Advancements in artificial micro/nanomotors for nucleic acid biosensing: a review of recent progress
Conghui
Liu
 ab,
Jingyu
Chen
ab,
Jingyu
Chen
 a,
Jiahui
Liang
a,
Tailin
Xu
a,
Jiahui
Liang
a,
Tailin
Xu
 *abc and
Xueji
Zhang
abc
*abc and
Xueji
Zhang
abc
aCollege of Chemistry and Environmental Engineering, Shenzhen University, Shenzhen 518060, China. E-mail: Tailin Xu xutailin@szu.edu.cn
bGuangdong Laboratory of Artificial Intelligence and Digital Economy (SZ), Shenzhen, 518060, China
cSchool of Biomedical Engineering, Health Science Center, Shenzhen University, Shenzhen 518060, China
First published on 1st August 2023
Abstract
Artificial micro/nanomotors represent a class of well-designed tools that exhibit dynamic motion and remote-control capabilities, endowing them with the capacity to perform complex tasks at the micro/nanoscale. Their utilization in nucleic acid biosensing has been paid significant attention, owing to their ability to facilitate targeted delivery of detection probes to designated sites and enhance hybridization between detection probes and target nucleic acids, thereby improving the sensitivity and specificity of biosensing. Within this comprehensive overview, we elucidate the advancement of nucleic acid biosensing through the integration of micro/nanomotors over the past decade. In particular, we provide an in-depth exploration of the diverse applications of micro/nanomotors in nucleic acid biosensing, including fluorescence recovery-based biosensing, velocity change-based biosensing, and aggregation-enhanced biosensing. Additionally, we outline the remaining challenges that impede the practical application of artificial micro/nanomotors in nucleic acid detection, and offer personal insights into prospective avenues for future development. By overcoming these obstacles, we anticipate that artificial micro/nanomotors will revolutionize conventional nucleic acid detection methodologies, providing enhanced sensitivity and reduced diagnostic timeframes, thereby facilitating more effective disease diagnosis.
1. Introduction
Nucleic acids are essential biomolecules for life, encompassing both deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). As they serve as storages, encoders, transmitters, carriers, and expressers of genetic information, nucleic acids play critical roles in various biological processes.1,2 Moreover, nucleic acids have demonstrated great potential as biomarkers for early diagnosis and disease screening.3 For example, circulating tumor DNA (ctDNA) is a type of cellular DNA that tumor cells release into the circulation, with significantly higher levels observed in cancer patients than in healthy individuals. This suggests that ctDNA holds great promise as a non-invasive biomarker for diagnostic and prognostic cancer tests.4,5 For another example, microRNAs (miRNAs), a class of non-protein-coding, short, single-stranded RNAs (19–23 nucleotides),6 have been associated with numerous diseases, including cancer,3 Alzheimer's disease7 and diabetes,8 due to their abnormal expression in bodily fluids. Therefore, nucleic acid analysis is crucial for predicting and monitoring several diseases. Traditional analytical methods of nucleic acids such as electrophoresis and blotting have great limitations in terms of sensitivity. Commonly used nucleic acid quantification techniques such as polymerase chain reaction (PCR) with high accuracy and wide dynamic range usually involve time-consuming and cumbersome operations. Additionally, the low abundance, sequence similarity, and susceptibility to degradation of nucleic acids pose significant challenges for their sensitive and selective detection.6,9Artificial micro/nanomotors are micro/nanoscale machines, prepared by mimicking natural micro/nanomotors such as kinesins and bacteria, and capable of converting energy (chemical, acoustic, thermal, light, magnetic, etc.) into autonomous motions and forces.10–14 Over the past twenty years, artificial micro/nanomotors have evolved into a cross-cutting frontier of materials, nano and biomedical sciences. Micro/nano dimensions, ease of modification and active movement ability enable the artificial micro/nanomotors to perform certain complex tasks including cargo delivery,15–19 noninvasive surgery,20–29 cell manipulation,30–32 bioimaging,18,33 and biosensing34–39 at the microscopic scale. One of the main applications of micro/nanomotors in biosensing is the detection of nucleic acids. It has been reported that artificial micro/nanomotors can enhance the transport of detection probes to the specific site to improve the sensitivity and specificity of nucleic acid biosensing. Additionally, the active motion of artificial micro/nanomotors can facilitate contact of substances and accelerate the hybridization reaction of nucleic acids in solution, providing a promising alternative tool for more sensitive, specific, and rapid nucleic acid biosensing.40
In the past decade, remarkable advancements have been made in the burgeoning field of artificial micro/nanomotors for biosensing, signifying its rapid growth and notable accomplishments.14,40–42 The objective of this review is to shed light on the progression of nucleic acid biosensing through the utilization of micro/nanomotors. Initially, we provide a concise overview of the prevailing techniques employed to propel micro/nanomotors for nucleic acid biosensing. Subsequently, we detailed discuss the specific applications of micro/nanomotors in nucleic acid biosensing, including fluorescence recovery-based biosensing, velocity change-based biosensing, and aggregation-enhanced biosensing (Fig. 1). Lastly, we present a comprehensive summary of the achievements thus far, identify the remaining challenges faced by micro/nanomotors in the realm of nucleic acid biosensing, and give a perspective on future development directions of the field.
2. Micro/nanomotors propulsion in nucleic acid sensing
The commonly used propulsion mechanism of artificial micro/nanomotors can be classified into chemically and physically driven methods.43 Chemically driven micro/nanomotors are self-propelled by microbubbles (O2,44–46 H2,47–50etc.) or a concentration gradient of molecules generated from local catalytic reactions with the presence of fuels (glucose,51,52 hydrogen peroxide,34,53 urea,19,54etc.) in fluids. Au is the common material used to prepare micro/nanomotors for nucleic acid sensing because the Au surface is easily anchored with thiolated DNA strands by forming a strong Au–S bond. Noble metals like Pt or natural enzymes (e.g., catalase, urease, glucose oxidase) are commonly used as catalysts, pre-modified or asymmetric immobilized on micro/nanomotors to break the symmetry of the system.55 For example, a tubular structure is commonly used for microbubble-propelled micro/nanomotors, which usually contain an outer Au layer and an inner Pt layer. The outer Au surface is used to modify the nucleic acid strand, and the inner Pt layer can catalyze hydrogen peroxide fuel to continuously generate O2 bubbles to propel the micro/nanomotors. In addition, Au–Pt nanowires are also commonly reported chemically driven nanomotors. Hydrogen peroxide fuel is oxidized by Pt to generate H+, resulting in the formation of an electric field gradient from Pt to Au, which in turn drives the nanomotors. However, heavy reliance on fuels results in the speed of micro/nanomotors being significantly influenced by fuel concentration. The low and limited concentration of bioavailable fuels in vivo may not continuously power the artificial micro/nanomotors, while high concentrations of fuel like hydrogen peroxide have harmful effects on the environment and human body, hindering further development in biomedical applications. Recently, it has been reported that apoptotic tumor DNA (nM–μM levels) can be used as fuel to power the DNase functionalized Janus nanomotors without external fuel,56 providing a new idea for self-propelled micro/nanomotor design.Compared to chemically driven micro/nanomotors, physically driven micro/nanomotors can realize continuous motion and remote directional control by applying physical stimuli such as optical, thermal, acoustic, electrical, or magnetic fields.57 Of these, magnetism and ultrasound are the popular driving methods for in vivo nucleic acid sensing due to their good biocompatibility and high precision. Magnetic propulsion is realized by introducing ferromagnetic nickel, iron or cobalt materials into micro/nanomotors by electrodeposition or physical vapor deposition. Then the magnetic micro/nanomotors can rotate or translate in magnetic field gradients at low strengths.58,59 Ultrasound propulsion is based on the acoustic radiation force exerted on the micro/nanomotors, which generate from a differential sound pressure field in the ultrasound setup at low amplitudes.60,61 Compared to magnetic propulsion, ultrasound control is a much simpler process and there are no specific requirements for the composition of micro/nanomotors. Au nanowires are the commonly used acoustically propelled nanorobots due to the facile fabrication progress of membrane template-assisted electrodeposition and the easily functionalized surface. In general, magnetic and ultrasound fields are harmless to cells and tissues, therefore they are the ideal propulsion and control methods for in vivo applications of micro/nanomotors.62
3. Applications of nucleic acid biosensing by micro/nanomotors
Currently, artificial micro/nanomotors with various functionalized modifications have been widely applied to nucleic acid biosensing due to their distinct advantages, including high sensitivity, non-invasive nature, and active-driven capabilities. The nucleic acid sensing strategies employed by micro/nanomotors can be classified into three principal categories based on the underlying sensing mechanisms, namely fluorescence recovery-based biosensing, velocity change-based biosensing, and aggregation-enhanced biosensing. A comprehensive discussion of these distinct methodologies will be presented in this section.3.1 Fluorescence recovery-based sensing method
Fluorescence signaling stands as a highly efficient method for nucleic acid detection with rapid responsiveness, simplified procedures, and unambiguous outcomes. By utilizing the micro/nanomotors modified with specific nucleic acid probes, these biosensors can serve as powerful tools, offering conditional recovery of fluorescence upon interaction with target nucleic acids. This innovative approach enables real-time nucleic acid sensing in human serum or within living cells.Wang's group proposed a novel intracellular fluorescence switching technique, referred to as an “OFF–ON” method, utilizing ultrasound-driven gold nanowires (AuNWs) coated with dye-labeled single-stranded DNA (ssDNA)/graphene oxide (GO).38 This groundbreaking approach enabled the penetration of intact cancer cells by the nanomotors, targeting the overexpressed microRNA-21 (miRNA-21) commonly found in tumor samples. The GO sheet, acting as a scaffold with a high surface area, absorbed FAM-ssDNA through π–π stacking interactions between nucleotide bases and carbon nanomaterial, leading to spontaneous quenching of fluorescence through fluorescence resonance energy transfer (FRET) mechanisms. To facilitate rapid migration towards the target cancer cells and enhance non-destructive internalization, an acoustic field was applied, propelling the nanomotors. This process promoted the hybridization of ssDNA with the endogenous content, as ssDNA had a stronger affinity for target RNA than for the GO surface. Consequently, the DNA–RNA duplex was released from the nanomotors, resulting in the recovery of detectable dye fluorescence signals upon separation from the GO quencher (Fig. 2a). Importantly, the use of ultrasound-based manipulation enables the design of specific motion trajectories, granting access to individual cancer cells while preserving their unique endogenous miRNA expression and eliminating interferences. Fig. 2b demonstrated that fluorescence intensity increases proportionally with the amplitude of applied ultrasound and over time. Notably, assays treated with the maximum voltage (9 V) exhibited a 17-fold higher fluorescence signal compared to the static condition. These results highlighted the effectiveness of ultrasound waves as regulators of nanomotor motion, significantly accelerating the cellular internalization process and facilitating sufficient contact and hybridization between ssDNA and target RNA at room temperature. This approach confirmed that ultrasound substantially enhanced detection sensitivity, enabling precise intracellular “on the move” detection of ultra-trace endogenous miRNA.
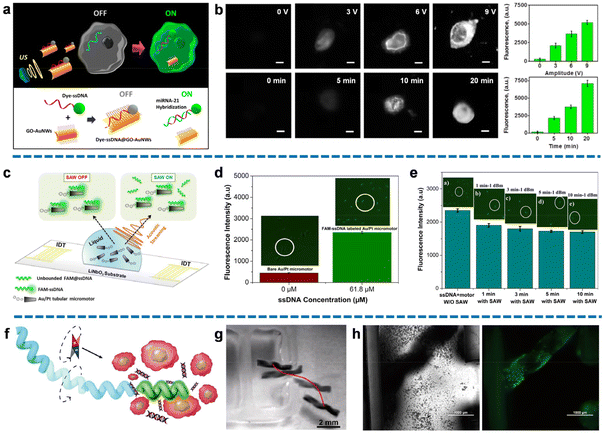 | ||
| Fig. 2 Fluorescence recovery-based nucleic acid biosensing. (a) Schematic of specifical reorganization of endogenous miRNA-21 and triggers fluorescence signal in intact cancer cells by the ultrasound-propelled ssDNA@GO-functionalized gold nanomotors. (b) Fluorescence recovery images of the modified gold nanomotor-treated MCF-7 cells using different voltages or different ultrasound propulsion times, and corresponding fluorescence intensity. Scale bar: 10 μm. Reproduced from ref. 38 with permission from the American Chemical Society, copyright 2015. (c) Schematic diagram of fluorescence detection of miRNA-21 by FAM-ssDNA@Au/Pt micromotors in the presence of surface acoustic wave (SAW). (d) Different fluorescence intensities correspond to micromotors with and without FAM-ssDNA immobilization. (e) The fluorescence intensities of micromotors under the same intensity of SAW treatment at different applied times correspond to the fluorescence intensities, confirming the capacity of SAW to remove unbound molecules. Reproduced from ref. 65 with permission from the American Chemical Society, copyright 2021. (f) Schematic representation of dye-labeled magnetic helical hydrogel micromotor for active detection of miRNA-21. (g) The trajectory of micromotor moving toward the tumor cells cluster under the rotating magnetic field. Scale bar: 2 mm. (h) Optical and fluorescent image of micromotor emitting a fluorescent signal by specifically recognizing the released miRNA-21 after moving to the cell-enriched area. Scale bar: 1000 μm. Reproduced from ref. 66 with permission from the Elsevier, copyright 2022. | ||
Moreover, these acoustically powered nanomotors have demonstrated successful extracellular and intracellular detection of HPV16 E6 mRNA transcripts.39 This “on the move” approach eliminates the need for target RNA extraction in each detection cycle, simplifying the assay procedure and offering a new perspective for downstream fluorescence-based nanomotor detection, enabling specific tracking of target cancer cells.
A catalytically self-propelled electrochemically reduced graphene oxide (erGO)/Pt microtubular motor exhibited a surprising performance for rapid movement and “OFF–ON” fluorescence detection of the gastric cancer biomarker RPRM.63 The study adopted the same mechanism of fluorescence quenching through interactions between graphene oxide (GO) and FAM-ssDNA, as well as fluorescence recovery resulting from the hybridization between dye-labeled DNA and target DNA to facilitate duplex release. However, the fluorescence intensity might diminish over time due to the gradual restoration of the new interaction, leading to a relatively high limit of detection (LOD) of 1.3 μM. Therefore, there remains a need to optimize the detection conditions and the geometric pattern of the micromotors to achieve stable, simple, and effective testing capabilities for these promising dual propulsion detection methods.
Cogal et al. introduced a dual-response detection method with dual motion propulsion. Their study employed radio frequency (RF) rotating plasma modification to achieve the rapid and uniform deposition of the W5O14/poly(3,4-ethylene dioxythiophene) (PEDOT) layer. Compared to other materials, PEDOT has a large surface area to modify more fluorescent probes, resulting in a stronger fluorescence signal and speed variation. The W5O14/PEDOT-Pt structure was synthesized using the magnetron sputtering technique and immobilized with the ssDNA probe to obtain the micromotors.64 These micromotors were powered by the catalytic decomposition of hydrogen peroxide, utilizing polymer microstructures with exceptional redox properties, high surface activity, and biocompatibility, thereby exhibiting significant potential in supporting detection. The micromotors were designed to provide sufficient binding sites with target miRNA-21 by both electrostatic interactions and hydrogen bonding. The formation of a duplex between the ssDNA probe and target miRNA-21 significantly affected the micromotors. The velocity of micromotors experienced a dramatic decrease due to the potential obstruction of catalytic sites caused by duplex formation. Meanwhile, the hybridization of the ssDNA probe with FAM-miRNA target enabled labeling of fluorescence intensity on the micromotors. Thus, the dual response of the specific recognition signals for the target RNA could be simultaneously detected, providing compelling evidence with a low LOD of 0.028 nM. However, it should be noted that the preparation process involving plasma modification may pose higher instrument requirements and limit the further application of these micromotors for simple and convenient detection.
The same research group also proposed an optimized approach that combines dual excitation-driven mechanisms with dual response characteristics, demonstrating the additional capabilities of this integrated scheme.65 The bimetallic tubular micromotors were fabricated through the electrodeposition of Au/Pt. Dye-labeled ssDNA was designed to be immobilized on the surface through van der Waals and hydrophobic interactions, enabling the recognition of target miRNA-21. By introducing an acoustic field to generate surface acoustic wave (SAW) within the system, both the speed and fluorescence intensity of the micromotors could be modulated by controlling the acoustic flow force. Firstly, SAW, as a component of the micromotor activator, could collaborate with chemical propulsion. SAW drove the emission of bubbles resulting from the decomposition of hydrogen peroxide fuel, leading to the faster movements of micromotors. Then, the scattering of SAW effectively removed the unbound miRNA-21, as the van der Waals adhesion forces between the hybridized duplex were affected by the acoustic streaming. Therefore, the application of ultrasound also reduced the fluorescence intensity as the binding rates decreased (Fig. 2c). Benefiting from the integration of dual-driven and dual-response characteristics, this detection scheme provided ultrasensitive and accurate information. Fig. 2d illustrated the enrichment of ssDNA, wherein an increased amount of immobilized FAM-ssDNA (61.8 μM) on the bimetallic surface resulted in a fluorescence signal about five times stronger than the control (0 μM). This result verified the hybridization of dye-labeled ssDNA with the micromotors, forming assemblies with the ability to recognize the target RNA. The feasibility of SAW-induced fluorescence intensity reduction was investigated in Fig. 2e. The fluorescence intensity exhibited a time-dependent property, with an increase in applied SAW resulting in a corresponding decrease in fluorescence intensity. This indicated that SAW effectively removed unbound ssDNA, eliminating potential interference signals for downstream miRNA-21 detection. By analyzing the degree of fluorescence intensity decrease and velocity decrease, quantitative information about the content of target RNA could be derived. Furthermore, SAW helped lower the LOD to 0.19 nM by reducing interference from false positive signals associated with unbound miRNA-21.
As mentioned earlier, magnetism is one of the most popular driving methods for in vivo nucleic acid sensing because the magnetic field guidance imparts active targeting capability to the micro/nanomotors. For example, Qin et al. constructed a magnetically driven helical hydrogel micromotor equipped with a dye-labeled SYBR Green I (SG) probe for DNA detection.66 The helical hydrogel matrix was formulated through the crosslinking of melamine and polyvinyl alcohol (PVA), followed by the loading of Fe3O4 nanoparticles and SG. The specific structure exhibited high biocompatibility, resistance to decomposition by organisms, and precise magnetic motion manipulation due to the modification of magnetic Fe3O4 nanoparticles. Fig. 2f demonstrates the manipulation of the helical hydrogel micromotor with the SG probe using a rotating magnetic field to migrate towards areas clustered with target tumor cells. The tumor cells were treated sequentially with NIR, hyperthermia, and further incubation, inducing cell apoptosis and releasing considerable endogenous DNA. Fig. 2g demonstrated the trajectory of the micromotor, highlighting its excellent ability to march directionally under magnetic actuation. The trajectory and speed of the micromotor could be regulated by adjusting the revolutions per minute (rpm), enabling not only a straight-line pattern but also the “V” and “J” trajectories. When the micromotor reached the area of enriched cells, the dissociated DNA spontaneously bound to SG in the fluorescent probe, triggering the appearance of an obvious green fluorescent signal for detection, with a mean fluorescence intensity of 55.339 (Fig. 2h). In comparison to traditional pathological examination methods, this study introduced a novel approach that advanced in directional DNA detection and hemocompatibility, offering promising prospects for in vivo detection. The formulation of a biocompatible helical hydrogel structure with magnetic nanoparticles allowed the micromotor with an appropriate diameter to navigate through vessels, and the trajectory could be easily controlled by manipulating the applied rotating magnetic field. This capability strongly supports in vivo directional DNA detection, enabling clinical analysis of relatively small sample volumes for accurate trace detection.
3.2 Velocity change-based nucleic acid biosensing
The velocity change-based biosensing method relies on the analysis of micro/nanomotor velocity and distance as crucial detection parameters, providing valuable analytical signals. By monitoring the changes in velocity and distance of the micro/nanomotor through microscopy, the concentration of the target nucleic acid can be directly and quantitatively inferred. Compared to fluorescence recovery-based biosensing, the velocity change-based biosensing method eliminates the need for precise, expensive, and cumbersome fluorescence-based microscopy. Instead, it leverages microscopy to monitor the motion of the micro/nanomotor, making it a more accessible and cost-effective technique. Furthermore, the motion signal in velocity change-based biosensing demonstrates enhanced stability over time when compared to the fluorescence signal. This attribute ensures reliable and consistent measurements throughout the detection process.Wu et al. pioneered the utilization of nanomotors for motion-based nucleic acid detection, presenting a platform that enables rapid, straightforward, and highly sensitive analysis of DNA and bacterial ribosomal RNA.34 Their approach converted probe–target hybridization events into measurable signals by monitoring the speed changes of nanomotors. As shown in Fig. 3a, thiolated capture probes were immobilized onto a gold electrode via Au–S bonds, and thiolated detector probes were anchored on the surface of Ag nanoparticles via the Ag–S bond. Upon the introduction of nucleic acid targets, stable sandwich duplexes were formed among the detector probes, capture probes, and target nucleic acids. The subsequent addition of a hydrogen peroxide solution resulted in the dissolution of Ag nanoparticles, releasing Ag+ ions that altered the speed of the unmodified magnetic nanomotors. The acceleration of nanomotors, indicative of target concentration, was captured as a long-distance signal (Fig. 3b). By recording the concentration-dependent distance signals using an optical microscope, the content of the target nucleic acid sample could be directly detected with high sensitivity, even at small concentration differences. Notably, the versatility of these nanomotors allowed for the detection of multiple nucleic acid targets through diverse functional modifications. As shown in Fig. 3c, the relationship between distance and target concentration held not only for DNA but also for different levels of 16S rRNA. The method demonstrated a low LOD of 40 amol for DNA and 7 × 103 CFU μL−1 for E. coli 16S rRNA, with satisfactory specificity (Fig. 3d). This study introduces a novel perspective on integrating nanomotors into nucleic acid detection, utilizing the speed and distance of the nanomotors as analytical signals.
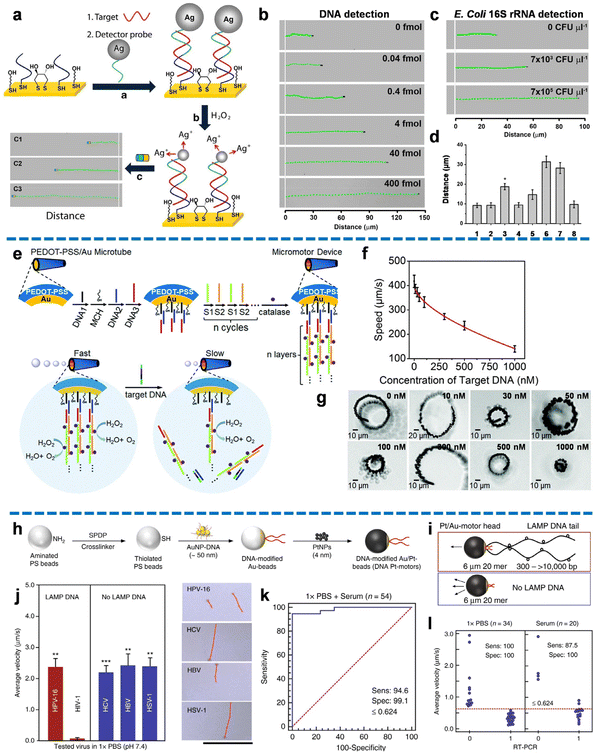 | ||
| Fig. 3 Velocity change-based nucleic acid biosensing. (a) The nucleic acid detection process of Ag NP-induced nanomotor acceleration. (b) The quantitative distance signals correspond to different target DNA concentrations. (c) The quantitative distance signals correspond to different concentrations of E. coli 16S rRNA (CFU, colony-forming units). (d) Specificity of Ag NP-induced nanomotor acceleration-based detection methods. Reproduced from ref. 34 with the permission from the Springer Nature, copyright 2010. (e) Schematic diagram of the preparation process of PEDOT-PSS/Au micromotors and the mechanism of speed reduction for target DNA detection by the conjugation and release of catalase decorations modified-multilayer DNA. (f) Diagram of the low power exponential function correlation between the nanomotor's speed and the concentration of the target DNA from 10 to 1000 nM. (g) The respective high kinematic behaviors of the micromotor at different concentrations of target DNA. Reproduced from the ref. 68 with permission from the Royal Society of Chemistry, copyright 2017. (h) Schematic of the preparation process of DNA Pt motors. (i) Altered structure and motion of the DNA Pt motors in the presence of LAMP DNA. (j) High specificity demonstrated by the mean speed of micromotors in the existence of amplification products of target and non-target viruses and their corresponding trajectories. Scale bar: 100 μm. (k) High sensitivity (sens) and specificity (spec) represented by the assay using 1× PBS (n = 35) and serum (n = 20) samples spiked with various HIV-1 concentrations for analyzing the receiver-operating characteristics (ROC) curve. (l) Real-time polymerase chain reaction (RT-PCR). Reproduced from ref. 35 with the permission from the Springer Nature, copyright 2018. | ||
Subsequently, Wu and coworkers proposed a series of bubble-driven micromotors for velocity decay-based DNA sensing based on DNA displacement hybridization-triggered catalase release.67–69 For example, a poly(3,4-ethylenedioxythiophene and sodium 4-styrenesulfonate)/Au (PEDOT-PSS/Au) microtube was used as micromotors, providing the inner surface of Au to form DNA sandwich hybridization (DNA1/2/3) as the sensing unit (Fig. 3e).68 Two assisting DNA strands (S1/S2) were attached to facilitate the binding of multi-catalase, creating multilayer DNA complex catalytic sites as the power unit. The motion of these tubular micromotors was driven by enzyme-catalyzed bubbles generated by hydrogen peroxide under optical microscopy. Upon the presence of target DNA, a velocity decay detection signal was observed as the hybridization of target DNA with DNA2 induced the release of catalase-decorated multilayer DNA. Fig. 3f showed the low power exponential function relationship between the speed of the micromotors and the concentration of target DNA from 10 to 1000 nM. The corresponding high-motion performance of the micromotor at different concentrations of target DNA was shown in Fig. 3g. To further improve the sensitivity, Wu and coworkers prepared a jellyfish-like micromotor consisting of a multimetallic (Au/Ag/Ni/Au) shell as the umbrella-shaped body and a DNA-catalase assembly as the muscle fibers of the jellyfish.69 This novel design exhibited a nearly fivefold reduction in micromotor speed upon the presence of target DNA, demonstrating a significantly enhanced sensitivity compared to the previous tubular micromotors with an inner surface. The umbrella-shaped structure of the jellyfish-inspired micromotor provided an open inner-shell space, facilitating efficient modification and hybridization efficiency of the concave surface with DNA probes. Furthermore, it promoted the diffusion of target DNA, resulting in higher catalytic activity for the detection of low concentrations of target DNA. Importantly, this detection method remained effective in various biomedia, including bovine serum albumin solution, serum, and cell culture. To ensure long-term detection, it is necessary to optimize the durability of the micromotors, as the catalytic activity of the enzymes decreased after 3 minutes of operation. Future research should focus on addressing this limitation to further improve the performance of these micromotors for sustained and reliable detection applications.
In contrast to the aforementioned approach, Nguyen et al. introduced a chemically propelled nanomotor based on specific substrate enzyme modifications, presenting a novel concept of “turn-on” motion-based biosensors triggered by target DNA.46 The specifically modified gold surface of microtubes with DNA1 probe acted as a catalytic site. The presence of target DNA induced the conjugation of unbound Pt NP-DNA2 assemblies to the obtrusive DNA1 probe, resulting in the capture of Pt nanoparticles that catalyzed the decomposition of the hydrogen peroxide to fuel nanomotors. The velocity of the nanomotors exhibited a quantitative correlation with the concentration of target DNA, achieving a LOD of 1 pmol. Although this detection methodology had inherent limitations in terms of specificity, catalytic activity, and detection threshold, it introduced a non-enzyme metal particle-catalyzed “turn-on” biosensing protocol triggered by the introduction of target DNA, providing valuable insights for subsequent investigations in this field.
The incorporation of velocity change-based biosensing into portable devices offers a rapid testing alternative to conventional nucleic acid detection methods, such as PCR. A noteworthy example involves the integration of cellphone-based optical biosensing with loop-mediated isothermal amplification (LAMP) and nanomotors for the detection of HIV-1.35 As shown in Fig. 3h, the specified DNA-modified Au/Pt-motor was constructed by assembling gold nanoparticles (Au NPs)-DNA conjugates into the polystyrene (PS) bead and then coating platinum nanoparticles (Pt NPs). Upon target HIV-1 RNA triggering LAMP amplification, a stem-looped amplicon with an increased size, owing to additional base pairs, was formed. The propulsion of nanomotors occurred through the generation of bubbles catalyzed by hydrogen peroxide. Notably, the hybridization of the DNA probe tail of the nanomotor with the amplicon led to a substantial decrease in velocity, generating a straightforward optical signal that could be accurately detected using handheld devices (Fig. 3i). In the absence of LAMP DNA, this optical signal was absent. Remarkably, exclusive velocity reduction of nanomotors was observed solely in the presence of HIV-1 DNA LAMP amplicons, illustrating the exceptional specificity of the method (Fig. 3j). Furthermore, the nanomotors maintained high sensitivity when applied to HIV-1 spiked serum and patient plasma samples, indicating the method's potential for broad application (Fig. 3k). The high accuracy of the method was illustrated in Fig. 3l, where the positive and negative rates were 100% and 90%, respectively, with a remarkable agreement (100%) with the traditional RT-PCR detection method. It is noteworthy that by monitoring the Pt NP coverage ratio on the bead surface, the spatial dispersion of DNA probes at the hybridization site was controlled, allowing for precise motion trajectory determination that could be tracked and imaged using optical sensing without the need for bulky instrumentation. The combination of these characteristics strongly suggests the prospective application of this method for rapid, sensitive, and simplified biosensing.
In addition to chemically driven nanomotors, Karaca et al. reported another type of nanomotor biosensing using magnetic propulsion.70 The nanomotors, comprised of chitosan-functionalized gold–nickel wire structures (CS@Au–Ni NMs), exhibited exceptional magnetic performance, owing to the presence of bimetallic layers. The chitosan layer, with a positive zeta-potential and ample surface area, effectively served as an electrostatic interaction site for negatively charged double-stranded DNA (dsDNA). Upon incubation for 30 minutes, the reduction in nanomotor speed correlated directly with the concentration increase of dsDNA, ranging from 0.01 mg L−1 to 10 mg L−1. This phenomenon was attributed to the immobilization of DNA onto chitosan through intense electrostatic interactions. In comparison to self-powered nanomotors, magnetic propulsion provided a more dynamic manipulation approach, enabling direct control over the motion trajectory of nanomotors and offering prospects for in vivo nanomotor detection.
3.3 Aggregation-enhanced nucleic acid biosensing
Ultrasound-induced aggregation of micro/nanomotors has emerged as a tremendously promising technique in the field of biosensing, owing to its exceptional attributes including rapid response, heightened maneuverability, and user-friendly operation. This approach primarily involves the adjustment of applied voltage and frequency to generate a pressure gradient, thereby inducing the formation of pressure nodes within the microcavity. As a consequence, micro/nanomotors or cells rapidly assemble and aggregate at these specific nodes, leading to amplified signals (such as fluorescence or Raman) at these localized sites. This technique enables enhanced nucleic acid detection, providing an innovative avenue for biosensing applications.The combination of ultrasound-induced aggregation of artificial motors with surface-enhanced Raman scattering (SERS) technique offers a precise analytical approach. Xu's group reported a method to achieve Raman enhancement by ultrasonic aggregation-induced enrichment of gold nanorods (Fig. 4a), enabling rapid and ultrasensitive detection of target DNA. This approach was successfully applied to detect target DNA at an astonishingly low concentration of 10−13 M in microliter-scale (10−6 L) serum samples.71 The gold nanorods were preliminarily modified with a DNA probe to capture the ROX-labeled target DNA. The assemblies were then dropped into a device integrated with the acoustic microfluidic instrument and Raman spectrometer (Fig. 4b). By capitalizing on the high acoustic response of gold nanorods, their motion could be controlled by adjusting the applied frequency and voltage to generate an asymmetric ultrasound field and exploit the acoustic radiation force, thereby promoting hybridization with the target DNA. Under ultrasound stimulation, the modified gold nanorods underwent enrichment, forming clusters of microparticles with numerous “hot spots” at the fixed pressure node within the microcavity within 25 seconds, leading to a significant enhancement of the Raman signal (Fig. 4c). The Raman intensity exhibited a strong quantitative relationship with the concentration of target DNA, demonstrating a LOD of 0.1 pM. Notably, this ultrasonic aggregation-induced enrichment of nanorods demonstrated an exceptionally low sample volume requirement of 1 μL to separate target DNA from microdroplets, making it highly promising for ultra-trace biomarker detection. However, it is worth mentioning that in this particular study, the Raman reporter ROX was utilized for modification in the target DNA, which may not be suitable for practical DNA detection.
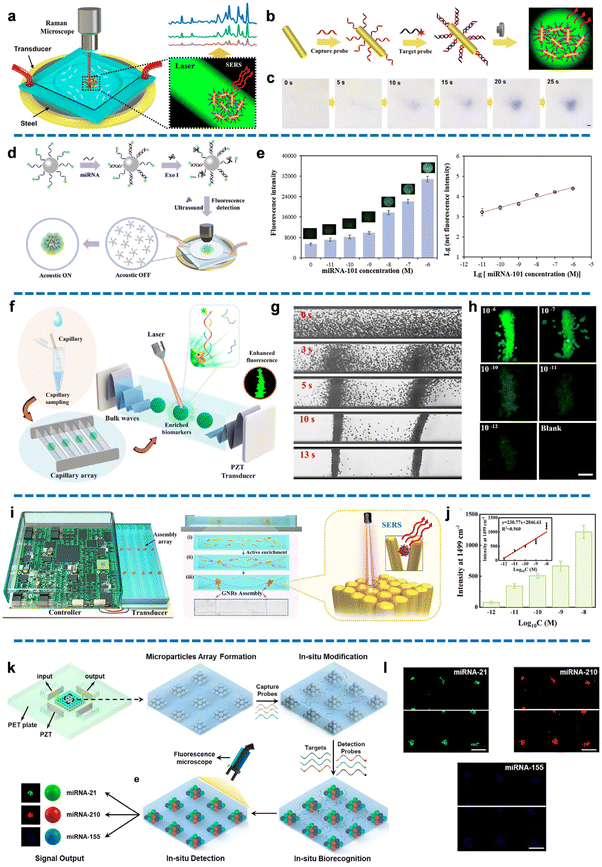 | ||
| Fig. 4 Aggregation-enhanced nucleic acid biosensing. (a) Schematic of ultrasound-induced aggregation of Au nanorod motors for SERS detection of DNA. (b) The modification processes of Au nanorod motors. (c) Actual time-lapse microscopic images of aggregation of Au nanorod motors. Scale bar: 10 μm. Reproduced from ref. 71 with permission from the American Chemical Society, copyright 2020. (d) Schematic of aggregation-induced separation and fluorescence enhancement for miRNA detection. (e) The fluorescence intensity at different concentrations of miRNA-101 and the corresponding linear relationship between the fluorescence intensity and miRNA-101 concentration. Reproduced from ref. 72 with permission from the Elsevier, copyright 2021. (f) Schematic diagram of ultra-trace enriching biosensing in nanoliter sample by integrating capillary with ultrasound-induced aggregation of micromotors. (g) The time-lapse images of the process of micromotor aggregation. Voltage: 1 V. (h) Fluorescence intensity at different concentrations of miRNA-21. Scale bar: 75 μm. Reproduced from ref. 73 with permission from the Elsevier, copyright 2022. (i) Schematic diagram of the portable active enrichment platform for highly sensitive and rapid SERS detection of COVID-19. (j) Raman intensity at different concentrations of target SARS-CoV-2N-Gene, and the corresponding linear relationship between the Raman intensity of 1499 cm−1 peak and logarithms of the target concentration. Reproduced from ref. 74 with permission from the American Chemical Society, copyright 2023. (k) Schematic illustration of ultrasound-induced programmable micromotor array for the enrichment and simultaneous detection of multiple miRNAs. (l) Fluorescence image of micromotor arrays for miRNAs detection. Scale bar: 200 μm. Reproduced from ref. 76 with permission from the American Chemical Society, copyright 2022. | ||
Building upon prior research, Sun et al. introduced a novel approach utilizing ultrasonic aggregation of micromotors for fluorescence-enhanced detection of target miRNA associated with Alzheimer's disease.72 Micromotors composed of polystyrene microparticles modified with FAM-labeled DNA probes were employed. A key innovation in this study was the incorporation of exonuclease I (Exo I) into the hybridization reaction, which facilitated the cleavage of unbound single-stranded DNA probes. Upon the presence of target miRNA-101, the DNA probes on the micromotor surface captured the miRNA-101, forming stable DNA/RNA double strands that resisted hydrolysis by Exo I. Conversely, the unhybridized DNA probes were sheared and released. Consequently, only the hybridized probes immobilized on the microspheres generated detectable fluorescence signals following ultrasonic aggregation (Fig. 4d). By manipulating the input voltage and frequency, the acoustic wave generated a fixed pressure node within the polydimethylsiloxane (PDMS) cavity, inducing the movement of micromotors towards the node and their immediate aggregation. Upon aggregation, the fluorescence intensity increased by approximately sixfold compared to the dispersed state. The method demonstrated good sensitivity, as evidenced by the linear relationship between the logarithm of fluorescence intensity and the logarithm of miRNA-101 concentration spanning from 10−11 to 10−6 M, with a LOD of 5 pM (Fig. 4e). Furthermore, successful application of this method was demonstrated in the detection of miRNA-101 in human serum, ranging from 10−10 to 10−6 M, underscoring its potential for in vivo detection of Alzheimer's disease.
Apart from the aforementioned studies, there are ultrasound-induced detection methods that integrate micro/nanomotor aggregation with imaging analysis within a single system, holding significant promise for the rapid detection of trace or ultra-trace samples in the future. An innovative approach introduced by Luo et al. involved a device for ultrasound-induced aggregation of polyethylene micromotors within a capillary, enabling fluorescence-enhanced biosensing of miRNA in nanoliter samples (Fig. 4f and g).73 Micromotors were constructed using polystyrene microspheres modified with Cy3-labeled capture probes. The gradient ultrasound field in the capillary enhanced the complete molecular interaction of the DNA probe-modified micromotors, target miRNA-21, and free FAM-labeled detection probes, which promoted the hybridization of the micromotors captured miRNA-21 and FAM-labeled detection probes to form a “sandwich” structure, thereby enriching the target miRNA-21 and intensifying the fluorescence signal. Notably, the fluorescence intensity of the aggregated micromotors exhibited a gradual decrease as the concentration of miRNA-21 decreased from 10−6 M to 10−12 M, enabling real-time visualized detection of miRNAs with a low LOD of 7.8 pM (Fig. 4h). Moreover, this work demonstrated an exceptionally low sample consumption in the nanoliter range (10−7 L) by ingeniously incorporating ultrasound-induced aggregation to expedite the recognition and hybridization of DNA probes and target miRNAs. This achievement holds considerable significance for the rapid detection of trace miRNAs in nanoliter biological samples, offering a practical pathway for future applications in biosensing.
Luo et al. extended their research to develop a portable active enrichment platform for highly sensitive and rapid SERS detection of COVID-19 using nanoliter samples (10−7 L) within a remarkably short time of 5 minutes (Fig. 4i).74 The experimental setup involved integrating a capillary as the experimental device, an ultrasound output circuit system, and a cell phone-based SERS detection system. While throat swabs were the prevailing sampling method, this platform enabled multiple samplings from the same sample, thereby reducing the potential for false-positive signals in COVID-19 detection. The device introduced a solution containing gold rod nanomotors, labeled probes, and ultra-trace target miRNAs into the nanocarrier system of the active enrichment platform. Through precise regulation of the applied frequency and voltage using an aggregation circuit system at the ultrasound output, the nanomotors autonomously migrated to specific nodes within the capillary for target capture. Leveraging the high acoustic response of gold nanorods, the aggregation points within the capillary created numerous “hot spots” under the influence of the acoustic field, leading to a remarkable 31.9-fold enhancement of the SERS signal compared to that of unaggregated nanomotors. The SERS signal was captured by the cell-phone-based SERS system, and its intensity exhibited a proportional relationship with the target miRNA concentration, yielding an impressive LOD of 6.15 × 10−13 M (Fig. 4j). The formation of stable gold nanorod assemblies required merely 1–2 seconds, and the detection process could be completed within 5 minutes, effectively minimizing the delay associated with current diagnostic methods. This innovative device effectively addressed the limitations of the prolonged nucleic acid detection period, cumbersome detection procedures, and low utilization efficiency of patient samples. It demonstrates the immense potential for achieving an ultrasensitive diagnosis of throat swab samples, thereby advancing the field of COVID-19 diagnostics.
Furthermore, the application of ultrasound-induced aggregation extends to the formation of micro/nanomotor arrays, facilitating the simultaneous in situ modification and detection of multiple samples within a single microcavity. This innovative approach enables efficient capture and aggregation of target nucleic acids, leading to enhanced signal intensity while minimizing the occurrence of false-positive signals.
Zhu et al. introduced an innovative acoustic chip that integrates two-dimensional micromotor arrays and fluorescence detection, utilizing piezoelectric transducers (PZTs) for ultrasound generation and PDMS for real-time visualization of the reaction process.75 This platform enabled in situ modification and detection, leading to enhanced performance. By leveraging dynamic ultrasound streaming, the microcavity generated well-organized ultrasound pressure nodes, facilitating rapid aggregation of particles and the formation of tissue-like arrays on these pressure nodes. This accelerated the efficiency of probe modification and miRNA detection. Furthermore, the acoustic wave allowed for the recycling of the target, enabling subsequent biological analyses. Although further advancements are needed to improve the capture efficiency of the micromotors on the target, the multi-locus reaction of the arrayed tissues opens up new possibilities for simultaneous in situ detection of multiple samples. This breakthrough greatly enhances the applicability and accuracy of disease diagnosis, presenting a promising direction for future diagnostic methodologies.
Subsequently, Zhu et al. developed an ultrasound-induced programmable micromotor array for the enrichment and simultaneous detection of multiple miRNAs in the sample solution (Fig. 4k).76 Through precise adjustment of the input frequency and voltage, a highly specific micromotor array was rapidly formed within the chamber, providing abundant reaction sites. The acoustic streaming further enhanced the contact between probes, facilitating subsequent modification processes. To enable multiplex detection, three distinct capture probes were immobilized on the microsphere surface using antigen–antibody interactions. Subsequently, miRNA-21 (FAM-labeled, red fluorescence), miRNA-210 (Cy3-labeled, green fluorescence), and miRNA-155 (Cy5-labeled, blue fluorescence) detection probes were introduced into the device. Upon the presence of target miRNAs, a sandwich structure formed, leading to the generation of multiple enhanced fluorescence signals, which were visualized using fluorescence microscopy (Fig. 4l). The fluorescence intensities exhibited a positive correlation with the corresponding miRNA concentrations. Remarkably, the limits of detection for miRNA-21, miRNA-210, and miRNA-155 probes in serum systems were achieved at 727.9 pM, 579.8 pM, and 584.3 pM, respectively. By engineering different protocols for the “sandwich” structure, this method enabled high-throughput, comprehensive, and cost-effective multi-miRNA detection. Notably, the approach demonstrated excellent specificity for different miRNAs and successfully demonstrated the feasibility of enriching and simultaneously detecting multiple miRNAs in real samples.
Compared with the chemically driven and magnetically driven methods, the ultrasound-induced aggregation technique emerges as a promising and versatile approach in the field of nucleic acid biosensing. This novel technique offers several advantages, including the ability to manipulate particle motion, integrate modification and detection processes, and accelerate the hybridization reactions between probes and micromotors within dedicated devices. Moreover, the ultrasound-induced aggregation method combines fluorescence and Raman enhancement, enabling precise quantitative detection of ultra-trace biomarkers. It successfully achieves nucleic acid enrichment and visual detection, demonstrating the significant potential for precise nucleic acid detection and early disease diagnosis. The integration of these capabilities positions ultrasound-induced aggregation as a valuable tool for advancing the field of diagnostic techniques.
Moreover, the integration of ultrasound-induced aggregation-enhanced biosensing with the lateral flow assay has been proposed by Huang et al. for the highly sensitive detection of ultra-trace proteins.78 Although this work is not exclusively focused on nucleic acid biosensing, its application can readily extend to the detection of DNA and RNA. The approach involved the utilization of ultrasound fields to facilitate the rapid capture of target proteins by antibody-modified gold nanoparticle nanomotors, which accumulated at specific ultrasound sites within the reaction cavity in just 20 seconds. Leveraging ultrasound-induced aggregation, a running solution was subsequently introduced into the cavity to remove any disturbances, such as blood cells and other proteins, present in the blood sample. The resulting complex, comprising gold nanoparticles, antibodies, and target proteins, was then transferred to the LFTS for protein detection. This innovative system enabled the specific detection of ultra-trace blood proteins within a linear range of 1–20 ng mL−1, achieving an impressive limit of detection (LOD) of 0.58 ng mL−1. This integrated platform demonstrated the synergistic combination of ultrasound-induced enrichment and the lateral flow assay, enhancing the detection capabilities for ultra-trace biomarkers in point-of-care clinical tests.
4. Conclusions and future perspectives
Herein, we have summarized micro/nanomotors based-biosensing strategies for the detection of nucleic acids, aiming to enable early disease diagnosis. The utilization of micro/nanomotors in these strategies offers unique advantages, including non-invasiveness, efficient target exposure, and the ability to perform tasks at an incredibly small scale. Traditional approaches have demonstrated the real-time detection of nucleic acids in human serum or living cells by surface modification of the micro/nanomotors with specific probes, which enable the capture of target nucleic acids and subsequent generation of detectable signals, such as quantitative fluorescence recovery or velocity changes. In the fluorescence recovery-based sensing method, the strong fluorescence signal can be imaged in real-time for precise analysis, and it can also be integrated with multiple external propulsions to achieve specific in vivo migration for directional detection. However, pretreatments involving modifications of probe and dye, complex hybridization reactions, and risks of fluorescence signal quenching are all reasons that hinder its further popularization. In addition, the velocity change-based method can quantitatively visualize the actual content of target nucleic acids into distinct range information, thus dramatically simplifying the detection procedure and stabilizing the signal. However, the reliance on chemical fuel for nano/micromotors propulsion limits its widely in vivo biosensing applications. Recently, researchers have introduced a novel technique called acoustic aggregation-enhanced nucleic acid biosensing, which exhibits distinctive advantages. This approach allows for the rapid in situ enrichment of target biomarkers and offers an ultrasensitive, fully integrated active platform with a simple structure and user-friendly operation. The acoustic field applied to the system can be adjusted to significantly amplify the signal generated by the aggregated target nucleic acids. As a result, the technique enables the ultrasensitive and real-time visualization of extremely low-abundance biomarkers, showcasing its exceptional capability for early-stage pathological examinations. Table 1 detailed summarized the artificial micro/nanomotors discussed in this review in terms of sensing mechanisms, materials, propulsion mode, and LOD.| Sensing mechanism | Materials | Propulsion mode | Analyte | LOD | Ref. |
|---|---|---|---|---|---|
| Fluorescence recovery-based | ssDNA/GO coated Au nanowires | Acoustic field | miRNA-21 and HPV16 E6 mRNA transcripts | — | 38 and 39 |
| erGO/Pt microtubes | Bubble propulsion | RPRM | 1.3 μM | 63 | |
| W5O14/PEDOT-Pt microwires | Bubble propulsion | miRNA-21 | 0.028 nM | 64 | |
| Au/Pt microtubes | SAW field | miRNA-21 | 0.19 nM | 65 | |
| PVA/MA/Fe3O4 | Magnetic field | Tumor DNA | — | 66 | |
| Velocity change-based | Au–Ni–Au–Pt nanowires | H2O2 decomposition | DNA | 40 aM | 34 |
| E. coli 16S rRNA | 7 × 103 CFU μL−1 | ||||
| Jellyfish-like Au/Ag/Ni/Au micromotors | Bubble propulsion | DNA | — | 69 | |
| DNA-modified Pt/Au NPs-PS beads | H2O2 decomposition | HIV-1 | ≥1000 virus particles per mL | 35 | |
| PEDOT-Au microtube | Bubble propulsion | DNA | 1 pmol | 46 | |
| Aggregation-enhanced nucleic acid biosensing | Gold nanorods | Acoustic field | DNA | 0.1 pM | 71 |
| FAM-labeled DNA probes-modified PS | Acoustic field | miRNA-101 | 5 pM | 72 | |
| Cy3-labeled capture probes modified PS | Acoustic field | miRNA-21 | 7.8 pM | 73 | |
| Gold nanorods | Acoustic field | COVID-19 | 6.15 × 10−13 M | 74 | |
| Cy3-labeled capture probes modified PS | Acoustic field | miRNA-141 | 107.9 pM (PBS) | 75 | |
| 768 pM (serum) | |||||
| Capture probes modified PS | Acoustic field | miRNA-21 | 727.9 pM | 76 | |
| miRNA-210 | 579.8 pM | ||||
| miRNA-155 | 584.3 pM | ||||
| Other sensing method | AuNR/PMO JNMs | NIR light propulsion | miRNA-21 | 18 fM | 77 |
Compared with traditional nucleic acid detection methods such as PCR and electrochemistry, micro/nanomotor assays undoubtedly have a huge advantage in terms of convenience and visualization with comparable detection limits and specificity. Despite the significant advancements made in micro/nanomotor-based detection methods, which have opened up new avenues for research and improved the sensitivity and selectivity of detection, there remain several challenges that need to be addressed for the successful implementation of portable in vivo diagnostic applications.
In the early stages of the disease, nucleic acid biomarkers are often present in ultra-trace amounts, posing a challenge for traditional qRT-PCR methods to achieve precise clinical diagnosis without significant misjudgment. Therefore, it is imperative to develop novel approaches that can enable rapid and accurate detection of extremely low volumes of target nucleic acids. This entails focusing on reducing the LOD and enhancing the sensitivity of biosensing techniques, ultimately paving the way for the development of promising and rapid ultra-sensitive biosensing platforms.
To further expand micro/nanomotors to practical large-scale nucleic acid biological detection, several factors require optimization:
(1) Expanding the possibilities of chemically driven fuels beyond the limited options like hydrogen peroxide is necessary. Innovative micro/nanomotor structures should be designed to autonomously identify and utilize bioavailable fuels in vivo for propulsion. This approach would eliminate the need for complex external fuels, enhancing the practicality of micro/nanomotors in biological systems.
(2) Passive micro/nanomotors, which solely rely on blood flow, may fail to reach the target area accurately, resulting in inefficient capture of target nucleic acids in complex biological samples. The inadequate diffusion also hampers the unambiguous identification of probes. This limitation can be overcome by precisely manipulating micro/nanomotors to the intended location using magnetic or acoustic driving techniques. However, it is essential to address the toxicity associated with magnetically modified micro/nanoparticles to ensure biocompatibility, allowing for harmless degradation or recycling during in vivo detection. Additionally, the limit of detection LOD of magnetic-based methods should be extended to enable large-scale detection of low-abundance target nucleic acids.
(3) Traditional qRT-PCR method involves time-consuming and inconvenient extraction and enrichment procedures before quantitative detection. Moreover, these methods exhibit poor responsiveness to ultra-trace biomarkers. To overcome these limitations, micro/nanomotor-based biosensing approaches should be economical, portable, and simple. They should be applicable in various analytical conditions and enable true real-time biosensing with affordable equipment, such as cell phone imaging. Liberating the detection process from bulky instruments and complex procedures would facilitate wider adoption and commercialization of micro/nanomotor-based biosensing.
(4) Ensuring high specificity is critical for removing false positive signals within heterogeneous environments. Micro/nanomotors should overcome the complex matrix effects within the serum, ensuring their successful arrival at designated areas for subsequent identification.
These challenges underscore the potential of micro/nanomotor-based nucleic acid biosensing methods. Further exploration is required to promote their widespread popularity, commercialization, and user-friendly application in medical diagnosis. Researchers are encouraged to dedicate more efforts toward addressing these challenges and unlocking the full potential of micro/nanomotor-based biosensing in the field of medical diagnostics.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge funding from National Natural Science Foundation of China (22104093 and 22234006), Guangdong Laboratory of Artificial Intelligence and Digital Economy (SZ) (Grant No. GML-KF-22-05), Shenzhen University 2035 Program for Excellent Research (86901-00000221), Shenzhen Stability Support Plan (20220809165141001), Joint Fund of the Ministry of Education for Equipment Pre-research (8091B022142), and Shenzhen Key Laboratory for Nano-Biosensing Technology (ZDSYS20210112161400001).References
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- Y. Zhao, X. Zuo, Q. Li, F. Chen, Y. R. Chen, J. Deng, D. Han, C. Hao, F. Huang, Y. Huang, G. Ke, H. Kuang, F. Li, J. Li, M. Li, N. Li, Z. Lin, D. Liu, J. Liu, L. Liu, X. Liu, C. Lu, F. Luo, X. Mao, J. Sun, B. Tang, F. Wang, J. Wang, L. Wang, S. Wang, L. Wu, Z. S. Wu, F. Xia, C. Xu, Y. Yang, B. F. Yuan, Q. Yuan, C. Zhang, Z. Zhu, C. Yang, X. B. Zhang, H. Yang, W. Tan and C. Fan, Sci. China: Chem., 2021, 64, 171–203 CrossRef CAS PubMed.
- L. Wu and X. Qu, Chem. Soc. Rev., 2015, 44, 2963–2997 RSC.
- P. Anker, H. Mulcahy, X. Q. Chen and M. Stroun, Cancer Metastasis Rev., 1999, 18, 65–73 CrossRef CAS PubMed.
- F. Diehl, K. Schmidt, M. A. Choti, K. Romans, S. Goodman, M. Li, K. Thornton, N. Agrawal, L. Sokoll, S. A. Szabo, K. W. Kinzler, B. Vogelstein and L. A. Diaz, Jr., Nat. Med., 2008, 14, 985–990 CrossRef CAS PubMed.
- H. Dong, J. Lei, L. Ding, Y. Wen, H. Ju and X. Zhang, Chem. Rev., 2013, 113, 6207–6233 CrossRef CAS PubMed.
- D. Guévremont, H. Tsui, R. Knight, C. J. Fowler, C. L. Masters, R. N. Martins, W. C. Abraham, W. P. Tate, N. Cutfield and J. M. Williams, Alzheimer's Dementia, 2020, 16, e04746 Search PubMed.
- C. Guay, E. Roggli, V. Nesca, C. Jacovetti and R. Regazzi, Transl. Res., 2011, 157, 253–264 CrossRef CAS PubMed.
- A. B. Chinen, C. M. Guan, J. R. Ferrer, S. N. Barnaby, T. J. Merkel and C. A. Mirkin, Chem. Rev., 2015, 115, 10530–10574 CrossRef CAS PubMed.
- C. Liu, T. Xu, L. P. Xu and X. Zhang, Micromachines, 2017, 9, 10 CrossRef PubMed.
- M. Wan, T. Li, H. Chen, C. Mao and J. Shen, Angew. Chem., Int. Ed., 2021, 60, 13158–13176 CrossRef CAS PubMed.
- K. Villa and M. Pumera, Chem. Soc. Rev., 2019, 48, 4966–4978 RSC.
- H. Zhao, Y. Zheng, Y. Cai, T. Xu, R. Dong and X. Zhang, Nano Today, 2023, 52, 101939 CrossRef CAS.
- W. Liu, Y. Liu, H. Li, H. M. Nie, M. Y. Tian and W. Long, Adv. Funct. Mater., 2023, 33, 2212452 CrossRef CAS.
- X. Y. Jiao, Z. M. Wang, J. D. Xiu, W. H. Dai, L. Zhao, T. L. Xu, X. Du, Y. Q. Wen and X. J. Zhang, Appl. Mater. Today, 2020, 18, 100504 CrossRef.
- Y. Xing, M. Y. Zhou, X. Du, X. Y. Li, J. Q. Li, T. L. Xu and X. J. Zhang, Appl. Mater. Today, 2019, 17, 85–91 CrossRef.
- X. Ma, K. Hahn and S. Sanchez, J. Am. Chem. Soc., 2015, 137, 4976–4979 CrossRef CAS PubMed.
- Z. Wu, L. Li, Y. Yang, P. Hu, Y. Li, S. Y. Yang, L. V. Wang and W. Gao, Sci. Robot., 2019, 4, eaax0613 CrossRef PubMed.
- S. Tang, F. Zhang, H. Gong, F. Wei, J. Zhuang, E. Karshalev, B. Esteban-Fernández de Ávila, C. Huang, Z. Zhou, Z. Li, L. Yin, H. Dong, R. H. Fang, X. Zhang, L. Zhang and J. Wang, Sci. Robot., 2020, 5, eaba6137 CrossRef PubMed.
- T. Cui, S. Wu, Y. Sun, J. Ren and X. Qu, Nano Lett., 2020, 20, 7350–7358 CrossRef CAS PubMed.
- M. Xuan, J. Shao, C. Gao, W. Wang, L. Dai and Q. He, Angew. Chem., 2018, 130, 12643–12647 CrossRef.
- Y. Xing, M. Zhou, T. Xu, S. Tang, Y. Fu, X. Du, L. Su, Y. Wen, X. Zhang and T. Ma, Angew. Chem., Int. Ed., 2020, 59, 14368–14372 CrossRef CAS PubMed.
- P. P. Yang, Y. G. Zhai, G. B. Qi, Y. X. Lin, Q. Luo, Y. Yang, A. P. Xu, C. Yang, Y. S. Li, L. Wang and H. Wang, Small, 2016, 12, 5423–5430 CrossRef CAS PubMed.
- J. Shao, M. Abdelghani, G. Shen, S. Cao, D. S. Williams and J. C. M. van Hest, ACS Nano, 2018, 12, 4877–4885 CrossRef CAS PubMed.
- M. Wan, Q. Wang, X. Li, B. Xu, D. Fang, T. Li, Y. Yu, L. Fang, Y. Wang, M. Wang, F. Wang, C. Mao, J. Shen and J. Wei, Angew. Chem., Int. Ed., 2020, 59, 14458–14465 CrossRef CAS PubMed.
- M. Wan, Q. Wang, R. Wang, R. Wu, T. Li, D. Fang, Y. Huang, Y. Yu, L. Fang, X. Wang, Y. Zhang, Z. Miao, B. Zhao, F. Wang, C. Mao, Q. Jiang, X. Xu and D. Shi, Sci. Adv., 2020, 6, eaaz9014 CrossRef CAS PubMed.
- Z. Sun, T. Wang, J. Wang, J. Xu, T. Shen, T. Zhang, B. Zhang, S. Gao, C. Zhao, M. Yang, F. Sheng, J. Yu and Y. Hou, J. Am. Chem. Soc., 2023, 145, 11019–11032 CrossRef CAS PubMed.
- G. Go, A. Yoo, H. W. Song, H. K. Min, S. Zheng, K. T. Nguyen, S. Kim, B. Kang, A. Hong, C. S. Kim, J. O. Park and E. Choi, ACS Nano, 2021, 15, 1059–1076 CrossRef CAS PubMed.
- X. Peng, S. Tang, D. Tang, D. Zhou, Y. Li, Q. Chen, F. Wan, H. Lukas, H. Han, X. Zhang, W. Gao and S. Wu, Sci. Adv., 2023, 9, eadh1736 CrossRef CAS PubMed.
- S. Sanchez, A. A. Solovev, S. Schulze and O. G. Schmidt, Chem. Commun., 2011, 47, 698–700 RSC.
- S. Balasubramanian, D. Kagan, C. M. Hu, S. Campuzano, M. J. Lobo-Castanon, N. Lim, D. Y. Kang, M. Zimmerman, L. Zhang and J. Wang, Angew. Chem., Int. Ed., 2011, 50, 4161–4164 CrossRef CAS PubMed.
- X. Ding, S. C. Lin, B. Kiraly, H. Yue, S. Li, I. K. Chiang, J. Shi, S. J. Benkovic and T. J. Huang, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11105–11109 CrossRef CAS PubMed.
- A. Aziz, J. Holthof, S. Meyer, O. G. Schmidt and M. Medina-Sanchez, Adv. Healthcare Mater., 2021, 10, e2101077 CrossRef PubMed.
- J. Wu, S. Balasubramanian, D. Kagan, K. M. Manesh, S. Campuzano and J. Wang, Nat. Commun., 2010, 1, 36 CrossRef PubMed.
- M. S. Draz, K. M. Kochehbyoki, A. Vasan, D. Battalapalli, A. Sreeram, M. K. Kanakasabapathy, S. Kallakuri, A. Tsibris, D. R. Kuritzkes and H. Shafiee, Nat. Commun., 2018, 9, 4282 CrossRef PubMed.
- Y. Wang, X. Liu, C. Chen, Y. Chen, Y. Li, H. Ye, B. Wang, H. Chen, J. Guo and X. Ma, ACS Nano, 2022, 16, 180–191 CrossRef CAS PubMed.
- V. V. Singh, K. Kaufmann, B. E.-F. de Ávila, E. Karshalev and J. Wang, Adv. Funct. Mater., 2016, 26, 6270–6278 CrossRef CAS.
- B. Esteban-Fernandez de Avila, A. Martin, F. Soto, M. A. Lopez-Ramirez, S. Campuzano, G. M. Vasquez-Machado, W. Gao, L. Zhang and J. Wang, ACS Nano, 2015, 9, 6756–6764 CrossRef CAS PubMed.
- J. R. Qualliotine, G. Bolat, M. Beltran-Gastelum, B. E. de Avila, J. Wang and J. A. Califano, Otolaryngol.–Head Neck Surg., 2019, 161, 814–822 CrossRef PubMed.
- M. Urso and M. Pumera, Adv. Funct. Mater., 2022, 32, 2200711 CrossRef CAS.
- S. Campuzano, D. Kagan, J. Orozco and J. Wang, Analyst, 2011, 136, 4621–4630 RSC.
- R. Maria-Hormigos, B. Jurado-Sanchez and A. Escarpa, Anal. Bioanal. Chem., 2022, 414, 7035–7049 CrossRef CAS PubMed.
- R. Dong, Y. Cai, Y. Yang, W. Gao and B. Ren, Acc. Chem. Res., 2018, 51, 1940–1947 CrossRef CAS PubMed.
- T. Xu, F. Soto, W. Gao, V. Garcia-Gradilla, J. Li, X. Zhang and J. Wang, J. Am. Chem. Soc., 2014, 136, 8552–8555 CrossRef CAS PubMed.
- T. Xu, F. Soto, W. Gao, R. Dong, V. Garcia-Gradilla, E. Magana, X. Zhang and J. Wang, J. Am. Chem. Soc., 2015, 137, 2163–2166 CrossRef CAS PubMed.
- K. Van Nguyen and S. D. Minteer, Chem. Commun., 2015, 51, 4782–4784 RSC.
- J. Li, S. Thamphiwatana, W. Liu, B. Esteban-Fernandez de Avila, P. Angsantikul, E. Sandraz, J. Wang, T. Xu, F. Soto, V. Ramez, X. Wang, W. Gao, L. Zhang and J. Wang, ACS Nano, 2016, 10, 9536–9542 CrossRef CAS PubMed.
- W. Gao, A. Pei and J. Wang, ACS Nano, 2012, 6, 8432–8438 CrossRef CAS PubMed.
- F. Mou, C. Chen, H. Ma, Y. Yin, Q. Wu and J. Guan, Angew. Chem., Int. Ed., 2013, 52, 7208–7212 CrossRef CAS PubMed.
- W. Gao, R. Dong, S. Thamphiwatana, J. Li, W. Gao, L. Zhang and J. Wang, ACS Nano, 2015, 9, 117–123 CrossRef CAS PubMed.
- Y. Ji, X. Lin, Z. Wu, Y. Wu, W. Gao and Q. He, Angew. Chem., Int. Ed., 2019, 58, 12200–12205 CrossRef CAS PubMed.
- H. Zhang, W. Duan, M. Lu, X. Zhao, S. Shklyaev, L. Liu, T. J. Huang and A. Sen, ACS Nano, 2014, 8, 8537–8542 CrossRef CAS PubMed.
- S. Sengupta, D. Patra, I. Ortiz-Rivera, A. Agrawal, S. Shklyaev, K. K. Dey, U. Cordova-Figueroa, T. E. Mallouk and A. Sen, Nat. Chem., 2014, 6, 415–422 CrossRef CAS PubMed.
- X. Ma, X. Wang, K. Hahn and S. Sanchez, ACS Nano, 2016, 10, 3597–3605 CrossRef CAS PubMed.
- S. Sanchez, L. Soler and J. Katuri, Angew. Chem., Int. Ed., 2015, 54, 1414–1444 CrossRef CAS PubMed.
- Y. Ye, F. Tong, S. Wang, J. Jiang, J. Gao, L. Liu, K. Liu, F. Wang, Z. Wang, J. Ou, B. Chen, D. A. Wilson, Y. Tu and F. Peng, Nano Lett., 2021, 21, 8086–8094 CrossRef CAS PubMed.
- T. Xu, W. Gao, L. P. Xu, X. Zhang and S. Wang, Adv. Mater., 2017, 29, 1603250 CrossRef PubMed.
- P. Mandal, G. Patil, H. Kakoty and A. Ghosh, Acc. Chem. Res., 2018, 51, 2689–2698 CrossRef CAS PubMed.
- H. Yu, W. Tang, G. Mu, H. Wang, X. Chang, H. Dong, L. Qi, G. Zhang and T. Li, Micromachines, 2018, 9, 540 CrossRef PubMed.
- T. Laurell, F. Petersson and A. Nilsson, Chem. Soc. Rev., 2007, 36, 492–506 RSC.
- T. Xu, L.-P. Xu and X. Zhang, Appl. Mater. Today, 2017, 9, 493–503 CrossRef.
- P. L. Venugopalan, B. Esteban-Fernandez de Avila, M. Pal, A. Ghosh and J. Wang, ACS Nano, 2020, 14, 9423–9439 CrossRef CAS PubMed.
- D. F. Báez, G. Ramos, A. Corvalán, M. L. Cordero, S. Bollo and M. J. Kogan, Sens. Actuators, B, 2020, 310, 127843 CrossRef.
- G. C. Cogal, G. Y. Karaca, E. Uygun, F. Kuralay, L. Oksuz, M. Remskar and A. U. Oksuz, Anal. Chim. Acta, 2020, 1138, 69–78 CrossRef CAS PubMed.
- G. C. Cogal, P. K. Das, G. Y. Karaca, V. R. Bhethanabotla and A. U. Oksuz, ACS Appl. Bio Mater., 2021, 4, 7932–7941 CrossRef PubMed.
- F. Y. Qin, J. Y. Wu, D. M. Fu, Y. Feng, C. Gao, D. Z. Xie, S. M. Fu, S. Y. Liu, D. A. Wilson and F. Peng, Appl. Mater. Today, 2022, 27, 101456 CrossRef.
- Y. Xie, S. Fu, J. Wu, J. Lei and H. Ju, Biosens. Bioelectron., 2017, 87, 31–37 CrossRef CAS PubMed.
- S. Fu, X. Zhang, Y. Xie, J. Wu and H. Ju, Nanoscale, 2017, 9, 9026–9033 RSC.
- X. Zhang, C. Chen, J. Wu and H. Ju, ACS Appl. Mater. Interfaces, 2019, 11, 13581–13588 CrossRef CAS PubMed.
- G. Y. Karaca, H. K. Kaya, F. Kuralay and A. U. Oksuz, Int. J. Biol. Macromol., 2021, 193, 370–377 CrossRef PubMed.
- T. Xu, Y. Luo, C. Liu, X. Zhang and S. Wang, Anal. Chem., 2020, 92, 7816–7821 CrossRef CAS PubMed.
- Y. Sun, Y. Luo, T. Xu, G. Cheng, H. Cai and X. Zhang, Talanta, 2021, 233, 122517 CrossRef CAS PubMed.
- Y. Luo, C. Fan, Y. Song, T. Xu and X. Zhang, Biosens. Bioelectron., 2022, 210, 114297 CrossRef CAS PubMed.
- Y. Luo, M. Zhou, C. Fan, Y. Song, L. Wang, T. Xu and X. Zhang, Anal. Chem., 2023, 95, 5316–5322 CrossRef CAS PubMed.
- Q. Zhu, T. Xu, Y. Song, Y. Luo, L. Xu and X. Zhang, Biosens. Bioelectron., 2020, 158, 112185 CrossRef CAS PubMed.
- Q. Zhu, T. Yan, Y. Yang, Y. Song, J. Lu, Y. Luo, L. P. Xu and T. Xu, ACS Sens., 2022, 7, 3654–3659 CrossRef CAS PubMed.
- M. Chen, E. Ma, Y. Xing, H. Xu, L. Chen, Y. Wang, Y. Zhang, J. Li, H. Wang and S. Zheng, ACS Sens., 2023, 8, 757–766 CrossRef CAS PubMed.
- Y. Huang, T. Xu, Y. Luo, C. Liu, X. Gao, Z. Cheng, Y. Wen and X. Zhang, Ultra-Trace Protein Detection by Integrating Lateral Flow Biosensor with Ultrasound Enrichment., Analytical Chemistry, 2021, 93(5), 2996–3001 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |