Nanoplastic at environmentally relevant concentrations activates a germline mir-240-rab-5 signaling cascade to affect the secreted ligands associated with transgenerational toxicity induction in C. elegans†
Xin
Hua
a,
Le
Zhang
a and
Dayong
Wang
 *ab
*ab
aKey Laboratory of Environmental Medicine Engineering of Ministry of Education, Medical School, Southeast University, Nanjing 210009, China. E-mail: dayongw@seu.edu.cn
bShenzhen Ruipuxun Academy for Stem Cell & Regenerative Medicine, Shenzhen, China
First published on 2nd July 2024
Abstract
Epigenetic regulation plays an important role in regulating the transgenerational toxicity of pollutants. However, the underlying mechanism of microRNAs (miRNAs) in regulating transgenerational nanoplastic toxicity remains largely unclear. We aimed to determine the miRNA-mediated mechanism for the induction of transgenerational nanoplastic toxicity. In Caenorhabditis elegans, although germline RNAi of both mir-240 and mir-36 suppressed polystyrene nanoparticle (PS-NP) toxicity, exposure to PS-NPs (1–100 μg L−1) only increased mir-240 expression. A transgenerational increase in mir-240 expression was observed after PS-NP exposure at P0 generation (P0-G), and the germline RNAi of mir-240 suppressed transgenerational PS-NP toxicity. Among the predicted target genes of mir-240 in the germline, the exposure to PS-NPs (1–100 μg L−1) decreased rab-5 and rab-6.2 expressions, whereas the germline RNAi of mir-240 only increased rab-5 expression in PS-NP exposed nematodes. A transgenerational decrease in rab-5 expression was detected after PS-NP exposure at P0-G, and the germline RNAi of rab-5 strengthened transgenerational PS-NP toxicity. Moreover, the resistance of mir-240(RNAi) to transgenerational PS-NP toxicity in inhibiting locomotion behavior and in reducing the brood size was inhibited by the germline RNAi of rab-5. Among the secreted ligands, the germline RNAi of rab-5 increased the expressions of genes encoding insulin peptides (ins-3, ins-39, and daf-28), FGF ligand (egl-17), and ephrin ligand (efn-3) in PS-NP exposed nematodes and their corresponding receptor genes (daf-2, egl-15, and vab-1) in the offspring of PS-NP exposed nematodes. Therefore, an increase in germline mir-240 mediated transgenerational PS-NP toxicity through insulin, FGF, and ephrin signals by affecting its target RAB-5. Our data demonstrated the important involvement of germline microRNA in mediating nanoplastic toxicity across multiple generations in organisms.
Environmental significanceThe release of nanoplastics is ubiquitous in the environment, which causes their exposure risk and poses health risk to humans. Caenorhabditis elegans is sensitive to pollutants at environmentally relevant concentrations (ERCs). Using C. elegans as the animal model, nanopolystyrene at ERCs induced a transgenerational increase in the expression of germline microRNA/mir-240, suggesting the activation of this epigenetic signal. Germline mir-240 mediated transgenerational nanoplastic toxicity through insulin, FGF, and ephrin signals by affecting its target RAB-5. Our findings demonstrated the transgenerational dysregulation of certain microRNAs by nanoplastics, which was associated with their toxicity induction across multiple generations. |
Introduction
In our daily lives, along with the tremendous applications of plastic-based products, plastic pollution has been considered as a global concern.1 This pollution is not only due to the existence of wide and vast waste plastic products, but also due to the limited biodegradation of waste plastics.2 Environmental degradation through microorganisms, photo-oxidation, and mechanical abrasion causes the formation of microplastics and even nanoplastics.3,4 In several ecosystems (such as the soil ecosystem), nanoplastics have been frequently detected.5,6 Besides this, various commercial applications result in the release of nanoplastics into the environment.7 Largely owing to these facts, the environmental fate, transport, and risk of microplastics and nanoplastics have received increasing attention.8–13Through the food chain, nanoplastics are bioavailable to humans.14,15 After ingestion, nanoparticles will encounter the intestinal barrier and then reach system circulation and other tissues.16,17 This form of translocation of nanoplastics leads to corresponding toxicity.18–20 Nanoplastic exposure results in the disruption of the normal reproductive function, development, nervous-system function, and immune response.21–24 In mammals, nanoplastics could induce genotoxicity, oxidative stress, and damage on biological membranes.25,26 Nanoplastic toxicity can be affected by several factors, such as dose, size, and charge.27,28
Caenorhabditis elegans have tiny size and high sensitivity to pollutants.29–34 Due to the short life-cycle, it is useful for the evaluation of transgenerational toxicity of pollutants.35,36 This animal model can be applied to assess the transgenerational toxicity of nanoplastics (such as polystyrene nanoparticle (PS-NP)),37–40 which was affected by the size and modification of PS-NP.41,42 Moreover, some secreted ligands, including insulin, Wnt, FGF, Notch, ephrin, and their receptors acted together to regulate transgenerational PS-NP toxicity.43–47
For underlying molecular basis, besides transgenerational signal communication, alteration in some histone methylation-related molecules were associated with transgenerational PS-NP toxicity.39,48 microRNA (miRNA) having 19–22 nucleotides can regulate the gene expression after binding to the 3′-UTR of targeted genes.49 At P0-generation (P0-G), some miRNAs have been identified to function in different tissues (intestine, neurons, and germline) to regulate the PS-NP toxicity.50–54 Moreover, the decrease in germline mir-38 controlled transgenerational PS-NP toxicity by inhibiting some targets, such as the hedgehog ligand of WRT-3.55 In the germline, some other miRNAs may also be involved in regulating the transgenerational PS-NP toxicity. Thus, we first identified other dysregulated germline miRNAs induced by PS-NP exposure. Moreover, we determined the underlying mechanism for candidate germline miRNA(s) in controlling the transgenerational PS-NP toxicity. Our results provided further epigenetic regulation mechanism for the transgenerational toxicity of nanoplastics in organisms.
Experimental
PS-NP properties
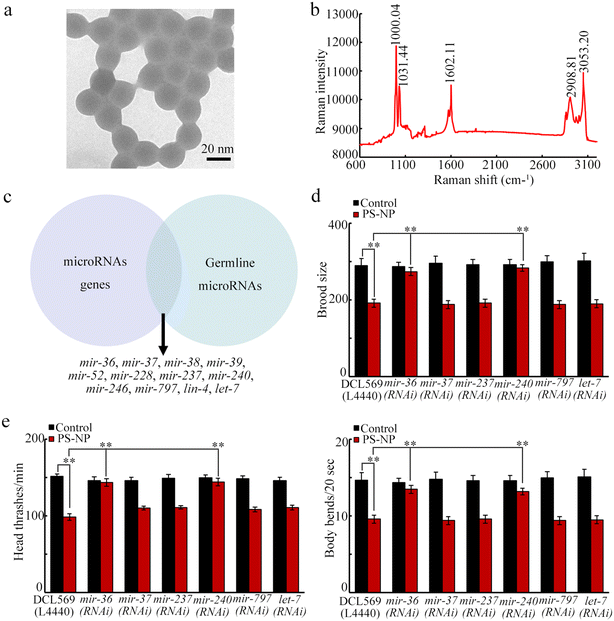
PS-NPs with a particle size of 20 nm was purchased from New-Materials Co., Ltd. Before the experiments, PS-NPs morphology was confirmed with a regular spherical shape and the particle size was 20.42 ± 2.4 nm using transmission electron microscopy (TEM) (Fig. 1a). Raman spectroscopy was used to reflect the plastic property of PS-NP. PS-NPs have three peaks at 1031.44 cm−1 (symmetric extension vibration of –C–C– in the benzene ring), 1000.04 cm−1 (indicating the respiratory vibration of the benzene ring), and 1602.11 cm−1 (indicating the asymmetric stretching vibration of the benzene ring carbon atoms) (Fig. 1b). The FTIR spectrum of PS-NP was described previously.42Maintenance of animals
Nematodes were cultured on nematode growth medium (NGM) fed with Escherichia coli (E. coli) OP50 at 20 °C.56 Strain information is provided in Table S1.† Eggs were isolated using lysis buffer from pregnant nematodes.57 After that, eggs were cultured to develop into L1-larvae nematodes for following experiments.PS-NP exposure
According to our previous study, the exposure concentrations of PS-NPs exposure were 1, 10, and 100 μg L−1.42 It was reported that the environmentally relevant concentrations (ERCs) of nanoplastics are in the range of ng L−1 to μg L−1.58 To evaluate the toxicity, animals were exposed to PS-NPs solutions at concentrations of 1–100 μg L−1 from L1-larvea to adult day-3, which is called P0-G. During the exposure process, PS-NPs exposure solutions were refreshed daily. After that, P0-G nematodes were removed, while their eggs were transferred to NGM plate to develop into adults, which is called F1-G. Using the same procedure, the following F2–F5 generation was obtained. All generations of nematodes were used for the subsequent determination of endpoints.Locomotion behavior and reproduction
To evaluate the toxicity, the locomotion behavior and reproduction were studied. Locomotion behavior was used to reflect the functional state of motor neurons.59 Brood size was used to reflect the reproductive capacity.60 In detail, locomotion behavior was evaluated by the frequencies of head thrashes and body bends.61 At the end of exposure, fifty nematodes were picked randomly in P0-G, and F1–F5 generations were used to count the number of head thrashes for 1 minute and number of body bends for 20 seconds under a stereomicroscope.62 The reproduction was evaluated using the endpoint of the brood size.63 The offspring number was counted until the end of spawning. Thirty animals were examined for the assessment of the brood size. Three independent experiments were conducted.Transcriptional expression analysis
Nematodes obtained from P0-G to F5-G were collected in centrifuge tubes, followed by the addition of Trizol to isolate the total RNA. The cDNA was synthesized using M-MuLV reverse transcriptase with the following reaction: 42 °C for 60 min and 70 °C for 10 min. Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted to assess the transcriptional expression of the examined genes. The expression of mir-240 was normalized by the expression of F35C11.9.64 For other genes' expression, tba-1 is the reference gene.65 Primers are given in Table S2.† Three replicates were performed.RNA interference (RNAi)
To analyze the function of the examined genes in transgenerational toxicity, nematodes were fed with E. coli HT115 expressing dsRNA.66 In detail, the gene construct for RNAi was generated using L4440, an empty vector, and subsequently transformed into HT115.67 Before the experiment, RNAi cells were cultured on NGM plates containing 1 mM isopropylthiogalactoside for 24 h to induce double-stranded RNA expression. For acquiring a higher RNAi efficiency, the F1-G of RNAi nematodes were collected for subsequent experiments. Meanwhile, the RNAi efficiency was determined to measure the expression of the determined gene or miRNAs by qRT-PCR, which were presented in Fig. S1.† The control consisted of HT115 expressing L4440.68Data analysis
Data were presented by mean ± standard deviation (SD). Significance was assessed by analysis of variance (ANOVA). In detail, one-way ANOVA was performed between the control and exposure groups. Two-way ANOVA was performed comparing multiple factors. The probability level threshold of 0.01 was used to define the statistical significance.Results
Identification of germline microRNAs required for the control of the PS-NP toxicity
Twelve miRNAs are expressed in the germline, including mir-38 (Fig. 1c) (https://wormbase.org/). Among these 12 miRNAs, mir-39, mir-52, mir-228, mir-246, and lin-4 were not required for controlling the toxicity of pollutants.64,69,70 Besides mir-38, we further identified whether some other miRNAs controlled the PS-NP toxicity. The germline RNAi of mir-37, mir-237, mir-797, and let-7 did not affect the PS-NP toxicity (Fig. 1d and e). However, the germline RNAi of mir-36 and mir-240 inhibited the PS-NP toxicity (Fig. 1d and e).Induction of transgenerational increase in the mir-240 expression after PS-NP exposure at P0-G
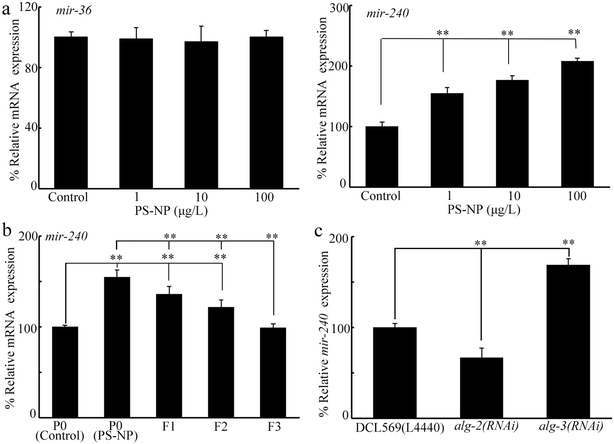
For candidate mir-36 and mir-240, we first investigated their expression at P0-G after PS-NP exposure. Exposure to PS-NP (1–100 μg L−1) did not affect the germline mir-36 expression but significantly increased germline mir-240 expression (Fig. 2a). Moreover, the increase in germline mir-240 expression was detected from P0-G to F2-G after PS-NP exposure (Fig. 2b). After PS-NP exposure at P0-G, the mir-240 expression was recovered to the control at F3-G (Fig. 2b).Previous study has indicated that the expression of certain miRNAs could be changed by the RNAi of genes (alg-1-5) encoding Argonaute proteins.55 In PS-NP exposed nematodes, alg-2 and alg-3 expressions could be affected by PS-NP.55 In PS-NP exposed nematodes, mir-240 expression was decreased by the germline RNAi of alg-2 and increased by the germline RNAi of alg-3 (Fig. 2c).
Germline RNAi of mir-240 confers resistance to transgenerational PS-NP toxicity
In nematodes, the germline RNAi of mir-240 inhibited transgenerational PS-NP toxicity in inhibiting the locomotion behavior and in reducing the brood size (Fig. 3a and b). Therefore, the resistance to transgenerational PS-NP toxicity could be observed in mir-240(RNAi).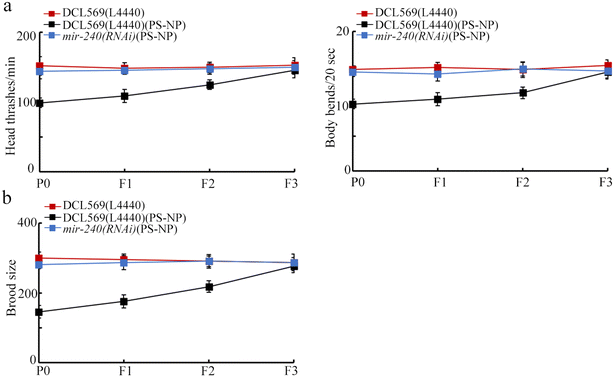 | ||
| Fig. 3 Effect of germline RNAi of mir-240 on the transgenerational toxicity of PS-NP on the locomotion behavior (a) and brood size (b). Exposure concentration of PS-NP was 1 μg L−1. | ||
Identification of targets of germline mir-240 in regulating transgenerational PS-NP toxicity
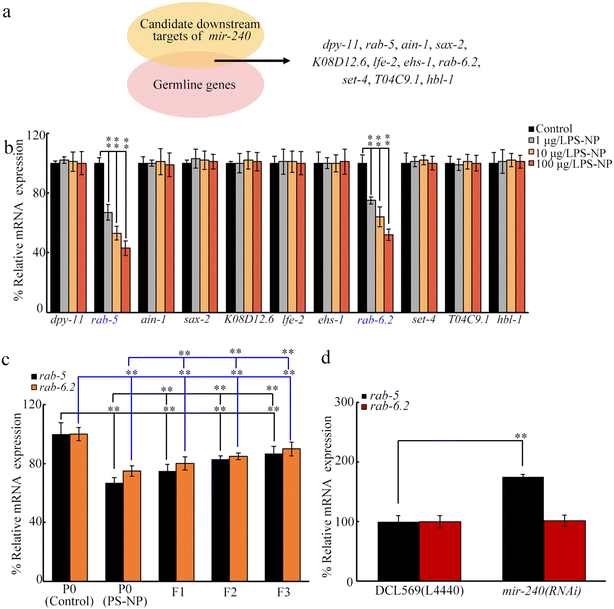
Using TargetScan (http://www.targetscan.org/worm_52/), 20 target genes are predicted for mir-240. Among these 20 genes, 11 genes are expressed in the germline (https://www.wormbase.org) (Fig. 4a). Among these 11 germline genes, exposure to PS-NP (1–100 μg L−1) decreased the germline expressions of rab-5 and rab-6.2 (Fig. 4b). After PS-NP exposure, the transgenerational decrease in germline expressions of rab-5 and rab-6.2 was further observed at F1-G and F2-G (Fig. 4c). Nevertheless, in PS-NP exposed nematodes, the germline RNAi of mir-240 did not affect rab-6.2 expression, whereas rab-5 expression was increased by the germline RNAi of mir-240 (Fig. 4d).Germline RNAi of rab-5 confers susceptibility to transgenerational PS-NP toxicity
Compared with PS-NP toxicity from P0-G to F2-G in DCL569(L4440), the PS-NP toxicity was detected from P0-G to F4-G in rab-5(RNAi) (Fig. 5a and b). This suggested the susceptibility of rab-5(RNAi) to transgenerational PS-NP toxicity.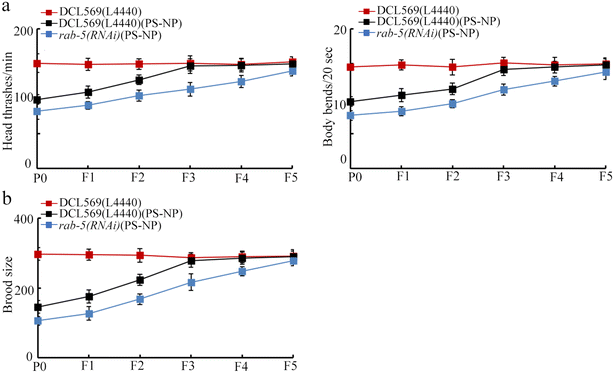 | ||
| Fig. 5 Effect of the germline RNAi of rab-5 on transgenerational PS-NP toxicity on the locomotion behavior (a) and brood size (b). Exposure concentration of PS-NP was 1 μg L−1. | ||
Genetic interaction between mir-240 and rab-5 in controlling transgenerational PS-NP toxicity
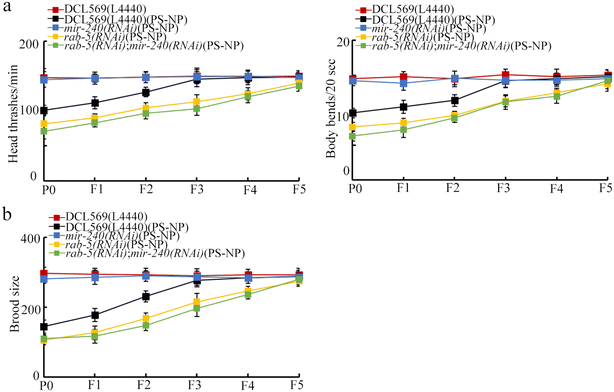
We performed double germline RNAi of mir-240 and rab-5 in PS-NP exposed nematodes. After PS-NP exposure, rab-5(RNAi); mir-240(RNAi) showed the similar phenotype of transgenerational PS-NP toxicity to that of rab-5(RNAi) (Fig. 6a and b). Therefore, the resistance of mir-240(RNAi) to transgenerational PS-NP toxicity in inhibiting the locomotion behavior and in reducing the brood size could be suppressed by the RNAi of rab-5.Identification of targets of germline RAB-5 in regulating transgenerational PS-NP toxicity
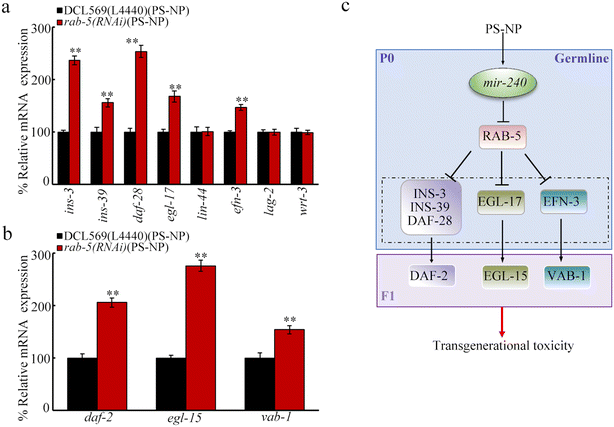
In C. elegans, RAB-5 regulates neuropeptide release,71 suggesting the potential of RAB-5 to regulate the secreted ligands. In the germline, insulin peptides (INS-3, INS-39, and DAF-28), Wnt ligand (LIN-44), FGF ligand (EGL-17), Notch ligand (LAG-2), hedgehog ligand (WRT-3), and ephrin ligand (EFN-3) were involved in controlling transgenerational PS-NP toxicity.43–48 Although the germline RNAi of rab-5 did not influence lin-44, lag-2, and wrt-3 expressions, ins-3, ins-39, daf-28, egl-17, and efn-3 expressions were increased by rab-5 RNAi at P0-G of the PS-NP exposed nematodes (Fig. 7a). DAF-2 is an insulin receptor, EGL-15 is an FGF receptor, and VAB-1 is an ephrin receptor.43,45,47 Moreover, after the RNAi of rab-5 at P0-G, at F1-G of PS-NP exposed rab-5(RNAi) nematodes, daf-2, egl-15, and vab-1 expressions were further increased (Fig. 7b).Discussion
Nanoplastics are widely distributed in the environment.72,73 Meanwhile, after being released or formed, nanoplastics are bioavailable to humans and organisms.74,75 After exposure, not only at P0-G, the nanoplastic toxicity could also be detected in the offspring of different organisms.76,77 The transgenerational toxicity of nanoplastics was further observed in C. elegans.78,79 Exposure to nanoplastics could cause epigenetic signatures associated with transgenerational impairment.80 Histone methylation regulation, a form of epigenetic regulation, was required for transgenerational toxicity induction of both pristine and aged nanoplastics.38,39 miRNA regulation is also a form of epigenetic regulation.81,82 We further found that the activation of miRNA could mediate the induction of transgenerational nanoplastic toxicity.We here found the role of activated germline mir-240 to mediate transgenerational PS-NP toxicity. Firstly, the germline RNAi of mir-240 conferred resistance to PS-NP toxicity at P0-G (Fig. 1d and e). Secondly, a transgenerational increase in the germline mir-240 expression was detected after PS-NP exposure (Fig. 2b). Thirdly, the germline RNAi of mir-240 caused resistance to transgenerational PS-NP toxicity (Fig. 3). Besides the involvement of the activated germline mir-240, the inhibited germline mir-38 also mediated transgenerational PS-NP toxicity. Germline mir-38 expression was decreased by PS-NP, and germline mir-38 overexpression inhibited transgenerational PS-NP toxicity.55 Therefore, nanoplastics can activate or inhibit certain germline miRNAs that lead to toxicity toward multiple generations. Our results demonstrated the crucial role of germline miRNAs in mediating an epigenetic mechanism to regulate the transgenerational toxicity of nanoplastics.
Besides germline mir-240, we also observed the resistance of mir-36(RNAi) to PS-NP toxicity (Fig. 1d and e). However, although PS-NP at 1–100 μg L−1 could induce the toxicity, the induced toxicity of PS-NP (1–100 μg L−1) was not associated with the germline mir-36. The reason was that the germline mir-36 expression was not affected by exposure to 1–100 μg L−1 PS-NP (Fig. 2a). In other words, PS-NP at higher concentrations or other pollutants with more severe toxicity may induce transgenerational toxicity by activating germline mir-36 in nematodes.
miRNAs biogenesis is governed by Argonaute proteins in C. elegans.83,84 Among genes encoding germline Argonaute proteins, alg-2 expression was increased by PS-NP and alg-3 expression was decreased by PS-NP.55 The alg-2(RNAi) animals showed resistance to PS-NP toxicity, and alg-3(RNAi) animals exhibited susceptibility to PS-NP toxicity.55 In PS-NP-exposed C. elegans, germline mir-240 expression was decreased by alg-2 RNAi and increased by alg-3 RNAi (Fig. 2c). The function of some C. elegans miRNAs in regulating stresses, such DNA damage and dauer formation, was also dependent on ALG-2.85,86 Additionally, germline mir-38 expression was decreased by the RNAi of C. elegans alg-3.55 Therefore, PS-NP exposure could activate germline mir-240 and inhibit germline mir-38 to induce transgenerational PS-NP toxicity by affecting the expressions of C. elegans germline Argonaute genes.
RAB-5 was further identified as the target of germline mir-240 (Fig. 7c). Firstly, PS-NP exposure caused transgenerational decrease in the expression of germline rab-5 (Fig. 4c). Secondly, germline rab-5 expression was decreased by the RNAi of mir-240 (Fig. 4d). Thirdly, the germline RNAi of rab-5 induced susceptibility to transgenerational PS-NP toxicity (Fig. 5). More importantly, the resistance of mir-240(RNAi) to transgenerational PS-NP toxicity was suppressed by the germline RNAi of rab-5 (Fig. 6). Therefore, activated germline mir-240 induced transgenerational nanoplastic toxicity by targeting and inhibiting RAB-5. RAB-5 is an endosomal Rab-type GTPase.87 RAB-5 was required for modulating the stress resistance and longevity.88
After PS-NP exposure, a transgenerational decrease in germline rab-6.2 was detected (Fig. 4c), suggesting that germline RAB-6.2 was also possibly required for controlling transgenerational PS-NP toxicity. Nevertheless, in PS-NP exposed nematodes, the RNAi of mir-240 did not alter the germline rab-6.2 expression (Fig. 4d). This implies that germline RAB-6.2 may participate in regulating the PS-NP toxicity under the control of other miRNAs.
Rab GTPases function as the master regulator for intracellular trafficking by regulating vesicle release.89,90 In C. elegans, Rab GTPases, including RAB-10 and RAB-5, control the release of certain ligands, such as neuropeptides.71 Among the identified secreted ligands controlling transgenerational PS-NP toxicity, insulin peptide genes (ins-3, ins-39, and daf-28), FGF ligand gene (egl-17), and ephrin ligand gene (efn-3) were increased by the germline RNAi of rab-5 after PS-NP exposure (Fig. 7a). Moreover, in the offspring of PS-NP-exposed nematodes, the expressions of corresponding receptor genes of these ligand genes were further increased by the germline RNAi of rab-5 (Fig. 7b). Thus, the decreased expression of germline rab-5 caused by PS-NP exposure was associated with the further activation of insulin, FGF, and ephrin ligands (Fig. 7c). In nematodes, the germline RNAi of these ligand genes resulted in resistance toward transgenerational PS-NP toxicity.43,45,47 In C. elegans, the decrease in germline mir-38 mediated transgenerational PS-NP toxicity by affecting WRT-3, a hedgehog ligand.55 The increase in the germline mir-240 mediated transgenerational PS-NP toxicity by affecting insulin peptides, FGF ligand, and ephrin ligand. These implied that germline mir-240 and mir-38 acted together to regulate the transgenerational toxicity of nanoplastics by affecting different ligands and their receptors. Unlike the alteration in insulin, FGF, and ephrin ligands, the expressions of ligand genes of Wnt, Notch, and hedgehog were not affected by the germline RNAi of rab-5 (Fig. 7a). In nematodes, these ligands in the germline also regulated transgenerational PS-NP toxicity.44,46,55 This implies that the expressions of Wnt, Notch, and hedgehog ligands may be under the control of other germline Rab GTPases in PS-NP exposed nematodes. Besides ligand release, RAB-5 is also required for endocytosis.71,91,92 The possible association of RAB-5 role in controlling the PS-NP toxicity with its function in modulating endocytosis needs to be further examined.
Conclusions
Together, we examined the role of germline miRNAs in mediating transgenerational nanoplastic toxicity. The increase in germline mir-240 expression mediated the transgenerational toxicity of PS-NP at ERCs. In the germline, RAB-5 acted as a downstream target of mir-240, and a decrease in germline rab-5 expression further mediated PS-NP toxicity across generations. Moreover, RAB-5 regulated transgenerational PS-NP toxicity by suppressing the expressions of ligands of insulin, FGF, and ephrin signals and expressions of their receptors in the offspring. Our results suggested the important involvement of activated germline miRNAs in mediating transgenerational nanoplastic toxicity by affecting the release of the secreted ligands across generations.Ethical statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Southeast University and approved by the Animal Care & Welfare Committee of Southeast University.Data availability
The data supporting this article have been included as part of ESI.†Conflicts of interest
There are no conflicts of interests.Acknowledgements
This study was supported by the grants from Natural Science Foundation of Guangdong Province (2024A1515011115) and Shenzhen Basic Research Project (JCYJ20220530163605011).References
- H.-T. Pinheiro, C. MacDonald, R.-G. Santos, R. Ali, A. Bobat, B.-J. Cresswell, R. Francini-Filho, R. Freitas, G.-F. Galbraith, P. Musembi, T.-A. Phelps, J.-P. Quimbayo, T. E. A. L. Quiros, B. Shepherd, P.-V. Stefanoudis, S. Talma, J.-B. Teixeira, L.-C. Woodall and L.-A. Rocha, Nature, 2023, 619, 311–316 CrossRef CAS PubMed
.
- J. Gigault, H. El Hadri, B. Nguyen, B. Grassl, L. Rowenczyk, N. Tufenkji, S. Feng and M. Wiesner, Nat. Nanotechnol., 2021, 16, 501–507 CrossRef CAS PubMed
.
- R. Jain, A. Gaur, R. Suravajhala, U. Chauhan, M. Pant, V. Tripathi and G. Pant, Sci. Total Environ., 2023, 905, 167098 CrossRef CAS PubMed
.
- S. Behera and S. Das, Chemosphere, 2023, 334, 138928 CrossRef CAS PubMed
.
- A. Wahl, C. Le Juge, M. Davranche, H. El Hadri, B. Grassl, S. Reynaud and J. Gigault, Chemosphere, 2021, 262, 127784 CrossRef CAS PubMed
.
- T. Li, L. Cui, Z. Xu, H. Liu, X. Cui and P. Fantke, Sci. Total Environ., 2023, 904, 166925 CrossRef CAS PubMed
.
- Y. Zhou, V. Ashokkumar, A. Amobonye, G. Bhattacharjee, R. Sirohi, V. Singh, G. Flora, V. Kumar, S. Pillai, Z. Zhang and M.-K. Awasthi, Environ. Pollut., 2023, 320, 121106 CrossRef CAS PubMed
.
- B. Zhang, J. Chao, L. Chen, L. Liu, X. Yang and Q. Wang, Sci. Total Environ., 2021, 757, 143791 CrossRef CAS PubMed
.
- X. Hua and D.-Y. Wang, Rev. Environ. Contam. Toxicol., 2022, 260, 12 CrossRef
.
- Y.-T. Shao, Y.-H. Li and D.-Y. Wang, Sci. Total Environ., 2024, 942, 173746 CrossRef CAS PubMed
.
- Z.-H. Zhuang, T.-W. Liu, Z.-Y. Liu and D.-Y. Wang, Ecotoxicol. Environ. Saf., 2024, 272, 116056 CrossRef CAS PubMed
.
- M.-F. Tang, G.-Y. Ding, L.-E. Li, G.-S. Xiao and D.-Y. Wang, Ecotoxicol. Environ. Saf., 2023, 262, 115131 CrossRef CAS PubMed
.
- M.-F. Tang, G.-Y. Ding, X.-Y. Lu, Q. Huang, H.-H. Du, G.-S. Xiao and D.-Y. Wang, Nanomaterials, 2022, 12, 4222 CrossRef CAS PubMed
.
- M. Carbery, W. O'Connor and T. Palanisami, Environ. Int., 2018, 115, 400–409 CrossRef PubMed
.
- M. Wang and W.-X. Wang, Chemosphere, 2023, 345, 140541 CrossRef CAS PubMed
.
- J.-J. Reineke, D.-Y. Cho, Y.-T. Dingle, A.-P. Morello, J. Jacob, C.-G. Thanos and E. Mathiowitz, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 13803–13808 CrossRef CAS PubMed
.
- T. Hong, W. Sun, Y. Deng, J.-D. Lyu, C.-H. Jin, Y.-L. Bai, J. Na, R. Zhang, Y. Gao, G.-W. Pan, Z.-S. Yang and L.-J. Yan, Biomed. Environ. Sci., 2024, 37, 31–41 Search PubMed
.
- Y.-H. Yang, W.-T. Dong, Q.-L. Wu and D.-Y. Wang, Nanoscale Adv., 2021, 3, 1997–2006 RSC
.
- J. Liang, F. Ji, A. L. B. Abdullah, W. Qin, T. Zhu, Y. J. Tay, Y. Li and M. Han, Sci. Total Environ., 2024, 942, 173770 CrossRef CAS PubMed
.
- N. Ali, J. Katsouli, E. L. Marczylo, T. W. Gant, S. Wright and J. Bernardino de la Serna, EBioMedicine, 2024, 99, 104901 CrossRef CAS PubMed
.
- D. Leistenschneider, A. Wolinski, J. Cheng, A. Ter Halle, G. Duflos, A. Huvet, I. Paul-Pont, F. Lartaud, F. Galgani, É. Lavergne, A.-L. Meistertzheim and J.-F. Ghiglione, Sci. Total Environ., 2023, 896, 164955 CrossRef CAS PubMed
.
- J. Bhagat, L. Zang, N. Nishimura and Y. Shimada, Sci. Total Environ., 2020, 728, 138707 CrossRef CAS PubMed
.
- Z.-Y. Liu, X. Hua, Y. Zhao, Q. Bian and D.-Y. Wang, Sci. Total Environ., 2024, 906, 167471 CrossRef CAS PubMed
.
- X. Hua and D.-Y. Wang, Sci. Total Environ., 2024, 918, 170760 CrossRef CAS PubMed
.
- W. Xu, Y. Yuan, Y. Tian, C. Cheng, Y. Chen, L. Zeng, Y. Yuan, D. Li, L. Zheng and T. Luo, J. Hazard. Mater., 2023, 454, 131470 CrossRef CAS PubMed
.
- A. Banerjee and W.-L. Shelver, Sci. Total Environ., 2021, 755, 142518 CrossRef CAS PubMed
.
- T. Kögel, Ø. Bjorøy, B. Toto, A.-M. Bienfait and M. Sanden, Sci. Total Environ., 2020, 709, 136050 CrossRef PubMed
.
- K. Pelegrini, T.-C.-B. Pereira, T.-G. Maraschin, L.-S. Teodoro, N.-R.-S. Basso, G.-L.-B. De Galland, R.-A. Ligabue and M.-R. Bogo, Sci. Total Environ., 2023, 878, 162954 CrossRef CAS PubMed
.
- D. Meyer and P. L. Williams, Toxicity testing of neurotoxic pesticides in Caenorhabditis elegans, J. Toxicol. Environ. Health, Part B, 2014, 17, 284–306 CAS
.
-
D.-Y. Wang, Exposure Toxicology in Caenorhabditis elegans, Springer Nature Singapore Pte Ltd., 2020 Search PubMed
.
- Y.-X. Wang, G.-Y. Liang, J. Chao and D.-Y. Wang, Sci. Total Environ., 2024, 927, 172306 CrossRef CAS PubMed
.
- X. Hua, G.-Y. Liang, J. Chao and D.-Y. Wang, J. Hazard. Mater., 2024, 472, 134598 CrossRef CAS PubMed
.
- X. Hua and D.-Y. Wang, Environ. Sci. Technol., 2023, 57, 19295–19303 CrossRef CAS PubMed
.
- Y.-X. Wang, X. Hua and D.-Y. Wang, Environ. Pollut., 2023, 333, 121937 CrossRef CAS PubMed
.
- Y. Zhao, J. Chen, R. Wang, X. Pu and D. Wang, J. Appl. Toxicol., 2023, 43, 122–145 CrossRef CAS PubMed
.
- Z.-Y. Liu, Y.-X. Wang, Q. Bian and D.-Y. Wang, Toxics, 2024, 12, 420 CrossRef PubMed
.
- H. Liu, Y. Wu and Z. Wang, J. Hazard. Mater., 2023, 459, 132124 CrossRef CAS PubMed
.
- H. Chen, Y. Gu, Y. Jiang, J. Yu, C. Chen, C. Shi and H. Li, Environ. Sci. Technol., 2023, 57, 19341–19351 CrossRef CAS PubMed
.
- C.-W. Yu, T.-C. Luk and V.-H. Liao, J. Hazard. Mater., 2021, 412, 125173 CrossRef CAS PubMed
.
- M. Qu, L. Miao, H. Chen, X. Zhang and Y. Wang, J. Hazard. Mater., 2023, 457, 131840 CrossRef CAS PubMed
.
- L.-M. Sun, K. Liao and D.-Y. Wang, Sci. Total Environ., 2021, 768, 144362 CrossRef CAS PubMed
.
- H.-L. Liu, L.-J. Tian, S.-T. Wang and D.-Y. Wang, Sci. Total Environ., 2021, 790, 148217 CrossRef CAS PubMed
.
- X. Hua, C. Cao, L. Zhang and D. Wang, J. Hazard. Mater., 2023, 451, 131174 CrossRef CAS PubMed
.
- W. He, A. Gu and D. Wang, Toxics, 2023, 11, 511 CrossRef CAS PubMed
.
- Y. Zhao, X. Hua, Q. Bian and D. Wang, Toxics, 2022, 10, 699 CrossRef CAS PubMed
.
- R. Xu, X. Hua, Q. Rui and D. Wang, NanoImpact, 2022, 28, 100425 CrossRef CAS PubMed
.
- H. Liu, Y. Zhao, X. Hua and D. Wang, Ecotoxicol. Environ. Saf., 2022, 243, 114022 CrossRef CAS PubMed
.
- L. Zhang, S.-T. Wang, Y. Zhao, K. Bi and D.-Y. Wang, Environ. Sci.: Nano, 2022, 9, 265–274 RSC
.
- H. Guo, N.-T. Ingolia, J.-S. Weissman and D.-P. Bartel, Nature, 2011, 466, 835–840 CrossRef PubMed
.
- H.-L. Liu, Y.-Y. Zhao, K. Bi, Q. Rui and D.-Y. Wang, Ecotoxicol. Environ. Saf., 2021, 212, 112018 CrossRef CAS PubMed
.
- D. Li, Y.-J. Yuan and D.-Y. Wang, Sci. Total Environ., 2020, 736, 139677 CrossRef CAS PubMed
.
- Y.-X. Qiu, Y.-Q. Liu, Y.-H. Li and D.-Y. Wang, Ecotoxicol. Environ. Saf., 2020, 201, 110857 CrossRef CAS PubMed
.
- S.-T. Wang, H.-L. Liu, Y.-Y. Zhao, Q. Rui and D.-Y. Wang, NanoImpact, 2020, 20, 100256 CrossRef
.
- Y.-H. Yang, Q.-L. Wu and D.-Y. Wang, Ecotoxicol. Environ. Saf., 2020, 206, 111404 CrossRef PubMed
.
- X. Hua, Y. Zhao, Y.-J. Yuan, L. Zhang, Q. Bian and D.-Y. Wang, J. Hazard. Mater., 2022, 437, 129302 CrossRef CAS PubMed
.
- S. Brenner, Genetics, 1974, 77, 71–94 CrossRef CAS PubMed
.
- C.-Y. Yuan, Y.-X. Wang, L. Zhang and D.-Y. Wang, Front. Pharmacol., 2024, 15, 1396733 CrossRef CAS PubMed
.
- R. Lenz, K. Enders and T. G. Nielsen, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E4121–E4122 CAS
.
- X. Wan, G.-Y. Liang and D.-Y. Wang, Chemosphere, 2024, 361, 142499 CrossRef CAS PubMed
.
- Z.-Y. Liu, Q. Bian and D.-Y. Wang, J. Hazard. Mater., 2024, 471, 134356 CrossRef CAS PubMed
.
- T.-W. Liu, Z.-H. Zhuang and D.-Y. Wang, Front. Pharmacol., 2023, 14, 1202379 CrossRef CAS PubMed
.
- X. Hua, X. Feng, Y.-S. Hua and D.-Y. Wang, Sci. Total Environ., 2023, 871, 162189 CrossRef CAS PubMed
.
- Y.-T. Shao, X. Hua, Y.-H. Li and D.-Y. Wang, J. Hazard. Mater., 2024, 466, 133545 CrossRef CAS PubMed
.
- M. Qu, L.-B. Luo, Y.-H. Yang, Y. Kong and D.-Y. Wang, Sci. Total Environ., 2019, 697, 134131 CrossRef CAS PubMed
.
- L. Zhang, Y.-X. Wang and D.-Y. Wang, Arch. Pharmacal Res., 2023, 46, 616–628 CrossRef CAS PubMed
.
- X. Hua and D.-Y. Wang, Environ. Pollut., 2023, 337, 122649 CrossRef CAS PubMed
.
- Y.-T. Shao, Y.-X. Wang, X. Hua, Y.-H. Li and D.-Y. Wang, Chemosphere, 2023, 336, 139193 CrossRef CAS PubMed
.
- Y.-X. Wang, L. Zhang, X.-A. Yuan and D.-Y. Wang, Front. Pharmacol., 2023, 14, 1114219 CrossRef CAS PubMed
.
- Y.-L. Zhao, Q.-L. Wu, Y.-P. Li, A. Nouara, R.-H. Jia and D.-Y. Wang, Nanoscale, 2014, 6, 4275–4284 RSC
.
- Q.-L. Wu, Y.-L. Zhao, G. Zhao and D.-Y. Wang, Nanomedicine, 2014, 10, 1401–1410 CrossRef CAS PubMed
.
- N. Sasidharan, M. Sumakovic, M. Hannemann, J. Hegermann, J.-F. Liewald, C. Olendrowitz, S. Koenig, B.-D. Grant, S.-O. Rizzoli, A. Gottschalk and S. Stefan Eimer, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 18944–18949 CrossRef CAS PubMed
.
- K. Mattsson, L.-A. Hansson and T. Cedervall, Environ. Sci.: Processes Impacts, 2015, 17, 1712–1721 RSC
.
- D. Huang, H. Chen, M. Shen, J. Tao, S. Chen, L. Yin, W. Zhou, X. Wang, R. Xiao and R. Li, J. Hazard. Mater., 2022, 438, 129515 CrossRef CAS PubMed
.
- J. Yi, Y. Ma, J. Ruan, S. You, J. Ma, H. Yu, J. Zhao, K. Zhang, Q. Yang, L. Jin, G. Zeng and D. Sun, Environ. Int., 2024, 183, 108432 CrossRef CAS PubMed
.
- J.-L. Xu, X. Lin, J.-J. Wang and A.-A. Gowen, Sci. Total Environ., 2022, 851, 158111 CrossRef CAS PubMed
.
- I.-C. Yeo, K.-Y. Shim, K. Kim and C.-B. Jeong, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2023, 269, 109635 CAS
.
- Y. He and R. Yin, J. Appl. Toxicol., 2024, 44, 66–85 CrossRef CAS PubMed
.
- M. C. López de Las Hazas, H. Boughanem and A. Dávalos, Adv. Nutr., 2022, 13, 1310–1323 CrossRef PubMed
.
- L. Zhao, M. Qu, G. Wong and D.-Y. Wang, Environ. Sci.: Nano, 2017, 4, 2356–2366 RSC
.
- Y. H. Lee, M. S. Kim, Y. Lee, D.-H. Kim and J.-S. Lee, J. Hazard. Mater., 2023, 449, 131037 CrossRef CAS PubMed
.
- V.-K. Gangaraju and H. Lin, Nat. Rev. Mol. Cell Biol., 2009, 10, 116–125 CrossRef CAS PubMed
.
- F. Sato, S. Tsuchiya, S. J. Meltzer and K. Shimizu, FEBS J., 2011, 278, 1598–1609 CrossRef CAS PubMed
.
- D.-G. Zisoulis, Z.-S. Kai, R.-K. Chang and A.-E. Pasquinelli, Nature, 2012, 486, 541–544 CrossRef CAS PubMed
.
- S. Bouasker and M.-J. Simard, Nucleic Acids Res., 2012, 40, 10452–10462 CrossRef CAS PubMed
.
- M.-A. Doll, N. Soltanmohammadi and B. Schumacher, Genetics, 2019, 213, 173–194 CrossRef CAS PubMed
.
- H. Roka Pun and X. Karp, G3: Genes, Genomes, Genet., 2024, 14, jkae007 CrossRef PubMed
.
- A. Audhya, A. Desai and K. Oegema, J. Cell Biol., 2007, 178, 43–56 CrossRef CAS PubMed
.
- A. Traa, S.-K. Soo, A. AlOkda, B. Ko, C.-E. Rocheleau and J.-M. Van Raamsdonk, Aging Cell, 2023, 22, e13762 CrossRef CAS PubMed
.
- M. Fukuda, Cell. Mol. Life Sci., 2008, 65, 2801–2813 CrossRef CAS PubMed
.
- H. Stenmark, Nat. Rev. Mol. Cell Biol., 2009, 10, 513–525 CrossRef CAS PubMed
.
- K. Smurova and B. Podbilewicz, Cell Rep., 2016, 14, 1517–1527 CrossRef CAS PubMed
.
- A. van der Vaart, S. Rademakers and G. Jansen, PLoS Genet., 2015, 11, e1005733 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4en00309h |
| This journal is © The Royal Society of Chemistry 2024 |