Extreme makeover: the incredible cell membrane adaptations of extremophiles to harsh environments
Archita
Maiti†
,
Shakkira
Erimban†‡
and
Snehasis
Daschakraborty
 *
*
Department of Chemistry, Indian Institute of Technology Patna, Bihar, 801106, India. E-mail: snehasis@iitp.ac.in
First published on 22nd August 2024
Abstract
The existence of life beyond Earth has long captivated humanity, and the study of extremophiles—organisms surviving and thriving in extreme environments—provides crucial insights into this possibility. Extremophiles overcome severe challenges such as enzyme inactivity, protein denaturation, and damage of the cell membrane by adopting several strategies. This feature article focuses on the molecular strategies extremophiles use to maintain the cell membrane's structure and fluidity under external stress. Key strategies include homeoviscous adaptation (HVA), involving the regulation of lipid composition, and osmolyte-mediated adaptation (OMA), where small organic molecules protect the lipid membrane under stress. Proteins also have direct and indirect roles in protecting the lipid membrane. Examining the survival strategies of extremophiles provides scientists with crucial insights into how life can adapt and persist in harsh conditions, shedding light on the origins of life. This article examines HVA and OMA and their mechanisms in maintaining membrane stability, emphasizing our contributions to this field. It also provides a brief overview of the roles of proteins and concludes with recommendations for future research directions.
1. Introduction
The question of whether life exists beyond Earth is a captivating inquiry that has intrigued humanity for centuries. While the vastness of the universe suggests the potential for extraterrestrial life, the immense distances involved in space exploration make direct contact seem dauntingly remote. However, the study of extremophiles presents a fascinating avenue for exploring this possibility.1–4 Extremophiles are organisms that survive and thrive in environments considered “extreme” by conventional standards, such as scorching heat, freezing cold, bone-crushing pressure, high salinity, high radiation, acidic or alkaline conditions, and low water conditions.5–14 By studying the survival strategies of extremophiles, scientists gain valuable insights into how life can adapt and thrive in seemingly inhospitable conditions, offering clues about the origins of life.The extremophiles face severe challenges at different extreme conditions, such as low enzyme activity, mechanical damage of cellular subunits by tiny ice crystals, drop down in the transcription and translation rate, cold and heat denaturation of proteins, disruption of the molecular structure of the cell membrane, reduction of cell membrane fluidity, loss of membrane barrier function, etc. To combat these challenges, the extremophiles must adapt to these extreme conditions. Two different types of adaptations are known: genotypic or phenotypic15–17 While genotypic adaptation occurs over an evolutionary timescale, phenotypic adaptation takes place within the lifetime of the organism and can have timescales ranging from minutes to days. Studying these strategies, which the extremophiles adopt to survive and thrive in extreme conditions, is important in various areas of science and technology, starting from the fundamental quest for the origins of life to the food processing and probiotic industries18–27
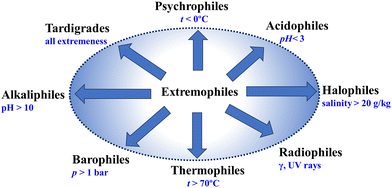
Fig. 1 presents a schematic representation showing different types of extremophiles, which survive and thrive in various extreme conditions. The extremophiles exhibit a diverse array of survival strategies tailored to their respective extreme environments. For instance, thermophiles have evolved specialized enzymes and proteins that remain stable at high temperatures, allowing them to thrive in hydrothermal vents or geothermal springs.28–31 Conversely, psychrophiles have mechanisms to prevent cellular freezing in frigid environments, such as polar regions or deep-sea trenches.32,33 Halophiles flourish in environments with high salt concentrations, employing adaptations to regulate osmotic pressure and mitigate the damaging effects of salt on cellular structures.34–36 Acidophiles21 and alkaliphiles37 adopt suitable strategies so that they are capable of surviving in environments with extreme pH levels, such as the use of proton efflux proteins.38–42 The barophiles, which thrive in high-pressure environments such as the deep sea, adopt strategies to combat high-pressure stress by morphological, physiological, and molecular evolutions.43–45 Some extremophiles even exhibit remarkable tolerance to radiation, earning them the label of radio-tolerant organisms.45–48 These organisms have mechanisms to repair DNA damage caused by high levels of radiation exposure, enabling them to persist in radioactive environments.
In this feature article, we focus on the molecular-level understanding of the adaptive strategies employed by extremophiles to maintain the packing density and fluidity of their cell membranes under various external stresses. The cell membrane, composed primarily of lipids and proteins, is a crucial component of the cell, providing essential functions. The packing density of lipids in the membrane indicates how closely the lipid molecules are arranged within the lipid bilayer, while fluidity refers to the ease with which these lipids can diffuse within the membrane. The packing density and fluidity of lipid membranes are severely challenged by extreme conditions. The extremophilic organisms must adopt effective strategies to combat the stress and retain the packing density and fluidity of the lipid membrane for several reasons:
(i) Barrier function: the appropriate barrier function of the cell membrane requires optimum fluidity, since higher or lower values of the latter may severely impact the barrier function.
(ii) Cell communication: some transmembrane proteins transmit signals between the exterior and interior of the cell, which requires appropriate fluidity of the cell membrane to enable these proteins to move and interact effectively with the signaling molecules.49–51
(iii) Transport of molecules: both active and passive transport of water and other molecules through the cell membrane are strongly impacted by a change of membrane fluidity.52–56
(iv) Cell shape and stability: the cell shape and stability are strongly affected by the packing density of the lipid membrane, preventing collapse or bursting due to osmotic pressure changes.57
(v) Cell recognition and adhesion: some membrane proteins aid in cell recognition and adhesion, essential for tissue formation and immune response.58–60 These are strongly influenced by the change of membrane packing density and fluidity.
Among various strategies that are adopted by extremophiles, we will primarily discuss here the two most important ones: (i) homeoviscous adaptation (HVA)61–65 and (ii) osmolyte-mediated adaptation (OMA)66–75 HVA is the regulation of the lipid composition of the cell membrane to keep the membrane structure and fluidity intact against external stress. HVA can occur via remodeling of both the lipid acyl chain and lipid head groups. On the other hand, OMA refers to the process by which osmolytes, small organic molecules that regulate osmotic pressure and maintain the structure and function of cellular components, stabilize lipid membranes by protecting their packing density and fluidity against environmental stress. We will also briefly discuss the direct and indirect roles of proteins in stabilizing lipid membranes under stressed conditions.
2. Homeoviscous adaptation (HVA) for protecting the cell membrane
HVA was first observed by Sinensky et al.76 in Escherichia coli (E. coli) bacteria surviving at higher growth temperatures. To date, several studies have shown a similar mode of adaptation in different classes of extremophiles and even in mammalian cell membranes.4,77–91 Therefore, adopting the theory of HVA is a universal paradigm of membrane adaptation to extreme growing environments. In this section, we detail the mechanism of HVA and its impact on the lipidomics of the cell membrane, providing sufficient examples from both simulation and experimental studies.Mechanism of HVA
The first step of HVA is sensing the changes in the cell membrane due to external stress with the help of sensor proteins.1,4,92–94 The changes in the local environment include the drastic alteration of the membrane fluidity, membrane curvature, and packing density of the lipids. Although significant progress has been made in recognizing potential sensory mechanisms, understanding their molecular mechanism and how they collaborate to uphold the physicochemical characteristics of cellular membranes remains largely elusive.95,96 There is a debate on what changes in membrane properties are sensed by sensor proteins. As per one school of thought, sensor proteins exhibit sensitivities to the membrane fluidity (measured by the lateral diffusion Dxy of the lipids).97–102 On the contrary, a more recent work4 proposed that instead of the fluidity of the membrane, the sensor protein can sense lipid-packing density to initiate a homeostatic response.103 Through the integration of molecular dynamics simulations with experimental approaches, the authors discovered a notable sensitivity of the transcriptional regulator Mga2 (a transcriptional regulator found in yeast, specifically Saccharomyces cerevisiae) to variations in the abundance, positioning, and arrangement of double bonds within lipid acyl chains. This revelation offers insights into the molecular principles governing membrane adaptation.The second step is as follows. Once the sensor proteins detect changes in membrane properties, they undergo conformational changes of the protein. These changes activate downstream signaling pathways, known as signal transduction. The nature of these pathways can vary depending on the organism and the specific sensor proteins involved. The signaling cascade often involves the activation of specific transcription factors.104 In the case of yeast (Saccharomyces cerevisiae), for example, the transcriptional regulator Mga2 is sensitive to changes in membrane lipid composition.105 Upon activation, these transcription factors move to the nucleus, where they bind to promoters of genes involved in lipid metabolism. This initiates transcription, producing mRNA, which is translated into proteins in the cytoplasm. These proteins include enzymes for lipid synthesis and modification, as well as other proteins crucial for membrane integrity.106
In the third step, newly synthesized enzymes and proteins adjust the cell membrane's lipid composition. They may increase unsaturated fatty acids for fluidity or alter lipid headgroups to adjust membrane curvature, restoring optimal membrane properties for proper cellular function.
The outcome of HVA on the lipid composition
The alterations in the fatty acid profile of the phospholipids in some of the extremophiles are shown in Fig. 2. Under subzero temperatures, high pressure, and dehydration the membrane undergoes a phase transition to an ordered gel state triggered by the tight packing of membrane lipids.68,107–116 This phase transition is fatal to the organism and is avoided by timely modifications in the lipid types. At low temperatures and high pressure, lipids with low melting temperatures (Tg) including monounsaturated fatty acids (MUFA), polyunsaturated fatty acids (PUFA), branched fatty acids (methyl branching), short-chain fatty acids, hydroxylated fatty acids, and polar headgroups are incorporated in a higher proportion.4,13,77–91,117–119 Unlike the trans double bond, the presence of kinks in the cis double bond pushes the lipids apart, decreasing the order of packing and avoiding the fluid-to-gel phase transition. For example, micrococcus cryophilus bacterium extracted from a temperature regime of −30 °C to −40 °C shows that 97% of the lipids are with unsaturated fatty acids (UFAs).120 Together with the changes in unsaturation, these species also set the best example for the shortening of fatty acids chain length with a lowering of growth temperature. The actual biochemical process of cold-induced shortening of the acyl chains is still not clearly understood. However, overactivity of some molecules, such as acetyl-CoA or malonyl-CoA in the cells during stress suggests their roles in the modification of the lipidome.121–124 Some psychrophiles show an increased abundance of lipids with short-chain (carbon length less than 12) and branched fatty acids.125–128 A study129 on Pseudomonas syringae (antarctic psychrotrophic bacterium) showed an increased level of hydroxylated fatty acids in the lipopolysaccharides (LPS) of the outer membrane when exposed to a temperature of 4 °C.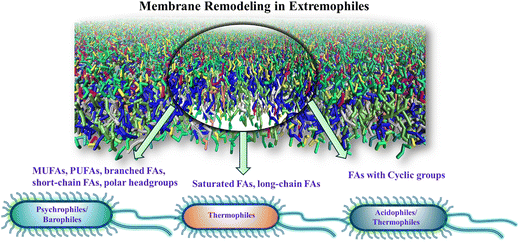 | ||
| Fig. 2 Schematic representation of the membrane remodeling occurring in the fatty acid profile of lipids in different classes of extremophiles. | ||
Remodeling of the lipid headgroup is also observed to be an effective adaptive strategy to maintain the structure and fluidity of the lipid membrane.130,131 A recent study by Chwastek et al.91 reported a headgroup-specific remodeling in Methylobacterium extorquens, a soil-based and plant-associated Gram-negative bacterium. The diurnal temperature drops to 6 °C and gradually increases the bacteria's PC/PE lipid ratio. An increased amount of lysophospholipids (LPS) is also reported in some psychrophilic yeast.121 These are inverted conical shapes and have been shown to disrupt the membrane packing and increase fluidity in response to cold.
Cyclisation of fatty acids in response to temperature and pH is also reported in some species although not widely documented to date.132 The cyclase enzyme synthesizes these derivatives from the unsaturated acyl chains. Like the MUFAs, the cyclic groups also maintain the membrane fluidity by introducing gauche defects and affect the packing of lipids. This is captured in the bacterial membrane of Lactococcus lactis when cultivated at lower fermentation temperature.133 The cyclopropanation of unsaturated fatty acids aids in perturbing the membrane rigidification upon freeze-drying and storage. The response of membrane lipids of R. erythropolis cells,134E. coli,135 and Salmonella enterica serovar typhimurium (S. typhimurium)136 with increased content of cyclopropane FAs follows the extremity in pH levels. Cyclopropane rings aid the survival of E. coli under acid shock and freeze-drying.
Along with the modifications in the lipid compositions, studies also report the role of sterols (cholesterol and hopanoids) in extremophiles.137,138 The increased expression of cholesterol biosynthetic genes in fishes is related to the temperature variations across seasons.139 At low temperatures, an increase in cholesterol can hinder the fluid-to-gel phase transition of the membrane through the cholesterol-induced condensation effect as in ‘lipid rafts’. The packing of cholesterol between the acyl chains of lipids perturbs the ordered packing of lipids and thereby the gel phase.
3. Protecting lipid membrane via HVA: insights from molecular dynamics simulations
Computer simulation studies can provide a molecular perspective on HVA under stress. However, these studies are less common than experimental ones. Here, we review the insights gained from simulation-based studies to date, including work from our own group.Role of unsaturation
The tuning of saturated/unsaturated lipids under sub-zero temperature on a bacteria Leeuwenhoekiella aequorea isolated from Antarctica was studied by Singh et al.90 The study reported a percentage profile of different fatty acid chains in bacteria over a temperature range of 15 to −20 °C involving a gradual reduction of temperature and freeze–thaw cycles. They analyzed the fatty acid profiles of three characteristic states of the bacteria: (i) I0, where the bacterial strains are incubated at standard growth conditions (15 °C) for 5 days, (ii) BFT, where the temperature is gradually reduced to 4 °C and (iii) PFT, where the system is quickly frozen to −20 °C and thawed back to 4 °C. In the I0 state, almost 70% of the lipids are saturated fatty acids (SFAs), while the remaining portion (∼30%) is unsaturated fatty acids (UFAs). This trend has reverted in both BFT and PFT systems, where more than 85% of the lipids are UFAs. Scrutinizing the profile, it was observed that the abundant 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 SFA gets converted to 18
0 SFA gets converted to 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 UFA at lower temperatures. To elucidate how the homeoviscous adaptation protects the cell membrane during cold stress, our group112 undertook a project to simulate a model lipid membrane of the bacteria in question. With some approximations, symmetric lipid bilayers were modeled and simulated over a temperature range from 250 K to 300 K using molecular dynamics (MD) simulation techniques, as depicted in Fig. 3. The study described the importance of the experimentally reported fatty acid remodeling with a reduction of temperature for the retention of membrane packing density and fluidity, the key for cell survival. This study provided a molecular basis of the observations that the I0 state of the lipid membrane, effective for cell survival at or above 280 K, becomes unsuitable below this temperature due to a fluid–gel phase transition. At temperatures near the phase transition, a phase separation occurs, with the unsaturated lipids forming a fluid-like phase and the saturated lipids forming a gel-like phase, along with a strong correlation between the leaflets. Our research also suggested an increasing role of the desaturase enzyme in cold adaptation as temperatures drop. By comparing the properties of five intermediate membranes, we outlined the gradual progression toward homeoviscous adaptation.
2 UFA at lower temperatures. To elucidate how the homeoviscous adaptation protects the cell membrane during cold stress, our group112 undertook a project to simulate a model lipid membrane of the bacteria in question. With some approximations, symmetric lipid bilayers were modeled and simulated over a temperature range from 250 K to 300 K using molecular dynamics (MD) simulation techniques, as depicted in Fig. 3. The study described the importance of the experimentally reported fatty acid remodeling with a reduction of temperature for the retention of membrane packing density and fluidity, the key for cell survival. This study provided a molecular basis of the observations that the I0 state of the lipid membrane, effective for cell survival at or above 280 K, becomes unsuitable below this temperature due to a fluid–gel phase transition. At temperatures near the phase transition, a phase separation occurs, with the unsaturated lipids forming a fluid-like phase and the saturated lipids forming a gel-like phase, along with a strong correlation between the leaflets. Our research also suggested an increasing role of the desaturase enzyme in cold adaptation as temperatures drop. By comparing the properties of five intermediate membranes, we outlined the gradual progression toward homeoviscous adaptation.
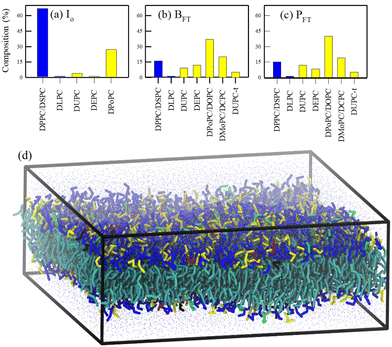 | ||
| Fig. 3 Lipid profiles (saturated lipids: blue; unsaturated lipids: yellow) of eight different lipid membranes (a)–(c). (d) A snapshot of the equilibrated I0 membrane. The head groups of 5 different lipids are color-coded as blue (DPPC/DSPC) red (DLPC), yellow (DPoPC), green (DUPC), and black (DEPC). The tails of all the lipids are represented by cyan color. CG water beads are represented by points. Reprinted with permission from ref. 112 Copyright {2020}American Chemical Society. | ||
The cold adaptation mechanism of marine algae has gained significant interest owing to their extensive applications. The aquatic environment's diurnal and seasonal temperature variations remarkably influence the lipid composition of marine algae's thylakoid membrane (thylakoid-LBM).140–144 These membranes are involved in photosynthesis and are composed of galacto- and sulfolipids with UFAs in their acyl chain. A recent study by Manna and coworkers145 revealed the purpose of membrane complexity in the thylakoid membrane of a commercially relevant Red algae Gracilaria corticata through atomistic simulations. The membrane composition at optimal growth conditions (25–30 °C) is modeled and further simulated over a temperature spectrum of 10 °C to 40 °C, at intervals of 5 °C. In between 25 °C and 30 °C they observed the ordered gel and disordered fluid domains of saturated and unsaturated lipids, respectively, in the multicomponent lipid mixture. Furthermore, a phase transition from fluid Lα phase to gel Lβ phase was observed below 10 °C. Fig. 4 shows the phase of the membrane across different temperatures they studied.
 | ||
| Fig. 4 Representative snapshots of side views of the thylakoid-LBM, the Lβ phase at 10 °C and the Lα phase at 40 °C. A snapshot with its periodic image shows the Lβ/Lα coexistence in the membrane with a wave-like surface at 30 °C. Reprinted with permission from ref. 145 Copyright {2023} American Chemical Society. | ||
This agrees with the phase separation observed in a binary/ternary/quaternary mixture of saturated and unsaturated lipids. The preferential interaction between lipids of the same acyl chain type drives the phase separation in the thylakoid-LBM. However, a segregation of lipids based on the acyl chain type is not observed in the membrane. The lipids with UFAs retain the fluidity of the membrane, whereas those with SFA get rigidified. The study concluded that the increased concentration of PUFAs can avoid the otherwise happening membrane rigidification under cold stress.
Role of polar headgroups
An experimental study by Chwastek et al.91 reported a headgroup-based lipidomic remodeling of a soil-based and plant-associated bacterium Methylobacterium extorquens under diurnal temperature variations. Almost 11 out of the total 25 lipid types undergo major remodeling across four temperatures: 30, 20, 13, and 6 °C. Lipids distributed across five headgroup types: phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), and cardiolipin (CL) are reported in the bacterium. An idiosyncratic adaptation strategy with tuning of the PE and PC class of lipids was reported remarkably at 6 °C. Under low temperatures (6 °C), the PE lipids are replaced with the PC class of lipids. Interestingly the bacterium did not have any saturated lipids even at the higher temperature regime (30 °C).Aiming to unravel the effect of headgroup-based membrane tuning from a molecular perspective, a study from our group146 modeled the membranes across all four temperatures 30, 20, 13, and 6 °C and named the membranes N30, N20, N13, and N6, respectively. These membranes are simulated at the respective temperatures. Fig. 5 shows the composition based on the headgroup for all four membranes. We performed cooling and heating simulations of the N30 and N6 membranes to understand how the membrane responds to abrupt temperature changes without an adaptation in its lipid profile. No phase transitions were marked in the membrane even at 6 °C. The study revealed that the membrane's properties are largely preserved through complex lipidome remodeling in response to both heat and cold stress. Specifically, the remodeling involves adjusting the acyl chain headgroups, fine-tuning the packing density and fluidity at various temperatures. At higher temperatures, lipids with headgroups that strongly interact become more prevalent, while at lower temperatures, lipids with headgroups that interact less strongly dominate the lipidome. This dynamic shift helps prevent disruptions to the lipid membrane caused by thermal stress. This study, therefore, unraveled the importance of headgroup remodeling during diurnal temperature variation of soil bacteria.
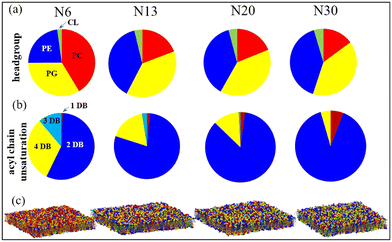 | ||
| Fig. 5 Pie charts showing the lipidome compositions of four native membranes N6, N13, N20, and N30 at four temperatures based on (a) headgroups (PC: phosphatidylcholine, PE: phosphatidylethanolamine, PG: phosphatidylglycerol, CL: cardiolipin) and (b) unsaturation level (DB: double bond) of the acyl chains. (c) Simulation snapshots of the equilibrated native membranes, where the lipids are color-coded the same as the pie charts representing four different headgroups. The phosphate beads of headgroups are represented by spherical beads. Reprinted with permission from ref. 146 Copyright {2022} American Chemical Society. | ||
Role of cyclic groups
Analogous to unsaturated fatty acids, studies show the incorporation of cyclic fatty acids to maintain membrane fluidity. Our group147 investigated the role of lipids with cyclopropane (CP) ring containing fatty acids on retaining the packing density of fluidity of the lipid membrane of E. coli bacteria with reduction of temperature. A realistic lipid bilayer of E. coli bacteria (containing 14 different types of lipids) was simulated using both coarse-grained and all-atom molecular dynamics simulations across a wide range of temperatures between 250 and 350 K. To address the question of why cyclopropane fatty acids are preferred over C–C single or C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C double bonds, a group of modified lipid membranes were also simulated by replacing cyclopropane (CP) groups with either single or double bonds. No differences were observed between the CP fatty acids with the unsaturated ones in coarse-grained resolution, suggesting that the CG model may not adequately represent the effects of cyclopropane-containing lipids. However, at the all-atom resolution, a noticeable difference emerged in membrane properties due to the presence of CPs and unsaturation, especially at lower temperatures. CPs are particularly effective at preventing close packing of membrane lipids, providing a rigid kink along the acyl chain that double bonds do not. Additionally, CPs not only inhibit the fluid-to-gel phase transition of the corresponding lipids but also prevent phase transitions in unsaturated lipids by interacting with various lipids in the membrane. This study helped in explaining why E. coli bacteria, which are vulnerable to freezing environments, utilize cyclopropanation of acyl chain double bonds. This explanation may also apply to other bacteria that adopt cyclopropanation as a survival strategy.
C double bonds, a group of modified lipid membranes were also simulated by replacing cyclopropane (CP) groups with either single or double bonds. No differences were observed between the CP fatty acids with the unsaturated ones in coarse-grained resolution, suggesting that the CG model may not adequately represent the effects of cyclopropane-containing lipids. However, at the all-atom resolution, a noticeable difference emerged in membrane properties due to the presence of CPs and unsaturation, especially at lower temperatures. CPs are particularly effective at preventing close packing of membrane lipids, providing a rigid kink along the acyl chain that double bonds do not. Additionally, CPs not only inhibit the fluid-to-gel phase transition of the corresponding lipids but also prevent phase transitions in unsaturated lipids by interacting with various lipids in the membrane. This study helped in explaining why E. coli bacteria, which are vulnerable to freezing environments, utilize cyclopropanation of acyl chain double bonds. This explanation may also apply to other bacteria that adopt cyclopropanation as a survival strategy.
Role of hydroxylation and methyl branching
The cell membrane of Archaea includes lipids with isoprenoid carbon chains connected to glycerol by ether bonds. Of these, isoprenoid glycerol dialkyl glycerol tetraethers (GDGTs) are the widely reported membrane lipids. The methylation of GDGTs at low temperatures is reported as the fundamental HVA strategy of the Archaea class of microorganisms. The methyl modification of GDGTs modulates the fluidity and packing of the membrane under cold stress. Recent MD simulation studies by Naafs et al.148 and Zhou et al.149 provided insights into the structural changes in the membrane of archaeal cells in response to cold environments. The methyl groups hinder the packing of lipids and attenuate the phase transition changes in membrane fluidity. Their study extensively detailed the structural features including area per lipid, core thickness, core order, hydrogen bonding between lipid with lipid and lipid with water, and mobility of lipids compared between membranes with and without the methyl modifications. The study148 compared tetra-, penta-, and hexa-methylated GDGTs with unmethylated ones at a constant temperature of 300 K and observed a consecutive increase in membrane fluidity and a decrease in membrane rigidity with the increase in degree of methylation. This observation supports the fact that in bacterial membranes the degree of methylation decreases with an increase in temperature owing to its cold adaptation.4. Protecting lipid membrane via OMA: molecular level insights
Osmolytes, which are low molecular weight organic solutes, help in maintaining cell volume under stress and stabilize the membrane structure. Some examples are amino acids, sugars, polyols, methylamines, and urea etc. Often functioning as antioxidants150 they support redox balance. These 'compatible solutes' do not interfere with macromolecules and can be regulated without disrupting cellular functions.151,152 At physiological pH, osmolytes are typically neutral but can be anionic153 in some bacteria and archaea, balanced by potassium ions. According to the compatibility, cells can use various osmolytes interchangeably for protection. Below, we discuss the use of various osmolytes for protecting the cell membrane under different stressed conditions and suggest possible mechanisms for the protection.Dehydration stress
Dehydration can cause phase transition, lateral segregation, and membrane fusion, disrupting the membrane barrier functions and leading to cell injury. Some organisms produce fructans,154 which stabilize the membranes by integrating into lipid headgroups, reducing leakage during freezing or drought. Cryptobiosis, another survival strategy, involves near-undetectable metabolic activity under extreme stress.155,156 Anhydrobiosis, a type of cryptobiosis, occurs in extreme dehydration, and involves the production of different osmolytes.Sugars are among the mostly abundant osmolytes observed in nematodes, tardigrades, bacteria, yeasts, mosses, fungi, pollens, seeds, and higher plants.157–159 Interactions between sugars and cell membranes are believed to play a vital role in preserving cells during desiccation. Experimental techniques and molecular dynamics (MD) simulations have been instrumental in uncovering lipid–sugar interactions. Together, these methods have led to the formulation of three main hypotheses regarding OMA, as discussed below.
(i) Water-replacement hypothesis (WRH): sugars replace water molecules by forming favorable hydrogen bonds around polar and charged groups of lipids, stabilizing the original self-assembly structure of the lipid bilayer even in the absence of water.68,158,160,161
(ii) Water-entrapment hypothesis: saccharides concentrate residual water molecules near the lipid bilayer and thereby maintain solvation, which is necessary for retaining the structural and dynamical properties.162–164
(iii) Vitrification hypothesis: sugars in anhydrobiotic systems act as vitrifying agents, forming amorphous glasses that protect biological structures.165–167 Vitrification reduces structural fluctuations and prevents mechanical disruption.
Note that the above three hypotheses are not only applicable for sugars but also valid for other osmolytes. Previous studies indicated that these hypotheses are not mutually exclusive and may collectively aid in protecting the cell membrane.70,168–174
Experiment and MD simulation suggested that the nature of interaction between lipid bilayers and sugars depends on the concentration of the sugar.175–179 The latter can either bind to or be expelled from a lipid bilayer, depending on their concentration. At low concentrations, small sugars bind strongly to the bilayer, causing the membrane to become thinner and laterally expanded due to the accumulation of sugar at the interface.176–178 At relatively larger concentration, sugars are gradually expelled from the membrane surface. This repulsive interaction helps counteract membrane thinning. This dual behavior of sugar–membrane interactions reconciles conflicting findings in earlier reports on how sugars modulate membrane properties.178,180 Ref. 169 and 181 showed that in multicomponent lipid bilayers, lipid mixing is linked to sugar concentration.
In an MD simulation study by our group70 we investigated how trehalose supports the survival of a realistic Escherichia coli (E. coli) bacterial membrane under severe dehydration. The hydration levels were varied from the fully hydrated state to severely dehydrated conditions. Fig. 6 illustrates that decreasing hydration levels lead to an increase in the packing density of the lipid bilayer measured by area per lipid and membrane thickness. However, unlike single-component lipid bilayers, the E. coli membrane exhibits only partial gel phase formation under reduced hydration. Trehalose plays a crucial role in stabilizing the lipid bilayer under low hydration conditions. By forming hydrogen bonds with lipids (WRH), trehalose prevents excessive lipid proximity and intra-lipid hydrogen bond formation. Importantly, trehalose does not necessarily enhance membrane fluidity but maintains the fluid phase by creating a viscous matrix at high sugar concentrations (vitrification hypothesis). In another work, we compared the effect of trehalose and sucrose on the dehydration-induced phase transition of the E. coli lipid membrane. The results show that both disaccharides exhibit nearly the same efficacy in stabilizing the membrane under severe desiccation.171
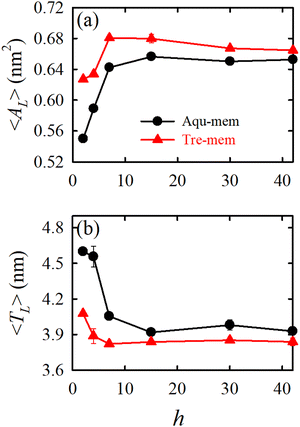 | ||
| Fig. 6 The packing density parameters: (a) average area per lipid 〈AL〉 and (b) average membrane thickness 〈TL〉 for different hydration levels (h) for the Aqu-mem (without trehalose) and Tre-mem (with trehalose) systems. Error bars are calculated over three independent trajectory segments of the production run. Reprinted with permission from ref. 70 Copyright {2023} American Chemical Society. | ||
In addition to sugar molecules, urea and trimethylamine-N-oxide (TMAO) serve as notable osmolytes that aid in stabilizing membranes during dehydration induced by osmotic stress, especially in marine organisms. Besides an experimental work by Schneck et al.,69 our simulation68 provided a thorough molecular level understanding on the impact of urea and TMAO on the dehydration-induced phase transition of lipid membranes from a fluid to a gel phase. TMAO counteracts urea's protein denaturation effects in deep-sea organisms. Fig. 7 shows that as the hydration level changes from fully hydrated (h = 35) to dehydrated (h = 4) conditions, the packing density of the membrane increases, leading to gel phase formation.
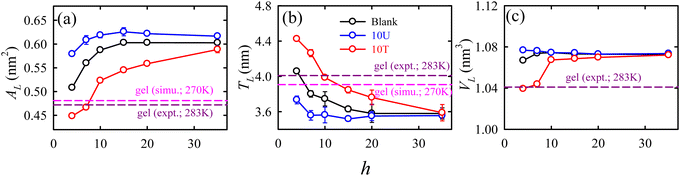 | ||
| Fig. 7 The hydration level-dependent packing density parameters: (a) area per lipid AL, (b) thickness TL, and (c) volume per lipid VL for different membrane systems: blank (black), 10U (blue), and 10T (red). The membrane systems with 10 wt% urea and 10 wt% TMAO are termed as 10U and 10T, respectively. “Blank” represents the membrane system without osmolytes. The error bars are the standard deviations over the average values calculated for three independent trajectory segments. The simulated113 and experimental182 values for the gel phase of DMPC, taken from the literature, are represented by horizontal dashed lines colour coded as pink and dark pink, respectively. Reprinted with permission from ref. 68 Copyright {2021} American Chemical Society. | ||
Urea prevents dehydration-induced changes, whereas TMAO induces a phase transition at lower hydration levels, causing a fluid-to-gel transition in the membrane. The normalized electron density profile (Fig. 8a) reveals that the TMAO density decreases earlier than urea, suggesting that urea penetrates the lipid headgroup region while TMAO stays excluded from the membrane interface. Radial distribution functions (Fig. 8b) and Kirkwood–Buff integrals (Fig. 8c) show that urea has a strong interaction with lipid headgroups, unlike TMAO, which has a weaker affinity. Hydrogen bonding analysis (Fig. 8d) indicated that urea (both an H-bond donor and acceptor) forms favourable H-bonds with lipids, maintaining membrane fluidity. On the contrary, TMAO, with only H-bond accepting ability, primarily accumulates in bulk water rather than the membrane interface. Osmolyte mixtures (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (urea
1 (urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TMAO)183) show similar membrane properties to the system without osmolyte, suggesting that TMAO counteracts urea's effects by removing some urea molecules from the lipid membrane.
TMAO)183) show similar membrane properties to the system without osmolyte, suggesting that TMAO counteracts urea's effects by removing some urea molecules from the lipid membrane.
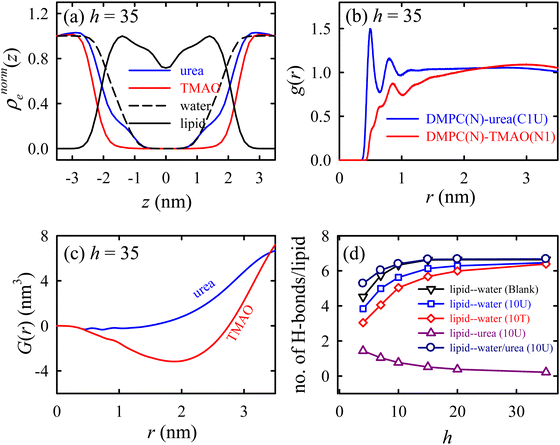 | ||
| Fig. 8 Specific interactions of the osmolytes with the lipid membrane. (a) The normalized EDP (ρnorme(z)) of the lipid, osmolytes, and water. (b) The radial distribution function g(r) between the lipid and the osmolytes. (c) The Kirkwood–Buff integral (KBI) for the osmolytes as a function of distance r. (d) The number of lipid–water and lipid–urea H-bonds per lipid as functions of the hydration level h for different membrane systems. These results are for the Blank (without osmolytes), 10U (with urea), and 10T (with TMAO) membrane systems. Reprinted with permission from ref. 68 Copyright {2021} American Chemical Society. | ||
Cold stress
We have already discussed the importance of HVA of the cell membrane of psychrophiles in previous sections. These organisms can also protect their cellular components under cold stress via cryoprotectants.184–187 Freeze-tolerant organisms synthesize various cryoprotectants, such as DMSO, glycerol, betaine, urea, TMAO, hydroxyectoine, sugars, proline, etc., to combat the cold shock.67,188–190 The impacts of these cryoprotectants on the lipid membrane at ambient conditions are also studied in detail using experiments and MD simulation techniques.67,154,191–197 For example, a simulation study by Smiatek et al.198 demonstrated that hydroxyectoine impacts the properties of aqueous solutions containing DPPC lipid bilayers. Hydroxyectoine, acting as a kosmotrope, was preferentially excluded from the membrane surface, leading to increased surface pressure and stabilization of the membrane structure.There are studies which focused on the protective action of these cryoprotectants on the lipid bilayer under cold stress.67,188–190 Our group113 investigated the effect of TMAO at different temperatures, particularly focusing on the fluid-to-gel phase transition of the lipid bilayer. Our findings indicate that TMAO has a more significant impact on the fluid phase of the membrane compared to the gel phase. Structural comparisons revealed that lipids with shorter acyl chains are more significantly influenced by TMAO. Furthermore, we assessed the gel-to-fluid phase transition temperature (Tm) using temperature annealing simulations. As shown in Fig. 9, Tm increases with higher TMAO concentrations, consistent with experimental observations.
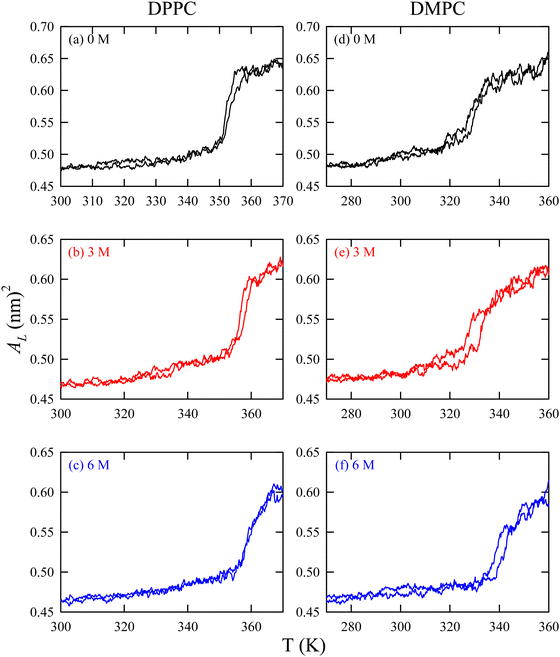 | ||
| Fig. 9 Area per lipid versus temperature for DPPC ((a) 0 M TMAO, (b) 3 M TMAO, and (c) 6 M TMAO) and DMPC ((d) 0 M TMAO, (e) 3 M TMAO, and (f) 6 M TMAO) lipid membranes during heating from gel to fluid phases. AL is different from 〈AL〉 in the sense that the latter is a time-averaged quantity of AL. Reprinted with permission from ref. 113 Copyright {2021} American Chemical Society. | ||
Heat stress
The majority of osmolytes amassed by thermophiles and hyperthermophiles are negatively charged,93,199–201 unlike mesophiles, which tend to accumulate neutral or zwitterionic compounds such as trehalose, glycerol, or proline.202 This observation suggests that charged solutes may play a crucial role in protecting cells from heat-induced damage. Studies have shown that negatively charged solutes, such as phosphodiester compounds or carboxylic acids,199 are particularly effective at enhancing protein stability. However, the use of these charged solutes requires the accumulation of positive counterions like potassium, which can be toxic to most mesophilic bacteria but not to hyperthermophiles, which have adapted to high salt concentrations.93,200,201 Xerophilic fungi, which survive and thrive in extreme dehydrated conditions, use glycerol to counterbalance external osmotic pressure, but during heat shock, they switch to accumulating trehalose.203,204 This switch is accompanied by changes in membrane lipids. After heat shock, fungi show greater thermotolerance in glycerol than in salt media, suggesting a synergistic effect between glycerol and trehalose and a crucial role of osmolyte composition and membrane lipid changes in heat stress adaptation.203,204Under heat stress, most genotypes displayed a notable accumulation of sugar and proline, alongside heightened activity of CAT, GPOX, and SOD.205 Trehalose has been demonstrated to effectively stabilize the native structure of biological systems in extreme environments. An MD simulation work by Liu et al.206 showed that as the temperature escalates from 303 K to 373 K, the presence of trehalose notably diminishes the disparity in membrane properties before and after heating, with this effect being more pronounced at higher trehalose concentrations. With an increase in trehalose concentration from 0 to 256, the relative variation upon heating decreases by approximately 46.6% for the area per lipid and 50.4% for bilayer thickness. This indicates that besides enhancing membrane structure at specific temperatures, trehalose also shields it from damage due to structural changes during abrupt temperature fluctuations. Furthermore, the underlying mechanism is attributed to the slow mobility and strong hydrogen bonding (H-bonding) ability of trehalose. These findings could offer valuable insights into the design of protective agents and processes.
Pressure stress
In the deep sea, high hydrostatic pressure disrupts the lipid membrane structure and fluidity. High pressure alters the lipid membrane's packing density, fluidity, and permeability, causing a phase transition from fluid to gel phase. To counteract this, certain solutes, termed ‘piezolytes,’207,208 accumulate under high-pressure conditions. In shallow-water marine organisms, trimethylamine-N-oxide (TMAO) is typically low or absent, except in ureosmotic fish like sharks. However, deep-sea teleost fishes and certain crustaceans and skates exhibit TMAO concentrations up to 300 mmol kg−1, increasing with depth.66,209,210 TMAO is known to stabilize the native structure of proteins. Deep-sea bacteria accumulate β-hydroxybutyrate in response to hydrostatic and osmotic pressure.Some experimental studies extensively investigated the effect of pressure on the structure and phase behavior of various lipid bilayers.211–213 The effect of osmolytes, such as TMAO and trehalose, on the lipid membrane under high pressure was studied by Winter and coworkers.214,215 Their findings reveal that under high hydrostatic pressure, the lipid order parameters in fluid membranes increase slightly, causing minor lipid elongation. However, TMAO's significant impact on interlamellar spacing in lipid bilayers is largely unaffected by temperature and high pressure. Additionally, TMAO alters the lateral organization of heterogeneous model membranes, promoting the coalescence of lipid domains. This coalescence is likely due to increased line tension between liquid-ordered and disordered domains within raft-like lipid bilayer structures.
These experimental studies motivated us to elucidate the observations using the MD simulation technique.216 We examined the pressure-induced phase transition of the lipid membranes from fluid-to-gel phase and the influence of two osmolytes, urea and TMAO. It was observed that neither solute can inhibit the pressure-induced phase transition of the membrane. Under high pressure, both solutes have negligible impact, and an interdigitated gel phase forms with reduced membrane thickness (Fig. 10). Urea maintains membrane fluidity at lower pressures through hydrogen bonds with lipid headgroups, while TMAO increases further dehydration. High pressure disrupts urea's hydrogen bonding, promoting the gel phase transition and expelling water and solutes from the membrane. These findings suggest that deep-sea organisms might use other strategies, like HVA, to protect their membranes under high pressure. Our findings align closely with the conclusions drawn by Schneck et al.69,116 in their combined experimental and simulation investigations into the impact of urea and TMAO on lipid membranes.
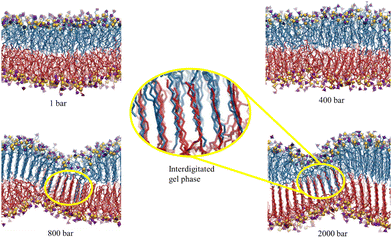 | ||
| Fig. 10 The final configurations of the simulation trajectories of the DMPC membrane in the Blank membrane system (without the osmolytes) at 1, 400, 800, and 2000 bar. The hydrogen-atoms of lipid, water and osmolyte molecules are not shown for clarity. The nitrogen and phosphorous atoms of the lipid headgroup are color-coded as violet and yellow, respectively. The magnified version of the interdigitated gel phase is shown. The opposite leaflets are shown in different colors. Reprinted with permission from ref. 216 Copyright {2022}American Chemical Society. | ||
Perturbing solutes
Certain organic osmolytes can counteract the disruptive effects of solutes that accumulate during osmotic stress and disrupt the macromolecules. Urea serves as one such disruptant and is counteracted by trimethylamine-N-oxide (TMAO).68,69,183 Elevation in the concentrations of specific inorganic ions (such as Na+ and K+) above their typical intracellular levels can negatively impact both the catalytic rate and the apparent Michaelis constant, Km of different enzymes found in plants and animals.202 Consequently, high intracellular salt concentrations during osmotic stress are likely to impair metabolic function and the maintenance of proper transmembrane potentials. Methylamines have been observed to mitigate the disruptive effects of salts.217 Methylated forms of glycine, such as sarcosine, dimethylglycine, and glycine betaine, can alleviate the inhibition of plant enzyme activity induced by NaCl, with the level of protection increasing in correlation with the degree of methylation. For instance, the introduction of exogenous glycine betaine (GB) significantly enhanced the salt tolerance of common bean plants.218,219 This enhancement was attributed to the augmentation of antioxidant defense mechanisms, encompassing both enzymatic (such as peroxidase, superoxide dismutase, and catalase) and non-enzymatic (like proline and glutathione) agents.202 The salt tolerance induced by GB in common bean plants primarily relies on its osmoregulatory effect, with its antioxidant capacity playing a secondary role. GB results in a considerable reduction in Na+ accumulation while concurrently promoting K+ uptake, thereby maintaining a higher K+/Na+ ratio.2195. Role of proteins in stabilizing lipid membranes under stress
Proteins play crucial roles in stabilizing lipid membranes under various stress conditions. In Section 2, we discussed how certain proteins aid in the homeoviscous adaptation of cell membranes, preventing the harmful effects of fluid-to-gel phase transitions in the lipid bilayer. Proteins also directly stabilize lipid membranes through interactions. Understanding these mechanisms provides insights into cellular adaptation and resilience. The protective roles of proteins on lipid membranes are summarized below with a few examples.At low temperatures, proteins protect lipid membranes both directly and indirectly. Cold shock proteins (CSPs) are expressed in response to low temperatures and maintain membrane fluidity by directly interacting with the lipid bilayer. The best-characterized CSP is CspA, which is the primary CSP expressed in Escherichia coli when the temperature decreases.220,221 CSPs facilitate the modification of lipid composition to ensure proper fluidity and functionality.221,222 On the other hand, cryoprotective proteins indirectly protect cell lipid membranes under cold stress. Antifreeze proteins, a type of cryoprotective protein, prevent ice formation within intracellular water.223–225 Experimental and simulation studies suggest that antifreeze proteins inhibit the growth and crystallization of ice by binding to ice crystals.226–230 This inhibition protects membrane integrity in freezing conditions.
The detrimental effect of heat stress on lipid membranes can be mitigated by proteins. The heat shock proteins (HSPs) can assist in refolding denatured proteins and stabilizing protein structures associated with the membrane, maintaining membrane integrity.231,232 HSP70, for example, binds to unfolding proteins and prevents aggregation, helping to preserve the overall stability of the membrane. Thermostable proteins are another class of proteins found in thermophiles that remain stable and functional at high temperatures. They can support membrane stability by maintaining their interactions with the lipid bilayer. These proteins often possess unique structural features that confer stability and enable them to operate under thermal stress.
Proteins play a crucial role in protecting lipid membranes under dehydration stress. Dehydrins, disordered proteins expressed in plants during embryogenesis and water-related stress, accumulate during dehydration and bind to phospholipids, preserving membrane structure. Acting as molecular chaperones, dehydrins stabilize macromolecules and membranes against dehydration-induced stress. Although the exact molecular function and structural mechanisms of dehydrins are not fully understood, evidence suggests that they protect cell membrane structure and dynamics.233,234 For example, the cold-induced dehydrin Lti30 binds to membranes via its conserved K segments. This binding, regulated by pH and phosphorylation, shifts the membrane phase transition to lower temperatures, aiding in cold stress adaptation. Several models propose that dehydrins stabilize plasma and organelle membranes,183,235–238 act as chaperones or cryoprotectants,239 are hygroscopic to prevent complete dehydration,240 and bind metal ions.241 Dehydrins contain a high proportion of hydrophilic and charged amino acids and are characterized by repetitive K-segments. Studies on dehydrin Lti30 (K6) from Arabidopsis thaliana show that it strongly associates with anionic lipid membranes, where the disordered K-segments fold into α-helices on the membrane surface.242–245 Aquaporins are the water channel proteins that regulate water transport, which can indirectly help in stabilizing membranes under desiccation. These channels manage water loss and maintain membrane hydration. They facilitate rapid water movement across the cell membrane, crucial for maintaining cell turgor and preventing desiccation damage.246
6. Concluding remarks
The study of extremophiles has unveiled remarkable adaptive strategies that enable organisms to thrive in some of Earth's most hostile environments. Through homeoviscous adaptation (HVA), these organisms meticulously regulate their lipid compositions to maintain the structural integrity and fluidity of their cell membranes amidst external stresses. By modulating lipid acyl chains and head groups, HVA ensures optimal barrier function, facilitates cell communication, regulates molecular transport, maintains cell shape and stability, and supports crucial cellular interactions such as recognition and adhesion. In parallel, osmolyte-mediated adaptation (OMA) employs small organic molecules to stabilize lipid membranes, protecting against fluctuations in osmotic pressure and environmental extremes. The insights gained from molecular-level studies of HVA and OMA not only deepen our understanding of extremophile biology but also offer insights into the broader potential for life in extreme environments, including extraterrestrial habitats.Looking forward, future research on extremophiles is poised to advance our understanding in several key areas. First, continued exploration of novel extremophiles and their adaptive mechanisms will expand our knowledge of the limits of life and its adaptation strategies under extreme conditions. This includes investigating extremophiles in unexplored terrestrial environments and potentially extraterrestrial habitats, leveraging advancements in technology and interdisciplinary approaches. Second, integrating omics technologies with structural biology and computational modeling will provide comprehensive insights into the molecular underpinnings of HVA and OMA. This holistic approach will elucidate how genetic, proteomic, and metabolomic adaptations synergistically contribute to membrane stability and function. Moreover, applying these insights to synthetic biology and biomaterials science holds promise for developing resilient biotechnological applications and materials that mimic extremophile strategies. Ultimately, by unraveling the adaptive strategies of extremophiles, we not only advance our scientific understanding but also inspire innovative solutions for sustainability, biotechnology, and the search for life beyond Earth.
Data availability
Data will be made available on request to the authors.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
A. M. acknowledges IIT Patna for funding. S. E. acknowledges the DST Inspire Fellowship (INSPIRE No. IF180090) for her research fellowships.References
- N. Merino, H. S. Aronson, D. P. Bojanova, J. Feyhl-Buska, M. L. Wong, S. Zhang and D. Giovannelli, Front. Microbiol., 2019, 10, 780 CrossRef.
- R. Cavicchioli, Astrobiology, 2002, 2, 281–292 CrossRef CAS PubMed.
- E. V. Pikuta, R. B. Hoover and J. Tang, Crit. Rev. Microbiol., 2007, 33, 183–209 CrossRef CAS PubMed.
- S. Ballweg, E. Sezgin, M. Doktorova, R. Covino, J. Reinhard, D. Wunnicke, I. Hänelt, I. Levental, G. Hummer and R. Ernst, Nat. Commun., 2020, 11, 756 CrossRef CAS PubMed.
- I. Kohli, N. C. Joshi, S. Mohapatra and A. Varma, Curr. Genomics, 2020, 21, 96–110 CAS.
- J. Schultz, A. dos Santos, N. Patel and A. S. Rosado, J. Indian Inst. Sci., 2023, 103, 721–737 CrossRef.
- N. Merino, H. S. Aronson, D. P. Bojanova, J. Feyhl-Buska, M. L. Wong, S. Zhang and D. Giovannelli, Front. Microbiol., 2019, 10, 780 CrossRef.
- N. Thakur, S. P. Singh and C. Zhang, Curr. Res. Microb. Sci., 2022, 3, 100141 CAS.
- L. J. Rothschild and R. L. Mancinelli, Nature, 2001, 409, 1092–1101 CrossRef CAS.
- R. D. MacElroy, BioSystems, 1974, 6, 74–75 CrossRef.
- H. B. Rappaport and A. M. Oliverio, Nat. Commun., 2023, 14, 4959 CrossRef CAS PubMed.
- A. Gree, Nature, 2020, 587, 165 CrossRef.
- P. H. Rampelotto, Life, 2013, 3, 482–485 CrossRef PubMed.
- S. Onofri, R. de la Torre, J.-P. de Vera, S. Ott, L. Zucconi, L. Selbmann, G. Scalzi, K. J. Venkateswaran, E. Rabbow and F. J. Sánchez Iñigo, Astrobiology, 2012, 12, 508–516 CrossRef PubMed.
- M. Ackermann, Nat. Rev. Microbiol., 2015, 13, 497–508 CrossRef CAS PubMed.
- B. Xue, P. Sartori and S. Leibler, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 13847–13855 CrossRef CAS.
- W. Johannsen, Am. Naturalist, 1911, 45, 129–159 CrossRef.
- M. Jin, Y. Gai, X. Guo, Y. Hou and R. Zeng, Mar. Drugs, 2019, 17, 656 CrossRef CAS PubMed.
- D. Zhu, W. A. Adebisi, F. Ahmad, S. Sethupathy, B. Danso and J. Sun, Front. Bioeng. Biotechnol., 2020, 8, 483 CrossRef PubMed.
- M. Khan and T. A. Sathya, Crit. Rev. Food Sci. Nutr., 2018, 58, 2017–2025 CrossRef CAS.
- A. Sharma, D. Parashar and T. Satyanarayana, Biotechnology of Extremophiles, Springer, Cham, 2016, pp. 215–241.
- P. Corral, M. A. Amoozegar and A. Ventosa, Mar. Drugs, 2019, 18, 33 CrossRef PubMed.
- R. Mehta, P. Singhal, H. Singh, D. Damle and A. K. Sharma, 3 Biotechnol., 2016, 6, 81 Search PubMed.
- J. A. Coker, F1000Research, 2016, 5, 396 Search PubMed.
- D. A. Cowan, Trends Biotechnol., 1992, 10, 315–323 CrossRef CAS PubMed.
- R. A. Herbert, Trends Biotechnol., 1992, 10, 395–402 CrossRef CAS.
- R. Margesin and F. Schinner, J. Biotechnol., 1994, 33, 1–14 CrossRef CAS.
- C. J. Reed, H. Lewis, E. Trejo, V. Winston and C. Evilia, Archaea, 2013, 2013, 373275 CrossRef.
- J. Pang and R. K. Allemann, Phys. Chem. Chem. Phys., 2007, 9, 711–718 RSC.
- A. Paz, D. Mester, I. Baca, E. Nevo and A. Korol, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 2951–2956 CrossRef CAS PubMed.
- K. E. Nelson, R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson and K. A. Ketchum, Nature, 1999, 399, 323–329 CrossRef CAS PubMed.
- S. D'Amico, T. Collins, J. C. Marx, G. Feller, C. Gerday and C. Gerday, EMBO Rep., 2006, 7, 385–389 CrossRef.
- G. Feller and C. Gerday, Nat. Rev. Microbiol., 2003, 1, 200–208 CrossRef CAS PubMed.
- Y.-H. Chen, C.-W. Lu, Y.-T. Shyu and S.-S. Lin, Sci. Rep., 2017, 7, 13037 CrossRef PubMed.
- P. Corral, M. A. Amoozegar and A. Ventosa, Mar. Drugs, 2019, 18, 33 CrossRef PubMed.
- A. Saini, A. Kumar, G. Singh and S. K. Giri, Microbial stress response: mechanisms and data science, ACS Publications, 2023, pp. 1–21 Search PubMed.
- K. Horikoshi, Alkaliphiles, Proc. Jpn. Acad., Ser. B, 2004, 80(4), 166–178 CrossRef CAS . Epub 2004 Apr 1.
- A. Sharma, Y. Kawarabayasi and T. Satyanarayana, Extremophiles, 2012, 16, 1–19 CrossRef CAS.
- K. Horikoshi, Proc. Jpn. Acad., Ser. B, 2004, 80, 166–178 CrossRef CAS.
- H. Richard and J. W. Foster, J. Bacteriol., 2004, 186, 6032–6041 CrossRef CAS PubMed.
- G. W. Tyson, J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar and J. F. Banfield, Nature, 2004, 428, 37–43 CrossRef CAS PubMed.
- D. Ma, P. Lu, C. Yan, C. Fan, P. Yin, J. Wang and Y. Shi, Nature, 2012, 483, 632–636 CrossRef CAS.
- C. Kato, L. Li, Y. Nogi, Y. Nakamura, J. Tamaoka and K. Horikoshi, Appl. Environ. Microbiol., 1998, 64, 1510–1513 CrossRef CAS PubMed.
- K. Wang, Y. Shen, Y. Yang, X. Gan, G. Liu, K. Hu, Y. Li, Z. Gao, L. Zhu and G. Yan, Nat. Ecol. Evol., 2019, 3, 823–833 CrossRef.
- K. Horikoshi, Curr. Opin. Microbiol., 1998, 1, 291–295 CrossRef CAS.
- M. J. Daly and K. W. Minton, J. Bacteriol., 1995, 177, 5495–5505 CrossRef CAS PubMed.
- M. J. Daly and K. W. Minton, Gene, 1997, 187, 225–229 CrossRef CAS PubMed.
- M. Cordovana, A. Deni, M. Kostrzewa, M. Abdalla and S. Ambretti, New Microbes New Infect., 2019, 30, 100546 CrossRef CAS PubMed.
- D. Kenzelmann, R. Chiquet-Ehrismann and R. Tucker, Cell. Mol. Life Sci., 2007, 64, 1452–1456 CrossRef CAS.
- K. Simons and D. Toomre, Nat. Rev. Mol. Cell Biol., 2000, 1, 31–39 CrossRef CAS.
- S. J. Singer and G. L. Nicolson, Science, 1972, 175, 720–731 CrossRef CAS.
- J. C. Mathai, S. Tristram-Nagle, J. F. Nagle and M. L. Zeidel, J. Gen. Physiol., 2008, 131, 69–76 CrossRef CAS.
- J. V. Vermaas, R. A. Dixon, F. Chen, S. D. Mansfield, W. Boerjan, J. Ralph, M. F. Crowley and G. T. Beckham, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 23117–23123 CrossRef CAS.
- A. Walter and J. Gutknecht, J. Membr. Biol., 1986, 90, 207–217 CrossRef CAS PubMed.
- R. M. Venable, A. Kramer and R. W. Pastor, Chem. Rev., 2019, 119, 5954–5997 CrossRef CAS PubMed.
- A. L. Lomize and I. D. Pogozheva, J. Chem. Inf. Model., 2019, 59, 3198–3213 CrossRef CAS.
- H. T. McMahon and J. L. Gallop, Nature, 2005, 438, 590–596 CrossRef CAS PubMed.
- B. Honig and L. Shapiro, Cell, 2020, 181, 520–535 CrossRef CAS.
- B. M. Gumbiner, Cell, 1996, 84, 345–357 CrossRef CAS PubMed.
- T. A. Springer, Nature, 1990, 346, 425–434 CrossRef CAS PubMed.
- R. Ernst, C. S. Ejsing and B. Antonny, J. Mol. Biol., 2016, 428, 4776–4791 CrossRef CAS PubMed.
- M. Sinensky, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 522–525 CrossRef CAS PubMed.
- S. Erimban and S. Daschakraborty, J. Phys. Chem. Lett., 2020, 11, 7709–7716 CrossRef CAS PubMed.
- M. H. Weber, W. Klein, L. Müller, U. M. Niess and M. A. Marahiel, Mol. Microbiol., 2001, 39, 1321–1329 CrossRef CAS PubMed.
- J. R. Hazel, Annu. Rev. Physiol., 1995, 57, 19–42 CrossRef CAS PubMed.
- P. H. Yancey, M. E. Clark, S. C. Hand, R. D. Bowlus and G. N. Somero, Science, 1982, 217, 1214–1222 CrossRef CAS.
- A. Nowacka, S. Douezan, L. Wadsö, D. Topgaard and E. Sparr, Soft Matter, 2012, 8, 1482–1491 RSC.
- A. Maiti and S. Daschakraborty, J. Phys. Chem. B, 2021, 125, 10149–10165 CrossRef CAS PubMed.
- A. Wolde-Kidan, Q. D. Pham, A. Schlaich, P. Loche, E. Sparr, R. R. Netz and E. Schneck, Phys. Chem. Chem. Phys., 2019, 21, 16989–17000 RSC.
- A. Maiti and S. Daschakraborty, J. Phys. Chem. B, 2023, 127, 4496–4507 CrossRef CAS PubMed.
- L. M. Crowe, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 2002, 131, 505–513 CrossRef PubMed.
- M. Reina-Bueno, M. Argandoña, J. J. Nieto, A. Hidalgo-García, F. Iglesias-Guerra, M. J. Delgado and C. Vargas, BMC Microbiol., 2012, 12, 1–17 CrossRef.
- Y. Newman, S. Ring and C. Colaco, Biotechnol. Genet. Eng. Rev., 1993, 11, 263–294 CrossRef CAS PubMed.
- J. A. Vriezen, F. J. De Bruijn and K. Nusslein, Appl. Environ. Microbiol., 2007, 73, 3451–3459 CrossRef CAS.
- O. Kandror, A. DeLeon and A. L. Goldberg, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9727–9732 CrossRef CAS PubMed.
- M. Sinensky, J. Bacteriol., 1971, 106, 449–455 CrossRef CAS PubMed.
- R. Evans, P. McClure, G. Gould and N. Russell, Int. J. Food Microbiol., 1998, 40, 159–167 CrossRef CAS.
- A. Kropinski, V. Lewis and D. Berry, J. Bacteriol., 1987, 169, 1960–1966 CrossRef CAS PubMed.
- C. Gill and J. Suisted, Microbiology, 1978, 104, 31–36 CrossRef CAS.
- W. C. Hazeleger, J. D. Janse, P. Koenraad, R. R. Beumer, F. M. Rombouts and T. Abee, Appl. Environ. Microbiol., 1995, 61, 2713–2719 CrossRef CAS.
- D. L. Macdonald and H. Goldfine, Appl. Environ. Microbiol., 1991, 57, 3517–3521 CrossRef CAS PubMed.
- M. Suutari and S. Laakso, Appl. Environ. Microbiol., 1992, 58, 2338–2340 CrossRef CAS.
- M. Suutari and S. Laakso, Biochim. Biophys. Acta, Lipids Lipid Metab., 1992, 1126, 119–124 CrossRef CAS PubMed.
- D. A. Los and N. Murata, Biochim. Biophys. Acta, Biomembr., 2004, 1666, 142–157 CrossRef CAS PubMed.
- C. E. Martin, C.-S. Oh and Y. Jiang, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2007, 1771, 271–285 CrossRef CAS.
- E. Saita, D. Albanesi and D. de Mendoza, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2016, 1861, 837–846 CrossRef CAS PubMed.
- D. Thewke, M. Kramer and M. S. Sinensky, Biochem. Biophys. Res. Commun., 2000, 273, 1–4 CrossRef CAS.
- P. Tiku, A. Gracey, A. Macartney, R. Beynon and A. Cossins, Science, 1996, 271, 815–818 CrossRef CAS.
- K. R. Levental, J. L. Symons, Y.-Y. Fan, R. Chapkin, R. Ernst and I. Levental, Biophys. J., 2020, 118, 77a CrossRef.
- A. Singh, K. P. Krishnan, D. Prabaharan and R. K. Sinha, J. Basic Microbiol., 2017, 57, 770–780 CrossRef CAS PubMed.
- G. Chwastek, M. A. Surma, S. Rizk, D. Grosser, O. Lavrynenko, M. Rucińska, H. Jambor and J. Sáenz, Cell Rep., 2020, 32, 108165 CrossRef CAS.
- E. A. Saita and D. de Mendoza, Biochim. Biophys. Acta, Biomembr., 2015, 1848, 1757–1764 CrossRef CAS.
- A. Somayaji, C. R. Dhanjal, R. Lingamsetty, R. Vinayagam, R. Selvaraj, T. Varadavenkatesan and M. Govarthanan, Microbiol. Res., 2022, 263, 127115 CrossRef CAS PubMed.
- G. Drin, Annu. Rev. Biochem., 2014, 83, 51–77 CrossRef CAS PubMed.
- R. Covino, G. Hummer and R. Ernst, Mol. Cell, 2018, 71, 458–467 CrossRef CAS PubMed.
- R. Ernst, S. Ballweg and I. Levental, Curr. Opin. Cell Biol., 2018, 53, 44–51 CrossRef CAS PubMed.
- D. de Mendoza and J. E. Cronan Jr, Trends Biochem. Sci., 1983, 8, 49–52 CrossRef CAS.
- D. Albanesi, M. C. Mansilla and D. de Mendoza, J. Bacteriol., 2004, 186, 2655–2663 CrossRef CAS.
- W. Fan and R. M. Evans, Cell, 2015, 161, 962–963 CrossRef CAS.
- L. E. Cybulski, D. Albanesi, M. C. Mansilla, S. Altabe, P. S. Aguilar and D. De Mendoza, Mol. Microbiol., 2002, 45, 1379–1388 CrossRef CAS.
- D. d Mendoza, Annu. Rev. Microbiol., 2014, 68, 101–116 CrossRef.
- M. Inaba, I. Suzuki, B. Z. Szalontai, Y. Kanesaki, D. A. Los, H. Hayashi and N. Murata, J. Biol. Chem., 2003, 278, 12191–12198 CrossRef CAS.
- D. Huster, S. Maiti and A. Herrmann, Adv. Mater., 2024, 36, 2312898 CrossRef CAS.
- T. Pawson and J. D. Scott, Science, 1997, 278, 2075–2080 CrossRef CAS PubMed.
- L. Klug and G. Daum, FEMS Yeast Res., 2014, 14, 369–388 CrossRef CAS PubMed.
- C. Barrero-Sicilia, S. Silvestre, R. P. Haslam and L. V. Michaelson, Plant Sci., 2017, 263, 194–200 CrossRef CAS.
- M. C. Mansilla, L. E. Cybulski, D. Albanesi and D. de Mendoza, J. Bacteriol., 2004, 186, 6681–6688 CrossRef CAS.
- G. M’Baye, Y. Mély, G. Duportail and A. S. Klymchenko, Biophys. J., 2008, 95, 1217–1225 CrossRef.
- W. Chen, F. Duša, J. Witos, S.-K. Ruokonen and S. K. Wiedmer, Sci. Rep., 2018, 8, 14815 CrossRef.
- E. L. Elson, E. Fried, J. E. Dolbow and G. M. Genin, Annu. Rev. Biophys., 2010, 39, 207–226 CrossRef CAS.
- A. Kumar and S. Daschakraborty, Phys. Chem. Chem. Phys., 2023, 25, 31431–31443 RSC.
- S. Erimban and S. Daschakraborty, J. Phys. Chem. Lett., 2020, 11, 7709–7716 CrossRef CAS.
- A. Maiti and S. Daschakraborty, J. Phys. Chem. B, 2021, 125, 1167–1180 CrossRef CAS PubMed.
- S. Malik and A. Debnath, J. Chem. Phys., 2021, 154, 174904 CrossRef CAS.
- M. J. Janiak, D. M. Small and G. G. Shipley, J. Biol. Chem., 1979, 254, 6068–6078 CrossRef CAS.
- Q. D. Pham, A. Wolde-Kidan, A. Gupta, A. Schlaich, E. Schneck, R. R. Netz and E. Sparr, J. Phys. Chem. B, 2018, 122, 6471–6482 CrossRef CAS PubMed.
- A. Cossins and C. Prosser, Proc. Natl. Acad. Sci. U. S. A., 1978, 75, 2040–2043 CrossRef CAS.
- J. R. Willdigg and J. D. Helmann, Front. Mol. Biosci., 2021, 8, 634438 CrossRef CAS PubMed.
- N. Russell, Philos. Trans. R. Soc., B, 1990, 326, 595–611 CAS.
- N. Russell, FEMS Microbiol. Lett., 1978, 4, 335–338 CrossRef CAS.
- G. Wu, R. Baumeister and T. Heimbucher, Cells, 2023, 12, 1353 CrossRef CAS PubMed.
- K. Zhu, X. Ding, M. Julotok and B. J. Wilkinson, Appl. Environ. Microbiol., 2005, 71, 8002–8007 CrossRef CAS.
- T. Hori, N. Nakamura and H. Okuyama, J. Biochem., 1987, 101, 949–956 CrossRef CAS PubMed.
- X. Bao, M. C. Koorengevel, M. J. Groot Koerkamp, A. Homavar, A. Weijn, S. Crielaard, M. F. Renne, J. H. Lorent, W. J. Geerts and M. A. Surma, EMBO J., 2021, 40, e107966 CrossRef CAS.
- S. E. Diomande, C. Nguyen-The, M.-H. Guinebretière, V. Broussolle and J. Brillard, Front. Microbiol., 2015, 6, 813 Search PubMed.
- Y.-M. Zhang and C. O. Rock, Nat. Rev. Microbiol., 2008, 6, 222–233 CrossRef CAS.
- K.-H. Choi, R. J. Heath and C. O. Rock, J. Bacteriol., 2000, 182, 365–370 CrossRef CAS.
- X. Qiu, A. E. Choudhry, C. A. Janson, M. Grooms, R. A. Daines, J. T. Lonsdale and S. S. Khandekar, Protein Sci., 2005, 14, 2087–2094 CrossRef CAS PubMed.
- G. S. Kumar, M. Jagannadham and M. Ray, J. Bacteriol., 2002, 184, 6746–6749 CrossRef CAS PubMed.
- A. Tamby, J. S. Sinninghe Damsté and L. Villanueva, Front. Mol. Biosci., 2023, 9, 1058381 CrossRef.
- I. A. Guschina and J. L. Harwood, FEBS Lett., 2006, 580, 5477–5483 CrossRef CAS.
- D. W. Grogan and J. E. Cronan Jr, Microbiol. Mol. Biol. Rev., 1997, 61, 429–441 CAS.
- H. Velly, M. Bouix, S. Passot, C. Pénicaud, H. Beinsteiner, S. Ghorbal, P. Lieben and F. Fonseca, Appl. Microbiol. Biotechnol., 2015, 99, 907–918 CrossRef CAS.
- C. C. de Carvalho, Res. Microbiol., 2012, 163, 125–136 CrossRef CAS PubMed.
- Y. Y. Chang and J. E. Cronan, Mol. Microbiol., 1999, 33, 249–259 CrossRef CAS.
- B. H. Kim, S. Kim, H. G. Kim, J. Lee, I. S. Lee and Y. K. Park, Microbiology, 2005, 151, 209–218 CrossRef CAS.
- M. Salvador-Castell, M. Golub, N. Erwin, B. Demé, N. J. Brooks, R. Winter, J. Peters and P. M. Oger, Commun. Biol., 2021, 4, 653 CrossRef CAS.
- M. Sugahara, M. Uragami, X. Yan and S. L. Regen, JACS, 2001, 123, 7939–7940 CrossRef CAS.
- G. N. Somero, B. L. Lockwood and L. Tomanek, Biochemical adaptation: response to environmental challenges, from life's origins to the Anthropocene, Sinauer Associates, Incorporated Publishers, 2016 Search PubMed.
- P. Kumari, M. Kumar, C. Reddy and B. Jha, Functional ingredients from algae for foods and nutraceuticals, Elsevier, 2013, pp. 87–134 Search PubMed.
- E. Susanto, A. S. Fahmi, M. Hosokawa and K. Miyashita, Mar. Drugs, 2019, 17, 630 CrossRef CAS PubMed.
- B. Narayan, K. Miyashita and M. Hosakawa, J. Aquat. Food Prod. Technol., 2005, 13, 53–70 CrossRef.
- M. Nomura, H. Kamogawa, E. Susanto, C. Kawagoe, H. Yasui, N. Saga, M. Hosokawa and K. Miyashita, J. Appl. Phycol., 2013, 25, 1159–1169 CrossRef CAS.
- K. Gounaris, A. Brain, P. Quinn and W. Williams, Biochim. Biophys. Acta, Bioenerg., 1984, 766, 198–208 CrossRef CAS.
- A. K. Rathod, D. Chavda and M. Manna, J. Chem. Inf. Model., 2023, 63, 3328–3339 CrossRef CAS PubMed.
- S. Erimban and S. Daschakraborty, J. Phys. Chem. B, 2022, 126, 7638–7650 CrossRef CAS.
- A. Maiti, A. Kumar and S. Daschakraborty, J. Phys. Chem. B, 2023, 127, 1607–1617 CrossRef CAS PubMed.
- B. Naafs, A. S. F. Oliveira and A. J. Mulholland, Geochim. Cosmochim. Acta, 2021, 312, 44–56 CrossRef CAS.
- J. Zhou and L. Dong, Chem. Geol., 2024, 644, 121844 CrossRef CAS.
- S. Ejaz, S. Fahad, M. A. Anjum, A. Nawaz, S. Naz, S. Hussain and S. Ahmad, in Sustainable Agriculture Reviews 39, ed. E. Lichtfouse, Springer International Publishing, Cham, 2020, pp. 95–117 DOI:10.1007/978-3-030-38881-2_4.
- P. S. Low, Molecular Basis of the Biological Compatibility of Nature’s Osmolytes, in Transport Processes, Iono- and Osmoregulation, ed. R. Gilles and M. Gilles-Baillien, Proceedings in Life Sciences, Springer, Berlin, Heidelberg, 1985, pp. 469–477 Search PubMed.
- P. H. Yancey, J. Exp. Biol., 2005, 208, 2819–2830 CrossRef CAS.
- Y. Zhang and P. S. Cremer, Annu. Rev. Phys. Chem., 2010, 61, 63–83 CrossRef CAS PubMed.
- I. J. Vereyken, V. Chupin, A. Islamov, A. Kuklin, D. K. Hincha and B. de Kruijff, Biophys. J., 2003, 85, 3058–3065 CrossRef CAS PubMed.
- C. Wang, M. A. Grohme, B. Mali, R. O. Schill and M. Frohme, PLoS One, 2014, 9, e92663 CrossRef PubMed.
- J. S. Clegg, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2001, 128, 613–624 CrossRef CAS PubMed.
- T. Grzyb and A. Skłodowska, Microorganisms, 2022, 10, 432 CrossRef CAS.
- L. Rebecchi, T. Altiero and R. Guidetti, Invertebrate Survival J., 2007, 4, 65–81 Search PubMed.
- J. H. Crowe, F. A. Hoekstra and L. M. Crowe, Annu. Rev. Physiol., 1992, 54, 579–599 CrossRef CAS.
- J. H. Crowe, J. S. Clegg and L. M. Crowe, Prop. Water Foods ISOPOW, 1998, 6, 440–455 Search PubMed.
- J. F. Carpenter, T. Arakawa and J. H. Crowe, Dev. Biol. Stand., 1992, 74, 225–238 CAS ; discussion 238-229.
- P. S. Belton and A. M. Gil, Biopolymers, 1994, 34, 957–961 CrossRef CAS PubMed.
- G. Cottone, G. Ciccotti and L. Cordone, J. Chem. Phys., 2002, 117, 9862–9866 CrossRef CAS.
- R. D. Lins, C. S. Pereira and P. H. Hünenberger, Proteins, 2004, 55, 177–186 CrossRef CAS PubMed.
- N. Grasmeijer, M. Stankovic, H. de Waard, H. W. Frijlink and W. L. Hinrichs, Biochim. Biophys. Acta, 2013, 1834, 763–769 CrossRef CAS.
- W. Q. Sun and A. C. Leopold, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 1997, 117, 327–333 CrossRef.
- W. Q. Sun, T. C. Irving and A. C. Leopold, Physiol. Plant., 1994, 90, 621–628 CrossRef CAS.
- C. S. Pereira and P. H. Hünenberger, J. Phys. Chem. B, 2006, 110, 15572–15581 CrossRef CAS.
- G. Moiset, C. A. López, R. Bartelds, L. Syga, E. Rijpkema, A. Cukkemane, M. Baldus, B. Poolman and S. J. Marrink, J. Am. Chem. Soc., 2014, 136, 16167–16175 CrossRef CAS.
- I. D. Pogozheva, G. A. Armstrong, L. Kong, T. J. Hartnagel, C. A. Carpino, S. E. Gee, D. M. Picarello, A. S. Rubin, J. Lee, S. Park, A. L. Lomize and W. Im, J. Chem. Inf. Model., 2022, 62, 1036–1051 CrossRef CAS.
- N. Rani, A. Maiti and S. Daschakraborty, Chem. Phys. Impact, 2024, 8, 100645 CrossRef.
- M. A. Villarreal, S. B. Díaz, E. A. Disalvo and G. G. Montich, Langmuir, 2004, 20, 7844–7851 CrossRef CAS.
- J. Kapla, J. Wohlert, B. Stevensson, O. Engström, G. Widmalm and A. Maliniak, J. Phys. Chem. B, 2013, 117, 6667–6673 CrossRef CAS.
- B. A. Horta, L. Perić-Hassler and P. H. Hünenberger, J. Mol. Graphics Modell., 2010, 29, 331–346 CrossRef CAS.
- J. Liu, C. Chen and W. Li, Fluid Phase Equilib., 2018, 474, 100–109 CrossRef CAS.
- S. Leekumjorn and A. K. Sum, J. Phys. Chem. B, 2008, 112, 10732–10740 CrossRef CAS.
- E. A. Golovina, A. Golovin, F. A. Hoekstra and R. Faller, Langmuir, 2010, 26, 11118–11126 CrossRef CAS.
- S. S. Stachura, C. J. Malajczuk and R. L. Mancera, Langmuir, 2019, 35, 15389–15400 CrossRef CAS.
- A. Roy, R. Dutta, N. Kundu, D. Banik and N. Sarkar, Langmuir, 2016, 32, 5124–5134 CrossRef CAS.
- E. A. Golovina, A. V. Golovin, F. A. Hoekstra and R. Faller, Biophys. J., 2009, 97, 490–499 CrossRef CAS.
- H. D. Andersen, C. Wang, L. Arleth, G. H. Peters and P. Westh, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1874–1878 CrossRef CAS.
- S. Tristram-Nagle, Y. Liu, J. Legleiter and J. F. Nagle, Biophys. J., 2002, 83, 3324–3335 CrossRef CAS.
- P. H. Yancey, M. E. Clark, S. C. Hand, R. D. Bowlus and G. N. Somero, Science, 1982, 217, 1214–1222 CrossRef CAS.
- B. J. Fuller, Cryo Lett., 2004, 25, 375–388 CAS.
- K. B. Storey and J. M. Storey, in Insects at Low Temperature, ed. R. E. Lee and D. L. Denlinger, Springer US, Boston, MA, 1991, pp. 64–93 DOI:10.1007/978-1-4757-0190-6_4.
- G. D. Elliott, S. Wang and B. J. Fuller, Cryobiology, 2017, 76, 74–91 CrossRef CAS.
- R. E. Watson, S. J. Wiegand, R. W. Clough and G. E. Hoffman, Peptides, 1986, 7, 155–159 CrossRef CAS.
- A. R. Mohammed, A. G. A. Coombes and Y. Perrie, Eur. J. Pharm. Sci., 2007, 30, 406–413 CrossRef CAS.
- J. L. MacCallum, W. F. D. Bennett and D. P. Tieleman, Biophys. J., 2008, 94, 3393–3404 CrossRef CAS.
- S. Björklund, J. Engblom, K. Thuresson and E. Sparr, Eur. J. Pharm. Sci., 2013, 50, 638–645 CrossRef.
- J. Lin, B. Novak and D. Moldovan, J. Phys. Chem. B, 2012, 116, 1299–1308 CrossRef CAS.
- M. Herzog, M. Dwivedi, R. K. Harishchandra, A. Bilstein, H.-J. Galla and R. Winter, Colloids Surf., B, 2019, 178, 404–411 CrossRef CAS.
- D. Harries and J. Rösgen, Methods Cell Biol., 2008, 84, 679–735 CAS.
- Q. D. Pham, A. Wolde-Kidan, A. Gupta, A. Schlaich, E. Schneck, R. R. Netz and E. Sparr, J. Phys. Chem. B, 2018, 122, 6471–6482 CrossRef CAS.
- F. Kosar, N. Aisha, M. Sadiq, F. Al-Qurainy and M. Ashraf, J. Plant Growth Regul., 2019, 38, 606–618 CrossRef CAS.
- K. N. Barton, M. M. Buhr and J. S. Ballantyne, Am. J. Physiol., 1999, 276, R397–406 CAS.
- O. Kukal, A. S. Serianni and J. G. Duman, J. Comput. Phys. B, 1988, 158, 175–183 CrossRef CAS.
- J. Smiatek, R. K. Harishchandra, H. J. Galla and A. Heuer, Biophys. Chem., 2013, 180–181, 102–109 CrossRef CAS.
- B. Kikani, R. Shukla and S. Singh, Curr. Res., Technol. Educ. Topics Appl. Microbiol. Microbial Biotechnol., 2010, 2, 1000–1007 Search PubMed.
- H. Santos, P. Lamosa and N. Borges, Methods in Microbiology, Academic Press, 2006, vol. 35, pp. 173–199 Search PubMed.
- C. Neves, S. da Costa Milton and H. Santos, Appl. Environ. Microbiol., 2005, 71, 8091–8098 CrossRef CAS.
- P. H. Yancey, J. Exp. Biol., 2005, 208, 2819–2830 CrossRef CAS.
- E. A. Ianutsevich, O. A. Danilova, A. B. Antropova and V. M. Tereshina, Fungal Biol., 2023, 127, 909–917 CrossRef CAS.
- S. Sarkar, N. A. Singh and N. Rai, in Extremophilic Fungi: Ecology, Physiology and Applications, ed. S. Sahay, Springer Nature Singapore, Singapore, 2022, pp. 253–270 DOI:10.1007/978-981-16-4907-3_13.
- A. Harsh, Y. K. Sharma, U. Joshi, S. Rampuria, G. Singh, S. Kumar and R. Sharma, Ann. Agric. Sci., 2016, 61, 57–64 CrossRef.
- J. Liu, C. Chen and W. Li, Mol. Simul., 2018, 44, 100–109 CrossRef CAS.
- F. Simonato, S. Campanaro, F. M. Lauro, A. Vezzi, M. D’Angelo, N. Vitulo, G. Valle and D. H. Bartlett, J. Biotechnol., 2006, 126, 11–25 CrossRef CAS.
- A. Scoma, P. Garrido-Amador, D. Nielsen Søren, H. Røy and U. Kjeldsen Kasper, Appl. Environ. Microbiol., 2019, 85, e00802–00819 CrossRef CAS.
- F. Meersman, D. Bowron, A. K. Soper and M. H. Koch, Biophys. J., 2009, 97, 2559–2566 CrossRef CAS.
- T.-Y. Lin and S. N. Timasheff, Biochemistry, 1994, 33, 12695–12701 CrossRef CAS.
- J. Eisenblätter and R. Winter, Biophys. J., 2006, 90, 956–966 CrossRef.
- K. Lai, B. Wang, Y. Zhang and Y. Zhang, Phys. Chem. Chem. Phys., 2012, 14, 5744–5752 RSC.
- S. Purushothaman, P. Cicuta, O. Ces and N. J. Brooks, J. Phys. Chem. B, 2015, 119, 9805–9810 CrossRef CAS.
- M. Manisegaran, S. Bornemann, I. Kiesel and R. Winter, Phys. Chem. Chem. Phys., 2019, 21, 18533–18540 RSC.
- S. R. Al-Ayoubi, P. K. F. Schinkel, M. Berghaus, M. Herzog and R. Winter, Soft Matter, 2018, 14, 8792–8802 RSC.
- A. Maiti and S. Daschakraborty, J. Phys. Chem. B, 2022, 126, 1426–1440 CrossRef CAS.
- S. Rahman, M. T. Rehman, L. R. Singh, M. Warepam, F. Ahmad and T. A. Dar, PLoS One, 2015, 10, e0119597 CrossRef.
- J. Giri, Plant Signaling Behav., 2011, 6, 1746–1751 CrossRef CAS.
- M. R. Sofy, N. Elhawat and A. Tarek, Ecotoxicol. Environ. Saf., 2020, 200, 110732 CrossRef CAS.
- W. Jiang, Y. Hou and M. Inouye, J. Biol. Chem., 1997, 272, 196–202 CrossRef CAS.
- M. B. Al-Fageeh and C. M. Smales, Biochem. J., 2006, 397, 247–259 CrossRef CAS.
- S. Phadtare, J. Alsina and M. Inouye, Curr. Opin. Microbiol., 1999, 2, 175–180 CrossRef CAS.
- A. L. DeVries and D. E. Wohlschlag, Science, 1969, 163, 1073–1075 CrossRef CAS.
- Z. Jia and P. L. Davies, Trends Biochem. Sci., 2002, 27, 101–106 CrossRef CAS.
- T. Sun, F. H. Lin, R. L. Campbell, J. S. Allingham and P. L. Davies, Science, 2014, 343, 795–798 CrossRef CAS.
- C. A. Knight, C. C. Cheng and A. L. DeVries, Biophys. J., 1991, 59, 409–418 CrossRef CAS.
- D. S. C. Yang, M. Sax, A. Chakrabartty and C. L. Hew, Nature, 1988, 333, 232–237 CrossRef CAS.
- D. Wen and R. A. Laursen, Biophys. J., 1992, 63, 1659–1662 CrossRef CAS.
- S. Chakraborty and B. Jana, Langmuir, 2017, 33, 7202–7214 CrossRef CAS.
- S. Chakraborty and B. Jana, J. Phys. Chem. B, 2018, 122, 3056–3067 CrossRef CAS.
- S. Lindquist and E. A. Craig, Annu. Rev. Genet., 1988, 22, 631–677 CrossRef CAS.
- F. U. Hartl and M. Hayer-Hartl, Science, 2002, 295, 1852–1858 CrossRef CAS.
- J. M. Andersson, Q. D. Pham, H. Mateos, S. Eriksson, P. Harryson and E. Sparr, J. Lipid Res., 2020, 61, 1014–1024 CrossRef CAS.
- S. Eriksson, N. Eremina, A. Barth, J. Danielsson and P. Harryson, Plant Physiol., 2016, 171, 932–943 CAS.
- P. Harryson, D. J. Morre and A. S. Sandelius, Plant Physiol., 1996, 110, 631–637 CrossRef.
- L. N. Rahman, F. McKay, M. Giuliani, A. Quirk, B. A. Moffatt, G. Harauz and J. R. Dutcher, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 967–980 CrossRef CAS.
- T. Furuki and M. Sakurai, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 2757–2766 CrossRef CAS.
- M. W. Clarke, K. F. Boddington, J. M. Warnica, J. Atkinson, S. McKenna, J. Madge, C. H. Barker and S. P. Graether, J. Biol. Chem., 2015, 290, 26900–26913 CrossRef CAS.
- M. Hara, S. Terashima and T. Kuboi, J. Plant Physiol., 2001, 158, 1333–1339 CrossRef CAS.
- T. J. Close, Physiol. Plant., 1996, 97, 795–803 CrossRef CAS.
- M. K. Alsheikh, B. J. Heyen and S. K. Randall, J. Biol. Chem., 2003, 278, 40882–40889 CrossRef CAS.
- A. Gupta, J. K. Marzinek, D. Jefferies, P. J. Bond, P. Harryson and T. Wohland, J. Biol. Chem., 2019, 294, 6468–6482 CrossRef CAS.
- S. K. Eriksson, M. Kutzer, J. Procek, G. Gröbner and P. Harryson, Plant Cell, 2011, 23, 2391–2404 CrossRef CAS.
- S. Eriksson, N. Eremina, A. Barth, J. Danielsson and P. Harryson, Plant Physiol., 2016, 171, 932–943 CAS.
- M.-C. Koag, R. D. Fenton, S. Wilkens and T. J. Close, Plant Physiol., 2003, 131, 309–316 CrossRef CAS.
- C. Maurel, L. Verdoucq, D.-T. Luu and V. Santoni, Annu. Rev. Plant Biol., 2008, 59, 595–624 CrossRef CAS.
Footnotes |
| † Equal contribution. |
| ‡ Present address: Department of Chemistry, The University of Utah, Salt Lake City, Utah 84112-0850, US. |
| This journal is © The Royal Society of Chemistry 2024 |