 Open Access Article
Open Access ArticleNatural deep eutectic solvents (NaDES): translating cell biology to processing
Miša Mojca
Cajnko
,
Filipa A.
Vicente
 *,
Uroš
Novak
*,
Uroš
Novak
 and
Blaž
Likozar
and
Blaž
Likozar

Department of Catalysis and Chemical Reaction Engineering, National Institute of Chemistry, Hajdrihova 19, 1000 Ljubljana, Slovenia. E-mail: filipa.andre.vicente@ki.si
First published on 26th September 2023
Abstract
This review examines the possible functional roles of liquid natural deep eutectic solvents (NaDES) in plants. Their cellular localization with biomolecular cell metabolites, a high chemical compound solubilizing capacity and a catalytic enzymatic activity indicate that they might form compartments, in which molecules with low water solubility are stored and/or converted. These environmental traits also make them an ideal reaction medium for interactions between molecules with opposite dissolution equilibria. By retaining water, stabilizing molecular structures, and yielding kinetics-determined product selectivity, they may prevent the degradation of active bonds, as in enzymes, under the conditions of high or low energy heat, drought and growth-inhibiting biomass production. Finally, the potential of NaDES has also been explored in a laboratory setting. Their ability to stabilize, catalyze and the overall substance toxicity makes NaDES an excellent candidate for (green) extraction, biocatalysis and cryopreservation, considering poorly soluble/thermally unstable intermediates.
Introduction
When NMR-based metabolomics were used to look at some of the major compounds present in cells,1 they showed that certain simple compounds like sugars, amino acids, choline and organic acids, are always present in cells in high amounts. Although sugars are a source of energy and their presence in high amounts is easily explained, other compounds do not have such a clear role in the cell metabolism. Thus, Choi et al.2 developed a novel theory about the role of these compounds in the biochemistry of living cells by hypothesizing that these cell metabolites may form a third liquid phase in cells, next to water and lipids. Their theory was based on the ability of greener solvents like ionic liquids (ILs) and deep eutectic solvents (DESs) to improve chemical and enzymatic reactions as well as enhance the extraction and dissolution of natural products. Even though these solvents share several properties, ILs and DES are still quite different solvents, starting with the fact that an IL is a single compound and a DES is a mixture of compounds. Typically, DESs are defined as a mixture of two or more pure compounds (usually in solid form), namely a hydrogen bond donor (HBD) and a hydrogen bond acceptor (HBA) that, upon mixing at specific molar ratios, establish hydrogen bonds and melt at lower temperatures, thus displaying a eutectic point temperature that is far below that of an ideal liquid mixture.3,4 Yet, the correct definition of a DES is that this is a eutectic mixture of Lewis or Brønsted acids and bases, in which it is not only required the presence of a hydrogen bond between a donor and an acceptor but there also must be a small difference in acidity between the HBD and the HBA.3,5 Choi et al.2 looked at some of these compounds and discovered more than 30 combinations in various molar ratios, that formed viscous liquids. Since these compounds were all of natural origin, they gave them a common term: natural deep eutectic solvents (NaDES).A good example of NaDES in nature are plant saps. The composition of maple (Acer spp.) syrup is mostly sucrose (Suc) with minor amounts of other sugars such as glucose (Glc) and malic acid (MalA). These compounds on their own have a melting point above 130 °C and are thus solid at room temperature, however, when mixed together, they form a viscous liquid. Another example of NaDES in nature is plant nectar, which is mostly composed of sugars and remains liquid even after freeze drying.2 This confirms the presence of NaDES mixtures in plants, but are they also present inside the cells and what is their role there? If NaDES are indeed the third liquid phase inside living cells, they may explain (i) how reactions between water-soluble and water-insoluble molecules take place, (ii) where molecules with low water solubility are stored at high concentrations and (iii) how certain organisms can survive in extreme environments. In order to answer these questions and further understand the underlying mechanisms of NaDES in nature, it is required to firstly understand them at the molecular level and in the laboratory.
As mentioned before, NaDES are mixtures of an HBA and an HBD that are generally cheap, easily available and non-toxic compounds with high solvability potential.6–11 As such, they represent good candidates for a new generation of green solvents with a wide range of interesting applications in the laboratory. This is due to their advantageous properties such as low volatility, high thermal and chemical stability, wide polarity range and adjustable viscosity. Several remain in a liquid state even at temperatures below 0 °C and possess a high degree of solubilization strength for a variety of different compounds. Furthermore, their numerous structural possibilities and the potential for adjustment and modeling of their physio-chemical properties make them ideal “designer solvents”. Some, if not most, NaDES compounds are high-purity components that are also safe for human consumption, hence making them good candidates for direct use in cosmetic, pharmaceutical and food applications without the need for their removal from the final product and/formulation.2,12–14 Consequently, this contributes to a preparation process that is waste-free.15
Among the different applications, (Na)DES have been extensively researched and reviewed,16–22 especially considering their exceptional aptitude in downstream processes23–27 and biomass processing.28–32 However, their role in living cells is still not entirely clear. Thus, in this review, we explore the potential roles of NaDES in natural environments (cells) by compiling the studies discussing phenomena that should not naturally occur if these solvents would not exist in plant cells. Herein, some laboratory studies are crucial to understand these phenomena. Additionally, we also briefly discuss the potential of NaDES in the laboratory as reaction media, extraction solvents and cryopreservation agents.
Localization and compound solubility – why NaDES are believed to be the third liquid phase in plants
NaDES are mixtures of sugars, organic acids, amino acids, choline salts and polyols, all of which can be found in living cells. However, their location and function are still unclear. Some clues have been provided by studies on the synthesis and localization of anthocyanins in cells. It was discovered that the levels of anthocyanins in cells can exceed their solubility in water,33 thus, anthocyanoplasts and anthocyanic vacuolar inclusions might be composed of NaDES. Additionally, these molecules are thought to be synthesized outside of the endoplasmic reticulum,34 which would fit the model suggested by Choi et al.2 that envisions NaDES being part of different organelles as well as attached to protein aggregates and cell membranes. It was also observed that in order to preserve liposomes upon dehydration and subsequent rehydration, sugar molecules were required on both sides of the membrane.35 Therefore, it was suggested that a dynamic NaDES layer could form around the membrane as a result of the interaction between the choline part of the membrane lipids and other NaDES components (acids, sugars…).2 Antioxidants like anthocyanins, glutathione, ascorbic acid and flavonoids can protect the membranes from oxidative damage by being present in high concentrations, dissolved in the membrane-associated NaDES. Georgieva et al.36 reported that a dense substance of unknown composition was formed in the thylakoid lumen of resurrection plant Haberlea rhodopensis chloroplasts upon dehydration. Previous studies assigned this substance a phenolic character.37 Thus, this substance may very well be the membrane-associated NaDES layer with dissolved flavonoids and/or anthocyanins that has the purpose of protecting the thylakoid membrane from oxidative damage.It is presumed that NaDES form a structure resembling liquid crystals in which the molecules are bound together through hydrogen bonds and other intermolecular interactions.8 In certain NaDES mixtures, water can be present as part of the solvent. Since this water is strongly bound to the liquid, it cannot be easily evaporated.2 The solubilizing effect that NaDES have on certain compounds is most likely due to the formation of H-bonds between NaDES components and the dissolved compound (Fig. 1). As the main intrinsic characteristic of NaDES is their H-bond network, these solvents can be fine-tuned to present a more or less extensive network, which, in turn, allows not only a better solubility of a wide range of compounds but also promotes different arrangements of the liquid crystals (steric effects).38 It was also implied that NaDES can induce some conformational changes in phenolic compounds dissolved in them, as it has been reported for curcumin38 and quercetin,39 resulting in higher stability of these compounds. These conformational changes may be the result of H-bond formation between the NaDES and the solute.39
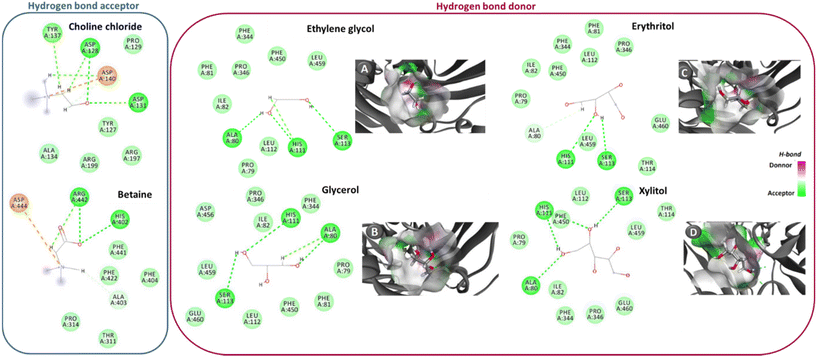 | ||
| Fig. 1 Hydrogen bonds (---, green interactions) between NaDES’ components and laccase amino acids. In the case of the hydrogen bond donors, there is also the establishment of electrostatic interactions with some of the enzyme amino acids (---, orange interactions). This data was acquired through simulation by molecular docking. Panels A–D show the docking pose with the lowest absolute value of affinity for laccase and the hydrogen bond acceptors. Adapted from ref. 40 with permission from American Chemical Society, copyright 2019. | ||
The high solubilizing capacity of numerous NaDES has previously been shown for a variety of poorly water-soluble macromolecules, small molecules, and synthetic drugs (Table 1). Dai et al.8 prepared over one hundred different NaDES mixtures and determined their viscosity, polarity, water activity, density and thermal properties as well as the effect of adding water on these physical properties. Afterwards, a few NaDES were selected and used to test their solubilizing potential on a range of compounds with poor or no water solubility. The results showed that in most cases the selected NaDES exhibited a significantly higher solubilization (18–460![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times) of the selected natural molecules that are poorly soluble or insoluble in water, like rutin, quercetin, ginkgolide B and cinnamic acid, compared to water. The best solubilizing potential was measured for the mixture of 1,2-propanediol
000 times) of the selected natural molecules that are poorly soluble or insoluble in water, like rutin, quercetin, ginkgolide B and cinnamic acid, compared to water. The best solubilizing potential was measured for the mixture of 1,2-propanediol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) choline chloride (ChCl)
choline chloride (ChCl)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), most likely because this NaDES was the least polar compared to others used. Furthermore, the addition of water to Glu
1), most likely because this NaDES was the least polar compared to others used. Furthermore, the addition of water to Glu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water DES mixture had a significant effect on the solubility of rutin, quercetin, cinnamic acid and carthamin and the amount of water was dependent on the solute.
water DES mixture had a significant effect on the solubility of rutin, quercetin, cinnamic acid and carthamin and the amount of water was dependent on the solute.
| Compound | Solubility in water | Solubility in NaDES | Ref. | |
|---|---|---|---|---|
| Abbreviations: MalA – malic acid; LacA – lactic acid; Glc – glucose; Pro – proline; ChCl – choline chloride; Xyl – xylose; Suc – sucrose; Fru – fructose; CitA – citric acid; MaleA – maleic acid; MaloA – malonic acid; Lys – lysine; AscA – ascorbic acid. 1,2PD – 1,2-propanediol; LevA – levulinic acid.a Values converted from g mol−1 to g mL−1 based on the measured NaDES density provided by the authors of the cited article.b The values were not converted to g mL−1 due to the fact that the authors did not provide any data on density of the selected NaDES. | ||||
| Macromolecules | ||||
| Salmon DNA | 26.9 × 10−3 g mL−1 | 39.4 × 10−3 g mL−1 | MalA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pro (1 Pro (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
2 |
| Starch | — | 17.2 × 10−3 g mL−1 | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (1 ChCl (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
2 |
| 0.034 g mL−1a | 1,2PD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
8 | ||
| 0.095 g mL−1a | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2 water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) 5) |
|||
| 0.027 g mL−1a | LacA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5 water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| DNA | 0.25 g mL−1a | 2.58 g mL−1a | LacA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5 water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
8 |
| Gluten | 0.0016 g mL−1a | 0.00083 g mL−1a | 1,2PD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
8 |
| 0.0012 g mL−1a | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2 water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) 5) |
|||
| 0.043 g mL−1a | LacA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5 water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| 0.0033 g mL−1a | Xyl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| Chitin | — | 102 × 10−3 g g−1b | ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LacA (1 LacA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
25 |
| 3.5 × 10−3 g g−1b | Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LacA (1 LacA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
|||
| Lignin | — | 252 × 10−3 g g−1b | 35% (w/w) Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Lys (1 Lys (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
41 |
| Small molecules | ||||
| Ginkgolide B | — | 5.85 × 10−3 g mL−1 | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (1 ChCl (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
2, 8 |
| 0.53 g mL−1a | 1,2PD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.082 g mL−1a | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2 water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) 5) |
|||
| 0.041 g mL−1a | LacA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5 water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| 0.17 g mL−1a | Xyl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| Quercetin | 0.02 × 10−3 g mL−1a | 1.63 g mL−1a | 1,2PD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
8 |
| 1.33 g mL−1a | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2 water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) 5) |
|||
| 0.042 g mL−1a | LacA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5 water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| 2.43 g mL−1a | Xyl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| Cinnamic acid | 0.0072 g mL−1a | 1.78 g mL−1a | 1,2PD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
8 |
| 0.51 g mL−1a | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2 water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) 5) |
|||
| 2.04 g mL−1a | LacA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5 water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| 0.64 g mL−1a | Xyl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| Oleanic acid | 0.017 × 10−3 g mL−1 | 3.4 × 10−3 g mL−1 | ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LevA (1 LevA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
27 |
| 1.02 × 10−3 g mL−1 | Acetamide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LacA (1 LacA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
|||
| Curcumin | 0.0006 × 10−3 g g−1b | 7.25–8.6 × 10−3 g g−1b | ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glyc (1 Glyc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
26 |
| 0.005 × 10−3 g mL−1 | 21.21 × 10−3 g mL−1 | ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LevA (1 LevA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
27 | |
| 0.88 × 10−3 g mL−1 | Acetamide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LacA (1 LacA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
|||
| 0.011 × 10−3 g mL−1 | 0.0051 × 10−3 g mL−1 | Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fru Fru![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc (1 Glc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
38, 42, 43 | |
| 0.0065 × 10−3 g mL−1 | CitA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc (1 Suc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.0521 × 10−3 g mL−1 | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc (1 Suc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.017 × 10−3 g mL−1 | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fru (1 Fru (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.0667 × 10−3 g mL−1 | MaleA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (1 ChCl (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| Benzoic acid | 3.0 × 10−3 g mL−1 | 35.0 × 10−3 g mL−1 | MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
44 |
| 18.0 × 10−3 g mL−1 | 75% (w/w) MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 11.0 × 10−3 g mL−1 | 50 wt% MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 229.0 × 10−3 g mL−1 | Urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 23 × 10−3 g mL−1 | 75% (w/w) urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 14 × 10−3 g mL−1 | 50% (w/w) urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| Rutin | 0.00055 g mL−1a | 1.48 g mL−1a | 1,2PD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
8 |
| 1.52 g mL−1a | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2 water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) 5) |
|||
| 0.33 g mL−1a | LacA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5 water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| 1.66 g mL−1a | Xyl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| 0.003 × 10−3 g mL−1 | 57.99 × 10−3 g mL−1 | ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
45 | |
| 59.3 × 10−3 g mL−1 | ChCl:1,2PD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| 4.26 × 10−3 g mL−1 | ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AscA AscA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2 water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) 5) |
|||
| 2.41 × 10−3 g mL−1 | ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MalA MalA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
|||
| Drugs | ||||
| Paclitaxel | — | 0.81 × 10−3 g mL−1 | Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (1 ChCl (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
2 |
| Danazol | <0.5 × 10−6 g mL−1 | 0.160 × 10−3 g mL−1 | MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
44 |
| 0.0044 × 10−3 g mL−1 | 75% (w/w) MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.002 × 10−3 g mL−1 | 50% (w/w) MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.048 × 10−3 g mL−1 | Urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.0061 × 10−3 g mL−1 | 75% (w/w) urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.002 × 10−3 g mL−1 | 50% (w/w) urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| Griseofulvin | 0.007 × 10−3 g mL−1 | 1 × 10−3 g mL−1 | MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
44 |
| 0.1 × 10−3 g mL−1 | 75% (w/w) MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.043 × 10−3 g mL−1 | 50% (w/w) MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.34 × 10−3 g mL−1 | Urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.016 × 10−3 g mL−1 | 75% (w/w) urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 0.015 × 10−3 g mL−1 | 50% (w/w) urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| Itraconazole | <1 × 10−6 g mL−1 | 22.0 × 10−3 g mL−1 | MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
44 |
| 6.6 × 10−3 g mL−1 | 75% (w/w) MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| 1.2 × 10−3 g mL−1 | 50% (w/w) MaloA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl (2 ChCl (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
The high solubilizing potential of NaDES was also shown in several studies. Paniwnyk et al.46 discovered that flowers of Sophora species contain high amounts of rutin. This flavonoid is poorly soluble in water but represents up to 10–30% of dry mass of the flower. This strongly suggests that rutin might only be present in such high amounts in a water-rich environment due to the presence of NaDES in plants. Paclitaxel and ginkgolide B, two compounds that do not dissolve in water, also showed high solubility in Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl-based NaDES. Wikene et al.38 tested the solubility of curcumin and showed that it was notably higher in selected NaDES compared to water.42,43 They also showed that, upon storage in airtight tubes for one month, the solubility increased up to 29% (v/v) in Glc
ChCl-based NaDES. Wikene et al.38 tested the solubility of curcumin and showed that it was notably higher in selected NaDES compared to water.42,43 They also showed that, upon storage in airtight tubes for one month, the solubility increased up to 29% (v/v) in Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc and maleic acid (MaleA)
Suc and maleic acid (MaleA)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl NaDES. Morrison et al.44 tested different poorly water-soluble model compounds in pure and diluted urea
ChCl NaDES. Morrison et al.44 tested different poorly water-soluble model compounds in pure and diluted urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl and malonic acid (MaloA)
ChCl and malonic acid (MaloA)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl mixtures and determined that the solubility in NaDES was up to 22
ChCl mixtures and determined that the solubility in NaDES was up to 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold higher compared to water. They also tested the same drugs in aqueous solutions of individual NaDES components and the results showed a significantly lower solubility than in the eutectic mixtures, proving that the combination of the HBA and HBD is crucial for improving solubility. Although these drugs were not of natural origin, the results of these studies show that the high solubilizing potential of certain NaDES extends beyond only natural compounds.
000-fold higher compared to water. They also tested the same drugs in aqueous solutions of individual NaDES components and the results showed a significantly lower solubility than in the eutectic mixtures, proving that the combination of the HBA and HBD is crucial for improving solubility. Although these drugs were not of natural origin, the results of these studies show that the high solubilizing potential of certain NaDES extends beyond only natural compounds.
The high solubilizing capacity of (Na)DES has many advantages in extraction and separation processes. Poorly water-soluble compounds can be extracted from plant biomass in high concentrations and afterwards separated from the (Na)DES by addition of water and subsequent compound precipitation. Thus, (Na)DES, alone or in combination with other methods, can be used to extract various compounds like polyphenols,47–51 pigments,52 enzymes53 and carbohydtrates.50 Their high solubilizing capacity can be exploited to overcome limited substrate solubility in enzymatic reactions54 as well as for separation techniques.55,56 In recent years, companies focused on natural ingredients have already turned their attention on using NaDES as extraction solvents for plant-based active compounds. They exploit the phenomenon they termed “eutectigenesis” which mimics the intracellular environment. The cosmetic products produced in this way were shown to have a wider range and amount of phytochemicals as well as higher biological activities compared to conventional extracts.57,58
Drought tolerance
Water is essential for cellular organization. If the cell is completely deprived of water, this organization is lost and cell death can occur. However, some organisms still manage to survive periods of severe water loss. Anhydrobiosis or “life without water” is the ability of some organisms to survive under almost complete or total water loss (dehydration), and is often used in nature to bridge periods of extreme conditions. Many organisms can survive dehydration to different extents. There are two types of dehydration tolerance: (1) drought tolerance, which is the tolerance to moderate dehydration where the moisture content is below the point where there is no bulk cytoplasmic water (∼23% water in fresh weight or ∼0.3 g H2O per g of dry weight), and (2) desiccation tolerance, which refers to further dehydration where the molecules gradually lose their hydration shells. The mechanisms of drought tolerance are based on stabilization of structures by preferential hydration, and the mechanisms of desiccation by replacing water with other molecules that are capable of H-bond formation.59 Desiccation tolerance is found throughout the plant kingdom (ferns, mosses, pollen and seeds of higher plants) as well as some other organisms such as fungi, protists, prokaryotes and animals such as crustaceans, nematodes and tardigrades.59The program for desiccation tolerance in a plant cell can be turned on by two factors, dehydration and a plant hormone called abscisic acid,59 which triggers gene expression associated with this phenomenon. It was discovered that drought tolerance is correlated with production of sugars and some amino acids like glutamate, glycine-betaine and proline, as well as molecules like compatible solutes and heat shock proteins.59 Many of these induced compounds have been shown to act as NaDES components and NMR-metabolomics of dry resurrection plants have shown the presence of some of these compounds inside the cells.2 By retaining water and preventing freezing, NaDES may help the living cells survive in drought conditions, which are the result of either high or low temperatures, by stabilizing membranes, enzymes and metabolites.2 This accumulation of metabolites has not only been shown for more extreme organisms that exhibit desiccation tolerance,59 but also for more moderate ones like the plant Arabidopsis, which showed increased levels of sugars, amino acids, organic acids and amines when exposed to water depletion.8
The effects of drought are very evident in enzymes, where the loss of water can cause enzyme denaturation and subsequent loss of function. This loss of function can not only lead to disruption of the cell's metabolic pathways, but also cause the accumulation of toxic compounds which would otherwise be inactivated with these enzymes. Knudsen and co-workers60 looked at how enzymes involved in the synthesis of a defense compound dhurrin were affected by drought. Dhurin is a natural insecticide that forms hydrogen cyanide gas upon hydrolysis61 and its synthesis is catalysed by a membrane multi-enzyme complex.62,63 The environment surrounding the enzyme complex and the produced dhurrin must be stable in order to promote the activity of the enzymes63 as well as prevent auto-hydrolysis of dhurrin at cytosolic pH.64 In drought conditions, the drought-tolerant sorghum (Sorghum bicolor) plant65 produces high amounts of dhurrin as well as NaDES forming compounds like organic acids, sugars and amino acids.66 Thus, Knudsen and co-workers60 hypothesized that dhurrin is stored in high-density NaDES-based condensates but had to show that the dhurrin metabolome is stable and/or active in such an environment (environment which mimics drought conditions). The researchers showed that, although the tested enzymes were not active in high concentrations of Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) tartrate (1
tartrate (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) NaDES, they regained a good portion of their initial activity upon dilution with water (rehydration). Similar results were shown by other groups as well.2,67 For example, it was shown that the enzyme laccase was completely dissolved in NaDES, yet it was not active. Nonetheless, when water was added to the mixture, the enzyme regained its activity.2 Similarly, Khodaverdian et al.67 used laccase from Bacillus HR03 and showed it retained about 92% and 82% of its initial activity in Sorb
1) NaDES, they regained a good portion of their initial activity upon dilution with water (rehydration). Similar results were shown by other groups as well.2,67 For example, it was shown that the enzyme laccase was completely dissolved in NaDES, yet it was not active. Nonetheless, when water was added to the mixture, the enzyme regained its activity.2 Similarly, Khodaverdian et al.67 used laccase from Bacillus HR03 and showed it retained about 92% and 82% of its initial activity in Sorb![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bet
Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and Glyc
1) and Glyc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bet (2
Bet (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), respectively, after water was added to the system. These studies all indicate that, if NaDES are present in the cell, they could act as stabilizing media in which enzymes and other active molecules are stored and protected until water is restored and their activity can be recovered.
1), respectively, after water was added to the system. These studies all indicate that, if NaDES are present in the cell, they could act as stabilizing media in which enzymes and other active molecules are stored and protected until water is restored and their activity can be recovered.
Overall, these findings suggest that NaDES could be used by the cell to stabilize enzymes and compounds when its water activity is low, such as under high environmental temperatures, drought, freezing and germination. The water shell around proteins could be substituted by the sugar components of NaDES providing an environment that protects against irreversible damage to these active molecules. Thus, the presence of NaDES in the cell in the form of the third liquid phase during desiccation, should prevent precipitation of proteins, polymers and metabolites while also preserving their function until water activity can be restored. Although the drought tolerance attributed to NaDES is not directly translatable, the stabilizing effects of (Na)DES discussed in this chapter can be exploited in a laboratory setting for storage of active molecules, thermal stabilization and reactions that require a water-free medium, which will be discussed in the following chapters.
Thermal tolerance
Water evaporation due to high temperatures leads to drought conditions – water loss in living cells. However, high temperatures also have other consequences like thermal deactivation of enzymes as a result of denaturation. Previous studies have shown that certain NaDES can provide favourable environments which preserve enzyme activities68 by stabilizing their secondary structures.69,70 For example, betaine (Bet), a common component of NaDES, has been shown to protect proteins against denaturation and aggregation.71 Thermal stability of laccase from Bacillus HR03 was tested at 80 °C in absence and presence of 20% (v/v) NaDES.67 The activity of the enzyme was markedly increased in 20% (v/v) sorbitol (Sorb)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bet
Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), where the half-life of the enzyme at 80 °C was 43 minutes, compared to only 7 minutes in aqueous buffer. The authors showed that Sorb
1), where the half-life of the enzyme at 80 °C was 43 minutes, compared to only 7 minutes in aqueous buffer. The authors showed that Sorb![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bet
Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water has 6 hydroxyl groups that bind to the enzyme through hydrogen bonds, making its structure more rigid and stable, thus, preventing it from unfolding (denaturation) at high temperatures. In another study, the thermal stability of laccase from Trametes versicolor was tested in various ChCl- and Bet-based aqueous NaDES.72 The enzyme was incubated at high temperatures in buffer or in 25% (w/w) aqueous solution of various NaDES. Although the relative activity of laccase did not increase, the thermal inactivation of laccase at 60, 70 and 90 °C was significantly slower in 25% (w/w) Bet
water has 6 hydroxyl groups that bind to the enzyme through hydrogen bonds, making its structure more rigid and stable, thus, preventing it from unfolding (denaturation) at high temperatures. In another study, the thermal stability of laccase from Trametes versicolor was tested in various ChCl- and Bet-based aqueous NaDES.72 The enzyme was incubated at high temperatures in buffer or in 25% (w/w) aqueous solution of various NaDES. Although the relative activity of laccase did not increase, the thermal inactivation of laccase at 60, 70 and 90 °C was significantly slower in 25% (w/w) Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylitol (1
xylitol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) NaDES compared to buffer. Furthermore, the half-life of the enzyme at 60 °C was increased from around 30 minutes in buffer to several hours in 25% (w/w) Bet
3) NaDES compared to buffer. Furthermore, the half-life of the enzyme at 60 °C was increased from around 30 minutes in buffer to several hours in 25% (w/w) Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylitol (1
xylitol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). The observed effects were shown to be due to the action of NaDES and not its individual components. The ChCl-based mixtures did not perform as well as Bet-based formulations, which shows the importance of the HBA selection. The choice of HBD, HBA
3). The observed effects were shown to be due to the action of NaDES and not its individual components. The ChCl-based mixtures did not perform as well as Bet-based formulations, which shows the importance of the HBA selection. The choice of HBD, HBA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HBD ratio and dilution factor also play an important role. The researchers showed that Bet
HBD ratio and dilution factor also play an important role. The researchers showed that Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sorb and Bet
Sorb and Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylitol significantly increased the thermostability, but exchanging the HBD for propanediol showed no effect compared to phosphate buffer. Different Bet
xylitol significantly increased the thermostability, but exchanging the HBD for propanediol showed no effect compared to phosphate buffer. Different Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylitol ratios and aqueous dilutions were also tried by the same group. They determined that the ratio 2
xylitol ratios and aqueous dilutions were also tried by the same group. They determined that the ratio 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 75% dilution (25% aqueous solution of NaDES) were the most advantageous. Delorme et al.72 and Toledo et al.40 both showed that the higher number of OH-groups of the HBD correlates with higher thermostability. Toledo et al.40 suggested that this could be due to the formation of H-bonds between OH-groups on the HBD and the amino acids of the enzyme, thus, a higher number of H-bonds would result in better stabilization of the protein structure (Fig. 2). Interestingly, one study also showed that some NaDES not only provide better thermostabilizing effects at high temperatures but also enable enzymatic reactions at low temperatures.6 The authors used horseradish peroxidase in enzyme-mediated free radical polymerization reactions and showed that the reactions take place in ChCl
1 and 75% dilution (25% aqueous solution of NaDES) were the most advantageous. Delorme et al.72 and Toledo et al.40 both showed that the higher number of OH-groups of the HBD correlates with higher thermostability. Toledo et al.40 suggested that this could be due to the formation of H-bonds between OH-groups on the HBD and the amino acids of the enzyme, thus, a higher number of H-bonds would result in better stabilization of the protein structure (Fig. 2). Interestingly, one study also showed that some NaDES not only provide better thermostabilizing effects at high temperatures but also enable enzymatic reactions at low temperatures.6 The authors used horseradish peroxidase in enzyme-mediated free radical polymerization reactions and showed that the reactions take place in ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol (1
glycerol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) even at 4 °C, while no polymer was obtained in water at the same temperature.
2) even at 4 °C, while no polymer was obtained in water at the same temperature.
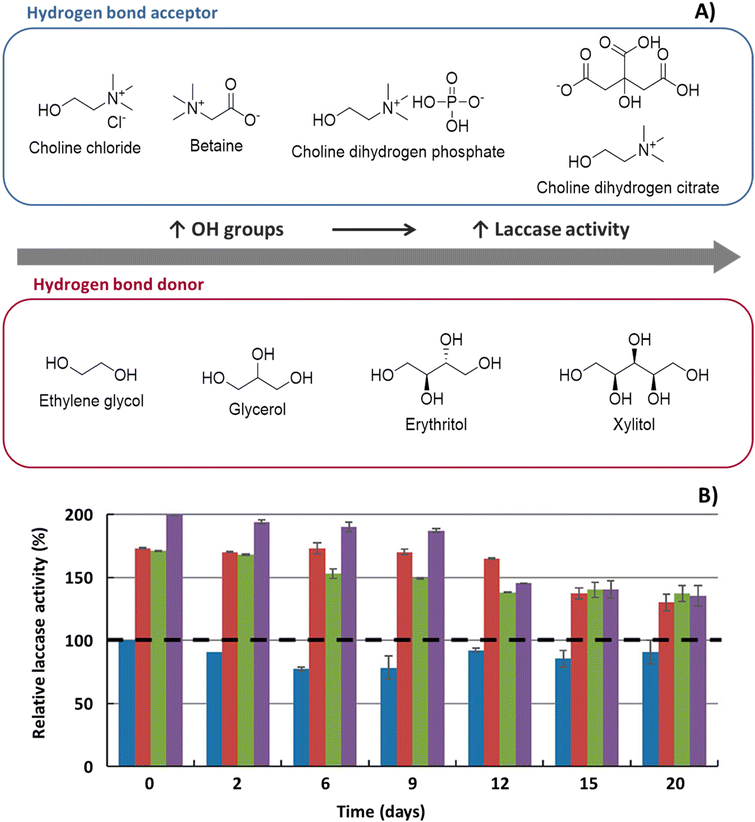 | ||
Fig. 2 A. Influence of the number of hydroxyl groups from the NaDES HBA and HBD upon laccase activity. B. Influence of NaDES over the thermal activity of laccase:  , control; , control;  , 10 wt% choline dihydrogen phosphate ([Ch]DHP) , 10 wt% choline dihydrogen phosphate ([Ch]DHP)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylitol (Xyl), (1 xylitol (Xyl), (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) molar ratio; 2) molar ratio;  , 25 wt% [Ch]DHP , 25 wt% [Ch]DHP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Xyl, (1 Xyl, (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) molar ratio; 2) molar ratio;  , 25 wt% choline dihydrogen citrate ([Ch]DHC) , 25 wt% choline dihydrogen citrate ([Ch]DHC)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylitol (Xyl), (1 xylitol (Xyl), (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) molar ratio. Adapted from ref. 40 with permission from American Chemical Society, copyright 2019. 2) molar ratio. Adapted from ref. 40 with permission from American Chemical Society, copyright 2019. | ||
NaDES were shown to also have a protective effect against other environmental effects that are associated with high temperatures, namely radiation. Dai et al.39 showed that natural colorants from safflower (Carthamus tinctorius) are more stable in sugar-based NaDES compared to water or 40% ethanol when exposed to both artificial light or sunlight. Carthamin, one of the major red pigments in safflower, has been shown to be very unstable in aqueous solutions.39,73,74 However, when carthamin, and other pigments extracted from safflower, were solubilized in selected NaDES (90% (v/v) Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2
water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) and 75% Suc
5) and 75% Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4)), their stability in sunlight conditions increased significantly compared to aqueous solution.
4)), their stability in sunlight conditions increased significantly compared to aqueous solution.
Not only high but also low temperatures can be detrimental to a living cell. Freezing can damage the cell mostly via mechanical damage caused by intra- and extracellular water crystals that form during the freezing process.75 It was shown that wheat (Triticum aestivum) tolerance to freezing was correlated with high proline (Pro) levels.76 This correlates with the results that show that proline is a good candidate for the formation of NaDES with organic acids and sugars.2 Freezing-related damage is not an issue only in nature but also in the laboratory when preservation of living cells or active molecules like enzymes (cryopreservation) is in question. In order to make the process of cryopreservation effective, cryoprotective agents (CPAs) have to be used. These CPAs prevent the formation of ice crystals via the formation of H-bonds with water.77 However, CPAs must be able to penetrate into the cell where the damage of ice crystal formation is the most detrimental, but at the same time not cause any significant toxic effects. CPAs like ethylene glycol, glycerol and DMSO penetrate cell membranes and increase cryopreservation, but can also cause cellular toxicity. On the other hand, non-toxic CPAs like sugars, amino acids and skim milk, have been studied and showed beneficial effects. However, they could not penetrate cell membranes, which diminishes their effectiveness as cryoprotectants.78,79 Because of their ability to form H-bonds with water as well as their low toxicity and favourable biodegradability,80 NaDES seem like promising candidates for novel and green cryopreservation solvents. Qiao et al.81 studied the effect of NaDES on cryopreservation of lactic acid bacteria, while using buffer as a control. Streptococcus thermophilus survival rate at −20 °C after 24 h in three different NaDES – ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylitol
xylitol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), ChCl
3), ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc
Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2
water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) and ChCl
5) and ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sorb
Sorb![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2
water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) – was 87–95%. The survival rate after 180 days was significantly higher in Glyc
6) – was 87–95%. The survival rate after 180 days was significantly higher in Glyc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pro (3
Pro (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) compared to buffer or glycerol alone, with only minimal disruption of cellular components and enzyme activities. The authors also determined that the bacterial suspension
1) compared to buffer or glycerol alone, with only minimal disruption of cellular components and enzyme activities. The authors also determined that the bacterial suspension![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaDES ratio is of vital importance and the optimal ratio was determined to be 1
NaDES ratio is of vital importance and the optimal ratio was determined to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v). Moreover, when the activity of individual enzymes, lactate dehydrogenase and β-galactosidase, was tested after 30 and 180 days of cryopreservation in selected NaDES, it too showed positive results compared to buffer. In certain cases, the enzyme activity was even increased. The authors also showed that, at freezing temperatures, NaDES were able to prevent the formation of ice crystals and, thus, prevent mechanical damage to the cell and its components. The cryoprotective effect was also shown on small molecules. Storage of carthamin and other pigments extracted from safflower (Carthamus tinctorius) at −20 °C and 4 °C resulted in significantly lower degradation in NaDES, especially in Suc
1 (v/v). Moreover, when the activity of individual enzymes, lactate dehydrogenase and β-galactosidase, was tested after 30 and 180 days of cryopreservation in selected NaDES, it too showed positive results compared to buffer. In certain cases, the enzyme activity was even increased. The authors also showed that, at freezing temperatures, NaDES were able to prevent the formation of ice crystals and, thus, prevent mechanical damage to the cell and its components. The cryoprotective effect was also shown on small molecules. Storage of carthamin and other pigments extracted from safflower (Carthamus tinctorius) at −20 °C and 4 °C resulted in significantly lower degradation in NaDES, especially in Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ChCl
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). This was most likely due to the high viscosity of this NaDES as it restricts the movement (reduced mass diffusivity) of molecules and allows more stable molecular interactions. This conclusion was reached since diluting the NaDES to lower the viscosity resulted in a decrease of their stabilizing effects.39
4). This was most likely due to the high viscosity of this NaDES as it restricts the movement (reduced mass diffusivity) of molecules and allows more stable molecular interactions. This conclusion was reached since diluting the NaDES to lower the viscosity resulted in a decrease of their stabilizing effects.39
Although NaDES have numerous potential applications and have already been used in food and pharmaceutical industries with beneficial safety and efficiency,80,82 not all NaDES are completely safe for all organisms.83,84 Therefore, it is evident that NaDES composition is of high importance and that not all NaDES are suitable for all purposes. Care must be taken when selecting the appropriate components for a desired purpose.
Based on the research presented in this section, the potential applications of (Na)DES is evident. The thermostabilizing effects of (Na)DES could allow enzymatic biocatalytic reactions at higher temperatures that subsequently result in higher reaction kinetics and higher product yields. Their ability to form H-bonds with water prevents ice crystal formation and consequent cell damage, making (Na)DES good candidates for cryopreservation. Additionally, their protective effects against radiation as well as thermostabilization, can be applied for storage of light and temperature-sensitive compounds like enzymes and pigments.
Medium for biocatalytic reactions
Generally, in enzyme-catalysed reactions, the reaction medium has two main functions: (i) provide a stable environment for the enzymes and (ii) enable the interaction between the enzyme and its substrate (i.e. the enzyme is active in the medium and both the enzyme and substrate are solubilized to allow for efficient mass transfer). It is thought that enzymatic reactions occur in an aqueous medium, but the question is, how these reactions function when molecules (reactants or products) with poor water-solubility are involved. One example is the synthesis of polymers like lignin, amylose and cellulose, which are not soluble in water, but likely need to be dissolved at some point of the reaction to allow further addition of monomers. In addition to the excellent solubilizing capacity of (Na)DES14,44 that would allow solubilization of these polymers and other compounds, research has shown that enzymes are also stable in synthetic ILs and DES and can be used in green chemistry.85–91 Therefore, it was implied that biosynthetic reactions between compounds of different water solubilities may occur in NaDES inside living cells. The model by Choi et al.2 mentioned in the section on Localization and compound solubility, envisions NaDES being part of different organelles as well as attached to protein aggregates and cell membranes where they act as media for reactions between enzymes and their poorly water-soluble substrates. Although this model or the general hypothesis of NaDES being the third liquid phase in cells has not yet been confirmed, there is strong evidence based on biocatalytic studies to suggest that this is the case.The field of biocatalytic reactions in (Na)DES has been extensively reviewed,16,17,92,93 with (Na)DES being able to act as (i) a co-solvent, (ii) a reaction medium, or (iii) a reaction medium and substrate, as shown in Fig. 3. Current research on enzymatic biocatalysis has shown significant differences in enzyme activities in aqueous buffer media compared to NaDES. A good example are laccases. Laccases are multi-copper enzymes, found in plants and fungi, that can oxidize a wide variety of aromatic substrates.94 However, not all of these substrates are soluble in water. Subsequently, organic solvents had to be used when performing laboratory tests.95 In order to find a more biocompatible approach, Khodaverdian et al.67 tested the activity of laccase from Bacillus HR03 in different ammonium-based (ChCl and Bet) NaDES with various HBDs and compared them to the activity in an aqueous buffer. Although ChCl-based NaDES resulted in a decrease in velocity of the reaction, likely due to an inhibitory effect of choline ions on laccase,96 some Bet-based mixtures resulted in an increase in laccase activity (150–300%) in 20% (v/v) NaDES.67 And a study by Toledo et al.40 showed that laccase is twice as active in 50% aqueous solution of choline dihydrogen citrate-based NaDES than in aqueous buffer. A very significant effect on enzyme activity was also measured for several lipase enzymes, where the researchers showed up to a 355% increase in ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc
Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (4
water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) and ChCl
4) and ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc
Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5
water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5).97 Another study showed a 156% increase in activity of lipase B from Candida antarctica when incubated in ChCl
5).97 Another study showed a 156% increase in activity of lipase B from Candida antarctica when incubated in ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol (1
glycerol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) with 0.3 mole fraction of water with a relatively high recycle stability (90.12%).98 A 55% increase in lipase activity was observed with ChCl
2) with 0.3 mole fraction of water with a relatively high recycle stability (90.12%).98 A 55% increase in lipase activity was observed with ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea
urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol (1
glycerol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) although the NaDES was used only as the co-solvent and was present in low (10%) concentrations.99 The authors also showed synergistic effects of NaDES components.
1) although the NaDES was used only as the co-solvent and was present in low (10%) concentrations.99 The authors also showed synergistic effects of NaDES components.
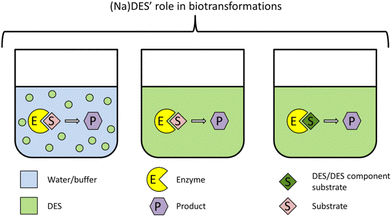 | ||
| Fig. 3 Role of (Na)DES in bio-transformations, namely as (i) a co-solvent, (ii) the reaction medium, or (iii) the reaction medium and substrate. | ||
Another interesting feature of NaDES was shown by Schweiger et al.54 on phenolic acid decarboxylase from Bacillus subtilis. The study not only showed increased reactivity against poorly water-soluble phenolic substrates but also that the choice of solvent strongly influences the choice of enzyme's substrates. Similarly, very low concentrations of NaDES showed a notable effect on laccase from Myceliophthora thermophila100 where the addition of only 8% (v/v) of Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lactic acid (LacA) (1
lactic acid (LacA) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) to sodium acetate buffer increased laccase activity by 300%.
2) to sodium acetate buffer increased laccase activity by 300%.
A marked effect was also observed for reaction kinetics, but, these effects varied considerably and did not always point to increased enzyme activity (Table 2). In the case of laccase from Bacillus HR03, enzyme kinetics in 20% (v/v) NaDES showed increased Michaelis constant (KM) and turnover numbers (kcat), which could be the result of high viscosity of eutectic mixtures containing sugars or carboxylic acids as HBDs.67 Also, interactions between the substrate and the HBD component of NaDES could affect binding affinity of the enzyme resulting in a higher KM value. As mentioned in section on Thermal tolerance, the hydroxyl groups of Sorb![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bet
Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water bind to the enzyme and make its structure more rigid and more stable. Although this effect increases the enzyme's thermal stability, it also limits the interactions between the enzyme and its substrate. An increase in KM value was also observed for potato epoxide hydrolase StEH1 in aqueous solutions of NaDES, though, kcat remained relatively unaffected.101 Thus, the loss of enzyme activity at higher NaDES concentrations was likely due to the destabilization of the enzyme/substrate or reaction intermediate complexes and not the enzyme denaturation. On the other hand, when Elgharbawy et al.97 performed lipase activity experiments in ChCl
water bind to the enzyme and make its structure more rigid and more stable. Although this effect increases the enzyme's thermal stability, it also limits the interactions between the enzyme and its substrate. An increase in KM value was also observed for potato epoxide hydrolase StEH1 in aqueous solutions of NaDES, though, kcat remained relatively unaffected.101 Thus, the loss of enzyme activity at higher NaDES concentrations was likely due to the destabilization of the enzyme/substrate or reaction intermediate complexes and not the enzyme denaturation. On the other hand, when Elgharbawy et al.97 performed lipase activity experiments in ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc
Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (4
water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) and 40% (v/v) ChCl
4) and 40% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc (4
Suc (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) NaDES, the results were somewhat different. They clearly showed a direct effect of the reaction medium on reaction kinetics with a reduction of KM and an increase of kcat. The extent of this effect was also dependent on the type of lipase. These results were in agreement with a previous study that showed that ChCl enhanced the activity of an alcohol dehydrogenase via increasing its substrate affinity (reduction of KM value).102 In contrast, the study by Chan et al.100 showed that ChCl-based NaDES had a notable inhibitory effect on laccase enzyme kinetics even at very low concentrations. When only 5% (v/v) of ChCl
1) NaDES, the results were somewhat different. They clearly showed a direct effect of the reaction medium on reaction kinetics with a reduction of KM and an increase of kcat. The extent of this effect was also dependent on the type of lipase. These results were in agreement with a previous study that showed that ChCl enhanced the activity of an alcohol dehydrogenase via increasing its substrate affinity (reduction of KM value).102 In contrast, the study by Chan et al.100 showed that ChCl-based NaDES had a notable inhibitory effect on laccase enzyme kinetics even at very low concentrations. When only 5% (v/v) of ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol (1
glycerol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) was added to sodium acetate buffer, the maximum rate of reaction (vmax) decreased to half whereas the KM increased 52%. Two more important NaDES features were presented in literature, amount of water and pH. Juneidi et al.103 used amano lipase from Burkholderia cepaci and tested its activity in pure, 96 and 40% (v/v) NaDES. The results showed an over 6-fold decrease in KM and a 5-fold increase in Kcat with the addition of only 4% of water (96% NaDES). Pure NaDES also resulted in a higher KM and lower Kcat compared to buffer. With the addition of 60% water (40% NaDES), the KM increased slightly, but was still much lower compared to pure NaDES. A very similar experiment was performed by Xu et al.104 with β-glicosidase with almost identical results, indicating that a small amount of water can have a very significant effect on reaction kinetics. Mamashli et al.105 went a step further and tested not only different amounts of water but also the pH of the medium. They used versatile peroxidase from Bjerkandera adusta and measured its activity in buffer and in 5, 10 and 25% (v/v) ChCl
2) was added to sodium acetate buffer, the maximum rate of reaction (vmax) decreased to half whereas the KM increased 52%. Two more important NaDES features were presented in literature, amount of water and pH. Juneidi et al.103 used amano lipase from Burkholderia cepaci and tested its activity in pure, 96 and 40% (v/v) NaDES. The results showed an over 6-fold decrease in KM and a 5-fold increase in Kcat with the addition of only 4% of water (96% NaDES). Pure NaDES also resulted in a higher KM and lower Kcat compared to buffer. With the addition of 60% water (40% NaDES), the KM increased slightly, but was still much lower compared to pure NaDES. A very similar experiment was performed by Xu et al.104 with β-glicosidase with almost identical results, indicating that a small amount of water can have a very significant effect on reaction kinetics. Mamashli et al.105 went a step further and tested not only different amounts of water but also the pH of the medium. They used versatile peroxidase from Bjerkandera adusta and measured its activity in buffer and in 5, 10 and 25% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Urea (1
Urea (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) at two different pH, 4.5 and 7. At pH 7, all of the NaDES dilutions had a 2 to 5-fold lower KM compared to buffer (the higher the amount of water, the lower the KM). At pH 4.5, a decrease in KM with the increase of the amount of water in NaDES was also measured, however, even at the highest water content, the KM value was still over 1.5-fold higher than in buffer. This indicates that the effect of pH is more significant than the effect of water content when comparing buffer to NaDES media. Taken together, it seems that the increase of enzyme activity in NaDES could be a result of better solubility of substrates and/or enzymes in this medium as well as due to stabilizing effects of NaDES on the enzyme structure. However, these results also indicate that specific NaDES mixtures may have different roles: the less viscous or more diluted can act as a reaction medium and the more viscous or less diluted serve as stabilizers at high/low temperatures and low water conditions.
2) at two different pH, 4.5 and 7. At pH 7, all of the NaDES dilutions had a 2 to 5-fold lower KM compared to buffer (the higher the amount of water, the lower the KM). At pH 4.5, a decrease in KM with the increase of the amount of water in NaDES was also measured, however, even at the highest water content, the KM value was still over 1.5-fold higher than in buffer. This indicates that the effect of pH is more significant than the effect of water content when comparing buffer to NaDES media. Taken together, it seems that the increase of enzyme activity in NaDES could be a result of better solubility of substrates and/or enzymes in this medium as well as due to stabilizing effects of NaDES on the enzyme structure. However, these results also indicate that specific NaDES mixtures may have different roles: the less viscous or more diluted can act as a reaction medium and the more viscous or less diluted serve as stabilizers at high/low temperatures and low water conditions.
| Enzyme | Medium | K M [mM] | k cat [s−1] | Ref. | |
|---|---|---|---|---|---|
| a The values were converted from min−1 to s−1. b The values refer to an esterification reaction where the concentration of one substrate was constant and the concentration of the other substrate was varied and vice versa. c The values refer to different substrates for the same enzyme. Abbreviations: Glyc – glycerol; Bet – betaine; MalA – malic acid; Sorb – sorbitol; ChCl – choline chloride; Suc – sucrose; EG – ethylene glycol; EAC – N,N-diethyl ethanol ammonium chloride; PG – propylene glycol; Chol DHP – choline dihydrogen phosphate; Fru – fructose; MTPB – methyltriphenylphosphonium bromide. | |||||
| Laccase from Bacillus HR03 | Citrate phosphate buffer | 76.0 × 10−3 ± 3.8 × 10−3 | 81.7 ± 4.0 | 67 | |
20% (v/v) Glyc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bet (2 Bet (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
649.0 × 10−3 ± 32 × 10−3 | 295.2 ± 14.5 | |||
20% (v/v) MalA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bet Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
870 × 10−3 ± 43 × 10−3 | 144.4 ± 5.8 | |||
20% (v/v) Sorb![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bet Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
861 × 10−3 ± 42 × 10−3 | 98.0 ± 4.9 | |||
Aqueous solution of Glyc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bet (2 Bet (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
280 × 10−3 ± 14.5 × 10−3 | 37.3 ± 1.8 | |||
| Potato epoxide hydrolase StEH1 | Phosphate buffer | 77 × 10−3 ± 10 × 10−3 | 63.0 ± 3.0 | 101 | |
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glyc (1 Glyc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
20% (v/v) | 120 × 10−3 ± 10 × 10−3 | 51 ± 2 | ||
| 40% (v/v) | 250 × 10−3 ± 40 × 10−3 | 57 ± 5 | |||
| 60% (v/v) | 530 × 10−3 ± 90 × 10−3 | 44 ± 5 | |||
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea (1 urea (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
20% (v/v) | 360 × 10−3 ± 40 × 10−3 | 65 ± 4 | ||
| 40% (v/v) | 1500 × 10−3 ± 30 × 10−3 | 92 ± 10 | |||
| Lipase from porcine pancreas | Phosphate buffer | 1.05 ± 0.52 | 0.085 ± 0.002a | 97 | |
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (4 water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
0.78 ± 0.09 | 0.26 ± 0.02a | |||
40% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc (4 Suc (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
0.53 ± 0.08 | 0.33 ± 0.04a | |||
| Amano lipase PS from Burkholderia cepacia | Phosphate buffer | 1.52 ± 0.25 | 0.12 ± 0.02a | ||
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (4 water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
0.68 ± 0.07 | 0.36 ± 0.04a | |||
40% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc (4 Suc (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
0.61 ± 0.08 | 0.41 ± 0.05a | |||
| Lipase from Rhizopus niveus | Phosphate buffer | 1.38 ± 0.92 | 0.084 ± 0.002a | ||
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (4 water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
0.66 ± 0.08 | 0.25 ± 0.03a | |||
40% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc (4 Suc (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
0.45 ± 0.05 | 0.30 ± 0.04a | |||
| Immobilized (acrylic resin) lipase from Candida antarctica | Phosphate buffer | 1.25 ± 0.25 | 0.08 ± 0.02a | ||
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (4 water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
1.08 ± 0.09 | 0.08 ± 0.03a | |||
40% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc (4 Suc (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
0.49 ± 0.05 | 0.23 ± 0.03a | |||
| Lipase from Candida rugosa | Phosphate buffer | 1.55 ± 0.35 | 0.089 ± 0.002a | ||
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (4 water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
0.57 ± 0.06 | 0.27 ± 0.03a | |||
40% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc (4 Suc (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
0.45 ± 0.07 | 0.31 ± 0.04a | |||
| Immobilized (Immobead) lipase B from Candida antarctica | Phosphate buffer | 0.61 ± 0.04 | 0.051 ± 0.002a | ||
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (4 water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
0.52 ± 0.03 | 0.17 ± 0.04a | |||
40% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc (4 Suc (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
0.42 ± 0.08 | 0.38 ± 0.05a | |||
| ω-Transaminase | Phosphate buffer | 8.00 ± 4.22c | 3.59 ± 1.5a,b | 106 | |
| 0.69 ± 0.13c | 1.25 ± 0.008a,c | ||||
10% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea (1 urea (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
3.27 ± 0.41b | 2.4 ± 0.2a,b | |||
| 0.49 ± 0.16c | 1.71 ± 0.2a,c | ||||
| Microreactor immobilized lipase from Candida antarctica | Phosphate buffer | 0.62 | — | 107 | |
10% (v/v) Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly |
0.3 | — | |||
| Amano lipase from Burkholderia cepaci | Phosphate buffer | 1.78 × 10−3 ± 0.04 | 0.48 ± 0.07a | 103 | |
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EG (1 EG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
Pure | 3.5 × 10−3 ± 1.2 | 0.22 ± 0.02a | ||
| 96% (v/v) | 0.53 × 10−3 ± 0.2 | 1.07 ± 0.1a | |||
| 40% (v/v) | 0.75 × 10−3 ± 0.21 | 0.9 ± 0.09a | |||
EAC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EG (1 EG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
Pure | 4.03 × 10−3 ± 1.1 | 0.059 ± 0.01a | ||
| 96% (v/v) | 0.61 × 10−3 ± 0.08 | 0.37 ± 0.06a | |||
| 40% (v/v) | 0.92 × 10−3 ± 0.4 | 0.31 ± 0.05a | |||
| β-Glucosidase | Phosphate buffer | 3.12 × 10−3 ± 1.40 | 0.38 ± 0.1a | 104 | |
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EG (1 EG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
Pure | 5.0 × 10−3 ± 0.08 | 0.12 ± 0.03a | ||
| 94% (v/v) | 0.52 × 10−3 ± 0.18 | 0.49 ± 0.1a | |||
| 40% (v/v) | 0.48 × 10−3 ± 0.02 | 0.56 ± 0.03a | |||
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PG (1 PG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
Pure | 5.08 × 10−3 ± 2.45 | 0.34 ± 0.1a | ||
| 94% (v/v) | 0.21 × 10−3 ± 0.04 | 0.58 ± 0.01a | |||
| 10% (v/v) | 0.49 × 10−3 ± 0.07 | 0.28 ± 0.03a | |||
| ω-Transaminase | Phosphate buffer | 0.64 ± 0.07b | 0.55 ± 0.0005a,b | 108 | |
| 0.65 ± 0.07b | 0.8 ± 0.0008a,b | ||||
10% ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EG (1 EG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
0.60 ± 0.06b | 0.68 ± 0.001a,b | |||
| 0.52 ± 0.04b | 1.59 ± 0.001a,b | ||||
10% ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EG:1,2-PD EG:1,2-PD |
0.51 ± 0.03b | 0.76 ± 0.0003a,b | |||
| 0.48 ± 0.02b | 1.71 ± 0.0006a,b | ||||
| Lipase from Rhizopus Niveus | Phosphate buffer | 1.27 | 0.211a | 109 | |
Menthol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) octanoic acid (1 octanoic acid (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
0.27 | 0.36a | |||
Menthol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) decanoic acid decanoic acid |
0.078 | 0.43a | |||
| Laccase from Myceliophthora thermophila | Sodium acetate buffer | 52.151 × 10−3 ± 2.0 | — | 100 | |
1% ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly (1 Gly (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
55.843 × 10−3 ± 2.8 | — | |||
5% ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly (1 Gly (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
79.011 × 10−3 ± 2.2 | — | |||
| Tyrosinase mCLEAs | Phosphate buffer | 9.7 ± 1.4 | — | 110 | |
10% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly (1 Gly (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
12.5 ± 1.9 | — | |||
10% (v/v) EAC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly (1 Gly (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5) 1.5) |
11.0 ± 1.6 | — | |||
10% (v/v) Bet![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly (1 Gly (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
9.0 ± 2.0 | — | |||
10% (v/v) Chol DHP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly (1 Gly (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
27.6 ± 5.2 | — | |||
10% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea (1 urea (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
15.2 ± 2.7 | — | |||
10% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly (1 Gly (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
12.1 ± 1.8 | — | |||
10% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BG (1 BG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
22.2 ± 4.2 | — | |||
10% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fru Fru![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5 water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) 5) |
9.9 ± 0.9 | — | |||
| Lipase from porcine pancreas | Phosphate buffer | 1.05 ± 0.52 | 0.084 ± 0.001a | 111 | |
ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.4 3.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
0.008 ± 0.001 | 0.014 ± 0.04a | |||
MTPB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.4 3.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
0.14 ± 0.05 | 0.038 ± 0.03a | |||
| β-Glucosidase | Acetate buffer | 10.31 ± 0.98c | — | 112 | |
| 13.35 ± 0.74c | |||||
| 17.32 ± 0.52c | |||||
30% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EG (2 EG (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
2.31 ± 0.38c | — | |||
| 4.96 ± 0.29c | |||||
| 5.76 ± 0.42c | |||||
| Versatile peroxidase from Bjerkandera adusta | Sodium tartrate buffer | pH 4.5 | 16.1 × 10−3 | 0.035 × 105a | 105 |
25% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea (1 urea (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
36.1 × 10−3 | 0.068 × 105a | |||
10% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea (1 urea (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
29.1 × 10−3 | 0.068 × 105a | |||
5% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea (1 urea (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
27.1 × 10−3 | 0.073 × 105a | |||
| Sodium tartrate buffer | pH 7 | 37.1 × 10−3 | 0.048 × 105a | ||
25% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea (1 urea (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
15.8 × 10−3 | 0.16 × 105a | |||
10% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea (1 urea (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
11.8 × 10−3 | 0.046 × 105a | |||
5% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea (1 urea (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
6.8 × 10−3 | 0.058 × 105a | |||
| β-D-Glucosidase | Phosphate buffer | 1.8 | — | 113 | |
30% (v/v) ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EG (2 EG (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
0.85 | — | |||
Despite the fact that the existence and/or function of NaDES in living cells has not yet been confirmed, the advantages that (Na)DES possess have been shown to be very beneficial in a laboratory setting.16,17,90 Biocatalytic reactions are reactions where enzymes or whole cells are used as catalysts for the conversion of various substrates (organic and inorganic).114,115 Enzymes have a highly catalytic and selective nature, are produced from renewable resources, are biodegradable and operate under mild conditions.114 All of these features make enzymes a very desirable option in green chemistry. However, the fact that some widely used enzymes like laccases have poor stability in commonly used process conditions (high temperatures, pH values outside of 6–7),116–118 presents a bottleneck for their widespread use. The high operating temperatures are often associated with higher reaction rates and yields but rapidly inactivate the enzyme.119,120 Chemical modification and immobilization are two common strategies for improving enzyme stability, yet, these techniques are often complex, non-sustainable and have a high cost.121,122 Due to their biocompatibility with enzymes, NaDES are promising non-toxic alternatives for biotechnological applications.123 Also, not all enzyme substrates are water soluble and, therefore, they either cannot be converted or organic solvents have to be used.95 Since NaDES have been shown to be excellent solvents for a variety of compounds, which are otherwise poorly soluble or not soluble in water (Table 1), they could expand the set of substrates that an enzyme can convert, thus, broadening an enzyme's use in technological processes. Furthermore, and as previously discussed in this section, some NaDES could enhance the enzyme's activity,40,67 which means that higher reaction rates can be achieved, thus, increasing the kinetics of the enzyme-catalysed reaction and subsequent product yields.
NaDES have also been shown to be beneficial in whole-cell biocatalysis. Yang et al.124 tested the conversion of isoeugenol to vanillin using Lysinibacillus fusiformis and showed that most of the NaDES tested improved product yields compared to aqueous buffer. The yields were improved up to 132% when 20% ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Raffinose (11
Raffinose (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) or ChCl
2) or ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Lactose (4
Lactose (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were added to the buffer. This was due to increased solubility of the substrate and increased permeability of the cell membrane, both of which increased the accessibility of the substrate to the enzymes. Baker's yeast have also been shown to be stable in selected NaDES (ChCl
1) were added to the buffer. This was due to increased solubility of the substrate and increased permeability of the cell membrane, both of which increased the accessibility of the substrate to the enzymes. Baker's yeast have also been shown to be stable in selected NaDES (ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol 1
glycerol 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) and were able to sustain ketone reduction for over 200 hours.125 Additionally, in these whole-cell reactions, HBDs can also play a role as co-substrates for cofactor regeneration or as a nutrition source for the cells.126,127
2) and were able to sustain ketone reduction for over 200 hours.125 Additionally, in these whole-cell reactions, HBDs can also play a role as co-substrates for cofactor regeneration or as a nutrition source for the cells.126,127
Another interesting and valuable feature of NaDES is their effect on reaction stereoselectivity.125,128–131 Since two enantiomers of the same compound can have different and sometimes even opposite activities, the synthesis of products with high enantiomeric purity is essential in the pharmaceutical industry.128 Pavoković et al.129 used a cell culture of sugar beet (Beta vulgaris) plant to produce the chiral alcohol (1R)-1-(3,4-dimethylphenyl)ethanol from prochiral 1-(3,4-dimethylphenyl)ethanone. The authors used aqueous solutions of ChCl-based (Na)DES containing either Glc or polyalcohols (glycerol and ethylene glycol) and determined that the (R)-alcohol configuration was predominant in most NaDES, compared to water, where enantioselectivity favoured the (S)-alcohol. An inversion in enantioselectivity was observed also in whole-cell biocatalysis with the use of baker's yeast for the reduction of ketones.125 By adding different amounts of ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol (1
glycerol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) to water, the enantioselectivity went from approx. 95% (S)-enantiomer in pure water to approx. 95% (R)-enantiomer in pure NaDES. The authors presumed that this was due to inhibitory effects of NaDES on some (S)-oxidoreductases.
2) to water, the enantioselectivity went from approx. 95% (S)-enantiomer in pure water to approx. 95% (R)-enantiomer in pure NaDES. The authors presumed that this was due to inhibitory effects of NaDES on some (S)-oxidoreductases.
NaDES have also been used as performance additives. Aqueous ChCl-based NaDES were used as alternative solvents for peroxygenase-catalysed oxy-functionalization reactions where they acted as solvent, enzyme stabilizer and as an electron donor for the generation of H2O2.132 Their results showed that, for every reaction or substrate, a different solvent composition was needed for an optimal result. Changes in stability, activity and selectivity in NaDES compared to aqueous solvents were also shown for an immobilized lipase B from Candida antarctica,133 epoxide hydrolase101 and peroxidase and cross-linked proteases.134,135 This stabilizing effect of NaDES was determined not only for enzymes but also for solutes such as phenolic compounds. When natural colorants from safflower were stored in sugar-based NaDES, their overall stability (thermal, storage at low temperatures, artificial and ambient light) was increased.39 Additionally, NaDES components can serve a dual purpose: as a reaction media and as a substrate for the reaction (cf.Fig. 3). In one study, menthol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) decanoic acid (1
decanoic acid (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was used in lipase-mediated production of glucose monodecanoate ester,136 and in another, menthol
1) was used in lipase-mediated production of glucose monodecanoate ester,136 and in another, menthol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lauric acid (2
lauric acid (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for the production of menthyl laurate ester.137 In both cases, one of the components (decanoic acid or menthol, respectively) was also used as a substrate for the esterification reaction. Another group went even further and used both NaDES components as reaction substrates.138 In a lipase catalysed esterification of menthol with dodecanoic acid these compounds first formed a NaDES reaction medium (menthol
1) for the production of menthyl laurate ester.137 In both cases, one of the components (decanoic acid or menthol, respectively) was also used as a substrate for the esterification reaction. Another group went even further and used both NaDES components as reaction substrates.138 In a lipase catalysed esterification of menthol with dodecanoic acid these compounds first formed a NaDES reaction medium (menthol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) decanoic acid 3
decanoic acid 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and when lipase was added, they were converted to menthyl decanoate ester.
1) and when lipase was added, they were converted to menthyl decanoate ester.
By using NaDES in biocatalytic reactions, we can not only improve the thermostability of the enzymes, but also increase their activity, stability, range of substrates (poorly water-soluble substrates) and direct their stereoselectivity. Furthermore, this stabilizing effect of NaDES can also be extended to solutes, i.e. reaction substrates, reducing their degradation under reaction conditions. However, comprehensive studies are needed in order to determine the best NaDES mixture and its optimal water dilution for a given reaction and its potential toxicity to the environment.
Critical perspective
Based on previous research, it is not so far-fetched to suggest that water and lipids are not the only liquid phases present in living systems and that NaDES are also one of them. The data indicates that NaDES may establish structurally confined high-density bio-condensates that serve a vital function in plants and other organisms. By having almost no water pressure and the ability to form hydrogen bonds with solutes, NaDES can retain water and stabilize proteins and other molecules during high heat and drought conditions, preventing their denaturation and loss of function. They may also be used for storage of bio-active compounds with poor water solubility and enable reactions between molecules with different water solubilities. NaDES-based condensates clearly have many advantageous features for living cells, and the presence of NaDES mixtures in plants has already been confirmed in the form of plant saps and nectars.2 However, that does not mean they actually exist inside the cells. So, what are the indicators that would prove or disprove the “NaDES as the third liquid phase” hypothesis?The first issue with current literature on NaDES that could disprove this hypothesis is in the NaDES preparation itself. Generally, NaDES are prepared using high temperatures (60 °C and above)67,72,81,97,124,129,132,139–141 which are not commonly found in a natural or cellular environment, raising the question if these mixtures can even be formed at temperatures that are more acceptable for living beings. Nevertheless, Dai et al.8 prepared different combinations of ChCl, Bet and amino acids with sugars (glucose, sucrose, maltose, fructose, etc.) and acids (citric, malic, malonic, maleic, lactic, etc.) at 50 °C, which is a temperature that is closer to some more extreme natural environments. In other studies, the combination of menthol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lauric acid137 and menthol
lauric acid137 and menthol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dodecanoic acid138 were prepared at 40 °C or below, hence, proving NaDES formation is not impossible at milder temperatures. Instead, it is only a matter of the HBA and HBD choice as well as the molar ratio. Moreover, NaDES are typically prepared at higher temperatures to avoid preliminary studies of the eutectic point of each mixture, which is only performed for a more in-depth understanding of the system. That said, just because most NaDES are reported to be formed at higher temperatures, it cannot be assumed that these will not be formed at lower temperatures as well without firstly studying the system eutectic point. More specific research is therefore needed, either by measuring the eutectic point of each mixture or simply by targeting NaDES formation at environmental and physiological temperatures. Furthermore, simulation studies could also be directed towards understanding the mechanisms of NaDES formation inside the cells since multiscale simulation has been gaining prominence for the study of complex physicochemical phenomena. Thereby, this field is still open for many interesting works that would be able to shed some light upon the hypothesis of NaDES being the “third liquid phase”.
dodecanoic acid138 were prepared at 40 °C or below, hence, proving NaDES formation is not impossible at milder temperatures. Instead, it is only a matter of the HBA and HBD choice as well as the molar ratio. Moreover, NaDES are typically prepared at higher temperatures to avoid preliminary studies of the eutectic point of each mixture, which is only performed for a more in-depth understanding of the system. That said, just because most NaDES are reported to be formed at higher temperatures, it cannot be assumed that these will not be formed at lower temperatures as well without firstly studying the system eutectic point. More specific research is therefore needed, either by measuring the eutectic point of each mixture or simply by targeting NaDES formation at environmental and physiological temperatures. Furthermore, simulation studies could also be directed towards understanding the mechanisms of NaDES formation inside the cells since multiscale simulation has been gaining prominence for the study of complex physicochemical phenomena. Thereby, this field is still open for many interesting works that would be able to shed some light upon the hypothesis of NaDES being the “third liquid phase”.
The second issue worth considering is the role of water in aqueous solutions of NaDES. Several studies addressed in the previous sections demonstrate the easier NaDES formation upon the addition of water as well as the use of aqueous solutions of NaDES to reduce the system viscosity. The former also favours the hypothesis here contested as NaDES would not require such high temperatures to be formed. But, more importantly, when water is added to the system other crucial questions are raised: what happens when you dilute a NaDES mixture with water beyond a certain point? Is the NaDES still a “true” NaDES? Vicente et al.142 addressed this issue in their work on chitin deacetylation in DES. They emphasized that it is important to evaluate how the HBA and HBD perform on their own since when using aqueous solutions of DES, we might end up with an aqueous solution of individual components instead of a “true” DES mixture due to a disruption of the DES H-bond network by the presence of water. A study by Hammond et al.143 indicates that the DES network is retained even at a remarkably high level of water, 42 wt% (58 wt% of DES). At 51 wt% of water (49 wt% DES), the DES network is disrupted, giving way to water–water and DES–water interactions. However, as some examples of enzyme-catalysed reactions from the previous section (medium for biocatalytic reactions) show that 40 or even 20% (v/v) DES still have an effect on reaction kinetics.67,97,101,124 Thus, it seems that even if the DES network is not completely intact, the presence of both HBA and HBD in the aqueous DES solution may have synergistic effects. It should be highlighted that this discussion is only relevant for hydrophilic (Na)DES since the moment water is added to hydrophobic (Na)DES, even at very small amounts, an emulsion will be formed.
The third issue, and possibly the strongest evidence pointing to the presence of NaDES in cells is related to drought and its associated high/low temperatures. Drought tolerance, as it turns out, is correlated with the production of not only compatible solutes (prevention of water loss) and heat-shock proteins (refolding of denatured proteins), but also with the production of sugars and amino acids59 which have been shown to act as NaDES components.2 The mechanisms of drought tolerance are based on stabilization of structures by preferential hydration, and the mechanisms of desiccation by replacing water with other molecules that are capable of H-bond formation.59 NaDES retain water2 and can thus either prevent dehydration or form a H-bond network with membranes and solutes in the absence of water or at high temperatures to stabilize them. The evidence in favor of this are the results that show high thermal stability of enzymes,59,67,72 re-activation of enzymes upon rehydration2,60,67 and stabilization of natural colorants which are regularly exposed to environmental conditions like high temperatures and radiation.39 A study also showed that in order to preserve liposomes after dehydration and subsequent rehydration, sugar molecules were needed on both sides of the membrane.35 In cells, these sugar molecules could form a membrane-associated NaDES layer with the choline-function of the membrane lipids.2
Another point favouring the NaDES presence inside cells is their high solubilizing capacity and the exceedingly high amounts of poorly water-soluble compounds like polyphenols inside plant cells.2,33,46 The antioxidants like anthocyanins, glutathione, ascorbic acid and flavonoids can protect cellular structures from oxidative stress, but some of them have very low or no water solubility. Thus, by being dissolved in membrane-associated NaDES, they may be present in high amounts and effectively protect membranes from oxidative damage. This is supported by a study that reported an unknown dense substance that was formed around the thylakoid lumen of chloroplasts upon dehydration.36 This observed substance was assigned a phenolic character,37 which could actually be membrane-bound NaDES with dissolved polyphenols that serve a purpose of protecting the membrane from oxidative damage.
Overall, and in spite of some research data supporting the presence of NaDES inside living cells, there is no direct evidence. Until we develop methods that can detect not only separate NaDES components but these components in a specific interaction (i.e. NaDES mixture) inside cells, it will be hard to definitively prove that NaDES are “the third liquid phase”. However, an increasing amount of evidence shows how NaDES could be useful to a cell and/or gives a possible explanation for processes like reactions between molecules of opposing water-solubilities, tolerance to high/low temperatures and unusually high amounts of poorly water-soluble compounds inside cells. Thus, despite the lack of direct evidence, the evidence we do have points to the “third liquid phase” hypothesis as being very plausible.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was funded by the Slovenian Research and Innovation Agency within the projects P2-0152 and NC-0024. We would also like to acknowledge Vid Urbančič for his invaluable help with some of the figures.References
- H. K. Kim, Y. H. Choi and R. Verpoorte, NMR-based metabolomic analysis of plants, Nat. Protoc., 2010, 5, 536–549 CrossRef CAS PubMed.
- Y. H. Choi, J. van Spronsen, Y. Dai, M. Verberne, F. Hollmann, I. W. C. E. Arends, G. J. Witkamp and R. Verpoorte, Are natural deep eutectic solvents the missing link in understanding cellular metabolism and physiology?, Plant Physiol., 2011, 156, 1701–1705 CrossRef CAS PubMed.
- A. P. Abbott, D. Boothby, G. Capper, D. L. Davies and R. K. Rasheed, Deep Eutectic Solvents formed between choline chloride and carboxylic acids: Versatile alternatives to ionic liquids, J. Am. Chem. Soc., 2004, 126, 9142–9147 CrossRef CAS PubMed.
- M. A. R. Martins, S. P. Pinho and J. A. P. Coutinho, Insights into the Nature of Eutectic and Deep Eutectic Mixtures, J. Solution Chem., 2019, 48, 962–982 CrossRef CAS.
- D. O. Abranches and J. A. P. Coutinho, Everything You Wanted to Know about Deep Eutectic Solvents but Were Afraid to Be Told, Annu. Rev. Chem. Biomol. Eng., 2023, 14, 141–163 CrossRef PubMed.
- R. J. Sánchez-Leija, J. R. Torres-Lubián, A. Reséndiz-Rubio, G. Luna-Bárcenas and J. D. Mota-Morales, Enzyme-mediated free radical polymerization of acrylamide in deep eutectic solvents, RSC Adv., 2016, 6, 13072–13079 RSC.
- Y. Dai, G. J. Witkamp, R. Verpoorte and Y. H. Choi, Tailoring properties of natural deep eutectic solvents with water to facilitate their applications, Food Chem., 2015, 187, 14–19 CrossRef CAS PubMed.
- Y. Dai, J. van Spronsen, G. J. Witkamp, R. Verpoorte and Y. H. Choi, Natural deep eutectic solvents as new potential media for green technology, Anal. Chim. Acta, 2013, 766, 61–68 CrossRef CAS.
- Y. P. Mbous, M. Hayyan, W. F. Wong, C. Y. Looi and M. A. Hashim, Unraveling the cytotoxicity and metabolic pathways of binary natural deep eutectic solvent systems, Sci. Rep., 2017, 7, 1–14 CrossRef.
- A. P. Abbott, G. Capper, D. L. Davies, R. K. Rasheed and V. Tambyrajah, Novel solvent properties of choline chloride/urea mixtures, Chem. Commun., 2003, 70–71 RSC.
- A. A. Quintana, A. M. Sztapka, V. de C. S. Ebinuma and C. Agatemor, Enabling Sustainable Chemistry with Ionic Liquids and Deep Eutectic Solvents: A Fad or the Future?, Angew. Chem., 2022, 134, e202205609 CrossRef.
- M. Cvjetko Bubalo, S. Vidović, I. Radojčić Redovniković and S. Jokić, Green solvents for green technologies, J. Chem. Technol. Biotechnol., 2015, 90, 1631–1639 CrossRef CAS.
- A. Paiva, R. Craveiro, I. Aroso, M. Martins, R. L. Reis and A. R. C. Duarte, Natural Deep Eutectic Solvents – Solvents for the 21st Century, ACS Sustainable Chem. Eng., 2014, 2, 1063–1071 CrossRef CAS.
- Y. Dai, G. J. Witkamp, R. Verpoorte and Y. H. Choi, Natural deep eutectic solvents as a new extraction media for phenolic metabolites in carthamus tinctorius L., Anal. Chem., 2013, 85, 6272–6278 CrossRef CAS PubMed.
- M. Jablonský and J. Šima, Phytomass Valorization by Deep Eutectic Solvents—Achievements, Perspectives, and Limitations, Crystals, 2020, 10, 800 CrossRef.
- Y. Liu, J. B. Friesen, J. B. McAlpine, D. C. Lankin, S. N. Chen and G. F. Pauli, Natural Deep Eutectic Solvents: Properties, Applications, and Perspectives, J. Nat. Prod., 2018, 81, 679–690 CrossRef CAS.
- A. Mišan, J. Nađpal, A. Stupar, M. Pojić, A. Mandić, R. Verpoorte and Y. H. Choi, The perspectives of natural deep eutectic solvents in agri-food sector, Crit. Rev. Food Sci. Nutr., 2019, 60, 2564–2592 CrossRef PubMed.
- F. M. Perna, P. Vitale and V. Capriati, Deep eutectic solvents and their applications as green solvents, Curr. Opin. Green Sustainable Chem., 2020, 21, 27–33 CrossRef CAS.
- O. V. Morozova, I. S. Vasil'eva, G. P. Shumakovich, E. A. Zaitseva and A. I. Yaropolov, Deep Eutectic Solvents for Biotechnology Applications, Biochemistry, 2023, 88, S150–S175 CAS.
- B. B. Hansen, S. Spittle, B. Chen, D. Poe, Y. Zhang, J. M. Klein, A. Horton, L. Adhikari, T. Zelovich, B. W. Doherty, B. Gurkan, E. J. Maginn, A. Ragauskas, M. Dadmun, T. A. Zawodzinski, G. A. Baker, M. E. Tuckerman, R. F. Savinell and J. R. Sangoro, Deep Eutectic Solvents: A Review of Fundamentals and Applications, Chem. Rev., 2021, 121, 1232–1285 CrossRef CAS PubMed.
- A. Azzouz and M. Hayyan, Potential applications of deep eutectic solvents in nanotechnology: Part II, Chem. Eng. J., 2023, 468, 143563 CrossRef CAS.
- M. Panić, M. Cvjetko Bubalo and I. Radojčić Redovniković, Designing a biocatalytic process involving deep eutectic solvents, J. Chem. Technol. Biotechnol., 2021, 96, 14–30 CrossRef.
- Y. Wang, K. H. Kim, K. Jeong, N. K. Kim and C. G. Yoo, Sustainable biorefinery processes using renewable deep eutectic solvents, Curr. Opin. Green Sustainable Chem., 2021, 27, 100396 CrossRef CAS.
- S. C. Cunha and J. O. Fernandes, Extraction techniques with deep eutectic solvents, TrAC, Trends Anal. Chem., 2018, 105, 225–239 CrossRef CAS.
- X. Wang, P. Zhou, X. Lv and Y. Liang, Insight into the structure-function relationships of the solubility of chitin/chitosan in natural deep eutectic solvents, Mater. Today Commun., 2021, 27, 102374 CrossRef CAS.
- T. Jeliński, M. Przybyłek and P. Cysewski, Natural Deep Eutectic Solvents as Agents for Improving Solubility, Stability and Delivery of Curcumin, Pharm. Res., 2019, 36, 1–10 CrossRef PubMed.
- J. Cao, J. Cao, H. Wang, L. Chen, F. Cao and E. Su, Solubility improvement of phytochemicals using (natural) deep eutectic solvents and their bioactivity evaluation, J. Mol. Liq., 2020, 318, 113997 CrossRef CAS.
- P. Kalhor and K. Ghandi, Deep Eutectic Solvents for Pretreatment, Extraction, and Catalysis of Biomass and Food Waste, Molecules, 2019, 24, 4012 CrossRef CAS PubMed.
- W. Wang and D. J. Lee, Lignocellulosic biomass pretreatment by deep eutectic solvents on lignin extraction and saccharification enhancement: A review, Bioresour. Technol., 2021, 339, 125587 CrossRef CAS PubMed.
- Y. Chen and T. Mu, Application of deep eutectic solvents in biomass pretreatment and conversion, Green Energy Environ., 2019, 4, 95–115 CrossRef.
- A. Procentese, F. Raganati, G. Olivieri, M. E. Russo, L. Rehmann and A. Marzocchella, Deep Eutectic Solvents pretreatment of agro-industrial food waste, Biotechnol. Biofuels, 2018, 11, 1–12 CrossRef.
- Z. Chen, A. Ragauskas and C. Wan, Lignin extraction and upgrading using deep eutectic solvents, Ind. Crops Prod., 2020, 147, 112241 CrossRef CAS.
- K. R. Markham, K. S. Gould, C. S. Winefield, K. A. Mitchell, S. J. Bloor and M. R. Boase, Anthocyanic vacuolar inclusions - Their nature and significance in flower colouration, Phytochemistry, 2000, 55, 327–336 CrossRef CAS PubMed.
- G. Hrazdina and R. A. Jensen, Spatial Organization of Enzymes in Plant Metabolic Pathways, Annu. Rev. Plant Physiol. Plant Mol. Biol., 2003, 43, 241–267 CrossRef.
- L. M. Crowe, Lessons from nature: the role of sugars in anhydrobiosis, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 2002, 131, 505–513 CrossRef.
- K. Georgieva, É. Sárvári and Á. Keresztes, Protection of thylakoids against combined light and drought by a lumenal substance in the resurrection plant Haberlea rhodopensis, Ann. Bot., 2010, 105, 117–126 CrossRef CAS PubMed.
- M. E. van Steveninck and R. F. M. van Steveninck, Plastids with densely staining thylakoid contents in Nymphoides indica, Protoplasma, 1980, 103, 333–342 CrossRef.
- K. O. Wikene, E. Bruzell and H. H. Tønnesen, Characterization and antimicrobial phototoxicity of curcumin dissolved in natural deep eutectic solvents, Eur. J. Pharm. Sci., 2015, 80, 26–32 CrossRef CAS PubMed.
- Y. Dai, R. Verpoorte and Y. H. Choi, Natural deep eutectic solvents providing enhanced stability of natural colorants from safflower (Carthamus tinctorius), Food Chem., 2014, 159, 116–121 CrossRef CAS PubMed.
- M. L. Toledo, M. M. Pereira, M. G. Freire, J. P. A. Silva, J. A. P. Coutinho and A. P. M. Tavares, Laccase Activation in Deep Eutectic Solvents, ACS Sustainable Chem. Eng., 2019, 7, 11806–11814 CrossRef CAS.
- Y. Liang, W. Duan, X. An, Y. Qiao, Y. Tian and H. Zhou, Novel betaine-amino acid based natural deep eutectic solvents for enhancing the enzymatic hydrolysis of corncob, Bioresour. Technol., 2020, 310, 123389 CrossRef CAS PubMed.
- H. H. Tønnesen, M. Másson and T. Loftsson, Studies of curcumin and curcuminoids. XXVII. Cyclodextrin complexation: solubility, chemical and photochemical stability, Int. J. Pharm., 2002, 244, 127–135 CrossRef PubMed.
- M. Esmaili, S. M. Ghaffari, Z. Moosavi-Movahedi, M. S. Atri, A. Sharifizadeh, M. Farhadi, R. Yousefi, J. M. Chobert, T. Haertlé and A. A. Moosavi-Movahedi, Beta casein-micelle as a nano vehicle for solubility enhancement of curcumin; food industry application, LWT–Food Sci. Technol., 2011, 44, 2166–2172 CrossRef CAS.
- H. G. Morrison, C. C. Sun and S. Neervannan, Characterization of thermal behavior of deep eutectic solvents and their potential as drug solubilization vehicles, Int. J. Pharm., 2009, 378, 136–139 CrossRef CAS.
- B. M. Popović, D. Uka, O. Alioui, R. Ždero Pavlović and Y. Benguerba, Experimental and COSMO-RS theoretical exploration of rutin formulations in natural deep eutectic solvents: Solubility, stability, antioxidant activity, and bioaccessibility, J. Mol. Liq., 2022, 359, 119266 CrossRef.
- L. Paniwnyk, E. Beaufoy, J. P. Lorimer and T. J. Mason, The extraction of rutin from flower buds of Sophora japonica, Ultrason. Sonochem., 2001, 8, 299–301 CrossRef CAS PubMed.
- L. M. de Souza Mesquita, F. H. B. Sosa, L. S. Contieri, P. R. Marques, J. Viganó, J. A. P. Coutinho, A. C. R. V. Dias, S. P. M. Ventura and M. A. Rostagno, Combining eutectic solvents and food-grade silica to recover and stabilize anthocyanins from grape pomace, Food Chem., 2023, 406, 135093 CrossRef CAS PubMed.
- L. M. de Souza Mesquita, L. S. Contieri, F. H. B. Sosa, R. S. Pizani, J. Chaves, J. Viganó, S. P. M. Ventura and M. A. Rostagno, Combining eutectic solvents and pressurized liquid extraction coupled in-line with solid-phase extraction to recover, purify and stabilize anthocyanins from Brazilian berry waste, Green Chem., 2023, 25, 1884–1897 RSC.
- E. El Maaiden, H. El Kahia, B. Nasser, K. Moustaid, N. Qarah, H. Boukcim, A. Hirich, L. Kouisni and Y. El Kharrassi, Deep eutectic solvent-ultrasound assisted extraction as a green approach for enhanced extraction of naringenin from Searsia tripartita and retained their bioactivities, Front. Nutr., 2023, 10, 1193509 CrossRef PubMed.
- A. Del-Castillo-Llamosas, F. Rodríguez-Rebelo, B. Rodríguez-Martínez, A. Mallo-Fraga, P. G. Del-Río and B. Gullón, Valorization of Avocado Seed Wastes for Antioxidant Phenolics and Carbohydrates Recovery Using Deep Eutectic Solvents (DES), Antioxidants, 2023, 12, 6 CrossRef PubMed.
- G. Zengin, M. D. Cádiz-Gurrea, Á. Fernández-Ochoa, F. J. Leyva-Jiménez, A. S. Carretero, M. Momotko, E. Yildiztugay, R. Karatas, S. Jugreet, M. F. Mahomoodally and G. Boczkaj, Selectivity Tuning by Natural Deep Eutectic Solvents (NADESs) for Extraction of Bioactive Compounds from Cytinus hypocistis—Studies of Antioxidative, Enzyme-Inhibitive Properties and LC-MS Profile, Molecules, 2022, 27, 18 Search PubMed.
- I. Fatima, M. Munir, R. Qureshi, U. Hanif, N. Gulzar and A. A. Sheikh, Advanced methods of algal pigments extraction: A review, Crit. Rev. Food Sci. Nutr., 2023, 1–18 CrossRef PubMed.
- J. Cao, R. Wu, Q. Dong, L. Zhao, F. Cao and E. Su, Effective Release of Intracellular Enzymes by Permeating the Cell Membrane with Hydrophobic Deep Eutectic Solvents, ChemBioChem, 2020, 21, 672–680 CrossRef CAS PubMed.
- A. K. Schweiger, N. Ríos-Lombardía, C. K. Winkler, S. Schmidt, F. Morís, W. Kroutil, J. González-Sabín and R. Kourist, Using Deep Eutectic Solvents to Overcome Limited Substrate Solubility in the Enzymatic Decarboxylation of Bio-Based Phenolic Acids, ACS Sustainable Chem. Eng., 2019, 7, 16364–16370 CrossRef CAS.
- U. G. Favero, N. Schaeffer, H. Passos, K. A. M. L. Cruz, D. Ananias, S. Dourdain and M. C. Hespanhol, Solvent extraction in non-ideal eutectic solvents – Application towards lanthanide separation, Sep. Purif. Technol., 2023, 314, 123592 CrossRef CAS.
- L. E. Meyer, M. B. Andersen and S. Kara, A Deep Eutectic Solvent Thermomorphic Multiphasic System for Biocatalytic Applications, Angew. Chem., Int. Ed., 2022, 61, e202203823 CrossRef CAS PubMed.
- F. Chemat, M. Abert Vian, H. K. Ravi, B. Khadhraoui, S. Hilali, S. Perino and A. S. Fabiano Tixier, Review of Alternative Solvents for Green Extraction of Food and Natural Products: Panorama, Principles, Applications and Prospects, Molecules, 2019, 24, 16 CrossRef PubMed.
- A. Lavaud, M. Laguerre, S. Bitric, A. S. Fabiano Tixier, M. Roller and F. Chemat, International. PatentWO2016/162703Al, 2015. Available online: https://worldwide.espacenet.com/publicationDetails/biblio?DB=EPODOC&II=4&ND=3&adjacent=true&locale=fr_EP&FT=D&date=20180406&CC=CN&NR=107889468A&KC=A# (accessed on 23 August 2023) Search PubMed.
- F. A. Hoekstra, E. A. Golovina and J. Buitink, Mechanisms of plant desiccation tolerance, Trends Plant Sci., 2001, 6, 431–438 CrossRef CAS PubMed.
- C. Knudsen, K. Bavishi, K. M. Viborg, D. P. Drew, H. T. Simonsen, M. S. Motawia, B. L. Møller and T. Laursen, Stabilization of dhurrin biosynthetic enzymes from Sorghum bicolor using a natural deep eutectic solvent, Phytochemistry, 2020, 170, 112214 CrossRef CAS PubMed.
- R. M. Gleadow and B. L. Møller, Cyanogenic Glycosides: Synthesis, Physiology, and Phenotypic Plasticity, Annu. Rev. Plant Biol., 2014, 65, 155–185 CrossRef CAS PubMed.
- J.-E. Bassard, B. L. Møller and T. Laursen, Assembly of Dynamic P450-Mediated Metabolons—Order Versus Chaos, Curr. Mol. Biol. Rep., 2017, 3(1), 37–51 CrossRef PubMed.
- T. Laursen, J. Borch, C. Knudsen, K. Bavishi, F. Torta, H. J. Martens, D. Silvestro, N. S. Hatzakis, M. R. Wenk, T. R. Dafforn, C. E. Olsen, M. S. Motawia, B. Hamberger, B. L. Møller and J. E. Bassard, Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum, Science, 2016, 354, 890–893 CrossRef CAS PubMed.
- C. Knudsen, N. J. Gallage, C. C. Hansen, B. L. Møller and T. Laursen, Dynamic metabolic solutions to the sessile life style of plants, Nat. Prod. Rep., 2018, 35, 1140–1155 RSC.
- O. I. Pavli, C. E. Vlachos, C. Kalloniati, E. Flemetakis and G. N. Skaracis, Metabolite profiling reveals the effect of drought on sorghum (Sorghum bicolor L. Moench) metabolism, Plant Omics, 2013, 6, 371–376 CAS.
- N. H. O'Donnell, B. L. Møller, A. D. Neale, J. D. Hamill, C. K. Blomstedt and R. M. Gleadow, Effects of PEG-induced osmotic stress on growth and dhurrin levels of forage sorghum, Plant Physiol. Biochem., 2013, 73, 83–92 CrossRef PubMed.
- S. Khodaverdian, B. Dabirmanesh, A. Heydari, E. Dashtban-moghadam, K. Khajeh and F. Ghazi, Activity, stability and structure of laccase in betaine based natural deep eutectic solvents, Int. J. Biol. Macromol., 2018, 107, 2574–2579 CrossRef CAS PubMed.
- E. Durand, J. Lecomte, B. Baréa, E. Dubreucq, R. Lortie and P. Villeneuve, Evaluation of deep eutectic solvent-water binary mixtures for lipase-catalyzed lipophilization of phenolic acids, Green Chem., 2013, 15, 2275–2282 RSC.
- Q. Zeng, Y. Wang, Y. Huang, X. Ding, J. Chen and K. Xu, Deep eutectic solvents as novel extraction media for protein partitioning, Analyst, 2014, 139, 2565–2573 RSC.
- A. Sanchez-Fernandez, K. J. Edler, T. Arnold, D. Alba Venero and A. J. Jackson, Protein conformation in pure and hydrated deep eutectic solvents, Phys. Chem. Chem. Phys., 2017, 19, 8667–8670 RSC.
- T. Caldas, N. Demont-Caulet, A. Ghazi and G. Richarme, Thermoprotection by glycine betaine and choline, Microbiology, 1999, 145, 2543–2548 CrossRef CAS.
- A. E. Delorme, J. M. Andanson and V. Verney, Improving laccase thermostability with aqueous natural deep eutectic solvents, Int. J. Biol. Macromol., 2020, 163, 919–926 CrossRef CAS PubMed.
- K. Saito, T. Yamamoto and K. I. Miyamoto, Isolation and partial purification of carthamine: an instrumentation manual, Z. Lebensm.-Unters. Forsch., 1992, 195, 550–554 CrossRef CAS.
- J. B. Kim and Y. S. Paik, Stability of carthamin from Carthamus tinctorius in aqueous solution:pH and temperature effects, Arch. Pharmacal Res., 1997, 20, 643–646 CrossRef CAS PubMed.
- B. Han and J. C. Bischof, Direct cell injury associated with eutectic crystallization during freezing, Cryobiology, 2004, 48, 8–21 CrossRef PubMed.
- Z. Kovács, L. Simon-Sarkadi, C. Sovány, K. Kirsch, G. Galiba and G. Kocsy, Differential effects of cold acclimation and abscisic acid on free amino acid composition in wheat, Plant Sci., 2011, 180, 61–68 CrossRef PubMed.
- N. Zhang, W. Li, C. Chen, J. Zuo and L. Weng, Molecular dynamics investigation of the effects of concentration on hydrogen bonding in aqueous solutions of methanol, ethylene glycol and glycerol, Bull. Korean Chem. Soc., 2013, 34, 2711–2719 CrossRef CAS.
- V. A. E. King and J. T. Su, Dehydration of Lactobacillus acidophilus, Process Biochem., 1993, 28, 47–52 CrossRef CAS.
- X. C. Meng, C. Stanton, G. F. Fitzgerald, C. Daly and R. P. Ross, Anhydrobiotics: The challenges of drying probiotic cultures, Food Chem., 2008, 106, 1406–1416 CrossRef CAS.
- Y. Huang, F. Feng, J. Jiang, Y. Qiao, T. Wu, J. Voglmeir and Z.-G. Chen, Green and efficient extraction of rutin from tartary buckwheat hull by using natural deep eutectic solvents, Food Chem., 2017, 221, 1400–1405 CrossRef CAS PubMed.
- Y. Qiao, H. L. Cai, X. Yang, Y. Y. Zang and Z. G. Chen, Effects of natural deep eutectic solvents on lactic acid bacteria viability during cryopreservation, Appl. Microbiol. Biotechnol., 2018, 102, 5695–5705 CrossRef CAS PubMed.
- B. Tang and K. H. Row, Recent developments in deep eutectic solvents in chemical sciences, Monatsh. Chem., 2013, 144, 1427–1454 CrossRef CAS.
- M. Hayyan, C. Y. Looi, A. Hayyan, W. F. Wong and M. A. Hashim, In Vitro and In Vivo Toxicity Profiling of Ammonium-Based Deep Eutectic Solvents, PLoS One, 2015, 10, e0117934 CrossRef.
- Q. Wen, J.-X. Chen, Y.-L. Tang, J. Wang and Z. Yang, Assessing the toxicity and biodegradability of deep eutectic solvents, Chemosphere, 2015, 132, 63–69 CrossRef CAS.
- J. T. Gorke, F. Srienc and R. J. Kazlauskas, Hydrolase-catalyzed biotransformations in deep eutectic solvents, Chem. Commun., 2008, 1235–1237 RSC.
- V. Gotor-Fernández and C. E. Paul, Deep eutectic solvents for redox biocatalysis, J. Biotechnol., 2019, 293, 24–35 CrossRef.
- M. Pätzold, S. Siebenhaller, S. Kara, A. Liese, C. Syldatk and D. Holtmann, Deep Eutectic Solvents as Efficient Solvents in Biocatalysis, Trends Biotechnol., 2019, 37, 943–959 CrossRef.
- J. A. Kist, H. Zhao, K. R. Mitchell-Koch and G. A. Baker, The study and application of biomolecules in deep eutectic solvents, J. Mater. Chem. B, 2021, 9, 536–566 RSC.
- R. A. Sheldon, M. L. Bode and N. Mathebula, Green and sustainable solvents for biocatalytic oxidations, Curr. Opin. Green Sustainable Chem., 2023, 39, 100741 CrossRef CAS.
- D. Arnodo, E. Maffeis, F. Marra, S. Nejrotti and C. Prandi, Combination of Enzymes and Deep Eutectic Solvents as Powerful Toolbox for Organic Synthesis, Molecules, 2023, 28, 516 CrossRef CAS PubMed.
- B. Nian and X. Li, Can deep eutectic solvents be the best alternatives to ionic liquids and organic solvents: A perspective in enzyme catalytic reactions, Int. J. Biol. Macromol., 2022, 217, 255–269 CrossRef CAS PubMed.
- S. Zhang, C. Ma, Q. Li, Q. Li and Y. C. He, Efficient chemoenzymatic valorization of biobased D-fructose into 2,5-bis(hydroxymethyl)furan with deep eutectic solvent Lactic acid:Betaine and Pseudomonas putida S12 whole cells, Bioresour. Technol., 2022, 344, 126299 CrossRef CAS PubMed.
- X. Han, W. Li, Z. Duan, X. Ma and D. Fan, Biocatalytic production of compound K in a deep eutectic solvent based on choline chloride using a substrate fed-batch strategy, Bioresour. Technol., 2020, 305, 123039 CrossRef CAS PubMed.
- E. I. Solomon, U. M. Sundaram and T. E. Machonkin, Multicopper oxidases and oxygenases, Chem. Rev., 1996, 96, 2563–2605 CrossRef CAS PubMed.
- B. Rasekh, K. Khajeh, B. Ranjbar, N. Mollania, B. Almasinia and H. Tirandaz, Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents, Eng. Life Sci., 2014, 14, 442–448 CrossRef CAS.
- C. Johannes and A. Majcherczyk, Laccase activity tests and laccase inhibitors, J. Biotechnol., 2000, 78, 193–199 CrossRef CAS PubMed.
- A. A. Elgharbawy, A. Hayyan, M. Hayyan, S. N. Rashid, M. R. M. Nor, M. Y. Zulkifli, Y. Alias and M. E. S. Mirghani, Shedding Light on Lipase Stability in Natural Deep Eutectic Solvents, Chem. Biochem. Eng. Q., 2018, 32, 359–370 CrossRef CAS.
- B. Nian, C. Cao and Y. Liu, Activation and stabilization of Candida antarctica lipase B in choline chloride-glycerol-water binary system via tailoring the hydrogen-bonding interaction, Int. J. Biol. Macromol., 2019, 136, 1086–1095 CrossRef CAS PubMed.
- S. H. Kim, S. Park, H. Yu, J. H. Kim, H. J. Kim, Y. H. Yang, Y. H. Kim, K. J. Kim, E. Kan and S. H. Lee, Effect of deep eutectic solvent mixtures on lipase activity and stability, J. Mol. Catal. B: Enzym., 2016, 128, 65–72 CrossRef CAS.
- J. C. Chan, B. Zhang, M. Martinez, B. Kuruba, J. Brozik, C. H. Kang and X. Zhang, Structural studies of Myceliophthora Thermophila Laccase in the presence of deep eutectic solvents, Enzyme Microb. Technol., 2021, 150, 109890 CrossRef CAS PubMed.
- D. Lindberg, M. de la Fuente Revenga and M. Widersten, Deep eutectic solvents (DESs) are viable cosolvents for enzyme-catalyzed epoxide hydrolysis, J. Biotechnol., 2010, 147, 169–171 CrossRef CAS PubMed.
- S. Weibels, A. Syguda, C. Herrmann and H. Weingärtner, Steering the enzymatic activity of proteins by ionic liquids. A case study of the enzyme kinetics of yeast alcohol dehydrogenase, Phys. Chem. Chem. Phys., 2012, 14, 4635–4639 RSC.
- I. Juneidi, M. Hayyan, M. A. Hashim and A. Hayyan, Pure and aqueous deep eutectic solvents for a lipase-catalysed hydrolysis reaction, Biochem. Eng. J., 2017, 117, 129–138 CrossRef CAS.
- W.-J. Xu, Y.-K. Huang, F. Li, D.-D. Wang, M.-N. Yin, M. Wang and Z.-N. Xia, Improving β-glucosidase biocatalysis with deep eutectic solvents based on choline chloride, Biochem. Eng. J., 2018, 138, 37–46 CrossRef CAS.
- F. Mamashli, J. Badraghi, B. Delavari, M. Sabbaghian, M. Hosseini and A. A. Saboury, Evaluation of Versatile Peroxidase's Activity and Conformation in the Presence of a Hydrated Urea Based Deep Eutectic Solvent, J. Solution Chem., 2019, 48, 689–701 CrossRef CAS.
- H. Wang, Y. Tao, M. V. Masuku, J. Cao, J. Yang, K. Huang, Y. Ge, Y. Yu, Z. Xiao, Y. Kuang, J. Huang and S. Yang, Effects of deep eutectic solvents on the biotransformation efficiency of ω-transaminase, J. Mol. Liq., 2023, 377, 121379 CrossRef CAS.
- M. G. Bellou, E. Gkantzou, A. Skonta, D. Moschovas, K. Spyrou, A. Avgeropoulos, D. Gournis and H. Stamatis, Development of 3D Printed Enzymatic Microreactors for Lipase-Catalyzed Reactions in Deep Eutectic Solvent-Based Media, Micromachines, 2022, 13, 1954 CrossRef PubMed.
- H. Wang, M. V. Masuku, Y. Tao, J. Yang, Y. Kuang, C. Lyu, J. Huang and S. Yang, Improved Stability and Catalytic Efficiency of ω-Transaminase in Aqueous Mixture of Deep Eutectic Solvents, Molecules, 2023, 28, 3895 CrossRef CAS.
- A. A. M. Elgharbawy, S. S. Syed Putra, H. W. Khan, N. A. Azmi, M. S. Sani, N. Ab llah, A. Hayyan, J. Jewaratnam and W. J. Basirun, Menthol and Fatty Acid-Based Hydrophobic Deep Eutectic Solvents as Media for Enzyme Activation, Processes, 2023, 11, 547 CrossRef CAS.
- M. G. Bellou, M. Patila, R. Fotiadou, K. Spyrou, F. Yan, P. Rudolf, D. P. Gournis and H. Stamatis, Tyrosinase Magnetic Cross-Linked Enzyme Aggregates: Biocatalytic Study in Deep Eutectic Solvent Aqueous Solutions, Biomolecules, 2023, 13, 643 CrossRef CAS PubMed.
- S. N. Rashid, A. Hayyan, M. Hayyan, M. A. Hashim, A. A. M. Elgharbawy, F. S. Sani, W. J. Basirun, V. S. Lee, Y. Alias, A. K. Mohammed, M. E. S. Mirghani, M. Y. Zulkifli and M. Rageh, Ternary glycerol-based deep eutectic solvents: Physicochemical properties and enzymatic activity, Chem. Eng. Res. Des., 2021, 169, 77–85 CrossRef CAS.
- Z. Ma, Y. Mi, X. Han, H. Li, M. Tian, Z. Duan, D. Fan and P. Ma, Transformation of ginsenoside via deep eutectic solvents based on choline chloride as an enzymatic reaction medium, Bioprocess Biosyst. Eng., 2020, 43, 1195–1208 CrossRef CAS PubMed.
- Q.-B. Cheng and L.-W. Zhang, Highly Efficient Enzymatic Preparation of Daidzein in Deep Eutectic Solvents, Molecules, 2017, 22, 186 CrossRef.
- G. Grogan, Practical biotransformations: a beginner's guide, Wiley, 2009 Search PubMed.
- J. B. va Beilen and Z. Li, Enzyme technology: an overview, Curr. Opin. Biotechnol., 2002, 13, 338–344 CrossRef.
- S. Rodríguez Couto and J. L. Toca Herrera, Industrial and biotechnological applications of laccases: A review, Biotechnol. Adv., 2006, 24, 500–513 CrossRef PubMed.
- A. Illanes, A. Cauerhff, L. Wilson and G. R. Castro, Recent trends in biocatalysis engineering, Bioresour. Technol., 2012, 115, 48–57 CrossRef CAS PubMed.
- C. Kang, D. Ren, S. Zhang, X. Zhang, X. He, Z. Deng, C. Huang and H. Guo, Effect of Polyhydroxyl Compounds on the ThermalStability and Structure of Laccase, Pol. J. Environ. Stud., 2019, 28, 3253–3259 CrossRef CAS.
- S. Kurniawati and J. A. Nicell, Characterization of Trametes versicolor laccase for the transformation of aqueous phenol, Bioresour. Technol., 2008, 99, 7825–7834 CrossRef CAS.
- D. M. Mate and M. Alcalde, Laccase engineering: From rational design to directed evolution, Biotechnol. Adv., 2015, 33, 25–40 CrossRef CAS.
- A. Wasak, R. Drozd, D. Jankowiak and R. Rakoczy, Rotating magnetic field as tool for enhancing enzymes properties - laccase case study, Sci. Rep., 2019, 9, 1–9 CrossRef CAS PubMed.
- L. Chen, M. Zou and F. F. Hong, Evaluation of fungal laccase immobilized on natural nanostructured bacterial cellulose, Front. Microbiol., 2015, 6, 1245 Search PubMed.
- B. Olivares, F. Martínez, L. Rivas, C. Calderón, J. M. Munita and P. R. Campodonico, A Natural Deep Eutectic Solvent Formulated to Stabilize β-Lactam Antibiotics, Sci. Rep., 2018, 8, 1–12 CAS.
- T. X. Yang, L. Q. Zhao, J. Wang, G. L. Song, H. M. Liu, H. Cheng and Z. Yang, Improving Whole-Cell Biocatalysis by Addition of Deep Eutectic Solvents and Natural Deep Eutectic Solvents, ACS Sustainable Chem. Eng., 2017, 5, 5713–5722 CrossRef CAS.
- Z. Maugeri and P. Domínguez De María, Whole-Cell Biocatalysis in Deep-Eutectic-Solvents/Aqueous Mixtures, ChemCatChem, 2014, 6, 1535–1537 CrossRef CAS.
- M. Cvjetko Bubalo, M. Mazur, K. Radošević and I. Radojčić Redovniković, Baker's yeast-mediated asymmetric reduction of ethyl 3-oxobutanoate in deep eutectic solvents, Process Biochem., 2015, 50, 1788–1792 CrossRef CAS.
- M. Panić, D. Delač, M. Roje, I. Radojčić Redovniković and M. Cvjetko Bubalo, Green asymmetric reduction of acetophenone derivatives: Saccharomyces cerevisiae and aqueous natural deep eutectic solvent, Biotechnol. Lett., 2019, 41, 253–262 CrossRef PubMed.
- R. N. Patel, Biocatalytic synthesis of chiral pharmaceutical intermediates, Food Technol. Biotechnol., 2004, 42, 305–325 CAS.
- D. Pavoković, K. Košpić, M. Panić, I. Radojčić Redovniković and M. Cvjetko Bubalo, Natural deep eutectic solvents are viable solvents for plant cell culture-assisted stereoselective biocatalysis, Process Biochem., 2020, 93, 69–76 CrossRef.
- C. R. Müller, I. Meiners and P. Domínguez De María, Highly enantioselective tandem enzyme–organocatalyst crossed aldol reactions with acetaldehyde in deep-eutectic-solvents, RSC Adv., 2014, 4, 46097–46101 RSC.
- A. Raczyńska, J. Jadczyk and M. Brzezińska-Rodak, Altering the Stereoselectivity of Whole-Cell Biotransformations via the Physicochemical Parameters Impacting the Processes, Catalysts, 2021, 11, 781 CrossRef.
- Y. Ma, Y. Li, S. Ali, P. Li, W. Zhang, M. C. R. Rauch, S. J. P. Willot, D. Ribitsch, Y. H. Choi, M. Alcalde, F. Hollmann and Y. Wang, Natural Deep Eutectic Solvents as Performance Additives for Peroxygenase Catalysis, ChemCatChem, 2020, 12, 989–994 CrossRef CAS.
- E. Durand, J. Lecomte, B. Baréa, G. Piombo, E. Dubreucq and P. Villeneuve, Evaluation of deep eutectic solvents as new media for Candida antarctica B lipase catalyzed reactions, Process Biochem., 2012, 47, 2081–2089 CrossRef CAS.
- H. Zhao, G. A. Baker and S. Holmes, Protease activation in glycerol-based deep eutectic solvents, J. Mol. Catal. B: Enzym., 2011, 72, 163–167 CrossRef CAS PubMed.
- B. P. Wu, Q. Wen, H. Xu and Z. Yang, Insights into the impact of deep eutectic solvents on horseradish peroxidase: Activity, stability and structure, J. Mol. Catal. B: Enzym., 2014, 101, 101–107 CrossRef CAS.
- R. Hollenbach, K. Ochsenreither and C. Syldatk, Enzymatic Synthesis of Glucose Monodecanoate in a Hydrophobic Deep Eutectic Solvent, Int. J. Mol. Sci., 2020, 21, 4342 CrossRef CAS PubMed.
- R. Craveiro, L. Meneses, L. Durazzo, Â. Rocha, J. M. Silva, R. L. Reis, S. Barreiros, A. R. C. Duarte and A. Paiva, Deep Eutectic Solvents for Enzymatic Esterification of Racemic Menthol, ACS Sustainable Chem. Eng., 2019, 7, 19943–19950 CrossRef CAS.
- M. Pätzold, B. O. Burek, A. Liese, J. Z. Bloh and D. Holtmann, Product recovery of an enzymatically synthesized (−)-menthol ester in a deep eutectic solvent, Bioprocess Biosyst. Eng., 2019, 42, 1385–1389 CrossRef PubMed.
- Z. Maugeri and P. Domínguez de María, Benzaldehyde lyase (BAL)-catalyzed enantioselective CC bond formation in deep-eutectic-solvents–buffer mixtures, J. Mol. Catal. B: Enzym., 2014, 107, 120–123 CrossRef CAS.
- A. Arıkaya, A. E. Ünlü and S. Takaç, Use of deep eutectic solvents in the enzyme catalysed production of ethyl lactate, Process Biochem., 2019, 84, 53–59 CrossRef.
- K. Radošević, N. Ćurko, V. Gaurina Srček, M. Cvjetko Bubalo, M. Tomašević, K. Kovačević Ganić and I. Radojčić Redovniković, Natural deep eutectic solvents as beneficial extractants for enhancement of plant extracts bioactivity, LWT–Food Sci. Technol., 2016, 73, 45–51 CrossRef.
- F. A. Vicente, M. Huš, B. Likozar and U. Novak, Chitin Deacetylation Using Deep Eutectic Solvents: Ab Initio-Supported Process Optimization, ACS Sustainable Chem. Eng., 2021, 9, 3874–3886 CrossRef CAS PubMed.
- O. S. Hammond, D. T. Bowron and K. J. Edler, The Effect of Water upon Deep Eutectic Solvent Nanostructure: An Unusual Transition from Ionic Mixture to Aqueous Solution, Angew. Chem., Int. Ed., 2017, 56, 9782–9785 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |
