DOI:
10.1039/D4DT01667J
(Paper)
Dalton Trans., 2024,
53, 14171-14181
A green approach to encapsulate proteins and enzymes within crystalline lanthanide-based Tb and Gd MOFs†
Received
8th June 2024
, Accepted 18th July 2024
First published on 19th July 2024
Abstract
In this work, bovine serum albumin (BSA) and Aspergillus sp. laccase (LC) were encapsulated in situ within two lanthanide-based MOFs (TbBTC and GdBTC) through a green one-pot synthesis (almost neutral aqueous solution, T = 25 °C, and atmospheric pressure) in about 1 h. Pristine MOFs and protein-encapsulated MOFs were characterized through wide angle X-ray scattering, scanning electron microscopy, thermogravimetric analysis, Fourier transform infrared and Raman spectroscopies. The location of immobilized BSA molecules, used as a model protein, was investigated through small angle X-ray scattering. BSA occurs both on the inner and on the outer surface of the MOFs. LC@TbBTC, and LC@GdBTC samples were also characterized in terms of specific activity, kinetic parameters, and storage stability both in water and acetate buffer. The specific activity of LC@TbBTC was almost twice that of LC@GdBTC (10.8 μmol min−1 mg−1vs. 6.6 μmol min−1 mg−1). Both biocatalysts showed similar storage stabilities retaining ∼60% of their initial activity after 7 days and ∼20% after 21 days. LC@TbBTC dispersed in acetate buffer exhibited a higher storage stability than LC@GdBTC. Additionally, terbium-based MOFs showed interesting luminescent properties. Together, these findings suggest that TbBTC and GdBTC are promising supports for the in situ immobilization of proteins and enzymes.
Introduction
Metal–organic frameworks (MOFs) are porous coordination networks constituted by metal ions bound together through organic linkers.1 To date, MOFs have been investigated for several applications such as gas adsorption,2 catalysis,3 sensing, drug delivery,4 removal of water pollutants,5 and other industrial applications.5–7 MOFs have also shown great potential as supports for enzyme immobilization.8–10 Among transition metals, lanthanides show some interesting properties like fluorescence and paramagnetism. Thus, lanthanide-based compounds have found applications in medicine for diagnostic and therapeutic purposes.11–15 This has suggested the potential use of lanthanide-based MOFs (Ln-MOFs) for the development of innovative biomedical applications.16 Recently, Zhang et al. developed a nanoprobe based on luminescent terbium MOF-coated gold nanorods for photo-stimulated thermal therapy and chemotherapy.17 Gadolinium-based complexes (i.e. gadopentetate dimeglumine) are widely used as contrast agents for magnetic resonance imaging (MRI).18 Gd-based MOFs showed a longer in vivo circulation time and a higher MRI relaxation time, due to Gd content, higher than conventional Gd-based contrast agents.19 Other recent works report the coupling of Ln-MOFs with biomolecules,20 such as enzymes/proteins or nucleic acids, to obtain hybrid biomaterials with improved properties for medical applications.21,22 Zhang et al. immobilized glucose oxidase (GOx) into an UiO type MOF modified with Eu3+ to obtain a fluorescence sensor for glucose detection in serum and urine.23 Gao et al. encapsulated GOx and carbon dots within a Tb3+ based composite obtaining a hybrid GOx&CDs@AMP/Tb biocatalyst which displayed about double catalytic activity than free GOx. Moreover, GOx&CDs@AMP/Tb was further modified with carboxyphenylboronic acid to obtain a dual-emissive GOx&CDs@AMP/Tb-CPBA composite for the ratiometric sensing of glucose.24 Although the coupling of biomolecules with Ln-MOFs is gaining interest, there are some synthetic issues to be solved. Indeed, enzymes and other biomolecules are generally immobilized post-synthesis by adsorption within the pores of solid carriers like ordered mesoporous silica and hierarchical zeolites.25–27 Since many MOFs are microporous (pore size < 2 nm), the post-synthesis immobilization would be only limited to the external surface of the material due to the larger size of enzyme molecules. Alternatively, the enzyme could be immobilized through encapsulation during the synthesis of the MOF.28 However, such in situ immobilization is not applicable when typical MOF synthesis methods are used. Indeed, they require harsh reaction conditions, like high temperatures or pressures, the use of organic solvents and long reaction times (minimum 1–3 days and up to 1–2 weeks for diffusion methods).29–32 These synthetic methods, besides resulting in enzyme inactivation, are not environmentally sustainable. Recently, in situ enzyme encapsulation during the MOF formation, via a ‘one pot’ approach under biocompatible synthetic conditions, has been proposed.33,34 Employing this strategy, enzymes larger than the average pore size of MOF materials can be embedded within the three-dimensional MOF structure thus preventing enzyme leaching, while retaining catalytic activity and allowing free substrate diffusion.35,36In situ encapsulation has been reported for both microporous MOFs, like ZIF-8,37,38 ZIF-90,39 HKUST-1@Fe3O4,40 and mesoporous MOFs, like ZIF-8,41 ZIF-67,42 and MIL-101(Cr).43 However, the number of MOFs obtained through sustainable synthetic methods which allows enzyme immobilization is still low and mainly focused on divalent metals-based MOFs which show lower thermal/chemical stability compared to trivalent metals based MOFs.44 On the other hand, the synthesis of trivalent metals-based MOFs, due to their higher reactivity (at room temperature) compared to divalent ones, might compromise the crystallinity of MOFs.44 For example, Sanchez-Sanchez et al. carried out the in situ encapsulation of enzymes in a non-crystalline MOF based on Fe3+ and the tridentate linker trimesic acid in aqueous medium and mild conditions (room temperature and almost neutral pH).45 Subsequently, the same procedure was used to encapsulate alcohol dehydrogenase, glucose oxidase, and lipase enzymes.9,10,46 Coming back to Ln-MOFs, the syntheses reported in the literature (i.e. for GdBTC and TbBTC MOFs) use microemulsion, mechanochemical, and solvothermal methods. All these approaches are not sustainable and incompatible with biomolecule encapsulation (i.e. solvothermal methods have long times and high reaction temperatures) or lead to the synthesis of amorphous materials (as in the cases of microemulsion and mechanochemical synthesis).47,48 Therefore, the sustainable synthesis and biomolecule encapsulation of trivalent (particularly lanthanides) metal ions-based crystalline MOFs is still a challenge.28 In this work, two lanthanide ions, Tb3+ and Gd3+, were used to synthesize crystalline TbBTC and GdBTC MOFs in a sustainable way, i.e. using water as the solvent, under mild reaction conditions (T = 25 °C and atmospheric pressure). The same synthetic strategy was used to encapsulate bovine serum albumin (BSA) and Aspergillus sp. laccase (LC) within TbBTC and GdBTC MOFs. The pristine MOFs and the hybrid protein@MOFs were characterized through wide-angle X-ray scattering (WAXS), scanning electron microscopy (SEM), Fourier transform infrared (FTIR) and Raman spectroscopies, and thermogravimetric analysis (TGA). The protein distribution after encapsulation within TbBTC and GdBTC MOFs was investigated through small angle X-ray scattering (SAXS) using BSA as a highly pure model protein. LC was used as a model enzyme to test the biological activity and stability of the LC@TbBTC and LC@GdBTC samples. To this purpose, LC@TbBTC and LC@GdBTC samples were characterized in terms of specific activity, kinetic parameters (KM and Vmax) and storage stability in water and acetate buffer. To the best of our knowledge, this is the first work regarding the in situ protein immobilization within crystalline lanthanide-based MOFs obtained through a sustainable synthetic approach.
Experimental section
Chemicals
Laccase from Aspergillus sp. (activity of ≥1000 LAMU g−1), 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonate) diammonium salt (ABTS) (≥98%), Bovine Serum Albumin (BSA) lyophilized powder (≥98%), Gd(NO3)3·6H2O (99.99%), Tb(NO3)3·6H2O (99.99%), sodium hydroxide, NaOH; trimesic acid, H3BTC (95%), sodium phosphate, monobasic NaH2PO4 (99%), sodium phosphate dibasic, Na2HPO4 (99%), Bradford reagent, HCl (37%), acetic acid (99%), sodium acetate (anhydrous) (≥99.0%), were purchased from Sigma-Aldrich. All reagents were used as received without further purification. Milli-Q water (≥18.2 MΩ cm) was used to prepare all aqueous solutions.
Synthesis of TbBTC, GdBTC, protein@TbBTC, and protein@GdBTC samples
A mass of 22 mg of trimesic acid (H3BTC) was dissolved in 5 mL of NaOH 1.06 M, then a solution containing 44 mg of Tb(NO3)3·6 H2O (or 60 mg of Gd(NO3)3·6H2O) in 5 mL of H2O was added dropwise under magnetic stirring. The obtained white suspension was maintained under stirring at 25 °C for 1 h. The precipitate, TbBTC (or GdBTC) MOF was recovered by centrifugation at 4500 rpm for 5 min (three times), then extensively washed with water, and dried at room temperature (25 °C) under vacuum. The synthesis of LC@GdBTC and LC@TbBTC, was carried out following the procedure to obtain TbBTC and GdBTC MOFs except for the addition of 2 mL of LC solution (4.5 mg mL−1) to the metal source solution. The resulting mix was added to the deprotonated BTC solution. The synthesis of BSA@TbBTC, and BSA@GdBTC were carried out as above by adding 1 mL of BSA solution (25 mg mL−1) to the MOFs reaction mixture.
Characterization of MOFs and protein@MOFs samples
SEM analysis was performed by using a Gemini Ultra 55 microscope (Carl-Zeiss) with an acceleration voltage of 1 kV. Fibrils cross-section in MOFs was measured through Image J software [https://www.nature.com/articles/nmeth.2019]. More specifically, the cross-section of 30 fibers for each MOF was measured on SEM micrographs with a 50–300 kX magnification. The average and standard deviation are reported in the Results and Discussion section. Attenuated total reflectance (ATR)-FTIR spectra were recorded using a Jasco FTIR 4100 instrument equipped with a PIKE GladiATR accessory with a single reflection diamond prism over the wavelength range, 4000–500 cm−1. An in Via Qontor confocal microRaman (Renishaw) was used to acquire Raman spectra. The confocal microRaman is equipped with a solid-state laser (785 nm, IPS R-type NIR785, 200 mW, 1200 l mm−1 grating), a front-illuminated CCD camera (655 × 1024 px, working temperature −70 °C) and a research-grade Leica DM 2700 microscope. TGA was carried out by Discovery SDT 650 (TA Instruments) in a temperature range from 25 to 700 °C and a heating ramp of 10 °C min−1, under continuous air flow (flow rate = 40 mL min−1). SAXS and WAXS data were acquired with a Xeuss 3.0 HR (Xenocs) apparatus, equipped with a GeniX 3D X-ray generator, containing a high brightness Cu X-ray tube (30 W per 40 μm, λx = 1.542 Å) and a FOX 3D single reflection multilayer optic. The detector used to detect the scattering signal was a Eiger 2R 1 M hybrid photon counting detector (Dectris, pixel dimension of 75 × 75 μm2). A powder/solid sample-holder was used, with Kapton tape as window (sample thickness: 1 mm). Measurements were performed at room temperature. 2D SAXS/WAXS images were collected at two sample-to-detector distances: 80 mm and 1800 mm to cover a scattering vector (Q) range of 0.0045–2.5 Å−1 and a maximum 2θ of about 35°. After circularly averaging 2D images, 1D curves (Intensity vs. Q) were obtained. Here the scattering vector modulus, Q, is defined as Q = (4π/λx) sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ, where 2θ is the scattering angle. The scattering camera was calibrated using silver behenate (d = 58.38 Å) as the ref. 49. Data were further corrected by empty cell subtraction and normalized, considering the measurement time and relative transmission factors. Finally, 1D SAXS/WAXS curves collected at the two different distances were merged in a single scattering profile and converted to absolute intensity, using glassy carbon as a secondary standard.50 SAXS/WAXS data treatment was performed by XSACT software (Xenocs). The materials crystallinity was evaluated by the WAXS technique and quantified by the following equation:
θ, where 2θ is the scattering angle. The scattering camera was calibrated using silver behenate (d = 58.38 Å) as the ref. 49. Data were further corrected by empty cell subtraction and normalized, considering the measurement time and relative transmission factors. Finally, 1D SAXS/WAXS curves collected at the two different distances were merged in a single scattering profile and converted to absolute intensity, using glassy carbon as a secondary standard.50 SAXS/WAXS data treatment was performed by XSACT software (Xenocs). The materials crystallinity was evaluated by the WAXS technique and quantified by the following equation:| | 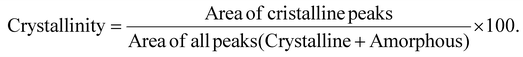 | (1) |
The fitting of SAXS curves was performed by using SASView software [https://www.sasview.org/]. The fitting model was chosen according to those reported in the literature for similar systems.51–53 More specifically, the fitting function is the sum of a 1/Qn term (with n Porod exponent in the low-Q region, used only for BSA@MOFs and calcinated BSA@MOFs), a Guinier–Porod model (IGP)54 and a background; the first two terms were weighted for two scaling factors, A and B, respectively:
| | 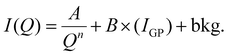 | (2) |
The Guinier–Porod model,
IGP, consists of two terms:
| | 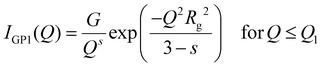 | (3) |
| | 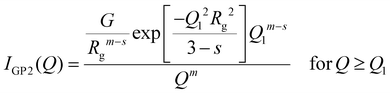 | (4) |
with
Rg the gyration radius of the scattering objects,
m Porod exponent at higher
Q,
s a dimensionality parameter, that equals 0 for spherical objects, 1 for rods and 2 for platelets or lamellae and:
| | 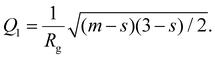 | (5) |
Immobilization efficiency and enzyme loading
Protein loading and immobilization efficiency of the immobilized LC@TbBTC and LC@GdBTC samples were obtained by means of the Bradford assay.55 Briefly, the protein content was determined using the Bradford reagent (Bio-Rad) and BSA as the protein standard. The protein concentration in the supernatant was evaluated by measuring the absorbance (λ = 595 nm) of a mixture containing 0.5 mL of supernatant and 0.5 mL of Bradford reagent after 10 min of incubation. The protein loading (Lprotein = [mgprotein gprotein@MOF−1]) was determined by the equation:| | 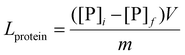 | (6) |
where, [P]i and [P]f are the initial and final protein concentrations (mg mL−1), respectively; V is the volume of the protein solution (mL), and m is the mass (g) of the obtained protein@MOF sample.
Encapsulation efficiency (EE%) is the percent ratio between the amount of immobilized protein and the amount of protein in the immobilizing solution:
| | | EE% = [([P]i − [P]f)/[P]i)] × 100%. | (7) |
Protein loading and encapsulation efficiency for the BSA modified MOFs, BSA@TbBTC and BSA@GdBTC, were obtained by TGA. The mass loss over the range from 260 °C and 430 °C is ascribed to the combustion of BSA.
Measurement of specific activity and kinetic parameters of free LC, LC@TbBTC and LC@GdBTC samples
The LC active site consists of four copper centres, classified as type I, type II, and type III. Substrate oxidation at the mononuclear site containing the type I Cu generates electrons that are transferred to the trinuclear site, which contains type II and type III copper ions, where O2 is reduced to H2O.56 The catalytic activity of free LC was determined through a Jasco 650 UV-Vis spectrophotometer by measuring the increase of absorbance, due the formation of the radical-ion ABTS˙+, at λ = 420 nm for 5 min. One unit (U) of laccase activity is defined as the amount of enzyme required to convert 1.0 μmol of ABTS to ABTS˙+ (εABTS˙+ = 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 M−1 cm−1) per minute at 25 °C. Briefly, 100 μL of commercially available Aspergillus sp. LC (diluted 1
700 M−1 cm−1) per minute at 25 °C. Briefly, 100 μL of commercially available Aspergillus sp. LC (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) were added to 2.7 mL of acetate buffer (pH 5, 100 mM) containing 150 μL of ABTS (5 mM) in a cuvette kept in the dark under stirring at T = 25 °C. The catalytic activity of LC@TbBTC and LC@GdBTC was carried out following the aforementioned procedure but adding 100 μL of biocatalyst dispersion (2 mg mL−1) instead of the free LC. The activity of TbBTC and GdBTC (with no LC) was carried out as a control experiment. No activity was detected for LC-free MOFs. The kinetic parameters, KM and Vmax, of free LC and LC@MOFs were obtained by measuring the initial reaction rate (V0) as a function of ABTS concentration in the range 5–250 μM. A plot of V0 (μM s−1) versus ABTS concentration was built and the experimental points were fitted using the Michaelis–Menten model by mean of KaleidaGraph (4.5.4) software. All measurements were carried out at least in triplicate.
500) were added to 2.7 mL of acetate buffer (pH 5, 100 mM) containing 150 μL of ABTS (5 mM) in a cuvette kept in the dark under stirring at T = 25 °C. The catalytic activity of LC@TbBTC and LC@GdBTC was carried out following the aforementioned procedure but adding 100 μL of biocatalyst dispersion (2 mg mL−1) instead of the free LC. The activity of TbBTC and GdBTC (with no LC) was carried out as a control experiment. No activity was detected for LC-free MOFs. The kinetic parameters, KM and Vmax, of free LC and LC@MOFs were obtained by measuring the initial reaction rate (V0) as a function of ABTS concentration in the range 5–250 μM. A plot of V0 (μM s−1) versus ABTS concentration was built and the experimental points were fitted using the Michaelis–Menten model by mean of KaleidaGraph (4.5.4) software. All measurements were carried out at least in triplicate.
LC@MOFs storage stability in water and acetate buffer
Two LC@TbBTC and LC@GdBTC dispersions (2 mg mL−1) in water and acetate buffer (pH 5, 100 mM) were prepared and stored in the fridge at 5 °C. At intervals of 7 days, the specific activity of both LC@TbBTC and LC@GdBTC samples was measured according to the procedure described above.
Results and discussion
Ln-MOF samples, i.e. TbBTC and GdBTC, were synthesized according to the “green” procedure described in the previous section. Then, two different model proteins, BSA and LC, were encapsulated in situ within TbBTC and GdBTC MOFs to obtain hybrid samples. Highly pure BSA (≥98%) was used to assess protein encapsulation feasibility and to investigate the location of the immobilized protein within the MOF structures. Since protein immobilization may result in the loss of the biological function, laccase was used as a model enzyme to test the catalytic activity and stability. All samples, pristine MOFs (TbBTC and GdBTC), BSA modified MOFs (BSA@TbBTC and BSA@GdBTC), and LC modified MOFs (LC@TbBTC and LC@GdBTC) were characterized using WAXS to quantify their crystallinity. The WAXS patterns show sharp reflections at 10.3°, 13.6°, 17.7°, 19.4°, 20.9°, and 25.5° in agreement with the crystal structure of Tb(BTC)(H2O)6 (Fig. 1A and B).47,57 A similar result was reported by Yang et al. who synthesized the Tb(BTC)(H2O)6, which is isostructural with La(BTC)(H2O)6 (CCDC 290771†).58–60 The slight shift of the peaks between MOF and protein@MOF samples can be attributed to the structure perturbation caused by the protein/enzyme encapsulated within the framework. A similar phenomenon was observed by Al-Harbi et al.61
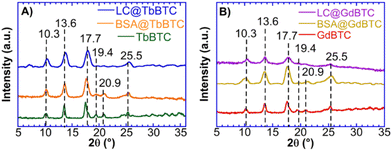 |
| | Fig. 1 WAXS patterns of (A) TbBTC, BSA@TbBTC and LC@TbBTC; (B) GdBTC, BSA@GdBTC and LC@GdBTC. | |
A rough estimation of the degree of crystallinity was obtained by mean of eqn (1).62 The enzyme/protein immobilization within TbBTC and GdBTC lead to a partial loss of crystallinity (Table 1). A similar phenomenon was observed by Qi et al. for the immobilization of Candida rugosa lipase within ZIF-8. Their data showed a decrease in ZIF-8 crystallinity with increasing enzyme loadings.63
Table 1 Crystallinity (%), encapsulation efficiency (EE%) and protein loading values of MOF and protein@MOF samples
| Samples |
Crystallinity (%) |
EE % |
Loadinga (mg g−1) |
Loadingb (mg g−1) |
|
Determined by Bradford assay.
Determined by TGA.
|
| TbBTC |
93 |
— |
— |
— |
| GdBTC |
85 |
— |
— |
— |
| BSA@TbBTC |
94 |
20.7 |
— |
127.9 |
| BSA@GdBTC |
50 |
30.0 |
— |
197.4 |
| LC@TbBTC |
70 |
70.2 |
59.3 |
73.2 |
| LC@GdBTC |
63 |
52.3 |
93.8 |
106.7 |
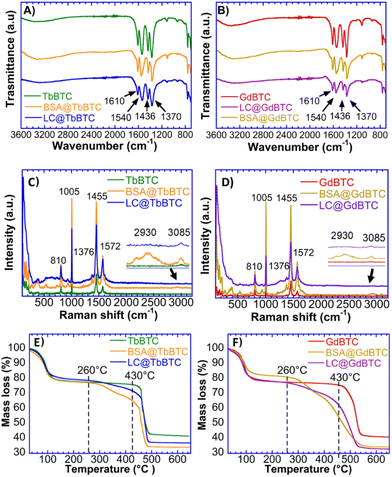
SEM images of TbBTC and GdBTC show a regular morphology with needle-shaped particles due to the preferred crystallization along one crystallographic direction (Fig. 2).64 Recently, Lo Presti et al. found that the morphology of the lanthanide-based MOF YBTC is affected by the reaction time and temperature of the synthesis. In particular, the needle morphology was obtained for the YBTC synthesized at room temperature.64 Owing to the encapsulation of BSA into TbBTC and GdBTC, the needle-shaped morphology changes to smaller spherical particles; the same phenomenon is observed for the immobilization of the laccase within GdBTC and TbBTC (Fig. 2). A change in morphology due to enzyme immobilization was previously obtained for Aspergillus sp. laccase immobilized within ZIF-zni MOF.8 This change might be due to the protein deposition on the MOF surface during the synthesis.52 However, it has also been observed that MOF morphology is affected by the type of immobilized protein. Indeed, Liang et al. observed the classical rhombic ZIF-8 dodecahedron crystal morphology for immobilized ribonuclease A, lipase, urease, and lysozyme, while the immobilization of ovalbumin, horseradish peroxidase, and trypsin led to leaves, flowers, and stars morphologies, respectively.51 FTIR spectra of pristine materials, BSA@MOF and LC@MOF samples are shown in Fig. 3A and B. The bands in the range of 1610 cm−1–1540 cm−1 and 1436 cm−1–1370 cm−1 are attributed to the asymmetric and symmetric C–O stretching vibration of the carboxylate group, respectively.65,66 The difference in wavenumber (Δν) between the asymmetric and symmetric stretching of the C–O gives information on the interaction between the carboxylate group and the metal ion.67 TbBTC and GdBTC showed a Δν value of ∼174 cm−1 and ∼170 cm−1, respectively, indicating that the organic linker and metal ions are coordinated via bidentate bridging mode.68–71 The two sharp peaks at 760 and 710 cm−1 correspond to the bending of aromatic C–H bonds.2,72 FTIR spectra obtained for BSA@TbBTC, BSA@GdBTC, LC@GdBTC and LC@TbBTC do not show alterations if compared to pristine lanthanide-based materials demonstrating that the inclusion of LC does not alter the functional groups of the hosting MOF. The Raman spectra of TbBTC, BSA@TbBTC, GdBTC and BSA@GdBTC samples are shown in Fig. 3C and D. Both TbBTC and GdBTC showed the same sharp bands at 536 cm−1, 810 cm−1, 1005 cm−1, 1376 cm−1, 1455 cm−1 and 1573 cm−1 assigned to the metal–O bond vibration,57 out-of-plane CH bending, ring stretching vibrations, symmetric C–O stretching of the carboxylic group, and the asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching, respectively.73 The Raman spectra obtained for BSA@TbBTC and BSA@GdBTC samples do not present substantial differences compared to the pristine Ln-MOFs, except for the Raman shift at 3085 cm−1 and 2930 cm−1 typically associated with the C–H stretching of aromatic amino acid residues, like phenylalanine or tyrosine and the symmetric stretching of CH2 groups in aliphatic amino acid residues such as leucine, isoleucine, and valine which confirms the presence of BSA immobilization in the materials.74 LC@TbBTC and LC@GdBTC spectra showed the same bands of BSA-modified MOFs except for the band at 2930 cm−1 which was not detected. This is likely due to the lower loading of laccase respect to BSA on the protein@MOF samples. Thermogravimetric profiles of TbBTC, GdBTC, BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC are shown in Fig. 3E and F. Both pristine materials and protein@MOF samples showed a mass loss of ∼15% over the ranges 25 °C–105 °C due to water desorption.TbBTC and GdBTC exhibited a good thermal stability in air, with only a slight mass decrease between 105 °C and 430 °C. These data agree with TGA profiles of lanthanide-based MOFs obtained by green methods.75 The final decomposition step (mass loss ∼35%) occurred from 430 °C to 650 °C likely due to the decomposition of the organic moiety (trimesic acid) of the TbBTC and GdBTC. In the range from 260 °C to 430 °C, BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC showed a mass loss of 12.8%, 19.7%, 7.4% and 10.7%, respectively, imputable to the protein mass loss. A similar mass loss profile was observed in previous works referred to Aspergillus sp. laccase immobilized within FeBTC MOF.8 The loading of BSA and LC in the BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC samples quantified by TGA (mass loss in the range 260–430 °C) were 127.9 mg g−1, 197.4 mg g−1, 73.2 mg g−1, and 106.7 mg g−1, respectively (Table 1). Since the commercial Aspergillus sp. LC used in this work showed a low degree of purity (1% ≤ LC concentration ≤ 10%) the enzymatic loading obtained by TGA could be overestimated due to the presence of organic additives present in the commercial LC preparation.
O stretching, respectively.73 The Raman spectra obtained for BSA@TbBTC and BSA@GdBTC samples do not present substantial differences compared to the pristine Ln-MOFs, except for the Raman shift at 3085 cm−1 and 2930 cm−1 typically associated with the C–H stretching of aromatic amino acid residues, like phenylalanine or tyrosine and the symmetric stretching of CH2 groups in aliphatic amino acid residues such as leucine, isoleucine, and valine which confirms the presence of BSA immobilization in the materials.74 LC@TbBTC and LC@GdBTC spectra showed the same bands of BSA-modified MOFs except for the band at 2930 cm−1 which was not detected. This is likely due to the lower loading of laccase respect to BSA on the protein@MOF samples. Thermogravimetric profiles of TbBTC, GdBTC, BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC are shown in Fig. 3E and F. Both pristine materials and protein@MOF samples showed a mass loss of ∼15% over the ranges 25 °C–105 °C due to water desorption.TbBTC and GdBTC exhibited a good thermal stability in air, with only a slight mass decrease between 105 °C and 430 °C. These data agree with TGA profiles of lanthanide-based MOFs obtained by green methods.75 The final decomposition step (mass loss ∼35%) occurred from 430 °C to 650 °C likely due to the decomposition of the organic moiety (trimesic acid) of the TbBTC and GdBTC. In the range from 260 °C to 430 °C, BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC showed a mass loss of 12.8%, 19.7%, 7.4% and 10.7%, respectively, imputable to the protein mass loss. A similar mass loss profile was observed in previous works referred to Aspergillus sp. laccase immobilized within FeBTC MOF.8 The loading of BSA and LC in the BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC samples quantified by TGA (mass loss in the range 260–430 °C) were 127.9 mg g−1, 197.4 mg g−1, 73.2 mg g−1, and 106.7 mg g−1, respectively (Table 1). Since the commercial Aspergillus sp. LC used in this work showed a low degree of purity (1% ≤ LC concentration ≤ 10%) the enzymatic loading obtained by TGA could be overestimated due to the presence of organic additives present in the commercial LC preparation.
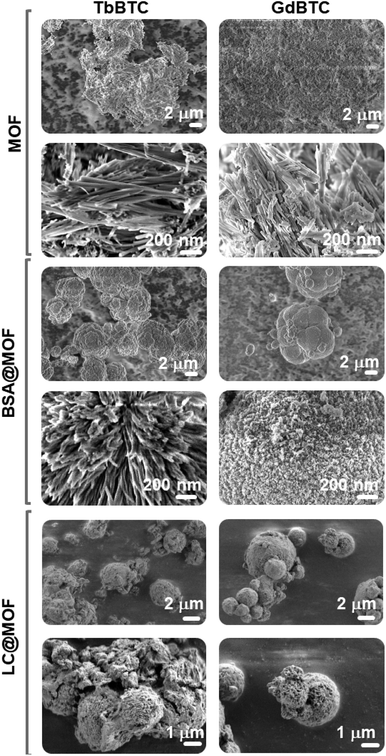 |
| | Fig. 2 SEM images of GdBTC, TbBTC, BSA@GdBTC, BSA@TbBTC LC@GdBTC and LC@TbBTC. | |
 |
| | Fig. 3 Samples characterizations. FTIR spectra of (A) TbBTC, BSA@TbBTC, and LC@TbBTC (B) GdBTC, BSA@GdBTC, and LC@GdBTC. Raman spectra of (C) TbBTC, BSA@TbBTC, and LC@TbBTC; (D) GdBTC, BSA@GdBTC, and LC@GdBTC. TGA curves from 25 °C to 650 °C of (E) TbBTC, BSA@TbBTC and LC@TbBTC; (F) GdBTC, BSA@GdBTC and LC@GdBTC. | |
Therefore, LC@GdBTC and LC@TbBTC loadings, were also quantified by mean of the Bradford assay. The results show a loading of 93.8 mg g−1 and 59.3 mg g−1 for the LC@GdBTC and LC@TbBTC samples respectively. The encapsulation efficiency (EE%), was 70.2% and 52.3% for LC@GdBTC and LC@ TbBTC, respectively (Table 1).
Location of encapsulated protein within MOFs
Although protein immobilization is gaining increasing attention, particularly regarding enzymes encapsulated within MOFs, the location of the protein within the material has not been thoroughly examined. This can be done by means of SAXS technique.52 However, that analysis can be complicated by the fact that commercial enzyme preparations often show very low purity. Indeed, they may contain other proteins, differing from the target enzyme, coming from the biological source. Moreover, commercial enzymes are formulated to optimize catalytic activity and stability by adding additives including, carbohydrates, salts, buffers etc. The result is that they may have a low enzyme content with the enzyme being only a low fraction of it.76 For these reasons, the SAXS analysis of the immobilized commercial enzymes within MOFs can be difficult. To solve these issues, BSA with high purity (>98%) was used as a model protein to investigate its location within the Ln-MOFs. More specifically, MOFs morphological changes induced by the protein loading and successive removal by calcination at 360 °C were investigated through SAXS. MOFs, BSA@MOFs and calcinated BSA@MOFs SAXS curves are shown in Fig. 4. SAXS curves were fitted according to the model reported in the Experimental section, and the fitting parameters are listed in Table 2. Pristine MOFs are characterized by inhomogeneities of 30–32 nm ca. (gyration radius, Rg, of 15–16 nm). Such objects size is attributable to the cross-section of MOFs fibrils, clearly visible in SEM micrographs (see Fig. 2). The average fibrils cross-section, measured through Image J software,77 is approximately 32 ± 7 nm in GdBTC and 36 ± 9 nm in TbBTC. This is further confirmed by the SAXS fitting parameter s, that indicates the shape of the scattering objects. Being s = 1, the scattering signal is produced by rod-like objects. BSA loading in the investigated MOFs was found to be 197.4 mg g−1 in BSA@GdBTC and 128 mg g−1 in BSA@TbBTC. Therefore, the scattering signal of BSA@MOFs is expected to be mainly produced by the MOF networks. More specifically, scattering is likely produced by mesopores in which the protein is located. As a matter of fact, protein is included within the MOFs during synthesis, inducing the formation of mesopores of about 8 nm, as previously reported for other coordination networks.51–53 This size is compatible with monomeric BSA, with each protein molecule occupying a single mesopore.51,63,78 In this case the parameter s indicates scattering from globular objects, being 0–0.4 as a result of the total or partial loss of the internal elongated structure (see Fig. 2). At low Q (length scales of 50–120 nm), the Porod exponent n indicates rough surfaces of BSA@MOFs and calcinated BSA@MOFs. Surface inhomogeneities are more pronounced in BSA@TbBTC (n = 3.3–3.4), where the scattering objects are globules containing radially-arranged needles (s = 0.44; see Fig. 2 SEM). At high Q (length scales of 3–10 nm), m = 3.80–4.00 indicates smoother surfaces in all MOFs.
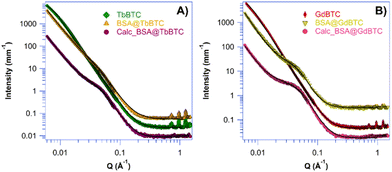 |
| | Fig. 4 SAXS profiles (full markers) and fitting (black lines) of the investigated MOFs. (A) TbBTC (green rhombus), BSA@ TbBTC (yellow triangle) and calcinated BSA@ TbBTC (purple circles). The curves have been shifted along the y axis for clarity. (B) GdBTC (red diamond), BSA@GdBTC (blue triangle) and calcinated BSA@GdBTC (pink circle). | |
Table 2 SAXS fitting parameters of MOFs, BSA@MOFs and calcinated (calc.) BSA@MOFs
| Samples |
A (scale), ×10−6 |
n (Porod low-Q) |
B (scale) |
R
g (nm) |
s
|
m (Porod high-Q) |
bkg × 10−2 |
| TbBTC |
— |
— |
21.4 ± 0.1 |
16.1 ± 0.2 |
1.04 ± 0.05 |
3.80 ± 0.05 |
1.0 ± 0.5 |
| BSA@TbBTC |
90.4 ± 0.6 |
3.34 ± 0.05 |
2.5 ± 0.1 |
4.1 ± 0.2 |
0.44 ± 0.02 |
4.00 ± 0.05 |
3.5 ± 0.5 |
| Calc. BSA@TbBTC |
57.6 ± 0.5 |
3.41 ± 0.05 |
22.4 ± 0.1 |
3.8 ± 0.2 |
0.00 ± 0.02 |
4.00 ± 0.05 |
7.0 ± 0.5 |
| GdBTC |
— |
— |
32.9 ± 0.1 |
16.6 ± 0.2 |
1.02 ± 0.05 |
3.80 ± 0.05 |
2.4 ± 0.5 |
| BSA@GdBTC |
6.49 ± 0.10 |
3.70 ± 0.05 |
14.2 ± 1.0 |
4.0 ± 0.2 |
0.00 ± 0.05 |
4.00 ± 0.06 |
13.8 ± 0.5 |
| Calc. BSA@GdBTC |
4.95 ± 0.15 |
3.60 ± 0.05 |
19.8 ± 2.0 |
3.7 ± 0.2 |
0.00 ± 0.02 |
4.00 ± 0.05 |
8.0 ± 0.5 |
Catalytic activity and stability of the LC@TbBTC and LC@GdBTC samples
Once demonstrated that the proteins are effectively encapsulated within MOFs, it is also important to verify if they keep their biological function. For this reason, the catalytic activity of LC@MOF samples was tested. The use of enzymes immobilized within MOFs obtained by green synthesis plays a key role in the carbon-neutral economy. Indeed biocatalytic processes are green and sustainable.79,80 Due to their low substrate specificity and high redox potential,81 LCs can degrade several compounds possessing a phenolic structure, including lignin.82 Thus, LCs are promising biocatalysts for biorefinery processes. Moreover, LCs are extensively used in bioremediation processes and for biosensing.6 Since the laccase kinetic parameters can be affected by the immobilization process as well as by the chosen support, the effect of immobilization on the kinetic parameters of Aspergillus sp. laccase, used as a model enzyme, was then investigated. More precisely, the Vmax and KM parameters were obtained from the Michaelis–Menten plot (Fig. 5A), measuring the initial rate, V0, as a function of ABTS concentration. Moreover, for a better comparison among free and immobilized enzymes, a plot specific activity vs. ABTS concentration is also shown (Fig. 5B). Both LC@GdBTC and LC@TbBTC showed an increase in KM value, that is 1.6 and 3.0 times higher than that of the free laccase, respectively, suggesting that ABTS substrate has a lower affinity once LC is immobilized in TbBTC and GdBTC. Aspergillus sp. LC showed a 3.4- and 2-fold loss in specific activity once immobilized within GdBTC and TbBTC, respectively (Table 3). LC@TbBTC showed a higher specific activity (10.8 μmol min−1 mg−1) than LC@GdBTC (6.6 μmol min−1 mg−1). Recently, Aspergillus sp. laccase immobilized in situ within FeBTC showed a specific activity of only 0.17 μmol mg−1 min−1.8 These data suggest that Aspergillus sp. laccase is better adapted to TbBTC and GdBTC MOFs rather than FeBTC. Similar modifications of kinetic parameters after enzyme immobilization have been previously observed. Indeed, Patil et al. obtained a 1.5 fold increase in KM value, and a 1.2 fold decrease in Vmax due to the immobilization of the LC from Trametes hirsuta within ZIF-8.83 Li et al. immobilized in situ a LC in a bimetallic CoCu-MOF finding that the Vmax decreased 3.86 times and the KM value increased of 6 times compared to the free LC.84 Zhang et al. immobilized in situ Bacillus subtilis laccase within CuBTC obtaining a KM increase from 0.108 mM to 0.159 mM due to the LC immobilization.85 Several factors such as the changes in protein conformation and the accessibility of the enzyme's active site, may lead to a change in kinetic parameters between free and immobilized enzymes.52
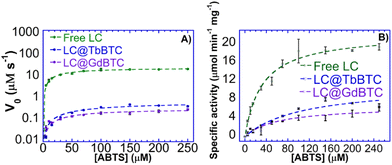 |
| | Fig. 5 Michaelis–Menten plots of the (A) V0vs. [ABTS] and and (B) specific activity vs. [ABTS] of free LC, LC@TbBTC and LC@GdBTC. | |
Table 3 Kinetic parameters of the free LC, LC@TbBTC and LC@GdBTC
| Samples |
K
M (μM) |
V
max (μM s−1) |
Specific activity (μmol min−1 mg−1) |
| Free LC |
32.1 |
27.2 |
22.3 |
| LC@TbBTC |
97.9 |
0.71 |
10.8 |
| LC@GdBTC |
59.7 |
0.34 |
6.6 |
The large-scale application of laccases is often hindered by their low stability, which increases their cost of use.86 Generally, compared with free laccases, immobilized laccases show higher operational and storage stabilities, moreover, immobilized LCs can be easily separated from the reaction medium.87 The storage stability of LC@TbBTC and LC@GdBTC samples dispersed in water were hence studied. LC@TbBTC storage stability (Fig. 6A) decreased up to ∼50% and the catalyst retained a constant activity up to 3 weeks. LC@GdBTC retained ∼60% of its initial activity at the 7th day and about ∼20% of its initial activity at the 21st day. A similar result was reported by Birhanlı et al. who encapsulated LC from T. trogii within cobalt- and copper-based MOFs retaining more than 58% of their initial activity over 28 days.88 A recent interesting work by Magner and coworkers showed that buffer composition and concentration play a key role in the stability of the MOFs.89 Their results indicated that the trimesic acid-based MOF (Fe-BTC, Cu-TMA, Co-TMA, Ni-TMA) are unstable in the presence of citrate buffer whilst the acetate is considered a promising storage medium. Hence, the stability of LC@TbBTC and LC@GdBTC stored in acetate buffer was also investigated. Both the LC@TbBTC stored in acetate buffer showed the same trend as the LC@TbBTC stored in water up to the 14th day. In the third week, LC@TbBTC stored in acetate buffer retained 37% of its initial activity, that is ∼10% of activity lower than LC@TbBTC stored in water (Fig. 6B) whilst the LC@GdBTC stored in acetate buffer (Fig. 6B) decreased its specific activity more rapidly than LC@GdBTC stored in water (30% vs. 24% after 1 week). From the 1st week it decreased up to ∼70% of its initial specific activity and retained a constant activity up to 3 weeks. LC@TbBTC showed a higher stability in acetate buffer rather than in water, whereas LC@GdBTC had an opposite behaviour. The in situ immobilization at room temperature within TbBTC and GdBTC MOFs, could be a valuable strategy to enhance the stability of Aspergillus sp. laccase. The results demonstrated that the MOF served as a protective host carrier that could maintain long storage stability.
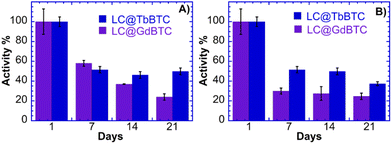 |
| | Fig. 6 Storage stability (A) LC@GdBTC and LC@TbBTC; (B) storage stability on acetate buffer LC@GdBTC and LC@TbBTC. | |
Luminescent properties of LC@TbBTC
Besides application for green processes, enzyme@MOFs could find application in medicine. Indeed, some Ln-based MOFs display luminescent properties that have interesting potential for sensing applications.90 Moreover some LCs (i.e. A. cylindracea and Pleurotus cornucopiae laccase) displayed antiproliferative activity toward cancer cells and inhibitory activity toward HIV-1 reverse transcriptase.91,92 Recently, Zhou et al. encapsulated a LC within ZIF-8 MOFs to obtain a laccase@ZIF-8-prodrug system with GSH redox cycle able to induce tumor cell apoptosis.93 Hence, the properties of laccases could be combined with those of Ln-MOFs for theranostic purposes.94,95 In this regards, the solid-state luminescence properties of TbBTC, BSA@TbBTC and LC@TbBTC were investigated at room temperature, whereas the GdBTC MOFs were not analyzed since they are not luminescent. Upon excitation at 366 nm, both pristine TbBTC and protein@TbBTC samples show emission peaks at 488, 544, 583, and 620 nm, which could be assigned to the 5D4 → 7FJ (J = 6, 5, 4, and 3) transitions of Tb3+ ions (Fig. 7). The strongest transition was the 5D4 → 7F5 band ca. 544 nm, which is responsible for the characteristic green color of Tb3+ emission.96 Protein@TbBTC could be used for sensing applications. For example, Wang et al. developed a biosensor based on a bimetallic Tb-Fe-MOF linked with an aptamer biomolecule able to bind to ovarian cancer biomarkers. The detection sensitivity of the obtained aptamer biosensor toward the targeted antigen was evaluated by electrochemical impedance spectroscopy, whereas the detection of living cancer cells was carried out by confocal laser spectroscopy due to the intrinsic fluorescence of Tb-Fe-MOF.97 Although the synthesis of luminescent MOFs is widely described in the literature, luminescent MOFs are typically obtained under harsh reaction conditions posing a challenge for the encapsulation of proteins within them. For example, Chen et al. synthesized a luminescent TbMOF capable of selectively detecting Al3+ and Fe3+ ions in water, however their synthesis was carried out at 80 °C for 72 h under autogenous pressure.98 The facile and green synthesis to obtain TbBTC reported in this work is more sustainable from the environmental and economic point of view compared to the conventional Tb-based MOFs synthesis.
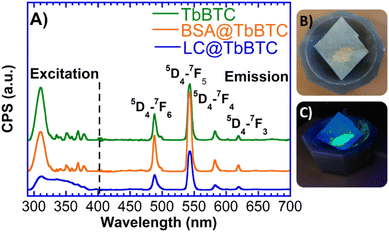 |
| | Fig. 7 (A) Solid state fluorescence spectra of TbBTC, BSA@TbBTC and LC@TbBTC samples. (B) LC@TbBTC sample and (C) LC@TbBTC sample under UV light (366 nm). | |
Conclusions
In this work, BSA protein and Aspergillus sp. LC enzyme were immobilized within two lanthanide-based crystalline MOFs in situ under mild reaction conditions, that is using water as the solvent in 1 h at 25 °C and atmospheric pressure, in agreement with the 3rd, 5th, and 6th principles of Green Chemistry.99 Pristine MOFs and protein@MOF samples were characterized employing WAXS, SEM, TGA, Raman and FTIR techniques. The enzyme location within MOFs significantly impacts their catalytic activity and stability. The immobilization of high-purity model proteins, like BSA, allows the investigation of the protein location in detail. SAXS analysis beside, confirming BSA encapsulation within MOFs structure, revealed the occurrence of mesopores of about 8 nm corresponding to the cavities created by encapsulated BSA molecules.52 SEM micrographs revealed a needle-shaped morphology of pristine MOFs with a cross-section of ca. 35 nm, also confirmed by SAXS data analysis. BSA loading changes MOFs morphology, and needle-shaped objects are partially retained only in BSA@TbBTC sample. Immobilized laccase samples, LC@TbBTC and LC@GdBTC, kept their catalytic activity although showing a slight a decrease in Vmax and an increase in KM compared to the free LC most likely due to the mass transfer or enzyme conformational change upon MOFs immobilization. Indeed, the Vmax as well the storage stability of Aspergillus sp. laccase was improved significantly upon immobilization within TbBTC and GdBTC in comparison with other MOFs like FeBTC.8 Furthermore, the immobilization of proteins/enzymes in a luminescent or a paramagnetic MOF, such as TbBTC and GdBTC, obtained through a ‘green’ approach, could lead to the sustainable development of biomedical devices. The luminescent properties of terbium-based MOFs combined with biomolecule activity suggest their applicability for theranostic purposes. For example, the laccase@ZIF-8-prodrug system with a GSH redox cycle developed by Zhou et al. to induce tumor cell apoptosis could be further improved by utilizing luminescent MOFs. Indeed, the intrinsic fluorescence of luminescent MOFs, compared to non-luminescent ones, could allow the detection of cancer cells by confocal laser spectroscopy, making the system a theranostic one.93 Further work is needed to investigate the paramagnetic features and theranostic potential of enzymes encapsulated within gadolinium-based MOFs. The immobilization of proteins within crystalline MOFs synthesized under mild reaction conditions proposed in this work, paves the way for the realization of innovative devices which could contribute to the sustainable evolution of biomedical technologies.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Data availability
The data supporting this article have been included as part of the ESI.†
Conflicts of interest
There are no conflicts of interest to declare.
Acknowledgements
DT thanks MIUR (PON RI 2014-2020, Azione l.1 “Dottorati Innovativi con Caratterizzazione industriale”-DOT1304455 2) for financing his Ph.D scholarship and Deutscher Akademische Austauschdienst (DAAD) for funding his research period abroad at FAU Erlangen-Nürnberg. CSGI (Florence) is kindly acknowledged for partial financial support. M.H. acknowledges financial support of this work by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), project-ID 416229255-SFB 1411.
References
- S. R. Batten, N. R. Champness, X.-M. Chen, J. Garcia-Martinez, S. Kitagawa, L. Öhrström, M. O’Keeffe, M. P. Suh and J. Reedijk, Pure Appl. Chem., 2013, 85, 1715–1724 CrossRef CAS.
- G. Autie-Castro, M. A. Autie, E. Rodríguez-Castellón, C. Aguirre and E. Reguera, Colloids Surf., A, 2015, 481, 351–357 CrossRef CAS.
- K. Hemmer, M. Cokoja and R. A. Fischer, ChemCatChem, 2021, 13, 1683–1691 CrossRef CAS.
- M.-X. Wu and Y.-W. Yang, Adv. Mater., 2017, 29, 1606134 CrossRef PubMed.
- G. R. Delpiano, D. Tocco, L. Medda, E. Magner and A. Salis, Int. J. Mol. Sci., 2021, 22, 788 CrossRef CAS PubMed.
- D. Tocco, C. Carucci, M. Monduzzi, A. Salis and E. Sanjust, ACS Sustainable Chem. Eng., 2021, 9, 2412–2432 CrossRef CAS.
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef PubMed.
- D. Tocco, C. Carucci, D. Todde, K. Shortall, F. Otero, E. Sanjust, E. Magner and A. Salis, Colloids Surf., B, 2021, 208, 112147 CrossRef CAS PubMed.
- V. Gascón, M. B. Jiménez, R. M. Blanco and M. Sanchez-Sanchez, Catal. Today, 2018, 304, 119–126 CrossRef.
- V. Gascón, C. Carucci, M. B. Jiménez, R. M. Blanco, M. Sánchez-Sánchez and E. Magner, ChemCatChem, 2017, 9, 1182–1186 CrossRef.
- S. Gulati, A. Choudhury, G. Mohan, R. Katiyar, M. A. Kurikkal M P, S. Kumar and R. S. Varma, J. Mater. Chem. B, 2023, 11, 6782–6801 RSC.
- L. Zhao, W. Zhang, Q. Wu, C. Fu, X. Ren, K. Lv, T. Ma, X. Chen, L. Tan and X. Meng, J. Nanobiotechnol., 2022, 20, 133 CrossRef CAS PubMed.
- S.-Y. Zhang, Z.-Y. Wang, J. Gao, K. Wang, E. Gianolio, S. Aime, W. Shi, Z. Zhou, P. Cheng and M. J. Zaworotko, Chem, 2019, 5, 1609–1618 CAS.
- K. Wang, Y.-L. Zhu, T.-F. Zheng, X. Xie, J.-L. Chen, Y.-Q. Wu, S.-J. Liu and H.-R. Wen, Anal. Chem., 2023, 95, 4992–4999 CrossRef CAS.
- Y. Li, B.-L. Chai, H. Xu, T.-F. Zheng, J.-L. Chen, S.-J. Liu and H.-R. Wen, Inorg. Chem. Front., 2022, 9, 1504–1513 RSC.
- S. Sahoo, S. Mondal and D. Sarma, Coord. Chem. Rev., 2022, 470, 214707 CrossRef CAS.
- T. Zhang, L. Qin, L. Liu, M. Zhang, T. Du, Y. Fan, H. Yan, P. Su, P. Zhou and Y. Tang, J. Rare Earths, 2022, 40, 1371–1381 CrossRef CAS.
- B. Ortega-Berlanga, L. Betancourt-Mendiola, C. del Angel-Olarte, L. Hernández-Adame, S. Rosales-Mendoza and G. Palestino, Crystals, 2021, 11, 1094 CrossRef CAS.
- W. Hatakeyama, T. J. Sanchez, M. D. Rowe, N. J. Serkova, M. W. Liberatore and S. G. Boyes, ACS Appl. Mater. Interfaces, 2011, 3, 1502–1510 CrossRef CAS PubMed.
- X. Wang, P. C. Lan and S. Ma, ACS Cent. Sci., 2020, 6, 1497–1506 CrossRef CAS PubMed.
- D. J. Tranchemontagne, J. L. Mendoza-Cortés, M. O'Keeffe and O. M. Yaghi, Chem. Soc. Rev., 2009, 38, 1257–1283 RSC.
- C. K. Oliveira, J. R. de Menezes Vicenti, R. A. Burrow, S. Alves, R. L. Longo and I. Malvestiti, Inorg. Chem. Commun., 2012, 22, 54–59 CrossRef CAS.
- Y. Zhang and B. Yan, Nanoscale, 2019, 11, 22946–22953 RSC.
- J. Gao, C. Wang and H. Tan, J. Mater. Chem. B, 2017, 5, 7692–7700 RSC.
- D. Tocco, D. Wisser, M. Fischer, W. Schwieger, A. Salis and M. Hartmann, Colloids Surf., B, 2023, 226, 113311 CrossRef CAS PubMed.
- A. Salis, M. Pisano, M. Monduzzi, V. Solinas and E. Sanjust, J. Mol. Catal. B: Enzym., 2009, 58, 175–180 CrossRef CAS.
- M. Hartmann and X. Kostrov, Chem. Soc. Rev., 2013, 42, 6277–6289 RSC.
- F. Lyu, Y. Zhang, R. N. Zare, J. Ge and Z. Liu, Nano Lett., 2014, 14, 5761–5765 CrossRef CAS PubMed.
- Y.-R. Lee, J. Kim and W.-S. Ahn, Korean J. Chem. Eng., 2013, 30, 1667–1680 CrossRef CAS.
- D. J. Tranchemontagne, J. R. Hunt and O. M. Yaghi, Tetrahedron, 2008, 64, 8553–8557 CrossRef CAS.
- X. Shi, G. Zhu, S. Qiu, K. Huang, J. Yu and R. Xu, Angew. Chem., Int. Ed., 2004, 43, 6482–6485 CrossRef CAS PubMed.
- Q. Xia, H. Wang, B. Huang, X. Yuan, J. Zhang, J. Zhang, L. Jiang, T. Xiong and G. Zeng, Small, 2019, 15, 1–25 Search PubMed.
- S. S. Nadar, L. Vaidya and V. K. Rathod, Int. J. Biol. Macromol., 2020, 149, 861–876 CrossRef CAS PubMed.
- R. J. Drout, L. Robison and O. K. Farha, Coord. Chem. Rev., 2019, 381, 151–160 CrossRef CAS.
- K. Liang, C. J. Coghlan, S. G. Bell, C. Doonan and P. Falcaro, Chem. Commun., 2016, 52, 473–476 RSC.
- S. S. Nadar and V. K. Rathod, Enzyme Microb. Technol., 2018, 108, 11–20 CrossRef CAS PubMed.
- M. Naseri, F. Pitzalis, C. Carucci, L. Medda, L. Fotouhi, E. Magner and A. Salis, ChemCatChem, 2018, 10, 5425 CrossRef CAS.
- Y. Cao, X. Li, J. Xiong, L. Wang, L. T. Yan and J. Ge, Nanoscale, 2019, 11, 22108–22117 RSC.
- F.-K. Shieh, S.-C. Wang, C.-I. Yen, C.-C. Wu, S. Dutta, L.-Y. Chou, J. V. Morabito, P. Hu, M.-H. Hsu, K. C. W. Wu and C.-K. Tsung, J. Am. Chem. Soc., 2015, 137, 4276–4279 CrossRef CAS PubMed.
- S. Chen, L. Wen, F. Svec, T. Tan and Y. Lv, RSC Adv., 2017, 7, 21205–21213 RSC.
- J. Cui, Y. Feng, T. Lin, Z. Tan, C. Zhong and S. Jia, ACS Appl. Mater. Interfaces, 2017, 9, 10587–10594 CrossRef CAS PubMed.
- X. Pei, Y. Wu, J. Wang, Z. Chen, W. Liu, W. Su and F. Liu, Nanoscale, 2020, 12, 967–972 RSC.
- E. Gkaniatsou, C. Sicard, R. Ricoux, L. Benahmed, F. Bourdreux, Q. Zhang, C. Serre, J. Mahy and N. Steunou, Angew. Chem., Int. Ed., 2018, 57, 16141–16146 CrossRef CAS PubMed.
- S. Dai, A. Tissot and C. Serre, Bull. Chem. Soc. Jpn., 2021, 94, 2623–2636 CrossRef CAS.
- M. Sanchez-Sanchez, I. de Asua, D. Ruano and K. Diaz, Cryst. Growth Des., 2015, 15, 4498–4506 CrossRef CAS.
- C. Carucci, L. Bruen, V. Gascón, F. Paradisi and E. Magner, Langmuir, 2018, 34, 8274–8280 CrossRef CAS PubMed.
- H. Brunckova, E. Mudra, L. Rocha, E. Nassar, W. Nascimento, H. Kolev, A. Kovalcikova, Z. Molcanova and M. Podobova, Appl. Surf. Sci., 2021, 542, 148731 CrossRef CAS.
- M. Oggianu, F. Manna, S. Ashoka Sahadevan, N. Avarvari, A. Abhervé and M. L. Mercuri, Crystals, 2022, 12, 763 CrossRef CAS.
- T. N. Blanton, T. C. Huang, H. Toraya, C. R. Hubbard, S. B. Robie, D. Louër, H. E. Göbel, G. Will, R. Gilles and T. Raftery, Powder Diffr., 1995, 10, 91–95 CrossRef CAS.
- F. Zhang, J. Ilavsky, G. G. Long, J. P. G. Quintana, A. J. Allen and P. R. Jemian, Metall. Mater. Trans. A, 2010, 41, 1151–1158 CrossRef.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Nat. Commun., 2015, 6, 7240 CrossRef CAS PubMed.
- D. Tocco, D. Chelazzi, R. Mastrangelo, A. Casini, A. Salis, E. Fratini and P. Baglioni, J. Colloid Interface Sci., 2023, 641, 685–694 CrossRef CAS PubMed.
- W. Liang, F. Carraro, M. B. Solomon, S. G. Bell, H. Amenitsch, C. J. Sumby, N. G. White, P. Falcaro and C. J. Doonan, J. Am. Chem. Soc., 2019, 141, 14298–14305 CrossRef CAS PubMed.
- B. Hammouda, J. Appl. Crystallogr., 2010, 43, 716–719 CrossRef CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- D. W. S. Wong, Appl. Biochem. Biotechnol., 2009, 157, 174 CrossRef CAS PubMed.
- H. Brunckova, E. Mudra, M. Streckova, L. Medvecky, T. Sopcak, I. Shepa, A. Kovalcikova, M. Lisnichuk and H. Kolev, Nanomaterials, 2022, 12, 2817 CrossRef CAS PubMed.
- K. Song, H. Yu, J. Zhang, Y. Bai, Y. Guan, J. Yu and L. Guo, Crystals, 2020, 10, 185 CrossRef CAS.
- K. Liu, Y. Zheng, G. Jia, M. Yang, Y. Song, N. Guo and H. You, J. Solid State Chem., 2010, 183, 2309–2316 CrossRef CAS.
- W. Yang, J. Feng and H. Zhang, J. Mater. Chem., 2012, 22, 6819–6823 RSC.
- S. A. Al-Harbi and Y. Q. Almulaiky, Heliyon, 2024, 10, e28396 CrossRef CAS.
- C. Nunes, A. Mahendrasingam and R. Suryanarayanan, Pharm. Res., 2005, 22, 1942–1953 CrossRef CAS PubMed.
- L. Qi, Z. Luo and X. Lu, ACS Sustainable Chem. Eng., 2019, 7, 7127–7139 CrossRef CAS.
- F. Lo, A. Borzì, A. Lucia, P. Rossi, P. Paoli and G. Malandrino, Results Chem., 2022, 4, 100640 CrossRef.
- Y. Yang, Y. Bai, F. Zhao, E. Yao, J. Yi, C. Xuan and S. Chen, RSC Adv., 2016, 6, 67308–67314 RSC.
- H. Lv, H. Zhao, T. Cao, L. Qian, Y. Wang and G. Zhao, J. Mol. Catal. A: Chem., 2015, 400, 81–89 CrossRef CAS.
- G. Deacon, Coord. Chem. Rev., 1980, 33, 227–250 CrossRef CAS.
- K. I. Hadjiivanov, D. A. Panayotov, M. Y. Mihaylov, E. Z. Ivanova, K. K. Chakarova, S. M. Andonova and N. L. Drenchev, Chem. Rev., 2021, 121, 1286–1424 CrossRef CAS PubMed.
- L. D. S. Flores, R. I. Rosa, J. D. S. Martins, R. R. Pinho, R. Diniz and C. C. Corrêa, Eur. J. Chem., 2019, 10, 180–188 CrossRef CAS.
- J. F. S. do Nascimento, A. M. U. de Araújo, J. Kulesza, A. F. de Farias Monteiro, S. A. Júnior and B. S. Barros, New J. Chem., 2018, 42, 5514–5522 RSC.
- E. R. Souza, I. G. N. Silva, E. E. S. Teotonio, M. C. F. C. Felinto and H. F. Brito, J. Lumin., 2010, 130, 283–291 CrossRef.
- B. Pangkumhang, P. Jutaporn, K. Sorachoti, P. Khamdahsag and V. Tanboonchuy, Sains Malays., 2019, 48, 199–208 CrossRef CAS.
- M. Zeng, A. Ren, W. Wu, Y. Zhao, C. Zhan and J. Yao, Chem. Sci., 2020, 11, 9154–9161 RSC.
- B. Fazio, C. D'Andrea, A. Foti, E. Messina, A. Irrera, M. G. Donato, V. Villari, N. Micali, O. M. Maragò and P. G. Gucciardi, Sci. Rep., 2016, 6, 26952 CrossRef CAS PubMed.
- Y. Li, S. Wu, Y. Zhang, Z. Ma, M. Zhu and E. Gao, Inorg. Chim. Acta, 2021, 528, 120632 CrossRef CAS.
- A. Salis, E. Sanjust, V. Solinas and M. Monduzzi, Biocatal. Biotransform., 2005, 23, 381–386 CrossRef CAS.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS.
- R. Murty, M. K. Bera, I. M. Walton, C. Whetzel, M. R. Prausnitz and K. S. Walton, J. Am. Chem. Soc., 2023, 145, 7323–7330 CrossRef CAS.
- A. R. Alcántara, P. Domínguez de María, J. A. Littlechild, M. Schürmann, R. A. Sheldon and R. Wohlgemuth, ChemSusChem, 2022, 15, e202102709 CrossRef.
- R. A. Sheldon and D. Brady, ChemSusChem, 2022, 15, e202102628 CrossRef CAS PubMed.
- S. Riva, Trends Biotechnol., 2006, 24, 219–226 CrossRef CAS PubMed.
- Y. Liu, G. Luo, H. H. Ngo, W. Guo and S. Zhang, Bioresour. Technol., 2020, 298, 122511 CrossRef CAS PubMed.
- P. D. Patil and G. D. Yadav, ChemistrySelect, 2018, 3, 4669–4675 CrossRef CAS.
- X. Li, Z. Wu, X. Tao, R. Li, D. Tian and X. Liu, J. Hazard. Mater., 2022, 438, 129525 CrossRef CAS.
- C. Zhang, S. You, J. Zhang, W. Qi, R. Su and Z. He, Bioresour. Technol., 2020, 308, 123271 CrossRef CAS.
- Z. Shokri, F. Seidi, S. Karami, C. Li, M. R. Saeb and H. Xiao, Carbohydr. Polym., 2021, 262, 117963 CrossRef CAS PubMed.
- I. Bassanini, E. E. Ferrandi, S. Riva and D. Monti, Catalysts, 2020, 11, 26 CrossRef.
- E. Birhanlı, S. A. A. Noma, F. Boran, A. Ulu, Ö. Yeşilada and B. Ateş, Chemosphere, 2022, 292, 133382 CrossRef PubMed.
- K. Shortall, F. Otero, S. Bendl, T. Soulimane and E. Magner, Langmuir, 2022, 38, 13382–13391 CrossRef CAS PubMed.
- P. Samanta, S. Let, W. Mandal, S. Dutta and S. K. Ghosh, Inorg. Chem. Front., 2020, 7, 1801–1821 RSC.
- D. D. Hu, R. Y. Zhang, G. Q. Zhang, H. X. Wang and T. B. Ng, Phytomedicine, 2011, 18, 374–379 CrossRef CAS PubMed.
- X. Wu, C. Huang, Q. Chen, H. Wang and J. Zhang, Biomed. Chromatogr., 2014, 28, 548–553 CrossRef CAS PubMed.
- Y. Zhou, C. Zhang, Y. Wang, J. Zhang, X. Yan, S. You, W. Qi, R. Su and Z. He, Colloids Surf., B, 2022, 220, 112853 CrossRef CAS PubMed.
- F. Wang, Y. Pu, X. Zhang, F. Zhang, H. Cheng and Y. Zhao, J. Lumin., 2019, 206, 192–198 CrossRef CAS.
- X. Ji, J. Hong and X. Guo, Analyst, 2012, 137, 710–715 RSC.
- K. P. Carter, S. J. A. Pope, M. Kalaj, R. J. Holmberg, M. Murugesu and C. L. Cahill, Z. Anorg. Allg. Chem., 2017, 643, 1948–1955 CrossRef CAS.
- M. Wang, M. Hu, Z. Li, L. He, Y. Song, Q. Jia, Z. Zhang and M. Du, Biosens. Bioelectron., 2019, 142, 111536 CrossRef PubMed.
- Y. Chen, J. Qiu, Z. Chen, Y. Zhao, B. Li and C. Zeng, Dyes Pigm., 2021, 194, 109671 CrossRef CAS.
- P. Anastas and N. Eghbali, Chem. Soc. Rev., 2010, 39, 301–312 RSC.
|
| This journal is © The Royal Society of Chemistry 2024 |
Click here to see how this site uses Cookies. View our privacy policy here.  c,
Andrea
Salis
c,
Andrea
Salis
 *a and
Martin
Hartmann
b
*a and
Martin
Hartmann
b
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ, where 2θ is the scattering angle. The scattering camera was calibrated using silver behenate (d = 58.38 Å) as the ref. 49. Data were further corrected by empty cell subtraction and normalized, considering the measurement time and relative transmission factors. Finally, 1D SAXS/WAXS curves collected at the two different distances were merged in a single scattering profile and converted to absolute intensity, using glassy carbon as a secondary standard.50 SAXS/WAXS data treatment was performed by XSACT software (Xenocs). The materials crystallinity was evaluated by the WAXS technique and quantified by the following equation:
θ, where 2θ is the scattering angle. The scattering camera was calibrated using silver behenate (d = 58.38 Å) as the ref. 49. Data were further corrected by empty cell subtraction and normalized, considering the measurement time and relative transmission factors. Finally, 1D SAXS/WAXS curves collected at the two different distances were merged in a single scattering profile and converted to absolute intensity, using glassy carbon as a secondary standard.50 SAXS/WAXS data treatment was performed by XSACT software (Xenocs). The materials crystallinity was evaluated by the WAXS technique and quantified by the following equation: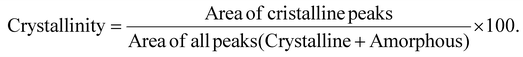
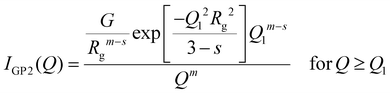
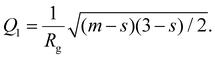
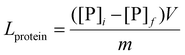
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 M−1 cm−1) per minute at 25 °C. Briefly, 100 μL of commercially available Aspergillus sp. LC (diluted 1
700 M−1 cm−1) per minute at 25 °C. Briefly, 100 μL of commercially available Aspergillus sp. LC (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) were added to 2.7 mL of acetate buffer (pH 5, 100 mM) containing 150 μL of ABTS (5 mM) in a cuvette kept in the dark under stirring at T = 25 °C. The catalytic activity of LC@TbBTC and LC@GdBTC was carried out following the aforementioned procedure but adding 100 μL of biocatalyst dispersion (2 mg mL−1) instead of the free LC. The activity of TbBTC and GdBTC (with no LC) was carried out as a control experiment. No activity was detected for LC-free MOFs. The kinetic parameters, KM and Vmax, of free LC and LC@MOFs were obtained by measuring the initial reaction rate (V0) as a function of ABTS concentration in the range 5–250 μM. A plot of V0 (μM s−1) versus ABTS concentration was built and the experimental points were fitted using the Michaelis–Menten model by mean of KaleidaGraph (4.5.4) software. All measurements were carried out at least in triplicate.
500) were added to 2.7 mL of acetate buffer (pH 5, 100 mM) containing 150 μL of ABTS (5 mM) in a cuvette kept in the dark under stirring at T = 25 °C. The catalytic activity of LC@TbBTC and LC@GdBTC was carried out following the aforementioned procedure but adding 100 μL of biocatalyst dispersion (2 mg mL−1) instead of the free LC. The activity of TbBTC and GdBTC (with no LC) was carried out as a control experiment. No activity was detected for LC-free MOFs. The kinetic parameters, KM and Vmax, of free LC and LC@MOFs were obtained by measuring the initial reaction rate (V0) as a function of ABTS concentration in the range 5–250 μM. A plot of V0 (μM s−1) versus ABTS concentration was built and the experimental points were fitted using the Michaelis–Menten model by mean of KaleidaGraph (4.5.4) software. All measurements were carried out at least in triplicate.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching, respectively.73 The Raman spectra obtained for BSA@TbBTC and BSA@GdBTC samples do not present substantial differences compared to the pristine Ln-MOFs, except for the Raman shift at 3085 cm−1 and 2930 cm−1 typically associated with the C–H stretching of aromatic amino acid residues, like phenylalanine or tyrosine and the symmetric stretching of CH2 groups in aliphatic amino acid residues such as leucine, isoleucine, and valine which confirms the presence of BSA immobilization in the materials.74 LC@TbBTC and LC@GdBTC spectra showed the same bands of BSA-modified MOFs except for the band at 2930 cm−1 which was not detected. This is likely due to the lower loading of laccase respect to BSA on the protein@MOF samples. Thermogravimetric profiles of TbBTC, GdBTC, BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC are shown in Fig. 3E and F. Both pristine materials and protein@MOF samples showed a mass loss of ∼15% over the ranges 25 °C–105 °C due to water desorption.TbBTC and GdBTC exhibited a good thermal stability in air, with only a slight mass decrease between 105 °C and 430 °C. These data agree with TGA profiles of lanthanide-based MOFs obtained by green methods.75 The final decomposition step (mass loss ∼35%) occurred from 430 °C to 650 °C likely due to the decomposition of the organic moiety (trimesic acid) of the TbBTC and GdBTC. In the range from 260 °C to 430 °C, BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC showed a mass loss of 12.8%, 19.7%, 7.4% and 10.7%, respectively, imputable to the protein mass loss. A similar mass loss profile was observed in previous works referred to Aspergillus sp. laccase immobilized within FeBTC MOF.8 The loading of BSA and LC in the BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC samples quantified by TGA (mass loss in the range 260–430 °C) were 127.9 mg g−1, 197.4 mg g−1, 73.2 mg g−1, and 106.7 mg g−1, respectively (Table 1). Since the commercial Aspergillus sp. LC used in this work showed a low degree of purity (1% ≤ LC concentration ≤ 10%) the enzymatic loading obtained by TGA could be overestimated due to the presence of organic additives present in the commercial LC preparation.
O stretching, respectively.73 The Raman spectra obtained for BSA@TbBTC and BSA@GdBTC samples do not present substantial differences compared to the pristine Ln-MOFs, except for the Raman shift at 3085 cm−1 and 2930 cm−1 typically associated with the C–H stretching of aromatic amino acid residues, like phenylalanine or tyrosine and the symmetric stretching of CH2 groups in aliphatic amino acid residues such as leucine, isoleucine, and valine which confirms the presence of BSA immobilization in the materials.74 LC@TbBTC and LC@GdBTC spectra showed the same bands of BSA-modified MOFs except for the band at 2930 cm−1 which was not detected. This is likely due to the lower loading of laccase respect to BSA on the protein@MOF samples. Thermogravimetric profiles of TbBTC, GdBTC, BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC are shown in Fig. 3E and F. Both pristine materials and protein@MOF samples showed a mass loss of ∼15% over the ranges 25 °C–105 °C due to water desorption.TbBTC and GdBTC exhibited a good thermal stability in air, with only a slight mass decrease between 105 °C and 430 °C. These data agree with TGA profiles of lanthanide-based MOFs obtained by green methods.75 The final decomposition step (mass loss ∼35%) occurred from 430 °C to 650 °C likely due to the decomposition of the organic moiety (trimesic acid) of the TbBTC and GdBTC. In the range from 260 °C to 430 °C, BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC showed a mass loss of 12.8%, 19.7%, 7.4% and 10.7%, respectively, imputable to the protein mass loss. A similar mass loss profile was observed in previous works referred to Aspergillus sp. laccase immobilized within FeBTC MOF.8 The loading of BSA and LC in the BSA@TbBTC, BSA@GdBTC, LC@TbBTC and LC@GdBTC samples quantified by TGA (mass loss in the range 260–430 °C) were 127.9 mg g−1, 197.4 mg g−1, 73.2 mg g−1, and 106.7 mg g−1, respectively (Table 1). Since the commercial Aspergillus sp. LC used in this work showed a low degree of purity (1% ≤ LC concentration ≤ 10%) the enzymatic loading obtained by TGA could be overestimated due to the presence of organic additives present in the commercial LC preparation.






