Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Peptide-based self-assembled monolayers (SAMs): what peptides can do for SAMs and vice versa
Carlos
Redondo-Gómez
ab,
Paula
Parreira
ab,
M. Cristina L.
Martins
abc and
Helena S.
Azevedo
 *ab
*ab
ai3S – Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Rua Alfredo Allen, 208, Porto, 4200-135, Portugal. E-mail: hazevedo@i3s.up.pt
bINEB – Instituto de Engenharia Biomédica, Universidade do Porto, Rua Alfredo Allen, 208, Porto, 4200-135, Portugal
cICBAS – Instituto de Ciências Biomédicas Abel Salazar, Universidade do Porto, Rua de Jorge Viterbo Ferreira, 4050-313 Porto, Portugal
First published on 8th March 2024
Abstract
Self-assembled monolayers (SAMs) represent highly ordered molecular materials with versatile biochemical features and multidisciplinary applications. Research on SAMs has made much progress since the early begginings of Au substrates and alkanethiols, and numerous examples of peptide-displaying SAMs can be found in the literature. Peptides, presenting increasing structural complexity, stimuli-responsiveness, and biological relevance, represent versatile functional components in SAMs-based platforms. This review examines the major findings and progress made on the use of peptide building blocks displayed as part of SAMs with specific functions, such as selective cell adhesion, migration and differentiation, biomolecular binding, advanced biosensing, molecular electronics, antimicrobial, osteointegrative and antifouling surfaces, among others. Peptide selection and design, functionalisation strategies, as well as structural and functional characteristics from selected examples are discussed. Additionally, advanced fabrication methods for dynamic peptide spatiotemporal presentation are presented, as well as a number of characterisation techniques. All together, these features and approaches enable the preparation and use of increasingly complex peptide-based SAMs to mimic and study biological processes, and provide convergent platforms for high throughput screening discovery and validation of promising therapeutics and technologies.
1. Introduction
Self-assembling monolayers (SAMs) are molecular monolayers typically formed spontaneously via adsorption onto solid substrates. This phenomenon was firstly described in 1983 by Nuzzo and Allara showing the assembly of dialkyl disulfides on gold (Au) surfaces.1 Conceptualisation of SAMs has evolved over time, with most works from the 1980s2 through mid-1990s, presenting them as ordered molecular assemblies formed by the adsorption of an active surfactant on a solid surface.3 Even though this seemed suitable for SAMs based on alkanethiols (ATs), greater potential for SAMs technologies was early identified, leading the field to striking growth and using adsorbates with increasing synthetic sophistication beyond AT long chain hydrocarbons.From that decade on, SAMs with electroconductive properties were increasingly explored, further moving into biosensing applications involving bioinspired building blocks, which paved the way for peptides to be incorporated into SAMs-based technologies.
SAMs are recognised as two-dimensional (2D) nanomaterials that exhibit thickness of one or few adsorbate molecules, in which the adsorbates organise spontaneously into highly ordered crystalline (or semicrystalline) structures in reproducible ways.4 Typically, molecules align vertically onto the surface with some tilt angle relative to the surface normal,5 enabling molecularly controlled nanostructured surfaces which are straightforward to prepare, easy to characterise, and that are also suitable for high-throughput screening of immobilised molecules.4,6,7 Moreover, SAMs are frequently used as proof-of-concept model surfaces, since information in terms of surface (bio)conjugation strategies to maintain the bioactivity of immobilised biomolecules8 is easily portable to “real world” biomaterials.
These features have made SAMs increasingly applied as biomimetic and biocompatible materials with defined compositions and architectures as part of biomedical applications, including cell adhesion, migration and differentiation, biosensing, antifouling and antimicrobial surfaces,9 among others that will be further described along this review article. This ample range of applications derives from the variety of chemical strategies that can be used to immobilise the adsorbates to different substrates, including chlorosilanes on quartz, silicon or glass substrates, carboxylic acids on metal oxides surfaces and organosulphur compounds on Au, the latter being by far the most explored substrate for fabricating SAMs.
Even though overall progress around SAMs has been extensively reviewed, particularly those involving ATs and dialkanethiols bound to Au surfaces,10 the present review will entirely focus on the structure, preparation, and applications of peptide-based SAMs formed from peptide-based building blocks on many other inorganic substrates. Although several multicomponent peptide assemblies11,12 and self-assembling peptide-based functional biomaterials13 have been reviewed in the literature, SAMs incorporating peptides as building blocks have not been looked in a systematic extent.
This review comprises advances made in peptide-based SAMs over the past 28 years (1995–2023), and its objectives are as follows:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (1) to review the structural, biochemical and functional features and advantages exhibited by SAMs formed by adsorption of peptides on inorganic surfaces; (2) to review the different fabrication approaches and techniques to prepare and characterise these molecularly engineered surfaces; (3) to illustrate applications of peptide-based SAMs interacting with (i) extracellular matrix components, (ii) mammalian cells, (iii) metal ions, (iv) biocatalytic targets, (v) biomarkers, and endorsing (vi) electron transfer, and (vii) antifouling properties; and (4) to sketch some of the challenges and opportunities to explore as future research avenues involving peptide-based SAMs.
(1) to review the structural, biochemical and functional features and advantages exhibited by SAMs formed by adsorption of peptides on inorganic surfaces; (2) to review the different fabrication approaches and techniques to prepare and characterise these molecularly engineered surfaces; (3) to illustrate applications of peptide-based SAMs interacting with (i) extracellular matrix components, (ii) mammalian cells, (iii) metal ions, (iv) biocatalytic targets, (v) biomarkers, and endorsing (vi) electron transfer, and (vii) antifouling properties; and (4) to sketch some of the challenges and opportunities to explore as future research avenues involving peptide-based SAMs.
In the hope of contributing to those scientific communities interested on self-assembling biomaterials and SAMs technologies, this review article intends to provide a comprehensive view of peptide-based SAMs platforms supported by selected examples, while placing the spotlight on peptide-based SAMs and their potential for high-throughput screening and analysis using methods established in many biomedical facilities. The enormous potential of synergies created by merging peptide science and SAMs technologies, i.e., what peptides can do for SAMs and vice versa, is the focus of this review article.
2. Peptides as self-assembled monolayers (SAMs)
Proteins are part of the molecular machinery produced by cells that are used to undertake a vast range of functions, from providing structure (e.g., collagen, actin) to biocatalysis (e.g., enzymes), from defence (e.g., antibodies) to regulation (e.g., hormones), from storage (e.g., ferritin) to transport (e.g., hemoglobin), and many others. Proteins constitute 17% (wt%) of the human body weight14 being essential building blocks of biological tissues.15 Extracellular proteins have received great attention by the biomaterials community focused on the design of synthetic extracellular matrices (ECMs). However, replicating the intricate organisation of the complex protein network in native ECMs remains a goal and challenge in biomaterials engineering.As highlighted in the comprehensive review by Ligorio![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) and Mata16 on function-encoding peptide epitopes, the function of many proteins is encoded by peptide epitopes located in specific regions of the proteins. Once identified, these epitopes can be isolated to derive peptide segments for undertaking mechanistic studies and yield improved understanding of their function, and reveal potentially new applications. Due to their small size, they can be easily synthesised in the mg–g scale, by well-established synthetic chemical methods (solid phase peptide synthesis, SPPS) using fully automated machines, and purified by standard reverse phase high performance liquid chromatography (RP-HPLC) to afford molecules with required grade to undertake reproducible structure–function relationship studies in a range of biological scenarios. Furthermore, they can be designed to contain inbuilt desirable functionalities (–NH2, –COOH, –SH, also present in the side chains of naturally occurring amino acids) to enable their direct immobilisation onto diverse inorganic substrates or conjugated onto pre-formed SAMs17via modification at N- or C-terminus (see Section 2.2).
and Mata16 on function-encoding peptide epitopes, the function of many proteins is encoded by peptide epitopes located in specific regions of the proteins. Once identified, these epitopes can be isolated to derive peptide segments for undertaking mechanistic studies and yield improved understanding of their function, and reveal potentially new applications. Due to their small size, they can be easily synthesised in the mg–g scale, by well-established synthetic chemical methods (solid phase peptide synthesis, SPPS) using fully automated machines, and purified by standard reverse phase high performance liquid chromatography (RP-HPLC) to afford molecules with required grade to undertake reproducible structure–function relationship studies in a range of biological scenarios. Furthermore, they can be designed to contain inbuilt desirable functionalities (–NH2, –COOH, –SH, also present in the side chains of naturally occurring amino acids) to enable their direct immobilisation onto diverse inorganic substrates or conjugated onto pre-formed SAMs17via modification at N- or C-terminus (see Section 2.2).
Advantages of peptide-based SAMs
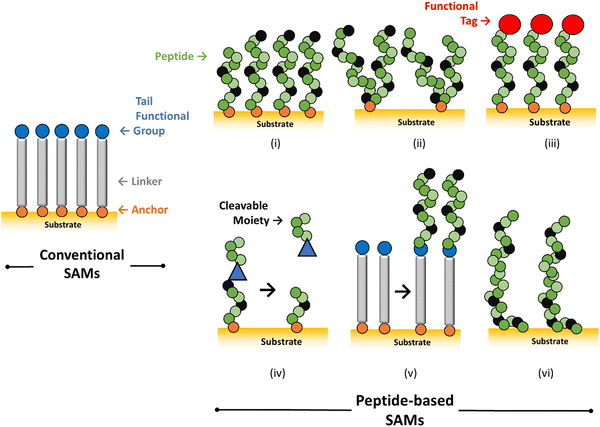
The use of peptides as self-assembling building blocks in SAMs provides several advantages:Molecules used in conventional SAMs usually consist of three segments: a head or anchor group (a thiol group), an alkyl chain (as linker), and a tail functional group (for instance, –COOH, –PO32−, or –OH) (Fig. 1).31 Peptide-based SAMs step-up this molecular design and allow for increasingly more complex nanoarchitectures. The most common strategy for the preparation of peptide-based SAMs involves direct tethering of peptide chains to the substrate via an anchor located either at the N- or C-terminus of the peptide (Fig. 1, panel i). These anchor functionalities comprise ATs, disulfide bonds, cysteine (Cys) residues, organosilanes, phosphonic acids, solid binding sequences, and other strategies (detailed in Section 2.2). The presence of built-in functionalities from amino acid side chains located in mid positions of the peptide sequence can also be exploited as anchors to the substrate (Fig. 1, panel ii). Either free terminus can be functionalised with a reporting tag (Fig. 1, panel iii). However, other strategies have been reported; peptides can be presented in dynamic/stimuli-responsive fashions (Fig. 1, panel iv), they can also be coupled to preformed reactive SAMs or be incorporated as part of AT-substituted peptides (Fig. 1, panel v), and solid-binding sequences can also be used to adsorb the peptide directly onto the solid substate without the need of a proper anchor as in the aforementioned systems (Fig. 1, panel vi).
The following subsections present a detailed description of substrates, anchoring functionalities, and peptide families, sequences and structural features displayed as part of selected peptide-based SAMs examples.
2.1 Substrates
Even though other noble metal surfaces have been studied,37 Au surfaces represent the most extensively studied members of the class of organic SAMs for several reasons. First, Au surfaces are reasonably inert, but bind to organosulphur species with high affinity10 (S–Au bound energy of ∼40 kcal mol−1), leading to Au–S bond formation4 as part of low cost surfaces with high structural order and packing.8,38 Second, Au thin films are easy to prepare through a number of deposition techniques, and they are easy to pattern via lithographic and chemical techniques. Au substrates are of common use in various analytical and spectroscopic techniques (see Section 4), such as ellipsometry, surface plasmon resonance (SPR) spectroscopy, and quartz crystal microbalance with dissipation (QCM-D), which facilitates their straightforward characterisation. Third, Au is compatible with cells and cell culture media conditions, allowing their use as biological interfaces with no evidence of toxicity.4Consequently, SAMs based on organosulphur compounds on Au are straightforward to prepare and exhibit good chemical and thermal stability suitable for a variety of applications.39 Peptide-based SAMs research has mostly focused on sulphur-mediated anchoring to Au substrates, including alkanethiols (ATs, R-SH), dialkyl disulphides (R–S–S–R) and dialkyl sulphides (R–S–R) and will be further described in Section 2.2. As thiols and other organosulphur functionalities can be easily incorporated in peptide sequences to enable immobilisation on Au, numerous Au–peptide-based SAMs have been prepared.
Apart from Au, peptide-based SAMs have also been prepared on other inorganic substrates, including metals such as Ti, Co, Cr, and stainless steels,22 metal oxides via phosphonic acids (R-PO3, R = alkyl chain),40 as well as other substrates such as quartz,35,41 silicon, glass,42 and modified glassy carbon electrodes (GCE), using different organosilane species (R–Si–X3, R2–Si–X2 or R3–Si–X, R = alkyl chain, X = Cl or alkoxy group)43 and other chemical surface reactions,17,44,45 as detailed in Sections 2.2.5 and 2.2.6.
2.2 Anchoring functionalities
Selection of anchor functionalities in peptide-based SAMs plays a pivotal role and must be made as a function of the substrate and application. For instance, the stability of organic SAMs on different substrates has been studied. Thiol-SAMs are not adequate for applications that require long term stability, since thiols tend to be oxidised and be displaced from the surface.46,47 SAMs of organosilanes also have limitations, as they can be hydrolytically unstable in aqueous systems and biological media.43,48 By contrast, phosphonate SAMs are more stable and are a promising system for applications that require long-term stability and have been studied as coatings for dental implants.40The bond between Au substrates and sulphur atoms represents the most frequently used in the formation of peptide-displaying SAMs (Tables 1–3, substrate column). It is important to note that anchor location plays a pivotal role in peptide orientation after tethering, as has been evidenced in the case of Au-bound helical peptides. Morita and collaborators have shown that N-terminally bound peptides generate more densely-packed SAMs than those from C-terminally bound peptides,49 these packing differences could be possibly attributed to several factors, including unfavourable electrostatic repulsion between the dipoles of the helical peptide directing toward the aqueous media and the dipoles of S–Au linkages (S−–Au+) opposing the dipoles from the helix, chain-length difference at the anchor segment of the molecules and different size and polarity of the protecting terminal groups. Therefore, attention must be paid to peptide molecular design and its further surface orientation before peptide-SAMs fabrication.
This section groups six main tethering strategies for designing peptide-displaying SAMs: peptide coupling to ATs, disulfide bonds, Cys residues, solid binding sequences, organosilanes and phosphonic acids, and others approaches including reactive SAMs.
| R–S–H + Au0n → R–S− Au+·Au0n + 1/2H2 |
| Application | Target | Peptide sequencea | Anchor | Substrate | Fabrication method | Comment | Ref. |
|---|---|---|---|---|---|---|---|
| a Amino acid residues are given in one-letter code, except for Aib (α-aminoisobutyric acid, a strongly helicogenic amino acid), and the ones employed as substrate anchors are highlighted in bold, and separated from the rest of the sequence. | |||||||
| Cell culture: binding to ECM components | Fibronectin (FN) | VPQIHGQNKGNQSFEEDTE-C (seven other mutant peptides were assayed) | Cys residue at C-terminus | Au | Immobilisation Au AFM tip | Pure SAM | 105 |
| Laminin (LN) | Very long sequence, 127 amino acids (named as rhNtA, recombinant, derived from human agrin) | N-terminus coupling to thiol-terminated poly(ethylene glycol) (PEG) | Au-coated substrates | Substrate immersion | Comprises residues Thr 30 – Pro 157 of human agrin N-terminal domain, mixed SAM with thiol-PEG-succinimidylglutaramide (HS-PEG-SGA) | 80 | |
| Collagen type II (Coll-II) | C-LRGRYW | Cys residue at N-terminus, Michael addition reaction to coumarin-bearing SAM | Glass substrates | Reactive μCP | Co-assembled fluorogenic SAM with TGB-β1-binding peptide | 42 | |
| Transforming growth factor β1 (TGB-β1) | C-LPLGNSH | Co-assembled fluorogenic SAM with Col II-binding peptide | |||||
| Hyaluronan (HA) | HS-GAHWQFNALTVR (Named as Pep-1, discovered by phage display) | Thiol from 3-mercaptopropionic acid (3-MPA) at N-terminus | Au-coated glass slides | Substrate immersion and μCP | Pure peptide-SAM | 33 | |
| Pure peptide-SAM | 31 | ||||||
| Substrate immersion | Mixed SAM, co-assembled SAM with 1-octanethiol | 34 | |||||
| Heparin (Hep) | GKKQRFRHRNRKG (Derived from VN) | N-terminus coupling to EG-AT linker | Au-coated glass slides and others | Substrate immersion | Employed for human pluripotent stem cell renewal | 51 | |
| GWQPPRARI (derived from FN) | |||||||
| FHRRIKA (derived from bone sialoprotein) | |||||||
| C-GKKQRFRHRNRKG (derived from vitronectin) | Cys residue at N-terminus, coupling to bromoacetamide-substituted surface | ||||||
| TYRSRKY | N-terminus coupling via amidation to carboxylate-terminated EG6-AT linker | Au | Substrate immersion | Hep/heparan sulfate-binding peptide, co-assembled SAM with RGDSP to test hMSCs adhesion | 106 | ||
| GGGKRTGQYKL | Au | Modulation of hMSCs behaviour | 73 | ||||
| KRTGQYKL | Au-coated glass | Hep proteoglycan binding peptide, synthesised with and without triglycine spacer, for hMSCs culture substrates | 25 | ||||
| poly(L-Lys, L-Leu) (named pKL, synthetic block co-polypeptide) | N-terminus coupling via 1,1′-carbonyldiimidazole (CDI) mediated amidation with EG4-AT linker | Au-coated silicon wafers | Substrate immersion | Culture substrates for hMSCs in vitro expansion | 91, 107 | ||
| KRSR | N-terminus coupling via CuCAA between peptide propargyl-PEG-NHS ester and azide-terminated AT linker | Au-coated coverslips | Microarray spotting | Pure and mixed SAMs with adhesion peptides (YIGSR, GRGDS), and KPSSAPTQLN (a BMP-7 derived peptide) | 108 | ||
| Cell culture: binding to cell membrane proteins | Integrins | cG![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) fE-(C12)2-C (underlined region derived from FN, C12 alkyl spacer) fE-(C12)2-C (underlined region derived from FN, C12 alkyl spacer) |
Cys residue via Michael addition reaction to maleimide-terminated SAM | Quartz substrates | Substrate immersion | Synergistic with C-GQGFSYPYKAVFSTQ in osteogenic differentiation of human mesenchymal stem cells (hMSCs) | 41 |
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) (underlined region derived from FN) (underlined region derived from FN) |
N-terminus coupling via CuAAC to azide-terminated EG6-AT linker | Au substrate | Co-assembled SAM with TYRSRKY to test hMSCs adhesion | 106 | |||
GGLWLGG![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) (underlined region derived from FN) (underlined region derived from FN) |
N-terminus coupling via amidation to carboxylate-terminated EG6-AT linker | Modulation of hMSCs behaviour | 73 | ||||
GG![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) (underlined region derived from FN) (underlined region derived from FN) |
N-terminus coupling via CuAAC to azide-terminated EG6-AT linker | Alkyne-terminated peptide (3-amino-5-hexynoyl group), to test modulation of hMSCs behaviour | |||||
G![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) (underlined region derived from FN) (underlined region derived from FN) |
N-terminus amine coupled to carboxylic acid-capped EG6-undecanethiol AT linker | Integrin binding peptide, for hMSCs culture substrates | 25 | ||||
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) (underlined region derived from FN, expressed in cutinase-FN fusion proteins) (underlined region derived from FN, expressed in cutinase-FN fusion proteins) |
Cutinase-mediated interaction with phosphonate presenting SAM, assembled from an EG3-terminated disulfide | Au substrates | Substrate immersion in bacterial lysate | Epitopes expressed as part of fusion proteins with cutinase, and immobilised to phosphonate presenting SAMs. Adhesion of baby hamster kidney (BHK) cells and 3T3 fibroblasts was studied | 72 | ||
![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) (Underlined region derived from FN, expressed in cutinase-FN fusion proteins) (Underlined region derived from FN, expressed in cutinase-FN fusion proteins) |
|||||||
c[![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) (DMNPB)f-K-] (A photoactivatable variant of c[RGDfK], DMNPB: 3-(4,5-dimethoxy-2-nitrophenyl)-2-butyl, a photocleavable group; underlined region derived from FN) (DMNPB)f-K-] (A photoactivatable variant of c[RGDfK], DMNPB: 3-(4,5-dimethoxy-2-nitrophenyl)-2-butyl, a photocleavable group; underlined region derived from FN) |
Lys residue via EDC/NHS coupling to carboxylate-terminated EG6-AT linker | Au-coated quartz sensors | Substrate incubation | Photodynamic peptide-SAM. Quantification of integrin activity in HUVECs | 109 | ||
c[![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) (DMNPB)f]-C (DMNPB: 3-(4,5-dimethoxy-2-nitrophenyl)-2-butyl, a photocleavable group; underlined region derived from FN) (DMNPB)f]-C (DMNPB: 3-(4,5-dimethoxy-2-nitrophenyl)-2-butyl, a photocleavable group; underlined region derived from FN) |
Cys residue coupling to maleimide terminated EG3-AT linker | Au substrates | Substrate incubation | Photodynamic peptide-SAM for HT-1080 and HUVECs adhesion, spreading, and migration studies | 110 | ||
C-![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) S (underlined section derived from FN) S (underlined section derived from FN) |
Cys residue at N-terminus via thiol–ene click reaction with vinyl terminated 5-hexenyldimethylchloro-silane SAM | Glass slides | Vapour deposition, then thiol–ene click reaction | Enhanced Schwann cell migration by concentration gradients on peptide-SAMs | 70 | ||
C-![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) (underlined section derived from LN) (underlined section derived from LN) |
|||||||
C-QAA![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) (underlined section derived from LN) (underlined section derived from LN) |
|||||||
C-![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) (underlined section derived from LN) (underlined section derived from LN) |
|||||||
C-![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) (underlined section derived from LN) (underlined section derived from LN) |
|||||||
C-SRARKQAAS![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) SADR (underlined section derived from LN) SADR (underlined section derived from LN) |
Cys residue at N-terminus via NHS/EDC coupling with maleimide AT linker | Au substrates | Substrate incubation, reactive SAM | Peptide binds to integrin α6β1 of embryonic mouse hippocampal neurons, known to promote neuronal attachment and neurite outgrowth | 111 | ||
G![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) S (underlined section derived from FN) S (underlined section derived from FN) |
N-terminus coupling via CuCAA between peptide propargyl-PEG-NHS ester and azide-terminated AT linker | Au-Coated coverslips | Microarray spotting | Adipose derived stem cells (ADSC) adhesion promoting, pure and mixed SAMs with Hep-binding and a BMP-7 derived peptide | 108 | ||
![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) (underlined region derived from LN) (underlined region derived from LN) |
|||||||
G![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) -K (underlined section derived from FN) -K (underlined section derived from FN) |
Lys sidechain coupled to carboxy AT linker (with and without EG4 moieties) | Au substrates | Chemical gradients created by computer-driven linear motion drive | Mixed SAMs, motogenic peptides to study migration of metastatic breast cancer cells (MDA-MB-231 line) | 27, 103 | ||
KG![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) S (underlined section derived from FN) S (underlined section derived from FN) |
N-terminus coupling to EG-AT linker | Au-coated glass slides and others | Substrate immersion | Long term culture of pluripotent stem cells, H1 and H9 cells were studied | 51 | ||
K![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) (derived from FN) (derived from FN) |
|||||||
GSDPG![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) (derived from LN) (derived from LN) |
|||||||
![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) (derived from LN γ-chain) (derived from LN γ-chain) |
N-terminus amine coupling to AT linker | Au-coated coverslips | Substrate immersion, manual spotting | Mixed SAMs as part of screening arrays. H1 and H9 cells were studied | 26 | ||
![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) (derived from LN γ-chain) (derived from LN γ-chain) |
|||||||
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) (derived from LN γ-chain) (derived from LN γ-chain) |
|||||||
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) (derived from LN γ-chain) (derived from LN γ-chain) |
|||||||
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) (derived from LN β-chain) (derived from LN β-chain) |
|||||||
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) (derived from LN α-chain) (derived from LN α-chain) |
|||||||
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) (derived from LN α-chain) (derived from LN α-chain) |
|||||||
![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) (derived from LN β-chain) (derived from LN β-chain) |
|||||||
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) (scrambled control) (scrambled control) |
|||||||
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) (derived from LN α-chain) (derived from LN α-chain) |
|||||||
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) (derived from LN α-chain) (derived from LN α-chain) |
|||||||
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) (derived from LN α-chain) (derived from LN α-chain) |
|||||||
![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) (derived from LN α-chain) (derived from LN α-chain) |
|||||||
![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) (derived from LN α-chain) (derived from LN α-chain) |
|||||||
![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) (derived from LN β-chain) (derived from LN β-chain) |
|||||||
![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) (derived from LN β-chain) (derived from LN β-chain) |
|||||||
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) (derived from LN γ-chain) (derived from LN γ-chain) |
|||||||
![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) (derived from LN γ-chain, modified) (derived from LN γ-chain, modified) |
|||||||
C-GGNGEP![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) TYRAYK-(FITC) (derived from bone sialoprotein, called bspRGD(15), bound to FITC fluorochrome) TYRAYK-(FITC) (derived from bone sialoprotein, called bspRGD(15), bound to FITC fluorochrome) |
Cys residue at N-terminus | Au-coated glass slides | Substrate immersion | bspRGD(15): 15 amino-acid ligand for αVβ3-integrin derived from bone sialoprotein. Peptide-SAMs supported hPSC expansion | 112 | ||
MUA-CGGNGEP![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) TYRAYK-(FITC) (derived from bone sialoprotein, called bspRGD(15)) TYRAYK-(FITC) (derived from bone sialoprotein, called bspRGD(15)) |
MUA moiety at N-terminus | ||||||
MUA-GGNGEP![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) TYRAYK (derived from bone sialoprotein, called 11-MUA-bspRGD(15)) TYRAYK (derived from bone sialoprotein, called 11-MUA-bspRGD(15)) |
Au-coated polystyrene PillarChips | Spot printing in 532-well microchip | |||||
C-EG-K![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) K-NMe3+ (underlined region derived from FN, named linear RGD when positive potential is applied, and cyclic RGD when negative potential is applied) K-NMe3+ (underlined region derived from FN, named linear RGD when positive potential is applied, and cyclic RGD when negative potential is applied) |
Cys residue attached to EG linker | Au-coated glass slides | Substrate incubation | Conformational modulation of RGD-NMe3via electric potential, able to undergo conformational changes with electric potential. NIH 3T3 fibroblasts migrated twice as fast in exposed conformation vs “cyclic” one | 113 | ||
C-EG-K![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) K-NH2 (underlined region derived from FN) K-NH2 (underlined region derived from FN) |
|||||||
C-GG![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) S (underlined region derived from FN) S (underlined region derived from FN) |
N-terminus coupling to maleimide-terminated AT disulfide linker | Au-coated glass coverslips and other substrates | Substrate immersion and μCP | Peptide and carbohydrate chips were prepared. Immobilisation of peptide to study adhesion of Swiss 3T3 fibroblasts, and quantitative enzyme inhibition assays | 78 | ||
![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) (underlined region derived from FN, expressed in cutinase-fibronectin fusion proteins) (underlined region derived from FN, expressed in cutinase-fibronectin fusion proteins) |
Cutinase-mediated interaction with phosphonate presenting SAM, assembled from an EG3-terminated disulfide | Au substrates | Substrate immersion in bacterial lysate | Epitopes expressed as part of fusion proteins with cutinase, and immobilised to phosphonate-presenting SAMs. Adhesion of baby hamster kidney (BHK) cells and 3T3 fibroblasts studied | 72 | ||
Cp-G![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) S (underlined region derived from FN, Cp: cyclopentadiene moiety, diene for cycloaddition coupling) S (underlined region derived from FN, Cp: cyclopentadiene moiety, diene for cycloaddition coupling) |
Cp moiety at N-terminus coupled via Diels–Alder reaction to benzoquinone-terminated EG3-AT linker | Au-coated glass coverslips | Substrate immersion | Linear or cyclic RGD-presenting peptides to study adhesion of Swiss 3T3 fibroblasts, cyclic peptide showed improved adhesion compared to linear one | 76 | ||
c![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) FK-Cp (underlined region derived from FN, Cp: cyclopentadiene moiety, diene for cycloaddition coupling) FK-Cp (underlined region derived from FN, Cp: cyclopentadiene moiety, diene for cycloaddition coupling) |
|||||||
G![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) SPG (underlined region derived from FN) SPG (underlined region derived from FN) |
C-terminus coupling to an EG3-AT linker | Au substrates | Substrate immersion, manual spotting | Mixed SAMs. Human melanoma cell line WM-115 was studied | 50 | ||
c![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) K-squarate (underlined region derived from FN) K-squarate (underlined region derived from FN) |
Coupling to an EG3-AT linker | ||||||
![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) Q-K (underlined section derived from FN) Q-K (underlined section derived from FN) |
Lys sidechain coupled to carboxy AT linker (with and without EG4 moieties) | Chemical gradients created by computer-driven linear motion drive | Mixed SAMs, motogenic peptides to study the migration of metastatic breast cancer cells (MDA-MB-231 line) | 27,103 | |||
| IDGQ-K (scrambled control) | Lys sidechain coupled to carboxy EG4-AT linker | ||||||
G![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) S (underlined section derived from FN) S (underlined section derived from FN) |
Electrostatic binding of ω-(GRGDS) bolaamphiphiles to oxoacid-terminated thiol SAMs | Au-coated well plates and coverslips | Multistep substrate incubation | Reversible peptide-SAM for the modulation of adhesion behaviour of MC3T3-E1 cells | 114 | ||
c[![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) fK] (underlined section derived from FN) fK] (underlined section derived from FN) |
Interfacial strain-promoted alkyne−azide cycloaddition (SPAAC) with cyclopropenone-displaying SAM | Au-coated glass slides | Multistep fabrication, cyclopropenone-displaying SAM, then UVA light irradiation | Peptide-SAMs characterized with polarization-modulation infrared reflection–absorption spectroscopy (PM-IRRAS) | 115 | ||
| Amidation via peptide amino groups and carboxylic groups from pre-formed MUA-displaying SAM | Au-coated QCM sensors | Substrate incubation | Interaction with αvβ3 integrins, peptide-SAM allowed to measure living chinese hamster ovary (CHO-K1) cell loading rate during adhesion and spreading | 116 | |||
| Supramolecular presentation in quatsomes (nonliposomal lipid-based unilamellar nanovesicles) | Au substrates | Multistep fabrication strategy | Hierarchical nanostructured SAM, displaying cell adhesion epitopes on cholesterol and surfactant assemblies, dramatic improvement of U2OS osteosarcoma cell adhesion compared to FN-coated Au substrates | 117 | |||
| Syndecan-1 & β1-integrins | MUA-GGRKLRQVQLSIRT (derived from Ag73: a syndecan-1 and β1-integrin binding peptide, called 11-MUA-Ag73) | MUA moiety at N-terminus | Au-coated polystyrene PillarChips | Spot printing in 532-well microchip | Peptide-SAMs supported hPSC expansion | 112 | |
| TGF-β receptors I and II | LTGKNFPMFHRN (identified by phage display) | C-terminus coupling to EG-AT linker | Au-coated glass slides | Spotting | Employed to exert activation of TGF-β signalling. Mouse mammary gland cells from the NmuMG line were studied | 20 | |
| MHRMPSFLPTTL (identified by phage display) | |||||||
| Fibroblast growth factor receptors | GGG-EVYVVAENQQGKSKA (spacer shown in italics) | N-terminus coupling to EG-AT linker | Au-coated glass slides and others | Substrate immersion | Studied in human embryonic stem cells (hESC) culture | 51 | |
| Bone morphogenetic protein 2 receptor (BMP2R) | KIPKASSVPTELSAISTLYL (derived from BMP2) | N-terminus amine coupled to carboxylic acid-capped EG6-undecanethiol AT linker | Au substrates | For hMSC culture substrates | 25 | ||
| Concanavalin A (mannose binding lectin) | 3-MUA-(Aib)n-NhtBu (n = 4–6) | MUA moiety at N-terminus | Au plates | Substrate incubation | Mixed SAM for the controlled display of bioactive carbohydrate ligands | 118 | |
| 3-MUA-(Aib)4-(D-mannose) | |||||||
| Other unspecified cell membrane proteins | KPSSAPTQLN (derived from BMP-7) | N-terminus coupling via CuCAA between peptide propargyl-PEG-NHS ester and azide-terminated AT linker | Au-coated coverslips | Microarray spotting | Pure and mixed SAMs with adhesion peptides (YIGSR, GRGDS), and a heparin-binding peptide (KRSR) mouse embryonic fibroblasts (MEFs) and ADSC were studied were studied | 108 | |
| C-GQGFSYPYKAVFSTQ (derived from BMP-7, named bone forming peptide-1 (BFP-1)) | Cys residue via Michael addition reaction to maleimide-terminated SAM | Quartz substrates | Substrate immersion | Synergistic effect with cG![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) fE-(C12)2-C in osteogenic differentiation of human mesenchymal stem cells (hMSCs) fE-(C12)2-C in osteogenic differentiation of human mesenchymal stem cells (hMSCs) |
41 | ||
G-C-![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) E (derived from bone marrow homing peptide 1 (BMHP1, underlined)) E (derived from bone marrow homing peptide 1 (BMHP1, underlined)) |
Cys residue close to N-terminus, coupling via Michael addition to maleimide surface | Substrate incubation | Pure SAMs to induce osteogenic differentiation of murine MSCs | 35 | |||
c[G-C-![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) E] (derived from bone marrow homing peptide 1 (BMHP1, underlined)) E] (derived from bone marrow homing peptide 1 (BMHP1, underlined)) |
Cys residue, coupling via Michael addition to maleimide-decorated surface | ||||||
![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) -C (derived from bone marrow homing peptide 1 (BMHP1, underlined)) -C (derived from bone marrow homing peptide 1 (BMHP1, underlined)) |
Cys residue at to C-terminus, coupling via Michael addition to maleimide surface | Peptide coupled to PEG5000 chain through a photoactivatable linker. Illumination-triggered MSCs osteogenic differentiation | 28 | ||||
| MUA-GKPLRAKREITKLKFG (derived from bone sialoprotein, called 11-MUA-linear (15–23)) | MUA moiety at N-terminus | Au-coated polystyrene PillarChips | Spot printing in 532-well microchip | Peptide-SAMs supported hPSC expansion | 112 | ||
| MUA-GG-PEG2-cyclo(DMGDGRPRK) (derived from bacterial peptide display, called 11-MUA-cyclic(7C-1)) | |||||||
| GGG-KLTWQELYQLKYKGI (de novo design, spacer shown in italics) | N-terminus amine coupled via EDC/NHS to carboxy-terminated EG6-AT linker | Au slides | SAMS array | Mixed SAMs, to target VEGF receptor (VEGFR) in HUVECs | 90 | ||
| TVKHRPDALHPQ (identified by phage display) | C-terminus coupling to an EG3-AT linker | Au-coated substrates | Two-step fabrication, spotting | Culture of pluripotent stem cells, H9 cells were studied | 79 | ||
| LTTAPKLPKVTR (identified by phage display) | |||||||
Thiol functionalities have also been incorporated to peptide building blocks via N-thiolation approaches without the need for a long alkyl chain. In this regard, Bilewicz et al. prepared a series of ferrocene (Fc) containing oligoglycine derivatives (Fc-CO(Gly)nNH-(CH2)2-SH) (Gly = glycine; n = 2–6) via SPPS techniques using a cysteamine 4-methoxytrityl resin.52 This same type of resin was also employed to prepare polyalanine derivatives R-(Ala)14NH-(CH2)2-SH (R = ferrocenecarbonyl or hydrogen) containing the same cysteamine linker (Table 2, peptide-SAMs for molecular electronics).53 Using a different approach, but a much shorter hydrocarbon chain, Azevedo and collaborators performed the 3-mercaptopropanoyl derivatisation at the N-terminus of the 12-mer GAHWQFNALTVR peptide before Au surface attachment (Table 1, peptide-SAMs binding to ECM components).31,33,34
| Application | Target | Peptide sequencea | Anchor | Substrate | Fabrication method | Comment | Ref. |
|---|---|---|---|---|---|---|---|
| a Amino acid residues are given in one-letter code, except for Aib (α-aminoisobutyric acid, a strongly helicogenic amino acid), and the ones employed as substrate anchors are highlighted in bold, and separated from the rest of the sequence. b Additional antifouling peptide-SAMs are presented in Table 3 as part of biosensing applications. c D-Amino acids are represented as DX. | |||||||
| Antimicrobial surfaces | Bacterial cell membranes | GKIIKLKASLKLL-C | Cys residue at C-terminus | Au-coated glass covers | Substrate immersion | Prevents biofilm formation against S. epidermidis and S. aureus. Antibiofilm properties translated to an elastin-like recombinamer (ELR) | 145 |
| KWIVWRWRFKR (de novo design, derived from antibacterial protein from crowberry) | Either-NH2 group from terminal amino acids, amidation with carboxylic acid from ((1-mercapto-11-undecyl))-EG4 linker activated with CDI | Au substrates | Multistep fabrication, substrate incubation, CDI activation, peptide incubation | Bactericidal activity against E. coli and S. aureus for the treatment of urinary catheter-associated infections | 128 | ||
| HS-Ahx-GIGKFLKKAKKFAKAFVKILKK (HS-MSI-78A (HS-pexiganan)) | Thiol from additional Cys residue and aminohexanoic acid (Ahx) spacer at either the N- or C-terminus coupling with maleimide-terminated surface | Au substrates | Multistep fabrication, neutravidin, biotin-PEG11-maleimide, peptide binding | MSI-78A bactericidal effect on H. pylori for gastric infection management | 129 | ||
| GIGKFLKKAKKFAKAFVKILKK-Ahx-C | |||||||
| KWKLFKKIGAVLKVL-C (derived from cecropin and melittin) | Cys residue at C-terminus that bind to Au NPs functionalised with-NH2 and –COOH terminated PEG spacers | Modified polyurethane substrates | Substrate incubation, Au NPs coating | Coating suitable to prevent bacterial infection on polyurethane devices | 146, 147 | ||
| KWKLFKKIGIGAVLKVLTTGLPALIS-C (hybrid peptide between cecropin (1–8) and melittin (1–18)) | Cys residue at C-terminus, first, an alkyne-terminated silane was used to grow the alkyne SAM, followed by conversion to maleimide terminated surface with azido-PEG3-maleimide linker (“click reaction”) | Silica surface | Multistep fabrication | Stable antimicrobial coating, tested against E. coli | 148 | ||
| Implants | Solid surfaces and bacterial cell membranes | MPA-(Ahx)3-GRRRRSVQWCA (hLf1–11, derived from lactoferrin; contains three 6-aminohexanoic acid units (Ahx)3) | Thiol from 3-mercaptopropionic acid (MPA) from N-terminus linker | Ti surface | Substrate grafting | Stable antimicrobial coating for dental implants | 84 |
| Reduction of S. sanguinis and L. salivarius adhesion to Ti implant, prevented early stages of microbial growth | 149 | ||||||
| KKGGGPPFLMLLKGSTRFC (derived from LN 332) | Onto silanised titanium (using 3-(chloropropyl)-triethoxysilane (CPTES)) | Grafted peptides promoted epithelial attachment around teeth and formation of hemidesmosomes | 150 | ||||
| KKKGGGVPIMDFADPQFPT (derived from ameloblastin, Ambn) | |||||||
| GKIIKLKASLKLL (derived from parotid secretory protein, named GL13K) | Silanised Ti disks using CPTES and diisopropyl-ethylamine (DIPEA), then grafting of GL13K AMP with a terminal amino group | Antibacterial dental implants | 151 | ||||
| LKLLKKLLKLLKKL (AMP, named E14LKK) | Solid binding sequences | Ti substrate | Substrate incubation | Combined via a triple-glycine linker with the metal binding sequences | 69 | ||
![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/b_char_0050_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/b_char_0045_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/b_char_004e_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/b_char_0045_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/b_char_004c_0332.gif) (TiBP1, underlined: metal binding sequence) (TiBP1, underlined: metal binding sequence) |
|||||||
![[S with combining low line]](https://www.rsc.org/images/entities/b_char_0053_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/b_char_0050_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/b_char_004e_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/b_char_0059_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/b_char_0053_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/b_char_0045_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/b_char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/b_char_0053_0332.gif) (TiBP2, underlined: metal binding sequence) (TiBP2, underlined: metal binding sequence) |
|||||||
| N 3-PEG12-KRWWKWWRR (named PEG-HHC36) | Terminal azide-functionality for a “click-reaction” | Substrate grafting | Antimicrobial Ti peptide-SAM | 152 | |||
| CMLPHHGAC (dual peptide construct, cHABP1: hydroxyapatite binding peptide-1, combined with an AMP: KRWWKWWRR) | HA coated nanotubular titania substrates | Substrate incubation | Hydroxyapatite binding peptide-1 and antimicrobial surface | 137 | |||
| FKRIVQRIKDFLRNLV (named FK-16) | OH groups silanised with APTES to generate surface amines, then, 6-maleimidohexanoic acid, was coupled to the APTES via EDC | Substrate grafting | Broad-spectrum activity against ESKAPE pathogens and anti-adhesion/biofilm against S. aureus and E. coli | 153 | |||
| Enzymes (for inhibition)c | DFPRPG | Free amine coupling to OH and COOH-terminated SAMs via CDI activation | Au substrate | Substrate incubation | Thrombin inhibition assays for blood contact implants applications | 154 | |
| Molecular electronics | Electron transfer | ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -(Ala)14 ( -(Ala)14 (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, redox tag) : ferrocene, redox tag) |
C-terminus coupling via AT short cysteamine linker | Au substrates | Substrate incubation | Mixed SAMs, observed ET liked to electric field generated by the molecular dipole of polyalanine helix | 53 |
| (Ala)14 | |||||||
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -KTAL10NP-C (Fc: ferrocene, redox moiety) -KTAL10NP-C (Fc: ferrocene, redox moiety) |
Cys residue at C-terminus | Au substrates | Substrate immersion | Redox-induced reorganisation of α-helical peptide-based SAMs of parallel and antiparallel dipole orientations, to study processes triggered by ET | 155 | ||
| Ac-KTAL10NP-C | |||||||
| Ac-C-TAL10NPK | |||||||
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -KTAL18NP-C (Fc: ferrocene, redox moiety) -KTAL18NP-C (Fc: ferrocene, redox moiety) |
Cys residue at C-terminus | Au substrates | Substrate immersion | Redox-active α-helical peptide-based SAMs, to study processes triggered by ET | 156 | ||
| Ac-KTAL18NP-C | |||||||
| Ac-C-TAL18NPK | |||||||
| Lipo-(γ-benzyl-L-Glu)86 | Lipoic acid at N-terminus | Au glass slides | Electric poling | Electric-field-enhanced self-assembly of α-helical polypeptides | 54 | ||
| SUA-(Glu)45-propyl (SUA: 11-(ethyldithio)undecanoic acid) | Disulfide group from AT at N-terminus | Au electrodes | Substrate immersion | Mixed SAMs, discrimination between N- and C-termini adsorption | 61 | ||
Pr-(Glu)45-C(O)CH2CH2C(O)-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ( (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, redox moiety) : ferrocene, redox moiety) |
Macrodipole interactions as guest peptide, Fc at N-terminus | ||||||
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -C(O)NHCH2CH2NH-(Glu)45 ( -C(O)NHCH2CH2NH-(Glu)45 (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, redox moiety) : ferrocene, redox moiety) |
Macrodipole interactions as guest peptide, Fc at C-terminus | ||||||
Boc-NHCH(CH2![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[z with combining low line]](https://www.rsc.org/images/entities/char_007a_0332.gif) )C(O)-(Ala-Aib)6-NH-Lipo (ECz: N-ethylcarbazoyl, photosensitizer) )C(O)-(Ala-Aib)6-NH-Lipo (ECz: N-ethylcarbazoyl, photosensitizer) |
Lipoic acid at C-terminus | Au-coated glass slides | Substrate incubation | Photocurrent generation in α-helical peptide-based SAMs, peptide-mediated electronic coupling is stronger than AT-mediated | 49 | ||
Lipo-(Ala-Aib)6-NHCH(CH2![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[z with combining low line]](https://www.rsc.org/images/entities/char_007a_0332.gif) )C(O)OEt (ECz: N-ethylcarbazoyl, photosensitiser) )C(O)OEt (ECz: N-ethylcarbazoyl, photosensitiser) |
Lipoic acid at N-terminus | ||||||
| Lipo-(Ala-Aib)6-OCH2Ph | Lipoic acid at N-terminus | ||||||
Boc-NHCH(CH2![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[z with combining low line]](https://www.rsc.org/images/entities/char_007a_0332.gif) )C(O)-(Ala-Aib)10-NH-Lipo (ECz: N-ethylcarbazoyl, photosensitiser) )C(O)-(Ala-Aib)10-NH-Lipo (ECz: N-ethylcarbazoyl, photosensitiser) |
Lipoic acid at C-terminus | ||||||
Boc-NH(CH2)2NHC(O)-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -C(O)-Ala(Leu-Aib)8-Lipo ( -C(O)-Ala(Leu-Aib)8-Lipo (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, redox moiety) : ferrocene, redox moiety) |
Lipoic acid at C-terminus | Au substrates | Substrate immersion | Fc moiety at N-terminus, helix dipole moment helix accelerated ET | 131 | ||
| Boc-(Leu-Aib)8-Lipo | Lipoic acid at C-terminus | — | |||||
Lipo-(Leu-Aib)8-NH(CH2)2NHC(O)-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -C(O)-Ala-OBzl ( -C(O)-Ala-OBzl (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, redox moiety) : ferrocene, redox moiety) |
Lipoic acid at N-terminus | Fc moiety at C-terminus, helix dipole moment helix accelerated ET | |||||
| Lipo-(Leu-Aib)8-OBzl | Lipoic acid at N-terminus | — | |||||
Lipo-(Leu-Aib)8-NHCH(CH2![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[z with combining low line]](https://www.rsc.org/images/entities/char_007a_0332.gif) )C(O)OEt ( )C(O)OEt (![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ![[z with combining low line]](https://www.rsc.org/images/entities/char_007a_0332.gif) : N-ethylcarbazoyl, photosensitiser) : N-ethylcarbazoyl, photosensitiser) |
Lipoic acid at N-terminus | Au substrates | Substrate immersion | Mixed SAMs, molecular photodiode, photocurrent direction can be switched by choosing the wavelength of irradiating light | 60 | ||
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) uC(O)-(DLeu-Aib)8-Lipo ( uC(O)-(DLeu-Aib)8-Lipo (![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) u: tris(2,2′-bipyridine)ruthenium(II)) u: tris(2,2′-bipyridine)ruthenium(II)) |
Lipoic acid at C-terminus | ||||||
Lipo-(Ala-Aib)2-Glu(OCH3)-(Ala-Aib)4-Glu(OCH3)-(Ala-Aib)2-NH(CH2)2NHC(O)-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ( (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, redox moiety) : ferrocene, redox moiety) |
Lipoic acid at N-terminus | Au substrates | Substrate incubation | Long-range ET through helical peptides | 157 | ||
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -C(O)NHCH(CH3)C(O)-(Ala-Aib)2-Glu(OCH3)-(Ala-Aib)4-Glu(OCH3)-(Ala-Aib)2-NH-Lipo ( -C(O)NHCH(CH3)C(O)-(Ala-Aib)2-Glu(OCH3)-(Ala-Aib)4-Glu(OCH3)-(Ala-Aib)2-NH-Lipo (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, redox tag) : ferrocene, redox tag) |
Lipoic acid at C-terminus | ||||||
Ac-S-Ph-C(O)-(Ala-Aib)2-Glu(OCH3)-(Ala-Aib)4-Glu(OCH3)-(Ala-Aib)2-NH(CH2)2NH-C(O)-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ( (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, redox moieties) : ferrocene, redox moieties) |
Thiophenyl group at N-terminus | ||||||
Ac-S-Ph-C(O)-(Ala-Aib)2-Glu(![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[r with combining low line]](https://www.rsc.org/images/entities/char_0072_0332.gif) )-(Ala-Aib)4-Glu( )-(Ala-Aib)4-Glu(![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[r with combining low line]](https://www.rsc.org/images/entities/char_0072_0332.gif) )-(Ala-Aib)2-NH(CH2)2NH-C(O)- )-(Ala-Aib)2-NH(CH2)2NH-C(O)-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ( (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene redox moieties, : ferrocene redox moieties, ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[r with combining low line]](https://www.rsc.org/images/entities/char_0072_0332.gif) : pyrenyl group as chromophore) : pyrenyl group as chromophore) |
Thiophenyl group at N-terminus | Inclusion of pyrenyl moieties did not disturb ET | |||||
| Lipo-(Ala-Aib)n-OBzl n = 6, 8, 12 | Lipoic acid at N-terminus | Au substrates | Substrate incubation | Lipo-(Ala-Aib)12-OBzl showed the smallest tilt angle and formed a nearly vertically oriented SAM with a parallel orientation of helices | 55 | ||
| Lipo-(Lys(CO2Bzl)-Aib)n-OCH3 | Lipoic acid at N-terminus | Role of helix-helix interaction to orient helical peptides in SAMs is greater than Au–S | |||||
| [tBoc-(Ala-Aib)8-NH(CH2)2-S-]2- | Disulfide group at C-terminus | ||||||
| -[-S-(CH2)10-C(O)-(Ala-Aib)8-OBzl]2 | Disulfide group from linker at N-terminus | ||||||
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -CO(Gly)nNH-(CH2)2-SH (n = 2, 3, 4, 5, 6, -CO(Gly)nNH-(CH2)2-SH (n = 2, 3, 4, 5, 6, ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene redox tag) : ferrocene redox tag) |
Thiol group from linker at C-terminus | Substrate incubation | ET rates through oligoglycine bridges decrease with distance | 52 | |||
| Lipo-Aib4-Trp-Aib | Lipoic acid at N-terminus | Substrate immersion | Pure SAM, short helical peptide aggregates horizontally onto Au surface | 38 | |||
| Substrate immersion | Pure SAM, Trp residue as photosensitiser to study the efficiency of photoinduced ET | 56 | |||||
Lipo-Aib3-Ala-![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -Ala ( -Ala (![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) : 2,2,6,6-tetramethylpiperidine-1-oxyl-4-amino-4-carboxylic acid, a redox active amino acid) : 2,2,6,6-tetramethylpiperidine-1-oxyl-4-amino-4-carboxylic acid, a redox active amino acid) |
Lipoic acid at N-terminus | Substrate immersion | Pure SAM, TOAC residue used to study the efficiency of ET | ||||
| C-Gn (n = 1–9) | Cys residue at N-terminus | Au and Ag substrates | Substrate incubation | Pronounced odd−even structural effect strongly affects peptide-SAM packing density and conformation. Promising elements of organic field-effect transistors (OFETs) | 158 | ||
| 3-MPA-E7 | 3-MPA linker at N-terminus | SAMs formed between Au layers | Substrate incubation | SAM conduction depends on peptide length, charge, amino acid, and secondary structure. Off-resonance tunneling as the dominant conduction mechanism | 159 | ||
| 3-MPA-An (n = 7, 20) | |||||||
| 3-MPA-Kn (n = 7, 20) | |||||||
| 3-MPA-Wn (n = 4–7) | |||||||
| 3-MPA-WA6 (named as W-1) | Au substrates | Solid-state electron transport with W residues as dopants, which increased the conductance when close to the electrode surface due to charge tunneling | 160 | ||||
| 3-MPA-A3WA3 (named as W-4) | |||||||
| 3-MPA-A6W (named as W-7) | |||||||
| 3-MPA-A7 (named as 7A) | |||||||
| Lipo-GGH | Lipoic acid residue at N-terminus | Au(111) surface | Substrate immersion, then copper complexation | Tripeptide GGH forms a complex with Cu2+ ions. Complexation highly dependent on peptide surface density and results in opposite trends: high-density SAMs are insulating, while lower density SAMs are more conductive | 161 | ||
| 3-MPA-GnK(Fc) (named series Gn, n = 1, 2, 3; Fc: ferrocene probe) | 3-MPA linker at N-terminus | Template stripped Au substrate | Substrate incubation | Relation between charge migration on solid state and in wet electrochemistry was investigated. Electron transfer rate in wet electrochemistry was F > G ≈ D, while charge transport rate on solid state was G ≈ F > D | 162 | ||
| 3-MPA-GnFK(Fc) (named series Fn+1, n = 1, 2, 3) | |||||||
| 3-MPA-GnDK(Fc) (named series Fn+1, n = 1, 2, 3) | |||||||
| (Boc)-Cys-(S-Acm)-(Ala-Leu)n-NH-(CH2)2-SH (n = 4–7, Acm: acetamidomethyl group) | Thiol group at C-terminus | Au substrates and AuNPs | Current-sensing atomic force microscopy (CS-AFM) showed ET due to possible tunneling mechanism between adjacent helices | 163 | |||
| Lipo-KL2HL6HL6 | Lipoic acid close to C-terminus | Au-coated PDMS/BaTiO3 NPs-based substrate | Multistep fabrication, including substrate incubation, then binding to Co(II) ions | Vertical α-helical peptide-SAM using piezoelectric field produced by a nanogenerator, able to rectify AC signal to DC signal | 164 | ||
| Lipo-(Aib)6-NHtBu | Lipoic acid at N-terminus | Au electrodes | Substrate immersion | Photoinduced ET remarkably improved by Pyr units, electronic flow through peptide helices is strongly asymmetric and favoured from the C-terminus to Au | 165 | ||
| Lipo-(Aib)6-Pyr (Pyr: pyrene, choromophore to enhance UV photon capture) | |||||||
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -KTALnNP-C ( -KTALnNP-C (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : redox tag, n = 10 (Fc10L), n = 14 (Fc14L), n = 18 (Fc18L)). : redox tag, n = 10 (Fc10L), n = 14 (Fc14L), n = 18 (Fc18L)). |
|||||||
| Cys residue at C-terminus | Au substrates | Substrate incubation | Fc labelled helical peptides. ET rates as a result of fluctuation-controlled tunneling mechanism | 166 | |||
| Thymine-(Aib-KL-Aib-KKL-Aib-KI-Lol)-adenine (helical undecapeptide analogue of the natural peptide trichogin GA IV, Lol: leucinol, a chiral (S) 1,2-aminoalcohol) | Watson–Crick base pairing between Adenine-displaying SAM and Thymine-functionalised peptide | Au electrodes | Multi-step fabrication, SAM assembly, then peptide, then porphyrin incubation | Peptide pH-driven 310- to α-helix conversion affects photocurrent generation, Zn porphyrin system attached to peptide | 167 | ||
| Probing protein and peptide folding and interactions | Peptide surface adsorption models | -[S-CH2CH2C(O)-(Ala-Aib)8-OCH2Ph]2 | Disulfide group from linker close to N-terminus | Au-coated glass slides | Substrate immersion | Model systems for helical protein binding. Pure SAMs evidence binding parallel to the surface, mixed SAMs evidence vertical orientation | 24 |
| [Boc-(Ala-Aib)8-NHCH2CH2-S-]2- | Disulfide group from linker close to C-terminus | ||||||
| LKKL-X-KKLLKKLLKK-X-LKKL, X = propargylglycine, referred to as α-LK2x20 | Two mid-sequence propargylglycine residues 1 coupling via CuAAC to azide-terminated AT linker | AuNPs | Substrate incubation | Click helical peptide attachment to further functionalise AuNPs with proteins and modulate their interaction with cells | 77, 168 | ||
| Peptide photo-isomerisation models | Boc-(DLeu-Aib)8-Ala-(azo)-C(O)CH2CH2C(O)-(DLeu-Aib)8-NH-Lipo (named as Nunchaku peptide) | Lipoic acid close to N-terminus | Au-coated glass slides | Photoresponsive peptides with two helical segments, change in orientation upon reversible isomerisation of azobenzene unit | 59 | ||
| Boc-(Leu-Aib)4-Ala-(azo)-C(O)CH2CH2C(O)-(Leu-Aib)8-NH-Lipo (named as Nunchaku Type-L) | Lipoic acid close to N-terminus | Au-coated silicon wafer | Model system for G-protein-coupled receptors (GPCRs) Adsorption of G- protein, human serum albumin (HSA) and human IgG is assessed | 62 | |||
| Protein–protein interaction studies | R-C | Cys residue at C-terminus | |||||
| Antifouling surfacesb | Protein adsorptionc | EKEKEKE-![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) |
Cys residue at C-terminus (underlined region defined as anchor by the authors) | Au-coated chips | Substrate incubation | Pure SAMs to assess fibrinogen (Fib) and lysozyme (Lyz) adsorption | 140 |
EKEKEKE-![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) |
|||||||
EKEKEKE-![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) |
|||||||
| EKEKEKE | C-terminus coupling to EG4-AT linker | Au-coated substrates | Substrate immersion and micro-contact printing | Resistant to Fib, Lyz, and albumin (Alb) adsorption | 169 | ||
| DKDKDKD-EG4-SH | |||||||
| ERERERE-EG4-SH | Not resistant to Fib, Lyz and Alb adsorption | ||||||
| DRDRDRD-EG4-SH | |||||||
| Propagyl-SGKGSSGSST | 11-Thioacetyl-undecanoic acid propargyl amide residue at N-terminus coupling via CuAAC to azide-terminated AT linker | Au-coated silicon wafer | Substrate incubation | Resistant to streptavidin (SA), bovine serum albumin (BSA), and FN adsorption | 170 | ||
| Propagyl-AAPAAAPAAL | Not resistant to streptavidin adsorption | ||||||
| C-Pn (n = 6 (named Pro6) and 9 (named Pro9), non-ionic anti-fouling peptides, promising for vascular devices) | Cys residue at N-terminus | Au-coated substrates | Substrate immersion | Adsorption of blood components and adhesion of blood cells strongly suppressed. Pro9-SAM inhibited thrombogenic response & platelet activation | 143 | ||
| Adsorption of HSA, Fib, bovine serum, and fatty acids was prevented. Adhesion of fibroblasts was suppressed | 142 | ||||||
EKEKEKE-![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) |
Cys residue at C-terminus (underlined region defined as anchor by the authors) | Au-coated silicon wafer | Pure SAMs resistant to fibrinogen and human platelets adsorption | 171 | |||
DKDKDKD-![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) |
|||||||
ERERERE-![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) |
Pure SAMs, adhered fibrinogen and platelets | ||||||
DRDRDRD-![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) |
|||||||
| (KE)4K | Thiol from MUA linker attached to N-terminus, unmodified free carboxylic acid group at C-terminus | Au-coated Nexterion B slides | Substrate incubation | Resist the nonspecific protein adsorption (Lyz and Fib). Further tested against attachment N. perminuta and C. marina & P. fluorescens | 172 | ||
| (DKDE)4DK | |||||||
| (HE)4H | |||||||
| (DHDE)4DH | |||||||
| Ion binding | Calcium phosphate mineralisation | C-DSSDSS | Cys residue at N-terminus | Au-coated silicon wafer | Substrate immersion | High affinity for Ca2+ ions, hydroxyapatite (HAp) deposition | 138 |
| C-DSSDSSDSS | |||||||
| C-DSSDSSG | |||||||
| C-DSSDSSDSSG | Highest capacity to induce HAp growth and nucleation | ||||||
| CMLPHHGAC (dual peptide construct, cHABP1: hydroxyapatite binding peptide-1, combined with an AMP: KRWWKWWRR) | HAp-coated nanotubular titania substrates | Ti substrate | Substrate incubation | Hydroxyapatite binding peptide-1 & antimicrobial surface | 137 | ||
| Other applications | Proof of concept of a characterisation technique | RRRVRRRGSGSRVTCDDYYYGFGCNKFCRPRGSGSGSGSGSK-biotin (de novo design) | Coupling to streptavidin-displaying mixed SAM | Au substrate | Substrate incubation | Mixed SAM, in situ underwater contact angle measurement model, electrically switched surfaces | 173 |
| RS–SR + Au0n → RS− Au+·Au0n |
Worley and collaborators coupled the rodlike polypeptide poly(γ-benzyl L-glutamate) (PBLG) to (±)-α-lipoic acid via its N-terminus (Table 2, peptide-SAMs for electron transfer). Peptide-SAMs were prepared by field-induced in situ molecular alignment by applying a voltage between two Au electrodes, this promoted higher lipoic acid-mediated chemisorption on the negative electrode compared with either the positive electrode or control samples without an applied voltage.54 Miura and collaborators prepared several helix-forming oligo- and polypeptide derivatives containing either lipoyl groups or disulfide functionalities (Table 2, peptide-SAMs for electron transfer). This report includes systematic variations in the solvent of choice, as well as the self-assembling peptide sequence, secondary structure (one or two helixes), length, and composition, leading to different tilt angles of the helix axis from the metal surface normal. It was also found that one-helix peptide with a lipoic acid group at the N-terminus showed a lower tilt than a two-helix peptide, in which the two helical peptides were connected by a disulfide linkage. These findings helped gaining a better mechanistic understanding of helix-forming peptide-SAMs. Initially, terminal lipoic acid groups cluster on one face of the helix bundle, then the cluster rapidly reacts with the Au substrate with the consequent fixation of the peptides with a vertical orientation, which is hindered by steric factors, as surface access of peptide clusters becomes statistically more unlikely.55
Venanzi and collaborators also employed lipoic acid coupling for preparing peptide-based SAMs. A helix-forming hexapeptide was N-terminus conjugated to (±)-α-lipoic acid (Table 2, peptide-SAMs for electron transfer). Self-assembling peptide units included a high content of 2-aminoisobutyric acid (Aib, a strongly helicogenic amino acid), which rendered a rigid helical conformation and contributed to the formation of a complex morphology made of ‘stripes’, which consisted on peptide aggregates horizontally layered on the Au substrate, and ‘holes’, that consisted on Au vacancy islands coated by the peptide monolayer.38 Lipoic acid conjugation has demonstrated to be friendly even with SAMs containing unnatural amino acids, as demonstrated by the work of Gatto and collaborators, who linked lipoic acid to a hexapeptide containing a 2,2,6,6-tetramethylpiperidine-1-oxyl-4-amino-4-carboxylic acid (TOAC) residue.56 TOAC is a rigid, paramagnetic, redox active amino acid, commonly used as a spin label in electron paramagnetic resonance (EPR) studies57 and as a fluorescence quencher.58
SAMs displaying two photoresponsive helical peptides attached to Au substrates via N-terminus lipoic acid coupling were prepared by Tada and collaborators (Table 2, peptide-SAMs for probing protein and peptide folding and interactions). Both peptides included two 16-mer helical segments with an intervening azobenzene at their C-terminus, both acquired vertical orientations, as well as exhibited cooperative conformational changes upon photoirradiation as a consequence of isomerisation of the azobenzene unit.59
Yasutomi and co-workers coupled lipoic acid moieties at either N- or C-termini of hexadecapeptides containing chromophores, this varying position permitted control over the helix dipole direction when immobilised as part of mixed SAMs on Au, allowing the fabrication of a molecular photodiode (Table 2, peptide-SAMs for electron transfer).60
The formation of helical poly(L-glutamic acid)-based amphiphile (PLGA) SAMs on Au substrates via interaction with a disulfide group at the N-terminus has also been reported. The source of choice for providing disulfide bonds was 11-(ethyldithio)undecanoic acid. Different packing densities were found as a function of adsorption rate and pH, and specific interactions with guest PLGAs containing a ferrocenyl group (as a redox active moiety) at the N- and C-termini were investigated, finding that guest helix PLGAs are captured through antiparallel, side-by-side helix-macrodipole interactions.61
The functionalised helix-forming 8-mer derivative Boc-(Ala-Aib)8-OCH2C6H5 with 3,3′-dithiodipropionic acid was described by Knoll and collaborators, rendering a peptide with two helices connected at the N-terminus via a short spacer containing a disulfide bond. Similarly, the 8-mer derivative Boc-(Ala-Aib)8-OH was made to react with cystamine dihydrochloride, generating a peptide with an equivalent disulfide bridge at the C-terminus. Both peptides were tethered to Au substrates to form either pure or mixed (multicomponent) SAMs. The thickness of the former suggested that helical peptides were adsorbed with a preferred orientation parallel to the surface. However, helix-forming peptides in an equimolar mixed SAM adopted a vertical orientation from the surface, as antiparallel helix packing is significantly more favourable than a parallel one, as suggested by the authors.24
Moreover, Cys residues have been employed for peptide tethering to other substrates apart from Au. For instance, Yin and co-workers anchored a bone morphogenetic protein-7 (BMP-7) derived and cyclic RGD peptides via a Michael addition reaction between peptide Cys residues and a maleimide terminated surface, achieving control over differentiation behaviours of mesenchymal stem cells (MSCs) (Fig. 2a).41 Another example based on organosilane-based peptide-SAM anchoring was reported by Cabanas-Danés and collaborators. They used a multi-step immobilisation strategy to tether Cys-terminated peptides to a modified glass surface. First, glass substrates were modified to include an azide-terminated SAM, these functionalities underwent a Huisgen 1,3-dipolar cycloaddition reaction with a triple bond linked to a coumarin derivative (via reactive μCP) which also carried a methyl-4-oxo-2-butenoate moiety, the latter was used to attach Cys-terminated peptides by means of Michael addition reactions (Fig. 2b).42
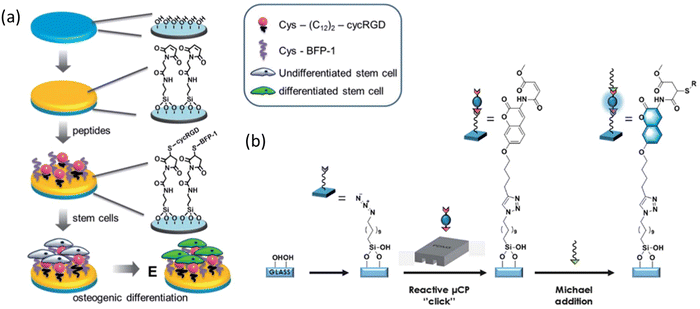 | ||
| Fig. 2 Preparation of peptide-based SAMs via Cys anchoring. (a) Multistep preparation of peptide-based SAMs on quartz substrates via a Michael addition reaction, starting with a blank quartz substrate, which is converted into a maleimide terminated substrate for peptide immobilisation, and then applied to culture MSCs and promote their osteogenic differentiation [adapted with permission from ref. 41].41 (b) Work-flow followed to prepare peptide-SAMs onto glass substrates via reactive μCP. Peptide immobilisation took place via the reactive μCP of a coumarin derivative on an azide-terminated SAM, followed by covalent immobilisation and detection of Cys-terminated bioactive peptides via a fluorogenic Michael addition reaction [adapted with permission from ref. 42].42 | ||
For instance, Lee and collaborators reported the Au-binding peptide WAGAKRLVLRRE (selected by directed evolution to specifically bind to Au substrates)67 as part of a chimeric biomolecule that also included a peptide nucleic acid domain that facilitates the anchoring of antisense oligonucleotide probes for the detection of nucleic acids (Table 3, biosensing of nucleic acids).68 SBP have also proved effective as part of coatings for Ti implants (Table 2, peptide-SAMs for osteo-integrative surfaces).69
| Class | Target/biomarker | Peptide sequencea | Anchor | Substrate/electrode modification | Fabrication method | Comment | Ref. |
|---|---|---|---|---|---|---|---|
| a Amino acid residues are given in one-letter code, except for non-proteinogenic amino acids, and the ones employed as substrate anchors are highlighted in bold, and separated from the rest of the sequence. b Related references regarding proteophobic peptide-SAMs are presented in Table 2. c |Indicates cleavage site by the corresponding enzyme or catalytic target. d Lower case letters correspond to nucleotide sequences in nucleic acid oligomers. | |||||||
| Metal ions | Copper(II) ions |
C-![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) -SIRKLEYEIEELRLRIG (underlined region: triglycine spacer) -SIRKLEYEIEELRLRIG (underlined region: triglycine spacer) |
Cys residue at N-terminus | Au chips | Substrate immersion | Specific detection of Cu2+ ions, metal binding induces conformational change from α-helix to β-sheet | 228 |
Ac-C-![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) -SIRKLEYEIEELRLRIG (underlined region: triglycine spacer) -SIRKLEYEIEELRLRIG (underlined region: triglycine spacer) |
Cys residue at N-terminus | Au NPs | Substrate incubation | Sensitive and selective colorimetric sensor for the determination of Cu2+ ions | 231 | ||
| Enzymes | Tyrosine kinase | I![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) GEFKKK-C (underlined residue acts as enzyme phosphorylation substrate) GEFKKK-C (underlined residue acts as enzyme phosphorylation substrate) |
Cys residue at C-terminus coupling to maleimide-terminated AT disulfide linker | Au-coated glass coverslips and other substrates | Substrate immersion and μCP | Immobilisation of peptide to study quantitative enzyme inhibition assays on chips. Antibody binding to phosphotyrosine-SAM-bound also took place. Peptide and carbohydrate chips were prepared | 78 |
| IFGEFKKK-C (negative control peptide, no phosphorylation takes place) | |||||||
| Protein kinase (PKA) | LRRA![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) LG-GGG-C (underlined residue acts as enzyme phosphorylation substrate) LG-GGG-C (underlined residue acts as enzyme phosphorylation substrate) |
Cys residue at C-terminus | Modified Au electrode | Multistep fabrication, substrate immersion, then passivation, then enzyme incubation, then redox tagging | Electrochemical sensor. Phosphorylated peptide binds to Zr4+, then 4-cyano-4-(phenylcarbonothioylthio)-pentanoic acid (CPAD) reacts at phosphorylated sites and served as RAFT agent for polymerisation of ferrocenylmethyl methacrylates (FcMMAs) as redox tags | 232 | |
| Protein tyrosine phosphatases (PTP) (22 kinds, including classical PTPs, regenerating liver phosphatases and one alkaline phosphatase (ALP)) | TRDX![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[p with combining low line]](https://www.rsc.org/images/entities/char_0070_0332.gif) ZT-C (X and Z comprise 19 canonical amino acids (except for Cys), originating an array of 361 peptide sequences. Underlined region: phosphotyrosine residue, test substrate for the assayed PTPs) ZT-C (X and Z comprise 19 canonical amino acids (except for Cys), originating an array of 361 peptide sequences. Underlined region: phosphotyrosine residue, test substrate for the assayed PTPs) |
Cys residue at C-terminus attached to a maleimide-displaying EG3 SAM | Array plate having 384 gold spots (microwell plate array) | Robotic liquid handler | Combinatorial approach using SAMs for matrix-assisted laser desorption/ionization mass spectrometry (SAMDI-MS) for the quantitative assay of phosphatase enzymes | 233 | |
| Phosphatases from cell lysates | GX-![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[p with combining low line]](https://www.rsc.org/images/entities/char_0070_0332.gif) / /![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[p with combining low line]](https://www.rsc.org/images/entities/char_0070_0332.gif) / /![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[p with combining low line]](https://www.rsc.org/images/entities/char_0070_0332.gif) -ZGR-C (X and Z comprise all natural amino acids except for Cys), either phosphoserine, phosphothreonine or phosphotyrosine residues were assayed in over 1000 peptide substrates -ZGR-C (X and Z comprise all natural amino acids except for Cys), either phosphoserine, phosphothreonine or phosphotyrosine residues were assayed in over 1000 peptide substrates |
Peptide arrays and SAMDI-MS to study phosphatase activity on Ser, Thr, and Tyr residues. Lysates from the following cell lines were studied: | 234 | ||||
| – NIH-3T3 (murine fibroblasts) | |||||||
| – HT-1080 (fibrosarcoma) | |||||||
| – MCF-7 (breast cancer) | |||||||
| – BT474 (ductal breast carcinoma) | |||||||
| – MDA-MB-231 (invasive ductal carcinoma) | |||||||
| β-1,4-Galactosyl transferase (βGalT, a glycosyl transferase) | GTTASN(βGlcNAc)YGTGFA | Carboxylic acid- or hydroxyl-terminated N-terminus and EG3-terminated AT linkers | Disposable Au slides | SPOT synthesis, peptide arrays | Peptide-SAMs used to evaluate peptides as enzyme substrates | 81 | |
| N-acetylgalactosaminyl transferase (ppGalNAcT, a glycosyl transferase) | AHGPTSAPA (derived from Mucin (MUC1), a transmembrane glycoprotein) | Carboxylic acid- or hydroxyl-terminated N-terminus and EG3-terminated AT linkers | Peptide-SAMs used to evaluate peptides as enzyme substrates | ||||
| Thermolysin (protease) | 56 tripeptide substrates were tested | Peptides were at-tached to the SAM surface through the C-terminal amino acid | Peptide-SAMs used to evaluate peptides as enzyme substrates | ||||
| Human immunodeficiency virus type-1 integrase (HIV-1 IN) |
Lipo-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -YQLLIRMIYKNI (derived from viral protein R (Vpr); -YQLLIRMIYKNI (derived from viral protein R (Vpr); ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : signalling redox tag) : signalling redox tag) |
Lipoic acid (thioctic acid) modified ferrocene (Thc-Fc). N-terminal of the peptides was attached to the activated carboxyl groups of Thc-Fc | Au electrodes | Multistep fabrication, substrate immersion | Electrochemical probing of HIV enzymes (that lack redox-active centres) using ferrocene-conjugated peptides | 85 | |
| Human immunodeficiency virus type-1 reverse transcriptase (HIV-1 RT) |
Lipo-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -VEAIIRILQQLLFIH (derived from viral protein R (Vpr); -VEAIIRILQQLLFIH (derived from viral protein R (Vpr); ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : signalling redox tag) : signalling redox tag) |
||||||
| Human immunodeficiency virus type-1 protease (HIV-1 PR) |
Lipo-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -VVStaASta (derived from pepstatine; -VVStaASta (derived from pepstatine; ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : signalling redox tag) : signalling redox tag) |
Lipoic acid (thioctic acid) acid modified ferrocene (Thc-Fc). N-Terminal of the peptide was attached to the activated carboxyl groups of Thc-Fc | Au electrodes | Multistep fabrication, substrate immersion | Electrochemical probing of HIV enzymes (that lack redox-active centres) using ferrocene-conjugated peptides | ||
(C-(NH-P4-C(O)-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -C(O)-VVStaASta-OH)2 (derived from pepstatine; -C(O)-VVStaASta-OH)2 (derived from pepstatine; ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : signalling redox tag; Sta: statine: (3S,4S)-4-amino-3-hydroxy-6-methylheptanoic acid, an unusual amino acid) : signalling redox tag; Sta: statine: (3S,4S)-4-amino-3-hydroxy-6-methylheptanoic acid, an unusual amino acid) |
Thiol from cystamine oligoproline Cys-(NH-Pro4-C(O)–Fc oligopeptide | Au electrode surfaces | Multistep fabrication. Modified with AuNPs or thiolated SWCNT/AuNPs | Impedance method for detecting HIV-1 protease and screening for its inhibitors. Peptide derived from pepstatin, a potent inhibitor of aspartyl proteases | 207 | ||
| Picomolar electrochemical detection of HIV-1 PR | 235 | ||||||
HS-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -VVStaASta (derived from pepstatin; -VVStaASta (derived from pepstatin; ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : signalling redox tag; Sta: see entry above) : signalling redox tag; Sta: see entry above) |
Screen-printed Au electrodes | Electrochemical approach for the detection of HIV-1 protease | 236 | ||||
| Caspase-3 (CASP3)c | ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -GDG -GDG![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) |G-C (CASP3 cleaves the N-terminus of the tetrapeptide motif |G-C (CASP3 cleaves the N-terminus of the tetrapeptide motif ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) , , ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : signalling redox tag) : signalling redox tag) |
Cys residue at C-terminus | Au electrode | Substrate immersion | Fc-tag is lost, resulting in the reduction of the redox current, evaluation of caspase activity in apoptosis | 237 | |
G![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) |-C (CASP3 cleaves the tetrapeptide motif |-C (CASP3 cleaves the tetrapeptide motif ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ) ) |
AuNPs deposited onto Au substrate | Substrate incubation | Detection of CASP3 using organic electrochemical transistors (OECT), detection limit 0.1 pM | 230 | |||
| Trypsinc | ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -Cter[G|RPS]Nter-PEG-disulfide (trypsin substrate, tetrapeptide, Fc: ferrocene at C terminus as signalling redox tag) -Cter[G|RPS]Nter-PEG-disulfide (trypsin substrate, tetrapeptide, Fc: ferrocene at C terminus as signalling redox tag) |
PEG7 disulfide (methoxy-terminated PEG6 disulfide, CH3O-EG6-S-)2) at the N terminus | Au surfaces | Substrate immersion | Mixed SAMs, for the rapid measurement of proteolytic activity | 238 | |
![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) - -![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) - -![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) |
Terminal Cys residues | QCM AT-cut quartz crystal with Au electrodes and Au NPs | Multistep fabrication, QCM electrode incubated with peptide, then AuNP deposition | Label-fee peptide based QCM biosensor for trypsin | 227 | ||
| C-RWEKIRLRWIKQ | |||||||
| Trypsinc | ![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) -FRR-PEGx-C ( -FRR-PEGx-C (![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) = methylene blue, redox tag, x = 2, 4, 6, 9, 12 ethylene glycol units) = methylene blue, redox tag, x = 2, 4, 6, 9, 12 ethylene glycol units) |
Cys residue from PEGx linker located at C-terminus | Au electrode | Substrate immersion, then MCH incubation | PEG-6 provided enhanced anti-fouling properties, limit of detection 200 pM for trypsin | 239 | |
| Papain |
Lipo-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -GGYR-OH (GGYR: papain-binding sequence, interacts with papain and acts as a competitive inhibitor, -GGYR-OH (GGYR: papain-binding sequence, interacts with papain and acts as a competitive inhibitor, ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : redox tag) : redox tag) |
Thiol from lipoic acid linker bound to amino group of ferrocene amino acid (1′-aminoferrocene-1-carboxylic acid) at N-terminus | Au surfaces | Substrate spotting | Interaction of a ferrocene-peptide conjugates with papain | 240 | |
| Au microelectrodes | Substrate immersion | Interaction of papain with peptide immobilised on Au surface altered the interfacial ET resistance | 241 | ||||
| Human neutrophil elastase (HNE)c | ![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) -APEEI|MRRQ-PEG2-C ( -APEEI|MRRQ-PEG2-C (![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) = methylene blue, redox tag; PEG2 = methylene blue, redox tag; PEG2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ethylene glycol units) 2 ethylene glycol units) |
Cys residue from PEG2 linker located at C-terminus | Au electrode | Substrate immersion, then MCH incubation | Bio-electrochemical sensing of HNE, produced during inflammation. Decrease of electrical signal after HNE cleavage measured by square wave voltammetry (SWV) | 242 | |
| Botulinum neurotoxin type A (BoNT/A, a bacterial enzyme) | GGGSNRTRIDEANQRATR{Nle}LGGG-C (derived from synaptosomal-associated protein (SNAP-25), involved in exocytosis of neurotransmitters, Nle: Norleucine) | Cys residue at C-terminus bound to amines (displayed by SAM) via sulfo-SMCC, (Succinimidyl-4-(N-maleimido-methyl)cyclohexane-1-carboxylate, a hetero-bifunctional cross-linker reactive for both thiols and amines) | Au slides | Multistep fabrication | Bacterial toxin sensing within arrayed microchannels, fluorescence detection as low as 3 pg mL−1. Peptide N-terminus tagged with fluorescein | 243 | |
| Legumain (cancer mediating protease)c |
H
2
N-(CH2)4-CO-AAN|L-NH-CH2-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ( (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene as signalling redox tag) : ferrocene as signalling redox tag) |
Amino groups from peptide linker at N terminus formed amide bond with substrate via EDC/sulfo-NHS coupling | Silicon chip modified with vertically aligned carbon nanofibers (VACNF) | Multistep fabrication, VACNF growth, then preconditioning, then passivation, then substrate immersion | Electrochemical protease biosensor for cancer diagnosis and treatment monitoring | 244 | |
| Cathepsin B (cancer mediating protease)c |
H
2
N-(CH2)4-CO-LR|FG-NH-CH2-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ( (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene as signalling redox tag) : ferrocene as signalling redox tag) |
Amino groups from peptide linker at N terminus formed amide bond with substrate via EDC/sulfo-NHS coupling | Silicon chip modified with vertically aligned carbon nanofibers (VACNF) | Multistep fabrication, VACNF growth, then pre-conditioning, then passivation, then substrate immersion | Electrochemical protease biosensor for cancer diagnosis and treatment monitoring | ||
| Matrix metalloprotease-2 (MMP-2)c | FGPLG|VRGKGGC-MUA (named Pep 1, binds to Pep 2 via host–guest interactions, cleaved by MMP-2) | Thiol from 11-mer-captoundecanoic acid (MUA) attached to Pep 1 assembles to Au surface, exposing Phe residue, which binds to Phe residue from Pep 2 via CB[8] non-covalent host–guest interactions | Au electrodes | Multistep approach, Ag NPs modification | Pep 1 having a preferred MMP-2 restriction site is firstly anchored on electrode, then it recruits peptide-templated Ag NPs through the CB[8] recognition, producing signal. In presence of MMP-2, peptide 1 is cleaved, with subsequent separation of Ag NP, reducing signal intensity | 74 | |
| FGGGASLWWSEKL (named Pep 2, templates Ag NPS, not cleaved by MMP-2, binds to Pap-1 via cucurbit[8]uril (CB[8]) non-covalent host–guest interactions) | |||||||
| C-SGGGPL|GVRG (MMP2 cleavage generates a free COOH group at C-terminus) | Cys residue at N-terminus | Multistep approach, substrate incubation, eRAFT polimerisation | Electrochemical detection of MMP-2. MMP2 cleavage generates a free COOH group at C-terminus that tethers CPAD via Zr4+ linkers, which is used for the eRAFT polymerisation of ferrocenylmethyl methacrylate as redox tags | 245 | |||
| Matrix metalloprotease-2 (MMP-2)c |
H
2
N-KG![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) GC (underlined region specific for MMP-2 recognition) GC (underlined region specific for MMP-2 recognition) |
Amino N-terminus group binds to GO via EDC/NHS coupling, Cys residue used for binding to Au-Pt nanorods | Modified glassy carbon electrode (GCE) | Multistep fabrication, chitosan-graphene quantum dots (CS-GQDs-COOH) | MMP-2 detection based on peptide-Au@Pt bimetallic nanorods | 246 | |
| Matrix metalloprotease-7 (MMP-7)c | KKKRPLALWRSCC-C | Cys residue at C-terminus | Au-rGO electrodes (reduced graphene oxide-Au) | Multi-step approach, electrode modified with Pd nanoparticles | Electrochemical detection of MMP-7, based on signal amplification by Pd nanoparticles dual catalytic reactions | 247 | |
| H2N-NAADLEKAIEA|LEKHLEAKGP-C-DAAQ|LEKQLEQAFEAFERAG-OH (two cleavage sites by MMP-7) | Cys-22 residue | Modified Au electrode | Multistep fabrication, Au NP/carbon nanotube electrochemical sensor | Electrochemical detection of MMP-7. After being cleaved by MMP7, both the size and net charge of the peptide remaining on the sensor surface are reduced, facilitating electron transfer between the electrode and the electrochemical redox probe (ferri/ferrocyanide) | 248 | ||
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -RPL|ALWRS-C ( -RPL|ALWRS-C (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, redox tag) : ferrocene, redox tag) |
Cys residue at C-terminus | Au electrode | Substrate incubation, then passivation | Fc-peptide is hydrolysed at the A–L bond. ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) redox tag is lost in the process from the electrode surface, leading to a large measurable signal decrease. Biosensor specificity tested against MMP-2 and MMP-3 redox tag is lost in the process from the electrode surface, leading to a large measurable signal decrease. Biosensor specificity tested against MMP-2 and MMP-3 |
249 | ||
| Matrix metalloprotease-9 (MMP-9)c | ![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) -GPLG|MWSR-C ( -GPLG|MWSR-C (![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) : methylene blue, redox tag) : methylene blue, redox tag) |
Cys residue at C-terminus, MB carboxylic acid N-hydroxysuccinimidyl ester (MB-NHS) conjugated at the peptide N-terminus | Au electrode, deposited onto Si/SiO2 substrates | Lithography, substrate incubation | Reference electrode-free electrochemical biosensor for detecting MMP-9 using a concentric electrode device | 225 | |
| Matrix metalloprotease-14 (MMP-14)c |
C-LPLRSWG|LK-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ( (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, signalling tag) : ferrocene, signalling tag) |
Cys residue at N-terminus | Au electrodes | Substrate incubation | Electrochemical biosensor for MMP-14 in overexpressed MCF-7 breast cancer cells. MMP-14 specifically cuts Pep-Fc to make Fc drop from electrode surface, causing signal reduction | 250 | |
| VMDGYPMP-(CH2)6-C (inhibitory peptide, interacts with hemopexin-like domain (PEX) of MMP-14 (PEX-14) preventing MMP-14 dimerisation and binding to CD44) | Cys residue from linker located at C-terminus | Modified Au electrode | EIS-based MMP-14 detection, based on binding interaction between MMP-14 and its inhibitory peptides | 208 | |||
| GYPKSALR-(CH2)6-C (inhibitory peptide, interacts with hemopexin-like domain (PEX) of MMP-14 (PEX-14) preventing MMP-14 dimerisation and binding to CD44) | |||||||
| Neuron specific enolase (NSE, a small cell lung cancer biomarker) | C-KGVLKAVDHINSTIAP-C (doubly Cys-modified peptide, used as template for molecular imprinting) | Cys residues located at N- and C-termini | Modified Au wire electrodes | Multistep fabrication, doubly Cys-modified peptide templates for subsequent molecular imprinting via electro-polymerization (EP) and target sensing | Detection of short 10-mer peptide and NSE protein via a novel epitope imprinting strategy, SAM epitope bridges on Au surfaces | 251 | |
| KAVDHINST (10-mer computationally derived from NSE) | Binds to templated electropolimerised surface via noncovalent interactions | ||||||
| Thrombin (TB) | G![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) DKDKDKDPPPP-C (thrombin recognising peptide region underlined, Fc is electrochemical tag) DKDKDKDPPPP-C (thrombin recognising peptide region underlined, Fc is electrochemical tag) |
Cys at the C-terminus binds to AuNPs, Fc-COOH was further reacted to the amine at the peptide N-terminus by EDC/NHS chemistry | Modified GCE | Multistep fabrication involving Nafion, MXene (a graphene-like transition metal carbon/nitride), MB and AuNPs decorated with the peptide | Anchor, antifouling, and recognition peptide. Thrombin detection with excellent electrochemical activity and antifouling capability | 252 | |
| α-Thrombinc | ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -Cter[RFS|RPQL]Nter-PEG-disulfide (α-Thrombin substrate, heptapeptide, -Cter[RFS|RPQL]Nter-PEG-disulfide (α-Thrombin substrate, heptapeptide, ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene as signalling redox tag) : ferrocene as signalling redox tag) |
PEG7 disulfide (methoxy-terminated PEG6 disulfide, CH3O-(CH2CH2O)6-S-)2) linker at the N terminus | Au surfaces | Substrate immersion | Mixed SAMs, for the rapid measurement of proteolytic activity | 238 | |
| Plasmin | ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -KTFK-GGGGGG-C ( -KTFK-GGGGGG-C (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene as signalling redox tag, KTFK: plasmin target sequence) : ferrocene as signalling redox tag, KTFK: plasmin target sequence) |
Cys residue at C-terminus | Au surfaces | Electrochemical-based sensor for plasmin activity (a trypsin-serine protease playing a critical role in blood hemostasis and fibrinolysis) | 253 | ||
| Aminopeptidase N (APN) | ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) -R-EKEKEKEK-AKAKAKA; (AKAKAKAK: anchoring and antifouling sequence; EKEKEKEK: antifouling sequence; R: linking which balanced the negative charge of the target recognising part; YVEYHLC: -R-EKEKEKEK-AKAKAKA; (AKAKAKAK: anchoring and antifouling sequence; EKEKEKEK: antifouling sequence; R: linking which balanced the negative charge of the target recognising part; YVEYHLC: ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[r with combining low line]](https://www.rsc.org/images/entities/char_0072_0332.gif) ![[e with combining low line]](https://www.rsc.org/images/entities/char_0065_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ![[o with combining low line]](https://www.rsc.org/images/entities/char_006f_0332.gif) ![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif) ![[n with combining low line]](https://www.rsc.org/images/entities/char_006e_0332.gif) ![[i with combining low line]](https://www.rsc.org/images/entities/char_0069_0332.gif) ![[s with combining low line]](https://www.rsc.org/images/entities/char_0073_0332.gif) ![[i with combining low line]](https://www.rsc.org/images/entities/char_0069_0332.gif) ![[n with combining low line]](https://www.rsc.org/images/entities/char_006e_0332.gif) ![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif) ![[s with combining low line]](https://www.rsc.org/images/entities/char_0073_0332.gif) ![[e with combining low line]](https://www.rsc.org/images/entities/char_0065_0332.gif) ![[1 with combining low line]](https://www.rsc.org/images/entities/char_0031_0332.gif) ![[u with combining low line]](https://www.rsc.org/images/entities/char_0075_0332.gif) ![[e with combining low line]](https://www.rsc.org/images/entities/char_0065_0332.gif) ![[n with combining low line]](https://www.rsc.org/images/entities/char_006e_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ![[e with combining low line]](https://www.rsc.org/images/entities/char_0065_0332.gif) ) ) |
Positively charged sequence AKAKAKAK can adsorb on the negatively charged poly(3,4-ethylenedioxythiophene) (PEDOT)-citrate film | Modified CGE | PEDOT-citrate films electro-deposited on GCEs | Three-in-one peptides with anchoring, antifouling, and recognising capabilities for highly sensitive and low-fouling electrochemical sensing. Target (APN) overexpressed in HepG2 cells | 254 | |
| Nucleic acids | DNA oligomersd |
![[W with combining low line]](https://www.rsc.org/images/entities/b_char_0057_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/b_char_004b_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/b_char_004c_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/b_char_0056_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/b_char_004c_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/b_char_0045_0332.gif) (named AuBP1, from a phage display combinatorial library, underlined: solid binding sequence) (named AuBP1, from a phage display combinatorial library, underlined: solid binding sequence) |
Solid-binding peptide (SBP) region anchors nucleic acid probes onto substrates | Au substrates | Injection onto substrates | Chimeric SBP-peptide nucleic acid bifunctional construct, quantitative nucleic acid detection | 68 |
tgaggct-![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/b_char_0057_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/b_char_004b_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/b_char_004c_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/b_char_0056_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/b_char_004c_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/b_char_0045_0332.gif) (AuBP1-GGG linker-PNA, underlined: solid binding sequence) (AuBP1-GGG linker-PNA, underlined: solid binding sequence) |
|||||||
tgaggct-(2-aminoethoxy-2-ethoxyacetic acid)![[W with combining low line]](https://www.rsc.org/images/entities/b_char_0057_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/b_char_004b_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/b_char_004c_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/b_char_0056_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/b_char_004c_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/b_char_0045_0332.gif) (AuBP1-O linker-PNA, unde (AuBP1-O linker-PNA, unde![[r with combining low line]](https://www.rsc.org/images/entities/char_0072_0332.gif) lined: solid binding sequence) lined: solid binding sequence) |
|||||||
| microRNAs | C-THNDRKQE | The peptide was covalently immobilised onto PANI surface via amino-thiol binding using SMCC bifunctional linker | Modified CGE | Multi-step modification, PANI/GCE incubated in SMCC, then peptide or DNA incubation | Ultrasensitive and low-fouling microRNA electrochemical biosensor, DNA-mediated RNA capture and sensing, detection sensitivity and linear range not negatively affected antifouling peptide | 255 | |
| microRNA21 (miRNA-21) | C-DSDS-PPPP-AEAKAEAK (multifunction peptide, doping sequence: DSDS; linking sequence: PPPP; antifouling sequence: AEAKAEAK) | Cys residue at N-terminus | Multistep fabrication, AuNPs electrodeposited on GCE | Aptamer biosensor, miRNA-21 is commonly used as a biomarker for disease diagnosis and therapy. Non-specific adsorption of Lyz, myoglobin (Mb), BSA, and fluorescein isothiocyanate-labelled BSA were tested | 256 | ||
| Peptides | β-Amyloid oligomer (AβO, Aβ(1–42)) | C-PPPPTHSQWNKPSKPKTNMK (capture peptide) | Cys residue at N-terminus | Au electrode | Substrate incubation with capture peptide and 6-mercaptohexane, then AβO capture, then with self-assembling signal probe, then signal enhancing | Signal amplification strategy based on peptide self-assembly for the identification of AβO | 257 |
C16-GGGTHSQWNKPSKPKTNMK-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) (Self-assembly signal probe, (Self-assembly signal probe, ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : redox tag) : redox tag) |
Non-covalent binding to AβO capturing peptide-SAM, forms fibrils | ||||||
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -THSQWNKPSKPKTNMK (signal probe, -THSQWNKPSKPKTNMK (signal probe, ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : redox tag) : redox tag) |
Non-covalent binding to fibril decorated peptide-SAM | ||||||
MUA-RGTWEGKWK-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ( (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : redox tag) : redox tag) |
Thiol from MUA linker at N-terminus | Au electrode | Substrate incubation, then passivation with 9-mercapto-1-nonanol (MNH) | Peptide-based electrochemical biosensor for Aβ(1–42) soluble oligomer, detection principle based on loss of peptide chain flexibility after Aβ(1–42) binding | 258 | ||
MUA-![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) EEEEEE-Y-GSNKGAIIGLM (bifunctional peptide, includes an amyloid β target sequence (underlined) and the 25 – 35 fragmented peptide, which induces release of amyloid β by platelets) EEEEEE-Y-GSNKGAIIGLM (bifunctional peptide, includes an amyloid β target sequence (underlined) and the 25 – 35 fragmented peptide, which induces release of amyloid β by platelets) |
Thiol from 11-MUA AT linker | Au slides | Multistep fabrication, substrate immersion, then poly-tyrosine is added for detection | Periphery platelets can secrete amyloid β and induce its cross-linking & aggregation to form a surface peptide nano-network, resulting in poly-tyrosine strands being covalently trapped in the network (to serve as a deficient signal amplifier, through Tyr electrochemical oxidation) | 259 | ||
| Micro-organisms | E. coli & Salmonella spp | GIGKFLHSAGKFGKAFVGEIMKS-C (derived from magainin I) | Cys residue at C-terminus | Au micro-electrodes | Substrate incubation | Sensor for pathogenic E. coli and Salmonella spp, detection limit approx. 1 bacterium/μL | 83 |
| C-GIGKFLHSAGKFGKAFVGEIMKS (derived from magainin I) | Cys residue at N-terminus | Reduced binding activity when immobilised via the N-terminus vs C-terminus | |||||
| Viruses | Norovirus | QHKMHKPHKNTK-![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) -C (Identified by phage display, flexible linker underlined) -C (Identified by phage display, flexible linker underlined) |
Cys residue at C-terminus with amines in PANI through the bifunctional sulfo-SMCC cross-linking | Modified Au electrode | Screen-printed electrode (SPE), then substrate incubation | Electrochemical biosensor for detection of human norovirus. Immersion of norovirus captured on the Au-working electrode in working buffer solution, which can be used for oxidation and reduction, affinity strength can be measured using EIS | 87 |
QHKMHKPHKNTK-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) )2-C (two repeats of flexible linkers underlined) )2-C (two repeats of flexible linkers underlined) |
|||||||
QHKMHKPHKNTK-![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) -C (rigid linker underlined) -C (rigid linker underlined) |
|||||||
QHKMHKPHKNTK-(![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) )2-C (two repeats of rigid linker underlined) )2-C (two repeats of rigid linker underlined) |
|||||||
QHKMHKPHKNTK-![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) -GGGGS-C (non-fouling sequence underlined, contains flexible linker) -GGGGS-C (non-fouling sequence underlined, contains flexible linker) |
|||||||
QHKMHKPHKNTK-![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) -(GGGGS)2-C (non-fouling sequence underlined, two repeats of flexible linker) -(GGGGS)2-C (non-fouling sequence underlined, two repeats of flexible linker) |
|||||||
QHKMHKPHKNTK-![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) -EAAAK-C (non-fouling sequence underlined, contains rigid linker) -EAAAK-C (non-fouling sequence underlined, contains rigid linker) |
|||||||
QHKMHKPHKNTK-![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) -(EAAAK)2-C (non-fouling sequence underlined, two repeats of rigid linker) -(EAAAK)2-C (non-fouling sequence underlined, two repeats of rigid linker) |
|||||||
| SARS-CoV-2 (coronavirus) | (C-KEKEKEKE)2KEPPPPKEKEKEKEK-biotin (Inverted Y-shaped peptide: 2 anchoring points) | Terminal Cys residues with amine groups from PANI (polyaniline, a conducting polymer) via SMCC crosslinker coupling | Modified GCE | Multistep fabrication, electrodeposition of PANI nanowires, then sulfo-SMCC, then peptide incubation | Enhanced antifouling capability for SARS-CoV-2 (COVID-19 causing agent) nucleic acid detection in complex biological media | 260 | |
| Proteins | Alpha fetoprotein (AFP) |
![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) - -![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) -EKEKEKEK (peptide modified with AFP aptamer) -EKEKEKEK (peptide modified with AFP aptamer) |
Cys residue at C-terminus with amine groups from PANI via SMCC bifunctional crosslinker coupling | Modified GCE | Conducting polymer PANI film electrodeposited onto GCE, then sulfo-SMCC, then peptide incubation, then aptamer incubation | Aptasensor. Hierarchically architectured zwitterionic peptide brushes, ultralow fouling detection of AFP in serum. False positive reactions caused by nonspecific protein adsorption prevented, highly sensitive detection of AFD was achieved | 261 |
![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) - -![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) -EKEKEK (peptide modified with AFP aptamer) -EKEKEK (peptide modified with AFP aptamer) |
|||||||
![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) - -![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) -EKEK (peptide modified with AFP aptamer) -EKEK (peptide modified with AFP aptamer) |
|||||||
![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) - -![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) -EK (peptide modified with AFP aptamer) -EK (peptide modified with AFP aptamer) |
|||||||
| Mucin 1 (Muc1) |
![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) - -![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) EK2(EK)4(EK) (branched presentation of the antifouling sequence EKEKEK) EK2(EK)4(EK) (branched presentation of the antifouling sequence EKEKEK) |
Cys residue at C-terminus, thiols were covalently bonded with PANI's (polyaniline) amino groups via sulfo-SMCC coupling | Modified GCE | Multistep electrode modification, galvanostatic PANI deposition on GCE, then substrate incubation with aptamers and SMCC | Electrochemical aptasensor for ultralow fouling cancer cell quantification. Peptide reduces unspecific protein adsorption for the detection MUC1-positive MCF-7 breast cancer cells in complex media (human serum) with high sensitivity and selectivity using aptamers as recognition elements | 262 | |
| C-Reactive protein (CRP) | ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -EA-C ( -EA-C (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : redox tag) : redox tag) |
Cys residue at C-terminus | Au electrodes | Multistep fabrication, substrate incubation with Fc tagged peptides, then incubation with polyclonal anti-CRP IgG antibodies | Immunodetection of C-reactive protein (CRP), a biomarker of inflammation, limit of detection 240 pM, ![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -EAA-C-Am peptide exhibited the best performance -EAA-C-Am peptide exhibited the best performance |
263 | |
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -EAA-C ( -EAA-C (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : redox tag) : redox tag) |
|||||||
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) -EAAA-C ( -EAAA-C (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : redox tag) : redox tag) |
|||||||
| Cytochrome c (Cyt c) | C-AE | Cys residue at N-terminus | Au film electrode | Substrate incubation | Mixed SAMs, peptide SAM as analogue of electron transfer protein partners. Cyt c adsorption via electrostatic interactions with surface | 264 | |
| Leucine-rich α-2-glycoprotein-1 (LRG1, biomarker for colorectal cancer) | ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) -GGGGS-C (bioactive sequence identified through phage display, underlined) -GGGGS-C (bioactive sequence identified through phage display, underlined) |
Cys residue at C-terminus | Au electrodes | Multistep fabrication, thiol functionalisation, then benzoquinone derivatisation, then peptide incubation | Electrochemical sensor for early diagnosis of adenoma-to-carcinoma progression based on rationally designed peptides | 265 | |
| Epidermal growth factor receptor (EGFR) |
HS-YHWYGYTPQNVI-![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ( (![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) : ferrocene, signalling redox tag) : ferrocene, signalling redox tag) |
Thiol from 9-mercapto-1-nonanol at N-terminus | Substrate incubation, then passivation with 6-MCH | Target receptor (EGFR) is overexpressed in several cancers | 266 | ||
| Lipopolysaccharide endotoxins (LPS, from Gram-negative bacteria) | ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) -C (underlined sequence discovered by phage display) -C (underlined sequence discovered by phage display) |
Cys residue at C-terminus binds to AuNPs | Modified CGE | Multistep fabrication, GO deposition, peptide incubation, exposure to endotoxin, then DNA-ferrocene modified AuNPs | Electrochemical method for bacterial endotoxin assay, combining peptide-modified graphene oxide and DNA-modified AuNPs | 88 | |
| Antibodies | Human immunoglobulin G (IgG) |
C-PPPP-EK![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) (Y-shaped peptide, underlined region recognises human IgG) (Y-shaped peptide, underlined region recognises human IgG) |
Cys residue at C terminus | GCE | Au NPs electrodeposited on GCE/PEDOT-citrate surface | Low fouling electrochemical biosensors for detection of IgG in human serum | 267 |
C-PPPP-EKEKEKE-![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) (linear peptide, underlined region recognises the cristallisable fragment (known as Fc) region of human IgG) (linear peptide, underlined region recognises the cristallisable fragment (known as Fc) region of human IgG) |
|||||||
(![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) D)4(KE)2KE-PPPP-DDDD-C (Doping sequence: DDDD; Linking sequence: PPPP; underlined region recognises the cristallisable fragment (known as Fc) region of human IgG) D)4(KE)2KE-PPPP-DDDD-C (Doping sequence: DDDD; Linking sequence: PPPP; underlined region recognises the cristallisable fragment (known as Fc) region of human IgG) |
Cys at the C-terminus | AuNPs modified electrode with electrodeposition of PEDOT (poly(3,4-ethylenedioxythiophene), a conducting polymer) | Sensitive antifouling biosensor for reliable IgG quantification in serum | 144 | |||
C-PPPP-NQNQNQNQD-![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ; (Anchoring domain: CPPPP; antifouling domain: NQNQNQNQ; underlined region recognises Fc region of human IgG) ; (Anchoring domain: CPPPP; antifouling domain: NQNQNQNQ; underlined region recognises Fc region of human IgG) |
Cys at the C-terminus via covalent bond formation with PANI mediated by sulfo-SMCC | Multistep fabrication, electro-polymerisation of PANI nanowire arrays on CGE, then substrate incubation and peptide immobilisation | IgG limit of detection of (0.26 ng mL−1) in neat serum and real clinical samples. | 268 | |||
| Human immunoglobulin E (IgE) | Ac-![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) (underlined hexapeptide exhibits high affinity and specificity to the Fc fragment of human IgG) (underlined hexapeptide exhibits high affinity and specificity to the Fc fragment of human IgG) |
C-Terminus coupling via HATU to an AT EG3-OH SAM presenting tris(2-aminoethyl)amine groups | Au-coated glass slides | Multistep fabrication, AT SAM assembly, then CDI activation, then tris(2-aminoethyl)-amine coupling, then peptide incubation | Peptide SAM with increased ligand density on branched amines. Increased sensitivity and selectivity for IgG adsorption from complex cell culture supernantants, regeneration-binding is demonstrated | 269 | |
Ac-![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) G (underlined hexapeptide exhibits high affinity and specificity to the Fc fragment of human IgG) G (underlined hexapeptide exhibits high affinity and specificity to the Fc fragment of human IgG) |
|||||||
| Human immunoglobulin E (IgE) | C-HHH-DDD | Amino groups from peptide covalently bind to carboxylic acid-decorated PABA (poly(m-aminobenzoic acid)) layer via NHS/EDC assisted coupling | Modified GCE | Multistep fabrication, electrochemical deposition of electroconductive PABA layer, then decoration with aptamer and antifouling peptide | Improve biosensing of Immunoglobulin E with aptamers using antifouling peptide. Cys residue from peptide apparently not involved in surface attachment | 270 | |
| Rituximab (chimeric monoclonal antibody for treating indolent B-cell non-Hodgkin's lymphoma) | C-GSGSGSWPRWLEN (cluster of differentiation 20 (CD20) mimetic peptide, named CN14) | Cys residue at N-terminus | Au electrode | Substrate immersion in Poly adenine (polyAn, n = 10–50, DNA sequence), then immersion in peptide solution | Rituximab detection in lymphoma patients’ plasma. Rituximab: a chimeric immunoglobulin (IgG) anti-CD20 antibody genetically engineered mAbs, approved by FDA | 271 | |
| Anti-DGP IgG monoclonal antibody (DGP: Alpha-2 deamidated gliadin peptide, derived from gliadin from gluten) | QLQPFPQPELPYPQPQLPYPQPQLPYPPQPQPF (recognition peptide: alpha-2 deamidated gliadin) | Recognition peptide coupled via COOH in Glu of previously anchored support peptide | Multistep fabrication, support helical peptide formation, then MB tagging, then attachment of recognition peptide, then binding of target antibody | Electrochemical peptide-based sensor for antibody detection. | 272 | ||
| Alpha-2 deamidated gliadin peptide (DGP, 33-mer peptide containing the 56–88 residues of alpha gliadin from gluten) | |||||||
Lipo-YAAAHAEAR(![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) ) (support helical peptide, labelled with methylene blue ( ) (support helical peptide, labelled with methylene blue (![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) ) as electrochemical tag) ) as electrochemical tag) |
Lipoic acid attached to N-terminus | ||||||
| HIV anti-p24 antibodies |
HS-C11-EAAEWDRVHP-K(![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) ) (derived from HIV-1 capsid protein p24, labelled with methylene blue ( ) (derived from HIV-1 capsid protein p24, labelled with methylene blue (![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) ) as redox label) ) as redox label) |
Thiol from modifier attached to N-terminus | Substrate incubation, then passivation | Electrochemical peptide-based biosensor for HIV detection, based on ET rate between MB and substrate after target binding | 273 | ||
| Autoantibodies involved in rheumatoid arthritis (RA) | ALQECit546DYIFGNYIECit556 (derived from inter-alpha-trypsin inhibitor-3 (ITIH3)). Cit: L-citrulline, a non-proteinogenic amino acid | N-terminus via NHS/EDC coupling to a pre-formed MUA-displaying SAM | Modified Au electrode | Multistep fabrication | EIS-based method for the detection of RA autoantibodies, better sensitivity than the current conventional ELISA method | 274 | |
| Antigens | PSA (prostate-specific antigen)c |
C-![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) | |![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) -Fc, (underlined sequence derived from semenogelin I and II, peptide tagged with ferrocene active moiety) -Fc, (underlined sequence derived from semenogelin I and II, peptide tagged with ferrocene active moiety) |
Cys at the N-terminus (peptide self-assembled onto AuNPs) | Modified GCE | Multistep fabrication, by casting a mixture of Nafion and AuNPs onto the surface of GCE | Electrogenerated chemiluminescence peptide-based biosensor for the determination of prostate-specific antigen. AuNPs were used as amplification platform. PSA is a serine protease secreted by both normal prostate glandular and prostate cancer cells | 275 |
| Cys residue at N-terminus | Au electrode | Multistep fabrication | Hydrolysis of S–S bond results in loss of the Fc-tag off the surface which in turn reduces the faradaic redox response of the system. Helical peptide | 276 | |||
Biotin-E![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) | |![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) -C (underlined sequence derived from semenogelin I and II) -C (underlined sequence derived from semenogelin I and II) |
Cys residue at C-terminus | AuNPs on 96-well plate | Peptide-based biosensor for prostate-specific antigen, based on electrochemical glucose readout | 82 | |||
4-Pentynoyl-GGGG-![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) L-OH; (underlined region corresponds to PSA-specific peptide) L-OH; (underlined region corresponds to PSA-specific peptide) |
4-Pentynoyl group at N-terminus coupling to azido groups immobilised on the electrode via CuAAC | GCE electrografted with 4-azidoaniline | Multistep fabrication, peptide grafting using diazonium electroreduction and click chemistry | Based on proteolytic activity of PSA towards grafted peptide carrying streptavidin at its distal end. Limit of quantification estimated in the nM to pM range | 75 | ||
4-Pentynoyl-GGGG-![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) L-(biotin)-OH (underlined region corresponds to PSA-specific peptide: HSSKLQ) L-(biotin)-OH (underlined region corresponds to PSA-specific peptide: HSSKLQ) |
|||||||
K![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) -DKKDKDPPPP-C (Co-assembled with ferrocene tagged peptide, PSA recognising peptide underlined, followed by region with antifouling properties) -DKKDKDPPPP-C (Co-assembled with ferrocene tagged peptide, PSA recognising peptide underlined, followed by region with antifouling properties) |
Cys residue at C-terminus | Modified Au electrode, electrodeposited onto ITO | Multistep fabrication, graphene oxide−Fe3O4−thionine probe | Mixed SAMs, dual mode biosensor, performs on human serum samples | 226 | ||
| Fc-DKDKDKDPPPP-C (Co-assembled, antifouling peptide) | |||||||
K![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) DKDKDKDPPPP-C (PSA recognising peptide underlined) DKDKDKDPPPP-C (PSA recognising peptide underlined) |
Cys at the C-terminus. Fc-COOH was further reacted to the amine at the peptide N-terminus by EDC/NHS chemistry | Modified GCE | Multistep fabrication involving Nafion, MXene (a graphene-like transition metal carbon/nitride), MB and AuNPs decorated with the peptide | PSA detection with excellent electrochemical activity and antifouling capability | 252 | ||
On the other hand, phosphonic acids (R-PO3H2, R = alkyl chain) can be incorporated onto hydroxylated substrates, such as Ti and silicone oxides, by thermal annealing, allowing the covalent binding between the phosphate group and the hydroxyl groups (P–O bound energy of ∼80 kcal mol−1).40 Other strategies involve surface decoration with phosphonate ligands, as described by Eisenberg and collaborators, presenting several epitopes on a SAM via attachment to a fusion construct containing cutinase, a serine esterase that forms a site-specific covalent adduct with phosphonate ligands (Table 1, peptide-SAMs binding to cell membrane proteins).72
Kato and collaborators presented peptide ligands containing either linear or cyclic RGD motifs attached to a benzoquinone-modified SAM. Ligands were immobilised to this surface through a quantitative Diels–Alder cycloaddition coupling between the benzoquinone groups presented by the SAM and either cyclopentadiene-conjugated linear GRGDS or cyclic RGDFK peptide building blocks. This Diels–Alder coupling was selective, high yield, ensured constant densities of ligand (even when different ligands were employed), and took place to completion as verified through cyclic voltammetry.76 This kind of method presents a synthetic setback, as each ligand-cyclopentadiene conjugate must be individually synthesised before immobilisation.78
2.3 Peptides
Most functional biomaterials are based on a rather limited number of peptide sequences derived from protein ligands for cell surface receptors. As few proteins possess short peptide sequences that alone can engage cell surface receptors, the repertoire of receptors that can be targeted using this approach is rather narrow.79 Peptide-based SAMs have benefited from this strategy, however, other approaches have nurtured the diversity of applications of peptide-based SAMs platforms.Table 1 shows selected peptide-based SAMs aimed at binding ECM components (including structural components like hyaluronan (HA), and soluble components like growth factors (GFs) and enzymes), and proteins located at the surface of mammalian cells (integrins, receptors, lectins), either derived from natural proteins or discovered by phage display (technologies).
Table 2 focuses on peptide-based SAMs applied in a variety of biomaterials science and engineering, including: AMPs (binding bacterial cell membranes), peptides designed for electron transfer (mostly helical and electroconductive ones), metal ion binding, and peptides with antifouling properties, including surface modification of implants and biomedical devices. Each system is presented with information on the displayed peptide sequence, anchoring mode, substrate, linker (if applicable), and fabrication method. Additionally, Table 3 outlines selected examples from the vast amount of peptide-based SAM combining several of these features (for instance, binding affinity and antifouling properties) as part of platforms devoted to biosensing applications. Here, the information is organised as a function of the binding target, including enzymes, antibodies, nucleic acids, and other clinically relevant biomarkers.
2.3.2.1 ECM-mimicking peptides. Cells interact with the ECM via multiple mechanisms using specialised transmembrane proteins. For example, cell adhesion to ECM proteins is generally mediated via integrins on the cell surface that bind to precise regions of the proteins, while binding to soluble ECM components is mediated by specific receptors. Peptides intended for controlling cellular functions include ECM-derived or ECM-mimicking peptides, as well as ECM-binding peptides. ECM proteins can be divided into insoluble macromolecules that provide physical support (collagens (Coll), elastin; glycoproteins, such as fibronectin (FN), vitronectin (VN), tenascin (TN), laminin (LN); proteoglycans (PGs)), and biochemical signals to cells as soluble proteins (ECM regulators, such as GFs, cytokines, chemokines; ECM-crosslinking and -degrading enzymes, like lysyl oxidases and transglutaminases, and matrix metalloproteinases (MMPs)).92
Polysaccharide-binding peptides. The ECM is also occupied with specific anionic polysaccharides (glycosaminoglycans, GAGs) that confer not only hydration to the matrix but are also implicated in the sequestration, retention, stabilisation and activation of GFs involved in many cellular activities (adhesion, proliferation, migration, differentiation and gene expression). GAGs include non-sulphated members (i.e., HA) and sulphated polysaccharides (heparin (Hep), heparan sulphate, chondroitin sulphate, dermatan sulphate, keratan sulphate). The formation of macromolecular complexes between sulphated GAGs and basic GFs are mediated by electrostatic interactions.93 Not surprisingly, peptide-based SAMs used to bind Hep are rich in basic amino acids (Table 1, peptide-SAMs for cell culture and binding ECM components). Non-sulphated GAGs of the ECM, being an exception, do not bind to GFs, but participate in the ECM organisation, through binding to certain ECM proteins (link protein, aggrecan, versican, TSG-6) and specific receptors on the cell surface (CD44, RHAMM).94
HA-Binding proteins, known as hyaladherins, interact with HA through a common domain (∼100 amino acids), called link module, which also contain clusters of basic amino acids.95 HA-binding peptides displaying binding motifs found in hyaladherins could be rationally designed to form SAMs for the subsequent supramolecular immobilisation of HA and studying its role on cellular activities.
On the other hand, phage display on HA as a target has rendered a 12-mer GAHWQFNALTVR peptide96 (Table 1, peptide-SAMs for cell culture and binding ECM components). This HA-binding peptide does contain clusters of basic amino acids seen in hyaladherins. Nonetheless, this HA-binding peptide, also known as HA inhibitor or HA blocking peptide, has been applied in a vast number of studies, from mechanistic studies to elucidate the role of endogenous HA to the functionalisation of biomaterials.86 It was found that the peptide binds strongly to cell surface HA and less strongly to HA in the ECM. Its HA-binding ability has been demonstrated in tumours97 and healthy tissues,98 and it inhibits a number of HA-mediated cell signalling pathways.99
Azevedo and collaborators reported the use of this oligopeptide presented via SAMs on Au surfaces via N-terminus thiolation (Fig. 3a) for the supramolecular immobilization of HA.31,33 They prepared patterned Pep-1 SAMs to investigate the contribution of HA to glycocalyx regulation of endothelial cells function and its effect to vascular integrity. SAMs surfaces allowed the non-covalent immobilisation of HA of different molecular sizes, and low molecular weight HA was shown to improve cell adhesion and stimulate the migration of human umbilical vein endothelial cells (HUVECs) cultured atop these bioengineered surfaces. Pep-1 has also been used to anchor HA at different densities.34 Different ratios of a thiolated peptide derivative and 1-octanethiol were tethered to Au substrates, originating mixed SAMs with different hydrophilicity, which achieved supramolecular immobilisation of varying size HA molecules, ranging from 5 to 700 kDa (Table 1, peptide-SAMs binding ECM components). Low molecular weight HA facilitated HUVECs alignment and elongation under laminar flow conditions and potentially drives their directional migration, not by the expression of CD44 (the major cell surface receptor for HA)100 but by the assembly of focal adhesions instead. This was apparently the result of higher expression of vinculin and stress fibres during cell–HA interaction, whereas soluble HA did not induce these effects in a significant manner (Fig. 3f).
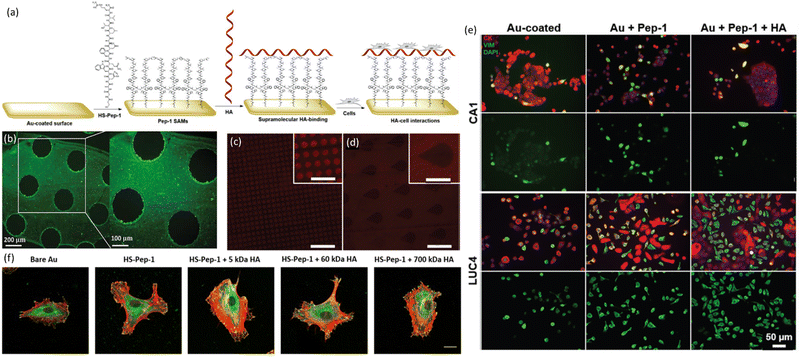 | ||
| Fig. 3 Supramolecular presentation of HA onto peptide-displaying SAMs. (a) Schematic illustration of supramolecular immobilisation of HA on Pep-1 SAM surfaces to study the role of HA in cancer stem cells and HUVECs. (b) Localisation of fluorescein-HA (green) on the SAM peptide areas. (c) Fluorescence images of surfaces patterned with a Pep-1-coated PDMS stamp, demonstrating the recognition of presented HA by an HA-binding protein, creating a spot pattern where HA is deposited in distinct foci or (d) a “negative” drop pattern in which HA forms a background coating (scale bar = 200 μm (large images) & 100 μm (inserts)) [reproduced from ref. 31 with permission from the Royal Society of Chemistry].31 (e) Fluorescence microscopy images of cultured CA1 and LUC4 cancer cells show an increased number of epithelial-to-mesenchymal transition cells bound to HA functionalised surfaces (scale bar is the same for all panels) [adapted with permission from ref. 33].33 (f) Confocal microscopy images demonstrating formation of focal adhesion of HUVECs seeded on bare Au and Pep-1 SAMs with or without HA for 24 h (scale bars = 20 μm; green, vinculin; red, F-actin) [adapted with permission from ref. 34. Copyright 2021, American Chemical Society].34 | ||
Cell adhesive and motogenic peptides. Bioactive surfaces involving peptide-based SAMs have demonstrated their efficacy to bind diverse classes of cell receptors, thus becoming important tools for the high-throughput identification of recognition elements for guiding cell growth and differentiation. Kiessling and co-workers presented the well-known integrin-binding RGD motif (linear and cyclic) as part of arrays atop Au substrates, which adequately supported adhesion of human melanoma cells.50
The interaction of cells with certain glycoproteins of the ECM has been elucidated to take place via dynamic binding events between integrins on the cell surface and specific domains or segments in the ECM protein, typically comprising short peptide sequences (FN: RGD (α5β1 integrin); LN: IKVAV (α6β1 integrin); Coll: GFOGER (α1β1, α2β1, α10β1, α11β1 integrins)). While the RGD and IKVAV sequences from FN and LN, respectively, have been widely applied for promoting cell adhesion onto surfaces, many other sequences have been also utilised (Table 1, peptide-SAMs binding to cell membrane proteins). In addition to cell-adhesive sequences, glycoproteins also contain additional peptide segments that either bind to ECM components (e.g., heparin, GFs) or are involved in other cell activities rather than cell adhesion. For example, the cryptic FN motif IGD, from the gelatin-binding domain in FN, was shown to possess motogenic activity in human dermal fibroblasts.101 This peptide motif can be applied to stimulate cell migration in biologically relevant scenarios, such as in cancer metastatic progression (Table 1, binding cell membrane proteins) (Fig. 5b).27,102,103
Peptide-SAMs have contributed greatly to develop ECM models. For instance, the work by Mrksich and collaborators has contributed to elucidate the role of peptide and protein ligands in cell–ECM interactions, by studying SAMs presenting RGD motifs with different packing densities and spacing to study the adhesion and spreading of different cell types.31 The work of Derda and co-workers demonstrated that peptide-SAMs enable the high-throughput discovery of sequences that support proliferation of pluripotent cells.79 For example, they have employed peptide-displaying SAMs to assess embryonic stem (ES) cell growth and renewal of 18 laminin-derived peptides.26 SAM-mediated display allowed for uniform density and defined peptide orientation. Five of these peptides proved effective at promoting ES cell proliferation in an undifferentiated state. This platform also offered the possibility to investigate on the mechanism and selectivity behind the receptor–ligand interaction, and also demonstrated the transferability of the information from the SAMs screen when presenting the sequence RNIAEIIKDI as part of peptide amphiphile molecules that self-assembled into nanofibers and generated 3D hydrogels.
In a remarkable study, phage display-based approaches were employed by Derda and collaborators to identify peptide sequences that bind specifically to the surface of pluripotent human embryonal carcinoma (EC) cells. The peptides were displayed by SAMs of ATs on Au substrates.79 These surfaces supported undifferentiated proliferation of human embryonic stem (ES) cells, and when cultured on SAMs presenting the sequence TVKHRPDALHPQ or LTTAPKLPKVTR they expressed markers of pluripotency at levels similar to those of cells cultured on Matrigel®. It is noteworthy that neither of these epitopes mediate cell adhesion via integrin binding, thus suggesting the combination of phage display technologies and SAMs approaches for investigating the mechanisms that govern cell growth and differentiation. Other known ECM-binding peptides (Coll-II, LN) can be used to retain nascent ECM proteins produced by cells known to impact cell fate.42
These studies showed the utility of peptides as recognition elements for elucidating cell−matrix interactions in 2D and their further translation into more complex scenarios, like the functionalisation of 3D biomaterials to control cell migration, proliferation, and differentiation. Peptide-displaying SAMs as a screening strategy represents a promising avenue for the generation of materials for cell-based therapeutics that control the fate of hematopoietic stem cells, induced pluripotent stem cells, and cancer stem cells, among others.
Growth factor-related peptides. The repertoire of peptides derived from ECM glycoproteins is larger when compared to sequences obtained from soluble ECM macromolecules, such as GFs. GFs are biologically relevant, as they are involved in numerous cellular processes, such as cell growth, differentiation, and migration.
Several sequences derived from GFs were tested in SAMs to discover peptide surfaces that guide cellular processes, such as cell differentiation (Table 1, peptide-SAMs binding to the cell surface). For example, using a rational design approach, a short peptide sequence (KLTWQELYQLKYKGI) able to recognise vascular endothelial GF (VEGF) receptor (VEGFR) has been reported.104 The peptide, designed based on the x-ray structure of VEGF bound to VEGFR reproducing a region of the VEGF binding interface, was further used to study its effects on endothelial cells when immobilised on surfaces via SAMs.90 Other GFR-binding peptides have been also tested in SAMs (Table 1) and a number of studies involving peptide-based SAMs have specifically targeted TGF-β, which controls cell proliferation, differentiation, adhesion, migration, apoptosis, and ECM deposition, thus playing fundamental roles in development, tissue homeostasis, and cancer. Signalling by this GF is characterised by its tight binding to its cell-surface receptor complex, but also by the fact that it also mediates the oligomerisation, assembly and activation of this complex, which is formed by two types of TGF-β receptors (TβRI-ED and TβRII-ED).
Kiessling and co-workers20 employed peptide-displaying SAMs as means to exert precise spatial control over activation of TGF-β signalling. In this study, two peptide sequences, LTGKNFPMFHRN and MHRMPSFLPTTL, which interact with both TGF-β receptors, are presented to NMuMG mouse mammary gland cells to investigate their attachment, which is mediated by the expression of such receptors. Cells adhered to SAMs presenting either peptide (even at peptide densities as low as 4%) and the functionalised SAMs also activated a Smad2/3 nuclear translocation mechanism (which constitutes a hallmark of TGF-β signalling). The work of Kiessling and co-workers is noteworthy, as it demonstrated that peptide-based SAMs approaches can help to pre-organise multivalent scaffolds at the cell-surface level by pre-organising the transmembrane receptors, to potentiating the amplification of specific GF signals.
Peptide-SAMs allow for the concomitant binding to GFs and other ECM components, as reported by Cabanas-Danés and co-workers, who used a fluorogenic peptide-based SAM surfaces for the simultaneous binding of transforming growth factor-β1 (TGF-β1) and Coll-II.42 In this work, the Coll-II-binding peptide CLRGRYW was patterned and co-immobilised onto a SAM substrate, allowing for the selective immobilisation and confinement of Coll-II molecules to patterned areas matching with the fluorogenic areas. Furthermore, surfaces of CLPLGNSH tethered to TGF-β1 were used to culture human articular chondrocytes (hACs), which retained their representative chondrogenic phenotype and exhibited increased GAG production after 7 days of culture compared to controls. This work also demonstrates the excellent capabilities of peptide-based SAMs for the simultaneous binding and display of different ECM components.42
These works are representative of GF-binding via peptide-display SAMs, and evidence the versatility of these platforms to create tailored and polyvalent surfaces that can deliver bio-signalling able to instruct cell behaviour with precise spatial control. Overall, peptide-based SAMs technologies as the ones described in this section represent attractive strategies to generate platforms combining tissue targeting and regeneration properties, which are in great demand in the regenerative medicine field.
2.3.2.2 Enzyme-responsive and high-affinity binding peptides. Enzyme overexpression is typically seen in different diseases (cancer, infection, pancreatitis) and also during inflammation. The enzyme landscape includes a range of biocatalysts that act on diverse substrates (proteases, such as MMPs, elastases, caspases and prostate-specific antigen (PSA); glycosidases, like hyaluronidase and β-galactosidase; esterases, such as lipases and phospholipases), from proteins to glycans and lipids. Therefore, enzyme levels are normally used as specific biomarkers for disease detection and therapy monitoring and their accurate measurement is paramount in routine assays (biosensing applications).
Quite often, the selection of peptide substrates has been based on the enzyme specificity and obtained through combinatorial design approaches119 or phage display,120 which can be further optimised to yield desirable enzyme kinetics. For example, MMPs are generally quite specific for the bonds they cleave. MMP-1 typically cleaves peptide bonds between glycine (G) and isoleucine (I), but peptide bonds between other amino acids have been reported for other MMPs. Single amino acid substitution in the neighbour amino acids were shown to alter the activity of various MMPs.121 Other proteases, like trypsin or papain, have broader specificity for peptide bonds and can accommodate greater diversity of peptide substrates. Measuring enzyme activity via SAMs requires conjugation to a fluorophore or redox tag (e.g., ferrocene (Fc) or methylene blue (MB)) for detection purposes (Table 3, peptide-based SAMs for biosensing of enzymes).
High-affinity peptide binders can also be selected from peptide libraries and used for detection of biomarkers of interest (Table 3, peptide-based SAMs for biosensing of antibodies).
2.3.2.3 Antimicrobial peptides (AMPs). AMPs have gained renewed interest fuelled by the antibiotic crisis that calls for the development of new therapeutic agents. AMPs have advantages over conventional antibiotics, namely broad-spectrum activity against several microorganisms (Gram-positive and Gram-negative bacteria, fungi and viruses), anti-biofilm and immunomodulatory effects, and slower emergence of resistance.122–124 AMPs (e.g., melittin, mastoparan, maculatin 1.1, aurein 1.2, magainins, indolicidin, myxinidin, LL-37, histatin 5, among others) are small (12–50 amino acid residues), cationic (carrying a net positive charge, +2 to +9), amphipathic (50% of hydrophobic amino acids) peptides widely distributed in living organisms.125
AMPs are typically derived from proteins and peptide involved in the host defence (e.g., human lactoferrin and its pepsin digested peptide lactoferricin, human cathelicidin, LL-37) originated from a range of organisms (e.g., animals, viruses, bacteria, insects, and amphibians). Their sequences have been further optimised to enhance their antimicrobial potency and stability, and are known to facilitate their initial binding to the negatively charged bacterial membranes.126 The overall amphipathic structure of AMPs enables their folding into an α-helix while partitioning into the hydrophobic cores of lipid bilayers. They can also be rationally designed (e.g., GALA peptide), following an amphipathic arrangement. More recently, machine-learning techniques have also generated libraries of AMPs.127
Structural and functional limitations, such as sensitivity to salt, serum, and pH, self-aggregation and proteolytic degradation have hampered their clinical translation as potential therapeutic options.128 An advocated strategy to bypass these drawbacks involves surface immobilisation of AMPs to confer protection against enzymatic degradation in vivo and to prevent aggregation as well. This might increase AMPs long-term stability, enhancing their activity and avoiding toxicity-related issues associated to the high concentrations used necessary to achieve the desired antimicrobial effect.123,125,129
SAMs are compatible with many techniques used for AMP grafting. As such, SAMs displaying AMPs are widely used in proof-of-concept studies to demonstrate their potential.129 AMPs’ ease of immobilisation via SAMs enables high-throughput studies to investigate their interaction with different types of bacteria and select the most potent molecules for cost-effective antimicrobial therapies. SAMs have been employed to screen AMPs for antimicrobial activity (Table 2, peptide-SAMs for antimicrobial surfaces and implants) as well as for the management of urinary and gastric infections.128,129 Further details on antimicrobial SAMs are detailed in Section 5.4.
2.3.2.4 Electrically-conductive peptides. Helical peptides are suitable model systems for protein secondary structure and ion-channel formation studies.130 The diameter of helical peptides ranges from 1.0 to 1.5 nm, as a function of the amino acid composition,60 and is larger than that of AT linker units commonly used in conventional SAMs. Helical peptides have been shown to act as excellent medium for electron transfer, as demonstrated by the work of Morita and collaborators, who found a stronger electronic coupling of N-ethylcarbazolyl groups and a gold surface in helical peptides compared to that of saturated AT hydrocarbon chains.49 This might be a consequence of the introduction of conformationally-constrained residues in the peptide sequence, which allows for control over the secondary structure, thus separating any electroactive group and the metal substrate (Table 2, peptide-SAMs for electron transfer).56
Additionally, helix-forming peptides exhibit a highly regular structure due to the formation of intramolecular hydrogen bonds between the amide proton at the 5th residue and the carbonyl oxygen at the 1st residue.59 This regular peptide bond alignment along the helix generates a large macrodipole moment,24 which, along with interactions among neighbouring helixes, is largely responsible for their long-range electron transfer131 and generation of surface potential.132 These features paved helical peptides’ way into SAMs to fabricate electroactive surfaces131 and molecular electronics,60 as will be further described in Section 5.3.
2.3.2.5 Metal ion-binding peptides. The ability of peptides to selectively bind metal ions, such as Ca2+ or Cu2+, has been exploited in the context of mineralisation studies, for the controlled formation of calcium phosphate mineral on surfaces, or for biosensing applications (detection of metal ions).
Peptide-mineralising sequences are typically composed of polar and acidic amino acids (e.g. EEEEEEEE)133 and inspired in non-collagenous proteins involved in biomineralisation,134 such as dentin matrix protein-1 (ESQES, QESQSEQDS),135 dentin phosphoprotein (DSS repeats), statherin136 (DDDEEKFLRRIGRFG). Hydroxyapatite-binding peptides have been also identified by phage display,86 but only few examples137,138 can be found as displayed on peptide-SAMs. In fact, the use of SAMs for the controlled display of known mineralising peptides has been reported to a less extent (Table 2, peptide-SAMs for biomineralisation).
A redox-sensitive peptide was rationally designed and used as Cu2+-binding peptide for detection purposes (Table 3, metal ion-binding peptide-SAMs). The designed 17-residue peptide adopts a helical conformation in the absence of metal cations and switches to a β-sheet secondary structure in the presence of Cu(II) ions, which then reverses to an α-helix upon electrochemical reduction of the metal species to Cu(I) (Fig. 4a).139
 | ||
| Fig. 4 Diverse peptide designs with inbuilt structural- and functional-encoded motifs. (a) Cu(II)-Binding redox-triggered switchable peptide. α-Helical peptide converts into a β-sheet assembly upon addition of cupric ions, while reduction of metal cations promotes reversal of conformation [adapted with permission from ref. 139].139 (b) Antifouling peptide-SAMs for biosensing applications. Schematic showing an all-in-one branched peptide including a anchoring, doping, linking, antifouling, and human IgG recognition domain for IgG quantification in serum samples [adapted with permission from ref. 144].144 (c) DNA target biosensing using chimeric SBPs assembled onto Au surfaces. Schematic shows the biofunctionalisation of gold substrates with a peptide-nucleic acid (PNA) chimera for further nucleic acid detection via complementary probes [adapted with permission from ref. 68. Copyright 2022, American Chemical Society].68 | ||
2.3.2.6 Antifouling and chimeric peptides. Antifouling surfaces have been developed to abolish or diminish nonspecific protein adsorption, mainly for cell adhesion studies, antibacterial surfaces and sensing applications.
While PEG has been widely utilised to create such type of antifouling surfaces, zwitterionic peptides can also provide antifouling properties, in addition to tuneable sequence and structure (α-helix or β-sheet). Zwitterionic peptides contain alternating positively (H, K, R,) and negatively charged (D, E) residues displayed in diverse orders (Fig. 4b).140 Being neutral and hydrophilic, they possess strong affinity for water, forming a surrounding water layer that prevents interactions with protein molecules. They are now seen as the next-generation of “stealth” molecules and their sequence can be rationally designed or generated by peptide computational design.141 However, not all antifouling peptide-SAMs contain charged residues. For example, the work of Noguchi ad co-workers on oligo-proline SAMs demonstrated the promising capabilities of this non-ionic antifouling peptides for the development of vascular devices with ultra-low fouling properties, including reduced thrombogenic response, and low protein and cell adhesion.142,143Antifouling peptides have been used in combination with other peptide monolayers (mixed SAMs) for improving specificity in biosensing, and preventing biofouling formation (Table 2, peptide-SAMs antibiofouling surfaces).
Chimeric peptide-DNA constructs have been reported in the context of biosensing applications. Particularly those involving SBPs, due to their modularity, self-organisation, ease of surface modification, and their control and optimisation over surface packing density.68 For instance, Lee and collaborators tethered the Au-binding peptide WAGAKRLVLRRE as part of chimeric peptide-nucleic acid biomolecules for immobilising nucleic acid probes to further detect DNA in aqueous media (Fig. 4c).67 The conformation adopted by this peptide sequence allowed strong anchoring of the chimeric molecule onto Au substrates, acted as a molecular erector for the proper display of the immobilised nucleic probes for target DNA recognition, and exhibited viscoelastic changes upon probe immobilisation, becoming more flexible and plausibly contributing to further DNA capture and detection from media (Table 3, peptide-SAMs for biosensing of nucleic acids).
3. Fabrication of peptide-based SAMs
Generally speaking, the structure and quality of SAMs are affected by factors such as surface roughness, concentration and purity of the self-assembled molecular building blocks, incubation/immersion time, choice of solvent, and temperature.4,31 Once in contact with the solid substrate the mechanism of SAMs formation includes two steps: the rapid and strong chemisorption between anchoring groups and substrates, and the subsequent slow reassembly due to van der Waals interactions between linker chains.31 These molecular events also take place in peptide-based SAMs, and a number of strategies have been developed in order to improve spatiotemporal display of peptide signals, including the formation of patterns,6 gradients, and dynamic surfaces.27,102,103,114,1743.1 Peptide layer deposition and pattern forming
Substrate immersion or incubation represents the amplest fabrication method for peptide-based SAMs. This technique is compatible with other methods, such as spotting for peptide array formation (Fig. 5a) and μCP, which can also be found in the literature. μCP is a soft non-photolithographic method which routinely renders SAMs containing regions terminated by different chemical functionalities with submicron lateral lengths.175 It consists on transferring an ink solution from a patterned elastomeric mould, or stamp, to a substrate by contact with its surface.176,177 The combination of μCP and SAMs is beneficial for obtaining good control over the surface chemistry and minimising defects due to peptide molecular self-organisation processes.175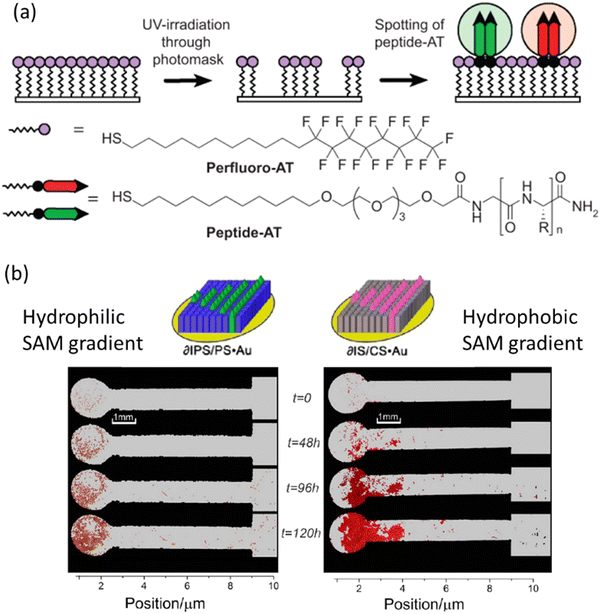 | ||
| Fig. 5 Fabrication methods available for precise spatial control of peptide-displaying SAMs. (a) Fabrication of peptide-based SAMs arrays via a two-step process: a pure perfluoro-AT SAM is photopatterned, followed by spotting of peptide-ATs onto the exposed areas to form peptide-terminated SAM array elements [adapted with permission from ref. 26. Copyright 2007. American Chemical Society].26 (b) Fabrication of peptide-SAM gradient surfaces exhibiting motogenic IGDQ-containing molecules: IPS (bearing an undecyl-PEG4 hydrophilic linker, green) and IS (bearing a hydrophobic octyl linker, pink) and time-dependent (t = 0–120 h) imaging of whole-population migration of metastatic breast cancer cells across the gradients (blue and grey blocks correspond to filler thiols) [adapted with permission from ref. 103. Copyright 2017. American Chemical Society].103 | ||
In the context of peptide-SAMs, μCP has been used to develop patterned surfaces for the controlled spatial presentation of HA via Pep-1-based SAMs on Au substrates (Fig. 3b–d),31,33 allowing precise spatial presentation of the HA, and combined with fluorescent and surface analysis techniques allowed for an adequate characterisation of the system and HUVECs response to it. Examples of reactive μCP can be found in peptide-SAMs systems,42 as patterns of SAMs generated by μCP provide a method for attaching cells on surfaces in a controlled fashion, thus indicating the potential of HA-patterned surfaces based on peptide-SAMs for cell sorting applications (Fig. 3c).4
Noteworthy, peptide-based SAMs used for biosensing applications tend to involve multistep fabrication processes, involving electrode modifications, passivation, and deposition of conducting polymers or nanostructures before depositing the peptide layer (Table 3, peptide-SAMs for biosensing applications).
3.2 Gradient and dynamic surfaces: controlled spatiotemporal display
As described in previous sections, homogenous and patterned peptide-SAMs can be formed to display peptide functionalities with some degree of spatial control, but they are based on non-reversible covalent bonds. These surfaces have been useful to study and control spatially defined cell adhesion and growth. However, natural ECMs are dynamic, being constantly remodelled by enzymes and its components subjected to post-translational modifications. In addition, soluble molecules like growth factors are secreted by cells in a time-regulated fashion necessary for coordinated signalling networks. In the human body, cells migrate in response to gradients of such soluble chemoattractants or to bound molecules. Replicating these temporally controlled events in 2D surfaces would be highly relevant to study cell migration in the context of cancer, for example, to identify key molecules involved in cancer invasion. Gradient surfaces display a steady variation of one or more physicochemical property in space which can also change over time.17Bonifazi and co-workers27,103 developed uniform peptide monolayers based on the IGD epitope of FN and gradients through one-step immersion and gradual immersion using a linear-motion drive,178 respectively. Gradients were formed through the co-assembly of thiol-containing IGDQ peptides anchored onto linkers of different length and hydrophilicity (e.g., hydrophilic undecyl-tetraethylene glycol (EG4) chain and hydrophobic n-octanethiol) and corresponding linkers as backfiller molecules. The approach enabled tuning of the surface polarity and variation of the peptide density in the SAMs. The IGDQ and GRGD SAM surfaces were then applied to study the migratory behaviour of metastatic breast cancer cells, revealing distinct cell subpopulations, with a “stationary” or a “migratory” phenotype, depending on the surface, and suggesting the use of IGDQ SAM gradients for the separation and characterisation of the highly motile cells to further study their metastatic phenotype.27 These investigations reveal that exists a mutual interaction among IGDQ-peptides, the surface fillers, and the substrate, controlling the structural features of the ECM-mimicking peptide-SAMs and dictating their motogenic potential (Fig. 5b).103
Building dynamic peptide-SAMs that change their properties controllably in time requires the inclusion of chemical moieties that are sensitive to specific stimuli. Light, electrochemical and enzyme activation of SAMs have been exploited to generate desired and controlled responses, either in cell culture or in biosensing applications. The use of electrical potential to generate oxidation and reduction reactions has been reported in SAMs functionalised with hydroquinone (HQ) and the oxidised reactive benzoquinone (BQ) pair.179 These dynamic surfaces can react with molecules functionalised either with ω-terminated-cyclopentadiene (Cp) or thiol groups via Diels–Alder reactions triggered by the application of low electrical potential. However, the reported SAMs did not integrate peptides in their composition. Zhao and co-workers reported the use electrical potentials to induce reversible conformation transitions (from linear to cyclic) in RGD-containing peptides and thus the ability to modulated cell adhesion and migration.113 The SAMs displayed RGD peptides containing a positively charged end group at distal end (a quaternary ammonium group, –NMe3). The application of a positive potential to the surface originated electrostatic repulsion of the positively charged distal end of the RGD-NMe3, encouraging a linear conformation. By contrast, when a negative potential is applied, electrostatic attraction flipped the positively charged distal end of the RGD-NMe3 towards the surface, causing bending of the RGD peptide backbone and resulting in a cyclic conformation.
An example of photoactivated peptide-SAMs employed SAMs of PEG-thiols functionalised with cyclic RGD modified with a 3-(4,5-dimethoxy-2-nitrophenyl)-2-butyl ester (DMNPB) (c[RGD(DMNPB)fK]) immobilised onto Au-coated surfaces (glass slides and quartz sensors) for triggering integrin binding and cell attachment onto these surfaces and for detection of these early events in cell culture experiments via QCM-D, respectively.109,180 DMNPB is a photolabile group that was attached via the COOH group of the aspartic acid (D) residue in the RGD peptide, as its acts as a ligand for one of the two bi-valent cations of the integrin αVβ3binding-site involved in the RGD–integrin. UV (at 365 nm) irradiation allows removal of the DMNPB cage and restoring the ability of cells to bind the RGD ligand (Fig. 6a). This photoactivable peptide SAM can be employed to create RGD patterns or gradients using a mask or a laser at different intensities. This report demonstrated that different cell types (HUVECs, NIH3T3 and CAL-72) respond differently to RGD ligand, and the QCM-D signal could be correlated with integrin levels expressed by cells. The results provide valuable information for biomaterial engineering and suggest the possibility for biosensing applications, by discriminating between abnormal and normal cells based on their integrin expression level.
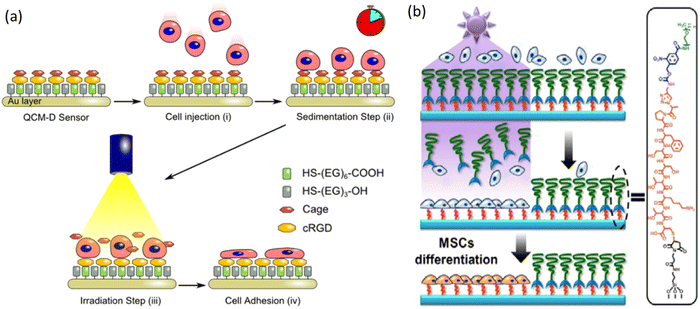 | ||
| Fig. 6 Fabrication methods for precise spatiotemporal control of peptide-displaying SAMs. (a) Synchronised cell adhesion and spreading based on photo-activatable adhesive peptide ligand c[RGD(DMNPB)fK] and the products generated upon photolysis [adapted from ref. 109 with permission from Springer Nature].109 (b) UV light triggered detachment of PEG shell via DMNPB cleaving, promoting MSCs osteogenic differentiation on functionalised quartz substrates displaying PFSSTKTC peptide-SAMs [adapted with permission from ref. 28. Copyright 2015. American Chemical Society].28 | ||
The use of photoactivable SAMs has also been reported in the context of stem cell differentiation.28 For instance, the PFSSTKT peptide (based on a phage display derived peptide) was shown to induce osteogenic differentiation of mesenchymal stem cells (MSCs) when displayed on peptide-SAMs. The peptide was synthesised with a Cys residue at the C-terminus and an azide group at the N-terminus (N3-PFSSTKT-C) to enable its tethering to maleimide modified quartz substrates (via a Michael addition reaction) and subsequent conjugation of alkynylated o-nitrobenzyl PEG5000 by click reaction at the N-terminus where the o-nitrobenzyl group acts as a photocleavable linker. The presence of the PEG shell prevents cell adhesion on the substrate, but local UV irradiation removes the PEG shell and expose PFSSTKT peptide and promotes the site-specific osteogenic differentiation of cultured MSCs (Fig. 6b).28
An elegant design of dynamic peptide-SAMs was reported by Huskens and co-workers using fluorogenic reactive SAMs for the signalled immobilisation of molecular thiols under physiological conditions, enabling direct visualisation of the localisation of the functionalised surface (Fig. 2b). Coumarin was used as fluorogenic probe and immobilised on an azide monolayer on glass through an alkyne moiety and reactive microcontact printing (μCP) with a Huisgen 1,3-dipolar cycloaddition.181 C-GRDS and M-GRDS (control sequence) peptides were applied on the fluorogenic reactive SAMs and only the C-GRDS peptide showed strong enhancement of fluorescence. Cell culture on SAMs with pre-immobilised C-RGDS showed co-localisation of mouse myoblast cells on fluorescently visualised regions with the peptide, especially when backfilled with hexa(ethylene glycol) (EG6).
Although the use of peptide sequences sensitive to enzyme activities has been largely explored in biosensing applications, it has not been reported for the display of peptide ligands overtime. Dynamic SAMs have been developed using dynamic chemistry for the selective and spatial-temporal control of the surface functionalisation. Guiseppone and collaborators described the use of dynamic covalent chemistry to generate functional gradients of pH-sensitive SAMs.182 Immersion of model surfaces (quartz, silicon) with immobilised aldehydes into solutions containing functional amines of various pKa values, enable the formation of chemical gradients over space and time upon removal from the solution at constant speed while the solution pH was varied in a time-dependent manner. The selective and reversible immobilisation of proteins on the aldehyde-coated surfaces was attempted through their lateral basic residues.
The development of dynamic, multi-responsive, adaptive and reversible surfaces is an important direction in SAMs research. Recently, reversible SAMs (rSAMs) with tuneable surface dynamics were reported for controlling cell adhesion using a supramolecular-based approach.114 Peptide-based SAMs were prepared on Au using oxoacid-terminated thiols in HEPES buffer (pH 8) containing various mole fractions of ω-(ethyleneglycol)2–4- and ω-(GRGDS)-, α-benzamidino bolaamphiphiles. Electrostatic interactions, formed between the amidine-functionalized amphiphiles and the oxoacid-displaying SAMs, enable molecular exchange, through the addition of inert filler amphiphiles to the RGD-functionalized rSAMs, for example, and thus modulation of the cell adhesion behaviour. This approach represents a breakthrough on dynamic SAMs surfaces, allowing the presentation of mobile bioactive ligands, capable of reversing cell adhesion in a non-invasive manner for cell harvesting, and the possibility of restoring surface functionality on demand.
4. Characterisation of peptide-based SAMs
Characterisation of peptide-SAMs can be done using several surface techniques, namely optical contact angle, X-ray photoelectron spectroscopy (XPS), ellipsometry, surface plasmon resonance (SPR), quartz crystal microbalance with dissipation (QCM-D), as well as routine scanning probe microscopy, vibrational spectroscopy, and electrochemical techniques. This section aims to describe their working principles and capabilities, referring the reader to specialised resources when necessary.4.1 Contact angle
Contact angle measurements are used to directly quantify the wettability of a surface and, indirectly, to calculate surface energy. In addition, it can also be applied to indirectly evaluate surface's roughness, heterogeneity, contamination, and molecular mobility.183 The contact angle (θ) is the angle at which the liquid drop encounters the solid surface (Fig. 7a). It is defined as the equilibrium of the drop under the action of three interfacial tensions: solid–liquid (γSL), solid–vapor (γSV), and liquid–vapor (γLV). The basic relationship describing this equilibrium is described by the Young's equation (eqn (1)):γSV = γSl + γlv![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ θ | (1) |
 | ||
| Fig. 7 Peptide-SAMs characterisation methods. (a) Schematic representation of an optical contact angle apparatus (not to scale) showing the sessile drop method. A drop of water is vertically positioned by a syringe onto sample surface, photographed by a high-resolution camera after which the contact angle (θ) is determined, via image analysis software by the angle formed between the tangent of the liquid drop and the solid surface [adapted with permission from ref. 183].183 (b) Schematic operation of a monochromatised XPS apparatus [adapted with permission from ref. 183].183 (c) Schematic showing the principle of ellipsometry, where Δ is the difference between the phase changes of the p and s components of the reflected light, and Ψ is an angle which tangent gives the ratio of amplitude changes for the p and s components of the reflective light [adapted with permission from ref. 188].188 | ||
SAMs displaying polar terminal functional groups, such as carboxylic acids and hydroxyls, are wetted by water. Those presenting non-polar organic groups, as methyl and trifluoromethyl, are hydrophobic and emerge dry from water. Moreover, alterations in surface wettability can be used to follow peptide bioconjugation on SAMs.91 The wettability of a surface can be precisely controlled using SAMs with different terminal functional groups.186,187 For instance, precise hydrophilic and hydrophobic gradient peptide-SAMs have been characterised by contact angle measurements.103 However, the technique presents limitations when it comes to track sequential binding events that do not significantly change surface hydrophilicity, like peptide-SAMs and HA.31
4.2 X-ray photoelectron spectroscopy (XPS)
XPS is a quantitative technique for surface chemical analysis.189 It measures the elemental composition and the chemical state of the atoms (except hydrogen and helium) within a material surface. This technique is based on the photoelectric effect. XPS measures the kinetic energy (keV) of the inner shell electrons ejected when the surface is irradiated with an X-ray beam in ultra-high vacuum. This process is described by eqn (2):| BE = hν − KE | (2) |
XPS is useful to analyse SAMs composition and peptide bioconjugation due to the appearance (or increase) of nitrogen content.128 Moreover, the use of different take-off angles allows the calculation of how deeply certain elements are positioned with respect to the monolayer surface.190 For SAMs of ATs on Au, sulphur high-resolution scans can provide information about bound and unbound thiol in a monolayer.191 They can also indicate if the thiolate head group in a monolayer was oxidised to a sulphonate group (RSO3−),46 this oxidation is also essential to produce patterned surfaces, where the UV exposed oxidised AT could be easily removed and replaced by other molecules.
4.3 Ellipsometry
Ellipsometry constitutes a non-destructive optical technique useful to characterise the thickness of thin organic/inorganic coatings (nanometer range, <100 nm) onto a reflective substrate.184,192 The most used reflective substrates are optically polished silicon wafers or flat wafers coated by sputtering with an optically reflecting metal film, such as Au, Ti, among others. Both monochromatic ellipsometry and spectroscopic ellipsometry are typically used to determine the thickness of the layers.184,193 Using a liquid cell, this technique can be used to measure the adsorption/desorption of molecules (e.g., peptides or proteins) onto a reflective surface in real-time and in a label-free environment.91,128,188The fundamental principles behind ellipsometry are based on the alteration of the polarisation state of light after reflection from a surface (Fig. 7c).194 The changes are different for both components of light polarised: parallel (p) and perpendicular (s) to the plane of incidence. Deposition of a thin film (like peptide-based SAMs) onto a reflective surface (like Au) shift the phase difference (Δ = §p − §s) and the amplitude ratio (tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ψ= rp/rs) of the reflected light. These coefficients depend on the wavelength, the angle of incidence, and the optical properties of the reflecting system.184
ψ= rp/rs) of the reflected light. These coefficients depend on the wavelength, the angle of incidence, and the optical properties of the reflecting system.184
4.4 Surface plasmon resonance (SPR)
SPR is an optical technique used to quantify molecular interactions in real time on a metal thin film due to alterations on its refractive index.195–197 SPR technique can quantify the binding of a soluble molecule (analyte) to a ligand bound on the metal thin film with a detection limit of around 0.5 ng cm−2.198 The metal thin film, usually Au, must be deposited on a glass prism with a thickness of ca. 40 nm. When the p-polarised light is reflected from the backside of the glass prism (Au-coated), an electromagnetic field component of the light (the evanescent wave) penetrates the metal layer. This evanescent wave is able to couple with the free oscillating electrons (plasmons) in the metal film at the specific angle of incidence (SPR angle or resonance angle). The SPR angle shift is proportional to the surface concentration of the adsorbed molecule.199 A schematic illustration of the operation of the SPR technique is shown in Fig. 8a.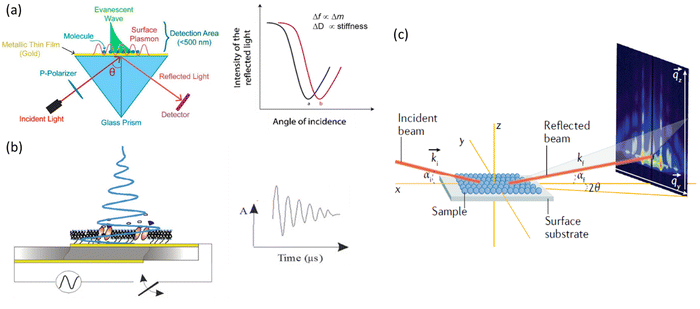 | ||
| Fig. 8 Peptide-SAMs characterisation methods. (a) Experimental set-up of a typical SPR experiment, depicting a dip in the intensity of the reflected light after surface plasmons excitation and an angular shift from trace a (black) to b (red) due to a change in the Au film refractive index (RI) [adapted from ref. 199 with permission from Multidisciplinary Digital Publishing Institute].199 (b) Schematic representation of a QCM-D. An alternating RF voltage is applied to a quartz crystal coated with metal (e.g., Au), the observed change in frequency (Δf) is proportional to the mass of adsorbed biomolecule species, whereas the change in dissipation (ΔD) is a function of the viscoelastic properties of the adsorbed layer [adapted with permission from ref. 200].200 (c) Schematic of a GISAXS experimental setup, showing the incident angle (αi) and the scattering angles along the horizontal (2θ) and vertical (αf) directions [adapted from ref. 201 with permission from Springer Nature].201 | ||
4.5 Quartz crystal microbalance with dissipation (QCM-D)
QCM-D represents a non-destructive technique able to quantify molecule (proteins, glycans and more) adsorption/binding with high sensitivity (in the nanogram range), under dynamic conditions (temperature and flow) and in real time.91,129,188,202Fig. 8b shows a schematic representation of a QCM-D. It uses an oscillating piezoelectric quartz crystal sensor at specific resonance frequencies to measure changes in frequency (Δf) and dissipation (ΔD).203 Changes in Δf result in ultra-sensitive determinations of absorbed or desorbed mass that can be quantified, while simultaneous information on the viscoelastic properties of the adsorbed molecules is given by ΔD: if the adsorbed mass is viscous and soft, it will not synchronise to the sensor oscillation leading to dissipation.204 For data modelling, the Sauerbrey equation (eqn (3)) or the Voigt model (eqn (4)) may be applied, with the choice being dependent on the dissipation values: for systems where ΔD/Δf is lower than 4 × 10−6 Hz−1 it is assumed that the adsorbed/bound film is rigid with no internal loss of energy, which is translated in low values of dissipation and the Sauerbrey equation remains valid. However, the Voigt model must be applied if the dissipation shifts are compatible with a viscoelastic layer, since this model takes in account the energy losses in the system (i.e., dissipation), correcting the deviation introduced by this factor.129,205,206| Δm = (Δf/n) × C | (3) |
| G* = GJ + GJJ = μ1 + i2πfη1 | (4) |
4.6 Electrochemical techniques
Measurements of electron transfer (ET) rates are routinely performed via electrochemical techniques,49,53 such as cyclic voltammetry, chronoamperometry, and photocurrent generation experiments,56 while electrochemical properties of modified electrodes for biosensing applications can be studied using electrochemical impedance spectroscopy (EIS).87,207,208 Tables 2 and 3 present numerous studies supported by these techniques.4.7 Infrared reflection–absorption spectroscopy (IRRAS)
A variety of vibrational spectroscopy techniques have been applied to peptide-SAMs characterisation. For instance, Fourier Transform Infrared Spectroscopy (FT-IR) has allowed to determine the tilt angle for helix-forming peptides on SAMs.53IRRAS represents another especially useful spectral technique to study structure and orientation of SAMs deposited on metallic, flat and reflective substrates, and are based on the frequencies and intensities of molecular vibrations.209 Radiation is used at grazing angles to maximize surface sensitivity. Since the incident and reflected component of the polarised light perpendicular to the plane of incidence combines to form a zero electric field at the surface, only molecular vibrations with dynamic dipole moment perpendicular to the surface will interact with light. This is particularly important to determine orientation relative to surface of the adsorbate chains in SAMs.210 Moreover, the C–H stretching regions can give information about the packing and orientation relative to surface of the adsorbate chains when compared with spectra of randomly oriented molecules in liquid/powder (KBr pellet).
IRRAS was recently used for the characterization of monolayers (packing density and conformation) produced by polypeptides composed of Gly and Cys on gold or silver substrates (GlynCys/Au(Ag), n = 1–9), based on the peptide amide I (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration) and amide II (N–H bending vibration) absorption bands (Table 2, peptide-SAMs for molecular electronics).158 The deconvolution of the amide bands is sensitive to both secondary structure and environmental factors of peptide-based SAMs. This technique was used to evaluate the secondary structure and the stability in aqueous solution of SAMs prepared with zwitterionic antifouling peptides composed of alternating K and E residues.211
O stretching vibration) and amide II (N–H bending vibration) absorption bands (Table 2, peptide-SAMs for molecular electronics).158 The deconvolution of the amide bands is sensitive to both secondary structure and environmental factors of peptide-based SAMs. This technique was used to evaluate the secondary structure and the stability in aqueous solution of SAMs prepared with zwitterionic antifouling peptides composed of alternating K and E residues.211
4.8 Grazing incidence small-angle X-ray scattering (GISAXS)
GISAXS is a powerful technique for the structural characterization (shape, size, and arrangement of molecules at the surface) of thin films (1–100 nm) deposited on flat substrates, and is an attractive tool for the characterization of peptide-based SAMs. This technique is based on the probability of each atom to scatter an incident X-ray photon at a particular energy. These scattering events depend of the X-ray wavelength, the absorption and the scattering properties of the sample, and the tilt angle of the incident beam regarding the substrate.201Fig. 8c shows a schematic view of the GISAXS apparatus. The incident X-ray beam reaches the nanofilm surface in a very small angle (typically <1°) and the scattered X-rays are recorded by a two-dimensional (2D) detector. GISAXS is based on the measurement of the intensity, I, of scattered X-rays as a function of scattering angle and its pattern comprises vertical (out-of-plane, qz) or lateral (in plane, qy) scattering intensities. Although GISAXS has not been applied for the characterization of peptide-SAMs, it was recently used to evaluate the structural properties of a lipid/AMP self-assembled layer on silicon substrate, based on different ratios of the glycerol monooleate (lipid) and the LL-37 AMP.2124.9 Scanning probe microscopy
The structural characterization of SAMs at atomic resolution often requires surface-sensitive techniques such as scanning tunneling microscopy (STM) and atomic force microscopy (AFM).STM principle of operation is based on the quantum tunneling effect. It uses a sharp metal tip (one electrode of the tunnel junction) placed close (0.3–1 nm) to the surface to be investigated, by applying an electrical voltage to the tip or to the sample (10 mV–1 V), the surface can be sensed/imaged.213 The STM is versatile enough to be used in a wide range of environments, from ultrahigh vacuum (UHV) to ambient air, water, and other liquids or gases, and temperatures down to millikelvin and as high as more than 1000 K. STM is distinguished from most other surface characterization methods by its unique capacity to probe the topographical and electrical features of plain surfaces with high spatial resolution, down to atomic scale.213 For instance, Gatto and co-workers used STM to characterize the topography of SAMs formed by a hexapeptide (including five Aib residues and a Trp unit functionalised at the N-terminus with a lipoic group for binding to an Au substrate).23
However, STM can only be applied to study surfaces that are electrically conductive. As such, AFM was later developed and is one of the foremost tools for surface characterization (topographic and force measurements) on the micro- to nanoscale of either electrically conductive or insulating samples.214 AFM uses a sharp tip (<10 nm in diameter) with the desired geometry (e.g., triangular, round, among others) that is attached to a flexible cantilever. This cantilever acts as the force sensor to scan the surface of a sample, recording the interaction between the tip and the sample to acquire images/perform force measurements (pN range) in real time.215
Compared to other microscopic techniques, such as optical microscopy and transmission electron microscopy, AFM has several advantages, including high spatial resolution (sub-nanometer laterally and sub-angstrom vertically), easy and minimal sample preparation, the ability to perform analysis at ambient conditions but also at different temperatures or in solution.216 Moreover, AFM can be coupled to other techniques to allow further in-depth characterization, namely with infrared spectroscope (AFM-IR) and with in situ high-temperature grazing incidence X-ray diffraction (GIWAXD). Recently, Wang and collaborators studied the melt recrystallization behaviour of nanoconfined polypeptoid films prepared on silicon substrates using GIWAXD in conjunction with AFM.217
5. Biomedical applications of peptide-based SAMs
This section provides a discussion on the main outcomes elicited by selected peptide-based SAMs use for cell culture, tissue engineering and regenerative medicine,42 nanoelectronics,56,131 protein folding and peptide aggregation,218,219 biosensing,220 antifouling,170,172,221 and antimicrobial129 and osteo-integrative surface applications.137,1495.1 Translational platforms for tissue engineering and regenerative medicine applications
As shown in Table 1, peptide-based SAMs have played a key role as translational platforms for the identification of biofunctionalities targeted for tissue engineering and regenerative medicine applications. They have been employed extensively in cell culture applications involving mammalian cells (including MSCs, HUVECs, ADSCs, human embryonic stem cells, kidney cells, Schwann cells, hippocampal cells, breast cancer and mammary gland cells, as well as fibroblasts and melanoma cells) for a number of purposes, as not only they allow to mimic the native ECM,32,222 but they can also provide insights into protein binding,4 cell adhesion, migration and differentiation,28,35,76,223 as well as contribute to elucidate the role of biomacromolecules in cancer progression and other diseases. For instance, Ramasubramanian and co-workers reported lately on the de novo high-throughput discovery of targeted peptide surfaces to promote in vitro self-renewal of human pluripotent stem cells (hPSC) and induced pluripotent stem cells (iPSC). The study describes several peptide-SAM agonists for α6-integrin (a laminin receptor and pluripotency regulator) that were prepared using AT-linkers on glass substrates and polystyrene microchips (PillarChips, Table 1, peptide-SAMs binding cell membrane proteins).112Another example is the HA-binding Pep-1, which has been used as part of a peptide-based SAM for unmodified HA immobilisation. μCP allowed to restrict the presentation of HA to specific regions, thereby creating patterned surfaces to examine in vitro adhesion and migration of human oral squamous cell carcinoma (SCC) derived immortalised keratinocyte CA1 and LUC4 cell lines.33 As LUC4 cells adhere to HA immobilised on surfaces, they remain attached and displayed constant shifting to and from an epithelial-to-mesenchymal transition phenotype, suggesting the use of the biomimetic surfaces as a platform for the development of cell sorting devices. Patterned Pep-1 SAMs were used to present HA in discrete regions, finding that low molecular weight HA improved the adhesion and stimulated HUVECs migration when cultured atop HA-presenting SAMs. This study provides insight into the use of SAMs for the controlled presentation of HA with defined molecular weight in cultures of HUVECs in order to better understand their functions and interactions with the ECM.31
Peptide-SAMs also generate results that can be translated as part of anti-angiogenic therapies, and as part of implantable endovascular devices, such as artificial vascular grafts and stents, to promote rapid endothelialisation and improve their performance.34 Lastly, peptide-SAMs have been used to drive MSCs differentiation224 by targeting different molecules, including GFs,42 BFP-1,41 as well as linear and cyclic BMHP1.35
Peptide-SAMs have also contributed to elucidate biomechanical aspects of cellular interactions with the ECM. A cyclic-RGDfK displayed as SAM was used to determine the cellular force applied on αvβ3 integrins during early stages of cell adhesion via QCM studies, demonstrating the potential of peptide-SAMs to contribute to rather fundamental aspects behind cell adhesion, spreading, and migration.116
5.2 Biosensing
Biological sensors, especially point-of-care sensing devices, have been pivotal for the early diagnosis of numerous diseases, as well as for the routine management of certain conditions (e.g., therapy monitoring). Different biomarkers, mainly enzymes, nucleic acids and antibodies, have been therefore used as target analytes in biosensing. Table 3 shows a larger number of biosensing applications utilising peptide-SAMs as the key element (ability to bind to or be hydrolysed/modified by the target analyte) in the biosensor design.Biosensing applications have benefited immensely from the progress made on the fabrication of SAMs, particularly on mixed SAMs, SAM gradients and dynamic SAMs. In addition, because many binding events involve protein–protein interactions, peptide-SAMs are also suitable choices for detection applications. In fact, highly sensitive detection of analytes in complex biological media (e.g., serum samples) requires surfaces able to reduce nonspecific protein adsorption to eliminate false positive reactions. Not surprisingly, zwitterionic peptides have been integrated in SAMs, either as mixed monolayers with the peptide ligand or as an integral part of the peptide monolayer (all-in-one peptide SAM, Fig. 4b). The design first reported by Nowinski et al.,140 attaching a linker made of four proline residues to the C-terminus of a nonfouling peptide (EKEKEKE-PPPP-C, Table 2, peptide-SAMs as antifouling surfaces) has been widely applied in the construction of biosensors (Table 3, peptide-SAMs for biosensing applications). The rigid, and hydrophobic linker enables the formation of well-ordered peptide-SAMs with high surface density.
The original zwitterionic peptide was shown to outperform PEG monolayers and has been applied as linear chain or in a branched fashion (see examples in Tables 2 and 3). In addition to provide antifouling property, the peptide ligand/substrate is typically conjugated with a redox tag, such as Fc or MB (Fig. 9b),225 to generate an electrochemical signal, upon binding or cleavage, to enable the detection of the target analyte by chronopotentiometry (CA) and voltammetry techniques (cyclic voltammetry (CV), differential pulse voltammetry (DPV), and square wave voltammetry (SWV)) (Fig. 9a).226 Nonetheless, label-free detection of trypsin using QCM has also been reported.227
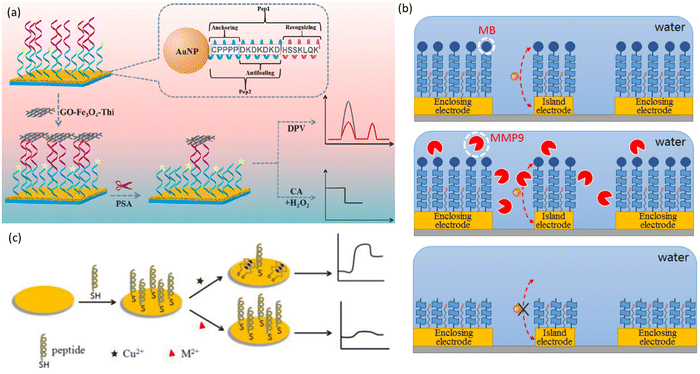 | ||
| Fig. 9 Peptide-SAMs for biosensing of enzymes, biomarkers and metal ions. (a) Working principle of an antifouling electrochemical biosensor for prostate-specific antigen (PSA) based on two antifouling peptides functionalised with either a graphene oxide–Fe3O4–thionine (GO–Fe3O4–Thi) probe (Pep1) or an internal reference ferrocene (Fc) probe (Pep2). Pep1 is recognised and cut by PSA, while Pep2 produced a differential pulse voltammetry (DPV) signal that remained constant and was independent of the presence of the PSA analyte, and also decreased the chronoamperometry (CA) signal of the reduction of H2O2 electrocatalysed by GO–Fe3O4 [adapted with permission from ref. 226. Copyright 2019. American Chemical Society].226 (b) Sensing mechanisms for the detection of MMP-9 via peptide substrate tagging with methylene blue (MB), which acts as a redox reporter, including: tunnelling of electrons in the presence of MB-peptide on Au electrode, peptide cleavage reaction of the MB-peptide with MMP-9, and decrease in the current of electron tunnelling after peptide cleavage, respectively [adapted with permission from ref. 225].225 (c) Experimental scheme for the detection of Cu(II) ions based on conformational change of peptide by SPR [adapted from ref. 228 with permission from the Royal Society of Chemistry].228 | ||
The integration of peptide substrates into microarrays like SAMs has attracted large interest, either for the detection of enzymes involved in numerous diseases (biosensing applications) or for determining enzyme substrate specificity for the identification of their natural substrates. Table 3 shows a clear predominance of enzymes as target analytes, in particular of proteolytic enzymes. Detection of MMP activity is typically performed using peptide-SAMs as substrates for enzyme cleavage at specific peptide bonds, but alternative approaches have also been proposed. For example, MMP-14 detection was also made by EIS and using SAMs of peptides that inhibit homodimerisation or heterodimerisation of MMP-14 (Table 3, peptide-SAMS for biosensing of enzyme targets).208 Prostate-specific antigen (PSA) is an enzyme (3.4.21.77) normally produced by the glandular tissue of the prostate and elevated levels of PSA in the blood are associated to abnormal conditions, such as prostate cancer. It recognises the HSSKLQK peptide, a specific and efficient peptide substrate for assaying the proteolytic activity of PSA,229 and cleaves the S|S bond. Several sensors have been developed based on this working principle (Fig. 9a and Table 3, peptide-SAMS for antigen detection). Recently, Yu and co-workers reported a label-free method to detect and quantify caspase-3 via organic electrochemical transistors (OECT)230 (Table 3, peptide-SAMS for enzyme detection).
Peptide-based biosensors have also been developed for detection of antibodies and other disease-relevant proteins. Here, the sensing principle is based on the binding of the target protein by the recognition peptides derived from epitopes in binding partners. More recently, virus detection via peptide-SAMs has also been attempted. For example, an electrochemical biosensor for detection of the human norovirus, known to cause acute foodborne gastroenteritis outbreaks worldwide, was reported using virus-specific peptides identified by phage display (Table 3, peptide-SAMS for virus detection). Interestingly, a redox-triggered peptide with potential Cu2+ binding sites (carboxylic acids of glutamate residues) was applied for the sensitive detection of Cu2+ ions by SPR228 or colorimetric assay231 (Table 3, metal–ion binding peptide-SAMs). The peptide reversibly switches from helical conformation to a β-sheet aggregate by reduction and oxidation of copper (Fig. 4a and 9c). Copper(II) is involved in gene expression and enzymatic reactions, playing important roles in maintaining the nervous and immune systems, but it can become detrimental at elevated concentrations. Its detection in different media (blood, drinking water) is, therefore, broadly useful.
Nucleic acid biomarkers hold great potential as key indicators for the diagnosis and monitoring of diseases. In a recent work by Lee and collaborators, Au-binding peptide-nucleic acid (PNA) chimeric molecules were used to immobilise nucleic acid probes to subsequently detect nucleic acid targets in aqueous environments (Fig. 4c).68 The researchers used SPR and QCM-D analyses to demonstrate the sequential assembly of the biomolecular constructs and the subsequent detection of DNA antisense oligomers from media under μM concentration regimes. Overall, Table 3 shows the broad spectrum of enzymes, cell bound receptors, nucleic acids, peptides, viruses, proteins, antibodies and antigens whose detection can be achieved by peptide-based SAMs.
5.3 Molecular electronics
As SAMs can be formed onto (semi-)conductor or dielectric surfaces, and their surface chemistry can be tailored, SAM-based platforms represent attractive candidates for molecular electronics.277 SAMs formed by conformationally constrained peptides functionalised with redox-active groups have shown unique electronic conduction properties in terms of long-range and directional electron transfer.56 As helical peptides are well known to act as good mediators for long-range electron transfer (ET),49 SAMs displaying helix forming peptides have been extensively studied due to their possible integration into (bio)electronic devices.278Morphological and peptide-packing modes studies have been carried out. For instance, Higashi and co-workers studied the capturing mechanism of guest Fc-containing helix peptides by PLGA SAMs on Au substrates. Different packing densities were found as a function of adsorption rate and pH, and specific interactions with guest PLGAs containing a ferrocenyl group (at either the N- and C-termini) were investigated, guest helix PLGAs were captured through an antiparallel, side-by-side helix-macrodipole interactions.61
In another study, Venanzi and colleagues studied the structural organisation of the short hexapeptide A4WA bound to Au substrates via a lipoyl group (Table 2, peptide-SAMs for electron transfer), providing insights on the packing modes of the peptide layer. The researchers found a peculiar peptide array made of densely packed nanometrically ordered structures (originated by a vertical peptide monolayer coating the substrate) and also a flat dimeric peptide layer arranged horizontally to the metal surface. This particular array was rationalised in terms of 3D conformational constrains of the peptide chains, as well as its helix-forming capacity, thus contributing to understand the competition between peptide/surface interactions and the strength of lateral interchain interactions.38,157
Using analogous Fc-containing systems, Gatto and collaborators studied the efficiency to mediate electron transfer and photocurrent generation capabilities of two hexamer-displaying SAMs. Both peptides were linked to Au substrates, exposing either 2,2,6,6-tetramethyl-N-oxyl-4-amino-4-carboxylic acid (TOAC) or tryptophan residues. For the former, long-range electron transport was found, consistent with a super-exchange mechanism from TOAC to the Au surface through the peptide spacer. Tryptophan-bearing SAMs showed superior response, sensitivity and reproducibility compared to conventional Au electrodes.56
Structure–function studies regarding electron transfer in peptide-based SAMs have been carried out. It is reported that electron conduction efficiency of peptide bridges, although higher than that of alkyl chains, decreases exponentially with peptide length, a factor that might be influenced as well as by the type of helical structure (for instance, having the same number of amino acid residues, 310-helices extend over longer distances than α-helices).38
Electron transfer rate distance dependence has been studied by Bilewicz and collaborators. This group has reported a series of Fc-containing oligoglycine spacers of different length (Table 2, peptide-SAMs for electron transfer). The rates of electron transfer decreased rapidly with distance only for short chain oligoglycine derivatives, while longer bridges exhibited a weaker distance dependence. This variation was plausibly attributed to differences in the secondary structure of the peptide bridges, and the change of the electron transfer mechanism.52
In another study, Sek and collaborators reported on Au electrode modification by forming SAMs of a helix-forming polyalanine Fc-containing derivative (Table 2, peptide-SAMs for electron transfer), to which a Fc moiety was attached to the N-terminus while the peptide C-terminus was thiolated with a cystamine functionality, thus the peptide acting as a bridge between redox active Fc units and metal surface. This study found a directional dependence of electron transfer and suggests that the solvation of amino acid residues by solvent molecules – water in this case – may play a role in dampening of the polyalanine helix dipole (Fig. 10a).53
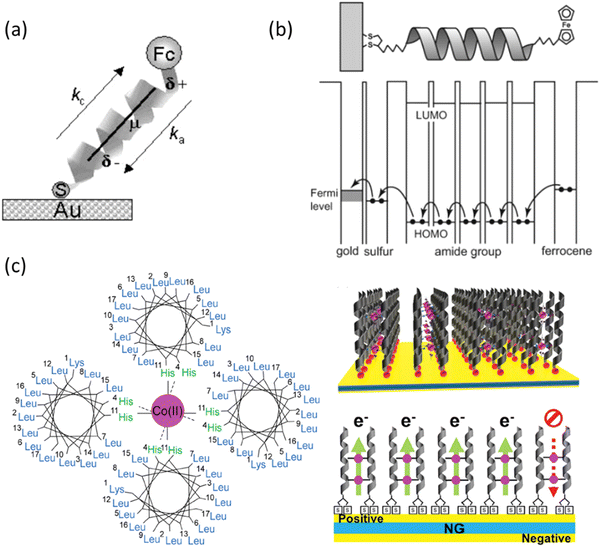 | ||
| Fig. 10 Peptide-SAMs for molecular electronics. (a) Schematics illustrating directional dependence of electron transfer (ET) through a helical polyalanine Fc-derivative [adapted with permission from ref. 53. Copyright 2005. American Chemical Society].53 (b) Energy diagram for the long-range electron transfer from a redox-active Fc moiety to Au substrate through the helical peptide by a hopping mechanism, using peptide amide groups as hopping sites [adapted with permission from ref. 157. Copyright 2005. American Chemical Society].157 (c) Piezoelectric-enhanced peptide-SAM formed by the binding of an α-helical peptide to Co(II) ions. Schematic diagrams of electron transfer of the Co(II)-peptide-SAMs when an input signal is applied. The arrows indicate the direction of the electron flow. [Adapted with permission from ref. 164].164 | ||
A hopping mechanism has been identified as responsible for electron transfer in helix peptide-based SAMs.131,157 Morita and collaborators prepared helical peptide-displaying SAMs carrying a Fc group either at the N- or C-termini, observing long-range electron transport over 4 nm accompanied by oxidation of the Fc tag. Electrochemical characterisation pointed that electron transport took place through an inelastic hopping mechanism with use of amide groups as hopping sites.131 Same as in the work of Watanabe and collaborators, who identified the same hopping mechanism, using the amide groups in the helical peptide as hopping sites on assembled Au SAMs of four kinds of Fc-containing helical octadecapeptides (Fig. 10b).157 This electron hopping mechanism is gaining increasing attention as it is present in biological system such as DNA double strands279 and its deeper understanding will play a role in the development of self-assembled nanoelectronics.280
Li and collaborators prepared an α-helical peptide-SAM (able to bind Co(II) ions via His residues) using a piezoelectric field produced by a BaTiO3-based nanogenerator (NG) (Fig. 10c). On the positive electrode of the bent NG, α-helical peptides led to electron transfer in the vertical direction through the SAM and along the macro-dipole moment, due to being able to unidirectionally enhance electron transfer and rectify an AC signal to a DC signal. This type of system holds great potential to be used in molecular memories, nonlinear molecular electronic devices, and biosensors.164
The remarkable work of Yasutomi and collaborators presents the fabrication of highly ordered bicomponent SAMs as part of a molecular photodiode system able to switch photocurrent direction by choosing the wavelength of irradiating light.60 This system includes two types of helical peptides with an alternating sequence of L- or D-leucine and α-aminoisobutyric acid carrying two different sensitisers: SSL16ECz carries a disulfide group at the N-terminus and a N-ethylcarbazolyl (ECz) group at the C-terminus; meanwhile, Rul16SS carries a tris(2,2′-bipyridine)ruthenium(II) complex (Ru group) at the N-terminus and a disulfide group at the C-terminus (Fig. 11). When both types of peptides were co-assembled on a gold substrate, the dipole moment of SSL16ECz oriented towards the Au surface, whereas that of Rul16SS oriented in the opposite direction, working as independent photodiodes with the help of their dipole moments.60
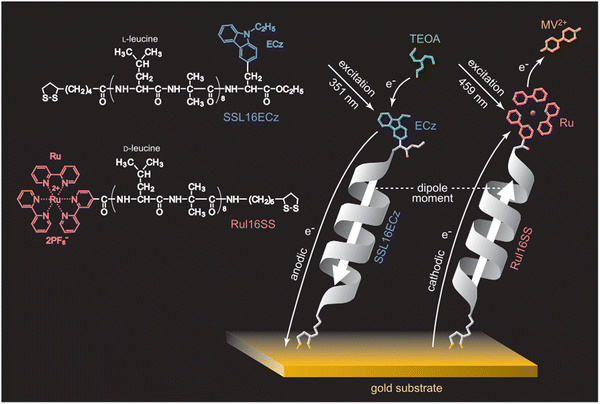 | ||
| Fig. 11 Peptide-SAMs for electron transport and molecular electronics. Preparation of a molecular photodiode system using two types of helical peptides carrying different chromophores (SSL16ECz and Rul16SS) and have different directions of dipole moments when immobilised onto Au substrates. The mixed peptide-SAM generated an anodic photocurrent when one of the two chromophores was photoexcited, which switched direction by selective chromophore excitation in presence of triethanolamine (TEOA), and/or 1-1′-dimethyl-4,4′-bypyridinium dichloride (methyl viologen; MV2+) [reproduced with permission from ref. 60].60 | ||
Altogether, these properties highlight the versatile role of peptide building blocks, whose rational design allow for their application as molecular wires,281 as part of biomolecular electronic devices,30,38,60 and even as part of potential platforms for the diagnosis of oxidative stress-related pathologies linked to reactive oxygen species (ROS).282
5.4 Antimicrobial surfaces and devices
This section is dedicated to peptide-based SAMs to assess antimicrobial and osteointegration performance of biomaterials surfaces. Biomaterials, such as cardiovascular grafts, urinary catheters, orthopaedic and dental implants, have improved the survival of patients as well as their quality of life. However, they remain as potential sources of infection, namely biomaterials-associated infections (BAI). This is a serious concern in modern healthcare, as bacteria become resistant to the most used antibiotics and few new ones have been discovered and commercialised in the last decades.283 Biomaterial coatings based on AMPs are exciting alternatives to fight BAI.In the context of urinary catheter-associated infections, which represent about 75% of hospital acquired infections, Monteiro and co-workers reported the use of a de novo designed AMP, Chain201D. Chain201D was successfully tethered to (1-mercapto-11-undecyl)-(tetra(ethylene glycol)) (EG4) terminated SAMs,8,128 which were able to bind and kill by contact a high percentage of adherent Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus), some of the most prevalent strains found in urinary catheter-associated infections. These results were obtained without any peptide modification (for chemoselective conjugation) and without the use of a spacer. In addition, increased amounts of immobilised AMP led to higher numbers of adhered/dead bacteria, showing a concentration-dependent behaviour, and demonstrating Chain201D's potential to develop antimicrobial urinary catheters.128
On the other hand, catheter-related blood stream infections are associated with the use of intravascular catheters and are mostly caused by bacteria present in the skin of patients or in the hospital environment, such as S. epidermidis.284 It has been described that a hybrid peptide combining cecropin and melittin peptide sequences (CM: KWKLFKKIGAVLKVLC) has better antimicrobial performance than the parent molecules.285,286
CM peptide was previously covalently immobilised onto glass and titanium surfaces by means of Au nanoparticles as a nanocoating for medical devices, and was described as active against different bacteria (E. coli, S. aureus, P. aeruginosa, and K. pneumoniae).146 To evaluate immobilised CM for intravascular catheters application, the peptide was synthesised with an extra Cys residue at its C-terminus and covalently immobilised onto medical grade polyurethane (the most common material used for intravascular devices) films. For that, polyurethane films were dip-coated with a layer of Au nanoparticles functionalised with –NH2 and –COOH terminated PEG spacers, as these improved CM peptide exposure/orientation during surface immobilisation and also avoided denaturation. S. epidermidis adhesion was significantly reduced (80%) accompanied by a 65% decrease on viable bacteria in comparison to the bare polyurethane surface. Moreover, the presence of human plasma did not impair the performance of the coating, showing the great potential of this CM-based strategy to prevent bacterial infection on polyurethane devices.147
Another C-terminus Cys-modified hybrid peptide, combining cecropin (1–8) and melittin (1–18) (KWKLFKKI-GIGAVLKVLTTGLPALIS-C) was studied when immobilised on a silica surface via a two step-reaction: first, an alkyne-terminated silane was used to grow the alkyne SAM, followed by preparation of a maleimide terminated surface by “clicking” the alkyne SAM with azido-PEG3-maleimide linker.148 These surfaces were able to kill substantial amounts of E. coli after 1 h. However, the number of bacteria killed diminished drastically with time, and no antimicrobial activity was observed after 5 days. This was attributed to the degradation of the silane, highlighting the short-term stability of this coating approach. It was postulated that the surface immobilised peptides may kill bacteria by charge interaction, instead of forming holes on the bacterial membrane (membrane disruption) as when free in solution. This may be related to the fact that immobilised AMPs (with no spacer such as a PEG linker as in the work of Querido et al.147) laydown on the surface instead of standing up and penetrating the bacterial cell membrane. This was also previously reported for surfaces with the immobilised alpha helical MSI-78 peptide (commercially known as Pexiganan, a 22-amino acid oligopeptide, designed through a series of amino acid substitutions and deletions to enhance the activity of the naturally occurring Magainin-2).287–289
Oral diseases affect about 3.5 billion people worldwide, with an estimated 267 million people suffering from tooth loss.290 Dental implants are medical devices surgically implanted into the jaw to recover function (chew) or aesthetics and also provide support for artificial teeth, such as crowns. Every year, since their introduction in the 1980s, thousands of dental implants are surgically implanted, with a vast majority of dental implant systems made of Ti or zirconium oxide (ZrO2). However, as it often happens with other implantable biomaterials, dental implants are prone to failure due to biofilm-associated/microbial infections291 caused by different bacteria, as Lactobacillus salivarius, Prevotella intermedia and Staphylococcus spp, among others.292
To reduce bacterial colonisation/biofilm formation on Ti implants, the lactoferrin-derived hLf1–11 (GRRRRSVQWCA) AMP was grafted onto organosilane SAMs. The Ti surface was amine functionalised using 3-aminopropyltriethoxysilane (APTES), through which the hLf1–11 peptide was grafted via thiol groups from 3-MPA residue bound to a three 6-aminohexanoic acid (Ahx) linker at the peptide N-terminus. hLf1–11-SAMs showed an outstanding reduction of S. sanguinis and L. salivarius adhesion to Ti implants and prevented early stages of microbial growth (Fig. 12a).149
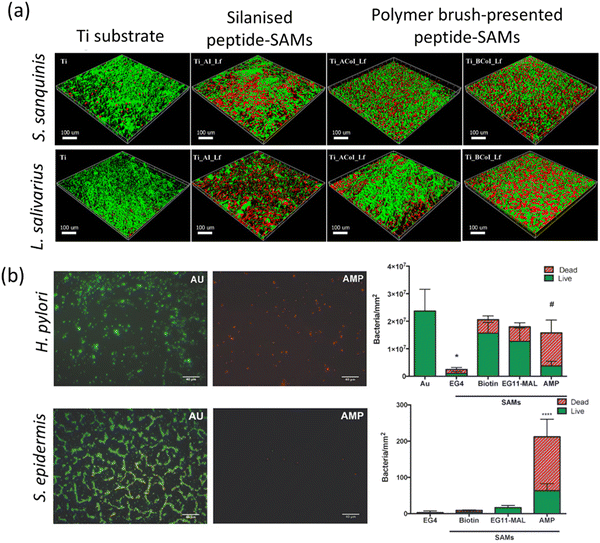 | ||
| Fig. 12 Peptide-SAMs for antimicrobial and osteo-integrative surface applications. (a) SAMs displaying the lactoferrin-derived antibacterial peptide hLf1–11 onto Ti osteo-integrative surfaces via silanisation with APTES, and polymer brush-based coatings with two different silanes. Images of live/dead staining of S. sanguinis and L. salivarius onto these substrates after 4 h incubation at 37 °C are shown (scale bars = 100 μm) [adapted with permission from ref. 150. Copyright 2015. American Chemical Society].149 (b) Live/dead staining of H. pylori J99 and S. epidermidis ATCC 35985 adhered to Au and the antimicrobial MSI-78A peptide, also antimicrobial activity assessment of AMP-SAMs is presented (scale bars = 40 μm) [adapted from ref. 129 with permission from Springer Nature].129 | ||
In addition to infection, most dental implant systems fail by lack of osteointegration. Therefore, to increase Ti implant osteointegration, the RGD peptide293 was grafted onto phosphonic acid SAMs on Ti surface.294 Phosphonic acids (like 11-hydroxyundecylphosphonic acid) self-assemble on oxide surfaces such as TiO2 or ZrO2, forming a strong P–O bond (energy of ∼80 kcal mol−1).40 These OH-terminated SAMs were then activated with a maleimide group that reacts with a thiol from the Cys residue at C-terminus in the RGD-C peptide. RGD-SAMs increased osteoblast adhesion and spreading on Ti with more than 90% of adherent cells well spread and with their actin filaments organized into robust stress fibers.
More recently, Hoyos-Nogués and co-workers prepared multifunctional coatings for bone regeneration by co-grafting the cell adhesive RGD sequence and the hLf1–11 AMP via a three-step synthesis approach: the Ti surface was first silanised, then cross-linked with N-succinimidyl-3-maleimidopropionate, followed by covalent peptide grafting. This coating demonstrated excellent ability to reduce colonisation by representative bacterial strains (S. aureus and S. sanguinis).71 Furthermore, Koidou and collaborators immobilised laminin 332- and ameloblastin-derived peptides, Lam (KKGGGPPFLMLLKGSTRFC) and Ambn (KKKGGGVPIMDFADPQFPT), respectively, onto silanised Ti (using CPTES), showing that the grafted peptides promoted epithelial attachment around teeth and formation of hemidesmosomes.150
The GL13K (GKIIKLKASLKLL) AMP features a modified 13-mer based on the sequence of parotid secretory protein. This AMP was bactericidal against P. gingivalis and prevented S. gordonii biofilm formation (putative pathogens of peri-implantitis), highlighting its potential for dental applications. Acosta et al. reported on a multifunctional modular design for SAMs based on recombinant coproduction of AMPs and elastin-like recombinamers (ELRs), which combine the antimicrobial properties of GL13K and low-fouling activity of an ELR in a synergistic manner (Fig. 13).
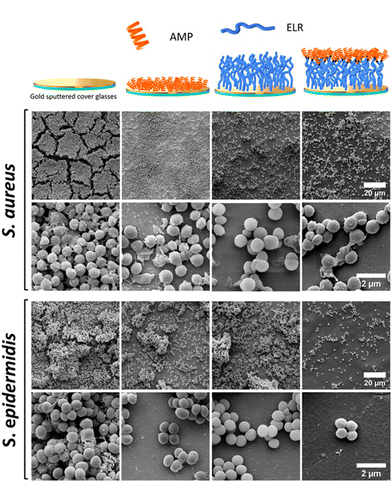 | ||
| Fig. 13 Peptide-SAMs for antimicrobial and osteo-integrative surface applications. SEM micrographs of biofilms formed by S. aureus and S. epidermidis onto the AMP, ELR (elastin-like recombiner), and AM-ELR coatings and control Au surfaces after 24 h incubation. All coated surfaces prevented formation of a mature biofilm [adapted with permission from ref. 145. Copyright 2019. American Chemical Society].145 | ||
Holmberg et al.151 developed an approach based on silanised Ti disks using CPTES and DIPEA for subsequent grafting of the GL13K AMP with a terminal amino group. The antibacterial properties of these GL13K-coated surfaces were tested against biofilms of P. gingivalis. Biofilms of this bacterium were cultured on GL13K-coated Ti discs and a pronounced reduction in the number of live cells (compared to control Ti surface without AMP) was observed by ATP content analysis and colony forming units (CFUs) counting. The antibacterial effect was attributed to electrostatic intrusion on the bacterial membrane by the positively charged lysine residues in the GL13K peptide, assisted by specific interaction with hydrophobic fimbrae of P. gingivalis.295 This coating was very stable, since GL13K was not significantly released from the Ti coated surfaces when immersed in saliva or serum for 1 week. Moreover, the cytocompatibility of the GL13K-functionalised Ti surfaces was assessed using human gingival fibroblasts and MC3T3-E1 murine osteoblasts. Nonetheless, it is noteworthy that this immobilisation process did not control peptide orientation and, although showing good performance in vitro, AMP covalent coupling can only be performed prior to dental implantation, as it is generally performed under harsh conditions (e.g., low pH). It was also demonstrated that the GL13K D-enantiomer grafted via “click-chemistry” on Ti has improved bacterial resistance296 compared to its enantiomer.
Several peptides with the ability to self-assemble on metal-, metal oxide-, mineral and polymer surfaces have been identified,297 allowing to overcome the “two/three-step” modification. This “surface-induced peptide self-assembly” strategy can be exemplified by Ti-binding peptides serving as an effective anchor for AMPs on implant surfaces.298 This approach is less complex and avoids adverse effects associated with introduction of additional molecules (e.g., linkers). However, it increases peptide length and cost.299 For instance, the E14LKK AMP (LKLLKKLLKLLKKL) was combined via a tri-glycine linker with metal binding sequences TiBP1 (RPRENRGRERGL) or TiBP2 (SRPNGYGGSESS).69 These peptides were then self-assembled onto Ti substrates and incubated with E. coli, S. mutans, and S. epidermidis. Significant differences were observed between adhered bacteria on the control surface (bare Ti) and peptide-functionalised surface. For S. epidermidis and S. mutans, 45-fold less bacteria adhered to the peptide-assembled on the Ti surface. Still, the surface with TiBP1-GGGAMP showed enhanced performance for S. mutans, while no differences on S. epidermidis adhesion were seen between TiBP1-GGGAMP and TiBP2-GGGAMP.
Ti-based biomaterials are also used in orthopaedics, due to their excellent mechanical strength, corrosion resistance, and biocompatibility.84 The main concern in orthopaedic infections includes ESKAPE microorganisms (E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa and E. cloacae), which account for over 90% of nosocomial infections.300
Chen and collaborators developed a strategy targeting ESKAPE bacteria based on silanised Ti discs with hydrolysed alkynyl-PEG3-triethoxysilane. The functionalised Ti substrates were then treated with a pegylated AMP, PEG-HHC36 (N3-PEG12-KRWWKWWRR), containing a terminal azide-functionality for a “click-reaction” to tether the peptide to the substrate. The surface yielded high AMP density and was able to inhibit 90.2% of S. aureus and 88.1% of E. coli after 2.5 h of incubation. Interestingly, after 4 days of degradation at 37 °C, the surface was still able to inhibit 69.5% of S. aureus.152 Using a mice model, the functionalised implant killed 78.8% of S. aureus after 7 days.
Yazici et al. resourced to electrochemically deposited calcium phosphate as the linking layer between nanotubular Ti surfaces and bound peptides.137 The cHABP-1-Spacer-AMP, consisting of the hydroxyapatite binding peptide-1 (cHABP1: CMLPHHGAC) and an AMP (tet-127: KRWWKWWRR),301 both combined with a flexible linker (GGG), were self-assembled by the calcium phosphate binding sequence on the metal surface. These chimeric peptides reduced bacterial adhesion of E. coli and S. mutans. On the same quest to challenge BAI infections, the FK-16 (FKRIVQRIKDFLRNLV) AMP was covalently attached onto silanised Ti substrates. Surfaces exhibited broad-spectrum activity against ESKAPE pathogens and anti-adhesion/biofilm against S. aureus and E. coli.153
Beyond prevention of BAI, SAMs functionalised with peptides have also been used as proof-of-concept for the development of novel strategies for infection management. Parreira and co-workers immobilised the MSI-78A AMP targeting the gastric pathogen H. pylori, a priority bacterium according to the World Health Organisation (WHO) in what concerns recent therapeutic options.129,302 MSI-78A is derived from the MSI-78 AMP by replacement of one amino acid (G13A, Table 2, peptide-SAMs as antimicrobial surfaces).8,129 This biotin-neutravidin strategy allowed controlling both the AMP orientation and exposure once grafted onto the surface. These MSI-78A functionalised SAMs were able to kill, by contact, 98% of planktonic H. pylori in only 2 h. Furthermore, these surfaces were H. pylori-specific since no effect was seen against the control bacteria (S. epidermidis). This proof-of-concept study highlighted the high bactericidal potential of grafted MSI-78A and established the foundations for the development of MSI-78A grafted nanoparticles for gastric infection management within a targeted nanomedicine concept (Fig. 12b).129
Thrombus formation, triggered by the activation of the coagulation cascade due to the adsorption of certain plasma proteins on the surface of biomaterials, is a severe clinical complication and a major concern for the success of blood-contacting medical devices.303,304 Hence, immobilisation of thrombin (a serine protease that plays a vital role on the coagulation system) inhibitors on the surface of biomaterials represents a viable strategy to prevent blood clotting and enhance their hemocompatibility. As a proof-of-concept, the thrombin inhibitor DFPRPG154 was grafted onto tetra(ethylene glycol) (EG4) terminated SAMs on Au surfaces (EG4-SAMs). Peptide-SAMs were found to inhibit the activity of adsorbed thrombin in a concentration-dependent manner. However, SAMs lost most of its thrombin-binding selectivity in the presence of plasma proteins, impeding their translation as thrombin inhibitor surface for blood contact implants.154
5.5 Enzyme assay platforms
In order to study enzyme activity, SAMs have been prepared by grafting peptides that can be degraded by the former. For instance, the work of Houseman and collaborators demonstrated the possibility to fabricate biochips suitable to study the phosphorylation of the immobilised peptide IYGEFKKKC by the enzyme tyrosine kinase c-src. Peptide-SAMs were incubated with this kinase and [γ-32P]ATP, incorporating nearly 500 cpm cm−2 of radiolabelled phosphate, unlike tyrosine-free and no peptide-containing controls. Additionally, presence of phosphotyrosine in modified SAMs substrates was examined via binding of an antibody monitored using SPR. SAMs presenting this tyrosine-containing peptide were then employed to study the noncompetitive inhibition of the enzyme in an array format. Mixtures containing the kinase, [γ-32P]ATP, and different inhibitor concentrations were incubated onto monolayers. Phosphorimager analysis allowed to determine concentration-dependent inhibition of enzyme activity, as well as an inhibition constant of 31 nM (Table 3, peptide-SAMs for biosensing of enzyme targets).78More recently, Mrksich and collaborators reported on the use of phosphotyrosine-containing peptide arrays based on SAMs for Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (SAMDI-MS) to profile phosphatase substrate selectivities.233,234 For instance, 22 protein tyrosine phosphatases were assessed against an array of 361 peptide sequences to give a profile of their specificities, providing valuable molecular insights into the biochemistry and biology of these clinically relevant enzyme family (Table 3, peptide-SAMs for biosensing of enzyme targets).233
6. Final overview, challenges, and prospects
The multiple inherent properties (e.g., chemical diversity and structural transitions, biocompatibility, biodegradability, selective recognition, conduction, ease of functionalization) of peptides have made this class of biomolecules highly attractive for R&D. Their integration in cell culture substrates is obvious, and the identification of novel peptide motifs from ECM proteins or by phage display has spurred the area of peptide-based SAMs in the early 2000s as screening platforms of peptide libraries. In fact, the majority of the applications in this area were reported between 2010 and 2015 (Table 1). Major advances were seen in the fabrication of anisotropic surfaces displaying peptide-SAMs with spatiotemporal control enabling the conceptualisation of more complex biological studies, such as the fabrication of SAM-based platforms for the hierarchical presentation of bioactive peptides.117 Current research in this applicational area is focused on the fabrication of dynamic surfaces, through supramolecular approaches114 or activation with external stimuli (e.g. light, electrical potentials) that are cell-compatible and allow multiple cycles of activation,110 as well as on the increasing sophistication of the SAMs platforms (e.g., automation, robotization), interfaced with mass spectrometry analytics,233 to support and advance high throughput screening (HTS) programs112 of large peptide libraries in drug discovery and diagnostics. The findings from these early studies revealed new potential applications, notably biosensing. In fact, the application of peptide-SAMs in biosensing has been an active area of research in recent years, as demonstrated in Table 3, integrating the knowledge and expertise generated from landmark studies on peptide-SAMs (e.g., zwitterionic peptides for antifouling surfaces). Another emerging applicational area of peptide-SAMs is in electron transfer with direct implications for the development of peptide-based bioelectronics, a research area still at its infancy.305 The advent of molecular and supramolecular electronics-based technologies will likely benefit by creating synergies with peptide-based platforms like SAMs, especially as SAMs-based technologies gain increasing attention including active molecular device layers and multifunctional SAMs which fulfil several layer functions of a device within one monolayer.306,307The diversity of peptide lengths and designs included in this review (Tables 1–3) attest the versatility of peptides for the development of SAMs and their adoption by scientific communities not specialized in peptide science. As a result, peptide sequences described in some published works have not been reported in the correct order (from the N- to C-terminus). In addition, the identification of the peptide anchoring mode and orientation, as well as the rationale behind the substrate selected, were sometimes not sufficiently clear, limiting the accurate reporting of details necessary to enhance the comprehension by readers less familiarised with the subject and facilitate the uptake of the peptide designs in further studies. Therefore, greater attention is recommended to ensure peptide sequences are presented in a consistent and accurate fashion.
Reflecting on what peptides can do for SAMs and vice versa, it is clear from this review that SAMs are a valuable platform for the controlled spatiotemporal and reproducible display of peptides on surfaces, either for screening purposes with a translational biomedical goal, or for answering biological questions, such as gaining understanding on peptide-target interactions. On the other hand, peptides offer chemical diversity and complexity, structural flexibility and dynamic behaviour, a set of traits not present in conventional SAMs, but crucial to meet the current demands for advanced cell-based screening devices, high-precision biological assays, anti-infective surfaces and sustainable bioelectronics. Although the field of SAMs seemed dormant, this review is a testimony that the field has not been stagnated, but rather deviated from its beginnings. With the discovery of novel peptide sequences by combinatorial, high-throughput, and machine learning methods (e.g., AMPs), and the emergence of applications using renewed peptide functions (e.g., peptide-assisted genome editing),308 as well as the need to provide hierarchical presentation of epitopes,117 peptide-SAMs will continue to provide a reliable platform for in vitro screening of artificial protein mimics that offers great flexibility for chemical processability, surface imaging and integration of sensors for detection. In fact, this review was conceived with the idea of reinvigorating the research on peptide-SAMs by showcasing the numerous transformative contributions made in the field and discussing emerging directions. While peptides offer an exquisite set of properties for applications in SAMs, there is still room for improvement and unforeseen opportunities to explore. For example, the fabrication of reversible surfaces using peptide-SAMs is an area not yet extensively exploited, but would pave the way for the creation of surfaces with tuneable fluidity and restorability (multiple uses) with great value for cell culture and biosensing applications, respectively. Another opportunity in peptide-SAMs would be the use of coiled peptides as molecular springs in sensors for measuring cell forces involved in cell adhesion.116 With active and eager researchers working across peptide and biomedical sciences, the field of peptide-SAMs is intriguing, with new developments and trends expected in the near future.
Abbreviations
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| ADSC | Adipose-derived stem cells |
| AFP | Alpha fetoprotein |
| Ahx | Aminohexanoic acid |
| Aib | 2-Aminoisobutyric acid |
| Ala, A | l-Alanine |
| Alb | Albumin |
| AMPs | Antimicrobial peptides |
| APN | Aminopeptidase N |
| APTES | 3-(Aminopropyl)-triethoxysilane |
| Arg, R | l-Arginine |
| Asn, N | l-Asparagine |
| Asp, D | l-Aspartic acid |
| ATP | Adenosine triphosphate |
| ATs | Alkanethiols |
| AβO, Aβ(1–42) | β-Amyloid oligomer |
| BAI | Biomaterials-associated infections |
| BFP-1 | Bone forming peptide-1 |
| BMHP1 | Bone marrow homing peptide 1 |
| BMP-7 | Bone morphogenetic protein-7 |
| Boc | Butyloxycarbonyl |
| BoNT/A | Botulinum neurotoxin type A |
| BQ | Benzoquinone |
| BSA | Bovine serum albumin |
| CA | Chronoamperometry |
| CASP3 | Caspase-3 |
| CB[8] | Cucurbit[8]uril |
| CM | Cecropin-mellitin peptide hybrid |
| Coll | Collagen |
| Coll-II | Collagen type II |
| Cp | Cyclopentadiene |
| CPAD | 4-Cyano-4-(phenylcarbonothioylthio)-pentanoic acid |
| CPTES | 3-(Chloropropyl)-triethoxysilane |
| CS-GQDs | Chitosan-graphene quantum dots |
| CuAAC | Copper(I)-catalysed azide–alkyne cycloaddition |
| CV | Cyclic voltammetry |
| Cys, C | l-Cysteine |
| Cyt c | Cytochrome c |
| DIPEA | Diisopropylethylamine |
| DMNPB | 3-(4,5-Dimethoxy-2-nitrophenyl)-2-butyl |
| DNA | Deoxyribonucleic acid |
| DPG | Alpha-2 deamidated gliadin peptide |
| DPV | Differential pulse voltammetry |
| EC | Human embryonal carcinoma cells |
| ECM | Extracellular matrix |
| ECz | N-Ethylcarbazoyl |
| EDC | 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide |
| EGFR | Epidermal growth factor receptor |
| EIS | Electrochemical impedance spectroscopy |
| ELR | Elastin-like recombinamer |
| EPR | Electron paramagnetic resonance |
| ES | Human embryonic stem cells |
| ET | Electron transfer |
| Fc | Ferrocene |
| FcMMAs | Ferrocenylmethylmethacrylates |
| Fib | Fibrinogen |
| FN | Fibronectin |
| FT-IR | Fourier Transform infrared spectroscopy |
| GAGs | Glycosaminoglycans |
| GCE | Glassy carbon electrode |
| GF | Growth factor |
| Gln, Q | l-Glutamine |
| Glu, E | l-Glutamic acid |
| Gly, G | Glycine |
| GO | Graphene oxide |
| GPCRs | G-Protein-coupled receptors |
| HA | Hyaluronan |
| HATU | 2-(1H-7-Azabenzotriazol-1-yl)-1,1,3,3-tetramethyl uranium hexafluorophosphate |
| hACs | Human articular chondrocytes |
| Hep | Heparin |
| HepG2 | Human hepatocellular carcinoma |
| His, H | l-Histidine |
| HIV-1 IN | Human immunodeficiency virus type-1 integrase |
| HIV-1 PR | Human immunodeficiency virus type-1 protease |
| HIV-1 RT | Human immunodeficiency virus type-1 reverse transcriptase |
| hMSCs | Human mesenchymal stem cells |
| HNE | Human neutrophil elastase |
| HQ | Hydroquinone |
| HSA | Human serum albumin |
| HTS | High-throughput screening |
| HUVECs | Human umbilical vein endothelial cells |
| IgE | Immunoglobulin E |
| IgG | Immunoglobulin G |
| Ile, I | l-Isoleucine |
| Leu, L | l-Leucine |
| Lipo | Lipoic acid |
| LN | Laminin |
| LRG1 | Leucine-rich α-2-glycoprotein-1 |
| Lys, K | l-Lysine |
| Lyz | Lysozyme |
| MB | Methylene blue |
| Mb | Mioglobin |
| Met, M | l-Methionine |
| MMP | Matrix metalloproteinase |
| MPA | 3-Mercaptopropionic acid |
| MSCs | Mesenchymal stem cells |
| MUA | 11-Mercaptoundecanoic acid |
| MV | Methyl viologen, 1-1′-dimethyl-4,4′-bypyridinium dichloride |
| NHS | N-Hydroxysuccinimide |
| NG | Nanogenerator |
| Nle | Norleucine |
| NPs | Nanoparticles |
| OFETs | Organic field-effect transistors |
| PANI | Polyaniline |
| PBLG | Poly(γ-benzyl l-glutamate) |
| PEG | Poly(ethylene glycol) |
| PGs | Proteoglycans |
| Phe, F | l-Phenylalanine |
| PKA | Protein kinase |
| PLGA | Poly(l-glutamic acid)-based amphiphile |
| PNA | Peptide-nucleic acid |
| GalNAcT | N-Acetylgalactosaminyl transferase |
| Pro, P | l-Proline |
| PSA | Prostate-specific antigen |
| Pyr | Pyrenyl |
| QCM-D | Quartz crystal microbalance with dissipation |
| RAFT | Reversible addition fragmentation chain-transfer polymerisation |
| RNA | Ribonucleic acid |
| ROS | Reactive oxygen species |
| RP-HPLC | Reverse phase high performance liquid chromatography |
| SA | Streptavidin |
| SAMs | Self-assembling monolayers |
| SBPs | Solid binding peptides |
| SCC | Squamous cell carcinoma |
| Ser, S | l-Serine |
| SMCC | Succinimidyl-4-(N-maleimido-methyl)cyclohexane-1-carboxylate |
| SPAAC | Strain-promoted alkyne–azide cycloaddition |
| SPE | Screen printed electrode |
| SPPS | Solid phase peptide synthesis |
| SPR | Surface plasmon resonance |
| SWV | Square wave voltammetry |
| TB | Thrombin |
| TEOA | Triethanolamine |
| TGF-β1 | Transforming growth factor-1 |
| Thc | Thioctic acid |
| Thr, T | l-Threonine |
| TN | Tenascin |
| TOAC | 2,2,6,6-Tetramethylpiperidine-1-oxyl-4-amino-4-carboxylic acid |
| Trp, W | l-Tryptophan |
| Tyr, Y | l-Tyrosine |
| VACNF | Vertically aligned carbon nanofibers |
| Val, V | l-Valine |
| VEGF | Vascular endothelial growth factor |
| VEGFR | VEGF receptor |
| VN | Vitronectin |
| WHO | World Health Organisation |
| XPS | X-ray photoelectron spectroscopy |
| βGalT | β-1,4-Galactosyl transferase |
| μCP | Micro-contact printing |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the MOBILIsE project funded by the European Union's Horizon 2020 research and innovation programme under Grant Agreement no. 951723. Authors also thank FCT (Fundação para a Ciência e a Tecnologia) for funding M. Cristina L. Martins (LA/P/0070/2020) and Paula Parreira (CEECIND/01210/2018).References
- R. G. Nuzzo and D. L. Allara, Adsorption of Bifunctional Organic Disulfides on Gold Surfaces, J. Am. Chem. Soc., 1983, 105(13), 4481–4483, DOI:10.1021/ja00351a063.
- C. D. Bain and G. M. Whitesides, Molecular-Level Control over Surface Order in Self-Assembled Monolayer Films of Thiols on Gold, Science, 1988, 240(4848), 62–63, DOI:10.1126/science.240.4848.62.
- A. Ulman, Formation and Structure of Self-Assembled Monolayers, Chem. Rev., 1996, 96(4), 1533–1554, DOI:10.1021/cr9502357.
- J. C. Love, L. A. Estroff, J. K. Kriebel, R. G. Nuzzo and G. M. Whitesides, Self-Assembled Monolayers of Thiolates on Metals as a Form of Nanotechnology, 2005, vol. 105 DOI:10.1021/cr0300789.
- S. Y. Kim, S. J. Cho, S. E. Byeon, X. He and H. J. Yoon, Self-Assembled Monolayers as Interface Engineering Nanomaterials in Perovskite Solar Cells, Adv. Energy Mater., 2020, 10(44), 1–21, DOI:10.1002/aenm.202002606.
- J. T. Koepsel and W. L. Murphy, Patterned Self-Assembled Monolayers: Efficient, Chemically Defined Tools for Cell Biology, ChemBioChem, 2012, 13(12), 1717–1724, DOI:10.1002/cbic.201200226.
- A. M. Ross and J. Lahann, Current Trends and Challenges in Biointerfaces Science and Engineering, Annu. Rev. Chem. Biomol. Eng., 2015, 6, 161–186, DOI:10.1146/annurev-chembioeng-060713-040042.
- V. Leiro, P. Parreira, S. C. Freitas, M. C. L. Martins and A. P. Pêgo, Conjugation Chemistry Principles and Surface Functionalization of Nanomaterials, Biomedical Applications of Functionalized Nanomaterials, Elsevier, 2018, pp. 35–66 DOI:10.1016/B978-0-323-50878-0.00002-1.
- P. Zou, W. T. Chen, T. Sun, Y. Gao, L. L. Li and H. Wang, Recent Advances: Peptides and Self-Assembled Peptide-Nanosystems for Antimicrobial Therapy and Diagnosis, Biomater. Sci., 2020, 8(18), 4975–4996, 10.1039/d0bm00789g.
- C. Vericat, M. E. Vela, G. Benitez, P. Carro and R. C. Salvarezza, Self-Assembled Monolayers of Thiols and Dithiols on Gold: New Challenges for a Well-Known System, Chem. Soc. Rev., 2010, 39(5), 1805–1834, 10.1039/b907301a.
- D. M. Raymond and B. L. Nilsson, Multicomponent Peptide Assemblies, Chem. Soc. Rev., 2018, 47(10), 3659–3720, 10.1039/c8cs00115d.
- B. O. Okesola, C. Redondo-Gómez and A. Mata, Multicomponent Self-Assembly: Supramolecular Design of Complex Hydrogels for Biomedical Applications, Self-assembling Biomaterials, Elsevier, 2018, pp. 371–397 DOI:10.1016/B978-0-08-102015-9.00019-8.
- Y. Huo, J. Hu, Y. Yin, P. Liu, K. Cai and W. Ji, Self-Assembling Peptide-Based Functional Biomaterials, ChemBioChem, 2023, 24(2), e202200582, DOI:10.1002/cbic.202200582.
- M. A. Meyers, P.-Y. Chen, A. Y.-M. Lin and Y. Seki, Biological Materials: Structure and Mechanical Properties, Prog. Mater. Sci., 2008, 53(1), 1–206, DOI:10.1016/j.pmatsci.2007.05.002.
- M. Eder, S. Amini and P. Fratzl, Biological Composites—Complex Structures for Functional Diversity, Science, 2018, 362(6414), 543–547, DOI:10.1126/science.aat8297.
- C. Ligorio and A. Mata, Synthetic Extracellular Matrices with Function-Encoding Peptides, Nat. Rev. Bioeng., 2023, 1(7), 518–536, DOI:10.1038/s44222-023-00055-3.
- C. Nicosia and J. Huskens, Reactive Self-Assembled Monolayers: From Surface Functionalization to Gradient Formation, Mater. Horiz., 2014, 1(1), 32–45, 10.1039/c3mh00046j.
- A. Adhikari, B. R. Bhattarai, A. Aryal, N. Thapa, P. Kc, A. Adhikari, S. Maharjan, P. B. Chanda, B. P. Regmi and N. Parajuli, Reprogramming Natural Proteins Using Unnatural Amino Acids, RSC Adv., 2021, 11(60), 38126–38145, 10.1039/d1ra07028b.
- X. Wang, X. Yang, Q. Wang and D. Meng, Unnatural Amino Acids: Promising Implications for the Development of New Antimicrobial Peptides, Crit. Rev. Microbiol., 2023, 49(2), 231–255, DOI:10.1080/1040841X.2022.2047008.
- N. Li, J. R. Klim, R. Derda, A. H. Courtney and L. L. Kiessling, Spatial Control of Cell Fate Using Synthetic Surfaces to Potentiate TGF-β Signaling, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(29), 11745–11750, DOI:10.1073/pnas.1101454108.
- P. M. Lundin, B. L. Fiser, M. S. Blackledge, H. L. Pickett and A. L. Copeland, Functionalized Self-Assembled Monolayers: Versatile Strategies to Combat Bacterial Biofilm Formation, Pharmaceutics, 2022, 14(8), 1613, DOI:10.3390/pharmaceutics14081613.
- G. T. L. Teixeira, J. P. L. do Nascimento, R. V. Gelamo, J. A. Moreto and N. B. L. Slade, Strategies for Functionalization of Metallic Surfaces with Bioactive Peptides: A Mini Review, Int. J. Pept. Res. Ther., 2023, 29(2), 1–12, DOI:10.1007/s10989-023-10497-3.
- E. Gatto and M. Venanzi, Self-Assembled Monolayers Formed by Helical Peptide Building Blocks: A New Tool for Bioinspired Nanotechnology, Polym. J., 2013, 45(5), 468–480, DOI:10.1038/pj.2013.27.
- T. Fujita, N. Bunjes, K. Nakajima, M. Hara, H. Sasabe and W. Knoll, Macrodipole Interaction of Helical Peptides in a Self-Assembled Monolayer on Gold Substrate, Langmuir, 1998, 14(21), 6167–6172, DOI:10.1021/la9801155.
- J. T. Koepsel, P. T. Brown, S. G. Loveland, W. J. Li and W. L. Murphy, Combinatorial Screening of Chemically Defined Human Mesenchymal Stem Cell Culture Substrates, J. Mater. Chem., 2012, 22(37), 19474–19481, 10.1039/c2jm32242k.
- R. Derda, L. Li, B. P. Orner, R. L. Lewis, J. A. Thomson and L. L. Kiessling, Defined Substrates for Human Embryonic Stem Cell Growth Identified from Surface Arrays, ACS Chem. Biol., 2007, 2(5), 347–355, DOI:10.1021/cb700032u.
- V. Corvaglia, R. Marega, F. De Leo, C. Michiels and D. Bonifazi, Unleashing Cancer Cells on Surfaces Exposing Motogenic IGDQ Peptides, Small, 2016, 12(3), 321–329, DOI:10.1002/smll.201501963.
- K. Han, W. N. Yin, J. X. Fan, F. Y. Cao and X. Z. Zhang, Photo-Activatable Substrates for Site-Specific Differentiation of Stem Cells, ACS Appl. Mater. Interfaces, 2015, 7(42), 23679–23684, DOI:10.1021/acsami.5b07455.
- A. Levin, T. A. Hakala, L. Schnaider, G. J. L. Bernardes, E. Gazit and T. P. J. Knowles, Biomimetic Peptide Self-Assembly for Functional Materials, Nat. Rev. Chem., 2020, 4(11), 615–634, DOI:10.1038/s41570-020-0215-y.
- N. S. Kehr, S. Atay and B. Ergün, Self-Assembled Monolayers and Nanocomposite Hydrogels of Functional Nanomaterials for Tissue Engineering Applications, Macromol. Biosci., 2015, 15(4), 445–463, DOI:10.1002/mabi.201400363.
- X. Pang, W. Li, E. Landwehr, Y. Yuan, W. Wang and H. S. Azevedo, Mimicking the Endothelial Glycocalyx through the Supramolecular Presentation of Hyaluronan on Patterned Surfaces, Faraday Discuss., 2019, 219, 168–182, 10.1039/C9FD00015A.
- G. A. Hudalla and W. L. Murphy, Chemically Well-Defined Self-Assembled Monolayers for Cell Culture: Toward Mimicking the Natural ECM, Soft Matter, 2011, 7(20), 9561–9571, 10.1039/c1sm05596h.
- X. Pang, C. O’Malley, J. Borges, M. M. Rahman, D. W. P. Collis, J. F. Mano, I. C. Mackenzie and H. S. Azevedo, Supramolecular Presentation of Hyaluronan onto Model Surfaces for Studying the Behavior of Cancer Stem Cells, Adv. Biosyst., 2019, 3(10), 1900017, DOI:10.1002/adbi.201900017.
- X. Pang, W. Li, L. Chang, J. E. Gautrot, W. Wang and H. S. Azevedo, Hyaluronan (HA) Immobilized on Surfaces via Self-Assembled Monolayers of HA-Binding Peptide Modulates Endothelial Cell Spreading and Migration through Focal Adhesion, ACS Appl. Mater. Interfaces, 2021, 13(22), 25792–25804, DOI:10.1021/acsami.1c05574.
- F. Y. Cao, W. N. Yin, J. X. Fan, R. X. Zhuo and X. Z. Zhang, A Novel Function of BMHP1 and CBMHP1 Peptides to Induce the Osteogenic Differentiation of Mesenchymal Stem Cells, Biomater. Sci., 2015, 3(2), 345–351, 10.1039/c4bm00300d.
- J. L. Gerardo-Nava, J. Jansen, D. Günther, L. Klasen, A. L. Thiebes, B. Niessing, C. Bergerbit, A. A. Meyer, J. Linkhorst and M. Barth, et al., Transformative Materials to Create 3D Functional Human Tissue Models In Vitro in a Reproducible Manner, Adv. Healthcare Mater., 2023, 2301030, 1–19, DOI:10.1002/adhm.202301030.
- P. E. Laibinis, G. M. Whitesides, D. L. Allara, Y. T. Tao, A. N. Parikh and R. G. Nuzzo, Comparison of the Structures and Wetting Properties of Self-Assembled Monolayers of n-Alkanethiols on the Coinage Metal Surfaces, Copper, Silver, and Gold, J. Am. Chem. Soc., 1991, 113(19), 7152–7167, DOI:10.1021/ja00019a011.
- M. Venanzi, G. Pace, A. Palleschi, L. Stella, P. Castrucci, M. Scarselli, M. De Crescenzi, F. Formaggio, C. Toniolo and G. Marletta, Densely-Packed Self-Assembled Monolayers on Gold Surfaces from a Conformationally Constrained Helical Hexapeptide, Surf. Sci., 2006, 600(2), 409–416, DOI:10.1016/j.susc.2005.10.040.
- L. Srisombat, A. C. Jamison and T. R. Lee, Stability: A Key Issue for Self-Assembled Monolayers on Gold as Thin-Film Coatings and Nanoparticle Protectants, Colloids Surf., A, 2011, 390(1–3), 1–19, DOI:10.1016/j.colsurfa.2011.09.020.
- M. Dubey, T. Weidner, L. J. Gamble and D. G. Castner, Structure and Order of Phosphonic Acid-Based Self-Assembled Monolayers on Si(100), Langmuir, 2010, 26(18), 14747–14754, DOI:10.1021/la1021438.
- W. N. Yin, F. Y. Cao, K. Han, X. Zeng, R. X. Zhuo and X. Z. Zhang, The Synergistic Effect of a BMP-7 Derived Peptide and Cyclic RGD in Regulating Differentiation Behaviours of Mesenchymal Stem Cells, J. Mater. Chem. B, 2014, 2(47), 8434–8440, 10.1039/c4tb01548g.
- J. Cabanas-Danés, C. Nicosia, E. Landman, M. Karperien, J. Huskens and P. Jonkheijm, A Fluorogenic Monolayer to Detect the Co-Immobilization of Peptides That Combine Cartilage Targeting and Regeneration, J. Mater. Chem. B, 2013, 1(14), 1903–1908, 10.1039/c3tb20109k.
- S. P. Pujari, L. Scheres, A. T. M. Marcelis and H. Zuilhof, Covalent Surface Modification of Oxide Surfaces, Angew. Chem., Int. Ed., 2014, 53(25), 6322–6356, DOI:10.1002/anie.201306709.
- L. Wang, U. S. Schubert and S. Hoeppener, Surface Chemical Reactions on Self-Assembled Silane Based Monolayers, Chem. Soc. Rev., 2021, 50(11), 6507–6540, 10.1039/d0cs01220c.
- C. Haensch, S. Hoeppener and U. S. Schubert, Chemical Modification of Self-Assembled Silane Based Monolayers by Surface Reactions, Chem. Soc. Rev., 2010, 39(6), 2323–2334, 10.1039/b920491a.
- J. Maciel, M. C. L. Martins and M. A. Barbosa, The Stability of Self-Assembled Monolayers with Time and under Biological Conditions, J. Biomed. Mater. Res., Part A, 2010, 94A, 833–843, DOI:10.1002/jbm.a.32746.
- N. T. Flynn, T. N. T. Tran, M. J. Cima and R. Langer, Long-Term Stability of Self-Assembled Monolayers in Biological Media, Langmuir, 2003, 19(26), 10909–10915, DOI:10.1021/la035331e.
- S.-J. Xiao, M. Textor, N. D. Spencer and H. Sigrist, Covalent Attachment of Cell-Adhesive, (Arg–Gly–Asp)-Containing Peptides to Titanium Surfaces, Langmuir, 1998, 14(19), 5507–5516, DOI:10.1021/la980257z.
- T. Morita, S. Kimura, S. Kobayashi and Y. Imanishi, Photocurrent Generation under a Large Dipole Moment Formed by Self-Assembled Monolayers of Helical Peptides Having an N-Ethylcarbazolyl Group, J. Am. Chem. Soc., 2000, 122(12), 2850–2859, DOI:10.1021/ja992769l.
- R. Derda, D. J. Wherritt and L. L. Kiessling, Solid-Phase Synthesis of Alkanethiols for the Preparation of Self-Assembled Monolayers, Langmuir, 2007, 23(22), 11164–11167, DOI:10.1021/la701386v.
- J. R. Klim, L. Li, P. J. Wrighton, M. S. Piekarczyk and L. L. Kiessling, A Defined Glycosaminoglycan-Binding Substratum for Human Pluripotent Stem Cells, Nat. Methods, 2010, 7(12), 989–994, DOI:10.1038/nmeth.1532.
- S. Sek, A. Sepiol, A. Tolak, A. Misicka and R. Bilewicz, Distance Dependence of the Electron Transfer Rate through Oligoglycine Spacers Introduced into Self-Assembled Monolayers, J. Phys. Chem. B, 2004, 108(24), 8102–8105, DOI:10.1021/jp049116y.
- S. Sek, A. Tolak, A. Misicka, B. Palys and R. Bilewicz, Asymmetry of Electron Transmission through Monolayers of Helical Polyalanine Adsorbed on Gold Surfaces, J. Phys. Chem. B, 2005, 109(39), 18433–18438, DOI:10.1021/jp052157p.
- C. G. Worley, R. W. Linton and E. T. Samulski, Electric-Field-Enhanced Self-Assembly of.Alpha.-Helical Polypeptides, Langmuir, 1995, 11(10), 3805–3810, DOI:10.1021/la00010a034.
- Y. Miura, S. Kimura, Y. Imanishi and J. Umemura, Formation of Oriented Helical Peptide Layers on a Gold Surface Due to the Self-Assembling Properties of Peptides, Langmuir, 1998, 14(24), 6935–6940, DOI:10.1021/la981296d.
- E. Gatto, L. Stella, F. Formaggio, C. Toniolo, L. Lorenzelli and M. Venanzi, Electroconductive and Photocurrent Generation Properties of Self-Assembled Monolayers Formed by Functionalized, Conformationally-Constrained Peptides on Gold Electrodes, J. Pept. Sci., 2008, 14(2), 184–191, DOI:10.1002/psc.973.
- T. I. Smirnova and A. I. Smirnov, Peptide–Membrane Interactions by Spin-Labeling EPR, 2015, pp. 219–258 DOI:10.1016/bs.mie.2015.08.018.
- K. Zamojć, W. Wiczk, B. Zaborowski, D. Jacewicz and L. Chmurzyński, Fluorescence Quenching of 7-Amino-4-Methylcoumarin by Different TEMPO Derivatives, Spectrochim. Acta, Part A, 2015, 136(PC), 1875–1880, DOI:10.1016/j.saa.2014.10.102.
- Y. Tada, T. Morita, J. Umemura, M. Iwamoto and S. Kimura, Photoresponsive Change of the Surface Potential Generated by Helical Peptide Self-Assembled Monolayers, Polym. J., 2005, 37(8), 599–607, DOI:10.1295/polymj.37.599.
- S. Yasutomi, T. Morita, Y. Imanishi and S. Kimura, A Molecular Photodiode System That Can Switch Photocurrent Direction, Science, 2004, 304(5679), 1944–1947, DOI:10.1126/science.1098489.
- M. Niwa, M. Morikawa and N. Higashi, Discrimination between N- and C-Termini of Polypeptides by a Two-Dimensional Array of Helical Poly(L-Glutamic Acid) Rods on Gold Surfaces, Langmuir, 1999, 15(15), 5088–5092, DOI:10.1021/la9817258.
- K. Uvdal and T. P. Vikinge, Chemisorption of the Dipeptide Arg-Cys on a Gold Surface and the Selectivity of G-Protein Adsorption, Langmuir, 2001, 17(6), 2008–2012, DOI:10.1021/la0009184.
- M. Sarikaya, C. Tamerler, A. K.-Y. Jen, K. Schulten and F. Baneyx, Molecular Biomimetics: Nanotechnology through Biology, Nat. Mater., 2003, 2(9), 577–585, DOI:10.1038/nmat964.
- C. Chen and N. L. Rosi, Peptide-Based Methods for the Preparation of Nanostructured Inorganic Materials, Angew. Chem., Int. Ed., 2010, 49(11), 1924–1942, DOI:10.1002/anie.200903572.
- L. Shao, J. Ma, J. L. Prelesnik, Y. Zhou, M. Nguyen, M. Zhao, S. A. Jenekhe, S. V. Kalinin, A. L. Ferguson and J. Pfaendtner, et al., Hierarchical Materials from High Information Content Macromolecular Building Blocks: Construction, Dynamic Interventions, and Prediction, Chem. Rev., 2022, 122(24), 17397–17478, DOI:10.1021/acs.chemrev.2c00220.
- N. Alvisi and R. de Vries, Biomedical Applications of Solid-Binding Peptides and Proteins, Mater. Today Bio, 2023, 19, 100580, DOI:10.1016/j.mtbio.2023.100580.
- M. Hnilova, E. E. Oren, U. O. S. Seker, B. R. Wilson, S. Collino, J. S. Evans, C. Tamerler and M. Sarikaya, Effect of Molecular Conformations on the Adsorption Behavior of Gold-Binding Peptides, Langmuir, 2008, 24(21), 12440–12445, DOI:10.1021/la801468c.
- R. V. Lee, H. M. Zareie and M. Sarikaya, Chimeric Peptide-Based Biomolecular Constructs for Versatile Nucleic Acid Biosensing, ACS Appl. Mater. Interfaces, 2022, 14(20), 23164–23181, DOI:10.1021/acsami.2c03341.
- H. Yazici, M. B. O’Neill, T. Kacar, B. R. Wilson, E. E. Oren, M. Sarikaya and C. Tamerler, Engineered Chimeric Peptides as Antimicrobial Surface Coating Agents toward Infection-Free Implants, ACS Appl. Mater. Interfaces, 2016, 8(8), 5070–5081, DOI:10.1021/acsami.5b03697.
- C. M. M. Motta, K. J. Endres, C. Wesdemiotis, R. K. Willits and M. L. Becker, Enhancing Schwann Cell Migration Using Concentration Gradients of Laminin-Derived Peptides, Biomaterials, 2019, 218(February), 119335, DOI:10.1016/j.biomaterials.2019.119335.
- M. Hoyos-Nogués, F. Velasco, M.-P. Ginebra, J. M. Manero, F. J. Gil and C. Mas-Moruno, Regenerating Bone via Multifunctional Coatings: The Blending of Cell Integration and Bacterial Inhibition Properties on the Surface of Biomaterials, ACS Appl. Mater. Interfaces, 2017, 9(26), 21618–21630, DOI:10.1021/acsami.7b03127.
- J. L. Eisenberg, J. L. Piper and M. Mrksich, Using Self-Assembled Monolayers to Model Cell Adhesion to the 9th and 10th Type III Domains of Fibronectin, Langmuir, 2009, 25(24), 13942–13951, DOI:10.1021/la901528c.
- G. A. Hudalla, N. A. Kouris, J. T. Koepsel, B. M. Ogle and W. L. Murphy, Harnessing Endogenous Growth Factor Activity Modulates Stem Cell Behavior, Integr. Biol., 2011, 3(8), 832–842, 10.1039/c1ib00021g.
- W. Cheng, J. Ma, D. Kong, Z. Zhang, A. Khan, C. Yi, K. Hu, Y. Yi and J. Li, One Step Electrochemical Detection for Matrix Metalloproteinase 2 Based on Anodic Stripping of Silver Nanoparticles Mediated by Host-Guest Interactions, Sens. Actuators, B, 2021, 330(September 2020), 129379, DOI:10.1016/j.snb.2020.129379.
- I. Strzemińska, S. Sainte Rose Fanchine, G. Anquetin, S. Reisberg, V. Noël, M. C. Pham and B. Piro, Grafting of a Peptide Probe for Prostate-Specific Antigen Detection Using Diazonium Electroreduction and Click Chemistry, Biosens. Bioelectron., 2016, 81, 131–137, DOI:10.1016/j.bios.2016.02.060.
- M. Kato and M. Mrksich, Using Model Substrates to Study the Dependence of Focal Adhesion Formation on the Affinity of Integrin-Ligand Complexes, Biochemistry, 2004, 43(10), 2699–2707, DOI:10.1021/bi0352670.
- L. M. Wilder, W. A. Fies, C. Rabin, L. J. Webb and R. M. Crooks, Conjugation of an α-Helical Peptide to the Surface of Gold Nanoparticles, Langmuir, 2019, 35(9), 3363–3371, DOI:10.1021/acs.langmuir.9b00075.
- B. T. Houseman, E. S. Gawalt and M. Mrksich, Maleimide-Functionalized Self-Assembled Monolayers for the Preparation of Peptide and Carbohydrate Biochips, Langmuir, 2003, 19(5), 1522–1531, DOI:10.1021/la0262304.
- R. Derda, S. Musah, B. P. Orner, J. R. Klim, L. Li and L. L. Kiessling, High-Throughput Discovery of Synthetic Surfaces That Support Proliferation of Pluripotent Cells, J. Am. Chem. Soc., 2010, 132(4), 1289–1295, DOI:10.1021/ja906089g.
- D. Barros, P. Parreira, J. Furtado, F. Ferreira-da-Silva, E. Conde-Sousa, A. J. García, M. C. L. Martins, I. F. Amaral and A. P. Pêgo, An Affinity-Based Approach to Engineer Laminin-Presenting Cell Instructive Microenvironments, Biomaterials, 2019, 192(July 2018), 601–611, DOI:10.1016/j.biomaterials.2018.10.039.
- N. Laurent, R. Haddoub, J. Voglmeir, S. C. C. Wong, S. J. Gaskell and S. L. Flitsch, SPOT Synthesis of Peptide Arrays on Self-Assembled Monolayers and Their Evaluation as Enzyme Substrates, ChemBioChem, 2008, 9(16), 2592–2596, DOI:10.1002/cbic.200800481.
- X. Hun, Y. Xu and X. Luo, Peptide-Based Biosensor for the Prostate-Specific Antigen Using Magnetic Particle-Bound Invertase and a Personal Glucose Meter for Readout, Microchim. Acta, 2015, 182(9–10), 1669–1675, DOI:10.1007/s00604-015-1483-y.
- M. S. Mannoor, S. Zhang, A. J. Link and M. C. McAlpine, Electrical Detection of Pathogenic Bacteria via Immobilized Antimicrobial Peptides, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(45), 19207–19212, DOI:10.1073/pnas.1008768107.
- M. Godoy-Gallardo, C. Mas-Moruno, M. C. Fernández-Calderón, C. Pérez-Giraldo, J. M. Manero, F. Albericio, F. J. Gil and D. Rodríguez, Covalent Immobilization of HLf1–11 Peptide on a Titanium Surface Reduces Bacterial Adhesion and Biofilm Formation, Acta Biomater., 2014, 10(8), 3522–3534, DOI:10.1016/j.actbio.2014.03.026.
- K. Kerman and H. B. Kraatz, Electrochemical Probing of HIV Enzymes Using Ferrocene-Conjugated Peptides on Surfaces, Analyst, 2009, 134(12), 2400–2404, 10.1039/b912083a.
- I. M. Martins, R. L. Reis and H. S. Azevedo, Phage Display Technology in Biomaterials Engineering: Progress and Opportunities for Applications in Regenerative Medicine, ACS Chem. Biol., 2016, 11(11), 2962–2980, DOI:10.1021/acschembio.5b00717.
- S. H. Baek, M. W. Kim, C. Y. Park, C. S. Choi, S. K. Kailasa, J. P. Park and T. J. Park, Development of a Rapid and Sensitive Electrochemical Biosensor for Detection of Human Norovirus via Novel Specific Binding Peptides, Biosens. Bioelectron., 2019, 123(June 2018), 223–229, DOI:10.1016/j.bios.2018.08.064.
- N. Yu, X. Zhang, Y. Gao, H. You, J. Zhang and P. Miao, Highly Sensitive Endotoxin Assay Combining Peptide/Graphene Oxide and DNA-Modified Gold Nanoparticles, ACS Omega, 2019, 4(10), 14312–14316, DOI:10.1021/acsomega.9b02013.
- G. P. Smith and V. A. Petrenko, Phage Display, Chem. Rev., 1997, 97(2), 391–410, DOI:10.1021/cr960065d.
- J. T. Koepsel, E. H. Nguyen and W. L. Murphy, Differential Effects of a Soluble or Immobilized VEGFR-Binding Peptide, Integr. Biol., 2012, 4(8), 914–924, 10.1039/c2ib20055d.
- M. Cimino, P. Parreira, V. Leiro, A. Sousa, R. M. Gonçalves, C. C. Barrias and M. C. L. Martins, Enhancement of HMSC In Vitro Proliferation by Surface Immobilization of a Heparin-Binding Peptide, Molecules, 2023, 28(8), 1–13, DOI:10.3390/molecules28083422.
- M. P. Lutolf and J. A. Hubbell, Synthetic Biomaterials as Instructive Extracellular Microenvironments for Morphogenesis in Tissue Engineering, Nat. Biotechnol., 2005, 23(1), 47–55, DOI:10.1038/nbt1055.
- H. S. Azevedo and I. Pashkuleva, Biomimetic Supramolecular Designs for the Controlled Release of Growth Factors in Bone Regeneration, Adv. Drug Delivery Rev., 2015, 94, 63–76, DOI:10.1016/j.addr.2015.08.003.
- J. Silva and H. S. Azevedo, Bioengineered Hyaluronan Hydrogels for the Delivery of Molecular and Cellular Therapies, Adv. Ther., 2023, 7, 2300182, DOI:10.1002/adtp.202300182.
- A. J. Day and G. D. Prestwich, Hyaluronan-Binding Proteins: Tying Up the Giant, J. Biol. Chem., 2002, 277(7), 4585–4588, DOI:10.1074/jbc.R100036200.
- M. E. Mummert, M. Mohamadzadeh, D. I. Mummert, N. Mizumoto and A. Takashima, Development of a Peptide Inhibitor of Hyaluronan-Mediated Leukocyte Trafficking, J. Exp. Med., 2000, 192(6), 769–780, DOI:10.1084/jem.192.6.769.
- S. R. Rudrabhatla, W. M. Petroll, C. L. Mahaffey and M. E. Mummert, Development of a Hyaluronan Targeted Contrast Reagent for the Demarcation of Melanoma Margins in Vivo [6], J. Invest. Dermatol., 2008, 128(3), 740–742, DOI:10.1038/sj.jid.5701097.
- J. M. Zmolik and M. E. Mummert, Pep-1 as a Novel Probe for the in Situ Detection of Hyaluronan, J. Histochem. Cytochem., 2005, 53(6), 745–751, DOI:10.1369/jhc.4A6491.2005.
- G. M. Campo, A. Micali, A. Avenoso, A. D’Ascola, M. Scuruchi, A. Pisani, A. Bruschetta, A. Calatroni, D. Puzzolo and S. Campo, Inhibition of Small HA Fragment Activity and Stimulation of A2A Adenosine Receptor Pathway Limit Apoptosis and Reduce Cartilage Damage in Experimental Arthritis, Histochem. Cell Biol., 2015, 143(5), 531–543, DOI:10.1007/s00418-014-1298-7.
- R. C. Savani, G. Cao, P. M. Pooler, A. Zaman, Z. Zhou and H. M. DeLisser, Differential Involvement of the Hyaluronan (HA) Receptors CD44 and Receptor for HA-Mediated Motility in Endothelial Cell Function and Angiogenesis, J. Biol. Chem., 2001, 276(39), 36770–36778, DOI:10.1074/jbc.M102273200.
- S. L. Schor, I. Ellis, J. Banyard and A. M. Schor, Motogenic Activity of IGD-Containing Synthetic Peptides, J. Cell Sci., 1999, 112(22), 3879–3888, DOI:10.1242/jcs.112.22.3879.
- S. Ayama-Canden, R. Tondo, L. Piñeros, N. Ninane, C. Demazy, M. Dieu, A. Fattaccioli, T. Tabarrant, S. Lucas and D. Bonifazi, et al., IGDQ Motogenic Peptide Gradient Induces Directional Cell Migration through Integrin (Av)B3 Activation in MDA-MB-231 Metastatic Breast Cancer Cells, Neoplasia, 2022, 31(C), 100816, DOI:10.1016/j.neo.2022.100816.
- F. De Leo, R. Marega, V. Corvaglia, R. Tondo, M. Lo Cicero, S. Silvestrini and D. Bonifazi, Unfolding IGDQ Peptides for Engineering Motogenic Interfaces, Langmuir, 2017, 33(30), 7512–7528, DOI:10.1021/acs.langmuir.6b04381.
- L. D. D’Andrea, G. Iaccarino, R. Fattorusso, D. Sorriento, C. Carannante, D. Capasso, B. Trimarco and C. Pedone, Targeting Angiogenesis: Structural Characterization and Biological Properties of a de Novo Engineered VEGF Mimicking Peptide, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(40), 14215–14220, DOI:10.1073/pnas.0505047102.
- N. N. Casillas-Ituarte, C. H. B. Cruz, R. D. Lins, A. C. DiBartola, J. Howard, X. Liang, M. Höök, I. F. T. Viana, M. R. Sierra-Hernández and S. K. Lower, Amino Acid Polymorphisms in the Fibronectin-Binding Repeats of Fibronectin-Binding Protein A Affect Bond Strength and Fibronectin Conformation, J. Biol. Chem., 2017, 292(21), 8797–8810, DOI:10.1074/jbc.M117.786012.
- G. A. Hudalla and W. L. Murphy, Immobilization of Peptides with Distinct Biological Activities onto Stem Cell Culture Substrates Using Orthogonal Chemistries, Langmuir, 2010, 26(9), 6449–6456, DOI:10.1021/la1008208.
- M. C. L. Martins, S. A. Curtin, S. C. Freitas, P. Salgueiro, B. D. Ratner and M. A. Barbosa, Molecularly Designed Surfaces for Blood Deheparinization Using an Immobilized Heparin-Binding Peptide, J. Biomed. Mater. Res., Part A, 2009, 88A(1), 162–173, DOI:10.1002/jbm.a.31849.
- D. Zhang and K. A. Kilian, Peptide Microarrays for the Discovery of Bioactive Surfaces That Guide Cellular Processes: A Single Step Azide-Alkyne “Click” Chemistry Approach, J. Mater. Chem. B, 2014, 2(27), 4280–4288, 10.1039/c4tb00375f.
- J. Iturri, L. García-Fernández, U. Reuning, A. J. García, A. Del Campo and M. J. Salierno, Synchronized Cell Attachment Triggered by Photo-Activatable Adhesive Ligands Allows QCM-Based Detection of Early Integrin Binding, Sci. Rep., 2015, 5, 1–8, DOI:10.1038/srep09533.
- P. Bugga and M. Mrksich, Sequential Photoactivation of Self-Assembled Monolayers to Direct Cell Adhesion and Migration, Langmuir, 2019, 35(17), 5937–5943, DOI:10.1021/acs.langmuir.8b04203.
- K. Jans, B. Van Meerbergen, G. Reekmans, K. Bonroy, W. Annaert, G. Maes, Y. Engelborghs, G. Borghs and C. Bartic, Chemical and Biological Characterization of Thiol SAMs for Neuronal Cell Attachment, Langmuir, 2009, 25(8), 4564–4570, DOI:10.1021/la802217r.
- A. Ramasubramanian, R. Muckom, C. Sugnaux, C. Fuentes, B. L. Ekerdt, D. S. Clark, K. E. Healy and D. V. Schaffer, High-Throughput Discovery of Targeted, Minimally Complex Peptide Surfaces for Human Pluripotent Stem Cell Culture, ACS Biomater. Sci. Eng., 2021, 7(4), 1344–1360, DOI:10.1021/acsbiomaterials.0c01462.
- J. Li, Y. Lei, C. Sun, W. Zheng, X. Jiang and H. Zhang, Rationally Designed Peptide Interface for Potential Modulated Cell Adhesion and Migration, Adv. Mater. Interfaces, 2015, 2, 1500335, DOI:10.1002/admi.201500335.
- S. Y. Yeung, Y. Sergeeva, G. Pan, S. Mittler, T. Ederth, T. Dam, P. Jönsson, Z. El-Schich, A. G. Wingren and A. Tillo, et al., Reversible Self-Assembled Monolayers with Tunable Surface Dynamics for Controlling Cell Adhesion Behavior, ACS Appl. Mater. Interfaces, 2022, 14(37), 41790–41799, DOI:10.1021/acsami.2c12029.
- W. Luo, S. M. Legge, J. Luo, F. Lagugné-Labarthet and M. S. Workentin, Investigation of Au SAMs Photoclick Derivatization by PM-IRRAS, Langmuir, 2020, 36(4), 1014–1022, DOI:10.1021/acs.langmuir.9b03782.
- K. Amar, I. I. Suni and F. Chowdhury, A Quartz Crystal Microbalance Based Study Reveals Living Cell Loading Rate via Avβ3 Integrins, Biochem. Biophys. Res. Commun., 2020, 524(4), 1051–1056, DOI:10.1016/j.bbrc.2020.01.149.
- M. Martínez-Miguel, M. Castellote-Borrell, M. Köber, A. R. Kyvik, J. Tomsen-Melero, G. Vargas-Nadal, J. Muñoz, D. Pulido, E. Cristóbal-Lecina and S. Passemard, et al., Hierarchical Quatsome-RGD Nanoarchitectonic Surfaces for Enhanced Integrin-Mediated Cell Adhesion, ACS Appl. Mater. Interfaces, 2022, 14(42), 48179–48193, DOI:10.1021/acsami.2c10497.
- J. M. Kaplan, J. Shang, P. Gobbo, S. Antonello, L. Armelao, V. Chatare, D. M. Ratner, R. B. Andrade and F. Maran, Conformationally Constrained Functional Peptide Monolayers for the Controlled Display of Bioactive Carbohydrate Ligands, Langmuir, 2013, 29(26), 8187–8192, DOI:10.1021/la4008894.
- B. E. Turk, L. L. Huang, E. T. Piro and L. C. Cantley, Determination of Protease Cleavage Site Motifs Using Mixture-Based Oriented Peptide Libraries, Nat. Biotechnol., 2001, 19(7), 661–667, DOI:10.1038/90273.
- S. Ohkubo, K. Miyadera, Y. Sugimoto, K. Matsuo, K. Wierzba and Y. Yamada, Substrate Phage as a Tool to Identify Novel Substrate Sequences of Proteases, Comb. Chem. High Throughput Screen., 2001, 4(7), 573–583, DOI:10.2174/1386207013330788.
- J. Patterson and J. A. Hubbell, Enhanced Proteolytic Degradation of Molecularly Engineered PEG Hydrogels in Response to MMP-1 and MMP-2, Biomaterials, 2010, 31(30), 7836–7845, DOI:10.1016/j.biomaterials.2010.06.061.
- M. Magana, M. Pushpanathan, A. L. Santos, L. Leanse, M. Fernandez, A. Ioannidis, M. A. Giulianotti, Y. Apidianakis, S. Bradfute and A. L. Ferguson, et al., The Value of Antimicrobial Peptides in the Age of Resistance, Lancet Infect. Dis., 2020, 20(9), e216–e230, DOI:10.1016/S1473-3099(20)30327-3.
- D. R. Fonseca, A. Moura, V. Leiro, R. Silva-Carvalho, B. N. Estevinho, C. L. Seabra, P. C. Henriques, M. Lucena, C. Teixeira and P. Gomes, et al., Grafting MSI-78A onto Chitosan Microspheres Enhances Its Antimicrobial Activity, Acta Biomater., 2022, 137, 186–198, DOI:10.1016/j.actbio.2021.09.063.
- C. Chen, J. Shi, D. Wang, P. Kong, Z. Wang and Y. Liu, Antimicrobial Peptides as Promising Antibiotic Adjuvants to Combat Drug-Resistant Pathogens, Crit. Rev. Microbiol., 2023, 1–18, DOI:10.1080/1040841X.2023.2186215.
- F. Costa, I. F. Carvalho, R. C. Montelaro, P. Gomes and M. C. L. Martins, Covalent Immobilization of Antimicrobial Peptides (AMPs) onto Biomaterial Surfaces, Acta Biomater., 2011, 7(4), 1431–1440, DOI:10.1016/j.actbio.2010.11.005.
- Y. Shi, D. W. Wareham, L. M. Phee and H. S. Azevedo, Self-Assembled Peptide Nanostructures for Antibacterial Applications, Peptide-based Biomaterials, The Royal Society of Chemistry, 2020, pp. 395–428 10.1039/9781839161148-00395.
- J. Huang, Y. Xu, Y. Xue, Y. Huang, X. Li, X. Chen, Y. Xu, D. Zhang, P. Zhang and J. Zhao, et al., Identification of Potent Antimicrobial Peptides via a Machine-Learning Pipeline That Mines the Entire Space of Peptide Sequences, Nat. Biomed. Eng., 2023, 7(6), 797–810, DOI:10.1038/s41551-022-00991-2.
- C. Monteiro, F. Costa, A. M. Pirttilä, M. V. Tejesvi and M. C. L. Martins, Prevention of Urinary Catheter-Associated Infections by Coating Antimicrobial Peptides from Crowberry Endophytes, Sci. Rep., 2019, 9(1), 1–14, DOI:10.1038/s41598-019-47108-5.
- P. Parreira, C. Monteiro, V. Graça, J. Gomes, S. Maia, P. Gomes, I. C. Gonçalves and M. C. L. Martins, Surface Grafted MSI-78A Antimicrobial Peptide Has High Potential for Gastric Infection Management, Sci. Rep., 2019, 9(1), 1–11, DOI:10.1038/s41598-019-53918-4.
- A. J. Scott, A. Niitsu, H. T. Kratochvil, E. J. M. Lang, J. T. Sengel, W. M. Dawson, K. R. Mahendran, M. Mravic, A. R. Thomson and R. L. Brady, et al., Constructing Ion Channels from Water-Soluble α-Helical Barrels, Nat. Chem., 2021, 13(7), 643–650, DOI:10.1038/s41557-021-00688-0.
- T. Morita and S. Kimura, Long-Range Electron Transfer over 4 Nm Governed by an Inelastic Hopping Mechanism in Self-Assembled Monolayers of Helical Peptides, J. Am. Chem. Soc., 2003, 125(29), 8732–8733, DOI:10.1021/ja034872n.
- Y. Miura, S. Kimura, S. Kobayashi, M. Iwamoto, Y. Imanishi and J. Umemura, Negative Surface Potential Produced by Self-Assembled Monolayers of Helix Peptides Oriented Vertically to a Surface, Chem. Phys. Lett., 1999, 315(1–2), 1–6, DOI:10.1016/S0009-2614(99)01191-4.
- T. He, G. Abbineni, B. Cao and C. Mao, Nanofibrous Bio-Inorganic Hybrid Structures Formed Through Self-Assembly and Oriented Mineralization of Genetically Engineered Phage Nanofibers, Small, 2010, 6(20), 2230–2235, DOI:10.1002/smll.201001108.
- D. K. Yarbrough, E. Hagerman, R. Eckert, J. He, H. Choi, N. Cao, K. Le, J. Hedger, F. Qi and M. Anderson, et al., Specific Binding and Mineralization of Calcified Surfaces by Small Peptides, Calcif. Tissue Int., 2010, 86(1), 58–66, DOI:10.1007/s00223-009-9312-0.
- H. Xu, B. Cao, A. George and C. Mao, Self-Assembly and Mineralization of Genetically Modifiable Biological Nanofibers Driven by β-Structure Formation, Biomacromolecules, 2011, 12(6), 2193–2199, DOI:10.1021/bm200274r.
- K. Shuturminska, N. V. Tarakina, H. S. Azevedo, A. J. Bushby, A. Mata, P. Anderson and M. Al-Jawad, Elastin-Like Protein, with Statherin Derived Peptide, Controls Fluorapatite Formation and Morphology, Front. Physiol., 2017, 8, 368, DOI:10.3389/fphys.2017.00368.
- H. Yazici, G. Habib, K. Boone, M. Urgen, F. S. Utku and C. Tamerler, Self-Assembling Antimicrobial Peptides on Nanotubular Titanium Surfaces Coated with Calcium Phosphate for Local Therapy, Mater. Sci. Eng., C, 2019, 94, 333–343, DOI:10.1016/j.msec.2018.09.030.
- S. Wang, Y. Yang, R. Wang, X. Kong and X. Wang, Mineralization of Calcium Phosphate Controlled by Biomimetic Self-Assembled Peptide Monolayers via Surface Electrostatic Potentials, Bioact. Mater., 2020, 5(2), 387–397, DOI:10.1016/j.bioactmat.2020.03.003.
- X. Wang, I. Bergenfeld, P. S. Arora and J. W. Canary, Reversible Redox Reconfiguration of Secondary Structures in a Designed Peptide, Angew. Chemie, 2012, 124(48), 12265–12267, DOI:10.1002/ange.201206009.
- A. K. Nowinski, F. Sun, A. D. White, A. J. Keefe and S. Jiang, Sequence, Structure, and Function of Peptide Self-Assembled Monolayers, J. Am. Chem. Soc., 2012, 134(13), 6000–6005, DOI:10.1021/ja3006868.
- C. Overby, S. Park, A. Summers and D. S. W. Benoit, Zwitterionic Peptides: Tunable next-Generation Stealth Nanoparticle Modifications, Bioact. Mater., 2023, 27, 113–124, DOI:10.1016/j.bioactmat.2023.03.020.
- Y. Noguchi, Y. Iwasaki, M. Ueda and S. Kakinoki, Surfaces Immobilized with Oligo-Prolines Prevent Protein Adsorption and Cell Adhesion, J. Mater. Chem. B, 2020, 8(11), 2233–2237, 10.1039/d0tb00051e.
- A. Mzyk, G. Imbir, Y. Noguchi, M. Sanak, R. Major, J. Wiecek, P. Kurtyka, H. Plutecka, K. Trembecka-Wójciga and Y. Iwasaki, et al., Dynamic in Vitro Hemocompatibility of Oligoproline Self-Assembled Monolayer Surfaces, Biomater. Sci., 2022, 10(19), 5498–5503, 10.1039/d2bm00885h.
- N. Liu, Y. Ma, R. Han, S. Lv, P. Wang and X. Luo, Antifouling Biosensors for Reliable Protein Quantification in Serum Based on Designed All-in-One Branched Peptides, Chem. Commun., 2021, 57(6), 777–780, 10.1039/D0CC07220F.
- S. Acosta, L. Quintanilla, M. Alonso, C. Aparicio and J. C. Rodríguez-Cabello, Recombinant AMP/Polypeptide Self-Assembled Monolayers with Synergistic Antimicrobial Properties for Bacterial Strains of Medical Relevance, ACS Biomater. Sci. Eng., 2019, 5(9), 4708–4716, DOI:10.1021/acsbiomaterials.9b00247.
- A. Rai, S. Pinto, M. B. Evangelista, H. Gil, S. Kallip, M. G. S. Ferreira and L. Ferreira, High-Density Antimicrobial Peptide Coating with Broad Activity and Low Cytotoxicity against Human Cells, Acta Biomater., 2016, 33, 64–77, DOI:10.1016/j.actbio.2016.01.035.
- M. M. Querido, H. P. Felgueiras, A. Rai, F. Costa, C. Monteiro, I. Borges, D. Oliveira, L. Ferreira and M. C. L. Martins, Cecropin–Melittin Functionalized Polyurethane Surfaces Prevent Staphylococcus Epidermidis Adhesion without Inducing Platelet Adhesion and Activation, Adv. Mater. Interfaces, 2018, 5, 1801390, DOI:10.1002/admi.201801390.
- M. Xiao, J. Jasensky, J. Gerszberg, J. Chen, J. Tian, T. Lin, T. Lu, J. Lahann and Z. Chen, Chemically Immobilized Antimicrobial Peptide on Polymer and Self-Assembled Monolayer Substrates, Langmuir, 2018, 34(43), 12889–12896, DOI:10.1021/acs.langmuir.8b02377.
- M. Godoy-Gallardo, C. Mas-Moruno, K. Yu, J. M. Manero, F. J. Gil, J. N. Kizhakkedathu and D. Rodriguez, Antibacterial Properties of HLf1–11 Peptide onto Titanium Surfaces: A Comparison Study Between Silanization and Surface Initiated Polymerization, Biomacromolecules, 2015, 16(2), 483–496, DOI:10.1021/bm501528x.
- V. P. Koidou, P. P. Argyris, E. P. Skoe, J. Mota Siqueira, X. Chen, L. Zhang, J. E. Hinrichs, M. Costalonga and C. Aparicio, Peptide Coatings Enhance Keratinocyte Attachment towards Improving the Peri-Implant Mucosal Seal, Biomater. Sci., 2018, 6(7), 1936–1945, 10.1039/C8BM00300A.
- K. V. Holmberg, M. Abdolhosseini, Y. Li, X. Chen, S.-U. Gorr and C. Aparicio, Bio-Inspired Stable Antimicrobial Peptide Coatings for Dental Applications, Acta Biomater., 2013, 9(9), 8224–8231, DOI:10.1016/j.actbio.2013.06.017.
- J. Chen, Y. Zhu, M. Xiong, G. Hu, J. Zhan, T. Li, L. Wang and Y. Wang, Antimicrobial Titanium Surface via Click-Immobilization of Peptide and Its in Vitro/Vivo Activity, ACS Biomater. Sci. Eng., 2019, 5(2), 1034–1044, DOI:10.1021/acsbiomaterials.8b01046.
- B. Mishra and G. Wang, Titanium Surfaces Immobilized with the Major Antimicrobial Fragment FK-16 of Human Cathelicidin LL-37 Are Potent against Multiple Antibiotic-Resistant Bacteria, Biofouling, 2017, 33(7), 544–555, DOI:10.1080/08927014.2017.1332186.
- S. C. Freitas, M. A. Barbosa and M. C. L. Martins, The Effect of Immobilization of Thrombin Inhibitors onto Self-Assembled Monolayers on the Adsorption and Activity of Thrombin, Biomaterials, 2010, 31(14), 3772–3780, DOI:10.1016/j.biomaterials.2010.01.097.
- A. J. Wain, H. N. L. Do, H. S. Mandal, H.-B. Kraatz and F. Zhou, Influence of Molecular Dipole Moment on the Redox-Induced Reorganization of α-Helical Peptide Self-Assembled Monolayers: An Electrochemical SPR Investigation, J. Phys. Chem. C, 2008, 112(37), 14513–14519, DOI:10.1021/jp804643c.
- H. S. Mandal and H.-B. Kraatz, Electron Transfer across α-Helical Peptides: Potential Influence of Molecular Dynamics, Chem. Phys., 2006, 326(1), 246–251, DOI:10.1016/j.chemphys.2006.01.010.
- J. Watanabe, T. Morita and S. Kimura, Effects of Dipole Moment, Linkers, and Chromophores at Side Chains on Long-Range Electron Transfer through Helical Peptides, J. Phys. Chem. B, 2005, 109(30), 14416–14425, DOI:10.1021/jp051592g.
- A. Grabarek, Ł. Walczak and P. Cyganik, Odd–Even Effect in Peptide SAMs—Competition of Secondary Structure and Molecule–Substrate Interaction, J. Phys. Chem. B, 2021, 125(39), 10964–10971, DOI:10.1021/acs.jpcb.1c06625.
- L. Sepunaru, S. Refaely-Abramson, R. Lovrinčić, Y. Gavrilov, P. Agrawal, Y. Levy, L. Kronik, I. Pecht, M. Sheves and D. Cahen, Electronic Transport via Homopeptides: The Role of Side Chains and Secondary Structure, J. Am. Chem. Soc., 2015, 137(30), 9617–9626, DOI:10.1021/jacs.5b03933.
- C. Guo, X. Yu, S. Refaely-Abramson, L. Sepunaru, T. Bendikov, I. Pecht, L. Kronik, A. Vilan, M. Sheves and D. Cahen, Tuning Electronic Transport via Hepta-Alanine Peptides Junction by Tryptophan Doping, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(39), 10785–10790, DOI:10.1073/pnas.1606779113.
- E. Mervinetsky, I. Alshanski, S. Lenfant, D. Guerin, L. Medrano Sandonas, A. Dianat, R. Gutierrez, G. Cuniberti, M. Hurevich and S. Yitzchaik, et al., Electron Transport through Self-Assembled Monolayers of Tripeptides, J. Phys. Chem. C, 2019, 123(14), 9600–9608, DOI:10.1021/acs.jpcc.9b01082.
- X. Zhu, Z. Xu, X. Li and C. Guo, Charge Migration of Ferrocene-Labeled Peptide Self-Assembled Monolayers at Various Interfaces: The Roles of Peptide Composition, Electrochim. Acta, 2023, 454, 142419, DOI:10.1016/j.electacta.2023.142419.
- J. Pawlowski, J. Juhaniewicz, D. Tymecka and S. Sek, Electron Transfer Across α-Helical Peptide Monolayers: Importance of Interchain Coupling, Langmuir, 2012, 28(50), 17287–17294, DOI:10.1021/la302716n.
- X. Wang, W. Jiang, Q. Zheng, L. Yan, Y. Jin, C. Han, J. Zhuang, H. Liu and Z. Li, Piezoelectric-Enhanced Oriented Cobalt Coordinated Peptide Monolayer with Rectification Behavior, Small, 2015, 11(37), 4864–4869, DOI:10.1002/smll.201500857.
- M. Venanzi, E. Gatto, M. Caruso, A. Porchetta, F. Formaggio and C. Toniolo, Photoinduced Electron Transfer through Peptide-Based Self-Assembled Monolayers Chemisorbed on Gold Electrodes: Directing the Flow-in and Flow-out of Electrons through Peptide Helices, J. Phys. Chem. A, 2014, 118(33), 6674–6684, DOI:10.1021/jp503791w.
- H. S. Mandal and H. B. Kraatz, Electron Transfer Mechanism in Helical Peptides, J. Phys. Chem. Lett., 2012, 3(6), 709–713, DOI:10.1021/jz300008s.
- S. Kubitzky, M. Venanzi, B. Biondi, R. Lettieri, M. De Zotti and E. Gatto, A PH-Induced Reversible Conformational Switch Able to Control the Photocurrent Efficiency in a Peptide Supramolecular System, Chem. – Eur. J., 2021, 27(8), 2810–2817, DOI:10.1002/chem.202004527.
- J. T. First and L. J. Webb, Agreement between Experimental and Simulated Circular Dichroic Spectra of a Positively Charged Peptide in Aqueous Solution and on Self-Assembled Monolayers, J. Phys. Chem. B, 2019, 123(21), 4512–4526, DOI:10.1021/acs.jpcb.9b02102.
- S. Chen, Z. Cao and S. Jiang, Ultra-Low Fouling Peptide Surfaces Derived from Natural Amino Acids, Biomaterials, 2009, 30(29), 5892–5896, DOI:10.1016/j.biomaterials.2009.07.001.
- R. Chelmowski, S. D. Köster, A. Kerstan, A. Prekelt, C. Grunwald, T. Winkler, N. Metzler-Nolte, A. Terfort and C. Wöll, Peptide-Based SAMs That Resist the Adsorption of Proteins, J. Am. Chem. Soc., 2008, 130(45), 14952–14953, DOI:10.1021/ja8065754.
- R. Chang, E. A. Quimada Mondarte, D. Palai, T. Sekine, A. Kashiwazaki, D. Murakami, M. Tanaka and T. Hayashi, Protein- and Cell-Resistance of Zwitterionic Peptide-Based Self-Assembled Monolayers: Anti-Biofouling Tests and Surface Force Analysis, Front. Chem., 2021, 9(October), 1–9, DOI:10.3389/fchem.2021.748017.
- C. D. Beyer, S. Thavalingam, T. Guseva, L. Schardt, R. Zimmermann, C. Werner, P. Dietze, J. E. Bandow, N. Metzler-Nolte and A. Rosenhahn, Zwitterionic Peptides Reduce Accumulation of Marine and Freshwater Biofilm Formers, ACS Appl. Mater. Interfaces, 2021, 13(42), 49682–49691, DOI:10.1021/acsami.1c13459.
- A. M. Ghafari, S. E. Domínguez, V. Järvinen, Z. Gounani, A. Schmit, M. Sjöqvist, C. Sahlgren, O. M. H. Salo-Ahen, C. Kvarnström and L. Torsi, et al., In Situ Coupled Electrochemical-Goniometry as a Tool to Reveal Conformational Changes of Charged Peptides, Adv. Mater. Interfaces, 2022, 9(4), 1–9, DOI:10.1002/admi.202101480.
- N. S. Kehr, K. Riehemann, J. El-Gindi, A. Schäfer, H. Fuchs, H. J. Galla and L. De Cola, Cell Adhesion and Cellular Patterning on a Self-Assembled Monolayer of Zeolite L Crystals, Adv. Funct. Mater., 2010, 20(14), 2248–2254, DOI:10.1002/adfm.201000205.
- Y. Xia and G. M. Whitesides, Soft Lithography, Angew. Chem., Int. Ed., 1998, 37(5), 550–575, DOI:10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G.
- D. Qin, Y. Xia and G. M. Whitesides, Soft Lithography for Micro- and Nanoscale Patterning, Nat. Protoc., 2010, 5(3), 491–502, DOI:10.1038/nprot.2009.234.
- S. Lamping, C. Buten and B. J. Ravoo, Functionalization and Patterning of Self-Assembled Monolayers and Polymer Brushes Using Microcontact Chemistry, Acc. Chem. Res., 2019, 52(5), 1336–1346, DOI:10.1021/acs.accounts.9b00041.
- S. Morgenthaler, S. Lee, S. Zürcher and N. D. Spencer, A Simple, Reproducible Approach to the Preparation of Surface-Chemical Gradients, Langmuir, 2003, 19(25), 10459–10462, DOI:10.1021/la034707l.
- A. R. Kyvik, C. Luque-Corredera, D. Pulido, M. Royo, J. Veciana, J. Guasch and I. Ratera, Stimuli-Responsive Functionalization Strategies to Spatially and Temporally Control Surface Properties: Michael vs Diels–Alder Type Additions, J. Phys. Chem. B, 2018, 122(16), 4481–4490, DOI:10.1021/acs.jpcb.8b01652.
- M. Wirkner, S. Weis, V. San Miguel, M. Álvarez, R. A. Gropeanu, M. Salierno, A. Sartoris, R. E. Unger, C. J. Kirkpatrick and A. del Campo, Photoactivatable Caged Cyclic RGD Peptide for Triggering Integrin Binding and Cell Adhesion to Surfaces, ChemBioChem, 2011, 12(17), 2623–2629, DOI:10.1002/cbic.201100437.
- C. Nicosia, J. Cabanas-Danés, P. Jonkheijm and J. Huskens, A Fluorogenic Reactive Monolayer Platform for the Signaled Immobilization of Thiols, ChemBioChem, 2012, 13(6), 778–782, DOI:10.1002/cbic.201200062.
- L. Tauk, A. P. Schröder, G. Decher and N. Giuseppone, Hierarchical Functional Gradients of PH-Responsive Self-Assembled Monolayers Using Dynamic Covalent Chemistry on Surfaces, Nat. Chem., 2009, 1(8), 649–656, DOI:10.1038/nchem.400.
- B. D. Ratner and D. G. Castner, Surface Properties and Surface Characterization of Biomaterials, in Biomaterials Science, ed. Buddy Ratner, Allan Hoffman, F. S. & J. L. on behalf of the S. for B., Elsevier, San Diego, CA, USA, 2020, pp. 53–75 DOI:10.1016/B978-0-12-816137-1.00006-4.
- W. Huaiyu and P. K. Chu, Surface Characterization of Biomaterials, in Characterization of Biomaterials, ed. A. Bandyopadhyay and S. Bose, Elsevier, 2013 DOI:10.1016/B978-0-12-415800-9.01001-9.
- A. Samanta, Q. Wang, S. K. Shaw and H. Ding, Roles of Chemistry Modification for Laser Textured Metal Alloys to Achieve Extreme Surface Wetting Behaviors, Mater. Des., 2020, 192, 108744, DOI:10.1016/j.matdes.2020.108744.
- C. D. Bain and G. M. Whitesides, Correlations between Wettability and Structure in Monolayers of Alkanethiols Adsorbed on Gold, J. Am. Chem. Soc., 1988, 110(11), 3665–3666, DOI:10.1021/ja00219a055.
- M. Cimino, P. Parreira, S. J. Bidarra, R. M. Gonçalves, C. C. Barrias and M. C. L. Martins, Effect of Surface Chemistry on HMSC Growth under Xeno-Free Conditions, Colloids Surf., B, 2020, 189, 110836, DOI:10.1016/j.colsurfb.2020.110836.
- M. C. L. Martins, S. R. Sousa, J. C. Antunes and M. A. Barbosa, Protein Adsorption Characterization, 2012, pp. 141–161 DOI:10.1007/978-1-61779-388-2_10.
- J. S. Apte, G. Collier, R. A. Latour, L. J. Gamble and D. G. Castner, XPS and ToF-SIMS Investigation of α-Helical and β-Strand Peptide Adsorption onto SAMs, Langmuir, 2010, 26(5), 3423–3432, DOI:10.1021/la902888y.
- M. C. L. Martins, E. Naeemi and B. D. Ratner, et al., Albumin Adsorption on Cibacron Blue F3G-A Immobilized onto Oligo(Ethylene Glycol)-Terminated Self-Assembled Monolayers, J. Mater. Sci.: Mater. Med., 2003, 14, 945–954, DOI:10.1023/A:1026394431100.
- S. Kumar, S. Soni, W. Danowski, C. L. F. van Beek, B. L. Feringa, P. Rudolf and R. C. Chiechi, Correlating the Influence of Disulfides in Monolayers across Photoelectron Spectroscopy Wettability and Tunneling Charge-Transport, J. Am. Chem. Soc., 2020, 142(35), 15075–15083, DOI:10.1021/jacs.0c06508.
- Y. Yang, A. M. Bittner, S. Baldelli and K. Kern, Study of Self-Assembled Triethoxysilane Thin Films Made by Casting Neat Reagents in Ambient Atmosphere, Thin Solid Films, 2008, 516(12), 3948–3956, DOI:10.1016/j.tsf.2007.07.208.
- H. Gu, S. Zhu, B. Song, M. Fang, Z. Guo, X. Chen, C. Zhang, H. Jiang and S. Liu, An Analytical Method to Determine the Complex Refractive Index of an Ultra-Thin Film by Ellipsometry, Appl. Surf. Sci., 2020, 507, 145091, DOI:10.1016/j.apsusc.2019.145091.
- K. Dorywalski, I. Maciejewski and T. Krzyżyński, Spectroscopic Ellipsometry Technique as a Materials Characterization Tool for Mechatronic Systems—The Case of Composition and Doping Concentration Monitoring in SBN Crystals, Mechatronics, 2016, 37, 33–41, DOI:10.1016/j.mechatronics.2015.11.005.
- J. Homola and M. Piliarik, in Surface Plasmon Resonance (SPR) Sensors, ed. J. Homola, Berlin, Heidelberg, 2006, pp. 45–67 DOI:10.1007/5346_014.
- R. Bakhtiar, Surface Plasmon Resonance Spectroscopy: A Versatile Technique in a Biochemist's Toolbox, J. Chem. Educ., 2013, 90(2), 203–209, DOI:10.1021/ed200549g.
- E. K. Hanson and R. J. Whelan, Application of the Nicoya OpenSPR to Studies of Biomolecular Binding: A Review of the Literature from 2016 to 2022, Sensors, 2023, 23(10), 4831, DOI:10.3390/s23104831.
- V. Silin, H. Weetall and D. J. Vanderah, SPR Studies of the Nonspecific Adsorption Kinetics of Human IgG and BSA on Gold Surfaces Modified by Self-Assembled Monolayers (SAMs), J. Colloid Interface Sci., 1997, 185(1), 94–103, DOI:10.1006/jcis.1996.4586.
- F. B. Kamal Eddin and Y. W. Fen, The Principle of Nanomaterials Based Surface Plasmon Resonance Biosensors and Its Potential for Dopamine Detection, Molecules, 2020, 25(12), 2769, DOI:10.3390/molecules25122769.
- B. Kasemo, Biological Surface Science, Surf. Sci., 2002, 500(1–3), 656–677, DOI:10.1016/S0039-6028(01)01809-X.
- C. M. Jeffries, J. Ilavsky, A. Martel, S. Hinrichs, A. Meyer, J. S. Pedersen, A. V. Sokolova and D. I. Svergun, Small-Angle X-Ray and Neutron Scattering, Nat. Rev. Methods Prim., 2021, 1(1), 70, DOI:10.1038/s43586-021-00064-9.
- L. Mousavifar, P. Parreira, A. Taponard, V. C. D. Graça, M. C. L. Martins and R. Roy, Validation of Selective Capture of Fimbriated Uropathogenic Escherichia Coli by a Label-Free Engineering Detection System Using Mannosylated Surfaces, ACS Appl. Bio Mater., 2022, 5(12), 5877–5886, DOI:10.1021/acsabm.2c00838.
- T. E. Alexander, L. D. Lozeau and T. A. Camesano, QCM-D Characterization of Time-Dependence of Bacterial Adhesion, Cell Surf., 2019, 5, 100024, DOI:10.1016/j.tcsw.2019.100024.
- A. Tarnapolsky and V. Freger, Modeling QCM-D Response to Deposition and Attachment of Microparticles and Living Cells, Anal. Chem., 2018, 90(23), 13960–13968, DOI:10.1021/acs.analchem.8b03411.
- E. Briand, V. Humblot, C.-M. Pradier, B. Kasemo and S. Svedhem, An OEGylated Thiol Monolayer for the Tethering of Liposomes and the Study of Liposome Interactions, Talanta, 2010, 81(4–5), 1153–1161, DOI:10.1016/j.talanta.2010.01.027.
- O. Azzaroni, M. Mir and W. Knoll, Supramolecular Architectures of Streptavidin on Biotinylated Self-Assembled Monolayers. Tracking Biomolecular Reorganization after Bioconjugation, J. Phys. Chem. B, 2007, 111(48), 13499–13503, DOI:10.1021/jp076707q.
- K. A. Mahmoud and J. H. T. Luong, Impedance Method for Detecting HIV-1 Protease and Screening for Its Inhibitors Using Ferrocene-Peptide Conjugate/Au Nanoparticle/Single-Walled Carbon Nanotube Modified Electrode, Anal. Chem., 2008, 80(18), 7056–7062, DOI:10.1021/ac801174r.
- F. Ma, J. Yan, L. Sun and Y. Chen, Electrochemical Impedance Spectroscopy for Quantization of Matrix Metalloproteinase-14 Based on Peptides Inhibiting Its Homodimerization and Heterodimerization, Talanta, 2019, 205(July), 120142, DOI:10.1016/j.talanta.2019.120142.
- A. Tricase, D. Blasi, A. Favia, A. Stefanachi, F. Leonetti, G. Colafemmina, L. Torsi and G. Scamarcio, Surface Composition of Mixed Self-Assembled Monolayers on Au by Infrared Attenuated Total Reflection Spectroscopy, Appl. Surf. Sci., 2021, 559, 149883, DOI:10.1016/j.apsusc.2021.149883.
- M. A. Ramin, G. Le Bourdon, N. Daugey, B. Bennetau, L. Vellutini and T. Buffeteau, PM-IRRAS Investigation of Self-Assembled Monolayers Grafted onto SiO2/Au Substrates, Langmuir, 2011, 27(10), 6076–6084, DOI:10.1021/la2006293.
- T. Ederth, M. Lerm, B. Orihuela and D. Rittschof, Resistance of Zwitterionic Peptide Monolayers to Biofouling, Langmuir, 2019, 35(5), 1818–1827, DOI:10.1021/acs.langmuir.8b01625.
- M. Zabara, Q. Ren, H. Amenitsch and S. Salentinig, Bioinspired Antimicrobial Coatings from Peptide-Functionalized Liquid Crystalline Nanostructures, ACS Appl. Bio Mater., 2021, 4(6), 5295–5303, DOI:10.1021/acsabm.1c00415.
- H. Wu, G. Li, J. Hou and K. Sotthewes, Probing Surface Properties of Organic Molecular Layers by Scanning Tunneling Microscopy, Adv. Colloid Interface Sci., 2023, 318, 102956, DOI:10.1016/j.cis.2023.102956.
- B. Bhushan and O. Marti, Scanning Probe Microscopy – Principle of Operation, Instrumentation, and Probes, Springer Handbook of Nanotechnology, Springer Berlin Heidelberg, Berlin, Heidelberg, 2007, pp. 591–636 DOI:10.1007/978-3-540-29857-1_22.
- J. Joshi, S. V. Homburg and A. Ehrmann, Atomic Force Microscopy (AFM) on Biopolymers and Hydrogels for Biotechnological Applications—Possibilities and Limits, Polymers, 2022, 14(6), 1267, DOI:10.3390/polym14061267.
- L. Kang, Q. Wang, L. Zhang, H. Zou, J. Gao, K. Niu and N. Jiang, Recent Experimental Advances in Characterizing the Self-Assembly and Phase Behavior of Polypeptoids, Materials, 2023, 16(11), 4175, DOI:10.3390/ma16114175.
- Q. Wang, L. Kang, X. Xu, M. Zhang, A. Chao, J. Chen, Z. Han, H. Yu, R. Li and Y. Zhao, et al., Multiscale Crystalline Structure of Confined Polypeptoid Films: The Effect of Alkyl Side Chain Branching, ACS Macro Lett., 2022, 11(9), 1060–1066, DOI:10.1021/acsmacrolett.2c00271.
- K. Nakano, J. Horiuchi, S. Hirata, M. Yamanaka, T. Himeno and R. Ishimatsu, Folding and Assembly of Vanilloid Receptor Secondary-Structure Peptide with Hexahistidine Linker at Nickel-Nitrilotriacetic Acid Monolayer for Capsaicin Recognition, Langmuir, 2019, 35(6), 2047–2054, DOI:10.1021/acs.langmuir.8b03202.
- C. Dey, M. Roy and S. G. Dey, Insights from Self-Assembled Aggregates of Amyloid β Peptides on Gold Surfaces, ACS Omega, 2022, 7(12), 9973–9983, DOI:10.1021/acsomega.1c06056.
- O. R. Bolduc, C. M. Clouthier, J. N. Pelletier and J. F. Masson, Peptide Self-Assembled Monolayers for Label-Free and Unamplified Surface Plasmon Resonance Biosensing in Crude Cell Lysate, Anal. Chem., 2009, 81(16), 6779–6788, DOI:10.1021/ac900956y.
- C. D. Beyer, M. L. Reback, S. M. Gopal, K. A. Nolte, J. A. Finlay, A. S. Clare, L. V. Schäfer, N. Metzler-Nolte and A. Rosenhahn, α-Aminoisobutyric Acid-Stabilized Peptide SAMs with Low Nonspecific Protein Adsorption and Resistance against Marine Biofouling, ACS Sustainable Chem. Eng., 2020, 8(7), 2665–2671, DOI:10.1021/acssuschemeng.9b05889.
- M. Mrksich, Using Self-Assembled Monolayers to Model the Extracellular Matrix, Acta Biomater., 2009, 5(3), 832–841, DOI:10.1016/j.actbio.2009.01.016.
- G. A. Hudalla and W. L. Murphy, Using “Click” Chemistry to Prepare SAM Substrates to Study Stem Cell Adhesion, Langmuir, 2009, 25(10), 5737–5746, DOI:10.1021/la804077t.
- L. S. Tew, J. Y. Ching, S. H. Ngalim and Y. L. Khung, Driving Mesenchymal Stem Cell Differentiation from Self-Assembled Monolayers, RSC Adv., 2018, 8(12), 6551–6564, 10.1039/c7ra12234a.
- J. Lee, J. Yeon, W. Cheol, S. Choi, J. Lim, H. Jeong, D. Shin and Y. June, Sensors and Actuators B: Chemical A Reference Electrode-Free Electrochemical Biosensor for Detecting MMP-9 Using a Concentric Electrode Device, Sens. Actuators, B, 2017, 240, 735–741, DOI:10.1016/j.snb.2016.09.026.
- C. Ding, X. Wang and X. Luo, Dual-Mode Electrochemical Assay of Prostate-Specific Antigen Based on Antifouling Peptides Functionalized with Electrochemical Probes and Internal References, Anal. Chem., 2019, 91(24), 15846–15852, DOI:10.1021/acs.analchem.9b04206.
- Z.-M. Dong, L. Cheng, P. Zhang and G.-C. Zhao, Label-Free Analytical Performances of a Peptide Based QCM Biosensor for Trypsin, Analyst, 2020, 145, 3329–3338, 10.1039/d0an00308e.
- H. Chen, S. Jia, J. Zhang, M. Jang, X. Chen, K. Koh and Z. Wang, Sensitive Detection of Copper(<scp > ii </Scp>) Ions Based on the Conformational Change of Peptides by Surface Plasmon Resonance Spectroscopy, Anal. Methods, 2015, 7(20), 8942–8946, 10.1039/C5AY02047F.
- S. R. Denmeade, W. Lou, J. Lövgren, J. Malm, H. Lilja and J. T. Isaacs, Specific and Efficient Peptide Substrates for Assaying the Proteolytic Activity of Prostate-Specific Antigen, Cancer Res., 1997, 57(21), 4924–4930 CAS.
- J. Yu, A. Yang, N. Wang, H. Ling, J. Song, X. Chen, Y. Lian, Z. Zhang, F. Yan and M. Gu, Highly Sensitive Detection of Caspase-3 Activity Based on Peptide-Modified Organic Electrochemical Transistor Biosensors, Nanoscale, 2021, 13(5), 2868–2874, 10.1039/d0nr08453k.
- H. Chen, J. Zhang, X. Liu, Y. Gao, Z. Ye and G. Li, Colorimetric Copper(II) Ion Sensor Based on the Conformational Change of Peptide Immobilized onto the Surface of Gold Nanoparticles, Anal. Methods, 2014, 6(8), 2580–2585, 10.1039/c3ay42211a.
- Q. Hu, J. Kong, D. Han, Y. Bao, X. Zhang, Y. Zhang and L. Niu, Ultrasensitive Peptide-Based Electrochemical Detection of Protein Kinase Activity Amplified by RAFT Polymerization, Talanta, 2020, 206(May 2019), 120173, DOI:10.1016/j.talanta.2019.120173.
- C. F. Huang and M. Mrksich, Profiling Protein Tyrosine Phosphatase Specificity with Self-Assembled Monolayers for Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry and Peptide Arrays, ACS Comb. Sci., 2019, 21(11), 760–769, DOI:10.1021/acscombsci.9b00152.
- L. C. Szymczak, D. J. Sykora and M. Mrksich, Using Peptide Arrays to Profile Phosphatase Activity in Cell Lysates, Chem. – Eur. J., 2020, 26(1), 165–170, DOI:10.1002/chem.201904364.
- K. A. Mahmoud, S. Hrapovic and J. H. T. Luong, Picomolar Detection of Protease Using Peptide/Single Walled Carbon Nanotube/Gold Nanoparticle-Modified Electrode, ACS Nano, 2008, 2(5), 1051–1057, DOI:10.1021/nn8000774.
- K. Kerman, K. A. Mahmoud and H. B. Kraatz, An Electrochemical Approach for the Detection of HIV-1 Protease, Chem. Commun., 2007,(37), 3829–3831, 10.1039/b707140j.
- H. Xiao, L. Liu, F. Meng, J. Huang and G. Li, Electrochemical Approach to Detect Apoptosis, Anal. Chem., 2008, 80(13), 5272–5275, DOI:10.1021/ac8005268.
- J. Adjémian, A. Anne, G. Cauet and C. Demaille, Cleavage-Sensing Redox Peptide Monolayers for the Rapid Measurement of the Proteolytic Activity of Trypsin and α-Thrombin Enzymes, Langmuir, 2010, 26(12), 10347–10356, DOI:10.1021/la100397g.
- E. González-Fernández, M. Staderini, N. Avlonitis, A. F. Murray, A. R. Mount and M. Bradley, Chemical Effect of Spacer Length on the Performance of Peptide-Based Electrochemical Biosensors for Protease Detection, Sens. Actuators, B, 2018, 255, 3040–3046, DOI:10.1016/j.snb.2017.09.128.
- K. A. Mahmoud and H. Kraatz, A Bioorganometallic Approach for the Electrochemical Detection of Proteins: A Study on the Interaction of Ferrocene–Peptide Conjugates with Papain in Solution and on Au Surfaces, Chem. – Eur. J., 2007, 13(20), 5885–5895, DOI:10.1002/chem.200601878.
- C. N. Tharamani, K. A. Mahmoud, G. R. Vasanthakumar and H. B. Kraatz, Studies into the Interaction of a Ferrocene-Conjugates of Gly-Gly-Arg-Tyr with Papain: AC Voltammetry, Impedance Spectroscopy and Surface Plasmon Resonance Studies, Sens. Actuators, B, 2009, 137(1), 253–258, DOI:10.1016/j.snb.2008.11.050.
- E. González-Fernández, M. Staderini, A. Yussof, E. Scholefield, A. F. Murray, A. R. Mount and M. Bradley, Electrochemical Sensing of Human Neutrophil Elastase and Polymorphonuclear Neutrophil Activity, Biosens. Bioelectron., 2018, 119(August), 209–214, DOI:10.1016/j.bios.2018.08.013.
- M. L. Frisk, W. H. Tepp, E. A. Johnson and D. J. Beebe, Self-Assembled Peptide Monolayers as a Toxin Sensing Mechanism within Arrayed Microchannels, Anal. Chem., 2009, 81(7), 2760–2767, DOI:10.1021/ac802707u.
- L. Z. Swisher, L. U. Syed, A. M. Prior, F. R. Madiyar, K. R. Carlson, T. A. Nguyen, D. H. Hua and J. Li, Electrochemical Protease Biosensor Based on Enhanced AC Voltammetry Using Carbon Nanofiber Nanoelectrode Arrays, J. Phys. Chem. C, 2013, 117(8), 4268–4277, DOI:10.1021/jp312031u.
- Q. Hu, L. Su, Y. Mao, S. Gan, Y. Bao, D. Qin, W. Wang, Y. Zhang and L. Niu, Electrochemically Induced Grafting of Ferrocenyl Polymers for Ultrasensitive Cleavage-Based Interrogation of Matrix Metalloproteinase Activity, Biosens. Bioelectron., 2021, 178(January), 113010, DOI:10.1016/j.bios.2021.113010.
- X. Xi, M. Wen, S. Song, J. Zhu, W. Wen, X. Zhang and S. Wang, A H2O2-Free Electrochemical Peptide Biosensor Based on Au@Pt Bimetallic Nanorods for Highly Sensitive Sensing of Matrix Metalloproteinase 2, Chem. Commun., 2020, 56(45), 6039–6042, 10.1039/d0cc01598a.
- Y. Zheng and Z. Ma, Dual-Reaction Triggered Sensitivity Amplification for Ultrasensitive Peptide-Cleavage Based Electrochemical Detection of Matrix Metalloproteinase-7, Biosens. Bioelectron., 2018, 108(November 2017), 46–52, DOI:10.1016/j.bios.2018.02.045.
- Q. Palomar, X. X. Xu, R. Selegård, D. Aili and Z. Zhang, Peptide Decorated Gold Nanoparticle/Carbon Nanotube Electrochemical Sensor for Ultrasensitive Detection of Matrix Metalloproteinase-7, Sens. Actuators, B, 2020, 325, 128789, DOI:10.1016/j.snb.2020.128789.
- G. Liu, J. Wang, D. S. Wunschel and Y. Lin, Electrochemical Proteolytic Beacon for Detection of Matrix Metalloproteinase Activities, J. Am. Chem. Soc., 2006, 128(38), 12382–12383, DOI:10.1021/ja0626638.
- L. Sun, Y. Chen, F. Chen and F. Ma, Peptide-Based Electrochemical Biosensor for Matrix Metalloproteinase-14 and Protein-Overexpressing Cancer Cells Based on Analyte-Induced Cleavage of Peptide, Microchem. J., 2020, 157(April), 105103, DOI:10.1016/j.microc.2020.105103.
- J. Drzazgowska, B. Schmid, R. D. Sussmuth and Z. Altintas, Self-Assembled Monolayer Epitope Bridges for Molecular Imprinting and Cancer Biomarker Sensing, Anal. Chem., 2020, 92(7), 4798–4806, DOI:10.1021/acs.analchem.9b03813.
- Y. Xu, X. Wang, C. Ding and X. Luo, Ratiometric Antifouling Electrochemical Biosensors Based on Multifunctional Peptides and MXene Loaded with Au Nanoparticles and Methylene Blue, ACS Appl. Mater. Interfaces, 2021, 13(17), 20388–20396, DOI:10.1021/acsami.1c04933.
- K. Ohtsuka, I. Maekawa, M. Waki and S. Takenaka, Electrochemical Assay of Plasmin Activity and Its Kinetic Analysis, Anal. Biochem., 2009, 385(2), 293–299, DOI:10.1016/j.ab.2008.11.006.
- Z. Song, M. Chen, C. Ding and X. Luo, Designed Three-in-One Peptides with Anchoring, Antifouling, and Recognizing Capabilities for Highly Sensitive and Low-Fouling Electrochemical Sensing in Complex Biological Media, Anal. Chem., 2020, 92(8), 5795–5802, DOI:10.1021/acs.analchem.9b05299.
- D. Wang, J. Wang, Z. Song and N. Hui, Highly Selective and Antifouling Electrochemical Biosensors for Sensitive MicroRNA Assaying Based on Conducting Polymer Polyaniline Functionalized with Zwitterionic Peptide, Anal. Bioanal. Chem., 2021, 413(2), 543–553, DOI:10.1007/s00216-020-03025-5.
- G. Wang, R. Han, Q. Li, Y. Han and X. Luo, Electrochemical Biosensors Capable of Detecting Biomarkers in Human Serum with Unique Long-Term Antifouling Abilities Based on Designed Multifunctional Peptides, Anal. Chem., 2020, 92(10), 7186–7193, DOI:10.1021/acs.analchem.0c00738.
- Y. Huang, B. Zhang, L. Yuan and L. Liu, A Signal Amplification Strategy Based on Peptide Self-Assembly for the Identification of Amyloid-β Oligomer, Sens. Actuators, B, 2021, 335, 129697, DOI:10.1016/j.snb.2021.129697.
- H. Li, Y. Cao, X. Wu, Z. Ye and G. Li, Peptide-Based Electrochemical Biosensor for Amyloid β 1 – 42 Soluble Oligomer Assay, Talanta, 2012, 93, 358–363, DOI:10.1016/j.talanta.2012.02.055.
- K. Zhang, Q. Yang, Z. Fan, J. Zhao and H. Li, Platelet-Driven Formation of Interface Peptide Nano-Network Biosensor Enabling a Non-Invasive Means for Early Detection of Alzheimer's Disease, Biosens. Bioelectron., 2019, 145(August), 111701, DOI:10.1016/j.bios.2019.111701.
- Z. Song, Y. Ma, M. Chen, A. Ambrosi, C. Ding and X. Luo, Electrochemical Biosensor with Enhanced Antifouling Capability for COVID-19 Nucleic Acid Detection in Complex Biological Media, Anal. Chem., 2021, 93(14), 5963–5971, DOI:10.1021/acs.analchem.1c00724.
- N. Liu, X. Fan, H. Hou, F. Gao and X. Luo, Electrochemical Sensing Interfaces Based on Hierarchically Architectured Zwitterionic Peptides for Ultralow Fouling Detection of Alpha Fetoprotein in Serum, Anal. Chim. Acta, 2021, 1146, 17–23, DOI:10.1016/j.aca.2020.12.031.
- N. Liu, J. Song, Y. Lu, J. J. Davis, F. Gao and X. Luo, Electrochemical Aptasensor for Ultralow Fouling Cancer Cell Quantification in Complex Biological Media Based on Designed Branched Peptides, Anal. Chem., 2019, 91(13), 8334–8340, DOI:10.1021/acs.analchem.9b01129.
- J. P. Piccoli, A. C. Soares, O. N. Oliveira and E. M. Cilli, Nanostructured Functional Peptide Films and Their Application in C-Reactive Protein Immunosensors, Bioelectrochemistry, 2021, 138, 107692, DOI:10.1016/j.bioelechem.2020.107692.
- T. M. Yawitz, K. S. Patterson, B. X. Onkst, F. Youmbi and R. A. Clark, Cytochrome c Electrochemistry on Peptide Self-Assembled Monolayers, J. Electroanal. Chem., 2018, 828, 59–62, DOI:10.1016/j.jelechem.2018.09.037.
- C. H. Cho, J. H. Kim, J. Kim, J. W. Yun, T. J. Park and J. P. Park, Re-Engineering of Peptides with High Binding Affinity to Develop an Advanced Electrochemical Sensor for Colon Cancer Diagnosis, Anal. Chim. Acta, 2021, 1146, 131–139, DOI:10.1016/j.aca.2020.11.011.
- R. Li, H. Huang, L. Huang, Z. Lin, L. Guo, B. Qiu and G. Chen, Electrochemical Biosensor for Epidermal Growth Factor Receptordetection with Peptide Ligand, Electrochim. Acta, 2013, 109, 233–237, DOI:10.1016/j.electacta.2013.07.151.
- M. Chen, Z. Song, R. Han, Y. Li and X. Luo, Low Fouling Electrochemical Biosensors Based on Designed Y-Shaped Peptides with Antifouling and Recognizing Branches for the Detection of IgG in Human Serum, Biosens. Bioelectron., 2021, 178, 113016, DOI:10.1016/j.bios.2021.113016.
- N. Liu, N. Hui, J. J. Davis and X. Luo, Low Fouling Protein Detection in Complex Biological Media Supported by a Designed Multifunctional Peptide, ACS Sens., 2018, 3(6), 1210–1216, DOI:10.1021/acssensors.8b00318.
- N. Islam, P. V. Gurgel, O. J. Rojas and R. G. Carbonell, Use of a Branched Linker for Enhanced Biosensing Properties in IgG Detection from Mixed Chinese Hamster Ovary Cell Cultures, Bioconjugate Chem., 2019, 30(3), 815–825, DOI:10.1021/acs.bioconjchem.8b00918.
- S. Wang, Y. Ma, Y. Wang, M. Jiao, X. Luo and M. Cui, One-Step Electrodeposition of Poly(m-Aminobenzoic Acid) Membrane Decorated with Peptide for Antifouling Biosensing of Immunoglobulin E, Colloids Surf., B, 2020, 186, 110706, DOI:10.1016/j.colsurfb.2019.110706.
- S. Huang, R. Tang, T. Zhang, J. Zhao, Z. Jiang and Q. Wang, Anti-Fouling Poly Adenine Coating Combined with Highly Specific CD20 Epitope Mimetic Peptide for Rituximab Detection in Clinical Patients’ Plasma, Biosens. Bioelectron., 2021, 171(September 2020), 112678, DOI:10.1016/j.bios.2020.112678.
- M. Puiu, A. Idili, D. Moscone, F. Ricci and C. Bala, A Modular Electrochemical Peptide-Based Sensor for Antibody Detection, Chem. Commun., 2014, 50(64), 8962, 10.1039/C4CC02858A.
- J. Y. Gerasimov and R. Y. Lai, An Electrochemical Peptide-Based Biosensing Platform for HIV Detection, Chem. Commun., 2010, 46(3), 395–397, 10.1039/b919070h.
- C. Y. Lin, U. T. Nhat Nguyen, H. Y. Hsieh, H. Tahara, Y. S. Chang, B. Y. Wang, B. C. Gu, Y. H. Dai, C. C. Wu and I. J. Tsai, et al., Peptide-Based Electrochemical Sensor with Nanogold Enhancement for Detecting Rheumatoid Arthritis, Talanta, 2022, 236(July 2021), 122886, DOI:10.1016/j.talanta.2021.122886.
- H. Qi, M. Li, M. Dong, S. Ruan, Q. Gao and C. Zhang, Electrogenerated Chemiluminescence Peptide-Based Biosensor for the Determination of Prostate-Specific Antigen Based on Target-Induced Cleavage of Peptide, Anal. Chem., 2014, 86(3), 1372–1379, DOI:10.1021/ac402991r.
- N. Zhao, Y. He, X. Mao, Y. Sun, X. Zhang, C. Li, Y. Lin and G. Liu, Electrochemical Assay of Active Prostate-Specific Antigen (PSA) Using Ferrocene-Functionalized Peptide Probes, Electrochem. Commun., 2010, 12(3), 471–474, DOI:10.1016/j.elecom.2010.01.022.
- S. Casalini, C. A. Bortolotti, F. Leonardi and F. Biscarini, Self-Assembled Monolayers in Organic Electronics, Chem. Soc. Rev., 2017, 46(1), 40–71, 10.1039/c6cs00509h.
- N. Amdursky, Electron Transfer across Helical Peptides, ChemPlusChem, 2015, 80(7), 1075–1095, DOI:10.1002/cplu.201500121.
- Y. A. Berlin, A. L. Burin and M. A. Ratner, Charge Hopping in DNA, J. Am. Chem. Soc., 2001, 123(2), 260–268, DOI:10.1021/ja001496n.
- X. Dai, Q. Li, A. Aldalbahi, L. Wang, C. Fan and X. Liu, DNA-Based Fabrication for Nanoelectronics, Nano Lett., 2020, 20(8), 5604–5615, DOI:10.1021/acs.nanolett.0c02511.
- X. Xiao, B. Xu and N. Tao, Conductance Titration of Single-Peptide Molecules, J. Am. Chem. Soc., 2004, 126(17), 5370–5371, DOI:10.1021/ja049469a.
- N. Song, Z. Zhou, Y. Song, M. Li, X. Yu, B. Hu and Z. Yu, In Situ Oxidation-Regulated Self-Assembly of Peptides into Transformable Scaffolds for Cascade Therapy, Nano Today, 2021, 38, 101198, DOI:10.1016/j.nantod.2021.101198.
- P. Li, R. Yin, J. Cheng and J. Lin, Bacterial Biofilm Formation on Biomaterials and Approaches to Its Treatment and Prevention, Int. J. Mol. Sci., 2023, 24(14), 11680, DOI:10.3390/ijms241411680.
- M. Van kerckhoven, A. Hotterbeekx, E. Lanckacker, P. Moons, C. Lammens, M. Kerstens, M. Ieven, P. Delputte, P. G. Jorens and S. Malhotra-Kumar, et al., Characterizing the in Vitro Biofilm Phenotype of Staphylococcus Epidermidis Isolates from Central Venous Catheters, J. Microbiol. Methods, 2016, 127, 95–101, DOI:10.1016/j.mimet.2016.05.009.
- A. Giacometti, O. Cirioni, W. Kamysz, G. D’Amato, C. Silvestri, M. S. Del Prete, J. Łukasiak and G. Scalise, Comparative Activities of Cecropin A, Melittin, and Cecropin A–Melittin Peptide CA(1–7)M(2–9)NH2 against Multidrug-Resistant Nosocomial Isolates of Acinetobacter Baumannii, Peptides, 2003, 24(9), 1315–1318, DOI:10.1016/j.peptides.2003.08.003.
- S. Ji, W. Li, L. Zhang, Y. Zhang and B. Cao, Cecropin A–Melittin Mutant with Improved Proteolytic Stability and Enhanced Antimicrobial Activity against Bacteria and Fungi Associated with Gastroenteritis in Vitro, Biochem. Biophys. Res. Commun., 2014, 451(4), 650–655, DOI:10.1016/j.bbrc.2014.08.044.
- M. Xiao, J. Jasensky, L. Foster, K. Kuroda and Z. Chen, Monitoring Antimicrobial Mechanisms of Surface-Immobilized Peptides in Situ, Langmuir, 2018, 34(5), 2057–2062, DOI:10.1021/acs.langmuir.7b03668.
- C. Monteiro, M. Fernandes, M. Pinheiro, S. Maia, C. L. Seabra, F. Ferreira-da-Silva, F. Costa, S. Reis, P. Gomes and M. C. L. Martins, Antimicrobial Properties of Membrane-Active Dodecapeptides Derived from MSI-78, Biochim. Biophys. Acta, Biomembr., 2015, 1848(5), 1139–1146, DOI:10.1016/j.bbamem.2015.02.001.
- X.-L. Zhang, A.-M. Jiang, Z.-Y. Ma, X.-B. Li, Y.-Y. Xiong, J.-F. Dou and J.-F. Wang, The Synthetic Antimicrobial Peptide Pexiganan and Its Nanoparticles (PNPs) Exhibit the Anti-Helicobacter Pylori Activity in Vitro and in Vivo, Molecules, 2015, 20(3), 3972–3985, DOI:10.3390/molecules20033972.
- L. L. Roberto, T. S. Crespo, R. S. Monteiro-Junior, A. M. E. B. L. Martins, A. M. B. De Paula, E. F. Ferreira and D. S. Haikal, Sociodemographic Determinants of Edentulism in the Elderly Population: A Systematic Review and Meta-analysis, Gerodontology, 2019, 36(4), 325–337, DOI:10.1111/ger.12430.
- S. Dhir, Biofilm and Dental Implant: The Microbial Link, J. Indian Soc. Periodontol., 2013, 17(1), 5, DOI:10.4103/0972-124X.107466.
- Å. Leonhardt, S. Renvert and G. Dahlén, Microbial Findings at Failing Implants, Clin. Oral. Implants Res., 1999, 10(5), 339–345, DOI:10.1034/j.1600-0501.1999.100501.x.
- P.-H. Chua, K.-G. Neoh, E.-T. Kang and W. Wang, Surface Functionalization of Titanium with Hyaluronic Acid/Chitosan Polyelectrolyte Multilayers and RGD for Promoting Osteoblast Functions and Inhibiting Bacterial Adhesion, Biomaterials, 2008, 29(10), 1412–1421, DOI:10.1016/j.biomaterials.2007.12.019.
- E. S. Gawalt, M. J. Avaltroni, M. P. Danahy, B. M. Silverman, E. L. Hanson, K. S. Midwood, J. E. Schwarzbauer and J. Schwartz, Bonding Organics to Ti Alloys: Facilitating Human Osteoblast Attachment and Spreading on Surgical Implant Materials, Langmuir, 2003, 19(1), 200–204, DOI:10.1021/la0203436.
- R. Chen, M. D. P. Willcox, N. Cole, K. K. K. Ho, R. Rasul, J. A. Denman and N. Kumar, Characterization of Chemoselective Surface Attachment of the Cationic Peptide Melimine and Its Effects on Antimicrobial Activity, Acta Biomater., 2012, 8(12), 4371–4379, DOI:10.1016/j.actbio.2012.07.029.
- S. Acosta, A. Ibañez-Fonseca, C. Aparicio and J. C. Rodríguez-Cabello, Antibiofilm Coatings Based on Protein-Engineered Polymers and Antimicrobial Peptides for Preventing Implant-Associated Infections, Biomater. Sci., 2020, 8(10), 2866–2877, 10.1039/D0BM00155D.
- A. Care, P. L. Bergquist and A. Sunna, Solid-Binding Peptides in Biomedicine, Adv. Exp. Med. Biol., 2017, 21–36, DOI:10.1007/978-3-319-66095-0_2.
- E. C. Wisdom, Y. Zhou, C. Chen, C. Tamerler and M. L. Snead, Mitigation of Peri-Implantitis by Rational Design of Bifunctional Peptides with Antimicrobial Properties, ACS Biomater. Sci. Eng., 2020, 6(5), 2682–2695, DOI:10.1021/acsbiomaterials.9b01213.
- A. Saha, S. Nir and M. Reches, Amphiphilic Peptide with Dual Functionality Resists Biofouling, Langmuir, 2020, 36(15), 4201–4206, DOI:10.1021/acs.langmuir.9b03997.
- M. S. Mulani, E. E. Kamble, S. N. Kumkar, M. S. Tawre and K. R. Pardesi, Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review, Front. Microbiol., 2019, 10, 539, DOI:10.3389/fmicb.2019.00539.
- J. Zhan, L. Wang, Y. Zhu, H. Gao, Y. Chen, J. Chen, Y. Jia, J. He, Z. Fang and Y. Zhu, et al., Temperature-Controlled Reversible Exposure and Hiding of Antimicrobial Peptides on an Implant for Killing Bacteria at Room Temperature and Improving Biocompatibility in Vivo, ACS Appl. Mater. Interfaces, 2018, 10(42), 35830–35837, DOI:10.1021/acsami.8b14534.
- B. N. Dang and D. Y. Graham, Helicobacter Pylori Infection and Antibiotic Resistance: A WHO High Priority?, Nat. Rev. Gastroenterol. Hepatol., 2017, 14(7), 383–384, DOI:10.1038/nrgastro.2017.57.
- M. F. Maitz, M. C. L. Martins, N. Grabow, C. Matschegewski, N. Huang, E. L. Chaikof, M. A. Barbosa, C. Werner and C. Sperling, The Blood Compatibility Challenge. Part 4: Surface Modification for Hemocompatible Materials: Passive and Active Approaches to Guide Blood-Material Interactions, Acta Biomater., 2019, 94, 33–43, DOI:10.1016/j.actbio.2019.06.019.
- J. Kuchinka, C. Willems, D. V. Telyshev and T. Groth, Control of Blood Coagulation by Hemocompatible Material Surfaces—A Review, Bioengineering, 2021, 8(12), 215, DOI:10.3390/bioengineering8120215.
- L. Zhang, J. R. Lu and T. A. Waigh, Electronics of Peptide- and Protein-Based Biomaterials, Adv. Colloid Interface Sci., 2021, 287, 102319, DOI:10.1016/j.cis.2020.102319.
- H. Chen and J. Fraser Stoddart, From Molecular to Supramolecular Electronics, Nat. Rev. Mater., 2021, 6(9), 804–828, DOI:10.1038/s41578-021-00302-2.
- T. Li, V. K. Bandari and O. G. Schmidt, Molecular Electronics: Creating and Bridging Molecular Junctions and Promoting Its Commercialization, Adv. Mater., 2023, 35(22), 2209088, DOI:10.1002/adma.202209088.
- Z. Zhang, A. E. Baxter, D. Ren, K. Qin, Z. Chen, S. M. Collins, H. Huang, C. A. Komar, P. F. Bailer and J. B. Parker, et al., Efficient Engineering of Human and Mouse Primary Cells Using Peptide-Assisted Genome Editing, Nat. Biotechnol., 2024, 42, 305–315, DOI:10.1038/s41587-023-01756-1.
| This journal is © The Royal Society of Chemistry 2024 |