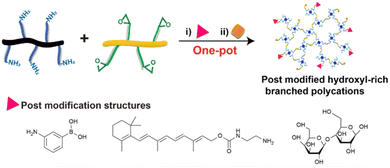
Hydroxyl-rich branched polycations for nucleic acid delivery
Mengrui
Su†
a,
Zichen
Hu†
a,
Yujie
Sun
a,
Yu
Qi
*bc,
Bingran
Yu
 *a and
Fu-Jian
Xu
*a and
Fu-Jian
Xu
 *a
*a
aKey Laboratory of Biomedical Materials of Natural Macromolecules (Beijing University of Chemical Technology, Ministry of Education), Beijing Laboratory of Biomedical Materials, Beijing University of Chemical Technology, Beijing 100029, PR China. E-mail: yubr@mail.buct.edu.cn; xufj@mail.buct.edu.cn
bChina Meat Food Research Center, Beijing Academy of Food Sciences, Beijing 100068, PR China. E-mail: qiyu_0501@163.com
cBeijing Forestry University, Beijing, 100083, PR China
First published on 17th November 2023
Abstract
Recently, nucleic acid delivery has become an amazing route for the treatment of various malignant diseases, and polycationic vectors are attracting more and more attention among gene vectors. However, conventional polycationic vectors still face many obstacles in nucleic acid delivery, such as significant cytotoxicity, high protein absorption behavior, and unsatisfactory blood compatibility caused by a high positive charge density. To solve these problems, the fabrication of hydroxyl-rich branched polycationic vectors has been proposed. For the synthesis of hydroxyl-rich branched polycations, a one-pot method is considered as the preferred method due to its simple preparation process. In this review, typical one-pot methods for fabricating hydroxyl-rich polycations are presented. In particular, amine-epoxide ring-opening polymerization as a novel approach is mainly introduced. In addition, various therapeutic scenarios of hydroxyl-rich branched polycations via one-pot fabrication are also generalized. We believe that this review will motivate the optimized design of hydroxyl-rich branched polycations for potential nucleic acid delivery and their bio-applications.
1. Introduction
Nucleic acid delivery is an approach that utilizes therapeutic nucleic acids, including DNA, RNA, and oligonucleotides, to treat conditions such as cancers, immunodeficiency, and many human malignancies.1–7 To achieve optimal therapeutic effect, the nucleic acids require efficient delivery to the targeted cells and efficient vectors are necessary. As we know, vectors can be mainly divided into two categories: viral and non-viral vectors. Viral vectors, such as adenoviruses and retroviruses, are restricted due to their uncontrolled bio-risks.8–10 Non-viral vectors mainly include liposomes,11,12 peptides,13–17 polycations,18–22 and organic–inorganic hybrid materials.7,23,24 Among non-viral vectors, polycations have attracted more attention due to their low manufacturing cost and robust modifiability. However, significant cytotoxicity and unsatisfactory transfection performance still hinder their applications.25–29 In addition, positively charged polycations can easily absorb negatively charged proteins in the extracellular fluid, which reduces the efficiency via which the vectors can complex with nucleic acids.30–32 Therefore, how to develop polycationic vectors for safe delivery and efficiently compressing nucleic acids has become a focus of research.Polycationic vectors mainly refer to polymers with positively charged groups, which rely on electrostatic interaction to complex nucleic acids.33–35 Conventional polycationic vectors such as poly(2-N,N-dimethylaminoethylmethacrylate) (PDMAEMA)36–38 and polyethyleneimine (PEI)31,39–41 have already been proven to have outstanding complexation capability due to their high positive charge density, while unignorable cytotoxicity, protein absorption behavior, and high hemolysis rate caused by the positive charges compromise their transfection efficiency and bio-compatibility. As a strategy to overcome these shortcomings, the fabrication of hydroxyl-rich polycationic vectors is suggested.42,43 Primarily, researchers mainly modify hydroxyl-rich structures, such as polyethylene glycol, on polycationic vectors.44,45 The obtained polycations were found to exhibit negligible cytotoxicity, low protein absorption capability, and good blood compatibility benefited from abundant hydroxyl groups. However, such a post-modification method usually requires two steps to complete, preparation of the polycations and post modification. The whole process is complicated and the yield of the final product is unacceptable. With the deepening of research, the “one-pot” method is gradually becoming a better choice. Instead of post-modification, the one-pot method focuses on the design and preparation of the building blocks, and hydroxyl structures can be obtained from efficient, green, and mild reactions of the building blocks.46–48 One example is the amine-epoxide ring-opening reaction, which is based on the reaction of primary amino groups and epoxy groups.49,50 With the opening of the epoxy groups, secondary amino groups and hydroxyl groups will occur simultaneously. Positively-charged secondary amino groups can complex with the negatively-charged nucleic acids, and hydroxyl groups provide hydrophilicity and biocompatibility.
In this brief review, we mainly introduce the recent progress in the fabrication of hydroxyl-rich polycations via a one-pot method. The typical one-pot methods for fabricating hydroxyl-rich polycations are presented. In particular, the amine-epoxide ring-opening reaction as a novel and efficient approach has been mainly highlighted. The therapeutic scenarios, including cancer, diabetes mellitus, liver fibrosis, antibacterial and wound healing treatments, of hydroxyl-rich branched polycations via the one-pot method are subsequently generalized. Hopefully, this review will inspire continuous endeavors in exploring more simple and efficient ways to develop various hydroxyl-rich branched polycations and enriching their biological applications for nucleic acid delivery.
2. One-pot method for fabricating hydroxyl-rich polycations
As an efficient and convenient synthetic strategy, the one-pot method can achieve a multi-step reaction process continuously without intermediate separation, which not only economizes cost, but also guarantees the yield of final product. In this section, many one-pot methods for the preparation of hydroxyl-rich polycations via ring-opening reactions of poly(glycidyl methacrylate) (PGMA), Michael addition reactions, and amine-epoxide ring-opening polymerization are generalized. Notably, amine-epoxide ring-opening polymerization is mainly introduced.2.1 Amino-epoxy group reaction of PGMA
PGMA is usually obtained by atom transfer radical polymerization (ATRP) or reversible addition–fragmentation chain transfer polymerization (RAFT) of glycidyl methacrylate monomers. Our previous work synthesized PGEA or PGED by using ring opening reactions of ethanolamine (EA) or ethylenediamine (ED) with epoxy groups in PGMA, which achieved poly-hydroxylation of PGMA successfully.51–55 Inspired by the synthetic methods of PGEA and PGED, many aminated building blocks were introduced into the preparation of hydroxyl-rich polycation carriers, together with EA or ED through ring-opening reactions to fabricate functionalized PGEA or PGED later. In this part, we summarize the one-pot methods for the fabrication of hydroxyl-rich polycationic vectors (Fig. 1).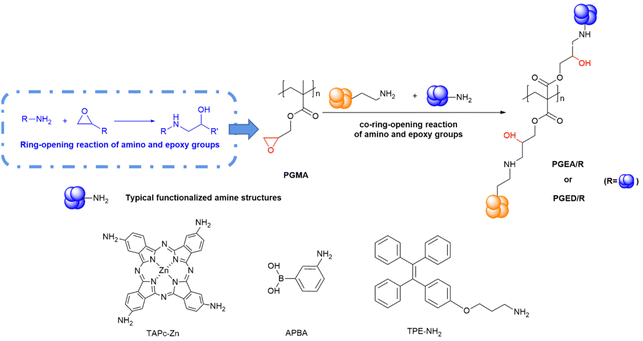 | ||
| Fig. 1 Strategies for the preparation of hydroxyl-rich polycations for co-ring-opening reactions of PGMA. | ||
For example, Li et al. used 3-aminophenyl boronic acid (APBA) and EA reacting with PGMA through the one-pot method to obtain functionalized PGEA, which lays the foundation for further works.56 Moreover, synthetic aminated building blocks, tetra-aminophthalocyanine (TAPc-Zn) and aminated tetraphenylethylene (TPE-NH2), were designed and constructed by Sun et al. and Qi et al.57,58 Then, PcPGEA and TPE-PGEA/TPE were obtained via a one-pot method by ring-opening reactions of PGMA, EA with TAPc-Zn and TPE-NH2, which conferred photodynamic therapy and fluorescent imaging capabilities on the PcPGEA and TPE-PGEA/TPE while delivering the nucleic acids.
Although such a one-pot method based on ring-opening reactions of PGMA could be utilized for the fabrication of hydroxyl-rich polycationic vectors, the degradability of the polycationic vectors has not been achieved owing to the backbone of PGMA. Therefore, considering the advantages of degradable biomaterials, novel strategies of one-pot methods for preparing hydroxyl-rich polycationic vectors are necessary.
2.2 Michael addition reactions
In order to achieve the degradability of polycations, researchers have made many attempts, including the design of polyamides and polyesters.59–61 However, the preparation of these polycations requires stringent reaction conditions, such as anhydrous and oxygen-free environments. Therefore, an easy-to-implement one-step method for the preparation of degradable polycations is urgently needed. Meanwhile, Michael addition reactions have attracted attention and proved to be a promising construction strategy.62–65 Specifically, a nucleophilic system, such as amino groups, could react with an electrophilic system, like vinyl groups, via a one-pot conjugate addition reaction under relaxed conditions.66,67 Polycations with flexible structures, including linear, branched, and three-dimensional structures, could be fabricated by Michael addition reactions. In addition, degradable groups are easy to obtain by Michael addition reactions to realize the degradability of the polycations (Fig. 2).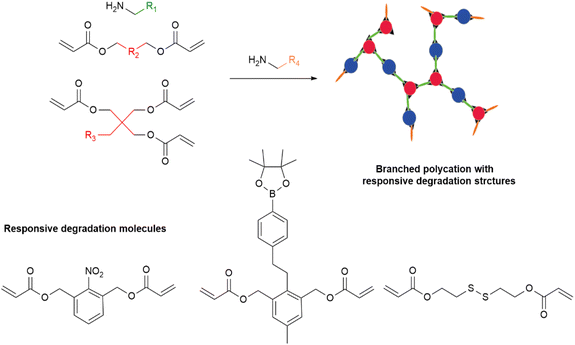 | ||
| Fig. 2 One-pot Michael addition reaction for the construction of degradable branched hydroxyl-rich polycations. | ||
Lu et al. synthesized a branched poly(β-amino ester) (PBAE) with backbone-embedded phenylboronic acid (PBA) via an amino-vinyl Michael addition reaction, which made the polycation degradable when driven by an oxidizing environment.68 Moreover, through an A2 + B3 + C3-type Michael addition reaction, Duan et al. synthesized a hydroxyl-rich polycation BPAE-NB by the use of 4-amino-1-butanol, trimethylolpropane triacrylate, and 2-nitro-1,3-phenylene bis(methylene) diacrylate. Owing to the introduction of 2-nitrobenzene moieties, the BPAE-NB backbone demonstrated good degradability when exposed to UV light, which promoted intracellular gene release and diminished the toxicity of the materials in the post-transfection state.69 Rui et al. used the bisulfide bond-containing monomer 2,2-disulfanediylbis(ethane-2,1-diyl) diacrylate (BR6) to create a series of reductively degradable quaternary polycations (rBEAQs) using a facile one-pot Michael addition reaction, and impressive nucleic acid binding affinity, cellular uptake efficiency, release performance, and functional nucleic acid delivery were proven.70
However, the Michael addition process does not yield hydroxyl groups, and hence requires the utilization of building blocks that contain hydroxyl groups, which severely limits the variety of building blocks.
In addition, many chemically active groups, such as hydroxyl, amino, and vinyl groups, are often involved in Michael addition, which means that side reactions and the production of by-products can easily occur, making the whole process inefficient. Therefore, how to expand the types of building blocks and improve the reaction efficiency has become a new problem.
2.3 Amine-epoxide ring-opening polymerization
Inspired by the Michael addition reaction, a one-pot method of amine-epoxide ring-opening polymerization was proposed.71 While retaining the amine-epoxy ring-opening reaction, this strategy abandons the polymerizations based on ATRP or RAFT, and designs building blocks containing a variety of responsive groups to achieve the degradability of the hydroxyl-rich polycation for nucleic acid delivery.72 Moreover, the terminal groups of the polycations are enriched, which is beneficial to the decoration of the vector and further improves the performance of the vectors.73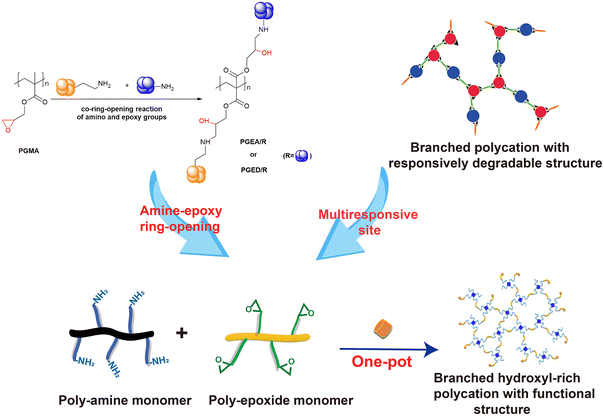 | ||
| Fig. 3 One-pot poly-amine and poly-epoxy ring-opening reaction for the construction of functionalized branched hydroxyl-rich polycations. | ||
The preparation of polycations through amine-epoxide ring-opening polymerization has been reported. Zeng et al. used the conventional aminoglycoside antibiotic gentamicin and poly(ethylene glycol) diglycidyl containing double epoxy groups via an amino-epoxy ring-opening polymerization to synthesize a branched polycation GPEG in one pot. ED as a capping agent was added when the polymerization was finished to consume the remaining epoxy groups.77 Moreover, the branched polycation TE was synthesized by the amine-epoxide ring-opening polymerization of ED and triglycidyl isocyanurate (TGIC) including three epoxy groups, which had good nucleic acid condensation ability and low cytotoxicity. Compared with the branched PEI, the optimal transfection efficiency of TE in the presence of serum was significantly higher.71
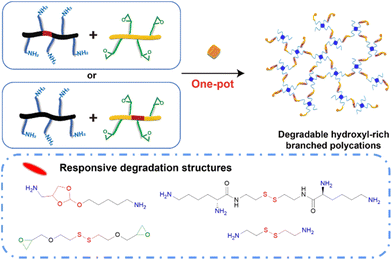 | ||
| Fig. 4 One-pot poly-amine and poly-epoxy ring-opening reaction for the construction of degradable branched hydroxyl-rich polycations. | ||
The progress of the typical responsive (redox-responsive and acid-responsive) degradable polycations for nucleic acid delivery will be introduced in detail. Huang et al. synthesized hydroxyl-rich branched polycation SS-HPs through amine-epoxide ring-opening polymerization involving disulfide-bonded HDDE and three polyaminoglycosides, which exhibited reductive degradability when used for gene therapy in glioma treatment.85 Chen et al. used lysine-decorated cystamine (CA) and TGIC to fabricate a hydroxyl-rich branched polycation via amine-epoxide ring-opening polymerization, which demonstrated reductive degradability when used for osteosarcoma therapy.86 Qi et al. used a synthetic poly-amine molecule involving an ortho ester linkage and TGIC to fabricate an acid-labile hydroxyl-rich branched polycation (ARP). Subsequently, 2-(2,2,3,3,4,4,5,5,5-nonafluoropentyl)oxirane, a kind of fluorinated epoxide molecule, was introduced to ARP via one-pot decoration to obtain ARP-F. Owing to the existence of fluorinated chains, ARP-F demonstrated impressive efficiencies of cellular uptake and transfection.87
 | ||
| Fig. 5 One-pot poly-amine and poly-epoxy ring-opening reaction for the construction of post modified branched hydroxyl-rich polycations. | ||
Several functional molecules have been used in the synthesis of hydroxyl-rich branched polycations through one-pot decoration. Wu et al. fabricated a hepatic stellate cell (HSC)-targeted hydroxyl-rich branched polycation named SS-HPVA by one-pot decoration of SS-HPT with the introduction of vitamin A (VA), leading to a significant accumulation of polycations within the liver.73 Shao et al. utilized two functional molecules, Rose Bengal (RB, an efficient photosensitizer) and APBA, to fabricate a bifunctional hydroxyl-rich branched polycation named HPT-RP. The introduction of RB and PBA could endow HPT-RP with the potential for receptor-mediated photodynamic therapy (PDT), which would offer potent antibacterial ability for infected wound treatment.88 Qi et al. created a reduction-responsive hydroxyl-rich branched polycation named LBP by using amine-epoxide ring-opening polymerization of poly-amine lactose with disulfide-bonded HDDE. Owing to the existence of lactose and disulfide groups, LBP possessed hepatocyte targeting ability and reduction-responsively degradability. In addition, the hydroxyl groups of LBP provided additional hydrophilicity for blood compatibility.89
In summary, amine-epoxide ring-opening polymerization can be carried out under mild reaction conditions, responsive moieties can be flexibly introduced into the building blocks through molecular design to achieve the degradability of the polycations, and the abundant active groups at the ends of the polycation provide a guarantee for further functionalization of the vector. Next, we summarize the different application scenarios of hydroxyl-rich polycations prepared by amine-epoxide ring-opening polymerization as nucleic acid delivery vectors.
3. Various scenarios for hydroxy-rich polycation application
With the continuous development of research, the application of nucleic acid delivery for disease treatment has gained widespread attention and shown good therapeutic effects. Based on the hydroxyl-rich polycations constructed by a one-pot method, researchers have innovatively designed several nucleic acid delivery systems with multiple functions against different application scenarios, such as malignant tumors, diabetes, and epidermal wounds. In this chapter, recent progress regarding the application of hydroxyl-rich polycations fabricated by a one-pot method is described.3.1 Degradable nucleic acid delivery systems for cancer therapy
As cancer is a fatal disease, researchers have been exploring different treatment methods for malignant tumors and gene therapy has been proven to be an effective strategy.90–94 Degradable hydroxy-rich polycations are widely used in nucleic acid delivery systems for tumor therapy due to their multiple responsiveness to the tumor microenvironment (TME).95–97 The TME is a special physiological environment required for tumor cell growth, and its salient features are higher glutathione (GSH) expression levels98,99 and a more acidic intracellular environment (pH: 5.0–5.5)100–102 compared with normal cell locations. As previously introduced, hydroxyl-rich polycations can be introduced with various responsive moieties via design of the building blocks, so responsive degradability can be successfully achieved influenced by the physiological microenvironment of tumor cells.103–105 The degradability of polycations is conducive to realizing the release of nucleic acids at the site of action, and greatly reduces the toxicity of their side effects on the organism.Reduction and acid-responsive degradable nucleic acid delivery systems have been mainly constructed. Huang et al. prepared several hydroxyl-rich branched polycations (SS-HPs) by amine-epoxide ring-opening reaction of disulfide-containing bis-epoxy compounds with aminoglycosides containing multiple amino groups (Fig. 6a).85 As verified by agarose gel electrophoresis assays, the SS-HPs showed an excellent degradation profile, which is attributed to the introduction of disulfide bonds (Fig. 6b). As shown in Fig. 6c, the SS-HPs demonstrated higher gene transfection efficiency in different cell lines than the un-disulfide HPs and branched PEI, which was mainly attributed to the reduction-responsive disulfide bonds in the SS-HP. Because of their reduction responsive degradability, SS-HPs can easily realize the release of nucleic acids. Subsequently, the classical anticancer gene p53 was further selected as the therapeutic nucleic acid to evaluate the in vivo antitumor performance of the SS-HPs (Fig. 6d). The results showed that the SS-HPs have the ability to efficiently deliver the p53 gene, and due to the reduction-sensitive property of the disulfide bonds, the p53 gene was effectively released into the cytoplasm. SS-HPs/p53 complexes significantly inhibited the growth of C6 glioma and did not produce significant systemic toxicity during treatment.
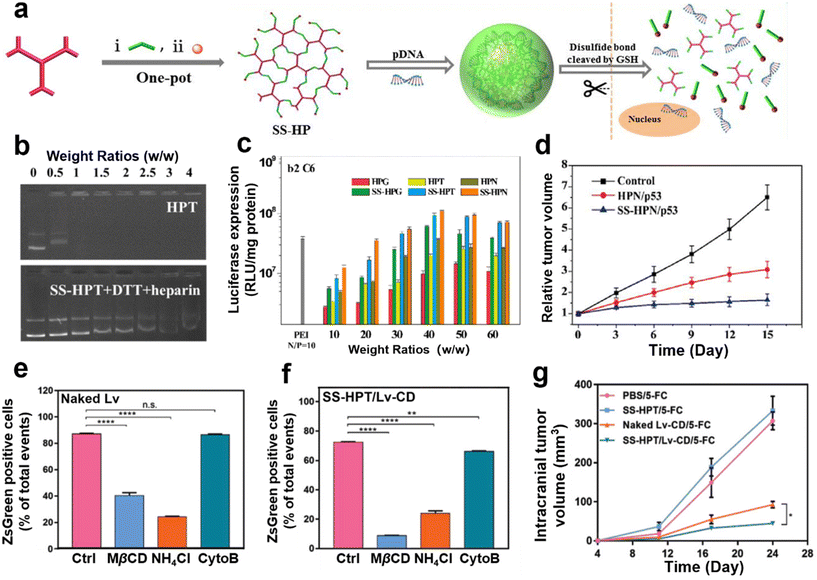 | ||
| Fig. 6 (a) Schematic of the preparation of multifunctional aminoglycoside degradable branched polycations (HPs). (b) Electrophoretic mobility of HPs/pDNA in a reduced environment with and without DTT simulation, exemplified by SS-HPN. (c) Transfection performance assay of HPs/pDNA in C6 cells. (d) Tumor volume curves in tumor-bearing mice after different groups of treatment. Reprinted with permission from ref. 85. Copyright 2016, Elsevier. (e) Quantitative FACS analysis of naked Lv and (f) SS-HPT/Lv-20 mediated reporter gene ZsGreen expression in C6 cells after treatment with different endocytosis inhibitors. (g) Tumor volume curves of tumor-bearing rats during different treatment groups. Reprinted with permission from ref. 106. Copyright 2019, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
Moreover, an SS-HP/Lv-CD system was constructed by coating CD suicide gene-engineered lentivirus (Lv-CD) with SS-HP for use against glioma by Shao et al.,106 which showed good biocompatibility. Cellular uptake assays by using C6 glioma cells showed that SS-HP/Lv could enter into the cells efficiently and release Lv particles in a wider range of pathways than exposed Lv (Fig. 6e and f). In addition, SS-HP/Lv demonstrated significantly higher transfection efficiency, which led to the expression of more CD proteins, further enabling SS-HPT/Lv-CD to significantly increase the sensitivity of glioma cells to 5-fluorocytosine (5-FC) (Fig. 6g). Furthermore, by evaluating the tumor volume and survival time of mice after treatment for glioma in situ, the results demonstrated that SS-HP/Lv-CD with 5-FC most effectively inhibited tumor growth and prolonged the survival time of the mice.
Encouraged by the above work, more versatile therapeutic nucleic acid delivery systems were constructed to achieve efficient treatment of cancer. Chen et al.86 modified biomolecule lysine with cystamine to synthesize SS-Lys, a polyamine compound with a disulfide bond. Following amine-epoxide ring-opening reaction of SS-Lys with TGIC, a reduction-responsive degradable hydroxyl-rich branched polycation named SS-LHP was fabricated. A branched polylysine gene vector (CC-HP) without disulfide bonds was used as a control. pKillerRed gene (pKR) was delivered by the polycation to estimate antitumor capability of the nucleic acid delivery system. KillerRed protein42,107 is a red fluorescent protein that can generate singlet oxygen (1O2) under visible light irradiation, which further enables the effects of PDT (Fig. 7a). Gel electrophoresis and atomic force microscopy were used to assess the degradation of the SS-LHP/pDNA complexes in a reducing environment. Compared to the non-degradable polycation CC-HP, SS-LHP was unable to encapsulate pDNA and SS-HP/pDNA did not exhibit a homogeneous and regular nanostructure in a reductive environment because of the existence of disulfide bonds. Owing to the degradability of polycations, the accelerated release of pDNA and good transfection efficiency were proved. Finally, SS-LHP carrying pKR plasmid was used for in vitro photodynamic anticancer evaluation. SS-HP/pKR transfected C6 glioma cells successfully expressed KillerRed protein and promoted apoptosis of tumor cells after light exposure (Fig. 7b).
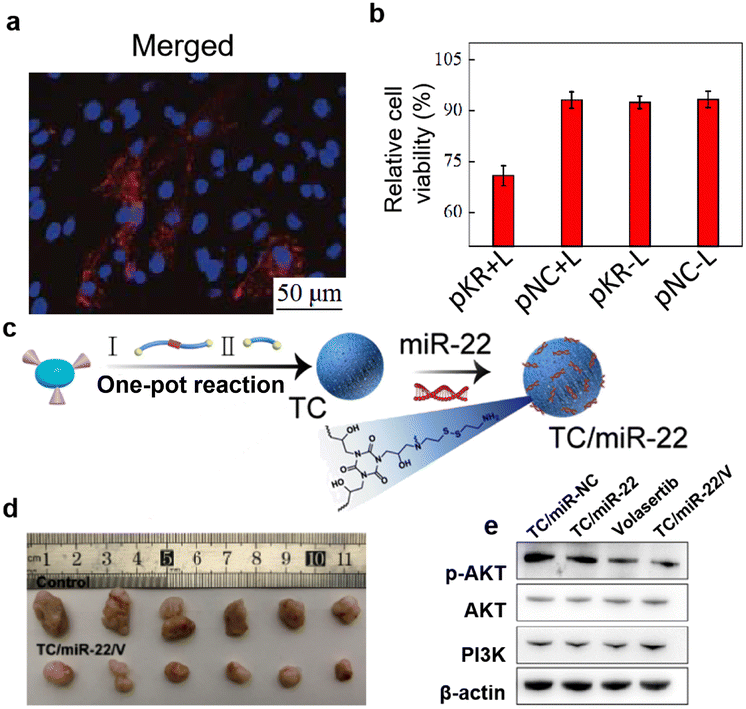 | ||
| Fig. 7 (a) Representative images of SS-HP-mediated KillerRed expression in C6 cells. (b) Relative cellular activity of SS-HP/pKR and SS-HP/pNC in C6 cells in the presence or absence of light. Reprinted with permission from ref. 86. Copyright 2019, Science Press. (c) Preparation process of reductively reactive polycationic TC with TC/miR-22. (d) Tumor images of drug-resistant PDX model mice after treatment with different treatment groups. (e) Relative expression of p-AKT protein in Saos-2 cells with different treatment groups. Reprinted with permission from ref. 108. Copyright 2023, KeAi Publishing. | ||
Osteosarcoma (OS) is the most common primary malignant bone tumor and is highly metastatic. Expression of miR-22 is significantly downregulated during OS development, making miR-22 a promising therapeutic target for OS. A novel class of reduction-sensitive hydroxyl-rich polycationic vector (TC) was constructed by a one-pot method through anime-epoxide ring-opening reaction of the epoxy group of cystamine with TGIC to deliver miR-22 and miR-30d, respectively, for the treatment of OS (Fig. 7c ).108 Due to its abundant secondary amino and hydroxyl groups, TC has good nucleic acid condensation ability and biocompatibility. The hydrated particle size of TC particles increased when treated with DTT, indicating that TC had a good reduction-responsive degradability. Moreover, TC showed good transfection performance in the OS cell line (Saos-2) and biocompatibility in the osteoblast cell line (MC3T3-E1). By inhibiting the PI3K/AKT pathway, the use of TC-mediated miR-22 transfection of Saos-2 cells achieved good anti-tumor effects in vitro, not only inhibiting the proliferation and migration of Saos-2 cells, but also inducing the promotion of apoptosis of Saos-2 cells. Combined with the anticancer drug Volasertib treatment, TC/miR-22 complexes were able to enhance the efficacy of treatment. In vivo results confirmed that TC/miR-22 achieved excellent anti-tumor effects and effectively reduced lung metastasis of tumor cells in both in situ OS and patient-derived xenograft (PDX) models. To investigate the improvement of the efficacy of chemotherapy-resistant OS, a drug-resistant PDX model was established. In the treatment of TC/miR-22/Volasertib, increasing the dosage of miRNA was not only able to inhibit the growth of drug-resistant tumors (Fig. 7d), but also did not produce significant cytotoxicity. At the end of treatment, in order to investigate the mechanism of action of TC/miR-22, western blot assay was used to analyze the regulation of AKT protein phosphorylation by TC/miR-22 in the PI3K/AKT pathway of Saos-2 cells, and it was found that both TC/miR-22 and Volasertib could effectively inhibit the expression of AKT protein, thus affecting the PI3K/AKT pathway (Fig. 7e).
3.2 Targeted nucleic acid delivery systems for disease therapy
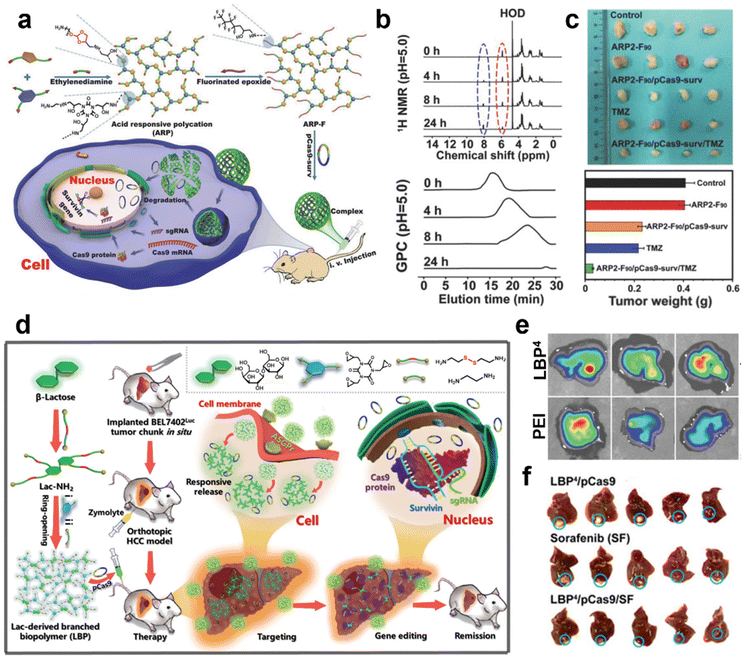 | ||
| Fig. 8 (a) Preparation of branched polycationic ARP-F with its in vivo delivery of nucleic acids with delivery of pCas9-surv for gene editing. (b) Degradation properties of ARP-F in vitro under simulated acidic conditions. (c) In vivo assay of ARP-F/pCas9-surv against mouse tumors. Reprinted with permission from ref. 87. Copyright 2018, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (d) Preparation of a lactose-derived branched biopolymer (LBP) and schematic representation of the delivery and gene editing process of pCas9-survivin for the treatment of HCC in situ. (e) Representative fluorescence images of the liver in the LBP-treated group. (f) Representative liver images for each treatment group. Reprinted with permission from ref. 89. Copyright 2020, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
In the process of gene therapy, how to make nucleic acids reach target cells without interfering with other normal cell physiological behaviors109–111 is a key scientific issue. Therefore, in addition to being degradable to ensure its biological safety, the nucleic acid delivery vector also needs to have the ability to target the cells of the lesion. As for the method of introducing targeting molecules into vectors, the aforementioned one-pot decoration method is widely used due to its simplicity of implementation and high efficiency of reaction.
According to previous research, the boronic acid group has affinity to the nucleus, showing a good tumor cell uptake efficiency and nucleus targeting ability. Shao et al. decorated a PBA moiety on a hydroxyl-rich branched polycation (SS-HPT) to get a PBA-functionalized branched polyaminoglycoside (SS-HPT-P).112 In the fluorescent imaging of tumor model animals, it was observed that the SS-HPT-P/pCas9-surv complexes were rapidly enriched into tumors due to the good targeting ability of PBA. The typical pCas9-surv delivery mediated by SS-HPT-P shows good gene editing performance. Due to the targeting ability of PBA, SS-HPT-P/pCas9-surv is significantly effective in inhibiting the proliferation and migration of lung tumor cells in vitro and in vivo when combined with TMZ.
In addition, lactose, as a common natural disaccharide, composed of one glucose molecule and one galactose molecule, is able to specifically bind to the asialoglycoprotein receptor (ASGPr) on the surface of hepatic carcinoma cells (HCCs). Qi et al.89 prepared a reduced-responsive hydroxyl-rich branched polycationic vector named LBP via amine-epoxide ring-opening polymerization of aminated lactose with TGIC for the treatment of in situ hepatic carcinoma (Fig. 8d). LBP demonstrated significant enrichment at the liver site via in vivo imaging characterization (Fig. 8e). Subsequently, in vitro and in vivo anti-tumor experiments demonstrated that the LBP-mediated CRISPR/Cas9 system (LBP/pCas9-survivin) not only effectively inhibits the proliferation of HCCs, but also accelerates cell apoptosis through effective gene editing. LBP/pCas9-survivin gene editing also enhances the anti-tumor effect and sensitizes the anticancer drugs (Fig. 8f).
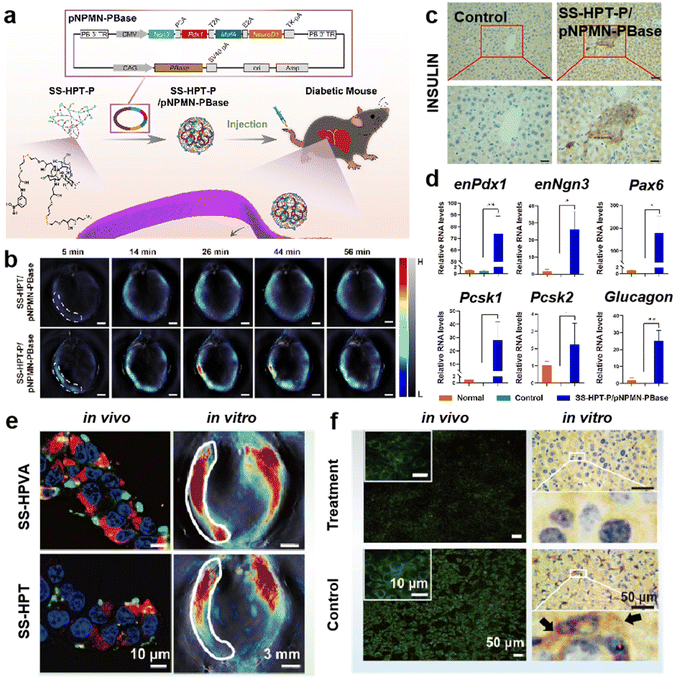 | ||
| Fig. 9 (a) Schematic illustration of the preparation process of hydroxyl-rich branched polycation SS-HPT-P, and SS-HPT-P/pNPMN-PBase targeting the liver of diabetic mice for gene reprogramming therapy. (b) Representative MSOT cross-sectional images of mice at different times. (c) Representative images of immunohistochemical staining of liver insulin. The images are observed upstream at 1000 times magnification and downstream at 2000 times magnification. (d) Quantitative real-time detection of mRNA expression of pancreatic transcription factors and insulin expression/secretion-related genes after treatment in different groups. Reprinted with permission from ref. 116. Copyright 2022, Elsevier. (e) Endocytosis fluorescence imaging of SS-HPVA/pDNA with the SS-HPT/pDNA complex in HSCs (left) and representative cross-sectional photoacoustic images of liver fibrosis model mice after tail vein injection of SS-HPT/pshRNA-TGFβ1 or SS-HPVA/pshRNA-TGFβ1. (f) Immunofluorescence imaging of TGFβ1 cells after different treatments (left) and immunohistochemical sections (right). Reprinted with permission from ref. 73. Copyright 2023, Elsevier. | ||
Liver fibrosis is a repair response to liver injury and is a key aspect in several liver diseases. Recent research studies revealed that the pro-fibrotic cytokine transforming growth factor (TGFβ1) is involved in almost all critical dimensions of liver fibrosis development. Therefore, down-regulating TGFβ1 expression has the potential to treat liver fibrosis. Wu et al.73 constructed a targeting hydroxyl-rich branched polycationic vector (SS-HPVA) based on the one-pot decoration of branched polyaminoglycoside (SS-HPT). As a HSC targeting molecule, VA was introduced to the SS-HPT. Multispectral photoacoustic tomography and in vivo fluorescent imaging verified that the SS-HPVA/pshRNA-TGFβ1 complexes demonstrated superior liver targeting ability, especially in the hepatic fibrosis model (Fig. 9e). On this basis, it was demonstrated that this system could exhibit good therapeutic effects on liver fibrosis by down-regulating TGFβ1 expression through non-invasive assays and histopathological analysis (Fig. 9f). Therefore, this gene silencing targeting delivery system is expected to provide a new strategy for the clinical treatment of liver fibrosis.
3.3 Nucleic acid delivery systems for antibacterials and wound healing
Growth factor delivery has attracted high attention for the treatment of severe skin wounds and the promotion of wound healing.117,118 Thus, Chen et al. developed a hydroxyl-rich branched polycation (SKP) via amine-epoxide polymerization of disulfide bond-modified lysine (SK) and TGIC.119 A transfection assay in the human umbilical vein endothelial cell line (HUVEC) indicated that SKP demonstrated good transfection efficiency compared with PEI. When vascular endothelial growth factor (VEGF) plasmid was delivered by SKP, it was further found that the SKP/pVEGF complexes successfully increase cell proliferation, and a western blot assay detected higher VEGF expression compared with the control group. Moreover, the SKP/pVEGF complexes could be coated on a pre-treated clinical stent to produce a pVEGF-eluting stent (S-SKP/pVEGF). The in vivo re-endothelialization and anti-restenosis performance of S-SKP/pVEGF were evaluated via implanting stents into rabbit abdominal aortas.Hydroxyl-rich branched polycations have also achieved antimicrobial activities and wound healing promotion. Aminoglycosides and their derivatives are active antimicrobial agents. A series of hydroxyl-rich reduction-responsive polycations (SS-HPG, SS-HPT and SS-HPN) synthesized based on aminoglycosides, including gentamicin, tobramycin, and neomycin, have been reported.85 Moreover, an Eosin Y (EY)-based antibacterial polycation (EY-QEGED-R, R = CH3 or C6H13) with versatile types of functional components including quaternary ammonium, photosensitizer, primary amino, and hydroxyl species was readily synthesized by Zhu et al. based on amine-epoxide ring-opening reactions (Fig. 10a).120 A remarkable synergistic antibacterial activity of the polycation owing to the combined photodynamic and quaternary ammonium antibacterial effects was proven (Fig. 10b). EY-QEGED-R also could be readily coated on nonwoven fabrics via an adhesive layer of polydopamine demonstrating antifouling capability against dead bacteria. Notably, EY-QEGED-R more significantly inhibited wound inflammation and promoted wound healing on an infected rat model under light conditions (Fig. 10c).
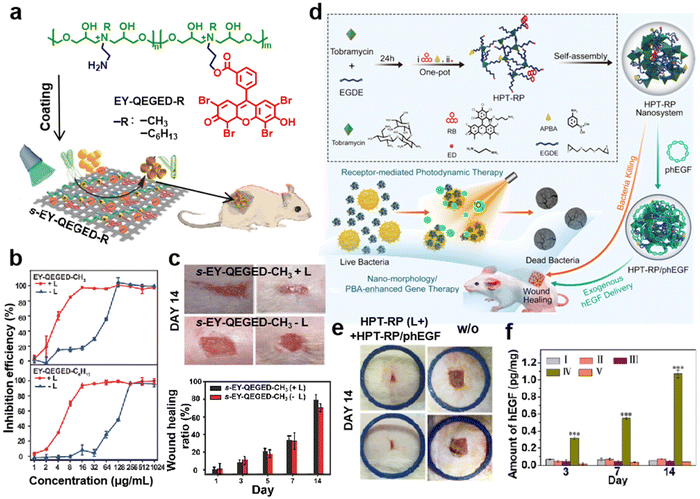 | ||
| Fig. 10 (a) Construction of EY-based antimicrobial polycations (EY-QEGED-R, R = CH3 or C6H13) by amino-epoxy ring-opening reaction and their antimicrobial properties after light exposure for wound dressings. (b) The inhibition effect of the antimicrobial polycations on S. aureus in the presence or absence of light. (c) Representative images and ratio of wound healing of a rat wound model on day 1 and day 14 after different groups of treatments. Reprinted with permission from ref. 120. Copyright 2018, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (d) Schematic of the preparation of HPT-RP/phEGF. (d) Bacteriostatic effect of HPT, HPT-R and HPT-RP in the presence or absence of light in the case of S. aureus. (e) Representative pictures of the rat infected-skin-defect model on day 14 after different groups of treatments. (f) Quantification of the expression levels F of the excised tissues around the wounds at day 3, 7 and 14 after treatment in different groups. Group I–V are: Control, HPT-RP(L-), HPT-RP(L+), HPT-RP(L+) + HPT-RP/phEGF and w/o. Reprinted with permission from ref. 88. Copyright 2021, Elsevier. | ||
Therefore, it is necessary to develop new delivery systems based on hydroxyl-rich branched polycations with a photosensitizer that have superiority for antibacterial, wound healing promotion and delivery performances. One bifunctional hydroxyl-rich branched polycation (HPT-RP) was constructed through the one-pot dual conjugation of a photosensitizer and bacterium/cell-affinity phenylboronic acid onto hyperbranched polyaminoglycosides, achieving antimicrobial properties while realizing its application to encoding epidermal growth factor (EGF) plasmid delivery (Fig. 10d).88 HPT-RP realized high anti-bacterial ability owing to the receptor-mediated photodynamic therapy, and impressive gene transfection efficiency because of phenylboronic acid conjugation. In a rat infected-skin-defect model, HPT-RP exhibited the remarkable performance of anti-infective/EGF-delivery and accelerated healing (Fig. 10e). More importantly, a new insight into rat-EGF production revealed the underlying mechanism of extraneous EGF delivery and endogenous EGF production (Fig. 10f).
4. Conclusions
Owing to their convenient fabrication process and flexible designability, one-pot methods have shown promising potential in the construction of hydroxyl-rich polycationic vectors for nucleic acid delivery. In this brief review, we mainly summarized three one-pot methods for the synthesis of hydroxyl-rich polycations for nucleic acid delivery: co-ring-opening reactions of PGMA, Michael addition reactions, and amine-epoxide ring-opening polymerization. Notably, amine-epoxide ring-opening polymerization as a novel approach was mainly introduced. In addition, various therapeutic scenarios used by multifunctional hydroxyl-rich polycation applications, including cancer, diabetes mellitus, liver fibrosis, antibacterial and wound healing treatments, were also generalized. We believe that this review will shed light on the design of hydroxyl-rich polycations and expand the biological applications of cationic gene carriers in the field of nucleic acid delivery.Author contributions
Mengrui Su and Zichen Hu performed the literature survey and edited the original draft; Yu Qi edited the original manuscript and made original draft revision; Bingran Yu and Fu-Jian Xu conceived the theme of this review, made the original draft revision, and supervised the work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (grant number 2021YFB3800900), National Natural Science Foundation of China (grant numbers 52073013, 22122501, 52221006, and 22005019), Beijing Natural Science Foundation (grant number 7222011), Beijing Outstanding Young Scientist Program (BJJWZYJH01201910010024), and Beijing Postdoctoral Research Foundation (2022-6ZZ-113).References
- X. M. Cai, R. Dou, C. Guo, J. R. Tang, X. J. Li, J. Chen and J. Y. Zhang, Pharmaceutics, 2023, 15, 1502 CrossRef CAS PubMed.
- S. A. Dilliard and D. J. Siegwart, Nat. Rev. Mater., 2023, 8, 282–300 CrossRef CAS PubMed.
- A. Gupta, J. L. Andresen, R. S. Manan and R. Langer, Adv. Drug Delivery Rev., 2021, 178, 113834 CrossRef CAS PubMed.
- Z. G. Lu, J. Shen, J. Yang, J. W. Wang, R. C. Zhao, T. L. Zhang, J. Guo and X. Zhang, Signal Transduction Targeted Ther., 2023, 8, 39 CrossRef CAS PubMed.
- A. S. Piotrowski-Daspit, A. C. Kauffman, L. G. Bracaglia and W. M. Saltzman, Adv. Drug Delivery Rev., 2020, 156, 119–132 CrossRef CAS PubMed.
- Y. Tian, M. V. Tirrell and J. L. LaBelle, Adv. Healthcare Mater., 2022, 11, 2102600 CrossRef CAS.
- Y. Y. Wang, P. K. Shahi, R. S. Xie, H. L. Zhang, A. A. Abdeen, N. Yodsanit, Z. Q. Ma, K. Saha, B. R. Pattnaik and S. Q. Gong, J. Controlled Release, 2020, 324, 194–203 CrossRef CAS PubMed.
- K. Isgrig, D. S. McDougald, J. L. Zhu, H. J. Wang, J. Bennett and W. W. Chien, Nat. Commun., 2019, 10, 427 CrossRef CAS.
- Y. Kim, D. Y. Lee, J. U. Choi, J. S. Park, S. M. L. ee, C. H. Kang and C. H. Park, Sci. Rep., 2023, 13, 12365 CrossRef CAS.
- H. E. Maunder, J. Wright, B. R. Kolli, C. R. Vieira, T. T. Mkandawire, S. Tatoris, V. Kennedy, S. Iqball, G. Devarajan, S. Ellis, Y. Lad, N. G. Clarkson, K. A. Mitrophanous and D. C. Farley, Nat. Commun., 2017, 8, 14838 CrossRef PubMed.
- Y. A. Aksoy, B. Y. Yang, W. J. Chen, T. Hung, R. P. Kuchel, N. W. Zammit, S. T. Grey, E. M. Goldys and W. Deng, ACS Appl. Mater. Interfaces, 2020, 12, 52433–52444 CrossRef CAS.
- J. W. Du, Y. Shao, Y. Hu, Y. W. Chen, J. H. Cang, X. Chen, W. Q. Pei, F. Q. Miao, Y. Q. Shen, M. Muddassir, Y. Zhang, J. Q. Zhang and G. J. Teng, ACS Appl. Mater. Interfaces, 2021, 13, 23396–23409 CrossRef CAS PubMed.
- Y. Cheng, R. C. Yumul and S. H. Pun, Angew. Chem., Int. Ed., 2016, 55, 12013–12017 CrossRef CAS.
- M. M. Khan, N. Filipczak and V. P. Torchilin, J. Controlled Release, 2021, 330, 1220–1228 CrossRef CAS.
- E. M. McErlean, M. Ziminska, C. M. McCrudden, J. W. McBride, S. P. Loughran, G. Cole, E. J. Mulholland, V. Kett, N. E. Buckley, T. Robson, N. J. Dunne and H. O. McCarthy, J. Controlled Release, 2021, 330, 1288–1299 CrossRef CAS PubMed.
- Y. Wang, Y. Qi, X. D. Guo, K. Zhang, B. R. Yu and F.-J. Xu, J. Mater. Chem. B, 2023, 11, 4121–4130 RSC.
- D. X. Yin, M. J. Zhang, J. X. Chen, Y. Y. Huang and D. H. Liang, Chin. Chem. Lett., 2021, 32, 1731–1736 CrossRef CAS.
- Q. Q. Fan, Z. Yang, Y. H. Li, Y. Y. Cheng and Y. W. Li, Adv. Funct. Mater., 2021, 31, 2101646 CrossRef CAS.
- J. H. Li, J. Qian, Y. Y. Xu, S. Yan, J. Shen and M. Z. Yin, ACS Sustainable Chem. Eng., 2019, 7, 6316 CrossRef CAS.
- N. Zhang, Y. Wang, R. Wu, C. Xu, J.-J. Nie, N. N. Zhao, B. R. Yu, Z. J. Liu and F.-J. Xu, Small, 2019, 15, 1904017 CrossRef CAS.
- Q. Zhang, C. N. Shen, N. Zhao and F.-J. Xu, Adv. Funct. Mater., 2017, 27, 1606229 CrossRef.
- J. Zhao, F. Han, P. Zhao, X. J. Wen and C. Lin, J. Mater. Chem. B, 2017, 5, 6119–6127 RSC.
- N. N. Zhao, W. L. Fan, X. Y. Zhao, Y. J. Liu, Y. Hu, F. Duan and F.-J. Xu, ACS Appl. Mater. Interfaces, 2020, 12, 11341–11352 CrossRef CAS PubMed.
- X. Y. Zhao, K. L. Guo, K. Zhang, S. Duan, M. W. Chen, N. N. Zhao and F.-J. Xu, Adv. Mater., 2022, 34, 2108263 CrossRef CAS PubMed.
- J. Chen, K. Wang, J. Y. Wu, H. Y. Tian and X. S. Chen, Bioconjugate Chem., 2019, 30, 338–349 CrossRef CAS.
- H. Fang, Z. Guo, L. Lin, J. Chen, P. Sun, J. Wu, C. Xu, H. Tian and X. Chen, J. Am. Chem. Soc., 2018, 140, 11992–12000 CrossRef CAS.
- H. M. Liu, H. Wang, W. J. Yang and Y. Y. Cheng, J. Am. Chem. Soc., 2012, 134, 17680–17687 CrossRef CAS.
- Y. Y. Pu, H. H. Yin, C. H. Dong, H. J. Xiang, W. C. Wu, B. G. Zhou, D. Du, Y. Chen and H. X. Xu, Adv. Mater., 2021, 33, 2104641 CrossRef CAS.
- C. N. Wu, J. H. Li, W. D. Wang and P. T. Hammond, ACS Nano, 2018, 12, 6504–6514 CrossRef CAS.
- Y. L. Cheng, D. L. Sellers, J. K. Y. Tan, D. J. Peeler, P. J. Horner and S. H. Pun, Biomaterials, 2017, 127, 89–96 CrossRef CAS.
- Q. Yang, S. Liu, X. Liu, Z. H. Liu, W. Xue and Y. Zhang, Acta Biomater., 2019, 96, 436–455 CrossRef CAS.
- C. Zhao, L. Shao, J. Lu, X. Deng and Y. Wu, ACS Appl. Mater. Interfaces, 2016, 8, 6400–6410 CrossRef CAS PubMed.
- Y. J. Feng, Z. P. Guo, J. Chen, S. J. Zhang, J. Y. Wu, H. Y. Tian and X. S. Chen, Biomater. Sci., 2022, 10, 6230–6243 RSC.
- W. C. Zhang, Y. X. Zhang, Y. L. Luo, S. B. Chen, Q. Y. Huang, Z. T. Cao, M. Liang and X. Z. Yang, Nano Today, 2022, 43, 101418 CrossRef CAS.
- N. Zheng, X. Q. Luo, Z. Y. Zhang, A. G. Wang and W. Z. Song, ACS Appl. Mater. Interfaces, 2021, 13, 27513–27521 CrossRef CAS PubMed.
- Y. L. Cheng, H. Wei, J. K. Y. Tan, D. J. Peeler, D. O. Maris, D. L. Sellers, P. J. Horner and S. H. Pun, Small, 2016, 12, 2750–2758 CrossRef CAS PubMed.
- S. T. G. Street, H. C. Parkin, L. Shopperly, J. Chrenek, K. Letwin, S. M. Willerth and I. Manners, Biomater. Sci., 2023, 11, 3512–3523 RSC.
- E. C. Tan, J. Lv, J. J. Hu, W. W. Shen, H. Wang and Y. Y. Cheng, J. Mater. Chem. B, 2018, 6, 7230–7238 RSC.
- B. C. Jiang, H. He, L. Yao, T. Zhang, J. P. Huo, W. Sun and L. C. Yin, Biomacromolecules, 2017, 18, 877–885 CrossRef CAS.
- Y. Jin, W. Yu, W. K. Zhang, C. Wang, Y. Liu, W. E. Yuan and Y. Feng, J. Colloid Interface Sci., 2023, 648, 287–298 CrossRef CAS.
- Z. W. Zhou, M. H. Zhang, Y. D. Liu, C. Z. Li, Q. Y. Zhang, D. Oupicky and M. J. Sun, Biomacromolecules, 2018, 19, 3776–3787 CrossRef CAS PubMed.
- C. Xu, W. T. Hu, N. Zhang, Y. Qi, J.-J. Nie, N. N. Zhao, B. R. Yu and F.-J. Xu, Biomaterials, 2020, 248, 120031 CrossRef CAS.
- J. Y. Zhu, X. Zeng, S. Y. Qin, S. S. Wan, H. Z. Jia, R. X. Zhuo, J. Feng and X. Z. Zhang, Biomaterials, 2016, 83, 79–92 CrossRef CAS.
- J. Kim, Y. C. Kang, S. Y. Tzeng and J. J. Green, Acta Biomater., 2016, 41, 293–301 CrossRef CAS PubMed.
- A. M. Martinez, R. H. F. V. de Souza, M. S. Petronio, G. O. Martins, J. C. Fernandes, M. Benderdour, V. A. O. de Tiera and M. J. Tiera, J. Nanostruct. Chem., 2022, 1–20 Search PubMed.
- G. Ahn, S. Kweon, C. Yang, J. E. Hwang, K. Kim and B. S. Kim, J. Polym. Sci., Part A: Polym. Chem., 2017, 55, 4013–4019 CrossRef CAS.
- X. F. Pan, Y. Y. Liu, Z. J. Li, S. D. Cui, H. Gebru, J. X. Xu, S. Q. Xu, J. Q. Liu and K. Guo, Macromol. Chem. Phys., 2017, 218, 1600483 CrossRef.
- P. F. Song, Y. L. Chen, Y. L. Li, J. P. Ma, L. Y. Wang and R. M. Wang, Macromol. Rapid Commun., 2020, 41, 2000436 CrossRef CAS.
- Y. Zhao, B. R. Yu, H. Hu, Y. Hu, N. N. Zhao and F.-J. Xu, ACS Appl. Mater. Interfaces, 2014, 6, 17911–17919 CrossRef CAS.
- Y. Zhi, C. Xu, D. D. Sui, J. Du, F.-J. Xu and Y. L. Li, Adv. Sci., 2019, 6, 1900023 CrossRef.
- Y. J. Huang, H. Hu, R. Q. Li, B. R. Yu and F.-J. Xu, ACS Appl. Mater. Interfaces, 2016, 8, 3919–3927 CrossRef CAS.
- J.-J. Nie, B. K. Qiao, S. Duan, C. Xu, B. Y. Chen, W. J. Hao, B. R. Yu, Y. L. Li, J. Du and F.-J. Xu, Adv. Mater., 2018, 30, 1801570 CrossRef.
- Y. L. Ren, R. Q. Li, Y. R. Cai, T. Xia, M. Yang and F.-J. Xu, Adv. Funct. Mater., 2016, 26, 7314–7325 CrossRef CAS.
- C. Xu, B. R. Yu, H. Hu, M. N. Nizam, W. Yuan, J. Ma and F.-J. Xu, Biomater. Sci., 2016, 4, 1233–1243 RSC.
- C. Xu, Y. Z. Z. Zhang, K. Xu, J.-J. Nie, B. R. Yu, S. J. Li, G. Cheng, Y. L. Li, J. Du and F.-J. Xu, Nat. Commun., 2019, 10, 3184 CrossRef PubMed.
- R. Q. Li, H. Q. Song and F.-J. Xu, Polym. Chem., 2015, 6, 6208–6218 RSC.
- Y. Qi, C. Xu, M. N. Nizam, Y. Li, B. R. Yu and F.-J. Xu, Polym. Chem., 2016, 7, 5630–5640 RSC.
- Y. Sun, H. Hu, N. N. Zhao, T. Xia, B. R. Yu, C. A. Shen and F.-J. Xu, Biomaterials, 2017, 117, 77–91 CrossRef CAS.
- L. Rotolo, D. Vanover, N. C. Bruno, H. E. Peck, C. Zurla, J. Murray, R. K. Noel, L. O'Farrell, M. Arainga, N. Orr-Burks, J. Y. Joo, L. C. S. Chaves, Y. H. Jung, J. Beyersdorf, S. Gumber, R. Guerrero-Ferreira, S. Cornejo, M. Thoresen, A. K. Olivier, K. T. M. Kuo, J. C. Gumbart, A. R. Woolums, F. Villinger, E. R. Lafontaine, R. J. Hogan, M. G. Finn and P. J. Santangelo, Nat. Mater., 2023, 22, 369 CrossRef CAS.
- L. Ye, H. M. Liu, X. Fei, D. Ma, X. Z. He, Q. Y. Tang, X. Zhao, H. B. Zou, X. J. Chen, X. M. Kong and P. F. Liu, Nano Res., 2022, 15, 1135–1144 CrossRef CAS.
- C. N. Zhang, R. Jin, P. Zhao and C. Lin, Acta Biomater., 2015, 22, 120–130 CrossRef CAS PubMed.
- D. M. Lynn and R. Langer, J. Am. Chem. Soc., 2000, 122, 10761–10768 CrossRef CAS.
- Y. Liu, Y. F. Li, D. Keskin and L. Q. Shi, Adv. Healthcare Mater., 2019, 8, 1801359 CrossRef PubMed.
- Y. Rui, D. R. Wilson, J. Choi, M. Varanasi, K. Sanders, J. Karlsson, M. Lim and J. J. Green, Sci. Adv., 2019, 5, eaay3255 CrossRef CAS.
- Q. Y. Zhu, H. M. Shen, Z. J. Yang and H. B. Ji, Chin. J. Catal., 2016, 37, 1227–1234 CrossRef CAS.
- W. R. Cheng, D. C. Wu and Y. Liu, Biomacromolecules, 2016, 17, 3115–3126 CrossRef CAS PubMed.
- B. D. Mather, K. Viswanathan, K. M. Miller and T. E. Long, Prog. Polym. Sci., 2006, 31, 487–531 CrossRef CAS.
- R. J. Lu, Y. J. Zheng, M. R. Wang, J. H. Lin, Z. Y. Zhao, L. Chen, J. H. Zhang, X. Liu, L. C. Yin and Y. B. Chen, Acta Biomater., 2022, 152, 355–366 CrossRef CAS PubMed.
- S. Z. Duan, D. S. Cao, X. D. Li, H. F. Zhu, M. Lan, Z. Z. Tan, Z. Y. Song, R. Y. Zhu, L. C. Yin and Y. B. Chen, Biomater. Sci., 2020, 8, 290–301 RSC.
- Y. Rui, D. R. Wilson, K. Sanders and J. J. Green, ACS Appl. Mater. Interfaces, 2019, 11, 10472–10480 CrossRef CAS.
- S. Duan, B. R. Yu, C. X. Gao, W. Yuan, J. Ma and F.-J. Xu, ACS Appl. Mater. Interfaces, 2016, 8, 29334–29342 CrossRef CAS.
- Q. F. Zhang, Q. Y. Yu, Y. Y. Geng, J. Zhang, W. X. Wu, G. Wang, Z. W. Gu and X. Q. Yu, ACS Appl. Mater. Interfaces, 2014, 6, 15733–15742 CrossRef CAS PubMed.
- T. Wu, Y. Qi, C. Xu, D. D. Sui and F.-J. Xu, Nano Today, 2023, 50, 101887 CrossRef CAS.
- Y. Qi, C. Xu and F.-J. Xu, WIREs Nanomed. Nanobiotechnol., 2020, 12, e1631 CrossRef CAS PubMed.
- F.-J. Xu, Prog. Polym. Sci., 2018, 78, 56–91 CrossRef CAS.
- Y. C. Zheng, S. P. Li, Z. L. Weng and C. Gao, Chem. Soc. Rev., 2015, 44, 4091–4130 RSC.
- Q. Zeng, Y. W. Zhu, B. R. Yu, Y. J. Sun, X. K. Ding, C. Xu, Y. W. Wu, Z. H. Tang and F.-J. Xu, Biomacromolecules, 2018, 19, 2805–2811 CrossRef CAS.
- Z. Guo, S. Li, M. Lv, Z. H. Liu and W. Xue, ACS Biomater. Sci. Eng., 2017, 3, 2420–2430 CrossRef CAS.
- C. R. Wang, X. X. Wang, L. L. Du, Y. L. Dong, B. Hu, J. H. Zhou, Y. L. Shi, S. P. Bai, Y. Y. Huang, H. Q. Cao, Z. C. Liang and A. J. Dong, ACS Appl. Mater. Interfaces, 2021, 13, 2218–2229 CrossRef CAS.
- B. Mondal, T. Dutta and S. S. Gupta, Chem. Commun., 2021, 57, 109–112 RSC.
- M. R. Schwartz, A. C. Debski and R. J. Price, J. Controlled Release, 2021, 339, 531–546 CrossRef CAS PubMed.
- C. Sardo, E. F. Craparo, B. Porsio, G. Giammona and G. Cavallaro, Polymers, 2019, 11, 889 CrossRef CAS PubMed.
- G. B. Yang, J. J. Liu, Y. F. Wu, L. Z. Feng and Z. Liu, Coord. Chem. Rev., 2016, 320, 100–117 CrossRef.
- R. L. Zhu, A. P. Qian, J. T. Yan, W. Li, K. Liu, T. Masuda and A. F. Zhang, ACS Appl. Mater. Interfaces, 2018, 10, 13258–13263 CrossRef CAS.
- Y. J. Huang, X. K. Ding, Y. Qi, B. R. Yu and F.-J. Xu, Biomaterials, 2016, 106, 134–143 CrossRef CAS PubMed.
- Y. M. Chen, Y. Qi, W. T. Hu, Z. Di, B. R. Yu and F.-J. Xu, Acta Polym. Sin., 2019, 50, 602–612 CAS.
- Y. Qi, H. Q. Song, H. H. Xiao, G. Cheng, B. R. Yu and F.-J. Xu, Small, 2018, 14, 1803061 CrossRef.
- M. Y. Shao, Y. Q. Fan, K. Zhang, Y. Hu and F.-J. Xu, Nano Today, 2021, 39, 101224 CrossRef CAS.
- Y. Qi, Y. L. Liu, B. R. Yu, Y. Hu, N. S. Zhang, Y. Zheng, M. Yang and F.-J. Xu, Adv. Sci., 2020, 7, 2001424 CrossRef CAS.
- J. L. Gao, T. Luo and J. K. Wang, Nat. Commun., 2021, 12, 5311 CrossRef CAS.
- R. W. Herzog and M. Suzuki, Cell, 2020, 186, 893–893 CrossRef.
- F. Li, N. C. Song, Y. H. Dong, S. Li, L. H. Li, Y. J. Liu, Z. M. Li and D. Y. Yang, Angew. Chem., Int. Ed., 2022, 61, e202116569 CrossRef CAS PubMed.
- S. Norberg, E. M. Von Euw, G. Parry, S. Highfill, Z. Franco, J. L. Gulley and C. S. Hinrichs, J. Clin. Oncol., 2022, 40, 16 CrossRef.
- T. Zheng, W. T. Wang, M. Mohammadniaei, J. Ashley, M. Zhang, N. L. Zhou, J. Shen and Y. Sun, Adv. Sci., 2022, 9, 2103812 CrossRef CAS.
- M. H. Andersen, Semin. Immunopathol., 2023, 45, 253–264 CrossRef CAS.
- T. Suwa, M. Kobayashi, J. M. Nam and H. Harada, Exp. Mol. Med., 2021, 53, 1029–1035 CrossRef CAS.
- Q. F. Zhang, G. Z. Kuang, W. Z. Li, J. L. Wang, H. Z. Ren and Y. J. Zhao, Nano-Micro Lett., 2023, 15, 44 CrossRef CAS.
- E. K. Bang, G. Gasparini, G. Molinard, A. Roux, N. Sakai and S. Matile, J. Am. Chem. Soc., 2013, 135, 2088–2091 CrossRef CAS.
- K. Luo, C. X. Li, L. Li, W. C. She, G. Wang and Z. W. Gu, Biomaterials, 2012, 33, 4917–4927 CrossRef CAS.
- E. Boedtkjer and S. F. Pedersen, Annu. Rev. Physiol., 2020, 82, 103–126 CrossRef CAS PubMed.
- S. Taylor, E. P. Spugnini, Y. G. Assaraf, T. Azzarito, C. Rauch and S. Fais, Drug Resistance Updates, 2015, 23, 69–78 CrossRef PubMed.
- C. C. Wang, P. R. Zhao, G. L. Yang, X. Y. Chen, Y. Q. Jiang, X. W. Jiang, Y. L. Wu, Y. Y. Liu, W. A. Zhang and W. B. Bu, Mater. Horiz., 2020, 7, 1180–1185 RSC.
- D. P. Feldmann, Y. L. Cheng, R. Kandil, Y. R. Xie, M. Mohammadi, H. Harz, A. Sharma, D. J. Peeler, A. Moszczynska, H. Leonhardt, S. H. Pun and O. M. Merkel, J. Controlled Release, 2018, 276, 50–58 CrossRef CAS.
- Y. H. Li, K. Tang, X. Zhang, W. Pan, N. Li and B. Tang, Chem. Commun., 2022, 58, 8754–8765 RSC.
- G. Yang, Y. Liu, J. Chen, J. Ding and X. Chen, Acc. Chem. Res., 2022, 3, 1232–1247 CAS.
- W. H. Fan, M. Y. Shao, J. W. Zhang, G. S. Jin, F.-J. Xu and F. S. Liu, Adv. Funct. Mater., 2019, 29, 1807104 CrossRef.
- Z. X. Liao, Y. C. Li, H. M. Lu and H. W. Sung, Biomaterials, 2014, 35, 500–508 CrossRef CAS PubMed.
- D. F. Chen, C. Y. Lei, W. F. Liu, M. Y. Shao, M. Z. Sun, J. X. Guo, J. J. Cao, P. Luo, Y. W. Luo, B. R. Yu, R. X. Wang, S. Duan and F.-J. Xu, Bioact. Mater., 2023, 28, 376–385 CAS.
- S. Padmakumar, A. D'Souza, N. N. Parayath, B. S. Bleier and M. M. Amiji, J. Controlled Release, 2022, 352, 121–145 CrossRef CAS PubMed.
- E. Wagner, R. Kircheis and G. F. Walker, Biomed. Pharmacother., 2004, 58, 152–161 CrossRef CAS PubMed.
- S. T. Xie, W. D. Sun, T. Fu, X. S. Liu, P. Chen, L. P. Qiu, F. L. Qu and W. H. Tan, J. Am. Chem. Soc., 2023, 145, 7677–7691 CrossRef CAS.
- M. Y. Shao, Y. Qi, D. D. Sui and F.-J. Xu, Biomater. Sci., 2021, 9, 7104–7114 RSC.
- K. Chevreul, K. B. Brigham and C. Bouche, Global Health, 2014, 10, 6 CrossRef PubMed.
- M. R. Khanna, J. Kovalevich, V. M. Y. Lee, J. Q. Trojanowski and K. R. Brunden, Alzheimers Dement., 2016, 12, 1051–1065 CrossRef PubMed.
- L. S. Satin, S. A. Soleimanpour, E. M. Walker and L. Isom, Pharmacol. Rev., 2021, 73, 1001–1015 CrossRef CAS PubMed.
- Y. Q. Pan, M. Y. Shao, P. Li, C. Xu, J.-J. Nie, K. Zhang, S. Wu, D. D. Sui and F.-J. Xu, J. Controlled Release, 2022, 343, 420–433 CrossRef CAS PubMed.
- H. Q. Song, M. Y. Shao, Y. Li, X. J. Ding and F.-J. Xu, Adv. Healthcare Mater., 2019, 8, 1800889 CrossRef.
- Y. W. Zhu, M. Y. Lin, W. T. Hu, J. K. Wang, Z. G. Zhang, K. Zhang, B. R. Yu and F.-J. Xu, Angew. Chem., Int. Ed., 2022, 61, e202200535 CrossRef CAS PubMed.
- W. J. Ye, Y. M. Chen, W. X. Tang, N. Zhang, Z. H. Li, Z. J. Liu, B. R. Yu and F.-J. Xu, ACS Appl. Mater. Interfaces, 2019, 11, 28307–28316 CrossRef CAS PubMed.
- Y. W. Zhu, C. Xu, N. Zhang, X. K. Ding, B. R. Yu and F.-J. Xu, Adv. Funct. Mater., 2018, 28, 1708709 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |






