Recent advances in nanocomposite-based delivery systems for targeted CRISPR/Cas delivery and therapeutic genetic manipulation
Muhammad Waseem
Ghani
 ab,
Ambreen
Iqbal
ab,
Ambreen
Iqbal
 ab,
Hammad
Ghani
ab,
Hammad
Ghani
 c,
Sidra
Bibi
c,
Sidra
Bibi
 d,
Zixun
Wang
d,
Zixun
Wang
 *ab and
Renjun
Pei
*ab and
Renjun
Pei
 *ab
*ab
aSchool of Nano-Tech and Nano-Bionics, University of Science and Technology of China (USTC), Hefei 230026, P. R. China
bCAS Key Laboratory for Nano-Bio Interface, Suzhou Institute of Nano-Tech and Nano-Bionics (SINANO), Chinese Academy of Science, Suzhou 215123, P. R. China. E-mail: zixunwang2021@sinano.ac.cn; rjpei2011@sinano.ac.cn
cBasic Health Unit Laleka, Primary and Secondary Healthcare Department, Bahawalngar, 62300, Punjab, Pakistan
dDepartment of Biology, The Islamia University of Bahawalpur, Bahawalnagar Campus 62300, Pakistan
First published on 13th February 2023
Abstract
CRISPR/Cas systems are novel gene editing tools with tremendous capacity and accuracy for gene editing and hold great potential for therapeutic genetic manipulation. However, the lack of safe and efficient delivery methods for CRISPR/Cas and its guide RNA hinders their wide adoption for therapeutic applications. To this end, there is an increasing demand for safe, efficient, precise, and non-pathogenic delivery approaches, both in vitro and in vivo. With the convergence of nanotechnology and biomedicine, functional nanocomposites have demonstrated unparalleled sophistication to overcome the limits of CRISPR/Cas delivery. The tunability of the physicochemical properties of nanocomposites makes it very easy to conjugate them with different functional substances. The combinatorial application of diverse functional materials in the form of nanocomposites has shown excellent properties for CRISPR/Cas delivery at the target site with therapeutic potential. The recent highlights of selective organ targeting and phase I clinical trials for gene manipulation by CRISPR/Cas after delivery through LNPs are at the brink of making it to routine clinical practice. Here we summarize the recent advances in delivering CRISPR/Cas systems through nanocomposites for targeted delivery and therapeutic genome editing.
10th Anniversary statementOver the past ten years, the studies on all aspects of materials chemistry, from chemistry to applications in biology and medicine, have been published in the Journal of Materials Chemistry B, which has earned a stellar reputation as a premier venue for publishing such work. We are honored to have our work published in this 10th anniversary themed issue. I would like to take this opportunity to congratulate everyone involved in the Journal of Materials Chemistry B and to offer my best wishes for the journal's continued success in the years to come, as it celebrates its tenth anniversary. |
1 Introduction
In the past decade, genetic manipulation in mammalian cells has become a routine practice for therapeutic ventures, given the promising advances in the safety and efficacy of gene-editing through clustered regularly interspaced short palindromic repeats (CRISPR) systems.1 The CRISPR-associated (Cas) enzymes, first discovered in Streptococcus thermophilus as a defensive shield against bacteriophages,2 have expanded into a large family regarding the functional capability for DNA editing through the CRISPR/Cas 9 system and RNA manipulation through the CRISPR/Cas 12 & 13 system. In brief, the CRISPR regions contain short repeats (called spacers) that come from the nucleic acid fragments of invading viruses. The spacers provide a sequence-specific memory and targeted defence against the invasion of exogenous infection. During the immune response, CRISPR will transcribe into precursor transcripts that subsequently produce CRISPR RNAs (called crRNAs) which can bind with Cas nucleases, thus directing the precise identification and cleavage of the invader's nucleic acids.1 Owing to its tremendous performance and fast iteration rate, the CRISPR/Cas system has recently been widely adopted in a variety of biomedical applications such as cancer therapy, AIDS, liver diseases, cardiovascular diseases, and eye-related congenital defects.3–11 In addition, the biomedical bids of the CRISPR/Cas system as biosensors have been adapted recently, given the specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) mechanism. The RNA-guided RNA endonuclease Cas13a could act as a nanosensor for the sensitive detection of canine parvovirus-2 DNA.12 Another example is the quick detection based on the CRISPR/Cas13a nanomachine to accurately detect the influenza virus through H7N9.13Given their incredible capacity and accuracy for genome manipulation, CRISPR/Cas systems hold great potential for therapeutic applications. As genetic manipulation should occur in living cells, the delivery of a fully functional CRISPR/Cas system into live cells and specified target regions of the animals has become a deterministic step. To serve this purpose, many viral and nonviral cargo delivery systems have been employed for delivering CRISPR tools such as plasmid DNA, mRNA, or protein along with guide RNA into the cell and animal models.14 The traditionally used viral delivery systems give promising results at the expense of toxicity issues, let alone their immunogenic and tumorigenic properties prompting the demand for the discovery of safe and less toxic nonviral delivery systems.15,16 Different nonviral delivery approaches, including microinjection,17 electroporation,18 sonoporation,19 liposomes,20 and CRISPR-delivering nanocomposites,21 have also been employed to overcome the issues related to viral delivery of CRISPR and to improve the efficiency and the targeted gene editing. Owing to the rapid development of nanotechnology and biomedical engineering techniques, nanocomposite-based nanocarriers have become one of the most promising strategies for CRISPR/Cas delivery due to their excellent biocompatibility, less toxicity, and enhanced delivery efficacy. Here, for the first time, we summarize the recent advances in the nanocomposite (including nanoparticle and nanostructure)-based delivery of CRISPR/Cas tools for targeted delivery and their respective therapeutic applications.
2 CRISPR/Cas systems and their therapeutic applications
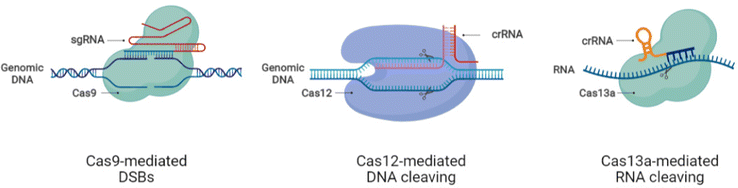
The discovery of CRISPR systems and their application for genetic manipulation in eukaryotic cells rendered the exploration of this system in other species of bacteria for broadening the repertoire of CRISPR systems based on the Cas enzymes. The current classification of CRISPR/Cas systems includes two classes, 1 and 2, and their six subtypes, I–VI, based on the conformation and functionality of the Cas protein.22,23 CRISPR/Cas9, a type II system from Streptococcus pyogenes (SpCas9), consists of Cas9 nuclease, crRNA, and transactivating crRNA (tracrRNA), mainly used for programmed genetic engineering in mammals.24,25 The ortholog of Cas9 found in Staphylococcus aureus Cas9 (SaCas9)26 and the recent discovery of other Cas enzymes (Cas12, Cas13, and Cas14) have added to the diversity of CRISPR/Cas systems.27,28 Further, the size of the Cas9 enzyme varies depending on the species of origin, as SaCas9, Campylobacter jejuni Cas9 (CjCas9),29 and Neisseria meningitidis Cas9 (NmeCas9)30 are more diminutive than SpCas9 easing the cargo loading and delivery.26 In addition, a recently reported subtype of Cas12, CasΦ enzyme, applicable for genome editing and DNA detection with a molecular weight half that of Cas9 and Cas12a, offers easy packaging and delivery.31,32 Xu et al. created a small Cas12f mutant CRISPR system called CasMINI, which is equally efficient as Cas12a in gene activation and gene editing.33 Similarly, the Cas14 enzyme, found in the archaea, capable of single-stranded DNA (ssDNA) cleavage in the target region, adds to easily deliverable CRISPR tools due to its compact size.342.1 The principles of operation of CRISPR/Cas systems
Mechanistically, CRISPR/Cas9 induces double-strand breaks (DSBs) at the genomic loci following the direction of designed guide RNA (gRNA) that is complementary to the target sequence. The presence of the protospacer adjacent motif (PAM) sequence ‘NGG’ at the end of the gRNA target region enhances the DNA binding efficiency and cleavability of SpCas9. The induction of DSBs activates the DNA repair system to mend the DNA damage either precisely by homology-directed repair (HDR) or by error-prone non-homologous end joining (NHEJ) or microhomologymediated end joining (MMEJ).35 The HDR mechanism permits only specific modifications, thus making it suitable for tailored genetic manipulation, while the NHEJ repair system works through insertions and deletions (indels), disrupting coding and noncoding sequences.35 The recently discovered Cas system, Cas13, allows editing at the RNA level providing a comprehensive application platform for research and medical drive.36–38 Hitherto, CRISPR/Cas systems have expanded into a large family with DNA and RNA editing capacity through CRISPR/Cas9 and CRISPR/Cas13, respectively.36,39–42The advances in CRISPR/Cas tools broadened the CRISPR-based repertoire of genome editing with different working modes. The variability in sequence recognition and respective functionality of different Cas enzymes increased the application range of CRISPR systems for biomedical applications and therapeutic purposes. The nucleotide base editing (BE) in DNA allows the conversion of C G to T A and A T to G C through changeover switches without generating DSBs.43,44 The function of prime editing (PE) could be achieved after linking Cas9H840A to an engineered reverse transcriptase enzyme, in combination with a PE gRNA (pegRNA) guiding the system to the required editing site.45 A modified form of dead Cas13 (dCas13b) upon tethering to human RNA adenosine deaminase can induce specific adenosine deamination in RNA.46 Other than the gene editing perks, CRISPR tools are also employed to repress gene transcription using an engineered form of Cas9, dead Cas9D10A/H840A (dCas9), which is unable to cleave DNA but can be used in CRISPR interference (CRISPRi) for gene knockdown through the fusion of dCas9 to the KRAB repressor.47,48 The gene activation role of CRISPR, termed CRISPR activation (CRISPRa), also involves dCas9 but in combination with the activator proteins such as VP64.39,49–51 The nucleic acid detection capacity of CRISPR/Cas12 and Cas13 systems based on SHERLOCK is being weighed for diagnostic and biosensor applications.52–54 With the evident advances in the CRISPR/Cas systems, the tailored genetic manipulation of most mammalian cell types is possible, and the efficacy of these systems is also being improved. Further, the presence of CRISPR systems in different species of bacteria provided the extensive functional diversity of genome editing.27,28,48 Despite the promising advances and improved functions of CRISPR/Cas systems, more research is essential for appropriately applying these systems to execute the required outcomes without harmful perturbations to the human body. Fig. 1 shows the schematic of the working principles of CRISPR/Cas tools for genetic manipulation.
2.2 Deliverable forms of CRISPR/Cas systems
CRISPR/Cas systems are delivered in multiple forms into the cells for genetic manipulation.55,56 Commonly, three delivery methods for the CRISPR/Cas system are implemented: plasmid DNA (pDNA) encoding the Cas enzyme and sgRNA, the RNA system with Cas mRNA and sgRNA, and the ribonucleoprotein (RNP) system with Cas protein and sgRNA.14,57,58 The Cas9 enzyme has a molecular weight of 160 kDa with a 7.5 nm diameter and a positive surface charge, making it difficult to deliver into the living systems.59,60 CRISPR pDNA transfection stands out for its superior stability and cost-effectiveness compared to the mRNA and RNP systems.61 However, practical pDNA-mediated genome editing faces unresolved delivery issues of compression, nuclear placement, and off-target risks.26 In the delivery of CRISPR/Cas tools based on the RNA system, CRISPR/Cas delivery vehicles should be equipped to transport and protect the highly unstable RNAs of the Cas enzyme and its guide simultaneously, as well as control their release in the cytoplasm. Thus, the selection of the CRISPR delivery strategy and the optimization of its use have a significant impact on the efficiency of therapeutic gene editing. To satisfy the requirements of clinical gene therapy, the delivery vehicle should be able to stay in circulation for prolonged periods, increasing the chances of reaching the targeted site and improving delivery efficiency.622.3 Therapeutic applications of CRISPR/Cas systems
The broadly spanning toolbox of CRISPR/Cas systems has been tested for therapeutic gene editing in different genetic diseases with promising results. The significant examples of therapeutic genetic manipulation in various genetic disorders include sickle cell anemia,4,5 thalassemia,6 cystic fibrosis disease,7 cancer,3 hereditary tyrosinemia type I8 alpha-1 antitrypsin deficiency,9 blindness,11 Huntington's disease,41,63 Duchenne muscular dystrophy,64 AIDS,10 and hepatitis B.65 The therapeutic functioning of CRISPR/Cas systems in the aforementioned genetic diseases has been investigated in cell models or animal models. Further, the human organoid models are currently being assessed as preclinical tools in gauging the safety, efficacy, and accuracy of CRISPR-based therapeutic applications such as for cystic fibrosis disease in intestinal organoids.66 In 2016, the first clinical trial of CRISPR/Cas9 mediated PD1 knockout (KO) T cells for treating lung cancer was carried out in China to evaluate the safety and efficacy.67 The results reported CRISPR/Cas9-mediated gene-edited T cells as a safe and feasible clinical option for lung cancer patients.68 Currently, ClinicalTrials.gov has listed more than 74 trials (https://clinicaltrials.gov/; accessed on 08/01/2023), including CRISPR/Cas9 and Ca12a, for treating genetic diseases, mainly cancer, hematological issues, and other genetic disorders. Recently, two trials for investigating the clinical diagnostic potential of CRISPR/Cas12a for pneumonia have also been listed.3 Nanoparticle (NP)-based nanocomposites for CRISPR/Cas delivery
Different delivery approaches have been developed for therapeutic genetic modification based on the prodigious therapeutic prospects of CRISPR/Cas systems.69–72 The safe and targeted delivery with sustained release (in some cases) of the CRISPR/Cas tools to the site of interest requires the carrying vehicle to deliver the cargo and protect it against enzymatic degradation and metabolic clearance. Therefore, the ideal advanced delivery systems should be equipped with efficient delivery in addition to their biocompatibility and biodegradation properties. With the convergence of biomaterials and nanotechnology, many attempts have been made to develop multifunctional nanoparticle-based nanocomposites for safe and feasible CRISPR/Cas delivery. This section will highlight the recent nanoparticle-based delivery systems, including lipid nanoparticles (LNPs), polymer-based nanoparticles, and hybrid nanoparticles, for targeted delivery of CRISPR/Cas tools and their therapeutic applications.3.1. Lipid nanoparticles for CRISPR/Cas delivery
Lipid-based nanoparticles for delivering the genetic scissors are made up of physiological lipids. Despite their biocompatibility and biodegradability, the traditionally used liposomes are not widely applicable for CRISPR/Cas systems due to their poor stability, low encapsulation efficiency, and short shelf life.73 The enormous success of mRNA vaccine technology in the fight against the COVID-19 pandemic has recently brought more attention to LNPs.74,75 LNPs are being widely developed and tested for delivering genetic materials to delineate their safety and efficacy to be considered as clinical delivery vehicles. To this end, the cationic carriers, DOTAP, DOPE, and cholesterol, are commonly used for shipping the negatively charged genetic material cargo after electrostatic interaction with the vehicle. Ionizable lipids are also used to carry negatively charged nucleic acids. LNPs containing ionizable lipids are typically used with conventional gears, such as cholesterol, phospholipid, and PEG derivatives, for particle, structural, and physiological stability, respectively.76,77Recently, Qiu et al. created an LNP-based method for delivering CRISPR/Cas9 and gRNA for Angptl3 in mouse hepatocytes.80 A single injection of LNPs containing Cas9 mRNA and Angptl3 gRNA effectively edited the Angptl3 gene in mice, lowering low-density lipoprotein (LDL) cholesterol by 57% and triglycerides by 29% for more than 100 days.80 Mutations in the Tymp gene encoding thymidine phosphorylase lead to mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). The LNPs delivered CRISPR/Cas9 and a Tymp cDNA to transport and activate a Tymp transgene into hepatocyte introns in a mouse model of MNGIE. The treated mice demonstrated a long-term (1-year) drop in plasma nucleoside levels associated with Tymp mRNA and effective enzymes in liver cells.81 Another recent study reported LNP-assisted delivery of the CRISPR base editor, ABE8.8, in cynomolgus monkeys as a preclinical demonstration. A single infusion of LNPs encapsulating ABE8.8 mRNA and sgPcsk9 achieved almost absolute knockdown (KD) of Pcsk9 in the liver with consequent reductions in PCSK9 and LDL cholesterol by about 90% and 60%, respectively. The persistence of the results for eight months after a single-dose treatment indicates the advancement of this approach towards clinical application.82 A biodegradable LNP (LNP INT01) delivered Cas9 mRNA along with end-modified sgRNA and entirely modified sgRNA targeting transthyretin (Ttr) to improve gene editing in vivo in mice. For 12 months after a single IV injection of LNP INT01, >97% of Ttr gene editing was achieved, decreasing the levels of the TTR serum protein by ∼70%.77 In 2020, the NTLA-2001 carrier system based on LNP, Cas9 mRNA, and sgTTR was formulated in an aqueous buffer for IV administration to demonstrate the clinical application.83 For assessing the targeting efficiency, NTLA-2001 underwent preclinical development that included simulation modeling and in vitro studies. NTLA-2001 was particularly potent and produced saturating levels of TTR editing (93.7%) in primary human hepatocytes, resulting in 91% drop in TTR mRNA and 95% drop in TTR protein levels. Preclinical investigations in mice and monkeys revealed that a single dose of NTLA-2001 or its substitute Cyn-LNP resulted in long-lasting TTR editing and near-complete eradication of blood TTR protein expression with no side effects.83 The LNP INT01 possessed the transient Ttr editing ability and was only tested in mouse and rat models. In contrast, the NTLA-2001 system for TTR gene editing was tested in human cell models, animal models, and Phase I clinical trials.
The advances in LNPs targeting liver cells for delivering CRISPR/Cas9 payloads have been effectively demonstrated in in vitro and in vivo preclinical models. To improve the delivery efficiency and gene therapy, the modification of LNPs and the delivery of different CRISPR systems have been tested. A significant advancement in this regard is the clinical trial of NTLA-2001 for TTR gene editing in liver cells. Systemic CRISPR/Cas delivery to non-liver organs remained a challenge until the recent discovery of LNPs with such delivery potential. Sago and colleagues developed a high throughput platform named FIND to discover LNPs. They found two LNPs, 7C2 and 7C3, for in vivo mRNA delivery to endothelial cell types in different organs. 7C3 efficiently transported RNAs of Cas9 and its guide to splenic endothelial cells, unlike LNPs, which target hepatocytes.84 In another recent study, Wei et al. created a modified LNP with a cationic lipid.85 The modified LNP delivered the CRISPR RNP complexes to specified organs like the lungs and liver of mice following IV injection (Fig. 5B). Furthermore, the tailored LNP transported Cas9 RNPs into the skeletal muscle of the MDX mouse model, restoring dystrophin expression.85 Mohanna et al. recently reported the delivery of LNP-RNPs for genetic correction in the small eye variant of the Pax6 gene in an aniridia mouse model. The ex vivo results demonstrated the successful modification of mutation in 17% of mouse cortical neurons; however, the mutation correction was not detected in corneal cells when LNP-RNPs were injected in the mouse model's cornea. However the LNPs delivered the RNPs to the target site highlighting the potential of this delivery approach for therapeutic gene editing related to genetic disorders of the eye.86
Despite the progress in delivering the genetic scissors to non-liver organs through LNPs, their targeted delivery to the specific site of genetic engineering is a big challenge following the systemic administration of LNPs. Recent research on LNP-CRISPR systems has mainly shifted towards selective organ targeting for target-specific genome editing to meet this challenge. Cheng et al.87 developed a selective organ targeting (SORT) technique in which an additional lipid, a SORT molecule, was placed on the surface of conventional LNPs to increase tissue targetability. The SORT-LNPs selectively target several organs, including epithelial cells, endothelial cells, immune cells, B cells, and T cells, in the liver, lungs, and spleen (Fig. 2A). As an example of gene editing, SORT-LNPs delivered Cas9 mRNA and sgPten or Cas9 protein/sgPten in mice upon injection leading to the successful Pten gene editing in liver and lung cells.87 Liu and colleagues synthesized multi-tailed ionizable phospholipids (iPhos) for in vivo gene editing in selective organs. iPhos synergized with helper lipids to form iPhos LNPs (iPLNPs) for specific organ targeting enabling tissue-selective gene editing through CRISPR/Cas delivery.88 Dual delivery of Cas9 mRNA and Tom1 sgRNA loaded in 9A1P9-5A2-SC8 iPLNPs specifically targeted the liver cells in mice upon IV injection. On the other hand, co-delivery of the same mRNA CRISPR system loaded in 9A1P9-DDAB iPLNPs targeted lung cells for gene editing. Similarly, Cas9 mRNA/sgPten co-delivery in mice by 9A1P9-5A2-SC8 and 9A1P9-DDAB iPLNPs presented efficient and targeted genetic manipulation in the liver and lungs, respectively.88
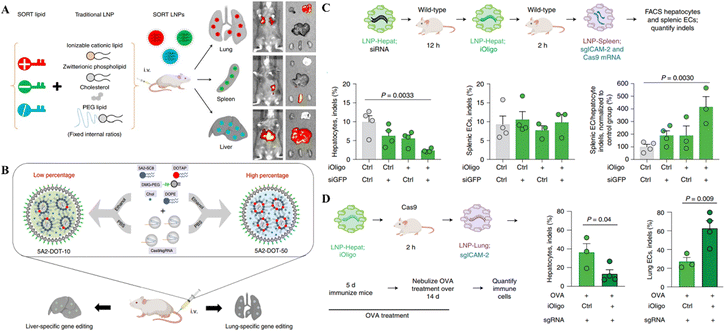 | ||
| Fig. 2 Targeted organ-specific CRISPR delivery through LNPs. (A) The schematic representation of the composition of SORT LNPs and their in vivo biodistribution. Reproduced with permission from (Cheng et al. 2020).87 (B) Graphical representation of particular site cargo delivery of CRISPR/Cas RNPs by chemically tailored LNPs for tissue-specific gene editing. Reproduced from (Wei et al. 2020).85 (C) Spleen-specific gene editing in mice through LNPs-Spleen. (D) Liver-specific gene editing in mice with LNP-Hepat. Reproduced with permission from (Sago et al. 2022).89 | ||
A recent study reported that the systemic administration of oligonucleotide blocking ICAM-2 sgRNA in SpCas9 mice bearing acute inflammation in the lungs reduced ICAM-2 indels in hepatocytes and enhanced them in lung endothelial cells. In wild-type animals, LNP-mediated transfer of a repressing oligonucleotide, trailed by Cas9-disrupting siRNA, reduced ICAM-2 genetic mutations in hepatocytes excluding splenic endothelial cells (Fig. 2C and D). This study demonstrated the tissue specific gene editing method following the delivery of CRISPR.89 Compared to the LNPs developed by Cheng et al. and Liu et al. for selective organ targeting, this study reported using siRNAs for selective gene editing in different organs instead of modifying the LNP composition as mentioned in the above two studies. This approach provides an additional way of targeting selective organs for genetic manipulation.
Overall, the selective organ delivery of CRISPR systems has been explored for assessing the effectiveness and safety of therapeutic gene editing in particular organs for targeted gene therapy. The selective delivery of CRISPR payloads in the liver, spleen, lungs, and tumor tissues has been accomplished hitherto.
The therapeutic potential of 5A2-DOT-X for in vivo gene editing through CRISPR/Cas was tested in two therapeutic animal models by Wei et al. Three times per week injections of 5A2-DOT-10 LNPs carrying Cas9/sgDmd RNPs into the anterior tibialis muscles of the Ex44 DMD mouse model led to the refurbishment of dystrophin expression. Similarly, the tail vein injection of 5A2-DOT-5 LNPs containing Cas9/sgPcsk9 RNPs in mice lowered the PCSK9 protein levels leading to the reduced level of harmful lipids associated with cardiovascular disorders. These two applications of 5A2-DOT-X LNPs have established the therapeutic efficacy of the LNP-CRISPR system for treating genetic illnesses.85 Another recent study tested the therapeutic application of SORT LNPs, resulting in 100% decline in PCSK9 protein expression in the liver and serum samples of treated mice delineating the strong clinical application potential of this system.87 Loss-of-function mutations in Angptl3 reduced lipid levels in the blood, highlighting it as a promising therapeutic target for lipoprotein metabolism disorders. The LNP delivery platform developed by Qiu et al. for CRISPR/Cas9-based KO of Angptl3 in the hepatic tissue of wild-type C57BL/6 mice reduced the levels of serum ANGPTL3, LDL cholesterol, and triglycerides (Fig. 3A and B). These therapeutic events lasted for 100 days following the single-dose treatment, and the efficiency of this LNP platform was higher in comparison to the FDA-approved MC-3 LNP. This research provides proof of concept for the therapeutic application of this platform in patients with lipoprotein disorders and atherosclerotic cardiovascular disease.80 In the clinical trial of LNP-CRISPR, a phase 1 clinical trial (ClinicalTrials.gov Identifier: NCT04601051) conducted by Intellia Therapeutics, NTLA-2001 led to a considerable decrease, 87%, of serum TTR protein content following the administration of a single dosage (0.30 mg kg−1) in patients of transthyretin amyloidosis, with no significant hostile effects noted. In six individuals of hATTR amyloidosis with polyneuropathy, systemic treatment of NTLA-2001 was linked with a sustained decrease in serum TTR protein concentrations. After 24 months of NTLA-2001 infusion, safety and therapeutic activity outcomes will be assessed.83 These results reveal the considerable potential of LNPs for conveying CRISPR/Cas9 to the human liver for therapeutic genetic manipulation. The targetable organs and genetic illnesses are anticipated to increase if LNP-CRISPR systems are improved further.
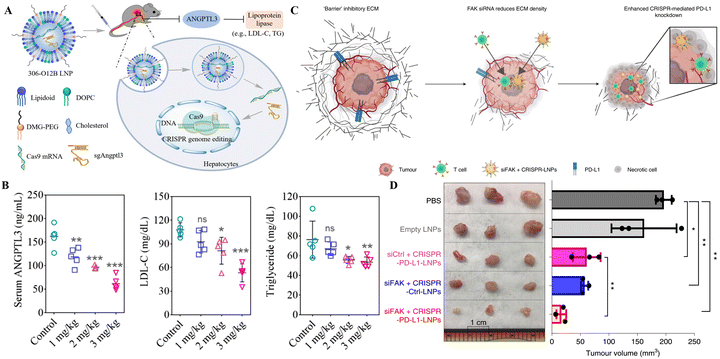 | ||
| Fig. 3 Therapeutic gene editing through LNP-based targeted delivery of CRISPR tools. (A) Schematic representation of LNP-based delivery of CRISPR/Cas9 in mice to drive loss-of-function mutations in Angptl3 to reduce lipid levels in the blood. (B) Serum ANGPTL3, LDL, and triglyceride levels in mice after 100 of LNP-CRISPR delivery. Adapted from (Qiu et al. 2021).80 Copyright (2021) National Academy of Sciences. (C) Schematic of addressing tumor mechanics to unlock a double-checkpoint cancer blockage to promote cancer therapy. (D) The excised tumor volume shows modified LNP-CRISPR's in vivo therapeutic efficiency. Reproduced with permission from (Zhang et al. 2022).93 | ||
Mutations in the kallikrein B1 (KLKB1) gene cause hereditary angioedema (HAE), characterized by swelling of the face, hands, and airways, which can be fatal in some cases. Intellia Therapeutics has started the phase 2 clinical trial of LNP-based NTLA-2002 (ClinicalTrials.gov Identifier: NCT05120830) for CRISPR/Cas9-based therapy of HEA by targeting KLKB1. The preclinical assessment of NTLA-2002 in cynomolgus monkeys and a humanized KLKB mouse model demonstrated that a single injection of NTLA-2002 resulted in 70% gene editing in KLKB1 and >90% decrease in kallikrein protein levels.90 Further, the single dose IV administration of NTLA-2002 at 0.3 mg kg−1 in a partial hepatectomy (PHx) humanized KLKB1 mouse model resulted in 74% KLKB1 gene editing and a 91% decrease in serum kallikrein before PHx and 71% gene editing and 94% decrease in serum kallikrein after 14 months of liver regeneration. These additional results delineate the permanence of NTLA-2002-based CRISPR/Cas9-induced genetic editing supporting the single-dose treatment for HAE patients.91
The LNP-CRISPR systems are on the verge of clinical application as cancer therapy based on the demonstrations in preclinical models thus far. In a mouse model of glioblastoma, a single intracerebral injection of sgPLK1-cLNPs killed tumor cells, slowed tumor development by 50%, and improved survival by 30%. Additionally, IP injections of EGFR-targeted sgPLK1-cLNPs permitted up to 80% editing of PLK1 in vivo, reduced tumor formation, and increased survival by 80% in the mice, opening new avenues for cancer treatment, research, and genetic editing of non-malignant tissues.92 The combination of SiFAK + CRISPR-PD-L1-LNP, created by Zhang et al.,93 inhibited the spread of cancer and the formation of tumors in four mouse liver cancer models. In mice with MYC-driven hepatocellular carcinoma, treatment with siFAK plus CRISPR-PD-L1-LNPs reduced the expression of PD-L1 and increased the elasticity of the extracellular matrix (ECM), resulting in a significant extension of the animal's survival time (Fig. 3C and D). This recent example points out that using CRISPR gene editing in a multiplexed and generalizable strategy presents an innovative method for treating cancer.93
Inclusively, the LNPs for CRISPR/Cas delivery have recently entered the clinical trials at Phase I and II levels for therapeutic genetic manipulation. NTLA-2001 and NTLA-2002 undergoing clinical trials are used for genetic illnesses, while the cancer treatment options have also been studied with promising results through LNP-CRISPR/Cas delivery systems.
3.2 Polymer-based nanoparticles for CRISPR/Cas delivery
Polymers are widely employed in medication delivery into cells and animal models. The polymeric nanocarriers include dendrimers, micelles, nanoconjugates, nanocapsules, and polyplexes.94 Akin to liposomes, the polymeric vectors are ideal for gene transfer since they have amino groups to form electrostatic complexes with negatively charged nucleotides without toxicity issues. This section will shed light on the recent advances of polymer-based NPs for CRISPR/Cas delivery.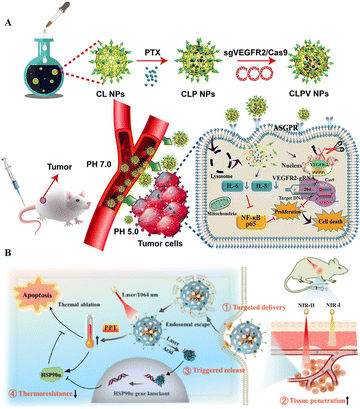 | ||
| Fig. 4 Polymeric NPs for targeted delivery of CRISPR/Cas tools. (A) Tumor targeted delivery of VEGFR2-gRNA/Cas9 in a mouse model. Adapted with permission from (Zhang et al. 2020).96 (B) Targeted delivery and anticancer gene-photothermal therapy by AP-RNP. Reproduced with permission from (Tao et al. 2022).98 | ||
The application of synthetic polymers like PEI and PC is less advisable due to their cytotoxic effects. Therefore, the application of natural polymers like chitosan, hyaluronic acid, and dextran with modifications is being investigated to safely deliver CRISPR/Cas systems. The polymer nanoparticle delivery systems allow the combined application of gene therapy and anticancer drugs or photothermal nano agents for PTT, increasing the targeted delivery chances and therapeutic potency of these systems.
The polymer-based NPs are mainly employed for tumor therapy and the tumor-targeted delivery of CRISPR/Cas tools. The combinatorial polymer-based NPs have been proven target specific, efficient, and safe given the coating of polymers or linking with target-specific peptides or linkers.
3.3 Hybrid nanoparticles for CRISPR/Cas delivery
To make the NPs more durable and biocompatible, lipids, particularly phospholipids, are used for coating, as these materials are unstable in aqueous solutions and can have harmful effects.100 Further, the formation of the hybrid nanocomposite with inorganic materials like calcium phosphate, carbon-based compounds, gold, silicon, and iron oxide has also been reported.101 The supramolecular formulations of these ions, in combination with CRISPR/Cas tools, are used as delivery vectors. For delivering the Cas9 and sgRNA plasmid into tumor cells, a functional lipid/gold nanocluster carrier was utilized to enhance the delivery efficiency. The surface modifications with DOPE, cholesterol and PEG are employed to improve cargo delivery efficacy.102 Coating NPs with cell membranes renders them biocompatible and increases the delivery efficiency of CRISPR/Cas tools.103 Recently designed metal-organic frameworks (MOFs) and metal-organic cages (MOCs) have also been used to deliver gene editing scissors.104 MOFs' high porosity and large capacity render them an auspicious tool for cellular CRISPR/Cas9 delivery.105,106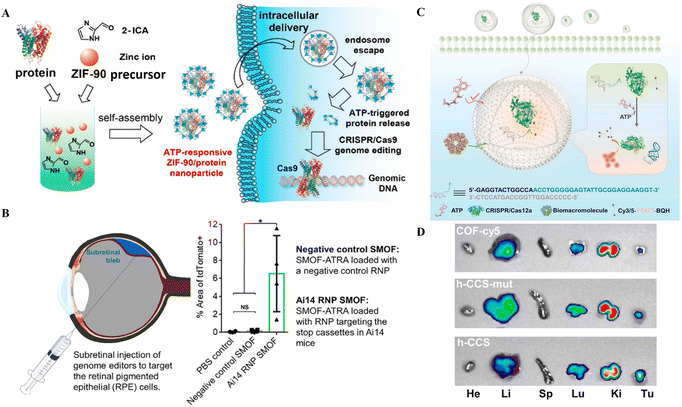 | ||
| Fig. 5 CRISPR/Cas delivery through hybrid NPs for therapeutic applications. (A) An illustration showing how ZIF-90 can self-assemble for intracellular transport and produce genome editing thereafter. Reproduced from (Yang et al. 2019).105 (B) Therapeutic application of the SMOF nanocomposite vehicle for CRISPR-based genome editing in retinal epithelial cells (Wang et al. 2020).108 (C) Schematic of the ATP-triggered CRISPR/aptamer-based nanosensor for ATP imaging. (D) Nanoreactor-based in vivo ATP imaging of different organs of tumor-bearing mice. Adapted with permission from (Pan et al. 2022).71 | ||
The ZIF-C approach by Poddar et al.107 used the combinatorial approach of RPSA manipulation at the RNA and DNA level to effectively treat PC. Further, the EGCG coating of NPs improved the delivery efficiency and acted as an antioxidant and anticancer agent. In contrast, the combination of silica with MOF allowed the addition of an ATRA targeting agent to the SMOF NPs, which helped in the RPE cell-targeted delivery of CRISPR/Cas9 RNPs. On the other hand, the coating of cell membranes onto NPs provides cell specificity and biomimetic properties. Overall, these studies provide the proof-of-concept for targeted delivery of CRISPR/Cas9 as well as other nucleotides and small molecule medicines through hybrid NPs.
The ZIF-C vehicles developed by Poddar et al.107 targeted PC resulting in retarded cell proliferation, hinting at the potential for PC treatment.107 ZIF-90, another type of ZIF formed through the self-assembly of zinc ions and 2-ICA, has a comparable porosity to ZIF-8 with the ability to deliver CRISPR/Cas9 RNPs into cells.113 ZIF-90, after entry, destroys ATP within the cell liberating fluorophores for real-time ATP imaging.113 ZIF-90's protein delivery and release efficacy are governed by ATP concentration in cells that is more in diseased cells (Fig. 5A). Thus, ATP-responsive ZIF-90 could be used for targeted gene therapy. More recently, Pan et al.71 reported a nanoreactor based on a covalent organic framework (COF) carrier packed with CRISPR/Cas12 and an aptamer-based nanosensor for ATP imaging. The hollow core of the COF encapsulated the nanoreactor based on CRISPR/Cas12, a fluorophore quencher and an ATP aptamer for in vivo imaging (Fig. 5C and D). The nanoreactor was suitable for ATP imaging in the cell and animal models.71
Zhang et al.114 developed a dual-locking nanoparticle (DLNP) capable of restricting the activation of CRISPR/Cas13a in tumor tissues. The polymer layer confers increased stability while circulating in the blood or when it is located in normal tissues. Additionally, it makes the endocytosis of the CRISPR/Cas13a system easier and activates gene editing when the DLNP is introduced into tumor tissue. In B16F10-bearing mice, DLNP demonstrates efficient activation of T cell-mediated antitumor immunity as well as immunosuppressive tumor microenvironment remodeling. This modification of the tumor microenvironment results in dramatically boosted and persistent antitumor effects.114
The hybrid combination of inorganic and polymeric nanomaterials improves the delivery of CRISPR/Cas tools. Additionally, the RNP-ZIF-90 NPs act as ATP-responsive agents for the intelligent delivery of CRISPR/Cas9 RNPs. More recently an ATP nanosensor based on a COF and an aptamer along with CRISPR/Cas12 was developed for ATP imaging. The RNP-ZIF-90 NPs used CRISPR/Cas9 for ATP-responsive genome editing in the target cells. In comparison, the ATP nanosensor based on the COF, aptamer, and CRISPR/Cas12 developed by Pan et al. can specifically be used for ATP imaging advocating the nanosensor potential of CRISPR/Cas tools for biomedical applications. The DLNP developed by Zhang et al.114 for delivering CRISPR/Cas13a for targeted tumor therapy also boosted the antitumor function of T-cells for effective cancer treatment. Altogether, the hybrid nanocomposites provide the additional capacity for linking, loading, and/or conjugating nucleotides, target specific molecules or small molecule medicines, and CRISPR/Cas tools for improved delivery and supporting genetic manipulation and treatment.
4 Nanostructure-based nanocomposites for delivery of CRISPR/Cas tools
The nanostructure-based nanocomposites are single or polycrystals with nanoscale grain size.115 The composition and size of nanostructures can vary depending on the type of nanostructure like extracellular vesicles, DNA nanostructures, and nanoneedles. Recently, the nanostructures have also been exploited to deliver CRISPR/Cas tools which will be discussed in this section.4.1 Extracellular vesicle-based nanostructures for delivery of CRISPR/Cas tools
Extracellular vesicles, often known as EVs, are diverse populations of membrane vesicles that are discharged from cells of various types. There are three distinct categories of EVs, namely, exosomes, microvesicles, and apoptotic bodies, characterized by 40 to 150 nm, 100–1000 nm, and 50 to 5000 nm size, respectively.116 These lipid bilayer membrane vesicles are oriented in the same topological fashion as the plasma membrane, act as cell-to-cell communication carriers, and are immune-privileged; therefore, they are rarely removed by the immune system and escape hypersensitive reactions.2,117 EVs appear to be capable of transporting their contents from one cell to another over extended distances.2 These properties make EVs suitable carrier vehicles for delivering CRISPR/Cas tools into the cell and animal models.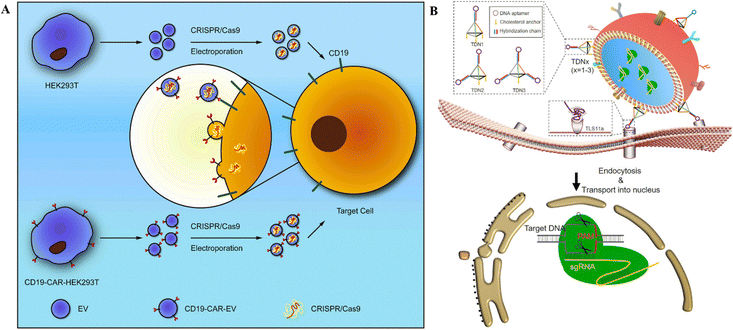 | ||
| Fig. 6 CRISPR/Cas RNPs’ delivery through EVs. (A) Schematic representation of anti-CD19-CAR-EV mediated cancer therapy. Electroporation loads MYC-targeting CRISPR/Cas9 into EVs. Anti-CD19-CAR-EVs boost the cargo's efficiency in CD19-positive tumor cells. Adapted with permission from (Xu et al. 2020).121 (B) Tumor targeted delivery of genetic scissors through TDNs. Reproduced with permission from (Zhuang et al. 2020).119 | ||
Burtkitt's lymphoma, diffused large cell lymphoma, multiple myeloma, and acute lymphocytic leukemia all have high levels of MYC overexpression.120 As a result, employing the CRISPR/Cas9 technology to target MYC could produce a favorable therapeutic effect in cancer treatment. Chimeric-antigen receptors (CAR) are used for tumor targeting.121 In this context, anti-CD19-CAR-EVs and unmodified EVs were electroporated with MYC-targeting sgRNA/Cas9 plasmids (Fig. 6A). Anti-CD19-CAR-EVs reduced CD19+ Raji and Nalm6 cell growth more than unmodified EVs. In mice with Raji xenografts, anti-CD19-CAR-EVs gathered in tumor areas, but unchanged EVs were identified in other tissues.121 Ye et al.122 created a modified exosome (M-CRISPR/Cas9 exosome) for improved encapsulation of CRISPR/Cas9 components. Researchers artificially generated exosomes by combining GFP and GFP nanobodies fused with CD63 and Cas9 proteins. Due to the selectivity of GFP-GFP nanobodies, Cas9 proteins can be precisely and effectively loaded into exosomes. Using A549stop-DsRed reporter cells, they found that CRISPR/Cas9-functionalized exosomes successfully suppressed the target gene in recipient cells.122
These studies highlight the targeted delivery of CRISPR/Cas tools after adding aptamers for target-specific binding. Moreover, the presence of CD proteins on the surface of EVs or exosomes stipulates the evasion of these delivery agents from the immune system and allows for long-term stay in circulation, increasing the chances for delivery to the target tissue. However, these studies only reported the encapsulation and delivery of CRISPR/Cas9 as RNPs rather than the mRNA form, given the unsuitability of SpCas9 mRNA encapsulation. All in all, the biomimetic, immune evasion, and target-specific delivery properties of EVs have added to the CRISPR/Cas delivery options for biomedical applications.
Gee et al. employed gene editing by developing an all-EV delivery system termed NanoMEDIC.125 HEK293T producer cells were lipofected with FKBP12 and FRB-Cas9 to produce EVs. After AP21967 treatment, EVs were introduced to HEK293T cells expressing a DMD1-targeting sgRNA. NanoMEDIC created DMD1 indels in up to 40% of human iPSCs. NanoMEDIC could target the dystrophin gene yielding 90% exon skipping in DMD patients’ skeletal muscle cells derived from the iPS cells (Fig. 7A and B). For in vivo testing on transgenic mice, the single IM injection of NanoMEDIC was associated with exon skipping in Mdx mice which lasted for 160 days.125 The NanoMEDIC system's stability is superior to existing in vivo genome editing technologies; DMD sufferers now have hope, thanks to the researchers for this development. HEK293-Exo and SKOV3-Exo were isolated by Kim et al.126 from HEK293 and SKOV3 cells, respectively. Electroporation was used for packaging exosomes with sgRNA/Cas9 targeting PARP-1. With cancer-derived exosomes, CRISPR/Cas9 inhibited PARP-1 in SKOV3 cells, and SKOV3-Exo induced indels with 27% efficacy in the PARP-1 gene. In mice, SKOV3-Exo injections (intravenous or intratumoral) reduced tumor weight and volume. The combined loading of cisplatin and CRISPR/Cas9 in SKOV3-Exo inhibited SKOV3 cell proliferation by 57% in vitro. The inhibition of PARP-1 sensitized the cancer cells to cisplatin, promoting its anticancer activity. In vivo, tagged exosomes were injected into the tail veins of SKOV3 xenograft mice, resulting in SKOV3-Exo accumulation at the tumor sites working as an anticancer agent (Fig. 7C and D).126
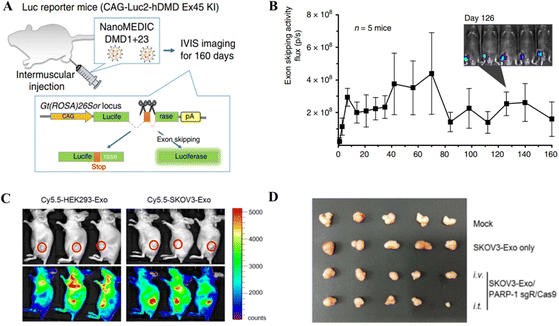 | ||
| Fig. 7 In vivo gene editing through EV-based CRISPR/Cas delivery. (A) Dystrophin gene editing in mice after IM injection of NanoMEDIC. (B) The exon skipping in mice treated with NanoMEDIC injection. Reproduced from (Gee et al. 2020).125 (C) Biodistribution of the exosome-mediated CRISPR nanocarrier targeting tumor cells in mice. (D) Effect of SKOV3-Exo-based delivery of CRISPR/Cas9 for gene editing a xenografted tumor in mice. Reproduced with permission from (Kim et al. 2017).126 | ||
The use of epithelial cells and cancer cells for the production of EVs encapsulating CRISPR/Cas tools diversifies the delivery function of EVs. In the case of GEDEX and NanoMEDIC, the EVs were produced from the epithelial cells with different applications depending upon the encapsulated CRISPR/Cas components. The GEDEX system contained dCas9, which was involved in HGF overexpression, while the NanoMEDIC system worked through indel generation for exon skipping in the target gene. In contrast, the SKOV3-Exo system developed by Kim et al. can be used explicitly for cancer treatment, given the dual function of genetic manipulation and chemotherapy. The encapsulation of CRISPR/Cas tools in EVs and exosomes added to the delivery options for therapeutic genetic manipulation with promising results.
4.2 DNA nanostructure-based delivery of CRISPR/Cas tools
DNA macromolecules are biocompatible and traceable; therefore, their use in nanomedicine is growing.127 DNA nanostructures serve as customizable CRISPR delivery systems protecting, targeting, and releasing the cargo.128 Recently, the use of DNA nanotechnology to deliver gene-editing tools has gained attention and is being widely researched for establishing an efficient and safe CRISPR/Cas delivery system. The recent developments in targeted delivery and therapeutic potential of DNA nanostructures as CRISPR/Cas delivery systems will be discussed in this section.Another recent study reported the DNA nano clews as a CRISPR/Cas delivery system. The sgRNA/Cas9 complexes were loaded in the nano clews, and loaded clews were coated with PEI for endosomal carrier escape. Besides, with the advancement of DNA nanotechnology, DNA nano clews are reported coated with polymers that are attached to positively charged carriers, which are released when the pH changes from physiological to acidic.130In vitro delivery of a Cas12a/CRISPR RNP system linked with decreased cholesterol blood levels in mice was achieved. Reduced PCSK9 expression was connected to PCSK9 disruption (48%) and lower cholesterol levels (45%).130 Ding et al.131 developed a similar delivery technique using a DNA-based nanogel. First, DNA-grafted polycaprolactone brushes (DNA-g-PCL) were loaded with sgRNA/Cas9 complexes and crosslinked using DNA linkers (Fig. 8A and B). The non-cationic nanogel protected sgRNA/Cas9 complexes from nuclease digestion and allowed for progressive release. The in vitro demonstration showed the knockdown of EGFP in HeLa cells after nanogel/Ca9/sgRNA delivery, demonstrating its targeted gene editing efficiency.131 Further research established that these nanogels could even be layered with PEI to enhance endosomal escape, similar to the nano clew coating with PEI. Poly-A mRNA tails hybridized with poly-T nanogel (poly-T20-grafted polycaprolactone) segments (T20-g-PCL). Packing and conveying Cas9-mRNA and EGFP-mRNA cargo confirmed their functionality in HeLa and HEK-293 cells.132 However, the efficacy of targeted gene editing after cargo delivery of CRISPR/Cas systems through nanogels in these two studies has only been demonstrated in vitro. Thus, the in vivo administration of these systems for clinical gene therapy is awaited. Wu et al.133 reported a broad technique for constructing a dsDNA-RNP fused nanostructure by folding dsDNA with a CRISPR/dCas9/12a system. They fused dCas9 and dCas12a via a stimuli-responsive peptide linker. Covalently bivalent dCas9-12a RNPs (staples) activated by gRNAs may recognize target sequences in the dsDNA scaffold and bind them to generate DNA-RNP hybrid nanostructures (Fig. 8C and D). The biologically derived hybrid nanostructure may shield genetic information when folded as well as induce stimuli-responsive gene transcription. This DNA folding and unfolding method enhances DNA nanotechnology for delivering the CRISPR system.133
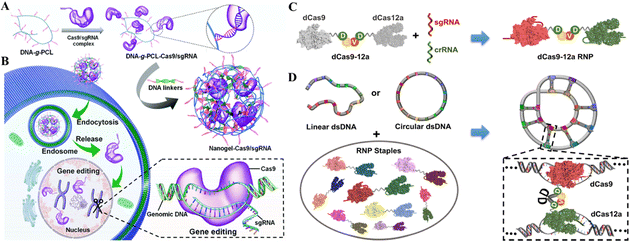 | ||
| Fig. 8 DNA nanostructures for CRISPR/Cas delivery. (A) Design of the DNA nanogel-based CRISPR carrier. (B) Gene editing mechanism after cell internalization of the nanocarrier. Adapted with permission from (Ding et al. 2019).131 (C) Fabrication of a dsDNA-RNP nanostructure using fused dCas9 and dCas12a. (D) Bivalent dCas9-12a RNP forms a highly condensed DNA-RNP hybrid nanostructure. Adapted with permission from (Wu et al. 2022).133 Copyright (2022). American Chemical Society. | ||
The targeted CRISPR/Cas delivery through DNA nanostructures is still in the early stages; therefore, their limited in vivo usage has been reported. However, the results of studies thus far indicate their efficacy to be considered as the clinical delivery vehicle for CRISPR/Cas delivery. On the other hand, the co-delivery of dCas9 and dCas12a allowed for multiplexed genetic manipulation, which stands out among the other CRISPR delivery systems.
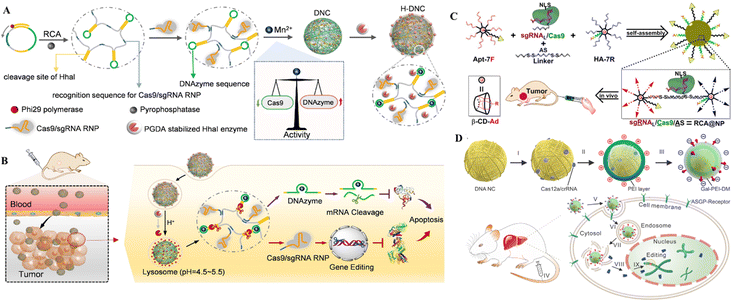 | ||
| Fig. 9 DNA nanostructures for delivering CRISPR/Cas9. (A) Formulation of DNCs. (B) In vivo gene editing in murine tumor cells. Adapted with permission from (Li et al. 2022).134 (C) DNA-based nanoplatforms to distribute sgRNA, Cas9, and antisense for combined tumor treatment. Adapted with permission from (Liu et al. 2019).129 Copyright 2019 American Chemical Society. (D) Charge-reversible DNA nano clews carrying Cas12a/crRNA wrapped in the Gal-PEI-DM polymer for in vivo CRISPR delivery. Adapted with permission from (Sun et al. 2020).130 | ||
The therapeutic genetic manipulation through CRISPR delivery using DNA nanostructures has not been extensively studied. A few studies have reported the preclinical demonstration in animal models, with a particular example of DNCs for treating breast cancer.
4.3 Nanoneedle-assisted delivery of CRISPR/Cas systems
Nanoneedles are vertical nanostructures with a high aspect ratio that may accurately influence complex interactions at the cell interface, allowing for active intracellular administration. This nanotechnological innovation has already permitted the establishment of effectual intracellular channels for delivering cell-impermeant payloads. A variety of one-dimensional (vertically arranged) nanostructures can be used as nanoneedles within these categories: nanowires,135,136 nanopillars,137,138 nanocones,139,140 nanostraws,141 and nanotubes.142Many ways of mechanically perforating cell membranes and delivering molecular payloads and Cas9 RNPs have been reported in nanoneedles and nanowires. Mechanistically, the nanoneedles are laid on the top of cells, and then the cells are centrifuged at a specified speed for a brief duration to poke the cells with nanoneedles, followed by removal of nanoneedles to allow the delivery of cargo into the cells. The centrifugation speed, time, acceleration and deceleration are critical for cell membrane penetration in these approaches. In a study by Kawamura et al., it was discovered that inserting 200 nm nanoneedles at a rate of 10 mm s−1 significantly increased the detection efficiency of cellular proteins due to the insertion efficacy.143 Because there is no substantial change beyond 10 mm s−1, a needle with a diameter of 200 nm should have a velocity of 10 mm s−1 or greater. They used silicon microfabrication technology to create monolithic nanoneedle arrays with 200 nm diameter and 40 mm length and nanoneedles spaced 10 mm from each other for molecular delivery and mechanically separating the cells targeting intracellular antigens. The introduction of Cas9 RNPs using this nanoneedle array resulted in a target gene KO efficacy of around 16 percent in mouse mammary cancer cells.144
Cell membrane penetration is effectively decreased if the nanoneedle array is submerged in cell-seeded media and permitted to come into contact with the cells via sedimentation. Chen et al.142 employed arrays of silicon nanotubes (300 nm inner tube diameter, 2 μm height, and 0.16 NT m−2 density) to administer Cas9 RNPs to murine fibroblasts, with a 6 percent indel efficiency for Hprt gene disruption. This could be because the needle-like substance stimulates membrane invagination and endocytosis when it touches the cells.145 Endocytosis is the mechanism behind the needles' sluggish approach to the cells. GFP-expressing B16F10 mouse melanoma cells were successfully wiped off by a gold nanowire driven by ultrasound to transduce Cas9 RNPs.146 According to this finding, the physical driving force imparted on nanoneedles appears critical for transmembrane transmission. In another study, silicon nanoblade chips (NB-chip) and the Cas9 RNP complex were integrated into a two-dimensional array of tens of thousands of nanoneedles to deliver Cas9-sgRNA into human cells (Fig. 10).144 FP10SC2 mouse breast cancer cells were used to prove the approach of delivering Cas9-sgRNA via a nanoneedle array, which knocked out nestin in FP10SC2 mouse breast cancer cells. This method's gene disruption efficiency in HeLa and murine mammary cancer cells proves its ex vivo gene-editing efficacy. Neuronal cells in 1 mm-thick brain tissues are too deep for typical nanoneedles (10 nm) to reach. Therefore Nomano et al. used nanoscale-tipped wire arrays with diameters of 100 nm and wire lengths of 200 nm to deliver biomolecules in slices of brain tissue. The nanotip arrays interact with cells in deep brain tissue to deliver FP expression vectors to target cells for electrophysiological monitoring and neuronal transfection. Pressing a nanotube wire array distributes plasmid DNA in multiple-neuronal cells in a brain slice with minor cell damage. A CRISPR/Cas9 genome-editing vector targeting Cetn1 can genetically modify the role of living neuronal cells in the brain and the pacemaker cells of mammalian circadian rhythms.147
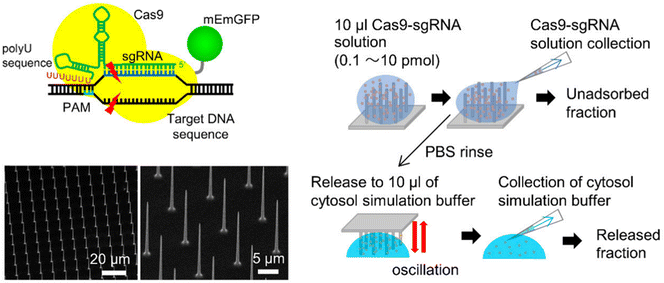 | ||
| Fig. 10 Gene editing through the use of mechanically responsive mechanisms. The mophology of nanoneedle array and the schematic of RNP loading and releasing mechanism from the nanoneedles. Adapted from (Yamagishi et al. 2019).144 © 2019 MDPI. | ||
The application of nanoneedles for CRISPR delivery has mainly been studied in vitro in the cell models, given their unsuitability for in vivo delivery in circulation owing to their structural configuration and coated CRISPR/Cas tools delivery method in the target cells. However, the nanoneedles can potentially deliver the CRISPR/Cas systems on the surface cells, such as skin and brain cells, after surgery.
5 Conclusion and outlook
Since their discovery, the CRISPR/Cas tools have been diversely weighed for therapeutic applications. CRISPR technology has already begun to answer many fundamental and complex concerns about gene therapy in numerous diseases. Albeit off-target activity and genome damage are potential challenges for CRISPR applications, the methods and technologies to assess and eliminate them are available and likely would not limit its clinical usage. An example of reduced off-target activity is the CRISPR/Cas12b system, which could be more effective for human genome editing than the CRISPR/Cas9 systems. The other likelihood to reduce off-target activity is the RNP system, which reduces the time limit for Cas enzyme activity and the chances of genome insertion. The application of CRISPR/Cas systems relies on safe, targeted, and efficient delivery. Despite being highly effective for transfection, the viral delivery vector is labor-intensive and expensive, and has repercussions to be used as the clinical CRISPR delivery system. Therefore, developing secure and reliable delivery systems presents a possible solution. In this regard, the recent advances in nanocomposite delivery systems, including nanoparticles and nanostructures, have proven effective for the targeted delivery of CRISPR/Cas systems and their respective therapeutic applications. However, further modification of these delivery systems is a requisite for enhanced cellular uptake, target site-specific delivery through blood vessel permeability, and addressing the major limitation of tissue penetration into deeper regions of different organs. The nanoneedle arrays with long-wired shapes could be the candidates for delivery into the deeper tissues of the body.147Comparatively, the wide variety of NPs and the extensive background research related to NPs and their drug or nucleotide delivery bids have made them more suitable candidates for CRISPR/Cas delivery. Further, the flexibility for chemical modification, composite formation capacity, and conjugation of target-specific linkers allow the effective loading and target-specific delivery of CRISPR/Cas. To this end, the chemical modification of LNPs rendered the delivery system more efficient and organ selective.87 The selective organ targeting for CRISPR/Cas delivery is mainly limited to organs like the liver, spleen, kidney, and lungs. The development of nanocomposite delivery systems to deliver CRISPR/Cas systems to other organs would increase the repertoire of therapeutic applications of CRISPR/Cas tools. In comparison, the delivery of CRISPR/Cas systems through nanostructures is still in its infancy, with some progress in recent years. The nanostructures have limitations, including maintaining conditions, physical/chemical parameter balancing, and diffusion issues, limiting the satisfactory CRISPR/Cas delivery through nanostructures. The delivery of CRISPR/Cas tools through nanostructures is mainly performed in the RNP form, while the NPs have been reported to deliver CRISPR/Cas tools through pDNA, mRNA, and RNP. Overall, the wide variety of nanocomposites, NPs, and nanostructures have been extrapolated recently for developing a safe and efficient CRISPR/Cas delivery system for clinical purposes.
The advanced CRISPR/Cas tools, such as the recently reported subtype of Cas12, CasΦ enzyme, Cas12f, with a molecular weight half that of Cas9 and Cas12a, would be easy to package and deliver.33–35 Similarly, the Cas14 enzyme, found in the archaea, capable of ssDNA cleavage in the target region, adds to easily deliverable CRISPR tools due to its compact size.34 Abbott et al. recently found CRISPR/Cas13-based therapy, PACMAN, against SARS-CoV-2 utilizing Ruminococcus flavefaciens Cas13d. PACMAN destroys SARS-CoV-2, disrupts IAV genomic sequences in human lung epithelial cells, and prevents viral replication. The authors claimed that PACMAN could be a pan-coronavirus inhibitory method to manage SARS-CoV-2 and its previous variants in animals and people.148 The nanocomposite-based delivery of PACMAN to the lungs could be its therapeutic application after developing such a delivery system.
The use of 3D cell models, primarily human tissue-derived organoids, has not been tested much for the delivery of CRISPR/Cas tools through nanocomposites. However, the organoids provide better preclinical modeling for their capacity to mimic patients' genetic setup and experiment with mutation correction through nanocomposite-based delivery of CRISPR/Cas tools.66,119 Alternatively, the nanocomposite-based delivery of CRISPR/Cas tools into the organoids from iPS cells or normal donor tissues can be used for modeling cancer and other genetic diseases for unraveling the underlying molecular mechanisms of genetic diseases or drug development.
Overall, the convergence of nanotechnology and biomedicine for CRISPR/Cas delivery has gained popularity and is being assessed for developing clinical-grade delivery systems. The LNPs and polymeric NPs, in addition to the nanostructure nanocomposites, are promising candidates winning the race towards clinical application as CRISPR delivery systems. The recent developments in CRISPR/Cas systems and delivery systems indicate that CRISPR will enter clinical practice in the near future.
Abbreviations
| ABE8.8 | Adenine base editor 8.8-m |
| ABP | Aptamer binding protein |
| Actc1 | Actin alpha cardiac muscle 1 |
| AIDS | Acquired immunodeficiency syndrome |
| Angptl3 | Angiopoietin-like 3 |
| ASGPR | Asialoglycoprotein receptor |
| ATP | Adenosine triphosphate |
| ATRA | All-retinoic acid |
| BE | Base editing |
| BP | Black phosphorus |
| CAR | Chimeric antigen receptor |
| Cas | CRISPR-associated |
| CD19 | CD19 molecule |
| CD63 | CD63 molecule |
| Cetn1 | Centrin 1 |
| COF | Covalent organic framework |
| COVID-19 | Coronavirus disease 2019 |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CRISPRa | CRISPR activation |
| CRISPRi | CRISPR interference |
| CjCas9 | Campylobacter jejuni Cas9 |
| crRNA | CRISPR RNA |
| CS | Chitosan |
| dCas | Dead Cas |
| DLNP | Dual-locking nanoparticle |
| DMD | Dystrophin muscular dystrophy |
| DNA-g-PCL | DNA-grafted polycaprolactone brushes |
| DOTAP | 1,2-Dioleoyl-3-trimethylammoniumpropane |
| DOPE | Dioleoylphosphatidylethanolamine |
| DSB | Double-strand break |
| ECM | Extracellular matrix |
| EGCG | Epigallocatechin gallate |
| EGFP | Enhanced green fluorescent protein |
| EGFR | Epidermal growth factor receptor |
| EV | Extracellular vesicle |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| FKBP12 | FKBP prolyl isomerase 12 |
| GEDEX | Genome editing with designed extracellular vesicles |
| GFP | Green fluorescent protein |
| GO | Graphene oxide |
| gRNA | Guide RNA |
| H7N9 | Influenza A virus subtype H7N9 |
| HAE | Hereditary angioedema |
| hATTR | Hereditary transthyretin-mediated |
| HCC | Hepatocellular carcinoma |
| HDR | Homology-directed repair |
| HEK293 cells | Human embryonic kidney 293 cells |
| HGF | Hepatocyte growth factor |
| HNF4a | Hepatocyte nuclear factor 4 alpha |
| Hprt | Hypoxanthine guanine phosphoribosyl transferase |
| HSCs | Hepatic stellate cells |
| HuR | Human antigen R |
| iPhos | Ionizable phospholipids |
| iPLNPs | iPhos NLPs |
| iPSCs | Induced pluripotent stem cells |
| ICAM-2 | Intercellular adhesion molecule 2 |
| Indels | Insertions and deletions |
| IP | Intraperitoneal |
| IV | Intravenous |
| KCNJ13 | Potassium inwardly rectifying channel subfamily J member 13 |
| KD | Knock down |
| KLB1 | kallikrein B1 |
| KO | Knockout |
| LCA16 | Leber congenital amaurosis |
| LDL | Low density lipoprotein |
| LNPs | Lipid nanoparticles |
| MDX | Duchenne muscular dystrophy |
| MMEJ | Microhomologymediated |
| MNGIE | Mitochondrial neurogastrointestinal encephalomyopathy |
| MOCs | Metal–organic cages |
| MOFs | Metal–organic frameworks |
| MYC | MYC proto-oncogene bHLH transcription factor |
| NHEJ | Non-homologous end joining |
| NmeCas9 | Neisseria meningitidis Cas9 |
| NP | Nanoparticle |
| PAM | Protospacer adjacent motif |
| PARP-1 | Poly(ADP-ribose) polymerase 1 |
| Pax6 | Paired box protein |
| PC | Prostate cancer |
| PD1 | Programmed cell death protein 1 |
| PE | Prime editing |
| pDNA | Plasmid DNA |
| PEG | Polyethylene glycol |
| PEI | Polyethyleneimine |
| pegRNA | PE gRNA |
| PHx | Partial hepatectomy |
| PLK1 | Polo like kinase 1 |
| Psck9 | Proprotein convertase subtilisin-like/kexin type 9 |
| Pten | Phosphatase and tensin homolog |
| PTT | Photothermal therapy |
| RCA@NP | sgRNA/Cas9/antisense-nanoparticle |
| RNP | Ribonucleoprotein |
| RPE | Retinal pigmented epithelium |
| RPSA | Ribosomal protein SA |
| SaCas9 | Staphylococcus aureus Cas9 |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SHERLOCK | Specific high-sensitivity enzymatic reporter unlocking |
| SMOF | Ailicon-MOF |
| SORT | Selective organ targeting |
| ssDNA | Single-stranded DNA |
| SpCas9 | Streptococcus pyogenes |
| TA | Tibialis anterior |
| tracrRNA | Transactivating crRNA |
| Ttr | Transthyretin |
| TYMP | Thymidine phosphorylase |
| VEGFR2 | Vascular endothelial growth factor receptor 2 |
| WNT10B | Wnt family member 10B |
| ZIF-8 | Zeolitic imidazolate framework-8. |
Author contributions
Muhammad Waseem Ghani perceived the conceptualization and wrote the original draft. Ambreen Iqbal helped in figure preparation and writing – review and editing. Hammad Ghani and Sidra Bibi helped in reviewing figures and writing – review and editing. Zixun Wang participated in conceptualization, writing – review and editing, and supervision. Renjun Pei acquired funding, helped with writing – review and editing, and supervised the manuscript preparation.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 32201176, 31971326, 22177129). During this work, Muhammad Waseem Ghani was supported by the Alliance of International Science Organizations (ANSO) fellowship program. Fig. 1 was designed on biorender (https://biorender.com/).References
- M. Chavez, X. Chen, P. B. Finn and L. S. Qi, Nat. Rev. Nephrol., 2023, 19, 9–22 CrossRef CAS PubMed
.
- B. Rodolphe, F. Christophe, D. Hélène, R. Melissa, B. Patrick, M. Sylvain, D. A. Romero and H. Philippe, Science, 2007, 315, 1709–1712 CrossRef PubMed
.
- D. Cyranoski, Nature, 2016, 539, 479 CrossRef CAS PubMed
.
- R. O. Bak, D. P. Dever and M. H. Porteus, Nat. Protoc., 2018, 13, 358–376 CrossRef CAS PubMed
.
- E. De Dreuzy, J. Heath, J. A. Zuris, P. Sousa, R. Viswanathan, S. Scott, J. Da Silva, T. Ta, S. Capehart, T. Wang, C. Fernandez, V. E. Myer, C. F. Albright, C. J. Wilson, S. Teixeira and K.-H. Chang, Blood, 2019, 134, 4636 CrossRef
.
- L. C. Cosenza, J. Gasparello, N. Romanini, M. Zurlo, C. Zuccato, R. Gambari and A. Finotti, Mol. Ther. – Methods Clin. Dev., 2021, 21, 507–523 CrossRef CAS PubMed
.
- J. Ruan, H. Hirai, D. Yang, L. Ma, X. Hou, H. Jiang, H. Wei, C. Rajagopalan, H. Mou, G. Wang, J. Zhang, K. Li, Y. E. Chen, F. Sun and J. Xu, Mol. Ther. – Nucleic Acids, 2019, 16, 73–81 CrossRef CAS PubMed
.
- H. Yin, C.-Q. Song, J. R. Dorkin, L. J. Zhu, Y. Li, Q. Wu, A. Park, J. Yang, S. Suresh, A. Bizhanova, A. Gupta, M. F. Bolukbasi, S. Walsh, R. L. Bogorad, G. Gao, Z. Weng, Y. Dong, V. Koteliansky, S. A. Wolfe, R. Langer, W. Xue and D. G. Anderson, Nat. Biotechnol., 2016, 34, 328–333 CrossRef CAS PubMed
.
- C.-Q. Song and W. Xue, Nat. Rev. Gastroenterol. Hepatol., 2018, 15, 251–252 CrossRef PubMed
.
- W. Zhu, R. Lei, Y. Le Duff, J. Li, F. Guo, M. A. Wainberg and C. Liang, Retrovirology, 2015, 12, 22 CrossRef PubMed
.
- S. Böhm, V. Splith, L. M. Riedmayr, R. D. Rötzer, G. Gasparoni, K. J. V. Nordström, J. E. Wagner, K. S. Hinrichsmeyer, J. Walter, C. Wahl-Schott, S. Fenske, M. Biel, S. Michalakis and E. Becirovic, Sci. Adv., 2020, 6, eaba5614 CrossRef PubMed
.
- H. Khan, A. Khan, Y. Liu, S. Wang, S. Bibi, H. Xu, Y. Liu, S. Durrani, L. Jin, N. He and T. Xiong, Chin. Chem. Lett., 2019, 30, 2201–2204 CrossRef CAS PubMed
.
- Y. Liu, H. Xu, C. Liu, L. Peng, H. Khan, L. Cui, R. Huang, C. Wu, S. Shen, S. Wang, W. Liang, Z. Li, B. Xu and N. He, J. Biomed. Nanotechnol., 2019, 15, 790–798 CrossRef CAS PubMed
.
- J. Yan, D. D. Kang and Y. Dong, Biomater. Sci., 2021, 9, 6001–6011 RSC
.
- C. L. Xu, M. Z. C. Ruan, V. B. Mahajan and S. H. Tsang, Viruses, 2019, 11, 28 CrossRef CAS PubMed
.
- M. A. Kotterman, T. W. Chalberg and D. V. Schaffer, Annu. Rev. Biomed. Eng., 2015, 17, 63–89 CrossRef CAS PubMed
.
- S. Z. Mirjalili Mohanna, J. W. Hickmott, S. L. Lam, N. Y. Chiu, T. C. Lengyell, B. M. Tam, O. L. Moritz and E. M. Simpson, Mol. Ther. – Methods Clin. Dev., 2020, 17, 478–490 CrossRef CAS PubMed
.
- A. Kagita, M. S. Y. Lung, H. Xu, Y. Kita, N. Sasakawa, T. Iguchi, M. Ono, X. H. Wang, P. Gee and A. Hotta, Stem Cell Rep., 2021, 16, 985–996 CrossRef CAS PubMed
.
- J. Cai, S. Huang, Y. Yi and S. Bao, J. Int. Med. Res., 2019, 47, 2199–2206 CrossRef CAS PubMed
.
- J. A. Zuris, D. B. Thompson, Y. Shu, J. P. Guilinger, J. L. Bessen, J. H. Hu, M. L. Maeder, J. K. Joung, Z.-Y. Chen and D. R. Liu, Nat. Biotechnol., 2015, 33, 73–80 CrossRef CAS PubMed
.
- Y. Lin, E. Wagner and U. Lächelt, Biomater. Sci., 2022, 10, 1166–1192 RSC
.
- E. V. Koonin, K. S. Makarova and F. Zhang, Curr. Opin. Microbiol., 2017, 37, 67–78 CrossRef CAS PubMed
.
- Z. Liu, H. Dong, Y. Cui, L. Cong and D. Zhang, Microb. Cell Fact., 2020, 19, 172 CrossRef PubMed
.
- F. A. Ran, P. D. Hsu, J. Wright, V. Agarwala, D. A. Scott and F. Zhang, Nat. Protoc., 2013, 8, 2281–2308 CrossRef CAS PubMed
.
- S. Shmakov, A. Smargon, D. Scott, D. Cox, N. Pyzocha, W. Yan, O. O. Abudayyeh, J. S. Gootenberg, K. S. Makarova, Y. I. Wolf, K. Severinov, F. Zhang and E. V. Koonin, Nat. Rev. Microbiol., 2017, 15, 169–182 CrossRef CAS PubMed
.
- F. A. Ran, L. Cong, W. X. Yan, D. A. Scott, J. S. Gootenberg, A. J. Kriz, B. Zetsche, O. Shalem, X. Wu, K. S. Makarova, E. V. Koonin, P. A. Sharp and F. Zhang, Nature, 2015, 520, 186–191 CrossRef CAS PubMed
.
- D. Hendriks, H. Clevers and B. Artegiani, Cell Stem Cell, 2020, 27, 705–731 CrossRef CAS PubMed
.
- A. Pickar-Oliver and C. A. Gersbach, Nat. Rev. Mol. Cell Biol., 2019, 20, 490–507 CrossRef CAS PubMed
.
- E. Kim, T. Koo, S. W. Park, D. Kim, K. Kim, H. Y. Cho, D. W. Song, K. J. Lee, M. H. Jung, S. Kim, J. H. Kim, J. H. Kim and J. S. Kim, Nat. Commun., 2017, 8, 1–12 CrossRef PubMed
.
- Y. Zhang, N. Heidrich, B. J. Ampattu, C. W. Gunderson, H. S. Seifert, C. Schoen, J. Vogel and E. J. Sontheimer, Mol. Cell, 2013, 50, 488–503 CrossRef CAS PubMed
.
- P. Pausch, B. Al-Shayeb, E. Bisom-Rapp, C. A. Tsuchida, Z. Li, B. F. Cress, G. J. Knott, S. E. Jacobsen, J. F. Banfield and J. A. Doudna, Science, 2020, 369, 333–337 CrossRef CAS PubMed
.
- B. Al-Shayeb, R. Sachdeva, L.-X. Chen, F. Ward, P. Munk, A. Devoto, C. J. Castelle, M. R. Olm, K. Bouma-Gregson, Y. Amano, C. He, R. Méheust, B. Brooks, A. Thomas, A. Lavy, P. Matheus-Carnevali, C. Sun, D. S. A. Goltsman, M. A. Borton, A. Sharrar, A. L. Jaffe, T. C. Nelson, R. Kantor, R. Keren, K. R. Lane, I. F. Farag, S. Lei, K. Finstad, R. Amundson, K. Anantharaman, J. Zhou, A. J. Probst, M. E. Power, S. G. Tringe, W.-J. Li, K. Wrighton, S. Harrison, M. Morowitz, D. A. Relman, J. A. Doudna, A.-C. Lehours, L. Warren, J. H. D. Cate, J. M. Santini and J. F. Banfield, Nature, 2020, 578, 425–431 CrossRef CAS PubMed
.
- X. Xu, A. Chemparathy, L. Zeng, H. R. Kempton, S. Shang, M. Nakamura and L. S. Qi, Mol. Cell, 2021, 81, 4333–4345.e4 CrossRef CAS PubMed
.
- L. B. Harrington, D. Burstein, J. S. Chen, D. Paez-Espino, E. Ma, I. P. Witte, J. C. Cofsky, N. C. Kyrpides, J. F. Banfield and J. A. Doudna, Science, 2018, 362, 839–842 CrossRef CAS PubMed
.
- A. Ventura and L. E. Dow, Annu. Rev. Cancer Biol., 2018, 2, 111–131 CrossRef
.
- D. B. T. Cox, J. S. Gootenberg, O. O. Abudayyeh, B. Franklin, M. J. Kellner, J. Joung and F. Zhang, Science, 2017, 358, 1019–1027 CrossRef CAS PubMed
.
- O. O. Abudayyeh, J. S. Gootenberg, P. Essletzbichler, S. Han, J. Joung, J. J. Belanto, V. Verdine, D. B. T. Cox, M. J. Kellner, A. Regev, E. S. Lander, D. F. Voytas, A. Y. Ting and F. Zhang, Nature, 2017, 550, 280–284 CrossRef PubMed
.
- A. East-Seletsky, M. R. O’Connell, S. C. Knight, D. Burstein, J. H. D. Cate, R. Tjian and J. A. Doudna, Nature, 2016, 538, 270–273 CrossRef CAS PubMed
.
- P. Mali, J. Aach, P. B. Stranges, K. M. Esvelt, M. Moosburner, S. Kosuri, L. Yang and G. M. Church, Nat. Biotechnol., 2013, 31, 833–838 CrossRef CAS PubMed
.
- L. Cong, F. Zhang, L. A. Marraffini, W. Jiang, X. Wu, P. D. Hsu, N. Habib, R. Barretto, S. Lin, D. Cox and F. A. Ran, Science, 2013, 339, 816–819 CrossRef PubMed
.
- J. E. Powell, C. K. W. Lim, R. Krishnan, T. X. McCallister, C. Saporito-Magriña, M. A. Zeballos, G. D. McPheron and T. Gaj, Sci. Adv., 2022, 8, 1–12 Search PubMed
.
- S. Konermann, P. Lotfy, N. J. Brideau, J. Oki, M. N. Shokhirev and P. D. Hsu, Cell, 2018, 173, 665–676.e14 CrossRef CAS PubMed
.
- A. C. Komor, Y. B. Kim, M. S. Packer, J. A. Zuris and D. R. Liu, Nature, 2016, 533, 420–424 CrossRef CAS PubMed
.
- N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson and D. R. Liu, Nature, 2017, 551, 464–471 CrossRef CAS PubMed
.
- A. V. Anzalone, P. B. Randolph, J. R. Davis, A. A. Sousa, L. W. Koblan, J. M. Levy, P. J. Chen, C. Wilson, G. A. Newby, A. Raguram and D. R. Liu, Nature, 2019, 576, 149–157 CrossRef CAS PubMed
.
- D. B. T. Cox, J. S. Gootenberg, O. O. Abudayyeh, B. Franklin, M. J. Kellner, J. Joung and F. Zhang, Science, 2017, 358, 1019–1027 CrossRef CAS PubMed
.
- L. S. Qi, M. H. Larson, L. A. Gilbert, J. A. Doudna, J. S. Weissman, A. P. Arkin and W. A. Lim, Cell, 2013, 152, 1173–1183 CrossRef CAS PubMed
.
- M. Adli, Nat. Commun., 2018, 9, 1911 CrossRef PubMed
.
- L. A. Gilbert, M. A. Horlbeck, B. Adamson, J. E. Villalta, Y. Chen, E. H. Whitehead, C. Guimaraes, B. Panning, H. L. Ploegh, M. C. Bassik, L. S. Qi, M. Kampmann and J. S. Weissman, Cell, 2014, 159, 647–661 CrossRef CAS PubMed
.
- M. L. Maeder, S. J. Linder, V. M. Cascio, Y. Fu, Q. H. Ho and J. K. Joung, Nat. Methods, 2013, 10, 977–979 CrossRef CAS PubMed
.
- P. Perez-Pinera, D. D. Kocak, C. M. Vockley, A. F. Adler, A. M. Kabadi, L. R. Polstein, P. I. Thakore, K. A. Glass, D. G. Ousterout, K. W. Leong, F. Guilak, G. E. Crawford, T. E. Reddy and C. A. Gersbach, Nat. Methods, 2013, 10, 973–976 CrossRef CAS PubMed
.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer, C. A. Freije, C. Myhrvold, R. P. Bhattacharyya, J. Livny, A. Regev, E. V. Koonin, D. T. Hung, P. C. Sabeti, J. J. Collins and F. Zhang, Science, 2017, 356, 438–442 CrossRef CAS PubMed
.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Science, 2018, 360, 439–444 CrossRef CAS PubMed
.
- S. Y. Li, Q. X. Cheng, X. Y. Li, Z. L. Zhang, S. Gao, R. B. Cao, G. P. Zhao, J. Wang and J. M. Wang, Cell Discov., 2018, 4, 18–21 Search PubMed
.
- S. W. Cho, S. Kim, Y. Kim, J. Kweon, H. S. Kim, S. Bae and J.-S. Kim, Genome Res., 2014, 24, 132–141 CrossRef CAS PubMed
.
- H. A. Han, J. K. S. Pang and B.-S. Soh, J. Mol. Med., 2020, 98, 615–632 CrossRef CAS PubMed
.
- N. Chang, C. Sun, L. Gao, D. Zhu, X. Xu, X. Zhu, J.-W. Xiong and J. J. Xi, Cell Res., 2013, 23, 465–472 CrossRef CAS PubMed
.
- C. A. Lino, J. C. Harper, J. P. Carney and J. A. Timlin, Drug Deliv., 2018, 25, 1234–1257 CrossRef CAS PubMed
.
- K. Schumann, S. Lin, E. Boyer, D. R. Simeonov, M. Subramaniam, R. E. Gate, G. E. Haliburton, C. J. Ye, J. A. Bluestone, J. A. Doudna and A. Marson, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 10437–10442 CrossRef CAS PubMed
.
- J. A. Zuris, D. B. Thompson, Y. Shu, J. P. Guilinger, J. L. Bessen, J. H. Hu, M. L. Maeder, J. K. Joung, Z. Y. Chen and D. R. Liu, Nat. Biotechnol., 2015, 33, 73–80 CrossRef CAS PubMed
.
- A. K. Levacic, S. Morys, S. Kempter, U. Lächelt and E. Wagner, Hum. Gene Ther., 2017, 28, 862–874 CrossRef CAS PubMed
.
- S. Mishra, P. Webster and M. E. Davis, Eur. J. Cell Biol., 2004, 83, 97–111 CrossRef CAS PubMed
.
- J. Wu, D. Ryskamp, L. Birnbaumer and I. Bezprozvanny, J. Huntingtons. Dis., 2018, 7, 35–50 CAS
.
- S.-M. Ryu, T. Koo, K. Kim, K. Lim, G. Baek, S.-T. Kim, H. S. Kim, D. Kim, H. Lee, E. Chung and J.-S. Kim, Nat. Biotechnol., 2018, 36, 536–539 CrossRef CAS PubMed
.
- Y.-C. Yang, Y.-H. Chen, J.-H. Kao, C. Ching, I.-J. Liu, C.-C. Wang, C.-H. Tsai, F.-Y. Wu, C.-J. Liu, P.-J. Chen, D.-S. Chen and H.-C. Yang, Mol. Ther.– Nucleic Acids, 2020, 20, 480–490 CrossRef CAS PubMed
.
- G. Schwank, B. K. Koo, V. Sasselli, J. F. Dekkers, I. Heo, T. Demircan, N. Sasaki, S. Boymans, E. Cuppen, C. K. Van Der Ent, E. E. S. Nieuwenhuis, J. M. Beekman and H. Clevers, Cell Stem Cell, 2013, 13, 653–658 CrossRef CAS PubMed
.
- D. Cyranoski, Nature, 2016, 535, 476–477 CrossRef CAS PubMed
.
- Y. Lu, J. Xue, T. Deng, X. Zhou, K. Yu, L. Deng, M. Huang, X. Yi, M. Liang, Y. Wang, H. Shen, R. Tong, W. Wang, L. Li, J. Song, J. Li, X. Su, Z. Ding, Y. Gong, J. Zhu, Y. Wang, B. Zou, Y. Zhang, Y. Li, L. Zhou, Y. Liu, M. Yu, Y. Wang, X. Zhang, L. Yin, X. Xia, Y. Zeng, Q. Zhou, B. Ying, C. Chen, Y. Wei, W. Li and T. Mok, Nat. Med., 2020, 26, 732–740 CrossRef CAS PubMed
.
- Y. Suzuki, H. Onuma, R. Sato, Y. Sato and A. Hashiba, J. Controlled. Release, 2021, 330, 61–71 CrossRef CAS PubMed
.
- L. Zhang, L. Wang, Y. Xie, P. Wang, S. Deng, A. Qin, J. Zhang, X. Yu, W. Zheng and X. Jiang, Angew. Chem., Int. Ed., 2019, 58, 12404–12408 CrossRef CAS PubMed
.
- Y. Pan, X. Luan, F. Zeng, Q. Xu, Z. Li, Y. Gao, X. Liu, X. Li, X. Han, J. Shen and Y. Song, Biosens. Bioelectron., 2022, 209, 114239 CrossRef CAS PubMed
.
- D. Huang, D. Zhao, J. Li, Y. Wu, L. Du, X.-H. Xia, X. Li, Y. Deng, Z. Li and Y. Huang, Curr. Drug Deliv., 2018, 16, 164–170 CrossRef PubMed
.
- P. Ghasemiyeh and S. Mohammadi-Samani, Res. Pharm. Sci., 2018, 13, 288–303 CrossRef PubMed
.
- M. J. Mulligan, K. E. Lyke, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, K. Neuzil, V. Raabe, R. Bailey, K. A. Swanson, P. Li, K. Koury, W. Kalina, D. Cooper, C. Fontes-Garfias, P.-Y. Shi, Ö. Türeci, K. R. Tompkins, E. E. Walsh, R. Frenck, A. R. Falsey, P. R. Dormitzer, W. C. Gruber, U. Şahin and K. U. Jansen, Nature, 2020, 586, 589–593 CrossRef CAS PubMed
.
- D. Chatzikleanthous, D. T. O’Hagan and R. Adamo, Mol. Pharm., 2021, 18, 2867–2888 CrossRef CAS PubMed
.
- Y. Granot and D. Peer, Semin. Immunol., 2017, 34, 68–77 CrossRef CAS PubMed
.
- J. D. Finn, A. R. Smith, M. C. Patel, L. Shaw, M. R. Youniss, J. van Heteren, T. Dirstine, C. Ciullo, R. Lescarbeau, J. Seitzer, R. R. Shah, A. Shah, D. Ling, J. Growe, M. Pink, E. Rohde, K. M. Wood, W. E. Salomon, W. F. Harrington, C. Dombrowski, W. R. Strapps, Y. Chang and D. V. Morrissey, Cell Rep., 2018, 22, 2227–2235 CrossRef CAS PubMed
.
- J. B. Miller, S. Zhang, P. Kos, H. Xiong, K. Zhou, S. S. Perelman, H. Zhu and D. J. Siegwart, Angew. Chem., Int. Ed., 2017, 56, 1059–1063 CrossRef CAS PubMed
.
- W. Cao, J. Liu, L. Wang, M. Li, M. M. A. Verstegen, Y. Yin, B. Ma, K. Chen, M. Bolkestein, D. Sprengers, L. J. W. van der Laan, M. Doukas, J. Kwekkeboom, R. Smits, M. P. Peppelenbosch and Q. Pan, Carcinogenesis, 2019, 40, 145–154 CrossRef CAS PubMed
.
- M. Qiu, Z. Glass, J. Chen, M. Haas, X. Jin, X. Zhao, X. Rui, Z. Ye, Y. Li, F. Zhang and Q. Xu, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2020401118 CrossRef CAS PubMed
.
- M. Parés, C. Fornaguera, F. Vila-Julià, S. Oh, S. H. Y. Fan, Y. K. Tam, N. Comes, F. Vidal, R. Martí, S. Borrós and J. Barquinero, Hum. Gene Ther., 2021, 32, 1210–1223 CrossRef PubMed
.
- K. Musunuru, A. C. Chadwick, T. Mizoguchi, S. P. Garcia, J. E. DeNizio, C. W. Reiss, K. Wang, S. Iyer, C. Dutta, V. Clendaniel, M. Amaonye, A. Beach, K. Berth, S. Biswas, M. C. Braun, H. M. Chen, T. V. Colace, J. D. Ganey, S. A. Gangopadhyay, R. Garrity, L. N. Kasiewicz, J. Lavoie, J. A. Madsen, Y. Matsumoto, A. M. Mazzola, Y. S. Nasrullah, J. Nneji, H. Ren, A. Sanjeev, M. Shay, M. R. Stahley, S. H. Y. Fan, Y. K. Tam, N. M. Gaudelli, G. Ciaramella, L. E. Stolz, P. Malyala, C. J. Cheng, K. G. Rajeev, E. Rohde, A. M. Bellinger and S. Kathiresan, Nature, 2021, 593, 429–434 CrossRef CAS PubMed
.
- J. D. Gillmore, E. Gane, J. Taubel, J. Kao, M. Fontana, M. L. Maitland, J. Seitzer, D. O’Connell, K. R. Walsh, K. Wood, J. Phillips, Y. Xu, A. Amaral, A. P. Boyd, J. E. Cehelsky, M. D. McKee, A. Schiermeier, O. Harari, A. Murphy, C. A. Kyratsous, B. Zambrowicz, R. Soltys, D. E. Gutstein, J. Leonard, L. Sepp-Lorenzino and D. Lebwohl, N. Engl. J. Med., 2021, 385, 493–502 CrossRef CAS PubMed
.
- C. D. Sago, M. P. Lokugamage, K. Paunovska, D. A. Vanover, C. M. Monaco, N. N. Shah, M. G. Castro, S. E. Anderson, T. G. Rudoltz, G. N. Lando, P. M. Tiwari, J. L. Kirschman, N. Willett, Y. C. Jang, P. J. Santangelo, A. V. Bryksin and J. E. Dahlman, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E9944–E9952 CrossRef CAS PubMed
.
- T. Wei, Q. Cheng, Y. L. Min, E. N. Olson and D. J. Siegwart, Nat. Commun., 2020, 11, 1–12 CrossRef PubMed
.
- S. Z. Mirjalili Mohanna, D. Djaksigulova, A. M. Hill, P. K. Wagner, E. M. Simpson and B. R. Leavitt, J. Controlled. Release, 2022, 350, 401–413 CrossRef CAS PubMed
.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Nat. Nanotechnol., 2020, 15, 313–320 CrossRef CAS PubMed
.
- S. Liu, Q. Cheng, T. Wei, X. Yu, L. T. Johnson, L. Farbiak and D. J. Siegwart, Nat. Mater., 2021, 20, 701–710 CrossRef CAS PubMed
.
- C. D. Sago, M. P. Lokugamage, D. Loughrey, K. E. Lindsay, R. Hincapie, B. R. Krupczak, S. Kalathoor, M. Sato, E. S. Echeverri, J. P. Fitzgerald, Z. Gan, L. Gamboa, K. Paunovska, C. A. Sanhueza, M. Z. C. Hatit, M. G. Finn, P. J. Santangelo and J. E. Dahlman, Nat. Biomed. Eng., 2022, 6, 157–167 CrossRef CAS PubMed
.
- J. Seitzer, J. Allergy Clin. Immunol., 2021, 147, AB147 CrossRef
.
- K. Walsh, P. Cottingham, C. Shaw, J. Phillips, J. Seitzer, C. Blanchard, N. Gardner, V. Doshi, K. Wood, S. Soukamneuth, B. Han, S. Jaligama, R. Cole, S. Parthasarathy, D. Kryukov and J. Butler, J. Allergy Clin. Immunol., 2022, 149, AB169 CrossRef
.
- D. Rosenblum, A. Gutkin, R. Kedmi, S. Ramishetti, N. Veiga, A. M. Jacobi, M. S. Schubert, D. Friedmann-Morvinski, Z. R. Cohen, M. A. Behlke, J. Lieberman and D. Peer, Sci. Adv., 2020, 6, eabc9450 CrossRef CAS PubMed
.
- D. Zhang, G. Wang, X. Yu, T. Wei, L. Farbiak, L. T. Johnson, A. M. Taylor, J. Xu, Y. Hong, H. Zhu and D. J. Siegwart, Nat. Nanotechnol., 2022, 7, 777–787 CrossRef PubMed
.
- A. S. Piotrowski-Daspit, A. C. Kauffman, L. G. Bracaglia and W. M. Saltzman, Adv. Drug Deliv. Rev., 2020, 156, 119–132 CrossRef CAS PubMed
.
- Z. Zhang, T. Wan, Y. Chen, Y. Chen, H. Sun, T. Cao, Z. Songyang, G. Tang, C. Wu, Y. Ping, F. J. Xu and J. Huang, Macromol. Rapid Commun., 2019, 40, 1–8 Search PubMed
.
- B.-C. Zhang, P.-Y. Wu, J.-J. Zou, J.-L. Jiang, R.-R. Zhao, B.-Y. Luo, Y.-Q. Liao and J.-W. Shao, Chem. Eng. J., 2020, 393, 124688 CrossRef CAS
.
- S. Abbasi, S. Uchida, K. Toh, T. A. Tockary, A. Dirisala, K. Hayashi, S. Fukushima and K. Kataoka, J. Controlled Release, 2021, 332, 260–268 CrossRef CAS PubMed
.
- W. Tao, X. Cheng, D. Sun, Y. Guo, N. Wang, J. Ruan, Y. Hu, M. Zhao, T. Zhao, H. Feng, L. Fan, C. Lu, Y. Ma, J. Duan and M. Zhao, Biomaterials, 2022, 287, 121621 CrossRef CAS PubMed
.
- M. Kabra, P. K. Shahi, Y. Wang, D. Sinha, A. Spillane, G. A. Newby, S. Saxena, A. A. Abdeen, K. L. Edwards, C. O. Theisen, D. M. Gamm, D. R. Liu, S. Gong, K. Saha and B. R. Pattnaik, bioRxiv, 2022, preprint DOI:10.1101/2022.07.12.499808.
- A. Luchini and G. Vitiello, Front. Chem., 2019, 7, 1–16 CrossRef PubMed
.
- Z. P. Xu, Q. H. Zeng, G. Q. Lu and A. B. Yu, Chem. Eng. Sci., 2006, 61, 1027–1040 CrossRef CAS
.
- P. Wang, L. Zhang, Y. Xie, N. Wang, R. Tang, W. Zheng and X. Jiang, Adv. Sci., 2017, 11, 1700175 CrossRef PubMed
.
- J.-Y. Zhu, D.-W. Zheng, M.-K. Zhang, W.-Y. Yu, W.-X. Qiu, J.-J. Hu, J. Feng and X.-Z. Zhang, Nano Lett., 2016, 16, 5895–5901 CrossRef CAS PubMed
.
- Z. Zhou, M. Vázquez-González and I. Willner, Chem. Soc. Rev., 2021, 50, 4541–4563 RSC
.
- X. Yang, Q. Tang, Y. Jiang, M. Zhang, M. Wang and L. Mao, J. Am. Chem. Soc., 2019, 141, 3782–3786 CrossRef CAS PubMed
.
- S. K. Alsaiari, S. Patil, M. Alyami, K. O. Alamoudi, F. A. Aleisa, J. S. Merzaban, M. Li and N. M. Khashab, J. Am. Chem. Soc., 2018, 140, 143–146 CrossRef CAS PubMed
.
- A. Poddar, S. Pyreddy, F. Carraro, S. Dhakal, A. Rassell, M. R. Field, T. S. Reddy, P. Falcaro, C. M. Doherty and R. Shukla, Chem. Commun., 2020, 56, 15406–15409 RSC
.
- Y. Wang, P. K. Shahi, R. Xie, H. Zhang, A. A. Abdeen, N. Yodsanit, Z. Ma, K. Saha, B. R. Pattnaik and S. Gong, J. Controlled Release, 2020, 324, 194–203 CrossRef CAS PubMed
.
- W. Berger, B. Kloeckener-Gruissem and J. Neidhardt, Prog. Retin. Eye Res., 2010, 29, 335–375 CrossRef CAS PubMed
.
- M. Z. Alyami, S. K. Alsaiari, Y. Li, S. S. Qutub, F. A. Aleisa, R. Sougrat, J. S. Merzaban and N. M. Khashab, J. Am. Chem. Soc., 2020, 142, 1715–1720 CrossRef CAS PubMed
.
- R. Shahbazi, G. Sghia-Hughes, J. L. Reid, S. Kubek, K. G. Haworth, O. Humbert, H. P. Kiem and J. E. Adair, Nat. Mater., 2019, 18, 1124–1132 CrossRef CAS PubMed
.
- H. Yue, X. Zhou, M. Cheng and D. Xing, Nanoscale, 2018, 10, 1063–1071 RSC
.
- J. Deng, K. Wang, M. Wang, P. Yu and L. Mao, J. Am. Chem. Soc., 2017, 139, 5877–5882 CrossRef CAS PubMed
.
- Z. Zhang, Q. Wang, Q. Liu, Y. Zheng, C. Zheng, K. Yi, Y. Zhao, Y. Gu, Y. Wang, C. Wang, X. Zhao, L. Shi, C. Kang and Y. Liu, Adv. Mater., 2019, 31, 1–10 Search PubMed
.
-
M. Rafique, M. B. Tahir, M. S. Rafique, N. Safdar and R. Tahir, Nanostructure materials and their classification by dimensionality, Elsevier Inc., 2020 Search PubMed
.
- R. Szatanek, M. Baj-Krzyworzeka, J. Zimoch, M. Lekka, M. Siedlar and J. Baran, Int. J. Mol. Sci., 2017, 18 Search PubMed
.
- F. J. M. Mojica, C. Díez-Villaseñor, J. García-Martínez and E. Soria, J. Mol. Evol., 2005, 60, 174–182 CrossRef CAS PubMed
.
- X. Yao, P. Lyu, K. Yoo, M. K. Yadav, R. Singh, A. Atala and B. Lu, J. Extracell. Vesicles, 2018, 10, e12076 Search PubMed
.
- J. Zhuang, J. Tan, C. Wu, J. Zhang, T. Liu, C. Fan, J. Li and Y. Zhang, Nucleic Acids Res., 2020, 48, 8870–8882 CrossRef CAS PubMed
.
- M. Kalkat, J. De Melo, K. A. Hickman, C. Lourenco, C. Redel, D. Resetca, A. Tamachi, W. B. Tu and L. Z. Penn, Genes, 2017, 8, 151 CrossRef PubMed
.
- Q. Xu, Z. Zhang, L. Zhao, Y. Qin, H. Cai, Z. Geng, X. Zhu, W. Zhang, Y. Zhang, J. Tan, J. Wang and J. Zhou, J. Controlled Release, 2020, 326, 455–467 CrossRef CAS PubMed
.
- Y. Ye, X. Zhang, F. Xie, B. Xu, P. Xie, T. Yang, Q. Shi, C. Y. Zhang, Y. Zhang, J. Chen, X. Jiang and J. Li, Biomater. Sci., 2020, 8, 2966–2976 RSC
.
- N. Luo, J. Li, Y. Chen, Y. Xu, Y. Wei, J. Lu and R. Dong, Drug Deliv., 2021, 28, 10–18 CrossRef CAS PubMed
.
- D. Lainšček, L. Kadunc, M. M. Keber, I. H. Bratkovič, R. Romih and R. Jerala, ACS Synth. Biol., 2018, 7, 2715–2725 CrossRef PubMed
.
- P. Gee, M. S. Y. Lung, Y. Okuzaki, N. Sasakawa, T. Iguchi, Y. Makita, H. Hozumi, Y. Miura, L. F. Yang, M. Iwasaki, X. H. Wang, M. A. Waller, N. Shirai, Y. O. Abe, Y. Fujita, K. Watanabe, A. Kagita, K. A. Iwabuchi, M. Yasuda, H. Xu, T. Noda, J. Komano, H. Sakurai, N. Inukai and A. Hotta, Nat. Commun., 2020, 11, 4–20 CrossRef PubMed
.
- S. M. Kim, Y. Yang, S. J. Oh, Y. Hong, M. Seo and M. Jang, J. Controlled Release, 2017, 266, 8–16 CrossRef CAS PubMed
.
- S. Jiang, Z. Ge, S. Mou, H. Yan and C. Fan, Chem, 2021, 7, 1156–1179 CAS
.
- V. Linko, A. Ora and M. A. Kostiainen, Trends Biotechnol., 2015, 33, 586–594 CrossRef CAS PubMed
.
- J. Liu, T. Wu, X. Lu, X. Wu, S. Liu, S. Zhao, X. Xu and B. Ding, J. Am. Chem. Soc., 2019, 141, 19032–19037 CrossRef CAS PubMed
.
- W. Sun, J. Wang, Q. Hu, X. Zhou, A. Khademhosseini and Z. Gu, Sci. Adv., 2020, 6, 1–12 Search PubMed
.
- F. Ding, X. Huang, X. Gao, M. Xie, G. Pan, Q. Li, J. Song, X. Zhu and C. Zhang, Nanoscale, 2019, 11, 17211–17215 RSC
.
- X. Huang, R. Zheng, F. Ding, J. Yang, M. Xie, X. Liu, J. Li, J. Feng, X. Zhu and C. Zhang, ACS Mater. Lett., 2020, 2, 1509–1515 CrossRef CAS
.
- T. Wu, Y. Cao, Q. Liu, X. Wu, Y. Shang, J. Piao, Y. Li, Y. Dong, D. Liu, H. Wang, J. Liu and B. Ding, J. Am. Chem. Soc., 2022, 144, 6575–6582 CrossRef CAS PubMed
.
- F. Li, N. Song, Y. Dong, S. Li, L. Li, Y. Liu, Z. Li and D. Yang, Angew. Chem.,
Int. Ed., 2022, 61, 1–10 Search PubMed
.
- A. K. Shalek, J. T. Robinson, E. S. Karp, J. S. Lee, D.-R. Ahn, M.-H. Yoon, A. Sutton, M. Jorgolli, R. S. Gertner, T. S. Gujral, G. MacBeath, E. G. Yang and H. Park, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1870–1875 CrossRef CAS PubMed
.
- H. Persson, C. Købler, K. Mølhave, L. Samuelson, J. O. Tegenfeldt, S. Oredsson and C. N. Prinz, Small, 2013, 9, 4006–4016 CrossRef CAS PubMed
.
- W. Zhao, L. Hanson, H.-Y. Lou, M. Akamatsu, P. D. Chowdary, F. Santoro, J. R. Marks, A. Grassart, D. G. Drubin, Y. Cui and B. Cui, Nat. Nanotechnol., 2017, 12, 750–756 CrossRef CAS PubMed
.
- J. Kim, S. Kim, J. Ahn, J. Lee and J. Nam, Adv. Mater., 2020, 32, 2001360 CrossRef CAS PubMed
.
- Y. Chen, S. Aslanoglou, G. Gervinskas, H. Abdelmaksoud, N. H. Voelcker and R. Elnathan, Small, 2019, 15, 1904819 CrossRef CAS PubMed
.
- H. Kim, H. S. Lee, Y. Jeon, W. Park, Y. Zhang, B. Kim, H. Jang, B. Xu, Y. Yeo, D. R. Kim and C. H. Lee, ACS Nano, 2020, 14, 7227–7236 CrossRef CAS PubMed
.
- E. Peer, A. Artzy-Schnirman, L. Gepstein and U. Sivan, ACS Nano, 2012, 6, 4940–4946 CrossRef CAS PubMed
.
- Y. Chen, S. Aslanoglou, G. Gervinskas, H. Abdelmaksoud, N. H. Voelcker and R. Elnathan, Small, 2019, 15, 1904819 CrossRef CAS PubMed
.
- R. Kawamura, K. Shimizu, Y. Matsumoto, A. Yamagishi, Y. R. Silberberg, M. Iijima, S. Kuroda, K. Fukazawa, K. Ishihara and C. Nakamura, J. Nanobiotechnol., 2016, 14, 12951 CrossRef PubMed
.
- A. Yamagishi, D. Matsumoto, Y. Kato, Y. Honda, M. Morikawa, F. Iwata, T. Kobayashi and C. Nakamura, Appl. Sci., 2019, 9, 965 CrossRef CAS
.
- S. Gopal, C. Chiappini, J. Penders, V. Leonardo, H. Seong, S. Rothery, Y. Korchev, A. Shevchuk and M. M. Stevens, Adv. Mater., 2019, 31, 1806788 CrossRef PubMed
.
- M. Hansen-Bruhn, B. E. F. de Ávila, M. Beltrán-Gastélum, J. Zhao, D. E. Ramírez-Herrera, P. Angsantikul, K. Vesterager Gothelf, L. Zhang and J. Wang, Angew. Chem., Int. Ed., 2018, 57, 2657–2661 CrossRef CAS PubMed
.
- R. Numano, A. Goryu, Y. Kubota, H. Sawahata, S. Yamagiwa, M. Matsuo, T. Iimura, H. Tei, M. Ishida and T. Kawano, FEBS Open Bio, 2022, 12, 835–851 CrossRef CAS PubMed
.
- T. R. Abbott, G. Dhamdhere, Y. Liu, X. Lin, L. Goudy, L. Zeng, A. Chemparathy, S. Chmura, N. S. Heaton, R. Debs, T. Pande, D. Endy, M. F. La Russa, D. B. Lewis and L. S. Qi, Cell, 2020, 181, 865–876 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2023 |