Open Access Article
Open Access ArticleAntibiotic resistance in urban stormwater: a review of the dissemination of resistance elements, their impact, and management opportunities
Kassidy
O'Malley
 ,
Walter
McDonald
and
Patrick
McNamara
,
Walter
McDonald
and
Patrick
McNamara
 *
*
Department of Civil, Construction and Environmental Engineering, Marquette University, 1637 W Wisconsin Ave, Milwaukee, WI 53233, USA
First published on 3rd August 2023
Abstract
The public health crisis of antibiotic resistance is a growing threat across the world that is only expected to intensify in the coming years. The cycling of antibiotic resistance genes (ARGs) in the environment via urban stormwater runoff is one means by which humans are exposed to resistant bacteria as traditional gray stormwater infrastructure facilitates the transport of resistance elements into water bodies utilized by the public. In this review, existing research on the occurrence of ARGs in urban stormwater runoff is critically reviewed with the goal of determining the role of stormwater in the dissemination and development of antibiotic resistance. From the research conducted, urban stormwater is concluded to be hotspot for antibiotic resistance due to the confluence of factors that contribute to an increase in the frequency of gene transfer including ARGs, antibiotic resistant bacteria, mobile genetic elements, and selective pressures such as metals. The long-term impact of ARG dissemination on downstream environments such as surface water bodies and green stormwater infrastructure (GSI) systems is also reviewed. GSI has the potential for ARG management within the urban water cycle while also presenting a threat for antibiotic resistance proliferation after collecting and possibly concentrating ARGs. More research is needed to understand how engineering decisions can be incorporated into GSI design to maximize the removal of ARGs from stormwater runoff while also minimizing ARG propagation and mobilization.
Water impactStormwater could be playing a crucial role in propagating and disseminating antibiotic resistance in the urban water cycle. Green stormwater infrastructure systems could serve as a hotspot for antibiotic resistance, or, pending their design, an opportunity to mitigate transport of antibiotic resistance genes into the urban environment. |
1. Introduction
1.1 The public health crisis of antibiotic resistance
The discovery and development of antibiotics in 1929 was revolutionary for public health. Bacterial infections that frequently led to illness and death became easily treatable.1 Now, such advancements are threatened by antibiotic resistance – a defense mechanism bacteria can acquire or develop to resist the lethal effects of antibiotics. As a result, there has been a severe increase in infections caused by antibiotic-resistant bacteria. In 2019 alone, 1.27 million deaths were caused by bacterial resistant infections, making antibiotic resistant infections the leading cause of death around the world that year.2 The consequences of antibiotic resistance also extend to the economy as the estimated cost in the United States was $55 billion per year through associated medical bills and loss in productivity.3 If actions are not taken to address this threat, by 2050 it is estimated that 10 million deaths per year globally will stem from antibiotic resistant infections and be associated with a total GDP loss of $100.2 trillion.4,5Antibiotic resistance is a natural phenomenon that predates the use of antibiotics by humans and animals.6 Resistance can be intrinsic to bacterial cells by ways of structural components, such as an impermeable outer membrane, and other chromosomally encoded elements (e.g., antibiotic inactivating enzymes and transposon insertion libraries).7,8 Resistance can also be acquired through a mutation in the chromosome or through horizontal gene transfer (HGT) of antibiotic resistance genes (ARGs).9 ARGs are the genetic material that provide bacteria the potential for resistance. Despite the natural occurrence of antibiotic resistance in the environment, the increased threat to public health seen today is due to a substantial increase in the rate of acquired resistance.10
One of the primary drivers of the increase in the acquisition of ARGs is the overuse and misuse of antibiotics in the human health and agrarian sectors. In the United States roughly 47 million prescriptions are unnecessarily prescribed to people every year.11 Moreover, the use of antibiotics in animals for growth promotion and prophylactic practices has led to the sale of antibiotics within the agricultural sector being four times that of which is sold to use in humans.12 Using medically important antibiotics in agriculture, moreover, has resulted in a causal relationship between antibiotic use in the agricultural sector and the exacerbation of antibiotic resistance in the clinic.13 The misuse and overuse of antibiotics, as well as improper disposal of antibiotics, has exacerbated this public health crisis because antibiotics exert selective pressures for antibiotic resistance, meaning that antibiotics can increase antibiotic resistance in a bacterial community.14,15 This selective pressure can occur in multiple ways. For instance, if a bacterial community is exposed to antibiotics, the antibiotics will inactivate or kill the susceptible bacteria, leaving the community dominated by antibiotic resistant bacteria (ARB).16 Additionally, the exposure of a bacterial community to antibiotics can trigger adaptive responses including the HGT of ARGs.17 This selective action for antibiotic resistance can also be driven by other chemical contaminants, such as many antimicrobials, heavy metals, disinfectants, pesticides, and biocides.18–20 As a result, an emerging concern in the fight against antibiotic resistance is the occurrence and possible proliferation of antibiotic resistance in environments plagued with these different contaminants and the subsequent connections and implications for human health.
1.2 The environmental occurrence of antibiotic resistance
To determine the role of the environment as a conduit of antibiotic resistance it is crucial to monitor present risks and classify future concerns. Several ARGs today though, are considered ubiquitous in the environment and are an inherent component of many bacterial communities, serving functions unrelated to antibiotics, and thus not posing an elevated risk to human health.21 So, to identify environments that warrant monitoring efforts, one approach is to classify environmental hotspots.22 Environmental hotspots can be classified by their ability to foster ARG dissemination and proliferation via HGT. Three HGT mechanisms have been identified: (1) conjugation, (2) transduction, and (3) transformation.23,24 Conjugation is gene transfer by cell-to-cell connection, transduction is transfer facilitated by bacteriophages, and transformation is the uptake and integration of extracellular DNA (eDNA).25 Water resource recovery facilities (WRRF) are an example of environmental hotspots for HGT due to the mixing of commensal and pathogenic bacteria with diverse waste streams containing ARGs, ARB, and selective pressures (e.g., antibiotics and metals). The frequency of ARG transfer via conjugation in activated sludge tanks has been shown to range between 10−2–10−7 transfers per recipient cell.26 The agriculture sector is another hotspot for ARGs dissemination with the rate of conjugation for ARGs in manure ranging from 10−3–10−8 transfers per recipient cell.26,27 Agriculture, moreover, has a direct link to public health as antibiotic resistant pathogens originating from food-animal production have been linked with foodborne outbreaks.28,29Additional considerations for characterizing environmental hotspots are the abundance of selective pressures and mobile genetic elements (MGEs). The hotspots discussed above, WRRF and the agricultural sector, are heavily polluted with selecting agents and thus can exert a pressure on microbial communities that select for ARB.27 In agriculture, for instance, the heavy metals copper, zinc, and mercury were correlated with the abundance of the ARGs sul1 and sulIII.30 Selective pressures can occur via co-resistance or cross-resistance. Co-resistance occurs when multiple resistance genes are acquired through the same genetic element (e.g., integrons gene cassettes) while cross-resistance is the result of a genetic determinant or biochemical systems (e.g., efflux pump) conferring resistance to multiple compounds.31 In either case, an increase in the abundance of selecting agents accelerates HGT, thereby diversifying the collection of resistance elements (e.g., the resistome) and increasing the risk of ARG dissemination to human pathogens.31,32
The mobility of ARGs is also known to impact its rate of transfer into bacterial cells. ARGs that are not associated with MGEs are not easily transferred between bacteria in the environment, consequently they are less likely to be transferred to clinically relevant species and are less of a concern for public health.33 MGEs, such as transposons, plasmids, integrons, gene cassettes, and bacteriophages, can carry a large number of ARGs and are able to move themselves within a cell and transfer to a recipient cell freely.34 Thus, the transfer of ARGs via MGE is found to be much more frequent and can enrich host cells with multidrug resistance.33,35,36
1.3 Urban Stormwater Runoff: An Emerging Area of Concern for Antibiotic Resistance
Stormwater runoff can be the cause of significant water quality impairments and lead to negative impacts for human and environmental health.37,38 In urban environments specifically, impervious surfaces are the primary exasperator of this impairment, as stormwater that has fallen on impervious surfaces, such as roofs, roads, sidewalks, and parking lots, can mobilize the pollutants found there.39 Stormwater can thus be a source and means of transport of diverse contaminants originating from anthropogenic activities into downstream environments. Numerous chemical contaminants, including heavy metals, polycyclic aromatic hydrocarbons, pesticides, and other organics, have been detected in stormwater as well as microbial contaminants, which are of notable concern for human health.40–42 Rain and snowmelt events play a role in the transport of bacteria, viruses, and protozoan oocysts and consequently, stormwater has also been linked to waterborne disease outbreaks through drinking water contamination.43 In an epidemiological study, it was found that people who swam in front of a stormwater discharge point were 50% more likely to develop broad range symptoms (i.e., upper respiratory and gastrointestinal) than those who swam further from the same discharge point.44 The distance from the drain was also found to be associated with the amount of bacterial indicator organisms and enteric pathogens. In numerous watersheds, the increased presence of pathogenic bacteria has been linked to rainfall events by ways of leaky sewer systems and septic tanks, combined sewer overflows (CSOs), agricultural runoff, and domestic animal waste.42 The introduction of bacterial populations of human, animal, and environmental origin paired with the various chemical selecting agents, makes stormwater a likely hotspot for antibiotic resistance.Urban stormwater runoff presents the unique risk of not only being a potential hotspot for the development of ARB but also being a dissemination route for ARGs across an urban watershed and into water bodies that humans directly interact with through fishing, recreation, or as a water supply. Stormwater has been found to be a source of ARGs; as stormwater travels over urban surfaces, it collects ARGs from diverse landscapes, such as streets, parking lots, urban green spaces, and stormwater sewers.45 In addition, stormwater can be a vehicle for selecting agents, such as pesticides and heavy metals, and MGEs.46 The confluence of resistance elements from different urban niches generates a unique resistance profile that, once disseminated into the environments that receive stormwater, has the potential to accumulate and proliferate. The resistome composition of stormwater and the downstream implications for the urban environment, however, has remained relatively unexplored in comparison to other environmental hotspots.47
1.4 Review Objectives
In this review, existing information on the occurrence of ARGs in urban stormwater runoff is critically reviewed with the goal of determining the role stormwater plays in the dissemination and development of ARGs, while also outlining future research and monitoring needs. The abundance and diversity of resistance elements in direct stormwater runoff will be summarized, with a focus on ARGs, MGEs, and selective pressures. Within this review a distinction is made between intracellular ARGs (iARGs) and extracellular ARGs (eARGs) and a discussion is provided on eARGs as an overlooked fraction of ARGs. The fate of resistance elements following transport into downstream environments is also presented to elucidate the long-term impacts of ARG contamination originating from stormwater. Green stormwater infrastructure (GSI) systems will be highlighted because they present the opportunity to manage the environmental occurrence of ARGs. The potential of GSI to capture and mitigate downstream transport of ARGs is critically reviewed in comparison to potential consequences of ARG accumulation in GSI soils. This review will ultimately introduce recommendations for future research directions with a focus on improving mitigation strategies for antibiotic resistance through urban stormwater runoff management.2. Antibiotic resistance in urban stormwater runoff
2.1 Resistance contaminants in direct stormwater runoff and stormwater outfalls
Stormwater is a transient and diffuse source of ARGs, making its characterization difficult. As a result, only a few studies have sampled ARGs in stormwater as direct runoff from urban surfaces (Table 1).48–50 Primarily, previous research has sought to describe the resistance elements in stormwater runoff by sampling at stormwater outfalls, where runoff from a larger urban area is piped and discharged into a water body (e.g., lake, river, reservoir) at one central point (Table 1).48–61 The stormwater disseminated from outfalls is mostly runoff collected via overland flow, but stormwater can include sewage inputs as it travels through a storm sewer system.56 The presence of sewage in stormwater can significantly alter the ARGs present and their concentration at an outfall.52,61 Primarily, sewage contamination is introduced into storm sewers through sanitary sewer cross-contamination or CSOs and is a sign of aging infrastructure.58 Stormwater contaminated with sewage of human origins is of great concern for the dissemination of ARGs. Wastewater, as discussed, is a known hotspot for antibiotic resistance proliferation due to the confluences of selective pressures and elevated concentrations of ARGs. The direct input of raw wastewater into the environment via stormwater has the potential to exacerbate the proliferation of resistance in downstream environments. Consequently, sources of sewage contaminants are an important consideration when characterizing the resistance elements at a stormwater outfall. The studies summarized in Table 1 specify whether the outfalls sampled were a part of a combined or separate sewer system. Such information indicates if sewage contamination is likely to be present, but it was rare for sewage inputs to be confirmed. Without confirmation it is challenging to determine if stormwater poses a risk to human health due to the ARGs and ARB mobilized in stormwater as it runs off urban surfaces or if the risk is due to the presence of sewage following transport through a storm sewer. Breaking down this distinction will indicate where management actions will be most effective.53 Research suggested that street cleaning efforts would limit the ARG contamination coming from stormwater outfalls because the ARGs were found to originate predominantly from urban surfaces and were not the result of sewage contamination. In the case though that sewage is introducing ARG contamination, updates to infrastructure systems would be more appropriate.53| Study Design | Analytical Methods | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study location | Number of sampling locations | Sampling location description | Sample collection description | Number of storm events | Sampled on temporal scale? | ARB analyzed | ARGs analyzed | ARG analysis method | Sequencing analysis | Selective pressures and other notable aspects analyzed | Mobile genetic elements | Citation |
| Tampa Bay, FL., USA | 12 | Separate storm sewer outfalls | Grab samples | 1 | No | NA | aadA5, acrD, ampC, blaKPC, blaNPS, blaOXA, blaSHV, cmlB, ereB, imp13, mefE, merA, mexB, nikA, qacF, qnrA, qnrB, rcnA, strB, sul1, sul2, sul3, tetA, tetL, tetM, tetS, tetW, tetX | Microfluidic qPCR | NA | Heavy metal resistance genes cadA, cadB8, chrA, copA | intl1, intl2, intl3 | 52 |
| Lincoln, NE., USA | 3 | Separate storm sewer outfalls | Grab samples | 2 per outfall; 4 events in total | Yes | NA | NA | NA | Whole genome shotgun metagenomic sequencing | NA | NA | 53 |
| Stockholm, Sweden | 5 | Separate storm sewer manholes | Grab samples | 6 | Yes | NA | NA | NA | Oxford Nanopore MinION sequencing | NA | Phage and plasmid identification | 57 |
| Natal, Rio Grande do Norte, Brazil | 3 | Separate storm and combined sewer outfalls | Grab samples | 9 | Yes | E. coli [ampicillin, cephalothin, chloramphenicol, nalidixic acid, nitrofurantoin, sulfamethoxazole, and tetracycline] | NA | NA | NA | E. coli [copper, zinc, and mercury] | NA | 51 |
| Sydney, New South Wales, Australia | 4 | Stormwater drains | Grab samples | 1 | No | All isolates [meropenem]; Rheinheimera and Variovorax isolates [meropenem, penicillin G, cefoxitin, kanamycin] | NA | NA | 16S rRNA amplicon sequencing | NA | Plasmid and transposon identification | 54 |
| New York City, NY., USA | 1 | Combined sewer outfall | Storm 1, two time points; storm 2 and 3, samples every 15–30 minutes | 3 | Yes | NA | sul1, tetG | qPCR | NA | NA | NA | 58 |
| Columbus, OH., USA | 4 | Separate storm sewer outfalls | Composite sample | 5 | Yes | NA | sul1, tetQ, KPC | ddPCR | Whole genome shotgun metagenomic sequencing (for only 1 storm event) | NA | MGE annotation | 55 |
| Munich, Germany | 2 | Stormwater quality improvement devices | Influent composite samples | 21 | Yes | NA | NA | NA | 16S rRNA amplicon sequencing and MinION sequencing | Chromium, copper, nickel, lead, zinc quantification; metal resistance gene annotation | intI1 quantification and inti1 gene cassette sequencing | 50 |
| Milwaukee, WI., USA | 1 | Combined sewer outfall | Grab samples | 4 | Yes | E. coli [ampicillin, chlortetracycline, kanamycin, nalidixic acid, neomycin, oxytetracycline, penicillin, streptomycin, sulfathiazole, tetracycline, and total resistance] | NA | NA | NA | NA | NA | 59 |
| Milwaukee, WI., USA | 2 | Separate storm sewer outfall and curbside storm drain | Grab samples | 3 | Yes | NA | blaTEM, ermF, sul1, tetC | ddPCR | NA | Lead, cadmium, arsenic, chromium, nickel, copper, zinc, iron quantification | intI1 quantification | 48 |
| Langfang and Beijing, China | 5 | Ground runoff and combined sewer outfall | Composite sample | 1 | No | NA | NA | NA | Whole genome shotgun metagenomic sequencing | NA | NA | 60 |
| Toronto, Ontario, Canada | 7 | Combined sewer outfall | Grab samples | 9–14 (outfall dependent) | Yes | E. coli [ampicillin] | NA | NA | NA | NA | NA | 56 |
| Central Coast, New South Wales, Australia | 3 | Separate storm sewer outfalls | Grab samples | 1-3 (outfall dependent) | Yes | NA | sul1, tetA, qnrS, dfrA1, vanB | qPCR | 16S rRNA amplicon sequencing | NA | intI1 quantification | 61 |
| Nanjing, Jiangsu, China | 3 | Urban underlying surface (i.e., curbside storm drain) | Grab samples | 11 | Yes | Heterotrophic plate counting [tetracycline, ampicillin, and sulfadiazine] | tetA-01, tetC-01, tetO-01, tetQ, tetW, tetM-01, blaTEM, blaOXA1/blaOXA30, blaSHV-01, blaSHV-02, sul1, sul2, sul3, ermB, ermC, ermF, qnrA | SmartChip Real-Time PCR | 16S rRNA amplicon sequencing | NA | intl1, intl2, trbC, tnpA-01 | 49 |
Without the dominant presence of sewage pollution, the composition and concentration of resistance elements at outfalls has been found to be dependent on the drainage area characteristics. Source tracking analysis of the sources of ARGs to a stormwater outfall revealed that impervious surfaces contributed 92–96% of the ARGs, indicating that the impervious nature of this outfall's drainage area is enabling stormwater to be a vector of ARG dissemination.53 Moreover, ARG concentrations were found to differ when four outfalls were comparatively analyzed across four different sewersheds that varied in land use, size, and imperviousness.55 In addition to watershed dynamics, hydrologic factors such as the antecedent dry weather period, rainfall intensity, and rainfall duration are important concerns for the profile of resistance elements observed at stormwater outfalls. Specifically, long antecedent dry weather periods enable the buildup of contaminants on impervious surfaces that are able to be picked up and transported by stormwater,48 while short, intense storms are able to mobilize a higher concentration of ARGs.55,58 Such research exemplifies the dynamic nature of ARGs in runoff collected from urban environments, in addition to highlighting the number of factors that must be considered when designing a study for sampling stormwater. The research papers summarized in Table 1 primarily did not report all the factors identified here as influencing resistance elements at outfalls. Moving forward it is critical that sewage contamination, drainage area characteristics, and hydrologic conditions preceding and during sampling are viewed as requirements in the conceptualization and reporting of antibiotic resistance in stormwater. This information will provide the appropriate information for characterizing the resistance elements in stormwater.
From other studies that reported variable ARG concentrations in stormwater runoff, the microbial community composition as well as the time of sampling were proposed as additional explanatory factors.58,61 For the time of sampling, specifically, there is a scarcity of data presenting ARG concentrations over the duration of a storm event at stormwater outfalls. Only one study presented data from two storms sampled in which ARG concentrations were quantified from samples collected every 15–30 minutes.58 The ARGs quantified displayed intra- and inter-storm variability with inconsistent timing of peak relative concentrations. In neither case did peak ARG concentration correspond with peak rainfall or microbial community signatures. While it is not clear from this study what dictates the timing of peak ARG concentrations, it is apparent that collecting one grab sample is not sufficient in characterizing the dynamics of ARG concentrations originating from stormwater outfalls. To identify the risk associated with the ARGs disseminated via stormwater, periods of peak ARG loading should be targeted in sampling, consequently time-series sampling of stormwater is necessary and further work should be dedicated to understanding how ARG contaminants partition across storm events.
What has been confirmed across studies is that stormwater contains a diverse resistome, with multidrug resistance (MDR) being the most common classification for the ARGs.53,57,60 MDR is a major concern for the public health crisis of antibiotic resistance because it indicates that the bacterium is able to confer resistance to more than one antibiotic. MDR can thus limit treatment options for bacterial infections and can ultimately lead to resistance to all antibiotics (i.e., pan-drug resistance).65 MDR in metagenomic datasets is frequently the result of an efflux pump (e.g., mexB and oprA), which is a cellular system that can extrude substrates from a cell, including antibiotics.66 Despite this ability, the presence of a MDR efflux pump does not imply phenotypic resistance to antibiotics as efflux pumps can serve many functional roles in environmental bacteria.66–68 As a result, the implications and relevancy of their presence in stormwater bacteria for public health is unclear.
Beyond efflux pumps, aminoglycoside and bacitracin resistance genes were the most frequently occurring ARGs in genome datasets.50,53,55,57,60 Through sequencing techniques, the microbial hosts of these ARGs have been identified. Aminoglycoside ARGs, for example, were found to be correlated with the Limnohabitans, Lefisonia, and Sphingomonas genera, which are associated with freshwater, soil and plant, and wastewater environments, respectively.57 This finding, again, emphasizes the numerous potential sources of ARGs in urban stormwater. Stormwater ARGs have also been linked with genera that contain pathogens. The Mycobacterium, Bordetella, Propionibacterium, Pseudomonas, and Corynebacterium genera have been documented in the research of stormwater outfalls, with further correlation analysis specifically finding the ARGs sul1, dfrA1 and qnrS correlated to Arcobacter sp., Comamonadaceae testosteroni, and Bacteroides fragilis.50,57,61
2.2 Extracellular ARGs: an underexplored antibiotic resistance element
Extracellular ARGs (eARGs) are crucial elements of the resistome that have been under investigated in urban stormwater runoff. eARGs can originate in the environment indirectly from input sources containing eARGs, or directly from the extrusion of iARGs from microbial cells or passive release as a result of cell lysis.72,73 In the environment, eARGs are a source of genetic material available for bacteria to acquire via horizontal gene transformation and thus, a means for antibiotic resistance propagation. The concentration of eARGs and the rate of transformation, moreover, can be influenced by abiotic and biotic factors differently in comparison to iARGs and conjugation.26 As such, to fully characterize the resistance elements in an environment as well as the risk of ARB proliferation by HGT mechanism, iARGs and eARGs must both be considered.The fate and stability of eARGs have been found to differ from iARGs.73–75 Crucially, eARGs have been found to persist under various environmental conditions longer than that of iARGs.76,77 Though the results are ranging, eARGs can persist in certain environments from a few days to years.78–81 The fate of eARGs is significantly influenced by temperature, salinity, radiation, solids, and DNA characteristics (e.g., size, concentration, and integrity).73,79 Soil colloids and particles specifically have been investigated for providing long term protection and stability to eARGs, and it has been confirmed that eARGs adsorbed to sediments remain bioavailable for transformation.75,82 Consequently, eARGs have been found to be the dominant fraction of ARGs in environments with elevated sediment and soil concentrations.83 Specific environments where eARGs have been quantified as a significant portion of the total ARGs include estuary sediments,83,84 marine sediments,78 airborne particulate matter,85 and municipal, hospital, and pharmaceutical industry wastewater effluents.86,87
Despite the significant and distinct role of eARGs in the proliferation of antibiotic resistance, their abundance in aquatic environments has not been well characterized. This gap in research exists in part due to the small size of eARGs and the large sample and chemical requirements for chemical precipitation methods that have been applied to soil-based samples.26,69,88 This made the extraction of eARG difficult, and only recently have methods been developed specifically to extract eARGs from aquatic samples with varying matrices and eARG concentrations.69 In stormwater, eARGs remain a significant gap in research, though the high concentrations of solids that frequently pollute urban runoff and the suspension of eARGs from riverbed sediments during rainfall events make their incidence likely. This research gap was recently addressed, in which it was found that stormwater runoff contributes to an increase in extracellular resistance elements in urban water bodies.48 Moreover, eARGs displayed different seasonal and temporal dynamics from that of iARGs, highlighting the importance of including eARGs in comprehensively characterizing the dynamics influencing the resistome of urban runoff.48 Beyond this one study, the fate and transport of eARGs in urban runoff and the impact of stormwater properties on the occurrence of eARGs and their rate of transformation remain unknown. As a result, future research should be dedicated to uncovering the specific role eARGs play in the development and propagation of antibiotic resistance in urban runoff.
3. The impact of antibiotic resistance dissemination via stormwater into downstream receiving environments
3.1 Resistance elements in downstream aquatic environments
The research targeting antibiotic resistance at stormwater outfalls has provided evidence that stormwater should be considered a hotspot for antibiotic resistance. Surface waters, however, are typically more connected to human health, as they are used as sources of drinking water and recreation. Consequently, researchers have focused on understanding the effects antibiotic resistance elements in stormwater can have on ecological functioning and human health by sampling water bodies receiving urban stormwater runoff (Table 2).48,51,53,54,56,58,59,61,89–101 Environments that receive direct inputs, such as rivers, estuaries, and lakes, have been described during, immediately after, and after an extended period following rainfall events. Conflicting results of ARG abundances during rainfall have been reported. Rainfall volume and depth have been observed to be correlated directly with ARG abundances,95,98 while inconsistent and variable relationships between ARG concentrations and hydrological conditions were also described.97 Specifically, it was found that only the sul2 relative concentration was greatest at the peak flow of the storms, while the sul1 relative concentration peaked in the rising limb and tetO, tetW, and ermF peaked with the falling limb of the storm events. Inter-storm variability was also reported, in which ARG concentrations were found to peak at different times across the three storms characterized.58 Such results can be explained by the several distinct sources of ARGs that can exist in a drainage area and the time required to transport peak concentrations across a watershed. Sources found to significantly impact the concentrations of ARGs over a storm event include agricultural runoff and sewage inputs from combined sewer overflows.58,59,92,96| Study Design | Analytical Methods | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study location | Downstream environment | Number of sampling sites | Baseflow sampling events | Stormwater sampling events | Stormwater sampling during, after, and/or extended period after rainfall | ARBs analyzed | ARGs analyzed | ARG analysis method | Sequencing analysis | Selective pressures and other notable aspects analyzed | Mobile genetic elements | Citation |
| Lincoln, NE., USA | Stream | 1 | Yes (n = 8) | 8 | During | NA | NA | NA | Metagenomic whole genome sequencing | NA | NA | 53 |
| Chicago, IL., USA | Beach water | 1 | Yes (n = 2) | 1 | After | Pseudomonas [fosfomycin, ciprofloxin, vancomycin] | NA | NA | 16S rRNA amplicon and Pseudomonas genome sequencing | Pseudomonas [mercury and copper] | Prophage detection | 99 |
| Natal, Rio Grande do Norte, Brazil | Beach water | 3 | Yes (n = 3) | 9 | After | E. coli [ampicillin, cephalothin, chloramphenicol, nalidixic acid, nitrofurantoin, sulfamethoxazole, tetracycline] | NA | NA | NA | E. coli [copper, zinc, and mercury] | NA | 51 |
| Sydney, North South Wales, Australia | Beach water | 2 | Yes | 16 | During, after, and after extended period (sampled weekly for two years) | NA | tetA, sul1, qnrS, dfrA1, vanBaac(6′), aac(6′)-cr, aac(3′)-II, aacA4(Amk), aacA4(Gen), aacA4cr, aacC1, aadB, armA, CMY-2, CTX-M, DHA, GES, IMP, KPC, NDM, OXA-10, OXA-23, OXA-30, OXA-48, panSHV, qnrA, qnrB, qnrS, rmtB, rmtC, rmtF, CHV nonESBL, SHV/ESBL, TEM, VEB, VIM | qPCR and multiplex PCR | 16S rRNA amplicon sequencing | NA | intI1 quantification | 93 |
| Chicago, IL., USA | Man-made stream | 1 | Yes (n = 3) | 2 | After (<24 hours) | NA | NA | NA | Metagenomic whole genome and 16S rRNA amplicon sequencing | NA | NA | 104 |
| Sydney, Australia | Seawater and freshwater stream | 3 each | Yes (n = 1) | 1 | After (<24 hours) | E. coli [meropenem, penicillin G, cefoxitin, kanamycin] | NA | NA | 16S rRNA amplicon sequencing | NA | Plasmid and transposon identification | 54 |
| Piedmont region, Italy | River | 1 | Yes (n = 1) | 1 | During | NA | tetA, ermB, blaCTX-M, sul1, qnrS | qPCR | NA | NA | NA | 95 |
| New York City, NY., USA | River | 3 | Yes (n = 2) | 1 | During or after (site dependent) | NA | sul1, sul2, tetG, tetO | qPCR | NA | NA | NA | 58 |
| Fort Collins, CO., USA | River | 5 | Yes (n = 1) | 4 | After extended period (3, 6, 8, 10 months) | NA | sul1, sul2, tetO, tetW, ermF | qPCR | 16S rRNA amplicon and metagenomic whole genome sequencing | Silver, arsenic, barium, cerium, chromium, copper, gadolinium, manganese, lead, platinum, strontium, thorium, uranium; anhydrotetracycline, azithromycin, clarithromycin, chlortetracycline, doxycycline, erythromycin, 4-epitetracycline, oxytetracycline, sulfadimethoxine, sulfamethoxazole, sulfamethazine, sulfadiazine, tetracycline, tylosin | NA | 96 |
| Blacksburg, VA., USA | Creek | 1 | Yes (n = 3) | 3 | During | NA | sul1, sul2, tetO, tetW, ermF | qPCR | Metagenomic whole genome sequencing | NA | NA | 97 |
| Xiamen City, China | Lagoon | 7 | Yes (n = 1) | 3 | After and after extended period (1, 2, and 11 days) | NA | 285 ARGs | HT-qPCR | 16S rRNA amplicon sequencing | NA | 10 MGEs (HT-qPCR) | 102 |
| Greenwood, NS., Canada | River | 8 | Yes (n = 4) | 1 | During | NA | bla CTX-M, blaTEM, ermB, qnrS, sul1, sul2, tetO, mecA, vanA | qPCR | NA | Amoxicillin, cefaclor, cefprozil, cefdinir, levofloxacin, ciprofloxacin, azithromycin, clindamycin, clarithromycin, triclocarbon | intI1 quantification | 98 |
| Milwaukee, WI., USA | Estuary, harbor, and lake | 3 each | Yes | 37 | During and after | E. coli [ampicillin, chlortetracycline, kanamycin, nalidixic acid, neomycin, oxytetracycline, penicillin, streptomycin, sulfathiazole, tetracycline, total resistance] | NA | NA | NA | NA | NA | 59 |
| Milwaukee, WI., USA | River and lake | 1 each | Yes (n = 3) | 3 | During | NA | bla TEM, ermF, sul1, tetC | ddPCR | NA | Lead, cadmium, arsenic, chromium, nickel, copper, zinc, iron | intI1 | 48 |
| Milwaukee, WI., USA | River | 10 | Yes (n = 3) | 5 | After and after extended period (<14 hours after 3 rainfall events; 18 and 42 hours after CSO event) | E. coli [ampicillin, streptomycin, chlorotetracycline, nalidixic acid, oxytetracycline, tetracyclinem, kanamycin, neomycin, sulfathiazole, penicillin] | NA | NA | NA | Arsenic, cadmium, chromium, copper, lead, magnesium, nickel, selenium, silver, zinc | NA | 91 |
| Adelaide, South Australia | Biofilms in a stormwater harvesting aquifer | 3 | No | 3 | After extended period (30, 60, and 90 days) | NA | 410 gene categories | Microarray | NA | Microarray of metal resistance genes | NA | 103 |
| Toronto, Ontario, Canada | River | 7 | Yes | 7–16 (site dependent) | Not specified | E. coli [ampicillin] | NA | NA | NA | NA | NA | 56 |
| Germany | Karst spring | 1 | Yes (n = 51) | 2 | During, after, and after extended period (weekly for 16 months) | NA | sul1, sul2, dfrA1, dfrA12, blaSHV, aadA, tetA, tetB, tetC, tetK, ermB | PCR and qPCR | NA | NA | NA | 101 |
| Central Coast, New South Wales, Australia | Lagoon and seawater | 5 (lagoon) and 27 (seawater) | Yes (n = 2) | 3 | After and after extended period (5 days) | NA | sul1, tetA, qnrS, dfrA1, vanB | qPCR | 16S rRNA amplicon sequencing | NA | intI1 quantification | 61 |
| New York City, NY., USA | Estuary | 10 | Yes (n = 6) | 17 | After | Heterotrophic bacteria [ampicillin and tetracycline] | NA | NA | NA | NA | NA | 92 |
| Nanjing City, Jiangsu, China | Lake | 6 | Yes (n = 1) | 1 | After | E. coli and S. aureus [levofloxacin, streptomycin, gentamicin, nalidixic acid, vancomycin, tetracycline, sulfamethoxazole, ceftriaxone, cefazolin] | tetE, tetM, tetO, sul1, sul2, sul3, sulA, strA, strB | PCR | NA | Aureomycin, chloramphenicol, ceftriaxone, cefazolin, doxycycline, erythromycin, florfenicol, gentamicin, levofloxacin, nalidixic acid, norfloxacin, ofloxacin, streptomycin, sulfadiazine, oxytetracycline, roxithromycin, sulfamethoxazole, sulfamethoxazole, tetracycline, thiamphenicol, vancomycin | intI1, intI2, intI3 | 89 |
Immediately following rainfall (∼24 hours) it was observed that the absolute abundance of ARGs increased and ARB phenotypes were more frequent in multiple water bodies, including streams, rivers, estuaries, and lakes.56,59,61,89–93,101 This result was reported consistently in watersheds of varying sizes and land uses, under different hydrologic conditions, in various seasons, and in several geographic locations. Accordingly, antibiotic resistance pollution originating from stormwater can be considered a widespread occurrence. Specifically, when the genes tetA, sul1, dfrA1, and qnrS were analyzed at an urban beach comparatively between baseflow and after rainfall the concentrations were found to increase 10- to 30-fold.93 A similar result was observed at a coastal lagoon where a 2–3 order of magnitude increase in the concentration of sul1 and dfrA1 was reported.61 The tetA gene, however, did not increase in concentration, indicating that individual ARGs can exhibit varying dynamics in downstream environments following rainfall.61 For ARB phenotypes after a storm event, resistance to ampicillin, oxytetracycline, penicillin, streptomycin, sulfathiazole, and tetracycline antibiotics increased in an urban river91 and resistance to levofloxacin, cefazolin, nalidixic acid, vancomycin, tetracycline, sulfamethoxazole, streptomycin increased in an urban lake.89
The impacts to downstream water bodies have also been shown to be persistent (>24 hours after rainfall). Concentrations of ARGs are typically highest one day following rainfall and decrease with time,100 but ARG abundances have been observed to remain elevated above baseline conditions for three weeks,95 one week,93 and five days.61 Moreover, following a significant flooding event, ARG concentrations remained elevated for 10-months while the profile of ARGs as determined through metagenomic sequencing never returned to a preflood state.96 Long-term alteration of the resistome of downstream environments has not been as regularly described as ARG concentrations and ARB phenotypic frequencies (Table 2). It is likely though that urban stormwater runoff can significantly alter the resistomes of urban water bodies long-term because stormwater can transport contaminants from across a drainage area, enabling the large-scale mixing of resistance elements in downstream water bodies from environments that are otherwise disconnected.105 Further evidence to this event is provided in a study of an urban stream after rain where it was found that the microbial community shifted significantly from a community associated with freshwater to one associated with urban-impacted waters. The phylum Actinobacteria and Bacteroidetes, specifically, decreased in relative abundance whereas the Gammaproteobacterial, Betaproteobacteria, and Chlamydia phylum increased long-term.90 Moreover, the waterborne pathogenic species Legionella pneumophila and Francisella tularensis increased 10- and 16-fold in relative abundance, respectively.90 Such a rise could also be due to an influx of sewage contaminated stormwater as rainfall has been observed to introduce wastewater associated genera, human sewage markers, and ARB displaying resistance patterns similar to WRRFs.56,59,93 A long-term concern that could thus arise from the introduction of urban stormwater into downstream water bodies is an increased connection between the ARGs and bacterial community of stormwater, including pathogenic bacteria, with the bacterial community of the downstream environment. Such connections could lead to an escalation in the rate of HGT and the generation of more and possible new superbugs in an environment with a closer proximity to human health.
Urban stormwater runoff, from these results, presents a clear impact to downstream environments in terms of the abundance and diversity of resistance elements. However, it remains unclear how the specific physical and chemical factors of stormwater runoff in combination with those of downstream waterbodies influence the transport and long-term fate of ARGs and ARB. For instance, in one study of a coastal lagoon, nutrient concentrations were found to be positively correlated with ARG relative concentration.100 Nutrient concentrations, as well as other pollutants such as metals, cations, solids, temperature, and conductivity, can be spatially and seasonally dependent. Further research should concentrate on investigating the influence of environmental factors and drainage area characteristics over the composition and persistence of resistance elements in downstream water bodies to provide perspective on the fate of ARGs under varying conditions.
3.2 Transport of antibiotic resistance elements into green stormwater infrastructure via urban stormwater runoff
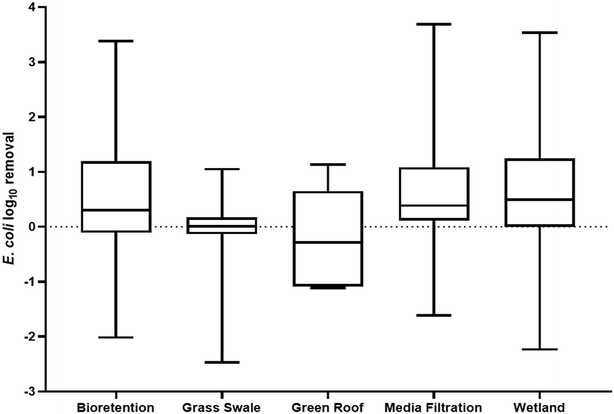
While many environments that receive urban runoff have been investigated as sinks of ARGs, GSI has remained significantly unexplored. GSI refers to the systems engineered to mimic natural hydrologic processes and infiltrate stormwater to reduce the volume of runoff and pollutant load to surface water bodies.106 In urban environments specifically, more impervious surfaces have led to a subsequent increase in water body impairment and thus, GSI systems have been presented as a solution to urban runoff by treating stormwater where it falls. GSI systems have been proven to reduce stormwater volumes, decrease flooding events, and remove stormwater pollutants, particularly sediments, metals, and nutrients, in urban watersheds.107 Moreover, in comparison to traditional gray infrastructure systems, GSI can provide co-environmental benefits such as groundwater recharge, carbon sequestration, and air quality improvement.108–110 Concerning the public health threat of antibiotic resistance, traditional gray stormwater infrastructure does not mitigate the transport of ARGs in the urban environment. Concentrations of ARGs and ARB at a storm drain and in a stormwater pond were found to be similar to those quantified in downstream rivers, indicating that gray infrastructure facilitates the movement of resistance elements into downstream environments.48,111 Consequently, GSI presents the possibility of removing ARGs from stormwater and mitigating the transport of ARGs in the urban water cycle. Examples of soil based GSI systems include bioretention cells, bioswales, green roofs, rain gardens, and constructed wetlands. GSI system such as these have been shown to remove stormwater contaminants by physical–chemical mechanisms, such as filtration, sedimentation, and adsorption.107,112–114 Despite the ability of GSI to remove stormwater contaminants, and stormwater being a known vector for antibiotic resistance elements, the occurrence of ARGs in GSI, the impact of GSI design, and the potential consequences of ARG accumulation in GSI is relatively unknown.GSI systems being able to remove ARGs from urban stormwater runoff indicates a potential management opportunity for the public health crisis of antibiotic resistance. The minimal research in this field however leads to no conclusive understanding as to the capability of GSI to serve a long-term role in mitigating the transport of ARGs via urban stormwater runoff and the governing design factors. Research on the removal of other biological contaminants through GSI however can provide some perspective into GSI's potential for ARG management, specifically hosts of ARGs like E. coli.Fig. 1 summarizes the reduction of E. coli concentrations from stormwater through various GSI systems. The results indicate that a positive removal rate can be achieved through all systems, but on average, grass swales and green roofs are not effective treatment systems for E. coli in stormwater and could even be leading to an enrichment of bacterial concentrations in the effluent of GSI. Rather, bioretention cells, media filtration systems, and wetlands provide an average positive removal rate at 0.476, 0.698, and 0.626-log, respectively. The difference in performance could be due to many different study and site-specific factors, but predominantly, soil media has been the focus of research into the performance of GSI as it dictates the primary removal mechanisms for biological contaminants: filtration and sorption.118
Sand-based media is the basis of most GSI systems as it promotes infiltration and can increase hydraulic conductivity, however its ability to remove biological contaminants is relatively poor.118 In fact, the amount of sand in the system relative to finer particles (e.g., silt and clay) has been found to be inversely related to bacterial removal as sand can only provide minimal physical straining and adsorption.119 This relationship is expected to remain true for ARGs, notably eARGs due to their small size. From these results, increasing the pore size of the soil media to increase the physical filtration capability of the system could increase bacterial removal, however this can inversely impact stormwater infiltration. As a result, increasing bacterial removal has been found to be most effective through an increase in the adsorptive capacity of the soil.119 Biochar is one of the most studied soil amendments in GSI for biological contaminant removal from stormwater and its performance has been reviewed previously.120,121 The addition of biochar can increase removal of E. coli in comparison to removal rates in sand-based systems due to an increase in attachment sites, surface roughness, and hydrophobicity.122,123 Specifically, E. coli removal increased from 0.3-log in a sand biofilter to 1.9-log when biochar was added to the system.124 The type of biochar can impact removal; woodchip-based biochar that was produced via pyrolysis at a low temperature and high temperature increased E. coli removal 3.33-log and 2.99-log, respectively, while a commercially purchased woodchip-based biochar achieved a 2.03-log increase in removal.122 Over a 5-month period, an average removal of 2.15-log was achieved for E. coli, indicating that removal is also able to be maintained long-term in biofilters amended with biochar.125 The effectiveness of biochar can be impeded however, specifically by influent natural organic matter competing for attachment sites as well as biofilm formation altering surface sites and hydraulic performance.123 Other amendments have thus been investigated as an alternative to biochar, including iron oxide coated sand, calcite, zeolite, iron fillings, and aluminum-magnesium graphene oxide coated sand.122,123,126–129 Similar removal capabilities were observed by these studies. Iron oxide coated sand increased the efficiency of E. coli removal from 82% to 99% due to an increase in surface charge and roughness.127 In addition, zeolite, iron fillings, and calcite increased removal of E. coli from approximately 20% to 99%, 85%, and 63%, respectively.
Such results indicate that GSI systems provide some reduction in bacterial contaminants from stormwater and can be engineered for further removal capabilities. As a result, it is likely that this ability could be extended for the removal of ARB and ARGs. However, the removal of ARGs in varying environmental treatment systems is not always as effective or correlated with the removal efficiency of different bacterium. For example, in a constructed wetland treating wastewater effluent, the removal of Enterococcus and human-associated Bacteroidetes ranged between 66.6–84.0%, while the ARG ermF only was reduced 13.1%.130 Moreover, an additional constructed wetland saw no reduction in ARG concentrations though E. coli concentrations decreased 22.3%.131 Such differences were linked to additional contaminant sources into the wetlands as well as factors that can have a contrasting impact on bacterial species, such as water temperature, turbidity, and sedimentation rates. Consequently, while GSI research does provide ample evidence of bacteria removal, research specifically investigating ARGs is needed to fully determine if there is an opportunity to mitigate a source and transport vector of ARGs and ARB into the urban environment. eARGs, moreover, are smaller in size and can have different surface properties than bacterial cells and thus should be investigated individually. Further, design factors that were also shown to influence the removal of bacteria from stormwater in GSI included plant selection, submerged zone depth, system size, and hydraulic conductivity.132,133 The influence such factors have on the removal of ARGs, in addition to soil media, are further avenues for future research.
HGT plays a crucial role in the public health crisis of antibiotic resistance because it facilitates the acquisition of new ARGs in bacteria. HGT can have an impact on the evolution of the microbial communities, including pathogenic bacteria, as ARGs, virulence factors, and diverse functions can be encoded on horizontally transferred genes.136,137 The carriage and maintenance of ARGs, however, is typically associated with a fitness cost to the cell.138 Bacteria are still able to maintain ARGs despite the assumed fitness cost because of the benefits ARGs can provide to cellular functioning in the face of selective pressures. Selective pressures are defined by their ability to stress bacterial communities and at low concentrations will not inhibit all cellular activity but trigger SOS responses. A specific SOS response to selective pressures that increases the fitness of the cell is HGT.139–142 Selective pressures, therefore, are one of the primary reasons why ARGs persist in the environment because they not only enrich resistomes with diverse mechanisms of resistance but can also reduce the fitness cost associated with resistance.143
An increase in the frequency of HGT has been directly linked with many selective pressures, including antibiotics,144–147 metals,71,148 disinfectants,149 disinfectant byproducts,150 biocides (e.g., triclosan, and chlorhexidine),145,151 and quaternary ammonium compounds.152 Environmental conditions are also known to impact the rate of HGT. The rate of conjugation is dependent upon abiotic factors including temperature, pH, nutrients, organic matter, oxygen, and moisture content.153,154 Transformation is similarly impacted by such factors, but interestingly the frequency of transformation is greater in environments lacking nutrients possibly as a result of nutrient deficiency causing cell lysis, leading to an increase in the concentration of eARGs available for uptake via transformation.26
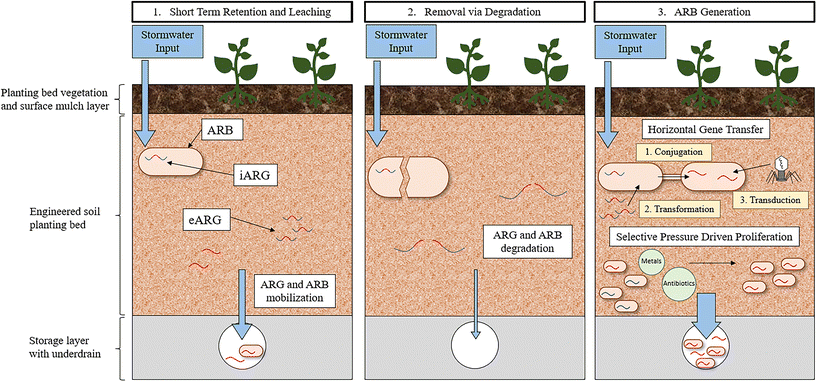
The rate of HGT in the soil environment of GSI has yet to be investigated. Determining the rate of HGT in environmental communities has been and remains a challenge due to a lack of methodology that can capture the inherent complexity of environmental systems while also providing direct quantification of observed transfers.155 For example, laboratory-based in vivo experiments that directly quantify HGT rates, often must simplify the bacterial recipient and transfer communities being evaluated to such a degree that meaningful conclusions are not possible. Moreover, in situ methods, often employing bioinformatic approaches, can capture environmental HGT dynamics but require that HGT is inferred and not quantified directly.155 The paucity of reliable methodologies to quantify HGT leaves a significant research gap in understanding the dissemination and evolution of ARGs in all environments. GSI environments specifically are likely a hotspot for HGT because it is a confluence of factors that contribute to an increase in frequency of gene transfer. Therefore, development of methods that can quantify HGT in these environments would help elucidate the role of GSI in the promotion of HGT. Heavy metals and biocides that select for resistance, nutrients that promote cellular growth, and solids that impart protection and stabilization are abundant in GSI systems. Moreover, GSI is plagued with anthropogenic pollutants from stormwater, including fecal contamination, pet waste, and other human associated bacteria, indicating that the rate of transfer to pathogens specifically, could be high in GSI. Taking all these conditions together, it is likely that the accumulation of ARGs in GSI could enrich the bacterial community with diverse functions and resistance traits, unique to the stormwater environment.
A further consequence of ARG accumulation in GSI is that, following the propagation of ARGs within the resistome, the bacterial community could be mobilized out of the GSI system during a subsequent storm event leading to diverse ARB entering the environment. ARG mobilization has been observed in GSI; a mixture of stormwater and untreated sewage was passed through the biofilter and then the biofilter was flushed with potable irrigation water.117 These experiments indicated that, for a minimum of two subsequent storm events, ARG concentrations increase in the effluent (1–2-log), implying that ARGs removed previously were mobilized out of the GSI system and into the effluent waters. More research will need to be dedicated to understanding how engineering decisions can be incorporated into GSI design to maximize the removal of ARGs while also minimizing ARG mobilization. A study investigating the fate of E. coli in GSI observed that, in a bioretention cell with conventional media, 99.98% die-off of the E. coli was achieved within one-week after a stormwater event primarily due to predation and competition in the bacterial community.127 No additional stormwater was passed through this bioretention cell however, and thus mobilization was not a possible fate. As a result, future research should investigate the long-term fate of ARGs in full-scale operating GSI systems to determine if GSI systems pose a risk through the propagation of resistance.
4. Conclusions and future research directions
Recent research has aimed to uncover the role of the environment in the exasperation of antibiotic resistance and the complex interrelationships the environment has with human health.22 Urban stormwater runoff has been identified as an area of concern for the introduction of antibiotic resistance into the environment, with elevated concentrations of iARGs and eARGs being transported via stormwater outfalls into downstream water bodies. Stormwater, moreover, can disseminate unique resistance elements from across an urban landscape, leading to the accumulation of ARGs and ARB in surface waters, diversifying the overall resistome.46 Following transport into downstream environments, research has concluded that the resistance elements from stormwater inputs can be persistent, presenting a long-term impact to water bodies utilized for drinking water, recreation, and fishing. Currently, there is limited research exploring ways to control the input of resistance elements via urban stormwater runoff. Though stormwater runoff is only one source of ARGs into urban water bodies, managing antibiotic resistance is required for all sources as they are all closely linked and interdependent. As such, action regarding stormwater runoff is key to advancing an overall concerted response to this public health threat.22 GSI are an existing infrastructure system installed in urban environments to manage stormwater volumes and thus present an opportunity to manage the cycling of ARGs in the urban environment. Research within this field is minimal, but does indicate that existing GSI can retain ARGs, and that engineered decisions can be implemented to enhance removal, with the soil media being a crucial component for ARG removal. Therefore, GSI could be a long-term tool for management of ARGs in urban stormwater runoff.There remains, however, significant research gaps in fully understanding this potential. Research gaps identified by the authors throughout this review that warrant future attention include:
• The influence of spatial and temporal dynamics on the composition of ARGs in stormwater runoff.
• The frequency of ARG co-location on MGEs in stormwater and implications for public health and ARG transfer.
• The rate of horizontal gene conjugation, transformation, and transduction in urban stormwater runoff and the impact of selective pressures on this rate.
• The fate and transport of eARG in urban runoff and the impact of stormwater properties on the occurrence of eARGs.
• The effect of GSI design factors on the removal, retention, and inactivation per cell lysis of ARGs and ARB from urban stormwater runoff.
• The long-term fate of ARGs in GSI in full-scale operating GSI systems and the risk of ARG propagation by HGT.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. 2041851. Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. This work was also supported by the Lafferty Family Charitable Foundation.Notes and references
- J. Davies, Where have all the antibiotics gone?, Can. J. Infect. Dis. Med. Microbiol., 2006, 17, 287–290 CrossRef PubMed.
- C. J. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. Robles Aguilar, A. Gray, C. Han, C. Bisignano, P. Rao, E. Wool, S. C. Johnson, A. J. Browne, M. G. Chipeta, F. Fell, S. Hackett, G. Haines-Woodhouse, B. H. Kashef Hamadani, E. A. P. Kumaran, B. McManigal, R. Agarwal, S. Akech, S. Albertson, J. Amuasi, J. Andrews, A. Aravkin, E. Ashley, F. Bailey, S. Baker, B. Basnyat, A. Bekker, R. Bender, A. Bethou, J. Bielicki, S. Boonkasidecha, J. Bukosia, C. Carvalheiro, C. Castañeda-Orjuela, V. Chansamouth, S. Chaurasia, S. Chiurchiù, F. Chowdhury, A. J. Cook, B. Cooper, T. R. Cressey, E. Criollo-Mora, M. Cunningham, S. Darboe, N. P. J. Day, M. De Luca, K. Dokova, A. Dramowski, S. J. Dunachie, T. Eckmanns, D. Eibach, A. Emami, N. Feasey, N. Fisher-Pearson, K. Forrest, D. Garrett, P. Gastmeier, A. Z. Giref, R. C. Greer, V. Gupta, S. Haller, A. Haselbeck, S. I. Hay, M. Holm, S. Hopkins, K. C. Iregbu, J. Jacobs, D. Jarovsky, F. Javanmardi, M. Khorana, N. Kissoon, E. Kobeissi, T. Kostyanev, F. Krapp, R. Krumkamp, A. Kumar, H. H. Kyu, C. Lim, D. Limmathurotsakul, M. J. Loftus, M. Lunn, J. Ma, N. Mturi, T. Munera-Huertas, P. Musicha, M. M. Mussi-Pinhata, T. Nakamura, R. Nanavati, S. Nangia, P. Newton, C. Ngoun, A. Novotney, D. Nwakanma, C. W. Obiero, A. Olivas-Martinez, P. Olliaro, E. Ooko, E. Ortiz-Brizuela, A. Y. Peleg, C. Perrone, N. Plakkal, A. Ponce-de-Leon, M. Raad, T. Ramdin, A. Riddell, T. Roberts, J. V. Robotham, A. Roca, K. E. Rudd, N. Russell, J. Schnall, J. A. G. Scott, M. Shivamallappa, J. Sifuentes-Osornio, N. Steenkeste, A. J. Stewardson, T. Stoeva, N. Tasak, A. Thaiprakong, G. Thwaites, C. Turner, P. Turner, H. R. van Doorn, S. Velaphi, A. Vongpradith, H. Vu, T. Walsh, S. Waner, T. Wangrangsimakul, T. Wozniak, P. Zheng, B. Sartorius, A. D. Lopez, A. Stergachis, C. Moore, C. Dolecek and M. Naghavi, Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, Lancet, 2022, 399(10325), 12–18 CrossRef PubMed.
- P. Dadgostar, Antimicrobial Resistance: Implications and Costs, Infect. Drug Resist., 2019, 12, 3903–3910 CrossRef CAS PubMed.
- IACG, No time to wait: securing the future from drug-resistant infections, No time to wait: securing the future from drug-resistant infections, 2016 Search PubMed.
- A. Chokshi, Z. Sifri, D. Cennimo and H. Horng, Global contributors to antibiotic resistance, J. Global Infect. Dis., 2019, 11, 36–42 CrossRef PubMed.
- V. M. D'Costa, C. E. King, L. Kalan, M. Morar, W. W. L. Sung, C. Schwarz, D. Froese, G. Zazula, F. Calmels, R. Debruyne, G. B. Golding, H. N. Poinar and G. D. Wright, Antibiotic resistance is ancient, Nature, 2011, 477, 457–461 CrossRef PubMed.
- G. Cox and G. D. Wright, Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions, Int. J. Med. Microbiol., 2013, 303, 287–292 CrossRef CAS PubMed.
- E. Peterson and P. Kaur, Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens, Front. Microbiol., 2018, 9, 1–21 CrossRef PubMed.
- J. M. A. Blair, M. A. Webber, A. J. Baylay, D. O. Ogbolu and L. J. V. Piddock, Molecular mechanisms of antibiotic resistance, Nat. Rev. Microbiol., 2015, 13, 42–51 CrossRef CAS PubMed.
- E. Goldman, Antibiotic abuse in animal agriculture: Exacerbating drug resistance in human pathogens, Hum. Ecol. Risk Assess., 2004, 10, 121–134 CrossRef.
- K. E. Fleming-Dutra, A. L. Hersh, D. J. Shapiro, M. Bartoces, E. A. Enns, T. M. File, J. A. Finkelstein, J. S. Gerber, Y. Hyun David, J. A. Linder, R. Lynfield, D. J. Margolis, L. S. May, D. Merenstein, J. P. Metlay, J. G. Newland, J. F. Piccirillo, R. M. Roberts, G. V. Sanchez, K. J. Suda, A. Thomas, T. M. Woo, R. M. Zetts and L. A. Hicks, Prevalence of Inappropriate Antibiotic Prescriptions Among US Ambulatory Care Visits, 2010-2011, JAMA, 2016, 315(17), 1864–1873 CrossRef CAS PubMed.
- B. Spellberg, G. R. Hansen, A. Kar, C. D. Cordova, L. B. Price and J. R. Johnson, Antibiotic Resistance in Humans and Animals, NAM Perspect., 2016 DOI:10.31478/201606d.
- W. Witte, Medical consequences of antibiotics use in agriculture, Science, 1998, 279, 996–997 CrossRef CAS PubMed.
- M. Anwar, Q. Iqbal and F. Saleem, Improper disposal of unused antibiotics: an often overlooked driver of antimicrobial resistance, Expert Rev. Anti-infect. Ther., 2020, 18, 697–699 CrossRef CAS PubMed.
- S. I. Polianciuc, A. E. Gurzău, B. Kiss, M. Georgia Ştefan and F. Loghin, Antibiotics in the environment: causes and consequences, Med. Pharm. Rep., 2020, 93, 231–240 Search PubMed.
- D. G. J. Larsson, Antibiotics in the environment, Upsala J. Med. Sci., 2014, 119, 108–112 CrossRef PubMed.
- A. Yakimov, I. Bakhlanova and D. Baitin, Targeting evolution of antibiotic resistance by SOS response inhibition, Comput. Struct. Biotechnol. J., 2021, 19, 777–783 CrossRef CAS PubMed.
- M. Zhuang, Y. Achmon, Y. Cao, X. Liang, L. Chen, H. Wang, B. A. Siame and K. Y. Leung, Distribution of antibiotic resistance genes in the environment, Environ. Pollut., 2021, 285, 117402 CrossRef CAS PubMed.
- M. Imran, K. R. Das and M. M. Naik, Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: An emerging health threat, Chemosphere, 2019, 215, 846–857 CrossRef CAS PubMed.
- A. D. Wales and R. H. Davies, Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens, Antibiotics, 2015, 4, 567–604 CrossRef PubMed.
- B. Berglund, Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics, Infect. Ecol. Epidemiol., 2015, 5, 28564 Search PubMed.
- United Nations Environment Programme, Bracing for Superbugs: Strengthening environmental action in the One Health response to antimicrobial resistance, Bracing for Superbugs: Strengthening environmental action in the One Health response to antimicrobial resistance, 2023 Search PubMed.
- C. J. H. Von Wintersdorff, J. Penders and J. M. Van Niekerk, Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer, Front. Microbiol., 2016, 7, 1–10 Search PubMed.
- S. B. Levy, Gene Transfer in the Environment, Gene Transfer in the Environment, McGraw-Hill, New York, 1989 Search PubMed.
- C. M. Thomas and K. M. Nielsen, Mechanisms of, and barriers to, horizontal gene transfer between bacteria, Nat. Rev. Microbiol., 2005, 3, 711–721 CrossRef CAS PubMed.
- A. Zarei-Baygi and A. L. Smith, Intracellular versus extracellular antibiotic resistance genes in the environment: Prevalence, horizontal transfer, and mitigation strategies, Bioresour. Technol., 2021, 319, 124181 CrossRef CAS PubMed.
- S. Kunhikannan, C. J. Thomas, A. E. Franks, S. Mahadevaiah, S. Kumar and S. Petrovski, Environmental hotspots for antibiotic resistance genes, MicrobiologyOpen, 2021, 10(3), e1197 CrossRef CAS PubMed.
- B. J. Koch, B. A. Hungate and L. B. Price, Food-animal production and the spread of antibiotic resistance: the role of ecology, Front. Ecol. Environ., 2017, 15, 309–318 CrossRef.
- E. A. Oniciuc, E. Likotrafiti, A. Alvarez-Molina, M. Prieto, J. A. Santos and A. Alvarez-Ordóñez, The present and future of whole genome sequencing (WGS) and whole metagenome sequencing (WMS) for surveillance of antimicrobial resistant microorganisms and antimicrobial resistance genes across the food chain, Genes, 2018, 9, 1–28 Search PubMed.
- X. Ji, Q. Shen, F. Liu, J. Ma, G. Xu, Y. Wang and M. Wu, Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China, J. Hazard. Mater., 2012, 235–236, 178–185 CrossRef CAS PubMed.
- C. Baker-Austin, M. S. Wright, R. Stepanauskas and J. V. McArthur, Co-selection of antibiotic and metal resistance, Trends Microbiol., 2006, 14, 176–182 CrossRef CAS PubMed.
- G. W. Sundin and C. L. Bender, Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants, Mol. Ecol., 1996, 5, 133–143 CrossRef CAS PubMed.
- K. Pärnänen, A. Karkman, M. Tamminen, C. Lyra, J. Hultman, L. Paulin and M. Virta, Evaluating the mobility potential of antibiotic resistance genes in environmental resistomes without metagenomics, Sci. Rep., 2016, 6, 35790 CrossRef PubMed.
- S. R. Partridge, S. M. Kwong, N. Firth and S. O. Jensen, Mobile Genetic Elements Associated with Antimicrobial Resistance, Clin. Microbiol. Rev., 2018, 31(4), e00088-17 CrossRef PubMed.
- J. L. Martínez, T. M. Coque and F. Baquero, What is a resistance gene? Ranking risk in resistomes, Nat. Rev. Microbiol., 2015, 13, 116–123 CrossRef PubMed.
- D. J. Rankin, E. P. C. Rocha and S. P. Brown, What traits are carried on mobile genetic elements, and why?, Heredity, 2011, 106, 1–10 CrossRef CAS PubMed.
- H. M. Hwang, M. J. Fiala, D. Park and T. L. Wade, Review of pollutants in urban road dust and stormwater runoff: part 1. Heavy metals released from vehicles, Int. J. Urban Sci., 2016, 20, 334–360 CrossRef.
- S. C. Jiang, K. Lim, X. Huang, D. McCarthy and A. J. Hamilton, Human and environmental health risks and benefits associated with use of urban stormwater, Wiley Interdiscip. Rev.: Water, 2015, 2, 683–699 CrossRef.
- L. Wang, J. Lyons, P. Kanehl and R. Bannerman, Impacts of urbanization on stream habitat and fish across multiple spatial scales, Environ. Manage., 2001, 28, 255–266 CrossRef CAS PubMed.
- E. Eriksson, A. Baun, P. S. Mikkelsen and A. Ledin, Chemical hazard identification and assessment tool for evaluation of stormwater priority pollutants, Water Sci. Technol., 2005, 51, 47–55 CrossRef CAS PubMed.
- M. Huber, A. Welker and B. Helmreich, Critical review of heavy metal pollution of traffic area runoff: Occurrence, influencing factors, and partitioning, Sci. Total Environ., 2016, 541, 895–919 CrossRef CAS PubMed.
- W. Ahmed, K. Hamilton, S. Toze, S. Cook and D. Page, A review on microbial contaminants in stormwater runoff and outfalls: Potential health risks and mitigation strategies, Sci. Total Environ., 2019, 692, 1304–1321 CrossRef CAS PubMed.
- S. J. Gaffield, R. L. Goo, L. A. Richards and R. J. Jackson, Public Health Effects of Inadequately Managed Stormwater Runoff, Am. J. Public Health, 2003, 93, 1527–1533 CrossRef PubMed.
- R. W. Haile, J. S. Witte, M. Gold, R. Cressey, C. McGee, R. C. Milikan, A. Glasser, N. Harawa, C. Ervin, P. Harmon, J. Harper, J. Dermand, J. Alamillo, K. Barrett, M. Nides and G. Wang, The Health Effects of Swimming in Ocean Water Contaminated by Storm Drain Runoff, Epidemiology, 1999, 10, 355–363 CrossRef CAS PubMed.
- A. Almakki, E. Jumas-Bilak, H. Marchandin and P. Licznar-Fajardo, Antibiotic resistance in urban runoff, Sci. Total Environ., 2019, 667, 64–76 CrossRef CAS PubMed.
- K. A. Hamilton, E. Garner, S. Joshi, W. Ahmed, N. Ashbolt, G. Medema and A. Pruden, Antimicrobial-resistant microorganisms and their genetic determinants in stormwater: A systematic review, Curr. Opin. Environ. Sci. Health, 2020, 16, 101–112 CrossRef.
- T. R. Burch, R. J. Newton, L. K. Kimbell, E. Lou LaMartina, K. O'Malley, S. M. Thomson, C. W. Marshall and P. J. McNamara, Targeting current and future threats: recent methodological trends in environmental antimicrobial resistance research and their relationships to risk assessment, J. Environ. Sci., 2022, 8, 1787–1802 CAS.
- K. O'Malley, P. J. McNamara and W. M. McDonald, Seasonal and spatial patterns differ between intracellular and extracellular antibiotic resistance genes in urban stormwater runoff, Env. Sci. Adv., 2022, 1, 380–390 RSC.
- X. Zuo, P. Suo, Y. Li and Q. Xu, Diversity and distribution of antibiotic resistance genes associated with road sediments transported in urban stormwater runoff ☆, Environ. Pollut., 2022, 292, 118470 CrossRef CAS PubMed.
- R. Liguori, S. H. Rommel, J. Bengtsson-Palme, B. Helmreich and C. Wurzbacher, Microbial retention and resistances in stormwater quality improvement devices treating road runoff, FEMS Microbiol. Ecol., 2021, 2, 1–13 Search PubMed.
- A. M. S. Cardonha, R. H. S. F. Vieira, D. P. Rodrigues, A. Macrae, G. Peirano and G. N. D. Teophilo, Fecal pollution in water from storm sewers and adjacent seashores in Natal, Rio Grande do Norte, Brazil, Int. Microbiol., 2004, 7, 213–218 Search PubMed.
- W. Ahmed, Q. Zhang, A. Lobos, J. Senkbeil, M. J. Sadowsky, V. J. Harwood, N. Saeidi, O. Marinoni and S. Ishii, Precipitation influences pathogenic bacteria and antibiotic resistance gene abundance in storm drain outfalls in coastal sub-tropical waters, Environ. Int., 2018, 116, 308–318 CrossRef CAS PubMed.
- D. Baral, B. I. Dvorak, D. Admiraal, S. Jia, C. Zhang and X. Li, Tracking the Sources of Antibiotic Resistance Genes in an Urban Stream during Wet Weather using Shotgun Metagenomic Analyses, Environ. Sci. Technol., 2018, 52, 9033–9044 CrossRef CAS PubMed.
- D. A. P. R. Dewi, B. Gotz and T. Thomas, Diversity and Genetic Basis for Carbapenem Resistance in a Coastal Marine Environment, Appl. Environ. Microbiol., 2020, 86(10), e02939-19 CrossRef PubMed.
- S. Lee, M. Suits, D. Wituszynski, R. Winston, J. Martin and J. Lee, Residential urban stormwater runoff: A comprehensive profile of microbiome and antibiotic resistance, Sci. Total Environ., 2020, 723, 138033 CrossRef CAS PubMed.
- Z. R. Staley, J. Grabuski, E. Sverko and T. A. Edge, Comparison of microbial and chemical source tracking markers to identify fecal contamination sources in the Humber River (Toronto, Ontario, Canada) and associated storm water outfalls, Appl. Environ. Microbiol., 2016, 82, 6357–6366 CrossRef CAS PubMed.
- M. Białasek and A. Miłobędzka, Revealing antimicrobial resistance in stormwater with MinION, Chemosphere, 2020, 258 DOI:10.1016/j.chemosphere.2020.127392.
- A. Eramo, H. D. Reyes and N. L. Fahrenfeld, Partitioning of antibiotic resistance genes and fecal indicators varies intra and inter-storm during combined sewer overflows, Front. Microbiol., 2017, 8, 1–13 Search PubMed.
- S. L. McLellan, E. J. Hollis, M. M. Depas, M. Van Dyke, J. Harris and C. O. Scopel, Distribution and fate of Escherichia coli in Lake Michigan following contamination with urban stormwater and combined sewer overflows, J. Great Lakes Res., 2007, 33, 566–580 CrossRef.
- X. Pan, L. Chen, L. Zhang and J. Zuo, Characteristics of antibiotic resistance gene distribution in rainfall runoff and combined sewer overflow, Environ. Sci. Pollut. Res., 2022, 30, 30766–30778 CrossRef PubMed.
- N. L. R. Williams, N. Siboni, S. L. McLellan, J. Potts, P. Scanes, C. Johnson, M. James, V. McCann and J. R. Seymour, Rainfall leads to elevated levels of antibiotic resistance genes within seawater at an Australian beach, Environ. Pollut., 2022, 307, 119456 CrossRef CAS PubMed.
- J. E. McLain, E. Cytryn, L. M. Durso and S. Young, Culture-based Methods for Detection of Antibiotic Resistance in Agroecosystems: Advantages, Challenges, and Gaps in Knowledge, J. Environ. Qual., 2016, 45, 432–440 CrossRef CAS PubMed.
- S. Nayfach and K. S. Pollard, Toward Accurate and Quantitative Comparative Metagenomics, Cell, 2016, 166, 1103–1116 CrossRef CAS PubMed.
- X. Yin, X. Chen, X. Jiang, Y. Yang, B. Li, M. H. Shum, T. T. Y. Lam, G. M. Leung, J. Rose, C. Sanchez-cid, T. M. Vogel, F. Walsh, T. U. Berendonk, J. Midega, C. Uchea, D. Frigon, G. D. Wright, S. Z. Ahammad, P. H. Nielsen, P. Hugenholtz, C. Bezuidenhout, R. C. Pica, N. J. Ashbolt, G. Corno, D. Fatta-kassinos, H. Bu, H. Schmitt, C. Cha, A. Pruden, K. Smalla, E. Cytryn, Y. Zhang, M. Yang, Y. Zhu, A. Dechesne, B. F. Smets, D. W. Graham, M. R. Gillings, W. H. Gaze, C. M. Manaia, M. C. M. Van Loosdrecht, P. J. J. Alvarez, M. J. Blaser, J. M. Tiedje, E. Topp and T. Zhang, Toward a Universal Unit for Quantification of Antibiotic Resistance Genes in Environmental Samples, Environ. Sci. Technol., 2023, 57, 9713–9721 CrossRef CAS PubMed.
- A. P. Magiorakos, A. Srinivasan, R. B. Carey, Y. Carmeli, M. E. Falagas, C. G. Giske, S. Harbarth, J. F. Hindler, G. Kahlmeter, B. Olsson-Liljequist, D. L. Paterson, L. B. Rice, J. Stelling, M. J. Struelens, A. Vatopoulos, J. T. Weber and D. L. Monnet, Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance, Clin. Microbiol. Infect., 2012, 18, 268–281 CrossRef CAS PubMed.
- P. Blanco, S. Hernando-Amado, J. A. Reales-Calderon, F. Corona, F. Lira, M. Alcalde-Rico, A. Bernardini, M. B. Sanchez and J. L. Martinez, Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants, Microorganisms, 2016, 4, 1–19 CrossRef PubMed.
- B. Youenou, S. Favre-Bonté, J. Bodilis, E. Brothier, A. Dubost, D. Muller and S. Nazaret, Comparative genomics of environmental and clinical Stenotrophomonas maltophilia strains with different antibiotic resistance profiles, Genome Biol. Evol., 2015, 7, 2484–2505 CrossRef CAS PubMed.
- K. T. Konstantinidis and J. M. Tiedje, Trends between gene content and genome size in prokaryotic species with larger genomes, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 3160–3165 CrossRef CAS PubMed.
- K. O'Malley, W. McDonald and P. McNamara, An extraction method to quantify the fraction of extracellular and intracellular antibiotic resistance genes in aquatic environments, J. Environ. Eng., 2022, 148, 04022017 CrossRef.
- M. R. Gillings, W. H. Gaze, A. Pruden, K. Smalla, J. M. Tiedje and Y. G. Zhu, Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution, ISME J., 2015, 9, 1269–1279 CrossRef CAS PubMed.
- Y. Zhang, A. Z. Gu, T. Cen, X. Li, M. He, D. Li and J. Chen, Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment, Environ. Pollut., 2018, 237, 74–82 CrossRef CAS PubMed.
- M. Nagler, H. Insam, G. Pietramellara and J. Ascher-Jenull, Extracellular DNA in natural environments: features, relevance and applications, Appl. Microbiol. Biotechnol., 2018, 102, 6343–6356 CrossRef CAS PubMed.
- K. M. Nielsen, P. J. Johnsen, D. Bensasson and D. Daffonchio, Release and persistence of extracellular DNA in the environment, Environ. Biosaf. Res., 2007, 6, 37–53 CrossRef CAS PubMed.
- D. Mao, Y. Luo, J. Mathieu, Q. Wang, L. Feng, Q. Mu, C. Feng and P. J. J. Alvarez, Persistence of extracellular DNA in river sediment facilitates antibiotic resistance gene propagation, Environ. Sci. Technol., 2014, 48, 71–78 CrossRef CAS PubMed.
- G. Pietramellara, J. Ascher, F. Borgogni, M. T. Ceccherini, G. Guerri and P. Nannipieri, Extracellular DNA in soil and sediment: Fate and ecological relevance, Biol. Fertil. Soils, 2009, 45, 219–235 CrossRef CAS.
- Y. Zhang, Z. Niu, Y. Zhang and K. Zhang, Occurrence of intracellular and extracellular antibiotic resistance genes in coastal areas of Bohai Bay (China) and the factors affecting them, Environ. Pollut., 2018, 236, 126–136 CrossRef CAS PubMed.
- Y. Yang, H. Li, Y. Wei, Z. Chen, T. Chen, Y. Liang, J. Yin, D. Yang, Z. Yang, D. Shi, S. Zhou, H. Wang, J. Li and M. Jin, Comprehensive insights into profiles and bacterial sources of intracellular and extracellular antibiotic resistance genes in groundwater, Environ. Pollut., 2022, 307, 119541 CrossRef CAS PubMed.
- C. Corinaldesi, R. Danovaro and A. Dell'Anno, Simultaneous recovery of extracellular and intracellular DNA suitable for molecular studies from marine sediments, Appl. Environ. Microbiol., 2005, 71, 46–50 CrossRef CAS PubMed.
- M. A. Barnes, C. R. Turner, C. L. Jerde, M. A. Renshaw, W. L. Chadderton and D. M. Lodge, Environmental conditions influence eDNA persistence in aquatic systems, Environ. Sci. Technol., 2014, 48, 1819–1827 CrossRef CAS PubMed.
- D. J. Levy-Booth, R. G. Campbell, R. H. Gulden, M. M. Hart, J. R. Powell, J. N. Klironomos, K. P. Pauls, C. J. Swanton, J. T. Trevors and K. E. Dunfield, Cycling of extracellular DNA in the soil environment, Soil Biol. Biochem., 2007, 39, 2977–2991 CrossRef CAS.
- B. Zhu, Degradation of plasmid and plant DNA in water microcosms monitored by natural transformation and real-time polymerase chain reaction ( PCR ), Water Res., 2006, 40, 3231–3238 CrossRef CAS PubMed.
- Y. Zhang, D. D. Snow, D. Parker, Z. Zhou and X. Li, Intracellular and Extracellular Antimicrobial Resistance Genes in the Sludge of Livestock Waste Management Structures, Environ. Sci. Technol., 2013, 47, 10206–10213 CrossRef CAS PubMed.
- Z. Zhao, K. Zhang, N. Wu, W. Li, W. Xu, Y. Zhang and Z. Niu, Estuarine sediments are key hotspots of intracellular and extracellular antibiotic resistance genes: A high-throughput analysis in Haihe Estuary in China, Environ. Int., 2020, 135, 105385 CrossRef CAS PubMed.
- Y. Zhang, Z. Niu, Y. Zhang and K. Zhang, Occurrence of intracellular and extracellular antibiotic resistance genes in coastal areas of Bohai Bay (China) and the factors affecting them, Environ. Pollut., 2018, 236, 126–136 CrossRef CAS PubMed.
- T. He, L. Jin, J. Xie, S. Yue, P. Fu and X. Li, Intracellular and Extracellular Antibiotic Resistance Genes in Airborne PM2.5for Respiratory Exposure in Urban Areas, Environ. Sci. Technol. Lett., 2021, 8, 128–134 CrossRef CAS.
- P. Dong, H. Wang, T. Fang, Y. Wang and Q. Ye, Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG, Environ. Int., 2019, 125, 90–96 CrossRef CAS PubMed.
- Y. Zhang, A. Li, T. Dai, F. Li, H. Xie, L. Chen and D. Wen, Cell-free DNA: A Neglected Source for Antibiotic Resistance Genes Spreading from WWTPs, Environ. Sci. Technol., 2018, 52, 248–257 CrossRef CAS PubMed.
- M. Liu, A. Hata, H. Katayama and I. Kasuga, Consecutive ultrafiltration and silica adsorption for recovery of extracellular antibiotic resistance genes from an urban river, Environ. Pollut., 2020, 260, 114062 CrossRef CAS PubMed.
- S. Zhang, S. Pang, P. F. Wang, C. Wang, N. Han, B. Liu, B. Han, Y. Li and K. Anim-Larbi, Antibiotic concentration and antibiotic-resistant bacteria in two shallow urban lakes after stormwater event, Environ. Sci. Pollut. Res., 2016, 23, 9984–9992 CrossRef CAS PubMed.
- A. Chaudhary, I. Kauser and A. Ray, Taxon-Driven Functional Shifts Associated with Storm Flow in an Urban Stream Microbial Community, Appl. Environ. Sci., 2018, 3, e00194-18 Search PubMed.
- A. K. Salmore, E. J. Hollis and S. L. McLellan, Delineation of a chemical and biological signature for stormwater pollution in an urban river, J. Water Health, 2006, 4, 247–262 CrossRef CAS PubMed.
- S. Young, A. Juhl and G. D. O'Mullan, Antibiotic-resistant bacteria in the Hudson River Estuary linked to wet weather sewage contamination, J. Water Health, 2013, 11, 297–310 CrossRef PubMed.
- R. L. Carney, M. Labbate, N. Siboni, K. A. Tagg, S. M. Mitrovic and J. R. Seymour, Urban beaches are environmental hotspots for antibiotic resistance following rainfall, Water Res., 2019, 167, 115081 CrossRef CAS PubMed.
- J. P. S. Sidhu, V. V. S. R. Gupta, C. Stange, J. Ho, N. Harris, K. Barry, D. Gonzalez, J. D. Van Nostrand, J. Zhou, D. Page, A. Tiehm and S. Toze, Prevalence of antibiotic resistance and virulence genes in the biofilms from an aquifer recharged with stormwater, Water Res., 2020, 185, 116269 CrossRef CAS PubMed.
- A. Di Cesare, E. M. Eckert, M. Rogora and G. Corno, Rainfall increases the abundance of antibiotic resistance genes within a riverine microbial community, Environ. Pollut., 2017, 226, 473–478 CrossRef CAS PubMed.
- E. Garner, J. S. Wallace, G. A. Argoty, C. Wilkinson, N. Fahrenfeld, L. S. Heath, L. Zhang, M. Arabi, D. S. Aga and A. Pruden, Metagenomic profiling of historic Colorado Front Range flood impact on distribution of riverine antibiotic resistance genes, Sci. Rep., 2016, 6, 1–10 CrossRef PubMed.
- E. Garner, R. Benitez, E. von Wagoner, R. Sawyer, E. Schaberg, W. C. Hession, L. A. H. Krometis, B. D. Badgley and A. Pruden, Stormwater loadings of antibiotic resistance genes in an urban stream, Water Res., 2017, 123, 144–152 CrossRef CAS PubMed.
- M. M. McConnell, L. T. Hansen, K. D. Neudorf, J. L. Hayward, R. C. Jamieson, C. K. Yost and A. Tong, Sources of Antibiotic Resistance Genes in a Rural River System, J. Environ. Qual., 2018, 47, 997–1005 CrossRef CAS PubMed.
- M. Batrich, L. Maskeri, R. Schubert, B. Ho, M. Kohout, M. Abdeljaber, A. Abuhasna, M. Kholoki, P. Psihogios, T. Razzaq, S. Sawhney, S. Siddiqui, E. Xoubi, A. Cooper, T. Hatzopoulos and C. Putonti, Pseudomonas diversity within urban freshwaters, Front. Microbiol., 2019, 10, 1–13 CrossRef PubMed.
- L. Hou, J. Li, H. Wang, Q. Chen, J. Q. Su, M. Gad, W. Ahmed, C. P. Yu and A. Hu, Storm promotes the dissemination of antibiotic resistome in an urban lagoon through enhancing bio-interactions, Environ. Int., 2022, 168, 107457 CrossRef CAS PubMed.
- C. Stange and A. Tiehm, Occurrence of antibiotic resistance genes and microbial source tracking markers in the water of a karst spring in Germany, Sci. Total Environ., 2020, 742 DOI:10.1016/j.scitotenv.2020.140529.
- L. Hou, J. Li, H. Wang, Q. Chen, J. Q. Su, M. Gad, W. Ahmed, C. P. Yu and A. Hu, Storm promotes the dissemination of antibiotic resistome in an urban lagoon through enhancing bio-interactions, Environ. Int., 2022, 168, 107457 CrossRef CAS PubMed.
- J. P. S. Sidhu, V. V. S. R. Gupta, C. Stange, J. Ho, N. Harris, K. Barry, D. Gonzalez, J. D. Van Nostrand, J. Zhou, D. Page, A. Tiehm and S. Toze, Prevalence of antibiotic resistance and virulence genes in the biofilms from an aquifer recharged with stormwater, Water Res., 2020, 185, 116269 CrossRef CAS PubMed.
- A. Chaudhary, I. Kauser and A. Ray, Taxon-Driven Functional Shifts Associated with Storm Flow in an Urban Stream Microbial Community, Appl. Environ. Sci., 2018, 3, e00194-18 Search PubMed.
- S. Saifur and C. M. Gardner, Loading, transport, and treatment of emerging chemical and biological contaminants of concern in stormwater, Water Sci. Technol., 2021, 83, 2863–2885 CrossRef CAS PubMed.
- V. Venkataramanan, A. I. Packman, D. R. Peters, D. Lopez, D. J. Mccuskey, R. I. Mcdonald, W. M. Miller and S. L. Young, A systematic review of the human health and social well-being outcomes of green infrastructure for stormwater and flood management, J. Environ. Manage., 2019, 246, 868–880 CrossRef PubMed.
- J. Sage, E. Berthier and M. C. Gromaire, Stormwater Management Criteria for On-Site Pollution Control: A Comparative Assessment of International Practices, Environ. Manage., 2015, 56, 66–80 CrossRef PubMed.
- V. M. Jayasooriya, A. W. M. Ng, S. Muthukumaran and B. J. C. Perera, Green infrastructure practices for improvement of urban air quality, Urban For Urban Green, 2017, 21, 34–47 CrossRef.
- T. A. M. Pugh, A. R. MacKenzie, J. D. Whyatt and C. N. Hewitt, Effectiveness of green infrastructure for improvement of air quality in urban street canyons, Environ. Sci. Technol., 2012, 46, 7692–7699 CrossRef CAS PubMed.
- K. V. Abhijith and P. Kumar, Field investigations for evaluating green infrastructure effects on air quality in open-road conditions, Atmos. Environ., 2019, 201, 132–147 CrossRef CAS.
- M. E. Delorenzo, B. Thompson, E. Cooper, J. Moore and M. H. Fulton, A long-term monitoring study of chlorophyll, microbial contaminants, and pesticides in a coastal residential stormwater pond and its adjacent tidal creek, Environ. Monit. Assess., 2012, 184, 343–359 CrossRef CAS PubMed.
- C. P. Muerdter, C. K. Wong and G. H. LeFevre, The Role of Vegetation in Bioretention for Stormwater Treatment in the Built Environment: Pollutant Removal , Hydrologic Function , and Ancillary Benefits, J. Environ. Sci., 2018, 4, 592–612 CAS.
- M. A. Gonzalez-Meler, L. A. Cotner, D. A. Massey, M. L. Zellner and E. S. Minor, The Environmental and Ecological Benefits of Green Infrastructure for Stormwater Runoff in Urban Areas, JSM Environ. Sci. Ecol., 2013, 1, 1007 Search PubMed.
- G. LeFevre, K. Paus, P. Natarajan, J. Gulliver, P. Novak and R. Hozalski, Review of dissolved pollutants in urban storm water and their removal and fate in bioretention cells, J. Environ. Eng., 2015, 141, 04014050 CrossRef CAS.
- X. Zuo, S. Zhang, F. Kong and Q. Xu, Application of electrochemical oxidation for the enhancement of antibiotic resistant bacteria removal in stormwater bioretention cells, Sci. Total Environ., 2023, 861 DOI:10.1016/J.SCITOTENV.2022.160477.
- X. Zuo, Q. Xu, Y. Li and K. Zhang, Antibiotic resistance genes removals in stormwater bioretention cells with three kinds of environmental conditions, J. Hazard. Mater., 2022, 429, 128336 CrossRef CAS PubMed.
- M. B. Rugh, S. B. Grant, W. C. Hung, J. A. Jay, E. A. Parker, M. Feraud, D. Li, S. Avasarala, P. A. Holden, H. Liu, M. A. Rippy, L. C. V. De Werfhorst, T. Kefela, J. Peng, S. Shao, K. E. Graham, A. B. Boehm, S. Choi, S. K. Mohanty and Y. Cao, Highly variable removal of pathogens, antibiotic resistance genes, conventional fecal indicators and human-associated fecal source markers in a pilot-scale stormwater biofilter operated under realistic stormflow conditions, Water Res., 2022, 219, 118525 CrossRef CAS PubMed.
- J. E. Grebel, S. K. Mohanty, A. A. Torkelson, A. B. Boehm, C. P. Higgins, R. M. Maxwell, K. L. Nelson and D. L. Sedlak, Engineered infiltration systems for urban stormwater reclamation, Environ. Eng. Sci., 2013, 30, 437–454 CrossRef CAS.
- T. K. Stevik, K. Aa, G. Ausland and J. F. Hanssen, Retention and removal of pathogenic bacteria in wastewater percolating through porous media: A review, Water Res., 2004, 38, 1355–1367 CrossRef CAS PubMed.
- A. B. Boehm, C. D. Bell, N. J. M. Fitzgerald, E. Gallo, C. P. Higgins, T. S. Hogue, R. G. Luthy, A. C. Portmann, B. A. Ulrich and J. M. Wolfand, Biochar-augmented biofilters to improve pollutant removal from stormwater-can they improve receiving water quality?, J. Environ. Sci., 2020, 6, 1520–1537 CAS.
- B. K. Biswal, K. Vijayaraghavan, D. L. Tsen-Tieng and R. Balasubramanian, Biochar-based bioretention systems for removal of chemical and microbial pollutants from stormwater: A critical review, J. Hazard. Mater., 2022, 422, 126886 CrossRef CAS PubMed.
- S. K. Mohanty, K. B. Cantrell, K. L. Nelson and A. B. Boehm, Efficacy of biochar to remove Escherichia coli from stormwater under steady and intermittent flow, Water Res., 2014, 61, 288–296 CrossRef CAS PubMed.
- A. R. M. N. Afrooz and A. B. Boehm, Escherichia coli removal in biochar-modified biofilters: Effects of biofilm, PLoS One, 2016, 11, 1–17 CrossRef PubMed.
- A. R. M. N. Afrooz, A. K. Pitol, D. Kitt and A. B. Boehm, Role of microbial cell properties on bacterial pathogen and coliphage removal in biochar-modified stormwater biofilters, Environ. Sci.: Water Res. Technol., 2018, 4, 2160–2169 RSC.
- A. R. M. Nabiul Afrooz and A. B. Boehm, Effects of submerged zone, media aging, and antecedent dry period on the performance of biochar-amended biofilters in removing fecal indicators and nutrients from natural stormwater, Ecol. Eng., 2017, 102, 320–330 CrossRef.
- C. T. Vu and T. Wu, Engineered multifunctional sand for enhanced removal of stormwater runoff contaminants in fixed-bed column systems, Chemosphere, 2019, 224, 852–861 CrossRef CAS PubMed.
- L. Zhang, E. A. Seagren, A. P. Davis and J. S. Karns, The Capture and Destruction of Escherichia coli from Simulated Urban Runoff Using Conventional Bioretention Media and Iron Oxide-coated Sand, Water Environ. Res., 2010, 82, 701–714 CrossRef CAS PubMed.
- G. Prabhukumar, G. S. Bhupal and K. R. Pagilla, Laboratory Evaluation of Sorptive Filtration Media Mixtures for Targeted Pollutant Removals from Simulated Stormwater, Water Environ. Res., 2015, 87, 789–795 CrossRef CAS PubMed.
- S. K. Mohanty, A. A. Torkelson, H. Dodd, K. L. Nelson and A. B. Boehm, Engineering solutions to improve the removal of fecal indicator bacteria by bioinfiltration systems during intermittent flow of stormwater, Environ. Sci. Technol., 2013, 47, 10791–10798 CrossRef CAS PubMed.
- J. G. Lamori, J. Xue, A. T. Rachmadi, G. U. Lopez, M. Kitajima, C. P. Gerba, I. L. Pepper, J. P. Brooks and S. Sherchan, Removal of fecal indicator bacteria and antibiotic resistant genes in constructed wetlands, Environ. Sci. Pollut. Res., 2019, 26, 10188–10197 CrossRef CAS PubMed.
- T. D. Hsu, W. J. Mitsch, J. F. Martin and J. Lee, Towards sustainable protection of public health : The role of an urban wetland as a frontline safeguard of pathogen and antibiotic resistance spread, Ecol. Eng., 2017, 108, 547–555 CrossRef.
- G. I. Chandrasena, T. Pham, E. G. Payne, A. Deletic and D. T. McCarthy, E. coli removal in laboratory scale stormwater biofilters: Influence of vegetation and submerged zone, J. Hydrol., 2014, 519, 814–822 CrossRef.
- C. P. Muerdter, C. K. Wong and G. H. LeFevre, Emerging investigator series: the role of vegetation in bioretention for stormwater treatment in the built environment: pollutant removal{,} hydrologic function{,} and ancillary benefits, Environ. Sci.: Water Res. Technol., 2018, 4, 592–612 RSC.
- W.-C. Hung, M. Rugh, M. Feraud, S. Avasarala, J. Kurylo, M. Gutierrez, K. Jimenez, N. Truong, P. A. Holden, S. B. Grant, H. Liu, R. F. Ambrose and J. A. Jay, Influence of soil characteristics and metal(loid)s on antibiotic resistance genes in green stormwater infrastructure in Southern California, J. Hazard. Mater., 2022, 424, 127469 CrossRef CAS PubMed.
- E. Gullberg, S. Cao, O. G. Berg, C. Ilbäck, L. Sandegren, D. Hughes and D. I. Andersson, Selection of resistant bacteria at very low antibiotic concentrations, PLoS Pathog., 2011, 7, 1–9 Search PubMed.
- M. Juhas, Horizontal gene transfer in human pathogens, Crit. Rev. Microbiol., 2015, 41, 101–108 CrossRef CAS PubMed.
- A. J. Lopatkin, T. A. Sysoeva and L. You, Dissecting the effects of antibiotics on horizontal gene transfer: Analysis suggests a critical role of selection dynamics, BioEssays, 2016, 38, 1283 CrossRef CAS PubMed.
- J. Bengtsson-Palme, E. Kristiansson and D. G. J. Larsson, Environmental factors influencing the development and spread of antibiotic resistance, FEMS Microbiol. Rev., 2018, 42, 68–80 CrossRef CAS PubMed.
- C. Torres-Barceló, M. Kojadinovic, R. Moxon and R. C. Maclean, The SOS response increases bacterial fitness, but not evolvability, under a sublethal dose of antibiotic, Proc. R. Soc. B: Biol. Sci., 2015, 282, 20150885 CrossRef PubMed.
- C. Miller, L. E. Thomsen, C. Gaggero, R. Mosseri, H. Ingmer and S. N. Cohen, SOS response induction by β-lactams and bacterial defense against antibiotic lethality, Science, 2004, 305, 1629–1631 CrossRef CAS PubMed.
- Z. Baharoglu and D. Mazel, Vibrio cholerae Triggers SOS and Mutagenesis in Response to a Wide Range of Antibiotics: a Route towards Multiresistance, Antimicrob. Agents Chemother., 2011, 55, 2438 CrossRef CAS PubMed.
- D. Hughes and D. I. Andersson, Selection of resistance at lethal and non-lethal antibiotic concentrations, Curr. Opin. Microbiol., 2012, 15, 555–560 CrossRef CAS PubMed.
- R. I. Aminov and R. I. Mackie, Evolution and ecology of antibiotic resistance genes, FEMS Microbiol. Lett., 2007, 271, 147–161 CrossRef CAS PubMed.
- S. Kim, Z. Yun, U. H. Ha, S. Lee, H. Park, E. E. Kwon, Y. Cho, S. Choung, J. Oh, C. A. Medriano and K. Chandran, Transfer of antibiotic resistance plasmids in pure and activated sludge cultures in the presence of environmentally representative micro-contaminant concentrations, Sci. Total Environ., 2014, 468–469, 813–820 CrossRef CAS PubMed.
- J. Jutkina, N. P. Marathe, C. F. Flach and D. G. J. Larsson, Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations, Sci. Total Environ., 2018, 616–617, 172–178 CrossRef CAS PubMed.
- R. Zhao, J. Feng, J. Liu, W. Fu, X. Li and B. Li, Deciphering of microbial community and antibiotic resistance genes in activated sludge reactors under high selective pressure of different antibiotics, Water Res., 2019, 151, 388–402 CrossRef CAS PubMed.
- H. Wu, D. Shi, D. Yang, J. Yin, Z. Yang, J. Li, W. Yang and M. Jin, Putative environmental levels of levofloxacin facilitate the dissemination of antibiotic-resistant Escherichia coli via plasmid-mediated transformability, Ecotoxicol. Environ. Saf., 2020, 195, 110461 CrossRef CAS PubMed.
- Y. Qingxiang, W. Qiang and Z. Kaiyue, Effects of Antibiotics and Metal Ions Exposure on the Natural Transformation Frequency of an Antibiotic Resistant Plasmid, Fresenius Environ. Bull., 2017, 26, 5732–5736 Search PubMed.
- Y. Zhang, A. Z. Gu, M. He, D. Li and J. Chen, Subinhibitory Concentrations of Disinfectants Promote the Horizontal Transfer of Multidrug Resistance Genes within and across Genera, Environ. Sci. Technol., 2017, 51, 570–580 CrossRef CAS PubMed.
- D. Li, S. Zeng, M. He and A. Z. Gu, Water Disinfection Byproducts Induce Antibiotic Resistance-Role of Environmental Pollutants in Resistance Phenomena, Environ. Sci. Technol., 2016, 50, 3193–3201 CrossRef CAS PubMed.
- J. Lu, Y. Wang, S. Zhang, P. Bond, Z. Yuan and J. Guo, Triclosan at environmental concentrations can enhance the spread of extracellular antibiotic resistance genes through transformation, Sci. Total Environ., 2020, 713, 136621 CrossRef CAS PubMed.
- Y. Han, Z. C. Zhou, L. Zhu, Y. Y. Wei, W. Q. Feng, L. Xu, Y. Liu, Z. J. Lin, X. Y. Shuai, Z. J. Zhang and H. Chen, The impact and mechanism of quaternary ammonium compounds on the transmission of antibiotic resistance genes, Environ. Sci. Pollut. Res., 2019, 26, 28352–28360 CrossRef CAS PubMed.
- R. I. Aminov and C. Knapp, Horizontal gene exchange in environmental microbiota, Front. Microbiol., 2011, 2, 1–19 Search PubMed.
- Y. Kohyama and S. Suzuki, Conjugative Gene Transfer between Nourished and Starved Cells of Photobacterium damselae ssp. damselae and Escherichia coli, Microbes Environ., 2019, 34, 388–392 CrossRef PubMed.
- J. Moralez, K. Szenkiel, K. Hamilton, A. Pruden and A. J. Lopatkin, Quantitative analysis of horizontal gene transfer in complex systems, Curr. Opin. Microbiol., 2021, 62, 103–109 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |