Open Access Article
Open Access ArticleEngineered aptamers for molecular imaging
Bingqian
Lin
 *,
Feng
Xiao
,
Jinting
Jiang
,
Zhengjia
Zhao
and
Xiang
Zhou
*,
Feng
Xiao
,
Jinting
Jiang
,
Zhengjia
Zhao
and
Xiang
Zhou

College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Department of Hematology of Zhongnan Hospital, Taikang Center for Life and Medical Sciences, Wuhan University, Wuhan 430072, China. E-mail: bingqianlin@whu.edu.cn
First published on 21st November 2023
Abstract
Molecular imaging, including quantification and molecular interaction studies, plays a crucial role in visualizing and analysing molecular events occurring within cells or organisms, thus facilitating the understanding of biological processes. Moreover, molecular imaging offers promising applications for early disease diagnosis and therapeutic evaluation. Aptamers are oligonucleotides that can recognize targets with a high affinity and specificity by folding themselves into various three-dimensional structures, thus serving as ideal molecular recognition elements in molecular imaging. This review summarizes the commonly employed aptamers in molecular imaging and outlines the prevalent design approaches for their applications. Furthermore, it highlights the successful application of aptamers to a wide range of targets and imaging modalities. Finally, the review concludes with a forward-looking perspective on future advancements in aptamer-based molecular imaging.
1. Introduction
Molecular imaging is a method that captures the molecular events happening inside the cells or organisms by visualizing and quantifying the functional bio-molecules.1,2 The studies of different molecular events, such as variation of molecular numbers, molecular mapping, and molecular interactions, promote a deep understanding of biological processes. Consequently, molecular imaging allows for the tracking of molecular abnormalities, which makes it different from other well-established diagnostic tests that measure the apparent symptoms. Thus, it is emerging as a powerful tool for early diagnosis, treatment evaluation, and prognostic monitoring.3In molecular imaging, recognition elements for various targets play a crucial role in high-performance imaging. Numerous molecules have been reported to be utilized as recognition elements, such as small organic molecules,4 peptides,5 antibodies,6 or functional nucleic acids (DNAzymes or aptamers7,8). Aptamers are single-stranded DNAs or RNAs that can fold into diverse secondary and tertiary structures for highly sensitive and specific binding with various target biomolecules.9 Compared to small organic molecules and peptides, aptamers possess a better programmability and biocompatibility. In molecular imaging, antibodies play a vital role in biomolecular recognition. Aptamers have a great superiority over antibodies as targeting ligands in molecular imaging owing to their intrinsic features. First, aptamers usually have lengths ranging 10–100 nucleotide residues (12–30 kDa), which are much smaller than antibodies (150–170 kDa).10 This characteristic enables aptamers to have a faster diffusion rate through tissues or any other biological barriers because of a minimized steric hindrance.11 It has been demonstrated that the glycosylation of proteins causes a spatial site blockage for antibody recognition, while the small size of aptamers allows for more efficient binding.12 Second, owing to the composition and small molecular weight of the aptamers, they exhibit low immunogenicity in vivo, allowing the potential for widespread use with low side effects.13 Third, the generation of aptamers towards different targets via the systematic evolution of ligands by exponential enrichment (SELEX) possesses the merits of being faster, reproducible, and more economical than the production of antibodies. Moreover, aptamers can be chemically synthesized and modified with various functional groups for a unique gain of functions.9
However, when compared to peptides and antibodies, aptamers as single-stranded oligonucleotides present distinct challenges in their in vivo imaging applications owing to their inherent characteristics, including flexibility and susceptibility to nucleases. Therefore, it is necessary to develop corresponding engineering methods to confer aptamers with higher resistance toward nucleases, improved stability, and enhanced specificity and sensitivity. Engineered aptamers can be achieved through various methods, including chemical modification, designed target activation strategies, and strategies involving nanostructures. First, aptamers can be easily cleared in vivo with a half-life of only a few minutes, especially for aptamers smaller than 5 nm in size, which are prone to degradation by nucleases or cleared through renal excretion.14 Consequently, the reduced circulation time results in a low efficiency of binding between aptamers and targets, thus limiting their application in molecular imaging. To improve the stability of aptamers in complex environments and to increase their half-life, researchers have developed many methods involving chemical modifications15 and the introduction of artificial bases.16
Additionally, in molecular imaging, aptamers can be strategically engineered to enable precise spatial–temporal imaging through various target activation approaches and nanostructure-assisted strategies. Specifically, aptamers efficiently identify the desired target, subsequently transducing it to the signal output module through diverse reporters, which enables detection through diverse imaging modalities. Depending on the imaging modality employed, such as fluorescence imaging (FL), magnetic resonance imaging (MRI), and computed tomography (CT), reporter probes can incorporate fluorescence moieties, radiolabels, and other appropriate functional groups.
In this review, we provide a summary of aptamers employed in molecular imaging and demonstrate rational design strategies for aptamer-based molecular imaging. Moreover, a comprehensive introduction is provided on the targets and imaging modalities of aptamers. Finally, the future prospects of aptamers in molecular imaging are discussed. Several recent reviews have addressed the use of aptamers in molecular imaging. For example, Yin et al. introduced aptamer-functionalized nanomaterials in molecular imaging.17 Chen et al. provided a general overview of design and application strategies in molecular imaging.8 In addition, Zhou et al. described the application of functional nucleic acids (including aptamers and DNAzymes) in cellular molecular imaging and manipulation.7 Compared to other reviews related to aptamer-based molecular imaging, this review focuses on strategies for engineering aptamers in molecular imaging and differs from other reviews because it specifically addresses relevant design strategies for aptamers in molecular imaging in a useful way. An overview of the engineered aptamer for molecular imaging is shown in Fig. 1.
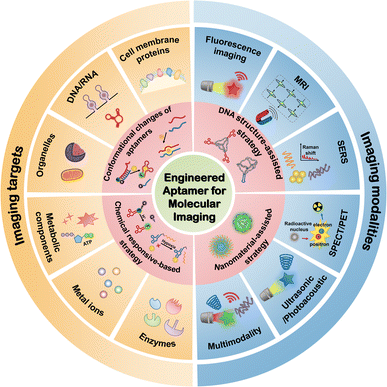 | ||
| Fig. 1 An overview of the engineered aptamer for molecular imaging, including design strategies, targets, and imaging modalities. | ||
2. Aptamers used in molecular imaging
Various aptamers have been applied in imaging, as summarized in Tables 1 and 2.| Aptamer name | Nature | Ligand/target | Imaging | Ref. |
|---|---|---|---|---|
| ATP aptamer | DNA | ATP | ATP | 18 and 19 |
| ATP aptamer | DNA | ATP | ATP | 20 |
| ATP aptamer | DNA | ATP | ATP | 21 |
| TD05 | DNA | Mucin 1 (MUC1) | Membrane proteins | 22 |
| AS1411 | DNA | Nucleolin | Membrane proteins | 23–27 |
| Sgc8c | DNA | Protein tyrosine kinase-7 (PTK7) | Membrane proteins | 28 and 29 |
| T-Met/T-TβRII | DNA | Hepatocyte growth factor (HGF)/Transforming growth factor-β type II receptor (TβRII) | Membrane proteins | 30 |
| Apt2 | DNA | Human epidermal growth factor receptor 2 (HER2) | Membrane proteins | 31 |
| SYL3C | DNA | Epidermal growth factor receptor (EpCAM) | Membrane proteins | 32 |
| MJ5C | DNA | Programmed cell death 1 ligand 1 (PD-L1) | Exosomes proteins | 33 |
| Neu5Ac | DNA | Glycan | Glycosylated RNAs | 34 |
| Aptamer name | Nature | Ligand/target | Imaging | Ref. |
|---|---|---|---|---|
| Broccoli | RNA | DFHBI/DFHBI-1T | RNA | 35 and 36 |
| SiRA | RNA | Silicon rhodamines | RNA | 37 |
| o-Coral | RNA | Gemini-561 | RNA | 38 |
| Mango II | RNA | To1-biotin | RNA | 39 |
| BeCA | RNA | BC fluorophores | RNA | 40 |
| degApt | RNA | csiFP | RNA | 41 |
| Inert pepper | RNA | HBC | RNA | 42 |
| RhoBAST | RNA | Rhodamine | RNA | 43 |
| Corn | RNA | DFHO | RNA | 44 |
| NapRA | RNA | Nap | RNA | 45 |
| Pepper | RNA | HBC | RNA | 46 |
| ATP aptamer | RNA | ATP | ATP | 47 |
| A9g | RNA | Prostate-specific membrane antigen (PSMA) | Membrane proteins | 48 |
| 5FU-EGFR aptamer | RNA | 5FU/epidermal growth factor receptor (EGFR) | Membrane proteins | 49 |
| MS2/PP7 aptamers | RNA | MS2/PP7 | Telomeres | 50 |
| DNB | RNA | NB | Adenine | 51 |
| MinE07 | RNA | EGFR | Membrane proteins | 52 |
| OTRS-FLAP | RNA | DFHBI/mitochondria | Mitochondria | 53 |
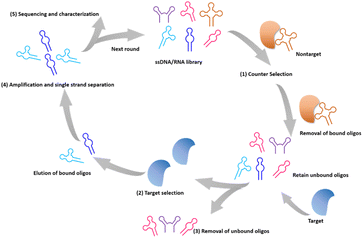
Advances in in vitro screening technologies have facilitated the versatility of aptamers for target recognition, allowing the screening of aptamers for different targets. Ellington54 and Tuerk55 introduced the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) in 1990, marking a significant milestone in the field of in vitro screening methods. As shown in Fig. 2, the typical SELEX contains a series of iterative selection rounds for aptamers with strong binding affinity. First, the initial library is designed with diverse sequences containing a random region of 20–50 bases, providing a vast repertoire ranging from 1013–1015.56 After a pre-treatment of counterselection with nontargets, this library is incubated with the targets under the defined conditions. When the unbound oligos are removed, the bound oligos are amplified to proceed to the next round of screening. This screening step is repeated until a collection of sequences with the desired affinity is obtained. These sequence sets are then subjected to sequencing, and the final sequence is obtained through synthesis and further optimization and identification processes.
Specifically, a technique known as Cell-SELEX allows the direct screening of aptamers by utilizing whole cells as the target for selection; it helps to screen for aptamers that specifically bind to cell surface proteins.57 This approach provides a screening solution for cell surface potential signature proteins and outperforms the strategy of screening for cell surface signature proteins alone, avoiding the problem of screening for aptamers that do not bind cells owing to incorrect target protein selection. Many tumor cell surface protein markers can be identified through Cell-SELEX screening, demonstrating high affinity and specificity.
The interactions between aptamers and ligands involve several effects, such as electrostatic forces,58 hydrogen bonds,59 hydrophobic effect,60 π–π stacking61 and van der Waals force.62,63 In addition, the structure of the aptamer can change with temperature and pH.64 Considering that aptamers are susceptible to degradation and removal in the physiological environment, researchers have developed various nucleic acid modification methods to improve the stability of aptamers in molecular imaging applications.15
The application of RNA aptamers in vivo is challenged by the inherent thermal and enzymatic instability of RNA. As shown in Fig. 3A, RNA thermal instability is due to the easy degradation of RNA, which starts with the deprotonation of the 2′ hydroxyl group attacking the adjacent 3′-phosphate, accompanied by the departure of the proximal 5′-hydroxyl group, resulting in the formation of a 2′3′-cyclic phosphate.65 However, the enzymatic fragility of RNA derives from the fact that most ribonucleases attack the phosphodiester bond by polarizing the 2′-hydroxyl group in the RNA aptamer. Consequently, the chemical substitution of the RNA 2′-hydroxyl group is an effective way to improve the bio-stability of RNA aptamers, such as 2′-fluoro group,66 2′-amino, and 2′-O-methyl,67–69 and locked nucleic acids (LNAs)70 (Fig. 3B(a)). In addition, the modification of the phosphodiester backbone by boranophosphate or phosphorothioate residues and nucleobases is effective because of the hydrolysis process (Fig. 3B(b and c)). In comparison to RNA aptamers, DNA aptamers exhibit superior stability owing to the absence of 2′-hydroxyl groups. Additionally, the stability of DNA aptamers can be further enhanced through various modifications, such as the incorporation of locking nucleic acids or the utilization of a phosphorothioate backbone. These modifications contribute to the increased stability and resilience of DNA aptamers in diverse biological contexts. Wang et al. prepared a single-molecule polymer–aptamer conjugate named PSP-pacDNA, which effectively suppressed nonspecific interactions between the aptamer and cells, leading to improved plasma pharmacokinetics.71
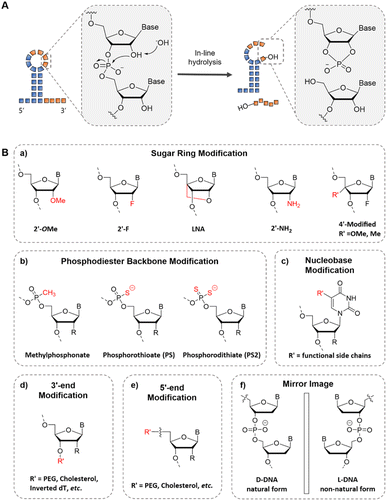 | ||
| Fig. 3 The chemical modification of aptamers for improved stability. (A) The mechanism for RNA thermal instability.65 (B) The chemical modification in different sites of aptamers: (a) the sugar ring modification, (b) phosphodiester backbone modification, (c) nucleobase modification, and (d and e) end modification. (f) The chiral form of DNA. R represents –H for DNA and –OH for RNA. | ||
Additionally, to address the issue of rapid clearance of aptamers in vivo, one of the most employed methods in molecular imaging is to conjugate polyethylene glycol (PEG) with aptamers at the 5′ or 3′ end (Fig. 3B(d and e)).71 This modification strategy increases the size and molecular weight of the aptamer, effectively alleviating renal clearance and prolonging the in vivo circulation time. Given that 3′ exonuclease activity primarily accounts for the degradation of deoxy oligonucleotides, implementing 3′-terminal modifications in DNA aptamers can significantly enhance their resistance against nucleases, such as 3′ inverted dT (Fig. 3B(d)).10 In addition, the L-form of DNA can confer resistance to enzymatic cleavage, thereby enhancing its stability (Fig. 3B(f)).72 The first application of L-aptamers in a living system was reported by Zhong et al.73 The structure of the block aptamer was formed using peptide nucleic acid (PNA) to form a heterochiral double strand with the L-Mango aptamer, thereby blocking the aptamer. In the presence of endogenous miRNA, complementary pairing with PNA releases the L-Mango aptamer, enabling it to bind with the dye and activate it. This allows for the imaging of miRNA-155 expression in living cells.
Although the aptamer is rapidly cleared from circulation, its rapid diffusion into the target tissue also allows the maximum signal-to-noise ratio to be reached in a short time. Considering the stability of the signal output and reproducibility, the stability of the aptamer is still the primary consideration.
3. Design strategies for aptamer-based molecular imaging
Aptamers can selectively bind to specific targets by adopting different structural conformations, such as pseudoknots, hairpins, G-quadruplexes, and convex rings.74 The binding modes between the aptamer and its target can be categorized into two types based on the spatial relationship between them. To bind small molecules, the targets are usually embedded in “pockets” folded by aptamer sequences, such as ATP,75 theophylline,76 cocaine,77 and K+.78 For large molecules, such as proteins with complex spatial structures, aptamers are usually adapted to the spatial structure of the target by folding into special shapes, such as thrombin79 and platelet-derived growth factor BB (PDGF-BB).80Because most aptamers undergo conformational changes during interaction with the target, this facilitates the signal output of various recognition activation methods. In addition, aptamers can achieve signal activation by modifying chemically responsive functional groups. DNA nanostructures can serve as carriers for aptamers and act as reactions for signal output. Furthermore, aptamers can be combined with nanomaterials as carriers or signal outputs for molecular imaging. This section aims to introduce various aptamer design strategies employed in molecular imaging (Fig. 4).
3.1 Design strategies based on conformational changes in aptamers
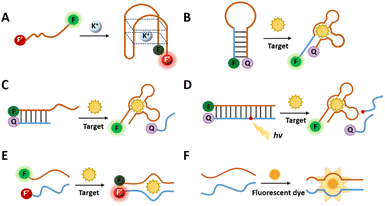
In general, probes for imaging usually comprise sensing and signal modules. The sensing module contains a recognition element for identifying the target. Upon target recognition, it transduces the signal to the signal module, which generates a signal that is detected by the corresponding imaging modality. Functionally modified aptamers can directly bind to targets for molecular imaging.81,82 For example, Wang et al. selected an aptamer yly12 that was against L1CAM.82 However, the direct binding mechanism of aptamers towards the target keeps the signal active, which leads to the low signal-to-noise, thus reducing the accuracy of the imaging.83 Considering this, researchers have developed several design strategies by utilizing the conformational change, making the signal output triggered conditionally.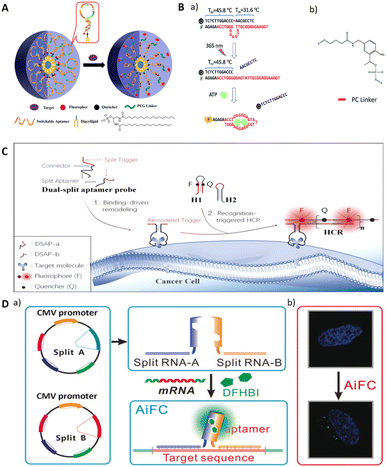 | ||
| Fig. 5 (A) The design of switchable aptamer micelle flares. Adapted with permission.85 Copyright 2013, American Chemical Society. (B) A schematic illustration of a photo-regulated aptamer sensor for ATP. Adapted with permission.75 Copyright 2020, Royal Society of Chemistry. (C) A schematic of the dual split aptamer strategy combined with the HCR for cell imaging. Adapted with permission.87 Copyright 2020, American Chemical Society. (D) An illustration of cell RNA imaging by the split light-up RNA aptamer. Adapted with permission.88 Copyright 2018 Wiley. | ||
Another approach to inducing conformational changes in aptamer sequences involves the incorporation of complementary sequences that bind to the aptamer region (Fig. 4C and D).89 The aptamer sequence and the complementary sequence are labeled with different reporter motifs at the adjacent ends of the sequence undergoing FRET. When the target molecule competes with the complementary sequence for binding to the aptamer, it can lead to dissociation of the complementary sequence, resulting in a detectable change in the fluorescence signal. This design provides a dynamic and responsive system for monitoring target binding events.
Lu's group reports an ATP detection device for mitochondria in living cells (Fig. 5B).75 The use of a photocleavable linker partially complementary to the aptamer sequence results in the functional inhibition of the aptamer, preventing it from binding to the target. Concurrently, the sequence with a photo-cleavable (PC) linker is labeled with FAM and quench groups at the same end of the complementary pairing with the aptamer, leading to fluorescence quenching. When the PC linker is disconnected by light, the aptamer binds to ATP, releasing the sequence with the quench group and restoring fluorescence. In addition to the photo-regulated method, Chai et al. designed a complementary sequence with a disulfide bond that binds to an ATP aptamer.90 In the presence of glutathione, the disulfide bond is cleaved, resulting in the conformational change in the aptamer and its subsequent binding to ATP.
One of the most commonly utilized types of split RNA aptamers is light-up RNA aptamers. This class of RNA aptamers includes Spinach,92 Broccoli,93 Pepper,94 and Mango95 which “turn on” fluorescence when combined with small molecule dyes. Split light-up aptamer designs typically contain two binding RNA aptamer fragments and a fluorescent dye. The binding of the two aptamer fragments enables the formation of dye binding sites, and the fluorescence of the dye is enhanced upon binding to the aptamer (Fig. 4F). When integrating such aptamers with target-specific sequences, different targets can be detected, including proteins, metabolites, and RNAs.96 Wang et al. reported an induced fluorescence complementation (AiFC) method for RNA imaging. Their method modifies the RNA aptamer Broccoli into two split fragments that can bind in tandem with the target mRNA. During gene coding in cells, endogenous mRNA molecules bind to each of the two fragments of split-Broccoli, bringing them in close spatial proximity to form in situ fluorophore binding sites and activate fluorescence (Fig. 5D).88
Designing split aptamers can be challenging because of the requirement to preserve the aptamer's binding capability. The insufficient understanding of aptamer's three-dimensional structures may result in incorrect division, thus causing the loss of aptamer binding ability. Another possibility is that the fragmented region does not affect the aptamer's binding, resulting in an ineffective split design. Therefore, more research needs to be invested in determining the three-dimensional structure of the different aptamers so that they can be designed accurately.
3.2 Chemical modification-based design strategies
The conformational change-dependent designs aforementioned may face problems such as incomplete noise suppression, poor stability, and the requirement for complex sequence design. An alternative approach involves activating aptamer signaling through a chemical modification that occurs via chemical removal in response to specific conditioning. Shi modified a fluorescent moiety near the aptamer tail, which is connected to the quenching moiety via an acid-unstable acetal linker 3,9-bis(3-aminopropyl)-2, 4, 8, 10-tetraoxaspiro[5.5]undecane (ATU) to construct a PH-responsive aptamer probe (pH-AAP) (Fig. 6A).97 Owing to the proximity of the two moieties, a 98% quenching rate was achieved with an ultra-low background. Fluorescence was restored by aptamer targeting the tumor site, while low pH caused acid hydrolysis of the acetal group. The probe maintained an ultra-low background in serum and mice in vivo, and it exhibited robust shelf stability.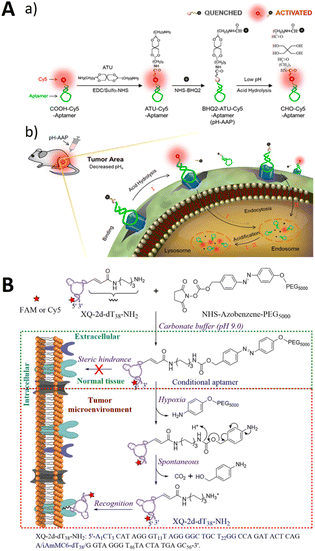 | ||
| Fig. 6 (A) The structure of the pH-activatable aptamer probe (a) and tumor imaging (b). Adapted with permission.97 Copyright 2019, American Chemical Society. (B) An illustration of the activatable aptamer recognition of XQ-2d-dT38-azo-PEG5000. Adapted with permission.98 Copyright 2019, American Chemical Society. | ||
In addition, Zhou et al. developed a chemically modified conditionally activated aptamer for cellular imaging (Fig. 6B).98 The aptamer XQ-2d-dT38-azo-PEG5000 used PEG5000-azobenzene-NHS as a conditionally recognized cage molecule. The caging molecule prevented target recognition. However, under special conditions, such as sodium disulfite treatment, hypoxia (<0.1% O2), or the tumor microenvironment, the caging molecule was removed, enabling the conditional recognition of the cellular target.
3.3 DNA structure-assisted design strategies
Owing to its inherent programmability, DNA can be precisely designed to form various intricate three-dimensional structures by utilizing complementary base pairings.99 With this precisely manipulated structure, aptamers can be precisely and quantitatively immobilized at specific positions by complementary pairing structures and are widely used in imaging. The use of DNA nanostructures can enhance the stability of DNA against nucleases and enable efficient delivery into target cells due to the properties of these nanostructures. Various DNA nanostructures have been applied to imaging, such as DNA tile,100 DNA triangular prism,101 DNA tetrahedron,102 and DNA octahedron.103 Furthermore, the assembly of DNA structures can be applied for signal amplification and enhancement of signals in imaging, such as hybrid chain reaction (HCR)104 and catalysed hairpin assembly (CHA)105,106 and rolling circle amplification (RCA).107 Dong et al. constructed an aptamer-functionalized cross DNA probe combined with CHA to achieve amplification imaging of multiple miRNAs in living cells.105Li et al. reported a tetrahedral probe that targets nucleolin with aptamer AS1411 modification on one of the vertices (DNA tetrahedron nanostructure, DTN).108 The other vertex modified two miRNA capture strands as well as two pairs of metastable catalytic hairpins located at different vertices. With aptamer targeting, the DTN probe was able to specifically enter tumor cells, in which the capture strands were able to hybridize to the miRNAs and initiate CHA only when both overexpressed miR-21 and miR-155 were present, resulting in a pronounced fluorescence resonance energy transfer signal. DNA dendrimer macromolecules were developed by applying a hairpin-less nonlinear HCR system, and the dendrimer-like structure makes its amplification efficiency significantly higher than that of linear HCR.109 Tang et al. devised a DNA dendrimer-assisted Förster resonance energy transfer (FRET) strategy to image cell-surface sialylation (Fig. 7B).110 Multiple FRET donors were labeled on target molecules by metabolic oligoengineering (MOE), and DNA dendrimers labeled the acceptors. Targeted recognition by aptamers significantly improved the FRET results. Using the DNA dendrimer-assisted FRET strategy, specific imaging of SMMC-7721 and CEM cell surface glycosylation was successfully achieved.
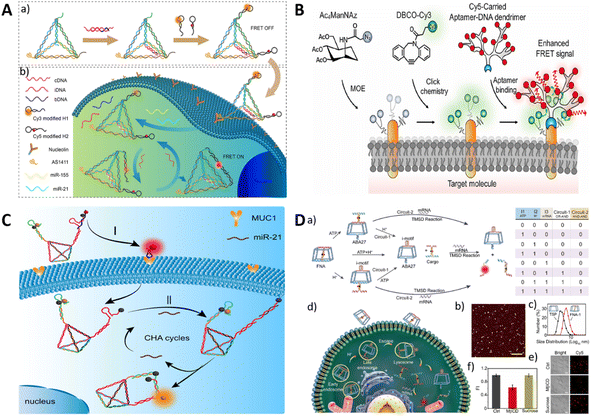 | ||
| Fig. 7 (A) The application of the DTN probe for miRNA imaging. Adapted with permission.108 Copyright 2022, American Chemical Society. (B) The aptamer-modified dye loaded with a DNA dendrimer for imaging cell surface sialylation. Adapted with permission.110 Copyright 2022, Elsevier. (C) The DNA logic device based on DNA tetrahedron by the dual recognition of MUC1 aptamer and miR-21. Adapted with permission.112 Copyright 2022, American Chemical Society. (D) A schematic of DNA computation circuits using a truncated square pyramid. Adapted with permission.113 Copyright 2020, Wiley. | ||
In addition to the addressable modifications of aptamers, DNA nanostructures exhibit a confinement effect, providing reaction cavities for molecules decorated on DNA nanostructures. DNA computation of multiple signal inputs improves reaction specificity. Hence, it is feasible to modify responsive probe molecules on DNA nanostructures, which achieve DNA molecular computing through the construction of diverse stimulus-responsive cooperative responses, thereby enhancing specificity. Gong et al. developed a “YES-AND” logic circuit-controlled smart DNA nano-assembly using two DNA tetrahedra modified with G-quadruplex (G4) and ATP aptamers.111 In the presence of both K+ and ATP, the output was a Förster resonance energy transfer (FRET) signal with high sensitivity and selectivity.
Apart from two-DNA structure boolean operations, most DNA-based boolean operations fix the input devices on a single structure. As shown in Fig. 7C, Li et al. constructed transmembrane DNA logic nanodevices (TDLNs) for the in situ sequential imaging and cellular recognition of the transmembrane glycoproteins mucin 1 (MUC1) and cytoplasmic microRNA-21 (miR-21).112 The TDLNs were designed by encoding two metastable hairpin DNAs in a DT scaffold, where the miR-21-triggered toe-in of H1 is capped by the MUC1-specific adaptor. After recognition by MUC1, the trigger strand of miR-21 was released, and in the presence of miR-21, the hairpin assembly (CHA) reaction was locally catalysed for signal amplification through the confinement effect of the DT structure. This method can be used to image Michigan Cancer Foundation-7 (MCF-7) cells expressing both MUC1 protein and miR-21.
Unlike the exogenous light-activated in vivo imaging methods reported above, auto-recognition responses triggered by abnormalities in the intracellular environment have been widely used. DNA nanostructures are biocompatible because of their nucleic acid nature. It was found to be rapidly internalized via a caveolin-mediated pathway and subsequently entered the lysosome while maintaining structural integrity for long periods in living cells.113 These properties make TDNs ideal nano scaffolds for dynamic studies in living cells. Li's group developed a series of DNA framework-based nanoplatforms for subcellular imaging in living cells.114 Wang et al. reported the construction of functional logic circuits for DNA using truncated square pyramid (TSP) cages as nanocarriers, as shown in Fig. 7D.113 The two edges of the TSP were modified with the ATP aptamer (ABA27) and an i-motif. Additionally, there is an interior double-stranded element containing an ASO used as a sensing component for TK1 mRNA. These two elements combine to create a cascade of logic gates, specifically an OR–AND gate and an AND–AND gate. When protons or ATP within acidic lysosomes activate the edge elements, the internal sensing element is released from the nanocage. Simultaneously, the presence of mRNA triggers the release of the fluorescent reporter strand.
3.4 Nanomaterial-assisted design strategies
The combination of aptamers with nanomaterials enhances the stability of aptamers. Moreover, nanomaterials serve as carriers for delivering aptamers and function as signal molecules in various imaging modalities. Currently, there are numerous applications of aptamer–nanomaterial conjugates in molecular imaging, including microspheres,115,116 nanoparticles (NPs),117 lipid nanoparticle formulations (LNPs),118,119 polymers,120–122 MOF123 and other nanomaterials.124Gold nanoparticles (AuNPs) have a wide range of applications in biosensors. Owing to their biocompatibility and non-toxicity, they have the advantage of intracellular application because they can be taken up by cells without the use of transfection reagents.125 In addition to being used as carriers for functional molecules, AuNPs can be used as signal quenching material. As shown in Fig. 8A, Yang et al. used AuNPs as fluorescent quenching material.126 When the aptamers recognized the targets, the AuNP-modified complementary sequences were released from the fluorescent-modified aptamers, thus recovering the signal.
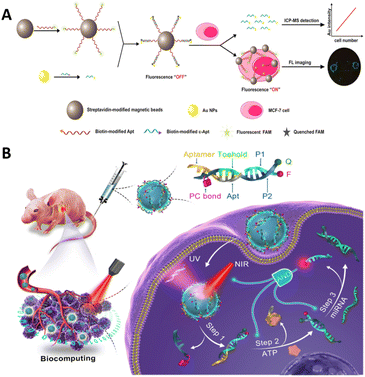 | ||
| Fig. 8 (A) A schematic of magnetic bead-based probes. Adapted with permission.126 Copyright 2018, American Chemical Society. (B) A schematic of a photo-triggered DNA nanocircuit. Adapted with permission.128 Copyright 2021, American Chemical Society. | ||
Upconversion nanoparticles have unique luminescence properties and can therefore be designed for multi-target detection.127 Mi et al. performed an “AND” boolean logic computation method for the simultaneous detection of two cancer biomarkers (Fig. 8B).128 This approach used a conceptual approach of photonic nanocircuits activated by spatially restricted near-infrared light that allows ATP to bind to its aptamer to stimulate structural transitions, which in turn led to the next step in the strand displacement reaction of miRNAs, releasing signaling molecules. The precise spatial accuracy of the signal output is achieved through the simultaneous input of three signals. The method was successfully imaged intracellularly and in vivo with the potential for spatiotemporal control. In addition, upconversion nanoparticles possess multicolor luminescence properties for multiplex imaging. Wu et al. used a dual metabolic labeling technique to label two fluorescent dye receptors on two cell surface monosaccharides, with aptamer-modified upconverted luminescent nanoparticles serving as donors.129
The exploration of aptamer-functionalized DNA hydrogels with favorable biocompatibility and responsiveness to stimuli has emerged as a novel research direction. Yang et al. introduced the development of a hydrogel-termed Apt-HEX/Cp-BHQ1 Gel (AHB Gel) for fluorescence-guided surgical resection.130 The AHB Gel exhibited rapid and intense fluorescence signal emission within 3 minutes when subjected to high ATP concentrations (ranging from 100 to 500 μM), a characteristic feature of the tumor microenvironment. Simultaneously, it displayed diminished fluorescence signals in response to low ATP concentrations (10–100 nM) typically found in normal tissues. In a mouse subcutaneous tumor xenograft model, the AHB Gel selectively displayed regional fluorescence signals limited to the areas covering the tumor tissue and residual tumor surfaces. The application of this hydrogel has promising potential for facilitating fluorescence-guided surgery.
4. Imaging targets of aptamer-based molecular imaging
In molecular imaging applications, aptamers are becoming effective recognition molecules for various types of targets, such as cell membrane proteins, nucleic acids, organelles, metabolites, and metal ions. This section introduces different targets for aptamer imaging.4.1 Cell membrane proteins
The visualization of tumor-specific protein biomarkers on cell membranes plays a pivotal role in detecting protein expression and categorizing tumors and holds promise for applications in basic biological research and clinical diagnosis.Wang et al. screened the DNA aptamer mApoc46 targeting patient-derived primary plasmacytoid ovarian cancer (pSOC) cells.131 The aptamer recognized pSOC cells with strong specificity while showing excellent selectivity for high-grade plasmacytoid ovarian cancer (HG-SOC) tissues and was expected to be applied to the identification of ovarian cancer histologic subtypes during surgery. It selectively accumulates in tumor regions within a patient-derived tumor xenograft NCG mouse model, making it a viable in vivo imaging method. For precise native-state imaging of tumor cell surface markers, Delcanale et al. introduced a point accumulation in a living cell (PAINT) method for nanoscale topological imaging using aptamers as probes (Fig. 9A).52 The localization and tracking of individual receptors rely on the localization and tracking of individual proteins, facilitated by random and transient binding interactions between aptamers and their respective targets. This method successfully performed single-molecule imaging of a tumor marker epidermal growth factor receptor (EGFR) on living cancer cells. In addition, by fine-tuning the entity sequence, the affinity to the EGFR can be adjusted to determine receptor motility and/or native receptor density.
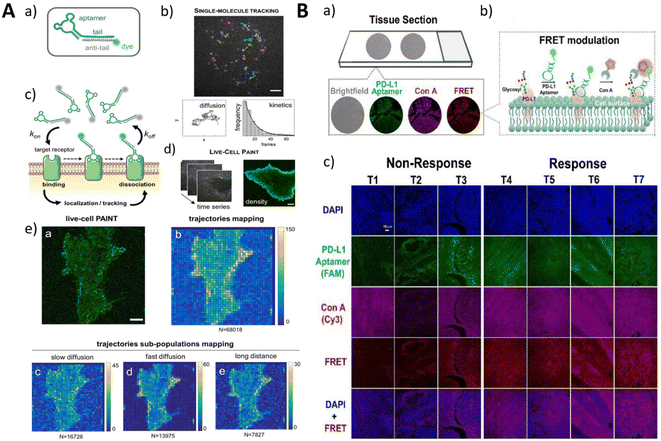 | ||
| Fig. 9 (A) The conjugation of the fluorescent aptamer probe and imaging mechanism (a–d). The reconstructed PAINT image and density of trajectories (e). Adapted with permission.52 Copyright 2020, Wiley. (B) The principle of the FLAG strategy for imaging glycosylated PD-L1 (a and b). The imaging of the tissue sections. Adapted with permission.134 Copyright 2021, American Chemical Society. | ||
An important advantage of aptamer imaging of cell surface proteins is the ability to distinguish distinctly characterized cell subpopulations using surface markers. Aptamers specific for multidrug resistance (MDR) provide effective molecular tools for the precise diagnosis and targeted therapy of drug-resistant tumors. Zhang et al. developed a phosphorothioate-modified PS-ZL-7c aptamer with high selectivity for multidrug-resistant hepatocellular carcinoma (HCC) cells (HepG2/MDR).81 The possibility that the PS-ZL-7c aptamer may target a generic drug resistance-associated protein opened up the possibility of developing generalized aptamer probes for drug resistance monitoring and targeted therapy. The labelling and analysis of different protein markers on the cell surface are important for the precise identification of target cell subtypes. Accurate fluorescent labelling of different protein markers on the cell surface can be achieved by multiple surface labelling DNA logic analysis methods.132
It has been found that specific glycosylation features found on tumor cells can be considered a novel immune checkpoint.133 Therefore, the glycosylation detection of tumor markers is of great significance. Huang et al. developed a FRET strategy (FLAG) based on glycolabeled lectins and programmed cell death 1 ligand 1 (PD-L1) recognition aptamers for the visualization of PD-L1-specific glycosylation (Fig. 9B).134 Through separate labeling of the concanavalin A (ConA) probe and the PD-L1 aptamer with fluorescent moieties, FRET occurs when both are simultaneously recognized, and the probe is close, generating a specific signal. This method was successfully applied to the in situ visualization of PD-L1-specific glycosylation in tissue biopsies. N-Azidoacetylneuraminic acid (SiaNAz) can be introduced into glycoproteins on the membrane surface of living cells using a cellular metabolic labelling assay,135 and the co-cultivation of cells with a cyclooctyne-functionalized aptamer allows the covalent attachment of the aptamer to the cell surface. Combined with signal amplification methods, it is possible to perform highly sensitive signal imaging.136–138
4.2 DNA/RNA
Specific sequences of DNA and RNA transcripts are important biomarkers in both basic biological research and clinical diagnosis. Aptamer-based DNA detection can be accomplished by applying gene editing technology, CRISPR-Cas9, which is based on the high affinity interaction between the MS2/PP7 RNA aptamer and its coat protein.139,140 The RNA aptamer can be integrated into the sgRNA of Cas9, thus accomplishing the imaging of the gene locus through the binding of the coat protein to the fluorescent protein.To improve the brightness and photostability of RNA aptamer lit fluorescent dyes, Bouhedda et al. developed a cell-permeable fluorescent dimer dye to improve the performance of RNA imaging methods.38,141 This method used a dimerized self-quenched sulforodamine B dye (Gemini-561) and a corresponding dimerized aptamer (o-Coral) to improve the brightness and high stability of the imaging. This approach allowed direct fluorescence imaging of RNA polymerase III transcription products and messenger RNA in living mammalian cells.
Genetically encoded RNA sensors based on lighted RNA aptamers are important tools for performing RNA imaging and can be used for live cell imaging. A compelling strategy for mitigating background fluorescence signals involves the use of aptamer probes with carefully designed fluorescence turn-on functionality.88,142 Fang et al. designed a series of fluorescence sensors based on the Pepper aptamer, exhibiting significant fluorescence signals only in the presence of the target molecule due to the ingenious sequence and structural design.143 Our research group developed an iPepper system, a tandem array based on the Pepper aptamer, which demonstrated excellent target-induced fluorescence turn-on capability and target specificity for real-time imaging of low-expressed mRNA in various cell lines (Fig. 10A).42 Jiang's group used genetically encoded RNA sensors to detect miRNAs and mRNAs. They first reported the use of two promoter plasmids expressing GFP and RNA sensors for the ratio detection of miRNAs;144 however, this method was not able to quantify RNAs. They then developed a single promoter plasmid expressing both a splittable fusion RNA sensor and GFP mRNA.145 The co-expressed RNA sensor and GFP mRNA could be split by applying the Dicer enzyme, which mitigated the interference of intramolecular hybridization on the correct folding of the RNA aptamer. This single promoter system was successfully applied to ratiometric imaging of survivin mRNA in tumor cells, providing a new paradigm for imaging mRNA in living cells.
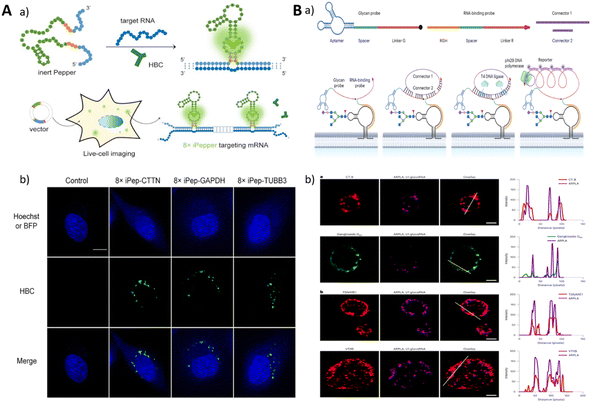 | ||
| Fig. 10 (A) A schematic of a genetically encoded system for endogenous RNA imaging. Adapted with permission.42 Copyright 2022, Oxford University Press. (B) A schematic illustration of ARPLA imaging for glycoRNA and the imaging of cells. Adapted with permission.34 Copyright 2023, Nature Research. | ||
In addition to imaging RNA with specific sequences, researchers have made advancements in imaging methods for glycosylated RNA (glycoRNAs), thereby expanding its detection capabilities in imaging. Imaging glycan RNAs can reveal their spatial distribution and expression levels, which are important for exploring their biological functions. Ma et al. reported an imaging method for glycosylated RNA profiling using an in situ hybridization DNA probe-mediated proximity ligation method (ARPLA) (Fig. 10B).34 The dual recognition of the probe containing N-acetylneuraminic acid (Neu5Ac) binding aptamer and RNA in situ hybridization (RISH) DNA probe led to the proximity to bind with connectors, generating fluorescent probes binding RCA products for imaging.34
4.3 Organelles and metabolic components
Organelles are specialized subunits within the cell that usually have a complete structure and perform important physiological activities. Mitochondria are critical to metabolism. Cellular metabolism is a complex life process, and the detection of metabolites therein is of great value in exploring cell-related activities. Hong et al. developed an aptamer sensor that binds to DQAsomes and targets mitochondria for spatiotemporally controlled monitoring of ATP.75 The light-activated construction prevented the sensor from being activated before it reached the mitochondria, and the sensor had a limit of ATP detection of 3.7 μM. This work represented the first successful delivery of a DNA aptamer sensor to mitochondria, providing a new platform for targeted delivery to subcellular organelles.The lysosome is an acidic environmental organelle with a pH ranging from 4.5–5.5 and is essential for maintaining cellular homeostasis. To characterize lysosomes, pH-responsive probes can be designed for lysosomal imaging. Du et al. reported a lysosome-recognized framework nucleic acid (FNA) nano-AND device. The device immobilized the i-motif and ATP junction appropriate body (ABA) at both ends of the DNA delta prism (DTP) as a logical control unit. The pH and ATP environment of the lysosome triggered structural changes in the FNA nanodevice, releasing reporter structures for subcellular imaging (Fig. 11A).146
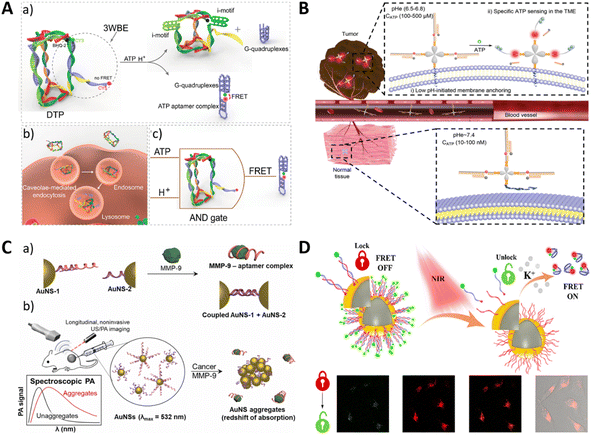 | ||
| Fig. 11 (A) A schematic of lysosome imaging by the cellular logic operation. Adapted with permission.146 Copyright 2019, American Chemical Society. (B) The imaging of extracellular ATP by acidic condition–driven DNA nanomachine. Adapted with permission.18 Copyright 2019, Wiley-Blackwell. (C) The aptamer-modified gold nanoparticle for the SERS imaging of MMP-9. Adapted with permission.147 Copyright 2022, Elsevier. (D) The imaging of potassium ions using an NIR remote-controlled “lock–unlock” nanosystem. Adapted with permission.148 Copyright 2020, American Chemical Society. | ||
In addition to metabolite detection in subcellular organelles, extracellular metabolites in the tumor microenvironment (TME) are also relevant targets for providing information about tumors. Di et al. designed an extra-TME pH-driven DNA nano-machine (Apt-LIP) to image extracellular ATP (Fig. 11B).18 Their work used a pH insertion peptide (pHLIP) module anchored to the tumor cell membrane. A DNA-responsive module linked to this system incorporated conformationally driven ATP aptamers. These aptamers released a fluorescent reporter sequence, thereby restoring fluorescence when exposed to the target and allowing the imaging of exogenous cellular ATP.
4.4 Enzymes
Enzymes constitute the essential components of living cells and play a critical role in sustaining life. Any dysregulation of enzymes can lead to various disorders.149 Kim et al. developed a photoacoustic-based biosensor for the detection of matrix metalloproteinase-9 (MMP-9) in the tumor microenvironment (Fig. 11C).147 The sensor utilized a DNA aptamer surface modified on plasmonic gold nanospheres, which are selectively bound to MMP-9, leading to aggregation of the nanospheres through DNA displacement and hybridization. The aggregation of the nanospheres enhanced the optical absorption in the near-infrared window, allowing for the detection of aggregated nanospheres within the tumor microenvironment using ultrasound-guided photoacoustic imaging. Experimental validation in solution, cell culture, and a xenograft mouse model of human breast cancer demonstrated the selective and sensitive detection of MMP-9. The development of this sensor provides feasibility for in vivo molecular-level photoacoustic imaging with potential applications in tumor monitoring and pathological studies for cancer treatment.4.5 Metal ions
Metals play key roles in fundamental biological processes, such as osmoregulation, biomineralization, metabolism, catalysis, and signalling.150 Yang et al. used an aptamer-based FRET nanofluorescent probe to detect intracellular K+. Dual fluorescently labeled G-quadruplexes were hybridized with short nucleotides immobilized on the surface of the nanoparticles.151 In the absence of the target K+, the donor FAM was far away from the acceptor TAMRA, and only the FAM signal was detected. In the presence of the target K+, the aptamer formed a G-quadruplex structure, which brought the donor and acceptor close, resulting in higher FRET efficiency and detectable fluorescence from TAMRA. The fluorescence emission ratio (A/D) of the acceptor to the donor can be used as a signal to detect target K+. For spatiotemporal modulation, Cui et al. integrated distant NIR excitation into a comparable strategy. In this approach, the aptamer thermally dissociated from its complementary chain, folded into a G-quadruplex structure when stimulated by K+, and underwent FRET, leading to a distinct signaling response (Fig. 11D).148Yu et al. developed a genetically encoded RNA-based sensor that can image silver ions in living bacterial cells.152 The silver ion signal was detected by introducing a cytosine-Ag+-cytosine metal base pair to the broccoli. Consequently, the binding of Ag+ induced the broccoli to fold and activate the fluorescent signal.
5. Modalities of aptamer-based molecular imaging and applications
5.1 Fluorescence imaging
Fluorescence imaging, as a robust and versatile tool, enables the visualization, localization, and quantification of specific target molecules, thereby facilitating the comprehension of intricate biological systems and driving advancements in drug discovery, diagnostics, and personalized medicine.153 The application of aptamers in fluorescence imaging confers several advantages, including non-invasiveness, high specificity, high affinity, and real-time imaging capability, leading to its widespread utilization in diverse domains, such as cellular investigations, animal models, and clinical diagnostics.9,154 Moreover, compared to certain alternative imaging modalities, fluorescence imaging generally reduces equipment costs and simplifies operations, enhancing its accessibility in laboratory and clinical environments.Nevertheless, fluorescence-based imaging methodologies are not exempted from inherent limitations when employed in in vivo experiments. For instance, the depth of fluorescence signal penetration within tissues is constrained by phenomena such as light scattering and absorption, thus circumscribing the imaging depth achievable using fluorescence techniques.155 In scenarios necessitating deep tissue imaging, alternative approaches, such as MRI or PET, may be more suitable alternatives. Additionally, fluorescence imaging in cells and tissues can be compromised by inherent spontaneous fluorescence and background noise, which may decrease the signal quality and specificity.156 Furthermore, the unfavorable pharmacokinetics and rapid degradation of unmodified aptamers in vivo pose significant obstacles to their clinical application in diagnostics.15
To overcome the aforementioned limitations, one strategy involves the utilization of near-infrared (NIR) fluorophores with longer excitation and emission wavelengths, enabling enhanced tissue penetration and reduced autofluorescence background.157–160 For example, Hu et al. developed and employed an integrated visible and NIR-I/II multispectral imaging system, which demonstrated enhanced contrast, sensitivity, and accuracy for the fluorescence imaging of deep tissues using the second NIR window (NIR-II, 1000–1700 nm).161 Sicco et al. utilized Alexa647-modified Sgc8c aptamers with far-red fluorescence to investigate the uptake characteristics of the probe in a murine metastatic melanoma model.162
Aptamers also play a pivotal role in enabling rapid ex vivo fluorescence imaging for tissue pathology diagnosis.164–167 For instance, Georges et al. conjugated a lymphoma-specific aptamer with Alexa Fluor 488 (TD05-488), enabling intraoperative tumor-specific diagnosis of CNS lymphoma within 11 minutes after biopsy.168 Chu et al. developed a fluorescent ATP probe comprising titanium carbide (TC) nanosheets modified with an ATP aptamer (TC/Apt) labeled with ROX.169 This probe facilitated the fluorescence detection and imaging of ATP in live cells and body fluids, such as urine and serum, and a mouse tumor model.
Moreover, simultaneous imaging of multiple targets and multicolor imaging represent future directions for aptamer-based fluorescence imaging. In the context of multicolor imaging, fluorophores can consist of diverse orthogonal aptamer–ligand complexes,46 fluorescent dyes conjugated to aptamers,163 or nanomaterials serving as carriers.23 Our research group has developed an intracellular RNA imaging method based on the CRISPR-dPspCas13b system (Fig. 12A).46 By linking sgRNAs with one or two aptamers, including Broccoli and Pepper, simultaneous imaging of two target RNAs can be achieved. By exchanging the ligand molecules in the imaging buffer, channel switching of the fluorescence signals during imaging can be achieved. This method has also been employed to study the interaction between long non-coding RNAs NEAT1 and MALAT1.
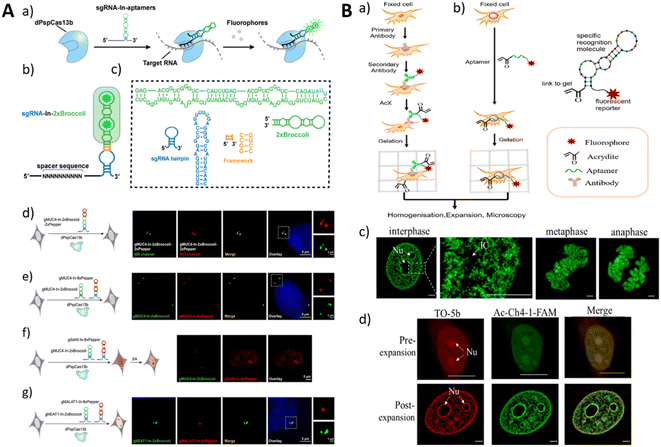 | ||
| Fig. 12 (A) Schematic illustration of RNA imaging using the CRISPR-dCas13 system with fluorescent RNA aptamer-modified sgRNAs (a–c) and RNA imaging in live cells (d–g). Adapted with permission.46 Copyright 2022 Royal Society of Chemistry. (B) The process of traditional ExM (a) and trifunctional aptamer-based ExM (b). Imaging of Ac-Ch4-1-FAM on different cell cycles (c) and cell nuclei stained with TO-5b and Ac-Ch4-1-FAM. Adapted with permission163 Copyright 2022 American Chemical Society. | ||
Furthermore, the advancement of aptamer-based fluorescence imaging encompasses novel imaging techniques and probe development, such as STROM Imaging and Expansion Microscopy.163,170 Expansion Microscopy (ExM) is an example of such techniques that enables nanoscale imaging using conventional microscopy. Sun et al. devised an ExM imaging method by employing aptamers as molecular probes (Fig. 12B).163 Through the binding of FAM and acridine-modified aptamer Ch4-1 to the cell nucleus, their participation in gelation, and their subsequent anchoring in a polyacrylamide gel, distinct chromatin morphologies at different stages of mitosis can be observed with high-resolution imaging using confocal microscopy. Recently, the development of single-molecule fluorescence imaging techniques has provided enhanced resolution and sensitivity for aptamer-based imaging. For instance, Sunbul et al. developed RhoBAST, an aptamer that combines fast binding and dissociation kinetics with fluorescence rhodamine dyes.43 This method enables subcellular imaging of RNA structures with a positioning accuracy of approximately 10 nm in live cells.
In conclusion, aptamer-based fluorescence imaging has been widely applied and developed in the realm of biomedical imaging. By harnessing the high specificity and affinity of aptamers and selectively labeling them with fluorescence probes, it becomes feasible to achieve high-resolution and real-time imaging of target molecules. We firmly believe that with continuous progress in novel imaging techniques and fluorescence probes, the application prospects of aptamer-based fluorescence imaging in biomedical research and clinical diagnostics will continue to expand.
5.2 SERS
Surface-Enhanced Raman Scattering (SERS) is a widely utilized technique that exploits the surface enhancement effect to amplify the Raman signals of target molecules through their interaction with nanostructured surfaces. This enables the highly sensitive detection and imaging of the target molecules. In contrast to fluorescence imaging, SERS provides Raman spectral information, allowing for comprehensive characterization and analysis of the molecules.171 SERS exhibits remarkable sensitivity and can detect target molecules even at low concentrations.172 Moreover, it enables the visualization of dynamic processes involving target molecules at the cellular or in vivo level, rendering it invaluable for studying biological processes, signal transduction, and molecular interactions.173 By employing diverse nanostructured surfaces or probes, simultaneous detection of multiple target molecules and multi-channel imaging can be achieved.174–176 However, SERS imaging encounters challenges associated with the stability of nanostructured surfaces. Prolonged laser irradiation or changes in experimental conditions can induce morphological alterations that may compromise the stability and accuracy of SERS signals.177–179 SERS typically relies on nanoparticles or other nanostructures as enhancers for target molecules, highlighting the importance of judiciously selecting and ensuring the stability of markers (e.g., nanoparticles) and aptamers. In the context of in vivo imaging, SERS also confronts limitations associated with optical imaging techniques, such as scattering and absorption in deep tissues, which can impede the intensity and depth penetration capacity of SERS signals, thereby restricting its application in deep tissue imaging.180 Furthermore, although SERS holds the potential for real-time imaging, practical implementation necessitates addressing challenges such as the need for rapid data acquisition and processing, as well as the accurate capture and analysis of dynamic processes.181–183As a molecular recognition element, aptamers exhibit high specificity in binding to specific target molecules. Through the conjugation of aptamers with SERS probes, the specific imaging of target molecules can be achieved. This molecular targeting imaging approach has broad applications in the detection and visualization of tumor markers, cell surface receptors, and other biomolecules, providing valuable insights into their presence and spatial distribution within biological systems.184,185 By utilizing aptamers that selectively bind to targets, such as tumor cells and vascular endothelial cells, real-time monitoring and imaging of dynamic biological processes can be achieved through SERS signals in live animal models. Aptamer-SERS probe conjugates offer potential applications in early disease diagnosis and therapeutic monitoring.186 By visualizing disease-related molecules, it becomes possible to detect and evaluate disease progression at an early stage and monitor the efficacy of treatments.187,188 Zou et al. fabricated isotopic cellular Raman-silent graphene-isolated-Au-nanocrystals (GIANs) as SERS-encoded nanoparticles, with the surface modification of aptamers for cancer cell pattern recognition. The 2D-band Raman shift of GIAN can be easily tuned through the fabrication of various isotopic forms. The GIAN tags showed Raman imaging capabilities in vivo and in vitro. With the conjugation of different aptamers targeting nucleolin, mucin and epithelial cell adhesion molecules, the isotopic CIANs achieved rapid imaging and pattern recognition of cancer cells (Fig. 13A).178 Similarly, Wang et al. fabricated a kind of SERS nanotags functionalized with aptamers and biorthogonal Raman reporters on the surface of gold nanorods (AuNR). The Raman reporter possessing a diynl group showed a Raman signal at 2205 cm−1 with a low background signal. With the targeting of aptamers (AS1411 and MUC1), the SERS nanotags demonstrated high-contrast imaging in vivo (Fig. 13B).177
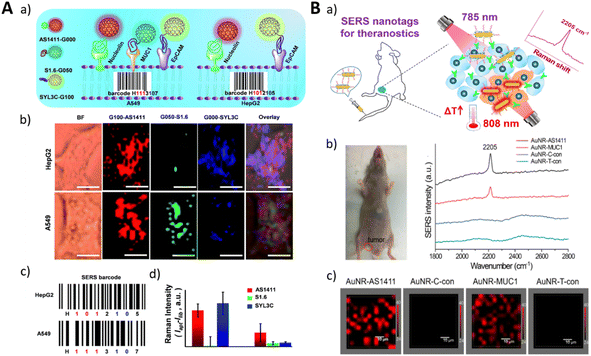 | ||
| Fig. 13 (A) A schematic illustration of the cancer cell pattern recognition using aptamer-functionalized isotopic GIAN-encoders and SERS images of cancer cells. Adapted with permission.178 Copyright 2018, Royal Society of Chemistry. (B) A schematic illustration of the precision treatment platform for mouse tumor detection and photothermal therapy based on biological orthogonal nanotags (a), a photograph of mouse bearing a tumor and SERS spectral detection of the tumor (b). SERS mapping images of the tumors (c). Adapted with permission.177 Copyright 2020, Elsevier, American Chemical Society. | ||
5.3 Nuclear imaging
Magnetic Resonance Imaging (MRI) is a widely used non-invasive imaging modality that offers high spatial resolution and unrestricted tissue penetration and does not involve radiation or radioactive exposure. It enables the visualization and characterization of tissue and organ structures, functions, and metabolic information within the human body.189,190 The introduction of exogenous contrast agents plays a vital role in enhancing image contrast and accuracy in MRI.191,192 In MRI imaging, contrast agents are commonly classified as T1 contrast agents and T2 contrast agents. T1 contrast agents shorten the tissue's T1 relaxation time, resulting in a high signal intensity contrast effect in T1-weighted images. Conversely, T2 contrast agents shorten the tissue's T2 relaxation time, leading to a low signal intensity contrast effect in T2-weighted images.193 These contrast agents can be administered intravenously or via other routes to enhance signal intensity in specific regions, thereby providing clearer visualization of anatomical structures or pathological lesions.194 However, conventional MRI contrast agents lack specificity for particular molecules or cells, and the development of aptamer-based MRI methods offers new prospects for molecular-level imaging.195 Aptamers, which interact with specific disease-associated molecular markers, such as tumor antigens, receptors, and enzymes, can be integrated with MRI contrast agents to achieve highly selective and sensitive imaging of these markers.196Yan et al. presented an aptamer (CD105) targeting endoglin, which exhibits high expression in both hepatocellular carcinoma (HCC) cells and endothelial cells of the neovasculature (Fig. 14A).197
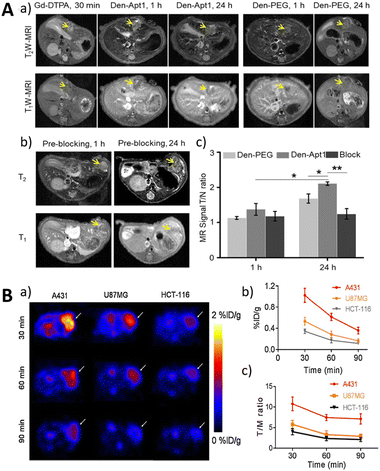 | ||
| Fig. 14 (A) The MRI imaging of Den-Apt1 in vivo. Adapted with permission197 Copyright 2018, American Chemical Society. (B) The PET imaging of the EGFR expression utilizing an 18F-labeled RNA aptamer. Adapted with permission.204 Copyright 2019, Springer. | ||
By conjugating the aptamer and imaging reporter onto a G5 dendrimer, a liver HCC-targeted nano-probe was generated. T1-weighted MRI exhibited a tumor-to-normal ratio of 2.1, demonstrating the successful visualization of in situ HCC xenografts as small as 1–4 mm in diameter. To enhance the MRI signal, Li et al. constructed a solid-phase synthesized nucleic acid aptamer, ApSC-Fc6, containing hydrophobic ferrocene artificial bases.16 The T1 MRI signal enhancement of pulmonary micrometastases was increased by 7–10 fold. To improve rapid clearance kinetics (t1/2, ∼20 h), Zhang et al. developed a series of perfluoropolyether (PFPE)-based hyperbranched (HBPFPE) nanoparticles for specific in vivo detection of breast cancer with high 19F MRI sensitivity.198
Zhao et al. developed an HCC-targeted magnetic resonance probe based on a glypican-3 (GPC3)-specific aptamer (AP613-1) with ultrasmall superparamagnetic iron oxide (USPIO).199 They observed specific uptake of Apt-USPIO in Huh-7 cells (human hepatocellular carcinoma cells, huh7), and in vivo T2-weighted MRI demonstrated significant tumor penetration ability of AP613-1 aptamer-conjugated NPs.
Although aptamer-based MRI methods hold promising potential, they still face several challenges, including aptamer design and synthesis, stability and biocompatibility of imaging agents, and sensitivity and resolution of imaging signals. Future research efforts will continue to address these issues and promote the application of aptamer-based MRI methods in clinical diagnosis and research.
Single Photon Emission Computed Tomography (SPECT) and Positron Emission Tomography (PET) are commonly used nuclear medicine imaging techniques with different principles, characteristics, and applications. Because nuclear imaging agents are radioactive and background radiation in living organisms or humans is minimal, only trace amounts of radioactive substances are required for nuclear imaging. This approach allows for high-sensitivity and high-specificity imaging of tissues without perturbing cellular biological processes, making nuclear imaging a promising tool for fundamental biological research. SPECT utilizes radiotracers to measure single gamma rays emitted by radioactive isotopes, providing functional information about organs and tissues within the human body.200 PET uses radioactive isotopes that undergo annihilation with positrons, measuring the resulting two opposing photons to obtain three-dimensional metabolic and functional information.201 Most PET scanners used in clinical practice have a resolution of approximately 2 millimeters, which enables accurate imaging of small tumor metastasis and has higher resolution and sensitivity compared to SPECT.202 SPECT and PET have the advantage of providing metabolic and molecular changes associated with different pathological conditions in the body, which is important for disease diagnosis, treatment evaluation, and efficacy research.203
Additionally, SPECT and PET offer high repeatability and quantifiability, allowing for dynamic studies and time-series data.205 In SPECT and PET imaging, aptamers serve as targeting probes, enabling targeted imaging of specific regions of interest. The introduction of aptamers enhances image contrast, improves the visibility of target areas, and helps with the precise localization and quantification of target molecules or pathological processes.
The earliest application of radiolabeled aptamers was reported by Charlton et al. in 1997 using technetium-99m (99mTc) labeled aptamers to target activated neutrophils for inflammation imaging in rats.206 Since then, various radiolabeling strategies for SPECT or PET have been developed, including the use of isotopes, such as 99mTc, 111In, and 123I for SPECT, and 11C, 18F, 68Ga, and 64Cu for PET.207–211 Bohrmann et al. developed an aptamer-targeted nanoplatform based on hyperbranched polyglycerol (HPG) to improve the stability and applicability of aptamer.210 Their study demonstrated that the Sgc8 aptamer exhibited highly specific binding to various cancer cells and displayed good binding characteristics and biocompatibility in vitro, with an extended half-life. However, in vivo SPECT/CT imaging showed no significant improvement in tumor uptake compared to non-targeted HPG or HPG conjugated with non-specific control ssDNA. This study highlights the importance of strict control and quantification in the evaluation of aptamer-targeted nanoplatforms and suggests further exploration of long-circulating polymer platform technologies in aptamer-targeted therapy.
Cheng et al. utilized 18F-fluorobenzoyl azide (FB) and click chemistry to modify the EGFR aptamer MinE07, resulting in the targeted EGFR PET tracer 18F-FB-ME07 (Fig. 14B).204 They investigated its cellular uptake and internalization properties. However, owing to rapid clearance in vivo, even with modifications to the aptamer backbone to increase resistance to enzymatic degradation, only a small amount of the probe remained intact in the bloodstream 20 minutes after injection.
As a low-cost alternative to radioactive isotopes, 68Ga has also been widely used in PET imaging studies. Choi et al. developed a 68Ga-labeled aptamer PD-L1 for targeting the programmed death-1 (PD-1) receptor.212 The binding affinity and targeting specificity of the aptamer in tumor cells were evaluated using flow cytometry, confocal microscopy, and PET imaging. The results showed strong binding of the PD-L1 aptamer to PD-L1 positive tumor cells and lower binding to PD-L1 negative tumor cells. PET imaging in mouse models demonstrated a higher uptake of 68Ga-NOTA-PD-L1 aptamer in PD-L1 positive tumors, suggesting that 68Ga-NOTA-PD-L1 aptamer may serve as an imaging tracer for PD-L1 positive tumors.
In summary, radiolabeled aptamers enable highly specific and quantitative SPECT/PET imaging, holding promise for their role in disease diagnosis and monitoring of treatment. Future developments will focus on optimizing the design and synthesis of aptamers, exploring applications of new radioactive isotopes and combining them with other imaging techniques.
5.4 Ultrasonic/photoacoustic
Photoacoustic imaging, also known as optoacoustic imaging or photoacoustic tomography, is an emerging non-invasive and non-ionizing biomedical imaging modality. When tissues absorb pulsed laser light, they undergo instantaneous heating, resulting in thermoelastic expansion and the generation of ultrasound waves.213 These ultrasound waves are then detected by ultrasound transducers and used to reconstruct tissue images. The intensity of the photoacoustic signal is directly proportional to local light absorption, enabling the visualization of functional and molecular information in deep tissues.213 This imaging modality provides high spatial resolution, excellent contrast, and the ability to image endogenous and exogenous contrast agents.214 In recent years, it has received significant attention because of its capability to provide high-resolution imaging with excellent optical contrast in deep tissues.The integration of aptamers with photoacoustic imaging technology has opened up new avenues for targeted imaging, which enables the specific detection and visualization of disease biomarkers or cells, thereby improving diagnostic accuracy and treatment outcomes. By conjugating aptamers with photoacoustic contrast agents, such as nanoparticles or nanobubbles, imaging specificity and sensitivity can be enhanced (Fig. 15A).215,216 These aptamer-conjugated contrast agents can selectively accumulate at the target site, enabling the visualization and characterization of diseases.
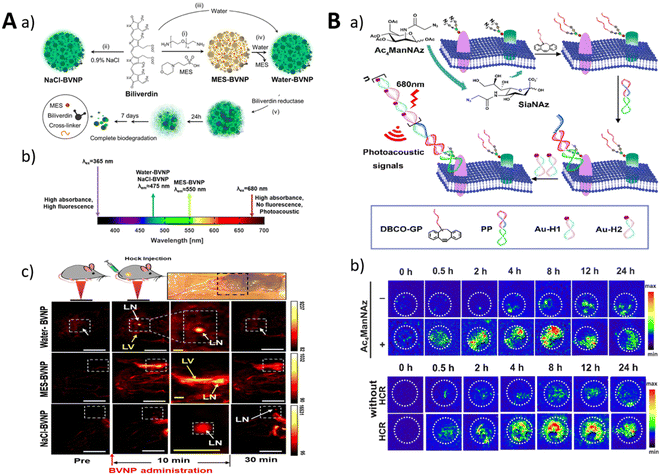 | ||
| Fig. 15 (A) The synthesis process for BVNP (a and b) and imaging of lymph nodes in mice (c). Adapted with permission.215 Copyright 2010, SPIE. (B) An illustration of the HCR-induced PA imaging system for MUC1-specific sialic acid (a) and PA imaging in BALB/c mice. Adapted with permission.137 Copyright 2021, American Chemical Society. | ||
Fan et al. developed an engineered cellular nanovesicle (ECNV)-based nanoplatform for photoacoustic (PA) imaging-guided precision chemoimmunotherapy.217 The ECNVs were loaded with gold nanorods (GNRs), gemcitabine (GEM), cytosine-phosphate-guanine oligodeoxynucleotides (CpG ODN), and a PD-L1 aptamer and surface-modified with M1-macrophage-derived cellular nanovesicles (CNVs). The combination of these components resulted in synergistic innate and adaptive immune responses, enhancing the antitumor effect of the ECNVs. In in vivo experiments, the ECNVs selectively accumulated in tumor tissues, and NIR irradiation triggered tumor ablation. The ECNVs exhibited excellent therapeutic efficacy, inhibiting primary and distal tumor growth and inducing changes in the tumor immune microenvironment. The nanoplatform demonstrated good biocompatibility and low systemic toxicity.
Liu et al. developed a photoacoustic (PA) nanoprobe based on the proximity-induced hybridization chain reaction (HCR) for amplified visual detection of protein-specific glycosylation in vivo (Fig. 15B).137 The nanoprobe consisted of glycan probes and protein probes, which were designed to selectively label glycans on target proteins and undergo proximity-induced hybridization. This led to the release of an HCR initiator domain, promoting the assembly of gold nanoparticles into aggregates. The aggregates produced PA signals that could be detected in vivo using photoacoustic imaging. The nanoprobe successfully enabled the in situ detection of MUC1-specific glycosylation in mice with breast cancer and allowed monitoring of dynamic changes in glycosylation during treatment. This approach provides a powerful platform for studying the effects of glycosylation on protein structure and function, thereby contributing to the understanding of glycosylation-related diseases.
The field of ligand-based photoacoustic imaging (PAI) holds promising prospects for future advancements. Potential directions for development include ligand-based molecular imaging, early disease diagnosis, guided therapies, and neuroscience research. However, several challenges need to be addressed for successful clinical translation, such as clinical validation, ligand selection and optimization, and the improvement of data processing and analysis methods. Through technological advancements and accumulated practical experience, ligand-based PAI approaches have the potential to revolutionize the field of biomedical research, providing more precise and personalized solutions.
5.5 Multimodality
Multimodality imaging is an advanced imaging approach that combines multiple imaging modalities to provide a more comprehensive and accurate assessment of biological processes and diseases. By integrating different imaging techniques, such as magnetic resonance imaging (MRI), positron emission tomography (PET), computed tomography (CT), single-photon emission computed tomography (SPECT), ultrasound (US), and optical imaging, multimodality imaging offers complementary information and synergistic advantages over individual modalities.The utilization of aptamers as probe molecules in multimodality imaging has several advantages. First, aptamers offer a high degree of target specificity, enabling precise molecular imaging. They can be designed to selectively bind to disease-related biomarkers, allowing for the visualization and quantification of specific molecular targets involved in various diseases. Second, aptamers exhibit excellent biocompatibility and low immunogenicity, making them suitable for in vivo imaging applications. Third, aptamers can serve as versatile modules for various imaging modalities. They can be easily conjugated with different imaging agents, including fluorescent dyes, radionuclides, paramagnetic agents, and nanoparticles, thereby enabling multimodal imaging approaches.218–220 This versatility allows for the combination of different imaging modalities to obtain complementary information and improve the accuracy and sensitivity of disease detection and monitoring.
Fluorescence imaging has low tissue penetration defects, which affect the depth and accuracy of imaging, and PAI compensates precisely for these defects. The combination of the two imaging modes can provide richer information for drug targeting and localization research. Zeng et al. developed aptamer-functionalized liposomes that can encapsulate temozolomide (TMZ) and the photothermal agent IR780 (Fig. 16A).221 These liposomes were designed to cross the blood–brain barrier and actively target gliomas. Owing to their stability and excellent photothermal conversion capabilities, the liposomes can be simultaneously employed for both fluorescence and photoacoustic imaging. The liposomes exhibited a chemo/photothermal synergistic therapeutic effect on gliomas, as evidenced by their ability to target orthotopic gliomas, alleviate tumor hypoxia, and reverse glioma cell resistance to TMZ.
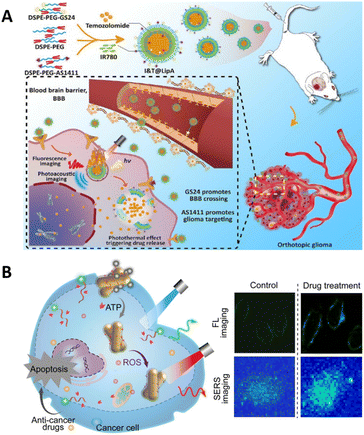 | ||
| Fig. 16 (A) The fluorescent imaging and photoacoustic imaging of aptamer-functionalized liposomes. Adapted with permission.221 Copyright 2023, Tsinghua University Press. (B) A schematic of the dual-modal imaging of ATP and ROS in living cells. Adapted with permission.223 Copyright 2022, American Chemical Society. | ||
Dai et al. developed an NIR-II light excitation phototheranostic nanomedicine for triple-negative breast cancer (TNBC).222 The nanomedicine, called Lip(PTQ/GA/AIPH), was designed by integrating a semiconducting polymer, an HSP inhibitor, and an azo compound into aptamer-modified liposomes. The nanomedicine exhibited excellent biocompatibility and tumor targeting properties. It provided dual-modal imaging capabilities of NIR-II fluorescence and photoacoustic imaging, as well as NIR-II photothermal therapy (PTT) and photonic thermodynamic therapy (PTDT). In vivo experiments demonstrated that nanomedicine achieved precise diagnosis and effective suppression of deep-seated TNBC, with enhanced PTT and PTDT effects.
Simultaneous multimodality imaging for multiple targets avoids mutual interference when a single mode is used for imaging multiple targets. To achieve duplex imaging of cellular ATP and reactive oxygen species (ROS), Jiang et al. engineered a dual-modal nanoprobe using metal–organic framework (MOF)-encapsulated AuNRs (MOF@AuNRs) for multimodality imaging (Fig. 16B).223 The fabricated MOF@AuNRs were modified with ROS-responsive SERS probe and FAM-labeled ATP aptamer. When recognized with ATP, the FAM-labeled aptamer can be released from MOF@AuNRs owing to an increased fluorescent signal. Moreover, the intracellular ROS changed the SERS probe for differential SERS signal. The proposed method achieved real-time monitoring of cancer cell apoptosis by simultaneously imaging the changes in ATP and ROS.
In summary, multimodality imaging, coupled with the utilization of aptamers as probe molecules, has great potential for advancing biomedical research and clinical diagnosis. The unique properties of aptamers, including their target specificity, versatility, and biocompatibility, make them valuable tools for visualizing and characterizing disease-related molecular targets. Further advancements in aptamer selection, probe design, and imaging technologies will continue to enhance the application of aptamer-based multimodality imaging, enabling more accurate disease detection, improved treatment planning, and personalized medicine approaches.
6. Summary and outlook
Aptamers have potential as recognition elements in molecular imaging because of their high affinity and specificity. Owing to their nucleic acid nature, aptamers have low immunogenicity and low toxicity in imaging, and they can be chemically synthesized with low cost and easy modification. Currently, researchers have developed a series of aptamers that bind to different targets and have been chemically modified to improve their stability against cleavage and their circulation time in vivo. Furthermore, the chemical modification-based response enhances the specificity of aptamer recognition, thus improving the spatiotemporal accuracy of imaging.Researchers have proposed different design strategies for the application of aptamers in imaging that have improved the signal-to-noise ratio, specificity, and spatiotemporal modulation of aptamer response, paving the way for aptamer-based molecular imaging in preclinical evaluation and clinical application. Additionally, through the design and computation of aptamers, the scope of imaging encompasses target types, target characteristics, multiple targets, and multimodality imaging, significantly expanding the application of aptamers in molecular imaging. By incorporating advanced imaging techniques, aptamer-based imaging has reached the level of single-molecule precision, providing tools for mapping molecular aberrations. However, there is still a long way to go for aptamer-based imaging applications, and many directions can be further optimized in the future.
First, despite the broad range of potential binding targets for aptamers, their application in real molecular imaging remains somewhat limited. Consequently, there is an urgent requirement to develop rapid and high-throughput methods for selecting efficient and highly specific aptamers that cater to a diverse array of clinical needs. Bioinformatics tools provide powerful assistance in aptamer screening, structure prediction, and affinity optimization. Using bioinformatics, precise prediction of aptamer structures and interaction with targets lays the foundation for designing strategies in subsequent applications in molecular imaging. In addition, bioinformatics assistance can lead to the development of advanced image analysis and processing tools, reducing noise and enhancing imaging quality.
Second, it has been demonstrated that aptamers can integrate with various imaging techniques, enhancing the specificity and resolution of molecular imaging. The development of multimodality aptamer imaging systems combined with clinical biomarkers has the potential to improve diagnostic and monitoring accuracy. Additionally, the creation of multi-target imaging systems provides valuable information for a comprehensive understanding of biological processes, clinical disease diagnosis, case analysis, and disease treatment. Furthermore, through the adoption of innovative labeling techniques utilizing low background noise and high-efficiency reporter molecules, imaging sensitivity can be enhanced, thus achieving high-resolution imaging of subcellular organelles to study cellular functions. Moreover, owing to the rapid binding and dissociation characteristics of aptamers, real-time imaging is expected to monitor biological processes or disease progression.
Third, the application of aptamers in clinical scenarios requires further investigation, such as the biological safety and toxicity of aptamers. Tan's group investigated for the first time the distribution and metabolism of radiolabeled Sgc8 aptamers in the human body by applying panoramic dynamic PET scanning and establishing relevant pharmacokinetic models.224 This work lays the foundation for the future development of more aptamer-based imaging. Therefore, in the future, more studies are required to explore additional applications of aptamers to foster their clinical utilization in medical imaging as well as to advance molecular medicine and translational medicine.
In conclusion, the use of aptamers as recognition elements in molecular imaging has the potential to be applied in various imaging modalities. It is hopeful that, in the near future, potent aptamer probes will become readily accessible, enabling their full-fledged application in clinical evaluation and diagnosis.
Author contributions
B. Lin conceived the outline. B. Lin, F. Xiao, J. Jiang and Z. Zhao wrote the manuscript. X. Zhou supervised the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (92153303, 21721005 and 91940000 to X. Z.) and China Postdoctoral Science Foundation (211000159 to B. L.).Notes and references
- T. F. Massoud and S. S. Gambhir, Genes Dev., 2003, 17, 545 CrossRef CAS PubMed.
- T. Hussain and Q. T. Nguyen, Adv. Drug Delivery Rev., 2014, 66, 90 CrossRef CAS PubMed.
- B. Soontornworajit and Y. Wang, Anal. Bioanal. Chem., 2011, 399, 1591 CrossRef CAS PubMed.
- H. W. Liu, L. L. Chen, C. Y. Xu, Z. Li, H. Y. Zhang, X. B. Zhang and W. H. Tan, Chem. Soc. Rev., 2018, 47, 7140 RSC.
- S. Lee, J. Xie and X. Y. Chen, Biochemistry, 2010, 49, 1364 CrossRef CAS PubMed.
- W. J. Wei, Z. T. Rosenkrans, J. J. Liu, G. Huang, Q. Y. Luo and W. B. Cai, Chem. Rev., 2020, 120, 3787 CrossRef CAS PubMed.
- Y. Zhou, Y. T. Zhuo, R. Z. Peng, Y. T. Zhang, Y. L. Du, Q. Zhang, Y. Sun and L. P. Qiu, Sci. China: Chem., 2021, 64, 1817 CrossRef CAS.
- L. L. Chen, Y. Lyu, X. Zhang, L. T. Zheng, Q. Q. Li, D. Ding, F. M. Chen, Y. H. Liu, W. Li, Y. T. Zhang, Q. L. Huang, Z. Q. Wang, T. T. Xie, Q. Zhang, Y. Sima, K. Li, S. Xu, T. B. Ren, M. Y. Xiong, Y. Wu, J. B. Song, L. Yuan, H. H. Yang, X. B. Zhang and W. H. Tan, Sci. China: Chem., 2023, 66, 1336 CrossRef CAS.
- L. L. Wu, Y. D. Wang, X. Xu, Y. L. Liu, B. Q. Lin, M. X. Zhang, J. L. Zhang, S. Wan, C. Y. Yang and W. H. Tan, Chem. Rev., 2021, 121, 12035 CrossRef CAS PubMed.
- L. Bohrmann, T. Burghardt, C. Haynes, K. Saatchi and U. O. Hafeli, Theranostics, 2022, 12, 4010 CrossRef CAS PubMed.
- S. Strauss, P. C. Nickels, M. T. Strauss, V. J. Sabinina, J. Ellenberg, J. D. Carter, S. Gupta, N. Janjic and R. Jungmann, Nat. Methods, 2018, 15, 685 CrossRef CAS PubMed.
- M. J. Huang, J. J. Yang, T. Wang, J. Song, J. L. Xia, L. L. Wu, W. Wang, Q. Y. Wu, Z. Zhu, Y. L. Song and C. Y. Yang, Angew. Chem., Int. Ed., 2020, 59, 4800 CrossRef CAS PubMed.
- A. D. Keefe, S. Pai and A. Ellington, Nat. Rev. Drug Discovery, 2010, 9, 537 CrossRef CAS PubMed.
- J. H. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181 CrossRef CAS PubMed.
- R. E. Wang, H. Wu, Y. Niu and J. Cai, Curr. Med. Chem., 2011, 18, 4126 CrossRef CAS PubMed.
- Y. Li, T. Li, H. Chen, L. Wang, Y. Xia, L. Zhang, Y. Xie, J. Li, C. Luo, Y. Xu, Y. Liu and W. Tan, ACS Appl. Mater. Interfaces, 2022, 14, 17032 CrossRef CAS PubMed.
- X. J. Yin, H. M. Zhao, Z. Q. He and X. Wang, Nanotechnol. Rev., 2023, 12, 20230107 CrossRef CAS.
- Z. H. Di, J. Zhao, H. Q. Chu, W. T. Xue, Y. L. Zhao and L. L. Li, Adv. Mater., 2019, 31, 1901885 CrossRef PubMed.
- Z. Xiang, J. Zhao, D. Yi, Z. Di and L. Li, Angew Chem. Int. Ed. Engl., 2021, 60, 22659 CrossRef CAS PubMed.
- Y. Pan, X. Luan, F. Zeng, Q. Xu, Z. Li, Y. Gao, X. Liu, X. Li, X. Han, J. Shen and Y. Song, Biosens. Bioelectron., 2022, 209, 114239 CrossRef CAS PubMed.
- L. Li, J. Wang, H. Jiang, X. Wen, M. Yang, S. Li, Q. Guo and K. Wang, Chin. Chem. Lett., 2023, 34, 107506 CrossRef CAS.
- B. Feng, Y. Xing, J. Lan, Z. Su and F. Wang, Talanta, 2020, 212, 120796 CrossRef CAS PubMed.
- Y. X. Wu, D. Zhang, X. Hu, R. Peng, J. Li, X. Zhang and W. Tan, Angew Chem. Int. Ed. Engl., 2021, 60, 12569 CrossRef CAS PubMed.
- Z. Xu, T. Shi, F. Mo, W. Yu, Y. Shen, Q. Jiang, F. Wang and X. Liu, Angew Chem. Int. Ed. Engl., 2022, 61, e202211505 CrossRef CAS PubMed.
- M. Jiao, Y. Wang, W. Wang, X. Zhou, J. Xu, Y. Xing, L. Chen, Y. Zhang, M. Chen, K. Xu and S. Zheng, Chem. Eng. J., 2022, 440, 135965 CrossRef CAS.
- Z. Xiang, J. Zhao, J. Qu, J. Song and L. Li, Angew Chem. Int. Ed. Engl., 2022, 61, e202111836 CrossRef CAS PubMed.
- Y. Jing, M. Cai, L. Zhou, J. Jiang, J. Gao and H. Wang, Talanta, 2020, 217, 121037 CrossRef CAS PubMed.
- S. Xie, Z. Wang, T. Fu, L. Zheng, H. Wu, L. He, H. Huang, C. Yang, R. Wang, X. Qian, L. Qiu and W. Tan, Angew Chem. Int. Ed. Engl., 2022, 61, e202201220 CrossRef CAS PubMed.
- S. Xu, T. Wu, N. Ren, J. Li, Y. Zhang, H. Wang, H. Liu and H. Liu, Biosens. Bioelectron., 2022, 213, 114478 CrossRef CAS PubMed.
- C. Dong, X. Fang, J. Xiong, J. Zhang, H. Gan, C. Song and L. Wang, ACS Nano, 2022, 16, 14055 CrossRef CAS PubMed.
- L. Xu, S. Liu, T. Yang, Y. Shen, Y. Zhang, L. Huang, L. Zhang, S. Ding, F. Song and W. Cheng, Theranostics, 2019, 9, 1993 CrossRef CAS PubMed.
- Y. Song, Z. Zhu, Y. An, W. Zhang, H. Zhang, D. Liu, C. Yu, W. Duan and C. J. Yang, Anal. Chem., 2013, 85, 4141 CrossRef CAS PubMed.
- L. Zhu, Y. Xu, X. Wei, H. Lin, M. Huang, B. Lin, Y. Song and C. Yang, Angew Chem. Int. Ed. Engl., 2021, 60, 18111 CrossRef CAS PubMed.
- Y. Ma, W. Guo, Q. Mou, X. Shao, M. Lyu, V. Garcia, L. Kong, W. Lewis, C. Ward, Z. Yang, X. Pan, S. S. Yi and Y. Lu, Nat. Biotechnol., 2023 DOI:10.1038/s41587-023-01801-z.
- X. Li, H. Kim, J. L. Litke, J. Wu and S. R. Jaffrey, Angew Chem. Int. Ed. Engl., 2020, 59, 4511 CrossRef CAS PubMed.
- Y. P. Zhang, Z. G. Wang, Y. F. Tian, L. H. Jiang, L. Zhao, D. M. Kong, X. Li, D. W. Pang and S. L. Liu, Angew Chem. Int. Ed. Engl., 2023, 62, e202217230 CrossRef CAS PubMed.
- R. Wirth, P. Gao, G. U. Nienhaus, M. Sunbul and A. Jaschke, J. Am. Chem. Soc., 2019, 141, 7562 CrossRef CAS PubMed.
- F. Bouhedda, K. T. Fam, M. Collot, A. Autour, S. Marzi, A. Klymchenko and M. Ryckelynck, Nat. Chem. Biol., 2020, 16, 69 CrossRef CAS PubMed.
- A. D. Cawte, P. J. Unrau and D. S. Rueda, Nat. Commun., 2020, 11, 1283 CrossRef CAS PubMed.
- J. Zhang, L. Wang, A. Jaschke and M. Sunbul, Angew Chem. Int. Ed. Engl., 2021, 60, 21441 CrossRef CAS PubMed.
- W. J. Zhou, H. Li, K. K. Zhang, F. Wang, X. Chu and J. H. Jiang, J. Am. Chem. Soc., 2021, 143, 14394 CrossRef CAS PubMed.
- Q. Wang, F. Xiao, H. Su, H. Liu, J. Xu, H. Tang, S. Qin, Z. Fang, Z. Lu, J. Wu, X. Weng and X. Zhou, Nucleic Acids Res., 2022, 50, e84 CrossRef CAS PubMed.
- M. Sunbul, J. Lackner, A. Martin, D. Englert, B. Hacene, F. Grun, K. Nienhaus, G. U. Nienhaus and A. Jaschke, Nat. Biotechnol., 2021, 39, 686 CrossRef CAS PubMed.
- Y. Gu, L. J. Huang, W. Zhao, T. T. Zhang, M. R. Cui, X. J. Yang, X. L. Zhao, H. Y. Chen and J. J. Xu, ACS Sens., 2021, 6, 2339 CrossRef CAS PubMed.
- C. Yan, L. Miao, Y. Zhang, X. Zhou, G. Wang, Y. Li, Q. Qiao and Z. Xu, Sens. Actuators, B, 2023, 386, 133731 CrossRef CAS.
- H. Tang, J. Peng, S. Peng, Q. Wang, X. Jiang, X. Xue, Y. Tao, L. Xiang, Q. Ji, S. M. Liu, X. Weng and X. Zhou, Chem. Sci., 2022, 13, 14032 RSC.
- G. Zheng, L. Zhao, D. Yuan, J. Li, G. Yang, D. Song, H. Miao, L. Shu, X. Mo, X. Xu, L. Li, X. Song and Y. Zhao, Biosens. Bioelectron., 2022, 198, 113827 CrossRef CAS PubMed.
- M. Omer, V. L. Andersen, J. S. Nielsen, J. Wengel and J. Kjems, Mol. Ther.--Nucleic Acids, 2020, 22, 994 CrossRef CAS PubMed.
- U. M. Mahajan, Q. Li, A. Alnatsha, J. Maas, M. Orth, S. H. Maier, J. Peterhansl, I. Regel, M. Sendler, P. R. Wagh, N. Mishra, Y. Xue, P. Allawadhi, G. Beyer, J. P. Kuhn, T. Marshall, B. Appel, F. Lammerhirt, C. Belka, S. Muller, F. U. Weiss, K. Lauber, M. M. Lerch and J. Mayerle, Gastroenterology, 2021, 161, 996 CrossRef CAS PubMed.
- S. Khosravi, P. Schindele, E. Gladilin, F. Dunemann, T. Rutten, H. Puchta and A. Houben, Front. Plant Sci., 2020, 11, 1254 CrossRef PubMed.
- R. Wu, A. Karunanayake Mudiyanselage, F. Shafiei, B. Zhao, Y. Bagheri, Q. Yu, K. McAuliffe, K. Ren and M. You, Angew Chem. Int. Ed. Engl., 2019, 58, 18271 CrossRef CAS PubMed.
- P. Delcanale, D. Porciani, S. Pujals, A. Jurkevich, A. Chetrusca, K. D. Tawiah, D. H. Burke and L. Albertazzi, Angew. Chem., Int. Ed., 2020, 59, 18546 CrossRef CAS PubMed.
- Y. Peng, X. Ai, Y. Yuan, J. Dong, X. Cui, F. Du, X. Huang and Z. Tang, Anal. Chim. Acta, 2022, 1209, 339816 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Sci., 1990, 249, 505 CrossRef CAS PubMed.
- T. Hianik and J. Wang, Electroanalysis, 2009, 21, 1223 CrossRef CAS.
- K. Sefah, D. Shangguan, X. L. Xiong, M. B. O'Donoghue and W. H. Tan, Nat. Protoc., 2010, 5, 1169 CrossRef CAS PubMed.
- X. Ma, A. Gosai and P. Shrotriya, J. Colloid Interface Sci., 2020, 559, 1 CrossRef CAS PubMed.
- S. Nonin-Lecomte, C. H. Lin and D. J. Patel, Biophys. J., 2001, 81, 3422 CrossRef CAS PubMed.
- N. A. Kolganova, V. B. Tsvetkov, I. P. Smirnov and E. N. Timofeev, Nucleic Acid Ther., 2019, 29, 208 CrossRef CAS PubMed.
- G. H. Xu, J. J. Zhao, N. Liu, M. H. Yang, Q. Zhao, C. G. Li and M. L. Liu, Nucleic Acids Res., 2019, 47, 5963 CrossRef CAS PubMed.
- L. H. Bie, Y. Wang, F. Z. Jiang, Z. Xiao, L. J. Zhang and J. Wang, Front. Mol. Biosci., 2022, 9 DOI:10.3389/fmolb.2022.1025313.
- S. D. Cai, J. H. Yan, H. J. Xiong, Y. F. Liu, D. M. Peng and Z. B. Liu, Analyst, 2018, 143, 5317 RSC.
- J. W. Liu and Y. Lu, J. Am. Chem. Soc., 2007, 129, 8634 CrossRef CAS PubMed.
- L. Fang, L. Xiao, Y. Jun, Y. Onishi and E. Kool, Nat. Chem., 2023, 15, 1296 CrossRef CAS PubMed.
- J. Aldag, T. Persson and R. K. Hartmann, Int. J. Mol. Sci., 2018, 19, 3883 CrossRef PubMed.
- M. Egli and M. Manoharan, Nucleic Acids Res., 2023, 51, 2529 CrossRef CAS PubMed.
- N. Freund, A. I. Taylor, S. Arangundy-Franklin, N. Subramanian, S.-Y. Peak-Chew, A. M. Whitaker, B. D. Freudenthal, M. Abramov, P. Herdewijn and P. Holliger, Nat. Chem., 2023, 15, 91 CrossRef CAS PubMed.
- J. Zhang, A. Zhu, M. Mei, J. Qu, Y. Huang, Y. Shi, M. Xue, J. Zhang, R. Zhang, B. Zhou, X. Tan, J. Zhao and Y. Wang, Adv. Sci., 2023, 10, e2300656 CrossRef PubMed.
- W. Ma, B. Chen, S. Zou, R. Jia, H. Cheng, J. Huang, H. Wang, X. He and K. Wang, Anal. Chem., 2019, 91, 12538 CrossRef CAS PubMed.
- Y. Wang, D. Wang, J. Lin, Z. Lyu, P. Chen, T. Sun, C. Xue, M. Mojtabavi, A. Vedadghavami, Z. Zhang, R. Wang, L. Zhang, C. Park, G. S. Heo, Y. Liu, S. Dong and K. Zhang, Angew. Chem., Int. Ed., 2022, 61, e202204576 CrossRef CAS PubMed.
- B. Yang, B. N. Zhou, C. F. Li, X. W. Li, Z. W. Shi, Y. X. Li, C. Y. Zhu, X. Li, Y. Hua, Y. F. Pan, J. He, T. Y. Cao, Y. W. Sun, W. L. Liu, M. Ge, Y. R. Yang, Y. C. Dong and D. S. Liu, Angew. Chem., Int. Ed., 2022, 61, e202202520 CrossRef CAS PubMed.
- W. R. Zhong and J. T. Sczepanski, ACS Sens., 2019, 4, 566 CrossRef CAS PubMed.
- T. N. Navien, R. Thevendran, H. Y. Hamdani, T. H. Tang and M. Citartan, Biochim, 2021, 180, 54 CrossRef CAS PubMed.
- S. Hong, X. Zhang, R. J. Lake, G. T. Pawel, Z. Guo, R. Pei and Y. Lu, Chem. Sci., 2020, 11, 713 RSC.
- E. E. Ferapontova, E. M. Olsen and K. V. Gothelf, J. Am. Chem. Soc., 2008, 130, 4256 CrossRef CAS PubMed.
- J. W. Liu and Y. Lu, Angew. Chem., Int. Ed., 2006, 45, 90 CrossRef CAS PubMed.
- W. A. Zhao, W. Chiuman, J. C. F. Lam, S. A. McManus, W. Chen, Y. G. Cui, R. Pelton, M. A. Brook and Y. F. Li, J. Am. Chem. Soc., 2008, 130, 3610 CrossRef CAS PubMed.
- Y. F. Bai, F. Feng, L. Zhao, C. Y. Wang, H. Y. Wang, M. Z. Tian, J. Qin, Y. L. Duan and X. X. He, Biosens. Bioelectron., 2013, 47, 265 CrossRef CAS PubMed.
- X. M. Ma, Z. H. Chen, J. Zhou, W. Weng, O. Zheng, Z. Y. Lin, L. H. Guo, B. Qiu and G. N. Chen, Biosens. Bioelectron., 2014, 55, 412 CrossRef CAS PubMed.
- L. Zhang, L. Zhou, H. Zhang, Y. Zhang, L. Li, T. Xie, Y. Chen, X. Li, N. Ling, J. Dai, X. Sun, J. Liu, J. Zhao, T. Peng and M. Ye, ACS Appl. Mater. Interfaces, 2021, 13, 54656 CrossRef CAS PubMed.
- L. Wang, T. Bing, Y. Liu, N. Zhang, L. Shen, X. Liu, J. Wang and D. Shangguan, J. Am. Chem. Soc., 2018, 140, 18066 CrossRef CAS PubMed.
- Z. H. Wang, M. Luo, C. Q. Mao, Q. Wei, T. Zhao, Y. Li, G. Huang and J. M. Gao, Angew. Chem., Int. Ed., 2017, 56, 1319 CrossRef CAS PubMed.
- H. Shi, X. He, K. Wang, X. Wu, X. Ye, Q. Guo, W. Tan, Z. Qing, X. Yang and B. Zhou, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3900 CrossRef CAS PubMed.
- C. Wu, T. Chen, D. Han, M. You, L. Peng, S. Cansiz, G. Zhu, C. Li, X. Xiong, E. Jimenez, C. J. Yang and W. Tan, ACS Nano, 2013, 7, 5724 CrossRef CAS PubMed.
- W. Ma, H. Sun, B. Chen, R. Jia, J. Huang, H. Cheng, X. He, M. Huang and K. Wang, Anal. Chem., 2021, 93, 14552 CrossRef CAS PubMed.
- J. Tang, Y. Lei, X. He, J. Liu, H. Shi and K. Wang, Anal. Chem., 2020, 92, 10839 CrossRef CAS PubMed.
- Z. J. Wang, Y. Luo, X. D. Xie, X. J. Hu, H. Y. Song, Y. Zhao, J. Y. Shi, L. H. Wang, G. Glinsky, N. Chen, R. Lal and C. H. Fan, Angew. Chem., Int. Ed., 2018, 57, 972 CrossRef CAS PubMed.
- Z. W. Tang, P. Mallikaratchy, R. H. Yang, Y. M. Kim, Z. Zhu, H. Wang and W. H. Tan, J. Am. Chem. Soc., 2008, 130, 11268 CrossRef CAS PubMed.
- X. Chai, Z. Fan, M.-M. Yu, J. Zhao and L. Li, Nano Lett., 2021, 21, 10047 CrossRef CAS PubMed.
- S. S. Shekhawat and I. Ghosh, Curr. Opin. Chem. Biol., 2011, 15, 789 CrossRef CAS PubMed.
- J. S. Paige, K. Y. Wu and S. R. Jaffrey, Sci, 2011, 333, 642 CrossRef CAS PubMed.
- G. S. Filonov, J. D. Moon, N. Svensen and S. R. Jaffrey, J. Am. Chem. Soc., 2014, 136, 16299 CrossRef CAS PubMed.
- X. Chen, D. Zhang, N. Su, B. Bao, X. Xie, F. Zuo, L. Yang, H. Wang, L. Jiang, Q. Lin, M. Fang, N. Li, X. Hua, Z. Chen, C. Bao, J. Xu, W. Du, L. Zhang, Y. Zhao, L. Zhu, J. Loscalzo and Y. Yang, Nat. Biotechnol., 2019, 37, 1287 CrossRef CAS PubMed.
- E. V. Dolgosheina, S. C. Y. Jeng, S. S. S. Panchapakesan, R. Cojocaru, P. S. K. Chen, P. D. Wilson, N. Hawkins, P. A. Wiggins and P. J. Unrau, ACS Chem. Biol., 2014, 9, 2412 CrossRef CAS PubMed.
- D. M. Kolpashchikov and A. A. Spelkov, Angew. Chem., Int. Ed., 2021, 60, 4988 CrossRef CAS PubMed.
- H. Shi, Y. Lei, J. Ge, X. He, W. Cui, X. Ye, J. Liu and K. Wang, Anal. Chem., 2019, 91, 9154 CrossRef CAS PubMed.
- F. Zhou, T. Fu, Q. Huang, H. Kuai, L. Mo, H. Liu, Q. Wang, Y. Peng, D. Han, Z. Zhao, X. Fang and W. Tan, J. Am. Chem. Soc., 2019, 141, 18421 CrossRef CAS PubMed.
- Z. Ge, H. Gu, Q. Li and C. Fan, J. Am. Chem. Soc., 2018, 140, 17808 CrossRef CAS PubMed.
- W. Ma, B. Chen, R. Jia, H. Sun, J. Huang, H. Cheng, H. Wang, X. He and K. Wang, Anal. Chem., 2021, 93, 10511 CrossRef CAS PubMed.
- X.-Q. Li, X.-N. Liu, Y.-L. Jia, X.-L. Luo, H.-Y. Chen and J.-J. Xu, Anal. Chem., 2021, 93, 14892 CrossRef CAS PubMed.
- R. Jia, X. He, W. Ma, Y. Lei, H. Cheng, H. Sun, J. Huang and K. Wang, Anal. Chem., 2019, 91, 15107 CrossRef CAS PubMed.
- L. Zhong, S. Cai, Y. Huang, L. Yin, Y. Yang, C. Lu and H. Yang, Anal. Chem., 2018, 90, 12059 CrossRef CAS PubMed.
- C. Yang, Y. Shi, Y. Zhang, J. He, M. Li, W. Huang, R. Yuan and W. Xu, Anal. Chem., 2023, 95, 10337 CrossRef CAS PubMed.
- Z. Dong, X. Xu, J. Ni, Y. Li, K. An, L. Meng and H. Wu, J. Mater. Chem. B, 2023, 11, 204 RSC.
- Z. Tian, P. Peng, H. Wang, J. Zheng, L. Shi and T. Li, Anal. Chem., 2020, 92, 10357 CrossRef CAS PubMed.
- Y. Fan, L. Li, M. Lu, H. Si and B. Tang, Chem. Commun., 2019, 55, 4043 RSC.
- C. H. Li, W. Y. Lv, F. F. Yang, S. J. Zhen and C. Z. Huang, ACS Appl. Mater. Interfaces, 2022, 14, 12059 CrossRef CAS PubMed.
- F. Xuan and I. M. Hsing, J. Am. Chem. Soc., 2014, 136, 9810 CrossRef CAS PubMed.
- J. Tang, B. Li, C. Qi, Z. Wang, K. Yin, L. Guo, W. Zhang and B. Yuan, Talanta, 2022, 243, 123399 CrossRef CAS PubMed.
- H. Gong, Q. Dai and P. Peng, ACS Appl. Mater. Interfaces, 2022, 14, 43026 CrossRef CAS PubMed.
- L. L. Li, W. Y. Lv, Y. T. Xu, Y. F. Li, C. M. Li and C. Z. Huang, Anal. Chem., 2022, 94, 4399 CrossRef CAS PubMed.
- H. H. Wang, P. Peng, Q. W. Wang, Y. Du, Z. J. Tian and T. Li, Angew. Chem., Int. Ed., 2020, 59, 6099 CrossRef CAS PubMed.
- P. Peng, Y. Du, J. Zheng, H. H. Wang and T. Li, Angew. Chem., Int. Ed., 2019, 58, 1648 CrossRef CAS PubMed.
- M. Y. Ali, I. Tariq, M. F. Sohail, M. U. Amin, S. Ali, S. R. Pinnapireddy, A. Ali, J. Schaefer and U. Bakowsky, Eur. J. Pharm. Biopharm., 2019, 145, 42 CrossRef CAS PubMed.
- W. Dai, Q. Liu, S. Li, Y. Gao, C. Feng, L. Guo, Y. Xiao, H. Lin, Y. Fan and X. Zhang, J. Mater. Chem. B, 2023, 11, 4050 RSC.
- M. Wei, X. Shen, X. Fan, J. Li and J. Bai, Front. Bioeng. Biotechnol., 2023, 11, 1224339 CrossRef PubMed.
- M. P. Lokugamage, C. D. Sago, Z. Can, B. R. Krupczak and J. E. Dahlman, Adv. Mater., 2019, 31, 1902251 CrossRef CAS PubMed.
- X. Li, K. Feng, L. Li, L. Yang, X. Pan, H. S. Yazd, C. Cui, J. Li, L. Moroz, Y. Sun, B. Wang, X. Li, T. Huang and W. Tan, Natl. Sci. Rev., 2020, 7, 1933 CrossRef CAS PubMed.
- M. Falsafi, N. H. Goji, A. S. Saljooghi, K. Abnous, S. M. Taghdisi, S. Nekooei, M. Ramezani and M. Alibolandi, Expert Opin. Drug Delivery, 2022, 19, 743 CrossRef CAS PubMed.
- S. Lu, B. Fu and Z. Zhang, Anal. Chem., 2022, 94, 14509 CrossRef CAS PubMed.
- F. Cheng, Y. Jiang, B. Kong, H. Lin, X. Shuai, P. Hu, P. Gao, L. Zhan, C. Huang and C. Li, Adv. Healthcare Mater., 2023, 12, 2300102 CrossRef CAS PubMed.
- X. Meng, H. Wang, M. Yang, J. Li, F. Yang, K. Zhang, H. Dong and X. Zhang, Anal. Chem., 2021, 93, 1693 CrossRef CAS PubMed.
- S. Fathi-Karkan, S. Mirinejad, F. Ulucan-Karnak, M. Mukhtar, H. G. Almanghadim, S. Sargazi, A. Rahdar and A. M. Diez-Pascual, Int. J. Biol. Macromol., 2023, 238, 124103 CrossRef CAS PubMed.
- A. Yañez-Aulestia, N. K. Gupta, M. Hernández, G. Osorio-Toribio, E. Sánchez-González, A. Guzmán-Vargas, J. L. Rivera, I. A. Ibarra and E. Lima, Chem. Commun., 2022, 58, 10886 RSC.
- B. Yang, B. Chen, M. He, X. Yin, C. Xu and B. Hu, Anal. Chem., 2018, 90, 2355 CrossRef CAS PubMed.
- Q. Su, S. Han, X. Xie, H. Zhu, H. Chen, C.-K. Chen, R.-S. Liu, X. Chen, F. Wang and X. Liu, J. Am. Chem. Soc., 2012, 134, 20849 CrossRef CAS PubMed.
- Y. Mi, J. Zhao, H. Chu, Z. Li, M. Yu and L. Li, Anal. Chem., 2021, 93, 2500 CrossRef CAS PubMed.
- N. Wu, L. Bao, L. Ding and H. Ju, Angew. Chem., Int. Ed., 2016, 55, 5220 CrossRef CAS PubMed.
- J. Yang, J. Qu, X. Teng, W. Zhu, Y. Xu, Y. Yang and X. Qian, Adv. Healthcare Mater., 2023, 12, 2301084 CrossRef CAS PubMed.
- J. Wang, X. Fang, C. Zhang, H. Ji, Q. Pang, X. Li, Z. Luo, Q. Wu and L. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 16118 CrossRef CAS PubMed.
- X. Chang, C. Zhang, C. Lv, Y. Sun, M. Zhang, Y. Zhao, L. Yang, D. Han and W. Tan, J. Am. Chem. Soc., 2019, 141, 12738 CrossRef CAS PubMed.
- E. RodrÍguez, S. Schetters and Y. van Kooyk, Nat. Rev. Immunol., 2018, 18, 204 CrossRef PubMed.
- M. Huang, L. Zhu, S. Kang, F. Chen, X. Wei, L. Lin, X. Chen, W. Wang, Z. Zhu, C. Yang and Y. Song, Anal. Chem., 2021, 93, 15958 CrossRef CAS PubMed.
- P. V. Robinson, G. de Almeida-Escobedo, A. E. de Groot, J. L. McKechnie and C. R. Bertozzi, J. Am. Chem. Soc., 2015, 137, 10452 CrossRef CAS PubMed.
- J. Li, S. Liu, L. Sun, W. Li, S.-Y. Zhang, S. Yang, J. Li and H.-H. Yang, J. Am. Chem. Soc., 2018, 140, 16589 CrossRef CAS PubMed.
- Z. Liu, Y. Liang, W. Cao, W. Gao and B. Tang, Anal. Chem., 2021, 93, 8915 CrossRef CAS PubMed.
- B. Yuan, Y. Chen, Y. Sun, Q. Guo, J. Huang, J. Liu, X. Meng, X. Yang, X. Wen, Z. Li, L. Li and K. Wang, Anal. Chem., 2018, 90, 6131 CrossRef CAS PubMed.
- S. Shao, W. Zhang, H. Hu, B. Xue, J. Qin, C. Sun, Y. Sun, W. Wei and Y. Sun, Nucleic Acids Res., 2016, 44, e86 CrossRef PubMed.
- Y. Fu, P. P. Rocha, V. M. Luo, R. Raviram, Y. Deng, E. O. Mazzoni and J. A. Skok, Nat. Commun., 2016, 7, 11707 CrossRef CAS PubMed.
- K. T. Fam, R. Pelletier, F. Bouhedda, M. Ryckelynck, M. Collot and A. S. Klymchenko, Anal. Chem., 2022, 94, 6657 CrossRef CAS PubMed.
- X. Liu, T. Wang, Y. Wu, Y. Tan, T. Jiang, K. Li, B. Lou, L. Chen, Y. Liu and Z. Liu, Biosens. Bioelectron., 2022, 208, 114231 CrossRef CAS PubMed.
- M. Fang, H. Li, X. Xie, H. Wang, Y. Jiang, T. Li, B. Zhang, X. Jiang, Y. Cao, R. Zhang, D. Zhang, Y. Zhao, L. Zhu, X. Chen and Y. Yang, Biosens. Bioelectron., 2023, 235, 115411 CrossRef CAS PubMed.
- Z.-M. Ying, Z. Wu, B. Tu, W. Tan and J.-H. Jiang, J. Am. Chem. Soc., 2017, 139, 9779 CrossRef CAS PubMed.
- Z.-M. Ying, Y.-Y. Yuan, B. Tu, L.-J. Tang, R.-Q. Yu and J.-H. Jiang, Chem. Sci., 2019, 10, 4828 RSC.
- Y. Du, P. Peng and T. Li, ACS Nano, 2019, 13, 5778 CrossRef CAS PubMed.
- J. Kim, A. M. Yu, K. P. Kubelick and S. Y. Emelianov, Photoacoustics, 2021, 25, 100307 CrossRef PubMed.
- M.-R. Cui, L.-X. Chen, X.-L. Li, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2020, 92, 4558 CrossRef CAS PubMed.
- J. Habchi, P. Tompa, S. Longhi and V. N. Uversky, Chem. Rev., 2014, 114, 6561 CrossRef CAS PubMed.
- K. P. Carter, A. M. Young and A. E. Palmer, Chem. Rev., 2014, 114, 4564 CrossRef CAS PubMed.
- Y. Yang, J. Huang, X. Yang, K. Quan, N. Xie, M. Ou, J. Tang and K. Wang, Chem. Commun., 2016, 52, 11386 RSC.
- Q. Yu, J. Shi, A. P. K. K. K. Mudiyanselage, R. Wu, B. Zhao, M. Zhou and M. You, Chem. Commun., 2019, 55, 707 RSC.
- R. R. Zhang, A. B. Schroeder, J. J. Grudzinski, E. L. Rosenthal, J. M. Warram, A. N. Pinchuk, K. W. Eliceiri, J. S. Kuo and J. P. Weichert, Nat. Rev. Clin. Oncol., 2017, 14, 347 CrossRef CAS PubMed.
- M. Mahmoudpour, S. Ding, Z. Lyu, G. Ebrahimi, D. Du, J. E. N. Dolatabadi, M. Torbati and Y. Lin, Nano Today, 2021, 39, 101177 CrossRef CAS.
- J. Hernandez-Gil, C. Y. Chow, H. Chatras, P. D. de Souza Franca, Z. V. Samuels, M. Cornejo, G. F. King, J. S. Lewis, T. Reiner and J. Gonzales, J. Am. Chem. Soc., 2023, 145, 14276 CrossRef CAS PubMed.
- K. E. Adams, S. Ke, S. Kwon, F. Liang, Z. Fan, Y. Lu, K. Hirschi, M. E. Mawad, M. A. Barry and E. M. Sevick-Muraca, J. Biomed. Opt., 2007, 12, 024017 CrossRef PubMed.
- D.-H. Li, R. S. Gamage, A. G. Oliver, N. L. Patel, S. M. Usama, J. D. Kalen, M. J. Schnermann and B. D. Smith, Angew. Chem., Int. Ed., 2023, 62, e202305062 CrossRef CAS PubMed.
- H. Shen, B. Wu, Q. Zhang, J. Ni, M. Liang, Y. Liu, X.-F. Zang, S. Wang, Y.-Y. Quan, X. Ye and Z.-S. Huang, Chem. Eng. J., 2023, 468, 143726 CrossRef CAS.
- S. Zhang, H. Yuan, S. Sun, C. Qin, Q. Qiu, Y. Feng, Y. Liu, Y. Li, L. Xu, Y. Ying, J. Qi and Y. Wang, Adv. Sci., 2023, e2207651, DOI:10.1002/advs.202207651.
- T. Zhang, Q. Cheng, J. H. Lei, B. Wang, Y. Chang, Y. Liu, G. Xing, C. Deng, Z. Tang and S. Qu, Adv. Mater., 2023, e2302705, DOI:10.1002/adma.202302705.
- Z. Hu, C. Fang, B. Li, Z. Zhang, C. Cao, M. Cai, S. Su, X. Sun, X. Shi, C. Li, T. Zhou, Y. Zhang, C. Chi, P. He, X. Xia, Y. Chen, S. S. Gambhir, Z. Cheng and J. Tian, Nat. Biomed. Eng., 2020, 4, 259 CrossRef PubMed.
- E. Sicco, A. Monaco, M. Fernandez, M. Moreno, V. Calzada and H. Cerecetto, Sci. Rep., 2021, 11, 19942 CrossRef CAS PubMed.
- Y. Sun, Z. Zhang, T. Bing, J. Liu, W. Li, X. Liu, N. Zhang, Y. Shu, J. Wang and D. Shangguan, Anal. Chem., 2022, 94, 6044 CrossRef CAS PubMed.
- T. Abraham, C. O. McGovern, S. S. Linton, Z. Wilczynski, J. H. Adair and G. L. Matters, Int. J. Nanomed., 2021, 16, 2297 CrossRef PubMed.
- L. Agnello, S. Tortorella, A. D’Argenio, C. Carbone, S. Camorani, E. Locatelli, L. Auletta, D. Sorrentino, M. Fedele, A. Zannetti, M. C. Franchini and L. Cerchia, J. Exp. Clin. Cancer Res., 2021, 40, 239 CrossRef CAS PubMed.
- A. P. Arevalo, R. Castelli, M. Ibarra, M. Crispo and V. Calzada, Int. J. Mol. Sci., 2022, 23, 2466 CrossRef CAS PubMed.
- H. Zheng, A. GhavamiNejad, P. GhavamiNejad, M. Samarikhalaj, A. Giacca and M. Poudineh, ACS Sens., 2022, 7, 2387 CrossRef CAS PubMed.
- J. Georges, X. D. Qi, X. W. Liu, Y. Zhou, E. C. Woolf, A. Valeri, Z. Al-Atrache, E. Belykh, B. G. Feuerstein, M. Preul, A. C. Scheck, M. Reiser, T. Anderson, J. Gopez, D. Appelt, S. Yocom, J. Eschbacher, H. Yan and P. Nakaji, J. Neurosurg., 2021, 134, 1783 CAS.
- B. Chu, A. Wang, L. Cheng, R. Chen, H. Shi, B. Song, F. Dong, H. Wang and Y. He, J. Nanobiotechnol., 2021, 19, 187 CrossRef CAS PubMed.
- J. Chen, H. Li, Q. Wu, Q. Yan, J. Sun, F. Liang, Y. Liu and H. Wang, Anal. Chem., 2021, 93, 936 CrossRef CAS PubMed.
- Kenry, F. Nicolson, L. Clark, S. R. Panikkanvalappil, B. Andreiuk and C. Andreou, Nanotheranostics, 2022, 6, 31 CrossRef CAS PubMed.
- A. Liang, H. Wang, D. Yao and Z. Jiang, Food Chem., 2019, 271, 39 CrossRef CAS PubMed.
- M. A. Tahir, N. E. Dina, H. Cheng, V. K. Valev and L. Zhang, Nanoscale, 2021, 13, 11593 RSC.
- M. Kamal Hossain, Chem. Rec., 2022, 22, e202200108 CrossRef CAS PubMed.
- X. Wang, Z. Xia, E. K. Fodjo, W. Deng and D. Li, J. Mater. Chem. B, 2022, 10, 3023 RSC.
- J. Zhang, X. Miao, C. Song, N. Chen, J. Xiong, H. Gan, J. Ni, Y. Zhu, K. Cheng and L. Wang, Biosens. Bioelectron., 2022, 212, 114379 CrossRef CAS PubMed.
- J. Wang, D. Liang, Q. Jin, J. Feng and X. Tang, Bioconjugate Chem., 2020, 31, 182 CrossRef CAS PubMed.
- Y. Zou, S. Huang, Y. Liao, X. Zhu, Y. Chen, L. Chen, F. Liu, X. Hu, H. Tu, L. Zhang, Z. Liu, Z. Chen and W. Tan, Chem. Sci., 2018, 9, 2842 RSC.
- D. Koukouzelis, A. R. N. Pontillo, S. Koutsoukos, E. Pavlatou and A. Detsi, J. Mol. Liq., 2020, 306, 112929 CrossRef CAS.
- Y. Zhang, R. Chen, F. Liu, P. Miao, L. Lin and J. Ye, Small Methods, 2023, 7, 2201334 CrossRef CAS PubMed.
- X. Qin, Y. Si, Z. Wu, K. Zhang, J. Li and Y. Yin, Anal. Chem., 2020, 92, 924 CrossRef CAS PubMed.
- Y. Wen, V. X. Truong and M. Li, Nano Lett., 2021, 21, 3066 CrossRef CAS PubMed.
- Y. Zhou, J. Liu, T. Zheng and Y. Tian, Anal. Chem., 2020, 92, 5910 CrossRef CAS PubMed.
- J. Wang, D. Liang, Q. Jin, J. Feng and X. Tang, Bioconjugate Chem., 2020, 31, 182 CrossRef CAS PubMed.
- Y. Zou, S. Huang, Y. Liao, X. Zhu, Y. Chen, L. Chen, F. Liu, X. Hu, H. Tu, L. Zhang, Z. Liu, Z. Chen and W. Tan, Chem. Sci., 2018, 9, 2842 RSC.
- J. He, J. Dong, Y. Hu, G. Li and Y. Hu, Nanoscale, 2019, 11, 6089 RSC.
- D. Liang, Q. Jin, N. Yan, J. Feng, J. Wang and X. Tang, Adv. Biosyst., 2018, 2, 1800100 CrossRef.
- J. Wang, D. Liang, J. Feng and X. Tang, Anal. Chem., 2019, 91, 11045 CrossRef CAS PubMed.
- M. Fritz, A. M. Klawonn and N. M. Zahr, J. Neurosci. Res., 2022, 100, 1140 CrossRef CAS PubMed.
- N. J. Stewart, L. J. Smith, H.-F. Chan, J. A. Eaden, S. Rajaram, A. J. Swift, N. D. Weatherley, A. Biancardi, G. J. Collier, D. Hughes, G. Klafkowski, C. S. Johns, N. West, K. Ugonna, S. M. Bianchi, R. Lawson, I. Sabroe, H. Marshall and J. M. Wild, Br. J. Radiol., 2022, 95, 20210207 CrossRef PubMed.
- A. E. Campbell-Washburn, R. Ramasawmy, M. C. Restivo, I. Bhattacharya, B. Basar, D. A. Herzka, M. S. Hansen, T. Rogers, W. P. Bandettini, D. R. McGuirt, C. Mancini, D. Grodzki, R. Schneider, W. Majeed, H. Bhat, H. Xue, J. Moss, A. A. Malayeri, E. C. Jones, A. P. Koretsky, P. Kellman, M. Y. Chen, R. J. Lederman and R. S. Balaban, Radiology, 2019, 293, 384 CrossRef PubMed.
- K. M. Jones, A. C. Pollard and M. D. Pagel, J. Magn. Reson. Imaging, 2018, 47, 11 CrossRef PubMed.
- L. M. De Leon-Rodriguez, A. F. Martins, M. C. Pinho, N. M. Rofsky and A. D. Sherry, J. Magn. Reson. Imaging, 2015, 42, 545 CrossRef PubMed.
- J. Pellico, C. M. Ellis and J. J. Davis, Contrast Media Mol. Imaging, 2019, 2019, 1845637 Search PubMed.
- M.-F. Bellin, Eur. J. Radiol., 2006, 60, 314 CrossRef PubMed.
- H. Bakhtiari, A. A. Palizban, H. Khanahmad and M. R. Mofid, Res. Pharm. Sci., 2020, 15, 107 CrossRef PubMed.
- H. Yan, X. Gao, Y. Zhang, W. Chang, J. Li, X. Li, Q. Du and C. Li, ACS Appl. Mater. Interfaces, 2018, 10, 17047 CrossRef CAS PubMed.
- C. Zhang, S. S. Moonshi, W. Wang, T. Hang Thu, Y. Han, F. Y. Han, H. Peng, P. Kral, B. E. Rolfe, J. J. Gooding, K. Gaus and A. K. Whittaker, ACS Nano, 2018, 12, 9162 CrossRef CAS PubMed.
- M. Zhao, Z. Liu, L. Dong, H. Zhou, S. Yang, W. Wu and J. Lin, Int. J. Nanomed., 2018, 13, 4433 CrossRef CAS PubMed.
- G. Mariani, L. Bruselli, T. Kuwert, E. E. Kim, A. Flotats, O. Israel, M. Dondi and N. Watanabe, Eur. J. Nucl. Med. Mol. Imaging, 2010, 37, 1959 CrossRef PubMed.
- R. Lecomte, Eur. J. Nucl. Med. Mol. Imaging, 2009, 36(Suppl 1), S69 CrossRef PubMed.
- L. W. Dobrucki and A. J. Sinusas, Nat. Rev. Cardiol., 2010, 7, 38 CrossRef PubMed.
- S. M. Park, S. Baek, J. H. Lee, S. K. Woo, T. S. Lee, H. S. Park, J. Lee, Y. K. Kang, S. Y. Kang, M. Y. Yoo, H. J. Yoon, B. S. Kim, K. P. Lee and B. S. Moon, Clin. Transl. Sci., 2023, 16, 1186 CrossRef CAS PubMed.
- S. Cheng, O. Jacobson, G. Zhu, Z. Chen, S. H. Liang, R. Tian, Z. Yang, G. Niu, X. Zhu and X. Chen, Eur. J. Nucl. Med. Mol. Imaging, 2019, 46, 948 CrossRef CAS PubMed.
- M. Muzi, F. O'Sullivan, D. A. Mankoff, R. K. Doot, L. A. Pierce, B. F. Kurland, H. M. Linden and P. E. Kinahan, Magn. Reson. Imaging, 2012, 30, 1203 CrossRef PubMed.
- J. Charlton, J. Sennello and D. Smith, Chem. Biol., 1997, 4, 809 CrossRef CAS PubMed.
- P. Li, C. Wang, W. Wang, X. Duan and J. Li, J. Radioanal. Nucl. Chem., 2023, 332, 2279 CrossRef CAS.
- A. V. Ozerskaya, T. N. Zamay, O. S. Kolovskaya, N. A. Tokarev, K. V. Belugin, N. G. Chanchikova, O. N. Badmaev, G. S. Zamay, I. A. Shchugoreva and R. V. Moryachkov, Mol. Ther.--Nucleic Acids, 2021, 26, 1159 CrossRef CAS PubMed.
- Y. Jiao, P. Xu, S. Luan, X. Wang, Y. Gao, C. Zhao and P. Fu, Nucl. Med. Biol., 2022, 104, 28 CrossRef PubMed.
- L. Bohrmann, T. Burghardt, C. Rodríguez-Rodríguez, M. M. Herth, K. Saatchi and U. O. Hafeli, ACS Omega, 2023, 8, 11003 CrossRef CAS PubMed.
- L. Filippi, O. Bagni and C. Nervi, Expert Rev. Med. Devices, 2020, 17, 751 CrossRef CAS PubMed.
- Y. J. Choi, J. Y. Park, Y. L. Cho, J. R. Chae, H. Cho and W. J. Kang, Biochem. Biophys. Res. Commun., 2022, 620, 105 CrossRef CAS PubMed.
- A. B. E. Attia, G. Balasundaram, M. Moothanchery, U. S. Dinish, R. Bi, V. Ntziachristos and M. Olivo, Photoacoustics, 2019, 16, 100144 CrossRef PubMed.
- J. Groehl, M. Schellenberg, K. Dreher and L. Maier-Hein, Photoacoustics, 2021, 22, 100241 CrossRef PubMed.
- C. Kim, R. Qin, J. S. Xu, L. V. Wang and R. Xu, J. Biomed. Opt., 2010, 15, 010510 CrossRef PubMed.
- P. Fathi, H. J. Knox, D. Sar, I. Tripathi, F. Ostadhossein, S. K. Misra, M. B. Esch, J. Chan and D. Pan, ACS Nano, 2019, 13, 7690 CrossRef CAS PubMed.
- Z. Fan, Y. Wang, L. Li, F. Zeng, Q. Shang, Y. Liao, C. Liang and L. Nie, ACS Nano, 2022, 16, 16177 CrossRef CAS PubMed.
- Z. Chen, Y. Peng, Y. Li, X. Xie, X. Wei, G. Yang, H. Zhang, N. Li, T. Li, X. Qin, S. Li, C. Wu, F. You, H. Yang and Y. Liu, ACS Nano, 2021, 15, 16683 CrossRef CAS PubMed.
- N. Yu, T. Huang, T. Duan, Y. Bao, R. Gao, X. Wang, K. Xu and C. Han, Chem. Eng. J., 2022, 440, 135801 CrossRef CAS.
- A. Najdian, M. Amanlou, D. Beiki, A. Bitarafan-Rajabi, M. Mirzaei and M. Shafiee Ardestani, Bioorg. Chem., 2022, 125, 105827 CrossRef CAS PubMed.
- Y. Zeng, L. Zhao, K. Li, J. Ma, D. Chen, C. Liu, W. Zhan and Y. Zhan, Nano Res., 2023, 9859 CrossRef CAS.
- Y. Dai, H. Zhao, K. He, W. Du, Y. Kong, Z. Wang, M. Li, Q. Shen, P. Sun and Q. Fan, Small, 2021, 17, 2102527 CrossRef CAS PubMed.
- L. Jiang, H.-Y. Chen, C.-H. He, H.-B. Xu, Z.-R. Zhou, M.-S. Wu, E. K. Fodjo, Y. He, M. E. Hafez, R.-C. Qian and D.-W. Li, Anal. Chem., 2023, 95, 3507 CrossRef CAS PubMed.
- D. Ding, H. Zhao, D. Wei, Q. Yang, C. Yang, R. Wang, Y. Chen, L. Li, S. An, Q. Xia, G. Huang, J. Liu, Z. Xiao and W. Tan, Research, 2023, 6, 0126 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |