV-2 infection, reducing virus replication and pathogenesis.35,71 Here, the ferritin and SARS-CoV-2 RBD were expressed in the same polypeptide chain. Second, a computationally designed 120-meric nanoparticle (named I53-50) displaying 60 copies of SARS-CoV-2 RBD (RBD-NP) induced 10 times more neutralizing antibodies in mice than the stabilized SARS-CoV-2 full-length S protein; it also protected NHPs from SARS-CoV-2 infection.72,73 Here, the nanoparticle protein and the RBD were also expressed in the same polypeptide chain. Third, a 60-meric lumazine synthase nanoparticle displaying 120 copies of SARS-CoV-2 RBD induced >5 times more neutralizing antibodies in mice than RBD by itself; it also conferred nearly complete protection to mice challenged by SARS-CoV-2 (mouse-adapted strain).37 The induced neutralizing antibodies lasted long time in mice. Moreover, the lumazine synthase nanoparticle vaccine was also effective against SARS-CoV-2 variants (B.1.1.7, B.1.351, and P.1), SARS-CoV-1, and SARS-related bat coronavirus. In this nanoparticle vaccine, Fc-tagged dimeric RBD is connected to protein A-tagged lumazine synthase through noncovalent Fc/protein A binding.37 Fourth, 60-meric nanoparticles (SpyCatcher-mi3 or SpyCatcher003-mi3) displaying SARS-CoV-2 RBD induced neutralizing antibodies in mice and pigs.74,75 These vaccines are temperature-resistant and maintains immunogenicity after lyophilization. Here, the RBD is connected to the nanoparticle protein through covalent SpyTag
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SpyCatcher binding. Overall, all of these nanoparticle-RBD vaccines prove the concept that target vaccines presented on the surface of nanoparticle scaffolds are significantly better recognized by mammalian immune systems than vaccines by themselves.
SpyCatcher binding. Overall, all of these nanoparticle-RBD vaccines prove the concept that target vaccines presented on the surface of nanoparticle scaffolds are significantly better recognized by mammalian immune systems than vaccines by themselves.
Nanoparticle vaccines displaying SARS-CoV-2 S protein or S1 fragment. Several nanoparticle vaccines presenting SARS-CoV-2 S protein or S1 subunit have been developed and tested in preclinical studies.36,39,45,76 The 60-mer trimeric lumazine synthase nanoparticle was used again to present the S protein, but the connection between lumazine synthase and the S protein was through the covalent SpyTag
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SpyCatcher binding.77 The 24-meric ferritin nanoparticle was also used to present several truncated versions of the S protein, and the connection between ferritin and the S protein was through the covalent SpyTag
SpyCatcher binding.77 The 24-meric ferritin nanoparticle was also used to present several truncated versions of the S protein, and the connection between ferritin and the S protein was through the covalent SpyTag![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SpyCatcher binding.77 These nanoparticle vaccines elicited neutralizing antibodies against pseudotyped SARS-CoV-2 and its variants (B.1.1.7, B.1.351, P.1, and B.1.617); they also demonstrated longer retention and stronger presentation on follicular dendritic cell dendrites, and induced higher germinal center B cell responses in immunized mice.39,77 Some of these nanoparticle vaccines protected hamsters against SARS-CoV-2 infection, with reduced disease and virus replication, as well as decreased weight loss, virus titers, and lung pathology.45 Bacteriophage AP205 nanoparticles presenting SARS-CoV-2 S1 resulted in strong neutralizing antibodies in immunized mice.76
SpyCatcher binding.77 These nanoparticle vaccines elicited neutralizing antibodies against pseudotyped SARS-CoV-2 and its variants (B.1.1.7, B.1.351, P.1, and B.1.617); they also demonstrated longer retention and stronger presentation on follicular dendritic cell dendrites, and induced higher germinal center B cell responses in immunized mice.39,77 Some of these nanoparticle vaccines protected hamsters against SARS-CoV-2 infection, with reduced disease and virus replication, as well as decreased weight loss, virus titers, and lung pathology.45 Bacteriophage AP205 nanoparticles presenting SARS-CoV-2 S1 resulted in strong neutralizing antibodies in immunized mice.76
Currently developed SARS-CoV-2 mRNA vaccines based on lipid nanoparticles
LNP-based SARS-CoV-2 mRNAs are among the most advanced COVID-19 vaccines under development. Vaccines of this class have been developed preclinically and clinically. Several of them have been approved for human use to prevent SARS-CoV-2 infection (Table 1).mRNA vaccines encoding SARS-CoV-2 RBD or S1 fragment. LNP-encapsulated non-replicating mRNA vaccines encoding SARS-CoV-2 RBD have been developed and shown protective efficacy in animal models against SARS-CoV-2 infection.47,81–84 An mRNA encoding the human Fc-tagged RBD (RBD-hFc) and formulated with LNPs elicited robust neutralizing IgG antibodies and Th1-based cellular immune responses in mice, partially protecting hACE2-transgenic (hACE2-Tg) mice from SARS-CoV-2 infection.85,86 mRNAs encoding the RBD without Fc (mRNA-RBD) also showed strong immunogenicity and elicited potent immune responses. Notably, a single-dose injection of an mRNA-RBD vaccine elicited sufficient neutralizing antibodies to protect hACE2-Tg mice from SARS-CoV-2 infection; passive transfer of sera of mice immunized with this mRNA inhibited SARS-CoV-2 infection of the same mouse model.83 An mRNA vaccine (named BNT162b1) encoding a trimerized SARS-CoV-2 RBD and formulated with LNPs also induced T-cell responses and neutralizing antibodies in mice and NHPs, protecting immunized NHPs from SARS-CoV-2 infection.84 It has been shown that SARS-CoV-2 RBD-specific Tfh, GC B, and memory B cell (MBC) responses were generated after mice were immunized with LNP-formulated mRNA-RBD vaccines.81,82 Compared to LNP-formulated RBD-mRNA vaccines, LNP-formulated mRNA-S1 vaccine induced significantly lower-titer neutralizing antibodies against SARS-CoV-2,47 suggesting that the RBD should play an more important role than other parts of the S1 fragment in the development of LNP-based SARS-CoV-2 mRNA vaccines.
mRNA vaccines encoding SARS-CoV-2 S protein. In addition to SARS-CoV-2 RBD, S protein is also a critical target for the development of mRNA vaccines. mRNA vaccines encoding SARS-CoV-2 S protein (mRNA-S) have been extensively studied, and two types of mRNA-S vaccines (self-amplifying and non-replicating) have been developed and tested for immunogenicity and protective efficacy in animal models.53,54,87,88
Several self-amplifying mRNA-S vaccines are being developed preclinically. These include an LNP-formulated, self-amplifying mRNA-S vaccine encoding alphavirus replicase which induced T-cell responses and neutralizing antibodies in mice.89 In addition, a replicon mRNA-S vaccine (LION/repRNA-CoV2S), which was derived from alphavirus and formulated with lipid inorganic nanoparticles (LIONs), also elicited specific neutralizing antibody responses in mice and NHPs.53 Furthermore, a self-replicating mRNA-S vaccine encoding alphavirus replicon (LUNAR-COV19) induced T-cell responses and neutralizing antibodies that protected hACE2-Tg mice from SARS-CoV-2.90
Non-replicating mRNAs encoding SARS-CoV-2 S protein have also been extensively studied. Several of them have demonstrated protective effective in animal models challenged by SARS-CoV-2. An LNP-formulated mRNA-S vaccine induced neutralizing antibody and specific T-cell, long-lived plasma cell (LLPC), and MBC responses.81,82 mRNA-1273, BNT162b2, and MRT5500 are three LNP-encapsulated mRNA-S vaccines encoding a stabilized prefusion SARS-CoV-2 S protein. In animal models (mice, hamsters, and NHPs), these mRNA-S vaccines induced neutralizing antibodies and specific T-cell responses, protecting immunized animal models against SARS-CoV-2 infection.84,88,91–93 Importantly, these mRNA-S vaccines did not cause vaccine-associated enhancement of disease after SARS-CoV-2 challenge.54,84,91 Other mRNA-S vaccines also elicited T-cell responses and neutralizing IgG antibodies specific to the RBD, S1, and S2 fragments; these immune responses lasted for 22 weeks and protected infant NHPs from SARS-CoV-2 infection.94
mRNA vaccines encoding SARS-CoV-2 therapeutics. In addition to expressing SARS-CoV-2-specific proteins as vaccine targets, mRNAs can also express SARS-CoV-2 therapeutics to directly inhibit SARS-CoV-2 entry. As the cellular receptor for SARS-CoV-2, ACE2 plays an essential role in SARS-CoV-2 entry.23,95 mRNAs encoding ACE2 receptor (mRNA-ACE2) produce recombinant ACE2 that directly binds to SARS-CoV-2 and blocks the virus from binding to cell-surface ACE2.96 It has been shown that after injection of mRNA-ACE2 into mice, secreted ACE2 reached a peak level in the blood circulation 6 hours and was detectable in bronchoalveolar lavage fluid 24 and 48 hours.96
In a Phase I clinical trial among people 18–55 years old, the mRNA-1273 vaccine elicited specific T-cell responses and IgG antibodies in a dose-dependent manner, neutralizing both pseudotyped and live SARS-CoV-2 infection.97 In another Phase I trial among older adults (56–70 or ≥71 years old), this vaccine also induced dose-dependent antibody responses with neutralizing activity, as well as CD4+ T-cell responses.98 The vaccine largely caused mild or moderate local and systemic adverse effects, including fatigue, chills, headache, myalgia, and pain at the injection site; only three participants at the highest injection dosage reported severe adverse effects.97,98 In addition, the vaccine induced durable antibodies that were maintained for 119 days after the first immunization but declined over time; these antibodies were able to neutralize both pseudotyped and authentic SARS-CoV-2 infections.99 In a Phase III trial, this mRNA vaccine was 94.1% effective in preventing COVID-19 illness in all participants, including those who were ≥65 years of age or had a SARS-CoV-2 infection at the time of vaccination.100
In a phase I clinical trial, BNT162b1 and BNT162b2 mRNA vaccines elicited similar dose-dependent SARS-CoV-2 neutralizing antibodies in adults 18–55 or 65–85 years old.101 Compared to BNT162b1, BNT162b2 was associated with fewer and milder systemic reactions. In phase I/II trials, BNT162b1 induced RBD-specific antibody responses, neutralizing antibodies, and T-cell responses in healthy adults aged 18–55 years, with mild to moderate local and systemic reactions.102,103 BNT162b2 exhibited 95% efficacy in prevention of COVID-19 in a phase III clinical trial, with mild side effects including pain at the injection site, fatigue, and headache.55 In a phase IV clinical trial, most of the participants developed at least one side effect, such as injection site pain, fatigue, headache, muscle pain, or chills.104
The BNT162b2 and mRNA-1273 vaccines are generally effective in inducing immune responses in individuals, particularly healthy people.113–115 It takes about two weeks after the second vaccination for immune responses to reach high levels.116 Vaccine-induced T cells, particularly memory T cells, were detectable 100 days after the second dose of the BNT162b2 vaccine.117 Overall, the immune responses can last up to 6 months.118 Reduced antibody or cellular immune responses were observed in elderly people, organ transplant patients, cancer patients, and patients with other immunosuppressed conditions.115,119–130 Between the two mRNA vaccines, mRNA-1273 led to better outcomes than BNT162b2 against COVID-19-related hospitalizations.107 Compared with an authorized viral vector vaccine (Ad26·COV2 from Johnson & Johnson), both of the mRNA vaccines induced more potent immune responses such as higher levels of S and RBD-specific antibodies.107
While overall both mRNA vaccines are safe to use in humans, some side effects have been identified. After the first or second dose of either of the two mRNA vaccines, regular local and systemic side reactions have been reported. These side effects include injection-site pain, muscle pain, joint pain, headache, fever, fatigue, malaise, chills, nausea, myalgia, and swelling.55,131–135 The side effects also include cardiovascular adverse events, such as tachycardia, flushing, hypertension, hypotension, and peripheral coldness. Allergic reactions were reported after receiving either of the two mRNA vaccines.136–138 Moreover, people >75 years old may also develop acute myocardial infarction, cardiac arrest, and circulatory collapse.139 For the BNT162b2 vaccine, some rare serious cases were reported in adults or elderly people, including Miller Fisher syndrome (a nerve disease related to Guillain-Barré syndrome), intracerebral hemorrhage due to vasculitis, and multisystem inflammation and organ dysfunction.140–142 Also for BNT162b2, myocarditis were reported in children, particularly in boys, after the second dose of the vaccine.143 It is believed that local or systemic adverse reactions are associated with increased SARS-CoV-2-specific IgG antibody responses.144
To further boost protection against COVID-19, the FDA has authorized a third dose of the two mRNA vaccines for individuals with immunocompromised diseases. The CDC recommended timing for the third dose for individuals aged 18 years or order is six months after the second dose. The third dose of BNT162b2 induced a moderate increase of S-specific antibody and/or cellular immune responses or neutralizing antibodies in some kidney transplant recipients and leukemia patients, which improved protection against SARS-CoV-2 infection.145–149 However, some organ (such as kidney) transplant recipients receiving immunosuppressive drugs did not develop effective S or RBD-specific antibody responses or neutralizing antibodies.150,151 Patients with solid tumors receiving chemotherapy produced moderately increased neutralizing antibodies, but not T-cell responses.152 The aforementioned side effects after the third dose of BNT162b2 were similar to, or better than, the second dose in immunocompromised individuals, the elderly, and other individuals.153,154
Currently developed nanobodies as anti-SARS-CoV-2 therapeutics
SARS-CoV-2 RBD is a key target for the development of potent nanobodies. Several SARS-CoV-2-specific nanobodies have been developed preclinically. Almost all of them target the RBD (Table 2).| Nanobody name | Target | Binding affinity and mechanism | Structure | Neutralizing activity | Protection and/or pharmacokinetics (PK) | Ref. |
|---|---|---|---|---|---|---|
| ACE2, angiotensin-converting enzyme 2; Cryo-EM: cryo-electron microscopy; EC50, half maximal effective concentration; hACE2-Tg, human ACE2-transgenic; IC50, half maximal inhibitory concentration; ND50, 50% neutralization dose; Kd, equilibrium dissociation constant; Nbs, nanobodies; RBD, receptor-binding domain; WT, wild-type. | ||||||
| 1E2 | RBD | Bound to RBD (EC50 0.996–35.52 nM), blocking the RBD-ACE2 binding | No | Neutralized pseudotyped (IC50 0.9–69 ng ml−1) and live (IC50 0.13–0.51 μg ml−1) SARS-CoV-2 infection | N/A | 156 |
| 4D8 | ||||||
| 2F2 | ||||||
| 3F11 | ||||||
| 5F8 | ||||||
| Ty1 | RBD | Bound to RBD (Kd 5–10 nM), preventing the RBD-ACE2 binding | Yes, cryo-EM structure (for Ty1) | Neutralized pseudotyped (IC50 0.77 μg ml−1 for Ty1; IC50 0.012 μg ml−1 for Ty1-Fc) SARS-CoV-2 infection | N/A | 67 |
| Ty1-Fc | ||||||
| Sb23 | RBD | Bound to RBD, competing its binding with ACE2 | Yes, crystal structure | Neutralized pseudotyped (IC50 0.6 μg ml−1) SARS-CoV-2 infection | N/A | 70 |
| H11-D4 | RBD | Bound to RBD (Kd 39 and 12 nM for H11-D4 and h11-H4), blocking the RBD-ACE2 interaction | Yes, crystal and cryo-EM structures (for H11-D4 and H11-H4) | Neutralized live (IC50 4-6 nM for H11-H4-Fc; 18 nM for H11-D4-Fc) SARS-CoV-2 infection | N/A | 155 |
| H11-H4 | ||||||
| H11-D4-Fc | ||||||
| H11-H4-Fc | ||||||
| Nb20 | RBD | Nbs 20 and 21 bound to RBD (Kd < 1 pM), overlapping with the ACE2 binding site | Yes, crystal structure (for Nb20) | Neutralized pseudotyped (IC50 0.045–0.133 nM) and live (IC50 0.022–0.154 nM) SARS-CoV-2 infection, with multimeric Nbs improving neutralizing activity against infection of pseudotyped (IC50 1.3–4.1 pM) and live SARS-CoV-2 | N/A | 68 |
| Nb21 | ||||||
| Nb89 | ||||||
| Nb203 | ||||||
| Nb213 | ||||||
| Nb3 | RBD | Bound to RBD by inhibiting the RBD-ACE2 interaction (via blocking the binding site or locking the RBD into an inactive conformation) | Yes, crystal (for mNb6) and cryo-EM (for Nb6, Nb11, and mNb6) structures | Neutralized pseudotyped (IC50 2.0–3.9 μM) SARS-CoV-2 infection, with trimeric or matured Nbs enhancing neutralizing activity against infection of pseudotyped (IC50 1.2–400 nM for tri-Nbs; 0.12 nM for mNb6-tri) and live (IC50 0.16–140 nM for tri-Nbs; 0.054 nM for mNb6-tri) SARS-CoV-2 | Prophylactically and therapeutically prevented hamsters from SARS-CoV-2 infection; inhalation treatment inhibited weight loss, with decreased lung viral titers and lung pathology | 69 and 159 |
| Nb6 | ||||||
| Nb11 | ||||||
| mNb6 | ||||||
| Nb3-tri | ||||||
| Nb6-tri | ||||||
| Nb11-tri | ||||||
| mNb6-tri | ||||||
| PiN-21 | ||||||
| C1 | RBD | Bound to WT or variant RBD (Kd 0.0003–2.523 nM) by overlapping with the ACE2 epitope (for H3 and C5) or a different epitope (for C1) | Yes, crystal (for C1, H3, and C5) and cryo-EM (for C5) structures | Neutralized (IC50 0.025–8.2 nM) infection of SARS-CoV-2 variants (B.1.1.7 or B.1.351) (for C1-trimer, C5-trimer, or H3-trimer) | C5-trimer or C5-Fc therapeutically prevented hamsters from SARS-CoV-2 infection | 158 |
| H3 | ||||||
| C5 | ||||||
| C1-trimer | ||||||
| H3-trimer | ||||||
| C5-trimer | ||||||
| C5-Fc | ||||||
| Nanosota-I | RBD | Bound to RBD (Kd 15.7 pg for Nanosota-1C-Fc), blocking the RBD-ACE2 binding | Yes, crystal structure (for Nanosota-1C) | Nanosota-IC-Fc inhibited infection of pseudotyped and/or live (ND50 0.16 μg ml−1) SARS-CoV-2 and variant (D614G) | Nanosota-IC-Fc prophylactically and therapeutically inhibited hamsters and hACE2-Tg mice from SARS-CoV-2 infection; >10-day half-life and high tissue bioavailability | 63 |
| Nanosota-IC | ||||||
| Nanosota-IC-Fc | ||||||
| K-874A | S1 (RBD-NTD) | Bound to S1 region (Kd 1.4 nM) between RBD and NTD of S protein | Cryo-EM structure | Blocked viral membrane fusion, neutralizing SARS-CoV-2 B.1.1.7 variant | Therapeutically protected hamsters from SARS-CoV-2 infection | 157 |
A number of nanobodies have been discovered from the camelid (e.g., llama and alpaca) immune system. In one approach, camelid animals were immunized with recombinant SARS-CoV-2 S1 or RBD protein, immunized nanobody libraries were established from the camelid immune cells, and these libraries were subsequently screened using proteomic strategy or phage display methods. Nanobodies that have been discovered using this approach include Nb20, Nb21, Nb89, Nb34, Nb95, and Ty1.67,68 In another approach, naïve nanobody libraries were established from naïve camelid immune cells and subsequently screened. Nanobodies screened from the naive library generally have low affinity for the target antigen and need to go through in vitro affinity maturation procedures. Nanobodies that have been discovered using this approach include Nb3, Nb6, Nb11, Sb23, 1E2, 4D8, H11-H4, H11-D4, and Nanosota-1C.63,69,70,155,156 For example, affinity-matured Nb6 and Nanosota-1 were both generated by mutating several amino acids at the antigen-binding sites and demonstrated much higher RBD-binding affinity and potent SARS-CoV-2 neutralizing potency than the respective nanobodies before the affinity maturation.63,69 Most nanobodies discovered from either of the above approach so far target SARS-CoV-2 RBD. Some of these nanobodies (such as Nb6, Nb11, Nb20, Nb21, Sb23, and Nanosota-1) recognize the RBD epitopes that overlap with the hACE2 binding site, whereas some other nanobodies (such as Nb34, Nb95, and K-874A) recognize the RBD epitopes that do not overlap with the ACE2 binding site.63,68–70,157 The former nanobodies directly block viral attachment to ACE2, whereas the latter may block the conformational changes of the S protein and hence inhibit the viral membrane fusion process.63,67–69,156,157
Multivalent (dimeric or trimeric) nanobodies have been constructed to improve the nanobodies’ target binding affinity, neutralizing potency, and in vivo half-life. One of the approaches is to link monomeric nanobodies linearly. These nanobodies include Nb3-tri, Nb6-tri, Nb11-tri, Nb203, Nb213, Nb20-20, Nb21-21, Nb21–Nb34, C1-trimer, H3-trimer, and C5-trimer. Compared to monomeric nanobodies, the linearly linked dimeric or trimeric nanobodies all neutralized pseudotyped SARS-CoV-2 infection more potently.63,68,69,158 In particular, compared to monomeric Nb6, its linearly linked dimer and trimer bound to SARS-CoV-2 RBD 750- and >20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times more strongly, respectively, and they neutralized pseudotyped SARS-CoV-2 infection 10 and 2000 times more potently respectively.69 These multivalent nanobodies maintain the high solubility, yield, and thermostability of monomeric nanobodies. Another approach to building multivalent nanobodies is to add a multivalent tag to the nanobodies. Dimeric Fc tags have been added to nanobodies, including Nanosota-1C-Fc 1E2-Fc, 4D8-Fc, 2F2-Fc, 3F11-Fc, 5F8-Fc, Ty1-Fc, and C5-Fc. Compared to monomeric nanobodies, the Fc-tagged dimeric nanobodies demonstrated binding to the RBD more tightly and neutralize SARS-CoV-2 infection more potently.63,67,156,158 A common misconception about building multivalent nanobodies is that multivalency makes nanobodies lose their advantages of being small. However, compared to conventional antibodies whose molecular weight is ∼160 kDa, Fc-tagged dimeric nanobodies are still much smaller with a molecular weight of ∼75–80 kDa. These multivalent nanobodies can still access cryptic epitopes on their targets because their antigen-binding sites are identical to monomeric nanobodies. Moreover, multivalent nanobodies maintain their good physical and chemical stabilities and production yields.63 It has been shown that some of the multivalent nanobodies even maintain their RBD-binding affinity and neutralizing activity after aerosolization, lyophilization, or heat treatment.69 Importantly, multivalent nanobodies have extended in vivo half-life because their molecular weight exceeds the molecular weight cutoff for kidney clearance (which is ∼60 kDa).63 Overall, multivalent nanobodies possess many advantages over monomeric nanobodies as antiviral therapeutics.
000 times more strongly, respectively, and they neutralized pseudotyped SARS-CoV-2 infection 10 and 2000 times more potently respectively.69 These multivalent nanobodies maintain the high solubility, yield, and thermostability of monomeric nanobodies. Another approach to building multivalent nanobodies is to add a multivalent tag to the nanobodies. Dimeric Fc tags have been added to nanobodies, including Nanosota-1C-Fc 1E2-Fc, 4D8-Fc, 2F2-Fc, 3F11-Fc, 5F8-Fc, Ty1-Fc, and C5-Fc. Compared to monomeric nanobodies, the Fc-tagged dimeric nanobodies demonstrated binding to the RBD more tightly and neutralize SARS-CoV-2 infection more potently.63,67,156,158 A common misconception about building multivalent nanobodies is that multivalency makes nanobodies lose their advantages of being small. However, compared to conventional antibodies whose molecular weight is ∼160 kDa, Fc-tagged dimeric nanobodies are still much smaller with a molecular weight of ∼75–80 kDa. These multivalent nanobodies can still access cryptic epitopes on their targets because their antigen-binding sites are identical to monomeric nanobodies. Moreover, multivalent nanobodies maintain their good physical and chemical stabilities and production yields.63 It has been shown that some of the multivalent nanobodies even maintain their RBD-binding affinity and neutralizing activity after aerosolization, lyophilization, or heat treatment.69 Importantly, multivalent nanobodies have extended in vivo half-life because their molecular weight exceeds the molecular weight cutoff for kidney clearance (which is ∼60 kDa).63 Overall, multivalent nanobodies possess many advantages over monomeric nanobodies as antiviral therapeutics.
Currently, only a few anti-SARS-CoV-2 nanobodies have been tested and shown to be efficacious in animal models. For example, single-dose injection of hamsters with Fc-fused Nanosota-1C, K-874A, C5-trimer, and C5-Fc protected the animals from SARS-CoV-2 infection both prophylactically and/or therapeutically.63,157,158 Inhalable treatment of hamsters with PiN-21 nanobody prevented SARS-CoV-2 infection with reduced weight loss and decreased viral titers and lung pathology.159 Fc-fused Nanosota-1C also protected hACE2-Tg mice from SARS-CoV-2 infection with reduced viral titers and/or absence of lung pathology in the lung.63 To date, however, no SARS-CoV-2-specific nanobody has been evaluated for efficacy in humans.
To identify the neutralizing epitopes of nanobodies, crystal structures of nanobody/RBD complexes and cryo-EM structures of nanobody/S complexes have been determined for the following nanobodies: Nb11, mNb6, C1, H3, C5, K-874A, Nb20, Nanosota-1C, H11-H4, H11-D4, Ty1, Sb23, and Nb6 (Fig. 5a–g).63,67–70,155,157,158 Whereas ACE2 only binds to SARS-CoV-2 RBD in the “up” position of the S protein, nanobodies can bind to SARS-CoV-2 RBD in the “up” or “down” conformation of the S protein due to their small size (Fig. 5d–g).67,70 Structural analysis of nanobodies in complex with the RBD or S protein is important for understanding the neutralizing mechanisms of the nanobodies and also provides a structural framework for optimizing the target-binding affinity and therapeutic potency of the nanobodies.
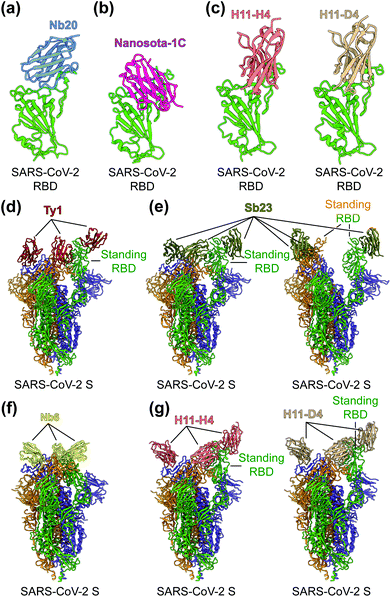 | ||
| Fig. 5 Structures of SARS-CoV-2 RBD or S protein trimer in complex with neutralizing nanobodies. (a–c) Crystal structures of SARS-CoV-2 receptor-binding domain (RBD) in complex with (a) neutralizing nanobodies Nb20 (PDB 7JVB), (b) Nanosota-1C (PDB 7KM5), (c) H11-H4 (PDB 6ZBP) or H11-D4 (PDB 6YZ5). SARS-CoV-2 RBD is colored in green and the four neutralizing nanobodies are colored in blue, magenta, salmon and tan, respectively. (d–g) Cryo-EM structures of SARS-CoV-2 spike (S) trimer in complex with neutralizing nanobodies (d) Ty1 with one RBD in the standing position (PDB 6ZXN), (e) Sb23 with one (PDB 7A25) and two (PDB 7A29) RBDs in the standing position, (f) Nb6 with all RBDs in the lying-down position (PDB 7KKK), (g) H11-H4 (PDB 6ZHD) and H11-D4 (PDB 6Z43) with one RBD in the standing position, respectively. Three copies of the S protein in the trimer are colored in green, orange and purple, respectively. Neutralizing nanobodies are colored in red (Ty1), olive (Sb23), yellow (Nb6), salmon (H11-H4), and tan (H11-D4), respectively. | ||
Potential challenges nanotechnology-based COVID-19 vaccines and therapeutics
SARS-CoV-2 mutates at fast paces, particularly in the RBD region of the S protein. Multiple mutant virus strains, including Omicron variant, have been identified all over the world, and they almost all harbor mutations in the RBD.160–164 Although the currently available COVID-19 mRNA-S vaccines are effective against original virus strain and some mutant variants, many vaccine-induced antibodies are less effective against mutant strains with the K417N/T, L452R, E484K, N501Y, K417N-E484K-N501Y, or T478K mutations in the RBD.72,165–169 Hence there is an urgent need for developing universal vaccines, particularly those that can induce protective T-cell responses against newly-emerged and future SARS-CoV-2 variant strains. In contrast to the fast-mutating RBD, the S2 region is more conserved among different SARS-CoV-2 strains and among SARS-CoV-2, SARS-CoV-1, and SARS-related beta-coronaviruses. Thus, the S2 region may serve as an important target for the development of pan-beta-coronavirus vaccines. Other conserved structural proteins such as N or M may also serve as targets for developing SARS-CoV-2 vaccines with broad efficacy. However, it remains to be seen whether vaccines targeting the S2 region of the S protein, N protein, and M protein can induce potent neutralizing or protective immune responses. If the answer is yes, both protein nanoparticle-based protein vaccines and lipid nanoparticle-based mRNA vaccines can incorporate these more conserved antigens as immunogens.Like vaccines, nanobody therapy also faces a similar challenge in neutralizing variant SARS-CoV-2 strains. Three potential strategies can be used. First, nanobodies can target different epitopes on SARS-CoV-2 S protein because mutations in multiple epitopes are less likely to occur than single-site mutations. To this end, multimeric nanobodies can be constructed by combining two or more nanobodies recognizing different epitopes on the same SARS-CoV-2 S protein. Alternatively, cocktails of nanobodies targeting different epitopes on the S protein may be used in therapies. Second, nanobodies can be developed to target conserved and essential epitopes on the S protein. Because of the high sequence conservation of the S2 region among SARS-CoV-2, SARS-CoV-1, and SARS-related coronaviruses, it is possible to develop broadly neutralizing nanobodies that target the S2 subunit. One advantage of nanobodies over conventional antibodies is that nanobodies can access cryptic epitopes that are essential for the function of the S protein. Third, nanobodies can be quickly adapted to new virus strains through in vitro affinity maturation using phage display. Overall, new nanobody therapies have the flexibility to be adapted to new SARS-CoV-2 variants.
While conventional antibodies have been developed for decades and technologies for developing them are much mature, the development of nanobodies as human therapeutics is still in its infancy. Nanobodies are generally produced by immunizing large camelid animals such as camels and llama, which can be difficult to access and also require very restricted regulatory procedures. Fortunately, recent development of naïve nanobody libraries makes it possible to rapidly screen target nanobodies quickly and conveniently. Nanobodies are generally safe to use as they have low immunogenicity and toxicity in humans.170–172 Nanobodies, such as Caplacizumab-yhdp (Cablivi), have been approved as therapeutic agents to treat human diseases, including thrombotic thrombocytopenic purpura (aTTP),173,174 confirming the safety of nanobodies.
Conclusion and future perspectives
Since the COVID-19 pandemic began, extensive global efforts and increased funds have been devoted to the development of effective vaccines and therapeutics to battle this pandemic. Using nanotechnology as a tool, protein nanoparticle-based and lipid nanoparticle-formulated mRNA vaccines have been developed rapidly and shown to be effective in animals. In particular, two mRNA vaccines have demonstrated both safety and high efficacy in preventing SARS-CoV-2 infection in humans. Future efforts will be needed to expand the spectrum of these vaccines against divergent virus variants. Notably, lipid nanoparticles have contributed substantially to the battle against COVID-19 by formulating with mRNA vaccines and delivering them into immune cells. It is worth noting that lipid nanoparticles can also deliver DNA to immune cells to treat non-viral diseases.175–177 Hence lipid nanoparticle-formulated DNA vaccines may be developed to battle COVID-19, and the result can be compared with the data from traditional approaches for DNA delivery.178–181 While several SARS-CoV-2-specific conventional antibodies have proceeded into human clinical trials,22,182,183 SARS-CoV-2-specific nanobodies are still being developed at the preclinical stage, and only a few of them have been tested for efficacy in animal models. We hope that some of the potent and cost-effective SARS-CoV-2-neutralizing nanobodies will move forward to human clinical trials where their safety and therapeutic efficacy can be tested in humans. Overall, nanotechnology has been and will continue to be one of the major scientific weapons in the global battle against the COVID-19 pandemic.Author contributions
L. D. and F. L. wrote and revised the manuscript. Y. Y and X. Z. prepared the figures.Conflicts of interest
The University of Minnesota has filed a patent on the lumazine synthase nanoparticle-based SARS-CoV-2 RBD vaccine with F. L and L. D. as two of the inventors and another patent on the Nanosota-1 candidate drugs with F. L. as one of the inventors. Other authors have declared that no competing interests exist.Acknowledgements
This study was supported by NIH grants (R01AI139092, R01AI137472, and R01AI157975).References
- K. Blecher, A. Nasir and A. Friedman, Virulence, 2011, 2, 395–401 CrossRef PubMed.
- L. Singh, H. G. Kruger, G. E. M. Maguire, T. Govender and R. Parboosing, Ther. Adv. Infect. Dis., 2017, 4, 105–131 CAS.
- M. S. Akilesh and A. Wadhwani, Curr. Drug Res. Rev., 2021, 13, 120–129 CrossRef PubMed.
- L. Milane and M. Amiji, Drug Delivery Transl. Res., 2021, 11, 1309–1315 CrossRef CAS PubMed.
- M. A. G. Hoffmann, Y. Bar-On, Z. Yang, H. B. Gristick, P. N. P. Gnanapragasam, J. Vielmetter, M. C. Nussenzweig and P. J. Bjorkman, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 18719–18728 CrossRef PubMed.
- M. Chakravarty and A. Vora, Drug Delivery Transl. Res., 2021, 11, 748–787 CrossRef CAS PubMed.
- Y. T. Lee, E. J. Ko, K. H. Kim, H. S. Hwang, Y. Lee, Y. M. Kwon, M. C. Kim, Y. N. Lee, Y. J. Jung and S. M. Kang, J. Biomed. Nanotechnol., 2017, 13, 84–98 CrossRef CAS PubMed.
- M. J. Rupar, P. Golusinski, W. Golusinski and M. M. Masternak, Rep. Pract. Oncol. Radiother., 2019, 24, 544–550 CrossRef PubMed.
- P. Zhou, X. L. Yang, X. G. Wang, B. Hu, L. Zhang, W. Zhang, H. R. Si, Y. Zhu, B. Li, C. L. Huang, H. D. Chen, J. Chen, Y. Luo, H. Guo, R. D. Jiang, M. Q. Liu, Y. Chen, X. R. Shen, X. Wang, X. S. Zheng, K. Zhao, Q. J. Chen, F. Deng, L. L. Liu, B. Yan, F. X. Zhan, Y. Y. Wang, G. F. Xiao and Z. L. Shi, Nature, 2020, 579, 270–273 CrossRef CAS PubMed.
- L. Du, Y. He, Y. Zhou, S. Liu, B. J. Zheng and S. Jiang, Nat. Rev. Microbiol., 2009, 7, 226–236 CrossRef CAS PubMed.
- A. M. Zaki, S. van Boheemen, T. M. Bestebroer, A. D. Osterhaus and R. A. Fouchier, N. Engl. J. Med., 2012, 367, 1814–1820 CrossRef CAS PubMed.
- F. Li, Annu. Rev. Virol., 2016, 3, 237–261 CrossRef CAS PubMed.
- L. Du, G. Zhao, Z. Kou, C. Ma, S. Sun, V. K. Poon, L. Lu, L. Wang, A. K. Debnath, B. J. Zheng, Y. Zhou and S. Jiang, J. Virol., 2013, 87, 9939–9942 CrossRef CAS PubMed.
- L. Du, W. Tai, Y. Zhou and S. Jiang, Expert Rev. Vaccines, 2016, 15, 1123–1134 CrossRef CAS PubMed.
- J. Cui, F. Li and Z. L. Shi, Nat. Rev. Microbiol., 2019, 17, 181–192 CrossRef CAS PubMed.
- Y. Zhou, S. Jiang and L. Du, Expert Rev. Vaccines, 2018, 17, 677–686 CrossRef CAS PubMed.
- N. Zhang, J. Shang, C. Li, K. Zhou and L. Du, Expert Rev. Vaccines, 2020, 19, 817–829 CrossRef CAS PubMed.
- L. Du, Y. Yang, Y. Zhou, L. Lu, F. Li and S. Jiang, Expert Opin. Ther. Targets, 2017, 21, 131–143 CrossRef CAS PubMed.
- N. Wang, J. Shang, S. Jiang and L. Du, Front. Microbiol., 2020, 11, 298 CrossRef PubMed.
- L. Du, Y. Yang and X. Zhang, Cell. Mol. Immunol., 2021, 18, 2293–2306 CrossRef CAS PubMed.
- Y. Yang and L. Du, Signal Transduction Targeted Ther., 2021, 6, 95 CrossRef CAS PubMed.
- S. Jiang, X. Zhang, Y. Yang, P. J. Hotez and L. Du, Nat. Biomed. Eng., 2020, 4, 1134–1139 CrossRef CAS PubMed.
- S. Jiang, X. Zhang and L. Du, Expert Opin. Ther. Targets, 2021, 25, 415–421 CrossRef CAS PubMed.
- S. Jiang, C. Hillyer and L. Du, Trends Immunol., 2020, 41, 355–359 CrossRef CAS PubMed.
- W. Tai, X. Zhang, Y. He, S. Jiang and L. Du, Antiviral Res., 2020, 179, 104820 CrossRef CAS PubMed.
- J. Shang, G. Ye, K. Shi, Y. Wan, C. Luo, H. Aihara, Q. Geng, A. Auerbach and F. Li, Nature, 2020, 581, 221–224 CrossRef CAS PubMed.
- J. Lan, J. Ge, J. Yu, S. Shan, H. Zhou, S. Fan, Q. Zhang, X. Shi, Q. Wang, L. Zhang and X. Wang, Nature, 2020, 581, 215–220 CrossRef CAS PubMed.
- Y. Wan, J. Shang, R. Graham, R. S. Baric and F. Li, J. Virol., 2020, 94, e00127-20 Search PubMed.
- F. Li, W. Li, M. Farzan and S. C. Harrison, Science, 2005, 309, 1864–1868 CrossRef CAS PubMed.
- F. Li, J. Virol., 2015, 89, 1954–1964 CrossRef PubMed.
- S. Xia, M. Liu, C. Wang, W. Xu, Q. Lan, S. Feng, F. Qi, L. Bao, L. Du, S. Liu, C. Qin, F. Sun, Z. Shi, Y. Zhu, S. Jiang and L. Lu, Cell Res., 2020, 30, 343–355 CrossRef CAS PubMed.
- J. Shang, Y. Wan, C. Luo, G. Ye, Q. Geng, A. Auerbach and F. Li, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 11727–11734 CrossRef CAS PubMed.
- S. Jiang, L. Du and Z. Shi, Emerging Microbes Infect., 2020, 9, 275–277 CrossRef CAS PubMed.
- A. C. Walls, Y. J. Park, M. A. Tortorici, A. Wall, A. T. McGuire and D. Veesler, Cell, 2020, 181, 281–292 CrossRef CAS PubMed.
- L. He, X. Lin, Y. Wang, C. Abraham, C. Sou, T. Ngo, Y. Zhang, I. A. Wilson and J. Zhu, Sci. Adv., 2021, 7, eabf1591 CrossRef PubMed.
- A. E. Powell, K. Zhang, M. Sanyal, S. Tang, P. A. Weidenbacher, S. Li, T. D. Pham, J. E. Pak, W. Chiu and P. S. Kim, ACS Cent. Sci., 2021, 7, 183–199 CrossRef CAS PubMed.
- Q. Geng, W. Tai, V. K. Baxter, J. Shi, Y. Wan, X. Zhang, S. A. Montgomery, S. A. Taft-Benz, E. J. Anderson, A. C. Knight, K. H. Dinnon, 3rd, S. R. Leist, R. S. Baric, J. Shang, S. W. Hong, A. Drelich, C. K. Tseng, M. Jenkins, M. Heise, L. Du and F. Li, PLoS Pathog., 2021, 17, e1009897 CrossRef CAS PubMed.
- C. Keech, G. Albert, I. Cho, A. Robertson, P. Reed, S. Neal, J. S. Plested, M. Zhu, S. Cloney-Clark, H. Zhou, G. Smith, N. Patel, M. B. Frieman, R. E. Haupt, J. Logue, M. McGrath, S. Weston, P. A. Piedra, C. Desai, K. Callahan, M. Lewis, P. Price-Abbott, N. Formica, V. Shinde, L. Fries, J. D. Lickliter, P. Griffin, B. Wilkinson and G. M. Glenn, N. Engl. J. Med., 2020, 383, 2320–2332 CrossRef CAS PubMed.
- Y. N. Zhang, J. Paynter, C. Sou, T. Fourfouris, Y. Wang, C. Abraham, T. Ngo, Y. Zhang, L. He and J. Zhu, Sci. Adv., 2021, 7, eabj3107 CrossRef CAS PubMed.
- R. Pati, M. Shevtsov and A. Sonawane, Front. Immunol., 2018, 9, 2224 CrossRef PubMed.
- V. Shinde, I. Cho, J. S. Plested, S. Agrawal, J. Fiske, R. Cai, H. Zhou, X. Pham, M. Zhu, S. Cloney-Clark, N. Wang, B. Zhou, M. Lewis, P. Price-Abbott, N. Patel, M. J. Massare, G. Smith, C. Keech, L. Fries and G. M. Glenn, Lancet Infect. Dis., 2021, 22, 73–84 CrossRef PubMed.
- D. M. Smith, J. K. Simon and J. R. Baker, Jr., Nat. Rev. Immunol., 2013, 13, 592–605 CrossRef CAS PubMed.
- S. Al-Halifa, L. Gauthier, D. Arpin, S. Bourgault and D. Archambault, Front. Immunol., 2019, 10, 22 CrossRef CAS PubMed.
- N. Zhang, C. Li, S. Jiang and L. Du, Vaccines, 2020, 8, 481 CrossRef CAS PubMed.
- K. M. Wuertz, E. K. Barkei, W. H. Chen, E. J. Martinez, I. Lakhal-Naouar, L. L. Jagodzinski, D. Paquin-Proulx, G. D. Gromowski, I. Swafford, A. Ganesh, M. Dong, X. Zeng, P. V. Thomas, R. S. Sankhala, A. Hajduczki, C. E. Peterson, C. Kuklis, S. Soman, L. Wieczorek, M. Zemil, A. Anderson, J. Darden, H. Hernandez, H. Grove, V. Dussupt, H. Hack, R. de la Barrera, S. Zarling, J. F. Wood, J. W. Froude, M. Gagne, A. R. Henry, E. B. Mokhtari, P. Mudvari, S. J. Krebs, A. S. Pekosz, J. R. Currier, S. Kar, M. Porto, A. Winn, K. Radzyminski, M. G. Lewis, S. Vasan, M. Suthar, V. R. Polonis, G. R. Matyas, E. A. Boritz, D. C. Douek, R. A. Seder, S. P. Daye, M. Rao, S. A. Peel, M. G. Joyce, D. L. Bolton, N. L. Michael and K. Modjarrad, npj Vaccines, 2021, 6, 129 CrossRef CAS PubMed.
- W. Tai, X. Zhang, Y. Yang, J. Zhu and L. Du, Transl. Res., 2021, S1931-5244(21)00280-2 Search PubMed.
- W. Tai, X. Zhang, A. Drelich, J. Shi, J. C. Hsu, L. Luchsinger, C. D. Hillyer, C. K. Tseng, S. Jiang and L. Du, Cell Res., 2020, 30, 932–935 CrossRef CAS PubMed.
- A. Wadhwa, A. Aljabbari, A. Lokras, C. Foged and A. Thakur, Pharmaceutics, 2020, 12, 102 CrossRef CAS PubMed.
- J. Kim, Y. Eygeris, M. Gupta and G. Sahay, Adv. Drug Delivery Rev., 2021, 170, 83–112 CrossRef CAS PubMed.
- N. A. C. Jackson, K. E. Kester, D. Casimiro, S. Gurunathan and F. DeRosa, npj Vaccines, 2020, 5, 11 CrossRef PubMed.
- M. C. Ballesteros-Briones, N. Silva-Pilipich, G. Herrador-Canete, L. Vanrell and C. Smerdou, Curr. Opin. Virol., 2020, 44, 145–153 CrossRef CAS PubMed.
- K. Bloom, F. van den Berg and P. Arbuthnot, Gene Ther., 2021, 28, 117–129 CrossRef CAS PubMed.
- J. H. Erasmus, A. P. Khandhar, M. A. O’Connor, A. C. Walls, E. A. Hemann, P. Murapa, J. Archer, S. Leventhal, J. T. Fuller, T. B. Lewis, K. E. Draves, S. Randall, K. A. Guerriero, M. S. Duthie, D. Carter, S. G. Reed, D. W. Hawman, H. Feldmann, M. Gale, Jr., D. Veesler, P. Berglund and D. H. Fuller, Sci. Transl. Med., 2020, 12, eabc9396 CrossRef CAS PubMed.
- K. S. Corbett, B. Flynn, K. E. Foulds, J. R. Francica, S. Boyoglu-Barnum, A. P. Werner, B. Flach, S. O'Connell, K. W. Bock, M. Minai, B. M. Nagata, H. Andersen, D. R. Martinez, A. T. Noe, N. Douek, M. M. Donaldson, N. N. Nji, G. S. Alvarado, D. K. Edwards, D. R. Flebbe, E. Lamb, N. A. Doria-Rose, B. C. Lin, M. K. Louder, S. O'Dell, S. D. Schmidt, E. Phung, L. A. Chang, C. Yap, J. M. Todd, L. Pessaint, A. Van Ry, S. Browne, J. Greenhouse, T. Putman-Taylor, A. Strasbaugh, T. A. Campbell, A. Cook, A. Dodson, K. Steingrebe, W. Shi, Y. Zhang, O. M. Abiona, L. Wang, A. Pegu, E. S. Yang, K. Leung, T. Zhou, I. T. Teng, A. Widge, I. Gordon, L. Novik, R. A. Gillespie, R. J. Loomis, J. I. Moliva, G. Stewart-Jones, S. Himansu, W. P. Kong, M. C. Nason, K. M. Morabito, T. J. Ruckwardt, J. E. Ledgerwood, M. R. Gaudinski, P. D. Kwong, J. R. Mascola, A. Carfi, M. G. Lewis, R. S. Baric, A. McDermott, I. N. Moore, N. J. Sullivan, M. Roederer, R. A. Seder and B. S. Graham, N. Engl. J. Med., 2020, 383, 1544–1555 CrossRef CAS PubMed.
- F. P. Polack, S. J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. Perez Marc, E. D. Moreira, C. Zerbini, R. Bailey, K. A. Swanson, S. Roychoudhury, K. Koury, P. Li, W. V. Kalina, D. Cooper, R. W. Frenck, Jr., L. L. Hammitt, O. Tureci, H. Nell, A. Schaefer, S. Unal, D. B. Tresnan, S. Mather, P. R. Dormitzer, U. Sahin, K. U. Jansen, W. C. Gruber and C. C. T. Group, N. Engl. J. Med., 2020, 383, 2603–2615 CrossRef CAS PubMed.
- A. M. Reichmuth, M. A. Oberli, A. Jaklenec, R. Langer and D. Blankschtein, Ther. Delivery, 2016, 7, 319–334 CrossRef CAS PubMed.
- A. K. Blakney, S. Ip and A. J. Geall, Vaccines, 2021, 9, 97 CrossRef CAS PubMed.
- S. Linares-Fernandez, C. Lacroix, J. Y. Exposito and B. Verrier, Trends Mol. Med., 2020, 26, 311–323 CrossRef CAS PubMed.
- R. J. Hoey, H. Eom and J. R. Horn, Exp. Biol. Med., 2019, 244, 1568–1576 CrossRef CAS PubMed.
- S. Muyldermans, FEBS J., 2021, 288, 2084–2102 CrossRef CAS PubMed.
- G. Zhao, L. He, S. Sun, H. Qiu, W. Tai, J. Chen, J. Li, Y. Chen, Y. Guo, Y. Wang, J. Shang, K. Ji, R. Fan, E. Du, S. Jiang, F. Li, L. Du and Y. Zhou, J. Virol., 2018, 92, e00837-18 Search PubMed.
- L. He, W. Tai, J. Li, Y. Chen, Y. Gao, J. Li, S. Sun, Y. Zhou, L. Du and G. Zhao, Viruses, 2019, 11, 166 CrossRef CAS PubMed.
- G. Ye, J. Gallant, J. Zheng, C. Massey, K. Shi, W. Tai, A. Odle, M. Vickers, J. Shang, Y. Wan, L. Du, H. Aihara, S. Perlman, A. LeBeau and F. Li, eLife, 2021, 10, e64815 CrossRef PubMed.
- Y. Wan, J. Shang, S. Sun, W. Tai, J. Chen, Q. Geng, L. He, Y. Chen, J. Wu, Z. Shi, Y. Zhou, L. Du and F. Li, J. Virol., 2020, 94, e02015-19 Search PubMed.
- I. Peyron, C. Kizlik-Masson, M. D. Dubois, S. Atsou, S. Ferriere, C. V. Denis, P. J. Lenting, C. Casari and O. D. Christophe, Res. Pract. Thromb. Haemostasis, 2020, 4, 1087–1110 CrossRef CAS PubMed.
- K. T. Xenaki, B. Dorrestijn, J. A. Muns, K. Adamzek, S. Doulkeridou, H. Houthoff, S. Oliveira and P. M. van Bergen En Henegouwen, Theranostics, 2021, 11, 5525–5538 CrossRef CAS PubMed.
- L. Hanke, L. Vidakovics Perez, D. J. Sheward, H. Das, T. Schulte, A. Moliner-Morro, M. Corcoran, A. Achour, G. B. Karlsson Hedestam, B. M. Hallberg, B. Murrell and G. M. McInerney, Nat. Commun., 2020, 11, 4420 CrossRef CAS PubMed.
- Y. Xiang, S. Nambulli, Z. Xiao, H. Liu, Z. Sang, W. P. Duprex, D. Schneidman-Duhovny, C. Zhang and Y. Shi, Science, 2020, 370, 1479–1484 CrossRef CAS PubMed.
- M. Schoof, B. Faust, R. A. Saunders, S. Sangwan, V. Rezelj, N. Hoppe, M. Boone, C. B. Billesbolle, C. Puchades, C. M. Azumaya, H. T. Kratochvil, M. Zimanyi, I. Deshpande, J. Liang, S. Dickinson, H. C. Nguyen, C. M. Chio, G. E. Merz, M. C. Thompson, D. Diwanji, K. Schaefer, A. A. Anand, N. Dobzinski, B. S. Zha, C. R. Simoneau, K. Leon, K. M. White, U. S. Chio, M. Gupta, M. Jin, F. Li, Y. Liu, K. Zhang, D. Bulkley, M. Sun, A. M. Smith, A. N. Rizo, F. Moss, A. F. Brilot, S. Pourmal, R. Trenker, T. Pospiech, S. Gupta, B. Barsi-Rhyne, V. Belyy, A. W. Barile-Hill, S. Nock, Y. Liu, N. J. Krogan, C. Y. Ralston, D. L. Swaney, A. Garcia-Sastre, M. Ott, M. Vignuzzi, Q. S. B. Consortium, P. Walter and A. Manglik, Science, 2020, 370, 1473–1479 CrossRef CAS PubMed.
- T. F. Custodio, H. Das, D. J. Sheward, L. Hanke, S. Pazicky, J. Pieprzyk, M. Sorgenfrei, M. A. Schroer, A. Y. Gruzinov, C. M. Jeffries, M. A. Graewert, D. I. Svergun, N. Dobrev, K. Remans, M. A. Seeger, G. M. McInerney, B. Murrell, B. M. Hallberg and C. Low, Nat. Commun., 2020, 11, 5588 CrossRef CAS PubMed.
- H. Li, L. Guo, H. Zheng, J. Li, X. Zhao, J. Li, Y. Liang, F. Yang, Y. Zhao, J. Yang, M. Xue, Y. Zuo, J. Zhou, Y. Chen, Z. Yang, Y. Li, W. Jin, H. Shi, Z. He, Q. Li and L. Liu, Bioconjugate Chem., 2021, 32, 1034–1046 CrossRef CAS PubMed.
- P. S. Arunachalam, A. C. Walls, N. Golden, C. Atyeo, S. Fischinger, C. Li, P. Aye, M. J. Navarro, L. Lai, V. V. Edara, K. Roltgen, K. Rogers, L. Shirreff, D. E. Ferrell, S. Wrenn, D. Pettie, J. C. Kraft, M. C. Miranda, E. Kepl, C. Sydeman, N. Brunette, M. Murphy, B. Fiala, L. Carter, A. G. White, M. Trisal, C. L. Hsieh, K. Russell-Lodrigue, C. Monjure, J. Dufour, S. Spencer, L. Doyle-Meyers, R. P. Bohm, N. J. Maness, C. Roy, J. A. Plante, K. S. Plante, A. Zhu, M. J. Gorman, S. Shin, X. Shen, J. Fontenot, S. Gupta, D. T. O'Hagan, R. Van Der Most, R. Rappuoli, R. L. Coffman, D. Novack, J. S. McLellan, S. Subramaniam, D. Montefiori, S. D. Boyd, J. L. Flynn, G. Alter, F. Villinger, H. Kleanthous, J. Rappaport, M. S. Suthar, N. P. King, D. Veesler and B. Pulendran, Nature, 2021, 594, 253–258 CrossRef CAS PubMed.
- A. C. Walls, B. Fiala, A. Schafer, S. Wrenn, M. N. Pham, M. Murphy, L. V. Tse, L. Shehata, M. A. O'Connor, C. Chen, M. J. Navarro, M. C. Miranda, D. Pettie, R. Ravichandran, J. C. Kraft, C. Ogohara, A. Palser, S. Chalk, E. C. Lee, K. Guerriero, E. Kepl, C. M. Chow, C. Sydeman, E. A. Hodge, B. Brown, J. T. Fuller, K. H. Dinnon, 3rd, L. E. Gralinski, S. R. Leist, K. L. Gully, T. B. Lewis, M. Guttman, H. Y. Chu, K. K. Lee, D. H. Fuller, R. S. Baric, P. Kellam, L. Carter, M. Pepper, T. P. Sheahan, D. Veesler and N. P. King, Cell, 2020, 183, 1367–1382 CrossRef CAS PubMed.
- Y. F. Kang, C. Sun, Z. Zhuang, R. Y. Yuan, Q. Zheng, J. P. Li, P. P. Zhou, X. C. Chen, Z. Liu, X. Zhang, X. H. Yu, X. W. Kong, Q. Y. Zhu, Q. Zhong, M. Xu, N. S. Zhong, Y. X. Zeng, G. K. Feng, C. Ke, J. C. Zhao and M. S. Zeng, ACS Nano, 2021, 15, 2738–2752 CrossRef CAS PubMed.
- T. K. Tan, P. Rijal, R. Rahikainen, A. H. Keeble, L. Schimanski, S. Hussain, R. Harvey, J. W. P. Hayes, J. C. Edwards, R. K. McLean, V. Martini, M. Pedrera, N. Thakur, C. Conceicao, I. Dietrich, H. Shelton, A. Ludi, G. Wilsden, C. Browning, A. K. Zagrajek, D. Bialy, S. Bhat, P. Stevenson-Leggett, P. Hollinghurst, M. Tully, K. Moffat, C. Chiu, R. Waters, A. Gray, M. Azhar, V. Mioulet, J. Newman, A. S. Asfor, A. Burman, S. Crossley, J. A. Hammond, E. Tchilian, B. Charleston, D. Bailey, T. J. Tuthill, S. P. Graham, H. M. E. Duyvesteyn, T. Malinauskas, J. Huo, J. A. Tree, K. R. Buttigieg, R. J. Owens, M. W. Carroll, R. S. Daniels, J. W. McCauley, D. I. Stuart, K. A. Huang, M. Howarth and A. R. Townsend, Nat. Commun., 2021, 12, 542 CrossRef CAS PubMed.
- L. van Oosten, J. J. Altenburg, C. Fougeroux, C. Geertsema, F. van den End, W. A. C. Evers, A. H. Westphal, S. Lindhoud, W. van den Berg, D. C. Swarts, L. Deurhof, A. Suhrbier, T. T. Le, S. Torres Morales, S. K. Myeni, M. Kikkert, A. F. Sander, W. A. de Jongh, R. Dagil, M. A. Nielsen, A. Salanti, M. Sogaard, T. M. P. Keijzer, D. Weijers, M. H. M. Eppink, R. H. Wijffels, M. M. van Oers, D. E. Martens and G. P. Pijlman, mBio, 2021, 12, e0181321 CrossRef PubMed.
- B. Zhang, C. W. Chao, Y. Tsybovsky, O. M. Abiona, G. B. Hutchinson, J. I. Moliva, A. S. Olia, A. Pegu, E. Phung, G. B. E. Stewart-Jones, R. Verardi, L. Wang, S. Wang, A. Werner, E. S. Yang, C. Yap, T. Zhou, J. R. Mascola, N. J. Sullivan, B. S. Graham, K. S. Corbett and P. D. Kwong, Sci. Rep., 2020, 10, 18149 CrossRef CAS PubMed.
- N. Formica, R. Mallory, G. Albert, M. Robinson, J. S. Plested, I. Cho, A. Robertson, F. Dubovsky, G. M. Glenn and V. S. G. nCo, PLoS Med., 2021, 18, e1003769 CrossRef CAS PubMed.
- P. T. Heath, E. P. Galiza, D. N. Baxter, M. Boffito, D. Browne, F. Burns, D. R. Chadwick, R. Clark, C. Cosgrove, J. Galloway, A. L. Goodman, A. Heer, A. Higham, S. Iyengar, A. Jamal, C. Jeanes, P. A. Kalra, C. Kyriakidou, D. F. McAuley, A. Meyrick, A. M. Minassian, J. Minton, P. Moore, I. Munsoor, H. Nicholls, O. Osanlou, J. Packham, C. H. Pretswell, A. San Francisco Ramos, D. Saralaya, R. P. Sheridan, R. Smith, R. L. Soiza, P. A. Swift, E. C. Thomson, J. Turner, M. E. Viljoen, G. Albert, I. Cho, F. Dubovsky, G. Glenn, J. Rivers, A. Robertson, K. Smith, S. Toback and V. S. G. nCo, N. Engl. J. Med., 2021, 385, 1172–1183 CrossRef CAS PubMed.
- V. Shinde, S. Bhikha, Z. Hoosain, M. Archary, Q. Bhorat, L. Fairlie, U. Lalloo, M. S. L. Masilela, D. Moodley, S. Hanley, L. Fouche, C. Louw, M. Tameris, N. Singh, A. Goga, K. Dheda, C. Grobbelaar, G. Kruger, N. Carrim-Ganey, V. Baillie, T. de Oliveira, A. Lombard Koen, J. J. Lombaard, R. Mngqibisa, A. E. Bhorat, G. Benade, N. Lalloo, A. Pitsi, P. L. Vollgraaff, A. Luabeya, A. Esmail, F. G. Petrick, A. Oommen-Jose, S. Foulkes, K. Ahmed, A. Thombrayil, L. Fries, S. Cloney-Clark, M. Zhu, C. Bennett, G. Albert, E. Faust, J. S. Plested, A. Robertson, S. Neal, I. Cho, G. M. Glenn, F. Dubovsky, S. A. Madhi and V. S. G. nCo, N. Engl. J. Med., 2021, 384, 1899–1909 CrossRef CAS PubMed.
- D. Laczkó, M. J. Hogan, S. A. Toulmin, P. Hicks, K. Lederer, B. T. Gaudette, D. Castaño, F. Amanat, H. Muramatsu, T. H. Oguin, 3rd, A. Ojha, L. Zhang, Z. Mu, R. Parks, T. B. Manzoni, B. Roper, S. Strohmeier, I. Tombácz, L. Arwood, R. Nachbagauer, K. Karikó, J. Greenhouse, L. Pessaint, M. Porto, T. Putman-Taylor, A. Strasbaugh, T. A. Campbell, P. J. C. Lin, Y. K. Tam, G. D. Sempowski, M. Farzan, H. Choe, K. O. Saunders, B. F. Haynes, H. Andersen, L. C. Eisenlohr, D. Weissman, F. Krammer, P. Bates, D. Allman, M. Locci and N. Pardi, Immunity, 2020, 53, 724–732 CrossRef PubMed.
- K. Lederer, D. Castano, D. Gomez Atria, T. H. Oguin 3rd, S. Wang, T. B. Manzoni, H. Muramatsu, M. J. Hogan, F. Amanat, P. Cherubin, K. A. Lundgreen, Y. K. Tam, S. H. Y. Fan, L. C. Eisenlohr, I. Maillard, D. Weissman, P. Bates, F. Krammer, G. D. Sempowski, N. Pardi and M. Locci, Immunity, 2020, 53, 1281–1295 CrossRef CAS PubMed.
- Q. Huang, K. Ji, S. Tian, F. Wang, B. Huang, Z. Tong, S. Tan, J. Hao, Q. Wang, W. Tan, G. F. Gao and J. Yan, Nat. Commun., 2021, 12, 776 CrossRef CAS PubMed.
- A. B. Vogel, I. Kanevsky, Y. Che, K. A. Swanson, A. Muik, M. Vormehr, L. M. Kranz, K. C. Walzer, S. Hein, A. Güler, J. Loschko, M. S. Maddur, A. Ota-Setlik, K. Tompkins, J. Cole, B. G. Lui, T. Ziegenhals, A. Plaschke, D. Eisel, S. C. Dany, S. Fesser, S. Erbar, F. Bates, D. Schneider, B. Jesionek, B. Sänger, A. K. Wallisch, Y. Feuchter, H. Junginger, S. A. Krumm, A. P. Heinen, P. Adams-Quack, J. Schlereth, S. Schille, C. Kröner, R. de la Caridad Güimil Garcia, T. Hiller, L. Fischer, R. S. Sellers, S. Choudhary, O. Gonzalez, F. Vascotto, M. R. Gutman, J. A. Fontenot, S. Hall-Ursone, K. Brasky, M. C. Griffor, S. Han, A. A. H. Su, J. A. Lees, N. L. Nedoma, E. H. Mashalidis, P. V. Sahasrabudhe, C. Y. Tan, D. Pavliakova, G. Singh, C. Fontes-Garfias, M. Pride, I. L. Scully, T. Ciolino, J. Obregon, M. Gazi, R. Carrion Jr., K. J. Alfson, W. V. Kalina, D. Kaushal, P. Y. Shi, T. Klamp, C. Rosenbaum, A. N. Kuhn, Ö. Türeci, P. R. Dormitzer, K. U. Jansen and U. Sahin, Nature, 2021, 592, 283–289 CrossRef CAS PubMed.
- U. Elia, S. Ramishetti, R. Rosenfeld, N. Dammes, E. Bar-Haim, G. S. Naidu, E. Makdasi, Y. Yahalom-Ronen, H. Tamir, N. Paran, O. Cohen and D. Peer, ACS Nano, 2021, 15, 9627–9637 CrossRef PubMed.
- U. Elia, S. Rotem, E. Bar-Haim, S. Ramishetti, G. S. Naidu, D. Gur, M. Aftalion, M. Israeli, A. Bercovich-Kinori, R. Alcalay, E. Makdasi, T. Chitlaru, R. Rosenfeld, T. Israely, S. Melamed, I. Abutbul Ionita, D. Danino, D. Peer and O. Cohen, Nano Lett., 2021, 21, 4774–4779 CrossRef CAS PubMed.
- N. van Doremalen, R. J. Fischer, J. E. Schulz, M. G. Holbrook, B. J. Smith, J. Lovaglio, B. Petsch and V. J. Munster, Viruses, 2021, 13, 1645 CrossRef CAS PubMed.
- K. S. Corbett, D. K. Edwards, S. R. Leist, O. M. Abiona, S. Boyoglu-Barnum, R. A. Gillespie, S. Himansu, A. Schäfer, C. T. Ziwawo, A. T. DiPiazza, K. H. Dinnon, S. M. Elbashir, C. A. Shaw, A. Woods, E. J. Fritch, D. R. Martinez, K. W. Bock, M. Minai, B. M. Nagata, G. B. Hutchinson, K. Wu, C. Henry, K. Bahl, D. Garcia-Dominguez, L. Ma, I. Renzi, W. P. Kong, S. D. Schmidt, L. Wang, Y. Zhang, E. Phung, L. A. Chang, R. J. Loomis, N. E. Altaras, E. Narayanan, M. Metkar, V. Presnyak, C. Liu, M. K. Louder, W. Shi, K. Leung, E. S. Yang, A. West, K. L. Gully, L. J. Stevens, N. Wang, D. Wrapp, N. A. Doria-Rose, G. Stewart-Jones, H. Bennett, G. S. Alvarado, M. C. Nason, T. J. Ruckwardt, J. S. McLellan, M. R. Denison, J. D. Chappell, I. N. Moore, K. M. Morabito, J. R. Mascola, R. S. Baric, A. Carfi and B. S. Graham, Nature, 2020, 586, 567–571 CrossRef CAS PubMed.
- P. F. McKay, K. Hu, A. K. Blakney, K. Samnuan, J. C. Brown, R. Penn, J. Zhou, C. R. Bouton, P. Rogers, K. Polra, P. J. C. Lin, C. Barbosa, Y. K. Tam, W. S. Barclay and R. J. Shattock, Nat. Commun., 2020, 11, 3523 CrossRef PubMed.
- R. de Alwis, E. S. Gan, S. Chen, Y. S. Leong, H. C. Tan, S. L. Zhang, C. Yau, J. G. H. Low, S. Kalimuddin, D. Matsuda, E. C. Allen, P. Hartman, K. J. Park, M. Alayyoubi, H. Bhaskaran, A. Dukanovic, Y. Bao, B. Clemente, J. Vega, S. Roberts, J. A. Gonzalez, M. Sablad, R. Yelin, W. Taylor, K. Tachikawa, S. Parker, P. Karmali, J. Davis, B. M. Sullivan, S. M. Sullivan, S. G. Hughes, P. Chivukula and E. E. Ooi, Mol. Ther., 2021, 29, 1970–1983 CrossRef CAS PubMed.
- A. T. DiPiazza, S. R. Leist, O. M. Abiona, J. I. Moliva, A. Werner, M. Minai, B. M. Nagata, K. W. Bock, E. Phung, A. Schafer, K. H. Dinnon, 3rd, L. A. Chang, R. J. Loomis, S. Boyoglu-Barnum, G. S. Alvarado, N. J. Sullivan, D. K. Edwards, K. M. Morabito, J. R. Mascola, A. Carfi, K. S. Corbett, I. N. Moore, R. S. Baric, B. S. Graham and T. J. Ruckwardt, Immunity, 2021, 54, 1869–1882 CrossRef CAS PubMed.
- K. V. Kalnin, T. Plitnik, M. Kishko, J. Zhang, D. Zhang, A. Beauvais, N. G. Anosova, T. Tibbitts, J. DiNapoli, G. Ulinski, P. Piepenhagen, S. M. Cummings, D. S. Bangari, S. Ryan, P. D. Huang, J. Huleatt, D. Vincent, K. Fries, S. Karve, R. Goldman, H. Gopani, A. Dias, K. Tran, M. Zacharia, X. Gu, L. Boeglin, J. Abysalh, J. Vargas, A. Beaulieu, M. Shah, T. Jeannotte, K. Gillis, S. Chivukula, R. Swearingen, V. Landolfi, T. M. Fu, F. DeRosa and D. Casimiro, npj Vaccines, 2021, 6, 61 CrossRef CAS PubMed.
- R. R. Ji, Y. Qu, H. Zhu, Y. Yang, A. B. Vogel, U. Sahin, C. Qin and A. Hui, Vaccines, 2021, 9, 324 CrossRef CAS PubMed.
- C. Garrido, A. D. Curtis 2nd, M. Dennis, S. H. Pathak, H. Gao, D. Montefiori, M. Tomai, C. B. Fox, P. A. Kozlowski, T. Scobey, J. E. Munt, M. L. Mallory, P. T. Saha, M. G. Hudgens, L. C. Lindesmith, R. S. Baric, O. M. Abiona, B. Graham, K. S. Corbett, D. Edwards, A. Carfi, G. Fouda, K. K. A. Van Rompay, K. De Paris and S. R. Permar, Sci. Immunol., 2021, 6, eabj3684 CrossRef PubMed.
- W. Tai, L. He, X. Zhang, J. Pu, D. Voronin, S. Jiang, Y. Zhou and L. Du, Cell. Mol. Immunol., 2020, 17, 613–620 CrossRef CAS PubMed.
- J. Kim, A. Mukherjee, D. Nelson, A. Jozic and G. Sahay, Preprint bioRxiv, 2020, DOI:10.1101/2020.07.24.205583.
- L. A. Jackson, E. J. Anderson, N. G. Rouphael, P. C. Roberts, M. Makhene, R. N. Coler, M. P. McCullough, J. D. Chappell, M. R. Denison, L. J. Stevens, A. J. Pruijssers, A. McDermott, B. Flach, N. A. Doria-Rose, K. S. Corbett, K. M. Morabito, S. O'Dell, S. D. Schmidt, P. A. Swanson, 2nd, M. Padilla, J. R. Mascola, K. M. Neuzil, H. Bennett, W. Sun, E. Peters, M. Makowski, J. Albert, K. Cross, W. Buchanan, R. Pikaart-Tautges, J. E. Ledgerwood, B. S. Graham and J. H. Beigel, N. Engl. J. Med., 2020, 383, 1920–1931 CrossRef CAS PubMed.
- E. J. Anderson, N. G. Rouphael, A. T. Widge, L. A. Jackson, P. C. Roberts, M. Makhene, J. D. Chappell, M. R. Denison, L. J. Stevens, A. J. Pruijssers, A. B. McDermott, B. Flach, B. C. Lin, N. A. Doria-Rose, S. O’Dell, S. D. Schmidt, K. S. Corbett, P. A. Swanson, II, M. Padilla, K. M. Neuzil, H. Bennett, B. Leav, M. Makowski, J. Albert, K. Cross, V. V. Edara, K. Floyd, M. S. Suthar, D. R. Martinez, R. Baric, W. Buchanan, C. J. Luke, V. K. Phadke, C. A. Rostad, J. E. Ledgerwood, B. S. Graham, J. H. Beigel and mRNA-1273 Study Group, N. Engl. J. Med., 2020, 383, 2427–2438 CrossRef CAS PubMed.
- A. T. Widge, N. G. Rouphael, L. A. Jackson, E. J. Anderson, P. C. Roberts, M. Makhene, J. D. Chappell, M. R. Denison, L. J. Stevens, A. J. Pruijssers, A. B. McDermott, B. Flach, B. C. Lin, N. A. Doria-Rose, S. O’Dell, S. D. Schmidt, K. M. Neuzil, H. Bennett, B. Leav, M. Makowski, J. Albert, K. Cross, V. V. Edara, K. Floyd, M. S. Suthar, W. Buchanan, C. J. Luke, J. E. Ledgerwood, J. R. Mascola, B. S. Graham, J. H. Beigel and mRNA-1273 Study Group, N. Engl. J. Med., 2021, 384, 80–82 CrossRef CAS PubMed.
- L. R. Baden, H. M. El Sahly, B. Essink, K. Kotloff, S. Frey, R. Novak, D. Diemert, S. A. Spector, N. Rouphael, C. B. Creech, J. McGettigan, S. Khetan, N. Segall, J. Solis, A. Brosz, C. Fierro, H. Schwartz, K. Neuzil, L. Corey, P. Gilbert, H. Janes, D. Follmann, M. Marovich, J. Mascola, L. Polakowski, J. Ledgerwood, B. S. Graham, H. Bennett, R. Pajon, C. Knightly, B. Leav, W. Deng, H. Zhou, S. Han, M. Ivarsson, J. Miller, T. Zaks and C. S. Group, N. Engl. J. Med., 2021, 384, 403–416 CrossRef CAS PubMed.
- E. E. Walsh, R. W. Frenck Jr., A. R. Falsey, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, K. Neuzil, M. J. Mulligan, R. Bailey, K. A. Swanson, P. Li, K. Koury, W. Kalina, D. Cooper, C. Fontes-Garfias, P. Y. Shi, O. Tureci, K. R. Tompkins, K. E. Lyke, V. Raabe, P. R. Dormitzer, K. U. Jansen, U. Sahin and W. C. Gruber, N. Engl. J. Med., 2020, 383, 2439–2450 CrossRef CAS PubMed.
- U. Sahin, A. Muik, E. Derhovanessian, I. Vogler, L. M. Kranz, M. Vormehr, A. Baum, K. Pascal, J. Quandt, D. Maurus, S. Brachtendorf, V. Lörks, J. Sikorski, R. Hilker, D. Becker, A. K. Eller, J. Grützner, C. Boesler, C. Rosenbaum, M. C. Kühnle, U. Luxemburger, A. Kemmer-Brück, D. Langer, M. Bexon, S. Bolte, K. Karikó, T. Palanche, B. Fischer, A. Schultz, P. Y. Shi, C. Fontes-Garfias, J. L. Perez, K. A. Swanson, J. Loschko, I. L. Scully, M. Cutler, W. Kalina, C. A. Kyratsous, D. Cooper, P. R. Dormitzer, K. U. Jansen and Ö. Türeci, Nature, 2020, 586, 594–599 CrossRef CAS PubMed.
- M. J. Mulligan, K. E. Lyke, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, K. Neuzil, V. Raabe, R. Bailey, K. A. Swanson, P. Li, K. Koury, W. Kalina, D. Cooper, C. Fontes-Garfias, P. Y. Shi, O. Tureci, K. R. Tompkins, E. E. Walsh, R. Frenck, A. R. Falsey, P. R. Dormitzer, W. C. Gruber, U. Sahin and K. U. Jansen, Nature, 2020, 586, 589–593 CrossRef CAS PubMed.
- A. Riad, B. Hockova, L. Kantorova, R. Slavik, L. Spurna, A. Stebel, M. Havrilak and M. Klugar, Pharmaceuticals, 2021, 14, 873 CrossRef CAS PubMed.
- S. E. Oliver, J. W. Gargano, M. Marin, M. Wallace, K. G. Curran, M. Chamberland, N. McClung, D. Campos-Outcalt, R. L. Morgan, S. Mbaeyi, J. R. Romero, H. K. Talbot, G. M. Lee, B. P. Bell and K. Dooling, MMWR Morb. Mortal. Wkly. Rep., 2020, 69, 1922–1924 CrossRef CAS PubMed.
- K. Dooling, J. W. Gargano, D. Moulia, M. Wallace, H. G. Rosenblum, A. E. Blain, S. C. Hadler, I. D. Plumb, H. Moline, J. Gerstein, J. P. Collins, M. Godfrey, D. Campos-Outcalt, R. L. Morgan, O. Brooks, H. K. Talbot, G. M. Lee, M. F. Daley and S. E. Oliver, MMWR Morb. Mortal. Wkly. Rep., 2021, 70, 1344–1348 CrossRef CAS PubMed.
- W. H. Self, M. W. Tenforde, J. P. Rhoads, M. Gaglani, A. A. Ginde, D. J. Douin, S. M. Olson, H. K. Talbot, J. D. Casey, N. M. Mohr, A. Zepeski, T. McNeal, S. Ghamande, K. W. Gibbs, D. C. Files, D. N. Hager, A. Shehu, M. E. Prekker, H. L. Erickson, M. N. Gong, A. Mohamed, D. J. Henning, J. S. Steingrub, I. D. Peltan, S. M. Brown, E. T. Martin, A. S. Monto, A. Khan, C. L. Hough, L. W. Busse, C. C. Ten Lohuis, A. Duggal, J. G. Wilson, A. J. Gordon, N. Qadir, S. Y. Chang, C. Mallow, C. Rivas, H. M. Babcock, J. H. Kwon, M. C. Exline, N. Halasa, J. D. Chappell, A. S. Lauring, C. G. Grijalva, T. W. Rice, I. D. Jones, W. B. Stubblefield, A. Baughman, K. N. Womack, C. J. Lindsell, K. W. Hart, Y. Zhu, L. Mills, S. N. Lester, M. M. Stumpf, E. A. Naioti, M. Kobayashi, J. R. Verani, N. J. Thornburg, M. M. Patel and I. V. Y. Network, MMWR Morb. Mortal. Wkly. Rep., 2021, 70, 1337–1343 CrossRef CAS PubMed.
- D. V. Parums, Med. Sci. Monit., 2021, 27, e934625 Search PubMed.
- M. Wallace, K. R. Woodworth, J. W. Gargano, H. M. Scobie, A. E. Blain, D. Moulia, M. Chamberland, N. Reisman, S. C. Hadler, J. R. MacNeil, D. Campos-Outcalt, R. L. Morgan, M. F. Daley, J. R. Romero, H. K. Talbot, G. M. Lee, B. P. Bell and S. E. Oliver, MMWR Morb. Mortal. Wkly. Rep., 2021, 70, 749–752 CrossRef CAS PubMed.
- K. R. Woodworth, D. Moulia, J. P. Collins, S. C. Hadler, J. M. Jones, S. C. Reddy, M. Chamberland, D. Campos-Outcalt, R. L. Morgan, O. Brooks, H. K. Talbot, G. M. Lee, B. P. Bell, M. F. Daley, S. Mbaeyi, K. Dooling and S. E. Oliver, MMWR Morb. Mortal. Wkly. Rep., 2021, 70, 1579–1583 CrossRef CAS PubMed.
- S. E. Oliver, J. W. Gargano, M. Marin, M. Wallace, K. G. Curran, M. Chamberland, N. McClung, D. Campos-Outcalt, R. L. Morgan, S. Mbaeyi, J. R. Romero, H. K. Talbot, G. M. Lee, B. P. Bell and K. Dooling, MMWR Morb. Mortal. Wkly. Rep., 2021, 69, 1653–1656 CrossRef CAS PubMed.
- A. P. Wodi, K. Ault, P. Hunter, V. McNally, P. G. Szilagyi and H. Bernstein, MMWR Morb. Mortal. Wkly. Rep., 2021, 70, 189–192 CrossRef PubMed.
- A. Glatman-Freedman, Y. Hershkovitz, Z. Kaufman, R. Dichtiar, L. Keinan-Boker and M. Bromberg, Emerging Infect. Dis., 2021, 27, 2919–2922 CrossRef CAS PubMed.
- J. G. Montoya, A. E. Adams, V. Bonetti, S. Deng, N. A. Link, S. Pertsch, K. Olson, M. Li, E. C. Dillon and D. L. Frosch, Microbiol. Spectrum, 2021, e0116221 CrossRef PubMed.
- L. Coppeta, G. Somma, C. Ferrari, A. Mazza, S. Rizza, M. Trabucco Aurilio, S. Perrone, A. Magrini and A. Pietroiusti, Vaccines, 2021, 9, 947 CrossRef CAS PubMed.
- S. Gil-Manso, D. Carbonell, L. Lopez-Fernandez, I. Miguens, R. Alonso, I. Buno, P. Munoz, J. Ochando, M. Pion and R. Correa-Rocha, Front. Immunol., 2021, 12, 726960 CrossRef CAS PubMed.
- C. Rossi, P. Lanuti, I. Cicalini, D. De Bellis, L. Pierdomenico, P. Del Boccio, M. Zucchelli, L. Natale, B. Sinjari, G. Catitti, S. Vespa, P. Simeone, G. Bologna, I. Bucci, K. Falasca, J. Vecchiet, L. Stuppia, V. De Laurenzi and D. Pieragostino, Vaccines, 2021, 9, 1164 CrossRef CAS PubMed.
- S. J. Thomas, E. D. Moreira Jr., N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. Perez Marc, F. P. Polack, C. Zerbini, R. Bailey, K. A. Swanson, X. Xu, S. Roychoudhury, K. Koury, S. Bouguermouh, W. V. Kalina, D. Cooper, R. W. Frenck Jr., L. L. Hammitt, O. Tureci, H. Nell, A. Schaefer, S. Unal, Q. Yang, P. Liberator, D. B. Tresnan, S. Mather, P. R. Dormitzer, U. Sahin, W. C. Gruber, K. U. Jansen and C. C. T. Group, N. Engl. J. Med., 2021, 385, 1761–1773 CrossRef CAS PubMed.
- D. Yahav, B. Rozen-Zvi, T. Mashraki, A. Atamna, H. Ben-Zvi, E. Bar-Haim and R. Rahamimov, BMJ Open, 2021, 11, e055611 CrossRef PubMed.
- C. Ammitzboll, L. E. Bartels, J. Bogh Andersen, S. Risbol Vils, C. Elbaek Mistegard, A. Dahl Johannsen, M. L. From Hermansen, M. Kragh Thomsen, C. Erikstrup, E. M. Hauge and A. Troldborg, ACR Open Rheumatol., 2021, 3, 622–628 CrossRef PubMed.
- M. Gavriatopoulou, E. Terpos, I. Ntanasis-Stathopoulos, A. Briasoulis, S. Gumeni, P. Malandrakis, D. Fotiou, M. Migkou, F. Theodorakakou, E. Eleutherakis-Papaiakovou, N. Kanellias, E. Kastritis, I. P. Trougakos and M. A. Dimopoulos, Blood Adv., 2021, 5, 4398–4405 CrossRef CAS PubMed.
- L. Rabinowich, A. Grupper, R. Baruch, M. Ben-Yehoyada, T. Halperin, D. Turner, E. Katchman, S. Levi, I. Houri, N. Lubezky, O. Shibolet and H. Katchman, J. Hepatol., 2021, 75, 435–438 CrossRef CAS PubMed.
- C. Ferri, F. Ursini, L. Gragnani, V. Raimondo, D. Giuggioli, R. Foti, M. Caminiti, D. Olivo, G. Cuomo, M. Visentini, F. Cacciapaglia, R. Pellegrini, E. Pigatto, T. Urraro, C. Naclerio, A. Tavoni, L. Puccetti, G. Varcasia, I. Cavazzana, M. L'Andolina, P. Ruscitti, M. Vadacca, P. Gigliotti, F. La Gualana, F. Cozzi, A. Spinella, E. Visalli, Y. Dal Bosco, G. Amato, F. Masini, G. Pagano Mariano, R. Brittelli, V. Aiello, R. Caminiti, D. Scorpiniti, G. Rechichi, T. Ferrari, M. Monti, G. Elia, F. Franceschini, R. Meliconi, M. Casato, F. Iannone, R. Giacomelli, P. Fallahi, S. A. Santini, A. L. Zignego and A. Antonelli, J. Autoimmun., 2021, 125, 102744 CrossRef CAS PubMed.
- Y. Davidov, K. Tsaraf, O. Cohen-Ezra, M. Likhter, G. Ben Yakov, I. Levy, E. G. Levin, Y. Lustig, O. Mor, G. Rahav and Z. Ben Ari, Liver Transpl., 2021 DOI:10.1002/lt.26366.
- D. Cucchiari, N. Egri, M. Bodro, S. Herrera, J. Del Risco-Zevallos, J. Casals-Urquiza, F. Cofan, A. Moreno, J. Rovira, E. Banon-Maneus, M. J. Ramirez-Bajo, P. Ventura-Aguiar, A. Perez-Olmos, M. Garcia-Pascual, M. Pascal, A. Vilella, A. Trilla, J. Rios, E. Palou, M. Juan, B. Bayes and F. Diekmann, Am. J. Transplant., 2021, 21, 2727–2739 CrossRef CAS PubMed.
- S. Herrera, J. Colmenero, M. Pascal, M. Escobedo, M. A. Castel, E. Sole-Gonzalez, E. Palou, N. Egri, P. Ruiz, M. Mosquera, A. Moreno, M. Juan, A. Vilella, A. Soriano, M. Farrero and M. Bodro, Am. J. Transplant., 2021, 21, 3971–3979 CrossRef CAS PubMed.
- V. Naranbhai, C. A. Pernat, A. Gavralidis, K. J. St Denis, E. C. Lam, L. M. Spring, S. J. Isakoff, J. R. Farmer, L. Zubiri, G. S. Hobbs, J. How, A. M. Brunner, A. T. Fathi, J. L. Peterson, M. Sakhi, G. Hambelton, E. N. Denault, L. J. Mortensen, L. A. Perriello, M. N. Bruno, B. Y. Bertaux, A. R. Lawless, M. A. Jackson, E. Niehoff, C. Barabell, C. N. Nambu, E. Nakajima, T. Reinicke, C. Bowes, C. J. Berrios-Mairena, O. Ofoman, G. E. Kirkpatrick, J. C. Thierauf, K. Reynolds, H. Willers, W. G. Beltran, A. S. Dighe, R. Saff, K. Blumenthal, R. J. Sullivan, Y. B. Chen, A. Kim, A. Bardia, A. B. Balazs, A. J. Iafrate and J. F. Gainor, J. Clin. Oncol., 2021, JCO2101891, DOI:10.1200/JCO.21.01891.
- T. Buttiron Webber, N. Provinciali, M. Musso, M. Ugolini, M. Boitano, M. Clavarezza, M. D'Amico, C. Defferrari, A. Gozza, I. M. Briata, M. Magnani, F. Paciolla, N. Menghini, E. Marcenaro, R. De Palma, N. Sacchi, L. Innocenti, G. Siri, O. D'Ecclesiis, I. Cevasco, S. Gandini and A. DeCensi, Eur. J. Cancer, 2021, 159, 105–112 CrossRef CAS PubMed.
- J. Melin, M. K. Svensson, B. Albinsson, O. Winqvist and K. Pauksens, BMC Immunol., 2021, 22, 70 CrossRef CAS PubMed.
- Y. Herishanu, I. Avivi, A. Aharon, G. Shefer, S. Levi, Y. Bronstein, M. Morales, T. Ziv, Y. Shorer Arbel, L. Scarfo, E. Joffe, C. Perry and P. Ghia, Blood, 2021, 137, 3165–3173 CrossRef CAS PubMed.
- M. Amanzio, D. D. Mitsikostas, F. Giovannelli, M. Bartoli, G. E. Cipriani and W. A. Brown, Lancet Reg. Health Eur., 2021, 100253 Search PubMed.
- A. Riad, A. Pokorna, J. Klugarova, N. Antalova, L. Kantorova, M. Koscik and M. Klugar, Pharmaceuticals, 2021, 14, 1049 CrossRef CAS PubMed.
- J. A. Esquivel-Valerio, C. M. Skinner-Taylor, I. A. Moreno-Arquieta, J. A. Cardenas-de la Garza, G. Garcia-Arellano, P. L. Gonzalez-Garcia, F. D. R. Almaraz-Juarez and D. A. Galarza-Delgado, Rheumatol. Int., 2021, 41, 2105–2108 CrossRef PubMed.
- M. Klugar, A. Riad, M. Mekhemar, J. Conrad, M. Buchbender, H. P. Howaldt and S. Attia, Biology, 2021, 10, 752 CrossRef CAS PubMed.
- F. B. S. Briggs, F. J. Mateen, H. Schmidt, K. M. Currie, H. M. Siefers, S. Crouthamel, B. F. Bebo, J. Fiol, M. K. Racke, K. C. O’Connor, L. G. Kolaczkowski, P. Klein, S. Loud and R. N. McBurney, Neurol. Neuroimmunol. Neuroinflamm., 2021, 9, e1104 CrossRef PubMed.
- B. Kaplan, S. Farzan, G. Coscia, D. W. Rosenthal, A. McInerney, A. M. Jongco, P. Ponda and V. R. Bonagura, Ann. Allergy, Asthma, Immunol., 2021, S1081-1206(21)01172-8 Search PubMed.
- L. Klimek, N. Novak, E. Hamelmann, T. Werfel, M. Wagenmann, C. Taube, A. Bauer, H. Merk, U. Rabe, K. Jung, W. Schlenter, J. Ring, A. Chaker, W. Wehrmann, S. Becker, N. Mulleneisen, K. Nemat, W. Czech, H. Wrede, R. Brehler, T. Fuchs, T. Jakob, T. Ankermann, S. M. Schmidt, M. Gerstlauer, C. Vogelberg, T. Zuberbier, K. Hartmann and M. Worm, Allergo J. Int., 2021, 30, 51–55 CrossRef PubMed.
- C. C.-R. Team Food and A. Drug, MMWR Morb. Mortal. Wkly. Rep., 2021, 70, 125–129 CrossRef PubMed.
- R. Jeet Kaur, S. Dutta, J. Charan, P. Bhardwaj, A. Tandon, D. Yadav, S. Islam and M. Haque, Int. J. Gen. Med., 2021, 14, 3909–3927 CrossRef PubMed.
- Y. Nishiguchi, H. Matsuyama, K. Maeda, A. Shindo and H. Tomimoto, BMC Neurol., 2021, 21, 452 CrossRef CAS PubMed.
- R. Takeyama, K. Fukuda, Y. Kouzaki, T. Koga, S. Hayashi, H. Ohtani and T. Inoue, Acta Neurochir., 2021 DOI:10.1007/s00701-021-05038-0.
- B. Kahn, S. A. Apostolidis, V. Bhatt, A. R. Greenplate, S. Kallish, A. LaCava, A. Lucas, N. J. Meyer, D. Negoianu, A. R. Ogdie, M. G. S. Shashaty, P. A. Takach, L. Zuroff, E. J. Wherry and G. L. Anesi, Crit. Care Explor., 2021, 3, e0578 CrossRef PubMed.
- A. Dionne, F. Sperotto, S. Chamberlain, A. L. Baker, A. J. Powell, A. Prakash, D. A. Castellanos, S. F. Saleeb, S. D. de Ferranti, J. W. Newburger and K. G. Friedman, JAMA Cardiol., 2021, 6, 1446–1450 CrossRef PubMed.
- Y. Rechavi, M. Shashar, J. Lellouche, M. Yana, D. Yakubovich and N. Sharon, Vaccines, 2021, 9, 977 CrossRef CAS PubMed.
- F. Massa, M. Cremoni, A. Gerard, H. Grabsi, L. Rogier, M. Blois, C. Couzin, N. B. Hassen, M. Rouleau, S. Barbosa, E. Martinuzzi, J. Fayada, G. Bernard, G. Favre, P. Hofman, V. L. M. Esnault, C. Czerkinsky, B. Seitz-Polski, N. Glaichenhaus and A. Sicard, EBioMedicine, 2021, 73, 103679 CrossRef CAS PubMed.
- N. Barda, N. Dagan, C. Cohen, M. A. Hernan, M. Lipsitch, I. S. Kohane, B. Y. Reis and R. D. Balicer, Lancet, 2021, 398, 2093–2100 CrossRef CAS.
- J. Marlet, P. Gatault, Z. Maakaroun, H. Longuet, K. Stefic, L. Handala, S. Eymieux, E. Gyan, C. Dartigeas and C. Gaudy-Graffin, Vaccines, 2021, 9, 1055 CrossRef CAS PubMed.
- E. Schrezenmeier, H. Rincon-Arevalo, A. L. Stefanski, A. Potekhin, H. Staub-Hohenbleicher, M. Choi, F. Bachmann, V. Pross, C. Hammett, H. Schrezenmeier, C. Ludwig, B. Jahrsdorfer, A. Lino, K. U. Eckardt, K. Kotsch, T. Doerner, K. Budde, A. Sattler and F. Halleck, J. Am. Soc. Nephrol., 2021, 32, 3027–3033 CrossRef PubMed.
- Y. Saciuk, J. Kertes, N. Shamir Stein and A. Ekka Zohar, J. Infect. Dis., 2021, 225, 30–33 CrossRef PubMed.
- N. Chavarot, A. Morel, M. Leruez-Ville, E. Vilain, G. Divard, C. Burger, A. Serris, R. Sberro-Soussan, F. Martinez, L. Amrouche, L. Bererhi, F. Lanternier, C. Legendre, J. Zuber, D. Anglicheau and A. Scemla, Am. J. Transplant., 2021, 21, 4043–4051 CrossRef CAS PubMed.
- A. H. Karaba, X. Zhu, T. Liang, K. H. Wang, A. G. Rittenhouse, O. Akinde, Y. Eby, J. E. Ruff, J. N. Blankson, A. T. Abedon, J. L. Alejo, A. L. Cox, J. R. Bailey, E. A. Thompson, S. L. Klein, D. S. Warren, J. M. Garonzik-Wang, B. J. Boyarsky, I. Sitaras, A. Pekosz, D. L. Segev, A. A. R. Tobian and W. A. Werbel, Preprint medRxiv, 2021, DOI:10.1101/2021.08.11.21261914.
- R. T. Shroff, P. Chalasani, R. Wei, D. Pennington, G. Quirk, M. V. Schoenle, K. L. Peyton, J. L. Uhrlaub, T. J. Ripperger, M. Jergovic, S. Dalgai, A. Wolf, R. Whitmer, H. Hammad, A. Carrier, A. J. Scott, J. Nikolich-Zugich, M. Worobey, R. Sprissler, M. Dake, B. J. LaFleur and D. Bhattacharya, Nat. Med., 2021, 27, 2002–2011 CrossRef CAS PubMed.
- S. Shapiro Ben David, N. Shamir-Stein, S. Baruch Gez, U. Lerner, D. Rahamim-Cohen and A. Ekka Zohar, Clin. Immunol., 2021, 232, 108860 CrossRef CAS PubMed.
- A. M. Hause, J. Baggs, J. Gee, P. Marquez, T. R. Myers, T. T. Shimabukuro and D. K. Shay, MMWR Morb. Mortal. Wkly. Rep., 2021, 70, 1379–1384 CrossRef CAS PubMed.
- J. Huo, A. Le Bas, R. R. Ruza, H. M. E. Duyvesteyn, H. Mikolajek, T. Malinauskas, T. K. Tan, P. Rijal, M. Dumoux, P. N. Ward, J. Ren, D. Zhou, P. J. Harrison, M. Weckener, D. K. Clare, V. K. Vogirala, J. Radecke, L. Moynie, Y. Zhao, J. Gilbert-Jaramillo, M. L. Knight, J. A. Tree, K. R. Buttigieg, N. Coombes, M. J. Elmore, M. W. Carroll, L. Carrique, P. N. M. Shah, W. James, A. R. Townsend, D. I. Stuart, R. J. Owens and J. H. Naismith, Nat. Struct. Mol. Biol., 2020, 27, 846–854 CrossRef CAS PubMed.
- X. Chi, X. Liu, C. Wang, X. Zhang, X. Li, J. Hou, L. Ren, Q. Jin, J. Wang and W. Yang, Nat. Commun., 2020, 11, 4528 CrossRef CAS PubMed.
- K. Haga, R. Takai-Todaka, Y. Matsumura, C. Song, T. Takano, T. Tojo, A. Nagami, Y. Ishida, H. Masaki, M. Tsuchiya, T. Ebisudani, S. Sugimoto, T. Sato, H. Yasuda, K. Fukunaga, A. Sawada, N. Nemoto, K. Murata, T. Morimoto and K. Katayama, PLoS Pathog., 2021, 17, e1009542 CrossRef CAS PubMed.
- J. Huo, H. Mikolajek, A. Le Bas, J. J. Clark, P. Sharma, A. Kipar, J. Dormon, C. Norman, M. Weckener, D. K. Clare, P. J. Harrison, J. A. Tree, K. R. Buttigieg, F. J. Salguero, R. Watson, D. Knott, O. Carnell, D. Ngabo, M. J. Elmore, S. Fotheringham, A. Harding, L. Moynie, P. N. Ward, M. Dumoux, T. Prince, Y. Hall, J. A. Hiscox, A. Owen, W. James, M. W. Carroll, J. P. Stewart, J. H. Naismith and R. J. Owens, Nat. Commun., 2021, 12, 5469 CrossRef CAS PubMed.
- S. Nambulli, Y. Xiang, N. L. Tilston-Lunel, L. J. Rennick, Z. Sang, W. B. Klimstra, D. S. Reed, N. A. Crossland, Y. Shi and W. P. Duprex, Sci. Adv., 2021, 7, eabh0319 CrossRef CAS PubMed.
- P. Mlcochova, S. A. Kemp, M. S. Dhar, G. Papa, B. Meng, I. A. T. M. Ferreira, R. Datir, D. A. Collier, A. Albecka, S. Singh, R. Pandey, J. Brown, J. Zhou, N. Goonawardane, S. Mishra, C. Whittaker, T. Mellan, R. Marwal, M. Datta, S. Sengupta, K. Ponnusamy, V. S. Radhakrishnan, A. Abdullahi, O. Charles, P. Chattopadhyay, P. Devi, D. Caputo, T. Peacock, C. Wattal, N. Goel, A. Satwik, R. Vaishya, M. Agarwal, Indian SARS-CoV-2 Genomics Consortium (INSACOG), Genotype to Phenotype Japan (G2P-Japan) Consortium, CITIID-NIHR BioResource COVID-19 Collaboration, A. Mavousian, J. H. Lee, J. Bassi, C. Silacci-Fegni, C. Saliba, D. Pinto, T. Irie, I. Yoshida, W. L. Hamilton, K. Sato, S. Bhatt, S. Flaxman, L. C. James, D. Corti, L. Piccoli, W. S. Barclay, P. Rakshit, A. Agrawal and R. K. Gupta, Nature, 2021, 599, 114–119 CrossRef CAS PubMed.
- G. French, M. Hulse, D. Nguyen, K. Sobotka, K. Webster, J. Corman, B. Aboagye-Nyame, M. Dion, M. Johnson, B. Zalinger and M. Ewing, MMWR Morb. Mortal. Wkly. Rep., 2021, 70, 1613–1616 CrossRef CAS PubMed.
- B. Venkatraja, G. Srilakshminarayana and B. Krishna Kumar, Am. J. Trop. Med. Hyg., 2021, tpmd210812 Search PubMed.
- H. Allen, A. Vusirikala, J. Flannagan, K. A. Twohig, A. Zaidi, D. Chudasama, T. Lamagni and C.-G. UK, Lancet Reg. Health Eur., 2021, 100252, DOI:10.1016/j.lanepe.2021.100252.
- M. Melloul, T. Chouati, M. Hemlali, S. Alaoui Amine, N. Touil, H. Elannaz, K. Ennibi, M. Youbi, M. Merabet, A. M. Bellefquih, J. Nourlil, A. Maaroufi and E. El Fahime, Microbiol. Resour. Announce., 2021, 10, e0072721 CrossRef PubMed.
- Z. Wang, F. Schmidt, Y. Weisblum, F. Muecksch, C. O. Barnes, S. Finkin, D. Schaefer-Babajew, M. Cipolla, C. Gaebler, J. A. Lieberman, T. Y. Oliveira, Z. Yang, M. E. Abernathy, K. E. Huey-Tubman, A. Hurley, M. Turroja, K. A. West, K. Gordon, K. G. Millard, V. Ramos, J. Da Silva, J. Xu, R. A. Colbert, R. Patel, J. Dizon, C. Unson-O'Brien, I. Shimeliovich, A. Gazumyan, M. Caskey, P. J. Bjorkman, R. Casellas, T. Hatziioannou, P. D. Bieniasz and M. C. Nussenzweig, Nature, 2021, 592, 616–622 CrossRef CAS PubMed.
- W. F. Garcia-Beltran, E. C. Lam, K. St Denis, A. D. Nitido, Z. H. Garcia, B. M. Hauser, J. Feldman, M. N. Pavlovic, D. J. Gregory, M. C. Poznansky, A. Sigal, A. G. Schmidt, A. J. Iafrate, V. Naranbhai and A. B. Balazs, Cell, 2021, 184, 2372–2383 CrossRef CAS PubMed.
- A. Kuzmina, Y. Khalaila, O. Voloshin, A. Keren-Naus, L. Boehm-Cohen, Y. Raviv, Y. Shemer-Avni, E. Rosenberg and R. Taube, Cell Host Microbe, 2021, 29, 522–528 CrossRef CAS PubMed.
- D. A. Collier, A. De Marco, I. A. T. M. Ferreira, B. Meng, R. P. Datir, A. C. Walls, S. A. Kemp, J. Bassi, D. Pinto, C. Silacci-Fregni, S. Bianchi, M. A. Tortorici, J. Bowen, K. Culap, S. Jaconi, E. Cameroni, G. Snell, M. S. Pizzuto, A. F. Pellanda, C. Garzoni, A. Riva, CITIID-NIHR BioResource COVID-19 Collaboration, A. Elmer, N. Kingston, B. Graves, L. E. McCoy, K. G. C. Smith, J. R. Bradley, N. Temperton, L. Ceron-Gutierrez, G. Barcenas-Morales, COVID-19 Genomics UK (COG-UK) Consortium, W. Harvey, H. W. Virgin, A. Lanzavecchia, L. Piccoli, R. Doffinger, M. Wills, D. Veesler, D. Corti and R. K. Gupta, Nature, 2021, 593, 136–141 CrossRef CAS PubMed.
- D. Zhou, W. Dejnirattisai, P. Supasa, C. Liu, A. J. Mentzer, H. M. Ginn, Y. Zhao, H. M. E. Duyvesteyn, A. Tuekprakhon, R. Nutalai, B. Wang, G. C. Paesen, C. Lopez-Camacho, J. Slon-Campos, B. Hallis, N. Coombes, K. Bewley, S. Charlton, T. S. Walter, D. Skelly, S. F. Lumley, C. Dold, R. Levin, T. Dong, A. J. Pollard, J. C. Knight, D. Crook, T. Lambe, E. Clutterbuck, S. Bibi, A. Flaxman, M. Bittaye, S. Belij-Rammerstorfer, S. Gilbert, W. James, M. W. Carroll, P. Klenerman, E. Barnes, S. J. Dunachie, E. E. Fry, J. Mongkolsapaya, J. Ren, D. I. Stuart and G. R. Screaton, Cell, 2021, 184, 2348–2361 CrossRef CAS PubMed.
- C. Ackaert, N. Smiejkowska, C. Xavier, Y. G. J. Sterckx, S. Denies, B. Stijlemans, Y. Elkrim, N. Devoogdt, V. Caveliers, T. Lahoutte, S. Muyldermans, K. Breckpot and M. Keyaerts, Front. Immunol., 2021, 12, 632687 CrossRef CAS PubMed.
- L. Broadbent, H. G. Parke, L. J. Ferguson, A. Millar, M. D. Shields, L. Detalle and U. F. Power, Antimicrob. Agents Chemother., 2020, 64, e02034-19 CrossRef PubMed.
- D. Svecova, M. W. Lubell, F. Casset-Semanaz, H. Mackenzie, R. Grenningloh and J. G. Krueger, J. Am. Acad. Dermatol., 2019, 81, 196–203 CrossRef CAS PubMed.
- C. Morrison, Nat. Rev. Drug Discovery, 2019, 18, 485–487 CrossRef CAS PubMed.
- S. Duggan, Drugs, 2018, 78, 1639–1642 CrossRef CAS PubMed.
- S. Kimura, I. A. Khalil, Y. H. A. Elewa and H. Harashima, J. Controlled Release, 2021, 330, 753–764 CrossRef CAS PubMed.
- J. E. Francis, I. Skakic, C. Dekiwadia, R. Shukla, A. C. Taki, A. Walduck and P. M. Smooker, Vaccines, 2020, 8, 551 CrossRef CAS PubMed.
- M. Maeta, N. Miura, H. Tanaka, T. Nakamura, R. Kawanishi, Y. Nishikawa, K. Asano, M. Tanaka, S. Tamagawa, Y. Nakai, K. Tange, H. Yoshioka, H. Harashima and H. Akita, Mol. Pharm., 2020, 17, 1237–1247 CrossRef CAS PubMed.
- F. Gambino Jr., W. Tai, D. Voronin, Y. Zhang, X. Zhang, J. Shi, X. Wang, N. Wang, L. Du and L. Qiao, Cell Rep., 2021, 35, 109107 CrossRef PubMed.
- D. Huang, Y. Huang and Z. Li, Methods Mol. Biol., 2020, 2050, 101–112 CrossRef CAS PubMed.
- M. A. Shah, Z. Ali, R. Ahmad, I. Qadri, K. Fatima and N. He, J. Nanosci. Nanotechnol., 2015, 15, 41–53 CrossRef CAS PubMed.
- M. Borggren, J. Nielsen, K. Bragstad, I. Karlsson, J. S. Krog, J. A. Williams and A. Fomsgaard, Hum. Vaccines Immunother., 2015, 11, 1983–1990 CrossRef PubMed.
- D. M. Weinreich, S. Sivapalasingam, T. Norton, S. Ali, H. Gao, R. Bhore, B. J. Musser, Y. Soo, D. Rofail, J. Im, C. Perry, C. Pan, R. Hosain, A. Mahmood, J. D. Davis, K. C. Turner, A. T. Hooper, J. D. Hamilton, A. Baum, C. A. Kyratsous, Y. Kim, A. Cook, W. Kampman, A. Kohli, Y. Sachdeva, X. Graber, B. Kowal, T. DiCioccio, N. Stahl, L. Lipsich, N. Braunstein, G. Herman, G. D. Yancopoulos and I. Trial, N. Engl. J. Med., 2021, 384, 238–251 CrossRef CAS PubMed.
- P. Chen, A. Nirula, B. Heller, R. L. Gottlieb, J. Boscia, J. Morris, G. Huhn, J. Cardona, B. Mocherla, V. Stosor, I. Shawa, A. C. Adams, J. Van Naarden, K. L. Custer, L. Shen, M. Durante, G. Oakley, A. E. Schade, J. Sabo, D. R. Patel, P. Klekotka, D. M. Skovronsky and B.- Investigators, N. Engl. J. Med., 2021, 384, 229–237 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |