Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dynamic self-assembling supramolecular dendrimer nanosystems as potent antibacterial candidates against drug-resistant bacteria and biofilms†
Dinesh
Dhumal
a,
Bar
Maron
b,
Einav
Malach
b,
Zhenbin
Lyu
a,
Ling
Ding
a,
Domenico
Marson
 c,
Erik
Laurini
c,
Aura
Tintaru
a,
Brigino
Ralahy
a,
Suzanne
Giorgio
a,
Sabrina
Pricl
c,
Erik
Laurini
c,
Aura
Tintaru
a,
Brigino
Ralahy
a,
Suzanne
Giorgio
a,
Sabrina
Pricl
 cd,
Zvi
Hayouka
cd,
Zvi
Hayouka
 *b and
Ling
Peng
*b and
Ling
Peng
 *a
*a
aAix Marseille Univ, CNRS, Centre Interdisciplinaire de Nanoscience de Marseille (CINaM), UMR 7325, Equipe Labelisée Ligue Contre le Cancer, Parc Scientifique et Technologique de Luminy, Marseille, France. E-mail: ling.peng@univ-amu.fr
bInstitute of Biochemistry, Food Science and Nutrition, The Robert H. Smith Faculty of Agriculture, Food and Environment, The Hebrew University of Jerusalem, Rehovot, Israel. E-mail: zvi.hayouka@mail.huji.ac.il
cMolecular Biology and Nanotechnology Laboratory (MolBNL@UniTS), DEA, University of Trieste, Trieste, Italy
dDepartment of General Biophysics, Faculty of Biology and Environmental Protection, University of Lodz, Lodz, Poland
First published on 1st June 2022
Abstract
The alarming and prevailing antibiotic resistance crisis urgently calls for innovative “outside of the box” antibacterial agents, which can differ substantially from conventional antibiotics. In this context, we have established antibacterial candidates based on dynamic supramolecular dendrimer nanosystems self-assembled with amphiphilic dendrimers composed of a long hydrophobic alkyl chain and a small hydrophilic poly(amidoamine) dendron bearing distinct terminal functionalities. Remarkably, the amphiphilic dendrimer with amine terminals exhibited strong antibacterial activity against both Gram-positive and Gram-negative as well as drug-resistant bacteria, and prevented biofilm formation. Multidisciplinary studies combining experimental approaches and computer modelling together demonstrate that the dendrimer interacts and binds via electrostatic interactions with the bacterial membrane, where it becomes enriched and then dynamically self-assembles into supramolecular nanoassemblies for stronger and multivalent interactions. These, in turn, rapidly promote the insertion of the hydrophobic dendrimer tail into the bacterial membrane thereby inducing bacterial cell lysis and constituting powerful antibacterial activity. Our study presents a novel concept for creating nanotechnology-based antibacterial candidates via dynamic self-assembly and offers a new perspective for combatting recalcitrant bacterial infection.
Introduction
Bacterial resistance to antibiotics has become a serious global threat with ever increasing prevalence of infections difficult to treat.1–3 This pressing public health crisis has been driving the development of new antibacterial agents to overcome drug resistance. Approaches for developing antibacterial agents include, to name but a few, the modification of existing antibiotics, the identification of active agents on novel targets against resistant bacteria, and the elaboration of antimicrobial peptides with dual antibacterial and immunomodulatory activities.4–7 Among these new strategies, the development of new antibacterial agents which are substantially different from conventional antibiotics is of particular interest. In this context, amphiphilic antibacterial agents that mimic natural antimicrobial peptides and amphiphilic antibacterial detergents constitute appealing candidates.8–11 Indeed, such agents are expected to harness the antibacterial features of both antimicrobial peptides and amphiphilic antibacterial detergents, while possessing the self-assembly capacity to form supramolecular nanostructures that further enforce the antibacterial activity via cooperative and multivalent interaction.7,12 Consequently, a myriad of amphiphilic molecules has been established as antibacterial candidates.13–15In an effort to meet the urgent need for new antibacterial agents, we steered our continuing efforts to develop amphiphilic dendrimers for use in biomedical applications16 as antibacterial candidates. Dendrimers are a unique family of synthetic molecules with precisely controlled radial architecture and special multivalent cooperativity confined within a small three-dimensional volume. Different dendrimers have been studied for their antimicrobial activity.17,18 Among them, the poly(amidoamine) (PAMAM) dendrimers are of special interest thanks to their excellent biocompatibility, owing to their peptide mimicry, and resistance to enzyme degradation by virtue of their dendritic structure. In addition, they are readily available via robust synthesis and commercial sources.19,20 Therefore, PAMAM dendrimers are considered as the ideal candidates to be elaborated as mimics of antimicrobial peptides. However, the most active PAMAM dendrimers are often high-generation molecules bearing numerous charged terminals that are unfortunately associated with considerable cytotoxicity at their MIC concentrations.21,22
Recently, small amphiphilic dendrimers composed of hydrophobic entities and hydrophilic dendrons were shown to have antibacterial activity that was frequently related to low dendrimer generation, terminal charge, and hydrophilicity/hydrophobicity balance.23–32 Specifically, various peptide, poly-lysine and poly(amide) dendrimers were synthesized to improve their antimicrobial activity,23–27 whereas polyester dendrimers were explored for imparting biodegradability while reducing toxicity.30–32 Also, amphiphilic dendrimers with carbosilane entities28 and poly(aryl ether) backbones29 were studied for antibacterial activity.
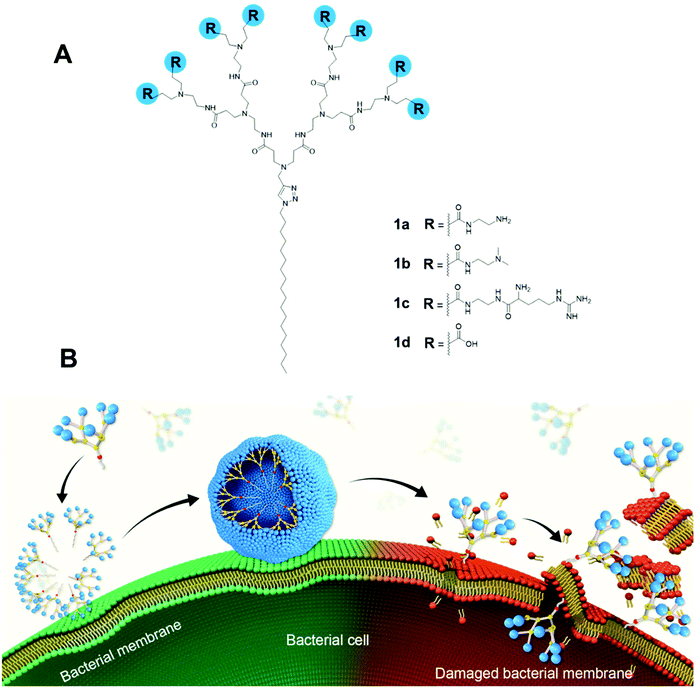
Here, we report the design, synthesis and evaluation of amphiphilic dendrimers composed of a long hydrophobic alkyl chain and a small hydrophilic PAMAM dendron carrying one of the following charged terminals: primary amine, tertiary amine, guanidine or carboxylate moieties for antibacterial activity (Fig. 1A). Importantly, we show that not only does the charge, the charge density and steric size associated with the terminal functionalities play crucial roles in the antibacterial activity, but the dynamic self-assembling feature of the amphiphilic dendrimers further enforces this antibacterial activity via membrane interaction and disruption (Fig. 1B). In particular, the amine-terminated dendrimer 1a exhibited the most potent antibacterial activity against both Gram-positive and Gram-negative bacteria as well as drug-resistant bacteria, and eradicated biofilm. Specifically, positively charged 1a dendrimers move towards and bind, via electrostatic interactions, to the negatively charged bacterial membrane, where they become enriched before dynamically self-assembling into supramolecular nanoassemblies that favor stronger and more multivalent interactions. These, in turn, rapidly promote the insertion of the hydrophobic dendrimer tail into the bacterial membrane and induce cell lysis, thereby generating powerful antibacterial activity. Here, we present our study in establishing amphiphilic dendrimer 1a as a promising antibacterial candidate, highlighting the importance of the self-assembly and surface terminal decoration of 1a in its antibacterial action.
Results and discussion
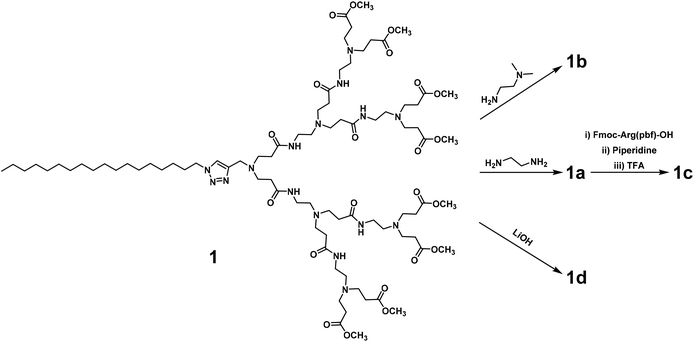
We first synthesized the amphiphilic dendrimers 1a–d using the ester-terminating dendrimer 1 as the common starting material (Scheme 1). The amidation of 1 with respectively ethylenediamine and N,N-dimethylethylenediamine, yielded 1a and 1b.33,34 Further conjugation of 1a with the protected arginine derivative followed by subsequent deprotection gave the arginine-terminated dendrimer 1c,35 whereas hydrolysis of 1 using LiOH furnished the acid-terminated dendrimer 1d. The structural integrity and purity of all the synthesized dendrimers were examined and confirmed using 1H- and 13C-NMR as well as high-resolution mass spectroscopic analysis (Fig. SA1–SA4, see ESI† Annex for details).All the dendrimers 1a–d were soluble in water, with concentrations up to 20 mM. By virtue of their amphiphilicity, these dendrimers self-assembled into small and spherical supramolecular nanomicelles ranging from 10 to 20 nm in size, as demonstrated by transmission electron microscopy (TEM) (Fig. 2, Fig. S1†). These data are in line with our previous results obtained for the amphiphilic dendrimers, which in water spontaneously form nanomicelles.34,36 In addition, NMR diffusion experiments (DOSY) were performed to estimate the average weights of the formed micelles (Fig. S2†).37 DOSY results confirmed the formation of homogeneous assemblies, which were composed of six molecules of 1a per micelle. Importantly, the nanomicelles formed by the dendrimers 1a–d had zeta potentials of +35 mV, +23 mV, +40 mV and −13 mV, respectively, the differences relating to the distinct chemical entities at their terminals. The absolute values of the zeta potentials were all over 10 mV, highlighting the colloidal stability of these nanomicelles.38
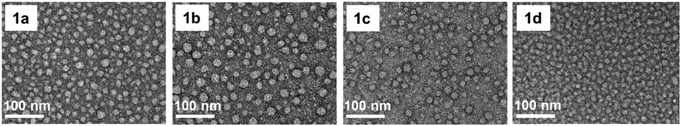 | ||
| Fig. 2 Transmission electron microscopy (TEM) images of the supramolecular nanomicelles formed by dendrimers 1a–d. | ||
To further assess the self-assembly of the amphiphilic dendrimers 1a–d, we determined their critical micelle concentration (CMC) using a fluorescence spectroscopic method based on the fluorescent dye pyrene.39 CMC is defined as the minimum concentration of an amphiphile above which micelles can form readily. 1d showed the lowest CMC value of 4.0 μM, whereas 1a and 1b had similar CMC values of 15 and 17 μM, and 1c had the highest CMC value of 35 μM (Table 1). The different CMC values for 1a–1d can reasonably be ascribed to their structural features: 1c has the largest hydrophilic entity and the greatest steric hindrance at its terminals which limit its packing and assembling ability and explain the highest CMC value; whereas dendrimer 1d has the smallest hydrophilic entity, hence the highest packing ability with the lowest CMC value.
| Dendrimer | CMCa (μM) | Particle sizeb (nm) | Zeta potential (mV) | Cytotoxicity IC50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c (μM) c (μM) |
Hemolysis IC50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) f (μM) f (μM) |
|
|---|---|---|---|---|---|---|
| L929d | HEK 293e | RBCg | ||||
| a CMC: critical micelle concentration. b Particle size: average values calculated with 300 nanoparticles measured in the TEM images. c Cytotoxicity IC50: concentration of a dendrimer required for 50% inhibition of cell growth. d L929: fibroblast cells. e HEK293: human embryonic kidney cell. f Hemolysis IC50: concentration required for lysis of 50% red blood cells. g RBC: red blood cells. | ||||||
| 1a | 15 | 14 ± 2 | +35 | >200 | 196 | 100 |
| 1b | 17 | 17 ± 4 | +23 | >200 | >200 | 100 |
| 1c | 35 | 15 ± 3 | +40 | 177 | 67 | 50 |
| 1d | 4 | 10 ± 2 | −13 | >200 | >200 | >250 |
To evaluate the safety of these dendrimers, we examined their cytotoxicity on fibroblast (L929) and kidney (HEK293) cell lines using the PrestoBlue assay. No notable adverse effect was observed for dendrimers 1a, 1b and 1d at concentrations up to 200 μM, whereas dendrimer 1c showed considerable toxicity towards both cell types (Table 1 and Fig. S3†). We also examined the hemolytic activity of these dendrimers on mouse red blood cells (RBCs). Similar to the trend observed for cytotoxicity, 1c had the highest hemolytic activity with a IC50 value of 50 μM compared to 100 μM for 1a and 1b; 1d showed no notable hemolysis, even at 250 μM (Table 1 and Fig. S4†). The observed toxicity of 1c may be ascribed to the arginine terminals, which interact and interfere strongly with the eukaryotic cell membrane via both electrostatic interaction and bivalent hydrogen bonds. Conversely, dendrimers 1a, 1b and 1d showed low cytotoxicity and good biocompatibility.
We then assessed the antibacterial activity of the dendrimers 1a–1d against the Gram-negative bacteria Escherichia coli (E. coli, K12) and Pseudomonas aeruginosa (P. aeruginosa, PAO1) as well as the Gram-positive bacteria Staphylococcus aureus (S. aureus, JLA512) and methicillin-resistant Staphylococcus aureus (MRSA, 1206). MRSA was isolated from a patient and is resistant to methicillin (MRSA), whereas PAO1 is an aerobe, mesophilic human pathogen that was isolated from infected wounds. The minimum inhibitory concentrations (MIC), i.e. the lowest concentrations required to inhibit 90% of bacteria, of these dendrimers against all the tested bacteria were determined using broth microdilution assay (Table 2). Remarkably, the amine-terminated dendrimer 1a exhibited strong antibacterial activity against both Gram-negative and Gram-positive bacteria, as well as the drug-resistant strain MRSA, with MIC values of 3.1 μM for all strains. Interestingly, the tertiary amine-terminated dendrimer 1b showed similar levels of antibacterial activity towards E. coli as 1a, but differed from 1a in that it was ineffective against all other tested bacteria; this would imply a possible narrow-spectrum antibacterial activity specific to certain bacteria. To our great surprise, the arginine-terminated dendrimer 1c, which showed the highest toxicity in eukaryotic cells, was inactive against all tested bacteria strains. The carboxylic acid-terminated dendrimer 1d was also devoid of notable antibacterial activity.
| Dendrimer | Minimum inhibition concentration (MIC) | |||||||
|---|---|---|---|---|---|---|---|---|
| Escherichia coli | Pseudomonas aeruginosa | Staphylococcus aureus | Methicillin resistant Staphylococcus aureus | |||||
| μM | μg mL−1 | μM | μg mL−1 | μM | μg mL−1 | μM | μg mL−1 | |
| MIC (minimum inhibitory concentration): lowest concentration required to inhibit 90% of bacterial growth, and value given in both μM and μg mL−1. | ||||||||
| 1a | 3.1 | 6.0 | 3.1 | 6.0 | 3.1 | 6.0 | 3.1 | 6.0 |
| 1b | 2.8 | 6.0 | 30 | 65 | 29 | 63 | 19 | 42 |
| 1c | 63 | 200 | 31 | 100 | 63 | 200 | >63 | >200 |
| 1d | 62 | 100 | >124 | >200 | 62 | 100 | 47 | 75 |
Noteworthy is the finding that the different antibacterial activities shown by the dendrimers could be ascribed to them having different terminal groups and thereby distinct interactions with bacteria. The inactivity shown by 1d can be readily explained by the repulsive forces in play between the negatively charged carboxylate terminals of the dendrimer and the negatively charged bacterial membrane. Although the dendrimers 1a, 1b and 1c all have positively charged terminals at neutral pH under physiological conditions, they showed different levels of antibacterial activity. The primary amine groups in 1a, by virtue of their small size and highly positive charge, could interact strongly with the bacterial membrane surface hence explaining the strong antibacterial activity. However, the arginine residues in 1c with the delocalized positive charges on the guanidine scaffolds, offered insufficient interaction with the bacterial cell membrane to allow any antibacterial activity. Similar observation was also reported where replacing ammonium functionalities by guanidiniums abolished antibacterial effect.32 Interestingly, dendrimer 1b bearing tertiary amine terminals, larger than the primary amine thus offering a lower charge density yet smaller than the guanidine moiety and hence providing higher charge density, generated rather variable activity toward different bacteria. This finding may highlight an adaptability of dendrimer 1b to be further exploited for developing selective and narrow-spectrum antibacterial agents.
Encouraged by the excellent antibacterial activity and safety profile of 1a, we further evaluated its activity against the biofilm formed by S. aureus.40,41 Remarkably, 1a retained its antibacterial activity with a MIC value maintained at 3.1 μM against the S. aureus biofilm (Fig. S5†), ultimately confirming the potential of 1a as a promising candidate in antibacterial treatment against drug-resistant bacteria and bacteria within biofilm matrix.
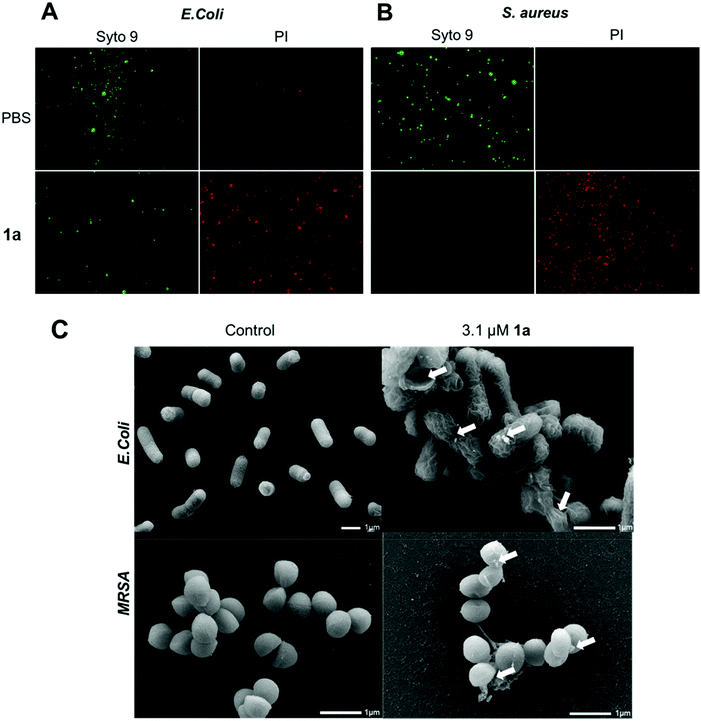
We next examined the ability of the most potent dendrimer 1a to disrupt the bacterial membrane and kill bacteria, using SYTO9 and propidium iodide (PI) dyes for staining the live and dead cells, respectively. SYTO9 is a universal dye that crosses intact membranes and stains nucleic acids of all live cells in green, whereas PI can only cross compromised bacterial membranes, and emits red fluorescence when binding to nucleic acids. The PI staining is therefore an indicator of membrane disintegration and cell death. Results from SYTO9 and PI staining (Fig. 3A/B) showed that both Gram-negative E. coli and Gram-positive S. aureus were damaged after treatment with 1a, highlighting the potent antibacterial activity of 1a, may kill bacterial cells by a mechanism most likely involving membrane damage and disruption.
To inspect and visualize the bacterial membrane integrity and morphology upon treatment with 1a, we performed scanning electron microscopic (SEM) studies. As shown in Fig. 3C and Fig. S6,† treatment with 1a induced obvious irregularities at the cell surface of both E. coli and S. aureus cells, with the appearance of blebbing and surface deformations indicating loss of membrane integrity. Also, drastic cell lysis was observed with triggered release of cellular materials. Untreated cells used as a control showed normal and smooth cell surface under SEM. These results highlight that bacterial membrane damaged by 1a may indeed contribute to its potent antibacterial activity.
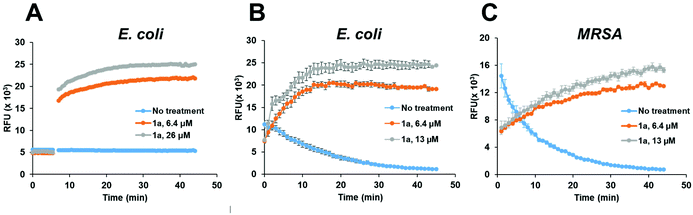
To confirm that the antibacterial mechanism of action used by 1a involved interaction with bacterial membrane, we further assessed the permeability and depolarization of the bacterial outer (OM) and inner membranes (IM). Membrane permeability was measured by monitoring the change in the fluorescent properties of the N-phenyl-1-naphthylamine (NPN) dye, a probe which displays increased fluorescence upon binding to hydrophobic membrane regions, whereas membrane depolarization was studied by recording the rapid increase in the fluorescence intensity of the 3,3′-dipropylthiadicarbocyanine iodide (diSC3(5)), a lipophilic potentiometric probe. As shown in Fig. 4, treatment with 1a led to significant outer membrane permeation in E. coli (Fig. 4A), and rapid inner membrane depolarization in both E. coli and MRSA (Fig. 4B/C). These results correlate well with the bacterial cell death observed by live/dead cell analysis and SEM imaging (Fig. 3), suggesting that the bacterial death was indeed associated with permeation and depolarization of both the outer and the inner membranes.
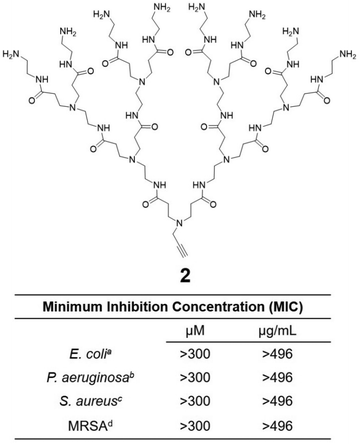
Cationic dendrimers of high generations have previously been reported to disrupt bacterial membrane by inducing pore formation via cooperative electrostatic interaction between the cationic dendrimer terminals and the anionic bacterial membrane surface.42,43 In our case however, dendrimer 1a showed strong antibacterial activity despite being of low generation. This is in strong contrast with the low-generation amine-terminating PAMAM dendrimer 2 (Fig. 5), the structural analogue of 1a but without the hydrophobic alkyl chain, which showed no activity at all against all bacteria tested in this study. Indeed, dendrimer 2 alone was insufficient to provide the strong electrostatic interaction with the bacterial membrane necessary for effective antibacterial activity, whereas the presence of the long and hydrophobic alkyl chain in 1a is clearly important for the observed strong antibacterial activity. Together the cationic terminals and the hydrophobic alkyl chain of 1a allow the formation of nanomicelles for more effective and multivalent interaction with bacterial membrane. This occurs via electrostatic interactions and the collective hydrophobic interactions between the alkyl chain of 1a and the hydrophobic region of the bacterial membrane, leading to strong inhibition of bacterial growth.
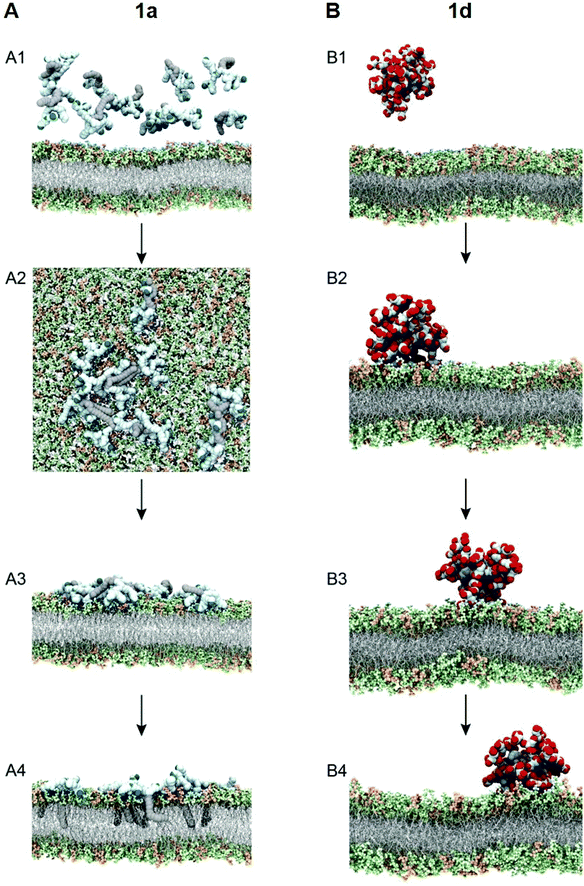
To test this notion, we investigated the interaction of 1a with bacterial membrane using molecular dynamics (MD) simulations of 1a on a bilayer model composed of mixed phospholipids representative of the E. coli membrane (Fig. 6A)44 (see ESI† for details). Starting from the dendrimer molecules randomly distributed in the water shell above the membrane (Fig. 6A1), the positively charged dendrimer molecules approach the bacterial membrane surface and accumulate on the bacterial surface owing to electrostatic interaction with the negatively charged bacterial membrane. Several clusters of small aggregates composed of 1a are detected on the bacterial surface at the end of the MD run (Fig. 6A2). The formation of aggregates shields the hydrophobic tails from water while allowing the positively-charged dendrons to bind strongly to bacterial membrane via cooperative and multivalent electrostatic interactions, as illustrated in Fig. 1B. A second round of atomistic MD simulations on these aggregates (Fig. 6A3) demonstrated complete disassembly and spreading of 1a across the upper membrane leaflet, along with full insertion of their hydrophobic tails into the bacterial membrane bilayer (Fig. 6A4). Findings of these in silico experiments support the proposed mechanistic notion of amphiphilic dendrimer molecules initially binding to and becoming enriched on the bacteria membrane via favorable electrostatic/polar interactions, before self-assembling into nanoclusters by virtue of their amphiphilicity. These clusters eventually rearrange, with the dendrimer molecules spreading along the bacterial surface allowing the long hydrophobic tails to fully insert into the lipid bilayer via collective hydrophobic interactions, hence generating strong antibacterial activity (Fig. 1B).
We also performed similar MD simulations of the least active dendrimer 1d for its interactions with the E. coli membrane model (Fig. 6B). Specifically, 1d has the lowest CMC which is also lower than its MIC value. Accordingly, 1d forms readily micelles in bulky solution (Fig. 6B1) and remains as micelles on the bacterial membrane (Fig. 6B2). These negatively charged micelles do not penetrate or induce damage to the bacterial membrane (Fig. 6B3 and B4) during the same time length of the simulation, mainly because of the repulsion generated by the negatively charged membrane. Consequently, 1d has no antibacterial activity. Collectively, these results highlight that the positive charge and the amphiphilicity alongside the dynamic self-assembling of 1a play the key role in the antibacterial activity.
Conclusion
Antibiotic-resistant bacterial infection is a pressing public health problem that is driving development of new and “outside of the box” antibacterial approaches. In this work, we have studied dynamical dendrimer nanosystems formed with amphiphilic dendrimers as potent antibacterial candidates. By virtue of their amphiphilic characteristics, these dendrimers all self-assembled spontaneously into small and stable supramolecular nanomicelles, and exhibited distinct antibacterial activity. The observed variant activity can be ascribed to the different charges and charge densities of the dendrimer terminals, which govern the interaction with the negatively charged bacterial membrane. The amine-terminated dendrimer 1a showed the most powerful activity against both Gram-positive and Gram-negative bacteria, as well as drug-resistant bacteria and allowed biofilm eradication, while having low toxicity and good safety profile. Most importantly, the self-assembling feature of the amphiphilic dendrimer 1a is responsible for and a decisive factor in its potent antibacterial activity, as the hydrophilic dendrimer counterpart without the alkyl chain showed no antibacterial activity at all. The dynamic feature of the self-assembling of 1a to form nanomicelles on the bacterial membrane surface can be further explored for nanotechnology-based delivery of antibiotics in order to impart synergic actions against recalcitrant bacterial infections. This study offers therefore a new perspective for the development of potent and effective supramolecular candidates as antibacterial nanodrugs.Author contributions
LP coordinated the project. DD, ZL and BR synthesized the agents; DD, ZL, LD, AT and SG characterized the agents; BM and EM carried out all experiments with bacteria; LD performed the toxicity studies; DM, EL, SP performed molecular simulation experiments; DD, ZL, SP, ZH and LP analyzed data; and LP wrote the paper with contributions from DD, ZH and SP. All authors proofed the manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the French National Research Agency under the framework of the ERA-NET EURONANOMED European Research projects ‘TARBRAINFECT’ (LP) and ‘antineuropatho’ (LP), the Chief Scientist office of the Israeli Ministry of Health grant number 3-17903 in the framework of EuroNanoMed 3 European Research project ‘antineuropatho’ (ZH), EU H2020 Research and Innovation program NMBP “SAFE-N-MEDTECH” (2019-2023) (grant agreement No. 814607, LP, BR), AMIDEX d'Aix-Marseille Université (LP), the Ligue Nationale Contre le Cancer (LP, ZL) and China Scholarship Council (LD). This article/publication is based upon work from COST Action CA 17140 “Cancer Nanomedicine from the Bench to the Bedside” supported by COST (European Cooperation in Science and Technology).References
- S. M. Schrader, J. Vaubourgeix and C. Nathan, Biology of antimicrobial resistance and approaches to combat it, Sci. Transl. Med., 2020, 12(549), eaaz6992, DOI:10.1126/scitranslmed.aaz6992
.
- J. M. A. Blair, M. A. Webber, A. J. Baylay, D. O. Ogbolu and L. J. V. Piddock, Molecular mechanisms of antibiotic resistance, Nat. Rev. Microbiol., 2015, 13, 42–51, DOI:10.1038/nrmicro3380
.
- A. H. Holmes, L. S. P. Moore, A. Sundsfjord, M. Steinbakk, S. Regmi and A. Karkey,
et al., Understanding the mechanisms and drivers of antimicrobial resistance, Lancet, 2016, 387, 176–187, DOI:10.1016/S0140-6736(15)00473-0
.
- Z. C. Wu and D. L. Boger, Maxamycins: Durable Antibiotics Derived by Rational Redesign of Vancomycin, Acc. Chem. Res., 2020, 53(11), 2587–2599, DOI:10.1021/acs.accounts.0c00569
.
- Z. D. Aron, A. Mehrani, E. D. Hoffer, K. L. Connolly, P. Srinivas, M. C. Torhan, J. N. Alumasa, M. Cabrera, D. Hosangadi, J. S. Barbor, S. C. Cardinale, S. M. Kwasny, L. R. Morin, M. M. Butler, T. J. Opperman, T. L. Bowlin, A. Jerse, S. M. Stagg, C. M. Dunham and K. C. Keiler, Translation inhibitors bind to a novel site on the ribosome and clear Neisseria gonorrhoeae in vivo, Nat. Commun., 2021, 12(1), 1799, DOI:10.1038/s41467-021-22012-7
.
- Y. Jiang, Y. Chen, Z. Song, Z. Tan and J. Cheng, Recent advances in design of antimicrobial peptides and polypeptides toward clinical translation, Adv. Drug Delivery Rev., 2021, 170, 261–280, DOI:10.1016/j.addr.2020.12.016
.
- J. M. V. Makabenta, A. Nabawy, C. H. Li, S. Schmidt-Malan, R. Patel and V. M. Rotello, Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections, Nat. Rev. Microbiol., 2021, 19, 23–36 CrossRef CAS PubMed
.
- N. Mookherjee, M. A. Anderson, H. P. Haagsman and D. J. Davidson, Antimicrobial host defence peptides: functions and clinical potential, Nat. Rev. Drug Discovery, 2020, 19(5), 311–332, DOI:10.1038/s41573-019-0058-8
.
- B. Findlay, G. G. Zhanel and F. Schweizer, Cationic amphiphiles, a new generation of antimicrobials inspired by the natural antimicrobial peptide scaffold, Antimicrob. Agents Chemother., 2010, 54(10), 4049–4058, DOI:10.1128/AAC.00530-10
.
- M. A. Rahman, M. Bam, E. Luat, M. S. Jui, M. S. Ganewatta, T. Shokfai, M. Nagarkatti, A. W. Decho and C. Tang, Macromolecular-clustered facial amphiphilic antimicrobials, Nat. Commun., 2018, 9(1), 5231, DOI:10.1038/s41467-018-07651-7
.
- A. G. Elliott, J. X. Huang, S. Neve, J. Zuegg, I. A. Edwards, A. K. Cain, C. J. Boinett, L. Barquist, C. V. Lundberg, J. Steen, M. S. Butler, M. Mobli, K. M. Porter, M. A. T. Blaskovich, S. Lociuro, M. Strandh and M. A. Cooper, An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria, Nat. Commun., 2020, 11(1), 3184, DOI:10.1038/s41467-020-16950-x
.
- A. R. Kirtane, M. Verma, P. Karandikar, J. Furin, R. Langer and G. Traverso, Nanotechnology approaches for global infectious diseases, Nat. Nanotechnol., 2021, 16, 369–384, DOI:10.1038/s41565-021-00866-8
.
- A. Levin, T. A. Hakala, L. Schnaider, G. J. Bernardes, E. Gazit and T. P. Knowles, Biomimetic peptide self-assembly for functional materials, Nat. Rev. Chem., 2020, 4(11), 615–634, DOI:10.1038/s41570-020-0215-y
.
- L. Schnaider, S. Brahmachari, N. W. Schmidt, B. Mensa, S. Shaham-Niv, D. Bychenko, L. Adler-Abramovich, L. J. Shimon, S. Kolusheva, W. F. DeGrado and E. Gazit, Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity, Nat. Commun., 2017, 8, 1365, DOI:10.1038/s41467-017-01447-x
.
- L. Liu, K. Xu, H. Wang, P. K. Tan, W. Fan, S. S. Venkatraman, L. Li and Y. Y. Yang, Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent, Nat. Nanotechnol., 2009, 4(7), 457–463, DOI:10.1038/nnano.2009.153
.
- Z. Lyu, L. Ding, A. Tintaru and L. Peng, Self-Assembling Supramolecular Dendrimers for Biomedical Applications: Lessons Learned from Poly(amidoamine) Dendrimers, Acc. Chem. Res., 2020, 53(12), 2936–2949, DOI:10.1021/acs.accounts.0c00589
.
- Y. Liu, L. Shi, L. Su, H. C. van der Mei, P. C. Jutte, Y. Ren and H. J. Busscher, Nanotechnology-based antimicrobials and delivery systems for biofilm-infection control, Chem. Soc. Rev., 2019, 48(2), 428–446, 10.1039/c7cs00807d
.
- M. A. Mintzer, E. L. Dane, G. A. O'Toole and M. W. Grinstaff, Exploiting dendrimer multivalency to combat emerging and re-emerging infectious diseases, Mol. Pharmaceutics, 2012, 9(3), 342–354 CrossRef CAS PubMed
.
- Z. Lyu, L. Ding, D. Dhumal, A. Y. Huang, C. L. Kao and L. Peng, Poly (amidoamine)(PAMAM) dendrimers: Synthesis and biological applications, Dendrimer Chem., 2020, 85–113, 10.1039/9781788012904-00085
.
- R. Duncan and L. Izzo, Dendrimer biocompatibility and toxicity, Adv. Drug Delivery Rev., 2005, 57(15), 2215–2237, DOI:10.1016/j.addr.2005.09.019
.
- A. Castonguay, E. Ladd, T. G. van de Ven and A. Kakkar, Dendrimers as bactericides, New J. Chem., 2012, 36, 199–204, 10.1039/C1NJ20481E
.
- K. Jain, P. Kesharwani, U. Gupta and N. K. Jain, Dendrimer toxicity: Let's meet the challenge, Int. J. Pharm., 2010, 394(1–2), 122–142, DOI:10.1016/j.ijpharm.2010.04.027
.
- M. Sowińska, A. Laskowska, A. Guśpiel, J. Solecka, M. Bochynska-Czyż, A. W. Lipkowski, K. Trzeciak and Z. Urbanczyk-Lipkowska, Bioinspired Amphiphilic Peptide Dendrimers as Specific and Effective Compounds against Drug Resistant Clinical Isolates of E. coli, Bioconjugate Chem., 2018, 29(11), 3571–3585, DOI:10.1021/acs.bioconjchem.8b00544
.
- T. N. Siriwardena, M. Stach, R. He, B. H. Gan, S. Javor, M. Heitz, L. Ma, X. Cai, P. Chen, D. Wei, H. Li, J. Ma, T. Köhler, C. van Delden, T. Darbre and J. L. Reymond, Lipidated Peptide Dendrimers Killing Multidrug-Resistant Bacteria, J. Am. Chem. Soc., 2018, 140(1), 423–432, DOI:10.1021/jacs.7b11037
.
- Z. Lai, Q. Jian, G. Li, C. Shao, Y. Zhu, X. Yuan, H. Chen and A. Shan, Self-Assembling Peptide Dendron Nanoparticles with High Stability and a Multimodal Antimicrobial Mechanism of Action, ACS Nano, 2021, 15(10), 15824–15840, DOI:10.1021/acsnano.1c03301
.
- M. Gide, A. Nimmagadda, M. Su, M. Wang, P. Teng, C. Li, R. Gao, H. Xu, Q. Li and J. Cai, Nano-Sized Lipidated Dendrimers as Potent and Broad-Spectrum Antibacterial Agents, Macromol. Rapid Commun., 2018, 39(24), e1800622, DOI:10.1002/marc.201800622
.
- W. Guo, Y. Wang, P. Wan, H. Wang, L. Chen, S. Zhang, C. Xiao and X. Chen, Cationic amphiphilic dendrons with effective antibacterial performance, J. Mater. Chem. B, 2022, 10, 456–467, 10.1039/D1TB02037D
.
- J. Fernandez, G. Acosta, D. Pulido, M. Malý, J. L. Copa-Patiño, J. Soliveri, M. Royo, R. Gómez, F. Albericio, P. Ortega and F. J. de la Mata, Carbosilane Dendron-Peptide Nanoconjugates as Antimicrobial Agents, Mol. Pharmaceutics, 2019, 16(6), 2661–2674, DOI:10.1021/acs.molpharmaceut.9b00222
.
- R. Kannan, P. Prabakaran, R. Basu, C. Pindi, S. Senapati, V. Muthuvijayan and E. Prasad, Mechanistic study on the antibacterial activity of self-assembled poly(aryl ether)-based amphiphilic dendrimers, ACS Appl. Bio Mater., 2019, 2, 3212–3224, DOI:10.1021/acsabm.9b00140
.
- S. R. Meyers, F. S. Juhn, A. P. Griset, N. R. Luman and M. W. Grinstaff, Anionic amphiphilic dendrimers as antibacterial agents, J. Am. Chem. Soc., 2008, 130, 14444–14445, DOI:10.1021/ja806912a
.
- A. Chen, A. Karanastasis, K. R. Casey, M. Necelis, B. R. Carone, G. A. Caputo and E. F. Palermo, Cationic Molecular Umbrellas as Antibacterial Agents with Remarkable Cell-Type Selectivity, ACS Appl. Mater. Interfaces, 2020, 12(19), 21270–21282, DOI:10.1021/acsami.9b19076
.
- A. Chen, E. Chen and E. F. Palermo, Guanidium-functionalized cationic molecular umbrellas as antibacterial agents, Polym. Chem., 2021, 12, 2374–2378, 10.1039/D1PY00071C
.
- T. Yu, X. Liu, A. L. Bolcato-Bellemin, Y. Wang, C. Liu, P. Erbacher, F. Qu, P. Rocchi, J. P. Behr and L. Peng, An amphiphilic dendrimer for effective delivery of small interfering RNA and gene silencing in vitro and in vivo, Angew. Chem., Int. Ed., 2012, 51(34), 8478–8484, DOI:10.1002/anie.201203920
.
- D. Dhumal, W. Lan, L. Ding, Y. Jiang, Z. Lyu, E. Laurini, D. Marson, A. Tintaru, N. Dusetti, S. Giorgio, J. Iovanna, S. Pricl and L. Peng, An ionizable supramolecular dendrimer nanosystem for effective siRNA delivery with a favorable safety profile, Nano Res., 2021, 14, 2247–2254, DOI:10.1007/s12274-020-3216-8
.
- X. Liu, C. Liu, J. Zhou, C. Chen, F. Qu, J. J. Rossi, P. Rocchi and L. Peng, Promoting siRNA delivery via enhanced cellular uptake using an arginine-decorated amphiphilic dendrimer, Nanoscale, 2015, 7(9), 3867–3875, 10.1039/c4nr04759a
.
- C. Chen, P. Posocco, X. Liu, Q. Cheng, E. Laurini, J. Zhou, C. Liu, Y. Wang, J. Tang, V. D. Col, T. Yu, S. Giorgio, M. Fermeglia, F. Qu, Z. Liang, J. J. Rossi, M. Liu, P. Rocchi, S. Pricl and L. Peng, Mastering Dendrimer Self-Assembly for Efficient siRNA Delivery: From Conceptual Design to In Vivo Efficient Gene Silencing, Small, 2016, 12(27), 3667–3676, DOI:10.1002/smll.201503866
.
- X. Liu, J. Wu, M. Yammine, J. Zhou, P. Posocco, S. Viel, C. Liu, F. Ziarelli, M. Fermeglia, S. Pricl, G. Victorero, C. Nguyen, P. Erbacher, J. P. Behr and L. Peng, Structurally flexible triethanolamine core PAMAM dendrimers are effective nanovectors for DNA transfection in vitro and in vivo to the mouse thymus, Bioconjugate Chem., 2011, 22(12), 2461–2473, DOI:10.1021/bc200275g
.
- S. Bhattacharjee, DLS and zeta potential–what they are and what they are not?, J. Controlled Release, 2016, 235, 337–351, DOI:10.1016/j.jconrel.2016.06.017
.
- J. Aguiar, P. Carpena, J. A. Molina-Bolıvar and C. C. Ruiz, On the determination of the critical micelle concentration by the pyrene 1:3 ratio method, J. Colloid Interface Sci., 2003, 258(1), 116–122, DOI:10.1016/S0021-9797(02)00082-6
.
- S. P. Hawser and L. J. Douglas, Biofilm formation by Candida species on the surface of catheter materials in vitro, Infect. Immun., 1994, 62(3), 915–921, DOI:10.1128/iai.62.3.915-921.1994
.
- E. A. Trafny, R. Lewandowski, I. Zawistowska-Marciniak and M. Stępińska, Use of MTT assay for determination of the biofilm formation capacity of microorganisms in metalworking fluids, World J. Microbiol. Biotechnol., 2013, 29(9), 1635–1643, DOI:10.1007/s11274-013-1326-0
.
- A. M. Holmes, J. R. Heylings, K. W. Wan and G. P. Moss, Antimicrobial efficacy and mechanism of action of poly (amidoamine)(PAMAM) dendrimers against opportunistic pathogens, Int. J. Antimicrob. Agents, 2019, 53(4), 500–507 CrossRef CAS PubMed
.
- M. A. Mintzer, E. L. Dane, G. A. O'Toole and M. W. Grinstaff, Exploiting dendrimer multivalency to combat emerging and re-emerging infectious diseases, Mol. Pharm., 2012, 9(3), 342–354, DOI:10.1021/mp2005033
.
- K. Murzyn, T. Róg and M. Pasenkiewicz-Gierula, Phosphatidylethanolamine-phosphatidylglycerol bilayer as a model of the inner bacterial membrane, Biophys. J., 2005, 88(2), 1091–1103, DOI:10.1529/biophysj.104.048835
.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S6 as well as all detailed experimental methods and protocols. See DOI: https://doi.org/10.1039/d2nr02305a |
| This journal is © The Royal Society of Chemistry 2022 |