Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The energy landscape of Aβ42: a funnel to disorder for the monomer becomes a folding funnel for self-assembly†
Moritz
Schäffler
ab,
David J.
Wales
 c and
Birgit
Strodel
c and
Birgit
Strodel
 *ab
*ab
aInstitute of Theoretical and Computational Chemistry, Heinrich Heine University Düsseldorf, 40225 Düsseldorf, Germany. E-mail: b.strodel@fz-juelich.de
bInstitute of Biological Information Processing, Structural Biochemistry (IBI-7), Forschungszentrum Jülich, 52428, Jülich, Germany
cYusuf Hamied Department of Chemistry, University of Cambridge, CB2 1EW Cambridge, UK
First published on 25th October 2024
Abstract
The aggregation of amyloid-β (Aβ) peptides, particularly Aβ1–42, plays a key role in Alzheimer's disease pathogenesis. In this study, we investigate how dimerisation transforms the free energy surface (FES) of the Aβ1–42 monomer when it interacts with another Aβ1–42 peptide. We find that the monomer FES is a structurally inverted funnel with a disordered state at the global minimum. However, in the presence of a second Aβ1–42 peptide, the landscape becomes a folding funnel, leading to a β-hairpin state. Using first passage time analysis, we analyse the pathway for the transition from disordered to the β-hairpin state, which highlights the initial formation of a D23–K28 salt bridge as the driving force, together with hydrophobic contacts.
Alzheimer's disease, a neurodegenerative disorder, is linked to the aggregation and misfolding of Aβ peptides, with Aβ42 being especially prone to forming insoluble fibrillar structures in the brain.1 As a monomer, Aβ is an intrinsically disordered peptide (IDP), lacking a stable secondary or tertiary structure.2,3 However, during the aggregation process, Aβ undergoes conformational changes, leading to more ordered structures.4 Understanding these structural transitions is crucial for elucidating the molecular mechanisms underlying Alzheimer's pathology, as they are pivotal in the formation of toxic oligomers and fibrils.5 Studying amyloid aggregation at the structural level in experiments is challenging due to the heterogeneity and transient nature of amyloid oligomers, as their diverse and short-lived states complicate detailed characterisation. Techniques such as nuclear magnetic resonance (NMR) spectroscopy, cryogenic electron microscopy (cryo-EM), and X-ray crystallography usually struggle to resolve these small, dynamic oligomeric intermediates.6 Molecular simulations can therefore be helpful, providing insights into the conformational dynamics and aggregation pathways of Aβ peptides.7
The energy landscapes of IDPs feature complex topographies with multiple minima representing various conformational states, which may correspond to an evolved intrinsically multifunctional capability.8 The energy landscapes for amyloid aggregation are even more heterogeneous, reflecting the diverse conformational and aggregation states amyloid proteins can adopt.9 In a previous study that applied NMR-based metadynamics simulations to Aβ40, the free energy landscape revealed what the authors called an ‘inverted landscape’, in which the global minimum consists of disordered structures, while higher energy regions contain transiently structured conformations, suggesting increased structuring at higher temperatures.10 This result contrasts with the multifunnel energy landscapes characterised for IDPs,8,11 which feature low-lying minima of similar energy separated by substantial energy barriers that trap the system in metastable states. In this study, we determine the free energy surface (FES) of Aβ42 as a monomer and for an initial aggregation step into β-sheet-rich dimers, offering fresh insights into the structural dynamics during Aβ42 aggregation. We suggest that the monomer landscape might be described as a ‘disordered funnel’, as explained below.
To obtain the FES of the monomer and the dimer, we performed molecular dynamics (MD) simulations using the all-atom force field CHARMM36m12 and sampled 6 and 24 μs for the Aβ42 monomer and dimer, respectively. The Aβ42 peptides in each frame of the MD simulations of both monomer and dimer are characterised by the distribution of reciprocal interatomic distance (DRID) metric, a structure-specific dimensonality reduction method known for preserving system kinetics effectively.13,14 We used the Cα atoms of structurally important residues of Aβ42, namely D1, F19, D23, K28, L34, and A42, as reference atoms to define the DRID metric. For each of the reference atoms, the first three moments of the DRID are used to define the state vector, resulting in a 3N → 3Nc dimensionality reduction, where N is the number of atoms in the molecule and Nc the number of reference atoms. After clustering, the trajectory of states is used to calculate the rate matrix of transitions between them, which is translated into free energies of the states (minima) and transition states that connect them.15,16 The resulting FESs are visualised using disconnectivity graphs,17,18 where the branches terminate at the energies of local minima, and the vertical scale is the free energy. These branches are joined at the threshold where the free energy exceeds the value for the highest transition state on the lowest path between them. Hence the branches correspond to a segregation of the minima into sets that can interconvert amongst themselves at any given level on the vertical scale. Similar states leading to the same local minimum are grouped in the same basin, facilitating intuitive analysis of the underlying structure of the landscape and identification of folding funnels. Central to our investigation is the application of first-passage time (FPT) distribution analysis to extract the kinetics of transitions between different states.16,19 This approach allows us to address the temporal aspects of the conformational changes, shedding light on the pathways and rates governing the transitions between disordered and β-sheet-rich conformations. Detailed information on the methods can be found in the ESI.†
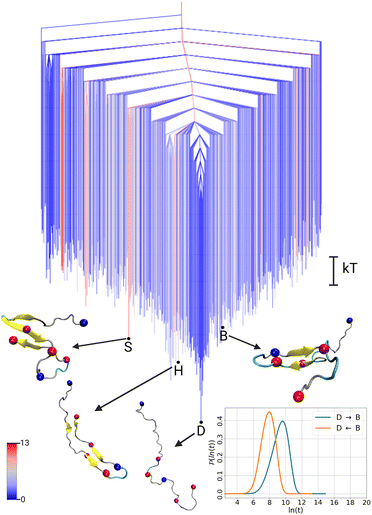
First, we discuss the free energy surface of the Aβ42 monomer, for which the disconnectivity graph is shown in Fig. 1. The FES exhibits one main funnel leading to the global minimum. In contrast to the usual landscapes of folded proteins, the global minimum corresponds to a disordered state of the Aβ42 monomer (labeled as state D for disorder). Conformations featuring partial secondary structure, such as a β-hairpin characteristic for Aβ oligomers20 or the S-shape conformation that is the usual building block of fibrils,4 are found as excited states in the FES of the monomer with energies of ΔFmonH = 2.3 kT and ΔFmonS = 3.2 kT respectively. Here, an excited state is defined as a higher free energy minimum, not to be confused with an electronic excited state. This organisation of the FES with (partially) folded states at the top of the funnel, and the the disordered states at the bottom, has previously been called an ‘inverted free energy landscape’.10 We suggest the nomenclature ‘structurally inverted funnel’ or simply ‘disordered funnel’, since it is the structure that is inverted, not the funnel.
To illustrate the FES of the Aβ42 dimer and allow direct comparison to the monomer, we treat the two chains in the dimer systems as individual peptides and present the disconnectivity graph of the dimer for an Aβ42 peptide in the environment of the second Aβ42 peptide. This representation further allows us to use the same DRID metric to calculate the states, highlighting the structural changes in the presence of another Aβ42 peptide. The resulting FES for the Aβ42 dimer, shown in Fig. 2, exhibits the topology of a folding funnel leading to a β-hairpin state with significant β-sheet content (hence labeled state B). Projecting the global minimum of the monomer FES (state D) onto the dimer FES, reveals that the disordered state appears as an excited state in the dimer FES, with a free energy difference of ΔFdimD = 3.9 kT. Similarly, the global minimum of the dimer FES (state B) can be projected onto the monomer FES (see Fig. 1), where it is likewise an excited state, with a free energy difference of ΔFmonB = 3.6 kT. We note that the D state in the dimer FES is not yet a dimer, but two disordered monomers, while the β-hairpin in the B state of the dimer is a result of interpeptide interactions due to dimerisation (Fig. S1, ESI†).
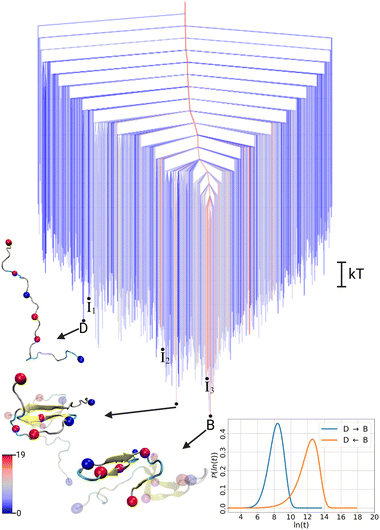 | ||
| Fig. 2 Free energy disconnectivity graph for the FES of the Aβ42 dimer. The branches are colored according to the average number of residues in β-sheet conformation in the ensemble of structures belonging to the respective minimum, ranging from blue (no β-sheets) to red (19 residues involved in β-sheets). For further details, see the caption of Fig. 1. The cartoons of all indicated dimer structures are shown in Fig. S1 (ESI†). | ||
To analyse the timescales associated with the interconversions between the disordered state D and the β-hairpin state B, we calculated the first passage time distribution for these transitions (see ESI† for details). The resulting FPTs for the monomer and dimer are shown in the lower right corner of Fig. 1 and 2, respectively. For the monomer, the transition times are τmonD→B ∼ 15 ns and τmonB→D ∼ 3 ns. While the transition time from the disordered state to the excited β-hairpin state is five times longer than the reverse transition, the transition to the excited state is still relatively fast. In contrast, for the dimer the transition times are τdimD→B ∼ 4 ns and τdimB→D ∼ 300 ns, with a ratio of about 75 for the transition from the global minimum to the excited disordered state with respect to the reverse transition. Thus, we observe a separation of timescales in the dimer, but not the monomer.
These results highlight the differences between the FES of the Aβ42 monomer and dimer. For the disordered funnel of the monomer, higher local minima are readily accessible and the time scales of interconversions between them are of similar magnitude, while transitions to excited states from the ordered global minimum of the dimer are relatively slow. Thus, while we characterise the monomer FES as a disordered funnel, the dimer FES is closer to the expected form for a folded protein. Our FES and timescale analysis of Aβ42 further confirms the predictions made for the energy landscapes of folded and disordered proteins based on the slope of the free energy funnels.21 Chong and Ham determined the FESs of two folded proteins, which exhibited steep folding funnels, and of an IDP, which has a gentler slope and led to similar transition times to and from the global minimum. A significant difference from our study is that the global minimum of the IDP they considered is a partially folded state, whereas at the global minimum of Aβ42 is essentially fully disordered.
To gain further insight into the structural transition from disordered state D to folded state B in the Aβ42 dimer, we extracted structural pathways associated with the fastest transition in the dimer FES (see ESI† for details). From the ensemble that includes all the possibilities D → B we identified three characteristic pathways (Fig. S2 in ESI†), by associating structures from the MD simulations with the states in the kinetic model. These pathways include three intermediate states (I1, I2, I3), which are marked on the FES in Fig. 2. Here, the most important of these three pathways, shown in Fig. 3, is discussed along with the FPTs associated with the fastest transitions between sequential states.
The initial transition D → I1 is characterised by a scissor motion, which closes the relatively extended conformation to a hairpin-like structure. The main driving force for this movement is the formation of a salt bridge between D23 and K28, which is then followed by the establishment of hydrophobic contacts between the strands on both sides of the turn, involving residues 17LVFFA21 and 30AIIGLMV36. The formation of these intrapeptide contacts occurs cooperatively with the establishment of interpeptide contacts between the hydrophobic regions of both peptides (Fig. S3 with in-depth discussion in the ESI†). In state I1, the most important hydrophobic contact is between F19 and L34, which is structurally suboptimal and therefore shifts to F19–I32 in state I2. In fact, the intermediate I1 is ∼ 1 kT higher in energy than state D, and there is a significant barrier of ∼ 9.4 kT that needs to be overcome for the D → I1 transition. This barrier is reflected in the four to nine times slower transition time for this first transition compared to the subsequent transitions. Nonetheless, the first passage time associated with the process τdimD→I1 ∼ 780 ps is still very fast compared to the height of the energy barrier, which can be explained with the FPT probability distribution (Fig. S4 in ESI†). This distribution features both fast and slow relaxation modes, resulting in a mean first passage time that appears slow, which is also reflected in the high free energy barrier. Hence, analysis of the full FPT distribution once again reveals important details that are not evident from average rates.19
After reaching I1, the system moves downhill in energy to the global minimum. After the positions of the hydrophobic contacts have been optimised in I2, hydrogen-bonds form between the strands, leading to a short antiparallel β-sheet in I3. Inspecting the dimer FES, we see that the intermediate I3 is already located in the central folding funnel, close to the global minimum. The final transition I3 → B, further stabilizes the β-sheet and involves a reorientation of the C-terminal.
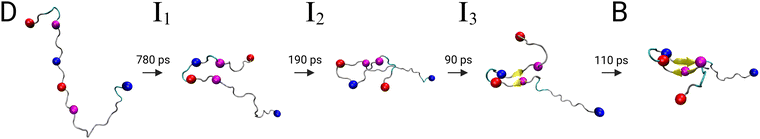 | ||
| Fig. 3 Fastest pathway from the intrinsically disordered state (D) to the global minimum (B) in the dimer free energy surface. The spheres represent the centroids used in the DRID metric. The FPTs associated with the fastest transition between the states along the pathway are shown above the arrows, which were calculated separately for each transition and therefore do not add up to the total transition time associated with the full pathway, where all possible recrossing events are included. Fig. S1 (ESI†) shows the pathway again, but with both peptides within the dimer. | ||
The elucidation of the pathway for structure formation during Aβ42 dimerisation confirms the relevance of the D23–K28 salt bridge for turn formation.22,23 Our results reveal that the creation of this salt bridge initiates the turn, leading to β-hairpin formation. A recent study demonstrated that Aβ oligomerisation depends on this specific β-hairpin motif, as variants of the Aβ peptide that cannot adopt this hairpin structure do not produce oligomers.20 In addition, this β-hairpin structure is crucial for mediating Aβ toxicity, as demonstrated by a study in which Aβ oligomers with a permanently present hairpin—achieved by introducing a disulfide bridge between residues 21 and 30—proved to be particularly toxic.24 Our observations of the hydrophobic contact formation are consistent with with a recent low-temperature solid-state NMR spectroscopy study of freeze-trapped Aβ40 oligomers.25 Tycko and coworkers identified β-hairpin conformations with contacts within the peptide and between the two main hydrophobic segments of Aβ40 that developed within 1 ms. The primary hydrophobic contact was located between the aromatic sidechain of F19 and aliphatic sidechains in residues 30–35, which fully agrees with the structures in the folding funnel of the dimer FES determined here.
In conclusion, our study provides new insight into the structural dynamics of Aβ42 with implications for Alzheimer's disease pathology. Through analysis of free energy landscapes and first-passage time distributions, we have elucidated key aspects of Aβ42 conformational transitions from a disordered to a β-sheet forming state. Our results reveal distinctive features in the free energy landscapes of Aβ42 monomers and dimers, highlighting the impact of the molecular environment on structural stability and propensity for aggregation. Specifically, we observe that the FES of the monomer resembles a structurally inverted folding funnel, where disordered states are lowest in energy. In contrast, the lowest minima for the dimer are characterised by more ordered structures, predominantly consisting of β-hairpins, which form as Aβ42 folds upon binding to the hydrophobic region of another Aβ42 peptide. Thus, the increase in hydrophobicity in the environment of Aβ42 appears to be crucial for its folding during self-assembly. Our analysis of first-passage times indicates that the FES of the monomer is relatively flat, with excited states (higher free energy minima) more readily accessible compared to the dimer. This organisation reflects the disordered nature of the low-lying states for the monomeric form of Aβ42.
Our pathway analysis identifies key intermediates and structural rearrangements involved in the transition from disordered to folded states, providing mechanistic insights into the initial aggregation steps for Aβ42. Starting from an extended conformation, the peptide chain undergoes a scissor-like motion, establishing a salt bridge between residues D23 and K28, followed by the formation of hydrophobic contacts. Subsequently, these contacts reorganise, leading to β-hairpin structures. Our results should contribute to a deeper understanding of the molecular mechanisms underlying Alzheimer's disease pathology, and may inform the development of targeted therapeutic interventions.
The authors gratefully acknowledge computing time on the supercomputer JURECA at Forschungszentrum Jülich under grant IDP, financial support from the German Academic Exchange Service, and valuable discussions with Prof. Debayan Chakraborty.
Data availability
The code for PATHSAMPLE can be found at https://www-wales.ch.cam.ac.uk/software.html. The data analysis scripts are available at https://github.com/strodel-group/ under ‘freeEnergyCalculation’ and ‘DRIDmetric’.Conflicts of interest
There are no conflicts to declare.References
- D. J. Selkoe and J. Hardy, EMBO Mol. Med., 2016, 8, 595–608 CrossRef CAS PubMed.
- J. Roche, Y. Shen, J. H. Lee, J. Ying and A. Bax, Biochemistry, 2016, 55, 762–775 CrossRef CAS PubMed.
- A. Paul, S. Samantray, M. Anteghini, M. Khaled and B. Strodel, Chem. Sci., 2021, 12, 6652–6669 RSC.
- D. Willbold, B. Strodel, G. F. Schröder, W. Hoyer and H. Heise, Chem. Rev., 2021, 121, 8285–8307 CrossRef CAS PubMed.
- U. Sengupta, A. N. Nilson and R. Kayed, EBioMedicine, 2016, 6, 42–49 CrossRef PubMed.
- P. H. Nguyen, A. Ramamoorthy, B. R. Sahoo, J. Zheng, P. Faller, J. E. Straub, L. Dominguez, J.-E. Shea, N. V. Dokholyan, A. De Simone, B. Ma, R. Nussinov, S. Najafi, S. T. Ngo, A. Loquet, M. Chiricotto, P. Ganguly, J. McCarty, M. S. Li, C. Hall, Y. Wang, Y. Miller, S. Melchionna, B. Habenstein, S. Timr, J. Chen, B. Hnath, B. Strodel, R. Kayed, S. Lesné, G. Wei, F. Sterpone, A. J. Doig and P. Derreumaux, Chem. Rev., 2021, 121, 2545–2647 CrossRef CAS PubMed.
- H. Fatafta, M. Khaled, B. Kav, O. O. Olubiyi and B. Strodel, WIREs Comput. Mol. Sci., 2024, 14, e1703 CrossRef CAS.
- Y. Chebaro, A. J. Ballard, D. Chakraborty and D. J. Wales, Sci. Rep., 2015, 5, 10386 CrossRef PubMed.
- B. Strodel, J. Mol. Biol., 2021, 433, 167182 CrossRef CAS PubMed.
- D. Granata, F. Baftizadeh, J. Habchi, C. Galvagnion, A. Simone, C. Camilloni, A. Laio and M. Vendruscolo, Sci. Rep., 2015, 5, 15449 CrossRef CAS PubMed.
- J. A. Joseph and D. J. Wales, J. Phys. Chem. B, 2018, 122, 11906–11921 CrossRef CAS PubMed.
- J. Huang, S. Rauscher, G. Nawrocki, R. Ting, M. Feig, B. de Groot, H. Grubmüller and A. MacKerell, Nat. Meth., 2017, 14, 71–73 CAS.
- T. Zhou and A. Caflisch, J. Chem. Theory Comput., 2012, 8, 2930–2937 CrossRef CAS PubMed.
- D. Chakraborty, J. E. Straub and D. Thirumalai, Sci. Adv., 2023, 9, eadd6921 CrossRef CAS PubMed.
- E. J. Woods, D. Kannan, D. J. Sharpe, T. D. Swinburne and D. J. Wales, Philos. Transact. A Math. Phys. Eng. Sci., 2023, 381, 20220245 Search PubMed.
- E. J. Woods and D. J. Wales, Phys. Chem. Chem. Phys., 2024, 26, 1640–1657 RSC.
- O. M. Becker and M. Karplus, J. Chem. Phys., 1997, 106, 1495–1517 CrossRef CAS.
- D. J. Wales, M. A. Miller and T. R. Walsh, Nature, 1998, 394, 758–760 CrossRef CAS.
- D. J. Wales, J. Phys. Chem. Lett., 2022, 13, 6349–6358 CrossRef CAS PubMed.
- M. Khaled, I. Rönnbäck, L. L. Ilag, A. Gräslund, B. Strodel and N. Österlund, J. Am. Chem. Soc., 2023, 145, 18340–18354 CrossRef CAS PubMed.
- S.-H. Chong and S. Ham, Sci. Rep., 2019, 9, 14927 CrossRef PubMed.
- G. Reddy, J. E. Straub and D. Thirumalai, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 11948–11953 CrossRef CAS PubMed.
- M. Schäffler, S. Samantray and B. Strodel, Int. J. Mol. Sci., 2023, 24, 11238 CrossRef PubMed.
- A. Sandberg, L. Luheshi, S. Söllvander, T. Pereira de Barros, B. Macao, T. Knowles, H. Biverstål, C. Lendel, F. Ekholm-Petterson, A. Dubnovitsky, L. Lannfelt, C. Dobson and T. Härd, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15595–15600 CrossRef CAS PubMed.
- J. Jeon, W.-M. Yau and R. Tycko, Nat. Commun., 2023, 14, 2964 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc02856b |
| This journal is © The Royal Society of Chemistry 2024 |